Abstract
MANF is an endoplasmic reticulum resident protein that plays a crucial role in attenuating ER stress responses. Although MANF is indispensable for the survival and function of mouse beta cells, its precise role in human beta cell development and function is unknown. Herein, we show that lack of MANF in humans results in diabetes due to increased ER stress leading to impaired beta cell function. We identified two patients from different families with childhood diabetes and a neurodevelopmental disorder associated with homozygous loss-of-function mutations in the MANF gene. To study the role of MANF in human beta cell development and function, we knocked out the MANF gene in human embryonic stem cells and differentiated them into pancreatic endocrine cells. Loss of MANF induced mild ER stress and impaired insulin processing capacity of beta cells in vitro. Upon implantation to immunocompromised mice, the MANF knockout grafts presented elevated ER stress and functional failure, particularly in diabetic recipients. By describing a new form of monogenic neurodevelopmental diabetes syndrome caused by disturbed ER function, we highlight the importance of adequate ER stress regulation for proper human beta cell function and demonstrate the crucial role of MANF in this process.
Introduction
Autosomal recessive causes of childhood-onset monogenic diabetes commonly present with syndromic diabetes (diabetes and additional non-autoimmune features). For instance, individuals carrying homozygous loss-of-function variants in WFS1, SLC19A2 and SLC29A3 develop diabetes in childhood with multiple non-autoimmune additional features (1–3). Autosomal recessive genetic diseases are enriched in populations with high rates of consanguinity (4). Genetic studies of children with syndromic diabetes who are born to consanguineous parents facilitates the identification of novel recessive causes of syndromic diabetes.
Recent studies highlight the important role of mesencephalic astrocyte-derived neurotrophic factor (MANF) in maintaining normal endoplasmic reticulum (ER) homeostasis (5). The major phenotype in juvenile MANF knockout mice is insulin-deficient diabetes, indicating the important role of MANF in the beta cell (6). Beta cells are under high demand for insulin synthesis, folding and secretion. ER is the intracellular compartment where proinsulin molecules fold to their native conformation before transport to Golgi for further granule packaging and secretion (7). Accumulation of unfolded and misfolded proteins in the ER leads to ER stress and activation of the unfolded protein response (UPR) (8). If prolonged, unresolved ER stress leads to cell death which is mainly mediated by DNA damage inducible transcript 3 (DDIT3/CHOP) (9). Therefore, any alterations in the mechanisms controlling protein folding may have a deleterious effect on beta cell function and survival (10).
In this study, we undertook whole genome sequencing for children with syndromic diabetes, aiming to identify novel autosomal recessive loss-of-function mutations (human knockout) causing monogenic diabetes. We show that homozygous loss-of-function variants in MANF cause a novel multisystem disorder characterised by childhood-onset diabetes, short stature, deafness, developmental delay and microcephaly. To study the role of MANF in human beta cell development and to create an experimental model for human diabetes caused by loss of MANF, we generated MANF knockout human embryonic stem cells (hESC) and differentiated them into pancreatic islet endocrine cells. Our results show that lack of MANF does not affect the development of human beta cells but increases their susceptibility to ER stress-induced functional failure. Our experimental findings provide a pathogenic explanation for the phenotype of the patients.
Research design and methods
Patient cohort
We investigated 32 children with syndromic diabetes with low likelihood of type 1 diabetes from 8 paediatric diabetes clinics across Turkey, a country with high rates of consanguinity (11). Syndromic diabetes was defined as the presence of any non-autoimmune features in addition to diabetes. All of the children were negative when tested for islet autoantibodies and their type 1 diabetes genetic risk score was <50th centile of type 1 diabetes population (12). Ethical approval for the study was from Kocaeli and Dokuz Eylul University ethics committee and Wales Research Ethics Committee. The study was conducted in accordance with the Declaration of Helsinki, and all subjects or their parents gave informed consent for genetic testing.
Molecular genetic analysis
The Molecular genetic analysis plan to identify novel homozygous predicted loss-of-function variant causing syndromic diabetes is described in supplementary figure 1.
We first performed targeted next-generation sequencing for all known monogenic diabetes genes as previously described (13). Individuals with likely pathogenic and pathogenic variants according to ACMG guideline were excluded (14). We calculated homozygosity from the off target reads of targeted next-generation sequencing gene panel using a SavvyHomozygosity tool (https://github.com/rdemolgen/SavvySuite) (15). We only took forward patients with calculated homozygosity >6.5% to increase the likelihood of identifying a novel homozygous pathogenic variant.
We performed gene discovery in the remaining eight individuals by whole genome sequencing. Genome sequencing was performed using BGI-Seg 500 on DNA extracted from peripheral blood leukocytes. The mean depth across these eight genomes were 35. The sequencing data were analysed using an approach based on the GATK best practice guidelines. GATK haplotype caller was used to identify variants which were annotated using Alamut batch version 1.11 (Sophia Genetics, Rouen, France) and variants which failed the QD2 VCF filter or had less than 5 reads supporting the variant allele were excluded. Copy number variants were called by SavvyCNV which uses read depth to judge copy number states (https://github.com/rdemolgen/SavvySuite) (15).
We prioritized homozygous protein truncating variants (frameshift, nonsense and splice acceptor/donor excluding last exon) in these genomes. These variants are predicted to cause loss of function by nonsense mediated mRNA decay (16). We investigated variants in the genes where there are no individuals with homozygous protein truncating variant in population and where homozygous protein truncating variants are not known to cause monogenic disease. We used multiple genome/exome population databases. This included gnomADv2.1 (n=135,743), gnomADv3 (n=71,702) (https://gnomad.broadinstitute.org) (17), TOPMed Freeze5 (n=62,784, https://bravo.sph.umich.edu/freeze5/hg38/) and two population with higher rates of consanguinity (The Greater Middle East (GME) Variome Project, n=2497)(http://igm.ucsd.edu/gme/index.php) (18) and Genes & Health, n=8921, Sep 2019 download (19).
Variant confirmation and co-segregation in family members were performed by Sanger sequencing (primer sequences available in Supplementary Table 1).
To identify additional patients with a variant in the putative candidate gene we used literature review, internet search (20) and Sanger sequencing of the candidate gene in patients selected based on homozygosity over the candidate gene and/or similar clinical features as a proband. The patients were referred for diagnostic genetic monogenic diabetes testing performed at the Department of Molecular Genetics, Royal Devon and Exeter NHS Trust, Exeter, UK and did not have a pathogenic variant in the known monogenic diabetes genes.
qRT-PCR analysis of MANF expression in human blood
Peripheral blood was collected in PAXgene Blood RNA tubes (PreAnalytix GmbH, Switzerland) and RNA extracted using the PAXgene Blood RNA kit (QIAGEN, Germany). Total RNA was DNase-treated using the TURBO DNA-free kit (Ambion, USA) and reverse transcribed using Superscript IV Reverse Transcriptase (Invitrogen, USA). Triplicate qPCR reactions (5μl) using TaqMan Gene Expression assays (Applied Biosystems, USA) were carried out on a QuantStudio 12K Flex (Thermo Fisher Scientific, USA). TaqMan Gene Expression assay IDs used are as follows: Hs00180640_m1 (MANF), Hs01060665_g1 (ACTB), Hs00831112_s1 (RPL11), Hs00984230_m1 (B2M), Hs00939627_m1 (GUSB) and Hs02786624_g1 (GAPDH). Expression of MANF was normalized using the geometric mean of five housekeeping genes (ACTB, RPL11, B2M, GUSB, GAPDH).
Cell culture and in vitro differentiation of human embryonic stem cells
Human embryonic stem cells (hESC) (WA01/H1 line, Wicell) were cultured on Matrigel-coated plates (BD Biosciences) in Essential 8 (E8) medium (Life technologies, A1517001). Differentiation was carried out following a seven-stage protocol as previously described with further modifications (21–23). The glucose concentration of each stage was as follow: S1-S4 cells were cultured in 10.5 mM glucose; S5-S6 cells were cultured in 20.5 mM glucose; and S7 cells were cultured in 5.5 mM glucose.
Genome editing
To recapitulate the gene mutations found in the patients, guide RNAs targeting the first exon of MANF gene for deletion were designed using Benchling (https://benchling.com). gRNAs with the highest quality score and lowest off-targets score were selected and purchased, alongside the RNP components (HiFi Cas9 protein, crRNA and tracrRNA), from Integrated DNA Technologies (IDT) and utilized according to the manufacturer’s recommended protocol. Two million undifferentiated stem cells were electroporated with the RNP complex using Neon Transfection system (Thermo Fisher, 1100 V, 20ms, two pulses), and plated on Matrigel-coated plates in E8 medium containing 10 μM ROCK inhibitor overnight. Afterwards, cells were single-cell sorted, expanded, and screened for the desired deletion using PCR. Positive clones were validated by Sanger sequencing and characterized for pluripotency and chromosomal integrity.
