SUMMARY
Microglial research has advanced considerably in recent decades yet has been constrained by a rolling series of dichotomies such as “resting versus activated” and “M1 versus M2.” This dualistic classification of good or bad microglia is inconsistent with the wide repertoire of microglial states and functions in development, plasticity, aging, and diseases that were elucidated in recent years. New designations continuously arising in an attempt to describe the different microglial states, notably defined using transcriptomics and proteomics, may easily lead to a misleading, although unintentional, coupling of categories and functions. To address these issues, we assembled a group of multidisciplinary experts to discuss our current understanding of microglial states as a dynamic concept and the importance of addressing microglial function. Here, we provide a conceptual framework and recommendations on the use of microglial nomenclature for researchers, reviewers, and editors, which will serve as the foundations for a future white paper.
NAMES, NAMES, NAMES
If the names are unknown, knowledge of the things also perishes.1
—Carolus Linnaeus
And yet, we humans instinctively tend to name things and use that name to define their properties. Biologists are no exception: from the time of 18th century father of taxonomy Carolus Linnaeus, the main purpose of biology has been categorizing the natural world as a way of understanding it. Naming species and grouping them together into taxa served to define evolutionary relationships; even today taxonomy and phylogeny are closely interrelated. But we must never forget that nomenclatures and categories are artificial constructs and that biology is seldom black and white but rather an extended continuum of greys. While giving names is natural and useful, we need to be aware that categorization constrains our thinking by forcing us to fit our observations into established classes. As sociologists say, “categorization spawns expectations.”2 This semantic issue has already been acknowledged by immunologists because, in fact, the given names have connotations that often imply a specific function.3 In this paper, we extend similar initiatives on macrophages,4 dendritic cells,3 interneurons,5 and astrocytes6 to discuss the widespread problems associated with categorization of microglia using outdated terms such as “resting versus activated” (Box 1) or “M1 versus M2” (Box 2).
Box 1. Resting versus activated microglia.
The development of specific silver staining techniques in 1919 allowed Río-Hortega to clearly identify microglia and study their response to experimental manipulations.7,145 Early on, Río-Hortega appreciated the striking morphological transformation of microglia following brain damage, but it was in the mid-1970s that the terms “resting” and “activated” microglia first appeared in the literature. These terms were used to morphologically describe cells with an affinity for silver staining that were observed in physiological (“resting”) versus pathological (“activated”) conditions. This nomenclature consolidated in the 1980s and became widely used during the 1990s,146 in parallel with the development and use of histochemical and immunohistochemical techniques, such as lectin staining,147 detection of phosphatases and phosphorylases,148 and antibodies against the complement receptor CR3.7 These techniques and nomenclature were pivotal in determining that “resting” microglia were unrelated to astrocytes, as some studies had wrongly concluded,149 and that “reactive” microglia shared many characteristics with the blood-borne monocytes.10 As shown by a PubMed search with microglia in all fields, there were only few papers published on the topic before the 1990s, and then a steady increase until the beginning of our century, followed by an exponential growth.150 There is a first inflexion point in 2005, with the seminal discovery using non-invasive two-photon in vivo imaging that microglia are extremely dynamic in the absence of pathological challenge, continuously surveying the parenchyma with their highly motile processes.55,56 The development of non-invasive methods was necessary for our understanding of microglial roles in the healthy brain (reviewed in Tremblay151). In 2005, microglial extreme dynamism in the intact brain was examined for the first time, through the skull of CX3CR1-GFP mice in which microglia are fluorescently labeled.55,56 As a result, microglia are now considered to be the most dynamic cells of the healthy mature brain.151 This seminal discovery prompted the renaming of quiescent or resting microglia as surveying56,152 or surveillant (from the verb to survey)153 microglia and also led to the proposal of the concept that microglia are never resting.154 Together, these and other in vivo two-photon imaging data put into serious doubt the concept of “activated” microglia, which suggests a unique form of response, as in fact microglia are always active, constantly responding (in different ways depending on the context) to the changes in their CNS environment, even under normal physiological conditions. Therefore, microglia do not switch from “resting” to “activated” in response to trauma, injury, infection, disease, and other challenges. Rather, microglia are continuously active and react to the stage of life, CNS region, species, sex, and context of health or disease by adopting different states and performing different functions. Thus, although still widely used, “resting” and “activated” microglia are labels that should be discontinued.
Box 2. M1 versus M2 microglia.
Another terminology emerged in the early 2000s from immunologists classifying macrophages based on findings obtained using in vitro models: “M1,” the classical activation, considered pro-inflammatory and neurotoxic, as well as closely related to the concept of “activated” microglia, and “M2,” or alternative activation, considered anti-inflammatory and neuroprotective.155 These responses were related to those of T helper lymphocytes (Th1 and Th2) based on their in vitro activation by specific immune stimuli that activated differential metabolic programs and changes in cytokine expression.156 An associated term is “M0” microglia, which describes their state when cultured in the presence of transforming growth factor β (TGFβ) and CSF1 to mimic in vivo counterparts.157 The terms became widely adopted in microglial research, and the 2010s saw a boom of papers phenotyping macrophages and microglia into “M1” and “M2” based on the expression of markers related to these categories, used to indirectly assume a detrimental (“M1”) or beneficial (“M2”) microglial role.156 In many cases, editors and reviewers have asked authors to comply with this nomenclature. However, it soon became evident that macrophage responses are more complex than simply “M1” and “M2.”158 In the case of microglia, the advent of single-cell technologies provided clear evidence that microglia in the living brain do not polarize to either of these categories, often co-expressing M1 and M2 markers,159 despite the continued use of M1 and M2 in the literature. We thus recommend strictly avoiding M1 and M2 labels and using more nuanced tools to investigate microglial function (reviewed in Devanney et al.160).
Dichotomic, rigid categories convey a dualistic idea of good versus bad microglia and may actually impede scientific advancement. Widely used terms, such as “neuroinflammation” as a synonym of microglial reactivity (Box 3) and naming a panoply of presumed microglial populations and assumed functions arising from single-cell transcriptomics, are misleading and increasingly problematic, especially to those entering the field of glial biology and neuroimmunology. This nomenclature does not address the important question: what are the specific functions of microglia in the contexts of development, health, aging, and disease? It is now clear that microglia exist in diverse, dynamic, and multidimensional states depending on the context, including local environment (Figure 1). We define dimensions as the key variables driving the phenotypic transformations of microglia. These variables are molecularly distinct signaling pathways regulated at multiple levels (e.g., transcriptional, epigenetic, translational, metabolic) that each give rise to distinct microglial functions or properties. In this manner, categorizing microglia based on a historical, one-dimensional nomenclature in the absence of functional data will constrain and stifle future progress and innovation.
Box 3. Microglial morphological responses across species.
Microglial cells display a profusion of morphologies that have fascinated researchers since the early days of Río-Hortega. Many were tempted to equate morphology with function. Ramified microglia were traditionally associated with the “resting” state, although we now know that ramified microglia actively play many functions during normal physiological conditions. In contrast, “reactive” microglia (rounder cell body, generally with fewer and shorter processes) were called “activated” and equated with an inflammatory response. Only recently, however, a mechanistic link between microglial reduced branching and increased release of the inflammatory cytokine interleukin 1β (IL-1β) was reported.161 Activation of P2YR12 by tissue damage signals potentiates the tonically active potassium THIK-1 channel, expressed in microglia, leading to both decreased microglial ramifications and activation of the inflammasome machinery processing IL-1β precursors into their mature form.161 Another morphology associated with functional changes is “ameboid” microglia, which were thought to be more “phagocytic,” but it is clear now that ramified microglia execute phagocytosis through their terminal or “en passant” branches notably during adult neurogenesis,162,163 while in disease conditions such as epilepsy, ameboid microglia can display reduced phagocytosis.164 Therefore, morphological changes should not be interpreted in functional terms but rather taken as a suggestion prompting further investigation of the relationship between microglial structure and function. While the categorization described above is now outdated, the analysis of microglial morphology is considered valuable and still often used across animal model and human postmortem brain studies. Studies in postmortem brain samples have revealed that human and mouse microglia can adopt similar morphologies. Using the now outdated terms “ramified,” “primed” (larger cell body, ramified processes), “reactive” (ameboid, few ramified processes), and “ameboid” (less than two unramified processes), microglia were described in middle-aged individuals.165 In addition, “rodshaped” microglia (elongated cell body, polarized processes) were found to become more abundant with aging.166 Similarly, “dystrophic” microglia, presenting apparently fragmented (but still intact at the ultrastructural level) processes were reported in aging.167,168 These different morphological types observed in humans were previously described in rodent models (reviewed in Savage et al.169). Nevertheless, a more sensitive quantitative microglial morphological assessment using a computational pipeline involving cluster analysis revealed differences between mouse and human, with distinct clusters found to be unique to each species.170 Subsequently, a high-throughput comparative morphology analysis revealed a generally conserved evolutionary pattern, with some intriguing differences observed between the leech, zebrafish, axolotl, turtle, chicken, gecko, snake, bearded dragon, bat, boar, sheep, whale, hamster, rat, mouse, marmoset, macaque, and human and across brain regions between mouse and human.76 While detailed comparative ultrastructural analyses of microglia between species are currently lacking, the state of “dark microglia” (named based on their increased electron density giving these cells a dark appearance, compared to other microglial states) discovered in 2016, which is defined using electron microscopy by its markers of cellular stress in contexts of aging and disease, was found to be conserved across mouse, rat, and human.171,172 New strategies are currently being developed to provide morphological data analyses based on automated pipeline, thus overcoming feature-selection-based biases.173 Future studies will show how these varied morphologies correlate with transcriptional and proteomic profiles and what they imply for the cell’s function. At the molecular level, recent single-cell transcriptome analyses also revealed that human microglia show multiple clusters that indicate a greater heterogeneity than in other mammalian species such as the mouse.76,91
Figure 1. Microglial nomenclatures: Past and future.
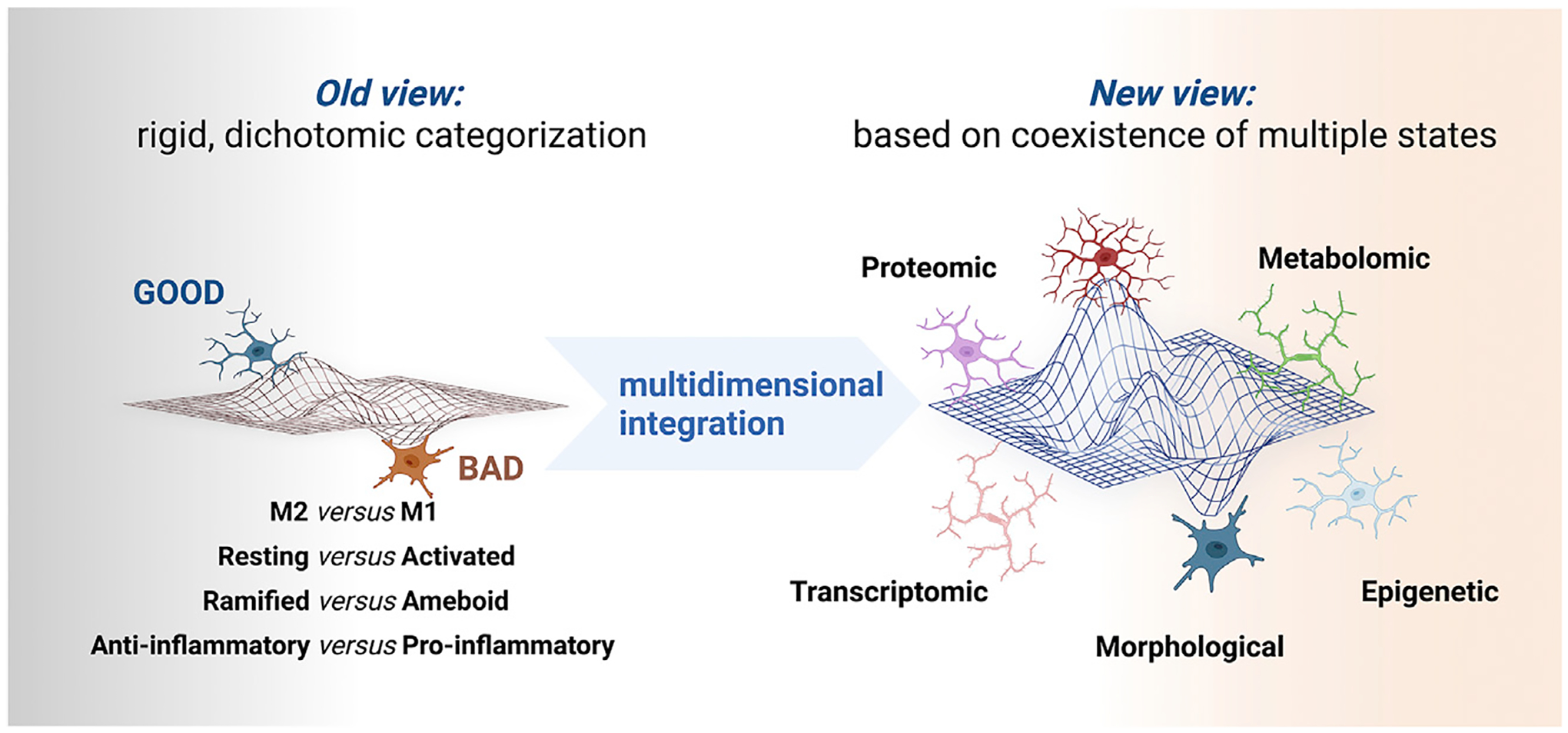
Microglia have been traditionally framed into dichotomic categories, but our current integration of epigenetic, transcriptomic, metabolomic, and proteomic data favors a multidimensional integration of coexisting states.
To examine and address these issues, we assembled a team of international experts who have made major contributions to microglia research, inclusive of various groups, and balancing gender, geographical distribution, and seniority. Authors from the fields of neuroscience, neurobiology, immunology, neuroimmunology, oncology, and neuropathology, from both academia and industry, discussed their perspectives on the current and future challenges in defining microglial states and nomenclature. A questionnaire (Data S1) was created to collect all the authors’ opinions on several nomenclature issues and the importance of directly addressing microglial function. The responses to the questionnaire, an online meeting held in June 2021, and an open session held at the EMBO meeting Microglia 2021 were used as a backbone to develop this paper.
Herein, we summarize our current knowledge about the identity of microglia and discuss best practices for how to define and study microglial state dynamics. We then outline “classical” microglial nomenclatures, highlighting some of the key discoveries that led to the above classifications and their limitations. We intentionally focus on citing studies related to the nomenclature rather than providing a comprehensive review of the history of microglial research, as it has been done elsewhere.7,8 We discuss the overall limitations and conclude with recommendations for the proper usage of microglial nomenclature as research evolves, provide a conceptual framework for discussing microglia, and offer perspectives on the future questions, gaps in knowledge, and challenges to tackle as a field.
MICROGLIAL IDENTITY: WHAT WE MEAN ABOUT WHEN WE TALK ABOUT MICROGLIA
The origin and identity of microglia was, for many years, a matter of debate. In the dim and distant past, Ramón y Cajal’s disciple, Pío del Río-Hortega, suggested that these cells were of mesodermal origin.9 However, over time, an ectodermal origin was also proposed,10 sparking controversy until the 1980s. The mesodermal origin took solid hold later with the advance of technical approaches revealing more similarities than differences with the functions and features of macrophages. In 1999, microglia were reported to appear in the brain rudiment as early as embryonic day 8 (E8) in mice and proposed to originate from yolk sac progenitors.11 The recent combination of fate mapping studies and transplantation approaches this debate, revealing key aspects of microglial identity and plasticity. In mice, unlike other model organisms such as zebrafish,12,13 microglia are now considered to originate from a pool of macrophages produced during primitive hematopoiesis in the yolk sac, which start invading the neuroepithelium at E8.5.14–17 In humans, microglial precursors invade the brain primordium around 4.5 to 5.5 gestational weeks.18
One key signaling pathway critical for microglial development and maintenance is the colony stimulating factor receptor (CSF1R). Ligands of CSF1R that sustain this pathway include two cytokines with different origins and primary sequences but similar tridimensional structures and binding to CSF1R: IL-34 and CSF1.19 IL-34 is produced by neurons, while CSF1 is secreted primarily by oligodendrocytes and astrocytes. Accordingly, the two ligands have distinct and non-overlapping functions in the establishment and maintenance of microglia within the gray and white matter.20 Microglia have the capacity for self-renewal in certain contexts, allowing them to repopulate the CNS within 1 week of depletion, even when more than 99% of microglia are ablated with CSF1R antagonists21,22 or diphtheria toxin.22 This process, termed “microglial repopulation” or “microglial self-renewal,”23–25 is different from “microglia replacement,” which, in contrast, occurs when endogenous microglia are replaced by exogenous cells that can include bone-marrow-derived myeloid cells,26–29 peripheral blood cells,28,30 stem-cell- or induced-pluripotent-stem-cell (iPSC)-derived peripheral blood cells,31 across various experimental or pathological conditions.31–33 Our current definition is that mammalian microglia are yolk-sac-derived, long-lived cells within the CNS parenchyma that persist into adulthood and self-renew without any contribution from bone-marrow-derived cells at a steady state.
The identification of microglia is currently based on the expression of specific genes highly enriched in microglia, which represent their transcriptional identity and are commonly employed as “microglial markers” (Table 1). However, the expression of each marker alone is not sufficient to define microglial identity, as levels of expression may change depending on microglial adaptation to local signals. The present consensus is that mammalian microglia can be identified by the expression of transcription factors like Pu.116, cytoplasmic markers such as ionized calcium-binding adapter molecule 1 (IBA1), and surface markers including the purinergic receptor P2YR12, trans-membrane protein 119 (TMEM119), and CSF1R.34 Based on these markers, genetic tools (such as Cx3cr1CreERT2, P2ry12CreERT2, Tmem119CreERT2, and HexbCreERT2 mouse lines) (Table 2) are available that allow for more specific manipulation or visualization of microglia, although they could also target other populations, including border-associated macrophages (BAMs), also named CNS-associated macrophages (CAMs), and other glial cells.35–40 Most recently, a new binary transgenic model relying on co-expression of Sall1 and Cx3cr1 has been introduced that specifically targets microglia in a non-inducible way.41
Table 1.
