Abstract
Observational studies exploring whether there is a non-linear effect of blood pressure on cardiovascular disease risk are hindered by confounding. This limitation can be overcome by leveraging randomly allocated genetic variants in ‘non-linear’ Mendelian randomization analyses. Based on their association with blood pressure traits in a genome-wide association study of 299,024 European ancestry individuals, we selected 253 genetic variants to proxy the effect of modifying systolic and diastolic blood pressure. Considering the outcomes of incident coronary artery disease, stroke and the combined outcome of cardiovascular disease, linear and non-linear Mendelian randomization analyses were performed on 255,714 European-ancestry participants without a history of cardiovascular disease or antihypertensive medication use. There was no evidence favouring non-linear relationships of genetically proxied systolic and diastolic blood pressure with the cardiovascular outcomes over linear relationships. For every 10mmHg increase in genetically proxied systolic blood pressure, risk of incident cardiovascular disease increased by 49% (hazard ratio 1.49, 95% confidence interval 1.38-1.61), with similar estimates obtained for coronary artery disease (hazard ratio 1.50, 95% confidence interval 1.38-1.63) and stroke (hazard ratio 1.44, 95% confidence interval 1.22-1.70). Genetically proxied blood pressure had a similar relationship with cardiovascular disease in males and females. These findings provide evidence to support that even for individuals that do not have elevated blood pressure, public health interventions achieving persistent blood pressure reduction will be of considerable benefit in the primary prevention of cardiovascular disease.
Keywords: blood pressure, coronary artery disease, stroke, Mendelian randomization
Introduction
More than one billion people worldwide suffer from hypertension (1), which is estimated to account for more than 20% of cardiovascular disease (CVD) (2). Meta-analyses of randomised controlled trials have shown that a 10mmHg reduction in systolic blood pressure (SBP) is associated with a 15-20% reduction in the risk of coronary artery disease (CAD) and a 25-30% reduction in the risk of stroke (3). As such, blood pressure lowering is one of the most effective strategies for reducing the burden of CVD (4, 5).
Large observational studies have previously explored the relationship between blood pressure and cardiovascular risk, potentially identifying linear associations in individuals free of CVD at baseline (6, 7), but J-shaped associations both in the general population (8), and in patients with a history of CAD (9) and stroke (10). However, it is difficult to make causal conclusions about the effects of altering blood pressure from such data because any identified associations may be susceptible to confounding from unknown or unmeasured factors. For the patients with elevated cardiovascular risk recruited to the SPRINT trial, SBP lowering to less than 120mmHg as compared to 140mmHg resulted in fewer major cardiovascular events (11). However, no high quality clinical trials have investigated the effect of blood pressure lowering below this level. Excessive blood pressure reduction in patients with atherosclerotic disease can reduce organ perfusion and increase CVD risk (12). Insight into the shape of the relationship between blood pressure and CVD risk is therefore critical for informing optimal prevention strategies.
In the Mendelian randomization (MR) paradigm, genetic variants can be used as proxies for studying the effect of varying blood pressure (13). In the same way as treatment allocation in a randomised controlled trial setting, random allocation of genetic variants means that they are unlikely to be affected by confounding from environmental factors (14). Recent methodological developments have allowed for MR investigation into the shape of the relationship between risk factors and outcomes (15–17). In this study, we employ MR to investigate the shape of the relationship between genetically proxied blood pressure and incident CVD in a general population without prior history of CVD or antihypertensive medication use. Our analyses aim to provide novel insight that can be used to inform public health strategies towards the primary prevention of CVD.
Methods
All data supporting the findings of this study are available from the corresponding author upon reasonable request. The UK Biobank study was approved by the North West Multicentre Research Ethics Committee, and all participants provided informed consent. All variants used as instruments in this study and their genetic association estimates are provided in the Online Supplement. All results from the analyses performed in this work are presented in the main manuscript or its supplementary files. This paper has been reported based on recommendations by the STROBE-MR Guidelines (Supplementary Research Checklist) (18). The study protocol and details were not pre-registered.
UK Biobank
The UK Biobank cohort is comprised of approximately 500,000 people (94% of self-reported European-ancestry) aged 40 to 69 at baseline and recruited between 2006 and 2010 at 22 assessment centres throughout the UK. Participants were followed up until January 1, 2018 or their date of death. Along with genotyping, the resource has information on clinical measurements, assays of biological samples, and self-reported health behaviour. Moreover, it is supplemented by linkage with electronic health records including hospital inpatient data, mortality data, and cancer registries (19).
For the exposures of interest, SBP and diastolic blood pressure (DBP), data were collected using an automated reading when participants attended the assessment centre for baseline measurements (UK Biobank fields 4080 for SBP and 4079 for DBP). When multiple baseline measurements were available, the mean of the measured values was used.
As our primary outcomes, we selected a combined incident cardiovascular endpoint of CAD and stroke (referred to hereafter as CVD), incident CAD, and incident stroke. We used hospitalization-based International Diagnostic Classification of Diseases and Health Related Problems version 10 (ICD-10) and Office of Population Censuses and Surveys (OPCS) Classification of Surgical Operations and Procedures (4th revision, OPCS-4) codes to identify events (Table S1). For individuals with multiple incident events (e.g. incident CAD and incident stroke), the first event recorded was used. Related individuals (kinship coefficient >0.0884) and those with prevalent CVD (identified through hospitalization-codes and self-report) were excluded from the analyses. Individuals taking antihypertensive medications at baseline (UK Biobank field 20003) were also excluded from the analyses because their observed blood pressure is not reflective of their genetically predicted blood pressure, thus introducing bias into the non-linear MR estimates (15, 17).
Candidate instrumental variables
For our primary analysis, we selected 253 uncorrelated (r2<0.1) single-nucleotide polymorphisms (SNPs) as candidate instrumental variables for SBP and DBP based on their previously published associations with blood pressure traits (20). Their associations with SBP and DBP were estimated in a genome-wide association study of 299,024 European ancestry individuals performed by the International Consortium of Blood Pressure study, which did not include UK Biobank participants (20). Using the coefficients for association with SBP and DBP (Table S2), a weighted allele score for each participant was created by multiplying the blood pressure-increasing allele dosage with the variant’s association with SBP or DBP respectively, and summing across the 253 variants. The above genetic association estimates were taken from a study that adjusted for body mass index. As this could theoretically bias the analyses (21), we further performed a sensitivity analysis that selected variants from a genome-wide association study meta-analysis of two non-UK Biobank cohorts that did not adjust for body mass index (N=122,361) (Supplementary Methods). Fixed-effects meta-analysis was performed using METAL (22), and variants reaching genome-wide significance (p<5x10-8) were clumped to correlation r2<0.01 using PLINK (23). We extracted 22 uncorrelated variants as instrumental variables for SBP and 27 uncorrelated variants as instrumental variables for DBP in this sensitivity analysis (Tables S3 and S4).
