Abstract
Children with B-cell non-Hodgkin lymphoma (B-NHL) have an excellent chance of survival, however, current clinical risk stratification places as many as half of patients in a high-risk group receiving very intensive chemo-immunotherapy. TP53 alterations are associated with adverse outcome in many malignancies; however, whilst common in paediatric B-NHL, their utility as a risk classifier is unknown. We evaluated the clinical significance of TP53 abnormalities (mutations, deletion and/or copy number neutral loss of heterozygosity) in a large UK paediatric B-NHL cohort and determined their impact on survival. TP53 abnormalities were present in 54.7% of cases and were independently associated with a significantly inferior survival compared to those without a TP53 abnormality (PFS 70.0% vs 100%, p<0.001, OS 78.0% vs 100%, p=0.002). Moreover, amongst patients clinically defined as high-risk (stage III with high LDH or stage IV), those without a TP53 abnormality have superior survival compared to those with TP53 abnormalities (PFS 100% vs 55.6%, p=0.005, OS 100% vs 66.7%, p=0.019). Biallelic TP53 abnormalities were either maintained from presentation or acquired at progression in all paired diagnosis/progression Burkitt lymphoma cases. TP53 abnormalities thus define clinical risk groups within paediatric B-NHL and offer a novel molecular risk stratifier, allowing more personalised treatment protocols.
Introduction
Current treatments for high-grade paediatric B-cell non-Hodgkin lymphoma (B-NHL) in resource-rich countries are extremely effective, with over 93% of children being cured 1–5 . Following the recent demonstration of the benefit of the anti-CD20 monoclonal antibody rituximab these same rates of cure are now achieved even in patients presenting with established high-risk clinical features (high lactate dehydrogenase (LDH), bone marrow (BM) and/or central nervous system (CNS) disease) 5 . This success, however, has required the use of intensive multi-agent chemo-immunotherapy regimens, associated with significant, predominantly acute, toxicity such as infection and mucositis as well as a small risk of long-term neurological side effects and second malignancy 6–8 . Accordingly, reduction in treatment intensity for at least some patients is a key objective. Unfortunately, previous attempts to reduce treatment intensity in high-risk bone marrow/CNS-positive patients resulted in unacceptable deterioration in survival, highlighting the need for biomarker-driven risk stratification to complement currently established clinical and laboratory risk features 2, 9 .
In contrast to the excellent overall survival currently achieved, the outcome for children with primary refractory or relapsed B-NHL is extremely poor, with fewer than 30% successfully salvaged despite the routine use of high-dose chemotherapy and stem cell rescue 10–13 . Mounting toxicity in this heavily pre-treated group means that escalating intensity alone cannot provide the solution to improving outcome in these patients 14 . Instead, a more comprehensive understanding of the biological drivers of therapy resistance is essential to support development of more effective and less toxic targeted therapies for this group of patients 15 .
Genomic studies have greatly improved our understanding of the pathogenesis and clinically-relevant heterogeneity of both Burkitt lymphoma (BL) and diffuse large B cell lymphoma (DLBCL) 16–28 . Amongst their findings is the frequent mutation in both diseases of the tumour suppressor gene TP53, inactivation of which has an established role in lymphomagenesis 29–33 . TP53 mutation, often accompanied by loss of heterozygosity, is present in 25-50% of sporadic BLs and approximately 20% of adult DLBCLs at diagnosis, while a recent report suggested a lower incidence amongst paediatric large B-cell lymphomas 16–22, 24–28, 34–36 . Consistently, TP53 mutations have been shown to be associated with adverse clinical outcomes in adult aggressive B-cell lymphoma, including DLBCL, and other lymphoid neoplasms 24, 34–38 . In chronic lymphocytic leukaemia (CLL), they are associated with a poor response to chemotherapy and detection of TP53 mutation and 17p deletion is used to guide therapeutic decisions 39 . However, to date no study has incorporated analysis of clinical outcome with a detailed characterisation of TP53 alterations in paediatric B-NHL, where BL predominates, and the potential utility of TP53 status as a clinical risk stratifier in this age group has remained unclear.
To address this question, we collected and analysed a clinically annotated cohort of 95 paediatric B-NHL patients from the UK. Using sequencing and copy number (CN) microarray data we show that TP53 abnormalities at presentation are associated with disease progression and poor outcome. Importantly, the absence of TP53 abnormalities is associated with an extremely low risk of relapse even in high-risk patients defined by high tumour stage and LDH. Moreover, we demonstrate that biallelic TP53 abnormalities are either maintained or acquired at the time of disease progression, implicating loss of TP53 function in the development of treatment resistance. Finally, we show that TP53 abnormalities are associated with complex chromosomal copy number profiles, identifying a potential mechanism underlying the evolution of chemo-resistant disease.
Methods
Patients and Clinical Samples
B-NHL samples from UK hospitals registered with the Children’s Cancer and Leukaemia Group (CCLG) Tissue Bank between 1993 and 2014 were obtained following informed consent from participants or their parent/guardian. A minimum of three-years follow-up was obtained for all survivors. Lymphomas were re-classified according to the World Health Organisation (WHO) Classification of Tumours of Haematopoietic and Lymphoid Tissues 40 . IG-MYC status was confirmed by fluorescence in situ hybridisation (FISH) as described in supplemental methods. The cohort comprised BL (n=64), DLBCL (n=19), Burkitt-like lymphoma with 11q aberration (BLL-11q, n=5) and remaining cases which could not be fully classified (B-NHL, NOS n=7).
TP53 Mutation and 17p Copy Number Analysis
TP53 mutational status was assessed using whole-exome sequencing (WES, n=90) or Sanger sequencing of exons 5 to 8 (n=5). WES data were generated using Illumina Nextera Exome enrichment (n=89) or TWIST Human Core Exome kit (n=1) and sequenced on an Illumina NovaSeq within the Newcastle University Genomics Core Facility or Illumina HiSeq by Eurofins Genomics (Germany). Data were analysed using the Genome Analysis Toolkit (GATK 3.7) and variants called using Mutect2. PCR products for Sanger sequencing were amplified using primers designed for TP53 (Supplemental Table 1) and sequenced by Eurofins Genomics. WES base calls were confirmed by Sanger sequencing in 39 cases, with 100% concordance between sequencing methods.
Copy number alterations (CNAs) of 17p and other chromosomes were identified using Affymetrix Cytoscan HD, Genome-wide Human SNP Array 6.0 or OncoScan arrays performed by Eurofins Genomics. Raw data were analysed and visualised in Nexus Copy Number 10.0 (BioDiscovery) to detect CNAs and copy number neutral loss of heterozygosity (CNN-LOH) in all samples.
Chromosomal Complexity Analysis
Complex patterns of chromosomal copy number abnormality were defined as those with a fluctuation between two or more copy number states involving two or more individual segments ≥100kb on a chromosome arm. These included chromothripsis-like patterns of alternation between two copy number states as well as stepwise increase in copy number of chromosome arms. This definition excludes regions with heterozygous deletion followed by homozygous deletion, single regions of gain, deletion or CNN-LOH, and abnormalities affecting alternate chromosome arms.
Statistical Analyses
Estimates of overall survival (OS) and progression free survival (PFS) were calculated and compared using Kaplan-Meier methods, log-rank tests and Cox-regression models. OS was defined as the time from diagnosis to death from any cause, with censoring at the date of last contact. PFS was defined as the date of diagnosis to the time of disease progression or death. We report 3-year OS and PFS survival rates. All variables conformed to the proportional hazards assumption. Other comparisons were performed using Fisher’s exact test. Analyses were performed using R Bioconductor packages ‘survival’ for univariate and multivariate analysis and ‘survminer’ for visualization of Kaplan-Meier survival curves.
