Abstract
Tristetraprolin (TTP) is an RNA binding protein that controls the inflammatory response by limiting the expression of several proinflammatory cytokines. TTP post-transcriptionally represses gene expression by interacting with AU-rich elements (AREs) in 3′UTRs of target mRNAs and subsequently engenders their deadenylation and decay. TTP accomplishes these tasks, at least in part, by recruiting the multi subunit CCR4–NOT deadenylase complex to the mRNA. Here we identify an evolutionarily conserved C-terminal motif in human TTP that directly binds to a central domain of CNOT1, a core subunit of the CCR4–NOT complex. A high-resolution crystal structure of the TTP-CNOT1 complex was determined, providing the first structural insight into an ARE-binding protein bound to the CCR4–NOT complex. Mutations at the CNOT1-TTP interface impair TTP-mediated deadenylation, demonstrating the significance of this interaction in TTP-mediated gene silencing.
Introduction
Mammalian gene expression is tightly regulated by an array of post-transcriptional control programs, including mRNA translation and stability. The mRNA 3′ untranslated region (UTR) plays a critical role in post-transcriptional control via cis-acting elements, such as micro (mi)RNA target sites and adenylate uridylate-rich elements (AREs) 1,2. These RNA elements are targeted by miRNAs and ARE-binding proteins (ARE-BPs). Tristetraprolin (TTP) is an ARE-BP, which represents the prototypical member of the TIS11 [TPA (12-O-tetradecanoylphorbol-13-acetate) inducible sequence-11] family of RNA-binding proteins that include the TTP-related proteins butyrate response factors (BRF)-1 and -2 3–5. TTP is an important regulator of the inflammatory response, and a bona fide tumor suppressor in lymphomas 6–9. TTP, along with BRF-1 and BRF-2, bind to AREs in the 3′UTRs of target mRNAs through their tandem zinc finger domains 10, to promote mRNA deadenylation and subsequent decay by recruiting the CCR4–NOT complex 11. The N-terminal and C-terminal domains of TTP effect deadenylation through the CCR4–NOT complex 11,12. However, controversy exists with respect to how TTP interacts with the deadenylase machinery. One report concluded that components of the CCR4–NOT complex interact only with the TTP N-terminal domain 11, whereas another only documented interactions with the TTP C-terminal domain 12. Thus, it is currently unclear how TTP physically interacts with the CCR4–NOT complex to bring about deadenylation.
CCR4–NOT is a multi-subunit protein complex originally described in Saccharomyces cerevisiae as a global regulator of transcription and the cell cycle 13. However, it has since gained prominence as a master regulator of mRNA stability that interfaces with several key post-transcriptional control programs, including those mediated by miRNAs and by TTP family proteins 14. The mammalian CCR4–NOT complex is comprised of multiple proteins, termed CCR4–NOT (CNOT) subunits. The CNOT1 subunit acts as a scaffold for other CCR4–NOT components, including the CNOT6 (CCR4) and CNOT7 (CAF1) deadenylases. In this study, we have identified a novel motif in TTP that binds CNOT1, elucidated the structural basis for this interaction, and established its significance for TTP-mediated gene silencing.
Results
TTP directly binds CNOT1
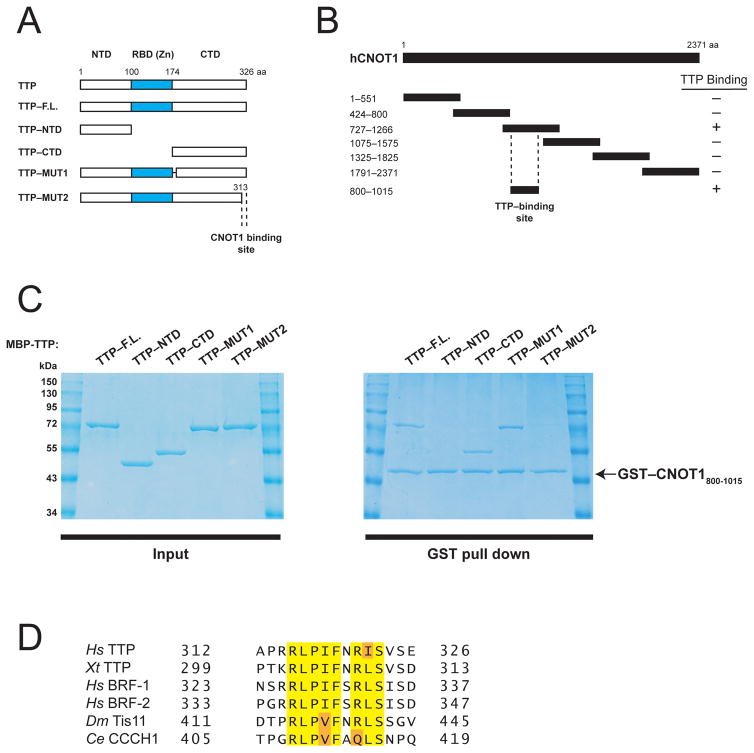
The human CNOT1 subunit is a large scaffolding protein that associates with TTP through unknown contacts 12. To demonstrate that CNOT1 directly interacts with TTP, as recently postulated 12, we performed in vitro co-precipitation experiments using recombinant MBP–fused TTP (Fig. 1A), and a series of GST–fused CNOT1 fragments covering the entire CNOT1 isoform C (2371 aa) protein (Fig. 1B). A CNOT1 fragment encompassing residues 727–1266 efficiently bound MBP–TTP, whereas all other CNOT1 fragments did not (Supplementary Fig. 1). We subsequently mapped the region of CNOT1 that binds TTP to residues 800–1015 (Fig. 1C and Supplementary Fig. 1).
Figure 1.
