Abstract
rRNAs are the central players in the reactions catalyzed by ribosomes, and the individual rRNAs are actively involved in different ribosome functions. Our previous demonstration that yeast 5S rRNA mutants (called mof9) can impact translational reading frame maintenance showed an unexpected function for this ubiquitous biomolecule. At the time, however, the highly repetitive nature of the genes encoding rRNAs precluded more detailed genetic and molecular analyses. A new genetic system allows all 5S rRNAs in the cell to be transcribed from a small, easily manipulated plasmid. The system is also amenable for the study of the other rRNAs, and provides an ideal genetic platform for detailed structural and functional studies. Saturation mutagenesis reveals regions of 5S rRNA that are required for cell viability, translational accuracy, and virus propagation. Unexpectedly, very few lethal alleles were identified, demonstrating the resilience of this molecule. Superimposition of genetic phenotypes on a physical map of 5S rRNA reveals the existence of phenotypic clusters of mutants, suggesting that specific regions of 5S rRNA are important for specific functions. Mapping these mutants onto the Haloarcula marismortui large subunit reveals that these clusters occur at important points of physical interaction between 5S rRNA and the different functional centers of the ribosome. Our analyses lead us to propose that one of the major functions of 5S rRNA may be to enhance translational fidelity by acting as a physical transducer of information between all of the different functional centers of the ribosome.
The ribosome is the central component of an extremely accurate cellular protein synthesis apparatus. Its function is to efficiently and accurately decode mRNAs. Eukaryotic ribosomes contain four rRNAs: three large-subunit-associated rRNAs (28S-25S in eukaryotes and 23S in prokaryotes, plus 5.8S and 5S) and the small-subunit rRNA (18S in eukaryotes and 16S in prokaryotes). Although these rRNAs were initially thought to provide the scaffolding for the enzymatic ribosomal proteins, early reconstitution and depletion experiments hinted at broader roles for these molecules (reviewed in references 37 and 39), and it is now clear that the rRNAs are the central players in the reactions catalyzed by ribosomes and that the individual rRNAs are actively involved in different ribosomal functions (reviewed in references 9, 30, 38, 41, and 63). Thus, understanding the molecular basis of rRNA structure and function is central to furthering our comprehension of the translational apparatus.
The 5S rRNA is a component of the large ribosomal subunit in all living organisms (with the exception of mitochondrial ribosomes) (see reference 27 for a review). In eukaryotic cells, 5S rRNA is synthesized in the nucleolus by RNA polymerase III, processed into its mature form, and then imported into the nucleus, where it associates with ribosomal protein L5. The 5S-L5 ribonuclear particle is reimported into the nucleolus, where it is assembled into the central protuberance as one of the last steps in the biogenesis of the 60S subunit (1, 3, 11, 12). The central protuberance lies opposite the head of the small subunit, and chemical probing and X-ray crystallographic analyses provide evidence to support the notion that 5S rRNA is in close contact with this subunit (56, 66). The crystal structures of the Haloarcula marismortui and Thermus thermophilus large subunits show that 5S rRNA also extends along the posterior face of the large subunit (2, 35). Chemical cross-linking, nucleolytic protection, pharmacological, and X-ray crystallographic studies show that 5S rRNA interacts with multiple functional regions of the large rRNA, including the A site region of the peptidyltransferase center and the GTPase-associated region of 23S rRNA (2, 18, 20, 36, 45, 52, 53, 56, 66). Numerous functions have been hypothesized for 5S rRNA, e.g., that it helps enhance aminoacyl-tRNA (aa-tRNA) binding to the ribosome (18, 21), that it assists in defining the topology of the peptidyltransferase center (25), and that it enhances peptidyltransferase activity (18, 20, 52).
Although 5S rRNA has been the subject of literally thousands of studies, how it works to ensure the proper function of the ribosome in vivo is still not clearly understood. Programmed ribosomal frameshifting, i.e., the ability of specific cis-acting viral signals to program ribosomes to shift translational reading frame by one base in either the 5′ (−1) or 3′ (+1) direction, provides a convenient probe of ribosome structure and function relationships in intact cells. A series of genetic screens of the yeast Saccharomyces cerevisiae that were designed to identify mutations that increased −1 programmed ribosomal frameshifting efficiencies revealed the presence of a family of genes called MOF, for maintenance of frame (reviewed in reference 13). The mof9 alleles were of particular interest because they were shown to be allelic to 5S rRNA, representing the first genetic phenotype associated with 5S rRNA (17). However, the presence of 100 to 200 tandemly arranged copies of rRNA genes on chromosome XII presented the major barrier to further genetic and functional studies of the function of 5S rRNA at the time. In the interim, in vivo systems for yeast which have progressively improved the prospects for this line of research have been developed (4, 10, 40, 60). Recently developed yeast strains called rdn1ΔΔ, in which the entire RDN1 locus, including all of the flanking 5S ribosomal DNA (rDNA) clusters have been deleted (40), are ideally suited for further studies on 5S rRNA because they allow examination of the effects of mutant 5S rRNAs without interference from wild-type sequences. Here we show that the rdn1ΔΔ strain background allows for both global and targeted mutagenesis of 5S rRNA in yeast. A total of 247 5S rRNA alleles were generated in the rdn1ΔΔ strain background and tested with regard to a series of translation-related phenotypes. The paucity of lethal 5S rRNA alleles and those responsible for ts− and cs− phenotypes supports other studies demonstrating the resiliency of the rDNAs (32, 43, 47, 58). Mapping the mutant 5S rRNA alleles with regard to termination suppression, nonsense-mediated mRNA decay, and maintenance of the killer virus phenotypes revealed functional regions of the molecule. The strongest global effects were seen in the most conserved regions of the molecule: the loop B→loop C region at the top of the central protuberance; the helix IV-loop E interface region, where 5S rRNA interacts with the A site finger (ASF); and the highly conserved G91 base in loop D, contacting helix 39 of the large subunit (25S) rRNA. Our findings lead us to propose an allosteric signaling role for 5S rRNA. We posit that, by providing a physical link between all of the different functional centers of the ribosome, it acts as a transducer of information, facilitating communication between the different functional centers and coordination of the multiple events catalyzed by the ribosome.
MATERIALS AND METHODS
Media, genetic methods, and enzymes.
Escherichia coli strains DH5α, CJ236, and MV1190 were used to amplify plasmids, and E. coli transformations were performed using the standard calcium chloride method as described previously (50). Yeasts were transformed using the alkali cation method (24). YPAD, YPG, SD, synthetic complete medium (H-), and 4.7-MB plates for testing the killer phenotype were used as described previously (62). Cytoduction of L-A and M1 from strain JD759 into rho-o strains was performed as previously described (16). Plasmid shuffle techniques using 5-flouroorotic acid (5-FOA) were as previously described (48). The sequences of the 5S rDNA mutants were determined using modified T7 DNA polymerase (57) (Sequenase, version 2.0; U.S. Biochemicals) using standard −20 and reverse primers (IDT).
Assays for killer virus maintenance and programmed ribosomal frameshifting followed previously described protocols (15). Briefly, strain 5X47 (MATa/MAT his1/+ trp1/+ ura3/+ K− R−) was used as the indicator strain to score for the presence of the killer virus. Yeast colonies containing either mutant or wild-type pJD209.TRP (control) plasmids were replica plated onto 4.7-MB plates seeded with a lawn of 5X47 cells and grown at 20°C for 2 to 3 days. Killer phenotypes were scored by either the lack of a halo of growth inhibition surrounding cells harboring 5S rDNA mutants (K−) or the lessening (Kw) of the diameter of the halo compared to the halo surrounding cells harboring the wild-type gene. Frameshifting efficiencies were calculated by dividing the beta-galactosidase activities obtained from cells harboring a −1 ribosomal frameshift reporter by those from cells harboring a 0-frameshift control. All assays were performed in triplicate, and standard errors were calculated as previously described (46).
Assays for cold and heat sensitivity and assays to monitor suppression of the ade2-1 and can1-100 alleles (at 60 μg of canavanine/ml) were performed as previously described (22). To monitor ade2-1 status, yeast colonies carrying either mutant or wild-type 5S rDNA plasmids were cultured overnight in synthetic liquid medium lacking leucine and tryptophan (H-Leu-Trp medium) and then used for the dilution spot assay on standard YPD plates. Serial dilutions of 2 × 105 to 2 × 10 CFU/5 μl were prepared in water, spotted onto the plates, and incubated at 30°C. Colonies harboring the wild-type gene were pink. Lack of color (white colonies) was interpreted as indicative of suppression of the ade2-1 nonsense mutation. Conversely, colonies that were bright red were indicative of high-fidelity mutants. A second serial dilution spot assay was used to score the nonsense suppression phenotypes of the 5S rRNA mutants with regard to the can1-100 allele, which confers resistance to canavanine in wild-type cells. Canavanine sensitivity was indicative of the ability of the mutant 5S rRNAs to act as nonsense suppressors. Scoring used a +/− system in which − was indicative of no growth, + indicated growth of the spot containing 2 × 105 CFU only, ++ indicated growth of the two densest spots, etc. Assays for heat and cold sensitivities were similarly performed and scored.
