Abstract
The rapid development of genome editing technology has brought major breakthroughs in the fields of life science and medicine. In recent years, the clustered regularly interspaced short palindromic repeats (CRISPR)-based genome editing toolbox has been greatly expanded, not only with emerging CRISPR-associated protein (Cas) nucleases, but also novel applications through combination with diverse effectors. Recently, transposon-associated programmable RNA-guided genome editing systems have been uncovered, adding myriads of potential new tools to the genome editing toolbox. CRISPR-based genome editing technology has also revolutionized cardiovascular research. Here we first summarize the advances involving newly identified Cas orthologs, engineered variants and novel genome editing systems, and then discuss the applications of the CRISPR-Cas systems in precise genome editing, such as base editing and prime editing. We also highlight recent progress in cardiovascular research using CRISPR-based genome editing technologies, including the generation of genetically modified in vitro and animal models of cardiovascular diseases (CVD) as well as the applications in treating different types of CVD. Finally, the current limitations and future prospects of genome editing technologies are discussed.
Keywords: Genome editing, CRISPR-Cas system, Base editing, Prime editing, Transposon-associated genome editing, Cardiovascular disease, Heart, Blood vessel, Gene therapy
Background
Genome editing technology refers to a series of technologies capable of manipulating cellular DNA sequences at desired genomic sites by generating altered DNA sequences through nuclease-mediated site-specific DNA breaks that are resolved through DNA repair pathways [1–3]. Among genome editing-associated nucleases, clustered regularly interspaced short palindromic repeats (CRISPR)/CRISPR-associated protein (Cas) nucleases are convenient, efficient, and precise, and are currently the most widely used [4–8]. After the CRISPR-Cas9 system was characterized and programmed to perform RNA-guided DNA cleavage at specific sites in prokaryotes, it was immediately proven to be an efficient tool for editing eukaryotic genomes [9]. Since then, CRISPR-based genome editing technology has drawn a worldwide attention and initiated extensive development. Emerging CRISPR-based tools with broadened targeting ranges, improved editing specificity and efficiency, and other distinct capabilities have facilitated eukaryotic genome editing by selecting optimal CRISPR-Cas tools. In addition to expanding the CRISPR-Cas nuclease arsenal, this system has also been applied to transcriptional regulation, epigenetic modification, and live-cell imaging by incorporation with other effector proteins [7, 10, 11].
The exponential development of genome editing technology has dramatically changed the landscape of biological and medical research, heralding a new era of precision medicine based on genome editing [12]. CRISPR-based nucleases are able to cut target DNA and generate double-strand breaks (DSBs), followed by the introduction of random mutations mediated by non-homologous end-joining (NHEJ), or make precise editing through homology-directed repair (HDR) [13]. The therapeutic potential of CRISPR-based tools has been investigated using the mouse models of various human diseases [7, 14]. However, precise gene correction for in vivo therapeutic utility remains challenging, which is partially due to the low efficiency of HDR-mediated DNA replacement. This strategy is usually not applicable to post-mitotic cells, as HDR occurs mainly in the S/G2 phase during cell division [15]. Precise genome editing tools have been developed and continuously optimized by fusing activity-impaired Cas nucleases with deaminases, called base editors, or with reverse transcriptases, called prime editors (PEs) [16–18]. Despite not achieving the goal of arbitrarily introducing any genetic substitutions at any targeted genomic site, we are now closer to this aspiration than ever.
Cardiovascular disease (CVD) is a group of disorders of the heart and blood vessels that has consistently been ranked as the leading threat to human health worldwide. Many gene mutations have been linked to CVD, and the number is still increasing [19, 20]. Loss-of-function studies in animals are required to address the causal relationship between these mutations and cardiovascular pathologies. With the help of the CRISPR-based toolbox, creating animal models of human diseases has become much easier, faster, and more flexible than ever before; these models will greatly advance our understanding of cardiovascular pathogenesis and the development of therapeutic strategies [14]. Furthermore, CRISPR-based genome editing technology holds promise for treating inherited CVD caused by rare mutations.
In this review, we highlight the recent advances in CRISPR-based genome editing technology, mainly in the past three years, and discuss the tremendous innovation this epoch-making technology has brought to the field of cardiovascular research.
Novel Cas orthologs and engineered variants
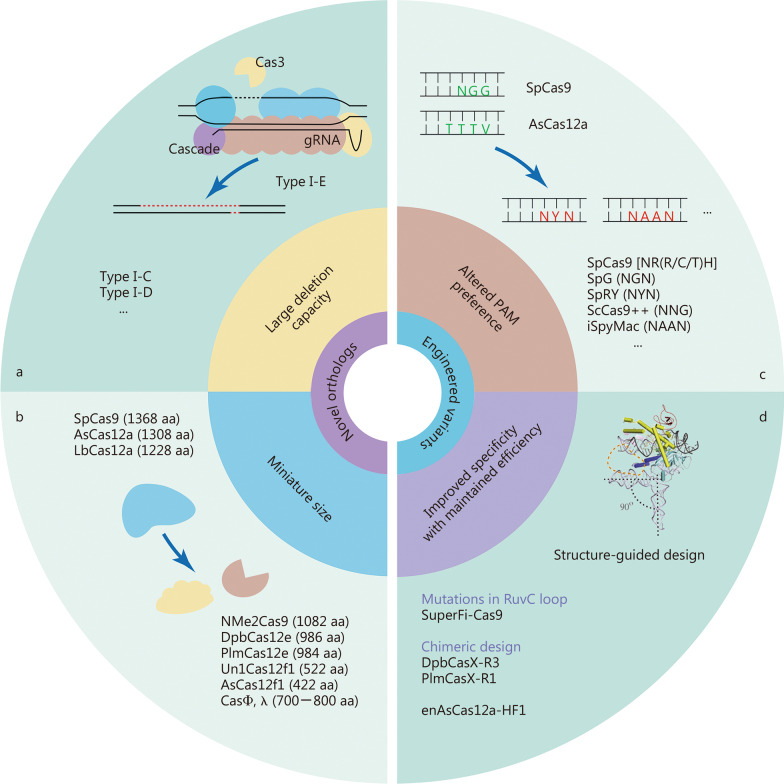
Natural CRISPR-Cas systems are originally identified as adaptive immune systems in bacteria and archaea, and can be divided into two classes based on their composition and mechanisms. These systems are further divided into six types (I–VI) and dozens of subtypes based on the characteristics and accessory genes flanking the CRISPR array [21]. The most widely used class 2 CRISPRs are characterized by their single effector proteins, including type II Cas9 and type V-A Cas12a. Although class 2 natural Cas nucleases have long been used for efficient genome editing, their applications are limited because of the requirement of specific protospacer adjacent motif (PAM) sequences, off-target DNA cleavage, and occasionally, large sizes. Class 1 CRISPR systems possess multiple effector molecules that have unique features, such as distinct PAM preferences, higher on-target specificity through longer target recognition, and production of long-range genomic deletion [22–25]. However, the requirement of multiple effectors and the relatively low editing efficiency must be improved before their widespread application. Continuous efforts have been made to characterize novel Cas orthologs and engineered Cas variants to improve genome editing efficiency and broaden compatibility (Fig. 1).
Fig. 1.
Characteristics of novel Cas orthologs and engineered variants. a Representative type I Cas orthologs capable of large-range deletions. b Representative Cas orthologs of miniature sizes. c Engineered Cas variants with diverse protospacer adjacent motif recognition capabilities. d Structure-guided strategies for improving DNA specificity without affecting the on-target cleavage efficiency
Characterizing novel Cas orthologs with distinctive features
The class 1 type I CRISPR system is the most prevalent CRISPR system, in which the multi-subunit CRISPR-associated complex for antiviral defense (Cascade) identifies DNA targets, and the helicase-nuclease enzyme Cas3 degrades DNA [26] (Fig. 1a). Several type I CRISPR systems have been characterized and applied to mammalian genome editing. Type I-E and type I-D systems have been used to induce unidirectional and bidirectional long-range deletions in human cells [22–24]. Recently, supplying Cas11 was shown to enable divergent I-C, I-D, and I-B CRISPR-Cas3 editors for eukaryotic applications, and efficiently produced large unidirectional deletions [25]. Therefore, type I CRISPR systems can greatly expand the genome editing toolbox owing to their unique mechanisms and advantages in deleting full-length genes, gene clusters, and non-coding sequences.
Recently, the IS200/605 transposon family encoded RNA-guided nucleases have been identified as ancestors of CRISPR-Cas nucleases [27, 28]. Cas9 endonucleases could likely have evolved from ancestral IscB proteins, whereas Cas12 endonucleases descended from TnpB proteins [27]. These transposon-encoded nucleases, together with the IsrB proteins, which are shorter IscB homologs also encoded in IS200/605 superfamily transposons, are called the obligate mobile element-guided activity (OMEGA) system [27]. IscB and TnpB are guided by non-coding RNAs called ωRNAs, which are derived from the left- or right-end elements of a transposon and combine the functions of CRISPR RNA (crRNA) and trans-activating CRISPR RNA (tracrRNA) [27–29]. Being only two-fifths the size of Cas9, IscB and TnpB can mediate double-strand DNA cleavage at the target sites with a 3′ or 5′ transposon-associated motif (TAM), and both have been adopted for genome editing in human cells [27, 28].
The in vivo application of most CRISPR systems is challenging because of their large size, especially when delivered by the widely used adeno-associated virus (AAV). As a result, the exploitation of miniature Cas proteins with high efficiency is in sustained demand (Fig. 1b). For instance, SaCas9 (1053 aa), CjCas9 (984 aa), and Nme1Cas9 (1082 aa) have been validated as mammalian genomic editors [30–32]. Recently, a compact Nme2Cas9 (1082 aa) recognizing an N4CC PAM was described with an identical target density as SpCas9 and few off-target effects [33]. In addition to the Cas9 orthologs mentioned above, several Cas12 nucleases with smaller sizes, such as Cas12e (or CasX, 986 aa), and Cas12f (400–700 aa), have also been identified as genome editing tools in mammalian cells [34–37]. The Un1Cas12f1 (522 aa) system was optimized to enable efficient genome editing in human cells [35]. Notably, with only a size of 422 aa, AsCas12f1 is currently the smallest RNA-guided Cas nuclease, and has been shown to be an effective programmed genome editing tool in both bacterial and human cells [36]. With approximately equal to or less than half the size of the widely used SpCas9, these miniature CRISPR tools facilitate the AAV-mediated all-in-one delivery of CRISPR components or catalytically inactive Cas variants fused with other functional proteins.
Very recently, CRISPR-Cas systems were found to be widely encoded in the genomes of diverse bacteriophages, where they are involved in competition with other viruses [38, 39]. The bacteriophage-encoded Cas proteins contain all known types of CRISPR-Cas systems, but have phage-specific properties. These Cas proteins, such as CasΦ [38] and Casλ [39], tend to have remarkably small sizes due to the compact viral genome. These hypercompact systems have been shown to edit the genomes of human and plant cells, indicating that viral Cas nucleases could serve as a new source of genome editing tools.
Expanding the range of genomic targets
Genomic targeting by Cas nucleases requires a PAM sequence near the site where the Cas nuclease binds DNA sequences complementary to the single guide RNA (sgRNA). The requirement of PAM is the gatekeeper for CRISPR-Cas mediated genome targeting, as whether a genomic sequence possesses a PAM for a certain Cas nuclease determines whether the site can be targeted and edited by CRISPR. Efforts have been made to develop Cas nucleases with broader PAM compatibility to pursue true PAM-free nucleases. However, PAM-free nucleases might have potential drawbacks, such as self-targeting of gRNA-expressing DNA constructs and reduced efficiency as more time is required for interrogating the whole genome. Therefore, it is better to develop an arsenal of divergent PAM-dependent Cas nucleases that collectively cover all genomic sequences [40] (Fig. 1c).
Using phage-assisted non-continuous and continuous evolution strategies, three new SpCas9 variants (SpCas9-NRRH, SpCas9-NRCH, and SpCas9-NRTH) were characterized to recognize most NR PAM sequences, together with SpCas9-NG (N = A/T/C/G, R = A/G, H = A/C/T) [41]. To relax the PAM preference of SpCas9, two SpCas9 variants, SpG (targeting NGN) and SpRY (targeting NYN), have been generated by structure-guided substitutions in several residues, making most of the genome targetable (Y = C/T) [42]. Structure-motivated engineering has also been used to expand targeting range of LbCas12a and AsCas12a [43, 44]. Chimeric Cas proteins created by exchanging PAM-interacting domains between naturally occurring Cas orthologs have also been applied to expand PAM recognition. Substituting the loop sequence of Cas9 from Streptococcus anginosus, together with the T1227K mutation, into the open reading frame (ORF) of ScCas9 generates ScCas9++ with NNG PAM compatibility [45]. A similar strategy was used to generate a variant iSpyMac by grafting the PAM-interacting domain of SmacCas9 into SpyCas9, which recognizes all adenine dinucleotide PAM (NAAN PAM) sequences [46]. The chimera generation approach has also been applied to replace PAM-interacting domains in SaCas9 [47] and Cas12a [48].
Improving the DNA specificity without affecting on-target cleavage efficiency
Off-target activity is a major challenge when using CRISPR tools for disease-related gene therapy. Over the years, continuous efforts have been made to construct high-fidelity Cas9 variants including eSpCas9(1.1) [49], SpCas9-HF1 [50], HypaCas9 [51], evoCas9 [52], HiFi Cas9 [53] and Sniper-Cas9 [54]. In addition, high-fidelity SaCas9s have been identified either by rational engineering [55] or directional screening [56]. The achievement of enhanced discrimination between on-target and off-target binding of these variants relies mainly on the energetic destabilization of the Cas9:sgRNA:DNA complex at off-target sites [57]. However, the improvement of these high-fidelity Cas9 variants seems to occur at the cost of decreased on-target efficiency [58, 59]. Recently, kinetics-guided cryo-electron microscopy was used to show that mismatches distal to PAM can be stabilized by a loop in the RuvC domain, allowing Cas9 activation [60]. Based on this observation, they designed a high-fidelity variant with mutations in the RuvC domain, named SuperFi-Cas9, which displayed significantly improved mismatch discrimination without compromising on-target DNA cleavage efficiency [60].
