Abstract
In Saccharomyces cerevisiae, in vitro mRNA cleavage and polyadenylation require the poly(A) binding protein, Pab1p, and two multiprotein complexes: CFI (cleavage factor I) and CPF (cleavage and polyadenylation factor). We characterized a novel essential gene, MPE1 (YKL059c), which interacts genetically with the PCF11 gene encoding a subunit of CFI. Mpe1p is an evolutionarily conserved protein, a homolog of which is encoded by the human genome. The protein sequence contains a putative RNA-binding zinc knuckle motif. MPE1 is implicated in the choice of ACT1 mRNA polyadenylation site in vivo. Extracts from a conditional mutant, mpe1-1, or from a wild-type extract immunoneutralized for Mpe1p are defective in 3′-end processing. We used the tandem affinity purification (TAP) method on strains TAP-tagged for Mpe1p or Pfs2p to show that Mpe1p, like Pfs2p, is an integral subunit of CPF. Nevertheless a stable CPF, devoid of Mpe1p, was purified from the mpe1-1 mutant strain, showing that Mpe1p is not directly involved in the stability of this complex. Consistently, Mpe1p is also not necessary for the processive polyadenylation, nonspecific for the genuine pre-mRNA 3′ end, displayed by the CPF alone. However, a reconstituted assay with purified CFI, CPF, and the recombinant Pab1p showed that Mpe1p is strictly required for the specific cleavage and polyadenylation of pre-mRNA. These results show that Mpe1p plays a crucial role in 3′ end formation probably by promoting the specific link between the CFI/CPF complex and pre-mRNA.
In eukaryotic cells, 3′-end cleavage and polyadenylation are essential steps in the synthesis of functional mRNAs. These processes are involved in transcription termination (21, 34) and the export of mature mRNAs from the nucleus (23). The poly(A) tail associated with the poly(A)-binding protein, Pab1p, is directly implicated in the regulation of gene expression, by stimulating the translation of mRNA and controlling its stability during deadenylation-dependent degradation (15, 32, 49). Prior to polyadenylation, the nascent transcripts are cleaved at a specific site. The cleavage and polyadenylation processes are coupled in vivo, and the specificity of the reaction is ensured by the recognition of cis-acting sequences on the transcript by a broad multiprotein complex. These two steps can be studied independently in cellular extracts. This led to the purification and characterization of many essential factors (reviewed in references 48 and 52). In the yeast Saccharomyces cerevisiae, two complexes, CFI (cleavage factor I) and CPF (cleavage and polyadenylation factor), plus Pab1p are responsible for the specific cleavage and polyadenylation of pre-mRNA. CFI was purified and shown to be composed of two subcomplexes, CFIA and CFIB (26). CFIA consists of Rna14p, Rna15p, Pcf11p, and Clp1p (26, 36). The phenotypes of Rna14p, Rna15p, and Pcf11p mutants confirmed that they are directly involved in 3′-end processing (4, 35). CFIB consists of a single protein, Hrp1p/Nab4p, which is specifically implicated in the correct choice of cleavage site (25, 33). Pab1p partially copurifies with CFI and is involved in the regulation of the length of the poly(A) tail but not in cleavage (3, 36). The 3′-end processing factors were initially identified by extensive chromatographic purification steps giving different subcomplexes, some of which contained common subunits (40, 53). More recently, affinity purification was used to study the proteins associated with a component of the polyadenylation complex, the protein A-tagged Pfs2p (38). This technique allowed the direct isolation of the CPF complex containing all of the previously identified proteins but not the CFI complex proteins, showing that CFI is a separate entity. The CPF complex contains the poly(A) polymerase enzyme and the Cft1p/Yhh1p, Cft2p/Ydh1p, Brr5p/Ysh1p, Pta1p, Fip1p, Yth1p, Pfs2p, and Pfs1p subunits (6, 38, 40). Only the Pfs1 protein remains uncharacterized. The exact functions of the different proteins in each complex are unknown, probably because they act together in a concerted manner. All of the subunits are essential, and the phenotypes of available conditional mutants show that, except for Fip1p, they are essential for both cleavage and polyadenylation (6, 38, 39, 52, 53). The protein directly involved in the endonucleolytic cleavage has not yet been identified. The Pap1 protein alone is known to catalyze polyadenylation. However, if CFI, CPF, and Pab1p are absent this polyadenylation occurs in an unregulated way (33, 38, 40).
The 3′-end processing apparatus is highly conserved between yeast and mammals, although there are differences in the sequences of the proteins and in the organization of the mRNA 3′-end processing signals (24, 44, 48, 52). Rna14p and Rna15p from yeast CFI have a high degree of identity with p77 and p64 from the mammalian cleavage stimulation factor, CstF. Similarly, Cft1p/Yhh1p, Cft2p/Ydh1p, Brr5p/Ysh1p, and Yth1p from CPF are the yeast homologues of the four subunits of the mammalian cleavage polyadenylation specificity factor (CPSF). Recently, the mammalian cleavage factor II, CFIIm, was purified and found to contain homologs to Pcf11p and Clp1p of yeast CFIA (12). A mammalian protein that interacts with CstF and seems to be involved in the assembly or the stability of the 3′-end processing complex shares similarities with Pta1p from yeast CPF (46). It is not yet clear whether the sequence homologies reflect functional identities. For example, the mammalian CFIIm is only required for cleavage in vitro, whereas the yeast CFI is necessary for both cleavage and polyadenylation. Nevertheless, it is probably mainly the protein-protein interactions that have acquired different affinities during evolution, resulting in a different repartition between subcomplexes, and the catalytic functions themselves have probably not been modified considerably.
An exhaustive study of the sequence of some genes has identified three domains defining the yeast polyadenylation signal: the efficiency element, the positioning element, and the cleavage site itself (reviewed in references 48 and 52). A recent statistical study on the 3′ end of a large number of genes suggested that two more signals exist, one on each side of the cleavage site (17, 47). The sequences of the efficiency and positioning elements are degenerate and redundant (47). Most of the genes contain many potential cleavage sites that are used by the 3′-end processing complex with different efficiencies (47). Modifications in the pattern of these signal sequences or mutations in the proteins constituting the 3′-end processing complex lead to identical alterations in the choice of cleavage site on mRNAs (14, 30, 31). This complexity offers a possibility for gene regulation by providing a choice of 3′ cleavage sites. Indeed, the distribution of different 3′-end transcripts of some genes has been found to be dependent on stress responses or growth conditions (22, 45).
We report here the characterization of MPE1, an essential yeast gene, which encodes a protein that is necessary for in vitro 3′-end processing and is a subunit of the CPF complex.
MATERIALS AND METHODS
Yeast strains generation of conditional mpe1 allele and alleles expressing tandem affinity purification (TAP)-tagged protein.
The pcf11-2, mpe1-S, mpe1-1, and mpe1-1-TAP strains were derived from the W303-1B strain (ura3-1 trp1-1 ade2-1 leu2-3,112 his3-11,15). The MPE1-TAP, MPE1ΔCterm-TAP, PFS2-TAP, and PCF11-TAP strains were derived from the BMA-64 strain (ura3-1 Δtrp1 ade2-1 leu2-3,112 his3-11,15).
The MPE1 gene was disrupted by replacing the coding region (amino acids 4 to 438) with the TRPI cassette in the diploid BMA64 strain according to the previously described PCR-based method (8). To generate mutants, the MPE1 gene was amplified with the oligonucleotides 5′-CCT TAA TCC ATT GCT GGT GC-3′, which begins 226 bp upstream of the start codon, and 5′-CGA CGG CTG CTA GAA TGG-3′, which begins 204 bp downstream of the stop codon. PCR was performed with Taq polymerase (Appligene) as described by the manufacturer except for the deoxynucleoside triphosphate concentration (200 mM concentrations of each except for either 50 mM dCTP or 50 mM dATP). The PCR products were cloned directly in vivo by gap repair of a pFL36 MPE1(LEU2) vector in a strain in which the MPE1 coding region had been deleted and that contained pFL38-MPE1 (URA3). The LEU2+ clones were then transferred onto 5-fluoro-orotic acid to lose the URA3+ marked plasmid and assayed for temperature sensitivity (10).
The strains expressing the TAP-tagged proteins were obtained by inserting the TAP-tag sequence immediately upstream of the stop codon of the corresponding gene as described by Puig et al. (41).
Most media and genetic methods were as described by Guthrie and Fink (19). Yeast strains were transformed by the lithium acetate method (16).
Sequence.