Western blot analysis
To extract whole cell protein from hESC-derived cells, cells were washed with ice-cold PBS and lysed with 300 μL Laemmli buffer (Bio-Rad) for 10 min on ice. The cells were then sonicated (3 x 15 s) and the homogenate centrifuged at 1000 rcf for 10 min at 4°C, heated at 95°C for 5 min. Equal amounts of supernatant were run on Any kD Mini-PROTEAN TGX gel (Bio-Rad, Hercules, CA, USA) at 100 V for 1 h in SDS running buffer (Bio-Rad). Proteins were then transferred to a nitrocellulose membrane (Invitrogen) using iBlot system (Invitrogen) for 5 min. The membrane was blocked in Tris-buffered saline containing 0.05% Tween (TBS-T) and 5% low fat dried milk for 90 min at 4°C. The membrane was then probed the primary antibody overnight at 4°C, washed twice with TBS-T, and incubated with the corresponding secondary antibody for 1 h at room temperature. Chemiluminescence detection was performed following Amersham ECL (Cytiva, RPN2235) exposure using Chemidoc XRS+ system and Image Lab Software (BioRad). Details of the used antibodies are listed in supplementary table 2.
Flow cytometry analysis
hESC-derived cells were dissociated into single cells by incubation with TrypLE for 7-10 min at 37°C and resuspended in cold FBS/PBS (5% v/v). For surface marker staining of CXCR4, 1 million cells were incubated with the directly conjugated antibody CD184/CXCR4 APC at a final dilution of 1:10 for 30 min at room temperature. For intracellular markers, 1 million cells were first fixed in 250 μl of Cytofix/Cytoprem Buffer (BD, No. 554722) for 20 min at 4°C, then washed twice with BD Perm/Wash Buffer Solution (BD, No. 554723). Cell pellet was resuspended in 80 μl FBS/BD Prem/Wash buffer (4% v/v) and incubated with the corresponding directly conjugated antibody at a final dilution of 1:80 overnight at 4°C. After incubation with the antibody, cells were washed twice and analyzed using FACSCalibur cytometer (BD Bioscience) and FlowJo software (Tree Star Inc.). Gating was defined using isotype antibodies. Details of the used antibodies are listed in supplementary table 2.
Quantitative RT-PCR
Gene expression was assessed at different stages of differentiation. Total RNA was extracted using NucleoSpin Plus RNA kit (Macherey-Nagel). Quantification and quality of the RNA preparations were measured using SimpliNano (General Electric) spectrophotometer. For cDNA synthesis, 1.5 μg RNA was first denatured for 1 min at 65°C, then reverse transcribed (RT) using GoScript™ Reverse Transcriptase kit (Promega). cDNA (50 ng) was amplified by qRT-PCR using 5x HOT FIREPol EvaGreen qPCR Mix Plus and 5 μL of forward and reverse primer mix at 2 μM each in a final volume of 20 μL. The reagents were pipetted by QIAgility (Quiagen) liquid handling system into 100 well disc and run in Rotor-Gene Q (Qiagen) thermo-cycler programmed as follows: 95°C for 15 min, followed by 40 cycles of 95°C for 25 s; 57°C for 25 s; 72°C for 25 s; and finished by a melting step. Rotor-Gene Q software (Qiagen) was used to calculate cycle threshold values (Ct). Cyclophilin G (PPIG) was used as an endogenous housekeeping gene to calculate ΔCt, and normalized to undifferentiated hESCs to calculate ΔΔCt values and relative gene expression. Exogenous positive control was used as a calibrator and RT-reaction without template was used as a negative control. Details of the used primers are listed in supplementary table 2.
Induction of endoplasmic reticulum stress
Fifty islet-like aggregates were manually picked and incubated in full S7 media supplemented with the corresponding concentration of ER-stress inducers. After the treatment, aggregates were collected, fixed with 4% PFA for 3 hours at room temperature, embedded in paraffin, and then processed for immunohistochemistry. 1 μM of thapsigargin (T9033, Sigma-Aldrich) and 5 μg/mL of tunicamycin (T7765, Sigma-Aldrich) were used for 48 hr, while 1 μg/ml of brefeldin A (B5936, Sigma-Aldrich) was used for 24 hr. DMSO was used as a vehicle control at 5 μL/mL.
Immunocytochemistry and histology
For adherent cultures, cells were fixed in 4% PFA for 15 min at RT, permeabilized with 0.5% triton-X100 in PBS, then blocked with UltraV block (ThermoFisher) for 10 min and incubated with primary antibodies diluted in 0.1% Tween in PBS overnight at 4°C. After incubation, cells were washed twice with PBS and incubated with corresponding secondary antibodies diluted in 0.1% Tween in PBS for 1 h at room temperature. For islet-like aggregates paraffin embedding, aggregates were fixed in 4% PFA for 3 h at RT, briefly stained with Eosin, then embedded in 2% low-melting agarose (Sigma-Aldrich) and transferred to paraffin blocks. For grafts, graft-containing kidney was retrieved, fixed in 4% PFA for 24 h at RT, and processed for paraffin embedding. Paraffin sections of 5 μm thickness were deparaffinised and antigen retrieval was carried out using the Decloaking Chamber (Biocare Medical) in citrate buffer at 95 °C for 12 min. Blocking and incubation with antibodies were performed as described above. For TUNEL assay, In Situ Cell Death Detection Fluorescein kit (Roche, #11684795910) was utilized according to manufacturer’s instruction. Details of the used antibodies are listed in supplementary table 3.
Static glucose stimulated insulin secretion (GSIS)
Fifty islet-like aggregates were manually picked and pre-incubated in 3.3 mM glucose-containing Krebs buffer in a 12-well plate placed on a rotating platform for 90 min at 37°C. Aggregates were then washed with Krebs buffer and sequentially incubated in Krebs buffer containing 3.3 mM glucose, 16.6 mM glucose, and 3.3 mM glucose + 30 mM KCL, for a period of 30 min. Samples of 300 μl were collected from the supernatant of each treatment and stored at -80°C for Insulin ELISA measurements. After the last incubation, aggregates were collected and lysed in acid ethanol for determination of total insulin content and DNA content. Stimulated insulin secretion results are presented as fractional release of total human insulin content after cell mass normalization using total DNA content.
Implantation of SC-islets
All experiments were approved by the Animal Welfare Committee of Southern Finland (ESAVI/14852/2018). NOD-SCID-gamma (NSG) (Jackson Laboratories, 0055577) were maintained at Biomedicum Helsinki animal facility, on a 12h light/dark cycle with ad libitum food. Mice were anesthesized with isoflurane, and subcutaneously injected with Carprofen (Rimadyl, 5 mg/Kg, Prizer, Helsinki, Finland) and Buprenorphine (Temgesic, 0.05 - 0.1 mg/Kg, RB pharmaceuticals Lmt, Berkshire, UK) as analgesics. Approximately 500 SC-islets were loaded on a PE-50 tubing and transplanted under the kidney capsule as previously described (24). Mouse blood samples were collected monthly from the saphenous vein using heparin-coated capillary tubes. Blood plasma was separated by centrifugation (5000 rcf, 5 min, RT) and stored at -80°C for human C-peptide analysis.
Streptozotocin treatment
After 3 months of implantation, mice were fasted for 4 h, then intraperitoneally injected with 130-150 mg/Kg streptozotocin (STZ, S0130, Sigma). STZ was solubilized in cold 0.1 M citrate buffer composing of 0.1 M citric acid monohydrate and 0.1 M trisodium citrate dehydrate. Drinking water was replaced with 10% sucrose-containing tap water for 48 h to avoid hypoglycemia caused by unregulated insulin release from dying β-cells.
Intraperitoneal glucose tolerance test (IPGTT)
After 6 h of fasting, mice were intraperitoneally injected with 2 g glucose/kg in the form of 30% glucose in saline. Blood glucose levels were measured using a glucometer (OneTouch Ultra, Lifescan, Milpitas, USA), and blood samples were collected for measuring human C-peptide levels at 0, 15, 30, 60, and 90 min after glucose injection.
ELISA
Insulin levels were measured from the in vitro samples with the Insulin ELISA kit (Mercodia, Sweden), and human-specific C-peptide was measured from plasma samples with Ultrasensetive C-PEPTIDE ELISA kit (Mercodia, Sweden).
Image acquisition and analysis
For adherent cultures, Evos inverted microscope (Life-Technologies) was used to image the cells. For paraffin sections, Zeiss Axio Observer Z1 with Apotome was used to image the cells, then images were processed with ZEN2 software. All paraffin sections were equally treated and imaged with the same microscope parameters. Image quantification were performed using CellProfiler software (25) and FIJI software (26).
Statistical analysis
All data are presented as individual points with their mean ± SEM. Statistical significance evaluation was conducted by unpaired, two-tailed Student t test with Welch’s correction using GraphPad Prism 8 software (GraphPad Software, La Jolla, CA). P-values under 0.05 were considered statistically significant and displayed as *, P-values under 0.001 were presented as **, and P-values under 0.0001 were presented as ***.
