Abstract

Mitragyna speciosa (“kratom”) is used as a natural remedy for pain and management of opioid dependence. The pharmacological properties of kratom have been linked to a complex mixture of monoterpene indole alkaloids, most notably mitragynine. Here, we report the central biosynthetic steps responsible for the scaffold formation of mitragynine and related corynanthe-type alkaloids. We illuminate the mechanistic basis by which the key stereogenic center of this scaffold is formed. These discoveries were leveraged for the enzymatic production of mitragynine, the C-20 epimer speciogynine, and fluorinated analogues.
Mitragyna speciosa (“kratom”) is a tree of the Rubiaceae family. Kratom consumption leads to stimulating effects at lower doses and opioid-like effects at higher doses.1 Manual workers have used it for centuries to endure heat and combat fatigue.2,3 Kratom is also consumed for the (self)treatment of pain, to mitigate opioid withdrawal symptoms, and to treat depression; however, rigorous clinical demonstration of kratom’s therapeutic efficacy is still lacking.4 Because of its purported analgesic properties, as well as for recreational purposes, kratom is increasingly used worldwide and is consumed by millions of people in the United States alone.5,6
The pharmacological effects of kratom have been linked to a mixture of >50 corynanthe- and oxindole-type alkaloids (Figure 1a,b).7 Most notable among these are the corynanthe-type alkaloid mitragynine (1) and the hydroxylated derivative 7OH-mitragynine (2). Both 1 and 2 are nanomolar partial agonists at the human μ-opioid receptor (hMOR), and 2 was found to be ∼10-fold more potent than morphine in mice.8,9 Intriguingly, speciogynine (3), the C-20 epimer of mitragynine (1), does not display agonist activity toward hMOR, though speciogynine (3), unlike mitragynine (1), is a smooth muscle relaxant. These differential bioactivities highlight the importance of the C-20 stereochemistry in the pharmacology of kratom alkaloids.10
Figure 1.
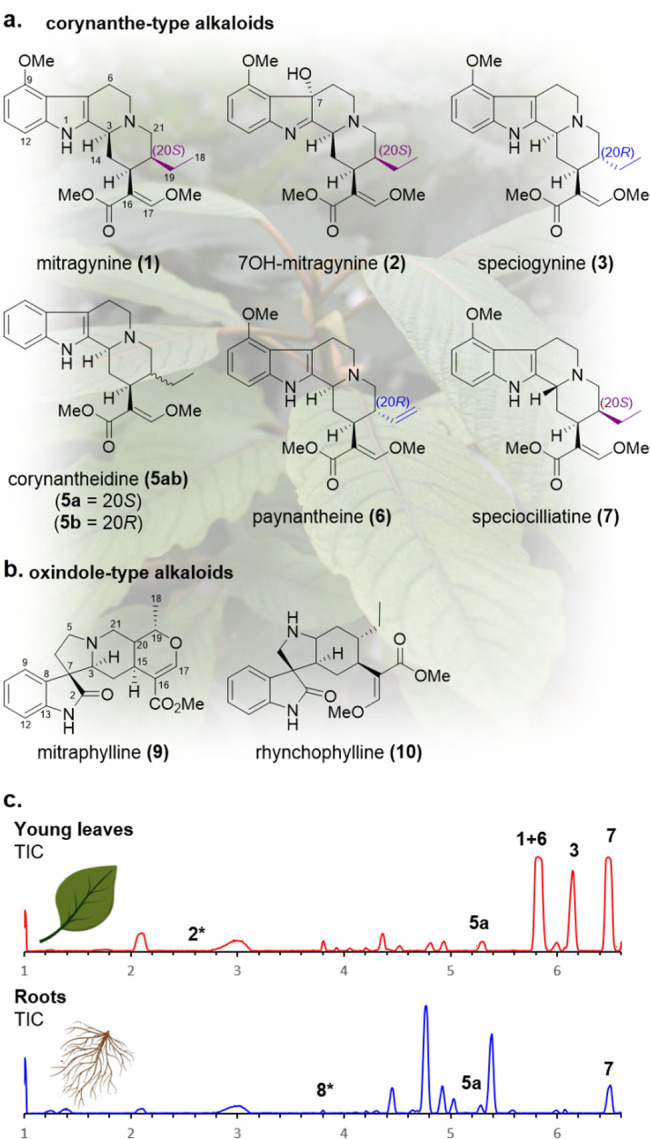
(a, b) Representative kratom alkaloids. (c) TIC of kratom leaf and root extracts. * observed in EIC.
