Abstract
Staphylococcus aureus and Staphylococcus epidermidis are frequently associated with medical device infections that involve establishment of a bacterial biofilm on the device surface. Staphylococcal surface proteins Aap, SasG, and Pls are members of the Periscope Protein class and have been implicated in biofilm formation and host colonization; they comprise a repetitive region (“B region”) and an N-terminal host colonization domain within the “A region,” predicted to be a lectin domain. Repetitive E-G5 domains (as found in Aap, SasG, and Pls) form elongated “stalks” that would vary in length with repeat number, resulting in projection of the N-terminal A domain variable distances from the bacterial cell surface. Here, we present the structures of the lectin domains within A regions of SasG, Aap, and Pls and a structure of the Aap lectin domain attached to contiguous E-G5 repeats, suggesting the lectin domains will sit at the tip of the variable length rod. We demonstrate that these isolated domains (Aap, SasG) are sufficient to bind to human host desquamated nasal epithelial cells. Previously, proteolytic cleavage or a deletion within the A domain had been reported to induce biofilm formation; the structures suggest a potential link between these observations. Intriguingly, while the Aap, SasG, and Pls lectin domains bind a metal ion, they lack the nonproline cis peptide bond thought to be key for carbohydrate binding by the lectin fold. This suggestion of noncanonical ligand binding should be a key consideration when investigating the host cell interactions of these bacterial surface proteins.
Keywords: adhesin, bacterial adhesion, biofilm, lectin, cell surface protein, Periscope proteins, glycomics
Abbreviations: Aap, accumulation associated protein; ConA, concanavalin A; nanoDSF, nanoscale differential scanning fluorimetry; Pls, plasmin sensitive protein; SasG, staphylococcal surface protein G; SEC-SAXS, size exclusion chromatography–small angle X-ray scattering; TLS, translation libration screw
The ability of bacteria to form biofilms on surfaces is a key factor in the establishment of infections on indwelling medical devices (1); thus, biofilms are involved in many nosocomial infections (2). Biofilm infections are difficult to treat, and infected devices often need to be removed to resolve the infection (3, 4); an improved understanding of adhesion and biofilm formation at the molecular level is required for the development of new preventative measures. Staphylococcus aureus and Staphylococcus epidermidis are gram-positive bacteria commonly isolated from device infections (5) and form biofilms through both exopolysaccharide and protein-mediated mechanisms (6, 7, 8). Accumulation associated protein (Aap), Staphylococcal surface protein G (SasG), and Plasmin sensitive protein (Pls) are homologous proteins from S. epidermidis (Aap) and S. aureus (SasG, Pls). Aap and SasG are visible as long fibrils on the bacterial cell surface (9, 10) and have been implicated in host cell binding and biofilm formation (8, 10, 11), while surface expression of Pls has been observed to reduce staphylococcal adhesion to fibronectin (12, 13, 14).
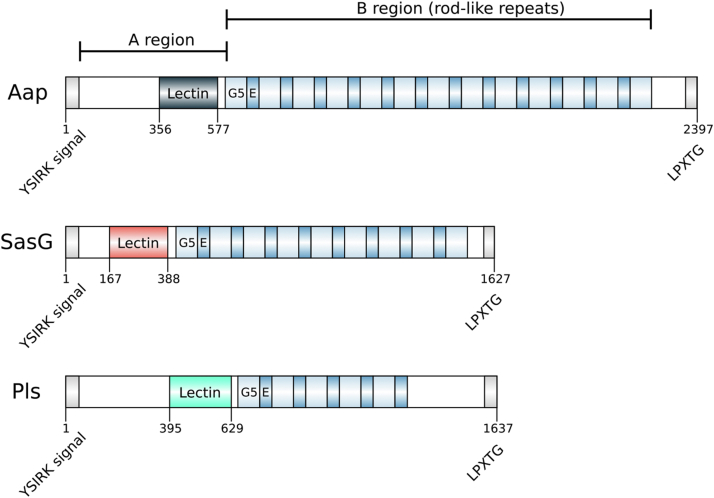
As well as having similar functions, Aap, SasG, and Pls have similar domain architectures (Fig. 1) including an N-terminal “A region” as a putative adhesin; however, the boundaries of the adhesin domain have yet to be defined (12, 15, 16). The A region in Aap contains a variable number (12 in S. epidermidis RP62A) of short (∼16 residue) repeats at the N terminus. Bowden and colleagues noted part of the A region of Aap is predicted to form an L-type lectin fold (17) a fold often associated with carbohydrate recognition. Indeed, National Center for Biotechnology Information (NCBI) Conserved Domain Database (CDD) (18) predicts a bacterial lectin (cl38937; pfam18483) for Aap residues 356 to 577, SasG residues 167 to 388, and Pls residues 395 to 629. The “B regions” of Aap, SasG, and Pls (Fig. 1) contain a variable number of highly conserved (∼128 residue) E-G5 repeats. In SasG and Aap these repeats together form an elongated rod-like structure (19, 20, 21, 22). Pls contains a serine-aspartate–rich region C-terminal of the E-G5 repeats, which has been shown to be glycosylated (23); all three proteins have a C-terminal LPXTG motif and thus are predicted to be attached covalently to the bacterial cell wall in a sortase-mediated process. For SasG, Aap, and Pls there is evidence for variation in E-G5 repeat number in close homologues, suggesting they are members of the Periscope Protein class we recently defined (24) that appear to play various roles in regulating the interactions of the bacterial surface with its environment (25).
Figure 1.
Domain schematics of bacterial surface proteins Aap, SasG and Pls. Schematic of Aap, SasG, and Pls (UniProt Q5HKE8, Q2G2B2, P80544) showing conserved functional and structural regions: N-terminal A region including YSIRK secretion signal and predicted lectin-like domain; B region featuring G5/E repeats, which form a rod-like structure; C-terminal LPXTG cell wall attachment motif. Lectin domain boundaries are annotated according to CDD predictions; see text and Table 2 for construct boundaries used in this work.
Many questions remain regarding the A region. Proteolytic processing of Aap and SasG to remove the A region promotes biofilm formation (8, 10), but unprocessed Aap has been linked to the attachment phase (26). The proteolytic processing site in Aap has been identified (27), but its structural context is unknown. The full A regions of SasG and Aap have previously been implicated in host cell binding (15, 28). Indeed, expression of SasG by the otherwise poorly adherent Lactococcus lactis enabled binding to desquamated nasal epithelial cells (15); binding was inhibited by the addition of recombinant intact A regions of both SasG and Aap, suggesting a common host cell receptor for the two proteins. However, a recombinant fragment of SasG comprising an ∼212-residue (predicted lectin-like) region conserved between SasG and Aap (see above) did not inhibit binding (15), suggesting it does not contain the complete functional region. More recently it was determined that Aap-mediated binding to corneocytes could be blocked by anti-AapA antisera, but not anti-AapB antisera, indicating that the Aap A region is important for this interaction (29). It has also been hypothesized that a catch-bond mechanism involving both the A and B domains could facilitate binding of Aap to von Willebrand Factor (30). However, the relationship between the structure and function of the A regions and their adhesive interactions remains elusive.
Here we redefine the structural and functional boundaries within the A regions of SasG, Aap, and Pls and solve the structures of the putative L-type lectin domains. The orientation of this domain with respect to the rod-like region formed by the E-G5 repeats reveal that the lectin fold is likely at the tip of the protein, in an optimal location for host–ligand binding. We identify a subtle but key structural difference when compared with other carbohydrate-binding L-type lectin folds, suggesting the Aap, SasG, and Pls lectin folds are likely to bind their ligand noncanonically. Our work thus provides important insights into the structure–function relationship of these biofilm-mediating proteins.
Results
Structure of the functional A domain
Previously described subdomain boundaries within the A regions of SasG and Aap highlighted a conserved region, but this region was nonfunctional in investigations of host cell binding by SasG (15). We investigated structural domain boundaries in SasG using limited proteolysis. Protease treatment of the full A region (52–420) identified a stable fragment of ∼32 kDa by SDS-PAGE and with an N terminus S145VDEG suggesting that, in SasG, Aap, and Pls, the N-terminal boundary is significantly N-terminal of both the predicted L-type lectin fold (Fig. 1) and the conserved region. In order to investigate the structure/function of the adhesin domains of these proteins, we first pursued structural characterization by X-ray crystallography.
