Abstract
Myelodysplastic syndrome (MDS) are clonal stem cell diseases characterized mainly by ineffective hematopoiesis. Here, we present an approach that enables robust long-term engraftment of primary MDS stem cells (MDS-SCs) in mice by implantation of human mesenchymal cell-seeded scaffolds. Critically for modelling MDS, where patient sample material is limiting, mononuclear bone marrow cells containing as few as 104 CD34+ cells can be engrafted and expanded by this approach with the maintenance of the genetic make-up seen in the patients. Non-invasive high-resolution ultrasound imaging shows that these scaffolds are fully perfused. Our data shows that human microenvironment but not mouse is essential to MDS-SCs homing and engraftment. Notably, the alternative niche provided by healthy donor MSCs enhanced engraftment of MDS-SCs. This study characterizes a new tool to model MDS human disease with the level of engraftment previously unattainable in mice, and offers insights into human-specific determinants of MDS-SC microenvironment.
Introduction
Myelodysplastic syndromes (MDS) are a group of hematopoietic stem cell (HSC) disorders (1,2), with a high propensity to transform to acute myeloid leukemia (AML). In a spectrum of myeloid disorders ranging from age-related clonal haematopoiesis (ARCH) to AML, MDS is mainly distinguished by the presence of peripheral blood cytopenias, dysplastic hematopoietic differentiation, and the lack of features that define acute leukemia. MDS disease is driven by a complex combination of somatic gene mutations and/or chromosomal abnormalities, particularly targeting the myeloid lineage (3–6).
Understanding the biology of MDS-SCs and decoding their interaction with the BM microenvironment remains a major challenge due to the lack of reliable in vivo disease models, therefore impeding the translational MDS research. Attempts to generate and use humanized transgenic mice have provided limited improvement in generating xenograft models for MDS (7,8). Although co-injection of MSCs along with MDS CD34+ cells into the murine bone marrow was initially suggested to help MDS engraftment in patient derived xenograft (PDX) models (7); however, subsequent studies showed no beneficial effect (8,9). These efforts to construct a model that replicates the cellular human BM niche have been limited until now, as they have been based on simple injection of MDS HSPCs into murine hematopoietic tissue. This has also restricted our ability to map the specific interactions that may exist in the human BM microenvironment (10–12). Being able to study these interactions in more physiological humanized conditions is essential that will allow us to better understand the intercellular signalling that we anticipate may be critical in the initiation, maintenance and progression of MDS. Recent advances in bioengineering have enabled the integration of novel biomaterials into developmental biology. These biomaterials provide a versatile tool to create a humanized microenvironment in immunodeficient mouse models (13). These ‘special niches’ are invaluable for providing architectural support for cell attachment, cellular differentiation and tissue development, therefore enabling key cell-cell biological interactions.
Results
MDS bone marrow stem cells robustly engraft in in vivo 3D humanized scaffolds
PDX mouse models have proven their reliability in recapitulating features of malignant hematopoiesis, particularly in acute leukemia (13–17). However, attempts to recapitulate this success in other more chronic hematopoietic malignancies such as MDS has yielded little or no success (3,6,8–12). This prompted us to develop an alternative in vivo system that enables rapid and reliable assessment of the hematopoietic stem and progenitor cells (HSPCs). This xenotransplantation system uses gelatin-based porous scaffolds (hereafter defined as humanized scaffolds) to generate niches in mice that mimic the human-specific microenvironment.
Our study is based on 37 patients (MDS-MLD =20, MDS-EB =4, MDS-U =2; MDS-SLD-RS=3, MDS-MLD-RS=4, CMML=2, MDS/MPN=1 and sAML, n=1) (Table S1–S3) and healthy donors (n=6). Whole-exome sequencing (WES) or myeloid-specific gene panel screening demonstrated a mutational distribution (Figure S1A) as previously reported (4,5). MDS patient BM MSCs isolated to be used for the in vivo experiments showed variable expansion levels, that is consistent with previous reports (18).
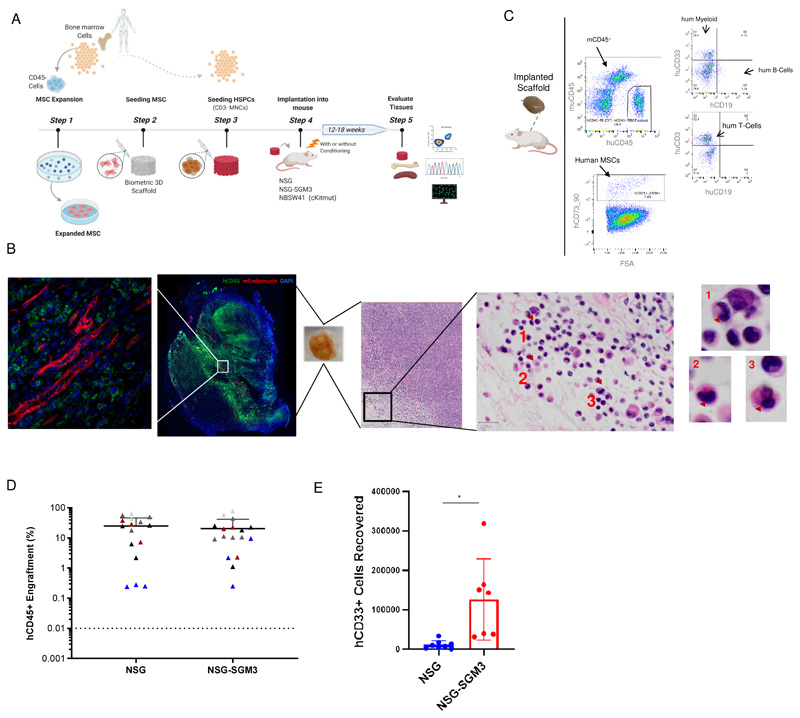
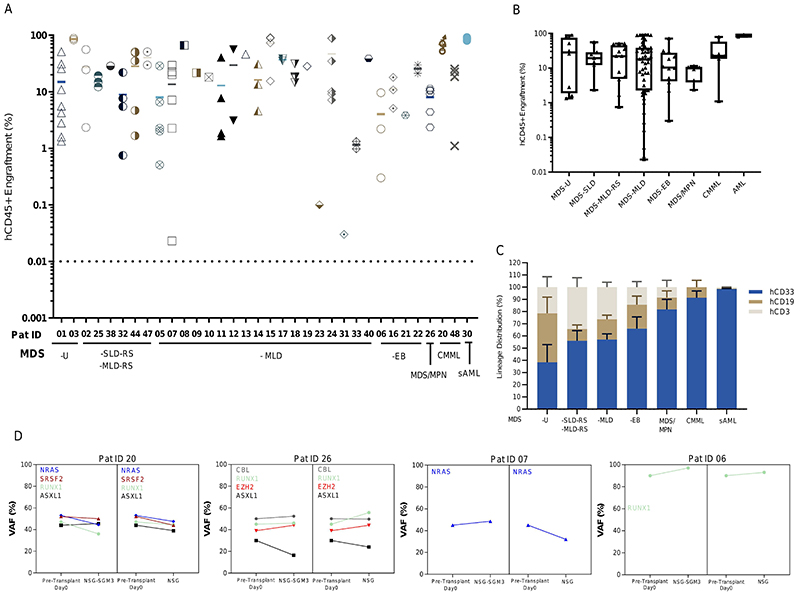
Initially, we chose to screen five patients in NSG and NSG-SGM3 mice by injecting BM CD3-depleted mononuclear cells (MNCs, hereafter defined as HSPCs) (0.25x106 to 0.75x106) into scaffolds that were pre-seeded with autologous (or allogenic MDS MSCs, where autologous MSCs were not available) patient derived MSCs (Figure 1A) and subcutaneously transplanted in mice. Humanized scaffold tissues recovered 12-18 weeks post-implantation showed uniform distribution of human CD45+ (hCD45+) cells throughout the scaffold as well as the presence of murine vasculature, with hCD45+ cells present adjacent to the murine vascular structures within the humanized scaffolds. Additionally, murine CD45+ cells are also observed in these humanized scaffolds but relatively fewer in numbers (Figure 1, B and C, Figure S1B). These humanized scaffolds act as a framework to support cell proliferation and differentiation, and not only maintains the primary cellular phenotype and function, but also the dysplastic morphology (Figure 1B, right panel; Figure S1C) typically observed in MDS bone marrow and blood. Although there was no significant difference in the hCD45+ cell engraftment between