Abstract
Objective:
Loss-of-function mutations in genes generating reactive oxygen species (ROS), such as NOX1, are associated with inflammatory bowel disease (IBD). Mechanisms whereby loss of ROS drive IBD are incompletely defined.
Design:
ROS measurements and single-cell transcriptomics were performed on colonoids stratified by NOX1 genotype and TNFα stimulation. Clustering of epithelial cells from human ulcerative colitis (UC, inflamed and uninflamed) scRNASeq was performed. Validation of M cell induction was performed by immunohistochemistry using UEA1 (Ulex Europaeus Agglutin-1 lectin) and in vivo with DSS injury.
Results:
TNFα induces ROS production more in NOX1-WT vs. NOX1-deficient murine colonoids under a range of Wnt- and Notch-mediated conditions. scRNASeq from inflamed and uninflamed human colitis versus TNFα-stimulated, in vitro colonoids defines substantially shared, induced transcription factors; NOX1-deficient colonoids express substantially lower levels of STAT3 (signal transducer and activator of transcription 3), CEBPD (CCAAT Enhancer Binding Protein Delta), DNMT1 (DNA methyltransferase) and HIF1A (hypoxia-inducible factor) baseline. Subclustering unexpectedly showed marked TNFα-mediated induction of M cells (sentinel cells overlying lymphoid aggregates) in NOX1-deficient colonoids. M cell induction by UEA1 staining is rescued with H2O2 and paraquat, defining extra- and intracellular ROS roles in maintenance of LGR5+ stem cells. DSS injury demonstrated GP2 (glycoprotein-2), basal lymphoplasmacytosis and UEA1 induction in NOX1-deficiency. Principal components analyses of M cell genes and decreased DNMT1 RNA velocity correlate with UC inflammation.
Conclusions:
NOX1 deficiency plus TNFα stimulation contribute to colitis through dysregulation of the stem cell niche and altered cell differentiation, enhancing basal lymphoplasmacytosis. Our findings prioritize ROS modulation for future therapies.
INTRODUCTION
The intestinal epithelium forms a key functional mediator between the intraluminal milieu and the mucosal immune system. Stress conditions enhance reactive oxygen species (ROS) production; loss-of-function mutations in genes of the NADPH oxidase complex in phagocytic leukocytes can result in chronic granulomatous disease (CGD) [1–4]. CGD typically presents in childhood with atypical infections, and approximately one-third to one-half of CGD patients have intestinal inflammation resembling polygenic inflammatory bowel disease (IBD) [5]. More recently, loss-of-function mutations in NOX1 [6,7] and DUOX2 (dual oxidase 2) [8], involved in epithelial production of ROS, have also been associated with IBD. The association of multiple loss-of-function genes expressed in diverse cellular sources underscores that loss of ROS represents a particularly essential pathway driving IBD; despite this, pathogenic mechanisms are incompletely defined.
The intestinal epithelium is constantly replenished by stem cells at the base of crypt structures, maintained by Wnt signaling [9]. Binary cell fate decisions between secretory and absorptive lineages are mediated via Notch signaling, with high Notch conditions favoring differentiation toward absorptive enterocytes. Nox1 knockout (KO) mice are characterized by an excess of cells of the secretory lineage [10]. Interspersed between LGR5+ (Leucine Rich Repeat Containing G Protein-Coupled Receptor 5) stem cells in the colon are deep crypt secretory cells (DCS), which highly express REG4 (regenerating family 4); ablation of DCS cells results in a loss of stem cells, with impaired epithelial differentiation [11]. Beyond the secretory vs. absorptive cell dichotomy, additional subspecialized cells include neuroendocrine cells, Tuft cells and M cells. In the small intestine, M cells overlie lymphoid aggregates and play a central role in mucosal immune surveillance[12]. In ulcerative colitis (UC), lymphoid aggregates and basal lymphoplasmacytosis[13] are frequently present[14], and can be induced in murine models via RANKL (Receptor Activator of Nuclear Factor Kappa-B Ligand)[15] or TNFα.
In this study, we explore the role of NOX1 and ROS through direct ex vivo studies of single cell UC transcriptomics (scRNAseq) and in vitro and in vivo murine studies. We show that under a variety of colonoid differentiation conditions, TNFα induced ROS secretion is dependent upon NOX1. We demonstrate that TNFα-induced transcription factors model very well differences observed direct ex-vivo comparing inflamed vs. uninflamed UC cells. Unexpectedly, we observed marked induction of M cells in Nox1KO enteroids with TNFα treatment. This induction was substantially rescued by exogenous H2O2. DSS injury demonstrate similar differential genotype effects in vivo. Taken together, these findings define a two-hit model (ROS deficiency plus TNFα) of M cell induction in colitis. The colonoid studies demonstrate lower baseline expression of multiple transcription factors with NOX1KO including DNMT1 (DNA methyltransferase 1) which plays a key role pluripotent somatic cell replication post-natally. The present study defines the role of ROS in maintaining intestinal stem cell niches during chronic inflammation.
METHODS
Additional methods can be found in online supplementary materials.
Human specimens
Human subject protocols were reviewed and approved by the Institutional Review Board (IRB) at the Icahn School of Medicine at Mount Sinai (HSM#14–00210). The demographic and clinical information of patients is in Supplementary table 1. Designation of active rectum and inactive sigmoid biopsies were determined endoscopically by Mayo Score and confirmed histologically. Biopsies were obtained from both the inflamed and non-inflamed regions and stored in RPMI-1640 (Corning), followed by incubation in 1mL dissociation medium (HBSS w/o Ca2+ Mg2+ with HEPES 10mM and EDTA 5mM, Life Technologies) at room temperature for 30 minutes. Following by two washes in RPMI, biopsies were digested in digestion medium (HBSS with Ca2+ Mg2+, FBS 2%, DNase I 0.5mg/mL (Sigma-Aldrich) and Collagenase IV 0.5mg/mL (Sigma-Aldrich)) for 40 min at 37°C under 100 rpm agitation.
Colonoid development and differentiation
Human and mice colonoid lines were generated according to the method from VanDussen et al. with the following modifications [16]. Human biopsies were obtained from IBD patients during routine colonoscopies. Full thickness murine colons (excluding cecum) were dissected and washed (DMEM/F12 medium with 15 mM HEPES, 2 mM L-glutamine, 100 U/ml of penicillin and 0.1 mg/ml of streptomycin) three times. Human biopsies and murine colons were dissociated by collagenase I (Thermo Fisher Scientific) incubation at 37°C for 30 min and vigorous pipetting of mixtures. Mixtures were passed through a 70-μm cell strainer (BD Biosciences) followed by two washes with wash medium. The crypt mixtures were suspended in Matrigel (Corning) and cultured in 50% L-WRN (L-cell expressing Wnt3a, Rspondin3 and Noggin) conditioned medium with 10 μM of Y-27632 (ROCK inhibitor; R&D Systems) and 10 μM SB-431542 (TGF-βR inhibitor; R&D Systems). L-WRN conditioned media was prepared as described, using cells generously provided by the Stappenbeck laboratory[17]. Cells were passaged every 4–6 days by trypsinization, and fresh 50% L-WRN was replaced every 2 days. After five passages, cells were frozen in 8.3% DMSO in wash media and kept in liquid nitrogen. Prior to differentiation, cells were thawed and expanded in 50% L-WRN with 10 μM of Y-27632 and 10 μM SB-43154. To induce differentiation, 2×105 of trypsinized spheroids were suspended in 30 ul of Matrigel (Corning, Cat#354234) precoated cultured in indicated differentiation medium for 2 (differentiation assays) or 3 days (ROS assays).
scRNAseq processing and analysis
UC epithelial clustering
Single cell data from inflamed rectum and uninflamed sigmoid from UC patients [18,19] (GEO GSE150516) were analyzed. Sequence reads were mapped using Cell Ranger (3.1.0). Raw matrices were analyzed using Seurat (v4_0_3). Epithelial cell doublets were removed using DoubletFinder. A mitochondrial percentage window of 2–50%, 6 PCs and 0.6 resolution were applied. Upon using unbiased markers for annotation, one of the clusters was identified as ‘apoptotic cells’, (marker list-MALAT1, MT-CYB, MT-ND2, HSPA1A, AKAP9, ARGLU1, FOSB, LYZ, IGLC3, MUC4) and was removed from downstream analysis. All heatmaps were generated using the Seurat wrapper “Scillus” (https://scillus.netlify.app/). All scripts used to generate corresponding figures can be found here: https://github.com/ChoBioLab/hsu-et-al-2022.