Results
Identification of individuals with homozygous protein truncating variants in MANF
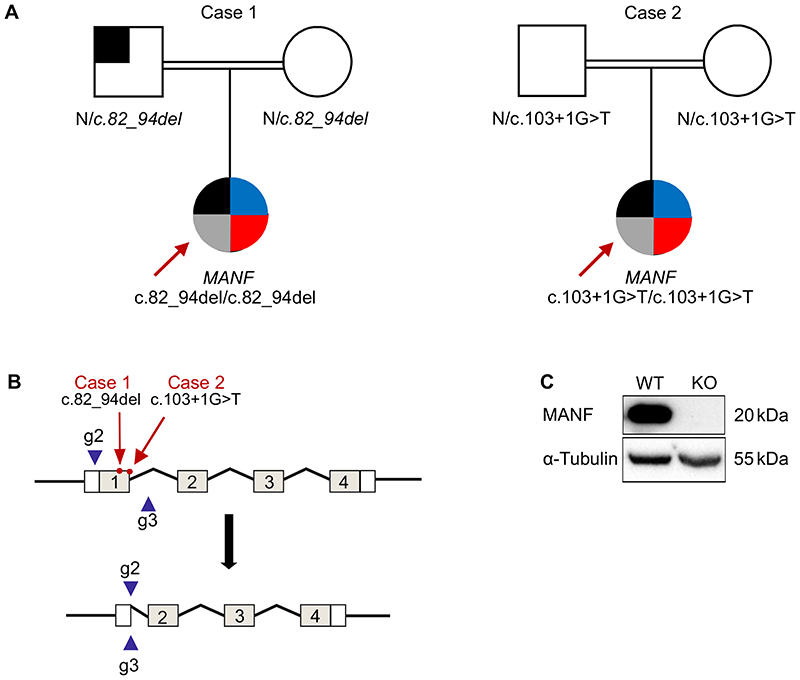
We performed gene discovery by whole genome sequencing on 8 subjects with non-autoimmune diabetes and additional extra-pancreatic features to identify novel homozygous loss of function cause of syndromic diabetes. The subjects did not have mutation in any of the known monogenic diabetes genes and had high genome wide homozygosity (>6.5%) (Supplementary figure 1A). Analysis of genome sequencing data identified one individual (case 1) with a novel homozygous frameshift variant in MANF (Chr3(GRCh37):g.51422846_51422858del, NM_006010.5:c.82_94del, p.Leu28Thrfs*33) (Fig 1) (Supplementary figure 1B). The variant was not present in 5 large independent population exome/genome databases (combined total n=281647) (17). None of these population databases had any homozygous protein truncating or rare homozygous missense (MAF <0.01) variants suggesting rare homozygous variants in MANF are highly intolerant to human.
Figure 1. Identification of homozygous MANF mutation in two patients with childhood diabetes.
A. Pedigrees of the two cases with homozygous MANF variants. Square symbolizes male and circle symbolizes female. Black shaded area indicates the presence of diabetes, Blue, and red area indicate the presence of short stature and deafness respectively whereas the grey shaded area indicates the presence of microcephaly and developmental delay.
B. Schematic presentation of the MANF gene knockout strategy utilizing CRISPR/Cas9. The positions of patient mutations are marked with red arrows, and the used guides to delete exon 1 are presented as blue arrow heads.
C. Representative immunoblot showing the absence of the MANF protein in the generated clone. α-tubulin used as a loading control.
Replication analysis identified a second independent individual (case 2) with a different homozygous protein truncating variant in MANF (NM_006010.5:c.103+1G>T, essential donor splice site). This case was partially described by Yavarna et al. (27). Case 2 was part of the exome study of individuals with clinically suspected mendelian disease (27). Similar to case 1, this variant is not seen in any population databases. The parents of both cases were heterozygous for MANF variants. Sanger sequencing of MANF from additional 187 patients presenting with diabetes, and microcephaly or developmental delay or deafness or short stature and/or homozygous for the region incorporating MANF, did not identify additional cases.
Both protein truncating variants are in exon 1. They generate premature termination codons and are predicted to cause loss of MANF due to classical nonsense-mediated mRNA decay of the mutant mRNA pathway (28). In line with this, MANF mRNA in peripheral blood of the case 1 was 5 fold lower compared to control individuals without damaging MANF variants (p=0.02) (Supplementary figure 2). These data suggest that both of these cases are lacking MANF and likely to represent a human knockout of MANF.
Lack of MANF causes diabetes, deafness, developmental delay, microcephaly and short stature
Both case 1 and case 2 present a similar clinical phenotype of childhood onset diabetes, short stature (<-3SD), bilateral sensorineural deafness, microcephaly and developmental delay (Fig. 1, Supplementary table 2). The diabetes was diagnosed at 10 and 17 years, respectively. Case 1 had brief treatment (10 days) with insulin followed by diet control diabetes for 1.5 y. Both patients are currently on insulin. They did not have autoantibodies associated with type 1 diabetes and have persistence of endogenous insulin secretion for ≥4 years post diagnosis, with C-peptide levels of 240 pmol/l and 330 pmol/l, respectively. Case 2 also has partial hypopituitarism with deficiency in growth hormone, thyroid stimulating hormone and gonadotropins. Hypopituitarism has not been detected in case 1 by 14y of age. Father of case 1 has a BMI of 29.4 and was diagnosed with type 2 diabetes at age 33. He is currently 41 and being treated with metformin. He does not present any other features as case 1.
Generation of a MANF knockout stem cell model
The homozygous mutations in both cases were found in exon 1 of the MANF gene (Fig. 1A and 1B). Therefore, we created an experimental model for dissecting the role of MANF in human beta cell development and function using CRISPR-Cas9 mediated genome editing to generate a deletion encompassing the first exon of MANF gene in H1 hESC. The genomic DNA sequencing of the generated clone showed the successful deletion of the targeted sequence which resulted in complete absence of MANF protein as shown by western blot analysis (Fig. 1C, Supplementary Fig. 2A). The generated clone was karyotypically normal and expressed markers of pluripotency (Supplementary Fig. 2B, 2C, and 2D). The KO clone, alongside its wild type counterpart, was differentiated into pancreatic islets for further studies.
MANF knockout stem cells differentiate normally to beta cells
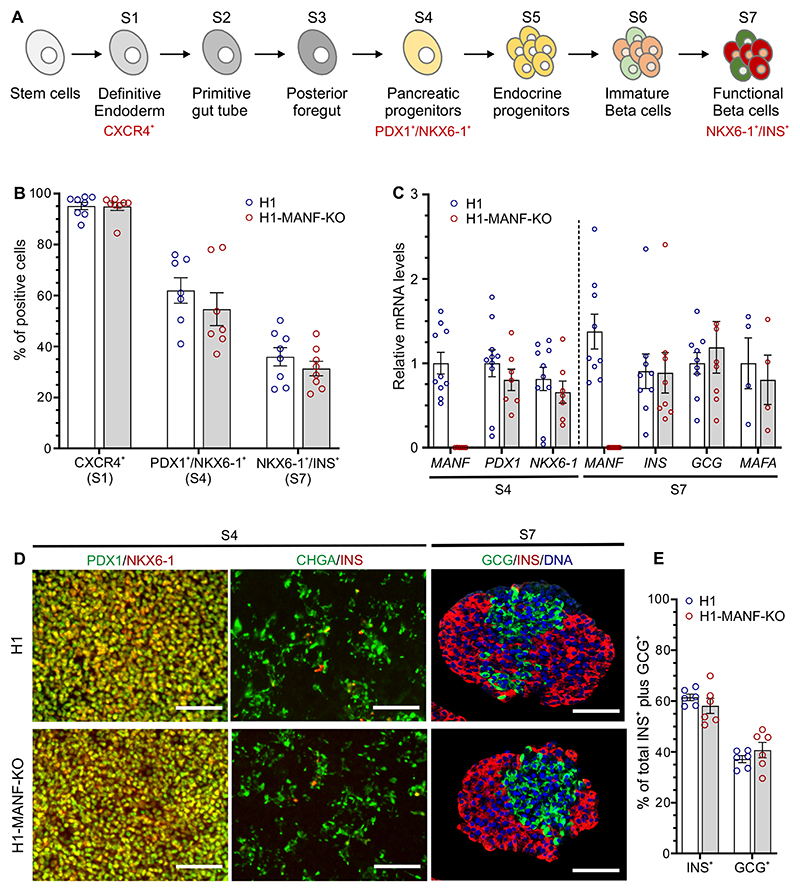
To study the role of MANF in pancreatic development, we differentiated MANF-KO H1 (H1-MANF-KO) and wild type H1 hESC into pancreatic islets following an optimized stepwise differentiation protocol (Fig. 2A). Both cell lines differentiated with similar efficiencies into definitive endoderm lineage, where approximately 95% of the cells expressed the definitive endoderm cell surface marker CXCR4 (Fig. 2B, Supplementary Fig. 3A) (29). After 12 days of differentiation (at S4, pancreatic progenitor stage), both cell lines were similarly co-expressing the pancreatic progenitor markers PDX1 and NKX6-1 in approximately 60% of the cells (Fig. 2B, Supplementary. Fig. 3A). This was further confirmed by their similar levels of PDX1 and NKX6-1 mRNA expression (Fig. 2C). Immunocytochemistry analysis at this stage showed similar expression pattern of pancreatic progenitor and endocrine markers in both cell lines (Fig. 2D). At the end stage (S7 SC-islets, islet endocrine cells), both cell lines generated approximately 35% insulin producing cells (Fig. 2B, Supplementary. Fig. 3A), and similar mRNA transcript levels of INS, GCG, and MAFA, a marker of mature beta cells (Fig. 2C) (22). Quantitative immunocytochemistry of these stem cell-derived islets (SC-islets) showed a similar ratio of monohormonal cells (INS+ and GCG+) (Fig. 2D and 2E). Taken together, these data indicate that MANF deficiency does not alter the differentiation of human pluripotent stem cells into pancreatic beta cells in vitro.
