Abstract
Background and Purpose
Metabolic traits affect ischemic stroke (IS) risk, but the degree to which this varies across different ethnic ancestries is not known. Our aim was to apply Mendelian randomization to investigate the causal effects of type 2 diabetes mellitus (T2DM) liability and lipid traits on IS risk in African ancestry individuals, and to compare them to estimates obtained in European ancestry individuals.
Methods
For African ancestry individuals, genetic proxies for T2DM liability and circulating lipids were obtained from a meta-analysis of the African Partnership for Chronic Disease Research study, the UK Biobank and the Million Veteran Program (total N=77,061). Genetic association estimates for IS risk were obtained from the Consortium of Minority Population Genome-Wide Association Studies of Stroke (3,734 cases and 18,317 controls). For European ancestry individuals, genetic proxies for the same metabolic traits were obtained from Million Veteran Program (lipids N=297,626, T2DM N=148,726 cases and 965,732 controls), and genetic association estimates for IS risk were obtained from the MEGASTROKE study (34,217 cases and 406,111 controls). Random-effects inverse-variance weighted Mendelian randomization was used as the main method, complemented with sensitivity analyses more robust to pleiotropy.
Results
Higher genetically proxied T2DM liability, low-density lipoprotein cholesterol (LDL-C), total cholesterol (TC) and lower genetically proxied high-density lipoprotein cholesterol (HDL-C) were associated with increased risk of IS in African ancestry individuals (odds ratio per doubling the odds of T2DM liability [95% confidence interval] 1.09 [1.07-1.11]; per standard-deviation increase in LDL-C: 1.12 [1.04-1.21]; TC: 1.23 [1.06-1.43]; HDL-C: 0.93 [0.89-0.99]). There was no evidence for differences in these estimates when performing analyses in European ancestry individuals.
Conclusions
Our analyses support a causal effect of T2DM liability and lipid traits on IS risk in African ancestry individuals, with MR estimates similar to those obtained in European ancestry individuals.
Keywords: ethnicity, Mendelian randomization, etiological risk factor, stroke
Introduction
Stroke is a major contributor to morbidity and mortality globally, responsible for over 5.5 million deaths per year1. The global burden of stroke disproportionately affects low and middle-income countries, with over 85% of all stroke deaths occurring in these nations2. While stroke was historically seen as a disease affecting affluent regions, Africa now reports the highest incidence of stroke and the highest case-fatality in the world3. There is thus a growing need to understand the risk factors for stroke in African ancestry individuals.
Large multi-national observational studies have established metabolic traits, such as dyslipidemia and type 2 diabetes mellitus (T2DM), as risk factors for stroke4. However, it is not clear how the effects of these risk factors vary between individuals of different genetic ancestries. Ethnic variation in stroke risk factors has previously been explored in observational studies, but these are liable to confounding and reverse causation, limiting the ability to make causal inferences5.
To address these issues, Mendelian randomization (MR) employs genetic variants as proxies for an exposure to study its effect on an outcome6. MR is analogous to a randomized controlled trial with individuals being randomly assigned genetic variants at conception, minimizing confounding and reverse causality. MR has been widely used to examine risk factors for stroke in European populations7–9. However, similar studies in other ethnic groups have not been undertaken, largely due to paucity of genetic data on individuals of non-European populations. The publication of the Consortium of Minority Population Genome-Wide Association Studies of Stroke (COMPASS)10 provides an opportunity to conduct MR studies in people of African ancestry.
Here, we used MR to investigate the causal effect of lipid traits and T2DM liability on ischemic stroke (IS) risk in African ancestry populations and compared estimates to those obtained in individuals of European ancestry.
Methods
Ethical approval, data availability and reporting
We used summary data from published studies that obtained relevant ethical approval and participant consent. These data are available on request to the original studies. The analysis codes are available on request to the corresponding author, and all results are presented in the main manuscript or its supplementary files.
Genetic association estimates
We used two-sample MR to investigate the associations of genetically proxied levels of five metabolic traits with IS risk: T2DM liability, high-density lipoprotein cholesterol (HDL-C), low-density lipoprotein cholesterol (LDL-C), total cholesterol (TC) and triglycerides (TG). Genetic association estimates were obtained from publicly available summary statistics of genome-wide association studies (GWAS) detailed in Supplementary Methods. Briefly, genetic associations for metabolic traits in African ancestry individuals were obtained from a meta-analysis of the African Partnership for Chronic Disease Research (APCDR), self-reported Black participants in the UK Biobank and African ancestry individuals in the Million Veteran Program (MVP), with a total of approximately 77,000 participants. The genetic associations with the risk of IS were obtained from COMPASS, a GWAS meta-analysis of 3,734 cases and 18,317 controls of African ancestry from 13 cohorts10.
