Abstract
Human society has been burdened by psychiatric disorders throughout the course of its history. The emergence and rapid advances of human brain organoid technology provide unprecedented opportunities for investigation of potential disease mechanisms and development of targeted or even personalized treatments for various psychiatric disorders. In this review, we summarize recent advances for generating organoids from human pluripotent stem cells to model distinct brain regions and diverse cell types. We also highlight recent progress, discuss limitations, and propose potential improvements in using patient-derived or genetically engineered brain region-specific organoids for investigating various psychiatric disorders.
Keywords: Patient iPSC, patterning, brain organoids, psychiatric disorders, cellular modeling
Introduction
Psychiatric disorders, including depression, bipolar disorder (BP), schizophrenia and other psychoses, cause great suffering to patients and their families, and impose a growing emotional and economic burden worldwide (1). Characterized by significant cognitive, emotional and behavioral disturbances, these disorders have been considered as developmental brain disorders (2, 3). Understanding underlying pathological mechanisms is crucial for developing effective treatments. Most psychiatric disorders are polygenic, and even mutations of a single gene can contribute to different symptoms. Mutations in DISC1, for instance, have been associated with autistic spectrum disorders (ASD), schizophrenia, BP, and major depression (4, 5). This makes psychiatric disorders difficult to recapitulate in traditional genetically modified animal models. Evolutionary increase in the complexity of human brain architecture and functions also makes it challenging to investigate the pathogenesis of human-specific psychiatric disorders with animal models, and many therapies developed based on animal studies have failed in clinical trials (6–8). Hence, utilization of patient brain tissues is important to overcome inter-species differences for both pathological research and therapeutic development. While the availability and usage of human fetal brain materials are subject to ethical and practical limitations, recent advancements in human stem cell-based models provide an alternative approach. For example, human induced pluripotent stem cells (hiPSCs) derived from patients with psychiatric disorders can be differentiated into neurons or glia in 2D cultures or 3D organoids for analyses (9, 10). Monolayer cultures usually possess limited cellular diversity, endogenous cell-cell interactions and tissue organization, whereas 3D brain organoids recapitulate several key characteristics of the developing human brain. Because psychiatric disorders often affect cellular architecture, functions, and interactions of multiple brain regions, brain organoids may provide new avenues for psychiatric research and drug development (11).
Growing numbers of organoid protocols have been developed to recapitulate distinct brain regions and subregions (12) and applied to psychiatric disorder research. In this review, we first briefly summarize current protocols for generating different brain region-specific organoids, then outline achievements and prospects for applying these models to investigate psychiatric disorders. We conclude with limitations and potential future improvements in using brain region-specific organoids for psychiatric disorder research.
Generation of brain region-specific organoids
Current brain region-specific organoid protocols are largely developed by following the in vivo developmental trajectory with temporally defined patterning by different chemical cues (Figure 1), providing an extensive and growing toolbox for psychiatric disorder research (Table 1).
Figure 1. Patient-derived brain region-specific organoids for studies of psychiatric disorders and therapeutic strategies.
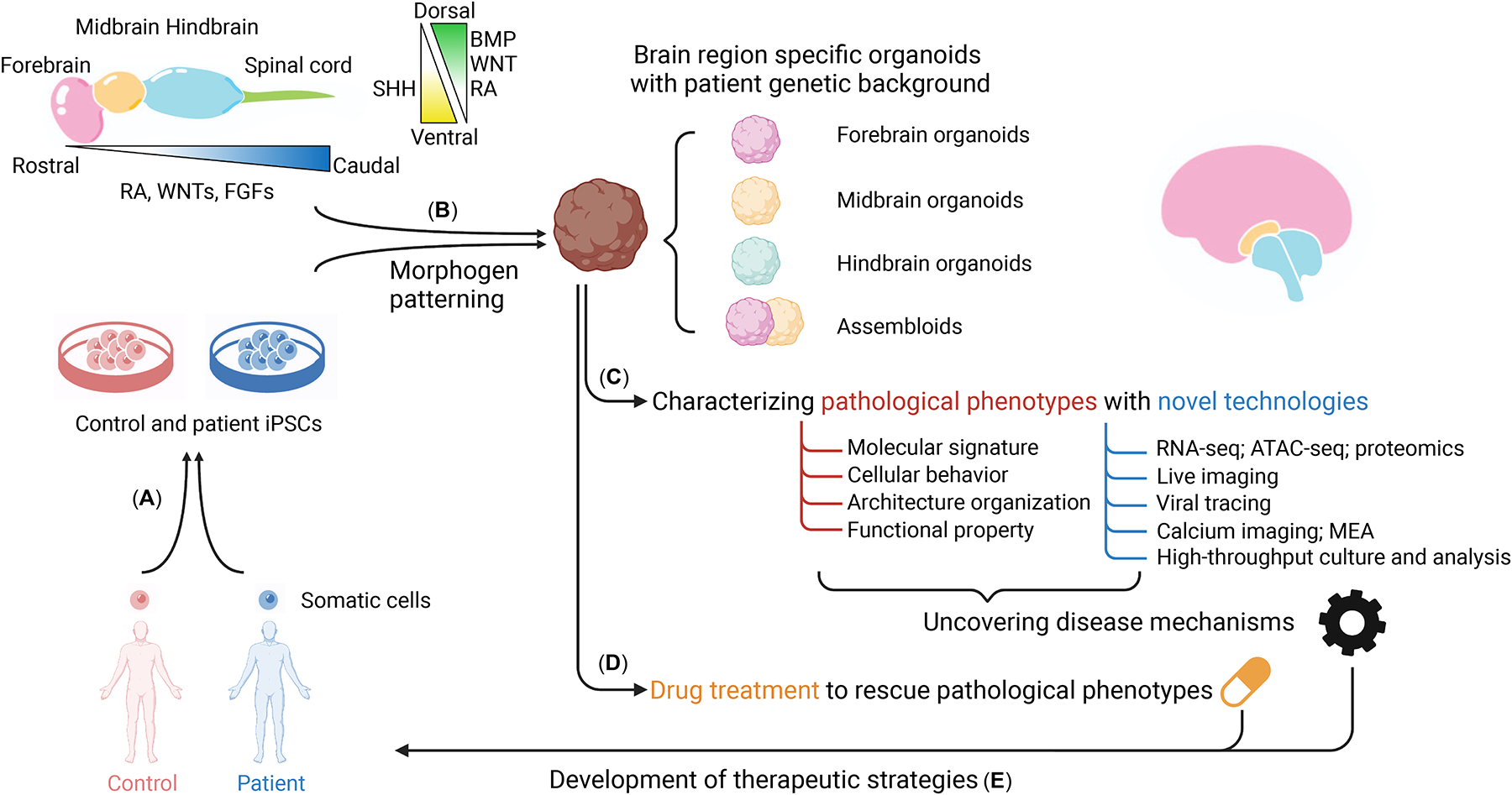
(a) Human iPSCs are established from donor tissue of patients or healthy family controls. (b) Distinct brain region-specific organoids are then generated by patterning with proper morphogens based on the endogenous human brain development process using these hiPSCs. (c) Comparative analysis between patient and control organoids can be performed to reveal pathological phenotypes and mechanisms at molecular, cellular, structural, and functional levels. (d) Based on the knowledge acquired from these studies, brain region-specific organoids can be used for testing drug responses and large-scale drug screening. (e) Eventually, brain region-specific organoids can be used to develop novel and personalized therapeutic strategies for patients.
Table 1.
Brain disorder modeling with brain region-specific organoids.