Mice colonoid data
The fastq files from 8 samples were aligned to mm10 using Cell Ranger software 3.1.0. The 8 samples include 2 different treatments (untreated and TNFα treated), 2 different genotypes (Nox1WT and Nox1KO) and duplication per treatment or genotype group. The resulting raw matrices were analyzed using Seurat (v4_0_3). We filtered out cells with less than 500 UMI and greater than 60k UMI. The mitochondrial percentage for each cell was calculated and analyzed to remove outlier cells with percentage mitochondrial gene > 30% or less than 5%. The cells were clustered iteratively examining the top unbiased markers using the ‘FindMarkers’ function in Seurat. We removed cells from clusters with most markers identified as mitochondrial genes indicative of apoptotic cells (markers genes-Cluster4: Gm42418, AY036118, Tff3, mt-Co2, mt-Atp6, mt-Nd2, mt-Co3, mt-Cytb, Pigr, mt-Nd5, Cluster11: Mki67, Top2a, mt-Atp6, mt-Nd2, mt-Co3, mt-Cytb, mt-Co2, Hsp90b1, mt-Nd3, mt-Nd1). The filtered set of 35355 cells were again clustered using the standardized pipeline. PCA was performed on the 2000 most variable genes and 12 PCs and 0.6 resolution were selected for the downstream clustering algorithm.
Given the marked genotype and TNFα-treated differences in the putative deep crypt secretory/Goblet cells cluster (1685 cells) in the mice colonoid data (Figure 2C), we performed further subclustering, iterating between expert annotation and Seurat-based marker selection. We identified some cells showing Secretory TA and Enteroendocrine markers incorrectly clustered with the Deep crypt secretory/Goblet cell cluster. These cells were re-annotated and the ‘Deep crypt secretory/Goblet’ cells were subclustered again with the remaining 1390 cells using 1000 top variable genes, 8 PC’s and 0.4 resolution. We removed clusters indicative of apoptotic state (mitochondrial gene markers) using the unbiased cluster markers and/or contributed only by one sample. The remaining clusters with 960 cells were again annotated using the unbiased marker and expert annotation approach. All heatmaps were generated using the Seurat wrapper “Scillus” (https://scillus.netlify.app/). All scripts used to generate corresponding figures can be found here: https://github.com/ChoBioLab/hsu-et-al-2022.
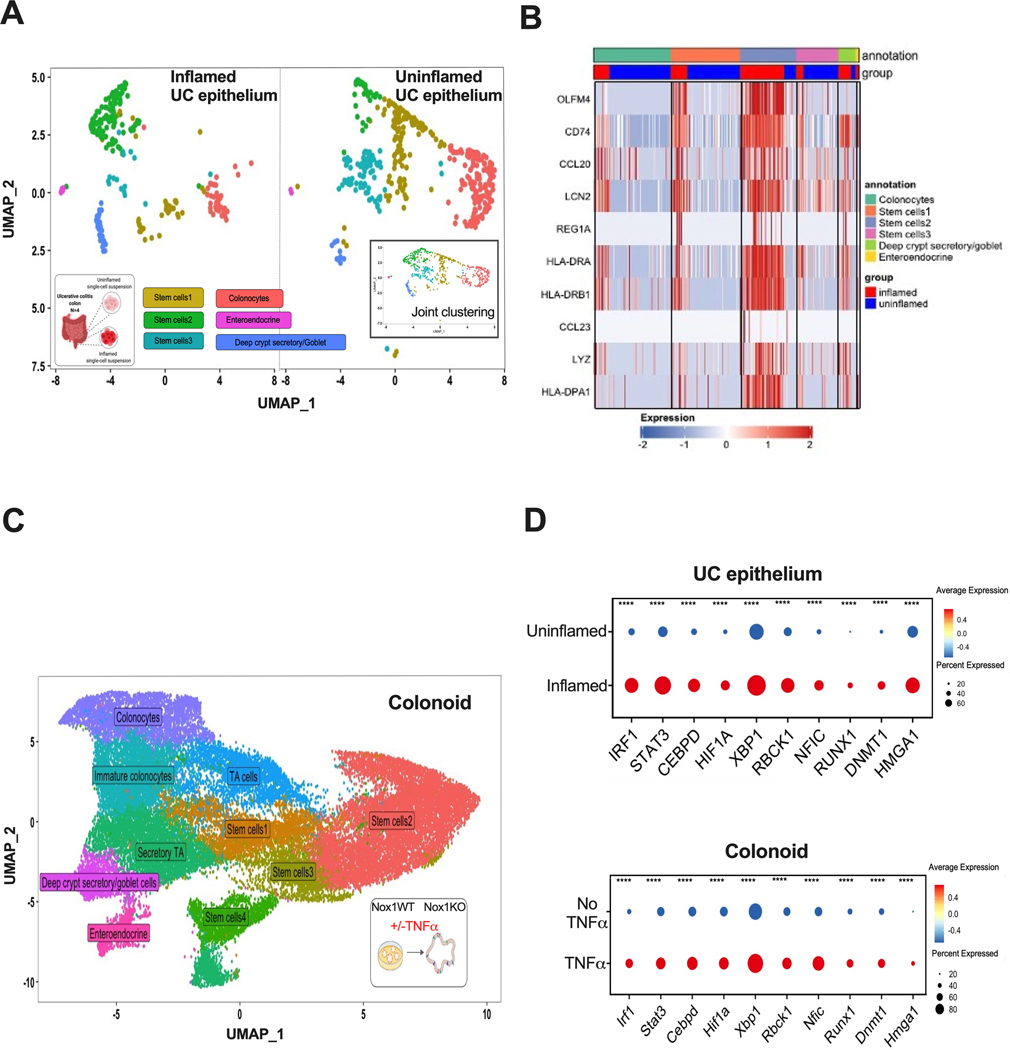
Figure 2. Single cell transcription factor gene expression comparing inflamed vs. uninflamed UC is similar in TNFα-induction in colonoids.
A. UMAP of joint clustering from epithelial cell clusters of UC scRNAseq stratified by inflammation status. The UMAP of the combined inflamed and uninflamed cells are shown in the inserted right panel. N=4 UC patients. B. Heatmap of the top 10 upregulated genes in the stem cell 2 cluster from UC epithelium scRNAseq. Expression is log2 normalized. C. UMAP showing joint clustering of colonoid cells from scRNAseq of untreated and TNFα-stimulated Nox1WT and Nox1KO cultured mice colonoids. D. Dot plots of inflammation or TNFα induced transcription factors in inflamed vs. uninflamed UC epithelial cells (top) similarly upregulated upon TNFα stimulation in colonoids (bottom). ****, P < 0.0001 (two-sided Wilcoxon signed-rank test).
Transcription Factor analysis
The list of transcription factors was taken from Lambert et al [20]. (http://humantfs.ccbr.utoronto.ca/download/v_1.01/TF_names_v_1.01.txt). The differential gene expression was analyzed using the “FindMarkers” function in Seurat for both colonoid and UC epithelium data sets. The colonoid data set was stratified by genotypes (Nox1WT vs Nox1KO) and treatments (untreated vs TNFα treated). UC epithelium data was stratified by disease conditions (UC uninflamed vs UC inflamed). The UC inflamed transcription-factors were generated via combination of transcription factor list and the most differential genes in UC inflamed vs uninflamed (P< 1X10−4). We further examined the conservation of UC inflamed and colonoid TNFα-induced transcription factors.
RESULTS
NOX1 is required for TNFα and RANKL induced ROS production
Because NOX1-deficiency favors secretory cell differentiation [10], we reasoned that NOX1 deficiency alters the stem cell niche and cellular differentiation under conditions of stress. To test this, we developed colonoid cell lines from wild-type and Nox1KO murine large intestine, evaluating the effects of TNFα and RANKL stimulation on ROS production. ROS production was measured by L012 enhanced chemiluminescence [21]. We observed a marked dependency of ROS secretion on NOX1 with both TNFα and RANKL, with dose-dependent induction of ROS observed in wild-type, but not Nox1KO enteroids (Figure 1A). Under these conditions (25% WRN, Wnt3a, R-spondin, Noggin) TNFα stimulation generated substantially higher ROS production than RANKL (Figure 1A).
