Abstract
To establish novel and effective treatment combinations for chronic myelomonocytic leukemia (CMML) preclinically, we hypothesized that supplementation of CMML cells with the human oncogene Meningioma 1 (MN1) promotes expansion and serial transplantability in mice, while maintaining the functional dependencies of these cells on their original genetic profile. Using lentiviral expression of MN1 for oncogenic supplementation and transplanting transduced primary mononuclear CMML cells into immunocompromised mice, we established three serially transplantable CMML-PDX models with disease-related gene mutations that recapitulate the disease in vivo. Ectopic MN1 expression was confirmed to enhance the proliferation of CMML cells, which otherwise did not engraft upon secondary transplantation. Furthermore, MN1-supplemented CMML cells were serially transplantable into recipient mice up to 5 generations. This robust engraftment enabled an in vivo RNA interference screening targeting CMML-related mutated genes including NRAS, confirming that their functional relevance is preserved in the presence of MN1. The novel combination treatment with azacitidine and the MEK-inhibitor trametinib additively inhibited ERK-phosphorylation and thus depleted the signal from mutated NRAS. The combination treatment significantly prolonged survival of CMML mice compared to single agent treatment. Thus, we identified the combination of azacitidine and trametinib as an effective treatment in NRAS-mutated CMML and propose its clinical development.
Introduction
Chronic myelomonocytic leukemia (CMML) is classified as a hematologic malignancy with myeloproliferative and myelodysplastic features(1) that is diagnosed in approximately 1000 patients per year in the United States.(2, 3) The prognosis varies by risk group and reaches from a median survival of 8 years in the favorable risk group to 16 months in the unfavorable risk group.(4) Similarly, the risk of transformation to acute myeloid leukemia (AML) varies from 0 to 48% among the favorable to high risk groups(5) and is expected in 16-19% of patients on average.(4, 6) Common cytogenetic aberrations are monosomy 7 and trisomy 8, and somatic mutations in genes that encode epigenetic modifiers (e.g. TET2, ASXL1, EZH2), splicing factors (e.g. SRSF2, SF3B1, ZRSF2), transcription factors (e.g. RUNX1), and signaling genes (e.g. NRAS, CBL, KRAS, JAK2) are commonly found in CMML patients.(5, 7–11)
Treatment options are limited in CMML and include observation, cytoreduction with hydroxyurea, treatment with hypomethylating agents azacitidine or decitabine and allogeneic hematopoietic cell transplantation (alloHCT) in eligible patients. Only alloHCT has a curative intent and resulted in a 3-year overall survival of 31% in single center experience.(12) Other treatment approaches aim at symptom relief in older patients. A matched-pair analysis of CMML patients treated with azacitidine (n=22) compared to patients treated with hydroxyurea (n=22) resulted in similar median overall survival (7.5 vs 6.2 months, respectively (p=0.251) and thus the benefit of hypomethylating agents over hydroxyurea is unclear.(13)
None of the mutations found in CMML patients is unique for the disease. However, ASXL1, TET2 and SRSF2 mutations occur more frequently in CMML patients (40-50% each) than in other myeloproliferative or myelodysplastic diseases.(14) Interestingly, mutations in signaling pathway genes like JAK2, NRAS and CBL are common in CMML and rare in MDS. CMML cells typically display hypersensitivity to granulocyte macrophage colony-stimulating factor (GM-CSF) in vitro, which likely contributes to the monocytic phenotype of the disease and is mediated by mutations in signaling genes.(15, 16)
In order to better understand the pathophysiology of CMML and to evaluate novel treatment combinations, patient-derived xenograft (PDX) models were developed by transplanting primary patient cells in immunocompromised mice. NOD.Cg-Prkdcscid Il2rgtm1Wjl/SzJ (NSG) mice transgenic for IL3, SCF and GM-CSF (NSGS) were shown to enhance engraftment of CMML cells likely through expression of GM-CSF.(17) Xenotransplantation of bone marrow or peripheral blood mononuclear cells from 18 CMML patients into NSGS mice gave rise to robust human hematopoietic cell engraftment in almost all cases. However, transplantation into secondary recipients succeeded only in 2 of 5 cases and further serial transplantation was not reported.(18) Similarly, cells from 15 of 16 CMML patients engrafted in primary NSG or NSGS recipient mice, but in secondary NSGS recipients only 3 of 10 samples engrafted with low levels of 2-9% human cells in bone marrow.(17)
Overexpression of the transcriptional co-factor MN1 has been observed in a broad spectrum of AML patients(19–23) and has been identified as an independent prognostic marker for AML with normal karyotype. High expression of MN1 is associated with poor prognosis, shorter overall and relapse-free survival, and poor response to treatment.(21) In murine cells human MN1 overexpression induces aggressive, fully penetrant AML through the promotion of leukemic cell self-renewal,(24–26) impairment of myeloid differentiation,(24, 25) and resistance to all trans retinoic acid-induced differentiation.(25) MN1-induced leukemias are also associated with upregulation of HOXA genes and MEIS1.(27) Moreover, functional assays have revealed the critical dependence of the MEIS1/AbdB-like HOX protein complex for MN1-induced transformation.(27)
We aimed to develop a xenograft model that allows expansion of CMML cells in vivo, serial transplantation for at least 3 generations of mice and functional manipulation of primary CMML cells for an in vivo shRNA screen. We hypothesized that supplementation of CMML cells with a human oncogene will promote expansion and serial transplantability while the functional dependencies of these cells on their genetic profile are maintained. We chose the MN1 oncogene for this approach as transduction of MN1 in CD34+ cord blood cells promotes self-renewal in vitro and engraftment in vivo, but does not induce AML in vivo nor does it enable serial transplantation.(28)
To establish a serially transplantable PDX model for CMML we used lentiviral expression of MN1 in primary CMML cells for oncogenic supplementation and evaluated the growth potential of these cells in vivo by serial transplantations, a functional in vivo shRNA screen and a novel combination treatment in vivo.
Materials and Methods
Patient samples and xenotransplantation
Diagnostic bone marrow or peripheral blood samples were collected after obtaining written informed consent from CMML patients diagnosed and treated at Hannover Medical School.(29, 30) Briefly, mononuclear cells were isolated from patient bone marrow aspirates or peripheral blood by density centrifugation using Biocoll Separating Solution (Biochrom, Berlin, Germany). The cell number was determined and depletion of CD3 positive cells was done following the manufacturer’s protocol (CD3 MicroBeads kit, Miltenyi Biotec, Bergisch Gladbach, Germany). Patient cells were analyzed according to the principles of the Declaration of Helsinki, and the study was approved by the institutional review board of Hannover Medical School. Patient characteristics are listed in Supplementary Table S1. Detailed methods for cell culture, viral vectors and infection of cells, clonogenic progenitor assays, immunoblotting and amplicon-based next generation sequencing(31, 32) of treated CMML cells are described in the Supplementary Methods.
For xenotransplantation studies, six- to eight-week old female NOD.Cg-Prkdcscid Il2rgtm1Wjl/SzJmice transgenic for human interleukin-3, granulocyte-macrophage-colony stimulating factor and stem cell factor (NSGS) were bred by the central animal laboratory of Hannover Medical School under pathogen-free conditions. All animal experiments were performed in accordance with the German animal protection law and approved by the Lower Saxony state office for consumer protection, Oldenburg, Germany. Xenotransplantation, engraftment monitoring, treatment and morphologic analysis of CMML cells are described in the Supplementary Methods. Technical details on serial transplantations are provided in Supplementary Table S2.
Quantitative RT-PCR
Total RNA was extracted from 1 million cells and reverse transcribed as previously described.(25) Quantitative reverse-transcriptase polymerase chain reaction (RT-PCR) was done as previously described using SYBR green (Invitrogen, ThermoFischer Scientific, Schwerte, Germany) for quantification of double stranded DNA on a StepOne Plus cycler (Applied Biosystems, Darmstadt, Germany). Relative expression was determined with the 2-△△CT method using ABL1 as the housekeeping gene, and expression values were normalized relative to control cells.(33) Used custom primers were shown in Supplementary Table S3.
Statistical analysis
Data are shown as mean ± standard error of mean (SEM). Student t test and Logrank test were used to assess differences between groups and survival curves, respectively. All statistical analysis were performed with GraphPad Prism Version 7 software. P < 0.05 was considered statistically significant. The size of the animal cohorts was based on our previous study.(34, 35) All in vitro experiments were performed at least 3 times unless otherwise stated and all attempts of replication were successful.