| Brain organoid subtypes | Psychiatric disorders | Genetic background | Findings | Ref | |
|---|---|---|---|---|---|
| Forebrain organoids | Cortical organoids | Angelman syndrome | Engineered UBE3A KO hESCs and isogenic controls | Hyperactivity of neurons and networks, which can be rescued by paxilline | (29) |
| Cortical organoids | Frontotempor al dementia | Patient iPSCs with tau-V337M and isogenic corrected iPSCs | Affected synaptic maturation and excitatory neuron survivability, which can be rescued by the PIKFYVE kinase inhibitor apilimod | (31) | |
| Cortical organoids | Autism spectrum disorder | Multiple patient iPSCs | Accelerated cell cycle and overproduction of inhibitory neurons | (16) | |
| Cortical organoids | Autism spectrum disorder | Engineered heterozygous mutation in SUV420H1, ARID1B or CHD8 with multiple parental iPSCs | Convergent phenotypes with asynchronous development of inhibitory and excitatory neurons | (25) | |
| Cortical organoids | Autism spectrum disorder | Patient iPSCs with a CNTNAP2 mutation and isogenic corrected iPSCs | Increased neural progenitor proliferation and organoid volume | (26) | |
| Cortical organoids | Schizophrenia | patient iPSCs with a DISC1 mutation | Disruption of cell cycle with delayed mitosis | (27) | |
| Cortical organoids | Schizophrenia | patient iPSCs with a DISC1 mutation | Deficits in cortical neuron fate specification | (19) | |
| Cortical organoids | Fragile X syndrome | Patient iPSCs and isogenic corrected iPSCs | Dysregulated neurogenesis, neuronal maturation and neuronal excitability | (28) | |
| Cortical organoids | Alzheimer’s Disease | Multiple patient iPSCs | Modeled amyloid aggregation and hyperphosphorylated tau protein, which is rescued by treatment with β- and γ-secretase inhibitors | (30) | |
| Cortical organoids | Lissencephaly / Miller-Dieker syndrome | Patient iPSCs with heterozygous deletion of chromosome 17p13.3 and rescue cell lines expressing LIS1 or YWHAE | Reduced organoid size caused by premature neurogenesis and can be rescued by Wnt activation | (24) | |
| Cortical organoids | Lissencephaly / Miller-Dieker syndrome | Patient iPSCs with heterozygous deletion of chromosome 17p13.3 and cell-autonomous corrected line | Reduced organoid size, increased apoptosis and horizontal divisions, and defects in neuronal migration and mitotic spindles | (23) | |
| Cortical organoids | 22q11.2 deletion syndrome | Patient iPSCs | Abnormal neuronal excitability-related gene expression, neuronal excitability and calcium signaling | (76) | |
| Cortical organoids | Microcephaly | Multiple engineered iPSCs with a WdR62 mutation | Reduction in organoid size due to decreased proliferation and premature differentiation of neural progenitor cells | (20) | |
| Cortical organoids | Microcephaly | Patient iPSCs with a ASPM mutation | Reduction in organoid size; neurogenesis defects; defective neuronal activity | (21) | |
| Cortical organoids | Microcephaly | Patient iPSCs from multiple families with missense or frameshift mutations in NARS1 | Disruption of protein synthesis; reduction in organoid size due to reduced RGC proliferation | (22) | |
| Cortical organoids | Microcephaly | Engineered iPSCs with a homozygous mutation in IER3IP1 | Reduced organoid size and rosette size which can be restored using ISRIB; increased unfolded protein response and ER stress; decreased extracellular matrix protein | (101) | |
| Cortical ventral forebrain organoids | Tuberous sclerosis complex | Patient iPSCs with mutations in TSC2 and isogenic corrected iPSC | Brain tumors and cortical malformations caused by over-proliferation of caudal late interneuron progenitor | (125) | |
| Cortical organoids | Rett syndrome | Patient iPSCs with mutations in MECP2 and isogenic corrected iPSCs | Cell migration deficits in distances, speeds, and radial trajectories | (84) | |
| Retinal organoids | Retinitis pigmentosa | Patient iPSCs with frameshift mutations in RPGR and isogenic corrected iPSCs | Photoreceptor defects in morphology, localization, transcriptional profiling, and electrophysiological activity | (126) | |
| ARC organoids | Prader-Willi syndrome | Patient iPSCs with major or minor deletion on the chromosome 15q11-q13 | Differentiation and functional deficits | (44) | |
| Midbrain organoids | Midbrain organoids | Parkinson’s disease | Gene edited iPSCs with LRRK2-G2019S mutation | Accumulation of a-synuclein; mimicked the gene expression profiles in patients | (48) |
| Midbrain organoids | Parkinson’s disease | Patient iPSCs with LRRK2-G2019S mutation | Decreased number and complexity in the expression of mDANs | (49) | |
| Midbrain rganoids | Parkinson’s disease | Patient iPSCs with SNCA triplication and CRISPR/Cas9 corrected isogenic control iPSCs | Elevated accumulation of a-synuclein and reduction in dopaminergic neurons | (50) | |
| Midbrain organoids | Parkinson’s disease | Patient iPSCs with a PINK1 mutation and CRISPR/Cas9 corrected isogenic control iPSCs | Imbalanced proliferation, apoptosis and mitophagy; reduced dopaminergic differentiation, which can be rescued by treatment with HP-β-CD | (52) | |
| Assembloids | Cortical organoids + Microglia assembloids | Alzheimer’s Disease | APP duplicated patient organoids cocultured with APOE3- or APOE4-carrying microglia-like cells | Increased Aβ aggregates and hyperphosphorylation of tau in organoids cocultured with APOE4-microglia | (127) |
| Cortical organoids + Blood vessels assembloids | Alzheimer’s Disease | A mixture of control and inducible ETV2-expressing hESCs | Aβ1–42-oligo treatment led to disruption of tight junctions | (62) | |
| Cortical-ventral forebrain assembloids | Timothy syndrome | Patient iPSCs with a CACNA1C mutation | Abnormal interneuron migration saltation. L-type calcium channel blocker rescued saltatory length | (39) | |
| Cortical-ventral forebrain assembloids | Timothy syndrome | Patient iPSCs with a CACNA1C mutation | Abnormal interneuron migration saltation. GABA-A receptor antagonism rescued saltatory frequency | (83) | |
| Cortical-striatal assembloids | Phelan-McDermid syndrome | Patient iPSCs with chromosome 22q13.3 deletion | Cortico-striatal connectivity defects with abnormal calcium activity | (42) | |
| Cerebral-ganglionic eminence assembloids | Rett syndrome | Patient iPSCs with mutations in MECP2 | Transcriptomic differences; epileptiform-like neural network activity that can be rescued by treatment with pifithrin-α | (90) | |
| Cortical ventral forebrain organoids and assembloids | Rett Syndrome | Patient iPSCs with mutations in MECP2 | Premature development of the deep-cortical layer; neural function defects; impaired interneuron migration | (86) | |
Cerebral cortex
The Sasai group developed the first guided cortical organoid protocol by inhibiting TGFβ and WNT signaling (13). Treatment with retinoic acid (RA) at a later culture stage leads to subregion-specificity resembling the human prefrontal cortex (14). Cortical organoids were also generated with SMAD inhibition, followed by a combination of early WNT inhibition and FGF2/EGF treatment (15, 16), or continuous TGFβ inhibition, and later WNT activation (17, 18). These protocols produce self-organized ventricular structures with cortical-specified neural progenitor cells and neuronal zones with different cortical neural types, with some containing cortical neurons of all six layers (17) and some exhibiting distinct superficial and deep cortical layers (19). These cortical organoids have been widely used to study various brain disorders, such as microcephaly (20–22), lissencephaly (23, 24), ASD (16, 25, 26), schizophrenia (19, 27), Fragile X syndrome (FXS) (28), Angelman syndrome (29), Alzheimer’s Disease (30) and frontotemporal dementia (31) (Table 1).
Hippocampus and choroid plexus
Hippocampal development is regulated by BMP signals secreted from the choroid plexus tissue (32). Hippocampal and choroid plexus organoids can be induced by combinational and timed treatment with BMP and WNT signaling (33–35). Choroid plexus organoids could provide a valuable tool to assess drug permeability into the brain.
Ganglionic eminence
Human cortical interneurons are mainly derived from the ganglionic eminence of the ventral pallium (36–38), which is patterned by high SHH and low WNT activity. Medial ganglionic eminence organoids have been generated by activating the SHH pathway (13, 39–41) and employed to model interneuron migration deficiency in Timothy syndrome. The lateral ganglionic eminence generates the striatum, which contains inhibitory medium spiny neurons that receive unidirectional inputs from excitatory cortical neurons. These features have been recapitulated in striatal organoids generated with a combination of TGFβ activation and WNT inhibition and later RA activation (42), which have been used to model 22q13.3 deletion syndrome (22q13.3DS).
Thalamus and hypothalamus
Thalamic and hypothalamic organoids can be generated using dual-SMAD inhibitors, followed by insulin treatment and then MEK-ERK inhibition for caudal forebrain fate induction (43), or SHH and WNT activation for ventral rostral forebrain induction (17). Organoids resembling the arcuate nucleus, a subregion of the hypothalamus, were generated using dual-SMAD inhibition followed by extended WNT inhibition and triple SHH activation, for modeling Prader-Willi syndrome (44).
Midbrain
Organoid protocols aimed at generating midbrain dopaminergic neurons have been developed using SHH activation (17, 45–50) or combined treatment with FGF8 (17, 45, 46, 50). Midbrain organoids have been used to study Parkinson’s disease (48–52) and treatments (52).
Hindbrain
Cerebellar organoids modeling the dorsal hindbrain have been generated using TGFβ inhibition, FGF2 and insulin for early cerebellar neuroepithelium induction, followed by FGF19 and SDF1 treatments for dorsal tissue differentiation (53). These organoids produced various cerebellar specific neural precursors and provide useful tools for studies of cerebellar-associated diseases. Ventral hindbrain organoids have been generated using SHH for ventralization and RA for caudalization (54) and contain various hindbrain specific cell types, including serotonin-synthesizing neurons, which are promising for studies and drug tests of human 5-HT-related diseases, such as depression.