Here, we leverage a multiomics approach to elucidate the key biosynthetic steps that form the corynanthe-type scaffold of kratom alkaloids. We report the discovery of two medium-chain alcohol dehydrogenases (MsDCS1 and MsDCS2) along with an enol O-methyltransferase (MsEnolMT) that converts strictosidine aglycone (4) to either (20S)-corynantheidine (5a) (the precursor to 1) or (20R)-corynantheidine (5b) (the precursor to 3). Rational mutagenesis of MsDCS1 revealed key amino acid residues that control the stereoselective reduction at C-20. A precursor directed biosynthesis approach was then used for the stereoselective production of 1 and 3, as well as fluorinated analogues.
We first identified where these alkaloids accumulate in planta by analyzing methanolic extracts of M. speciosa root, stem, bark, and leaf tissue using targeted metabolomics (Figure 1c; Figures S1–S6). Consistent with literature reports11,12 the alkaloid content in the leaves was higher than other organs, with mitragynine (1), paynantheine (6), speciogynine (3), and speciocilliatine (7) being the dominant products. Low levels of 7OH-mitragynine (2), (20S)-corynantheidine (5a), and strictosidine (8) were also observed. Stem and bark showed similar metabolic profiles, with 7 as the dominant alkaloid and low quantities of 1 and 3 also observed. Notably, root tissue was completely lacking in 1 and 3, with only 7, 8, and 5a detected.
Strictosidine aglycone (4) is the central intermediate for most monoterpene indole alkaloids, including 1, 3, and other kratom-derived alkaloids. The biosynthetic pathway for 4 has been elucidated in Catharanthus roseus,13 and we identified orthologues of these biosynthetic genes in the kratom transcriptome (Scheme 2a). Notably, although 1 and 3 accumulate primarily in leaf and stem, the strictosidine aglycone (4) biosynthetic genes were preferentially expressed in roots, suggesting that this organ is the primary site of biosynthesis for the early pathway steps toward 1. Therefore, either 1 and 3 are produced in the root and subsequently transported to leaf/stem, or alternatively, a biosynthetic intermediate of 1 and 3 is transported to the leaf/stem where the final biosynthetic steps would take place.
Scheme 2. (a) Expression Profiles of Identified Genes in Kratom; (b) Proposed Mechanism for the Formation of the Corynanthe-Type Skeleton; (c) EIC (m/z = 355) of Assays Featuring Combinations of 8, CrSGD, and MsDCS1/MsDCS2/CpDCS; (d) EIC (m/z = 369) of Assays Featuring Combinations of 8, CrSGD, MsEnolMT, and MsDCS1/MsDCS2/CpDCS; and (e) EIC (m/z 369) Corresponding to Transient Expression of CrSTR, CrSGD, MsDCS1/CpDCS, and MsEnolMT in Tobacco.
Expression levels are represented as FPKM of the M. speciosa transcriptome.
Strictosidine aglycone (4) is a reactive intermediate that can be reductively trapped into numerous isomers.14,15 One isomer, dihydrocorynantheine (11ab), has the same scaffold as 1 and 3, and is therefore a likely biosynthetic intermediate for these alkaloids. Recently, we reported the discovery of a medium-chain alcohol dehydrogenase from Cinchona pubescens, dihydrocorynantheine synthase (CpDCS), that converts strictosidine aglycone (4) to (20R)-dihydrocorynantheine (11b) during the biosynthesis of quinine (12) (Scheme 1a).16 The structure of 11b was inferred based on (HR)MS/MS experiments and by NMR characterization of the decarboxylated product (20R)-dihydrocorynantheal (Figure S7).16 It seemed logical that orthologous enzymes should catalyze the reduction of strictosidine aglycone to the dihydrocorynantheine scaffold in kratom (Scheme 1b).16,17 Moreover, given the presence of dihydrocorynantheine-like alkaloids with both (20S)- and (20R)-stereochemistry in kratom, we further hypothesized that kratom would have multiple DCS orthologues with differing stereoselectivity.
Scheme 1. (a) Reduction of 4 by CpDCS and (b) Proposed Pathway Toward Major Kratom Alkaloids.
