This letter to the editor is a response by a large number of investigators in the field of protein synthesis to the minireview published by Dr. Kozak in Molecular and Cellular Biology (9). This minireview attempts to create significant doubts regarding the published literature that we believe are unwarranted and to bolster Dr. Kozak's own point of view regarding translation initiation. We therefore take serious issue with the scholarliness of the Kozak minireview. As will be shown, the Kozak minireview contains numerous distortions of fact and of published data and selectively utilizes the published literature. In every field of research there are legitimate concerns regarding the interpretation of results and the reproducibility of certain published data. Several of the issues raised by Dr. Kozak are legitimate in this regard, but they are not new and have hardly gone unnoticed, having been raised in scholarly and critical reviews elsewhere. At issue here is not the right to critically question results and interpretations but rather whether the Kozak minireview is scholarly and its tone is professional.
We point out that much of the work challenged in the Kozak minireview was published in Molecular and Cellular Biology, as well as other leading peer-reviewed journals, and forms a mainstream of research on protein synthesis which is taking place in scores of laboratories around the world. In this minireview, Dr. Kozak dismisses three novel mechanisms for translation initiation which have now been well studied and extensively documented. One mechanism is internal ribosome entry, which she rejects in favor of ribosome scanning, a mechanism for translation initiation which she proposed over 20 years ago. In ribosome scanning, it is proposed that the 40S small ribosome subunit enters the mRNA from the 5′ cap and undergoes a linear 5′-to-3′ search for the initiation codon, which is typically an AUG. Internal ribosome entry involves the internal association of ribosome subunits at or near the initiation codon without the need for entry from the 5′ end of the mRNA. A second mechanism opposed by Dr. Kozak is the initiation of protein synthesis without Met-tRNA, a universal and key component, as shown for several insect virus mRNAs. The ability to carry out translation without this initiator tRNA, and from the A site of the ribosome, has enormous implications for our understanding of protein synthesis and its evolution. A third mechanism of translation initiation which Dr. Kozak takes issue with is known as ribosome shunting or discontinuous scanning, which combines features of 5′ entry of ribosomes by scanning and the internal translocation of ribosome subunits without further scanning to the initiation codon. It is clear to a great many researchers, as represented by the signatory list below, that the initiation of protein synthesis in eukaryotes is dynamic and flexible, involving a variety of mechanisms that have evolved to meet the complex demands of eukaryotic cells and viruses.
Dr. Kozak has spent more than 10 years in strenuous opposition to the evidence for viral internal ribosome entry and the recognition of specific viral cis-acting internal ribosome entry site (IRES) elements. The minireview now attempts to use almost entirely the same kinds of arguments against cellular IRESs and other means of nonscanning translation initiation that Dr. Kozak used previously in her unsuccessful efforts to disprove viral IRESs. Whether Dr. Kozak explicitly acknowledges internal ribosome entry as an established fact, at least for viruses, is not at all clear in the minireview, although she does compare translation functions to the encephalomyocarditis virus (EMCV) IRES, but without comment or acceptance. It would be fairer to the reader and more intellectually honest to explicitly acknowledge internal ribosome entry as an established fact, at least for viruses, or—if she still wishes to oppose the idea—to do so openly. Needless to say, it is now difficult to mount a convincing case for blanket repudiation of IRESs in the face of overwhelming data, including elegant and compelling evidence from viral IRES-dependent translation of a circular RNA (3), which was not cited in the Kozak review.
It is not practical to document here all of the examples in which published results were inappropriately presented in the minireview by Dr. Kozak. We refer readers to a recent comprehensive review which summarizes the current evidence in support of viral and cellular IRES elements and alternate mechanisms of translation initiation in eukaryotes and briefly overviews some of the key techniques which were questioned by Dr. Kozak (7). Consequently, we list below just several specific examples which are emblematic of the serious issues which are of concern to us.