Assembloids
Various brain region-specific organoid subtypes can be assembled (assembloids) to model crossregional interactions, such as cell migration, axonal projections and synaptic formation, as well as functional interactions, for relevant brain disorders. Many cortical interneurons are derived from the ventral forebrain and migrate into the dorsal cortex during development. Several studies using dorsal-ventral forebrain assembloids show the recapitulation of the saltatory (39) and tangential (40, 41) migration of interneurons and its regulation by neurotransmitter signaling (55). Cortical-striatal assembloids form unidirectional axonal projections and functional connections from excitatory cortical neurons to striatal GABAergic medium spiny neurons (42), resembling the in vivo network connectivity in forebrains. Thalamic-cortical assembloids recapitulate reciprocal thalamic-cortical axonal projections (43), and cortico-motor assembloids model cortical-spinal-muscle circuit connections that can functionally control muscle contraction (56). Hypothalamic-pituitary projections and functional units were also modeled with assembloids (57). We expect many more region-specific protocols and assembly approaches to be developed, which will help to build organoids with more complex cellular populations and regional interactions for studies of relevant psychiatric disorders.
Co-cultured organoids
Patterning methods produce cell populations with similar regional identities and reduce the variability of organoids, however, this consistency comes at the cost of restricted cellular and brain regional diversity. Neuroectoderm derived region-specific organoids rarely produce tissues with non-neuroectodermal origin, such as microglia and vasculature. A co-culture strategy can overcome this caveat and enhance cellular diversity. For example, hiPSC-derived microglia or macrophage precursors readily integrate into dorsal and ventral forebrain (58, 59), or midbrain organoids (60). Co-culturing primitive neural and macrophage progenitors produced cortical organoids with a controllable ratio of microglia (61). Brain organoids with microglia showed improved synaptic pruning, tissue maturation, and functionality. Cortical organoids with vascular-like structures can be generated by co-culturing human stem cells with inducible hETV2-expressing cells (62) or human umbilical vein endothelial cells (63). Blood vessel-associated endothelial cells and pericytes are rarely produced in current brain organoid protocols and have not yet been modeled in co-culture. Transplanting organoids into rodent brains also results in host integrated functional vessels with circulation (64–67).
Taken together, progress in patterning methods and co-culture strategies provides a versatile toolbox of brain organoid protocols that allow heterogeneous cellular diversity and multiple brain region interactions. With these advances, brain organoid technology is moving toward making a significant impact on psychiatric disorder research.
Patient iPSC-derived brain region-specific organoids for psychiatric disorder research
Brain region-specific organoids derived from patients with clinical profiles provide a powerful platform to maintain the patient genomic context to recapitulate known disease phenotypes, investigate underlying mechanisms, and develop rational drug treatments (Figure 1). These studies can provide insights at multiple levels for psychiatric disorders.
At the molecular level
Brain development is dynamically governed by comprehensive molecular processes, which are ultimately coded in the genome. Rapid advances in high-throughput molecular profiling technologies make it possible to systematically compare molecular signatures of human fetal brain tissue with organoids at the bulk or single-cell level. Numerous studies showed that (epi)transcriptomic and epigenomic profiles of brain region-specific organoids reliably reflected the developmental trajectory of the human fetal brain (14, 68–75), laying the foundation for applying patient-derived brain organoids to study pathological molecular events of psychiatric disorders within patient-specific genomic contexts.
Brain region-specific organoids derived from patients with monogenic etiology have been extensively used to investigate downstream molecular consequences. For example, cortical organoids derived from patients with a homozygous mutation in CNTNAP2 revealed differentially expressed genes for cell proliferation and neuronal differentiation, which are enriched for ASD-associated genes (26). Organoids generated from isogenic corrected patient iPSCs or isogenic mutant iPSCs with various genetic backgrounds can be further used to determine causality and understand mechanisms underlying disease phenotypes.
Brain region-specific organoids derived from patients with known large-scale genomic variants allow identification of causal gene(s) and relevant mechanisms for certain pathological phenotypes. For example, cortical organoids derived from patients with heterozygous deletion of chromosome 17p13.3 showed a reduced expansion rate caused by increased asymmetric cell division in ventricular neural stem cells (23, 24). Re-expression of LIS1 or YWHAE, two genes within 17p13.3, in patient-derived organoids partially restored the expansion and cell division defects, revealing their causal roles in controlling ventricular neurogenesis and cortical expansion. Similarly, deletion of the DGCR8 gene was identified as the cause for abnormal neuronal excitability and associated gene expression of 22q11DS with patient-derived cortical organoids (76).
Brain region-specific organoids derived from idiopathic patients can also be used to identify common molecular mechanisms of pathogenesis. Compared with those from unaffected family relatives, cortical organoids derived from multiple idiopathic ASD patients with macrocephaly showed upregulated expression of genes involved in cell proliferation, neuronal differentiation, synaptic assembly, as well as overproduction of GABAergic neurons (16). FOXG1 is among the top upregulated genes and downregulation of FOXG1 in patient-derived organoids rescued the dysregulation of GABAergic neuronal fate. This study highlights the importance of brain region-specific organoids for revealing convergent molecular pathways for psychiatric disorders with unidentified genomic variations.
With advances in multi-omics approaches, including at the single cell level, we expect that many omics datasets, including transcriptomic, epigenomic, proteomic and metabolomic signatures of human brain tissues and organoids, will help to decipher and cross-validate mechanisms of psychiatric disorders at the molecular level.
At the cellular level
Deficits in cellular development processes, such as proliferation, differentiation, migration, axonal projections, and maturation, have been broadly linked to various psychiatric disorders (16, 77–80). Several studies have demonstrated the utility of brain organoids in recapitulating these developmental processes and elucidating their pathological roles in psychiatric disorders. For example, FXS-derived cortical organoids exhibit dysregulated neural progenitor proliferation, neural differentiation and maturation (28). Prader-Willi syndrome patient-derived arcuate organoids also exhibit aberrant neural differentiation and selective loss of POMC-expressing neurons (44). Cortical organoids from Miller-Dieker Syndrome (MDS) patients with lissencephaly showed a reduced organoid expansion rate due to premature neural differentiation (24), increased apoptosis, and defects in cell division and neuronal migration (23). Excitation/inhibition imbalance is known to be associated with psychiatric disorders (81, 82). Overproduction of inhibitory neurons caused by increased inhibitory neuron differentiation and early precursor proliferation was identified as a common feature in ASD patient-derived cortical organoids (16). Dorsal-ventral forebrain assembloids derived from Timothy syndrome patients exhibit abnormal interneuron migration (39, 83).
Together, these studies demonstrated that patient-derived brain region-specific organoids can reveal cellular mechanisms for psychiatric disorders. Leveraging various co-culture or multi-region organoids and innovative imaging technologies, such as label-free live imaging (84), will allow more complex cellular diversity and interactions to be visualized in organoids, which will provide novel insights into cytopathological mechanisms of psychiatric disorders.
At the architectural level
With highly ordered spatial organization and interactions of various cell types, organoids can model many architectural features of the developing human brain and disease-associated deficits. Cortical organoids faithfully resemble early cerebral development by displaying polarized laminar structures, including the subventricular zone (SVZ) harboring proliferating apical radial glia cells, the outer subventricular zone (oSVZ) harboring outer radial glia cells, and the cortical plate (CP) harboring cortical neurons (85). Altered thickness or areas of these laminar structures have been shown in patient organoids with Rett syndrome (RTT) (86), microcephaly (22), and periventricular heterotopia (87). The CP expands in an inside-out manner and separates into six cortical layers. A slicing method developed to bypass diffusion limitations and allow for long-term maintenance generates cortical organoids with laminar expansion and segregation into distinct cortical layers (19). This study further showed that sliced cortical organoids from psychiatric patients with a DISC1 mutation exhibited aberrant cortical layer distribution due to deficits in cortical fate specification (19). At later stages of development, the human cerebral cortex folds and generates gyrifications. PTEN mutations have been demonstrated to expand SVZ/oSVZ layers and promote surface folding in unpatterned brain organoids (88), however, the developmental process could be different in this system because this folding is absent in organoids without the PTEN mutation. A recent on-chip approach to generate cortical folding-like structures by growing organoids in Matrigel-filled thin microchannels showed that a LIS1 heterozygous mutation resulted in lissencephalic organoids with reduced convolutions and cell elasticity (89).
Accumulating evidence is redefining psychiatric disorders as brain circuit disorders (1). Incorporating circuit abnormalities into phenotypic assessments and further tuning these aberrant circuits will substantially contribute to precision diagnostics and therapeutics for psychiatric disorders. Synaptic deficits have been widely identified in patient-derived brain organoids (28, 29, 31, 86). While abnormal long-distance projection synaptic connections were only recently modeled with patient-derived dorsal-ventral forebrain assembloids (90), other types of assembloids also demonstrated reciprocal thalamic-cortical (43) and unidirectional cortical-striatal projections (42). Furthermore, xenotransplantation could promote circuit maturation and incorporate human organoids within host brain circuits (65, 67). In the future, more advanced organoid cultures, combined with trans-synaptic viral tracers, such as rabies and herpes simplex virus, can be used to reveal long-distance projection circuits communicating among various brain regions, or even sensory organs to model pathological defects in psychiatric disorders.