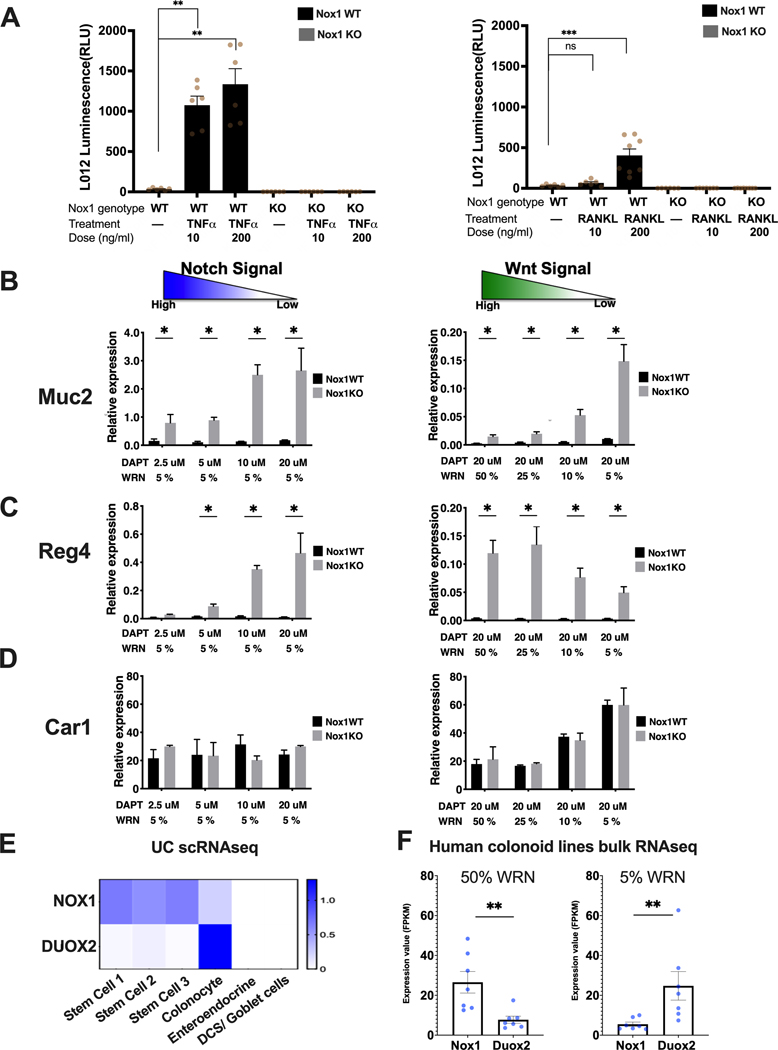
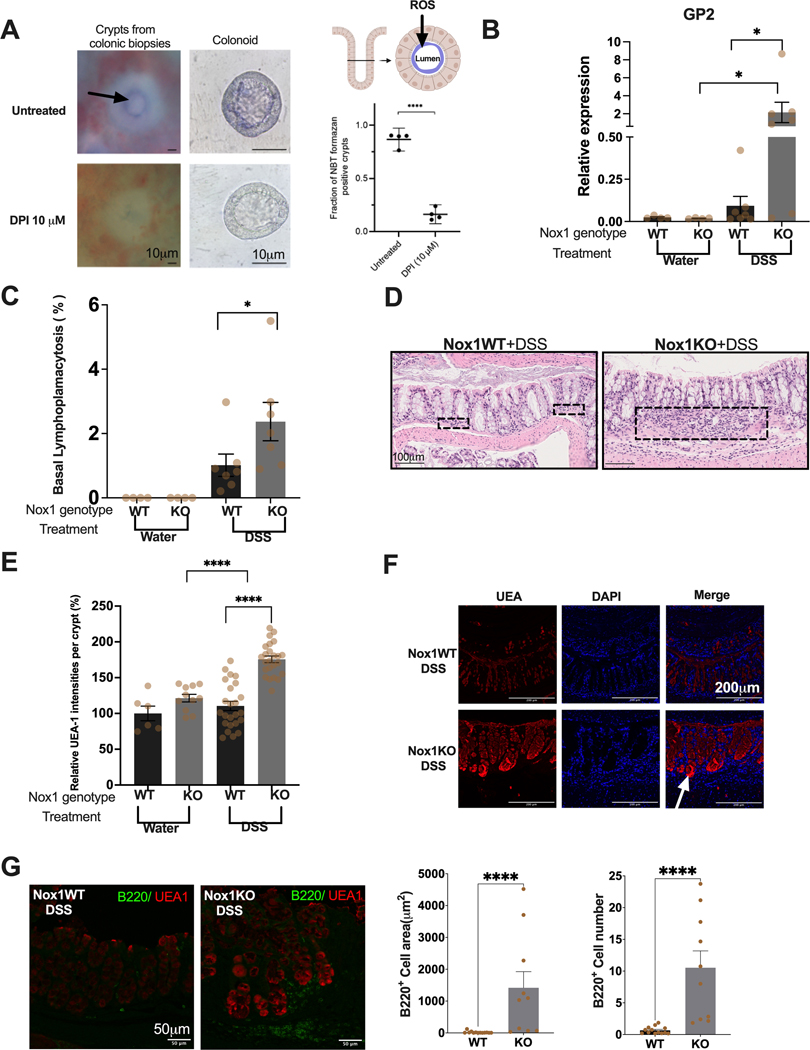
Figure 1. Stem cell expressed NOX1 is required for TNFα induced ROS production and NOX1 deficiency results in secretory precursor enhancement.
A. TNFα (left) and RANKL (right) induced ROS levels in Nox1WT and Nox1KO cells (25% WRN, 20 μM DAPT, measured 3 days after plating). Ns, non-significant. **, P < 0.01. ***, P < 0.001. (Mann-Whitney test, n = 6 to 8 per group). B, C, D. Cell type specific gene expression changes with decreasing Notch (left) and Wnt (right) culture conditions stratified by Nox1 genotype. RT-PCR results for Muc2 (B--goblet cells), Reg4 (C--deep crypt secretory cells) and Car1 (D--colonocytes) for Nox1WT (black) and Nox1KO (gray) colonoids normalized to a house keeping gene, Hprt1 (n=4 per group). *, P < 0.05 (Mann-Whitney test) E. Heatmap of average NOX1 and DUOX2 expression from uninflamed biopsy human (scRNAseq) of UC. F. NOX1 and DUOX2 expression from uninflamed human colonoid lines bulk RNA seq data. human colonoid lines (n = 7 lines) were cultured under 50%WRN (stem cells are the major cell type) or 5% WRN (colonocytes are major cell type). **, P < 0.01 (Mann-Whitney test).
We next evaluated marker gene expression under conditions of decreasing Notch or Wnt signaling after 48 hours, keeping the other signaling pathway fixed at a low activity (Figure 1B-D). Marker gene expression was normalized to a house keeping gene, Hprt1 (hypoxanthine phosphoribosyltransferase 1). Systematic decreases of both Notch or Wnt signaling resulted in a marked enhancement of Muc2 (mucin 2, goblet cell marker) in Nox1KO compared to wild-type colonoids, consistent with prior reports with NOX1 knockouts[10] (Figure 1B). Similarly, with low Wnt (Figure 1C, left panel), progressive decreases of Notch signaling also resulted in progressive increases in Reg4 in Nox1KO compared to wild-type cells. At fixed, low Notch conditions (Figure 1C, right panel), decreasing Wnt (50% to 5% WRN) results in lowered Reg4 gene expression in Nox1KO cells; this models the gradient effect of high Wnt conditions at the base of intestinal crypts enhancing Reg4 expression marking deep crypt secretory cells[11,22]. No genotype differences were observed for Car1 (carbonic anhydrase 1, absorptive colonocyte marker) gene expression generally (Figure 1D), with increasing Car1 expression observed with progressively decreasing Wnt dosage (Figure 1D, right panel). Finally, the highest ROS production was observed in wild-type cells in low Wnt (5% WRN), high Notch conditions (Supplementary Figure 1).