Statistical analyses
All statistical analyses were performed using R (version 3.6.2). Differences in characteristics between UK Biobank population subgroups were assessed using a Student’s t-test, Wilcoxon rank-sum test, Fisher’s exact test or Chi-square test as appropriate. We performed MR analyses investigating the association between genetically proxied blood pressure (either SBP or DBP) and incident CVD, CAD, and stroke risk. Analyses were performed by modelling a linear relationship between genetically proxied blood pressure and the outcomes (‘linear MR’) (14, 24), and also using the fractional polynomial method to test for a non-linear relationship between genetically proxied blood pressure and the outcomes (‘non-linear MR’) (15, 17).
Linear MR
We used the ratio of coefficients method to perform MR analyses that assumed a linear association of genetically proxied blood pressure with the risk of incident CVD, CAD, and stroke (25). This represents the association of the allele score with the cardiovascular outcome (incident CVD, CAD or stroke) divided by the association of the allele score with the blood pressure trait (either SBP or DBP) (26). Linear regression was used to estimate the association of the allele score with blood pressure, incorporating age, sex, principal components 1-10 of genetic ancestry, genotyping chip and assessment centre as covariates. The proportion of blood pressure variance explained by the allele score and its F-statistic were calculated to estimate instrument strength (27). Cox proportional hazard regression was used to estimate the association of the allele score with the outcomes, incorporating age, sex, principal components 1-10 of genetic ancestry, genotyping chip and assessment centre as covariates. As sensitivity analyses, we considered each variant in the allele score separately and performed MR methods that differ in their requisite assumptions regarding the inclusion of pleiotropic variants: random-effects inverse-variance weighted MR, MR-Egger, weighted median MR and MR-PRESSO (28). An intercept term in MR-Egger differing from zero can be used to evidence the presence of directional pleiotropy (29), and MR-PRESSO is able to identify variants with outlying estimates that may in turn be excluded from analyses (30).
Non-linear MR
We applied the fractional polynomial method to investigate for evidence of a non-linear relationship between genetically proxied blood pressure and risk of incident CVD, CAD, and stroke. This approach has been described in detail previously (15–17), and is outlined in the Supplementary Methods. Briefly, we stratified the population into centiles based on residual blood pressure, defined as a participant’s blood pressure minus the genetic contribution to blood pressure from the allele score. By doing this, we aimed to compare individuals in the population who would have similar blood pressure values (values in the same centile) if they had the same genetic predisposition. Stratifying on blood pressure directly would introduce collider bias to distort estimates, as blood pressure is on the causal pathway from the genetic variants to CVD (17, 31). For each centile, we calculated a linear MR estimate for the association of genetically proxied blood pressure with the outcome using the ratio of coefficients method, as described above (26). Using a flexible semiparametric framework, we then performed a meta-regression of the linear MR estimates obtained for each centile against the mean blood pressure in that centile (16, 17). A fractional polynomial test was used to investigate whether a non-linear model fit this meta-regression better than a linear model (further detailed in the Supplementary Methods). A Bonferroni correction was applied to account for multiple testing of the two blood pressure traits and three outcomes, with p<8x10-3 representing statistical significance. We further conducted a priori specified subgroup analyses considering males and females separately to investigate potential sex-specific effects.
Individuals with elevated blood pressure are more likely to be prescribed antihypertensive medications, and therefore exclusion of these individuals from the main analysis could potentially distort MR estimates due to selection effects and introduction of collider bias. Inverse-probability weighting was therefore performed in a sensitivity analysis to investigate this, as described in the Supplementary Methods.
Results
A total of 255,714 participants were included in analyses, after excluding 66,011 individuals with a history of antihypertensive medication use and 6,506 individuals with a history of CVD (but not on antihypertensive medications). There were 10,606 incident CVD events, including 8,430 incident CAD events (68.1% ICD-10 based) and 2,176 incident stroke events. The allele score explained 4.8% and 4.5% of the variance for SBP and DBP respectively, corresponding to F-statistics of 58.6 and 54.1 and low risk of substantial weak instrument bias. The distribution of CVD risk factors for individuals in the analysed population that had a weighted allele score for SBP and DBP above and below the population median in the main and sensitivity analyses are provided in Table 1. Table S5 provides these data for individuals in the top and bottom decile of residual blood pressure in the main analysis.
Table 1.
Distribution of risk factors for individuals in the analysed population that had a weighted allele score for systolic blood pressure (SBP) and diastolic blood pressure (DBP) above and below the population median in the main and sensitivity analyses. BMI: body mass index; LDL-C: low-density lipoprotein cholesterol; SD: standard deviation.