Figure 2. MANF knockout hESC differentiate normally to beta cells.
A. Schematic presentation of the seven-stage differentiation protocol (for further details, see research design and methods).
B. Graphical presentation of flow cytometry analysis of the differentiating hESC for specific relevant markers of each stage in H1 (wild type) and H1-MANF-KO (N=8 for S1 and S7 cells; N=7 For S4 cells). For the definitive endoderm stage, CXCR4 was used as a surface marker; PDX1 and NKX6-1 double positive staining was used as a marker of the pancreatic progenitor population (S4); while NKX6-1 and INS double positive staining was used as a marker of end stage β-like cells (S7).
C. Relative gene expression levels of pancreatic genes analyzed by RT-qPCR in H1 and H1-MANF-KO (N=4 - 9).
D. Immunocytochemistry analysis of S4 monolayer cells for PDX1, NKX6-1, chromogranin A (CHGA), and insulin (Scale bar=100 μm); and S7 aggregates for insulin, glucagon, and the nuclear stain Hochest (N=6, scale bar=100 μm).
E. Quantification of the percentage of insulin positive- and glucagon positive cells in H1 and H1-MANF-KO SC-islets.
H1 is presented as blue circles and H1-MANF-KO is presented as red circles. Data is presented as mean ± SEM
MANF knockout sensitizes beta cells to ER stress
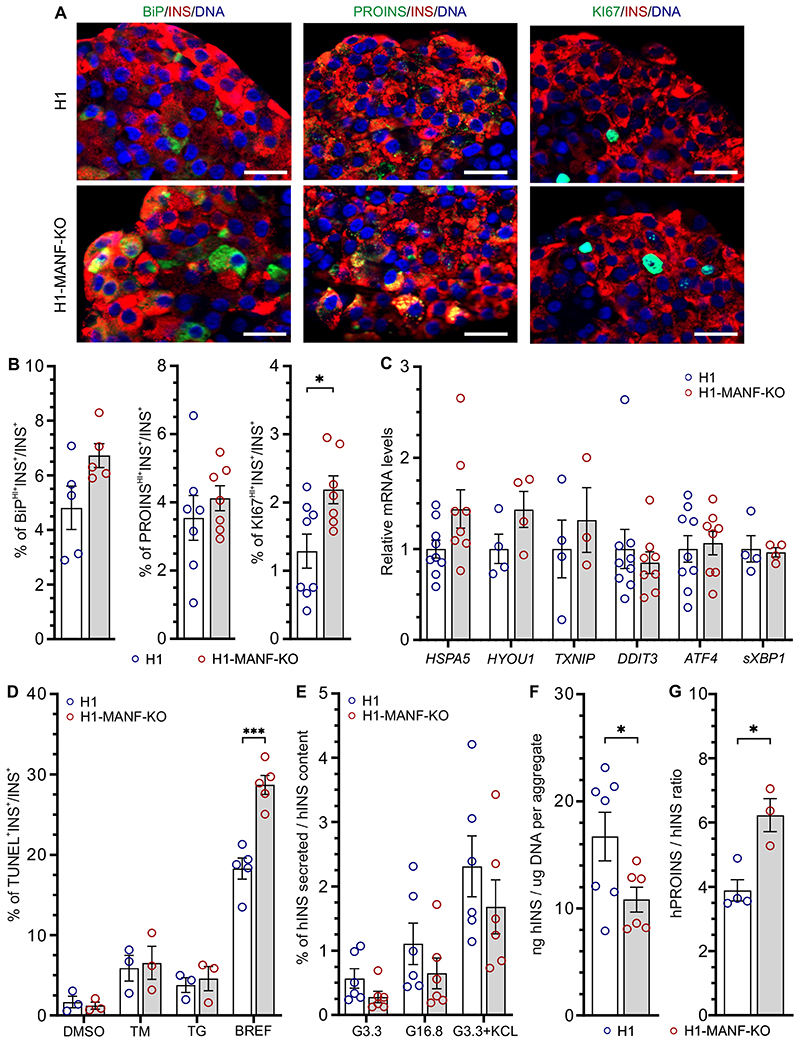
To assess the effect of MANF-KO on the expression levels of ER stress markers we employed immunocytochemistry on S7 SC-islets. MANF-KO beta-like cells showed marginally higher levels of BiP when compared to WT (Fig. 3A and B). This might indicate a higher level of ER stress in the MANF-KO beta-like cells, which could be associated with the higher accumulation of proinsulin in these cells (Fig. 3A and B). Interestingly, MANF-KO cells displayed significantly higher number of KI67+ proliferative beta-like cells when compared to the wild type (Fig. 3A and B). Furthermore, we confirmed our findings by quantifying the mRNA levels of ER stress markers using RT-qPCR, which showed an increasing trend in the expression levels of BiP (HSPA5), GRP170 (HYOU1), and TXNIP in MANF-KO cells (Fig. 3C, Supplementary Fig. 3B). However, both genotypes showed similar very low basal mRNA transcript levels of the inflammatory markers TNFα, CXCL-10, IL-6, and IFNα (data not shown).
Figure 3. MANF knockout Beta cells tend to have induced ER stress and significantly reduced insulin processing capacity.
A. Immunocytochemistry analysis of H1 and H1-MANF-KO S7 aggregates for β-like cells (INS+) which express ER stress marker BiP (N=5), proinsulin (PROINS) (N=6), and proliferation marker (Ki67) (N=8). (Scale bar=20 μM)
B. Quantified relative amounts of BiP, proinsulin and Ki67 positive beta cells from (A).
C. Relative gene expression levels of ER stress genes analyzed by RT-qPCR in H1 and H1-MANF-KO (N= 4 - 9).
D. ER stress-induced apoptosis of S7 aggregates. Quantification of the percentage of insulin positive cell which are labeled by TUNEL assay after treating the aggregates with the following chemical ER stress inducers: tunicamycin (TM), thapsigargin (TG) (N=3), and Brefaldin A (BREF) (N=5). DMSO was used as a vehicle control.
E. Static glucose stimulated insulin secretion of S7 SC-islets. Values are presented as fractional secretion of human insulin over total insulin content. (N=6)
F. Human insulin content of S7 SC-islets. Cell mass is normalized by average DNA content of each aggregate. (N=6)
G. Ratio of human proinsulin content to human insulin content per aggregate. (N=4 for H1, and 3 for H1-MANF-KO).
H1 is presented as blue circles and H1-MANF-KO is presented as red circles. Data is presented as mean ± SEM. P-values under 0.05 are displayed as *, and P-values under 0.0001 are displayed as ***.
To evaluate the role of MANF in beta cell survival, we quantified apoptotic cells using the TUNEL assay on S7 SC-islets treated with different ER stressors. In the basal condition, approximately 1.5% of the insulin positive cells were TUNEL+ in both MANF-KO and WT (Fig. 3D). Upon the induction of experimental ER stress using tunicamycin (TM), an inhibitor of N-linked glycosylation, or thapsigargin (TG), which depletes ER calcium (30), the percentage of apoptotic beta-like cells increased similarly in both genotypes (~ 8% with TM, and 5% with TG). However, upon the treatment with brefeldin A, an inhibitor of ER-to-Golgi cargo transfer (31), the percentage of MANF-KO apoptotic beta-like cells was significantly higher than WT (29% vs. 18%) (Fig. 3D, Supplementary Fig. 3C). These findings suggest that depletion of MANF results in mildly increased levels of ER stress in stem cell-derived beta cells in vitro, making them more sensitive to additional ER stress.