Main antibody markers used to visualize microglia in rodents and humans from early embryonic development to adulthood and aging Other proteins expressed by microglia but whose specificity is not confirmed include APOE, CLEC7A, ITGAX, and LPL.
| Marker | Specificity | Labeled states | Staining patterns | Main applications | Reference |
|---|---|---|---|---|---|
| F4/80 (EMR1) | macrophages including microglia | homeostatic conditions and disease associated. expressed in rodents but presence not yet confirmed in human. |
does not provide a detailed cellular visualization, especially in homeostatic conditions, because of its low basal expression. its expression varies significantly between species and is low in human macrophages. |
brightfield or fluorescence analysis of microglial density, distribution, and categorization into morphological states. | Lawson et al.;175 Gautier et al.;176 Waddell et al.177 |
| CX3CR1 | macrophages including microglia | homeostatic conditions and disease associated, but downregulated by the DAMs, MGnD, dark microglia, and other pathological states. | CX3CR1-GFP reporter line generally used for visualization, with or without GFP immunostaining. | brightfield or fluorescence analysis of microglial density, distribution, and categorization into morphological states. | Keren-Shaul et al.;58 Krasemann et al.;59 Jung et al.;178 Wolf et al.;179 Bisht et al.180 |
| IBA1 | macrophages including microglia | homeostatic conditions and disease associated. used to study microglia in early embryonic and postnatal development. conserved across several species including human. |
provides exceptional visualization of microglial cell body and processes, including distal extremities. diffuses throughout the cytoplasm. staining can, however, be discontinuous in aging. |
brightfield or fluorescence analysis of microglial density, distribution, and morphology. ultrastructural studies. |
Keren-Shaul et al.;58 Geirsdottir et al.;76 Tischer et al.;168 Imai et al.;181 Ito et al.;182 Shapiro et al.;183 Wake et al.;184 Tremblay et al.;185 Lier et al.186 |
| MerTK | macrophages including microglia | homeostatic conditions and disease associated. expressed in health and across various contexts of disease, notably in association with the phagocytosis of newborn neurons, amyloid, and myelin. |
partial visualization of microglial cell bodies and diffuse staining of their processes preventing a complete morphological visualization. | brightfield or fluorescence analysis of microglial density, distribution. morphological analysis or categorization into morphological states possible in combination with IBA1. |
Fourgeaud et al.;187 Savage et al.;188 Healy et al.;189 Huang et al.190 |
| CD11b/c | macrophages including microglia | homeostatic conditions and disease associated. used to study microglia in early postnatal development. conserved across species including human. |
visualization of microglial cell body and processes. low basal expression in adult microglia. staining is mainly restricted to the plasma membrane. |
brightfield or fluorescence analysis of microglial density, distribution, and morphology ultrastructural studies of subsets downregulating IBA1. | Bisht et al.;180 Robinson et al.;191 Milligan et al.;192 McKay et al.;193 Blackbeard et al.;194 Marshall et al.195 |
| P2RY12 | largely microglia specific (not expressed by monocytes), but state dependent | homeostatic marker. strongly downregulated in disease-associated and reactive states (but upregulated in status epilepticus). used to study microglia in early postnatal development. conserved across several species including human. |
visualization of microglial cell body and processes. staining can localize to the plasma membrane or diffuse throughout the cytoplasm and can be more profuse than IBA1 depending on staining conditions. |
brightfield or fluorescence analysis of microglial density, distribution, and morphology. ultrastructural studies. |
Avignone et al.;117 Peng et al.;196 Haynes et al.;197 Sipe et al.198 |
| TMEM119 | largely microglia specific, but state dependent | homeostatic conditions and disease associated, but downregulated on reactive microglia in some contexts (e.g., traumatic brain injury and ischemia, MS). developmentally regulated. conserved across species including human. |
partial visualization of microglial cell bodies and diffuse staining of their processes preventing a complete morphological visualization. | brightfield or fluorescence analysis of microglial density, distribution. morphological analysis or categorization into morphological states possible in combination with IBA1. |
Kanamoto et al.;199 Bennett et al.;200 Satoh et al.;201 van Wageningen et al.;202 Gonzalez Ibanez203 |
| TREM2 | macrophages including microglia, state dependent | microglial subsets in early postnatal development, aging, and disease conditions (e.g., microglia involved in synaptic pruning or associated with amyloid plaques in AD pathology). shown to label monocytes or neurons instead of microglia in human. |
visualization of microglial cell body and processes. staining diffuses throughout the cytoplasm. |
brightfield or fluorescence analysis of microglial density, distribution, and categorization into morphological states. ultrastructural studies of pathological states downregulating IBA1. |
Bisht et al.;180 Savage et al.;188 Satoh et al.;201 Chertoff et al.;204 Fahrenhold et al.205 |
Table 2.
Main mouse lines used to visualize microglia from early embryonic development to adulthood and aging Other proteins expressed by microglia but whose specificity is not confirmed include APOE, CLEC7A, ITGAX, and LPL.
| Mouse line | Specificity | Labeled states | Expression patterns | Main applications | Reference |
|---|---|---|---|---|---|
| CX3CR1-GFP | macrophages including microglia | homeostatic conditions and disease associated, but downregulated in DAM, MGnD, dark microglia, and other pathological states. | visualization of microglial cell body and processes. fluorescence diffuses throughout the cytoplasm. bright enough for two-photon in vivo imaging. a limitation is that the heterozygous mice used for in vivo imaging are partially deficient in fractalkine signaling, with possible outcomes on the brain and behavior.206 The homozygous mice are knockout for CX3CR1 and used to study the outcomes of fractalkine receptor deficiency. |
two-photon in vivo imaging or fluorescence analysis of microglial density, distribution, dynamics, interactions with other parenchymal elements, and categorization into morphological states. ultrastructural studies using staining against GFP. |
Davalos et al.;55 Nimmerjahn et al.;56 Jung et al.;178 Bisht et al.;180 Tremblay et al.;185 Paolicelli et al.207 |
| Iba1-EGFP | macrophages including microglia | homeostatic conditions and disease associated. downregulated in some contexts (e.g., obesity and aging) and in some pathological states (e.g., DAM, dark microglia). used to study microglia in early embryonic and postnatal development. conserved across several species including human. |
visualization of microglial cell body and processes. fluorescence diffuses throughout the cytoplasm. less bright than fluorescence in CX3CR1-GFP mice but generally sufficient for two-photon in vivo imaging of cell body and proximal processes. these mice are not partially deficient in IBA1 in their heterozygous state, which is a main advantage. |
two-photon in vivo imaging or fluorescence analysis of microglial density, distribution, dynamics, interactions with other parenchymal elements, and categorization into morphological states. ultrastructural studies using staining against GFP. |
Bisht et al.;180 Wake et al.;184 Hirasawa et al.208 |
| Fms-EGFP or CSF1R-EGFP; CSF1R-FusionRed | Macrophages including microglia. CSF1R is expressed by most microglia. |
Homeostatic conditions and disease-associated, but considered to be downregulated in DAM and other pathological states. | Fluorescence is less bright than in CX3CR1-GFP mice, and generally sufficient for two-photon in vivo imaging. It also allows for fluorescence-activated cell sorting and fluorescence imaging when combined with immunostaining. These mice are not partially deficient in CSF1R in their heterozygous state, which is a main advantage. |
Fluorescence-activated cell sorting and fluorescence analysis of microglial density, distribution, dynamics, interactions with other parenchymal elements, and categorization into morphological states when combined with immunostaining. | Grabert et al.;34 Sierra et al.;162 Sasmono et al.209 |
| HEXB-TdTomato | largely overlaps with IBA1 staining but restricted to microglia. Does not label CAMs and other border-associated macrophage populations. | expression appears stable in homeostatic conditions and disease-associated states. The labeled microglia are also depleted by CSF1R inhibition. |
visualization of microglial cell body and processes. fluorescence diffuses throughout the cytoplasm. bright enough for two-photon in vivo imaging. a limitation is that the heterozygous mice used for in vivo imaging are partially deficient in HEXB. However, their microglial gene expression patterns do not appear affected. |
two-photon in vivo imaging or fluorescence analysis of microglial density, distribution, dynamics, interactions with other parenchymal elements, and categorization into morphological states. | Masuda et al.38 |
Nonetheless, many of these markers are downregulated in pathological states and can be expressed by other brain macrophage populations such as BAMs residing in the perivascular space and leptomeninges,42,43 which also derive from the yolk sac.44 In addition, caution must be exercised, because many classical microglial markers can also be expressed by cells originating from monocytes or iPSCs, and therefore their presence does not imply bona fide microglia. These cells should be more accurately described as monocyte-derived microglia-like or iPSC-derived microglia-like cells (iMGL cells).
As resident macrophages of the brain parenchyma, microglia participate in many critical CNS functions ranging from glio-, vasculo-, and neurogenesis to synaptic and myelination through their process motility, release of soluble factors, and capacity for phagocytosis (Figure 2). These functions have been revealed using several constitutive and inducible knockout models for microglial-specific genes45 and by microglial-depletion paradigms in animal models,46 particularly rodents and zebrafish.
Figure 2. Microglial core properties and functions.
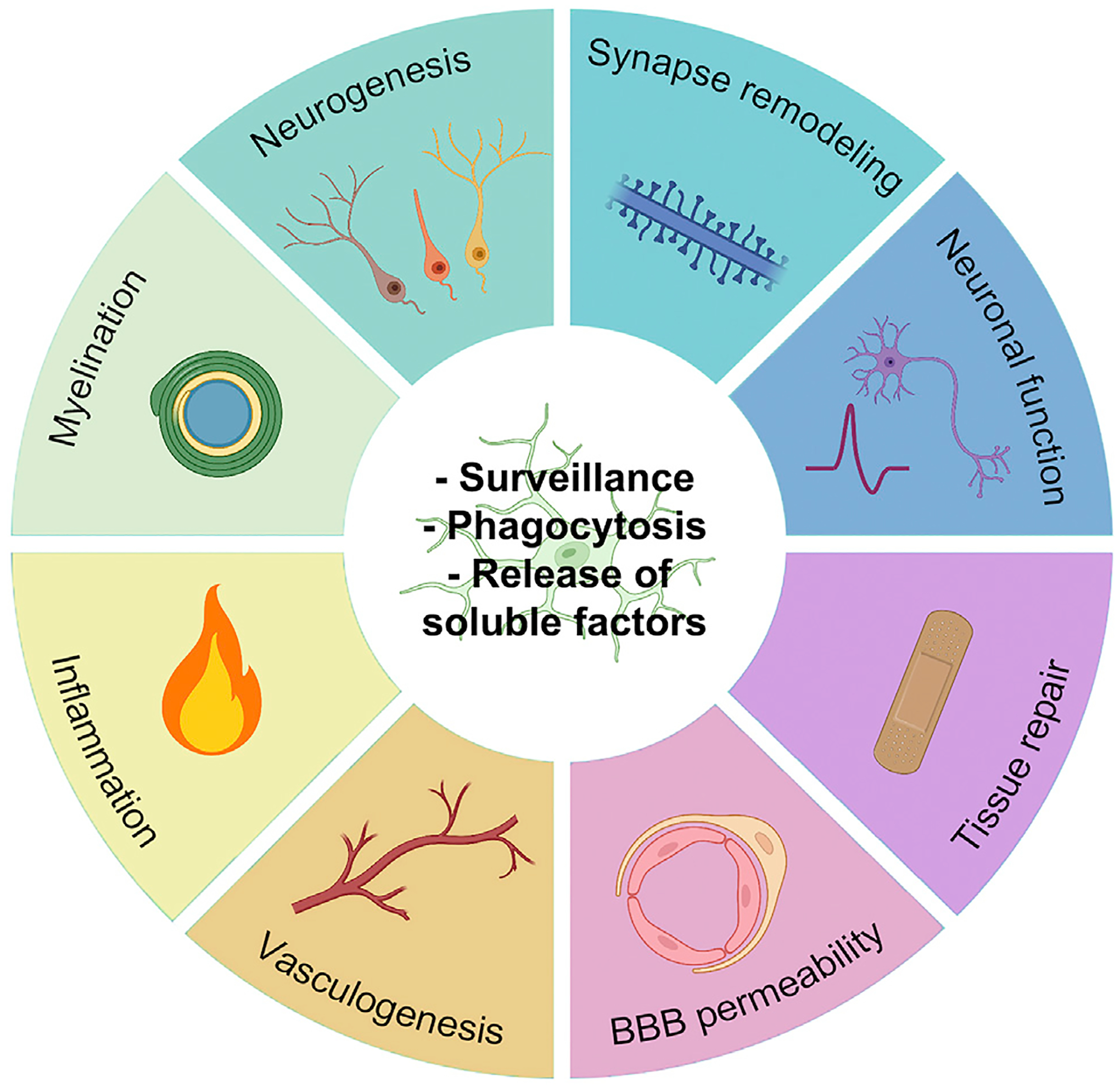
Phagocytosis, surveillance, and capacity for releasing soluble factors (inner circle) are core properties through which microglia contribute to key biological functions (outer circle). Created with BioRender.com.
The key role of microglia in maintaining CNS health is also supported by the severe phenotype displayed by patients lacking microglia due to loss-of-function CSF1R mutations. Heterozygous mutations, particularly in the kinase domain of CSF1R, are associated with adult-onset leukoencephalopathy with axonal spheroids and pigmented glia (ALSP; OMIM: 221820) characterized by reduced microglial numbers and white matter atrophy that result in progressive cognitive and motor impairment, dementia, and early death.47 Additionally, biallelic mutations are reported to cause complete absence of microglia with developmental brain malformation, hydrocephalus, bony lesions, and early death.48,49 This phenotype, however, seems in apparent contradiction with the reported absence of gross neurological abnormalities at birth observed in mice with genomic deletion of FIRE, an intra-intronic super enhancer in the Csf1r gene enhancer region, whose brains lack microglia,50 though more nuanced analyses are needed. Nonetheless, FIRE mice have premature lethality and increased amyloid pathology as early as 5 months of age.51 The source of discrepancy between the developmental impact of CSF1R mutations in humans and mice is not yet fully understood. One possibility is that microglial developmental functions are partly redundant, modified by other environmental factors, or compensated in their absence by other cell types, such as astrocytes.52 It will be important to determine how microglia communicate with other glial cells and immune cell populations to support CNS maturation and function in the future.
(RE)DEFINING MICROGLIAL STATES: DAMs, HAMs, WAMs, AND MORE
Core markers of cellular identity are useful to identify microglia but are not necessarily informative about the functional “state” of microglia, which depends on the context (i.e., the physiological conditions in which microglia are found at any given CNS region and time). Microglia have a complex “sensome,”53 a series of surface receptors that allow them to detect changes in their environment. Microglial states are thus dynamic, and the outcome of the cell’s epigenome, transcriptome, proteome, and metabolome yields discrete morphological, ultrastructural, and/or functional outputs (Figure 3). Microglia are anything but static, as they are exceptionally responsive to alterations in their local environment. In the mature healthy CNS, the distribution of microglia is largely uniform and generally regular with little overlap between adjacent territories.54 The cell bodies are largely sessile, but their processes are constantly moving and scanning the brain parenchyma.55,56 Microglial functions adapt to their location and reciprocal interactions with nearby cells and structures. Their morphology, ultrastructure, and molecular profile are similarly dynamic and plastic, resulting in many different cell states. As Conrad H. Waddington, founding father of systems biology, eloquently described: “Cells are residents of a vast ‘landscape’ of possible states, over which they travel during development and in disease”.57
Figure 3. Microglial identity and states.
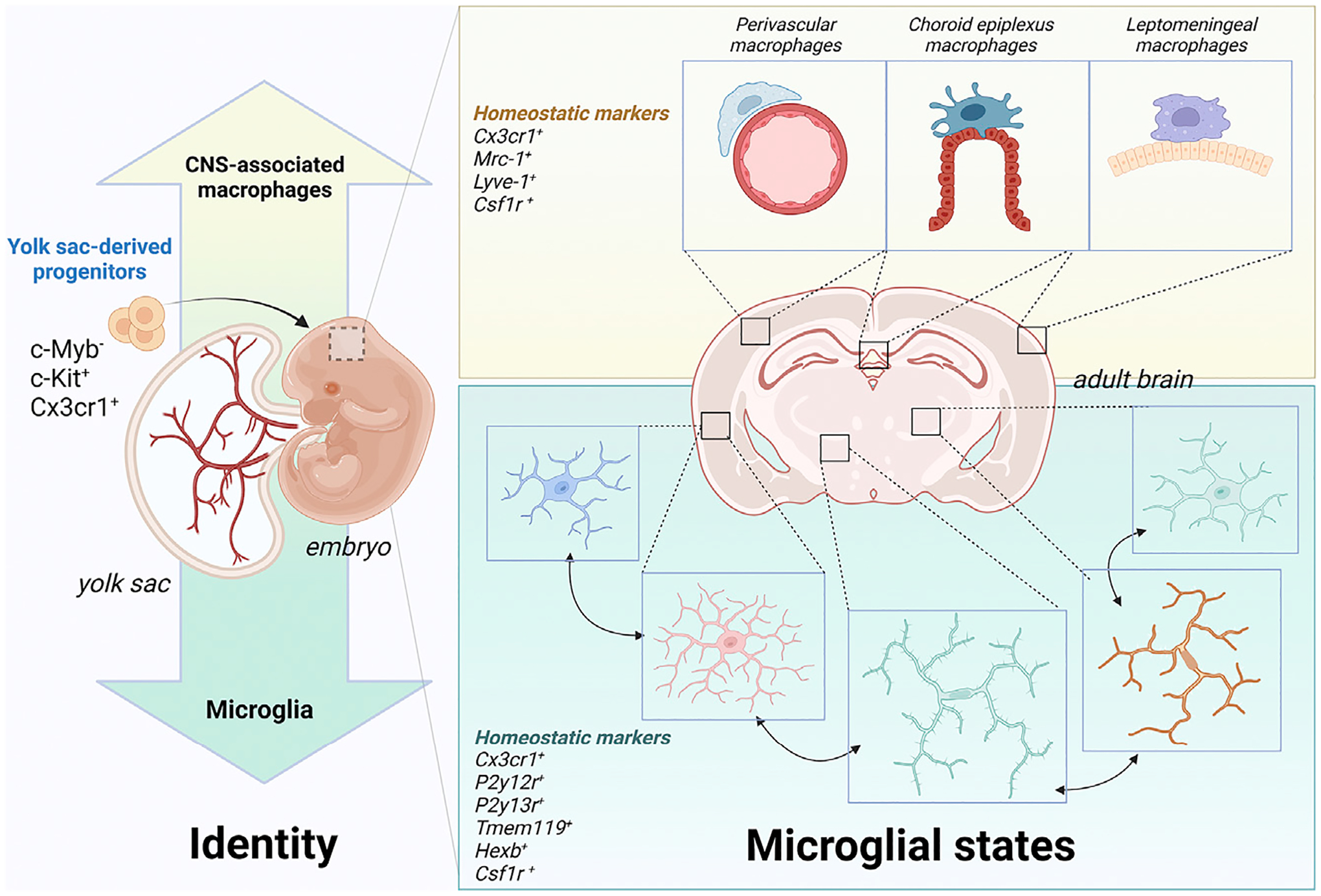
The identity of microglia, compared to other CNS-associated macrophages in the perivascular space, choroid plexus, and leptomeninges, is established early on from yolk-sac-derived progenitors. Once they colonize the brain parenchyma and differentiate, they can adopt multiple states depending on the particular spatiotemporal context, as shown in more detail in Figure 5. Created with BioRender.com.
