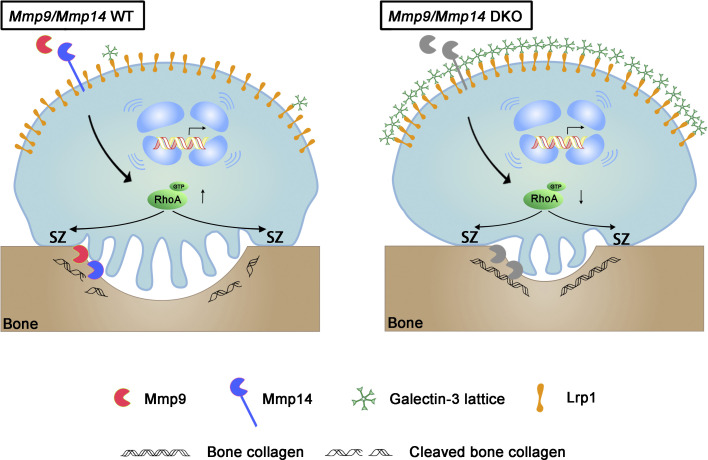
Zhu et al. identify a critical role for a heretofore undescribed interaction between a dual set of metalloproteins, Mmp9 and Mmp14, and the carbohydrate-binding protein galectin-3, whose binding interaction with the transmembrane receptor Lrp-1 shapes the transcriptional programs and signaling cascades directing osteoclast-mediated bone resorption.
Abstract
Bone-resorbing osteoclasts mobilize proteolytic enzymes belonging to the matrix metalloproteinase (MMP) family to directly degrade type I collagen, the dominant extracellular matrix component of skeletal tissues. While searching for additional MMP substrates critical to bone resorption, Mmp9/Mmp14 double-knockout (DKO) osteoclasts—as well as MMP-inhibited human osteoclasts—unexpectedly display major changes in transcriptional programs in tandem with compromised RhoA activation, sealing zone formation and bone resorption. Further study revealed that osteoclast function is dependent on the ability of Mmp9 and Mmp14 to cooperatively proteolyze the β-galactoside–binding lectin, galectin-3, on the cell surface. Mass spectrometry identified the galectin-3 receptor as low-density lipoprotein-related protein-1 (Lrp1), whose targeting in DKO osteoclasts fully rescues RhoA activation, sealing zone formation and bone resorption. Together, these findings identify a previously unrecognized galectin-3/Lrp1 axis whose proteolytic regulation controls both the transcriptional programs and the intracellular signaling cascades critical to mouse as well as human osteoclast function.
Graphical Abstract
Introduction
As a metabolically dynamic tissue, bone is continuously remodeled through repetitive phases of bone resorption and formation (Teitelbaum and Ross, 2003; Zaidi, 2007). Osteoclasts are giant multinucleated cells (MNCs) that are specialized in their ability to resorb bone (Teitelbaum and Ross, 2003; Zaidi, 2007). Although their activity is normally integrated with requirements for skeletal morphogenesis and remodeling, excessive bone resorption gives rise to bone-wasting diseases ranging from osteoporosis and rheumatoid arthritis to metastasis-associated osteolysis (Compston et al., 2019; Teitelbaum and Ross, 2003; Zaidi, 2007). Consequently, deciphering the molecular mechanisms governing osteoclast activity is critical to our understanding of skeletal remodeling under both physiologic and pathologic conditions.
Osteoclasts are derived from the monocyte/macrophage lineage cells of both embryonic erythro-myeloid lineage progenitors and hematopoietic stem cell precursors (Jacome-Galarza et al., 2019; Teitelbaum and Ross, 2003; Yahara et al., 2020). In response to the cytokines, receptor activator of NF-κB ligand (RANKL) and macrophage colony-stimulating factor (M-CSF), monocyte precursors differentiate into pre-osteoclasts that ultimately fuse to form polykaryons (Compston et al., 2019; Jacome-Galarza et al., 2019; Teitelbaum and Ross, 2003; Yahara et al., 2020; Zaidi, 2007). Upon attachment to bone, osteoclasts then polarize and undergo extensive morphologic changes to generate a prominent actin ring at their basal surface, known as the sealing zone, which in turn, surrounds the ruffled border, a differentiated region of the plasma membrane where protons, chloride ions, and proteases are secreted into the underlying resorption lacunae (Blangy et al., 2020; Teitelbaum and Ross, 2003). To degrade type I collagen, the dominant extracellular matrix component found in bone, osteoclasts mobilize triple-helical collagenases belonging to the cysteine proteinase family, i.e., cathepsin K (CTSK) as well as a functionally redundant network of the secreted matrix metalloproteinase (MMP), MMP9, and the membrane-anchored MMP, MMP14 (Brömme et al., 2016; Zhu et al., 2020). Whereas MMP14 functions as a collagenase by hydrolyzing acid-soluble type I collagen within its triple-helical domain (Ohuchi et al., 1997; Tang et al., 2013), MMP9 was revealed to depolymerize and cleave the non-helical telopeptide domains that contain the covalent cross-links that maintain the type I collagen fibrillar network in decalcified bone (Okada et al., 1995; Parikka et al., 2001; Zhu et al., 2020). As the bone-resorptive defects observed in Mmp9/Mmp14 double-knockout (DKO) osteoclasts could be rescued by transducing cells with either proteinase alone, these data were most consistent with a model wherein the metalloproteinases controlled bone resorption by “simply” acting as bone type I collagenases (Zhu et al., 2020).
Nevertheless, an increasing body of evidence indicates that MMP activity is not necessarily confined to matrix-degradative activity (Alabi et al., 2021; Alonso-Herranz et al., 2020; Hui et al., 2020; Jin et al., 2011; Kelly et al., 2018; Nishida et al., 2012; Orgaz et al., 2014; Sakamoto and Seiki, 2010; Shimizu-Hirota et al., 2012; Wong et al., 2016). Hence, we designed a series of experiments to test the possibility that the MMP9/MMP14 axis exerts additional, but collagenase-independent, control over osteoclast activity. Indeed, transcriptional profiling of Mmp9/Mmp14 DKO osteoclasts uncovered remarkable changes in gene expression relative to wild-type cells in the absence of an extracellular matrix substratum. In searching for potential MMP targets responsible for these effects, our attention turned to galectin-3, a chimeric member of the β-galactoside–binding galectin family, whose association with cell surface carbohydrate-containing molecules and glycoproteins can modulate intracellular signaling pathways as well as cell function (Nabi et al., 2015; Thiemann and Baum, 2016). Herein, we report that MMP9 and MMP14 directly cleave endogenously derived galectin-3 on the osteoclast surface, thereby controlling osteoclast gene expression, RhoA activation, sealing zone formation and bone resorption in vitro as well as in vivo. Further, we demonstrated that galectin-3 mediates its intracellular effects on osteoclast activity through its binding to the low-density lipoprotein receptor-related protein-1 (Lrp1), a multi-functional member of the low-density lipoprotein receptor family of lipoprotein receptors (Yang and Williams, 2017). Taken together, our results identify a novel MMP9/MMP14–galectin-3/Lrp1 axis that functions as an auto-regulating proteolytic rheostat of osteoclast activity and function.
Results
Mmp9/Mmp14 DKO osteoclasts display major changes in gene expression programs
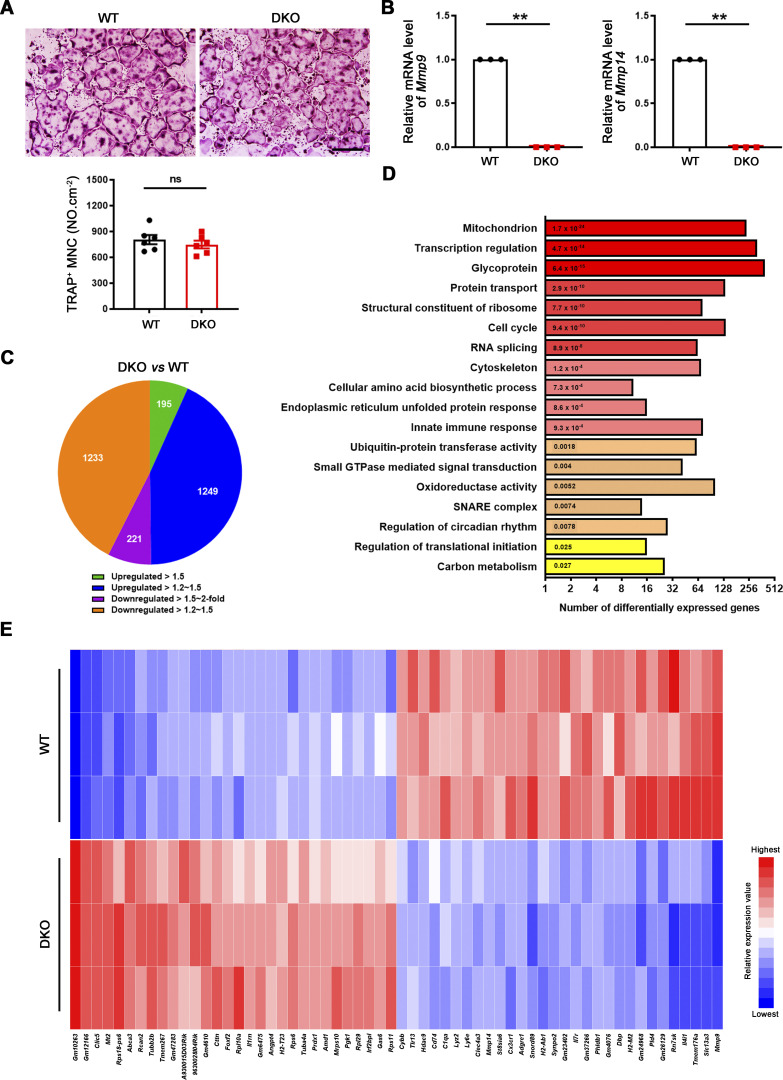
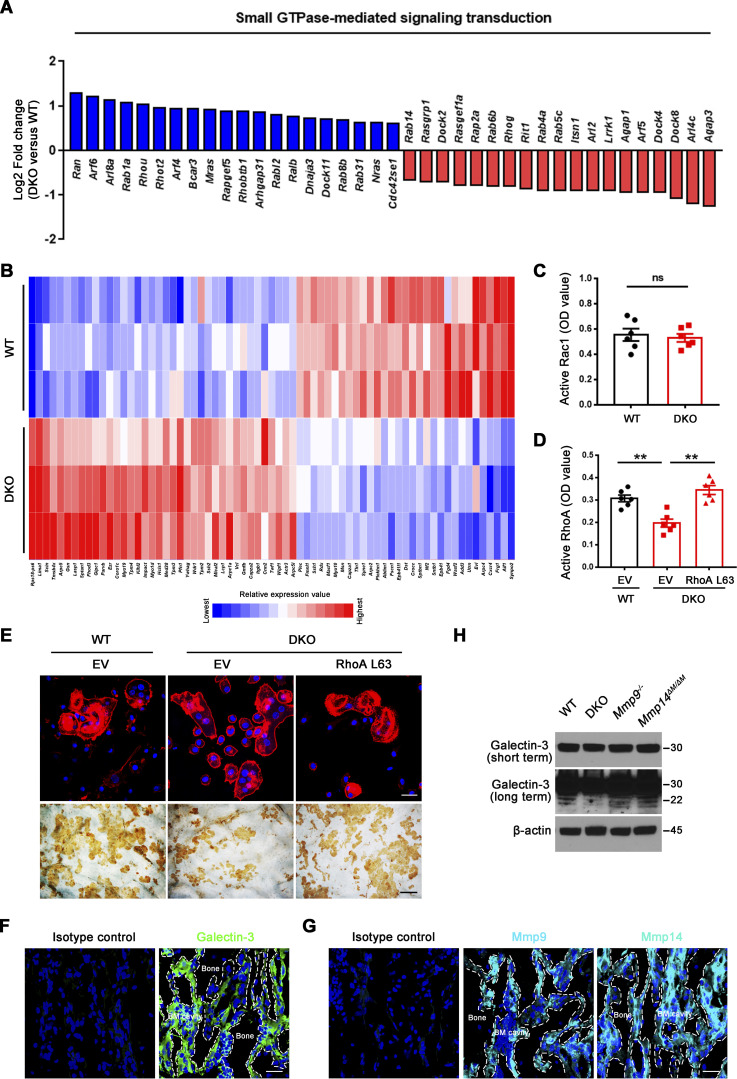
As Mmp9 and Mmp14 potentially exert pleiotropic effects on cell function independent of their matrix-degradative activities (Alabi et al., 2021; Alonso-Herranz et al., 2020; Hui et al., 2020; Jin et al., 2011; Kelly et al., 2018; Nishida et al., 2012; Orgaz et al., 2014; Sakamoto and Seiki, 2010; Shimizu-Hirota et al., 2012; Wong et al., 2016), we sought to first screen for global effects on osteoclast activity in an unbiased fashion by interrogating gene expression programs. To circumvent the potential confounding effects of differences in collagenolytic activity and the generation of bioactive matrikines during bone resorption, we performed unbiased transcription profiling of osteoclasts fully differentiated atop plastic substrata using bone marrow–derived macrophages (BMDMs) generated from Csf1r-Cre/Mmp14f/f/Mmp9−/− conditional knockout mice and littermate controls. As described previously (Zhu et al., 2020), Mmp9/Mmp14 DKO BMDMs stimulated with RANKL undergo fusion into osteoclasts comparably to controls (Fig. 1, A and B). Three biological replicates were performed for each group to assess the reproducibility of RNA sequencing (RNA-seq) data. Remarkably, however, using a minimum of 1.2-fold change as a cutoff and an adjusted P value of <0.05, dual Mmp9/Mmp14 targeting alters the expression of 2,898 unique transcripts (Fig. 1, C–E). On the basis of DAVID Gene Ontology (GO) analysis, we found that several important biological processes, such as mitochondrial, cytoskeletal, small GTPase-mediated signaling transduction, carbon metabolism, and glycoprotein pathways are enriched among the pattern of differentially expressed genes (Fig. 1 D).
Figure 1.
Genome wide analysis of gene expression programs in Mmp9/Mmp14 DKO osteoclasts. (A) BMDMs were isolated from wild-type or DKO mice and cultured on plastic substrata with 20 ng/ml M-CSF and 30 ng/ml RANKL for 5 d, stained with TRAP, and the number of TRAP+ MNCs was determined. Scale bar, 500 μm. Data are presented as mean ± SEM (n = 6 biological replicates). (B) The relative mRNA levels of Mmp9 and Mmp14 in DKO osteoclasts were assessed by qPCR compared to wild-type osteoclasts (normalized to 1). Data are presented as mean ± SEM (n = 3 biological replicates). (C) Pie chart depicts the distribution of total transcripts changed in DKO osteoclasts relative to wild-type osteoclasts (n = 3 biological replicates). (D) DAVID GO analysis of differentially expressed genes from Mmp9/Mmp14-deleted versus control osteoclasts (n = 3 biological replicates). (E) The 30 most highly upregulated and downregulated transcripts in DKO osteoclasts as compared with wild-type osteoclasts are presented (n = 3 biological replicates; color bar, relative expression value). **P < 0.01. Statistical significance was assessed using unpaired two-sided Student’s t test.
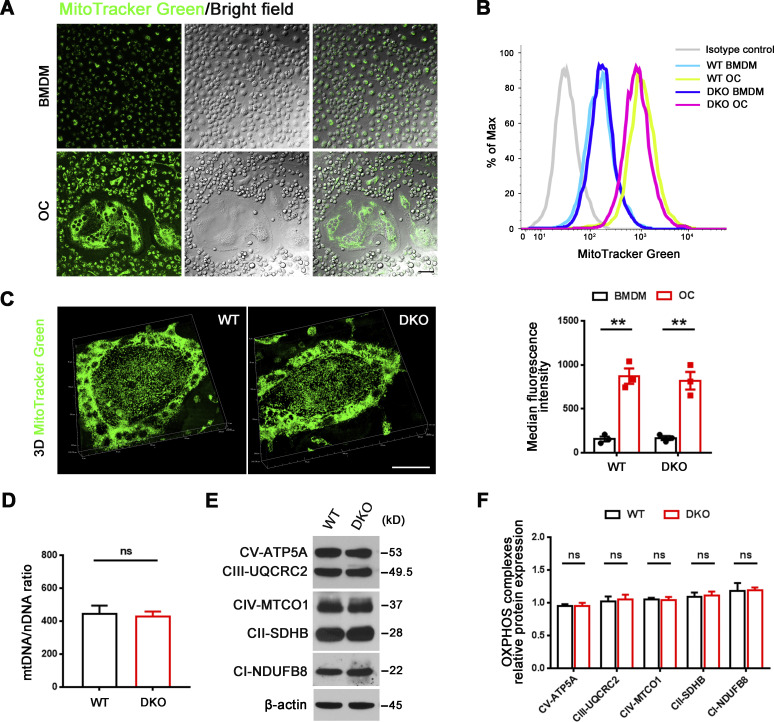
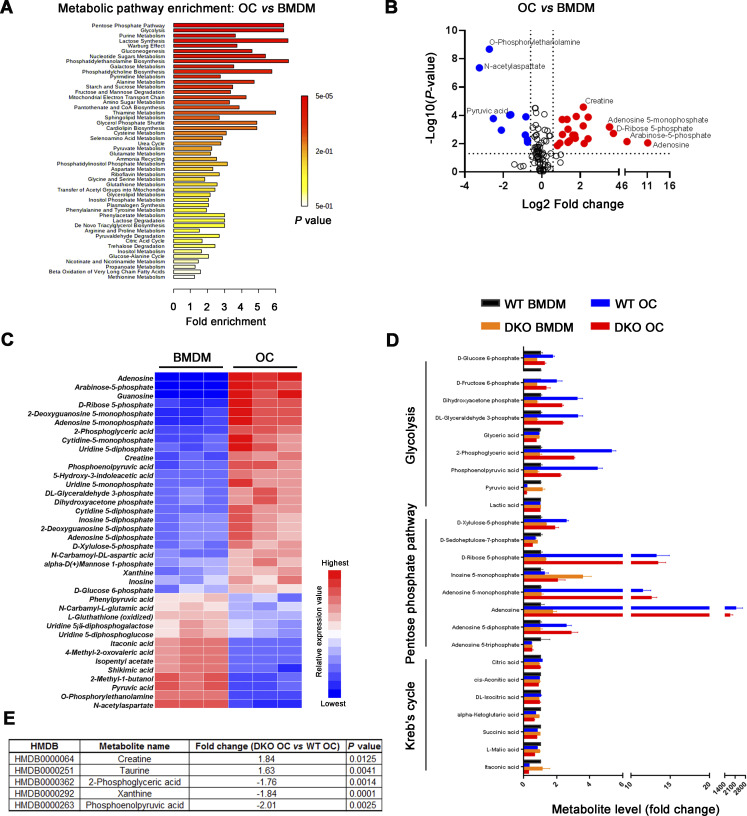
As differentially expressed genes included almost 250 mitochondrial-associated transcripts, our attention first focused on recent findings describing functional connections between mitochondrial metabolism and dynamic gene expression during osteoclast differentiation (Arnett and Orriss, 2018; Bae et al., 2017; Nishikawa et al., 2015) along with complementary reports linking Mmp9 and Mmp14 to mitochondrial activity and energy metabolism (Kowluru et al., 2011; Mori et al., 2016; Sakamoto and Seiki, 2010). As expected, mitochondrial mass, as assessed by microscopy and flow cytometry after MitoTracker Green staining, increases significantly as BMDMs are induced to differentiate into mature osteoclasts (Fig. S1, A and B). However, despite the changes in gene expression, the RANKL-induced shift in mitochondrial mass and morphology is indistinguishable between control and Mmp9/Mmp14-deficient osteoclasts (Fig. S1, B and C). Further, mitochondrial DNA (mtDNA) copy number is also unaltered between wild-type and DKO osteoclasts (Fig. S1 D), as are protein levels of mitochondrial complexes I, II, III, IV, and V relative to control cells (Fig. S1, E and F). To probe for possible effects of dual Mmp9/Mmp14 targeting at the metabolite level, we performed quantitative metabolomic profiling of BMDM and osteoclasts derived from both wild-type and DKO mice. Using a minimum of 1.5-fold change as a cutoff and an adjusted P value of <0.05, 37 metabolites are changed during the BMDM–osteoclast transition in wild-type cells with 25 metabolites consistently upregulated and 13 metabolites downregulated (Fig. S2, A–C). A search of the differentially expressed metabolites by MetaboAnalyst (Xia and Wishart, 2016) indicates that multiple metabolic pathways, including pentose phosphate pathway, glycolysis, purine metabolism, gluconeogenesis, phosphatidylethanolamine biosynthesis, galactose metabolism, and alanine metabolism, are all enriched, underlining the complex metabolic reprogramming that occurs during the BMDM–osteoclast transition (Fig. S2 A). However, no major changes in these pathways are detected between wild-type and DKO osteoclasts, with only small changes detected in five metabolites, including creatine, taurine, 2-phosphoglyceric acid, xanthine, and phosphoenolpyruvic acid (Fig. S2, D and E). Of note, the remarkable drop in itaconic acid levels, a negative regulator of osteoclast differentiation and function (Sun et al., 2019), that occurs during the wild-type BMDM–osteoclast transition proceeds normally in DKO cells (Fig. S2 D). Taken together, the detected changes in mitochondrial-associated gene expression appear unlikely to cause major changes in DKO osteoclast metabolism.