Plasmids.
pNOY290 is a 2μm plasmid containing both the URA3 and leu2d selectable markers and a complete copy of an rDNA repeat that carries the hygromycin resistance (hygr) allele of 25S rRNA (40). pNOY353 is a 2μm plasmid containing the TRP1 and leu2d markers and a complete copy of an rDNA repeat in which transcription of the 35S operon is driven from a GAL7 promoter (40). The pJD180 series of plasmids (pJD180.URA and pJD180.TRP) are pRS400-series 2μm vectors (5) with a 9,082-bp DNA fragment that contains a complete copy of an rDNA repeat from pRDN1-wt (kindly provided by Y. O. Chernoff) (4) inserted into the SmaI restriction site. Digestion of pJD180.TRP with BstEI removed a 0.8-kb fragment containing 5S rDNA, and subsequent self-ligation was used to produce pJD210.TRP. Digestion of this plasmid with SalI and NotI produced a 7.5-kb fragment containing the complete 35S rDNA operon, which was inserted into similarly restricted pRS425 to create pJD211.LEU.
Plasmids expressing 5S rRNA alone were constructed as follows. High-copy-number URA3-selectable vector pJD106.URA was made by inserting the 2.1-kb EcoRI fragment containing a copy of the 5S rDNA gene into pRS426, and pJD106.TRP was constructed by cloning the insert from pJD106.URA into pRS424 (17). pJD209.TRP was prepared by excising the 424-bp SmaI/SalI fragment containing 5S rDNA from pJD116Y5 (17) and subcloning it into SmaI/SalI-restricted pRS424.
Yeast strains.
HFY870 (MATa ade2-1 his3-11,15 leu2-3,112 trp1-1 ura3-1 can1-100 upf1::HIS3 UPF2 UPF3) was a kind gift from the laboratory of A. Jacobson. The rdn1ΔΔ strain NOY891 (MATa ade2-1 ura3-1 leu2-3 his3-11 trp1 can1-100 rdn1ΔΔ::HIS3 plus pNOY353) was kindly provided by M. Nomura (40). This strain is an adaptation of previous rdn1 deletion strains in which the entire RDN1 locus and flanking regions of chromosome XII, including an unspecified number of 5S rDNA genes, have been deleted. In this strain background, all of the cellular rRNAs are produced from pNOY353.
To obtain strains that were more amenable to genetic analyses, a series of strains was constructed so that the final killer+ strain harbored the 35S rDNA operon on a high-copy-number LEU2 vector and the 5S rDNA gene on a 2μm URA3 vector, thus allowing us to transform cells with mutant 5S rDNA alleles on a high-copy-number TRP1-based vector and screen for loss of the wild-type plasmid using 5-FOA. The genealogies of the strains are as follows. JD1089 was constructed by introducing pJD180.URA into NOY891 by transformation, and strains that had lost pNOY353 were identified by their Ura+ Trp− Leu− phenotypes. The killer virus was introduced into the resulting strain by cytoduction from strain JD759 (MATα kar1-1 arg1 thr[i,x] [L-A HN M1]), and the killer+ phenotype was confirmed as described below. Transformation of JD1089 with pJD180.TRP and subsequent identification of colonies that had lost pJD180.URA by their ability to grow in the presence of 5-FOA were used to construct strain JD1110. Plasmids pJD106.TRP and pJD211.LEU were subsequently cotransformed into JD1110, and cells that had lost pJD180.TRP were identified by their Ura+ Leu+ Trp− killer+ phenotypes. A single colony having this phenotype was selected to be JD1111. The lack of chromosomal copies of rDNA genes was confirmed by DNA blot analysis using 5S and 25S rDNA-specific probes.
Mutagenesis of 5S rDNA. pJD209.TRP was used to make a systematic collection of mutants covering the entire 121-bp sequence of 5S rDNA. pJD209.TRP was introduced into E. coli strain CJ236, and uracil-containing single-stranded DNA was obtained by infection with the R408 helper phage (Promega, Madison, Wis.). Site-directed oligonucleotide mutagenesis using T4 DNA polymerase and subsequent transformation into MV1190 were performed in accordance with standard methods (28). To mutate 5S rDNA to saturation, a series of 121 mutagenic antisense 31-mers were designed to “walk along” the (plus strand) single-stranded DNA (ssDNA) 5S rDNA sequence of uracil containing ssDNA of pJD209.TRP. The identity of the 16th base of each synthetic oligonucleotide was randomized so that it contained a mixture of all three possible mutant bases for each of the 121 different positions. The 5′ and 3′ 15 nucleotides flanking the mutagenic base perfectly complemented the 5S rDNA sequences present on the plasmid. The identity of each mutant was confirmed by DNA sequence analysis. On a technical note, since the position but not the identity of a particular mutant was known beforehand, we were able to multiplex sequence using up to five different clones (mutated at different positions) per sequencing reaction.
Generation of the yeast mutant collection.
Strain JD1111 was grown in H-Leu-Ura synthetic complete medium, transformed with mutant pJD209.TRP plasmids, and plated onto H-Leu-Trp. After 5 to 6 days of growth at 30°C, the yeast colonies were replica plated onto H-Leu-Trp medium supplemented with 1 g of 5-FOA/liter and incubated for 6 days at 30°C. The resultant yeast colonies were replica plated onto H-Ura medium to test for the absence of the initial pJD106.URA plasmid containing wild-type 5S rDNA. For each variant, at least three colonies were selected and stored at −80°C as stocks for all further work. To confirm that the process was not selecting for reversion or second-site mutations, mutant 5S rRNAs were amplified from cell lysates from 10 selected strains by direct sequencing of reverse transcription products in accordance with previously described protocols (65) using primer 5′ AGGTTGCGGCCATATG 3′ (complementary to the 3′ end of 5S rRNA) and the products were analyzed by sequencing.
RNA blot analyses.
RNA blot analyses to monitor for the presence of the L-A and M1 double-stranded RNAs (dsRNAs) and to monitor the endogenous Cyh2 pre-mRNA and Cyh2 mRNA species were performed as previously described (7, 16). The L-A and M1 blots were normalized for genomic DNA concentrations by using standardized samples. For the Cyh2 blots, equal units of optical density at 260 nm, indicative of cellular RNA, were used. The images were visualized and quantitated by phosphorimagery using a Typhoon 8600 phosphorimager and ImageQuant, version 5.2, software (Molecular Dynamics).
RESULTS
Construction of a system for mutagenesis of 5S rRNA.
The present study was made possible by the initial development of a yeast strain lacking the entire RDN1 locus, as well as flanking sequences that contain an unknown number of 5S rDNA genes (40). The rRNAs were originally supplied to this rdn1ΔΔ strain by a large plasmid (pNOY353) that contains a single rDNA repeat encoding all four rRNAs. Due to its size, this plasmid is rather unstable in E. coli making it difficult to manipulate. To solve this problem, we replaced pNOY353 with two plasmids: pJD106.URA, which carried only the 5S rRNA gene, and pJD211.LEU, which carried the 35S rRNA operon (the product of which is processed into the 25S, 18S, and 5.8S rRNAs). The resultant strain, JD1111, had no discernible growth defects compared to the parental strain and was able to stably support the yeast killer virus. This strain served as the basis for the subsequent phenotypic characterization of a saturation library of mutant 5S rRNAs.
