Abstract
A key challenge for the discovery of novel molecular targets and therapeutics against pediatric bone metastatic disease is the lack of bona fide in vitro cell models. Here, we show that a beta-tricalcium phosphate (β-TCP) multicellular 3D in vitro bone microtissue model reconstitutes key phenotypic and transcriptional patterns of native metastatic tumor cells while promoting their stemness and proinvasive features. Comparing planar with interconnected channeled scaffolds, we identified geometry as a dominant orchestrator of proangiogenic traits in neuroblastoma tumor cells. On the other hand, the β-TCP-determined gene signature was DNA replication related. Jointly, the geometry and chemical impact of β-TCP revealed a prometastatic landscape of the engineered tumor microenvironment. The proposed 3D multicellular in vitro model of pediatric bone metastatic disease may advance further analysis of the molecular, genetic and metabolic bases of the disease and allow more efficient preclinical target validations.
Keywords: Metastatic bone model, β-TCP ceramics Application, 3D bioengineering, Tumor microenvironment, In vitro biological models
Graphical abstract

1. Introduction
Recent advances in the field of bioengineering have opened a new perspective for artificially reproducing biological moieties with high levels of complexity while reducing the current limitations of conventional two-dimensional (2D) in vitro experimental models [1]. For instance, in the case of bone replicates, a combination of diverse types of biomaterials with mesenchymal stromal (stem) cells (MSCs) was successful in achieving proper cell differentiation and extracellular matrix (ECM) production and mineralization as hub processes engaged in forming the bone microenvironment [2]. The addition of endothelial cells to that system is a necessary step toward warranting the vascularization of this three-dimensional (3D) bone system. Together, these components are essential for assembly into a microtissue [3]. The bioengineering strategies that reconstruct bone microtissues were initially recognized as highly relevant only for regenerative medicine. Recently, other scientific fields, including oncology, have shown interest in adopting these microtissues for the 3D reproduction of specific nonphysiological conditions, e.g., in vitro cancer models [4]. Their introduction enabled more systematic investigations of the biological processes that endure tumor cell behavior under in vivo-approaching growth conditions [5]. In addition, by more closely recapitulating the geometric intricacy of native tissues, bone microtissues are suitable for delineating the coordinative activities of malignant and nonmalignant compartments within the third dimension [6].
Among the biomaterials that are applied for bone microtissue assembly, beta-tricalcium phosphate (β-TCP) has been extensively tested since it displays required osteoconductive and osteoinductive properties while maintaining a superior degradation rate compared to hydroxyapatite (HA) in vivo [7]. Importantly, β-TCP releases Ca2+ ions that are essential for tempering MSC proliferation and differentiation toward osteoblast-like cells [8], but also for driving the neovascularization process [9]. In addition, biochemical (such as growth factors, cytokines, DNA and RNA) or biophysical (such as stiffness, porosity, elasticity and pore size) cues of the bone also strongly influence the molecular landscape of the cells, impacting their function [10]. Thus, it is important to consider each of these aspects when choosing proper 3D structures to accurately reflect their impacts on cell (malignant and/or nonmalignant) morphology, transcriptional background and overall behavior [11].
In the case of metastatic solid tumors, 3D structures have been confirmed as exceedingly valuable models for studying either the premetastatic niche or disseminated disease [12]. Their application is particularly useful in malignancies for which tumor material is not easily accessible to biologists and biomedical experts. Moreover, 3D tumor models are especially acknowledged in recognizing more informative biological and pharmacological features, either individual or combinatorial, that govern metastatic tumor cell behavior. In the case of neuroblastoma (NB), the most common and most aggressive pediatric extracranial solid tumor with a high tendency for metastatic spread [13], the introduction of 3D disease models would open new opportunities for reliable basic research. As an alternative to the scarce tumor-derived specimens, biologically reliable 3D models that recap substantial phenotypical and molecular heterogeneity found in neoplastic tissues would ultimately serve as a guide toward defining more effective therapeutic interventions [14]. The most frequent distant metastasis of NB develops in the bones [15]. Bone metastasis is a negative prognostic marker and determines unfavorable outcomes of patients affected by NB for whom new therapeutic policies are still required in the era of substantial advances in the molecular and genetic characterization of disease [16]. Currently, there are few in vitro alternatives to patient-derived material for studying metastatic NB [17]. At the same time, in vivo mouse models of NB disease involving bones are still very limited [18]. Bone microtissues might therefore be considered a promising starting point for engineering metastatic tumors in vitro by artificially recreating the bone microenvironment. In the following, these models could be investigated regarding the types of interactions in place between tumor and adjacent nonmalignant cellular and acellular components (e.g., ECM) while explaining how these interfaces shape the molecular landscape of NB cells. In terms of medical translation, it is expected that the experimental readouts may set a base for more trustworthy preclinical screenings of targeted or biological therapies [19] against NB or against the tumor microenvironment (TME) [20].
Herein, we used β-TCP-derived NB metastatic bone microtissue with increased cellular heterogeneity to better approach its native state. In doing so, along with the stromal component of the TME, we explored the possibility of introducing endothelial (nonmalignant) cells and zoomed out their organization in two geometrically distinct β-TCP scaffolds, with interconnected channels (CSc) and planar (PSc) structure. Finally, the influence of β-TCP, geometry and coculturing complexity over tumorigenic features of NB cells were comprehensively inspected at the molecular level.
2. Materials and methods
2.1. Cell lines, primary cells and culturing conditions
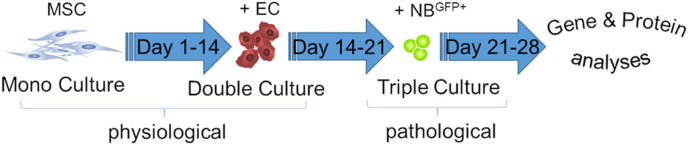
The human NB tumor cell line (SH-SY5Y) was obtained from the Deutsche Sammlung von Mikroorganismen und Zellkulturen (DSMZ, Braunschweig, Germany). To generate stably-expressing enhanced green fluorescent protein (eGFP) reporter cells, cells were transfected with the GFP-plasmid construct (GeneCopoeia, Vienna, Austria) using Effectene® Transfection Reagent [21]. The NBGFP + cells were grown in DMEM supplemented with 10% FCS and 100 U/mL penicillin/streptomycin (all from PAN-Biotech, Aidenbach, Germany). The use of human primary multipotent mesenchymal stromal cells (MSCs) and human umbilical vein endothelial cells (ECs) was approved by the local ethics committee (EK 424/19). Primary cells were obtained and cultured up to the 5th passage as described elsewhere [21]. Mono-, double- and triple-cultures of MSC, EC and NB tumor (SH-SY5Y) cells were obtained by a time-dependent seeding schedule (Fig. 1). Briefly, β-TCP structures were placed in a 48-well plate and equilibrated for 15 min with MSC growth medium (Mesenpan, PAN Biotech, Aidenbach, Germany) before cell seeding. Following this, 30,000 MSCs suspended in 30 μL of growth medium were equally distributed inside the channels (3D channeled β-TCP scaffolds – CSc) or seeded on the top surface of planar β-TCP counterparts (PSc). The same approach was adopted for seeding 60,000 EC on day 14 and 10,000 tumor cells on day 21. A triple culture was maintained in endothelial basal medium enriched with EGM-2 growth supplements (PromoCell, Heidelberg, Germany) until day 28.
Fig. 1.
A 3D metastatic bone model – cell seeding time frame scheme. MSC – mesenchymal stromal cells, EC – endothelial cells, NBGFP + - green fluorescent protein expressing neuroblastoma cells. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
2.2. Scaffold manufacturing
Using tailored Computer Aided Design (CAD) models as a starting point, wax negative molds were printed to create the 3D scaffolds with a network of the interconnected microchannels with 500 μm in diameter. The molds were made of a building wax (Indura®Cast) and supporting wax (Indura®Fill, Solidscape, New Hampshire, USA). Layer by layer, the molds were printed utilizing drop on demand technology. Following printing, the support wax was dissolved in petroleum (Merck 1.09718, Merck, Darmstadt, Germany) to produce the wax mold. Next, the mold was fixed to the base using melted building wax and filled with the slurry made of organic solvent and β-TCP powder (Budenheim Pharmaceutical Technology, Germany). Sintering temperature was set to around 1200 °C for 3 h to avoid phase decomposition.
2.3. Lactate dehydrogenase (LDH) activity measurement and live/dead staining
The percentage of damaged or injured cells in our samples was measured indirectly by colorimetric LDH assay (Abcam, Cambridge, UK). For this purpose, the supernatants were collected on days 10, 14, 21 and 28 and processed according to the manufacturer's protocol for determining LDH activity. The absorbance intensity was measured at 490 nm by SpectraMax, and enzymatic activity was presented as mU/mL. In addition, a two-color fluorescence (calcein AM and ethidium homodimer; Invitrogen, Karlsruhe, Germany) cell viability assay was used for the simultaneous determination of live (green) and dead (red) cells; data were analyzed by two-photon microscopy.