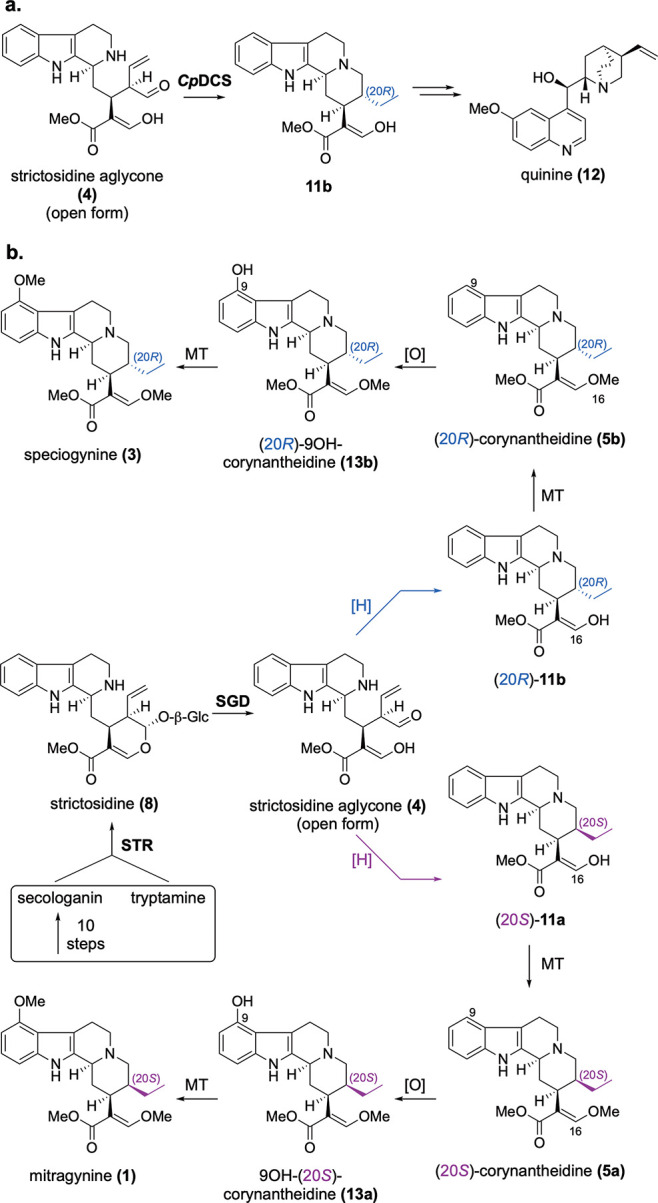
To identify enzyme candidates from kratom that catalyze formation of 11a or 11b from strictosidine aglycone (4), we used the protein sequence of CpDCS to mine the kratom transcriptome. From this process, 27 candidates that showed homology to CpDCS and/or coexpressed with genes involved in (8)-formation were expressed in Escherichia coli (Figures S8 and S9). To assay for enzymatic activity, strictosidine (8) was deglycosylated in situ with strictosidine glucosidase from C. roseus (CrSGD) and incubated with kratom reductase candidates and NADPH. CpDCS, which afforded (20R)-dihydrocorynantheine (11b),16 was used as a positive control (Scheme 2c; TR = 4.4 min; [M + H]+ calcd for C21H27N2O3, 355.2022; found, 355.2014).16 Two of the tested kratom candidates, MsDCS1 and MsDCS2, also produced 11b (Scheme 2c; Figure S10). Intriguingly, MsDCS1 also yielded a second product with the same HRMS ([M + H]+ calcd for C21H27N2O3, 355.2022; found, 355.2015) and MS/MS fragmentation pattern as 11b, but a different retention time (TR = 3.9 min; Scheme 2c; Figure S10). We assumed that this product was (20S)-dihydrocorynantheine (11a), but due to poor stability, this compound could not be characterized.
Subsequent O-methylation at C-16 of 11ab would yield corynantheidine (5ab), which is the next predicted intermediate in the biosynthesis of 1 and 3. To identify gene candidates that catalyze O-methylation at C-16 of 11ab, we identified annotated methyltransferase genes that coexpress with MsDCS1 (r > 0.8, Pearson correlation coefficient). Five purified enzyme candidates were assayed in vitro with strictosidine (8), strictosidine glucosidase (CrSGD), NADPH, SAM, and either CpDCS, MsDCS1, or MsDCS2. A single enzyme, MsEnolMT (r = 0.96), showed methyltransferase activity in all enzyme assays (Scheme 2d). Coincubation of MsEnolMT with MsDCS1 afforded two products with the expected nominal mass of 368 corresponding to the methylated product of 11ab (HRMS: [M + H]+ calcd for C22H28N2O3, 369.2178; found, 369.2164 and 369.2167). The minor product (TR = 4.9 min) eluted at the same retention time and displayed identical MS/MS spectra compared to an authentic standard of 5a (Figures S11 and S12),18 validating that MsDCS1 generates (20S)-dihydrocorynantheine (11a). The major product 5b (TR = 5.4 min) displayed identical MS/MS patterns to 5a (Figure S12), but different retention time. This product has the same retention time and MS/MS pattern as the product of CpDCS, which had been previously established to have R stereoselectivity (Scheme 2c,d).16 Moreover, the MS/MS and the retention time of the decarboxylated major MsDCS1 product matched an authentic standard of (20R)-dihydrocorynantheal (Figure S7).16MsDCS1 also showed R stereoselectivity (Scheme 2c).