Results
Patient Demographics and Clinical Characteristics
The cohort consisted of the diagnostic tumour samples from 95 UK paediatric B-NHL patients. In total 89 of the 95 (94%) cases were uniformly treated on FAB/LMB 96 protocols (trial or interim guidelines) and had complete follow-up with a median follow-up of 66.2 months (1-270.4 months) (Table 1). In keeping with previously published clinical trial cohorts 4, 41 , the 89 FAB/LMB96-treated cases in the present study demonstrated a median age of 8 years, a male predominance of 3.4:1, a predominance (67%) of Burkitt lymphoma, a majority of high stage (III/IV) patients (73%), bone marrow disease in 18% and CNS disease in 6% of cases. MYC status was available for 84/89 samples (five cases failed FISH or had no available material), amongst which 63 (75%) had an IG-MYC translocation, including 56/58 BL, 4/16 DLBCL, 3/5 B-NHL, NOS and 0/5 BLL-11q cases. Survival estimates at 3 years for PFS and OS were 83.1% (95% CI 75.7-91.3) and 87.6% (95% CI 81.1-94.8), respectively. For those patients with disease progression (primary refractory or relapsed disease), the median time from initial diagnosis to progression was 4.5 months (range 2.8-7.7 months). The additional 6 of the 95 cases (4 BL, 1 DLBCL and 1 B-NHL, NOS) were included in the genomic analysis only.
Genomic Analysis of the TP53 locus
TP53 mutations were found in 46/95 (48.4%) cases: 37 cases had a single non-synonymous somatic mutation, eight had two different somatic mutations and one had a germline mutation (DLBCL with Li-Fraumeni syndrome) (Figure 1A, Supplemental Table 2). TP53 mutations were found in 37/64 (57.8%) BL, 4/19 (21.0%) DLBCL, 4/7 (57.1%) B-NHL, NOS cases and 1/5 (20.0%) BLL-11q cases. All three DLBCL cases with somatic TP53 mutations and all four mutated B-NHL, NOS cases carried IG-MYC rearrangements. The 35 distinct mutations included 33 missense mutations and two deletions leading to frameshifts. All but two mutations involved the DNA binding domain and experimental data from the UMD and IARC TP53 databases showed all but two of the mutations to be functionally deleterious 42, 43 . As seen in other cancers 42, 44 , the most commonly mutated residues were R175 (n=6), G245 (n=5), R248 (n=7) and R273 (n=4).
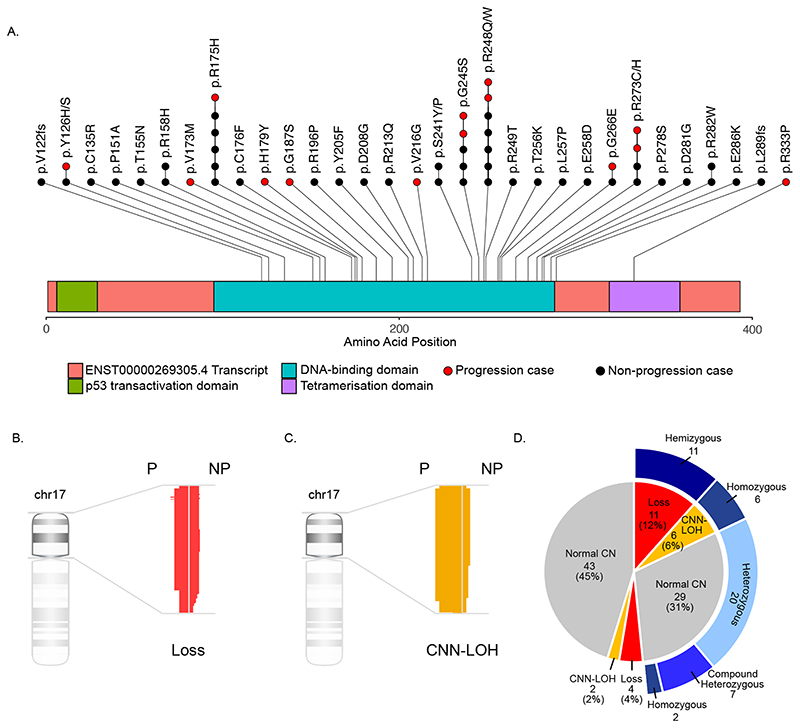
Figure 1. TP53 mutations and genomic copy number abnormalities are detected in a high proportion of paediatric B-NHL cases.
(A) A lolliplot showing 53 somatic and one germline mutation (R248Q detected in a Li-Fraumeni syndrome case). (B) Deletion of the TP53 locus was detected in 3/14 patients with disease progression (P) and 12/78 with no disease progression (NP). (C) CNN-LOH of the TP53 locus was detected in 5/14 patients with disease progression (P) and 3/78 with no disease progression (NP). (D) Representation of co-occurrence of TP53 abnormalities. Inner circle represents TP53 copy number status; outer ring represents TP53 mutation status.
Next, we analysed 17p copy number alteration and identified deletions in 15/95 (15.8%) cases: 10/64 (15.6%) BL, 4/19 (21.0%) DLBCL and 1/7 (14.3%) B-NHL, NOS. The median region of deletion was 20.6 Mb (range 17.9-22.2 Mb), resulting in deletion of >80% of 17p (Figure 1B). Additionally, 8/95 cases (8.4%) showed CNN-LOH: 7/64 (10.9%) BL and 1/19 (5.3%) DLBCL. The median region of CNN-LOH was 19.5 Mb (range 12.9-22.3 Mb), again covering >80% of 17p (Figure 1C). All 17p deletions and CNN-LOH involved the TP53 locus. The presence or absence of TP53 copy number deletion was assessed by FISH in 76/95 cases: 49/64 BL, 16/19 DLBCL, 5/5 BLL-11q and 6/7 B-NHL, NOS (Supplemental Table 2). These included 12/15 cases with TP53 deletion, 5/8 with CNN-LOH and 59/72 with neither. In each case the FISH results were concordant with the copy number array findings.
Combining the mutation and CN data for the TP53 locus showed that abnormalities are common at presentation with 52/95 (54.7%) cases harbouring at least one abnormality and functionally inactivating biallelic events present in 26/95 (27.4%) cases (Figure 1D; Supplemental Table 2). Of the 26 cases with monoallelic TP53 abnormalities, 20 had a single somatic heterozygous mutation (median variant allele frequency 39%, range 18-50%), four had a deletion and two had CNN-LOH. Among the 26 cases with biallelic abnormalities, 17 had TP53 mutation together with a deletion or CNN-LOH, two had a homozygous mutation and seven had compound heterozygous mutations. The correlation between p53 protein expression and TP53 status was assessed by p53 immunohistochemistry (IHC) in a subset of cases. In most cases TP53 mutation was associated with overexpression of p53 protein but the correlation between p53 expression levels and TP53 status was variable and IHC could not reliably detect the range of genomic alterations (Supplemental Table 3).