At the functional level
Brain organoids not only recapitulate many cellular and architectural features of the developing brain, but also exhibit some functionalities, such as releasing various signaling molecules (33, 91), producing neural and synaptic activities (15, 17), generating complex network dynamics (29, 90, 92), and even sensing light stimuli (91) and controlling muscle movement (56). Recent advances have just started to realize the potential of organoids for studying functional deficiencies in psychiatric disorders. Whole-cell patch-clamp recording or calcium imaging has revealed altered neural activities at the single cell level from patient organoids in various diseases, such as FXS (28), TS (39), microcephaly (21) and 22q11DS (76). Complex neural network activity involves interactions among distinct brain regions and cell types. Medium spiny neurons in cortical-striatal assembloids from 22q13.3DS patients exhibited elevated spontaneous calcium activity, but recued global network synchronization (42). TS patient-derived cortical organoids showed hypersynchronous network calcium dynamics, which was further exacerbated in cortical-subpallial assembloids (83), consistent with clinical features of frequent recurrent seizures in TS patients (93). Similarly, dorsal-ventral forebrain assembloids derived from RTT patients displayed hypersynchrony and epileptiform-like network activities, consistent with the common symptom of epilepsy in patients (90).
Collectively, patient-derived brain region-specific organoids are shedding light on functional abnormalities in psychiatric disorders. Nevertheless, sophisticated brain functions are built on highly organized tissue with diverse cell types, fully mature cells and complex circuit connections, and shaped through interactions with sensory, motor and other systems. Future efforts, such as long-term cultures and xenotransplantation strategies, will hopefully enhance the potential of brain organoids in functional studies of psychiatric disorders.
Responses to drug treatment
Patient-derived brain region-specific organoids are becoming increasingly important for developing therapeutic strategies. So far, progress has been made by testing specific drugs to restore phenotypic abnormalities observed in patient-derived organoids. MECP2 deficiency has been reported to activate the TP53 pathway and promote neuronal senescence (94). The key physiological phenotype of epileptiform-like network activity in RTT patient assembloids with MECP2 mutations can be restored by a putative TP53 inhibitor, pifithrin-α, and a commonly used anti-seizure medication, sodium valproate (90). Tissue growth defects caused by impaired N-Cadherin/β-Catenin signaling in MDS patient-derived cortical organoids can be ameliorated by the GSK3β inhibitor CHIR99021 (24).
Scalability and high-throughput phenotypic analyses are major roadblocks that hinder the effective applications of brain organoids in drug discovery. Renner et al. (95, 96) developed a robotic technology to control initial cell dispensing, organoid patterning, feeding, staining and analysis, thus creating a scalable workflow for generation and maintenance of human midbrain organoids that is amenable to automatic high-content optical analysis.
Human brain organoids could also play invaluable roles for testing drug toxicity. Due to the inaccessibility of human embryos, certain approved drugs, such as thalidomide and isotretinoin, used by pregnant women resulted in severe birth defects, although they were preclinically tested to be non-teratogenic in animal models (97). Thus, fully exploiting the potential of brain organoids for screening teratogens or drug toxicity will be crucial to avoid similar outcomes in the future.
Genetically engineered brain region-specific organoids for psychiatric disorder research
Human iPSCs and derived brain organoids provide remarkable accessibility for genetic engineering. In addition to revealing disease phenotypes in patient-derived organoids, engineered brain organoids from healthy individuals are increasingly used in investigating roles of genetic risk variants linked to human brain disorders (Figure 2).
Figure 2. Genetically engineered brain organoids derived from healthy donors for functional studies of risk gene variants.
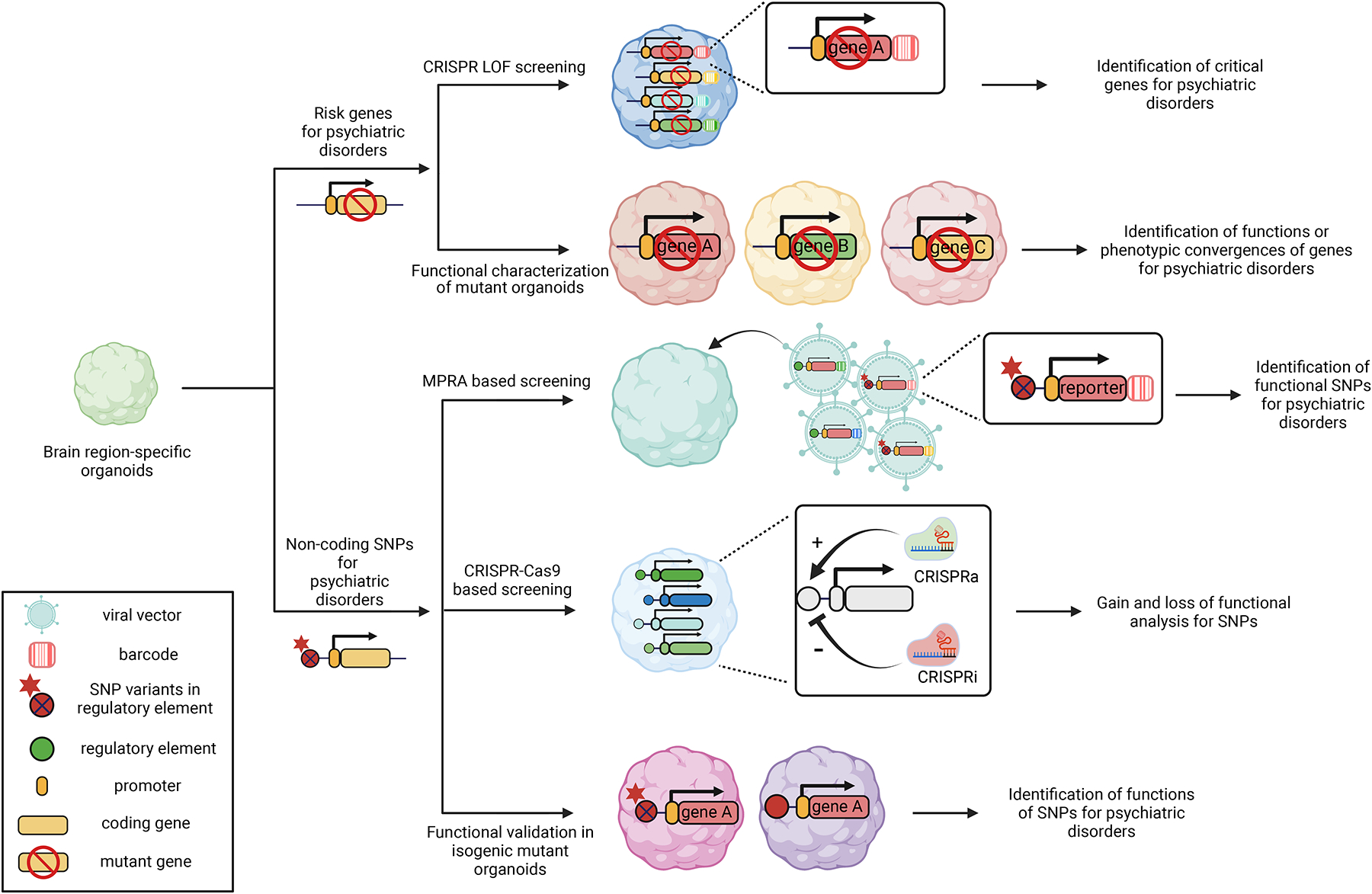
Engineered brain organoids can be used for highly efficient functional screening and in-depth phenotypic convergence studies of protein-coding risk genes and non-coding SNP variants.
Analyses of functions of psychiatric disorder risk genes in human brain development
Advancements in human genetics have led to the identification of an ever increasing number of genes linked to psychiatric disorders. Brain organoids provide powerful platforms for analyzing risk gene functions in diverse cell types at single-cell resolution in a human-tissue context. CRISPR technology not only permits genomic editing of a single gene, but also enables systematic assays of many candidate genes in a multiplex fashion (98, 99). A recent study highlighted how brain organoids could provide insight into phenotypic convergences of multiple risk genes (25). Heterozygous mutations of SUV420H1, ARID1B or CHD8, top ASD risk genes linked to different biological functions (100), were introduced to iPSCs from different healthy donors. Analysis of proteomic and single-cell transcriptomic profiles from generated cortical organoids showed that although the three genes function through distinct molecular pathways, they exhibited common phenotypes of asynchronous development of inhibitory and excitatory cortical neurons. Scalability was further demonstrated in another study, in which 173 microcephaly candidate genes were screened for their effects on cell proliferation using a CRISPR-Cas9-mediated loss-of-function assay in healthy human cortical organoids (101). This study applied a dual barcoding strategy to link individual lineages with a given gene deletion to their lineage sizes, thus overcoming the impact of inherently unequal proliferation dynamics.
