Abstract
Non-coding RNAs (ncRNAs) are a newly discovered functional RNA different from messenger RNA, which can participate in regulating the occurrence and development of tumors. More and more research results show that ncRNAs can participate in the regulation of gastric cancer (GC) radiotherapy response, and its mechanism may be related to its effect on DNA damage repair, gastric cancer cell stemness, cell apoptosis, activation of epidermal growth factor receptor signaling pathway, etc. This article summarizes the relevant mechanisms of ncRNAs regulating the response to radiotherapy in gastric cancer, which will be directly important for the introduction of ncRNAs particularly microRNAs (miRNAs), long non-coding RNAs (lncRNAs) and circular RNAs (circRNAs) into clinical medicine as biomarkers and therapeutic targets.
Keywords: non-coding RNA, radiotherapy, oncogenesis, therapeutic targets, biomarkers, gastric cancer, biofluids
1 Introduction
The incidence of gastric cancer (GC) ranks fifth in the world, and its death rate ranks third in the world (Smyth et al., 2020). Because the early symptoms of GC are often hidden and atypical, many patients usually present with advanced disease when they see a doctor. Radiation therapy (radiotherapy), as one of the main treatment methods for GC, has shown the advantages of reducing the recurrence rate and prolonging the survival of patients (Leong, 2005; Foo et al., 2014). However, due to the low sensitivity of GC to radiotherapy, an important problem remains the resistance of this tumor and, in particular, what mechanisms are involved in this (Pasechnikov et al., 2014; Ruan et al., 2020). Therefore, it is necessary to develop tumor-targeted drugs or radiosensitizers to enhance the radiosensitivity of GC and improve the radiotherapy efficacy of GC patients. Studies have found that the aberrant expression of non-coding RNAs (ncRNAs) are involved in regulating the radiotherapy sensitivity of various tumors such as nasopharyngeal carcinoma (NPC), non-small cell lung cancer (NSCLC), colorectal cancer (CC), GC and significantly affects the radiotherapy efficacy of tumors (Slack and Chinnaiyan, 2019; Gareev et al., 2020; Machlowska et al., 2020; Yan and Bu, 2021). NcRNAs are usually divided into basic structure type and regulatory type according to different functions. Regulatory ncRNAs are mainly composed of long non-coding RNAs (lncRNAs), microRNAs (miRNAs) and circular RNAs (circRNAs) (Beylerli et al., 2021) (Figure 1). This article reviews the dysregulation of ncRNAs in GC, summarizes and analyzes the research results of ncRNAs related to GC radiotherapy sensitivity, and explores new directions for improving the prognosis of GC patients after radiotherapy.
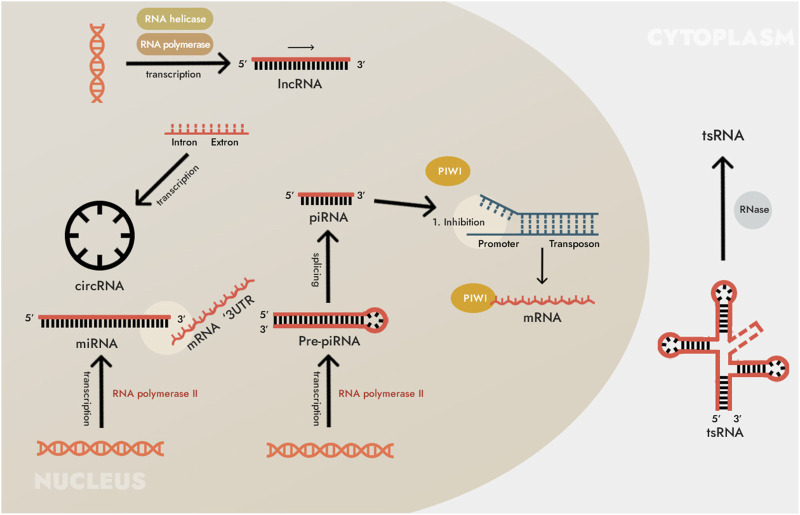
FIGURE 1.
Biosynthesis of non-coding RNAs (ncRNAs). The main types of ncRNAs are microRNAs (miRNAs), long non-coding RNAs (lncRNAs) and circular RNAs (circRNAs). In addition to those listed, many other types of RNA are also included in ncRNAs. For instance, small ncRNAs include RNAs that interact with PIWI proteins (piRNA, piwi-interacting RNA, piwiRNA), transfer RNA (tRNA)-derived small non-coding RNA (tsRNA), and others. However, these types of ncRNAs have not been found in human cells, and/or their pathogenetic and diagnostic (as well as therapeutic) significance has not yet been shown, so we do not consider them here.
2 Regulation of DNA damage by ncRNAs in GC cells
Damage to DNA and cell membranes is considered to be the main cause of radiation-induced cancer cell death (Beylerli et al., 2021). Normal and cancer cells have the ability to sense DNA damage and initiate DNA damage repair, the DNA damage response. The DNA damage response plays an important role in sensing DNA double-strand breaks, inducing cell cycle arrest, and initiating DNA repair (Wang and Xie, 2022). The DNA damage response manifests as a signaling cascade in which DNA-damaging factors first activate telangiectatic ataxia mutant factors, which subsequently activate cellular checkpoint kinases, phosphorylate histone 2A variants, and inhibit cell entry into S and M phases, leading to cell cycle arrest and initiation of damage repair to maintain genome stability. The study by Hu et al. confirmed that, under X-ray irradiation, hsa-let-7 g can significantly increase the radiosensitivity of GC by reducing the expression of telangiectasia ataxia mutation factor in GC cells and indirectly inhibiting the activation of DNA damage response (Hu et al., 2015).
DNA double-strand breaks are the most prevalent and potent type of damage induced by radiation therapy. The role of ncRNA in DNA double-strand damage repair has been confirmed (Nickoloff et al., 2020). In SNU-638 GC cells, ectopically expressed miR-196b can reduce the expression of DNA repair protein RAD23B, leading to the blockage of DNA damage repair mechanism in GC cells, inducing cell death, and increasing the radiosensitivity of GC cells (Tsai et al., 2010). Tissue-based GC-related ncRNAs are listed in Table 1, focusing particularly on their involvement in DNA damage (Qin et al., 2018; Huang et al., 2019; Manoel-Caetano et al., 2019; Zhang et al., 2019; Zhang et al., 2020; Guo et al., 2022; Gupta et al., 2022)
TABLE 1.
Some non-coding RNAs (ncRNAs) that regulate DNA damage in gastric cancer (GC) cells under the influence of radiotherapy.
| NcRNA | Model study | Expression | Target gene/pathway | Type of ncRNA | Biological function | References |
|---|---|---|---|---|---|---|
| miR-21, miR-24 and miR-421 | In silico and in vitro | Up | ATM/ATR/H2AX | Oncogene | Recognition and repair of DNA damage | Manoel-Caetano et al. (2019) |
| miR-192 and miR-215 | In vivo and in vitro | Up | SET8 and p53 | Oncogene | Promotes DNA damage repair and promotes progression of GC | Zhang et al. (2020) |
| miR-129-3p | In vitro | Down | SUMO-activating enzyme subunit 1 (SAE1) | Tumor suppressor | Induces more DNA damage and cell apoptosis, and inhibits GC cell proliferation, migration and invasion | Zhang et al. (2019) |
| lncRNA GAS5 | Mathematical model | Down | ATM/p38 MAPK and miR-34c | Tumor suppressor | Stimulate of DNA damage and enhances the radiosensitivity of GC | Gupta et al. (2022) |
| lncRNA FOXD2-AS1 | In vitro | Up | SETD1A | Oncogene | Promotes DNA damage repair and reduces the radiosensitivity of GC | Guo et al. (2022) |
| lncRNA MDC1-AS | In vivo and in vitro | Up | MDC1 | Oncogene | Promotes DNA damage repair and significantly inhibits cell proliferation and metastasis | Qin et al. (2018) |
| Circular RNA AKT3 | In vivo and in vitro | Up | PIK3R1 | Oncogene | Promotes DNA damage repair and inhibits the apoptosis of GC cells | Huang et al. (2019) |
Abbreviations: ATM, Ataxia-telangiectasia mutated; ATR, Ataxia telangiectasia and Rad3-related protein; SAE1, SUMO-activating enzyme subunit one; MAPK, Mitogen-activated protein kinase; MDC, Macrophage-derived/CCL22 chemokine; PIK3R1, Phosphatidylinositol 3-kinase regulatory subunit alpha; GAS5, Growth arrest-specific five; FOXD2-AS1, Forkhead box D2 adjacent apposite strand RNA, one; MDC1-AS, Mediator of DNA, damage checkpoint protein one; AKT3, AKT, serine/threonine kinase 3.