| Variable | Main analysis (allele score adjusted for BMI) | Sensitivity analysis (allele score not adjusted for BMI) | ||||||
|---|---|---|---|---|---|---|---|---|
| SBP weighted allele score | DBP weighted allele score | SBP weighted allele score | DBP weighted allele score | |||||
| Below median | Above median | Below median | Above median | Below median | Above median | Below median | Above median | |
| Age, mean (SD), y | 55.9 (8.0) | 55.4 (8.1) | 55.8 (8.0) | 55.4 (8.1) | 55.7 (8.0) | 55.6 (8.0) | 55.7 (8.0) | 55.5 (8.0) |
| Sex, N (%) | ||||||||
| Male | 54642 (43.0) | 53959 (42.4) | 53842 (42.5) | 53533 (42.3) | 54549 (42.9) | 54052 (42.5) | 53770 (42.5) | 53605 (42.3) |
| Female | 72563 (57.0) | 73246 (57.6) | 72740 (57.5) | 73049 (57.7) | 72653 (57.1) | 73156 (57.5) | 72812 (57.5) | 72977 (57.7) |
| Socioeconomic status, N (%)* | ||||||||
| Quintile 1 | 27823 (21.9) | 28066 (22.1) | 27742 (21.9) | 27953 (22.1) | 27744 (21.8) | 28145 (22.1) | 27819 (22.0) | 27876 (22.0) |
| Quintile 2-4 | 78655 (61.8) | 78287 (61.5) | 78210 (61.8) | 77999 (61.6) | 78562 (61.8) | 78380 (61.6) | 78243 (61.8) | 77966 (61.6) |
| Quintile 5 | 20727 (16.3) | 20852 (16.4) | 20630 (16.3) | 20630 (16.3) | 20896 (16.4) | 20683 (16.3) | 20520 (16.2) | 20740 (16.4) |
| Smoking index, mean (SD)† | 0.4 (0.7) | 0.4 (0.7) | 0.4 (0.7) | 0.4 (0.7) | 0.4 (0.7) | 0.4 (0.7) | 0.4 (0.7) | 0.4 (0.7) |
| BMI, mean (SD), kg/m2 | 26.9 (4.5) | 26.7 (4.4) | 26.9 (4.5) | 26.7 (4.4) | 26.9 (4.5) | 26.9 (4.4) | 26.9 (4.5) | 26.9 (4.4) |
| SBP, mean (SD), mmHg | 134.6 (17.8) | 138.3 (18.6) | 135.0 (18.0) | 137.9 (18.5) | 135.5 (18.0) | 137.4 (18.5) | 135.7 (18.2) | 137.3 (18.4) |
| DBP, mean (SD), mmHg | 80.9 (9.9) | 82.7 (10.1) | 80.7 (9.9) | 82.9 (10.1) | 81.4 (10.0) | 82.2 (10.1) | 81.3 (10.0) | 82.4 (10.1) |
| Diabetes diagnosed, N (%) | 2738 (2.1) | 2583 (2.0) | 2610 (2.1) | 2487 (2.0) | 2954 (2.3) | 2567 (2.0) | 2685 (2.1) | 2612 (2.1) |
| LDL-C, mean (SD), mmol/L | 3.7 (0.8) | 3.7 (0.8) | 3.7 (0.8) | 3.7 (0.8) | 3.7 (0.8) | 3.7 (0.8) | 3.7 (0.8) | 3.7 (0.8) |
Socioeconomic status quintiles according to Townsend deprivation index combining information on social class, employment, car availability and housing.
Lifetime smoking index, as detailed in Wootton et al. (39).
Linear MR
Linear MR analyses identified a strong association of both genetically proxied SBP and DBP with the cardiovascular outcomes. For a 10mmHg increase in genetically proxied SBP, the hazard ratio (HR) of incident CVD was 1.49 (95% confidence interval [CI] 1.38-1.61; p=7x10-25), incident CAD was 1.50 (95% CI 1.38-1.63; p=2x10-21) and incident stroke was 1.44 (95% CI 1.22- 1.70; p=1x10-5). For a 5mmHg increase in genetically proxied DBP, the HR of incident CVD was 1.35 (95% CI 1.29-1.42; p=5x10-34), incident CAD was 1.36 (95% CI 1.26-1.47; p=1x10-15) and incident stroke was 1.39 (95% CI 1.20-1.62; p=2x10-5). The MR-Egger test did not detect significant directional pleiotropy (Table S6), and MR-PRESSO only identified 16 SNPs as outliers in the analysis of genetically proxied SBP and CAD (Table S2). Similar MR estimates were obtained in sensitivity analyses (Table S6, Figures S1 and S2).
Non-linear MR
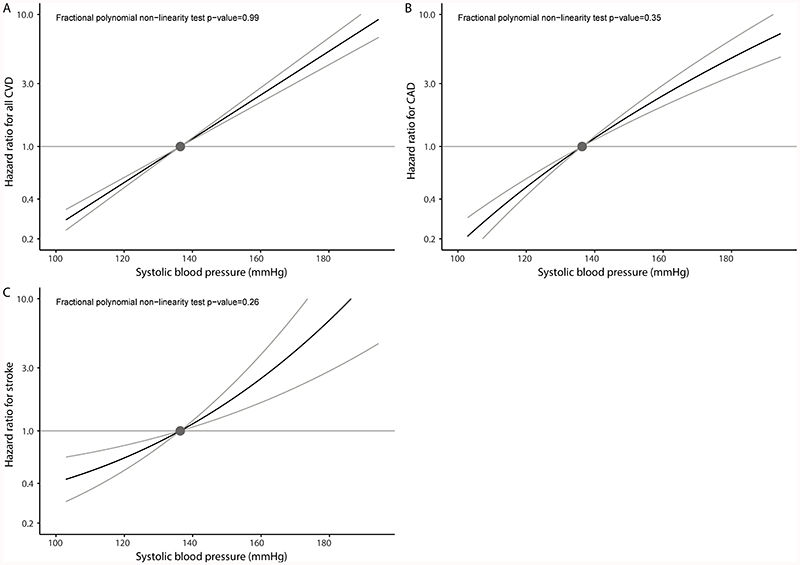
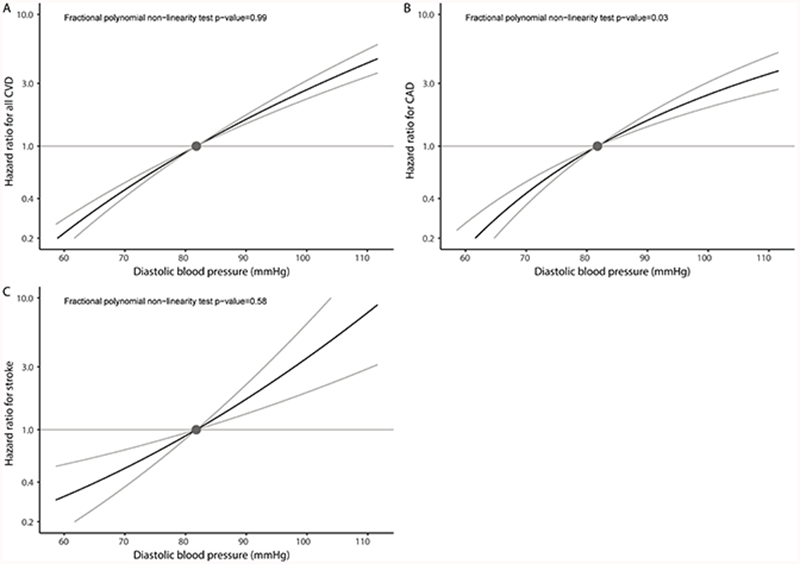
While in some cases the best-fitting fractional polynomial was a non-linear function, we observed no evidence favouring a non-linear relationship between genetically proxied blood pressure and the cardiovascular outcomes over a linear one (Figures 1 and 2). This means that any departure from linearity was no greater than would be expected by chance due to random variability. Compared to the population mean SBP of 137mmHg, individuals with a genetically proxied SBP of 120mmHg had a 47% lower risk of incident CVD (HR 0.53, 95% CI 0.49-0.58) (Table 2). Compared to the population mean DBP of 82mmHg, individuals with a genetically proxied DBP of 70mmHg had a 53% lower risk of incident CVD (HR 0.47, 95% CI 0.41-0.53) (Table 2). MR estimates for population subgroups based on stratification into SBP and DBP centiles are provided in Tables S7 and S8, respectively.