NOX1 and DUOX2 (dual oxidase 2) are two major epithelial NADPH oxidase proteins both enhancing ROS production. Paired biopsies from inflamed rectum and uninflamed sigmoid regions from four UC patients for scRNASeq were analyzed for comparative NOX1 vs. DUOX2 expression. NOX1 is expressed in all UC stem cell clusters; in contrast, DUOX2 was mainly found in differentiated colonocytes (Figure 1E). Similar trends were observed in bulk RNA sequencing in human colonoids. Higher NOX1 mRNA was observed with 50% WRN, with the converse (i.e., higher DUOX2 than NOX1) expression observed with 5% WRN (Figure 1F). Taken together, these data establish that NOX1 is required for TNFα-induction of ROS and highlight an essential role of NOX1 in the maintenance of the stem cell niche, via inhibition of secretory cell differentiation.
TNFα-stimulated colonoids models a common set of transcription factors present direct ex-vivo in inflamed UC
We next restricted inflamed vs. uninflamed UC scRNASeq analyses to epithelia (Figure 2A, Supplementary Figure 2A), with a particular focus on transcription factor gene expression. Clustering all epithelial cells together, we observed a relative decrease in mature absorptive colonocytes in inflamed compared to uninflamed regions (Figure 2A, red cluster). Among stem cell subclusters, we observed differences in relative fractions of inflamed vs. uninflamed regions, with increases in the stem cell 2 (SC2) cluster in inflamed tissues observed in all patients (Figure 2A, green cluster). Inspection of genes differentially expressed between the different stem cell clusters (Figure 2B) showed that, similar to prior reports [23], under conditions of stress, expression of genes involved in antigen processing (MHC class II, PSMB9, proteasome 20S subunit Beta 9—involved in peptide processing) was observed. In addition, SC2 in UC is enhanced in genes involved in cellular killing (LYZ, lysozyme, LCN1, lipocalin), anti-apoptosis (OLFM4, olfactomedin 4), lymphocyte recruitment (CCL20, C-C motif chemokine ligand 20) and cellular regeneration (REG1A, regenerating family member 1A).
We cultured Nox1WT and Nox1KO colonoids at intermediate, 25% WRN conditions and performed scRNAseq (Figure 2C, Supplementary Figure 2B). We assessed to what extent the inflammation-induced transcription factors observed in UC epithelium scRNASeq were also induced in TNFα-treated colonoids. To a substantial extent, the most induced transcription factors observed direct ex vivo in UC epithelium scRNAseq, were similarly induced by TNFα stimulated colonoids in vitro, reflecting the key role of TNFα in IBD pathogenesis (Figure 2D). Comparing Nox1WT vs. Nox1KO colonoids, the large majority of transcription factors demonstrated similar directional effects (Supplementary table 2), with similar induction of numerous transcription factors observed in inflamed (UC) and TNFα-treated (colonoid) cells.
Baseline and TNFα stimulated NOX1-dependent differences in transcription factor gene expression expressed in UC SC2 cluster
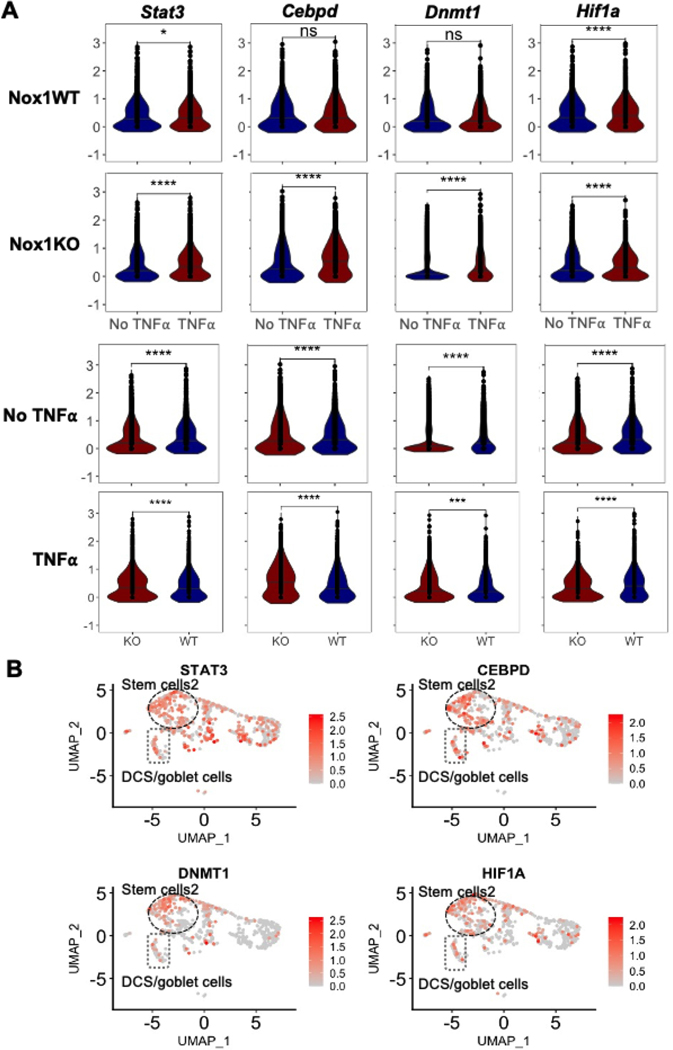
Genotype-dependent differences (either in untreated, or with TNFα induction) included Stat3 (Signal Transducer and Activator of Transcription 3), Cebpd (CCAAT enhancer binding protein delta), Dnmt1 (DNA methyltransferase 1), and Hif1a (Hypoxia induced factor 1A) (Figure 3A). For these four genes, generally greater induction was observed in Nox1KO colonoids with TNFα stimulation compared to wild-type colonoids (Figure 3A, first two rows). Substantially, this reflects lower unstimulated expression of these transcription factors in NOX1-deficiency (Figure 3A, third row). For example, substantially lower Dnmt1 is observed in baseline NOX1KO compared to wild-type cells (P-value = 9.48 × 10−40); DNMT1 plays a key role in maintaining maintenance progenitor function in self-renewing somatic tissues[24]. Hif1a is induced in both wild-type and Nox1KO colonoids treated with TNFα; higher levels of Hif1a are expressed in untreated wild-type colonoids, with induced expression observed with TNFα treatment (P-value = 2.50 × 10−4). This observation is consistent with prior literature demonstrating that ROS transcriptionally stimulates HIF1A expression via AMPK (adenosine activated protein kinase, catalytic subunit) [25]. Projection of these genes onto direct ex vivo human UC scRNAseq data (Figure 3B) demonstrated predominant gene expression in the SC2 cluster. Marked induction of HIF1A is expressed in inflamed rectum compared to uninflamed bulk mRNA sigmoid (log2 = 0.93, P-value = 8.6 × 10−13, adjusted), with more modest induction present with DNMT1 (log2 = 0.30, P-value = 8.5 × 10−4, adjusted).
Figure 3. NOX1- and TNFα-modulated transcription factor expression is predominant in UC stem cells.
A. Violin plot of transcription factors showing differential expression in inflamed vs. uninflamed tissue and being differentially expressed (without TNFα) by genotype. TNFα-stimulated differences for Nox1WT (first row) and Nox1KO (second row) colonoids. Genotype dependent differences without (third row) and with (forth row) TNFα stimulation. Ns, non-significant. *, P<0.05. ****, P < 0.0001 (two-sided Wilcoxon signed-rank test). B. Feature plot of induced transcription factors projected onto UMAP from UC epithelial cells (see Figure 2A for cell annotation).
