Diffuse large B cell lymphoma (DLBCL) is an aggressive non-Hodgkin lymphoma that is molecularly and clinically heterogeneous. Gene expression studies have revealed how DLBCL can be divided into germinal center (GC) and activated B cell (ABC) subtypes. The ABC subtype is associated with constitutive activation of the NF-kB pathway, commonly as a consequence of genetic activation of the B cell receptor (BCR) pathway1. Components of the BCR pathway that are activated by mutation include CD79B, MYD88 and CARD11. Chronic stimulation of the BCR in ABC DLBCL may also result from engagement of the BCR by self-antigens in the tumor microenvironment. These preclinical observations suggest a role for the targeted inhibitors of the BCR pathway in the treatment of DLBCL1.
DLBCL most commonly presents as malignant infiltration of lymph nodes. However, extranodal disease is seen in one third of cases. Some forms of DLBCL have an apparent tissue-specific restriction that allows them to be defined as specific disease subtypes. These “special site” forms of DLBCL include primary central nervous system lymphoma (PCNSL), primary testicular lymphoma (PTL) and primary cutaneous DLBCL. For reasons that remain mysterious, primary cutaneous DLBCL most commonly involves the lower leg and Primary Cutaneous DLBCL-leg type (PCDLBCL-LT) is recognized as a specific entity in the WHO classification of lymphoma2. It predominantly affects older patients and is associated with poor response to conventional therapy3. The vast majority of PCDLBCL-LT exhibit an ABC DLBCL gene expression profile and are strongly enriched for genetic activation of the BCR pathway, suggesting a pathogenesis shared with nodal ABC DLBCL4, 5.
Drugs that target the BCR pathway include inhibitors of the downstream kinases BTK, SYK and P110delta. Inhibitors of BTK have led to dramatic responses in patients with chronic lymphocytic leukemia (CLL) and mantle cell lymphoma. The use of BTK inhibitors is now incorporated into standard therapy for these conditions and the genetic basis of acquired resistance in CLL, dominated by acquired mutation of BTK or PLCG2, has been well-studied6, 7. In contrast, the role of BCR inhibition in DLBCL remains less clear and little is known about the genetics of acquired resistance.
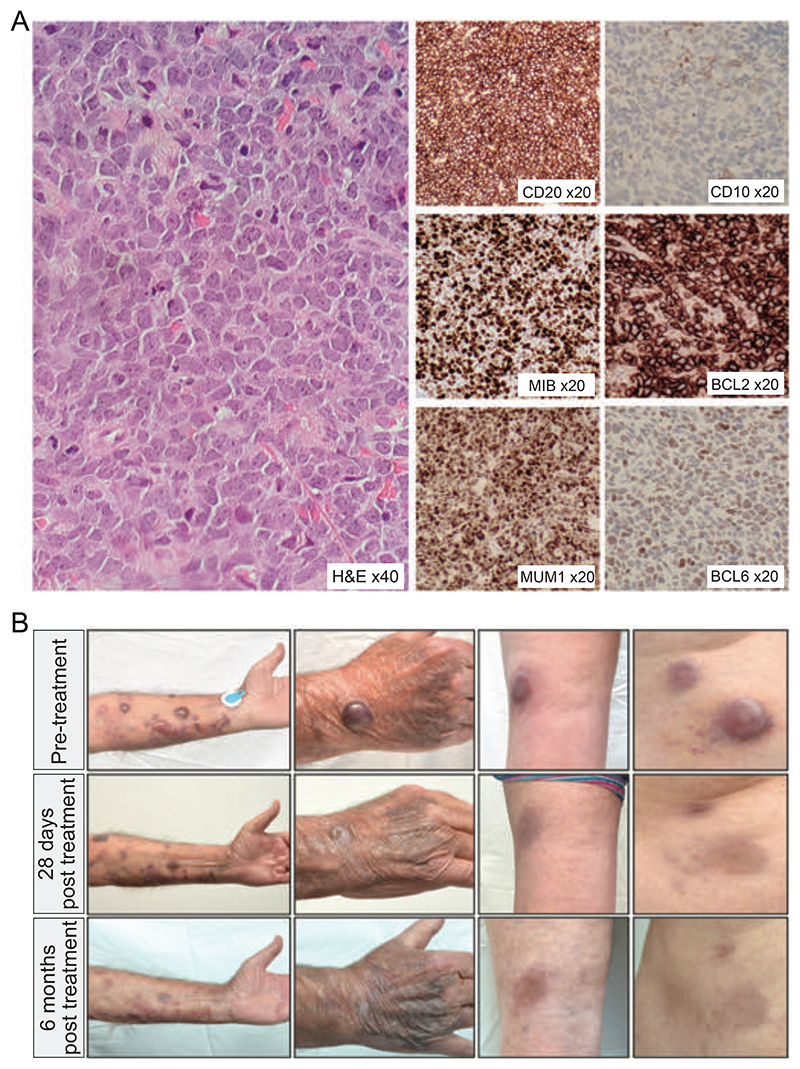
