Abstract
AlgX is a periplasmic protein required for the production of the exopolysaccharide alginate in Pseudomonas sp. and Azotobacter vinelandii. AlgX has been overexpressed and purified and diffraction-quality crystals have been grown using iterative seeding and the hanging-drop vapor-diffusion method. The crystals grew as flat plates with unit-cell parameters a = 46.4, b = 120.6, c = 86.9 Å, β = 95.7°. The crystals exhibited the symmetry of space group P21 and diffracted to a minimum d-spacing of 2.1 Å. On the basis of the Matthews coefficient (VM = 2.25 Å3 Da−1), two molecules were estimated to be present in the asymmetric unit.
1. Introduction
The later stages of cystic fibrosis (CF) disease are characterized by chronic respiratory infections caused by the opportunistic pathogen Pseudomonas aeruginosa. The establishment of these infections is an indicator of poor prognoses ( ▸) and P. aeruginosa infections remain the leading cause of death and morbidity in CF patients ( ▸). Treatment of the chronic infections is ineffective owing to the intrinsic resistance of the bacteria ( ▸; ▸), which arises at least in part from the conversion of the bacteria from a nonmucoid to a mucoid phenotype ( ▸; ▸). The mucoid phenotype is characterized by the production of large quantities of the exopolysaccharide alginate.
Alginate is synthesized as a linear polymer of β-1,4-linked D-mannuronic acid from the activated sugar-nucleotide precursor GDP-mannuronic acid ( ▸). Polymerization is believed to occur on the cytoplasmic face of the inner membrane and appears to require not only Alg8, a putative CAZy (carbohydrate-active enzymes) family 2 glycosyltransferase ( ▸; ▸; ▸), but also the bis-(3′,5′)-cyclic GMP-binding PilZ-domain-containing Alg44 ( ▸; ▸; ▸; ▸). The exact mechanism by which the growing homopolymeris exported across the inner membrane into the periplasm is not known but is thought to involve the transmembrane-spanning domains of Alg8. In the periplasm the D-mannuronate residues can be either epimerized to α-L-guluronic acid by AlgG ( ▸), selectively acetylated at the O2′ and/or O3′ hydroxyls by the concerted action of AlgI, AlgJ and AlgF ( ▸)or remain unaltered ( ▸). As the modified alginatepolymer traverses the periplasm, its proper export is dependent upon the remaining proteins of the putative alginate-biosynthetic complex: AlgK, AlgE, AlgL and AlgX ( ▸, ▸; ▸; ▸). The AlgK/E proteins have been proposed to form the basis of a novel class of exopolysaccharide secretin ( ▸). AlgE bears a resemblance to outer-membrane β-barrel porins and has been proposed to form a pore that allows the polymer to traverse the outer membrane ( ▸; ▸; ▸). Recent structural and functional characterization of AlgK has shown that this protein is composed of at least 9.5 tetratricopeptide (TPR) protein–protein interaction motifs and may form the scaffold for the assembly of the periplasmic alginate-biosynthetic complex. Indeed, AlgK has been shown to facilitate the correct localization of AlgE ( ▸) and may also be involved in the coordination of AlgL and AlgX. AlgL appears to have a dual role involving both the degradation of alginate within the periplasm and as a protein within these cretion complex ( ▸; ▸), while the exact role of AlgX is currently unknown ( ▸; ▸).