Figure 1.
Non-linear Mendelian randomization considering genetically proxied systolic blood pressure (SBP) and incident cardiovascular outcomes: (A) all incident cardiovascular disease (CVD) events, (B) incident coronary artery disease (CAD), and (C) incident stroke. Displayed on the x-axis are SBP values in mmHg. The y-axis shows the hazard ratio for the respective incident cardiovascular event. Reference is set to a population mean SBP value of 136.5mmHg. Grey lines depict the 95% confidence interval. Fractional polynomial test is a goodness-of-fit test assessing whether any improvement of fit using a non-linear function to model the data compared with a linear function is greater than expected due to chance alone.
Figure 2.
Non-linear Mendelian randomization considering genetically proxied diastolic blood pressure (DBP) and incident cardiovascular outcomes: (A) all incident cardiovascular disease (CVD) events, (B) incident coronary artery disease (CAD), and (C) incident stroke. Displayed on the x-axis are DBP values in mmHg. The y-axis shows the hazard ratio for the respective incident cardiovascular event. Reference is set to a population mean DBP value of 81.8mmHg. Grey lines depict the 95% confidence interval.
Table 2.
Non-linear Mendelian randomization estimates for the association between systolic blood pressure and incident cardiovascular outcomes. Reference is made to a population mean systolic blood pressure value of 136.5mmHg and a population mean diastolic blood pressure value of 81.8mmHg.
| Blood pressure | Cardiovascular disease | Coronary artery disease | Stroke | ||||
|---|---|---|---|---|---|---|---|
| Hazard ratio | 95% confidence interval | Hazard ratio | 95% confidence interval | Hazard ratio | 95% confidence interval | ||
| Systolic blood pressure (mmHg) | min (102.9) | 0.28 | 0.23-0.33 | 0.21 | 0.15-0.29 | 0.42 | 0.29-0.63 |
| 110 | 0.36 | 0.32-0.42 | 0.3 | 0.24-0.39 | 0.49 | 0.36-0.68 | |
| 120 | 0.53 | 0.49-0.58 | 0.49 | 0.43-0.57 | 0.62 | 0.50-0.77 | |
| 130 | 0.78 | 0.75-0.81 | 0.77 | 0.73-0.81 | 0.82 | 0.75-0.90 | |
| 140 | 1.14 | 1.12-1.17 | 1.16 | 1.12-1.20 | 1.13 | 1.07-1.19 | |
| 150 | 1.67 | 1.56-1.80 | 1.7 | 1.52-1.90 | 1.63 | 1.30-2.05 | |
| 160 | 2.45 | 2.16-2.78 | 2.43 | 2.02-2.93 | 2.49 | 1.64-3.79 | |
| 170 | 3.59 | 2.99-4.30 | 3.41 | 2.64-4.39 | 4.02 | 2.12-7.61 | |
| 180 | 5.25 | 4.16-6.64 | 4.68 | 3.40-6.44 | 6.87 | 2.84-16.63 | |
| 190 | 7.69 | 5.76-10.26 | 6.32 | 4.31-9.26 | 12.55 | 3.93-40.10 | |
| max (194.6) | 9.15 | 6.69-12.52 | 7.16 | 4.76-10.76 | 16.65 | 4.58-60.56 | |
| Diastolic blood pressure (mmHg) | min (58.6) | 0.2 | 0.15-0.25 | 0.14 | 0.09-0.23 | 0.3 | 0.16-0.52 |
| 60 | 0.22 | 0.17-0.28 | 0.17 | 0.11-0.26 | 0.31 | 0.18-0.54 | |
| 65 | 0.32 | 0.27-0.39 | 0.28 | 0.21-0.38 | 0.39 | 0.25-0.62 | |
| 70 | 0.47 | 0.41-0.53 | 0.44 | 0.36-0.54 | 0.51 | 0.37-0.70 | |
| 75 | 0.65 | 0.61-0.70 | 0.64 | 0.58-0.72 | 0.67 | 0.55-0.81 | |
| 80 | 0.9 | 0.88-0.91 | 0.9 | 0.87-0.92 | 0.9 | 0.85-0.95 | |
| 85 | 1.21 | 1.17-1.24 | 1.21 | 1.15-1.26 | 1.23 | 1.11-1.35 | |
| 90 | 1.6 | 1.48-1.72 | 1.57 | 1.41-1.75 | 1.71 | 1.32-2.21 | |
| 95 | 2.08 | 1.85-2.35 | 1.99 | 1.68-2.35 | 2.42 | 1.58-3.70 | |
| 100 | 2.68 | 2.28-3.14 | 2.45 | 1.97-3.06 | 3.5 | 1.92-6.39 | |
| 105 | 3.4 | 2.78-4.15 | 2.97 | 2.28-3.88 | 5.16 | 2.35-11.34 | |
| max (111.7) | 4.6 | 3.58-5.89 | 3.72 | 2.70-5.14 | 8.88 | 3.11-25.27 | |
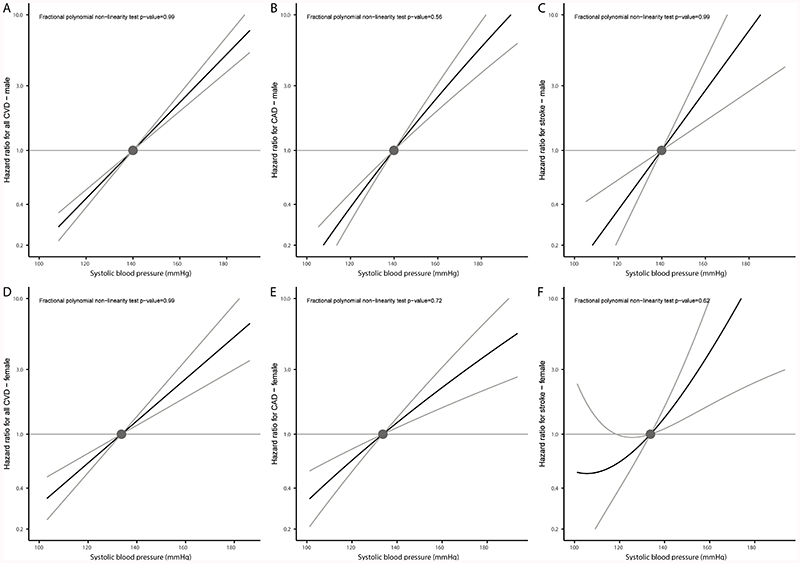
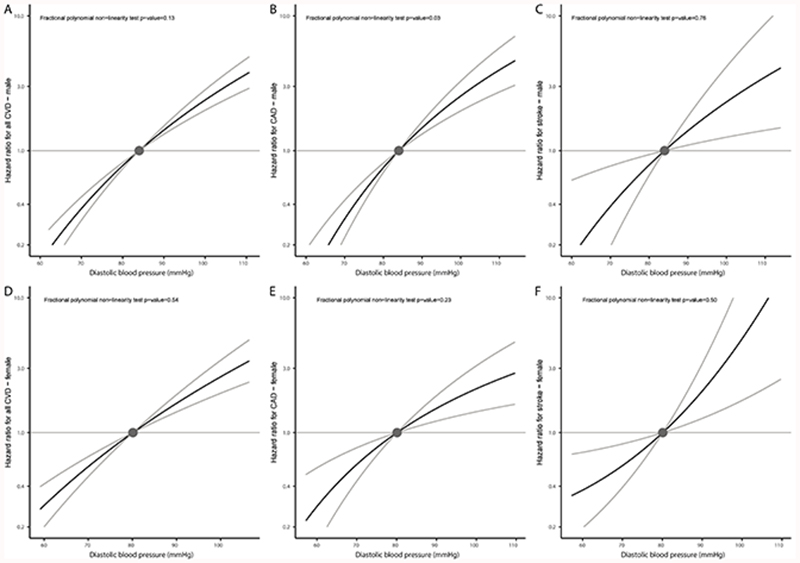
Subgroup analyses considering males and females separately produced similar results to the main analyses (Figures 3 and 4). Findings were also similar in the two sensitivity analyses: i) using inverse-probability weighting to correct for potential selection bias related to exclusion of individuals taking antihypertensive medications at baseline (Figures S3 and S4) and ii) using a different set of variants as instruments, which were obtained from studies not including UK Biobank participants, and without adjustment for body mass index (Figures S5 and S6, and Table S9).
Figure 3.
Non-linear Mendelian randomization considering genetically proxied systolic blood pressure (SBP) and incident cardiovascular outcomes split by sex: (A) all incident cardiovascular disease (CVD) events in males, (B) incident coronary artery disease (CAD) in males, and (C) incident stroke in males. Sections D-F show the equivalent analyses in females. Displayed on the x-axis are SBP values in mmHg. The y-axis shows the hazard ratio for the respective incident cardiovascular event. Reference is set to a mean SBP value of 136.5mmHg. Grey lines depict the 95% confidence interval.
Figure 4.
