Abstract
Discovery of deafness genes and elucidating their functions have substantially contributed to our understanding of hearing physiology and its pathologies. Here we report on DNA variants in MINAR2, encoding membrane integral NOTCH2-associated receptor 2, in four families underlying autosomal recessive non-syndromic deafness. Neurologic evaluation of affected individuals at ages ranging from 4 to 80 years old does not show additional abnormalities. MINAR2 is a recently annotated gene with limited functional understanding. We detected three MINAR2 variants, c.144G>A (p.Trp48*), c.412_419delCGGTTTTG (p.Arg138Valfs*10), and c.393G>T, in 13 individuals with congenital or prelingual-onset severe to profound sensorineural hearing loss. The c.393G>T variant is shown to disrupt a splice donor site. We show that Minar2 is expressed in the mouse inner ear, with the protein localizing mainly in the hair cells, spiral ganglia, the spiral limbus, and the stria vascularis. Mice with loss-of-function of the Minar2 protein (Minar2tm1b/tm1b) present with rapidly progressive sensorineural hearing loss associated with a reduction in outer hair cell stereocilia in the shortest row and degeneration of hair cells at a later age. We conclude that MINAR2 is essential for hearing in humans and mice and its disruption leads to sensorineural hearing loss. Progressive hearing loss observed in mice and in some affected individuals and as well as relative preservation of hair cells provides an opportunity to interfere with hearing loss using genetic therapies.
Keywords: Autosomal recessive, deafness, hearing loss, MINAR2, NOTCH2
Classification: Biological Sciences (Major), Genetics (Minor)
Introduction
Hearing loss (HL) is one of the most common sensory deficits, affecting ~1 in 500 newborns (1). Genetic factors are implicated in the majority of cases, with more than 80% of the inherited form exhibiting autosomal recessive transmission (2). No additional findings are present in over 70% of the cases, which are then classified as non-syndromic HL (Hereditary Hearing Loss Homepage, https://hereditaryhearingloss.org/) (2, 3). Genetic testing for etiologic evaluation has become a standard of care in people with congenital or childhood-onset sensorineural HL, which is caused by pathologies of the inner ear and auditory nerve (4, 5). Recent studies have shown that screening all recognized HL genes for variants reveals underlying cause in about half of the affected individuals, leaving a significant portion of people with HL with an unknown etiology (6–9). In the era of emerging genetic therapies for HL, finding the etiology of HL in affected individuals has become a critical task. This is especially relevant for progressive HL, as genetic therapies may potentially stop progression while cochlear hair cells are still alive (10–12).
MINAR2 (previously known as uncharacterized protein KIAA1024L and mouse gene A730017C20Rik) has recently been identified, and based on its structural similarity to MINAR1, named as major intrinsically disordered NOTCH2-associated receptor 2 or membrane integral NOTCH2-associated receptor 2 (13). A mutant mouse model of Minar2 showed motor deficits similar to those seen in Parkinson disease, with no information about hearing abnormalities (13). A Minar2 mutant mouse line, Minar2tm1b, has also recently been reported to show no auditory brainstem responses at 14 weeks old as part of a large HL screen (Mouse Genome Informatics; MGI: 2442934) (11). Functional aspects of MINAR2 and consequences of its dysfunction in humans remain unknown.
In this study, to better map the landscape of hereditary HL, we sought DNA variants underlying deafness in 13 affected individuals from four families. We identified three different MINAR2 variants in the families co-segregating with HL. We further showed that homozygous Minar2tm1b mutant mice develop rapidly progressive HL associated with changes in outer hair cell stereocilia. Finally, via in vitro studies we demonstrated that MINAR2 suppresses NOTCH2, suggesting that notch signaling might play a role in pathogenesis.