A 75 year old male presented with rapidly enlarging cutaneous nodules. These initially involved the feet and legs, but subsequently emerged on the arms and abdomen. A CT scan showed no lymph node enlargement. Biopsy revealed sheets of large lymphoid cells expressing PAX5, CD20, BCL2, MUM1 and weak BCL6. CD10 staining was negative. The proliferation fraction was 90%. A diagnosis of primary cutaneous DLBCL was made. Because of comorbidity, he was initially treated with rituximab, gemcitabine, cyclophosphamide, vincristine and prednisolone with partial response. At relapse two years later, he was retreated with rituximab and gemcitabine, and subsequently oral etoposide with minimal response to either therapy. He was referred to our tertiary lymphoma service for further management. Clinical examination revealed cutaneous nodules up to 25mm in diameter affecting the feet, legs, arms and abdomen. A repeat biopsy showed features identical to his original diagnostic biopsy (Figure 1A) and CT scan confirmed exclusively cutaneous disease. He was commenced on oral therapy with BTK and SYK inhibitors. By day 28, he had a near complete resolution of his cutaneous lesions, which further improved by 6 months (Figure 1B). With ongoing therapy, his remission lasted for thirteen months before the recurrence of rapidly growing cutaneous nodules and lymph node involvement. A repeat biopsy confirmed recurrent DLBCL with immunophenotype identical to his diagnostic biopsy. The patient was managed with palliative radiotherapy.
Figure 1.
(A) Histology and immunohistochemistry from tumor biopsy showing a diffuse infiltration by large atypical cells with predominantly centroblast-like morphology expressing CD20, MUM1, BCL2, weak BCL6, no CD10 and MIB1 proliferation faction 90% (B) Images of selected lesions on the forearm, hand, popliteal fossa and abdomen taken prior to therapy (top row), at 28 days (middle row) and at 6 months (bottom row) of BCR-targeted therapy.
The rapid clinical response and prolonged remission, followed by later reemergence of tumor suggested the acquisition of new genetic alterations driving resistance to BCR inhibition. We therefore performed whole-exome sequencing using DNA extracted from biopsies at initial diagnosis, immediately prior to BCR inhibition and at relapse. The patient provided written informed consent for genetic analysis and publication of clinical photographs. The study was approved by the East of England Cambridge South Research Ethics Committee (approval reference number 07/MRE05/44).