The mature form of AlgX is a 49.9 kDa periplasmic protein ( ▸; ▸). At the amino-acid level, the protein shares 31% and 69% sequence identity and similarity, respectively, to AlgJ, a protein that is required for alginateacetylation ( ▸, ▸). At the tertiary level, the Phyre structure-prediction server ( ▸)suggests that the N-terminal region of the protein, residues 50–200, shares structural homology to members of the SGNH hydrolyase superfamily of enzymes, which typically remove acyl groups from carbohydrates and other compounds ( ▸; ▸). The closest match is to an Enterococcus faecalis hydrolase(PDB code 1yzf; R. Zhang, C. Hatzos, S. Clancy, F. Collart & A. Joachimiak, unpublished work), which has 9% sequence identity to AlgX over this region. At present no enzymatic activity has been demonstrated for AlgX; while sequence analysis suggests thatAsp174 and His176 could form part of the signature Ser-His-Aspcatalytic triad of SGNH hydrolases, no candidate for the catalyticserine can be readily identified. The C-terminal region of the protein, residues 201–474, shows no similarity to any protein of known structure and there are no clearly conserved residues that would define a putative function for this region of the protein. The identification of an interaction between AlgX and MucD ( ▸), which is required for temperature resistance and alginate-gene regulation and is a close homologue of the Escherichia coli periplasmicserine protease HtrA ( ▸), further complicates our understanding of the role that AlgX may play in alginate production. Therefore, in order to obtain further insight in to the mechanism by which alginate is synthesized and exported and the role of AlgX in this process we have undertaken structural studies of AlgX and describe here the overexpression, purification and crystallization of the protein.
2. Materials and methods
2.1. Cloning and expression
The nucleotide sequence of algX from P. aeruginosa PAO1 was codon-optimized for expression in E. coli and the synthetic gene was prepared by GenScript Corporation (Piscataway, New Jersey, USA). Care was taken to ensure that the changes resulted only in silent mutations of the coding sequence so that the expressed protein would be identical at the amino-acid level to P. aeruginosa PAO1-encodedAlgX (GenBank/EMBL protein accession No. NP_252236; ▸). Unique restriction sites were chosen for the N- and C-termini (NdeI and XhoI, respectively) to facilitate cloning of the synthesized segment into the pET24b(+) vector (Novagen). This construct (pPLHJTWX) facilitates the expression of the entire AlgX protein including its signal sequence and a C-terminal His6 tag under the control of an isopropyl β-D-1-thiogalactopyranoside (IPTG)inducible promoter.
For the expression of high levels of protein, E. coli BL21 Codon-Plus (λDE3) cells (Stratagene) transformed with the AlgX expression vector pPLHJTWX were grown in 1 l rich medium (32 g tryptone, 20 g yeast extract, 5 g NaCl) containing 50 μg ml−1 kanamycin at 310 K until the OD600 of the cell culture reached 0.6, at which point protein expression was induced by the addition of IPTG to a final concentration of 1 mM. After induction, the cells were incubated at 298 K for 16 h prior to being harvested by centrifugation at 5000g for 15 min at 277 K. The resulting cell pellet was stored at 253 K until required.
2.2. Purification
To purify the AlgX His6-tagged protein, the cell pellet from 1 l of bacterial culture was thawed and resuspended in 40–50 ml buffer A[500 mM NaCl, 50 mM Tris–HCl pH 7.8, 2%(v/v) glycerol, 2 mM DTT] containing 1 mg ml−1 lysozyme, 0.5 mM EDTA and one tablet of Roche Complete protease-inhibitor cocktail (EDTA-free). This suspension was incubated at 277 K for 30 min before being subjected to lysis by sonication (Misonix Sonicator 3000) with cooling on ice for 3 min at power 5 (10 s on/10 s off) or until the lysate was translucent. Unlysed cells were removed by centrifugation at 5000g for 15 min at 277 K. The supernatant was subsequently loaded onto a 5 ml Ni2+–NTA Superflow Cartridge (Qiagen) pre-equilibrated with buffer A. The column was washed with at least 15 column volumes of buffer A to remove contaminating proteins. Bound protein was recovered from the column using a gradient from 0 to 150 mM imidazole (in buffer A) at a flow rate of 1 ml min−1. The AlgX-containing fractions, which appeared at ~75–90 mM imidazole, were pooled and concentrated by centrifugation (2200g at 277 K) using a Millipore concentrator with a 30 kDa molecular-weight cutoff. The protein was subsequently further purified and buffer-exchanged into buffer B[50 mM Tris–HCl pH 7.8, 2%(v/v) glycerol, 2 mM DTT] by size-exclusion chromatography using a HiLoad 16/60 Superdex 200 prep-grade gel-filtration column (GE Healthcare). The purity of theAlgX-His6 protein was monitored at all stages of the purification process using SDS–PAGE ( ▸). The purified protein was analyzed by MALDI–TOF mass spectrometry at the Advanced Protein Technology Centre at The Hospital for Sick Children and revealed a single peak corresponding to the expected molecular weight of the mature AlgX protein (i.e. minus its signal sequence but containing the C-terminal His6 tag). Following purification, samples could be stored at 277 K for up to one week at concentrations below 4 mg ml−1 with out visible precipitation or loss of the ability to readily form crystals.