Impaired insulin processing in MANF knockout beta cells
Next, we examined the functionality of the SC-islets by performing static insulin secretion tests. Both genotypes responded to high glucose with a two-fold increase in human insulin secretion (Fig. 3E, Supplementary. Fig. 3D). Additionally, KCl induced a four-fold increase of insulin release in both genotypes, indicating normal insulin secretion response to depolarization (Fig 3E). Although both genotypes showed a similar pattern of insulin secretion, the absolute amount of insulin secreted by the MANF-KO SC-islets was lower and their insulin content was significantly lower than in wild type (Fig. 3F). Additionally, MANF-KO cells showed a significantly higher ratio of proinsulin to insulin content (Fig 3G) even though proinsulin content was similar (Supplementary Fig. 3E). Thus, MANF-KO had a minor effect on beta cell function but a significant effect on the insulin processing capacity of the beta cells in vitro.
Impaired function of MANF knockout beta cells after implantation
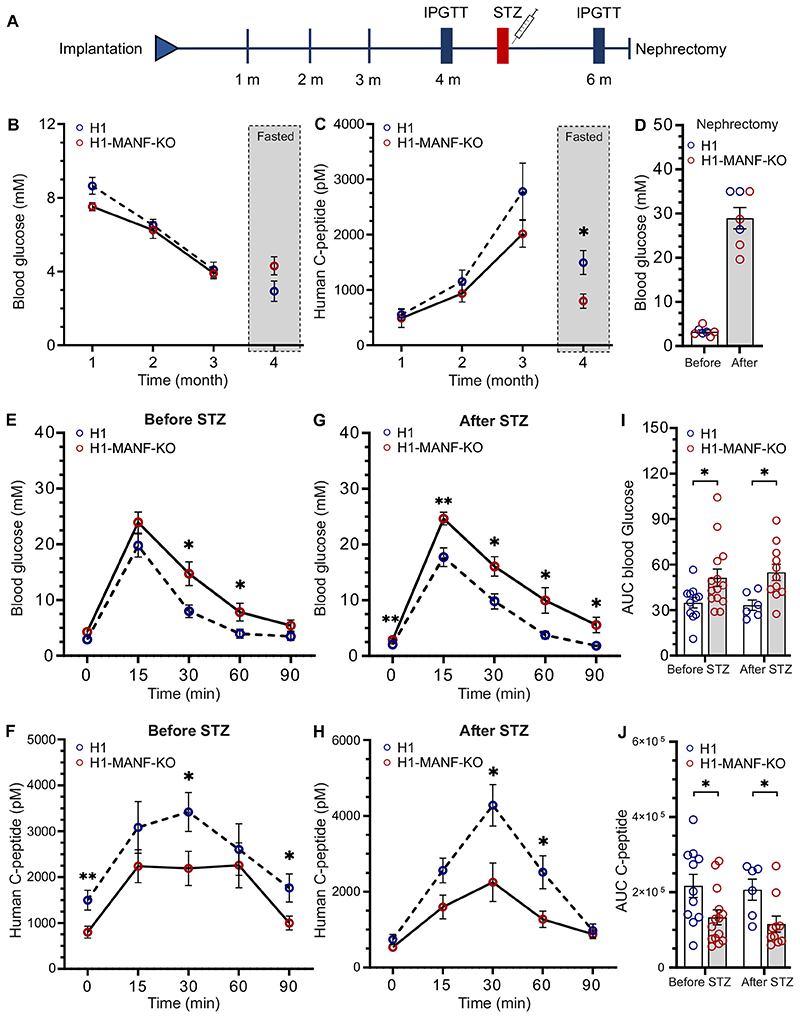
To evaluate the functionality of SC-islets in vivo, we implanted MANF-KO and WT SC-islets under the kidney capsule of immunocompromised (NSG) mice. We followed the functionality of the grafted cells for 6 months by measuring the circulating glucose and human C-peptide levels (Fig. 4A). Blood glucose levels of the mice implanted with both MANF-KO and WT cells decreased over time from usual normoglycemic levels for mice (8 mM) to human levels (4 mM) by 3 months, suggesting that the grafted cells were able to efficiently control the blood glucose of transplanted mice (Fig. 4B). This was consistent with a gradual increase in the levels of circulating human C-peptide in randomly fed mice, reaching up to 2800 pM after 3 months (Fig. 4C). At 4 months post-implantation, the levels of C-peptide in fasted mice were significantly higher in mice implanted with WT cells compared to MANF-KO (Fig. 4C).
Figure 4. Impaired response to glucose of MANF knockout cells in vivo.
A. Schematic presentation of the in vivo workflow.
B. Monthly follow-up of H1 and H1-MANF-KO in vivo functionality by measuring the blood glucose levels of implanted mice. Measurements of the first three months were done on randomly fed mice; while for the last month, mice were fasted. (N= 11 for H1 and 14 for H1-MANF-KO)
C. Monthly follow-up of H1 and H1-MANF-KO in vivo functionality by measuring the circulating human C-peptide levels of implanted mice. Measurements were done on randomly fed mice for the first three months; while for the last month, mice were fasted. (STZ). (N= 11 for H1 and 14 for H1-MANF-KO)
D. Blood glucose measurements before and after removing the implanted kidney of STZ-induced diabetic mice by surgical unilateral nephrectomy. (N=7)
E. Blood glucose measurements during an intraperitoneal glucose tolerance test of fasted H1 and H1-MANF-KO implanted mice before STZ treatment. (N= 11 for H1 and 14 for H1-MANF-KO)
F. Levels of circulating human C-peptide during an intraperitoneal glucose tolerance test of fasted H1 and H1-MANF-KO implanted mice before STZ treatment. (N= 11 for H1 and 14 for H1-MANF-KO)
G. Blood glucose measurements during an intraperitoneal glucose tolerance test of fasted H1 and H1-MANF-KO implanted mice after treatment. (N= 6 for H1 and 11 for H1-MANF-KO)
H. Levels of circulating human C-peptide during an intraperitoneal glucose tolerance test of fasted H1 and H1-MANF-KO implanted mice after STZ treatment. (N= 6 for H1 and 11 for H1-MANF-KO)
I. Quantification of the efficiency of the implanted grafts to regulate the mice blood glucose during IPGTT by measuring the area under the curve of (E and G).
J. Quantification of the ability of the implanted grafts to secrete human C-peptide by measuring the area under the curve of (F and H).
H1 is presented as blue circles and H1-MANF-KO is presented as red circles. Data is presented as mean ± SEM. P-values under 0.05 are displayed as *, and P-values under 0.001 are displayed as **.
Four months after implantation, the mice were challenged with an intraperitoneal glucose tolerance test. Blood glucose excursions were significantly higher in mice transplanted with MANF-KO grafts (Fig. 4 E and I). This was associated with lower secretion of human C-peptide (Fig. 4F and 4J). The glucose tolerance test was repeated after eradication of mouse beta cells using a single high dose of streptozotocin (STZ). The mice carrying WT grafts maintained normal glucose tolerance and high human C-peptide secretion in this test. However, the mice transplanted with MANF-KO grafts showed reduced glucose clearance (Fig. 4G and 4I) and a substantially lower increase in human C-peptide secretion (area under the curve, AUC, p= 0.02) (Fig. 4H and 4J). To confirm the functionality of the grafts and the success of the STZ treatment in eradicating the mouse beta cells, grafts were retrieved after 6 months by unilateral nephrectomy, which resulted in immediate hyperglycemia in all mice (Fig. 4D). Taken together, these data show impaired functionality of implanted MANF-KO beta cells.