3 Regulation of ncRNA on tumor cell stemness in GC
It's no secret that, cancer stem cells are more resistant to radiation than mature cancer cells. Studies have shown that cancer stem cells have a strong ability to scavenge or reduce the level of reactive oxygen species (ROS), resulting in less DNA damage than mature cancer cells (Gareev et al., 2021). Currently, the Wnt/β-catenin pathway is considered to be one of the main targets of anti-tumor stem cell therapy. Studies have found that lncRNA HNF1A antisense RNA 1 (HNF1A-AS1), miR-501-5, and circFAM73A can promote the stemness of GC cells by activating the downstream Wnt/β-catenin pathway (Fan et al., 2016; Liu et al., 2018; Xia et al., 2021).
In addition, lncRNA metastasis-associated lung adenocarcinoma transcript 1 (MALAT1) promotes the stemness of GC cells by combining with sex determining region Y (SRY)-related HMG-box 2 (SOX) mRNA, Sox2 SRY (sex determining region Y)-box 2 mRNA, and knockdown of lncRNA MALAT1 can enhance the radiosensitivity of GC cells (Xu et al., 2021). In summary, some ncRNAs may be potential targets for GC radiotherapy by promoting the stem cell-like characteristics of GC, enhancing the scavenging ability of reactive ROS, reducing the damage effect of radiation on GC cells, and reducing the radiosensitivity of GC. A variety of ncRNAs have been reported to be involved in tumor cell stemness in GC (Table 2) (Song et al., 2018; Hong et al., 2019; Song et al., 2019; Sun et al., 2019; Zhao et al., 2020; Ni et al., 2021; Taghehchian et al., 2022; Xing et al., 2022; Zhang et al., 2022; Liu et al., 2023).
TABLE 2.
Some non-coding RNAs (ncRNAs) that regulate tumor cell stemness in gastric cancer (GC) under the influence of radiotherapy.
| NcRNA | Model study | Expression | Target gene/pathway | Type of ncRNA | Biological function | References |
|---|---|---|---|---|---|---|
| miR-375 | In vivo and in vitro | Down | SLC7A11 | Tumor suppressor | Inhibits the stemness and can induce ferroptosis of GC cells | Ni et al. (2021) |
| miR-18 | In vitro | Up | Meis2 and HMGB3 | Oncogene | Promotes the stemness of GC cells | Zhang et al. (2022) |
| miR-216a-3p | In vitro | Down | BRD4/Wnt/β-catenin pathway | Tumor suppressor | Promotes the stemness of GC cells | Song et al. (2019) |
| lncRNA PTCSC3 | In vitro | Down | PTCSC3 | Tumor suppressor | Inhibits the stemness and GC cells proliferation | Hong et al. (2019) |
| lncRNA THOR | In vitro | Down | SOX9 | Tumor suppressor | Inhibits the stemness | Song et al. (2018) |
| lncRNA LOXL1-AS1 | In vitro | Up | miR‐708‐5p and USF1 | Oncogene | Promotes the stemness and contributes to GC cells proliferation, migration and EMT. Reflects poor prognosis | Sun et al. (2019) |
| lncRNA LINC00332 | In silico, in vitro and bioinformatics | Down | MMP-13 | Tumor suppressor | Inhibits the stemness and proliferation, migration, and invasion of GC cells | Taghehchian et al. (2022) |
| circ-NOTCH1 | In vivo and in vitro | Up | miR-449c-5p/MYC/NOTCH1 axis | Oncogene | Promotes metastasis and stemness in GC | Zhao et al. (2020) |
| circRPPH1 | In vitro | Up | SLC7A11 | Oncogene | Promotes the stemness and can regulate ferroptosis of GC cells | Liu et al. (2023) |
| circ0007360 | In vivo and in vitro | Up | miR-762/IRF7 axis | Tumor suppressor | Inhibitory effects of circ0007260 on the survival, migration, invasion, and stemness of GC cells | Xing et al. (2022) |
Abbreviations: PTCSC3, Papillary thyroid carcinoma susceptibility candidate three; LOXL1-AS1, LOXL1 Antisense RNA, one; LINC00332, Long intergenic non-protein coding RNA, 332; SLC7A11, Meis2, Meis homeobox two; HMGB3, High-mobility group protein B3; BRD4, Bromodomain-containing protein 4; PTCSC3, Papillary thyroid carcinoma susceptibility candidate three; SOX9, SRY-Box transcription factor 9; USF1, Upstream stimulatory factor 1; MMP-13, Matrix metalloproteinase 13; NOTCH1, Neurogenic locus notch homolog protein one; SLC7A11, Solute carrier family 7, membrane 11; IRF7, Interferon regulatory factor 7; EMT, Epithelial-mesenchymal transition.
4 Regulation of ncRNA on apoptosis of GC cells
When repair of DNA damage caused by radiation fails, cells initiate automatic death programs (apoptosis) to maintain genome stability. For radiation-induced cell damage, whether tumor cells choose to repair the damage or initiate apoptosis is of great significance to the prognosis of tumors.
The p53 gene is currently the most widely studied cell regulatory gene. Studies have shown that in p53-deficient GC cells, miR-34 can restore p53 function and induce cell apoptosis (Xiong et al., 2019). miR-375 can directly interact with the 3′-untranslated region (3′-UTR) mRNA of the p53 gene, negatively regulate p53 expression and downstream pathways, and reduce the radiosensitivity of GC cells by inhibiting apoptosis and causing cell cycle arrest (Xu et al., 2011). In addition, the study found that in GC cells after radiation exposure, inhibition of the miR-221/222 cluster can upregulate the expression of phosphatase and tensin homolog deleted on chromosome 10 (PTEN) in GC cells, activate phosphatidylinositol-3-hydroxykinase (PI3K)/Akt signaling pathway, induce apoptosis, and enhance the radiosensitivity of GC cells (Chun-Zhi et al., 2010). LncRNA growth arrest-specific 5 (GAS5) significantly inhibits GC cell proliferation, promotes apoptosis, and enhances radiosensitivity by targeting miR-196a (Li et al., 2016). It can be seen that the abnormal expression of ncRNA is closely related to the apoptosis of GC cells, which largely determines the sensitivity of GC cells to radiotherapy (Table 3) (He et al., 2014; Zhang et al., 2014; Mao et al., 2019; Wei et al., 2019; Liu et al., 2021a; Wang et al., 2021a; Qin et al., 2022; Wang et al., 2022; Zhou et al., 2022).
TABLE 3.
Some non-coding RNAs (ncRNAs) that regulate stability to apoptosis in gastric cancer (GC) cells under the influence of radiotherapy.