Figure 1.
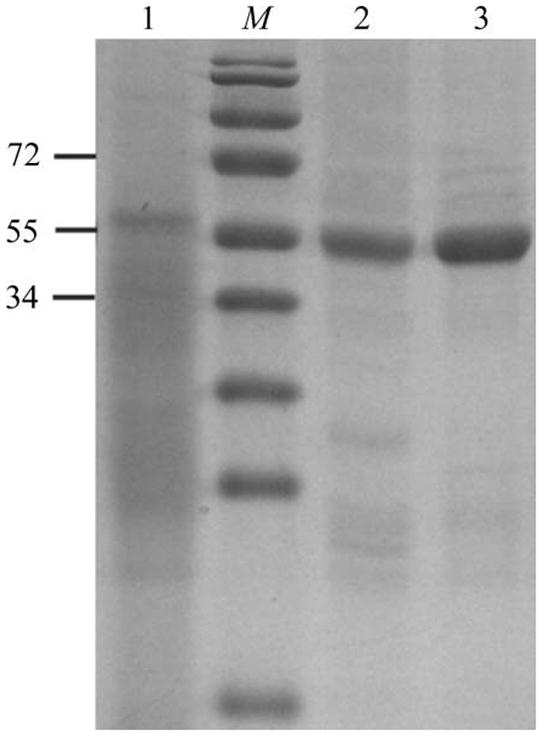
SDS–PAGE analysis of AlgX-His6 during expression and purification. Lane M, molecular-weight markers (kDa); lane 1, whole cell lysate; lane 2, AlgX after Ni-affinity chromatography; lane 3, AlgX after size-exclusion chromatography.
2.3. Crystallization
Preparations of purified protein at 4 and 8 mg ml−1 were used for the initial crystallization trials with ten different sparse-matrix crystal screens from Emerald BioSystems and Qiagen. These trials were setup in 48-well VDX plates (Hampton Research) by hand with 2 μl drops containing different ratios (1:1, 2:1 and 1:2) of protein solution and mother liquor over a reservoir containing 0.2 ml mother liquor. There were a number of hits from these trials, but the best-looking crystals always contained a low-molecular-weight polyethylene glycol(PEG) as the precipitant. Extensive optimization was carried out on condition No. 6 [100 mM sodium citrate pH 5.5, 20%(w/v) PEG 3000]from the Wizard I random sparse-matrix crystallization screen(Emerald BioSystems) by setting up approximately 150 variations of these conditions using the hanging-drop vapour-diffusion method in 24-well VDX plates (Hampton Research) with 4 μl drops containing a 1:1 ratio of protein solution (3 mg ml−1) and mother liquor over are servoir containing 0.5 ml mother liquor. This yielded single crystals that were small in size (50 × 20 × 20 μm) and took up to two weeks to grow at 293 K ( ▸). Further optimization involved the use of at least two successive rounds of seeding with seed beads and a streak-seeding tool (Hampton Research). Briefly, 4–8 crystals were extracted from a drop using a pipette and resuspended in 50 μl reservoir solution containing an additional 2–5%(w/v) PEG 3000. These crystals were crushed by vortexing them for 30 s in the presence of a seed bead and diluted by 20–50-fold. Streak-seeding from these diluted stocks was performed by dipping the streak-seeding tool into the diluted seed-stock solution and then dragging it once through prepared drops. This procedure yielded a variety of crystals from 100 mM sodium citrate pH 5.2, 14–18%(w/v) PEG 3000 that took only a few days to grow to maximum dimensions of 200 × 100 × 100 μm ( ▸).