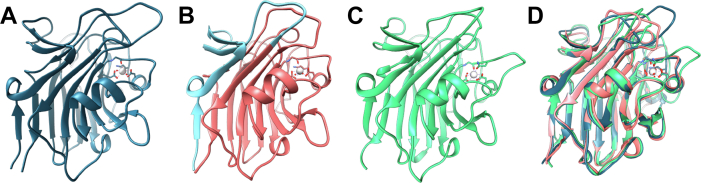
Based on the N-terminal boundaries suggested by limited proteolysis results, constructs of Aap338–608, SasG144–423, and Pls391–656 were recombinantly expressed and crystal structures (Fig. 2) determined at resolutions of 1.30, 1.65, and 1.21 Å (Table 1 and Figs. S1 and S2), respectively, in the presence of Ca2+. Notably, for SasG144–423, residues 144 to 162 were not resolved in the electron density map; thus, this region was excluded from further analysis. A new expression construct was designed comprising SasG163–421 for analysis of metal binding, host cell binding, and glycan array screening. The Aap, SasG, and Pls structures are very similar; alignment of Aap, SasG, and Pls using PDBeFold (31), matching chain A to chain A, reveals a root-mean-squared deviation (RMSD) of 1.06 Å over 242 residues for Aap compared with SasG; 1.18 Å over 235 residues for SasG against Pls; and 1.26 Å over 240 residues for Aap against Pls; the areas of weakest sequence conservation are the loops (Fig. 2D). PDBeFold (31) analyses show Aap, SasG, and Pls domain structures fit most closely with superfamily b.29.1 (Concanavalin A-like lectins/glucanases) and subfamily b.29.1.1 (Legume lectin). The overall fold consists of two antiparallel β-sheets, one concave sheet of seven strands (known as the front face) and a flat six-stranded sheet, known as the back face. Dali (32) found the closest structural matches for the SasG, Aap, and Pls structures to be the L-type lectin domain from S. aureus SraP (4M01; PF18483) (Fig. S3). L-type lectin domains typically bind divalent cations, enabling carbohydrate binding; thus, we investigated the impact of divalent cation binding on domain stability.
Figure 2.
Structures of L-type lectin domains from Aap, SasG and Pls. X-ray crystal structures of L-type lectin domains from Aap (A), SasG (B), and Pls (C), and the aligned structures (D). The lectin domains of Aap338–608, SasG144–423, and Pls391–656 were crystallized and experimental phases determined for SeMet-labeled Aap; subsequently, native data acquired for all three proteins were phased by molecular replacement (Table 1). The structures describe L-type lectin domains (ribbon representation) with a single coordinated Ca2+ ion (gray sphere). The blue colored region shown in (B) indicates the portion of the protein that was missing in a previous nonfunctional construct (see main text).
Table 1.
Data collection and refinement statistics
| Data collection | Aap338–608 native | Aap338–608 SeMet | Aap351–813 | SasG144–423 | Pls391–656 |
|---|---|---|---|---|---|
| Beamline | I03 | I03 | I02 | I02 | I04-1 |
| Space group | P212121 | P212121 | P1 | P41 | P21 |
| Cell dimensions | |||||
| a, b, c (Å) | 35.8, 68.7, 109.2 | 35.9, 68.5, 109.3 | 58.5, 66.6, 66.9 | 63.2, 63.2, 273.4 | 52.2, 66.7, 71.5 |
| α, β, γ (°) | 90, 90, 90 | 90, 90, 90 | 84.7, 80.5, 89.7 | 90, 90, 90 | 90, 110.3, 90 |
| Peak | |||||
| Wavelength (Å) | 0.9762 | 0.9793 | 0.9795 | 0.9795 | 0.9159 |
| Resolution (Å) | 16.5–1.30 (1.32–1.30) | 24.2–1.35 (1.37–1.35) | 39.7–2.30 (2.38–2.30) | 57.4–1.65 (1.68–1.65) | 67.2–1.21 (1.23–1.21) |
|
Rmerge (%) <I%σI)> |
4.0 (6.6) 34.1 (21.8) |
6.1 (10.8) 28.3 (10.7) |
8.0 (58.6) 4.8 (1.3) |
10.0 (63.2) 14.0 (3.5) |
10.6 (89.8) 5.8 (1.8) |
| CC(1/2) | 99.8 (99.6) | 99.8 (97.6) | 98.9 (76.4) | 99.8 (80.8) | 98.9 (39.1) |
| Completeness (%) | 99.5 (98.7) | 99.5 (93.6) | 97.5 (96.9) | 99.8 (99.4) | 99.9 (99.8) |
| Redundancy | 6.5 (6.6) | 5.9 | 2.2 (2.1) | 11.4 (10.7) | 3.9 (3.6) |
| No. unique reflections | 66,831 | 60,071 | 42,896 | 127,888 | 139,706 |
| No. molecules/AU | 1 | 1 | 2 | 4 | 2 |
| Solvent content (%) | 46 | 46 | 52 | 49 | 40 |
| Refinement | |||||
| Resolution (Å) | 1.30 | 2.30 | 1.65 | 1.21 | |
| No. reflections | 63,480 | 40,775 | 120,320 | 132,407 | |
| Rwork/Rfree (%) | 10.0/12.4 | 19.8/24.3 | 18.5/22.6 | 12.0/15.8 | |
| No. atoms | |||||
| Protein | 2124 | 6704 | 7918 | 4499 | |
| Ligand/ion | 2 | 4 | 16 | 2 | |
| Water | 385 | 168 | 354 | 748 | |
| B-Factors (Å2) | |||||
| Protein | 9.3 | 58.0 | 17.7 | 14.4 | |
| Ligand/ion | 6.4 | 37.8 | 21.6 | 9.2 | |
| Water | 24.0 | 39.2 | 16.5 | 28.2 | |
| R.m.s deviations | |||||
| Bond lengths (Å) | 0.020 | 0.008 | 0.014 | 0.018 | |
| Bond angles (°) | 2.07 | 1.41 | 1.92 | 2.00 | |
| Ramachandran (%) | |||||
| Favored | 95.7 | 96.3 | 94.3 | 94.4 | |
| Allowed | 4.3 | 3.4 | 5.2 | 5.4 | |
| Outliers | 0 | 0.2 | 0.6 | 0.2 | |
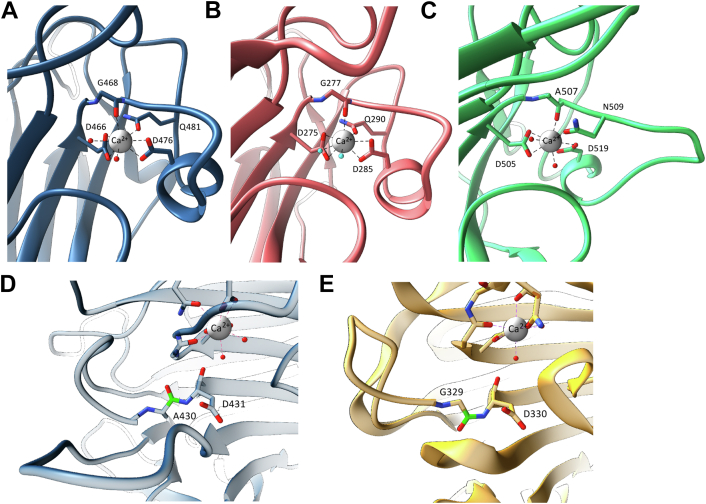
Similar to the SasG, Aap, and Pls domains, the SraP domain is monomeric and binds only a single Ca2+ (33). Many lectin domains bind divalent cations (usually Ca2+ and Mn2+); we determined the effect of Ca2+ and Mn2+ on the thermally induced unfolding of A domain constructs of Aap, SasG, and Pls using nanoscale differential scanning fluorimetry (nanoDSF). The thermal stability of Aap353–608, Aap351–813 (lectin-G51-E1-G52), SasG163–421, SasG163–629, and Pls391–656 was measured in the presence and absence (EDTA control) of Ca2+ and Mn2+. Consistent with the presence of a Ca2+ ion in the crystal structures, nanoDSF indicated that, while Mn2+ and Ca2+ binding both increase melting temperature (Tm) relative to the Tm in the presence of EDTA, Ca2+ binding has the most significant effect (Fig. S4). In Aap and SasG the binding site for the single Ca2+ is conserved (Figs. 3, A and B and S2); octahedral geometry is created by two water molecules, bidentate coordination by the side chains of two Asp residues (466 and 476 in Aap; and 275 and 285 in SasG), by the side chain oxygen of a Gln residue (Aap481 and SasG290), and the backbone carbonyl of a Gly residue (Aap468 and SasG277). In Pls, there is an insertion in the loop near the Ca2+-binding site; the ion is coordinated through two water molecules, bidentate coordination by Asp505 (homologous to Aap466 and SasG275) and Asp519 (homologous to Aap476 and SasG285), the backbone carbonyl of Ala507 (homologous to the Gly residues Aap468 and SasG277) and the side chain oxygen of Asn509 (homologous to glutamine residues Aap481 and SasG290) (Fig. 3C). Having established that the A domains of Aap, SasG, and Pls have an L-type lectin fold and are stabilized by divalent cation binding, we next sought to investigate the host cell interactions and carbohydrate binding of these domains.
Figure 3.
Metal coordination and the notable absence of a nonproline cis peptide bond in L-type lectin domains of Aap, SasG and Pls. Ca2+ coordination in (A) Aap, (B) SasG, and (C) Pls. (D) In Aap (also in SasG and Pls; data not shown) the unusual nonproline cis-peptide bond, observed in other sugar-binding lectin domains such as concanavalin A and (E) SraP (gold), is in the trans orientation (D). This is consistent with the lack of carbohydrate binding observed for Aap, SasG, and Pls (Fig. S5). Protein structures are illustrated as ribbon diagrams, with highlighted residues shown in cylinder representation coordinating Ca2+ (gray sphere) and water (red spheres, cyan for clarity in (B)). Atoms are colored by element (O, red; N, blue), and peptide bond in trans (D) and cis (E) is highlighted in green.