| NcRNA | Model study | Expression | Target gene/pathway | Type of ncRNA | Biological function | References |
|---|---|---|---|---|---|---|
| miR-4537 | In vitro | Down | ZNF587 | Tumor suppressor | Inhibits the ability of cell proliferation, but on the contrary, it promotes the ability of cell apoptosis and improves radiosensitivity of GC cells | Liu et al. (2021a) |
| miR-4766-5p | In vitro | Down | NKAP | Tumor suppressor | Induces GC cell apoptosis | Wei et al. (2019) |
| miR-300 and miR-642 | In vitro and bioinformatics | Down | BCL2L11, GAS2, CASP8AP2, APAF1, DLC1, TP53, CASPS2, CASPS7, CASPS9, CASPS10, and BCL2L11 | Tumor suppressor | Regulate cellular radiation response by modulating apoptosis and cell cycle regulation | He et al. (2014) |
| lncRNA LINC00152 | In vitro and in vivo | Up | Bcl-2 | Oncogene | Activates cell cycle signaling, promotes migration and invasion, and suppress apoptosis | Mao et al. (2019) |
| lncRNA OGFRP1 | In vitro | Up | miR-149-5p/MAP3K3 axis | Oncogene | Promotes proliferation and suppresses GC cells radiosensitivity | Qin et al. (2022) |
| lncRNA CARLo-5 | In vitro | Up | ERK/MAPK pathway | Oncogene | Promotes the GC cells proliferation and inhibits apoptosis | Zhang et al. (2014) |
| lncRNA SLC25A21-AS1 | In vitro | Down | miR-15a-5p | Tumor suppressor | Inhibits cell malignant behaviors (e.g., promotes to apoptosis) and enhances cell radiosensitivity in GC | Wang et al. (2022) |
| circ_0003506 | In vitro and in vivo | Up | miR-1256/BMPR2 axis | Oncogene | Downregulation of circ_0003506 inhibits radioresistance to repress proliferation, migration and invasion but increase apoptosis in radioresistant GC cells | Zhou et al. (2022) |
| circ_HN1 | In vitro and in vivo | Up | miR-302b-3p/ROCK2 axis | Oncogene | Promotes tumor growth, cell proliferation, migration, invasion, and inhibit cell apoptosis in GC cells | Wang et al. (2021a) |
Abbreviations: OGFRP1, Opioid growth factor receptor pseudogene one; SLC25A21-AS1, SLC25A21 antisense RNA, one; circ_HN1, circRNA, Jupiter microtubule associated homolog one; ZNF587, Zinc finger protein 587; NKAP, NF-kappa-B-activating protein; BCL2L11, recombinant human Bcl-2-like protein 11; GAS2, Growth arrest-specific protein two; CASP8AP2, Caspase eight associated protein two; APAF1, Apoptotic protease activating factor 1; DLC1, Deleted in liver cancer one; TP53, Tumor protein P53; CASPS2, Caspase two; CASPS7, Caspase seven; CASPS9, Caspase nine; CASPS10, Caspase 10; BCL2L11, Proapoptotic member of the B-cell CLL/lymphoma two; Bcl-2, B-cell lymphoma two MAP3K3, Mitogen-activated protein kinase kinase kinase three; ERK, Extracellular signal-regulated kinase; MAPK, Mitogen-activated protein kinase; BMPR2, Bone morphogenetic protein receptor type 2; ROCK2, Rho associated coiled-coil containing protein kinase two.
5 Activation of epidermal growth factor receptor (EGFR) signaling pathway by ncRNA
Radiation can cause abnormal expression of various genes in tumor cells, including epidermal growth factor receptor (EGFR). Overexpression of EGFR is related to lymphatic metastasis of GC, and can lead to growth and invasion of GC cells through the Akt pathway (Chen et al., 2021). As a stress response to radiation, EGFR is rapidly activated and induces the mitogen-activated protein kinase (MAPK)/extracellular signal-regulated kinase (ERK) and PI3K/Akt signaling pathways (Lei et al., 2022). Activation of these signaling pathways may repair radiation-induced DNA damage, evade apoptosis, and promote cell proliferation through homologous and non-homologous recombination (Molina-Castro et al., 2017).
Previous studies have shown that anti-EGFR-targeted therapy is an effective radiosensitizer for EGFR-overexpressing GC cells and xenografts. This radiosensitization is associated with inhibition of GC cell proliferation and promotion of apoptosis (Dragovich and Campen, 2009). Recent studies have shown that a variety of ncRNAs mediate the expression of EGFR in GC, so these ncRNAs can be used as a medium to target and regulate the expression of EGFR, and then promote the apoptosis of GC cells in the process of radiotherapy, thereby improving the sensitivity of GC to radiotherapy (Carlomagno et al., 2017; D'Souza et al., 2020; Kong et al., 2021; Ye et al., 2022; Lazăr et al., 2016) (Figure 2). Therefore, analyzing the regulatory mechanism of ncRNA on EGFR expression is a new direction worth exploring to improve the radiosensitivity of GC, and the EGFR inhibitor derived from this is expected to be a selective and effective radiosensitizer for GC.
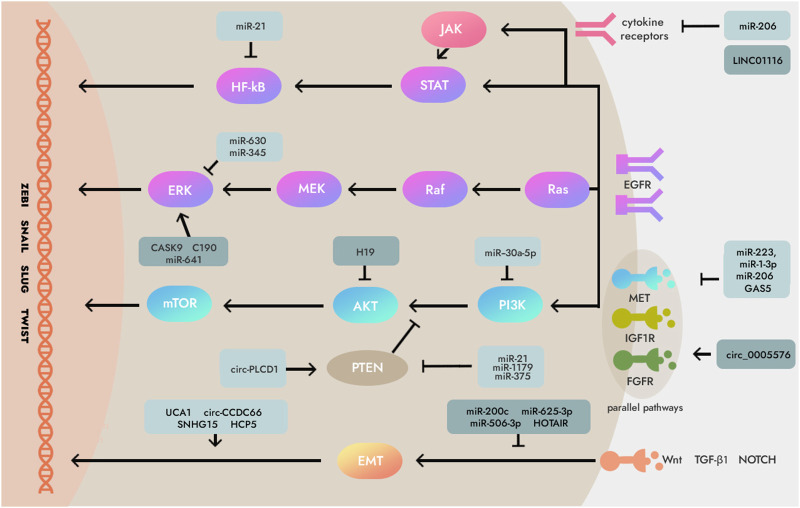
FIGURE 2.
Mechanisms of non-coding RNAs (ncRNAs) which involved in essential signaling pathways downstream and parallel pathways of epidermal growth factor receptor (EGFR) in some human cancers including gastric cancer (GC). Several critical ncRNAs can regulate EGFR signaling pathways like Ras/Raf/MEK/ERK axis, PI3K/AKT/mTOR axis, JAK/STAT/NF‐κB, PTEN, and Wnt/TGF‐β1/NOTCH. Note: MEK, Mitogen-activated protein kinase; ERK, Extracellular signal-regulated kinase; PI3K, Phosphoinositide 3-kinases; AKT, Serine/threonine-protein kinase; mTOR, Mammalian target of rapamycin; JAK, Janus kinase two; STAT, Signal transducer and activator of transcription; NF‐κB, Nuclear factor kappa-light-chain-enhancer of activated B cells; NOTCH, Neurogenic locus notch homolog protein; EMT, epithelial‐mesenchymal transition; IGF1R, Insulin‐like growth factor 1 receptor; PTEN, Phosphatase and ten sin homolog deleted on chromosome 10; TGF‐β1, Transforming growth factor‐β1; FGFR, Fibroblast growth factor receptor.
6 Regulation of epithelial-mesenchymal transition (EMT) by ncRNAs
A phenotypic change in tumor cells that may result in enhanced tumor cell motility and invasiveness, increased metastatic potential, and radiotherapy resistance (Lu et al., 2022). LncRNA HOX transcript antisense RNA (HOTAIR) can bind to miR-331-3p and inhibit its function, leading to upregulation of human epidermal growth factor receptor 2 (HER2) expression, promoting epithelial-mesenchymal transition (EMT) through HER2/Akt/HSF-1/slug signaling pathway. This may be related to the radiotherapy resistance of GC cells (Wang et al., 2015; Abdi et al., 2020).
MiR-544a-5p can act on cadherin E and Wnt/β-catenin to induce GC cell EMT through two independent pathways, and lncRNA RP11-789C1.1 inhibits EMT in GC via the RP11-789C1.1/miR-5003/cadherin E axis (Chen et al., 2018). LncRNA-h19 can combine with miR-141-5p to promote the EMT process of GC cells by up-regulating the expression of zinc finger E-box binding homeobox 1 (ZEB1) (Liu et al., 2022). Therefore, there may be a special signaling pathway between ncRNA and EMT, and through the regulation of EMT expression, it can affect the curative effect of GC cell radiotherapy (Figure 3) (Yang et al., 2015; Feng et al., 2019; Liu et al., 2021b; Beilerli et al., 2022).