Several reasons are described by Dr. Kozak for dismissing reports of cellular IRESs. Dr. Kozak argues that because cellular IRESs often represent modest translation increases over background levels, they result from fortuitous positioning of RNA sequences in experimental constructs. This argument ignores the evidence that IRESs have been shown to represent a range of activities from weak to strong and to function by a variety of mechanisms. Indeed, the expression of many cellular genes encoding regulatory proteins is often tightly controlled at multiple steps to guarantee that correct protein levels are achieved, which is not generally equivalent to high protein levels. An IRES may therefore be relatively weak, but in combination with other levels of gene control, it achieves significant or correct protein expression levels under different physiological conditions. One example is the IRES of the proto-oncogene c-sis, which encodes platelet-derived growth factor 2 (PDGF2). This IRES is activated severalfold during megakaryocyte differentiation, in conjunction with induction of PDGF2/c-sis gene expression during differentiation (1, 16). The modest translation stimulation directed by the cellular PDGF2 IRES fine tunes PDGF2/c-sis gene expression during differentiation. Similar mechanisms are likely employed by other critical regulatory genes and may have widespread implications for cellular growth and development. Thus, it is arbitrary to dismiss cellular IRES elements as physiologically irrelevant artifacts merely because their effects on translation are moderate. A more considered view is that regulatory elements act at all levels of gene control, including transcription, mRNA transport, and mRNA stability and translation, and permit exquisite control precisely because they involve multiple and modest additive effects which can be independently combined and regulated.
There are several functional ways to study IRESs. Construction of a dicistronic mRNA containing an internal downstream second open reading frame that is ordinarily not translated is typically used to detect IRES activity. Other approaches include insertion of very stable, translation-blocking hairpin structures upstream of the IRES and biochemical detection of IRES interaction with initiation factors and ribosome subunits. Important control studies must be performed to validate the integrity of the dicistronic mRNA and to exclude the presence of cryptic promoters or aberrant splicing that could lead to production of subgenomic mRNAs or removal of intervening RNA sequences that would normally prevent internal translation initiation by ribosome scanning. With this in mind, apart from one “potential” cellular IRES, Dr. Kozak dismisses all other published reports as artifacts arising from improper experimental methodologies, a lack of proper control studies, or poor experimental design. However, most but not all cellular IRES studies included the use of other RNA segments that did not contain IRES elements or IRES sequences with defined mutations, which failed to mediate internal ribosome initiation. Thus, selective translation by internal ribosome entry was in fact shown to be specific for a small number of RNA elements. These controls were largely ignored in the Kozak minireview, inappropriately casting doubt on the integrity of the conclusions from these reports. Dr. Kozak is particularly critical of cellular IRES reports because the background control level of translation in the absence of the IRES varies between different constructs and because it is not zero. This argument can be misapplied to most molecular systems. For instance, deletion of all transcription elements seldom completely abolishes activity, and the basal activities of different control constructs typically vary. As proof for this view, Dr. Kozak points out that an antisense version of a putative cellular IRES directed translation at 40% of the level of the sense form (13). However, in other examples the antisense verification did not function as an IRES. In other cases, Dr. Kozak asserts (in the absence of any evidence to support her view) that a control RNA sequence has depressed translation, making it only appear that the cellular IRES element directs translation initiation. As one example, the Kozak minireview inappropriately compares experiments described in two papers (11, 21). In the Nature paper (11), the BiP IRES mediated translation of the second cistron 15-fold over the Antp control sequences. Importantly, introduction of a hairpin at the 5′ end of the dicistronic mRNA completely abolished translation of the second cistron (Fig. 2 in reference 11), demonstrating that the BiP sequence has IRES activity. In the Nucleic Acids Research paper (21), the BiP IRES was stimulated 10-fold over the Antp sequence; as Dr. Kozak pointed out, translation was lower (2.5-fold) compared to the “empty” vector control. This was interpreted by the authors as readthrough mediated by the 30-nucleotide sequence located between the two cistrons. This does not constitute a serious “discrepancy of results,” in contrast to its presentation by Dr. Kozak. In addition, while studies have shown that varying the length of the intercistronic region influences translation initiation frequency (5), a potential confounding problem, the effect acts predominantly on scanning-dependent rather than internal initiation of translation.