DpbCas12e has been validated as a naturally occurring high-fidelity Cas nuclease with striking avoidance of off-target activity [61]. In a recent cryo-EM-based structural engineering study, unique nucleotide-binding loops within Cas12e were found to be important for DNA cleavage efficacy. Based on this finding, newly designed chimeric Cas12e proteins (DpbCasX-R3 and PlmCasX-R1) and sgRNA (sgRNAv2) exhibited substantially improved DNA editing efficiency in mammalian cells [62]. Structure-guided protein engineering has also been used to improve the performance of AsCas12a [43]. E174R/S542R/K548R substitutions were introduced into AsCas12a to construct a variant called enAsCas12a that possesses an expanded targeting range and increased cleavage activity. A high-fidelity version of enAsCas12a (enAsCas12a-HF1) with an additional N282A substitution was also engineered to reduce off-target effects [43] (Fig. 1d).
Advances in precise genome editing
Precise genome editing is essential for preclinical research and clinical gene therapy, and HDR-mediated gene editing has long been the only option. Efforts have been made to improve HDR efficiency, such as the use of rationally designed single-stranded oligodeoxynucleotide (ssODN) templates instead of double-stranded DNA [63] or the addition of NHEJ chemical inhibitors [64]. The delivery of Cas9 and HDR templates by AAVs has accomplished precise genome editing in post-mitotic hippocampal neurons and cardiomyocytes in mice [65–67]. However, the efficiency of HDR-mediated editing is still relatively low compared to that of the predominant NHEJ repair pathway, and DSBs made by this conventional method may introduce undesired damage to the genome [68–70]. Its clinical application is hampered by the need for additional DNA templates and HDR-promoting chemical agents with potential cytotoxicity, such as SCR7 [64], azidothymidine, trifluridine [71], NU7026, and NU7441 [72]. Motivated by these problems, novel precise genome editing tools that do not require DSBs or exogenous DNA templates have been developed (Fig. 2).
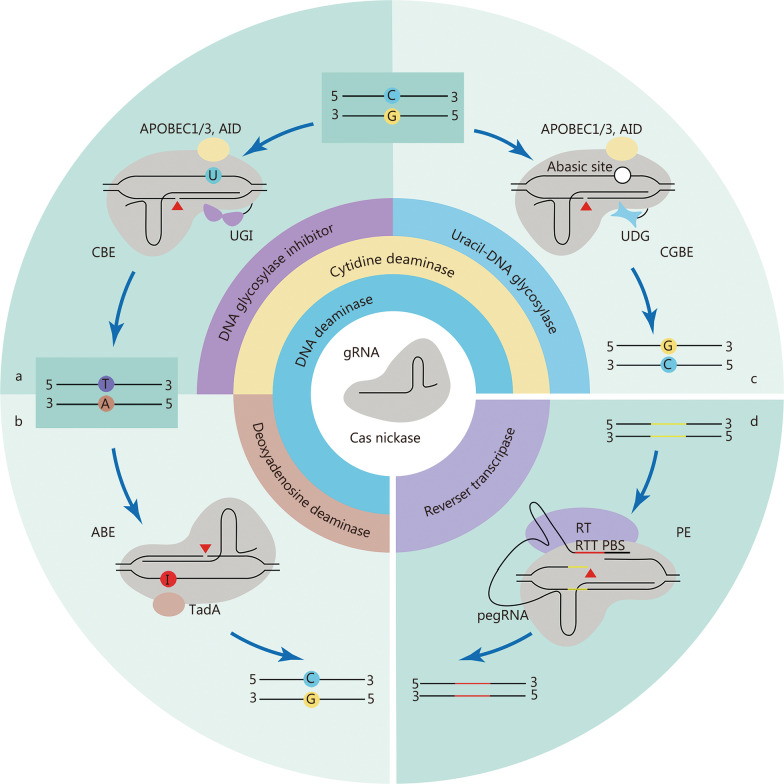
Fig. 2.
CRISPR-Cas-based DNA base editing tools. a-c Schematic diagrams of CBE (a), ABE (b), and CGBE (c). d Schematic of PE. UGI uracil-DNA glycosylase inhibitor, AID activation-induced cytidine deaminase, UNG uracil-DNA glycosylase, TadA deoxyadenosine deaminases, RT reverse transcriptase, PBS primer binding site, RTT RT template, CBE cytosine base editor, ABE adenine base editor, CGBE C-to-G base editor, PE Prime editor
Base editors
Base editors can make precise base substitutions without requiring DSBs or donor DNA templates, and are independent of HDR, providing a promising therapeutic tool for human genetic diseases in which the most relevant variants are single nucleotide mutations [73, 74]. Current base editors are constructed by fusing DNA deaminase enzymes to catalytically impaired Cas nucleases, which can precisely change a single base in a targeted sequence [75]. Base editing jointly harnesses the genome-targeting function of a Cas protein and the DNA base modification role of a deaminase, and sometimes additional regulatory elements are also required to achieve the desired performance. To date, base editors can be used not only for precise genome editing at specific single loci, but also for large-scale functional screening of genetic variants or key amino acid residues [76–78].
The cytosine base editor (CBE) and adenine base editor (ABE) are widely used base editors that enable the editing of all four types of base transitions (C-to-T, A-to-G, T-to-C, and G-to-A) [16, 17] (Fig. 2a, b). Since these two editors were developed in 2016 and 2017, subsequent efforts have significantly expanded their genome-targeting range and improved their efficiency and product purity. The use of Cas nickase, fusion of a second uracil-DNA glycosylase inhibitor (UGI) domain, the addition of a nuclear localization sequence, and linker and codon optimization have greatly increased the editing efficiencies of base editors [79–81]. To maximize the editing scope of base editors, diverse base editors with natural, engineered, or evolved Cas variants that recognize alternative PAMs and various deaminases have been created [82–88].
Programmable C-to-G base editors (CGBEs) that can achieve targeted C-to-G and G-to-C base transversions have recently been developed [89, 90]. CGBEs originate from CBE by replacing the UGI with a uracil-DNA glycosylase (UNG), which excises the U base generated by cytosine deaminase, resulting in an abasic site followed by the preferential installation of a G base through the DNA repair mechanism (Fig. 2c). Although CGBEs can provide efficient C-to-G and G-to-C editing, very few sites are suitable for CGBE editing. Several studies have used machine learning to optimize CGBEs to improve editing efficiency and product purity by changing the species origin, modifying the relative positions of UNG and deaminase, and optimizing codons [91, 92].
The therapeutic applications of base editors have been hampered by their genome-wide off-target effects. Recent studies have shown that cytidine deaminases used in CBE induce genome-wide off-target editing independently of sgRNA or Cas9 [93, 94]. In addition, both ABE and CBE can cause transcriptome-wide mutations [95, 96]. Continuous efforts have been made to reduce off-target effects by engineering the DNA- [97, 98] or RNA-binding domain [95, 96, 99], thereby bringing base editors closer to clinical applications.
PEs
Prime editing is a newly developed precise genome editing technology that enables all types of base conversion, small deletions, and insertions, as desired. PEs consist of a prime editor protein and prime editing gRNA (pegRNA). The PE protein is constructed by fusing an engineered Cas9 nickase (H840A) with reverse transcriptase, which can be targeted to the genomic locus by pegRNA [18]. The pegRNA combines a gRNA recognizing the target genomic sequence, a reverse transcriptase template encoding the desired edits, and a primer binding site to initiate reverse transcription [18] (Fig. 2d). The newly synthesized edited DNA strand is incorporated into the target locus to generate heteroduplex DNA, in which the non-edited strand is eventually replaced by an edited strand through DNA repair. Compared to base editing, which often introduces bystander editing of extra bases in an activity window, prime editing is more versatile and precise.
A series of PE systems, namely PE2, PE3b, PE4, and PE5b, have been developed and are most widely used. All these systems share a common PE2 protein with an engineered Moloney murine leukemia virus (M-MLV) reverse transcriptase instead of the wild-type M-MLV reverse transcriptase in PE1 to increase editing efficiency [18, 100]. The PE3 system contains an additional sgRNA that targets the non-edited strand to increase the editing efficiency [18]. The PE2 and PE3 systems were further optimized by introducing a DNA mismatch repair-inhibiting domain MLH1dn to generate PE4 and PE5 systems, respectively [18, 100]. Systems ending in “b”, namely PE3b and PE5b, use an edit-specific nicking sgRNA to reduce indel levels [18, 100]. Constant efforts are being devoted to optimizing PEs, with a primary focus on improving their editing efficiency. Optimization of PE2 protein architecture by codon optimization, SpCas9 mutation, and alterations of the nuclear localization signal and peptide linker sequence results in PEmax protein architecture, which greatly enhances editing efficiency [100]. They also constructed two types of engineered pegRNAs (epegRNAs) by incorporating 3′ structural motifs, which stabilize pegRNA and increase prime editing efficiency [101]. Many other groups have adopted similar strategies by optimizing either PE proteins [102–104] or pegRNAs [105].
In addition, a dual pegRNA strategy has been used to improve editing efficiency, which can also achieve programmable insertion, deletion, and replacement of large genomic sequences at specific genomic sites [106–110]. Using a pair of pegRNAs, each of which targets a different DNA strand and template the synthesis of complementary DNA flaps, endogenous targeted DNA sequence between the PE-induced nick sites is successfully replaced. This strategy also achieves targeted insertion of gene-sized DNA plasmids (> 5 kb) and targeted inversions of 40 kb in human cells when co-expressing a site-specific serine recombinase, Bxb1 integrase [108]. The dual pegRNA strategy with expanded capabilities of precision genome editing provides new possibilities for treating genetic disorders caused by large DNA deletions or complex structural mutations.
CRISPR-associated transposon (CAST) systems for large DNA insertion
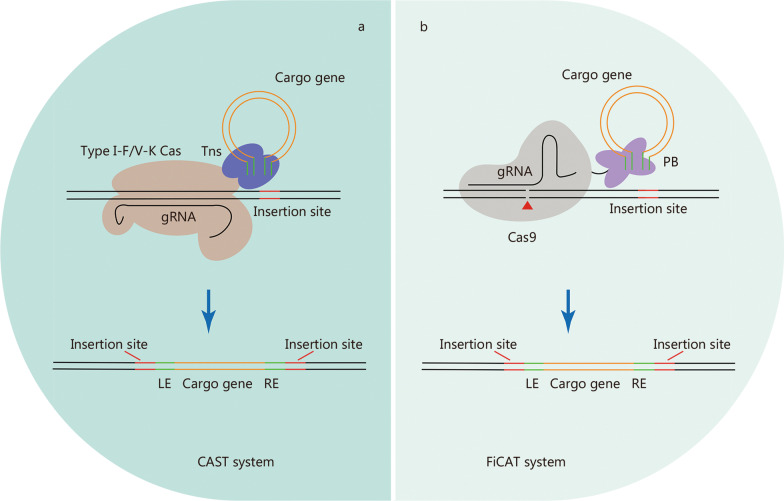
CAST systems, consisting of transposase subunits and CRISPR effectors, facilitate the RNA-guided transposition of mobile genetic elements, making it a promising system for targeted, precise, and efficient insertion of large DNA segments. Most identified CASTs are derived from Tn7-like transposons that retain the core genes of the transposition machinery, but have no genes for target selection [111, 112]. Instead, CASTs co-opt nuclease-deficient CRISPR-Cas proteins to induce RNA-guided transposition [111, 112]. Several CAST systems have been experimentally or bioinformatically characterized, including type I-B, type I-C, type I-F, type IV, and type V-K CAST systems [111, 113–115]. Bioinformatic analysis of the metagenomic database also revealed a non-Tn7 CAST system that co-opts a nuclease-inactive Cas12 and type I-E cascade [111].
Type I-F and type V-K CAST systems have been successfully reconstituted to achieve the integration of donor DNA into specific bacterial genome sites [113, 114] (Fig. 3a). An improved version of the type I-F CAST system enables highly specific and effective integration of up to 10 kb DNA fragments in the bacterial genome [116]. However, the application of these two systems in mammalian cells has not been reported. A very recent study developed an artificial transposon-associated CRISPR-Cas system named find and cut-and-transfer (FiCAT) system by coupling a SpCas9 protein with an engineered piggyBac (PB) transposase (Fig. 3b), which achieved the targeted integration of multi-kilobase DNA fragments into the genomes of mammalian cell lines and mouse liver [117]. The discovery of CAST systems has expanded the genome editing toolkit, although CAST systems still require extensive modification and optimization until they can be conveniently and effectively applied to biomedical research.
Fig. 3.
CRISPR-Cas-based transposon systems. Schematic of CRISPR-based transposon systems, CAST system (a) and FiCAT system (b), which mediate site-specific DNA integration. Tns Tn7-like transposases, PB piggyBac transposase, LE transposon left end sequences, RE transposon right end sequences, CAST CRISPR-associated transposon, FiCAT find and cut-and-transfer
Delivery systems for CRISPRs
The safe, effective, and tissue-specific delivery of CRISPR-Cas tools in vivo determines whether CRISPR-based gene therapy can be used for this tissue. Therapeutic in vivo delivery systems for CRISPR-Cas have recently been discussed [118, 119]. CRISPR-Cas tools can be delivered in the form of DNA, mRNA, or ribonucleoprotein complexes (RNP) through ex vivo or in vivo approaches. Various robust methods have been established to deliver genome editing reagents ex vivo, some of which have been used in multiple clinical trials involving different types of diseases [120–122]. The most efficient method of in vivo delivery of editors reported so far is the use of AAV, which can deliver editor-encoding DNA to target tissues and has been applied in clinical trials [118, 123, 124]. However, AAV-based delivery of DNA-encoding editing agents has a number of disadvantages, such as the possibility of viral vector integration into the transduced cell genome and increased frequency of off-target editing due to prolonged expression [75, 119, 125, 126], which limits its clinical application. Therefore, safer alternative strategies for in vivo delivery of genome editors must be developed.
As a gene therapy delivery system approved by the Food and Drug Administration (FDA), lipid nanoparticles (LNP) have been demonstrated to safely deliver therapeutic small molecules and nucleic acid drugs to hepatocytes and antigen-presenting cells via systemic administration or intramuscular injection. The LNP system was used to deliver gene editing tools in the first clinical trial involving human gene editing in vivo [123]. However, because intravenously delivered LNP showed liver tropism, delivering editors to non-hepatocytes has been a huge challenge. Recent studies have shown that high-throughput screening identifies nanoparticles targeting non-hepatocytes, including endothelial cells (ECs) and spleen immune cells [127, 128]. In addition, cell-type specificity of LNP-mediated Cas9 therapies could be modified by reducing Cas9-mediated insertions and deletions in hepatocytes using inhibitory oligonucleotides and siRNAs [129].
