Abstract
The role of Programmed Cell Death Ligand 1 (PD-L1) expression in predicting epidermal growth factor receptor tyrosine kinase inhibitor (EGFR-TKIs) efficacy remains controversial. Recent studies have highlighted that tumor-intrinsic PD-L1 signaling can be modulated by STAT3, AKT, MET oncogenic pathway, epithelial–mesenchymal transition, or BIM expression. This study aimed to investigate whether these underlying mechanisms affect the prognostic role of PD-L1. We retrospectively enrolled patients with EGFR mutant advanced stage NSCLC who received first-line EGFR-TKI between January 2017 and June 2019, the treatment efficacy of EGFR-TKI was assessed. Kaplan–Meier analysis of progression-free survival (PFS) revealed that patients with high BIM expression had shorter PFS, regardless of PD-L1 expression. This result was also supported by the COX proportional hazard regression analysis. In vitro, we further proved that the knockdown of BIM, instead of PDL1, induced more cell apoptosis following gefitinib treatment. Our data suggest that among the pathways affecting tumor-intrinsic PD-L1 signaling, BIM is potentially the underlying mechanism that affects the role of PD-L1 expression in predicting response to EGFR TKI and mediates cell apoptosis under treatment with gefitinib in EGFR-mutant NSCLC. Further prospective studies are required to validate these results.
Subject terms: Oncology, Cancer therapy, Lung cancer
Introduction
Epidermal growth factor receptor (EGFR) mutation is the most common oncogenic driver gene in advanced non-small cell lung cancer (NSCLC), accounting for approximately 50% of patients1. The use of EGFR-tyrosine kinase inhibitors (TKIs) provides better progression-free survival (PFS) and objective response rate (ORR), it has become the mainstay treatment strategy in these patient populations2. However, approximately 20–30% of patients receiving EGFR-TKIs remain unresponsive3. The mechanism of primary resistance includes the activation of bypass or downstream signaling pathways4, presence of resistant kinase domain mutations (T790M mutation)4, histological transformation5, and deletion polymorphism of the Bcl-2 family member (BIM)6. Recently, there have been increasing studies regarding the role of the tumor microenvironment in the treatment efficacy of EGFR-TKIs, including the programmed death 1 (PD-1) and programmed death-ligand 1 (PD-L1) pathways7.
The role of PD-L1 in predicting treatment outcomes to EGFR-TKIs remains controversial; some cohort studies have demonstrated that high PD-L1 expression is associated with poor treatment efficacy of EGFR-TKIs8–15. Other studies have shown that PD-L1 expression levels do not affect the efficacy of EGFR-TKIs16–19. Current studies had highlighted the multiple aspects of tumor PD-L1 signaling, including cell-intrinsic effects and cell-extrinsic PD-L1 signaling20. The cell-extrinsic PD-L1, high expressed PD-L1 on the surface of cancer or immune cells, is pathogenic primarily by inhibiting CD8+ cytotoxic T cells and contributes to the development of immune checkpoint blockade in treating NSCLC. In contrast, cell-intrinsic PD-L1 signals originate from PD-L1 in the surface, cytosol, and nucleus by post-translational modifications and can be elicited by a PD-1-independent pathway. It can be upregulated by activated EGFR through the interleukin 6 (IL6)/Janus kinase (JAK)/signal transducer and activator of transcription 3 (STAT3) signaling pathway in NSCLC cells20,21. Moreover, the downstream pathway of EGFR and STAT3 not only directly binds to the promoter to upregulate the transcriptional expression of PD-L122 but also mediates resistance to EGFR-TKI via the Zeb1 activity of the EMT pathway23. AKT activation also triggers PD-L1 expression in EGFR-mutant lung cancer cell lines24 and becomes a convergent feature of acquired resistance to EGFR-TKIs25. Additionally, epithelial-mesenchymal transition (EMT) could upregulate PD-L1 expression via an NF-κB-dependent mechanism26. Moreover, the expression of PD-L1 was also associated with increased BIM expression27, which is an important factor in EGFR-TKI-mediated apoptosis28.
Since these pathways both mediate PD-L1 expression and are associated with primary resistance to EGFR-TKI, we conducted this study to identify the key pathway that may impact the inconsistency in the prognostic value of PD-L1 in advanced non-small cell lung cancer patients treated with EGFR-TKIs.
Methods
Patient characteristics
Patients with EGFR-mutant advanced-stage NSCLC who received first-line EGFR-TKIs at a tertiary referral center were retrospectively reviewed from January 2017 to June 2019. At the initial diagnosis, all patients underwent imaging examinations for complete staging, including computed tomography of the chest, brain magnetic resonance imaging, and bone scan. Staging was performed according to the American Joint Committee on Cancer 8th edition. Baseline characteristics, including age, sex, stage, performance status, presence of brain metastasis, EGFR mutation subtype, and choice of first-line EGFR-TKI therapy, were recorded. After the initiation of EGFR-TKI treatment, all patients underwent computed tomography (CT) of the chest to evaluate treatment response every 3 months until disease progression. The radiological response was defined using the Response Evaluation Criteria in Solid Tumors (RECIST) version 1.129. The study protocol was reviewed and approved by the Review Board and Ethics Committee of National Cheng Kung University Hospital (NCKUH B-ER-107-374). Given the retrospective nature of the current study, the need of informed consent was waived by the Review Board and Ethics Committee of National Cheng Kung University Hospital. All the study protocol of this study followed the Declaration of Helsinki guidelines.
Immunohistochemical staining and scoring for PD-L1
Immunohistochemical (IHC) staining of PD-L1 expression on tumor cells was assessed using the PD-L1 Clone 22C3 pharmDx kit (Dako; Agilent Technologies, Inc., Santa Clara, CA, USA) and the Automated Link 48 platform (Dako, Carpinteria, CA). The percentage of positive membrane staining of PD-L1 was calculated after the evaluation of at least 100 viable cells, which was referred to as the tumor proportional score (TPS)30. TPS was categorized into three groups (no expression with TPS of < 1%, low expression with TPS of 1–49%, and high expression with TPS of 50–100%), which used 22C3 antibodies as a companion diagnostic test31,32. The stained tissue sections were independently scored by Dr. Chang-Yao Chu at Chi-Mei Medical Center and other pathologists at NCKUH who were blinded to the patients’ clinical characteristics and outcomes.
Immunohistochemical staining of other biomarkers
Immunohistochemical staining for p-STAT3, p-AKT, MET, E-cadherin, and BIM was performed using the streptavidin-peroxidase method. Briefly, 4-μm-thick sections were deparaffinized, rehydrated, and treated with 0.3% H2O2 to block endogenous peroxidase activity. Following rehydration through graded concentrations of ethanol and autoclave, nonspecific binding sites were blocked with 10% normal goat serum. The sections were then incubated at 4 °C overnight with the following antibodies: rabbit antibodies against human p-STAT3 (Cell Signaling Technology, #4060, dilution 1:100), p-AKT (Cell Signaling Technology, catalog #9145, dilution 1:100), MET (Abcam, catalog #Ab227637, dilution 1:30), E-cadherin (Cell Signaling Technology, catalog #3195, dilution 1:50), and BIM (Cell Signaling Technology, catalog #2933, dilution 1:100). The sections were then incubated with biotinylated peroxidase-labeled anti-rabbit antibody (DakoCytomation, catalog #K4003) for 30 min, followed by incubation with streptavidin–biotin peroxidase complex solution. The chromogen 3,3′-diaminobenzidine tetrahydrochloride was used. The expression of p-STAT3, p-AKT, MET, and BIM was defined in samples as any intensity of antibody staining and ≥ 1% of the tumor33–36. Samples with no E-cadherin membranous staining in any percentage of the tumor were categorized as a loss of expression, while those with E-cadherin membranous staining were categorized as having preserved expression37. Finally, the sections were lightly counterstained with hematoxylin. The stained tissue sections were independently scored by Dr. Chang-Yao Chu at Chi-Mei Medical Center and other pathologists at NCKUH who were blinded to the patients’ clinical characteristics and outcomes.
