A transcriptional rheostat orchestrated by RELA is found to control the expression of type I/III interferon in T cells. Unlocking IFN-I/III expression in T cells confers self-defense against HIV infection and increases the antitumor activity of CAR T cells.
Abstract
In innate immune cells, intracellular sensors such as cGAS-STING stimulate type I/III interferon (IFN) expression, which promotes antiviral defense and immune activation. However, how IFN-I/III expression is controlled in adaptive cells is poorly understood. Here, we identify a transcriptional rheostat orchestrated by RELA that confers human T cells with innate-like abilities to produce IFN-I/III. Despite intact cGAS-STING signaling, IFN-I/III responses are stunted in CD4+ T cells compared with dendritic cells or macrophages. We find that lysine residues in RELA tune the IFN-I/III response at baseline and in response to STING stimulation in CD4+ T cells. This response requires positive feedback driven by cGAS and IRF7 expression. By combining RELA with IRF3 and DNA demethylation, IFN-I/III production in CD4+ T cells reaches levels observed in dendritic cells. IFN-I/III production provides self-protection of CD4+ T cells against HIV infection and enhances the elimination of tumor cells by CAR T cells. Therefore, innate-like functions can be tuned and leveraged in human T cells.
Introduction
The classic paradigm of immunology describes a division of labor between the innate and adaptive immune cell types (Iwasaki and Medzhitov, 2010). Innate immune cells armed with germ-line encoded pathogen recognition receptors (PRRs) sense the presence of pathogens or danger and respond by producing cytokines, upregulating costimulatory molecules, and presenting antigens. Adaptive immune cells are subsequently clonally stimulated by antigenic peptide presentation to mediate helper and effector functions. Even though it is now recognized that PRRs are also expressed in adaptive cells, these cells are not generally recognized for having the capacity to efficiently detect and signal pathogens or danger the way innate immune cells do. The mechanisms that differentiate innate and adaptive cells downstream of PRR are elusive.
Type I and type III interferons (IFN) are crucial cytokines that activate antiviral defenses and contribute to inflammation downstream of PRR. IFN-I/III expression is controlled by the transcription factors NF-κB, IRF3, IRF7, ATF2, and c-Jun, which are tightly controlled by multiple posttranslational modifications (PTM) to ensure directed and timely expression of target genes. Detection of intracellular nucleic acids by PRRs—dsRNA by RIG-I and MDA5, and dsDNA by the cGAS–cGAMP–STING pathway—potently stimulates IFN-I/III expression. IFN-I is also expressed constitutively at low levels, resulting in tonic IFN signaling that is crucial for antiviral defenses (Gough et al., 2012; Schoggins et al., 2014). cGAS activation by endogenous DNA has been implicated in constitutive IFN production (Härtlova et al., 2015; Gentili et al., 2019). RELA (p65 subunit of NF-κB) is also required for autocrine IFN-I production at baseline (Basagoudanavar et al., 2011).
While antigen-presenting cells readily produce IFN-I/III in response to PRR stimulation, lymphocytes are not generally considered a significant source of IFN-I/III. For example, CD4+ T cells are not able to produce IFN-I in response to HIV infection, resulting in their inability to control virus infection, while monocyte-derived dendritic cells do so in similar conditions (Manel et al., 2010; Elsner et al., 2020). However, IFN production by T cells has been associated with desirable functional outcomes, including spontaneous resistance to HIV infection (Angin et al., 2019) and antitumor activity of chimeric antigen receptor (CAR) T cells (Zhao et al., 2015).
T cells express a wide range of IFN-inducing PRR at the protein level, including cGAS, STING, and RIG-I (Li et al., 2016; Cerboni et al., 2017). Infection with Sendai virus that activates RIG-I leads to detectable levels of IFN-I protein production by activated T cells (Manel et al., 2010). In one study, IFN-I production has been detected following cGAS–STING stimulation by electroporation of DNA or cGAMP, or following infection with a herpes simplex virus type 1 mutant (Elsner et al., 2020), while another study failed to detect IFN-I production and IFN responses after DNA transfection (Berg et al., 2014). IFN production by T cells has also been observed in pathogenic conditions. Splenic T cells with a pathogenic mutation in TREX1 produce elevated levels of IFN-I (Simpson et al., 2019). IFN-I expression in T cells has also been described in the context of cancer but the upstream signaling was not evaluated (Kagamu et al., 2020; Chen et al., 2021). PRR activation in T cells additionally leads to IFN-independent responses in T cells. Endogenous cGAS–STING-driven IFN-I signaling contributes to stemness maintenance in CD8+ T cells in the context of cancer immune responses (Li et al., 2020), while STING activation by exogenous agonists leads to cell death in murine T cells (Cerboni et al., 2017; Gulen et al., 2017; Wu et al., 2019). There exist important species-specific differences in activation and response to STING signaling between murine and human T cells. For instance, pathogenic variants in STING lead to T cell imbalances in peripheral naive and memory compartments and a block in proliferation in humans, whereas the mouse model develops a severe combined immunodeficiency phenotype (Jeremiah et al., 2014; Liu et al., 2014; Cerboni et al., 2017; Bouis et al., 2019; Luksch et al., 2019). Individually, these findings illustrate the notion that CD4+ T cells have the capacity to produce IFN-I/III, but a general mechanism to explain the regulation of IFN-I/III expression in these cells is lacking. As a result, this ability has yet to be exploited for functional applications.
Results
Restricted IFN-I/III production in human CD4+ T cells benchmarked to dendritic cells
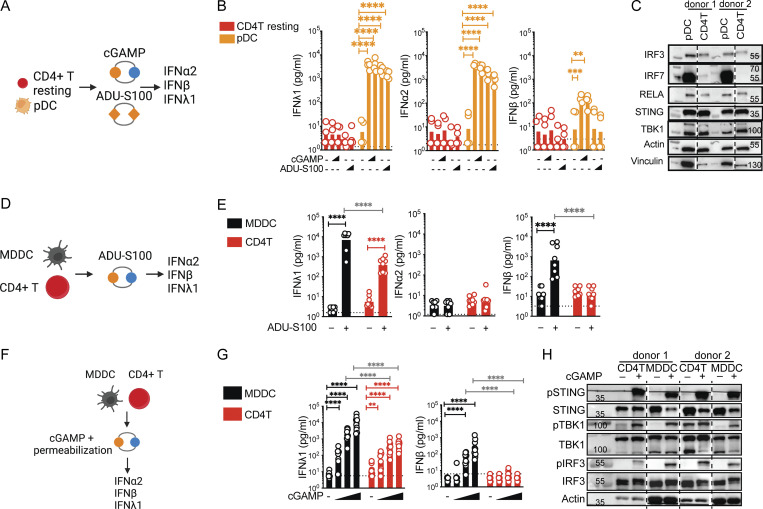
To assess the IFN response of human blood CD4+ T cells to STING stimulation, we benchmarked their response to donor-matched human blood plasmacytoid dendritic cell (pDC) subsets (Fig. 1 A). Stimulation with soluble synthetic STING ligands, namely 2′3′-cGAMP (cGAMP) or the cyclic dinucleotide (CDN) analog and clinical candidate ADU-S100 (Corrales et al., 2015), revealed that pDC produces significant amounts of IFN-III and IFN-I, while the IFN-I/III response to STING ligands in resting CD4+ T cells was undetectable (Fig. 1 B). Key signaling proteins STING, TBK1, IRF3, and RELA were expressed at similar levels in both cell types (Fig. 1 C). We noted that IRF7 was highly expressed in pDCs as reported (Honda et al., 2005), while CD4+ T cells barely expressed it. It has been previously proposed that T cell receptor (TCR) stimulation is required to detect IFN-I expression following STING stimulation of murine CD4+ T cells (Imanishi et al., 2019). Stimulation of TCR-activated human cells with ADU-S100 induced IFN-III but no detectable IFN-I (Fig. 1, D and E), while donor-matched monocyte-derived dendritic cells (MDDCs) produced both a 10-fold more IFNλ1 and high levels of IFNβ. Therefore, TCR stimulation is not sufficient to enable DC-like IFN-I production in human CD4+ T cells.
Figure 1.
IFN-I/III production is stunted in CD4+ T cells compared to dendritic cells. (A) Stimulation with extracellular STING ligands in primary blood cells, experimental outline. (B) IFN-I/III concentrations after treatment of enriched total resting CD4+ T subsets and FACS-sorted pDC, with cGAMP or ADU-S100 (1 or 2.5 µg) (n = 4 donors combined from two independent experiments). (C) Western blot of key signaling proteins involved in STING signaling and control proteins in the indicated resting primary cell types (representative of two independent experiments). (D) Delivery of STING ligands in media, experimental design. (E) IFN-I/III concentrations after stimulation of TCR-activated CD4+ T cells (CD4T) and MDDCs with ADU-S100 (2.5 µg) stimulation (n = 8 donors combined from four independent experiments). (F) Delivery of STING ligands by permeabilization, experimental design. (G) IFN-I/III concentrations after treatment of TCR-activated CD4+ T cells and MDDCs with cGAMP (0.06, 0.6, and 6 µg/ml) delivered by digitonin-mediated membrane permeabilization (n = 10 donors combined from five independent experiments). (H) Western blot of key signaling proteins involved in STING signaling and control proteins 6 h following cGAMP (6 µg/ml) stimulation in TCR-activated CD4+ T cells and MDDCs (representative of three independent experiments). Each symbol represents one donor, bars represent geometric mean, paired one-way ANOVA with Tukey’s multiple comparison test, **P ≤ 0.01, ****P ≤ 0.0001. Figure was generated using Biorender. Source data are available for this figure: SourceData F1.
DCs might be more efficient at taking up synthetic ligands than CD4+ T cells. We forced STING ligand entry from the media into cells using a well-established digitonin-mediated membrane permeabilization protocol throughout the rest of the study (Woodward et al., 2010; Fig. 1 F). Using this method, we found that CD4+ T cells remained 10- to 100-fold less capable of producing IFNλ1 mRNA and protein than MDDC, and IFNβ protein remained undetectable in CD4+ T cells (Fig. 1 G and Fig. S1 A). The phosphorylated levels of STING, TBK1, and IRF3 were identical between CD4+ T cells and MDDCs in this assay. While we do not exclude kinetic signaling differences between the cell types, these results indicate that there is no general defect in the activation of these key signaling proteins by cGAMP in CD4+ T cells (Fig. 1 H).
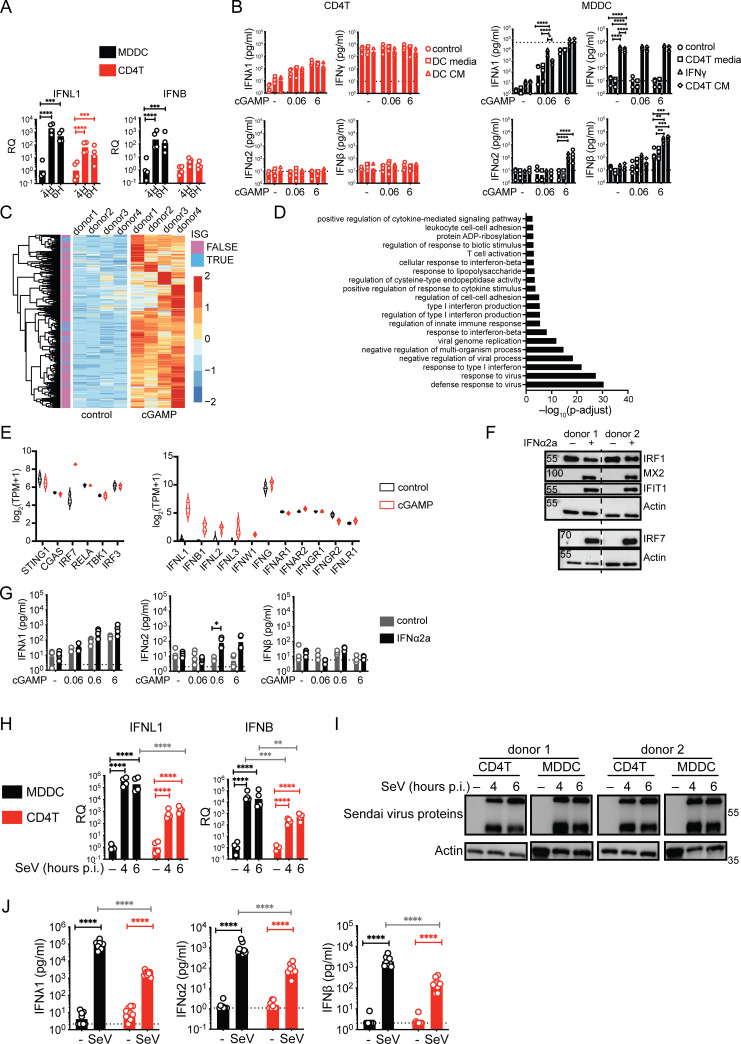
Figure S1.
Characterization of the IFN response of T cells to cGAMP stimulation, IFNα2a pre-treatment, and Sendai virus infection. (A) RT-qPCR of IFNL1 and IFNB expression following cGAMP (6 µg/ml) stimulation of TCR-activated CD4+ T cells or MDDCs at indicated time points (n = 4 donors from two independent experiments). Relative quantities (RQ) normalized to the average of controls, each symbol represents one donor, geometric mean, paired one-way ANOVA with Tukey’s multiple comparison test. (B) IFN-I/II/III concentrations following cGAMP (0.06, 6 µg/ml) stimulation of TCR-activated CD4+ T cells and MDDCs. Cells were cultured for 24 h prior to stimulation with the indicated fresh culture media, conditioned media (CM), or treated with IFNγ (10 ng/ml; n = 4 donors from two independent experiments). (C) Heat-map of upregulated genes 5 h following cGAMP (6 µg/ml) stimulation of TCR-activated CD4+ T cells. Data are shown as color-coded Z-scores of transcripts per million (TPM) values (n = 4 donors combined from two independent experiments). Annotation of ISGs is indicated. (D) Enriched biological processes among upregulated genes following cGAMP (6 µg/ml) stimulation of CD4+ T cells (n = 4 donors combined from two independent experiments). (E) Violin plots of expression levels of key proteins involved in STING-IFN signaling pathway (lines denote median and quartiles, n = 4 donors combined from two independent experiments). (F) Western blot of IFIT1, MX2, IRF1, IRF7, and actin 18 h following IFNα2a (1,000 U/ml) treatment of CD4+ T cells (representative of two independent experiments). (G) IFN-I/III concentrations following cGAMP (6 µg/ml) stimulation of CD4+ T cells pre-treated for 18 h with IFNα2a (n = 4 donors from two independent experiments, bars represent geometric mean, paired one-way ANOVA with Tukey’s multiple comparison test). (H) RT-qPCR of IFNL1 and IFNB expression following Sendai virus (SeV 200 HA/ml) infection of CD4+ T cells and MDDCs at indicated time points post-infection (p.i.; n = 4 donors from two independent experiments). RQ normalized to the average of controls, each symbol represents one donor, bars represent geometric mean, paired one-way ANOVA with Tukey’s multiple comparison test. (I) Western blot of Sendai virus proteins and actin following Sendai virus (SeV 200 HA/ml) infection of CD4+ T cells and MDDCs at indicated time points (representative of two independent experiments). (J) IFN-I/III concentrations after infection of TCR-activated CD4+ T cells and MDDCs with Sendai virus (SeV 200 HA/ml; n = 9 donors combined from five independent experiments). *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001, ****P ≤ 0.0001. Source data are available for this figure: SourceData FS1.
We also asked if different ex vivo culture conditions between the donor-matched CD4+ T cells and MDDCs could impact the IFN-I/III response. To test this possibility, STING stimulation was carried out in activated CD4+ T cells exposed to MDDC-conditioned media or in MDDCs exposed to CD4+ T cell-conditioned media. The IFN-I and IFN-III responses to cGAMP in CD4+ T cells did not change, while they further increased in MDDCs exposed to CD4+ T conditioned media (Fig. S1 B). Type II IFNγ is produced by activated CD4+ T cells, and we found that treatment of MDDCs with IFNγ was sufficient to enhance their IFN-I/III response to cGAMP. This indicates that the environment of CD4+ T cells does not contain suppressive but rather stimulatory signals for IFN-I/III production in other cells, indicative of a cell-intrinsic restriction in CD4+ T cells.
To identify intrinsic factors that could regulate the IFN-I/III response of T cells in response to cGAMP, we performed RNA-seq analysis. This revealed the enrichment of antiviral and IFN-I–related gene sets in the upregulated genes, indicating that the low IFN-I/III production by CD4+ T cells is biologically active (Fig. S1, C and D and Table S1). The transcripts for IFNL1, IFNL2, IFNL3, and IFNB were detected, but counts were low relative to other transcripts (Fig. S1 E). We did not detect changes induced by cGAMP for key signaling components or IFN receptor genes, except the IFN-stimulated gene IRF7 expression that was induced by cGAMP (Fig. S1 E). In pDCs, IRF7 dictates their ability to produce a strong IFNα response (Honda et al., 2005). To test if IRF7 induction could be sufficient to lift the restriction of IFN-I/III production in CD4+ T cells, we treated cells with IFNα2a prior to cGAMP stimulation in CD4+ T cells (Fig. S1, F and G). This treatment led to an increase in IFNα2 production by CD4+ T cells in response to high doses of cGAMP, but no induction of IFNβ and no increase of IFNλ1. Therefore, while increasing IRF7 can have a positive impact, it does not lift the restriction.