The ratios of 11a and 11b produced by MsDCS1 varied considerably among in vitro assays, suggesting that assay conditions affect the stereochemical outcome. To corroborate MsDCS1 activity, we transiently expressed MsDCS1 together with CrSTR, CrSGD, and MsEnolMT in Nicotiana benthamiana leaves. Infiltration with tryptamine and secologanin (19) [the precursors to strictosidine (8)] afforded reproducible ratios of 5a and 5b, with 5a as the dominant product (Scheme 2e). Exchange of MsDCS1 with CpDCS afforded solely 5b, in agreement with previous observations.
Formation of 11ab may likely proceed via an initial 1,4-reduction of the α,β-unsaturated iminium dehydrogeissoschizine (15), which can form in situ upon deglycosidation of 8 (Scheme 2b).16 The reduced intermediate 16 tautomerizes to the iminium form, 17ab, upon protonation at C-20, after which a second 1,2-reduction would occur at C-21. Protonation at C-20 during tautomerization would therefore define the stereochemical outcome. We hypothesized that differences within the active site of these enzymes controlled the face of protonation.
To identify candidate amino acid residues that direct the C-20 stereochemistry, we generated a structural model of MsDCS1 (Figure 2a–c).19,20 Seven amino acids in the binding pocket differentiate MsDCS1 from MsDCS2/CpDCS (Figure 2c,d; Figure S13). These residues from MsDCS2/CpDCS were introduced into MsDCS1 to swap stereoselectivity at C-20. Assays were performed by transient expression of the resulting mutants in tobacco leaves (together with CrSTR, CrSGD, MsEnolMT, tryptamine, and secologanin; Figure 2e; Figure S14). Mutagenesis of residues 295–298 (SGAS to ATGG) was sufficient to invert the ratio between 5a and 5b (from ∼74% 5a in wild-type MsDCS1 to <35% 5a in the mutant). In a septuple mutant of MsDCS1 (T53F, I100M, S116N, SGAS295–298ATGG), formation of the (20S)-isomer 5a was nearly abolished (<5%). Similar results were observed in analogous mutations in CpDCS, which forms the (20R) product 5b (Figure 2f). In the septuple CpDCS mutant (F53T, M100I, N116S, ATGG295–298SGAS), the amount of 5a changed to ∼45% in the mutant compared to 0% in the wild type enzyme (Figure 2f; compare to Figure S14 for additional mutations). Mining of the kratom genome revealed that MsDCS1 is the only homologue harboring these residues at these positions, so we speculate that MsDCS1 is solely responsible for production of corynanthe-type alkaloids with (20S)-stereochemistry in kratom (Figure S15).
Figure 2.
(a, b) Structural model of MsDCS1 in complex with NADPH and dehydrogeissoschizine (15). (c) Key active site residues directing the stereoselectivity. (d) Sequence alignment between MsDCS1, MsDCS2, and CpDCS. (e, f) Levels of (20S)-5a and (20R)-5b in mutants of MsDCS1/CpDCS; displayed are EICs (m/z 369) corresponding to transient expression of CrSTR, CrSGD, and ADH mutants and MsEnolMT in Nicotiana benthamiana.
These seven amino acids may collectively affect the orientation in which dehydrogeissoschizine (15) binds in the enzyme active site, which would in turn control tautomerization and protonation of 16 to either (20S)-17a or (20R)-17b (Scheme 2b; Figure S16). The model of the active site did not contain an amino acid that would be appropriately positioned to catalyze this stereoselective protonation, suggesting that a bound water molecule may be responsible, as previously proposed for other monoterpene indole alkaloid reductases.21 This mutational analysis lays the foundation for metabolic engineering strategies to improve production of mitragynine (1); for example, MsDCS2 could be knocked out or mutated in kratom to generate plants with increased levels of alkaloids with (20S)-stereoconfiguration.
Completion of mitragynine (1) and speciogynine (3) biosynthesis requires methoxylation at C-9 of 5ab (Scheme 1b).22 Since early pathway genes are expressed in roots, while 1 is found exclusively in leaves, it is difficult to predict where the genes responsible for methoxylation would be located. Therefore, we screened oxidases with a variety of expression profiles. However, although 172 candidate oxidase genes were assayed, none showed activity toward either 5a or 5b (Figures S17 and S18). Attempts to use the fungal cytochrome P450 monooxygenase PsiH, another oxidase known to hydroxylate this position of the indole moiety, also failed to hydroxylate 5a or 5b (Figure S19).23
Therefore, we switched to a mutasynthetic strategy to reconstitute mitragynine (1) biosynthesis. N. benthamiana leaves were transiently expressed with CrSTR, CrSGD, MsDCS1, and MsEnolMT and infiltrated with 4-methoxy-tryptamine (18) and secologanin (19). Consistent with the previously observed stereoselectivity of MsDCS1, this afforded a mixture of 1 and 3, with 1 as the dominant product (Scheme 3a,b). Exchange of MsDCS1 with CpDCS solely afforded speciogynine (3). In vitro assays yielded similar results (Figure S20).