The Mpe1p sequence was analyzed with the PSI-BLAST and CDD programs (1). The S. cerevisiae sequence (GenBank accession no. 6322791) was lined up by CLUSTALX with corresponding motifs found in genes from Shizosaccharomyces pombe (GenBank accession no. 7490387), Arabidopsis thaliana (GenBank accession no. 10178219) and Drosophila melanogaster (GenBank accession no. 4972740) and in three cDNAs from Homo sapiens (H.s.1, GenBank accession no. 13295901; H.s.2, GenBank accession no. 10991320; and H.s.3, GenBank accession no. 12002024). The putative human open reading frame (ORF) was obtained with the GENSCANW and GeneMark programs. It consists of 16 exons in the contig NT_010604.3 (c836544 to 799544) on chromosome 16 in the following positions: 1, c834386 to 833812 with the start codon at 833977; 2, c828441 to 828342; 3, c825659 to 825510 (GENSCANW) or c822228 to 822117 (GeneMark); 4, c819257 to 819169; 5, c819051 to 818955; 6, c818564 to 818425; 6′, c816451 to 816338 (GENSCANW); 7, c815367 to 815195; 8, c813260 to 813157; 9, c813048 to 812711; 10, c811673 to 811577; 11, c811451 to 811371; 12, c811262 to 811208; 12′, c810075 to 810007 (GeneMark); 13, c807079 to 806978; 14, c806126 to 804372; and 15, c803995 to 802056, with the stop codon at 802424 and the AATAAA polyadenylation signal at c802035.
mRNA analysis.
RNase H treatment and Northern blots were carried out as previously described (2). The ACT1 mRNA contained in 20 μg of total RNA was cleaved by RNase H in the presence of both oligo(dT) and the oligonucleotide 5′-GAAGATGGAGCCAAAGC-3′, which is complementary to nucleotides 158 to 174 upstream of the ACT1 stop codon.
Recombinant proteins, antibodies, and immunoblots.
Purified recombinant Pab1p was obtained as described previously (3). The C-terminal region of Mpe1p (amino acids 264 to 441) was expressed as a His6 fusion peptide after being cloned into pET22b(+) (Novagen). The recombinant polypeptide was produced in Escherichia coli BL21(DE3) and purified on nickel-nitriloacetic acid agarose (Qiagen) according to the manufacturer's instructions. Polyclonal anti-Mpe1p antiserum was obtained by immunizing rabbits. Immunoblots were processed, and signals were detected by chemiluminescence (SuperSignal West Pico Chemiluminescent Substrate; Pierce) according to standard procedures (20).
In vitro 3′-end processing assays.
32P-labeled CYC1 RNA and precleaved CYC1 RNA were synthesized as runoff transcripts from linearized plasmids as described previously (35, 40). Proteins were extracted and fractionated by centrifugation and ammonium sulfate precipitation as previously described (11).
The CFI and CPF complexes were purified by the TAP method (42) starting from strains expressing the different TAP-tagged proteins. The complexes were eluted from immunoglobulin G (IgG) agarose by a tobacco etch virus proteolytic treatment, dialyzed against 20 mM Tris-HCl (pH 7.9)–0.2 mM EDTA–50 mM KCl–0.5 mM dithiothreitol–20% glycerol, and kept at −80°C.
In vitro polyadenylation assays were carried out in the presence of ATP and magnesium acetate. Specific cleavage was assayed in the presence of CTP and EDTA to inhibit the formation of poly(A) tails, as previously described (35).
TAP.
The TAP-tag contains the calmodulin-binding peptide (CBP), the specific cleavage site of the TEV protease, and protein A and is a 25-kDa peptide. It allows the complex associated with a TAP-tagged protein to bind to IgG agarose and to be eluted by TEV proteolytic treatment. The complex is then repurified on calmodulin agarose and finally eluted off with EGTA. After this procedure, the tagged protein still contains the CBP domain, the molecular mass of which is increased by 5 kDa.
Complexes were purified from 1 liter of yeast cells expressing the TAP-tagged proteins that had been grown to an optical density at 600 nm of 2 to 4 in rich medium. Complexes were purified according to the standard TAP procedure, except that after adsorption of IgG agarose the matrix was washed twice at a higher stringency with a 400 mM NaCl washing buffer for 5 min (42). Purified proteins were precipitated with trichloroacetic acid, separated on a sodium dodecyl sulfate (SDS)-10% polyacrylamide gel, and stained with Coomassie blue.
Mass-spectrometric identification.
Stained bands were excised and subjected to proteolytic digestion with endoproteinase Lys-C (Roche Diagnostics, Meylan, France) as described elsewhere (18) A microchromatographic separation of proteolytic digests was performed on C18 ZipTips (Millipore, Bedford, Mass.). Two fractions were eluted with 3 μl of 30 and 70% acetonitrile in 0.1% trifluoroacetic acid, respectively.
Matrix-assisted laser desorption ionization (MALDI)-mass spectrometry was performed with a Bruker (Bremen, Germany) Reflex III instrument with a 20-kV acceleration voltage and a 24-kV reflector voltage. External calibration was achieved with a mixture of eight peptides of from 942 to 3,495 Da. Alpha-cyano-4-hydroxy-cinnamic acid was used as a matrix, prepared as a saturated solution in 50% acetonitrile–0.1% trifluoroacetic acid. Samples were prepared with the dried droplet method on a stainless steel target for 26 samples.
Measured monoisotopic mass values were used with a 0.15-Da tolerance to search the NCBI database with the Protein Prospector MSFit search engine (http://falcon.ludwig.ucl.ac.uk) by restricting the species to S. cerevisiae.
RESULTS
The product of the essential MPE1 gene interacts functionally with Pcf11p.
The yeast PCF11 gene encodes a CFIA subunit required both for cleavage and polyadenylation of the 3′ end of mRNA. A collection of conditional thermosensitive PCF11 mutants, including the pcf11-2 strain, was previously selected by PCR mutagenesis (4). The pcf11-2 strain grows slowly at 34°C and is unable to grow at 37°C (Fig. 1). Extracts from the mutant are deficient in vitro both for cleavage and polyadenylation (4). To find new genes that interact functionally with the CFIA complex, we selected extragenic suppressors of the thermosensitivity of the pcf11-2 mutant after UV mutagenesis. A semidominant suppressor was found, and the corresponding gene was named MPE1 for mutant PCF11 extragenic suppressor 1. The suppressor allele, mpe1-s, has no phenotype by itself (Fig. 1). It suppressed pcf11-3 mutant but not pcf11-1 and pcf11-9 mutants. It was also found to suppress rna15-2 weakly but not rna14-1 and rna14-5 mutants (data not shown).
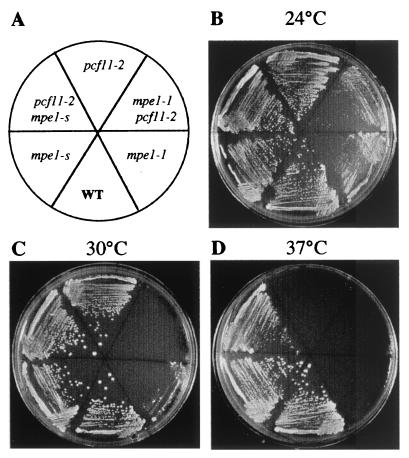
FIG. 1.
Growth characteristics of mpe1-1, pcf11-2, and pcf11-2 mpe1-1 mutant strains. Positions of the wild-type, mutant, and suppressor strains are indicated in the diagram. Cells were grown at the indicated temperatures for 2 days on minimal medium supplemented for the auxotrophies of the W303-1B strain.
To clone the MPE1 gene, we used DNA isolated from a pcf11-2 mpe1-s strain to synthesize a genomic library in the pFL38 centromeric vector. After transformation of the thermosensitive pcf11-2 strain and selection of thermoresistant clones, we found that the essential ORF, YKL059c (50), contained the suppressor mpe1-s. The corresponding protein was 441 amino acids long, with a predicted molecular mass of 49.5 kDa. To determine the function of the protein, we used PCR to replace its ORF with the TRP1 gene in the diploid strain, BMA64. (8). Southern blot analysis confirmed that MPE1 had been deleted and that TRP1 had been inserted into the locus (data not shown). After sporulation, only the two spores with a tryptophan auxotrophic phenotype were viable in each tetrad. The lethal deletion was perfectly complemented by the MPE1 gene on a centromeric vector. Thus, MPE1 encodes for the suppressor and is essential for cell viability like most of the genes involved in mRNA 3′-end processing.
We obtained a conditional allele of MPE1 by PCR mutagenesis. After integration to the locus, this stable thermosensitive mpe1-1 mutant grew slowly at 30°C but was unable to grow at 37°C (Fig. 1). The double mutant (pcf11-2 mpe1-1) strain showed a stronger phenotype of slow growth at 25°C than each single mutant strain and is unable to grow at 30°C (Fig. 1). The sequence of Mpe1-1p revealed four point mutations compared to the wild-type protein (Fig. 2). A nonsense mutation at position 354 eliminates 87 amino acids from the C-terminal end of Mpe1p and results in the synthesis of a shortened protein (see Fig. 6A, lane 4). Nevertheless, this truncation alone probably does not determine the mutant phenotype because the introduction of a TAP-tag into a wild-type strain at the position of the nonsense codon also resulted in a shortened protein (Fig. 6A, lane 3) without a detectable phenotypic change (data not shown).