Figure S1.
Mitochondrial abundance and OXPHOS expression in Mmp9/Mmp14 DKO osteoclasts. (A) MitoTracker Green (green) immunofluorescence and phase contrast morphology of BMDMs and osteoclasts generated from wild-type mice. Scale bar, 20 μm. Results are representative of three independent experiments. (B) Measurements of mitochondrial mass of BMDMs and osteoclasts (OC) generated from wild-type or DKO mice using MitoTracker Green with flow cytometry. Data are presented as mean ± SEM (n = 3 biological replicates). (C) MitoTracker Green (green) immunofluorescence of wild-type or DKO osteoclasts. Scale bar, 50 μm. Results are representative of three independent experiments. (D) Relative mtDNA copy number per nuclear genome in osteoclasts generated from wild-type or DKO mice. Data are presented as mean ± SEM (n = 3 biological replicates). (E and F) OXPHOS protein expression as assessed by Western blot (E) in osteoclasts generated from wild-type or DKO mice and quantified (F). Data are presented as mean ± SEM (n = 3 biological replicates). **P < 0.01. Statistical significance was assessed using two-way ANOVA with Bonferroni correction (B) and unpaired two-sided Student’s t test (D and F). Source data are available for this figure: SourceData FS1.
Figure S2.
Identification and quantitative analysis of the metabolome in Mmp9/Mmp14 DKO osteoclasts. (A and B) MetaboAnalyst analysis (A) and volcano plots (B) of differentially expressed metabolites from wild-type osteoclasts versus BMDMs (n = 3 biological replicates). (C) Heat map representation of metabolomics profile data highlights differentially expressed metabolites from wild-type osteoclasts (OC) versus BMDMs (n = 3 biological replicates; color bar, relative expression value). (D) Relative abundance of key metabolites from pentose phosphate pathway, glycolysis, and Kreb’s cycle in BMDMs and osteoclasts generated from wild-type or DKO mice. Data are presented as mean ± SEM (n = 3 biological replicates). (E) Lists of five differentially expressed metabolites from DKO osteoclasts versus wild-type osteoclasts (n = 3 biological replicates). Statistical significance was assessed using unpaired two-sided Student’s t test.
Mmp9/Mmp14 co-dependent control of osteoclast RhoA activation and cytoskeletal organization
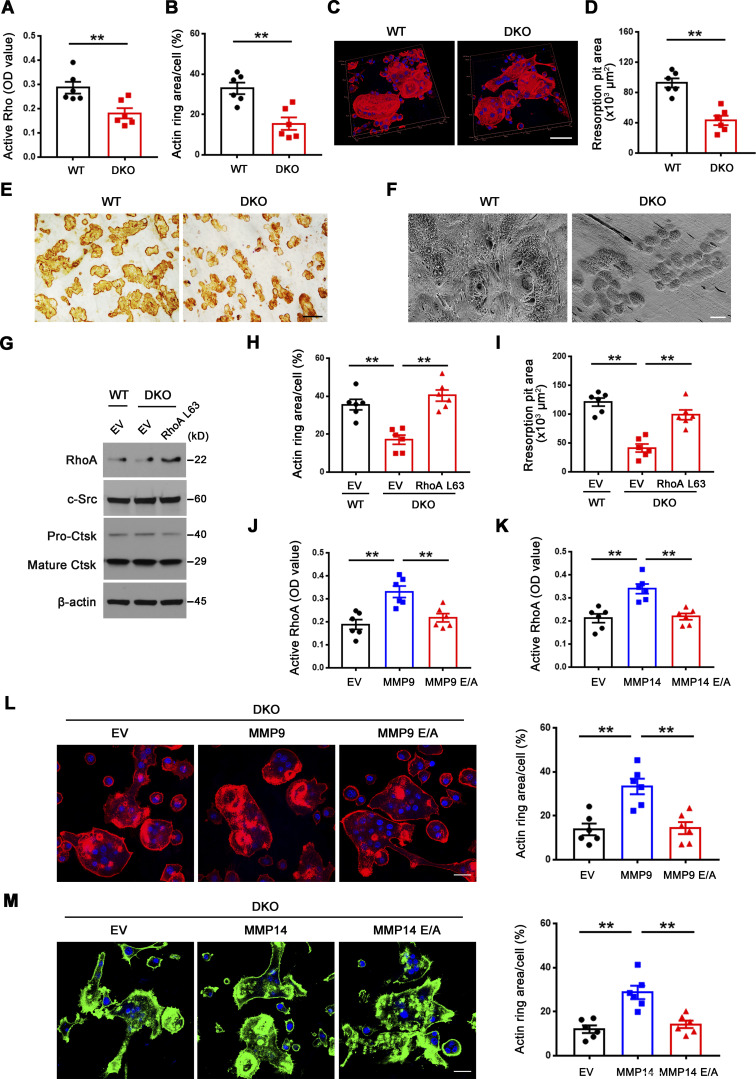
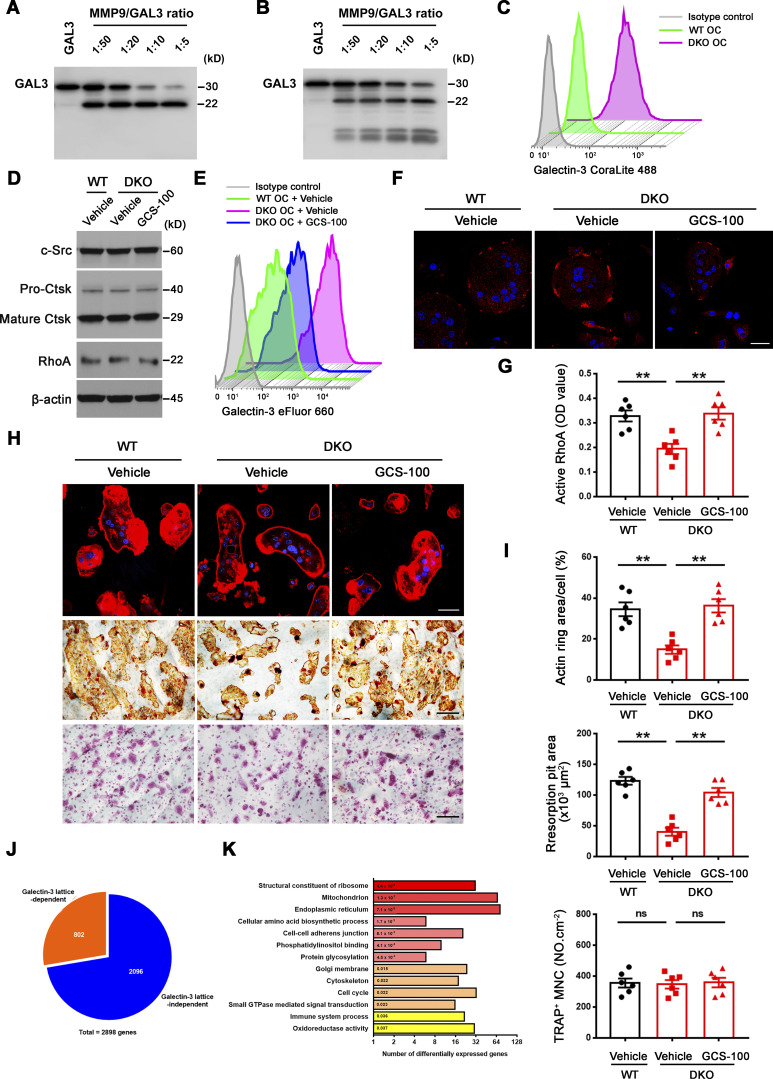
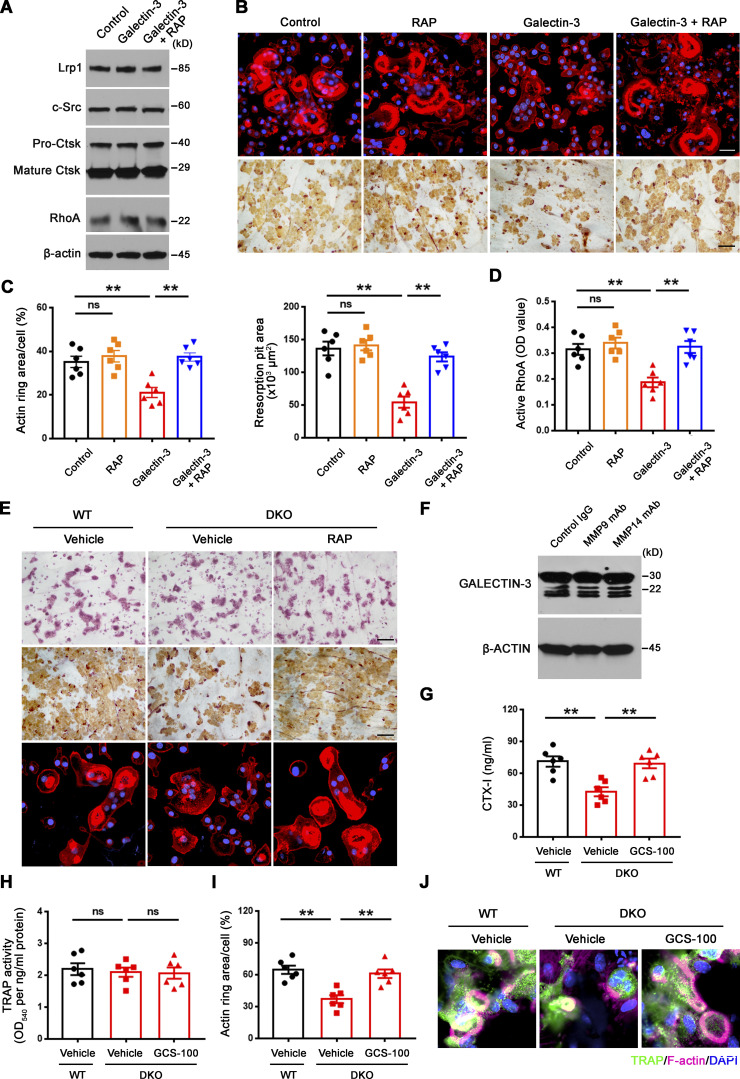
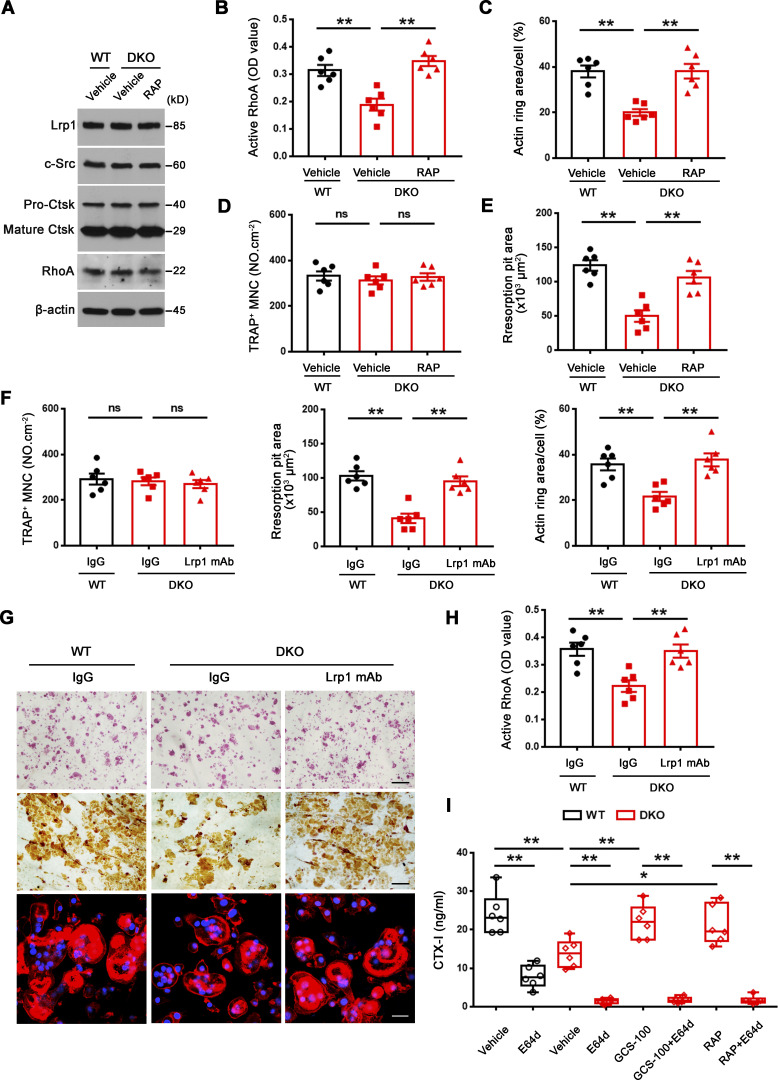
Small GTPase-mediated signaling transduction and cytoskeleton-associated pathways arise among the most altered GO categories in DKO osteoclasts (Fig. 1 D; and Fig. S3, A and B) and Rho GTPase–dependent activation of cytoskeletal reorganization and sealing zone formation play critical roles in supporting osteoclast-mediated bone resorption (Blangy et al., 2020; Teitelbaum, 2011; Touaitahuata et al., 2014). While RhoA and Rac1 remain unaltered, we noted changes in the expression of a series of small GTPase-mediated signaling transduction-related transcripts, including Arf6, Rhou, Bcar3, Rasgrp1, Arl4c, Dock4, Arl2, and Rap2a (Fig. S3 A), that have been previously linked to the differential regulation of RhoA or Rac1 activation (Boshans et al., 2000; Matsumoto et al., 2014; Meng et al., 2018; Salzer et al., 2016; Slaymi et al., 2019; Taniuchi et al., 2011; Wilson et al., 2013; Yazbeck et al., 2022). In this regard, relative to wild-type osteoclasts, GTP-loaded active RhoA, but not Rac1, levels are suppressed significantly in Mmp9/Mmp14-targeted osteoclasts cultures (Fig. 2 A and Fig. S3 C). Consistent with this finding, sealing zone-associated actin ring formation is likewise repressed when DKO cells are cultured atop bone in tandem with striking decreases in bone resorption as assessed by wheat germ agglutinin-diaminobenzidine acid (WGA-DAB) staining or scanning electron microscopy (Fig. 2, B–F), though DKO osteoclasts formed a morphologically normal podosome belt when cultured on glass surfaces (Zhu et al., 2020). To assess the relative importance of RhoA activation in controlling osteoclast function in DKO osteoclasts, cells were next transduced with a constitutively active form of RhoA (ca-RhoA; Fig. 2 G and Fig. S3 D). While protein expression of osteoclast maturation markers such as c-Src and pro/mature-Ctsk are unaltered, RhoA activity, sealing zone formation and bone-resorptive activity are each upregulated to wild-type levels in DKO osteoclasts (Fig. 2, G–I; and Fig. S3, D and E).
Figure S3.
Active RhoA rescues defects in sealing zone formation and bone resorption in DKO osteoclasts. (A) Relative expression levels of small GTPase-mediated signal transduction-related transcripts retrieved from DAVID GO categories were assessed in DKO osteoclasts as compared with wild-type osteoclasts (normalized to 1; n = 3). (B) The differentially expressed transcripts of cytoskeleton-associated pathways in DKO osteoclasts as compared with wild-type osteoclasts are presented (n = 3; color bar, relative expression value). (C) Rac1 activity of wild-type and DKO osteoclasts cultured on plastic upon activation with 20 ng/ml M-CSF and 30 ng/ml RANKL for 15 min. Data are presented as mean ± SEM (n = 6 biological replicates). (D) EV-transduced wild-type BMDMS, and EV- or ca-RhoA–transduced DKO BMDMs were differentiated into osteoclasts on plastic, and RhoA activity determined upon activation 20 ng/ml M-CSF and 30 ng/ml RANKL for 15 min. Data are presented as mean ± SEM (n = 6 biological replicates). (E) EV-transduced wild-type pre-osteoclasts, and EV- or ca-RhoA–transduced DKO pre-osteoclasts were cultured atop bone slices for 3 d and stained with phalloidin (red). Osteoclasts were removed and resorption pits visualized by WGA-DAB staining (n = 6 biological replicates). Scale bar, upper 20 μm, lower 100 μm. (F and G) Immunofluorescence staining of a femur section from a wild-type mouse with anti-galectin-3 (#125401; Biolegend; clone M3/38; F; green), anti-Mmp9, anti-Mmp14 antibodies (G; cyan), or corresponding isotype controls. BM cavity, bone marrow cavity. Scale bar, 30 μm. Results are representative of three independent experiments. (H) Galectin-3 expression and cleavage as assessed by Western blot in osteoclasts generated from wild-type, Mmp9−/−, Mmp14ΔM/ΔM, or DKO mice. Results are representative of three independent experiments. **P < 0.01. Statistical significance was assessed using unpaired two-sided Student’s t test (C) and one-way ANOVA with Bonferroni correction (D). Source data are available for this figure: SourceData FS3.
Figure 2.
Mmp9/Mmp14-dependent RhoA activation and cytoskeletal organization. (A) RhoA activity of wild-type and DKO osteoclasts cultured on plastic upon activation with 20 ng/ml M-CSF and 30 ng/ml RANKL for 15 min (n = 6 biological replicates). (B and C) After a 6-d culture atop bone slice, phalloidin staining (red) was performed in wild-type versus DKO osteoclasts (C), and actin ring area per cell quantified (B). Scale bar, 50 μm. Data are presented as mean ± SEM (n = 6 biological replicates). (D and E) Wild-type or DKO osteoclasts were removed from bone slices, resorption pits visualized by WGA-DAB staining (E), and resorption pit area quantified (D). Scale bar, 100 μm. Data are presented as mean ± SEM (n = 6 biological replicates). (F) Resorption pits generated on bone slice by wild-type and DKO osteoclasts were imaged by scanning electron microscopy. Scale bar, 10 μm. Results are representative of three independent experiments. (G) RhoA, c-Src, and Ctsk expression in empty vector (EV)–transduced wild-type osteoclasts, and EV- or ca-RhoA–transduced DKO osteoclasts, as assessed by Western blot. Results are representative of three independent experiments. (H and I) EV-transduced wild-type pre-osteoclasts, and EV- or ca-RhoA–transduced DKO pre-osteoclasts were cultured atop bone slices for 3 d and stained by phalloidin along with actin ring area per cell quantified (H). Data are presented as mean ± SEM (n = 6 biological replicates). Osteoclasts were removed and resorption pits visualized by WGA-DAB staining and resorption pit area quantified (I). Data are presented as mean ± SEM (n = 6 biological replicates). (J) DKO BMDMs were transduced with lentiviral vectors expressing full-length MMP9, an MMP9E/A mutant, or an empty control and differentiated into osteoclasts, and RhoA activity assessed upon activation with 20 ng/ml M-CSF and 30 ng/ml RANKL for 15 min. Data are presented as mean ± SEM (n = 6 biological replicates). (K) DKO BMDMs were transduced with lentiviral vectors expressing full-length MMP14, MMP14E/A, or an empty control and differentiated into osteoclasts, and RhoA activity assessed upon activation with 20 ng/ml M-CSF and 30 ng/ml RANKL for 15 min. Data are presented as mean ± SEM (n = 6 biological replicates). (L) MMP9- or MMP9E/A-transduced pre-osteoclasts were cultured atop bone slices for 3 d, and F-actin stained with phalloidin (red), and actin ring area per cell quantified. Data are presented as mean ± SEM (n = 6 biological replicates). Scale bar, 20 μm. (M) MMP14- or MMP14E/A-transduced pre-osteoclasts were cultured atop bone slices for 3 d, and F-actin staining (green) and quantification assessed as described in L. Scale bar, 20 μm. Data are presented as mean ± SEM (n = 6 biological replicates). **P < 0.01. Statistical significance was assessed using unpaired two-sided Student’s t test (A, B, and D) and one-way ANOVA (H–M) with Bonferroni correction (H–M). Source data are available for this figure: SourceData F2.
As MMPs are able to regulate cell function by both proteinase-dependent and -independent mechanisms (Orgaz et al., 2014; Sakamoto and Seiki, 2010; Shimizu-Hirota et al., 2012; Zhu et al., 2020), we sought to determine whether MMP9/MMP14-dependent regulation of RhoA activity does—or does not—require proteolytic activity. As such, DKO osteoclasts were transduce with lentiviral expression vectors encoding wild-type MMP9 or MMP14 versus catalytically inactive forms of MMP9 (MMP9E/A) or MMP14 (MMP14E/A; Zhu et al., 2020). Under these conditions, only wild-type MMP9 or MMP14, but not the MMPE/A mutants, rescue RhoA activation and sealing zone formation in DKO osteoclasts (Fig. 2, J–M). Together, these studies indicate that a proteolytically active MMP9/MMP14 axis controls RhoA activation along with downstream control of osteoclast cytoskeletal organization and bone resorption.
