Abstract
The control of rRNA transcription, tightly coupled to the cell cycle and growth state of the cell, is a key process for understanding the mechanisms that drive cell proliferation. Here we describe a novel protein, ribin, found in rodents, that binds to the rRNA promoter and stimulates its activity. The protein also interacts with the basal rRNA transcription factor UBF. The open reading frame encoding ribin is 96% complementary to a central region of the large rRNA. This demonstrates that ribosomal DNA-related sequences in higher eukaryotes can be expressed as protein-coding messages. Ribin contains two predicted nuclear localization sequence elements, and green fluorescent protein-ribin fusion proteins localize in the nucleus. Cell lines overexpressing ribin exhibit enhanced rRNA transcription and faster growth. Furthermore, these cells significantly overcome the suppression of rRNA synthesis caused by serum deprivation. On the other hand, the endogenous ribin level correlates positively with the amount of serum in the medium. The data show that ribin is a limiting stimulatory factor for rRNA synthesis in vivo and suggest its involvement in the pathway that adapts ribosomal transcription and cell proliferation to physiological changes.
A major class of housekeeping genes, highly repeated and clustered in the nucleolus in eukaryotes, encode rRNA. The transcription of these genes, executed by RNA polymerase I (Pol I) and assisting factors, is strictly cell cycle and growth regulated (reviewed in references15, 32, and 34). It has been demonstrated that both activating factors and repressors are involved in a rapid adjustment of Pol I activity to the growth state of the cell (9, 16, 23, 39). A Pol I-associated factor, TIF-IA/C*, implicated in the growth regulation of rRNA transcription in mammals, has recently been cloned (8). The activity of this factor fluctuates with the growth conditions, and its binding to the polymerase is required for a transcription-competent complex to form on the rRNA promoter (9, 39). Recent studies have shown that other Pol I transcription factors are also targets of regulation, finding that the phosphorylation status and activity of UBF, SL1, and TTFI are cell cycle controlled through cyclin-dependent kinases (20, 24, 41, 44). Ribosomal transcription is a target of variety of factors affecting cell proliferation, such as hormones and phorbol esters, viral antigens, and the tumor suppressor proteins p53 and Rb (10, 15, 16, 48, 49).
Pol I transcription machinery is generally species specific (15, 32). The structure divergence found in rRNA promoters of different species could partially account for this. SL1 has been shown to play a major role in selection of the rRNA promoter by homologous Pol I transcription apparatus, although UBF and Pol I can also contribute to this selectivity (5, 15, 37). The DNA-protein and protein-protein aspects of specific recognition of the rRNA promoter are not yet clear. Previous data demonstrated that UBF and SL1 can interact with the rRNA promoter. This interaction is synergistic over an extended rDNA region, spanning the core promoter and an upstream control element (6, 34). It has also been shown that the core promoter sequence alone is sufficient to inhibit PolI transcription in trans, presumably competing for a factor(s) binding to it, and detection of such binding activity in cells supported this finding (23, 30). A p70 protein that requires SL1 to bind the rRNA promoter has recently been isolated (47).
In an attempt to extend these studies, we used a Southwestern binding assay (SWA) to identify and clone a polypeptide that interacts with the core element of the rRNA promoter. The coding region of this novel protein revealed 96% complementarity to rRNA. This protein was capable of modulating ribosomal transcription and cell proliferation, and its cellular level correlated with the growth state of the cells.
MATERIALS AND METHODS
Cells and extracts.
Hamster BHK-21 cells, green monkey Vero cells, and rat hepatoma N1S1 cells were obtained from the American Type Culture Collection (ATCC, Rockville, Md.). BHK and Vero cells were grown in alpha minimal essential medium (α-MEM) supplemented with 10% (or less, where indicated) fetal bovine serum, nonessential amino acids, and MEM vitamins, plus 100 U of penicillin and 100 μg of streptomycin per ml. Medium for N1S1 cells was Swims S-77 supplemented as above with addition of pluronic F68 to 0.1%. Whole and nuclear cell extracts and a transcriptionally competent 175 mM ammonium sulfate-DEAE fraction (DEAE-175) were prepared according to published procedures (12, 26, 46).
Expression vectors and plasmids.
Sindbis virus-based expression vector pSinRep21 (2), generously provided by C. Rice, was used for establishing cell lines overexpressing ribin. A 990-bp BglI-EcoRI fragment of cDNA containing the ribin open reading frame (ORF) was ligated at the PmlI cloning site of this vector in the sense and antisense orientation with respect to the viral subgenomic promoter. The same cDNA was used to generate ribin-green fluorescent protein (Rib/GFP) and GFP/Rib fusion constructs by ligating it in frame to the N or C terminus of the GFP ORF, provided in the pEGFP-N1 and pEGFP-C1 vectors from Clontech. In the glutathione S-transferase (GST)-ribin fusion construct (pGST/Rib), ribin cDNA was ligated to the C terminus of the GST ORF in the SalI site of a pGEX2T-derived pT7-GT expression vector, kindly provided by I. Verma. DNA manipulation steps were performed according to conventional procedures (3, 38).
Transfection and selection of cell lines.
Subconfluent (50 to 60%) BHK or Vero cells seeded in 35-mm dishes were transfected with 1 μg of plasmid DNA from pSinRep21, pSin21/RibS, pSin21/RibAS, pEGFP-N1 or -C1, pRib/GFP, or pGFP/Rib. Transfection was done with 6 μl of Lipofectamine in Opti-MEM (Life Technologies) according to the manufacturer's recommendations for 5 h at 37°C. At 24 to 48 h posttransfection, cells were seeded in selective medium containing 2 μg of puromycin or 300 μg of gentamicin per ml. After about a week of selection, when no control (mock-transfected) cells survived, the transformed cells were pooled and expanded for stock freezing and experiments. For expression analysis of the GFP fusion constructs, transfected cells were processed for GFP detection after 6 to 8 days of growth in selective medium, or the selection was omitted and cells were analyzed 42 h posttransfection.
Expression screening of cDNA library.
The Southwestern technique, probing phage plaques with radiolabeled DNA (3, 40), was used to screen a rat liver cDNA expression library (Stratagene; Lambda ZAPII vector) with a labeled double-stranded oligonucleotide probe derived from rat ribosomal DNA (rDNA) core promoter (rCPr) and spanning nucleotides −36 to +18 with respect to the transcription initiation site (36) (sequence given below). The probe was 3′-end labeled by Klenow fill-in reaction (38), gel purified, and used in binding reactions at 4 × 105 to 8 × 105 cpm/ml. The lysed colonies were transferred to nitrocellulose filters and subjected to stepwise renaturation with guanidine hydrochloride as described (40), followed by blocking in 5% nonfat milk. Filters were preincubated for 30 min in binding buffer containing 25 mM Tris-HCl (pH 7.5), 50 mM KCl, 5 mM MgCl2, 0.2 mM EDTA, 0.04% NP-40, 5% glycerol, and 2 μg of poly(dI:dC) per ml. The binding reaction was started by addition of the labeled probe and incubated for 2 h at 25°C. The filters were finally washed three to four times in the binding buffer containing 100 mM KCl, dried, and exposed to X-ray film. About 1.5 × 106 plaques were screened for DNA-binding activity through several cycles, ending with three identical positive cDNA clones. The cDNA sequence was determined following the USB Sequenase kit protocol and confirmed in some regions by fluorescent DNA sequencing (ABI Prism 377; Perkin Elmer).