Thus, multiplexed CRISPR-based gene editing in brain organoids may provide a framework for high-throughput causative screens, in-depth functional characterization of risk genes, and mechanism discovery for psychiatric disorders.
Linking functional SNPs to target genes in specific cell types
Most psychiatric disease-associated variants reside in non-coding regions (102, 103). Chromatin structure mapping studies suggest that human psychiatric disorder-associated non-coding single-nucleotide polymorphisms (SNPs) are enriched in enhancer-promoter interactions in a cell type-specific manner (104, 105), which can regulate target genes over a long distance via 3D chromatin structures (106). Functions of non-coding SNP candidates have been explored in hiPSCs and derived brain cell cultures by leveraging CRISPR-based genetic engineering (107, 108). High-throughput massively parallel reporter assays (MPRAs) have been widely used for large-scale screening of enhancer–promoter interactions in mammalian brain cells (109–113). An MPRA library can be designed by fusing DNA-barcoded reporter genes with candidate regulatory elements with either reference or risk SNP variants and then integrated into brain organoids. By quantifying barcoded transcripts through single-cell sequencing, MPRA can provide biological relevance of SNPs to disease-related gene expression at a large scale and in specific cell types. However, MPRA is limited in recapitulating the size and endogenous chromatin structural context of regulatory elements. CRISPR-Cas9-based in situ SNP editing (114) overcomes this limitation and can be used for in-depth functional validation of candidate SNPs identified from MPRA or other screening models. This method, however, is low throughput and lacks scalability (115). CRISPR-based tiling deletion (116) or activation/interference (CRISPRa/i) (114, 117, 118) in regulatory elements can be an alternate for increasing the throughput for functional validations of risk SNPs, but with a lower depth. Taken together, non-coding, disorder-associated SNP functions can be potentially studied in-depth with high throughput by integrating these genetic engineering approaches in human iPSC-derived organoids, although currently these technologies have only been applied to 2D cultures.
Current limitations and potential improvement
The field of applying brain organoids for psychiatric research is still nascent and current technologies have considerable limitations. First, human brain organoids can only mimic fetal or, at most, early postnatal brain development rather than mature brains (119), whereas most psychiatric disorders manifest only in postnatal or adult stages. Sustaining long-term healthy cultures through methods such as slicing (19) and air-liquid interface (120), or xenotransplantation, can promote maturation to a limited degree. Recently, a cocktail of pharmacological compounds targeting several signaling pathways significantly accelerates functional and structural maturation in both human cortical organoids and various hiPSC-derived cell types (121), providing a new strategy for promoting organoid maturation. Second, organoids only recapitulate limited human brain architecture. Although layer segregation has been reported, a fully defined six-layer structure with distinct radial unit/column distributions and phenotypic normal gyrification has not been demonstrated in human cortical organoids. Further efforts are needed to improve organoid cultures to show these key architectural features, potentially with bioengineering approaches. Third, despite the emergence of various brain region-specific organoid and assembloid methods, brain organoids with multiple well-organized regions have not been reported. Assembloids do not model the intrinsic multi-region developmental process that generates a smooth continuum and structural intermediaries among different regions, and thus are unable to recapitulate neural circuit formation dynamics during brain region development and could potentially yield artificial connections. A recent study using bioengineering approaches to apply spatial gradient morphogens generated neural tube-like organoids with precursor cells representing multiple brain regions (12). Future studies aiming to maintain these multi-regional neural tubes will potentially produce multi-regional or whole brain organoids, which could provide better insight into endogenous multi-region brain development and interactions as well as disease processes. Fourth, upregulated cellular stress has been reported in brain organoids, which may hinder cell-type specification (72, 119, 122). Cellular stress can be reduced by improving culture conditions, such as using sliced cultures (19), or by xenotransplantation (122). On the other hand, a recent study revealed that cellular stress is restricted to a specific subpopulation and can be removed using bioinformatic analysis (123), and some studies argue that it could reflect a distinct in vitro homeostatic metabolic state (119), and does not affect most cellular identity acquisition process in organoids (124). Lastly, we need to be cautious in the interpretation of data coming just from organoid models. For instance, the high concentration of patterning molecules used in organoid protocols could mask the pathology caused by altered patterning during development in some disease conditions. Organoids lack certain cell types, which are critical for developing psychiatric disorders. A combinational use of organoids as well as in vivo animal models could help to address this concern. Together, limitations and challenges exist and future improvements are needed to make brain organoids a better model for psychiatric research.
Summary
Various methods have been developed to model distinct brain (sub)regions with diverse resident cell types using organoids. Both patient-derived and genetically engineered brain region-specific organoids have been applied in pathological and pharmacological studies of psychiatric disorders and novel insights have been gained in the understanding of disease pathogenesis. Only by fully acknowledging the limitations can we properly interpret data obtained from organoid studies. We envision that future improvements will further unlock the potential of this technology and leaps toward a deeper understanding of psychiatric disorders and develop better and more rational therapeutics.
Acknowledgements
We thank members of Song and Ming laboratories for comments and suggestions. The research in the authors’ laboratories were supported by grants from the National Institutes of Health (R35NS116843 and RF1AG079557 to H.S, and R35NS097370, RF1MH123979, and R01MH125528 to G-l.M.) and the Adelson Medical Research Foundation (to G-l.M.)
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of interest statement
The authors report no biomedical financial interests or potential conflicts of interest.
References
- 1.Insel TR, Cuthbert BN (2015): Medicine. Brain disorders? Precisely. Science. 348:499–500. [DOI] [PubMed] [Google Scholar]
- 2.Lewis DA, Levitt P (2002): Schizophrenia as a Disorder of Neurodevelopment. Annual Review of Neuroscience. 25:409–432. [DOI] [PubMed] [Google Scholar]
- 3.Raedler TJ, Knable MB, Weinberger DR (1998): Schizophrenia as a developmental disorder of the cerebral cortex. Current Opinion in Neurobiology. 8:157–161. [DOI] [PubMed] [Google Scholar]
- 4.Blackwood DH, Fordyce A, Walker MT, St Clair DM, Porteous DJ, Muir WJ (2001): Schizophrenia and affective disorders--cosegregation with a translocation at chromosome 1q42 that directly disrupts brain-expressed genes: clinical and P300 findings in a family. Am J Hum Genet. 69:428–433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brandon NJ, Sawa A (2011): Linking neurodevelopmental and synaptic theories of mental illness through DISC1. Nat Rev Neurosci. 12:707–722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schnoll JG, Temsamrit B, Zhang D, Song H, Ming G-l, Christian KM (2021): Evaluating Neurodevelopmental Consequences of Perinatal Exposure to Antiretroviral Drugs: Current Challenges and New Approaches. Journal of Neuroimmune Pharmacology. 16:113–129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.van der Worp HB, Howells DW, Sena ES, Porritt MJ, Rewell S, O’Collins V, et al. (2010): Can Animal Models of Disease Reliably Inform Human Studies? PLOS Medicine. 7:e1000245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dawson TM, Golde TE, Lagier-Tourenne C (2018): Animal models of neurodegenerative diseases. Nat Neurosci. 21:1370–1379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wen Z, Christian KM, Song H, Ming G-l (2016): Modeling psychiatric disorders with patient-derived iPSCs. Current Opinion in Neurobiology. 36:118–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang DY, Song H, Ming G-l (2021): Modeling neurological disorders using brain organoids. Seminars in Cell & Developmental Biology. 111:4–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Qian XY, Song HJ, Ming GL (2019): Brain organoids: advances, applications and challenges. Development. 