Statistical analysis
The frequencies and descriptive statistics of the demographic and clinical variables were calculated. Categorical variables were compared using the chi-square test or Fisher’s exact test, whereas continuous variables were compared using the Student’s t-test or Wilcoxon rank-sum test. PFS was determined as the time period from initiation of EGFR-TKI therapy to radiological progression or death, which was estimated using the Kaplan–Meier method and compared with the log-rank test. The p-value was used to indicate statistical significance (P < 0.05). The Cox proportional hazards regression analysis was also performed to determine the determinants of PFS. The Cox proportional hazards were estimated using the formula:
where t represents survival time and is the expected hazard at time t. The hazard function was determined by a set of determinants (X1, X2,…,Xp). The coefficients (b1, b2,…,bp) represent the impact of each determinants. The is the baseline hazard and represents the hazard when all the determinants are equal to zero. The possible determinants included prognostic factors based on prior studies38,39 and the IHC staining results of the current study. Statistical Analysis System® software (version 9.4; SAS Institute, Cary, NC, USA) was used to perform analyses.
Cell culture and transfection of siRNA
The HCC827 cell line was grown in RPMI-1640 medium (Thermo Fisher Scientific, Waltham, MA, USA) with supplement of 10% (v/v) fetal bovine serum (FBS) (Gibco; Life Technologies, Carlsbad, CA, USA) at 37 °C in an incubator with 5% carbon dioxide. The cells were maintained by monolayer culture and passaged twice or thrice per week. For siRNA transfection, HCC827 cells were cultured in a 6-cm dish. Thereafter, the cultured cells were washed once with sterile phosphate-buffered saline and the culture medium was supplemented with Lipofectamine (Dharmacon, Horizon Discovery, Lafayette, CO, USA) plus 100 nM of the ON-TARGETplus siRNA targeting CD274 or BCL2L11 (Dharmacon, Horizon Discovery, Lafayette, CO, USA). For drug treatment, gefitinib (Selleck Chemicals LLC, Houston, TX, USA) was dissolved in DMSO and diluted to the desired final concentrations with a growth medium immediately before administration. After 24-h transfection, the culture medium was replaced with a medium containing 1 μM or 5 μM gefitinib for the following 24 h.
Assessing apoptotic ratio via flow cytometry
The flow cytometry was performed to assess the apoptotic fractions of cancer cell line. After 24-h drug treatment, the cells were trypsinized and stained with 5 μL FITC-Annexin V stock solution (#556547; BD Biosciences Pharmingen, CA, USA) and 5 μL propidium iodide (PI) stock solution (#550825; BD Biosciences Pharmingen, CA, USA). Then, the samples were incubated in the dark for 15 min, followed by the addition of a 400 μL Annexin V Binding Buffer (#422201, BioLegend, CA, USA) for the subsequent measurement. Both fluorochromes were excited by an argon-ion laser at 488 nm. FITC-Annexin V was measured on FL1 channel (filter: 530/30 nm) and PI was measured on the FL2 channel (filter: 585/42 nm). Total 1 × 104 events were evaluated in each sample via flow cytometry (CytoFLEX Flow Cytometry, Beckman Coulter, Brea, CA, USA). Further analysis of the apoptotic ratios was implemented by CytExpert 2.2 (Beckman Coulter, CA, USA).
Genomic testing of the BIM intronic deletion polymorphism
In patients with sufficient residual tissue, the genomic DNA was extracted from formalin-fixed paraffin-embedded (FFPE) biopsy slides using iCatcher FFPE Tissue DNA Kit (CatchGene Co., Ltd.). BIM polymorphic genotyping was performed as previously described40. Briefly, two separate polymerase chain reactions (PCRs) were performed for each sample, to determine the presence of the wild-type and deletion alleles using the forward primer 5′-CCACCAATGGAAAAGGTTCA-3′ for both wild-type and deletion alleles, the reverse primers 5′-CTGTCATTTCTCCCCACCAC-3′ and 5′-GGCACAGCCTCTATGGAGAA-3′ for the wild-type and deletion alleles, respectively. The PCRs were performed with the following thermo cycling conditions: 95 °C for 5 min; 39 cycles of 95 °C for 50 s, 58 °C for 50 s, and 72 °C for 1 min; and 72 °C for 10 min. Finally, the PCR products were analyzed on a 2% agarose gel, with 284 bp for the deletion alleles and 362 bp for the wild-type alleles.
Results
Patient demographics
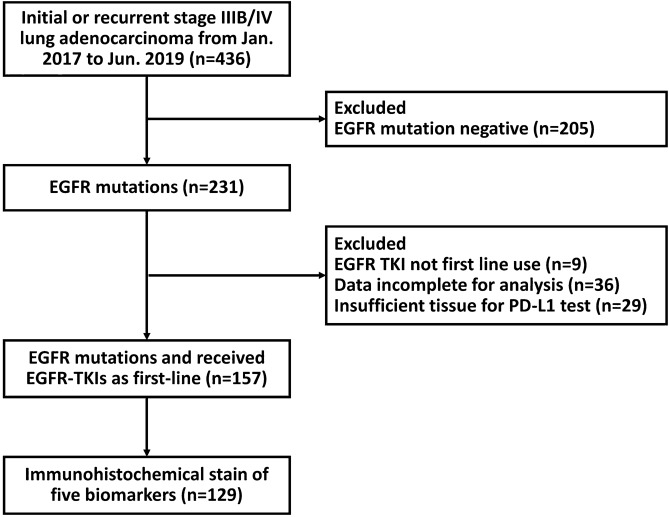
Of the 231 patients with advanced EGFR-mutated NSCLC recruited between January 2017 and June 2019, 157 had sufficient tissue for PD-L1 evaluation. Among these patients, 129 underwent immunohistochemical staining for an additional five biomarkers. Figure 1 shows the flowchart for enrolling the subjects. The Table 1 had summarized the baseline characteristics of 157 patients. All the patients had histological adenocarcinomas and advanced-stage disease. The median age of the patients was 66 years (range, 61–75 years), and there were 61 men (38.9%) and 96 women (61.1%). Sixty-eight patients (43.3%) had brain metastases. The EGFR genotyping test revealed exon 19 deletion mutations in 74 (47.1%), L858R mutations in 74 (47.1%), uncommon mutations in 6 (3.8%), and complex mutations in 3 (2.0%) patients with NSCLC. Immunohistochemical staining of PD-L1 revealed no expression (< 1%) in 95 patients (60.5%), low expression (1–49%) in 42 patients (26.8%), and high expression (≥ 50%) in 20 patients (12.7%).
Figure 1.
Patient flowchart.
Table 1.
Patient characteristics.
| Characteristic | Total patients (%) N = 157 |
|---|---|
| Age | |
| Median (range), years | 66 (61–75) |
| < 65 years | 67 (42.7%) |
| > 65 years | 90 (57.5%) |
| Sex | |
| Female | 96 (61.1%) |
| Male | 61 (38.9%) |
| Brain metastasis | 68 (43.3%) |
| Stage | |
| 3B | 8 (5.1%) |
| 4A | 40 (25.5%) |
| 4B | 109 (69.4) |
| Tumor size | |
| < 3 cm | 32 (20.4%) |
| ≥ 3 cm | 125 (79.6%) |
| Nodal stage | |
| N0-2 | 48 (30.6%) |
| N3 | 109 (69.4%) |
| Performance status | |
| ECOG 0–1 | 138 (87.9%) |
| ECOG ≥ 2 | 19 (12.1%) |
| EGFR mutation | |
| Exon 19 deletion | 74 (47.1%) |
| Exon 21 L858R substitution | 74 (47.1%) |
| Uncommon mutation | 6 (3.8%) |
| Complex mutation | 3 (2.0%) |
| EGFR-TKI | |
| Gefitinib | 22 (14.0%) |
| Erlotinib | 67 (42.7%) |
| Afatinib | 68 (43.3%) |
EGFR, epidermal growth factor receptor; TKI, tyrosine kinase inhibitor.