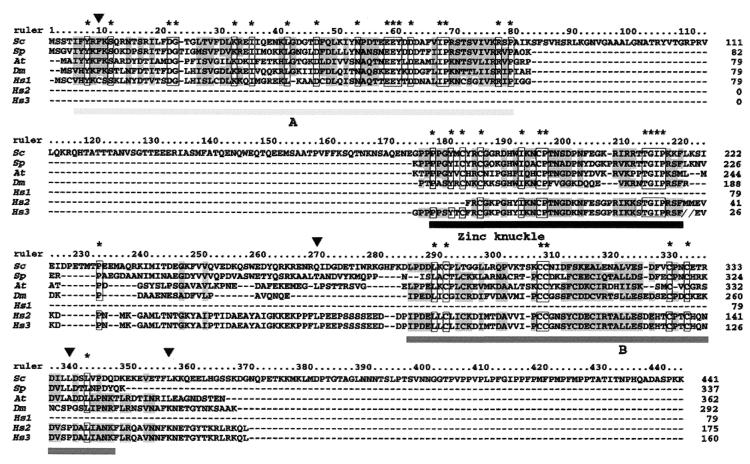
FIG. 2.
The human genome encodes an Mpe1p homolog. The sequence of three conserved motifs were compared for S. cerevisiae (Sc), S. pombe (Sp), A. thaliana (At), and D. melanogaster (Dm) and for polypeptides encoded by three cDNAs from H. sapiens (Hs1 to -3). The three conserved motives are designated A, zinc knuckle, and B. Identical residues are in frames and marked with asterisks, and similar residues are indicated by gray shading. The identity scores between S. cerevisiae and H. sapiens are 35% for A, 65% for the zinc knuckle domain, and 29% for B. The arrows show the positions of the mutations in the mpe1-1 mutant (amino acid 9, phenylalanine changed to a serine; amino acid 268, glutamine changed to a lysine; amino acid 337, lysine changed to a phenylalanine; and amino acid 354, lysine changed to a stop codon).
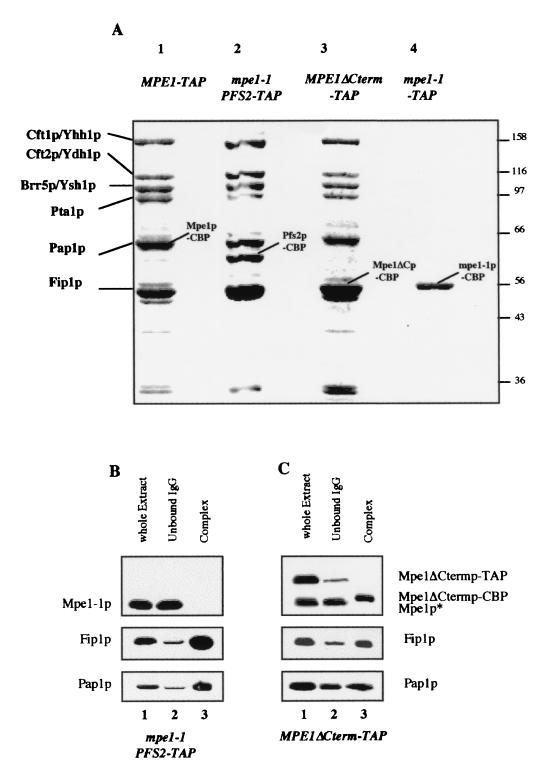
FIG. 6.
The mutant protein, Mpe1-1p, is not stably associated to the CPF complex. (A) Analysis of the protein complexes associated with the mutant proteins Mpe1-1p and Mpe1ΔCtermp. The different complexes were purified and analyzed as described in Fig. 5A either from the wild-type strain MPE1-TAP (lane 1) or from the mpe1-1 PFS2-TAP (lane 2), MPE1ΔCterm-TAP (lane 3), and mpe1-1–TAP (lane 4) mutant strains. (B and C) Western blot analysis of the TAP from the mpe1-1 PFS2-TAP (B) or MPE1ΔCterm-TAP (C) strains as described in Fig. 5B and C.
Mpe1p is an evolutionarily conserved protein.
A search for known protein motifs in Mpe1p revealed a zinc knuckle (CX2 CX4 HX4 C) between amino acids 182 and 195. This motif has been implicated in the interactions between proteins and single-stranded nucleic acids (9, 28). A homology search based on the zinc knuckle identified genes from S. pombe, A. thaliana, and D. melanogaster in which the Mpe1p domain between amino acids 176 and 216 is conserved. When these protein sequences were aligned with the Mpe1p sequence, two more regions, A and B, were found to contain similarities. The sequence comparison of the three conserved domains is shown in Fig. 2. The A region encompasses amino acids 5 to 78 of Mpe1p and does not contain any known motifs. The B domain extends from amino acids 284 to 343 of Mpe1p; it is cysteine-rich and presents some of the characteristics of a RING finger. RING fingers are often implicated in protein-protein interactions, notably in the ubiquitination pathway (5). These three conserved domains are also found in nonoverlapping human cDNA's, which all belong to the same region of chromosome 16. Based on these data and on the sequence of the human genome, we selected a set of exons and reconstituted the most probable ORF containing the three conserved motifs (see Materials and Methods). Like the A. thaliana and D. melanogaster genes, this human ORF contains a much longer C-terminal polypeptide downstream of the B motif than the MPE1 gene. These results strongly indicate that a human Mpe1p homolog exists.
The mpe1-1 mutation modifies the choice of the mRNA polyadenylation site in vivo.
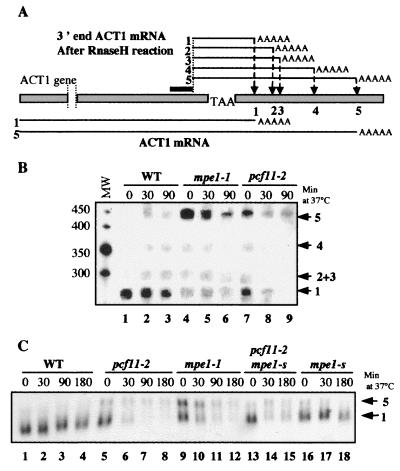
To determine whether Mpe1p is directly involved in mRNA 3′-end processing, we studied the cleavage of ACT1 mRNA in the mpe1-1 mutant strain in vivo. This transcript, encoding the yeast actin, contains five potential cleavage sites in its 3′ untranslated region (Fig. 3A). They are used at variable frequencies in vivo depending on the efficiency of the cleavage and polyadenylation complex. Mutations in the subunits of CFI, such as Rna14p or Rna15p, or in the poly(A) polymerase modify the sites used (2, 31). We studied the in vivo distribution of ACT1 mRNA polyadenylation sites in pcf11-2 and mpe1-1 mutants according to the temperature. The mRNAs were treated with RNase H after hybridization to both a specific oligonucleotide designed to cut the transcript near its 3′ end and oligo(dT) to degrade the heterogeneous poly(A) tails. After separation on a polyacrylamide gel, the ACT1 mRNA ends were revealed by Northern blotting (Fig. 3B). As already described, in the wild-type strain most of the transcripts were cleaved at the proximal site and the other sites were of minor importance regardless of temperature (lanes 1 to 3). Interestingly, the situation was reversed in the mpe1-1 and pcf11-2 mutants, in which the most distal site was the major site cleaved at permissive temperatures and the only site used at 37°C (lanes 4 to 6 and lanes 7 to 9, respectively). This demonstrates that the mpe1-1 and pcf11-2 mutations alter the choice of cleavage site of the ACT1 transcripts in the same way as has already been observed for mutants of the polyadenylation complex (2, 31).
FIG. 3.
MPE1 is required for the choice of ACT1 mRNA polyadenylation site in vivo. (A) Structure of the 3′ end of the ACT1 gene. Arrows indicate the potential polyadenylation sites numbered 1 to 5 from 5′ to 3′. The position of the specific oligonucleotide used for RNase H treatment is indicated by a black bar. (B) Comparison of the polydenylation sites of ACT1 mRNA. Total RNA was extracted at time zero and at various times after a shift to 37°C. Equal quantities (20 μg) of each sample were treated with RNase H after hybridization to the complementary oligonucleotide and oligo(dT) to shorten the ACT1 mRNA and to eliminate the poly(A) tails. After PAGE and Northern blotting, the amount of each different ACT1 mRNA was estimated. The marker sizes are indicated in nucleotides on the left. (C) Analysis of ACT1 mRNA decay after a shift to 37°C. Equal quantities of total RNA (10 μg) extracted at the indicated times at 37°C were separated on a 1.2% agarose gel in the presence of 5% formaldehyde, and ACT1 mRNA was visualized by Northern blotting. The polyadenylation sites are indicated on the right.