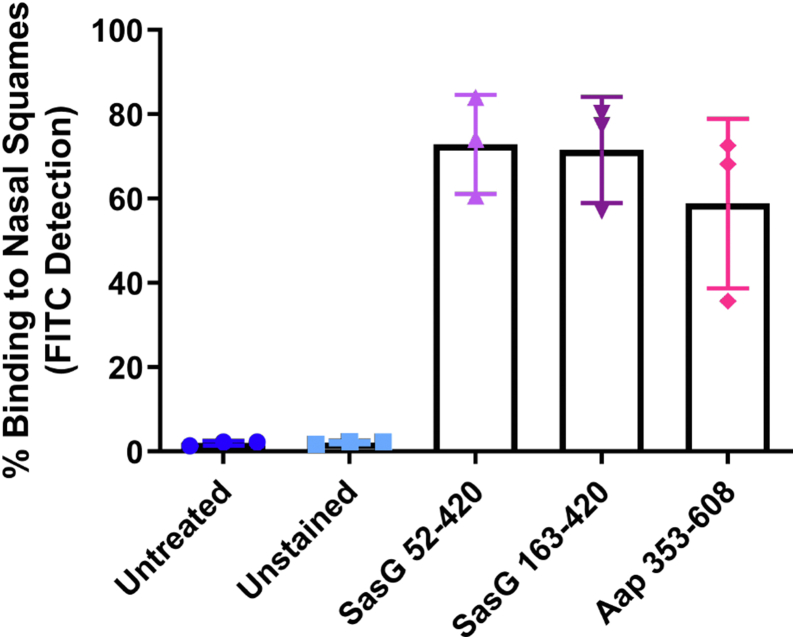
The structures explain the previously observed lack of activity of the SasG(207–428) construct in functional assays (15); Figure 2B indicates key structural elements that would have been lacking. Thus, based on the SasG A domain structure, two SasG and one Aap constructs were tested in a cell binding assay (Fig. 4); the full A domain (SasG52–420) and the lectin domains (SasG163–421 and Aap353–608) all bound to desquamated nasal epithelial cells. The ligand for a canonical lectin fold is a carbohydrate; however, SasG163–421 and Aap353–608 were inactive in screening against a microarray containing 492 glycan probes (Fig. S5 and Table S1) including a variety of mammalian type sequences, representative of N-glycans, peripheral regions of O-glycans; blood group antigen-related sequences on linear or branched backbones and their sialylated and/or sulfated analogs; linear and branched poly-N-acetyllactosamine sequences; gangliosides, oligosaccharide fragments of glycosaminoglycans, and polysialic acid. The arrays also included microbial and plant-derived homo-oligomers of glucose and of other monosaccharides (Table S1). The lack of binding in the glycan arrays suggests that the binding to host cells may not be mediated by a carbohydrate interaction, although it remains possible that the carbohydrate ligands were not included in the arrayed probe library. Intriguingly, but consistent with the lack of carbohydrate binding activity, Figure 3D clearly shows that a nonproline cis peptide bond, first identified in the structure of the plant lectin Concanavalin A (ConA) and subsequently found conserved in legume and legume-like lectin protein folds and conserved in SraP (4M01; Fig. 3E), is in the trans conformation in Aap; the homologous peptide bond is also in the trans conformation in SasG and Pls lectin folds (data not shown). It is worth noting that the ConA lectin included as a control protein in the glycan microarray analyses showed strong binding to the N-glycan probes, especially to the mannose-rich oligo/high-mannose and hybrid-type N-glycans, as predicted (34, 35) (Fig. S5). L-type lectins typically bind carbohydrate ligands near to the bound cation, at the top of the “front face” of the fold. Thus, we investigated the structural relationship between the lectin fold and the rod-like repeat region of Aap to determine the potential exposure of the L-type lectin ligand-binding site.
Figure 4.
Binding of recombinant proteins containing the lectin domains of SasG and Aap to desquamated nasal epithelial cells. FITC-labeled SasG52–420 (lilac triangles), SasG163–421 (purple inverted triangles), and Aap353–608 (pink diamonds) were incubated with human desquamated nasal epithelial cells, and cell-associated fluorescence was measured by flow cytometry. Binding is expressed as the percentage of FITC-positive cells in the population. Bars show the means of three separate experiments ± standard deviation. Untreated cells (dark blue circles) were not incubated with recombinant protein and unstained cells (light blue squares) were incubated with the buffer used for dialysis after FITC labeling.
Orientation of the lectin domain
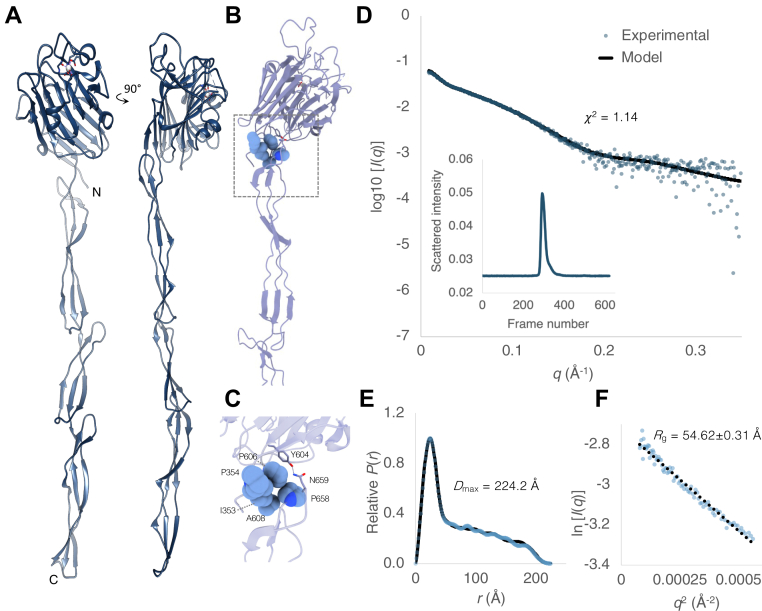
We solved the structure of Aap351–813, which contains the lectin domain followed by the G51-E1-G52 domains (Fig. 5, A–C). The L-type lectin fold is largely unaffected by the addition of G51-E1-G52 (superposition with the isolated domain results in a Cα RMSD of 0.4 Å over 245 residues). The presence of an ordered loop in Aap353–608 (residues 581–589), which is disordered in Aap351–813, is likely a result of packing interactions in the crystal lattice of the isolated lectin fold. G51-E1-G52 forms an elongated array of planar β-sheet modules as determined previously for the homologous regions of SasG (19, 20) and for G512-E12-G513 from Aap of S. epidermidis RP62a (22). The N terminus of the lectin domain (I353-P354) forms predominantly van der Waals interactions at the N terminus of the G51 domain (P606, A608) and the G51 loop (P658), with formation of a hydrogen bond between Y604 (lectin domain) and N659 (G51 loop), incurring a defined orientation of the lectin domain relative to the rod-like G51-E1-G52 array (Fig. 5, B and C).
Figure 5.
TheX-ray crystal structure and SAXS analysis of a contiguous adhesin domain and rod-like B region are consistent with exposure of the Aap ligand binding site at the cell surface. (A) Crystal structure of Aap351–813 showing rod-like B region and lectin-like A region with N and C termini annotated. A disordered loop is illustrated as a dashed line. B residues forming a van der Waals interface between the lectin domain and B repeat region G51 are illustrated as spheres, and hydrogen bonding side chains are shown as cylinders. These residues are annotated in a magnified image in (C). D, fit of the Aap351–813 model to experimental scattering data. The data are accounted for by a single model of Aap351–813. Experimental data (blue) and calculated (black) scattering curves are displayed to a maximal momentum transfer of q = 0.35 Å, with fit value (χ2) annotated. D (inset): size exclusion chromatography trace of Aap351–813 sample injection. E, normalized pair-distance distribution function (P(r)) with calculated maximum intraparticle diameter (Dmax) displayed. Experimental data (blue) and calculated (black) curves are displayed. Real space Rg = 64.55 ± 0.9 Å; I(0) = 0.069. F, guinier region; linearity confirms monodispersity (q∗Rg range 0.48–1.29; Rg = 54.62 ± 0.31 Å; I(0) = 0.065). The dotted line shows linear regression with an R2 value of 0.9772.
The solution structures of Aap353–608, Aap351–813, SasG163–421, SasG163–629, and Pls391–656 were analyzed using size exclusion chromatography coupled to small-angle X-ray scattering (SEC-SAXS) (Fig. 5, D–F). Experimental scattering curves produced by SEC-SAXS were then compared with predicted scattering curves calculated for the crystal structures (or a model of SasG163–629 based on the crystal structure of Aap351–813; Figs. 5, D and E and S6 and S7). This analysis shows close agreement (χ2 value ≤ 1.25) between experimental and theoretical scattering curves, indicating that the crystal structures are representative of the protein solution structures. The theoretical scattering curve calculated for Aap351–813 also fits well to the SasG163–629 experimental scattering data (χ2 value 1.02), suggesting Aap351–813 and SasG163–629 adopt similar solution structures.