Western and Southwestern blotting
Immunoblotting was performed following established procedure (38). Equal amounts of protein from total cell extracts, determined by the Bio-Rad protein microassay kit, were electrophoresed in sodium dodecyl sulfate–10% polyacrylamide gel electrophoresis (SDS-PAGE) and transferred to nitrocellulose filters. A polyclonal antibody raised in a rabbit against bacterially expressed and gel-purified protein was used at a 1:500 to 1:1,000 dilution, in 1 h of incubation at 25°C. For immunodetection of ribin, the Amersham ECL kit including horseradish peroxidase-conjugated anti-rabbit immunoglobulin G (IgG) was used according to the manufacturer's protocol. In some experiments, cell extracts were immunoprobed in parallel with a mouse monoclonal antiactin antibody (1:2,000 dilution), as a normalization control.
In Southwestern analysis, after the protein-blotting step, the filter membranes were processed for DNA binding as described for library screening, including a prolonged prebinding reaction of 1 h and 150 mM KCl in the filter-washing steps. The rCPr probe or an analogous mouse core promoter probe (mCPr [23]), spanning nucleotides −43 to +13 with respect to transcription initiation (22), was end labeled, purified, and used as described above. In some experiments, a mutant variant of the rCPr probe, with a G to A change at the −16 position was used (rCPrMut). For testing bacterially expressed protein for promoter binding, the GST-ribin fusion was induced with IPTG (isopropylthiogalactopyranoside) as described below, and 10 to 20 μg of cell lysates was subjected to SWA.
The rRNA promoter-binding probes (noncoding strand) were rCPr (CTTTGC TATC TCTCC T TAT TGTACC TGGAGATATATGC TGACACGCTG TCC TTT), rCPrMut (same as rCPr but with the italic G changed to A), and mCPr (TTGTGATCTTTTCTATCTGTTCCTATTGGACCTGGAGATAGGTACTGACACG).
GST fusion protein purification and pull-down binding assay.
Escherichia coli BL-21(DE3) was transformed with the pGST/Rib fusion construct or with the parental vector expressing GST alone, and protein expression was induced with 1 mM IPTG for 4 h. To obtain bacterial extracts, cells were suspended in ice-cold extraction buffer (50 mM Tris-HCl [pH 7.5], 2 mM EDTA, 1 mM dithiothreitol [DTT], 2 mM phenylmethylsulfonyl fluoride [PMSF], and 10 μg/ml each of pepstatin A, leupeptin, and aprotinin), followed by rapid freezing and thawing. Cells were then lysed with 0.5 mg/ml lysozyme for 15 min at 4°C, NaCI was added to 1 M for an additional 15 min, and the cell debris was removed by centrifugation. Lysate proteins were precipitated with 65% ammonium sulfate and suspended in phosphate-buffered saline (PBS) buffer containing 0.5% NP-40 and protease inhibitors as above. The GST fusion was affinity purified by glutathione-Sepharose beads (Pharmacia), as described (1). Protein bound to beads was washed five times and suspended in HEN buffer (10 mM HEPES-KOH [pH 7.9], 100 mM KCl, 0.5 mM EDTA, 1 mM DTT, 0.5 mM PMSF, 0.25% NP-40, 10% glycerol).
Nuclear extracts from BHK cells used in the binding assay were precleared with glutathione-Sepharose beads equilibrated with the above buffer by rocking for 1 h at 4°C and brief centrifugation. Protein-protein interaction was performed with 5 μl of protein-loaded beads and 20 μl of nuclear extract for 1 h at 4°C in a final volume of 100 μl, adjusted with HEN buffer. Bead-bound complexes were precipitated by brief centrifugation, and the pellet was washed five times with HEN buffer and resuspended in 60 μl of the same buffer. Then 20-μl samples were taken from the binding reaction before precipitation (total), from the supernatant (unbound), and from the suspended pellet (bound), boiled in SDS sample buffer, and analyzed by Western blotting with an antiserum (1:1,000) raised against Xenopus laevis UBF (kindly provided by C. Pikaard).
PCR amplification.
From 60 to 80 ng of rat, mouse, or human genomic DNA (Novagen) was amplified with primers A plus B or A plus C, 10 pmol of each (see Fig. 1b for primer locations). Primer A was TTAGAGCCAATCCTTATCCCGAAGTTACG, primer B was ATCGAAAGGGAGTCGGGTTCAGATCT, and primer C was TGAGAGATGGGCGAGTGCCGTTCCGAA. KlentaqLA enzyme (DNA Polymerase Technology) was used with the manufacturer's buffer in 50-μl reactions, including 1.3 M betaine (Sigma). Samples were heated for 4 min at 94°C, followed by 35 to 38 amplification cycles of 50 s at 94°C and 2 min at 68°C in a RoboCycler (Stratagene). Amplified products were analyzed in ethidium bromide-stained 2% agarose gel, along with a 100-bp DNA ladder.
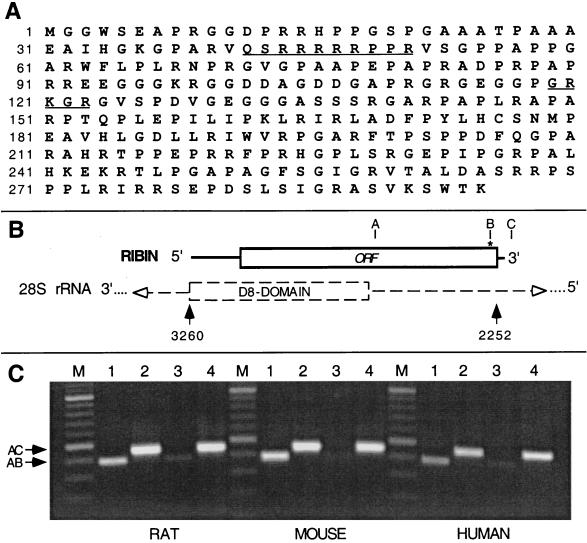
FIG. 1.
Amino acid sequence of ribin ORF, rDNA homology region, and PCR detection of the cloned gene. (A) Amino acid sequence of the 295-amino-acid ribin ORF. NLS-type sequences are underlined. (B) The ribin gene is homologous to an internal region of 28S rDNA; the ribin ORF is oriented opposite to rRNA polarity. About half of this ORF overlaps the D8 variable domain of the rRNA gene. (C) Rat, mouse, and human DNAs are PCR amplified with primers A plus B (lanes 1 and 3) or A plus C (lanes 2 and 4) (primer positions are shown in panel B). Primers A and C are derived from rDNA, where A is within the ribin coding region, while C is located beyond the 3′ end of ribin cDNA. Primer B ends in a ribin-specific BglII site at the 3′ terminus of the gene (shown by the asterisk in panel B). The AB and AC products should therefore represent ribin and the bulk rRNA genes, respectively. In lanes 3 and 4, the template DNA was digested with BglII prior to PCR. Amplified products (386 and 450 bp, shown by arrows) were resolved in a 2% ethidium bromide-stained agarose gel, along with a 100-bp DNA ladder (lanes M).