Non-linear Mendelian randomization considering genetically proxied diastolic blood pressure (DBP) and incident cardiovascular outcomes split by sex: (A) all incident cardiovascular disease (CVD) events in males, (B) incident coronary artery disease (CAD) in males, and (C) incident stroke in males. Sections D-F show the equivalent analyses in females. Displayed on the x-axis are DBP values in mmHg. The y-axis shows the hazard ratio for the respective incident cardiovascular event. Reference is set to a mean DBP value of 81.8mmHg. Grey lines depict the 95% confidence interval.
Discussion
By applying non-linear MR methods in the UK Biobank, we were able to examine the shape of the relationship between genetically proxied blood pressure and incident CVD in a population without a prior history of CVD or antihypertensive medication use. We found no evidence favouring non-linear relationships between genetically proxied SBP or DBP and risk of the cardiovascular outcomes over linear ones. Similar results were obtained when considering males and females separately.
Blood pressure control represents a global health challenge (32), and hypertension thresholds have been lowered in recent consensus guidelines (33). The MR estimates obtained in this study may be used to quantify the effect of a persistent, life-long reduction in blood pressure on the primary prevention of CVD, and highlight the potential gains of clinical and public health interventions that achieve this. Importantly, they support the notion that for a population without history of CVD or antihypertensive medication use, a similar relative reduction in CVD risk will be observed irrespective of baseline blood pressure, including for individuals that have normal blood pressure (34). This means that fixed changes in blood pressure will lead to similar changes in CVD risk on the hazard ratio scale. On the absolute scale, risk reduction will be greater for those with higher baseline blood pressure. This finding is consistent with previous large-scale observational analyses performed in individuals free of CVD at baseline (6, 7). In contrast, excessive blood pressure reduction in patients with atherosclerotic disease can reduce organ perfusion and increase CVD risk (12), and it is therefore important that our findings are not extrapolated to infer the effect of blood pressure lowering in individuals with pre-existing CVD. It is also important to appreciate that absolute risk reduction conferred from blood pressure lowering will remain greatest for those with highest blood pressure. Our current data support the concept that risk factor targeting in low- and medium-risk individuals on a population-wide level is likely to also substantially contribute to reducing the burden of CVD (35, 36). Dietary modification and reduced sodium consumption represent examples of public health strategies that can be adopted to achieve this (37, 38).
We found no evidence for a J-shaped association of either genetically proxied SBP or DBP with any of the outcomes. This contrasts the findings of a recent observational study using data from 1.3 million general outpatients with a low prevalence of CAD (8), which identified a J-shaped association of blood pressure with the composite outcome of myocardial infarction and stroke. This J-shape was only partially attenuated after adjusting for age, ethnicity and co-morbidities (8), and there remains the possibility that residual unknown or unmeasured confounding factors are responsible for the discrepancy with our findings. A systematic review and meta-analysis of blood pressure lowering trials considering 613,815 participants from 123 studies found no trend for CVD risk reduction per 10mmHg lower SBP when stratifying trials by mean baseline SBP (3). In the SPRINT trial, SBP lowering to less than 120mmHg as compared to 140mmHg resulted in fewer major cardiovascular events (11). The findings from our current MR study additionally support a relative CVD risk reduction from blood pressure lowering below this level in patients without a history of CVD.
Our study has a number of strengths. By employing randomly allocated genetic variants as proxies for the effect of modifying blood pressure, we were able to use the MR paradigm to overcome the environmental confounding bias that can limit causal inference in observational association studies. The implementation of both linear and non-linear MR methods within the comprehensive UK Biobank resource enabled us to efficiently study the relationships of genetically proxied SBP and DBP with incident CVD, CAD, and stroke, including in sex-stratified analyses. Importantly, the fractional polynomial method allowed us to investigate for evidence of non-linear associations.