2.4. Cytokine profiling
Relative cytokine expression was measured in the supernatants collected on day 28 using the samples from day 21 for normalization. Several approaches were used: interleukin 6 (IL-6) and vascular endothelial growth factor (VEGF) concentrations were measured on an Applied Biosystems ABI 7900HT Real Time Thermo Cycler using ProQuantum High-Sensitivity Immunoassays kits (Thermo Fisher, Karlsruhe, Germany) following the manufacturer's instructions. The results are presented as the mean concentration (pg/mL) of the protein obtained from the experimental triplicates. Angiogenic factors (including VEGF, IL-6, basic fibroblast growth factor - FGFb, epidermal growth factor - EGF, transforming growth factor-beta - TGF-b, tumor necrosis factor α - TNFα, insulin-like growth factor 1 - IGF-1, and leptin) profiling was assessed simultaneously using Angiogenesis ELISA Strip I Profiling Assay (Signosis, BioCat GmbH, Heidelberg, Germany). The results are presented as the mean absorbance obtained from experimental quadruplicates. Measurements were performed on a SpectraMax spectrophotometer at 450 nm.
2.5. Two-photon analysis and scanning electron microscopy (SEM)
For intrachannel cell visualization, immunostaining followed by two-photon microscopy was adopted. Details of the procedure are described elsewhere [22]. Briefly, the samples were fixed with 4% PFA for 3 h, washed 3x with PBS and permeabilized with 0.25% Triton X-100 buffer to allow intracellular protein staining. Blocking was performed in 3% BSA solution, and antibody dilutions were prepared following the manufacturer's recommendations (phalloidin-TRITC #P1951 and phalloidin-FITC #P5282 for F-actin visualization, Sigma-Aldrich, 1:400; anti-VE-cadherin #sc-9989 and anti-IGF2 #sc-515805 primary antibodies, SC Biotechnology, Heidelberg, Germany, 1:400). Alexa Fluor-594 (Abcam #ab150120; 1:2000) was used as a secondary antibody for protein visualization. DAPI (Thermo Fisher #62248; 1:10,000) was used for nuclear labeling. Imaging was performed by a two-photon microscope (FV1000MPE, Olympus, Tokyo, Japan) with a 25x NA 1.05 water dipping objective. The excitation wavelength was mode-locked to 800 nm using a pulsed Ti:Sapphire laser (Mai Tai DeepSee, Spectra Physics, Santa Clara, CA, USA). Single images (approximately 20–30 for PSc, and 100–150 for CSc) were recorded in the X–Y plane and collected at different 1.0 μm Z heights. Image processing, 3D reconstruction of the samples, and directionality histograms were made using specific plugins provided by Fiji [23]. For SEM, fixation with 3% glutaraldehyde (Agarscientific, Wetzlar, Germany) was performed, followed by washing in 0.1 M Soerensen's Phosphate Buffer (Merck, Darmstadt, Germany) for 15 min, dehydration in an ascending dilution of acetone and drying with a critical point drying method in liquid CO2 (CriticalPointDryer, Polaron, QuorumTechnologies Ltd., Ashford, Kent, England). The samples were then coated with 10 nm gold/palladium film (Sputter Coater EM SCD500, Leica, Wetzlar, Germany), and microscopy was performed in a high-vacuum environment (ESEM XL30 FEG, FEI, Eindhoven, The Netherlands) with a 10 kV acceleration voltage.
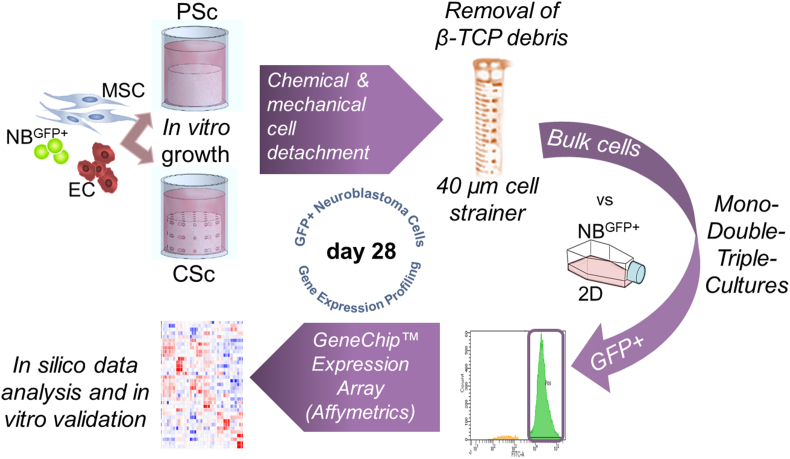
2.6. Cell sorting, RNA extraction and gene expression array performance
After 28 days of growth, cells were removed from the scaffolds by trypsinization sustained by slight vortexing. Recovery of GFP + tumor cells from bulk material was performed, after passing the material through 40 μm EASYstrainer™ filters, by sorting on a BD FACS Aria II instrument equipped with 3 lasers (Violet 407 nm, Blue 488 nm, Red 633 nm). After sorting, the cells were washed once with PBS, pelleted, resuspended in TRIzol reagent (Invitrogen; Thermo Fisher), and stored at −80 °C until RNA isolation. Total RNA was extracted using columns from Direct-zol RNA Purification Kits (Zymo Research) according to the manufacturer's recommendations. RNA concentration was detected with Qubit (Invitrogen, Thermo Fisher), while quality and integrity were estimated with an RNA 6000 PICO Assay conducted on an Agilent 2100 Bioanalyzer (Agilent Technologies, Tokyo, Japan). One nanogram of total RNA was used for performing in vitro transcription, hybridization and biotin labeling with the GeneChip™ Expression Arrays 3′ IVT Pico Kit (Affymetrics, Thermo Fisher Scientific). The experiments were run as experimental duplicates and technical triplicates. The hybridization was performed on the Human Clariom™ S Gene Chip cartridge array.
2.7. Differentially expressed genes (DEGs), functional enrichment analysis and Protein‒Protein interaction (PPI) network construction
Raw data (CEL files) were processed and normalized using the Robust Multichip Average (RMA) algorithm (rma function of the R package oligo (v1.58.0)) with standard parameters. Annotation of the resulting expression set was conducted using the annotateEset function of the R package affycoretools (v1.66.0). All rows with ‘NA’ entries were omitted. Probes for the same gene of a specific array were aggregated using the mean value. The design matrix contains the batch effect introduced by performing two separate experiments in addition to the experimental conditions as a factor of interest. The differential expression analyses were performed following the standard limma workflow using the lmFit, contrasts. fit, and eBayes methods from the limma package (v350.3). Correction for multiple tests was performed using the Benjamini Hochberg adjustment method. To compute the specific marker genes of each experimental condition, we used the one-against-all approach, where the gene expression from one condition is compared to the average expression from the other conditions. In that case, we selected robust = True and trend = True to account for hypervariable or hypovariable average expression of genes and the mean-variance relationship, respectively. For downstream analysis, genes with adjusted P value≤0.05 and |log2FC| ≥ 0.56 were selected. Signals shown in heatmaps are row-scaled expression values. Hierarchical clustering was performed using the Euclidean distance and the complete linkage method of the function “hclust” from the R stats package (v4.1.3). The heatmaps were generated using the R package pheatmap (v1.012). GO functional and KEGG pathway enrichment analyses for the intersected or unique genes were run online using the Enrichr v2021 web server. QuickGo v2022 was used to compare connections between GO terms. The protein interaction data were initially analyzed by the STRING database (https://string-db.org/) and imported into Cytoscape v3.9.1 for PPI network construction and module definition. Biological networks defined by DEGs were inspected by the NDEx network viewer [24]. Patient-derived material from the SEQC-498 cohort was obtained from the publicly available GSE49710 dataset and analyzed on the R2 platform (https://r2.amc.nl).
2.8. Statistical analysis
In vitro data were plotted as the mean values with standard deviation. Unless stated otherwise, three experimental replicates (n = 3) were performed. Statistical significance was determined by a two-tailed Student's t-test and one-way ANOVA with a post hoc Tukey test using GraphPad Prism, and significantly different values were considered for p value < 0.05 (∗).
3. Results
3.1. The engineered β-TCP scaffold microenvironment resembles a native bone metastatic niche
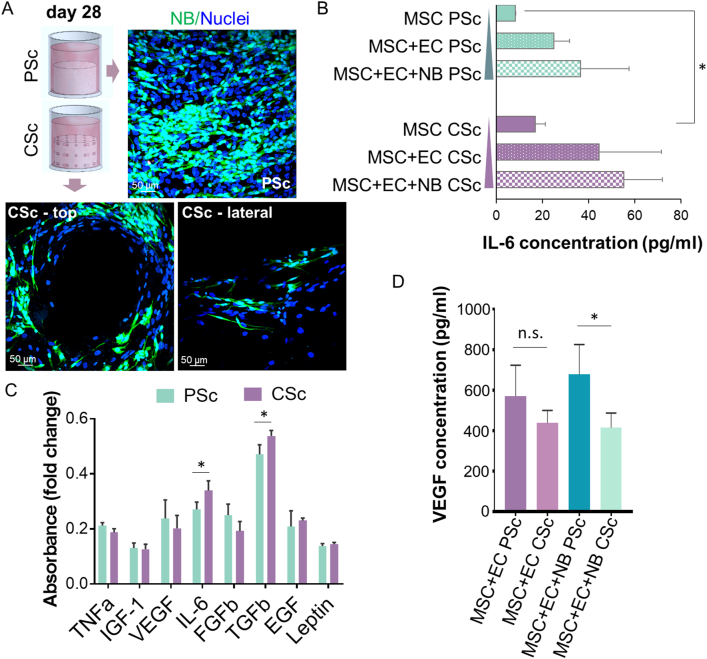
The objective of this study was to develop 3D microtissue of NB bone metastasis recapitulating the in vivo organization and morphology of disseminated tumor cells. 3D-printed β-TCP scaffolds with either planar surface (PSc) or channeled structure (CSc) were used to investigate the effects of geometry on cell behavior. In our previous work, the stromal component was confirmed as a substantial element of 3D models for enabling meshwork-like cellular organizations and formation of the TME necessary for sustaining tumor cell aggregation in the forms of rosettes [25]. Here, an endothelial cell (EC) component has been introduced to more closely mimic the microtissue composition of a native bone metastatic niche [26]. We compared how this triple culture system (mesenchymal stromal cells - MSC, EC, and NB cells) behaves microscopically when grown either on the planar surface or inside the channels of the engineered β-TCP scaffolds (Fig. 2A; PSc and CSc, respectively). Similarly to what we have reported recently, a dense meshwork-like organization of nonmalignant cells was detected [22]. However, more disseminated pattern of NBGFP + cells was found only inside the CSc structures. A low activity of LDH (Fig. 2B) was measured at different time points in both types of scaffolds (PSc and CSc) and for all ranges of cellular complexity (mono-, double- and triple-culture), therefore excluding the cytotoxic impact of the structure on any of the applied cell types. In addition, optimal cell viability (>90%) was maintained until day 28 in both of the β-TCP scaffolds, PSc or CSc, as confirmed by live/dead staining (Fig. 2C). Together, these data imply that the adopted 3D growth conditions successfully nourished the coculture of all three implemented cell types, confirming the validity of the system for further biological investigations.