Figure 2.
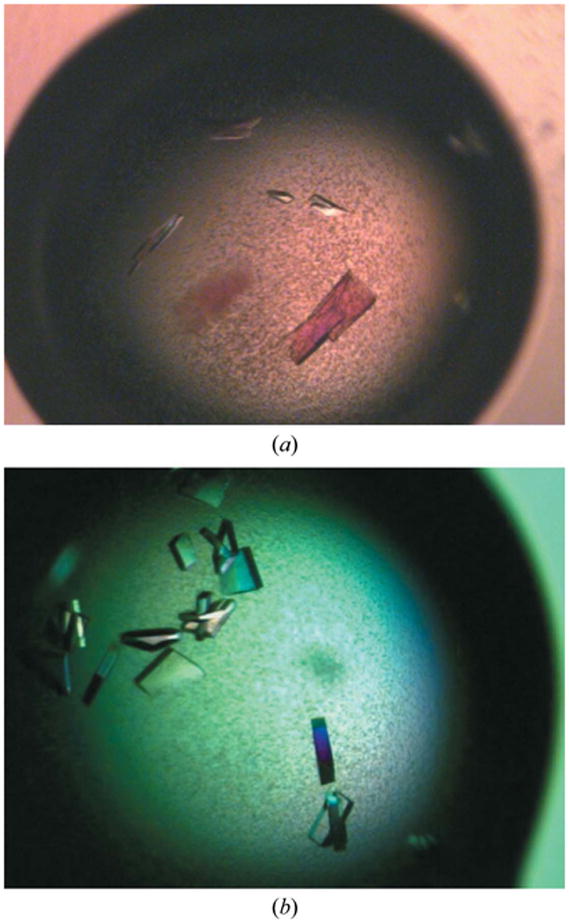
Crystals of AlgX. (a) Crystals of the AlgX protein from the initial crystallization hit; the average dimensions are 50 × 20 × 20 μm. (b) Crystals of AlgX following optimization and successive rounds of seeding. The crystal dimensions are approximately 200 × 100 × 100 μm.
2.4. Data collection
In preparation for data collection, two different cryosolutions were tested. The first consisted of adding glycerol directly to the reservoirsolution of the crystal to a final concentration of 25%(v/v). However, the crystals fared much better in cryoprotectants that consisted of exact concentrations of 100 mM sodium citrate pH 5.2, 0.5 mM DTT, 25%(v/v) glycerol and a PEG 3000 concentration that was 2–5%(w/v)higher than the reservoir solution of the crystal. Crystals were soaked for 2–3 min in cryoprotectants prior to vitrification in liquid nitrogen. To reduce the cracking of the crystals when immersed directly in the cryosolution, the cryosolution was first added directly to the crystallization drop. After 1–2 min soaking in this solution, the crystals were then moved with a loop into fresh cryosolution prior to vitrification in liquid nitrogen. Owing to variation in the diffraction quality, crystals were first screened on our in-house X-ray diffraction facilities at The Hospital for Sick Children (Cu Kα X-ray radiation from anRU-H3R rotating-anode generator with R-AXIS IV++ image-platedetector) prior to shipment to the National Synchrotron Light Source(NSLS). A complete set of data was collected at Station X29 at NSLS at 100 K. A 0.16 mm collimator was used to collect a total of 450 images of 1° Δϕ oscillations on an ADSC Quantum-315 detector with a 240 mm crystal-to-detector distance and an exposure time of 0.5 s per image. The data were integrated, reduced and scaled using HKL-2000 v. 0.95 ( ▸).