The PFS based on different PD-L1 and biomarker expression
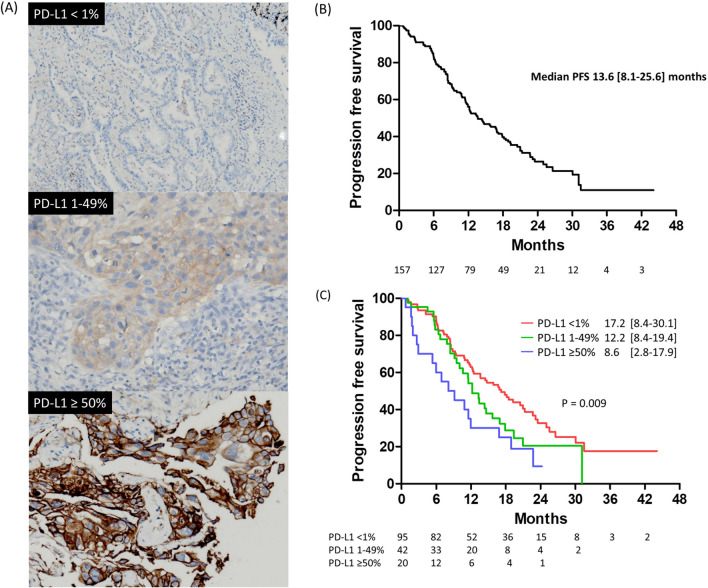
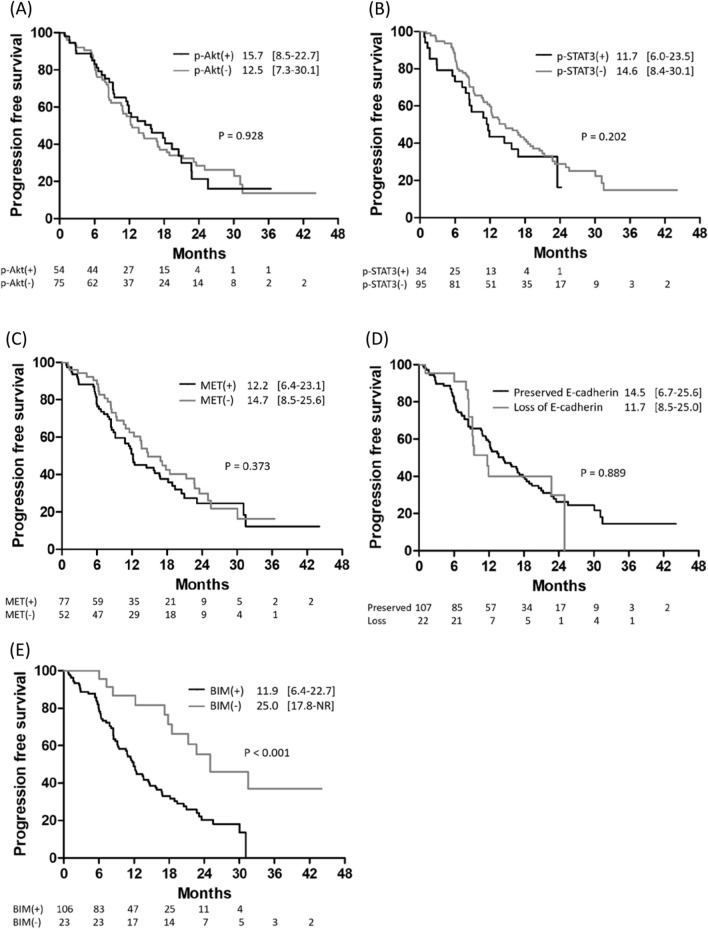
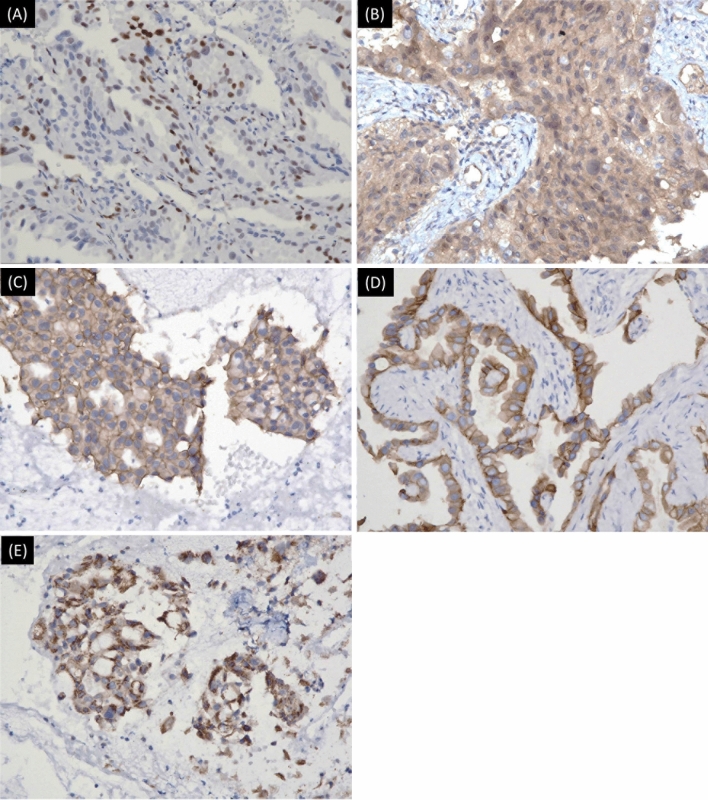
The immunohistochemical staining of different PD-L1 expression levels is presented in Fig. 2A. The median follow-up period was 19.1 months (interquartile range: 11.6–25.8). The median PFS among the 157 patients was 13.6 months (interquartile range: 8.1–25.6) (Fig. 2B). When patients were classified based on PD-L1 expression level, the patients with no PD-L1 expression had PFS of 17.2 months (interquartile range: 8.4–30.1), which was significantly longer than that of patients with low PD-L1 expression (median PFS 12.2 months, interquartile range: 8.4–19.4) and patients with high PD-L1 expression (median PFS 8.6 months, interquartile range, 2.8–17.9) (log-rank p = 0.009, Fig. 2C). Moreover, we evaluated the differences in PFS between patients with different biomarker expression levels. However, patients who had increased expression of phosphor-STAT3 (p-STAT3), phosphor-AKT (p-AKT), or MET had similar PFS to those who did not (Fig. 3A–C). Moreover, patients with decreased E-cadherin expression had similar PFS to patients with preserved E-cadherin expression (Fig. 3D). Finally, the patients without BIM expression had PFS of 25.0 months (interquartile range: 18.7–not reached) than patients with BIM expression (11.9 months, interquartile range: 6.4–22.7) (p < 0.001, Fig. 3E). Immunohistochemical staining of the five biomarkers is presented in Fig. 4.
Figure 2.
(A) The immunohistochemical stain of different PD-L1 expression levels. (B) Progression-free survival among EGFR-mutant NSCLC patients receiving first-line EGFR-TKI. (C) Kaplan–Meier analysis of progression-free survival among patients with different PD-L1 expression levels.
Figure 3.
Kaplan–Meier analysis of progression-free survival among patients with the different expression levels of p-AKT (A), p-STAT3 (B), MET (C), E-cadherin (D), and BIM (E).
Figure 4.
The immunohistochemical stain of p-AKT (1:100) (A), p-STAT3 (1:100) (B), MET (1:30) (C), E-cadherin (1:50) (D), and BIM (1:100) (E).