Virus-like particle (VLP) systems, which combine the advantages of viral and non-viral delivery systems, are another promising in vivo gene editing delivery vehicle [126]. VLPs can package genome editing agents in the forms of mRNA or RNP. The short cellular lifespan of RNPs effectively restricts off-target editing. Almost all current VLPs are derived from retroviruses and contain most viral components but no viral genome [118, 130–133]. Very recently, fourth-generation engineered VLPs (eVLPs) based on M-MLV have been developed to deliver Cas9 or base editor RNPs both in vitro and in vivo [126]. A single intravenous injection of eVLPs carrying a base editor targeting proprotein convertase subtilisin/kexin type 9 (Pcsk9) can achieve base editing in multiple tissues, reduce serum PCSK9 levels by 78%, and partially restore visual function when designed for retinal editing in a mouse model of blindness [126]. The mammalian endogenous retrovirus-like protein PEG10 has also been programmed as a VLP system called selective endogenous encapsidation for cellular delivery (SEND) platform, which can package and deliver mRNA encoding Cas9 in vivo [134]. Based on endogenous mammalian proteins, the SEND system may be less immunogenic than bona fide retrovirus-based VLP systems.
Applications of genome editing in modeling and treating CVD
CVD, including heart and vascular diseases, are leading causes of morbidity and mortality at different ages [135, 136]. In recent years, a tremendous amount of new genetic information related to CVD has been identified using next-generation sequencing technologies [19, 20]. Owing to the advent and development of the CRISPR-Cas system, we can now handle this information and determine CVD-related functions much more easily than ever. The CRISPR-Cas system also provides more possibilities for treating inherited CVD by correcting disease-causing mutations in the patient genome. As the most commonly used Cas proteins, SpCas9 and SaCas9 have been broadly applied in CVD-related modeling and therapeutic purposes, both in vitro [137–140] and in vivo [141–144]. Newly developed base editing and prime editing systems have also been used [145, 146]. Additionally, delivering CRISPR-Cas components to the cardiovascular system remains challenging, and AAV-based systems are currently the most widely used methods [147–150].
Modeling CVD using CRISPR
Genetic studies have identified various pathogenic genetic variants associated with the occurrence of CVD [151, 152]. Revealing the consequences of specific mutations in CVD-related genes is important for CVD genetic diagnosis and precise medicine. CVD models have played a critical role in establishing causal links between genetic variants and CVD, dissecting the molecular mechanisms underlying CVD, validating therapeutic targets, and preclinical evaluation of therapeutic agents. At present, multiple gene editing tools have been applied to create in vitro and in vivo models of CVD [151].
In vitro models of CVD
Human induced pluripotent stem cells (hiPSCs) are promising for modeling human cardiomyopathies in vitro because they can differentiate into cardiomyocytes [153]. Through genome editing of hiPSCs followed by their differentiation into cardiomyocytes (hiPSC-CMs), isogenic hiPSC-CMs have been broadly used to verify causative genes or mutations in cardiomyopathies. Gene disruption induced by CRISPR-Cas9 in hiPSC-CMs is straightforward and suitable for determining the role of a gene in CVD. For example, DNA methyltransferase 3A (DNMT3A) gene-deleted hiPSC-CMs generated using CRISPR-Cas9 gene editing showed altered contraction kinetics and impaired glucose/lipid metabolism, suggesting an important role of DNA methylation in cardiac diseases [137]. The homozygous SCN10A gene (encoding NaV1.8) knockout hiPSC-CMs help demonstrate that the voltage-gated sodium channel Nav1.8 contributes to late Na+ current (INaL) formation and displays a harmful proarrhythmogenic function [138].
The precise introduction of point mutations into hiPSC-CMs facilitates the determination of the causal relationship between genetic mutations and heart diseases. Striated muscle-enriched protein kinase (SPEG) E1680K homozygous mutant hiPSC-CMs recapitulate the hallmarks of dilated cardiomyopathy (DCM), confirming that SPEG E1680K is a novel DCM-causing mutation [139]. Genome editing of hiPSCs has also been used to identify several causative mutations of arrhythmias. hiPSC-CMs expressing an R211H substitution in the Ras-related associated with diabetes (RRAD) gene mimic the single-cell electrophysiological characteristics of Brugada syndrome, a disorder predisposing the patient to ventricular arrhythmias, indicating that RRAD is possibly a novel susceptibility gene for Brugada syndrome [154]. CRISPR-Cas9-engineered hiPSC-CMs carrying three different mutations of ryanodine receptor 2 (RyR2), R420Q, Q4201R, or F2483I, exhibit various pathological features of catecholaminergic polymorphic ventricular tachycardia 1 (CPVT1)-associated arrhythmia, suggesting that different RyR2 mutations cause varied Ca2+ signaling consequences and drug sensitivities [155]. Genome-edited hiPSC-CMs can also be used as high-throughput platforms for scalable functional validation of the pathogenicity and pathophysiology of genetic variants identified in the human population. To determine the functional significance of cardiac troponin T (TNNT2) variants, the endogenous TNNT2 gene was knocked out in hiPSCs using CRISPR-Cas9, and 51 different TNNT2 variants were expressed using lentivirus in differentiated TNNT2 knockout hiPSC-CMs. The results revealed that various TNNT2 variants exhibit different pathogenic mechanisms, greatly expanding the knowledge of which and how TNNT2 variants cause hypertrophic cardiomyopathy (HCM) and DCM [156].
Genome editing has been used to correct mutations to generate optimal isogenic controls for patient-derived iPSC-CMs, enabling the determination of genotype–phenotype relationships more precisely. Genome editing has been performed to correct the missense mutation T618I in the potassium channel gene KCNH2 in short QT syndrome patience-specific hiPSC-CMs to elucidate the single-cell phenotype of short QT syndrome [157]. Using iPSC-CMs derived from doxorubicin-treated pediatric patients, cytosine base editing has helped identify the single nucleotide polymorphism rs11140490 in the SLC28A3 locus, which is a novel protector against doxorubicin-induced cardiotoxicity [145]. Isogenic hiPSC-CM controls generated by CRISPR-based gene correction have also been used as platforms to evaluate other therapeutic methods. Type 1 long QT syndrome is caused by loss-of-function variants in the KCNQ1-encoded Kv7.1 potassium channel α-subunit. iPSC-CMs generated from patients with KCNQ1-V254M and -A344A/spl mutations have recently been corrected using CRISPR-Cas9 to act as isogenic controls, which have been used to evaluate a dual-component suppression-and-replacement gene therapy method [158]. Base editing and prime editing could possibly be widely used in establishing hiPSC-CM-based CVD models in the near future.
Animal models of CVD
CRISPR-based germline genome editing tools have revolutionized the generation of genetically modified animal models of CVD. Compared to conventional gene targeting technologies using embryonic stem cells, CRISPR-based gene editing technologies are easier to operate, faster, and applicable to most species. One strategy for generating animal models of CVD is to introduce targeted point mutations, insertions, or deletions using HDR-mediated germline genome editing. A mouse model of HCM with a Myh6 R404Q mutation was generated using SpCas9/ssODN-mediated directed genomic DNA editing, and heterozygous mice developed a typical HCM phenotype [141]. A similar approach was also utilized to insert an additional adenine nucleotide into the lysosomal acid alpha-glucosidase (Gaa) gene at the c.1826 locus and generate a novel mouse model of infantile-onset Pompe disease (IOPD), which recapitulates HCM and the skeletal muscle weakness of human IOPD [142]. A CRISPR-Cas9-generated rat model, with a 9 bp deletion within the hotspot analogous to the novel mutation of the human PDE3A gene, recapitulates arterial hypertension with brachydactyly, demonstrating that mutant PDE3A causes arterial hypertension [143]. Recently, a 94 bp out of frame deletion was generated in exon 1 of Kcnk3 using SpCas9/ssODN-mediated genome editing, creating a novel rat model of pulmonary arterial hypertension [159]. Another strategy is to delete exon(s) using two sgRNAs flanking specific exon(s). Exon deletion mutations in the dystrophin are among the most common causes of Duchenne muscular dystrophy (DMD). Several mouse models of DMD have been generated using CRISPR-Cas9 genome editing [144, 160], which are discussed further in the next section. CRISPR-Cas9 mediated mosaic inactivation of zebrafish ccm2 led to a lethal multi-cavernous lesion that histologically mimics the typical human hemorrhagic cerebral cavernous malformation [161].
Compared with germline genome editing, somatic genome editing is a more flexible method for obtaining CVD models, which overcomes the challenges of germline modification, such as embryonic lethality and the cost and time required to establish, reproduce, and maintain these models. It is also suitable for rapid and relatively high-throughput studies on the functions of CVD-related genes. As early as 2016, a cardiomyocyte-specific SpCas9 transgenic mouse model was successfully generated to achieve somatic editing in the heart [162]. Following this study, intraperitoneal injection of AAV9 encoding sgRNA against three genes critical for the heart, Myh6, Sav1, and Tbx20, in postnatal cardiomyocyte-Cas9 transgenic mice caused a similar degree of DNA disruption and subsequent mRNA downregulation, but only Myh6 disruption induced HCM and heart failure, suggesting that the effect of postnatal cardiac genome editing is target-dependent [147]. Mouse models can also be generated by activating endogenous gene expression through CRISPR-mediated genome editing in the postnatal heart [163]. CRISPR-mediated endogenous activation of myocyte enhancer factor 2D (Mef2d) leads to cardiac hypertrophy in mice, indicating that CRISPR-mediated genome editing can be used to generate CVD mouse models by controlling transcription in the postnatal heart [163]. Several recent studies have shown that CRISPR-Cas9 can be used to edit endothelial genes in vivo to obtain vascular disease models and enable reverse genetic studies of gene function in the mammalian vascular endothelium. Co-injection of an adenovirus harboring sgRNAs targeting the Alk1 gene and AAV1-VEGF successfully induced mutations in Alk1 in brain ECs and generated brain arteriovenous malformations in adult mice [164]. We recently generated a blood–brain barrier (BBB) breakdown mouse model by AAV-BR1-CRISPR mediated somatic genome editing. A single intravenous administration of brain microvascular EC targeting AAV-BR1 encoding sgRNA against the β-catenin (Ctnnb1) gene resulted in a mutation of 36.1% of the Ctnnb1 alleles and dramatically decreased levels of CTNNB1 in brain ECs, leading to BBB breakdown in EC-restricted Tie2Cas9 mice [148]. The AAV-BR1-CRISPR system established in this study allowed for the rapid construction of BBB perturbation models in vivo and may be helpful for developing drug delivery systems in the central nervous system. Recently, the nanoparticle-mediated delivery of CRISPR plasmid DNA expressing Cas9 under the control of the Cdh5 promoter resulted in efficient genome editing in the ECs of the peripheral vasculature in adult mice, which provides a powerful tool to construct animal models of peripheral vascular diseases [165].
Genome editing in CVD treatment
Therapeutic genome editing can be used to treat monogenic CVD, and the technology could permanently correct mutations and eventually eradicate specific CVD. Programmed edits were introduced into the human germline genome [166–169]. However, human germline genome editing faces significant ethical concerns and is prohibited in most countries [170]. Somatic editing is a promising technology for editing CVD-causing mutations without the risk of passing genomic changes to the offspring. Table 1 summarizes the latest applications of genome editing in treating different types of CVD.
Table 1.
Recent applications of genome editing in the treatment of CVD
| Diseases | Models | Mutations | Strategies | Outcomes | References |
|---|---|---|---|---|---|
| DMD |
Mice hiPSC-CMs |
Deletion of Dmd exon44 | Skipping and/or reframing of exon43 or 45 by single sgRNA targeting splice acceptor or donor site | Restore dystrophin expression and improve muscle function | [144] |
|
Mice hiPSC-CMs |
Deletion of Dmd exon43, 45 or 52 | Skipping and/or reframing of exon44 or 53 by single sgRNA targeting splice acceptor or donor site | Rescue the dystrophic phenotype, reduce necrotic cells and centralized nuclei | [171] | |
|
Pigs hiPSC-CMs |
Deletion of Dmd exon52 | Deletion of exon51 by dual sgRNAs targeting flanking sequences | Improve muscle function and mobility and prevent malignant arrhythmias | [172] | |
| Mice | A nonsense mutation in Dmd exon23 | Deletion of exon23 or 21–23 by dual sgRNAs targeting flanking sequences | Restore dystrophin expression and improve cardiac function without inducing serious adverse effects | [149, 150] | |
|
Mice hiPSC-CMs |
Deletion of Dmd exon51 | Skipping of exon50 by destroying splice donor site using ABEmax in vivo; reframing of exon52 by PE in vitro | Restore dystrophin protein expression and normalize contractile abnormalities | [146] | |
| Mice | A 4 bp deletion in Dmd exon4 | Skipping of exon4 by destroying splice donor site using CBE | Improve cardiac function and increase life span | [160] | |
| Mice | A nonsense mutation in Dmd exon53 | Correction of the stop codon by ABE | A near complete rescue of dystrophin in hearts without obvious toxicity | [173] | |
| HCM | Mouse embryos | A missense mutation in Myh6 c.1211C > T (p. Arg404Trp) | Correction of the mutant nucleotide by ABEmax | Abolish HCM phenotypes in mice born from edited embryos | [141] |
| hiPSC-CMs | Biallelic variants in LZTR1 (paternal c.1943-256C > T create a new splice donor site and maternal c.27dupG) | Disruption of the SNP-induced splice donor site in the paternal allele by single sgRNA | Reverse the NS-associated hypertrophic phenotype including cell size, contractile and electrophysiological properties | [174] | |
| Human pronuclear stage zygotes | A dominant 4 bp deletion in MYBPC3 exon16 | HDR by microinjection of sgRNA, Cas9 protein and ssODN | Heterozygous gene mutations can be corrected in human gametes or early embryos | [175] | |
| DCM | hiPSC-CMs | A 2 bp insertion in TTN exon327 c.70692_70693insAT | HDR by nucleofection of CRISPR/Cas9 RNP complex targeting the mutated allele and ssODN | Normalize titin protein level and contractile function | [140] |
| hiPSC-CMs | A 1 bp deletion in TTN exon276 | Reframing of the mutant allele by single sgRNA targeting the mutant site | Increase full-length TTN protein levels and sarcomere function | [176] | |
| hiPSC-CMs | Missense mutations in RBM20 c.1901G > A (p. Arg634Gln) and c.1906C > A (p. Arg636Ser) | Correction of the mutant nucleotide by ABEmax and PE | Restore RBM20 nuclear localization and eliminate RNP granules in the cytoplasm | [177] | |
| Mice | A missense mutation in Rbm20 c.1907C > A (p. Arg636Gln) | Correction of the mutant nucleotide by ABEmax | Restore RBM20 nuclear localization, eliminate RNP granules, restore cardiac size and function, and increase life span | [177] | |
| Cardiac arrhythmia | Humanized mice | A 3 bp deletion in PLN c.40_42AGA (p.Arg14del) | Disruption of the mutant allele by single sgRNA targeting the mutant site | Improve cardiac function and reduce sustained ventricular tachycardia susceptibility | [178] |
| Atherosclerosis | Mice | – | Disrupting liver Pcsk9 gene by adenovirus/AAV-delivered sgRNA targeting coding sequences | Disrupt Pcsk9 genes and chronically decrease serum cholesterol levels | [30, 179] |
|
Mice macaques cynomolgus monkeys |
– | Disruption of the splice donor site of liver PCSK9 exon1 by AAV/LNP/eVLP-delivered ABE | Reduce plasma PCSK9 and LDL levels | [180, 181] |
DMD Duchenne muscular dystrophy, hiPSC-CMs human induced pluripotent stem cell-derived cardiomyocytes, sgRNA single guide RNA, ABE adenine base editor, PE prime editor, CBE cytosine base editor, HCM hypertrophic cardiomyopathy, SNP single nucleotide polymorphism, NS Noonan syndrome, HDR homology-directed repair, Cas9 CRISPR-associated nuclease 9, ssODN single-stranded oligodeoxynucleotide, DCM dilated cardiomyopathy, CRISPR clustered regularly interspaced short palindromic repeats, RNP ribonucleoprotein complexes, AAV adeno-associated virus, LNP lipid nanoparticle, eVLP engineered virus-like particle, LDL low-density lipoprotein
DMD
DMD is an X-linked disorder characterized by proximal muscle weakness and cardiomyopathy caused by mutations in the largest human gene, dystrophin (DMD) [182]. A variety of mutations exist throughout the DMD gene, most of which are located in the regions crossing exons 43 to 53 and disrupt the ORF, resulting in non-functional truncated proteins.
