Follicular T helper (Tfh) cells coordinate optimal germinal center responses, but abnormal Tfh cell function contributes to autoantibody-dependent autoimmunity. Seth et al. demonstrate that AP-1–independent NFAT gene expression, typically associated with hyporesponsive T cells, is required for Tfh cell maintenance and is a therapeutic target in murine lupus.
Abstract
Coordinated gene expression programs enable development and function of T cell subsets. Follicular helper T (Tfh) cells coordinate humoral immune responses by providing selective and instructive cues to germinal center B cells. Here, we show that AP-1–independent NFAT gene expression, a program associated with hyporesponsive T cell states like anergy or exhaustion, is also a distinguishing feature of Tfh cells. NFAT signaling in Tfh cells, maintained by NFAT2 autoamplification, is required for their survival. ICOS signaling upregulates Bcl6 and induces an AP-1–independent NFAT program in primary T cells. Using lupus-prone mice, we demonstrate that genetic disruption or pharmacologic inhibition of NFAT signaling specifically impacts Tfh cell maintenance and leads to amelioration of autoantibody production and renal injury. Our data provide important conceptual and therapeutic insights into the signaling mechanisms that regulate Tfh cell development and function.
Introduction
Follicular helper T (Tfh) cells are necessary for orchestrating normal humoral immune responses elicited by pathogens or immunizations (Crotty, 2011; Seth and Craft, 2019; Webb and Linterman, 2017). The central transcriptional regulator of Tfh cells is Bcl6, which promotes Tfh cell development by suppressing the expression of transcriptional regulators that either promote non-Tfh cell fates or repress Tfh cell commitment (Choi and Crotty, 2021; Johnston et al., 2009; Nurieva et al., 2009; Yu et al., 2009). Unlike other Th effector cells, which leave their site of activation in secondary lymphoid organs and migrate to effector locations in the periphery, Tfh cells largely remain in the lymphoid organs, upregulating proteins such as CXCR5, which allow them to migrate to the B cell follicle and ultimately germinal centers (GCs) alongside maturing B cells (Song and Craft, 2019). Tfh cells express a number of costimulatory and coinhibitory receptors including CD28, ICOS, and PD1, which are necessary for their development, maintenance, and function (Crotty, 2011). ICOS signals are particularly important early after naive T cell activation to induce Bcl6, which is necessary for Tfh cell differentiation (Choi et al., 2011; Johnston et al., 2009; Nurieva et al., 2009; Yu et al., 2009). ICOS-specific costimulatory pathways, many of which have yet to be delineated, are likely to be important for Tfh cell development and function (Pedros et al., 2016; Wikenheiser and Stumhofer, 2016). Tfh cells promote follicular and ultimately GC B cell proliferation and maturation through contact-dependent and soluble factors, including CD40L (CD154) and the cytokine IL-21. In return, their maintenance is dependent upon B cell provision of antigen, thereby providing a self-contained mechanism for shutting down Tfh cell responses once antigen is cleared (Barnett et al., 2014; Baumjohann et al., 2013; Deenick et al., 2010; Johnston et al., 2009). Abnormal expansion and/or function of Tfh cells has been associated with human autoimmune diseases, such as systemic lupus erythematosus (SLE, lupus; Craft, 2012). We and others have demonstrated that targeting Tfh cell function in mouse models of autoimmune disease can lead to therapeutic effects and disease amelioration (Bubier et al., 2009; Choi et al., 2017; Herber et al., 2007; McPhee et al., 2013; Rankin et al., 2012; Yan et al., 2017; Yin et al., 2015). Determining the unique features of Tfh cell gene expression is essential for understanding how these cells differ from other T helper subsets. This knowledge will guide development of therapeutic strategies to target these cells in vivo, either to augment humoral immunity, as in a vaccine response, or to inhibit it in autoantibody-dependent autoimmune diseases.
The NFAT family of transcription factors is rapidly activated upon T cell stimulation with redundant and specific functional effects in different T cell subsets (Macián, 2005). Calcium influx triggered by TCR activation leads to activation of the phosphatase calcineurin, which directly acts to dephosphorylate NFAT family members, resulting in the unmasking of nuclear localization signals allowing NFAT proteins to translocate to the nucleus and regulate gene expression. Calcineurin inhibitors (CNIs), which inhibit NFAT signaling, are used routinely to prevent allo-transplant rejection and have recently experienced revived interest for the treatment of autoimmune diseases including SLE (Chang et al., 2021; Park et al., 2020). In canonical NFAT signaling, NFAT family members cooperate with AP-1 family proteins to coordinately regulate gene expression of T cell activation programs (Hogan, 2017; Rao et al., 1997). An alternate NFAT signaling pathway, termed AP-1–independent signaling, has also been described and is associated with hyporesponsive T cell states including anergy and exhaustion (Macián et al., 2001). Thus, NFAT signaling can exist in various modes depending on the biological context.
Our group and others have demonstrated the central role of NFAT2 for the development and function of Tfh cells that arise during viral infection in mice (Martinez et al., 2016; Ray et al., 2015). We also found upregulation of specific genes usually associated with AP-1–independent NFAT gene expression, the module previously described in hyporesponsive T cells (Ray et al., 2015), but the specific function of NFAT2 in Tfh cells remains elusive. NFAT1 and NFAT2, the most well-studied NFAT family members, are induced following T cell activation and are thought to have largely overlapping functions in most Th effector cells (Dietz et al., 2015; Srinivasan and Frauwirth, 2007; Xu et al., 2019a). A unique feature of NFAT2 signaling, autoregulation of its expression via an NFAT-mediated, stimulation-dependent positive-feedback loop, enables high-level expression of NFAT2 protein and is important for osteoclast development, endocardial cell development, and protection of T cells from activation-induced apoptosis in vitro (Serfling et al., 2006). Blockade of NFAT2 autoregulation by CNIs has been used to monitor their therapeutic efficacy (Kannegieter et al., 2018), but the physiologic role of NFAT2 autoregulation in T cells has yet to be established.
The specific role of NFAT2 or its autoregulation in Tfh cell development and function is unknown. We analyzed the transcriptomes of murine Tfh cells following pathogen challenge and in murine lupus and found upregulated expression of AP-1–independent NFAT genes. The global transcriptomes of Tfh cells, relative to other Th effector cells, exhibited greater similarity to anergic or exhausted T cells, suggesting that AP-1–independent NFAT gene expression may be an essential feature of Tfh cell identity, distinguishing them from other Th cell subsets. Using mutants of NFAT2 unable to interact with AP-1, we established that the AP-1–independent NFAT gene expression program is sufficient for development of Tfh cells in vivo and necessary for their survival. In contrast to hyporesponsive cells, which rely on impaired costimulation to generate AP-1–independent signaling, we hypothesized that the Tfh cell transcriptional regulator, Bcl6, previously shown to interact with and repress AP-1 function in different cell types, including Tfh cells (Hatzi et al., 2015; Tunyaplin et al., 2004; Vasanwala et al., 2002), may enforce AP-1–independent NFAT signaling by sequestering AP-1 from NFAT2. Consistent with this idea, we found that ICOS costimulation of naive T cells in vitro, with TCR signaling and CD28 costimulation, led to robust Bcl6 expression and induction of an AP-1–independent mode of NFAT gene expression. We also found that T cell overexpression of the AP-1 factor, cFos, resulted in impaired Tfh cell development in vivo, while suppression of AP-1 function had the opposite effect, suggesting that the stoichiometry of NFAT:AP-1 may be important for AP-1–independent NFAT gene expression in Tfh cells. Using cyclosporine A (CsA), a prototypic CNI, as a pharmacologic probe of NFAT signaling, we made the unexpected discovery that CNIs selectively deplete Tfh cells in vivo during acute viral infection in mice. We exploited this finding to demonstrate that NFAT signaling, generally, and NFAT2, specifically, are critical for development of murine lupus, a systemic illness reliant upon persistent autoantigenic T cell stimulation (Riemekasten and Hahn, 2005). Our findings suggest that the ability of CNIs to specifically deplete Tfh cells underlies their therapeutic efficacy in autoantibody-driven autoimmunity.
Results
Tfh cells are characterized by an AP-1–independent NFAT gene expression signature
During acute lymphocytic choriomeningitis virus (LCMV) infection, activated CD4+CD44hi T cells initially regulate the expression of Ly6c and P-selectin glycoprotein ligand-1 (PSGL-1), with the activated CD44hiPSGL1loLy6clo population representing pre-Tfh cells and CD44hiPSGL1hiLy6chi cells becoming their Th1 counterparts (Iyer et al., 2013; Marshall et al., 2011; Odegard et al., 2008; Poholek et al., 2010). The former upregulate CXCR5 and PD1 becoming PSGL1loLy6cloPD1+CXCR5+ upon further differentiation and B cell interaction (Kerfoot et al., 2011; Poholek et al., 2010; Weinstein et al., 2014), with subsequent follicular entry and then GC migration as GC Tfh cells (Yusuf et al., 2010). Accordingly, we identified CD4+ T cells by these surface markers.
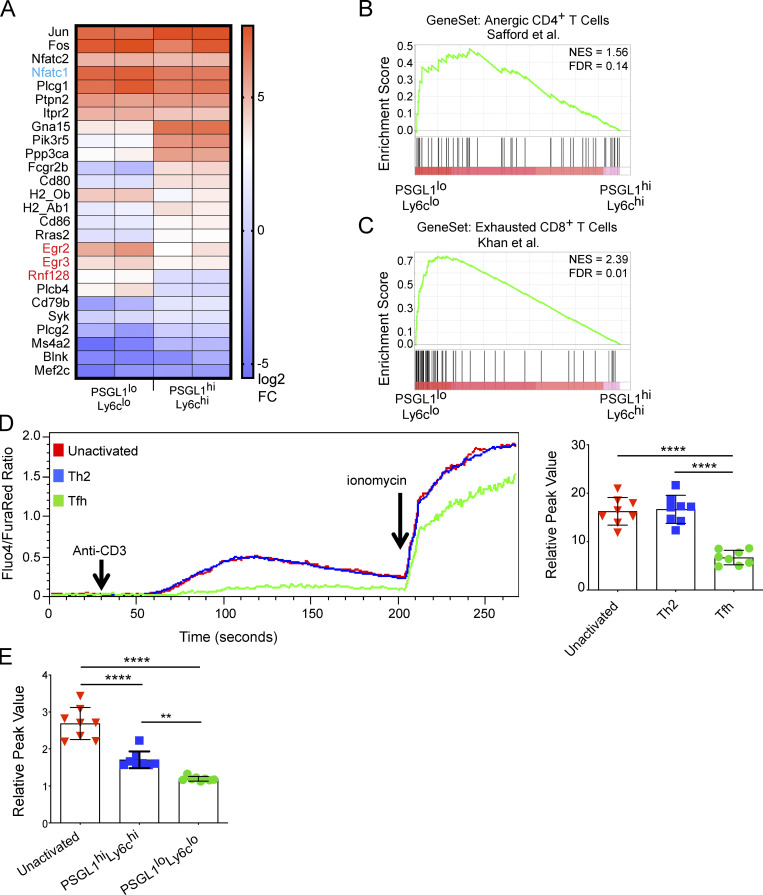
PSGL1loLy6clo T cells from LCMV-infected mice have reduced expression of many NFAT-dependent genes, but a small subset, including Egr2, Egr3, and Rnf128 are upregulated relative to PSGL1hiLy6chi T cells (Ray et al., 2015; Fig. 1 A). This transcriptional profile is consistent with AP-1–independent NFAT gene expression, a mode of NFAT signaling typically associated with hyporesponsive T cell states such as anergy or exhaustion and that is distinct from other effector T cell populations (Rao et al., 1997). We used gene set enrichment analysis (GSEA), an algorithm for interpreting gene expression data at the genome-wide level, to better characterize the Tfh cell transcriptional program (Subramanian et al., 2005). GSEA showed similarity between PSGL1loLy6clo T cells from acute LCMV infection and anergic mouse CD4+ T cells (Fig. 1 B; Safford et al., 2005), with false discovery rate well below the 0.25 cutoff, recommended for this type of exploratory discovery analysis (Subramanian et al., 2005). Similarly, GSEA using two different gene sets derived from exhausted CD8+ T cells (Khan et al., 2019; Wherry et al., 2007) exhibited similarity to PSGL1loLy6clo cells from acute LCMV infection (Fig. 1 C and Fig. S1 A), suggesting that an AP-1–independent gene signature, as occurs in T cell hyporesponsive states, could be a feature of the Tfh cell developmental program. Consistent with this idea, we found the same AP-1–independent NFAT gene signature in CD4+CD44hiCXCR5+PD1+IL-21+ T cells following infection with the helminth Nippostrongylus brasiliensis (Weinstein et al., 2016) and in CD4+CD44+ PD1+CXCR5+ T cells that promote aberrant B cell maturation and autoantibody production from lupus-prone mice, respectively (Dong et al., 2021; Fig. S1, B and D). GSEA of these respective Tfh cell populations also demonstrated similarity to anergic CD4+ cells and exhausted CD8+ T cells (Fig. S1, C and E). The conserved presence of an AP-1–independent NFAT gene signature along with global transcriptomic similarity to anergic/exhausted T cell states in Tfh cells derived from different biological settings prompted us to further examine the functional significance of these findings.
Figure 1.
Tfh cells are characterized by an AP-1–independent NFAT gene expression signature. (A) NFAT gene signature from RNA-Seq of PSGL1hiLy6chi and PSGL1loLy6clo CD4+ T cells (Ray et al., 2015). Nfatc1, the gene encoding NFAT2 is shown in blue. Egr2, Egr3, and Rnf128, canonical AP-1–independent NFAT gene transcripts, are highlighted in red. (B) GSEA of PSGL1hiLy6chi and PSGL1loLy6clo CD4+ T cells (Ray et al., 2015) using gene set from anergic CD4+ T cells (Safford et al., 2005). (C) GSEA using gene set derived from exhausted CD8+ T cells in mice chronically infected with LCMV (Khan et al., 2019). (D) TCR stimulation mediated in vitro calcium flux after papain/NP-ova immunization. CD4+ T cell populations were defined as unactivated (CD4+CD44lo), Th2 (CD4+CD44hiPD1−CXCR5−), or Tfh (CD4+CD44hiPD1+CXCR5+). The left panel shows a representative time course for one experiment. The Fluo4/FuraRed ratio is plotted over time and reports cytosolic calcium in response to the indicated agents. The right panel shows cumulative data for multiple samples and plots the relative calcium peak (calculated as a ratio of peak Fluo4/FuraRed value to mean baseline Fluo4/FuraRed value) for the different T cell subsets. (E) TCR stimulation mediated in vitro calcium flux after acute infection with LCMV. Data in D and E are representative of at least three independent experiments with three to five mice per experiment for D and five to 10 mice per experiment for E. One-way ANOVA with post-hoc Tukey’s test used for analysis of D and E. See also Fig. S1. **** = P ≤ 0.0001, ** = P ≤ 0.01. NES, normalized enrichment score; FDR, false discovery rate.
Figure S1.
Tfh cells are characterized by an AP-1–independent NFAT gene expression signature. Related to Fig. 1. (A) GSEA of PSGL1hiLy6chi and PSGL1loLy6clo CD4+ T cells (Ray et al., 2015) using gene set from exhausted CD8+ T cells in mice chronically infected with LCMV (Wherry et al., 2007). (B) NFAT gene signature from RNA-Seq of Tfh (CD4+CD44hiCXCR5+PD1+IL-21+) cells and Th2 (CD4+CD44hiCXCR5−PD1−IL-4+) cells following infection with N. brasiliensis (Weinstein et al., 2016), average of duplicate data. Nfatc1, the gene encoding NFAT2 is shown in blue. Egr2, Egr3, and Rnf128, canonical AP-1–independent NFAT gene transcripts, are highlighted in red. (C) GSEA of Tfh and Th2 cells following infection with N. brasiliensis using gene sets from anergic CD4+ (Safford et al., 2005) or exhausted CD8+ T cells (Khan et al., 2019). (D) NFAT gene signature from RNA-Seq of Tfh (CD4+CD44hiLy6cloPSGL1loCXCR5+PD1+) cells and Th1 (CD4+CD44hiLy6chiPSGL1hi) cells from murine lupus (Dong et al., 2021), average of duplicate data. (E) GSEA of Tfh and Th1 cells from 2-mo-old lupus-prone mice using gene sets from anergic CD4+ (Safford et al., 2005) or exhausted CD8+ T cells (Khan et al., 2019). (F) Inhibitory gene signature from PSGL1hiLy6chi and PSGL1loLy6clo CD4+ T cells isolated 8 d after infection with LCMV (Ray et al., 2015). (G) Flow cytometric measurement of inhibitory proteins from STG+ T cells isolated 8 d after infection with LCMV. Data in G are representative of at least three independent experiments with three to five mice per experiment. **** = P ≤ 0.0001, *** = P ≤ 0.001, ** = P ≤ 0.01, * = P ≤ 0.05, and ns P > 0.05. NES, normalized enrichment score; FDR, false discovery rate.