Single-cell technologies, multi-omics, and integrative analyses of gene and protein expression have helped to not only locate cells on this landscape but also provide new insight into the molecular mechanisms that shape the landscape and regulate specific cell states in a given context (e.g., development, adult, disease, or injury model, etc.). Many diverse and context-dependent microglial states have been observed across species and models. Some examples of these states are the disease-associated microglia (DAMs), originally associated with Alzheimer’s disease (AD) pathology models;58 microglial neurodegenerative phenotype (MGnD) documented across several disease models;59 activated response microglia (ARMs) and interferon-responsive microglia (IRMs) in an AD pathology mouse model;60 human AD microglia (HAMs);61 microglia inflamed in multiple sclerosis (MS) (MIMS);62 and lipid-droplet-accumulating microglia (LDAMs) in aging mice and humans,63 brain tumors (glioma-associated microglia, GAMs),64 amyotrophic lateral sclerosis (ALS)-associated signature,65 and Parkinson disease (PD) microglial signature.66 In the developing and aging brain, the white matter-associated microglia (WAMs),67 axon tract-associated microglia (ATMs),68 and proliferative-region-associated microglia (PAMs, related to phagocytosis of developing oligodendrocytes)69 may share some features with the core DAM signature. In the developing human CNS, microglia also express some of the DAM/MGnD/ARM-like profiles.70
While gene expression signatures indicate biological pathways, the functional implications of these states and relationship to one another remain unclear. In fact, the ever-growing list of branding clusters in single-cell RNA sequencing (scRNA-seq) experiments and use of acronyms is not consistent across research groups and could hinder future advance of the field without validation and functional experiments to understand their meaning. Moreover, transcriptomic signatures depend on tissue dissection and gating strategies that can lead to isolation artifacts,71–74 which, when layered with the technical limitations of single-cell sequencing, can make it difficult to assign state identity across different studies. Another source of complexity comes from evident interspecies differences,75–77 which can further hamper comparisons. Advances in computational tools and approaches, which enable the alignment and integration of single-cell datasets, can help solve some of these issues, providing a powerful way to determine microglial-state similarities across contexts.78,79
A practical limitation of solely defining functional states by their transcriptional signature is that mRNA expression may not directly predict protein levels.80 Protein expression signatures obtained by methods, such as single-cell mass cytometry, have their own technical limitations81 but may better represent true cell states.82,83 Importantly, mRNA or protein expression alone does not necessarily predict microglial function, although they can be used to generate functional hypotheses that need to be experimentally tested. There are many methods that allow for the classification of microglia based on their constituent states, including gene expression, protein expression, post-translational modifications, mRNA profiling, morphology, and ultrastructure. All these approaches can vary in coverage (e.g., expression of a single-cell versus whole-transcriptome profiling), which has created overall confusion and mislabeling in the field. Presumably, each microglial state is associated with unique or specialized functions, although the unique roles of any observed state have so far remained elusive. Thus, it is critical that we begin to define microglial states taking into account their specific context within and between species, across sex, space, and time (e.g., CNS region and biological age) as well as layers of complexity (e.g., epigenetic, transcriptional, translational, and metabolic signatures), which ultimately determine together the cell’s phenome (i.e., motility, morphology, and ultrastructure) and function (Figure 5).
Figure 5. Microglial states defined by their intrinsic and extrinsic determinants, spatiotemporal context, and layers of complexity.
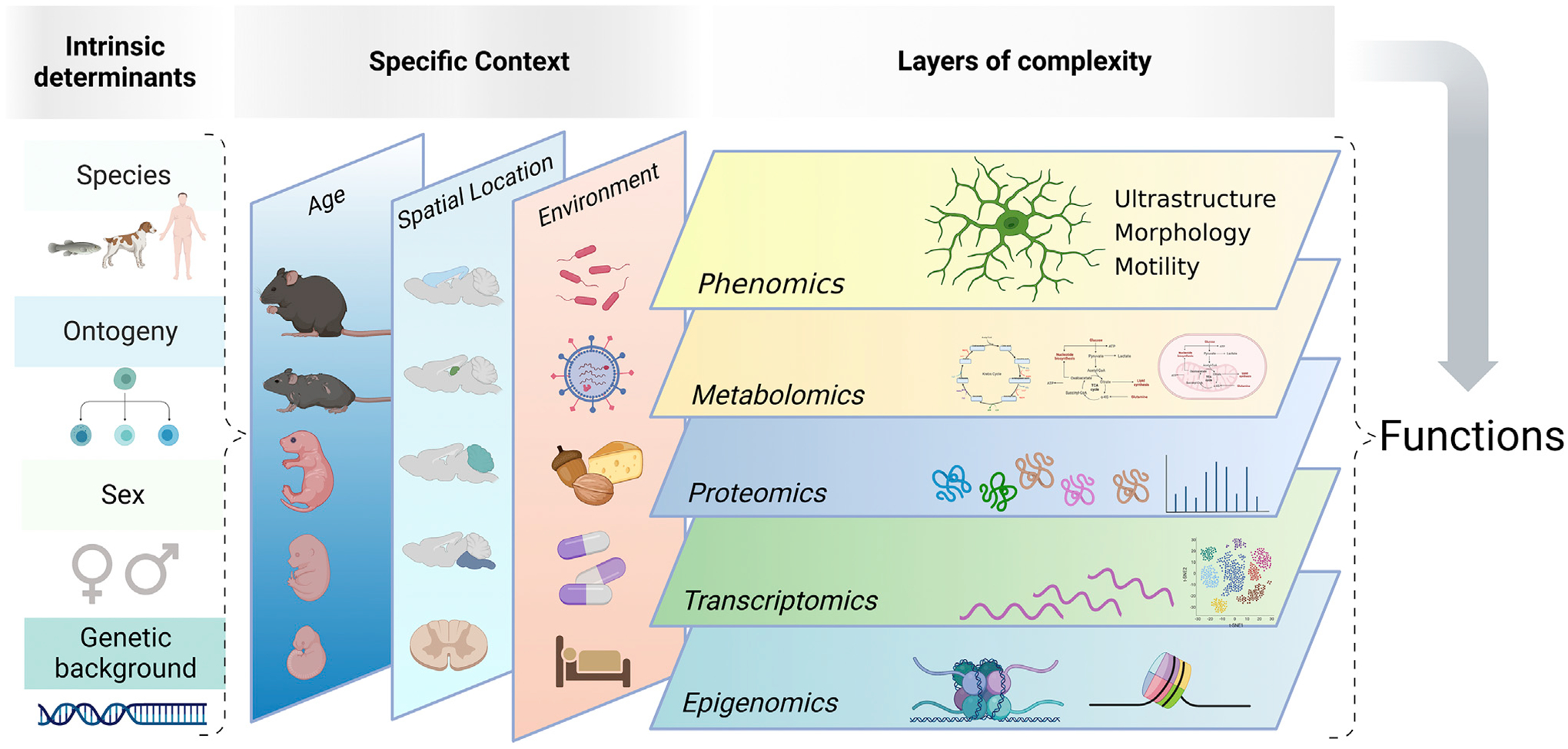
Microglial states depend on intrinsic determinants (such as species, ontogeny, sex, or genetic background) as well as the specific context they inhabit, including age, spatial location, and environmental factors (such as nutrition, microbiota, pathogens, drugs, etc.). All together, these factors impinge on microglia at multiple levels (i.e., epigenomic, transcriptomic, proteomic, metabolomics, ultrastructural, and phenomic), which ultimately determine microglial functions. Created with https://BioRender.com
One major conceptual limitation of the various “one-off” microglial acronyms (e.g., DAMs, MGnD, etc.) is that they suggest stable states or phenotypes of microglia associated with a disease context, such as neurodegeneration. Intuitively, this classification system is similar to the concept of neuronal cell types, where neurons cluster into distinct subtypes based on their gene expression or neuroanatomy. However, contrary to microglia, neuronal groupings are considered fixed and terminally differentiated.5 We do not know how temporally or spatially dynamic microglial states may be, as microglia are remarkably heterogeneous and plastic. Therefore, these cells are probably not permanently “locked” into any single functional state. From the evidence available so far, microglial states appear dynamic and plastic, possibly transitory, and strongly dependent on the context.84 New tools including imaging reporters for microglial states are needed to track transitions within individual cells over time and across the lifespan, following different challenges and perturbations, as well as in response to treatment.
MICROGLIAL HETEROGENEITY: IT ALL DEPENDS ON THE CONTEXT
The term “homeostatic” is used to refer to microglia in physiological conditions, but there are different interpretations of this nomenclature when describing microglia in health and disease. While homeostatic relates to the “physiological” context assessed in space and time, it does not necessarily correspond to a unique molecular profile because, even without any perturbation, microglia display diverse morphological and functional states depending on the signals from the CNS microenvironment. This continuous microglial sensing results in multiple transcriptional signatures from development to aging depending on the specific local signals or challenges to the brain at each developmental stage.53 A less responsive microglial state, which in other contexts would be considered more homeostatic, might be less effective at responding to damage or pathological cues in aging and disease contexts. For example, in aging and neurodegenerative disease, microglia may have reduced ability to rapidly respond to brain challenges (i.e., removing toxic amyloid, infected, damaged, or degenerating neurons), leading to CNS dysfunction and disease progression. Microglia from adult TREM2 knockout mice have been described as “locked in a homeostatic state” as they are less responsive to challenges (such as amyloid) and do not adopt a transcriptional DAM signature in disease contexts.85,86 From this example, the term “homeostatic” is not informative if not well defined and placed in the context of function.
Key modifying factors that lead to microglial heterogeneous states include age, sex, circadian time, local CNS signals, and peripheral cues, such as the changes in the microbiota87,88 or other systemic diseases (e.g., asthma)89 in addition to the pathophysiological state of the CNS and overall organism (discussed in more depth in the next section). Age, indeed, has a key influence on the microglial homeostatic state, which goes through several distinct temporal stages (embryonic, perinatal, adult, and aging microglia), each notably characterized by an enrichment of defined regulatory factors and gene expression profiles.68,90 After the initial establishment of microglial identity by a network of developmentally programmed and environment-dependent transcription factors,75,90 microglia become extremely heterogeneous in their transcriptome during early postnatal development, as determined by scRNA-seq.68,69,91 In contrast, microglia display a more limited transcriptomic heterogeneity in the adult CNS, where the different microglial scRNA-seq clusters fall into a transcriptional continuum instead of representing distinct states.68,69,91 Relatively small transcriptional differences may, however, lead to relevant functional differences, as exemplified by the functional variations between hippocampal and cerebellar microglia.92,93
Sex differences due to sex chromosomes and/or gonadal hormones may also impact microglial states in different contexts. A growing body of evidence shows that male and female microglia differ in their transcriptomic, proteomic, and morphological profiles across brain colonization, maturation, and function in health and disease.88,94–96 Of note, the microglial sex-specific transcriptomic signatures appear to be intrinsically determined, being maintained when microglia are transplanted into the brains of mice from the other sex.96 Sexually differentiated roles of microglia could critically influence a variety of biological processes, in a time-dependent manner, and, thus, emerge as key disease modifiers across various pathological conditions with sexual dimorphism in prevalence, manifestation, and response to treatment.97 A well-characterized example for sex-specific divergence is the purinergic receptor P2X4R, identified as the male-biased microglial mediator of chronic pain.98 Sex differences in sexually dimorphic responses in physiology and pathology likely arise from a combination of Y-chromosome-specific genes, sex hormones, neuronal circuit-related factors, and epigenetic mechanisms.99
Regardless of the reduced heterogeneity in the mature adult (compared to embryonic) CNS,7,68,90 microglia do differ among CNS areas in terms of their morphology and ultrastructure, transcriptional, proteomic, epigenetic profiles, and functional specialization, suggesting that microglial states are modulated by local cues.83,100,101 However, local CNS signals are not sufficient to determine microglial identity because macrophages engrafted in the brain parenchyma can acquire a microglia-like morphology without reaching a transcriptomic signature identical to host microglia, even after prolonged CNS residence,26,102,103 supporting the idea that microglia are distinct from peripherally derived macrophages, even when they colonize a similar niche. In addition, these findings suggest that once their identity is established, microglia assume different functional states in response to local CNS signals. Therefore, both the developmental genetic programs and CNS environment (nature and nurture) collaborate to dynamically determine microglial functional states.
Microglia not only respond to local cues within the brain, but they also receive continuous inputs from the periphery, including signals from the gastrointestinal tract.104 In this context, the role of the host microbiota is gaining momentum in controlling microglial maturation and function in the CNS,88 with growing evidence that microbiota-derived short-chain fatty acids represent major mediators of the gut-brain axis.87,105 Another example of crosstalk between microglia and the periphery is the so called “sickness behavior,” as a result of the central response to peripherally released cytokines produced by peripheral immune cells and tissue-resident macrophages detecting specific pathogen-associated molecular patterns (PAMPs).106 This complex and coordinated response, in which the functional role of microglia remains poorly understood, gives rise to adaptive behavioral strategies, including lethargy. Acute systemic inflammation, nevertheless, was extensively shown to impact on microglia107,108 and induce a microglial state associated with robust IL-1β production.109
The concept of the brain as an immune-privileged organ has been challenged and definitely revisited in recent years. Indeed, peripherally produced cytokines and immune cells access the CNS and patrolthe perivascularspace in disease but alsoin health, thus playing important roles in coordinating central and peripheral immune responses.110 It was also suggested that microglia require resident CD4+ T cells in the healthy developing brain for proper maturation and complete fetal-to-adult transition.111 Microglia and T cell crosstalk was shown to help maintain homeostasis in the CNS, with dysfunctional regulation occurring in diseases, such as MS,112 ALS,113 AD,114 and encephalitis.115 It will be important to continue investigating the influence of the peripheral immune system, including B cells, natural killer cells, and other cells, on microglial states and function in both health and disease.
MICROGLIAL STATES IN THE DISEASED CNS
Microglia are keen responders and critical players in numerous neurodevelopmental, neurological, and neurodegenerative conditions, as thoroughly reviewed elsewhere. Altered microglial states have been described in the diseased human brain and across various animal models of disease pathology based on morphology and gene expression signature. In addition, these states also differ depending on the timing (i.e., disease stage), genetic background, and local environment. Context-dependent signals vary dramatically during disease progression; they range from apoptotic cells, extracellular debris, toxic proteins (i.e., amyloid, α-synuclein), and signals resulting from blood-brain barrier disruption and altered function of neurons and other glial cells. Microglia respond to these challenges by changing their molecular profile, morphology, and ultrastructure (Box 3), as well as motility and function.
The expression of core microglial markers is also altered over the course of disease, including downregulation of the homeostatic microglial signature. A prototypical example is P2RY12, one of the most widely used markers to discriminate microglia from other macrophages, with its reduced expression being one of the salient features of the microglial response to AD pathology and other disease conditions,116 as shown in several mouse models of disease (Figure 4). The apparent contradiction that core markers do not have a steady expression, as could perhaps be expected, is likely reflecting the functions those proteins have and how they change in the diseased brain. For instance, P2RY12 upregulation in epilepsy may relate to microglial sensing ATP and nucleotides released during seizures.117 This seeming paradox strengthens the fact that determining microglial expression profile is far from attributing any function to microglia, as it may only be suggestive of a potential functional identity, which, with unanimous consensus from all the authors, requires experimental validation using appropriate animal models and mutagenesis while using analyses that preserve the environmental influences shaping microglial function.
Figure 4. Microglial transcriptomic signatures.
