Highlights
-
•
An association between Alzheimer’s disease genetic rinks with static and dynamic functional network connectivity was observed.
-
•
Participants with higher risk of Alzheimer’s disease showed less visual sensory connectivity.
-
•
Participants with lower risk of Alzheimer’s disease showed more cognitive control network connectivity.
-
•
Sex has a significant contribution in the link between Alzheimer’s disease genetic risks with static and dynamic functional network connectivity.
Keywords: Alzheimer's disease, Genetic risk resting-state fMRI static functional network connectivity, Dynamic functional network connectivity sex difference
Abstract
Apolipoprotein E (APOE) polymorphic alleles are genetic factors associated with Alzheimer’s disease (AD) risk. Although previous studies have explored the link between AD genetic risk and static functional network connectivity (sFNC), to the best of our knowledge, no previous studies have evaluated the association between dynamic FNC (dFNC) and AD genetic risk. Here, we examined the link between sFNC, dFNC, and AD genetic risk with a data-driven approach. We used rs-fMRI, demographic, and APOE data from cognitively normal individuals (N = 886) between 42 and 95 years of age (mean = 70 years). We separated individuals into low, moderate, and high-risk groups. Using Pearson correlation, we calculated sFNC across seven brain networks. We also calculated dFNC with a sliding window and Pearson correlation. The dFNC windows were partitioned into three distinct states with k-means clustering. Next, we calculated the proportion of time each subject spent in each state, called occupancy rate or OCR and frequency of visits. We compared both sFNC and dFNC features across individuals with different genetic risks and found that both sFNC and dFNC are related to AD genetic risk. We found that higher AD risk reduces within-visual sensory network (VSN) sFNC and that individuals with higher AD risk spend more time in a state with lower within-VSN dFNC. We also found that AD genetic risk affects whole-brain sFNC and dFNC in women but not men. In conclusion, we presented novel insights into the links between sFNC, dFNC, and AD genetic risk.
1. Introduction
Alzheimer’s disease (AD) is the most prevalent age-related dementia in individuals above 65 years of age (Masters et al., 2015). While global biomedical research efforts for AD prevention have expanded, the number of individuals affected by AD is still growing significantly every year. Even though there is no effective AD therapy to date, some medications can slow down disease progression (Yiannopoulou and Papageorgiou, 2020). It has been hypothesized that AD progression begins affecting brain functional connectivity many years prior to disease onset (Agosta et al., 2012, Demirtaş et al., 2017). As such, knowing how AD risk alters brain connectivity in cognitively normal individuals might shed light on the mechanisms associated with AD development later in life.
While previous studies showed that environmental factors such as diet, living in rural versus urban areas, smoking, not exercising, and infections are risk factors for AD, genetic factors are believed to contribute 70% to AD risk (Elsheikh et al., 2020, Van Cauwenberghe et al., 2016). Apolipoprotein E polymorphic alleles are genetic factors linked to Alzheimer’s disease (AD). There are three common alleles, including ɛ2, ɛ3, and ɛ4, that can produce six genotypes such as ɛ2/ɛ2, ɛ2/ɛ3, ɛ2/ɛ4, ɛ3/ɛ3, ɛ3/ɛ4, and ɛ4/ɛ4 in which one gene is inherited from the father and the other is from the mother (Yamazaki et al., 2019). Individuals carrying the ɛ4 allele have the highest risk of AD and younger mean age at dementia onset compared to those carrying the ɛ3 and ɛ2 alleles, whereas individuals with the ɛ2 allele have the lowest chance and older mean age of dementia onset (Bird, 2008).
Previous studies explored the link between AD's genetic risk and static functional network connectivity or sFNC (Axelrud et al., 2019, Chiesa et al., 2017, McKenna et al., 2016, Turney et al., 2020). For example, a recent study found that individuals carrying ɛ4 have lower temporal default mode network (DMN) functional connectivity than those without ɛ4 (Turney et al., 2020). Another study showed an increase in functional connectivity between the hippocampus and prefrontal/parietal/temporal cortex in healthy individuals carrying ɛ4 (Zheng et al., 2018). Despite extensive research on the effect of ɛ4 on rs-fMRI sFNC, the effects of other alleles (i.e., ɛ2 and ɛ3) on sFNC have not been explored. Additionally, previous literature that studied the APOE effect on FNC assumed that FNC is static over time and ignored its dynamics. However, recent studies have shown that FNC is highly dynamic during both task and resting conditions (Allen et al., 2014, Sendi et al., 2021a, Sendi et al., 2021b). Therefore, we hypothesized here that studying the dynamics of whole-brain FNC might give insight into how the APOE could disrupt FNC in AD.
This study aimed to explore how dFNC and sFNC differ among individuals with different genetic risks for AD using a relatively large dataset (N >850). To model the dFNC of each participant, we utilized a sliding window approach followed by k-means clustering to estimate a set of connectivity states (Allen et al., 2014). Next, we modeled the temporal changes by calculating the occupancy rate (OCR) and frequency of visits of each state from the dFNC. Next, we explored the difference between cognitively normal participants with different AD risk levels via statistical analysis of the estimated OCR features and the frequency of visits. In addition, we compared sFNC cell-wise differences between cognitively normal individuals differing in genetic risk of AD.
2. Methods and materials
2.1. Participants and dataset
Neuroimaging data of 886 cognitively normal brains (354 females) and their associated demographic information from the longitudinal Open Access Series of Imaging Studies (OASIS)-3 cohort was used in this study (LaMontagne et al., 2019). The participants' cognitive functionality at the time of scanning was evaluated by the clinical dementia rating scale (CDR) scores, and the CDR scores were equal to 0. The participants' age at scanning time ranged from 42.46 years to 95.39 years, with a mean of 70.13 years. We divided the data into three groups, including the low genetic risk of AD or LGR_AD (N = 127, 55 females), consisting of all individuals with at least one ɛ2 allele (i.e., ɛ2/ɛ2 and ɛ2/ɛ3), moderate genetic risk of AD or MGR_AD (N = 558, 219 females), containing all individuals with only ɛ3 allele (i.e., ɛ3/ɛ3), and high genetic risk of AD or HGR_AD (N = 201, 80 females) consisting of all individuals with at least one ɛ4 allele (i.e., ɛ3/ɛ4 and ɛ4/ɛ4). The demographic and clinical information of each group is shown in Table 1. No significant age, gender, and mini-mental state examination differences were observed between any pair of groups (p > 0.05).
Table 1.