146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang Z, O’Laughlin R, Song H, Ming GL (2022): Patterning of brain organoids derived from human pluripotent stem cells. Curr Opin Neurobiol. 74:102536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kadoshima T, Sakaguchi H, Nakano T, Soen M, Ando S, Eiraku M, et al. (2013): Selforganization of axial polarity, inside-out layer pattern, and species-specific progenitor dynamics in human ES cell–derived neocortex. Proceedings of the National Academy of Sciences. 110:20284–20289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ziffra RS, Kim CN, Ross JM, Wilfert A, Turner TN, Haeussler M, et al. (2021): Single-cell epigenomics reveals mechanisms of human cortical development. Nature. 598:205–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Paşca AM, Sloan SA, Clarke LE, Tian Y, Makinson CD, Huber N, et al. (2015): Functional cortical neurons and astrocytes from human pluripotent stem cells in 3D culture. Nature Methods. 12:671–678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mariani J, Coppola G, Zhang P, Abyzov A, Provini L, Tomasini L, et al. (2015): FOXG1Dependent Dysregulation of GABA/Glutamate Neuron Differentiation in Autism Spectrum Disorders. Cell. 162:375–390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Qian X, Nguyen Ha N, Song Mingxi M, Hadiono C, Ogden Sarah C, Hammack C, et al. (2016): Brain-Region-Specific Organoids Using Mini-bioreactors for Modeling ZIKV Exposure. Cell. 165:1238–1254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Qian X, Jacob F, Song MM, Nguyen HN, Song H, Ming G-l (2018): Generation of human brain region–specific organoids using a miniaturized spinning bioreactor. Nature Protocols. 13:565–580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Qian X, Su Y, Adam CD, Deutschmann AU, Pather SR, Goldberg EM, et al. (2020): Sliced Human Cortical Organoids for Modeling Distinct Cortical Layer Formation. Cell Stem Cell. 26:766–781.e769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang W, Yang SL, Yang M, Herrlinger S, Shao Q, Collar JL, et al. (2019): Modeling microcephaly with cerebral organoids reveals a WDR62-CEP170-KIF2A pathway promoting cilium disassembly in neural progenitors. Nat Commun. 10:2612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li R, Sun L, Fang A, Li P, Wu Q, Wang X (2017): Recapitulating cortical development with organoid culture in vitro and modeling abnormal spindle-like (ASPM related primary) microcephaly disease. Protein Cell. 8:823–833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang L, Li Z, Sievert D, Smith DEC, Mendes MI, Chen DY, et al. (2020): Loss of NARS1 impairs progenitor proliferation in cortical brain organoids and leads to microcephaly. Nat Commun. 11:4038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bershteyn M, Nowakowski TJ, Pollen AA, Di Lullo E, Nene A, Wynshaw-Boris A, et al. (2017): Human iPSC-Derived Cerebral Organoids Model Cellular Features of Lissencephaly and Reveal Prolonged Mitosis of Outer Radial Glia. Cell Stem Cell. 20:435–449 e434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Iefremova V, Manikakis G, Krefft O, Jabali A, Weynans K, Wilkens R, et al. (2017): An Organoid-Based Model of Cortical Development Identifies Non-Cell-Autonomous Defects in Wnt Signaling Contributing to Miller-Dieker Syndrome. Cell Reports. 19:50–59. [DOI] [PubMed] [Google Scholar]
- 25.Paulsen B, Velasco S, Kedaigle AJ, Pigoni M, Quadrato G, Deo AJ, et al. (2022): Autism genes converge on asynchronous development of shared neuron classes. Nature. 602:268–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.de Jong JO, Llapashtica C, Genestine M, Strauss K, Provenzano F, Sun Y, et al. (2021): Cortical overgrowth in a preclinical forebrain organoid model of CNTNAP2-associated autism spectrum disorder. Nat Commun. 12:4087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ye F, Kang E, Yu C, Qian X, Jacob F, Yu C, et al. (2017): DISC1 Regulates Neurogenesis via Modulating Kinetochore Attachment of Ndel1/Nde1 during Mitosis. Neuron. 96:1041–1054 e1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kang Y, Zhou Y, Li Y, Han Y, Xu J, Niu W, et al. (2021): A human forebrain organoid model of fragile X syndrome exhibits altered neurogenesis and highlights new treatment strategies. Nat Neurosci. 24:1377–1391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sun AX, Yuan Q, Fukuda M, Yu W, Yan H, Lim GGY, et al. (2019): Potassium channel dysfunction in human neuronal models of Angelman syndrome. Science. 366:1486–1492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Raja WK, Mungenast AE, Lin YT, Ko T, Abdurrob F, Seo J, et al. (2016): Self-Organizing 3D Human Neural Tissue Derived from Induced Pluripotent Stem Cells Recapitulate Alzheimer’s Disease Phenotypes. PLoS One. 11:e0161969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bowles KR, Silva MC, Whitney K, Bertucci T, Berlind JE, Lai JD, et al. (2021): ELAVL4, splicing, and glutamatergic dysfunction precede neuron loss in MAPT mutation cerebral organoids. Cell. 184:4547–4563 e4517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Emerich DF, Skinner SJ, Borlongan CV, Vasconcellos AV, Thanos CG (2005): The choroid plexus in the rise, fall and repair of the brain. Bioessays. 27:262–274. [DOI] [PubMed] [Google Scholar]
- 33.Pellegrini L, Bonfio C, Chadwick J, Begum F, Skehel M, Lancaster MA (2020): Human CNS barrier-forming organoids with cerebrospinal fluid production. Science. 369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sakaguchi H, Kadoshima T, Soen M, Narii N, Ishida Y, Ohgushi M, et al. (2015): Generation of functional hippocampal neurons from self-organizing human embryonic stem cell-derived dorsomedial telencephalic tissue. Nat Commun. 6:8896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jacob F, Pather SR, Huang W-K, Zhang F, Wong SZH, Zhou H, et al. (2020): Human Pluripotent Stem Cell-Derived Neural Cells and Brain Organoids Reveal SARS-CoV-2 Neurotropism Predominates in Choroid Plexus Epithelium. Cell Stem Cell. 27:937–950.e939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wonders CP, Anderson SA (2006): The origin and specification of cortical interneurons. Nat Rev Neurosci. 7:687–696. [DOI] [PubMed] [Google Scholar]
- 37.Ma T, Wang C, Wang L, Zhou X, Tian M, Zhang Q, et al. (2013): Subcortical origins of human and monkey neocortical interneurons. Nat Neurosci. 16:1588–1597. [DOI] [PubMed] [Google Scholar]
- 38.Hansen DV, Lui JH, Flandin P, Yoshikawa K, Rubenstein JL, Alvarez-Buylla A, et al. (2013): Non-epithelial stem cells and cortical interneuron production in the human ganglionic eminences. Nat Neurosci. 16:1576–1587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Birey F, Andersen J, Makinson CD, Islam S, Wei W, Huber N, et al. (2017): Assembly of functionally integrated human forebrain spheroids. Nature. 545:54–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bagley JA, Reumann D, Bian S, Lévi-Strauss J, Knoblich JA (2017): Fused cerebral organoids model interactions between brain regions. Nature Methods. 14:743–751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Xiang Y, Tanaka Y, Patterson B, Kang Y-J, Govindaiah G, Roselaar N, et al. (2017): Fusion of Regionally Specified hPSC-Derived Organoids Models Human Brain Development and Interneuron Migration. Cell Stem Cell. 21:383–398.e387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Miura Y, Li MY, Birey F, Ikeda K, Revah O, Thete MV, et al. (2020): Generation of human striatal organoids and cortico-striatal assembloids from human pluripotent stem cells. Nat Biotechnol. 38:1421–1430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Xiang Y, Tanaka Y, Cakir B, Patterson B, Kim K-Y, Sun P, et al. (2019): hESC-Derived Thalamic Organoids Form Reciprocal Projections When Fused with Cortical Organoids. Cell Stem Cell. 24:487–497.e487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Huang WK, Wong SZH, Pather SR, Nguyen PTT, Zhang F, Zhang DY, et al. (2021): Generation of hypothalamic arcuate organoids from human induced pluripotent stem cells. Cell Stem Cell. 28:1657–1670 e1610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tieng V, Stoppini L, Villy S, Fathi M, Dubois-Dauphin M, Krause K-H (2014): Engineering of Midbrain Organoids Containing Long-Lived Dopaminergic Neurons. Stem Cells and Development. 23:1535–1547. [DOI] [PubMed] [Google Scholar]
- 46.Jo J, Xiao Y, Sun Alfred X, Cukuroglu E, Tran H-D, Göke J, et al. (2016): Midbrain-like Organoids from Human Pluripotent Stem Cells Contain Functional Dopaminergic and Neuromelanin-Producing Neurons. Cell Stem Cell. 19:248–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Monzel AS, Smits LM, Hemmer K, Hachi S, Moreno EL, van Wuellen T, et al. (2017): Derivation of Human Midbrain-Specific Organoids from Neuroepithelial Stem Cells. Stem Cell Reports. 8:1144–1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kim H, Park HJ, Choi H, Chang Y, Park H, Shin J, et al. (2019): Modeling G2019S-LRRK2 Sporadic Parkinson’s Disease in 3D Midbrain Organoids. Stem Cell Reports. 12:518–531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Smits LM, Reinhardt L, Reinhardt P, Glatza M, Monzel AS, Stanslowsky N, et al. (2019): Modeling Parkinson’s disease in midbrain-like organoids. npj Parkinson’s Disease. 5:5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mohamed NV, Sirois J, Ramamurthy J, Mathur M, Lepine P, Deneault E, et al. (2021): Midbrain organoids with an SNCA gene triplication model key features of synucleinopathy. Brain Commun. 3:fcab223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jo J, Yang L, Tran HD, Yu W, Sun AX, Chang YY, et al. (2021): Lewy Body-like Inclusions in Human Midbrain Organoids Carrying Glucocerebrosidase and alpha-Synuclein Mutations. Ann Neurol. 90:490–505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jarazo J, Barmpa K, Rosety I, Smits LM, Arias-Fuenzalida J, Walter J, et al. (2019): Parkinson’s disease phenotypes in patient specific brain organoids are improved by HP-β-CD treatment. bioRxiv. 813089. [Google Scholar]