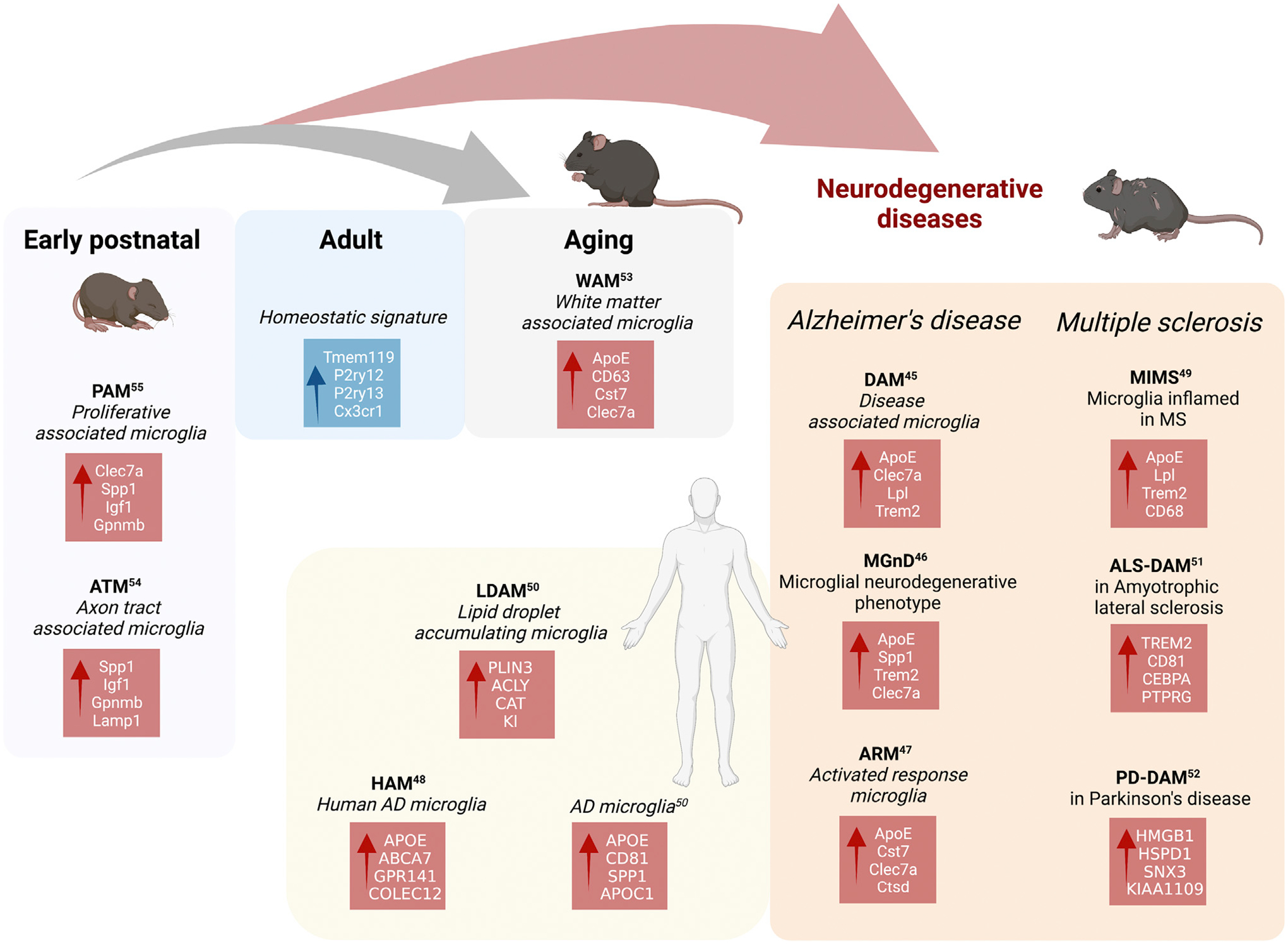
Recent scRNA-seq studies have identified many microglial transcriptional signatures including, but not limited to, PAM and ATM in development; DAM, MgnD, ARM, and MIMS in disease models of AD, MS, ALS, and PD; and WAM, LDAM, and HAM in aging, in both mice and human. The key upregulated (red) and downregulated (blue) genes in each signature are indicated. Created with BioRender.com.
A microglial state that has received particular focus is the one denoted by the DAM signature, initially identified in a mouse model with mutations within five AD genes (5XFAD)58 and later detected in other AD mouse models and samples from human AD (reviewed in Chen and Colonna116) and MS patients.62,118 Single-cell transcriptomic profiling of human microglial nuclei revealed a tau-associated microglia cluster that had not been identified in mice,119 reinforcing the idea that more human studies are needed. The shared DAM signature includes downregulation of CX3CR1 and P2RY12 and upregulation of APOE, AXL, SPP1, and TREM2,116 and it has been recently shown that it comprises two ontogenetically different cell lineages, both expressing TREM2, resident microglia and invading monocyte-derived cells (termed disease inflammatory macrophages, DIMs) that accumulate during aging.120 Many questions remain open regarding the functional significance of the DAM signature.
Are DAMs beneficial, detrimental, or both? Several studies, in both mouse and human stem-cell-differentiated microglia, demonstrated that the transition to a DAM state is dependent on TREM2.58,59,85,121 How the TREM2 receptor drives the DAM transcriptional phenotype remains unclear, although the TREM2-ApoE signaling pathway is necessary for the switch from homeostatic to MGnD.59 Further investigations are required to fully elucidate the role of TREM2. For instance, is TREM2 a key sensor for amyloid-β and other AD-related pathology, or does its loss of function cause developmental defects in microglia that render them unable to change state? Is TREM2 controlling the microglial state by regulating their energetic and anabolic metabolism?122,123
New bulk and single-cell epigenetic approaches75,124–129 will help answer these questions and ultimately may provide a means to toggle microglial states at will, enabling the field to finally understand the function of distinct microglial states and their impact in different contexts. Additionally, many genes of the DAM signature were identified across various contexts. For example, a common set of markers including (but not limited to) an upregulation of TREM2, APOE, CD11c, CLEC7A, and LPL and downregulation of TGFβ, CSF1R, P2RY12, and TMEM119 has been recently used to denote a microglial state that associates with myelinating areas in the developing brain but also with aging and several models of degenerative diseases, such as AD, ALS,130 and MS.58,67,131 These observations raise the question as to whether the DAM is a signature strictly associated with certain diseases, as the name implies, or perhaps represents a more universal core signature that appears in response to various challenges and may differ between the young/developing versus aged/diseased CNS and across distinct regions. Most likely, the same states that are beneficial in certain contexts may be detrimental in others, strictly depending on the complex interactions between microglia and their surrounding environment. One of the most relevant questions to be addressed is to which extent microglial states identified in the mouse brain are conserved and functionally relevant in the human brain.
NOMENCLATURE TROUBLES
Our current understanding of the plasticity of microglial states is at odds with the simplistic scenario established using outdated microglial nomenclature (resting versus activated and M1 versus M2; Boxes 1 and 2). Thus, a systematic, careful naming approach would greatly benefit microglial biology. As a first step to guide the field regarding the use of nomenclature, we generated a questionnaire (Data S1) and collected the responses from the co-authors.
Surprisingly, there was more consensus than disagreement that the current nomenclature has severe limitations, and a more useful conceptual framework is needed to properly understand microglial states. There is also agreement that this framework is a first important step to guide the field and should be revisited every 5 to 10 years by an international panel of experts as new discoveries are made. There is also a broad agreement that microglial responses should be framed in a multidimensional space and should not be simplified as dichotomic good versus bad (Figure 1). Another point of strong agreement: abandon M1/M2 (and similar) nomenclature once and for all and generally avoid using the vague term “neuroinflammation.” Most agree that inflammation is not always detrimental but, instead, represents an adaptive response to damage that can sometimes get out of control (Box 4). Quite importantly, a vast majority of authors support the use of “markers” (genes or proteins) to identify cell populations, but not as a readout of cell functions, which need to be addressed directly.
Box 4. Microglia and the term “neuroinflammation”.
There is a long historical literature stating that inflammation is an important part of recovery from infection, injury, and disease, and it is the lack of resolution of this inflammatory response that is problematic in the context of CNS cell “reactivity.” Therefore, when the term “neuroinflammation” is encountered in the literature, the reader must be aware that it means different things depending on the context.
While the term “neuroinflammation” is widely used in the field as a synonym of microglial “activation,”174 its definition also varies dramatically among authors, according to our survey. Below are representative definitions which are currently used by the authors:
Neuroinflammation is inflammation of neural tissue particularly mediated by glial cells.
Neuroinflammation is strictly limited to conditions in which leukocytes enter CNS, e.g., in stroke and MS.
Neuroinflammation is a mixed cellular response to brain infection or damage involving innate and adaptive responses of resident brain cells and circulating immune cells.
The term neuroinflammation is too unclear and imprecise and should be avoided.
Considering that different definitions are used across authors, our main recommendation for the field is to liberate neuroinflammation from microglia and microglia from neuroinflammation and to use both terms rigorously. The consensus among authors is 4-fold. First, protection against tissue damage and extreme departures from homeostasis as well as repair (i.e., “inflammation”) encompasses, in the CNS, a highly complex set of local responses and equally complex interactions with circulating immune cells or with immune cells residing in brain-blood and brain-cerebrospinal fluid interphases. In other words, “neuroinflammation” is not a substitute for “microglial reaction.” Second, there are numerous transcriptional states of microglia, astrocytes, and oligodendrocytes. The functional outcomes of cells undergoing these transcriptional states remain incompletely understood. Furthermore, it is uncertain which transcriptional states are transient or represent durable cell-fate choices. It is also unknown whether changes in states during diseases are “inflammatory” or dedicated to maintaining microglial homeostatic functions. Taking these considerations together, one should exercise extreme caution in simplifying these phenomena as “neuroinflammation,” as at least some of these phenomena may represent alternative homeostatic or non-inflammatory reactive states. Third, it is not appropriate to imply that neuroinflammation is invariably deleterious. Rather, it should be recognized that each inflammatory response may exert adaptive or maladaptive effects, contingent on context. To be more specific, research is necessary to explore functions and distinct actions of cytokine-enriched microglia secretomes beyond binary characterizations such as “pro-inflammatory” and “anti-inflammatory.” Fourth, with regards to nomenclature, we recommend the use of modest and precise terms to describe specific phenomena such as: microglial reaction, astrocytic reaction, molecules involved, loss of barrier function at the blood-brain barrier, etc. All in all, the main message we wish to convey is that inflammation associated with the CNS follows unique rules that need to be fully discerned experimentally and not simply extrapolated from observations in non-nervous tissue.
Nonetheless, there were a few points that are still under intense debate. The term “resting” microglia is strongly avoided by some authors, whereas others acknowledge that they still use it even with its limitations for lack of a better term. “Homeostatic” has more acceptance, although it is recognized that it is based on a very particular gene signature not shared by microglia across all physiological contexts, such as embryonic and postnatal development, and that several homeostatic states likely exist. Thus, the term “homeostatic” should always be accompanied by an accurate description of the context.
The opinion on use of the term “DAM,” on the other hand, is highly polarized. Many authors consider that a core set of transcripts in this signature is common to several pathological conditions and some physiological processes, including the development of white matter, whereas an equal number of authors state there is not enough evidence for “DAM” to be a universal signature of microglial response to damage. Finally, the extent to which microglia are unique or similar to other brain-associated or tissue macrophages is evolving with new data and profiling methods: most agree that because of their lineage, microglia are, to some extent, similar to other macrophages but have unique functions resulting from their longer residence in the CNS environment.
RECOMMENDATIONS: DO’S AND DON’TS
Based on the collective opinions from the authors, we provide a series of recommendations for researchers, reviewers, and editors. As the field has not yet reached a consensus on several nomenclature topics, including the appropriate use of descriptors for microglial states, it is premature to provide clearer recommendations. Nevertheless, we aim to raise awareness on these issues and stimulate the launch of further initiatives that will guide the field and allow to develop more specific guidelines.
Classic nomenclature
Consider microglia as highly dynamic and plastic cells that display multivariate morphological/ultrastructural, transcriptional, metabolic, and functional states in both the healthy and pathological CNS.
Describe microglia using as many layers of complexity as possible: ontogeny, morphology/ultrastructure, motility, -omics, and function, always placing them into a species and spatiotemporal context (Figure 5).
Refer to microglia in basal conditions as “homeostatic” instead of “resting” microglia, considering the limitations discussed above (i.e., that these terms refer to microglia under physiological conditions and not to the function of microglia). Use the term “surveillant/surveilling” to refer to microglia that are engaged in surveillance, but not as a synonym of microglia under normal physiological conditions.
Refer to microglia in your experimental condition as “reactive to” or “responding to” while describing the particular signals they respond to (i.e., the context) instead of using the widely used broad term “activated,” as microglia are active in both health and disease.
Disregard simplistic, dichotomic categorizations by providing the observed data and its context.
Describe profiles of cytokine expression, considering that microglial complexity cannot be reduced to oversimplified and polarized “pro-inflammatory” versus “anti-inflammatory” categories. Similarly, do not use M1 versus M2 classification.
When using the term “DAM,” do not use it as a universal term applicable to all diseases, models, or challenges. The jury is still out to test whether its full or core signature is common to all or a subset of pathologies, particularly in the human brain.
Introducing new terminology
Until a consensus is reached about true subtype(s) of microglia, with defined ontogeny, physical niches, functions, and transcriptional profiles (whether permanent or transient), use the term “state” rather than “subpopulation.”
Use combinations of gene or protein “markers” to identify putative subpopulations but be aware that their expression is plastic and may change over time and under different experimental conditions. Use fate mapping approaches with lineage tracing to track individual microglial cells and assess possible intrinsic differences as well as changes in their state over time.84,132
In scRNA-seq studies, describe the transcriptional signatures (sets or modules of expressed genes) that can be compared with other studies.116,133 To describe groups of transcriptionally similar cells in terms of signature, use the term “cluster.”
Avoid the use of acronyms wherever possible, and only use these once multiple laboratories have defined a stable state with a clearly defined functional role.
If new terminology needs to be introduced, follow FAIR principles: findable, accessible, interoperable, and reusable (https://neuronline.sfn.org/professional-development/data-sharing-principles-to-promote-open-science). An example of naming cell lines following these principles can be found here.134
Microglial markers and function
Use integrative methodological approaches that allow probing of microglia using different levels of analysis (Figure 5).
Follow updated consensus guidelines when using methodologies such as scRNA-seq,135 qRT-PCR,136 or digital PCR.137
Do not use morphology or gene/protein expression as a substitute for directly assessing cell function. Morphology and expression can be used to generate hypotheses about function that need to be specifically tested.
Grammar quandary
“Microglia” as a population is a plural noun in English but a singular noun in Latin-derived languages, which occasionally causes confusion. In English texts, microglial cells should always be referred to in the plural form unless referring to an individual cell. For example, “microglia are brain cells” but “this microglia is adjacent to a neuron”.
FUTURE QUESTIONS AND CHALLENGES
From words to action
A key challenge in the field is to match microglial morphological, ultrastructural, transcriptomic, proteomic, metabolomics, and emerging lipidomic changes with functional responses (Figure 3). In the current single-cell era, an overwhelming wealth of data has been generated, profiling the expression of millions of microglia in different organisms, at different ages, across diverse brain regions. Yet, such “omics” identities are not necessarily linked to functional states and often lack spatial resolution. Additionally, many widely used microglial markers are sensome genes, whose expression and activity at the microglial membrane may reflect functional adaptations to a changing environment and are possibly more indicative of the microglial functional state than the transcription profile.
Transcriptional analysis will benefit from ribosome profiling by RiboSeq138 and from gene-trap insertion profiling by TRAP-Seq.139 Proteomic approaches combined with in situ studies will provide better information in this respect, bridging the gap between expression and function. Further integration of complementary approaches, such as spatial transcriptomics, imaging mass cytometry, and correlative or conjugate electron microscopy in combination with other single-cell approaches, will provide a more comprehensive characterization of microglia. Ultimately, functional studies using specific pharmacological and transgenic approaches in animal models, as well as human-derived cells and organoids, are indispensable to understand the multiple roles of microglia within specific spatiotemporal contexts of health and disease.
How are microglial states coordinated?
Even as we acquire more data about microglial states, there are still key questions remaining unanswered. To which extent are microglial states plastic and reversible? What is the relationship between microglial state and cellular function? These varied single-cell characterizations ultimately need to be linked to particular functions to become relevant to development, health, and diseases. How do these states come about? How do signals from the CNS environment get integrated in microglia to produce specific states? New imaging tools and reporters that enable tracking and manipulation of specific microglial states are needed to address these questions.
How similar are peripherally derived macrophages and microglia?
A burning question that surely requires further investigation is related to the identity and function of microglia versus other brain macrophages. Although recent studies have provided evidence for an intrinsic unique core signature of microglia, their functional resemblances and differences remain undetermined. For instance, could engrafted parenchymal macrophages functionally replace the resident microglia despite having a different molecular identity, and could they serve as therapeutic vectors?
The devil is in the details
Another major caveat is that microglia are incredibly reactive cells, and evidence indicates that artifacts are often introduced during sample processing for a variety of methodologies, such as RNA profiling, immunohistochemistry, fluorescence-activated cell sorting, in vivo imaging, and so on. Hence, we may be missing or confounding important pieces of information because we unintentionally introduce changes in the parameters we are trying to measure. In addition, these artifacts are likely to generate variability across laboratories using different protocols. A future challenge is to increase reproducibility of data across laboratories by coordinating a shared database of protocols and analysis pipelines curated using STAR Methods guidelines. In addition, in the current single-cell multi-omics era, the challenges in big data analysis are exponentially growing.140 Statistical methods (including multivariate statistics)141 and artificial-intelligence-based data mining approaches (such as machine learning)142 will have to be introduced to uniformly process and integrate large datasets, as well as extract the biological relevance of the findings.
Diversity as a source of richness
Many transcriptional states have been reported during embryonic development, aging, and disease. How many different microglial states can be identified? Within the homeostatic microglia, how many states exist? How do microglia navigate among their many states? Are they related through a transcriptional continuum or perhaps as a hub-and-spoke set of states, as has been proposed for macrophages?4 How dynamic are these states? And how spatially defined are they? Future research will need to address these important questions.
Male versus female microglia
Sex differences have been reported to affect the brain colonization, maturation, structure, transcriptomic, proteomic, and functional profiles of microglia in a time-dependent manner. To what extent these differences may regulate the susceptibility to neurological diseases remains a fascinating question that urgently awaits answers. Investigating the molecular and cellular mechanisms underlying sex-mediated differences in microglial states would advance our understanding of microglial implication in diseases with clear sex-related differences in their prevalence, symptoms, and progression, as well as response to treatments.
