Abstract
De novo mutations (DNMs) are important in Autism Spectrum Disorder (ASD), but so far analyses have mainly been on the ~1.5% of the genome encoding genes. Here, we performed whole genome sequencing (WGS) of 200 ASD parent-child trios and characterized germline and somatic DNMs. We confirmed that the majority of germline DNMs (75.6%) originated from the father, and these increased significantly with paternal age only (p=4.2×10−10). However, when clustered DNMs (those within 20kb) were found in ASD, not only did they mostly originate from the mother (p=7.7×10−13), but they could also be found adjacent to de novo copy number variations (CNVs) where the mutation rate was significantly elevated (p=2.4×10−24). By comparing DNMs detected in controls, we found a significant enrichment of predicted damaging DNMs in ASD cases (p=8.0×10−9; OR=1.84), of which 15.6% (p=4.3×10−3) and 22.5% (p=7.0×10−5) were in the non-coding or genic non-coding, respectively. The non-coding elements most enriched for DNM were untranslated regions of genes, boundaries involved in exon-skipping and DNase I hypersensitive regions. Using microarrays and a novel outlier detection test, we also found aberrant methylation profiles in 2/185 (1.1%) of ASD cases. These same individuals carried independently identified DNMs in the ASD risk- and epigenetic- genes DNMT3A and ADNP. Our data begins to characterize different genome-wide DNMs, and highlight the contribution of non-coding variants, to the etiology of ASD.
Introduction
Autism Spectrum Disorder (ASD), a neurobehavioral condition characterized by atypical development of social-communication, and the presence of restrictive interests and repetitive behaviors, can have a genetic basis1. ASD exhibits extensive clinical and genetic heterogeneity with high heritability2 and recurrence risk3, and males are affected more often than girls (~4:1)4. Copy number variations (CNVs)5,6, insertion-deletions (indels)7,8, single nucleotide mutations6,9 have implicated >100 ASD susceptibility genes5,6,10 of variable penetrance and expressivity, some of which are making their way into clinical genetic testing11,12, but most of which are still to be defined10. Functionally, ASD risk genes often converge in pathways that modulate synaptic transmission, chromatin remodeling and transcriptional regulation5,9. Common genetic variants may also contribute to ASD13.
With over a decade of experience in genomic studies of ASD, the approach of searching for de novo mutations (DNMs) continually emerges as an effective method to initially sort through increasingly more complex datasets14–17. Due to previous technology limitations in resolution and cost, the vast majority of studies have interrogated the small (~1.5%) gene-coding segments of the genome. In a recent study, penetrant DNMs in genes were estimated to contribute to ASD in ~11% of parent-child trios (simplex) families6. Even our own research using whole genome sequencing (WGS)7,18 focused only on annotating genes, since sample sizes were insufficient to discern statistically relevant data from the larger non-coding regions (~98.5% of the genome).
Here, we developed new approaches to characterize de novo mutations from WGS data, with an emphasis on determining their origin and functional impact on non-coding DNA in ASD. Our most compelling data found that clustered DNMs in ASD mostly originated from the mother, and are often found adjacent to de novo CNVs. In addition, we found that coding and non-coding de novo point mutations in ASD are enriched in genes that are responsible for synaptic, translational and chromatin remodeling function. We have also demonstrated that these DNMs may have deleterious effects on the epigenetic profiles of individuals with ASD. Somatic mutations potentially relevant to ASD were also detectable in the WGS data.
Results
Detection of genome-wide de novo mutations
We performed WGS in 200 unrelated idiopathic ASD trio families (600 individuals) using the Illumina Hiseq 2000 technology. The families were selected based on the fact that the index case (proband) was the only individual in the family affected with ASD. Subjects met criteria for ASD based on the Autism Diagnostic Interview-Revised (ADI-R), the Autism Diagnostic Observation Schedule-Generic (ADOS) plus clinical evaluation. All probands were genotyped for CNVs using high-resolution microarrays (Supplementary information).
Of the 200 probands, genomic DNA was obtained for 192, four and four subjects from whole blood, lymphocyte cell-line (LCL), and leucocytes, respectively. The average coverage relative to the hg19 reference sequence (non-N bases) was 99.7% or 32x (Supplementary Table 1). Using an improved DNM detection approach7, we identified 9,774 germline DNMs. This represents 50.9 de novo single nucleotide variants (SNVs), 3.9 de novo indels and 0.052 de novo CNVs (defined as unbalanced changes >10kb) per genome, and their validation rates were 95.7% (377 of 396), 100% (21 of 21) and 62.5% (10 of 16), respectively (Supplementary Figure 1; Supplementary Table 2 and 3). In the exonic regions, there were 0.99 de novo SNVs, 0.1 de novo indels and 0.03 de novo CNVs (Supplementary Table 2 and 3) per individual. We found an unusually high number of DNMs in four of the LCL samples (Supplementary Table 2), consistent with previous observations18,19. We also found a shift of the allelic fraction (alternate reads over total reads) supporting the variant towards the lower end (Supplementary Figure 2), confirming that most of the DNMs were cell-line derived mutations of a mosaic nature18. These 8 samples (including the four from leucocytes) were therefore removed from our analysis.
Origin of de novo mutations
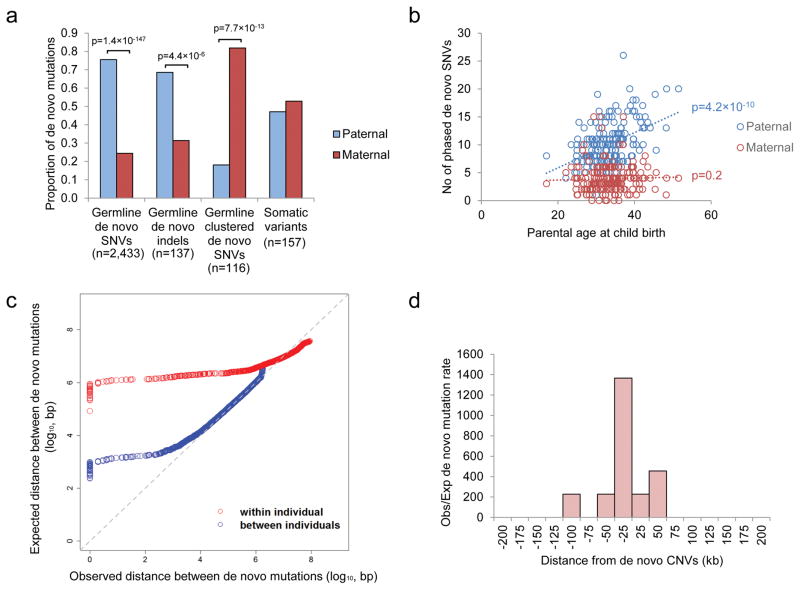
We performed phasing to determine the chromosome of origin of the DNM (Supplementary information) and determined that 75.6% of the de novo SNVs and 68.6% of the de novo indels originated from the father (Figure 2a). Consistent with previous reports7,20, the number of germline DNMs was found to increase with paternal age (Pearson correlation test, r=0.4; p=4.2×10−10; Figure 2b), which is mostly attributed to the higher number of replication events in the older paternal gamete20. However, we found no correlation between the number of de novo SNVs on the maternal allele and the maternal age, suggesting few de novo mutations were accumulated throughout life in female. The number of phased de novo indels was insufficient for robust statistical analysis, but we could demonstrate the total aggregate number of de novo indels was more significantly correlated with paternal rather than maternal age (Poisson regression beta coefficient based on Student’s t distribution, p=6.4×10−3 for paternal age and p=0.74 for maternal age; Supplementary Figure 3).