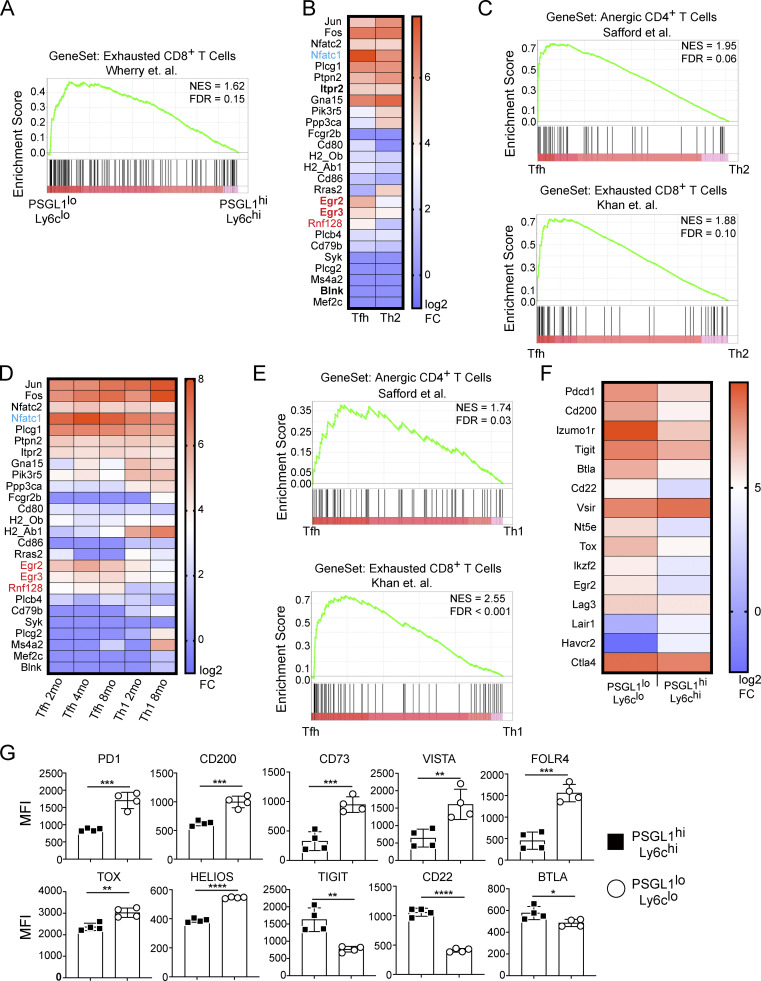
Hyporesponsive T cells express a variety of inhibitory receptors and transcriptional regulators (Nurieva et al., 2011; Pereira et al., 2017; Schietinger and Greenberg, 2014; Shin and Wherry, 2007). The inhibitory receptor PD1 is used to identify Tfh cells and is critical for their maintenance and function (Crotty, 2011; Haynes et al., 2007; Yusuf et al., 2010). Other inhibitory proteins, such as CD200, FolR4, and CD73, have also been found to be highly expressed by Tfh cells (Chtanova et al., 2004; Iyer et al., 2013). Tox family transcription factors, including Tox and Tox2, are important for Tfh cell development (Xu et al., 2019b) and are key regulators of anergy and exhaustion pathways (Alfei et al., 2019; Khan et al., 2019; Pereira et al., 2017; Scott et al., 2019; Seo et al., 2019). Analysis of our prior RNA sequencing (RNA-Seq) data revealed that transcripts of several inhibitory receptors and signaling molecules were upregulated in PSGL1loLy6clo T cells compared with their PSGL1hiLy6chi counterparts (Fig. S1 F). Accordingly, we also found increased expression at the protein level for many of these inhibitory molecules (Fig. S1 G).
We have previously reported that Tfh cells flux cytosolic calcium when treated with ionomycin (Ray et al., 2015); however, a functional consequence of AP-1–independent NFAT signaling in anergic cells is reduced intracellular calcium flux in response to TCR stimulation. CD4+CD44+PD1+CXCR5+ T cells generated after immunization with papain/NP-ova of B6 mice exhibited decreased calcium flux in response to TCR stimulation compared with unactivated CD4+CD44lo or to CD4+CD44hiPD1−CXCR5− Th2 cells (Fig. 1 D). PSGL1loLy6clo CD4+ T cells generated after acute LCMV infection exhibited decreased TCR-stimulation-mediated calcium flux compared with unactivated CD4+CD44lo T cells and PSGL1hiLy6chi CD4+ T cells (Fig. 1 E). Decreased calcium flux in the PSGL1loLy6clo CD4+ T cells was not due to decreased expression of calcium flux genes, including Stim1/Stim2/Ora1, based on our previously published transcriptome data (data not shown; Ray et al., 2015). The association of an AP-1–independent NFAT signaling pathway with Tfh cells is also consistent with their production of IL-21, a cytokine regulated by NFAT independently of AP-1 (Kim et al., 2005). Taken together, our genetic, phenotypic, and functional profiling suggested that AP-1–independent NFAT signaling is operational in Tfh cells. We next sought to understand if this transcriptional program has physiologic significance for Tfh cell development and/or function.
AP-1–independent NFAT signaling is sufficient for Tfh cell development
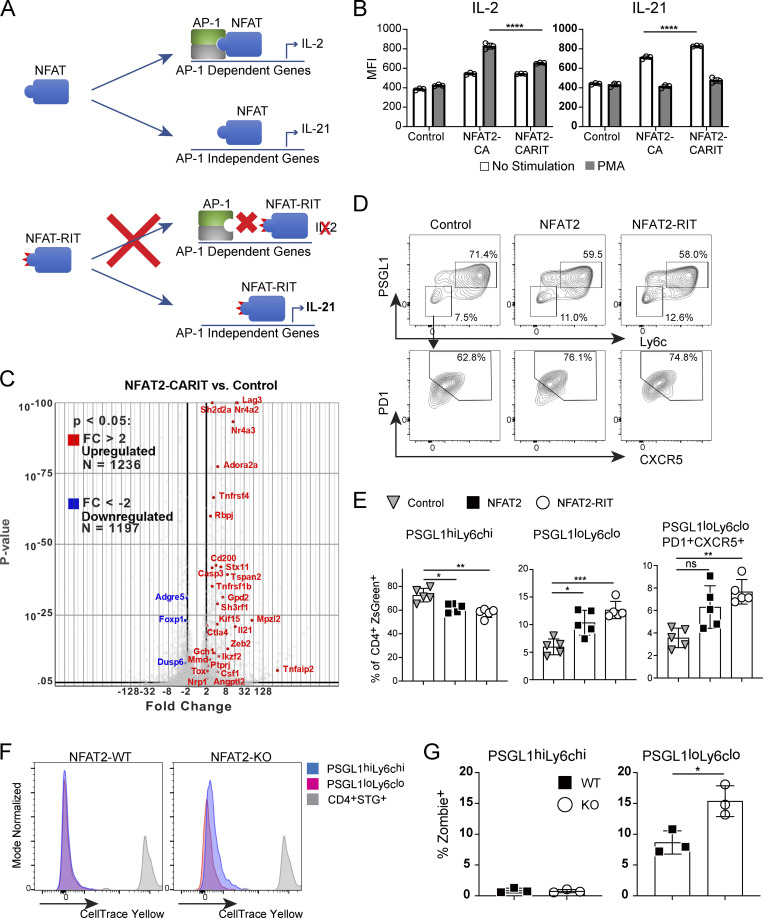
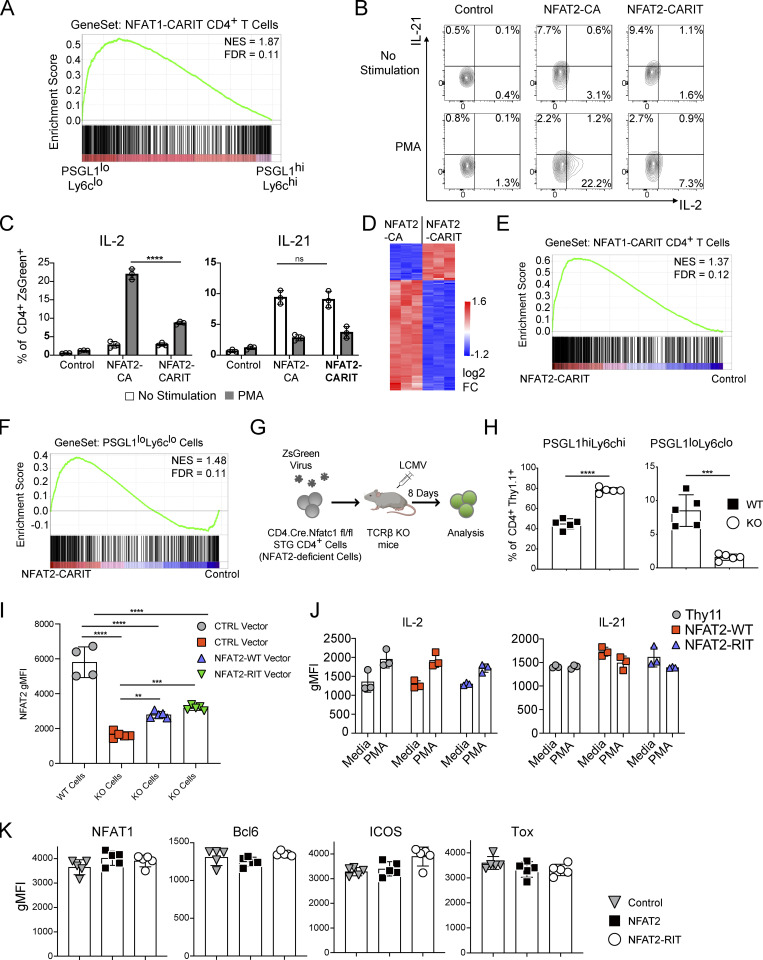
NFAT1 (gene name Nfatc2) and NFAT2 (gene name Nfatc1) are highly homologous proteins and are therefore largely redundant in terms of their downstream transcriptional targets (Rao et al., 1997); however, NFAT2 is selectively activated and required in Tfh cells (Martinez et al., 2016; Ray et al., 2015). Mutation of three conserved residues in NFAT1 (R468A, I469A, and T535G), generating the mutant protein NFAT1-RIT, abolish its ability to interact with AP-1 family members (Fig. 2 A; Macián et al., 2000, 2002). Overexpression of NFAT1-CARIT, a constitutively active mutant of NFAT that is unable to engage AP-1–dependent pathways, in primary T cells induces an anergy gene signature (Martinez et al., 2015). This NFAT1-CARIT signature is also enriched in PSGL1loLy6clo CD4+ T cells (Fig. S2 A). To better understand the role of AP-1–independent NFAT signaling in Tfh cells, we introduced the corresponding NFAT1 RIT point mutations (R471A, L472A, and T538G) into retroviral expression constructs encoding WT NFAT2 or constitutively active NFAT2 (NFAT2-CA). To validate that the RIT triple mutation hinders AP-1 binding in the context of NFAT2, we expressed NFAT2-CA or NFAT2-CARIT in primary CD4+ T cells and assessed the capacity of these mutants to support production of IL-2, an AP-1–dependent cytokine, or IL-21, an AP-1–independent cytokine (Kim et al., 2005). When incubated in media alone, cells expressing either NFAT2-CA or NFAT2-CARIT upregulated IL-21 production (Fig. 2 B; and Fig. S2, B and C), consistent with this condition favoring AP-1–independent NFAT signaling and with both mutants capable of supporting AP-1–independent signaling, as is observed when T cells are stimulated only with ionomycin (Macián et al., 2002). When stimulated with PMA, which would be expected to pharmacologically restore stimulation akin to that through TCR and CD28 favoring canonical AP-1–dependent NFAT signaling (Macián et al., 2002), cells expressing NFAT2-CA, capable of AP-1–dependent and AP-independent signaling, upregulated IL-2 production, while cells expressing NFAT2-CARIT, capable of AP-1–independent signaling only, were significantly impaired (Fig. 2 B; and Fig. S2, B and C). To further demonstrate that NFAT2-CARIT expression can support an AP-1–independent program, we performed transcriptome analysis of CD4+ T cells transduced with control vector or NFAT2-CARIT. Similar to expression of NFAT1-CARIT, expression of NFAT2-CARIT led to differential expression of ∼2,000 genes compared with control cells, about half of which are upregulated and include many anergy-associated genes (Fig. 2 C; Martinez et al., 2015; Safford et al., 2005). There were ∼200 differentially expressed genes between NFAT2-CARIT and NFAT2-CA transduced cells (Fig. S2 D), consistent with the prediction that these two mutants would have distinct capacities to support NFAT-mediated signaling pathways, i.e., NFAT2-CA being competent to participate in both AP-1–dependent and AP-1–independent signaling and NFAT2-CARIT being competent to support only the latter. We would anticipate even greater gene expression differences between these mutants under conditions actively promoting AP-1–dependent NFAT signaling. The gene signature of NFAT2-CARIT expressing cells was very similar to NFAT1-CARIT expressing cells (Fig. S2 E), and, accordingly, showed similarity to PSGL1loLy6clo T cells from acute LCMV infection (Fig. S2 F). Together, these data validated that the RIT triple mutant, which previously has been shown to impair AP-1–dependent signaling in the context of NFAT1, had a similar effect when introduced into NFAT2.
Figure 2.
AP-1–independent NFAT signaling is sufficient for Tfh cell development. (A) Schematic representation of WT NFAT and NFAT-RIT mutant proteins and their differential abilities to engage AP-1–dependent or AP-1–independent gene expression programs. (B) CD4+ T cells were transduced with pMIZ control, pMIZ-NFAT2, or pMIZ-NFAT2-CARIT retroviral vectors. After 72 h, cells were either incubated with fresh media or stimulated with PMA and then analyzed by flow cytometry for expression of ZsGreen (to mark transduced cells), IL-2, and IL-21. One-way ANOVA with post-hoc Tukey’s test used for analysis. Representative of three independent experiments. (C) Volcano plot of gene expression from RNA-Seq of CD4+ T cells transduced with control or NFAT2-CARIT. Selected anergy-associated genes (Martinez et al., 2015; Safford et al., 2005) have been annotated. (D) Representative flow plots of PSGL1hiLy6chi, PSGL1loLy6clo, and PSGL1loLyc6loPD1+CXCR5+ CD4+ T cell populations following acute LCMV Armstrong infection of mice receiving NFAT2-deficient STG+ T cells transduced with control, NFAT2, or NFAT2-RIT retroviral expression vectors. (E) Quantification of data from D. One-way ANOVA with post-hoc Tukey’s test used for analysis. (F) Flow cytometry measurement CellTrace Yellow cell proliferation dye in the indicated cell populations. (G) Viability of the indicated cell populations assessed by the Zombie cell death dye. Data are representative of at least three replicates and two independent experiments for D and E (at least five animals per group) and two independent experiments for G and H (three animals per group). See also Fig. S2. **** = P ≤ 0.0001, *** = P ≤ 0.001, ** = P ≤ 0.01, * = P ≤ 0.05, and ns P > 0.05.
Figure S2.
AP-1–independent NFAT signaling is sufficient for Tfh cell development. Related to Fig. 2. (A) GSEA demonstrating PSGL1loLy6clo CD4+ T cells from acute LCMV infection (Ray et al., 2015) exhibit greater similarity to NFAT1-CARIT expressing cells (Martinez et al., 2015) than PSGL1hiLy6chi CD4+ T cells. (B) Representative flow cytometry plot showing IL-21 or IL-2 intracellular staining for control, NFAT2, or NFAT-CARIT transduced CD4+ T cells stimulated with PMA or media control (related to Fig. 2 B). (C) IL-2 or IL-21–producing cells as a percentage of the transduced cells (related to Fig. 2 B and Fig. S2 B). One-way ANOVA with post-hoc Tukey’s test used for analysis. (D) Differentially expressed genes between NFAT-CA and NFAT2-CARIT transduced CD4+ T cells. (E) GSEA demonstrating similarity of NFAT2-CARIT transduced cells to NFAT1-CARIT transduced cells (Martinez et al., 2015). (F) GSEA demonstrating similarity of NFAT2-CARIT transduced cells to Tfh cells at day 8 after LCMV Armstrong infection (Ray et al., 2015). (G) Schematic of adoptive transfer experimental strategy. NFAT2-deficient (CD4Cre+.Nfatc1fl/fl) STG+ CD4+ T cells were transduced with retroviruses carrying ZsGreen-tagged vectors. Cells were transferred into TCRβ− mice that were then acutely infected with LCMV Armstrong. 8 d after infection, splenocytes were isolated for analysis. (H) NFAT2-deficient (CD4Cre+.Nfatc1fl/fl) or NFAT2-sufficient CD4Cre-.Nfatc1fl/fl) Thy1.1 STG+ T cells were adoptively transferred into recipient mice that were then infected with LCMV Armstrong. 8 d after infection, splenocytes were isolated and analyzed by flow cytometry. (I) Mean NFAT2 expression in splenic CD4+ZsGreen+ T cells after isolation. One-way ANOVA with post-hoc Tukey’s test used for analysis. (J) CD4+ T cells were transduced with retroviral vectors encoding Thy1.1 (CD90.1, control), NFAT2-WT, or NFAT2-RIT, and IL-2 and IL-21 were measured as described in Fig. 2 B. (K) MFI of selected markers in ZsGreen+PSGL1loLy6cloPD1+CXCR5+ CD4+ T cells. Results are representative of three experiments with at least three replicates for B and C and with three to five animals per group for H, I, and K. For J, results are representative of at least two independent experiments. **** = P ≤ 0.0001, *** = P ≤ 0.001, ** = P ≤ 0.01, and ns P > 0.05. NES, normalized enrichment score; FDR, false discovery rate.
NFAT2-deficient CD4+ (CD4Cre+.Nfatc1fl/fl) T cells exhibit impaired Tfh cell development in vivo during acute LCMV infection of B6 mice (Martinez et al., 2016). To determine whether AP-1–independent NFAT signaling is sufficient for Tfh cell development in vivo, we developed an adoptive transfer system using SMARTA TCR transgenic (STG+) CD4+ T cells, specific for the immunodominant I-Ab-restricted LCMV glycoprotein peptide 66–77 (Fig. S2 G; Oxenius et al., 1998), crossed to CD4Cre+.Nfatc1fl/fl B6 mice.
Consistent with previous results using a similar system (Martinez et al., 2016), NFAT2-deficient (CD4Cre+.Nfatc1fl/fl) STG+ T cells had impaired PSGL1loLy6clo T cell development compared with control (CD4Cre-.Nfatc1fl/fl) STG+ T cells (Fig. S2 H). We next tested the ability of different NFAT2 constructs to rescue Tfh cell development of the NFAT2-deficient cells in this system. NFAT2-deficient (CD4Cre+.Nfatc1fl/fl) STG+ T cells transduced with control vector had impaired development of PSGL1loLy6clo and PSGL1loLy6cloPD1+CXCR5+ T cells compared with cells transduced with NFAT2 or NFAT2-RIT (Fig. 2, D and E), the latter impaired in AP-1–dependent NFAT signaling but competent to drive AP-1–independent signaling. Conversely, as expected there was reduction in PSGL1hiLy6chi T cells upon transduction with NFAT2 or NFAT2-RIT. The degree of NFAT2 or NFAT2-RIT overexpression in transduced cells was equivalent and was less than physiologic expression of NFAT2 in WT cells, possibly due to lack of positive feedback mechanisms in the transduced vector-mediated overexpression of NFAT2, suggesting our results are not due to supraphysiologic expression of NFAT2 in the transduction system (Fig. S2 I). In contrast to the constitutively active CA mutants used to validate the RIT mutations in vitro (Fig. 2 A), neither NFAT2 nor NFAT2-RIT had any significant effect on baseline IL-2 or IL-21 production in transduced cells (Fig. S2 J), suggesting the in vivo results are the result of physiologic NFAT2 activation and signaling. Rescuing NFAT2-deficient CD4+ T cells with either NFAT2 or NFAT2-RIT had a quantitative effect on Tfh cell development but not a qualitative one in that the expression of key molecules such as NFAT1, Bcl6, ICOS, or Tox were not different in the PSGL1loLy6cloPD1+CXCR5+ Tfh cells generated in the different experimental groups (Fig. S2 K).