Results
Non-syndromic sensorineural HL is diagnosed in 13 individuals from four unrelated families
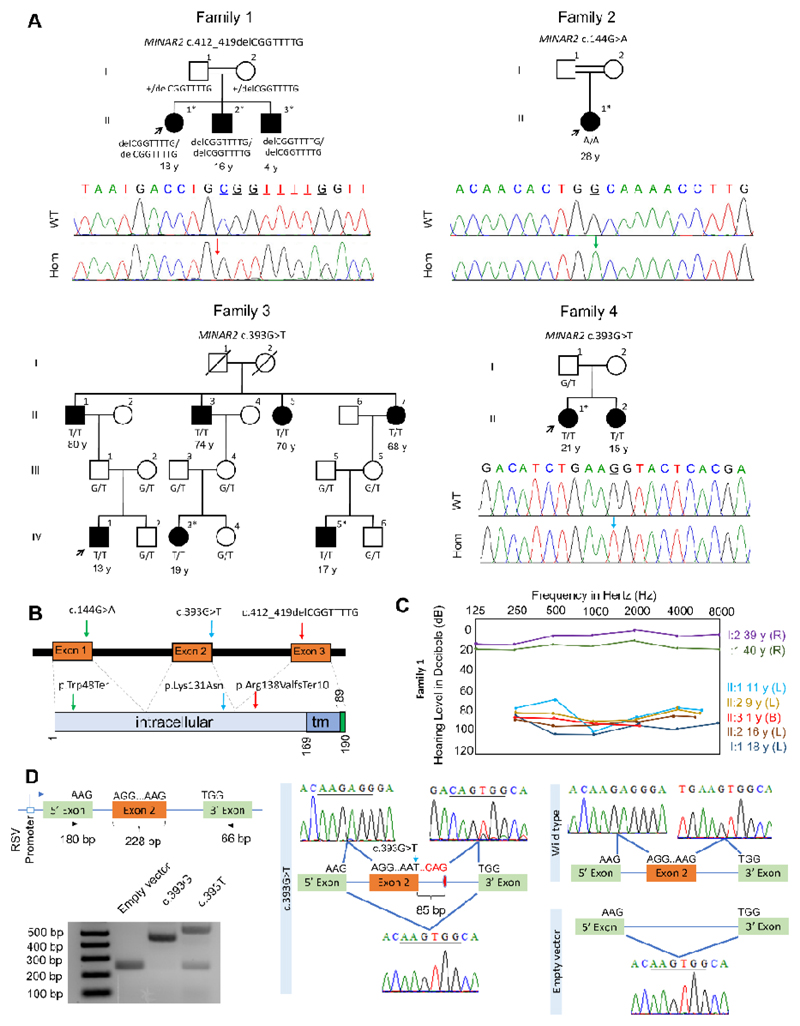
A summary of the auditory phenotype is shown in SI Appendix, Table S1. Ages ranged from 4 to 80 years old at the last examination. Each affected individual was diagnosed with HL either at birth or during infancy. Families 1 and 2 were of Turkish ancestry (Fig. 1). Parents of Family 1 stated that in II:1 and II:2, HL was milder in younger ages and progressed to severe or profound degree by around age 10. Otoacoustic emissions were absent in these individuals. These siblings received unilateral cochlear implants at ages 12 and 10, respectively, which improved their oral communication. Individual II:3 in Family 1 was diagnosed with profound sensorineural HL after failing the newborn hearing screening test. Otoacoustic emissions were absent at diagnosis. He received a unilateral cochlear implant at age 1 and communicates orally. Pure tone audiograms in parents showed normal thresholds (SI Appendix, Fig. S1).
Figure 1. Families, MINAR2 variants, and the effects of the c.393G>T variant on splicing.
(A) Pedigrees and segregation of the MINAR2 variants in families. Filled symbols denote affected individuals and double lines indicate first cousin consanguinity. Electropherograms show the identified variant. The wildtype traces are from an unrelated individual. WT: Wildtype, Hom: Homozygous mutant; exome/genome sequencing was performed in individuals marked with an (*) asterisk. (B) Locations of the identified variants. tm: transmembrane (C) Audiogram of the Family 1 (R: Right ear, L: Left ear, B: Bilateral). (D) MINAR2 exon 2 inserted into a vector consisting of 5’ and 3’ exons in the exon trap assay is shown. There are larger and smaller PCR products in the c.393G>T sample compared to wildtype. Sanger sequencing confirms insertion of 85 bp at the donor site of exon 2 and skipping of exon 2.
Families 3 and 4 were of Indian ancestry. While there is no known consanguinity in any of the marriages in these two pedigrees, they are all from the same small town belonging to the same ethnic background, i.e Hindu (religion) and Mali (Caste). All affected individuals in Family 3 were born deaf and used signs, simple words, or sounds to be able to communicate. Severity of HL appeared to have remained the same in all the affected individuals from the beginning of life. Individuals IV:1 and IV:2 in Family 4 were diagnosed with severe to profound sensorineural HL at the age of 3 years via auditory brainstem response (ABR) studies. Parents indicated a progression in the severity of HL. Their mode of communication at that age was predominantly non-verbal.