Figure 2.
Origins of de novo mutations in ASD. (a) Parent-of-origin of germline and somatic variants. Number of germline de novo SNVs and de novo indels derived from the father were significantly higher than that from the mother. On the other hand, there are significantly more clustered (within 20kb) germline de novo mutations originated from the mother than from the father, while somatic mutations can be found in similar proportion from both parents. (b) Number of de novo SNVs found on the paternal allele increases with the age of father, but there is no correlation between the number of de novo SNVs found on the maternal allele and the age of the mother. (c) Distance between de novo mutations is shorter than expected for a subset of de novo mutations both between and within individuals. (d) Mutation rate is significantly higher than the background within 100kb flanking the de novo CNVs.
We also found a substantial portion of DNMs clustered (two or more mutations occurring within a 20kb segment) in the same individual (239 DNMs in Supplementary Table 6) (Figure 2c). This phenomenon has been described previously in Dutch population controls21, and similarly we found that clustered DNMs have different sequence signatures than non-clustered ones (Supplementary Figure 4)21. Remarkably, 43.9% of them (105 out of 239 DNMs) clustered within 200bp (Supplementary Table 6). One such cluster of DNMs was found in a known ASD-risk gene, SYNGAP15,6,9; two de novo events were identified in the coding region of the gene in ASD case 3-0438-000. These mutations result in a 12bp to 7bp substitution that removes a core splice site of the exon (Supplementary Figure 5).
Contrary to what was observed in the Dutch population controls where fathers contribute a majority of clustered DNMs (Supplementary Figure 6), in our ASD families, we found that the majority of the clustered DNMs originated on the maternal lineage (Fisher’s exact test, p=7.7×10−13; Figure 2a). We also validated this finding upon re-analysis of our previously reported ASD WGS data (Supplementary Figure 6)18. In search of an explanation, we found that mutation rates have been reported to be increased near CNVs22. Indeed, we found that the DNMs near the 10 de novo CNVs (+/− 100kb) found in our sample are significantly higher than the expected genome background (Binominal test, p=2.4×10−24; Figure 2d). This involved 11 DNMs (7 of the 11 DNMs were clustered DNMs described above) in 5 de novo CNVs, and they were all separated over 1kb (Supplementary Table 7). Interestingly, there is a significant reduction of maternal contribution in DNMs separated >200bp (68% maternal) than those separated <200bp (88% maternal) (Fisher’s exact test, p=0.01). No significant difference was found for the origin of de novo CNVs (or rare-inherited) from the parents5 (Supplementary Table 7), so it is unlikely that maternal enrichment of clustered DNMs can be explained due to a higher de novo rate of CNV from the mother. Instead, it may be caused by sex-based differences on DNA repair mechanisms during gametogenesis23 (Supplementary Table 7). Also, the fact that not all of the clustered de novo point mutations were found in de novo CNVs may be partially due to the false negative rate of CNV detection from current WGS technology18,24.
Somatic mutations
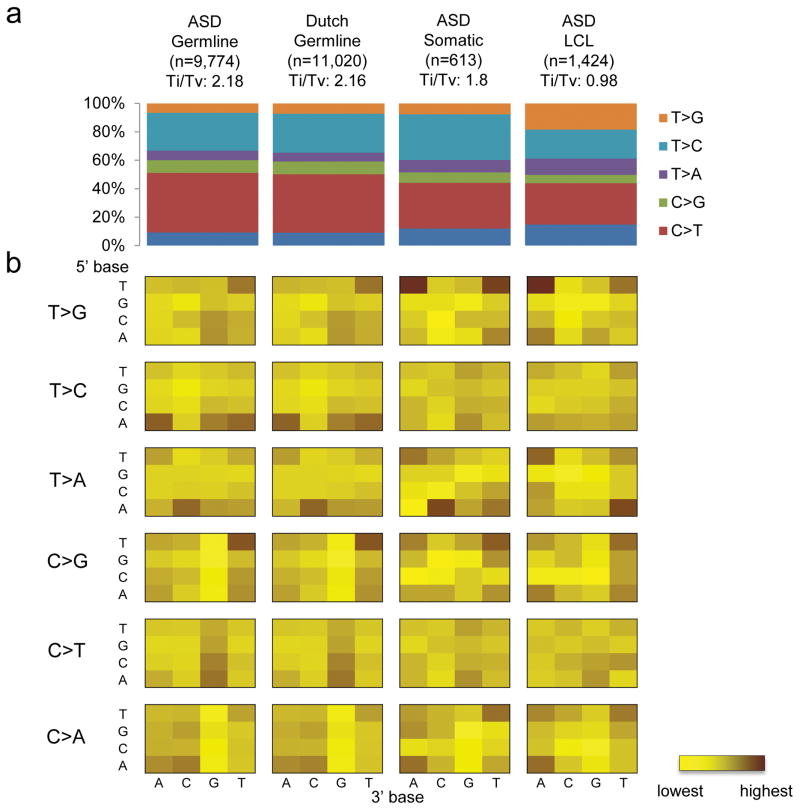
Among the DNMs, we found that there is a substantial portion of variants with a lower allelic fraction (33%, 2 S.D. from the mean). We compared the sequence context of the DNMs with less than 33% allelic fraction to the rest of the variants (Supplementary Figure 7). We found that their sequence context is similar to that of the LCL derived variants (Figure 1), suggesting that they may be generated by a similar mechanism. Therefore, most of these DNMs are likely to be somatic in origin, which is supported by the fact they were found almost equally from both maternal and paternal alleles (Figure 2a), and differ from what is seen in the constitutional genome. These correspond to 3.19 somatic mutations per genome and 0.036 per exome (Supplementary Table 2). At least one of these somatic mutations affects the NRXN1, a known ASD known ASD-risk gene17,25 (Supplementary Figure 8; Supplementary Table 3). Although the status of these mutations in the brain of carrier individuals would not be known, the relatively high allelic representation (16%) suggest they arose early in post-zygotic development and therefore may be extensively represented in cells throughout the body and therefore have phenotypic consequence.
Figure 1.
Sequence context of regions with de novo mutations. (a) Transition (Ti) to transversion (Tv) ratio of different kinds of de novo mutations found in: germline of ASD, germline of Dutch population controls, somatic events of ASD and lymphocyte derived cell line (LCL) of ASD. (b) Sequence context of the base substitution mutation spectra for different de novo mutations. Each of the 96 mutated trinucleotides (mutated position at center) from each cohort is represented in a heatmap (intensity of color correspond to frequency of each mutation). The 5′ base to the mutated site is shown on the vertical axis, while the 3′ base is shown on the horizontal axis.
Functional characteristics of de novo mutations
To assess the potential functional effect of the DNMs identified in the ASD cohort, we compared them with the DNMs detected in a Dutch control population, in which the genomes of 258 parent-child trios (250 families) were sequenced with the same platform21,26. These samples were collected without ascertaining on the basis of disease26. We compared only the autosomal de novo germline SNVs from the ASD data because they were the only DNMs that were reported from that control population. While there is a difference in the sequence depth between our cohort (32×) and the control cohort (13.3×), we found that the sequence context associated with the DNMs were similar between the two (Pearson correlation test, p=5.8×10−61; Figure 1), which is not observed when using different sequencing platforms or DNM detection methods (Supplementary Figure 9). This observation suggests that there is no significant sequencing or detection bias between the cases and controls in this study. The high validation rate of our DNM detected (95.7%) is also comparable to that of the controls (94.6% specificity). The difference in sequence coverage, however, can lead to variant detectability in regions with extreme GC content. Indeed, we found a minor difference in GC content between our cases and controls (Supplementary Figure 10). Therefore, we used a logistic regression test with GC content as a covariate to correct for this potential confounding effect (see Methods).