TP53 Abnormalities are Associated with Chromosomal Complexity
Genomic TP53 alterations have been associated with genomic complexity in DLBCL and other tumours 21, 33, 44–46 . In this study, TP53 abnormalities were not associated with the number of CNAs or the percentage of genome altered (Supplemental Table 4). However, TP53 abnormalities were associated with complex CN profiles of specific chromosomes. (Figure 2; Supplemental Table 5). Excluding BLL-11q, which is defined by a complex 11q rearrangement, chromosomes with complex CNAs were present in 30/51 cases with TP53 abnormality but only in 5/39 cases with wild-type TP53 (p<0.001). This association was particularly strong for BL, in which 24/40 (60%) TP53 abnormal cases harboured a complex chromosome compared to only 1/24 (4.2%) TP53 wild-type cases (p=0.001). The Li-Fraumeni syndrome case (DLBCL with TP53 biallelic abnormality) demonstrated the highest number of complex CNAs within the whole cohort, but otherwise there was no association between TP53 abnormalities and complex CNAs in DLBCL cases.
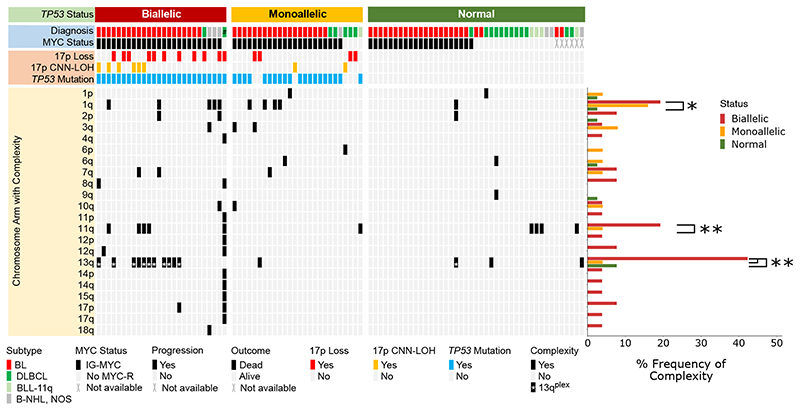
Figure 2. A pattern of complex chromosomal abnormalities in paediatric B-NHL is associated with TP53 status.
Oncoplot showing TP53 status (upper panel) and associated complex chromosomes (lower panel). Histogram displaying the frequency of complex abnormalities of each chromosome arm in biallelic, monoallelic and TP53 normal groups. As a diagnosis of BLL-11q is determined by the presence of a complex 11q rearrangement, these five cases were excluded from this analysis. TP53 Biallelic with 1q complexity vs TP53 Normal with 1q complexity, p=0.026; TP53 Biallelic with 11q complexity vs TP53 Normal with 11q complexity, p=0.006. TP53 Biallelic with 13q complexity vs TP53 Normal with 13q complexity, p=0.001; TP53 Biallelic with 13q complexity vs TP53 Monoallelic with 13q complexity, p=0.002. Fisher’s Exact test *= p<0.05, **=p<0.01; # Li-Fraumeni syndrome case.
Most notably, complex CNAs of 1q, 11q and 13q were associated with biallelic TP53 abnormalities (p<0.05) (Figure 2). Complexity of 1q typically involved stepwise increases in copy number, with only two cases having telomeric deletion (Supplemental Figure 1). Both chromosome arms 11q and 13q recurrently showed a pattern of a region of gain adjacent to telomeric deletion, similar to the profile defining BLL-11q cases 47 (Supplemental Figures 2-3). All ten cases with this characteristic gain-telomeric deletion pattern of 13q (here termed 13qplex) were BL (Supplemental Table 5). The MIR17HG locus mapped within the region of 13q CN gain/amplification, notably located in the highest peak for 9/10 (90%) 13qplex cases (Supplemental Figure 3).
Survival Analysis of TP53 Abnormalities in Paediatric B-NHL
The clinical characteristics of cases with or without a TP53 abnormality are presented in Supplemental Table 6. Univariate survival analyses identified a significantly adverse PFS for patients with high disease stage, bone marrow involvement or high LDH (Supplemental Table 7). CNS involvement was associated with a hazard ratio of 3.0 but did not reach significance, likely due to the low number of cases (n=5). Those with mutations had significantly inferior 3-year survival compared to those without (PFS 66.7% (95% CI 54.2-82.0) vs 100% (95% CI 100-100), p<0.001 and OS 75.6% (95% CI 64.0-89.2) vs 100% (95% CI 100-100), p<0.001) (Supplemental Figure 4A-B), although no hazard ratio could be reported due to the absence of events in cases without TP53 mutation. Likewise, 17p CNN-LOH involving TP53 was also associated with worse outcome (PFS HR 4.9 (95% CI 1.6-15.6), p=0.007; OS HR 4.7 (95% CI 1.2-17.7), p=0.023) (Supplemental Figure 4C-D, Supplemental Table 7). In contrast, as previously reported in the context of the FAB/LMB96 trial, univariate analysis showed that, agnostic of TP53 mutation status, cases with deletion of 17p including the TP53 locus did not have inferior outcome (Supplemental Figure 4E-F, Supplemental Table 7) 2 .
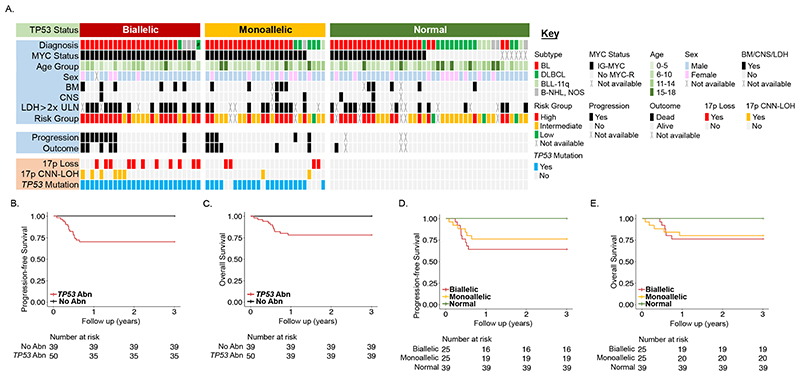
Combining the TP53 locus alterations (Figure 3A), patients with at least one TP53 abnormality had a significantly inferior 3-year survival compared to those with no abnormality (PFS 70.0% (95% CI 58.4-83.9) vs 100 (95% CI 100-100), p<0.001 and OS 78.0% (95% CI 67.3-90.4) vs 100 (95% CI 100-100), p=0.002) (Figure 3B-C). Importantly, those patients without any TP53 abnormality at initial diagnosis had a PFS of 100% (95% CI 100-100) and an OS of 100% (95% CI 100-100). Most TP53 abnormalities were seen in patients diagnosed with BL and the presence of any TP53 abnormality remained adversely prognostic within this subgroup (PFS 67.5% (95% CI 54.4-83.7) vs 100% (95% CI 100-100), p=0.005 and OS: 75% (95% CI 62.7-89.7) vs 100 (95% CI 100-100, p=0.017) (Supplemental Figure 5). Both monoallelic and biallelic TP53 abnormalities were associated with adverse outcome, when compared with TP53 wild-type cases (monoallelic PFS 76.0% (95% CI 61.0-94.7) vs 100% (95% CI 100-100), p=0.001 and OS 80.0% (95% CI 65.8-97.3) vs 100% (95% CI 100-100), p=0.004; biallelic PFS 64.0% (95% CI 47.7-85.9) vs 100% (95% CI 100-100), p<0.001 and OS 76.0% (95% CI 61.0-94.7) vs 100 (95% CI 100-100) p=0.001), but with no significant difference identified between these two groups (Figure 3D-E). There was no association between prognosis and either complex CNA patterns of 1q, 11q or 13q (Supplemental Table 8) or gain of MIR17HG. Neither was there a difference in outcome for patients with or without any complex CNA (PFS HR 1.7 (95% CI 0.6-4.7), p=0.295; OS HR 1.8 (95% CI 0.5-5.9), p=0.332).