To determine if the IFN-I/III restriction was specific to STING activation, activated CD4+ T cells were infected with Sendai virus sensed by the RIG-I-MAVS pathway. Sendai virus induced both IFN-III and IFN-I in T cells; however, the magnitude was 10- to 100-fold lower in comparison to donor-matched MDDCs expressing similar levels of viral proteins (Fig. S1, H–J). These results indicate that human CD4+ T cells have the potential to produce IFN-I/III, but that it is restricted in response to external stimuli compared with DCs. This restriction occurs downstream of the phosphorylation events of STING–TBK1–IRF3 proteins.
RELA is a key regulator of IFN expression in CD4+ T cells
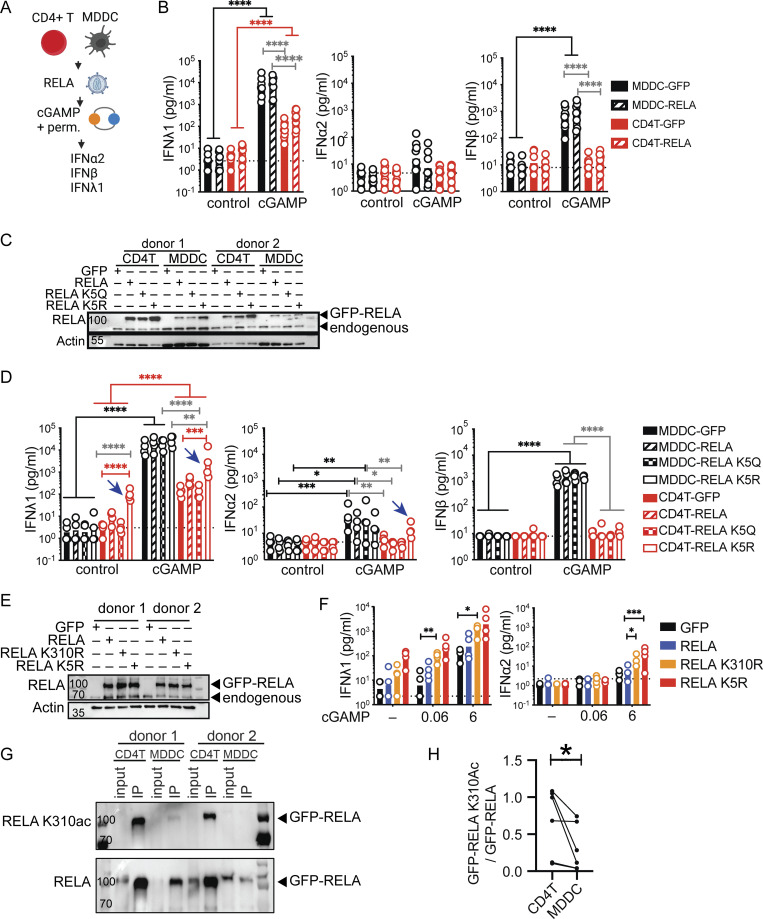
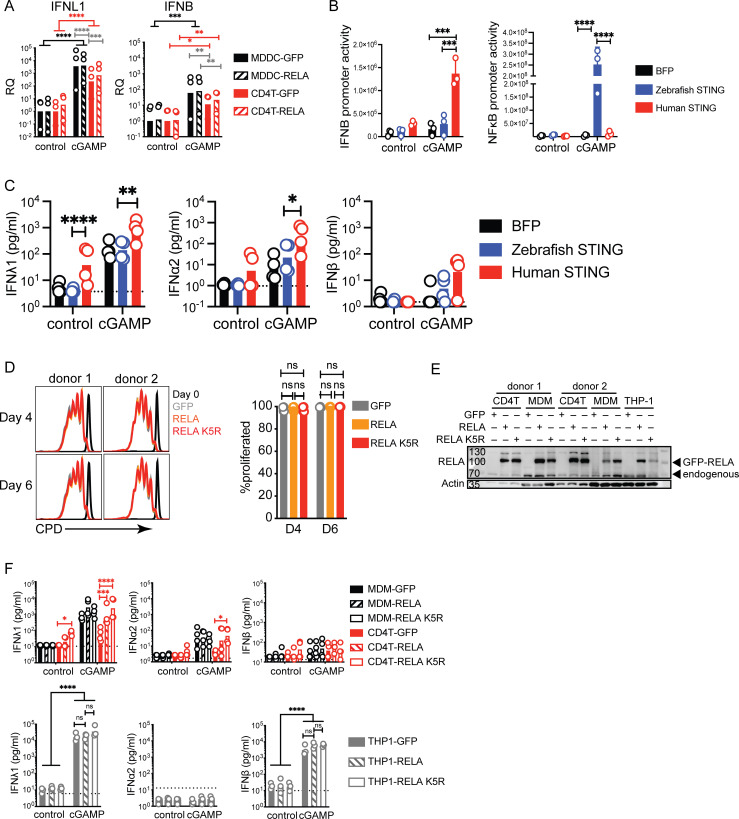
We hypothesized that the regulation or availability of transcription factors could be restricting IFN expression in CD4+ T cells. In model systems, overexpression of NF-κB subunits is sufficient to induce the IFN-β promoter (Apostolou and Thanos, 2008). Therefore, we increased the availability of RELA, a key NF-κB subunit that directly binds DNA implicated in IFN-I expression, through overexpression in CD4+ T cells and MDDCs (Freaney et al., 2013; Fig. 2 A). However, simply increasing RELA in CD4+ T cells and MDDCs did not significantly enhance the IFN-I/III response to cGAMP neither at RNA nor protein levels (Fig. 2 B and Fig. S2 A). It was recently demonstrated that the expression of zebrafish STING strongly activates NF-κB when expressed in human cells (de Oliveira Mann et al., 2019). We, therefore, tested if zebrafish STING would enhance IFN-I/III expression by CD4+ T cells. We confirmed that zebrafish STING strongly activates an NF-κB reporter in response to cGAMP but not an IFNB reporter (Fig. S2 B). In CD4+ T cells, expression of human STING enhanced IFN-I/III production, in agreement with previous results (Cerboni et al., 2017; Fig. S2 C). In contrast, zebrafish STING, however, did not enhance IFN-I/III production, despite the presence of endogenous human STING. Therefore, increasing NF-κB activation by STING is not sufficient to enhance the production of IFN-I/III by T cells in response to cGAMP.
Figure 2.
RELA enhances IFN-I/III expression in CD4+ T cells. (A) STING ligand stimulation in cells transduced with RELA, experimental outline. (B) IFN-I/III concentration following cGAMP (6 µg/ml) stimulation of TCR-activated CD4+ T cells and MDDCs transduced with control (GFP) or RELA lentivectors (n = 8 donors combined from four individual experiments). (C) Western blot of RELA and actin in CD4+ T cells and MDDCs transduced with either control, RELA, RELA K5Q, or RELA K5R (representative of two independent experiments). (D) IFN-I/III concentration after cGAMP (6 µg/ml) stimulation of CD4+ T and MDDCs transduced with control (GFP), RELA, RELA K5Q, or RELA K5R (n = 4 donors combined from two individual experiments). (E) Western blot of RELA and actin 4 d after lentiviral transduction of CD4+ T cells as indicated (representative of two independent experiments). (F) IFN-I/III concentration following cGAMP (0.06, 6 µg/ml) stimulation of CD4+ T cells transduced with control (GFP), RELA, RELA K310R, or RELA K5R (n = 4 donors combined from two independent experiments). (G) Western blot of RELA K310ac and total RELA in CD4+ T cells and MDDCs following immunoprecipitation of GFP-RELA 4 or 5 d after lentiviral transduction (representative of three independent experiments). (H) Quantification of the ratio of GFP-RELA K310ac over total GPP-RELA (n = 6 donors combined from three independent experiments, Wilcoxon test). Each symbol represents one donor, bars represent geometric mean, paired one-way ANOVA with Tukey’s multiple comparison test except indicated otherwise, *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001, ****P ≤ 0.0001. Figure was generated using Biorender. Source data are available for this figure: SourceData F2.
Figure S2.
Impact of RELA on IFN response to cGAMP in CD4+ T cells MDDCs, MDMs, and THP-1 cells. (A) RT-qPCR of IFNL1 and IFNB expression 5 h following stimulation with cGAMP (6 µg/ml) in CD4+ T cells and MDDCs transduced with either control or RELA (n = 3 donors combined from two independent experiments). RQ normalized to average of controls, each symbol represents one donor, bars represent geometric mean, paired one-way ANOVA with Tukey’s multiple comparison test. (B) Activity of IFNB promoter and NF-κB promoter luciferase reporter in 293FT following transfection with BFP, human STING or zebrafish STING and stimulation with cGAMP (4 µg/ml). Each symbol represents one experiment (n = 3 independent experiments). (C) IFN-I/III concentration following cGAMP (6 µg/ml) stimulation of CD4+ T cells, transduced with control (BFP), human STING, or zebrafish STING (n = 4 donors combined from two independent experiments). Each symbol represents one donor, bars represent mean, paired one-way ANOVA with Tukey’s multiple comparison test. (D) Proliferation measured using a cell proliferation dye (CPD) of CD4+ T cells transduced with either control (GFP) RELA or RELA K5R lentivector on day 4 and 6 post TCR stimulation (n = 4 donors combined from two independent experiments). Each symbol represents one donor, bars represent mean + SEM, paired one-way ANOVA with Tukey’s multiple comparison test. (E) Western blot of RELA and actin in CD4+ T cells, MDMs, and THP-1 transduced with either control, RELA, or RELA K5R (representative of two individual experiments). (F) IFN-I/III concentration following cGAMP (6 µg/ml) stimulation of (top) CD4+ T cells, MDMs, and (bottom) THP-1 transduced with control (GFP), RELA, or RELA K5R (n = 4 donors combined from two independent experiments, for THP-1 combined from three independent experiments). Each symbol represents one donor, bars represent geometric mean, paired one-way ANOVA with Tukey’s multiple comparison test, *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001, ****P ≤ 0.0001. Source data are available for this figure: SourceData FS2.
These results suggested that RELA might be regulated in T cells to limit IFN promoter induction at the posttranslational level. PTM of lysine residues in RELA such as acetylation and methylation are reported to impact transcriptional activity (Chen et al., 2002; Li et al., 2012; Ziesché et al., 2013). We focused on two RELA mutants converting five lysine residues distributed in the REL homology domain (K122, K123, K310, K314, and K315) either to glutamine (RELA K5Q) or arginine (RELA K5R). The RELA K5Q is predicted to mimic acetylation while RELA K5R is predicted to maintain a non-acetylated basic state, and both are reported to impact TNFα- and IL1α-driven gene expression in cell lines (Buerki et al., 2008; Li et al., 2012; Ziesché et al., 2013). Overexpression of RELA, RELA K5R, or RELA K5Q had no impact on endogenous RELA expression (Fig. 2 C). The IFN-I/III response to cGAMP in MDDCs remained unchanged following RELA, RELA K5Q, and RELA K5R overexpression compared with the control condition (Fig. 2 D). In contrast, in CD4+ T cells, the non-acetylated RELA K5R increased baseline IFNλ1 expression, and this response was augmented in response to cGAMP (Fig. 2 D). This baseline increase in IFNλ1 was not related to increased proliferation rate or survival of CD4+ T cells expressing RELA K5R (Fig. S2 D). Notably, IFNα2 but not IFNβ was now detectable in response to cGAMP when CD4+ T cells expressed RELA K5R. The increase in IFNλ1 at baseline upon RELA K5R expression was not observed in MDDCs, monocyte-derived macrophages (MDMs), or THP-1 cells (Fig. 2 D and Fig. S2, E and F). It was also not observed with RELA K5Q in CD4+ T cells. Together, this data suggests that endogenous RELA regulation in activated CD4+ T cells differs from monocyte-derived cells in a manner that negatively impacts IFN-I/III expression. Notably, the lysine residues K122, 123, 310, 314, and 315 in RELA are important in determining the magnitude of IFN expression, specifically in CD4+ T cells.
K310 has been individually identified to modulate the transcriptional activity of RELA and can also impact the PTM of surrounding lysine residues (Yang et al., 2010). This raised the possibility that K310 might be non-redundant among the five lysine residues responsible for controlling IFN-I/III expression in CD4+ T cells. To determine its importance, IFN response to cGAMP was compared between RELA K310R and RELA K5R in CD4+ T cells. K310R overexpression in CD4+ T cells increased baseline IFNλ1 similar to RELA K5R and the response was augmented in the presence of cGAMP (Fig. 2, E and F). IFNα2 was also detected in K310R overexpressing cells stimulated with cGAMP, similar to RELA K5R overexpressing cells. We hypothesized that the pool of acetylated RELA K310 might differ between CD4+ T cells and MDDCs. Pull-down experiments of GFP-RELA in donor-matched CD4+ T cells and MDDCs revealed a significant increase in K310-acetylated RELA (K310ac) in CD4+ T cells (Fig. 2, G and H). This data highlights the crucial role of K310 in RELA in tuning not only cGAMP-driven but also tonic IFN-I/III expression in CD4+ T cells.
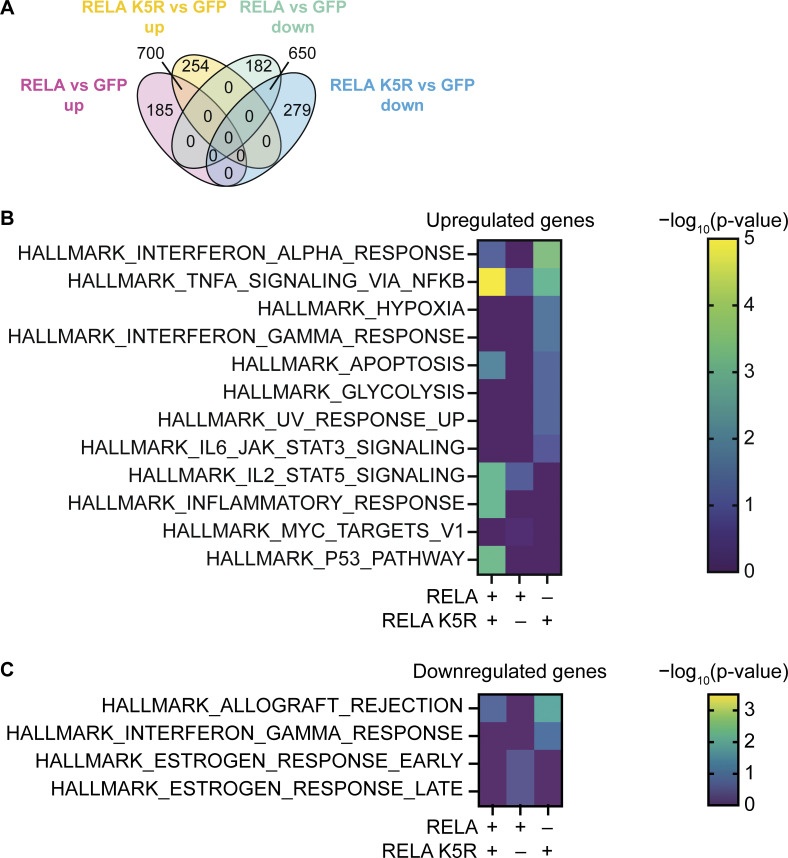
To assess the relative impact of RELA vs. RELA K5R on NF-κB–driven transcription, we analyzed the transcriptome of CD4+ T cells expressing RELA or RELA K5R. We detected upregulated or downregulated genes that were either unique or shared between RELA and RELA K5R (Fig. 3 A and Table S1). Accordingly, enrichment analyses revealed hallmark gene sets that were overlapping or exclusive to RELA or RELA K5R (Fig. 3, B and C). Gene sets annotated as the NF-κB response to TNFα, apoptosis, and p53 were most enriched in the upregulated genes overlapping for RELA and RELA K5R. The IFN-I response gene set was more strongly enriched in RELA K5R. Additionally, RELA K5R-expressing cells induced genes related to the UV response, IFNγ signaling, the JAK–STAT3 pathway, glycolysis, and hypoxia. Altogether, these data indicate that RELA and RELA K5R are both active and induce a common NF-κB–related gene program, but they also regulated distinct genes, characterized by a heightened IFN-I signature for RELA K5R.
Figure 3.
Transcriptional profile of CD4+ T cells expressing RELA or RELA K5R. (A) Venn diagram of differentially expressed genes common or exclusive to CD4+ T cells transduced with either RELA or RELA K5R (n = 4 donors from three independent experiments). (B) Enrichment of MySigDB Hallmark gene sets in upregulated genes of CD4+ T cells transduced with RELA or RELA K5R (n = 4 donors from three independent experiments). (C) Enrichment of gene sets in downregulated genes.
IRF3 and 5-azacytidine synergize with RELA K5R to unlock IFN-I/III production by CD4+ T cells
Since RELA K5R partially unlocks the restriction to IFN-I/III production in a T cell–specific manner, we asked whether additional factors that generally control IFN-I/III production could synergize to fully unlock the system.