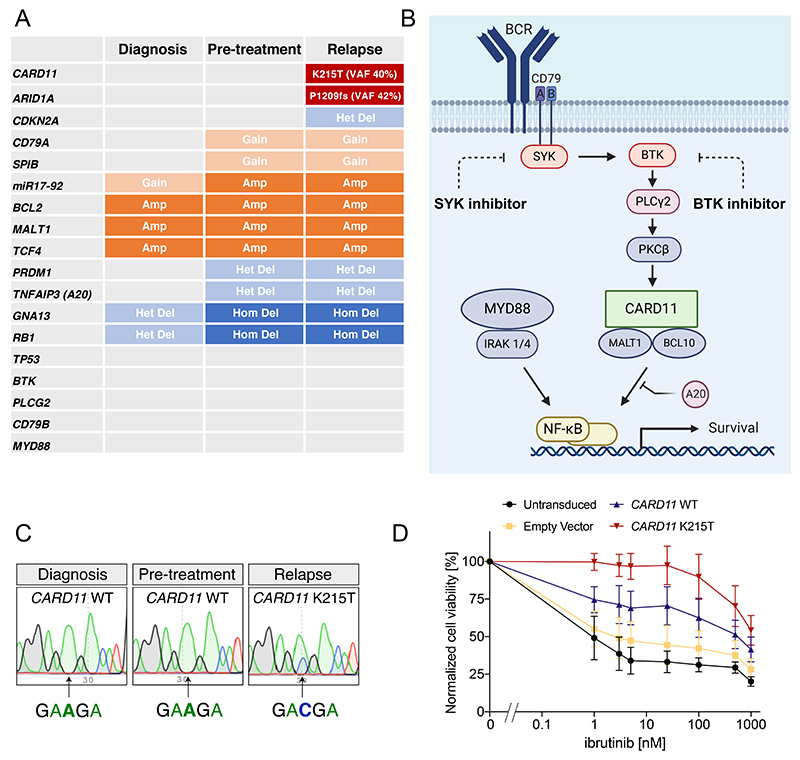
Full variant and copy number data are presented in Supplementary Data 1&2. Notable genetic alterations detected prior to BCR inhibition included amplification of Chr18q21 (including BCL2, MALT1 and TCF4), amplification of Chr13q31 (including miR17-92), and homozygous deletions of GNA13 and RB1 (Figure 2A and Supplementary Figure 1) We observed gains of CD79A and SPIB and single copy loss of PRDM1 and TNFAIP3 (A20). We did not identify mutation of MYD88 or CD79B (Figure 2A & Supplementary Figure 2). Importantly, many of the identified genetic alterations are predicted to enhance activity of the BCR and NF-kB pathways1 (Figure 2B). These data suggest that this patient’s tumor matched the biology of ABC DLBCL, and relied on chronic active BCR signaling to maintain NF-kB activity.
Figure 2.
Summary of whole exome sequencing of samples taken from three different timepoints; diagnosis, prior to BCR-targeted therapy and at the time of relapse. Mutations and copy number changes are indicated within each cell. Grey indicates no mutation/copy number change identified. (B) A simplified schematic showing the critical signaling components of the BCR pathway that converge onto activation of canonical NFKB. Components targeted pharmacologically in this patient are indicated. Created with BioRender.com. (C) Sanger sequencing from samples at the indicated timepoints confirm the acquisition of an A to C mutation in CARD11, which results in the activating, coiled-coil domain K215T mutation. (D) Ibrutinib sensitivity assay in the ABC DLBCL line U2932 transduced with either empty vector or CARD11 WT or K215T. Data show mean and SEM of 5 independent experiments.
We then screened all genetic alterations acquired during BCR inhibition to identify those that might drive resistance. We identified 34 protein-altering mutations that arose during exposure to BCR inhibition, including the known DLBCL driver genes – ARID1A and CARD11 (Figure 2A&B & Supplementary Data 1). Copy number analysis showed a new single copy loss of the CDKN2A locus (Supplementary Data 2). We manually examined the loci of PLCG2 and BTK, mutations which are commonly acquired in BTKi-resistant CLL, and confirmed both genes to be wild type at all timepoints. Alteration of TP53 was not detected. The majority of altered genes lacked any known association with BCR signaling. CDKN2A loss is common in ABC DLBCL, however we considered it unlikely that single copy loss could mediate resistance to BCR inhibition. In contrast, CARD11 is a critical scaffold protein which, together with BCL10 and MALT1, functions downstream of SYK and BTK in the BCR pathway to activate canonical NF-kB signaling1. Activating mutation in the coiled-coil domain of CARD11 was previously proposed as a mechanism of resistance to BCR inhibition in mantle cell lymphoma8 and primary resistance in DLBCL9. The acquired K215T mutation in our patient is located in the coiled-coil domain and has been previously shown to activate NF-kB10. We confirmed the near-clonal presence of the CARD11 K215T mutation at relapse (VAF 40%) but zero mutant reads at this position in either of the two pretreatment biopsies (read depth 180 and 210, Supplementary Figure 3). Sanger sequencing verified the acquisition of the activating CARD11 K215T mutation in the relapse sample but not at diagnosis or in the pre-treatment sample (Figure 2C). To establish the ability of this mutation to mediate resistance, we expressed CARD11 WT or K215T in the ABC DLBCL line U2932 and treated cells with the BTK inhibitor ibrutinib. CARD11 K215T induced drug resistance, with IC50 increased by three orders of magnitude (Figure 2D). Taken together, these data strongly suggest that CARD11 mutation was the genetic driver of the acquired resistance in this case.