For European ancestry individuals, genetic associations for the metabolic traits were obtained from the MVP (T2DM liability, 148,726 cases, 965,732 controls; lipids N= 297,626) via dbGaP11,12. The genetic associations with the risk of IS were obtained from the MEGASTROKE consortium (34,217 cases, 406,111 controls, Supplementary Methods)13. All genetic associations for both ancestries were adjusted for age, sex and population stratification.
Mendelian randomization analysis
For each exposure, we identified ancestry-specific instrumental variables for MR, based on GWAS on the exposure in the relevant ethnic group: variants that associated with the exposure at p < 5×10−8 and were available in the outcome dataset were clumped at r2 < 0.01 within ± 500 kb, using the corresponding reference ancestry in 1000 Genomes Project. The remaining variants were used as instrumental variables for MR.
To measure instrument strength, we calculated the variance explained and F statistics for the individual variants. To evaluate statistical power, we calculated the minimum detectable odds ratio (OR) for each exposure at power = 0.8, given the exposure GWAS sample size, total variance explained by the genetic instruments (calculated as the sum of the variances explained by each individual instrument) and type I error rate = 0.0514.
The main analyses estimating the association of genetically proxied levels of each exposure with risk of IS were performed using the random-effects inverse-variance weighted (IVW) method15. We examined the differences in the MR estimates between populations of European and African ancestries using the propagation of error method. Further sensitivity analyses – namely MR-Egger, weighted median, weighted mode and contamination mixture method – were conducted to assess the robustness of the results to violations in instrumental variable assumptions (Supplementary Methods)15. MR effect estimates are expressed as ORs per standard deviation (SD) increase in genetically predicted levels of the exposure for continuous traits, and per doubling the odds (log-OR per unit change in exposure log-odds multiplied by loge(2)) in the exposure for T2DM.
Results
The demographics for African ancestry individuals in UK Biobank are given in Table 1, and the demographics for other considered populations can be found in their original publications10–13,16. Table 2 provides minimum detectable ORs for MR analyses, and the association estimates for the variants used as instrumental variables and their individual F statistics are given in Supplementary Table I.
Table 1. Demographics for African ancestry individuals in UKBiobank, N = 6,614.
| Variable | Median (interquartile range) or percentage |
|---|---|
| Age at recruitment in years | 51.6 (46.3-58.9) |
| Female sex | 59.0% |
| Type 2 diabetes | 9.0% |
| Body mass index in kg/m2 | 28.8 (25.9-32.4) |
| Low-density lipoprotein cholesterol in mmol/L | 3.3 (2.7-3.8) |
| High-density lipoprotein cholesterol in mmol/L | 1.4 (1.2-1.7) |
| Triglycerides in mmol/L Total Cholesterol in mmol/L |
1.0 (0.7-1.4) 5.2 (4.5-5.9) |
Table 2. Exposure summary data and statistical power calculations.
| Ancestry | Trait | Sample size | Variants (N) | Variance explained (%) |
Detectable odds ratio† |
|---|---|---|---|---|---|
| African | Type 2 diabetes mellitus | 24,646 cases, 31,446 controls | 22 | NA* | NA* |
| High-density lipoprotein cholesterol | 77,060 | 41 | 9.9 | 0.85 | |
| Low-density lipoprotein cholesterol | 77,060 | 71 | 21.4 | 1.11 | |
| Total cholesterol | 77,061 | 70 | 16.2 | 1.13 | |
| Triglycerides | 77,061 | 27 | 7.3 | 1.20 | |
| European | Type 2 diabetes mellitus | 148,726 cases, 965,732 controls | 557 | NA* | NA* |
| High-density lipoprotein cholesterol |
215,551 | 225 | 12.4 | 0.96 | |
| Low-density lipoprotein cholesterol |
215,551 | 145 | 10.4 | 1.05 | |
| Total cholesterol | 215,551 | 161 | 8.5 | 1.06 | |
| Triglycerides | 215,551 | 200 | 11.9 | 1.05 |
The sources for the summary data are detailed in the Supplementary Material.
Minimum detectable odds ratio per 1 standard deviation change in the exposure, at 80% power and Type I error rate = 0.05.