The peak coordinates of components and labels.
| Component Name |
Peak Coordinate (mm) |
||||
|---|---|---|---|---|---|
| 1 | SCN | Caudate (69) | 6.5 | 10.5 | 5.5 |
| 2 | Subthalamus/hypothalamus (53) | −2.5 | −13.5 | −1.5 | |
| 3 | Putamen (98) | −26.5 | 1.5 | −0.5 | |
| 4 | Caudate (99) | 21.5 | 10.5 | −3.5 | |
| 5 | Thalamus (45) | −12.5 | −18.5 | 11.5 | |
| 6 | AUN | Superior temporal gyrus ([STG], 21) | 62.5 | –22.5 | 7.5 |
| 7 | Middle temporal gyrus ([MTG], 56) | −42.5 | −6.5 | 10.5 | |
| 8 | SMN | Postcentral gyrus ([PoCG], 3) | 56.5 | −4.5 | 28.5 |
| 9 | Left postcentral gyrus ([L PoCG], 9) | −38.5 | –22.5 | 56.5 | |
| 10 | Paracentral lobule ([ParaCL], 2) | 0.5 | –22.5 | 65.5 | |
| 11 | Right postcentral gyrus ([R PoCG], 11) | 38.5 | −19.5 | 55.5 | |
| 12 | Superior parietal lobule ([SPL], 27) | −18.5 | −43.5 | 65.5 | |
| 13 | Paracentral lobule ([ParaCL], 54) | −18.5 | −9.5 | 56.5 | |
| 14 | Precentral gyrus ([PreCG], 66) | −42.5 | −7.5 | 46.5 | |
| 15 | Superior parietal lobule ([SPL], 80) | 20.5 | −63.5 | 58.5 | |
| 16 | Postcentral gyrus ([PoCG], 72) | −47.5 | −27.5 | 43.5 | |
| 17 | VSN | Calcarine gyrus ([CalcarineG], 16) | −12.5 | −66.5 | 8.5 |
| 18 | Middle occipital gyrus ([MOG], 5) | –23.5 | −93.5 | −0.5 | |
| 19 | Middle temporal gyrus ([MTG], 62) | 48.5 | −60.5 | 10.5 | |
| 20 | Cuneus (15) | 15.5 | −91.5 | 22.5 | |
| 21 | Right middle occipital gyrus ([R MOG], 12) | 38.5 | −73.5 | 6.5 | |
| 22 | Fusiform gyrus (93) | 29.5 | −42.5 | −12.5 | |
| 23 | Inferior occipital gyrus ([IOG], 20) | −36.5 | −76.5 | −4.5 | |
| 24 | Lingual gyrus ([LingualG], 8) | −8.5 | −81.5 | −4.5 | |
| 25 | Middle temporal gyrus ([MTG], 77) | −44.5 | −57.5 | −7.5 | |
| 26 | CCN | Inferior parietal lobule ([IPL], 68) | 45.5 | −61.5 | 43.5 |
| 27 | Insula (33) | −30.5 | 22.5 | −3.5 | |
| 28 | Superior medial frontal gyrus ([SMFG], 43) | −0.5 | 50.5 | 29.5 | |
| 29 | Inferior frontal gyrus ([IFG], 70) | −48.5 | 34.5 | −0.5 | |
| 30 | Right inferior frontal gyrus ([R IFG], 61) | 53.5 | 22.5 | 13.5 | |
| 31 | Middle frontal gyrus ([MiFG], 55) | −41.5 | 19.5 | 26.5 | |
| 32 | Inferior parietal lobule ([IPL], 63) | −53.5 | −49.5 | 43.5 | |
| 33 | Left inferior parietal lobue ([R IPL], 79) | 44.5 | −34.5 | 46.5 | |
| 34 | Supplementary motor area ([SMA], 84) | −6.5 | 13.5 | 64.5 | |
| 35 | Superior frontal gyrus ([SFG], 96) | −24.5 | 26.5 | 49.5 | |
| 36 | Middle frontal gyrus ([MiFG], 88) | 30.5 | 41.5 | 28.5 | |
| 37 | Hippocampus ([HiPP], 48) | 23.5 | −9.5 | −16.5 | |
| 38 | Left inferior parietal lobue ([L IPL], 81) | 45.5 | −61.5 | 43.5 | |
| 39 | Middle cingulate cortex ([MCC], 37) | −15.5 | 20.5 | 37.5 | |
| 40 | Inferior frontal gyrus ([IFG], 67) | 39.5 | 44.5 | −0.5 | |
| 41 | Middle frontal gyrus ([MiFG], 38) | −26.5 | 47.5 | 5.5 | |
| 42 | Hippocampus ([HiPP], 83) | −24.5 | −36.5 | 1.5 | |
| 43 | DMN | Precuneus (32) | −8.5 | −66.5 | 35.5 |
| 44 | Precuneus (40) | −12.5 | −54.5 | 14.5 | |
| 45 | Anterior cingulate cortex ([ACC], 23) | −2.5 | 35.5 | 2.5 | |
| 46 | Posterior cingulate cortex ([PCC], 71) | −5.5 | −28.5 | 26.5 | |
| 47 | Anterior cingulate cortex ([ACC], 17) | −9.5 | 46.5 | −10.5 | |
| 48 | Precuneus (51) | −0.5 | −48.5 | 49.5 | |
| 49 | Posterior cingulate cortex ([PCC], 94) | −2.5 | 54.5 | 31.5 | |
| 50 | CB | Cerebellum ([CB], 13) | −30.5 | −54.5 | −42.5 |
| 51 | Cerebellum ([CB], 18) | –32.5 | −79.5 | −37.5 | |
| 52 | Cerebellum ([CB], 4) | 20.5 | −48.5 | −40.5 | |
| 53 | Cerebellum ([CB], 7) | 30.5 | −63.5 | −40.5 | |
2.2. Imaging acquisition protocol
The T2*-weighted functional images were collected via echoplanar imaging (EPI) using Trio 3 T scanners with 20-channel head coils (Siemens Medical Solutions USA, Inc.). The data collecting protocol includes TE = 27 ms, TR = 2.2 s, flip angle = 90˚, slice thickness = 4 mm, slice gap (center-to-center) = 4 mm, matrix = 64×64, number of slices = 32, and field of view = 256×256 mm2. The duration of the scanning was 6 min.
2.3. Preprocessing
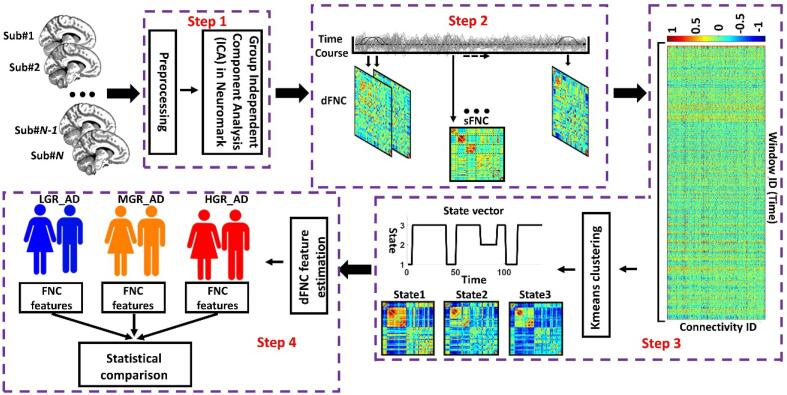
Fig. 1 shows the analytic pipeline that we used in this study. The following steps describe the details of our method. The first step (Step 1 in Fig. 1) removed the first five dummy scans before the preprocessing. We used statistical parametric mapping (SPM12, https://www.fil.ion.ucl.ac.uk/spm/) default slice timing routines. In this method, we used the slice acquired in the middle of the sequence as the reference slice. We applied rigid body motion correction to adjust for participant head movement. Next, we normalized the imaging data to the standard Montreal Neurological Institute (MNI) space using the echo-planar imaging (EPI) template. Finally, we smoothed the images by applying a Gaussian kernel with a full width at half maximum (FWHM) of 6 mm.
Fig. 1.