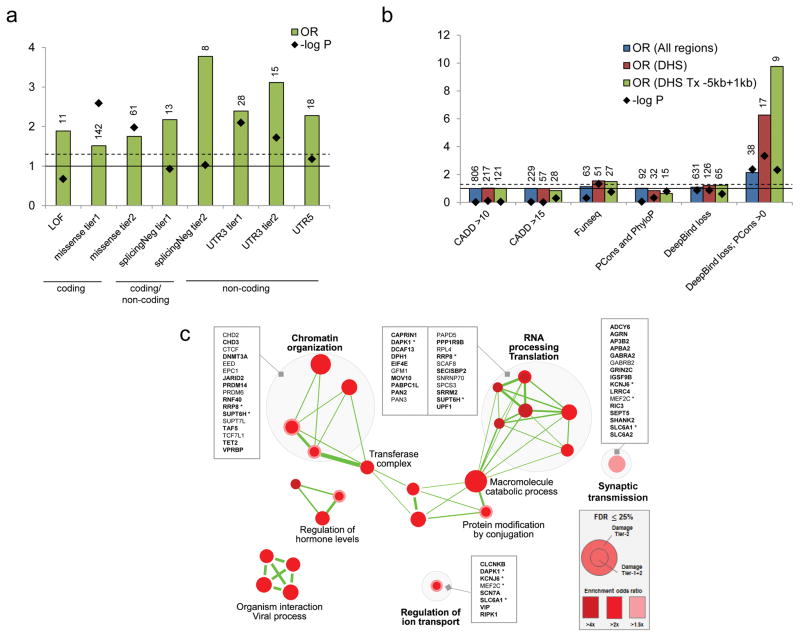
Comparing the 9,774 germline DNMs from our ASD cohort (192 trios) with 11,020 DNMs from the Dutch control cohort at different genomic regions, we found that the DNM rate is higher at the 5′ untranslated region (5′-UTR) and the coding exons in ASD (Figure 3a). We further examined the in silico predicted effects of the DNMs. While loss-of-function (LOF) DNMs have a higher odds ratio compared to the control sample, they are not significantly enriched because of the small number of LOF mutations involved (Figure 3; Supplementary Table 8). On the other hand, we found a significant enrichment of de novo missense mutations in the ASD sample compared to controls (Figure 3a). This was not previously observed in the simplex proband-sibling comparison27. Perhaps some of the supposedly unaffected siblings in families with ASD children were in fact at risk of ASD or other developmental phenotypes15, a phenomenon that we have found previously7,18.
Figure 3.
Functional impact of genome-wide damaging de novo mutations. (a) Damaging de novo mutations are significantly enriched in both coding (missense) and non-coding regions (splicingNeg, UTR3 and UTR5) in ASD compare to population controls. Definition of damaging tiers can be found in the Methods. LOF: loss of function mutations; missense: missense mutations; splicingNeg: exon-skipping mutations predicted by SPANR; UTR: untranslated regions. Number of variants is indicated above each bar. Solid horizontal line indicates OR=1, and dash horizontal line represents p=0.05. (b) Non-coding de novo mutations in non-genic regions are significantly enriched in DNase I hypersensitive regions (DHS). Damaging de novo mutations predicted by “Deepbind loss; PCons >0” are significantly enriched in ASD in general (All regions), but further enriched in DNase I hypersensitive regions and regions proximal to genes. DHS: DNase I hypersensitive sites; Tx: transcript; PCons: PhastCons. Number of variants is indicated above each bar. Solid horizontal line indicates OR=1, and dash horizontal line represents p=0.05. (c) Damaging de novo mutations are significantly enriched (FDR ≤0.25) in Gene Ontology defined pathways that are related to chromatin organization, RNA processing and translation, synaptic transmission, and others. Genes involved in the pathways are listed. Genes with DNMs in coding region are bolded. Asterisk represents gene that is found in more than one gene-pathway.
Beyond the coding region, we found that, in addition to the 5′-UTR, the 3′ untranslated region (3′-UTR) was significantly enriched with DNMs when we restricted our analysis to the conserved regions (Figure 3a). Although there is some enrichment of DNMs at the conserved long non-coding RNA, the difference did not reach the statistical significance (Supplementary Table 8). We have also applied a variant effect prediction tool we developed, SPANR28, to annotate the effect of the variants at predicted splice sites (both exonic and intronic regions). We also found that the variants predicted with an exon-skipping effect (splicingNeg)28 represent the highest significant enrichment of variants in ASD compared to the controls (Figure 3a), while no significant enrichment was found in predicted benign missense (OR=1.2; p=0.27), synonymous (OR=1.19; p=0.65) and intronic (excluding predicted damaging. OR=1; p=0.98) DNMs (Supplementary Table 8). For variants in the non-genic regions, we applied four different prediction tools and examined the burden of these ASD variants in different chromatin states from ENCODE29 and Epigenomic Roadmap30 (see Methods and Supplementary Table 9). We found that DNMs were significantly predicted to lead to loss of transcriptional binding factors (Figure 3b). They were enriched in DNase I hypersensitive regions and proximal to genes. For example, a loss of KDM5B binding was found at the promoter of a candidate autism-risk gene, EFR3A (Supplementary Figure 11). Comparing 71 different human primary cell types or tissues, the effect of transcriptional binding factor loss was enriched in quiescent states of different brain regions (Supplementary Figure 12). Selecting a set of brain-specific enhancers without applying prediction algorithms, we also found a trend of enrichment in cases (OR=1.7, p=0.07). Taking together, putative non-coding DNMs that were significantly enriched in ASD represent 38% (93 out of 244) of the variants considered to be damaging (Table 1; Supplementary Table 10).
Table 1.
Summary of de novo SNVs contribution*
| ASD (n=192) | Control (n=258) | odds ratio (p) | ||
|---|---|---|---|---|
|
|
||||
| Germline | All | 9,774a | 11,020 | - |
| Coding | 193 (1.98%) | 136 (1.23%) | 1.38 (5.0×10−3) | |
|
| ||||
| Predicted damaging | All | 244 | 141 | 1.84 (8.0×10−9) |
| Coding | 151 (61.9%) | 97 (68.8%) | 1.53 (1.3×10−3)b | |
| Genic non-coding | 55 (22.5%) | 23 (16.3%) | 2.59 (7.0×10−5)b | |
| Non-genic non-coding | 38 (15.6%) | 21 (14.9%) | 2.14 (4.3×10−3)c | |
Comparison was based on a logistic regression model with GC content correction (see Methods);
Somatic mutations (n=613) were removed.
All exonic and intronic variants as the universe;
All non-exonic variants as the universe.
To evaluate the functional relevance of the predicted damaging DNMs, we compared the mutation burden between the ASD cases and the controls in the gene-sets previously shown to be involved in ASD5,18. Since it is still challenging to elucidate the target genes for linked noncoding variants in the non-genic regions, we focused our analyses on the DNMs found in gene-encompassing regions (exonic and intronic regions; 206 DNMs in total). Consistent with previous findings5,9, we found that the predicted damaging DNMs in ASD samples have a significantly higher mutation burden in genes that are expressed in the brain, are FMRP targets, and other genes that are known to be involved in neurodevelopmental or behavioral phenotypes (Supplementary Figure 13).