Adjusting the possible confounders by multivariate Cox proportional hazards regression analysis, both the presence of brain metastasis (hazard ratio 1.69, 95% confidence interval: 1.06–2.70; P = 0.028) and BIM expression (hazard ratio 2.94, 95% confidence interval: 1.41–6.14; P = 0.004) were independent poor prognostic factors for PFS (Table 2). Among the 23 patients without BIM expression, 20 were also negative for PD-L1 expression, which was significantly correlated (p = 0.021). Therefore, we further analyzed the PFS of subgroups of patients with BIM expression and found that the PFS was similar among patients with different PD-L1 expression levels (Fig. 5). Moreover, all subgroups of patients with BIM expression had significantly shorter PFS than patients without BIM expression, regardless of PD-L1 expression (p = 0.004) (Fig. 5).
Table 2.
Cox proportional hazards regression analysis of progression-free survival.
| HR (95% CI) | P-value | ||
|---|---|---|---|
| Age (years) | > 65 years vs. < 65 years | 1.09 (0.63–1.87) | 0.759 |
| Sex | Male vs. Female | 0.99 (0.63–1.55) | 0.963 |
| Brain metastasis | Presence vs. Absence | 1.69 (1.06–2.70) | 0.028 |
| Performance status | ECOG ≥ 2 vs. 0–1 | 1.66 (0.85–3.25) | 0.138 |
| EGFR mutation | Exon 19 deletion vs. Others | 0.97 (0.60–1.57) | 0.908 |
| PD-L1 level | ≥ 50% vs. < 1% | 1.42 (0.73–2.76) | 0.300 |
| 1–49% vs. < 1% | 1.17 (0.68–2.01) | 0.576 | |
| E-cadherin | Loss vs. preserved | 1.33 (0.69–2.56) | 0.393 |
| MET | Presence vs. absence | 1.00 (0.62–1.62) | 0.989 |
| p-STAT3 | Presence vs. absence | 0.78 (0.48–1.29) | 0.337 |
| p-AKT | Presence vs. absence | 1.08 (0.64–1.83) | 0.767 |
| BIM | Presence vs. absence | 2.94 (1.41–6.14) | 0.004 |
EGFR, epidermal growth factor receptor.
Figure 5.
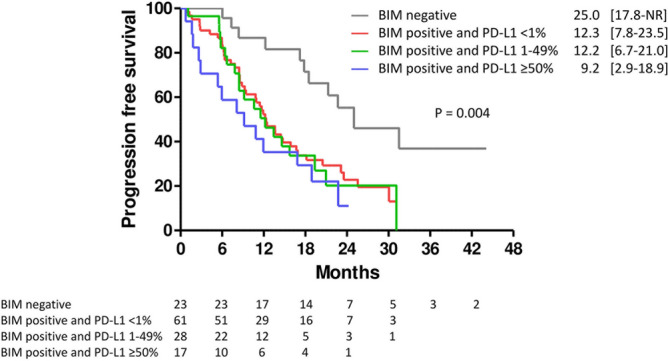
Kaplan–Meier analysis of progression-free survival among patients with different BIM and PD-L1 expression levels. The patients with BIM expression had shorter progression-free survival than those without BIM expression, regardless of PD-L1 expression.
We found that PD-L1 expression was associated with shorter PFS, but with only borderline statistical significance in patients who received first-generation EGFR-TKI (11.5 versus 14.5 months, p = 0.076, Supplementary Fig. 1A). In contrast, BIM expression was associated with shorter PFS (10.9 versus 22.0 months, p = 0.030, Supplementary Fig. 1B) in this patient subgroup. Additionally, both PD-L1 (12.2 versus 25.6 months, p = 0.049) and BIM (12.2 versus NR, p = 0.002) expression was associated with significantly shorter PFS in patients who received second-generation EGFR-TKI (Supplementary Fig. 2A,B). We further adjusted for possible confounding factors using a multivariate Cox proportional hazard regression model. The presence of BIM expression was a poor prognostic factor for PFS, with borderline statistical significance (p = 0.070, Supplementary Table 1) in patients who received first-generation EGFR-TKI; however, it was an independent poor prognostic factor for PFS in patients who received second-generation EGFR-TKI (p = 0.002, Supplementary Table 2). In contrast, high PD-L1 expression was not associated with shorter PFS in patients who received first-generation (p = 0.665) or second-generation (p = 0.887) EGFR-TKI. Although the use of second-generation EGFR-TKI had been associated with a better treatment outcome, BIM expression was still an independent prognostic factor in this patient subgroup in our study (Supplementary Fig. 2B, Supplementary Table 2).
Assessing apoptotic ratio via flow cytometry
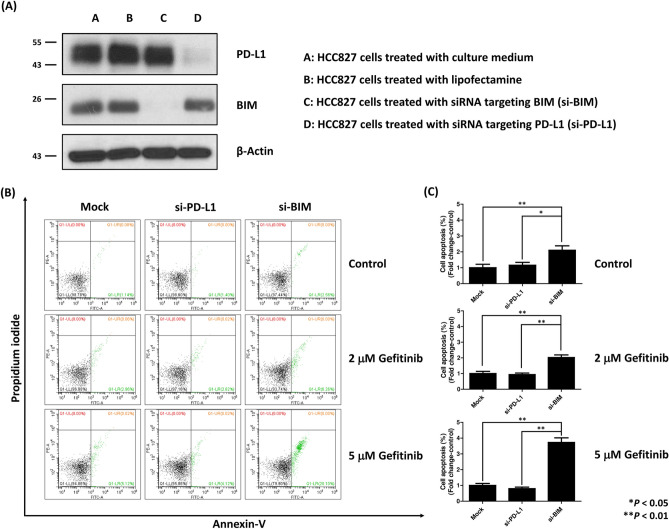
To investigate the role of BIM knockdown on drug sensitivity, flow cytometry of apoptosis was performed after transfecting HCC827 cells with siRNA targeting PD-L1 or BIM. With transfection of siRNA targeting PD-L1 and BIM, the western blotting of PD-L1 and BIM decreased significantly (Fig. 6A). The original blots are presented in Supplementary Fig. 3. After 24 h of siRNA transfection, HCC827 cells were treated with gefitinib for 24 h. The apoptotic rate of HCC827 cells was determined using flow cytometry with annexin V/PI staining (Fig. 6B). The apoptosis rates in cells transfected with siRNA targeting PD-L1 were similar to those in the control group (Fig. 6C), whereas the apoptosis rates of cells treated with siRNA targeting BIM were significantly higher than those in the control group or cells treated with siRNA targeting PD-L1 (Fig. 6C). These results suggested that the knockdown of BIM expression enhanced the drug sensitivity of HCC827 cells to gefitinib.
Figure 6.
The BIM silencing increased lung cancer cell apoptosis compared to the PD-L1 silencing after treatment with gefitinib. (A) The western blot confirmed the silencing of PD-L1 and BIM of HCC827 cells by transfecting siRNA. (B) After transfecting with siRNA targeting PD-L1 and BIM, the HCC827 cells were cultured in control conditions (DMSO) or in the presence of the indicated doses of gefitinib, for 24 h. Flow cytometry was performed after staining cells with Annexin V-FITC and propidium iodide (PI). The cells in the Mock group were treated with transfecting vector only. (C) Quantitative analysis of Annexin V-FITC flow cytometry showed an increase in apoptotic cells in HCC827 cells with BIM silencing. *P < 0.05, **P < 0.01.
Genomic testing of the BIM intronic deletion polymorphism
A previous study demonstrated that the BIM deletion polymorphism reduced the treatment efficacy of EGFR-TKI40. To better clarify the role of BIM protein expression on EGFR-TKI treatment efficacy, we collected residual tissue to perform BIM polymorphic genotyping and investigate the association between BIM deletion polymorphism and the protein expression of BIM. There were 42 patients with lung cancer who had sufficient residual tumor tissue for genomic testing. Among them, DNA from six samples did not qualify for subsequent PCR testing. Of the remaining 36 samples, 4 had the BIM deletion polymorphism and 32 had only the wild-type allele (Supplementary Fig. 4). The association between the BIM deletion polymorphism and BIM protein expression is summarized in Supplementary Table 3. Although there was no statistical significance, all the individuals with the polymorphism also had BIM protein expression.