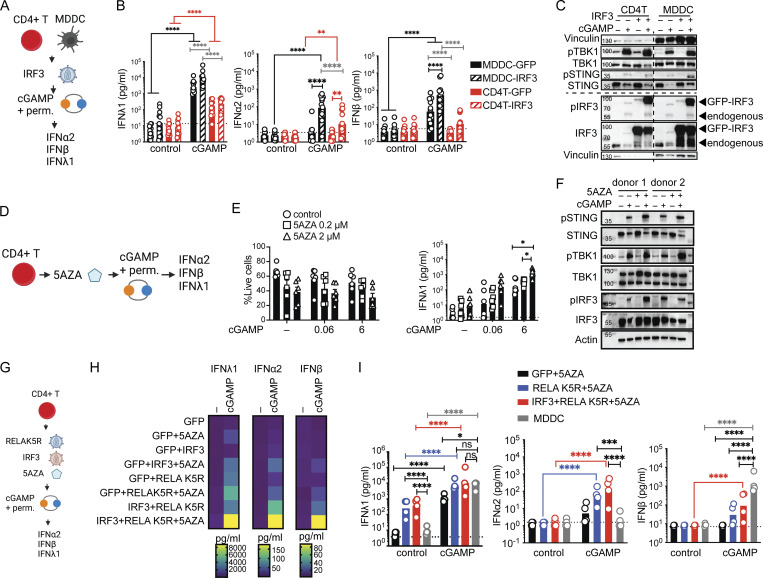
We first overexpressed IRF3 alone in CD4+ T cells and MDDCs (Fig. 4 A). Increasing IRF3 augmented cGAMP-mediated IFNλ1, IFNα2, and IFNβ production by 10-fold in MDDCs. In contrast, IRF3 only increased the IFNα2 response to a small extent in CD4+ T cells and IFNβ remained undetectable (Fig. 4 B). IFNL1 and IFNB transcripts were increased by IRF3 in CD4+ T cells in response to cGAMP (Fig. S3 A), indicating that the promoters were responsive, but insufficiently. IRF3 overexpression resulted in a massive increase in the phospho-IRF3 levels in response to cGAMP that were equivalent in CD4+ T cells and MDDCs, and other components of the pathway remained unchanged (Fig. 4 C). These results show that IRF3 is a limiting factor for IFN-I/III production, but its full potential observed in MDDCs is restricted in CD4+ T cells.
Figure 4.
IRF3 and a DNA methylation inhibitor synergize with RELA K5R to fully lift the IFN-I/III restriction in CD4+ T cells. (A) cGAMP stimulation in CD4+ T cells and MDDCs transduced with IRF3 lentivectors, outline of the experiment. (B) IFN-I/III concentration after cGAMP (6 µg/ml) stimulation of CD4+ T cells and MDDCs transduced with either control or IRF3 (n = 12 donors combined from six independent experiments). (C) Western blot of key signaling proteins involved in STING signaling and actin 6 h following cGAMP (6 µg/ml) stimulation of CD4+ T cells and MDDCs transduced with either control or IRF3 lentivectors (representative of two independent experiments). (D) cGAMP stimulation in CD4+ T cells treated with a DNA methylation inhibitor (5AZA), outline of the experiment. (E) Viability (mean + SEM) and IFNλ1 concentration (geometric mean) following 5AZA (2 µM) treatment and cGAMP (0.06, 6 µg/ml) stimulation of CD4+ T cells (n = 6 donors from three independent experiments). (F) Western blot of key signaling proteins involved in STING signaling and actin 6 h following cGAMP (6 µg/ml) stimulation of CD4+ T cells pretreated for 48 h with 5AZA (2 µM; representative of two independent experiments). (G) cGAMP stimulation in CD4+ T cells treated with a DNA methylation inhibitor (5AZA) and transduced with RELA K5R and IRF3 lentivectors, outline of the experiment. (H) Heat-map showing IFN-I/III concentration following cGAMP (6 µg/ml) stimulation of CD4+ T cells transduced with control (GFP), IRF3, RELA K5R, and pretreated for 48 h with 5AZA (2 µM; n = 8 donors combined from four independent experiments, geometric mean). (I) IFN-I/III concentration following cGAMP (6 µg/ml) stimulation of untransduced MDDCs and CD4+ T cells transduced with control (GFP), IRF3, RELA K5R and treated for 48 h with 5AZA (2 µM; n = 4 donors combined from two independent experiments). Each symbol represents one donor, bars represent geometric mean, paired one-way ANOVA with Tukey’s multiple comparison test, *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001, ****P ≤ 0.0001. Figure was generated using Biorender. Source data are available for this figure: SourceData F4.
Figure S3.
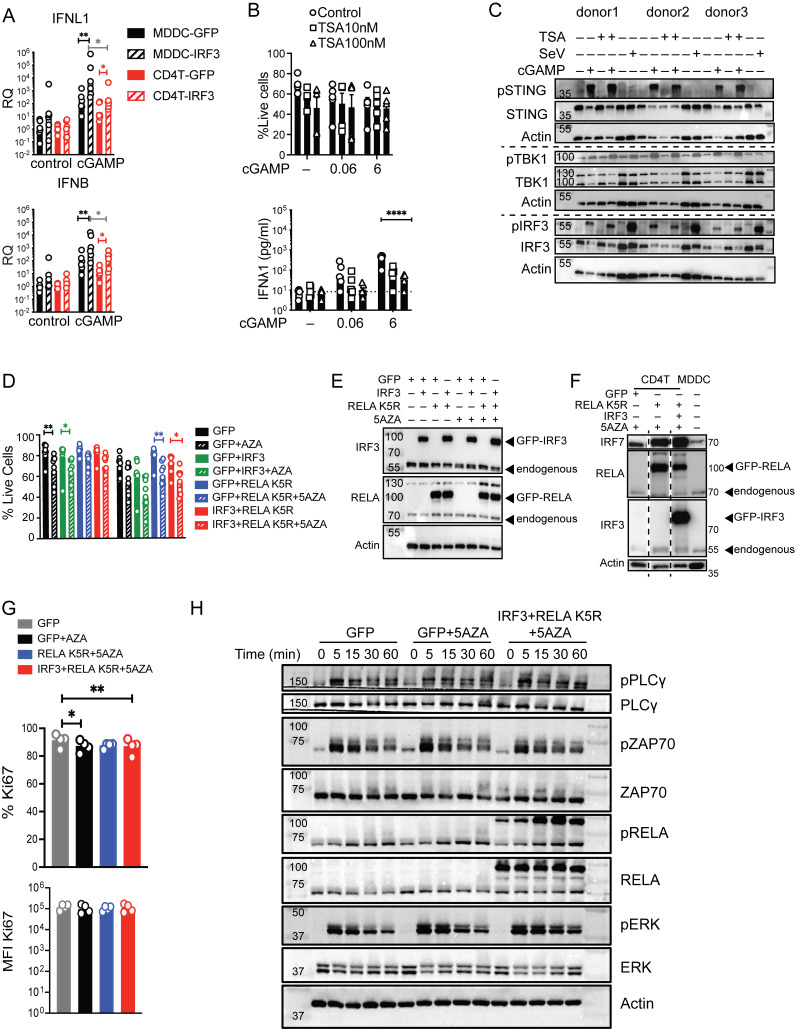
Impact of IRF3, TSA, 5AZA, and RELA K5R on IFN response to cGAMP, viability, and signaling in CD4+ T cells. (A) RT-qPCR of IFNL1 and IFNB 6 h following stimulation with cGAMP (6 µg/ml) of CD4+ T cells and MDDCs transduced with control (GFP) or IRF3 lentivectors (n = 6 donors combined from three independent experiments). RQ normalized to average of controls, each symbol represents one donor, bar represents geometric mean, paired one-way ANOVA with Tukey’s multiple comparison test. (B) Viability (mean + SEM) and IFNl1 concentration following TSA (100 nM) treatment and cGAMP (0.06, 6 µg/ml) stimulation of CD4+ T cells (n = 6 donors combined from three independent experiments). (C) Western blot of key signaling proteins involved in STING signaling and actin, 6 h following cGAMP (6 µg/ml) stimulation or Sendai virus (SeV 200 HA/ml) infection of CD4+ T cells in combination with TSA (100 nM; representative of two independent experiments). (D) Viability (mean + SEM) of CD4+ T cells transduced with control or IRF3 and RELA K5R lentivectors stimulated with cGAMP (6 µg/ml; n = 6 donors combined from three independent experiments). Each symbol represents one donor, bars represent mean + SEM, paired one-way ANOVA with Tukey’s multiple comparison test. (E) Western blot of IRF3, RELA, and actin in CD4+ T cells transduced as indicated (representative of n = 8 independent donors in two experiments). (F) Western blot of IRF3, RELA, and actin in CD4+ T cells transduced as indicated and untransduced MDDCs (representative of n = 4 independent donors in two experiments). (G) Percentage and MFI of Ki67 in CD4+ T cells transduced either with control (GFP) IRF3 and RELA K5R and pretreated with 5AZA (2 µM; n = 4 donors combined from two independent experiments). Each symbol represents one donor, bars represent mean + SEM, paired one-way ANOVA with Tukey’s multiple comparison test. (H) Western blot of indicated TCR signaling response proteins in CD4+ T cells transduced either with control (GFP) IRF3 and RELA K5R and pretreated with 5AZA (2 µM), harvested at the indicated times (representative of n = 4 independent donors in two experiments). *P ≤ 0.05, **P ≤ 0.01, ****P ≤ 0.0001. Source data are available for this figure: SourceData FS3.
IFN expression is also subject to epigenetic regulation by histone modifications, DNA methylation, and acetyl transferases (Salvi et al., 2010; Chen et al., 2012; Chiappinelli et al., 2015; Marié et al., 2018). A counter-intuitive feature of IFN and IFN-stimulated gene expression is a requirement for histone deacetylase (HDAC) activity, commonly associated with gene expression silencing (Marié et al., 2018).To identify the dominant class of epigenetic regulation in CD4+ T cells, the IFN response to cGAMP was screened in the context of histone deacetylase inhibition by Trichostatin A (TSA) and DNA methylation inhibition by 5′azacytidine (5AZA; Fig. 4 D and Fig. S3 B). TSA reduced the already low IFNλ1 production in CD4+ T cells (Fig. S3 B). cGAMP-mediated STING signaling was intact in TSA-treated cells, consistent with an effect at the transcriptional level (Fig. S3 C). In contrast, 5AZA enhanced the IFNλ1 response to cGAMP (Fig. 4 E). This enhancement did not occur through a detectable increase in STING signaling, in accordance with a transcriptional effect (Fig. 4 F).
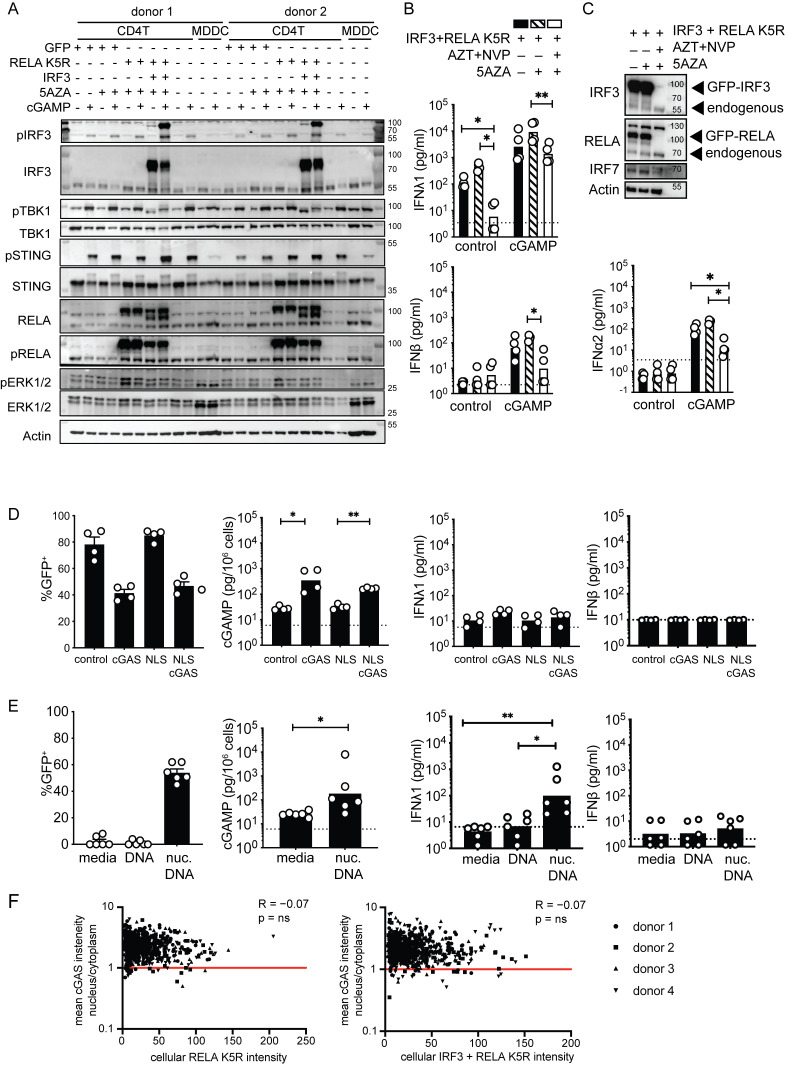
These results revealed that IRF3 and 5AZA can enhance IFN-I/III expression in response to cGAMP stimulation but not at baseline, like RELA K5R. Next, we combined IRF3 and 5AZA with RELA K5R to assess synergy in IFN-I/III at the basal level and after cGAMP stimulation (Fig. 4, G and H). RELA K5R and IRF3 expression did not compromise cell viability, while 5AZA significantly reduced it (Fig. S3, D and E). Combining RELA K5R, IRF3, and 5AZA gradually increased the production of all tested IFN-I/III in CD4+ T cells in response to cGAMP (Fig. 4 H). IFNλ1 was produced at similar levels to the MDDCs benchmark in response to cGAMP (Fig. 4 I). IFNα2a was also induced by >100-fold by CD4+ T cells, while it remained undetectable in MDDCs (Fig. 4 I). This difference could be associated with a difference in levels of IRF7 between MDDCs and CD4+ T cells in these conditions (Fig. S3 F). IFNβ reached significant levels of production in CD4+ T cells, although they remain below MDDCs (Fig. 4 I). The expression of endogenous and overexpressed RELA and IRF3 was similar in all conditions evaluated (Fig. S3 F). Expression of Ki67, a marker of cell proliferation, was not impacted by RELA K5R, but was reduced under 5AZA treatment (Fig. S3 G). The signaling response to TCR stimulation, as measured by phospho-PLCγ, phospho-ZAP70, phospho-ERK, and phospho-RELA, was not altered by 5AZA, IRF3, or RELA K5R (Fig. S3 H). The levels and phosphorylation of endogenous IRF3, TBK1, STING, RELA, and ERK in response to cGAMP were also unaffected by 5AZA, RELA K5R, and IRF3 (Fig. S4 A). Of note, ERK1 (p44) was more phosphorylated in CD4+ T cells, while ERK2 (p42) was more phosphorylated in MDDCs. As a control, we verified using reverse transcriptase inhibitors azidothymidine and nevirapine that the synergistic effects of IRF3 and RELA K5R expression in CD4+ T cells were specifically due to the lentivirus-mediated overexpression and not to confounding factors associated with the viral lentivirus particles in the first place, such as cGAMP transferred in viral particles (Fig. S4, B and C; Bridgeman et al., 2015; Gentili et al., 2015). Altogether, these results establish that the combination of RELA K5R, IRF3, and 5AZA in CD4+ T cells, which will be referred to as type I/III-IFN producing CD4+ T cells (iCD4+ T cells), unlocks their ability to produce type I and type III IFN to the same level as the dendritic cell benchmark in response to cGAMP.
Figure S4.
Impact of IRF3, 5AZA, and RELA K5R on IFN response to cGAMP in CD4+ T cells. (A) Western blot of STING signaling response and ERK1/2 in CD4+ T cells transduced and treated as indicated, and untransduced MDDCs (representative of n = 2 independent experiments). (B) IFN-I/III concentration following cGAMP (6 µg/ml) stimulation and 5AZA (2 µM) treatment of CD4+ T cells transduced with control or IRF3 and RELA K5R lentivectors in the presence of AZT (25 µM) and NVP (10 µM; n = 4 donors, combined from two independent experiments). Each symbol represents one donor, bars represent geometric mean, paired one-way ANOVA with Tukey’s multiple comparison test. (C) Western blot of IRF3, RELA, IRF7, and actin proteins in CD4+ T cells transduced as indicated and treated with AZT (25 µM), NVP (10 µM), and 5AZA (2 µM; representative of 4 n = 2 independent donors in two experiments). (D) Percentage of GFP expression, and cGAMP and IFN-I/III quantification in CD4+ T cells transduced either with control, cGAS, NLS, or NLS-cGAS (n = 4 donors combined from two independent experiments). Each symbol represents one donor, bars represent mean + SEM (percentage of GFP) or geometric mean (concentrations), paired one-way ANOVA with Tukey’s multiple comparison test. (E) Percentage of GFP expression, and cGAMP and IFN-I/III quantification in CD4+ T cells nucleofected with a control GFP plasmid DNA (n = 6 donors combined from three independent experiments). Each symbol represents one donor, bars represent mean + SEM (percentage of GFP) or geometric mean (concentrations), paired one-way ANOVA with Tukey’s multiple comparison test or paired t test. (F) Pearson correlation of GFP intensity with the ratio of average intensity of nuclear to cytoplasmic cGAS signal in CD4+ T cells transduced with either RELA K5R or IRF3 and RELA K5R. Each point represents a single cell of a donor (n = 4 donors combined from two independent experiments). *P ≤ 0.05, **P ≤ 0.01. Source data are available for this figure: SourceData FS4.