M cell induction in Nox1 knockout with TNFα stimulation
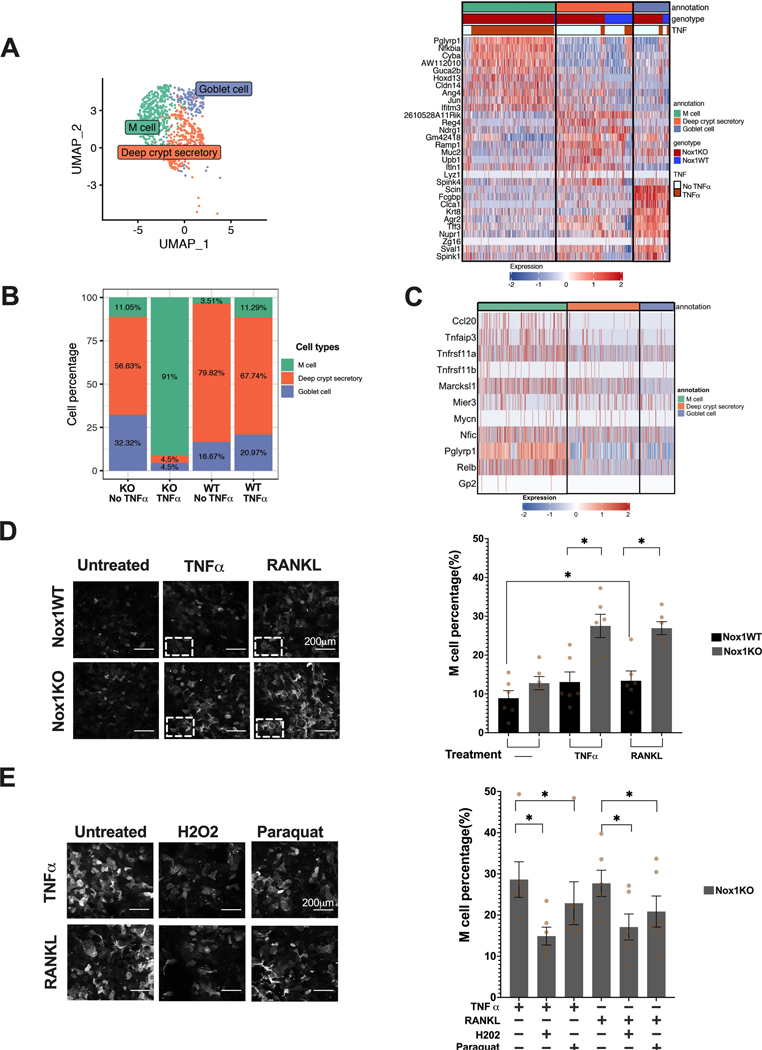
We observed marked genotype differences in the putative deep crypt secretory/goblet cells and therefore performed subclustering of the putative DCS/goblet cell cluster (Figure 4A). Unexpectedly, within the TNFα-treated, Nox1KO colonoids, we observed marked induction of M (microfold) cells (Figure 4B), characterized by high Pglyrp1 (Peptidoglycan recognition protein 1), Nfkbia (Nuclear factor of kappa light polypeptide gene enhancer in B cells inhibitor) and Ccl20 gene expression (Figure 4C). M cells serve as key sentinels for antigen presentation within the gut and are classically described as being induced by RANKL. Spi-B, the key transcription factor linked with M cells, was not observed in the scRNASeq data [15]. Under these conditions, the M cell induction disproportionately observed with the combination of NOX1-deficiency plus TNFα treatment, thereby defining a two-hit model for M cell induction. (Figure 4B).
Figure 4. scRNASeq identifies marked M cell induction with Nox1-deficiency plus TNFα stimulation, rescued by H2O2 and paraquat.
A. UMAP of joint subclustering (left) of putative colonoid DCS/goblet cells clustering. Heatmap (Right) of top 10 upregulated genes in each cluster from sub-clustering of putative DCS/goblet cells from colonoid scRNAseq shows unexpected presence of M cells. B. Stacked bar graph of subcluster percentages stratified by Nox1 genotype and TNFα treatment. C. Heatmap of known M cell markers of putative colonoid DCS/goblet cells. D. UEA1 staining of Nox1WT and deficient monolayer cells stimulated with TNFα or RANKL for 72 hours. Representative images of each group are shown. Scale bar denotes 200μm. Quantification of M cell percentage was performed on 42 images for each treatment group. *, P < 0.05 (two-sided Wilcoxon signed-rank test). E. Representative UEA1 staining of TNFα or RANKL stimulated NOX1KO colonoids co-treated with H2O2 or Paraquat. M cell percentages were quantitated on 42 images per treatment group. *, P <0.05 (two-sided Wilcoxon signed-rank test).
Validation of M cell induction with NOX1 deficiency and rescue by exogenous H2O2 validating a two-hit model
We validated the induction of M cells by testing for induction of UEA1-positive cells [26],[27] with both TNFα and RANKL treatment in Nox1KO compared to wild-type colonoids (Figure 4D, Supplementary table 3). Enhanced UEA1-positive cells were observed with Nox1KO colonoids with either TNFα (average = 27.5%) or RANKL (average = 26.9%) treatment. Finally, we observed significant rescue of M cell induction with treatment with both H2O2 or paraquat, with a trend toward greater rescue with H2O2 (Figure 4E, Supplementary table 4). Exogenous administration of H2O2 models paracrine effects to a greater extent than paraquat, which enhances mitochondrial ROS production [28]. This demonstrates a two-hit model (TNFα plus NOX1-driven ROS deficiency) for induction of M cells, involving TNFα-induction of ROS, which normally serves to maintain stem cells. Absent ROS plus TNFα results in marked induction of M cells (Figure 4B, 4D), which is substantially rescued by exogenous H2O2 administration (Figure 4E, Supplementary table 4).
In vivo validation of NOX1-mediated M cell induction, RNA velocity and gene signature correlation in ulcerative colitis implicate an altered stem cell niche in chronic inflammation
To explore localization of ROS, we isolated crypts from healthy human colon stimulated with TNF, with and without diphenyleneiodonium chloride (DPI) treatment, which inhibits ROS formation (Figure 5A). In the absence of DPI, most crypts demonstrate nitroblue tetrazolium (NBT) chloride staining at the apical surface, consistent with secretion and functional effects throughout the crypt lumens. We next sought to validate genotype-dependent differences in vivo by treating NOX1 KO and wild-type mice with DSS 5% for 5 days. In our single cell data, GP2 was exclusively observed in M cells, with no expression observed in deep crypt secretory or goblet cells. We further validated by RT-PCR on GP2 (Fig 5B) (P-value = 0.03). GP2 functions as an M cell transcytotic receptor for type 1 piliated bacteria to elicit immune responses [29]. Importantly, we observed significant induction (P-value < 0.0001) of basal lymphoplasmacytosis in NOX1 KO compared to wild-type mice (Figure 5C-D). The recruitment of lymphocytes and plasma cells between the crypt bases and muscularis mucosae reflects chronic inflammation. We observed particularly marked UEA1 staining of crypts (Figure 5E), most marked at crypt bases (Figure 5F), implicating altered stem cell niche function in NOX1-deficient mice with DSS injury. To validate the relationship between UEA1 positive cells and basal lymphoplasmacytosis, we performed immunofluorescence staining of the murine colonic tissue with the B lymphocyte marker (B220). B220 cells were localized to the crypt base and in close proximity to UEA1 positive cells (Figure 5G left panel). We showed significantly increased of B220 positive cell numbers and surface areas in Nox1KO under DSS treatment (Figure 5G, right panel). Together with increase of M cells in Nox1KO during DSS injury (Figure 5E), Our results indicate the importance of ROS in maintaining stem cell niche via inhibiting M cell differentiation thus preventing the formation of basal lymphoplasmacytosis.
Figure 5. Isolated crypt ROS secretion and in vivo validation of NOX1 genotype effects.
A. Representative images of ROS production via NBT formazan (blue) from direct ex vivo colonic biopsies and colonic organoids (left and middle panels). The black arrows indicate NBT formazan. Scale bars 10 μm. ROS (Blue) is localized to the lumen of crypt. Quantification of NBT positive crypts fraction of untreated and DPI treated direct ex vivo colonic biopsies from 4 biopsies (901 crypts in total) (Right Panel, Bottom). ****, p<0.0001 (Paired t-test). B. RT-PCR results of GP2 from 5% DSS or water-treated mice for 5 days (n=4 Nox1WT+water; n=4 Nox1KO+water; n=7 Nox1WT+ 5%DSS; n=7 Nox1KO+5%DSS). *, p<0.05 (Mann-Whitney test). C. Fraction of colonic area with basal lymphoplasmacytosis of 5% DSS or water treated mice. (n=4 Nox1WT+water; n=4 Nox1KO+water; n=7 Nox1WT+ 5%DSS; n=7 Nox1KO+5%DSS). * p<0.05 (Mann-Whitney test). D. Representative H&E images of Nox1WT+DSS and Nox1KO+DSS. Black rectangles demonstrate expansion of basal lymphoplasmacytosis with the Nox1KO. Scale bars 100 μm. E. Quantitation of UEA1 intensity of 65 crypts (n=6 Nox1WT+water; n=11 Nox1KO+water; n=24 Nox1WT+5%DSS; n=24 Nox1KO+ 5%DSS). ****, p<0.0001 (Mann-Whitney test) F. Representative immunofluorescence images of UEA1 in Nox1WT-DSS and Nox1KO-DSS. Scale bars 200 μm. White arrow indicates crypt base. G. Representative immunofluorescence images of B220 and UEA1 (Left Panel). Scale bars 50 μm. Quantification of B220 positive area and cell number (Middle and Right Panel) from 23 images (n=13 Nox1WT+5%DSS; n=10 Nox1KO+5%DSS). ****, p<0.0001 (Mann-Whitney test).