In vitro translation and transcription.
“Capped” RNA transcripts of the ribin cDNA clone in the BlueScript II vector (Stratagene) were synthesized in vitro according to the Promega Guide protocols. From 1 to 2 μg of RNA template was then used in the translation reaction (25 to 50 μl) with rabbit reticulocyte lysate (Promega) containing unlabeled (1 mM) or 35S-labeled (20 μCi) methionine for 90 min at 30°C. In mock translation controls, the RNA template was omitted.
For testing the transcriptional activity of the synthesized protein, aliquots of unlabeled translation reactions were added directly to the transcription assay. pB7-2.0 plasmid (kindly provided by L. Rothblum), encompassing the transcription initiation site of the rat rRNA gene in a 2-kb SalI fragment, was used as the runoff template. It was linearized with HindIII to produce 124-nucleotide (nt) transcripts (36). Transcription conditions were essentially as described previously (23). rDNA template (50 ng) was transcribed in a 25-μl reaction with 4 μl of DEAE-175 fraction obtained from rat N1S1 cells, with addition of 2 to 4 μl of ribin or mock translation reactions in the presence of α-amanitin (100 μg/ml). The labeled transcripts were purified and analyzed in 5% polyacrylamide–7 M urea gels.
Nucleus isolation, run-on transcription, and dot hybridization.
Control or ribin-overexpressing BHK or Vero cell lines were grown to strictly equal density (60 to 70% confluency), and the same amounts of cells were then harvested for nucleus isolation. Transcriptionally active nuclei were prepared by the NP-40 lysis method, and run-on transcription was performed in the presence of [α-32P]UTP as described for the basic protocol (3). The amount of nuclei used in the reactions was equalized based on the 280 nm absorbance of the total nuclear lysate. Where indicated, α-amanitin was present at 150 μg/ml in order to block Pol II and Pol III transcription.
The transcription reaction, carried out for 30 min at 30°C, was stopped, and the radiolabeled nuclear RNA was isolated following the abovementioned protocol except using acidic (100 mM Tris-acetate [pH 5.2] saturated) phenol for RNA extraction. RNA preparations were checked in an agarose gel for absence of DNA, and the specific activity of RNA (disintegrations per minute [dpm] per microgram) was estimated after trichloroacetic acid filter precipitation of RNA aliquots. For identification of the newly transcribed RNA, 1 to 3 μg of the following plasmid DNAs was denatured and immobilized on nitrocellulose filters (3) as dot hybridization samples: pBS-28SBE plasmid, containing a 2,029-bp BglII-EcoRI fragment of mouse 28S rRNA (obtained from a pGEM2-28S plasmid [18], a kind gift from J.-P. Bachellerie); and pTri-GAPDH plasmid (Ambion), containing a 316-bp fragment of human glyceraldehyde-3-phosphate dehydrogenase (GAPDH) cDNA. pBS/SK vector DNA (Stratagene) was used as a negative hybridization control. In some experiments 0.1 to 0.2 μg of sense RNA transcript of the ribin cDNA clone in the pBS/SK vector was used as an additional rRNA hybridization substrate.
Nitrocellulose strips containing identical dotted DNAs were UV cross-linked and hybridized with equal amounts (usually 15 μg) of radiolabeled nuclear RNA probes. After 6 h of prehybridization, 24 to 36 h of hybridization was carried out at 68°C in 6× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate)–2× Denhardt's reagent–0.2% SDS–100 μg of tRNA per ml. Filters were washed twice with 2× SSC for 1 h at 65°C, treated with 2× SSC containing RNase A (10 μg/ml) for 30 min at 37°C, and washed additionally with fresh 2× SSC for 1 h at 37°C. After drying, filters were exposed to x-ray film or a phosphorimager screen. Hybridization dot intensities were quantified using the Image J Program (National Institutes of Health).
Cellular localization of ribin-GFP fusion proteins.
BHK cells were analyzed for GFP fluorescence 42 h or 7 days posttransfection with Rib/GFP or GFP/Rib fusion constructs. Control cells were transfected with the parental plasmid pEGFP. Transfected cells were processed for GFP fluorescent detection basically as recommended in the Clontech protocol. Cells were seeded on cover slips and allowed to attach and grow for 24 h. Cells were then washed with PBS, fixed with 4% paraformaldehyde–5% sucrose–PBS for 30 min, and mounted on microscope slides. Samples were examined on Zeiss Axioplan fluorescence microscope equipped with the Bio-Rad MRC 1024 laser confocal scanning system. Digital images were processed using Adobe Photoshop (Adobe Systems).
Nucleotide sequence accession number.
The GenBank accession number for the cDNA sequence reported in this work is U77931.
RESULTS
Detection and cloning of an rRNA core promoter binding protein, ribin, present in rodent cells.
An rRNA core promoter binding activity was previously detected in mouse cells by a gel mobility shift assay, where the specificity of binding was demonstrated by nonspecific competitors (22, 23). To test if this activity resides in a monomer polypeptide and estimate the size of the potential promoter binding protein(s), we made use of the SWA. This technique, successfully used in studies on the Pol I transcription factor UBF, proved useful in expression cloning of DNA-binding proteins and testing the specificity of DNA-protein interaction (31, 40).
Applying a high-stringency SWA protocol which eliminates UBF binding, we detected a 32-kDa protein in rodent cell extracts that binds to the core rRNA promoter (illustrated in Fig. 2A and 3C). This binding was sensitive to a G to A mutation in the probe at position −16 (Fig. 2A), known to significantly inactivate the rRNA promoter (6, 30, 46).
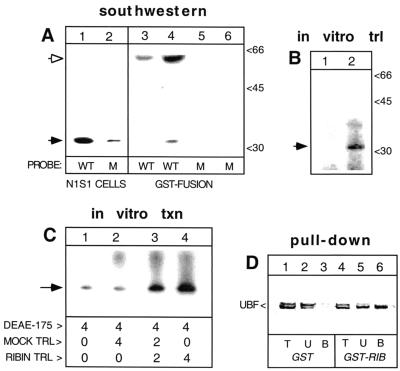
FIG. 2.