A high-resolution temporal bone CT scan or MRI was normal in at least one affected member of each family (SI Appendix, Fig. S1). None of the affected individuals had additional clinical findings for a syndromic form of deafness. Their neurodevelopmental skills were on target except for speech delay. None of the affected individuals showed impaired balance on tandem walking and Romberg test. Neurological examination was normal except for hearing loss in six affected individuals with ages ranging from 4 to 80 years. No affected individual was noted to have bradykinesia, tremor, or rigidity similar to those seen in Parkinson disease (SI Appendix, Table S2).
MINAR2 variants co-segregate with autosomal recessive deafness
We performed genome sequencing in four affected individuals in Families 1 and 2 and exome sequencing in three affected individuals in Families 3 and 4 (Fig. 1A; individuals marked with an asterisk, SI Appendix, Table S3). Sequencing data in affected individuals were first analyzed for variants (SNVs: Single nucleotide variants, indels: insertions and/or deletions, and CNVs: Copy number variants) in recognized HL genes retrieved from the Hereditary Hearing Loss Homepage (https://hereditaryhearingloss.org/) (3), Online Mendelian Inheritance in Man (OMIM: http://omim.org/), University of Miami Molecular Genetics Laboratory HL gene panel, and a virtual gene panel for HL (v2.176) from PanelApp (https://www.genomicsengland.co.uk ). Minor allele frequency (MAF) thresholds of 0.005 for recessive and 0.001 for dominant variants were used. Population allele frequencies were obtained from genome aggregation database (gnomAD: http://gnomad.broadinstitute.org/) (14) and the single nucleotide polymorphism database (dbSNP: https://www.ncbi.nlm.nih.gov/projects/SNP/), as well as from our internal exome/genome database that includes > 7,000 samples from different ethnicities. American College of Medical Genetics (ACMG) and ClinGen HL expert panel guidelines were followed for variant interpretation (15, 16). This analysis did not reveal a plausible variant under any inheritance model.
After excluding variants in previously recognized deafness genes, in Family 1 we filtered shared homozygous coding and splice variants (SNVs, indels, and CNVs) in three affected siblings with a MAF of <0.005 in dbSNP, gnomAD highest sub-ethnicity, and our internal control database. This filter reveals only a MINAR2 (GeneBank: NM_001257308.2) c.412_419delCGGTTTTG (p.Arg138Valfs*10) variant in the family. The variant is located within a 9.4 MB shared homozygous run in three siblings, which is the only homozygous region >1 MB (SI Appendix, Tables S4 and S5). This frame-shift variant is predicted to introduce a premature stop codon and lead to early truncation of MINAR2.
In the proband of Family 2, MINAR2 is located within the second longest homozygous run at chr5:128,253,080-141,730,596 (hg19) (SI Appendix, Table S6). She is homozygous for the c.144G>A (p.Trp48*) variant in MINAR2. This nonsense variant is predicted to cause an early stop codon and result in truncation of MINAR2.
In the exome data of two affected individuals from Family 3, filtering variants via the same criteria used in Family 1 reveals only one variant for which both individuals are homozygous: MINAR2 c.393G>T (p.Lys131Asn). SNP arrays show that this variant is located within the only shared homozygous run, >1 MB, in all seven affected individuals in Family 3. This homozygous run is flanked by markers rs13174854 and rs377767449, which is 2.96 MB on chr5:128,738,407-131,705,915 (hg19). In Family 4, two affected siblings share a 5.76 MB homozygous run on chr5: 126,978,108-132,742,450 (hg19), flanked by markers rs11241936 and rs11242152. Exome sequencing showed that the proband in Family 4 is homozygous for the same MINAR2 variant detected in Family 3.
None of the detected MINAR2 variants is listed in dbSNP or gnomAD databases and all variants are predicted to be deleterious (SI Appendix, Table S7). Sanger sequencing confirmed all three MINAR2 variants and showed that each variant co-segregates with autosomal recessive HL in all families (Fig. 1A).
MINAR2 c.393G>T (p.Lys131Asn) leads to aberrant splicing
While it is a missense change, the c.393G>T (p.Lys131Asn) variant substitutes the last nucleotide of exon 2 and is predicted to abolish a splice donor site (SI Appendix, Table S7 and Fig. S2). Via exon trapping experiments, we show that this variant leads to an addition of 85 intronic nucleotides into exon 2, which alters the amino acid composition of the rest of the protein leading to a premature stop codon (Fig. 1D and SI Appendix, Fig. S2). The same variant also leads to skipping of exon 2 entirely (Fig. 1D).