Northern blotting analysis revealed that pcf11-2 and mpe1-1 mutants are defective in the accumulation of actin mRNA when shifted to restrictive temperatures (Fig. 3C, lanes 5 to 8 and lanes 9 to 12, respectively); the same decrease was also seen for CYC1, URA1, and URA2 mRNAs (data not shown). This defect has already been observed in many thermosensitive mutants of the 3′-end processing complex and reflects their inability to synthesize mature mRNAs after a temperature shift (4, 37, 39). Two ACT1 mRNAs were separated in these conditions, as a result of cleavage at polyadenylation sites 1 and 5 (2). The amounts of both mRNAs found in the different strains confirm the results shown in Fig. 3B. Indeed, the frequency of cleavage at the distal site 5 was 0.08 in the wild-type strain and 0.70 and 0.60, respectively, in the pcf11-2 and mpe1-1 strains after 180 min at 37°C. Moreover, it can be seen that the suppression of pcf11-2 by mpe1-s partially restores the accumulation of actin mRNA and the correct choice of cleavage site (Fig. 3C, lanes 13 to 15).
We can conclude from these results that MPE1 is required for the specificity of mRNA 3′-end cleavage in vivo.
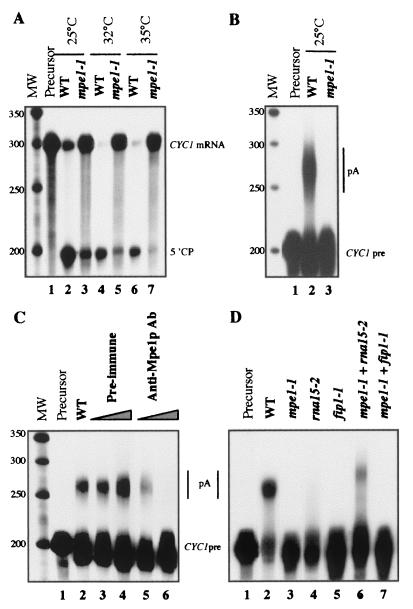
Mpe1p is essential for in vitro cleavage and polyadenylation.
Extracts from the mpe1-1 mutant strain were used to test the role of Mpe1p in specific cleavage and polyadenylation in vitro. The cleavage reaction was done on a synthetic pre-mRNA containing the 3′ end of CYC1 and showed that the activity of the mpe1-1 extract was reduced at 25 and 32°C and was very weak at 35°C (Fig. 4A). At these three temperatures the percentages of 5′ cleaved product were respectively 67, 96, and 98% in the wild-type strain (lanes 2, 4, and 6) and 22, 15, and 7% in the mpe1-1 mutant strain (lanes 3, 5, and 7) The polyadenylation reaction was carried out on a precleaved CYC1 synthetic mRNA and showed that specific polyadenylation was completely abolished even at 25°C (Fig. 4B, lane 3). These results were confirmed in similar assays carried out on wild-type extracts in the presence of increasing quantities of specific anti-Mpe1p antibodies. Polyadenylation was completely blocked by immunoneutralizing Mpe1p (Fig. 4C, lanes 5 and 6), whereas the preimmune serum had no effect (Fig. 4C, lanes 3 and 4). Cleavage was only slightly modified in these conditions (data not shown)
FIG. 4.
Mpe1p is necessary for in vitro 3′-end processing. (A) Specific in vitro cleavage assay. Equal quantities of wild-type extract (lanes 2, 4, and 6) or mpe1-1 mutant extract (lanes 3, 5, and 7) were added to a cleavage reaction mix containing the CYC1 pre-mRNA (lane 1, precursor alone). The temperature of the reaction is indicated at the top. The positions of the substrate (CYC1 mRNA) and of the 5′ cleavage product (5′CP) are indicated on the right. The positions and sizes of the markers are indicated on the left in nucleotides. (B) Specific in vitro polyadenylation assay. The experiment was carried out as in panel A except that a reaction mix specific for polyadenylation was used in the presence of precleaved CYC1 mRNA (CYC1 pre) to obtain polyadenylated products (pA). Lane 1, precursor alone; lane 2, wild-type extract; lane 3, mutant mpe1-1 extract. Markers are as in panel A. (C) Mpe1p immunoneutralization inhibits in vitro polyadenylation. In vitro polyadenylation was carried out as in panel B at 25°C. Lane 1, precursors alone; lane 2, wild-type extract alone; lanes 3 and 4, wild-type extract with increasing amounts of preimmune serum; lanes 5 and 6, wild-type extract with increasing amounts of anti-Mpe1p antibodies raised against the C-terminal domain of the protein. Markers are as described in panel A. (D) Complementation of polyadenylation activity in the mpe1-1 mutant extract. In vitro polyadenylation was carried out as in panel B at 25°C. Extracts were assayed alone or combined as indicated at the top of each lane.
A rna15-2 mutant extract was inactive alone due to the loss of the CFIA function (Fig. 4D, lane 4) (35). Nevertheless, it could complement the mpe1-1 extract (Fig. 4D, lane 6). Identical results were obtained with an extract of rna14-1 mutant (data not shown). Similar complementations have already been shown between mutant extracts of CFI subunits and mutant extracts of CPF subunits (fip1-1, yth1-1, pta1-1, and pfs2-1) (7, 38, 39, 53). Conversely, the fip1-1 mutant extract inactivated for CPF function (Fig. 4D, lane 5) did not complement the polyadenylation deficiency of the mpe1-1 extract (Fig. 4D, lane 7).
This leads us to conclude that Mpe1p has an important function in the 3′-end formation likely at the level of the CPF complex because the mpe1-1 extract contains an active CFI complex but an inactive CPF complex.
Mpe1p is an integral subunit of the CPF complex.
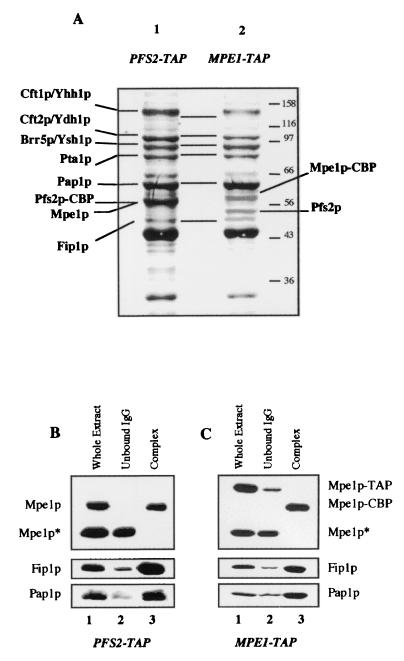
TAP was used as a mild method for purifying the proteins associated with either the Mpe1p or the Pfs2p subunit of the CPF complex (42). The TAP-tag sequence was inserted immediately upstream of the MPE1 or PFS2 stop codon in a wild-type strain (see Materials and Methods). This insertion did not affect the growth rate of the corresponding strains at any temperature (data not shown). Pfs2p was chosen as a control because the protein A-tagged Pfs2p had already been successfully used to purify the CPF complex (38).
We used SDS-polyacrylamide gel electrophoresis (PAGE) to separate the purified proteins from the PFS2-TAP strain and then stained them with Coomassie blue (Fig. 5A, lane 1). The pattern was identical to those previously published for the CPF purified either by affinity with the protein A-tagged Pfs2p or Yth1p or by biochemical techniques (6, 38, 40). The purified Mpe1p-associated proteins are shown in Fig. 5A, lane 2. It can be seen that the profile is identical to the one obtained with Pfs2p (lane 1). In addition, we used MALDI-mass spectrometry to identify the components of the CPF complex after in-gel proteolytic digestion with endoproteinase LysC. Seven peptide masses matched the values calculated for a theoretical (in silico) Mpe1p digest (data not shown). Thus, Mpe1p is clearly one of the proteins associated with Pfs2p (lane 1). Mpe1p migrates on gels with an apparent molecular mass of 58 kDa, although its calculated mass is 49.5 kDa (data not shown). Hence, Mpe1p comigrates with Pfs2p, which remains associated with the calmodulin-binding peptide (Pfs2p-CBP) after the TAP purification and the molecular mass of which was increased by 5 kDa.
FIG. 5.