Human TTP directly binds a central fragment of CNOT1. (A) Schematic diagram of full-length TTP and TTP fragments used in co-precipitation experiments (Fig. 1C). Dashed lines indicate the region required for CNOT1 binding (Fig. 1C). (B) Schematic diagram of full-length CNOT1 and CNOT1 fragments used in co-precipitation experiments (Fig. 1C and Supplemental Fig. 1). Coordinates are marked to the left of each fragment. Dashed lines indicate the region required for TTP binding (Figure 1C). (C) Right panel: Recombinant glutathione-S-transferase (GST)–tagged CNOT1800–1015 (Figure 1B) was immobilized on glutathione-Sepharose beads and incubated with (MBP)–tagged full-length TTP, or TTP fragments (see left panel). Precipitated proteins were separated by SDS-PAGE and visualized by Coomassie staining. (D) Sequence alignment of conserved amino acids within the TTP C-terminus that binds CNOT1 of human (Hs) TTP, BRF-1, and BRF-2, Xenopus tropicalis (Xt) TTP, Drosophila melanogaster (Dm), Tis11 and Caenorhabditis elegans (Ce) CCCH1. Highlighted are amino acids identical in 100% of proteins (yellow) or conservative substitutions by related amino acids (orange).
Next, we set out to identify what region of TTP binds to the CNOT1800–1015 fragment. To this end, we generated MBP-fused TTP fragments containing either the TTP N-terminal (residues 1–99) or C-terminal (residues 174–326) regions (Fig. 1A), which have been reported to associate with the CCR4–NOT 11,12. GST-CNOT1800–1015 did not interact with the TTP N-terminal domain in binding assays (Fig. 1C). In contrast, it bound the TTP C-terminal domain very efficiently (~50% of input) (Fig. 1C). Sequence alignment of the TTP C-terminal domain (CTD) and its related proteins, BRF-1 and BRF-2, revealed two conserved patches of amino acids in the TTP CTD: residues 179–192 and 314–326 (Supplementary Fig. 2). GST–CNOT1800–1015 bound MBP-TTP lacking amino acids 179–192 [(TTP-MUT1) Figs. 1A and C]. However, MBP–TTP lacking the terminal 13 amino acids (TTP–MUT2) failed to bind GST–CNOT1800–1015 (Figs. 1A and C). Importantly, the TTP C-terminal amino acid sequence is highly conserved in Tis11 homologs, including in Xenopus tropicalis, Drosophila melanogaster and Caenorhabditis elegans (Fig. 1D). Taken together, these results demonstrate that CNOT1 interacts with an evolutionarily conserved amino acid motif at the C-terminus of TTP.
Structural determination of the TTP–CNOT1 interaction
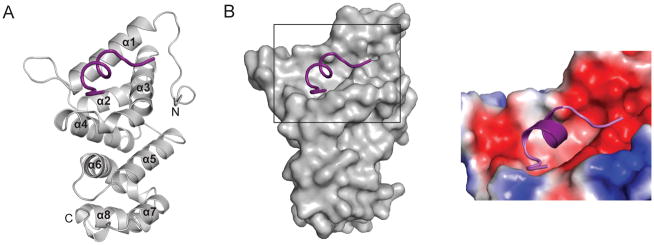
To gain structural insight into the CNOT1–TTP interaction we solved the crystal structure of a TTP-interacting fragment of CNOT1 (residues 800–999) in complex with a peptide covering residues Ala312 to Glu326 of hTTP (Fig. 1D) at 1.5 Å resolution (Fig. 2A and Table 1). The N-terminal 20 residues (800–819) of the CNOT1 fragment are disordered, whereas TTP could be modeled from residues Arg314 to Ser325. CNOT1820–999 consists of eight α-helices stacked as a series of helix-turn-helix motifs in an arrangement similar to the HEAT repeats of the Middle domain of Initiation Factor 4G (MIF4G) 15 (Supplementary Fig. S3A). The MIF4G domain is generally utilized as a protein-protein interaction domain and is found in numerous scaffolding proteins involved in the regulation of translation and RNA metabolism 16. The main difference between the CNOT1 domain and the MIF4G domain resides at the N-terminal region, where the MIF4G domain forms the first HEAT repeat. In CNOT1, this region forms a long loop held in place by Trp828, which folds over α3 and contributes to a highly acidic surface adjacent to the TTP binding site (not shown in figure).
Figure 2.
Crystal structure of the TTP–CNOT1 complex. (A) Cartoon representation of the crystal structure of human CNOT1820–999 (grey) in complex with TTP peptide (purple). The eight CNOT1 helices are numbered starting from the N-terminus. (B) Left panel: Surface representation of CNOT1820–999. The bound TTP312–326 peptide (purple) is shown as a cartoon representation. Right panel: magnification of TTP binding site on CNOT1820–999 colored according to electrostatic potential, from dark red (−kT/e) to white (0 kT/e) to dark blue (+5 kT/e).
Table 1.