Minar2tm1b homozygous mutant mice show sensorineural HL
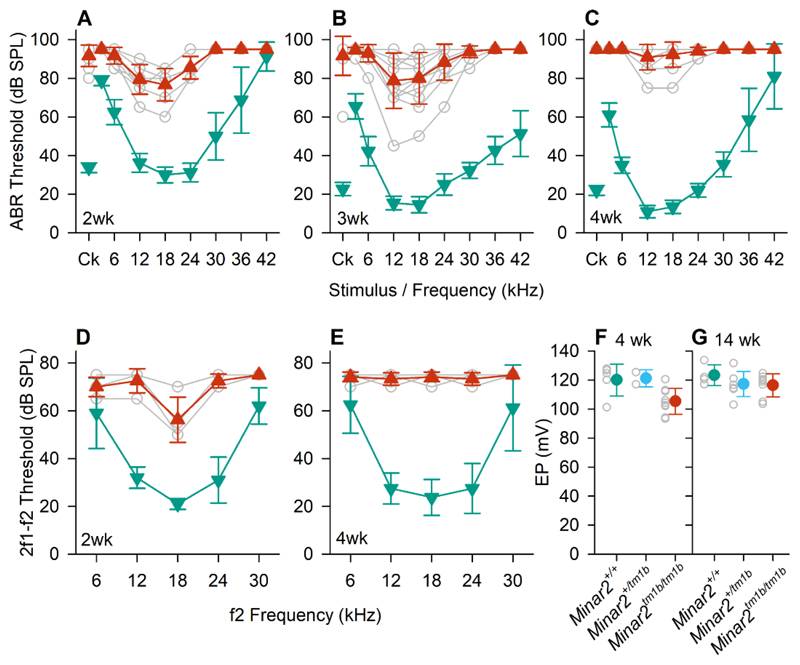
To prove causality of disruption of Minar2 in sensorineural HL, we evaluated hearing in Minar2 mutant mice. Hearing sensitivity of Minar2tm1b mutant mice (SI Appendix, Fig. S3) was assessed using two methods: ABR, a measure of neural activity in the auditory nerve and brainstem, and Distortion Product Otoacoustic Emission (DPOAE), a measure of outer hair cell electromotility and resulting nonlinearities in the cochlea. Thresholds for both ABRs and DPOAEs were raised in homozygous mutants compared with wildtype littermates from the earliest age studied, postnatal day (P) 14, only two days after the usual onset of hearing (Fig. 2). By four weeks old, the mutant mice showed severe elevations in threshold or no response at the highest sound levels used. Endocochlear potential was only slightly reduced from a mean of 120 mV to 105 mV in Minar2tm1b/tm1b mice, with most mutant mice exhibiting an EP in the normal physiological range for mice of over 100mV (Fig. 2).
Figure 2. Auditory studies in Minar2 mutant mice.
(A-C) Auditory Brainstem Response (ABR) thresholds. Mean (±SD) ABR thresholds are plotted against stimulus for Minar2+/+ mice (teal down-triangles) and Minar2tm1b/tm1b mice (red up-triangles). Open grey circles indicate thresholds for individual Minar2tm1b/tm1b mice. Results are plotted for data obtained at P14 (A) (n=4 Minar2+/+, 9 Minar2tm1b/tm1b); P21. (B) (n=11 Minar2+/+, 12 Minar2tm1b/tm1b and P27-28 (C) (n=10 Minar2+/+, 11 Minar2tm1b/tm1b). Ck, click. (D-E) Distortion Product OtoAcoustic Emission (DPOAE) thresholds. Mean (±SD) DPOAE thresholds are plotted against f2 stimulus frequency for Minar2 control mice (teal down-triangles) and Minar2tm1b/tm1b mice (red up-triangles). Open grey circles indicate thresholds for individual Minar2tm1b/tm1b mice. Results are plotted for data obtained at P14 (D) (n=4 controls, comprising 1 wildtype and 3 heterozygotes, n=4 mutants) and P27-28 (E) (n=4 wildtypes, n=6 mutants). (F, G) Endocochlear Potential (EP) in Minar2 mutant mice aged 4 weeks (F) and 14 weeks old (G). Colored circles indicate the mean (±SD) EP measurement in Minar2+/+ (teal), Minar2+/tm1b (cyan) Minar2tm1b/tm1b (red) mice. Open grey circles indicate the EP values recorded in individual mice. At 4 weeks old (P30-32) Minar2+/+ n=5, range 101.2 – 127.8 mV; Minar2+/tm1b n=2 range 117.0 – 125.4 mV; Minar2tm1b/tm1b n=9, range 93.4 – 120.5 mV. At 14 weeks old Minar2+/+ n=4, range 117 – 133.6 mV; Minar2+/tm1b n=7, range 113.8 – 131.6 mV; Minar2tm1b/tm1b n=8, range 103.5 – 124.7 mV.