Fig. 2.
Biocompatibility of planar scaffolds (PSc) and 3D scaffolds with an interconnected channel structure (CSc). A) Two-photon fluorescence microscopy graphs represent the organization of the MSC, EC and NB cells on the PSc and inside the CSc. Labeling: red – F-actin (phalloidin-TRITC); enhanced green protein (eGFP) – NB reporter cells; blue – DAPI (nuclear staining). Scale bar: 50 μm. Insets represent the macroscopic view of the β-TCP scaffolds: PSc and CSc cut laterally. A red asterisk indicates the cell positioning (on top of PSc, and in the inner channel after cutting the CSc). B) Lactate dehydrogenase (LDH) activity measurement for i) MSCs alone, ii) cocultures of MSCs and ECs, and iii) cocultures of MSCs, ECs and NBs. Data are presented as the mean ± SD, n ≥ 3. P value – n. s. (nonsignificant) by one-way ANOVA. C) Live/dead staining for cell viability evaluation in PSc and CSc after 28 days of in vitro growth in 48-well plates. Data are presented as the mean ± SD, n = 3. P value – n. s. (nonsignificant) by Student's t-test. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
Electron microscopy micrographs displayed compact and uniform cell spreading on the top surface of PSc and meshwork-like cell organization inside the CSc (Fig. 3A(i-iii) and 3B(i-iii)). To distinguish the morphology and organization of different cell types, immunostaining analysis was implemented (Fig. 3A(iv-vi) and 3B(iv-vi)). In particular, investigation of VE-cadherin, an EC-specific adhesion protein that favors cell-to-cell contact, in combination with F-actin (used for cytoskeleton labeling), documented a domination of the continuous monolayer of EC in either PSc or CSc that is indicative of vascular integrity maintenance and barrier function [27]. These findings validate the biocompatibility of the β-TCP material and adopted growth conditions for all cell types, including EC. Importantly, in some parts of the scaffolds, the initiation of EC reorganization into elongated, structures was captured recalling a beginning of the microcapillary-like self-assembly (Fig. 3A(v-vi) and 3B(v-vi)). This outcome supports the potential generation of better-defined microvessels upon longer incubation times, as reported previously [28]. Interestingly, the directionality histogram of EC grown on PSc showed their uniform distribution on the surface of the scaffold without any preferred angle, while the directionality histogram of the same cells grown inside the CSc was impacted by the channel presence (Fig. 3C). This is an indication of the fact that the channeled structure potentially affects the morphology (directionality) of the EC.
Fig. 3.
Multicell organization in the β-TCP scaffolds. An overview of the cell morphology on A) the PSc and inside B) the CSc. Scanning electron microscope images (i-ii) and two-photon imaging (iv-vi) of MSC/EC/NB-cell coculture. Dashed squares indicate magnification zones. Red squares indicate the focus of the two-photon imaging. Labeling: red – VE-cadherin; green – F-actin (phalloidin-FITC); blue – DAPI (nuclear staining). Scale bar: 50 μm. C) The favored direction of the EC in the two different β-TCP scaffolds is presented. The analysis was done for the entire population of the cells in the proximity of the scaffold surface. The insets with VE-cadherin staining (red signal) are representative magnifications of the analyzed sections. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
3.2. Scaffold geometry affects the production of IL-6 and TGF-β1 cytokines
After one week of growth in the triple culture conditions, we detected a directional elongation of NB cells: a change in their phenotype that was particularly evident in CSc (Fig. 4A). This observation implied that likely some paracrine mediators were released in the surrounding microenvironment, prompting us to assess the profiles of the most relevant inflammatory and angiogenic cytokines frequently found in the metastatic niche [29]. Similar to our previous report [22], an increase in the amount of the pleiotropic cytokine IL-6 was confirmed for MSCs (Fig. 4B) and NB cells (Fig. 4C) grown in CSc. The addition of EC resulted in an overall increase in IL-6 levels in cocultures, although it was not significantly different between the compared conditions. The majority of other angiogenic factors, such as TNFα, IGF-1, FGFb, EGF, and leptin, did not show significant fluctuations between tumor cells originating from the two different geometrical growth conditions, except for TGF-β1, which increased in CSc-derived samples (Fig. 4C). Remarkably, a tendency toward a lower amount of VEGF was revealed in the supernatants isolated from the CSc double-culture system (EC and MSC) with respect to PSc (Fig. 4D). This difference was confirmed to be significant in the triple culture within the CSc, implying a projected increase in VEGF consumption by ECs, as reported previously [30].
Fig. 4.
Cytokine production in the PSc and CSc. A) Two-photon immunofluorescence analysis of NB-cell morphology in the coculture with MSCs and ECs on top of PSc or inside CSc (for the latter, a top and lateral view after CSc cutting is shown). Labeling: eGFP – NB reporter cells; blue – DAPI (nuclear staining). Scale bar: 50 μm. B) The concentration of IL-6 (pg/mL) was measured in the supernatants of MSCs alone, MSCs/ECs and MSCs/ECs/NBs. Data are presented as the mean ± SD, n = 3, P∗ value < 0.05 by one-way ANOVA. C) The absorbance fold changes for TNFa, IGF-1, VEGF, IL-6, FGFb, TGFb, EGF and leptin were calculated for NB cells grown in PSc and CSc. Data are presented as the mean ± SD, n = 3, P∗ value < 0.05 by one-way ANOVA. D) The concentration of VEGF (pg/mL) was measured for the coculture of MSC/EC and MSC/EC/NB cells. Data are presented as the mean ± SD, n = 3, P∗ value < 0.05 by one-way ANOVA. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
3.3. Geometry shows prevalence over culturing complexity in determining the activation of unique gene sets in tumor cells
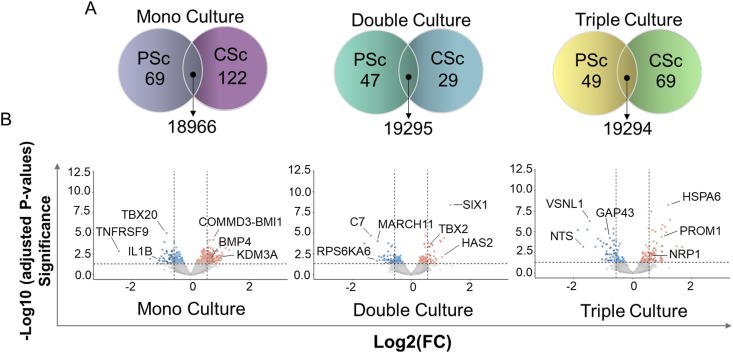
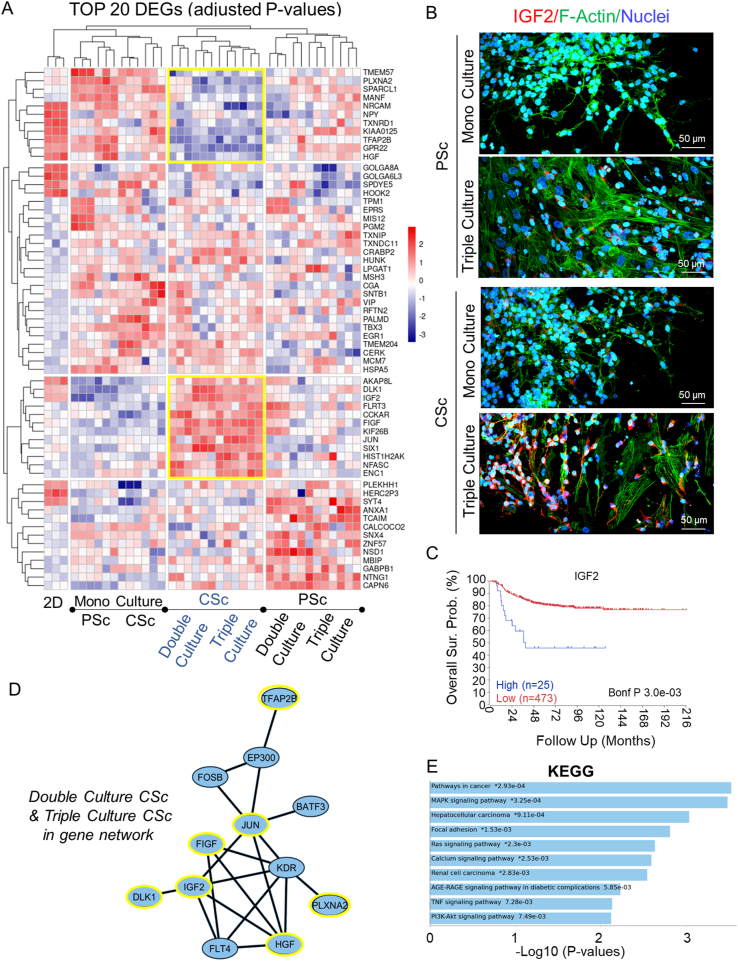
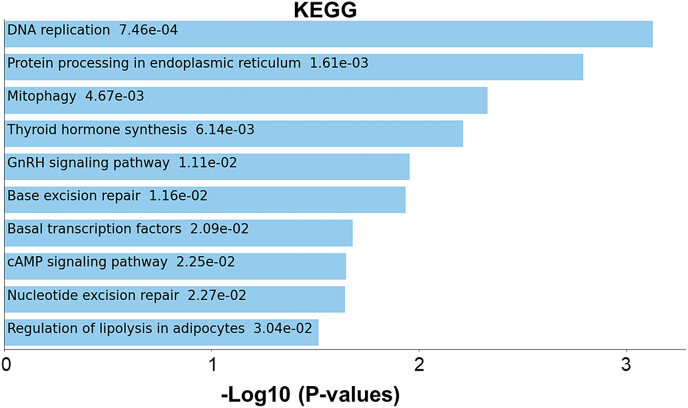
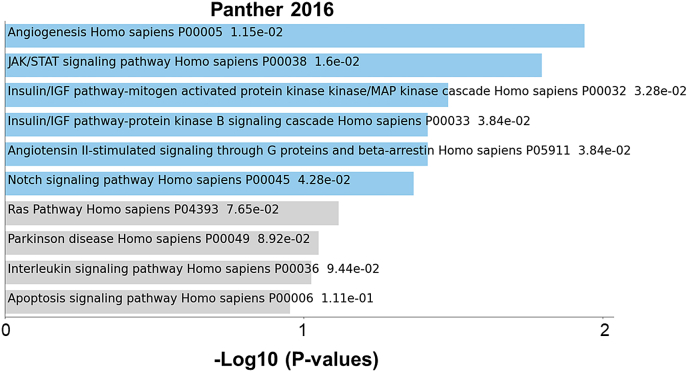