An unexpected link between MMP9/MMP14-mediated osteoclast activation and galectin-3 lattice remodeling
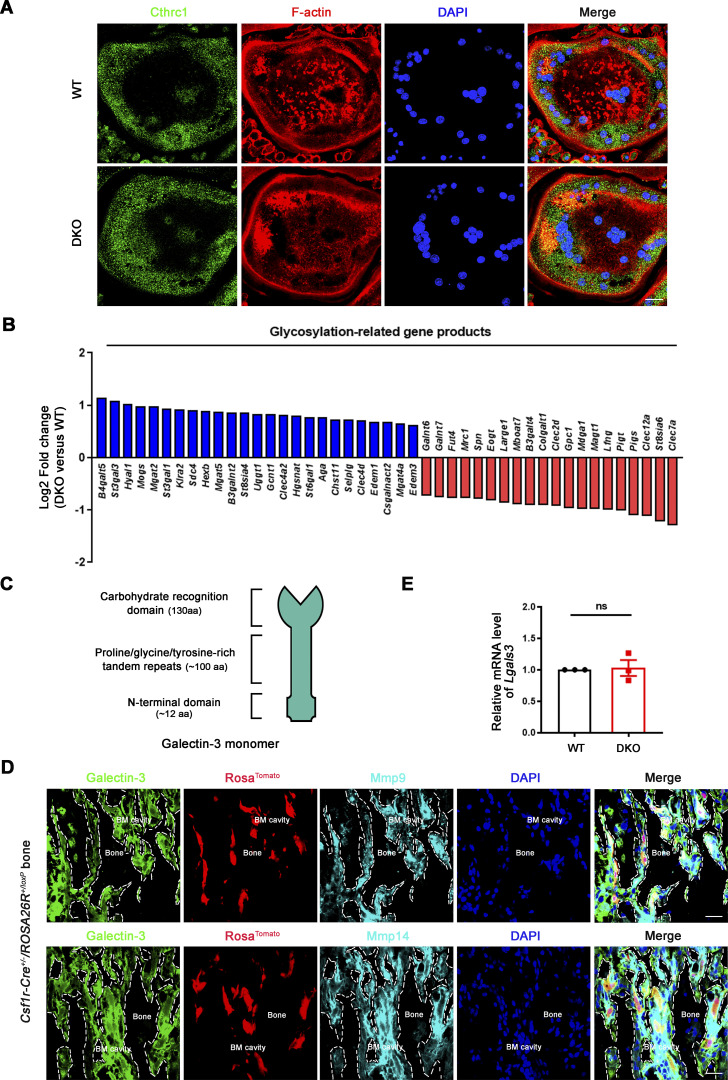
Given that Mmp9/Mmp14 DKO osteoclasts exhibit defects in RhoA activation and cytoskeletal organization independent of direct bone collagenolysis (i.e., defects are observed when cells are cultured atop a plastic substratum), we considered the existence of additional, but cell-intrinsic target substrates, susceptible to cleavage by both MMP9 and MMP14 that serve to regulate RhoA activation. Osteoclasts are known to secrete collagen triple helix repeat containing 1, Cthrc1, a molecule that not only regulates collagen expression and bone formation, but also is sensitive to collagenase-mediated hydrolysis (Takeshita et al., 2013). Nevertheless, no differences are observed in Cthrc1 levels between wild-type and DKO osteoclasts (Fig. 3 A). Alternatively, our transcriptome analysis of DKO osteoclasts highlighted major changes in glycosylation-related gene products, including Mgat1, Mgat4, and Mgat5 (Fig. 3 B). N-acetylglucosaminyltransferases encoded by Mgat1, Mgat2, Mgat4, and Mgat5 act sequentially to generate the N-acetylglucosamine branched complex type N-glycans on glycoprotein receptors, thereby increasing their affinities for galectin binding (Lau et al., 2007; Nabi et al., 2015; Nielsen et al., 2018; Smith et al., 2018). As such, our attention shifted to the carbohydrate-binding protein, galectin-3, as it (i) is highly expressed in osteoclasts (Colnot et al., 1999), (ii) under cell-free conditions, is sensitive to both MMP14- as well as MMP9-dependent hydrolysis (Ochieng et al., 1998; Toth et al., 2005), and (iii) interacts with Magt1/Mgat4/Mgat5-modified N-glycans on various cell surface receptors and proteins to form a multivalent, biologically active lattice (Lau et al., 2007; Nabi et al., 2015; Nielsen et al., 2018; Smith et al., 2018; Taniguchi et al., 2017). Further, cleavage of the N-terminal proline, glycine, and tyrosine-rich tandem repeats found within galectin-3 alters its affinity for glycoconjugates as well as inhibits its ability to self-associate into an interlocking, lattice-like network on cell surfaces (Nabi et al., 2015; Fig. 3 C). Immunofluorescence staining (along with isotype controls) was next conducted to interrogate galectin-3 and Mmp9/Mmp14 expression in vivo. In the bone marrow compartment, galectin-3 is ubiquitously distributed intracellularly, on the cell membrane, as well as in the extracellular milieu (Fig. S3 F; Elola et al., 2015; Nabi et al., 2015; Thiemann and Baum, 2016). In turn, Mmp9 and Mmp14 signals are found expressed in multiple cell types within the bone marrow (Fig. S3 G), potentially including osteoclasts, macrophages, endothelial cells, osteoblasts, mesenchymal stem cells, and hematopoietic stem cells (Bonnans et al., 2014; Rowe and Weiss, 2009). To further confirm that galectin-3 is expressed in Mmp9/Mmp14-positive osteoclasts in vivo, bone marrow samples were harvested from Csf1r-Cre/RosaTomato/+ transgenic mice wherein the Tomato red signal marks macrophage/osteoclast lineage cells (Dallas et al., 2018) and triple-stained for Mmp9 or Mmp14 with galectin-3. As shown in Fig. 3 D, bone-adherent osteoclasts score positive for all three markers.
Figure 3.
Dual Mmp9/Mmp14 deletion does not affect Cthrc1 levels or galectin-3 expression in osteoclasts. (A) Cthrc1 (green) immunofluorescence stained with an anti-Cthrc1 polyclonal antibody (ab85739; Abcam) and F-actin (red) imaged following phalloidin staining of wild-type and DKO osteoclasts. Results are representative of three independent experiments. Scale bar, 20 μm. (B) Relative expression levels of glycosylation-related transcripts retrieved from DAVID GO categories were assessed in DKO osteoclasts as compared with wild-type osteoclasts (normalized to 1; n = 3 biological replicates). (C) Schematics of galectin-3 monomer domains are presented. (D) Galectin-3 (green) stained with an anti–galectin-3 monoclonal antibody (#125401; Biolegend; clone M3/38) and Mmp9/Mmp14 (cyan) immunofluorescence (upper and lower panels, respectively) of a femur section from a Csf1r-Cre+/−/RosaTomato/+ mouse. BM cavity, bone marrow cavity. Scale bar, 30 μm. Results are representative of three independent experiments. (E) The relative mRNA levels of Lgals3 in DKO osteoclasts were assessed by qPCR compared to wild-type osteoclasts (normalized to 1). Data are presented as mean ± SEM (n = 3 biological replicates). Statistical significance was assessed using unpaired two-sided Student’s t test.
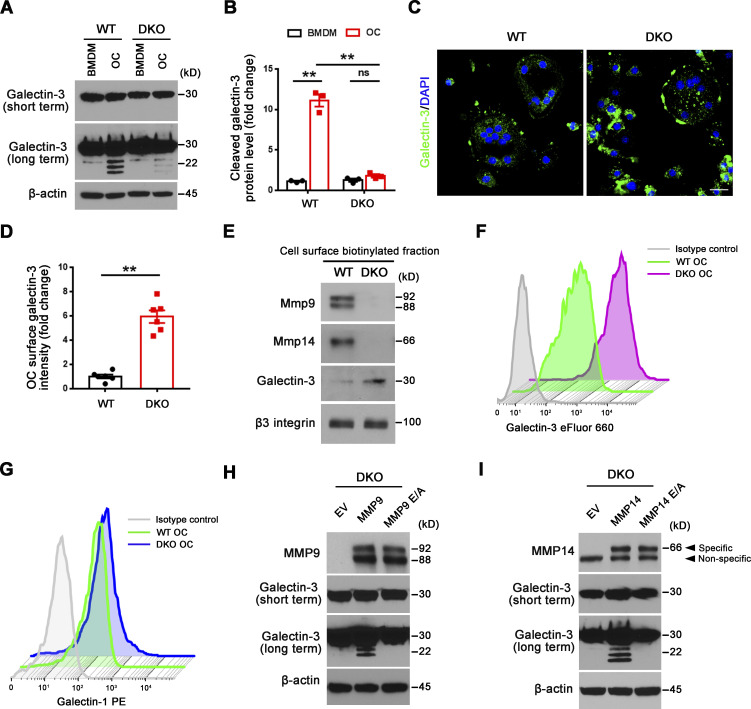
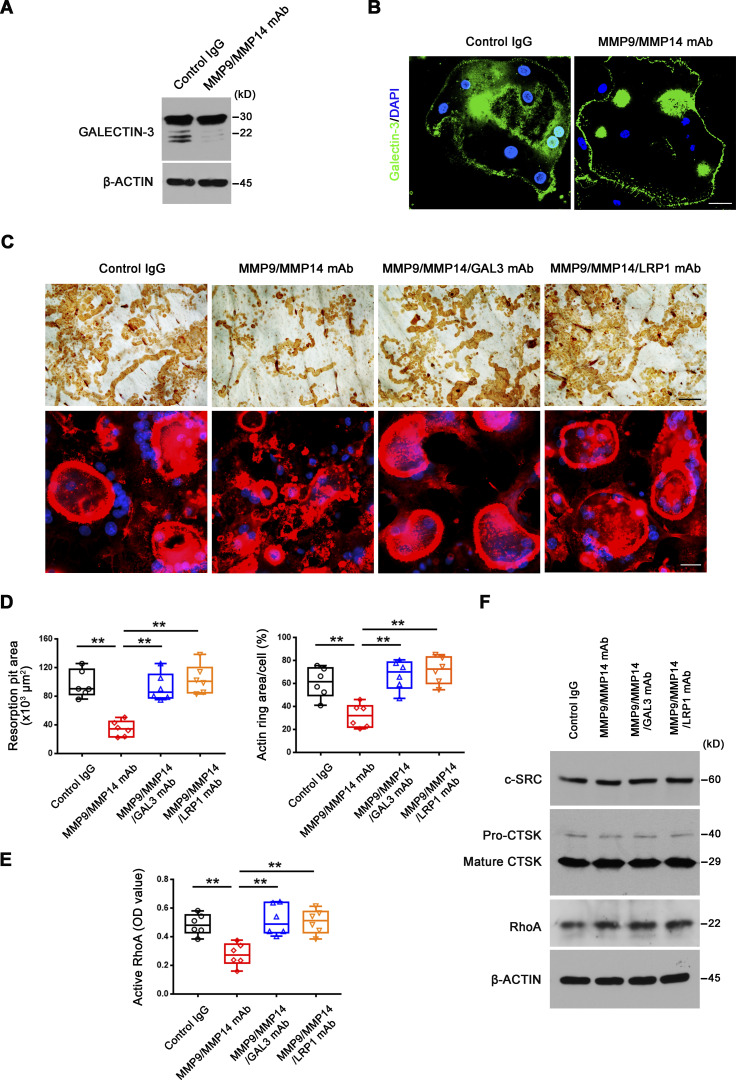
Dual deficiency of Mmp14 and Mmp9 had no effect on Lgals3 mRNA levels in isolated osteoclasts (Fig. 3 E). Further, Western blot analyses of galectin-3 levels using a monospecific monoclonal antibody in wild-type versus DKO osteoclasts did not, at first glance, suggest significant changes in expression (Fig. 4 A). However, intracellular levels of galectin-3 can overwhelm the surface pool, potentially obfuscating extracellular processing. Indeed, following longer exposure, a series of galectin-3 degradation products are detected in wild-type, Mmp9−/−/Mmp14f/f, and Mmp9+/+/Csf1r-Cre/Mmp14f/f osteoclasts, but not DKO cells (Fig. 4, A and B; and Fig. S3 H). Consistent with the proposition that the detected cleavage products accumulate in a pericellular pool, surface-associated galectin-3 levels were assessed in wild-type and DKO osteoclasts by immunofluorescence staining and flow cytometry. Indeed, immunostaining of cell surface galectin-3 confirms dramatically increased levels in DKO osteoclasts relative to wild-type cells (Fig. 4, C and D). Further, following cell surface biotinylation and capture with streptavidin magnetic beads (Maupin et al., 2022; Shiratori et al., 2018), only low levels of galectin-3 are detected in wild-type cells as opposed to DKO osteoclasts (Fig. 4 E), a finding corroborated by flow cytometry analysis (Fig. 4 F). MMP9 and MMP14 cleave galectin-3 at the Ala62-Tyr63 peptide bond generating a ∼22-kD fragment with an intact carbohydrate recognition domain (McClung et al., 2007; Ochieng et al., 1994; Ochieng et al., 1998; Toth et al., 2005). While the galectin-3 monoclonal antibody used here detects the ∼22 kD fragment (Fig. S4 A), the N-terminal cleavage product is not recognized, raising the possibility that wild-type osteoclast may yet be decorated with this smaller fragment. However, using a second galectin-3 polyclonal antibody that can detect both the ∼22 kD and smaller ∼9 kD cleavage products (Fig. S4 B), we again found that cell surface galectin-3 staining confirms dramatically increased levels in DKO osteoclasts relative to wild-type cells (Fig. S4 C). Interestingly, galectin-1-null osteoclasts have been reported to display increased resorbing activity (Muller et al., 2019) and likewise can be cleaved by MMPs (Butler et al., 2008; Prudova et al., 2010). However, Mmp9/Mmp14 DKO osteoclasts display unaltered surface galectin-1 level (Fig. 4 G), excluding galectin-1 as the potential downstream target of Mmp9/Mmp14 in regulating osteoclast activity. Finally, to assess the relative abilities of MMP9 and MMP14 to cleave endogenously expressed galectin-3 in osteoclasts, DKO BMDMs were transduced with lentiviral constructs expressing wild-type MMP9 versus MMP9E/A, or wild-type MMP14 versus MMP14E/A, and differentiated into osteoclasts (Zhu et al., 2020). Having confirmed equivalent expression of wild-type and mutant MMP9/MMP14 in the transduced cells, only wild-type MMP9- or MMP14-transduced osteoclasts regain galectin-3–degrading activity (Fig. 4, H and I). Together, these studies support a model wherein MMP9 and/or MMP14 serve as the principal proteinases responsible for cell surface galectin-3 lattice remodeling in osteoclasts.
Figure 4.
Mmp9/Mmp14 regulates galectin-3 lattice remodeling in osteoclasts. (A and B) Galectin-3 expression and cleavage as assessed with anti–galectin-3 monoclonal antibody (ab2785; Abcam; epitopes mapped against N-terminal region) by Western blot (A) and quantification (B) in BMDMs and osteoclasts (OC) generated from wild-type or DKO mice. Data are presented as mean ± SEM (n = 3 biological replicates). (C and D) Galectin-3 (green) immunofluorescence of non-permeabilized wild-type or DKO osteoclasts stained with an anti–galectin-3 monoclonal antibody (#125401; Biolegend; clone M3/38, epitopes mapped against N-terminal region; C), and surface galectin-3 level quantified (D). Scale bar, 20 μm. Data are presented as mean ± SEM (n = 3 biological replicates with two technical replicates each). (E) Following cell surface biotinylation and capture with streptavidin magnetic beads, Mmp9, Mmp14, galectin-3, and β3 integrin expression in the membrane fraction of wild-type and DKO osteoclasts as assessed by Western blot. Results are representative of three independent experiments. (F) Measurements of surface galectin-3 in wild-type and DKO osteoclasts with eFluor 660–conjugated anti–galectin-3 monoclonal antibody (#50-5301-82; Thermo Fisher Scientific; clone M3/38, epitopes mapped within the N-terminal domain) by flow cytometry. Results are representative of three independent experiments. (G) Measurements of surface galectin-1 in wild-type and DKO osteoclasts by flow cytometry. Results are representative of three independent experiments. (H) DKO BMDMs were transduced with lentiviral vectors expressing full-length MMP9, an MMP9E/A mutant, or an empty control, and differentiated into osteoclasts. Cell lysates were collected for MMP9 and galectin-3 expression and cleavage as assessed by Western blot. Results are representative of three independent experiments. (I) DKO BMDMs were transduced with lentiviral vectors expressing full-length MMP14, MMP14E/A, or an empty control and differentiated into osteoclasts. Cell lysates were collected for MMP14 and galectin-3 expression and cleavage as assessed by Western blot. Results are representative of three independent experiments. **P < 0.01. Statistical significance was assessed using two-way ANOVA with Bonferroni correction (B) and unpaired two-sided Student’s t test (D). Source data are available for this figure: SourceData F4.
Figure S4.
Galectin-3 surface binding antagonist reverses functional defects in DKO osteoclasts. (A and B) Galectin-3 cleavage by Western blot using either an anti–galectin-3 full-length polyclonal antibody (Proteintech, 14979-1-AP; A) or an anti–galectin-3 monoclonal antibody (ab2785; Abcam; epitope mapped within the N-terminal region; B), following co-incubation with 3 μg recombinant human GALECTIN-3 and activated recombinant MMP9 at molar ratios from 1:50 to 1:5 (enzyme to substrate ratio) at 37°C for 30 min. Results are representative of three independent experiments. (C) Detection of surface galectin-3 in wild-type and DKO osteoclasts (OC) with the CoraLite 488–conjugated galectin-3 full-length polyclonal antibody (CL488-14979; Proteintech) as detected by flow cytometry. Results are representative of three independent experiments. (D) c-Src, Ctsk, and RhoA expression in wild-type osteoclasts differentiated and treated with vehicle, and DKO osteoclasts treated with vehicle or 20 μg/ml GCS-100 for 5 d at 37°C as assessed by Western blot. Results are representative of three independent experiments. (E and F) Surface galectin-3 levels were determined in wild-type osteoclasts treated with vehicle, and DKO osteoclasts treated with vehicle or 20 μg/ml GCS-100 for 2 h at 37°C, followed by flow cytometry stained with an eFluor 660–conjugated anti–galectin-3 monoclonal antibody (#50-5301-82; Thermo Fisher Scientific; clone M3/38, epitope mapped within the N-terminal region; E) or immunofluorescence stained with an anti–galectin-3 monoclonal antibody (#125401; Biolegend; clone M3/38; F; red). Scale bar, 20 μm. Results are representative of three independent experiments. (G) Wild-type osteoclasts were treated with vehicle, and DKO osteoclasts were treated with vehicle or 20 μg/ml GCS-100 for 2 h at 37°C, and RhoA activity determined following activation with 20 ng/ml M-CSF and 30 ng/ml RANKL for 15 min. Data are presented as mean ± SEM (n = 6 biological replicates). (H and I) Phalloidin staining (red), resorption pits visualized with WGA-DAB staining, and TRAP staining in wild-type pre-osteoclasts treated with vehicle, and DKO pre-osteoclasts treated with vehicle or 20 μg/ml GCS-100 cultured on bone slices for 3 d at 37°C (H) with actin ring area per cell, resorption pit area, and the number of TRAP+ MNCs quantified (I). Scale bar, upper 20 μm, middle and lower 100 μm. Data are presented as mean ± SEM (n = 6 biological replicates). (J and K) Transcriptional profiling analysis of cultured wild-type osteoclasts treated with vehicle, and DKO osteoclasts differentiated and treated with vehicle or 20 μg/ml GCS-100 for 5 d at 37°C (n = 3 biological replicates). Pie chart depicts the distribution of reversed transcripts by GCS-100 in DKO osteoclasts as compared with wild-type osteoclasts (J; n = 3 biological replicates). DAVID GO analysis of reversed expressed genes from DKO osteoclasts treated with GCS-100 versus DKO osteoclasts (K; n = 3 biological replicates). **P < 0.01. Statistical significance was assessed using one-way ANOVA with Bonferroni correction. Source data are available for this figure: SourceData FS4.