Minar2 is expressed in the mouse cochlea
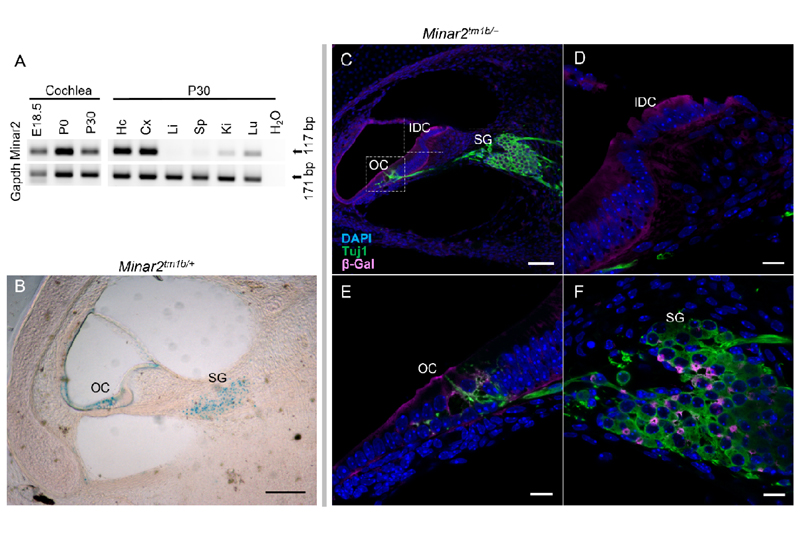
To further understand the role of Minar2 in hearing, we assessed the presence of Minar2 transcript in different mouse tissues, including the inner ear, and specifically the cochlea. Total RNA was isolated from wildtype mice at embryonic day (E)18.5, P0, and P30. RT-PCR with a forward primer located in exon 2 and a reverse primer in exon 3 of the Minar2 gene (GenBank: NM_173759) produced a unique band of 171 bp corresponding to the wildtype mRNA in all analyzed tissues, with the exception of the liver. We find that Minar2 is highly expressed in the inner ear, and specifically in the cochlea, at E18.5, P0, and P30 (Fig. 3A). To determine specific locations of Minar2 expression within the cochlea, we proceeded to study Minar2tm1b heterozygous mutant mice taking advantage of the reporter gene. β-galactosidase staining of P1 cross sections and whole mount organ of Corti preparations reveals that Minar2 is mainly localized in the hair cells, the supporting cells, spiral ganglion (including the nerve fibers), the spiral limbus, and the stria vascularis (Fig. 3B and SI Appendix, Fig. S4 A and B). Of note, the gene expression analysis resource (gEAR; https://umgear.org/ ) portal shows Minar2 expression both in hair cells and supporting cells (SI Appendix, Fig. S4C) (17, 18). Double-labelling for Minar2 and Tuj1 in heterozygous mutant mice show that the two are expressed in different cell types, Minar2 is not present in the neurons of the spiral ganglion that were marked with Tuj1, suggesting that Minar2 is located in spiral ganglion glial cells (Fig. 3 C, E and F).
Figure 3.
Expression of Minar2, localization of the protein in the cochlea and innervation of cochlear hair cells in Minar2 heterozygous mice, using the LacZ reporter gene component of the inserted cassette in the mutant allele which expresses β galactosidase (see Appendix SI, Fig. S3). (A) RT-PCR of Minar2 expression in the cochlea at embryonic 18.5 (E18.5), postnatal day 0 (P0), and day 30 (P30). Also, expression in different mouse tissues at P30 like Hc: hippocampus, Cx: cortex, Li: liver, Sp: spleen, Ki: kidney, Lu: lung. Gapdh was used as a control. (B) Localization of Minar2 using the reporter gene LacZ of the mutant allele and in β-gal staining shown in blue. Note the localization of Minar2 at the SG: spiral ganglion, OC: organ of Corti. Scale bar: 70 µm. (C) Cross section from 24 kHz region of P1 mutant inner ear was labelled with anti-β gal (magenta) to detect Minar2 localization; and Tuj1 (green) to label neurons. Bar=70µm in C and a zoom in is shown in (D), (E), and (F) scale bar=10µm. SG; Spiral ganglion, OC: organ of Corti, IDC: Interdental cells.