An IRF7-positive feedback drives IFN-I/III production in iCD4+ T cells
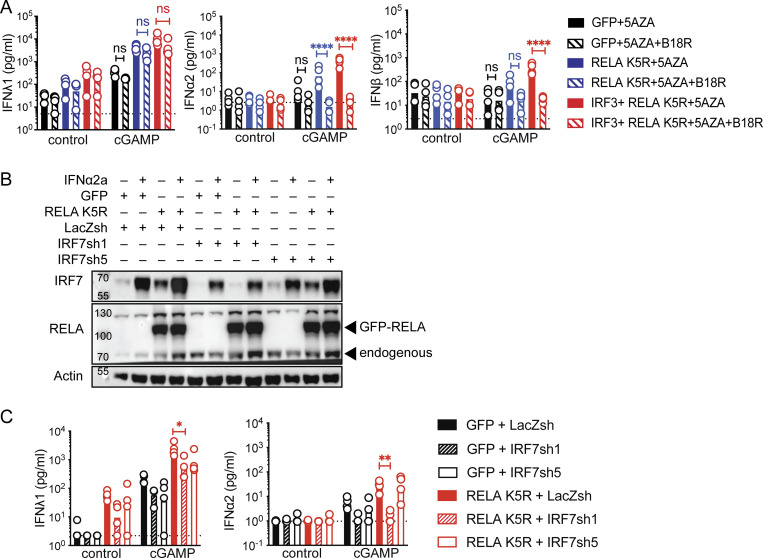
The enhanced expression of IRF7 in iCD4+ T by RELA K5R (Fig. S3 F) suggested that positive feedback from IFN-I signaling might be important for IFN-I production in these cells. To test this, iCD4+ T cells were treated with decoy IFN-I receptor B18R protein prior to cGAMP treatment (Hernáez et al., 2018). Adding B18R to iCD4+ T cells blocked cGAMP-dependent IFN-I response (Fig. 5 A). The IFNλ1 response was not significantly impacted by B18R exposure. To test if IRF7 was required for the positive feedback loop of IFN-I, IRF7 was knocked down using two different shRNA in the context of CD4+ T cells expressing only RELA K5R. As the basal level of IRF7 is low in CD4+ T cells, IFNα2a was added to the culture media to visualize IRF7 expression. This allowed a validation of IRF7 knock-down in CD4+ T cells (Fig. 5 B). Both shRNA reduced IRF7 expression, and shRNA1 was more potent (Fig. 5 B). shRNA1 abolished the induction of IFNα2 by cGAMP stimulation (Fig. 5 C). The IRF7 shRNAs also reduced the production of IFNλ1 at baseline and after cGAMP stimulation. Notably, in the absence of RELA K5R, increasing IRF7 was not sufficient to enhance the IFNλ1 and IFNβ response in CD4+ T cells (Fig. S1, F and G). Therefore, IRF7 mediates positive feedback to enable IFN-I and maximize IFN-III production by iCD4+ T cells.
Figure 5.
IRF7 positive feedback is required for IFN-I/III expression in iCD4+ T cells. (A) IFN-I/III concentration following cGAMP (6 µg/ml) stimulation of CD4+ T cells transduced with control (GFP), RELA K5R, IRF3, in the presence of control or B18R and pretreated for 48 h 5AZA (2 µM; n = 4 donors combined from two independent experiments). (B) Western blot of RELA, IRF7, and actin in CD4+ T cells transduced with either LacZsh, IRF7sh1, or IRF7sh5 and treated with IFNα2a (1,000 U/ml) for 18–24 h (n = 4 independent donors in two experiments). (C) IFN-I/III concentration following cGAMP (6 µg/ml) stimulation of CD4+ T cells cotransduced with control (GFP) or RELA K5R and LacZsh, IRF7sh1, or IRF7sh5 (n = 4 independent donors in two experiments). Each symbol represents one donor, bars represent geometric mean, paired one-way ANOVA with Tukey’s multiple comparison test, ****P ≤ 0.0001; ns, not significant. Source data are available for this figure: SourceData F5.
cGAS drives the IRF7 positive feedback in iCD4+ T cells
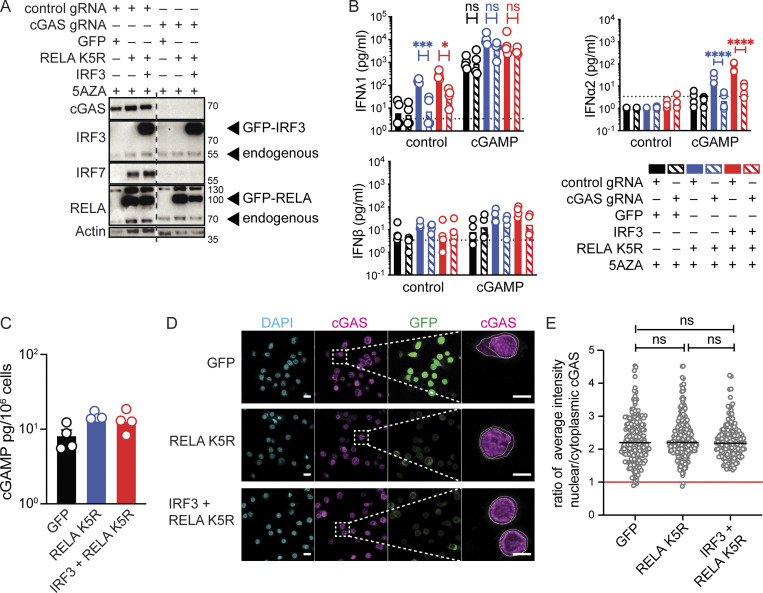
The expression of RELA K5R in CD4+ T cells was consistently sufficient to induce the expression of IFNλ1 and IRF7. This raised the possibility that an upstream innate sensor could be active in CD4+ T cells to activate RELA K5R. We first tested the functionality of cGAS in T cells. cGAS overexpression is sufficient for cGAMP production due to the promiscuous detection of endogenous nucleic acids, in particular in the nucleus (Gentili et al., 2019). CD4+ T cells contained a low but detectable quantity of endogenous cGAMP (Fig. S4 D). Overexpression of cGAS or nuclear localization signal (NLS)-cGAS in CD4+ T cells increased intracellular cGAMP levels by 10–20-fold (Fig. S4 E). IFN-I/III remained undetectable (Fig. S4 D). We next examined the functionality of endogenous cGAS and other DNA sensors by nucleofecting a plasmid DNA coding for GFP. Intracellular cGAMP increased by 50-fold and minimal levels of IFNλ1 were induced by DNA transfection, while IFN-β remained undetected (Fig. S4 E). Therefore, endogenous cGAS is functional in T cells but not sufficient, even after overexpression, to unlock IFN-I/III expression, consistent with a downstream bottleneck at the level of RELA. Next, we evaluated the role of cGAS in the context of iCD4+ T cells. Knock-out (KO) of cGAS had no impact on IRF3 or RELA expression (Fig. 6 A). However, cGAS KO iCD4+ T cells were unable to upregulate IRF7 (Fig. 6 A). cGAS KO also inhibited the basal production of IFNλ1 and the IFNα2 response to cGAMP (Fig. 6 B). To evaluate if cGAS activity was enhanced in cells expressing RELA K5R or IRF3, cellular cGAMP was quantified, but no significant increase was observed, indicating that cGAS is activated upstream of RELA K5R and IRF3 (Fig. 6 C). To understand how cGAS was activated, we examined its localization since it determines its ability to be activated by endogenous DNA ligands (Barnett et al., 2019; Gentili et al., 2019; Li et al., 2021). In activated CD4+ T cells, we found that cGAS is predominantly nuclear and this remained unchanged in cells overexpressing IRF3 or RELA K5R (Fig. 6, D and E and Fig. S4 F). These results show that tonic cGAS activation by endogenous ligands is required to drive the IRF7 loop leading to IFN-I/III expression in iCD4+ T cells.
Figure 6.
Tonic cGAS activity is required for IFN expression in iCD4+ T cells. (A) Western blot of cGAS, IRF3, IRF7, RELA, and actin in CD4+ T cells nucleofected with either control or cGAS targeting guides and subsequently transduced with control (GFP), RELA K5R or IRF3 lentivectors (n = 4 independent donors in two experiments). (B) IFN-I/III concentration following cGAMP (6 µg/ml) stimulation of CD4+ T cells nucleofected with control or cGAS gRNA and transduced with control, RELA K5R, or IRF3 lentivectors. Cells were pretreated with 5AZA (2 µM) for 48 h prior to cGAMP stimulation (n = 4 donors combined from two independent experiments). Each symbol represents one donor, bars represent geometric mean, paired one-way ANOVA with Tukey’s multiple comparison test. (C) cGAMP quantification by ELISA in CD4+ T cells transduced with either control (GFP), RELA K5R, or IRF3 lentivectors (n = 4 donors combined from two independent experiments). (D) DAPI, cGAS staining, and GFP localization by confocal microscopy in CD4+ T cells transduced with control (GFP), RELA K5R, or IRF3 lentivectors. Scale bar at 10 µm. Right: Magnification of cGAS channel showing contours used to delineate nuclear and cellular contents, scale bar at 5 µm (n = 4 donors combined from two independent experiments). (E) Quantification of the ratio of average intensity of nuclear to cytoplasmic cGAS signal in CD4+ T cells transduced with either control (GFP), RELA K5R, or IRF3 lentivectors (n = 4 donors combined from two independent experiments). Each symbol represents one cell, line represents median, mixed-effect analysis, *P ≤ 0.05, ***P ≤ 0.001, ****P ≤ 0.0001, ns, not significant. Source data are available for this figure: SourceData F6.
RELA K5R increases the antitumor activity of CAR T cells
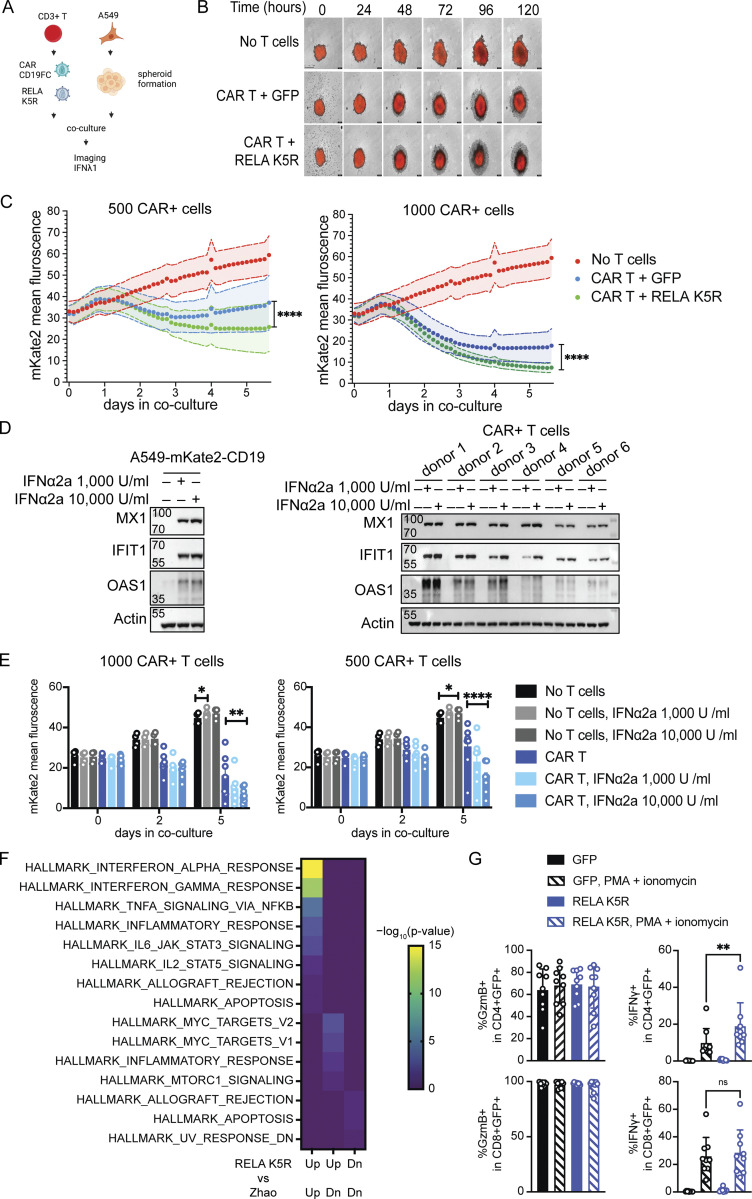
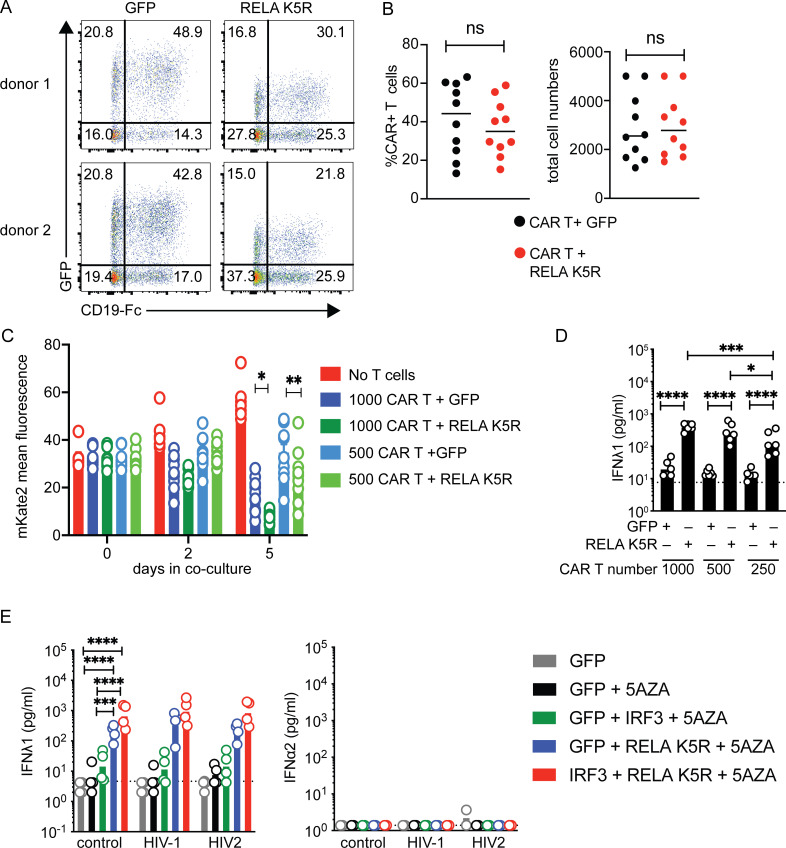
We next aimed to assess the functional impact of endowing CD4+ T cells with strong IFN-I/III responses. Transient induction of IRF7-dependent IFN induction has been previously shown to be required for the optimal antitumor activity of CD19-targeting CAR T cells (Zhao et al., 2015). Since IFN-I/III enhancement by RELA K5R was dependent on IRF7, we evaluated if RELA K5R could improve CAR T-mediated tumor killing using a spheroid tumor model using A549 cells expressing CD19 (Fig. 7 A). T cells transduced with CD19-targeting CART with control GFP or RELA K5R were added to tumors and tumor growth was monitored for 5–6 d. Since the rate of CAR transduction varied slightly when cotransduced with GFP or RELA K5R vectors, the cell numbers were adjusted to normalize the CAR-positive cells (Fig. S5, A and B). The spheroids grew with time when cultured alone and reduced in size partially when cocultured with CD19-targeting CAR T cells (Fig. 7, B and C). When CAR T cells coexpressed RELA K5R, tumor cells were significantly more efficiently eliminated when compared with CAR T cells alone (Fig. 7 C and Fig. S5C). Notably, the RELA K5R delay of tumor growth was also observed with lower numbers of CAR T cells (Fig. 7 C, left panel). IFNλ1 was detected at the end of the 6-d coculture in all dilutions of CART and RELA K5R cells (Fig. S5 D). Since RELA K5R T cells are characterized by a IFN-I signature (Fig. 3 B), we next sought to test the contribution of IFN-I in the elimination of tumor cells by CAR T cells, specifically its action on tumor cells vs. T cells. The addition of IFNα induced IFN-stimulated gene (ISG) expression in both A549 and CAR+ T cells (Fig. 7 D). Interestingly, however, IFNα enhanced the elimination of tumor cells only in the presence of CAR+ T cells (Fig. 7 E). To further substantiate the significance of RELA K5R expression in CAR T cells, we determined the overlap of gene expression signatures between our data set of CD4+ T cells expressing RELA K5R and a published dataset of CAR T cells where IRF7 and IFN-I expression was crucial for optimal antitumor efficacy (Zhao et al., 2015). We found that gene signatures of IFN-I and IFN-II responses were shared and were the most significantly overlapping among the upregulated genes (Fig. 7 F). Next, we examined the expression of T cell effector proteins Granzyme B and IFNγ. RELA K5R significantly increased the expression of IFNγ in CD4+ T cells (Fig. 7 G). Altogether, these results demonstrate that RELA K5R enhances the antitumor activity of CAR T cells and they establish a concordance with IFN-I activity in T cells.
Figure 7.
RELA K5R enhances CAR-mediated tumor killing. (A) CAR-T tumor spheroid killing assay, experimental outline. (B) Representative images of mKate2+CD19+ A549 cells (red) alone or in coculture with CAR+ GFP+ or CAR+ RELA K5R+ T cells acquired over 5 d. (C) Fluorescence intensity of mKate2+CD19+ A549 cells over time during coculture with CAR+ GFP+ or CAR+ RELA K5R+ T cells (n = 10 donors combined from five independent experiments). (D) Western blot of MX1, IFIT1, OAS1, and actin expression following overnight treatment of A549-CD19-mKate2 cells or CAR-expressing CD3+ T cells with IFNα2a (representative of n = 3 independent experiments for A549 cells). (E) Fluorescence intensity of mKate2+CD19+ A549 cells over time during coculture with CAR+ T cells treated or not with IFNα2a at the time of coculture (n = 7 donors combined from four independent experiments). (F) Enrichment of MySigDB Hallmark gene sets in shared differentially expressed genes between CD4+ T cells expressing RELA K5R and the comparison of CAR constructs 1928z-4-1-BBL vs. 1928z in Zhao et al. (2015). (G) Percentage of IFNγ and Granzyme B (GzmB) expressing cells in GFP+ CD4+ and CD8+ T cells also transduced with the CAR construct (n = 9 donors combined from five independent experiments). Each symbol represents one donor, bars represent mean, paired one-way ANOVA with Tukey’s multiple comparison test, except if indicated otherwise, *P ≤ 0.05, ***P ≤ 0.001, ****P ≤ 0.0001, ns, not significant. Figure was generated using Biorender. Source data are available for this figure: SourceData F7.