Osteoclast MMP9/MMP14-dependent galectin-3 lattice remodeling controls RhoA activation, sealing zone formation and bone resorption
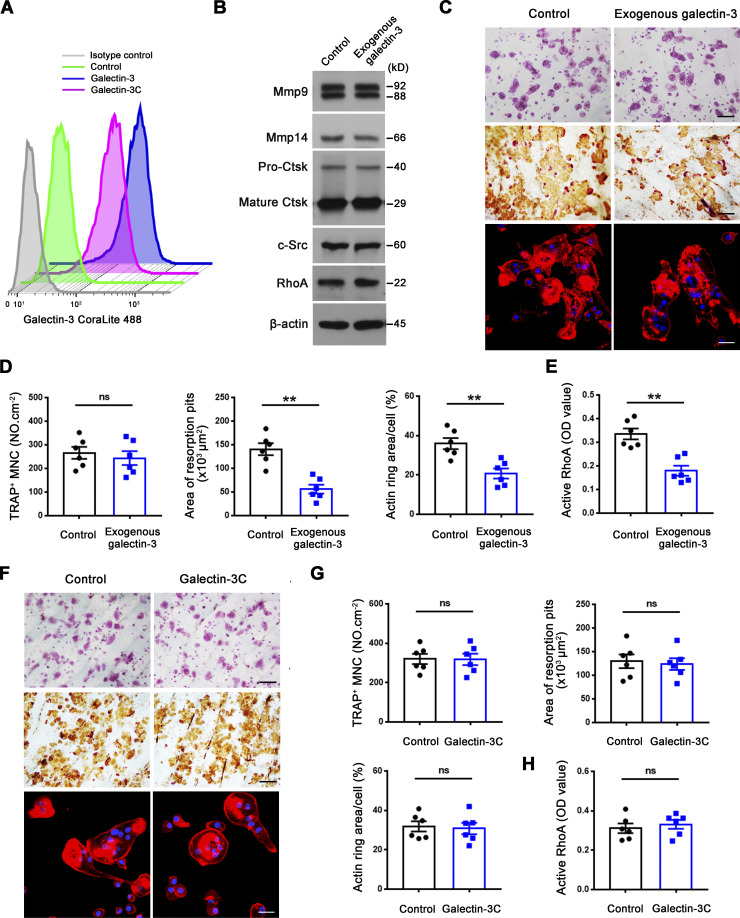
As global galectin-3 knockout mice display decreased bone mass coupled with increased osteoclast activity (Iacobini et al., 2018; Simon et al., 2017), we considered the possibility that membrane-bound galectin-3 serves as an endogenous regulator of osteoclast activity. Consistent with this premise, while exogenously supplied galectin-3 does not affect osteoclastogenesis of wild-type cells, bone-resorptive activity is significantly inhibited in tandem with large increases in surface galectin-3 exposure and repressed RhoA activation as well as sealing zone formation, a phenotype similar, if not identical, to that observed in DKO osteoclasts (Fig. 5, A–E). Multimerization of galectin-3 often leads to cross-linking of its saccharide ligands and formation of lattice-like structures on plasma membranes, events critical to transducing biological activity (Farhadi et al., 2021; Nabi et al., 2015; Thiemann and Baum, 2016; Zhao et al., 2021). As the self-association of galectin-3 into cell surface networks requires its N-terminal domain, cleaved galectin-3 would be unable to form the galectin-3 latticework (Ochieng et al., 1998; Zhao et al., 2021). However, several studies indicate that galectin-3C, a construct containing only the C-terminal carbohydrate recognition domain, may attach to cell surface in the absence of self-association while retaining biological activity (Nieminen et al., 2007; Sundqvist et al., 2018; Yang et al., 1998). Consequently, to determine whether the osteoclast-inhibitory effects exerted by galectin-3 requires multimerization, osteoclasts were cultured with galectin-3C, a peptide containing amino acids 108–250 which includes the C-terminal carbohydrate recognition domain of galectin-3 (John et al., 2003; Massa et al., 1993). Despite the fact that either exogenously applied galectin-3 or galectin-3C associate strongly with the wild-type osteoclast surface (Fig. 5 A), only galectin-3, and not galectin-3C, inhibits sealing zone formation and bone resorption (Fig. 5, B–H), indicating that galectin-3 binding and multimerization are necessary to support osteoclast-inhibitory activity.
Figure 5.
Exogenous galectin-3, but not galectin-3C, regulates osteoclast RhoA GTPase activation, sealing zone formation and bone resorptive activity. (A) Surface galectin-3 levels in wild-type osteoclasts exogenously treated with 0.1 μM galectin-3 or galectin-3C (corresponds to amino acids 108–250 containing the C-terminal carbohydrate recognition domain of galectin-3) for 30 min at 37°C (John et al., 2003; Massa et al., 1993), followed by staining with the CoraLite 488–conjugated galectin-3 polyclonal antibody (CL488-14979; Proteintech; epitopes mapped against full-length) as detected by flow cytometry. Results are representative of three independent experiments. (B) Mmp9, Mmp14, Ctsk, c-Src, and RhoA expression in wild-type osteoclasts treated with exogenous 0.1 μM galectin-3 as assessed by Western blot. Results are representative of three independent experiments. (C and D) TRAP (red), WGA-DAB, and phalloidin staining (red) of wild-type pre-osteoclasts cultured atop bone slices in the presence of exogenous 0.1 μM galectin-3 for 3 d at 37°C (C), with the number of TRAP+ MNCs, resorption pit area, and actin ring area per cell quantified (D). Scale bar, upper and middle 100 μm, lower 20 μm. Data are presented as mean ± SEM (n = 6 biological replicates). (E) Wild-type osteoclasts were cultured and treated with exogenous 0.1 μM galectin-3 for 2 h at 37°C and RhoA activity determined upon activation with 20 ng/ml M-CSF and 30 ng/ml RANKL for 15 min. Data are presented as mean ± SEM (n = 6 biological replicates). (F and G) Wild-type pre-osteoclasts were cultured atop bone slices and treated with exogenous 0.1 μM galectin-3C for 3 d at 37°C (John et al., 2003; Massa et al., 1993). The number of TRAP+ MNCs, resorption pit area, and actin ring area per cell were quantified (G) following TRAP, WGA-DAB, and phalloidin staining (red) (F), respectively. Scale bar, upper and middle 100 μm, lower 20 μm. Data are presented as mean ± SEM (n = 6 biological replicates). (H) Wild-type osteoclasts were cultured and treated with exogenous 0.1 μM galectin-3C for 2 h at 37°C and RhoA activity determined upon activation with 20 ng/ml M-CSF and 30 ng/ml RANKL for 15 min. Data are presented as mean ± SEM (n = 6 biological replicates). **P < 0.01. Statistical significance was assessed using unpaired two-sided Student’s t test. Source data are available for this figure: SourceData F5.
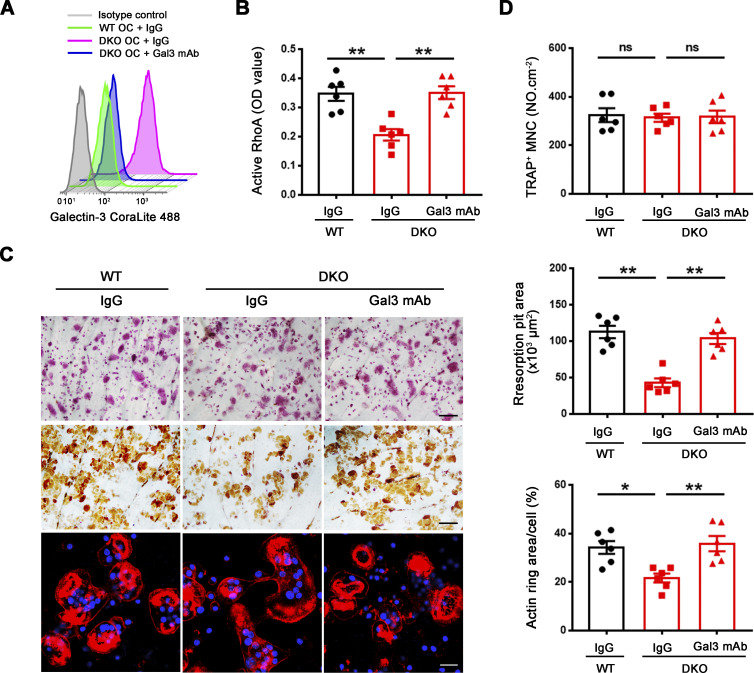
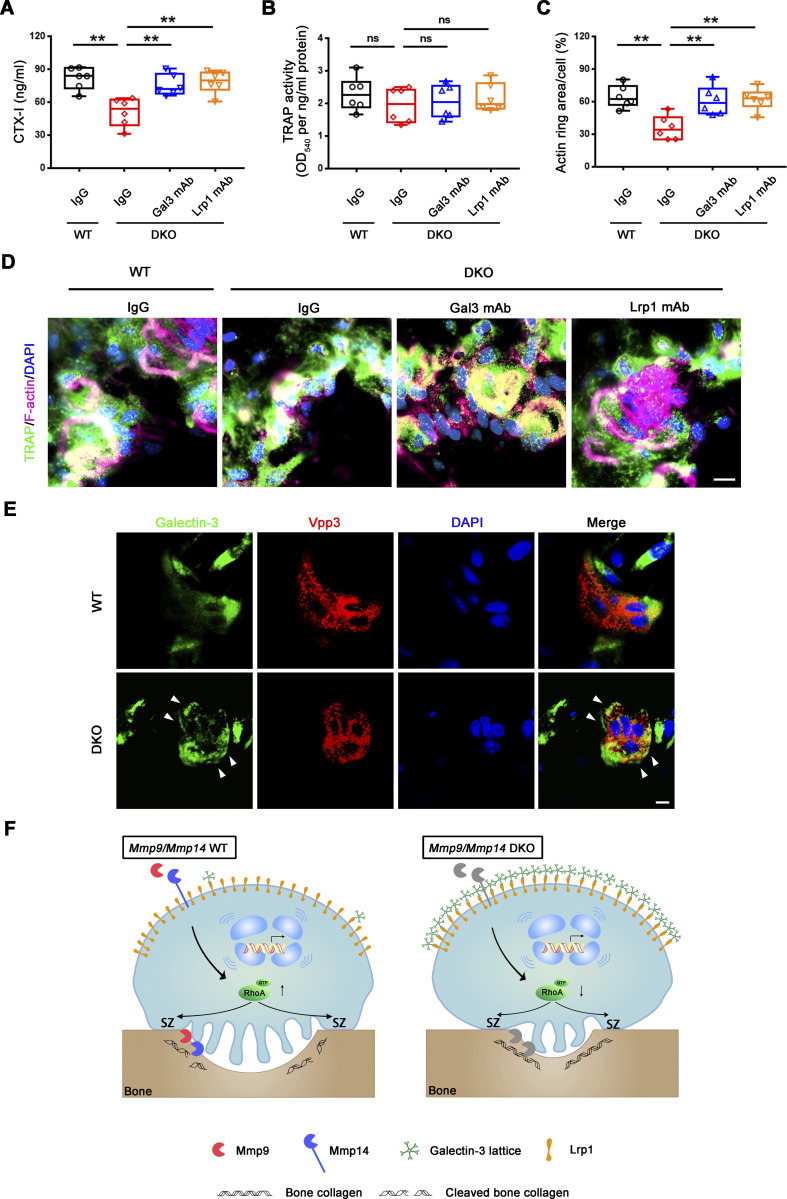
Given these findings, we sought to establish whether depleting endogenously derived galectin-3 from the surface of DKO osteoclasts would rescue bone-resorbing activity. Hence, we cultured DKO osteoclasts in the presence of the galectin-3 neutralizing antibody, B2C10 (Chen et al., 2015), that binds the N-terminal domain of galectin-3, thereby inhibiting its self-association without affecting carbohydrate-binding activity (Liu et al., 1996). Following treatment of DKO osteoclasts with B2C10 for 2 h, galectin-3 surface exposure in DKO osteoclasts is inhibited without affecting osteoclast differentiation or the expression of osteoclast maturation markers (Fig. 6 A). By contrast, the galectin-3 neutralizing antibody reverses the impaired RhoA activation as well as the defective cytoskeletal organization and bone resorption observed in DKO osteoclasts (Fig. 6, B–D). Alternatively, and consistent with these findings, treatment with the galectin-3 binding inhibitor, GCS-100, a modified pectin carbohydrate (Demotte et al., 2010; Seguin et al., 2017), likewise inhibits galectin-3 surface exposure and restores the bone resorptive program in DKO osteoclasts (Fig. S4, D–I). In support of galectin-3 functioning as a general gatekeeper of osteoclast function and transcription, when DKO osteoclasts are treated with GCS-100 and transcriptionally profiled, 770 unique transcripts are normalized to wild-type expression levels (Fig. S4 J). Further, consistent with the ability of GCS-100 to rescue RhoA activity and sealing zone formation in DKO osteoclasts, GO analysis reveals that categories of cytoskeleton and small GTPase-mediated signaling transduction are specifically recovered, including expression of 30–40% of small GTPase-mediated signaling transduction-related genes (Fig. S4 K). Hence, depleting galectin-3 from the cell surface partially restores the gene expression profile of DKO osteoclasts in tandem with an almost complete rescue of bone-resorptive activity.
Figure 6.
Mmp9/Mmp14-dependent galectin-3 lattice remodeling controls osteoclast RhoA activation, sealing formation and bone resorptive activity. (A) Surface galectin-3 levels were determined in wild-type osteoclasts (OC) treated with control IgG, and DKO osteoclasts treated with control IgG or galectin-3 function-blocking mAb at 25 μg/ml for 2 h at 37°C, followed by staining with the CoraLite 488–conjugated galectin-3 full-length polyclonal antibody (CL488-14979; Proteintech) as detected by flow cytometry. Results are representative of three independent experiments. (B) Wild-type osteoclasts were treated with control IgG, and DKO osteoclasts were treated with control IgG or galectin-3 function-blocking mAb at 25 μg/ml for 2 h at 37°C, and RhoA activity determined upon activation with 20 ng/ml M-CSF and 30 ng/ml RANKL for 15 min. Data are presented as mean ± SEM (n = 6 biological replicates). (C and D) TRAP (red), WGA-DAB, and phalloidin staining (red) of wild-type pre-osteoclasts cultured atop bone slices treated with control IgG, and DKO pre-osteoclasts treated with control IgG or galectin-3 function-blocking mAb (Gal3 mAb; 25 μg/ml) for 3 d at 37°C (C), and the number of TRAP+ MNCs, resorption pit area, and actin ring area per cell quantified (D). Scale bar, upper and middle 100 μm, lower 20 μm. Data are presented as mean ± SEM (n = 6 biological replicates). *P < 0.05, **P < 0.01. Statistical significance was assessed using one-way ANOVA with Bonferroni correction.
MMP9/MMP14 co-dependent regulation of osteoclast function through the galectin-3–centric control of Lrp1 activation
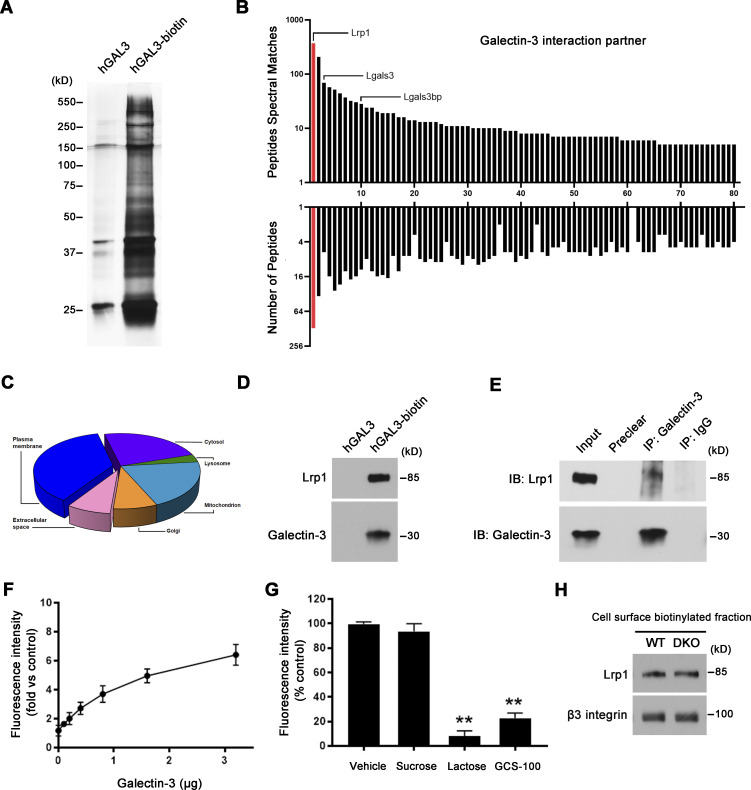
To define the mechanisms by which MMP9/MMP14-dependent galectin-3 remodeling affects osteoclast activity, we sought to identify its lectin-binding targets on the cell surface. To this end, biotinylated galectin-3 was added to osteoclasts and galectin-3–interacting proteins at the cell surface identified following streptavidin magnetic bead pulldown and mass spectrometry (Hönig et al., 2018; Fig. 7 A). As expected, the list of galectin-3 binders contained mostly glycoproteins from the plasma membrane, extracellular space, or intracellular compartments, including endogenous galectin-3 (Fig. 7, B and C), corroborating the fact that the recombinant lectin oligomerizes with the endogenously expressed orthologue into larger lattice networks (Hönig et al., 2018; Nabi et al., 2015).
Figure 7.
Mmp9/Mmp14 co-dependent regulation of osteoclast function through the galectin-3–centric control of Lrp1 activation. (A–C) Pre-osteoclasts were incubated with either human GALECTIN-3 or GALECTIN-3-biotin (hGAL3-biotin; 1.5 µM) for 30 min at 4°C. Biotinylated GALECTIN-3 was precipitated with streptavidin magnetic beads, separated by SDS-PAGE (A), and analyzed by mass spectrometry (B). Hits were classified according to the peptides spectral matches and the number of peptides per protein. Red columns: the top hit of GALECTIN-3 binding partner. The numbering on the x axis relates to Table S3. Pie chart depicts GALECTIN-3 interactors based on the GO annotation “cellular component” in FunRich (C). (D) Lrp1 and galectin-3 expression from the above streptavidin magnetic beads–precipitated lysates as assessed by Western blot. Results are representative of three independent experiments. (E) Protein lysates from wild-type osteoclasts were immunoprecipitated with the galectin-3 antibody, and Lrp1 and galectin-3 levels assessed by Western blot. Results are representative of three independent experiments. (F and G) Cell-free binding assay demonstrating direct binding between 0.1 and 3.2 μg GALECTIN-3 and 0.5 μg LRP1 for 4 h at 30°C (F), and the competitive inhibition of binding between GALECTIN-3 and LRP1 (1.6 versus 0.5 μg) by 50 mM sucrose, 50 mM lactose, or 10 mg/ml GCS-100 (G). Data are presented as mean ± SEM (n = 3 biological replicates). (H) Following cell surface biotinylation and capture with streptavidin magnetic beads, Lrp1 and β3 integrin expression in the membrane fraction of wild-type and DKO osteoclasts as assessed by Western blot. Results are representative of three independent experiments. **P < 0.01. Statistical significance was assessed using one-way ANOVA with Bonferroni correction. Source data are available for this figure: SourceData F7.
Interestingly, the top hit from this screen is Lrp1 (Fig. 7 B and Table S3), a heavily glycosylated cell surface protein capable of binding other galectin family members and previously implicated in regulating osteoclast function as well as small GTPase activity in other cell systems (Lu et al., 2018; Mantuano et al., 2010; Obermann et al., 2017; Vinik et al., 2015; Yang and Williams, 2017). Interactions between both exogenous and endogenous galectin-3 with Lrp1 are validated by pulldown and co-immunoprecipitation assays, respectively (Fig. 7, D and E). To confirm a direct interaction between these molecules, we constructed a cell-free protein-binding assay using recombinant galectin-3 and an Lrp1 cluster II Fc chimeric construct that covers ∼10% of the full-length Lpr1 protein sequence, including three N-glycosylation sites as well as a key ligand binding domain (Herz and Strickland, 2001). Under these conditions, galectin-3 binds to the extracellular domain of the Lrp1 construct through a process that is blocked by either GCS-100 or the small competitive binding inhibitor, lactose (Demotte et al., 2010; Seguin et al., 2017; Fig. 7, F–H).
To functionally assess the relative importance of Lrp1 as a galectin-3 signaling intermediate capable of regulating osteoclast activity, we first cultured wild-type cells in the presence of inhibitory concentrations of exogenous galectin-3 in the presence of the Lrp1 antagonist, receptor-associated protein (RAP; Lee et al., 2007). Under these conditions, RAP significantly increases RhoA activation, sealing zone formation and bone resorptive activity of the galectin-3–treated osteoclasts (Fig. S5, A–D). Importantly, DKO osteoclasts cultured in the presence either RAP or an Lrp1 blocking antibody (Lee et al., 2007; Mantuano et al., 2016), likewise recover RhoA activation, sealing zone formation and bone resorptive activity (Fig. 8, A–H and Fig. S5 E). By contrast, wild-type osteoclasts that display only low levels of surface-bond galectin-3 are largely unaffected when cultured in the presence of RAP (Fig. S5, B–D). Of note, the recovery of bone resorption in DKO osteoclasts occurs despite the absence of MMP9 and MMP14, previously established as functionally important type I collagenases that operate in concert with CTSK (Zhu et al., 2020). However, whereas wild-type osteoclasts retain significant bone collagenolytic activity in the absence of CTSK activity, the cysteine proteinase inhibitor E-64d almost completely inhibits the collagen-degradative activity of GCS-100– or RAP-treated DKO osteoclasts as assessed by the release of the type I collagen cross-linked telopeptide CTX-1 (Zhu et al., 2020; Fig. 8 I). Finally, though Lrp1 has been reported to be sensitive to MMP14-dependent processing (Alabi et al., 2021), both intracellular and membrane Lrp1 protein levels are unchanged between wild-type and DKO osteoclasts in our system (Fig. 7 H and Fig. 8 A). Together, these studies indicate that MMP9/MMP14–galectin-3 axis regulates Lrp1-mediated RhoA signaling, thereby controlling osteoclast activation and bone resorption.
Figure S5.