We sought to determine how different microenvironment complexities affect the expression of protein coding genes in NB cells. Considering that our two systems also differ in geometry, in addition to the chemical impact of β-TCP, we explored how each factor is reflected in the set of active genes in NB cells. Therefore, we sorted NBGFP + reporter cells from both PSc and CSc repeatedly for the mono-, double-, and triple-cultures to investigate the set of differentially expressed genes (DEGs) in these cells compared to the 2D monoculture counterpart (Fig. 5). The NDEx Integrated Query (v1.3.1) algorithm was applied to the lists of DEGs in PSc-derived cells and identified a group of genes preferentially affecting cell proliferation and growth suppression escape pathways (Fig. 6A). In contrast, in CSc-derived NB cells, the most evident influence was observed on a group of genes known to regulate cell invasion and metastases (full lists of deregulated genes in each group are provided in Supplementary Table S1). This result was observed regardless of mono- (Fig. 6A(i)), double- (Fig. 6A(ii)) or triple-culture (Fig. 6A(iii)). Next, when DEGs from all the experimental conditions were compared together, a total of 99 genes were found to be specifically deregulated in all β-TCP models (Fig. 6B, shared area between all conditions; Supplementary Fig. S1), allowing us to attribute to them a chemistry-determined signature (Supplementary Table S1). The genes were linked in 3 main modules (Fig. 6C) that were associated principally with unfolded protein response (n = 10; GO:0006986; PPI enrichment p value: 3e-15; FDR = 0.0130), nucleosome assembly (n = 5; GO:0006334; PPI enrichment p value: 7.48e-06; FDR = 0.00016) and DNA replication (n = 4; GO:0006271; PPI enrichment p value: 5.79e-06; FDR = 5.47e-05). The same pathways were found among the top 10 enriched terms when a complete set of 99 shared DEGs was inspected by Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analysis (Supplementary Fig. S2). These results imply that each of the two variables, geometry and chemistry, determine the gene expression patterns in NB cells. Next, we assessed the most specific markers for each experimental condition by comparing the gene expression within each condition to the average gene expression in all other conditions. As expected, the top 15 marker genes of each β-TCP-dependent condition were significantly different from a conventional 2D derived gene set (Fig. 6D). Surprisingly, in the CSc culture system, 23 upregulated genes found in NB cells originating from double culture were also confirmed in triple culture (Fig. 6D; cluster rows 4 and 6). When these 23 upregulated DEGs were inspected by the PANTHER pathway database, significantly enriched terms acquired for the input gene set showed overlap with the proangiogenic processes (Supplementary Fig. S3). More importantly, among them, 10 (43.5%) genes (HIST1H2AK, FIGF, DLK1, ASCL1, LAYN, EBF3, MFAP4, HSAP6, SERPINF1 and IGF2; the latter gene will be discussed more in details further on) showed prognostic value when overexpressed (Supplementary Fig. S4). The obtained results indicate that geometry might have prevalence over culturing complexity in determining the activation of this specific gene set and molecular pathway.
Fig. 5.
Gene expression assessment – the experimental workflow. A multistep preparative procedure for cell isolation and gene expression array performance is displayed.
Fig. 6.
Gene expression profile analyses in mono-, double-, and triple-culture. A) Numbers indicate central common DEGs or unique DEGs in i) mono-, ii) double-, and iii) triple-cultures with respect to NB cells from 2D cell growth. The genes from the most significant biological processes in each of the 3 conditions delineated by NDEx and the genes regulating them are listed. B) The number of common genes between different experimental setups is presented by a Venn diagram generated by Python 3.10. C) PPI network analysis of 99 central DEGs shared across all experimental β-TCP conditions when compared to the wild type (from Panel B) using a combination of STRING online tools and AutoAnnotation App from Cytoscape 3.9.1 software for protein interaction and main modules (proteins that contribute jointly to a shared function) definition, respectively. Module 1 – unfolded protein response; Module 2 – nucleosome assembly; Module 3 – DNA replication. D) Heatmap showing the 15 top upregulated DEGs for each experimental condition compared to all others. Genes were sorted based on the levels of significance (adjusted P value). Only genes with|logFC ≥0.56|and adjusted P value of ≤0.05 were selected for mono-, double-, triple- and 2D culture.
3.4. A bone microenvironment promotes pro-malignant tumor phenotypes
To scrutinize in more detail the contribution of the geometry in defining the molecular landscape of NB cells, we continued by assessing unique DEGs characterizing the cells isolated from the CSc vs. PSc model, regardless of the conventional 2D system (Fig. 7; Supplementary Table S2). As shown in Fig. 7A, a large number of genes were shared between the cells obtained from the three different coculturing modalities, while only a few genes were uniquely expressed in only one of the comparison groups. Among them, TBX20, XPNPEP3, IL-1β, COMMD3-BMI1, BMP4, and KDM3A were deregulated in monoculture; C7, MARCH11, RPS6KA6, TBX2, and HAS2 were deregulated in double culture; and GAP43, VSNL1, NTS, PROM1, NRP1, and HSPA6 [31] were deregulated in triple-culture (Fig. 7B; Supplementary Table S2). These genes were reported previously to be involved in cell function regulation and maintenance of the bone-like niche or have been correlated with tumor progression [28,[32], [33], [34], [35], [36]]. Particularly, in the triple-culture CSc system, a stem feature of NB cells was elicited through sustained overexpression of PROM1 (human prominin 1; a CD133 protein coding gene) and a decrease in GAP43 (neuromodulin; an NB differentiation marker gene) [37]. In fact, CD133 has been associated with stem-like phenotypes in numerous solid tumors, including NB, where its expression has been reported in tumors with aggressive clinical profiles [38]. Next, the functional connection of those DEGs (Supplementary Table S2) was explored. More than one module was recognized for all conditions except for PSc double-culture with no specific functional gene module (Supplementary Fig. S5). In monoculture, a core position for the CSc condition (hub module; Supplementary Fig. S5A(i)) was occupied by master regulators of ECM remodeling (TIMP3, IGF2, BMP4) and tumor growth and metastatic progression (DLK1, IGF2, NTRK2, SIX1, ALX1). Other modules were composed of the genes regulating cell proliferation and chromatin organization (Supplementary Fig. S5B). Under PSc conditions, a hub module accounted for the genes regulating cytokine signaling (PTGS2, RELB, ICAM1, BIRC3), the inflammatory response (TLR4), or both (CCL2, CCL20, IL1B). In double-culture, a hub module was accountable for the metabolic activity of CSc-derived NB cells (Supplementary Fig. S5A(ii)). The triple-culture system favored the expression of genes regulating cell differentiation (CREM, ATF3, NRP1, BHLHE40, PROM1, DLK1, ASCL1, LHX8, NDNF, SIX1, BMP7) and proliferation (FIGF, PDGFRA, MAP2K1, RPS6KA2) and has already been described to sustain pro-malignant phenotypes (SLC2A1, PDGFRA, FIGF, AGT, MAP2K1, ASCL1, BMP7, PROM1; Supplementary Fig. S5A(iii)) [[38], [39], [40]]. Additional modules have been related to cell adhesion, synaptic plasticity and growth-promoting processes (Supplementary Fig. S5C). In the case of PSc, cholesterol biosynthesis and metabolism played a hub role in sustaining the growth of NB cells (Supplementary Fig. S5A(iii)), highlighting the relevance of this pathway in providing growth advantages to malignant cell compartments [41].