Discussion
The work presented here resolves some important questions regarding the structure, function, and potential activation of the homologous Periscope Proteins Aap, SasG, and Pls. Previously it was shown that expression of SasG by L. lactis enabled binding to desquamated nasal epithelial cells (15); this was inhibited by the addition of full-length recombinant A region (SasG52-428) but not by two A region fragments SasG52-207 and SasG207-428; thus, the functional region was not identified. Here we provide the probable solution to this puzzle. Figure 2B shows that SasG207-428 would lack key structural features at the N terminus and is unlikely to adopt a stable fold.
In both Aap (8) and SasG (10), proteolytic processing (to remove the A domain) is necessary to induce biofilm formation and the addition of the protease inhibitor α2-macroglobulin inhibits biofilm formation. An S. epidermidis protease was recently shown to cleave at two positions Aap335 and Aap601 (Fig. S2) (27). Rohde et al. (8) reported that Aap detected on the surface of a biofilm-positive strain of S. epidermidis (5179-R1) had a different distribution of sizes compared with the biofilm-negative strain (5179). In S. epidermidis 5179-R1, an additional shorter (140 kDa) protein was detected in addition to the 220 kDa and 180 kDa forms. This apparent proteolytic processing of the 220 kDa form was abolished in both strains on addition of α2-macroglobulin, with only the 220 kDa band being detected. The Aap sequences between the two strains differ within the lectin fold, where 5179-R1 is missing ∼30 aa (corresponding to residues 401–430 in Aap from strain RP62A); we predict this deletion would likely destabilize the domain structure (Fig. S2). Thus, it is possible that the R1-deletion and proteolytic processing act in the same way to promote biofilm formation, that is, by removing the functional lectin domain.
Aap, SasG, and Pls contain differing amounts of primary sequence N-terminal to the folded lectin domain (Fig. 1). Typical of the L-type lectin fold, in SasG, Aap, and Pls, the N and C termini are proximal, capping the six-stranded antiparallel β-sheet of the lectin domain “back face” (Fig. 2). The lower-resolution structure of Aap351–813 (lectin_G51-E-G52; Fig. 5) confirms that, when connected to the rod-like domain, the N terminus remains oriented back toward the bacterial cell surface. This is important because it suggests that, although the lectin domain is not at the N-terminal tip of the primary sequence, it is at the tip of the rod-like “Periscope.” This is somewhat reminiscent of the N-terminal N2N3 domain arrangement in FnBPA (36), in which the orientation of the N3 domain C terminus back toward the N2 domain might result in the N3 domain, which contains the majority of the fibrinogen-binding residues, being furthest from the bacterial surface despite it not being the most N-terminal region in the primary sequence.
The previous prediction that the A region contains a lectin fold would lead, naturally, to the prediction that this host colonization domain binds carbohydrates; however, no definitive ligand has been identified to date. The His-tagged versions of Aap353–608 and SasG163–423 were screened against a broad-spectrum array and no definitive “hits” were identified. A DALI (32) analysis identified SraP sialic acid (N-acetylneuraminic acid)-binding protein from S. aureus (33) as the closest structural homologue to the Aap, SasG, and Pls lectin domains. Like SasG, Aap, and Pls, the SraP L-type lectin fold is monomeric and binds a single Ca2+. Fig. S3 shows a sequence alignment of SasG, Aap, and Pls domains with SraP. The primary carbohydrate-binding site in legume lectin domains contains conserved Asp, Asn, and Gly residues involved in interactions (direct or water mediated) with the Ca2+ (37) and a highly conserved hydrophobic (frequently aromatic) residue. Typically, the conserved aromatic residue backbone carbonyl acts as a Ca2+ ligand, while the side chain interacts with the sugar ligand. This hydrophobic residue is conserved in SraP but is replaced with a Gly in SasG and Aap and by an Ala in Pls (Figs. 3 and S2).
Most published structures of the leguminous lectin folds that bind carbohydrates have a nonproline cis peptide bond near the carbohydrate-binding site. Such bonds are usually highly thermodynamically unfavorable compared with the trans conformation and are thus rare, with an incidence in proteins of ∼0.03% (38); it has been suggested that they are found near functionally important regions of a protein structure (39). In fact, the mannose-binding lectin ConA was the first protein structure in which a nonproline cis peptide bond was identified (40, 41); in SraP, the homologous peptide bond is also in the cis conformation. Thus, the relaxation of the bond to the trans conformation in SasG, Aap, and Pls, and the resultant remodeling of what would normally be the carbohydrate-binding site, is consistent with these legume lectin folds having noncanonical ligand binding activity.
Experimental procedures
Sequences and cloning
Translated nucleotide sequences were obtained from GenBank: Aap from S. epidermidis RP62A (AAW53239.1), SasG from S. aureus NCTC 8325 (ABD31801.1), Pls from S. aureus (AAD09131.1). Residue numbering for all constructs was derived from these sequences. Sequence alignments (Figs. S1 and S2) were performed using Clustal Omega (42). The coding sequences of Aap, SasG, and Pls were amplified by PCR (see Table 2 for list of constructs) and cloned into the pETFPP_1 vector, a derivative of the pET-YSBLIC3C vector (43) using a recombination-based method (Takara InFusion).
Table 2.
Vectors and constructs
| Construct | Description |
|---|---|
| pETFPP_1 | Modified pET expression vector encoding a 3C-cleavable N-terminal hexa-histidine tag |
| SasG 52–420 | SasG A region |
| SasG 144–423 | SasG lectin domain |
| SasG 163–421 | SasG lectin domain |
| SasG 207–428 | SasG lectin domain (predicted) |
| SasG 163–629 | SasG lectin domain and G5-E-G5 |
| Aap 338–608 | Aap lectin domain |
| Aap 353–608 | Aap lectin domain |
| Aap 351–813 | Aap lectin domain and G5-E-G5 |
| Pls 391–656 | Pls lectin domain |
Protein expression and purification
All protein constructs (Table 2) were expressed in Escherichia coli BL21 DE3 with 3C protease cleavable N-terminal hexa-histidine tags. LB cultures were inoculated and incubated at 37 ˚C with shaking at 180 rpm; cell growth was monitored to A600 0.6, and protein expression was induced with addition of 0.4 mM IPTG for ∼16 h at 17 ˚C with shaking. Cells were harvested by centrifugation and resuspended in lysis buffer comprising 20 mM Tris-HCl, 150 mM NaCl, 20 mM Imidazole, 10 mM CaCl2, pH 7.5 (Aap351–813), or 20 mM Tris pH 7.5, 500 mM NaCl, 30 mM Imidazole, 10 mM CaCl2, pH 7.5 (SasG52–420, SasG144–423, SasG163–421, Aap338–608, Aap353–608, Pls391–656, and SasG163–629) and lysed by sonication on ice. The lysate was clarified by centrifugation and purified by immobilized Ni affinity chromatography (IMAC, 5 ml HisTrap-HP, GE Healthcare) by washing with lysis buffer to remove unbound contaminants and eluted with a gradient (0–50%) of lysis buffer supplemented with 500 mM Imidazole. Peak fractions were dialyzed overnight into buffer comprising 50 mM Tris-HCl, 150 mM NaCl, 10 mM CaCl2, 1 mM TCEP, pH 7.5 at 4 °C. For crystallization samples, 3C protease was added at a ratio of 1:50 w/w (3C:eluant) and incubated for 20 h at 4 °C. Following proteolysis, the protein was applied to a Ni-charged IMAC column equilibrated in dialysis buffer; the flow-through was collected and concentrated to ∼50 mg/ml and separated by size exclusion chromatography (Superdex 75 16/600 HiLoad column, GE Healthcare [SasG52–420, SasG144–423, SasG163–421, Aap338–608, Aap353–608, and Pls391–656] and Superdex 200 16/600 HiLoad, GE Healthcare [Aap351–813 and SasG163–629]) in 20 mM Tris-HCl, 150 mM NaCl, 10 mM CaCl2, pH 7.5; or 20 mM Tris-HCl, 150 mM NaCl, 1 mM CaCl2, pH 7.5 for Pls391–656. Selenomethionine was incorporated into Aap338–608 as described in (44) with protein expression induced in E. coli B834 (DE3) cells in minimal media with 40 μg/ml L-selenomethionine and protein purified as above as a heavy atom derivative to acquire phases for X-ray crystallographic structure determination.
Limited proteolysis
Limited proteolysis of the previously predicted lectin domain (SasG207-428) and of the complete A domain (A52–428) was used to identify the structural domain boundaries within the A domain of SasG (and thus by homology) of Aap. The 0.5 ml cleavage reaction contained 50 mM Tris-HCl pH 7.8, 10 mM CaCl2, and 0.08 μg of trypsin and was left for 2 h at room temperature. The reaction was stopped using 10 μl of Protease Inhibitor Cocktail 1 (Calbiochem). The sample was then dialyzed against H2O overnight. Following SDS-PAGE analysis the major band was excised for N-terminal sequencing (AltaBioscience).