Scheme 3. (a) N. benthamiana Infiltration Strategy; and (b, c) EICs of Methanolic Extracts from the Transient Expression of CrSTR, CrSGD, MsEnolMT, and Indicated ADH Enzymes with Different Tryptamine Analogues.
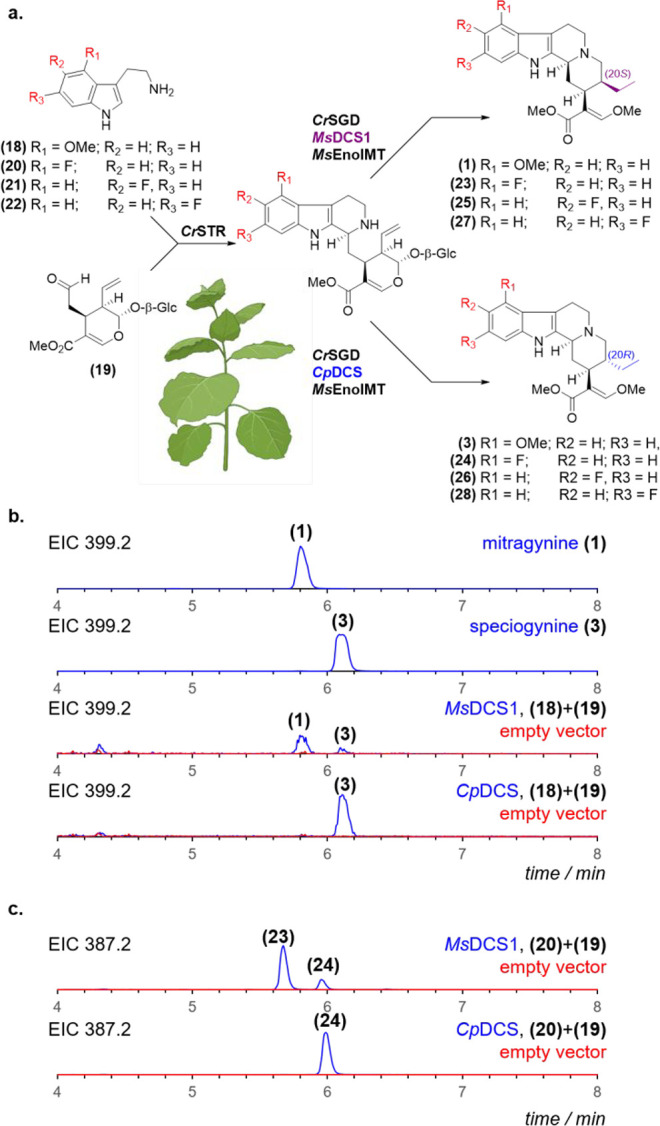
Notably, fluorinated mitragynine analogues have enhanced pharmacological activity.24 Therefore, we assessed the potential for the biocatalytic production of fluorinated analogues of 1 and 3. Infiltration of secologanin (19) and either 4F-, 5F-, or 6F-tryptamine (20–22), along with CrSTR, CrSGD, MsDCS1, and MsEnolMT in N. benthamiana, afforded compounds that corresponded to the expected fluorinated analogues (23–28) as evidenced by HRMS (Scheme 3a,c; Figures S21–S26). Although attempts to isolate these compounds in quantities sufficient for NMR analysis failed, this sets the stage for exploring more efficient yeast-based strategies for mitragynine analogue engineering.
In conclusion, we elucidated the key enzymatic steps for the production of corynanthe-type alkaloids in kratom. Mutagenesis experiments suggest a mechanism that is responsible for the control of the stereochemistry at the crucial C-20 position. These discoveries will enable targeted genome editing in kratom to fine-tune alkaloid profiles. Given the recent advent of yeast expression systems for the production of monoterpene indole alkaloids,25 we anticipate that these enzymes will enable development of robust production platforms for mitragynine, speciogynine, and related analogues.
Acknowledgments
We gratefully acknowledge Delia Ayled Serna Guerrero, Sarah Heinicke, and Maritta Kunert for assistance with mass spectrometry. Jens Wurlitzer is thanked for help with molecular cloning. Eva Rothe and the MPI-CE greenhouse team is thanked for taking care of plants. Carlos E. Rodríguez López is kindly thanked for help with bioinformatics. Maite Colinas, Prashant Sonawane, Chloe Langley, and Matilde Florean are kindly thanked for helpful discussion and advice on methodology. This work was supported by grants from the European Research Council (788301) and the Max Planck Society. A portion of this study was supported by UG3DA048353 and R01DA047855 grants from the National Institute on Drug Abuse and the University of Florida Clinical and Translational Science Institute, which is supported in part by the NIH National Center for Advancing Translational Sciences under award number UL1TR001427. The plant art in Figures 1 and 2 and Schemes 2 and 3 was created with BioRender.com. The heatmap in Scheme 2 was created with Morpheus (https://software.broadinstitute.org/morpheus).