The above results supported the idea that AP-1–independent NFAT2 signaling is sufficient to restore Tfh cell development in the absence of WT NFAT2. We next sought to understand whether NFAT2 deficiency impaired Tfh cell survival and/or proliferation. NFAT2-deficient (CD4Cre+.Nfatc1fl/fl) STG+ T cells (NFAT2-KO) and control (CD4Cre-.Nfatc1fl/fl) STG+ T cells (NFAT2-WT) cells were labeled in vitro with the proliferation tracking dye CellTrace-Yellow and transferred into host mice that were then infected with LCMV. Both WT and KO PSGL1hiLy6chi T cells and PSGL1loLy6clo T cells had near complete dilution of the proliferation tracking dye compared with STG+ T cells transferred into uninfected hosts, indicating that NFAT2-deficiency has no significant effect on the proliferation of the T helper cells (Fig. 2 F). Using the Zombie cell viability dye, we also found near complete viability of PSGL1hiLy6chi T cells with no significant difference between NFAT2-WT and NFAT2-KO cells (Fig. 2 G). However, we did find decreased cell viability of PSGL1loLy6clo T cells (Fig. 2 G), consistent with our previously reported results of enhanced apoptosis of Tfh cells (Ray et al., 2015), with deficiency of NFAT2 approximately doubling the percentage of non-viable PSGL1loLy6clo T cells (Fig. 2 G), suggesting that NFAT2 acts as a survival factor for Tfh cells in vivo.
ICOS costimulation drives Bcl6 expression and induces an AP-1–independent NFAT gene signature in T cells
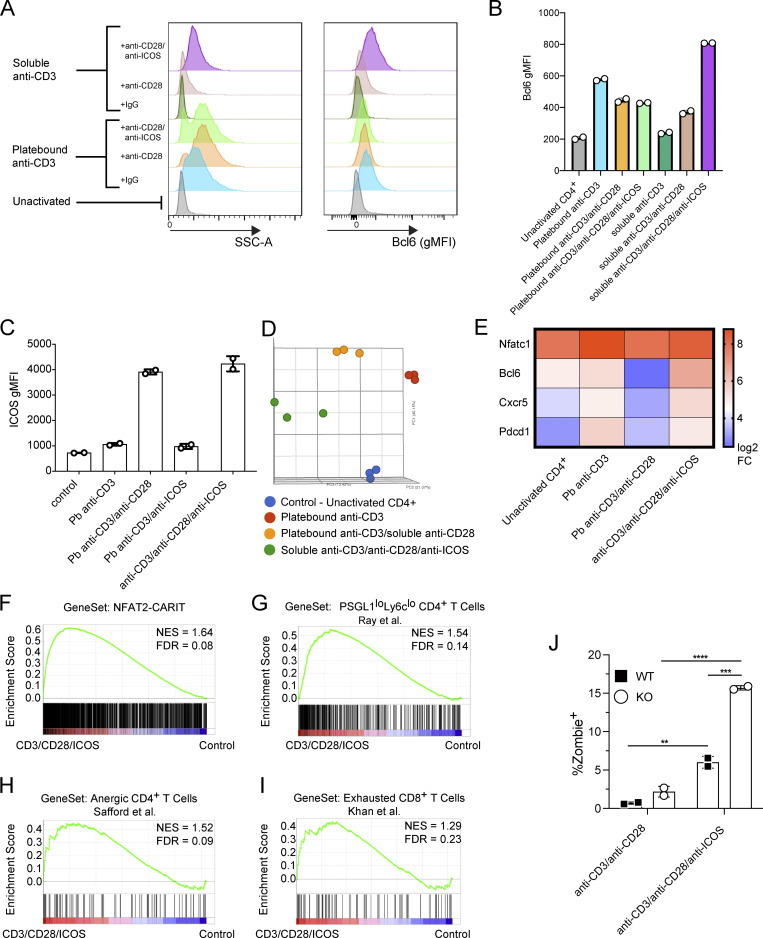
We next sought to understand the signaling mechanisms that promote AP-1–independent NFAT signaling in Tfh cells. In anergic cells, AP-1–independent NFAT signaling is established by TCR stimulation in the absence of CD28 costimulation (Schwartz, 2003), while in exhausted cells, PD1 antagonism of CD28 costimulation is one route toward AP-1–independent NFAT signaling (Hui et al., 2017; Pereira et al., 2017). We reasoned that these mechanisms were unlikely to be relevant for differentiation of Tfh cells given that CD28 costimulation is required not just for their priming, but also promotes their function (Linterman et al., 2014; Weber et al., 2015). Bcl6 is the subset-defining transcriptional regulator of Tfh cells (Johnston et al., 2009; Nurieva et al., 2009; Yu et al., 2009), antagonizing AP-1 activity via physical sequestration (Hatzi et al., 2015). In the absence of Bcl6, AP-1 would therefore be free to bind activated NFAT and engage canonical NFAT signaling, but in its presence, the relative amount of AP-1 would be reduced and therefore, NFAT signaling would be skewed toward an AP-1–independent pathway. As ICOS signaling is required for Bcl6 upregulation in developing Tfh cells (Choi et al., 2011), we reasoned it should induce AP-1–independent NFAT signaling in activated T cells. Thus, we activated naive CD4+ T cells in vitro with anti-ICOS, in combination with agonistic anti-CD3 and anti-CD28. Activation in the presence of a soluble mixture of all three antibodies led to robust upregulation of Bcl6 as well as increased side scatter (Fig. 3, A and B). By contrast, in the presence of platebound anti-CD3 and soluble costimulation signals, the ability of ICOS costimulation to upregulate Bcl6 in CD4+ T cells was masked (Fig. 3, A and B). CD3 and ICOS costimulation were insufficient to upregulate Bcl6, likely because CD28 signaling is required for ICOS upregulation (McAdam et al., 2000; Fig. 3 C). The transcriptome of ICOS-activated CD4+ T cells formed a discrete cell cluster by principal component analysis compared with control naive CD4+ T cells, anti-CD3 stimulated cells, or anti-CD3/anti-CD28 stimulated cells (Fig. 3 D), with ICOS costimulation leading to upregulation of key Tfh cell genes including Nfatc1, Bcl6, Cxcr5, and Pdcd1 (Fig. 3 E), suggesting that combination costimulation conditions led to a distinct T cell program. Moreover, GSEA showed similarity between the anti-CD3/anti-CD28/anti-ICOS stimulated T cell program and cell states associated with AP-1–independent NFAT signaling including NFAT2-CARIT overexpressing cells (Fig. 3 F), PSGL1loLy6clo CD4+ T cells from acute LCMV Armstrong infection (Fig. 3 G), anergic CD4+ T cells (Fig. 3 H), and exhausted CD8+ T cells (Fig. 3 I). These results indicated that T cell activation in the presence of combination costimulation through CD28 and ICOS enhanced Bcl6 expression, and accordingly, skewed activated T cells toward an AP-1–independent NFAT gene expression profile.
Figure 3.
ICOS costimulation induces an AP-1–independent NFAT signature. (A) Representative histograms showing Side Scatter (SSC-A; left panel) or Bcl6 geometric MFI (gMFI; right panel) as measured by flow cytometry of cells stimulated for 72 h under the indicated conditions. (B) Cumulative data for Bcl6 gMFI. Results are representative of three independent experiments with two replicates each. (C) ICOS gMFI of naive CD4+ T cells stimulated with the indicated antibodies. Results are representative of two independent experiments with two replicates each. (D) Principal component analysis of transcriptome data of in vitro activated or control (naive) CD4+ T cells. (E) Heat map depicting expression of indicated genes for each stimulation condition. Each sample represents average of three independent experiments. (F–I) GSEA of CD4+ T cells activated in the presence of anti-CD3/anti-CD28/anti-ICOS vs. control cells (naive CD4+ T cells) using gene set from NFAT2-CARIT overexpressing CD4+ T cells (this publication; F), PSGL1loLy6clo CD4+ T cells isolated on day 8 after LCMV Armstrong infection (Ray et al., 2015; G), anergic CD4+ T cells (Safford et al., 2005; H) or exhausted CD8+ T cells in mice chronically infected with LCMV (Khan et al., 2019; I). (J) Viability of in vitro stimulated CD4+ T cells by the Zombie cell death dye. Results are representative of three independent experiments with two replicates each. Data were analyzed using unpaired t tests. **** = P ≤ 0.0001, *** = P ≤ 0.001, ** = P ≤ 0.01. NES, normalized enrichment score; FDR, false discovery rate.
Our in vivo results showed that NFAT2 operates in an AP-1–independent mode in Tfh cells and is required for Tfh cell survival. We next assessed whether T cells receiving combination costimulation in vitro, which adopted an AP-1–independent transcriptional program, were also dependent upon NFAT2 for survival. T cells stimulated with conventional plate-bound anti-CD3 and soluble anti-CD28 were largely viable and exhibited no significant differences in viability whether or not they were NFAT2-sufficient or deficient (Fig. 3 J). However, T cells stimulated with soluble anti-CD3 and combination costimulation with soluble anti-CD28 and anti-ICOS were less viable than T cells receiving plate-bound anti-CD3 and soluble anti-CD28 (Fig. 3 J). Further, as we saw with in vivo NFAT2-KO Tfh cells (Fig. 2 G), NFAT2-KO T cells stimulated with soluble anti-CD3 and combination costimulation were significantly less viable than their NFAT2-WT counterparts (Fig. 3 J). Taken together, these results suggested that our in vitro ICOS costimulation protocol recapitulates the AP-1–independent mode of NFAT signaling and dependence on NFAT2 for survival that we have observed in Tfh cells in vivo.
Perturbing NFAT2–AP-1 balance impairs Tfh cell development
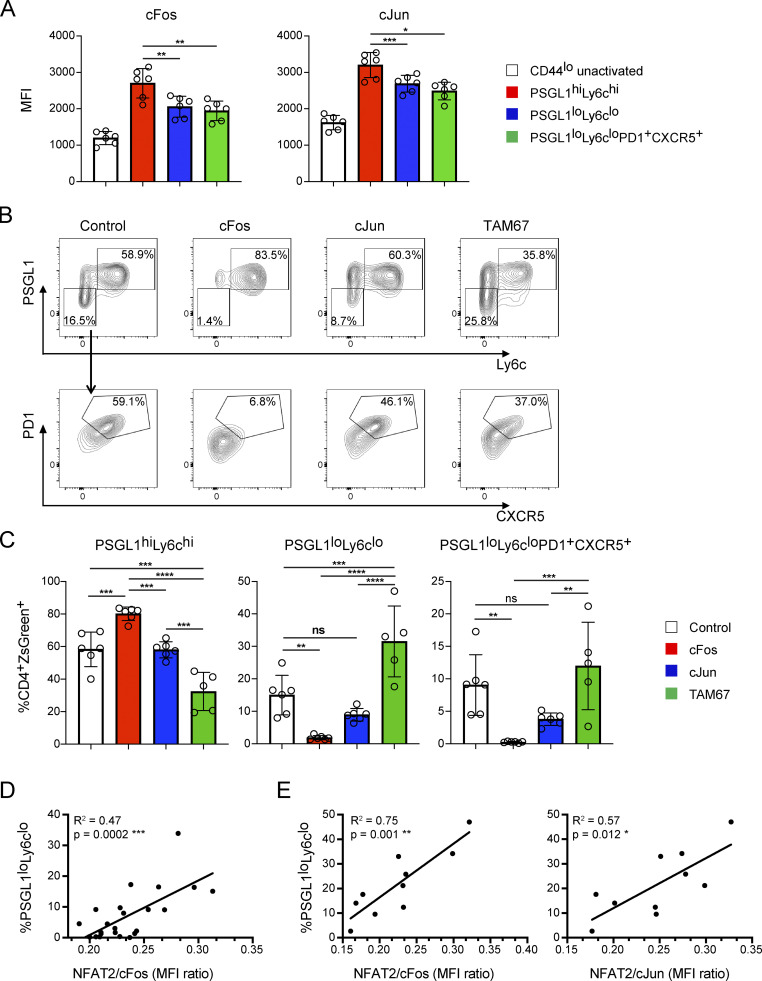
We next asked if increasing AP-1 availability in Tfh cells would antagonize their development and whether reducing AP-1 availability would potentiate it, as would be predicted, since Bcl6 acts as a competitive inhibitor of NFAT and AP-1 interactions (Hatzi et al., 2015). AP-1 proteins are comprised of Fos and Jun family members with a varying potential to homo- and hetero-dimerize to drive gene transcription (Curran and Franza, 1988). We focused on cFos and cJun as they are the AP-1 family members that have been most widely studied in AP-1–dependent NFAT signaling (Jain et al., 1993; McCaffrey et al., 1993; Peterson et al., 1996). Both cFos and cJun were expressed in PSGL1loLy6clo and PSGL1loLy6cloPD1+CXCR5+ T cells on day 8 following LCMV infection compared with control cells, albeit to a lesser extent than in PSGL1hiLy6chi cells (Fig. 4 A). We transduced STG+CD4+ T cells with ZsGreen expressing vectors encoding cFos or cJun and adoptively transferred them into OT-II TCR transgenic recipients that were then infected with LCMV, with the use of TCR transgenic recipients enabling expansion of adoptively transferred LCMV-specific cells (Marshall et al., 2015; Fig. S3 A). Transferred cells transduced with control vector exhibited PSGL1loLy6clo and PSGL1loLy6cloPD1+CXCR5+ T cell development (Fig. 4, B and C). Transduction of cFos led to significant cFos overexpression (Fig. S3 B) and was associated with decreased PSGL1loLy6clo and PSGL1loLy6cloPD1+CXCR5+ T cell development and with increased PSGL1hiLy6chi cell development (Fig. 4, B and C). In a like manner, transduction of cJun led to a trend decrease of PSGL1loLy6clo and PSGL1loLy6cloPD1+CXCR5+ T cell development, even though cJun expression was relatively minimally increased (Fig. S3 B). Expression of neither cFos nor cJun significantly changed NFAT2 nor NFAT1 expression in the remaining PSGL1loLy6cloPD1+CXCR5+ T cells (Fig. S3 C). Bcl6 was reduced in cFos and cJun transduced cells (Fig. S3 C), suggesting that Bcl6 and these factors work antagonistically in Tfh cells, consistent with previous work (Hatzi et al., 2015; Tunyaplin et al., 2004; Vasanwala et al., 2002). Tox was also reduced and T-bet increased in cFos transduced cells, consistent with the changes in PSGL1loLy6clo and PSGL1loLy6cloPD1+CXCR5+, and in PSGL1hiLy6chi cells, respectively (Fig. S3 C). Moreover, the NFAT2/cFos mean fluorescence intensity (MFI) ratio correlated with the PSGL1loLy6clo T cell frequency (Fig. 4 D). A correlation with cJun was not observed, likely due to relative inefficiency of its expression (not shown).
Figure 4.
AP-1–independent NFAT signaling is necessary for Tfh cell development. (A) cFos or cJun MFI in CD4+ T cell subsets 8 d after infection with LCMV. (B) Representative flow cytometry plots of splenic STG+CD4+ T cells 8 d after acute infection with LCMV Armstrong. Cells were transduced with ZsGreen expressing retroviral vectors encoding the indicated gene prior to adoptive transfer and infection. (C) Percentages of PSGL1hiLy6chi, PSGL1loLy6clo, and PSGL1loLyc6loPD1+CXCR5+ CD4+ T cells for each experimental condition. (D) Linear regression plot of PSGL1loLy6clo CD4+ T cell frequency vs. NFAT2/cFos gMFI ratio in cFos transduced cells. (E) Linear regression plot of PSGL1loLy6clo CD4+ T cell frequency vs. NFAT2/cFos or NFAT2/cJun gMFI ratio in TAM67 transduced cells. Results are representative of three independent experiments with three to five animals per experimental group. One-way ANOVA with post-hoc Tukey’s test used for analysis of A and C. See also Fig. S3. **** = P ≤ 0.0001, *** = P ≤ 0.001, ** = P ≤ 0.01, * = P ≤ 0.05, and ns P > 0.05.
Figure S3.
AP-1–independent NFAT signaling is necessary for Tfh cell development. Related to Fig. 4. (A) Schematic of experimental strategy. STG+CD4+ T cells were transduced with retroviruses carrying control, cFos, cJun, or TAM67 expression vectors. Cells were adoptively transferred into OTII mice which were then infected with LCMV. 8 d after infection, splenocytes were isolated for analysis. (B) cFos and cJun expression in STG+CD4+ZsGreen+ T cells for each transduction condition. (C) MFI of the indicated markers in STG+CD4+ZsGreen+PSGL1loLyc6loPD1+CXCR5+ T cells. Results are representative of three independent experiments with three to five animals per group. One-way ANOVA with post-hoc Tukey’s test used for analysis of the data. **** = P ≤ 0.0001, *** = P ≤ 0.001, ** = P ≤ 0.01, * = P ≤ 0.05, and ns P > 0.05.
Since we were not able to substantially overexpress cJun in these experiments, we employed an alternative strategy to assess its effects, transducing STG+CD4+ T cells with a vector encoding TAM67, a dominant negative mutant of cJun, which antagonizes AP-1 function (Brown et al., 1993; Chen et al., 1996; Domann et al., 1994; Dong et al., 1994; Ham et al., 1995; Petrak et al., 1994), which we predicted would increase the effective NFAT:AP-1 ratio. We found that PSGL1loLy6clo T cell development was enhanced with a trend toward increased PSGL1loLy6cloPD1+CXCR5+ T cells (Fig. 4, B and C) and preserved Bcl6 in the latter (Fig. S3 C). In the TAM67 transduced cells, both the NFAT2/cFos MFI and NFAT2/cJun MFI correlated with PSGL1loLy6clo T cell frequency (Fig. 4 E). Together, these results are consistent with a model in which AP-1–independent NFAT signaling in Tfh cells depends on the balance of NFAT2 and available AP-1.