NSG and NSG-SGM3 mice (Figure 1D), humanized scaffolds harvested from the NSG-SGM3 mice appeared larger compared to the ones obtained from NSG mice (Figure S1D) and the recovery of human myeloid cells (CD45+CD33+) was significantly higher from NSG-SGM3 mice (Figure 1E). We did not observe any significant increase in the engraftment levels in the humanized scaffolds when mice were kept alive for up to 24 weeks (NSG-SGM3 12 weeks median hCD45+ cells = 10.6%, 24 weeks hCD45+ cells = 4.4%; P value = 0.1) (Figure S1E). We therefore opted to use our 3D humanized scaffold system in NSG-SGM3 mice to study a cohort additional MDS patients, covering a range of MDS risk groups and closely related myeloid malignancies (Table S2). We observed persistent long-term engraftment of hCD45+ cells within the humanized scaffolds ranging from 0.2% to 86%, with 82% of cases having ≥20% hCD45+ cells (Figure 2A). There was no significant difference in the engraftment levels between low-risk cases (mean hCD45+ cells, MDS-U = 35.5%, MDS-SLD = 22.5%, MDS-MLD-RS = 30.4%, MDS-MLD = 30.7%) and high-risk MDS cases (mean hCD45+ cells, MDS-EB = 21.88%) (Figure 2, A and B). In some cases, such as patients 23 and 31, the average engraftment of MDS cells was low (hCD45+ cells <1%). The humanized scaffolds in these mice showed multi-lineage engraftment of human hematopoiesis. Notably, mice that received HSPCs from MDS-EB cases had higher myeloid lineage engraftment compared to low-risk MDS cases (MDS-SLD-RS hCD33+ 44%; MDS-MLD hCD33+ 46.6%; MDS-EB hCD33+ 65%) (Figure 2C). Similarly, myeloid bias was also observed in mice transplanted with MDS/MPN, CMML and sAML BM HSPCs. Taken together, our data provides a reliable in vivo model system that can be used to robustly engraft MDS HSPCs in preclinical studies.
Figure 1. Humanized scaffold system in NSG and NSG-SGM3 immunodeficient mice.
(A) Schematic representation of the in vivo protocol used for generating humanized scaffolds. Illustration was created with BioRender.com (B) Representative example of Immunofluorescence and H&E (Hematoxylin and eosin) staining for the scaffold retrieved following the xenotransplantation. H&E staining of a section of the engrafted scaffold shows multiple neutrophils with hypolobated nuclei (examples indicated with arrows), consistent with dysplasia in the granulocytic lineage. (C) Representative flow cytometry plot of the cells retrieved from the humanized scaffolds following the xenotransplantation. (D) Comparison of hCD45+ cells engraftment in humanized scaffolds retrieved from NSG and NSG-SGM3 mice. (E) Absolute cell counts of human myeloid (hCD45+hCD33+) cells retrieved from humanized scaffolds implanted in NSG and NSG-SGM3 mice. BM- Bone marrow cells, MSC- Mesenchymal stromal cells.
Figure 2. Humanized niches in immunodeficient mice enable the robust engraftment of MDS HSPCs.
(A) Total hCD45+ cell engraftment in humanized scaffolds implanted in NSG-SGM3 mice. (B) Comparison of total hCD45+ cell engraftment in humanized scaffolds following xenotransplantation in various MDS WHO subtypes and other related myeloid malignancies. (C) Lineage distribution within the hCD45+ cells recovered from the humanized scaffolds implanted in NSG-SGM3 mice. (D) Mutational spectrum of the MDS HSPCs in primary pre-transplanted cells and post-transplanted cells from humanized MDS scaffolds in NSG and NSG-SGM3 mice. VAF for xenografted samples is the average between 2-3 mice (where applicable). VAF- Variant allele frequency.
HSPCs carrying MDS associated gene mutations constitute a reservoir of pre-leukemic stem cells (3,5,6) that undergo evolution acquiring additional mutations, leading to transformation to acute myeloid leukemia (AML). We used DNA molecular testing to compare the mutations present in the engrafted cells harvested from the humanized scaffolds in the mice with those that were initially identified in the patients’ primary bone marrow HSPCs (Table S3). Patient-specific targeted mutation screening was performed on the harvested hCD45+hCD33+ cells by next-generation MiSeq sequencing. Our analysis showed that in the implanted humanized scaffolds, variant allele frequency of the mutations was largely maintained in both NSG as well as NSG-SGM3 mouse models (Figure 2D, Figure S2 and Table S4). Interestingly, mutations that have been previously described as disease initiating such as SF3B1, DNMT3A, SRSF2 and TET2, as well as those associated with disease progression such as ASXL1, NRAS, RUNX1, were all maintained in the humanized scaffolds in all the cases, indicating that these humanized niches in mice maintain the original clonal MDS architecture.
Humanized scaffolds enable the maintenance of long-term self-renewal capacity of MDS-SCs
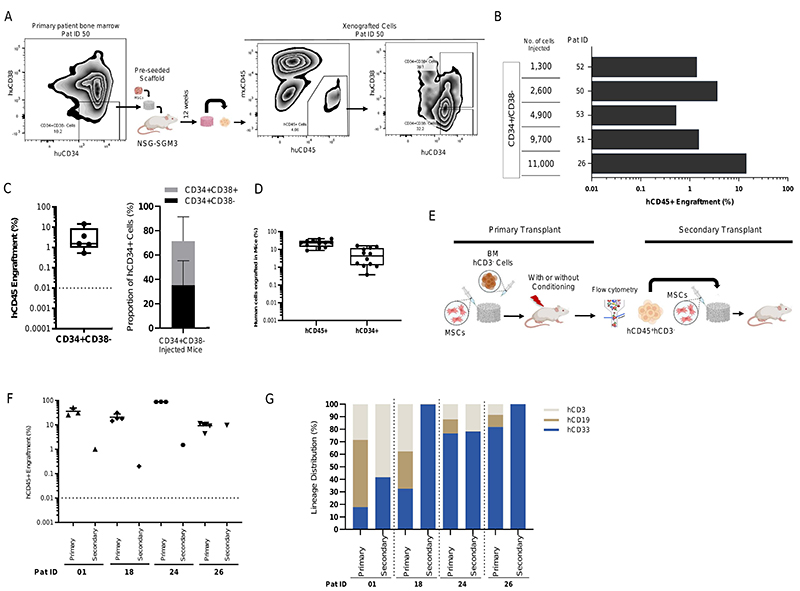
Long-term self-renewal ability has been traditionally used to define stemness of the HSCs. We first sought to determine if the humanized niches implanted in the immunodeficient mice were able to maintain the long-term MDS-SC. MDS CD34+CD38- stem cells were flow sorted, seeded into the humanized niches and then transplanted into the NSG-SGM3 mice. Even though we transplanted variable number of CD34+CD38- stem cells (range 1300 to 11000 CD34+CD38-), engraftment of these MDS stem cells was observed in all the cases (Figure 3, A, B and C). Interestingly, MDS CD34+ HSPCs were detected at high frequency in CD34+CD38- injected mice, (mean hCD34+CD38- = 35.24%; hCD34+CD38+ = 36.26) than in BM MNC CD3- injected mice, (mean hCD34+=6.1%) in primary transplants even up to 18 weeks post xenotransplantation (Figure 3C-3D). In addition, CD34+CD38- also differentiated to mature progeny (CD34+C38+) within these humanized scaffolds.