Variance explained and the minimal detectable odds ratio not estimated for type 2 diabetes mellitus because it is a binary exposure.
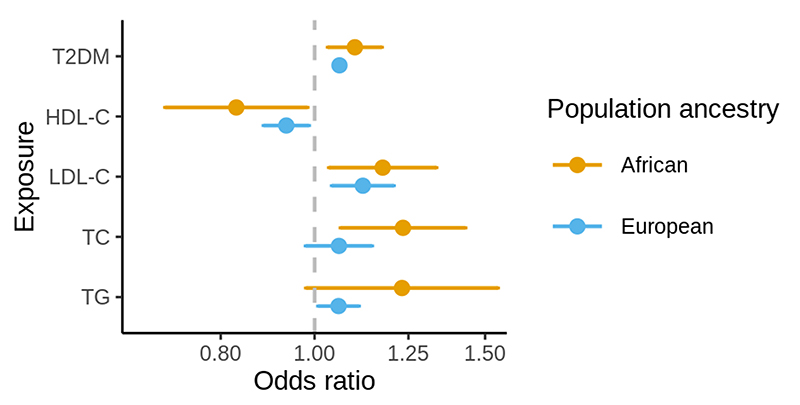
In the MR analysis of African ancestry populations, higher genetically proxied T2DM liability, LDL-C, and TC and lower genetically proxied HDL-C were associated with increased risk of IS (Figure, Supplementary Table II). The MR estimate for TG was similar in magnitude to the estimates for TC, LDL-C and inversely to HDL-C, however with 95% CI for OR overlapping the null.
Figure. Forest plot showing the main Mendelian randomization estimates for the association between genetically proxied lipid traits and type 2 diabetes mellitus liability with risk of ischemic stroke in African and European ancestry populations.
Estimates represent odds ratios and their 95% confidence intervals for ischemic stroke risk per 1 standard deviation increase in genetically-predicted levels of the exposure. T2DM = type 2 diabetes mellitus; HDL-C = high-density lipoprotein cholesterol; LDL-C = low density lipoprotein cholesterol; TC = total cholesterol; TG = triglycerides.
In European ancestry individuals, higher genetically proxied T2DM liability, LDL-C, and lower genetically proxied HDL-C were associated with increased risk of IS (Figure, Supplementary Table II). The effect estimate for genetically proxied TC was similar in the absolute value to other traits, however with 95% CI for the OR marginally overlapping the null.
The comparison of MR estimates between European and African ancestry populations showed no strong evidence for differences in the MR estimates. The point estimates were marginally larger in African ancestry individuals for all traits (Supplementary Table III). The associations between genetically proxied metabolic traits with the risk of IS were mostly consistent in the sensitivity analysis, apart from the estimates for HDL-C and TG in European ancestry populations which were shrunk towards the null in the sensitivity analyses, implying some degree of horizontal pleiotropy (Supplementary Table II, Supplementary Figures I-VI).
Discussion
This MR study found evidence for causal effects of lipid traits and T2DM liability on IS risk for African ancestry individuals. When compared to the estimates obtained in European ancestry individuals, there was no evidence for marked differences in the effects.
These findings are of direct clinical relevance, as they support that optimization of these risk factors will be of benefit in reducing IS for all individuals, irrespective of ethnic background. However, while the effect of dyslipidemia and T2DM on IS risk may be similar, it is also important to appreciate that the prevalence of these metabolic traits does vary considerably between different ethnic groups17, resulting in marked differences in the proportion of stroke that can be attributed to these risk factors18.
By leveraging large-scale genetic association data from African and European populations, we were able to investigate the comparative effects of T2DM liability and lipid traits on stroke risk in these ethnic groups. The use of genetically proxied metabolic traits in MR approach offers robustness against environmental confounding and reverse causation that can hinder causal inference in observational studies. The findings were mostly consistent in sensitivity analyses more robust to the inclusion of pleiotropic variants, suggesting that this is unlikely to be a major source of bias.