A single-cut genome editing strategy was applied in both iPSC-CMs and mouse models of DMD bearing an exon 44 deletion mutation (Δ44), one of the most common causative mutations of DMD. The ORF can be restored by disrupting the exon splice site to skip the adjacent exon, inserting one nucleotide, or deleting two nucleotides in exon 44 [144]. Systemic delivery of gene editing components by a single dose of AAV9 restores ~ 90% dystrophin protein expression in the hearts of Δ44 mice within 4 weeks [144]. This approach also helps to correct DMD models bearing deletions of exons 43, 45, and 52 (Δ43, Δ45, and Δ52) both in vitro and in vivo [171]. A dual-AAV system was used to deliver SpCas9 and a single sgRNA targeting the splice donor site of exon 44 (for Δ43 or Δ45 mice) or splice acceptor site of exon 53 (for Δ52 mice) in vivo to restore dystrophin expression. Both exon skipping and reframing were induced in Δ45 and Δ52 mice, and the efficacy of dystrophin in these two models was higher than that in Δ43 mice, in which only exon skipping was generated [171]. Restoration of dystrophin has also been achieved in hiPSC-CMs from these DMD models [171]. However, this study did not specify whether exon skipping and/or reframing could subsequently rescue the cardiac phenotypes of DMD models [171]. Another strategy is to delete exon(s) using two sgRNAs flanking on either side, thus restoring the ORF of the DMD gene. The systemic application of AAV9 carrying an intein-split SpCas9 and a pair of sgRNAs targeting sequences flanking exon 51 in a pig model of DMD lacking exon 52 induced dystrophin expression in the heart and reduced arrhythmogenic vulnerability [172]. The long-term efficacy and safety of therapeutic editing for DMD have also been studied [149, 150]. AAV vectors carrying SaCas9 and a pair of sgRNAs targeting exon 23 or exons 21–23 were administrated for one year or 19 months, respectively, to mdx mice. Cardiac functions were improved without serious adverse effects, indicating that in vivo CRISPR genome editing may be a safe therapeutic strategy for DMD [149, 150].
Base editing and prime editing show great promise for treating DMD. Both ABE and PE can restore dystrophin protein expression by inducing exon skipping or exon reframing to correct the Dmd exon 51 deletion mutation in iPSC-CMs, and intramuscular delivery of AAV9 encoding ABE components amends the mutation in ∆E51 DMD mice [146]. CBE has been shown to rescue dystrophic cardiomyopathy in DmdE4* mice, which harbor a 4 bp deletion in exon 4 of the Dmd gene and recapitulate many characteristics of human DMD [160]. A single-dose administration of AAV9-eTAM encoding a fused nuclease-defective SaCas9 (KKH) with activation-induced cytidine deaminase (AID) and UGI, together with AAV9-sgRNA, efficiently induced splice site mutation and exon 4 skipping of the Dmd gene and restored up to 90% of dystrophin proteins in the heart of DmdE4* mice, resulting in improved cardiac function and an increased life span [160]. Alternatively, a dual AAV-mediated protein trans-splicing approach was used to deliver a modified ABE-NG to an mdx4cv mouse model carrying a premature stop codon (CAA-to-TAA) in exon 53 of the Dmd gene. After 10 months of treatment, a near-complete rescue of dystrophin was found in the hearts of mdx4cv mice without obvious toxicity [173].
HCM and DCM
Inherited cardiomyopathies, including HCM and DCM, are candidate genetic disorders that are suitable for genome editing-related treatment. ABEmax-NG has been shown to correct a pathogenic R404Q/+ mutation in embryos of the HCM mouse model [141]. Administration of ABEmax-NG mRNA to Myh6R404Q/+ embryos corrects the mutant allele at a rate of 62.5% to 70.8%, abolishing the HCM phenotype in postnatal mice and their progeny. Moreover, in utero delivery of intein-split ABEmax-NG induced a high correction rate without introducing indels or off-target editing in Myh6R404Q/+ fetuses [141]. Intronic CRISPR repair has been demonstrated as efficient in a preclinical iPSC-CM model of Noonan syndrome-associated HCM [174]. CRISPR-Cas9-mediated destruction of the mutation-induced additional intronic donor splice site can reverse the hypertrophic phenotypes in Noonan syndrome patient-derived iPSC-CMs carrying biallelic mutations in intron 16 of the leucine zipper-like transcription regulator 1 (LZTR1) gene, indicating new possibilities for personalized therapeutic genome editing in HCM patients [174]. Notably, CRISPR-based genome editing has been shown to have potential to correct a well-documented heterozygous dominant 4 bp deletion in exon 16 of MYBPC3, which causes familial HCM, in human embryos [175]. Co-injection of Cas9 proteins, mutation-specific sgRNAs, and mutant sperm into healthy metaphase II oocytes corrected the deletion by wild-type maternal allele-mediated HDR, resulting in a high yield of homozygous embryos carrying the wild-type MYBPC3 gene without mosaicism or off-target mutations [175].
Genome editing has also been used to correct DCM-causing mutations. In addition to previous studies using hiPSC-CMs showing that truncated titin (TTNtv) mutations are the most common causes of DCM [183–185], recently, the pathological mechanisms of TTNtv-associated DCM have been highlighted and a new genome editing strategy has been developed to treat TTNtv-associated DCM. iPSC-CMs with patient-derived or CRISPR-Cas9-generated TTN mutations were corrected using SpCas9/ssODN. Engineered heart muscle generated from corrected hiPSC-CMs shows normalized titin protein levels and contractile function [140]. Genome editing using SpCas9 and A-band TTNtv-specific sgRNA was also shown to restore the reading frame of TTN protein in hiPSC-CMs, leading to increased full-length TTN protein levels and normalized sarcomere function [176]. More recently, the application of precise genome editing technology for treating DCM caused by mutations in RBM20 has been reported [177]. The RBM20R634Q and RBM20R636S mutant iPSCs were corrected by ABE and PE, with the efficiency of 92% and 40%, respectively. In addition, AAV9-mediated systemic delivery of ABE components corrected 66% of the RBM20 transcripts expressed in cardiomyocytes of postnatal RBMR636Q/R636Q mice. The corrected mice showed restored cardiac size and function, and prolonged life span [177].
Cardiac arrhythmia
Cardiac arrhythmia caused by autosomal-dominant mutations can be treated with CRISPR-mediated specific disruption of the mutant allele, which has been validated in several mouse models [186, 187]. Recently, humanized mice expressing a human mutant PLN (hPLN-R14del) demonstrated bi-ventricular dilation and a higher propensity for sustained ventricular tachycardia. Disruption of the hPLN-R14del allele by AAV9-CRISPR-Cas9 improved cardiac function and reduced sustained ventricular tachycardia susceptibility in young adult humanized PLN-R14del mice, providing a potential therapeutic strategy for the arrhythmogenic phenotype in human patients with the PLN-R14del mutation [178].
Atherosclerosis
Atherosclerosis is a chronic disease that refers to the formation of fibrofatty lesions in the arterial wall, and causes ischemic stroke, ischemic cardiomyopathy, myocardial infarction, and peripheral arterial disease. Blood concentration of low-density lipoprotein cholesterol (LDL-C) is one of the best-established causal risk factors for atherosclerosis.
The lipid metabolism-related gene, Pcsk9, is specifically expressed in the liver and functions primarily as an antagonist to the LDL receptor. Disruption of PCSK9 activity can reduce circulating LDL-C levels, thereby lowering the risk of atherosclerosis [188]. Several clinical trials have investigated monoclonal antibodies targeting PCSK9. However, even if these antibody-based drugs are effective, their effect on LDL-C is short-lived. Genome editing using CRISPR systems provides an alternative method for reducing PCSK9 levels. A single administration of adenovirus co-expressing SpCas9 and sgRNA targeting exon 1 of the mouse Pcsk9 gene can efficiently introduce loss-of-function mutations into endogenous Pcsk9 genes in vivo and chronically decrease plasma cholesterol levels in the blood [179]. The AAV-SaCas9 system has also been proven to be effective in editing the Pcsk9 gene in vivo, leading to significantly decreased serum PCSK9 and cholesterol levels [30]. Single injections of engineered DNA-free VLPs targeting the Pcsk9 gene into adult mice demonstrated 63% base editing in the liver, resulting in 78% reduction in serum PCSK9 levels [126]. In addition to these studies carried out in rodents, somatic Pcsk9 gene editing has also been validated in nonhuman primates [180, 181]. CRISPR base editors delivered using LNPs proved highly effective in editing the Pcsk9 gene in the liver of macaques and cynomolgus monkeys. A single-dose treatment of LNPs carrying CRISPR base editors leads to stable Pcsk9 knockdown in the liver and a 60% reduction in blood LDL-C for at least 8 months [180]. To ensure the safety of gene editing in comparatively more proliferative organs, such as the liver, the proportion of edited cells that remain stable over time must be investigated. All these genome editing approaches offer the potential for once-and-done therapies for the lifelong treatment of atherosclerosis-associated CVD.
Perspectives
CRISPR-based genome editing technology has been rapidly applied in almost all fields, from basic biology to translational medicine. The development of novel systems and tools for more accurate, efficient, and faster genome editing and tighter control of the duration, efficiency, and specificity of genome editors will further benefit their translational applications. Newly uncovered thousands of phage-encoded CRISPR systems provide a valuable resource for searching novel miniature single-effector CRISPR-Cas systems [39]. In addition, newly developed Cas13a-based RNA editing tools can achieve RNA knockdown and precise base editing of mammalian transcripts without causing DNA damage, providing a promising potential therapeutic strategy in translational cardiovascular medicine [11, 189]. Notably, type III-E CRISPR-Cas7-11 effector has recently been shown to cleavage protein under target RNA guidance [190, 191], bringing new potential CRISPR tools for CVD diagnosis and treatment.
Genome editing technologies have been successfully translated into human clinical trials for enhanced chimeric antigen receptor (CAR) T-cell therapy, cell-based regenerative medicine, and treatment of monogenic diseases, such as transfusion-dependent beta thalassemia (TDT) and sickle cell disease (SCD) [192, 193]. Researchers have used in vivo genome editing to target the transthyretin (TTR) gene to treat transthyretin amyloidosis and have achieved very encouraging results in phase 1 clinical trials, taking the most critical step towards applying CRISPR-based genome editing technology to treat human genetic diseases [123]. Taking the most optimistic view, CVD with known causal genes can theoretically be treated with CRISPR technology. However, there are still several important challenges. Recently, CRISPR-based genome editing in human embryos was shown to cause unpredictable genomic alterations, including DNA rearrangements, large deletions, and even loss of allele-specific chromosomes [168, 194, 195]. Therefore, potential technical safety concerns, including mosaicism, off-target effects, and long-term risks caused by genome editing, need to be addressed before the therapeutic applications of CRISPR technology in treating CVD. Perhaps striking a balance between the efficiency and safety of genome editing is crucial. At present, efficient delivery of CRISPR-Cas systems to human cardiovascular system remains a challenge. In addition, the efficacy and safety of each therapeutic gene editing strategy for each CVD need to be confirmed by clinical trials. Although this paper uses the CVD as example to illustrate the progress of CRISPR-based genome editing in modeling and treating diseases, the same strategies could also be used for numerous diseases in other tissues.
Like any cutting-edge technology, gene editing technology could be a double-edged sword. Genome editing has been listed as a potential weapon of mass destruction in the 2016 annual worldwide threat assessment report of the U.S. intelligence community, indicating a high risk of extreme misuse. Recent rapid advances have made genome editing technologies more accessible and difficult to control, which may further lower the threshold for genome editing misuse and increase biosecurity threats. The possible misuse of genome editing technology and biosecurity risks may include, but are not limited to creating (1) pathogens with increased virulence, (2) new pathogens and biotoxins, and (3) gene-driven animals that may have irreversible effects on specific populations and the environment. Regulations and guidelines should be developed after extensive consultation to ensure that the development of gene editing technologies will not harm living organisms, including humans, or the environment.