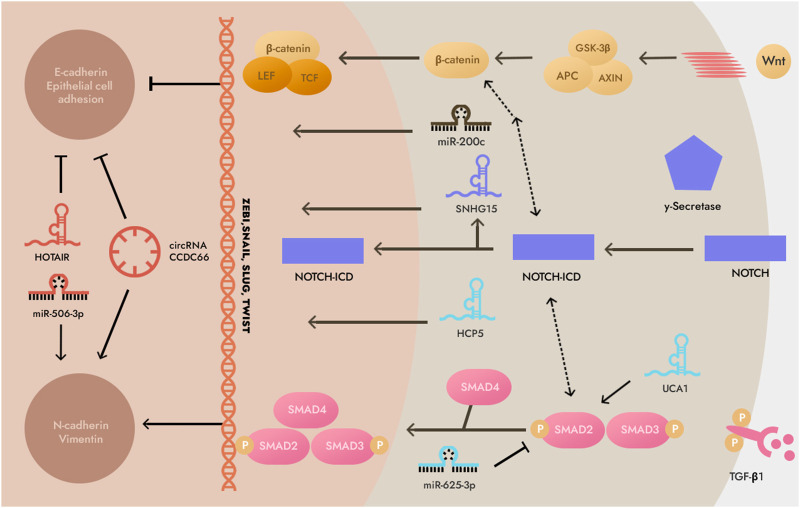
FIGURE 3.
Non-coding RNAs (ncRNAs) involved in epithelial-mesenchymal transition (EMT). This illustration demonstrates that the EMT process is predominantly composed of transforming growth factor-β1 (TGF-β), WNT and neurogenic locus notch homolog protein (NOTCH) signaling pathways. Note: SMAD2,3,4, Mothers against decapentaplegic homolog 2,3,4; GSK3β, glycogen synthase kinase-3β; LEF, Lymphoid enhancer-binding factor; N-cadherin, Neural cadherin; TCF, T cell factor; NOTCH-ICD, intracellular domain of the NOTCH receptor.
7 Regulation of oxygen adaptive metabolism by ncRNAs
The most striking feature of tumor cells is the ability to metabolize energy by glycolysis even in the presence of sufficient oxygen, which is known as the Warburg effect. Studies have shown that aerobic glycolysis in malignant tumors is closely related to tumor radiotherapy resistance (Yuan et al., 2022). ROS play an important role in radiation-induced DNA damage. The generation of reactive oxygen species is mainly in the process of oxidative phosphorylation in cells. Electron leakage during mitochondrial electron transport is the main source of electrons for the generation of intracellular ROS, and glycolysis leads to reduced generation of ROS, which seriously affects ROS-induced radiation damage (Tanprasert et al., 2022).
It is evident from many recent studies that when NSCLC, cervical cancer (CC), and glioma cells are exposed to radiation, various ncRNAs such as miR-449a and lncRNA urothelial carcinoma-associated 1 (UCA1) in the cells target multiple functions in the glycolytic metabolic pathway, specifically by decreasing the rate of a key rate-limiting enzyme, leading to an increase in the sensitivity of tumor cells to radiation (Yao et al., 2015; Nie et al., 2016; Wang et al., 2019). This phenomenon also occurs in GC cells, indicating that ncRNAs may play a role in altering the metabolic mode of GC and influencing its radiation sensitivity. MiR-4290 inhibits pyruvate dehydrogenase kinase 1 (PDK1), inhibiting glycolysis (Qian et al., 2021). MiR-7 can inhibit the glycolysis, cell proliferation and colony formation of GC cells by regulating the expression of lactate dehydrogenase (Xie et al., 2014). Therefore, ncRNA may enhance the curative effect of GC radiotherapy by regulating the glycolysis process of GC cells, changing the metabolic mode of cells, and increasing the level of ROS in cells.
Hypoxia is a pathophysiological feature of solid malignancies. Under hypoxic conditions, hypoxia-inducible factor-1α (HIF-1α) is upregulated, activating hypoxic adaptation pathways, including angiogenesis, erythropoiesis, and glycolysis (Li et al., 2019). HIF-1α protects blood vessels after radiotherapy and regulates glycolysis and pentose phosphate pathways, which increases the antioxidant capacity of tumors, thereby counteracting the oxidative stress caused by radiotherapy and affecting the radiosensitivity of tumors (Yasui et al., 2008). Previous studies have found that ncRNAs can regulate tumor glycolysis by regulating HIF-1α and its downstream glycolysis-related enzymes, thereby affecting tumor radiosensitivity (Zheng et al., 2021; Xu et al., 2022). The high expression of lncRNA ZNFX1 antisense RNA 1 (ZFAS1) in gastric cardia adenocarcinoma (GCA) assists EPAS1 to enhance the epigenetic silencing of HIF-1α and promote the proliferation and metastasis of cancer cells (Zhu et al., 2020). MiR-376a binds lncRNA NUTM2A Antisense RNA 1 (NUTM2A-AS1) and negatively regulates HIF-1α to inhibit the invasion of GC (Wang et al., 2020). Therefore, the ncRNA/HIF-1α/glycolysis-related enzyme signaling pathway may be a potential target for regulating the radiosensitivity of GC, and it may become a new direction to improve the radiosensitivity of GC by regulating tumor glucose metabolism and tumor microenvironment. The ncRNAs that have been associated with the oxygen adaptive metabolism and their molecular pathways are listed in Table 4 (Tu et al., 2014; Hong et al., 2016; Fang et al., 2020; Wang et al., 2021b; Deng et al., 2021; Sun et al., 2021; Li et al., 2022; Tan et al., 2022; Yang et al., 2022).
TABLE 4.
Some non-coding RNAs (ncRNAs) that regulate oxygen adaptive metabolism in gastric cancer (GC) cells.
| NcRNA | Model study | Expression | Target gene/pathway | Type of ncRNA | Biological function | References |
|---|---|---|---|---|---|---|
| miR-21 | In vitro | Up | PDCD4 | Oncogene | Participates in balance of oxidation and antioxidant system in patients with GC | Tu et al. (2014) |
| miR-622 | Bioinformatics, in vitro and in vivo | Down | NUAK1/p-protein kinase B (Akt) axis | Tumor suppressor | Decreases GC cell proliferation and migration but increases oxidative stress and inhibits the development of tumor | Yang et al. (2022) |
| miR-448 | In vitro | Up | KDM2B | Oncogene | Promotes glycolytic metabolism of GC. Significantly associated with poor clinical outcomes of GC patients | Hong et al. (2016) |
| THUMPD3-AS1 | In vitro | Down | miR-1252-3p and CXCL17 | Tumor suppressor | Inhibits proliferation, migration, invasion and ROS accumulation of GC cells | Tan et al. (2022) |
| lncRNA LINC00242 | In vitro and in vivo | Up | miR-1-3p/G6PD axis | Oncogene | Promotes cell proliferation and aerobic glycolysis and relieve the tumorigenesis | Deng et al. (2021) |
| lncRNA H19 | Bioinformatics, in vitro and in vivo | Up | miR-519d-3p/LDHA axis | Oncogene | Promotes aerobic glycolysis, proliferation, and immune escape of GC cells | Sun et al. (2021) |
| circSLAMF6 | In vitro and in vivo | Up | miR-204-5p/MYH9 axis | Oncogene | Promotes cell glycolysis, migration, and invasion of GC cells | Fang et al. (2020) |
| circDNMT1 | In vitro and in vivo | Up | miR-576-3p/HIF-1α axis | Oncogene | Promotes the proliferation, migration, invasion and glycolysis of GC cells. Promotes malignant behaviors and metabolic reprogramming of GC | Li et al. (2022) |
| circBFAR | In vitro and in vivo | Up | miR-513a-3p/HK2 axis | Oncogene | Promotes proliferation and glycolysis in GC | Wang et al. (2021b) |
Abbreviations: THUMPD3-AS1, THUMPD3 antisense RNA, one; LINC00242, Long intergenic non-protein coding RNA, 242; circDNMT1, circRNA DNA, methyltransferase one; circBFAR, circular RNA, bifunctional apoptosis regulator (circBFAR); PDCD4, Programmed cell death protein 4; NUAK1, Novel (nua) kinase family one; KDM2B, Human Lysine-specific demethylase 2B; CXCL17, Chemokine (C-X-C motif) ligand 17; G6PD, Glucose-6-phosphate dehydrogenase; LDHA, Lactate dehydrogenase A; MYH9, Myosin heavy chain nine; HIF-1α, Hypoxia-inducible factor 1-alpha.