3. Results
The periplasmic protein AlgX from P. aeruginosa has been expressed and purified to near-homogeneity (98%; ▸). Approximately 8 mg of purified AlgX-His6 could be routinely obtained per litre of cell culture. Diffraction-quality crystals have been grown and optimized with successive rounds of seeding. The crystals diffracted to 2.1 Å resolution with no significant evidence of radiation decay over the collected images. The crystals belonged to space group P21, with unit-cell parameters a = 46.4, b = 120.6, c = 86.9 Å, β = 95.7°. Data-collection statistics are summarized in ▸. On the basis of density calculations ( ▸), each asymmetric unit is predicted to contain two monomers of AlgX (VM = 2.25Å3 Da−1). We are currently in the process of determining the structure of this protein using selenomethionine incorporation and the anomalous diffraction technique ( ▸).
Table 1. Data-collection statistics.
Values in parentheses are for the highest resolution shell.
| Wavelength (Å) | 1.0 |
| Temperature (K) | 100 |
| Space group | P21 |
| Unit-cell parameters (Å, °) | a = 46.4, b = 120.6, c = 86.9, β = 95.7 |
| Resolution (Å) | 50–2.1 (2.18–2.10) |
| Total No. of reflections | 334105 (26390) |
| No. of unique reflections | 53287 (5075) |
| Redundancy | 6.27 (5.20) |
| Completeness (%) | 97.2 (95.7) |
| Average I/σ(I) | 13.9 (2.70) |
| Rmerge† (%) | 11.1 (55.1) |
Rmerge = Σhkl Σi |Ii(hkl) − 〈I(hkl)〉|/ΣhklΣi Ii(hkl), where Ii(hkl) and 〈I(hkl)〉 represent the diffraction-intensity values of the individual measurements and the corresponding mean values, respectively.
Acknowledgments
The authors thank the Advanced Protein Technology Centre at The Hospital for Sick Children for assistance with the mass spectrometry. This work was supported by research grants from the Canadian Institutes of Health Research (CIHR No. MT13337) to PLH and the National Institutes of Health Research (GM081419) to PAT. JTW was funded in part by postdoctoral fellowships from The Hospital for Sick Children and the Natural Sciences and Engineering Research Council (NSERC) of Canada. PLH is the recipient of a Canada Research Chair.
References
- Bakkevig K, Sletta H, Gimmestad M, Aune R, Ertesvag H, Degnes K, Christensen BE, Ellingsen TE, Valla S. J Bacteriol. 2005;187:8375–8384. doi: 10.1128/JB.187.24.8375-8384.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conway SP, Brownlee KG, Denton M, Pekham DG. Am J Respir Med. 2003;2:321–322. doi: 10.1007/BF03256660. [DOI] [PubMed] [Google Scholar]
- Evans LR, Linker A. J Bacteriol. 1973;116:915–924. doi: 10.1128/jb.116.2.915-924.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franklin MJ, Chitnis CE, Gacesa P, Sonesson A, White DC, Ohman DE. J Bacteriol. 1994;176:1821–1830. doi: 10.1128/jb.176.7.1821-1830.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franklin MJ, Ohman DE. J Bacteriol. 1996;178:2186–2195. doi: 10.1128/jb.178.8.2186-2195.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franklin MJ, Ohman DE. J Bacteriol. 2002;184:3000–3007. doi: 10.1128/JB.184.11.3000-3007.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Govan JRW, Deretic V. Microbiol Rev. 1996;60:539–574. doi: 10.1128/mr.60.3.539-574.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutsche J, Remminghorst U, Rehm BH. Biochimie. 2006;88:245–251. doi: 10.1016/j.biochi.2005.06.003. [DOI] [PubMed] [Google Scholar]
- Hancock RE. Clin Infect Dis. 1998;27:S93–S99. doi: 10.1086/514909. [DOI] [PubMed] [Google Scholar]