Figure 3. MDS stem cells engraft in humanized niches in NSG-SGM3 mice and demonstrate long-term self-renewal capacity.
(A) Representative flow cytometry plot showing the day 0 patient bone marrow CD34/CD38 phenotype and engraftment of hCD45+ cells in humanized scaffolds as well as the hCD34/hCD38 phenotype of these xenografted cells in NSG-SGM3 mice. (B) Total hCD45+ cell engraftment in humanized scaffolds that were seeded with CD34+CD38- MDS stem cells in NSG-SGM3 mice. (C) Percentage of hCD45+ cells (left panel) in mice injected with CD34+CD38- MDS cells and proportion of CD34+C38-/CD34+CD38+ cells within hCD34+ HSPCs in these humanized scaffolds retrieved from NSG-SGM3 mice. (D) Percentage of hCD45+ cells and proportion of hCD34+ cells within the hCD45+ cell fraction in the humanized scaffolds that were seeded with MDS CD3- MNCs. (E) Schematic representation of the in vivo protocol for secondary transplantation. (F) Percentage of total hCD45+ cells engrafted in humanized niches from primary and secondary NSG-SGM3 mice. (G) Lineage distribution within the hCD45+ cells in humanized niches implanted in primary and secondary NSG-SGM3 mice.
Next, we tested whether the MDS-SCs present in the humanized scaffolds in the primary mice had self-renewal capacity by performing secondary transplantation assays. Following the primary xenotransplantation, hCD45+CD3- cells were isolated by fluorescence-activated cell sorting (FACS) from mice previously engrafted with MDS BM CD3- cells (Figure 3E). Four patients were used for serial transplantation. Variable number of hCD45+CD3- cells were transplanted into the 3D scaffolds in the mice (Table S5). Twelve weeks following xenotransplantation in the secondary recipients, all mice demonstrated human cell engraftment in the humanized niches (Figure 3F). For patient 26, engraftment in the secondary recipient (hCD45+ =10%) was similar to that observed in the primary transplants. However, for the other three patients, which were all high-engrafters in the primary transplant settings, we observed hCD45+ cell engraftment levels between 0.1% to 1.5% in the secondary recipients. Furthermore, even though T cell depleted (hCD45+CD3-) MDS HSPCs were injected into the secondary recipients, we observed mixed lineage engraftment in two out of four cases (Figure 3G). Patients 18 and 26 displayed exclusive myeloid lineage engraftment in the secondary recipients. Although the engraftment in some secondary recipients was relatively reduced compared to the corresponding primary transplants, this could be due to the intrinsic ageing of the BM HSCs, as similar results have also been observed in normal aged mouse HSCs elsewhere (19,20), but also clearly depend on patient samples. Therefore, our data demonstrates that irrespective of the disease risk-group and the genomic mutational architecture, the humanized scaffold in vivo system can support long-term self-renewal activity of the malignant MDS-SCs.
MDS HSPCs are highly dependent on humanized niches
Previous studies have shown that MDS patient BM HSPCs can engraft in the immunodeficient murine BM microenvironment (3,8,9), albeit at a low level. Therefore, we wanted to determine if MDS HSPCs engrafted in the humanized niches could colonize the murine bone marrow microenvironment. In the majority of the cases, MDS HSPCs were not detected in the BM of sub-lethally irradiated NSG or NSG-SGM3 mice (Figure S3A). Only three mice had low levels of hCD45+ cells engrafted in the BM. In order to determine whether the humanized scaffolds had sufficiently perfused vasculature to allow migration of human HSPCs to the mouse BM, we next assessed the perfusion of the scaffolds by high-resolution ultrasound imaging of live mice following injection of micro-bubbles via the tail vein. The flow of the micro-bubbles through the mouse vasculature system demonstrated that these humanized scaffolds are fully perfused (Supplementary video 1). The photoacoustic imaging of biological tissue-engineered scaffolds in live mice provides a novel and promising, non-invasive and non-destructive method for assessing vascular and endothelial alterations that have been previously associated with myeloid malignancies, particularly AML (21).
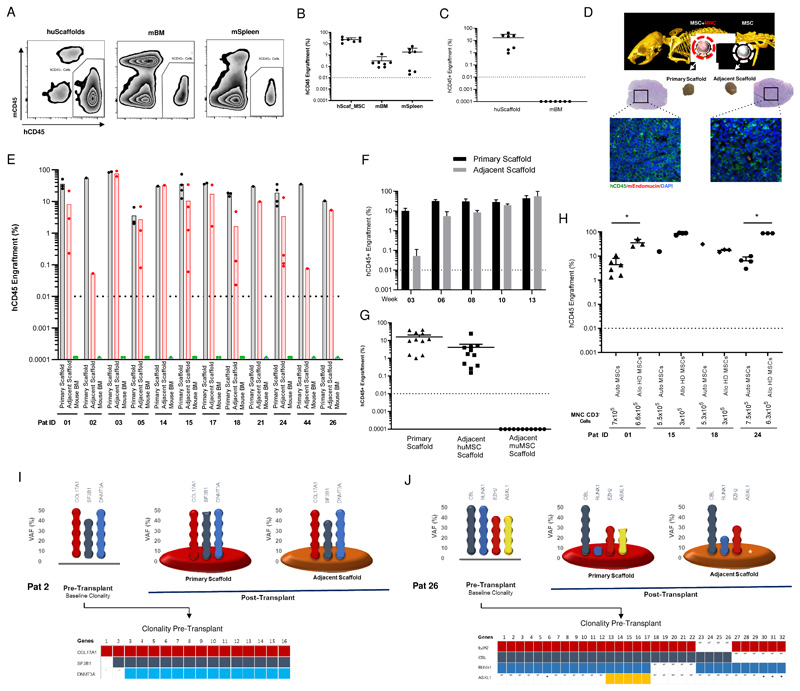
These data prompted us to use a more permissive mouse model (NSGW41) (22) where the endogenous murine HSCs have no advantage, thus enabling engraftment of human HSPCs without myeloablative irradiation. We first used NSGW41 and NSG-SGM3 immunodeficient mice to test if BM cells from healthy adult individuals (n=6 adult donors) can engraft in the humanized scaffolds and whether these cells can both migrate and colonize murine hematopoietic tissues. Following xenotransplantation, healthy adult donor HSPCs injected into the humanized scaffolds were able to engraft not only in the humanized niches but also in the murine BM and spleen at 12 weeks (Figure 4, A and B). The migratory behavior of healthy stem cells has been reported previously in healthy animals and humans (23–25). In contrast, transplantation of MDS HSPCs in NSGW41 mice yielded hCD45+ cell engraftment only in the humanized biometric scaffolds (Figure 4C). This behavior of MDS HSPCs led us to design an experiment to assess whether the MDS-SCs were unable to migrate through the vasculature or specifically failed in colonizing the murine hematopoietic tissues. In addition to the primary humanized scaffold containing MDS HSPCs, we implanted an adjacent MSC seeded ‘empty scaffold’ to provide an uncolonized humanized niche in NSG-SGM3 and NSGW41 mice (Figure 4D). Following xenotransplantation, hCD45+ engraftment was quantified in the primary scaffold and adjacent human MSC-seeded but not to the murine bone marrow. Our data clearly demonstrate that MDS HSPCs do migrate out of the primary scaffold and colonize the adjacent humanized scaffolds (Figure 4, D and E; Figure S3B-E). It is noteworthy that human hematopoietic cells start migrating from primary to adjacent scaffold as early as week 3 (Figure 4F), before reaching a plateau at week 10-13. This migratory property of MDS-SCs was observed in all the cases studied here, irrespective of the disease risk groups. Migration of the MDS-SC occurred regardless of the location or distance between the primary and adjacent scaffolds. Notably, when NSG-SGM3 murine MSCs were used in adjacent scaffolds along with human MSCs in another adjacent scaffold, MDS HSPCs substantially migrated and homed only in adjacent niche with human MSCs and not in adjacent niche seeded with murine MSCs (Figure 4G). This suggest that in a more permissive NSGW41 mouse model, healthy donor HSPCs can relatively compete with murine counterparts in the murine niche, however this potent fitness is lacking in MDS HSPCs. Altogether, our data demonstrate that human MDS-SCs are entirely dependent on a humanized microenvironment in order to home and reconstitute, therefore implying a cross-talk between the MDS-SCs and the humanized niche supporting cells (in this case hMSCs) that may be critical for establishment and progression of the disease.