Conclusions
The emerging novel Cas nucleases and their extended applications have greatly expanded the CRISPR-based genome editing toolbox and promoted the development of life science and medicine. CRISPR-based genome editing technology has also revolutionized cardiovascular research, accelerating the generation of genetically modified models of CVD and its application in the treatment of different types of CVD. However, this technology may also bring huge potential biological threats, which should be strictly controlled to prevent its abuse.
Abbreviations
- AAV
Adeno-associated virus
- ABE
Adenine base editor
- BBB
Blood–brain barrier
- CRISPR
Clustered regularly interspaced short palindromic repeats
- Cas
CRISPR-associated protein
- CVD
Cardiovascular disease
- Cascade
CRISPR-associated complex for antiviral defense
- crRNA
CRISPR RNA
- CBE
Cytosine base editor
- CGBE
C-to-G base editor
- CAST
CRISPR-associated transposon
- CAR
Chimeric antigen receptor
- DCM
Dilated cardiomyopathy
- DMD
Duchenne muscular dystrophy
- DSB
Double-strand break
- EC
Endothelial cell
- FiCAT
Find and cut-and-transfer
- hiPSC
Human induced pluripotent stem cell
- HCM
Hypertrophic cardiomyopathy
- HDR
Homology-directed repair
- LNP
Lipid nanoparticle
- LDL-C
Low-density lipoprotein cholesterol
- M-MLV
Moloney murine leukemia virus
- NHEJ
Non-homologous end-joining
- ORF
Open reading frame
- OMEGA
Obligate mobile element-guided activity
- PAM
Protospacer adjacent motif
- PE
Prime editor
- pegRNA
Prime editing gRNA
- RNP
Ribonucleoprotein complexes
- sgRNA
Single guide RNA
- ssODN
Single-stranded oligodeoxynucleotide
- SCD
Sickle cell disease
- tracrRNA
Trans-activating CRISPR RNA
- TAM
Transposon-associated motif
- TDT
Transfusion-dependent beta thalassemia
- UGI
Uracil-DNA glycosylase inhibitor
- UNG
Uracil-DNA glycosylase
- VLP
Virus-like particle
Author contributions
ZHL, JW, JW and XY drafted the manuscript. ZHL and JPX prepared figures. JW and XY conceived and supervised the study. All authors have read and approved the final manuscript.
Funding
This work was supported by the National Natural Science Foundation of China (82270355, 82270354, 81970134, 82030011, 31630093), and the National Key Research and Development Program of China (2019YFA0801601, 2021YFA1101801).
Availability of data and materials
Not applicable.
Declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Zhen-Hua Li and Jun Wang contributed equally to this work
Contributor Information
Zhen-Hua Li, Email: lzhh639@163.com.
Jun Wang, Email: wangjun227@126.com.
Jing-Ping Xu, Email: xujping@126.com.
Jian Wang, Email: wangjian7773@126.com.
Xiao Yang, Email: yangx@bmi.ac.cn.
References
- 1.Doudna JA, Charpentier E. Genome editing. The new frontier of genome engineering with CRISPR-Cas9. Science. 2014;346(6213):1258096. doi: 10.1126/science.1258096. [DOI] [PubMed] [Google Scholar]
- 2.Doudna JA. The promise and challenge of therapeutic genome editing. Nature. 2020;578(7794):229–236. doi: 10.1038/s41586-020-1978-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nambiar TS, Baudrier L, Billon P, Ciccia A. CRISPR-based genome editing through the lens of DNA repair. Mol Cell. 2022;82(2):348–388. doi: 10.1016/j.molcel.2021.12.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hsu PD, Lander ES, Zhang F. Development and applications of CRISPR-Cas9 for genome engineering. Cell. 2014;157(6):1262–1278. doi: 10.1016/j.cell.2014.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Komor AC, Badran AH, Liu DR. CRISPR-based technologies for the manipulation of eukaryotic genomes. Cell. 2017;169(3):559. doi: 10.1016/j.cell.2017.04.005. [DOI] [PubMed] [Google Scholar]
- 6.Knott GJ, Doudna JA. CRISPR-Cas guides the future of genetic engineering. Science. 2018;361(6405):866–869. doi: 10.1126/science.aat5011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li G, Li X, Zhuang S, Wang L, Zhu Y, Chen Y, et al. Gene editing and its applications in biomedicine. Sci China Life Sci. 2022;65(4):660–700. doi: 10.1007/s11427-021-2057-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yang X. Applications of CRISPR-Cas9 mediated genome engineering. Mil Med Res. 2015;2:11. doi: 10.1186/s40779-015-0038-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, et al. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339(6121):819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu G, Lin Q, Jin S, Gao C. The CRISPR-Cas toolbox and gene editing technologies. Mol Cell. 2022;82(2):333–347. doi: 10.1016/j.molcel.2021.12.002. [DOI] [PubMed] [Google Scholar]
- 11.Pickar-Oliver A, Gersbach CA. The next generation of CRISPR-Cas technologies and applications. Nat Rev Mol Cell Biol. 2019;20(8):490–507. doi: 10.1038/s41580-019-0131-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Porteus MH. A new class of medicines through DNA editing. N Engl J Med. 2019;380(10):947–959. doi: 10.1056/NEJMra1800729. [DOI] [PubMed] [Google Scholar]
- 13.Yeh CD, Richardson CD, Corn JE. Advances in genome editing through control of DNA repair pathways. Nat Cell Biol. 2019;21(12):1468–1478. doi: 10.1038/s41556-019-0425-z. [DOI] [PubMed] [Google Scholar]
- 14.Clark JF, Dinsmore CJ, Soriano P. A most formidable arsenal: genetic technologies for building a better mouse. Genes Dev. 2020;34(19–20):1256–1286. doi: 10.1101/gad.342089.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nishiga M, Liu C, Qi LS, Wu JC. The use of new CRISPR tools in cardiovascular research and medicine. Nat Rev Cardiol. 2022;19(8):505–521. doi: 10.1038/s41569-021-00669-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Komor AC, Kim YB, Packer MS, Zuris JA, Liu DR. Programmable editing of a target base in genomic DNA without double-stranded DNA cleavage. Nature. 2016;533(7603):420–424. doi: 10.1038/nature17946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gaudelli NM, Komor AC, Rees HA, Packer MS, Badran AH, Bryson DI, et al. Programmable base editing of A·T to G·C in genomic DNA without DNA cleavage. Nature. 2017;551(7681):464–471. doi: 10.1038/nature24644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Anzalone AV, Randolph PB, Davis JR, Sousa AA, Koblan LW, Levy JM, et al. Search-and-replace genome editing without double-strand breaks or donor DNA. Nature. 2019;576(7785):149–157. doi: 10.1038/s41586-019-1711-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Watkins WS, Hernandez EJ, Wesolowski S, Bisgrove BW, Sunderland RT, Lin E, et al. De novo and recessive forms of congenital heart disease have distinct genetic and phenotypic landscapes. Nat Commun. 2019;10(1):4722. doi: 10.1038/s41467-019-12582-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jin SC, Homsy J, Zaidi S, Lu Q, Morton S, Depalma SR, et al. Contribution of rare inherited and de novo variants in 2,871 congenital heart disease probands. Nat Genet. 2017;49(11):1593–1601. doi: 10.1038/ng.3970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Makarova KS, Wolf YI, Alkhnbashi OS, Costa F, Shah SA, Saunders SJ, et al. An updated evolutionary classification of CRISPR-Cas systems. Nat Rev Microbiol. 2015;13(11):722–736. doi: 10.1038/nrmicro3569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dolan AE, Hou Z, Xiao Y, Gramelspacher MJ, Heo J, Howden SE, et al. Introducing a spectrum of long-range genomic deletions in human embryonic stem cells using type I CRISPR-Cas. Mol Cell. 2019;74(5):936–50.e5. doi: 10.1016/j.molcel.2019.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Morisaka H, Yoshimi K, Okuzaki Y, Gee P, Kunihiro Y, Sonpho E, et al. CRISPR-Cas3 induces broad and unidirectional genome editing in human cells. Nat Commun. 2019;10(1):5302. doi: 10.1038/s41467-019-13226-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Osakabe K, Wada N, Murakami E, Miyashita N, Osakabe Y. Genome editing in mammalian cells using the CRISPR type I-D nuclease. Nucleic Acids Res. 2021;49(11):6347–6363. doi: 10.1093/nar/gkab348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tan R, Krueger RK, Gramelspacher MJ, Zhou X, Xiao Y, Ke A, et al. Cas11 enables genome engineering in human cells with compact CRISPR-Cas3 systems. Mol Cell. 2022;82(4):852–67.e5. doi: 10.1016/j.molcel.2021.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu TY, Doudna JA. Chemistry of class 1 CRISPR-Cas effectors: binding, editing, and regulation. J Biol Chem. 2020;295(42):14473–14487. doi: 10.1074/jbc.REV120.007034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Altae-Tran H, Kannan S, Demircioglu FE, Oshiro R, Nety SP, Mckay LJ, et al. The widespread IS200/IS605 transposon family encodes diverse programmable RNA-guided endonucleases. Science. 2021;374(6563):57–65. doi: 10.1126/science.abj6856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Karvelis T, Druteika G, Bigelyte G, Budre K, Zedaveinyte R, Silanskas A, et al. Transposon-associated TnpB is a programmable RNA-guided DNA endonuclease. Nature. 2021;599(7886):692–696. doi: 10.1038/s41586-021-04058-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schuler G, Hu C, Ke A. Structural basis for RNA-guided DNA cleavage by IscB-ωRNA and mechanistic comparison with Cas9. Science. 2022;376(6600):1476–1481. doi: 10.1126/science.abq7220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ran FA, Cong L, Yan WX, Scott DA, Gootenberg JS, Kriz AJ, et al. In vivo genome editing using Staphylococcus aureus Cas9. Nature. 2015;520(7546):186–191. doi: 10.1038/nature14299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hou Z, Zhang Y, Propson NE, Howden SE, Chu LF, Sontheimer EJ, et al. Efficient genome engineering in human pluripotent stem cells using Cas9 from Neisseria meningitidis. Proc Natl Acad Sci U S A. 2013;110(39):15644–15649. doi: 10.1073/pnas.1313587110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kim E, Koo T, Park SW, Kim D, Kim K, Cho HY, et al. In vivo genome editing with a small Cas9 orthologue derived from Campylobacter jejuni. Nat Commun. 2017;8:14500. doi: 10.1038/ncomms14500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Edraki A, Mir A, Ibraheim R, Gainetdinov I, Yoon Y, Song CQ, et al. A compact, high-accuracy Cas9 with a dinucleotide PAM for in vivo genome editing. Mol Cell. 2019;73(4):714–26 e4. doi: 10.1016/j.molcel.2018.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liu JJ, Orlova N, Oakes BL, Ma E, Spinner HB, Baney KLM, et al. CasX enzymes comprise a distinct family of RNA-guided genome editors. Nature. 2019;566(7743):218–223. doi: 10.1038/s41586-019-0908-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kim DY, Lee JM, Moon SB, Chin HJ, Park S, Lim Y, et al. Efficient CRISPR editing with a hypercompact Cas12f1 and engineered guide RNAs delivered by adeno-associated virus. Nat Biotechnol. 2022;40(1):94–102. doi: 10.1038/s41587-021-01009-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wu Z, Zhang Y, Yu H, Pan D, Wang Y, Wang Y, et al. Programmed genome editing by a miniature CRISPR-Cas12f nuclease. Nat Chem Biol. 2021;17(11):1132–1138. doi: 10.1038/s41589-021-00868-6. [DOI] [PubMed] [Google Scholar]
- 37.Xu X, Chemparathy A, Zeng L, Kempton HR, Shang S, Nakamura M, et al. Engineered miniature CRISPR-Cas system for mammalian genome regulation and editing. Mol Cell. 2021;81(20):4333–45.e4. doi: 10.1016/j.molcel.2021.08.008. [DOI] [PubMed] [Google Scholar]
- 38.Pausch P, Al-Shayeb B, Bisom-Rapp E, Tsuchida CA, Li Z, Cress BF, et al. CRISPR-CasΦ from huge phages is a hypercompact genome editor. Science. 2020;369(6501):333–337. doi: 10.1126/science.abb1400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Al-Shayeb B, Skopintsev P, Soczek KM, Stahl EC, Li Z, Groover E, et al. Diverse virus-encoded CRISPR-Cas systems include streamlined genome editors. Cell. 2022;185(24):4574–86.e16. doi: 10.1016/j.cell.2022.10.020. [DOI] [PubMed] [Google Scholar]
- 40.Collias D, Beisel CL. CRISPR technologies and the search for the PAM-free nuclease. Nat Commun. 2021;12(1):555. doi: 10.1038/s41467-020-20633-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Miller SM, Wang T, Randolph PB, Arbab M, Shen MW, Huang TP, et al. Continuous evolution of SpCas9 variants compatible with non-G PAMs. Nat Biotechnol. 2020;38(4):471–481. doi: 10.1038/s41587-020-0412-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Walton RT, Christie KA, Whittaker MN, Kleinstiver BP. Unconstrained genome targeting with near-PAMless engineered CRISPR-Cas9 variants. Science. 2020;368(6488):290–296. doi: 10.1126/science.aba8853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kleinstiver BP, Sousa AA, Walton RT, Tak YE, Hsu JY, Clement K, et al. Engineered CRISPR-Cas12a variants with increased activities and improved targeting ranges for gene, epigenetic and base editing. Nat Biotechnol. 2019;37(3):276–282. doi: 10.1038/s41587-018-0011-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tóth E, Varga É, Kulcsár PI, Kocsis-Jutka V, Krausz SL, Nyeste A, et al. Improved LbCas12a variants with altered PAM specificities further broaden the genome targeting range of Cas12a nucleases. Nucleic Acids Res. 2020;48(7):3722–3733. doi: 10.1093/nar/gkaa110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chatterjee P, Jakimo N, Lee J, Amrani N, Rodriguez T, Koseki SRT, et al. An engineered ScCas9 with broad PAM range and high specificity and activity. Nat Biotechnol. 2020;38(10):1154–1158. doi: 10.1038/s41587-020-0517-0. [DOI] [PubMed] [Google Scholar]