Divalent cation binding
The thermal stability of Aap353–608, Aap351–813, SasG163–421, SasG163–629, and Pls391–656 was measured in the presence and absence of divalent cations. Protein was dialyzed against 20 mM Tris-HCl, 150 mM NaCl, 1 mM EDTA, pH 7.5 to remove bound cations. A sample was then redialyzed against 20 mM Tris-HCl, 150 mM NaCl, pH 7.5 supplemented with 1 mM CaCl2 or 1 mM MnCl2. Protein was diluted in the relevant buffer to a concentration ≤ 1 mg/ml and loaded into nanoDSF grade standard capillaries. The ratio of integrated fluorescence was measured using a Prometheus NanoTemper Differential Scanning Fluorimeter at a temperature ramp speed of 1 °C/min and monitored at wavelengths of 330 and 350 nm.
Cell binding assays
Binding of SasG and Aap to desquamated nasal epithelial cells was determined using flow cytometry. SasG52–420, SasG163–421, and Aap353–608 were labeled by incubation with fluorescein isothiocyanate (FITC; 0.1 mg/ml, Sigma) for 1 h at room temperature. The sample was dialyzed into 20 mM Tris-HCl, 150 mM NaCl, 10 mM CaCl2, pH 7.5 overnight at 4 °C. Desquamated nasal epithelial cells were collected from healthy human volunteers as described (45). Briefly the nasal septum was swabbed and the swab was transferred to a tube containing PBS and agitated vigorously to release the cells. The cells were washed with PBS and adjusted to a density of 1 × 105 cells per mL and incubated with FITC-labeled protein (1 μM) at 37 °C for 1 h with occasional shaking. Labeled proteins were detected with a blue laser at an excitation of 488 nm using a filter of 530/30 using a BD Accuri C6. Fluorescent cells were detected by initially gating cell population using unstained cells and then gating FITC-positive cells in this population. A control was carried out to ensure that excess FITC in the buffer did not stain the cells by incubating cells with dialysis buffer (unstained). Analysis was performed using the BD Accuri C6 Analysis software and plotted with GraphPad Prism v8.0.
Glycan microarray binding assays
Microarray screening analyses were carried out using the neoglycolipid-based microarray system (46). The microarray binding assay of the recombinant His-tagged constructs Aap353–608 and SasG163–421 was performed at 20 °C, essentially as described (46). In brief, after blocking the microarray slides with 0.02% v/v Casein (Pierce) and 1% w/v bovine serum albumin (Sigma A8577) diluted in 5 mM Hepes-buffered saline, pH 7.4, 150 mM NaCl) with 5 mM CaCl2, the microarrays were overlaid with the His-tagged-proteins precomplexed with mouse monoclonal anti-poly-histidine (Ab1) and biotinylated anti-mouse IgG antibodies (Ab2; both from Sigma) in a ratio of 1:2:2 (by weight). The protein–antibody precomplexes were prepared by preincubating Ab1 with Ab2 for 15 min, followed by addition of the His-tagged protein and incubation for an additional 15 min. The protein–antibody complexes were diluted in the blocking solution, to give a 50 μg/ml final concentration of the protein, and then overlaid onto the arrays for 1.5 h. Binding was detected with Alexa Fluor 647–labeled streptavidin (Molecular Probes, 1 μg/ml). The microarray data of biotinylated Concanavalin A (Vector Laboratories) was included as a control for the microarray (Fig. S5). For this lectin, a different blocking solution was used: 3% w/v bovine serum albumin in Hepes-buffered saline with 5 mM CaCl2, and the biotinylated lectin was overlaid at 0.5 μg/ml before detection using Alexa Fluor 647–labeled streptavidin. Details of the glycan probes included in the study, the generation of the microarrays, imaging, and data analysis are in the Supplementary glycan microarray document (Table S2) in accordance with the Minimum Information Required for A Glycomics Experiment (MIRAGE) guidelines for reporting glycan microarray-based data (47).
Size exclusion chromatography–small-angle X-ray scattering
SEC-SAXS was performed on Aap353–608, Aap351–813, SasG163–421, SasG163–629, and Pls391–656 using the B21 beam line at Diamond Light Source. Protein samples were injected at a concentration of 9.5 mg/ml (SasG163–421), 9 mg/ml (Aap353–608, Pls391–656), or 8 mg/ml (SasG163–629, Aap351–813) into a Shodex KW402.5-4F column equilibrated in 20 mM Tris, 150 mM NaCl, 10 mM CaCl2, pH 7.5. SAXS was performed over a momentum transfer range (q) of 0.0032 to 0.38 Å-1. Scattering intensity was collected using an Eiger 4M detector with an incident beam energy of 12.4 keV and a beam-to-detector distance of 4014 mm.
Data processing was performed using the ATSAS program suite as described (48): briefly, data reduction and merging was performed using Chromixs; determination of Guinier region and estimation of Rg and distance distribution function were performed using PRIMUS (49). Useful angular data range was estimated using Shanum (50). All-atom ensembles were generated with Allosmod (51). For each example, 50 independent pools of 100 models were created. Calculation and fitting of theoretical scattering curves from models to experimental data was performed in FoxS (52). This process was automated using Allosmod-FoxS. Plots were generated in Excel.
Crystallization
Aap338–608 crystals were grown by sitting drop vapor diffusion at 4 ˚C, screening for initial hits using PEG/Ion I&II, Index, PACT, Morpheus, and Hampton I&II. Native protein in 50 mM Tris-HCl, 150 mM NaCl, 5 mM CaCl2, pH 7.5 was concentrated to 41 mg/ml. Conditions were set up using a Mosquito liquid handling robot in MRC-Wilden 96-well plates, mixing protein in a 1:1 ratio with well solution. Large single crystals were obtained from a range of screen conditions, with the best from 0.2 M LiCl, 10% (v/v) PEG 6000, 0.1 M sodium acetate, pH 5.0 in the PACT screen (53). Crystals were cryoprotected with 20% (v/v) ethylene glycol and flash cooled in liquid nitrogen prior to data collection. Selenomethionine-labeled Aap338–608 was crystallized at 4 ˚C in the same conditions as the native protein at a concentration of 43 mg/ml. Aap351–813 was crystallized at 20 ˚C, 26 mg/ml by hanging drop vapor diffusion in a 2-uL drop at 1:1 (v/v) protein/well solution comprising 20% PEG 550 MME, 10% PEG 20K, 100 mM Mops/Hepes (Morpheus buffer system 2) pH 7.0 (54) and vitrified directly from the drop without addition of further cryoprotectant.
SasG144–423 crystals were also grown by sitting drop vapor diffusion at 4 °C, screening using the same conditions as for Aap338–608. The best crystals were obtained from the Peg Ion screen in 20% (v/v) PEG 3350, 0.1 M sodium malonate pH 5.0. Crystals were cryoprotected with 20% (v/v) ethylene glycol and flash cooled in liquid nitrogen prior to data collection.
Pls391–656 crystals were grown by sitting drop vapor diffusion at 20 °C, screening for initial hits using JCSG+, Ammonium sulfate, and Hampton I&II. Native protein was at a concentration of 17.6 mg/ml in 20 mM Tris, 100 mM NaCl, 1 mM CaCl2. Conditions were set up using a Mosquito liquid handling robot in MRC-Wilden 96-well plates, mixing protein in a 1:1 and 1:2 ratio with well solution. Crystals were obtained from a number of conditions, with the best from 25% (w/v) PEG 3350, 0.1 M Bis-Tris pH 5.5 in the JSCG+ screen (53). Crystals were cryoprotected with 20% (v/v) PEG 3350 and flash cooled in liquid nitrogen prior to data collection.
Data collection and refinement
Both the selenomethionine derivative and native data for Aap338–608 were collected from a single crystal at the Diamond Light Source, beamline I03. Images were indexed and integrated with XDS (55), then scaled using AIMLESS (56). The structure was determined by phasing from SeMet data using ShelxCDE (57), by building the main chain backbone using Buccaneer (58) and then filling gaps manually in Coot (59). The model was then used to provide phases for high-resolution native data, refining using REFMAC (60) and building in Coot (59). Waters were added using Arp/Warp (61) and manually checked in Coot (59). Density for a divalent metal ion was modeled with Ca2+ and restraints were added in REFMAC (60), with the refined model having one molecule in the asymmetric unit comprising residues 353 to 607. Notably, no density was evident for residues 338 to 352, and thus crystals were washed in well solution and subjected to electrospray mass spectrometric analysis (data not shown), which revealed that residues 338 to 346 were absent from crystallized protein.
Native data for Aap351–813 were collected from a single crystal at Diamond Light Source, beamline I02. Images were indexed and integrated with XDS (62) scaled with AIMLESS (56) and phases determined using MOLREP (63) with the Aap338–608 structure as the search model, which defined two molecules in the asymmetric unit in space group P1. The model of Aap351–813 was built in Coot and refined in REFMAC, with noncrystallographic symmetry restraints and ProSMART (64) external restraints to the Aap338–608 model imposed, and translation libration screw (TLS) refinement with four groups per chain (351–612, 613–685, 686–737, 738–813) determined using the TLS motion determination server (TLSMD) (65). The model includes two molecules comprising residues 351 to 580 and 590 to 813 with a single Ca2+ atom bound per molecule; the region including residues 581 to 589 was not visible in the electron density.