Figure 4. MDS HSPCs are highly dependent on the human MSC niche.
(A) Representative flow cytometry plot showing the engraftment of healthy hCD45+ cells in humanized niches, murine bone marrow and spleen in NSGW41 mice. (B) Percentage of healthy donor hCD45+ cells in humanized niche, murine bone marrow and murine spleen in NSGW41 and/or NSG-SGM3 mice. (C) Percentage of MDS hCD45+ cells in humanized scaffolds and murine bone marrow in NSGW41 mice. (D) Representative plot (Immunofluorescence and H&E staining) of the primary scaffold and adjacent scaffold after xenotransplantation in NSG-SGM3 mice. (E) Percentage of total hCD45+ cells in primary scaffold, adjacent scaffold and murine bone marrow after xenotransplantation in NSG-SGM3 mice. (F) In vivo migration kinetics of hCD45+ cells in the primary and adjacent scaffolds in NSG-SGM3 mice at various time points. (G) Percentage of MDS hCD45+ cells in humanized primary niche, adjacent niche with human MSCs and adjacent niche with murine MSCs in NSG-SGM3 mice. (H) Percentage of total hCD45+ cells in the humanized scaffolds that were seeded either with autologous MDS MSCs or allogenic healthy donor MSCs. (I, J) MDS patient clonal distribution in the primary pre-transplanted HSPCs, primary and adjacent scaffolds following the xenotransplantation. Bottom part of each figure shows the clonal spectrum at single cell level (single cell clonogenic assay) of primary day 0 pre-transplanted HSPCs for each patient. (A-H) Mice were injected with CD3- MNCs. huScaffold- humanized scaffold, mBM- Mouse bone marrow, mSpleen- Mouse Spleen. huMSC-human mesenchymal stromal cells, muMSC-Murine mesenchymal stromal cells. * P value <0.05.
Healthy donor MSCs could also provide niche support for MDS stem cells
Emerging evidence from our data and others suggests that there are functional deficits in the proliferation and differentiation of MDS MSCs (18). It is also becoming increasingly clear that alterations in the ageing BM microenvironment can induce changes in hematopoiesis and that this effect may be exacerbated among MDS BM components due to the dysregulated inflammatory microenvironment (26–28). This led us to assess whether MSCs derived from BM of the MDS patients are best to provide support to malignant HSPCs. Therefore, we compared the supportive capabilities of autologous MSCs (4 patients) with allogenic healthy donor MSCs using our humanized scaffold system in NSG-SGM3 mice. Both types of scaffolds were seeded with the same number of MSCs (≈5x105 cells per scaffold) and similar numbers of MDS HSPCs were injected into all of these scaffolds. Notably, the engraftment of MDS CD45+ cells was similar and in 3 out of 4 cases even higher in mice that were implanted with scaffolds seeded with healthy donor MSCs (Figure 4H). In patient 01, engraftment of hCD45+ cells increased from 4.4% (in autologous seeded MSCs) to 35.9% (in healthy donor seeded MSCs). In patient 24, this increase was significantly higher: from 6.9% (in autologous seeded MSCs) to 88.7% (in healthy donor seeded MSCs). Our data suggest that MSCs derived from healthy donors are functionally able to support MDS-SCs, even possibly better than autologous MDS-MSCs. Given the importance of this preliminary observation, further studies with larger patient cohorts and investigation of the clonal distribution in the engrafted cells will be needed to confirm this behavior.
MDS HSPC clones migrate between the humanized niches
Having observed the migratory behavior of MDS HSPCs in in vivo humanized scaffolds, we next studied the clonal composition of the cells from the primary scaffold and those cells that had migrated into the adjacent scaffold. DNA sequencing analysis of the xenografted cells revealed that the mutant clones that migrate to the adjacent scaffolds are genetically similar (Figure 4, I and J, Figure S3F, Table S4). For example, the clonality of the engrafted HSPCs in patient 02 was maintained in both primary and adjacent scaffolds (Figure 4I). Interestingly, this patient had comparatively few subclones in the pre-transplanted bone marrow HSPCs at day 0 (Figure 4I). In contrast, a multiclonal pattern was seen in the BM HSPCs of patient 26 prior to xenotransplantation (day 0) (Figure 4J). Our single cell clonogenic assay revealed that at day 0 BM HSPCs from patient 26 had multiple subclones with EZH2 as the founding mutation while the other gene aberrations were acquired in a sequential order (Figure 4J). Interestingly, a clonal fluctuation within the HSPCs was observed in the engrafted cells in the primary scaffolds. On the other hand, HSPC clones that had migrated into the adjacent scaffold lacked ASXL1 mutations that was detected in both the primary scaffold and day 0 HSPCs. This data demonstrates the migratory behavior of malignant HSPCs and of potential differential motility behavior in in vivo settings of some subclones.
Discussion
Understanding the biology of MDS-SCs and their interaction with the BM microenvironment remains a major challenge due to the lack of reliable in vivo disease models, therefore impeding the translational MDS research. Here, for the first time, we provide an in vivo modelling approach to create a humanized microenvironment for MDS-SCs. These humanized scaffolds act as a framework to support cell proliferation and differentiation, and not only maintains the primary cellular phenotype and function, but also the dysplastic morphology typically observed in MDS bone marrow and blood. MDS patient derived MSCs were essential to create a stromal layer on the carrier biomaterial (or humanized scaffold) that acts as a ‘HSPC supportive niche’, when the scaffolds are implanted into immunodeficient mice. These extramedullary humanized niches supported both human normal and malignant hematopoietic cells. Using high resolution ultrasound imaging, we were able to show that these biometric scaffolds are vascularized as well as fully perfused. Remarkably, this humanized scaffold approach enabled engraftment of HSCs from ≈94% of the MDS patients tested here, independent of MDS subtypes. The disease/age-associated lineage skewing that is often observed clinically and in MDS mouse models (29–32) was also captured in this in vivo system, in which we observed a myeloid differentiation bias that was particularly pronounced in high-risk cases. We were also able to show that MDS-SCs engraft and remained in primary transplants. Using secondary transplantation assays, we were able to functionally demonstrate their self-renewal and differentiation capacity within the in vivo humanized niches, thereby proving their stem cell capability. Although the engraftment in some secondary recipients was relatively reduced compared to the corresponding primary transplants, this could be due to the intrinsic ageing of the BM HSCs, as similar results have also been observed in normal aged mouse HSCs elsewhere (19,20). Furthermore, in the knock-in MISTRG mouse model, similar engraftment in secondary transplants for low-risk MDS cases has been reported (33). The heterogeneity present amongst MDS patients is also reflected in engraftment levels in our secondary transplants. Given the robustness of our scaffold-based system in generating primary as well as secondary transplants, it will be interesting to test more samples and also evaluate our scaffold model in MISTRG mouse knock-in strain, and investigate whether further humanization of our scaffolds via the use of human endothelial cells for example could further improve the maintenance of long-term self-renewing MDS-SCs. Our quantitative targeted mutational analysis demonstrated the maintenance of the mutations that were present in the pre-transplanted cells, indicating that these humanized niches in immunodeficient mice retain the original subclonal architecture of the MDS patients.