- 46.Chatterjee P, Lee J, Nip L, Koseki SRT, Tysinger E, Sontheimer EJ, et al. A Cas9 with PAM recognition for adenine dinucleotides. Nat Commun. 2020;11(1):2474. doi: 10.1038/s41467-020-16117-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ma D, Xu Z, Zhang Z, Chen X, Zeng X, Zhang Y, et al. Engineer chimeric Cas9 to expand PAM recognition based on evolutionary information. Nat Commun. 2019;10(1):560. doi: 10.1038/s41467-019-08395-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Liu RM, Liang LL, Freed E, Chang H, Oh E, Liu ZY, et al. Synthetic chimeric nucleases function for efficient genome editing. Nat Commun. 2019;10(1):5524. doi: 10.1038/s41467-019-13500-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Slaymaker IM, Gao L, Zetsche B, Scott DA, Yan WX, Zhang F. Rationally engineered Cas9 nucleases with improved specificity. Science. 2016;351(6268):84–88. doi: 10.1126/science.aad5227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kleinstiver BP, Pattanayak V, Prew MS, Tsai SQ, Nguyen NT, Zheng Z, et al. High-fidelity CRISPR-Cas9 nucleases with no detectable genome-wide off-target effects. Nature. 2016;529(7587):490–495. doi: 10.1038/nature16526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chen JS, Dagdas YS, Kleinstiver BP, Welch MM, Sousa AA, Harrington LB, et al. Enhanced proofreading governs CRISPR-Cas9 targeting accuracy. Nature. 2017;550(7676):407–410. doi: 10.1038/nature24268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Casini A, Olivieri M, Petris G, Montagna C, Reginato G, Maule G, et al. A highly specific SpCas9 variant is identified by in vivo screening in yeast. Nat Biotechnol. 2018;36(3):265–271. doi: 10.1038/nbt.4066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Vakulskas CA, Dever DP, Rettig GR, Turk R, Jacobi AM, Collingwood MA, et al. A high-fidelity Cas9 mutant delivered as a ribonucleoprotein complex enables efficient gene editing in human hematopoietic stem and progenitor cells. Nat Med. 2018;24(8):1216–1224. doi: 10.1038/s41591-018-0137-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lee JK, Jeong E, Lee J, Jung M, Shin E, Kim YH, et al. Directed evolution of CRISPR-Cas9 to increase its specificity. Nat Commun. 2018;9(1):3048. doi: 10.1038/s41467-018-05477-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tan Y, Chu AHY, Bao S, Hoang DA, Kebede FT, Xiong W, et al. Rationally engineered Staphylococcus aureus Cas9 nucleases with high genome-wide specificity. Proc Natl Acad Sci U S A. 2019;116(42):20969–20976. doi: 10.1073/pnas.1906843116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Xie H, Ge X, Yang F, Wang B, Li S, Duan J, et al. High-fidelity SaCas9 identified by directional screening in human cells. PLoS Biol. 2020;18(7):e3000747. doi: 10.1371/journal.pbio.3000747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bak SY, Jung Y, Park J, Sung K, Jang HK, Bae S, et al. Quantitative assessment of engineered Cas9 variants for target specificity enhancement by single-molecule reaction pathway analysis. Nucleic Acids Res. 2021;49(19):11312–11322. doi: 10.1093/nar/gkab858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kim N, Kim HK, Lee S, Seo JH, Choi JW, Park J, et al. Prediction of the sequence-specific cleavage activity of Cas9 variants. Nat Biotechnol. 2020;38(11):1328–1336. doi: 10.1038/s41587-020-0537-9. [DOI] [PubMed] [Google Scholar]
- 59.Schmid-Burgk JL, Gao L, Li D, Gardner Z, Strecker J, Lash B, et al. Highly parallel profiling of Cas9 variant specificity. Mol Cell. 2020;78(4):794–800.e8. doi: 10.1016/j.molcel.2020.02.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bravo JPK, Liu MS, Hibshman GN, Dangerfield TL, Jung K, Mccool RS, et al. Structural basis for mismatch surveillance by CRISPR-Cas9. Nature. 2022;603(7900):343–347. doi: 10.1038/s41586-022-04470-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhang L, Rube HT, Vakulskas CA, Behlke MA, Bussemaker HJ, Pufall MA. Systematic in vitro profiling of off-target affinity, cleavage and efficiency for CRISPR enzymes. Nucleic Acids Res. 2020;48(9):5037–5053. doi: 10.1093/nar/gkaa231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Tsuchida CA, Zhang S, Doost MS, Zhao Y, Wang J, O’brien E, et al. Chimeric CRISPR-CasX enzymes and guide RNAs for improved genome editing activity. Mol Cell. 2022;82(6):1199–209.e6. doi: 10.1016/j.molcel.2022.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Richardson CD, Ray GJ, Dewitt MA, Curie GL, Corn JE. Enhancing homology-directed genome editing by catalytically active and inactive CRISPR-Cas9 using asymmetric donor DNA. Nat Biotechnol. 2016;34(3):339–344. doi: 10.1038/nbt.3481. [DOI] [PubMed] [Google Scholar]
- 64.Maruyama T, Dougan SK, Truttmann MC, Bilate AM, Ingram JR, Ploegh HL. Increasing the efficiency of precise genome editing with CRISPR-Cas9 by inhibition of nonhomologous end joining. Nat Biotechnol. 2015;33(5):538–542. doi: 10.1038/nbt.3190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Nishiyama J, Mikuni T, Yasuda R. Virus-mediated genome editing via homology-directed repair in mitotic and postmitotic cells in mammalian brain. Neuron. 2017;96(4):755–68.e5. doi: 10.1016/j.neuron.2017.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ishizu T, Higo S, Masumura Y, Kohama Y, Shiba M, Higo T, et al. Targeted genome replacement via homology-directed repair in non-dividing cardiomyocytes. Sci Rep. 2017;7(1):9363. doi: 10.1038/s41598-017-09716-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zheng Y, Vandusen NJ, Butler CE, Ma Q, King JS, Pu WT. Efficient in vivo homology-directed repair within cardiomyocytes. Circulation. 2022;145(10):787–789. doi: 10.1161/CIRCULATIONAHA.120.052383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kosicki M, Tomberg K, Bradley A. Repair of double-strand breaks induced by CRISPR-Cas9 leads to large deletions and complex rearrangements. Nat Biotechnol. 2018;36(8):765–771. doi: 10.1038/nbt.4192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Cullot G, Boutin J, Toutain J, Prat F, Pennamen P, Rooryck C, et al. CRISPR-Cas9 genome editing induces megabase-scale chromosomal truncations. Nat Commun. 2019;10(1):1136. doi: 10.1038/s41467-019-09006-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Liu M, Zhang W, Xin C, Yin J, Shang Y, Ai C, et al. Global detection of DNA repair outcomes induced by CRISPR-Cas9. Nucleic Acids Res. 2021;49(15):8732–8742. doi: 10.1093/nar/gkab686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Yu C, Liu Y, Ma T, Liu K, Xu S, Zhang Y, et al. Small molecules enhance CRISPR genome editing in pluripotent stem cells. Cell Stem Cell. 2015;16(2):142–147. doi: 10.1016/j.stem.2015.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Riesenberg S, Maricic T. Targeting repair pathways with small molecules increases precise genome editing in pluripotent stem cells. Nat Commun. 2018;9(1):2164. doi: 10.1038/s41467-018-04609-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Genomes Project C. Auton A, Brooks LD, Durbin RM, Garrison EP, Kang HM, et al. A global reference for human genetic variation. Nature. 2015;526(7571):68–74. doi: 10.1038/nature15393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Eichler EE. Genetic variation, comparative genomics, and the diagnosis of disease. N Engl J Med. 2019;381(1):64–74. doi: 10.1056/NEJMra1809315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Anzalone AV, Koblan LW, Liu DR. Genome editing with CRISPR-Cas nucleases, base editors, transposases and prime editors. Nat Biotechnol. 2020;38(7):824–844. doi: 10.1038/s41587-020-0561-9. [DOI] [PubMed] [Google Scholar]
- 76.Despres PC, Dube AK, Seki M, Yachie N, Landry CR. Perturbing proteomes at single residue resolution using base editing. Nat Commun. 2020;11(1):1871. doi: 10.1038/s41467-020-15796-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Hanna RE, Hegde M, Fagre CR, Deweirdt PC, Sangree AK, Szegletes Z, et al. Massively parallel assessment of human variants with base editor screens. Cell. 2021;184(4):1064–80.e20. doi: 10.1016/j.cell.2021.01.012. [DOI] [PubMed] [Google Scholar]
- 78.Cuella-Martin R, Hayward SB, Fan X, Chen X, Huang JW, Taglialatela A, et al. Functional interrogation of DNA damage response variants with base editing screens. Cell. 2021;184(4):1081–97.e19. doi: 10.1016/j.cell.2021.01.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Komor AC, Zhao KT, Packer MS, Gaudelli NM, Waterbury AL, Koblan LW, et al. Improved base excision repair inhibition and bacteriophage Mu Gam protein yields C:G-to-T: A base editors with higher efficiency and product purity. Sci Adv. 2017;3(8):eaao4774. doi: 10.1126/sciadv.aao4774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Zafra MP, Schatoff EM, Katti A, Foronda M, Breinig M, Schweitzer AY, et al. Optimized base editors enable efficient editing in cells, organoids and mice. Nat Biotechnol. 2018;36(9):888–893. doi: 10.1038/nbt.4194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Koblan LW, Doman JL, Wilson C, Levy JM, Tay T, Newby GA, et al. Improving cytidine and adenine base editors by expression optimization and ancestral reconstruction. Nat Biotechnol. 2018;36(9):843–846. doi: 10.1038/nbt.4172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Kim YB, Komor AC, Levy JM, Packer MS, Zhao KT, Liu DR. Increasing the genome-targeting scope and precision of base editing with engineered Cas9-cytidine deaminase fusions. Nat Biotechnol. 2017;35(4):371–376. doi: 10.1038/nbt.3803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Li X, Wang Y, Liu Y, Yang B, Wang X, Wei J, et al. Base editing with a Cpf1-cytidine deaminase fusion. Nat Biotechnol. 2018;36(4):324–327. doi: 10.1038/nbt.4102. [DOI] [PubMed] [Google Scholar]
- 84.Thuronyi BW, Koblan LW, Levy JM, Yeh WH, Zheng C, Newby GA, et al. Continuous evolution of base editors with expanded target compatibility and improved activity. Nat Biotechnol. 2019;37(9):1070–1079. doi: 10.1038/s41587-019-0193-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Huang TP, Zhao KT, Miller SM, Gaudelli NM, Oakes BL, Fellmann C, et al. Circularly permuted and PAM-modified Cas9 variants broaden the targeting scope of base editors. Nat Biotechnol. 2019;37(6):626–631. doi: 10.1038/s41587-019-0134-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Richter MF, Zhao KT, Eton E, Lapinaite A, Newby GA, Thuronyi BW, et al. Phage-assisted evolution of an adenine base editor with improved Cas domain compatibility and activity. Nat Biotechnol. 2020;38(7):883–891. doi: 10.1038/s41587-020-0453-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Gaudelli NM, Lam DK, Rees HA, Sola-Esteves NM, Barrera LA, Born DA, et al. Directed evolution of adenine base editors with increased activity and therapeutic application. Nat Biotechnol. 2020;38(7):892–900. doi: 10.1038/s41587-020-0491-6. [DOI] [PubMed] [Google Scholar]
- 88.Tan J, Zhang F, Karcher D, Bock R. Expanding the genome-targeting scope and the site selectivity of high-precision base editors. Nat Commun. 2020;11(1):629. doi: 10.1038/s41467-020-14465-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Kurt IC, Zhou R, Iyer S, Garcia SP, Miller BR, Langner LM, et al. CRISPR C-to-G base editors for inducing targeted DNA transversions in human cells. Nat Biotechnol. 2021;39(1):41–46. doi: 10.1038/s41587-020-0609-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Zhao D, Li J, Li S, Xin X, Hu M, Price MA, et al. Glycosylase base editors enable C-to-A and C-to-G base changes. Nat Biotechnol. 2021;39(1):35–40. doi: 10.1038/s41587-020-0592-2. [DOI] [PubMed] [Google Scholar]
- 91.Koblan LW, Arbab M, Shen MW, Hussmann JA, Anzalone AV, Doman JL, et al. Efficient C·G-to-G·C base editors developed using CRISPRi screens, target-library analysis, and machine learning. Nat Biotechnol. 2021;39(11):1414–1425. doi: 10.1038/s41587-021-00938-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Yuan T, Yan N, Fei T, Zheng J, Meng J, Li N, et al. Optimization of C-to-G base editors with sequence context preference predictable by machine learning methods. Nat Commun. 2021;12(1):4902. doi: 10.1038/s41467-021-25217-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Zuo E, Sun Y, Wei W, Yuan T, Ying W, Sun H, et al. Cytosine base editor generates substantial off-target single-nucleotide variants in mouse embryos. Science. 2019;364(6437):289–292. doi: 10.1126/science.aav9973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Mcgrath E, Shin H, Zhang L, Phue JN, Wu WW, Shen RF, et al. Targeting specificity of APOBEC-based cytosine base editor in human iPSCs determined by whole genome sequencing. Nat Commun. 2019;10(1):5353. doi: 10.1038/s41467-019-13342-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Grunewald J, Zhou R, Garcia SP, Iyer S, Lareau CA, Aryee MJ, et al. Transcriptome-wide off-target RNA editing induced by CRISPR-guided DNA base editors. Nature. 2019;569(7756):433–437. doi: 10.1038/s41586-019-1161-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Zhou C, Sun Y, Yan R, Liu Y, Zuo E, Gu C, et al. Off-target RNA mutation induced by DNA base editing and its elimination by mutagenesis. Nature. 2019;571(7764):275–278. doi: 10.1038/s41586-019-1314-0. [DOI] [PubMed] [Google Scholar]
