Abstract
The fucose α(1→2) galactose β structure is expressed by uterine epithelial cells in the mouse and has been implicated in blastocyst adhesion events thought to be required for murine implantation. Fucα(1→2)Galβ moieties and cognate fucosyltransferases are also expressed by epithelial cells of the male reproductive tract and have been implicated in sperm maturation events that may contribute to fertilization. To determine directly if Fucα(1→2)Galβ moieties are required for fertility, we have generated strains of mice that are deficient in genes encoding FUT1 and FUT2, a pair of GDP-l-fucose:β(1→4)-d-galactosyl-R 2-α-l-fucosyltransferase enzymes (EC 2.4.1.69) responsible for Fucα(1→2)Galβ synthesis and expression. FUT1 null mice and FUT2 null mice develop normally and exhibit no gross phenotypic abnormalities. The Fucα(1→2)Galβ epitope is absent from the uterine epithelia of FUT2 null mice and from the epithelia of the epididymis of FUT1 null mice. Fully normal fertility is observed in FUT1 null intercrosses and in FUT2 null intercrosses. These observations indicate that Fucα(1→2)Galβ moieties are not essential to blastocyst-uterine epithelial cell interactions required for implantation and are not required for sperm maturation events that permit fertilization and that neither the FUT loci nor their cognate fucosylated glycans are essential to normal development.
In mammals, the large number of discrete cell surface glycan structures includes many that are decorated with fucose (42). Fucose is found on these molecules in direct linkage to some serine residues, for example, where they may contribute to Notch-dependent intracellular signal transduction events (3, 32). Fucose also decorates some asparagine- and serine/threonine-linked glycans, by attachment in an α(1,3) linkage. Subsets of the latter molecules contribute to selectin-dependent leukocyte adhesion (45). Fucose has also been described for the α(1,6) linkage exclusively on the N-acetylglucosamine residue attached to asparagine on N-linked glycans (31), though the function of this modification remains unknown. Some of the major blood group antigens, including the ABO and Lewis blood group antigens, also correspond to fucosylated glycan epitopes (28). These are composed of α(1,4)-linked and/or α(1,2)-linked fucose, some additionally modified by terminal galactose or N-acetylgalactosamine moieties, and are displayed by glycoproteins and glycolipids on erythrocytes and many types of epithelium. In the stomach, for example, gastric epithelial cells can express the Lewis b antigen, an epitope corresponding to an α(1,4)- and α(1,2)fucosylated glycan, which has been assigned a role in the attachment and pathogenesis of Helicobacter pylori infection (11). Interactions between microbes and fucosylated glycans on mammalian host cells extend to the small intestine in rodents, where host-derived α(1,2)fucosylated glycans support the growth of and are in turn regulated by Bacteroides thetaiotaomicron (4). In each of these contexts, expression of cell surface fucosylated glycans is directed in part by one or more of the cognate fucosyltransferases responsible for their synthesis (24, 25).
Cell surface fucosylated glycans are also implicated in reproductive physiology. For example, α(1,3)-fucosylated glycans are associated with the process of morula compaction (1, 13) and may contribute to sperm-egg interactions in some species (7). There is substantial experimental support for the hypothesis that in rodents, α(1,2)fucosylated glycans contribute to the process of blastocyst attachment to the uterine epithelial wall in the context of implantation (23). During the rodent reproductive cycle, α(1,2)fucosylated glycan expression by the uterine epithelium is dynamically regulated, in concert with hormonal changes that account for the physiology of the estrous cycle, at a time that correlates with endometrial receptivity for blastocyst implantation (21, 22). Coculture of blastocysts with uterine epithelia supports blastocyst attachment (27). Attachment is inferred to be dependent on α(1,2)fucosylated glycans, since attachment is inhibited by the addition of α(1,2)fucosylated glycoconjugates, but not by control fucosylated glycans, and by monoclonal antibodies (MAbs) specific for α(1,2)fucosylated epitopes (27). Intrauterine injection of a MAb directed against the α(1,2)fucosylated α(1,3)fucosylated glycan Lewis Y inhibited blastocyst implantation in a dose-dependent manner (50). Similarly, this antibody, but not control antibodies, inhibited implantation in an embryo transfer model and in a uterine epithelial cell-blastocyst coculture (46). Furthermore, fluoresceinated probes prepared from α(1,2)fucosylated glycans localize to the mural trophectoderm on hatched blastocysts and thus colocalize with the site of blastocyst attachment to the receptive uterine wall (26, 48). Considered together, these observations imply an important or even essential role for α(1,2)fucosylated glycans in the implantation process.
Control of cell surface fucosylation in this context is mediated by cycle-dependent modulation of α(1,2)fucosyltransferase activity in association with estrogen and progesterone levels (47). Dynamic control of fucosyltransferase activity is in turn apparently controlled by the dynamic modulation of the level of transcripts derived from one or more α(1,2)fucosyltransferase genes (9, 40). In the uterine epithelium, control of α(1,2)fucosylated glycan expression largely correlates with cycle-dependent modulation of expression of the FUT2 α(1,2)fucosyltransferase locus (9). In oophorectomized mice, estrogen activates the accumulation of uterine epithelial cell α(1,2)fucosyltransferase activity and its mRNA, whereas progesterone inhibits accumulation (19, 40).
Fucosylated glycans are also implicated in the maturation of sperm in the rodent (30, 34). Fucosyltransferase activities, including α(1,2)fucosyltransferase activities, are expressed in the rodent seminiferous tubules and epididymis (37, 38). A gradient of fucosyltransferase activity exists from the caput epididymis to the cauda epididymis (6, 34, 36) and is apparently elaborated by the epithelial cells that line the epididymis. This gradient aligns with the gradient of expression of the FUT1 locus observed by in situ hybridization in the mouse epididymal epithelia (8). The gradient of fucosyltransferase expression parallels the apparent acquisition by spermatozoa of cell surface α(1,2)fucosylated glycans and correlates with acquisition of the ability to fertilize that occurs during passage of spermatozoa through the epididymis in the mouse (5, 43). α(1,2)Fucosylated glycans may also be involved in sperm capacitation. Incubation of spermatozoa from the cauda epididymis of mice with exogenous fucose displaces a decapacitation factor, accelerating capacitation, while incubation with the α(1,2)fucose-specific lectin Ulex europaeus agglutinin-I (UEA-I) binds to the postacrosomal region and blocks decapacitation factor reassociation (14). These observations suggest an important or perhaps essential role for epididymal α(1,2)fucosylation events in fertility.
To directly determine if α(1,2)fucosylated glycans contribute essentially to the blastocyst implantation events or sperm maturation process discussed above, and to begin to define the functions of cell surface α(1,2)fucosylated glycans and their fucosyltransferases in mammalian physiology, we carried out the construction and initial characterization of strains of mice with induced null mutations in either the FUT2 locus or the FUT1 locus. Homozygously FUT2 null mice are viable, appear healthy, no longer exhibit the wild-type expression pattern of α(1,2)fucosylated glycans in the uterine epithelium, and are fertile. Homozygously FUT1 null mice are also viable and healthy, are deficient in epididymal cell surface α(1,2)fucosylated glycans expressed by wild-type mice, yet are also fertile. These observations imply that α(1,2)fucosylated glycans play nonessential roles in blastocyst implantation or sperm function in mice.
MATERIALS AND METHODS
Targeting constructs.