To identify novel gene pathways that were enriched in the genes disrupted by the DNMs, we tested the mutation burden in all the gene-sets listed in the Gene Ontology. We found a significant enrichment of variants in pathways involved in “Chromatin Organization”, “RNA Processing Translation” and “Synaptic Transmission” among others (Figure 3c), which is largely consistent with the previous findings5,9. These included many genes that are known to be involved in ASD, for example, SHANK2, EIF4E and DAPK1, further supporting the critical role of these pathways in ASD.
Importantly, we applied our previously developed tools to identify damaging non-coding variants. We showed that these predicted damaging non-coding variants were enriched in the splicing, 5′ and 3′-UTR, and together contribute 22.5% of the potential damaging DNMs examined (Table 1). Indeed, from the gene sets that were enriched with the pathways mentioned above, 29% (16 out of 56) of the genic variants involved were non-coding (Figure 3c), supporting the hypothesis in ASD that damaging non-coding variants may affect gene function in a manner similar to coding variants. We estimated that the damaging DNMs in genic regions (including coding and non-coding variants) contribute ~45% of the ASD cases in simplex families, which is largely consistent with that previously estimated27.
Mutations altering epigenetic profiles
Given that DNMs in ASD can affect chromatin organization, we performed DNA methylation profiling using Illumina Infinium array of 185 probands for which whole blood DNA was available to assess for epigenetic aberrations that might be mapped to the genomic sequence. Since mutations in ASD are highly heterogeneous, we speculated that samples having extreme epigenetic aberration would be rare. Therefore, we sought to identify samples with “rare methylation signatures”31 by detecting outliers from the overall DNA methylation pattern. To capture this effect, we developed a new approach called Methylation Outlier Sample Test (MOST)(see Methods).
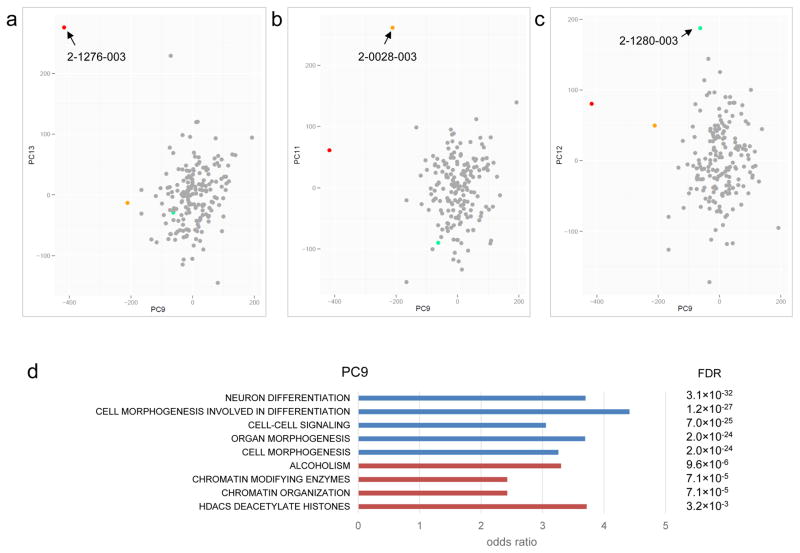
After normalization and the removal of problematic probe array data, we performed principal component (PC) analysis on the samples. We generated up to 20 PCs and used the Grubbs test for the detection of outliers (see Methods). For each PC, we also adjusted for covariates such as gender, ethnicity, age, blood cell composition, batch effects and array chip orders32. After correcting for covariates, we identified three significant outlier samples (from 185, 1.6%) from five different PCs (Supplementary Table 11): 2-0028-003 was identified from 3 PCs, 2-1276-003 was identified from 2 PCs, and 2-1280-003 was identified from 1 PC (Figure 4). Interestingly, 2-0028-003 carries a de novo damaging missense mutation at DNMT3A, a gene involved in de novo DNA methylation33, which is also a risk factor for ASD7,34. The other outlier, 2-1276-003, carries a de novo frameshift deletion at ADNP, known to be involved in chromatin remodeling35 and ASD-risk36. Both 2-0028-003 and 2-1276-003 were outliers in PC9, which captured genes enriched for function in neuron differentiation, cell morphogenesis and chromatin organization (Figure 3c and Figure 4c). The third outlier, 2-1280-003, did not carry a detectable predicted damaging DNM in a gene related to epigenetic regulation, but instead a maternally inherited mutation predicted to be damaging was detected in KMT5C, a gene function as a histone methyltransferase37. It is not clear if this inherited mutation in KMT5C would lead to the aberrant DNA methylation profile, but the PC data may guide additional genetic or functional testing (see Supplementary Figure 14).
Figure 4.
Sample outliers identified by the Methylation Outlier Sample Test (MOST). (a) Sample 2-1276-003, carrying a de novo damaging heterozygous mutation at DNMT3A (p.R635W), was identified as an outlier sample in principle component (PC) 9 and 13. (b) Sample 2-0028-003, which carries a de novo frameshift mutation at ADNP (p.Q345fs), was identified as an outlier sample in PC 11 and 14. (c) Sample 2-1280-003 was identified as an outlier sample in PC 12. No de novo mutation in known epigene was found, but there is a maternal inherited rare damaging missense mutation at KMT5C (p.R205Q). (d) Functional enrichment of genes involved in the PC9 responsible for the sample outliers. Functions from negative loadings are in blue and that from positive loadings are in red.
Discussion
We have conducted a comprehensive analysis of the distribution of DNM across the entire genome in ASD cases and controls and discovered a cadre of new germline rare genetic variants of relevance to ASD. Lower-resolution microarray and targeted sequencing studies have implicated rare mutations in non-coding genes like PTCHD1AS138,39, 5′-UTR of MBD540, introns of NRXN141, and more complex regulatory structural variants39,42, but here our unbiased WGS assessment of germline mutations implicate numerous functional elements involved in regulating gene expression and chromatin organization (Figure 3 and Figure 4). We also found that a proportion (1.1%) of DNMs previously thought to be germline in origin7 were in fact likely somatic events. So far, there are no genome-wide somatic mutation profiles in controls that we can compare our data against, but our findings of somatic rates in ASD are comparable to a study of intellectual disability43. Moreover, using targeted genes, it has recently been shown that there is an excess rate of somatic mutation rate found in the coding regions of ASD probands compared to their unaffected siblings44.
Our most surprising observation was the clustering of germline DNMs arising on the maternal chromosome. We hypothesize that the generation of a de novo CNVs might disturb DNA repair22, and this entire process may be influenced in a sex-dependent manner both in gametogenesis23 and ultimately in post-natal phenotypic expression5. While the overall contribution of this novel mechanism of mutation to ASD needs to be determined through much larger studies, we did find one ASD case (3-0438-000) having two de novo events impacting the coding region of the known ASD-risk gene, SYNGAP1 (Supplementary Table 5). The finding of only one such example from 192 cases studied (0.5%), suggests like all other known genetic mechanism involved in ASD is rare, but it could increase when the mutational impact on the non-coding genome is better understood. Our data also suggest, in the clinical genetics setting characterizing de novo CNVs alone may be insufficient when attempting to understand a full genotype and phenotype correlation sequencing near the breakpoints or better yet WGS may be required12,42,45. Applying other improved WGS technologies, such as long-read and linked-read sequencing, for SV identification implied that our previous knowledge of SVs was rather primitive. Our understanding of the genetics of ASD may further improve as these methods start to be widely used46–48.