8 Clinical perspective of ncRNAs in GC radiotherapy
There are suggestions that the direct involvement of miRNAs, lncRNAs, and circRNAs in GC radiosensitivity is likely to be applied in clinical practice in the near future. And this possibility may involve many steps. First, before radiotherapy is given to patients with GC, it will be necessary to assess the range of expression changes in radiodependent miRNAs, lncRNAs, and circRNAs in human biofluids (e.g., whole blood, plasma/serum, or gastric juice), to in order to: 1) predict the response to radiation of each specific patient, 2) determine the individual radiation dose, and 3) minimize acute and latent damage to normal cells/tissues (Das et al., 2019; May et al., 2021). Secondly, in the course of radiotherapy, checking the expression change in radiodependent miRNAs, lncRNAs, and circRNAs in biological fluids and changing the expression of a number of certain miRNAs, lncRNAs, and circRNAs among radiodependent ncRNAs can help to effectively achieve the desired effect of radiation therapy and further increase radiosensitivity of GC. Thirdly, during the period of radiotherapy, radiation therapy itself can be combined with chemotherapy drugs (e.g., oxaliplatin (FLO) or cisplatin (FLP)), small molecule inhibitors (e.g., tyrosine kinase inhibitors) and drugs that target specific miRNAs, lncRNAs, and circRNAs to enhance the genetic instability of cancer cells, increase the rate of destruction of radiation, and enhance the overall effect of radiotherapy (Song et al., 2017; Patel and Cecchini, 2020; Fong et al., 2022). And fourth, when radiotherapy is completed, determining the expression of so-called predictive miRNAs, lncRNAs, and circRNAs in body fluids can help control the therapeutic effect of radiation and reduce the risk of metastasis and recurrence of GC (Figure 4) (Wei et al., 2020). In addition, the discovery of the role of miRNAs, lncRNAs, and circRNAs, as well as their interaction with each other, in the regulation of GC radiosensitivity increases the likelihood that these ncRNAs, in particular radio-dependent ones, will provide a promising direction in the clinical practice of prevention, diagnosis, prognosis and treatment of GC.
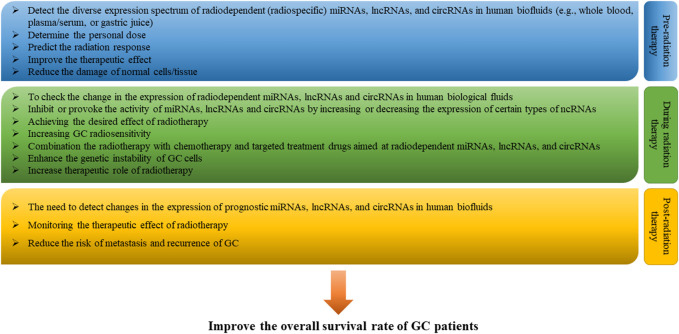
FIGURE 4.
Clinical perspective of microRNAs (miRNAs), long non-coding RNAs (lncRNAs) and circular RNAs (circRNAs) in gastric cancer (GC) radiotherapy. In the future, in clinical practice with the use of radiotherapy, it is possible to assess and monitor the range of changes in the expression of radiodependent miRNAs, lncRNAs, and circRNAs in human biofluids (e.g., whole blood, plasma/serum, or gastric juice) pre-radiation therapy, during radiation therapy, and post-radiation therapy periods in order to enhance the therapeutic effect of radiotherapy and improve the overall survival of GC patients.
Advances in fluid biopsy, that is, the non-invasive detection of radiospecific miRNAs, lncRNAs and circRNAs as biomarkers in biological fluids to assess response to GC radiotherapy, are entirely possible. In addition, the potential to improve the radiotherapeutic effect by activating or inhibiting the expression of certain miRNAs, lncRNAs and circRNAs and downstream target genes is extremely promising.
9 Conclusion
To sum up, ncRNAs may play a role in regulating the sensitivity of GC to radiation therapy by impacting important biological processes such as DNA damage response, cell stemness, apoptosis, EGFR activation, EMT, and oxygen adaptive metabolism. This suggests new opportunities for research to further investigate the impact of ncRNA dysregulation on the radiation sensitivity of GC. Further studies are needed to explore the potential role of ncRNA in regulating GC radiation sensitivity in-depth, and to develop a prediction model and screening system for ncRNAs to regulate radiation sensitivity of GC, which can bring new hope to improve the prognosis of GC patients. Overall, the role of ncRNA in the development and progression of tumors is a current area of focus in tumor biology research. Targeting ncRNA may be an effective method to reduce the resistance of GC to radiation therapy, which can help improve the effectiveness of radiation therapy for GC patients and provide new ideas and strategies for GC radiation therapy.
Funding Statement
This work was supported by the Bashkir State Medical University Strategic Academic Leadership Program (PRIORITY-2030) and The Science and Technology Bureau of Banan District, Chongqing [grant number 2021-36].
Author contributions
MU: Conceptualization and writing—Original draft. IG: Writing—Review and editing, investigation, project administration, and resources.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The reviewer GY declared a past co-authorship with the authors AB, AS, TI, IG to the handling editor.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
- Abdi E., Latifi-Navid S., Abdi F., Taherian-Esfahani Z. (2020). Emerging circulating MiRNAs and LncRNAs in upper gastrointestinal cancers. Expert Rev. Mol. Diagn 20 (11), 1121–1138. 10.1080/14737159.2020.1842199 [DOI] [PubMed] [Google Scholar]
- Beilerli A., Gareev I., Beylerli O., Yang G., Pavlov V., Aliev G., et al. (2022). Circular RNAs as biomarkers and therapeutic targets in cancer. Semin. Cancer Biol. 83, 242–252. 10.1016/j.semcancer.2020.12.026 [DOI] [PubMed] [Google Scholar]
- Beylerli O., Khasanov D., Gareev I., Valitov E., Sokhatskii A., Wang C., et al. (2021). Differential non-coding RNAs expression profiles of invasive and non-invasive pituitary adenomas. Noncoding RNA Res. 6 (3), 115–122. 10.1016/j.ncrna.2021.06.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlomagno N., Incollingo P., Tammaro V., Peluso G., Rupealta N., Chiacchio G., et al. (2017). Diagnostic, predictive, prognostic, and therapeutic molecular biomarkers in third millennium: A breakthrough in gastric cancer. Biomed. Res. Int. 2017, 7869802. 10.1155/2017/7869802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen N., Han X., Yin B., Bai X., Wang Y. (2021). FGD5 facilitates tumor growth by regulating EGFR ubiquitination in gastric cancer. Biochem. Biophys. Res. Commun. 562, 43–49. 10.1016/j.bbrc.2021.04.106 [DOI] [PubMed] [Google Scholar]
- Chen Z., Wu J., Huang W., Peng J., Ye J., Yang L., et al. (2018). Long non-coding RNA RP11-789C1.1 suppresses epithelial to mesenchymal transition in gastric cancer through the RP11-789C1.1/MiR-5003/E-cadherin Axis. Cell Physiol. Biochem. 47 (6), 2432–2444. 10.1159/000491617 [DOI] [PubMed] [Google Scholar]
- Chun-Zhi Z., Lei H., An-Ling Z., Yan-Chao F., Xiao Y., Guang-Xiu W., et al. (2010). MicroRNA-221 and microRNA-222 regulate gastric carcinoma cell proliferation and radioresistance by targeting PTEN. BMC Cancer 10, 367. 10.1186/1471-2407-10-367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Souza L. C., Mishra S., Chakraborty A., Shekher A., Sharma A., Gupta S. C. (2020). Oxidative stress and cancer development: Are noncoding RNAs the missing links? Antioxid. Redox Signal 33 (17), 1209–1229. 10.1089/ars.2019.7987 [DOI] [PubMed] [Google Scholar]