FUT1 and FUT2 genomic DNA clones were isolated and mapped from a commercially available P1 genomic library (strain 129P2/OlaHsd; Incyte Genomics, St. Louis, Mo.) as previously described (9). Targeting vectors were derived from the β-galactoside-containing nucleus-localizing plasmid pnlacF (49). An existing SacI site within the 5′ polylinker site of pnlacF was modified into a NotI restriction site by utilizing two overlapping 15-mer oligonucleotides: 5′-CGC GGC CGC AGA GCT-3′ and 5′-CTG CGG CCG CGA GCT-3′ (NotI site is underlined; overhangs for ligation into SacI at the 3′ end). The 15-mer oligonucleotides were heated together to 65°C, cooled, ligated with partially SacI-digested full-length pnlacF vector (6.3 kb), and electroporated (Gene Pulser; Bio-Rad Laboratories) into Escherichia coli JM109 (Promega Corp.), and blue colonies were isolated by blue-white selection on agar plates containing X-Gal (5′-bromo-4-chloro-3-indolyl-β-d-galactopyranoside; Roche Molecular Biochemicals). Insertion of the new NotI site maintained in-frame translation of the lacZ coding sequence and was confirmed by DNA sequencing. The resulting vector, pnlacF NotI, was modified by ligation with a PGK-neomycin resistance cassette (44) into an EcoRI site in the 3′ polylinker sequence of partially digested full-length pnlacF. Orientation of the neomycin cassette in the resulting 7.9-kb vector (pnlacF NotI Neo) was determined by sequencing from the 3′ end using the M13 forward sequencing primer and from the opposite end using a 19-mer, 5′-TTG ACT ACC AAG CCA CCT G-3′, from the vector sequence within the 3′ pnlacF polylinker. Oligonucleotides were designed using the program MacVector (Oxford Molecular Group) and synthesized on Applied Biosystems DNA synthesizers in the University of Michigan DNA Synthesis Core. All sequencing was performed in the University of Michigan DNA Sequencing Core Facility on Applied Biosystems DNA sequencers.
The FUT2 targeting vector was made by ligation of a 2.1-kb HindIII genomic fragment 3 kb 3′ to the translation stop codon of the FUT2 open reading frame into a unique HindIII site within the 3′ polylinker sequence of linearized pnlacF NotI Neo. The orientation of the fragment was determined by sequencing. A second ligation was then performed with a KpnI partial digestion fragment that spans 3.5 kb of DNA 5′ to the FUT2 open reading frame and ends after the first six codons of the FUT2 open reading frame. This fragment was ligated into a unique KpnI site within the 5′ polylinker sequence of linearized pnlacF NotI Neo containing the correctly oriented 3′ fragment and sequenced for orientation. Maintenance of the translational reading frame of the FUT2-LacZ fusion protein at the KpnI site was ensured by sequencing with a FUT2 open reading frame 20-mer forward primer, 5′-GGC ACA ATG CAG ATG ATT AG-3′. The resulting 13.4-kb targeting vector was linearized with NotI prior to electroporation in embryonic stem (ES) cells.
The FUT1 targeting vector was made by isolation of a 2.4-kb EcoRV/NcoI genomic fragment 0.14 kb 3′ to the translation stop codon of the FUT1 open reading frame. The fragment was blunted and ligated to HindIII phosphorylated linkers (New England Biolabs), restricted, and ligated into a unique HindIII site within the 3′ polylinker sequence of linearized pnlacF NotI Neo. Orientation of the 3′ genomic fragment was determined by sequencing. A second ligation utilized a 4.4-kb PCR fragment of genomic DNA that spans 4.4 kb 5′ to the FUT1 open reading frame and ends at an inserted NcoI site after the first five codons of the FUT1 open reading frame. The PCR amplification used a 46-mer forward PCR primer, 5′-ATA AGA ATG CGG CCG CAG GGG CAG GTC ATG GGC TTC AGA CTG GTA C-3′ (NotI site underlined), and a 42-mer reverse PCR primer, 5′-CAT GCC ATG GGA GTC CAC ATA GCT ATG GGG AGG AAA GAC GAG-3′ (NcoI site underlined), and was done with the Extend PCR kit (Roche Molecular Biochemicals) using buffer 3 containing 2.25 mM MgCl2 at an annealing temperature of 65°C for 25 cycles. The 4.4-kb PCR product was subcloned into pGEM-T (Promega Corp). Sequencing from each end of the insert with T7 and SP6 primers demonstrated the correct FUT1 sequence, although the NotI site in the 46-mer was not present in the isolated clones. However, a NotI site within the pGEM-T multiple cloning site was immediately 5′ to the cloned 4.4-kb fragment and allowed restriction of the fragment with NotI and NcoI and directional ligation into NotI-linearized, partially NcoI-digested pnlacF NotI Neo containing the correctly oriented 3′ fragment. Correct ligation into the 5′ polylinker site was confirmed by DNA sequencing. Maintenance of the translational reading frame of the FUT1-LacZ fusion protein at the NcoI site was ensured by sequencing with a FUT1 open reading frame 19-mer forward primer, 5′-TGA CCT CAC TGA AAA TCT C-3′. The resulting 14.7-kb targeting vector was linearized with NotI prior to electroporation in ES cells.
ES cell manipulation.
A mouse ES cell line (designated Pat5) was prepared from mouse strain 129X1/SvJ (stock no. 000691; Jackson Laboratory) according to a published immunosurgery protocol (41). Briefly, blastocysts obtained by mating 129X1/SvJ mice were subjected to immunosurgery to eliminate trophectoderm cells, as described previously (41). The inner cell mass cells were established in tissue culture on feeder cells. Primary mouse embryo fibroblasts (MEF) were prepared from mice transgenic for the neo gene and used as feeder cells. ES cells were cultured in high-glucose Dulbecco's minimal essential medium supplemented with 15% fetal bovine serum, 1 μM β-mercaptoethanol, 4 mM glutamine, 50 IU of penicillin per ml, 50 μg of streptomycin per ml, and 1,000 U of recombinant leukemia inhibitory factor (ESGRO; Chemicon International, Inc) per ml. In addition, R1 ES cells were independently used for FUT1 and FUT2 targeting (33).
The EndoFree plasmid maxi-kit (Qiagen) was used for the purification of the targeting vectors prior to ES electroporation. Plasmid DNA (200 μg) was linearized with NotI, extracted with phenol-chloroform and then with chloroform, ethanol precipitated, and resuspended in sterile Tris-EDTA at 2 mg/ml. Electroporation of Pat5 and R1 ES cells with targeting vector and selection for clones were carried out essentially as described by Kendall et al. (20). Six electroporations were performed for each construct. Briefly, 107 ES cells were electroporated with 30 μg of linearized construct (0.4-cm electroporation cuvette; 0.3 kV; 250 μF; time constant, approximately 4 s) and then diluted onto feeder layers of mitomycin C-treated neomycin-resistant MEF in media containing leukemia inhibitory factor. G418-resistant transfectant ES cell colonies were picked and plated on six 96-well MEF plates. The cells were grown to confluence and split among three 96-well plates. Two sets of plates were stored at −80°C. The cells in the third set of plates were grown to high density and split between two 96-well plates. The cells in the final duplicate plates were grown to high density, and genomic DNA was extracted (35) for Southern blot analysis. Clones that were positive in the initial screening were expanded, and their genotypes were confirmed by additional Southern blot analyses.
Southern blot screening of targeted ES cells.