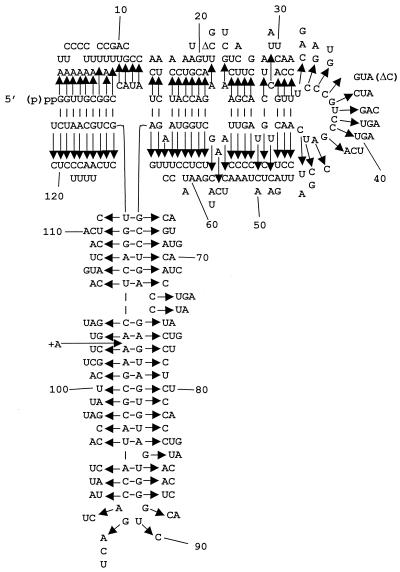
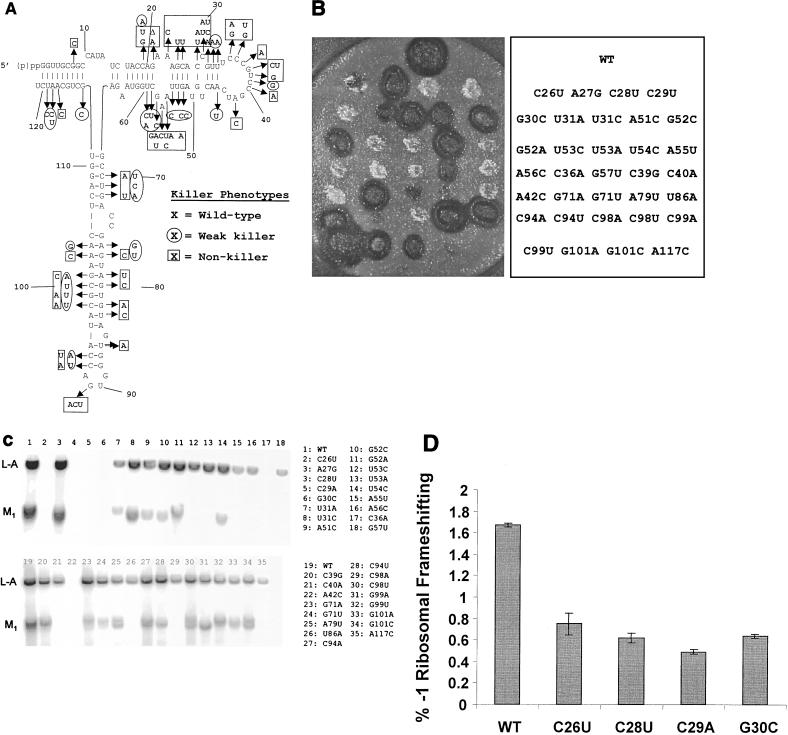
Oligonucleotide site-directed mutagenesis was used to create the library of 5S rRNA mutants because it allowed the location of each mutation to be known in advance and also because it ensured against biases for or against specific nucleotides or regions of the molecule. The 5S rRNA-encoding plasmid pJD209.TRP was the target for mutagenesis. This plasmid contains a short (424-bp) fragment of rDNA harboring the 121-bp 5S rDNA plus 5′ and 3′ flanking regions of 147 and 156 bp, respectively. Each position of the 5S rDNA was mutated using a specific synthetic oligonucleotide reagent consisting of a mixture of three oligonucleotides that were identical except for a mutation of the corresponding wild-type base into any of the three possible substitutions. Mutagenesis reactions were performed separately for all 121 positions of 5S rDNA. Mutant plasmids were isolated from E. coli clones and checked by sequencing for the presence of desired mutations. In total, 1,024 plasmid samples were sequenced, yielding 246 unique point mutants at each of the 121 positions of 5S rDNA. These are shown in Fig. 1. Only one allele was generated for 24 positions, two alleles were generated for 69 positions, and all three possible mutants were generated for 28 positions. Two mutant 5S rDNA clones harbored deletions (marked Δ), and one contained an additional A residue between positions 103 and 104 (marked +A104). Substitutions of pyrimidines greatly outnumbered those of purines (especially G residues), presumably because the smaller size of the pyrimidines favored their incorporation during the chemical synthesis of the mutagenic oligonucleotides. Standard plasmid shuffle methods were used to replace the wild-type 5S rDNA plasmid with those harboring mutants. Direct reverse transcription sequencing of cellular 5S rRNAs harvested from 10 different mutants was used to confirm that all 5S rRNAs in the cells corresponded to single clonal mutants.
FIG. 1.
Map of yeast 5S rRNA. The wild-type Saccharomyces cerevisiae 5S rRNA sequence is shown. Arrows indicate the 246 mutants with point mutations. Δ, deletion allele (e.g., deletion of one nucleotide at positions 21 and 36); +A, insertion of an extra A residue between positions 103 and 104.
Phenotypic characterization of the 5S rRNA mutants.
Endogenous genetic markers were used to assess the effects of the 5S rRNA alleles on ribosome-related functions. These are summarized in Table 1. In general, transversions (Pu↔Py) tended to be twice as likely to produce mutant phenotypes as transitions (Pu↔Pu and Py↔Py). This was especially apparent in replacements of pyrimidines by purines, indicating that the phenotype might be more affected by the size of a substituting nucleotide rather than by the precise nature of the substitution. The effects of the 5S rRNA mutants on specific phenotypes are discussed in greater detail below.
TABLE 1.
Summary of properties of 5S rRNA mutants
| Sequence typeb | No. of alleles (%) conferring indicated phenotypec
|
|||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Growtha
|
Nonsense suppression
|
Killer (229)
|
||||||||||||
| Lethal (246 alleles) | 13°C (239)
|
37°C (236)
|
ade2-1 (239)
|
can1-100 (223)
|
||||||||||
| WT | cs− | WT | ts− | WT | Hyperacc. | Supps | WT | Suppw | Supps | WT | Kw | K− | ||
| Consensus | 2 | 118 (92) | 1 (1) | 90 (69) | 2 (1) | 94 (73) | 2 (1) | 34 (26) | 85 (70) | 1 (1) | 36 (29) | 72 (58) | 21 (16) | 33 (26) |
| Noncons. | 2 | 107 (98) | 0 | 88 (84) | 2 (2) | 94 (86) | 3 (3) | 12 (11) | 90 (88) | 1 (1) | 10 (11) | 76 (73) | 12 (12) | 15 (15) |
| Total | 4 | 225 (94) | 1 (<1) | 178 (75) | 4 (2) | 188 (79) | 5 (2) | 46 (19) | 173 (78) | 2 (1) | 47 (21) | 148 (65) | 33 (14) | 48 (21) |
General growth phenotypes.
Consensus and noncons., consensus and nonconsensus eukaryotic 5S rRNA sequences, respectively, of mutant alleles.
Total numbers of alleles tested are in parentheses next to phenotypes and allele designations. WT, wild type; Hyperacc., hyperaccurate; Supps, strong suppression; Suppw, weak suppression.
Effects of the 5S rRNA mutants on cell growth and viability.
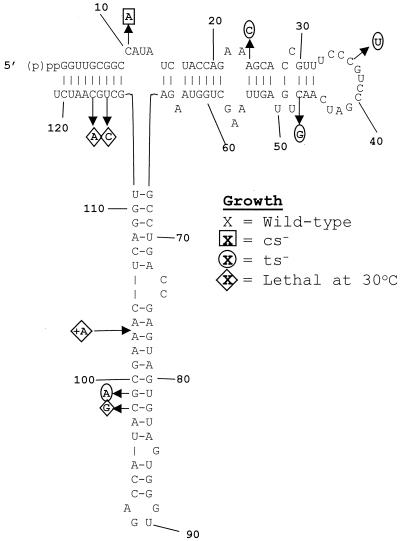
Although 5S rRNA is highly conserved, only four of the mutants were lethal: C98G, U114C, and C116A alleles and the +A104 allele (Fig. 2). To test whether a compensatory mutation would rescue the C116A allele, we constructed a 5S rDNA clone harboring the C116A plus G5U allele. Expression of this 5S rRNA species had no discernible effect on cell growth, nor did it produce a result in any other obvious mutant phenotype.
FIG. 2.
Growth phenotypes of the 5S rRNA mutants. The sequence of wild-type S. cerevisiae 5S rRNA is in the center. Alleles conferring specific growth-related phenotypes are in boldface. Diamonds, lethal alleles; circles and squares, alleles conferring lethality due to temperature sensitivity (37°C) and cold sensitivity, respectively.
Since many mutant alleles in yeast confer sensitivity to extremes in temperature, we assayed the effects of the library of 5S rRNA mutants at both 13 and 37°C (Fig. 2). Because cold sensitivity tends to be associated with macromolecule assembly (e.g., ribosome biogenesis) defects, we monitored cell growth at 13°C. Only one cs−-conferring allele was identified, the C10A allele. This was the only allele in the highly conserved loop A region conferring any detectable phenotype, and the C10U allele had no such effect. The paucity of cs−-conferring alleles is consistent with the fact that the 5S-L5 particle is assembled into the ribosome late in the ribosomal biogenesis program, i.e., that 5S rRNA does not provide a critical foundation for assembly of subsequent essential ribosomal structural features. Four ts−-conferring alleles were also identified: the A24C, C36U, C47G, and G99A alleles.
Nonsense suppression and nonsense-mediated mRNA decay-associated phenotypes.
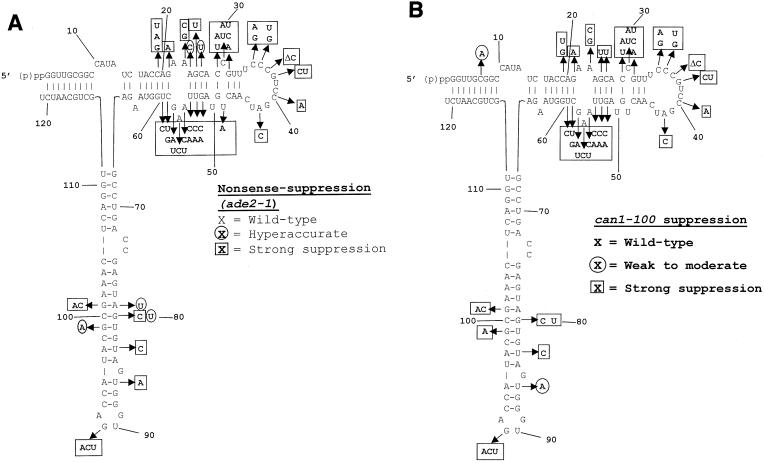
The presence of endogenous nonsense-containing alleles (ade2-1 and can1-100) provided inbuilt monitors of the effects of the 5S rRNA mutants on the ability of ribosomes to recognize termination codons. The ade2-1 allele of the phosphoribosylaminoimidazole carboxylase (AIR decarboxylase) gene harbors a nonsense (ochre) codon, and cells harboring this allele that are wild type with respect to most other genes are pink when grown in the absence of exogenous adenine as a result of the accumulation of a red pigment (AIR). Cells that were able to suppress this mutation grew as either light pink or white colonies, indicating increased readthrough of the nonsense codon. In contrast, cells in which a mutation causes the translational apparatus to be hyperaccurate generated bright red colonies in this strain background. Of the entire collection of 242 viable mutants, 239 were assayed regarding their Ade phenotypes (Fig. 3A). A total of 44 (19%) of the alleles were strong suppressors. There was almost a threefold-greater frequency of this class of mutants occurring at a conserved nucleotide than at a nonconserved one (32 to 12). Of the five alleles (2% of the total) that conferred hyperaccurate phenotypes, there was an even split between mutations at conserved versus nonconserved positions (three to two).