The earliest defect in Minar2tm1b mutant mice appears in stereocilia bundles
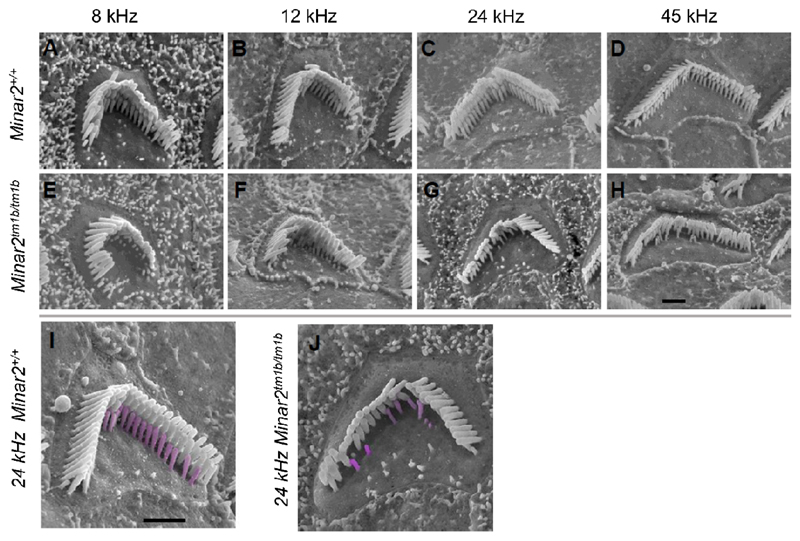
We used scanning electron microscopy (SEM) (Fig. 4) to examine the cochlea of mice aged 14 days old because at this age the ABR thresholds were raised but synapses appeared normal in numbers (Fig. 5). The organization of the organ of Corti appeared normal and there were very few missing stereocilia bundles in mutants, suggesting that hair cell degeneration cannot explain the raised ABR thresholds at this age (SI Appendix, Fig. S5). However, when we examined the stereocilia bundles of outer hair cells, we saw the shortest row was depleted, with reduced numbers and abnormally short stereocilia (Fig. 4). Outer hair cells along the entire length of the cochlear duct showed this feature. Inner hair cells appeared close to normal, although the middle row of stereocilia occasionally appeared more irregular than in the control littermates. Heterozygotes looked the same as wildtype littermates at this age.
Figure 4.
Scanning electron microscopy reveals stereocilia defects. Outer hair cells at 8kHz (85% of distance from base), 12kHz (70%), 24kHz (40%) and 45kHz (20%) best frequency locations in wildtype mice (A-D) and Minar2tm1b homozygotes (E-H). Higher magnification images with the shortest row colored in magenta in a wildtype (I) and a mutant (J) outer hair cell, showing the reduction in numbers. Scale bars (in H and I) indicate 1 μm.
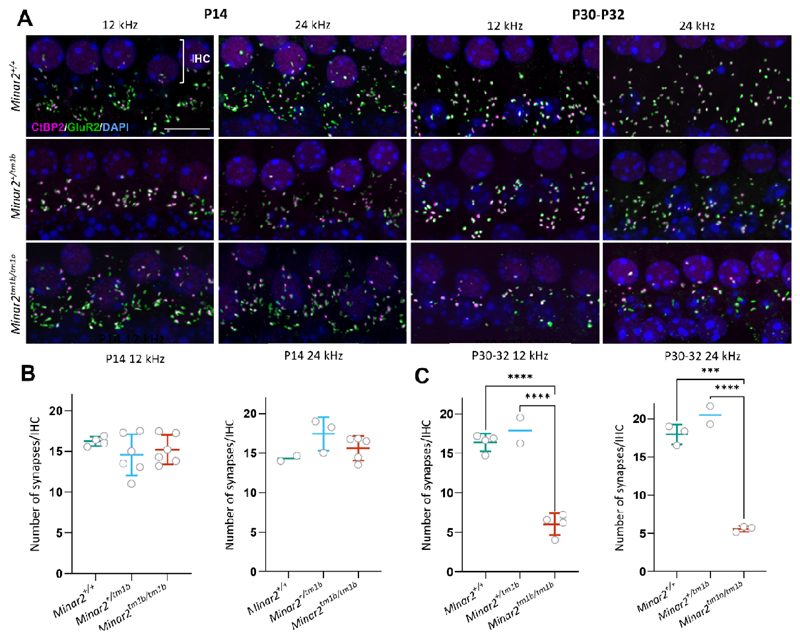
Figure 5. Synaptic abnormalities of cochlear hair cells in Minar2 mutant mice.
