Abstract
Kombucha beverage produced through fermentation of sugared tea using bacteria and yeast has gained attention for its beneficial health benefits. However, the cost linked to the raw materials often increases the upstream process expenses, thereby the overall operating expenditures. Thus, there is a need to explore alternative waste and cost-effective raw materials for Kombucha fermentation. The present study, compared the physico-chemical and microbial growth pattern of Kombucha beverage production using tea waste from the tea processing industries with that of the green/black tea, reporting similar trends irrespective of its type. Further, the amplicon sequencing of 16S rRNA showed dominant presence of Komagataeibacter rhaeticus and high throughput sequencing of ITS1 confirmed the presence of yeast species similar to Brettanomyces bruxellensis in the tea waste based Kombucha beverage. Appreciable amount of carbohydrates (8.5/100 g) and energy (34 kcal/100 g) with appropriate organoleptic properties favourable for human consumption were also observed during the nutritional content and qualitative property assessment. The overall study showed a broad taxonomic and functional diversity existing during Kombucha fermentation process with tea waste to maintain a sustained eco-system to facilitate cost-effective beverage production with desired properties for safe consumption.
Supplementary Information
The online version contains supplementary material available at 10.1007/s13197-022-05476-3.
Keywords: Fermentation, Kombucha, Metagenomics, Microbial diversity, Probiotic beverages, Physicochemical analysis
Introduction
Microorganisms, since time-immemorial have played a pivotal role in nature as they produce various secondary metabolites that have numerous social implications. Various food and beverages produced are supposedly being fermented by these microorganisms and are of immense beneficial for the mankind (Ray et al. 2014).
Kombucha is a traditional beverage obtained from the fermentation of black or green tea (Camellia sinensis) sweetened with sugar by a symbiotic microbial consortium, which is mainly composed of acetic acid bacteria (AAB) (Gluconacetobacter sp.) and osmophilic yeasts (Zygosaccharomyces sp.) (El Sheikha et al. 2018). The Kombucha tea is a socially accepted beverage for human consumption (El Sheikha and Hu 2020) and has been proposed to pose remarkable health benefits owing to the diverse metabolic content produced by the variability in substrate composition and different operational processing conditions (Laavanya et al. 2021). Kombucha tea production involves inoculation of the culture medium with 5–20% of the previously brewed tea having the probiotic bacterial and yeast communities in an active state to disperse homogenously and propagate the fermentation process. Often the growth parameters and the chemical constituents in the medium determines the microbiome ecology (Villarreal-Soto et al. 2020).
Several researchers (Marsh et al. 2014; Coton et al. 2017) have analysed the microbiome proposing the presence of Komagataeibacter sp., Acetobacter sp., and Gluconobacter sp., as the dominant bacterial genera and Brettanomyces sp., and Zygosaccharomyces sp. as the dominant fungal genera. The symbiotic metabolism of these microbes helps in the organoleptic properties, enhancing the taste of the beverage and decides the yield of cellulosic biofilm (Villarreal-Soto et al. 2020). The yeast metabolizes the complex sugar (sucrose) into glucose and fructose, which thereby is metabolized into ethanol (Laavanya et al. 2021). The glucose molecules are thereby metabolized into organic acids like acetic, gallic, succinic, gluconic and glucuronic acid by the glucose oxidase enzyme activity of AAB. Thereafter, Komagataeibacter sp., and Gluconobacter sp., utilizes cellulose synthase enzyme during oxidative fermentation producing extracellularly a cellulosic biofilm. Autolysis of yeast also has been proposed to provide vitamins that promotes the growth of acetic acid bacteria, thereby the cellulosic biofilms (Chakravorty et al. 2016). The interaction between the microbes in the culture medium determines the quantitative and qualitative properties of the products formed. Therefore, it is essential to understand and study the microbiome diversity during the variable conditions of Kombucha fermentation process in-order to obtain the desirable properties.
Most of the microbiological studies related to identification of the bacterial and yeast genera are culture based, mostly relied on the variation of the phenotypic traits (Coton et al. 2017). These methods often have a low throughput and leads to misinterpretation of the strains. Marsh et al. (2014) conducted the first high-throughput sequencing analysis reporting the presence of Gluconobacter sp., in > 85% of the samples and Zygosaccharomyces sp., as the dominant yeast in > 95% of the samples. Chakravorty et al. (2016) utilized terminal restriction fragment length polymorphism and high throughput sequencing reporting the presence of Komagataeibacter and Candida sp., as the dominant bacteria and yeast respectively. A recent study by Villarreal-Soto et al. (2020) utilized short-gun metagenomic sequencing revealing the presence of the bacterial and fungal genera and their direct linkage with a number of bioactive metabolites. Most of the above-mentioned studies on genome analysis for Kombucha beverage has been done utilizing the commercial black tea/green tea as the nitrogenous source. Since, the commercial scale production of beverage is influenced by the raw material costs (Behera et al. 2022), it is essential to explore the cost-effective substrates without much influence on the underlying microbial ecology.