Dr. Kozak asserts that cryptic promoters or cryptic splicing of RNAs cannot be excluded as a source of smaller mRNAs that could be translated from truncated positions, providing the false impression of internal ribosome initiation or initiation by ribosome shunting. Dr. Kozak is not alone in expressing concern regarding some claims for internal ribosome entry, particularly when there is no accompanying data verifying the integrity or size of the mRNA species. Indeed, a few studies noted unanticipated smaller transcripts and noted that they likely arose from splicing, generally at low levels, from a few of the dicistronic constructs (e.g., see reference 6). However, this study demonstrated that the translation of the second cistron could not have occurred from the low-abundance monocistronic mRNA. Dr. Kozak cites the fact that unanticipated splicing was sometimes detected but does not present the data fully and accurately. Additionally, many studies involved in vitro-synthesized mRNAs that were monitored in cell-free systems or examined after expression in cultured cells, and the RNAs were found to be intact. While Dr. Kozak highlighted instances in which important RNA structural data were absent, she often failed to reference studies in which it was included and the RNAs were found to be intact. In some other cases she inappropriately dismisses the data as of poor quality or not sufficiently sensitive. For example, Dr. Kozak criticized published work on the vascular endothelial growth factor (VEGF) IRES as not convincing because of the presence of an internal promoter but failed to cite another paper which showed that translation initiation from an internal promoter cannot account for VEGF translation results (8). Again, it is not appropriate to assume that moderate translation effects, which can be quite important biologically, are artifacts because they do not conform to an arbitrary value. Internal initiation of c-Myc2 protein synthesis was similarly dismissed by Dr. Kozak despite evidence for only a single mRNA because transfection of the mRNA itself into cells, compared to its expression from a DNA vector, failed to lead to translation (17, 18). It was suggested by the authors of these two papers that the c-myc IRES might require nuclear binding proteins to function, which is reasonable given the importance of noncanonical factors for the activity of certain viral IRESs (7). This was rejected by Dr. Kozak, and other well-controlled studies which demonstrated c-Myc IRES function were not cited (e.g., reference 12). Thus, the minireview provides the false impression that only limited and poorly controlled research has been performed on cellular IRESs.
Dr. Kozak also asserts that biochemical studies have never been conducted to show that initiation factor 4G (eIF4G), a key factor that promotes ribosome binding, can associate with sufficient affinity to a natural IRES so as to mediate internal ribosome entry. This conclusion is meant to cast doubt on the validity of translation by internal ribosome entry in eukaryotic cells. In fact, Lomakin, Hellen, and Pestova (10) directly measured the affinity of the central domain of eIF4G alone and as a complex with eIF4A for the EMCV IRES and for β-globin mRNA. They found that the eIF4G/4A complex binds the EMCV IRES with an affinity sufficient for the IRES to be able to compete with cellular capped mRNAs for eIF4F, a complex of factors which contains eIF4G and helps to direct ribosome binding to capped mRNA. While these data do not prove a mechanistic function, they account for a critical first step. This reference was not cited by Dr. Kozak, nor in fact was any of the literature that analyzed the functional, specific interactions of eIF4G/4A/4F with EMCV-like IRESs and of eIF3, another essential initiation factor that binds to the 40S small ribosome subunit, with hepatitis C virus-like IRESs. True, these are viral IRESs. However, this is a well-known literature published in leading journals, and it provides a quantitative and partial mechanistic understanding of IRES function that may be applied to cellular IRESs.