A galectin-3–Lrp1 axis regulates RhoA activation and sealing zone formation in osteoclasts. (A) Lrp1, c-Src, Ctsk, and RhoA expression in wild-type osteoclasts differentiated and treated with exogenous 0.1 μM galectin-3 in the presence or absence of 50 nM RAP for 5 d at 37°C as assessed by Western blot. Results are representative of three independent experiments. (B and C) Phalloidin (red) and WGA-DAB staining of wild-type pre-osteoclasts cultured atop bone slices treated with exogenous 0.1 μM galectin-3 in the presence or absence of 50 nM RAP for 3 d at 37°C (B), and actin ring area per cell and resorption pit area quantified (C). Scale bar, upper 20 μm, lower 100 μm. Data are presented as mean ± SEM (n = 6 biological replicates). (D) Wild-type osteoclasts were cultured and treated with exogenous 0.1 μM galectin-3 in the presence or absence of 50 nM RAP for 2 h at 37°C, and RhoA activity determined upon activation with 20 ng/ml M-CSF and 30 ng/ml RANKL for 15 min. Data are presented as mean ± SEM (n = 6 biological replicates). (E) TRAP (red), WGA-DAB, and phalloidin staining (red) of wild-type pre-osteoclasts cultured atop bone slices treated with vehicle, and DKO osteoclasts treated with vehicle or 50 nM RAP for 3 d at 37°C (n = 6 biological replicates). Scale bar, upper and middle 100 μm, lower 20 μm. (F) GALECTIN-3 expression and cleavage as assessed by Western blot in human osteoclasts differentiated from macrophages in the presence or absence of either an MMP9 function-blocking mAb (25 μg/ml) or MMP14 function-blocking antibody (DX-2400; 100 μg/ml). Results are representative of three independent experiments. (G and H) Calvaria isolated from wild-type and DKO mice were cultured in the presence or absence of 20 μg/ml GCS-100 for 5 d at 37°C, and supernatants or whole cell lysates collected for CTX-I ELISA (G) and TRAP activity (H), respectively. Data are presented as mean ± SEM (n = 6 biological replicates). (I and J) Actin ring area per cell (I) and phalloidin staining (magenta) with TRAP (green) immunofluorescence (J) are shown of wild-type and DKO calvaria explants cultured in the presence or absence of 20 μg/ml GCS-100 for 5 d at 37°C. Scale bar, 10 μm. Data are presented as mean ± SEM (n = 6 biological replicates). **P < 0.01. Statistical significance assessed using one-way ANOVA with Bonferroni correction. Source data are available for this figure: SourceData FS5.
Figure 8.
Inhibition of Lrp1 function reverses defects in RhoA activation, sealing zone formation and bone resorption in DKO osteoclasts. (A) c-Src, Ctsk, and RhoA expression in wild-type osteoclasts treated with vehicle, and DKO osteoclasts treated with vehicle or 50 nM RAP as assessed by Western blot. Results are representative of three independent experiments. (B) Wild-type osteoclasts were treated with vehicle, and DKO osteoclasts were treated with vehicle or 50 nM RAP for 2 h at 37°C, and RhoA activity determined upon activation with 20 ng/ml M-CSF and 30 ng/ml RANKL for 15 min. Data are presented as mean ± SEM (n = 6 biological replicates). (C–E) Phalloidin staining of wild-type pre-osteoclasts treated with vehicle, and DKO pre-osteoclasts treated with vehicle or 50 nM RAP cultured on bone slices for 3 d at 37°C, and actin ring area per cell quantified (C). Cells were stained for TRAP activity and the number of TRAP+ MNCs quantified (D). Osteoclasts were removed and resorption pits visualized by WGA-DAB staining and resorption pit area quantified (E). Data are presented as mean ± SEM (n = 6 biological replicates). (F and G) TRAP (red), WGA-DAB, and phalloidin staining (red) of wild-type pre-osteoclasts cultured atop bone slices treated with control IgG, and DKO pre-osteoclasts treated with control IgG or Lrp1 function-blocking mAb (25 μg/ml) for 3 d at 37°C (G), and the number of TRAP+ MNCs, actin ring area per cell, and resorption pit area quantified (F). Scale bar, upper and middle 100 μm, lower 20 μm. Data are presented as mean ± SEM (n = 6 biological replicates). (H) Wild-type osteoclasts were treated with control IgG, and DKO osteoclasts were treated with control IgG or Lrp1 function-blocking mAbs at 25 μg/ml for 2 h, and RhoA activity determined upon activation with 20 ng/ml M-CSF and 30 ng/ml RANKL for 15 min. Data are presented as mean ± SEM (n = 6 biological replicates). (I) Pre-osteoclasts differentiated from wild-type or DKO BMDMs were cultured atop cortical bone slices with or without 20 μM E64d, 20 μg/ml GCS-100, or 50 nM RAP for 3 d at 37°C, and supernatants collected for CTX-I ELISA. Data are presented as mean ± SEM (n = 6 biological replicates). *P < 0.05, **P < 0.01. Statistical significance was assessed using one-way ANOVA (B–F, H) and two-way ANOVA (I) with Bonferroni correction. Source data are available for this figure: SourceData F8.
Finally, given potential disparities in extrapolating findings from murine to human cell populations (Ginhoux and Jung, 2014), we examined the role of MMP9 and MMP14 in directing the proteolytic control of the GALECTIN-3/LRP1 axis in human osteoclasts. The GALECTIN-3 cleavage products are similarly present when human osteoclasts are treated with either function-blocking MMP9 or MMP14 mAbs alone, but disappear when the antibodies are used in combination (Fig. 9 A and Fig. S5 F). As described in mouse osteoclasts, cell surface GALECTIN-3 levels dramatically increase in human osteoclasts cultured in the presence of MMP9/MMP14 dual function-blocking antibodies, in tandem with decreased RhoA activation, defective cytoskeletal organization, and bone resorption (Fig. 9, B–F). Likewise, in the presence of MMP9/MMP14 dual function-blocking antibodies, either GALECTIN-3 or LRP1 neutralizing antibodies rescue RhoA activation, actin ring formation and bone resorptive activity (Fig. 9, C–F), supporting the role of MMP9/MMP14–GALECTIN-3/LRP1 axis as a gatekeeper of human osteoclast-mediated bone resorption.
Figure 9.
MMP9/MMP14–GALECTIN-3–LRP1 axis regulates human osteoclast activity. (A) GALECTIN-3 expression and cleavage as assessed with anti–galectin-3 monoclonal antibody (ab2785; Abcam; epitopes mapped against N-terminal region) by Western blot in human osteoclasts cultured in the presence or absence of MMP9 and MMP14 function-blocking mAbs. Results are representative of three independent experiments. (B) GALECTIN-3 (green) immunofluorescence stained with an anti-galectin-3 monoclonal antibody (#125401; Biolegend; clone M3/38, epitopes mapped against N-terminal region) of non-permeabilized human osteoclasts differentiated from macrophages with M-CSF/RANKL for 9 d at 37°C in the presence or absence of MMP9 and MMP14 function-blocking antibodies. Scale bar, 20 μm. Results are representative of three independent experiments. (C and D) Osteoclastic resorption pits and phalloidin staining (red) of human osteoclasts cultured atop bone slices with MMP9 and MMP14 function-blocking mAbs in the presence or absence of a GALECTIN-3 function-blocking mAb (GAL3 mAb) or LRP1 function-blocking mAb (25 μg/ml) for 9 d at 37°C (C) with resorption pit and actin ring areas quantified (D). Scale bar, upper 100 μm, lower 20 μm. Data are presented as mean ± SEM (n = 6 biological replicates). (E) Human osteoclasts were differentiated from macrophages with M-CSF/RANKL for 9 d in the presence or absence of MMP9 and MMP14 function-blocking mAbs with or without GAL3 mAb or LRP1 function-blocking mAb at 25 μg/ml, and RhoA activity determined upon activation with 20 ng/ml M-CSF and 20 ng/ml RANKL for 15 min. Data are presented as mean ± SEM (n = 6 biological replicates). (F) c-Src, Ctsk, and RhoA expression in human osteoclasts differentiated from macrophages with M-CSF/RANKL for 9 d in the presence or absence of MMP9 and MMP14 function-blocking mAbs with or without GAL3 mAb or LRP1 function-blocking mAb (25 μg/ml) as assessed by Western blot. Results are representative of three independent experiments. **P < 0.01. Statistical significance was assessed using one-way ANOVA with Bonferroni correction. Source data are available for this figure: SourceData F9.
Ex vivo control of osteoclast activity by an MMP9/MMP14–galectin-3/Lrp1 axis
To finally assess the role of the MMP9/MMP14–galectin-3/Lrp1 axis in regulating osteoclast activity in a natural bone microenvironment, we characterized bone resorption in calvarial organ cultures prepared from wild-type and DKO mice (Inada et al., 2004). Consistent with our in vitro results, DKO calvaria explants exhibit a decrease in the release of bone collagen turnover as assessed by CTX-I relative to wild-type controls despite similar osteoclast numbers as defined by tartrate-resistant acid phosphatase (TRAP) activity level (Fig. 10, A and B). Of note, defects in bone resorption correlate with increased galectin-3 staining at the cell surface of DKO osteoclasts in tandem with a striking decrease in sealing zone formation (Fig. 10, C–E). However, when DKO explants are cultured in the presence of anti–galectin-3 or anti-Lrp1 blocking antibodies, both sealing zone formation and bone resorption increase to wild-type levels (Fig. 10, A–D). Consistent with these findings, treatment with GCS-100 likewise restores sealing zone formation and the bone resorptive program of DKO osteoclasts in calvaria explants (Fig. S5, G–J). Together, these data demonstrate that the MMP9/MMP14-regulated galectin/Lrp1 axis functions as a physiologically relevant control mechanism of osteoclast activation in situ.
Figure 10.
Mmp9/Mmp14–galectin-3–Lrp1 axis controls osteoclast activity in a calvaria explant model. (A and B) Calvaria isolated from wild-type and DKO mice were cultured in the presence or absence of either the galectin-3 or Lrp1 function-blocking mAbs (25 μg/ml) for 5 d at 37°C, and supernatants or whole cell lysates collected for CTX-I ELISA (A) and TRAP activity (B), respectively. Data are presented as mean ± SEM (n = 6 biological replicates). (C and D) Actin ring area per cell (C) and phalloidin staining (magenta) with TRAP (green) immunofluorescence with an anti-TRAP polyclonal antibody (sc-30833; Santa Cruz; D) are shown of wild-type and DKO calvaria explants cultured in the presence or absence of either galectin-3 or Lrp1 function-blocking mAbs (25 μg/ml) for 5 d at 37°C. Scale bar, 10 μm. Data are presented as mean ± SEM (n = 6 biological replicates). **P < 0.01. Statistical significance was assessed using one-way ANOVA with Bonferroni correction. (E) Galectin-3 (green) and Vpp3 (red) immunofluorescence stained with an anti–galectin-3 monoclonal antibody (#125401; Biolegend; clone M3/38) and an anti-Vpp3 polyclonal antibody (Abcam, ab200839) of wild-type and DKO calvarial organ. White arrowhead, galectin-3 lattice. Scale bar, 10 μm. Results are representative of three independent experiments. (F) Schematic depicting dual roles for MMP9 and MMP14 in osteoclast-mediated bone resorption by both modulating the galectin-3/Lrp1-dependent control of RhoA-GTPase activity, but also serving as direct-acting bone type I collagenolysis (Zhu et al., 2020). Galectin-3 is depicted as a pentamer only as an example of a potential multimer complex.
Discussion
The unique capacity of osteoclasts to resorb bone, in both physiological and pathological contexts, highlights the importance of defining the molecular mechanisms controlling their development and activity (Compston et al., 2019; Teitelbaum and Ross, 2003; Zaidi, 2007). Using conditional knockout mouse models that target both Mmp9 and Mmp14 in tandem, we previously reported that wild-type osteoclasts are able to use either metalloenzyme to degrade bone type I collagen in vitro or in vivo, thereby requiring their dual knockout in order to uncover a combined role in bone resorption (Zhu et al., 2020). Applying Occam’s razor, i.e., with multiple explanations in hand, the simplest model is the most preferred, we assumed that osteoclasts reserve metalloenzymes for collagenolytic activity alone and predicted that DKO cells would display normal patterns of gene expression as well as intact function when cultured atop plastic surfaces in the absence of a type I collagen substratum. Unexpectedly, DKO osteoclasts display major shifts in gene expression, precipitating efforts to identify new MMP9/MMP14-directed functions and target substrates that might account for these global effects. Herein, we report that MMP9/MMP14 exert protease-dependent control over RhoA activation, sealing zone formation and bone resorption by modulating a previously unidentified galectin-3/Lrp1 signaling axis in mouse as well as human osteoclasts.
Given the range of observed changes in gene expression that followed Mmp9/Mmp14 targeting, we first searched for functional clues by focusing on affected GO categories. In this regard, our attention first centered on mitochondrial function given the number of differentially expressed transcripts and the known importance of mitochondrial function in regulating osteoclast activity (Arnett and Orriss, 2018; Bae et al., 2017; Nishikawa et al., 2015). This focus was further rationalized on a complementary body of literature linking MMP9 and MMP14 to metabolic control in other cell systems (de Castro Brás et al., 2014; Kowluru et al., 2011; Mori et al., 2016; Sakamoto and Seiki, 2010). However, from a functional perspective, we were unable to detect major changes in mitochondrial activity or metabolomics in DKO osteoclasts. Alternatively, we noted multiple effects on gene families related to small GTPases, proteins known to play key roles in regulating osteoclast–bone adhesive interactions (Blangy et al., 2020; Teitelbaum, 2011; Touaitahuata et al., 2014). Whereas both Rac1 and Rac2 as well as RhoA can regulate osteoclast activity (Blangy et al., 2020; Teitelbaum, 2011; Touaitahuata et al., 2014), only active RhoA-GTP levels were affected with significant decreases noted in DKO cells. While actin ring formation atop plastic substrata remains intact in DKO osteoclasts (Zhu et al., 2020), DKO cells display major defects when cultured atop bone, both with regard to sealing zone formation and resorptive activity, phenotypes pathognomonic of RhoA inhibition (Chellaiah et al., 2000; Uehara et al., 2017). Underlining the functional significance of defects in osteoclast RhoA activation, sealing zone formation and bone resorptive activity are reversed in DKO osteoclasts following transduction with a constitutively active form of the small GTPase.
In searching for substrates whose cleavage by either MMP9 or MMP14 could potentially regulate osteoclast function, we elected to search for bioactive targets known to contain collagenase-cleavable sequences. The galectins are a large family of β-galactoside–binding proteins that share considerable homology in their respective carbohydrate recognition domains, but only galectin-3 contains an N-terminal domain that supports multimer formation (Nabi et al., 2015; Thiemann and Baum, 2016). Consequently, upon binding to cell surface ligands, galectin-3 lattice-like networks may be formed, enabling the modulation of a wide variety of intracellular signaling pathways that, in turn, alter cell function (Nabi et al., 2015; Thiemann and Baum, 2016). Importantly, cleavage within the N-terminal proline, glycine, and tyrosine-rich tandem repeats of galectin-3 by either MMP9 or MMP14 has previously been shown to alter its affinity for glycoconjugates as well as inhibit galectin-3 self-association into cell surface networks (McClung et al., 2007; Ochieng et al., 1994; Ochieng et al., 1998; Toth et al., 2005; Zhao et al., 2021). Nevertheless, the ability of these metalloproteinases to hydrolyze galectin-3 in an intact cell system under physiologic conditions, including the presence of potent plasma-derived proteinase inhibitors, has not been described previously. We now find that osteoclasts constitutively proteolyze endogenously derived surface galectin-3 by mobilizing either MMP9 or MMP14, and that this process is only abrogated following their dual targeting.
Strengthening a potential link between galectin-3 levels and osteoclast function is the fact that Lgal3−/−/galectin-3-null mice often, but not always, display decreased bone mass coupled with increased osteoclast activity in vivo (Iacobini et al., 2018; Simon et al., 2017). Complicating efforts to extrapolate these results to our model system, it is noteworthy that all cell populations are targeted in these global knockouts, including osteoblasts, and that galectin-3 has both extracellular and intracellular functions (Iacobini et al., 2018; Simon et al., 2017; Thiemann and Baum, 2016). These caveats notwithstanding, when we cultured wild-type osteoclasts with exogenous galectin-3 at levels designed to overwhelm proteolytic processing, RhoA activation, sealing zone formation and bone resorption were inhibited in a fashion similar to that observed in Mmp9/Mmp14 DKO cells. More importantly, the functional defects displayed by DKO osteoclasts were largely reversed following the displacement of surface-bound galectin-3. Interestingly, preventing galectin-3 exposure rescued close to 30% (i.e., 770 unique genes) of the differentially expressed transcripts impacted in Mmp9/Mmp14 DKO osteoclasts. The incomplete rescue of the entire transcript profile indicates that MMP9 and MMP14 likely target additional substrates beyond extracellular galectin-3, but the effectiveness of galectin-3 neutralizing antibody and a galectin-3 binding inhibitor in rescuing RhoA-dependent sealing zone formation and bone resorption in DKO osteoclasts strongly supports the contention that the galectin-3 lattice serves as the preeminent negative regulator of osteoclast activity. We do note that non-specific inhibitory effects have been assigned to GCS-100 and other similarly modified pectin carbohydrate compounds (Nishikawa and Suzuki, 2019; Pedrosa et al., 2022; Stegmayr et al., 2016). Indeed, the ability of pectins to act as competitive inhibitors of galectin-3 binding interactions has been questioned, though GCS-100 was not specifically studied here (Stegmayr et al., 2016). Despite these concerns, the Cheresh group similarly used GCS-100 to block galectin-3/αvβ3 interactions in both cell-free and intact cell systems (Seguin et al., 2017). Nevertheless, it remains possible that GCS-100 may have other mechanisms here for affecting osteoclast function in addition to disrupting surface exposure of galectin-3. We stress, however, that effects observed here with GCS-100 duplicated those observed with a galectin-3 neutralizing antibody and that the modified pectin rescued, rather than inhibited, DKO osteoclast function.
The striking biological activity of extracellular galectin-3 on osteoclast function spurred efforts to identify cognate receptors. Though galectin-3 is known to bind to numerous targets in other cell systems (Joeh et al., 2020; Obermann et al., 2017), using mass spectrometry, we identified a cohort of potentially important receptors, ranging from vacuolar ATPases to Piezo channels whose functional role as galectin-3 targets remain to be determined, but the dominant glycoprotein detected was Lrp1 (Yang and Williams, 2017). Lrp1 is a heavily glycosylated glycoprotein composed of a 515 kD extracellular ligand-binding subunit (α-chain) that contains four clusters of ligand binding domains linked non-covalently to an 85 kD transmembrane subunit encompassing a short cytoplasmic tail (β-chain; Yang and Williams, 2017). This transmembrane receptor is a multi-functional member of the low-density lipoprotein receptor family of lipoprotein receptors, best characterized as an endocytic receptor that recognizes over 30 targets, including a broad range of proteases, including MMP family members, as well as protease inhibitors (Yang and Williams, 2017). Independent of Lrp1’s role in regulating the extracellular concentration of specific cargoes, the receptor also modulates a number of cellular signaling pathways, including those controlled by Rho-GTPase activity through as yet-to-be-defined mechanisms (Appert-Collin et al., 2017; Gao et al., 2016; Mantuano et al., 2010). While MMP14 has been reported to shed Lrp1 from cell surfaces in other systems (Alabi et al., 2021; Lehti et al., 2009), Lrp1 receptor levels were indistinguishable between wild-type and DKO osteoclasts. Despite its possible role in regulating galectin-3–dependent osteoclast activity, only a single recent report identified Lrp1 as a potential galectin-3 receptor in retinal pigment epithelial cells (Obermann et al., 2017), and the functional impact of this interaction has not been examined in this or any other cell population. Remarkably, we found that RAP, an Lrp1-binding chaperone that blocks all cargo interactions with the receptor (Lee et al., 2007), fully restored bone resorption in DKO osteoclasts. As predicted, RAP exerts little effect on wild-type osteoclasts wherein surface levels of galectin-3 are maintained at low levels secondary to MMP/MMP14-dependent proteolysis. In considering the pathophysiological ramifications of our identification of Lrp1 as the downstream effector of the MMP9/MMP14–galectin-3 proteolytic axis, it is interesting to note that targeting Lrp1 in mouse osteoclasts gives rise to a dramatically decreased trabecular bone mass in vivo in tandem with increased osteoclast activity (Lu et al., 2018). Further, in humans, LRP1 single nucleotide polymorphisms are associated with low bone mineral density (Sims et al., 2008). Future studies are yet needed to identify Lrp1-interacting partner(s) that allow galectin-3 to modulate RhoA activity at either—or both—the transcriptional or/and post-transcriptional levels. Interestingly, our mass spectrometry–based analysis of galectin-3 binding partners also included a number of potential targets that are capable of depressing RhoA activity by enhancing RhoGAP activity, including Rap1a, moesin, and Slc3a2 (Boulter et al., 2013; Neisch et al., 2013; Post et al., 2015). Finally, independent of Lrp1’s ability to transduce intracellular signaling through extracellular galectin-3 binding, we noted that binding to cell surface glycoproteins can trigger galectin-3 oligomerization through a recently described process similar to liquid–liquid phase separation wherein galectin-3 valency scales with both its binding affinity and biological activity (Chiu et al., 2020; Farhadi et al., 2021; Zhao et al., 2021). Nevertheless, the oligomerization status of galectin-3 at the osteoclast surface remains to be determined.