Analytic pipeline: In Step 1, fifty-three time-course components of the whole brain were identified using group-independent component analysis (ICA). In Step 2, a taper sliding window was used to segment the time-course signals and calculate the functional network connectivity (FNC). After vectorizing the FNC matrixes, we concatenated them and used k-means clustering, k = 3, to group them into three distinct states (Step 3). The elbow criteria were used to find the optimal k. In addition, the correlation distance metric was used for the clustering. Then, based on the state vector of each subject, the occupancy rate or OCR, in total 3 features, and the frequency of visits of each state, in total 3 features, were calculated from the state vector of each subject. We next compared the OCR among groups with a two-sample t-test. We adjusted all p values by the Benjamini-Hochberg false discovery rate (FDR) correction in each analysis (Step 4). LGR: LGR-AD: Low genetic risk of AD, MGR-AD: Moderate genetic risk of AD, HGR-AD: High genetic risk of AD.
2.4. Extracting independent components using Neuromark
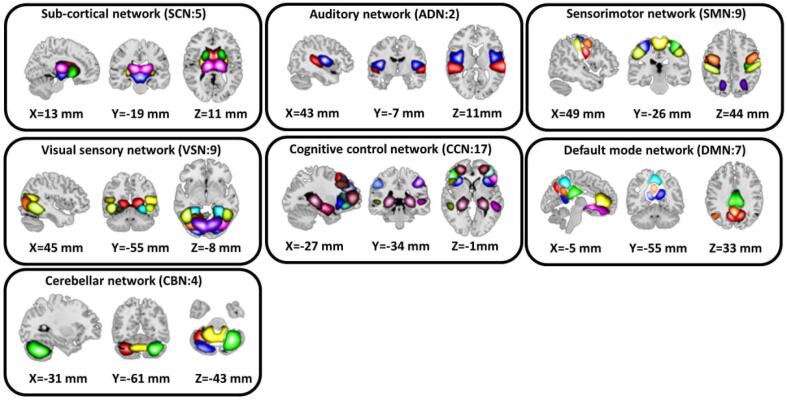
This study used a set of robust network priors to extract comparable components across subjects from the OASIS dataset. The network priors were extracted via the Neuromark pipeline (Du et al., 2020). This framework performed group ICA with model order as 100 on two healthy controls datasets, human connectome project (HCP: https://www.humanconnectome.org/study/hcp-young-adult/document/1200-subjects-data-release, 823 subjects after the subject selection) and genomics superstruct project (GSP: https://dataverse.harvard.edu/dataverse/GSP, 1005 subjects after the subject selection) for creating the network priors. The extracted ICs from the two datasets were matched by comparing the corresponding group-level spatial maps. If they showed a higher spatial correlation than a given threshold of 0.4, we considered that the IC pairs were reproducible. The reproducible ICs pairs were further evaluated by examining their spatial activations and low-frequency fluctuations of their corresponding time courses (TCs). 53 pairs of ICs were identified as meaningful and were arranged into seven functional domains based on anatomic and functional prior knowledge. These seven functional domains were the subcortical network (SCN), auditory network (ADN), sensorimotor network (SMN), visual network (VSN), cognitive control network (CCN), default-mode network (DMN), and cerebellar network (CBN) as shown in Fig. 2. The peak coordinates of components are shown in Table 1.
Fig. 2.
We used the Neuromark pipeline to extract reliable intrinsic connectivity networks (ICNs, in total, 53 components) for the whole brain that are replicated across independent datasets. We put them into seven networks, including subcortical network (SCN), auditory network (AND), visual sensory network (VSN), sensorimotor network (SMN), cognitive control network (CCN), default mode network (DMN), and cerebellar network (CBN).
Finally, after obtaining the subject-specific time courses, we did additional post-processing on the time courses to remove the noise, including 1) detrending linear, quadratic, and cubic trends, 2) multiple regression of the 6 realignment parameters and their derivatives, 3) removal of detected outliers, and 4) low-pass filtering with a cutoff frequency of 0.15 Hz. Our filtering was performed on the TC of ICs, not on the voxel-based fMRI data because we want to retain more information on fMRI for ICA decomposition. This strategy has been successfully applied in a wide range of previous ICA-based dFNC studies (Dini et al., 2021, Du et al., 2020, Sendi et al., 2021a, Sendi et al., 2021b).
2.5. Dynamic and static functional network connectivity estimation
The sliding window approach with a tapered window (size of 44 s and standard deviation of 3 s) was used to estimate the whole-brain dFNC. We used Pearson correlation to assess the functional network connectivity among all 53 ICs within each window. With 53 ICNs, the symmetric matrix size is 53×53. Additionally, with 53 ICNs, we had = 1378 connectivity features within each window. Next, we concatenated the functional network connectivity estimates of each window for each individual to form an (F × T) array (where F = 1378 denotes the number of connectivity features and T = 139 is the number of windows for each participant), which represented the changes in brain connectivity between ICNs as a function of time. Next, we concatenated all matrices across all participants (Step 2 in Fig. 1).
2.6. Dynamic functional network connectivity clustering
In the next step, we used the k-means clustering approach to assign the windowed FNCs of all participants into a set of clusters (or states). The optimal number of clusters (k) was set to three based upon the elbow criterion, e.g., “the ratio of within-cluster to between cluster distance.” Pearson correlation was used as a distance metric, and 1000 iterations were used. Even though we randomly set the maximum number of iterations to 1000 times, we did not observe any failure in convergence with this number. The output of k-means clustering was three states for the entire group of all 886 participants and a state vector for each individual. The state vector represents changes in whole-brain FNC over time. For example, in the state vector shown in Fig. 1, the participant is in state1, then state 3, state1, state3, state2, state3, state1, and finally, state 3. This pattern would be different for another participant. Next, we calculated the proportion of each participant's time spent in each state, called occupancy rate (OCR) hereafter. To calculate OCR in state i for each participant, we counted all windows in state i belonging to that participant and then divided it by 139 (the entire number of windows). Having three states, we estimated three OCRs for each individual. Additionally, we calculated the number of transitions to state i as the frequency of visits for that state. With three states, we calculated three frequencies of visits for each participant.
2.7. Statistical analysis
A two-sample t-test was used to compare the OCR (number of null hypotheses or N = 3) and the frequency of visits (number of null hypotheses or N = 3) of each pair of groups. Similarly, a two-sample t-test was used to compare the sFNC (number of null hypotheses or N = 1387) of each pair of groups. We adjusted all p values with Benjamini-Hochberg false discovery rate (FDR) correction in both dFNC and sFNC analyses (Yoav Benjamini and Yosef Hochberg, 1995).
3. Results
3.1. Clinical and demographic information
The demographic and clinical information of each group is shown in Table 2. No significant age, gender, and mini-mental state examination differences were observed between any pair of groups (p > 0.05).
Table 2.
Demographic and clinical information of subjects.
| LGR-AD | MGR-AD | HGR-AD | |
|---|---|---|---|
| Number | 127 | 558 | 201 |
| Age | 71.22±8.42 | 70.14±8.58 | 69.37±8.59 |
| Gender (M/F) | 72/55 | 339/219 | 121/80 |
| MMSE | 28.98±1.17 | 29.15±1.11 | 29.12±1.30 |
Note: LGR-AD: Low genetic risk for AD, MGR-AD: Moderate genetic risk for AD, HGR-AD: High genetic risk for AD, MMSE: mini-mental state examination.