TCR stimulation-mediated NFAT2 autoregulation is required for Tfh cell maintenance and survival
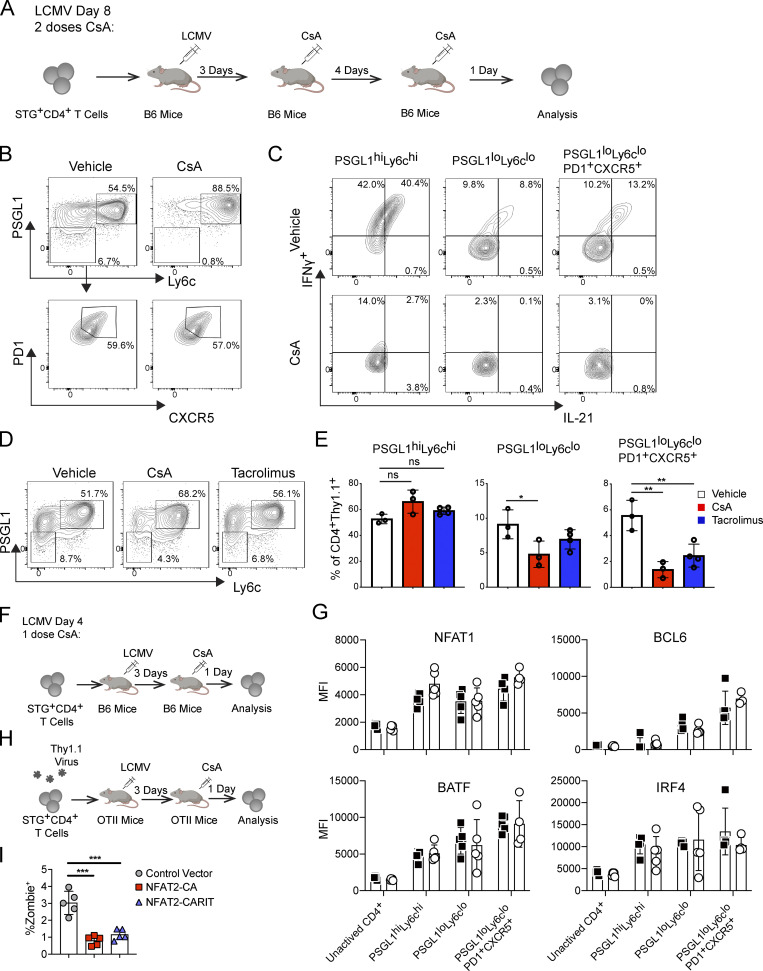
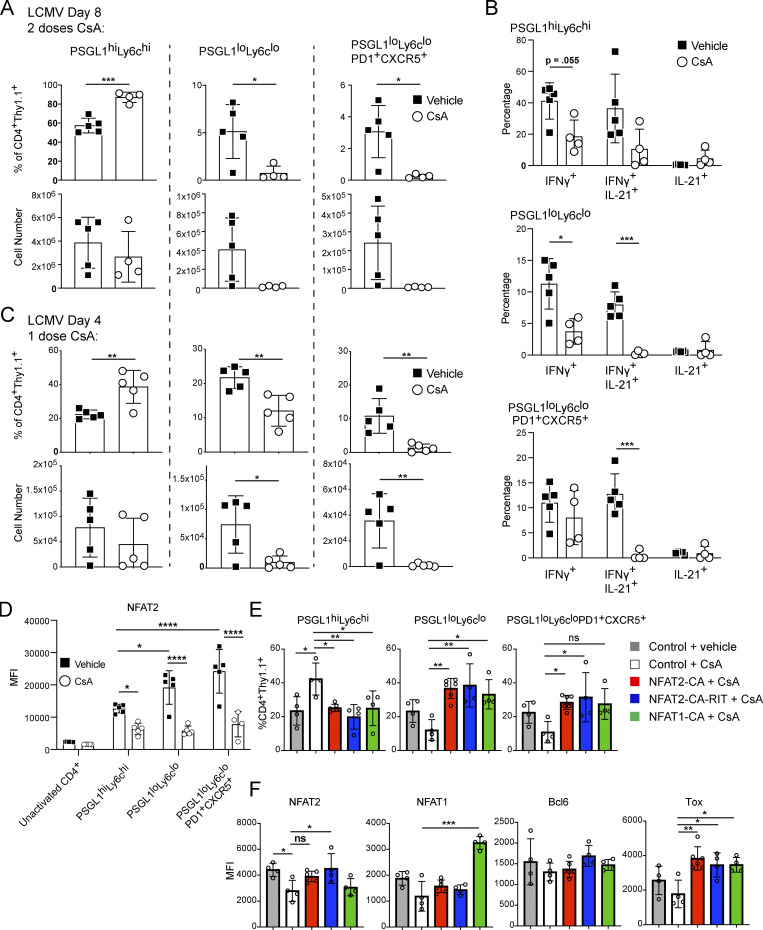
We next employed a pharmacologic approach using CNIs to further understand the role of NFAT signaling in Tfh cells in vivo. We transferred Thy1.1 (CD90.1) STG+ T cells into congenically marked WT B6 recipients and infected them with LCMV. We administered 80 mg/kg CsA or vehicle i.p. at days 3 and 7 after infection, isolating splenocytes for analysis on day 8 after infection (Fig. S4 A). The CsA-treated animals exhibited reduced frequency and number of PSGL1loLy6clo and PSGL1loLy6cloPD1+CXCR5+ CD4+ T cells relative to the control group (Fig. 5 A and Fig. S4 B). The frequency of PSGL1hiLy6chi CD4+ T cells was reciprocally increased, but their numbers were not statistically different between the groups (Fig. 5 A). As expected, CsA treatment affected the functional ability of all activated CD4+ T cells to produce effector cytokines when stimulated ex vivo (Fig. 5 B and Fig. S4 C). While the ability of CsA to impair cytokine production was expected, as NFAT signaling is universally required for activated T cell function, the selective ability of CsA to deplete PSGL1loLy6clo and PSGL1loLy6cloPD1+CXCR5+ CD4+ T cells was unexpected, but perhaps not surprising given our finding above that NFAT2 signaling is required for Tfh cell survival. We confirmed this selective depletion of Tfh cells was due to calcineurin inhibition and not a CsA-specific effect by using tacrolimus instead of CsA in parallel experiments (Fig. S4, D and E).
Figure S4.
TCR stimulation-mediated NFAT2 autoregulation is required for Tfh cell maintenance and survival. Related to Fig. 5. (A) Schematic of experimental strategy. STG+CD4+ T cells were adoptively transferred into B6 mice that were then acutely infected with LCMV Armstrong. Mice were treated with vehicle or 80 mg/kg CsA i.p. on day 3 and 7 following infection. 8 d after infection, splenocytes were isolated for analysis. (B) Representative flow cytometry plots of splenic STG+CD4+ T cells from vehicle control or CsA-treated animals, related to Fig. 5 A. (C) Representative flow cytometric analysis of ex vivo cytokine production after day 8 LCMV infection, related to Fig. 5 B. (D) Representative flow cytometry plots of splenic STG+CD4+ T cells from mice treated with vehicle control, CsA, or tacrolimus. (E) Percentages of PSGL1hiLy6chi, PSGL1loLy6clo, and PSGL1loLyc6loPD1+CXCR5+ T cells from panel D. One-way ANOVA with post-hoc Tukey’s test used for analysis. (F) Schematic of experimental strategy for single-dose CsA treatment. STG+CD4+ T cells were adoptively transferred into B6 mice that were then acutely infected with LCMV Armstrong. Mice were treated with vehicle or 80 mg/kg CsA i.p. on day 3. On the following day, splenocytes were isolated for analysis. (G) MFI of NFAT1, Bcl6, BATF, or IRF4 in the indicated cell subsets after day 4 LCMV infection, related to Fig. 5 C. (H) Schematic of experimental strategy. STG+CD4+ T cells were transduced with retroviruses carrying Thy1.1-tagged vectors and transferred into OT-II TCR transgenic mice, which were then acutely infected with LCMV Armstrong. 3 d after infection, CsA or vehicle was given i.p. After 24 h, splenocytes were isolated for analysis. (I) CD4+ T cells were transduced with retroviral vectors encoding Thy1.1 (control), NFAT2-CA, or NFAT2-CARIT. After 72 h, cells were labeled with the Zombie cell viability dye and analyzed by flow cytometry. One-way ANOVA with post-hoc Tukey’s test used for analysis. B, C, and G are representative of three experiments with three to five animals per group. D and E are representative of one experiment with three animals per group. I is representative of two independent experiments. *** = P ≤ 0.001, ** = P ≤ 0.01, * = P ≤ 0.05, and ns P > 0.05.
Figure 5.
TCR stimulation-mediated NFAT2 autoregulation is required for Tfh cell maintenance and survival. (A) Percentages and numbers of PSGL1hiLy6chi, PSGL1loLy6clo, and PSGL1loLyc6loPD1+CXCR5+ CD4+ T cells after day 8 LCMV infection with vehicle or CsA treatment. (B) Percentages of IFNγ+, IL-21+IFNγ+, and IL-21+ CD4+ T cells within each CD4+ T cell subset. (C) Percentages and numbers of PSGL1hiLy6chi, PSGL1loLy6clo, and PSGL1loLyc6loPD1+CXCR5+ CD4+ T cells after day 4 LCMV infection with vehicle or CsA treatment. (D) MFI of NFAT2 after day 4 LCMV infection in the indicated cell subsets. Data were analyzed with two-way ANOVA. (E) Percentages of PSGL1hiLy6chi, PSGL1loLy6clo, and PSGL1loLyc6loPD1+CXCR5+ CD4+ T cells for each indicated condition. (F) MFI of NFAT2, NFAT1, Tox or Bcl6 in PSGL1loLyc6loPD1+CXCR5+ CD4+ T cells. One-way ANOVA was used to analyze E and F. Results are representative of three independent experiments with three to five mice per experimental group. See also Fig. S4. **** = P ≤ 0.0001, *** = P ≤ 0.001, ** = P ≤ 0.01, * = P ≤ 0.05, and ns P > 0.05.
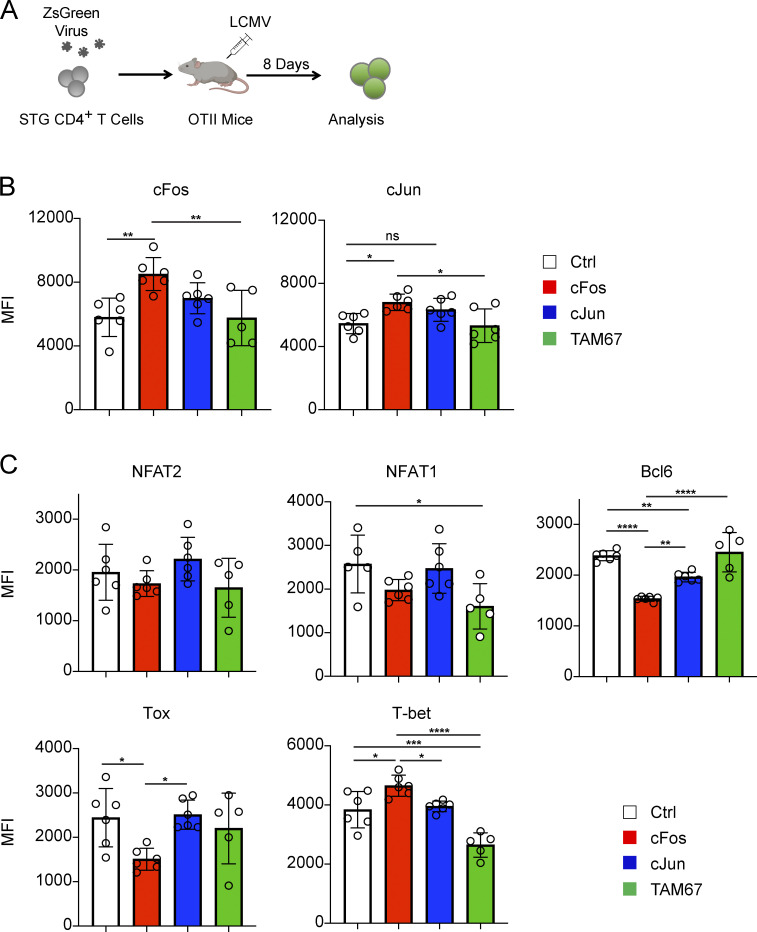
To further characterize the effect of CsA on Tfh cell development, we performed a shorter time course infection wherein CsA was administered 72 h after LCMV infection and cell populations were analyzed 24 h after CsA treatment (Fig. S4 F). We again observed a marked reduction in PSGL1loLy6clo and PSGL1loLy6cloPD1+CXCR5+ CD4+ T cell frequency and numbers, while the number of PSGL1hiLy6chi cells was relatively preserved (Fig. 5 C). We next investigated whether single-dose CsA-treatment affected key transcription factor expression. We found that PSGL1loLy6clo and PSGL1loLy6cloPD1+CXCR5+ CD4+ T cells in the control animals upregulated NFAT2 by day 4, more than PSGL1hiLy6chi CD4+ T cells, consistent with our previous findings on NFAT2 activation on day 8 after LCMV infection (Ray et al., 2015); however, this upregulation was abrogated in all cell populations in the animals that received a single dose of CsA 24 h earlier (Fig. 5 D). In contrast, the relative expression of NFAT1, Bcl6, BATF, and IRF4 were not significantly different between control and CsA-treated animals (Fig. S4 G). These results suggested that NFAT2 upregulation is a key early event in Tfh cell development that is dependent on calcium flux downstream of TCR stimulation. Supporting this idea was our observation that NFAT2 upregulation was detectable in PSGL1loLy6clo T cells that had not yet highly upregulated PD1 and CXCR5 (Fig. 5 D). The CsA sensitivity of NFAT2 upregulation at an early stage of Tfh cell development suggested that NFAT2 upregulation is NFAT signaling dependent. Indeed, NFAT-mediated upregulation of NFAT2 is a well-documented positive-feedback loop, termed NFAT autoregulation, which has been shown to protect T cells from activation-induced apoptosis in vitro (Chuvpilo et al., 2002), but has not been clearly associated with survival of a particular T helper cell subset, including Tfh cells, in vivo.
The ability of CsA to inhibit Tfh cell development allowed us to further probe the requirement for AP-1–independent NFAT signaling in these cells. We transduced STG+CD4+ T cells with constructs encoding different constitutively active NFAT mutants and transferred them into OT-II TCR transgenic mice which were then infected with LCMV and subsequently treated with CsA to block endogenous NFAT signaling (Fig. S4 H). Since all the mutants were constitutively active and, therefore, insensitive to the presence of CsA, Tfh cell persistence would depend on the functionality of the transduced NFAT mutant. While STG+CD4+ T cells transduced with control vector had impaired development of PSGL1loLy6clo and PSGL1loLy6cloPD1+CXCR5+ CD4+ T cells in the presence of CsA (Fig. 5 E), both NFAT2-CA and NFAT2-CARIT transduced cells were able to develop into PSGL1loLy6clo and PSGL1loLy6cloPD1+CXCR5+ CD4+ T cells under the same conditions, as expected, since both mutants can support AP-1–independent signaling (Fig. 5 E). Neither NFAT2-CA nor NFAT2-CARIT had any detrimental effect on transduced cell viability (Fig. S4 I). These results supported the idea that AP-1–independent NFAT signaling was sufficient for Tfh cell production in vivo. NFAT1-CA was also able to rescue PSGL1loLy6clo and PSGL1loLy6cloPD1+CXCR5+ CD4+ T cell development in vivo (Fig. 5 E), suggesting that total NFAT activity rather than a specific NFAT family member is required for Tfh cell development. All of the tested NFAT constructs decreased the relative proportion of PSGL1hiLy6chi T cells (Fig. 5 E). While Bcl6 was not differentially regulated in the presence or absence of any of the NFAT constructs, Tox was specifically induced in the conditions with NFAT overexpression (Fig. 5 F), supporting the idea that the role of NFAT signaling in Tfh cells is to engage AP-1–independent pathways, of which Tox is a marker (Pereira et al., 2017; Seo et al., 2019), rather than engage Tfh-defining pathways, of which Bcl6 is a marker.
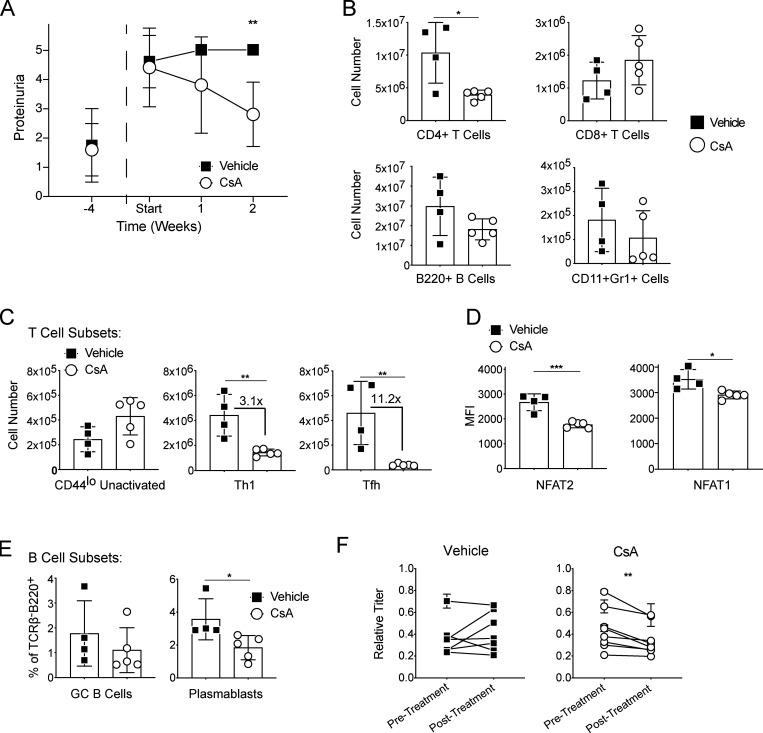
Calcineurin inhibition depletes Tfh cells and ameliorates disease in lupus-prone mice
We sought to investigate the role of NFAT signaling in Tfh cells in murine lupus, given their role in driving autoreactive B cell maturation with autoantibody production (Choi et al., 2017). To assess whether pharmacologic inhibition of NFAT signaling could reverse established disease, we administered CsA to NZBxW F1 lupus-prone mice. Starting at 15 wk age of age, we measured proteinuria weekly until all mice in our cohort developed > 3+ proteinuria as evidence of inflammatory renal disease (lupus nephritis; Casey, 1968). We then split the mice into two groups with the experimental mice receiving i.p. CsA 80 mg/kg three times per week and the control mice receiving vehicle control. Over the course of 2 wk, CsA-treated animals had improved proteinuria while the control animals had persistent, elevated proteinuria (Fig. 6 A). Upon sacrifice after 2 wk of treatment, CsA-treated animals had fewer total splenic CD4+ T cells than control animals, while CD8+ T cells, B220+ B cells, and TCRβ−B220−Gr1+CD11b+ myeloid cells were similar between the two groups (Fig. 6 B). While CsA-treated animals had decreased Th1 cells compared with controls, Tfh cells were the most affected T cell subtype with their numbers markedly decreased (Fig. 6 C). Within the Tfh cell subset, NFAT1 and NFAT2 were both reduced, with NFAT2 being more affected (Fig. 6 D), as we observed in acute LCMV infection. While we did not observe significant differences in GC B cells (Fig. 6 E), likely a consequence of the relatively short duration of CsA treatment, plasmablasts and anti-dsDNA (double-stranded DNA) titers were reduced (Fig. 6, E and F).