- 97.Doman JL, Raguram A, Newby GA, Liu DR. Evaluation and minimization of Cas9-independent off-target DNA editing by cytosine base editors. Nat Biotechnol. 2020;38(5):620–628. doi: 10.1038/s41587-020-0414-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Zuo E, Sun Y, Yuan T, He B, Zhou C, Ying W, et al. A rationally engineered cytosine base editor retains high on-target activity while reducing both DNA and RNA off-target effects. Nat Methods. 2020;17(6):600–604. doi: 10.1038/s41592-020-0832-x. [DOI] [PubMed] [Google Scholar]
- 99.Wang L, Xue W, Zhang H, Gao R, Qiu H, Wei J, et al. Eliminating base-editor-induced genome-wide and transcriptome-wide off-target mutations. Nat Cell Biol. 2021;23(5):552–563. doi: 10.1038/s41556-021-00671-4. [DOI] [PubMed] [Google Scholar]
- 100.Chen PJ, Hussmann JA, Yan J, Knipping F, Ravisankar P, Chen PF, et al. Enhanced prime editing systems by manipulating cellular determinants of editing outcomes. Cell. 2021;184(22):5635–52.e29. doi: 10.1016/j.cell.2021.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Nelson JW, Randolph PB, Shen SP, Everette KA, Chen PJ, Anzalone AV, et al. Engineered pegRNAs improve prime editing efficiency. Nat Biotechnol. 2022;40(3):402–410. doi: 10.1038/s41587-021-01039-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Song M, Lim JM, Min S, Oh JS, Kim DY, Woo JS, et al. Generation of a more efficient prime editor 2 by addition of the Rad51 DNA-binding domain. Nat Commun. 2021;12(1):5617. doi: 10.1038/s41467-021-25928-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Park SJ, Jeong TY, Shin SK, Yoon DE, Lim SY, Kim SP, et al. Targeted mutagenesis in mouse cells and embryos using an enhanced prime editor. Genome Biol. 2021;22(1):170. doi: 10.1186/s13059-021-02389-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Zong Y, Liu Y, Xue C, Li B, Li X, Wang Y, et al. An engineered prime editor with enhanced editing efficiency in plants. Nat Biotechnol. 2022;40(9):1394–1402. doi: 10.1038/s41587-022-01254-w. [DOI] [PubMed] [Google Scholar]
- 105.Jiang YY, Chai YP, Lu MH, Han XL, Lin Q, Zhang Y, et al. Prime editing efficiently generates W542L and S621I double mutations in two ALS genes in maize. Genome Biol. 2020;21(1):257. doi: 10.1186/s13059-020-02170-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Choi J, Chen W, Suiter CC, Lee C, Chardon FM, Yang W, et al. Precise genomic deletions using paired prime editing. Nat Biotechnol. 2022;40(2):218–226. doi: 10.1038/s41587-021-01025-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Lin Q, Jin S, Zong Y, Yu H, Zhu Z, Liu G, et al. High-efficiency prime editing with optimized, paired pegRNAs in plants. Nat Biotechnol. 2021;39(8):923–927. doi: 10.1038/s41587-021-00868-w. [DOI] [PubMed] [Google Scholar]
- 108.Anzalone AV, Gao XD, Podracky CJ, Nelson AT, Koblan LW, Raguram A, et al. Programmable deletion, replacement, integration and inversion of large DNA sequences with twin prime editing. Nat Biotechnol. 2021;40(5):731–740. doi: 10.1038/s41587-021-01133-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Jiang T, Zhang XO, Weng Z, Xue W. Deletion and replacement of long genomic sequences using prime editing. Nat Biotechnol. 2022;40(2):227–234. doi: 10.1038/s41587-021-01026-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Zhuang Y, Liu J, Wu H, Zhu Q, Yan Y, Meng H, et al. Increasing the efficiency and precision of prime editing with guide RNA pairs. Nat Chem Biol. 2022;18(1):29–37. doi: 10.1038/s41589-021-00889-1. [DOI] [PubMed] [Google Scholar]
- 111.Rybarski JR, Hu K, Hill AM, Wilke CO, Finkelstein IJ. Metagenomic discovery of CRISPR-associated transposons. Proc Natl Acad Sci U S A. 2021;118(49):e2112279118. doi: 10.1073/pnas.2112279118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Klompe SE, Jaber N, Beh LY, Mohabir JT, Bernheim A, Sternberg SH. Evolutionary and mechanistic diversity of type I-F CRISPR-associated transposons. Mol Cell. 2022;82(3):616–28.e5. doi: 10.1016/j.molcel.2021.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Klompe SE, Vo PLH, Halpin-Healy TS, Sternberg SH. Transposon-encoded CRISPR-Cas systems direct RNA-guided DNA integration. Nature. 2019;571(7764):219–225. doi: 10.1038/s41586-019-1323-z. [DOI] [PubMed] [Google Scholar]
- 114.Strecker J, Ladha A, Gardner Z, Schmid-Burgk JL, Makarova KS, Koonin EV, et al. RNA-guided DNA insertion with CRISPR-associated transposases. Science. 2019;365(6448):48–53. doi: 10.1126/science.aax9181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Saito M, Ladha A, Strecker J, Faure G, Neumann E, Altae-Tran H, et al. Dual modes of CRISPR-associated transposon homing. Cell. 2021;184(9):2441–53.e18. doi: 10.1016/j.cell.2021.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Vo PLH, Ronda C, Klompe SE, Chen EE, Acree C, Wang HH, et al. CRISPR RNA-guided integrases for high-efficiency, multiplexed bacterial genome engineering. Nat Biotechnol. 2021;39(4):480–489. doi: 10.1038/s41587-020-00745-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Pallarès-Masmitjà M, Ivančić D, Mir-Pedrol J, Jaraba-Wallace J, Tagliani T, Oliva B, et al. Find and cut-and-transfer (FiCAT) mammalian genome engineering. Nat Commun. 2021;12(1):7071. doi: 10.1038/s41467-021-27183-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Raguram A, Banskota S, Liu DR. Therapeutic in vivo delivery of gene editing agents. Cell. 2022;185(15):2806–2827. doi: 10.1016/j.cell.2022.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Wang D, Zhang F, Gao G. CRISPR-based therapeutic genome editing: strategies and in vivo delivery by AAV vectors. Cell. 2020;181(1):136–150. doi: 10.1016/j.cell.2020.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Newby GA, Yen JS, Woodard KJ, Mayuranathan T, Lazzarotto CR, Li Y, et al. Base editing of haematopoietic stem cells rescues sickle cell disease in mice. Nature. 2021;595(7866):295–302. doi: 10.1038/s41586-021-03609-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Lattanzi A, Camarena J, Lahiri P, Segal H, Srifa W, Vakulskas CA, et al. Development of β-globin gene correction in human hematopoietic stem cells as a potential durable treatment for sickle cell disease. Sci Transl Med. 2021;13(598):eabf2444. doi: 10.1126/scitranslmed.abf2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Zhang J, Hu Y, Yang J, Li W, Zhang M, Wang Q, et al. Non-viral, specifically targeted CAR-T cells achieve high safety and efficacy in B-NHL. Nature. 2022;609(7926):369–374. doi: 10.1038/s41586-022-05140-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Gillmore JD, Gane E, Taubel J, Kao J, Fontana M, Maitland ML, et al. CRISPR-Cas9 in vivo gene editing for transthyretin amyloidosis. N Engl J Med. 2021;385(6):493–502. doi: 10.1056/NEJMoa2107454. [DOI] [PubMed] [Google Scholar]
- 124.Mendell JR, Al-Zaidy SA, Rodino-Klapac LR, Goodspeed K, Gray SJ, Kay CN, et al. Current clinical applications of in vivo gene therapy with AAVs. Mol Ther. 2021;29(2):464–488. doi: 10.1016/j.ymthe.2020.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Chandler RJ, Sands MS, Venditti CP. Recombinant adeno-associated viral integration and genotoxicity: insights from animal models. Hum Gene Ther. 2017;28(4):314–322. doi: 10.1089/hum.2017.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Banskota S, Raguram A, Suh S, Du SW, Davis JR, Choi EH, et al. Engineered virus-like particles for efficient in vivo delivery of therapeutic proteins. Cell. 2022;185(2):250–65.e16. doi: 10.1016/j.cell.2021.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Sago CD, Lokugamage MP, Paunovska K, Vanover DA, Monaco CM, Shah NN, et al. High-throughput in vivo screen of functional mRNA delivery identifies nanoparticles for endothelial cell gene editing. Proc Natl Acad Sci U S A. 2018;115(42):E9944–E9952. doi: 10.1073/pnas.1811276115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Ni H, Hatit MZC, Zhao K, Loughrey D, Lokugamage MP, Peck HE, et al. Piperazine-derived lipid nanoparticles deliver mRNA to immune cells in vivo. Nat Commun. 2022;13(1):4766. doi: 10.1038/s41467-022-32281-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Sago CD, Lokugamage MP, Loughrey D, Lindsay KE, Hincapie R, Krupczak BR, et al. Augmented lipid-nanoparticle-mediated in vivo genome editing in the lungs and spleen by disrupting Cas9 activity in the liver. Nat Biomed Eng. 2022;6(2):157–167. doi: 10.1038/s41551-022-00847-9. [DOI] [PubMed] [Google Scholar]
- 130.Lyu P, Wang L, Lu B. Virus-like particle mediated CRISPR/Cas9 delivery for efficient and safe genome editing. Life (Basel) 2020;10(12):366. doi: 10.3390/life10120366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Lu Z, Yao X, Lyu P, Yadav M, Yoo K, Atala A, et al. Lentiviral capsid-mediated streptococcus pyogenes Cas9 ribonucleoprotein delivery for efficient and safe multiplex genome editing. CRISPR J. 2021;4(6):914–928. doi: 10.1089/crispr.2020.0106. [DOI] [PubMed] [Google Scholar]
- 132.Hamilton JR, Tsuchida CA, Nguyen DN, Shy BR, Mcgarrigle ER, Sandoval Espinoza CR, et al. Targeted delivery of CRISPR-Cas9 and transgenes enables complex immune cell engineering. Cell Rep. 2021;35(9):109207. doi: 10.1016/j.celrep.2021.109207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Lyu P, Lu Z, Cho SI, Yadav M, Yoo KW, Atala A, et al. Adenine base editor ribonucleoproteins delivered by lentivirus-like particles show high on-target base editing and undetectable RNA off-target activities. CRISPR J. 2021;4(1):69–81. doi: 10.1089/crispr.2020.0095. [DOI] [PubMed] [Google Scholar]
- 134.Segel M, Lash B, Song J, Ladha A, Liu CC, Jin X, et al. Mammalian retrovirus-like protein PEG10 packages its own mRNA and can be pseudotyped for mRNA delivery. Science. 2021;373(6557):882–889. doi: 10.1126/science.abg6155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Roth GA, Mensah GA, Johnson CO, Addolorato G, Ammirati E, Baddour LM, et al. Global burden of cardiovascular diseases and risk factors, 1990–2019: update from the GBD 2019 study. J Am Coll Cardiol. 2020;76(25):2982–3021. doi: 10.1016/j.jacc.2020.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Leopold JA, Loscalzo J. Emerging role of precision medicine in cardiovascular disease. Circ Res. 2018;122(9):1302–1315. doi: 10.1161/CIRCRESAHA.117.310782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Madsen A, Höppner G, Krause J, Hirt MN, Laufer SD, Schweizer M, et al. An important role for DNMT3A-mediated DNA methylation in cardiomyocyte metabolism and contractility. Circulation. 2020;142(16):1562–1578. doi: 10.1161/CIRCULATIONAHA.119.044444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Bengel P, Dybkova N, Tirilomis P, Ahmad S, Hartmann N, Mohamed BA, et al. Detrimental proarrhythmogenic interaction of Ca2+/calmodulin-dependent protein kinase II and NaV1.8 in heart failure. Nat Commun. 2021;12(1):6586. doi: 10.1038/s41467-021-26690-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Levitas A, Muhammad E, Zhang Y, Perea Gil I, Serrano R, Diaz N, et al. A novel recessive mutation in SPEG causes early onset dilated cardiomyopathy. PLoS Genet. 2020;16(9):e1009000. doi: 10.1371/journal.pgen.1009000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Fomin A, Gärtner A, Cyganek L, Tiburcy M, Tuleta I, Wellers L, et al. Truncated titin proteins and titin haploinsufficiency are targets for functional recovery in human cardiomyopathy due to TTN mutations. Sci Transl Med. 2021;13(618):eabd3079. doi: 10.1126/scitranslmed.abd3079. [DOI] [PubMed] [Google Scholar]
- 141.Ma S, Jiang W, Liu X, Lu WJ, Qi T, Wei J, et al. Efficient correction of a hypertrophic cardiomyopathy mutation by ABEmax-NG. Circ Res. 2021;129(10):895–908. doi: 10.1161/CIRCRESAHA.120.318674. [DOI] [PubMed] [Google Scholar]
- 142.Huang JY, Kan SH, Sandfeld EK, Dalton ND, Rangel AD, Chan Y, et al. CRISPR-Cas9 generated Pompe knock-in murine model exhibits early-onset hypertrophic cardiomyopathy and skeletal muscle weakness. Sci Rep. 2020;10(1):10321. doi: 10.1038/s41598-020-65259-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Ercu M, Markó L, Schächterle C, Tsvetkov D, Cui Y, Maghsodi S, et al. Phosphodiesterase 3A and arterial hypertension. Circulation. 2020;142(2):133–149. doi: 10.1161/CIRCULATIONAHA.119.043061. [DOI] [PubMed] [Google Scholar]
- 144.Min YL, Li H, Rodriguez-Caycedo C, Mireault AA, Huang J, Shelton JM, et al. CRISPR-Cas9 corrects Duchenne muscular dystrophy exon 44 deletion mutations in mice and human cells. Sci Adv. 2019;5(3):eaav4324. doi: 10.1126/sciadv.aav4324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Magdy T, Jouni M, Kuo HH, Weddle CJ, Lyra-Leite D, Fonoudi H, et al. Identification of drug transporter genomic variants and inhibitors that protect against doxorubicin-induced cardiotoxicity. Circulation. 2022;145(4):279–294. doi: 10.1161/CIRCULATIONAHA.121.055801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Chemello F, Chai AC, Li H, Rodriguez-Caycedo C, Sanchez-Ortiz E, Atmanli A, et al. Precise correction of Duchenne muscular dystrophy exon deletion mutations by base and prime editing. Sci Adv. 2021;7(18):eabg4910. doi: 10.1126/sciadv.abg4910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Johansen AK, Molenaar B, Versteeg D, Leitoguinho AR, Demkes C, Spanjaard B, et al. Postnatal cardiac gene editing using CRISPR/Cas9 with AAV9-mediated delivery of short guide RNAs results in mosaic gene disruption. Circ Res. 2017;121(10):1168–1181. doi: 10.1161/CIRCRESAHA.116.310370. [DOI] [PubMed] [Google Scholar]