Native data for SasG144–423 and Pls391–656 were collected from single crystals at Diamond Light Source, beamlines I02 and I04-1, respectively. Images were indexed and integrated with XDS (55) and scaled with AIMLESS (56). L-test data indicated twinning for SasG144–423. Phases were determined for both using MOLREP (63), using the Aap338–608 structure as the search model. For Pls391–656, the asymmetric unit contains two molecules, which were built in Coot (59) and refined by REFMAC (60), comprising residues 391 to 656 (chain A) with one bound Ca2+ ion per chain. For SasG144–423, two molecules were found in the asymmetric unit with space group P41212. After the first round of twinned refinement, R-factors were above 60% indicating an incorrect solution. Inspection of pseudotranslation vectors in the Patterson map revealed pseudotranslation vectors at fractions 0.5, 0.5, 0.0. Space group validation of the molecular replacement solution was carried out using Zanuda (66). PDBset (67) was used to generate a P41 model from the P41212 Zanuda output model with symmetry operator y, x, -z, then these coordinates moved to the P41 origin with the symmetry operator x, y-1/2, z. FreeR flags were assigned to the integrated P41 data making sure that the twinned pairs h, k, l and k, h, -l have the same flag set, using CAD (68). Model building and twinned refinement were carried out with Coot (59) and REFMAC (60), generating a model with four molecules in the asymmetric unit comprising residues 163 to 419, with one Ca2+ ion bound per chain.
Data collection and refinement statistics for all structures are detailed in Table 1; all were validated using MolProbity (69). For Aap338–608, 242 residues (95.7%) were in the favored region and 11 residues (4.3%) in the allowed region of the Ramachandran plot. For SasG144–423, 952 residues (94.3%) in favored, 52 residues (5.2%) in allowed and 6 residues (0.6%) were outliers in the Ramachandran plot. For Pls391–656, 492 residues (94.4%) in favored conformations, 28 (5.4%) allowed, and 1 (0.2%) was outlier in the Ramachandran plot. For Aap351–813, 867 residues (96.3%) were in favored conformations, 31 allowed (3.4%), and 2 (0.2%) were outliers in the Ramachandran plot.
Structural analysis
PISA (70) analysis of any interfaces was used to predict the oligomeric state of the domains. Structures were visualized and interatomic distances analyzed using Pymol (The PyMOL Molecular Graphics System, Schrödinger, LLC) or UCSF Chimera (71). Electron density (Fig. S3) was rendered with ccp4mg (72).
Data availability
The coordinates of structures reported here have been deposited in the Protein Data Bank (PDB) under accession codes Aap338–608 7sie, Aap351–813 8deo, SasG144–423 7smh, and Pls391–656 7sjk.
Supporting information
This article contains supporting information (73, 74, 75).
Conflict of interest
The authors declare that they have no conflicts of interest with the contents of this article.
Acknowledgments
Jean Whittingham and Judith Hawkhead assisted with structural biology and protein production, respectively. Eleanor Dodson assisted with structure solution of twinned data. Dominika Gruszka assisted with experimental design, implementation, and analysis. We acknowledge Diamond Light Source for time on Beamlines I02, I03, and I04-1 under Proposals MX7864 and MX18598, and the University of York Bioscience Technology Facility for technical assistance. The glycan microarray studies were performed in the Carbohydrate Microarray Facility at the Imperial College Glycosciences Laboratory. The sequence-defined glycan microarrays contain many saccharides provided by collaborators whom we thank as well as members of the Glycosciences Laboratory for their contribution in the establishment of the neoglycolipid-based microarray system. The glycan microarray studies were supported by the Wellcome Trust Biomedical Resource Grants (WT099197/Z/12/Z, 108430/Z/15/Z, and 218304/Z/19/Z) and in part by the March of Dimes Prematurity Research Center grant 22-FY18-82. This work was funded by BHF grant number PG/17/19/32862.
Author contributions
L. C. C., K. E. A., A. S. B., G. H., A. M. T. conceptualization; L. C. C., K. E. A., A. S. B., G. H., A. M. T., J. P. T. methodology; Y. L. validation; L. C. C., K. E. A., F. W., A. S. B., G. H., A. M. T., J. P. T., Y. L., S. C. G., J. R. P. formal analysis; L. C. C., K. E. A., F. W., A. S. B., G. H., A. M. T., J. P. T., Y. L., S. C. G., J. R. P. investigation; J. P. T., Y. L. data curation; L. C. C., F. W., J. R. P. writing – original draft; L. C. C., F. W., G. H., Y. L., T. F., S. C. G., J. A. G., J. R. P. writing – review & editing; L. C. C., F. W., Y. L., J. R. P. visualization; T. F., J. A. G., J. R. P. supervision; Y. L., T. F., J. A. G., J. R. P. project administration; Y. L., T. F., J. R. P. funding acquisition.
Funding and additional information
F. W. is supported by the Ramsay Fellowship in Applied Science. A. M. T. and J. A. G. were supported by a British Skin Foundation Research Project Grant 7013s.
Reviewed by members of the JBC Editorial Board. Edited by Chris Whitfield
Contributor Information
Fiona Whelan, Email: fiona.whelan@adelaide.edu.au.
Jennifer R. Potts, Email: jennifer.potts@sydney.edu.au.
Supporting information
References
- 1.Götz F., Peters G. In: Infections associated with indwelling medical devices. Third edition. Waldvogel Francis A., Bisno Alan L., editors. American society of microbiology; Washington, D.C.: 2000. Colonization of medical devices by coagulase-negative staphylococci. [Google Scholar]
- 2.Darouiche R.O. Treatment of infections associated with surgical implants. N. Engl. J. Med. 2004;350:1422–1429. doi: 10.1056/NEJMra035415. [DOI] [PubMed] [Google Scholar]
- 3.Di Domenico E.G., Rimoldi S.G., Cavallo I., D'Agosto G., Trento E., Cagnoni G., et al. Microbial biofilm correlates with an increased antibiotic tolerance and poor therapeutic outcome in infective endocarditis. BMC Microbiol. 2019;19:228. doi: 10.1186/s12866-019-1596-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hellmark B., Soderquist B., Unemo M., Nilsdotter-Augustinsson Å. Comparison of Staphylococcus epidermidis isolated from prosthetic joint infections and commensal isolates in regard to antibiotic susceptibility, agr type, biofilm production, and epidemiology. Int. J. Med. Microbiol. 2013;303:32–39. doi: 10.1016/j.ijmm.2012.11.001. [DOI] [PubMed] [Google Scholar]
- 5.Percival S.L., Suleman L., Vuotto C., Donelli G. Healthcare-associated infections, medical devices and biofilms: risk, tolerance and control. J. Med. Microbiol. 2015;64:323–334. doi: 10.1099/jmm.0.000032. [DOI] [PubMed] [Google Scholar]
- 6.Otto M. Staphylococcal biofilms. Curr. Top Microbiol. Immunol. 2008;322:207–228. doi: 10.1007/978-3-540-75418-3_10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Götz F. Staphylococcus and biofilms. Mol. Microbiol. 2002;43:1367–1378. doi: 10.1046/j.1365-2958.2002.02827.x. [DOI] [PubMed] [Google Scholar]
- 8.Rohde H., Burdelski C., Bartscht K., Hussain M., Buck F., Horstkotte M.A., et al. Induction of Staphylococcus epidermidis biofilm formation via proteolytic processing of the accumulation-associated protein by staphylococcal and host proteases. Mol. Microbiol. 2005;55:1883–1895. doi: 10.1111/j.1365-2958.2005.04515.x. [DOI] [PubMed] [Google Scholar]
- 9.Banner M.A., Cunniffe J.G., Macintosh R.L., Foster T.J., Rohde H., Mack D., et al. Localized tufts of fibrils on Staphylococcus epidermidis NCTC 11047 are comprised of the accumulation-associated protein. J. Bacteriol. 2007;189:2793–2804. doi: 10.1128/JB.00952-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Corrigan R.M., Rigby D., Handley P., Foster T.J. The role of Staphylococcus aureus surface protein SasG in adherence and biofilm formation. Microbiology. 2007;153:2435–2446. doi: 10.1099/mic.0.2007/006676-0. [DOI] [PubMed] [Google Scholar]
- 11.Reiter K.C., Villa B., Paim T., G. d S., Sambrano G.E., de Oliveira C.F., et al. Enhancement of antistaphylococcal activities of six antimicrobials against sasG-negative methicillin-susceptible Staphylococcus aureus: an in vitro biofilm model. Diagn. Microbiol. Infect. Dis. 2012;74:101–105. doi: 10.1016/j.diagmicrobio.2012.05.034. [DOI] [PubMed] [Google Scholar]