Glossary
Abbreviations
- ADH
alcohol dehydrogenase
- Cr
Catharanthus roseus
- Cp
Cinchona pubescens
- DCS
dihydrocorynantheine synthase
- EIC
extracted ion chromatogram
- FPKM
fragments per kilobase of exon per million of mapped fragments
- GES
geraniol synthase
- G8H
geraniol 8-hydroxylase
- GOR
8-hydroxygeraniol oxidoreductase
- GPP
geranylpyrophosphate
- ISY
iridoid synthase
- hMOR
human μ-opioid receptor
- HRMS
high-resolution mass spectrometry
- Ms
Mitragyna speciosa
- NADPH
nicotinamide adenine dinucleotide phosphate
- SAM
S-adenosyl-methionine
- SGD
strictosidine glucosidase
- STR
strictosidine synthase
- TIC
total ion chromatogram
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/jacs.2c13644.
Detailed methods and materials and additional figures and data including chemical structures, LCMS results, extracted ion chromatograms, sequence alignments, pairwise sequence identities, MS/MS data, and Geneious results (PDF)
Author Present Address
# Y.J.: CAS Key Laboratory of Quantitative Engineering Biology, Shenzhen Institute of Synthetic Biology, Shenzhen Institute of Advanced Technology, Chinese Academy of Sciences, Shenzhen, China
Author Present Address
∇ T.-T.T.D.: Department of Chemistry, Irving K. Barber Faculty of Science, University of British Columbia, Kelowna, British Columbia, Canada
Author Present Address
○ F.L.: Department of Drug Discovery and Biomedical Sciences, College of Pharmacy, University of South Carolina, Columbia, SC 29208, USA
Author Present Address
◆ M.M.: Laboratory for Neglected Disease Drug Discovery, College of Science, Department of Chemistry and Chemical Biology, Northeastern University, 02115 Boston, USA
Author Contributions
⊥ C.S. and Y.J. contributed equally.
Open access funded by Max Planck Society.
The authors declare no competing financial interest.
This paper was published on February 22, 2023. Scheme 2 and Figure 2 have been updated. The revised version of the paper was re-posted on March 8, 2023.
Supplementary Material
References
- Han C.; Schmitt J.; Gilliland K. M. DARK Classics in Chemical Neuroscience: Kratom. ACS Chem. Neurosci. 2020, 11 (23), 3870–3880. 10.1021/acschemneuro.9b00535. [DOI] [PubMed] [Google Scholar]
- Assanangkornchai S.; Muekthong A.; Sam-angsri N.; Pattanasattayawong U. The Use of Mitragynine Speciosa (“Krathom”), an Addictive Plant, in Thailand. Subst. Use Misuse 2007, 42 (14), 2145–2157. 10.1080/10826080701205869. [DOI] [PubMed] [Google Scholar]
- Cinosi E.; Martinotti G.; Simonato P.; Singh D.; Demetrovics Z.; Roman-Urrestarazu A.; Bersani F. S.; Vicknasingam B.; Piazzon G.; Li J.-H.; Yu W.-J.; Kapitány-Fövény M.; Farkas J.; Di Giannantonio M.; Corazza O. Following “the Roots” of Kratom (Mitragyna Speciosa): The Evolution of an Enhancer from a Traditional Use to Increase Work and Productivity in Southeast Asia to a Recreational Psychoactive Drug in Western Countries. BioMed. Res. Int. 2015, 2015, 1–11. 10.1155/2015/968786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grundmann O. Patterns of Kratom Use and Health Impact in the US—Results from an Online Survey. Drug Alcohol Depend. 2017, 176, 63–70. 10.1016/j.drugalcdep.2017.03.007. [DOI] [PubMed] [Google Scholar]
- Swogger M. T.; Smith K. E.; Garcia-Romeu A.; Grundmann O.; Veltri C. A.; Henningfield J. E.; Busch L. Y. Understanding Kratom Use: A Guide for Healthcare Providers. Front. Pharmacol. 2022, 13, 801855. 10.3389/fphar.2022.801855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schimmel J.; Amioka E.; Rockhill K.; Haynes C. M.; Black J. C.; Dart R. C.; Iwanicki J. L. Prevalence and Description of Kratom (Mitragyna Speciosa) Use in the United States: A Cross-sectional Study. Addiction 2021, 116 (1), 176–181. 10.1111/add.15082. [DOI] [PubMed] [Google Scholar]