Thus, the present study utilized tea leaves discarded as waste from the tea processing industry as the nitrogen source during the Kombucha beverage production. The biochemical composition and the microbial load during the fermentation with waste tea leaves has been compared with that of the black and green tea leaves essentially utilized as nitrogenous substrate during the commercial production of Kombucha beverage. The 16S rRNA sequencing and high throughput internal transcribed sequencing (ITS) was utilized to analyse the bacterial and yeast diversity in the Kombucha beverage produced from tea waste. The physicochemical and organoleptic properties were also evaluated to confirm its suitability for human consumption. The study will facilitate the use of waste and cost-efficient resources for Kombucha beverage production, thereby bringing down the production costs during commercialization.
Methodology
Kombucha culture and maintenance
The Kombucha beverage was produced via the process of fermentation with the tea (C. sinensis) discarded as waste from the tea processing industry (Parry Agro, Tamil Nadu, India) and sucrose (common sugar) as substrate. Fermentation was carried out over a period of 14 days with 8% w/v sucrose, 1.2% w/v tea, along with 10% v/v of previously fermented Kombucha tea and 5% w/v of inoculum/symbiotic culture of bacteria and yeast [SCOBY] at 30 ± 5 °C. For comparative purpose, fermentation under the similar process conditions were also carried with black tea and green tea. Samples were collected from the day of inoculation till the 14th day at an interval of 2 days for analysing the pH and glucose via spectrophotometric determination using dinitrosalicylic acid assay (Miller 1959). Sucrose (°Brix) and potential alcohol (%v/v) were also estimated using refractometer (Hanna Instruments, Model HI96813). The radical scavenging activity of Kombucha beverage samples were measured using 1,1-Diphenyl-2-picrylhydrazyl (DPPH) assay (Soni 2014). The growth pattern of bacteria and yeast during fermentation with different tea substrates were determined by inoculating the samples drawn at specific time-intervals in Glucose chalk yeast extract broth and potato dextrose broth respectively. For metagenomics study, Kombucha beverage samples prepared from the waste tea leaves were centrifuged after the completion of 8 days of fermentation and the pellets obtained was stored in − 20 °C until further analysis.
Analysis of microbiome in Kombucha beverage
DNA isolation from Kombucha culture
Deoxyribonucleic acid (DNA) was extracted using the PSP Spin DNA Plus Kit (Invitek Inc.) as per the given standard protocol. Using 1 μl DNA sample, the isolated metagenomic DNA was tested for purity by A260/280 ratio using a NanoDrop (Eppendorf, BioPhotometer). Qubit was used to assess the DNA concentration (ThermoFisher Scientific). DNA was thereafter resolved on a 0.8% agarose gel at 80 V for around 30 min to assess the purity and integrity of the DNA.
16S rRNA sequencing of bacterial isolates
The 16S rRNA amplicon data was generated using Oxford Nanopore Technologies MinION platform's long read sequencing method (Jain et al. 2016). Universal primer sets 27F (5′-AGAGTTTGATCCTGGCTCAG-3′) and 1492R (5′-GGTTACCTTGTTACGACTT-3′) were utilised to produce 16S rRNA gene amplicons from quality control (QC) passed genomic DNA (gDNA). Thermal cycling was initiated with a 3 min denaturation step at 94 °C, followed by 25 cycles of denaturation at 94 °C for 30 s, primer annealing at 55 °C for 30 s, and elongation at 72 °C for 1 min. Cycling was finished with a 10 min elongation step at 72 °C. The PCR amplicons were bead filtered to eliminate the polymerase chain reaction (PCR) chemicals and other contaminants before being utilized for library preparation, which included end prep, barcode and adaptor ligation, according to the protocol for the native barcoding of amplicons as prescribed by the Oxford Nanopore technologies. The library was processed with the SQL-LSK 109 sequencing kit before being put onto the MinION Mk1B sequencer at the appropriate concentration. About 90,000–1,10,000 reads were generated after sequencing, which was subjected to quality control using the FastQC software and further bioinformatics analysis was carried out to taxonomically identify the bacterial genera.
High-throughput internal transcribed sequencing (ITS) of yeast isolates
The quality of DNA for yeast genera identification was assessed by running the sample in 1% agarose gel resulting in a single high molecular weight DNA band. PCR was used to amplify a fragment of the ITS region. When resolved on agarose, a single distinct PCR amplicon band of around 700 bp was seen. The ITS1 (5′-TCCGTAGGTGAACCTGCGG-3′) and ITS4 (5′-TCCTCCGCTTATTGATATGC-3′) primers were used as forward and reverse primers respectively for DNA sequencing reaction of PCR amplicon on an ABI 3730xl genetic analyzer using the BDT v3.1 cycle sequencing kit. Using aligner software, a consensus sequence of the PCR amplicon was constructed from the forward and reverse sequencing data.