Southern blot probes derived from positions 5′ and 3′ to the borders of the targeting constructs were used in primary and secondary screening of ES colonies. For FUT2, a 5′ 400-bp BamHI/KpnI probe and 3′ 390-bp KpnI/SacI probe were derived from genomic clones previously mapped 5′ and 3′ to FUT2 (9). For FUT1, PCR was used to generate a 3′ 355-bp probe (25-mer forward primer, 5′-CGC TTG TCT ATT TAG GAC AGG AAC C-3′; 25-mer reverse primer, 5′-GAT TCA GGA GCC AGA ACT GAT TCT C-3′) and a 5′ 238-bp probe (22-mer forward primer, 5′-GTG GAA ACC CAA AAA GAA ACA G-3′; 20-mer reverse primer, 5′-CGT CTG CCT CAG AAG GAC TC-3). These probes and Southern blot analyses identified four homologously recombinant FUT2-LacZ clones and more than 12 homologously recombinant FUT1-LacZ clones.
Blastocyst injection of homologously targeted ES cells.
Clones with the modal number of 40 chromosomes were injected into host C57BL/6J blastocysts by the Transgenic Animal Model Core using standard protocols (2). The resulting male ES cell-mouse chimeras were crossed with wild-type female C57BL/6J mice to obtain germ line transmission of the targeted alleles. Two FUT2-LacZ clones and four FUT1-LacZ clones yielded germ line transmission.
PCR genotyping of progeny from chimeric mice.
Southern blotting and PCR genotyping were used to genotype the first 100 progeny derived from FUT2-LacZ and FUT1-LacZ germ line-transmitting chimeric mice. The Southern blot probes are described above. Tail DNA was purified by proteinase K digestion, NaCl precipitation, and spooling on glass pipettes (29). PCR genotyping of the FUT2-LacZ locus used a forward primer whose sequence is present in both the wild-type and the FUT2-LacZ allele (21-mer primer a, 5′-CCT GCC ATG CTT TCT TTC CTG-3′). Two reverse primers were used with primer a to discriminate between the wild-type and the FUT2-LacZ allele. Reverse primer b corresponds to a sequence in the wild-type allele (23-mer, 5′-ATT CCT TCT CTG ACA GGG TTT GG-3′). Reverse primer c corresponds to the lacZ sequence in the FUT2-LacZ allele (20-mer, 5′-TGG GTA ACG CCA GGG TTT TC-3′). Primer pairs a and b yield a 154-bp fragment from the wild-type allele. Primer pairs a and c yield a 191-bp fragment from the FUT2-LacZ null allele.
PCR genotyping of the FUT1-LacZ locus used a forward primer whose sequence is present in the wild-type and the FUT1-LacZ allele (24-mer primer d, 5′-CGG TGG CTT AAT CTG TGT GTC TTC-3′). Two reverse primers were used with primer d to discriminate between the wild-type and the FUT1-LacZ allele. Reverse primer e corresponds to the wild-type sequence in both the wild-type and FUT1-LacZ alleles (23-mer, 5′-GCA ATG GAT GAG GTA GGC ATA CC-3′). Reverse primer f corresponds to the lacZ sequence in the FUT1-LacZ allele (24-mer, 5′-CCA GTC ACG ACG TTG TAA AAC GAC-3′). Primer pairs d and e yield a 239-bp fragment from the wild-type allele. Primer pairs d and f yield a 130-bp fragment from the FUT1-LacZ null allele.
The DNA template for PCR genotyping used either 0.75 μg of purified tail DNA as described above or 2-μl aliquots of a crude proteinase K digestion of mouse tails: the distal 5 mm of a tail tip biopsy sample was digested overnight in 50 μl of digestion buffer (10 mM Tris-HCl [pH 9.0], 50 mM KCl, 0.1% Triton X-100, 0.4 mg of proteinase K/ml) at 56°C and then heated to 94°C for 10 min to denature proteinase K prior to PCR. All PCRs used primers at 0.4 μM (except primer c, which was used at 0.8 μM), 1.5 mM MgCl2, Taq polymerase (Life Technologies, Gibco BRL), and buffer conditions supplied by the manufacturer. Annealing was done at 55°C for FUT2 and 60°C for FUT1. PCR fragments were fractionated though 2% agarose gels containing 0.5% NuSieve agarose (FMC BioProducts).
Lectin histochemistry.
The α(1,2)fucose-specific lectin UEA-I conjugated to biotin (EY Laboratories Inc.) and blood group MAbs BG-4 and BG-1 (Signet Pathology Systems Inc.) were used in automated immunoperoxidase staining. BG-4 is specific for type I H antigen; BG-1 is specific for nonfucosylated type I lacto-N-biose disaccharide. Adult wild-type, FUT2 null, and FUT1 null female mice were monitored through the estrous cycle by vaginal washings. Uteri from mice in estrus were fixed in 10% buffered formalin at 4°C overnight, embedded in paraffin, and processed for immunohistochemistry. Epididymides from adult male mice were processed in the same manner. Immunohistochemistry was done in the University of Michigan Comprehensive Cancer Center Research Histology and Immunoperoxidase Laboratory using a Dako Corp. autostainer immunostaining system and the manufacture's protocol. No antigen retrieval step was necessary. With mouse MAbs (BG-4 and BG-1), preincubation with an immunoglobulin blocking reagent was performed according to the manufacturer's protocol (Mouse-on-Mouse immunodetection kit peroxidase; Vector Laboratories). Control sections processed for immunohistochemistry with all reagents except the primary reagents showed virtually no staining.
Breeding of F1 and F2 generations.
Four male-female F1 FUT1 heterozygous (+/−) and FUT2 heterozygous (+/−) pairs were mated. In excess of 160 progeny were sexed and genotyped to ascertain any sex ratio distortion or decreased fetal or postnatal viability as a function of FUT1 or FUT2 genotype. For the F2 crosses, gestational duration and enumeration of litter size were done with male-female pairs that were continuously housed. Progeny were immediately removed after birth to ensure that the female would be continuously fertile.
RESULTS AND DISCUSSION
Construction of mice with targeted deletions of the FUT2 and FUT1 α(1,2)fucosyltransferase genes.
A targeting vector for homologous recombination-mediated inactivation of the FUT2 locus was constructed from a mouse FUT2 genomic fragment (Fig. 1). This vector deletes the majority of the single coding exon of the FUT2 locus, including all of the part of the exon that corresponds to the enzyme's catalytic domain (9). The E. coli β-galactosidase coding sequence was fused in frame to the amino-terminal six residues proximal to the transmembrane segment of FUT2, for eventual use in immunohistochemical detection of the cell type-specific expression patterns of this locus. Chimeric males obtained from blastocyst injection of two independent heterozygous ES clones derived with this construct gave rise to heterozygous progeny when crossed with wild-type females. Heterozygous progeny were intercrossed to generate mice homozygous for the FUT2 locus. A Mendelian distribution of genotypes was observed among the progeny, males and females were present in equal numbers regardless of genotype, and no gross behavioral or morphological abnormalities were observed among heterozygous or homozygous null progeny.
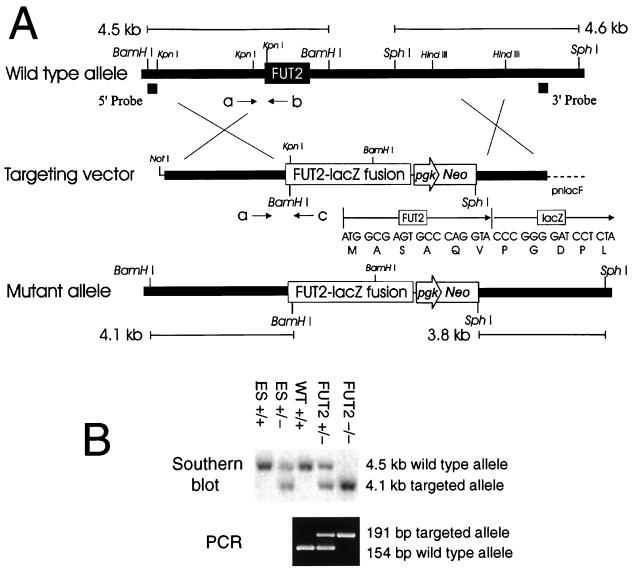
FIG. 1.