Binding of ribin to rRNA core promoter, effect on Pol I transcription, and interaction of the protein with UBF. (A) SWA of extracts from rat N1S1 cells (lanes 1 and 2) or bacterial cells induced with IPTG to express the GST-ribin fusion (lanes 4 and 6) or uninduced (lanes 3 and 5). Duplicate samples of each extract were probed with the wild-type (WT; rCPr, lanes 1, 3, and 4) or point-mutated (M; rCPrM, lanes 2, 5, and 6) rat rRNA core promoter probe. The binding signals corresponding to ribin and the GST fusion are marked by black and white arrowheads, respectively, and the positions of protein standards are shown (in kilodaltons) on the right. (B) Ribin was translated (trl) in the presence of [35S]methionine, and the radiolabeled product was analyzed by SDS–10% PAGE (lane 2, shown by arrowhead). In mock translation, the RNA template is omitted (lane 1). (C) In vitro-translated (unlabeled) ribin was tested directly in a cell-free transcription assay. pB7-2.0/HindIII-cut rat rDNA template (50 ng) was transcribed with 4 μl of DEAE-175 fraction alone (lane 1) or supplemented with 4 μl of mock translation reaction (lane 2), 4 μl of ribin translation (txn) reaction (lane 4), or 2 μl of each (lane 3). The specific transcript of 124 nt is marked by an arrow. (D) Nuclear extract from BHK cells was incubated with GST-ribin fusion protein (lanes 4 to 6) or GST alone (lanes 1 to 3), followed by sedimentation of the products formed on glutathione-Sepharose bead complexes. Aliquots taken from the total reaction mix before precipitation (T), from the supernatants (unbound fraction, U), and from the resuspended pellets (bound fraction, B) were subjected to immunoblotting with an anti-UBF antibody.
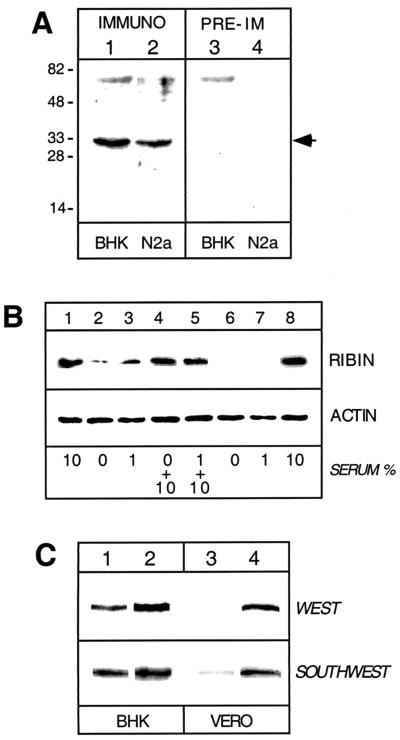
FIG. 3.
Ribin expression levels upon serum depletion and after transfection. (A) Total cell extracts from hamster BHK cells and mouse N2a cells were probed in Western blotting with antiribin serum (lanes 1 and 2) or preimmune serum (lanes 3 and 4). In both types of cells, endogenous ribin (approximately 32 kDa) was detected. The positions of protein standards are shown (in kilodaltons) on the left. (B) BHK cells were cultured for 22 h with 10, 0, or 1% serum (lanes 1 to 3). Portions of the last two (serum deficient) samples were cultured for an additional 18 h with (lanes 4 and 5) or without (lanes 6 and 7) addition of serum back to 10%. Lane 8, control cells incubated continuously with 10% serum. Equal amounts of total cell protein from each variant were used for immunoblotting with antiribin antibody. The same cell extracts were also immunoprobed for actin as a reference control. (C) BHK and Vero cells stably transfected with pSinRep21 expression vector carrying ribin cDNA (lanes 2 and 4) or with blank vector (lanes 1 and 3) were probed for ribin by immunoblotting or Southwestern technique. In the latter, a mouse rDNA core promoter fragment, mCPr, was used as a binding probe.
The same SWA protocol was then applied to screen a rat cDNA expression library. After multiple rounds of screening three identical clones expressing core promoter binding activity were obtained. The cDNA (GenBank U77931) reveals a 1,008-bp sequence that is 96% similar to an internal region of the 28S rRNA gene (positions 2252 to 3260, Fig. 1). This sequence contains a 295-amino-acid open reading frame oriented opposite the direction of rRNA and is followed by a 30-nt nonribosomal sequence matching the 3′-end polyadenylation signal of ferritin mRNA (GenBank J04716). Thus, the message of the cloned gene is essentially an antisense transcript of rRNA (giving the name ribin for the encoded protein). Interestingly, the 5′ half of the rRNA homology region (nt 2 to 611) corresponds to the largest variable domain in rRNA, D8, and shows the best match to a D8 sequence found in the mouse (18).
We were able to amplify the cloned sequences from genomic DNA using combinations of an rDNA-derived primer and a selective primer from the 3′ terminus of ribin cDNA that covers a specific BglII site present in the clone. As expected, the amplified product was sensitive to BglII digestion (Fig. 1C, lanes 3), while amplification of the corresponding region of rRNA genes with two bona fide ribosomal primers was not (lanes 4). This shows that the conditions used favor selective amplification of the ribin sequence, minimizing the potential background from the ribosomal genes. The data also suggest that despite its homology to 28S rDNA, the ribin gene is in lower copy number than the rRNA genes. Identical PCR results were obtained with rat, mouse, and human DNA (Fig. 1C), suggesting the existence of ribin-type rDNA sequences in mammals.
The deduced amino acid sequence of ribin revealed two motifs, QSRRRRRPPR and GRKGR (Fig. 1A, positions 42 to 51 and 119 to 123), homologous to the nuclear localization signal (NLS) of the Tat and Rev proteins (19, 35). A PGAPAGXXG sequence present in the enhancer-binding protein C/EBP (25) was located at positions 248 to 256. Multiple putative protein kinase C and casein kinase II phosphorylation sites were found clustered at the C-terminal portion of the protein.
We generated a GST fusion of the cloned protein and assessed its rRNA promoter binding activity. As in SWA performed with the rat cell extract, the point mutation at position −16 in the promoter probe abolished binding (Fig. 2A). A minor binding signal corresponding to about 32 kDa was also observed. It too was sensitive to that mutation and most probably reflected partial cleavage of the fusion.
We also detected ribin in cells by using a polyclonal antibody raised against the cloned protein (Fig. 3A). The apparent mass of 32 kDa of the reacting protein corresponded to the binding signal obtained in SWA (Fig. 2). Thus, both Western and Southwestern assays confirmed the ORF predicted in the cDNA clone. With the antiribin antibody, we have been able to detect related proteins in rat, mouse, and hamster cells. In SWAs a mouse rDNA core promoter probe, shown earlier to inhibit Pol I transcription in trans (23, 30), produced a stronger binding signal with rodent than primate cells (Fig. 3C). These results suggest that divergent ribin orthologues may exist in more distant species.
Ribin activates rRNA transcription in vitro and interacts with the basal Pol I transcription factor UBF.