3.2. The genetic risk associated with sFNC
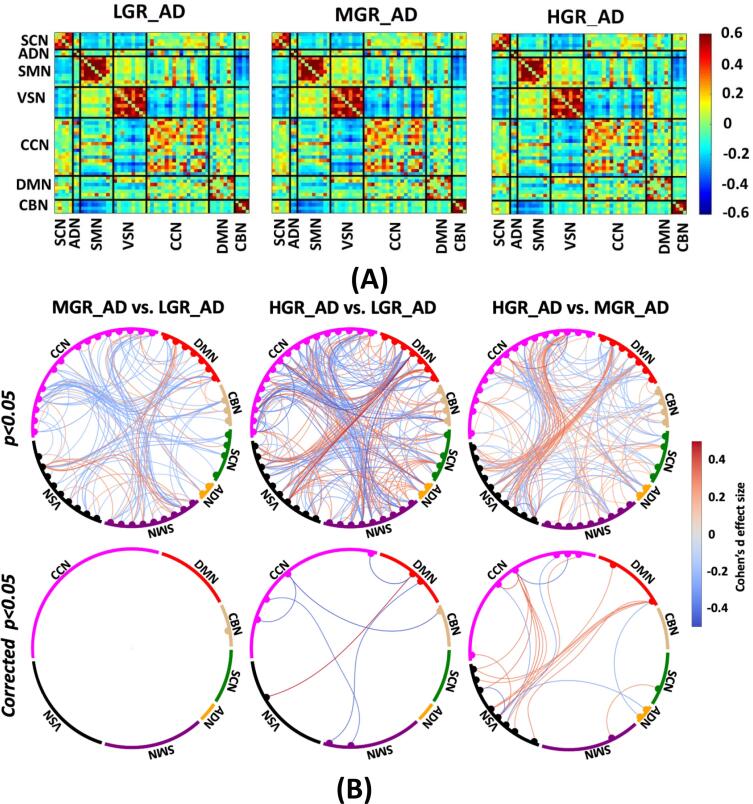
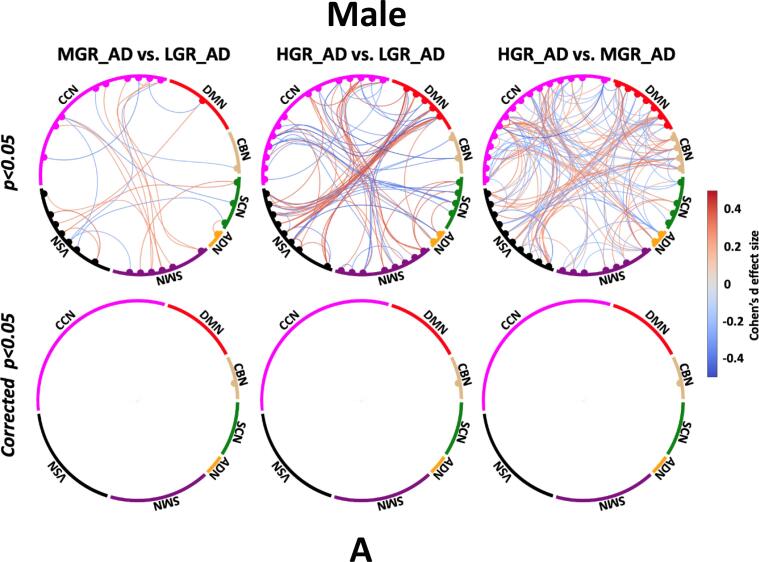
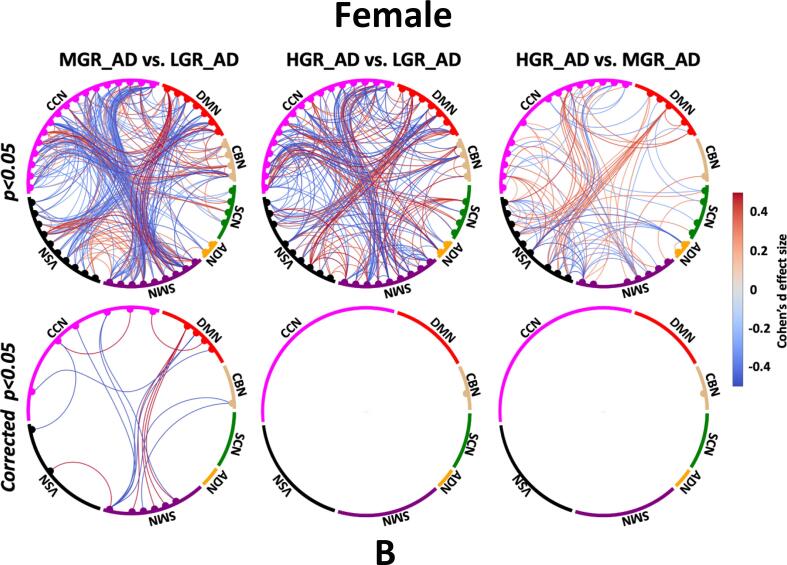
The average sFNC of each group is shown in Fig. 3A. Also, Fig. 3B shows the Cohen’s d effect size when we compared the cell-wise FNC between each pair of groups using a two-sample t-test. Those significant group differences with p < 0.05 and corrected p < 0.05 are shown in Fig. 3B top and bottom panels, respectively. The red and blue colors in this figure show positive and negative differences, respectively. Fig. 3B (left panel) shows the Cohen’s d effect size while we compared LGR_AD with MGR_AD (MGR_AD vs. LGR_AD). We did not observe a significant sFNC difference between LGR_AD and MGR_AD groups after FDR correction. While the difference between LGR_AD and HDR_AD (or HGR_AD vs. LGR_AD) was significant in some networks, as shown in Fig. 3B (middle panel). As Fig. 3B shows, we see significant differences in SMN/CCN (connectivity between SMN and CCN), VSN/DMN, CCN/DBN, and within CCN. Therefore, a pattern of internetwork differences spanning multiple resting-state networks across the whole brain was observed.
Fig. 3.
Estimated sFNC for each group. A) Estimated sFNC for LGR-AD (left panel), MGR-AD (middle panel), and HGR-AD (right panel). The colorbar shows the intensity of sFNC values, B) Top panel: The Cohen’s d effect size of sFNC difference between each pair of groups (uncorrected p < 0.05). Bottom panel: Significant group differences passing the multiple comparison thresholds (false discovery rate [FDR] corrected, p < 0.05). The colorbar shows Cohen’s d effect size calculated using MATLAB toolbox (https://www.mathworks.com/matlabcentral/fileexchange/62957-computecohen_d-x1-x2-varargin.). SCN: subcortical network SCN, ADN: auditory network, SMN: sensory motor network, VSN: visual sensory network, CCN: cognitive control network, DMN: default mode network, and CBN: cerebellar network. LGR-AD: Low genetic risk of AD, MGR-AD: Moderate genetic risk of AD, HGR-AD: High genetic risk of AD. The chord graphs are generated using the NiChord toolbox in Python (https://github.com/paulcbogdan/NiChord).
In contrast, the cell-wise FNC difference between MGR_AD and HDR_AD (or HGR_AD vs. MGR_AD) was more focused on VSN, as shown in Fig. 3B (right panel). As shown in this figure, we found that individuals with a higher risk of AD have less within VSN connectivity than those with a moderate risk of AD. In comparison, the connectivity between VSN and CCN and between VSN and DMN was higher for individuals with higher AD risk.
3.3. Dynamic functional network connectivity states
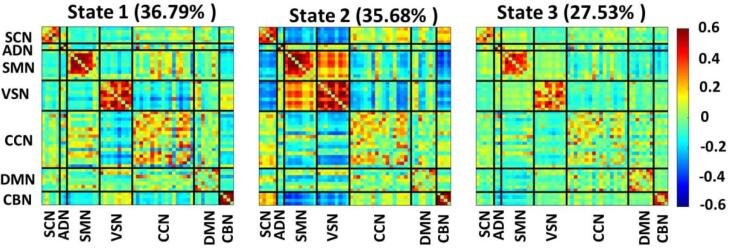
Fig. 4 shows three distinct dFNC states estimated by k-means clustering. State1 and state 2 show more positive connectivity within CCN, within CBN, within SMN, and within VSN relative to state 3. State 2 offers the most positive connectivity among sensory domains (i.e., ADN, SMN, and VSN). Meanwhile, the connectivity between these three domains with the rest of the brain is negative in this state. Additionally, the connectivity between SMN and the rest of the brain is relatively high in state1.