Targeting constructs of the murine α(1,2)fucosyltransferase gene FUT2. (A) FUT2 targeting construct. Genomic DNA fragments 5′ and 3′ to the open reading frame of FUT2 were serially ligated into a targeting vector (pnlacF NotI Neo) as described in Materials and Methods. Southern blot analysis using the 5′ probe and BamHI digests identified ES clones with homologously recombinant targeting events. Southern blot analysis using the 3′ probe and SphI digestion confirmed the fidelity of recombination events at the 3′ end of the locus. The positions of PCR primers a, b, and c (see Materials and Methods) used to genotype the wild-type and targeted alleles are shown below each schematic. The DNA and protein sequences of the fusion between the first six amino acids of FUT2 and β-galactosidase are presented above the schematic of the mutant allele. (B) Genotyping of tail DNA from wild-type (WT +/+), FUT2+/−, and FUT2−/− mice generated in crosses between FUT2 heterozygotes. (Top) Southern blot of BamHI digests of wild-type ES cells (ES +/+), FUT2-targeted ES cells (ES +/−), and cells from littermates of three different genotypes probed with the 5′ probe; (bottom) corresponding genotype data from the same tail DNA samples using PCR primers a and c (targeted allele) or a and b (wild-type allele).
A FUT1 targeting vector was constructed in a similar manner, deleting most of the single coding segment of this gene, including the part corresponding to the catalytic domain, while simultaneously fusing the E. coli β-galactosidase coding sequence in frame to the amino-terminal five residues proximal to the transmembrane segment of FUT1 (Fig. 2). Chimeric males obtained from blastocyst injection of four independent heterozygous ES clones derived with this construct gave rise to heterozygous progeny when crossed with wild-type females. Heterozygous progeny were intercrossed to generate mice homozygous for the FUT1 locus. The genotypes of the progeny were Mendelian in distribution, males and females were present in equal numbers in all genotypes, and no obvious behavioral or morphological abnormalities were observed in any of the progeny.
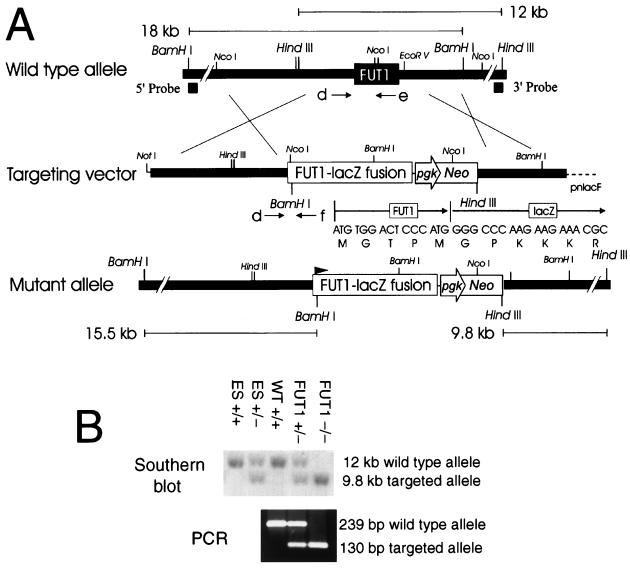
FIG. 2.
Targeting constructs of the murine α(1,2)fucosyltransferase gene FUT1. (A) FUT1 targeting construct. Genomic DNA fragments 5′ and 3′ to the open reading frame of FUT1 were serially ligated into a targeting vector (pnlacF NotI Neo) as described in Materials and Methods. Southern blot analysis using the 3′ probe and HindIII digests identified ES clones with homologously recombinant targeting events. Southern blot analysis using the 5′ probe and BamHI digestion confirmed the fidelity of recombination events at the 5′ end of the locus. The positions of PCR primers d, e, and f (see Materials and Methods) used to genotype the wild-type and targeted alleles are shown below each schematic. The DNA and protein sequences of the fusion between the first five amino acids of FUT1 and β-galactosidase are presented above the schematic of the mutant allele. (B) Genotyping of tail DNA from wild-type (WT +/+) mice, FUT1+/−, or FUT1−/− mice generated in crosses between FUT1 heterozygotes. (Top) Southern blot of HindIII digests of wild-type ES cells (ES +/+), FUT1-targeted ES cells (ES +/−), and cells from littermates of three different genotypes probed with the 3′ probe; (bottom) corresponding genotype data from the same tail DNA samples using PCR primers d and e (wild-type allele) or d and f (targeted allele).
Expression of α(1,2)fucosylated glycans in uterine epithelia requires FUT2 and is exclusive of a contribution by FUT1.
Hybridization in situ and biochemical analyses are generally if not completely consistent with an assignment to the FUT2 locus, and not the FUT1 locus, in controlling expression of α(1,2)fucosylated glycans in uterine epithelium during the estrous cycle (9, 40). To confirm these assignments, and to verify that deletion of the FUT2 locus abolishes expression of α(1,2)fucosylated glycans in this context, expression of H antigen was assessed on uterine epithelia in FUT2−/− mice, FUT1−/− mice, and wild-type controls (Fig. 3) at estrus, where, in wild-type mice, expression of these glycans is prominent (22, 47) and implicated in implantation (21, 27). H antigen is clearly evident on wild-type epithelial cells, as detected with a lectin with specificity for α(1,2)fucosylated glycans (UEA-I), and with a MAb with specificity for type I H blood group structure (Fig. 3A). H-reactive glycans localize primarily to the glycocalyx on the luminal side of the cells, where they are eligible for interaction with blastocysts susceptible to implantation. By contrast, the uterine epithelium in FUT2−/− mice is devoid of detectable H structures when probed with these reagents. Deletion of α(1,2)fucosylated glycans in the FUT2−/− uterine epithelia is not consequent to deletion of glycans that may serve as H-antigen precursors (28), since such unmodified type I glycans continue to be expressed by these cells (Fig. 3B). Small amounts of FUT1 transcripts are detected in the mouse uterus at estrus, though in situ hybridization analyses do not clearly indicate that they are localized to the uterine epithelia (9). These transcripts and their cognate fucosyltransferase do not detectably contribute to α(1,2)fucosylated glycans in uterine epithelia, since such glycans are fully deleted in FUT2 null epithelia. These observations confirm that the FUT2 mutant allele is null, definitively assign the FUT2 locus to the control of α(1,2)fucosylated glycan expression in uterine epithelia, and indicate that FUT2-dependent control of α(1,2)fucosylated glycans in uterine epithelia is exclusive of a contribution by the FUT1 locus, since such glycans are fully deleted in FUT2 null epithelia yet are retained in the FUT1−/− epithelia (Fig. 3).