Bioinformatic analysis of the bacterial and yeast microbiome
Bioinformatics analysis was performed to identify the bacterial and yeast genera utilizing a systematic procedure. In case of bacteria, after removing the adaptor and overrepresented sequences, the high-quality sequences were submitted to taxonomy categorization using the Kraken2 suite (Wood et al. 2019) with the SILVA database (Quast et al. 2013) as the target. For taxonomic annotation, the high-quality reads were blasted with the NCBI Reference sequence curated database using more than 97% identity and 90% query coverage.
In case of yeast, the ITS region sequence was utilized to perform Basic Local Alignment Search Tool (BLAST) against the National Centre for Biotechnology Information (NCBI) Genbank database. Alignment was carried out using nucleotide BLAST (BLASTN) suite with reference database as nucleotide collection (nr/nt). The blast program was chosen as megablast with scoring of 1/− 2 (match/mismatch) which is specific for obtaining highly similar sequences. The top 10 sequences chosen based on maximum identity score were aligned using Clustal W [a multiple alignment software programme] (Larkin et al. 2007). The distance matrix was obtained, and the phylogenetic tree was generated using MEGA 7 (Kumar et al. 2016).
Biochemical analysis of Kombucha beverage
The quality of water used for Kombucha beverage production using tea waste along with its nutrition profile comprising carbohydrate, protein, total fats, and energy content was measured by following the standard operating procedure (SOP) as outlined in the manual of methods for analysis of foods by the Food Safety and Standard Authority of India (Food Safety and Standard Authority of India (FSSAI) 2016). The physiochemical properties pH, acidity, total soluble solids along with the presence of metals like arsenic, copper, lead and zinc were analysed as per the FSSAI regulations using the standard protocols (Food Safety and Standard Authority of India (FSSAI) 2016).
Organoleptic study was conducted to understand the consumer acceptability of the fermented Kombucha and its flavored (apple, orange, kiwi, pomegranate and lemon) prototypes. Sensory evaluation is a scientific method for evaluating a product where research is in the form of preference or hedonic tests for untrained participants are utilized. Around 150 untrained people, consisting of mostly students and staff from the Department of Biotechnology and Medical Engineering, National Institute of Technology (NIT) Rourkela, participated in this sensory analysis study. The participants were asked to fill out a questionnaire that requested their basic demographic information (age, gender) as well as their familiarity with the beverage, along with a consent form. Consumer acceptance was determined using the 9-point hedonic scale and the food action rating scale test. For sensory evaluation, participants were asked to rate the prototype on a 9-point hedonic scale on attributes such as sweetness, aroma, flavor, tartness, mouthfeel and overall preference. The food action rating scale test measured the incidences of drinking using a 9-point scale. The data collected was then assessed to find consumer acceptability.
Results and discussion
Variation in microbial load during Kombucha fermentation with different tea substrates
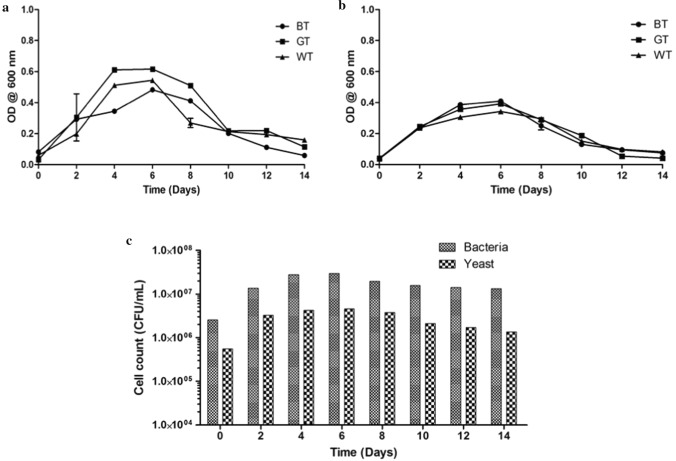
The growth rate of microorganisms depicts the progress of Kombucha fermentation and thereby the nutrient consumption and product yield. The growth rate of bacteria and yeasts sampled from Kombucha beverage cultured with different tea substrates over the period of 14 days are represented in Fig. 1a, b. For all the tea substrates, the bacteria and yeast growth were found to rapidly increase up to 6th day of fermentation, beyond which a decline was observed in the present study. Thus, it was confirmed that the tea waste provided the requisite biochemical components for the growth of desired bacteria and yeast. Thus, to further confirm the capacity of the waste tea leaves to support microbial growth, the plate count was carried out as shown in Fig. 1c. Maximum cell density of 4 × 106 CFU mL−1 and 3 × 107 CFU mL−1 was observed for yeast and bacteria respectively after the 6th day in case of tea waste based Kombucha beverage. Similar results were also obtained by Tran et al. (2022) reporting the maximum cell density of 6 × 106 CFU mL−1 and 1 × 107 CFU mL−1 for yeast and bacteria respectively in Kombucha beverage prepared from black tea. Comparable to the present study, the authors have also reported a decline in growth of microorganism after 7 days due to a decline in pH.