Discussion
In the present study, we first proved that both BIM and PDL1 expression was correlated with the PFS of EGFR-TKI treatment in EGFR-mutant NSCLC. Using COX proportional hazard regression analysis, we further demonstrated that BIM expression, instead of PD-L1 expression, was an independent poor prognostic factor for PFS (hazard ratio 2.94, 95% confidence interval, 1.41–6.14; p = 0.004). We further verified the proof-of-concept in vitro; the knockdown of BIM instead of PD-L1 using siRNA enhanced gefitinib-induced apoptosis. Our study is the first to globally identify the effectors mediating the predicting role of PD-L1 expression in EGFR-mutant advanced NSCLC patients treated with EGFR-TKIs, which explains the inconsistency of the prognostic value of PD-L1 in previous studies.
Activation of the EGFR pathway has been shown to increase the expression of PD-L1 and other immunosuppressive factors, suggesting a role for EGFR signaling pathway in remodeling the tumor microenvironment. Tumors from patients with EGFR-mutant NSCLC were also found to be PD-L1 positive by IHC analysis28,41–43. The association between PD-L1 expression and EGFR-TKI treatment efficacy has been widely studied, but the results have been inconsistent (Table 3)8–15,19,41–44. In a retrospective study conducted by Matsumoto et al., high PD-L1 expression deteriorated the treatment efficacy of EGFR-TKIs. This phenomenon was more significant when higher CD8-positive T cell infiltration was observed in the tumor microenvironment10, which implied that PD-1/PD-L1 inhibitors might provide clinical benefits in patients with EGFR-mutant NSCLC and high PD-L1 expression. However, the phase 3 open-label CAURAL trial investigated osimertinib plus durvalumab versus osimertinib monotherapy in patients with EGFR-TKI sensitizing and EGFR T790M mutation-positive advanced NSCLC and disease progression after EGFR-TKI therapy45. Although the study was terminated early because of high interstitial lung disease in the separate phase Ib TATTON trial, and the objective response rates were 80% in the osimertinib arm and only 64% in the combination arm. The study not only demonstrated no additional benefit of combination immunotherapy in patients with EGFR-mutant NSCLC but also implied the contradictory role of PD-L1 expression in predicting response to EGFR-TKI.
Table 3.
Summary of data regarding the prognostic role of PD-L1 in treatment efficacy of EGFR-TKIs.
| Author | Year | Patient number | PD-L1 antibody | Outcome associated with high PD-L1 expression |
|---|---|---|---|---|
| D’Incecco et al.42 | 2015 | 95 | Ab58810 | Better RR and longer TTP |
| Lin et al.43 | 2015 | 56 | Ab58810 | Better DCR and longer PFS |
| Tang et al.41 | 2015 | 64 | E1L3N | No association |
| Soo et al.11 | 2017 | 90 | SP142 | Shorter PFS |
| Yoneshima et al.14 | 2018 | 71 | Dako 22C3 | Shorter PFS |
| Su et al.12 | 2018 | 101 | SP142 | Poor RR, shorter PFS, higher primary resistance rate |
| Hsu et al.8 | 2019 | 123 | SP263 | Shorter PFS, higher primary resistance rate |
| Matsumoto et al.10 | 2019 | 52 | 28-8 | Shorter PFS |
| D7U8C | ||||
| Yang et al.13 | 2020 | 153 | Dako 22C3 | Poor RR, shorter PFS, higher primary resistance rate |
| Yoon et al.15 | 2020 | 131 | Dako 22C3 | Poor RR and shorter PFS |
| Kim et al.44 | 2020 | 69 | SP142 | No association |
| SP263 | ||||
| Dako 22C3 | ||||
| Chang et al.19 | 2021 | 114 | Dako 22C3 | No association |
| Liu et al.9 | 2021 | 186 | SP263 | Shorter PFS |
| Present study | 2022 | 157 | Dako 22C3 | Shorter PFS |
| Associated with BIM expression |
DCR, disease control rate; EGFR, epidermal growth factor receptor; PFS, progression-free survival; RR, response rate; TKIs, tyrosine kinase inhibitors; TTP, time to progression.
The recent studies have disclosed cancer cell-intrinsic PD-L1 signals, including the control of tumor growth and survival pathways, stemness, immune effects, DNA damage responses, and gene expression regulation, especially those that are PD-1-independent20. Many studies have provided evidence for oncogenic pathway activation and EMT in modulating cancer cell-intrinsic PD-L1 signals. In a cell line study, constitutive activation of the EGFR signaling pathway induced upregulation of PD-L1 expression through the p-ERK1/2/p-c-Jun pathway46. The similar phenomenon was observed in the activation of other oncogenic pathways, including ALK47, KRAS48, and MET49. Moreover, the inhibition of downstream AKT and STAT3 signaling can decrease PD-L1 expression in PC9 cells50. Similarly, lung cancer samples with EMT phenotypes, defined by decreased E-cadherin expression, also had significantly higher PD-L1 expression than those with epithelial phenotypes51. The suppression of EMT by siRNA-mediated ZEB1 knockdown can also suppress PD-L1 expression52. Another mechanism associated with cell-intrinsic PD-L1 expression, which may also affect the treatment response to EGFR-TKIs, is the expression of BIM53. BIM is a BH3-only protein of the Bcl-2 family that promotes apoptosis by regulating antiapoptotic Bcl-2 proteins54 and is an important factor in EGFR-TKI-mediated apoptosis28. BIM upregulation is also associated with the expression of PD-L1 in melanoma55. However, data regarding the role of BIM expression in the treatment efficacy of EGFR-TKIs remains controversial27,56–59. In the post-hoc analysis of the EURTAC trial, patients with high BIM expression had significantly shorter PFS when receiving erlotinib (9.2 vs. 15.0 months, p = 0.02)56. This result was further supported by the other two cohort studies27,57. In contrast, in an in vitro cell line study, the apoptotic ratio after gefitinib treatment was positively correlated with BIM expression58. Moreover, cells treated with shRNA-mediated BIM knockdown were more resistant to gefitinib therapy59. To the best of our knowledge, no prior studies globally examined the impact of these pathways (AKT, STAT3, MET, BIM, and EMT), especially their interaction with PD-L1 expression, on the treatment efficiency of EGFR-TKI. Using IHC to detect the activity of these signaling pathways, we found that activation of the oncogenic pathway (AKT, STAT3, and MET) and EMT pathway (loss of E-cadherin) did not affect PFS. (Fig. 3A–D). However, patients with BIM expression had a shorter PFS than those without BIM expression (Fig. 3E), regardless of PD-L1 expression (Fig. 5). COX proportional hazard regression analysis further confirmed that BIM expression is an independent prognostic factor, whereas the expression level of PD-L1 did not affect PFS in multivariate analysis (Table 2). Furthermore, although the use of second-generation EGFR-TKI is associated with a better treatment outcome60, BIM expression is still an independent prognostic factor in patients who received second-generation EGFR-TKI (Supplementary Fig. 2B, Supplementary Table 2). To verify the significant role of BIM in mediating the sensitivity to gefitinib, we transfected EGFR-mutant lung cancer cells (HCC827) with siRNA targeting BIM or PD-L1. We found only the knockdown of BIM increased apoptotic cells following gefitinib treatment (Fig. 6), which is compatible with the IHC finding that only BIM expression has an impact on PFS in patients with EGFR-mutant NSCLC receiving EGFR-TKI treatment.
The current study had some limitations. First, this was a retrospective study of patients from a single center. The baseline characteristics of patients with different PD-L1 or BIM expressions were imbalanced. To eliminate potential confounding factors, we performed COX proportional hazard regression analysis to confirm that BIM expression is an independent prognostic factor. The prospective study with balanced characteristics is required to confirm the results of this study. Second, we used IHC staining to evaluate the role of BIM in the treatment efficacy of EGFR-TKIs instead of the BIM deletion polymorphism. The relationship between BIM expression levels and BIM deletion polymorphisms requires further investigation. We discovered that the patients with the BIM deletion polymorphism also expressed BIM, but there was no statistical significance of this correlation due to the limited patient number. However, previous studies regarding the role of BIM deletion polymorphism in EGFR-TKI treatment still had inconclusive results6,61–63. Moreover, we have used the HCC827 cell line with an siRNA-mediated knockdown to confirm the role of BIM expression in the treatment efficacy of EGFR-TKI.