Data collection and refinement statistics (molecular replacement)
| 4J8S | |
|---|---|
| Data collection | |
| X-ray source | Rotating Cu anode |
| Wavelength (Å) | 1.5418 |
| Resolution (Å) | 30–1.55(1.61–1.55) |
| Space group | P23 |
| Cell parameters (Å, °) | a=b=c=80.07, α=β=γ=90.00 |
| Molecules per ASU | 1 |
| Mosaicity (°) | 0.45 |
| No. reflections | 57193 |
| Redundancy | 11.6(3.5) |
| I/σ(I) | 28.8(2.4) |
| Completeness (%) | 99.0(90.2) |
| Rsym‡ | 0.065(0.45) |
| Rwork/Rfree | 16.1/18.3 |
| Refinement | |
| Number of atoms, all | 1886 |
| Protein | 1645 |
| Water | 241 |
| R.m.s deviations | |
| Bond lengths (Å) | 0.012 |
| Bond angles (°) | 1.313 |
| B-factors (Å2), all | 19.75 |
| Protein | 18.06 |
| Water | 31.23 |
| Rwork/Rfree | 16.1/18.3 |
The TTP binding site is located close to the N-terminus of CNOT1820–999, and is formed by a highly conserved hydrophobic groove between helices α1 and α3 that is flanked by negatively charged patches of amino acids (Fig. 2B). The central portion of the TTP peptide forms a short, two-turn amphipathic α-helix resulting in the insertion of Leu316, Ile318, Phe319, and Ile322 into the hydrophobic groove of CNOT1 (Fig. 3A). The bottom of the groove is lined with aromatic residues Phe847, Tyr851, and Tyr900 (Fig. 3B). A network of electrostatic interactions between several polar residues located at the termini of the TTP peptide and the surrounding negatively charged residues in CNOT1 is also formed. The most prominent of these interactions is a salt bridge between Glu893 of CNOT1 and Arg315 of TTP at the edge of the groove (Fig. 3B). Hydrogen bonding between Glu893 and Tyr900 of CNOT1 with residues Arg315 and Ser323 of TTP, respectively, generates a stable closed-loop conformation in the TTP peptide (Fig. 3B). Additional contacts stabilizing the complex are provided by hydrogen bonds on the solvent exposed side of the TTP helix between residues Pro317 and Arg321 of TTP with Asn844 and Gln848 of CNOT1, respectively (Fig. 3B). The sum of these interactions confers an affinity (Kd) of ~2μM between the TTP peptide and CNOT1820–999 as measured by isothermal titration calorimetry (ITC) experiments (Supplementary Fig. S3B).
Figure 3.
Analysis of TTP–CNOT1 interaction. (A) Electron density (grey) of the TTP312–326 peptide (shown as purple sticks). Density (Fo-Fc of a map calculated by omitting the TTP peptide) is contoured at 2.5σ. (B) A view of the interface between the CNOT1 domain (grey) and TTP peptide (purple). Interacting side chains of CNOT1 (blue) and TTP (purple) are shown as sticks. The backbone amide of P317 is shown as a blue sphere. Dashed lines indicate hydrogen-bonding interactions. (C) Recombinant glutathione-S-transferase (GST)–tagged CNOT1800–1015 (Figure 1B) was immobilized on glutathione-Sepharose beads and incubated with (MBP)–tagged TTP-CTD, or TTP-CTD mutants. Precipitated proteins were separated by SDS-PAGE and visualized by Coomassie staining.
We generated mutants of TTP to biochemically validate the structure. Alanine substitutions of Arg315, which makes a salt bridge with Glu893 of CNOT1, and Phe319, which is at the center of the hydrophobic interaction site and forms Van der Waals contacts with the CNOT1 surface (Supplementary Fig. 4A), in the TTP-CTD abolished its interaction with CNOT1800–1015 in vitro (Fig. 3C). In addition, mutating Phe319 to alanine in the TTP peptide disrupted its binding to CNOT1820–999 [Kd = ~7mM (Supplementary Fig. S3C)]. Mutation of Asn320, which does not make contact with CNOT1, to alanine failed to disrupt the TTP-CNOT1 interaction (Fig. 3C). Taken together, these data demonstrate that the TTP–CNOT1 interaction requires several invariant residues in the C-terminus of TTP, a sequence we now refer to as a TTP–CCR4–NOT-Interaction Motif (TTP-CIM).
TTP–CNOT1 contact promotes mRNA deadenylation in vitro
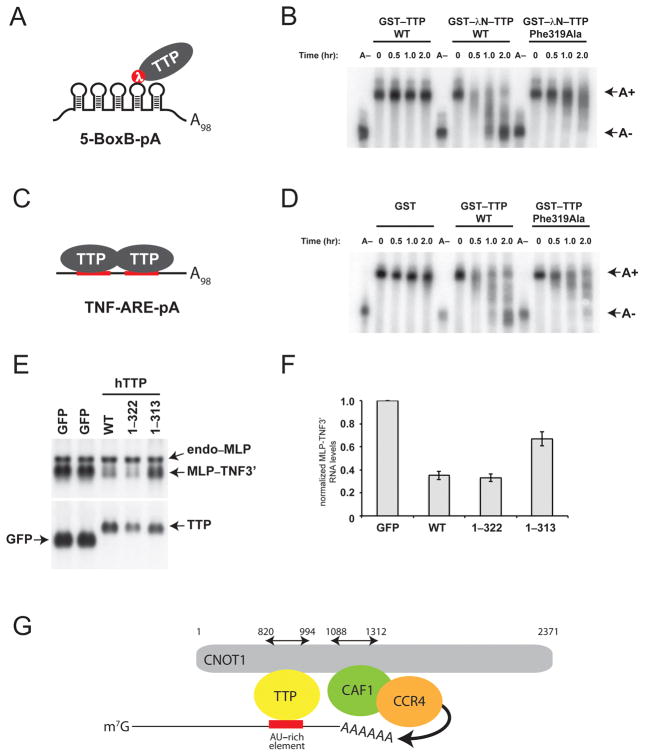
TTP requires its CTD to effect maximal deadenylation of target mRNAs 11,12. To examine the functional significance of the interaction between CNOT1 and TTP, we established an in vitro assay for TTP-mediated deadenylation. Using the λN–BoxB system 17, we artificially tethered recombinant TTP proteins to an RNA reporter containing 5 BoxB stem-loops and a 98-nucleotide poly(A) tail (5-BoxB-pA), which was added to a cell-free extract derived from mouse Krebs-2 ascites (termed Krebs extract) (Fig. 4A). This strategy has been used successfully to biochemically dissect the mechanics of miRNA-mediated deadenylation by the CCR4–NOT complex 18. Recombinant wild-type GST–TTP was unable to mediate deadenylation, whereas wild-type GST–λNHA–TTP engendered complete deadenylation of 5-BoxB-pA RNA in the Krebs extract within 2 hours (Fig. 4B). In contrast, a TTP mutant that could not bind to CNOT1 (TTP-Phe319Ala) displayed severely impaired deadenylation, with deadenylation barely detectable by 2 h (Fig. 4B, right panel). Similar results were obtained when using a polyadenylated reporter containing two TTP-binding sites derived from the tumor necrosis factor 3′UTR (TNF-ARE-pA), a bona fide TTP mRNA target (Fig. 4C). Wild-type GST–TTP engendered complete deadenylation of TNF-ARE-pA by 2 hours, whereas GST-TTP-Phe319Ala displayed partial and incomplete deadenylation (Fig. 4D). Taken together, these results demonstrate that TTP-mediated deadenylation in vitro requires the TTP-CTD–CNOT1 interaction, irrespective of whether TTP is directly binding or being artificially tethered to a target RNA.