Mpe1p belongs to the CPF complex. (A) Analysis of the multiprotein complex associated with the TAP-tagged Pfs2p or the TAP-tagged Mpe1p. After purification by the TAP procedure starting from strains PFS2-TAP (lane 1) or MPE1-TAP (lane 2), the proteins were separated on a 10% polyacrylamide-SDS gel and then stained with Coomassie blue. The indicated proteins were identified by mass spectrometry from bands excised from the gel. CBP indicates the TAP-tagged proteins that remained fused with the CBP after purification. (B and C) Western blot analysis of the TAP. Mpe1p, Fip1p, and Pap1 were detected by specific antibodies in the whole extract (lane 1), in the material not bound to IgG agarose (lane 2), and in the purified complex (lane 3), starting either from the PFS2-TAP strain (B) or from the MPE1-TAP strain (C). The fractions contain identical quantities of cells. TAP and CBP indicate that proteins are fused to the TAP peptide or to the CBP; Mpe1p* is a truncated form of Mpe1p revealed by the anti-Mpe1p antibodies and detected in variable quantities in all the extracts. It was never found in the purified complexes and was not fused with the C-terminal TAP-tag.
The proteins present in the complexes were subjected to immunoblotting. The repartition of Mpe1p, Fip1p, and Pap1p was monitored throughout the purification of the CPF obtained from the strains PFS2-TAP (Fig. 5B) and MPE1-TAP (Fig. 5C) in the whole-cell extract (lane 1), in the effluent of the IgG agarose column (lane 2), and in the purified complex (lane 3). The three proteins were mainly found in both purified complexes and were present at very low level in the effluent. It is noteworthy that Fip1p and Pap1p were mainly copurified with the MPE1-TAP-purified CPF, suggesting that most of the CPF complex contains the Mpe1p protein. The CFIA proteins (Rna14p, Rna15p, Pcf11p, and Clp1p) (36) and Pab1p were not detected on blots in the purified CPFs (data not shown). Moreover, in coimmunoprecipitation experiments no interaction was obtained between recombinant Pcf11p and in vitro-translated Mpe1p or between both in vitro-translated proteins Rna14p and Mpe1p (data not shown), indicating that there would not be a direct and stable interaction between Mpe1p and Pcf11p.
All these data clearly show the association between Mpe1p and the CPF complex. This is a strong association because the IgG agarose was washed with a buffer with a higher ionic strength than those usually used (400 mM NaCl instead of 150 mM NaCl). Mpe1p does not appear to belong to any other complexes because only the CPF complex proteins were purified with the Mpe1p-TAP. The lack of complementation between mpe1-1 and fip1-1 (see Fig. 4D) shows that once Mpe1p is assembled in CPF, it cannot diffuse from one complex to another in vitro as already shown for several components of the cleavage and polyadenylation complex (35, 40). Therefore, Mpe1p can be considered an integral subunit of CPF, one required both for cleavage and polyadenylation in vitro.
The mutant protein, Mpe1-1p, is not tightly associated with the CPF complex.
To investigate whether the composition of CPF is modified by the mpe1-1 mutation, this complex was purified through a TAP-tagged Pfs2p in the mpe1-1 mutant background. The pattern obtained on gel (Fig. 6A, lane 2) was identical to the wild-type CPF patterns obtained with the MPE1-TAP strain (lane 1) and with the PFS2-TAP strain (Fig. 5A, lane 1). The apparent stoichiometry of Cft1p/Yhh1p, Cft2p/Ydh1p, Brr5p/Ysh1p, and Pta1 was similar in both the wild-type and the mutant backgrounds. Since Mpe1-1p is truncated at its C terminus, its apparent molecular mass on a gel was very close to that of Fip1p (data not shown). Thus, we used immunodetection to reveal the presence or absence of Mpe1-1p in the PFS2-TAP mpe1-1 strain (Fig. 6B). Mpe1-1p was present in the whole cellular extract; thus, it is stable and accumulates within cells. However, it is only found in the effluent from the IgG agarose column and not in the isolated CPF complex. This result was strengthened by purification from a strain in which the mutant Mpe1-1p protein was TAP tagged at the stop codon generated by the nonsense mutation. Only the mutant protein that fused with the CBP, Mpe1-1p–CBP, could be purified without the other proteins from the CPF complex (Fig. 6A, lane 4). The same result was obtained even when the stringency of the washing step of IgG agarose was reduced (150 mM NaCl instead of 400 mM NaCl [data not shown]). The mutant phenotype was not essentially due to the Mpe1-1p truncation. Indeed, we constructed a strain in which the wild-type ORF was TAP tagged at the position of the nonsense codon. In this strain the CPF complex was still copurified with the shortened Mpe1ΔCterm protein (Fig. 6A, lane 3), and Fip1p was mainly detected in the purified complex by immunoblotting (Fig. 6C).
The Mpe1-1p mutant protein largely lost its affinity for CPF. Nevertheless, the presence and the relative levels of the other components were not modified considerably in the complex lacking Mpe1p, which was prepared from the (PFS2-TAP mpe1-1) mutant extract. Thus, we can conclude that, although Mpe1p is essential for 3′-end processing, it is not implicated in the overall stability of CPF.
Mpe1p is required in vitro for the specificity of mRNA 3′-end processing.
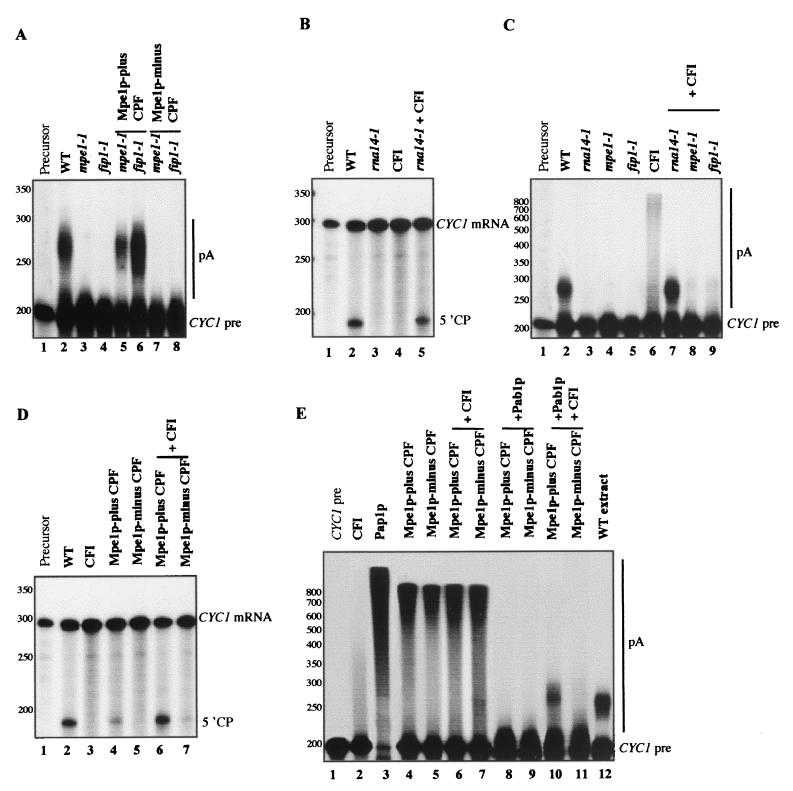
The purification of a Mpe1p-devoided CPF allowed us to analyze the role of Mpe1p in 3′-end processing. We tested the cleavage and polyadenylation activities in reconstituted assays with the purified factors CFI and CPF, either with or without Mpe1p, and the recombinant Pab1p. We purified the complexes only to the first step of the TAP procedure after elution by the TEV protease. CPF were isolated either from MPE1-TAP or from mpe1-1 PFS2-TAP strains. In these conditions PAGE did not reveal any major contaminants after Coomassie blue staining, and no CFIA proteins were detected by immunodetection in either CPF (data not shown). As before, the CPF isolated from the wild-type MPE1-TAP strain contained Mpe1p and was named Mpe1p-plus CPF, whereas the complex purified from the mpe1-1 PFS2-TAP mutant strain did not contain mutant Mpe1-1p. It was named Mpe1p-minus CPF.
Mutant mpe1-1 or fip1-1 extracts were inactive for polyadenylation due to the loss of CPF function (Fig. 7A, lanes 3 and 4, respectively) (39). Mpe1p-plus CPF was functional and complemented the polyadenylation deficiency of both mutants (Fig. 7A, lanes 5 and 6), whereas the Mpe1p-minus complex was unable to provide this complementation (Fig. 7A, lanes 7 and 8). This confirms the essential role of Mpe1p in the specific processing activity of CPF. Neither CPF complemented the cleavage defect of the mutant rna14-1 extract (data not shown).
FIG. 7.