Results
Supplementation of primary CMML cells with MN1 promotes engraftment in vivo
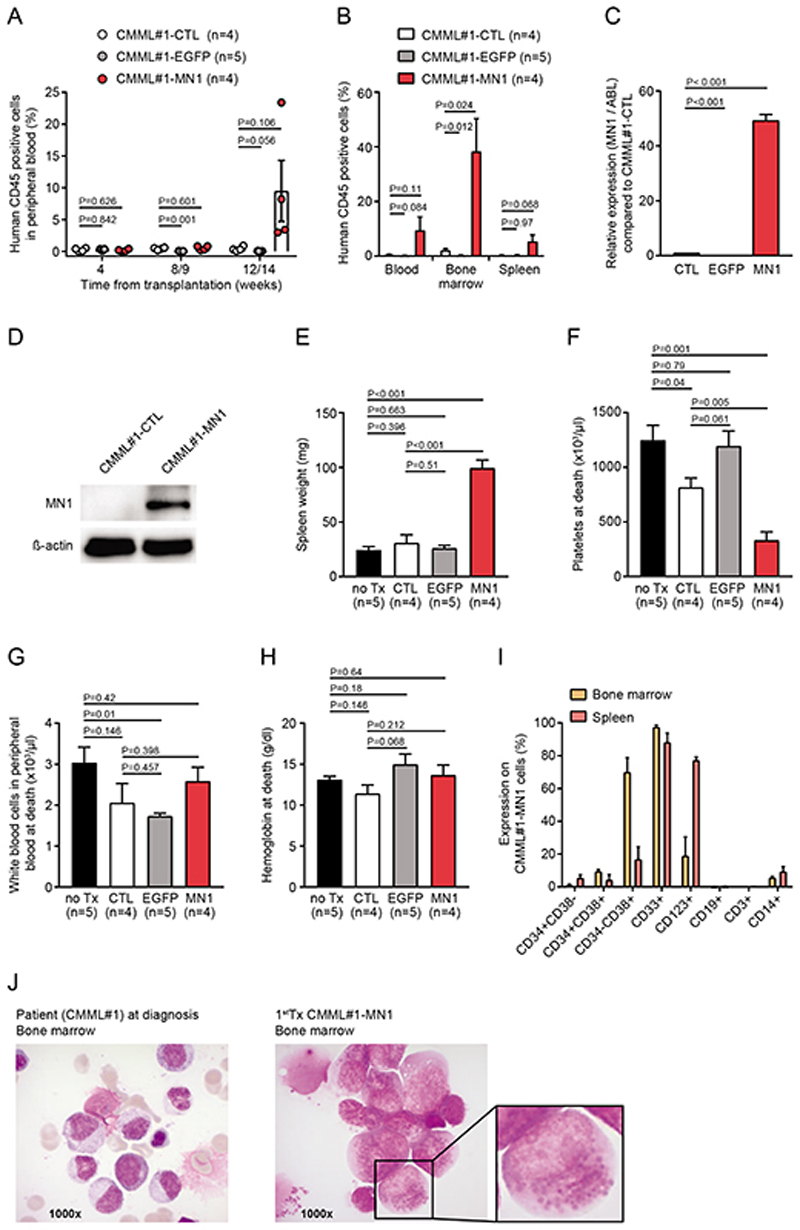
We evaluated the ability of oncogenic supplementation of patient CMML cells with the oncogene MN1 to enhance robust engraftment of CMML in vivo and enable serial transplantation.(27, 28) Transduction of mononuclear cells from bone marrow aspirate of a patient diagnosed with CMML-1 with a control vector (CMML#1-EGFP) or MN1 (CMML#1-MN1) resulted in 77% and 16% efficacy in vitro, respectively (Supplementary Figure S1A). Intravenous injection of untransduced (CMML#1-CTL) and EGFP-transduced (CMML#1-EGFP) primary CMML#1 cells into sublethally irradiated NSGS recipient mice resulted in minimal or undetectable engraftment of human CD45+ cells for CMML#1-CTL and CMML#1-EGFP cells in peripheral blood 12 weeks after transplantation (Figure 1A), and in bone marrow 22 weeks after transplantation (0,8% to 3% for CMML#1-CTL, 0,6% for CMML#1-EGFP) and spleen (0.5% for CMML#1-CTL, 0,6% for CMML#1-EGFP) (Figure 1B). In contrast, MN1- transduced CMML#1 cells initiated robust human hematopoietic engraftment ranging from 3% to 23% in peripheral blood, 21% to 73% in bone marrow and 2% to 12% in spleen 13 weeks after transplantation (Figure 1A and 1B). In CMML#1-MN1 cells all human cells expressed EGFP (Supplementary Figure S1B), and MN1 overexpression was confirmed by RNA and protein expression in engrafted PDX cells compared to primary untransduced CMML#1 cells of the corresponding patient (Figure 1C and 1D), suggesting that MN1 was required for engraftment. CMML#1-MN1 mice had enlarged spleens (Figure 1E) and reduced platelet counts (Figure 1F), while white blood cell (WBC) counts and hemoglobin levels were similar (Figure 1G and 1H) when compared with CMML#1-CTL, CMML#1-EGFP or non-engrafted control (no Tx) groups.
Figure 1. Supplementation of primary CMML cells with MN1 promotes engraftment in vivo.
A) Engraftment of human hematopoietic cells (CD45 positive) in the peripheral blood of primary recipient NSGS mice monitored at the indicated time points. Recipient mice were transplanted with untransduced CMML#1 cells (CMML#1-CTL), EGFP- (control vector) transduced CMML#1 cells (CMML#1-EGFP) or MN1-transduced CMML#1 cells (CMML#1-MN1), (number of analyzed mice is indicated in the figure; mean ± SEM).
B) Engraftment of human CD45 positive cells in peripheral blood, bone marrow and spleen of primary recipient NSGS mice at sacrifice (number of analyzed mice is indicated in the figure; mean ± SEM).
C) Expression levels of MN1 in untransduced CMML#1 patient cells (CMML#1-CTL), EGFP-transduced CMML cells (CMML#1-EGFP) and MN1-transduced CMML cells (CMML#1-MN1). Gene expression was determined by quantitative RT-PCR relative to the housekeeping gene ABL1 and normalized to gene expression in untransduced CMML#1 patient cells (n=3), (mean ± SEM).
D) Western blot showing MN1 expression in primary bone marrow mononuclear cells from a patient with CMML (CMML#1-CTL) and in their MN1-transduced counterparts isolated from bone marrow of engrafted recipient mice. ß-actin was used in the same blot as loading control.
E) Spleen weight of primary recipient NSGS mice at sacrifice (number of analyzed mice is indicated in the figure; mean ± SEM).
F) Platelet count in peripheral blood of primary recipient NSGS mice at sacrifice (number of analyzed mice is indicated in the figure; mean ± SEM).
G) White blood cell count in peripheral blood of primary recipient NSGS mice at sacrifice (number of analyzed mice is indicated in the figure; mean ± SEM).
H) Hemoglobin level in peripheral blood of primary recipient NSGS mice at sacrifice (number of analyzed mice is indicated in the figure; mean ± SEM).
I) Immunophenotype of engrafted CMML#1 cells from bone marrow and spleen of primary recipient NSGS mice at sacrifice (gated on human CD45+EGFP+ CMML#1-MN1 cells; n=3; mean ± SEM).
J) Morphology from bone marrow smears of the CMML#1 patient at diagnosis and from a transplanted mouse at sacrifice. The enlarged cell shows granules in the cytoplasm that can be identified as lipid granules in cells from later transplantations (see Figure 3B).
The immunophenotype of engrafted human CD45 and EGFP positive CMML#1-MN1 cells showed a myelo-monocytic differentiation expressing CD33, CD38 and CD14, including progenitor markers CD38 and in few cells CD34 (Figure 1I). Morphologic analysis of bone marrow from CMML#1-MN1 mice showed myeloid progenitors and monocytic cells similarly to bone marrow smears from the CMML#1 patient at diagnosis and revealed MN1 expression by EGFP fluorescence (Figure 1J and Supplementary Figure S1C). In summary, oncogenic supplementation of CMML cells with MN1 enabled engraftment of these cells that otherwise did not engraft in vivo.