Fig. 1.
Optical density (OD) at 600 nm for a bacterial and b yeast cultures during Kombucha beverage fermentation c cell count of bacterial and yeast during tea waste based Kombucha beverage fermentation
Variation in biochemical constituents during Kombucha fermentation with different tea substrates
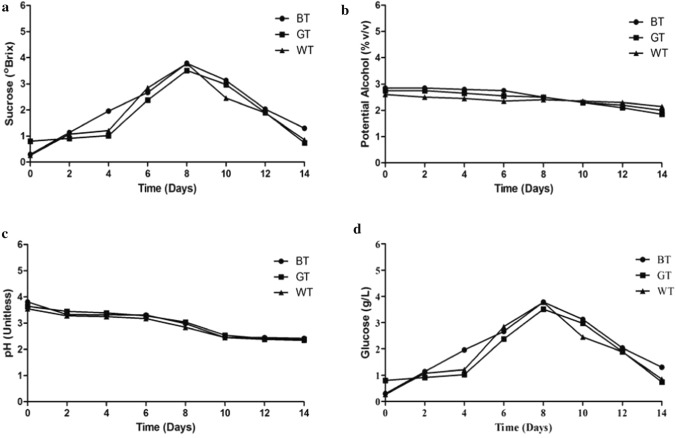
The nitrogenous compounds in tea influences the metabolic activity of the symbiotic culture of bacteria and yeast, which thereby affects the biochemical profile of Kombucha beverage. Figure 2a–d showed the variation in pH, sugar content, glucose concentration and potential alcohol in Kombucha beverage at an interval of two days. The initial sucrose content (apparent brix) and potential alcohol was found to be similar in case of all the tea substrates. Over a period of 14 days, the sucrose content (apparent brix) declined from 5° Brix, by almost 30% in all the cases (Fig. 2a). The potential alcohol content in all tea samples fairly remained constant and a slight decline was observed after 14 days (Fig. 2b). An increase in acid concentration with decline in pH value was also observed over the given time-period as evident in Fig. 2c. pH of the Kombucha beverage was found to decline from 4 gradually to 2, for all the tea substrates after a period of 14 days This could be corroborated to the reason that sucrose is metabolized by the yeast to produce glucose which is then used by the acetic acid bacteria producing cellulose and acids like glucuronic and gluconic acid etc. As evident in Fig. 2d, the glucose concentration was found to increase gradually until the 8th day reaching the maximum value of 3.5–3.8 g/L, beyond which it was found to decline for all the tea substrates. Gaggìa et al. (2019) reported a variation in glucose concentration of 8–11 g/L after 7 days of fermentation depending on the type of tea used i.e., black tea, green tea or rooibos tea. Since, the nitrogenous components and vitamins play an essential role in governing the metabolism of yeast, thereby its potential to breakdown sucrose into glucose and fructose, the concentration of glucose in Kombucha beverage might vary with the tea substrate. Kallel et al. (2012) also project a relative variation in the biochemical profile and the rate of glucose consumption with the changes in type of tea used during Kombucha fermentation. A decrease in pH of Kombucha beverage samples were also observed by Zubaidah et al. (2018) due to the metabolic conversion of glucose by Acetobacter into organic acids. pH also influences the sensory property of Kombucha beverage as increase in acidity might make the product too sour for consumption (Jayabalan et al. 2010). Thus, an incubation time of 8 days with a glucose content of 3.8 g/L with pH of 3 is good enough for consumption.
The radical scavenging activity of different Kombucha samples determined by DPPH assay are shown in Table 1. The DPPH radical scavenging activity was found to increase with time for all the tea substrates. Maximum radical scavenging activity of 56.27 ± 1.1% was obtained with green tea after a period of 14 days. Kombucha beverage produced with tea waste also showed a comparable antioxidant potential as that of the black and green tea with radical scavenging activity of 40.44 ± 4.38%. Degirmencioglu et al. (2020) have also studied the Kombucha fermentation with different tea substrate and found the green tea Kombucha had maximum antioxidant properties. Muhialdin et al. (2019) showed that the Kombucha culture exhibited good antioxidant properties throughout the fermentation period of 14 days (Fig. 2).
Table 1.