Role of Mpe1p in in vitro 3′-end processing reactions reconstituted from purified CFI and CPF complexes. (A) Complementation of polyadenylation activity by purified CPF. The reactions were performed at 25°C with precleaved CYC1 mRNA as described in Fig. 4B. The extracts were incubated alone or in combination with purified Mpe1p-plus CPF or Mpe1p-minus CPF as indicated at the top of the figure. The positions of the precleaved CYC1 mRNA (CYC1 pre) and of the polyadenylated products (pA) are shown on the right. The positions and the sizes of the molecular mass markers are shown on the left in nucleotides. (B) Complementation of cleavage activity by purified CFI. The reactions were performed at 25°C with the CYC1 pre-mRNA as described in Fig. 4A. The extracts were incubated alone or in combination with purified CFI as indicated at the top of the figure. The positions of the substrate (CYC1 mRNA) and of the 5′ cleavage product (5′CP) are shown on the right. Molecular mass markers are as described in panel A. (C) Complementation of polyadenylation activity by purified CFI. The reactions were performed as described in panel A. The extracts were incubated alone or in combination with purified CFI, as indicated at the top of the figure. Molecular mass markers are as described in panel A. (D) Cleavage activity reconstituted from purified complexes. The reactions were performed as described in panel B with CYC1 pre-mRNA. Purified CFI, Mpe1p-plus CPF, and Mpe1p-minus CPF were associated as indicated at the top of the figure. Molecular mass markers are as described in panel A. (E) Polyadenylation activity reconstituted from purified complexes. The reactions were performed as described in panel A with pre-cleaved CYC1 mRNA. Purified CFI, Mpe1p-plus CPF, Mpe1p-minus CPF, recombinant Pap1p (USB, Cleveland, Ohio), and recombinant Pab1p (125 ng/assay) were associated as indicated at the top of the figure. Molecular mass markers are as described in panel A.
We constructed a strain expressing the TAP-tagged Pcf11p to isolate wild-type CFI by the TAP method. An rna14-1 extract, mutated in a CFI subunit, was deficient in cleavage and polyadenylation (Fig. 7B, lane 3, and Fig. 7C, lane 3, respectively). The purified CFI complemented both 3′-end processing steps in the rna14-1 mutant extract (Fig. 7B, lane 5, and Fig. 7C, lane 7, respectively). Nevertheless, it could not rescue the polyadenylation defect of the mpe1-1 and fip1-1 mutant extracts (Fig. 7C, lanes 8 and 9). Thus, this fraction essentially contained the active CFI factor and was used in the reconstituted 3′-end processing assays.
The purified factors still showed residual processing activities. There was a very low level of polyadenylation activity in CFI (Fig. 7C, lane 6). This is certainly due to the presence of a small amount of Pap1p found in the purified fraction by immunoblotting, whereas Mpe1p was never detected in the same conditions (data not shown). Indeed, Pap1p has already been shown to be associated to purified CFI (27). As previously shown (38), a low level of cleavage activity was associated with the wild-type Mpe1p-plus CPF (Fig. 7D, lane 4). However, full cleavage activity was only restored when CFI and Mpe1p-plus CPF were associated (Fig. 7D, lane 6). In the same conditions this activity could not be reconstituted in the presence of Mpe1p-minus CPF (Fig. 7D, lane 7). This definitely confirms that Mpe1p is necessary for cleavage.
Pap1p alone is known to catalyze the 3′-end polyadenylation of RNA substrates without specificity and regardless of the length of the poly(A) tails (29, 40). We confirmed that all RNA molecules were simultaneously polyadenylated during this reaction, showing that the polymerization was distributive (Fig. 7E, lane 3). A number of conclusions can be made. (i) As previously shown for the CPFs purified from wild-type strains (38, 40), the polyadenylation of precleaved mRNA by Mpe1p-plus CPF was processive. Only a small amount of the substrate RNA was elongated (Fig. 7E, lane 4). (ii) The length of the poly(A) tails was not regulated. (iii) This activity was completely blocked by excess recombinant Pab1p (Fig. 7E, lane 8). Remarkably identical results were obtained with the purified Mpe1p-minus CPF (Fig. 7E, lanes 5 and 9). Thus, in this complex devoided of Mpe1p the unspecific polyadenylation activity was maintained, and poly(A) addition remained processive (lane 5), suggesting that the interactions between CPF and the RNA substrate were preserved in the absence of Mpe1p (40). The polyadenylation remained totally inhibited by Pab1p (lane 9). These data indicate that Mpe1p would not be directly implicated in the modulation of the Pap1p activity by CPF. This is consistent with the observations presented here showing that the composition and the stability of CPF are not affected significantly in the absence of Mpe1p.
As previously shown (33, 38), the specific polyadenylation took place when CFI, the wild-type Mpe1p-plus CPF, and Pab1p were combined (Fig. 7E, line 10). CFI was necessary to prevent the inhibition of polyadenylation by Pab1p. The length of the synthesized poly(A) tails was regulated and very similar to this obtained in a wild-type extract (Fig. 7E, lane 12). It has been shown that Pab1p is directly involved in the regulation of this process (3, 36). Indeed, long poly(A) tails are synthesized in a mutant extract for Pab1p or in an extract immunodepleted for this protein. In the absence of Pab1p, long poly(A) tails were also observed in a reconstituted assay with CFI and Mpe1p-plus CPF (Fig. 7E, lane 6). In this assay the poly(A) tail profile was identical to that obtained with Mpe1p-plus CPF alone (Fig. 7E, compare lanes 4 and 6). The only evidence for the role of CFI is that the activity of Mpe1p-plus CPF is totally inhibited by Pab1p (Fig. 7E, lane 8), whereas this CPF combined with CFI and Pab1p results in a regulated poly(A) addition (Fig. 7E, lane 10). The polyadenylation could not be reconstituted in an assay containing Mpe1p-minus CPF, CFI, and Pab1p (Fig. 7E, lane 11), demonstrating that Mpe1p is required in vitro for the specific poly(A) addition. The polyadenylation activity found in a mixture of CFI and Mpe1p-minus CPF (Fig. 7E, lane 7) remained inhibited by Pab1p (Fig. 7E, lane 11).
DISCUSSION
Our results demonstrate that a zinc knuckle protein, Mpe1p, is required for both steps of the mRNA 3′-end processing in S. cerevisiae. Mpe1p is essential for cell viability. In vitro assays demonstrated that an extract from the conditionally lethal mpe1-1 mutant strain is impaired for cleavage and polyadenylation and that antibodies raised against recombinant Mpe1p inhibit polyadenylation in a wild-type strain extract. By using the TAP method, we found that Mpe1p copurifies with the CPF isolated with a TAP-tagged Pfs2p without leaving significant amounts of soluble Mpe1p in the extract depleted from the complex. Conversely, CPF alone was purified with a TAP-tagged Mpe1p. The protein profile of this complex was identical to that found in the complexes purified with tagged forms of different genuine CPF components (6, 38, 40). Thus, Mpe1p is a novel integral subunit of CPF. Consistent with these results, a zinc knuckle protein, Pfs1p, with a molecular mass similar to that of Mpe1p has been repeatedly copurified with CPF, although the corresponding gene has not yet been characterized (6, 38, 40, 53).
In the mutant mpe1-1 strain, Mpe1p was not associated with CPF. Nevertheless, it was possible to isolate the other CPF subunits that formed a stable structure devoid of Mpe1p. We can conclude that (i) the mpe1-1 mutation drastically decreases the affinity of the protein for the CPF complex and (ii) Mpe1p is not essential for the overall stability of CPF. Thus, the processing defect displayed by the mpe1-1 mutant is not the result of a CPF destabilization, in contrast to what has been observed in strains mutated for other subunits of CPF (6, 38, 53). We also showed that CPF, with or without Mpe1p, displays its own polyadenylation activity. It is able to catalyze the processive poly(A) addition on any RNA substrate, whereas the poly(A) polymerase, Pap1p, alone had a distributive polyadenylation (40). This implies that Mpe1p is not involved in the modulation of Pap1p activity by the CPF complex. CPF, in particular, could form more stable complex with RNA than free Pap1p does, even in the absence of Mpe1p. However, the binding of CPF to the substrate is not specific, and the CFI factor is required for proper substrate recognition and for accurate 3′-end processing (6, 33, 38). Reconstituted assays with purified factors demonstrated that Mpe1p is strictly required for the cleavage of a pre-mRNA and for the poly(A) addition at the genuine polyadenylation site. This indicates that Mpe1p is involved in the mechanism by which CFI and CPF function synergistically to form the correct 3′ ends. Several observations suggest a role of Mpe1p in this mechanism. (i) Mpe1p may be required to stabilize the interactions between CFI and CPF, leading to the formation of a stable specific processing complex. Consistent with this hypothesis, some pcf11 mutations were suppressed by a Mpe1p mutant, suggesting that these two proteins interact closely when the CFI and CPF are associated. (ii) Mpe1p contains a zinc knuckle protein and is therefore a putative RNA-binding protein (9, 28). This is supported by the fact that the zinc knuckle motif present in the CPSF 30K subunit of the mammalian cleavage and polyadenylation specificity factor is involved in the interactions of this protein with the pre-mRNA (7). Mpe1p may be one of the components required to anchor the processing machinery to the correct location on RNA. (iii) Mpe1p is involved in the recognition of the correct ACT1 mRNA cleavage site in vivo. Like ACT1, a large number of yeast genes contain multiple potential polyadenylation sites (47). Some of them produce transcripts of different lengths due to the choice between alternative poly(A) sites in response to modifications of physiological state of the cells (22, 45). These observations suggest that regulated 3′-end processing is one of the mechanisms of the regulation of gene expression in which components of polyadenylation complex, such as Mpe1p, might be implicated.