- 53.Muguruma K, Nishiyama A, Kawakami H, Hashimoto K, Sasai Y (2015): Self-Organization of Polarized Cerebellar Tissue in 3D Culture of Human Pluripotent Stem Cells. Cell Reports. 10:537–550. [DOI] [PubMed] [Google Scholar]
- 54.Valiulahi P, Vidyawan V, Puspita L, Oh Y, Juwono VB, Sittipo P, et al. (2021): Generation of caudal-type serotonin neurons and hindbrain-fate organoids from hPSCs. Stem Cell Reports. 16:1938–1952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bajaj S, Bagley JA, Sommer C, Vertesy A, Nagumo Wong S, Krenn V, et al. (2021): Neurotransmitter signaling regulates distinct phases of multimodal human interneuron migration. EMBO J. 40:e108714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Andersen J, Revah O, Miura Y, Thom N, Amin ND, Kelley KW, et al. (2020): Generation of Functional Human 3D Cortico-Motor Assembloids. Cell. 183:1913–1929 e1926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kasai T, Suga H, Sakakibara M, Ozone C, Matsumoto R, Kano M, et al. (2020): Hypothalamic Contribution to Pituitary Functions Is Recapitulated In Vitro Using 3D-Cultured Human iPS Cells. Cell Reports. 30:18–+. [DOI] [PubMed] [Google Scholar]
- 58.Song L, Yuan X, Jones Z, Vied C, Miao Y, Marzano M, et al. (2019): Functionalization of Brain Region-specific Spheroids with Isogenic Microglia-like Cells. Sci Rep. 9:11055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Popova G, Soliman SS, Kim CN, Keefe MG, Hennick KM, Jain S, et al. (2021): Human microglia states are conserved across experimental models and regulate neural stem cell responses in chimeric organoids. Cell Stem Cell. 28:2153–2166 e2156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sabate-Soler S, Nickels SL, Saraiva C, Berger E, Dubonyte U, Barmpa K, et al. (2022): Microglia integration into human midbrain organoids leads to increased neuronal maturation and functionality. Glia. 70:1267–1288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Xu R, Boreland AJ, Li X, Erickson C, Jin M, Atkins C, et al. (2021): Developing human pluripotent stem cell-based cerebral organoids with a controllable microglia ratio for modeling brain development and pathology. Stem Cell Reports. 16:1923–1937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Cakir B, Xiang Y, Tanaka Y, Kural MH, Parent M, Kang YJ, et al. (2019): Engineering of human brain organoids with a functional vascular-like system. Nat Methods. 16:1169–1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Shi Y, Sun L, Wang M, Liu J, Zhong S, Li R, et al. (2020): Vascularized human cortical organoids (vOrganoids) model cortical development in vivo. PLoS Biol. 18:e3000705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Shi YC, Sun L, Wang MD, Liu JW, Zhong SJ, Li R, et al. (2020): Vascularized human cortical organoids (vOrganoids) model cortical development in vivo. Plos Biology. 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Mansour AA, Goncalves JT, Bloyd CW, Li H, Fernandes S, Quang D, et al. (2018): An in vivo model of functional and vascularized human brain organoids. Nat Biotechnol. 36:432–441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Daviaud N, Friedel RH, Zou HY (2018): Vascularization and Engraftment of Transplanted Human Cerebral Organoids in Mouse Cortex. Eneuro. 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Revah O, Gore F, Kelley KW, Andersen J, Sakai N, Chen X, et al. (2022): Maturation and circuit integration of transplanted human cortical organoids. Nature. 610:319–326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Camp JG, Badsha F, Florio M, Kanton S, Gerber T, Wilsch-Brauninger M, et al. (2015): Human cerebral organoids recapitulate gene expression programs of fetal neocortex development. P Natl Acad Sci USA. 112:15672–15677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Luo CY, Lancaster MA, Castanon R, Nery JR, Knoblich JA, Ecker JR (2016): Cerebral Organoids Recapitulate Epigenomic Signatures of the Human Fetal Brain. Cell Reports. 17:3369–3384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Amiri A, Coppola G, Scuderi S, Wu F, Roychowdhury T, Liu FC, et al. (2018): Transcriptome and epigenome landscape of human cortical development modeled in organoids. Science. 362:1268–+. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Velasco S, Kedaigle AJ, Simmons SK, Nash A, Rocha M, Quadrato G, et al. (2019): Individual brain organoids reproducibly form cell diversity of the human cerebral cortex. Nature. 570:523–+. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Pollen AA, Bhaduri A, Andrews MG, Nowakowski TJ, Meyerson OS, Mostajo-Radji MA, et al. (2019): Establishing Cerebral Organoids as Models of Human-Specific Brain Evolution. Cell. 176:743–+. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kanton S, Boyle MJ, He ZS, Santel M, Weigert A, Sanchis-Calleja F, et al. (2019): Organoid single-cell genomic atlas uncovers human-specific features of brain development. Nature. 574:418+. [DOI] [PubMed] [Google Scholar]
- 74.Trevino AE, Sinnott-Armstrong N, Andersen J, Yoon SJ, Huber N, Pritchard JK, et al. (2020): Chromatin accessibility dynamics in a model of human forebrain development. Science. 367:404–+. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Yoon KJ, Ringeling FR, Vissers C, Jacob F, Pokrass M, Jimenez-Cyrus D, et al. (2017): Temporal Control of Mammalian Cortical Neurogenesis by m(6)A Methylation. Cell. 171:877–+. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Khan TA, Revah O, Gordon A, Yoon SJ, Krawisz AK, Goold C, et al. (2020): Neuronal defects in a human cellular model of 22q11.2 deletion syndrome. Nature Medicine. 26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Marin O (2012): Interneuron dysfunction in psychiatric disorders. Nat Rev Neurosci. 13:107–120. [DOI] [PubMed] [Google Scholar]
- 78.Zhang ZN, Freitas BC, Qian H, Lux J, Acab A, Trujillo CA, et al. (2016): Layered hydrogels accelerate iPSC-derived neuronal maturation and reveal migration defects caused by MeCP2 dysfunction. P Natl Acad Sci USA. 113:3185–3190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Marchetto MC, Belinson H, Tian Y, Freitas BC, Fu C, Vadodaria KC, et al. (2017): Altered proliferation and networks in neural cells derived from idiopathic autistic individuals. Molecular Psychiatry. 22:820–835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Windrem MS, Osipovitch M, Liu ZS, Bates J, Chandler-Militello D, Zou L, et al. (2017): Human iPSC Glial Mouse Chimeras Reveal Glial Contributions to Schizophrenia. Cell Stem Cell. 21:195–+. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Rubenstein JLR, Merzenich MM (2003): Model of autism: increased ratio of excitation/inhibition in key neural systems. Genes Brain Behav. 2:255–267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Marin O, Rubenstein JLR (2003): Cell migration in the forebrain. Annual Review of Neuroscience. 26:441–483. [DOI] [PubMed] [Google Scholar]
- 83.Birey F, Li MY, Gordon A, Thete MV, Valencia AM, Revah O, et al. (2022): Dissecting the molecular basis of human interneuron migration in forebrain assembloids from Timothy syndrome. Cell Stem Cell. 29:248–+. [DOI] [PubMed] [Google Scholar]
- 84.Yildirim M, Delepine C, Feldman D, Pham VA, Chou S, Ip J, et al. (2022): Label-free threephoton imaging of intact human cerebral organoids for tracking early events in brain development and deficits in Rett syndrome. Elife. 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Yang Q, Hong Y, Zhao T, Song HJ, Ming GL (2022): What Makes Organoids Good Models of Human Neurogenesis? Front Neurosci-Switz. 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Gomes AR, Fernandes TG, Vaz SH, Silva TP, Bekman EP, Xapelli S, et al. (2020): Modeling Rett Syndrome With Human Patient-Specific Forebrain Organoids. Front Cell Dev Biol. 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Klaus J, Kanton S, Kyrousi C, Ayo-Martin AC, Di Giaimo R, Riesenberg S, et al. (2019): Altered neuronal migratory trajectories in human cerebral organoids derived from individuals with neuronal heterotopia. Nature Medicine. 25:561–+. [DOI] [PubMed] [Google Scholar]