Fig. 7.
Analysis of a unique set of genes for different coculturing complexities. A) Venn diagrams show a number of unique and common DEGs for two conditions (CSc and PSc) in mono-, double-, and triple-cultures. B) Volcano plots indicate the number of negative (blue) and positive (red) DEGs and the positions of some of the most biologically significant DEGs (manual annotation). (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
3.5. Multicellular 3D bone microtissue favors the expression of proangiogenic markers in neuroblastoma
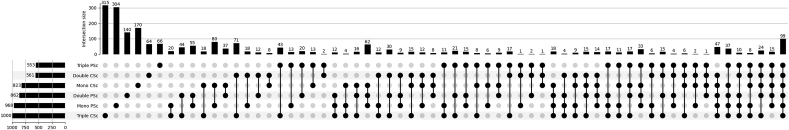
To more closely address the most relevant modules of gene expression patterns in NB cells, the 20 most significant DEGs among designated growth conditions were scrutinized. The unsupervised hierarchical clustering (Fig. 8A) confirmed a similar molecular background when NB cells were grown as monocultures in PSc and CSc, indicating that the role of geometry, as a variable, in this case is marginal. In contrast, in the case of introducing stromal (double-culture) and EC (triple-culture) components to the system, a major effect of geometry on the gene transcripts of NB was revealed; hence, a combination of the two variables, multiple-culturing conditions and geometry, plays a determinant role in defining which gene transcripts will be activated in NB cells. More importantly, tumor cells from double- and triple-cultures isolated from CSc were enriched in multiple factors with known proangiogenic features in tumors, such as DLK1, IGF2, JUN and SIX1 [42,43]. Finally, to confirm the results obtained from in silico analysis, the expression of IGF2 protein was validated (Fig. 8B). As evident from these two-photon microscopy images, IGF2 protein was abundantly expressed only in the triple-culture CSc system. The clinical significance of the IGF2 level was then supported by comparing the overall survival for IGF2-high NB and IGF2-low NB (Fig. 8C). The two groups of patients presented significantly different survival rates (P = 0.003), which was attributed to IGF2 as a putative prognostic marker. Based on functional connectivity, the abovementioned proangiogenic genes create a network with other transcriptional regulators of angiogenesis, such as KDR, FLT4 and FOSB (Fig. 8D) [44]. On the other hand, the triple-culture CSc system revealed the downregulation of several tumor suppressor genes, including SPARCL1, NRCAM, and KIAA0125 [[45], [46], [47]], hence securing proliferative advantages for tumor cells. These genes, inspected by KEGG analysis, defined several pathways that are indicative of tumor progression, including MAPK and PI3K-AKT signaling (Fig. 8E). Gene Ontology (GO) functional enrichment analysis based on unique DEGs for double- or triple-culture CSc-derived NB cells (Supplementary Fig. 6A, Supplementary Table S3) highlighted the top 10 biological processes (Supplementary Fig. 6B(i)), molecular functions (Supplementary Fig. 6B(ii)), and cellular components (Supplementary Fig. 6B(iii)) that were favored in each of these two conditions. A more complex (triple-) culture system preferentially sustained genes involved in regulating physical contacts between tumor cells and ECM and biochemical signaling while supporting promigratory processes in CSc. Finally, GO annotation for CSc triple-culture confirmed our previous observation, which was attributed to our bone metastatic microtissue model serving a supportive role in regulating NB-cell differentiation, locomotion and signaling (Fig. 9A, Supplementary Table S5). From the viewpoint of molecular function, vascular endothelial growth factor-activated receptor activity (GO:0005021) and insulin-like growth factor binding (GO:0031,994) were highlighted (Fig. 9B, Supplementary Table S4; DEGs), whereas the plasma membrane was the most pertinent cellular component affected by the 3D geometry of microtissue (Fig. 9C, Supplementary Table S5). Together, these processes represent part of a composite signal transduction pathway (Supplementary Fig. S7), which explains our system as a complex and dynamic composition of tumor cells inside the multicellular bone metastatic niche.
Fig. 8.
Screening of NB differentially expressed genes (DEGs) and network analysis in a multiparametric cell system. A) Unsupervised heatmap showing the 20 top DEGs for each experimental condition based on the levels of significance (adjusted P value). Only genes with |logFC ≥0.56| and adjusted P value of ≤0.05 were selected. Yellow squares highlight the CSc-selective DEG signature for higher-complexity systems (double- and triple-cultures). B) Two-photon fluorescence microscopy analysis of IGF2 protein (red fluorescence) levels in two experimental conditions, mono- and triple-culture, and in the PSc and CSc systems. Labeling: green – F-actin (phalloidin-FITC) – to distinguish morphology of nontumor cells; eGFP – NB reporter cells; blue – DAPI (nuclear staining). Scale bar: 50 μm. C) Kaplan‒Meier survival curve showing the overall survival of patients affected by NB with IGF2-low expression (n = 473; red) or IGF2-high expression (n = 25; blue). P values from a Cox regression analysis between IGF2-high and IGF2-low NB are indicated. Significance is adjusted after Bonferroni correction (P value < 0.05). D) Combination of STRING and Cytoscape 3.9.1 software to determine the extended PPI network for CSc-selective DEGs. Yellow circles from the extended gene network highlight the genes extrapolated form the heatmap (yellow squares). E) KEGG pathway analysis for CSc-selective DEGs by Enrichr. The y-axis refers to pathway terms, and the x-axis refers to significance (-log10(P value)). (∗) - indicates the terms with a significant adjusted P value < 0.05. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
Fig. 9.
A GO function enrichment analysis specific for NB cells from a triple culture. A) Distribution of the most significant biological processes enriched specifically in triple-culture CSc-derived NB cells (vs. PSc). B) Scatter plot of GO_Molecular_Function_2021 gene set library terms obtained by the Enrichr gene set enrichment analysis tool. From a total of 16 clusters from a database (Supplementary Table S4), clusters 0–5 are highlighted having the ten most significant GO terms. The terms are plotted based on the first two UMAP dimensions and automatically assigned to a specific cluster (computed with the Leiden algorithm). C) Bar graph indicates the number of genes defining the five most significantly enriched cellular components.
4. Discussion
The bone marrow (BM) microenvironment in close proximity to the bone surface is defined by a sophisticated interaction between multicellular components such as osteoblasts, osteoclasts, endothelial, hematopoietic and immune cells, as well as acellular (ECM) substrates that provide biomechanical (e.g., collagen I and fibronectin) and biochemical support (e.g., mineralized bone components and growth factors) to the system [48]. Based on the type of residing cells, diverse areas (niches) inside the BM can be defined, including osteoblastic (endosteal) [49] and perivascular niches [50]. Under physiological conditions, they balance the processes of cell proliferation and differentiation while supporting the stemness of hematopoietic and nonhematopoietic cell progenitors [51]. In the case of malignancies, the niches are instrumental for provisioned tumor grafting as well as tumor growth and progression [52]. The biological processes behind these actions are intensively under investigation with the goal of isolating precise cellular and molecular events (such as genes, proteins and soluble factors) responsible for tumor evolution. In vitro cellular models of metastatic disease have become indispensable for unraveling the essential biophysical and biomolecular mechanisms assumed by tumor cells and for validating their clinical relevance [53]. However, recapitulating such complex structures in vitro is certainly a challenge and requires multidisciplinary approaches to define optimal biochemical, molecular and physical features that shape the TME. However, progress in manufacturing bone tumor models is evident and proceeds from a conventional 2D monolayer cell culture system toward more sophisticated and functional three-dimensional (3D) multicellular models [54]. Starting from this context, we engineered a bone metastatic microtissue in which genetic circuits have been studied depending on the chemical, microenvironmental and geometric complexity. We then investigated the morphological and transcriptional characteristics of tumor cells grown in β-TCP in vitro models with a special focus on the deadliest and highly metastatic pediatric tumor known as neuroblastoma (NB).