(A) Synapses were examined using anti-CtBP2 antibody to mark pre-synaptic ribbons (magenta) and anti-GluR2 antibody to mark postsynaptic densities (green). Nuclei are shown in blue (DAPI). Scale bar = 5 µm. The images correspond to the cochlear best-frequency regions of 12 kHz and 24 kHz. The row of nuclei at the top of each image corresponds to the IHCs. (B, C) Quantification of ribbon synapses per IHC at P14 (B) and P30-P32 (C). Colocalised pre and postsynaptic components were defined as a synapse. The average for each mouse is plotted as a circle. All data are shown as mean ± SD and statistically analyzed by one way ANOVA with multiple comparisons. At P14, no significant difference was seen between homozygotes (n = 6 at 12 kHz and n = 5 at 24 kHz) and heterozygotes (n = 6 at 12 kHz and n = 3 at 24 kHz) or wildtypes (n=4 at 12 kHz and n = 2 at 24 kHz). At P30-P32, there are significantly fewer colocalised synapses in Minar2tm1b/tm1b (n = 4 for 12 kHz and n = 3 for 24 kHz) compared to wildtypes (n = 4 at 12 kHz and n = 3 at 24 kHz) and compared to heterozygotes (n = 2). *** p =0.0001 and **** p < 0.0001
Mechanoelectrical transduction (MET) channels are functional in Minar2tm1b/tm1b mice
To study the MET, we used the FM1-43 FX dye, a molecule commonly used to assay the MET channels function, since functional MET channels are required for the uptake of this fluorescent dye into the hair cells (19). We perfused cochleae from P14 mice and observed FM1-43 FX dye uptake by both inner and outer hair cells in Minar2+/+ and Minar2tm1b/tm1b mice. Heterozygotes looked the same as wildtype littermates. While the intensity of the staining varied from mouse to mouse, all the samples showed uptake of the molecule specifically in hair cells, indicating that the MET channels are active in the Minar2 mutants (SI Appendix, Fig. S6).
Hair cell degeneration is a secondary feature in Minar2tm1b mutant mice
Hair cell degeneration is a common feature in HL, and extensive loss of hair cells may preclude some approaches to treatments. Therefore, we examined older Minar2tm1b mutant mice, aged 14 weeks, to assess the loss of hair cells. We have previously reported that these mutants show no auditory brainstem responses at 14 weeks old (11) and the EP is normal at this age (Fig. 2G). At earlier ages we see minimal evidence of any hair cell degeneration (SI Appendix, Fig. S5). However, by 14 weeks of age, there is extensive loss of hair cells, with some discrete patches distributed along the length of the cochlear duct where the entire organ of Corti appears to have degenerated while other regions show scattered missing hair cells, both inner and outer, within the organ of Corti (Fig. 6). Heterozygotes had a similar appearance to the wildtype mice.
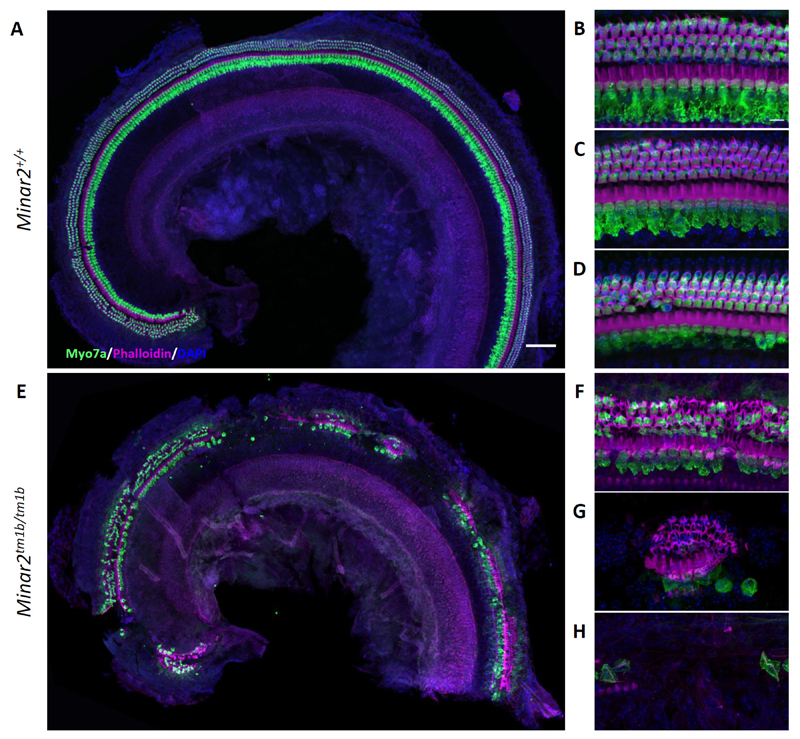
Figure 6. Hair cell degeneration pattern in Minar2tm1b mutant mice at 14 weeks old.