CMML-MN1 cells engraft secondary recipient mice and are serially transplantable
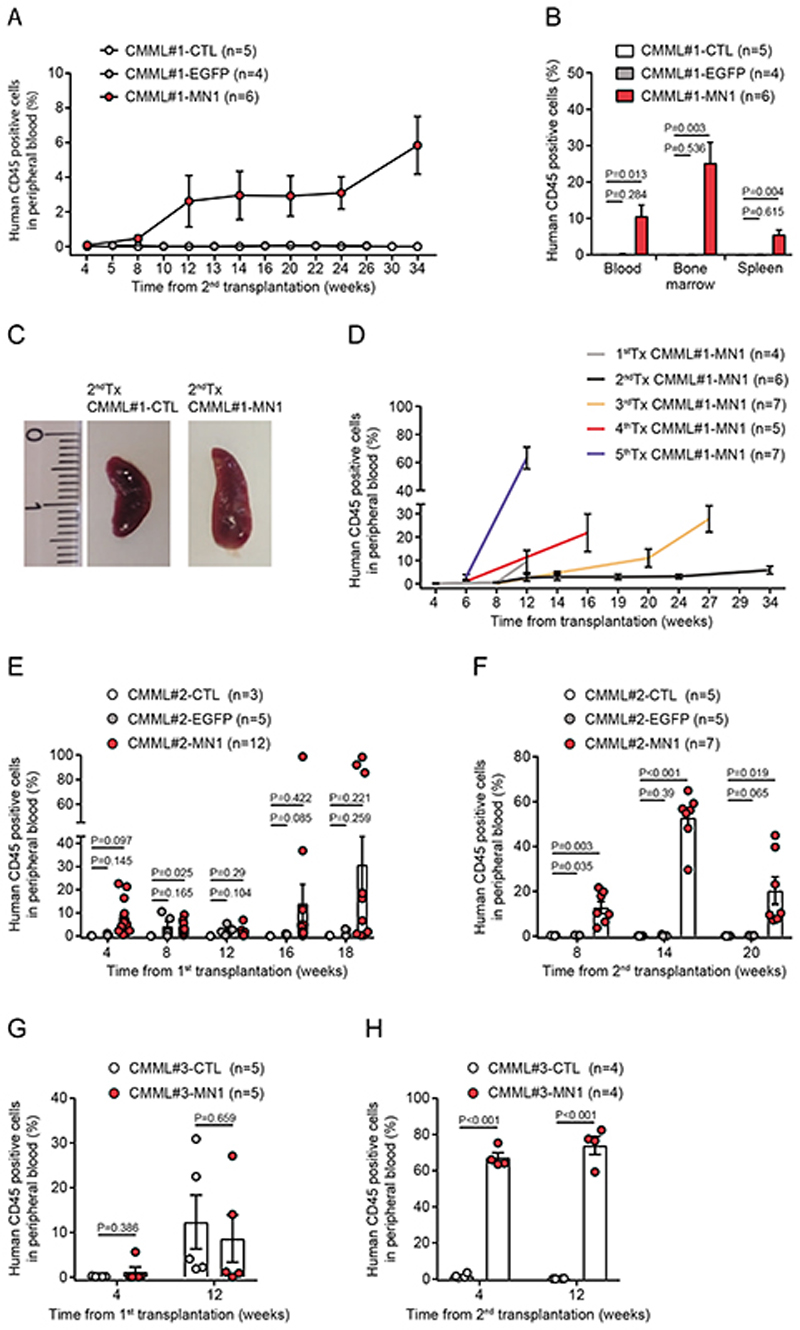
To evaluate whether CMML#1-MN1 cells can be serially transplanted, CMML#1-CTL, CMML#1-EGFP and CMML#1-MN1 cells from bone marrow of primary recipients were isolated and intravenously transplanted into sublethally irradiated secondary NSGS recipient mice. Only CMML#1-MN1 cells engrafted in secondary recipients (Figure 2A), and showed mean engraftment of 10%, 25% and 5% of human CD45+ cells in peripheral blood, bone marrow and spleen, respectively, at the time of sacrifice (Figure 2B). Engrafted CMML#1-MN1 cells had a myeloid phenotype expressing CD33, CD38, and CD123, but lacked expression of lymphoid markers CD3 and CD19 (Supplementary Figure S1D), and developed splenomegaly (Figure 2C).
Figure 2. CMML-MN1 cells engraft secondary recipient mice and are serially transplantable.
A) Engraftment of human hematopoietic cells (CD45 positive) in the peripheral blood of secondary recipient NSGS mice monitored at the indicated time points (number of analyzed mice is indicated in the figure; mean ± SEM).
B) Engraftment of CMML#1-MN1 cells in peripheral blood, bone marrow and spleen of secondary recipient NSGS mice at sacrifice (number of analyzed mice is indicated in the figure; mean ± SEM).
C) Representative photographs of spleens from secondary recipient mice transplanted with bone marrow from primary recipient mice which had received either untransduced CMML#1 cells (CMML#1-CTL) or MN1-transduced CMML#1 cells (CMML#1-MN1) (scale bar indicates millimeters).
D) Engraftment of CMML#1-MN1 cells in serial transplantations shown as percentage of human CD45 positive cells detected in peripheral blood at the indicated time points (number of analyzed mice is indicated in the figure; mean ± SEM).
E) Engraftment of CMML#2 cells in the peripheral blood of primary recipient NSGS mice transplanted with untransduced CMML#2 (CMML#2-CTL), EGFP (control vector) transduced CMML#2 (CMML#2-EGFP) or MN1-transduced CMML#2 cells (CMML#2-MN1), (number of analyzed mice is indicated in the figure; mean ± SEM).
F) Engraftment of CMML#2 cells (from Figure 2E) in the peripheral blood of secondary recipient NSGS mice at the indicated time points (number of analyzed mice is indicated in the figure; mean ± SEM).
G) Engraftment of CMML#3 cells in the peripheral blood of primary recipient NSGS mice transplanted with untransduced CMML#3 (CMML#3-CTL) or MN1-transduced CMML#3 cells (CMML#3-MN1), (number of analyzed mice is indicated in the figure; mean ± SEM).
H) Engraftment of CMML#3 cells (from Figure 2G) in the peripheral blood of secondary recipient NSGS mice at the indicated time points (number of analyzed mice is indicated in the figure; mean ± SEM).
Cryopreserved CMML#1-MN1 cells from bone marrow and spleen of secondary transplanted mice were used as source to initiate serial transplantations in NSGS recipients up to 5 times (Figure 2D). Serial transplantation resulted in higher engraftment levels in peripheral blood and shorter latency, suggesting that the cells underwent clonal selection.
Cells from two additional CMML patients were used to confirm MN1-enhanced serial engraftment. Detailed patient characteristics are provided in Supplementary Table S1. In the CMML#2 model MN1-transduced cells showed higher engraftment levels in several mice at 16 and 18 weeks after transplantation compared to CTL mice (Figure 2E). Upon secondary transplantation only MN1-transduced cells engrafted (7% to 44% human CD45+ in peripheral blood, 11% to 98% human CD45+ in bone marrow and 18% to 36% human CD45+ in spleen), while CTL and EGFP transduced cells did not engraft (Figure 2F and Supplementary Figure S2A). In the CMML#3 model we observed comparable robust engraftment of both CMML#3-MN1 and CMML#3-CTL cells at week 12 after transplantation in primary recipient NSGS mice (Figure 2G). Upon secondary transplantation of spleen and bone marrow cells (65% to 85% human CD45+ cells in 1stTx CMML#3-MN1 and 26% to 47% human CD45+ cells in 1stTxCMML#3-CTL mice) only MN1-transduced cells engrafted at high levels (59% to 82% human CD45+ cells in peripheral blood, 93% to 96% in bone marrow and 74% to 88% in spleen), while the CTL group did not engraft secondary recipient NSGS mice (Figure 2H and Supplementary Figure S3A). CMML#2-MN1 and CMML#3-MN1 secondary recipient mice had enlarged spleens (Supplementary Figures S2B and S3B), reduced platelet counts and similar white blood cell counts and hemoglobin levels (Supplementary Figures S2C-E and S3C-E) when compared with CMML#2-CTL and CMML#2-EGFP, or CMML#3-CTL mice, respectively. Morphology of bone marrow cells isolated from CMML#2 and CMML#3 secondary recipient mice showed myeloid progenitors and monocytic cells similar to bone marrow smears from the CMML#2 and CMML#3 patients (Supplementary Figures S2F and S3F). Thus, oncogenic supplementation enabled serial transplantation of CMML cells and set the stage for functional in vivo studies in primary patient cells.