Figure S5.
Functional impact of iCD4+ T cells in the context of HIV infection and anti-tumor response. (A) Representative cotransduction rates of control (GFP) or RELA K5R and CAR (CD19-Fc staining) lentivectors in T cells. (B) Rates of CAR+ cells in control (GFP) or RELA K5R transduced T cells (n = 10 donors combined from five independent experiments). Each symbol represents one donor, bar indicates mean, paired one-way ANOVA with Tukey’s multiple comparison test. (C) Bar plots representation of data shown in Fig. 7 C on days 0, 2, and 5 after coculture of CAR+ GFP+ or CAR+ RELA K5R+ T cells with mKate2+CD19+ A549 cells (n = 10 donors combined from five independent experiments). (D) IFNλ1 concentration at the end of the coculture period with 1,000, 500, and 250 CAR+ GFP+ or CAR+ RELA K5R+ T cells (n = 6 donors combined from three independent experiments). (E) IFN-I/III concentration 48 h following HIV-1 or HIV-2 infection of CD4+ T cells transduced with the indicated lentivectors and pretreated with 5AZA (2 µM) for 48 h (n = 4 donors combined from two independent experiments). Each symbol represents one donor, bar represents geometric mean, paired one-way ANOVA with Tukey’s multiple comparison test, *P ≤ 0.05, **P ≤0.01, ***P ≤ 0.001, ****P ≤ 0.0001, ns, not significant.
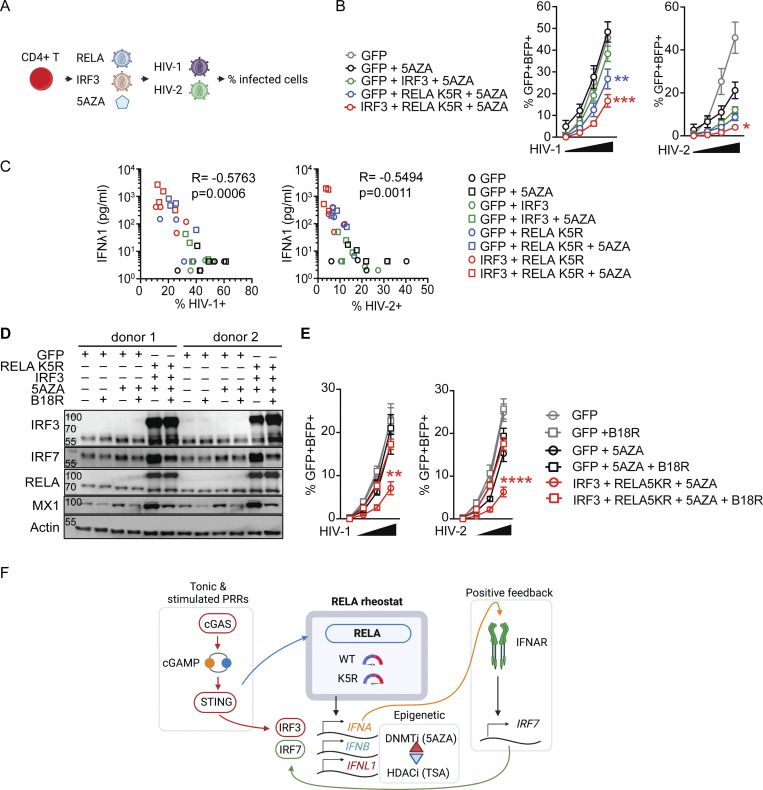
iCD4+ T cells autonomously resist HIV infection
To extend these findings, we moved to HIV infection. CD4+ T cells are the primary targets of HIV infection. CD4+ T cells are unable to mount an antiviral IFN-I response following infection, therefore failing to protect themselves (Manel et al., 2010). We tested if iCD4+ T cells could now autonomously resist HIV-1 infection. Activated CD4+ T cells transduced with either control, IRF3, RELA K5R, IRF3, or RELA K5R in the presence of 5AZA were challenged with single-round HIV-1 and HIV-2 reporter viruses (Fig. 8 A). After 48 h, infection rates and IFN levels were quantified. The rates of HIV-1 and HIV-2 infections were the lowest in iCD4+ T cells (IRF3+RELA K5R+5AZA; Fig. 8 B). Notably, the levels of infection negatively correlated with baseline IFNλ1 levels detected across conditions (Fig. 8 C and Fig. S5 E). There was no further detectable increase in IFN-I/III response upon infection (Fig. S5 E). Blocking type I IFN with B18R abrogated the antiviral state of iCD4+ T cells against HIV-1 and HIV-2 (Fig. 8, D and E). This data provides evidence for the superior self-defense of iCD4+ T cells in the context of HIV infection.
Figure 8.
iCD4+ T cells have increased resistance to HIV infection. (A) HIV infection challenge in iCD4+ T cells, experimental outline. (B) Rate of HIV-1 or HIV-2 infection, 48 h after infection of CD4+ T cells transduced with control (GFP), IRF3, and RELA K5R lentivectors. Cells were transduced, pretreated with 5AZA (2 µM) for 48 h, and subsequently infected with HIV-1 or HIV-2 single-round virus (n = 4 donors combined from two independent experiments). Each symbol represents one donor, bars represent mean ± SEM of four donors, paired one-way ANOVA with Tukey’s multiple comparison test on the highest dose of virus. (C) Pearson correlation of infection rates with IFNλ1 concentration of CD4+ T cells transduced and treated with 5AZA as indicated. (D) Western blot of IRF3, IRF7, RELA, MX1, and Actin in CD4+ T cells transduced with either GFP or RELA K5R and IRF3, and pretreated with 5AZA (2 µM), control or B18R supernatant for 3 d (representative of two independent experiments). (E) Rate of HIV-1 or HIV-2 infection, 48 h after infection of CD4+ T cells transduced with control (GFP), IRF3, and RELA K5R lentivectors. Cells were transduced, pretreated with 5AZA (2 µM), control, or B18R supernatants for 48 h and subsequently infected with HIV-1 or HIV-2 single-round virus (n = 4 donors combined from two independent experiments). Each symbol represents one donor, bars represent mean ± SEM of four donors, paired one-way ANOVA with Tukey’s multiple comparison test on the highest dose of virus. (F) Working model. RELA functions as a rheostat to control IFN-I/III expression levels in CD4+ T cells. IFN-I/III expression requires tonic cGAS activity or PRR stimulation and positive feedback from IRF7 signaling. *P ≤ 0.05, **P ≤0.01, ***P ≤ 0.001, ****P ≤ 0.0001. Figure was generated using Biorender. Source data are available for this figure: SourceData F8.
Discussion
In this study, we addressed the functionality of the cGAS–STING–IFN innate sensing pathway in human CD4+ T cells. We identified that the production of IFN-III and to a much larger extent IFN-I is tightly regulated in CD4+ T cells with several restrictions at the level of both transcription (RELA, IRF3) and epigenetic factors (DNA methylation). To understand the basis of low IFN-I/III responses, we systematically compared the IFN-I/III response of CD4+ T cells and dendritic cells. We find that upstream phosphorylation-based signaling by cGAS–STING–TBK1–IRF3 is intact in CD4+ T cells. Instead, we identified RELA as a limiting factor of both tonic and inducible IFN-I/III expression in T cells. IRF3 expression and DNA methylation inhibition synergize with RELA K5R to enhance IFN expression, but unlike RELA K5R, they are not able to promote spontaneous IFN-III expression. The crucial role of RELA is further supported by our finding that five lysines (122, 123, 310, 314, and 315), in particular K310, and not the strength of NF-κB stimulation per se, as demonstrated with zebrafish STING, are key determinants of IFN-I/III expression in CD4+ T cells but not in dendritic cells. Therefore, we propose that RELA is a rheostat for IFN-I/III expression in lymphocytes (Bardwell, 2008).
The PTM of each of the five lysines of RELA and their mutants have been reported to have variable effects depending on the target gene and the stimulus. Mutations to acetyl-mimic glutamine blunted the enhancement of IFNλ1 by RELA, while mutations to arginine, which mimic a lack of PTM, enhanced IFN-I/III production. Both RELA K5R and RELA K5Q mutations have been previously shown to induce p65-dependent genes in response to stimuli such as TNFα and IL1α (Buerki et al., 2008; Li et al., 2012). However, the overlap of differentially expressed genes is low suggesting the differential affinity for cofactors rather than DNA binding affinity or transactivation potential (Buerki et al., 2008; Li et al., 2012). In T cells, RELA is activated downstream of the TCR but the PTM state of these lysines has not been extensively studied, with the exception of K310. K310 is reported to be acetylated in response to TCR stimulation in mouse primary T cells (Clavijo and Frauwirth, 2012) and in response to p300 expression in the human Jurkat T cell line (Kwon et al., 2008). In non-immune cell lines, K310 can be deacetylated by SIRT1 (Yeung et al., 2004; Kwon et al., 2008). In cell lines, while acetylation of RELA at K310 and the other lysine residues has been associated with both activation and repression of gene expression (Kiernan et al., 2003; Li et al., 2012; Ziesché et al., 2013), their methylation is reported to negatively impact NF-κB–mediated gene expression (Yang et al., 2009; Yang et al., 2010; Levy et al., 2011). Further studies are required to appreciate the roles of the various RELA PTM in the control of IFN-I/III production by T cells. Interestingly, RELA in MDDCs, MDMs, and THP-1 cells did not appear to be a limiting factor with neither RELA lysine mutants impacting IFN-I/III expression at baseline nor in response to cGAMP. We also identified differences in the pool of cellular acetylated RELA K310 between CD4+ T cells and MDDCs. It is thus tempting to speculate that, through PTM, the pool of endogenous RELA in T cells may be preferentially associated with T-cell–specific gene programs, such as TCR signaling-induced genes, as opposed to PRR-induced IFN-I/III genes. Accordingly, the RNA-seq analysis revealed both shared and specific gene signatures associated with RELA and RELA K5R.
We found that unlike RELA, overexpression of IRF3 enhances IFN-I expression in both MDDCs and CD4+ T cells, suggesting that IRF3 is a limiting factor in both cell types tested. We also demonstrate that IRF7 induction by RELA is crucial for inducing IFN-I and enhancing IFN-III expression in T cells. Importantly, however, IRF3 and IRF7 are individually insufficient to promote IFNL1 and IFNB expression in T cells. We also show that treatment of T cells with 5AZA has a positive impact on IFN expression in T cells. A recent study identified a single cytosine in the IFNB promoter that, when methylated, negatively impacts IRF3 recruitment and thus IFN expression in murine macrophages (Nishioka et al., 2019). Alternatively, inhibition of DNA methyltransferases could indirectly enhance IFN-I expression through the reexpression of endogenous retroelements and activation of innate sensors (Chiappinelli et al., 2015). We also found that TSA inhibits IFN expression at baseline with RELA K5R and after cGAMP stimulation. HDAC inhibition has been shown to reactivate HIV expression from latent integrated proviruses in vitro and advanced to clinical trials in vivo (Lehrman et al., 2005). We propose that in addition to activating the viral promoters, HDAC inhibition also diminishes IFN expression in vivo, thereby promoting viral expression. Altogether, our findings highlight an emerging regulation of IFN gene expression by epigenetic regulation.
We describe tonic IFN production in T cells that is dependent on cGAS, RELA, and IRF7 expression. In the assays we used, baseline IFN production is detectable at the protein level for IFNλ1 when unlocked by RELA K5R in T cells, but IFN-I remains below detection limits in accordance with tonic signaling (Gough et al., 2012). We find that cGAS is largely nuclear in T cells, therefore its association with nuclear DNA could permit low levels of cGAS activity as described in other cells (Gentili et al., 2019). Alternatively, cGAS may be activated by mitochondrial DNA. Supporting this, we observed the upregulation of the TNF signaling pathway using RNA-seq, and TNF was recently shown to release mitochondrial DNA activate cGAS–STING (Willemsen et al., 2021). Notably, neither the activity nor cellular localization of cGAS is altered upon RELA K5R overexpression.
Consistent with the restriction of IFN-I/III in T cells, human T cells have not been previously identified as a cellular source of IFN-I or IFN-III in patients with interferonopathies so far (Rodero et al., 2017). Intriguingly, patients carrying mutations in RELA have been described to have elevated IFN-I and an ISG signature at a steady state (Barnabei et al., 2020 Preprint). However, the mechanism of how these RELA mutants might induce unchecked IFN-I expression, the requirement for upstream intracellular sensors and cellular subtypes involved in the IFN-I induction in the pathology of these patients, is currently unclear.
We noted qualitative differences in ERK1 and ERK2 phosphorylation between CD4+ T cells and MDDCs. Namely, ERK2 was more phosphorylated at baseline in MDDCs than in T cells, while ERK1 was less phosphorylated in MDDCs over T cells. In the Huh7 hepatocyte cell line, ERK1 and ERK2 were found to be collectively required for type I (not type III) IFN induction in response to Sendai virus infection (Odendall et al., 2014). It is therefore possible that relative differences between ERK1 and ERK2 phosphorylation contribute to the regulation of IFN-I vs. IFN-III in CD4+ T cells vs. MDDCs.
We provide evidence that intrinsic induction of IFN-I/III in T cells can be beneficial in two contexts. First, it increases resistance to HIV infection. Second, RELA K5R is able to improve tumor elimination by CAR T cells robustly in vitro. Importantly, RELA K5R does not reduce cell viability or proliferation. It would be interesting to evaluate the relevance of RELA K5R CAR+ T cells in tumor models in vivo. Given that RELA lies downstream of several receptors and is involved in the protection from apoptosis and the production of IL-2 (Oh and Ghosh, 2013), the impact of RELA K5R may also be broader than IFN-I/III expression in T cells. Therefore, RELA emerges as a target for enhancing IFN-I/III expression by T cells in these therapeutic contexts. Expressing RELA K5R may constitute a viable path for the improvement of CAR-based therapies. Additionally, combining RELA K5R with IRF3 and 5AZA provides maximal IFN-I/III expression, but the practical use of this combination may be more challenging.
Our findings show that T cells, unlike antigen-presenting cells, are specifically programmed to limit IFN-I/III expression, even if their PRRs get activated. This implies that the ability of CD4+ T cells to produce IFN-I/III production has been counterselected during evolution. In contrast to T cells, pDC are well appreciated as a major producer of IFN-I in infections and pathological conditions, and they constitute a minor fraction of circulating and tissue-resident immune cells. We thus propose that the restriction of IFN-I/III expression in T cells is a mechanism to avoid pathogenic levels of a toxic cytokine from a cell type that constitutes a large proportion of immune cells.
Materials and methods
Primary cells isolation and culture
Plasmapheresis blood pockets were obtained from healthy adult volunteers after informed consent from the Establishment of French blood collection (approved by the Institut National de la Santé et de la Recherche Médicale ethics committee). Peripheral blood mononuclear cells were isolated using Ficoll-Paque Plus (GE-17144003) and left overnight at 4°C in RPMI (#61870-010; Thermo Fisher Scientific) with 10% heat-decomplemented fetal bovine serum (FBS; #CVFSVF00-01; Eurobio). CD14+ cells were enriched by positive selection (#130-050-201; Miltenyi) and the CD14 negative fraction was subsequently used to isolate total CD4+ T cells by negative selection (#17952; Stem Cell). CD14+ cells were cultured in RPMI containing 10% FBS, 5% penicillin-streptomycin (PS; #10378-016; Thermo Fisher Scientifc), 50 μg/ml Gentamicin (#15750-045; Thermo Fisher Scientific), and 10 mM HEPES (#15630-056; Thermo Fisher Scientific). CD14+ cells were differentiated ex vivo into MDDCs by supplementing the culture media with 50 ng/ml IL-4 (#130-093-922; Miltenyi) and 10 ng/ml GM-CSF (#130-093-867; Miltenyi). MDDCs was used for experiments 3 or 4 d following the start of the culture. CD14+ cells were differentiated ex vivo into macrophages by adding 50 ng/ml MCSF (#130-096-492; Miltenyi) to the culture media containing RPMI with 1% PS, 5% FBS, and 5% human serum (#H4522; Sigma-Aldrich). MDMs were used 7 d following culture. Freshly isolated CD4+ T cells were cultured in X-VIVO 15 (#BE02-060F; Lonza) and activated using anti-CD3 and anti-CD28 Dynabeads (#10548353; Thermo Fisher Scientific) at a ratio of 1:5 (bead:cell). 100 U/ml human IL-2 (#1134002; Immunotools) was added to cultures 2 d following TCR stimulation and media with IL-2 was replenished every 48 h. To assess proliferation, freshly isolated CD4+ T cells were stained with cell proliferation dye (#65-0840-85; Thermo Fisher Scientific) prior to TCR stimulation.
Peripheral pDCs were enriched using pan-DC enrichment kit (#19251; Stem Cell) and subsequently stained with anti-HLA-DR APCeFluor780, anti-CD123 Viogreen, anti-CD45RA Vioblue (Miltenyi), anti-AXL PE (Clone #108724; R&D Systems), anti-CD33 PE-CF594, and with a cocktail of FITC conjugated antibodies against lineage markers CD19 (Miltenyi), CD3, CD14, CD16, and CD34 (BD), and sorted on a FACSAria as previously described (Ruffin et al., 2019). pDCs were defined as follows: Lin− HLA-DR+ CD33− CD45RA+ CD123+ AXL−, Lin− corresponds to CD19−CD16−CD3−CD34−CD14−. Sorted cells were cultured in X-VIVO 15 with 5% PS and 3 μg/ml GM-CSF.