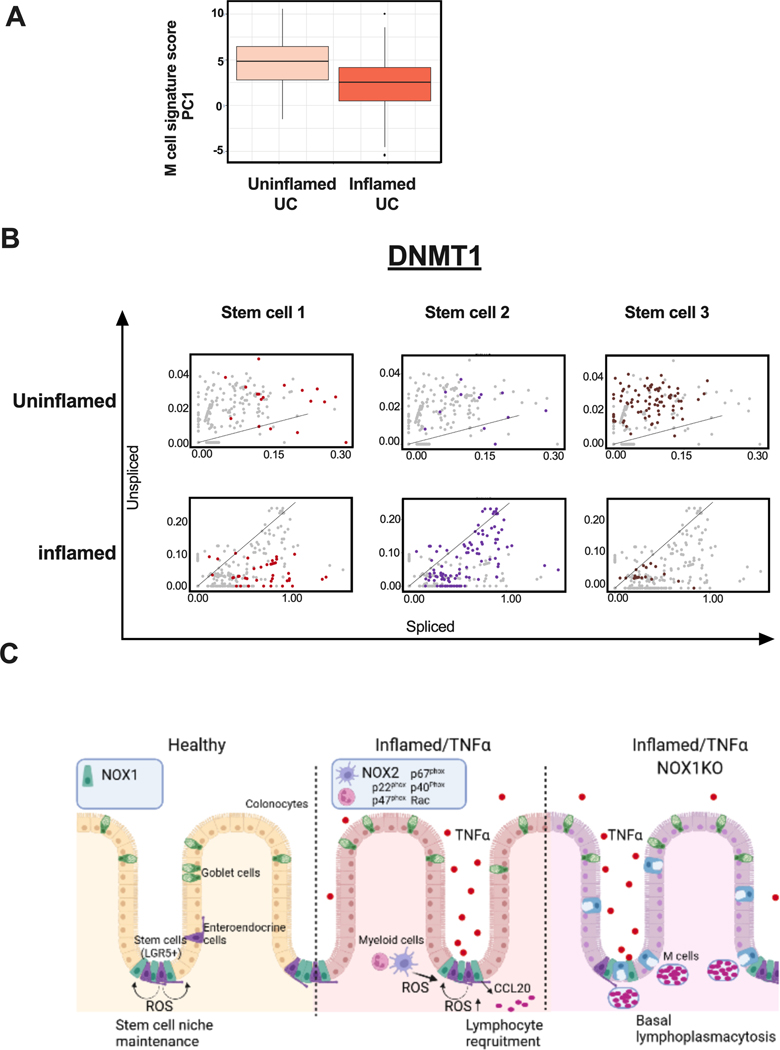
To determine whether this M cell gene signature is correlated with ulcerative colitis generally, we performed principal components analysis of the 111 genes defined by our single cell enteroid data on a bulk UC dataset comparing inflamed vs. uninflamed UC (supplementary table 5). PC1 (Figure 6A) demonstrated significant correlation of this M cell gene module (P-value = 0.0002). Finally, to define more specific transcription factor mechanisms, we performed RNA velocity analysis[30] of the direct ex-vivo data from human UC data. scVelo estimates steady-state transcription for individual transcripts (Figure 6B, diagonals), with increased unspliced (Y-axis) and spliced (X-axis) gene expression indicating increasing and decreasing RNA velocity, respectively. Of the 4 transcription factors, DNMT1 showed the most consistent trends in SC1–3. Specifically, we observed increasing RNA velocity in uninflamed UC and decreasing RNA velocity in uninflamed UC in all stem cell clusters. SC3 is characteristic of uninflamed stem cells, is more abundant in uninflamed tissues, and contains the most cells with increasing RNA velocity (Figure 6B), reflecting baseline ROS secretion present. SC2 is common in inflamed regions, and the decreasing RNA velocity of DNMT1 would infer that suppression of differentiation that DNMT1 mediates in health is being released in inflamed tissue, resulting in aberrant differentiation (e.g. Paneth cell metaplasia) commonly observed.
Figure 6. M cell signatures and DNMT1 RNA velocity correlated to UC inflammation status, implicating ROS-mediated mechanisms driving chronic inflammation.
A. Box whisker plot of UC patients’ M cell gene signature scores comparing bulk RNASeq (144 inflamed vs. 167 uninflamed rectal UC)-derived composite scores for PC1 (36.7% variance explained). Supplementary table 5 includes scores for the first 10 PCs. B. RNA velocity of DNMT1 in UC stem cells. C. Model for ROS-mediated, genotype-dependent differences in generating lymphoid aggregates, contributing to UC inflammation. i) Healthy: wild-type NOX1-mediated maintenance of the stem cell niche, ii) Inflamed/TNFα: expansion of CCL20+ cells generally, recruiting CCR6+ lymphocytes, and iii) Inflamed/TNFα/Nox1KO: ROS deficiency, together with TNFα treatment, induces M cells expansion and leads to basal lymphoplasmacytosis.
DISCUSSION
The association to IBD of loss-of-function mutations in multiple ROS-generating genes expressed in myriad cells (epithelium, myeloid cells, endothelial cells [31] traditionally, with recent reports also including myofibroblasts [32]) indicates that ROS deficiency is a particularly important pathogenic mechanism. ROS deficient mutations lead to increased colitis susceptibility and loss of epithelial barrier function in mice [33]. However, depending on the context, ROS generation can have inflammatory effects as well, driven by local hypoxia, microbiota and redox states[34]. Major therapies for moderate to severe IBD currently include monoclonal antibodies blocking proinflammatory cytokines (anti-TNFα, anti-IL12/23), and agents altering leukocyte trafficking [35]. Given the critical importance of epithelial barrier and complete mucosal healing in long-term IBD outcomes [36], there is a paucity of therapies that specifically target epithelial cells. NOX1, expressed most highly in intestinal LGR5+ stem cells, therefore represents a particularly powerful pathway to examine IBD-relevant (e.g., TNFα stimulation) and fundamental mechanisms of epithelial stress responses.
Single-cell transcriptomics have highlighted the critical role of heterotypic cell interactions in homeostasis and disease [37]. These interactions affect cellular differentiation from stem and other pluripotent cells, tissue development (e.g., Wnt pathways), and mechanisms of tissue repair and regeneration. We have leveraged both direct ex vivo intestinal biopsies and in vitro colonoid studies stratified on NOX1 genotype to define general and genotype-specific insight into epithelial differentiation. NOX1 mutations have been described with both subtypes of IBD, namely Crohn’s disease and UC [6]; we chose to focus on UC given the generally greater role for epithelial cell differentiation [38] and its more uniform, general nature (Crohn’s disease is more genetically and phenotypically variable than UC ). We focused on complete murine knockouts, as many of the human-associated mutations demonstrate only nominal decreases in ROS production with limited genetic penetrances [6]; most of the pathways implicated here are highly conserved in murine models. It is possible that loss-of-function mutations in other NADPH oxidase genes mediate similar effects, meriting further study.
Central among these conserved pathways is stem cell maintenance in somatic tissues, which has been previously reported to be altered permanently in UC[39]. In UC, CCL20 is enriched in inflammation-induced SC2 stem cells (Supplementary Fig 2A) and TNFα induces CCL20 in colonoids (Supplementary Figure 3). CCL20 recruits CCR6-expressing B and T cell lymphocytes. These results indicate a potential role of SC2-like stem cells in recruiting lymphocytes, observed in greater fractions in DSS-treated NOX1 knockouts compared to wild-type mice (Figure 5C). In the absence of stress, intact ROS production by epithelial, immune and stromal cells serves to maintain stem cells, with a role for DNMT1 in maintaining progenitor function during replication [24]. That DNMT1 is reduced in Nox1ko compared to Nox1 wild-type colonoids (P-value = 9.48 × 10−40) is consistent with a role for ROS maintenance of stem cells. Anti-TNFα therapy is the primary agent used to treat moderate to severe IBD [40]. TNFα has marked gut pleiotropic effects, reflecting the broad distribution of its receptors in multiple cell clusters (Supplementary Figure 4A). Notably, the putative DCS/goblet cell cluster observed in colonoids (Figure 4C) contains substantial M cells (likely derived from LGR5+ stem cells [15]) by mRNA and UEA1 staining, most marked in TNFα-treated Nox1KO colonoids (Figure 4B, 4D). M cells are typically described in the small intestine, but have also been described throughout the gastrointestinal tract [12,41]. RANKL is classically regarded as being essential to M cell development [15], with prior reports demonstrating an augmenting effect of TNFα [42]. RANKL expression in ILC3 cells (Supplementary Figure 4B) contributes to developmental gastrointestinal lymphoid tissue formation, with innate immunity developing first; importantly, gut developmental pathways are co-opted with inflammation and stress responses [43].