In normal adults, HSCs continuously migrate between the BM and blood, as such they are always available to exit circulation in order to fill empty BM HSC niches (23,25). Our data also demonstrates that HSPCs from healthy individuals migrated from human to human niche as well as to the murine hematopoietic niches. This behavior was not seen in MDS-SCs. Interestingly, MDS-SCs were able to migrate out of the primary niche but were subsequently homing and engrafting only in niches seeded with human MSCs. This preferential human niche requirement was observed even when these mice were maintained for 6 months after implantation of scaffolds. The preferential migratory behavior of primary MDS malignant stem cells in vivo setting is consistent with some previous reports in AML, ALL (34,35) and further studies are needed to understand the dynamics of the migration and if this is associated with more aggressive MDS-SC properties. Our model provides a unique opportunity to study the clonal behavior of the cancers in general and to understand how these cancerous cells interact with different types of niches (for example, healthy vs malignant MSCs, young vs aged MSCs and role of endothelial cells).
It is becoming increasingly clear that alterations in the ageing BM microenvironment can induce changes in hematopoiesis and that this effect may be exacerbated among MDS BM components due to the dysregulated inflammatory microenvironment (26–28). The change in MDS microenvironment was noticeable from our in vivo data, as the alternative niche provided by HD MSCs seems to enhance engraftment of MDS-SCs. This indicates the existence of a ‘dysplasia-like’ defect in the MDS MSCs, which are abnormal compared to their healthy counterparts but still provide enough support for the maintenance of the disease. Further studies are needed to confirm these preliminary findings.
Altogether, we believe that this new in vivo model is going to be critical for future investigations of the pathogenesis of MDS. It provides a robust preclinical system that will enable the development of new approaches to the treatment of MDS disease. Targeting the HSC-niche specific interactions might not only prevent disease progression from precursor states such as age-related clonal haematopoiesis (ARCH), and from MDS to AML, but will also enhance the effectiveness of current therapies directed at the malignant cells in a humanized microenvironment. Future functional studies are required to explore the receptor-ligand interacting signalling networks that may be essential for the MDS HSPC-niche interactions. In general, this will ultimately lead to the identification of rational therapeutic targets and the development of optimal strategies for intervention.
Material and Methods
Patient samples
Patient sample (n=37) were received from King’s College Haemato-oncology tissue bank under research ethics protocol (08/H0906/94). Patient demographic and clinical characteristics are detailed in supplementary table S2. The clinical variables for all patients were ascertained at the time of sample collection. Whole-exome sequencing data was available for 25 patients and myeloid-associated gene panel data was available for 2 patients (Table S1–S3).
Targeted DNA mutation sequencing
Targeted mutational analysis was performed on xenografted cells retrieved from the mice as previously described (3). Polymerase chain reaction (PCR) primers for patient specific mutations were designed using Primer3 (RRID:SCR_003139) program (36,37). Here, PCR amplified amplicons were normalized, mixed and then processed using transposon-based Nextera XT technology (Illumina, Cat FC-131-1096, Cat FC-131-2001). Libraries were sequenced on the Illumina MiSeq platform. VCF and BAM data files were visualized using variant studio (Illumina) and integrated genome viewer (IGV, RRID:SCR_011793), respectively.
MNC, CD34+ and CD3- isolation from the human bone marrow cells
MNCs were isolated from the bone marrow cells by centrifugation using Ficoll-PaqueTM PLUS (GE Healthcare Life Sciences). CD34+ and/or CD3- cell enrichment was performed using Easysep Human CD3 positive selection kit (StemCell Technologies, Cat 18051) and Easysep Human CD34 positive selection kit (StemCell Technologies, Cat 18056), respectively, along with Easysep magnet (StemCell Technologies, Cat 18000) according to the manufacturer’s instructions.
Human MSC expansion
Human bone marrow MNCs were used for CD45 selection. CD45 selection was performed using Easysep Human CD45 positive selection kit (StemCell Technologies, Cat 18259) and Easysep magnet (StemCell Technologies, Cat 18000) according to the manufacturer’s instructions. CD45- cells were seeded at a density of 1x106/cm2/0.2 - 0.3 mL of MSC culture media [MEM Alpha Medium (1X) + GlutaMAX-1 (Gibco, Cat 32571-029),1% penicillin/streptomycin (Sigma-Aldrich, Cat P4333) and 10% human MSC-FBS (Gibco, Cat 12662-029)]. Cell culture media was removed and replenished 24 hours after seeding, and then once every week. Expanded MSCs were frozen as viable cells at passage 1 or 2.
Mouse models
NOD/SCID/IL2rγ−/− (NSG, RRID:IMSR_JAX:005557) mice and NOD/SCID/IL2rγ−/−/IL-3/GM/SF (NSG-SGM3, RRID:IMSR_JAX:013062) mice were originally obtained from Leonard Shultz (The Jackson Laboratory, Bar Harbor, Maine, USA). NOD/SCID/ IL2rγ−/−/Tyr+/Kit W41J (NBSGW, RRID:IMSR_JAX:026622) mice were purchased from the Jackson laboratory. All three strains of mice (male/female) aged between 8 to 12 weeks used in this study have been bred at the Francis Crick Institute Biological Resource facility. All animal experiments were performed at the Francis Crick Institute in accordance with UK Home Office and CRICK guidelines and were undertaken under the Home Office project license PLL 70/8904.
Mouse MSC expansion
NSG-SGM3 mice (age, up to 4 weeks; RRID:IMSR_JAX:013062) were sacrificed and bones (femurs, tibias and pelvis) were recovered. Mouse bone were cut longitudinally and then placed (inner bone marrow tissue facing downwards) onto the surface of a 10cm culture dish. Culture dish was incubated for 10 minutes at room temperature. 10mls of mouse MSC media (MesenCult™ Expansion Kit, StemCell technologies, Cat 05514; 1% penicillin/streptomycin, Sigma-Aldrich, Cat P4333) was added to the culture dish. After 3 days, media was collected and centrifuged at 300xg for 6 minutes. Culture dish with mouse bones attached was washed 3 times with PBS without disturbing the bones. Fresh mouse MSC media (mixed with conditioned media, ratio 1:4) with was added to the culture dish. At day 14, attached bones were removed using sterile forceps. 75% of the media was replaced every week with fresh mouse MSC media. Cells were frozen at passage 1 and used later for in vivo experiments.
Cell seeding of scaffolds
All procedures were done in sterile conditions in a Class II biological safety cabinet. Human (and/or murine) MSCs were plated 7-10 days prior to seeding into the biometric scaffolds. Gelatin-based sponges (Gelfoam hemostatic agent absorbable gelatin sponge, Pfizer, Cat 00300090315085) were sectioned into 24 similar slices (6.6 mm x 7.5 mm x 7 mm), washed once with 70% ethanol, once with sterile PBS and finally rehydrated with sterile PBS. MSCs (5x104 – 1x105, Passage P2-P3) in 50μl of respective MSC culture media were injected into each scaffold using a sterile insulin syringe. Seeded scaffolds were transferred to polystyrene ultra-low attachment multi-well plate (Corning). Seeded scaffolds were incubated for up to 2 hours in a humidified cell culture incubator that was maintained at 37°C and 5% CO2. Following on, MSC culture media was slowly added to the wells without disturbing scaffolds and incubated for additional 48 hours.
CD3 depleted human bone marrow MNCs (2.5x105 to 7.5X105) from patient or healthy donors were resuspended into MyeloCult H5100 (StemCell technologies, Cat 05150; supplemented with 1% penicillin/streptomycin, Sigma-Aldrich, Cat P4333) supplemented with cytokines (20ng/mL G-CSF, PeproTech Cat 300-23; 20ng/mL IL-3, PeproTech Cat 200-03; 20ng/mL TPO, PeproTech Cat AF-300-18). CD3 depleted bone marrow MNCs were then injected into the scaffolds that were pre-seeded with MSCs. For experiments where CD34+CD38- cells were injected into the scaffolds, 3x105 CD34- accessory cells (irradiated 15 Gys) were also seeded alongside into each scaffold. Scaffolds were then incubated for up to 2 hours in a humidified cell culture incubator that was maintained at 37°C and 5% CO2. Following on, MyeloCult H5100 (StemCell technologies, Cat 05150; supplemented with 1% penicillin/streptomycin, Sigma-Aldrich, Cat P4333) supplemented with cytokines (20ng/mL G-CSF, PeproTech Cat 300-23; 20ng/mL IL-3, PeproTech Cat 200-03; 20ng/mL TPO, PeproTech Cat AF-300-18) was added slowly to the wells and incubated for additional 24 hours.