Previously, we confirmed an excellent biocompatibility of the proposed β-TCP bioceramic (both, PSc and CSc) while evaluating our simplified in vitro bone models before and after the transformation step [22,25]. Biocompatibility is an essential characteristic of biomaterials, necessary for the role and function required in the intended application [55]. It has also been verified in the in vitro multicellular metastatic bone model (composed of the stromal, endothelial and NB tumor cells) described here. Along with the optimal biocompatibility of either planar (PSc) or 3D printed β-TCP scaffolds with interconnected channels (CSc), we also detected vigorous and consistent cell-to-cell interactions within a third dimension. Communication between NB and stromal cells in CSc β-TCP scaffolds was enabled through focal adhesion and connexin complexes, confirming previous findings [22]. Similarly, we observed that even in the absence of dynamic cell cultivation, e.g., when using a perfusion bioreactor, diverse geometrical conditions substantially affected cellular organization and orientation (Fig. 2, Fig. 3, Fig. 4A). Likewise, secreted bioactive factors, such as transforming growth factor-β (TGF-β) and interleukin-6 (IL-6), were conditioned by geometry (Fig. 4), suggesting that an osteolytic-like process might be active in our 3D cell system while preserving the aggressive behavior of tumor cells [56]. These data corroborate the roles of TGF-β and IL-6 as the main molecular modulators of the metastatic TME in 3D conditions, closely approaching previous in vivo findings [57]. Moreover, geometry also strongly impacted the transcriptome landscape of tumor cells (Fig. 6, Fig. 7). Modulation of a number of genes that have been previously described in more severe NB tumor phenotypes, including TBX2 [58], PROM1 [38], NRP1, and KDM3A, was particularly marked in 3D CSc-derived NB cells. Importantly, the genes involved in the regulation of angiogenesis (FIGF, IGF2, DLK1) [59] and cell proliferation (TFAP2B, JUN, IGFBP5) [60,61] were more potentiated in the 3D microtissue models (Fig. 6, Fig. 8A). Consistently, biological pathways, such as MAPK and PI3K-AKT, i.e., those pertinent to bone microenvironment remodeling, metastatic disease spread and tumor progression [62,63], were triggered under more complex geometric and coculture growth conditions (Fig. 8E). Each of the latter pathways has already been recognized as an IGF-governed downstream effector [64] that sustains the IL-6-dependent inflammatory and immunomodulatory microenvironment in bone metastases [65]. Additionally, a functional relevance of IGF2 protein that has been previously correlated with tumor colonization, survival inside the bone microenvironment and disease progression [66] was evidenced in our 3D model (Fig. 8B), implying the pertinence of IGF2 to bone metastatic NB-cell growth.
Overall, the observed transcriptional regulation underlined in our 3D in vitro tumor model supported the migratory/invasive pathways of NB tumor cells while sustaining cell stemness (Fig. 6, Fig. 7B). This highlights the specific contributions of chemistry (β-TCP), coculturing complexity and geometry in defining aggressive tumor cell behavior [67,68]. Despite this potential, we are aware that additional features of our 3D model remain to be addressed to achieve the complexity of TME found in vivo. This includes a functional vasculature of the system, dynamic interactions between bone, tumor, and nerve cells, but also the inclusion of immune cells (e.g., T cells and macrophages). Once this level of complexity is secured, it will open a prospective for further understanding of the inflammatory processes governing the metastatic niche (e.g. secretion of inflammatory and neurotrophic factors, cytokines, and chemokines) [69], the release of neurochemicals and neurotransmitters by nerve cells [70], and the dynamic of metabolic rewiring in tumor cells (e.g. serine metabolism) [71]. Together, these comprehensive mechanisms not only promote tumor initiation but also support tumor growth, invasion, and metastasis formation.
Nevertheless, our findings corroborate that the 3D-printed β-TCP multicellular in vitro metastatic model, even if simplified with respect to native tissue, is sufficiently complex to recapitulate key biological processes that have been defined in patient-derived specimens. Hence, it can become a valuable low-cost in vitro tool for studying molecular pathways sustaining bone metastatic processes that are still largely unknown [57,72]. Moreover, it opens new horizons for the preclinical evaluation of innovative therapeutic strategies against specific biological effectors or pathways active in the bone microenvironment.
Author contributions
SA, HF – conceptualization; SA – methodology and work design; ATA, MP, MV, SR, CZ – data acquisition and analysis; RN, SA, ATA – data interpretation; RN, SA – original draft preparation; ATA – software; HF, ATA, CZ – manuscript editing and reviewing. All authors read and approved the current version of the manuscript.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
The authors acknowledge the financial support from the Fondazione Cassa di Risparmio di Padova e Rovigo (CARIPARO Foundation) - project ID 20/11(SA), the Italian Association for Cancer Research (AIRC) - project ID 26484 (SA), and the Deutsche Forschungsgemeinschaft (DFG, German Research Foundation) - 363055819/GRK2415 (HF). This work is done in cooperation with: the “Flow Cytometry”, the “Two-Photon Imaging” and the “Genomics” core facilities of the Interdisciplinary Center for Clinical Research (IZKF) Aachen within the Faculty of Medicine at RWTH Aachen University; the Electron Microscopy Facility of the Institute of Pathology, RWTH University Hospital, Aachen, Germany. The authors would like to thank Roswitha Davtalab and Michael Weber from the Department of Dental Materials and Biomaterials Research, RWTH Aachen University Hospital, for their technical support.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.mtbio.2023.100596.
Appendix A. Supplementary data
The following are the Supplementary data to this article.
figs1.
figs2.
figs3.
figs4.
figs5.
figs6.
figs7.
Data availability
Data will be made available on request.
References
- 1.Martinez E., St-Pierre J.-P., Variola F. Advanced bioengineering technologies for preclinical research. Adv. Phys. X. 2019;4 doi: 10.1080/23746149.2019.1622451. [DOI] [Google Scholar]
- 2.Yu X., Tang X., Gohil S.V., Laurencin C.T. Biomaterials for bone regenerative engineering. Adv. Healthc. Mater. 2015;4:1268–1285. doi: 10.1002/adhm.201400760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Krishnan L., Willett N.J., Guldberg R.E. Vascularization strategies for bone regeneration. Ann. Biomed. Eng. 2014;42:432–444. doi: 10.1007/s10439-014-0969-9. [DOI] [PubMed] [Google Scholar]
- 4.Sitarski A.M., Fairfield H., Falank C., Reagan M.R. 3D tissue engineered in vitro models of cancer in bone. ACS Biomater. Sci. Eng. 2018;4:324–336. doi: 10.1021/acsbiomaterials.7b00097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Marturano-Kruik A., Nava M.M., Yeager K., Chramiec A., Hao L., Robinson S., Guo E., Raimondi M.T., Vunjak-Novakovic G. Human bone perivascular niche-on-a-chip for studying metastatic colonization. Proc. Natl. Acad. Sci. USA. 2018;115:1256–1261. doi: 10.1073/pnas.1714282115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Othman Z., Mohren R.J.C., Cillero-Pastor B., Shen Z., Lacroix Y.S.N.W., Guttenplan A.P.M., Tahmasebi Birgani Z., Eijssen L., Luider T.M., van Rijt S., Habibovic P. Comparative proteomic analysis of human mesenchymal stromal cell behavior on calcium phosphate ceramics with different osteoinductive potential. Mater. Today Bio. 2020;7 doi: 10.1016/j.mtbio.2020.100066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lu H., Zhou Y., Ma Y., Xiao L., Ji W., Zhang Y., Wang X. Current application of beta-tricalcium phosphate in bone repair and its mechanism to regulate osteogenesis. Front. Mater. 2021;8 doi: 10.3389/fmats.2021.698915. [DOI] [Google Scholar]
- 8.Maeno S., Niki Y., Matsumoto H., Morioka H., Yatabe T., Funayama A., Toyama Y., Taguchi T., Tanaka J. The effect of calcium ion concentration on osteoblast viability, proliferation and differentiation in monolayer and 3D culture. Biomaterials. 2005;26:4847–4855. doi: 10.1016/j.biomaterials.2005.01.006. [DOI] [PubMed] [Google Scholar]