Figure 4.
Disruption of the TTP–CNOT1 interaction impairs mRNA deadenylation in vitro and mRNA stability in vivo. (A) Schematic depiction of GST–λNHA–TTP tethered to 5-BoxB-pA RNA. (B) 5-BoxB-pA RNA deadenylation in Krebs extract in the presence of recombinant wild-type GST–TTP, or wild-type or mutant (Phe319Ala) GST–λNHA–tagged TTP. A(−) RNA was prepared by incubating 5-BoxB-pA with Oligo(dT) and RNase H. Polyadenylated and deadenylated mRNAs are marked on the right of the figure. (C) Schematic depiction of TTP associated with TNF-ARE-pA RNA. (D) TNF-ARE-pA RNA deadenylation in Krebs extract in the presence of recombinant GST, or wild-type or mutant (Phe319Ala) GST–tagged TTP. A(−) RNA was prepared by incubating TNF-ARE-pA with Oligo(dT) and RNase H. Polyadenylated and deadenylated mRNAs are marked on the right of the figure. (E) HEK-293 cells were co-transfected with the reporter construct pMLP-TNF3′ and DNA encoding GFP or TTP protein constructs. Total cellular RNA was harvested and subjected to electrophoresis and Northern blotting. In the upper panel, the Northern blot was probed with a 32P-labeled MLP probe; in the lower panel, the blot was probed with a 32P-labeled TTP probe. In the upper panel, the endogenous MLP mRNA (endo-MLP) the reporter MLP-TNF3′ species are indicated by arrowheads. In the lower panel, the expression levels of transcripts for GFP alone and GFP–TTP are indicated. (F) Quantification of MLP-TNF3′ RNA levels (see Figure 4E), normalized to those in GFP-expressing cells, in the presence of GFP, TTP WT, TTP 1–322 and TTP 1–313. Mean values ±s.e. from seven independent experiments are shown. (G) Model for structural organization of mRNA-bound TTP in complex with CNOT1 and CAF1 and CCR4 deadenylases. The cartoon summarizes structural data reported in this manuscript, combined with data from crystal structures of yeast 20 and human 21 NOT1 proteins in complex with CAF1 and CCR4 deadenylases. TTP and CAF1 binding domains in CNOT1 (820–999 and 1088–1312, respectively) refer to coordinates of human CNOT1 isoform c. Other CCR4–NOT subunits (i.e. CNOT2, 3, 9 and 10) are not shown.
TTP–CNOT1 contact promotes mRNA decay in vivo
We also examined the significance of the TTP–CNOT1 interaction on the stability of a TTP-targeted reporter mRNA in vivo. We employed a plasmid containing the mouse MARCKS-like protein (MLP) promoter and coding region that is fused to the TTP-targeted TNFα 3′UTR (MLP-TNF3′), which has previously served as a reporter to assay TTP function in cell cultures 19. HEK-293 cells were co-transfected with the MLP-TNF3′ reporter plasmid, along with a plasmid coding for (i) green fluorescent protein (GFP), (ii) wild-type TTP or (iii) various TTP mutants (Figs. 4E and F). We observed significantly less MLP-TNF3′ RNA in wild-type TTP-expressing cells as compared to GFP-expressing cells, indicative of an increase in reporter mRNA degradation. Similar effects on MLP-TNF3′ RNA stability were observed in cells expressing a TTP mutant that contains the TTP-CIM but lacks four C-terminal amino acids (1–322) (lane 4). In contrast, MLP-TNF3′ RNA was significantly stabilized in cells expressing a TTP mutant where an additional 9 amino acids have been deleted, thereby removing the TTP-CIM (1–313) (lane 5). Thus, these results strongly suggest that TTP requires its C-terminal CNOT1-interacting motif to bring about efficient decay of a target mRNA in vivo.
Discussion
We have identified a novel CCR4–NOT-interaction motif (CIM) at the C-terminus of human TTP. The TTP-CIM directly binds the CNOT1 subunit of the CCR4–NOT complex. Importantly, this represents the first structure of the interface between an ARE-BP and the CCR4–NOT complex. Furthermore, our data demonstrate that optimal TTP-mediated deadenylation requires the TTP-CIM to be bound to CNOT1, as disrupting this interaction impairs, but does not completely abolish, deadenylation in vitro (Figs. 4A and B) and mRNA decay in vivo (Fig. 4C). These data fit well with previous studies demonstrating that the TTP N-terminal domain facilitated deadenylation of target mRNAs in a CCR4–NOT complex-dependent manner. How the deadenylase machinery interacts with the TTP N-terminal domain remains to be determined. Interestingly, the TTP-binding domain of CNOT1 is N-terminally adjacent to the CNOT1 domain that binds the CAF1 deadenylase 20,21. It is therefore plausible that TTP binding next to CAF1 on CNOT1 may help position the CAF1 deadenylase such that it has unimpeded access to the mRNA poly(A) tail (Fig. 4G).