Figure 3. Identification of patient risk groups based on TP53 status.
(A) Oncoplot showing TP53 status with clinical and molecular parameters as described in the key. Data is plotted from left to right according to TP53 status. Kaplan-Meier plots showing (B) progression free and (C) overall survival for any TP53 abnormality (deletion, CNN-LOH and/or mutation), and (D) progression-free and (E) overall survival according to normal (green), monoallelic (amber) or biallelic (red) TP53 status. # Li-Fraumeni syndrome case.
Multivariate analysis was performed to assess the impact of TP53 abnormalities alongside established high-risk clinical factors (BM and CNS involvement, stage and high LDH) (Supplemental Table 7). Due to the lack of an event amongst the TP53 wild-type patients included in this analysis we are unable to report a hazard ratio for TP53 status; however, having included this strong prognostic factor in the model, those established high-risk clinical factors were not independently significant.
TP53 status risk stratifies patients with clinically defined high-risk disease
The Inter-B-NHL ritux 2010 trial (NCT01516580) has recently demonstrated the survival benefit of adding rituximab to modified FAB/LMB96 chemotherapy for patients with high-risk disease (stage III and LDH > 2x ULN, stage IV or Burkitt leukaemia) 5 . Nevertheless, rituximab-chemotherapy was associated with increased incidence of prolonged hypogammaglobulinaemia and may be associated with a greater number of infections 5 . Given the inferior outcome of these patients prior to the use of rituximab and the unacceptable deterioration in that outcome with a previous attempt to reduce therapy intensity 9 , they represent a key group for further biomarker-driven stratification. We therefore sought to understand the prognostic impact of TP53 abnormalities specifically in this group. High-risk patients had a higher rate of TP53 abnormality (27/41, 66%) than low/intermediate-risk groups (20/42, 48%), but here this association was specifically within the TP53 biallelic group (p=0.037) (Supplemental Table 6). As expected, the high-risk group had an inferior survival compared to the intermediate and low-risk groups (PFS 70.7% (95% CI 58.1-86.1) vs 94.7% (95% CI 87.9-100.0) vs 100.0% (95% CI 100.0-100.0), p=0.013) and OS 78.6% (95% CI 66.4-91.8) vs 94.7% (95% CI 87.9-100.0) vs 100.0% (95% CI 100.0-100.0), p=0.067) (Figure 4A-B). Strikingly, despite the substantial clinical value of stage and LDH, the absence of any TP53 abnormality in high-risk patients remained strongly associated with an excellent clinical outcome compared to high-risk patients with any TP53 abnormality (PFS 100% (95% CI 100-100) vs 55.6% (95% CI 39.7-77.9), p=0.005 and OS 100% (95% CI 100-100) vs 66.7% (95% CI 51.1-87.0), p=0.019) (Figure 4C-D). Stratification of the low/intermediate-risk group identified an inferior outcome for patients with any TP53 abnormality but this did not reach significance (PFS 100 % (95% CI 100-100) vs 90% (95% CI 77.8%-100%) vs, p=0.133 and OS 100% (95% CI 100-100) vs 90% (95% CI 70.7%-100%), p=0.133) (Supplemental Figure 6).
Figure 4. TP53 status differentiates patients in the clinically defined high-risk group in paediatric B-NHL.
Kaplan-Meier plots showing (A) progression-free and (B) overall survival for high, intermediate and low clinical risk groups, and (C) progression-free and (D) overall survival in high-risk patients according to the presence of any TP53 abnormality.
Biallelic TP53 Abnormalities Evolve during Disease Progression
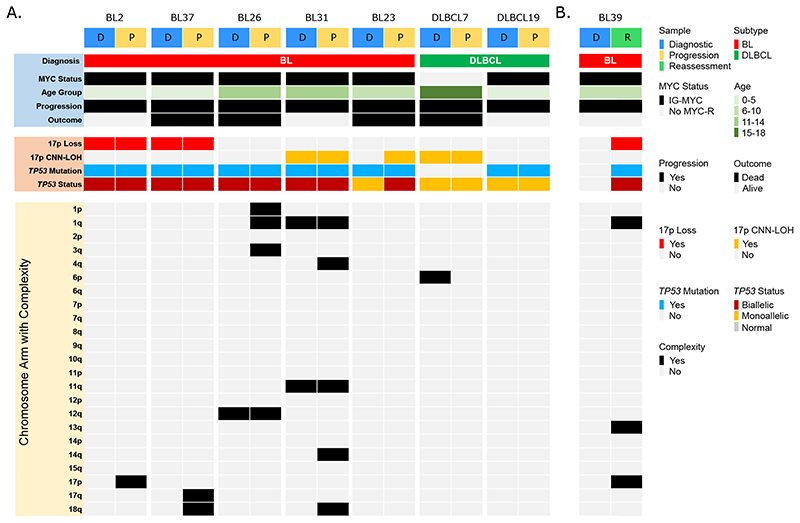
Very little is known about the genomic changes associated with therapy resistance and disease progression in BL or other paediatric B-NHL 48 . To understand the evolution of TP53 abnormalities associated with therapy resistance we investigated the paired biopsies taken at the time of disease progression for 7 cases and one further paired biopsy (patient BL39) taken from a viable residual tumour mass detected at routine reassessment following the CYM-1 component of FAB/LMB96 therapy. Amongst six BL cases, four had major clonal biallelic TP53 abnormalities at the time of the original diagnosis (Figure 5 and Supplemental Table 9) and these same biallelic abnormalities were maintained at the time of disease progression. Interestingly, the other two BL cases showed acquisition of biallelic TP53 abnormalities in the second biopsies, further supporting a role for TP53 in treatment resistance. The first, BL23, had only a monoallelic clonal G245S mutation at diagnosis but developed CNN-LOH at relapse, rendering this mutation biallelic. The other, BL39, lacked a detectable TP53 abnormality at initial diagnosis but the residual tumour had both a deletion of chromosome 17p and a classical pathogenic R248W mutation. Analysis of WES data failed to identify even a minor clonal R248W mutation at presentation (0/124 reads). In contrast to the BL cases, both DLBCL cases with paired material harboured a monoallelic TP53 abnormality at both diagnosis and progression. In five of the six BL cases, therapy resistant disease was associated with an increase in the number of chromosomes showing a complex copy number profile (Figure 5).
Figure 5. Biallelic TP53 abnormalities are either maintained or acquired at the time of progression of BL.
(A) Overview of clinical and molecular parameters for 7 patients with matched samples taken at initial diagnosis (D) and at the time of progression (P). (B) One patient had matched samples from initial diagnosis and the time of reassessment.
Discussion
The addition of rituximab to first-line therapy for children with high-risk B-NHL has resulted in ≥93% event-free survival (EFS) in all risk groups 5 . This success requires intensive multi-agent chemotherapy and comes at the cost of significant acute toxicity and there now exists a growing understanding of the long-term sequelae 6–8 . However, with very few relapse events occurring, identifying the prognostic biomarkers required for improved risk stratification in this rare patient group has become extremely challenging. Consequently, we undertook an analysis of the prognostic significance of TP53 status in a large national cohort of paediatric mature B-NHL cases diagnosed prior to the routine introduction of rituximab therapy. This showed that the presence or absence of TP53 abnormalities defines two patient groups with markedly different progression-free and overall survival rates, adverse and favourable respectively.