Relevance to humans
It will be imperative to study developmental and functional differences between human and animal model microglia. To date, most of the studies on microglia were conducted in mice, and a direct comparison among brain regions is still missing. Whether microglial states identified in mice also exist in humans is still under debate. Translating and validating these findings across species is critical and will help prevent failure of clinical trials that stem from animal model limitations. In addition, most human microglial studies were performed in Caucasians, and only recently data from other groups, such as African American individuals, are becoming available.143
Toward a unified nomenclature
The conclusion of this paper is that the community has not yet reached an agreement on what defines microglial identity compared to other cell types, nor consensus on the number, dynamic nature, or definition of microglial states. The community advocates for creating harmonized, curated databases and guidelines for introducing novel terminology; following STAR methods; and sharing data as early as possible. Until such consensus is reached, the community urges all microglial studies to present data with all their layers of complexity and carefully define the context examined to offer clarity instead of confusion, thereby contributing to a more thorough understanding of the many facets of microglial biology. To establish new guidelines for microglial states and nomenclature, we call for a community-based approach, whereby the issues and progress are discussed openly in workshops and meetings, with input from diverse researchers across fields and career stages. A useful model to look after are the 10 Human Leukocyte Differentiation Antigen workshops that have taken place since 1982, in charge of renaming cluster of differentiation (CD) antigens (https://www.sinobiological.com/research/cd-antigens/hlda1). We lastly advocate for the creation of an international panel/committee of experts in charge of overseeing the guidelines and establishing a specific roadmap to write a white paper in the nearest future.
We would like to conclude with the words of Río-Hortega, who sarcastically identified the problems of microglial nomenclature already 100 years ago: “If we were fond of introducing new nomenclature to describe microglia, as many modern histologists are, who think that enriching nomenclature resolves problems, we would find for microglia names that would indicate their origin, or morphology, or function, in addition to classify all the shapes that acquire when moving and evolving—resulting in the same absurdity that occurs in some branches of Histology and, particularly, Hematology.”144
Supplementary Material
ACKNOWLEDGMENTS
The authors are deeply grateful to Richard Ransohoff, Monica Carson, and Elena Galea, who contributed the section on neuroinflammation. We would also like to express our gratitude for our lab members, who contributed with fruitful discussions. We are particularly grateful to Sol Beccari (who conceived Figure 1), Lasse Dissing-Olesen, Alec Walker, Martine Therrien, and Yvanka de Soysa. We are very thankful for the technical support of Diane Hirshon during the preparation of the manuscript. We are grateful for the help of all the student hosts who contributed to the virtual workshop held in June 2021: Ifoeluwa Awogbindin, Elisa Gonçalves de Andrade, Fernando Gonzalez Ibanez, Mohammadparsa Khakpour, Torin Halvorson, Victor Lau, Sophia Loewen, Chloe McKee, Jared VanderZwaag, and Haley Vecchiarelli (Tremblay lab); An Buckinx, Anne-Claire Compagnion, and Fanny Martineau (Paolicelli lab); Sol Beccari, Alice Louail, and Noelia Rodriguez-Iglesias (Sierra lab); and Martine Therrien, Yvanka De-Soysa, and Anna Kane (Stevens lab). Finally, we are grateful for the input we received from young trainees during the EMBO 2021 Workshop on Microglia and for our lab members who helped with the organization. We would like to thank the creativity of Sophie Robinson, who proposed the term “homeodynamic” to refer to the dynamic nature of microglia, although its similar pronunciation with the current term hemodynamic prevented us from recommending its use in this paper.
All authors in the author list are listed in alphabetical order. This work was supported by grants from the Dementia Research Switzerland – Synapsis Foundation, Swiss National Science Foundation (SNSF 310030_197940), and European Research Council (ERC StGrant REMIND 804949) to R.C.P.; the Spanish Ministry of Science and Innovation Competitiveness MCIN/AEI/10.13039/501100011033 and FEDER “A way to make Europe” (RTI2018-099267-B-I00 and RYC-2013-12817), a Tatiana Foundation award (P-048-FTPGB 2018), and a Basque Government Department of Education project (PIBA 2020_1_0030) to A.S.; Cure Alzheimer’s Fund and Alzheimer’s Association to B.S.; the Canadian Institutes of Health Research (foundation grant 341846, project grant 461831) and Natural Sciences and Engineering Research Council of Canada (discovery grant RGPIN-2014-05308) to M.E.T. M.E.T. is a Tier II Canada Research Chair in Neurobiology of Aging and Cognition. This work was also funded by DFG CRC/TRR167 “NeuroMac” to I.A., J.P., M.P., and S.J. and by DFG SFB 1052, Project 209933838 to I.B. Australian Research Council support for project DP150104472 to M.B.G. is gratefully acknowledged.
DECLARATION OF INTERESTS
B.A. is the shareholder and member of scientific advisory board of Tranquis Therapeutics. K.B. is an employee and shareholder of AbbVie. M.C. receives research support from Vigil, is a member of the scientific advisory board of Vigil, and has a patent on TREM2. S.C. is a recipient of research funding from Eli Lilly and Company. C.C. is a member of the advisory board of Exalys Therapeutics and is the recipient of a research grant from IONIS therapeutics. B.D.S. is occasionally consulting for different companies. He is founding scientist of Augustin TX and of Muna TX. He is also shareholder of Muna TX. C.H. collaborates with Denali Therapeutics. C.H. is chief advisor of ISAR Bioscience and a member of the advisory board of AviadoBio. J.K. is a scientific advisor and collaborator with PureTech. T.M. is a cofounder of REGAIN Therapeutics, owner of a provisional patent on compositions and methods for treatment and/or prophylaxis of proteinopathies, and owner of a provisional patent on preventing or reverting abnormal amyloid deposition. R.M. has scientific collaborations with Alector, Nodthera, and Alchemab and is a consultant for Sanofi. B.M. has received consultancy fees from AstraZeneca. A. Sierra is a recipient of a research grant from Hoffmann La Roche.
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental information can be found online at https://doi.org/10.1016/j.neuron.2022.10.020.
REFERENCES
- 1.Stafleu FA (1971). Linnaeus and the Linnaeans: The Spreading of their Ideas in Systematic Botany, 1735–1789 (A. Oosthoek’s Uitgeversmaat-schappij; ). [Google Scholar]
- 2.Charmaz K (2006). The power of names. J. Contemp. Ethnogr 35, 396–399. 10.1177/0891241606286983. [DOI] [Google Scholar]
- 3.Guilliams M, Ginhoux F, Jakubzick C, Naik SH, Onai N, Schraml BU, Segura E, Tussiwand R, and Yona S (2014). Dendritic cells, monocytes and macrophages: a unified nomenclature based on ontogeny. Nat. Rev. Immunol 14, 571–578. 10.1038/nri3712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Murray PJ, Allen J, Biswas S, Fisher E, Gilroy D, Goerdt S, Gordon S, Hamilton J, Ivashkiv L, Lawrence T, et al. (2014). Macrophage activation and polarization: nomenclature and experimental guidelines. Immunity 41, 14–20. 10.1016/j.immuni.2014.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yuste R, Hawrylycz M, Aalling N, Aguilar-Valles A, Arendt D, Armananzas R, Ascoli GA, Bielza C, Bokharaie V, Bergmann TB, et al. (2020). A community-based transcriptomics classification and nomenclature of neocortical cell types. Nat. Neurosci 23, 1456–1468. 10.1038/s41593-020-0685-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Escartin C, Galea E, Lakatos A, O’Callaghan JP, Petzold GC, Serrano-Pozo A, Steinhauser C, Volterra A, Carmignoto G, Agarwal A, et al. (2021). Reactive astrocyte nomenclature, definitions, and future directions. Nat. Neurosci 24, 312–325. 10.1038/s41593-020-00783-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sierra A, Paolicelli RC, and Kettenmann H (2019). Cien anos de microglia: Milestones in a century of microglial research. Trends Neurosci 42, 778–792. 10.1016/j.tins.2019.09.004. [DOI] [PubMed] [Google Scholar]
- 8.Rezaie P, and Hanisch U-K (2014). Historical context. In Microglia in Health and Disease, Tremblay ME and Sierra A, eds. (Springer; ), pp. 7–46. [Google Scholar]
- 9.Río-Hortega P (1919). El tercer elemento de los centros nerviosos. III. Naturaleza probable de la microglía. Bol. Soc. Esp. Biol 9, 108–120. [Google Scholar]
- 10.Oehmichen M (1982). Are resting and/or reactive microglia macrophages? Immunobiology 161, 246–254. 10.1016/S0171-2985(82)80080-6. [DOI] [PubMed] [Google Scholar]
- 11.Alliot F, Godin I, and Pessac B (1999). Microglia derive from progenitors, originating from the yolk sac, and which proliferate in the brain. Brain Res. Dev. Brain Res 117, 145–152. 10.1016/s0165-3806(99)00113-3. [DOI] [PubMed] [Google Scholar]
- 12.Xu J, Zhu L, He S, Wu Y, Jin W, Yu T, Qu J, and Wen Z (2015). Temporal-spatial resolution fate mapping reveals distinct origins for embryonic and adult microglia in zebrafish. Dev. Cell 34, 632–641. 10.1016/j.devcel.2015.08.018. [DOI] [PubMed] [Google Scholar]
- 13.Ferrero G, Mahony CB, Dupuis E, Yvernogeau L, Di Ruggiero E, Miserocchi M, Caron M, Robin C, Traver D, Bertrand JY, and Wittamer V (2018). Embryonic microglia derive from primitive macrophages and are replaced by cmyb-dependent definitive microglia in zebrafish. Cell Rep 24, 130–141. 10.1016/j.celrep.2018.05.066. [DOI] [PubMed] [Google Scholar]
- 14.Ginhoux F, Greter M, Leboeuf M, Nandi S, See P, Gokhan S, Mehler MF, Conway SJ, Ng LG, Stanley ER, et al. (2010). Fate mapping analysis reveals that adult microglia derive from primitive macrophages. Science 330, 841–845. 10.1126/science.1194637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schulz C, Perdiguero EG, Chorro L, Szabo-Rogers H, Cagnard N, Kierdorf K, Prinz M, Wu B, Jacobsen SEW, Pollard JW, et al. (2012). A lineage of myeloid cells independent of Myb and hematopoietic stem cells. Science 336, 86–90. 10.1126/science.1219179. [DOI] [PubMed] [Google Scholar]
- 16.Kierdorf K, Erny D, Goldmann T, Sander V, Schulz C, Perdiguero EG, Wieghofer P, Heinrich A, Riemke P, Holscher C, et al. (2013). Microglia emerge from erythromyeloid precursors via Pu.1- and Irf8-dependent pathways. Nat. Neurosci 16, 273–280. 10.1038/nn.3318. [DOI] [PubMed] [Google Scholar]
- 17.Stremmel C, Schuchert R, Wagner F, Thaler R, Weinberger T, Pick R, Mass E, Ishikawa-Ankerhold HC, Margraf A, Hutter S, et al. (2018). Yolk sac macrophage progenitors traffic to the embryo during defined stages of development. Nat. Commun 9, 75. 10.1038/s41467-017-02492-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Andjelkovic AV, Nikolic B, Pachter JS, and Zecevic N (1998). Macrophages/microglial cells in human central nervous system during development: an immunohistochemical study. Brain Res 814, 13–25. 10.1016/s0006-8993(98)00830-0. [DOI] [PubMed] [Google Scholar]
- 19.Chitu V, Gokhan S, Nandi S, Mehler MF, and Stanley ER (2016). Emerging roles for CSF-1 receptor and its ligands in the nervous system. Trends Neurosci 39, 378–393. 10.1016/j.tins.2016.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Easley-Neal C, Foreman O, Sharma N, Zarrin AA, and Weimer RM (2019). CSF1R ligands IL-34 and CSF1 are differentially required for microglia development and maintenance in white and gray matter brain regions. Front. Immunol 10, 2199. 10.3389/fimmu.2019.02199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ajami B, Bennett JL, Krieger C, Tetzlaff W, and Rossi FM (2007). Local self-renewal can sustain CNS microglia maintenance and function throughout adult life. Nat. Neurosci 10, 1538–1543. 10.1038/nn2014. [DOI] [PubMed] [Google Scholar]
- 22.Bruttger J, Karram K, Wortge S, Regen T, Marini F, Hoppmann N, Klein M, Blank T, Yona S, Wolf Y, et al. (2015). Genetic cell ablation reveals clusters of local self-renewing microglia in the mammalian central nervous system. Immunity 43, 92–106. 10.1016/j.immuni.2015.06.012. [DOI] [PubMed] [Google Scholar]
- 23.Huang Y, Xu Z, Xiong S, Qin G, Sun F, Yang J, Yuan TF, Zhao L, Wang K, Liang YX, et al. (2018). Dual extra-retinal origins of microglia in the model of retinal microglia repopulation. Cell Discov 4, 9. 10.1038/s41421-018-0011-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Huang Y, Xu Z, Xiong S, Sun F, Qin G, Hu G, Wang J, Zhao L, Liang YX, Wu T, et al. (2018). Repopulated microglia are solely derived from the proliferation of residual microglia after acute depletion. Nat. Neurosci 21, 530–540. 10.1038/s41593-018-0090-8. [DOI] [PubMed] [Google Scholar]
- 25.Zhan L, Krabbe G, Du F, Jones I, Reichert MC, Telpoukhovskaia M, Kodama L, Wang C, Cho S, Sayed F, et al. (2019). Proximal recolonization by self-renewing microglia re-establishes microglial homeostasis in the adult mouse brain. PLoS Biol 17, e3000134. 10.1371/journal.pbio.3000134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cronk JC, Filiano AJ, Louveau A, Marin I, Marsh R, Ji E, Goldman DH, Smirnov I, Geraci N, Acton S, et al. (2018). Peripherally derived macrophages can engraft the brain independent of irradiation and maintain an identity distinct from microglia. J. Exp. Med 215, 1627–1647. 10.1084/jem.20180247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Priller J, Flugel A, Wehner T, Boentert M, Haas CA, Prinz M, Fernandez-Klett F, Prass K, Bechmann I, de Boer BA, et al. (2001). Targeting gene-modified hematopoietic cells to the central nervous system: use of green fluorescent protein uncovers microglial engraftment. Nat. Med 7, 1356–1361. 10.1038/nm1201-1356. [DOI] [PubMed] [Google Scholar]
- 28.Xu Z, Rao Y, Huang Y, Zhou T, Feng R, Xiong S, Yuan TF, Qin S, Lu Y, Zhou X, et al. (2020). Efficient Strategies for Microglia Replacement in the Central Nervous System. Cell Rep 32, 108041. 10.1016/j.celrep.2020.108041. [DOI] [PubMed] [Google Scholar]
- 29.Xu Z, Zhou X, Peng B, and Rao Y (2021). Microglia replacement by bone marrow transplantation (Mr BMT) in the central nervous system of adult mice. STAR Protoc 2, 100666. 10.1016/j.xpro.2021.100666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xu Z, Rao Y, and Peng B (2021). Protocol for microglia replacement by peripheral blood (Mr PB). STAR Protoc 2, 100613. 10.1016/j.xpro.2021.100613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xu R, Li X, Boreland AJ, Posyton A, Kwan K, Hart RP, and Jiang P (2020). Human iPSC-derived mature microglia retain their identity and functionally integrate in the chimeric mouse brain. Nat. Commun 11, 1577. 10.1038/s41467-020-15411-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hasselmann J, Coburn MA, England W, Figueroa Velez DX, Kiani Shabestari S, Tu CH, McQuade A, Kolahdouzan M, Echeverria K, Claes C, et al. (2019). Development of a Chimeric Model to Study and Manipulate Human Microglia In Vivo. Neuron 103, 1016–1033.e10. 10.1016/j.neuron.2019.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mancuso R, Van Den Daele J, Fattorelli N, Wolfs L, Balusu S, Burton O, Liston A, Sierksma A, Fourne Y, Poovathingal S, et al. (2019). Stem-cell-derived human microglia transplanted in mouse brain to study human disease. Nat. Neurosci 22, 2111–2116. 10.1038/s41593-019-0525-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Grabert K, Sehgal A, Irvine KM, Wollscheid-Lengeling E, Ozdemir DD, Stables J, Luke GA, Ryan MD, Adamson A, Humphreys NE, et al. (2020). A Transgenic Line That Reports CSF1R Protein Expression Provides a Definitive Marker for the Mouse Mononuclear Phagocyte System. J. Immunol 205, 3154–3166. 10.4049/jimmunol.2000835. [DOI] [PubMed] [Google Scholar]
- 35.Kaiser T, and Feng G (2019). Tmem119-EGFP and Tmem119-CreERT2 Transgenic Mice for Labeling and Manipulating Microglia. eNeuro 6, ENEURO.0448–18.2019. 10.1523/ENEURO.0448-18.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chappell-Maor L, Kolesnikov M, Kim J, Shemer A, Haimon Z, Grozovski J, Boura-Halfon S, Masuda T, Prinz M, and Jung S (2020). Comparative analysis of CreER transgenic mice for the study of brain macrophages: A case study. Eur. J. Immunol 50, 353–362. 10.1002/eji.201948342. [DOI] [PubMed] [Google Scholar]