Figure 6.
Calcineurin inhibition depletes Tfh cells and ameliorates disease in lupus-prone mice. (A) Proteinuria measurements in NZBXW F1 mice. CsA treatment was started at age 15 wk (labeled Start). (B) Flow cytometric analysis of splenic immune cell populations in vehicle control and CsA-treated mice. (C) Numbers of CD4+CD44lo unactivated, Th1 (CD4+CD44hiPSGL1hi), and Tfh (CD4+CD44hiPSGL1loPD1+CXCR5+) T cells. (D) MFI of NFAT transcription factors in Tfh cell populations. (E) Percentage of splenic GC B cells (TCRβ−B220+CD138−CD95+GL7+) or plasmablasts (TCRβ−B220+CD138+). (F) dsDNA antibody titers at the beginning and the end of the experiment in the vehicle and CsA-treated groups. Statistical significance was determined using a paired t test of the pre- and posttreatment mean values within each cohort. Results are representative of two independent experiments with three to five animals per experimental group. *** = P ≤ 0.001, ** = P ≤ 0.01, * = P ≤ 0.05.
NFAT2 is required in CD4+ T cells for development of murine lupus
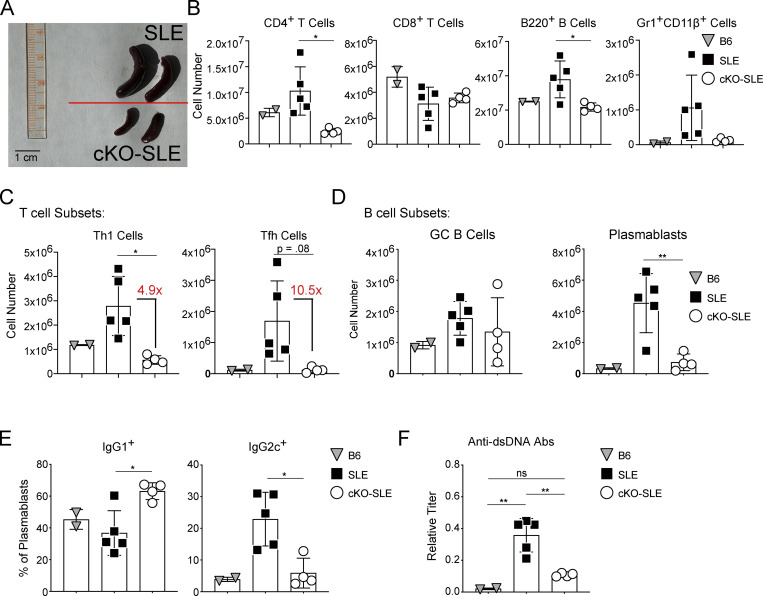
The experiments above suggested that intact NFAT signaling is required for the development of Tfh cells and the manifestations of autoimmune disease in a lupus-prone mouse model. To look at the specific role of NFAT2 in T cells in a model of autoimmunity, we turned to a genetically tractable murine lupus strain in a B6 strain background, B6.Sle1.Yaa (Deane et al., 2007; Murphy and Roths, 1979; Pisitkun et al., 2006; Subramanian et al., 2006). We bred lupus-prone B6.Sle1.Yaa mice to CD4Cre+.Nfatc1fl/fl.B6 mice with screening to ensure intact Sle1 locus, known for its recombination hotspots (Brunschwig et al., 2012). We use SLE to refer to CD4Cre-.Nfatc1fl/fl.B6.Sle1.Yaa and conditional KO-SLE (cKO-SLE) to CD4Cre+.Nfatc1fl/fl.B6.Sle1.Yaa mice. We aged SLE and cKO-SLE mice to 14–20 wk and analyzed splenic immune cell populations, with 15-wk-old B6 mice as additional controls, as previously described (Choi et al., 2017). The cKO-SLE mice exhibited reduced splenomegaly compared with SLE controls and reduced splenic cellularity (Fig. 7, A and B). Overall, cKO-SLE mice had decreased total CD4+ T, B220+ B, and Gr1+CD11β+ myeloid cells, while CD8+ T cells were similar (Fig. 7 B). These results suggested that T cell–restricted NFAT2 deficiency interferes with the immune cell expansion associated with autoimmunity.
Figure 7.
NFAT2 is required in CD4+ T cells for development of murine lupus. (A) Photograph of spleens isolated from aged SLE or NFAT2 cKO-SLE mice. Scale bar = 1 cm. (B) Numbers of splenic immune cell populations measured by flow cytometry comparing age-matched B6, SLE, and cKO-SLE mice. (C) Numbers of splenic T cell subsets. (D) Numbers of subtypes of splenic B cells. (E) Percentage of IgG1+ or IgG2c+ positive plasmablasts. (F) Serum dsDNA autoantibody titers from B6, SLE, or cKO-SLE mice measured by ELISA. T and B cell subsets are gated as described in Fig. 6. Results representative of three independent experiments with two to five animals per group. One-way ANOVA was used to analyze the data. ** = P ≤ 0.01, * = P ≤ 0.05, and ns P > 0.05.
We further characterized the T cell subsets in SLE vs. cKO-SLE animals and found that while Th1 effector cells were reduced approximately fivefold, there was a >10-fold reduction in Tfh cells (Fig. 7 C). While GC B cells were not significantly different between SLE and cKO-SLE animals, plasmablasts were significantly reduced (Fig. 7 D). The small number of remaining plasmablasts in the cKO-SLE animals were predominantly skewed to IgG1+, while SLE plasmablasts were significantly more IgG2c+, the pathogenic IgG isotype in B6.Sle1.Yaa mice (Fairhurst et al., 2008; Subramanian et al., 2006; Fig. 7 E). These results indicated a disproportionate effect of NFAT2 deficiency on the development and function of Tfh cells, leading to diminished pathogenic T–B collaboration, an essential contributor to autoimmunity in this model (Choi et al., 2017). Consistent with reduced splenomegaly and immune cell expansion, cKO-SLE mice had reduced anti-dsDNA antibody titers, relative to SLE controls, that were not statistically different from age-matched B6 controls suggesting that T cell NFAT2 is the primary driver of autoimmunity in this genetic model (Fig. 7 F).
Discussion
Using pharmacologic, genetic, and bioinformatic approaches, we have dissected the unique role of NFAT signaling in Tfh cells and demonstrated its importance to their development in autoimmunity. We found that an AP-1–independent NFAT gene program is essential for the proper development and maintenance of Tfh cells. This signature was found in acute infection and autoimmunity, suggesting that this gene program is central to their identity. Canonical NFAT signaling underlies T cell activation following TCR stimulation (Rao et al., 1997). Heretofore, the non-canonical AP-1–independent NFAT pathway has been described in hyporesponsive T cell states, such as anergy or exhaustion, and has been thought to be incompatible with effector T cell states (Hogan, 2017). We posited that AP-1–independent NFAT signaling plays an important role in Tfh cell development given their biological function to provide supportive and instructive cues to guide B cell development, selection, and maturation in the face of multiple successive rounds of antigen stimulation (Crotty, 2011; Liu et al., 2015). To test our hypothesis, we searched for evidence of overlap between Tfh cells and hyporesponsive T cell states. We have previously reported that Tfh cells are smaller, less proliferative, and less metabolically active compared with Th1 cells during acute viral infection, thus lending credence to the idea of viewing the former through the prism of hyporesponsiveness—at least relative to Th1 cells (Ray et al., 2015). The transcriptome of Tfh cells showed similarity to the gene signatures of anergic CD4+ and exhausted CD8+ T cells, findings consistent with previous observations of their phenotypic similarity to exhausted CD4+ cells in chronic viral infection (Crawford et al., 2014; Fahey et al., 2011). Abundant expression of inhibitory receptors and signaling proteins is characteristic of hyporesponsive cells. Tfh cells are also enriched for these proteins, such as PD1, although studies will be needed to focus on the specific role of individual inhibitory proteins in contributing to AP-1–independent NFAT signaling in these cells. Functionally, like hyporesponsive T cells, Tfh cells exhibited reduced calcium flux to TCR stimulation in vitro. As NFAT2 is their primary activated NFAT family member, we assessed the role of AP-1–independent NFAT signaling using NFAT2-RIT mutant proteins defective in AP-1 binding, designed based on published studies using NFAT1 (Macián et al., 2000; Martinez et al., 2015). Such mutants exhibited impaired activation of canonical NFAT pathways but retained the ability to engage AP-1–independent NFAT signaling. Our results showed that NFAT2 likely does not play a role in orchestrating the global transcriptional program of Tfh cells. Rather they suggest that NFAT2 has a critical role in maintaining their survival in vivo. Using the NFAT2-RIT mutants, we showed that AP-1–independent NFAT signaling was sufficient to restore Tfh cell development either when CD4+ T cells lacked NFAT2 expression or when endogenous NFAT2 activation was blocked pharmacologically. Together, these observations argue that AP-1–independent NFAT signaling, a driver of the hyporesponsiveness gene program, plays a fundamental role in Tfh cell survival. A similar AP-1–independent mode of NFAT2 signaling has also been described as critical for the survival of pathologic, but indolent B cells in chronic lymphocytic leukemia, arguing that our findings may represent a broader biological role for NFAT2 across cell types (Apollonio et al., 2013; Märklin et al., 2017). The specific molecular mechanisms by which AP-1–independent NFAT signaling promotes cell survival generally, and in Tfh cells in particular, are important areas for further investigation.
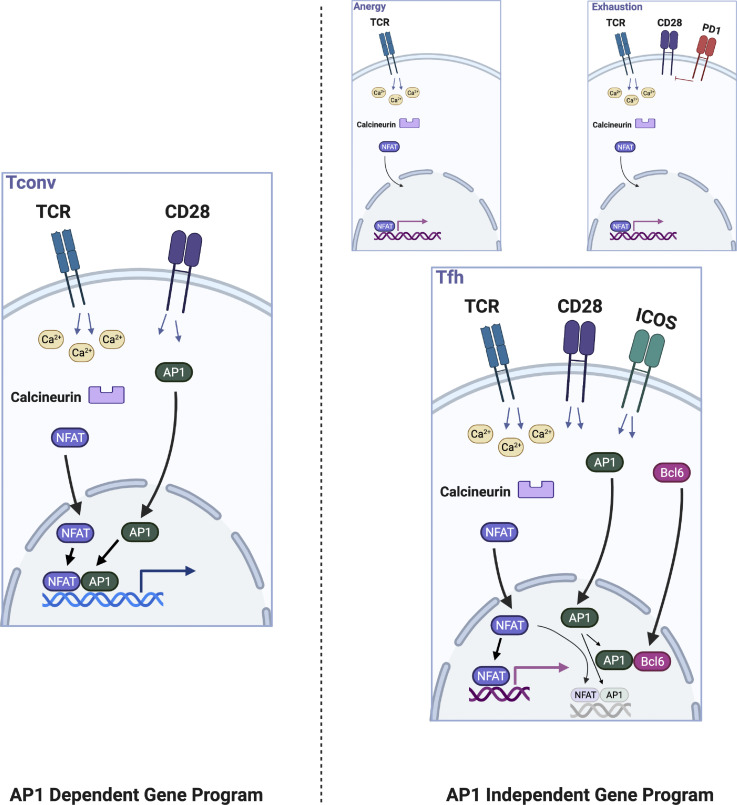
As effector Th cells regulating B cell maturation and selection, why might Tfh cells employ a non-canonical NFAT gene expression program previously described in the context of hyporesponsive T cell states? A systemic immune response entails the concerted action of distinct cell types and subsets, each genetically programmed to fulfill its physiological function in its biological niche. Upon activation in lymphoid tissues, Th cells, such as Th1, Th2, or Th17, rapidly expand and then migrate to peripheral sites of infection where they assist in pathogen clearance, but may also promote collateral inflammatory damage (Medzhitov et al., 2012). By contrast, Tfh cells arguably need to be more controlled in their effective promotion of B cell maturation. For example, disruption of splenic architecture, including disrupted GCs, is attributed to excessive inflammatory cytokine production and is postulated to impair adaptive immune responses to malaria (Lewis et al., 2019). Heightened inflammatory signals during chronic viral infection drive GC B cell dysregulation and impaired antibody development (Staupe et al., 2019 Preprint). Further, Tfh cell numbers are actively limited during normal immune responses (Pratama and Vinuesa, 2014) with their excessive accumulation associated with development of autoimmunity (Linterman et al., 2009; Vinuesa et al., 2005). Tfh cells are also known as frugal cytokine producers (Dan et al., 2016; Havenar-Daughton et al., 2016). All of these features, consistent with their biological function to facilitate GC B cell selection and competition in order to produce clones of optimal affinity, have been postulated to be regulated by Bcl6, but elucidation of specific molecular mechanisms has been elusive (Crotty, 2019; Qi, 2016). We provide evidence that ICOS costimulation via Bcl6 upregulation is key to allowing T cells to engage an AP-1–independent NFAT program in which their survival becomes dependent on intact NFAT2 signaling. In turn, this mode of NFAT gene expression likely enables Tfh cells to provide help in an effective and productive, yet restrained manner. Considering the ability of Bcl6 to physically sequester and antagonize AP-1 (Hatzi et al., 2015), we postulated that a specific and critical role of Bcl6 would be to reduce the pool of AP-1 available to interact with NFAT in developing Tfh cells, effectively reducing the NFAT:AP-1 ratio, enforcing an AP-1–independent NFAT gene expression program in Tfh cells. Consistent with this idea, we found that overexpression of cFos inhibited, and overexpression of an AP-1 inhibitor augmented, respectively, Tfh cell development in vivo. Our results suggest that they access AP-1–independent NFAT signaling using a molecular pathway distinct from that used by anergic or exhausted cells, likely explaining the differences that exist between these disparate populations of T cells (Fig. S5, model). Future studies will be needed to define the precise molecular interactions allowing Bcl6 to regulate AP-1 directly, i.e., specific effects on cJun or cFos, and identify additional mechanisms that may contribute to enforcing AP-1–independent NFAT signaling in Tfh cells.
Figure S5.
Model of AP-1–independent NFAT signaling in Tfh cells. Tfh cells exhibit AP-1–independent NFAT signaling as do anergic and exhausted cells. Our data suggest a distinct molecular mechanism underlies this gene expression program in Tfh cells. Canonical T helper cells utilize AP-1–dependent NFAT signaling as a consequence of signaling pathways activated downstream of TCR and CD28 engagement. Anergic cells receive TCR signal in the absence of CD28 costimulation. One mechanism in exhausted cells may be the blunting of CD28 signaling through the action of inhibitory receptors such as PD-1. In Tfh cells, our data suggest that ICOS signaling, which upregulates Bcl6, an AP-1 antagonist, is responsible for skewing NFAT signaling toward an AP-1–independent mode. Thus, while Tfh cells share some features with anergic and exhausted cells, they also have distinct functional capabilities.
CNIs depress humoral responses, but their specific effects on Tfh cells have not been reported (Goodlad and Macartney, 1995; Legrand et al., 2013; Moriyama et al., 2012). We found that cytokine production by both Tfh and Th1 cells was reduced in mice treated with CsA during acute LCMV infection, as expected. However, Tfh cell numbers were decimated, while Th1 cell numbers were relatively unchanged, similar to the effect of T cell–restricted NFAT2 genetic deficiency. The observation that CsA treatment phenocopied the effect of NFAT2 deficiency suggested we could use it as a pharmacological probe of the role of NFAT2 in Tfh cell development and maintenance. Administration of a single dose of CsA 72 h after LCMV infection dramatically reduced the Tfh cell population within 24 h, with relatively minimal effect on Th1 cell numbers. At this time point, Tfh cells from control animals exhibited a significant upregulation of NFAT2 protein relative to naive T cells. Upregulation was not simply a consequence of activation as NFAT2 protein expression in Tfh cells was higher compared with Th1 cells. The relative upregulation of NFAT2 in Tfh cells was completely blocked by CsA. In contrast, Bcl6 expression was relatively unchanged. As CsA blocks the transmission of TCR signals through the calcium-regulated phosphatase calcineurin, it effectively abolishes autoregulation of NFAT2 (Chuvpilo et al., 2002). The exquisite sensitivity of Tfh cell numbers to interrupted TCR signaling is reminiscent of the TCR “addiction” observed in exhausted CD8+ cells during chronic viral infection (Scott et al., 2019; Shin and Wherry, 2007; Wherry, 2011) and could explain our previously reported observation that Tfh cells are more susceptible to apoptotic cell death ex vivo compared with Th1 cells (Ray et al., 2015) and could be related to the heightened cell death we observed in vitro when NFAT2-deficient CD4+ T cells were stimulated in the presence of CD28 and ICOS combination costimulation. The inhibition of NFAT2 autoregulation as one of the first observable effects of CsA suggests autoregulation of NFAT2 as the molecular mediator of this addiction in vivo and provides a molecular explanation for the observed dependence of Tfh cells on antigen persistence (Baumjohann et al., 2013; Deenick et al., 2010).