Given the increasingly appreciated importance of chromatin remodeling function in the pathology of ASD5,6, we established a general method to connect aberrant methylation profiles detected by microarrays to DNMs in WGS data (which can also act as a functional evaluation of the DNMs). Here, we found DNMs directly affecting the coding regions of two (of 185 or 1.1%) known genes, DNMT3A34 and ADNP36, which control the epigenetic cascade. This same approach should be equally amenable to implicate non-coding regulatory elements, and downstream target regions or genes for a particular epigene, as well as, to confirm the damaging effects of mutations. Moreover, environmental influences on the epigenome in ASD biospecimens could be monitored using this strategy.
Our study provides a framework of how to use WGS in the study of ASD. The early data arising lends further support for a multifactorial threshold model underlying ASD49–51 with all types of variation (SNV/indel/CNV in coding and non-coding DNA, germ-line, somatic, epigenetic) involved. Here, we focused on studying the impact of DNMs as an entry point into WGS data, but similar studies of rare and common-inherited genetic variants52, as well as non-genetic factors, will now need to be assessed in larger cohorts in order to quantitate relative risks for ASD.
Methods
Samples for whole-genome sequencing
We selected 200 unrelated trio families from a cohort of Canadian ASD families, based on the fact that the index case (proband) was the only affected individual in the family at the time of proband’s diagnosis (simplex families). Diagnosis was based on using the ADI-R, ADOS plus clinical evaluation7. We also considered the availability of genomic DNA from whole blood and completeness of phenotype information. We obtained informed consent from all participants, as approved by the Research Ethics Boards at The Hospital for Sick Children, McMaster University and Memorial Hospital. We genotyped all the samples using high-resolution microarray platforms for the detection of CNVs.
Whole-genome sequencing and variant detection
We sequenced trio families (two parents and one proband). We extracted genomic DNA from all samples and sequenced them with Illumina Hiseq2000 technology (Illumina, San Diego, CA). We ligated the purified DNA fragments with adaptor oligonucleotides to form pair-end DNA libraries with an insert size of 500bp. Sequencing depth and coverage for each sample is summarized in Supplementary Table 1. We aligned the filtered reads to the reference genome (build GRCh37) with the Burrows-Wheeler Aligner as a sorted binary format (BAM). We performed local realignment and quality recalibration with the Genome Analysis Toolkit for each genome. Details of the procedure can be found in Supplementary Information.
De novo SNV and indel detection
We considered the variants in the proband to be a candidate de novo SNV if it was not present at the same position in both of his/her parents. We used ForestDNM method detect de novo SNV calls and filtering method for indels in all trios as previously described7. We validated all the exonic and a subset of non-coding de novo SNVs and indels using Sanger sequencing. Details can be found in Supplementary Information.
De novo CNV and detection
We used Segseq53 and ERDS54 to detect potential de novo CNVs. We also used Meerkat55 to detect potential de novo SVs. Details of the procedures can be found in Supplementary Information. We validated all the detected putative de novo CNV and SV by quantitative-PCR (qPCR) and/or Sanger sequencing.
Functional annotation of de novo mutations
We annotated the vcf using a custom pipeline based on Annovar (November 2014 version)56. We defined genes from RefSeq gene models (hg19 genome build; downloaded from UCSC 2013 February 12). We annotated the genomic conservation at the variant position using UCSC PhyloP and phastCons for placental mammals and 100 vertebrates57.
For the functional impact of genic variants, we used predictors including SIFT58, PolyPhen259, Mutation Assessor60, Mutation Taster61 and CADD62. We also expanded the annotation of non-coding regulatory sequence through implementation of splicing exon inclusion/exclusion predictions28.
We created filtering tiers and annotated each variant based on conservation and predicted impact on coding and non-coding sequence. Damaging tier 1 defined as having odds ratio higher than 1.5. Damaging tier 2 is defined as variants having odds ratio higher than 2.5 (except LOF) (Supplementary Table 8).
Damaging tier 1 genic variants include: 1) all LOF (stop gain + core splice site) variants; 2) all the missense (including stoploss) variants; 3) splicing (both intronic and exonic, excluding stop-gain) negative variants, as predicted by SpliceDx with dPSI<-3.5; 4) all 5′ UTR variants and 5) 3′ UTR variants with PhastCons>0.
Damaging tier 2 genic variants include: 1) all LOF (stop gain + core splice site) variants; 2) missense variants with at least 5 out of 7 predictive programs meeting damaging criteria: PhyloPMam ≥2.30, phyloVert100 ≥4.0, SIFT <0.05, Polyphen2 ≥0.90, Mutation Assessor ≥1.9, Mutation Taster ≥0.5, CADD phred ≥15; 3) splicing (both intronic and exonic, excluding stop-gain) negative variants, as predicted by SpliceDx with dPSI<−5; 4) all 5′ UTR variants; 5) 3′ UTR variants with PhastCons>0 and PhylopP>=1.5.
For non-genic variants, we annotated the vcf by overlapping the DNase I hypersensitive sites and chromatin states extracted from FANTOM enhancers63 and enhancers in developing fetal brain64, ENCODE29 and Epigenomic Roadmap30. Details of tracks extracted can be found in Supplementary Table 12. For the functional impact of variants, we used predictors including CADD, DeepBind65, FunSeq66 and conservation score (PhastCons and PhyloP).
We annotated each variant based on the overlap of chromatin states, conservation and predicted impact on the non-coding sequence. We assigned damaging tiers based on their high burden in ASD cohort (294 combinations; FDR<0.2). Damaging tier 1 defined as having odds ratio higher than 1.5. Damaging tier 2 is defined as variants having odds ratio higher than 2 (Supplementary Table 9).
Damaging tier 1 non-genic variants include:
DeepBind loss; PCons >0 – DeepBind predicted loss of transcriptional binding factor (wild type reference binding score >= 99.9% percentile of genome background distribution and variant binding score <= 99% percentile of genome background distribution, additionally requiring placental mammal, PhastCons>0).
Damaging tier 2 non-genic variants include:
DeepBind loss; PCons >0 (DHS) – DeepBind predicted loss of transcriptional binding factor in DNase I hypersensitive regions (ENCODE).
GC content correction
Coverage difference is known to affect the number of variants detected (sensitivity), in which we have adjusted it from our statistics (using total overall variants detected). We have also showed that there is no bias on the underlying sequence context. The non-linear regional variability is known to affect regions in extreme GC content and repetitive regions. Both our data and the control data have the repetitive regions masked for de novo variant detection using Machine-learning. For GC content, we indeed found minor but statistically significant difference between ours and the control data. By comparing the GC content flanking the de novo SNVs (50bp, 200bp, 500bp) between our cases and the controls, we found a minor but statistical significant difference in GC content (with 50bp having the highest bias) (Supplementary Figure 10). Therefore, we used a logistic regression test and used the 50bp flanking GC content as a covariate to correct for the confounding effect:
where
x1 = GC content
x2 = membership to variant set (based on impact prediction and region-of-interest)
c = intercept
Burden and pathway analysis of annotated de novo mutations
We performed logistic regression test to determine the significance of higher burden of de novo mutations in cases than controls67. We used all de novo mutations in exonic coding, UTR and intronic regions as the universe for genic variant comparison. We used all de novo mutations, except exonic and predicted splicing mutations, as the universe for non-genic variant comparison. We only counted once if a variant appeared more than once in different annotated categories (to avoid double-count for variants with multiple annotations).