Variation in DPPH radical scavenging activity during Kombucha fermentation with variation in tea substrate
| Tea Substrate | Day 0 | Day 8 | Day 14 |
|---|---|---|---|
| Black Tea | 29.41 ± 0.095% | 35.52 ± 9.3% | 48.68 ± 2.13% |
| Green Tea | 46.35 ± 5.37% | 54.67 ± 2.22% | 56.27 ± 1.1% |
| Waste Tea | 32.03 ± 4.17% | 36.3 ± 6.8% | 40.44 ± 4.38% |
*Other conditions of fermentation comprising 8% w/v sucrose, 1.2% w/v tea, along with 10% v/v of previously fermented Kombucha tea and 5% w/v of inoculum/symbiotic culture of bacteria and yeast [SCOBY] at 30 ± 5 °C were kept constant during the entire period of study
Fig. 2.
a Sucrose content, b Potential alcohol, c pH, d Glucose Concentration during the fermentation of Kombucha beverage with different tea substrates
Metagenomic study of isolates of Kombucha beverage produced from tea waste
Microbial taxonomy of bacterial isolates
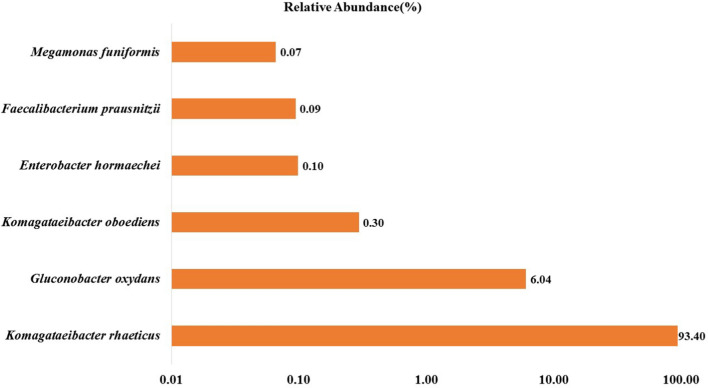
Sequencing was done to generate 90,000–1,10,000 reads per sample and were analysed for quality using FastQC. A total of 113,448 sequences of varying length between 360 and 1646 bp was obtained from sequencing. After the removal of primers and adapter sequences, the GC content was found to be 55%. rRNA gene-based results showed that there are two different bacterial phyla in the Kombucha beverage e.g. Firmicutes, and Proteobacteria. Among these phyla, Proteobacteria was dominant (> 99%) in the sample throughout the fermentation process. Amplicon analyses showed that 93.4% of the sequences assigned to the Komagataeibacter rhaeticus belonged to Proteobacteria phylum (Fig. 3).
Fig. 3.
Relative abundance (%) of different bacterial genera in Kombucha beverage produced from the tea waste
Komagataeibacter rhaeticus and Gluconobacter oxydans were observed to be the two main bacterial species present in the Kombucha beverage prepared from the tea waste in the present study. More specifically, at the species level, Komagataeibacter rhaeticus was the dominant bacteria (93%), followed by Gluconacetobacter sp. (6%) as evident from Fig. 3. Similar results were obtained by Villarreal et al. (2020) showing the presence of 80% of Acetobacteraceae, with the dominant genera being Komagataeibacter, Gluconocetobacter and Gluconobacter in Kombucha beverage prepared from black tea. Authors reported the presence of Komagataeibacter rhaeticus as the dominant bacteria followed by Gluconabacter sp. SXCCI at the species level. Gaggìa et al. (2019) projected that while beverage prepared from black tea and green tea had Komagataeibacter sp. in dominance, however, Gluconobacter sp., was found to be dominant in Kombucha beverage prepared from rooibos tea. The bacterial genera obtained is in agreement with the studies done by most authors (Marsh et al. 2014; Chakravorty et al. 2016) on Kombucha tea using black tea as substrate. Thus, it could be projected that tea waste could act as cheaper substrate to produce beverage with microbiome and quality similar to the commercially marketed beverage from black/green tea. Further, the presence of Komagataeibacter and Gluconabacter is also expected to form a symbiotic relationship with the microbes present in the gut, thereby promoting the probiotic effect.