TTP residues that interact with CNOT1 are phylogenetically conserved in Tis11 proteins, including the Drosophila melanogaster Tis11 homolog and the Caenorhabditis elegans protein CCCH-1 (Fig. 1D). Moreover, CNOT1 residues that form a groove, which contains amino acids that interact with the TTP-CIM (Phe847, Tyr851, Glu893 and Tyr900) are highly conserved across these species as well (Supplementary Fig. 4B). A recent study reported the crystal structure of the yeast Not1 N-terminal region (residues 154–753) 20. It consists of 13 HEAT repeats, of which repeats 10–13 (residues 571–746) correspond to the 4 HEAT repeats found in human CNOT1820–999. Although yeast Not1 and human CNOT1 proteins share only 35% sequence identity in this region, the structures are very similar with a Cα-RMSD of 1.19 Å (Supplementary Figs. 4C and D). Interestingly, yeast Not1 HEAT repeats 10–13 form a separate unit within the protein in that they are arranged in a parallel fashion while there is a rotation by 90° with respect to repeats 1–9 20. This suggests that HEAT repeats 10–13 form a separate domain. Our results confirm this notion since we show that this domain is stable and can carry out its function of TTP recruitment in the absence of the more N-terminal HEAT repeats. The residues in human CNOT1 that form the TTP docking site are also conserved in yeast Not1, suggesting that they may also act as a protein binding site. However, the yeast Tis11 homolog CTH1 (Cysteine Three Histidine 1) does not contain a sequence that shares any detectable homology to the hTTP-CIM. Nevertheless, it is conceivable that ancient NOT1 proteins maintained this protein-interaction groove, which was exploited during the course of eukaryotic evolution by proteins such as TTP. CNOT1 binds the miRNA-associated GW182 protein through two conserved CIMs and additional tryptophan-containing sequences 18,22. However, the TTP-CIM does not share homology with the GW182 CIMs, suggesting that TTP and GW182 bind to different regions of CNOT1.
Mouse TTP activity is controlled by phosphorylation on a number of residues, including Ser52 and Ser178 (Ser60 and Ser186 in human TTP, respectively) by the p38 MAPK-activated protein kinase 2 (MK2) 23. Serines 52 and 178, when phosphorylated, act as substrates for 14-3-3 adaptor proteins that bind to and stabilize TTP 23,24. Phosphorylation of TTP via MK2 also impairs TTP recruitment of the CCR4–NOT complex and TTP-mediated deadenylation 25,26. Based on these data, it has been postulated that 14-3-3 binding to TTP impairs deadenylase recruitment 26. However, our data demonstrate that 14-3-3 binding sites (Ser52 and Ser178) do not overlap with the C-terminal TTP CIM. Notwithstanding these data, MK2-induced TTP phosphorylation partially inhibits CCR4–NOT association even when these serines are mutated in tandem to alanines, thereby abolishing 14-3-3 binding 26. It has therefore been suggested that additional phosphorylation sites act to inhibit deadenylase recruitment to TTP 26. Interestingly, MK2 has also been reported to phosphorylate TTP and BRF-1 on a highly conserved serine residue residing within the TTP-CIM [Ser323 in hTTP (Fig. 1D)] 23. Our structural data indicate that Ser323 forms multiple contacts at the TTP-CNOT1 interface, both within the TTP peptide and with CNOT1 (Fig 3B). Thus, the addition of a phosphate group to Ser323 would likely perturb TTP–CNOT1 binding. Indeed, a TTP-CIM peptide containing a phospho-Ser323 binds with a significantly lower affinity to CNOT1820–999 [Kd = ~625μM (Supplementary Fig. S3D)] as compared to wild-type. Taken together, these data suggest that the ability of the TTP-CIM to bind CNOT1 is regulated through the p38-signaling pathway. This model may help explain how MK2-phosphorylation regulates TTP recruitment of the CCR4–NOT complex. It is possible that 14-3-3 proteins and the CCR4–NOT complex compete for TTP binding. Phosphorylation of Ser323 may therefore aid the TTP transition from being bound to the CCR4–NOT complex to interacting with 14-3-3 proteins. Whether this post-translational modification plays a role in regulating TTP-mediated silencing, however, remains to be determined.
ONLINE METHODS
DNA constructs
For preparation of CNOT1 recombinant proteins, human CNOT1 fragments were PCR amplified from CNOT1 isoform c (accession: NP_001252541) and subcloned into pGEX-6P1 (GE Healthcare) with six C-terminal histidines. Human TTP fragments were PCR amplified and subcloned into pMAL-c5x (New England Biolabs) with six C-terminal histidines, or subcloned into pGEX-6P1 (GE Healthcare) in frame with or without an upstream sequence encoding a λN-HA peptide, and with six C-terminal histidines. Multiple CNOT1 fragments were cloned into the EcoRI and NotI sites of the pSMT3 vector 1, which contains an N-terminal Ulp1 cleavable His6-SUMO tag. These CNOT1 fragments were tested for crystallization, with CNOT1800–999 yielding diffractable crystals. RL-5BoxB-pA has previously been described 2. RL-TNF-pA was generated by inserting TTP binding sites from the TNF alpha 3′UTR into RL-pA using site-directed mutagenesis. Expression plasmids CMV.mTNFα [127–1325], CMV.hTTP.tag (WT) and its zinc finger mutant (C124R) were constructed as described (Lai et al., 1999); plasmid pMlp.TNF-3′UTR was made as described 3. A fusion plasmid, CMV.GFP.hTTP97-end, that expresses the human TTP protein from the tandem zinc finger to the C-terminus was constructed by fusing EGFP in frame with hTTP residues 97–326 (of GenBank accession number NM_003398.1), and ligating the two fragments with the HindIII and XhoI digested vector CMV.BGH3′/BS+ 4. Carboxyl terminal (C-term) mutants of CMV.hTTP, or CMV.GFP.hTTP97-end, were made using the PCR primer-overlapping mutagenesis technique. The EGFP expression construct CMV.EGFP BGH3′ was made by releasing the HindIII-NotI fragment, containing the EGFP coding sequence, from pEGFP-N1 (Clontech), and ligating it with the HindIII and NotI digested vector CMV.BGH3′/BS+.