Fig. 4.
Three dFNC states were identified with the k-means clustering method. Each state is a 53×53 matrix in which positive connectivity is shown in red and negative connectivity is shown in blue. We put all 53 components in 7 domains, including the subcortical network (SCN), auditory network (ADN), sensory motor network (SMN), visual sensory network (VSN), cognitive control network (CCN), default mode network (DMN), and cerebellar network (CBN). We found individuals spent 36.79 %, 35.68 %, and 27.53 % in state 1, state 2, and state 3, respectively. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
3.4. Genetics risk associated with dFNC features
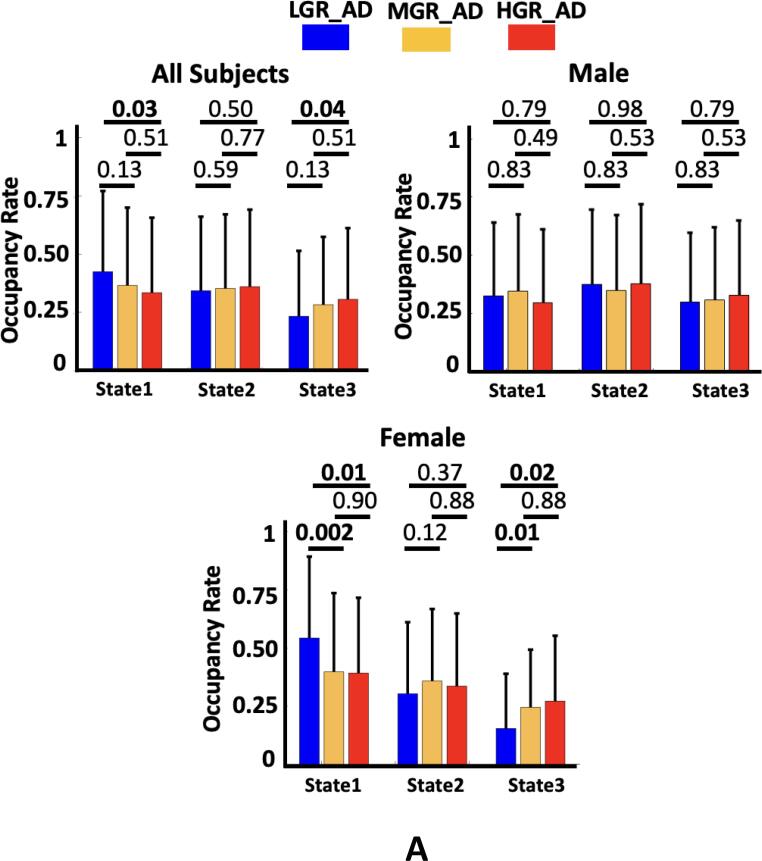
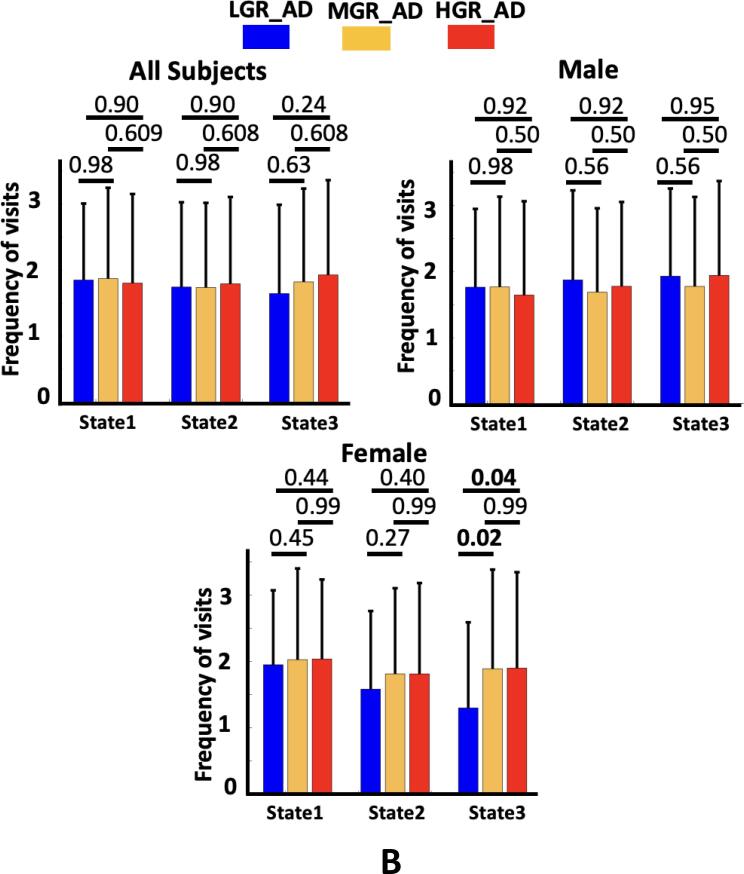
We compared the OCR and frequency of visits of each state across three groups of individuals. The results are shown in Fig. 5A, and Supplementary Table 1 shows the Cohen’s d effect size of each pair of group comparisons. As this figure shows, we found that the OCR of state1 was significantly less for HGR_AD than for LGR_AD (corrected p = 0.03). In contrast, HGR_AD had higher OCR than LGR_AD in state 3 (corrected p = 0.04). Additionally, we did not find any significant OCR difference across groups in state 2. Besides, no significant difference was observed between MGR_AD and LGR_AD and between HGR_AD and MGR_AD. Also, we did not observe any significant difference in the frequency of visits, as shown in Fig. 5B (All subjects).
Fig. 5.
Genetic risk effects on dFNC features. A) The occupancy rate (OCR) of each group in state 1, state 2, and state 3. All individuals: The individuals with the ɛ2 allele spent more time in state 1 than those individuals without the ɛ2 allele (corrected p < 0.05). Male: The occupancy rate (OCR) of each group in state 1, state 2, and state 3. No significant difference was observed among the groups. Female: The occupancy rate (OCR) of each group in state 1, state 2, and state 3. The individuals without the ɛ2 allele spent more time in state 3 than those individuals with the ɛ2 allele (corrected p < 0.05). B) The frequency of visits to states 1, state 2, and state 3. All individuals: No significant difference was observed across the group. Male: No significant difference was observed among the groups. Female: participants with the ɛ2 allele have fewer visits to state 3 than those with the ɛ3 and ɛ4 alleles (corrected p < 0.05). LGR-AD: Low genetic risk of AD, MGR-AD: Moderate genetic risk of AD, HGR-AD: High genetic risk of AD. The error bars represent the standard deviation.