- 9.Moccia, Negri Shekha, Faris, Guerra Endothelial Ca2+ signaling, angiogenesis and vasculogenesis: just what it takes to make a blood vessel. Int. J. Mol. Sci. 2019;20:3962. doi: 10.3390/ijms20163962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li J., Liu Y., Zhang Y., Yao B., Enhejirigala, Li Z., Song W., Wang Y., Duan X., Yuan X., Fu X., Huang S. Biophysical and biochemical cues of biomaterials guide mesenchymal stem cell behaviors. Front. Cell Dev. Biol. 2021;9 doi: 10.3389/fcell.2021.640388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tekin H., Simmons S., Cummings B., Gao L., Adiconis X., Hession C.C., Ghoshal A., Dionne D., Choudhury S.R., Yesilyurt V., Sanjana N.E., Shi X., Lu C., Heidenreich M., Pan J.Q., Levin J.Z., Zhang F. Effects of 3D culturing conditions on the transcriptomic profile of stem-cell-derived neurons. Nat. Biomed. Eng. 2018;2:540–554. doi: 10.1038/s41551-018-0219-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Aguado B.A., Bushnell G.G., Rao S.S., Jeruss J.S., Shea L.D. Engineering the pre-metastatic niche. Nat. Biomed. Eng. 2017;1 doi: 10.1038/s41551-017-0077. 0077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shimada H., Ikegaki N. Genetic and histopathological heterogeneity of neuroblastoma and precision therapeutic approaches for extremely unfavorable histology subgroups. Biomolecules. 2022;12:79. doi: 10.3390/biom12010079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kock A., Bergqvist F., Steinmetz J., Elfman L.H.M., Korotkova M., Johnsen J.I., Jakobsson P., Kogner P., Larsson K. Establishment of an in vitro 3D model for neuroblastoma enables preclinical investigation of combined tumor-stroma drug targeting. Faseb. J. 2020;34:11101–11114. doi: 10.1096/fj.202000684R. [DOI] [PubMed] [Google Scholar]
- 15.Whittle S.B., Smith V., Doherty E., Zhao S., McCarty S., Zage P.E. Overview and recent advances in the treatment of neuroblastoma. Expert Rev. Anticancer Ther. 2017;17:369–386. doi: 10.1080/14737140.2017.1285230. [DOI] [PubMed] [Google Scholar]
- 16.Rifatbegovic F., Frech C., Abbasi M.R., Taschner-Mandl S., Weiss T., Schmidt W.M., Schmidt I., Ladenstein R., Ambros I.M., Ambros P.F. Neuroblastoma cells undergo transcriptomic alterations upon dissemination into the bone marrow and subsequent tumor progression. Int. J. Cancer. 2018;142:297–307. doi: 10.1002/ijc.31053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Almici E., Caballero D., Montero J., Samitier J. Biomater. 3D Tumor Model. Elsevier; 2020. 3D neuroblastoma in vitro models using engineered cell-derived matrices; pp. 107–130. [DOI] [Google Scholar]
- 18.Seong B.K.A., Fathers K.E., Hallett R., Yung C.K., Stein L.D., Mouaaz S., Kee L., Hawkins C.E., Irwin M.S., Kaplan D.R. A metastatic mouse model identifies genes that regulate neuroblastoma metastasis. Cancer Res. 2017;77:696–706. doi: 10.1158/0008-5472.CAN-16-1502. [DOI] [PubMed] [Google Scholar]
- 19.Falzone L., Salomone S., Libra M. Evolution of cancer pharmacological treatments at the turn of the third millennium. Front. Pharmacol. 2018;9 doi: 10.3389/fphar.2018.01300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Saw P.E., Chen J., Song E. Targeting CAFs to overcome anticancer therapeutic resistance. Trends in Cancer. 2022;8:527–555. doi: 10.1016/j.trecan.2022.03.001. [DOI] [PubMed] [Google Scholar]
- 21.Duarte Campos D.F., Bonnin Marquez A., O'Seanain C., Fischer H., Blaeser A., Vogt M., Corallo D., Aveic S., Campos D.F.D., Marquez A.B., O'Seanain C., Fischer H., Blaeser A., Vogt M., Corallo D., Aveic S., Duarte Campos D.F., Bonnin Marquez A., O'Seanain C., Fischer H., Blaeser A., Vogt M., Corallo D., Aveic S. Exploring cancer cell behavior in vitro in three-dimensional multicellular bioprintable collagen-based hydrogels. Cancers. 2019;11:180. doi: 10.3390/cancers11020180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Aveic S., Janßen S., Nasehi R., Seidelmann M., Vogt M., Pantile M., Rütten S., Fischer H. A 3D printed in vitro bone model for the assessment of molecular and cellular cues in metastatic neuroblastoma. Biomater. Sci. 2021;9:1716–1727. doi: 10.1039/D0BM00921K. [DOI] [PubMed] [Google Scholar]
- 23.Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., Preibisch S., Rueden C., Saalfeld S., Schmid B., Tinevez J.-Y., White D.J., Hartenstein V., Eliceiri K., Tomancak P., Cardona A. Fiji: an open-source platform for biological-image analysis. Nat. Methods. 2012;9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pillich R.T., Chen J., Churas C., Liu S., Ono K., Otasek D., Pratt D. NDEx: accessing network models and streamlining network biology workflows. Curr. Protoc. 2021;1 doi: 10.1002/cpz1.258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Aveic S., Davtalab R., Vogt M., Weber M., Buttler P., Tonini G.P.G.P., Fischer H. Calcium phosphate scaffolds with defined interconnecting channel structure provide a mimetic 3D niche for bone marrow metastasized tumor cell growth. Acta Biomater. 2019;88:527–539. doi: 10.1016/j.actbio.2019.02.030. [DOI] [PubMed] [Google Scholar]
- 26.Grosso A., Burger M.G., Lunger A., Schaefer D.J., Banfi A., Di Maggio N. It takes two to tango: coupling of angiogenesis and osteogenesis for bone regeneration. Front. Bioeng. Biotechnol. 2017;5 doi: 10.3389/fbioe.2017.00068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Giannotta M., Trani M., Dejana E. VE-cadherin and endothelial adherens junctions: active guardians of vascular integrity. Dev. Cell. 2013;26:441–454. doi: 10.1016/j.devcel.2013.08.020. [DOI] [PubMed] [Google Scholar]
- 28.Cenni E., Perut F., Baldini N. In vitro models for the evaluation of angiogenic potential in bone engineering. Acta Pharmacol. Sin. 2011;32:21–30. doi: 10.1038/aps.2010.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.A. Eggert, J.H. Maris, Angiogenic Cytokines: Quantitative and Functional Analysis , in: Cytokines Colony Stimul. Factors, Humana Press, New Jersey, n.d.: pp. 117–134. 10.1385/1-59259-345-3:117. [DOI] [PubMed]
- 30.Inomata K., Honda M. Co-culture of osteoblasts and endothelial cells on a microfiber scaffold to construct bone-like tissue with vascular networks. Materials. 2019;12:2869. doi: 10.3390/ma12182869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dikshit B., Irshad K., Madan E., Aggarwal N., Sarkar C., Chandra P.S., Gupta D.K., Chattopadhyay P., Sinha S., Chosdol K. FAT1 acts as an upstream regulator of oncogenic and inflammatory pathways, via PDCD4, in glioma cells. Oncogene. 2013;32:3798–3808. doi: 10.1038/onc.2012.393. [DOI] [PubMed] [Google Scholar]
- 32.Jubb A.M., Strickland L.A., Liu S.D., Mak J., Schmidt M., Koeppen H. Neuropilin-1 expression in cancer and development. J. Pathol. 2012;226:50–60. doi: 10.1002/path.2989. [DOI] [PubMed] [Google Scholar]
- 33.Hu T., Kitano A., Luu V., Dawson B., Hoegenauer K.A., Lee B.H., Nakada D. Bmi1 suppresses adipogenesis in the hematopoietic stem cell niche. Stem Cell Rep. 2019;13:545–558. doi: 10.1016/j.stemcr.2019.05.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang W., Wang B. KDM3A-mediated SP1 activates PFKFB4 transcription to promote aerobic glycolysis in osteosarcoma and augment tumor development. BMC Cancer. 2022;22:562. doi: 10.1186/s12885-022-09636-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Okuda H., Kobayashi A., Xia B., Watabe M., Pai S.K., Hirota S., Xing F., Liu W., Pandey P.R., Fukuda K., Modur V., Ghosh A., Wilber A., Watabe K. Hyaluronan synthase HAS2 promotes tumor progression in bone by stimulating the interaction of breast cancer stem–like cells with macrophages and stromal cells. Cancer Res. 2012;72:537–547. doi: 10.1158/0008-5472.CAN-11-1678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu X., Li K., Wang L., Zhang M., Wang X. Variations of human heat shock proteins in multiple cancers. Clin. Transl. Med. 2021;11 doi: 10.1002/ctm2.320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Edsjö A., Holmquist L., Påhlman S. Neuroblastoma as an experimental model for neuronal differentiation and hypoxia-induced tumor cell dedifferentiation. Semin. Cancer Biol. 2007;17:248–256. doi: 10.1016/j.semcancer.2006.04.005. [DOI] [PubMed] [Google Scholar]
- 38.Corallo D., Zanon C., Pantile M., Tonini G.P., Zin A., Francescato S., Rossi B., Trevisson E., Pinato C., Monferrer E., Noguera R., Aliño S.F., Herrero M.J., Biffi A., Viscardi E., Aveic S. Integrated CGH/WES analyses advance understanding of aggressive neuroblastoma evolution: a case study. Cells. 2021;10:2695. doi: 10.3390/cells10102695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang L., Tan T.K., Durbin A.D., Zimmerman M.W., Abraham B.J., Tan S.H., Ngoc P.C.T., Weichert-Leahey N., Akahane K., Lawton L.N., Rokita J.L., Maris J.M., Young R.A., Look A.T., Sanda T. ASCL1 is a MYCN- and LMO1-dependent member of the adrenergic neuroblastoma core regulatory circuitry. Nat. Commun. 2019;10:5622. doi: 10.1038/s41467-019-13515-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lebedev T., Vagapova E., Spirin P., Rubtsov P., Astashkova O., Mikheeva A., Sorokin M., Vladimirova U., Suntsova M., Konovalov D., Roumiantsev A., Stocking C., Buzdin A., Prassolov V. Growth factor signaling predicts therapy resistance mechanisms and defines neuroblastoma subtypes. Oncogene. 2021;40:6258–6272. doi: 10.1038/s41388-021-02018-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Huang B., Song B., Xu C. Cholesterol metabolism in cancer: mechanisms and therapeutic opportunities. Nat. Metab. 2020;2:132–141. doi: 10.1038/s42255-020-0174-0. [DOI] [PubMed] [Google Scholar]