The limitations of this work should be acknowledged. The statistical power may have been insufficient to identify small differences in the MR estimates between European and African ancestry populations. The binary categorization of individuals as either of European or African ancestry is an over-simplification and will not capture the wider genetic diversity of individuals within each group. Furthermore, there may also be a population effect that impacts the genetic associations for individuals of the same ancestry when considered in different contexts19. Genetic association estimates were pooled from studies of heterogeneous populations with varying demographics. Despite the adjustments for age, sex and population stratification, population heterogeneity may introduce bias to the MR estimates. Summary statistics for the MVP data were publicly available via dbGaP only for variants with p<10-4, and therefore we were not able to conduct multivariable MR to investigate the mutually adjusted, direct effect of each considered cardiometabolic trait. Similarly, nor could we perform bi-directional MR to explore for reverse causality. We could not expand our analyses to other cardiometabolic traits, such as blood pressure or obesity, as sufficiently large GWAS summary statistics on these traits in African ancestry populations were not available to us. Finally, we were not able to examine the associations across different stroke subtypes, as subtype-specific GWAS summary statistics were not available in COMPASS.
In conclusion, our results are consistent with T2DM liability and lipid traits having a similar effect on IS risk in both African and European ancestry populations. Optimization of these risk factors will be of benefit for reducing the population burden of IS.
Supplementary Material
Acknowledgements
The COMPASS project would like to thank the staff and participants of the ARIC (Atherosclerosis Risk in Communities), CHS (Cardiovascular Health Study), VISP (Vitamin Intervention for Stroke Prevention), HANDLS (Healthy Aging in Neighborhoods of Diversity Across the Life Span), INTERSTROKE, ISGS (Ischemic Stroke Genetics Study), JHS (Jackson Heart Study), SIGNET-REGARDS (Sea Islands Genetics Network–Reasons for Geographic and Racial Differences in Stroke), GEOS (Genetics of Early Onset Stroke), SiGN (Stroke Genetics Network), SLESS (South London Ethnicity and Stroke Study), SWISS (Siblings With Ischemic Stroke Study), and WHI (Women’s Health Initiative) studies for their dedication, and willingness to participate in the respective research studies, which made this work possible. The MEGASTROKE project received funding from sources specified at http://www.megastroke.org/acknowledgments.html.
Funding
SF is funded by the Wellcome International Intermediate fellowship (220740/Z/20/Z) at the MRC/UVRI and LSHTM. DG is supported by the Wellcome 4i Program (203928/Z/16/Z) and by a National Institute for Health Research Clinical Lectureship (CL-2020-16-001) at St. George’s, University of London. VK and DG are supported by British Heart Foundation Centre of Research Excellence (RE/18/4/34215) at Imperial College London. TC is an international training fellow supported by the Wellcome Trust grant (214205/Z/18/Z). BU and MN are supported by the Wellcome Trust. MN and S.F. received support from the Makerere University-Uganda Virus Research Institute Centre of Excellence for Infection and Immunity Research and Training (MUII). MUII is supported through the DELTAS Africa Initiative (grant 107743). The DELTAS Africa Initiative is an independent funding scheme of the African Academy of Sciences (AAS), Alliance for Accelerating Excellence in Science in Africa (AESA), and supported by the New Partnership for Africa’s Development Planning and Coordinating Agency (NEPAD Agency) with funding from the Wellcome Trust (107743) and the U.K. government. MN acknowledges the support of MacNCD. This research is made possible by the Makerere University Non-Communicable Diseases (MakNCD) Research Training Program: NIH 1D43TW011401-01 through the Fogarty International Center (FIC). CZ, BU and SF acknowledge H3Africa Bioinformatics Network (H3ABioNet) Node, National Biotechnology Development Agency (NABDA) and the Center for Genomics Research and Innovation (CGRI) Abuja, Nigeria. SMD was supported by the Department of Veterans Affairs Office of Research and Development (IK2-CX001780). AMM is funded by the EC-Innovative Medicines Initiative (BigData@Heart). SB is supported by a Sir Henry Dale Fellowship jointly funded by the Wellcome Trust and the Royal Society (204623/Z/16/Z). This research was funded by UKRI Medical Research Council (MC_UU_00002/7) and supported by the NIHR Cambridge Biomedical Research Centre (BRC-1215-20014). The views expressed are those of the author(s) and not necessarily those of the NIHR or the Department of Health and Social Care. The funding sources did not have any role in designing the study, performing analysis or communicating findings.