The specific interaction of ribin with the rRNA promoter raised the obvious question of whether the protein can also affect promoter function. We tested the transcriptional effect of the in vitro-translated protein directly in a cell-free assay. As shown in Fig. 2B, the ribin ORF can be efficiently translated in a rabbit reticulocyte lysate. A rat rDNA template, pB7-2.0, was transcribed with a DEAE-175 cell fraction that contains basal Pol I transcription activity (46). When portions of the ribin translation reaction were added to this assay, a significant dose-dependent increase in the specific 124-nt transcripts was obtained, while a mock translation reaction had no effect (Fig. 2C). These results show that ribin is capable of enhancing the rRNA promoter-driven transcription in vitro. Furthermore, a pull-down assay performed with highly purified GST fusion protein showed that ribin interacts with UBF (Fig. 2D). The latter is known to bind to some Pol I factors, and it is also a target of repressors of rRNA transcription (reviewed in reference 15).
Cellular level of ribin correlates with amount of serum in medium.
We next determined whether the expression of the cloned protein changes with the growth state of the cells. Immunoblotting was performed with cells subjected to serum variations. Serum and growth factor deficiencies are known to arrest cell division at the G0/G1 phase of the cell cycle, while readdition of serum stimulates quiescent cells to progress through G1 (reference 15 and references therein). The ribin level was found to correlate with the serum level. When the serum amount was highly reduced or completely withdrawn, the ribin was barely or not detectable, while when cells were growth stimulated by readding serum, the protein level was restored close to normal (Fig. 3B). This implies a possible link of ribin to the serum-triggered regulatory pathways.
Overexpression of ribin in mammalian cells results in enhanced ribosomal transcription and proliferation rate.
It is well documented that rRNA synthesis fluctuates with the serum changes to adapt ribosome biogenesis to the growth rate of the cells (15, 34). Since ribin was capable of stimulating Pol I transcription in vitro, we entertained the possibility that a target of ribin function in vivo might be rRNA synthesis. We explored a Sindbis virus-based expression system (2) to establish cell lines overexpressing ribin and examine the protein's effect in vivo. As shown in Fig. 3C, the ribin level, as detected by Western or Southwestern techniques, was either increased (more than twofold) or only detectable in these cells. This test also demonstrates that the cloned protein, expressed upon transfection, comigrates with the endogenous one.
The rRNA transcription rate in these cell lines was then analyzed by nuclear run-on/dot hybridization assays. In the Vero cell line overexpressing ribin (Fig. 3C, lane 4), the estimated rRNA transcription, performed in either the presence or absence of α-amanitin, was increased by 62 to 68% compared to the vector-transfected control cell line, as shown in Fig. 4A. The data indicate that ribin can act as an activator of rRNA synthesis in vivo. Consistent with this, expression of ribin antisense RNA had a reverse effect on ribosomal transcription, reducing it by one third of the control value. Thus, the ribin-dependent modulation of cellular rRNA synthesis ranged to 2.3- to 2.6-fold. These values, obtained from the dot hybridization signals, were in agreement with the specific activity of the nuclear RNA reached after run-on transcription. Since, in the presence of α-amanitin, Pol II and Pol III polymerase activities should be negligible, if present at all, the incorporated radiolabel will mostly reflect the amount of newly synthesized rRNA.
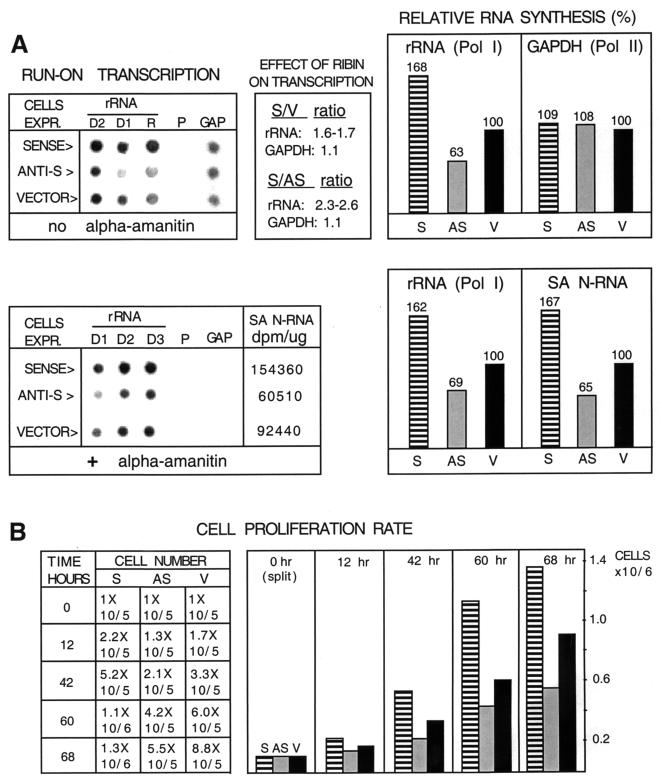
FIG. 4.
Effect of ribin overexpression on rRNA synthesis and cell proliferation. (A) Nuclei isolated from Vero cells stably transfected with pSinRep21 expression vector harboring ribin cDNA in the sense (S) or antisense (AS) orientation, or with blank vector (V), were used for a run-on transcription assay in the absence or presence of α-amanitin (150 μg/ml, lower panel). The samples in lanes V and S are the same cell lines shown in Fig. 3C, lanes 3 and 4, respectively. Radiolabeled nuclear RNA was then isolated and used in dot hybridization. Dot samples: D1 to D3, 1, 2, and 3 μg, respectively, of pBS-28SBE plasmid DNA, containing a 2,029-bp fragment of mouse 28S rDNA; R, 0.1 μg of sense RNA transcript of ribin cDNA; P, 3 μg of pBS vector DNA; GAP, 1 μg of plasmid pTRI-GAPDH, containing a 316-bp fragment of the GAPDH gene (note that P and GAP dots were exposed eight times longer than rDNA dots). SA N-RNA, specific activity of the radiolabeled nuclear RNA obtained after run-on transcription. Reactions were normalized by using equal amounts of nuclei for transcription and of nuclear RNA for hybridization. The panels on the right show a densitometry quantitation of the hybridization signals and the relative effect of ribin overexpression on Pol I and Pol II RNA transcription. (B) Growth rate of the cell transfectants. Equal starting amounts of the three cell variants were grown for 68 h (still subconfluent), and cell number was counted in the time intervals.
In contrast, ribin overexpression had a minor effect on a Pol II-transcribed reference gene, GAPDH, suggesting that the Pol I machinery can be selectively activated by the cloned protein. Furthermore, the three cell variants also exhibited proliferation rates proportional to their rRNA transcription, highest with the cells overproducing ribin and lowest with the ones expressing its antisense RNA (Fig. 4B). The initial indication of this growth difference was the fastest recovery of the ribin-overexpressing cells after selection, ahead of the rest of the transfectants in establishing single-cell colonies (not illustrated).
These effects of ribin in vivo suggest that the protein is a limiting stimulatory factor for rRNA synthesis and cell proliferation. The estimated enhancing effect of ribin on endogenous rRNA transcription is comparable to the increase in Pol I activity observed earlier in serum-stimulated resting cells (33) and in cells overexpressing UBF (17).
Ribin can counteract serum-mediated suppression of rRNA transcription.