- 148.Song X, Cui Y, Wang Y, Zhang Y, He Q, Yu Z, et al. Genome editing with AAV-BR1-CRISPR in postnatal mouse brain endothelial cells. Int J Biol Sci. 2022;18(2):652–660. doi: 10.7150/ijbs.64188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Xu L, Lau YS, Gao Y, Li H, Han R. Life-long AAV-mediated CRISPR genome editing in dystrophic heart improves cardiomyopathy without causing serious lesions in mdx mice. Mol Ther. 2019;27(8):1407–1414. doi: 10.1016/j.ymthe.2019.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Nelson CE, Wu Y, Gemberling MP, Oliver ML, Waller MA, Bohning JD, et al. Long-term evaluation of AAV-CRISPR genome editing for Duchenne muscular dystrophy. Nat Med. 2019;25(3):427–432. doi: 10.1038/s41591-019-0344-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Liu N, Olson EN. CRISPR modeling and correction of cardiovascular disease. Circ Res. 2022;130(12):1827–1850. doi: 10.1161/CIRCRESAHA.122.320496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Heimlich JB, Bick AG. Somatic mutations in cardiovascular disease. Circ Res. 2022;130(1):149–161. doi: 10.1161/CIRCRESAHA.121.319809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Strong A, Musunuru K. Genome editing in cardiovascular diseases. Nat Rev Cardiol. 2017;14(1):11–20. doi: 10.1038/nrcardio.2016.139. [DOI] [PubMed] [Google Scholar]
- 154.Belbachir N, Portero V, Al Sayed ZR, Gourraud JB, Dilasser F, Jesel L, et al. RRAD mutation causes electrical and cytoskeletal defects in cardiomyocytes derived from a familial case of Brugada syndrome. Eur Heart J. 2019;40(37):3081–3094. doi: 10.1093/eurheartj/ehz308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Zhang XH, Wei H, Xia Y, Morad M. Calcium signaling consequences of RyR2 mutations associated with CPVT1 introduced via CRISPR/Cas9 gene editing in human-induced pluripotent stem cell-derived cardiomyocytes: comparison of RyR2-R420Q, F2483I, and Q4201R. Heart Rhythm. 2021;18(2):250–260. doi: 10.1016/j.hrthm.2020.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Pettinato AM, Ladha FA, Mellert DJ, Legere N, Cohn R, Romano R, et al. Development of a cardiac sarcomere functional genomics platform to enable scalable interrogation of uhman TNNT2 variants. Circulation. 2020;142(23):2262–2275. doi: 10.1161/CIRCULATIONAHA.120.047999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Guo F, Sun Y, Wang X, Wang H, Wang J, Gong T, et al. Patient-specific and gene-corrected induced pluripotent stem cell-derived cardiomyocytes elucidate single-cell phenotype of short QT syndrome. Circ Res. 2019;124(1):66–78. doi: 10.1161/CIRCRESAHA.118.313518. [DOI] [PubMed] [Google Scholar]
- 158.Dotzler SM, Kim CSJ, Genfrom WAC, Zhou W, Ye D, Bos JM, et al. Suppression-replacement KCNQ1 gene therapy for type 1 long QT syndrome. Circulation. 2021;143(14):1411–1425. doi: 10.1161/CIRCULATIONAHA.120.051836. [DOI] [PubMed] [Google Scholar]
- 159.Lambert M, Capuano V, Boet A, Tesson L, Bertero T, Nakhleh MK, et al. Characterization of Kcnk3-mutated rat, a novel model of pulmonary hypertension. Circ Res. 2019;125(7):678–695. doi: 10.1161/CIRCRESAHA.119.314793. [DOI] [PubMed] [Google Scholar]
- 160.Li J, Wang K, Zhang Y, Qi T, Yuan J, Zhang L, et al. Therapeutic exon skipping through a CRISPR-guided cytidine deaminase rescues dystrophic cardiomyopathy in vivo. Circulation. 2021;144(22):1760–1776. doi: 10.1161/CIRCULATIONAHA.121.054628. [DOI] [PubMed] [Google Scholar]
- 161.Li W, Tran V, Shaked I, Xue B, Moore T, Lightle R, et al. Abortive intussusceptive angiogenesis causes multi-cavernous vascular malformations. Elife. 2021;10:e62155. doi: 10.7554/eLife.62155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Carroll KJ, Makarewich CA, Mcanally J, Anderson DM, Zentilin L, Liu N, et al. A mouse model for adult cardiac-specific gene deletion with CRISPR/Cas9. Proc Natl Acad Sci U S A. 2016;113(2):338–343. doi: 10.1073/pnas.1523918113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Schoger E, Carroll KJ, Iyer LM, Mcanally JR, Tan W, Liu N, et al. CRISPR-mediated activation of endogenous gene expression in the postnatal heart. Circ Res. 2020;126(1):6–24. doi: 10.1161/CIRCRESAHA.118.314522. [DOI] [PubMed] [Google Scholar]
- 164.Zhu W, Saw D, Weiss M, Sun Z, Wei M, Shaligram S, et al. Induction of brain arteriovenous malformation through CRISPR/Cas9-mediated somatic Alk1 gene mutations in adult mice. Transl Stroke Res. 2019;10(5):557–565. doi: 10.1007/s12975-018-0676-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Zhang X, Jin H, Huang X, Chaurasiya B, Dong D, Shanley TP, et al. Robust genome editing in adult vascular endothelium by nanoparticle delivery of CRISPR-Cas9 plasmid DNA. Cell Rep. 2022;38(1):110196. doi: 10.1016/j.celrep.2021.110196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Liang P, Xu Y, Zhang X, Ding C, Huang R, Zhang Z, et al. CRISPR/Cas9-mediated gene editing in human tripronuclear zygotes. Protein Cell. 2015;6(5):363–372. doi: 10.1007/s13238-015-0153-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Kang X, He W, Huang Y, Yu Q, Chen Y, Gao X, et al. Introducing precise genetic modifications into human 3PN embryos by CRISPR/Cas-mediated genome editing. J Assist Reprod Genet. 2016;33(5):581–588. doi: 10.1007/s10815-016-0710-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Zuccaro MV, Xu J, Mitchell C, Marin D, Zimmerman R, Rana B, et al. Allele-specific chromosome removal after Cas9 cleavage in human embryos. Cell. 2020;183(6):1650–64.e15. doi: 10.1016/j.cell.2020.10.025. [DOI] [PubMed] [Google Scholar]
- 169.Turocy J, Adashi EY, Egli D. Heritable human genome editing: research progress, ethical considerations, and hurdles to clinical practice. Cell. 2021;184(6):1561–1574. doi: 10.1016/j.cell.2021.02.036. [DOI] [PubMed] [Google Scholar]
- 170.Baylis F, Darnovsky M, Hasson K, Krahn TM. Human germ line and heritable genome editing: the global policy landscape. CRISPR J. 2020;3(5):365–377. doi: 10.1089/crispr.2020.0082. [DOI] [PubMed] [Google Scholar]
- 171.Min YL, Chemello F, Li H, Rodriguez-Caycedo C, Sanchez-Ortiz E, Mireault AA, et al. Correction of three prominent mutations in mouse and human models of Duchenne muscular dystrophy by single-cut genome editing. Mol Ther. 2020;28(9):2044–2055. doi: 10.1016/j.ymthe.2020.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Moretti A, Fonteyne L, Giesert F, Hoppmann P, Meier AB, Bozoglu T, et al. Somatic gene editing ameliorates skeletal and cardiac muscle failure in pig and human models of Duchenne muscular dystrophy. Nat Med. 2020;26(2):207–214. doi: 10.1038/s41591-019-0738-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Xu L, Zhang C, Li H, Wang P, Gao Y, Mokadam NA, et al. Efficient precise in vivo base editing in adult dystrophic mice. Nat Commun. 2021;12(1):3719. doi: 10.1038/s41467-021-23996-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Hanses U, Kleinsorge M, Roos L, Yigit G, Li Y, Barbarics B, et al. Intronic CRISPR repair in a preclinical model of noonan syndrome-associated cardiomyopathy. Circulation. 2020;142(11):1059–1076. doi: 10.1161/CIRCULATIONAHA.119.044794. [DOI] [PubMed] [Google Scholar]
- 175.Ma H, Marti-Gutierrez N, Park SW, Wu J, Lee Y, Suzuki K, et al. Correction of a pathogenic gene mutation in human embryos. Nature. 2017;548(7668):413–419. doi: 10.1038/nature23305. [DOI] [PubMed] [Google Scholar]
- 176.Romano R, Ghahremani S, Zimmerman T, Legere N, Thakar K, Ladha FA, et al. Reading frame repair of TTN truncation variants restores titin quantity and functions. Circulation. 2022;145(3):194–205. doi: 10.1161/CIRCULATIONAHA.120.049997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Nishiyama T, Zhang Y, Cui M, Li H, Sanchez-Ortiz E, Mcanally JR, et al. Precise genomic editing of pathogenic mutations in RBM20 rescues dilated cardiomyopathy. Sci Transl Med. 2022;14(672):eade1633. doi: 10.1126/scitranslmed.ade1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Dave J, Raad N, Mittal N, Zhang L, Fargnoli A, Oh JG, et al. Gene editing reverses arrhythmia susceptibility in humanized PLN-R14del mice: modelling a European cardiomyopathy with global impact. Cardiovasc Res. 2022;118(15):3140–3150. doi: 10.1093/cvr/cvac021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Ding Q, Strong A, Patel KM, Ng SL, Gosis BS, Regan SN, et al. Permanent alteration of PCSK9 with in vivo CRISPR-Cas9 genome editing. Circ Res. 2014;115(5):488–492. doi: 10.1161/CIRCRESAHA.115.304351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Musunuru K, Chadwick AC, Mizoguchi T, Garcia SP, Denizio JE, Reiss CW, et al. In vivo CRISPR base editing of PCSK9 durably lowers cholesterol in primates. Nature. 2021;593(7859):429–434. doi: 10.1038/s41586-021-03534-y. [DOI] [PubMed] [Google Scholar]
- 181.Rothgangl T, Dennis MK, Lin PJC, Oka R, Witzigmann D, Villiger L, et al. In vivo adenine base editing of PCSK9 in macaques reduces LDL cholesterol levels. Nat Biotechnol. 2021;39(8):949–957. doi: 10.1038/s41587-021-00933-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Hoffman EP, Brown RH, Jr, Kunkel LM. Dystrophin: the protein product of the Duchenne muscular dystrophy locus. Cell. 1987;51(6):919–928. doi: 10.1016/0092-8674(87)90579-4. [DOI] [PubMed] [Google Scholar]
- 183.Hinson JT, Chopra A, Nafissi N, Polacheck WJ, Benson CC, Swist S, et al. Heart disease. Titin mutations in iPS cells define sarcomere insufficiency as a cause of dilated cardiomyopathy. Science. 2015;349(6251):982–986. doi: 10.1126/science.aaa5458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Chopra A, Kutys ML, Zhang K, Polacheck WJ, Sheng CC, Luu RJ, et al. Force generation via β-cardiac myosin, titin, and α-actinin drives cardiac sarcomere assembly from cell-matrix adhesions. Dev Cell. 2018;44(1):87–96.e5. doi: 10.1016/j.devcel.2017.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Zaunbrecher RJ, Abel AN, Beussman K, Leonard A, Von Frieling-Salewsky M, Fields PA, et al. Cronos titin is expressed in human cardiomyocytes and necessary for normal sarcomere function. Circulation. 2019;140(20):1647–1660. doi: 10.1161/CIRCULATIONAHA.119.039521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Xie C, Zhang YP, Song L, Luo J, Qi W, Hu J, et al. Genome editing with CRISPR/Cas9 in postnatal mice corrects PRKAG2 cardiac syndrome. Cell Res. 2016;26(10):1099–1111. doi: 10.1038/cr.2016.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Pan X, Philippen L, Lahiri SK, Lee C, Park SH, Word TA, et al. In vivo Ryr2 editing corrects catecholaminergic polymorphic ventricular tachycardia. Circ Res. 2018;123(8):953–963. doi: 10.1161/CIRCRESAHA.118.313369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Libby P. The changing landscape of atherosclerosis. Nature. 2021;592(7855):524–533. doi: 10.1038/s41586-021-03392-8. [DOI] [PubMed] [Google Scholar]
- 189.Kannan S, Altae-Tran H, Jin X, Madigan VJ, Oshiro R, Makarova KS, et al. Compact RNA editors with small Cas13 proteins. Nat Biotechnol. 2022;40(2):194–197. doi: 10.1038/s41587-021-01030-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Kato K, Okazaki S, Schmitt-Ulms C, Jiang K, Zhou W, Ishikawa J, et al. RNA-triggered protein cleavage and cell growth arrest by the type III-E CRISPR nuclease-protease. Science. 2022;378(6622):882–889. doi: 10.1126/science.add7347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Strecker J, Demircioglu FE, Li D, Faure G, Wilkinson ME, Gootenberg JS, et al. RNA-activated protein cleavage with a CRISPR-associated endopeptidase. Science. 2022;378(6622):874–881. doi: 10.1126/science.add7450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Frangoul H, Altshuler D, Cappellini MD, Chen YS, Domm J, Eustace BK, et al. CRISPR-Cas9 gene editing for sickle cell disease and beta-thalassemia. N Engl J Med. 2021;384(3):252–260. doi: 10.1056/NEJMoa2031054. [DOI] [PubMed] [Google Scholar]
- 193.Stadtmauer EA, Fraietta JA, Davis MM, Cohen AD, Weber KL, Lancaster E, et al. CRISPR-engineered T cells in patients with refractory cancer. Science. 2020;367(6481):eaba7365. doi: 10.1126/science.aba7365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Grunwald HA, Gantz VM, Poplawski G, Xu XS, Bier E, Cooper KL. Super-Mendelian inheritance mediated by CRISPR-Cas9 in the female mouse germline. Nature. 2019;566(7742):105–109. doi: 10.1038/s41586-019-0875-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Alanis-Lobato G, Zohren J, McCarthy A, Fogarty NME, Kubikova N, Hardman E, et al. Frequent loss of heterozygosity in CRISPR-Cas9-edited early human embryos. Proc Natl Acad Sci U S A. 2021;118(22):e2004832117. doi: 10.1073/pnas.2004832117. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.