- Hay ID, Rehman ZU, Rehm BH. Appl Environ Microbiol. 2009;75:1110–1120. doi: 10.1128/AEM.02416-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendrickson WA. Science. 1991;254:51–58. doi: 10.1126/science.1925561. [DOI] [PubMed] [Google Scholar]
- Jain S, Ohman DE. J Bacteriol. 1998;180:634–641. doi: 10.1128/jb.180.3.634-641.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain S, Ohman DE. In: Pseudomonas. Ramos J-L, editor. Vol. 3. New York: Kluwer Academic/Plenum; 2004. pp. 53–81. [Google Scholar]
- Jain S, Ohman DE. Infect Immun. 2005;73:6429–6436. doi: 10.1128/IAI.73.10.6429-6436.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keiski C, Harwich M, Jain S, Neculai AM, Yip P, Robinson H, Whitney JC, Riley L, Burrows LL, Ohman DE, Howell PL. Structure. 2010;18:265–273. doi: 10.1016/j.str.2009.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley LA, Sternberg MJ. Nature Protoc. 2009;4:363–371. doi: 10.1038/nprot.2009.2. [DOI] [PubMed] [Google Scholar]
- Koch C, Høiby N. Respiration. 2000;67:239–247. doi: 10.1159/000029503. [DOI] [PubMed] [Google Scholar]
- Leid JG, Willson CJ, Shirtliff ME, Hassett DJ, Parsek MR, Jeffers AK. J Immunol. 2005;175:7512–7518. doi: 10.4049/jimmunol.175.11.7512. [DOI] [PubMed] [Google Scholar]
- Lo Y, Lin S, Shaw J, Liaw Y. J Mol Biol. 2003;330:539–551. doi: 10.1016/s0022-2836(03)00637-5. [DOI] [PubMed] [Google Scholar]
- Maharaj R, May TB, Wang SK, Chakrabarty AM. Gene. 1993;136:267–269. doi: 10.1016/0378-1119(93)90477-k. [DOI] [PubMed] [Google Scholar]
- Matthews BW. J Mol Biol. 1968;33:491–497. doi: 10.1016/0022-2836(68)90205-2. [DOI] [PubMed] [Google Scholar]
- Merighi M, Lee VT, Hyodo M, Hayakawa Y, Lory S. Mol Microbiol. 2007;65:876–895. doi: 10.1111/j.1365-2958.2007.05817.x. [DOI] [PubMed] [Google Scholar]
- Mølgaard A, Kauppinen S, Larsen S. Structure. 2000;8:373–383. doi: 10.1016/s0969-2126(00)00118-0. [DOI] [PubMed] [Google Scholar]
- Monday SR, Schiller NL. J Bacteriol. 1996;178:625–632. doi: 10.1128/jb.178.3.625-632.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oglesby LL, Jain S, Ohman DE. Microbiology. 2008;154:1605–1615. doi: 10.1099/mic.0.2007/015305-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otwinowski Z, Minor W. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- Pier GB, Coleman F, Grout M, Franklin M, Ohman DE. Infect Immun. 2001;69:1895–1901. doi: 10.1128/IAI.69.3.1895-1901.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rehm BH, Boheim G, Tommassen J, Winkler UK. J Bacteriol. 1994;176:5639–5647. doi: 10.1128/jb.176.18.5639-5647.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remminghorst U, Rehm BH. Appl Environ Microbiol. 2006a;72:298–305. doi: 10.1128/AEM.72.1.298-305.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remminghorst U, Rehm BH. FEBS Lett. 2006b;580:3883–3888. doi: 10.1016/j.febslet.2006.05.077. [DOI] [PubMed] [Google Scholar]
- Robles-Price A, Wong TY, Sletta H, Valla S, Schiller NL. J Bacteriol. 2004;186:7369–7377. doi: 10.1128/JB.186.21.7369-7377.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitney JCC, Neculai AM, Ohman DE, Howell PL. Acta Cryst. 2009;F65:463–466. doi: 10.1107/S174430910901094X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winsor GL, Lo R, Sui SJ, Ung KS, Huang S, Cheng D, Ching WK, Hancock RE, Brinkman FS. Nucleic Acids Res. 2005;33:D338–D343. doi: 10.1093/nar/gki047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood LF, Ohman DE. J Bacteriol. 2006;188:3134–3137. doi: 10.1128/JB.188.8.3134-3137.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]