In another set of experiments, we asked of whether the Pol I-stimulatory effect of ribin could interfere with the known suppression of rRNA transcription upon serum starvation. BHK cell lines overexpressing ribin were cultured along with control cells in serum-deficient medium (0.5% serum for 12 h), and rRNA transcription was analyzed again by nuclear run-on assay. As shown in Fig. 5, this serum deficiency caused 68% inhibition of rRNA transcription in the control cells, while the cells overexpressing ribin were significantly less inhibited (by 24%). Remarkably, the Pol I transcription rate in the latter was above the control value even upon serum starvation. As with Vero cells, the dot hybridization values were consistent with the relative amounts of rRNA synthesized, as deduced from the specific activity of the nuclear RNA. These results show that by enhancing rRNA transcription, ribin can significantly relieve the serum-triggered inhibition of the Pol I machinery.
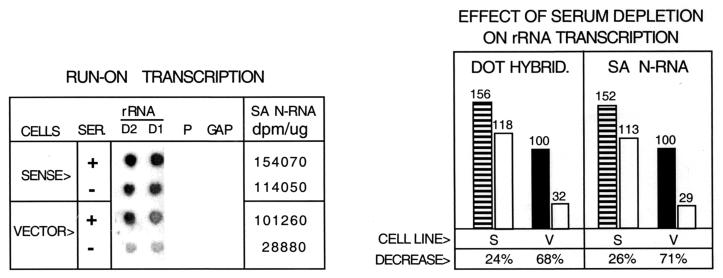
FIG. 5.
Effect of serum starvation on rRNA synthesis in cells overexpressing ribin. BHK cells stably transfected with the pSinRep21 expression vector carrying ribin cDNA (S) or with the blank vector (V) were cultured with a control amount (10%, SER+) or a reduced amount (SER−) of serum (0.5% for 12 h). Nuclei isolated from these cells were used for run-on transcription in the presence of 150 μg of α-amanitin, followed by dot hybridization, as in Fig. 2. The dot samples D1, D2, P, and GAP are also as in Fig. 2. The specific activity of the radiolabeled nuclear RNA is given as in Fig. 4 (SA N-RNA). A densitometric quantitation of the average transcriptional signals is shown on the right. The values for the control (V, SER+) cells were taken as 100%, and the SER− values are presented by open bars.
Ribin-GFP fusion proteins localize in the nucleus.
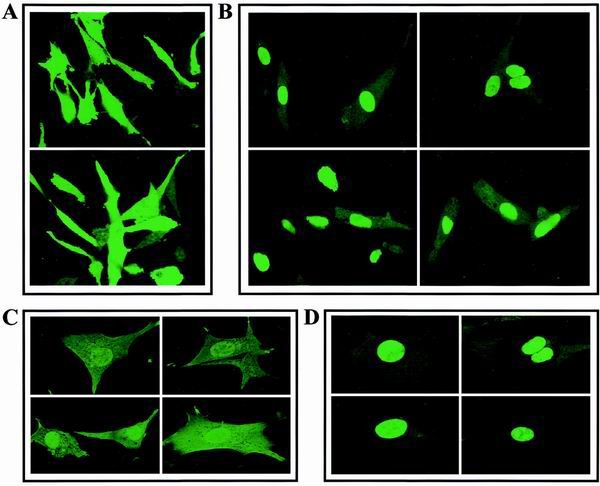
Two NLS are apparent in the amino acid sequence of ribin. To test the functionality of these elements, we transfected hamster BHK cells with GFP fusion constructs having GFP at either the N or C terminus of the ribin ORF. As illustrated in Fig. 6, in both cases the expressed fusion protein localized clearly in the nucleus, and this nuclear accumulation was observed with both short- and long-term-transfected cells. Some cells showed dominant nucleolar staining, and the subnuclear distribution of ribin remains to be established.
FIG. 6.
Cellular localization of ribin. BHK cells were transfected with GFP (A and C) or ribin-GFP fusion constructs (B and D). Identical accumulation of ribin in the nucleus was observed whether the GFP moiety was N or C terminal to the ribin ORF, as illustrated with the upper and lower images in each panel. Fluorescent images were taken by confocal microscope 42 h (A and B) or 7 days (C and D) after transfection.
DISCUSSION
We isolated a cDNA clone expressing a 32-kDa rRNA core promoter binding protein of rodent cells. The protein interacted with the Pol I factor UBF and was capable of enhancing rRNA transcription and cell proliferation.
The exact role of ribin in Pol I transcription activation remains to be elucidated. Our data favor a model in which ribin, by interacting with the rDNA promoter and UBF, could play a “bridging” role in the formation and/or stability of the transcription initiation complex. Since only a fraction of the eukaryotic rRNA genes are transcriptionally active, rRNA synthesis could be enhanced either through activating of silent genes or by increased transcription of the established active ones (34 and references therein). Although our run-on assay cannot discriminate between these possibilities, it is unlikely that ribin affects the transcription elongation rate, because the incubation time used should be sufficient for all nascent rRNA transcripts to be completed.
Our in vivo data suggest that the ribin level can modulate rRNA synthesis and cell proliferation. Moreover, ribin can significantly relieve the serum-triggered repression of rRNA synthesis, rendering cells less serum dependent. This is of particular interest, as tolerance to serum deprivation is a hallmark of tumor cells. Furthermore, the findings that the cellular level of ribin both depends on the serum and affects rRNA synthesis suggest a mediator role for this protein in serum-responsive rRNA regulation.
As the ribin-overexpressing cells manifest both enhanced rRNA transcription and accelerated growth, an intriguing question is whether the sole effect of this protein is activation of rRNA synthesis. If so, this would be a key step in triggering the cascade of events leading to cell division. Yet our data cannot rule out another function of the protein related eventually to cell division.
It remains to be understood whether the ribin-mediated effect on rRNA synthesis is linked to the pathway(s) controlling the activity of some Pol I factors, such as TIF-IA and UBF, in a growth- and cell cycle-dependent mode (8, 24, 44).
A remarkable aspect of the ribin gene is its extensive homology to rDNA. Transcripts with sequence similarity to rRNA are known to exist, such as small nucleolar RNAs (4) and some mRNAs that contain relatively short segments similar to rRNA (27). With its 1-kb complementarity to rRNA, the ribin coding region is an extreme example of that kind, demonstrating a heretofore unknown genetic potential of rDNA-related sequences in higher eukaryotes to express protein-coding messages.