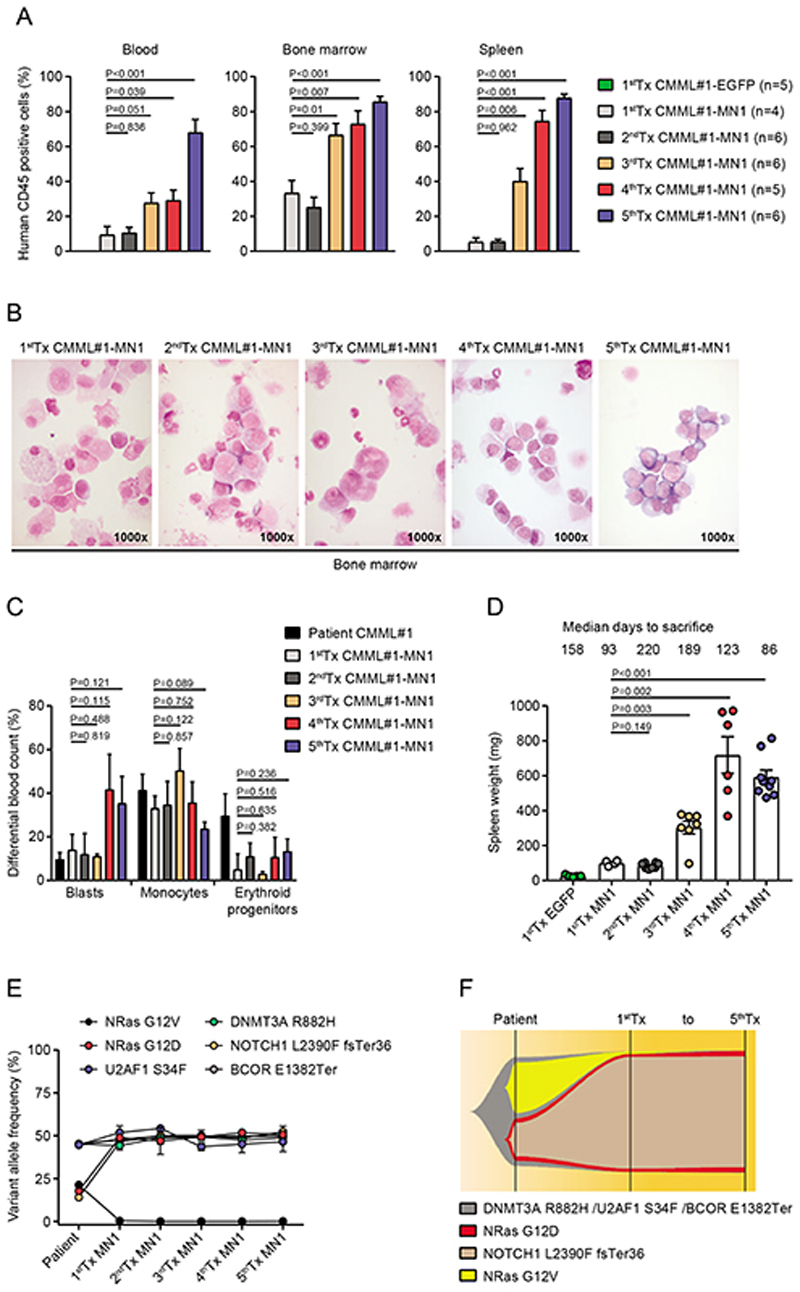
Clonal dominance of an NRAS/NOTCH1 mutated subclone during serial transplantation of CMML#1-MN1 cells
We evaluated the biologic and genetic evolution of CMML#1-MN1 cells during serial transplantation from first to fifth generation. Up to the third transplantation CMML#1-MN1 cells showed preferential engraftment in bone marrow over peripheral blood and spleen (Figure 3A). In the fourth transplantation engraftment in bone marrow and spleen was comparable and in the fifth transplantation engraftment was equally high in blood, bone marrow and spleen (Figure 3A). Bone marrow morphology showed a similar monocyte dominated picture compared to the patients’ bone marrow from first to third transplantation, while the blast count increased in the fourth and fifth transplantations above 20%, while the monocytes remained at high levels (Figure 3B-C). This indicates that upon serial transplantation the most aggressive clones prevail and the disease is more related to AML, while it still reflects the same genetics that were found in the patients’ cells. Spleen weight in sacrificed mice increased over time consistent with engraftment levels in the spleen (Figure 3D). Genetic profiling of the patient`s cells identified a founding clone characterized by DNMT3A, U2AF1 and BCOR mutations and two subclones with additional mutations. The first subclone was characterized by a NRAS G12D mutation and a NOTCH1 mutation, and the second subclone included a NRAS G12V mutation (Figure 3E). Upon transduction with MN1 and transplantation in vivo only the NRAS G12D/NOTCH1 subclone containing also the DNMT3A, U2AF1 and BCOR mutations engrafted in PDX mice and persisted without further changes up to the fifth transplantation (Figure 3F and Supplementary Table S4). In summary, serial transplantation led to the selection of a more aggressive leukemic clone that was characterized by mutations recurrently found in CMML and AML patients. Only the NRAS G12D/NOTCH1 mutated subclone stably engrafted in vivo, and its consistent representation in all transplantations suggests that mutations in DNMT3A, U2AF1, BCOR, NRAS and NOTCH1 were required for in vivo proliferation despite the presence of MN1. We found that clonal selection of MN1 transduced CMML cells occured during engraftment in vivo and not during the ex vivo transduction procedure as shown for the CMML#3-MN1 model (Supplementary Figure S4).
Figure 3. Clonal dominance of an NRAS mutated subclone during serial transplantation of CMML#1-MN1 cells.
A) Engraftment of human hematopoietic cells in peripheral blood, bone marrow and spleen of NSGS recipient mice from first to fifth serial transplantation (number of analyzed mice is indicated in the figure, mean ± SEM).
B) Morphology from bone marrow smears of engrafted CMML#1-MN1 cells of NSGS recipient mice from first to fifth serial transplantation.
C) Differential blood count quantification of bone marrow smears from the patient (CMML#1) at diagnosis and engrafted CMML#1-MN1 cells in mice from first to fifth transplantation (n=3; mean ± SEM).
D) Spleen weight of primary CMML#1-EGFP recipient mice and CMML#1-MN1 recipient mice from first to fifth serial transplantation at the time of sacrifice (each dot represents one mouse; mean ± SEM).
E) Variant allele frequencies of mutated genes in bone marrow cells of the CMML#1 patient and corresponding CMML#1-MN1 cells engrafted in the bone marrow and spleen of NSGS recipient mice at sacrifice from first to fifth serial transplantation (n=3; mean ± SEM).
F) Graphical representation of clonal evolution of CMML#1 cells from the patient and its corresponding CMML#1-MN1 cells from first to fifth serial transplantation in NSGS recipient mice.
The known GM-CSF/CSF2 dependency of CMML cells was evaluated for CMML#1-MN1 cells by transplanting cells from the fourth recipient generation into either NSG or NSGS mice. The latter mouse strain produces human GM-CSF besides IL3 and SCF. Upon transplantation of equal cell numbers CMML#1-MN1 cells engrafted much faster in NSGS compared to NSG recipient mice, suggesting that these MN1-transduced cells retained their GM-CSF sensitivity (Supplementary Figure S5).