Elevated ER stress and proinsulin accumulation in MANF knockout grafts
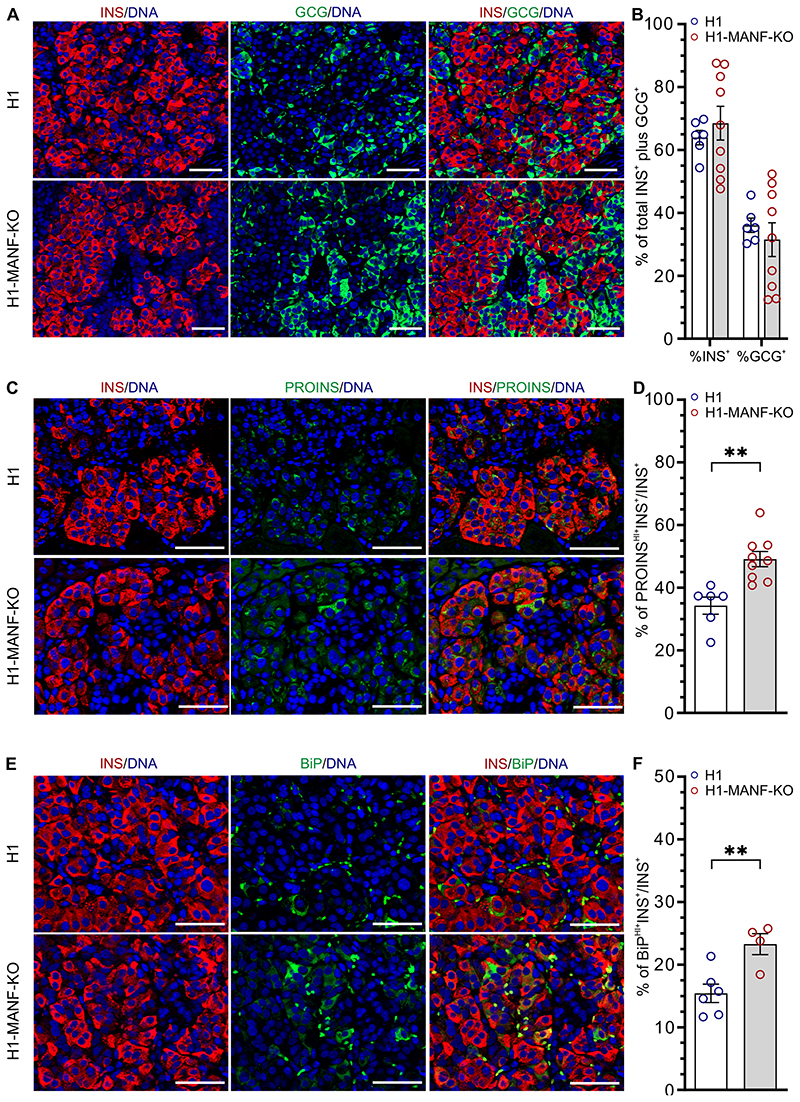
To further evaluate the effect of MANF-KO on the implanted SC-islets, grafts were retrieved at 1, 2, 4, and 6 months and examined by immunohistochemistry. Staining for INS and GCG in 6 months old grafts showed no differences in the proportions of beta and alpha cells between the genotypes, further indicating that MANF depletion did not affect pancreatic endocrine differentiation (Figure 5A and B). The percentage of INS+ cells in the grafts increased gradually with time (Supplementary Fig. 4E), while the number of proliferating beta cells gradually decreased (Supplementary. Fig. 4C and 4D). Interestingly, MANF-KO beta cells showed significantly higher levels of proinsulin (50% PROINSHI+INS+/INS+) compared to their wild type counterparts (35% PROINSHI+INS+/INS+) in the 6-month-old grafts (Fig 5C and D). A similar pattern was observed in the grafts retrieved at 2, and 4 months, suggesting an important defect in proinsulin processing in the ER (Supplementary Fig. 4G). In addition, immunohistochemistry analysis for ER stress markers in 6-month grafts revealed significantly higher levels of the ER stress markers BiP (Fig 5E, 5F, Supplementary Fig. 4H) and GRP170 (Supplementary Fig 4A and 4B) in MANF-KO grafts compared to the wild type grafts (23.3% vs. 15.4% BiPHI+INS+/INS+ cells and 27.9% vs. 10.2% GRP170HI+INS+/INS+ cells. Finally, TUNEL assay for apoptotic INS+ cells revealed an absence of beta cell death in the grafts of both genotypes (Supplementary Fig. 4F).
Figure 5. Increased ER stress and accumulation of PROINS in MANF knockout grafts.
A. Immunohistochemistry analysis of 6 months old grafts for insulin (INS) and glucagon (GCG). Scale bar=50 μM.
B. Quantification of the percentage of insulin and glucagon positive cells from (A) (N= 6 for H1 and 9 for H1-MANF-KO)
C. Immunohistochemistry analysis of 6 months old grafts for insulin (INS) and proinsulin (PROINS). Scale bar=50 μM.
D. Quantification of the relative number of insulin and proinsulin positive cells from (C) (N= 6 for H1 and 9 for H1-MANF-KO)
E. Immunohistochemistry analysis of 6 months old grafts for insulin (INS) cells which are expressing the ER stress marker BiP. Scale bar=50 μM.
F. Quantification of the relative number of BiP positive beta cells from (E) (N= 6 for H1 and 4 for H1-MANF-KO)
H1 is presented as blue circles and H1-MANF-KO is presented as red circles. Data is presented as mean ± SEM. P-values under 0.05 are displayed as *, and P-values under 0.001 are displayed as **.
Discussion
In this study we report MANF homozygous loss-of-function mutations as a novel genetic cause of childhood onset diabetes, deafness, developmental delay, microcephaly and short stature. We employed differentiation of hESCs lacking MANF to the pancreatic islets to model the consequences of these genetic defects. This revealed the crucial role of MANF in mitigating beta cell ER stress, and how its absence leads to impaired proinsulin processing and impaired beta cell function.
Strong genetic, clinical and functional data suggest that lack of MANF is indeed the cause of diabetes in these patients. The childhood progressive diabetes is unlikely to be type 1 diabetes as they lacked islet autoantibodies, had low genetic risk of type 1 diabetes and significant residual C-peptide 4 years after diagnosis (32). Both patients with different homozygous mutations have strikingly similar extra pancreatic phenotypes with microcephaly, developmental delay, sensorineural deafness and short stature. The brain is sensitive to ER stress mediated damage and almost always affected along with pancreas in other ER stress mediated monogenic diabetes forms (33). This strongly suggests that MANF deficiency is aetiological also for these features.
Both global and beta-cell specific loss of MANF results in severe early onset diabetes in transgenic mice (6,34). The two patients identified in this study were diagnosed with diabetes at the age of 10 and 17 years, suggesting a slower progression of the beta-cell failure in humans. In our MANF-KO hESC model, obvious signs of severe ER stress and functional deterioration were only evident in the in vivo model when the stem-cell derived beta cells became responsible for the insulin secretion of the diabetic host, suggesting again that loss of MANF leads to progressively impaired beta cell function and diabetes.
MANF-KO mice present neurological phenotypes that resemble those in the human MANF-KO patients. MANF inactivation in the mouse inner ear leads to a progressive death of outer hair cells of the cochlea accompanied with hearing loss (35). Similarly to the patients, MANF-KO mice show a growth defect and short stature that is independent of diabetes (6).
The ER stress response is a crucial adaptive pathway for beta cell function and survival. ER stress can be triggered by the accumulation of misfolded protein that is unable to exit the ER, as in the case of the dominant INS mutations causing severe forms of diabetes in Akita mice and humans (23,36). Pathogenic mutations in genes involved in the UPR also lead to neonatal diabetes, as in the case of EIF2AK3 mutations in Wolcott-Rallison syndrome (37), IER3IP1 mutations (38) or YIPF5 mutations (39). Childhood onset diabetes has been described in individuals carrying WFS1 and CISD2 mutations, leading to Wolfram syndrome (1), and recessive mutations in the DNAJC3 (40) and PPP1R15B genes (41). Indeed, the MANF syndrome clinical features of diabetes, microcephaly and other neurological features are overlapping with those seen in patients with above mutations (33,42). Thus, homozygous MANF mutations constitute a novel form of childhood onset diabetes caused by dysregulated ER stress and therefore MANF should be included in the gene panels testing for monogenic diabetes.
Similar to Wolfram syndrome, the MANF syndrome has childhood-onset diabetes with progressive beta cell dysfunction, developmental delay, and deafness. However, it lacks optic atrophy, diabetes insipidus, and urological defects which are common in Wolfram syndrome. Furthermore, the short stature and microcephaly seen in MANF syndrome are not reported in Wolfram syndrome. The difference in the clinical features indicates some differences in the pathogenesis. Further work with additional patients and animal models will be needed to fully understand these differences.
High expression of ER-stress markers in human adult beta cells has been linked to higher tendency of proliferation (43). Additionally, induction of mild ER stress by hyperglycemia has been reported to induce beta cell proliferation (44,45). Conversely, sub-threshold chronic ER stress has been shown to inhibit the proliferation of beta cells (46). We observed a similar pattern in our model where the in vitro mild ER stress significantly induced the proliferation of MANF-KO beta cells, while after implantation and exposure to more severe ER stress, there was a trend of decreased proliferation (Figure 3, Supplementary Fig. 4D).
As a compensatory response to ER stress, UPR tends to attenuate protein translation to decrease the folding load on the ER (8). This may result in decreased insulin synthesis and processing capacity in ER-stressed beta cells. A previous study on Wolfram syndrome, which is caused by ER dysfunction (47), reported that although the low levels of UPR signaling decreased the insulin content of beta cells, it did not alter their glucose stimulated insulin secretion in vitro (48). These results are well in line with the findings of our study, since we can detect a lower level of insulin content and secretion in the MANF-KO beta cells compared to wild type cells, while the insulin response to glucose stimulation was preserved. Additionally, the significantly higher ratio of proinsulin to insulin content in the MANF-KO cells, observed both in vitro and in vivo, indicates impaired insulin processing capacity, also reported in human prediabetes (49,50)
In conclusion, our findings indicate the essential role of MANF in protecting beta cells from ER stress-induced failure and apoptosis in humans. Our study also highlights the potential of the modelling platform which enables detailed studies of isogenic stem-cell derived beta cells both in vitro and in vivo. The mild phenotype observed in vitro, representing an immature beta cell population, followed by an exacerbated phenotype in more actively functioning beta cells in vivo, is likely to recapitulate the pathogenic evolution of the diabetic phenotype in the patients with MANF mutation.