- 42.Xu W.W., Li B., Guan X.Y., Chung S.K., Wang Y., Yip Y.L., Law S.Y.K., Chan K.T., Lee N.P.Y., Chan K.W., Xu L.Y., Li E.M., Tsao S.W., He Q.-Y., Cheung A.L.M. Cancer cell-secreted IGF2 instigates fibroblasts and bone marrow-derived vascular progenitor cells to promote cancer progression. Nat. Commun. 2017;8 doi: 10.1038/ncomms14399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Grassi E.S., Pietras A. Emerging roles of DLK1 in the stem cell niche and cancer stemness. J. Histochem. Cytochem. 2022;70:17–28. doi: 10.1369/00221554211048951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hamik A., Wang B., Jain M.K. Transcriptional regulators of angiogenesis. Arterioscler. Thromb. Vasc. Biol. 2006;26:1936–1947. doi: 10.1161/01.ATV.0000232542.42968.e3. [DOI] [PubMed] [Google Scholar]
- 45.Li T., Liu X., Yang A., Fu W., Yin F., Zeng X. Associations of tumor suppressor SPARCL1 with cancer progression and prognosis. Oncol. Lett. 2017;14:2603–2610. doi: 10.3892/ol.2017.6546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wachowiak R., Mayer S., Suttkus A., Martynov I., Lacher M., Melling N., Izbicki J.R., Tachezy M. CHL1 and NrCAM are primarily expressed in low grade pediatric neuroblastoma. Open Med. 2019;14:920–927. doi: 10.1515/med-2019-0109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yang Y., Zhao Y., Hu N., Zhao J., Bai Y. lncRNA KIAA0125 functions as a tumor suppressor modulating growth and metastasis of colorectal cancer via Wnt/β-catenin pathway. Cell Biol. Int. 2019;43:1463–1470. doi: 10.1002/cbin.11196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lee-Thedieck C., Schertl P., Klein G. The extracellular matrix of hematopoietic stem cell niches. Adv. Drug Deliv. Rev. 2022;181 doi: 10.1016/j.addr.2021.114069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Calvi L.M., Adams G.B., Weibrecht K.W., Weber J.M., Olson D.P., Knight M.C., Martin R.P., Schipani E., Divieti P., Bringhurst F.R., Milner L.A., Kronenberg H.M., Scadden D.T. Osteoblastic cells regulate the haematopoietic stem cell niche. Nature. 2003;425:841–846. doi: 10.1038/nature02040. [DOI] [PubMed] [Google Scholar]
- 50.Morrison S.J., Scadden D.T. The bone marrow niche for haematopoietic stem cells. Nature. 2014;505:327–334. doi: 10.1038/nature12984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tekinturhan F., Zimmerlin L., Donnenberg V.S., Pfeifer M.E., Monlish D.A., Guvenc B., Bradley J., Donnenberg A.D. Characterization of hematopoietic and non-hematopoietic stem/progenitor cells in freshly isolated adult human bone marrow using an 8-color flow cytometric assay. Blood. 2007;110 doi: 10.1182/blood.V110.11.4045.4045. 4045–4045. [DOI] [Google Scholar]
- 52.Zhang W., Bado I., Wang H., Lo H.-C., Zhang X.H.-F. Bone metastasis: find your niche and fit in. Trends in Cancer. 2019;5:95–110. doi: 10.1016/j.trecan.2018.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.González Díaz E.C., Sinha S., Avedian R.S., Yang F. Tissue-engineered 3D models for elucidating primary and metastatic bone cancer progression. Acta Biomater. 2019;99:18–32. doi: 10.1016/j.actbio.2019.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Duval K., Grover H., Han L.-H., Mou Y., Pegoraro A.F., Fredberg J., Chen Z. Modeling physiological events in 2D vs. 3D cell culture. Physiology. 2017;32:266–277. doi: 10.1152/physiol.00036.2016. [DOI] [PMC free article] [PubMed] [Google Scholar] [Research Misconduct Found]
- 55.Donaruma L.G. In: J. Polym. Sci. Polym. Lett. Ed. 26. Williams D.F., editor. Elsevier; Amsterdam: 1988. Definitions in biomaterials. 1987, 72 414–414. [DOI] [Google Scholar]
- 56.Mundy G.R. Metastasis to bone: causes, consequences and therapeutic opportunities. Nat. Rev. Cancer. 2002;2:584–593. doi: 10.1038/nrc867. [DOI] [PubMed] [Google Scholar]
- 57.Weilbaecher K.N., Guise T.A., McCauley L.K. Cancer to bone: a fatal attraction. Nat. Rev. Cancer. 2011;11:411–425. doi: 10.1038/nrc3055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Decaesteker B., Denecker G., Van Neste C., Dolman E.M., Van Loocke W., Gartlgruber M., Nunes C., De Vloed F., Depuydt P., Verboom K., Rombaut D., Loontiens S., De Wyn J., Kholosy W.M., Koopmans B., Essing A.H.W., Herrmann C., Dreidax D., Durinck K., Deforce D., Van Nieuwerburgh F., Henssen A., Versteeg R., Boeva V., Schleiermacher G., van Nes J., Mestdagh P., Vanhauwaert S., Schulte J.H., Westermann F., Molenaar J.J., De Preter K., Speleman F. TBX2 is a neuroblastoma core regulatory circuitry component enhancing MYCN/FOXM1 reactivation of DREAM targets. Nat. Commun. 2018;9:4866. doi: 10.1038/s41467-018-06699-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Dallinga M.G., Yetkin-Arik B., Kayser R.P., Vogels I.M.C., Nowak-Sliwinska P., Griffioen A.W., van Noorden C.J.F., Klaassen I., Schlingemann R.O. IGF2 and IGF1R identified as novel tip cell genes in primary microvascular endothelial cell monolayers. Angiogenesis. 2018;21:823–836. doi: 10.1007/s10456-018-9627-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ikram F., Ackermann S., Kahlert Y., Volland R., Roels F., Engesser A., Hertwig F., Kocak H., Hero B., Dreidax D., Henrich K.-O., Berthold F., Nürnberg P., Westermann F., Fischer M. Transcription factor activating protein 2 beta (TFAP2B) mediates noradrenergic neuronal differentiation in neuroblastoma. Mol. Oncol. 2016;10:344–359. doi: 10.1016/j.molonc.2015.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Dong C., Zhang J., Fang S., Liu F. IGFBP5 increases cell invasion and inhibits cell proliferation by EMT and Akt signaling pathway in Glioblastoma multiforme cells. Cell Div. 2020;15:4. doi: 10.1186/s13008-020-00061-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Corallo D., Donadon M., Pantile M., Sidarovich V., Cocchi S., Ori M., De Sarlo M., Candiani S., Frasson C., Distel M., Quattrone A., Zanon C., Basso G., Tonini G.P.G.P., Aveic S. LIN28B increases neural crest cell migration and leads to transformation of trunk sympathoadrenal precursors. Cell Death Differ. 2020;27:1225–1242. doi: 10.1038/s41418-019-0425-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Micalizzi D.S., Wang C.-A., Farabaugh S.M., Schiemann W.P., Ford H.L. Homeoprotein Six1 increases TGF-β type I receptor and converts TGF-β signaling from suppressive to supportive for tumor growth. Cancer Res. 2010;70:10371–10380. doi: 10.1158/0008-5472.CAN-10-1354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Mancarella C., Morrione A., Scotlandi K. Novel regulators of the IGF system in cancer. Biomolecules. 2021;11:273. doi: 10.3390/biom11020273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yin J.J., Pollock C.B., Kelly K. Mechanisms of cancer metastasis to the bone. Cell Res. 2005;15:57–62. doi: 10.1038/sj.cr.7290266. [DOI] [PubMed] [Google Scholar]
- 66.Mu Q., Wang L., Yu F., Gao H., Lei T., Li P., Liu P., Zheng X., Hu X., Chen Y., Jiang Z., Sayari A.J., Shen J., Huang H. Imp2 regulates GBM progression by activating IGF2/PI3K/Akt pathway. Cancer Biol. Ther. 2015;16:623–633. doi: 10.1080/15384047.2015.1019185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Enderling H., Park D., Hlatky L., Hahnfeldt P. The importance of spatial distribution of stemness and proliferation state in determining tumor radioresponse. Math. Model Nat. Phenom. 2009;4:117–133. doi: 10.1051/mmnp/20094305. [DOI] [Google Scholar]
- 68.Futrega K., Mosaad E., Chambers K., Lott W.B., Clements J., Doran M.R. Bone marrow-derived stem/stromal cells (BMSC) 3D microtissues cultured in BMP-2 supplemented osteogenic induction medium are prone to adipogenesis. Cell Tissue Res. 2018;374:541–553. doi: 10.1007/s00441-018-2894-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Sharma G.P., Frei A.C., Narayanan J., Gasperetti T., Veley D., Amjad A., Albano K., Fish B.L., Himburg H.A. Brain-derived neurotrophic factor promotes immune reconstitution following radiation injury via activation of bone marrow mesenchymal stem cells. PLoS One. 2021;16 doi: 10.1371/journal.pone.0259042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Maryanovich M., Takeishi S., Frenette P.S. Neural regulation of bone and bone marrow. Cold Spring Harb. Perspect. Med. 2018;8:a031344. doi: 10.1101/cshperspect.a031344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Amelio I., Cutruzzolá F., Antonov A., Agostini M., Melino G. Serine and glycine metabolism in cancer. Trends Biochem. Sci. 2014;39:191–198. doi: 10.1016/j.tibs.2014.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hernandez R.K., Maegbaek M.L., Liede A., Sørensen H.T., Ehrenstein V. Bone metastases, skeletal-related events, and survival among children with cancer in Denmark. J. Pediatr. Hematol. Oncol. 2014;36:528–533. doi: 10.1097/MPH.0000000000000106. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data will be made available on request.