Dr. Kozak questioned the quality and integrity of work which demonstrated the possibility of initiator-independent translation from the ribosome A site, as shown to occur in the cricket paralysis virus (CrPV) mRNA (19). This is a recent seminal finding in the field of protein synthesis. Importantly, a landmark paper (15), which demonstrated that a related insect virus IRES is also translated without tRNA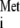 and is therefore highly supportive, was not cited in the minireview. While this paper is included in a review cited by Dr. Kozak, that review was referenced in a manner so as to cast doubt on these findings. Figure 3 in the PNAS paper (15) provides unambiguous data demonstrating that the CrPV IRES does not use initiator tRNA to initiate translation, strongly arguing that general rules of scanning-dependent initiation do not apply in this case. The Cell CrPV paper (19) challenged by Dr. Kozak confirmed these earlier findings from a related insect virus genome and disclosed an important and unexpected alternate molecular mechanism for protein synthesis. Dr. Kozak claims that the IRES-ribosome complexes that were reported in reference 19 are merely aggregates that are not translationally active complexes and ignores the fact that the authors examined the oligomeric state of the RNA molecules in these studies. Results also showed that 80S ribosome/CrPV IRES formation does not need GTP hydrolysis and is quite insensitive to the addition of l-methioninol (an approach also used by Dr. Kozak). Notwithstanding this evidence for unconventional initiation, Dr. Kozak questions the validity of the formation of initiator tRNAMet-independent 80S ribosome/CrPV complexes on the basis of the concentrations of edeine used in the experiments. It is true that edeine at 1 to 10 μM inhibits translation initiation at the 40S ribosome-AUG recognition step. These concentrations of edeine will inhibit the pausing of 40S subunits at the AUG initiation codon on all mRNAs examined so far, except the CrPV IRES. A 40S ribosome can be detected at the CrPV-IRES initiation codon by toeprinting analysis. While CrPV IRES-mediated translation is unaffected in the presence of 0.5 μM edeine, translation is inhibited by 80% at 1 μM. This finding could be explained if edeine has an affect on a step in translation that is subsequent to the 40S subunit-start codon recognition step. Indeed, it has been shown (2) that the enzymatic association (aided by eEF1 and GTP) of Phe-tRNA to the ribosomal A site is abolished by 80% in the presence of 1 μM edeine. The sucrose gradient-toeprinting data in reference 19 support the hypothesis that edeine inhibits a postinitiation step in CrPV IRES-mediated translation. As this concentration of edeine inhibits the AUG recognition by 40S ribosome subunits in all examined mRNAs, a subsequent affect of edeine on elongation would not be trivial to detect. It is therefore difficult to understand Dr. Kozak's claim to have disproven that the CrPV-like IRESs have an unusual mechanism of initiator tRNA-independent translation initiation, which does not use the ribosomal P site. Dr. Kozak also states that CrPV may synthesize subgenomic mRNAs that are translated, providing initiation from 5′-truncated transcripts that only appear to constitute internal translation initiation. This claim ignores compelling and rigorous literature (none of which was cited) demonstrating that CrPV does not produce subgenomic mRNAs in infected cells and that full-length genomic RNA extracted from virions is directly translatable to yield the protein in question from the downstream open reading frame (4, 14, 20).