In conclusion, we use retrospective cohort study to reveal that patients with high BIM expression had significantly shorter PFS, regardless of PD-L1 expression. Subsequent in vitro studies also demonstrated an increased apoptotic ratio of HCC827 cells to gefitinib therapy after transfection with siRNA targeting BIM. Our data suggests that, among the pathways associated with cell-intrinsic PD-L1 signaling, BIM is potentially the underlying mechanism that affects the prognostic role of PD-L1 and mediates cell apoptosis when receiving gefitinib treatment in EGFR-mutant NSCLC. Further prospective studies are required to validate the results of this study.
Supplementary Information
Acknowledgements
This study was based in part on data sourced from the Cancer Data Bank of National Cheng Kung University Hospital.
Author contributions
C.-Y.C., C.-Y.L., C.-C.L., and P.-L.S. designed the study. C.-Y.C., C.-C.L., and P.-L.S. were responsible for the biomarker measurements. S.-C.Y. and P.-L.S. performed the statistical analysis. J-S.T., C.-W.C., C.-Y.L., C.-C.C., Y.-T.Y., Y.-L.T., and W.-C.S. collected the clinical data. C.-Y.C., C.-Y.L., C.-C.L., and P.-L.S. participated in the manuscript preparation and revised the manuscript. C.-F.L. the performed BIM deletion polymorphism analysis. All authors read and approved the final version of the manuscript.
Funding
This research was funded by the National Science and Technology Council of Taiwan (NSTC 110-2314-B-006-098-MY3 and NSTC 111-2314-B-006-092-MY3). This research was also funded by the National Cheng Kung University Hospital, NCKUH-11102019.
Data availability
The raw data supporting the conclusions of this article will be made available on reasonable request by the corresponding author, Dr. Po-Lan Su.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Chang-Yao Chu, Chien-Yu Lin and Chien-Chung Lin.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-023-30565-4.
References
- 1.Lynch TJ, Bell DW, Sordella R, Gurubhagavatula S, Okimoto RA, Brannigan BW, et al. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N. Engl. J. Med. 2004;350:2129–2139. doi: 10.1056/NEJMoa040938. [DOI] [PubMed] [Google Scholar]
- 2.Lee CK, Davies L, Wu YL, Mitsudomi T, Inoue A, Rosell R, et al. Gefitinib or erlotinib vs chemotherapy for EGFR mutation-positive lung cancer: Individual patient data meta-analysis of overall survival. J. Natl. Cancer Inst. 2017 doi: 10.1093/jnci/djw279. [DOI] [PubMed] [Google Scholar]
- 3.Soria JC, Ohe Y, Vansteenkiste J, Reungwetwattana T, Chewaskulyong B, Lee KH, et al. Osimertinib in untreated EGFR-mutated advanced non-small-cell lung cancer. N. Engl. J. Med. 2018;378:113–125. doi: 10.1056/NEJMoa1713137. [DOI] [PubMed] [Google Scholar]
- 4.Reita D, Pabst L, Pencreach E, Guérin E, Dano L, Rimelen V, et al. Molecular mechanism of EGFR-TKI resistance in EGFR-mutated non-small cell lung cancer: Application to biological diagnostic and monitoring. Cancers. 2021;13:4926. doi: 10.3390/cancers13194926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rotow J, Bivona TG. Understanding and targeting resistance mechanisms in NSCLC. Nat. Rev. Cancer. 2017;17:637–658. doi: 10.1038/nrc.2017.84. [DOI] [PubMed] [Google Scholar]
- 6.Isobe K, Hata Y, Tochigi N, Kaburaki K, Kobayashi H, Makino T, et al. Clinical significance of BIM deletion polymorphism in non-small-cell lung cancer with epidermal growth factor receptor mutation. J. Thorac. Oncol. 2014;9:483–487. doi: 10.1097/JTO.0000000000000125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hanahan D, Weinberg RA. Hallmarks of cancer: The next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 8.Hsu KH, Huang YH, Tseng JS, Chen KC, Ku WH, Su KY, et al. High PD-L1 expression correlates with primary resistance to EGFR-TKIs in treatment naïve advanced EGFR-mutant lung adenocarcinoma patients. Lung Cancer. 2019;127:37–43. doi: 10.1016/j.lungcan.2018.11.021. [DOI] [PubMed] [Google Scholar]
- 9.Liu J, Itchins M, Nagrial A, Cooper WA, De Silva M, Barnet M, et al. Relationship between PD-L1 expression and outcome in EGFR-mutant lung cancer patients treated with EGFR tyrosine kinase inhibitors. Lung Cancer. 2021;155:28–33. doi: 10.1016/j.lungcan.2021.03.004. [DOI] [PubMed] [Google Scholar]
- 10.Matsumoto Y, Sawa K, Fukui M, Oyanagi J, Izumi M, Ogawa K, et al. Impact of tumor microenvironment on the efficacy of epidermal growth factor receptor-tyrosine kinase inhibitors in patients with EGFR-mutant non-small cell lung cancer. Cancer Sci. 2019;110:3244–3254. doi: 10.1111/cas.14156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Soo RA, Kim HR, Asuncion BR, Fazreen Z, Omar MFM, Herrera MC, et al. Significance of immune checkpoint proteins in EGFR-mutant non-small cell lung cancer. Lung Cancer. 2017;105:17–22. doi: 10.1016/j.lungcan.2017.01.008. [DOI] [PubMed] [Google Scholar]
- 12.Su S, Dong ZY, Xie Z, Yan LX, Li YF, Su J, et al. Strong programmed death ligand 1 expression predicts poor response and de novo resistance to EGFR tyrosine kinase inhibitors among NSCLC patients with EGFR mutation. J. Thorac. Oncol. 2018;13:1668–1675. doi: 10.1016/j.jtho.2018.07.016. [DOI] [PubMed] [Google Scholar]
- 13.Yang CY, Liao WY, Ho CC, Chen KY, Tsai TH, Hsu CL, et al. Association between programmed death-ligand 1 expression, immune microenvironments, and clinical outcomes in epidermal growth factor receptor mutant lung adenocarcinoma patients treated with tyrosine kinase inhibitors. Eur. J. Cancer. 2020;124:110–122. doi: 10.1016/j.ejca.2019.10.019. [DOI] [PubMed] [Google Scholar]
- 14.Yoneshima Y, Ijichi K, Anai S, Ota K, Otsubo K, Iwama E, et al. PD-L1 expression in lung adenocarcinoma harboring EGFR mutations or ALK rearrangements. Lung Cancer. 2018;118:36–40. doi: 10.1016/j.lungcan.2018.01.024. [DOI] [PubMed] [Google Scholar]
- 15.Yoon BW, Chang B, Lee SH. High PD-L1 expression is associated with unfavorable clinical outcome in EGFR-mutated lung adenocarcinomas treated with targeted therapy. Onco. Targets Ther. 2020;13:8273–8785. doi: 10.2147/OTT.S271011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hsu P-C, Wang C-W, Kuo SC-H, Lin S-M, Lo Y-L, Huang AC-C, et al. The co-expression of programmed death-ligand 1 (PD-L1) in untreated EGFR-mutated metastatic lung adenocarcinoma. Biomedicines. 2020;8:36. doi: 10.3390/biomedicines8020036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bai Y, Chen X, Hou L, Qian J, Jiang T, Zhou C, et al. PD-L1 expression and its effect on clinical outcomes of EGFR-mutant NSCLC patients treated with EGFR-TKIs. Cancer Biol. Med. 2018;15:434–442. doi: 10.20892/j.issn.2095-3941.2018.0223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Brown H, Vansteenkiste J, Nakagawa K, Cobo M, John T, Barker C, et al. Programmed cell death ligand 1 expression in untreated EGFR mutated advanced NSCLC and response to osimertinib versus comparator in FLAURA. J. Thorac. Oncol. 2020;15:138–143. doi: 10.1016/j.jtho.2019.09.009. [DOI] [PubMed] [Google Scholar]