3.5. Gender effect on sFNC and dFNC features
To consider sex effects on the results, we separated men and women in each group of individuals and repeated our analysis. The Cohen’s d effect size group sFNC difference for each sex is shown in Fig. 6. Fig. 6A shows the sFNC difference for each male participant group, and Fig. 6B shows similar results for female. Interestingly, we did not find any significant sFNC difference across groups for males after FDR correction. In contrast, the sFNC difference between LGR_AD and HGR_AD was significant for women, as shown in Fig. 6B (left panel). We did not observe a significant difference between women LGR_AD and MGR_AD and between women MGR_AD and HGR_AD, as shown in the middle and right panel of Fig. 6B. We also studied group differences in dFNC features for both men and women. We did not observe a significant group difference in the OCR and frequency of visits to each state (Fig. 5B) for men like we did for the sFNC analysis. In contrast, the group difference in dFNC features was significant for women. In more detail, we found a significant OCR difference between LGR_AD and MGR_AD and between LGR_AD and HGR_AD in state 1 and state 3 (corrected p < 0.05). Besides, the frequency of visits in state 3 was significantly higher in MGR_AD (corrected p = 0.02) and HGR_AD (corrected p = 0.04) compared with LGR_AD.
Fig. 6.
Sex effects on sFNC. A) Top panel: sFNC differences between pairs of groups for males (uncorrected p < 0.05). Bottom panel: sFNC differences between pairs of groups for males (corrected p < 0.05). B) Top panel: sFNC differences between pairs of groups for females (uncorrected p < 0.05). Bottom panel: sFNC differences between pairs of groups for females (corrected p < 0.05). The colorbar shows Cohen’s d effect size calculated using MATLAB toolbox (https://www.mathworks.com/matlabcentral/fileexchange/62957-computecohen_d-x1-x2-varargin.). SCN: subcortical network SCN, ADN: auditory network, SMN: sensory motor network, VSN: visual sensory network, CCN: cognitive control network, DMN: default mode network, and CBN: cerebellar network. LGR-AD: Low genetic risk of AD, MGR-AD: Moderate genetic risk of AD, HGR-AD: High genetic risk of AD. The chord graphs are generated using the NiChord toolbox in Python (https://github.com/paulcbogdan/NiChord).
4. Discussion
Previous studies showed that FNC estimated from rs-fMRI is highly dynamic even without external input (Allen et al., 2014, Sendi et al., 2021c). Therefore, here we hypothesized that the genetic risk of AD not only alters the strength of the functional connectivity between pairs of brain networks, as would be shown in sFNC, but also the dynamic fluctuations of connectivity among those networks, as would be shown in dFNC. To the best of our knowledge, the present study is the first to report a link between AD genetic risk and dFNC estimated from rs-fMRI recorded of cognitively normal participants. We also compared the results obtained from both sFNC and dFNC data as the measures are complementary and could provide distinct insights into the relationship between brain network connectivity and the genetic risk of AD. Lastly, we examined the effects of an individual’s sex on the degree to which sFNC, dFNC, and the genetic risk of AD are associated.
We observed that AD genetic risk causes a different pattern that spans multiple inter-network connections across the resting-state networks in cognitively normal individuals, which is consistent with previous studies (Sheline et al., 2010, Trachtenberg et al., 2012). We found that genetic risk affects the VSN significantly more than other brain networks. In more detail, we found that individuals carrying ɛ4 have less within-VSN functional connectivity compared to those individuals carrying ɛ3 and not ɛ2. A decrease in VSN activity during a visual task has been reported in individuals carrying ɛ4 (Smith et al., 1999). A recent study reported functional connectivity decreases in the primary, secondary, and associative visual cortices for cognitively normal individuals with the ɛ4 allele compared with ɛ4 allele non-carriers (McKenna et al., 2016). We also found that cognitively normal individuals carrying ɛ4 have higher VSN/CCN than individuals with moderate AD risk and more increased VSN/DMN connectivity relative to individuals with moderate and lower AD risk. We hypothesize that this higher VSN/CCN connectivity in ɛ4 carriers could be a compensatory mechanism to offset the VSN FC reduction in these individuals. A similar compensatory mechanism in functional connectivity has been reported in individuals with ɛ4, enabling them to achieve the same performance level as individuals with ɛ3 in a memory task (Bondi et al., 2005). It is worth mentioning that we did not find a significant sFNC difference between the individuals with the lowest risk and the moderate risk of AD.
We also explored the whole-brain dFNC across cognitively normal individuals with different genetic risks for AD. We observed a significant difference in the OCR of individuals carrying ɛ4 and individuals having ɛ2. At the same time, we did not observe any significant differences between those with ɛ2 and ɛ3 and those with ɛ3 and ɛ4. In more detail, we found that individuals carrying ɛ4 spend more time than those having ɛ2 in state3 with lower within-VSN functional connectivity (relative to state1 and state2). This result is consistent with the result obtained from the sFNC data, which showed that individuals with higher AD genetic risk have less within-VSN functional connectivity. That supported our hypothesis about the genetic risk effects on both the strength and the temporal pattern of FNC estimated from rs-fMRI.
Additionally, we found that individuals with ɛ4 spend more time in state3 with lower SMN, lower CCN, and lower CBN functional connectivity (relative to state1 and state2), while we did not observe a significant difference in those networks by analyzing sFNC data. These results might reveal new evidence of the CCN, VSN, and CBN’s role in differentiating individuals with different genetic risks for AD. We did not observe a significant difference in OCRs of the individuals with a moderate and lower risk of AD, consistent with the sFNC analysis results. These pieces of evidence might suggest that a moderate genetic risk of AD does not affect either brain sFNC or dFNC.
We observed a considerable difference in sFNC when comparing the individuals having ɛ3 with those having ɛ4. However, no significant difference between the two groups was observed by looking at OCR. While previous studies only analyzed sFNC (Axelrud et al., 2019, Chiesa et al., 2017, McKenna et al., 2016, Turney et al., 2020), the results reported above demonstrated the importance of examining both sFNC and dFNC data to differentiate individuals with different AD risks.
We also separated men and women to examine the effect of sex on our results. We did not observe a significant difference across the three groups of men for either sFNC or dFNC data. While a significant difference between women carrying ɛ4 and ɛ2 and between women carrying ɛ4 and ɛ3 was observed in the dFNC analysis, we only found a significant difference between the women with ɛ2 and ɛ4 in the sFNC analysis. We did not observe a significant difference between individuals having ɛ3 and ɛ4 in either analysis. A recent study analyzed 5496 healthy individuals carrying ɛ4 and showed that AD's conversion rate is significantly higher for women (Altmann et al., 2014). A study showed that ɛ4 could be used to predict mild cognitive impairment (MCI) to AD transition, while ɛ4 affects the transition speed in women but not men (Kim et al., 2015). Another study showed a significant reduction in DMN connectivity of females with ɛ4 than females with ɛ3, while this difference was not substantial in males with ɛ4 versus those with ɛ3 (Damoiseaux et al., 2012). Our results showing that differences in sFNC across female groups are significant and that differences in sFNC across male groups are not significant is consistent with these studies mentioned above. Additionally, our current study provided new information about how sex differences significantly contribute to the differentiation of the dFNC of cognitively normal individuals with different AD genetic risks.
A recent study on the Alzheimer's Disease Neuroimaging Initiative dataset found that p-tau levels in cerebrospinal fluid grow faster in female ɛ4 carriers than in noncarriers and males, indicating that the ɛ4 allele has a sex-specific impact (Buckley et al., 2019). This might explain why we see a significant difference in the FNC of female individuals with higher AD genetic risk compared to those with lower AD genetic risk in our study. It could also explain why this pattern has not been observed in male individuals due to slower tau growth in this group. Similarly, we compared sFNC and dFNC feature across ɛ4 carriers and noncarriers and found a significant sFNC difference in females but not in males (Supplementary Fig. 1 and Fig. 2). Similarly, we found a significant sFNC difference between ɛ4 carriers and noncarriers in females but not males. Although previous studies considered the ɛ3 allele to be a neutral factor in AD, a recent study claimed that the ɛ3 allele might be a protective factor rather than a neutral one (de-Almada et al., 2012). Our results may suggest that the ɛ3 allele is a more significant risk factor for AD in women than in men.