The sequence data show that Mpe1p homologues exist in S. pombe, A. thaliana, and D. melanogaster. The zinc knuckle-containing domain and two other regions are well conserved in these organisms. Since the polyadenylation complex has not yet been thoroughly studied in these species, we do not yet know whether these putative homologues are involved in 3′-end processing. In mammals, the CPSF 30K subunit contains five zinc finger repeats besides the zinc knuckle motif, which all participate in the interactions of this protein with pre-mRNA. The yeast homolog of CPSF 30K, Yth1p, contains five zinc fingers but no zinc knuckle (7). Since this motif is found in Mpe1p, we can speculate that the role of the mammalian CPSF 30K protein is assumed in yeast by two proteins, Yth1p and Mpe1p, whereas the function of Mpe1p has been incorporated into CPSF 30K in higher eukaryotes. However, our results strongly suggest that Mpe1p is a highly conserved protein and that a true human homolog exists. It remains to be determined whether this human protein is the functional homolog of Mpe1p that has not yet been identified in the mammalian polyadenylation complex or if this protein assumes a different function in higher eukaryotic cells. Interestingly, one of the human cDNAs, which contains a domain conserved in Mpe1p, codes for the Rbbp6 protein, and it has been shown that this protein interacts with the tumor suppressor pRB1 (43). The pRB1 protein is involved in cell differentiation and is localized to centers of mRNA processing in the nucleus (13, 51). This support the idea that the human Mpe1 homolog could participate in 3′-end processing.
ACKNOWLEDGMENTS
We are grateful to Bertrand Seraphin, Lionel Minvielle-Sebastia, Yves D'Aubenton, and Claude Thermes for helpful discussions. We are indebted to Marc Bonneu and Katell Bathany for contributing to this work. We thank Marie Elisabeth Dufour for her technical assistance. We also thank Bertrand Seraphin and Lionel Minvielle-Sebastia for providing so many plasmids and antibodies.
This work was supported by the Centre National de la Recherche Scientifique, the Association pour la Recherche sur le Cancer (grant 9922), and the Fondation pour la Recherche Médicale.
REFERENCES
- 1.Altschul S F, Madden T L, Schaffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Amrani N, Dufour M E, Bonneaud N, Lacroute F. Mutations in STS1 suppress the defect in 3′ mRNA processing caused by the rna15–2 mutation in Saccharomyces cerevisiae. Mol Gen Genet. 1996;252:552–562. doi: 10.1007/BF02172401. [DOI] [PubMed] [Google Scholar]
- 3.Amrani N, Minet M, Le Gouar M, Lacroute F, Wyers F. yeast Pab1 interacts with Rna15 and participates in the control of the poly(A) tail length in vitro. Mol Cell Biol. 1997;17:3694–3701. doi: 10.1128/mcb.17.7.3694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Amrani N, Minet M, Wyers F, Dufour M E, Aggerbeck L P, Lacroute F. PCF11 encodes a third protein component of yeast cleavage and polyadenylation factor I. Mol Cell Biol. 1997;17:1102–1109. doi: 10.1128/mcb.17.3.1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Aravind L, Koonin E V. The U box is a modified RING finger—a common domain in ubiquitination. Curr Biol. 2000;10:R132–134. doi: 10.1016/s0960-9822(00)00398-5. [DOI] [PubMed] [Google Scholar]
- 6.Barabino S M, Ohnacker M, Keller W. Distinct roles of two Yth1p domains in 3′-end cleavage and polyadenylation of yeast pre-mRNAs. EMBO J. 2000;19:3778–3787. doi: 10.1093/emboj/19.14.3778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Barabino S M L, Hub N, Jenny W A, Minvielle-Sebastia L, Keller W. The 30-kD subunit of mammalian cleavage and polyadenylation specificity factor and its yeast homolog are RNA-binding zinc finger proteins. Genes Dev. 1997;11:1703–1716. doi: 10.1101/gad.11.13.1703. [DOI] [PubMed] [Google Scholar]
- 8.Baudin A, Ozier-Kalogeropoulos O, Denouel A, Lacroute F, Cullin C. A simple and efficient method for direct gene deletion in Saccharomyces cerevisiae. Nucleic Acids Res. 1993;21:3329–3330. doi: 10.1093/nar/21.14.3329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Berg J M, Shi Y. The galvanization of biology: a growing appreciation for the roles of zinc. Science. 1996;271:1081–1085. doi: 10.1126/science.271.5252.1081. [DOI] [PubMed] [Google Scholar]
- 10.Boeke J D, Lacroute F, Fink G R. A positive selection for mutants lacking orotidine-5′-phosphate decarboxylase activity in yeast: 5-fluoro-orotic acid resistance. Mol Gen Genet. 1984;197:345–346. doi: 10.1007/BF00330984. [DOI] [PubMed] [Google Scholar]
- 11.Butler J S, Sadhale P P, Platt T. RNA processing in vitro produces mature 3′ ends of a variety of Saccharomyces cerevisiae mRNAs. Mol Cell Biol. 1990;10:2599–2605. doi: 10.1128/mcb.10.6.2599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.de Vries H, Ruegsegger U, Hubner W, Friedlein A, Langen H, Keller W. Human pre-mRNA cleavage factor II(m) contains homologs of yeast proteins and bridges two other cleavage factors. EMBO J. 2000;19:5895–5904. doi: 10.1093/emboj/19.21.5895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Durfee T, Mancini M A, Jones D, Elledge S J, Lee W H. The amino-terminal region of the retinoblastoma gene product binds a novel nuclear matrix protein that co-localizes to centers for RNA processing. J Cell Biol. 1994;127:609–622. doi: 10.1083/jcb.127.3.609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Duvel K, Braus G H. Different positioning elements select poly(A) sites at the 3′-end of GCN4 mRNA in the yeast Saccharomyces cerevisiae. Nucleic Acids Res. 1999;27:4751–4758. doi: 10.1093/nar/27.24.4751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gallie D R. A tale of two termini: a functional interaction between the termini of an mRNA is a prerequisite for efficient translation initiation. Gene. 1998;216:1–11. doi: 10.1016/s0378-1119(98)00318-7. [DOI] [PubMed] [Google Scholar]
- 16.Gietz R D, St. Jean A, Woods R A, Schiestl R H. Improved method for high efficiency transformation of intact yeast cells. Nucleic Acids Res. 1992;20:1425. doi: 10.1093/nar/20.6.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Graber J H, Cantor C R, Mohr S C, Smith T F. Genomic detection of new yeast pre-mRNA 3′ end processing signals. Nucleic Acids Res. 2001;27:888–894. doi: 10.1093/nar/27.3.888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Grandier-Vazeille X, Bathany K, Chaignepain S, Camougrand N, Manon S, Schmitter J. Yeast mitochondrial dehydrogenases are associated in a supramolecular complex. Biochemistry. 2001;40:9758–9769. doi: 10.1021/bi010277r. [DOI] [PubMed] [Google Scholar]
- 19.Guthrie C, Fink G R. Guide to yeast genetics and molecular biology. Vol. 194. New York, N.Y: Academic Press, Inc.; 1991. [Google Scholar]
- 20.Harlow E, Lane D. Antibodies: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1988. [Google Scholar]
- 21.Hirose Y, Manley J L. RNA polymerase II and the integration of nuclear events. Genes Dev. 2000;14:1415–1429. [PubMed] [Google Scholar]
- 22.Hoopes B C, Bowers G D, DiVisconte M J. The two Saccharomyces cerevisiae SUA7 (TFIIB) transcripts differ at the 3′-end and respond differently to stress. Nucleic Acids Res. 2000;28:4435–4443. doi: 10.1093/nar/28.22.4435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huang Y, Carmichael G G. Role of polyadenylation in nucleocytoplasmic transport of mRNA. Mol Cell Biol. 1996;16:1534–1542. doi: 10.1128/mcb.16.4.1534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Keller W, Minvielle-Sebastia L. A comparison of mammalian and yeast pre-mRNA 3′-end processing. Curr Opin Cell Biol. 1997;9:329–336. doi: 10.1016/s0955-0674(97)80004-x. [DOI] [PubMed] [Google Scholar]