- 37.McKinsey GL, Lizama CO, Keown-Lang AE, Niu A, Santander N, Larpthaveesarp A, Chee E, Gonzalez FF, and Arnold TD (2020). A new genetic strategy for targeting microglia in development and disease. Elife 9, e54590. 10.7554/eLife.54590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Masuda T, Amann L, Sankowski R, Staszewski O, Lenz M, d’Errico P, Snaidero N, Costa Jordao MJ, Bottcher C, Kierdorf K, et al. (2020). Novel Hexb-based tools for studying microglia in the CNS. Nat. Immunol 21, 802–815. 10.1038/s41590-020-0707-4. [DOI] [PubMed] [Google Scholar]
- 39.Parkhurst C, Yang G, Ninan I, Savas J, Yates J, Lafaille J, Hempstead B, Littman D, and Gan WB (2013). Microglia promote learning-dependent synapse formation through brain-derived neurotrophic factor. Cell 155, 1596–1609. 10.1016/j.cell.2013.11.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yona S, Kim KW, Wolf Y, Mildner A, Varol D, Breker M, Strauss-Ayali D, Viukov S, Guilliams M, Misharin A, et al. (2013). Fate mapping reveals origins and dynamics of monocytes and tissue macrophages under homeostasis. Immunity 38, 79–91. 10.1016/j.immuni.2012.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kim JS, Kolesnikov M, Peled-Hajaj S, Scheyltjens I, Xia Y, Trzebanski S, Haimon Z, Shemer A, Lubart A, Van Hove H, et al. (2021). A Binary Cre Transgenic Approach Dissects Microglia and CNS Border-Associated Macrophages. Immunity 54, 176–190.e177. 10.1016/j.immuni.2020.11.007. [DOI] [PubMed] [Google Scholar]
- 42.Goldmann T, Wieghofer P, Jordao MJC, Prutek F, Hagemeyer N, Frenzel K, Amann L, Staszewski O, Kierdorf K, Krueger M, et al. (2016). Origin, fate and dynamics of macrophages at central nervous system interfaces. Nat. Immunol 17, 797–805. 10.1038/ni.3423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Van Hove H, Martens L, Scheyltjens I, De Vlaminck K, Pombo Antunes AR, De Prijck S, Vandamme N, De Schepper S, Van Isterdael G, Scott CL, et al. (2019). A single-cell atlas of mouse brain macrophages reveals unique transcriptional identities shaped by ontogeny and tissue environment. Nat. Neurosci 22, 1021–1035. 10.1038/s41593-019-0393-4. [DOI] [PubMed] [Google Scholar]
- 44.Masuda T, Amann L, Monaco G, Sankowski R, Staszewski O, Krueger M, Del Gaudio F, He L, Paterson N, Nent E, et al. (2022). Specification of CNS macrophage subsets occurs postnatally in defined niches. Nature 604, 740–748. 10.1038/s41586-022-04596-2. [DOI] [PubMed] [Google Scholar]
- 45.Paolicelli RC, and Ferretti MT (2017). Function and Dysfunction of Microglia during Brain Development: Consequences for Synapses and Neural Circuits. Front. Synaptic Neurosci 9, 9. 10.3389/fnsyn.2017.00009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Green KN, Crapser JD, and Hohsfield LA (2020). To Kill a Microglia: A Case for CSF1R Inhibitors. Trends Immunol 41, 771–784. 10.1016/j.it.2020.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chitu V, Gokhan S, and Stanley ER (2021). Modeling CSF-1 receptor deficiency diseases - how close are we? FEBS J 289, 5049–5073. 10.1111/febs.16085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Oosterhof N, Chang IJ, Karimiani EG, Kuil LE, Jensen DM, Daza R, Young E, Astle L, van der Linde HC, Shivaram GM, et al. (2019). Homozygous Mutations in CSF1R Cause a Pediatric-Onset Leukoence-phalopathy and Can Result in Congenital Absence of Microglia. Am. J. Hum. Genet 104, 936–947. 10.1016/j.ajhg.2019.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Guo L, Bertola DR, Takanohashi A, Saito A, Segawa Y, Yokota T, Ishibashi S, Nishida Y, Yamamoto GL, Franco JFS, et al. (2019). Bi-allelic CSF1R Mutations Cause Skeletal Dysplasia of Dysosteosclerosis-Pyle Disease Spectrum and Degenerative Encepha-lopathy with Brain Malformation. Am. J. Hum. Genet 104, 925–935. 10.1016/j.ajhg.2019.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rojo R, Raper A, Ozdemir DD, Lefevre L, Grabert K, Wollscheid-Lengeling E, Bradford B, Caruso M, Gazova I, Sanchez A, et al. (2019). Deletion of a Csf1r enhancer selectively impacts CSF1R expression and development of tissue macrophage populations. Nat. Commun 10, 3215. 10.1038/s41467-019-11053-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kiani Shabestari S, Morabito S, Danhash EP, McQuade A, Sanchez JR, Miyoshi E, Chadarevian JP, Claes C, Coburn MA, Hasselmann J, et al. (2022). Absence of microglia promotes diverse pathologies and early lethality in Alzheimer’s disease mice. Cell Rep 39, 110961. 10.1016/j.celrep.2022.110961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Konishi H, Okamoto T, Hara Y, Komine O, Tamada H, Maeda M, Osako F, Kobayashi M, Nishiyama A, Kataoka Y, et al. (2020). Astrocytic phagocytosis is a compensatory mechanism for microglial dysfunction. EMBO J 39, e104464. 10.15252/embj.2020104464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hickman SE, Kingery ND, Ohsumi TK, Borowsky ML, Wang L, Means TK, and El Khoury J (2013). The microglial sensome revealed by direct RNA sequencing. Nat. Neurosci 16, 1896–1905. 10.1038/nn.3554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hume DA, Perry VH, and Gordon S (1983). Immunohistochemical localization of a macrophage-specific antigen in developing mouse retina: phagocytosis of dying neurons and differentiation of microglial cells to form a regular array in the plexiform layers. J. Cell. Biol 97, 253–257. 10.1083/jcb.97.1.253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Davalos D, Grutzendler J, Yang G, Kim JV, Zuo Y, Jung S, Littman DR, Dustin ML, and Gan WB (2005). ATP mediates rapid microglial response to local brain injury in vivo. Nat. Neurosci 8, 752–758. 10.1038/nn1472. [DOI] [PubMed] [Google Scholar]
- 56.Nimmerjahn A, Kirchhoff F, and Helmchen F (2005). Resting microglial cells are highly dynamic surveillants of brain parenchyma in vivo. Science 308, 1314–1318. 10.1126/science.1110647. [DOI] [PubMed] [Google Scholar]
- 57.Wang J, Zhang K, Xu L, and Wang E (2011). Quantifying the Waddington landscape and biological paths for development and differentiation. Proc. Natl. Acad. Sci. USA 108, 8257–8262. 10.1073/pnas.1017017108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Keren-Shaul H, Spinrad A, Weiner A, Matcovitch-Natan O, Dvir-Szternfeld R, Ulland TK, David E, Baruch K, Lara-Astaiso D, Toth B, et al. (2017). A Unique Microglia Type Associated with Restricting Development of Alzheimer’s Disease. Cell 169. 1276–1290 e1217. 10.1016/j.cell.2017.05.018. [DOI] [PubMed] [Google Scholar]
- 59.Krasemann S, Madore C, Cialic R, Baufeld C, Calcagno N, El Fatimy R, Beckers L, O’Loughlin E, Xu Y, Fanek Z, et al. (2017). The TREM2-APOE Pathway Drives the Transcriptional Phenotype of Dysfunctional Microglia in Neurodegenerative Diseases. Immunity 47, 566–581.e9. 10.1016/j.immuni.2017.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sala Frigerio C, Wolfs L, Fattorelli N, Thrupp N, Voytyuk I, Schmidt I, Mancuso R, Chen WT, Woodbury ME, Srivastava G, et al. (2019). The Major Risk Factors for Alzheimer’s Disease: Age, Sex, and Genes Modulate the Microglia Response to Aβ Plaques. Cell Rep 27, 1293–1306.e6. 10.1016/j.celrep.2019.03.099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Srinivasan K, Friedman BA, Etxeberria A, Huntley MA, van der Brug MP, Foreman O, Paw JS, Modrusan Z, Beach TG, Serrano GE, and Hansen DV (2020). Alzheimer’s Patient Microglia Exhibit Enhanced Aging and Unique Transcriptional Activation. Cell Rep 31, 107843. 10.1016/j.celrep.2020.107843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Absinta M, Maric D, Gharagozloo M, Garton T, Smith MD, Jin J, Fitzgerald KC, Song A, Liu P, Lin JP, et al. (2021). A lymphocyte-microglia-astrocyte axis in chronic active multiple sclerosis. Nature 597, 709–714. 10.1038/s41586-021-03892-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Marschallinger J, Iram T, Zardeneta M, Lee SE, Lehallier B, Haney MS, Pluvinage JV, Mathur V, Hahn O, Morgens DW, et al. (2020). Lipid-droplet-accumulating microglia represent a dysfunctional and proinflammatory state in the aging brain. Nat. Neurosci 23, 194–208. 10.1038/s41593-019-0566-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.De Andrade Costa A, Chatterjee J, Cobb O, Sanapala S, Scheaffer S, Guo X, Dahiya S, and Gutmann DH (2022). RNA sequence analysis reveals ITGAL/CD11A as a stromal regulator of murine low-grade glioma growth. Neuro Oncol 24, 14–26. 10.1093/neuonc/noab130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Limone F, Mordes D, Couto A, Pietiläinen O, Joseph BJ, Burberry A, Dia Ghosh S, Meyer D, Goldman M, Bortolin L, et al. (2021). Single-nucleus sequencing reveals enriched expression of genetic risk factors sensitises Motor Neurons to degeneration in ALS. Preprint at bioRxiv. 10.1101/2021.07.12.452054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Smajic S, Prada-Medina CA, Landoulsi Z, Ghelfi J, Delcambre S, Dietrich C, Jarazo J, Henck J, Balachandran S, Pachchek S, et al. (2022). Single-cell sequencing of human midbrain reveals glial activation and a Parkinson-specific neuronal state. Brain 145, 964–978. 10.1093/brain/awab446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Safaiyan S, Besson-Girard S, Kaya T, Cantuti-Castelvetri L, Liu L, Ji H, Schifferer M, Gouna G, Usifo F, Kannaiyan N, et al. (2021). White matter aging drives microglial diversity. Neuron 109, 1100–1117.e10. 10.1016/j.neuron.2021.01.027. [DOI] [PubMed] [Google Scholar]
- 68.Hammond TR, Dufort C, Dissing-Olesen L, Giera S, Young A, Wysoker A, Walker AJ, Gergits F, Segel M, Nemesh J, et al. (2019). Single-Cell RNA Sequencing of Microglia throughout the Mouse Lifespan and in the Injured Brain Reveals Complex Cell-State Changes. Immunity 50, 253–271.e6. 10.1016/j.immuni.2018.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Li Q, Cheng Z, Zhou L, Darmanis S, Neff NF, Okamoto J, Gulati G, Bennett ML, Sun LO, Clarke LE, et al. (2019). Developmental Heterogeneity of Microglia and Brain Myeloid Cells Revealed by Deep Single-Cell RNA Sequencing. Neuron 101, 207–223.e10. 10.1016/j.neuron.2018.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kracht L, Borggrewe M, Eskandar S, Brouwer N, Chuva de Sousa Lopes SM, Laman JD, Scherjon SA, Prins JR, Kooistra SM, and Eggen BJL (2020). Human fetal microglia acquire homeostatic immune-sensing properties early in development. Science 369, 530–537. 10.1126/science.aba5906. [DOI] [PubMed] [Google Scholar]
- 71.Wu YE, Pan L, Zuo Y, Li X, and Hong W (2017). Detecting Activated Cell Populations Using Single-Cell RNA-Seq. Neuron 96, 313–329.e6. 10.1016/j.neuron.2017.09.026. [DOI] [PubMed] [Google Scholar]
- 72.Marsh SE, Walker AJ, Kamath T, Dissing-Olesen L, Hammond TR, de Soysa TY, Young AMH, Murphy S, Abdulraouf A, Nadaf N, et al. (2022). Dissection of artifactual and confounding glial signatures by single-cell sequencing of mouse and human brain. Nat. Neurosci 25, 306–316. 10.1038/s41593-022-01022-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Mattei D, Ivanov A, van Oostrum M, Pantelyushin S, Richetto J, Mueller F, Beffinger M, Schellhammer L, vom Berg J, Wollscheid B, et al. (2020). Enzymatic Dissociation Induces Transcriptional and Proteotype Bias in Brain Cell Populations. Int. J. Mol. Sci 21, 7944. 10.3390/ijms21217944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Summers KM, Bush SJ, and Hume DA (2020). Network analysis of transcriptomic diversity amongst resident tissue macrophages and dendritic cells in the mouse mononuclear phagocyte system. PLoS Biol 18, e3000859. 10.1371/journal.pbio.3000859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Gosselin D, Skola D, Coufal NG, Holtman IR, Schlachetzki JCM, Sajti E, Jaeger BN, O’Connor C, Fitzpatrick C, Pasillas MP, et al. (2017). An environment-dependent transcriptional network specifies human microglia identity. Science 356, eaal3222. 10.1126/science.aal3222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Geirsdottir L, David E, Keren-Shaul H, Weiner A, Bohlen SC, Neuber J, Balic A, Giladi A, Sheban F, Dutertre CA, et al. (2019). Cross-Species Single-Cell Analysis Reveals Divergence of the Primate Microglia Program. Cell 179, 1609–1622.e16. 10.1016/j.cell.2019.11.010. [DOI] [PubMed] [Google Scholar]
- 77.Kolodziejczyk AA, Kim JK, Svensson V, Marioni JC, and Teichmann SA (2015). The technology and biology of single-cell RNA sequencing. Mol. Cell 58, 610–620. 10.1016/j.molcel.2015.04.005. [DOI] [PubMed] [Google Scholar]
- 78.Welch JD, Kozareva V, Ferreira A, Vanderburg C, Martin C, and Macosko EZ (2019). Single-Cell Multi-omic Integration Compares and Contrasts Features of Brain Cell Identity. Cell 177, 1873–1887.e17. 10.1016/j.cell.2019.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Stuart T, Butler A, Hoffman P, Hafemeister C, Papalexi E, Mauck WM, Hao Y, Stoeckius M, Smibert P, and Satija R (2019). Comprehensive Integration of Single-Cell Data. Cell 177, 1888–1902.e21. 10.1016/j.cell.2019.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Koussounadis A, Langdon SP, Um IH, Harrison DJ, and Smith VA (2015). Relationship between differentially expressed mRNA and mRNA-protein correlations in a xenograft model system. Sci. Rep 5, 10775. 10.1038/srep10775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Fernandez-Zapata C, Leman JKH, Priller J, and Bottcher C (2020). The use and limitations of single-cell mass cytometry for studying human microglia function. Brain Pathol 30, 1178–1191. 10.1111/bpa.12909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Ajami B, Samusik N, Wieghofer P, Ho PP, Crotti A, Bjornson Z, Prinz M, Fantl WJ, Nolan GP, and Steinman L (2018). Single-cell mass cytometry reveals distinct populations of brain myeloid cells in mouse neuroinflammation and neurodegeneration models. Nat. Neurosci 21, 541–551. 10.1038/s41593-018-0100-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Bottcher C, Schlickeiser S, Sneeboer MAM, Kunkel D, Knop A, Paza E, Fidzinski P, Kraus L, Snijders GJL, Kahn RS, et al. (2019). Human microglia regional heterogeneity and phenotypes determined by multiplexed single-cell mass cytometry. Nat. Neurosci 22, 78–90. 10.1038/s41593-018-0290-2. [DOI] [PubMed] [Google Scholar]
- 84.Tay TL, Mai D, Dautzenberg J, Fernandez-Klett F, Lin G, Sagar, Datta M, Drougard A, Stempfl T, Ardura-Fabregat A, et al. (2017). A new fate mapping system reveals context-dependent random or clonal expansion of microglia. Nat. Neurosci 20, 793–803. 10.1038/nn.4547. [DOI] [PubMed] [Google Scholar]
- 85.McQuade A, Kang YJ, Hasselmann J, Jairaman A, Sotelo A, Coburn M, Shabestari SK, Chadarevian JP, Fote G, Tu CH, et al. (2020). Gene expression and functional deficits underlie TREM2-knockout microglia responses in human models of Alzheimer’s disease. Nat. Commun 11, 5370. 10.1038/s41467-020-19227-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Mazaheri F, Snaidero N, Kleinberger G, Madore C, Daria A, Werner G, Krasemann S, Capell A, Trumbach D, Wurst W, et al. (2017). TREM2 deficiency impairs chemotaxis and microglial responses to neuronal injury. EMBO Rep 18, 1186–1198. 10.15252/embr.201743922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Erny D, Hrabe de Angelis AL, Jaitin D, Wieghofer P, Staszewski O, David E, Keren-Shaul H, Mahlakoiv T, Jakobshagen K, Buch T, et al. (2015). Host microbiota constantly control maturation and function of microglia in the CNS. Nat. Neurosci 18, 965–977. 10.1038/nn.4030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Thion MS, Low D, Silvin A, Chen J, Grisel P, Schulte-Schrepping J, Blecher R, Ulas T, Squarzoni P, Hoeffel G, et al. (2018). Microbiome Influences Prenatal and Adult Microglia in a Sex-Specific Manner. Cell 172, 500–516.e16. 10.1016/j.cell.2017.11.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Chatterjee J, Sanapala S, Cobb O, Bewley A, Goldstein AK, Cordell E, Ge X, Garbow JR, Holtzman MJ, and Gutmann DH (2021). Asthma reduces glioma formation by T cell decorin-mediated inhibition of microglia. Nat. Commun 12, 7122. 10.1038/s41467-021-27455-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Matcovitch-Natan O, Winter DR, Giladi A, Vargas Aguilar S, Spinrad A, Sarrazin S, Ben-Yehuda H, David E, Zelada Gonzalez F, Perrin P, et al. (2016). Microglia development follows a stepwise program to regulate brain homeostasis. Science 353, aad8670. 10.1126/science.aad8670. [DOI] [PubMed] [Google Scholar]
- 91.Masuda T, Sankowski R, Staszewski O, Bottcher C, Amann L, Sagar, Scheiwe C, Nessler S, Kunz P, van Loo G, et al. (2019). Spatial and temporal heterogeneity of mouse and human microglia at single-cell resolution. Nature 566, 388–392. 10.1038/s41586-019-0924-x. [DOI] [PubMed] [Google Scholar]