Surgical implantation of pre-seeded scaffold into mice
Surgical implantation of the pre-seeded scaffolds was performed by following local named veterinary surgeon (NVS) guidelines for aseptic techniques. Mice surgical area was prepared (shaved) 24 hours prior to the surgery. 24-48 hours prior to procedure, NSG and NSG-SGM3 mice received a sub-lethal dose of radiation (375 cGy) from a cesium-137 source. Mice were given analgesic (Carprofen in drinking water, 0.1mg/mL of water) 24 hours before the surgery. On the day of the surgery, mice were anesthetized in a chamber filled with 0.5% isoflurane and 2 L/min O2. Analgesia (Buprenorphin, 0.1 mg/kg and Meloxicam, 10 mg/kg) was administered via the subcutaneous route. Then the skin around the surgical area was sterilized using 10% chlorhexidine solution with a clean swab. 0.5cm anterior-to-posterior incision of the skin was created and then a pocket was made under the skin by using a sterile round ended scissors (or forceps). Pre-seeded scaffold(s) was inserted into the incision, making sure it was placed deep within the pocket. Up to 3 pre-seeded (MSC+CD3 depleted MNCs) scaffolds were implanted into each mouse. Scaffolds seeded with only MSCs were also implanted (when necessary). Incision site(s) was closed with surgical staples. Mice were maintained on analgesia (Carprofen in drinking water, 0.1mg/mL of water) for 48 hours post-surgery. Surgical staples were removed after 7 days (no more than 10 days) of the surgical procedure. Mice were administered with OTK3 (BioXCell, RRID:AB_1107632) (38) via the intraperitoneal route on weekly basis for up to 4 weeks post-surgery.
Engraftment was assessed in the scaffolds, bone marrow (pooled femurs, tibias, pelvis) and spleen at the time of sacrifice (12-18 weeks, or otherwise stated).
H&E and Immunofluorescence of biometric scaffolds
Scaffolds retrieved from mice were fixed overnight in 10% neutral buffered formalin (NBF). Then, scaffolds were processed, paraffin embedded and sectioned (5μm) for histological analysis. Hematoxylin/eosin (H&E) staining was performed on the scaffolds. For immunofluorescence (IF) analysis heat antigen retrieval was performed. Primary unconjugated antibodies used were specific for the following proteins: human CD45 (Agilent, RRID:AB_2314143) and Endomucin (Santa Cruz Biotechnology, RRID:AB_2100037). Secondary fluorescent antibodies (Invitrogen) were also used in this protocol. Images were captured using Zeiss Axio Scan Z1 slice scanner and Zen blue edition software.
Tissue digestion
Scaffolds retrieved from mice were excised into small pieces and transferred into a 1.5mL microcentrifuge tube that contained 1mL of tissue digestion solution (Dispase, Sigma-Aldrich; 2mg/mL Collagen Type I, StemCell Technologies Cat 04902; 1 mg/mL DNase I, Sigma-Aldrich Cat D4527-500KU; 10% FBS, Sigma-Aldrich). Tubes were incubated for up to a maximum of 1 hour in a water bath at 37°C. Digested cell suspension was filtered through a sterile 5mL tube with a cell strainer cap. Cells were washed twice with washing buffer (PBS, 2% FBS and 1% penicillin/streptomycin).
Cell sorting
MDS patient bone marrow cells were stained with antibodies specific for human antigens (CD34 and CD38). DAPI (4,6, diamidino-2-phenylindole, Sigma-Aldrich, Cat D9542) staining was used to exclude dead cells and debris from the analysis. CD34+CD38- cells were FACS sorted and then used for injections into the scaffold pre-seeded with MSC, that were then used in in vivo experiments.
For xenografted cells, cells were stained with antibodies specific for human or murine antigens (mCD45, Thermo Fisher Scientific, RRID:AB_1107002; hCD45, Thermo Fisher Scientific RRID:AB_1944375, Agilent RRID:AB_2314143; hCD33, BD Biosciences RRID:AB_395843, RRID:AB_398502, Thermo Fisher Scientific RRID:AB_1907380; hCD19, Thermo Fisher Scientific RRID:AB_1272053, BD Biosciences RRID:AB_398597, RRID:AB_395812; hCD3, BD Biosciences RRID:AB_398591 RRID:AB_395740; hCD73, BD Biosciences RRID:AB_2738063; hCD90, BD Biosciences RRID:AB_2872219; hCD34, Thermo Fisher Scientific RRID:AB_1963576, BD Biosciences Cat 555824, RRID:AB_398614, RRID:AB_2868843; hCD38 Thermo Fisher Scientific RRID:AB_2573346). DAPI or PI (Propidium iodide, BD Biosciences RRID:AB_2869075) staining was used to exclude dead cells and debris from the analysis. Cells were sorted as follows, myeloid cells (mCD45-hCD45+hCD33+), lymphoid T cells (mCD45-hCD45+hCD3+), lymphoid B cells (mCD45-hCD45+hCD19+) and MSCs (mCD45-hCD45-hCD73+hCD90+).
Cell sorting for was performed using a FACS Aria SORP (BD Biosciences, Oxford, UK). Sorted cells were washed in PBS and harvested in order to later perform further analysis (where needed).
Flow cytometry analysis
Following the tissue digestion of the scaffolds, xenografted cells were washed with PBS (2% fetal bovine serum, 1% P/S) and used for flow analysis. In addition, cells were also recovered from the bones (femurs, tibias and pelvis) and spleen of these mice. Cells were stained with antibodies specific for human or murine antigens (mCD45, Thermo Fisher Scientific, RRID:AB_1107002; hCD45, Thermo Fisher Scientific RRID:AB_1944375, Agilent RRID:AB_2314143; hCD33, BD Biosciences RRID:AB_395843, RRID:AB_398502, Thermo Fisher Scientific RRID:AB_1907380;hCD19, Thermo Fisher Scientific RRID:AB_1272053, BD Biosciences RRID:AB_398597, RRID:AB_395812; hCD3, BD Biosciences RRID:AB_398591 RRID:AB_395740; hCD73, BD Biosciences RRID:AB_2738063; hCD90, BD Biosciences RRID:AB_2872219; hCD34 Thermo Fisher Scientific RRID:AB_1963576, BD Biosciences Cat 555824, RRID:AB_398614, RRID:AB_2868843; hCD38 Thermo Fisher Scientific RRID:AB_2573346), DAPI or PI (Propidium iodide, BD Biosciences RRID:AB_2869075) staining was used to exclude dead cells and debris from the analysis. Cells were immunophenotyped by using Fortessa flow cytometer (BD Biosciences, Oxford, UK). Human myeloid, lymphoid and MSCs were detected as depicted in flow cytometry plot (Figure 1C).
High-resolution ultrasound perfusion imaging of the scaffold in in vivo
In order to assess the blood perfusion of the humanized scaffolds, non-linear contrast-enhanced ultrasound (CEUS) imaging was performed using a Vevo 3100 system with a MX550D transducer (VisualSonics, Toronto, Canada) and Vevo MicroMarker non-targeted contrast agent (VisualSonics). The contrast agent was reconstituted according to the manufacturers protocol at a concentration of 2x109 microbubbles/ml. Regular b-mode ultrasound imaging was used to position the imaging plane through the centre of the scaffold before switching to non-linear contrast (NLC) mode. Mice were anesthetized in a chamber filled with 0.5% isoflurane and 2 L/min O2. Imaging area around the implanted scaffold was shaved. While recording, a bolus injection of 50μl contrast agent was administered via a tail vein catheter at a flow rate of 300 μl/min using a Vevo infusion pump (VisualSonics). Mice were maintained on 0.5% isoflurane and 2 L/min O2 during the imaging process. At the end of the procedure, mice recovered to normality. Data analysis was performed using Vevo lab software (version 3.2.6, VisualSonics).