Microbial taxonomy analysis of yeast genera
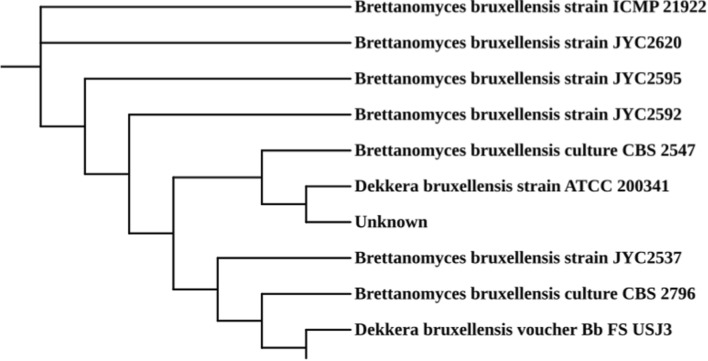
The BLAST results showed the yeast strain in Kombucha beverage produced from tea waste had high similarity with the Brettanomyces bruxellensis species (Supplementary Table S1). Two genera of yeast i.e. Brettanomyces bruxellensis and Dekkera bruxellensis were identified in the Kombucha beverage (Fig. 4). Villarreal-Soto et al. (2020) reported the abundance of 29.56% of B. bruxellensis and 16.11% of S. pombe. Contrary to the present study, Marsh et al. (2014) have reported the presence of Zygosaccharomyces sp., in Kombucha beverage obtained from black tea. Similar results were obtained by Coton et al. (2017), suggesting that Kombucha beverage from green tea showed the presence of Dekkera bruxellensis, D. anomala, Zygosaccharomyces bailii and Hanseniaspora valbyensis. Thus, as evident from the literature comparisons, the type of tea might influence the presence of yeast genera. The presence of Dekkera sp., in the present study is also expected to provide a pleasant flavour to beverage as projected in the study by De Keersmaecke, (1996), thereby improving its organoleptic properties.
Fig. 4.
Phylogenetic tree generated using neighbor joining algorithm for identification of the closely related evolutionary strains in Kombucha beverage produced from the tea waste
Nutrition content, physicochemical and organoleptic analysis of Kombucha beverage prepared from tea waste
Nutritional and physiochemical properties of Kombucha beverage plays an essential role in influencing the health aspects during consumption. The nutritional content, trace metals as well as the microbial and physico-chemical properties of the Kombucha beverage (light brown in colour) prepared from tea waste and the water utilized in analysis was evaluated through the manual of methods for analysis of foods by the Food Safety and Standard Authority of India (Food Safety and Standard Authority of India (FSSAI) 2016). The quality of water analysed showed no load of microbial contaminants, and physical as well as chemical parameters within the agreeable limits, thus ensuring the safety of product during the regular consumption (Supplementary Table S2). The beverage had significant quantity of carbohydrates (8.5%) and energy (32 kcal/100 g) with minimal protein and fat content as provided in Table 2. The pH of the beverage was 3.4, with total acidity (in terms of citric acid content) of 0.012% weight. 100% raw Kombucha contained 12 kcal of energy with 3.33 g carbohydrates in 100 ml of sample (U.S. Department of Agriculture 2019). Kaashyap et al. (2021) reported a pH of 3.0, with 1.87 g/L glucose, 1.11 g/L sucrose and 3.31 µg/ml in Kombucha beverage prepared from 75% green tea and 25% oolong tea. Very minimal concentration of trace metals was observed in Kombucha beverage, showing its suitability for consumption. Bauer-Petrovska and Petrushevska-Tozi (2000) also showed negligible presence of trace elements in Kombucha beverage. Jakubczyk et al. (2020) projected that the biochemical and nutritional composition of Kombucha beverage is influenced by the tea type, since it provides the major source of nitrogenous components for promoting the growth of the microbial community.
Table 2.
Nutritional and trace metal content of Kombucha beverage produced from the tea waste
| Parameters | Unit | Result | Test method |
|---|---|---|---|
| Carbohydrates | g/100 g | 8.5 | KLPL/SOP/FOOD-139 |
| Energy | Kcal/100 g | 34.0 | KLPL/SOP/FOOD-139 |
| Protein | g/100 g | < 0.4 | KLPL/SOP/FOOD-160 |
| Total fat | g/100 g | < 0.1 | KLPL/SOP/FOOD-139 |
| Total soluble solids | % by mass | 8.2 | Manual of methods, metals FSSAI,2016 |
| Acidity as citric acid | % by mass | 0.012 | FSSAI Manual 2016 |
| Arsenic (As) | mg/kg | < 0.001 | Manual of methods, metals FSSAI,2016 |
| Copper (Cu) | mg/kg | < 0.5 | Manual of methods, metals FSSAI,2016 |
| Lead (P) | mg/kg | < 0.02 | Manual of methods, metals FSSAI,2016 |
| Zinc (Zn) | mg/kg | < 0.1 | Manual of methods, metals FSSAI,2016 |
Evaluation of the sensory value of Kombucha tea showed maximum acceptability for the natural brew, followed by moderate acceptability for other flavours. On the Hedonics scale from 1 to 9, the Kombucha beverage overall was rated with a scale of 7.17 ± 1.76. The ratings for other attributes were highest in flavour (7.47 ± 1.34), followed by mouthfeel (7.13 ± 1.76), tartness (7.01 ± 1.77), sweetness (6.99 ± 1.60) and aroma (6.84 ± 1.80). On the Food Action Rating Scale Test (1 to 9), the average rating for the beverage was 6.50 ± 1.76 referring to “I like this and would drink it now and then”. Thus, it could be concluded that the Kombucha beverage produced from the tea waste had the desirable nutritional qualities and organoleptic properties for consumption.