- 12.Savolainen K., Paulin L., Westerlund-Wikström B., Foster T.J., Korhonen T.K., Kuusela P. Expression of pls, a gene closely associated with the mecA gene of methicillin-resistant Staphylococcus aureus, prevents bacterial adhesion in vitro. Infect. Immun. 2001;69:3013–3020. doi: 10.1128/IAI.69.5.3013-3020.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Juuti K.M., Sinha B., Werbick C., Peters G., Kuusela P.I. Reduced adherence and host cell invasion by methicillin-resistant Staphylococcus aureus expressing the surface protein Pls. J. Infect. Dis. 2004;189:1574–1584. doi: 10.1086/383348. [DOI] [PubMed] [Google Scholar]
- 14.Hussain M., Schafer D., Juuti K.M., Peters G., Haslinger-Loffler B., Kuusela P.I., et al. Expression of Pls (plasmin sensitive) in Staphylococcus aureus negative for pls reduces adherence and cellular invasion and acts by steric hindrance. J. Infect. Dis. 2009;200:107–117. doi: 10.1086/599359. [DOI] [PubMed] [Google Scholar]
- 15.Roche F.M., Meehan M., Foster T.J. The Staphylococcus aureus surface protein SasG and its homologues promote bacterial adherence to human desquamated nasal epithelial cells. Microbiology. 2003;149:2759–2767. doi: 10.1099/mic.0.26412-0. [DOI] [PubMed] [Google Scholar]
- 16.Hussain M., Herrmann M., von Eiff C., Perdreau-Remington F., Peters G. A 140-kilodalton extracellular protein is essential for the accumulation of Staphylococcus epidermidis strains on surfaces. Infect. Immun. 1997;65:519–524. doi: 10.1128/iai.65.2.519-524.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bowden M.G., Chen W., Singvall J., Xu Y., Peacock S.J., Valtulina V., et al. Identification and preliminary characterization of cell-wall-anchored proteins of Staphylococcus epidermidis. Microbiology. 2005;151:1453–1464. doi: 10.1099/mic.0.27534-0. [DOI] [PubMed] [Google Scholar]
- 18.Marchler-Bauer A., Derbyshire M.K., Gonzales N.R., Lu S., Chitsaz F., Geer L.Y., et al. Cdd: NCBI's conserved domain database. Nucl. Acids Res. 2015;43:D222–D226. doi: 10.1093/nar/gku1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gruszka D.T., Wojdyla J.A., Bingham R.J., Turkenburg J.P., Manfield I.W., Steward A., et al. Staphylococcal biofilm-forming protein has a contiguous rod-like structure. Proc. Natl. Acad. Sci. U. S. A. 2012;109:E1011–E1018. doi: 10.1073/pnas.1119456109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gruszka D.T., Whelan F., Farrance O.E., Fung H.K.H., Paci E., Jeffries C.M., et al. Cooperative folding of intrinsically disordered domains drives assembly of a strong elongated protein. Nat. Commun. 2015;6:7271. doi: 10.1038/ncomms8271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gruszka D.T., Mendonça C.A.T.F., Paci E., Whelan F., Hawkhead J., Potts J.R., et al. Disorder drives cooperative folding in a multidomain protein. Proc. Natl. Acad. Sci. U. S. A. 2016;113:11841–11846. doi: 10.1073/pnas.1608762113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Conrady D.G., Wilson J.J., Herr A.B. Structural basis for Zn2+-dependent intercellular adhesion in staphylococcal biofilms. Proc. Natl. Acad. Sci. U. S. A. 2013;110:E202–211. doi: 10.1073/pnas.1208134110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bleiziffer I., Eikmeier J., Pohlentz G., McAulay K., Xia G., Hussain M., et al. The plasmin-sensitive protein pls in methicillin-resistant Staphylococcus aureus (MRSA) is a Glycoprotein. PLoS Pathog. 2017;13:e1006110. doi: 10.1371/journal.ppat.1006110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Whelan F., Lafita A., Gilburt J., Dégut C., Griffiths S.C., Jenkins H.T., et al. Periscope Proteins are variable-length regulators of bacterial cell surface interactions. Proc. Natl. Acad. Sci. U. S. A. 2021;118 doi: 10.1073/pnas.2101349118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Whelan F. The long and the short of Periscope Proteins. Biochem. Soc. Trans. 2022;50:1293–1302. doi: 10.1042/BST20220194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Conlon B.P., Geoghegan J.A., Waters E.M., McCarthy H., Rowe S.E., Davies J.R., et al. Role for the A domain of unprocessed accumulation-associated protein (Aap) in the attachment phase of the Staphylococcus epidermidis biofilm phenotype. J. Bacteriol. 2014;196:4268–4275. doi: 10.1128/JB.01946-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Paharik A.E., Kotasinska M., Both A., Hoang T.-M.N., Büttner H., Roy P., et al. The metalloprotease SepA governs processing of accumulation-associated protein and shapes intercellular adhesive surface properties in Staphylococcus epidermidis. Mol. Microbiol. 2017;103:860–874. doi: 10.1111/mmi.13594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Macintosh R.L., Brittan J.L., Bhattacharya R., Jenkinson H.F., Derrick J., Upton M., et al. The terminal A domain of the fibrillar accumulation-associated protein (Aap) of Staphylococcus epidermidis mediates adhesion to human corneocytes. J. Bacteriol. 2009;191:7007–7016. doi: 10.1128/JB.00764-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Roy P., Horswill A.R., Fey P.D. Glycan-dependent corneocyte adherence of Staphylococcus epidermidis mediated by the lectin subdomain of Aap. mBio. 2021;12:e0290820. doi: 10.1128/mBio.02908-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chantraine C., Mathelié-Guinlet M., Pietrocola G., Speziale P., Dufrêne Y.F. AFM identifies a protein complex involved in pathogen adhesion which ruptures at three nanonewtons. Nano Lett. 2021;21:7595–7601. doi: 10.1021/acs.nanolett.1c02105. [DOI] [PubMed] [Google Scholar]
- 31.Krissinel E., Henrick K. In: Computational Life Sciences: First International Symposium, CompLife 2005, Konstanz, Germany. R. Berthold M., Glen R.C., Diederichs K., Kohlbacher O., Fischer I., editors. Springer Berlin Heidelberg; Berlin, Heidelberg: 2005. Multiple Alignment of Protein Structures in Three Dimensions; pp. 67–78. September 25-27, 2005. Proceedings. [Google Scholar]
- 32.Holm L., Rosenström P. Dali server: conservation mapping in 3D. Nucl. Acids Res. 2010;38:W545–W549. doi: 10.1093/nar/gkq366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yang Y.H., Jiang Y.L., Zhang J., Wang L., Bai X.H., Zhang S.J., et al. Structural insights into SraP-mediated Staphylococcus aureus adhesion to host cells. PLoS Pathog. 2014;10:e1004169. doi: 10.1371/journal.ppat.1004169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Moothoo D.N., Naismith J.H. Concanavalin A distorts the beta-GlcNAc-(1-->2)-Man linkage of beta-GlcNAc-(1-->2)-alpha-Man-(1-->3)-[beta-GlcNAc-(1-->2)-alpha-Man- (1-->6)]-Man upon binding. Glycobiology. 1998;8:173–181. doi: 10.1093/glycob/8.2.173. [DOI] [PubMed] [Google Scholar]
- 35.Naismith J.H., Field R.A. Structural basis of trimannoside recognition by concanavalin A. J. Biol. Chem. 1996;271:972–976. doi: 10.1074/jbc.271.2.972. [DOI] [PubMed] [Google Scholar]
- 36.Stemberk V., Jones R.P., Moroz O., Atkin K.E., Edwards A.M., Turkenburg J.P., et al. Evidence for steric regulation of fibrinogen binding to Staphylococcus aureus fibronectin-binding protein A (FnBPA) J. Biol. Chem. 2014;289:12842–12851. doi: 10.1074/jbc.M113.543546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Loris R., Hamelryck T., Bouckaert J., Wyns L. Legume lectin structure. Biochim. Biophys. Acta (Bba) - Protein Struct. Mol. Enzymol. 1998;1383:9–36. doi: 10.1016/s0167-4838(97)00182-9. [DOI] [PubMed] [Google Scholar]
- 38.Jabs A., Weiss M.S., Hilgenfeld R. Non-proline Cis peptide bonds in proteins 1. J. Mol. Biol. 1999;286:291–304. doi: 10.1006/jmbi.1998.2459. [DOI] [PubMed] [Google Scholar]
- 39.Weiss M.S., Metzner H.J., Hilgenfeld R. Two non-proline cis peptide bonds may be important for factor XIII function. FEBS Lett. 1998;423:291–296. doi: 10.1016/s0014-5793(98)00098-2. [DOI] [PubMed] [Google Scholar]
- 40.Reeke G.N., Jr., Becker J.W., Edelman G.M. The covalent and three-dimensional structure of concanavalin A. IV. Atomic coordinates, hydrogen bonding, and quaternary structure. J. Biol. Chem. 1975;250:1525–1547. [PubMed] [Google Scholar]
- 41.Garcia-Pino A., Buts L., Wyns L., Loris R. Interplay between metal binding and cis/trans isomerization in legume lectins: structural and thermodynamic study of P. Angolensis lectin. J. Mol. Biol. 2006;361:153–167. doi: 10.1016/j.jmb.2006.06.006. [DOI] [PubMed] [Google Scholar]