Supplementary Material
Acknowledgements
HM, KP, DB, ATH, JSV, and TO conceived the project and planned the experiments. KAP, MW, KC, KD, TB and JL analyzed the genetic and clinical data. MW developed the genome sequencing and homozygosity mapping pipelines. DB performed genome editing and edited the manuscript. HI, VL, AN, and JSV assisted in data acquisitions. ML, and VC edited the manuscript. The first two authors have equal contribution: HM performed SC-islet differentiation, acquired and analyzed the data and wrote the related first draft of the manuscript, and KAP identified the causative gene in the patients and wrote the related first draft of the manuscript. All the authors reviewed and edited the manuscript. TO is the guarantor of this work. There are no conflicts of interest.
We thank the families for participating in the study. The authors are grateful to Solja Eurola, Jarkko Ustinov, Heli Grym and Anni Laitinen for expert technical assistance. We are also grateful to Rebecca Ward for her help with Sanger sequencing. The genetic studies were funded by Wellcome Trust awarded to Kashyap Patel (grant 110082/Z/15/Z). Kashyap Patel is a Wellcome trust Fellow (grant 219606/Z/19/Z). A.T.H. is a Wellcome Trust Senior Investigator (WT098395/Z/12/Z) and is also supported by a National Institute for Health Research (NIHR Senior Investigator award). The NIHR Exeter Clinical Research Facility is a partnership between the University of Exeter Medical School, College of Medicine and Health, and Royal Devon and Exeter NHS Foundation Trust. The views expressed are those of the author(s) and not necessarily those of the NIHR, Wellcome Turst or the Department of Health and Social Care, UK. The experimental studies were funded by the Academy of Finland (grant 297466 and MetaStem Center of Excellence grant 312437), the Sigrid Jusélius Foundation, the Novo Nordisk Foundation and the JDRF (Grant 2-SRA-2018-496-A-B, PI Mart Saarma, co-PI TO). HM is a member of the Doctoral Program in Integrative Life Science at University of Helsinki.
Footnotes
Prior presentation
Preliminary results from this study were presented at the European Association for the Study of Diabetes 55th annual meeting (16 – 20 September 2019) in Barcelona, Spain.
Data and resource availability
The data generated and analyzed during this study are included in the published article and its online supplementary files. Additional data and resources are available from the corresponding author upon reasonable request.
References
- 1.Vilan A, Faria O, Campos MM. Wolfram syndrome. Rev Neurol. 2009;49(4):221–2. [PubMed] [Google Scholar]
- 2.Labay V, Raz T, Baron D, Mandel H, Williams H, Barrett T, et al. Mutations in SLC19A2 cause thiamine-responsive megaloblastic anaemia associated with diabetes mellitus and deafness. Nat Genet. 1999 Jul;22(3):300–4. doi: 10.1038/10372. [DOI] [PubMed] [Google Scholar]
- 3.Noavar S, Behroozi S, Tatarcheh T, Parvini F, Foroutan M, Fahimi H. BMC Medical Genetics. Vol. 20. BioMed Central Ltd; 2019. A novel homozygous frame-shift mutation in the SLC29A3 gene: A new case report and review of literature. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fareed M, Afzal M. Genetics of consanguinity and inbreeding in health and disease. [cited 2020 Oct 31];Ann Hum Biol [Internet] 2017 Feb;44(2):99–107. doi: 10.1080/03014460.2016.1265148. [DOI] [PubMed] [Google Scholar]
- 5.Lindahl M, Saarma M, Lindholm P. Neurobiology of Disease. Vol. 97. Academic Press Inc; 2017. Unconventional neurotrophic factors CDNF and MANF: Structure, physiological functions and therapeutic potential; pp. 90–102. [DOI] [PubMed] [Google Scholar]
- 6.Lindahl M, Danilova T, Palm E, Lindholm P, Võikar V, Hakonen E, et al. MANF Is Indispensable for the Proliferation and Survival of Pancreatic βCells. Cell Rep. 2014 Apr;7(2):366–75. doi: 10.1016/j.celrep.2014.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fonseca SG, Gromada J, Urano F. Trends in Endocrinology and Metabolism. Vol. 22. Elsevier Inc; 2011. Endoplasmic reticulum stress and pancreatic β-cell death; pp. 266–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ron D, Walter P. Signal integration in the endoplasmic reticulum unfolded protein response. Nat Rev Mol Cell Biol. 2007;8(7):519–29. doi: 10.1038/nrm2199. [DOI] [PubMed] [Google Scholar]
- 9.Oyadomari S, Koizumi A, Takeda K, Gotoh T, Akira S, Araki E, et al. Targeted disruption of the Chop gene delays endoplasmic reticulum stress-mediated diabetes. J Clin Invest. 2002 Feb 15;109(4):525–32. doi: 10.1172/JCI14550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Eizirik DL, Cardozo AK, Cnop M. Endocrine Reviews. Vol. 29. Oxford Academic; 2008. The role for endoplasmic reticulum stress in diabetes mellitus; pp. 42–61. [DOI] [PubMed] [Google Scholar]
- 11.Koc I. Prevalence and sociodemographic correlates of consanguineous marriages in Turkey. J Biosoc Sci. 2008;40(1):137–48. doi: 10.1017/S002193200700226X. [DOI] [PubMed] [Google Scholar]
- 12.Patel KA, Oram RA, Flanagan SE, De Franco E, Colclough K, Shepherd M, et al. Type 1 diabetes genetic risk score: A novel tool to discriminate monogenic and type 1 diabetes. Diabetes. 2016 Jul 1;65(7):2094–9. doi: 10.2337/db15-1690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ellard S, Lango Allen H, De Franco E, Flanagan SE, Hysenaj G, Colclough K, et al. Improved genetic testing for monogenic diabetes using targeted next-generation sequencing. Diabetologia. 2013;56(9):1958–63. doi: 10.1007/s00125-013-2962-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J, et al. Standards and guidelines for the interpretation of sequence variants: A joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015 May 8;17(5):405–24. doi: 10.1038/gim.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Laver TW, De Franco E, Johnson MB, Patel K, Ellard S, Michael N. SavvyCNV: genome-wide CNV calling from off-target reads. 2019:1–23. doi: 10.1371/journal.pcbi.1009940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rivas MA, Pirinen M, Conrad DF, Lek M, Tsang EK, Karczewski KJ, et al. Effect of predicted protein-truncating genetic variants on the human transcriptome. Science (80-) 2015 May 8;348(6235):666–9. doi: 10.1126/science.1261877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Karczewski KJ, Francioli LC, Tiao G, Cummings BB, Alföldi J, Wang Q, et al. The mutational constraint spectrum quantified from variation in 141,456 humans. Nature. 2020 May 28;581(7809):434–43. doi: 10.1038/s41586-020-2308-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Scott EM, Halees A, Itan Y, Spencer EG, He Y, Azab MA, et al. Nature Genetics. Vol. 48. Nature Research; 2016. Characterization of greater middle eastern genetic variation for enhanced disease gene discovery; pp. 1071–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Narasimhan VM, Hunt KA, Mason D, Baker CL, Karczewski KJ, Barnes MR, et al. Health and population effects of rare gene knockouts in adult humans with related parents. Science (80-) 2016 Apr 22;352(6284):474–7. doi: 10.1126/science.aac8624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sobreira N, Schiettecatte F, Valle D, Hamosh A. GeneMatcher: A Matching Tool for Connecting Investigators with an Interest in the Same Gene. Hum Mutat. 2015 Oct 1;36(10):928–30. doi: 10.1002/humu.22844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pagliuca FW, Millman JR, Gürtler M, Segel M, Van Dervort A, Ryu JH, et al. Generation of functional human pancreatic β cells in vitro. Cell. 2014 Oct 9;159(2):428–39. doi: 10.1016/j.cell.2014.09.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rezania A, Bruin JE, Arora P, Rubin A, Batushansky I, Asadi A, et al. Reversal of diabetes with insulin-producing cells derived in vitro from human pluripotent stem cells. Nat Biotechnol. 2014 Sep 11;32(11):1121–33. doi: 10.1038/nbt.3033. [DOI] [PubMed] [Google Scholar]