Colony-forming cell assay
Five hundred primary bone marrow CD34+ HSPCs were plated in 0.5mL in 24 well plate with MethoCult H4434 (StemCell Technologies, Cat 04434) supplemented with 1% penicillin/streptomycin (Sigma-Aldrich, Cat P4333). Assay was performed under hypoxic conditions (37°C and 3% O2). After 14 days of culture, the numbers of colonies were counted and subsequently picked individually. Cells were washed with PBS and then frozen as cell pellet to be used for genomic sequencing analysis.
Genotyping of colonies derived from single cells
Single cell colonies (CFU-GM and BFU-E) were harvested at day 14, and washed twice with PBS. All single cell derived colonies were individually subjected to whole-genome amplification (GenomePlex® Single Cell Whole Genome Amplification Kit, Sigma-Aldrich, Cat WGA4). Single cell colonies were screened for patient specific mutations. Gene mutation specific amplicons were amplified through PCR using the primers as described in earlier section. All amplified libraries were subjected to Nextera XT (Illumina, Cat FC-131-1096, Cat FC-131-2001) based library preparation and subsequently sequenced using MiSeq sequencing platform (Illumina).
Statistical analysis
Prism Version 6 software (GraphPad, RRID:SCR_002798) was used for statistical analysis. Data is presented as the mean ± S.E.M (where applicable). Statistical analysis was performed using the unpaired t-test for comparison of two groups to determine the level of significance. All the significant P values are described in the legends of the figures (where applicable).
Supplementary Material
Statement of significance.
These findings are significant for understanding the niche dependence of MDS. This report provides the evidence of the migratory behavior of HSCs in myeloid cancers. Our model offers a unique opportunity to study the clonal behavior of the myeloid/lymphoid cancers and delineate how cancer cells interact with different niches.
Acknowledgments
We would like to thank King’s College London and Kings College Hospital NHS Foundation Trust for funding the Kings College London Haemato-oncology Tissue Bank. We would like to thank Rajani Chelliah for assisting with patient sample processing. We would also like to thank the Biological Resource Facility, Histopathology, In Vivo Imaging Facility and the Flow Cytometry Core Facility teams at the Francis Crick Institute. This work was supported by The Francis Crick Institute, which receives its core funding from Cancer Research UK (FC001045), The UK Medical Research Council (FC001045), and the Welcome Trust (FC001045) as well by the Blood Cancer UK program grant support (to Kings College London and the Francis Crick Institute).
Footnotes
Conflict of interest statement
The authors have declared that no conflict of interest exists.
Author Contributions
S.A.M designed the study, performed experiments, analyzed and interpreted data, and wrote the manuscript; A.A, K.L.K, C.O, A.E.S, A.B helped with the experiments; K.R.P reviewed the manuscript and helped with the experiments; A.E.S helped with the sequencing data analysis; H.W provided the clinical data and reviewed the manuscript; P.J. helped with the in vivo imaging; G.J.M and D.B supervised the project, interpreted data, and contributed to writing of the paper.
References
- 1.Mufti GJ, Bennett JM, Goasguen J, Bain BJ, Baumann I, Brunning R, et al. Diagnosis and classification of myelodysplastic syndrome: International Working Group on Morphology of myelodysplastic syndrome (IWGM-MDS) consensus proposals for the definition and enumeration of myeloblasts and ring sideroblasts. Haematologica. 2008;93:1712–7. doi: 10.3324/haematol.13405. [DOI] [PubMed] [Google Scholar]
- 2.Steensma DP. Myelodysplastic syndromes current treatment algorithm 2018. Blood Cancer J. 2018;8:47. doi: 10.1038/s41408-018-0085-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mian SA, Rouault-Pierre K, Smith AE, Seidl T, Pizzitola I, Kizilors A, et al. SF3B1 mutant MDS-initiating cells may arise from the haematopoietic stem cell compartment. Nat Commun. 2015;6:10004. doi: 10.1038/ncomms10004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mian SA, Smith AE, Kulasekararaj AG, Kizilors A, Mohamedali AM, Lea NC, et al. Spliceosome mutations exhibit specific associations with epigenetic modifiers and proto-oncogenes mutated in myelodysplastic syndrome. Haematologica. 2013;98:1058–66. doi: 10.3324/haematol.2012.075325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Haferlach T, Nagata Y, Grossmann V, Okuno Y, Bacher U, Nagae G, et al. Landscape of genetic lesions in 944 patients with myelodysplastic syndromes. Leukemia. 2014;28:241–7. doi: 10.1038/leu.2013.336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Woll PS, Kjallquist U, Chowdhury O, Doolittle H, Wedge DC, Thongjuea S, et al. Myelodysplastic syndromes are propagated by rare and distinct human cancer stem cells in vivo. Cancer Cell. 2014;25:794–808. doi: 10.1016/j.ccr.2014.03.036. [DOI] [PubMed] [Google Scholar]
- 7.Medyouf H, Mossner M, Jann JC, Nolte F, Raffel S, Herrmann C, et al. Myelodysplastic cells in patients reprogram mesenchymal stromal cells to establish a transplantable stem cell niche disease unit. Cell Stem Cell. 2014;14:824–37. doi: 10.1016/j.stem.2014.02.014. [DOI] [PubMed] [Google Scholar]
- 8.Rouault-Pierre K, Mian SA, Goulard M, Abarrategi A, Di Tulio A, Smith AE, et al. Preclinical modeling of myelodysplastic syndromes. Leukemia. 2017;31:2702–8. doi: 10.1038/leu.2017.172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Krevvata M, Shan X, Zhou C, Dos Santos C, Habineza Ndikuyeze G, Secreto A, et al. Cytokines increase engraftment of human acute myeloid leukemia cells in immunocompromised mice but not engraftment of human myelodysplastic syndrome cells. Haematologica. 2018;103:959–71. doi: 10.3324/haematol.2017.183202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pang WW, Pluvinage JV, Price EA, Sridhar K, Arber DA, Greenberg PL, et al. Hematopoietic stem cell and progenitor cell mechanisms in myelodysplastic syndromes. Proc Natl Acad Sci U S A. 2013;110:3011–6. doi: 10.1073/pnas.1222861110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Benito AI, Bryant E, Loken MR, Sale GE, Nash RA, John Gass M, et al. NOD/SCID mice transplanted with marrow from patients with myelodysplastic syndrome (MDS) show long-term propagation of normal but not clonal human precursors. Leuk Res. 2003;27:425–36. doi: 10.1016/s0145-2126(02)00221-7. [DOI] [PubMed] [Google Scholar]
- 12.Thanopoulou E, Cashman J, Kakagianne T, Eaves A, Zoumbos N, Eaves C. Engraftment of NOD/SCID-beta2 microglobulin null mice with multilineage neoplastic cells from patients with myelodysplastic syndrome. Blood. 2004;103:4285–93. doi: 10.1182/blood-2003-09-3192. [DOI] [PubMed] [Google Scholar]