- 92.Grabert K, Michoel T, Karavolos MH, Clohisey S, Baillie JK, Stevens MP, Freeman TC, Summers KM, and McColl BW (2016). Microglial brain region-dependent diversity and selective regional sensitivities to aging. Nat. Neurosci 19, 504–516. 10.1038/nn.4222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Kana V, Desland FA, Casanova-Acebes M, Ayata P, Badimon A, Nabel E, Yamamuro K, Sneeboer M, Tan IL, Flanigan ME, et al. (2019). CSF-1 controls cerebellar microglia and is required for motor function and social interaction. J. Exp. Med 216, 2265–2281. 10.1084/jem.20182037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Hanamsagar R, Alter MD, Block CS, Sullivan H, Bolton JL, and Bilbo SD (2017). Generation of a microglial developmental index in mice and in humans reveals a sex difference in maturation and immune reactivity. Glia 65, 1504–1520. 10.1002/glia.23176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Guneykaya D, Ivanov A, Hernandez DP, Haage V, Wojtas B, Meyer N, Maricos M, Jordan P, Buonfiglioli A, Gielniewski B, et al. (2018). Transcriptional and Translational Differences of Microglia from Male and Female Brains. Cell Rep 24, 2773–2783.e6. 10.1016/j.celrep.2018.08.001. [DOI] [PubMed] [Google Scholar]
- 96.Villa A, Gelosa P, Castiglioni L, Cimino M, Rizzi N, Pepe G, Lolli F, Marcello E, Sironi L, Vegeto E, and Maggi A (2018). Sex-Specific Features of Microglia from Adult Mice. Cell Rep 23, 3501–3511. 10.1016/j.celrep.2018.05.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Lynch MA (2022). Exploring Sex-Related Differences in Microglia May Be a Game-Changer in Precision Medicine. Front. Aging Neurosci 14, 868448. 10.3389/fnagi.2022.868448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Halievski K, Ghazisaeidi S, and Salter MW (2020). Sex-Dependent Mechanisms of Chronic Pain: A Focus on Microglia and P2X4R. J. Pharmacol. Exp. Ther 375, 202–209. 10.1124/jpet.120.265017. [DOI] [PubMed] [Google Scholar]
- 99.Han J, Fan Y, Zhou K, Blomgren K, and Harris RA (2021). Uncovering sex differences of rodent microglia. J. Neuroinflammation 18, 74. 10.1186/s12974-021-02124-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.De Biase LM, Schuebel KE, Fusfeld ZH, Jair K, Hawes IA, Cimbro R, Zhang HY, Liu QR, Shen H, Xi ZX, et al. (2017). Local Cues Establish and Maintain Region-Specific Phenotypes of Basal Ganglia Microglia. Neuron 95, 341–356.e6. 10.1016/j.neuron.2017.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Ayata P, Badimon A, Strasburger HJ, Duff MK, Montgomery SE, Loh YHE, Ebert A, Pimenova AA, Ramirez BR, Chan AT, et al. (2018). Epigenetic regulation of brain region-specific microglia clearance activity. Nat. Neurosci 21, 1049–1060. 10.1038/s41593-018-0192-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Bennett FC, Bennett ML, Yaqoob F, Mulinyawe SB, Grant GA, Hayden Gephart M, Plowey ED, and Barres BA (2018). A Combination of Ontogeny and CNS Environment Establishes Microglial Identity. Neuron 98, 1170–1183.e8. 10.1016/j.neuron.2018.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Shemer A, Grozovski J, Tay TL, Tao J, Volaski A, SuB P, Ardura-Fabregat A, Gross-Vered M, Kim JS, David E, et al. (2018). Engrafted parenchymal brain macrophages differ from microglia in transcriptome, chromatin landscape and response to challenge. Nat. Commun 9, 5206. 10.1038/s41467-018-07548-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Abdel-Haq R, Schlachetzki JCM, Glass CK, and Mazmanian SK (2019). Microbiome-microglia connections via the gut-brain axis. J. Exp. Med 216, 41–59. 10.1084/jem.20180794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Erny D, Dokalis N, Mezo C, Castoldi A, Mossad O, Staszewski O, Frosch M, Villa M, Fuchs V, Mayer A, et al. (2021). Microbiota-derived acetate enables the metabolic fitness of the brain innate immune system during health and disease. Cell Metab 33, 2260–2276.e7. 10.1016/j.cmet.2021.10.010. [DOI] [PubMed] [Google Scholar]
- 106.Dantzer R (2009). Cytokine, sickness behavior, and depression. Immunol. Allergy Clin. North Am 29, 247–264. 10.1016/j.iac.2009.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Shemer A, Scheyltjens I, Frumer GR, Kim JS, Grozovski J, Ayanaw S, Dassa B, Van Hove H, Chappell-Maor L, Boura-Halfon S, et al. (2020). Interleukin-10 Prevents Pathological Microglia Hyperactivation following Peripheral Endotoxin Challenge. Immunity 53, 1033–1049.e7. 10.1016/j.immuni.2020.09.018. [DOI] [PubMed] [Google Scholar]
- 108.Sousa C, Golebiewska A, Poovathingal SK, Kaoma T, Pires-Afonso Y, Martina S, Coowar D, Azuaje F, Skupin A, Balling R, et al. (2018). Single-cell transcriptomics reveals distinct inflammation-induced microglia signatures. EMBO Rep 19, e46171. 10.15252/embr.201846171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Cunningham C, Wilcockson DC, Campion S, Lunnon K, and Perry VH (2005). Central and systemic endotoxin challenges exacerbate the local inflammatory response and increase neuronal death during chronic neurodegeneration. J. Neurosci 25, 9275–9284. 10.1523/JNEUROSCI.2614-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Louveau A, Harris TH, and Kipnis J (2015). Revisiting the Mechanisms of CNS Immune Privilege. Trends Immunol 36, 569–577. 10.1016/j.it.2015.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Pasciuto E, Burton OT, Roca CP, Lagou V, Rajan WD, Theys T, Mancuso R, Tito RY, Kouser L, Callaerts-Vegh Z, et al. (2020). Microglia Require CD4 T Cells to Complete the Fetal-to-Adult Transition. Cell 182, 625–640.e24. 10.1016/j.cell.2020.06.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Dong Y, and Yong VW (2019). When encephalitogenic T cells collaborate with microglia in multiple sclerosis. Nat. Rev. Neurol 15, 704–717. 10.1038/s41582-019-0253-6. [DOI] [PubMed] [Google Scholar]
- 113.Beers DR, Henkel JS, Zhao W, Wang J, and Appel SH (2008). CD4+ T cells support glial neuroprotection, slow disease progression, and modify glial morphology in an animal model of inherited ALS. Proc. Natl. Acad. Sci. USA 105, 15558–15563. 10.1073/pnas.0807419105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Mittal K, Eremenko E, Berner O, Elyahu Y, Strominger I, Apelblat D, Nemirovsky A, Spiegel I, and Monsonego A (2019). CD4 T Cells Induce A Subset of MHCII-Expressing Microglia that Attenuates Alzheimer Pathology. iScience 16, 298–311. 10.1016/j.isci.2019.05.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Di Liberto G, Pantelyushin S, Kreutzfeldt M, Page N, Musardo S, Coras R, Steinbach K, Vincenti I, Klimek B, Lingner T, et al. (2018). Neurons under T Cell Attack Coordinate Phagocyte-Mediated Synaptic Stripping. Cell 175, 458–471.e19. 10.1016/j.cell.2018.07.049. [DOI] [PubMed] [Google Scholar]
- 116.Chen Y, and Colonna M (2021). Microglia in Alzheimer’s disease at single-cell level. Are there common patterns in humans and mice? J. Exp. Med 218, e20202717. 10.1084/jem.20202717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Avignone E, Ulmann L, Levavasseur F, Rassendren F, and Audinat E (2008). Status epilepticus induces a particular microglial activation state characterized by enhanced purinergic signaling. J. Neurosci 28, 9133–9144. 10.1523/JNEUROSCI.1820-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Zrzavy T, Hametner S, Wimmer I, Butovsky O, Weiner HL, and Lassmann H (2017). Loss of ‘homeostatic’ microglia and patterns of their activation in active multiple sclerosis. Brain 140, 1900–1913. 10.1093/brain/awx113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Gerrits E, Brouwer N, Kooistra SM, Woodbury ME, Vermeiren Y, Lambourne M, Mulder J, Kummer M, Moller T, Biber K, et al. (2021). Distinct amyloid-beta and tau-associated microglia profiles in Alzheimer’s disease. Acta Neuropathol 141, 681–696. 10.1007/s00401-021-02263-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Silvin A, Uderhardt S, Piot C, Da Mesquita S, Yang K, Geirsdottir L, Mulder K, Eyal D, Liu Z, Bridlance C, et al. (2022). Dual ontogeny of disease-associated microglia and disease inflammatory macrophages in aging and neurodegeneration. Immunity 55, 1448–1465.e6. 10.1016/j.immuni.2022.07.004. [DOI] [PubMed] [Google Scholar]
- 121.Zhou Y, Song WM, Andhey PS, Swain A, Levy T, Miller KR, Poliani PL, Cominelli M, Grover S, Gilfillan S, et al. (2020). Human and mouse single-nucleus transcriptomics reveal TREM2-dependent and TREM2-independent cellular responses in Alzheimer’s disease. Nat. Med 26, 131–142. 10.1038/s41591-019-0695-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Ulland TK, Song WM, Huang SCC, Ulrich JD, Sergushichev A, Beatty WL, Loboda AA, Zhou Y, Cairns NJ, Kambal A, et al. (2017). TREM2 Maintains Microglial Metabolic Fitness in Alzheimer’s Disease. Cell 170, 649–663.e13. 10.1016/j.cell.2017.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Xiang X, Wind K, Wiedemann T, Blume T, Shi Y, Briel N, Beyer L, Biechele G, Eckenweber F, Zatcepin A, et al. (2021). Microglial activation states drive glucose uptake and FDG-PET alterations in neurodegenerative diseases. Sci. Transl. Med 13, eabe5640. 10.1126/scitranslmed.abe5640. [DOI] [PubMed] [Google Scholar]
- 124.Ma S, Zhang B, LaFave LM, Earl AS, Chiang Z, Hu Y, Ding J, Brack A, Kartha VK, Tay T, et al. (2020). Chromatin Potential Identified by Shared Single-Cell Profiling of RNA and Chromatin. Cell 183, 1103–1116.e20. 10.1016/j.cell.2020.09.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Buenrostro JD, Wu B, Chang HY, and Greenleaf WJ (2015). ATAC-seq: A Method for Assaying Chromatin Accessibility Genome-Wide. Curr. Protoc. Mol. Biol 109, 21.29.1–21.29.9. 10.1002/0471142727.mb2129s109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.van Galen P, Viny AD, Ram O, Ryan RJ, Cotton MJ, Donohue L, Sievers C, Drier Y, Liau BB, Gillespie SM, et al. (2016). A Multi-plexed System for Quantitative Comparisons of Chromatin Landscapes. Mol. Cell 61, 170–180. 10.1016/j.molcel.2015.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Bartosovic M, Kabbe M, and Castelo-Branco G (2021). Single-cell CUT&Tag profiles histone modifications and transcription factors in complex tissues. Nat. Biotechnol 39, 825–835. 10.1038/s41587-021-00869-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Schaafsma W, Zhang X, van Zomeren K, Jacobs S, Georgieva P, Wolf S, Kettenmann H, Janova H, Saiepour N, Hanisch UK, et al. (2015). Long-lasting pro-inflammatory suppression of microglia by LPS-preconditioning is mediated by RelB-dependent epigenetic silencing. Brain Behav. Immun 48, 205–221. 10.1016/j.bbi.2015.03.013. [DOI] [PubMed] [Google Scholar]
- 129.Wendeln AC, Degenhardt K, Kaurani L, Gertig M, Ulas T, Jain G, Wagner J, Hasler LM, Wild K, Skodras A, et al. (2018). Innate immune memory in the brain shapes neurological disease hallmarks. Nature 556, 332–338. 10.1038/s41586-018-0023-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Chiu I, Morimoto E, Goodarzi H, Liao J, O’Keeffe S, Phatnani H, Muratet M, Carroll M, Levy S, Tavazoie S, et al. (2013). A neurode-generation-specific gene-expression signature of acutely isolated microglia from an amyotrophic lateral sclerosis mouse model. Cell Rep 4, 385–401. 10.1016/j.celrep.2013.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Sobue A, Komine O, Hara Y, Endo F, Mizoguchi H, Watanabe S, Murayama S, Saito T, Saido TC, Sahara N, et al. (2021). Microglial gene signature reveals loss of homeostatic microglia associated with neurodegeneration of Alzheimer’s disease. Acta Neuropathol. Commun 9, 1. 10.1186/s40478-020-01099-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Jordao MJC, Sankowski R, Brendecke SM, Sagar, Locatelli G, Tai YH, Tay TL, Schramm E, Armbruster S, Hagemeyer N, et al. (2019). Single-cell profiling identifies myeloid cell subsets with distinct fates during neuroinflammation. Science 363, eaat7554. 10.1126/science.aat7554. [DOI] [PubMed] [Google Scholar]
- 133.Olah M, Menon V, Habib N, Taga MF, Ma Y, Yung CJ, Cimpean M, Khairallah A, Coronas-Samano G, Sankowski R, et al. (2020). Single cell RNA sequencing of human microglia uncovers a subset associated with Alzheimer’s disease. Nat. Commun 11, 6129. 10.1038/s41467-020-19737-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Kurtz A, Seltmann S, Bairoch A, Bittner MS, Bruce K, Capes-Davis A, Clarke L, Crook JM, Daheron L, Dewender J, et al. (2018). A Standard Nomenclature for Referencing and Authentication of Pluripotent Stem Cells. Stem Cell Rep 10, 1–6. 10.1016/j.stemcr.2017.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Luecken MD, and Theis FJ (2019). Current best practices in single-cell RNA-seq analysis: a tutorial. Mol. Syst. Biol 15, e8746. 10.15252/msb.20188746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Bustin SA, Benes V, Garson JA, Hellemans J, Huggett J, Kubista M, Mueller R, Nolan T, Pfaffl MW, Shipley GL, et al. (2009). The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin. Chem 55, 611–622. 10.1373/clinchem.2008.112797. [DOI] [PubMed] [Google Scholar]
- 137.dMIQE Group, and Huggett JF (2020). The Digital MIQE Guidelines Update: Minimum Information for Publication of Quantitative Digital PCR Experiments for 2020. Clin. Chem 66, 1012–1029. 10.1093/clinchem/hvaa125. [DOI] [PubMed] [Google Scholar]
- 138.Ingolia NT, Brar GA, Rouskin S, McGeachy AM, and Weissman JS (2012). The ribosome profiling strategy for monitoring translation in vivo by deep sequencing of ribosome-protected mRNA fragments. Nat. Protoc 7, 1534–1550. 10.1038/nprot.2012.086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Mayor-Ruiz C, Dominguez O, and Fernandez-Capetillo O (2017). Trap(Seq): An RNA Sequencing-Based Pipeline for the Identification of Gene-Trap Insertions in Mammalian Cells. J. Mol. Biol 429, 2780–2789. 10.1016/j.jmb.2017.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Rautenstrauch P, Vlot AHC, Saran S, and Ohler U (2022). Intricacies of single-cell multi-omics data integration. Trends Genet 38, 128–139. 10.1016/j.tig.2021.08.012. [DOI] [PubMed] [Google Scholar]
- 141.Paczkowska M, Barenboim J, Sintupisut N, Fox NS, Zhu H, Abd-Rabbo D, Mee MW, Boutros PC, and Reimand J (2020). Integrative pathway enrichment analysis of multivariate omics data. Nat. Commun 11, 735. 10.1038/s41467-019-13983-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Reel PS, Reel S, Pearson E, Trucco E, and Jefferson E (2021). Using machine learning approaches for multi-omics data analysis: A review. Biotechnol. Adv 49, 107739. 10.1016/j.biotechadv.2021.107739. [DOI] [PubMed] [Google Scholar]
- 143.Kunkle BW, Schmidt M, Klein HU, Naj AC, Hamilton-Nelson KL, Larson EB, Evans DA, De Jager PL, Crane PK, Buxbaum JD, et al. (2021). Novel Alzheimer Disease Risk Loci and Pathways in African American Individuals Using the African Genome Resources Panel: A Meta-analysis. JAMA Neurol 78, 102–113. 10.1001/jamaneurol.2020.3536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Río-Hortega P.d.R. (1921). Histogenesis and normal evolution: exodus and regional distribution of microglia. Mem. R. Soc. Esp. Hist. Nat 11, 213–268. [Google Scholar]
- 145.Sierra A, de Castro F, del Rio-Hortega J, Rafael Iglesias-Rozas J, Garrosa M, and Kettenmann H (2016). The “Big-Bang” for modern glial biology: Translation and comments on Pio del Rio-Hortega 1919 series of papers on microglia. Glia 64, 1801–1840. 10.1002/glia.23046. [DOI] [PubMed] [Google Scholar]
- 146.Streit WJ, Graeber MB, and Kreutzberg GW (1988). Functional plasticity of microglia: a review. Glia 1, 301–307. 10.1002/glia.440010502. [DOI] [PubMed] [Google Scholar]
- 147.Acarin L, Vela JM, Gonzalez B, and Castellano B (1994). Demonstration of poly-N-acetyl lactosamine residues in ameboid and ramified microglial cells in rat brain by tomato lectin binding. J. Histochem. Cytochem 42, 1033–1041. 10.1177/42.8.8027523. [DOI] [PubMed] [Google Scholar]
- 148.Castellano B, Gonzalez B, Jensen MB, Pedersen EB, Finsen BR, and Zimmer J (1991). A double staining technique for simultaneous demonstration of astrocytes and microglia in brain sections and astroglial cell cultures. J. Histochem. Cytochem 39, 561–568. 10.1177/39.5.1707903. [DOI] [PubMed] [Google Scholar]
- 149.Kitamura T, Miyake T, and Fujita S (1984). Genesis of resting microglia in the gray matter of mouse hippocampus. J. Comp. Neurol 226, 421–433. 10.1002/cne.902260310. [DOI] [PubMed] [Google Scholar]