Our study also has limitations. This work only considered participants without a prior history of CVD or antihypertensive medication use, and its findings should not be extrapolated to populations with established CVD (9, 10). Individuals that reported taking antihypertensive medications were excluded in order to allow for meaningful stratification into blood pressure quantiles, and as such there is the possibility that ascertainment bias may have been introduced. Reassuringly, similar findings were obtained in inverse-probability weighting sensitivity analyses, suggesting that any such bias is unlikely to be affecting our conclusions. The employed MR approach assumes that the genetic variants utilized as proxies for blood pressure do not affect CVD risk through alternative (pleiotropic) pathways, an assumption that cannot be tested and if violated could introduce bias to the obtained estimates. Our employed MR method also explores the effects of lifelong changes in blood pressure and its estimates should therefore not be extrapolated to quantify the effect of blood pressure modification in adult life, such as through use of anti-hypertensive medications. Finally, there were differences in the distribution of risk factors between individuals in the highest and lowest decile of residual blood pressure (Table S5), suggesting that this MR analysis may still be vulnerable to environmental confounding.
Perspectives
For a population without history of CVD or antihypertensive medication use, genetically proxied blood pressure reduction was associated with lower CVD risk at all levels of blood pressure. These findings provide evidence to support that public health interventions achieving persistent, population-wide blood pressure reduction will be of considerable benefit in the primary prevention of CVD.
Supplementary Material
Novelty and Significance.
What is new?
Recent methodological developments have enabled randomly allocated genetic variants to be leveraged in ‘non-linear’ Mendelian randomization analyses that explore the shape of the relationship between a risk factor and an outcome.
Performing linear and non-linear Mendelian randomization analyses in 255,714 European-ancestry UK Biobank participants without a history of cardiovascular disease or antihypertensive medication use, this study found no evidence favouring non-linear relationships of genetically proxied systolic and diastolic blood pressure with incident coronary artery disease, stroke or a combined endpoint.
What is relevant?
At all levels of blood pressure, public health interventions achieving persistent blood pressure reduction are likely to be of considerable benefit in the primary prevention of cardiovascular disease.
Summary
For a population without history of cardiovascular disease or antihypertensive medication use, genetically proxied blood pressure reduction was associated with lower cardiovascular disease risk at all levels of blood pressure.
Acknowledgements
This research has been conducted using the UK Biobank Resource (UK Biobank application 2532). UK Biobank data is available on application at https://www.ukbiobank.ac.uk/register-apply. The Trøndelag Health Study (The HUNT Study) is a collaboration between HUNT Research Centre, (Faculty of Medicine and Health Sciences, NTNU, Norwegian University of Science and Technology), Nord-Trøndelag County Council, Central Norway Regional Health Authority, and the Norwegian Institute of Public Health. The genotype quality control and imputation in HUNT has been conducted by the K.G. Jebsen Center for Genetic Epidemiology, Department of Public Health and Nursing, Faculty of Medicine and Health Sciences, NTNU, Norwegian University of Science and Technology.
Sources of Funding
This work was supported by the UK National Institute for Health Research Cambridge Biomedical Research Centre. MKG is funded by a scholarship from the Onassis Foundation. SMD was supported by the Department of Veterans Affairs Office of Research and Development (IK2-CX001780). PE acknowledges support from the British Heart Foundation (RE/18/4/34215), the Medical Research Council (MR/S019669/1), the National Institute for Health Research Imperial Biomedical Research Centre, Imperial College London (RDF03), the UK Dementia Research Institute (DRI) at Imperial College London funded by UK DRI Ltd (funded by Medical Research Council, Alzheimer’s Society, Alzheimer’s Research UK), and Health Data Research (HDR) UK London funded by HDR UK Ltd (funded by a consortium led by the Medical Research Council 1004231). JNH is supported by K12 HD04348. MMS is funded by the National Institutes of Health (DK108444). SB is supported by a Sir Henry Dale Fellowship jointly funded by the Wellcome Trust and the Royal Society (204623/Z/16/Z). MD acknowledges funding from the European Union’s Horizon 2020 research and innovation programme (666881), SVDs@target (667375), CoSTREAM, SyNergy (EXC 2145 SyNergy – ID 390857198), the CRC 1123 (B3 and project DI 722/13-1), the Corona Foundation, the LMUexcellent fond, the e:Med program (e:AtheroSysMed) and the FP7/2007-2103 European Union project CVgenes@target (Health-F2-2013-601456). DG is supported by the Wellcome Trust 4i Programme (203928/Z/16/Z) and British Heart Foundation Centre of Research Excellence (RE/18/4/34215) at Imperial College London, and a National Institute for Health Research Clinical Lectureship at St. George’s, University of London (CL-2020-16-001). The BioVU dataset used for the analyses described were obtained from Vanderbilt University Medical Center’s BioVU which is supported by numerous sources: institutional funding, private agencies, and federal grants. These include the National Institutes of Health (NIH) funded Shared Instrumentation Grant S10RR025141; and CTSA grants UL1TR002243, UL1TR000445, and UL1RR024975. Genomic data are also supported by investigator-led projects that include U01HG004798, R01NS032830, RC2GM092618, P50GM115305, U01HG006378, U19HL065962, R01HD074711; and additional funding sources listed at https://victr.vumc.org/biovu-funding/. The genotyping in HUNT was financed by the NIH; University of Michigan; The Research Council of Norway; The Liaison Committee for Education, Research and Innovation in Central Norway; and the Joint Research Committee between St. Olavs hospital and the Faculty of Medicine and Health Sciences, NTNU.
Footnotes
Contributors
DG, RM, MV, SB and MKG designed the study. RM, MV, JHN, MMS, TLE, BMB, TR, CF, AG analysed the data. RM, DG, JHN, BMB and MKG drafted the manuscript. All authors interpreted the results and critically revised the manuscript for intellectual content.
Disclosures
DG is employed part-time by Novo Nordisk and has received consultancy fees from Abbott Laboratories. SMD has received grants from the U.S. Department of Veterans Affairs, Calico Labs, and Renalytix AI plc outside the submitted work. The remaining authors have no conflicts of interest to declare.