For curated ASD-related gene-sets, we used a Fisher’s Exact Test on the contingency table defined by the case and control variants groups intersected by the damaging and not-damaging variant groups (self-contained). For Gene-Ontology Function gene-sets, we used the same approach by including gene-sets with gene number between 50 and 1200. The rows where all intersections were equal to 0 were removed (gene-sets that do not satisfy this condition are not considered to be interesting). We extracted the Gene-Ontology gene sets from the National Cancer Institute at the US National Institutes of Health (NCI-NIH). We computed the FDR using the Benjamini-Hochberg procedure.
DNA methylation array
We bisulfite-converted the genomic DNA from 185 samples using the EpiTect PLUS Bisulfite Kit (Qiagen, CA, USA) according to the manufacturer’s instructions. We eliminated probes: 1) on the sex chromosomes, 2) containing SNPs, and 3) with detection P values >0.05 in any of the samples from the study. We performed background subtraction using the ‘noob’ method from Methylumi68 and normalization by ‘SWAN’69. Details of the procedures can be found in the Supplementary Information.
Methylation Outlier Sample Test
We performed principal component (PC) analysis using the R stats package function procomp (with scaling and centering). We generated 20 PCs from the DNA methylation beta values, the same was repeated on a randomized matrix to determine 20 sets of eigenvalues. We analyzed the top 14 PCs, which have eigenvalues higher the maximum of each randomized eigenvalue (2637.107). The corrected and uncorrected PC values for the samples follow a normal distribution (Supplementary Figure 15). We then performed Grubbs test for outlier detection. For each PC, we adjusted for covariates including gender, ethnicity, age, blood cell composition, batch effects and array chip orders. We estimated the blood cell composition based on the beta values using celltypes450 (version 0.1) from R70. We consider a sample being an outlier when it has a Grubbs test FDR<0.1 both before and after covariate correction.
Supplementary Material
Acknowledgments
We thank the families for their participation in the study, BGI-Shenzhen and The Centre for Applied Genomics analytical and technical support. This work was funded by Autism Speaks, Autism Speaks Canada, the Canadian Institutes for Advanced Research, the University of Toronto McLaughlin Centre, Genome Canada/Ontario Genomics Institute, the government of Ontario, the Canadian Institutes of Health Research (CIHR), and The Hospital for Sick Children Foundation. R.K.C.Y. holds CIHR Postdoctoral Fellowship, NARSAD Young Investigator award and Thrasher Early Career Award. K.T. holds a fellowship from the Swedish Research Council. M.U. holds the Banting Postdoctoral Fellowship. P.S. holds the Patsy and Jamie Anderson Chair in Child and Youth Mental Health. S.W.S. holds the GlaxoSmithKline-CIHR Chair in Genome Sciences at the University of Toronto and The Hospital for Sick Children.
Footnotes
Contributions
R.K.C.Y., D.M. and S.W.S. conceived and designed the experiments. R.K.C.Y., D.M., H.C., B.T., X. T., Y. S., D.C., T.Z., X.W., X.J., Z.Z., X.L., T.N. processed and analyzed the whole genome sequencing data. S.W., K.T., A.C. and L.D. designed and performed experiments for variants characterization and validation. G.P., B.A., Z.W., J.R.M., E.D., M.T.S., M.U., M.Z., M.B., J. B., S.S.G., D.L. and C.R.M. helped perform different components of analysis and validation experiments. R.K.C.Y., H.C., J.L.H., M.W., Y.L., J.W., J.W., H.Y., X.X., S.W.S. coordinated the whole genome sequencing experiments. R.K.C.Y., D.M., M.P., R.H.R. and S.W.S. conceived and coordinated the project. E.A., L.Z., R.W., B.A.F., W.R., P.S. recruited, diagnosed and examined the recruited participants. R.K.C.Y. and S.W.S. wrote the manuscript.
Competing Interests
The authors declare no conflict of interest.
Availability of data and materials
The sequence data can be accessed through the MSSNG database on Google Genomics (for access see https://research.mss.ng).
References
- 1.Anagnostou E, Zwaigenbaum L, Szatmari P, Fombonne E, Fernandez BA, Woodbury-Smith M, et al. Autism spectrum disorder: advances in evidence-based practice. CMAJ. 2014;186:509–519. doi: 10.1503/cmaj.121756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Colvert E, Tick B, McEwen F, Stewart C, Curran SR, Woodhouse E, et al. Heritability of Autism Spectrum Disorder in a UK Population-Based Twin Sample. JAMA Psychiatry. 2015;72:415–423. doi: 10.1001/jamapsychiatry.2014.3028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ozonoff S, Young GS, Carter A, Messinger D, Yirmiya N, Zwaigenbaum L, et al. Recurrence risk for autism spectrum disorders: a Baby Siblings Research Consortium study. Pediatrics. 2011;128:e488–495. doi: 10.1542/peds.2010-2825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fombonne E. Epidemiology of pervasive developmental disorders. Pediatr Res. 2009;65:591–598. doi: 10.1203/PDR.0b013e31819e7203. [DOI] [PubMed] [Google Scholar]
- 5.Pinto D, Delaby E, Merico D, Barbosa M, Merikangas A, Klei L, et al. Convergence of genes and cellular pathways dysregulated in autism spectrum disorders. American journal of human genetics. 2014;94:677–694. doi: 10.1016/j.ajhg.2014.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sanders SJ, He X, Willsey AJ, Ercan-Sencicek AG, Samocha KE, Cicek AE, et al. Insights into Autism Spectrum Disorder Genomic Architecture and Biology from 71 Risk Loci. Neuron. 2015;87:1215–1233. doi: 10.1016/j.neuron.2015.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jiang YH, Yuen RK, Jin X, Wang M, Chen N, Wu X, et al. Detection of Clinically Relevant Genetic Variants in Autism Spectrum Disorder by Whole-Genome Sequencing. American journal of human genetics. 2013;93:249–263. doi: 10.1016/j.ajhg.2013.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dong S, Walker MF, Carriero NJ, DiCola M, Willsey AJ, Ye AY, et al. De novo insertions and deletions of predominantly paternal origin are associated with autism spectrum disorder. Cell Rep. 2014;9:16–23. doi: 10.1016/j.celrep.2014.08.068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.De Rubeis S, He X, Goldberg AP, Poultney CS, Samocha K, Cicek AE, et al. Synaptic, transcriptional and chromatin genes disrupted in autism. Nature. 2014;515:209–215. doi: 10.1038/nature13772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Buxbaum JD, Daly MJ, Devlin B, Lehner T, Roeder K, State MW, et al. The autism sequencing consortium: large-scale, high-throughput sequencing in autism spectrum disorders. Neuron. 2012;76:1052–1056. doi: 10.1016/j.neuron.2012.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Carter MT, Scherer SW. Autism spectrum disorder in the genetics clinic: a review. Clin Genet. 2013;83:399–407. doi: 10.1111/cge.12101. [DOI] [PubMed] [Google Scholar]
- 12.Tammimies K, Marshall CR, Walker S, Kaur G, Thiruvahindrapuram B, Lionel AC, et al. Molecular Diagnostic Yield of Chromosomal Microarray Analysis and Whole-Exome Sequencing in Children With Autism Spectrum Disorder. Jama. 2015;314:895–903. doi: 10.1001/jama.2015.10078. [DOI] [PubMed] [Google Scholar]