- 13.Abarrategi A, Mian SA, Passaro D, Rouault-Pierre K, Grey W, Bonnet D. Modeling the human bone marrow niche in mice: From host bone marrow engraftment to bioengineering approaches. J Exp Med. 2018;215:729–43. doi: 10.1084/jem.20172139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Taussig DC, Vargaftig J, Miraki-Moud F, Griessinger E, Sharrock K, Luke T, et al. Leukemia-initiating cells from some acute myeloid leukemia patients with mutated nucleophosmin reside in the CD34(-) fraction. Blood. 2010;115:1976–84. doi: 10.1182/blood-2009-02-206565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Borgmann A, Baldy C, von Stackelberg A, Beyermann B, Fichtner I, Nurnberg P, et al. Childhood all blasts retain phenotypic and genotypic characteristics upon long-term serial passage in NOD/SCID mice. Pediatr Hematol Oncol. 2000;17:635–50. doi: 10.1080/08880010050211349. [DOI] [PubMed] [Google Scholar]
- 16.Rombouts WJ, Martens AC, Ploemacher RE. Identification of variables determining the engraftment potential of human acute myeloid leukemia in the immunodeficient NOD/SCID human chimera model. Leukemia. 2000;14:889–97. doi: 10.1038/sj.leu.2401777. [DOI] [PubMed] [Google Scholar]
- 17.Nijmeijer BA, Mollevanger P, van Zelderen-Bhola SL, Kluin-Nelemans HC, Willemze R, Falkenburg JH. Monitoring of engraftment and progression of acute lymphoblastic leukemia in individual NOD/SCID mice. Exp Hematol. 2001;29:322–9. doi: 10.1016/s0301-472x(00)00669-x. [DOI] [PubMed] [Google Scholar]
- 18.Geyh S, Oz S, Cadeddu RP, Frobel J, Bruckner B, Kundgen A, et al. Insufficient stromal support in MDS results from molecular and functional deficits of mesenchymal stromal cells. Leukemia. 2013;27:1841–51. doi: 10.1038/leu.2013.193. [DOI] [PubMed] [Google Scholar]
- 19.Dykstra B, Olthof S, Schreuder J, Ritsema M, de Haan G. Clonal analysis reveals multiple functional defects of aged murine hematopoietic stem cells. J Exp Med. 2011;208:2691–703. doi: 10.1084/jem.20111490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yamamoto R, Wilkinson AC, Ooehara J, Lan X, Lai CY, Nakauchi Y, et al. Large-Scale Clonal Analysis Resolves Aging of the Mouse Hematopoietic Stem Cell Compartment. Cell Stem Cell. 2018;22:600–7 e4. doi: 10.1016/j.stem.2018.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Passaro D, Di Tullio A, Abarrategi A, Rouault-Pierre K, Foster K, Ariza-McNaughton L, et al. Increased Vascular Permeability in the Bone Marrow Microenvironment Contributes to Disease Progression and Drug Response in Acute Myeloid Leukemia. Cancer Cell. 2017;32:324–41 e6. doi: 10.1016/j.ccell.2017.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cosgun KN, Rahmig S, Mende N, Reinke S, Hauber I, Schafer C, et al. Kit regulates HSC engraftment across the human-mouse species barrier. Cell Stem Cell. 2014;15:227–38. doi: 10.1016/j.stem.2014.06.001. [DOI] [PubMed] [Google Scholar]
- 23.Wright DE, Wagers AJ, Gulati AP, Johnson FL, Weissman IL. Physiological migration of hematopoietic stem and progenitor cells. Science. 2001;294:1933–6. doi: 10.1126/science.1064081. [DOI] [PubMed] [Google Scholar]
- 24.Abkowitz JL, Robinson AE, Kale S, Long MW, Chen J. Mobilization of hematopoietic stem cells during homeostasis and after cytokine exposure. Blood. 2003;102:1249–53. doi: 10.1182/blood-2003-01-0318. [DOI] [PubMed] [Google Scholar]
- 25.Bixel MG, Kusumbe AP, Ramasamy SK, Sivaraj KK, Butz S, Vestweber D, et al. Flow Dynamics and HSPC Homing in Bone Marrow Microvessels. Cell Rep. 2017;18:1804–16. doi: 10.1016/j.celrep.2017.01.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ganan-Gomez I, Wei Y, Starczynowski DT, Colla S, Yang H, Cabrero-Calvo M, et al. Deregulation of innate immune and inflammatory signaling in myelodysplastic syndromes. Leukemia. 2015;29:1458–69. doi: 10.1038/leu.2015.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Haas S, Hansson J, Klimmeck D, Loeffler D, Velten L, Uckelmann H, et al. Inflammation-Induced Emergency Megakaryopoiesis Driven by Hematopoietic Stem Cell-like Megakaryocyte Progenitors. Cell Stem Cell. 2015;17:422–34. doi: 10.1016/j.stem.2015.07.007. [DOI] [PubMed] [Google Scholar]
- 28.Zambetti NA, Ping Z, Chen S, Kenswil KJG, Mylona MA, Sanders MA, et al. Mesenchymal Inflammation Drives Genotoxic Stress in Hematopoietic Stem Cells and Predicts Disease Evolution in Human Pre-leukemia. Cell Stem Cell. 2016;19:613–27. doi: 10.1016/j.stem.2016.08.021. [DOI] [PubMed] [Google Scholar]
- 29.Sun D, Luo M, Jeong M, Rodriguez B, Xia Z, Hannah R, et al. Epigenomic profiling of young and aged HSCs reveals concerted changes during aging that reinforce self-renewal. Cell Stem Cell. 2014;14:673–88. doi: 10.1016/j.stem.2014.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Challen GA, Sun D, Mayle A, Jeong M, Luo M, Rodriguez B, et al. Dnmt3a and Dnmt3b have overlapping and distinct functions in hematopoietic stem cells. Cell Stem Cell. 2014;15:350–64. doi: 10.1016/j.stem.2014.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Moran-Crusio K, Reavie L, Shih A, Abdel-Wahab O, Ndiaye-Lobry D, Lobry C, et al. Tet2 loss leads to increased hematopoietic stem cell self-renewal and myeloid transformation. Cancer Cell. 2011;20:11–24. doi: 10.1016/j.ccr.2011.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Balderman SR, Li AJ, Hoffman CM, Frisch BJ, Goodman AN, LaMere MW, et al. Targeting of the bone marrow microenvironment improves outcome in a murine model of myelodysplastic syndrome. Blood. 2016;127:616–25. doi: 10.1182/blood-2015-06-653113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Song Y, Rongvaux A, Taylor A, Jiang T, Tebaldi T, Balasubramanian K, et al. A highly efficient and faithful MDS patient-derived xenotransplantation model for pre-clinical studies. Nat Commun. 2019;10:366. doi: 10.1038/s41467-018-08166-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Antonelli A, Noort WA, Jaques J, de Boer B, de Jong-Korlaar R, Brouwers-Vos AZ, et al. Establishing human leukemia xenograft mouse models by implanting human bone marrow-like scaffold-based niches. Blood. 2016;128:2949–59. doi: 10.1182/blood-2016-05-719021. [DOI] [PubMed] [Google Scholar]
- 35.Reinisch A, Thomas D, Corces MR, Zhang X, Gratzinger D, Hong WJ, et al. A humanized bone marrow ossicle xenotransplantation model enables improved engraftment of healthy and leukemic human hematopoietic cells. Nat Med. 2016;22:812–21. doi: 10.1038/nm.4103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Untergasser A, Cutcutache I, Koressaar T, Ye J, Faircloth BC, Remm M, et al. Primer3--new capabilities and interfaces. Nucleic Acids Res. 2012;40:e115. doi: 10.1093/nar/gks596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Koressaar T, Lepamets M, Kaplinski L, Raime K, Andreson R, Remm M. Primer3_masker: integrating masking of template sequence with primer design software. Bioinformatics. 2018;34:1937–8. doi: 10.1093/bioinformatics/bty036. [DOI] [PubMed] [Google Scholar]
- 38.Wunderlich M, Brooks RA, Panchal R, Rhyasen GW, Danet-Desnoyers G, Mulloy JC. OKT3 prevents xenogeneic GVHD and allows reliable xenograft initiation from unfractionated human hematopoietic tissues. Blood. 2014;123:e134–44. doi: 10.1182/blood-2014-02-556340. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.