Conclusion
Tea waste produced from the tea processing industries could act as an alternative to the commonly utilized black and green tea leaves during Kombucha beverage production. The microbial load, nutrient profile along with the product yield and antioxidant scavenging potential of the Kombucha beverage produced from the tea waste were comparable to that of the green and black tea. The present study also reported the presence of Komagataeibacter rhaeticus (93% abundance), while the yeast species was found to have 98% similarity with Brettanomyces bruxellensis in Kombucha beverage prepared from tea waste. The nutritional content analysis revealed the presence of significant amount of carbohydrates (8.5 g/100 g) and energy (34 kcal/100 g) with organoleptic properties suitable for human consumption. Even-though the symbiotic presence of the identified bacterial and yeast microbiota is expected to favourably influence the gut microbiota, detailed probiotic assays are essential to confirm their health impacts. More such studies linked with metabolomics are required to establish the linkage between the diversity in microbiome with the changes in the bioprocessing conditions influencing the functional properties of the beverage, promoting its beneficial applications.
Supplementary Information
Below is the link to the electronic supplementary material.
Acknowledgements
The authors thank the Department of Biotechnology and Medical Engineering of National Institute of Technology Rourkela for providing the research facility.
Abbreviations
- AAB
Acetic acid bacteria
- rRNA
Ribosomal ribonucleic acid
- ITS
Internal transcribed sequencing
- SCOBY
Symbiotic culture of bacteria and yeast
- DNA
Deoxyribonucleic acid
- BLAST
Basic local alignment search tool
- NCBI
National Centre for Biotechnology Information
- SOP
Standard operating procedure
- FSSAI
Food Safety and Standard Authority of India
Author’s Contributions
SP: Conceptualization, Data Curation, Writing-Original draft preparation. MRP: Conceptualization, Data Curation, Writing-Original draft preparation. KR: Investigation, Writing—Reviewing and Editing. BD: Organoleptic analysis, Writing—Reviewing and Editing. SJ: Organoleptic analysis, Writing—Reviewing and Editing. BB: Conceptualization, Data Curation, Writing—Reviewing and Editing. BP: Conceptualization, Funding acquisition, Writing- Reviewing and Editing, Final Approval.
Funding
This research was funded by the Biotechnology Ignition Grant (BIG), Biotechnology Industry Research Assistance Council (BIRAC), Department of Biotechnology, Government of India, for BIRAC/KIIT0471/BIG-13/18.
Declarations
Conflict of interest
Authors declare no conflict of interest.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- Bauer-Petrovska B, Petrushevska-Tozi L. Mineral and water soluble vitamin content in the Kombucha drink. Int J Food Sci Technol. 2000;35:201–205. doi: 10.1046/J.1365-2621.2000.00342.X. [DOI] [Google Scholar]
- Behera B, Laavanya D, Balasubramanian P. Techno-economic feasibility assessment of bacterial cellulose biofilm production during the Kombucha fermentation process. Bioresour Technol. 2022;346:126659. doi: 10.1016/J.BIORTECH.2021.126659. [DOI] [PubMed] [Google Scholar]
- Belloso-Morales G, Hernández-Sánchez H. Manufacture of a beverage from cheese whey using a “tea fungus” fermentation. Rev Latinoam Microbiol. 2003;45:1–2. [PubMed] [Google Scholar]
- Chakravorty S, Bhattacharya S, Chatzinotas A, et al. Kombucha tea fermentation: microbial and biochemical dynamics. Int J Food Microbiol. 2016;220:63–72. doi: 10.1016/J.IJFOODMICRO.2015.12.015. [DOI] [PubMed] [Google Scholar]
- Coton M, Pawtowski A, Taminiau B, et al. Unraveling microbial ecology of industrial-scale Kombucha fermentations by metabarcoding and culture-based methods. FEMS Microbiol Ecol. 2017 doi: 10.1093/FEMSEC/FIX048. [DOI] [PubMed] [Google Scholar]
- De Keersmaecke J. The Mystery of Lambic Beer. Sci Am. 1996;275:74–80. doi: 10.1038/scientificamerican0896-74. [DOI] [Google Scholar]
- Değirmencioğlu N, Yildiz E, Guldas M, Gurbuz O. Health benefits of Kombucha tea enriched with olive leaf and honey. J Obes Chronic Dis. 2020 doi: 10.17756/jocd.2020-031. [DOI] [Google Scholar]
- El Sheikha AF, Hu DM. Molecular techniques reveal more secrets of fermented foods. Crit Rev Food Sci Nutr. 2020;60:11–32. doi: 10.1080/10408398.2018.1506906. [DOI] [PubMed] [Google Scholar]
- El Sheikha AF, Levin R, Xu J. Revolution in fermented foods: From artisan household technology to the era of biotechnology. Mol Techniq Food Biol Safety Biotechnol Authent Traceability. 2018 doi: 10.1002/9781119374633.ch10. [DOI] [Google Scholar]