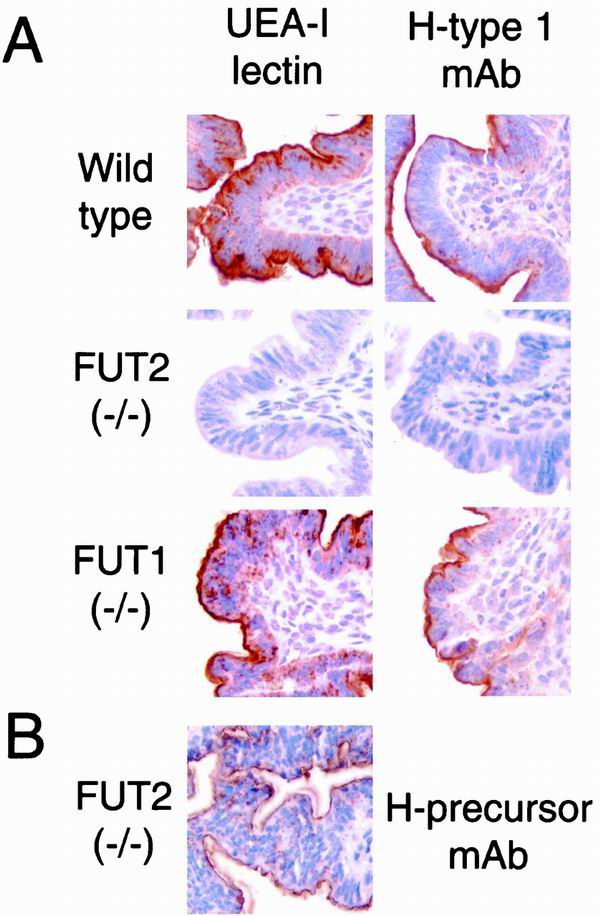
FIG. 3.
Lectin and blood group immunohistochemistry of the uterus in wild-type, FUT2 null, and FUT1 null mice. Adult female mice were sacrificed on the first day of estrus and processed for immunohistochemistry (see Materials and Methods). (A) Uteri stained with the α(1,2)fucosylated glycan-specific lectin UEA-I or H type I-specific MAb. The brown staining identifies H antigen-reactive glycans within the glycocalyx on the apical surfaces of luminal uterine epithelial cells from wild-type mice and FUT1−/− mice. Staining is absent from the glycocalyx of FUT2−/− uterine epithelia. (B) A FUT2−/− uterus was stained with a MAb (H-precursor MAb; BG-1) specific for nonfucosylated type I glycans that serve as precursors for H type I blood group synthesis (see Materials and Methods).
Expression of α(1,2)fucosylated glycans in epididymal epithelia requires FUT1 and is exclusive of a contribution by FUT2.
In the murine epididymis, a gradient of fucosyltransferase expression associated with spermatozoa increases from the caput, near the seminiferous tubules, to the cauda, near the junction with the vas deferens (6, 34, 36). This gradient parallels the acquisition by spermatozoa of α(1,2)fucosylated glycans, correlates with acquisition by spermatozoa of the ability to fertilize (5, 43), and implies a requirement for these epididymis-associated α(1,2)fucosylation events in fertility. In situ hybridization analyses define a gradient of expression of FUT1 transcripts in the epithelia along the length of the epididymis (8) but do not detect FUT2 transcripts in this organ, implying that the FUT1 locus controls epididymal α(1,2)fucosylation events associated with maturation of spermatozoa. To confirm these assignments and to verify that deletion of the FUT1 locus disables expression of α(1,2)fucosylated glycans, H-antigen expression was assessed on the epithelia of the proximal and mid-caput portions of the epididymis and on the corpus of the epididymis in FUT1−/− mice, FUT2−/− mice, and wild-type controls (Fig. 4). In wild-type mice, H antigen, detected with the lectin UEA-I, is prominently expressed in the mid-caput of the epididymis but is absent from the proximal segment of the organ (Fig. 4). In the mid-caput, H antigen localizes to the luminal surface of the epididymal epithelia and is also associated with spermatozoa and molecules within the tubules.
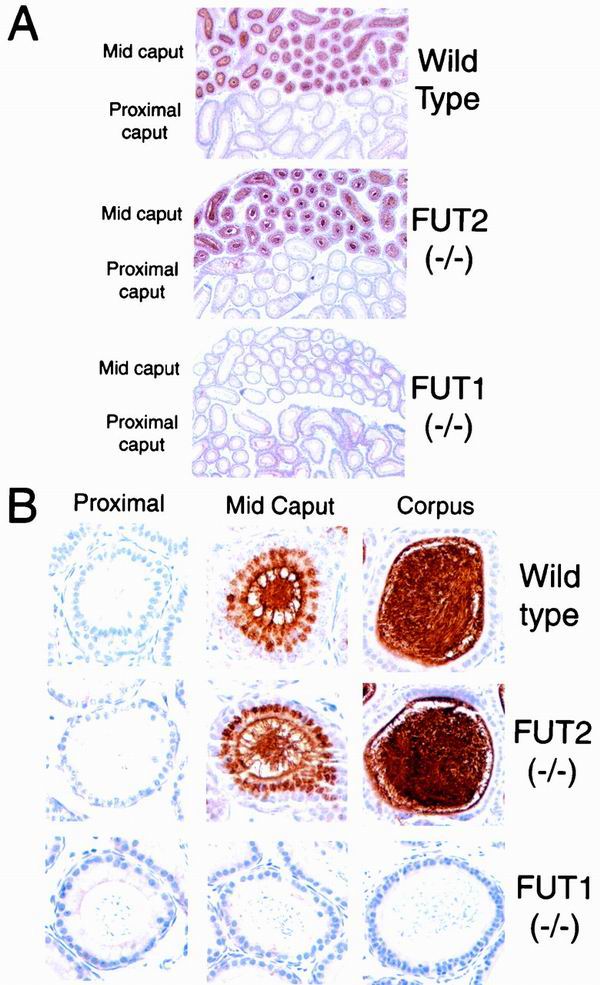
FIG. 4.
Lectin histochemistry of the epididymis in wild-type, FUT2 null, and FUT1 null mice. Epididymides from adult wild-type, FUT2 null, and FUT1 null male mice were stained with UEA-I (see Materials and Methods). (A) Low magnification (×50) of epididymis UEA-I histochemistry. UEA-I staining is prominent in the mid-caput but absence from the proximal caput in wild-type mice and FUT2−/− mice. UEA-I does not stain the mid-caput or proximal caput in FUT1−/− mice. (B) High magnification (×500) of epididymis UEA-I histochemistry. Single epididymal tubules are shown from proximal caput, mid-caput, and corpus from adult wild-type, FUT2 null, and FUT1 null mice. Mid-caput epithelial cells and adjacent spermatozoa show specific UEA-I staining in wild-type and FUT2 null mice but not FUT1 null mice. Spermatozoa from the corpus epididymis display dense UEA-I staining in wild-type and FUT2 null mice but not FUT1 null mice.
Epithelia in the mid-caput also display prominent intracellular expression of H antigen (Fig. 4B). Intracellular staining is most evident in a supranuclear position suggestive of Golgi localization, which implies that the mid-caput epididymal epithelia are highly active in the elaboration of cell surface and secreted H-active glycans. H-antigen expression is also observed in the corpus and is associated with the surface of the epithelia and with the spermatozoa and other tubular contents. The prominent intracellular staining observed in mid-caput epithelia is not apparent in the epithelia of the corpus. This observation implies that epithelia in the corpus maintain little if any H-glycan synthetic activity and suggests that much or even all of the apparent association of H antigen with the luminal surface of the corpus epithelium may be accounted for by adsorption of H-active molecules present in the lumen but synthesized more proximally in the epididymis.
FUT2−/− mice are indistinguishable from wild-type mice with respect to the density of H-antigen expression on epididymal epithelia and in the luminal contents, to the gradient of H expression along the length of the epididymis, to the extent and location of intracellular expression in mid-caput epithelia, and to the overall microscopic morphology of the tubules (Fig. 4). These observations indicate that FUT2 is not required for H expression in these contexts or for the organization or structural integrity of the tissue. By contrast, the epididymal epithelia in FUT1−/− mice are devoid of detectable H structures. This observation demonstrates that the FUT1 mutant allele is null and assigns the FUT1 locus, exclusive of FUT2, to the control of α(1,2)fucosylated glycan expression in the epididymis. Tubular and epithelial morphology and the overall organization of the epididymal structures are normal in FUT1−/− mice, indicating that FUT1 is not required for the development of this tissue.