FIG. 3.
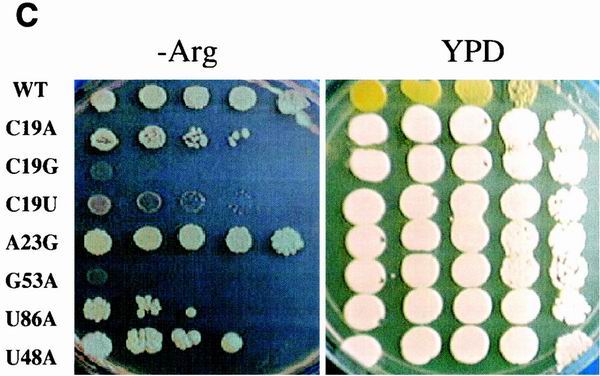
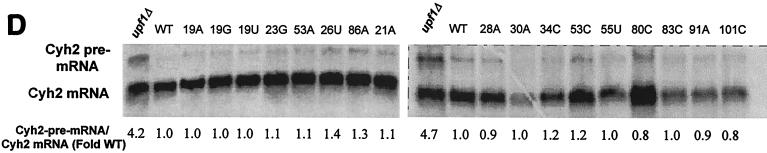
Nonsense-suppression phenotypes of the 5S rRNA mutants. The endogenous ade2-1 (A) and can1-100 (B) nonsense-containing alleles were used to monitor the effects of all of the 5S rRNA mutants on the ability of the ribosomes to recognize termination codons. Scoring for degree of accuracy, e.g., hyperaccurate, wild-type, and weak and strong suppression, is described in Materials and Methods. (C) The C19A, A23G, U86A and U48A alleles of 5S rRNA can suppress ade2-1 but not can1-100. Serial dilutions of cells harboring wild-type 5S rRNA or the indicated 5S rRNA alleles were spotted onto H-Arg medium containing 60 μg of canavanine/ml or onto YPD medium lacking adenine and incubated at 30°C for 3 days. Suppression of can1-100 is indicated by the inability of cells to grow in the presence of canavanine. Suppression of ade2-1 is indicated by the formation of white colonies. (D) Mutations in 5S rRNA do not affect the nonsense-mediated mRNA decay pathway. Total RNAs (10 μg) extracted from JD1111 cells harboring wild-type (WT) 5S rRNA or selected 5S rRNA mutant alleles or from the upf1Δ strain HFY870 were separated through a 1% denaturing agarose gel, transferred to nitrocellulose, and probed with a 32P-labeled Cyh2 fragment as previously described (6). The abundances of the mature Cyh2 mRNA and the Cyh2 pre-mRNA were determined using a Typhoon 8600 phosphorimager and ImageQuant, version 5.2, software (Molecular Dynamics). The pre-Cyh2/Cyh2 ratios for all of the mutants were normalized to that from cells harboring wild-type 5S rRNA.
Similarly, the can1-100 allele harbors an ochre mutation in the CAN1-encoded arginine permease that prevents transport of canavanine, a toxic arginine homolog, into the cell. Thus, the parental JD1111 strain is Canr, whereas suppression of this allele renders cells CanS. We tested 223 mutants for growth in the presence of 60 μg of canavanine/ml (Fig. 3B). The results closely paralleled the ade2-1 tests: 47 total alleles conferred canavanine sensitivity, with a threefold greater chance of this occurring at a conserved nucleotide. Interestingly, though four of the 5S rRNA mutants were able to suppress the ade2-1 nonsense-containing allele, they were unable to suppress the can1-100 allele (C19A, A23G, U86A and U48A alleles; Fig. 3C). We suggest that this is reflective of quantitative differences in their abilities of suppress nonsense alleles.
The presence of premature termination codons in mRNAs, as a consequence of point mutations or frameshift mutations, or of those contained in the introns of unspliced mRNAs that escape to the cytoplasm could result in production of toxic truncated protein products. It is therefore important that cells be able to identify and rapidly rid themselves of nonsense-containing mRNAs. A complex, yet evolutionarily conserved, apparatus involving trans-acting factors and cis-acting signals called the nonsense-mediated mRNA decay (NMD) pathway has evolved to handle this problem (reviewed in references 23 and 49). Since nonsense suppression defects have been linked to defects in the NMD pathway, we surveyed the NMD phenotypes in a selected subset of mutants. The inefficiently spliced endogenous Cyh2 precursor mRNA was used to monitor the status of the NMD pathway in cells expressing both wild-type and mutant 5S rRNAs. The results from this partial survey of the mutants show that none of the strong ade2-1 or can1-100 suppressors confer defects in NMD (Fig. 3D). Probing these blots for the Can1 transcript revealed that this was also not stabilized by any of the 5S rRNA mutants (data not shown). The fact that none of the nonsense suppressors had NMD defects is consistent with previous work demonstrating that, although the two mechanisms are linked, they are also separable (reviewed in reference 8).
Killer virus maintenance and programmed −1 ribosomal frameshifting.
Propagation of the yeast killer virus is extremely sensitive to the status of the host translational apparatus, providing an inbuilt indicator of defects in ribosome function (44). Figure 4A shows the summary results for the mutants for the killer phenotype. Of 229 alleles examined, over one-third had discernible effects on the killer. Virus-infected yeast cells can be further divided into those that completely lost the killer phenotype (K−) and those around which the zone of growth inhibition was reduced, i.e., weak killers (Kw). Examples of these different phenotypes are shown in Fig. 4B. The 5S rRNA alleles tended to favor production of the K− over the Kw phenotype by a ratio of approximately 3:2 (Table 1).
FIG. 4.
Killer phenotypes of the 5S rRNA mutants. (A) Effects of each of the mutants on the killer virus phenotype. The map of 5S rRNA and mutants is as described for Fig. 1, and killer phenotypes are shown. (B) Representative killer plate assay. Colonies of JD1111 cells harboring various 5S rRNA alleles were replica plated onto a lawn of cells that are sensitive to the secreted killer toxin produced by the M1 satellite virus of L-A. Killer activity was observed as a zone of growth inhibition around the colonies. (Left) Photograph of the killer plate assay. (Right) Key indicating 5S rRNA genotypes. WT, wild type. (C) Northern blot probing for L-A and M1 viral RNAs. Total RNAs were isolated from the indicated strains and analyzed by Northern blotting for the presence of the L-A and M1 viral RNAs as described in Materials and Methods. (D) Programmed −1 ribosomal frameshifting phenotypes of selected mutants. JD1111 cells harboring wild-type, C26U, C28U, C29A, or G30C 5S rRNA alleles were transformed with either p-1 frameshift indicator or p0 control plasmids, and efficiencies of L-A virus-promoted programmed −1 ribosomal frameshifting were monitored as previously described (17). Assays were performed in triplicate, and bars denote percent error.
A number of factors can contribute to an observed killer phenotype. These include loss or decreased copy numbers of the helper L-A virus and/or the killer toxin-producing M1 satellite virus, defects in the gene products that are responsible for processing the killer toxin precursor into the mature toxin, and defects in the apparatus responsible for secretion of the toxin out of the cell. It has been shown that two types of flaws in the translational apparatus, general 60S ribosomal subunit biogenesis defects (44) and those that promote increased or decreased programmed −1 ribosomal frameshift efficiencies on the L-A viral mRNA (reviewed in reference 14), can lead to total or partial loss of L-A and/or M1. In light of the data suggesting a role for 5S rRNA in enhancing translational fidelity, the effects of the 5S rRNA mutants on the ability of cells to maintain these viral dsRNA genomes was examined (Fig. 4C). RNA hybridization analyses of a selected subset of the alleles revealed that loss of the killer phenotype could result from either (i) complete loss of both L-A and M1 (e.g., C28U; sample 4 in Fig. 4C), (ii) loss of M1 with or without attendant decreases in L-A copy number (e.g., compare U55A [no. 15]) with U53A [no. 13], or (3) the production of defective-interfering M1 deletion mutants (e.g., G99A [no. 31] and G101C [no. 34]). The Kw phenotype was found to be due to decreases in M1 copy numbers either with or without accompanying decreases in L-A concentrations (e.g., compare U31A [no. 7]) with U54C [no. 14]). In no cases were effects on the killer phenotype found associated with wild-type M1 levels, suggesting that all the translational defects of the 5S rRNA mutants affected virus propagation rather than processing and export of the toxin.