and is therefore highly supportive, was not cited in the minireview. While this paper is included in a review cited by Dr. Kozak, that review was referenced in a manner so as to cast doubt on these findings. Figure 3 in the PNAS paper (15) provides unambiguous data demonstrating that the CrPV IRES does not use initiator tRNA to initiate translation, strongly arguing that general rules of scanning-dependent initiation do not apply in this case. The Cell CrPV paper (19) challenged by Dr. Kozak confirmed these earlier findings from a related insect virus genome and disclosed an important and unexpected alternate molecular mechanism for protein synthesis. Dr. Kozak claims that the IRES-ribosome complexes that were reported in reference 19 are merely aggregates that are not translationally active complexes and ignores the fact that the authors examined the oligomeric state of the RNA molecules in these studies. Results also showed that 80S ribosome/CrPV IRES formation does not need GTP hydrolysis and is quite insensitive to the addition of l-methioninol (an approach also used by Dr. Kozak). Notwithstanding this evidence for unconventional initiation, Dr. Kozak questions the validity of the formation of initiator tRNAMet-independent 80S ribosome/CrPV complexes on the basis of the concentrations of edeine used in the experiments. It is true that edeine at 1 to 10 μM inhibits translation initiation at the 40S ribosome-AUG recognition step. These concentrations of edeine will inhibit the pausing of 40S subunits at the AUG initiation codon on all mRNAs examined so far, except the CrPV IRES. A 40S ribosome can be detected at the CrPV-IRES initiation codon by toeprinting analysis. While CrPV IRES-mediated translation is unaffected in the presence of 0.5 μM edeine, translation is inhibited by 80% at 1 μM. This finding could be explained if edeine has an affect on a step in translation that is subsequent to the 40S subunit-start codon recognition step. Indeed, it has been shown (2) that the enzymatic association (aided by eEF1 and GTP) of Phe-tRNA to the ribosomal A site is abolished by 80% in the presence of 1 μM edeine. The sucrose gradient-toeprinting data in reference 19 support the hypothesis that edeine inhibits a postinitiation step in CrPV IRES-mediated translation. As this concentration of edeine inhibits the AUG recognition by 40S ribosome subunits in all examined mRNAs, a subsequent affect of edeine on elongation would not be trivial to detect. It is therefore difficult to understand Dr. Kozak's claim to have disproven that the CrPV-like IRESs have an unusual mechanism of initiator tRNA-independent translation initiation, which does not use the ribosomal P site. Dr. Kozak also states that CrPV may synthesize subgenomic mRNAs that are translated, providing initiation from 5′-truncated transcripts that only appear to constitute internal translation initiation. This claim ignores compelling and rigorous literature (none of which was cited) demonstrating that CrPV does not produce subgenomic mRNAs in infected cells and that full-length genomic RNA extracted from virions is directly translatable to yield the protein in question from the downstream open reading frame (4, 14, 20).
A number of studies have identified yet another alternate mechanism for translation initiation known as ribosome shunting. As it occurs in adenovirus mRNAs expressed during the late stage of infection, ribosome shunting was shown to involve sequences in the viral 5′ noncoding region that are complementary to 18S rRNA (22). Dr. Kozak's minireview misrepresents the central conclusion of this paper, falsely stating that this study claims to have demonstrated direct interaction between mRNA and 18S rRNA for initiation of translation by ribosome shunting. In fact, this study concluded that ribosome shunting on adenovirus late mRNAs might occur by any of several mechanisms that involve sequences complementary to 18S rRNA, including but not limited to structural RNA mimicry or direct interaction with 18S rRNA. Dr. Kozak also asserts that only rudimentary mapping, large deletions, and a failure to conduct mRNA integrity analysis underlie the conclusion that 5′ noncoding sequences in adenovirus late mRNAs facilitate ribosome initiation by utilizing sequences complementary to 18S rRNA. This assertion ignores control Northern mRNA analyses presented in this paper and elsewhere, and it improperly represents the size of deletions introduced in the 5′ noncoding region in such a way as to leave the impression that they are nonspecific.
The examples cited above represent only a sampling of numerous significant errors in the minireview published by Dr. Kozak. Careful inspection of this minireview reveals a lack of scholarly accuracy that will only serve to confuse and mislead readers. While Dr. Kozak is entitled to her opinions, we believe very strongly that only manuscripts of acceptable scholarly standards should be published in Molecular and Cellular Biology.