While pharmacologic treatment with CsA and genetic disruption had similar effects on Tfh cell development, the CsA effect was consistently much more pronounced. As CsA would be expected to block NFAT1 and NFAT2 signaling in T cells, this observation suggests that NFAT1 also has a role in Tfh cell development. This is consistent with a previous study demonstrating that while Tfh numbers are markedly reduced in NFAT2 single KOs, doubly deficient NFAT1/2 KO T cells have greater defects in Tfh cell PD1 expression and GC B cell percentage (Martinez et al., 2016). Our data that NFAT1-CA overexpression can rescue NFAT2 deficiency argues that, with sufficient expression and activation, NFAT1 can substitute qualitatively for NFAT2. This is consistent with the idea that NFAT proteins exhibit biological redundancy (Macián, 2005; Rao et al., 1997), but our data (this study) and previously published data (Martinez et al., 2016) also suggest that, under physiologic conditions, NFAT1 cannot compensate for the loss of NFAT2. Toward this point, while autoregulation and positive feedforward amplification of NFAT2 are well described (Chuvpilo et al., 2002; Serfling et al., 2006), NFAT1 is not regulated in a similar manner (Hogan et al., 2003). Perhaps these intrinsic differences in the biological regulation of NFAT1 vs. NFAT2 underlie their different functional roles in Tfh cells. Thus, the residual Tfh cell population in NFAT2-deficient animals may represent functional Tfh cells that have arisen due to partial but not complete compensation by NFAT1, i.e., they are programmed like Tfh cells but are decreased in number because they do not have sufficient NFAT signaling to prevent cell death. In support of this idea, we have found the transcriptome of NFAT2-deficient Tfh cells to be nearly identical to that of WT Tfh cells (data not shown). Overall, our observations are consistent with the notion that the specific role of NFAT2 in Tfh cells is a consequence of its unique regulation—relatively robust accumulation due to positive feedback—rather than engagement of an NFAT2-specific transcriptional program. Whether there are active mechanisms that restrict NFAT1 function in Tfh cells or whether the physiological amount of NFAT1 is simply insufficient to meet the NFAT signaling demand of Tfh cells is not clear but would be an important area for future investigation.
Our results further our understanding of the Tfh transcriptional program and elucidate the importance of ICOS, Bcl6, and NFAT2 in Tfh cell survival and development. But future studies are required to rigorously test our model of Tfh cell development (Fig. S5). First, we have been unable to directly measure NFAT2:AP-1 complexes in vivo or in vitro. This is due to technical limitations as we have found, as previously reported (Klein-Hessling et al., 2017; Minami et al., 2013), that commercially available NFAT2 antibodies are not reliable for immunoprecipitation in our hands (data not shown). Second, while the results with cFos and TAM67 are consistent with the idea that NFAT:AP-1 stoichiometry is important for Tfh cell development, our ability to draw stronger conclusions was limited by the lack of an ability to titrate AP-1 levels consistently over a broad concentration range. Developing the technological capability to finely regulate NFAT:AP-1 stoichiometry will enable direct assessment of AP-1–independent NFAT signaling in the development of autoimmune disease, where we have shown genetically and pharmacologically that NFAT signaling is required for the abnormal persistence of Tfh cells in two different models of murine lupus.
Finally, recent advances in development of CNIs have led to a rekindled interest in their deployment in treatment of autoimmune diseases (Lee et al., 2018), increasing the imperative to understand how they exert therapeutic benefits. Our results show that Tfh cells are a major target of CNIs in mouse models of lupus. Further, our finding that Tfh cells are rapidly depleted following CsA administration raises the intriguing possibility that CNI treatment of human lupus need not be given continuously—rather intermittent boluses may be sufficient to keep Tfh cell expansion and autoimmune disease manifestations in check—thereby lowering cumulative drug exposure and potentially minimizing medication side effects. While NFAT family members are the primary targets of calcineurin inhibition, other substrates also exist and likely mediate some of the therapeutic benefits of CNIs (Faul et al., 2008; Spurney, 2014). However, these other substrates may also contribute to the most limiting side effects of CNIs, i.e., nephrotoxicity (Cheriyan et al., 2021; Gooch et al., 2007; Ume et al., 2021). This has led to interest in developing therapies that directly target NFAT family members. Our results suggest that NFAT2-specific inhibitors could selectively target Tfh cells and, therefore, might have clinical utility in autoantibody-mediated autoimmune disease.
Materials and methods
Mice
Mice were housed in specific pathogen–free conditions in accordance with U.S. federal regulations and approved by Yale University’s Institutional Animal Care and Use Committee. Unless otherwise noted, experiments were conducted on male and female mice, aged 6–8 wk.
C57BL/6 (B6) mice were purchased from Charles River Laboratories. SMARTA Transgenic (TG[TCRLCMV]1Aox; STG) mice were obtained from Hans Hengartner at the University of Zürich, Switzerland (Oxenius et al., 1998). NZBWF1/J (NZBXW F1), Tg(CD4-Cre)1Cwi/BfluJ (CD4Cre+), B6.Cg-Tg(TcraTcrb)425Cbn/J (OT-II), B6.129P2-Tcrbtm1Mom/J (TCRβ−), and B6(Cg)-Nfatc1<tm3Glm>/AoaJ (NFATc1 flox) mice were purchased from The Jackson Laboratory. B6.Sle1.Yaa mice were provided by E. Wakeland (University of Texas Southwestern Medical School, Dallas, TX, USA). CD4Cre+ mice were crossed with Nfatc1 flox mice to generate CD4Cre+.Nfatc1fl/fl mice. CD4Cre+.Nfatc1fl/fl were crossed with B6.Sle1.Yaa mice to generate CD4Cre+.Nfatc1fl/f.B6.Sle1.Yaa mice. The presence of the intact Sle1 locus was confirmed by microsatellite screening within the locus including D1Mit15, D1Mit17, D1Mit47, D1Mit202, D1Mit113, D1Mit206, D1Mit353, D1Mit407, D1Mit105, D1Mit274, D1Mit400, and D1Mit541 (Morel et al., 2000).
Cell culture
HEK293T cells (ATCC), used for retroviral packaging, were passaged in DMEM supplemented with 10% FBS and penicillin/streptomycin. Primary murine T cells were cultured in RPMI-1640 supplemented with 10% FBS, penicillin/streptomycin, L-glutamate, and 2-mercaptoethanol (T cell media). All cells were maintained in a humidified incubator with 5% CO2 at 37°C.
DNA constructs
The pMIGR1 vector (Pear et al., 1998) was modified to encode either Thy1.1 or ZsGreen (Matz et al., 1999) downstream of the IRES site instead of GFP. Constructs encoding NFAT1, NFAT2, cJun, cFos, or their mutants were cloned into the multiple cloning site using the In-Fusion HD Cloning Kit (Clontech Laboratories). Vectors encoding NFAT1 (gift from Anjana Rao, ID 11791; Addgene), NFAT1-CA (gift from Anjana Rao, ID 11792; Addgene), NFAT2 (gift from Anjana Rao, ID 11101; Addgene; Monticelli and Rao, 2002), NFAT2-CA (gift from Anjana Rao, ID 11793; Addgene), cJun (gift from Axel Behrens, ID 47443; Addgene; Aguilera et al., 2011), and cFos (MC203181; Origene) were used as cDNA templates. Based on the sequence alignment of NFAT1 and NFAT2, the R471A/L472A/T538G point mutations, designed to mimic the R468A/I469A/T535G point mutations in NFAT1-CARIT that impair AP-1 binding (Macián et al., 2000), were introduced into NFAT2 and NFAT2-CA using PCR-directed site mutagenesis to create NFAT2-RIT and NFAT2-CARIT, respectively. TAM67, a dominant negative mutant of cJun lacking residues 3–122 (Brown et al., 1993), was cloned from the cJun vector.
LCMV infection
The Armstrong strain of LCMV was grown as previously described (Ahmed and Oldstone, 1988). Mice were injected i.p. with 2 × 105 plaque-forming units of LCMV for each infection.
Immunization
Mice were immunized in bilateral foot pads with 100 μl of a mixture of 100 μg papain (Sigma-Aldrich) and 100 μg 4-hydroxy-3-nitrophenylacetyl conjugated to ovalbumin (NP-ova; Sigma-Aldrich). After 5 d, inguinal and popliteal lymph nodes were harvested. Lymphocytes were prepared for calcium flux analysis as described below.
Retroviral transduction followed by in vitro culture
On day 1, HEK293T cells were transfected with 1 μg of vector plus 0.5 μg of pCL-Eco plasmid (Naviaux et al., 1996) using X-tremeGene 9 DNA transfection reagent (Roche). On day 2, the media was exchanged with 1 ml fresh T cell media. CD4+ T cells were isolated from the spleens of B6 mice using an EasySep Mouse Naive CD4+ T cell isolation kit (Stemcell Technologies) and cultured in 96-well plates coated with anti-CD3 antibody (10 μg/ml; BD Biosciences) at a density of 1–2 × 106 cells per well in 0.1 ml of T cell media with soluble anti-CD28 antibody (10 μg/ml; BD Biosciences). On day 3, the activated T cells were transferred to 24-well plates at a density of 1–2 × 106 cells per well, washed, and then resuspended in virus-containing supernatants from the HEK293T cell cultures. Polybrene was added to a final concentration of 8 μg/ml. The cell–virus mixture was then centrifuged at 3,500 rpm for 90 min at 32°C to achieve transduction. Following transduction, cells were cultured in 96-well plates coated with anti-CD3 antibody (1 μg/ml) at a density of 5 × 105 cells per well in T cell media with soluble anti-CD28 antibody (1 μg/ml). On the following day, cells were washed and cultured in fresh 96-well plates in T cell media with IL-2 (20 ng/ml) until ready for further analysis.
Retroviral transduction followed by adoptive transfer
HEK293T cells were transfected and treated as described above. On day 2, splenocytes were prepared from STG mice and cultured in 10-cm plates at a density of 5–10 × 107 cells in 15 ml T cell media with soluble LCMV peptide GP61-80 (1 μg/ml; AnaSpec). The following day, activated STG+ T cells were isolated using an EasySep Mouse CD4+ T cell isolation kit (Stemcell Technologies) and plated in 24-well plates at a density of 1–2 × 106 cells per well. Cells were transduced with viral supernatants as described above. Following transduction, cells were washed and resuspended in PBS for adoptive transfer via retro-orbital injection into recipient mice.
In vitro T cell stimulation
CD4+ T cells were isolated from the spleens of B6 mice using an EasySep Mouse Naive CD4+ T cell isolation kit (Stemcell Technologies) and cultured in 96-well plates at a density of 1 × 106 cells per well in 0.2 ml of T cell media. For plate-bound stimulation, wells were precoated with anti-CD3 antibody (10 μg/ml; BD Biosciences, Purified NA/LE anti-mouse CD3 clone 145-2C11, Catalog Number 553057), and soluble anti-CD28 antibody (10 μg/ml; BD Biosciences, Purified NA/LE anti-mouse CD28 clone 37.51, Catalog Number 553294) was added at the time of cell plating. For ICOS stimulation, anti-CD3 antibody (1 μg/ml; BD Biosciences), anti-CD28 antibody (1 μg/ml), anti-ICOS antibody (2 μg/ml; Biolegend, Ultra-LEAF anti-ICOS clone C398.4A, Catalog Number 313540), and anti-hamster crosslinking antibody (25 μg/ml; Jackson ImmunoResearch, AffiniPure Goat Anti-Armenian Hamster IgG (H+L), Catalog Number 127-005-099) were added at the time of cell plating. Cells were incubated for 72 h and then processed for subsequent analysis.
Calcium flux assay
Cell preparations were suspended in 1 ml loading buffer (DPBS supplemented with 1 mM CaCl2, 1 mM MgSO4, and 1% FBS) at a density of 1 × 107 cells per ml and loaded with the calcium indicator dyes Fluo-4 (Thermo Fisher Scientific) and FuraRed (Thermo Fisher Scientific) at 37°C according to manufacturer’s directions. Cells were then washed and stained for flow cytometry at room temperature. Immediately before measurement, cells were incubated at 37°C for 5 min and then transferred to the flow cytometer for continuous monitoring. After obtaining a baseline measurement for 30 s, anti-CD3 antibodies (5 μg/ml) and IgG crosslinking antibodies (25 μg/ml) were added to the sample. After 180 s, ionomycin (1 μg/ml) was added to stimulate maximal cytosolic calcium influx.
Pharmacologic agents
For in vitro T cell stimulation assays, PMA (Sigma-Aldrich) was used at 50 ng/ml and ionomycin (Sigma-Aldrich) was used at 1 μg/ml final concentrations. For CsA (Calbiochem) treatment, 10x stock solutions were prepared in EtOH. CsA stocks were diluted with olive oil (Sigma-Aldrich) and, after thorough mixing, were injected i.p. at a dose of 80 mg/kg. For tacrolimus (Calbiochem), the final treatment dose was 2.5 mg/kg.
Flow cytometry
For flow cytometric analysis of splenocytes, single-cell suspensions were prepared. In general, 3 × 106 cells were surface stained in ice-cold PBS for 1 h at 4°C. CXCR5 staining was performed at room temperature for 1 h to optimize detection. After staining, cells were washed and analyzed on an LSRII Multilaser Cytometer (BD Biosciences). Intracellular staining for the indicated transcription factors was performed using the FoxP3/Transcription Factor Fixation/Permeabilization Kit (eBioscience) according to the manufacturer’s directions. Cell number was quantified using a reference number of microsphere beads (Spherotech ACBP70-10) added to specific samples. For intracellular cytokine staining, cells were plated in 96-well plates in T cell media. Cells were stimulated with PMA/ionomycin or media control. After 1 h, GolgiPlug (BD Biosciences) was added to each well. After 3 h, cells were harvested and surface stained as described above. Cells were then fixed with BD Cytofix (BD Biosciences) for 15 min followed by permeabilization with Permeabilization Buffer (Invitrogen) for 1 h. IL-21 staining was performed sequentially using an IL-21R-Fc chimeric protein (R&D Systems), followed by PE-labeled anti-human IgG (Jackson ImmunoResearch Laboratories) as previously described (Ray et al., 2015). IFN-γ antibody (Biolegend) was added at the initial step of IL-21 staining to simultaneously detect intracellular IFN-γ. For cell proliferation assays, CellTrace Yellow was purchased from ThermoFisher and used according to the manufacturer’s direction. For cell viability, Zombie Fixable Viability Kit was purchased from Biolegend and used according to the manufacturer’s directions.
RNA-Seq
Primary CD4+ T cells were isolated from the spleens of B6 mice using an EasySep Mouse Naive CD4+ T cell isolation kit (Stemcell Technologies). 1.5 × 106 cells were transduced with pMIT (control vector) or pMIT-NFAT2-CARIT as described above. 3 d after transduction, CD4+ T cells were sorted and RNA was isolated using an RNeasy mini kit (Qiagen). Purified RNA was submitted to the Yale Center for Genome Analysis where paired-end polyA RNA-Seq was performed. Gene analysis was performed using Partek Flow software, version 10. GSEA analysis was conducted using the GenePattern website (Reich et al., 2006). All sequencing data have been submitted to GEO and are available via SuperSeries accession number GSE223978.
Analysis of lupus-prone mice
Anti-dsDNA antibody titers were determined using ELISA as previously described (Choi et al., 2017). Briefly, ELISA plates were coated with methylated BSA (Sigma-Aldrich) followed by calf thymus DNA. Diluted sera were added to the plates and incubated overnight at 4°C. Anti-dsDNA antibodies were detected with goat anti-mouse IgG conjugated to HRP (Southern Biotechnology). Plates were developed with TMB Microwell peroxidase Substrate (SureBlue; KPL) according to the manufacturer’s directions and read at 450 nm using a microplate reader. Proteinuria was measured using Uristix urinalysis reagent strips (Siemens). Splenocyte preparations were made and stained for flow cytometry as described above.
Statistical analysis
Unless otherwise indicated in the figure legends, data were analyzed using unpaired t tests. Statistical analysis was done with GraphPad Prism version 9.0.0 for MacOS (GraphPad Software, http://www.graphpad.com). Statistical significance was defined as ****P ≤ 0.0001, ***P ≤ 0.001, **P ≤ 0.01, *P ≤ 0.05, and ns P > 0.05. Error bars represent SD.
Online supplementary material
Fig. S1 shows additional supporting data regarding the presence of an AP-1–independent NFAT gene program in Tfh cells. Fig. S2 provides additional data on the comparison of signaling programs induced by NFAT2-CA vs. NFAT2-CARIT in vitro and between NFAT2 vs. NFAT2-RIT in vivo. Fig. S3 provides additional characterization of Tfh cells generated in the context of AP-1 protein overexpression. Fig. S4 provides supporting data on the experiments used to probe the effects of CNIs on Tfh cell development and function. Fig. S5 provides a model of NFAT-signaling in Tfh cells as distinct from other types of CD4+ T cell subsets.
Acknowledgments
We thank Jason Weinstein and past and current Craft lab members for helpful discussions. Fig. S5 and mouse images in Figs. S2, S3, and S4 were created with Biorender.com.
This work was supported by grants from the National Institutes of Health (R37AR40072 and R01AR074545) and from the Lupus Research Alliance (J. Craft), Patterson Trust (A. Seth), and Daiichi Sankyo Co. Ltd. (Y. Yokokura).
Author contributions: Conceptualization: A. Seth and J. Craft; Methodology: A. Seth, Y. Yokokura, and J. Craft; Investigation: A. Seth, Y. Yokokura, J.-Y. Choi, J.A. Shyer, A. Vidyarthi; Writing—Original Draft, Revised Draft: A. Seth; Figures: A. Seth, Y. Yokokura; Writing—Review and Editing: A. Seth, Y. Yokokura, and J. Craft; Funding Acquisition: A. Seth, Y. Yokokura, and J. Craft; Supervision: J. Craft.