- 13.Anney R, Klei L, Pinto D, Almeida J, Bacchelli E, Baird G, et al. Individual common variants exert weak effects on the risk for autism spectrum disorderspi. Hum Mol Genet. 2012;21:4781–4792. doi: 10.1093/hmg/dds301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sebat J, Lakshmi B, Malhotra D, Troge J, Lese-Martin C, Walsh T, et al. Strong association of de novo copy number mutations with autism. Science. 2007;316:445–449. doi: 10.1126/science.1138659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ronemus M, Iossifov I, Levy D, Wigler M. The role of de novo mutations in the genetics of autism spectrum disorders. Nat Rev Genet. 2014;15:133–141. doi: 10.1038/nrg3585. [DOI] [PubMed] [Google Scholar]
- 16.Uddin M, Tammimies K, Pellecchia G, Alipanahi B, Hu P, Wang Z, et al. Brain-expressed exons under purifying selection are enriched for de novo mutations in autism spectrum disorder. Nat Genet. 2014;46:742–747. doi: 10.1038/ng.2980. [DOI] [PubMed] [Google Scholar]
- 17.Autism Genome Project C. Szatmari P, Paterson AD, Zwaigenbaum L, Roberts W, Brian J, et al. Mapping autism risk loci using genetic linkage and chromosomal rearrangements. Nat Genet. 2007;39:319–328. doi: 10.1038/ng1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yuen RK, Thiruvahindrapuram B, Merico D, Walker S, Tammimies K, Hoang N, et al. Whole-genome sequencing of quartet families with autism spectrum disorder. Nat Med. 2015;21:185–191. doi: 10.1038/nm.3792. [DOI] [PubMed] [Google Scholar]
- 19.Awadalla P, Gauthier J, Myers RA, Casals F, Hamdan FF, Griffing AR, et al. Direct measure of the de novo mutation rate in autism and schizophrenia cohorts. American journal of human genetics. 2010;87:316–324. doi: 10.1016/j.ajhg.2010.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kong A, Frigge ML, Masson G, Besenbacher S, Sulem P, Magnusson G, et al. Rate of de novo mutations and the importance of father’s age to disease risk. Nature. 2012;488:471–475. doi: 10.1038/nature11396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Francioli LC, Polak PP, Koren A, Menelaou A, Chun S, Renkens I, et al. Genome-wide patterns and properties of de novo mutations in humans. Nat Genet. 2015;47:822–826. doi: 10.1038/ng.3292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Carvalho CM, Pehlivan D, Ramocki MB, Fang P, Alleva B, Franco LM, et al. Replicative mechanisms for CNV formation are error prone. Nat Genet. 2013;45:1319–1326. doi: 10.1038/ng.2768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Baarends WM, van der Laan R, Grootegoed JA. DNA repair mechanisms and gametogenesis. Reproduction. 2001;121:31–39. doi: 10.1530/rep.0.1210031. [DOI] [PubMed] [Google Scholar]
- 24.Pang AW, Macdonald JR, Yuen RK, Hayes VM, Scherer SW. Performance of High-Throughput Sequencing for the Discovery of Genetic Variation Across the Complete Size Spectrum. G3 (Bethesda) 2014;4:63–65. doi: 10.1534/g3.113.008797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kim HG, Kishikawa S, Higgins AW, Seong IS, Donovan DJ, Shen Y, et al. Disruption of neurexin 1 associated with autism spectrum disorder. American journal of human genetics. 2008;82:199–207. doi: 10.1016/j.ajhg.2007.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Genome of the Netherlands C. Whole-genome sequence variation, population structure and demographic history of the Dutch population. Nat Genet. 2014;46:818–825. doi: 10.1038/ng.3021. [DOI] [PubMed] [Google Scholar]
- 27.Iossifov I, O’Roak BJ, Sanders SJ, Ronemus M, Krumm N, Levy D, et al. The contribution of de novo coding mutations to autism spectrum disorder. Nature. 2014;515:216–221. doi: 10.1038/nature13908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xiong HY, Alipanahi B, Lee LJ, Bretschneider H, Merico D, Yuen RK, et al. RNA splicing. The human splicing code reveals new insights into the genetic determinants of disease. Science. 2015;347:1254806. doi: 10.1126/science.1254806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Consortium EP. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Roadmap Epigenomics C. Kundaje A, Meuleman W, Ernst J, Bilenky M, Yen A, et al. Integrative analysis of 111 reference human epigenomes. Nature. 2015;518:317–330. doi: 10.1038/nature14248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Choufani S, Cytrynbaum C, Chung BH, Turinsky AL, Grafodatskaya D, Chen YA, et al. NSD1 mutations generate a genome-wide DNA methylation signature. Nat Commun. 2015;6:10207. doi: 10.1038/ncomms10207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Harper KN, Peters BA, Gamble MV. Batch effects and pathway analysis: two potential perils in cancer studies involving DNA methylation array analysis. Cancer Epidemiol Biomarkers Prev. 2013;22:1052–1060. doi: 10.1158/1055-9965.EPI-13-0114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Okano M, Bell DW, Haber DA, Li E. DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell. 1999;99:247–257. doi: 10.1016/s0092-8674(00)81656-6. [DOI] [PubMed] [Google Scholar]
- 34.Tatton-Brown K, Seal S, Ruark E, Harmer J, Ramsay E, Del Vecchio Duarte S, et al. Mutations in the DNA methyltransferase gene DNMT3A cause an overgrowth syndrome with intellectual disability. Nat Genet. 2014;46:385–388. doi: 10.1038/ng.2917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mandel S, Gozes I. Activity-dependent neuroprotective protein constitutes a novel element in the SWI/SNF chromatin remodeling complex. J Biol Chem. 2007;282:34448–34456. doi: 10.1074/jbc.M704756200. [DOI] [PubMed] [Google Scholar]
- 36.Helsmoortel C, Vulto-van Silfhout AT, Coe BP, Vandeweyer G, Rooms L, van den Ende J, et al. A SWI/SNF-related autism syndrome caused by de novo mutations in ADNP. Nat Genet. 2014;46:380–384. doi: 10.1038/ng.2899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schotta G, Lachner M, Sarma K, Ebert A, Sengupta R, Reuter G, et al. A silencing pathway to induce H3-K9 and H4-K20 trimethylation at constitutive heterochromatin. Genes Dev. 2004;18:1251–1262. doi: 10.1101/gad.300704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Noor A, Whibley A, Marshall CR, Gianakopoulos PJ, Piton A, Carson AR, et al. Disruption at the PTCHD1 Locus on Xp22.11 in Autism spectrum disorder and intellectual disability. Sci Transl Med. 2010;2:49ra68. doi: 10.1126/scitranslmed.3001267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Marshall CR, Noor A, Vincent JB, Lionel AC, Feuk L, Skaug J, et al. Structural variation of chromosomes in autism spectrum disorder. American journal of human genetics. 2008;82:477–488. doi: 10.1016/j.ajhg.2007.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hodge JC, Mitchell E, Pillalamarri V, Toler TL, Bartel F, Kearney HM, et al. Disruption of MBD5 contributes to a spectrum of psychopathology and neurodevelopmental abnormalities. Mol Psychiatry. 2014;19:368–379. doi: 10.1038/mp.2013.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Duong LT, Hoeffding LK, Petersen KB, Knudsen CD, Thygesen JH, Klitten LL, et al. Two rare deletions upstream of the NRXN1 gene (2p16.3) affecting the non-coding mRNA AK127244 segregate with diverse psychopathological phenotypes in a family. Eur J Med Genet. 2015;58:650–653. doi: 10.1016/j.ejmg.2015.11.004. [DOI] [PubMed] [Google Scholar]