Non-standard Abbreviations and Acronyms:
- APCDR
African Partnership for Chronic Disease Research
- COMPASS
Consortium of Minority Population Genome-Wide Association Studies of Stroke
- dbGaP
database of Genotypes and Phenotypes
- HDL-C
High Density Lipoprotein Cholesterol
- IS
Ischemic stroke
- LDL-C
Low Density Lipoprotein Cholesterol
- MR
Mendelian randomization
- MVP
Million Veteran Program
- T2DM
Type 2 Diabetes Mellitus
- TC
Total Cholesterol
- TG
Triglycerides
Footnotes
Contributors
SF and DG designed the study. VK and SF performed statistical analyses. SF, VK, DKR, AT, DG and drafted the manuscript. All authors interpreted results, edited the manuscript for intellectual content, and take responsibility for the integrity of the study. SF and VK had full access to all the data in the study and take responsibility for the integrity of the data and the accuracy of the data analysis.
Disclosures
SMD has received grants from the U.S. Department of Veterans Affairs, Calico Labs, and Renalytix AI plc outside the submitted work. DG is employed part-time by Novo Nordisk. TS reports grants and personal fees from NIH Cooperative Agreement 1 U2R TW010673-01 for West African Center of Excellence for Global Health Bioinformatics Research Training during the conduct of the study; personal fees from NIH Cooperative Agreement 1 U2R TW010673-01 for West African Center of Excellence for Global Health Bioinformatics Research Training outside the submitted work. AMM reports grants from National Institute for Health Research [Cambridge Biomedical Research Centre at the Cambridge University Hospitals NHS Foundation Trust] and grants from EC-Innovative Medicines Initiative (BigData@Heart) during the conduct of the study. The remaining authors have no conflicts of interest to declare.?
References
- 1.Donkor ES. Stroke in the 2 1 s t Century: A Snapshot of the Burden, Epidemiology, and Quality of Life. Stroke Research and Treatment. 2018;2018:1–10. doi: 10.1155/2018/3238165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Owolabi M, Akarolo-Anthony S, Akinyemi R, Arnett D, Gebregziabher M, Jenkins C, Tiwari H, Arulogun O, Akpalu A, Sarfo F, et al. The burden of stroke in Africa: a glance at the present and a glimpse into the future: review article. CVJA. 2015;26:S27–S38. doi: 10.5830/CVJA-2015-038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Owolabi M, Olowoyo P, Popoola F, Lackland D, Jenkins C, Arulogun O, Akinyemi R, Akinyemi O, Akpa O, Olaniyan O, et al. The epidemiology of stroke in Africa: A systematic review of existing methods and new approaches. J Clin Hypertens. 2018;20:47–55. doi: 10.1111/jch.13152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.O’Donnell MJ, Chin SL, Rangarajan S, Xavier D, Liu L, Zhang H, Rao-Melacini P, Zhang X, Pais P, Agapay S, et al. Global and regional effects of potentially modifiable risk factors associated with acute stroke in 32 countries (INTERSTROKE): a case-control study. The Lancet. 2016;388:761–775. doi: 10.1016/S0140-6736(16)30506-2. [DOI] [PubMed] [Google Scholar]
- 5.Agyemang C, Addo J, Bhopal R, de Graft Aikins A, Stronks K. Cardiovascular disease, diabetes and established risk factors among populations of sub-Saharan African descent in Europe: a literature review. Global Health. 2009;5:7. doi: 10.1186/1744-8603-5-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Davies NM, Holmes MV, Davey Smith G. Reading Mendelian randomisation studies: a guide, glossary, and checklist for clinicians. BMJ. 2018:k601. doi: 10.1136/bmj.k601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hopewell JC, Clarke R. Emerging Risk Factors for Stroke: What Have We Learned From Mendelian Randomization Studies? Stroke. 2016;47:1673–1678. doi: 10.1161/STROKEAHA.115.010646. [DOI] [PubMed] [Google Scholar]