Recombinant protein expression and purification
His-tagged recombinant proteins were expressed in Rosetta-2(DE3) E. coli cells (EMD Biosciences) and purified using Ni-NTA resin (Qiagen). CNOT1800–999 was subsequently treated with Ulp1 to cleave the SUMO-his tag and dialyzed into buffer containing 25 mM Tris-HCl pH 8.0, 100 mM NaCl. The sample was diluted twice to obtain a final NaCl concentration of 50 mM. The sample was loaded onto a Mono Q (GE Healthcare) anion-exchange column. Fractions containing the CNOT1 protein were collected, concentrated and further purified over a Superdex-75 size-exclusion column in 25 mM MES-HCl, pH 6.5, and 200 mM NaCl.
GST pull-down experiments
35 pmol of various MBP-TTP fragments was incubated with 20 μl of packed glutathione Sepharose-4B and 35 pmol of GST, or various recombinant GST-CNOT1 fragments in binding buffer (25 mM Tris, pH 7.5; 150 mM NaCl; 0.1% NP-40) for 2 hours at 4°C with gentle rocking. Beads were then washed five times with 1 ml of binding buffer, and proteins were eluted by boiling the beads with 40 μl of Laemmli sample buffer at 95°C for 8 minutes. Proteins were separated by SDS-PAGE and visualized by Coomassie staining.
Peptide synthesis and purification
Peptides corresponding to the wild-type (312-APRRLPIFNRISVSE-326) and Phe319Ala mutant (312-APRRLPIANRISVSE-326) CNOT1 binding site in TTP (accession GI:136471) were synthesized by Fmoc N-(9-fluorenyl)-methoxycarbonyl solid-phase peptide synthesis (Sheldon Biotechnology Center) and purified by reverse-phase chromatography on a Vydac C18 column. Peptides was verified by ion-spray quadrapole mass spectroscopy. The pSer323 peptide (312-APRRLPIFNRIpSVSE-326) was generated and purified by Canpeptide (Montreal, Canada).
Crystallization, data collection, and structure determination
TTP peptide (concentration ~7 mg/mL in buffer containing 25 mM MES-HCl pH 6.5, 200 mM NaCl) was mixed with CNOT1800–999 (concentration 15 mg/mL) at a 1.5:1 molar ratio of peptide to protein and crystals were grown by hanging-drop vapor diffusion. The protein-peptide complex was mixed 1:1 with well solution containing 0.2 M MgCl2, 0.1 M Bis-Tris pH 5.5, and 25% PEG3350 at room temperature. Crystals diffracting to 1.55 Å with excellent quality appeared after 7 days. Native data for 360 degrees collected in 0.5 degree images on a rotating anode Cu Kα home source (λ=1.54 Å) at 100 K were processed with the HKL2000 software package 5. CNOT1 in complex with TTP peptide crystallized in space group P23 with cell dimensions a=b=c=80.07 Å. The structure of CNOT1800–999 was solved using sulfur-single wavelength anomalous dispersion (S-SAD). The protein contains 9 sulfur atoms (6 from Met and 3 from Cys). The sulfur substructure was solved using the program SHELXD 6, which found 8 of 9 sites. Data up to 2.5 Å were used for the search where the signal to noise was ~55 σ in that shell. Phases were calculated using SHELXE with the −e1.0 flag (free lunch algorithm 7) for data up to 1.55 Å, which produced electron density maps of extremely high quality that were almost completely traced by Arp/Warp 8 including the ligand peptide. The structure was further refined using the PHENIX program 9. Statistics are described in Table 1. 99.5 % of residues were found in the Ramachandran favored regions and 0 % outliers were observed.
In vitro deadenylation assays
5-BoxB-pA and TNF-pA RNA transcripts were generated from a PCR product derived from RL-5-BoxB-pA and RL-TNF-pA, respectively. PCR product was digested with AgeI (New England Biolabs). To synthesize radiolabeled mRNA, [α-32P]UTP (800 Ci/mmol, 10 mCi/ml; PerkinElmer) was used according to the manufacturer’s protocol. The mRNA was loaded on a mini Quick Spin RNA Column (Roche) to remove unincorporated nucleotides. In vitro deadenylation assays were carried out as described 10 in the presence of 172 nM GST, GST–TTP and GST–λNHA–TTP proteins.
In vivo mRNA stability assays
HEK 293 cells were transiently transfected with CMV.mTNFα or other constructs in calcium-phosphate precipitates as described previously 4. Each 100-mm dish of cells (0.8 × 106) was co-transfected with 3 μg of pLMP-TNF3′ and 10 ng of hTTP expression plasmid, or 5 ng of plasmid CMV.EGFP.BGH3′ together with 5 ng of vector CMV.BGH3′/BS+ as a control. Vector plasmid (BS+) was added to make the total amount of co-transfected DNA 5 μg per dish in all cases. In every experiment, three dishes of cells were pooled for each co-transfection condition. Total cellular RNA was harvested from the cells using the illustra RNAspin Mini RNA Isolation Kit (GE Healthcare). Each gel lane for Northern blotting contained 10 μg of RNA. Northern blots were hybridized to a random-primed, α-32P-labeled Mlp MscI fragment, or a mouse TTP cDNA together with the EGFP coding cDNA 3,4.
Isothermal titration calorimetry (ITC)
Software for data acquisition and instrument control was from MicroCal Inc. (Northampton, MA). The ITC buffer used contained 20 mM Tris pH 7.2, 150 mM NaCl, and 200 μM TCEP. The reaction cell contained 1.8 ml of 25–30 μM CNOT1800–999 at 20°C. The sample cell was titrated with 29 injections of 10 μl peptide (300 μM-1mM) with a 4 minute time interval between injections. The isotherm was fitted with a 1:1 binding model to determine the thermodynamic binding constant and stoichiometry.