- Das S., Ansel K. M., Bitzer M., Breakefield X. O., Charest A., Galas D. J., et al. Extracellular RNA Communication Consortium (2019). The extracellular RNA communication consortium: Establishing foundational knowledge and technologies for extracellular RNA research. Cell 177 (2), 231–242. 10.1016/j.cell.2019.03.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng P., Li K., Gu F., Zhang T., Zhao W., Sun M., et al. (2021). LINC00242/miR-1-3p/G6PD axis regulates Warburg effect and affects gastric cancer proliferation and apoptosis. Mol. Med. 27 (1), 9. 10.1186/s10020-020-00259-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dragovich T., Campen C. (2009). Anti-EGFR-targeted therapy for esophageal and gastric cancers: An evolving concept. J. Oncol. 2009, 804108. 10.1155/2009/804108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan D., Ren B., Yang X., Liu J., Zhang Z. (2016). Upregulation of miR-501-5p activates the wnt/β-catenin signaling pathway and enhances stem cell-like phenotype in gastric cancer. J. Exp. Clin. Cancer Res. 35 (1), 177. 10.1186/s13046-016-0432-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang X., Bai Y., Zhang L., Ding S. (2020). Silencing circSLAMF6 represses cell glycolysis, migration, and invasion by regulating the miR-204-5p/MYH9 axis in gastric cancer under hypoxia. Biosci. Rep. 40 (6), BSR20201275. 10.1042/BSR20201275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng W., Ding Y., Zong W., Ju S. (2019). Non-coding RNAs in regulating gastric cancer metastasis. Clin. Chim. Acta 496, 125–133. 10.1016/j.cca.2019.07.003 [DOI] [PubMed] [Google Scholar]
- Fong C., Johnston E., Starling N. (2022). Neoadjuvant and adjuvant therapy approaches to gastric cancer. Curr. Treat. Options Oncol. 23 (9), 1247–1268. 10.1007/s11864-022-01004-9 [DOI] [PubMed] [Google Scholar]
- Foo M., Crosby T., Rackley T., Leong T. (2014). Role of (chemo)-radiotherapy in resectable gastric cancer. Clin. Oncol. R. Coll. Radiol. 26 (9), 541–550. 10.1016/j.clon.2014.06.004 [DOI] [PubMed] [Google Scholar]
- Gareev I., Beylerli O., Liang Y., Xiang H., Liu C., Xu X., et al. (2021). The role of MicroRNAs in therapeutic resistance of malignant primary brain tumors. Front. Cell Dev. Biol. 9, 740303. 10.3389/fcell.2021.740303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gareev I., Beylerli O., Yang G., Sun J., Pavlov V., Izmailov A., et al. (2020). The current state of MiRNAs as biomarkers and therapeutic tools. Clin. Exp. Med. 20 (3), 349–359. 10.1007/s10238-020-00627-2 [DOI] [PubMed] [Google Scholar]
- Guo F., Guo R., Zhang L. (2022). Downregulation of lncRNA FOXD2-AS1 confers radiosensitivity to gastric cancer cells via miR-1913/setd1a Axis. Cytogenet Genome Res. 162 (1-2), 10–27. 10.1159/000522653 [DOI] [PubMed] [Google Scholar]
- Gupta S., Panda P. K., Luo W., Hashimoto R. F., Ahuja R. (2022). Network analysis reveals that the tumor suppressor lncRNA GAS5 acts as a double-edged sword in response to DNA damage in gastric cancer. Sci. Rep. 12 (1), 18312. 10.1038/s41598-022-21492-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- He J., Hua J., Ding N., Xu S., Sun R., Zhou G., et al. (2014). Modulation of microRNAs by ionizing radiation in human gastric cancer. Oncol. Rep. 32 (2), 787–793. 10.3892/or.2014.3246 [DOI] [PubMed] [Google Scholar]
- Hong L., Wang H., Wang J., Wei S., Zhang F., Han J., et al. (2019). LncRNA PTCSC3 inhibits tumor growth and cancer cell stemness in gastric cancer by interacting with lncRNA linc-pint. Cancer Manag. Res. 11, 10393–10399. 10.2147/CMAR.S231369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong X., Xu Y., Qiu X., Zhu Y., Feng X., Ding Z., et al. (2016). MiR-448 promotes glycolytic metabolism of gastric cancer by downregulating KDM2B. Oncotarget 7 (16), 22092–22102. 10.18632/oncotarget.8020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu H., Zhao X., Jin Z., Hou M. (2015). Hsa-let-7g miRNA regulates the anti-tumor effects of gastric cancer cells under oxidative stress through the expression of DDR genes. J. Toxicol. Sci. 40 (3), 329–338. 10.2131/jts.40.329 [DOI] [PubMed] [Google Scholar]
- Huang X., Li Z., Zhang Q., Wang W., Li B., Wang L., et al. (2019). Circular RNA AKT3 upregulates PIK3R1 to enhance cisplatin resistance in gastric cancer via miR-198 suppression. Mol. Cancer 18 (1), 71. 10.1186/s12943-019-0969-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong W., Yin G., Zheng S., Liu X., Zhu A., Yu P., et al. (2021). Long noncoding RNA (lncRNA) HOTAIR: Pathogenic roles and therapeutic opportunities in gastric cancer. Genes Dis. 9 (5), 1269–1280. 10.1016/j.gendis.2021.07.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazăr D. C., Tăban S., Cornianu M., Faur A., Goldiş A. (2016). New advances in targeted gastric cancer treatment. World J. Gastroenterol. 22 (30), 6776–6799. 10.3748/wjg.v22.i30.6776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei Z. N., Teng Q. X., Tian Q., Chen W., Xie Y., Wu K., et al. (2022). Signaling pathways and therapeutic interventions in gastric cancer. Signal Transduct. Target Ther. 7 (1), 358. 10.1038/s41392-022-01190-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leong T. (2005). Chemotherapy and radiotherapy in the management of gastric cancer. Curr. Opin. Gastroenterol. 21 (6), 673–678. 10.1097/01.mog.0000179833.28158.b7 [DOI] [PubMed] [Google Scholar]
- Li H., Cao B., Zhao R., Li T., Xu X., Cui H., et al. (2022). circDNMT1 promotes malignant progression of gastric cancer through targeting miR-576-3p/hypoxia inducible factor-1 alpha Axis. Front. Oncol. 12, 817192. 10.3389/fonc.2022.817192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Jia Y., Wang Y. (2019). Targeting HIF-1α signaling pathway for gastric cancer treatment. Pharmazie 74 (1), 3–7. 10.1691/ph.2019.8674 [DOI] [PubMed] [Google Scholar]
- Li T., Mo X., Fu L., Xiao B., Guo J. (2016). Molecular mechanisms of long noncoding RNAs on gastric cancer. Oncotarget 7 (8), 8601–8612. 10.18632/oncotarget.6926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H. T., Liu S., Liu L., Ma R. R., Gao P. (2018). EGR1-Mediated transcription of lncRNA-hnf1a-AS1 promotes cell-cycle progression in gastric cancer. Cancer Res. 78 (20), 5877–5890. 10.1158/0008-5472.CAN-18-1011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J., Wang G., Zhao J., Liu X., Zhang K., Gong G., et al. (2022). LncRNA H19 promoted the epithelial to mesenchymal transition and metastasis in gastric cancer via activating wnt/β-catenin signaling. Dig. Dis. 40 (4), 436–447. 10.1159/000518627 [DOI] [PubMed] [Google Scholar]
- Liu J., Yan S., Hu J., Ding D., Liu Y., Li X., et al. (2021). MiRNA-4537 functions as a tumor suppressor in gastric cancer and increases the radiosensitivity of gastric cancer cells. Bioengineered 12 (1), 8457–8467. 10.1080/21655979.2021.1982843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J., Yang H., Deng J., Jiang R., Meng E., Wu H. (2023). CircRPPH1 promotes the stemness of gastric cancer cells by targeting miR-375/SLC7A11 axis. Environ. Toxicol. 38 (1), 115–125. 10.1002/tox.23668 [DOI] [PubMed] [Google Scholar]