Plasmids
pSIV3+, psPAX2, HXB2 env, CMV-VSVG, pTRIP-SFFV-GFP, and pTRIP-SFFV-GFPIRF3 were previously described (Manel et al., 2010). pTRIP-SFFV-GFP-RELA was obtained by PCR cloning of RELA (#23255; Addgene) into the pTRIP-SFFV-GFP backbone. RELA K5R and RELA K5Q mutants were generated by subcloning DNA fragments (Twist Biosciences) into the pTRIP-SFFV-GFP-RELA plasmid, resulting in pTRIP-SFFV-GFP-RELA K5R and pTRIP-SFFV-GFP-RELA K5Q. RELA K310R mutant was generated by overlapping PCR mutagenesis in pTRIP-SFFV-GFPRELA plasmid resulting in pTRIP-SFFV-GFP-RELA K310R. pLKO.1-puro-IRF7sh1 (IRF7sh1, 5′-CCCGAGCTGCACGTTCCTATA-3′), pLKO.1-puroIRF7sh5 (IRF7sh5, 5′-CGCAGCGTGAGGGTGTGTCTT-3′), and pLKO.1puro-shLacZ (LacZsh) were previously described (Döring et al., 2021). HIV-mTagBFP2 and HIV-2 ROD9 ∆env∆nef mTagBFP2+ were previously described (Bhargava et al., 2021). pTRIP-SFFV-GFP (control), pTRIP-SFFV-EGFP-FLAG-cGAS (cGAS), pTRIP-SFFV-EGFP-NLS (NLS), and pTRIP-SFFV-EGFP-NLS-FLAG-cGAS (NLS-cGAS) were previously described (Gentili et al., 2019). pTRIP-SFF-tagBFP-2A, pTRIP-SFFV–tagBFP-2A-STING, IFNB-pGL3, and pTRH1-NFkB-dscGFP have been previously described (Cerboni et al., 2017). pTRIP-SFFV-tagBFP-2A-ZebrafishSTING was subcloned from pCOM37_pcDNA4 ZFish STING (de Oliveira Mann et al., 2019). Human CD19 cDNA (NCBI accession no. NM_001178098; Genescript) was cloned into the pCDH-CMV-MCS-EF1-Puro plasmid (System Biosciences) to create pCDH-CMV-CD19 puro.
Lentivirus production
293FT cells were cultured in DMEM (#61965026; Thermo Fisher Scientific) with 5% PS and 10% FBS. 293FT cells were plated at 0.8 million cells in a 6-well plate and transfected with 3 μg of DNA complexed in 8 μl of TransIT-293 (#MIR2706; Mirus Bio) per well. The ratio of DNA used for transfections was as follows: 0.2 μg HXB2 env, 0.2 μg CMV-VSVG, 1 μg psPAX2, and 1.6 μg of pTRIP-SFFV or pLKO.1 lentivector. The ratio of plasmids for the production of HIV-1 and HIV-2 BFP single-round reporter viruses was 0.2 μg HXB2 env, 0.2 μg CMV-VSVG, and 2.6 μg HIV plasmid. SIV-VLPs were produced using 0.4 μg CMV-VSVG with 2.6 μg pSIV3+. Lentiviruses for MDMs were prepared by plating 7 million 293FT cells in T75 flasks and transfected with 8.35 μg DNA complexed in 116 µl PEImax (0.75 mM; #24765; Polyscience) per flask. The ratio of DNA used for transfection includes 3 μg psPAX2, 1.25 μg CMV-VSVG, and 4.10 µg pTRIP-SFFV-GFP. For SIV-VLP production for MDMs, 2.5 μg CMV-VSVG with 8.2 μg pSIV3+ was used. 18 h following transfection, media was removed and replenished with fresh media (3 ml for T cells and MDDCs or 8.5 ml for MDMs). 24–26 h later, viral supernatants were harvested, filtered using 0.45 μM filters, and used fresh or stored at −80°C.
For CAR expression, rLV.EFA.19BBz CAR lentivirus was produced using pLV, pHIV-GagPol, and pEnv plasmids and concentrated by ultracentrifugation (Flash Therapeutics). Titer was determined by serial dilution on activated human T cells.
CRISPR-Cas9 nucleofections
24 h after TCR stimulation, 2 million cells were washed with PBS and nucleofected using a Lonza kit (#V4XP-3032). Cr-RNA and TracrRNA (#1072534; IDT) were annealed by heating at 95°C for 5 min. Two gRNAs targeting cGAS were used and 100 pmol of each gRNA was incubated separately with 66 pmol of Cas9 protein (#1081059; IDT) for 10 mins at room temperature (RT) to form complexes. The two complexes were combined and a final volume of 5 μl containing 200 pmol of guides and 135 pmol of Cas9 was used per reaction. Nucleofection was performed using EH-100 program using the 4D-Nucelofector (Lonza). Following nucleofection cells were cultured for 3 d in 300 U/ml IL-2. 72 h following nucleofection, cells were harvested to assess the efficiency of gene knockout. cGAS gRNA targeted the following genomic sequences 5′-AGACTCGGTGGGATCCATCG-3′ (IDT# Hs.Cas9.MB21D1.1.AA) and 5′-CGAAGCCAAGACCTCCGCCC-3′ (IDT# Hs.Cas9.MB21D1.1.AL). Nucleofections were performed 5–6 h prior to lentivirus transductions.
Lentivirus transductions
CD14+ cells were plated at 1 million cells/ml and transduced with equal volumes of freshly harvested SIV-VLPs and pTRIP-SFFV vectors in the presence of 8 μg/ml protamine (#P4020; Sigma-Aldrich). CD4+ T cells were transduced with lentivectors 24 h post-TCR stimulation in 100 μl of cells (0.2 million cells) and 100 μl of freshly harvested lentivirus in the presence of 8 μg/ml protamine. T cells were spinoculated at 1,200 g for 2 h at 25°C. 0.5 million THP-1 cells in 500 μl media (RPMI, 10% FBS, and 1% PS) were transduced with 500 μl freshly harvested lentivirus in the presence of 8 μg/ml protamine. 1 μg/ml Puromycin (#ant-pr-1; Invivogen) was added 2 d after transduction when cells were transduced with pLKO.1-puro plasmid.
Primary cell stimulations and treatments
For stimulation of STING by extracellular ligands, 2.5 μg of ADUS100 (#tlrl-nacda2r-01; Invivogen) or 2′3′-cGAMP (#tlrl-nacga23-02; cGAMP; Invivogen ) was added to 100,000 cells in 100 µl media either on freshly isolated primary cells or 3–4 d after differentiation for MDDCs and 3 d after TCR stimulation for CD4+ T cells. Cells and supernatant were harvested 18–24 h after stimulation.
For delivery of STING ligands after permeabilization, T cells were used 3 or 4 d after TCR stimulation, and MDDCs were used 3–4 d following differentiation. 150,000 cells were plated in round-bottom 96-well plate. Media was removed by centrifugation and 20 µl 2× permeabilization buffer (100 mM HEPES, 200 mM KCl, 6 mM MgCl2, 0.2 mM DTT, 170 mM sucrose, 2 mM ATP, 2 mM GTP, 0.4% bovine serum albumin [BSA], and 20 μg/ml digitonin) and 20 μl cGAMP (12 μg/ml) was added to cells. 18–20 h following stimulation supernatants and cells were harvested. Cells were stained with a fixable viability dye (#65-0865-14; Invitrogen) and acquired on a FACSVerse (BD).
For cGAS stimulation, 2 µg of control GFP plasmid DNA (provided in the kit) was nucelofected in 2 million T cells 72 h after TCR stimulation (#VPA-1002; Lonza) using the program T020 (#Nucleofector 2b; Lonza). Cells were harvested at 4 and 24 h after nucleofection for analysis.
For experiments with conditioned media, CD4+ T cell– and MDDC-conditioned media refer to 0.45 µM filtered conditioned media from cultures of activated CD4+ T cell and MDDCs, respectively. CD4+ T cells were pretreated with 100 μl of MDDC-conditioned media or fresh MDDC culture media, 24 h prior to cGAMP stimulation. MDDCs were pretreated with 100 μl of CD4+ T cell-conditioned media or fresh CD4+ T cell culture media, 24 h prior to cGAMP stimulation. Where indicated, 10 ng/ml recombinant IFN-γ1b (#130-096-484; Miltenyi) was added.
Where indicated, cells were treated with 5AZA (# A2385; Sigma-Aldrich) 48 h prior to cGAMP stimulation. Fresh 5AZA was added daily to cells, and media was replenished with 5AZA following cGAMP stimulation. 100 nM TSA (# T1952; Sigma-Aldrich) was added to cells following cGAMP stimulation and left overnight. 25 µM Azidothymidine (AZT; #A2169; Sigma-Aldrich) and 10 µM nevirapine (NVP; #SML0097; Sigma-Aldrich) were added prior to lentiviral transduction and replenished when cells were expanded. B18R supernatant and control supernatant were produced from baculovirus-infected cells as described (Fernández de Marco et al., 2010). 10% of supernatants were added to T cell culture media 2 d after transduction and fresh supernatants were added every day for the next 2 d and maintained overnight after cGAMP stimulation. Where indicated, 1,000 U/ml IFNα2a (#11343506; Immunotools) was added 20–24 h prior to cGAMP stimulation.
To study TCR signaling response in iCD4+ T cells, 0.8 to 1 million CD4+ T cells were washed, resuspended in 150 µl of RPMI, and rested for 20 min at 37°C. Cells were left unstimulated or stimulated by adding Dynabeads at a ratio of 1:1 for the indicated time points. After activation, cells were centrifuged at 2,000 rpm for 3 min at RT and lysed in 40–50 µl of RIPA buffer (89901; Thermo Fisher Scientific) supplemented with a protease inhibitor cocktail (11873580001; Roche) and a phosphatase inhibitor cocktail (815-968-0747; Thermo Fisher Scientific). Lysates were processed as described in the protein isolation section.
Luciferase reporter assays
45,000 293FT cells were plated in a 24-well plate and transfected the following day with 200 ng IFNB-pGL3 or pTRH1-NFkB-dscGFP and 300 ng of pTRIP-SFFV-tagBFP-2A, pTRIP-SFFV-tagBFP-2A STING, pTRIP-SFFV-tagBFP-2A-ZebrafishSTING with TransIT-293 (Mirus). The next day the media was removed, and cells were stimulated with 4 µg/ml cGAMP (Invivogen) complexed with Lipofectamine 2000. 24 h later cells were lysed with passive lysis buffer (Promega) and 10 µl of the lysate was used for measuring luciferase activity using Luciferase Assay Reagent (E1483; Promega). Luminescence was acquired on a FLUOstar OPTIMA microplate reader (BMG Labtech).
IFN quantification
Culture supernatants were stored at −80°C and thawed prior to quantification. Concentrations of IFNα2, IFNβ, IFNγ, IFNλ1, and IFNλ2/3 were measured using LEGENDplex Human Type 1/2/3 Interferon Panel (#740396; BioLegend) according to the manufacturer’s protocol. Data were acquired on a BD FACSVerse (BD) and analyzed with LEGENDplex Software (BioLegend). In figures, dotted lines indicate limits of detection.
Gene expression analysis
RNA was extracted from 0.4 to 1 million cells using the Nucleospin RNA II kits (#740955.50; Macherey-Nagel). cDNA was synthesized using random primers (#48190-011; Invitrogen) from 0.1 μg RNA using Superscript III Reverse transcriptase (#18080044; Thermo Fisher Scientific). Real-time qPCR (RT-qPCR) was carried out in 20 μl reactions using SYBR Green I Master (#4887352001; Roche). Primers used were as follows: IFNL1 (Fwd: 5′-GGTGACTTTGGTGCTAGGCT, Rev: TGAGTGACTCTTCCAAGGCG-3′), IFNB (Fwd: 5′-GTCTCCTCCAAATTGCTCTC, Rev: ACAGGAGCTTCTGACACTGA-3′), ACTB (Fwd: 5′-GGACTTCGAGCAAGAGATGG, Rev: AGCACTGTGTTGGCGTACAG-3′), B2M (Fwd: 5′-TCTCTGCTGGATGACGTGAG, Rev: TAGCTGTGCTCGCGCTACT-3′).
Bulk RNA-seq data analysis of CD4+ T cells stimulated with cGAMP
5 h after cGAMP (6 µg/ml) stimulation of activated CD4+ T cells, total RNA was extracted (#740955.50; Macherey-Nagel). RNA integrity was verified using Agilent Bioanalyzer (#5607-1511; Agilent RNA 6000 Nano kit) and all samples had a RIN >9. RNA sequencing libraries were prepared from 500 ng of total RNA using the Illumina TruSeq Stranded mRNA Library preparation kit. A first step of polyA selection using magnetic beads was performed to focus sequencing on polyadenylated transcripts. After fragmentation, cDNA synthesis was performed and resulting fragments were used for dA-tailing and ligated to the TruSeq-indexed adapters. PCR amplification was performed to create the final cDNA library (with 13 cycles). After quantification of PCR products, sequencing was carried out using 2 × 100 cycles (paired-end reads, 100 nucleotides) on a Novaseq 6000 instrument, targeting 25 M clusters. The data were aligned to the hg19 (ENSEMBL annotation v.74) genome using the RNA-seq pipeline of the Curie bioinformatics platform, rnaseq v3.1.1. Reads were trimmed with TrimGalore (v.0.6.2) and aligned on the reference genome using STAR (v 2.6.1; Dobin et al., 2013). Quality control was done with picard (v.2.18.15), RSeQC (v.2.6.4), and preseq (v.2.0.3; Wang et al., 2012). Read counts were generated with STAR. Quality reports were generated with MultiQC (v.1.7). We filtered the count matrix only keeping genes that have in at least one sample a transcripts per million value of 1, this strategy left us with 10,671 genes tested of a total of 57,820 genes in the count matrix. Differential expression analysis was performed using DESeq2 (1.26.0), and a complete list of differentially expressed genes is provided in Table S1 (Love et al., 2014). A gene was designated as differentially expressed with an adjusted P value of <0.05 and an absolute log fold change >1. A list of 625 ISGs was used for annotation (Silvin et al., 2017; Cerboni et al., 2021). Additionally, Bioconductor package clusterProfiler (3.14.3) was used for the pathway over-representation analysis using public databases GO and Kegg (Yu et al., 2012). Upregulated and downregulated genes were analyzed separately. Pathways with an adjusted P value <0.05 and that contained at least five genes from our dataset were considered significant. Gene expression data have been deposited at GEO (accession no. GSE182647).
Bulk RNA-seq data analysis of CD4+ T cells expressing RELA or RELA K5R
CD4+ T cells transduced with either pTRIP-SFFV-GFP or pTRIP-SFFV-GFP-RELA or pTRIP-SFFV-GFP-RELA K5R and RNA was isolated 4 d after transduction. The analysis was performed by GenoSplice. Analysis of sequencing data quality, reads repartition (e.g., for potential ribosomal contamination), inner distance size estimation, genebody coverage, and strand-specificity of the library were performed using FastQC v0.11.2, Picard-Tools v1.119, Samtools v1.0, and RSeQC v2.3.9. Reads were mapped using STAR v2.7.5a on the human hg38 genome assembly and read count was performed using featureCount from SubRead v1.5.0. Gene expression was estimated as described previously (Paillet et al., 2021) using Human FAST DB v2022_1 annotations. Only genes expressed in at least one of the two compared conditions were analyzed further. Genes were considered as expressed if their FPKM value was greater than the FPKM of 96% of the intergenic regions (background). Analysis at the gene level was performed using DESeq2 using experiment ID in the DESeq2 GLM model. Genes were considered differentially expressed for fold changes ≥1.5 and P values ≤0.05. Pathway analyses were performed using WebGestalt v0.4.4 merging results from upregulated and downregulated genes only, as well as all regulated genes. Pathways and networks were considered significant with P values ≤0.05. The results of this analysis were compared to those from Zhao et al. (2015) corresponding to the GSE68329 GEO dataset ID. Regulated genes from Zhao et al. (2015) were retrieved using GEO2R using adjusted P value ≤0.05 and fold change ≥ 1.5. Gene expression data have been deposited at GEO (accession no. GSE182647).