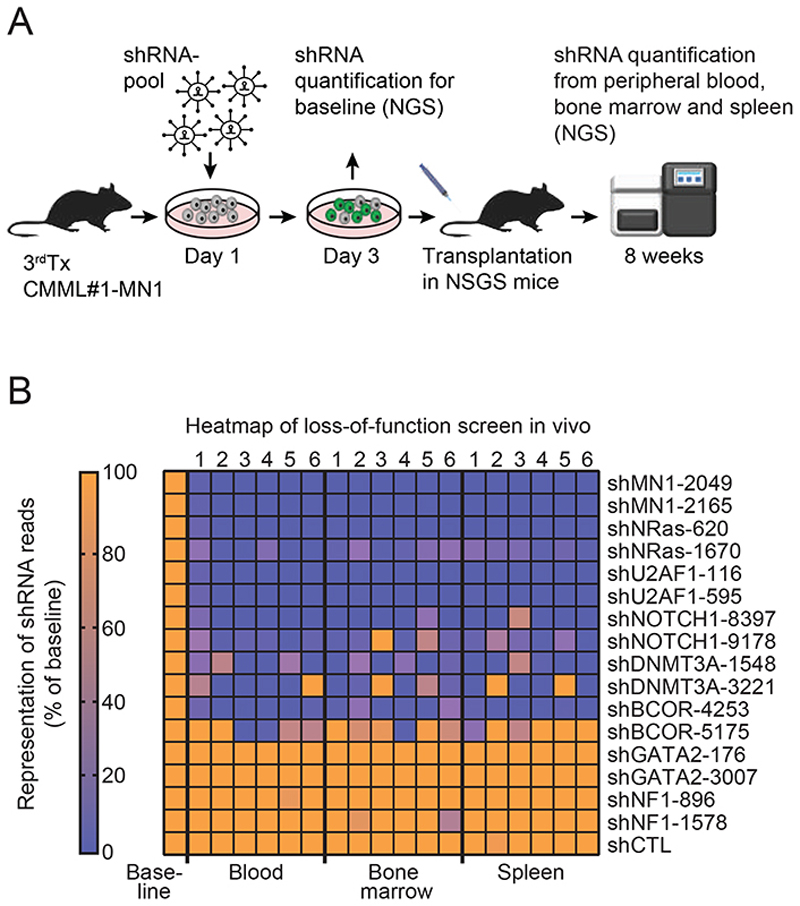
Loss-of-function shRNA-screening identifies functional dependencies in CMML#1-MN1 cells
To evaluate the functional importance of the mutated genes and MN1 we performed a targeted shRNA screening in vivo. Bone marrow cells from the third generation of CMML#1-MN1 mice were harvested and transduced with a shRNA pool consisting of 2 independent shRNAs against 8 genes and 1 control shRNA against luciferase. Six shRNAs targeted the mutated genes in this patient or MN1, while 2 shRNAs targeted 2 control genes that are recurrently mutated in myeloid disease (GATA2 and NF1). 48 hours after transduction the cells were either transplanted in NSGS mice or harvested for shRNA quantification. Eight weeks after transplantation the cells were harvested from blood, bone marrow and spleen and DNA was extracted from unsorted cells to quantify shRNA copies in total DNA by NGS (Figure 4A). The shRNAs against the 8 genes were validated in vitro and induced knockdown usually by more than 80% (Supplementary Figure S6 and Supplementary Table S5). shRNAs against the control genes luciferase, GATA2 and NF1 were not depleted after transplantation in mice compared to the cells harvested after transduction in vitro (Figure 4B). Interestingly, the shRNAs against the mutated genes and MN1 were strongly depleted in most mice and cell sources (blood, bone marrow, spleen) after in vivo transplantation except one of the two shRNAs against BCOR (Figure 4B). This analysis suggests that at least four mutated genes are required for the survival of CMML-MN1 cells in vivo in addition to MN1 and shows that our model can be used to evaluate the specific contribution and biology of each mutated gene despite the ectopic expression of the MN1 oncogene.
Figure 4. Loss-of-function shRNA-screening identifies functional dependencies in CMML#1-MN1 cells.
A) Experimental outline of loss-of-function shRNA screening showing lentiviral transduction of CMML#1-MN1 cells with a shRNA pool, transplantation into NSGS recipient mice, cell harvest after eight weeks and quantification of shRNAs by next-generation sequencing.
B) Representation of shRNAs in blood, bone marrow and spleen of NSGS recipient mice at eight weeks after transplantation compared to baseline shRNA representation (baseline: transduced cells kept in vitro and quantified on day 3), (each column represents one mouse, n=6).
The MEK-inhibitor trametinib prolongs survival of CMML-MN1 mice when combined with azacitidine
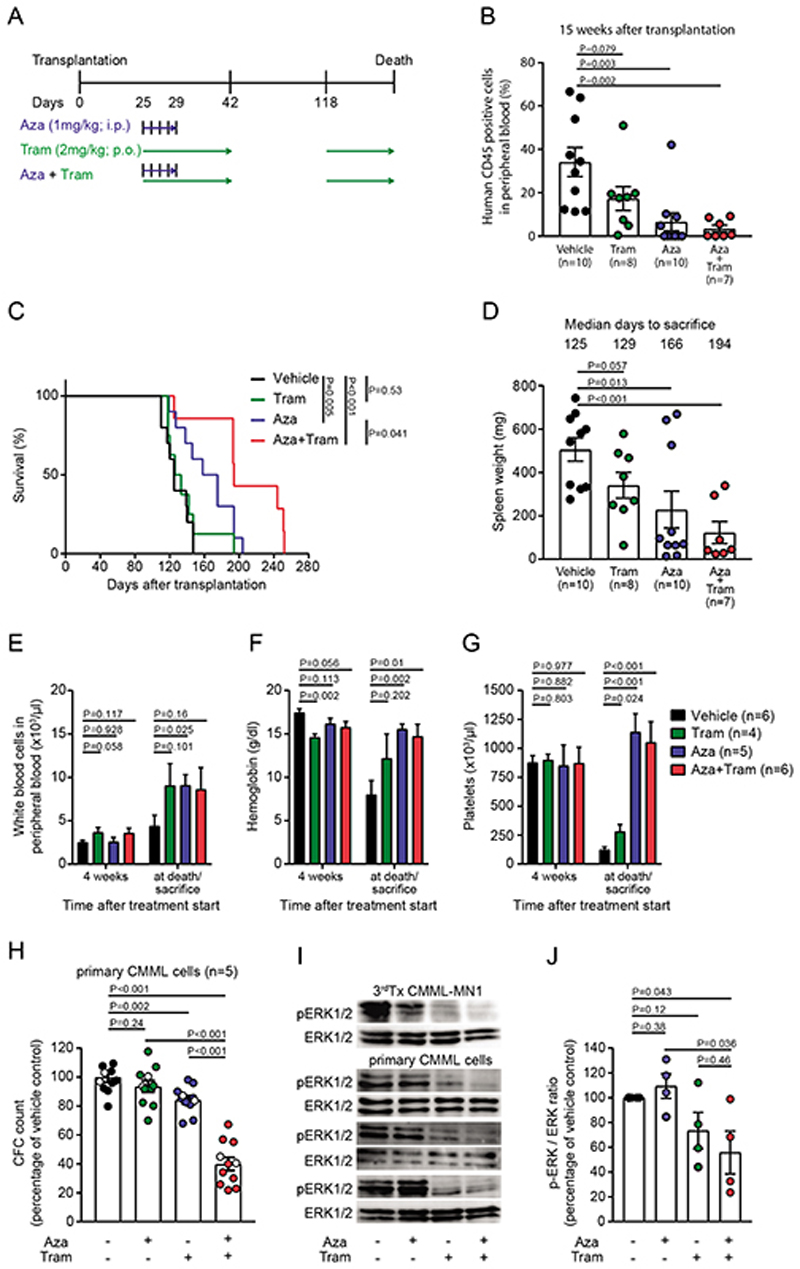
Hypomethylating agents are used in CMML patients with proliferative disease who are not eligible for alloHCT. As the NRAS mutated subclone engrafted efficiently in NSGS mice and knockdown of NRAS depleted CMML#1 cells, we hypothesized that the combination of the MEK inhibitor trametinib, which inhibits the signaling pathway downstream of NRAS, with azacitidine more efficiently inhibits CMML development than either drug alone. We transplanted CMML#1-MN1 cells from the fifth transplantation in NSGS mice and confirmed similar engraftment of human CD45+EGFP+ cells in peripheral blood 2 weeks after transplantation (Supplementary Figure S7A). Treatment was started four weeks after transplantation with azacitidine (1 mg/kg i.p. for 5 days) and/or trametinib (2 mg/kg p.o. for 18 days). At this time we assume similar engraftment levels between the indicated groups, as three weeks after the start of treatment engraftment was still comparable between the treatment groups (Supplementary Figure S7B). Trametinib treatment was interrupted until day 118 and then continued until the end of the experiment (Figure 5A). Engraftment monitoring at week 15 after transplantation revealed diminished engraftment in peripheral blood of mice treated with azacitidine (6% CD45+ cells) and the combination of azacitidine+trametinib (3% CD45+ cells) as compared to trametinib treated (17% CD45+ cells) and vehicle treated (34% CD45+ cells) mice (Figure 5B and Supplementary Figure S7B). Trametinib treated mice had similar survival as vehicle treated mice, while azacitidine treated mice showed improved survival (Figure 5C). Interestingly, mice treated with the combination of trametinib/azacitidine survived significantly longer than vehicle treated, azacitidine treated or trametinib treated mice alone (median survival for vehicle 125 days, azacitidine 166 days, trametinib 129 days and trametinib/azacitidine 194 days, Figure 5C, P=0.041). Engraftment of CMML-MN1 cells was significantly lower in azacitidine and trametinib/azacitidine treated groups compared to vehicle control (Supplementary Figure S7C). The weight of the spleens was lower in the single and combination treatment groups compared to the vehicle group despite a longer latency to sacrifice (Figure 5D). At the end of the treatment, WBC counts and hemoglobin counts were decreased in the vehicle group but remained in the normal range for azacitidine and trametinib/azacitidine treated groups. Platelets were increased in azacitidine and trametinib/azacitidine treated groups when compared with vehicle or trametinib treated mice (Figure 5E-G).