In sum, our work defines new roles for osteoclast MMP9 and MMP14 that extend beyond their function as collagenolysis. That is, rather than confine their functions to matrix degradation, MMP9/MMP14 also serve as upstream regulators of osteoclast activity by modulating the galectin-3/Lrp1-dependent control of active RhoA levels (Fig. 10 F). Therefore, our findings define a new model wherein a previously unrecognized quartet of interacting proteases, a carbohydrate-binding lectin, and a transmembrane protein together serve as a rheostat that autoregulates osteoclast activity in vitro and in vivo. Nevertheless, given our findings, previously documented defects in MMP9- or MMP14-targeted macrophages, lymphocytes, endothelial cells, and hematopoietic stem cells (Jin et al., 2011; Kelly et al., 2018; Miloudi et al., 2016; Nishida et al., 2012; Shimizu-Hirota et al., 2012) may well require re-evaluation within the context of previously unappreciated changes in galectin-3 proteolytic regulation and function.
Materials and methods
Mouse strains
Mice with Mmp14-floxed alleles (Mmp14f/f) were generated in our laboratory (Tang et al., 2013; Zhu et al., 2020), while Mmp9−/− mice and Csf1r-Cre mice were obtained from Jackson Laboratories. Mmp14f/f mice were bred with Csf1r-Cre transgenic mice to obtain Mmp14ΔM/ΔM (Csf1r-Cre/Mmp14f/f) mice. To generate Csf1r-Cre/Mmp9−/−/Mmp14f/f (DKO) mice, Csf1r-Cre/Mmp9+/−/Mmp14f/+ mice were crossed with Mmp9+/−/Mmp14f/+ mice (Zhu et al., 2020). Csf1r-Cre expressing mice were crossed onto a ROSA26R+/loxP background to obtain the osteoclast and myeloid lineage-labeled Csf1r-Cre/ROSA26R+/loxP mice. For genotyping primers, see Table S1.
All mouse work was performed under the guidelines and approval of the Animal Welfare and Ethics Committee (School and Hospital of Stomatology, Wuhan University) and the University of Michigan Institutional Animal Care & Use Committee.
In vitro osteoclast differentiation
Bone marrow cells extracted from femurs and tibiae of 10–12-wk old mice were cultured in α-MEM containing 10% FBS, 100 IU/ml penicillin, and 100 μg/ml streptomycin with 20 ng/ml recombinant mouse M-CSF (416-ML; R&D Systems) in plastic petri dishes. Cells were incubated at 37°C in 95% air/5% CO2 for 4 d and then lifted with 5 mM EDTA in PBS. Recovered BMDMs were cultured in α-MEM containing 10% FBS supplemented with 20 ng/ml mouse M-CSF and 30 ng/ml mouse RANKL (462-TEC; R&D Systems) for 5 d in tissue-culture dishes to induce osteoclast formation (Tang et al., 2009; Zhu et al., 2020). Mature osteoclasts were characterized by staining for TRAP activity using an Acid Phosphatase Leukocyte Kit (387A; Sigma-Aldrich) and TRAP-positive MNCs (>3 nuclei/cell) counted.
RNA extraction, quantitative PCR (qPCR), and RNA-seq analysis
RNA was isolated from cultured cells with TRIzol reagent (Invitrogen) and purified using QIAGEN RNeasy Mini-kit columns (QIAGEN). RNA quality was confirmed using an Agilent 2100 Bioanalyzer. Mouse cDNA was reverse transcribed from 1 μg total RNA with SuperScript VILO Master Mix (Life Technologies). qPCR was performed in triplicate samples using SYBR Green PCR master mix (Applied Biosystems) according to the manufacturer’s instructions. For each transcript examined, raw data (CT) were obtained by qPCR, and the ΔΔCT method (ΔΔCT = ΔCT (experimental gene) − ΔCT (controlled gene)) was used to calculate the relative fold of gene expression (fold = 2ΔΔCT). ΔCT was calculated using a housekeeping gene (Gapdh; whose equivalent expression in wild-type and DKO cells was confirmed in our transcriptional profiling) and averaged (ΔCT = CTGAPDH − CTgene). For qPCR primers, see Table S2.
For RNA-seq, the construction of libraries was generated using QuantSeq 3′ mRNA-Seq Library Prep Kit (Lexogen) according to the manufacturer’s instructions. High-throughput sequencing was performed as single-end 75 sequencing using NextSeq 500 (Illumina). Each sample was analyzed at the University of Michigan Advanced Genomics Core. Data were aligned using the STAR aligner and featureCounts v1.6.4 software, and reads per kilobase of transcript per million mapped read values on gene level were estimated for ensemble transcriptome (Dobin et al., 2013; Liao et al., 2014). DESeq2 was used to estimate significance between any two experimental groups (Love et al., 2014). Principal component analysis was performed on the RNA-seq data to visualize sample-to-sample variance. Differentially expressed genes were analyzed using the DAVID Bioinformatics Resources 6.8 (Jiao et al., 2012).
mtDNA quantitation
mtDNA copy number per nuclear genome in osteoclasts was quantitated (Nakahira et al., 2011). Total DNA purification was performed using the DNeasy kit (Qiagen). The abundance of nuclear and mitochondrial DNA, reflected by 18s ribosomal RNA and cytochrome c oxidase I, was quantified by real-time PCR under standard conditions using a SYBR Green PCR kit (Qiagen). The ratios between mtDNA and nuclear DNA concentrations were calculated.
Snapshot metabolomics
Cells for snapshot metabolomics were lysed in cold (−80°C) 80% methanol. After centrifuging, samples were normalized and lyophilized by speed vac. Dried pellets were resuspended in 1:1 of methanol:H2O before liquid chromatography/mass spectrometry–mass spectrometry analysis (Bian et al., 2020). Samples were run in triplicate on an Agilent QQQ 6470 liquid chromatography/mass spectrometry–mass spectrometry with ion pairing chromatography acquiring dynamic multiple reaction monitoring for 226 metabolites with a delta retention time window of 1 min. Data were preprocessed by applying a threshold area of 3,000 ion counts and a coefficient of variation of 0.5 among triplicates. All chromatography analysis was performed with Agilent MassHunter Quantitative Analysis 9.0.647.0. Heatmaps were generated and data clustered using Morpheus Matrix Visualization and analysis tool (https://software.broadinstitute.org/morpheus). Pathway analyses were conducted using MetaboAnalyst (https://www.metaboanalyst.ca).
In vitro bone resorbing activity
For bone resorption assays, BMDMs or pre-osteoclasts were seeded and cultured on bovine cortical bone slices (DT-1BON1000-96; Immunodiagnostic Systems) with 20 ng/ml M-CSF and 30 ng/ml RANKL (R&D Systems; Wu et al., 2017; Zhang et al., 2018; Zhu et al., 2020), in the presence or absence of galectin-3 (8259-GA; R&D Systems), galectin-3C (10110-GA; R&D Systems), GCS-100 (La Jolla Pharmaceutical), RAP (4480-LR; R&D Systems), an anti–galectin-3 blocking antibody (sc-32790L; Santa Cruz), or an anti-Lrp1 blocking antibody (MA1-27198; Thermo Fisher Scientific; Chen et al., 2015; Demotte et al., 2010; John et al., 2003; Moxon et al., 2015; Seguin et al., 2017). After the indicated culture period, bone samples were sonicated in PBS, stained with 20 μg/ml WGA-lectin (L3892; Sigma-Aldrich) for 45 min and then incubated with DAB tablets (D4418; Sigma-Aldrich) for 15 min. Image J software was used to quantify the resorbed area. The concentration of the CTX-I was measured using the CrossLaps for Culture CTX-I ELISA kit (AC-07F1; Immunodiagnostic Systems) according to the manufacturer’s instructions.
Scanning electron microscopy analysis
After removing cells, bone slices were fixed overnight in 2.5% glutaraldehyde in 0.1 M Sorensen’s phosphate buffer (pH 7.4). Samples were then dehydrated in ascending ethanol series, processed for critical point drying (Balzers Union) and Au/Pg sputtered (E-5100, Polaron Equipment Ltd.). Samples were imaged on an AMRAY 1910 field emission scanning electron microscope.
Rho GTPase activity assay
Osteoclasts were starved for 4 h in 2% FBS containing α-MEM, and then stimulated with 20 ng/ml M-CSF and 30 ng/ml RANKL for 15 min and lysed. Cell lysates were harvested, and Rho GTPase activity analyzed using RhoA/Rac1 G-LISA Activation Assay kits (BK135; Cytoskeleton) according to the manufacturer’s instructions.
Lentiviral or retroviral gene transduction
Where indicated, BMDMs were transduced with either a control vector, wild-type human MMP9, a catalytically inactive MMP9 E402/A mutant (Orgaz et al., 2014), wild-type human MMP14, a catalytically inactive MMP14 E240/A mutant expression vector (Shimizu-Hirota et al., 2012), or a constitutively active human RhoA L63 expression vector (RTV-204; Cell Biolabs). The MMP9 and MMP14 constructs were cloned into the pLentilox-IRES-GFP lentiviral vector, while the RhoA constructs were inserted into pBABEhygro retroviral vector. Expression of the recombinant proteins was confirmed by Western blot.
Western blot and immunoprecipitation analysis
Cell lysate preparation, SDS-PAGE, and Western blotting were carried out according to standard protocol. Proteins were harvested in cell lysis buffer supplemented with proteinase inhibitor cocktail (P8340; Sigma-Aldrich) and phosphatase inhibitor cocktail 2 (P5726; Sigma-Aldrich). Antigen detection was performed using antibodies directed against c-Src (rabbit anti-mouse/human antibody; #2109; Cell Signaling), Ctsk (mouse anti-mouse/human antibody; sc-48353; Santa Cruz), Rho (mouse anti-mouse/human antibody; #05-778; Millipore), galectin-3 (mouse anti-mouse/human antibody; ab2785; Abcam; epitopes mapped within the N-terminal region), Lrp1 (mouse anti-mouse antibody; MABN1796; Millipore), Mmp9 (rabbit anti-mouse antibody; ab38898; Abcam), Mmp14 (rabbit anti-mouse antibody; ab53712; Abcam), OXPHOS (rabbit anti-mouse antibody; ab110413; Abcam), vinculin (mouse anti-mouse antibody; V9131; Sigma-Aldrich), β3 integrin (rabbit anti-mouse antibody; #4702; Cell Signaling), or β-actin (rabbit anti-mouse antibody; #4970; Cell Signaling). Goat anti-rabbit IgG horseradish peroxidase (#65-6120; Thermofisher Scientific) or goat anti-mouse IgG horseradish peroxidase (#32430; Thermofisher Scientific) were used as secondary antibody. Bound primary antibodies (diluted to 1:1,000) were detected with horseradish peroxidase–conjugated species-specific secondary antibodies (Santa Cruz; diluted to 1:2,000) using the Super Signal Pico system (Thermo Fisher Scientific).
For immunoprecipitation analysis, cells were solubilized in IP Lysis Buffer (#87788; Thermo Fisher Scientific) supplemented with complete protease inhibitor cocktail (Roche). Immunoprecipitation was performed by incubation with a mouse monoclonal IgG (#5415; Cell Signaling) or anti–galectin-3 antibody (sc-32790; Santa Cruz) followed by the addition of Protein A/G Magnetic Beads (#88803; Thermo Fisher Scientific). Immune complexes were separated by electrophoresis followed by blotting with antibodies directed against Lrp1 (MABN1796; Millipore) and galectin-3 (ab2785; Abcam).
Immunofluorescence staining
For immunofluorescence analyses, osteoclasts were fixed with 4% paraformaldehyde, non-permeabilized and blocked for 1 h prior to an overnight incubation at 4°C with primary antibodies directed against galectin-3 (1:500; rat anti-mouse antibody; #125401; Biolegend; clone M3/38, epitopes mapped within the N-terminal region) or Cthrc1 (1:500; rabbit anti-mouse antibody; ab85739; Abcam). For F-actin staining, osteoclasts were permeabilized with 0.1% Triton X-100 and incubated with TRITC- or FITC-conjugated phalloidin (P1951 or P5282; Sigma-Aldrich). For Mitotracker Green staining, non-fixed cells were incubated with 100 nM Mitotracker Green (M7514; Invitrogen) in HBSS for 20 min. For bone tissue immunofluorescence, femurs from 8-wk-old Csf1r-Cre+/−/ROSA26R+/loxP mice were fixed in 4% paraformaldehyde at 4°C overnight, decalcified in 10% EDTA, and transferred to 30% sucrose in PBS for embedding in OCT (Fisher Healthcare). Frozen sections were permeabilized with 0.3–0.5% Triton X-100 in PBS and blocked for 2 h prior to adding primary antibodies directed against Mmp9 (1:500; rabbit anti-mouse antibody; ab38898; Abcam) or Mmp14 (1:200; rabbit anti-mouse antibody; ab53712; Abcam) with galectin-3 (1:500; rat anti-mouse antibody; #125401; Biolegend; clone M3/38, epitopes mapped within the N-terminal region). Following primary antibody incubations, sections were incubated with donkey anti-rat Alexa 488–, donkey anti-rabbit Alexa 488–, or donkey anti-rabbit Alexa 647–conjugated secondary antibodies (A-21208; A-21206; A-31573; Invitrogen Molecular Probes). Isotype control antibodies, including normal rabbit IgG (#31235; Invitrogen) and normal rat IgG (#31933; Invitrogen), were used as negative controls to confirm the specificity of immunofluorescence staining. Nuclei were visualized by staining with 1 μg/ml DAPI (D9542). Fluorescence images were acquired with a Nikon A1 laser-scanning confocal microscope using a 60× oil immersion objective (NA 1.4) at room temperature and analyzed with NIS Elements software and Image J software.
Cell surface biotinylation
Cell surface biotinylation was conducted as described (Maupin et al., 2022; Shiratori et al., 2018). In brief, cells were rinsed with PBS and surface proteins were biotinylated by incubating cells with 0.25 mg/ml EZ-Link Sulfo-NHS-SS-Biotin (A39258; Thermo Fisher Scientific) in PBS for 30 min with horizontal motion at 4°C. After labeling, plates were washed with quenching buffer (PBS containing 100 mM glycine) for 20 min at 4°C, then rinsed once with PBS. Cells were then lysed in IP Lysis Buffer and lysates were cleared by centrifugation. Cell lysates of equivalent amounts of protein were equilibrated overnight with Pierce Streptavidin Magnetic Beads (#88817; Thermo Fisher Scientific) at 4°C. After repeated washing steps, biotinylated proteins were released from the beads by heating to 95°C with 2× Laemmli buffer, separated by SDS-PAGE, and analyzed by Western blotting.
Flow cytometry analysis
Single-cell suspensions of osteoclasts were collected after passing through a 40 µm cell strainer (BD Bioscience), incubated with Fc receptor block (#101319; Biolegend) for 10 min, and then incubated with CoraLite 488–conjugated galectin-3 polyclonal antibody (rabbit anti-mouse antibody; CL488-14979; Proteintech; epitopes mapped throughout the full-length protein), eFluor 660–conjugated anti–galectin-3 monoclonal antibody (rabbit anti-mouse antibody; #50-5301-82; Thermo Fisher Scientific; clone M3/38, epitopes mapped within the N-terminal region), PE-conjugated anti–galectin-1 antibody (goat anti-mouse antibody; IC1245P; R&D), or the corresponding rabbit and goat isotype control (#31235; #31245; Invitrogen) in flow cytometry staining buffer (eBioscience) for 30 min at 4°C. For Mitotracker Green staining, cells were incubated with 100 nM Mitotracker Green (M7514; Invitrogen) in HBSS for 45 min. Then, cells were subjected to flow cytometry analysis on a FACS Canto II (BD Bioscience). Data analysis was carried out using FlowJo software.
Proteolytic cleavage of GALECTIN-3
Recombinant human GALECTIN-3 protein (A13506; Abclonal) was incubated with activated recombinant MMP9 (RP00103; Abclonal) by adding 5 mM p-aminophenylmercuric acetate (A9563; Sigma-Aldrich) at 37°C for 30 min in 50 mM Tris-HCl buffer containing 150 mM NaCl and 10 mM CaCl2 (pH 7.5). The incubation was stopped by adding with a volume of 4× Laemmli sample buffer, and the sample was separated on a 12% SDS-PAGE and analyzed by Western blot analysis using either anti–galectin-3 polyclonal antibody (rabbit anti-mouse antibody; 14979-1-AP; Proteintech) or anti–galectin-3 monoclonal antibody (mouse anti-mouse antibody; ab2785; Abcam).
Mass spectroscopy–based proteomics
Conjugation of recombinant human GALECTIN-3 protein (hGAL3; #774408; Biolegend) to EZ-Link Sulfo-NHS-LC-LC-Biotin (A35358; Thermo Fisher Scientific) was performed (Hönig et al., 2018). 1.5 µM hGAL3-biotin was added to osteoclast culture for 30 min at 4°C. Cells were then lysed with IP Lysis Buffer (Thermo Fisher Scientific) and biotinylated hGAL3, and associated proteins were isolated with Pierce Streptavidin Magnetic Beads (Thermo Fisher Scientific) and separated by SDS-PAGE. Excised gel slices were washed and reduced with tris(2-carboxyethyl)phosphine and alkylated in the dark with iodoacetamide prior to treatment with trypsin (Promega). The resulting peptides were analyzed by capillary high-performance liquid chromatography (HPLC)–electrospray ionization tandem mass spectrometry on a Thermo Fisher Scientific LTQ Orbitrap Velos Pro mass spectrometer. On-line HPLC separation was accomplished with an RSLC NANO HPLC system (Thermofisher Scientific/Dionex): column, PicoFrit (New Objective; 75 μm i.d.) packed to 15 cm with C18 adsorbent (Vydac; 218MS 5 μm, 300 Å). Precursor ions were acquired in the Orbitrap in centroid mode at 60,000 resolution (m/z 400); data-dependent collision-induced dissociation spectra of the six most intense ions in each precursor scan were acquired at the same time in the linear trap (30% normalized collision energy). Mascot (v2.7.0; Matrix Science) was used to search the spectra against a combination of the mouse and human subsets of the UniProt database plus a database of common contaminants (UniProt_Mouse 20161004 [57,951 sequences; 26,573,730 residues]; UniProt Human 20181204 [95,936 sequences; 38,067,061 residues]; contaminants 20120713 [247 sequences; 128,130 residues]). Cysteine carbamidomethylation was set as a fixed modification and methionine oxidation and deamidation of glutamine and asparagine were considered as variable modifications; trypsin was specified as the proteolytic enzyme, with one missed cleavage allowed. The 13C setting was 1. Subset search of the identified proteins by X! Tandem, cross-correlation with the Mascot results, and determination of protein and peptide identity probabilities were assessed by Scaffold (v4.9.0; Proteome Software). The thresholds for acceptance of peptide and protein assignments in Scaffold were 95 and 99%, respectively, resulting in ≤1% protein false discovery rate. Proteins identified in the control were subtracted from the proteins identified in the hGal3-associated proteins and a shortened list interrogated for potential galectin-3 receptor.
Plate-based cell-free protein binding assay
96-well plates were coated with recombinant human IgG1.Fc (A42561; Invitrogen) or human LRP-1 Cluster II Fc Chimera Protein covering ∼10% of the full-length Lpr1 protein sequence, including 3 N-glycosylation sites (#2368-L2-050; R&D Systems) at 0.5 μg in 100 μl PBS, incubated at 4°C overnight, and blocked with 50 mg/ml BSA for 90 min at 30°C. Serial concentrations ranging from 0.1 to 3.2 μg of recombinant human GALECTIN-3 (#774408; Biolegend), and test agents were added in a total volume of 50 μl and then incubated for 4 h at 30°C. Wells were washed, fixed with 2% PFA in PBS for 15 min at room temperature, washed, and incubated with rat anti-mouse monoclonal galectin-3 antibody (#125401; Biolegend; clone M3/38, epitopes mapped within the N-terminal region) for 30 min on ice. After washing, wells were incubated with AF488-conjugated donkey anti-rat secondary antibody (A-21208; Invitrogen Molecular Probes) for 30 min on ice. After washing, fluorescence was determined using a SpectraMax L (Molecular Devices) plate reader (excitation 485, emission 538) to quantify galectin-3–Lrp1 binding.
Human osteoclast culture
Normal human CD14+ monocytes (Lonza) were cultured in α-MEM supplemented with 10% FBS, 2 mM L-glutamine, 100 U/ml penicillin-streptomycin, and 20 ng/ml M-CSF for 6 d to expand the precursor population and differentiate into macrophages. Cells were then detached with Accutase (A1110501; Gibco), seeded on tissue-culture plastic dishes, and cultured with M-CSF (20 ng/ml) and RANKL (20 ng/ml) for 9 d to induce multinuclear osteoclast formation, or reseeded on bone slices for bone resorption assay (Raynaud-Messina et al., 2018; Zhu et al., 2020). Human osteoclasts were cultured with isotype control IgG, MMP9 function-blocking monoclonal antibody (sc-12759L; Santa Cruz), and a function-blocking antibody anti-MMP14 (DX-2400) at 100 μg/ml (Ager et al., 2015; Marshall et al., 2015; Zhu et al., 2020), in the presence or absence of an anti–GALECTIN-3 blocking antibody (sc-32790L; Santa Cruz) or an anti-LRP1 blocking antibody (MA1-27198; Thermo Fisher Scientific) at 25 μg/ml (Chen et al., 2015; Moxon et al., 2015; Seguin et al., 2017). DX-2400 was provided by the Kadmon Corporation.