- Liu X., Ma R., Yi B., Riker A. I., Xi Y. (2021). MicroRNAs are involved in the development and progression of gastric cancer. Acta Pharmacol. Sin. 42 (7), 1018–1026. 10.1038/s41401-020-00540-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu E., Gareev I., Yuan C., Liang Y., Sun J., Chen X., et al. (2022). The mechanisms of current platinum anticancer drug resistance in the glioma. Curr. Pharm. Des. 28 (23), 1863–1869. 10.2174/1381612828666220607105746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machlowska J., Baj J., Sitarz M., Maciejewski R., Sitarz R. (2020). Gastric cancer: Epidemiology, risk factors, classification, genomic characteristics and treatment strategies. Int. J. Mol. Sci. 21 (11), 4012. 10.3390/ijms21114012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manoel-Caetano F. S., Rossi A. F. T., Calvet de Morais G., Severino F. E., Silva A. E. (2019). Upregulation of the APE1 and H2AX genes and miRNAs involved in DNA damage response and repair in gastric cancer. Genes Dis. 6 (2), 176–184. 10.1016/j.gendis.2019.03.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao Y., Tie Y., Du J., He J. (2019). Linc00152 promotes the proliferation of gastric cancer cells by regulating b-cell lymphoma-2. J. Cell Biochem. 120 (3), 3747–3756. 10.1002/jcb.27655 [DOI] [PubMed] [Google Scholar]
- May J. M., Bylicky M., Chopra S., Coleman C. N., Aryankalayil M. J. (2021). Long and short non-coding RNA and radiation response: A review. Transl. Res. 233, 162–179. 10.1016/j.trsl.2021.02.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molina-Castro S., Pereira-Marques J., Figueiredo C., Machado J. C., Varon C. (2017). Gastric cancer: Basic aspects. Helicobacter 22, e12412. 10.1111/hel.12412 [DOI] [PubMed] [Google Scholar]
- Ni H., Qin H., Sun C., Liu Y., Ruan G., Guo Q., et al. (2021). MiR-375 reduces the stemness of gastric cancer cells through triggering ferroptosis. Stem Cell Res. Ther. 12 (1), 325. 10.1186/s13287-021-02394-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nickoloff J. A., Sharma N., Taylor L. (2020). Clustered DNA double-strand breaks: Biological effects and relevance to cancer radiotherapy. Genes (Basel) 11 (1), 99. 10.3390/genes11010099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nie W., Ge H. J., Yang X. Q., Sun X., Huang H., Tao X., et al. (2016). LncRNA-UCA1 exerts oncogenic functions in non-small cell lung cancer by targeting miR-193a-3p. Cancer Lett. 371 (1), 99–106. 10.1016/j.canlet.2015.11.024 [DOI] [PubMed] [Google Scholar]
- Pasechnikov V., Chukov S., Fedorov E., Kikuste I., Leja M. (2014). Gastric cancer: Prevention, screening and early diagnosis. World J. Gastroenterol. 20 (38), 13842–13862. 10.3748/wjg.v20.i38.13842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel T. H., Cecchini M. (2020). Targeted therapies in advanced gastric cancer. Curr. Treat. Options Oncol. 21 (9), 70. 10.1007/s11864-020-00774-4 [DOI] [PubMed] [Google Scholar]
- Qian Y., Song W., Wu X., Hou G., Wang H., Hang X., et al. (2021). DLX6 antisense RNA 1 modulates glucose metabolism and cell growth in gastric cancer by targeting microRNA-4290. Dig. Dis. Sci. 66 (2), 460–473. 10.1007/s10620-020-06223-4 [DOI] [PubMed] [Google Scholar]
- Qin H., Li X., Zhang W., Ding Z. (2022). LncRNA OGFRP1 promotes cell proliferation and suppresses cell radiosensitivity in gastric cancer by targeting the miR-149-5p/MAP3K3 axis. J. Mol. Histol. 53 (2), 257–271. 10.1007/s10735-022-10058-w [DOI] [PubMed] [Google Scholar]
- Qin Y., Zhuang S., Wen J., Zheng K. (2018). Long non-coding RNA MDC1-AS inhibits human gastric cancer cell proliferation and metastasis through an MDC1-dependent mechanism. Exp. Ther. Med. 15 (1), 191–197. 10.3892/etm.2017.5370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruan T., Liu W., Tao K., Wu C. (2020). A review of research progress in multidrug-resistance mechanisms in gastric cancer. Onco Targets Ther. 13, 1797–1807. 10.2147/OTT.S239336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slack F. J., Chinnaiyan A. M. (2019). The role of non-coding RNAs in oncology. Cell 179 (5), 1033–1055. 10.1016/j.cell.2019.10.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyth E. C., Nilsson M., Grabsch H. I., van Grieken N. C., Lordick F. (2020). Gastric cancer. Lancet 396 (10251), 635–648. 10.1016/S0140-6736(20)31288-5 [DOI] [PubMed] [Google Scholar]
- Song H., Shi L., Xu Y., Xu T., Fan R., Cao M., et al. (2019). BRD4 promotes the stemness of gastric cancer cells via attenuating miR-216a-3p-mediated inhibition of Wnt/β-catenin signaling. Eur. J. Pharmacol. 852, 189–197. 10.1016/j.ejphar.2019.03.018 [DOI] [PubMed] [Google Scholar]
- Song H., Xu Y., Shi L., Xu T., Fan R., Cao M., et al. (2018). LncRNA THOR increases the stemness of gastric cancer cells via enhancing SOX9 mRNA stability. Biomed. Pharmacother. 108, 338–346. 10.1016/j.biopha.2018.09.057 [DOI] [PubMed] [Google Scholar]
- Song Z., Wu Y., Yang J., Yang D., Fang X. (2017). Progress in the treatment of advanced gastric cancer. Tumour Biol. 39 (7), 1010428317714626. 10.1177/1010428317714626 [DOI] [PubMed] [Google Scholar]
- Sun L., Li J., Yan W., Yao Z., Wang R., Zhou X., et al. (2021). H19 promotes aerobic glycolysis, proliferation, and immune escape of gastric cancer cells through the microRNA-519d-3p/lactate dehydrogenase A axis. Cancer Sci. 112 (6), 2245–2259. 10.1111/cas.14896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Q., Li J., Li F., Li H., Bei S., Zhang X., et al. (2019). LncRNA LOXL1-AS1 facilitates the tumorigenesis and stemness of gastric carcinoma via regulation of miR-708-5p/USF1 pathway. Cell Prolif. 52 (6), e12687. 10.1111/cpr.12687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taghehchian N., Alemohammad R., Farshchian M., Asoodeh A., Abbaszadegan M. R. (2022). Inhibitory role of LINC00332 in gastric cancer progression through regulating cell EMT and stemness. Life Sci. 305, 120759. 10.1016/j.lfs.2022.120759 [DOI] [PubMed] [Google Scholar]
- Tan Y., Liu L., Zhang X., Xue Y., Gao J., Zhao J., et al. (2022). THUMPD3-AS1 is correlated with gastric cancer and regulates cell function through miR-1252-3p and CXCL17. Crit. Rev. Eukaryot. Gene Expr. 32 (8), 69–80. 10.1615/CritRevEukaryotGeneExpr.2022042848 [DOI] [PubMed] [Google Scholar]
- Tanprasert P., Limpakan Yamada S., Chattipakorn S. C., Chattipakorn N., Shinlapawittayatorn K. (2022). Targeting mitochondria as a therapeutic anti-gastric cancer approach. Apoptosis 27 (3-4), 163–183. 10.1007/s10495-022-01709-0 [DOI] [PubMed] [Google Scholar]
- Tsai K. W., Hu L. Y., Wu C. W., Li S. C., Lai C. H., Kao H. W., et al. (2010). Epigenetic regulation of miR-196b expression in gastric cancer. Genes Chromosom. Cancer 49 (11), 969–980. 10.1002/gcc.20804 [DOI] [PubMed] [Google Scholar]
- Tu H., Sun H., Lin Y., Ding J., Nan K., Li Z., et al. (2014). Oxidative stress upregulates PDCD4 expression in patients with gastric cancer via miR-21. Curr. Pharm. Des. 20 (11), 1917–1923. 10.2174/13816128113199990547 [DOI] [PubMed] [Google Scholar]
- Wang D., Jiang X., Liu Y., Cao G., Zhang X., Kuang Y. (2021). Circular RNA circ_HN1 facilitates gastric cancer progression through modulation of the miR-302b-3p/ROCK2 axis. Mol. Cell Biochem. 476 (1), 199–212. 10.1007/s11010-020-03897-2 [DOI] [PubMed] [Google Scholar]
- Wang J., Song Y. X., Wang Z. N. (2015). Non-coding RNAs in gastric cancer. Gene 560 (1), 1–8. 10.1016/j.gene.2015.02.004 [DOI] [PubMed] [Google Scholar]