Protein isolation, immunoprecipitations, and Western blot
0.5 million cells were lysed for 30 min on ice in 50 µl RIPA buffer (50 mM Tris HCl, 150 mM NaCl, 0.1% SDS, 0.5% DOC, and 1% NP-40) with Protease inhibitor (#1187358001; Roche). To evaluate, phospho-proteins 50 mM NaF and 1 mM sodium orthovanadate were added to the lysis buffer. Protein lysates were cleared by centrifugation at 12,000 g for 10–12 min at 4°C. 10 µl of 6× Laemmli buffer (#J61337; Thermo Fisher Scientific) was added to 50 μl protein lysates and samples were boiled at 95°C for 15 min. For immunoprecipitations, 4 or 5 d after transduction equal cell numbers were harvested from donor-matched CD4+ T cells and MDDCs transduced with pTRIP-SFFV-GFP-RELA. 5 to 7 million cells were lysed in 250–300 µl of RIPA with protease inhibitors, 50 mM NaF, and 1 mM sodium orthovanadate for 15 mins on ice and sonicated for 15 min at 4°C. Protein lysates were cleared by centrifugation at 12,000 g for 10–12 min at 4°C. 10% of lysates were harvested for input and the rest were incubated for 2.5 h with 15 µl of GFP-trap magnetic agarose beads (Chromotek # gtma-20) at 18 rpm. Beads were washed thrice with PBS prior to incubation with protein lysates. At the end of the incubation period, beads were washed thrice with RIPA lysis buffer using a magnet to remove unbound proteins, resuspended in 30 µl of lysis buffer and 5 µl of 6× Laemmli buffer, denatured at 95°C and stored at −20°C. Protein samples were resolved on 4–20% BioRad precast SDS-PAGE gels (#5671125) and transferred onto PVDF membranes (#1704157; BioRad). Membranes were blocked in 5% non-fat dry milk in PBS 0.1% Tween or in TBS 5% BSA 0.1% Tween to detect phosphorylated proteins. Primary antibodies were used at 1:1,000: phospho-STING (S366; clone-D7C3S), STING (clone-D2P2F), phospho-TBK1 (S172; clone-D52C2), TBK1 (clone-D1B4), phospho IRF3 (S396; clone-D6O1M), IRF3 (clone-D83B9), RELA (clone C22B4, clone D14E12 used for immunoprecipitation analysis, and clone C-20 used for T cell signaling analysis), acetyl-NFκB (K310; CST #3045), phospho p65 (S536; clone-93H1), IRF7 (clone-12G9A36), cGAS (clone-D1D3G), Actin (clone-C4), Sendai viral proteins (# MBL-PD029; Biozol), phospho-ERK1/2 (T202/Y204; clone-E10), ERK1/2 (clone-137F5), phospho-PLCγ1 (Y783; clone-D6M9S), PLCγ1 (CST# 2822S), phospho-Zap-70 (Y319)/Syk (Y352; CST#2701S), and Zap-70 (clone-D1C10E). Secondary anti-rabbit or anti-mouse antibodies were used at 1:10,000 (#sc-2357, #sc-516102; Santa Cruz), secondary antibodies for T cell signaling (#111-036-046 or # 115-035-146; Jackson ImmunoResearch) and secondary antibody for immunoprecipitation analysis (Ebioscience 18-8816 Rabbit Trueblot) were used at 1:3,000. ECL signal (#1705061 and #1705062; BioRad) was recorded on a ChemiDoc Touch BioRad Imager. Data were analyzed and quantified with Image Lab software (BioRad). In figures, dotted lines indicate the reordering of lanes from a single original membrane for visualization purpose.
Confocal microscopy and analysis
4–5 d following transduction, 500,000 cells were plated on 0.1% poly-L-lysine in water-coated (#8920; Sigma-Aldrich) coverslips and left for 30 min to attach. Cells were fixed in PBS 4% paraformaldehyde for 20 min at RT. Coverslips were washed twice in PBS and quenched with 500 μl of freshly prepared PBS glycine (375 mg glycine in 50 ml of PBS) for 10 min at RT. Cells were then permeabilized and simultaneously blocked with PBS, 0.2 % BSA, 0.05% saponin, and 1% goat serum (#G9023; Sigma-Aldrich) for 30 min at RT. Coverslips were incubated with primary antibody against cGAS (clone D1D3G) or isotype control (clone DA1E) at a concentration of 0.085 μg/ml in PBS, 0.2% BSA, and 0.05% saponin, and left overnight at 4°C in a moist chamber. Coverslips were washed five times with PBS, 0.2% BSA, and 0.05% saponin, and incubated in secondary antibody goat anti-rabbit IgG at 1:400 (#A21246; Invitrogen) for 45 min at RT. Coverslips were washed five times with PBS, 0.2% BSA, and 0.05% saponin, rinsed in water, and mounted on a glass slide using 10 µl fluoromount-G with DAPI (#00-4959-520; Invitrogen). Glass slides were allowed to dry in a 37°C chamber for 30 min and stored at 4°C. Images were acquired on a Leica DmI8 inverted microscope equipped with an SP8 confocal unit using a 40× (1.3 NA) oil objective. Image analysis was performed using Fiji software (Schindelin et al., 2012). Homemade scripts were used to analyze cGAS localization. All images were smoothened using a filtering of mean radius 1 pixel. For each Z-stacks, an optical section in the middle of the nuclei was chosen. Then, for each condition, a binary mask of the nuclei and a mask of the cell were obtained by applying a threshold respectively on the DAPI signal or the cGAS signal to define nuclear and cytosolic regions. To avoid out-of-focus cells and to be sure that measurement will be done inside nuclei, a filter was applied to keep only cells with a section larger than 20 µm2 containing a nucleus section larger than 16 μm2. Finally, average cGAS and GFP intensities were measured in the whole cell, the nuclei, and the cytosol (defined by the whole cell excluding the nuclear region).
cGAMP isolation and quantification
Cell pellets were stored at −80°C until extraction. Frozen pellets were fixed in 80% cold methanol and subjected to five freeze-thaw cycles in liquid nitrogen. Samples were then centrifuged at 16,000 g for 20 min at 4°C. The supernatants were further subjected to drying in Savant DNA Speed Vac DNA 110 at 65°C for 2 h. The pellets were resuspended in 100 μl RNase- and DNase-free water and cGAMP was quantified by ELISA as per manufacturer’s protocol (#501700; Interchim). cGAMP quantity was normalized to cell numbers and represented as picograms/million cells.
Viral infections
For HIV infections, 4 d after lentivirus transduction and 2 d following 5AZA treatment, 0.07 million cells in 70 μl media were infected with 70 μl BFP-reporter single-round HIV-1 or HIV-2 with 8 μg/ml protamine and fresh 5AZA (2 μM). Cells were pretreated with B18R or control supernatants (10%) for 48 h prior to viral infections. Fresh B18R or control supernatants were added at the time of infection and left untouched for 48 h. Serial dilutions of viruses at 1/3 were performed. GHOST X4R5 cells were infected in parallel to control the viral titer of the inoculum. Cells and supernatants were harvested 48 h after infection.
For Sendai virus infections, 100,000 cells in 100 μl were infected with 100 μl of Sendai virus 200 HA/ml (Charles River, Cantell Strain) with 8 μg/ml protamine. Culture supernatants for IFN quantification were harvested 18–24 h after infection. Cells for RNA and protein extraction were harvested 6 h following infection.
CAR-T tumor spheroid killing assay
CD19+ mKate2+ A549 cells were produced by lentiviral transduction using pCDH-CMV-CD19 puro and ILV-EF1a-mKate-9-X01(Flash Therapeutics) lentivectors. 1,000 cells were plated 4 d prior to coculture in low-cluster 96-well plates (#7007; Costar) and allowed to form spheroids in RPMI, 10% FBS, and 5% PS. Total CD3+ T cells were isolated by negative selection from peripheral blood mononuclear cells (#17951; Stem Cell) and stimulated with Dynabeads at a ratio of 1:3 (beads:cell). Cells were cultured at 1 million per ml in X-VIVO 15 with 5% PS, 5% decomplemented human serum (Sigma-Aldrich), and 50 mM 2-mercaptoethanol (#31350-010; GIBCO). 24 h following TCR stimulation, 100,000 cells in 100 μl were cotransduced with 100 μl of lentivector viral particles (pTRIP-SFFV-GFP or pTRIP-SFFV-GFPRELAK5R) combined with the CAR lentivirus (rLV.EFA.19BBz; Flash Therapeutics) at a multiplicity of infection (MOI) of 10 (corresponding usually to <1 µl). Spinoculation was carried out at 1,200 g for 2 h at 25°C. Cells were expanded for 48 h with fresh media and 300 U/ml IL-2. 3 d following transduction, T cells were stained with CD19 CAR detection reagent (130-115-965; Miltenyi) for 15 min at 4°C, washed twice, and subsequently with streptavidin A647 conjugate (#S21374; Invitrogen) for 30 min at 4°C. Cells were fixed and acquired on a FACSVerse (BD). The next day, Dynabeads were removed by magnetic separation, and 250, 500, or 1,000 CAR+ cells were added per well of A549 spheroids in quadruplicates. To evaluate the impact of IFN-I on CART function, IFNα2a (10,000 U/ml or 1,000 U/ml) was added at the time of coculture. Plates were then placed in an IncuCyteS3 and images were acquired using a 10× objective (1.24 μm/pixel) every 3 h for 5–6 d. Images were analyzed using the IncuCyte S3 software. At the end of the culture, supernatants were harvested and assayed for IFN concentrations.
Cell stimulation and intracellular staining
Total CD3+ T cells were cotransduced with CAR lentivirus and either pTRIP-SFFV-GFP or pTRIP-SFFV-GFP-RELA K5R for 4 d prior to staining. 0.15 million cells were treated with cell-stimulation cocktail and protein transport inhibitors (#15516286 or 15172069; Thermo Fisher Scientific) for 4 h. Cells were stained with Fixable Viability Dye eFluor 780 (#15383562; Thermo Fisher Scientific), CD4 BV510 (#562971; Thermo Fisher Scientific), and CD8a PE (#15506706; Thermo Fisher Scientific), and then fixed and permeabilized using the eBioscience FOXP3 transcription factor staining buffer set (#11500597; Thermo Fisher Scientific). Permeabilized cells were stained with antibodies against IFNγ PeCy7 (#13417646; Thermo Fisher Scientific) and Granzyme B APC (#515405; Biolegend). Samples were acquired on a NovoCyte (Agilent). Isotype controls were used to define gates.
Statistical analysis
Data were analyzed using PRISM software (version 9, GraphPad Prism). Statistical tests and parameters including statistical significance are reported in figure legends.
Online supplemental material
Fig. S1 shows the characterization of the IFN response of T cells to cGAMP stimulation, IFNα2a pretreatment, and Sendai virus infection. Fig. S2 shows the impact of RELA on IFN response to cGAMP in CD4+ T cells MDDCs, MDMs, and THP-1 cells. Fig. S3 shows the impact of IRF3, TSA, 5AZA, and RELA K5R on IFN response to cGAMP, viability, and signaling in CD4+ T cells. Fig. S4 shows the impact of IRF3, 5AZA, and RELA K5R on IFN response to cGAMP in CD4+ T cells. Fig. S5 shows the functional impact of iCD4+ T cells in the context of HIV infection and anti-tumor response. Table S1 provides a complete list of differentially expressed genes.
Supplementary Material
provides a complete list of differentially expressed genes.
is the source file for Fig. 1.
is the source file for Fig. 2.
is the source file for Fig. 4.
is the source file for Fig. 5.
is the source file for Fig. 6.
is the source file for Fig. 7.
is the source file for Fig. 8.
is the source file for Fig. S1.
is the source file for Fig. S2.
is the source file for Fig. S3.
is the source file for Fig. S4.
Acknowledgments
We thank Jon Kagan and the other anonymous reviewers. We thank U932 members and in particular Manel lab members for sharing their expertise and for insightful scientific discussions, Claire Hivroz for critical reading of the manuscript, Frédéric Rieux-Laucat (Institut Imagine, Paris, France) for sharing the GFP-RELA plasmid, Franck Perez (Institut Curie, Paris, France) for the ILV-EF1a-mKate-9-X01 plasmid, Carina de Oliveira Mann (Technical University of Munich, Munich, Germany) for the pCOM37_pcDNA4 ZFish STING plasmid, Flavien Brouiller (Institut Curie) for kindly providing sorted primary dendritic cell subsets, Sheila Lopez-Cobo and Sebastian Amigorena (Institut Curie) for providing the CAR lentivirus and sharing their expertise on CAR T cell generation and culture, and Maria C. Fernández-Moyano and Antonio Alcamí (Centro de Biología Molecular Severo Ochoa, Madrid, Spain) for providing the recombinant B18 protein from vaccinia virus. High-throughput sequencing was performed by the ICGex NGS platform of Institut Curie (Equipex ANR-10-EQPX-03 and France Génomique Consortium ANR-10-INBS-09-08).
This work was supported by Institute Curie, INSERM, Agence Nationale de la Recherche (ANR-17-CE15-0025-01, ANR-19-CE15-0018-01, ANR-18-CE92-0022-01, 11-LABX-0043), Fondation pour la Recherche Médicale (EQU202103012774), Agence Nationale de Recherche sur le Sida et les Hépatites (ANRS, ECTZ36691, ECTZ25472, ECTZ71745, ECTZ118797), Région Ile-de-France (DIM Virofector), Sidaction (20-2-AEQ-12822-2, 17-1-AAE-11097-2, VIH2016126002) Fondation Chercher et Trouver, and Fondation pour la Recherche Médicale (EQU202103012774 to N. Manel). N. Jeremiah was supported by the Institute Curie International Post-doc fellowship and ANRS post-doctoral Fellowship.
Author contributions: N. Jeremiah designed, performed experiments, and wrote the manuscript. N. Manel conceptualized the study, designed experiments, and wrote the manuscript. H. Ferran and K. Antoniadou provided assistance with experiments. K. De Azevedo performed the RNA-seq data analysis of the cGAMP response. J. Nikolic and P. Benaroch provided expertise on CART Tumor spheroid killing assay. M. Maurin provided expertise in image acquisition and analysis.