Ex vivo calvarial organ cultures
Calvariae from 5-d-old wild-type or DKO mice were isolated aseptically, cleaned, and cultured for 16 h at 37°C in 0.5 ml of BGJb medium (Life Technologies) containing 1 mg/ml BSA (fraction V; Sigma-Aldrich; Moxon et al., 2015). Half calvariae were transferred to fresh medium with 0.1 μM parathyroid hormone (H-4835.0005; Bachem) in the presence or absence of an anti–galectin-3 monoclonal antibody (sc-32790L; Santa Cruz) or an anti-Lrp1 monoclonal antibody (MA1-27198; Thermo Fisher Scientific), and cultured for an additional 5 d. Culture supernatant was collected for bone resorption marker CTX-I level detection using RatLaps CTX-I EIA kit (AC-06F1; Immunodiagnostic Systems). The half calvariae were either fixed for immunostaining with an anti-TRAP polyclonal antibody (sc-30833; Santa Cruz), an anti–galectin-3 monoclonal antibody (#125401; Biolegend; clone M3/38), or an anti-V-type proton pump-3 (Vpp3) polyclonal antibody (ab200839; Abcam) as describe above or snap-frozen for TRAP activity assay.
Calvarial tissue lysate preparation and TRAP activity assay
The snap-frozen calvariae were extracted and TRAP activity was detected (Cackowski et al., 2010). After homogenization using a ground glass homogenizer in 200 μl of NP-40 lysis buffer (1% NP-40, 150 mM NaCl, 50 mM Tris, pH 8), each sample was put through five freeze–thaw cycles, kept on ice for 1 h followed by centrifugation to remove calvarial bone and debris, and bone tissue extracts stored at −70°C until required. For TRAP activity detection, 35 μl of bone tissue lysates was added to 200 μl of TRAP substrate (50 mM sodium acetate, pH 5, 25 mM sodium tartrate, 0.4 mM MnCl2, 0.4% N,N,-dimethylformamide, 0.2 mg/ml fast red violet, 0.5 mg/ml naphthol AS-MX phosphate), then incubated 3 h at 37°C, and the absorbance read at 540 nm. Values were normalized for the protein concentration of the calvarial tissue lysates.
Statistical analysis
Statistical analyses were carried out using GraphPad Prism 7. All values were expressed as mean ± SEM. Unpaired two-sided Student’s t test was used to analyze the differences between two groups, while one- or two-way ANOVA with Bonferroni correction was used to evaluate differences among multiple comparisons. Data distribution was assumed to be normal, but this was not formally tested. A P value of <0.05 was considered statistically significant. All representative experiments shown were repeated three or more times.
Online supplemental material
Fig. S1 displays mitochondrial abundance and OXPHOS expression in Mmp9/Mmp14 DKO osteoclasts. Fig. S2 shows the identification and quantitative analysis of the metabolome in Mmp9/Mmp14 DKO osteoclasts. Fig. S3 depicts that active RhoA rescues defects in sealing zone formation and bone resorption in DKO osteoclasts. Fig. S4 shows that galectin-3 surface binding antagonist reverses functional defects in DKO osteoclasts. Fig. S5 depicts a galectin-3–Lrp1 axis regulates RhoA activation and sealing zone formation in osteoclasts. Table S1 is the list of the genotyping PCR primers. Table S2 lists the quantitative real-time PCR primers. Table S3 shows the list of galectin-3 interacting partners through mass spectrometry.
Supplementary Material
lists genotyping PCR primers.
lists quantitative real-time PCR primers.
shows listing of galectin-3 interacting partners.
is the source file for Fig. 2.
is the source file for Fig. 4.
is the source file for Fig. 5.
is the source file for Fig. 7.
is the source file for Fig. 8.
is the source file for Fig. 9.
is the source file for Fig. S1.
is the source file for Fig. S3.
is the source file for Fig. S4.
is the source file for Fig. S5.
Acknowledgments
We thank H.A. Remmer (University of Michigan Proteomics and Peptide Synthesis Core) for analysis of proteomics data, and S. Pardo and D. Molleur (University of Texas Health Science Center at San Antonio Institutional Mass Spectrometry Laboratory) for expert technical assistance of mass spectrometry.
This study was supported by National Institutes of Health grants (R01-105068, R01-CA071699 and R01-AR075168), the Breast Cancer Research Foundation, and the Margolies Family Discovery Fund for Cancer Research (S.J. Weiss), the National Natural Science Foundation of China 81970919, and the Fundamental Research Funds for the Central Universities 2042022kf1207 (L. Zhu).
Author contributions: L. Zhu and S.J. Weiss designed and supervised the project, analyzed data, wrote the manuscript, and approved the final version. L. Zhu, Y. Tang, X.-Y. Li, S.A. Kerk, C.A. Lyssiotis, X. Sun, Z. Wang, J.-S. Cho, and J. Ma performed experiments, analyzed data, and provided relevant advice.
Data availability
The RNA-seq data have been deposited to the Gene Expression Omnibus database at accession number GSE220835 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE220835). The metabolomics data have been deposited to Mendeley (https://doi.org/10.17632/8h7vsykvpd.1). The mass spectrometry–based proteomics data have been deposited to Mendeley (https://doi.org/10.17632/pdhxwr8jc8.2).
References
- Ager, E.I., Kozin S.V., Kirkpatrick N.D., Seano G., Kodack D.P., Askoxylakis V., Huang Y., Goel S., Snuderl M., Muzikansky A., et al. 2015. Blockade of MMP14 activity in murine breast carcinomas: Implications for macrophages, vessels, and radiotherapy. J. Natl. Cancer Inst. 107:djv017. 10.1093/jnci/djv017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alabi, A., Xia X.D., Gu H.M., Wang F., Deng S.J., Yang N., Adijiang A., Douglas D.N., Kneteman N.M., Xue Y., et al. 2021. Membrane type 1 matrix metalloproteinase promotes LDL receptor shedding and accelerates the development of atherosclerosis. Nat. Commun. 12:1889. 10.1038/s41467-021-22167-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonso-Herranz, L., Sahún-Español Á., Paredes A., Gonzalo P., Gkontra P., Núñez V., Clemente C., Cedenilla M., Villalba-Orero M., Inserte J., et al. 2020. Macrophages promote endothelial-to-mesenchymal transition via MT1-MMP/TGFβ1 after myocardial infarction. Elife. 9:e57920. 10.7554/eLife.57920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Appert-Collin, A., Bennasroune A., Jeannesson P., Terryn C., Fuhrmann G., Morjani H., and Dedieu S.. 2017. Role of LRP-1 in cancer cell migration in 3-dimensional collagen matrix. Cell Adhes. Migr. 11:316–326. 10.1080/19336918.2016.1215788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnett, T.R., and Orriss I.R.. 2018. Metabolic properties of the osteoclast. Bone. 115:25–30. 10.1016/j.bone.2017.12.021 [DOI] [PubMed] [Google Scholar]
- Bae, S., Lee M.J., Mun S.H., Giannopoulou E.G., Yong-Gonzalez V., Cross J.R., Murata K., Giguère V., van der Meulen M., and Park-Min K.H.. 2017. MYC-dependent oxidative metabolism regulates osteoclastogenesis via nuclear receptor ERRα. J. Clin. Invest. 127:2555–2568. 10.1172/JCI89935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bian, Y., Li W., Kremer D.M., Sajjakulnukit P., Li S., Crespo J., Nwosu Z.C., Zhang L., Czerwonka A., Pawłowska A., et al. 2020. Cancer SLC43A2 alters T cell methionine metabolism and histone methylation. Nature. 585:277–282. 10.1038/s41586-020-2682-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blangy, A., Bompard G., Guerit D., Marie P., Maurin J., Morel A., and Vives V.. 2020. The osteoclast cytoskeleton: Current understanding and therapeutic perspectives for osteoporosis. J. Cell Sci. 133:jcs244798. 10.1242/jcs.244798 [DOI] [PubMed] [Google Scholar]
- Bonnans, C., Chou J., and Werb Z.. 2014. Remodelling the extracellular matrix in development and disease. Nat. Rev. Mol. Cell Biol. 15:786–801. 10.1038/nrm3904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boshans, R.L., Szanto S., van Aelst L., and D’Souza-Schorey C.. 2000. ADP-ribosylation factor 6 regulates actin cytoskeleton remodeling in coordination with Rac1 and RhoA. Mol. Cell. Biol. 20:3685–3694. 10.1128/MCB.20.10.3685-3694.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boulter, E., Estrach S., Errante A., Pons C., Cailleteau L., Tissot F., Meneguzzi G., and Féral C.C.. 2013. CD98hc (SLC3A2) regulation of skin homeostasis wanes with age. J. Exp. Med. 210:173–190. 10.1084/jem.20121651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brömme, D., Panwar P., and Turan S.. 2016. Cathepsin K osteoporosis trials, pycnodysostosis and mouse deficiency models: Commonalities and differences. Expert Opin. Drug Discov. 11:457–472. 10.1517/17460441.2016.1160884 [DOI] [PubMed] [Google Scholar]
- Butler, G.S., Dean R.A., Tam E.M., and Overall C.M.. 2008. Pharmacoproteomics of a metalloproteinase hydroxamate inhibitor in breast cancer cells: Dynamics of membrane type 1 matrix metalloproteinase-mediated membrane protein shedding. Mol. Cell. Biol. 28:4896–4914. 10.1128/MCB.01775-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cackowski, F.C., Anderson J.L., Patrene K.D., Choksi R.J., Shapiro S.D., Windle J.J., Blair H.C., and Roodman G.D.. 2010. Osteoclasts are important for bone angiogenesis. Blood. 115:140–149. 10.1182/blood-2009-08-237628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chellaiah, M.A., Soga N., Swanson S., McAllister S., Alvarez U., Wang D., Dowdy S.F., and Hruska K.A.. 2000. Rho-A is critical for osteoclast podosome organization, motility, and bone resorption. J. Biol. Chem. 275:11993–12002. 10.1074/jbc.275.16.11993 [DOI] [PubMed] [Google Scholar]
- Chen, S.S., Sun L.W., Brickner H., and Sun P.Q.. 2015. Downregulating galectin-3 inhibits proinflammatory cytokine production by human monocyte-derived dendritic cells via RNA interference. Cell. Immunol. 294:44–53. 10.1016/j.cellimm.2015.01.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu, Y.P., Sun Y.C., Qiu D.C., Lin Y.H., Chen Y.Q., Kuo J.C., and Huang J.R.. 2020. Liquid-liquid phase separation and extracellular multivalent interactions in the tale of galectin-3. Nat. Commun. 11:1229. 10.1038/s41467-020-15007-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colnot, C., Sidhu S.S., Poirier F., and Balmain N.. 1999. Cellular and subcellular distribution of galectin-3 in the epiphyseal cartilage and bone of fetal and neonatal mice. Cell. Mol. Biol. 45:1191–1202 [PubMed] [Google Scholar]
- Compston, J.E., McClung M.R., and Leslie W.D.. 2019. Osteoporosis. Lancet. 393:364–376. 10.1016/S0140-6736(18)32112-3 [DOI] [PubMed] [Google Scholar]
- Dallas, S.L., Xie Y., Shiflett L.A., and Ueki Y.. 2018. Mouse Cre models for the study of bone diseases. Curr. Osteoporos. Rep. 16:466–477. 10.1007/s11914-018-0455-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Castro Brás, L.E., Cates C.A., DeLeon-Pennell K.Y., Ma Y., Iyer R.P., Halade G.V., Yabluchanskiy A., Fields G.B., Weintraub S.T., and Lindsey M.L.. 2014. Citrate synthase is a novel in vivo matrix metalloproteinase-9 substrate that regulates mitochondrial function in the postmyocardial infarction left ventricle. Antioxid. Redox Signal. 21:1974–1985. 10.1089/ars.2013.5411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demotte, N., Wieërs G., Van Der Smissen P., Moser M., Schmidt C., Thielemans K., Squifflet J.L., Weynand B., Carrasco J., Lurquin C., et al. 2010. A galectin-3 ligand corrects the impaired function of human CD4 and CD8 tumor-infiltrating lymphocytes and favors tumor rejection in mice. Cancer Res. 70:7476–7488. 10.1158/0008-5472.CAN-10-0761 [DOI] [PubMed] [Google Scholar]
- Dobin, A., Davis C.A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., and Gingeras T.R.. 2013. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics. 29:15–21. 10.1093/bioinformatics/bts635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elola, M.T., Blidner A.G., Ferragut F., Bracalente C., and Rabinovich G.A.. 2015. Assembly, organization and regulation of cell-surface receptors by lectin-glycan complexes. Biochem. J. 469:1–16. 10.1042/BJ20150461 [DOI] [PubMed] [Google Scholar]
- Farhadi, S.A., Liu R., Becker M.W., Phelps E.A., and Hudalla G.A.. 2021. Physical tuning of galectin-3 signaling. Proc. Natl. Acad. Sci. USA. 118:e2024117118. 10.1073/pnas.2024117118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao, Q., Xia Y., Liu L., Huang L., Liu Y., Zhang X., Xu K., Wei J., Hu Y., Mu Y., and Li K.. 2016. Galectin-3 enhances migration of minature pig bone marrow mesenchymal stem cells through inhibition of RhoA-GTP activity. Sci. Rep. 6:26577. 10.1038/srep26577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ginhoux, F., and Jung S.. 2014. Monocytes and macrophages: Developmental pathways and tissue homeostasis. Nat. Rev. Immunol. 14:392–404. 10.1038/nri3671 [DOI] [PubMed] [Google Scholar]
- Herz, J., and Strickland D.K.. 2001. LRP: A multifunctional scavenger and signaling receptor. J. Clin. Invest. 108:779–784. 10.1172/JCI200113992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hönig, E., Ringer K., Dewes J., von Mach T., Kamm N., Kreitzer G., and Jacob R.. 2018. Galectin-3 modulates the polarized surface delivery of β1-integrin in epithelial cells. J. Cell Sci. 131:jcs213199. 10.1242/jcs.213199 [DOI] [PubMed] [Google Scholar]
- Hui, L., Nie Y., Li S., Guo M., Yang W., Huang R., Chen J., Liu Y., Lu X., Chen Z., et al. 2020. Matrix metalloproteinase 9 facilitates Zika virus invasion of the testis by modulating the integrity of the blood-testis barrier. PLoS Pathog. 16:e1008509. 10.1371/journal.ppat.1008509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iacobini, C., Blasetti Fantauzzi C., Bedini R., Pecci R., Bartolazzi A., Amadio B., Pesce C., Pugliese G., and Menini S.. 2018. Galectin-3 is essential for proper bone cell differentiation and activity, bone remodeling and biomechanical competence in mice. Metabolism. 83:149–158. 10.1016/j.metabol.2018.02.001 [DOI] [PubMed] [Google Scholar]
- Inada, M., Wang Y., Byrne M.H., Rahman M.U., Miyaura C., López-Otín C., and Krane S.M.. 2004. Critical roles for collagenase-3 (Mmp13) in development of growth plate cartilage and in endochondral ossification. Proc. Natl. Acad. Sci. USA. 101:17192–17197. 10.1073/pnas.0407788101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacome-Galarza, C.E., Percin G.I., Muller J.T., Mass E., Lazarov T., Eitler J., Rauner M., Yadav V.K., Crozet L., Bohm M., et al. 2019. Developmental origin, functional maintenance and genetic rescue of osteoclasts. Nature. 568:541–545. 10.1038/s41586-019-1105-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao, X., Sherman B.T., Huang da W., Stephens R., Baseler M.W., Lane H.C., and Lempicki R.A.. 2012. DAVID-WS: A stateful web service to facilitate gene/protein list analysis. Bioinformatics. 28:1805–1806. 10.1093/bioinformatics/bts251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin, G., Zhang F., Chan K.M., Xavier Wong H.L., Liu B., Cheah K.S., Liu X., Mauch C., Liu D., and Zhou Z.. 2011. MT1-MMP cleaves Dll1 to negatively regulate Notch signalling to maintain normal B-cell development. EMBO J. 30:2281–2293. 10.1038/emboj.2011.136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joeh, E., O’Leary T., Li W., Hawkins R., Hung J.R., Parker C.G., and Huang M.L.. 2020. Mapping glycan-mediated galectin-3 interactions by live cell proximity labeling. Proc. Natl. Acad. Sci. USA. 117:27329–27338. 10.1073/pnas.2009206117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- John, C.M., Leffler H., Kahl-Knutsson B., Svensson I., and Jarvis G.A.. 2003. Truncated galectin-3 inhibits tumor growth and metastasis in orthotopic nude mouse model of human breast cancer. Clin. Cancer Res. 9:2374–2383 [PubMed] [Google Scholar]
- Kelly, A., Gunaltay S., McEntee C.P., Shuttleworth E.E., Smedley C., Houston S.A., Fenton T.M., Levison S., Mann E.R., and Travis M.A.. 2018. Human monocytes and macrophages regulate immune tolerance via integrin αvβ8-mediated TGFβ activation. J. Exp. Med. 215:2725–2736. 10.1084/jem.20171491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kowluru, R.A., Mohammad G., dos Santos J.M., and Zhong Q.. 2011. Abrogation of MMP-9 gene protects against the development of retinopathy in diabetic mice by preventing mitochondrial damage. Diabetes. 60:3023–3033. 10.2337/db11-0816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau, K.S., Partridge E.A., Grigorian A., Silvescu C.I., Reinhold V.N., Demetriou M., and Dennis J.W.. 2007. Complex N-glycan number and degree of branching cooperate to regulate cell proliferation and differentiation. Cell. 129:123–134. 10.1016/j.cell.2007.01.049 [DOI] [PubMed] [Google Scholar]
- Lee, D., Walsh J.D., Migliorini M., Yu P., Cai T., Schwieters C.D., Krueger S., Strickland D.K., and Wang Y.X.. 2007. The structure of receptor-associated protein (RAP). Protein Sci. 16:1628–1640. 10.1110/ps.072865407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehti, K., Rose N.F., Valavaara S., Weiss S.J., and Keski-Oja J.. 2009. MT1-MMP promotes vascular smooth muscle dedifferentiation through LRP1 processing. J. Cell Sci. 122:126–135. 10.1242/jcs.035279 [DOI] [PubMed] [Google Scholar]
- Liao, Y., Smyth G.K., and Shi W.. 2014. featureCounts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 30:923–930. 10.1093/bioinformatics/btt656 [DOI] [PubMed] [Google Scholar]
- Liu, F.T., Hsu D.K., Zuberi R.I., Hill P.N., Shenhav A., Kuwabara I., and Chen S.S.. 1996. Modulation of functional properties of galectin-3 by monoclonal antibodies binding to the non-lectin domains. Biochemistry. 35:6073–6079. 10.1021/bi952716q [DOI] [PubMed] [Google Scholar]
- Love, M.I., Huber W., and Anders S.. 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15:550. 10.1186/s13059-014-0550-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu, D., Li J., Liu H., Foxa G.E., Weaver K., Li J., Williams B.O., and Yang T.. 2018. LRP1 suppresses bone resorption in mice by inhibiting the RANKL-stimulated NF-κB and p38 pathways during osteoclastogenesis. J. Bone Miner. Res. 33:1773–1784. 10.1002/jbmr.3469 [DOI] [PubMed] [Google Scholar]
- Mantuano, E., Brifault C., Lam M.S., Azmoon P., Gilder A.S., and Gonias S.L.. 2016. LDL receptor-related protein-1 regulates NFκB and microRNA-155 in macrophages to control the inflammatory response. Proc. Natl. Acad. Sci. USA. 113:1369–1374. 10.1073/pnas.1515480113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mantuano, E., Jo M., Gonias S.L., and Campana W.M.. 2010. Low density lipoprotein receptor-related protein (LRP1) regulates Rac1 and RhoA reciprocally to control Schwann cell adhesion and migration. J. Biol. Chem. 285:14259–14266. 10.1074/jbc.M109.085126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall, D.C., Lyman S.K., McCauley S., Kovalenko M., Spangler R., Liu C., Lee M., O’Sullivan C., Barry-Hamilton V., Ghermazien H., et al. 2015. Selective allosteric inhibition of MMP9 is efficacious in preclinical models of ulcerative colitis and colorectal cancer. PLoS One. 10:e0127063. 10.1371/journal.pone.0127063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massa, S.M., Cooper D.N., Leffler H., and Barondes S.H.. 1993. L-29, an endogenous lectin, binds to glycoconjugate ligands with positive cooperativity. Biochemistry. 32:260–267. 10.1021/bi00052a033 [DOI] [PubMed] [Google Scholar]