- Wang J., Yu Z., Wang J., Shen Y., Qiu J., Zhuang Z. (2020). LncRNA NUTM2A-AS1 positively modulates TET1 and HIF-1A to enhance gastric cancer tumorigenesis and drug resistance by sponging miR-376a. Cancer Med. 9 (24), 9499–9510. 10.1002/cam4.3544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Wang Z., Wang L. (2022). Long noncoding RNA Solute carrier family 25 member 21 antisense RNA 1 inhibits cell malignant behaviors and enhances radiosensitivity of gastric cancer cells by upregulating synuclein gamma expression. Tohoku J. Exp. Med. 257 (3), 225–239. 10.1620/tjem.2022.J029 [DOI] [PubMed] [Google Scholar]
- Wang L., Zhao Y., Xiong W., Ye W., Zhao W., Hua Y. (2019). MicroRNA-449a is downregulated in cervical cancer and inhibits proliferation, migration, and invasion. Oncol. Res. Treat. 42 (11), 564–571. 10.1159/000502122 [DOI] [PubMed] [Google Scholar]
- Wang M., Xie C. (2022). DNA damage repair and current therapeutic approaches in gastric cancer: A comprehensive review. Front. Genet. 13, 931866. 10.3389/fgene.2022.931866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D., Cao B., Zhao R., Li H., Wei B., Dai G. (2021). Knockdown of circBFAR inhibits proliferation and glycolysis in gastric cancer by sponging miR-513a-3p/hexokinase 2 axis. Biochem. Biophys. Res. Commun. 560, 80–86. 10.1016/j.bbrc.2021.04.131 [DOI] [PubMed] [Google Scholar]
- Wei L., Sun J., Zhang N., Zheng Y., Wang X., Lv L., et al. (2020). Noncoding RNAs in gastric cancer: Implications for drug resistance. Mol. Cancer 19 (1), 62. 10.1186/s12943-020-01185-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei Y., Wang Y., Zang A., Wang Z., Fang G., Hong D. (2019). MiR-4766-5p inhibits the development and progression of gastric cancer by targeting NKAP. Onco Targets Ther. 12, 8525–8536. 10.2147/OTT.S220234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia Y., Lv J., Jiang T., Li B., Li Y., He Z., et al. (2021). CircFAM73A promotes the cancer stem cell-like properties of gastric cancer through the miR-490-3p/HMGA2 positive feedback loop and HNRNPK-mediated β-catenin stabilization. J. Exp. Clin. Cancer Res. 40 (1), 103. 10.1186/s13046-021-01896-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie J., Chen M., Zhou J., Mo M. S., Zhu L. H., Liu Y. P., et al. (2014). miR-7 inhibits the invasion and metastasis of gastric cancer cells by suppressing epidermal growth factor receptor expression. Oncol. Rep. 31 (4), 1715–1722. 10.3892/or.2014.3052 [DOI] [PubMed] [Google Scholar]
- Xing Y., Chen H., Guo Z., Zhou X. (2022). Circular RNA circ0007360 attenuates gastric cancer progression by altering the miR-762/IRF7 Axis. Front. Cell Dev. Biol. 10, 789073. 10.3389/fcell.2022.789073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong S., Hu M., Li C., Zhou X., Chen H. (2019). Role of miR-34 in gastric cancer: From bench to bedside (Review). Oncol. Rep. 42 (5), 1635–1646. 10.3892/or.2019.7280 [DOI] [PubMed] [Google Scholar]
- Xu W., Ding M., Wang B., Cai Y., Guo C., Yuan C. (2021). Molecular mechanism of the canonical oncogenic lncRNA MALAT1 in gastric cancer. Curr. Med. Chem. 28 (42), 8800–8809. 10.2174/0929867328666210521213352 [DOI] [PubMed] [Google Scholar]
- Xu X., Liang Y., Gareev I., Liang Y., Liu R., Wang N., et al. (2022). LncRNA as potential biomarker and therapeutic target in glioma. Mol. Biol. Rep. 50, 841–851. 10.1007/s11033-022-08056-y [DOI] [PubMed] [Google Scholar]
- Xu Y., Deng Y., Yan X., Zhou T. (2011). Targeting miR-375 in gastric cancer. Expert Opin. Ther. Targets 15 (8), 961–972. 10.1517/14728222.2011.581232 [DOI] [PubMed] [Google Scholar]
- Yan H., Bu P. (2021). Non-coding RNA in cancer. Essays Biochem. 65 (4), 625–639. 10.1042/EBC20200032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J., Lu J., Yin N., Sun J., Pu J., Zang J. (2022). miR-622 counteracts the NUAK1-induced gastric cancer cell proliferation and the antioxidative stress. Dis. Markers 2022, 9616764. 10.1155/2022/9616764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Q., Zhang R. W., Sui P. C., He H. T., Ding L. (2015). Dysregulation of non-coding RNAs in gastric cancer. World J. Gastroenterol. 21 (39), 10956–10981. 10.3748/wjg.v21.i39.10956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao Y., Ma J., Xue Y., Wang P., Li Z., Li Z., et al. (2015). MiR-449a exerts tumor-suppressive functions in human glioblastoma by targeting Myc-associated zinc-finger protein. Mol. Oncol. 9 (3), 640–656. 10.1016/j.molonc.2014.11.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasui H., Ogura A., Asanuma T., Matsuda A., Kashiwakura I., Kuwabara M., et al. (2008). Inhibition of HIF-1alpha by the anticancer drug TAS106 enhances X-ray-induced apoptosis in vitro and in vivo . Br. J. Cancer 99 (9), 1442–1452. 10.1038/sj.bjc.6604720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye M., Xiu L. J., Ji Q. Q., Zhang Y. C., Sun Y. W., Zhao Y., et al. (2022). Research progress in targeted therapies for gastric cancer. Int. J. Clin. Pharmacol. Ther. 60 (12), 509–514. 10.5414/CP204250 [DOI] [PubMed] [Google Scholar]
- Yuan Y., Li H., Pu W., Chen L., Guo D., Jiang H., et al. (2022). Cancer metabolism and tumor microenvironment: Fostering each other? Sci. China Life Sci. 65 (2), 236–279. 10.1007/s11427-021-1999-2 [DOI] [PubMed] [Google Scholar]
- Zhang M., Jiang D., Xie X., He Y., Lv M., Jiang X. (2019). miR-129-3p inhibits NHEJ pathway by targeting SAE1 and represses gastric cancer progression. Int. J. Clin. Exp. Pathol. 12 (5), 1539–1547. [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Peng Y., Yuan Y., Gao Y., Hu F., Wang J., et al. (2020). Histone methyltransferase SET8 is regulated by miR-192/215 and induces oncogene-induced senescence via p53-dependent DNA damage in human gastric carcinoma cells. Cell Death Dis. 11 (10), 937. 10.1038/s41419-020-03130-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Lin W., Jiang W., Wang Z. (2022). MicroRNA-18 facilitates the stemness of gastric cancer by downregulating HMGB3 though targeting Meis2. Bioengineered 13 (4), 9959–9972. 10.1080/21655979.2022.2062529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Ma M., Liu W., Ding W., Yu H. (2014). Enhanced expression of long noncoding RNA CARLo-5 is associated with the development of gastric cancer. Int. J. Clin. Exp. Pathol. 7 (12), 8471–8479. [PMC free article] [PubMed] [Google Scholar]
- Zhao X., Zhong Q., Cheng X., Wang S., Wu R., Leng X., et al. (2020). miR-449c-5p availability is antagonized by circ-NOTCH1 for MYC-induced NOTCH1 upregulation as well as tumor metastasis and stemness in gastric cancer. J. Cell Biochem. 121 (10), 4052–4063. 10.1002/jcb.29575 [DOI] [PubMed] [Google Scholar]
- Zheng F., Chen J., Zhang X., Wang Z., Chen J., Lin X., et al. (2021). The HIF-1α antisense long non-coding RNA drives a positive feedback loop of HIF-1α mediated transactivation and glycolysis. Nat. Commun. 12 (1), 1341. 10.1038/s41467-021-21535-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou K., Zhang J., Song S., Xie K., Hu X. (2022). Knockdown of Circ_0003506 impedes radioresistance, cell growth, migration and invasion in gastric cancer. Dig. Dis. Sci. 68, 128–137. 10.1007/s10620-022-07534-4 [DOI] [PubMed] [Google Scholar]
- Zhu T., Wang Z., Wang G., Hu Z., Ding H., Li R., et al. (2020). Long non-coding RNA ZFAS1 promotes the expression of EPAS1 in gastric cardia adenocarcinoma. J. Adv. Res. 28, 7–15. 10.1016/j.jare.2020.06.006 [DOI] [PMC free article] [PubMed] [Google Scholar]