Northern blot analyses identify transcripts derived from the SEC1 locus in testes and epididymis (9). The SEC1 locus is predicted to encode a polypeptide with substantial primary sequence similarity with FUT1 and FUT2, directs low-level expression of α(1,2)fucosylated glycans in transfected mammalian cells (9), and is therefore a candidate for control of expression of α(1,2)fucosylated glycans in the epididymis. However, SEC1 does not contribute in this context, since α(1,2)fucosylated epitopes are fully deleted from the glycans in the FUT2 null epididymis.
Uterine epithelial α(1,2)fucosylated glycans are dispensable for fertility.
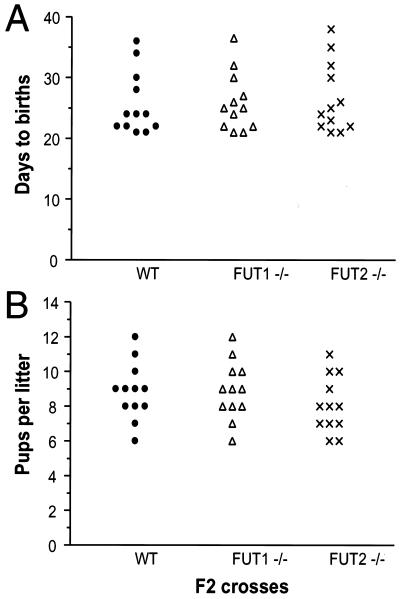
Blastocyst attachment to uterine epithelia in vitro and blastocyst implantation in vivo are inhibited by blockade of α(1,2)fucosylated glycans corresponding to the H and Lewis Y epitopes (27, 46, 50). Blockade correlates with binding of α(1,2)fucosylated glycans to the mural trophectoderm on hatched blastocysts, the site of blastocyst attachment to the receptive uterine wall (26, 48). These observations, together with cycle-dependent expression if α(1,2)fucosylated glycans by the uterine epithelia, have suggested an essential role for uterine α(1,2)fucosylated glycan-dependent adhesive interactions in fertility. To directly examine this issue, fertility was assessed in FUT2 null mice, whose uterine epithelia are genetically deficient in expression of α(1,2)fucosylated glycans. In intercrosses of FUT2 null adults, elapsed time between the first day when a male is housed with a receptive female and the day of birth (mean ± standard deviation, 26.6 ± 5.8 days) does not differ from elapsed time to birth in wild-type intercrosses (25.7 ± 5.2 days) (Fig. 5). FUT2 null intercrosses also yield litter sizes (8.1 ± 1.6 pups) equivalent to the sizes of litters born in wild-type intercrosses (8.8 ± 1.6 pups). These observations indicate that FUT2 null females are as competent as wild-type females in initiating and sustaining pregnancy and excludes a requirement for uterine epithelial cell surface α(1,2)fucosylated glycans in the support of blastocyst adhesion or implantation or in the maintenance of pregnancy. These observations also show that the FUT2 locus is dispensable for fertility in males.
FIG. 5.
Frequencies of births and litter sizes in wild-type, FUT1−/−, and FUT2−/− intercrosses. (A) Days to birth. The duration of gestation is defined as the time elapsed between the first day when a male was housed with a receptive female to the day of birth. Each data point corresponds to a single litter and derives from matings between four breeding pairs within each genotype. (B) Number of pups per litter at birth. The number of live pups from each birth was recorded.
Normal fertility in the absence of epididymal α(1,2)fucosylated glycans.
During passage from the caput to the cauda of the epididymis, spermatozoa acquire α(1,2)fucosylated glycans during a maturational translocation from the testes that is associated with acquisition of competence for fertility (5, 43). To determine if epididymal acquisition of α(1,2)fucosylated glycans is a prerequisite for the fertilization competence of spermatozoa, fertility was assessed in FUT1 null mice, wherein α(1,2)fucosylated glycans are absent from the epididymal epithelia. In FUT1 null intercrosses, elapsed time to birth (26.0 ± 4.8 days) is equivalent to that in wild-type intercrosses (Fig. 5), as is the average litter size (8.9 ± 1.7 pups) (Fig. 5). FUT1 null males are therefore as fertile as wild-type males, excluding a requirement for FUT1-dependent epididymal fucosylation events in the spermatozoan maturation process. These observations also exclude a requirement for FUT1-dependent fucosylation in postcoital fertilization events, where fucosylated glycans have been implicated in sperm capacitation (14), and exclude a requirement for FUT1 in female fertility.
These studies exclude an essential role in fertility for the α(1,2)fucosylated glycans elaborated by FUT1 and FUT2 but leave open the possibility that such glycans contribute to other functions in the reproductive tract, including protection from pathogens. The lineage-restricted expression of FUT1 and FUT2 characteristic of the reproductive tract extends to other tissues in the mouse, inferring potential functions for their cognate glycans in other biological contexts. Aside from the uterus, expression of the FUT2 locus is restricted to the stomach and colon among the major organs in an adult mouse (9). Extraepididymal expression of the FUT1 locus has also been assigned to the stomach and colon; in those locales, the two corresponding fucosyltransferases may contribute to host-microbe interactions that involve fucosylated glycans (18). Efforts are in progress with FUT1−/− and FUT2−/− mice to determine if the absence of α(1,2)fucosylation in the gastrointestinal tract modulates their susceptibility to microbial symbiosis, commensalism, or pathogenicity and to more carefully define expression patterns in these contexts using the β-galactosidase reporter installed in each locus.
These mice are also being used to determine if thymus-specific expression of FUT1 (8) controls expression of α(1,2)fucosylated glycans displayed by the medullary epithelial cells in this organ (12) and to identify functional correlates for such expression. Similar considerations apply, for example, to the intestinal M cell, an H-antigen-positive epithelial cell that overlies mucosa-associated lymphoid tissues (17), to the α(1,2)fucosylated olfactory epithelium (10), to rare α(1,2)fucosylated enteroendocrine cells of the gastrointestinal tract (15, 16), and to other cell types revealing restricted expression of α(1,2)fucosylated glycans in development and differentiation (39).
ACKNOWLEDGMENTS
This work was supported by a fellowship grant from the Reproductive Scientist Development Program and NIH K08 HD01195 (S.E.D), P01 CA71932 (J.B.L), University of Michigan Cancer Research Committee John S. and Suzanne C. Munn Endowed Research Fund (S.E.D.), and University of Michigan Phoenix Memorial Laboratory Michigan Memorial-Phoenix Project no. 856 (S.E.D.). J.B.L. is an Investigator of the Howard Hughes Medical Institute.
We are grateful to the University of Michigan Transgenic Animal Core (L. Samuelson, S. Camper, E. Hughes, M. Berand, and M. Van Keuren), DNA Sequencing Core (R. Lyons, S. Genik, and C. Esposito), DNA Synthesis Core (C. Wong), and Comprehensive Cancer Center Research Histology and Immunoperoxidase Laboratory (M. Rubin, M. LeBlanc, and N. McAnsh). We acknowledge R. Palmiter for plasmid pnlacF, T. Doetschman for mice transgenic for the neo gene, and J. Rossant and J. C. Roder for the R1 cell line made available through the Transgenic Animal Model Core.