Figure 5. The MEK-inhibitor trametinib prolongs survival of CMML#1-MN1 mice when combined with azacitidine.
A) Schematic representation of the treatment regimens (Aza, azacitidine; Tram, trametinib).
B) Monitoring of CMML burden in recipient mice at week 15 after transplantation (each dot represents one mouse; mean ± SEM)
C) Survival of NSGS recipient mice engrafted with CMML#1-MN1 cells and treated with vehicle (n=10), trametinib (n=8), azacitidine (n=10) and the combination of trametinib+azacitidine (n=7).
D) Spleen weight of NSGS recipient mice engrafted with CMML#1-MN1 cells and treated with vehicle, trametinib, azacitidine and the combination of trametinib+azacitidine at sacrifice (each dot represents one mouse; mean ± SEM).
E) White blood cell count in peripheral blood of CMML#1-MN1 bearing mice treated with vehicle, trametinib, azacitidine and the combination of trametinib+azacitidine 4 weeks after treatment start and at sacrifice (number of individual mice is indicated in the figure; mean ± SEM).
F) Hemoglobin levels in peripheral blood of CMML#1-MN1 bearing mice treated with vehicle, trametinib, azacitidine and the combination of trametinib+azacitidine 4 weeks after treatment start and at sacrifice (number of individual mice is indicated in the figure; mean ± SEM).
G) Platelet count in peripheral blood of CMML#1-MN1 bearing mice treated with vehicle, trametinib, azacitidine and the combination of trametinib+azacitidine 4 weeks after treatment start and at sacrifice (number of individual mice is indicated in the figure; mean ± SEM).
H) Effect of the combination treatment in five different CMML patients (unrelated to the CMML#1-MN1 cells). Primary CMML cells were treated with vehicle, azacitidine (500 nM), trametinib (20 nM) or the combination of azacitidine (500 nM) + trametinib (20 nM) and plated in duplicate in CFC media. After 20 days colonies were counted and expressed as percentage of the vehicle treated cells. White dots within all groups represent cells from a patient without mutation in a signaling gene (n=5, mean ± SEM).
I) Immunoblotting for p-ERK and ERK in azacitidine/trametinib- and vehicle-treated CMML patients (n=3) and CMML#1-MN1 (n=1) cells from third recipient mice at 6h after treatment.
J) Average p-ERK-to-ERK ratio from immunoblots shown in Figure 5I as percentage of vehicle treated CMML cells (n=4, mean ± SEM).
To evaluate whether the additive effect of trametinib and azacitidine was also found in primary cells from CMML patients we performed CFC assays from peripheral blood or bone marrow cells of five CMML patients (Supplementary Table S1). The combination treatment reduced the CFC count significantly stronger compared to either azacitidine or trametinib monotherapy (Figure 5H). In line with this finding, phosphorylated ERK (pERK) was most effectively inhibited in CMML#1-MN1 cells harvested from NSGS mice as well as primary CMML cells by the combination treatment compared to single agent azacitidine, trametinib or vehicle treatment (Figure 5I-J). In summary, the combination of the MEK-inhibitor trametinib and a hypomethylating agent prolonged survival in the CMML#1-MN1 model by additively inhibiting pERK signaling.
Discussion
We developed patient-derived CMML models that recapitulate the disease in vivo and allowed us to demonstrate that (1) ectopic MN1 expression enhances the engraftment and proliferation of cells that otherwise do not engraft in mice, (2) the functional relevance of CMML-related gene mutations is preserved in the presence of MN1, (3) MEK/ERK signaling is important for in vivo proliferation of CMML cells, (4) the serial transplantability of the MN1-CMML cells enable functional RNAi in vivo and (5) the combination of azacitidine with trametinib is an effective combination treatment in CMML#1-MN1 cells in vivo and primary CMML cells in vitro.
It was shown in recent years that CMML cells readily engraft in immunocompromised mice by intrafemoral or intravenous injection and are transplantable in secondary recipient mice in some cases.(17, 18) However, transplantation in secondary recipients was inconsistent and no tertiary transplantation was reported. We chose the MN1 oncogene to enhance the engraftment of CMML cells as MN1 enhances self-renewal and blocks differentiation of CD34+ cord blood cells in vitro and induces a myeloproliferative disease, but not AML, in vivo. Only by combining MN1 with another oncogene, NUP98-HOXD13, could cord blood cells be transformed to AML and serially transplanted.(28) Here we confirm our hypothesis that the mutations present in CMML cells collaborate with MN1 and maintain their functional relevance. So far, only expression of the fusion genes MLL-AF9 and MLL-ENL were able to fully transform normal human hematopoietic stem cells.(36–39) The incomplete transformative capacity of MN1 may be ideally suited to enhance the proliferative capacity of malignant cells and enable their investigation in vivo.
The robust engraftment of MN1-CMML cells allowed us to perform in vivo RNA interference targeting the mutated genes in our patient. The cells were clearly dependent on MN1, but also on NRAS, NOTCH1, U2AF1 and DNMT3A, while shRNAs against the non-mutated genes GATA2 and NF1 and the control shRNA were not depleting. The results for BCOR were inconsistent between the two targeting shRNAs. Mutations in signaling gens like NRAS are found in 30% of CMML patients (7, 9) and the NRAS/NOTCH1 mutated subclone in our patient was selected during the transplantation procedure and persisted throughout all 5 serial transplantations. Ricci et al. (40) underscored the importance of RAS pathway mutations by showing that they induce a CMML-like disease in mice. NOTCH1 mutations have not been reported in CMML, but mutations in NOTCH pathway genes were found in 5 of 47 CMML patients.(41) Klinakis et al. showed that deletion of at least NOTCH1 and NOTCH2 induced a CMML-like disease in mice.(41) It remains unclear whether the NOTCH1 mutation in our model was a loss-of-function or rather gain-of-function mutation, as inhibiting NOTCH1 in our experiments by shRNA depleted the CMML cells in vivo. U2AF1 mutations occur in approximately 7% of CMML patients.(42) U2AF1 mutated cells had a competitive disadvantage in mouse models compared to U2AF1 wildtype cells. The combination of mutated U2AF1 with RUNX1 induced AML in mice, but 2 of 3 established AMLs lost the U2AF1 mutation.(43) It is thus surprising that the U2AF1 mutation was maintained in our model over 5 generations, which represents a faithful model to study the impact of U2AF1 mutations in vivo. Knockdown of U2AF1 resulted in depletion of CMML cells in vivo in our study. A gain-of-function has been suspected for U2AF1 mutations, as they occur only at 2 hotspots in this gene, but catastrophic depletion of the splicing factor SRSF2 has also led to cell depletion (44), and thus the nature of our U2AF1 mutation remains unclear. DNMT3A mutations occur in approximately 5% of CMML patients.(45) The Arg882His mutation found in our patient indicates an origin of the disease in clonal hematopoiesis of indeterminate potential.(46) This mutation impairs the methyltransferase activity of DNMT3A (47) and loss of DNMT3A leads to enhanced self-renewal of hematopoietic stem cells and expansion of the DNMT3A null clone.(48) It was therefore unexpected that shRNA-mediated knockdown of DNMT3A depleted CMML cells in vivo. However, DNMT3A depletion in hepatocellular carcinoma cells resulted in reduced proliferation.(49)
As RAS-pathway mutations are frequent in CMML and the MEK-inhibitor trametinib is approved for melanoma and shows single agent clinical activity in RAS mutated patients with myeloid malignancies including CMML(50), we tested the combination treatment of trametinib and azacitidine in vivo. The reduced engraftment at 15 weeks, reduced spleen weight at death and the significantly prolonged survival compared to azacitidine monotherapy suggests that this combination is at least additive in NRAS mutated CMML cells. The additive effect was confirmed in CFC assays with primary, non-transduced patient cells from five CMML patients and by the strong synergistic inhibition of phosphorylated ERK (p-ERK) in MN1-CMML and primary patient cells. All five patients included in the CFC assay lacked mutations in NRAS, but four of the five patients had a mutation in signaling genes that regulate the same pathway (2 patients with CBL mutations, one patient with a cKit mutation and one patient with a JAK2 mutation). This suggests that CMML cells may be sensitive to trametinib independent of NRAS mutations, as they have frequent comutations in other signaling genes. So far, the combination of azacitidine with trametinib has not been tested in CMML patients with or without RAS-pathway mutations, but our data suggest that this combination should be evaluated in a phase II clinical trial.
In summary, we established a transplantable CMML model with NRAS, NOTCH1, DNMT3A, U2AF1 and BCOR mutations and validated the functional importance of mutated NRAS. We identified the combination of azacitidine and trametinib as an effective treatment in NRAS mutated CMML and suggest its clinical development to eventually improve the outcome of CMML patients.