- 25.Kessler M M, Henry M F, Shen E, Zhao J, Gross S, Silver P A, Moore C L. Hrp1, a sequence specific RNA-binding protein that shuttles between the nucleus and the cytoplasm, is required for mRNA 3′-end formation in yeast. Genes Dev. 1997;11:2545–2556. doi: 10.1101/gad.11.19.2545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kessler M M, Zhao J, Moore C L. Purification of the Saccharomyces cerevisiae cleavage/polyadenylation factor I. J Biol Chem. 1996;271:27167–27175. doi: 10.1074/jbc.271.43.27167. [DOI] [PubMed] [Google Scholar]
- 27.Kessler M M, Zhelkovsky A M, Skvorak A, Moore C L. Monoclonal antibodies to yeast poly(A) polymerase (PAP) provide evidence for association of PAP with cleavage factor I. Biochemistry. 1995;34:1750–1759. doi: 10.1021/bi00005a032. [DOI] [PubMed] [Google Scholar]
- 28.Laity J H, Lee B M, Wright P E. Zinc finger proteins: new insights into structural and functional diversity. Curr Opin Struct Biol. 2001;11:39–46. doi: 10.1016/s0959-440x(00)00167-6. [DOI] [PubMed] [Google Scholar]
- 29.Lingner J, Radkte I, Wahle E, Keller W. Purification and characterization of poly(A) polymerase from Saccharomyces cerevisiae. J Biol Chem. 1991;266:8741–8746. [PubMed] [Google Scholar]
- 30.Mandart E. Effect of mutations in the Saccharomyces cerevisiae RNA14 gene on the abundance and polyadenylation of its transcripts. Mol Gen Genet. 1998;258:16–25. doi: 10.1007/s004380050702. [DOI] [PubMed] [Google Scholar]
- 31.Mandart E, Parker R. Effects of mutations in the Saccharomyces cerevisiae RNA14, RNA15, and PAP1 genes on polyadenylation in vivo. Mol Cell Biol. 1995;15:6979–6986. doi: 10.1128/mcb.15.12.6979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McCarthy J E G. Posttranscriptional control of gene expression in yeast. Microbiol Mol Biol Rev. 1998;62:1492–1553. doi: 10.1128/mmbr.62.4.1492-1553.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Minvielle-Sebastia L, Beyer K, Krecic A M, Hector R E, Swanson M S, Keller W. Control of cleavage site selection during mRNA 3′ end formation by a yeast hnRNP. EMBO J. 1998;17:7454–7468. doi: 10.1093/emboj/17.24.7454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Minvielle-Sebastia L, Keller W. mRNA polyadenylation and its coupling to other RNA processing reactions and to transcription. Curr Opin Cell Biol. 1999;11:352–357. doi: 10.1016/S0955-0674(99)80049-0. [DOI] [PubMed] [Google Scholar]
- 35.Minvielle-Sebastia L, Preker P J, Keller W. RNA14 and RNA15 proteins as components of a yeast pre-mRNA 3′-end processing factor. Science. 1994;266:1702–1705. doi: 10.1126/science.7992054. [DOI] [PubMed] [Google Scholar]
- 36.Minvielle-Sebastia L, Preker P J, Wiederkehr T, Strahm Y, Keller W. The major yeast poly(A)-binding protein is associated with cleavage factor IA and functions in premessager RNA3′-end formation. Proc Natl Acad Sci USA. 1997;94:7897–7902. doi: 10.1073/pnas.94.15.7897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Minvielle-Sebastia L, Winsor B, Bonneaud N, Lacroute F. Mutations in the yeast RNA14 and RNA15 genes result in an abnormal mRNA decay rate: sequence analysis reveals an RNA-binding domain in the RNA15 protein. Mol Cell Biol. 1991;11:3075–3087. doi: 10.1128/mcb.11.6.3075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ohnacker M, Barabino S M, Preker P J, Keller W. The WD-repeat protein pfs2p bridges two essential factors within the yeast pre-mRNA 3′-end-processing complex. EMBO J. 2000;19:37–47. doi: 10.1093/emboj/19.1.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Preker P J, Lingner J, Minvielle-Sebastia L, Keller W. The FIP1 gene encodes a component of a yeast pre-mRNA polyadenylation factor that directly interacts with poly(A) polymerase. Cell. 1995;81:379–389. doi: 10.1016/0092-8674(95)90391-7. [DOI] [PubMed] [Google Scholar]
- 40.Preker P J, Ohnaker M, Minvielle-Sebastia L, Keller W. A multisubunit 3′ end processing factor from yeast containing poly(A) polymerase and homologues of the subunits of mammalian cleavage and polyadenylation specificity factor. EMBO J. 1997;16:4727–4737. doi: 10.1093/emboj/16.15.4727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Puig O, Rutz B, Luukkonen B G, Kandels-Lewis S, Bragado-Nilsson E, Seraphin B. New constructs and strategies for efficient PCR-based gene manipulations in yeast. Yeast. 1998;14:1139–1146. doi: 10.1002/(SICI)1097-0061(19980915)14:12<1139::AID-YEA306>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- 42.Rigaut G, Shevchenko A, Rutz B, Wilm M, Mann M, Seraphin B. A generic protein purification method for protein complex characterization and proteome exploration. Nat Biotechnol. 1999;17:1030–1032. doi: 10.1038/13732. [DOI] [PubMed] [Google Scholar]
- 43.Sakai Y, Saijo M, Coelho K, Kishino T, Niikawa N, Taya Y. cDNA sequence and chromosomal localization of a novel human protein, RBQ-1 (RBBP6), that binds to the retinoblastoma gene product. Genomics. 1995;30:98–101. doi: 10.1006/geno.1995.0017. [DOI] [PubMed] [Google Scholar]
- 44.Shatkin A J, Manley J L. The ends of the affair: capping and polyadenylation. Nat Struct Biol. 2000;7:838–842. doi: 10.1038/79583. [DOI] [PubMed] [Google Scholar]
- 45.Sparks K A, Dieckmann C L. Regulation of poly(A) site choice of several yeast mRNAs. Nucleic Acids Res. 1998;26:4676–4687. doi: 10.1093/nar/26.20.4676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Takagaki Y, Manley J L. Complex protein interactions within the human polyadenylation machinery identify a novel component. Mol Cell Biol. 2000;20:1515–1525. doi: 10.1128/mcb.20.5.1515-1525.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.van Helden J, del Olmo M, Perez-Ortin J E. Statistical analysis of yeast genomic downstream sequences reveals putative polyadenylation signals. Nucleic Acids Res. 2000;28:1000–1010. doi: 10.1093/nar/28.4.1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wahle E, Ruegsegger U. 3′-End processing of pre-mRNA in eukaryotes. FEMS Microbiol Rev. 1999;23:277–295. doi: 10.1111/j.1574-6976.1999.tb00400.x. [DOI] [PubMed] [Google Scholar]
- 49.Wilusz C J, Wormington M, Peltz S W. The cap-to-tail guide to mRNA turnover. Nat Rev Mol Cell Biol. 2001;2:237–246. doi: 10.1038/35067025. [DOI] [PubMed] [Google Scholar]
- 50.Winzeler E A, Shoemaker D D, Astromoff A, Liang H, Anderson K, Andre B, Bangham R, Benito R, Boeke J D, Bussey H, Chu A M, Connelly C, Davis K, Dietrich F, Dow S W, El Bakkoury M, Foury F, Friend S H, Gentalen E, Giaever G, Hegemann J H, Jones T, Laub M, Liao H, Davis R W, et al. Functional characterization of the S. cerevisiae genome by gene deletion and parallel analysis. Science. 1999;285:901–906. doi: 10.1126/science.285.5429.901. [DOI] [PubMed] [Google Scholar]
- 51.Witte M M, Scott R E. The proliferation potential protein-related (P2P-R) gene with domains encoding heterogeneous nuclear ribonucleoprotein association and Rb1 binding shows repressed expression during terminal differentiation. Proc Natl Acad Sci USA. 1997;94:1212–1217. doi: 10.1073/pnas.94.4.1212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhao J, Hyman L, Moore C. Formation of mRNA 3′ ends in eukaryotes: mechanism, regulation, and interrelationships with other steps in mRNA synthesis. Microbiol Mol Biol Rev. 1999;63:405–445. doi: 10.1128/mmbr.63.2.405-445.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhao J, Kessler M, Helmling S, O'Connor J P, Moore C. Pta1, a component of yeast CF II, is required for both cleavage and poly(A) addition of mRNA precursor. Mol Cell Biol. 1999;19:7733–7740. doi: 10.1128/mcb.19.11.7733. [DOI] [PMC free article] [PubMed] [Google Scholar]