- 23.Balboa D, Saarimäki-Vire J, Borshagovski D, Survila M, Lindholm P, Galli E, et al. Insulin mutations impair beta-cell development in a patient-derived iPSC model of neonatal diabetes. Elife. 2018 Nov 1;7 doi: 10.7554/eLife.38519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Saarimäki-Vire J, Balboa D, Russell MA, Saarikettu J, Kinnunen M, Keskitalo S, et al. An Activating STAT3 Mutation Causes Neonatal Diabetes through Premature Induction of Pancreatic Differentiation. Cell Rep. 2017 Apr 11;19(2):281–94. doi: 10.1016/j.celrep.2017.03.055. [DOI] [PubMed] [Google Scholar]
- 25.McQuin C, Goodman A, Chernyshev V, Kamentsky L, Cimini BA, Karhohs KW, et al. CellProfiler 3.0: Next-generation image processing for biology. PLoS Biol. 2018 Jul 3;16(7) doi: 10.1371/journal.pbio.2005970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rueden CT, Schindelin J, Hiner MC, Dezonia BE, Walter AE, Arena ET, et al. ImageJ2: ImageJ for the next generation of scientific image data. BMC Bioinformatics. 2017;18:529. doi: 10.1186/s12859-017-1934-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yavarna T, Al-Dewik N, Al-Mureikhi M, Ali R, Al-Mesaifri F, Mahmoud L, et al. High diagnostic yield of clinical exome sequencing in Middle Eastern patients with Mendelian disorders. Hum Genet. 2015 Sep 10;134(9):967–80. doi: 10.1007/s00439-015-1575-0. [DOI] [PubMed] [Google Scholar]
- 28.Khajavi M, Inoue K, Lupski JR. Nonsense-mediated mRNA decay modulates clinical outcome of genetic disease. Eur J Hum Genet. 2006;14(10):1074–81. doi: 10.1038/sj.ejhg.5201649. [DOI] [PubMed] [Google Scholar]
- 29.D’Amour KA, Agulnick AD, Eliazer S, Kelly OG, Kroon E, Baetge EE. Efficient differentiation of human embryonic stem cells to definitive endoderm. Nat Biotechnol. 2005 Dec 28;23(12):1534–41. doi: 10.1038/nbt1163. [DOI] [PubMed] [Google Scholar]
- 30.Abdullahi A, Stanojcic M, Parousis A, Patsouris D, Jeschke MG. Modelin acute ER stress in vivo and in vitro. Shock. 2017 Apr 1;47(4):506–13. doi: 10.1097/SHK.0000000000000759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tran K, Li Y, Duan H, Arora D, Lim HY, Wang W. Identification of small molecules that protect pancreatic β cells against endoplasmic reticulum stress-induced cell death. ACS Chem Biol. 2014 Dec 19;9(12):2796–806. doi: 10.1021/cb500740d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jones AG, Hattersley AT. Diabetic Medicine. Vol. 30. Wiley-Blackwell; 2013. The clinical utility of C-peptide measurement in the care of patients with diabetes; pp. 803–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cnop M, Toivonen S, Igoillo-Esteve M, Salpea P. Molecular Metabolism. Vol. 6. Elsevier GmbH; 2017. Endoplasmic reticulum stress and eIF2α phosphorylation: The Achilles heel of pancreatic β cells; pp. 1024–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Danilova T, Belevich I, Li H, Palm E, Jokitalo E, Otonkoski T, et al. MANF Is Required for the Postnatal Expansion and Maintenance of Pancreatic b-Cell Mass in Mice. Diabetes. 2019;68:66–80. doi: 10.2337/db17-1149. [DOI] [PubMed] [Google Scholar]
- 35.Herranen A, Ikäheimo K, Lankinen T, Pakarinen E, Fritzsch B, Saarma M, et al. Deficiency of the ER-stress-regulator MANF triggers progressive outer hair cell death and hearing loss. Cell Death Dis. 2020 Feb 1;11(2):1–12. doi: 10.1038/s41419-020-2286-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mizobuchi N, Hoseki J, Kubota H, Toyokuni S, Nozaki JI, Naitoh M, et al. ARMET is a soluble ER protein induced by the unfolded protein response via ERSE-II element. Cell Struct Funct. 2007;32(1):41–50. doi: 10.1247/csf.07001. [DOI] [PubMed] [Google Scholar]
- 37.Delépine M, Nicolino M, Barrett T, Golamaully M, Mark Lathrop G, Julier C. EIF2AK3, encoding translation initiation factor 2-α kinase 3, is mutated in patients with Wolcott-Rallison syndrome. Nat Genet. 2000;25(4):406–9. doi: 10.1038/78085. [DOI] [PubMed] [Google Scholar]
- 38.Shalev SA, Tenenbaum-Rakover Y, Horovitz Y, Paz VP, Ye H, Carmody D, et al. Microcephaly, Epilepsy and Neonatal Diabetes Due to Compound Heterozygous Mutations in IER3IP1: Insights into the Natural History of a Rare Disorder. Pediatr Diabetes. 2014;15(3):252–6. doi: 10.1111/pedi.12086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.De Franco E, Lytrivi M, Ibrahim H, Montaser H, Wakeling MN, Fantuzzi F, et al. YIPF5 mutations cause neonatal diabetes and microcephaly through endoplasmic reticulum stress. J Clin Invest. 2020 Nov 9;130(12) doi: 10.1172/JCI141455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Synofzik M, Haack TB, Kopajtich R, Gorza M, Rapaport D, Greiner M, et al. Absence of BiP Co-chaperone DNAJC3 causes diabetes mellitus and multisystemic neurodegeneration. Am J Hum Genet. 2014 Dec 4;95(6):689–97. doi: 10.1016/j.ajhg.2014.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Abdulkarim B, Nicolino M, Igoillo-Esteve M, Daures M, Romero S, Philippi A, et al. A missense mutation in PPP1R15B causes a syndrome including diabetes, short stature, and microcephaly. Diabetes. 2015 Nov 1;64(11):3951–62. doi: 10.2337/db15-0477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stone SI, Abreu D, McGill JB, Urano F. Monogenic and syndromic diabetes due to endoplasmic reticulum stress. J Diabetes Complications. 2020 May 8;:107618. doi: 10.1016/j.jdiacomp.2020.107618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Xin Y, Gutierrez GD, Okamoto H, Kim J, Lee AH, Adler C, et al. Pseudotime ordering of single human B-cells reveals states of insulin production and unfolded protein response. Diabetes. 2018 Sep 1;67(9):1783–94. doi: 10.2337/db18-0365. [DOI] [PubMed] [Google Scholar]
- 44.Porat S, Weinberg-Corem N, Tornovsky-Babaey S, Schyr-Ben-Haroush R, Hija A, Stolovich-Rain M, et al. Control of pancreatic β cell regeneration by glucose metabolism. Cell Metab. 2011 Apr 6;13(4):440–9. doi: 10.1016/j.cmet.2011.02.012. [DOI] [PubMed] [Google Scholar]
- 45.Sharma RB, Arvan P, Alonso LC, Sharma RB, Donnell ACO, Stamateris RE, et al. Insulin demand regulates b cell number via the unfolded protein response Find the latest version: Insulin demand regulates β cell number via the unfolded protein response. J Clin Invest. 2015;125(10):3831–46. doi: 10.1172/JCI79264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Szabat M, Page MM, Panzhinskiy E, Skovsø S, Mojibian M, Fernandez-Tajes J, et al. Reduced Insulin Production Relieves Endoplasmic Reticulum Stress and Induces β Cell Proliferation. Cell Metab. 2016 Jan 12;23(1):179–93. doi: 10.1016/j.cmet.2015.10.016. [DOI] [PubMed] [Google Scholar]
- 47.Riggs AC, Bernal-Mizrachi E, Ohsugi M, Wasson J, Fatrai S, Welling C, et al. Mice conditionally lacking the Wolfram gene in pancreatic islet beta cells exhibit diabetes as a result of enhanced endoplasmic reticulum stress and apoptosis. Diabetologia. 2005 Nov 8;48(11):2313–21. doi: 10.1007/s00125-005-1947-4. [DOI] [PubMed] [Google Scholar]
- 48.Shang L, Hua H, Foo K, Martinez H, Watanabe K, Zimmer M, et al. β-cell dysfunction due to increased ER stress in a stem cell model of wolfram syndrome. Diabetes. 2014 Mar 1;63(3):923–33. doi: 10.2337/db13-0717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.MykkÄnen L, Haffner SM, Kuusisto J, PyörÄlÄ K, Hales CN, Laakso M. Serum proinsulin levels are disproportionately increased in elderly prediabetic subjects. Diabetologia. 1995;38(10):1176–82. doi: 10.1007/BF00422366. [DOI] [PubMed] [Google Scholar]
- 50.Rodriguez-Calvo T, Zapardiel-Gonzalo J, Amirian N, Castillo E, Lajevardi Y, Krogvold L, et al. Increase in pancreatic proinsulin and preservation of β-cell mass in autoantibody-positive donors prior to type 1 diabetes onset. Diabetes. 2017 May 1;66(5):1334–45. doi: 10.2337/db16-1343. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data generated and analyzed during this study are included in the published article and its online supplementary files. Additional data and resources are available from the corresponding author upon reasonable request.