Supplementary Material
Acknowledgments
We would like to thank all participating patients and contributing nurses and doctors, the staff of the Central Animal Facility of Hannover Medical School, and Martin Wichmann, Kerstin Görlich and Karin Battmer for their support. We acknowledge the assistance of the Cell Sorting Core Facility of Hannover Medical School supported in part by the Braukmann-Wittenberg-Herz-Stiftung and the Deutsche Forschungsgemeinschaft. This work was supported by an ERC grant under the European Union’s Horizon 2020 research and innovation program (No. 638035), by grants 109714 and 70112697 from Deutsche Krebshilfe; and DFG grants HE 5240/5-1, HE 5240/5-2, HE 5240/6-1, and HE 5240/6-2.
Footnotes
Competing Interests
The authors declare no competing financial interests.
References
- 1.Arber DA, Orazi A, Hasserjian R, Thiele J, Borowitz MJ, Le Beau MM, et al. The 2016 revision to the World Health Organization classification of myeloid neoplasms and acute leukemia. Blood. 2016;127(20):2391–405. doi: 10.1182/blood-2016-03-643544. [DOI] [PubMed] [Google Scholar]
- 2.Rollison DE, Howlader N, Smith MT, Strom SS, Merritt WD, Ries LA, et al. Epidemiology of myelodysplastic syndromes and chronic myeloproliferative disorders in the United States, 2001-2004, using data from the NAACCR and SEER programs. Blood. 2008;112(1):45–52. doi: 10.1182/blood-2008-01-134858. [DOI] [PubMed] [Google Scholar]
- 3.Williamson PJ, Kruger AR, Reynolds PJ, Hamblin TJ, Oscier DG. Establishing the incidence of myelodysplastic syndrome. British journal of haematology. 1994;87(4):743–5. doi: 10.1111/j.1365-2141.1994.tb06733.x. [DOI] [PubMed] [Google Scholar]
- 4.Wassie EA, Itzykson R, Lasho TL, Kosmider O, Finke CM, Hanson CA, et al. Molecular and prognostic correlates of cytogenetic abnormalities in chronic myelomonocytic leukemia: a Mayo Clinic-French Consortium Study. American journal of hematology. 2014;89(12):1111–5. doi: 10.1002/ajh.23846. [DOI] [PubMed] [Google Scholar]
- 5.Elena C, Galli A, Such E, Meggendorfer M, Germing U, Rizzo E, et al. Integrating clinical features and genetic lesions in the risk assessment of patients with chronic myelomonocytic leukemia. Blood. 2016;128(10):1408–17. doi: 10.1182/blood-2016-05-714030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Onida F, Kantarjian HM, Smith TL, Ball G, Keating MJ, Estey EH, et al. Prognostic factors and scoring systems in chronic myelomonocytic leukemia: a retrospective analysis of 213 patients. Blood. 2002;99(3):840–9. doi: 10.1182/blood.v99.3.840. [DOI] [PubMed] [Google Scholar]
- 7.Itzykson R, Kosmider O, Renneville A, Gelsi-Boyer V, Meggendorfer M, Morabito M, et al. Prognostic score including gene mutations in chronic myelomonocytic leukemia. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2013;31(19):2428–36. doi: 10.1200/JCO.2012.47.3314. [DOI] [PubMed] [Google Scholar]
- 8.Meggendorfer M, Roller A, Haferlach T, Eder C, Dicker F, Grossmann V, et al. SRSF2 mutations in 275 cases with chronic myelomonocytic leukemia (CMML) Blood. 2012;120(15):3080–8. doi: 10.1182/blood-2012-01-404863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kohlmann A, Grossmann V, Klein HU, Schindela S, Weiss T, Kazak B, et al. Next-generation sequencing technology reveals a characteristic pattern of molecular mutations in 72.8% of chronic myelomonocytic leukemia by detecting frequent alterations in TET2, CBL, RAS, and RUNX 1. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2010;28(24):3858–65. doi: 10.1200/JCO.2009.27.1361. [DOI] [PubMed] [Google Scholar]
- 10.Gambacorti-Passerini CB, Donadoni C, Parmiani A, Pirola A, Redaelli S, Signore G, et al. Recurrent ETNK1 mutations in atypical chronic myeloid leukemia. Blood. 2015;125(3):499–503. doi: 10.1182/blood-2014-06-579466. [DOI] [PubMed] [Google Scholar]
- 11.Itzykson R, Kosmider O, Rensneville A, Morabito M, Preudhomme C, Berthon C, et al. Clonal architecture of chronic myelomonocytic leukemias. Blood. 2013;121(12):2186–98. doi: 10.1182/blood-2012-06-440347. [DOI] [PubMed] [Google Scholar]
- 12.Krishnamurthy P, Lim ZY, Nagi W, Kenyon M, Mijovic A, Ireland R, et al. Allogeneic haematopoietic SCT for chronic myelomonocytic leukaemia: a single-centre experience. Bone marrow transplantation. 2010;45(10):1502–7. doi: 10.1038/bmt.2009.375. [DOI] [PubMed] [Google Scholar]
- 13.Pleyer L, Germing U, Sperr WR, Linkesch W, Burgstaller S, Stauder R, et al. Azacitidine in CMML: matched-pair analyses of daily-life patients reveal modest effects on clinical course and survival. Leukemia research. 2014;38(4):475–83. doi: 10.1016/j.leukres.2014.01.006. [DOI] [PubMed] [Google Scholar]
- 14.Patnaik MM, Tefferi A. Chronic myelomonocytic leukemia: 2018 update on diagnosis, risk stratification and management. American journal of hematology. 2018;93(6):824–40. doi: 10.1002/ajh.25104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Padron E, Painter JS, Kunigal S, Mailloux AW, McGraw K, McDaniel JM, et al. GM-CSF-dependent pSTAT5 sensitivity is a feature with therapeutic potential in chronic myelomonocytic leukemia. Blood. 2013;121(25):5068–77. doi: 10.1182/blood-2012-10-460170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kotecha N, Flores NJ, Irish JM, Simonds EF, Sakai DS, Archambeault S, et al. Single-cell profiling identifies aberrant STAT5 activation in myeloid malignancies with specific clinical and biologic correlates. Cancer cell. 2008;14(4):335–43. doi: 10.1016/j.ccr.2008.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang Y, He L, Selimoglu-Buet D, Jego C, Morabito M, Willekens C, et al. Engraftment of chronic myelomonocytic leukemia cells in immunocompromised mice supports disease dependency on cytokines. Blood advances. 2017;1(14):972–9. doi: 10.1182/bloodadvances.2017004903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yoshimi A, Balasis ME, Vedder A, Feldman K, Ma Y, Zhang H, et al. Robust patient-derived xenografts of MDS/MPN overlap syndromes capture the unique characteristics of CMML and JMML. Blood. 2017;130(4):397–407. doi: 10.1182/blood-2017-01-763219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu PP, Hajra A, Wijmenga C, Collins FS. Molecular pathogenesis of the chromosome 16 inversion in the M4Eo subtype of acute myeloid leukemia. Blood. 1995;85(9):2289–302. [PubMed] [Google Scholar]
- 20.Valk PJ, Verhaak RG, Beijen MA, Erpelinck CA, Barjesteh van Waalwijk van Doorn Khosrovani S, Boer JM, et al. Prognostically useful gene-expression profiles in acute myeloid leukemia. The New England journal of medicine. 2004;350(16):1617–28. doi: 10.1056/NEJMoa040465. [DOI] [PubMed] [Google Scholar]
- 21.Heuser M, Beutel G, Krauter J, Dohner K, von Neuhoff N, Schlegelberger B, et al. High meningioma 1 (MN1) expression as a predictor for poor outcome in acute myeloid leukemia with normal cytogenetics. Blood. 2006;108(12):3898–905. doi: 10.1182/blood-2006-04-014845. [DOI] [PubMed] [Google Scholar]
- 22.Langer C, Marcucci G, Holland KB, Radmacher MD, Maharry K, Paschka P, et al. Prognostic importance of MN1 transcript levels, and biologic insights from MN1-associated gene and microRNA expression signatures in cytogenetically normal acute myeloid leukemia: a cancer and leukemia group B study. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2009;27(19):3198–204. doi: 10.1200/JCO.2008.20.6110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Metzeler KH, Dufour A, Benthaus T, Hummel M, Sauerland MC, Heinecke A, et al. ERG expression is an independent prognostic factor and allows refined risk stratification in cytogenetically normal acute myeloid leukemia: a comprehensive analysis of ERG, MN1, and BAALC transcript levels using oligonucleotide microarrays. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2009;27(30):5031–8. doi: 10.1200/JCO.2008.20.5328. [DOI] [PubMed] [Google Scholar]