Experiments were carried out on a MicroCal VP-ITC titration calorimeter using the VPViewer software for data acquisition and instrument control (MicroCal Inc., Northampton, MA). The ITC buffer contained 20 mM Tris pH 7.2, 150 mM NaCl, and 200 μM TCEP. The reaction cell contained 1.8 ml of 25–30 μM CNOT1800–999 and was thermostated to 20°C. The sample cell was titrated with 29 injections of 10 μl peptide (300 μM-1mM) with a 4 min time interval between injections. The isotherm for the wild-type titration was fitted with a 1:1 binding model to determine the thermodynamic binding constant and stoichiometry. The number of binding sites was set to 1 for calculating the binding constants of the mutant and phosphorylated peptides.
Supplementary Material
Acknowledgments
This work was supported by Canadian Institutes of Health Research grants to N.S. (MOP-93607) and to B.N (MOP-82929). N.S. was also supported by a Howard Hughes Medical Institute (HHMI) Senior International Scholarship. P.B. was supported by the Intramural Research Program of the US National Institutes of Health, National Institute of Environmental Health Sciences. N.Siddiqui holds a fellowship from the Cole Foundation.
Footnotes
ACCESSION CODES
Structure factors and coordinates for the TTP-CNOT1 complex have been deposited in the Protein Data Bank with accession the code 4J8S.
AUTHOR CONTRIBUTIONS
M.R.F., F.F., C.R. and N. Siddiqui designed experiments. M.R.F. and C.R. performed binding assays and in vitro deadenylation assays. N. Siddiqui performed ITC experiments. F.F. crystallized the CNOT1-TTP complex and B.N. and F.F. determined its structure. W.S.L. and P.J.B. performed in vivo stability assays. A.K. provided technical support. M.R.F., F.F., B.N. and N.S. wrote the manuscript.
References
- 1.Fabian MR, Sonenberg N, Filipowicz W. Regulation of mRNA translation and stability by microRNAs. Annual review of biochemistry. 2010;79:351–79. doi: 10.1146/annurev-biochem-060308-103103. [DOI] [PubMed] [Google Scholar]
- 2.von Roretz C, Di Marco S, Mazroui R, Gallouzi IE. Turnover of AU-rich-containing mRNAs during stress: a matter of survival. Wiley interdisciplinary reviews. RNA. 2011;2:336–47. doi: 10.1002/wrna.55. [DOI] [PubMed] [Google Scholar]
- 3.Sanduja S, Blanco FF, Dixon DA. The roles of TTP and BRF proteins in regulated mRNA decay. Wiley interdisciplinary reviews. RNA. 2011;2:42–57. doi: 10.1002/wrna.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blackshear PJ. Tristetraprolin and other CCCH tandem zinc-finger proteins in the regulation of mRNA turnover. Biochem Soc Trans. 2002;30:945–52. doi: 10.1042/bst0300945. [DOI] [PubMed] [Google Scholar]
- 5.Lai WS, Stumpo DJ, Blackshear PJ. Rapid insulin-stimulated accumulation of an mRNA encoding a proline-rich protein. The Journal of biological chemistry. 1990;265:16556–63. [PubMed] [Google Scholar]
- 6.Sanduja S, Blanco FF, Young LE, Kaza V, Dixon DA. The role of tristetraprolin in cancer and inflammation. Frontiers in bioscience : a journal and virtual library. 2012;17:174–88. doi: 10.2741/3920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rounbehler RJ, et al. Tristetraprolin impairs myc-induced lymphoma and abolishes the malignant state. Cell. 2012;150:563–74. doi: 10.1016/j.cell.2012.06.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Carballo E, Lai WS, Blackshear PJ. Feedback inhibition of macrophage tumor necrosis factor-alpha production by tristetraprolin. Science. 1998;281:1001–5. doi: 10.1126/science.281.5379.1001. [DOI] [PubMed] [Google Scholar]
- 9.Taylor GA, et al. A pathogenetic role for TNF alpha in the syndrome of cachexia, arthritis, and autoimmunity resulting from tristetraprolin (TTP) deficiency. Immunity. 1996;4:445–54. doi: 10.1016/s1074-7613(00)80411-2. [DOI] [PubMed] [Google Scholar]
- 10.Lai WS, Carballo E, Thorn JM, Kennington EA, Blackshear PJ. Interactions of CCCH zinc finger proteins with mRNA. Binding of tristetraprolin-related zinc finger proteins to Au-rich elements and destabilization of mRNA. The Journal of biological chemistry. 2000;275:17827–37. doi: 10.1074/jbc.M001696200. [DOI] [PubMed] [Google Scholar]
- 11.Lykke-Andersen J, Wagner E. Recruitment and activation of mRNA decay enzymes by two ARE-mediated decay activation domains in the proteins TTP and BRF-1. Genes & development. 2005;19:351–61. doi: 10.1101/gad.1282305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sandler H, Kreth J, Timmers HT, Stoecklin G. Not1 mediates recruitment of the deadenylase Caf1 to mRNAs targeted for degradation by tristetraprolin. Nucleic acids research. 2011;39:4373–86. doi: 10.1093/nar/gkr011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Collart MA, Panasenko OO. The Ccr4--not complex. Gene. 2012;492:42–53. doi: 10.1016/j.gene.2011.09.033. [DOI] [PubMed] [Google Scholar]
- 14.Goldstrohm AC, Wickens M. Multifunctional deadenylase complexes diversify mRNA control. Nature reviews. Molecular cell biology. 2008;9:337–44. doi: 10.1038/nrm2370. [DOI] [PubMed] [Google Scholar]
- 15.Schutz P, et al. Crystal structure of the yeast eIF4A-eIF4G complex: an RNA-helicase controlled by protein-protein interactions. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:9564–9. doi: 10.1073/pnas.0800418105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ponting CP. Novel eIF4G domain homologues linking mRNA translation with nonsense-mediated mRNA decay. Trends Biochem Sci. 2000;25:423–6. doi: 10.1016/s0968-0004(00)01628-5. [DOI] [PubMed] [Google Scholar]