REFERENCES
- 1.Bird J M, Kimber S J. Oligosaccharides containing fucose linked alpha(1-3) and alpha(1-4) to N-acetylglucosamine cause decompaction of mouse morulae. Dev Biol. 1984;104:449–460. doi: 10.1016/0012-1606(84)90101-5. [DOI] [PubMed] [Google Scholar]
- 2.Bradley A. Production and analysis of chimeric mice. In: Robertson E J, editor. Teratocarcinomas and embryonic stem cells. Oxford, United Kingdom: IRL Press at Oxford University Press; 1987. [Google Scholar]
- 3.Bruckner K, Perez L, Clausen H, Cohen S. Glycosyltransferase activity of Fringe modulates Notch-Delta interactions. Nature. 2000;406:411–415. doi: 10.1038/35019075. [DOI] [PubMed] [Google Scholar]
- 4.Bry L, Falk P, Midtvedt T, Gordon J. A model of host-microbial interactions in an open mammalian ecosystem. Science. 1996;273:1380–1383. doi: 10.1126/science.273.5280.1380. [DOI] [PubMed] [Google Scholar]
- 5.Cardullo R A, Armant D R, Millette C F. Characterization of fucosyltransferase activity during mouse spermatogenesis: evidence for a cell surface fucosyltransferase. Biochemistry. 1989;28:1611–1617. doi: 10.1021/bi00430a028. [DOI] [PubMed] [Google Scholar]
- 6.Cossu G, Boitani C. Lactosaminoglycans synthesized by mouse male germ cells are fucosylated by an epididymal fucosyltransferase. Dev Biol. 1984;102:402–408. doi: 10.1016/0012-1606(84)90204-5. [DOI] [PubMed] [Google Scholar]
- 7.Dell A, Morris H R, Easton R L, Patankar M, Clark G F. The glycobiology of gametes and fertilization. Biochim Biophys Acta. 1999;1473:196–205. doi: 10.1016/s0304-4165(99)00179-8. [DOI] [PubMed] [Google Scholar]
- 8.Domino S E, Hiraiwa N, Lowe J B. Molecular cloning and tissue-specific expression of a murine α(1,2)fucosyltransferase expressed in thymic and epididymal epithelial cells. Biochem J. 1997;327:105–115. doi: 10.1042/bj3270105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Domino S E, Zhang L, Lowe J B. Molecular cloning, genomic mapping, and expression of two secretor blood group α(1,2)fucosyltransferase genes differentially regulated in mouse uterine epithelium and gastrointestinal tract. J Biol Chem. 2001;276:23748–23756. doi: 10.1074/jbc.M100735200. [DOI] [PubMed] [Google Scholar]
- 10.Ducray A, Propper A, Kastner A. Detection of alpha-L fucose containing carbohydrates in mouse immature olfactory neurons. Neurosci Lett. 1999;274:17–20. doi: 10.1016/s0304-3940(99)00674-6. [DOI] [PubMed] [Google Scholar]
- 11.Falk P, Bry L, Holgersson J, Gordon J. Expression of a human alpha-1,3/4-fucosyltransferase in the pit cell lineage of FVB/N mouse stomach results in production of Leb-containing glycoconjugates: a potential transgenic mouse model for studying Helicobacter pylori infection. Proc Natl Acad Sci USA. 1995;92:1515–1519. doi: 10.1073/pnas.92.5.1515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Farr A, Hosier S, Nelson A, Itohara S, Tonegawa S. Distribution of thymocytes expressing gamma delta receptors in the murine thymus during development. J Immunol. 1990;144:492–498. [PubMed] [Google Scholar]
- 13.Fenderson B A, Zehavi U, Hakomori S. A multivalent lacto-N-fucopentaose III-lysyllysine conjugate decompacts preimplantation mouse embryos, while the free oligosaccharide is ineffective. J Exp Med. 1984;160:1591–1596. doi: 10.1084/jem.160.5.1591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fraser L R. Interactions between a decapacitation factor and mouse spermatozoa appear to involve fucose residues and a GPI-anchored receptor. Mol Reprod Dev. 1998;51:193–202. doi: 10.1002/(SICI)1098-2795(199810)51:2<193::AID-MRD9>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- 15.Gebert A, Cetin Y. Expression of fucose residues in entero-endocrine cells. Histochem Cell Biol. 1998;109:161–165. doi: 10.1007/s004180050213. [DOI] [PubMed] [Google Scholar]
- 16.Gebert A, Posselt W. Glycoconjugate expression defines the origin and differentiation pathway of intestinal M-cells. J Histochem Cytochem. 1997;45:1341–1350. doi: 10.1177/002215549704501003. [DOI] [PubMed] [Google Scholar]
- 17.Giannasca P J, Giannasca K T, Falk P, Gordon J I, Neutra M R. Regional differences in glycoconjugates of intestinal M cells in mice: potential targets for mucosal vaccines. Am J Physiol. 1994;267:G1108–G1121. doi: 10.1152/ajpgi.1994.267.6.G1108. [DOI] [PubMed] [Google Scholar]
- 18.Hooper L V, Gordon J I. Glycans as legislators of host-microbial interactions: spanning the spectrum from symbiosis to pathogenicity. Glycobiology. 2001;11:1R–10R. doi: 10.1093/glycob/11.2.1r. [DOI] [PubMed] [Google Scholar]
- 19.Illingworth I M, Kimber S J. Demonstration of oestrogenic control of H-type-1 carbohydrate antigen in the murine endometrial epithelium by use of ICI 182,780. J Reprod Fertil. 1999;117:89–95. doi: 10.1530/jrf.0.1170089. [DOI] [PubMed] [Google Scholar]
- 20.Kendall S K, Samuelson L C, Saunders T L, Wood R I, Camper S A. Targeted disruption of the pituitary glycoprotein hormone alpha-subunit produces hypogonadal and hypothyroid mice. Genes Dev. 1995;9:2007–2019. doi: 10.1101/gad.9.16.2007. [DOI] [PubMed] [Google Scholar]
- 21.Kimber S J, Lindenberg S. Hormonal control of a carbohydrate epitope involved in implantation in mice. J Reprod Fertil. 1990;89:13–21. doi: 10.1530/jrf.0.0890013. [DOI] [PubMed] [Google Scholar]
- 22.Kimber S J, Lindenberg S, Lundblad A. Distribution of some Galβ1-3(4)GlcNAc related carbohydrate antigens on the mouse uterine epithelium in relation to the peri-implantational period. J Reprod Immunol. 1988;12:297–313. doi: 10.1016/0165-0378(88)90015-0. [DOI] [PubMed] [Google Scholar]
- 23.Kimber S J, Spanswick C. Blastocyst implantation: the adhesion cascade. Semin Cell Dev Biol. 2000;11:77–92. doi: 10.1006/scdb.2000.0154. [DOI] [PubMed] [Google Scholar]
- 24.Lin B, Hayashi Y, Saito M, Sakakibara Y, Yanagisawa M, Iwamori M. GDP-fucose: β-galactoside α1,2-fucosyltransferase, MFUT-II, and not MFUT-I or -III, is induced in a restricted region of the digestive tract of germ-free mice by host-microbe interactions and cycloheximide. Biochim Biophys Acta. 2000;1487:275–285. doi: 10.1016/s1388-1981(00)00103-7. [DOI] [PubMed] [Google Scholar]