Footnotes
Vadim I. Agol, Raul Andino, Francis Bayard, Douglas R. Cavener, Stephen A. Chappell, Jane-Jane Chen, Jean-Luc E. Darlix, Asim Dasgupta, Olivier Donzé, Roger Duncan, Orna Elroy-Stein, Philip J. Farabaugh, Witold Filipowicz, Michael Gale, Jr., Lee Gehrke, Emanuel Goldman, Yoram Groner, Joe B. Harford, Maria Hatzoglou, Bin He, Christopher U. T. Hellen, Matthias W. Hentze, John Hershey, Panda Hershey, Thomas Hohn, Martin Holcik, Craig P. Hunter, Kazuei Igarashi, Richard Jackson, Rosemary Jagus, Leonard S. Jefferson, Bhavesh Joshi, Raymond Kaempfer, Michael G. Katze, Randal J. Kaufman, Megerditch Kiledjian, Scot R. Kimball, Adi Kimchi, Karla Kirkegaard, Antonis E. Koromilas, Robert M. Krug, Veronique Kruys, Barry J. Lamphear, Stanley Lemon, Richard E. Lloyd, Lynne E. Maquat, Encarnacion Martinez-Salas, Michael B. Mathews, Vincent P. Mauro, Suzanne Miyamoto, Ian Mohr, David R. Morris, Eric G. Moss, Nobuhiko Nakashima, Ann Palmenberg, Neil T. Parkin, Tsafi Pe'ery, Jerry Pelletier, Stuart Peltz, Tatyana V. Pestova, Evgeny V. Pilipenko, Anne-Catherine Prats, Vincent Racaniello, G. Sullivan Read, Robert E. Rhoads, Joel D. Richter, Rolando Rivera-Pomar, Tracey Rouault, Alan Sachs, Peter Sarnow, Gert C. Scheper, Leslie Schiff, Daniel R. Schoenberg, Bert L. Semler, Aleem Siddiqui, Tim Skern, Nahum Sonenberg, Wayne Sossin, Nancy Standart, Stanley M. Tahara, Adri A. M. Thomas, Jean-Jacques Toulmé, Jeff Wilusz, Eckard Wimmer, Gary Witherell, and Michael Wormington
REFERENCES
- 1.Bernstein J, Sella O, Le S Y, Elroy-Stein O. PDGF2/c-sis mRNA leader contains a differentiation-linked internal ribosomal entry site (D-IRES) J Biol Chem. 1997;272:9356–9362. doi: 10.1074/jbc.272.14.9356. [DOI] [PubMed] [Google Scholar]
- 2.Carrasco L, Battaner E, Vazquez D. The elongation step in protein synthesis in eukaryotic ribosomes: effects of antibiotics. Methods Enzymol. 1974;30:282–289. doi: 10.1016/0076-6879(74)30031-6. [DOI] [PubMed] [Google Scholar]
- 3.Chen C Y, Sarnow P. Initiation of protein synthesis by the eukaryotic translational apparatus on circular RNAs. Science. 1995;268:415–417. doi: 10.1126/science.7536344. [DOI] [PubMed] [Google Scholar]
- 4.Eaton B J, Steacie A D. Cricket paralysis virus RNA has a 3′ terminal poly A. J Gen Virol. 1980;50:167–171. [Google Scholar]
- 5.Gallie D R, Ling J, Niepel M, Morley S J, Pain V M. The role of 5′-leader length, secondary structure and PABP concentration on cap and poly(A) tail function during translation in Xenopus oocytes. Nucleic Acids Res. 2000;28:2943–2953. doi: 10.1093/nar/28.15.2943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Grundhoff A, Ganem D. Mechanisms governing expression of the v-FLIP gene of Kaposi's sarcoma-associated herpesvirus. J Virol. 2001;75:1857–1863. doi: 10.1128/JVI.75.4.1857-1863.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hellen C U, Sarnow P. Internal ribosome entry sites in eukaryotic mRNA molecules. Genes Dev. 2001;15:1593–1612. doi: 10.1101/gad.891101. [DOI] [PubMed] [Google Scholar]