The principal clinical challenges in paediatric B-NHL have, for some years, been: 1) improving survival in high-risk patients; 2) reducing therapy intensity without increasing risk of relapse 9 ; 3) identifying mechanisms of therapy resistance which result in extremely poor survival following relapse. Addressing the first of these challenges, the international collaborative trial Inter-B-NHL ritux 2010 (NCT01516580) recently demonstrated an 11% increase in EFS in high-risk patients (defined as stage III with LDH greater than twice the upper limit of normal, stage IV or Burkitt leukaemia) with the addition of six doses of rituximab to a modified FAB/LMB96 chemotherapy schedule 5 . This contrasts with the unacceptable reduction in survival seen in high-risk bone marrow/central nervous system positive patients randomised to reduced intensity therapy within the FAB/LMB96 study 9 . Nevertheless, within the reduced intensity arm of the high risk FAB/LMB96 study, an 80% EFS was seen, implying that only a minority of patients benefit from intensification of treatment. The inability to identify those with a low risk of treatment failure has hampered risk stratification beyond clinicopathological features. Here, we show that within the high-risk patient group, (as defined in the Inter-B-NHL trial), TP53 wild-type patients have a very low risk of relapse, despite the absence of rituximab therapy. If validated in international trial cohorts, analysis of TP53 status may allow identification of a subset of patients currently considered high-risk for whom chemo-immunotherapy can be de-intensified without compromising efficacy. Initially this could involve omitting rituximab but further reduction in chemotherapy similar to that attempted within FAB/LMB96, could also be considered. Our finding provides a potential biomarker platform for future trials of therapy reduction, albeit that a large number of patients and a more effective salvage strategy would be necessary for such a trial.
Critical to the better treatment of progressive or relapsed disease is a much deeper understanding of the drivers of therapy resistance. The finding that TP53 abnormalities are associated with increased risk of disease progression, and our analysis of paired Burkitt lymphoma samples showing that all cases either maintained or developed biallelic TP53 abnormalities at progression, suggest a key role for TP53 loss of function in this process. In support of this assertion, Reutter et al 49 found multiple TP53 abnormalities at diagnosis and relapse in each of 5 relapsed BL cases studied. Given the many biological pathways impacted by p53, the mechanisms by which TP53 abnormalities promote therapy resistance merits further investigation in support of developing an effective salvage therapy. It is notable, therefore, that we found a strong association between TP53 abnormalities and MYC rearrangements in our cohort. Since these two cancer genes cooperate to drive experimental lymphomagenesis in mice and have been associated with a particularly poor prognosis when concurrently mutated and translocated, respectively, in adult DLBCL, a deeper mechanistic understanding of the interactions between p53 and MYC in paediatric B-NHL may be particularly informative in this regard 29, 32, 38 .
Mutations, present in half of cases, were the most frequent TP53 abnormalities detected in our cohort. As expected, TP53 deletions and CNN-LOH were less prevalent and were mostly present in tumours with TP53 mutation 2, 50–53 . Overall, half of all cases with a TP53 abnormality had biallelic alterations expected to abrogate wild-type TP53 functions at diagnosis and biallelic events were present in all BL tumours at the time of progression. However, while the presence of any TP53 abnormality was associated with an increased risk of disease progression and death, there was no difference in PFS or OS between cases with monoallelic or biallelic alterations at diagnosis. This contrasts with recent findings in myelodysplastic syndrome and plasma cell myeloma, in which specifically biallelic TP53 alterations at diagnosis are associated with poor survival 54–56 . The increasing evidence that many mutant p53 proteins exhibit gain of function or dominant negative properties may provide a partial explanation but our analysis of paired diagnosis/progression BL samples suggests the alternative explanation that clones/subclones with monoallelic TP53 abnormalities may evolve to a biallelic state during disease progression under the selective pressure of therapy 32, 33, 57 . Interesting in this regard is the recent report of a case of relapsed BL with expansion of a low level (2% VAF) TP53 R248Q mutation at diagnosis to a 93% VAF at relapse, secondary to a combination of clonal expansion and CNN-LOH 58 .
TP53 mutations are associated with several types of genomic instability, including aneuploidy and chromothripsis, and thus increased genome complexity across the cancer spectrum 33, 44, 46 . Notably, a recently described subset of adult DLBCL carrying frequent biallelic inactivation of TP53 by mutation and 17p deletion is selectively associated with an increase in small and large CNAs 21, 45 . Similarly, an association between TP53 abnormalities and chromothripsis-like changes, including of 13q, has been reported in adult DLBCL 59, 60 . Although we did not see an effect on global genomic complexity in our predominantly BL paediatric cohort, we did observe the correlation of TP53 abnormalities with complex copy number patterns involving 1q, 11q and 13q. The 13q gains centred on the MIR17HG gene, in keeping with a previous report of an association between 17p deletion and MIR17HG gain in a small sample set 50 . That report speculated that MIR17HG gain was associated with relapse but our data suggest that this probably results indirectly from the association of MIR17HG gain with TP53 abnormalities 50 .
We have demonstrated the clinical importance of TP53 abnormalities in paediatric B-NHL, identifying them as a potential biomarker capable of further risk-stratifying patients currently considered high-risk. These findings now need to be validated in a large international trial cohort. For those children without a TP53 abnormality, the risk of disease progression is extremely low and stratified trials examining therapy reduction should be considered. Evolution of biallelic abnormalities at relapse implicates TP53 biology as a driver of therapy resistance and relapse and further understanding of the underlying mechanisms could lead to new strategies to prevent or treat therapy resistant disease.
Supplementary Material
Table 1. Clinical and cytogenetic characteristics of the FAB/LMB96-treated paediatric B-NHL cases.
| FAB/LMB96-treated Cohort | |||
|---|---|---|---|
| Total Cases | 89 | ||
| Diagnosis | BL | 60 (67.4%) | |
| DLBCL | 18 (20.2%) | ||
| BLL-11q | 5 (5.6%) | ||
| B-NHL, NOS | 6 (6.7%) | ||
| Median Age at Diagnosis (range), years | 8 (0.5-17) | ||
| Sex | Male | 68 (76.4%) | |
| Female | 20 (22.5%) | ||
| Not available | 1 (1.1%) | ||
| Tumour Stage | Stage I or II | 24 (27.0%) | |
| Stage III or IV | 65 (73.0%) | ||
| Not available | 0 (0.0%) | ||
| BM Involvement | Y | 16 (18.0%) | |
| N | 71 (79.8%) | ||
| Not available | 2 (2.2%) | ||
| CNS Involvement* | Y | 5 (5.6%) | |
| N | 83 (93.3%) | ||
| Not available | 1 (1.1%) | ||
| LDH > 2x ULN | Y | 38 (42.7%) | |
| N | 38 (42.7%) | ||
| Not available | 13 (14.6%) | ||
| MYC Translocation | Y | 63 (70.7%) | |
| N | 21 (23.6%) | ||
| Not available | 5 (5.6%) | ||
| Risk Group | High | 41 (46.1%) | |
| Intermediate | 38 (42.7%) | ||
| Low | 4 (4.5%) | ||
| Not available | 6 (6.7%) | ||
| Treatment Group | Group A | 4 (4.5%) | |
| Group B | 68 (76.4%) | ||
| Group C | 15 (16.9%) | ||
| Group unknown | 2 (2.2%) | ||
| Rituximab Added** | 2 (2.2%) | ||
| No Rituximab | 84 (94.4%) | ||
| Rituximab unknown | 3 (3.4%) | ||
| Outcome | PFS | Progression / relapse | 15 (16.9%) |
| Median time to event (range), months | 4.5 (0.9-7.7) | ||
| OS | Deaths | 11 (12.4%) | |
| Median time to death (range), months | 6.5 (0.9-11.1) | ||
BL = Burkitt lymphoma; DLBCL = diffuse large B-cell lymphoma; BLL-11q = Burkitt-like lymphoma with 11q aberrations; B-NHL, NOS = B-cell non-Hodgkin lymphoma not otherwise specified; Y = Yes; N= No; “-” = no event, hazard ratio not reported; BM = bone marrow; CNS = central nervous system; CSF = cerebrospinal fluid; LDH = lactate dehydrogenase; ULN = upper limit of normal. PFS = progression-free survival. OS = overall survival.