- 24.Carella C, Bonten J, Sirma S, Kranenburg TA, Terranova S, Klein-Geltink R, et al. MN1 overexpression is an important step in the development of inv(16) AML. Leukemia. 2007;21(8):1679–90. doi: 10.1038/sj.leu.2404778. [DOI] [PubMed] [Google Scholar]
- 25.Heuser M, Argiropoulos B, Kuchenbauer F, Yung E, Piper J, Fung S, et al. MN1 overexpression induces acute myeloid leukemia in mice and predicts ATRA resistance in patients with AML. Blood. 2007;110(5):1639–47. doi: 10.1182/blood-2007-03-080523. [DOI] [PubMed] [Google Scholar]
- 26.Liu T, Jankovic D, Brault L, Ehret S, Baty F, Stavropoulou V, et al. Functional characterization of high levels of meningioma 1 as collaborating oncogene in acute leukemia. Leukemia. 2010;24(3):601–12. doi: 10.1038/leu.2009.272. [DOI] [PubMed] [Google Scholar]
- 27.Heuser M, Yun H, Berg T, Yung E, Argiropoulos B, Kuchenbauer F, et al. Cell of origin in AML: susceptibility to MN1-induced transformation is regulated by the MEIS1/AbdB-like HOX protein complex. Cancer cell. 2011;20(1):39–52. doi: 10.1016/j.ccr.2011.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Imren S, Heuser M, Gasparetto M, Beer PA, Norddahl GL, Xiang P, et al. Modeling de novo leukemogenesis from human cord blood with MN1 and NUP98HOXD13. Blood. 2014;124(24):3608–12. doi: 10.1182/blood-2014-04-564666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jyotsana N, Sharma A, Chaturvedi A, Budida R, Scherr M, Kuchenbauer F, et al. Lipid nanoparticle-mediated siRNA delivery for safe targeting of human CML in vivo. Annals of hematology. 2019;98(8):1905–18. doi: 10.1007/s00277-019-03713-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chaturvedi A, Araujo Cruz MM, Jyotsana N, Sharma A, Goparaju R, Schwarzer A, et al. Enantiomer-specific and paracrine leukemogenicity of mutant IDH metabolite 2-hydroxyglutarate. Leukemia. 2016;30(8):1708–15. doi: 10.1038/leu.2016.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Thol F, Gabdoulline R, Liebich A, Klement P, Schiller J, Kandziora C, et al. Measurable residual disease monitoring by NGS before allogeneic hematopoietic cell transplantation in AML. Blood. 2018;132(16):1703–13. doi: 10.1182/blood-2018-02-829911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Heuser M, Gabdoulline R, Loffeld P, Dobbernack V, Kreimeyer H, Pankratz M, et al. Individual outcome prediction for myelodysplastic syndrome (MDS) and secondary acute myeloid leukemia from MDS after allogeneic hematopoietic cell transplantation. Annals of hematology. 2017;96(8):1361–72. doi: 10.1007/s00277-017-3027-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(T)(-Delta Delta C) method. Methods. 2001;25(4):402–8. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 34.Chaturvedi A, Araujo Cruz MM, Jyotsana N, Sharma A, Yun H, Gorlich K, et al. Mutant IDH1 promotes leukemogenesis in vivo and can be specifically targeted in human AML. Blood. 2013;122(16):2877–87. doi: 10.1182/blood-2013-03-491571. [DOI] [PubMed] [Google Scholar]
- 35.Chaturvedi A, Herbst L, Pusch S, Klett L, Goparaju R, Stichel D, et al. Pan-mutant-IDH1 inhibitor BAY1436032 is highly effective against human IDH1 mutant acute myeloid leukemia in vivo. Leukemia. 2017;31(10):2020–8. doi: 10.1038/leu.2017.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Reimer J, Knoss S, Labuhn M, Charpentier EM, Gohring G, Schlegelberger B, et al. CRISPRCas9-induced t(11;19)/MLL-ENL translocations initiate leukemia in human hematopoietic progenitor cells in vivo. Haematologica. 2017;102(9):1558–66. doi: 10.3324/haematol.2017.164046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Barabe F, Kennedy JA, Hope KJ, Dick JE. Modeling the initiation and progression of human acute leukemia in mice. Science. 2007;316(5824):600–4. doi: 10.1126/science.1139851. [DOI] [PubMed] [Google Scholar]
- 38.Barabe F, Gil L, Celton M, Bergeron A, Lamontagne V, Roques E, et al. Modeling human MLL-AF9 translocated acute myeloid leukemia from single donors reveals RET as a potential therapeutic target. Leukemia. 2017;31(5):1166–76. doi: 10.1038/leu.2016.302. [DOI] [PubMed] [Google Scholar]
- 39.Wei J, Wunderlich M, Fox C, Alvarez S, Cigudosa JC, Wilhelm JS, et al. Microenvironment determines lineage fate in a human model of MLL-AF9 leukemia. Cancer cell. 2008;13(6):483–95. doi: 10.1016/j.ccr.2008.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ricci C, Fermo E, Corti S, Molteni M, Faricciotti A, Cortelezzi A, et al. RAS mutations contribute to evolution of chronic myelomonocytic leukemia to the proliferative variant. Clinical cancer research : an official journal of the American Association for Cancer Research. 2010;16(8):2246–56. doi: 10.1158/1078-0432.CCR-09-2112. [DOI] [PubMed] [Google Scholar]
- 41.Klinakis A, Lobry C, Abdel-Wahab O, Oh P, Haeno H, Buonamici S, et al. A novel tumour-suppressor function for the Notch pathway in myeloid leukaemia. Nature. 2011;473(7346):230–3. doi: 10.1038/nature09999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tefferi A, Mudireddy M, Finke CM, Nicolosi M, Lasho TL, Hanson CA, et al. U2AF1 mutation variants in myelodysplastic syndromes and their clinical correlates. American journal of hematology. 2018;93(6):E146–E8. doi: 10.1002/ajh.25084. [DOI] [PubMed] [Google Scholar]
- 43.Fei DL, Zhen T, Durham B, Ferrarone J, Zhang T, Garrett L, et al. Impaired hematopoiesis and leukemia development in mice with a conditional knock-in.allele of a mutant splicing factor gene U2af 1. Proceedings of the National Academy of Sciences of the United States of America; 2018. pp. E10437–E46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lee SC, Dvinge H, Kim E, Cho H, Micol JB, Chung YR, et al. Modulation of splicing catalysis for therapeutic targeting of leukemia with mutations in genes encoding spliceosomal proteins. Nature medicine. 2016;22(6):672–8. doi: 10.1038/nm.4097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Patnaik MM, Barraco D, Lasho TL, Finke CM, Hanson CA, Ketterling RP, et al. DNMT3A mutations are associated with inferior overall and leukemia-free survival in chronic myelomonocytic leukemia. American journal of hematology. 2017;92(1):56–61. doi: 10.1002/ajh.24581. [DOI] [PubMed] [Google Scholar]
- 46.Steensma DP, Bejar R, Jaiswal S, Lindsley RC, Sekeres MA, Hasserjian RP, et al. Clonal hematopoiesis of indeterminate potential and its distinction from myelodysplastic syndromes. Blood. 2015;126(1):9–16. doi: 10.1182/blood-2015-03-631747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Russler-Germain DA, Spencer DH, Young MA, Lamprecht TL, Miller CA, Fulton R, et al. The R882H DNMT3A mutation associated with AML dominantly inhibits wild-type DNMT3A by blocking its ability to form active tetramers. Cancer cell. 2014;25(4):442–54. doi: 10.1016/j.ccr.2014.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Challen GA, Sun D, Jeong M, Luo M, Jelinek J, Berg JS, et al. Dnmt3a is essential for hematopoietic stem cell differentiation. Nature genetics. 2011;44(1):23–31. doi: 10.1038/ng.1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhao Z, Wu Q, Cheng J, Qiu X, Zhang J, Fan H. Depletion of DNMT3A suppressed cell proliferation and restored PTEN in hepatocellular carcinoma cell. Journal of biomedicine & biotechnology. 2010:737535. doi: 10.1155/2010/737535. 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Borthakur G, Popplewell L, Boyiadzis M, Foran J, Platzbecker U, Vey N, et al. Activity of the oral mitogen-activated protein kinase kinase inhibitor trametinib in RAS-mutant relapsed or refractory myeloid malignancies. Cancer. 2016;122(12):1871–9. doi: 10.1002/cncr.29986. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.