- 8.Huez I, Creancier L, Audigier S, Gensac M C, Prats A C, Prats H. Two independent internal ribosome entry sites are involved in translation initiation of vascular endothelial growth factor mRNA. Mol Cell Biol. 1998;18:6178–6190. doi: 10.1128/mcb.18.11.6178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kozak M. New ways of initiating translation in eukaryotes? Mol Cell Biol. 2001;21:1899–1907. doi: 10.1128/MCB.21.6.1899-1907.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lomakin I B, Hellen C U, Pestova T V. Physical association of eukaryotic initiation factor 4G (eIF4G) with eIF4A strongly enhances binding of eIF4G to the internal ribosomal entry site of encephalomyocarditis virus and is required for internal initiation of translation. Mol Cell Biol. 2000;20:6019–6029. doi: 10.1128/mcb.20.16.6019-6029.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Macejak D G, Sarnow P. Internal initiation of translation mediated by the 5′ leader of a cellular mRNA. Nature. 1991;353:90–94. doi: 10.1038/353090a0. [DOI] [PubMed] [Google Scholar]
- 12.Nanbru C, Lafon I, Audigier S, Gensac M C, Vagner S, Huez G, Prats A C. Alternative translation of the proto-oncogene c-myc by an internal ribosome entry site. J Biol Chem. 1997;272:32061–32066. doi: 10.1074/jbc.272.51.32061. [DOI] [PubMed] [Google Scholar]
- 13.Negulescu D, Leong L E, Chandy K G, Semler B L, Gutman G A. Translation initiation of a cardiac voltage-gated potassium channel by internal ribosome entry. J Biol Chem. 1998;273:20109–20113. doi: 10.1074/jbc.273.32.20109. [DOI] [PubMed] [Google Scholar]
- 14.Reavy B, Moore N F. In vitro translation of cricket paralysis virus RNA. Arch Virol. 1981;67:175–180. doi: 10.1007/BF01318602. [DOI] [PubMed] [Google Scholar]
- 15.Sasaki J, Nakashima N. Methionine-independent initiation of translation in the capsid protein of an insect RNA virus. Proc Natl Acad Sci USA. 2000;97:1512–1515. doi: 10.1073/pnas.010426997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sella O, Gerlitz G, Le S Y, Elroy-Stein O. Differentiation-induced internal translation of c-sis mRNA: analysis of the cis elements and their differentiation-linked binding to the hnRNP C protein. Mol Cell Biol. 1999;19:5429–5440. doi: 10.1128/mcb.19.8.5429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stoneley M, Paulin F E, Le Quesne J P, Chappell S A, Willis A E. c-Myc 5′ untranslated region contains an internal ribosome entry segment. Oncogene. 1998;16:423–428. doi: 10.1038/sj.onc.1201763. [DOI] [PubMed] [Google Scholar]
- 18.Stoneley M, Subkhankulova T, Le Quesne J P, Coldwell M J, Jopling C L, Belsham G J, Willis A E. Analysis of the c-myc IRES; a potential role for cell-type specific trans-acting factors and the nuclear compartment. Nucleic Acids Res. 2000;28:687–694. doi: 10.1093/nar/28.3.687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wilson J E, Pestova T V, Hellen C U, Sarnow P. Initiation of protein synthesis from the A site of the ribosome. Cell. 2000;102:511–520. doi: 10.1016/s0092-8674(00)00055-6. [DOI] [PubMed] [Google Scholar]
- 20.Wilson J E, Powell M J, Hoover S E, Sarnow P. Naturally occurring dicistronic cricket paralysis virus RNA is regulated by two internal ribosome entry sites. Mol Cell Biol. 2000;20:4990–4999. doi: 10.1128/mcb.20.14.4990-4999.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yang Q, Sarnow P. Location of the internal ribosome entry site in the 5′ non-coding region of the immunoglobulin heavy-chain binding protein (BiP) mRNA: evidence for specific RNA-protein interactions. Nucleic Acids Res. 1997;25:2800–2807. doi: 10.1093/nar/25.14.2800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yueh A, Schneider R J. Translation by ribosome shunting on adenovirus and Hsp70 mRNas facilitated by complementarity to 18S rRNA. Genes Dev. 2000;14:414–421. [PMC free article] [PubMed] [Google Scholar]