It should be noted that our study does have some limitations. In particular, previous studies have indicated that other risk factors, in addition to genetic risk, can lead to AD (Durazzo et al., 2014, Rahman et al., 2020, Thomas et al., 2020). For example, a recent study showed that individuals with diabetes and the ɛ4 allele demonstrated a faster functional decline than those without diabetes (Thomas et al., 2020). Other confounding factors like smoking (Durazzo et al., 2014), physical activity (Meng et al., 2020), and education levels (Meng and Arcy, 2012) could introduce some bias into our results. This information was not included in the dataset. Future studies are needed to explore AD genetic risk factors combined with other potential risk factors in both sFNC and dFNC data.
In conclusion, by analyzing the link between AD genetic risk with sFNC and dFNC for the first time, we found that AD genetic risk affects both sFNC and dFNC and that each analysis provides information about different aspects of the effects of AD risk on brain connectivity. When analyzing sFNC data, it was possible to differentiate people with lower risk from those with higher risk and people with moderate risk from those with higher risk. An analysis of dFNC showed that people with low risk could be discriminated from those with higher risk and that the SMN and CBN helped differentiate the two groups. When analyzing sFNC and dFNC from individuals of both sexes, we found that a higher risk of AD is associated with a reduction in within-VSN connectivity and an increase in VSN/DMN connectivity, potentially as a compensatory mechanism. However, when analyzing only women, we did not observe a similar compensatory mechanism. The lack of a compensatory mechanism in women could explain the higher AD conversion rate in women that has been identified in previous studies (Altmann et al., 2014). Additionally, our findings suggested that having only an ɛ3 allele could be more of a risk factor in women than in men. Our results shed new light on the genetic risk interactions for AD and brain connectivity in cognitively normal individuals and could assist future diagnostic and treatment efforts.
Declaration of Competing Interest
Elizabeth Mormino provides consulting services for Neurotrack and Eli Lilly. David Salat is a founder and has equity interest in Niji Corp. The remaining authors declare no competing interests.
Acknowledgments
Acknowledgments
We thank those who collected the data and the participants of this study.
Funding
This work was supported by the NIH grants: R01AG063153, R01EB020407, R01MH094524, R01MH119069, R01MH118695, and R01MH121101. Time spent preparing the revised manuscript for publication was supported by T32MH125786 (to W. Carlezon/K. Ressler, MPIs).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.nicl.2023.103363.
Contributor Information
Mohammad S.E. Sendi, Email: msendi@mclean.harvard.edu.
Vince D. Calhoun, Email: vcalhoun@gsu.edu.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
Data availability
Data will be made available on request.
References
- Agosta F., Pievani M., Geroldi C., Copetti M., Frisoni G.B., Filippi M. Resting state fMRI in Alzheimer’s disease: Beyond the default mode network. Neurobiology of Aging. 2012;33:1564–1578. doi: 10.1016/j.neurobiolaging.2011.06.007. [DOI] [PubMed] [Google Scholar]
- Allen E.A., Damaraju E., Plis S.M., Erhardt E.B., Eichele T., Calhoun V.D. Tracking whole-brain connectivity dynamics in the resting state. Cerebral Cortex. 2014;24:663–676. doi: 10.1093/cercor/bhs352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altmann A., Tian L., Henderson V.W., Greicius M.D. Sex modifies the APOE-related risk of developing Alzheimer disease. Annals of Neurology. 2014;75:563–573. doi: 10.1002/ana.24135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Axelrud L.K., Sato J.R., Santoro M.L., Talarico F., Pine D.S., Rohde L.A., Zugman A., Junior E.A., Bressan R.A., Grassi-Oliveira R., Pan P.M., Hoffmann M.S., Simioni A.R., Guinjoan S.M., Hakonarson H., Brietzke E., Gadelha A., Pellegrino da Silva R., Hoexter M.Q., Miguel E.C., Belangero S.I., Salum G.A. Genetic risk for Alzheimer’s disease and functional brain connectivity in children and adolescents. Neurobiology of Aging. 2019;82:10–17. doi: 10.1016/j.neurobiolaging.2019.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bird T.D. Genetic aspects of Alzheimer disease. Genetics in Medicine. 2008;10:231–239. doi: 10.1097/GIM.0b013e31816b64dc. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bondi M.W., Houston W.S., Eyler L.T., Brown G.G. fMRI evidence of compensatory mechanisms in older adults at genetic risk for Alzheimer disease. Neurology. 2005;64:501–508. doi: 10.1212/01.WNL.0000150885.00929.7E. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckley R.F., Mormino E.C., Chhatwal J., Schultz A.P., Rabin J.S., Rentz D.M., Acar D., Properzi M.J., Dumurgier J., Jacobs H., Gomez-Isla T., Johnson K.A., Sperling R.A., Hanseeuw B.J. Associations between baseline amyloid, sex, and APOE on subsequent tau accumulation in cerebrospinal fluid. Neurobiology of Aging. 2019;78:178–185. doi: 10.1016/j.neurobiolaging.2019.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiesa P.A., Cavedo E., Lista S., Thompson P.M., Hampel H. Revolution of Resting-State Functional Neuroimaging Genetics in Alzheimer’s Disease. Trends in Neurosciences. 2017;40:469–480. doi: 10.1016/j.tins.2017.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damoiseaux J.S., Seeley W.W., Zhou J., Shirer W.R., Coppola G., Karydas A., Rosen H.J., Miller B.L., Kramer J.H., Greicius M.D. Gender modulates the APOE ε4 effect in healthy older adults: Convergent evidence from functional brain connectivity and spinal fluid tau levels. Journal of Neuroscience. 2012;32:8254–8262. doi: 10.1523/JNEUROSCI.0305-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de-Almada, B.V.P., de-Almeida, L.D., Camporez, D., de-Moraes, M. V.D., Morelato, R.L., Perrone, A.M.S., Belcavello, L., Louro, I.D., de-Paula, F., 2012. Protective effect of the APOE-e3 allele in Alzheimer’s disease. Brazilian Journal of Medical and Biological Research 45, 8–12. https://doi.org/10.1590/S0100-879X2011007500151. [DOI] [PMC free article] [PubMed]
- Demirtaş M., Falcon C., Tucholka A., Gispert J.D., Molinuevo J.L., Deco G. A whole-brain computational modeling approach to explain the alterations in resting-state functional connectivity during progression of Alzheimer’s disease. NeuroImage: Clinical. 2017;16:343–354. doi: 10.1016/j.nicl.2017.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dini H., Sendi M.S.E., Sui J., Fu Z., Espinoza R., Narr K.L., Qi S., Abbott C.C., van Rooij S.J.H., Riva-Posse P., Bruni L.E., Mayberg H.S., Calhoun V.D. Dynamic Functional Connectivity Predicts Treatment Response to Electroconvulsive Therapy in Major Depressive Disorder. Front Hum Neurosci. 2021;15:1–11. doi: 10.3389/fnhum.2021.689488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du Y., Fu Z., Sui J., Gao S., Xing Y., Lin D., Salman M., Abrol A., Rahaman M.A., Chen J., Hong L.E., Kochunov P., Osuch E.A., Calhoun V.D. NeuroMark: An automated and adaptive ICA based pipeline to identify reproducible fMRI markers of brain disorders. Neuroimage Clin. 2020;28 doi: 10.1016/j.nicl.2020.102375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durazzo T.C., Mattsson N., Weiner M.W. Smoking and increased Alzheimer ’ s disease risk : A review of. Alzheimer’s & Dementia. 2014;10:S122. doi: 10.1016/j.jalz.2014.04.009. S145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elsheikh S.S.M., Chimusa E.R., Mulder N.J., Crimi A. Genome-Wide Association Study of Brain Connectivity Changes for Alzheimer’s Disease. Scientific Reports. 2020;10:1–16. doi: 10.1038/s41598-020-58291-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S., Kim M.J., Kim S., Kang H.S., Lim S.W., Myung W., Lee Y., Hong C.H., Choi S.H., Na D.L., Seo S.W., Ku B.D., Kim S.Y., Kim S.Y., Jeong J.H., Park S.A., Carroll B.J., Kim D.K. Gender differences in risk factors for transition from mild cognitive impairment to Alzheimer’s disease: A CREDOS study. Comprehensive Psychiatry. 2015;62:114–122. doi: 10.1016/j.comppsych.2015.07.002. [DOI] [PubMed] [Google Scholar]
- LaMontagne, P.J., Benzinger, T.L.S., Morris, J.C., Keefe, S., Hornbeck, R., Xiong, C., Grant, E., Hassenstab, J., Moulder, K., Vlassenko, A., Raichle, M.E., Cruchaga, C., Marcus, D., 2019. OASIS-3: Longitudinal Neuroimaging, Clinical, and Cognitive Dataset for Normal Aging and Alzheimer Disease. medRxiv. https://doi.org/10.1101/2019.12.13.19014902.