One case had CSF involvement.
Two cases received rituximab from the start of first-line therapy, two other patients received rituximab following switch to group C therapy following post-CYM-1 reassessment.
Acknowledgements
This study was supported by a Blood Cancer UK Senior Bennett Fellowship (#12005), the Newcastle upon Tyne Hospitals NHS Charity, North East Promenaders Against Cancer (NEPAC), the JGW Patterson Foundation, the Little Princess Trust, the North of England Children’s Cancer Research Fund (NECCR), the Good Will Cause, Cancer Research North East, the National Institute for Health Research (NIHR) Newcastle In Vitro Diagnostics Co-operative, the MRC/EPSRC Newcastle Molecular Pathology Node (NovoPath) and MRC Clinician Scientist Fellowship (MR/S021590/1). We thank all the patients, clinicians and staff who participated in the collection of material and clinical data at the Children’s Cancer and Leukaemia Group (CCLG) Tissue Bank. The CCLG Tissue Bank is funded by Cancer Research UK.
Footnotes
Authorship Contributions
V.R. conceived the study, secured funding and finalized the manuscript with editorial assistance from all authors. V.R., S.B. and C.M.B. designed the study, directed the research and wrote the manuscript; V.R., A.M.N., S.B. and C.M.B. co-ordinated and participated in data collection, analysis and interpretation; V.R., M.Z., A.E.B and A.M.N. processed and analysed the copy number and whole exome sequencing data; P.Z., A.Er., A.Ba., R.E.C, and S.W. did laboratory experiments and analysis. V.R., A.M.N., M.Z. and A.E. performed statistical analysis; V.R., C.M.B, S.B, A.B., S.T., M.T. and C.H. gathered samples and patient data, and provided clinical interpretation. C.M.B., K.W. and D.T. conducted the pathological review of the cases. All authors approved the manuscript.
Disclosure of Conflicts of Interest
The authors declare no competing financial interests.
References
- 1.Patte C, Auperin A, Gerrard M, Michon J, Pinkerton R, Sposto R, et al. Results of the randomized international FAB/LMB96 trial for intermediate risk B-cell non-Hodgkin lymphoma in children and adolescents: it is possible to reduce treatment for the early responding patients. Blood. 2007 Apr 1;109(7):2773–2780. doi: 10.1182/blood-2006-07-036673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Poirel HA, Cairo MS, Heerema NA, Swansbury J, Auperin A, Launay E, et al. Specific cytogenetic abnormalities are associated with a significantly inferior outcome in children and adolescents with mature B-cell non-Hodgkin’s lymphoma: results of the FAB/LMB 96 international study. Leukemia. 2009 Feb;23(2):323–331. doi: 10.1038/leu.2008.312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Minard-Colin V, Brugieres L, Reiter A, Cairo MS, Gross TG, Woessmann W, et al. Non-Hodgkin Lymphoma in Children and Adolescents: Progress Through Effective Collaboration, Current Knowledge, and Challenges Ahead. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2015 Sep 20;33(27):2963–2974. doi: 10.1200/JCO.2014.59.5827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cairo MS, Sposto R, Gerrard M, Auperin A, Goldman SC, Harrison L, et al. Advanced stage, increased lactate dehydrogenase, and primary site, but not adolescent age (>/= 15 years), are associated with an increased risk of treatment failure in children and adolescents with mature B-cell non-Hodgkin’s lymphoma: results of the FAB LMB 96 study. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2012 Feb 1;30(4):387–393. doi: 10.1200/JCO.2010.33.3369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Minard-Colin V, Auperin A, Pillon M, Burke GAA, Barkauskas DA, Wheatley K, et al. Rituximab for High-Risk, Mature B-Cell Non-Hodgkin’s Lymphoma in Children. N Engl J Med. 2020 Jun 4;382(23):2207–2219. doi: 10.1056/NEJMoa1915315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ehrhardt MJ, Chen Y, Sandlund JT, Bluhm EC, Hayashi RJ, Becktell K, et al. Late Health Outcomes After Contemporary Lymphome Malin de Burkitt Therapy for Mature B-Cell Non-Hodgkin Lymphoma: A Report From the Childhood Cancer Survivor Study. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2019 Oct 1;37(28):2556–2570. doi: 10.1200/JCO.19.00525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Attarbaschi A, Carraro E, Ronceray L, Andres M, Barzilai-Birenboim S, Bomken S, et al. Second malignant neoplasms after treatment of non-Hodgkin’s lymphoma-a retrospective multinational study of 189 children and adolescents. Leukemia. 2020 May 11; doi: 10.1038/s41375-020-0841-x. [DOI] [PubMed] [Google Scholar]
- 8.Srinivasan S, Roy Moulik N, Kc A, Narula G, Sankaran H, Prasad M, et al. Increased toxicities in children with Burkitt lymphoma treated with rituximab: Experience from a tertiary cancer center in India. Pediatr Blood Cancer. 2020 Aug 31;:e28682. doi: 10.1002/pbc.28682. [DOI] [PubMed] [Google Scholar]
- 9.Cairo MS, Gerrard M, Sposto R, Auperin A, Pinkerton CR, Michon J, et al. Results of a randomized international study of high-risk central nervous system B non-Hodgkin lymphoma and B acute lymphoblastic leukemia in children and adolescents. Blood. 2007 Apr 1;109(7):2736–2743. doi: 10.1182/blood-2006-07-036665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Anoop P, Sankpal S, Stiller C, Tewari S, Lancaster DL, Khabra K, et al. Outcome of childhood relapsed or refractory mature B-cell non-Hodgkin lymphoma and acute lymphoblastic leukemia. Leukemia & lymphoma. 2012 Oct;53(10):1882–1888. doi: 10.3109/10428194.2012.677534. [DOI] [PubMed] [Google Scholar]
- 11.Jourdain A, Auperin A, Minard-Colin V, Aladjidi N, Zsiros J, Coze C, et al. Outcome of and prognostic factors for relapse in children and adolescents with mature B-cell lymphoma and leukemia treated in three consecutive prospective “Lymphomes Malins B” protocols. A Societe Francaise des Cancers de l’Enfant study. Haematologica. 2015 Jun;100(6):810–817. doi: 10.3324/haematol.2014.121434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rigaud C, Auperin A, Jourdain A, Haouy S, Couec ML, Aladjidi N, et al. Outcome of relapse in children and adolescents with B-cell non-Hodgkin lymphoma and mature acute leukemia: A report from the French LMB study. Pediatr Blood Cancer. 2019 Sep;66(9):e27873. doi: 10.1002/pbc.27873. [DOI] [PubMed] [Google Scholar]
- 13.Woessmann W, Zimmermann M, Meinhardt A, Muller S, Hauch H, Knorr F, et al. Progressive or relapsed Burkitt lymphoma or leukemia in children and adolescents after BFM-type first-line therapy. Blood. 2020 Apr 2;135(14):1124–1132. doi: 10.1182/blood.2019003591. [DOI] [PubMed] [Google Scholar]