The limited information about acquired resistance to BCR inhibition in DLBCL suggests potential differences compared to that of CLL. The latter is predominantly associated with mutation of BTK and PLCG26,7. CARD11 mutation is identified in 10-15% of DLBCL at presentation11 but is rarely identified (<1%) in CLL12. This suggests that the BCR signal in CLL and DLBCL may be qualitatively different and therefore that genetic mechanisms of resistance to BCR inhibition may also differ. However, establishing the genetic basis of resistance to BCR inhibition in DLBCL presents specific challenges that contrast with the situation in CLL. Firstly, DLBCL patients receiving BCR inhibitors are typically treated simultaneously with multiagent immunochemotherapy regimens making it hard to establish which mutations provide resistance to which agent, or indeed if sensitivity to the BCR inhibitor ever existed in the first place. Secondly, relapsing DLBCL frequently presents as a rapidly progressive disease that may preclude the opportunity for a repeat biopsy in patients who are often treated palliatively. These factors make analysis of rare, exceptional responders especially important for DLBCL. The genetics of our case recapitulate the biology of ABC DLBCL and importantly, we observed clear initial sensitivity to therapy targeted exclusively to the BCR pathway. Combined with the biopsy-accessible cutaneous location of disease this provided a rare opportunity to study genetic mechanisms of resistance to BCR inhibition in ABC DLBCL.
Indeed, we have found only one other case describing the genetic basis of acquired resistance to BCR inhibition in DLBCL. This also involved a case of PCDLBCL-LT and also reported acquisition of CARD11 mutation (interesting also K215 mutant) in a patient who relapsed following an initial response to a BTK inhibitor13. However, their findings of NFKBIE mutation and IgH-IRF8 translocation may also have contributed to resistance, leaving the role of the CARD11 mutation uncertain. Our finding of acquired activating CARD11 mutation, the absence of any likely alternative genetic explanation and the demonstrated ability of the K215T mutation to effect BTKi resistance in vitro strongly suggest that CARD11 mutation is the dominant driver of BCR independence in our case.
As we move towards the introduction of precision medicine in DLBCL and the real time monitoring of clonal evolution, it will become increasingly important to understand genetic mechanisms of drug resistance. Our study uses the unique features of this case of cutaneous DLBCL to highlight the importance of CARD11 mutation as a driver of acquired resistance to BCR inhibition in DLBCL. We speculate that such cases might be targeted successfully in the future by inhibition of downstream targets such as MALT1 or NF-kB14, 15.
Supplementary Material
Acknowledgements
D.H. was supported by a Clinician Scientist Fellowship from the Medical Research Council (MR/M008584/1) and a project grant from the Kay Kendall Leukaemia Fund. The Hodson laboratory receives core funding from Wellcome and MRC to the Wellcome-MRC Cambridge Stem Cell Institute and from the CRUK Cambridge Cancer Centre.
Footnotes
Author Contributions
RC designed and performed experiments. JM analyzed whole exome sequencing data. JG performed drug resistance experiments. IW analyzed sequencing data. NS provided clinical expertise. LRB provided histopathology expertise. SA analyzed and discussed data. DH designed experiments, provided clinical expertise, directed the research and wrote the manuscript with contributions from RC and IW.
Conflicts of Interest
RC - Consultancy for Karus Therapeutics. DH - Research funding from Gilead Sciences. The remaining authors declare no competing interests.