References
- Aguilera, C., Nakagawa K., Sancho R., Chakraborty A., Hendrich B., and Behrens A.. 2011. c-Jun N-terminal phosphorylation antagonises recruitment of the Mbd3/NuRD repressor complex. Nature. 469:231–235. 10.1038/nature09607 [DOI] [PubMed] [Google Scholar]
- Ahmed, R., and Oldstone M.B.. 1988. Organ-specific selection of viral variants during chronic infection. J. Exp. Med. 167:1719–1724. 10.1084/jem.167.5.1719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alfei, F., Kanev K., Hofmann M., Wu M., Ghoneim H.E., Roelli P., Utzschneider D.T., von Hoesslin M., Cullen J.G., Fan Y., et al. 2019. TOX reinforces the phenotype and longevity of exhausted T cells in chronic viral infection. Nature. 571:265–269. 10.1038/s41586-019-1326-9 [DOI] [PubMed] [Google Scholar]
- Apollonio, B., Scielzo C., Bertilaccio M.T.S., Ten Hacken E., Scarfò L., Ranghetti P., Stevenson F., Packham G., Ghia P., Muzio M., and Caligaris-Cappio F.. 2013. Targeting B-cell anergy in chronic lymphocytic leukemia. Blood. 121:3879–3888. 10.1182/blood-2012-12-474718 [DOI] [PubMed] [Google Scholar]
- Barnett, L.G., Simkins H.M.A., Barnett B.E., Korn L.L., Johnson A.L., Wherry E.J., Wu G.F., and Laufer T.M.. 2014. B cell antigen presentation in the initiation of follicular helper T cell and germinal center differentiation. J. Immunol. 192:3607–3617. 10.4049/jimmunol.1301284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumjohann, D., Preite S., Reboldi A., Ronchi F., Ansel K.M., Lanzavecchia A., and Sallusto F.. 2013. Persistent antigen and germinal center B cells sustain T follicular helper cell responses and phenotype. Immunity. 38:596–605. 10.1016/j.immuni.2012.11.020 [DOI] [PubMed] [Google Scholar]
- Brown, P.H., Alani R., Preis L.H., Szabo E., and Birrer M.J.. 1993. Suppression of oncogene-induced transformation by a deletion mutant of c-jun. Oncogene. 8:877–886 [PubMed] [Google Scholar]
- Brunschwig, H., Levi L., Ben-David E., Williams R.W., Yakir B., and Shifman S.. 2012. Fine-scale maps of recombination rates and hotspots in the mouse genome. Genetics. 191:757–764. 10.1534/genetics.112.141036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bubier, J.A., Sproule T.J., Foreman O., Spolski R., Shaffer D.J., Morse H.C. III, Leonard W.J., and Roopenian D.C.. 2009. A critical role for IL-21 receptor signaling in the pathogenesis of systemic lupus erythematosus in BXSB-Yaa mice. Proc. Natl. Acad. Sci. USA. 106:1518–1523. 10.1073/pnas.0807309106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casey, T.P. 1968. Immunosuppression by cyclophosphamide in NZB X NZW mice with lupus nephritis. Blood. 32:436–444. 10.1182/blood.V32.3.436.436 [DOI] [PubMed] [Google Scholar]
- Chang, A., Clark M.R., and Ko K.. 2021. Cellular aspects of the pathogenesis of lupus nephritis. Curr. Opin. Rheumatol. 33:197–204. 10.1097/BOR.0000000000000777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, T.K., Smith L.M., Gebhardt D.K., Birrer M.J., and Brown P.H.. 1996. Activation and inhibition of the AP-1 complex in human breast cancer cells. Mol. Carcinog. 15:215–226. [DOI] [PubMed] [Google Scholar]
- Cheriyan, A.M., Ume A.C., Francis C.E., King K.N., Linck V.A., Bai Y., Cai H., Hoover R.S., Ma H.P., Gooch J.L., et al. 2021. Calcineurin A-alpha suppression drives Nuclear Factor κB-mediated NADPH oxidase-2 upregulation. Am. J. Physiol. Renal. Physiol. 320:F336–F341. 10.1152/ajprenal.00254.2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi, J., and Crotty S.. 2021. Bcl6-Mediated transcriptional regulation of follicular helper T cells (TFH). Trends Immunol. 42:336–349. 10.1016/j.it.2021.02.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi, J.-Y., Seth A., Kashgarian M., Terrillon S., Fung E., Huang L., Wang L.C., and Craft J.. 2017. Disruption of pathogenic cellular networks by IL-21 blockade leads to disease amelioration in murine lupus. J. Immunol. 198:2578–2588. 10.4049/jimmunol.1601687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi, Y.S., Kageyama R., Eto D., Escobar T.C., Johnston R.J., Monticelli L., Lao C., and Crotty S.. 2011. ICOS receptor instructs T follicular helper cell versus effector cell differentiation via induction of the transcriptional repressor Bcl6. Immunity. 34:932–946. 10.1016/j.immuni.2011.03.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chtanova, T., Tangye S.G., Newton R., Frank N., Hodge M.R., Rolph M.S., and Mackay C.R.. 2004. T follicular helper cells express a distinctive transcriptional profile, reflecting their role as non-Th1/Th2 effector cells that provide help for B cells. J. Immunol. 173:68–78. 10.4049/jimmunol.173.1.68 [DOI] [PubMed] [Google Scholar]
- Chuvpilo, S., Jankevics E., Tyrsin D., Akimzhanov A., Moroz D., Jha M.K., Schulze-Luehrmann J., Santner-Nanan B., Feoktistova E., König T., et al. 2002. Autoregulation of NFATc1/A expression facilitates effector T cells to escape from rapid apoptosis. Immunity. 16:881–895. 10.1016/S1074-7613(02)00329-1 [DOI] [PubMed] [Google Scholar]
- Craft, J.E. 2012. Follicular helper T cells in immunity and systemic autoimmunity. Nat. Rev. Rheumatol. 8:337–347. 10.1038/nrrheum.2012.58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawford, A., Angelosanto J.M., Kao C., Doering T.A., Odorizzi P.M., Barnett B.E., and Wherry E.J.. 2014. Molecular and transcriptional basis of CD4⁺ T cell dysfunction during chronic infection. Immunity. 40:289–302. 10.1016/j.immuni.2014.01.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crotty, S. 2011. Follicular helper CD4 T cells (TFH). Annu. Rev. Immunol. 29:621–663. 10.1146/annurev-immunol-031210-101400 [DOI] [PubMed] [Google Scholar]
- Crotty, S. 2019. T follicular helper cell biology: A decade of discovery and diseases. Immunity. 50:1132–1148. 10.1016/j.immuni.2019.04.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curran, T., and Franza B.R. Jr. 1988. Fos and jun: The AP-1 connection. Cell. 55:395–397. 10.1016/0092-8674(88)90024-4 [DOI] [PubMed] [Google Scholar]
- Dan, J.M., Lindestam Arlehamn C.S., Weiskopf D., da Silva Antunes R., Havenar-Daughton C., Reiss S.M., Brigger M., Bothwell M., Sette A., and Crotty S.. 2016. A cytokine-independent approach to identify antigen-specific human germinal center T follicular helper cells and rare antigen-specific CD4+ T cells in blood. J. Immunol. 197:983–993. 10.4049/jimmunol.1600318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deane, J.A., Pisitkun P., Barrett R.S., Feigenbaum L., Town T., Ward J.M., Flavell R.A., and Bolland S.. 2007. Control of toll-like receptor 7 expression is essential to restrict autoimmunity and dendritic cell proliferation. Immunity. 27:801–810. 10.1016/j.immuni.2007.09.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deenick, E.K., Chan A., Ma C.S., Gatto D., Schwartzberg P.L., Brink R., and Tangye S.G.. 2010. Follicular helper T cell differentiation requires continuous antigen presentation that is independent of unique B cell signaling. Immunity. 33:241–253. 10.1016/j.immuni.2010.07.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietz, L., Frommer F., Vogel A.L., Vaeth M., Serfling E., Waisman A., Buttmann M., and Berberich-Siebelt F.. 2015. NFAT1 deficit and NFAT2 deficit attenuate EAE via different mechanisms. Eur. J. Immunol. 45:1377–1389. 10.1002/eji.201444638 [DOI] [PubMed] [Google Scholar]
- Domann, F.E., Levy J.P., Birrer M.J., and Bowden G.T.. 1994. Stable expression of a c-JUN deletion mutant in two malignant mouse epidermal cell lines blocks tumor formation in nude mice. Cell Growth Differ. 5:9–16 [PubMed] [Google Scholar]
- Dong, X., Antao O.Q., Song W., Sanchez G.M., Zembrzuski K., Koumpouras F., Lemenze A., Craft J., and Weinstein J.S.. 2021. Type I interferon-activated STAT4 regulation of follicular helper T cell-dependent cytokine and immunoglobulin production in lupus. Arthritis Rheumatol. 73:478–489. 10.1002/art.41532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong, Z., Birrer M.J., Watts R.G., Matrisian L.M., and Colburn N.H.. 1994. Blocking of tumor promoter-induced AP-1 activity inhibits induced transformation in JB6 mouse epidermal cells. Proc. Natl. Acad. Sci. USA. 91:609–613. 10.1073/pnas.91.2.609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fahey, L.M., Wilson E.B., Elsaesser H., Fistonich C.D., McGavern D.B., and Brooks D.G.. 2011. Viral persistence redirects CD4 T cell differentiation toward T follicular helper cells. J. Exp. Med. 208:987–999. 10.1084/jem.20101773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fairhurst, A.M., Hwang S.H., Wang A., Tian X.H., Boudreaux C., Zhou X.J., Casco J., Li Q.Z., Connolly J.E., and Wakeland E.K.. 2008. Yaa autoimmune phenotypes are conferred by overexpression of TLR7. Eur. J. Immunol. 38:1971–1978. 10.1002/eji.200838138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faul, C., Donnelly M., Merscher-Gomez S., Chang Y.H., Franz S., Delfgaauw J., Chang J.M., Choi H.Y., Campbell K.N., Kim K., et al. 2008. The actin cytoskeleton of kidney podocytes is a direct target of the antiproteinuric effect of cyclosporine A. Nat. Med. 14:931–938. 10.1038/nm.1857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gooch, J.L., Roberts B.R., Cobbs S.L., and Tumlin J.A.. 2007. Loss of the alpha-isoform of calcineurin is sufficient to induce nephrotoxicity and altered expression of transforming growth factor-beta. Transplantation. 83:439–447. 10.1097/01.tp.0000251423.78124.51 [DOI] [PubMed] [Google Scholar]
- Goodlad, J.R., and Macartney J.C.. 1995. Regulation of murine germinal centre cell proliferation in vivo: A stathmokinetic study examining the effect of differently timed doses of cyclosporin A. J. Pathol. 176:87–97. 10.1002/path.1711760113 [DOI] [PubMed] [Google Scholar]
- Ham, J., Babij C., Whitfield J., Pfarr C.M., Lallemand D., Yaniv M., and Rubin L.L.. 1995. A c-Jun dominant negative mutant protects sympathetic neurons against programmed cell death. Neuron. 14:927–939. 10.1016/0896-6273(95)90331-3 [DOI] [PubMed] [Google Scholar]
- Hatzi, K., Nance J.P., Kroenke M.A., Bothwell M., Haddad E.K., Melnick A., and Crotty S.. 2015. BCL6 orchestrates Tfh cell differentiation via multiple distinct mechanisms. J. Exp. Med. 212:539–553. 10.1084/jem.20141380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Havenar-Daughton, C., Reiss S.M., Carnathan D.G., Wu J.E., Kendric K., Torrents de la Peña A., Kasturi S.P., Dan J.M., Bothwell M., Sanders R.W., et al. 2016. Cytokine-independent detection of antigen-specific germinal center T follicular helper cells in immunized nonhuman primates using a live cell activation-induced marker technique. J. Immunol. 197:994–1002. 10.4049/jimmunol.1600320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haynes, N.M., Allen C.D.C., Lesley R., Ansel K.M., Killeen N., and Cyster J.G.. 2007. Role of CXCR5 and CCR7 in follicular Th cell positioning and appearance of a programmed cell death gene-1high germinal center-associated subpopulation. J. Immunol. 179:5099–5108. 10.4049/jimmunol.179.8.5099 [DOI] [PubMed] [Google Scholar]
- Herber, D., Brown T.P., Liang S., Young D.A., Collins M., and Dunussi-Joannopoulos K.. 2007. IL-21 has a pathogenic role in a lupus-prone mouse model and its blockade with IL-21R.Fc reduces disease progression. J. Immunol. 178:3822–3830. 10.4049/jimmunol.178.6.3822 [DOI] [PubMed] [Google Scholar]
- Hogan, P.G. 2017. Calcium-NFAT transcriptional signalling in T cell activation and T cell exhaustion. Cell Calcium. 63:66–69. 10.1016/j.ceca.2017.01.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogan, P.G., Chen L., Nardone J., and Rao A.. 2003. Transcriptional regulation by calcium, calcineurin, and NFAT. Genes Dev. 17:2205–2232. 10.1101/gad.1102703 [DOI] [PubMed] [Google Scholar]
- Hui, E., Cheung J., Zhu J., Su X., Taylor M.J., Wallweber H.A., Sasmal D.K., Huang J., Kim J.M., Mellman I., et al. 2017. T cell costimulatory receptor CD28 is a primary target for PD-1-mediated inhibition. Science. 355:1428–1433. 10.1126/science.aaf1292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyer, S.S., Latner D.R., Zilliox M.J., McCausland M., Akondy R.S., Penaloza-Macmaster P., Hale J.S., Ye L., Mohammed A.U.R., Yamaguchi T., et al. 2013. Identification of novel markers for mouse CD4(+) T follicular helper cells. Eur. J. Immunol. 43:3219–3232. 10.1002/eji.201343469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain, J., McCaffrey P.G., Miner Z., Kerppola T.K., Lambert J.N., Verdine G.L., Curran T., and Rao A.. 1993. The T-cell transcription factor NFATp is a substrate for calcineurin and interacts with Fos and Jun. Nature. 365:352–355. 10.1038/365352a0 [DOI] [PubMed] [Google Scholar]
- Johnston, R.J., Poholek A.C., DiToro D., Yusuf I., Eto D., Barnett B., Dent A.L., Craft J., and Crotty S.. 2009. Bcl6 and Blimp-1 are reciprocal and antagonistic regulators of T follicular helper cell differentiation. Science. 325:1006–1010. 10.1126/science.1175870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kannegieter, N.M., Hesselink D.A., Dieterich M., de Graav G.N., Kraaijeveld R., and Baan C.C.. 2018. Analysis of NFATc1 amplification in T cells for pharmacodynamic monitoring of tacrolimus in kidney transplant recipients. PLoS One. 13:e0201113. 10.1371/journal.pone.0201113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerfoot, S.M., Yaari G., Patel J.R., Johnson K.L., Gonzalez D.G., Kleinstein S.H., and Haberman A.M.. 2011. Germinal center B cell and T follicular helper cell development initiates in the interfollicular zone. Immunity. 34:947–960. 10.1016/j.immuni.2011.03.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan, O., Giles J.R., McDonald S., Manne S., Ngiow S.F., Patel K.P., Werner M.T., Huang A.C., Alexander K.A., Wu J.E., et al. 2019. TOX transcriptionally and epigenetically programs CD8+ T cell exhaustion. Nature. 571:211–218. 10.1038/s41586-019-1325-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, H.P., Korn L.L., Gamero A.M., and Leonard W.J.. 2005. Calcium-dependent activation of interleukin-21 gene expression in T cells. J. Biol. Chem. 280:25291–25297. 10.1074/jbc.M501459200 [DOI] [PubMed] [Google Scholar]
- Klein-Hessling, S., Muhammad K., Klein M., Pusch T., Rudolf R., Flöter J., Qureischi M., Beilhack A., Vaeth M., Kummerow C., et al. 2017. NFATc1 controls the cytotoxicity of CD8+ T cells. Nat. Commun. 8:511. 10.1038/s41467-017-00612-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, J.U., Kim L.K., and Choi J.M.. 2018. Revisiting the concept of targeting NFAT to control T cell immunity and autoimmune diseases. Front. Immunol. 9:2747. 10.3389/fimmu.2018.02747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legrand, J.J., Bouchez C., Mimouni C., N’Guyen A., Bouchard J., Ameller T., and Descotes J.. 2013. Immunotoxic effects of cyclophosphamide and cyclosporine in the dog. J. Immunotoxicol. 10:90–95. 10.3109/1547691X.2012.723766 [DOI] [PubMed] [Google Scholar]
- Lewis, S.M., Williams A., and Eisenbarth S.C.. 2019. Structure and function of the immune system in the spleen. Sci. Immunol. 4:eaau6085. 10.1126/sciimmunol.aau6085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linterman, M.A., Rigby R.J., Wong R.K., Yu D., Brink R., Cannons J.L., Schwartzberg P.L., Cook M.C., Walters G.D., and Vinuesa C.G.. 2009. Follicular helper T cells are required for systemic autoimmunity. J. Exp. Med. 206:561–576. 10.1084/jem.20081886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linterman, M.A., Denton A.E., Divekar D.P., Zvetkova I., Kane L., Ferreira C., Veldhoen M., Clare S., Dougan G., Espéli M., and Smith K.G.C.. 2014. CD28 expression is required after T cell priming for helper T cell responses and protective immunity to infection. Elife. 3:e03180. 10.7554/eLife.03180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, D., Xu H., Shih C., Wan Z., Ma X., Ma W., Luo D., and Qi H.. 2015. T-B-cell entanglement and ICOSL-driven feed-forward regulation of germinal centre reaction. Nature. 517:214–218. 10.1038/nature13803 [DOI] [PubMed] [Google Scholar]
- Macián, F. 2005. NFAT proteins: Key regulators of T-cell development and function. Nat. Rev. Immunol. 5:472–484. 10.1038/nri1632 [DOI] [PubMed] [Google Scholar]
- Macián, F., García-Rodríguez C., and Rao A.. 2000. Gene expression elicited by NFAT in the presence or absence of cooperative recruitment of Fos and Jun. EMBO J. 19:4783–4795. 10.1093/emboj/19.17.4783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macián, F., López-Rodríguez C., and Rao A.. 2001. Partners in transcription: NFAT and AP-1. Oncogene. 20:2476–2489. 10.1038/sj.onc.1204386 [DOI] [PubMed] [Google Scholar]
- Macián, F., García-Cózar F., Im S.H., Horton H.F., Byrne M.C., and Rao A.. 2002. Transcriptional mechanisms underlying lymphocyte tolerance. Cell. 109:719–731. 10.1016/S0092-8674(02)00767-5 [DOI] [PubMed] [Google Scholar]