References
- 1.NCD Risk Factor Collaboration. Worldwide trends in blood pressure from 1975 to 2015: a pooled analysis of 1479 population-based measurement studies with 19.1 million participants. Lancet. 2017;389(10064):37–55. doi: 10.1016/S0140-6736(16)31919-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yusuf S, Joseph P, Rangarajan S, Islam S, Mente A, Hystad P, et al. Modifiable risk factors, cardiovascular disease, and mortality in 155 722 individuals from 21 high-income, middle-income, and low-income countries (PURE): a prospective cohort study. Lancet. 2020;395(10226):795–808. doi: 10.1016/S0140-6736(19)32008-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ettehad D, Emdin CA, Kiran A, Anderson SG, Callender T, Emberson J, et al. Blood pressure lowering for prevention of cardiovascular disease and death: a systematic review and meta-analysis. Lancet. 2016;387(10022):957–67. doi: 10.1016/S0140-6736(15)01225-8. [DOI] [PubMed] [Google Scholar]
- 4.Arnett DK, Blumenthal RS, Albert MA, Buroker AB, Goldberger ZD, Hahn EJ, et al. 2019 ACC/AHA Guideline on the Primary Prevention of Cardiovascular Disease: Executive Summary: A Report of the American College of Cardiology/American Heart Association Task Force on Clinical Practice Guidelines. Circulation. 2019;140(11):e563–e95. doi: 10.1161/CIR.0000000000000677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Piepoli MF, Hoes AW, Agewall S, Albus C, Brotons C, Catapano AL, et al. 2016 European Guidelines on cardiovascular disease prevention in clinical practice: The Sixth Joint Task Force of the European Society of Cardiology and Other Societies on Cardiovascular Disease Prevention in Clinical Practice (constituted by representatives of 10 societies and by invited experts)Developed with the special contribution of the European Association for Cardiovascular Prevention & Rehabilitation (EACPR) Eur Heart J. 2016;37(29):2315–81. doi: 10.1093/eurheartj/ehw106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rapsomaniki E, Timmis A, George J, Pujades-Rodriguez M, Shah AD, Denaxas S, et al. Blood pressure and incidence of twelve cardiovascular diseases: lifetime risks, healthy life-years lost, and age-specific associations in 1.25 million people. Lancet. 2014;383(9932):1899–911. doi: 10.1016/S0140-6736(14)60685-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lewington S, Clarke R, Qizilbash N, Peto R, Collins R, Collaboration. PS Age-specific relevance of usual blood pressure to vascular mortality: a meta-analysis of individual data for one million adults in 61 prospective studies. Lancet. 2002;360(9349):1903–13. doi: 10.1016/s0140-6736(02)11911-8. [DOI] [PubMed] [Google Scholar]
- 8.Flint AC, Conell C, Ren X, Banki NM, Chan SL, Rao VA, et al. Effect of Systolic and Diastolic Blood Pressure on Cardiovascular Outcomes. N Engl J Med. 2019;381(3):243–51. doi: 10.1056/NEJMoa1803180. [DOI] [PubMed] [Google Scholar]
- 9.Vidal-Petiot E, Ford I, Greenlaw N, Ferrari R, Fox KM, Tardif JC, et al. Cardiovascular event rates and mortality according to achieved systolic and diastolic blood pressure in patients with stable coronary artery disease: an international cohort study. Lancet. 2016;388(10056):2142–52. doi: 10.1016/S0140-6736(16)31326-5. [DOI] [PubMed] [Google Scholar]
- 10.Ovbiagele B, Diener HC, Yusuf S, Martin RH, Cotton D, Vinisko R, et al. Level of Systolic Blood Pressure Within the Normal Range and Risk of Recurrent Stroke. JAMA. 2011;306(19):2137–44. doi: 10.1001/jama.2011.1650. [DOI] [PubMed] [Google Scholar]
- 11.Sprint Research Group. Wright JT, Jr, Williamson JD, Whelton PK, Snyder JK, Sink KM, et al. A Randomized Trial of Intensive versus Standard Blood-Pressure Control. N Engl J Med. 2015;373(22):2103–16. doi: 10.1056/NEJMoa1511939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mancia G, Grassi G. Aggressive Blood Pressure Lowering Is Dangerous: The J-Curve Pro Side of the Argument. Hypertension. 2014;63(1):29–36. doi: 10.1161/01.hyp.0000441190.09494.e9. [DOI] [PubMed] [Google Scholar]
- 13.Nazarzadeh M, Pinho-Gomes AC, Smith Byrne K, Canoy D, Raimondi F, Ayala Solares JR, et al. Systolic Blood Pressure and Risk of Valvular Heart Disease: A Mendelian Randomization Study. JAMA Cardiol. 2019;4(8):788–95. doi: 10.1001/jamacardio.2019.2202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Burgess S, Butterworth A, Malarstig A, Thompson SG. Use of Mendelian randomisation to assess potential benefit of clinical intervention. BMJ. 2012:345. doi: 10.1136/bmj.e7325. [DOI] [PubMed] [Google Scholar]
- 15.Sun YQ, Burgess S, Staley JR, Wood AM, Bell S, Kaptoge SK, et al. Body mass index and all cause mortality in HUNT and UK Biobank studies: linear and non-linear mendelian randomisation analyses. BMJ. 2019;364:l1042. doi: 10.1136/bmj.l1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Staley JR, Burgess S. Semiparametric methods for estimation of a nonlinear exposure-outcome relationship using instrumental variables with application to Mendelian randomization. Genet Epidemiol. 2017;41(4):341–52. doi: 10.1002/gepi.22041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Burgess S, Davies NM, Thompson SG, Consortium EP-I. Instrumental variable analysis with a nonlinear exposure-outcome relationship. Epidemiology. 2014;25(6):877–85. doi: 10.1097/EDE.0000000000000161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Davey Smith G, Davies NM, Dimou N, Egger M, Gallo V, Golub R, et al. STROBE-MR: Guidelines for strengthening the reporting of Mendelian randomization studies. PeerJ Preprints. 2019;7:e27857v1. doi: 10.7287/peerj.preprints.27857v1. [DOI] [Google Scholar]