- 88.Li Y, Muffat J, Omer A, Bosch I, Lancaster MA, Sur M, et al. (2017): Induction of Expansion and Folding in Human Cerebral Organoids. Cell Stem Cell. 20:385–+. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Karzbrun E, Kshirsagar A, Cohen SR, Hanna JH, Reiner O (2018): Human brain organoids on a chip reveal the physics of folding. Nat Phys. 14:515–+. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Samarasinghe RA, Miranda OA, Buth JE, Mitchell S, Ferando I, Watanabe M, et al. (2021): Identification of neural oscillations and epileptiform changes in human brain organoids. Nature Neuroscience. 24:1488–1500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Quadrato G, Nguyen T, Macosko EZ, Sherwood JL, Yang SM, Berger DR, et al. (2017): Cell diversity and network dynamics in photosensitive human brain organoids. Nature. 545:48–+. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Trujillo CA, Gao R, Negraes PD, Gu J, Buchanan J, Preissl S, et al. (2019): Complex Oscillatory Waves Emerging from Cortical Organoids Model Early Human Brain Network Development. Cell Stem Cell. 25:558–569.e557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Splawski I, Timothy KW, Sharpe LM, Decher N, Kumar P, Bloise R, et al. (2004): Ca(V)1.2 calcium channel dysfunction causes a multisystem disorder including arrhythmia and autism. Cell. 119:19–31. [DOI] [PubMed] [Google Scholar]
- 94.Ohashi M, Korsakova E, Allen D, Lee P, Fu K, Vargas BS, et al. (2018): Loss of MECP2 Leads to Activation of P53 and Neuronal Senescence. Stem Cell Reports. 10:1453–1463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Renner H, Grabos M, Becker KJ, Kagermeier TE, Wu J, Otto M, et al. (2020): A fully automated high-throughput workflow for 3D-based chemical screening in human midbrain organoids. Elife. 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Renner H, Grabos M, Scholer HR, Bruder JM (2021): Generation and Maintenance of Homogeneous Human Midbrain Organoids. Bio-Protocol. 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Shafique S (2022): Stem cell-based region-specific brain organoids: Novel models to understand neurodevelopmental defects. Birth Defects Res. [DOI] [PubMed] [Google Scholar]
- 98.Cong L, Ran FA, Cox D, Lin SL, Barretto R, Habib N, et al. (2013): Multiplex Genome Engineering Using CRISPR/Cas Systems. Science. 339:819–823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Li C, Fleck JS, Martins-Costa C, Burkard TR, Stuempflen M, Vertesy Á, et al. (2022): Single-cell brain organoid screening identifies developmental defects in autism. bioRxiv.2022.2009.2015.508118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Sanders SJ, Murtha MT, Gupta AR, Murdoch JD, Raubeson MJ, Willsey AJ, et al. (2012): De novo mutations revealed by whole-exome sequencing are strongly associated with autism. Nature. 485:237–U124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Esk C, Lindenhofer D, Haendeler S, Wester RA, Pflug F, Schroeder B, et al. (2020): A human tissue screen identifies a regulator of ER secretion as a brain-size determinant. Science. 370:935–+. [DOI] [PubMed] [Google Scholar]
- 102.Gusev A, Lee SH, Trynka G, Finucane H, Vilhjalmsson BJ, Xu H, et al. (2014): Partitioning Heritability of Regulatory and Cell-Type-Specific Variants across 11 Common Diseases. American Journal of Human Genetics. 95:535–552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Edwards SL, Beesley J, French JD, Dunning AM (2013): Beyond GWASs: Illuminating the Dark Road from Association to Function. American Journal of Human Genetics. 93:779–797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Lu LN, Liu XX, Huang WK, Giusti-Rodriguez P, Cui J, Zhang SS, et al. (2020): Robust Hi-C Maps of Enhancer-Promoter Interactions Reveal the Function of Non-coding Genome in Neural Development and Diseases. Mol Cell. 79:521–+. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Song M, Yang XY, Ren XJ, Maliskova L, Li BK, Jones IR, et al. (2019): Mapping cis-regulatory chromatin contacts in neural cells links neuropsychiatric disorder risk variants to target genes. Nat Genet. 51:1252–+. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Sey NYA, Hu BX, Mah W, Fauni H, McAfee JC, Rajarajan P, et al. (2020): A computational tool (H-MAGMA) for improved prediction of brain-disorder risk genes by incorporating brain chromatin interaction profiles. Nature Neuroscience. 23:583–+. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Townsley KG, Brennand KJ, Huckins LM (2020): Massively parallel techniques for cataloguing the regulome of the human brain. Nature Neuroscience. 23:1509–1521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Fujita Y, Pather SR, Ming GL, Song H (2022): 3D spatial genome organization in the nervous system: From development and plasticity to disease. Neuron. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Shen SQ, Myers CA, Hughes AEO, Byrne LC, Flannery JG, Corbo JC (2016): Massively parallel cis-regulatory analysis in the mammalian central nervous system. Genome Res. 26:238255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Inoue F, Kreimer A, Ashuach T, Ahituv N, Yosef N (2019): Identification and Massively Parallel Characterization of Regulatory Elements Driving Neural Induction. Cell Stem Cell. 25:713+. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Melnikov A, Zhang XL, Rogov P, Wang L, Mikkelsen TS (2014): Massively Parallel Reporter Assays in Cultured Mammalian Cells. Jove-J Vis Exp. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Inoue F, Kircher M, Martin B, Cooper GM, Witten DM, McManus MT, et al. (2017): A systematic comparison reveals substantial differences in chromosomal versus episomal encoding of enhancer activity. Genome Res. 27:38–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Uebbing S, Gockley J, Reilly SK, Kocher AA, Geller E, Gandotra N, et al. (2021): Massively parallel discovery of human-specific substitutions that alter enhancer activity. P Natl Acad Sci USA. 118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Schrode N, Ho SM, Yamamuro K, Dobbyn A, Huckins L, Matos MR, et al. (2019): Synergistic effects of common schizophrenia risk variants. Nat Genet. 51:1475–+. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Sternberg SH, Redding S, Jinek M, Greene EC, Doudna JA (2014): DNA interrogation by the CRISPR RNA-guided endonuclease Cas9. Nature. 507:62–+. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Diao YR, Fang RX, Li B, Meng ZP, Yu JT, Qiu YJ, et al. (2017): A tiling-deletion-based genetic screen for cis-regulatory element identification in mammalian cells. Nature Methods. 14:629–+. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Tian RL, Gachechiladze MA, Ludwig CH, Laurie MT, Hong JY, Nathaniel D, et al. (2019): CRISPR Interference-Based Platform for Multimodal Genetic Screens in Human iPSC-Derived Neurons. Neuron. 104:239–+. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Tian RL, Abarientos A, Hong JS, Hashemi SH, Yan R, Drager N, et al. (2021): Genomewide CRISPRi/a screens in human neurons link lysosomal failure to ferroptosis. Nature Neuroscience. 24:1020–1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Gordon A, Yoon SJ, Tran SS, Makinson CD, Park JY, Andersen J, et al. (2021): Long-term maturation of human cortical organoids matches key early postnatal transitions. Nature Neuroscience. 24:331–342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Giandomenico SL, Sutcliffe M, Lancaster MA (2021): Generation and long-term culture of advanced cerebral organoids for studying later stages of neural development. Nature Protocols. 16:579–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Hergenreder E, Zorina Y, Zhao Z, Munguba H, Calder EL, Baggiolini A, et al. (2022): Combined small molecule treatment accelerates timing of maturation in human pluripotent stem cell-derived neurons. bioRxiv.2022.2006.2002.494616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Bhaduri A, Andrews MG, Leon WM, Jung D, Shin D, Allen D, et al. (2020): Cell stress in cortical organoids impairs molecular subtype specification. Nature. 578:142–+. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Vértesy Á, Eichmueller OL, Naas J, Novatchkova M, Esk C, Balmaña M, et al. (2022): Cellular stress in brain organoids is limited to a distinct and bioinformatically removable subpopulation. bioRxiv.2022.2003.2011.483643. [Google Scholar]
- 124.Uzquiano A, Kedaigle AJ, Pigoni M, Paulsen B, Adiconis X, Kim K, et al. (2022): Proper acquisition of cell class identity in organoids allows definition of fate specification programs of the human cerebral cortex. Cell. 185:3770–3788 e3727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Eichmuller OL, Corsini NS, Vertesy A, Morassut I, Scholl T, Gruber VE, et al. (2022): Amplification of human interneuron progenitors promotes brain tumors and neurological defects. Science. 375:eabf5546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Deng WL, Gao ML, Lei XL, Lv JN, Zhao H, He KW, et al. (2018): Gene Correction Reverses Ciliopathy and Photoreceptor Loss in iPSC-Derived Retinal Organoids from Retinitis Pigmentosa Patients. Stem Cell Reports. 10:1267–1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Lin YT, Seo J, Gao F, Feldman HM, Wen HL, Penney J, et al. (2018): APOE4 Causes Widespread Molecular and Cellular Alterations Associated with Alzheimer’s Disease Phenotypes in Human iPSC-Derived Brain Cell Types. Neuron. 98:1141–1154 e1147. [DOI] [PMC free article] [PubMed] [Google Scholar]