- Food Safety and Standard Authority of India (FSSAI) (2016) Manual of Methods of Analysis of Food
- Gaggìa F, Baffoni L, Galiano M, et al. Kombucha beverage from green, black and rooibos teas: a comparative study looking at microbiology, chemistry and antioxidant activity. Nutrition. 2018;1(11):1. doi: 10.3390/NU11010001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain M, Olsen HE, Paten B, Akeson M. The Oxford Nanopore MinION: delivery of nanopore sequencing to the genomics community. Genome Biol. 2016;171(17):1–11. doi: 10.1186/S13059-016-1103-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakubczyk K, Kałduńska J, Kochman J, Janda K. Chemical profile and antioxidant activity of the kombucha beverage derived from white, green, black and red tea. Antioxidants. 2020;447(9):447. doi: 10.3390/ANTIOX9050447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jayabalan R, Baskaran S, Marimuthu S, et al. Effect of Kombucha tea on Aflatoxin B 1 induced acute hepatotoxicity in albino rats-prophylactic and curative studies. J Korean Soc Appl Biol Chem. 2010;53:407–416. doi: 10.3839/jksabc.2010.063. [DOI] [Google Scholar]
- Kaashyap M, Cohen M, Mantri N, et al. Microbial diversity and characteristics of Kombucha as revealed by metagenomic and physicochemical analysis. Nutrition. 2021;4446(13):4446. doi: 10.3390/NU13124446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kallel L, Desseaux V, Hamdi M, et al. Insights into the fermentation biochemistry of Kombucha teas and potential impacts of Kombucha drinking on starch digestion. Food Res Int. 2012;49:226–232. doi: 10.1016/J.FOODRES.2012.08.018. [DOI] [Google Scholar]
- Kumar S, Stecher G, Tamura K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 2016;33:1870–1874. doi: 10.1093/MOLBEV/MSW054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laavanya D, Shirkole S, Balasubramanian P. Current challenges, applications and future perspectives of SCOBY cellulose of Kombucha fermentation. J Clean Prod. 2021;295:126454. doi: 10.1016/J.JCLEPRO.2021.126454. [DOI] [Google Scholar]
- Larkin MA, Blackshields G, Brown NP, et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/BIOINFORMATICS/BTM404. [DOI] [PubMed] [Google Scholar]
- Marsh AJ, O’Sullivan O, Hill C, et al. Sequence-based analysis of the bacterial and fungal compositions of multiple kombucha (tea fungus) samples. Food Microbiol. 2014;38:171–178. doi: 10.1016/J.FM.2013.09.003. [DOI] [PubMed] [Google Scholar]
- Miller GL. Use of dinitrosalicylic acid reagent for determination of reducing sugar. Anal Chem. 1959;31:426–428. doi: 10.1021/AC60147A030. [DOI] [Google Scholar]
- Muhialdin BJ, Osman FA, Muhamad R, et al (2019) Effects of sugar sources and fermentation time on the properties of tea fungus (kombucha) beverage. Int Food Res J. 26
- Quast C, Pruesse E, Yilmaz P, et al. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 2013;41:D590–D596. doi: 10.1093/NAR/GKS1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray RC, El Sheikha AF, Kumar S. Microorganisms and fermentation of traditional foods. Boca Raton: Food biology series. Science Publishers Inc.; 2014. Oriental fermented functional (probiotic) foods; pp. 283–311. [Google Scholar]
- Soni NO. Antioxidant assay in vivo and vitro. Int J Phytopharm. 2014;5:58. [Google Scholar]
- Tran T, Grandvalet C, Verdier F, et al. Microbial dynamics between yeasts and acetic acid bacteria in Kombucha: impacts on the chemical composition of the beverage. Foods. 2020;9:963. doi: 10.3390/foods9070963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- U.S. Department of Agriculture (2019) Food Data Central. In: 100% Raw Kombucha. https://fdc.nal.usda.gov/fdc-app.html#/food-details/556430/nutrients. Accessed 15 Mar 2022
- Villarreal-Soto SA, Bouajila J, Pace M, et al. Metabolome-microbiome signatures in the fermented beverage. Kombucha Int J Food Microbiol. 2020;333:108778. doi: 10.1016/J.IJFOODMICRO.2020.108778. [DOI] [PubMed] [Google Scholar]
- Wood DE, Lu J, Langmead B. Improved metagenomic analysis with Kraken 2. Genome Biol. 2019;20:1–13. doi: 10.1186/S13059-019-1891-0/FIGURES/2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zubaidah E, Yurista S, Rahmadani NR. Characteristic of physical, chemical, and microbiological kombucha from various varieties of apples. IOP Conf Ser Earth Environ Sci. 2018;131:012040. doi: 10.1088/1755-1315/131/1/012040. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.