- 19.Chang CY, Lai YC, Wei YF, Chen CY, Chang SC. PD-L1 expression and outcome in patients with metastatic non-small cell lung cancer and EGFR mutations receiving EGFR-TKI as frontline treatment. Onco. Targets Ther. 2021;14:2301–2309. doi: 10.2147/OTT.S290445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kornepati AVR, Vadlamudi RK, Curiel TJ. Programmed death ligand 1 signals in cancer cells. Nat. Rev. Cancer. 2022;22:174–189. doi: 10.1038/s41568-021-00431-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang N, Zeng Y, Du W, Zhu J, Shen D, Liu Z, et al. The EGFR pathway is involved in the regulation of PD-L1 expression via the IL-6/JAK/STAT3 signaling pathway in EGFR-mutated non-small cell lung cancer. Int. J. Oncol. 2016;49:1360–1368. doi: 10.3892/ijo.2016.3632. [DOI] [PubMed] [Google Scholar]
- 22.Luo F, Luo M, Rong Q-X, Zhang H, Chen Z, Wang F, et al. Niclosamide, an antihelmintic drug, enhances efficacy of PD-1/PD-L1 immune checkpoint blockade in non-small cell lung cancer. J. Immunother. Cancer. 2019;7:245. doi: 10.1186/s40425-019-0733-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu Z, Ma L, Sun Y, Yu W, Wang X. Targeting STAT3 signaling overcomes gefitinib resistance in non-small cell lung cancer. Cell Death Dis. 2021;12:561. doi: 10.1038/s41419-021-03844-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lastwika KJ, Wilson W, Li QK, Norris J, Xu H, Ghazarian SR, et al. Control of PD-L1 expression by oncogenic activation of the AKT–mTOR pathway in non-small cell lung cancer. Cancer Res. 2016;76:227–238. doi: 10.1158/0008-5472.CAN-14-3362. [DOI] [PubMed] [Google Scholar]
- 25.Jacobsen K, Bertran-Alamillo J, Molina MA, Teixidó C, Karachaliou N, Pedersen MH, et al. Convergent Akt activation drives acquired EGFR inhibitor resistance in lung cancer. Nat. Commun. 2017;8:410. doi: 10.1038/s41467-017-00450-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xu D, Li J, Li RY, Lan T, Xiao C, Gong P. PD-L1 expression is regulated by NF-κB during EMT signaling in gastric carcinoma. Onco. Targets Ther. 2019;12:10099–10105. doi: 10.2147/OTT.S224053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Isobe K, Kakimoto A, Mikami T, Kaburaki K, Kobayashi H, Yoshizawa T, et al. PD-L1 mRNA expression in EGFR-mutant lung adenocarcinoma. Oncol. Rep. 2018;40:331–338. doi: 10.3892/or.2018.6442. [DOI] [PubMed] [Google Scholar]
- 28.Gong Y, Somwar R, Politi K, Balak M, Chmielecki J, Jiang X, et al. Induction of BIM is essential for apoptosis triggered by EGFR kinase inhibitors in mutant EGFR-dependent lung adenocarcinomas. PLoS Med. 2007;4:e294. doi: 10.1371/journal.pmed.0040294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tirkes T, Hollar MA, Tann M, Kohli MD, Akisik F, Sandrasegaran K. Response criteria in oncologic imaging: Review of traditional and new criteria. Radiographics. 2013;33:1323–1341. doi: 10.1148/rg.335125214. [DOI] [PubMed] [Google Scholar]
- 30.Garon EB, Rizvi NA, Hui R, Leighl N, Balmanoukian AS, Eder JP, et al. Pembrolizumab for the treatment of non-small-cell lung cancer. N. Engl. J. Med. 2015;372:2018–2028. doi: 10.1056/NEJMoa1501824. [DOI] [PubMed] [Google Scholar]
- 31.Herbst RS, Baas P, Kim DW, Felip E, Pérez-Gracia JL, Han JY, et al. Pembrolizumab versus docetaxel for previously treated, PD-L1-positive, advanced non-small-cell lung cancer (KEYNOTE-010): A randomised controlled trial. Lancet. 2016;387:1540–1550. doi: 10.1016/S0140-6736(15)01281-7. [DOI] [PubMed] [Google Scholar]
- 32.Reck M, Rodríguez-Abreu D, Robinson AG, Hui R, Csőszi T, Fülöp A, et al. Pembrolizumab versus chemotherapy for PD-L1-positive non-small-cell lung cancer. N. Engl. J. Med. 2016;375:1823–1833. doi: 10.1056/NEJMoa1606774. [DOI] [PubMed] [Google Scholar]
- 33.Lee J, Jiffar T, Kupferman ME. A novel role for BDNF-TrkB in the regulation of chemotherapy resistance in head and neck squamous cell carcinoma. PLoS ONE. 2012;7:e30246. doi: 10.1371/journal.pone.0030246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lin WH, Chang YW, Hong MX, Hsu TC, Lee KC, Lin C, et al. STAT3 phosphorylation at Ser727 and Tyr705 differentially regulates the EMT-MET switch and cancer metastasis. Oncogene. 2021;40:791–805. doi: 10.1038/s41388-020-01566-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Castiglione R, Alidousty C, Holz B, Wagener S, Baar T, Heydt C, et al. Comparison of the genomic background of MET-altered carcinomas of the lung: Biological differences and analogies. Mod. Pathol. 2019;32:627–638. doi: 10.1038/s41379-018-0182-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lu J, Zang H, Zheng H, Wagener S, Baar T, Heydt C, et al. Overexpression of p-Akt, p-mTOR and p-eIF4E proteins associates with metastasis and unfavorable prognosis in non-small cell lung cancer. PLoS ONE. 2020;15:e0227768. doi: 10.1371/journal.pone.0227768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yoo JY, Yang SH, Lee JE, Cho DG, Kim HK, Kim SH, et al. E-cadherin as a predictive marker of brain metastasis in non-small-cell lung cancer, and its regulation by pioglitazone in a preclinical model. J. Neurooncol. 2012;109:219–227. doi: 10.1007/s11060-012-0890-8. [DOI] [PubMed] [Google Scholar]
- 38.Su PL, Wu YL, Chang WY, Ho CL, Tseng YL, Lai WW, et al. Preventing and treating brain metastases with three first-line EGFR-tyrosine kinase inhibitors in patients with EGFR mutation-positive advanced non-small cell lung cancer. Ther. Adv. Med. Oncol. 2018;10:1758835918797589. doi: 10.1177/1758835918797589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Båtevik R, Grong K, Segadal L, Stangeland L. The female gender has a positive effect on survival independent of background life expectancy following surgical resection of primary non-small cell lung cancer: A study of absolute and relative survival over 15 years. Lung Cancer. 2005;47:173–181. doi: 10.1016/j.lungcan.2004.08.014. [DOI] [PubMed] [Google Scholar]
- 40.Ng KP, Hillmer AM, Chuah CT, Juan WC, Ko TK, Teo AS, et al. A common BIM deletion polymorphism mediates intrinsic resistance and inferior responses to tyrosine kinase inhibitors in cancer. Nat. Med. 2012;18:521–528. doi: 10.1038/nm.2713. [DOI] [PubMed] [Google Scholar]