Here, we demonstrate that TNFα effects on epithelia are, in part, indirectly mediated ROS effects on differentiation of the stem cell niche. Direct effects of TNFα on ROS secretion are myriad and include RIP1-contatining signaling complexes[44]. RANKL is dispensable for M cell differentiation and CCL20 induction in a model involving TNFα stimulation combined with decreased ROS. As with IBD generally, an unpredictable subset of NOX1 mutant carriers with IBD respond to anti-TNFα treatment [6], reflecting a complex pathogenesis and therapeutic response [37,45]. Given the present paucity of therapies targeting the epithelium, new approaches targeting oxidative stress and redox states of epithelial stem cells should be prioritized. Furthermore, given the key role of ROS in microbiome interactions[34] and perianal diseases [46], our findings of key transcription factors driving aberrant stem cell maintenance and differentiation may provide insight into the prevention and treatment of perianal fistulae.
Supplementary Material
Significance of this Study.
1. What is already known on this topic
Loss-of-function mutations in multiple epithelial (in particular NOX1) or myeloid cell-expressed genes producing reactive oxygen species (ROS) are associated with inflammatory bowel disease (IBD).
NOX1 is expressed in epithelial cells, and NOX1-deficiency favors secretory cell differentiation.
Interactions between CCL20 chemokine and CCR6-expressing lymphocytes contribute to chronic gut inflammation
Single-cell transcriptomics is a powerful tool to dissect heterotypic cell-cell interactions and uncover novel disease mechanisms.
2. What this study adds
NOX1 is expressed in stem cells, and Nox1 is essential for TNFα induced ROS production.
Transcription factor induction observed in inflamed vs. uninflamed human ulcerative colitis are mirrored by TNFα stimulated colonoids. Unstimulated NOX1-deficient colonoids express lower levels of many inflammation-induced transcription factors which play a key role in stem cell maintenance.
Marked induction of M cells in Nox1-deficient, TNFα-treated colonoids expressing CCL20 can potentially lead to subsequent lymphocyte recruitment.
Exogenous H2O2 rescues M cell induction in Nox1-deficient, TNFα-treated colonoids
With DSS injury, NOX1-deficient mice demonstrate increased M cell (GP2 and UEA1) and B220 positive basal lymphoplasmacytosis compared to wild-type mice, highlighting markers of chronic inflammation.
3. How this study might affect research, practice or policy
ROS secretion within intestinal crypt bases is required for optimal stem cell niche function. ROS-based and epithelial stem cell gene expression may serve as discriminating biomarkers of epithelial wound repair at various stages of disease and treatment.
Epithelial stem cell function may be optimized by therapies directed at oxidative states (e.g. hyperbaric oxygen therapy), which have shown some benefit in IBD patients.
ACKNOWLEDGEMENTS
We gratefully acknowledge the patients who participating in these studies. Microscopy were performed at the Microscopy CoRE at the Icahn School of Medicine at Mount Sinai with assistance from Nikos Tzavaras and Shilpa Dilipkumar. Single cell seq experiments were performed at the Human Immune Monitor Center at the Icahn School of Medicine at Mount Sinai with assistance from Travis Dawson and Laura A. Walker. Histology processing were performed at Mount Sinai biorepository and pathology core with the assistance from Alan Soto.
FUNDING
This study was supported by U01 DK062422, U24 DK052429, R01 DK123758 (JHC), R01 DK106593 (JHC). AA is supported by Deutsche Forschungsgemeinschaft (AZ 167/1-1).
Footnotes
COMPETING INTERESTS STATEMENT
The authors declare no competing financial interests.
PATIENT AND PUBLIC INVOLVEMENT
Patients and/or the public were not involved in the design, or conduct, or reporting, or dissemination plans of this research.
PATIENT CONSENT FOR PUBLICATION Not required.
ETHICS APPROVAL This study was approved by Institutional Animal Care and Use Committee Institutional Review Board at Mount Sinai school of medicine.
REFERENCES
- 1.Rae J, Newburger PE, Dinauer MC, et al. X-Linked chronic granulomatous disease: mutations in the CYBB gene encoding the gp91-phox component of respiratory-burst oxidase. Am J Hum Genet 1998;62:1320–31. doi: 10.1086/301874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Matute JD, Arias AA, Wright NAM, et al. A new genetic subgroup of chronic granulomatous disease with autosomal recessive mutations in p40 phox and selective defects in neutrophil NADPH oxidase activity. Blood 2009;114:3309–15. doi: 10.1182/blood-2009-07-231498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Winkelstein JA, Marino MC, Johnston RB, et al. Chronic granulomatous disease. Report on a national registry of 368 patients. Medicine (Baltimore) 2000;79:155–69. doi: 10.1097/00005792-200005000-00003 [DOI] [PubMed] [Google Scholar]
- 4.Segal BH, Leto TL, Gallin JI, et al. Genetic, biochemical, and clinical features of chronic granulomatous disease. Medicine (Baltimore) 2000;79:170–200. doi: 10.1097/00005792-200005000-00004 [DOI] [PubMed] [Google Scholar]
- 5.Huang C, De Ravin SS, Paul AR, et al. Genetic Risk for Inflammatory Bowel Disease Is a Determinant of Crohn’s Disease Development in Chronic Granulomatous Disease. Inflamm Bowel Dis 2016;22:2794–801. doi: 10.1097/MIB.0000000000000966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schwerd T, Bryant RV, Pandey S, et al. NOX1 loss-of-function genetic variants in patients with inflammatory bowel disease. Mucosal Immunol 2018;11:562–74. doi: 10.1038/mi.2017.74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hayes P, Dhillon S, O’Neill K, et al. Defects in NADPH Oxidase Genes NOX1 and DUOX2 in Very Early Onset Inflammatory Bowel Disease. Cell Mol Gastroenterol Hepatol 2015;1:489–502. doi: 10.1016/j.jcmgh.2015.06.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Grasberger H, Magis AT, Sheng E, et al. DUOX2 variants associate with preclinical disturbances in microbiota-immune homeostasis and increased inflammatory bowel disease risk. J Clin Invest 2021;131:141676. doi: 10.1172/JCI141676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Karin M, Clevers H. Reparative inflammation takes charge of tissue regeneration. Nature 2016;529:307–15. doi: 10.1038/nature17039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Coant N, Ben Mkaddem S, Pedruzzi E, et al. NADPH oxidase 1 modulates WNT and NOTCH1 signaling to control the fate of proliferative progenitor cells in the colon. Mol Cell Biol 2010;30:2636–50. doi: 10.1128/MCB.01194-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sasaki N, Sachs N, Wiebrands K, et al. Reg4 + deep crypt secretory cells function as epithelial niche for Lgr5 + stem cells in colon. Proceedings of the National Academy of Sciences 2016;113:E5399–407. doi: 10.1073/pnas.1607327113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dillon A, Lo DD. M Cells: Intelligent Engineering of Mucosal Immune Surveillance. Front Immunol 2019;10:1499. doi: 10.3389/fimmu.2019.01499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.DeRoche TC, Xiao S-Y, Liu X. Histological evaluation in ulcerative colitis. Gastroenterology Report 2014;2:178–92. doi: 10.1093/gastro/gou031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yeung MM, Melgar S, Baranov V, et al. Characterisation of mucosal lymphoid aggregates in ulcerative colitis: immune cell phenotype and TcR-gammadelta expression. Gut 2000;47:215–27. doi: 10.1136/gut.47.2.215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.de Lau W, Kujala P, Schneeberger K, et al. Peyer’s patch M cells derived from Lgr5(+) stem cells require SpiB and are induced by RankL in cultured “miniguts.” Mol Cell Biol 2012;32:3639–47. doi: 10.1128/MCB.00434-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.VanDussen KL, Liu T-C, Li D, et al. Genetic variants synthesize to produce paneth cell phenotypes that define subtypes of Crohn’s disease. Gastroenterology 2014;146:200–9. doi: 10.1053/j.gastro.2013.09.048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Miyoshi H, Stappenbeck TS. In vitro expansion and genetic modification of gastrointestinal stem cells in spheroid culture. Nat Protoc 2013;8:2471–82. doi: 10.1038/nprot.2013.153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen E, Chuang L-S, Giri M, et al. Inflamed Ulcerative Colitis Regions Associated With MRGPRX2-Mediated Mast Cell Degranulation and Cell Activation Modules, Defining a New Therapeutic Target., Gene Expression Omnibus, GEO# GSE150516, April 1,2021, 10.1053/j.gastro.2020.12.076 [DOI] [PMC free article] [PubMed]