- 14.Burke GAA, Beishuizen A, Bhojwani D, Burkhardt B, Minard-Colin V, Norris RE, et al. Ibrutinib plus CIT for R/R mature B-NHL in children (SPARKLE trial): initial safety, pharmacokinetics, and efficacy. Leukemia. 2020 Feb 18;34(8):2271–2275. doi: 10.1038/s41375-020-0749-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pearson ADJ, Scobie N, Norga K, Ligas F, Chiodin D, Burke A, et al. ACCELERATE and European Medicine Agency Paediatric Strategy Forum for medicinal product development for mature B-cell malignancies in children. Eur J Cancer. 2019 Mar;110:74–85. doi: 10.1016/j.ejca.2019.01.013. [DOI] [PubMed] [Google Scholar]
- 16.Love C, Sun Z, Jima D, Li G, Zhang J, Miles R, et al. The genetic landscape of mutations in Burkitt lymphoma. Nat Genet. 2012 Dec;44(12):1321–1325. doi: 10.1038/ng.2468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schmitz R, Young RM, Ceribelli M, Jhavar S, Xiao W, Zhang M, et al. Burkitt lymphoma pathogenesis and therapeutic targets from structural and functional genomics. Nature. 2012 Oct 4;490(7418):116–120. doi: 10.1038/nature11378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Panea RI, Love CL, Shingleton JR, Reddy A, Bailey JA, Moormann AM, et al. The whole genome landscape of Burkitt lymphoma subtypes. Blood. 2019 Sep 26;134(19):1598–1607. doi: 10.1182/blood.2019001880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Grande BM, Gerhard DS, Jiang A, Griner NB, Abramson JS, Alexander TB, et al. Genome-wide discovery of somatic coding and non-coding mutations in pediatric endemic and sporadic Burkitt lymphoma. Blood. 2019 Jan 7;133(12):1313–1324. doi: 10.1182/blood-2018-09-871418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lopez C, Kleinheinz K, Aukema SM, Rohde M, Bernhart SH, Hubschmann D, et al. Genomic and transcriptomic changes complement each other in the pathogenesis of sporadic Burkitt lymphoma. Nat Commun. 2019 Mar 29;10(1):1459. doi: 10.1038/s41467-019-08578-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chapuy B, Stewart C, Dunford AJ, Kim J, Kamburov A, Redd RA, et al. Molecular subtypes of diffuse large B cell lymphoma are associated with distinct pathogenic mechanisms and outcomes. Nat Med. 2018 May;24(5):679–690. doi: 10.1038/s41591-018-0016-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schmitz R, Wright GW, Huang DW, Johnson CA, Phelan JD, Wang JQ, et al. Genetics and Pathogenesis of Diffuse Large B-Cell Lymphoma. N Engl J Med. 2018 Apr 12;378(15):1396–1407. doi: 10.1056/NEJMoa1801445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Reddy A, Zhang J, Davis NS, Moffitt AB, Love CL, Waldrop A, et al. Genetic and Functional Drivers of Diffuse Large B Cell Lymphoma. Cell. 2017 Oct 5;171(2):481–494. doi: 10.1016/j.cell.2017.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lacy SE, Barrans SL, Beer PA, Painter D, Smith AG, Roman E, et al. Targeted sequencing in DLBCL, molecular subtypes, and outcomes: a Haematological Malignancy Research Network report. Blood. 2020 May 14;135(20):1759–1771. doi: 10.1182/blood.2019003535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lohr JG, Stojanov P, Lawrence MS, Auclair D, Chapuy B, Sougnez C, et al. Discovery and prioritization of somatic mutations in diffuse large B-cell lymphoma (DLBCL) by whole-exome sequencing. Proc Natl Acad Sci U S A. 2012 Mar 6;109(10):3879–3884. doi: 10.1073/pnas.1121343109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang J, Grubor V, Love CL, Banerjee A, Richards KL, Mieczkowski PA, et al. Genetic heterogeneity of diffuse large B-cell lymphoma. Proc Natl Acad Sci U S A. 2013 Jan 22;110(4):1398–1403. doi: 10.1073/pnas.1205299110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Morin RD, Mungall K, Pleasance E, Mungall AJ, Goya R, Huff RD, et al. Mutational and structural analysis of diffuse large B-cell lymphoma using whole-genome sequencing. Blood. 2013 Aug 15;122(7):1256–1265. doi: 10.1182/blood-2013-02-483727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pasqualucci L, Trifonov V, Fabbri G, Ma J, Rossi D, Chiarenza A, et al. Analysis of the coding genome of diffuse large B-cell lymphoma. Nat Genet. 2011 Jul 31;43(9):830–837. doi: 10.1038/ng.892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Eischen CM, Weber JD, Roussel MF, Sherr CJ, Cleveland JL. Disruption of the ARF-Mdm2-p53 tumor suppressor pathway in Myc-induced lymphomagenesis. Genes Dev. 1999 Oct 15;13(20):2658–2669. doi: 10.1101/gad.13.20.2658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rowh MA, DeMicco A, Horowitz JE, Yin B, Yang-Iott KS, Fusello AM, et al. Tp53 deletion in B lineage cells predisposes mice to lymphomas with oncogenic translocations. Oncogene. 2011 Nov 24;30(47):4757–4764. doi: 10.1038/onc.2011.191. [DOI] [PubMed] [Google Scholar]
- 31.Xu-Monette ZY, Medeiros LJ, Li Y, Orlowski RZ, Andreeff M, Bueso-Ramos CE, et al. Dysfunction of the TP53 tumor suppressor gene in lymphoid malignancies. Blood. 2012 Apr 19;119(16):3668–3683. doi: 10.1182/blood-2011-11-366062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Aubrey BJ, Janic A, Chen Y, Chang C, Lieschke EC, Diepstraten ST, et al. Mutant TRP53 exerts a target gene-selective dominant-negative effect to drive tumor development. Genes Dev. 2018 Nov 1;32(21–22):1420–1429. doi: 10.1101/gad.314286.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kastenhuber ER, Lowe SW. Putting p53 in Context. Cell. 2017 Sep 7;170(6):1062–1078. doi: 10.1016/j.cell.2017.08.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Xu-Monette ZY, Wu L, Visco C, Tai YC, Tzankov A, Liu WM, et al. Mutational profile and prognostic significance of TP53 in diffuse large B-cell lymphoma patients treated with R-CHOP: report from an International DLBCL Rituximab-CHOP Consortium Program Study. Blood. 2012 Nov 8;120(19):3986–3996. doi: 10.1182/blood-2012-05-433334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zenz T, Kreuz M, Fuge M, Klapper W, Horn H, Staiger AM, et al. TP53 mutation and survival in aggressive B cell lymphoma. Int J Cancer. 2017 Oct 1;141(7):1381–1388. doi: 10.1002/ijc.30838. [DOI] [PubMed] [Google Scholar]