References
- Angin, M., Volant S., Passaes C., Lecuroux C., Monceaux V., Dillies M.A., Valle-Casuso J.C., Pancino G., Vaslin B., Le Grand R., et al. 2019. Metabolic plasticity of HIV-specific CD8+ T cells is associated with enhanced antiviral potential and natural control of HIV-1 infection. Nat. Metab. 1:704–716. 10.1038/s42255-019-0081-4 [DOI] [PubMed] [Google Scholar]
- Apostolou, E., and Thanos D.. 2008. Virus Infection Induces NF-κB-dependent interchromosomal associations mediating monoallelic IFN-β gene expression. Cell. 134:85–96. 10.1016/j.cell.2008.05.052 [DOI] [PubMed] [Google Scholar]
- Bardwell, L. 2008. Signal transduction: Turning a switch into a rheostat. Curr. Biol. 18:R910–R912. 10.1016/j.cub.2008.07.082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnabei, L., Lamrini H., Castela M., Jeremiah N., Stolzenberg M.C., Chentout L., Jacques S., Bouafia A., Magérus-Chatinet A., Moncan M., et al. 2020. Heterozygous RELA mutations cause early-onset systemic lupus erythematosus by hijacking the NF-κB pathway towards transcriptional activation of type-I Interferon genes. bioRxiv. 10.1101/2020.04.27.046102 [DOI] [Google Scholar]
- Barnett, K.C., Coronas-Serna J.M., Zhou W., Ernandes M.J., Cao A., Kranzusch P.J., and Kagan J.C.. 2019. Phosphoinositide interactions position cGAS at the plasma membrane to ensure efficient distinction between self- and viral DNA. Cell. 176:1432–1446.e11. 10.1016/j.cell.2019.01.049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basagoudanavar, S.H., Thapa R.J., Nogusa S., Wang J., Beg A.A., and Balachandran S.. 2011. Distinct roles for the NF-κ B RelA subunit during antiviral innate immune responses. J. Virol. 85:2599–2610. 10.1128/JVI.02213-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berg, R.K., Rahbek S.H., Kofod-Olsen E., Holm C.K., Melchjorsen J., Jensen D.G., Hansen A.L., Jørgensen L.B., Ostergaard L., Tolstrup M., et al. 2014. T cells detect intracellular DNA but fail to induce type I IFN responses: Implications for restriction of HIV replication. PLoS One. 9:e84513. 10.1371/journal.pone.0084513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhargava, A., Williart A., Maurin M., Davidson P.M., Jouve M., Piel M., Lahaye X., and Manel N.. 2021. Inhibition of HIV infection by structural proteins of the inner nuclear membrane is associated with reduced chromatin dynamics. Cell Rep. 36:109763. 10.1016/j.celrep.2021.109763 [DOI] [PubMed] [Google Scholar]
- Bouis, D., Kirstetter P., Arbogast F., Lamon D., Delgado V., Jung S., Ebel C., Jacobs H., Knapp A.M., Jeremiah N., et al. 2019. Severe combined immunodeficiency in stimulator of interferon genes (STING) V154M/wild-type mice. J. Allergy Clin. Immunol. 143:712–725.e5. 10.1016/j.jaci.2018.04.034 [DOI] [PubMed] [Google Scholar]
- Bridgeman, A., Maelfait J., Davenne T., Partridge T., Peng Y., Mayer A., Dong T., Kaever V., Borrow P., and Rehwinkel J.. 2015. Viruses transfer the antiviral second messenger cGAMP between cells. Science. 349:1228–1232. 10.1126/science.aab3632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buerki, C., Rothgiesser K.M., Valovka T., Owen H.R., Rehrauer H., Fey M., Lane W.S., and Hottiger M.O.. 2008. Functional relevance of novel p300-mediated lysine 314 and 315 acetylation of RelA/p65. Nucleic Acids Res. 36:1665–1680. 10.1093/nar/gkn003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerboni, S., Jeremiah N., Gentili M., Gehrmann U., Conrad C., Stolzenberg M.C., Picard C., Neven B., Fischer A., Amigorena S., et al. 2017. Intrinsic antiproliferative activity of the innate sensor STING in T lymphocytes. J. Exp. Med. 214:1769–1785. 10.1084/jem.20161674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerboni, S., Marques-Ladeira S. and Manel N.. 2021. Virus-stimulated dendritic cells elicit a T antiviral transcriptional signature in human CD4+ lymphocytes. J. Mol. Biol. 436:167389. 10.1016/j.jmb.2021.167389 [DOI] [PubMed] [Google Scholar]
- Chen, J., Sun H.W., Yang Y.Y., Chen H.T., Yu X.J., Wu W.C., Xu Y.T., Jin L.L., Wu X.J., Xu J., and Zheng L.. 2021. Reprogramming immunosuppressive myeloid cells by activated T cells promotes the response to anti-PD-1 therapy in colorectal cancer. Signal Transduct.Target.Ther. 6:4. 10.1038/s41392-020-00377-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, L.F., Mu Y., and Greene W.C.. 2002. Acetylation of RelA at discrete sites regulates distinct nuclear functions of NF-κB. EMBO J. 21:6539–6548. 10.1093/emboj/cdf660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, X., Barozzi I., Termanini A., Prosperini E., Recchiuti A., Dalli J., Mietton F., Matteoli G., Hiebert S., and Natoli G.. 2012. Requirement for the histone deacetylase Hdac3 for the inflammatory gene expression program in macrophages. Proc. Natl. Acad. Sci. USA. 109:E2865–E2874. 10.1073/pnas.1121131109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiappinelli, K.B., Strissel P.L., Desrichard A., Li H., Henke C., Akman B., Hein A., Rote N.S., Cope L.M., Snyder A., et al. 2015. Inhibiting DNA methylation causes an interferon response in cancer via dsRNA including endogenous retroviruses. Cell. 162:974–986. 10.1016/j.cell.2015.07.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clavijo, P.E., and Frauwirth K.A.. 2012. Anergic CD8+ T lymphocytes have impaired NF-κB activation with defects in p65 phosphorylation and acetylation. J. Immunol. 188:1213–1221. 10.4049/jimmunol.1100793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corrales, L., Glickman L.H., McWhirter S.M., Kanne D.B., Sivick K.E., Katibah G.E., Woo S.R., Lemmens E., Banda T., Leong J.J., et al. 2015. Direct activation of STING in the tumor microenvironment leads to potent and systemic tumor regression and immunity. Cell Rep. 11:1018–1030. 10.1016/j.celrep.2015.04.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin, A., Davis C.A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., and Gingeras T.R.. 2013. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics. 29:15–21. 10.1093/bioinformatics/bts635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Döring, M., De Azevedo K., Blanco-Rodriguez G., Nadalin F., Satoh T., Gentili M., Lahaye X., De Silva N.S., Conrad C., Jouve M., et al. 2021. Single-cell analysis reveals divergent responses of human dendritic cells to the MVA vaccine. Sci. Signal. 14:eabd9720. 10.1126/scisignal.abd9720 [DOI] [PubMed] [Google Scholar]
- Elsner, C., Ponnurangam A., Kazmierski J., Zillinger T., Jansen J., Todt D., Döhner K., Xu S., Ducroux A., Kriedemann N., et al. 2020. Absence of cGAS-mediated type I IFN responses in HIV-1-infected T cells. Proc. Natl. Acad. Sci. USA. 117:19475–19486. 10.1073/pnas.2002481117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernández de Marco, M.d. M., Alejo A., Hudson P., Damon I.K., and Alcami A.. 2010. The highly virulent variola and monkeypox viruses express secreted inhibitors of type I interferon. FASEB J. 24:1479–1488. 10.1096/fj.09-144733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freaney, J.E., Kim R., Mandhana R., and Horvath C.M.. 2013. Extensive cooperation of immune master regulators IRF3 and NFκB in RNA Pol II recruitment and pause release in human innate antiviral transcription. Cell Rep. 4:959–973. 10.1016/j.celrep.2013.07.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gentili, M., Kowal J., Tkach M., Satoh T., Lahaye X., Conrad C., Boyron M., Lombard B., Durand S., Kroemer G., et al. 2015. Transmission of innate immune signaling by packaging of cGAMP in viral particles. Science. 349:1232–1236. 10.1126/science.aab3628 [DOI] [PubMed] [Google Scholar]
- Gentili, M., Lahaye X., Nadalin F., Nader G.P.F., Puig Lombardi E., Herve S., De Silva N.S., Rookhuizen D.C., Zueva E., Goudot C., et al. 2019. The N-terminal domain of cGAS determines preferential association with centromeric DNA and innate immune activation in the nucleus. Cell Rep. 26:2377–2393.e13. 10.1016/j.celrep.2019.01.105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gough, D.J., Messina N.L., Clarke C.J.P., Johnstone R.W., and Levy D.E., 2012. Constitutive type I interferon modulates homeostatic balance through tonic signaling. Immunity. 36:166–174. 10.1016/j.immuni.2012.01.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gulen, M.F., Koch U., Haag S.M., Schuler F., Apetoh L., Villunger A., Radtke F., and Ablasser A.. 2017. Signalling strength determines proapoptotic functions of STING. Nat. Commun. 8:427. 10.1038/s41467-017-00573-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Härtlova, A., Erttmann S.F., Raffi F.A., Schmalz A.M., Resch U., Anugula S., Lienenklaus S., Nilsson L.M., Kröger A., Nilsson J.A., et al. 2015. DNA damage primes the type I interferon system via the cytosolic DNA sensor STING to promote anti-microbial innate immunity. Immunity. 42:332–343. 10.1016/j.immuni.2015.01.012 [DOI] [PubMed] [Google Scholar]
- Hernáez, B., Alonso-Lobo J.M., Montanuy I., Fischer C., Sauer S., Sigal L., Sevilla N., and Alcamí A.. 2018. A virus-encoded type I interferon decoy receptor enables evasion of host immunity through cell-surface binding. Nat. Commun. 9:5440. 10.1038/s41467-018-07772-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honda, K., Yanai H., Negishi H., Asagiri M., Sato M., Mizutani T., Shimada N., Ohba Y., Takaoka A., Yoshida N., and Taniguchi T.. 2005. IRF-7 is the master regulator of type-I interferon-dependent immune responses. Nature. 434:772–777. 10.1038/nature03464 [DOI] [PubMed] [Google Scholar]
- Imanishi, T., Unno M., Kobayashi W., Yoneda N., Matsuda S., Ikeda K., Hoshii T., Hirao A., Miyake K., Barber G.N., et al. 2019. Reciprocal regulation of STING and TCR signaling by mTORC1 for T-cell activation and function. Life Sci. Alliance. 2:e201800282. 10.26508/lsa.201800282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwasaki, A., and Medzhitov R.. 2010. Regulation of adaptive immunity by the innate immune system. Science. 327:291–295. 10.1126/science.1183021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeremiah, N., Neven B., Gentili M., Callebaut I., Maschalidi S., Stolzenberg M.C., Goudin N., Frémond M.L., Nitschke P., Molina T.J., et al. 2014. Inherited STING-activating mutation underlies a familial inflammatory syndrome with lupus-like manifestations. J. Clin. Invest. 124:5516–5520. 10.1172/JCI79100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kagamu, H., Kitano S., Yamaguchi O., Yoshimura K., Horimoto K., Kitazawa M., Fukui K., Shiono A., Mouri A., Nishihara F., et al. 2020. CD4+ T-cell immunity in the peripheral blood correlates with response to anti-PD-1 therapy. Cancer Immunol. Res. 8:334–344. 10.1158/2326-6066.CIR-19-0574 [DOI] [PubMed] [Google Scholar]
- Kiernan, R., Brès V., Ng R.W.M., Coudart M.P., El Messaoudi S., Sardet C., Jin D.Y., Emiliani S., and Benkirane M.. 2003. Post-activation turn-off of NF-κB-dependent transcription is regulated by acetylation of p65. J. Biol. Chem. 278:2758–2766. 10.1074/jbc.M209572200 [DOI] [PubMed] [Google Scholar]
- Kwon, H.-S., Brent M.M., Getachew R., Jayakumar P., Chen L.F., Schnolzer M., McBurney M.W., Marmorstein R., Greene W.C., and Ott M.. 2008. Human immunodeficiency virus type 1 Tat protein inhibits the SIRT1 deacetylase and induces T cell hyperactivation. Cell Host Microbe. 3:158–167. 10.1016/j.chom.2008.02.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehrman, G., Hogue I.B., Palmer S., Jennings C., Spina C.A., Wiegand A., Landay A.L., Coombs R.W., Richman D.D., Mellors J.W., et al. 2005. Depletion of latent HIV-1 infection in vivo: A proof-of-concept study. Lancet. 366:549–555. 10.1016/S0140-6736(05)67098-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy, D., Kuo A.J., Chang Y., Schaefer U., Kitson C., Cheung P., Espejo A., Zee B.M., Liu C.L., Tangsombatvisit S., et al. 2011. Lysine methylation of the NF-κB subunit RelA by SETD6 couples activity of the histone methyltransferase GLP at chromatin to tonic repression of NF-κB signaling. Nat. Immunol. 12:29–36. 10.1038/ni.1968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, H., Wittwer T., Weber A., Schneider H., Moreno R., Maine G.N., Kracht M., Schmitz M.L., and Burstein E.. 2012. Regulation of NF-κB activity by competition between RelA acetylation and ubiquitination. Oncogene. 31:611–623. 10.1038/onc.2011.253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, P., Kaiser P., Lampiris H.W., Kim P., Yukl S.A., Havlir D.V., Greene W.C., and Wong J.K.. 2016. Stimulating the RIG-I pathway to kill cells in the latent HIV reservoir following viral reactivation. Nat. Med. 22:807–811. 10.1038/nm.4124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, T., Huang T., Du M., Chen X., Du F., Ren J., and Chen Z.J.. 2021. Phosphorylation and chromatin tethering prevent cGAS activation during mitosis. Science. 371:eabc5386. 10.1126/science.abc5386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, W., Lu L., Lu J., Wang X., Yang C., Jin J., Wu L., Hong X., Li F., Cao D., et al. 2020. cGAS-STING-mediated DNA sensing maintains CD8+ T cell stemness and promotes antitumor T cell therapy. Sci. Transl. Med. 12:eaay9013. 10.1126/scitranslmed.aay9013 [DOI] [PubMed] [Google Scholar]
- Liu, Y., Jesus A.A., Marrero B., Yang D., Ramsey S.E., Sanchez G.A.M., Tenbrock K., Wittkowski H., Jones O.Y., Kuehn H.S., et al. 2014. Activated STING in a vascular and pulmonary syndrome. N. Engl. J. Med. 371:507–518. 10.1056/NEJMoa1312625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love, M.I., Huber W., and Anders S.. 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15:550. 10.1186/s13059-014-0550-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luksch, H., Stinson W.A., Platt D.J., Qian W., Kalugotla G., Miner C.A., Bennion B.G., Gerbaulet A., Rösen-Wolff A., and Miner J.J.. 2019. STING-associated lung disease in mice relies on T cells but not type I interferon. J. Allergy Clin. Immunol. 144:254–266.e8. 10.1016/j.jaci.2019.01.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manel, N., Hogstad B., Wang Y., Levy D.E., Unutmaz D., and Littman D.R.. 2010. A cryptic sensor for HIV-1 activates antiviral innate immunity in dendritic cells. Nature. 467:214–217. 10.1038/nature09337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marié, I.J., Chang H.-M., and Levy D.E.. 2018. HDAC stimulates gene expression through BRD4 availability in response to IFN and in interferonopathies. J. Exp. Med. 215:3194–3212. 10.1084/jem.20180520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishioka, K., Daidoji T., and Nakaya T.. 2019. Demethylation around the transcriptional start site of the IFN-β gene induces IFN-β production and protection against influenza virus infection. Biochem. Biophys. Res. Commun. 520:269–276. 10.1016/j.bbrc.2019.09.136 [DOI] [PubMed] [Google Scholar]
- Odendall, C., Dixit E., Stavru F., Bierne H., Franz K.M., Durbin A.F., Boulant S., Gehrke L., Cossart P., and Kagan J.C.. 2014. Diverse intracellular pathogens activate type III interferon expression from peroxisomes. Nat. Immunol. 15:717–726. 10.1038/ni.2915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh, H., and Ghosh S.. 2013. NF-κB: Roles and regulation in different CD4+ T-cell subsets. Immunol. Rev. 252:41–51. 10.1111/imr.12033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Oliveira Mann, C.C., Orzalli M.H., King D.S., Kagan J.C., Lee A.S.Y., and Kranzusch P.J.. 2019. Modular architecture of the STING C-terminal tail allows interferon and NF-κB signaling adaptation. Cell Rep. 27:1165–1175.e5. 10.1016/j.celrep.2019.03.098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paillet, J., Plantureux C., Lévesque S., Le Naour J., Stoll G., Sauvat A., Caudana P., Tosello Boari J., Bloy N., Lachkar S., et al. 2021. Autoimmunity affecting the biliary tract fuels the immunosurveillance of cholangiocarcinoma. J. Exp. Med. 218:e20200853. 10.1084/jem.20200853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodero, M.P., Decalf J., Bondet V., Hunt D., Rice G.I., Werneke S., McGlasson S.L., Alyanakian M.A., Bader-Meunier B., Barnerias C., et al. 2017. Detection of interferon alpha protein reveals differential levels and cellular sources in disease. J. Exp. Med. 214:1547–1555. 10.1084/jem.20161451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruffin, N., Gea-Mallorquí E., Brouiller F., Jouve M., Silvin A., See P., Dutertre C.A., Ginhoux F., and Benaroch P.. 2019. Constitutive Siglec-1 expression confers susceptibility to HIV-1 infection of human dendritic cell precursors. Proc. Natl. Acad. Sci. USA. 116:21685–21693. 10.1073/pnas.1911007116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salvi, V., Bosisio D., Mitola S., Andreoli L., Tincani A., and Sozzani S.. 2010. Trichostatin A blocks type I interferon production by activated plasmacytoid dendritic cells. Immunobiology. 215:756–761. 10.1016/j.imbio.2010.05.023 [DOI] [PubMed] [Google Scholar]
- Schindelin, J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., Preibisch S., Rueden C., Saalfeld S., Schmid B., et al. 2012. Fiji: An open-source platform for biological-image analysis. Nat. Methods. 9:676–682. 10.1038/nmeth.2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoggins, J.W., MacDuff D.A., Imanaka N., Gainey M.D., Shrestha B., Eitson J.L., Mar K.B., Richardson R.B., Ratushny A.V., Litvak V., et al. 2014. Pan-viral specificity of IFN-induced genes reveals new roles for cGAS in innate immunity. Nature:505:691–695. 10.1038/nature12862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silvin, A., Yu C.I., Lahaye X., Imperatore F., Brault J.B., Cardinaud S., Becker C., Kwan W.H., Conrad C., Maurin M., et al. 2017. Constitutive resistance to viral infection in human CD141+ dendritic cells. Sci. Immunol. 2:eaai8071. 10.1126/sciimmunol.aai8071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson, S.R., Rego S.L., Harvey S.E., Liu M., Hemphill W.O., Venkatadri R., Sharma R., Grayson J.M., and Perrino F.W.. 2020. T cells produce IFN-α in the TREX1 D18N model of lupus-like autoimmunity. J. Immunol. 204:348–359. 10.4049/jimmunol.1900220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, L., Wang S., and Li W.. 2012. RSeQC: Quality control of RNA-seq experiments. Bioinformatics. 28:2184–2185. 10.1093/bioinformatics/bts356 [DOI] [PubMed] [Google Scholar]
- Willemsen, J., Neuhoff M.T., Hoyler T., Noir E., Tessier C., Sarret S., Thorsen T.N., Littlewood-Evans A., Zhang J., Hasan M., et al. 2021. TNF leads to mtDNA release and cGAS/STING-dependent interferon responses that support inflammatory arthritis. Cell Rep. 37:109977. 10.1016/j.celrep.2021.109977 [DOI] [PubMed] [Google Scholar]
- Woodward, J.J., Iavarone A.T., and Portnoy D.A.. 2010. c-di-AMP secreted by intracellular Listeria monocytogenes activates a host type I interferon response. Science. 328:1703–1705. 10.1126/science.1189801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, J., Chen Y.J., Dobbs N., Sakai T., Liou J., Miner J.J., and Yan N.. 2019. STING-mediated disruption of calcium homeostasis chronically activates ER stress and primes T cell death. J. Exp. Med. 216:867–883. 10.1084/jem.20182192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, X.-D., Huang B., Li M., Lamb A., Kelleher N.L., and Chen L.F.. 2009. Negative regulation of NF-κB action by Set9-mediated lysine methylation of the RelA subunit. EMBO J. 28:1055–1066. 10.1038/emboj.2009.55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, X.-D., Tajkhorshid E., and Chen L.-F.. 2010. Functional interplay between acetylation and methylation of the RelA subunit of NF-κB. Mol. Cell. Biol. 30:2170–2180. 10.1128/MCB.01343-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeung, F., Hoberg J.E., Ramsey C.S., Keller M.D., Jones D.R., Frye R.A., and Mayo M.W.. 2004. Modulation of NF-κB-dependent transcription and cell survival by the SIRT1 deacetylase. EMBO J. 23:2369–2380. 10.1038/sj.emboj.7600244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, G., Wang L.G., Han Y., and He Q.Y.. 2012. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS. 16:284–287. 10.1089/omi.2011.0118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, Z., Condomines M., van der Stegen S.J.C., Perna F., Kloss C.C., Gunset G., Plotkin J., and Sadelain M.. 2015. Structural design of engineered costimulation determines tumor rejection kinetics and persistence of CAR T cells. Cancer Cell. 28:415–428. 10.1016/j.ccell.2015.09.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziesché, E., Kettner-Buhrow D., Weber A., Wittwer T., Jurida L., Soelch J., Müller H., Newel D., Kronich P., Schneider H., et al. 2013. The coactivator role of histone deacetylase 3 in IL-1-signaling involves deacetylation of p65 NF-κB. Nucleic Acids Res. 41:90–109. 10.1093/nar/gks916 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
provides a complete list of differentially expressed genes.
is the source file for Fig. 1.
is the source file for Fig. 2.
is the source file for Fig. 4.
is the source file for Fig. 5.
is the source file for Fig. 6.
is the source file for Fig. 7.
is the source file for Fig. 8.
is the source file for Fig. S1.
is the source file for Fig. S2.
is the source file for Fig. S3.
is the source file for Fig. S4.