- 42.Talkowski ME, Rosenfeld JA, Blumenthal I, Pillalamarri V, Chiang C, Heilbut A, et al. Sequencing chromosomal abnormalities reveals neurodevelopmental loci that confer risk across diagnostic boundaries. Cell. 2012;149:525–537. doi: 10.1016/j.cell.2012.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Campbell IM, Yuan B, Robberecht C, Pfundt R, Szafranski P, McEntagart ME, et al. Parental somatic mosaicism is underrecognized and influences recurrence risk of genomic disorders. American journal of human genetics. 2014;95:173–182. doi: 10.1016/j.ajhg.2014.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.D’Gama AM, Pochareddy S, Li M, Jamuar SS, Reiff RE, Lam AT, et al. Targeted DNA Sequencing from Autism Spectrum Disorder Brains Implicates Multiple Genetic Mechanisms. Neuron. 2015;88:910–917. doi: 10.1016/j.neuron.2015.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Stavropoulos DJ, Merico D, Jobling R, Meyn MS, Bowdin S, Monfared N, et al. Whole Genome Sequencing Expands Diagnostic Utility and Improves Clinical Management in Paediatric Medicine. npj Genomic Medicine. 2016;1:15012. doi: 10.1038/npjgenmed.2015.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zheng GX, Lau BT, Schnall-Levin M, Jarosz M, Bell JM, Hindson CM, et al. Haplotyping germline and cancer genomes with high-throughput linked-read sequencing. Nat Biotechnol. 2016;34:303–311. doi: 10.1038/nbt.3432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.English AC, Salerno WJ, Hampton OA, Gonzaga-Jauregui C, Ambreth S, Ritter DI, et al. Assessing structural variation in a personal genome-towards a human reference diploid genome. BMC Genomics. 2015;16:286. doi: 10.1186/s12864-015-1479-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Noll AC, Miller NA, Smith LD, Yoo B, Fiedler S, Cooley LD, et al. Clinical detection of deletion structural variants in whole genome sequences. npj Genomic Medicine. doi: 10.1038/npjgenmed.2016.26. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bailey A, Phillips W, Rutter M. Autism: towards an integration of clinical, genetic, neuropsychological, and neurobiological perspectives. J Child Psychol Psychiatry. 1996;37:89–126. doi: 10.1111/j.1469-7610.1996.tb01381.x. [DOI] [PubMed] [Google Scholar]
- 50.Bourgeron T. From the genetic architecture to synaptic plasticity in autism spectrum disorder. Nat Rev Neurosci. 2015;16:551–563. doi: 10.1038/nrn3992. [DOI] [PubMed] [Google Scholar]
- 51.Devlin B, Scherer SW. Genetic architecture in autism spectrum disorder. Current opinion in genetics & development. 2012;22:229–237. doi: 10.1016/j.gde.2012.03.002. [DOI] [PubMed] [Google Scholar]
- 52.He X, Sanders SJ, Liu L, De Rubeis S, Lim ET, Sutcliffe JS, et al. Integrated model of de novo and inherited genetic variants yields greater power to identify risk genes. PLoS Genet. 2013;9:e1003671. doi: 10.1371/journal.pgen.1003671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chiang DY, Getz G, Jaffe DB, O’Kelly MJ, Zhao X, Carter SL, et al. High-resolution mapping of copy-number alterations with massively parallel sequencing. Nat Methods. 2009;6:99–103. doi: 10.1038/nmeth.1276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhu M, Need AC, Han Y, Ge D, Maia JM, Zhu Q, et al. Using ERDS to infer copy-number variants in high-coverage genomes. American journal of human genetics. 2012;91:408–421. doi: 10.1016/j.ajhg.2012.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yang L, Luquette LJ, Gehlenborg N, Xi R, Haseley PS, Hsieh CH, et al. Diverse mechanisms of somatic structural variations in human cancer genomes. Cell. 2013;153:919–929. doi: 10.1016/j.cell.2013.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38:e164. doi: 10.1093/nar/gkq603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pollard KS, Hubisz MJ, Rosenbloom KR, Siepel A. Detection of nonneutral substitution rates on mammalian phylogenies. Genome Res. 2010;20:110–121. doi: 10.1101/gr.097857.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ng PC, Henikoff S. Predicting deleterious amino acid substitutions. Genome Res. 2001;11:863–874. doi: 10.1101/gr.176601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Adzhubei IA, Schmidt S, Peshkin L, Ramensky VE, Gerasimova A, Bork P, et al. A method and server for predicting damaging missense mutations. Nat Methods. 2010;7:248–249. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Reva B, Antipin Y, Sander C. Predicting the functional impact of protein mutations: application to cancer genomics. Nucleic Acids Res. 2011;39:e118. doi: 10.1093/nar/gkr407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Schwarz JM, Rodelsperger C, Schuelke M, Seelow D. MutationTaster evaluates disease-causing potential of sequence alterations. Nat Methods. 2010;7:575–576. doi: 10.1038/nmeth0810-575. [DOI] [PubMed] [Google Scholar]
- 62.Kircher M, Witten DM, Jain P, O’Roak BJ, Cooper GM, Shendure J. A general framework for estimating the relative pathogenicity of human genetic variants. Nat Genet. 2014;46:310–315. doi: 10.1038/ng.2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Andersson R, Gebhard C, Miguel-Escalada I, Hoof I, Bornholdt J, Boyd M, et al. An atlas of active enhancers across human cell types and tissues. Nature. 2014;507:455–461. doi: 10.1038/nature12787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Visel A, Taher L, Girgis H, May D, Golonzhka O, Hoch RV, et al. A high-resolution enhancer atlas of the developing telencephalon. Cell. 2013;152:895–908. doi: 10.1016/j.cell.2012.12.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Alipanahi B, Delong A, Weirauch MT, Frey BJ. Predicting the sequence specificities of DNA- and RNA-binding proteins by deep learning. Nat Biotechnol. 2015;33:831–838. doi: 10.1038/nbt.3300. [DOI] [PubMed] [Google Scholar]
- 66.Khurana E, Fu Y, Colonna V, Mu XJ, Kang HM, Lappalainen T, et al. Integrative annotation of variants from 1092 humans: application to cancer genomics. Science. 2013;342:1235587. doi: 10.1126/science.1235587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Merico D, Isserlin R, Stueker O, Emili A, Bader GD. Enrichment map: a network-based method for gene-set enrichment visualization and interpretation. PLoS One. 2010;5:e13984. doi: 10.1371/journal.pone.0013984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Triche TJ, Jr, Weisenberger DJ, Van Den Berg D, Laird PW, Siegmund KD. Low-level processing of Illumina Infinium DNA Methylation BeadArrays. Nucleic Acids Res. 2013;41:e90. doi: 10.1093/nar/gkt090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Maksimovic J, Gordon L, Oshlack A. SWAN: Subset-quantile within array normalization for illumina infinium HumanMethylation450 BeadChips. Genome Biol. 2012;13:R44. doi: 10.1186/gb-2012-13-6-r44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Houseman EA, Accomando WP, Koestler DC, Christensen BC, Marsit CJ, Nelson HH, et al. DNA methylation arrays as surrogate measures of cell mixture distribution. BMC Bioinformatics. 2012;13:86. doi: 10.1186/1471-2105-13-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.