- 36.Intlekofer AM, Joffe E, Batlevi CL, Hilden P, He J, Seshan VE, et al. Integrated DNA/RNA targeted genomic profiling of diffuse large B-cell lymphoma using a clinical assay. Blood Cancer J. 2018 Jun 12;8(6):60. doi: 10.1038/s41408-018-0089-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Xu P, Liu X, Ouyang J, Chen B. TP53 mutation predicts the poor prognosis of non-Hodgkin lymphomas: Evidence from a meta-analysis. PLoS One. 2017;12(4):e0174809. doi: 10.1371/journal.pone.0174809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Clipson A, Barrans S, Zeng N, Crouch S, Grigoropoulos NF, Liu H, et al. The prognosis of MYC translocation positive diffuse large B-cell lymphoma depends on the second hit. J Pathol Clin Res. 2015 Jul;1(3):125–133. doi: 10.1002/cjp2.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Campo E, Cymbalista F, Ghia P, Jager U, Pospisilova S, Rosenquist R, et al. TP53 aberrations in chronic lymphocytic leukemia: an overview of the clinical implications of improved diagnostics. Haematologica. 2018 Dec;103(12):1956–1968. doi: 10.3324/haematol.2018.187583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Swerdlow SH, Campo E, Harris NL, Jaffe E, Pileri SA, Stein H, Thiele J, Arber DA, Hasserjian RP, Le Beau MM, Orazi A, et al. WHO Classification of Tumours of Haematopoietic and Lymphoid Tissues. Revised 4th Edition edn. International Agency for Research on Cancer (IARC); 2017. [Google Scholar]
- 41.Woessmann W, Seidemann K, Mann G, Zimmermann M, Burkhardt B, Oschlies I, et al. The impact of the methotrexate administration schedule and dose in the treatment of children and adolescents with B-cell neoplasms: a report of the BFM Group Study NHL-BFM95. Blood. 2005 Feb 1;105(3):948–958. doi: 10.1182/blood-2004-03-0973. [DOI] [PubMed] [Google Scholar]
- 42.Bouaoun L, Sonkin D, Ardin M, Hollstein M, Byrnes G, Zavadil J, et al. TP53 Variations in Human Cancers: New Lessons from the IARC TP53 Database and Genomics Data. Hum Mutat. 2016 Sep;37(9):865–876. doi: 10.1002/humu.23035. [DOI] [PubMed] [Google Scholar]
- 43.Leroy B, Anderson M, Soussi T. TP53 mutations in human cancer: database reassessment and prospects for the next decade. Hum Mutat. 2014 Jun;35(6):672–688. doi: 10.1002/humu.22552. [DOI] [PubMed] [Google Scholar]
- 44.Donehower LA, Soussi T, Korkut A, Liu Y, Schultz A, Cardenas M, et al. Integrated Analysis of TP53 Gene and Pathway Alterations in The Cancer Genome Atlas. Cell Rep. 2019 Jul 30;28(5):1370–1384. doi: 10.1016/j.celrep.2019.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Monti S, Chapuy B, Takeyama K, Rodig SJ, Hao Y, Yeda KT, et al. Integrative analysis reveals an outcome-associated and targetable pattern of p53 and cell cycle deregulation in diffuse large B cell lymphoma. Cancer Cell. 2012 Sep 11;22(3):359–372. doi: 10.1016/j.ccr.2012.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Eischen CM. Genome Stability Requires p53. Cold Spring Harb Perspect Med. 2016 Jun 1;6(6):a026096. doi: 10.1101/cshperspect.a026096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Salaverria I, Martin-Guerrero I, Wagener R, Kreuz M, Kohler CW, Richter J, et al. A recurrent 11q aberration pattern characterizes a subset of MYC-negative high-grade B-cell lymphomas resembling Burkitt lymphoma. Blood. 2014 Feb 20;123(8):1187–1198. doi: 10.1182/blood-2013-06-507996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Aukema SM, Theil L, Rohde M, Bauer B, Bradtke J, Burkhardt B, et al. Sequential karyotyping in Burkitt lymphoma reveals a linear clonal evolution with increase in karyotype complexity and a high frequency of recurrent secondary aberrations. Br J Haematol. 2015 Sep;170(6):814–825. doi: 10.1111/bjh.13501. [DOI] [PubMed] [Google Scholar]
- 49.Reutter K, Sandmann S, Rohde J, Muller S, Woste M, Khanam T, et al. Reconstructing clonal evolution in relapsed and non-relapsed Burkitt lymphoma. Leukemia. 2020 May 14; doi: 10.1038/s41375-020-0862-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schiffman JD, Lorimer PD, Rodic V, Jahromi MS, Downie JM, Bayerl MG, et al. Genome wide copy number analysis of paediatric Burkitt lymphoma using formalin-fixed tissues reveals a subset with gain of chromosome 13q and corresponding miRNA over expression. Br J Haematol. 2011 Nov;155(4):477–486. doi: 10.1111/j.1365-2141.2011.08883.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Scholtysik R, Kreuz M, Klapper W, Burkhardt B, Feller AC, Hummel M, et al. Detection of genomic aberrations in molecularly defined Burkitt’s lymphoma by array-based, high resolution, single nucleotide polymorphism analysis. Haematologica. 2010 Dec;95(12):2047–2055. doi: 10.3324/haematol.2010.026831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Havelange V, Pepermans X, Ameye G, Theate I, Callet-Bauchu E, Barin C, et al. Genetic differences between paediatric and adult Burkitt lymphomas. Br J Haematol. 2016 Apr;173(1):137–144. doi: 10.1111/bjh.13925. [DOI] [PubMed] [Google Scholar]
- 53.Scholtysik R, Kreuz M, Hummel M, Rosolowski M, Szczepanowski M, Klapper W, et al. Characterization of genomic imbalances in diffuse large B-cell lymphoma by detailed SNP-chip analysis. Int J Cancer. 2015 Mar 1;136(5):1033–1042. doi: 10.1002/ijc.29072. [DOI] [PubMed] [Google Scholar]
- 54.Thanendrarajan S, Tian E, Qu P, Mathur P, Schinke C, van Rhee F, et al. The level of deletion 17p and bi-allelic inactivation of TP53 has a significant impact on clinical outcome in multiple myeloma. Haematologica. 2017 Sep;102(9):e364–e367. doi: 10.3324/haematol.2017.168872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Walker BA, Mavrommatis K, Wardell CP, Ashby TC, Bauer M, Davies F, et al. A high-risk, Double-Hit, group of newly diagnosed myeloma identified by genomic analysis. Leukemia. 2019 Jan;33(1):159–170. doi: 10.1038/s41375-018-0196-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bernard E, Nannya Y, Hasserjian RP, Devlin SM, Tuechler H, Medina-Martinez JS, et al. Implications of TP53 allelic state for genome stability, clinical presentation and outcomes in myelodysplastic syndromes. Nature Medicine. 2020;26(10):1549–1556. doi: 10.1038/s41591-020-1008-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Boettcher S, Miller PG, Sharma R, McConkey M, Leventhal M, Krivtsov AV, et al. A dominant-negative effect drives selection of TP53 missense mutations in myeloid malignancies. Science. 2019 Aug 9;365(6453):599–604. doi: 10.1126/science.aax3649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wever CM, Geoffrion D, Grande BM, Yu S, Alcaide M, Lemaire M, et al. The genomic landscape of two Burkitt lymphoma cases and derived cell lines: comparison between primary and relapse samples. Leukemia & lymphoma. 2018 Sep;59(9):2159–2174. doi: 10.1080/10428194.2017.1413186. [DOI] [PubMed] [Google Scholar]
- 59.Karube K, Enjuanes A, Dlouhy I, Jares P, Martin-Garcia D, Nadeu F, et al. Integrating genomic alterations in diffuse large B-cell lymphoma identifies new relevant pathways and potential therapeutic targets. Leukemia. 2018 Mar;32(3):675–684. doi: 10.1038/leu.2017.251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Rausch T, Jones DT, Zapatka M, Stutz AM, Zichner T, Weischenfeldt J, et al. Genome sequencing of pediatric medulloblastoma links catastrophic DNA rearrangements with TP53 mutations. Cell. 2012 Jan 20;148(1–2):59–71. doi: 10.1016/j.cell.2011.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.