Functional polypeptides encoded by transcripts complementary to the large rRNA or within 16S rRNA have been found in some primitive eukaryotes and bacteria (7, 21, 42, 43). This is in accord with the hypothesis that rRNA was originally an agglomeration of individually transcribed genomic sequences, where structural and protein-coding sequences might have been intermingled (11). Interestingly, the large rRNA genes ranging from Drosophila to humans all reveal inverted ORFs overlapping the ribin homology region (M. Kermekchiev, unpublished analysis). This observation is rather unexpected, given the fact that this ORF region includes one of the largest rRNA variable domains, D8. It has been assumed that in rRNA evolution, selective constraints operate to preserve the secondary structure rather than the nucleotide sequence of the variable domains (13, 14, 28). This concept, established for the bulk of rRNA genes, would presumably not apply to the ribin gene. Furthermore, if the complementarity of ribin transcripts to rRNA is significant for the control of ribosome synthesis, the ribin gene must be maintained by more stringent selective pressure. It remains to be determined if the cloned gene is positioned near the rDNA repeats or if it is located outside the nucleolus, similar to some “jumped” rRNA pseudogenes and orphan rDNA sequences (29, 45). This gene might have originated by duplication of the above-mentioned portion of rDNA with protein-coding potential.
ACKNOWLEDGMENTS
We thank W. Barnes for offering the ABI Prism sequencer and S. Jacob and S. Schlesinger for their support in preparation of antiribin antibody and establishing stably transfected cell lines. We are also grateful to S. Schlesinger, M. Johnston, and S. Gerbi for helpful comments and suggestions on the manuscript, as well as to lab members and colleagues for numerous stimulating discussions.
REFERENCES
- 1.Abath F, Simpson A. A simple method for the recovery of purified recombinant peptides cleaved from glutathione-S-transferase fusion proteins. Peptide Res. 1990;3:167–168. [PubMed] [Google Scholar]
- 2.Agapov E, Frolov I, Lindenbach B, Pragai B, Schlesinger S, Rice C. Noncytopathic Sindbis virus RNA vectors for heterologous gene expression. Proc Natl Acad Sci USA. 1998;95:12989–12994. doi: 10.1073/pnas.95.22.12989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ausubel F, Brent R, Kingston R, Moore D, Seidman J, Smith J, Struhl K, editors. Current protocols in molecular biology. New York, N.Y: John Wiley & Sons; 1992. [Google Scholar]
- 4.Bachellerie J-P, Michot B, Nicoloso M, Balakin A, Ni J, Fournier M. Antisense snoRNAs: a family of nucleolar RNAs with long complementarities to RNA. Trends Biochem Sci. 1995;20:261–264. doi: 10.1016/s0968-0004(00)89039-8. [DOI] [PubMed] [Google Scholar]
- 5.Beckmann H, Chen J-L, O'Brien T, Tjian R. Coactivator and promoter-selective properties of RNA polymerase I TAFs. Science. 1995;270:1506–1509. doi: 10.1126/science.270.5241.1506. [DOI] [PubMed] [Google Scholar]
- 6.Bell S, Jantzen H-M, Tjian R. Assembly of alternative multiprotein complexes directs rRNA promoter selectivity. Genes Dev. 1990;4:943–954. doi: 10.1101/gad.4.6.943. [DOI] [PubMed] [Google Scholar]
- 7.Berg K, Squires C L, Squires C. In vivo translation of a region within the rrn 16S rRNA gene of Escherichia coli. J Bacteriol. 1987;169:1691–1701. doi: 10.1128/jb.169.4.1691-1701.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bodem J, Dobreva G, Hoffmann-Rohrer U, Iben S, Zentgraf H, Delius H, Vingron M, Grummt I. TIF-IA, the factor mediating growth-dependent control of ribosomal RNA synthesis, is the mammalian homolog of yeast Rrn3p. EMBO Rep. 2000;1:171–175. doi: 10.1093/embo-reports/kvd032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brun R, Rayan K, Sollner-Web B. Factor C*, the specific initiation component of the mouse RNA polymerase I holoenzyme, is inactivated early in the transcription process. Mol Cell Biol. 1994;14:5010–5021. doi: 10.1128/mcb.14.7.5010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Budde A, Grummt I. p53 represses ribosomal gene transcription. Oncogene. 1999;18:1119–1124. doi: 10.1038/sj.onc.1202402. [DOI] [PubMed] [Google Scholar]
- 11.Clark C G. On the evolution of ribosomal RNA. J Mol Evol. 1987;25:343–350. doi: 10.1007/BF02603119. [DOI] [PubMed] [Google Scholar]
- 12.Dignam J, Lebovitz R, Roeder R. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 1983;11:1475–1489. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gerbi S. Expansion segments of ribosomal RNA. In: Zimmermann R, Dahlberg A, editors. Ribosomal RNA: structure, evolution, processing and function in protein biosynthesis. Boca Raton, Fla: CRC Press; 1996. pp. 71–87. [Google Scholar]
- 14.Gorski J, Gonzalez I, Schmickel R. The secondary structure of human 28S rRNA: the structure and evolution of a mosaic rRNA gene. J Mol Evol. 1987;24:236–251. doi: 10.1007/BF02111237. [DOI] [PubMed] [Google Scholar]
- 15.Grummt I. Regulation of mammalian ribosomal gene transcription by RNA polymerase I. Prog Nucleic Acid Res Mol Biol. 1999;62:109–153. doi: 10.1016/s0079-6603(08)60506-1. [DOI] [PubMed] [Google Scholar]
- 16.Hannan K, Kennedy B, Cavanaugh A, Hannan R, Hirschler-Laszkiewicz I, Jefferson L, Rothblum L. RNA polymerase transcription in confluent cells: Rb downregulates rDNA transcription during confluence-induced cell-cycle arrest. Oncogene. 2000;19:3487–3497. doi: 10.1038/sj.onc.1203690. [DOI] [PubMed] [Google Scholar]
- 17.Hannan R, Stefanovsky V, Arino T, Rothblum L, Moss T. Cellular regulation of ribosomal DNA transcription: both rat and Xenopus UBF1 stimulate rDNA transcription in 3T3 fibroblasts. Nucleic Acids Res. 1999;27:1205–1213. doi: 10.1093/nar/27.4.1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hassouna N, Michot B, Bachellerie J-P. The complete nucleotide sequence of mouse 28S rRNA gene. Implications for the process of size increase of the large subunit rRNA in higher eukaryotes. Nucleic Acids Res. 1984;12:3563–3583. doi: 10.1093/nar/12.8.3563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hauber J, Malim M, Cullen B. Mutational analysis of the conserved basic domain of human immunodeficiency virus Tat protein. J Virol. 1989;63:1181–1187. doi: 10.1128/jvi.63.3.1181-1187.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Heix J, Vente A, Voit R, Budde A, Michaelidis T, Grummt I. Mitotic silencing of human rRNA synthesis: inactivation of the promoter selectivity factor SL1 by cdc2/cyclin B-mediated phosphorylation. EMBO J. 1998;17:7373–7381. doi: 10.1093/emboj/17.24.7373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jansson A, Gillin F, Kagardt U, Hagblom P. Coding of hemolysins within the ribosomal RNA repeat on a plasmid in Entamoeba histolytica. Science. 1994;263:1440–1443. doi: 10.1126/science.8128227. [DOI] [PubMed] [Google Scholar]