- Märklin, M., Heitmann J.S., Fuchs A.R., Truckenmüller F.M., Gutknecht M., Bugl S., Saur S.J., Lazarus J., Kohlhofer U., Quintanilla-Martinez L., et al. 2017. NFAT2 is a critical regulator of the anergic phenotype in chronic lymphocytic leukaemia. Nat. Commun. 8:755. 10.1038/s41467-017-00830-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall, H.D., Chandele A., Jung Y.W., Meng H., Poholek A.C., Parish I.A., Rutishauser R., Cui W., Kleinstein S.H., Craft J., and Kaech S.M.. 2011. Differential expression of Ly6C and T-bet distinguish effector and memory Th1 CD4(+) cell properties during viral infection. Immunity. 35:633–646. 10.1016/j.immuni.2011.08.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall, H.D., Ray J.P., Laidlaw B.J., Zhang N., Gawande D., Staron M.M., Craft J., and Kaech S.M.. 2015. The transforming growth factor beta signaling pathway is critical for the formation of CD4 T follicular helper cells and isotype-switched antibody responses in the lung mucosa. Elife. 4:e04851. 10.7554/eLife.04851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez, G.J., Pereira R.M., Äijö T., Kim E.Y., Marangoni F., Pipkin M.E., Togher S., Heissmeyer V., Zhang Y.C., Crotty S., et al. 2015. The transcription factor NFAT promotes exhaustion of activated CD8⁺ T cells. Immunity. 42:265–278. 10.1016/j.immuni.2015.01.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez, G.J., Hu J.K., Pereira R.M., Crampton J.S., Togher S., Bild N., Crotty S., and Rao A.. 2016. Cutting edge: NFAT transcription factors promote the generation of follicular helper T cells in response to acute viral infection. J. Immunol. 196:2015–2019. 10.4049/jimmunol.1501841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matz, M.V., Fradkov A.F., Labas Y.A., Savitsky A.P., Zaraisky A.G., Markelov M.L., and Lukyanov S.A.. 1999. Fluorescent proteins from nonbioluminescent Anthozoa species. Nat. Biotechnol. 17:969–973. 10.1038/13657 [DOI] [PubMed] [Google Scholar]
- McAdam, A.J., Chang T.T., Lumelsky A.E., Greenfield E.A., Boussiotis V.A., Duke-Cohan J.S., Chernova T., Malenkovich N., Jabs C., Kuchroo V.K., et al. 2000. Mouse inducible costimulatory molecule (ICOS) expression is enhanced by CD28 costimulation and regulates differentiation of CD4+ T cells. J. Immunol. 165:5035–5040. 10.4049/jimmunol.165.9.5035 [DOI] [PubMed] [Google Scholar]
- McCaffrey, P.G., Luo C., Kerppola T.K., Jain J., Badalian T.M., Ho A.M., Burgeon E., Lane W.S., Lambert J.N., Curran T., et al. 1993. Isolation of the cyclosporin-sensitive T cell transcription factor NFATp. Science. 262:750–754. 10.1126/science.8235597 [DOI] [PubMed] [Google Scholar]
- McPhee, C.G., Bubier J.A., Sproule T.J., Park G., Steinbuck M.P., Schott W.H., Christianson G.J., Morse H.C. III, and Roopenian D.C.. 2013. IL-21 is a double-edged sword in the systemic lupus erythematosus-like disease of BXSB.Yaa mice. J. Immunol. 191:4581–4588. 10.4049/jimmunol.1300439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medzhitov, R., Schneider D.S., and Soares M.P.. 2012. Disease tolerance as a defense strategy. Science. 335:936–941. 10.1126/science.1214935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minami, T., Jiang S., Schadler K., Suehiro J.I., Osawa T., Oike Y., Miura M., Naito M., Kodama T., and Ryeom S.. 2013. The calcineurin-NFAT-angiopoietin-2 signaling axis in lung endothelium is critical for the establishment of lung metastases. Cell Rep. 4:709–723. 10.1016/j.celrep.2013.07.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monticelli, S., and Rao A.. 2002. NFAT1 and NFAT2 are positive regulators of IL-4 gene transcription. Eur. J. Immunol. 32:2971–2978. [DOI] [PubMed] [Google Scholar]
- Morel, L., Croker B.P., Blenman K.R., Mohan C., Huang G., Gilkeson G., and Wakeland E.K.. 2000. Genetic reconstitution of systemic lupus erythematosus immunopathology with polycongenic murine strains. Proc. Natl. Acad. Sci. USA. 97:6670–6675. 10.1073/pnas.97.12.6670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moriyama, A., Maeda H., Hirai T., and Yamaguchi R.. 2012. Pathological effects in lymphoid tissues of the spleen, lymph nodes, and Peyer’s patches in cyclosporin-treated cynomolgus monkeys. J. Vet. Med. Sci. 74:1487–1491. 10.1292/jvms.12-0155 [DOI] [PubMed] [Google Scholar]
- Murphy, E.D., and Roths J.B.. 1979. A Y chromosome associated factor in strain BXSB producing accelerated autoimmunity and lymphoproliferation. Arthritis Rheum. 22:1188–1194. 10.1002/art.1780221105 [DOI] [PubMed] [Google Scholar]
- Naviaux, R.K., Costanzi E., Haas M., and Verma I.M.. 1996. The pCL vector system: Rapid production of helper-free, high-titer, recombinant retroviruses. J. Virol. 70:5701–5705. 10.1128/jvi.70.8.5701-5705.1996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nurieva, R.I., Chung Y., Martinez G.J., Yang X.O., Tanaka S., Matskevitch T.D., Wang Y.H., and Dong C.. 2009. Bcl6 mediates the development of T follicular helper cells. Science. 325:1001–1005. 10.1126/science.1176676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nurieva, R.I., Liu X., and Dong C.. 2011. Molecular mechanisms of T-cell tolerance. Immunol. Rev. 241:133–144. 10.1111/j.1600-065X.2011.01012.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odegard, J.M., Marks B.R., DiPlacido L.D., Poholek A.C., Kono D.H., Dong C., Flavell R.A., and Craft J.. 2008. ICOS-dependent extrafollicular helper T cells elicit IgG production via IL-21 in systemic autoimmunity. J. Exp. Med. 205:2873–2886. 10.1084/jem.20080840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oxenius, A., Bachmann M.F., Zinkernagel R.M., and Hengartner H.. 1998. Virus-specific MHC-class II-restricted TCR-transgenic mice: Effects on humoral and cellular immune responses after viral infection. Eur. J. Immunol. 28:390–400. [DOI] [PubMed] [Google Scholar]
- Park, Y.J., Yoo S.A., Kim M., and Kim W.U.. 2020. The role of calcium-calcineurin-NFAT signaling pathway in health and autoimmune diseases. Front. Immunol. 11:195. 10.3389/fimmu.2020.00195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pear, W.S., Miller J.P., Xu L., Pui J.C., Soffer B., Quackenbush R.C., Pendergast A.M., Bronson R., Aster J.C., Scott M.L., and Baltimore D.. 1998. Efficient and rapid induction of a chronic myelogenous leukemia-like myeloproliferative disease in mice receiving P210 bcr/abl-transduced bone marrow. Blood. 92:3780–3792. 10.1182/blood.V92.10.3780 [DOI] [PubMed] [Google Scholar]
- Pedros, C., Zhang Y., Hu J.K., Choi Y.S., Canonigo-Balancio A.J., Yates J.R. III, Altman A., Crotty S., and Kong K.F.. 2016. A TRAF-like motif of the inducible costimulator ICOS controls development of germinal center TFH cells via the kinase TBK1. Nat. Immunol. 17:825–833. 10.1038/ni.3463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira, R.M., Hogan P.G., Rao A., and Martinez G.J.. 2017. Transcriptional and epigenetic regulation of T cell hyporesponsiveness. J. Leukoc. Biol. 102:601–615. 10.1189/jlb.2RI0317-097R [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson, B.R., Sun L.J., and Verdine G.L.. 1996. A critical arginine residue mediates cooperativity in the contact interface between transcription factors NFAT and AP-1. Proc. Natl. Acad. Sci. USA. 93:13671–13676. 10.1073/pnas.93.24.13671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrak, D., Memon S.A., Birrer M.J., Ashwell J.D., and Zacharchuk C.M.. 1994. Dominant negative mutant of c-Jun inhibits NF-AT transcriptional activity and prevents IL-2 gene transcription. J. Immunol. 153:2046–2051. 10.4049/jimmunol.153.5.2046 [DOI] [PubMed] [Google Scholar]
- Pisitkun, P., Deane J.A., Difilippantonio M.J., Tarasenko T., Satterthwaite A.B., and Bolland S.. 2006. Autoreactive B cell responses to RNA-related antigens due to TLR7 gene duplication. Science. 312:1669–1672. 10.1126/science.1124978 [DOI] [PubMed] [Google Scholar]
- Poholek, A.C., Hansen K., Hernandez S.G., Eto D., Chandele A., Weinstein J.S., Dong X., Odegard J.M., Kaech S.M., Dent A.L., et al. 2010. In vivo regulation of Bcl6 and T follicular helper cell development. J. Immunol. 185:313–326. 10.4049/jimmunol.0904023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pratama, A., and Vinuesa C.G.. 2014. Control of TFH cell numbers: Why and how? Immunol. Cell Biol. 92:40–48. 10.1038/icb.2013.69 [DOI] [PubMed] [Google Scholar]
- Qi, H. 2016. T follicular helper cells in space-time. Nat. Rev. Immunol. 16:612–625. 10.1038/nri.2016.94 [DOI] [PubMed] [Google Scholar]
- Rankin, A.L., Guay H., Herber D., Bertino S.A., Duzanski T.A., Carrier Y., Keegan S., Senices M., Stedman N., Ryan M., et al. 2012. IL-21 receptor is required for the systemic accumulation of activated B and T lymphocytes in MRL/MpJ-Fas(lpr/lpr)/J mice. J. Immunol. 188:1656–1667. 10.4049/jimmunol.1003871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao, A., Luo C., and Hogan P.G.. 1997. Transcription factors of the NFAT family: Regulation and function. Annu. Rev. Immunol. 15:707–747. 10.1146/annurev.immunol.15.1.707 [DOI] [PubMed] [Google Scholar]
- Ray, J.P., Staron M.M., Shyer J.A., Ho P.C., Marshall H.D., Gray S.M., Laidlaw B.J., Araki K., Ahmed R., Kaech S.M., and Craft J.. 2015. The interleukin-2-mTORc1 kinase Axis defines the signaling, differentiation, and metabolism of T helper 1 and follicular B helper T cells. Immunity. 43:690–702. 10.1016/j.immuni.2015.08.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reich, M., Liefeld T., Gould J., Lerner J., Tamayo P., and Mesirov J.P.. 2006. GenePattern 2.0. Nat. Genet. 38:500–501. 10.1038/ng0506-500 [DOI] [PubMed] [Google Scholar]
- Riemekasten, G., and Hahn B.H.. 2005. Key autoantigens in SLE. Rheumatology 44:975–982. 10.1093/rheumatology/keh688 [DOI] [PubMed] [Google Scholar]
- Safford, M., Collins S., Lutz M.A., Allen A., Huang C.T., Kowalski J., Blackford A., Horton M.R., Drake C., Schwartz R.H., and Powell J.D.. 2005. Egr-2 and Egr-3 are negative regulators of T cell activation. Nat. Immunol. 6:472–480. 10.1038/ni1193 [DOI] [PubMed] [Google Scholar]
- Schietinger, A., and Greenberg P.D.. 2014. Tolerance and exhaustion: Defining mechanisms of T cell dysfunction. Trends Immunol. 35:51–60. 10.1016/j.it.2013.10.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz, R.H. 2003. T cell anergy. Annu. Rev. Immunol. 21:305–334. 10.1146/annurev.immunol.21.120601.141110 [DOI] [PubMed] [Google Scholar]
- Scott, A.C., Dündar F., Zumbo P., Chandran S.S., Klebanoff C.A., Shakiba M., Trivedi P., Menocal L., Appleby H., Camara S., et al. 2019. TOX is a critical regulator of tumour-specific T cell differentiation. Nature. 571:270–274. 10.1038/s41586-019-1324-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo, H., Chen J., González-Avalos E., Samaniego-Castruita D., Das A., Wang Y.H., López-Moyado I.F., Georges R.O., Zhang W., Onodera A., et al. 2019. TOX and TOX2 transcription factors cooperate with NR4A transcription factors to impose CD8+ T cell exhaustion. Proc. Natl. Acad. Sci. USA. 116:12410–12415. 10.1073/pnas.1905675116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serfling, E., Chuvpilo S., Liu J., Höfer T., and Palmetshofer A.. 2006. NFATc1 autoregulation: A crucial step for cell-fate determination. Trends Immunol. 27:461–469. 10.1016/j.it.2006.08.005 [DOI] [PubMed] [Google Scholar]
- Seth, A., and Craft J.. 2019. Spatial and functional heterogeneity of follicular helper T cells in autoimmunity. Curr. Opin. Immunol. 61:1–9. 10.1016/j.coi.2019.06.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin, H., and Wherry E.J.. 2007. CD8 T cell dysfunction during chronic viral infection. Curr. Opin. Immunol. 19:408–415. 10.1016/j.coi.2007.06.004 [DOI] [PubMed] [Google Scholar]
- Song, W., and Craft J.. 2019. T follicular helper cell heterogeneity: Time, space, and function. Immunol. Rev. 288:85–96. 10.1111/imr.12740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spurney, R.F. 2014. Non-immunologic actions of calcineurin inhibitors in proteinuric kidney diseases. Front. Endocrinol. 5:181. 10.3389/fendo.2014.00181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinivasan, M., and Frauwirth K.A.. 2007. Reciprocal NFAT1 and NFAT2 nuclear localization in CD8+ anergic T cells is regulated by suboptimal calcium signaling. J. Immunol. 179:3734–3741. 10.4049/jimmunol.179.6.3734 [DOI] [PubMed] [Google Scholar]
- Staupe, R.P., Vella L.A., Manne S., Giles J.R., Meng W., Herati R.S., Khan O., Wu J.E., Baxter A.E., Luning Prak E.T., et al. 2019. Chronic viral infection promotes early germinal center exit of B cells and impaired antibody development. bioRxiv. (Preprint posted November 21, 2019). 10.1101/849844 [DOI] [Google Scholar]
- Subramanian, A., Tamayo P., Mootha V.K., Mukherjee S., Ebert B.L., Gillette M.A., Paulovich A., Pomeroy S.L., Golub T.R., Lander E.S., and Mesirov J.P.. 2005. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA. 102:15545–15550. 10.1073/pnas.0506580102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian, S., Tus K., Li Q.Z., Wang A., Tian X.H., Zhou J., Liang C., Bartov G., McDaniel L.D., Zhou X.J., et al. 2006. A Tlr7 translocation accelerates systemic autoimmunity in murine lupus. Proc. Natl. Acad. Sci. USA. 103:9970–9975. 10.1073/pnas.0603912103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tunyaplin, C., Shaffer A.L., Angelin-Duclos C.D., Yu X., Staudt L.M., and Calame K.L.. 2004. Direct repression of prdm1 by Bcl-6 inhibits plasmacytic differentiation. J. Immunol. 173:1158–1165. 10.4049/jimmunol.173.2.1158 [DOI] [PubMed] [Google Scholar]
- Ume, A.C., Wenegieme T.-Y., and Williams C.R.. 2021. Calcineurin inhibitors: A double-edged sword. Am. J. Physiol. Renal. Physiol. 320:F336–F341. 10.1152/ajprenal.00262.2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasanwala, F.H., Kusam S., Toney L.M., and Dent A.L.. 2002. Repression of AP-1 function: A mechanism for the regulation of blimp-1 expression and B lymphocyte differentiation by the B cell lymphoma-6 protooncogene. J. Immunol. 169:1922–1929. 10.4049/jimmunol.169.4.1922 [DOI] [PubMed] [Google Scholar]
- Vinuesa, C.G., Tangye S.G., Moser B., and Mackay C.R.. 2005. Follicular B helper T cells in antibody responses and autoimmunity. Nat. Rev. Immunol. 5:853–865. 10.1038/nri1714 [DOI] [PubMed] [Google Scholar]
- Webb, L.M.C., and Linterman M.A.. 2017. Signals that drive T follicular helper cell formation. Immunology. 152:185–194. 10.1111/imm.12778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber, J.P., Fuhrmann F., Feist R.K., Lahmann A., Al Baz M.S., Gentz L.J., Vu Van D., Mages H.W., Haftmann C., Riedel R., et al. 2015. ICOS maintains the T follicular helper cell phenotype by down-regulating Krüppel-like factor 2. J. Exp Med. 212:217–233. 10.1084/jem.20141432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinstein, J.S., Bertino S.A., Hernandez S.G., Poholek A.C., Teplitzky T.B., Nowyhed H.N., and Craft J.. 2014. B cells in T follicular helper cell development and function: Separable roles in delivery of ICOS ligand and antigen. J. Immunol. 192:3166–3179. 10.4049/jimmunol.1302617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinstein, J.S., Herman E.I., Lainez B., Licona-Limón P., Esplugues E., Flavell R., and Craft J.. 2016. TFH cells progressively differentiate to regulate the germinal center response. Nat. Immunol. 17:1197–1205. 10.1038/ni.3554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wherry, E.J. 2011. T cell exhaustion. Nat. Immunol. 12:492–499. 10.1038/ni.2035 [DOI] [PubMed] [Google Scholar]
- Wherry, E.J., Ha S.J., Kaech S.M., Haining W.N., Sarkar S., Kalia V., Subramaniam S., Blattman J.N., Barber D.L., and Ahmed R.. 2007. Molecular signature of CD8+ T cell exhaustion during chronic viral infection. Immunity 27:824. 10.1016/j.immuni.2007.09.006 [DOI] [PubMed] [Google Scholar]
- Wikenheiser, D.J., and Stumhofer J.S.. 2016. ICOS Co-Stimulation. Front. Immunol. 7:304. 10.3389/fimmu.2016.00304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, T., Keller A., and Martinez G.J.. 2019a. NFAT1 and NFAT2 differentially regulate CTL differentiation upon acute viral infection. Front. Immunol. 10:184. 10.3389/fimmu.2019.00184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, W., Zhao X., Wang X., Feng H., Gou M., Jin W., Wang X., Liu X., and Dong C.. 2019b. The transcription factor Tox2 drives T follicular helper cell development via regulating chromatin accessibility. Immunity. 51:826–839.e5. 10.1016/j.immuni.2019.10.006 [DOI] [PubMed] [Google Scholar]
- Yan, L., de Leur K., Hendriks R.W., van der Laan L.J.W., Shi Y., Wang L., and Baan C.C.. 2017. T follicular helper cells as a new target for immunosuppressive therapies. Front. Immunol. 8:1510. 10.3389/fimmu.2017.01510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin, Y., Choi S.C., Xu Z., Perry D.J., Seay H., Croker B.P., Sobel E.S., Brusko T.M., and Morel L.. 2015. Normalization of CD4+ T cell metabolism reverses lupus. Sci. Transl. Med. 7:274ra18. 10.1126/scitranslmed.aaa0835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, D., Rao S., Tsai L.M., Lee S.K., He Y., Sutcliffe E.L., Srivastava M., Linterman M., Zheng L., Simpson N., et al. 2009. The transcriptional repressor Bcl-6 directs T follicular helper cell lineage commitment. Immunity. 31:457–468. 10.1016/j.immuni.2009.07.002 [DOI] [PubMed] [Google Scholar]
- Yusuf, I., Kageyama R., Monticelli L., Johnston R.J., Ditoro D., Hansen K., Barnett B., and Crotty S.. 2010. Germinal center T follicular helper cell IL-4 production is dependent on signaling lymphocytic activation molecule receptor (CD150). J. Immunol. 185:190–202. 10.4049/jimmunol.0903505 [DOI] [PMC free article] [PubMed] [Google Scholar]