- 25.Lin B, Saito M, Sakakibara Y, Hayashi Y, Yanagisawa M, Iwamori M. Characterization of three members of murine α1,2-fucosyltransferases: change in the expression of the Se gene in the intestine of mice after administration of microbes. Arch Biochem Biophys. 2001;388:207–215. doi: 10.1006/abbi.2001.2303. [DOI] [PubMed] [Google Scholar]
- 26.Lindenberg S, Kimber S J, Kallin E. Carbohydrate binding properties of mouse embryos. J Reprod Fertil. 1990;89:431–439. doi: 10.1530/jrf.0.0890431. [DOI] [PubMed] [Google Scholar]
- 27.Lindenberg S, Sundberg K, Kimber S J, Lundblad A. The milk oligosaccharide, lacto-N-fucopentaose I, inhibits attachment of mouse blastocysts on endometrial monolayers. J Reprod Fertil. 1988;83:149–158. doi: 10.1530/jrf.0.0830149. [DOI] [PubMed] [Google Scholar]
- 28.Lowe J B. The blood group-specific human glycosyltransferases. Bailliere's Clin Haematol. 1993;6:465–492. doi: 10.1016/s0950-3536(05)80155-6. [DOI] [PubMed] [Google Scholar]
- 29.Miller S A, Dykes D D, Polesky H F. A simple salting out procedure for extracting DNA from human nucleated cells. Nucleic Acids Res. 1988;16:1215. doi: 10.1093/nar/16.3.1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Millette C F, Cardullo R A, Armant D R, Gerton G L. Fucosylation events during mammalian spermatogenesis. Ann N Y Acad Sci. 1987;513:58–73. doi: 10.1111/j.1749-6632.1987.tb24998.x. [DOI] [PubMed] [Google Scholar]
- 31.Miyoshi E, Noda K, Yamaguchi Y, Inoue S, Ikeda Y, Wang W, Ko J H, Uozumi N, Li W, Taniguchi N. The α1-6-fucosyltransferase gene and its biological significance. Biochim Biophys Acta. 1999;1473:9–20. doi: 10.1016/s0304-4165(99)00166-x. [DOI] [PubMed] [Google Scholar]
- 32.Moloney D J, Panin V M, Johnston S H, Chen J, Shao L, Wilson R, Wang Y, Stanley P, Irvine K D, Haltiwanger R S, Vogt T F. Fringe is a glycosyltransferase that modifies Notch. Nature. 2000;406:369–375. doi: 10.1038/35019000. [DOI] [PubMed] [Google Scholar]
- 33.Nagy A, Rossant J, Nagy R, Abramow-Newerly W, Roder J C. Derivation of completely cell culture-derived mice from early-passage embryonic stem cells. Proc Natl Acad Sci USA. 1993;90:8424–8428. doi: 10.1073/pnas.90.18.8424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ram P A, Cardullo R A, Millette C F. Expression and topographical localization of cell surface fucosyltransferase activity during epididymal sperm maturation in the mouse. Gamete Res. 1989;22:321–332. doi: 10.1002/mrd.1120220309. [DOI] [PubMed] [Google Scholar]
- 35.Ramirez-Solis R, Rivera-Perez J, Wallace J D, Wims M, Zheng H, Bradley A. Genomic DNA microextraction: a method to screen numerous samples. Anal Biochem. 1992;201:331–335. doi: 10.1016/0003-2697(92)90347-a. [DOI] [PubMed] [Google Scholar]
- 36.Raychoudhury S S, Millette C F. Glycosidic specificity of fucosyltransferases present in rat epididymal spermatozoa. J Androl. 1995;16:448–456. [PubMed] [Google Scholar]
- 37.Raychoudhury S S, Millette C F. Multiple fucosyltransferases and their carbohydrate ligands are involved in spermatogenic cell-Sertoli cell adhesion in vitro in rats. Biol Reprod. 1997;56:1268–1273. doi: 10.1095/biolreprod56.5.1268. [DOI] [PubMed] [Google Scholar]
- 38.Raychoudhury S S, Millette C F. Presence of multiple fucosyltransferases in rat Sertoli cells and spermatogenic cells. Biol Reprod. 1994;51:1006–1013. doi: 10.1095/biolreprod51.5.1006. [DOI] [PubMed] [Google Scholar]
- 39.Sato M, Yonezawa S, Uehara H, Arita Y, Sato E, Muramatsu T. Differential distribution of receptors for two fucose-binding lectins in embryos and adult tissues of the mouse. Differentiation. 1986;30:211–219. doi: 10.1111/j.1432-0436.1986.tb00783.x. [DOI] [PubMed] [Google Scholar]
- 40.Sidu S S, Kimber S J. Hormonal control of H-type alpha(1-2)fucosyltransferase messenger ribonucleic acid in the mouse uterus. Biol Reprod. 1999;60:147–157. doi: 10.1095/biolreprod60.1.147. [DOI] [PubMed] [Google Scholar]
- 41.Solter D, Knowles B B. Immunosurgery of mouse blastocyst. Proc Natl Acad Sci USA. 1975;72:5099–5102. doi: 10.1073/pnas.72.12.5099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Staudacher E, Altmann F, Wilson I B, Marz L. Fucose in N-glycans: from plant to man. Biochim Biophys Acta. 1999;1473:216–236. doi: 10.1016/s0304-4165(99)00181-6. [DOI] [PubMed] [Google Scholar]
- 43.Tulsiani D R, Skudlarek M D, Holland M K, Orgebin-Crist M C. Glycosylation of rat sperm plasma membrane during epididymal maturation. Biol Reprod. 1993;48:417–428. doi: 10.1095/biolreprod48.2.417. [DOI] [PubMed] [Google Scholar]
- 44.Tybulewicz V L, Crawford C E, Jackson P K, Bronson R T, Mulligan R C. Neonatal lethality and lymphopenia in mice with a homozygous disruption of the c-abl proto-oncogene. Cell. 1991;65:1153–1163. doi: 10.1016/0092-8674(91)90011-m. [DOI] [PubMed] [Google Scholar]
- 45.Vestweber D, Blanks J E. Mechanisms that regulate the function of the selectins and their ligands. Physiol Rev. 1999;79:181–213. doi: 10.1152/physrev.1999.79.1.181. [DOI] [PubMed] [Google Scholar]
- 46.Wang X Q, Zhu Z M, Fenderson B A, Zeng G Q, Cao Y J, Jiang G T. Effects of monoclonal antibody directed to LeY on implantation in the mouse. Mol Hum Reprod. 1998;4:295–300. doi: 10.1093/molehr/4.3.295. [DOI] [PubMed] [Google Scholar]
- 47.White S, Kimber S J. Changes in alpha(1-2)fucosyltransferase activity in the mouse endometrial epithelium during the estrous cycle, early pregnancy, and after ovariectomy and hormone replacement. Biol Reprod. 1994;50:73–81. doi: 10.1095/biolreprod50.1.73. [DOI] [PubMed] [Google Scholar]
- 48.Yamagata T, Yamazaki K. Implanting mouse embryo stain with a LNF-I bearing fluorescent probe at their mural trophectodermal side. Biochem Biophys Res Commun. 1991;181:1004–1009. doi: 10.1016/0006-291x(91)92036-j. [DOI] [PubMed] [Google Scholar]
- 49.Zambrowicz B, Zimmermann J, Harendza C, Simpson E, Page D, Brinster R, Palmiter R. Expression of a mouse Zfy-1/lacZ transgene in the somatic cells of the embryonic gonad and germ cells of the adult testis. Development. 1994;120:1549–1559. doi: 10.1242/dev.120.6.1549. [DOI] [PubMed] [Google Scholar]
- 50.Zhu Z M, Kojima N, Stroud M R, Hakomori S, Fenderson B A. Monoclonal antibody directed to Le(y) oligosaccharide inhibits implantation in the mouse. Biol Reprod. 1995;52:903–912. doi: 10.1095/biolreprod52.4.903. [DOI] [PubMed] [Google Scholar]