- 150.Tremblay ME, Lecours C, Samson L, Sanchez-Zafra V, and Sierra A (2015). From the Cajal alumni Achucarro and Rio-Hortega to the rediscovery of never-resting microglia. Front. Neuroanat 9, 45. 10.3389/fnana.2015.00045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Tremblay ME (2011). The role of microglia at synapses in the healthy CNS: novel insights from recent imaging studies. Neuron Glia Biol 7, 67–76. 10.1017/S1740925X12000038. [DOI] [PubMed] [Google Scholar]
- 152.Hanisch UK, and Kettenmann H (2007). Microglia: active sensor and versatile effector cells in the normal and pathologic brain. Nat. Neurosci 10, 1387–1394. 10.1038/nn1997. [DOI] [PubMed] [Google Scholar]
- 153.Tremblay ME, Madore C, Bordeleau M, Tian L, and Verkhratsky A (2020). Neuropathobiology of COVID-19: The Role for Glia. Front. Cell. Neurosci 14, 592214. 10.3389/fncel.2020.592214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Sierra A, Tremblay ME, and Wake H (2014). Never-resting microglia: physiological roles in the healthy brain and pathological implications. Front. Cell. Neurosci 8, 240. 10.3389/fncel.2014.00240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Michelucci A, Heurtaux T, Grandbarbe L, Morga E, and Heuschling P (2009). Characterization of the microglial phenotype under specific pro-inflammatory and anti-inflammatory conditions: Effects of oligomeric and fibrillar amyloid-beta. J. Neuroimmunol 210, 3–12. 10.1016/j.jneuroim.2009.02.003. [DOI] [PubMed] [Google Scholar]
- 156.Mills CD, Kincaid K, Alt JM, Heilman MJ, and Hill AM (2000). M-1/M-2 macrophages and the Th1/Th2 paradigm. J. Immunol 164, 6166–6173. 10.4049/jimmunol.164.12.6166. [DOI] [PubMed] [Google Scholar]
- 157.Butovsky O, Jedrychowski MP, Moore CS, Cialic R, Lanser AJ, Gabriely G, Koeglsperger T, Dake B, Wu PM, Doykan CE, et al. (2014). Identification of a unique TGF-beta-dependent molecular and functional signature in microglia. Nat. Neurosci 17, 131–143. 10.1038/nn.3599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Martinez FO, and Gordon S (2014). The M1 and M2 paradigm of macrophage activation: time for reassessment. F1000Prime Rep 6, 13. 10.12703/P6-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Ransohoff RM (2016). A polarizing question: do M1 and M2 microglia exist? Nat. Neurosci 19, 987–991. 10.1038/nn.4338. [DOI] [PubMed] [Google Scholar]
- 160.Devanney NA, Stewart AN, and Gensel JC (2020). Microglia and macrophage metabolism in CNS injury and disease: The role of immuno-metabolism in neurodegeneration and neurotrauma. Exp. Neurol 329, 113310. 10.1016/j.expneurol.2020.113310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Madry C, Kyrargyri V, Arancibia-Carcamo IL, Jolivet R, Kohsaka S, Bryan RM, and Attwell D (2018). Microglial Ramification, Surveillance, and Interleukin-1β Release Are Regulated by the Two-Pore Domain K+ Channel THIK-1. Neuron 97, 299–312.e6. 10.1016/j.neuron.2017.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Sierra A, Encinas JM, Deudero JJ, Chancey JH, Enikolopov G, Overstreet-Wadiche LS, Tsirka SE, and Maletic-Savatic M (2010). Microglia shape adult hippocampal neurogenesis through apoptosis-coupled phagocytosis. Cell Stem Cell 7, 483–495. 10.1016/j.stem.2010.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.VanRyzin JW, Marquardt AE, Argue KJ, Vecchiarelli HA, Ashton SE, Arambula SE, Hill MN, and McCarthy MM (2019). Microglial Phagocytosis of Newborn Cells Is Induced by Endocannabinoids and Sculpts Sex Differences in Juvenile Rat Social Play. Neuron 102, 435–449.e6. 10.1016/j.neuron.2019.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Abiega O, Beccari S, Diaz-Aparicio I, Nadjar A, Laye S, Leyrolle Q, Gomez-Nicola D, Domercq M, Perez-Samartin A, Sanchez-Zafra V, et al. (2016). Neuronal Hyperactivity Disturbs ATP Microgradients, Impairs Microglial Motility, and Reduces Phagocytic Receptor Expression Triggering Apoptosis/Microglial Phagocytosis Uncoupling. PLoS Biol 14, e1002466. 10.1371/journal.pbio.1002466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Torres-Platas SG, Cruceanu C, Chen GG, Turecki G, and Mechawar N (2014). Evidence for increased microglial priming and macrophage recruitment in the dorsal anterior cingulate white matter of depressed suicides. Brain Behav. Immun 42, 50–59. 10.1016/j.bbi.2014.05.007. [DOI] [PubMed] [Google Scholar]
- 166.Bachstetter AD, Ighodaro ET, Hassoun Y, Aldeiri D, Neltner JH, Patel E, Abner EL, and Nelson PT (2017). Rod-shaped microglia morphology is associated with aging in 2 human autopsy series. Neurobiol. Aging 52, 98–105. 10.1016/j.neurobiolaging.2016.12.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Streit WJ, Sammons NW, Kuhns AJ, and Sparks DL (2004). Dystrophic microglia in the aging human brain. Glia 45, 208–212. 10.1002/glia.10319. [DOI] [PubMed] [Google Scholar]
- 168.Tischer J, Krueger M, Mueller W, Staszewski O, Prinz M, Streit WJ, and Bechmann I (2016). Inhomogeneous distribution of Iba-1 characterizes microglial pathology in Alzheimer’s disease. Glia 64, 1562–1572. 10.1002/glia.23024. [DOI] [PubMed] [Google Scholar]
- 169.Savage JC, Carrier M, and Tremblay ME (2019). Morphology of Microglia Across Contexts of Health and Disease. Methods Mol. Biol 2034, 13–26. 10.1007/978-1-4939-9658-2_2. [DOI] [PubMed] [Google Scholar]
- 170.Salamanca L, Mechawar N, Murai KK, Balling R, Bouvier DS, and Skupin A (2019). MIC-MAC: An automated pipeline for high-throughput characterization and classification of three-dimensional microglia morphologies in mouse and human postmortem brain samples. Glia 67, 1496–1509. 10.1002/glia.23623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Stratoulias V, Venero JL, Tremblay MÈ, and Joseph B (2019). Microglial subtypes: diversity within the microglial community. EMBO J 38, e101997. 10.15252/embj.2019101997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.St-Pierre MK, Carrier M, Gonzalez Ibanez F, Simoncicova E, Wallman MJ, Vallieres L, Parent M, and Tremblay ME (2022). Ultra-structural characterization of dark microglia during aging in a mouse model of Alzheimer’s disease pathology and in human post-mortem brain samples. J Neuroinflammation 19, 235. 10.1186/s12974-022-02595-8 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Colombo G, Cubero RJA, Kanari L, Venturino A, Schulz R, Scolamiero M, Agerberg J, Mathys H, Tsai LH, Chacholski W, et al. (2021). Microglial MorphOMICs unravel region- and sex-dependent morphological phenotypes from postnatal development to degeneration. Preprint at bioRxiv. 10.1101/2021.11.30.470610. [DOI] [Google Scholar]
- 174.Graeber MB (2010). Changing face of microglia. Science 330, 783–788. 10.1126/science.1190929. [DOI] [PubMed] [Google Scholar]
- 175.Lawson LJ, Perry VH, Dri P, and Gordon S (1990). Heterogeneity in the distribution and morphology of microglia in the normal adult mouse brain. Neuroscience 39, 151–170. 10.1016/0306-4522(90)90229-w. [DOI] [PubMed] [Google Scholar]
- 176.Gautier EL, Shay T, Miller J, Greter M, Jakubzick C, Ivanov S, Helft J, Chow A, Elpek KG, Gordonov S, et al. (2012). Gene-expression profiles and transcriptional regulatory pathways that underlie the identity and diversity of mouse tissue macrophages. Nat. Immunol 13, 1118–1128. 10.1038/ni.2419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Waddell LA, Lefevre L, Bush SJ, Raper A, Young R, Lisowski ZM, McCulloch MEB, Muriuki C, Sauter KA, Clark EL, et al. (2018). ADGRE1 (EMR1, F4/80) Is a Rapidly-Evolving Gene Expressed in Mammalian Monocyte-Macrophages. Front. Immunol 9, 2246. 10.3389/fimmu.2018.02246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Jung S, Aliberti J, Graemmel P, Sunshine MJ, Kreutzberg GW, Sher A, and Littman DR (2000). Analysis of fractalkine receptor CX(3)CR1 function by targeted deletion and green fluorescent protein reporter gene insertion. Mol. Cell Biol 20, 4106–4114. 10.1128/mcb.20.11.4106-4114.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Wolf Y, Yona S, Kim KW, and Jung S (2013). Microglia, seen from the CX3CR1 angle. Front. Cell. Neurosci 7, 26. 10.3389/fncel.2013.00026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Bisht K, Sharma KP, Lecours C, Gabriela Sanchez M, El Hajj H, Milior G, Olmos-Alonso A, Gomez-Nicola D, Luheshi G, Vallieres L, et al. (2016). Dark microglia: A new phenotype predominantly associated with pathological states. Glia 64, 826–839. 10.1002/glia.22966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Imai Y, Ibata I, Ito D, Ohsawa K, and Kohsaka S (1996). A novel gene iba1 in the major histocompatibility complex class III region encoding an EF hand protein expressed in a monocytic lineage. Biochem. Biophys. Res. Commun 224, 855–862. 10.1006/bbrc.1996.1112. [DOI] [PubMed] [Google Scholar]
- 182.Ito D, Imai Y, Ohsawa K, Nakajima K, Fukuuchi Y, and Kohsaka S (1998). Microglia-specific localisation of a novel calcium binding protein, Iba1. Brain Res. Mol. Brain Res 57, 1–9. 10.1016/s0169-328x(98)00040-0. [DOI] [PubMed] [Google Scholar]
- 183.Shapiro LA, Perez ZD, Foresti ML, Arisi GM, and Ribak CE (2009). Morphological and ultrastructural features of Iba1-immunolabeled microglial cells in the hippocampal dentate gyrus. Brain Res 1266, 29–36. 10.1016/j.brainres.2009.02.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Wake H, Moorhouse AJ, Jinno S, Kohsaka S, and Nabekura J (2009). Resting microglia directly monitor the functional state of synapses in vivo and determine the fate of ischemic terminals. J. Neurosci 29, 3974–3980. 10.1523/JNEUROSCI.4363-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Tremblay ME, Lowery RL, and Majewska AK (2010). Microglial interactions with synapses are modulated by visual experience. PLoS Biol 8, e1000527. 10.1371/journal.pbio.1000527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Lier J, Winter K, Bleher J, Grammig J, Mueller WC, Streit W, and Bechmann I (2019). Loss of IBA1-Expression in brains from individuals with obesity and hepatic dysfunction. Brain Res 1710, 220–229. 10.1016/j.brainres.2019.01.006. [DOI] [PubMed] [Google Scholar]
- 187.Fourgeaud L, Traves PG, Tufail Y, Leal-Bailey H, Lew ED, Burrola PG, Callaway P, Zagorska A, Rothlin CV, Nimmerjahn A, and Lemke G (2016). TAM receptors regulate multiple features of microglial physiology. Nature 532, 240–244. 10.1038/nature17630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Savage JC, Jay T, Goduni E, Quigley C, Mariani MM, Malm T, Ransohoff RM, Lamb BT, and Landreth GE (2015). Nuclear receptors license phagocytosis by trem2+ myeloid cells in mouse models of Alzheimer’s disease. J. Neurosci 35, 6532–6543. 10.1523/JNEUROSCI.4586-14.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Healy LM, Perron G, Won SY, Michell-Robinson MA, Rezk A, Ludwin SK, Moore CS, Hall JA, Bar-Or A, and Antel JP (2016). MerTK Is a Functional Regulator of Myelin Phagocytosis by Human Myeloid Cells. J. Immunol 196, 3375–3384. 10.4049/jim-munol.1502562. [DOI] [PubMed] [Google Scholar]
- 190.Huang Y, Happonen KE, Burrola PG, O’Connor C, Hah N, Huang L, Nimmerjahn A, and Lemke G (2021). Microglia use TAM receptors to detect and engulf amyloid beta plaques. Nat. Immunol 22, 586–594. 10.1038/s41590-021-00913-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Robinson AP, White TM, and Mason DW (1986). Macrophage heterogeneity in the rat as delineated by two monoclonal antibodies MRC OX-41 and MRC OX-42, the latter recognizing complement receptor type 3. Immunology 57, 239–247. [PMC free article] [PubMed] [Google Scholar]
- 192.Milligan CE, Cunningham TJ, and Levitt P (1991). Differential immunochemical markers reveal the normal distribution of brain macrophages and microglia in the developing rat brain. J. Comp. Neurol 314, 125–135. 10.1002/cne.903140112. [DOI] [PubMed] [Google Scholar]
- 193.McKay SM, Brooks DJ, Hu P, and McLachlan EM (2007). Distinct types of microglial activation in white and grey matter of rat lumbosacral cord after mid-thoracic spinal transection. J. Neuropathol. Exp. Neurol 66, 698–710. 10.1097/nen.0b013e3181256b32. [DOI] [PubMed] [Google Scholar]
- 194.Blackbeard J, O’Dea K, Wallace V, Segerdahl A, Pheby T, Takata M, Field M, and Rice A (2007). Quantification of the rat spinal microglial response to peripheral nerve injury as revealed by immunohistochemical image analysis and flow cytometry. J. Neurosci. Methods 164, 207–217. 10.1016/j.jneumeth.2007.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Marshall SA, McClain JA, Kelso ML, Hopkins DM, Pauly JR, and Nixon K (2013). Microglial activation is not equivalent to neuroinflammation in alcohol-induced neurodegeneration: The importance of microglia phenotype. Neurobiol. Dis 54, 239–251. 10.1016/j.nbd.2012.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Peng J, Liu Y, Umpierre AD, Xie M, Tian DS, Richardson JR, and Wu LJ (2019). Microglial P2Y12 receptor regulates ventral hippocampal CA1 neuronal excitability and innate fear in mice. Mol. Brain 12, 71. 10.1186/s13041-019-0492-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Haynes SE, Hollopeter G, Yang G, Kurpius D, Dailey ME, Gan WB, and Julius D (2006). The P2Y12 receptor regulates microglial activation by extracellular nucleotides. Nat. Neurosci 9, 1512–1519. 10.1038/nn1805. [DOI] [PubMed] [Google Scholar]
- 198.Sipe GO, Lowery RL, Tremblay ME, Kelly EA, Lamantia CE, and Majewska AK (2016). Microglial P2Y12 is necessary for synaptic plasticity in mouse visual cortex. Nat. Commun 7, 10905. 10.1038/ncomms10905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Kanamoto T, Mizuhashi K, Terada K, Minami T, Yoshikawa H, and Furukawa T (2009). Isolation and characterization of a novel plasma membrane protein, osteoblast induction factor (obif), associated with osteoblast differentiation. BMC Dev. Biol 9, 70. 10.1186/1471-213X-9-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Bennett ML, Bennett FC, Liddelow SA, Ajami B, Zamanian JL, Fernhoff NB, Mulinyawe SB, Bohlen CJ, Adil A, Tucker A, et al. (2016). New tools for studying microglia in the mouse and human CNS. Proc. Natl. Acad. Sci. USA 113, E1738–E1746. 10.1073/pnas.1525528113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Satoh J.i., Kino Y, Asahina N, Takitani M, Miyoshi J, Ishida T, and Saito Y (2016). TMEM119 marks a subset of microglia in the human brain. Neuropathology 36, 39–49. 10.1111/neup.12235. [DOI] [PubMed] [Google Scholar]
- 202.van Wageningen TA, Vlaar E, Kooij G, Jongenelen CAM, Geurts JJG, and van Dam AM (2019). Regulation of microglial TMEM119 and P2RY12 immunoreactivity in multiple sclerosis white and grey matter lesions is dependent on their inflammatory environment. Acta Neuropathol. Commun 7, 206. 10.1186/s40478-019-0850-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Gonzalez Ibanez F, Picard K, Bordeleau M, Sharma K, Bisht K, and Tremblay ME (2019). Immunofluorescence Staining Using IBA1 and TMEM119 for Microglial Density, Morphology and Peripheral Myeloid Cell Infiltration Analysis in Mouse Brain. J. Vis. Exp 10.3791/60510. [DOI] [PubMed] [Google Scholar]
- 204.Chertoff M, Shrivastava K, Gonzalez B, Acarin L, and Gimenez-Llort L (2013). Differential modulation of TREM2 protein during postnatal brain development in mice. PLoS One 8, e72083. 10.1371/journal.pone.0072083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Fahrenhold M, Rakic S, Classey J, Brayne C, Ince PG, Nicoll JAR, and Boche D (2018). TREM2 expression in the human brain: a marker of monocyte recruitment? Brain Pathol 28, 595–602. 10.1111/bpa.12564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Rogers JT, Morganti JM, Bachstetter AD, Hudson CE, Peters MM, Grimmig BA, Weeber EJ, Bickford PC, and Gemma C (2011). CX3CR1 deficiency leads to impairment of hippocampal cognitive function and synaptic plasticity. J. Neurosci 31, 16241–16250. 10.1523/JNEUROSCI.3667-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Paolicelli RC, Bisht K, and Tremblay ME (2014). Fractalkine regulation of microglial physiology and consequences on the brain and behavior. Front. Cell. Neurosci 8, 129. 10.3389/fncel.2014.00129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Hirasawa T, Ohsawa K, Imai Y, Ondo Y, Akazawa C, Uchino S, and Kohsaka S (2005). Visualization of microglia in living tissues using Iba1-EGFP transgenic mice. J. Neurosci. Res 81, 357–362. 10.1002/jnr.20480. [DOI] [PubMed] [Google Scholar]
- 209.Sasmono RT, Oceandy D, Pollard JW, Tong W, Pavli P, Wain-wright BJ, Ostrowski MC, Himes SR, and Hume DA (2003). A macrophage colony-stimulating factor receptor-green fluorescent protein transgene is expressed throughout the mononuclear phagocyte system of the mouse. Blood 101, 1155–1163. 10.1182/blood-2002-02-0569. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