- 19.Gettler K, Levantovsky R, Moscati A, et al. Common and rare variant prediction and penetrance of IBD in a large, multi-ethnic, health system-based biobank cohort. Gene Expression Omnibus, GEO# GSE150516, April 1,2021, 10.1053/j.gastro.2020.12.034 [DOI] [PMC free article] [PubMed]
- 20.Lambert SA, Jolma A, Campitelli LF, et al. The Human Transcription Factors. Cell 2018;172:650–65. doi: 10.1016/j.cell.2018.01.029 [DOI] [PubMed] [Google Scholar]
- 21.Patel KK, Miyoshi H, Beatty WL, et al. Autophagy proteins control goblet cell function by potentiating reactive oxygen species production. EMBO J 2013;32:3130–44. doi: 10.1038/emboj.2013.233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Beumer J, Clevers H. Cell fate specification and differentiation in the adult mammalian intestine. Nat Rev Mol Cell Biol 2021;22:39–53. doi: 10.1038/s41580-020-0278-0 [DOI] [PubMed] [Google Scholar]
- 23.Smillie CS, Biton M, Ordovas-Montanes J, et al. Intra- and Inter-cellular Rewiring of the Human Colon during Ulcerative Colitis. Cell 2019;178:714–730.e22. doi: 10.1016/j.cell.2019.06.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sen GL, Reuter JA, Webster DE, et al. DNMT1 maintains progenitor function in self-renewing somatic tissue. Nature 2010;463:563–7. doi: 10.1038/nature08683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jung S-N, Yang WK, Kim J, et al. Reactive oxygen species stabilize hypoxia-inducible factor-1 alpha protein and stimulate transcriptional activity via AMP-activated protein kinase in DU145 human prostate cancer cells. Carcinogenesis 2008;29:713–21. doi: 10.1093/carcin/bgn032 [DOI] [PubMed] [Google Scholar]
- 26.Jang MH, Kweon M-N, Iwatani K, et al. Intestinal villous M cells: An antigen entry site in the mucosal epithelium. Proceedings of the National Academy of Sciences 2004;101:6110–5. doi: 10.1073/pnas.0400969101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Giannasca PJ, Giannasca KT, Leichtner AM, et al. Human intestinal M cells display the sialyl Lewis A antigen. Infect Immun 1999;67:946–53. doi: 10.1128/IAI.67.2.946-953.1999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rodríguez-Colman MJ, Schewe M, Meerlo M, et al. Interplay between metabolic identities in the intestinal crypt supports stem cell function. Nature 2017;543:424–7. doi: 10.1038/nature21673 [DOI] [PubMed] [Google Scholar]
- 29.Hase K, Kawano K, Nochi T, et al. Uptake through glycoprotein 2 of FimH(+) bacteria by M cells initiates mucosal immune response. Nature 2009;462:226–30. doi: 10.1038/nature08529 [DOI] [PubMed] [Google Scholar]
- 30.Bergen V, Lange M, Peidli S, et al. Generalizing RNA velocity to transient cell states through dynamical modeling. Nat Biotechnol 2020;38:1408–14. doi: 10.1038/s41587-020-0591-3 [DOI] [PubMed] [Google Scholar]
- 31.Pendyala S, Gorshkova IA, Usatyuk PV, et al. Role of Nox4 and Nox2 in hyperoxia-induced reactive oxygen species generation and migration of human lung endothelial cells. Antioxid Redox Signal 2009;11:747–64. doi: 10.1089/ars.2008.2203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wu N, Sun H, Zhao X, et al. MAP3K2-regulated intestinal stromal cells define a distinct stem cell niche. Nature Published Online First: 3 March 2021. doi: 10.1038/s41586-021-03283-y [DOI] [PubMed]
- 33.Aviello G, Singh AK, O’Neill S, et al. Colitis susceptibility in mice with reactive oxygen species deficiency is mediated by mucus barrier and immune defense defects. Mucosal Immunol 2019;12:1316–26. doi: 10.1038/s41385-019-0205-x [DOI] [PubMed] [Google Scholar]
- 34.Aviello G, Knaus U. ROS in gastrointestinal inflammation: Rescue Or Sabotage?: ROS and GI inflammation. British Journal of Pharmacology 2017;174:1704–18. doi: 10.1111/bph.13428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Neurath MF. Current and emerging therapeutic targets for IBD. Nat Rev Gastroenterol Hepatol 2017;14:269–78. doi: 10.1038/nrgastro.2016.208 [DOI] [PubMed] [Google Scholar]
- 36.Pineton de Chambrun G, Peyrin-Biroulet L, Lémann M, et al. Clinical implications of mucosal healing for the management of IBD. Nat Rev Gastroenterol Hepatol 2010;7:15–29. doi: 10.1038/nrgastro.2009.203 [DOI] [PubMed] [Google Scholar]
- 37.Nayar S, Morrison JK, Giri M, et al. A myeloid-stromal niche and gp130 rescue in NOD2-driven Crohn’s disease. Nature Published Online First: 31 March 2021. doi: 10.1038/s41586-021-03484-5 [DOI] [PMC free article] [PubMed]
- 38.Mokry M, Middendorp S, Wiegerinck CL, et al. Many inflammatory bowel disease risk loci include regions that regulate gene expression in immune cells and the intestinal epithelium. Gastroenterology 2014;146:1040–7. doi: 10.1053/j.gastro.2013.12.003 [DOI] [PubMed] [Google Scholar]
- 39.Dotti I, Mora-Buch R, Ferrer-Picón E, et al. Alterations in the epithelial stem cell compartment could contribute to permanent changes in the mucosa of patients with ulcerative colitis. Gut 2017;66:2069–79. doi: 10.1136/gutjnl-2016-312609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Danese S, Vuitton L, Peyrin-Biroulet L. Biologic agents for IBD: practical insights. Nat Rev Gastroenterol Hepatol 2015;12:537–45. doi: 10.1038/nrgastro.2015.135 [DOI] [PubMed] [Google Scholar]
- 41.Bennett KM, Parnell EA, Sanscartier C, et al. Induction of Colonic M Cells during Intestinal Inflammation. Am J Pathol 2016;186:1166–79. doi: 10.1016/j.ajpath.2015.12.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wood MB, Rios D, Williams IR. TNF-α augments RANKL-dependent intestinal M cell differentiation in enteroid cultures. Am J Physiol Cell Physiol 2016;311:C498–507. doi: 10.1152/ajpcell.00108.2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Elmentaite R, Ross ADB, Roberts K, et al. Single-Cell Sequencing of Developing Human Gut Reveals Transcriptional Links to Childhood Crohn’s Disease. Dev Cell 2020;55:771–783.e5. doi: 10.1016/j.devcel.2020.11.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kim Y-S, Morgan MJ, Choksi S, et al. TNF-induced activation of the Nox1 NADPH oxidase and its role in the induction of necrotic cell death. Mol Cell 2007;26:675–87. doi: 10.1016/j.molcel.2007.04.021 [DOI] [PubMed] [Google Scholar]
- 45.Martin JC, Chang C, Boschetti G, et al. Single-Cell Analysis of Crohn’s Disease Lesions Identifies a Pathogenic Cellular Module Associated with Resistance to Anti-TNF Therapy. Cell 2019;178:1493–1508.e20. doi: 10.1016/j.cell.2019.08.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Denson LA, Jurickova I, Karns R, et al. Clinical and Genomic Correlates of Neutrophil Reactive Oxygen Species Production in Pediatric Patients With Crohn’s Disease. Gastroenterology 2018;154:2097–110. doi: 10.1053/j.gastro.2018.02.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.