- 17.Baron-Benhamou J, Gehring NH, Kulozik AE, Hentze MW. Using the lambdaN peptide to tether proteins to RNAs. Methods in molecular biology. 2004;257:135–54. doi: 10.1385/1-59259-750-5:135. [DOI] [PubMed] [Google Scholar]
- 18.Fabian MR, et al. miRNA-mediated deadenylation is orchestrated by GW182 through two conserved motifs that interact with CCR4–NOT. Nature structural & molecular biology. 2011;18:1211–7. doi: 10.1038/nsmb.2149. [DOI] [PubMed] [Google Scholar]
- 19.Blackshear PJ, et al. Zfp36l3, a rodent X chromosome gene encoding a placenta-specific member of the Tristetraprolin family of CCCH tandem zinc finger proteins. Biol Reprod. 2005;73:297–307. doi: 10.1095/biolreprod.105.040527. [DOI] [PubMed] [Google Scholar]
- 20.Basquin J, et al. Architecture of the Nuclease Module of the Yeast CCR4–NOT Complex: the Not1-Caf1-Ccr4 Interaction. Molecular cell. 2012 doi: 10.1016/j.molcel.2012.08.014. [DOI] [PubMed] [Google Scholar]
- 21.Petit AP, et al. The structural basis for the interaction between the CAF1 nuclease and the NOT1 scaffold of the human CCR4–NOT deadenylase complex. Nucleic acids research. 2012 doi: 10.1093/nar/gks883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chekulaeva M, et al. miRNA repression involves GW182-mediated recruitment of CCR4–NOT through conserved W-containing motifs. Nature structural & molecular biology. 2011;18:1218–26. doi: 10.1038/nsmb.2166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chrestensen CA, et al. MAPKAP kinase 2 phosphorylates tristetraprolin on in vivo sites including Ser178, a site required for 14-3-3 binding. The Journal of biological chemistry. 2004;279:10176–84. doi: 10.1074/jbc.M310486200. [DOI] [PubMed] [Google Scholar]
- 24.Stoecklin G, et al. MK2-induced tristetraprolin:14-3-3 complexes prevent stress granule association and ARE-mRNA decay. The EMBO journal. 2004;23:1313–24. doi: 10.1038/sj.emboj.7600163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Marchese FP, et al. MAPKAP kinase 2 blocks tristetraprolin-directed mRNA decay by inhibiting CAF1 deadenylase recruitment. The Journal of biological chemistry. 2010;285:27590–600. doi: 10.1074/jbc.M110.136473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Clement SL, Scheckel C, Stoecklin G, Lykke-Andersen J. Phosphorylation of tristetraprolin by MK2 impairs AU-rich element mRNA decay by preventing deadenylase recruitment. Molecular and cellular biology. 2011;31:256–66. doi: 10.1128/MCB.00717-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nguyen MN, Tan KP, Madhusudhan MS. CLICK--topology-independent comparison of biomolecular 3D structures. Nucleic acids research. 2011;39:W24–8. doi: 10.1093/nar/gkr393. [DOI] [PMC free article] [PubMed] [Google Scholar]
ONLINE METHODS REFERENCES
- 1.Mossessova E, Lima CD. Ulp1-SUMO crystal structure and genetic analysis reveal conserved interactions and a regulatory element essential for cell growth in yeast. Molecular cell. 2000;5:865–76. doi: 10.1016/s1097-2765(00)80326-3. [DOI] [PubMed] [Google Scholar]
- 2.Jinek M, Fabian MR, Coyle SM, Sonenberg N, Doudna JA. Structural insights into the human GW182-PABC interaction in microRNA-mediated deadenylation. Nature structural & molecular biology. 2010;17:238–40. doi: 10.1038/nsmb.1768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blackshear PJ, et al. Zfp36l3, a rodent X chromosome gene encoding a placenta-specific member of the Tristetraprolin family of CCCH tandem zinc finger proteins. Biol Reprod. 2005;73:297–307. doi: 10.1095/biolreprod.105.040527. [DOI] [PubMed] [Google Scholar]
- 4.Lai WS, et al. Evidence that tristetraprolin binds to AU-rich elements and promotes the deadenylation and destabilization of tumor necrosis factor alpha mRNA. Molecular and cellular biology. 1999;19:4311–23. doi: 10.1128/mcb.19.6.4311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Minor W, Cymborowski M, Otwinowski Z, Chruszcz M. HKL-3000: the integration of data reduction and structure solution--from diffraction images to an initial model in minutes. Acta crystallographica. Section D, Biological crystallography. 2006;62:859–66. doi: 10.1107/S0907444906019949. [DOI] [PubMed] [Google Scholar]
- 6.Schneider TR, Sheldrick GM. Substructure solution with SHELXD. Acta crystallographica. Section D, Biological crystallography. 2002;58:1772–9. doi: 10.1107/s0907444902011678. [DOI] [PubMed] [Google Scholar]
- 7.Uson I, Stevenson CE, Lawson DM, Sheldrick GM. Structure determination of the O-methyltransferase NovP using the ‘free lunch algorithm’ as implemented in SHELXE. Acta crystallographica. Section D, Biological crystallography. 2007;63:1069–74. doi: 10.1107/S0907444907042230. [DOI] [PubMed] [Google Scholar]
- 8.Langer G, Cohen SX, Lamzin VS, Perrakis A. Automated macromolecular model building for X-ray crystallography using ARP/wARP version 7. Nature protocols. 2008;3:1171–9. doi: 10.1038/nprot.2008.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Adams PD, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta crystallographica. Section D, Biological crystallography. 2010;66:213–21. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fabian MR, et al. miRNA-mediated deadenylation is orchestrated by GW182 through two conserved motifs that interact with CCR4–NOT. Nature structural & molecular biology. 2011;18:1211–7. doi: 10.1038/nsmb.2149. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.