- 19.Bycroft C, Freeman C, Petkova D, Band G, Elliott LT, Sharp K, et al. The UK Biobank resource with deep phenotyping and genomic data. Nature. 2018;562(7726):203–9. doi: 10.1038/s41586-018-0579-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Evangelou E, Warren HR, Mosen-Ansorena D, Mifsud B, Pazoki R, Gao H, et al. Genetic analysis of over 1 million people identifies 535 new loci associated with blood pressure traits. Nat Genet. 2018;50(10):1412–25. doi: 10.1038/s41588-018-0205-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Holmes MV, Davey Smith G. Problems in interpreting and using GWAS of conditional phenotypes illustrated by ‘alcohol GWAS’. Mol Psychiatry. 2018 doi: 10.1038/s41380-018-0037-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Willer CJ, Li Y, Abecasis GR. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics. 2010;26(17):2190–1. doi: 10.1093/bioinformatics/btq340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81(3):559–75. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Burgess S, Dudbridge F, Thompson SG. Combining information on multiple instrumental variables in Mendelian randomization: comparison of allele score and summarized data methods. Stat Med. 2016;35(11):1880–906. doi: 10.1002/sim.6835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Burgess S, Small DS, Thompson SG. A review of instrumental variable estimators for Mendelian randomization. Stat Methods Med Res. 2017;26(5):2333–55. doi: 10.1177/0962280215597579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Burgess S, Thompson SG. Use of allele scores as instrumental variables for Mendelian randomization. Int J Epidemiol. 2013;42(4):1134–44. doi: 10.1093/ije/dyt093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Palmer TM, Lawlor DA, Harbord RM, Sheehan NA, Tobias JH, Timpson NJ, et al. Using multiple genetic variants as instrumental variables for modifiable risk factors. Stat Methods Med Res. 2012;21(3):223–42. doi: 10.1177/0962280210394459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Slob EAW, Burgess S. A comparison of robust Mendelian randomization methods using summary data. Genet Epidemiol. 2020;44(4):313–29. doi: 10.1002/gepi.22295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bowden J, Davey Smith G, Burgess S. Mendelian randomization with invalid instruments: effect estimation and bias detection through Egger regression. Int J Epidemiol. 2015;44(2):512–25. doi: 10.1093/ije/dyv080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Verbanck M, Chen CY, Neale B, Do R. Detection of widespread horizontal pleiotropy in causal relationships inferred from Mendelian randomization between complex traits and diseases. Nat Genet. 2018;50(5):693–8. doi: 10.1038/s41588-018-0099-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Didelez V, Sheehan N. Mendelian randomization as an instrumental variable approach to causal inference. Stat Methods Med Res. 2007;16(4):309–30. doi: 10.1177/0962280206077743. [DOI] [PubMed] [Google Scholar]
- 32.Chow CK, Gupta R. Blood pressure control: a challenge to global health systems. Lancet. 2019;394(10199):613–5. doi: 10.1016/S0140-6736(19)31293-0. [DOI] [PubMed] [Google Scholar]
- 33.Whelton PK, Carey RM, Aronow WS, Casey DE, Jr, Collins KJ, Dennison Himmelfarb C, et al. 2017 ACC/AHA/AAPA/ABC/ACPM/AGS/APhA/ASH/ASPC/NMA/PCNA Guideline for the Prevention, Detection, Evaluation, and Management of High Blood Pressure in Adults: Executive Summary: A Report of the American College of Cardiology/American Heart Association Task Force on Clinical Practice Guidelines. Hypertension. 2018;71(6):1269–324. doi: 10.1161/HYP.0000000000000066. [DOI] [PubMed] [Google Scholar]
- 34.Rose G. Sick individuals and sick populations. Int J Epidemiol. 1985;14(1):32–8. doi: 10.1093/ije/14.1.32. [DOI] [PubMed] [Google Scholar]
- 35.Emberson J, Whincup P, Morris R, Walker M, Ebrahim S. Evaluating the impact of population and high-risk strategies for the primary prevention of cardiovascular disease. Eur Heart Js. 2004;25(6):484–91. doi: 10.1016/j.ehj.2003.11.012. [DOI] [PubMed] [Google Scholar]
- 36.Hardy ST, Loehr LR, Butler KR, Chakladar S, Chang PP, Folsom AR, et al. Reducing the Blood Pressure-Related Burden of Cardiovascular Disease: Impact of Achievable Improvements in Blood Pressure Prevention and Control. J Am Heart Assoc. 2015;4(10):e002276. doi: 10.1161/JAHA.115.002276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.He FJ, Brinsden HC, MacGregor GA. Salt reduction in the United Kingdom: a successful experiment in public health. J Hum Hypertens. 2014;28(6):345–52. doi: 10.1038/jhh.2013.105. [DOI] [PubMed] [Google Scholar]
- 38.Xu A, Ma J, Guo X, Wang L, Wu J, Zhang J, et al. Association of a Province-Wide Intervention With Salt Intake and Hypertension in Shandong Province, China, 2011-2016. JAMA Intern Med. 2020;180(6):877–86. doi: 10.1001/jamainternmed.2020.0904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wootton RE, Richmond RC, Stuijfzand BG, Lawn RB, Sallis HM, Taylor GMJ, et al. Evidence for causal effects of lifetime smoking on risk for depression and schizophrenia: a Mendelian randomisation study. Psychol Med. 2019:1–9. doi: 10.1017/S0033291719002678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Roden DM, Pulley JM, Basford MA, Bernard GR, Clayton EW, Balser JR, et al. Development of a large-scale de-identified DNA biobank to enable personalized medicine. Clin Pharmacol Ther. 2008;84(3):362–9. doi: 10.1038/clpt.2008.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Das S, Forer L, Schonherr S, Sidore C, Locke AE, Kwong A, et al. Next-generation genotype imputation service and methods. Nat Genet. 2016;48(10):1284–7. doi: 10.1038/ng.3656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.McCarthy S, Das S, Kretzschmar W, Delaneau O, Wood AR, Teumer A, et al. A reference panel of 64,976 haplotypes for genotype imputation. Nat Genet. 2016;48(10):1279–83. doi: 10.1038/ng.3643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Krokstad S, Langhammer A, Hveem K, Holmen TL, Midthjell K, Stene TR, et al. Cohort Profile: the HUNT Study, Norway. Int J Epidemiol. 2013;42(4):968–77. doi: 10.1093/ije/dys095. [DOI] [PubMed] [Google Scholar]
- 44.Loh PR, Danecek P, Palamara PF, Fuchsberger C, Reshef YA, Finucane HK, et al. Reference-based phasing using the Haplotype Reference Consortium panel. Nat Genet. 2016;48(11):1443–8. doi: 10.1038/ng.3679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Loh PR, Tucker G, Bulik-Sullivan BK, Vilhjalmsson BJ, Finucane HK, Salem RM, et al. Efficient Bayesian mixed-model analysis increases association power in large cohorts. Nat Genet. 2015;47(3):284–90. doi: 10.1038/ng.3190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Royston P, Altman DG. Regression Using Fractional Polynomials of Continuous Covariates - Parsimonious Parametric Modeling. J R Stat Soc C-Appl. 1994;43(3):429–67. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.