- a Flores-Bocanegra L.; Raja H. A.; Graf T. N.; Augustinović M.; Wallace E. D.; Hematian S.; Kellogg J. J.; Todd D. A.; Cech N. B.; Oberlies N. H. The Chemistry of Kratom [Mitragyna Speciosa]: Updated Characterization Data and Methods to Elucidate Indole and Oxindole Alkaloids. J. Nat. Prod. 2020, 83 (7), 2165–2177. 10.1021/acs.jnatprod.0c00257. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Nguyen T.-A. M.; Grzech D.; Chung K. C.; Xia Z.; Nguyen T.-D.; Dang T. T. T. Discovery of a Cytochrome P450 Enzyme Catalyzing the Formation of Spirooxindole Alkaloid Scaffold in Kratom. Front. Plant Sci. 2023, 14, 1125158. 10.3389/fpls.2023.1125158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumoto K.; Horie S.; Ishikawa H.; Takayama H.; Aimi N.; Ponglux D.; Watanabe K. Antinociceptive Effect of 7-Hydroxymitragynine in Mice: Discovery of an Orally Active Opioid Analgesic from the Thai Medicinal Herb Mitragyna Speciosa. Life Sci. 2004, 74, 2143–2155. 10.1016/j.lfs.2003.09.054. [DOI] [PubMed] [Google Scholar]
- Takayama H.; Ishikawa H.; Kurihara M.; Kitajima M.; Aimi N.; Ponglux D.; Koyama F.; Matsumoto K.; Moriyama T.; Yamamoto L. T.; Watanabe K.; Murayama T.; Horie S. Studies on the Synthesis and Opioid Agonistic Activities of Mitragynine-Related Indole Alkaloids: Discovery of Opioid Agonists Structurally Different from Other Opioid Ligands. J. Med. Chem. 2002, 45 (9), 1949–1956. 10.1021/jm010576e. [DOI] [PubMed] [Google Scholar]
- Kruegel A. C.; Gassaway M. M.; Kapoor A.; Váradi A.; Majumdar S.; Filizola M.; Javitch J. A.; Sames D. Synthetic and Receptor Signaling Explorations of the Mitragyna Alkaloids: Mitragynine as an Atypical Molecular Framework for Opioid Receptor Modulators. J. Am. Chem. Soc. 2016, 138 (21), 6754–6764. 10.1021/jacs.6b00360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takayama H. Chemistry and Pharmacology of Analgesic Indole Alkaloids from the Rubiaceous Plant, Mitragyna Speciosa. Chem. Pharm. Bull. 2004, 52 (8), 916–928. 10.1248/cpb.52.916. [DOI] [PubMed] [Google Scholar]
- Todd D. A.; Kellogg J. J.; Wallace E. D.; Khin M.; Flores-Bocanegra L.; Tanna R. S.; McIntosh S.; Raja H. A.; Graf T. N.; Hemby S. E.; Paine M. F.; Oberlies N. H.; Cech N. B. Chemical Composition and Biological Effects of Kratom (Mitragyna Speciosa): In Vitro Studies with Implications for Efficacy and Drug Interactions. Sci. Rep. 2020, 10 (1), 19158. 10.1038/s41598-020-76119-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miettinen K.; Dong L.; Navrot N.; Schneider T.; Burlat V.; Pollier J.; Woittiez L.; van der Krol S.; Lugan R.; Ilc T.; Verpoorte R.; Oksman-Caldentey K.-M.; Martinoia E.; Bouwmeester H.; Goossens A.; Memelink J.; Werck-Reichhart D. The Seco-Iridoid Pathway from Catharanthus Roseus. Nat. Commun. 2014, 5 (1), 3606. 10.1038/ncomms4606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatsis E. C.; Carqueijeiro I.; Dugé de Bernonville T.; Franke J.; Dang T.-T. T.; Oudin A.; Lanoue A.; Lafontaine F.; Stavrinides A. K.; Clastre M.; Courdavault V.; O’Connor S. E. A Three Enzyme System to Generate the Strychnos Alkaloid Scaffold from a Central Biosynthetic Intermediate. Nat. Commun. 2017, 8 (1), 316. 10.1038/s41467-017-00154-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stavrinides A.; Tatsis E. C.; Foureau E.; Caputi L.; Kellner F.; Courdavault V.; O’Connor S. E. Unlocking the Diversity of Alkaloids in Catharanthus Roseus: Nuclear Localization Suggests Metabolic Channeling in Secondary Metabolism. Chem. Biol. 2015, 22 (3), 336–341. 10.1016/j.chembiol.2015.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trenti F.; Yamamoto K.; Hong B.; Paetz C.; Nakamura Y.; O’Connor S. E. Early and Late Steps of Quinine Biosynthesis. Org. Lett. 2021, 23 (5), 1793–1797. 