The inability to maintain both L-A and M1 is unusual: to date, mutants from only three other complementation groups, MAK3, MAK10, and PET18, have been shown to confer this phenotype (reviewed in reference 61). A preliminary characterization of the programmed −1 ribosomal frameshifting phenotypes of four examples of this class of 5S rRNA mutants revealed that they all promoted strong decreases in frameshifting efficiencies (Fig. 4D). We suggest that the reason for L-A loss in these mutants is due to decreased availability of the Gag-Pol dimer, excluding encapsidation of the L-A dsRNA into nascent viral particles.
DISCUSSION
A powerful system for rRNA structure and function studies of a model eukaryotic organism.
The results presented here demonstrate the strength of the rdn1ΔΔ strains as a tool to address issues related to rRNA structure and function in a model eukaryotic system. We have introduced modifications to the original strain and plasmids that make such analyses more practical, specifically stabilizing the clones by separating the RNA polymerase I and III transcribed operons onto two plasmids and making LEU2 available as a selectable marker. Although this report focuses on 5S rRNA for both historical and scientific reasons, the results presented here provide proof of the principle that this system is amenable for mutagenesis-based structure and function studies of all of the yeast rRNAs. It should also be possible to use this system for large-scale substitution studies, e.g., replacement of either entire yeast rRNA genes or regions thereof with homologous sequences from other organisms.
Genotype, phenotype, structure, and function: clustering of sequences responsible for mutant phenotypes at the critical contacts between 5S rRNA and the major functional regions of the ribosome.
Overall, just 4 of the 246 alleles were lethal, and only 5 more conferred either cold or heat sensitivity (Table 1). These findings support suggestions from other investigators that the rRNAs are resilient molecules that can retain their functions despite mutations at universally conserved bases within critical regions, e.g., the peptidyltransferase center or the sarcin-ricin loop (31, 32, 47, 58). The presence of multiple endogenous genetic markers in this strain enabled us to qualitatively address 5S rRNA function by examining the effects of the 5S rRNA mutants on other phenotypes. At a general level, nucleotides at 64 of the 121 positions of yeast 5S rRNA correspond to the eukaryotic consensus 5S rRNA sequence, and mutations introduced into these evolutionarily conserved positions were approximately twice as likely to promote mutant phenotypes than those at the nonconserved positions. There are notable exceptions to this trend: mutations of some highly conserved bases did not elicit phenotypes (e.g., U33, G49, G85, G87, etc.), while some mutations of some of the nonconserved bases promoted strong mutant phenotypes (e.g., nucleotide C19).
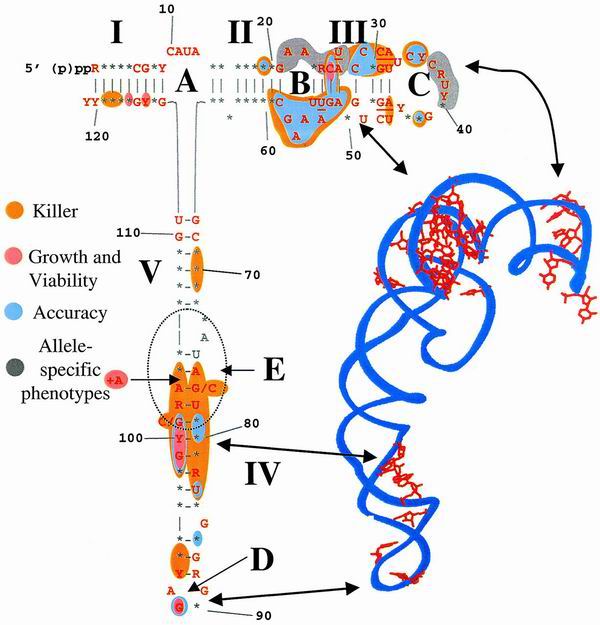
Charting the various phenotypes of the 5S rRNA mutants onto a consensus map of the eukaryotic 5S rRNA and projecting these onto a topological map of the H. marismortui 5S rRNA crystal structure (2) reveal that the phenotypes tended to cluster into two minor and three major discrete regions (Fig. 5). The minor regions were (i) bases 69 to 71 and (ii) helix I. The contribution of bases 69 to 71 was minor, only affecting the maintenance of the killer virus. The helix I alleles tended either to be lethal or to only affect the killer phenotype. The importance of this region of the gene for correct posttranscriptional 3′ end processing of 5S rRNA may provide an explanation for the lethal phenotypes produced by the U114C and C116A alleles. With regard to nucleotide 116, whereas the C116A allele exhibited a strong dominant lethal phenotype, cells harboring either the C116U allele or the C116A plus G5U double mutant appeared to be defect free. Though a likely conclusion from these data suggests that, while the identity of the base at position 116 is important, the effects of changing it can be overcome by ensuring base pairing with the nucleotide at position 5. Overexpression of the C116G allele from a high-copy-number plasmid in an RDN1 wild-type background also produced no discernible defect (29). Thus, the answer to this quandary awaits more detailed biochemical and molecular analyses. Mutations in the highly conserved loop A region (and adjacent sequences, notably the conserved G67-U111 and C68-G110 base pairs) also had no significant effects. Thus, it appears either that specific nucleotides in this region do not play a major role in the function of 5S rRNA in the context of translational fidelity or that, if this arm of the molecule has a specific function, our analysis was not sufficient to elucidate it.
FIG. 5.
Mapping the different phenotypes of the 5S rRNA mutants onto the eukaryotic 5S rRNA consensus sequence and H. marismortui 5S rRNA crystal structure reveals phenotypic clustering. (Left) Eukaryotic 5S rRNA consensus sequence. R, purine; Y, pyramidine; ∗, any base. Red bases are conserved in the yeast 5S rRNA sequence. Classes of phenotypes are colored as indicated. Killer, impact on the ability of cells harboring mutant 5S rRNA alleles to maintain the killer phenotype as shown in Fig. 4; growth and viability, impact of the mutants on cell viability (lethality at 13, 25, or 37°C) as shown in Fig. 2; accuracy, nonsense-suppression phenotypes as shown in Fig. 3; allele-specific phenotypes, regions in which individual mutant alleles promote specific phenotypic defects. (Right) Crystal structure of H. marismortui 5S rRNA (2). Red, bases from this study corresponding to suppressors of the ade2-1 allele; arrows, analogous regions of the molecule as depicted by the two different topological representations.
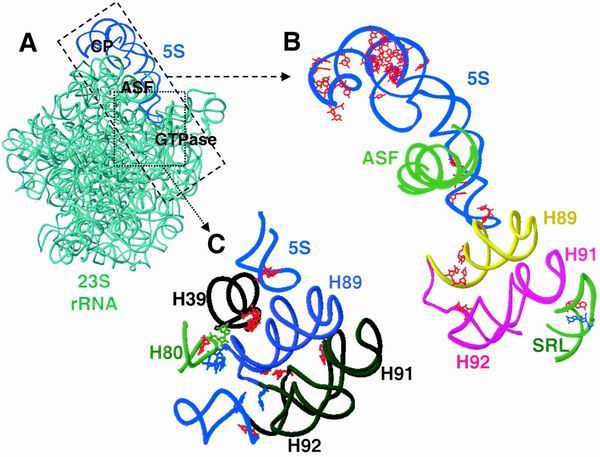
In contrast, mutations in three defined regions of the molecule had very significant impacts on ribosome function. These were (i) the loop B→loop C arm, (ii) the loop E-helix IV interface, and (iii) the bottom of helix IV and the highly conserved G91 base in loop D. These regions are topologically located at the head, middle, and tail of the molecule, respectively (Fig. 5). With regard to the atomic scale structures of H. marismortui and T. thermophilus ribosomes, 5S rRNA appears to form a chain running from the top of the central protuberance of the large subunit, down through the ASF and ending at the GTPase-associated center (Fig. 6) (2, 66). Mutations in the loop B→loop C arm tended to produce a broad range of defects. While the most severe of them appear to map along a helical face of the molecule from nucleotide 19, crossing over to positions 59 to 51 and back again to nucleotides 28 to 35, those in the bases on the other face of this helix tended to have fewer severe consequences (Fig. 5 and 6B). The yeast loop B→loop C 5S rRNA mutants also coincide with regions from E. coli 5S rRNA that were previously shown to be affected by the binding of the small subunit (56). This region of the molecule interacts with several ribosomal proteins (in T. thermophilus, these are L5 [yeast L11] and L18 [yeast L5]) and helps to form a critical contact between the large and small ribosomal subunits (66). This region of 5S rRNA also interacts indirectly with the peptidyl-tRNA (66), and we have demonstrated that mutant alleles encoding the yeast ribosomal protein L5 have 5S rRNA-dependent peptidyl-tRNA binding defects, promoting increased rates of programmed ribosomal frameshifting, which in turn inhibit propagation of both the M1 killer and Ty1 viruses (34). The existence of this phenotypic cluster suggests that loop B→loop C region of 5S rRNA may be involved both in communicating with the small subunit and in maintaining translational reading frame at the P site.