- 41.Tang Y, Fang W, Zhang Y, Hong S, Kang S, Yan Y, et al. The association between PD-L1 and EGFR status and the prognostic value of PD-L1 in advanced non-small cell lung cancer patients treated with EGFR-TKIs. Oncotarget. 2015;6:14209–14219. doi: 10.18632/oncotarget.3694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.D'Incecco A, Andreozzi M, Ludovini V, Rossi E, Capodanno A, Landi L, et al. PD-1 and PD-L1 expression in molecularly selected non-small-cell lung cancer patients. Br. J. Cancer. 2015;112:95–102. doi: 10.1038/bjc.2014.555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lin C, Chen X, Li M, Liu J, Qi X, Yang W, et al. Programmed death-ligand 1 expression predicts tyrosine kinase inhibitor response and better prognosis in a cohort of patients with epidermal growth factor receptor mutation-positive lung adenocarcinoma. Clin. Lung Cancer. 2015;16:e25–35. doi: 10.1016/j.cllc.2015.02.002. [DOI] [PubMed] [Google Scholar]
- 44.Kim T, Cha YJ, Chang YS. Correlation of PD-L1 expression tested by 22C3 and SP263 in non-small cell lung cancer and its prognostic effect on EGFR mutation-positive lung adenocarcinoma. Tuberc. Respir. Dis. 2020;83:51–60. doi: 10.4046/trd.2019.0026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yang JC, Shepherd FA, Kim DW, Lee GW, Lee JS, Chang GC, et al. Osimertinib plus durvalumab versus osimertinib monotherapy in EGFR T790M-positive NSCLC following previous EGFR TKI therapy: CAURAL brief report. J. Thorac. Oncol. 2019;14:933–939. doi: 10.1016/j.jtho.2019.02.001. [DOI] [PubMed] [Google Scholar]
- 46.Chen N, Fang W, Zhan J, Hong S, Tang Y, Kang S, et al. Upregulation of PD-L1 by EGFR activation mediates the immune escape in EGFR-driven NSCLC: Implication for optional immune targeted therapy for NSCLC patients with EGFR mutation. J. Thorac. Oncol. 2015;10:910–923. doi: 10.1097/JTO.0000000000000500. [DOI] [PubMed] [Google Scholar]
- 47.Ota K, Azuma K, Kawahara A, Hattori S, Iwama E, Tanizaki J, et al. Induction of PD-L1 expression by the EML4-ALK oncoprotein and downstream signaling pathways in non-small cell lung cancer. Clin. Cancer Res. 2015;21:4014–4021. doi: 10.1158/1078-0432.CCR-15-0016. [DOI] [PubMed] [Google Scholar]
- 48.Glorieux C, Xia X, He YQ, Hu Y, Cremer K, Robert A, et al. Regulation of PD-L1 expression in K-ras-driven cancers through ROS-mediated FGFR1 signaling. Redox Biol. 2021;38:101780. doi: 10.1016/j.redox.2020.101780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Saigi M, Alburquerque-Bejar JJ, Mc Leer-Florin A, Pereira C, Pros E, Romero OA, et al. MET-oncogenic and JAK2-inactivating alterations are independent factors that affect regulation of PD-L1 expression in lung cancer. Clin. Cancer Res. 2018;24:4579–4587. doi: 10.1158/1078-0432.CCR-18-0267. [DOI] [PubMed] [Google Scholar]
- 50.Abdelhamed S, Ogura K, Yokoyama S, Saiki I, Hayakawa Y. AKT-STAT3 pathway as a downstream target of EGFR signaling to regulate PD-L1 expression on NSCLC cells. J. Cancer. 2016;7:1579–1586. doi: 10.7150/jca.14713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kim S, Koh J, Kim MY, Kwon D, Go H, Kim YA, et al. PD-L1 expression is associated with epithelial-to-mesenchymal transition in adenocarcinoma of the lung. Hum. Pathol. 2016;58:7–14. doi: 10.1016/j.humpath.2016.07.007. [DOI] [PubMed] [Google Scholar]
- 52.Tsutsumi S, Saeki H, Nakashima Y, Ito S, Oki E, Morita M, et al. Programmed death-ligand 1 expression at tumor invasive front is associated with epithelial-mesenchymal transition and poor prognosis in esophageal squamous cell carcinoma. Cancer Sci. 2017;108:1119–1127. doi: 10.1111/cas.13237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Costa DB, Halmos B, Kumar A, Schumer ST, Huberman MS, Boggon TJ, et al. BIM mediates EGFR tyrosine kinase inhibitor-induced apoptosis in lung cancers with oncogenic EGFR mutations. PLoS Med. 2007;4:1669–1679. doi: 10.1371/journal.pmed.0040315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Youle RJ, Strasser A. The BCL-2 protein family: Opposing activities that mediate cell death. Nat. Rev. Mol. Cell Biol. 2008;9:47–59. doi: 10.1038/nrm2308. [DOI] [PubMed] [Google Scholar]
- 55.Gibbons RM, Liu X, Pulko V, Harrington SM, Krco CJ, Kwon ED, et al. B7–H1 limits the entry of effector CD8(+) T cells to the memory pool by upregulating Bim. Oncoimmunology. 2012;1:1061–1073. doi: 10.4161/onci.20850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Karachaliou N, Codony-Servat J, Teixidó C, Pilotto S, Drozdowskyj A, Codony-Servat C, et al. BIM and mTOR expression levels predict outcome to erlotinib in EGFR-mutant non-small-cell lung cancer. Sci. Rep. 2015;5:17499. doi: 10.1038/srep17499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Incharoen P, Charonpongsuntorn C, Saowapa S, Sirachainan E, Dejthevaporn T, Kampreasart K, et al. Role of BIM deletion polymorphism and BIM expression as predictive biomarkers to maximize the benefit of EGFR-TKI treatment in EGFR-positive NSCLC. Asian Pac. J. Cancer Prev. 2019;20:3581–3589. doi: 10.31557/APJCP.2019.20.12.3581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Faber AC, Corcoran RB, Ebi H, Sequist LV, Waltman BA, Chung E, et al. BIM expression in treatment-naive cancers predicts responsiveness to kinase inhibitors. Cancer Discov. 2011;1:352–365. doi: 10.1158/2159-8290.CD-11-0106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lin PL, Wu TC, Wu DW, Wang L, Chen CY, Lee H. An increase in BAG-1 by PD-L1 confers resistance to tyrosine kinase inhibitor in non-small cell lung cancer via persistent activation of ERK signalling. Eur. J. Cancer. 2017;85:95–105. doi: 10.1016/j.ejca.2017.07.025. [DOI] [PubMed] [Google Scholar]
- 60.Park K, Tan EH, O'Byrne K, Zhang L, Boyer M, Mok TSK, et al. Afatinib versus gefitinib as first-line treatment of patients with EGFR mutation-positive non-small-cell lung cancer (LUX-Lung 7): A phase 2B, open-label, randomised controlled trial. Lancet Oncol. 2016;17:577–589. doi: 10.1016/S1470-2045(16)30033-X. [DOI] [PubMed] [Google Scholar]
- 61.Lee JY, Ku BM, Lim SH, Lee MY, Kim H, Kim M, et al. The BIM deletion polymorphism and its clinical implication in patients with EGFR-mutant non-small-cell lung cancer treated with EGFR tyrosine kinase inhibitors. J. Thorac. Oncol. 2015;10:903–909. doi: 10.1097/JTO.0000000000000535. [DOI] [PubMed] [Google Scholar]
- 62.Nakagawa T, Takeuchi S, Yamada T, Ebi H, Sano T, Nanjo S, et al. EGFR-TKI resistance due to BIM polymorphism can be circumvented in combination with HDAC inhibition. Cancer Res. 2013;73:2428–2434. doi: 10.1158/0008-5472.CAN-12-3479. [DOI] [PubMed] [Google Scholar]
- 63.Zhao M, Zhang Y, Cai W, Li J, Zhou F, Cheng N, et al. The Bim deletion polymorphism clinical profile and its relation with tyrosine kinase inhibitor resistance in Chinese patients with non-small cell lung cancer. Cancer. 2014;120:2299–2307. doi: 10.1002/cncr.28725. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The raw data supporting the conclusions of this article will be made available on reasonable request by the corresponding author, Dr. Po-Lan Su.