References
- 1.Young RM, Shaffer AL, 3rd, Phelan JD, Staudt LM. B-cell receptor signaling in diffuse large B-cell lymphoma. Seminars in hematology. 2015 Apr;52(2):77–85. doi: 10.1053/j.seminhematol.2015.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Swerdlow SH, Campo E, Pileri SA, Harris NL, Stein H, Siebert R, et al. The 2016 revision of the World Health Organization classification of lymphoid neoplasms. Blood. 2016 May 19;127(20):2375–2390. doi: 10.1182/blood-2016-01-643569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Grange F, Beylot-Barry M, Courville P, Maubec E, Bagot M, Vergier B, et al. Primary cutaneous diffuse large B-cell lymphoma, leg type: clinicopathologic features and prognostic analysis in 60 cases. Archives of dermatology. 2007 Sep;143(9):1144–1150. doi: 10.1001/archderm.143.9.1144. [DOI] [PubMed] [Google Scholar]
- 4.Mareschal S, Pham-Ledard A, Viailly PJ, Dubois S, Bertrand P, Maingonnat C, et al. Identification of Somatic Mutations in Primary Cutaneous Diffuse Large B-Cell Lymphoma, Leg Type by Massive Parallel Sequencing. The Journal of investigative dermatology. 2017 Sep;137(9):1984–1994. doi: 10.1016/j.jid.2017.04.010. [DOI] [PubMed] [Google Scholar]
- 5.Pham-Ledard A, Prochazkova-Carlotti M, Andrique L, Cappellen D, Vergier B, Martinez F, et al. Multiple genetic alterations in primary cutaneous large B-cell lymphoma, leg type support a common lymphomagenesis with activated B-cell-like diffuse large B-cell lymphoma. Modern pathology : an official journal of the United States and Canadian Academy of Pathology, Inc. 2014 Mar;27(3):402–411. doi: 10.1038/modpathol.2013.156. [DOI] [PubMed] [Google Scholar]
- 6.Woyach JA, Ruppert AS, Guinn D, Lehman A, Blachly JS, Lozanski A, et al. BTK(C481S)-Mediated Resistance to Ibrutinib in Chronic Lymphocytic Leukemia. J Clin Oncol. 2017 May 1;35(13):1437–1443. doi: 10.1200/JCO.2016.70.2282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ahn IE, Underbayev C, Albitar A, Herman SE, Tian X, Maric I, et al. Clonal evolution leading to ibrutinib resistance in chronic lymphocytic leukemia. Blood. 2017 Mar 16;129(11):1469–1479. doi: 10.1182/blood-2016-06-719294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wu C, de Miranda NF, Chen L, Wasik AM, Mansouri L, Jurczak W, et al. Genetic heterogeneity in primary and relapsed mantle cell lymphomas: Impact of recurrent CARD11 mutations. Oncotarget. 2016 Jun 21;7(25):38180–38190. doi: 10.18632/oncotarget.9500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wilson WH, Young RM, Schmitz R, Yang Y, Pittaluga S, Wright G, et al. Targeting B cell receptor signaling with ibrutinib in diffuse large B cell lymphoma. Nat Med. 2015 Aug;21(8):922–926. doi: 10.1038/nm.3884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lenz G, Davis RE, Ngo VN, Lam L, George TC, Wright GW, et al. Oncogenic CARD11 mutations in human diffuse large B cell lymphoma. Science. 2008 Mar 21;319(5870):1676–1679. doi: 10.1126/science.1153629. [DOI] [PubMed] [Google Scholar]
- 11.Lacy SE, Barrans SL, Beer PA, Painter D, Smith AG, Roman E, et al. Targeted sequencing in DLBCL, molecular subtypes, and outcomes: a Haematological Malignancy Research Network report. Blood. 2020 May 14;135(20):1759–1771. doi: 10.1182/blood.2019003535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Puente XS, Beà S, Valdés-Mas R, Villamor N, Gutiérrez-Abril J, Martín-Subero JI, et al. Non-coding recurrent mutations in chronic lymphocytic leukaemia. Nature. 2015 Oct 22;526(7574):519–524. doi: 10.1038/nature14666. [DOI] [PubMed] [Google Scholar]
- 13.Fox LC, Yannakou CK, Ryland G, Lade S, Dickinson M, Campbell BA, et al. Molecular Mechanisms of Disease Progression in Primary Cutaneous Diffuse Large B-Cell Lymphoma, Leg Type during Ibrutinib Therapy. International journal of molecular sciences. 2018 Jun 13;19(6) doi: 10.3390/ijms19061758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fontan L, Yang C, Kabaleeswaran V, Volpon L, Osborne MJ, Beltran E, et al. MALT1 small molecule inhibitors specifically suppress ABC-DLBCL in vitro and in vivo. Cancer cell. 2012 Dec 11;22(6):812–824. doi: 10.1016/j.ccr.2012.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nagel D, Spranger S, Vincendeau M, Grau M, Raffegerst S, Kloo B, et al. Pharmacologic inhibition of MALT1 protease by phenothiazines as a therapeutic approach for the treatment of aggressive ABC-DLBCL. Cancer cell. 2012 Dec 11;22(6):825–837. doi: 10.1016/j.ccr.2012.11.002. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.