FIG. 6.
Structure and function mapping of the yeast 5S rRNA mutants onto the H. marismortui large-subunit rRNA. (A) Ribbon diagram of the H. marismortui large-subunit rRNAs. Blue, 5S rRNA; aqua, 23S rRNA. CP, central protuberance; GTPase, GTPase-associated center. (B) 5S rRNA as a bridge to mediate communication between the decoding center and the GTPase-associated center. Dashed box and arrow, region of the large subunit enhanced. Red nucleotides map mutations in 5S rRNA which suppress the ade2-1 allele of yeast (this study) and those in 23S rRNA which also decrease translational accuracy in E. coli. Nucleotides in which substitutions lead to the hyperaccurate phenotype are blue. Shown are 5S rRNA (blue); the ASF (green); helices 89 (yellow), 91 (magenta), and 92 (magenta); and the sarcin-ricin loop (SRL) (green) of the 23S rRNA of H. marismortui (2). (C) 5S rRNA as a bridge to mediate communication between the peptidyltransferase center and the elongation factor binding center. Dotted box and arrow, region of the large subunit enhanced. Nucleotides increasing their reactivity upon mutation at E. coli 23S rRNA residue 960 are in red, and residues promoting decreased reactivity are in blue (54). The latter nucleotides coincide with the residues protected by the P site-bound tRNA and may influence helix 89 (indigo) and 5S rRNA. Mutations in 5S rRNA at positions 87 and 91 or 5S rRNA (red) also affect translational accuracy. Helices 39 (black) and 80 (green) of the large-subunit rRNA are located in immediate proximity to the peptidyltransferase center. We suggest that an allosteric signal transmission pathway through 5S rRNA could serve to coordinate the peptidyltransferase reaction with subsequent EF-2 binding and GTP hydrolysis.
The loop E-helix IV interface is the site of the second important cluster of 5S rRNA alleles: these map to a point of contact between 5S rRNA and helix 38, also known as the ASF (Fig. 6B). The +A104 allele is of particular interest because the associated region of 5S rRNA contains an A minor motif that makes the most significant contact with 23 S rRNA through an “A patch” (36). We speculate that the addition of an extra nucleotide into this region interfered with formation of this critical structural determinant to an extent sufficient to prevent viability. The C98G and G99A alleles are also of special interest in that they were previously shown to promote a significant conformational change in the 5S rRNA molecule (59), and their overexpression in a wild-type background promoted increases in both −1 and +1 programmed ribosomal frameshifting (17). The lethality of the C98G mutation is suggestive of ribosomes with catastrophic translational fidelity defects, while the temperature sensitivity-conferring G99A allele likely has a similar but less severe impact. The existence of this phenotypic cluster suggests that this region of 5S rRNA is involved in helping the ribosome monitor the status of the aa-tRNA in the A site.
The third cluster of mutations, those at the bottom of helix IV, tended to be allele (compare U86A to U86C) and phenotype (e.g., C93 and C94) specific. The highly conserved G91 was the only base in loop D with any phenotypic effects. The yeast G91 is located at the same structural region of the molecule as the E. coli U89, which has been shown to make multiple contacts with the large subunit rRNA (reviewed in reference 35). These findings suggest that specific base contacts are important for function in this region of the 5S rRNA molecule. Structurally, the helix IV-loop D region is where 5S interacts with helices 89 and 39 of the large-subunit rRNA: mutants in this region lie at crucial contact points between 5S rRNA, the GTPase-associated center, and the peptidyltransferase center, respectively (Fig. 6B and C). Thus, this phenotypic cluster links 5S rRNA to the two remaining functional centers of the ribosome.
5S rRNA as a physical transducer of information within the ribosome.
We propose that a major function of 5S rRNA could be to enhance translational fidelity by acting as a physical transducer of information between all of the different functional centers of the ribosome. For example, though the decoding of the genetic information encoded in the mRNA is considered to be the function of the small ribosomal subunit, the genetic and biochemical evidence leads us to hypothesize that 5S rRNA may help to enhance translational fidelity by coordinating the transfer of aa-tRNA from eukaryotic elongation factor 1A to the ribosomal A site by linking this small-subunit functional center with the large-subunit GTPase center. In support of this, random-mutagenesis approaches have demonstrated that several components of the 23S rRNA are involved in maintaining translational fidelity (42). One of these, the loop of helix 92, interacts with the A site-bound aa-tRNA (26). This loop also interacts with helix 89, which in turn contacts 5S rRNA (Fig. 6B). Thus, 5S rRNA could communicate with helix 92 through helix 89. There is also a link from the tip of 5S rRNA through helix 89 to the sarcin-ricin loop via helices 92 and 91. Both error-prone and hyperaccurate mutants are known to occur at the sarcin-ricin loop (33, 43), and thus it is possible that upon the binding of cognate aa-tRNA an allosteric signal could be transduced from the small subunit to the large subunit to 5S rRNA through the intersubunit contact through L5 (yeast L11). From there, the signal would be transduced through 5S rRNA to the GTPase-associated center, activating the GTPase activity of eEF-1A. By our model, the translational fidelity phenotypes associated with the mutant alleles at position 91 in particular are a consequence of defects in their abilities to efficiently transmit information across the gap between the tip of 5S rRNA and helix 89, thus inhibiting communication between the small subunit and the GTPase-associated center.
The grouping of mutants in the loop E-helix IV region of 5S rRNA suggests the presence of another critical contact between the aa-tRNA and the GTPase-associated center. We propose that the reason that these mutant 5S rRNAs affect translational fidelity is that they inefficiently convey information along this transmission line. The existence of this phenotypic cluster suggests that 5S rRNA may also be involved in monitoring the transfer of aa-tRNA from eEF-1A to the ribosomal A site. Thus, accommodation of the CCA end of the aa-tRNA could be monitored via transmission of a signal through this contact to the other functional centers of the ribosome. It is important to note that we have drawn the 5S rRNA loop E as a base-paired region because the high-resolution crystal structures show it as such (2, 36, 66). However, most depictions of 5S rRNA prior to elucidation of the crystal structures show this region as a loop. As has been noted elsewhere, many different crystal forms of ribosomes can be obtained, each of which diffract X rays with various degrees of resolution, presumably because of differences in the extent of order or disorder among them (51, 64). Thus, the reason that loop E is closed in these crystal structures may simply be that this conformation contributes to the overall order of the crystals, i.e., those that have been used in the analyses because they diffract X rays with the highest resolution. Viewed in this light, it is possible that the loop E region may indeed alternate between open and closed conformations, contributing to the allosteric signaling potential of 5S rRNA.
Another putative signal transmission chain could coordinate the activities of the peptidyltransferase and elongation factor binding centers (Fig. 6C). The existence of this chain and the involvement of 5S rRNA in this pathway were first postulated in cross-linking experiments (19, 20, 55) and were further supported by site-directed mutagenesis of helix 39, which is in contact with the D loop of the 5S rRNA (54). These mutants specifically alter the chemical reactivity of nucleotides that have been implicated in binding of the CCA end of the peptidyl-tRNA at the P site. Helix 39 interacts with helix 80, which base pairs with the P site-bound peptidyl-tRNA; thus the interaction of 5S rRNA with helix 39 means that 5S rRNA may influence the interactions between the ribosome and peptidyl-tRNAs and vice versa. The third critical cluster of 5S rRNA mutants, those at the tip of helix IV of 5S rRNA, and G91 lie in close proximity to helix 39 (Fig. 6C). As noted above, this phenotypic cluster is also a site of interaction between 5S rRNA and the functional region formed by helices 89, 92, and 91 and the sarcin-ricin loop. Thus, this cluster points to a functional connection through 5S rRNA between the peptidyltransferase center and the major sites of interaction between the elongation factors and the ribosome. This suggests that 5S rRNA is also involved in the communication between the peptidyltransferase center and the elongation factor binding center. An allosteric signal transmission pathway through 5S rRNA could thus serve to coordinate the peptidyltransferase reaction with subsequent eEF-2 binding and GTP hydrolysis.
In conclusion, we propose that all of the different functional centers of the ribosome are able to coordinate their activities by transmission of allosteric signals through 5S rRNA. In this manner, 5S rRNA could serve to ensure the fidelity of each step in the translation program. The 5S rRNA mutants described in this study will enable us to perform meaningful biochemical tests of this model. Additionally, in light of this model it is interesting that the assembly of the 5S rRNA into the ribosome is one of the last steps in ribosome biogenesis. One can speculate that the translational apparatus has evolved so as to ensure that ribosomes are only functionally activated at a late stage in the biogenesis program by addition of 5S rRNA to ensure protein synthesis in the appropriate cellular compartments.
ACKNOWLEDGMENTS
We extend our warmest thanks to M. Nomura for his kind gift of the rdn1ΔΔ yeast strain and to A. Jacobson for the upf1Δ strain. We also thank Manan Patel and Deepu Abraham for technical help and Jason Harger, Kristi Muldoon, Ewan Plant, and Gary Brewer for critical reviews of the manuscript.