- 42.Madeira F., Pearce M., Tivey A.R.N., Basutkar P., Lee J., Edbali O., et al. Search and sequence analysis tools services from EMBL-EBI in 2022. Nucl. Acids Res. 2022;50:W276–W279. doi: 10.1093/nar/gkac240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fogg M.J., Wilkinson A.J. Higher-throughput approaches to crystallization and crystal structure determination. Biochem. Soc. Trans. 2008;36:771–775. doi: 10.1042/BST0360771. [DOI] [PubMed] [Google Scholar]
- 44.Hendrickson W.A., Horton J.R., LeMaster D.M. Selenomethionyl proteins produced for analysis by multiwavelength anomalous diffraction (MAD): a vehicle for direct determination of three-dimensional structure. EMBO J. 1990;9:1665–1672. doi: 10.1002/j.1460-2075.1990.tb08287.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.O'Brien L.M., Walsh E.J., Massey R.C., Peacock S.J., Foster T.J. Staphylococcus aureus clumping factor B (ClfB) promotes adherence to human type I cytokeratin 10: implications for nasal colonization. Cell Microbiol. 2002;4:759–770. doi: 10.1046/j.1462-5822.2002.00231.x. [DOI] [PubMed] [Google Scholar]
- 46.Liu Y., Childs R.A., Palma A.S., Campanero-Rhodes M.A., Stoll M.S., Chai W., et al. Neoglycolipid-based oligosaccharide microarray system: preparation of NGLs and their noncovalent immobilization on nitrocellulose-coated glass slides for microarray analyses. Met. Mol. Biol. 2012;808:117–136. doi: 10.1007/978-1-61779-373-8_8. [DOI] [PubMed] [Google Scholar]
- 47.Liu Y., McBride R., Stoll M., Palma A.S., Silva L., Agravat S., et al. The minimum information required for a glycomics experiment (MIRAGE) project: improving the standards for reporting glycan microarray-based data. Glycobiology. 2017;27:280–284. doi: 10.1093/glycob/cww118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Whelan F., Lafita A., Griffiths S.C., Cooper R.E.M., Whittingham J.L., Turkenburg J.P., et al. Defining the remarkable structural malleability of a bacterial surface protein Rib domain implicated in infection. Proc Natl Acad Sci U. S. A. 2019;116:26540–26548. doi: 10.1073/pnas.1911776116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Konarev P.V., Volkov V.V., Sokolova A.V., Koch M.H.J., Svergun D.I. Primus: a windows PC-based system for small-angle scattering data analysis. J. Appl. Crystallogr. 2003;36:1277–1282. [Google Scholar]
- 50.Franke D., Petoukhov M.V., Konarev P.V., Panjkovich A., Tuukkanen A., Mertens H.D.T., et al. Atsas 2.8: a comprehensive data analysis suite for small-angle scattering from macromolecular solutions. J. Appl. Crystallogr. 2017;50:1212–1225. doi: 10.1107/S1600576717007786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Weinkam P., Pons J., Sali A. Structure-based model of allostery predicts coupling between distant sites. Proc. Natl. Acad. Sci. U. S. A. 2012;109:4875–4880. doi: 10.1073/pnas.1116274109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Schneidman-Duhovny D., Hammel M., Sali A. FoXS: a web server for rapid computation and fitting of SAXS profiles. Nucl. Acids Res. 2010;38:W540–544. doi: 10.1093/nar/gkq461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Newman J., Egan D., Walter T.S., Meged R., Berry I., Ben Jelloul M., et al. Towards rationalization of crystallization screening for small- to medium-sized academic laboratories: the PACT/JCSG+ strategy. Acta Crystallogr. D Biol. Crystallogr. 2005;61:1426–1431. doi: 10.1107/S0907444905024984. [DOI] [PubMed] [Google Scholar]
- 54.Gorrec F. The MORPHEUS protein crystallization screen. J. Appl. Crystallogr. 2009;42:1035–1042. doi: 10.1107/S0021889809042022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kabsch W. Evaluation of single-crystal X-ray diffraction data from a position-sensitive detector. J. Appl. Crystallogr. 1988;21:916–924. [Google Scholar]
- 56.Evans P.R., Murshudov G.N. How good are my data and what is the resolution? Acta Crystallogr. Sect. D, Biol. Crystallogr. 2013;69:1204–1214. doi: 10.1107/S0907444913000061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sheldrick G. Experimental phasing with SHELXC/D/E: combining chain tracing with density modification. Acta Crystallogr. Sect. D. 2010;66:479–485. doi: 10.1107/S0907444909038360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Cowtan K. The Buccaneer software for automated model building. 1. Tracing protein chains. Acta Crystallogr. Sect. D. 2006;62:1002–1011. doi: 10.1107/S0907444906022116. [DOI] [PubMed] [Google Scholar]
- 59.Emsley P., Lohkamp B., Scott W.G., Cowtan K. Features and development of Coot. Acta Crystallogr. Sect. D. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Murshudov G.N., Skubak P., Lebedev A.A., Pannu N.S., Steiner R.A., Nicholls R.A., et al. REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallogr. Sect. D. 2011;67:355–367. doi: 10.1107/S0907444911001314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Perrakis A., Morris R., Lamzin V.S. Automated protein model building combined with iterative structure refinement. Nat. Struct. Mol. Biol. 1999;6:458–463. doi: 10.1038/8263. [DOI] [PubMed] [Google Scholar]
- 62.Kabsch W. Xds. Acta Crystallogr. Sect. D. 2010;66:125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Vagin A., Teplyakov A. Molrep: an automated program for molecular replacement. J. Appl. Crystallogr. 1997;30:1022–1025. [Google Scholar]
- 64.Nicholls R.A., Fischer M., McNicholas S., Murshudov G.N. Conformation-independent structural comparison of macromolecules with ProSMART. Acta Crystallogr. D Biol. Crystallogr. 2014;70:2487–2499. doi: 10.1107/S1399004714016241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Painter J., Merritt E.A. Optimal description of a protein structure in terms of multiple groups undergoing TLS motion. Acta Crystallogr. D Biol. Crystallogr. 2006;62:439–450. doi: 10.1107/S0907444906005270. [DOI] [PubMed] [Google Scholar]
- 66.Lebedev A.A., Isupov M.N. Space-group and origin ambiguity in macromolecular structures with pseudo-symmetry and its treatment with the program Zanuda. Acta Crystallogr. Sect. D. 2014;70:2430–2443. doi: 10.1107/S1399004714014795. [DOI] [PubMed] [Google Scholar]
- 67.Collaborative Computational Project, N The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D Biol. Crystallogr. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 68.Abagyan R.A., Totrov M.M. Contact area difference (CAD): a robust measure to evaluate accuracy of protein models11Edited by B. Honig. J. Mol. Biol. 1997;268:678–685. doi: 10.1006/jmbi.1997.0994. [DOI] [PubMed] [Google Scholar]
- 69.Chen V.B., Arendall W.B., III, Headd J.J., Keedy D.A., Immormino R.M., Kapral G.J., et al. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. Sect. D. 2010;66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Krissinel E., Henrick K. Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 2007;372:774–797. doi: 10.1016/j.jmb.2007.05.022. [DOI] [PubMed] [Google Scholar]
- 71.Pettersen E.F., Goddard T.D., Huang C.C., Couch G.S., Greenblatt D.M., Meng E.C., et al. UCSF Chimera–a visualization system for exploratory research and analysis. J. Comput. Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 72.McNicholas S., Potterton E., Wilson K.S., Noble M.E. Presenting your structures: the CCP4mg molecular-graphics software. Acta Crystallogr. D Biol. Crystallogr. 2011;67:386–394. doi: 10.1107/S0907444911007281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Chai W., Stoll M.S., Galustian C., Lawson A.M., Feizi T. Neoglycolipid Technology: Deciphering Information Content of Glycome. Meth. Enzymol. 2003;362:160–195. doi: 10.1016/S0076-6879(03)01012-7. [DOI] [PubMed] [Google Scholar]
- 74.Liu Y., Feizi T., Campanero-Rhodes M.A., Childs R.A., Zhang Y., Mulloy B., et al. Neoglycolipid probes prepared via oxime ligation for microarray analysis of oligosaccharide-protein interactions. Chem. Biol. 2007;14:847–859. doi: 10.1016/j.chembiol.2007.06.009. [DOI] [PubMed] [Google Scholar]
- 75.Palma A.S., Liu Y., Childs R.A., Herbert C., Wang D., Chai W., et al. The human epithelial carcinoma antigen recognized by monoclonal antibody AE3 is expressed on a sulfoglycolipid in addition to neoplastic mucins. Biochem. Biophys. Res. Commun. 2011;408:548–552. doi: 10.1016/j.bbrc.2011.04.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The coordinates of structures reported here have been deposited in the Protein Data Bank (PDB) under accession codes Aap338–608 7sie, Aap351–813 8deo, SasG144–423 7smh, and Pls391–656 7sjk.