- Masters C.L., Bateman R., Blennow K., Rowe C.C., Sperling R.A., Cummings J.L. Alzheimer’s disease. Nature Reviews Disease Primers. 2015;1:1–18. doi: 10.1038/nrdp.2015.56. [DOI] [PubMed] [Google Scholar]
- McKenna F., Koo B.B., Killiany R. For the Alzheimer’s Disease Neuroimaging Initiative, Comparison of ApoE-related brain connectivity differences in early MCI and normal aging populations: an fMRI study. Brain Imaging and Behavior. 2016;10:970–983. doi: 10.1007/s11682-015-9451-z. [DOI] [PubMed] [Google Scholar]
- Meng X., Arcy C.D. Education and Dementia in the Context of the Cognitive Reserve Hypothesis : A Systematic Review with Meta- Analyses and Qualitative Analyses. PLoS ONE. 2012;7 doi: 10.1371/journal.pone.0038268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng Q., Lin M., Tzeng I. Relationship Between Exercise and Alzheimer ’ s Disease : A Narrative Literature Review. Frontiers in Neuroscience. 2020;14:1–6. doi: 10.3389/fnins.2020.00131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahman M., White E.M., Thomas K.S., Jutkowitz E. Assessment of Rural-Urban Differences in Health Care Use and Survival Among Medicare Beneficiaries With Alzheimer Disease and Related Dementia. JAMA Network Open. 2020;3:1–13. doi: 10.1001/jamanetworkopen.2020.22111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sendi M.S.E., Pearlson G.D., Mathalon D.H., Ford J.M., Preda A., van Erp T.G.M., Calhoun V.D. Multiple overlapping dynamic patterns of the visual sensory network in schizophrenia. Schizophr Res. 2021;228:103–111. doi: 10.1016/j.schres.2020.11.055. [DOI] [PubMed] [Google Scholar]
- Sendi M.S.E., Zendehrouh E., Fu Z., Liu J., Du Y., Mormino E., Salat D.H., Calhoun V.D., Miller, Robyn L. Disrupted Dynamic Functional Network Connectivity Among Cognitive Control Networks in the Progression of Alzheimer’s Disease. Brain Connect. 2021;1–10 doi: 10.1089/brain.2020.0847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sendi M.S.E., Zendehrouh E., Miller R.L., Fu Z., Du Y., Liu J., Mormino E.C., Salat D.H., Calhoun V.D. Alzheimer’s Disease Projection From Normal to Mild Dementia Reflected in Functional Network Connectivity: A Longitudinal Study. Frontiers in Neural Circuits. 2021;14 doi: 10.3389/fncir.2020.593263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheline Y.I., Morris J.C., Snyder A.Z., Price J.L., Yan Z., D’Angelo G., Liu C., Dixit S., Benzinger T., Fagan A., Goate A., Mintun M.A. APOE4 allele disrupts resting state fMRI connectivity in the absence of amyloid plaques or decreased CSF Aβ42. Journal of Neuroscience. 2010;30:17035–17040. doi: 10.1523/JNEUROSCI.3987-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith C.D., Andersen A.H., Kryscio R.J., Schmitt F.A., Kindy M.S., Blonder L.X., Avison M.J. Altered brain activation in cognitively intact individuals at high risk for Alzheimer’s disease. Neurology. 1999;53:1391–1396. doi: 10.1212/wnl.53.7.1391. [DOI] [PubMed] [Google Scholar]
- Thomas K.R., Bangen K.J., Weigand A.J., Edmonds E.C., Sundermann E., Wong C.G., Joel S., Werhane M.L., Delano-wood L., Mark W. Type II diabetes interacts with Alzheimer’s disease risk factors to predict functional decline. Alzheimer Disease and Associated Disorders. 2020;34:10–17. doi: 10.1097/WAD.0000000000000332.Type. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trachtenberg A.J., Filippini N., Ebmeier K.P., Smith S.M., Karpe F., Mackay C.E. The effects of APOE on the functional architecture of the resting brain. NeuroImage. 2012;59:565–572. doi: 10.1016/j.neuroimage.2011.07.059. [DOI] [PubMed] [Google Scholar]
- Turney I.C., Chesebro A.G., Rentería M.A., Lao P.J., Beato J.M., Schupf N., Mayeux R., Manly J.J., Brickman A.M. APOE ε 4 and resting-state functional connectivity in racially/ethnically diverse older adults. Alzheimer’s & Dementia: Diagnosis, Assessment & Disease Monitoring. 2020;1–8 doi: 10.1002/dad2.12094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Cauwenberghe C., Van Broeckhoven C., Sleegers K. The genetic landscape of Alzheimer disease: Clinical implications and perspectives. Genetics in Medicine. 2016;18:421–430. doi: 10.1038/gim.2015.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamazaki Y., Zhao N., Caulfield T.R. Apolipoprotein E and Alzheimer disease: pathobiology and targeting strategies. Nature Reviews Neurology. 2019;15 doi: 10.1038/s41582-019-0228-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yiannopoulou K.G., Papageorgiou S.G. Current and Future Treatments in Alzheimer Disease: An Update. Journal of Central Nervous System Disease. 2020;12 doi: 10.1177/1179573520907397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoav Benjamini, Yosef Hochberg Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. Royal Statistical Society. Series B (Methodological) 1995;57:289–300. [Google Scholar]
- Zheng L.J., Su Y.Y., Wang Y.F., Schoepf U.J. Different Hippocampus Functional Connectivity Patterns in Healthy Young Adults with Mutations of APP/Presenilin-1/2 and APOE ε 4. Molecular Neurobiology. 2018;55:3439–3450. doi: 10.1007/s12035-017-0540-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data will be made available on request.