- 8.Larsson SC, Scott RA, Traylor M, Langenberg CC, Hindy G, Melander O, Orho-Melander M, Seshadri S, Wareham NJ, Markus HS, et al. Type 2 diabetes, glucose, insulin, BMI, and ischemic stroke subtypes: Mendelian randomization study. Neurology. 2017;89:454–460. doi: 10.1212/WNL.0000000000004173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hindy G, Engström G, Larsson SC, Traylor M, Markus HS, Melander O, Orho-Melander M. Role of Blood Lipids in the Development of Ischemic Stroke and its Subtypes: A Mendelian Randomization Study. Stroke. 2018;49:820–827. doi: 10.1161/STROKEAHA.117.019653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Keene KL, Hyacinth HI, Bis JC, Kittner SJ, Mitchell BD, Cheng Y-C, Pare G, Chong M, O’Donnell M, Meschia JF, et al. Genome-Wide Association Study Meta-Analysis of Stroke in 22 000 Individuals of African Descent Identifies Novel Associations With Stroke. Stroke. 2020;51:2454–2463. doi: 10.1161/STROKEAHA.120.029123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Global Lipids Genetics Consortium, Myocardial Infarction Genetics (MIGen) Consortium, The Geisinger-Regeneron DiscovEHR Collaboration, The VA Million Veteran Program. Klarin D, Damrauer SM, Cho K, Sun YV, Teslovich TM, Honerlaw J, et al. Genetics of blood lipids among ~300,000 multi-ethnic participants of the Million Veteran Program. Nat Genet. 2018;50:1514–1523. doi: 10.1038/s41588-018-0222-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vujkovic M, Keaton JM, Lynch JA, Miller DR, Zhou J, Tcheandjieu C, Huffman JE, Assimes TL, Lorenz K, Zhu X, et al. Discovery of 318 new risk loci for type 2 diabetes and related vascular outcomes among 1.4 million participants in a multi-ancestry meta-analysis. Nature Genetics. 2020;52:680–691. doi: 10.1038/s41588-020-0637-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Malik R, Chauhan G, Traylor M, Sargurupremraj M, Okada Y, Mishra A, Rutten-Jacobs L, Giese A-K, van der Laan SW, Gretarsdottir S, et al. Multiancestry genome-wide association study of 520,000 subjects identifies 32 loci associated with stroke and stroke subtypes. Nature Genetics. 2018;50:524–537. doi: 10.1038/s41588-018-0058-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Burgess S. Sample size and power calculations in Mendelian randomization with a single instrumental variable and a binary outcome. International Journal of Epidemiology. 2014;43:922–929. doi: 10.1093/ije/dyu005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Slob EAW, Burgess S. A comparison of robust Mendelian randomization methods using summary data. Genetic Epidemiology. 2020;44:313–329. doi: 10.1002/gepi.22295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gurdasani D, Carstensen T, Fatumo S, Chen G, Franklin CS, Prado-Martinez J, Bouman H, Abascal F, Haber M, Tachmazidou I, et al. Uganda Genome Resource Enables Insights into Population History and Genomic Discovery in Africa. Cell. 2019;179:984–1002.:e36. doi: 10.1016/j.cell.2019.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cruz-Flores S, Rabinstein A, Biller J, Elkind MSV, Griffith P, Gorelick PB, Howard G, Leira EC, Morgenstern LB, Ovbiagele B, et al. Racial-Ethnic Disparities in Stroke Care: The American Experience: A Statement for Healthcare Professionals From the American Heart Association/American Stroke Association. Stroke. 2011;42:2091–2116. doi: 10.1161/STR.0b013e3182213e24. [DOI] [PubMed] [Google Scholar]
- 18.Hajat C, Tilling K, Stewart JA, Lemic-Stojcevic N, Wolfe CDA. Ethnic Differences in Risk Factors for Ischemic Stroke: A European Case–Control Study. Stroke. 2004;35:1562–1567. doi: 10.1161/01.STR.0000131903.04708.b8. [DOI] [PubMed] [Google Scholar]
- 19.Gola D, Erdmann J, Läll K, Mägi R, Müller-Myhsok B, Schunkert H, König IR. Population Bias in Polygenic Risk Prediction Models for Coronary Artery Disease. [cited 2021 Mar 28];Circ: Genomic and Precision Medicine. 2020 :13. doi: 10.1161/CIRCGEN.120.002932. [Internet]. Available from: [DOI] [PubMed] [Google Scholar]
- 20.Bycroft C, Freeman C, Petkova D, Band G, Elliott LT, Sharp K, Motyer A, Vukcevic D, Delaneau O, O’Connell J, et al. The UK Biobank resource with deep phenotyping and genomic data. Nature. 2018;562:203–209. doi: 10.1038/s41586-018-0579-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gaziano JM, Concato J, Brophy M, Fiore L, Pyarajan S, Breeling J, Whitbourne S, Deen J, Shannon C, Humphries D, et al. Million Veteran Program: A mega-biobank to study genetic influences on health and disease. Journal of Clinical Epidemiology. 2016;70:214–223. doi: 10.1016/j.jclinepi.2015.09.016. [DOI] [PubMed] [Google Scholar]
- 22.Wasserstein RL, Schirm AL, Lazar NA. Moving to a World Beyond “p < 0.05.”. The American Statistician. 2019;73:1–19. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.