- 22.Kato H, Nagamine M, Kominami R, Muramatsu M. Formation of the transcription initiation complex on mammalian rDNA. Mol Cell Biol. 1986;6:3418–3427. doi: 10.1128/mcb.6.10.3418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kermekchiev M, Muramatsu M. Presence of an inhibitor of RNA polymerase I mediated transcription in extracts from growth arrested mouse cells. Nucleic Acids Res. 1993;21:447–453. doi: 10.1093/nar/21.3.447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Klein J, Grummt I. Cell cycle-dependent regulation of RNA polymerase I transcription: The nucleolar transcription factor UBF is inactive in mitosis and early G1. Proc Natl Acad Sci USA. 1999;96:6096–6101. doi: 10.1073/pnas.96.11.6096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Landshulz W, Johnson P, Adashi E, Graves B, McKnight S. Isolation of a recombinant copy of the gene encoding C/EBP. Genes Dev. 1988;2:786–800. doi: 10.1101/gad.2.7.786. [DOI] [PubMed] [Google Scholar]
- 26.Manley J, Fire A, Cano A, Sharp P, Gefter M. DNA-dependent transcription of adenovirus genes in a soluble whole-cell extract. Proc Natl Acad Sci USA. 1980;77:3855–3859. doi: 10.1073/pnas.77.7.3855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mauro V, Edelman G. rRNA-like sequences occur in diverse primary transcripts: implication for the control of gene expression. Proc Natl Acad Sci USA. 1997;94:422–427. doi: 10.1073/pnas.94.2.422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Michot B, Bachellerie J-P. Comparisons of large subunit rRNAs reveal some eukaryote-specific elements of secondary structure. Biochemie. 1987;69:11–23. doi: 10.1016/0300-9084(87)90267-7. [DOI] [PubMed] [Google Scholar]
- 29.Munro J, Burdon R, Leader D. Characterization of a human orphan 28S ribosomal DNA. Gene. 1986;48:65–70. doi: 10.1016/0378-1119(86)90352-5. [DOI] [PubMed] [Google Scholar]
- 30.Nagamine M, Kishimoto T, Aono J, Kato H, Kominami R, Muramatsu M. Sequestration analysis for RNA polymerase I transcription factors with various deletion and point mutations reveals different functional regions of the mouse rRNA gene promoter. Mol Cell Biol. 1987;7:1486–1495. doi: 10.1128/mcb.7.4.1486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.O'Mahony D, Smith D, Xie W, Rothblum L. Analysis of the phosphorylation, DNA-binding and dimerization properties of the RNA polymerase I transcription factors UBF1 and UBF2. Nucleic Acids Res. 1992;20:1301–1308. doi: 10.1093/nar/20.6.1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Paule M R, White R J. Survey and summary: transcription by RNA polymerase I and III. Nucleic Acids Res. 2000;28:1283–1298. doi: 10.1093/nar/28.6.1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Perrone-Bizzozero N, Lapalucci-Espinoza S, Medrano E, Franze-Fernandez M. Transcription of rRNA is differentially controlled in resting and growing BALB/c 3T3 cells. J Cell Physiol. 1985;124:160–164. doi: 10.1002/jcp.1041240125. [DOI] [PubMed] [Google Scholar]
- 34.Reeder R. Regulation of RNA polymerase I transcription in yeast and vertebrates. Prog Nucleic Acid Res Mol Biol. 1999;62:293–328. doi: 10.1016/s0079-6603(08)60511-5. [DOI] [PubMed] [Google Scholar]
- 35.Rosen C. Regulation of HIV gene expression by RNA-protein interaction. Trends Genet. 1991;7:9–13. doi: 10.1016/0168-9525(91)90015-i. [DOI] [PubMed] [Google Scholar]
- 36.Rothblum L, Reddy R, Cassidy B. Transcription initiation site of rat ribosomal DNA. Nucleic Acids Res. 1982;10:7345–7356. doi: 10.1093/nar/10.22.7345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rudloff U, Eberhard D, Tora L, Stunnenberg H, Grummt I. TBP-associated factors interact with DNA and govern species specificity of RNA polymerase I transcription. EMBO J. 1994;13:2611–2616. doi: 10.1002/j.1460-2075.1994.tb06551.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sambrook J, Fritsch E, Maniatis T. Molecular cloning: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 39.Schnapp A, Pfleiderer C, Rosenbauer H, Grummt I. A growth-dependent transcription initiation factor (TIF-IA) interacting with RNA polymerase I regulates mouse ribosomal RNA synthesis. EMBO J. 1990;9:2857–2863. doi: 10.1002/j.1460-2075.1990.tb07475.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Singh H, LeBowitz J, Baldwin A, Sharp P. Molecular cloning of an enhancer binding protein: isolation by screening of an expression library with a recognition site DNA. Cell. 1988;52:415–423. doi: 10.1016/s0092-8674(88)80034-5. [DOI] [PubMed] [Google Scholar]
- 41.Sirri V, Roussel P, Hernandez-Verdun D. The mitotically phosphorylated form of the transcription termination factor TTF-1 is associated with the repressed rDNA transcription. J Cell Sci. 1999;112:3259–3268. doi: 10.1242/jcs.112.19.3259. [DOI] [PubMed] [Google Scholar]
- 42.Tenson T, De Blasio A, Mankin A. A functional peptide encoded in the E. coli 23S rRNA. Proc Natl Acad Sci USA. 1996;93:5641–5646. doi: 10.1073/pnas.93.11.5641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Upcroft J, Healey A, Mitchell R, Boreham P, Upcroft P. Antigen expression from the ribosomal DNA repeat unit of Giardia intestinalis. Nucleic Acids Res. 1990;18:7077–7081. doi: 10.1093/nar/18.23.7077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Voit R, Hoffman M, Grummt I. Phosphorylation by G1-specific cdk-cyclin complexes activates the nucleolar transcription factor UBF. EMBO J. 1999;18:1891–1899. doi: 10.1093/emboj/18.7.1891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang S, Pritle I, Pritle R. A human 28S ribosomal RNA retropseudogene. Gene. 1997;196:105–111. doi: 10.1016/s0378-1119(97)00214-x. [DOI] [PubMed] [Google Scholar]
- 46.Xie W, Rothblum L. Domains of the rat rDNA promoter must be aligned stereospecifically. Mol Cell Biol. 1992;12:1266–1275. doi: 10.1128/mcb.12.3.1266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yamamoto K, Koga A, Yamamoto M, Nishi Y, Tamura T, Nogi Y, Muramatsu M. Identification of a novel 70 kDa protein that binds to the core promoter element and is essential for ribosomal DNA transcription. Nucleic Acids Res. 2000;28:1199–1205. doi: 10.1093/nar/28.5.1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhai W, Comai L. Repression of RNA polymerase I transcription by the tumor suppressor p53. Mol Cell Biol. 2000;20:5930–5938. doi: 10.1128/mcb.20.16.5930-5938.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhai W, Tuan A, Comai L. SV40 large T antigen binds to the TBP-TAF(I) complex SLI and coactivates ribosomal RNA transcription. Genes Dev. 1997;11:1605–1617. doi: 10.1101/gad.11.12.1605. [DOI] [PubMed] [Google Scholar]