This work was supported by grants to J.D.D. from the National Institutes of Health (R01 GM58859 and R01 GM62143).
REFERENCES
- 1.Allison L A, North M T, Murdoch K J, Romaniuk P J, Deschamps S, Le Marie M. Structural requirements of 5S rRNA for nuclear transport, 7S ribonucleoprotein particle assembly, and 60S ribosomal subunit assembly in Xenopus oocytes. Mol Cell Biol. 1993;13:6819–6831. doi: 10.1128/mcb.13.11.6819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ban N, Nissen P, Hansen J, Moore P B, Steitz T A. The complete atomic structure of the large ribosomal subunit at 2.4 D resolution. Science. 2000;289:905–920. doi: 10.1126/science.289.5481.905. [DOI] [PubMed] [Google Scholar]
- 3.Brow D A, Geiduschek E P. Modulation of yeast 5S rRNA synthesis in vitro by ribosomal protein YL3. J Biol Chem. 1987;262:13953–13958. [PubMed] [Google Scholar]
- 4.Chernoff Y O, Vincent A, Liebman S W. Mutations in eukaryotic 18S ribosomal RNA affect translational fidelity and resistance to aminoglycoside antibiotics. EMBO J. 1994;13:906–913. doi: 10.1002/j.1460-2075.1994.tb06334.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Christianson T W, Sikorski R S, Dante M, Shero J H, Hieter P. Multifunctional yeast high-copy-number shuttle vectors. Yeast. 1992;110:119–122. doi: 10.1016/0378-1119(92)90454-w. [DOI] [PubMed] [Google Scholar]
- 6.Cui Y, Dinman J D, Peltz S W. mof4-1 is an allele of the UPF1/IFS2 gene which affects both mRNA turnover and −1 ribosomal frameshifting efficiency. EMBO J. 1996;15:5726–5736. doi: 10.1002/j.1460-2075.1996.tb00956.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cui Y, Kinzy T G, Dinman J D, Peltz S W. Mutations in the MOF2/SUI1 gene affect both translation and nonsense-mediated mRNA decay. RNA. 1998;5:794–804. doi: 10.1017/s1355838299982055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Czaplinski K, Ruiz-Echevarria M J, Gonzalez C I, Peltz S W. Should we kill the messenger? The role of the surveillance complex in translation termination and mRNA turnover. Bioessays. 1999;21:685–696. doi: 10.1002/(SICI)1521-1878(199908)21:8<685::AID-BIES8>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- 9.Dahlberg A E. The functional role of ribosomal RNA in protein synthesis. Cell. 1989;57:525–529. doi: 10.1016/0092-8674(89)90122-0. [DOI] [PubMed] [Google Scholar]
- 10.Dechampesme A M, Koroleva O, Leger-Silvestre I, Gas N, Camier S. Assembly of 5S ribosomal RNA is required at a specific step of the pre-rRNA processing pathway. J Cell Biol. 1999;145:1369–1380. doi: 10.1083/jcb.145.7.1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Deshmukh M, Stark J, Yeh L C, Lee J C, Woolford J L., Jr Multiple regions of yeast ribosomal protein L1 are important for its interaction with 5 S rRNA and assembly into ribosomes. J Biol Chem. 1995;270:30148–30156. doi: 10.1074/jbc.270.50.30148. [DOI] [PubMed] [Google Scholar]
- 12.Deshmukh M, Tsay Y F, Paulovich A G, Woolford J L., Jr Yeast ribosomal protein L1 is required for the stability of newly synthesized 5S rRNA and the assembly of 60S ribosomal subunits. Mol Cell Biol. 1993;13:2835–2845. doi: 10.1128/mcb.13.5.2835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dinman J D. Ribosomal frameshifting in yeast viruses. Yeast. 1995;11:1115–1127. doi: 10.1002/yea.320111202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dinman J D, Ruiz-Echevarria M J, Peltz S W. Translating old drugs into new treatments: identifying compounds that modulate programmed −1 ribosomal frameshifting and function as potential antiviral agents. Trends Biotechnol. 1998;16:190–196. doi: 10.1016/S0167-7799(97)01167-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dinman J D, Wickner R B. Ribosomal frameshifting efficiency and Gag/Gag-Pol ratio are critical for yeast M1 double-stranded RNA virus propagation. J Virol. 1992;66:3669–3676. doi: 10.1128/jvi.66.6.3669-3676.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dinman J D, Wickner R B. Translational maintenance of frame: mutants of Saccharomyces cerevisiae with altered −1 ribosomal frameshifting efficiencies. Genetics. 1994;136:75–86. doi: 10.1093/genetics/136.1.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dinman J D, Wickner R B. 5S rRNA is involved in fidelity of translational reading frame. Genetics. 1995;141:95–105. doi: 10.1093/genetics/141.1.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dohme F, Nierhaus K H. Role of 5S RNA in assembly and function of the 50S subunit from Escherichia coli. Proc Natl Acad Sci USA. 1976;73:2221–2225. doi: 10.1073/pnas.73.7.2221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dokudovskaya S, Dontsova O, Shpanchenko O, Bogdanov A, Brimacombe R. Loop IV of 5S ribosomal RNA has contacts both to domain II and to domain V of the 23S RNA. RNA. 1996;2:146–152. [PMC free article] [PubMed] [Google Scholar]
- 20.Dontsova O A, Tishkov V, Dokudovskaya S, Bogdanov A, Doring T, Rinke-Appel J, Thamm S, Greuer B, Brimacombe R. Stem-loop IV of 5S rRNA lies close to the peptidyltransferase center. Proc Natl Acad Sci USA. 1994;91:4125–4129. doi: 10.1073/pnas.91.10.4125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Goringer H U, Bertram S, Wagner R. The effect of tRNA binding on the structure of 5 S RNA in Escherichia coli. A chemical modification study. J Biol Chem. 1984;259:491–496. [PubMed] [Google Scholar]
- 22.Hampsey M. A review of phenotypes in Saccharomyces cerevisiae. Yeast. 1997;13:1099–1133. doi: 10.1002/(SICI)1097-0061(19970930)13:12<1099::AID-YEA177>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- 23.Hentze M W, Kulozik A E. A perfect message: RNA surveillance and nonsense-mediated decay. Cell. 1999;96:307–310. doi: 10.1016/s0092-8674(00)80542-5. [DOI] [PubMed] [Google Scholar]
- 24.Ito H, Fukuda Y, Murata K, Kimura A. Transformation of intact yeast cells treated with alkali cations. J Bacteriol. 1983;153:163–168. doi: 10.1128/jb.153.1.163-168.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Khaitovich P, Mankin A S. Effect of antibiotics on large ribosomal subunit assembly reveals possible function of 5 S rRNA. J Mol Biol. 1999;291:1025–1034. doi: 10.1006/jmbi.1999.3030. [DOI] [PubMed] [Google Scholar]
- 26.Kim D F, Green R. Base-pairing between 23S rRNA and tRNA in the ribosomal A site. Mol Cell. 1999;4:859–864. doi: 10.1016/s1097-2765(00)80395-0. [DOI] [PubMed] [Google Scholar]
- 27.Kitakawa M, Isono K. The mitochondrial ribosomes. Biochimie. 1991;73:813–825. doi: 10.1016/0300-9084(91)90061-5. [DOI] [PubMed] [Google Scholar]
- 28.Kunkel T. Rapid and efficient site-specific mutagenesis without phenotype selection. Proc Natl Acad Sci USA. 1985;82:488–492. doi: 10.1073/pnas.82.2.488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lee Y, Nazar R N. Ribosomal 5 S rRNA maturation in Saccharomyces cerevisiae. J Biol Chem. 1997;272:15206–15212. doi: 10.1074/jbc.272.24.15206. [DOI] [PubMed] [Google Scholar]
- 30.Lieberman K R, Dahlberg A E. Ribosome-catalyzed peptide-bond formation. Prog Nucleic Acid Res Mol Biol. 1995;50:1–23. [PubMed] [Google Scholar]
- 31.Macbeth M R, Wool I G. Characterization of in vitro and in vivo mutations in non-conserved nucleotides in the ribosomal RNA recognition domain for the ribotoxins ricin and sarcin and the translation elongation factors. J Mol Biol. 1999;285:567–580. doi: 10.1006/jmbi.1998.2337. [DOI] [PubMed] [Google Scholar]
- 32.Macbeth M R, Wool I G. The phenotype of mutations of G2655 in the sarcin/ricin domain of 23 S ribosomal RNA. J Mol Biol. 1999;285:965–975. doi: 10.1006/jmbi.1998.2388. [DOI] [PubMed] [Google Scholar]