- Matsumoto, S., Fujii S., Sato A., Ibuka S., Kagawa Y., Ishii M., and Kikuchi A.. 2014. A combination of Wnt and growth factor signaling induces Arl4c expression to form epithelial tubular structures. EMBO J. 33:702–718. 10.1002/embj.201386942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maupin, K.A., Diegel C.R., Stevens P.D., Dick D., Williams B.O., Transgenic C., Williams B.O., and VAI Vivarium and Transgenic Core . 2022. Mutation of the galectin-3 glycan-binding domain (Lgals3-R200S) enhances cortical bone expansion in male mice and trabecular bone mass in female mice. FEBS Open Bio. 12:1717–1728. 10.1002/2211-5463.13483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClung, H.M., Thomas S.L., Osenkowski P., Toth M., Menon P., Raz A., Fridman R., and Rempel S.A.. 2007. SPARC upregulates MT1-MMP expression, MMP-2 activation, and the secretion and cleavage of galectin-3 in U87MG glioma cells. Neurosci. Lett. 419:172–177. 10.1016/j.neulet.2007.04.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng, Z., Qiu Y., Lin K.C., Kumar A., Placone J.K., Fang C., Wang K.C., Lu S., Pan M., Hong A.W., et al. 2018. RAP2 mediates mechanoresponses of the Hippo pathway. Nature. 560:655–660. 10.1038/s41586-018-0444-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miloudi, K., Binet F., Wilson A., Cerani A., Oubaha M., Menard C., Henriques S., Mawambo G., Dejda A., Nguyen P.T., et al. 2016. Truncated netrin-1 contributes to pathological vascular permeability in diabetic retinopathy. J. Clin. Invest. 126:3006–3022. 10.1172/JCI84767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori, H., Bhat R., Bruni-Cardoso A., Chen E.I., Jorgens D.M., Coutinho K., Louie K., Bowen B.B., Inman J.L., Tecca V., et al. 2016. New insight into the role of MMP14 in metabolic balance. PeerJ. 4:e2142. 10.7717/peerj.2142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moxon, J.V., Behl-Gilhotra R., Morton S.K., Krishna S.M., Seto S.W., Biros E., Nataatmadja M., West M., Walker P.J., Norman P.E., and Golledge J.. 2015. Plasma low-density lipoprotein receptor-related protein 1 concentration is not associated with human abdominal aortic aneurysm presence. Eur. J. Vasc. Endovasc. Surg. 50:466–473. 10.1016/j.ejvs.2015.06.023 [DOI] [PubMed] [Google Scholar]
- Muller, J., Duray E., Lejeune M., Dubois S., Plougonven E., Léonard A., Storti P., Giuliani N., Cohen-Solal M., Hempel U., et al. 2019. Loss of stromal galectin-1 enhances multiple myeloma development: Emphasis on a role in osteoclasts. Cancers. 11:261. 10.3390/cancers11020261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nabi, I.R., Shankar J., and Dennis J.W.. 2015. The galectin lattice at a glance. J. Cell Sci. 128:2213–2219. 10.1242/jcs.151159 [DOI] [PubMed] [Google Scholar]
- Nakahira, K., Haspel J.A., Rathinam V.A., Lee S.J., Dolinay T., Lam H.C., Englert J.A., Rabinovitch M., Cernadas M., Kim H.P., et al. 2011. Autophagy proteins regulate innate immune responses by inhibiting the release of mitochondrial DNA mediated by the NALP3 inflammasome. Nat. Immunol. 12:222–230. 10.1038/ni.1980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neisch, A.L., Formstecher E., and Fehon R.G.. 2013. Conundrum, an ARHGAP18 orthologue, regulates RhoA and proliferation through interactions with Moesin. Mol. Biol. Cell. 24:1420–1433. 10.1091/mbc.e12-11-0800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen, M.I., Stegmayr J., Grant O.C., Yang Z., Nilsson U.J., Boos I., Carlsson M.C., Woods R.J., Unverzagt C., Leffler H., and Wandall H.H.. 2018. Galectin binding to cells and glycoproteins with genetically modified glycosylation reveals galectin-glycan specificities in a natural context. J. Biol. Chem. 293:20249–20262. 10.1074/jbc.RA118.004636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nieminen, J., Kuno A., Hirabayashi J., and Sato S.. 2007. Visualization of galectin-3 oligomerization on the surface of neutrophils and endothelial cells using fluorescence resonance energy transfer. J. Biol. Chem. 282:1374–1383. 10.1074/jbc.M604506200 [DOI] [PubMed] [Google Scholar]
- Nishida, C., Kusubata K., Tashiro Y., Gritli I., Sato A., Ohki-Koizumi M., Morita Y., Nagano M., Sakamoto T., Koshikawa N., et al. 2012. MT1-MMP plays a critical role in hematopoiesis by regulating HIF-mediated chemokine/cytokine gene transcription within niche cells. Blood. 119:5405–5416. 10.1182/blood-2011-11-390849 [DOI] [PubMed] [Google Scholar]
- Nishikawa, H., and Suzuki H.. 2019. Response by Nishikawa and Suzuki to letter regarding article, “modified citrus pectin prevents blood-brain barrier disruption in mouse subarachnoid hemorrhage by inhibiting galectin-3”. Stroke. 50:e137. 10.1161/STROKEAHA.119.025133 [DOI] [PubMed] [Google Scholar]
- Nishikawa, K., Iwamoto Y., Kobayashi Y., Katsuoka F., Kawaguchi S., Tsujita T., Nakamura T., Kato S., Yamamoto M., Takayanagi H., and Ishii M.. 2015. DNA methyltransferase 3a regulates osteoclast differentiation by coupling to an S-adenosylmethionine-producing metabolic pathway. Nat. Med. 21:281–287. 10.1038/nm.3774 [DOI] [PubMed] [Google Scholar]
- Obermann, J., Priglinger C.S., Merl-Pham J., Geerlof A., Priglinger S., Götz M., and Hauck S.M.. 2017. Proteome-wide identification of glycosylation-dependent interactors of galectin-1 and galectin-3 on mesenchymal retinal pigment epithelial (RPE) cells. Mol. Cell. Proteomics. 16:1528–1546. 10.1074/mcp.M116.066381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ochieng, J., Fridman R., Nangia-Makker P., Kleiner D.E., Liotta L.A., Stetler-Stevenson W.G., and Raz A.. 1994. Galectin-3 is a novel substrate for human matrix metalloproteinases-2 and -9. Biochemistry. 33:14109–14114. 10.1021/bi00251a020 [DOI] [PubMed] [Google Scholar]
- Ochieng, J., Green B., Evans S., James O., and Warfield P.. 1998. Modulation of the biological functions of galectin-3 by matrix metalloproteinases. Biochim. Biophys. Acta. 1379:97–106. 10.1016/S0304-4165(97)00086-X [DOI] [PubMed] [Google Scholar]
- Ohuchi, E., Imai K., Fujii Y., Sato H., Seiki M., and Okada Y.. 1997. Membrane type 1 matrix metalloproteinase digests interstitial collagens and other extracellular matrix macromolecules. J. Biol. Chem. 272:2446–2451. 10.1074/jbc.272.4.2446 [DOI] [PubMed] [Google Scholar]
- Okada, Y., Naka K., Kawamura K., Matsumoto T., Nakanishi I., Fujimoto N., Sato H., and Seiki M.. 1995. Localization of matrix metalloproteinase 9 (92-kilodalton gelatinase/type IV collagenase = gelatinase B) in osteoclasts: Implications for bone resorption. Lab. Invest. 72:311–322 [PubMed] [Google Scholar]
- Orgaz, J.L., Pandya P., Dalmeida R., Karagiannis P., Sanchez-Laorden B., Viros A., Albrengues J., Nestle F.O., Ridley A.J., Gaggioli C., et al. 2014. Diverse matrix metalloproteinase functions regulate cancer amoeboid migration. Nat. Commun. 5:4255. 10.1038/ncomms5255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parikka, V., Lehenkari P., Sassi M.L., Halleen J., Risteli J., Härkönen P., and Väänänen H.K.. 2001. Estrogen reduces the depth of resorption pits by disturbing the organic bone matrix degradation activity of mature osteoclasts. Endocrinology. 142:5371–5378. 10.1210/endo.142.12.8533 [DOI] [PubMed] [Google Scholar]
- Pedrosa, L.F., Raz A., and Fabi J.P.. 2022. The complex biological effects of pectin: Galectin-3 targeting as potential human health improvement? Biomolecules. 12:289. 10.3390/biom12020289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Post, A., Pannekoek W.J., Ponsioen B., Vliem M.J., and Bos J.L.. 2015. Rap1 spatially controls ArhGAP29 to inhibit Rho signaling during endothelial barrier regulation. Mol. Cell. Biol. 35:2495–2502. 10.1128/MCB.01453-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prudova, A., auf dem Keller U., Butler G.S., and Overall C.M.. 2010. Multiplex N-terminome analysis of MMP-2 and MMP-9 substrate degradomes by iTRAQ-TAILS quantitative proteomics. Mol. Cell. Proteomics. 9:894–911. 10.1074/mcp.M000050-MCP201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raynaud-Messina, B., Bracq L., Dupont M., Souriant S., Usmani S.M., Proag A., Pingris K., Soldan V., Thibault C., Capilla F., et al. 2018. Bone degradation machinery of osteoclasts: An HIV-1 target that contributes to bone loss. Proc. Natl. Acad. Sci. USA. 115:E2556–E2565. 10.1073/pnas.1713370115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowe, R.G., and Weiss S.J.. 2009. Navigating ECM barriers at the invasive front: The cancer cell-stroma interface. Annu. Rev. Cell Dev. Biol. 25:567–595. 10.1146/annurev.cellbio.24.110707.175315 [DOI] [PubMed] [Google Scholar]
- Sakamoto, T., and Seiki M.. 2010. A membrane protease regulates energy production in macrophages by activating hypoxia-inducible factor-1 via a non-proteolytic mechanism. J. Biol. Chem. 285:29951–29964. 10.1074/jbc.M110.132704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salzer, E., Cagdas D., Hons M., Mace E.M., Garncarz W., Petronczki O.Y., Platzer R., Pfajfer L., Bilic I., Ban S.A., et al. 2016. RASGRP1 deficiency causes immunodeficiency with impaired cytoskeletal dynamics. Nat. Immunol. 17:1352–1360. 10.1038/ni.3575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seguin, L., Camargo M.F., Wettersten H.I., Kato S., Desgrosellier J.S., von Schalscha T., Elliott K.C., Cosset E., Lesperance J., Weis S.M., and Cheresh D.A.. 2017. Galectin-3, a druggable vulnerability for KRAS-addicted cancers. Cancer Discov. 7:1464–1479. 10.1158/2159-8290.CD-17-0539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu-Hirota, R., Xiong W., Baxter B.T., Kunkel S.L., Maillard I., Chen X.W., Sabeh F., Liu R., Li X.Y., and Weiss S.J.. 2012. MT1-MMP regulates the PI3Kδ·Mi-2/NuRD-dependent control of macrophage immune function. Genes Dev. 26:395–413. 10.1101/gad.178749.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiratori, T., Kyumoto-Nakamura Y., Kukita A., Uehara N., Zhang J., Koda K., Kamiya M., Badawy T., Tomoda E., Xu X., et al. 2018. IL-1β induces pathologically activated osteoclasts bearing extremely high levels of resorbing activity: A possible pathological subpopulation of osteoclasts, accompanied by suppressed expression of Kindlin-3 and Talin-1. J. Immunol. 200:218–228. 10.4049/jimmunol.1602035 [DOI] [PubMed] [Google Scholar]
- Simon, D., Derer A., Andes F.T., Lezuo P., Bozec A., Schett G., Herrmann M., and Harre U.. 2017. Galectin-3 as a novel regulator of osteoblast-osteoclast interaction and bone homeostasis. Bone. 105:35–41. 10.1016/j.bone.2017.08.013 [DOI] [PubMed] [Google Scholar]
- Sims, A.M., Shephard N., Carter K., Doan T., Dowling A., Duncan E.L., Eisman J., Jones G., Nicholson G., Prince R., et al. 2008. Genetic analyses in a sample of individuals with high or low BMD shows association with multiple Wnt pathway genes. J. Bone Miner. Res. 23:499–506. 10.1359/jbmr.071113 [DOI] [PubMed] [Google Scholar]
- Slaymi, C., Vignal E., Crès G., Roux P., Blangy A., Raynaud P., and Fort P.. 2019. The atypical RhoU/Wrch1 Rho GTPase controls cell proliferation and apoptosis in the gut epithelium. Biol. Cell. 111:121–141. 10.1111/boc.201800062 [DOI] [PubMed] [Google Scholar]
- Smith, L.K., Boukhaled G.M., Condotta S.A., Mazouz S., Guthmiller J.J., Vijay R., Butler N.S., Bruneau J., Shoukry N.H., Krawczyk C.M., and Richer M.J.. 2018. Interleukin-10 directly inhibits CD8+ T cell function by enhancing N-glycan branching to decrease antigen sensitivity. Immunity. 48:299–312.e5. 10.1016/j.immuni.2018.01.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stegmayr, J., Lepur A., Kahl-Knutson B., Aguilar-Moncayo M., Klyosov A.A., Field R.A., Oredsson S., Nilsson U.J., and Leffler H.. 2016. Low or No inhibitory potency of the canonical galectin carbohydrate-binding site by pectins and galactomannans. J. Biol. Chem. 291:13318–13334. 10.1074/jbc.M116.721464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun, X., Zhang B., Pan X., Huang H., Xie Z., Ma Y., Hu B., Wang J., Chen Z., and Shi P.. 2019. Octyl itaconate inhibits osteoclastogenesis by suppressing Hrd1 and activating Nrf2 signaling. FASEB J. 33:12929–12940. 10.1096/fj.201900887RR [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundqvist, M., Welin A., Elmwall J., Osla V., Nilsson U.J., Leffler H., Bylund J., and Karlsson A.. 2018. Galectin-3 type-C self-association on neutrophil surfaces; the carbohydrate recognition domain regulates cell function. J. Leukoc. Biol. 103:341–353. 10.1002/JLB.3A0317-110R [DOI] [PubMed] [Google Scholar]
- Takeshita, S., Fumoto T., Matsuoka K., Park K.A., Aburatani H., Kato S., Ito M., and Ikeda K.. 2013. Osteoclast-secreted CTHRC1 in the coupling of bone resorption to formation. J. Clin. Invest. 123:3914–3924. 10.1172/JCI69493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang, Y., Rowe R.G., Botvinick E.L., Kurup A., Putnam A.J., Seiki M., Weaver V.M., Keller E.T., Goldstein S., Dai J., et al. 2013. MT1-MMP-dependent control of skeletal stem cell commitment via a β1-integrin/YAP/TAZ signaling axis. Dev. Cell. 25:402–416. 10.1016/j.devcel.2013.04.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang, Y., Wu X., Lei W., Pang L., Wan C., Shi Z., Zhao L., Nagy T.R., Peng X., Hu J., et al. 2009. TGF-beta1-induced migration of bone mesenchymal stem cells couples bone resorption with formation. Nat. Med. 15:757–765. 10.1038/nm.1979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taniguchi, T., Woodward A.M., Magnelli P., McColgan N.M., Lehoux S., Jacobo S.M.P., Mauris J., and Argüeso P.. 2017. N-Glycosylation affects the stability and barrier function of the MUC16 mucin. J. Biol. Chem. 292:11079–11090. 10.1074/jbc.M116.770123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taniuchi, K., Iwasaki S., and Saibara T.. 2011. BART inhibits pancreatic cancer cell invasion by inhibiting ARL2-mediated RhoA inactivation. Int. J. Oncol. 39:1243–1252. 10.3892/ijo.2011.1156 [DOI] [PubMed] [Google Scholar]
- Teitelbaum, S.L. 2011. The osteoclast and its unique cytoskeleton. Ann. N. Y. Acad. Sci. 1240:14–17. 10.1111/j.1749-6632.2011.06283.x [DOI] [PubMed] [Google Scholar]
- Teitelbaum, S.L., and Ross F.P.. 2003. Genetic regulation of osteoclast development and function. Nat. Rev. Genet. 4:638–649. 10.1038/nrg1122 [DOI] [PubMed] [Google Scholar]
- Thiemann, S., and Baum L.G.. 2016. Galectins and immune responses-just how do they do those things they do? Annu. Rev. Immunol. 34:243–264. 10.1146/annurev-immunol-041015-055402 [DOI] [PubMed] [Google Scholar]
- Toth, M., Osenkowski P., Hesek D., Brown S., Meroueh S., Sakr W., Mobashery S., and Fridman R.. 2005. Cleavage at the stem region releases an active ectodomain of the membrane type 1 matrix metalloproteinase. Biochem. J. 387:497–506. 10.1042/BJ20041324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Touaitahuata, H., Blangy A., and Vives V.. 2014. Modulation of osteoclast differentiation and bone resorption by Rho GTPases. Small GTPases. 5:e28119. 10.4161/sgtp.28119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uehara, S., Udagawa N., Mukai H., Ishihara A., Maeda K., Yamashita T., Murakami K., Nishita M., Nakamura T., Kato S., et al. 2017. Protein kinase N3 promotes bone resorption by osteoclasts in response to Wnt5a-Ror2 signaling. Sci. Signal. 10:eaan0023. 10.1126/scisignal.aan0023 [DOI] [PubMed] [Google Scholar]
- Vinik, Y., Shatz-Azoulay H., Vivanti A., Hever N., Levy Y., Karmona R., Brumfeld V., Baraghithy S., Attar-Lamdar M., Boura-Halfon S., et al. 2015. The mammalian lectin galectin-8 induces RANKL expression, osteoclastogenesis, and bone mass reduction in mice. Elife. 4:e05914. 10.7554/eLife.05914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson, A.L., Schrecengost R.S., Guerrero M.S., Thomas K.S., and Bouton A.H.. 2013. Breast cancer antiestrogen resistance 3 (BCAR3) promotes cell motility by regulating actin cytoskeletal and adhesion remodeling in invasive breast cancer cells. PLoS One. 8:e65678. 10.1371/journal.pone.0065678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong, H.L., Jin G., Cao R., Zhang S., Cao Y., and Zhou Z.. 2016. MT1-MMP sheds LYVE-1 on lymphatic endothelial cells and suppresses VEGF-C production to inhibit lymphangiogenesis. Nat. Commun. 7:10824. 10.1038/ncomms10824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, M., Chen W., Lu Y., Zhu G., Hao L., and Li Y.P.. 2017. Gα13 negatively controls osteoclastogenesis through inhibition of the Akt-GSK3β-NFATc1 signalling pathway. Nat. Commun. 8:13700. 10.1038/ncomms13700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia, J., and Wishart D.S.. 2016. Using MetaboAnalyst 3.0 for comprehensive metabolomics data analysis. Curr. Protoc. Bioinformatics. 55:14.10.1–14.10.91. 10.1002/cpbi.11 [DOI] [PubMed] [Google Scholar]
- Yahara, Y., Barrientos T., Tang Y.J., Puviindran V., Nadesan P., Zhang H., Gibson J.R., Gregory S.G., Diao Y., Xiang Y., et al. 2020. Erythromyeloid progenitors give rise to a population of osteoclasts that contribute to bone homeostasis and repair. Nat. Cell Biol. 22:49–59. 10.1038/s41556-019-0437-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, R.Y., Hill P.N., Hsu D.K., and Liu F.T.. 1998. Role of the carboxyl-terminal lectin domain in self-association of galectin-3. Biochemistry. 37:4086–4092. 10.1021/bi971409c [DOI] [PubMed] [Google Scholar]
- Yang, T., and Williams B.O.. 2017. Low-density lipoprotein receptor-related proteins in skeletal development and disease. Physiol. Rev. 97:1211–1228. 10.1152/physrev.00013.2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yazbeck, P., Cullere X., Bennett P., Yajnik V., Wang H., Kawada K., Davis V.M., Parikh A., Kuo A., Mysore V., et al. 2022. DOCK4 regulation of Rho GTPases mediates pulmonary vascular barrier function. Arterioscler. Thromb. Vasc. Biol. 42:886–902. 10.1161/ATVBAHA.122.317565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaidi, M. 2007. Skeletal remodeling in health and disease. Nat. Med. 13:791–801. 10.1038/nm1593 [DOI] [PubMed] [Google Scholar]
- Zhang, Y., Rohatgi N., Veis D.J., Schilling J., Teitelbaum S.L., and Zou W.. 2018. PGC1β organizes the osteoclast cytoskeleton by mitochondrial biogenesis and activation. J. Bone Miner. Res. 33:1114–1125. 10.1002/jbmr.3398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, Z., Xu X., Cheng H., Miller M.C., He Z., Gu H., Zhang Z., Raz A., Mayo K.H., Tai G., and Zhou Y.. 2021. Galectin-3 N-terminal tail prolines modulate cell activity and glycan-mediated oligomerization/phase separation. Proc. Natl. Acad. Sci. USA. 118:e2021074118. 10.1073/pnas.2021074118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu, L., Tang Y., Li X.Y., Keller E.T., Yang J., Cho J.S., Feinberg T.Y., and Weiss S.J.. 2020. Osteoclast-mediated bone resorption is controlled by a compensatory network of secreted and membrane-tethered metalloproteinases. Sci. Transl. Med. 12:eaaw6143. 10.1126/scitranslmed.aaw6143 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
lists genotyping PCR primers.
lists quantitative real-time PCR primers.
shows listing of galectin-3 interacting partners.
is the source file for Fig. 2.
is the source file for Fig. 4.
is the source file for Fig. 5.
is the source file for Fig. 7.
is the source file for Fig. 8.
is the source file for Fig. 9.
is the source file for Fig. S1.
is the source file for Fig. S3.
is the source file for Fig. S4.
is the source file for Fig. S5.
Data Availability Statement
The RNA-seq data have been deposited to the Gene Expression Omnibus database at accession number GSE220835 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE220835). The metabolomics data have been deposited to Mendeley (https://doi.org/10.17632/8h7vsykvpd.1). The mass spectrometry–based proteomics data have been deposited to Mendeley (https://doi.org/10.17632/pdhxwr8jc8.2).