10.1021/acs.orglett.1c00206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brose J.; Lau K. H.; Dang T. T. T.; Hamilton J. P.; Martins L. do V.; Hamberger B.; Hamberger B.; Jiang J.; O’Connor S. E.; Buell C. R. The Mitragyna Speciosa (Kratom) Genome: A Resource for Data-Mining Potent Pharmaceuticals That Impact Human Health. G3 GenesGenomesGenetics 2021, 11 (4), jkab058. 10.1093/g3journal/jkab058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obeng S.; Kamble S. H.; Reeves M. E.; Restrepo L. F.; Patel A.; Behnke M.; Chear N. J.-Y.; Ramanathan S.; Sharma A.; León F.; Hiranita T.; Avery B. A.; McMahon L. R.; McCurdy C. R. Investigation of the Adrenergic and Opioid Binding Affinities, Metabolic Stability, Plasma Protein Binding Properties, and Functional Effects of Selected Indole-Based Kratom Alkaloids. J. Med. Chem. 2020, 63 (1), 433–439. 10.1021/acs.jmedchem.9b01465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baek M.; DiMaio F.; Anishchenko I.; Dauparas J.; Ovchinnikov S.; Lee G. R.; Wang J.; Cong Q.; Kinch L. N.; Schaeffer R. D.; Millán C.; Park H.; Adams C.; Glassman C. R.; DeGiovanni A.; Pereira J. H.; Rodrigues A. V.; van Dijk A. A.; Ebrecht A. C.; Opperman D. J.; Sagmeister T.; Buhlheller C.; Pavkov-Keller T.; Rathinaswamy M. K.; Dalwadi U.; Yip C. K.; Burke J. E.; Garcia K. C.; Grishin N. V.; Adams P. D.; Read R. J.; Baker D. Accurate Prediction of Protein Structures and Interactions Using a Three-Track Neural Network. Science 2021, 373 (6557), 871–876. 10.1126/science.abj8754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kochnev Y.; Hellemann E.; Cassidy K. C.; Durrant J. D. Webina: An Open-Source Library and Web App That Runs AutoDock Vina Entirely in the Web Browser. Bioinformatics 2020, 36 (16), 4513–4515. 10.1093/bioinformatics/btaa579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stavrinides A.; Tatsis E. C.; Caputi L.; Foureau E.; Stevenson C. E. M.; Lawson D. M.; Courdavault V.; O’Connor S. E. Structural Investigation of Heteroyohimbine Alkaloid Synthesis Reveals Active Site Elements That Control Stereoselectivity. Nat. Commun. 2016, 7 (1), 12116. 10.1038/ncomms12116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franke J.; Kim J.; Hamilton J. P.; Zhao D.; Pham G. M.; Wiegert-Rininger K.; Crisovan E.; Newton L.; Vaillancourt B.; Tatsis E.; Buell C. R.; O’Connor S. E. Gene Discovery in Gelsemium Highlights Conserved Gene Clusters in Monoterpene Indole Alkaloid Biosynthesis. ChemBioChem. 2019, 20 (1), 83–87. 10.1002/cbic.201800592. [DOI] [PubMed] [Google Scholar]
- Fricke J.; Blei F.; Hoffmeister D. Enzymatic Synthesis of Psilocybin. Angew. Chem., Int. Ed. 2017, 56 (40), 12352–12355. 10.1002/anie.201705489. [DOI] [PubMed] [Google Scholar]
- Takayama H.; Misawa K.; Okada N.; Ishikawa H.; Kitajima M.; Hatori Y.; Murayama T.; Wongseripipatana S.; Tashima K.; Matsumoto K.; Horie S. New Procedure to Mask the 2,3-π Bond of the Indole Nucleus and Its Application to the Preparation of Potent Opioid Receptor Agonists with a Corynanthe Skeleton. Org. Lett. 2006, 8 (25), 5705–5708. 10.1021/ol062173k. [DOI] [PubMed] [Google Scholar]
- Zhang J.; Hansen L. G.; Gudich O.; Viehrig K.; Lassen L. M. M.; Schrübbers L.; Adhikari K. B.; Rubaszka P.; Carrasquer-Alvarez E.; Chen L.; D’Ambrosio V.; Lehka B.; Haidar A. K.; Nallapareddy S.; Giannakou K.; Laloux M.; Arsovska D.; Jørgensen M. A. K.; Chan L. J. G.; Kristensen M.; Christensen H. B.; Sudarsan S.; Stander E. A.; Baidoo E.; Petzold C. J.; Wulff T.; O’Connor S. E.; Courdavault V.; Jensen M. K.; Keasling J. D. A Microbial Supply Chain for Production of the Anti-Cancer Drug Vinblastine. Nature 2022, 609 (7926), 341–347. 10.1038/s41586-022-05157-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.