- 33.Melancon P, Tapprich W E, Brakier-Gingras L. Single-base mutations at position 2661 of Escherichia coli 23S rRNA increase efficiency of translational proofreading. J Bacteriol. 1992;174:7896–7901. doi: 10.1128/jb.174.24.7896-7901.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Meskauskas A, Dinman J D. Ribosomal protein L5 helps anchor peptidyl-tRNA to the P-site in Saccharomyces cerevisiae. RNA. 2001;7:1084–1096. doi: 10.1017/S1355838201001480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mueller F, Sommer I, Baranov P, Matadeen R, Stoldt M, Wohnert J, Gorlach M, van Heel M, Brimacombe R. The 3D arrangement of the 23 S and 5 S rRNA in the Escherichia coli 50 S ribosomal subunit based on a cryo-electron microscopic reconstruction at 7.5 D resolution. J Mol Biol. 2000;298:35–59. doi: 10.1006/jmbi.2000.3635. [DOI] [PubMed] [Google Scholar]
- 36.Nissen P, Ippolito J A, Ban N, Moore P B, Steitz T A. RNA tertiary interactions in the large ribosomal subunit: the A-minor motif. Proc Natl Acad Sci USA. 2001;98:4899–4903. doi: 10.1073/pnas.081082398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Noller H F. Ribosomal RNA and translation. Annu Rev Biochem. 1991;60:191–227. doi: 10.1146/annurev.bi.60.070191.001203. [DOI] [PubMed] [Google Scholar]
- 38.Noller H F. Peptidyltransferase: protein, ribonucleoprotein, or RNA? J Bacteriol. 1993;175:5297–5300. doi: 10.1128/jb.175.17.5297-5300.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Noller H F. Ribosomes and translation. Annu Rev Biochem. 1997;66:679–716. doi: 10.1146/annurev.biochem.66.1.679. [DOI] [PubMed] [Google Scholar]
- 40.Oakes M, Aris J P, Brockenbrough J S, Wai H, Vu L, Nomura M. Mutational analysis of the structure and localization of the nucleolus in the yeast Saccharomyces cerevisiae. J Cell Biol. 1998;143:23–34. doi: 10.1083/jcb.143.1.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.O'Connor M, Brunelli C A, Firpo M A, Gregory S T, Lieberman K R, Lodmell J S, Moine H, Van Ryk D I, Dahlberg A E. Genetic probes of ribosomal RNA function. Biochem Cell Biol. 1995;73:859–868. doi: 10.1139/o95-093. [DOI] [PubMed] [Google Scholar]
- 42.O'Connor M, Dahlberg A E. The involvement of two distinct regions of 23 S ribosomal RNA in tRNA selection. J Mol Biol. 1995;254:838–847. doi: 10.1006/jmbi.1995.0659. [DOI] [PubMed] [Google Scholar]
- 43.O'Connor M, Dahlberg A E. The influence of base identity and base pairing on the function of the alpha-sarcin loop of 23S rRNA. Nucleic Acids Res. 1996;24:2701–2705. doi: 10.1093/nar/24.14.2701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ohtake Y, Wickner R B. Yeast virus propagation depends critically on free 60S ribosomal subunit concentration. Mol Cell Biol. 1995;15:2772–2781. doi: 10.1128/mcb.15.5.2772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Osswald M, Brimacombe R. The environment of 5S rRNA in the ribosome: cross-links to 23S rRNA from sites within helices II and III of the 5S molecule. Nucleic Acids Res. 1999;27:2283–2290. doi: 10.1093/nar/27.11.2283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Peltz S W, Hammell A B, Cui Y, Yasenchak J, Puljanowski L, Dinman J D. Ribosomal protein L3 mutants alter translational fidelity and promote rapid loss of the yeast killer virus. Mol Cell Biol. 1999;19:384–391. doi: 10.1128/mcb.19.1.384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Polacek N, Gaynor M, Yassin A, Mankin A S. Ribosomal peptidyl transferase can withstand mutations at the putative catalytic nucleotide. Nature. 2001;411:498–501. doi: 10.1038/35078113. [DOI] [PubMed] [Google Scholar]
- 48.Rose M D, Winston F, Hieter P. Methods in yeast genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Press; 1990. [Google Scholar]
- 49.Ruiz-Echevarria M J, Czaplinski K, Peltz S W. Making sense of nonsense in yeast. Trends Biochem Sci. 1996;21:433–438. doi: 10.1016/s0968-0004(96)10055-4. [DOI] [PubMed] [Google Scholar]
- 50.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 51.Schluenzen F, Tocilj A, Zarivach R, Harms J, Gluehmann M, Janell D, Bashan A, Bartels H, Agmon I, Franceschi F, Yonath A. Structure of functionally activated small ribosomal subunit at 3.3 angstroms resolution. Cell. 2000;102:615–623. doi: 10.1016/s0092-8674(00)00084-2. [DOI] [PubMed] [Google Scholar]
- 52.Schulze H, Nierhaus K H. Minimal set of ribosomal components for reconstitution of the peptidyltransferase activity. EMBO J. 1982;1:609–613. doi: 10.1002/j.1460-2075.1982.tb01216.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sergiev P, Dokudovskaya S, Romanova E, Topin A, Bogdanov A, Brimacombe R, Dontsova O. The environment of 5S rRNA in the ribosome: cross-links to the GTPase-associated area of 23S rRNA. Nucleic Acids Res. 1998;26:2519–2525. doi: 10.1093/nar/26.11.2519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sergiev P V, Bogdanov A A, Dahlberg A E, Dontsova O. Mutations at position A960 of E. coli 23 S ribosomal RNA influence the structure of 5 S ribosomal RNA and the peptidyltransferase region of 23 S ribosomal RNA. J Mol Biol. 2000;299:379–389. doi: 10.1006/jmbi.2000.3739. [DOI] [PubMed] [Google Scholar]
- 55.Sergiev P V, Lavrik I N, Dokudovskaya S S, Dontsova O A, Bogdanov A A. Structure of the decoding center of the ribosome. Biochemistry (Moscow) 1998;63:963–976. [PubMed] [Google Scholar]
- 56.Shpanchenko O V, Dontsova O A, Bogdanov A A, Nierhaus K H. Structure of 5S rRNA within the Escherichia coli ribosome: iodine-induced cleavage patterns of phosphorothioate derivatives. RNA. 1998;4:1154–1164. doi: 10.1017/s1355838298980359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tabor S, Richardson C C. DNA sequence analysis with a modified bacteriophage T7 DNA polymerase. Proc Natl Acad Sci USA. 1987;84:4767–4771. doi: 10.1073/pnas.84.14.4767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Thompson J, Kim D F, O'Connor M, Lieberman K R, Bayfield M A, Gregory S T, Green R, Noller H F, Dahlberg A E. Analysis of mutations at residues A2451 and G2447 of 23S rRNA in the peptidyltransferase active site of the 50S ribosomal subunit. Proc Natl Acad Sci USA. 2001;98:9002–9007. doi: 10.1073/pnas.151257098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Van Ryk D I, Nazar R N. Effect of sequence mutations on the higher order structure of the yeast 5S rRNA. J Mol Biol. 1992;226:1027–1035. doi: 10.1016/0022-2836(92)91050-y. [DOI] [PubMed] [Google Scholar]
- 60.Venema J, Dirks-Mulder A, Faber A W, Raue H A. Development and application of an in vivo system to study yeast ribosomal RNA biogenesis and function. Yeast. 1995;11:145–156. doi: 10.1002/yea.320110206. [DOI] [PubMed] [Google Scholar]
- 61.Wickner R B. Double-stranded RNA viruses of Saccharomyces cerevisiae. Microbiol Rev. 1996;60:250–265. doi: 10.1128/mr.60.1.250-265.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wickner R B, Leibowitz M J. Two chromosomal genes required for killing expression in killer strains of Saccharomyces cerevisiae. Genetics. 1976;82:429–442. doi: 10.1093/genetics/82.3.429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Woolford J L, Warner J R. The ribosome and its synthesis. In: Broach J R, Pringle J R, Jones E W, editors. The molecular and cellular biology of the yeast Saccharomyces. Vol. 1. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1991. pp. 587–626. [Google Scholar]
- 64.Yonath A, Harms J, Hansen H A, Bashan A, Schlunzen F, Levin I, Koelln I, Tocilj A, Agmon I, Peretz M, Bartels H, Bennett W S, Krumbholz S, Janell D, Weinstein S, Auerbach T, Avila H, Piolleti M, Morlang S, Franceschi F. Crystallographic studies on the ribosome, a large macromolecular assembly exhibiting severe nonisomorphism, extreme beam sensitivity and no internal symmetry. Acta Crystallogr. 1998;54:945–955. doi: 10.1107/s010876739800991x. [DOI] [PubMed] [Google Scholar]
- 65.Yoshioka K, Kanda H, Takamatsu N, Togashi S, Kondo S, Miyake T, Sakaki Y, Shiba T. Efficient amplification of Drosophila simulans copia directed by high-level reverse transcriptase activity associated with copia virus-like particles. Gene. 1992;120:191–196. doi: 10.1016/0378-1119(92)90093-5. [DOI] [PubMed] [Google Scholar]
- 66.Yusupov M M, Yusupova G Z, Baucom A, Lieberman K, Earnest T N, Cate J H, Noller H F. Crystal structure of the ribosome at 5.5 D resolution. Science. 2001;292:883–896. doi: 10.1126/science.1060089. [DOI] [PubMed] [Google Scholar]