Confocal maximum intensity projection images of the whole-mount organ of Corti of 14-week old mice. Hair cells were examined using anti-Myo7a antibody (green) and phalloidin (magenta). Nuclei are shown in blue (DAPI). Scale bar= 200 µm in A and 10µm in B. The images are representative examples of Minar2+/+ (A, B, C, D) and Minar2tm1b/tm1b (E, F, G, H) mice. (A, E) Overview of the apical half of the cochlea. (B, C, D) High magnification images corresponding to the 12 kHz, 24 kHz and 36 kHz best-frequency regions respectively. (F, G, H) High magnification images of three different patterns of hair cell loss observed in Minar2tm1b mutant mice. All 5 homozygotes that were studied at 14 weeks old showed similar patches of loss of the organ of Corti at varying locations along the length of the cochlear duct interspersed with regions that showed only scattered hair cell loss. Minar2+/+ (n = 4), Minar2+/tm1b (n = 5) and Minar2tm1b/tm1b (n = 5).
Minar2tm1b mutant mice show reduction in the number of inner hair cell ribbon synapses at P30
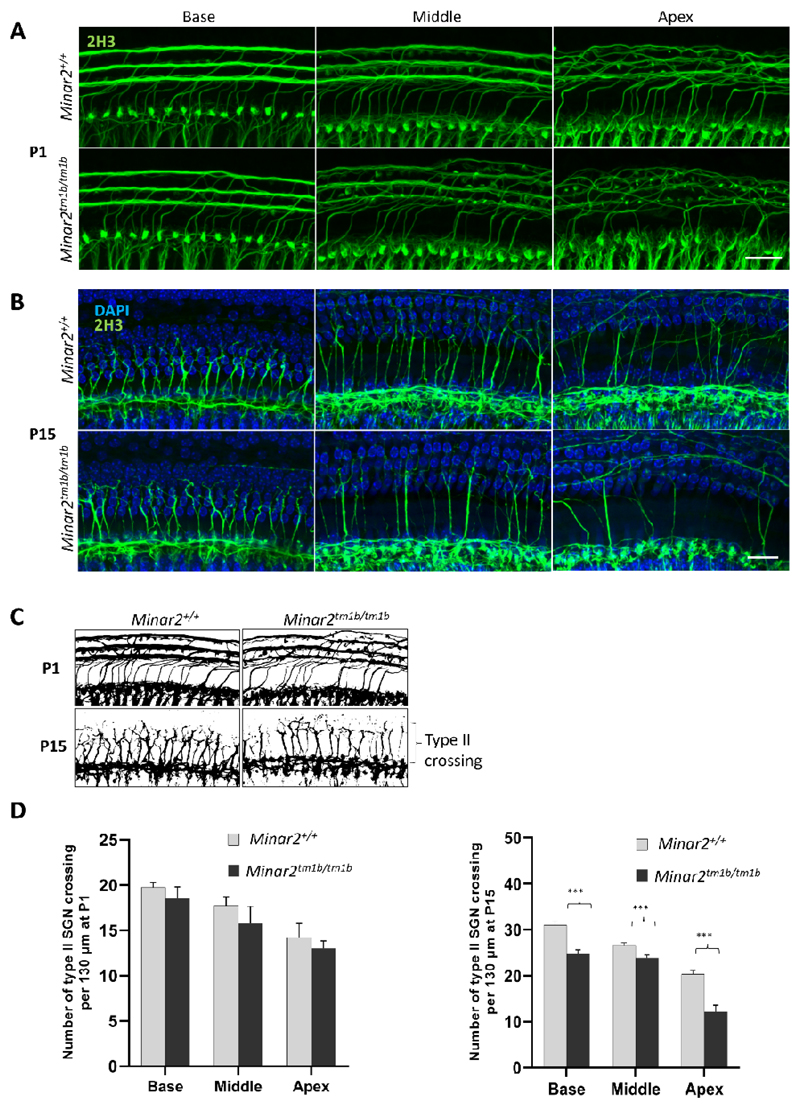
To understand underlying pathophysiology further, we evaluated the innervation pattern of hair cells by staining neurons with an anti-Neurofilament antibody in the organ of Corti at P1 (SI Appendix, Fig. S7). Homozygous mutant mice show an altered innervation pattern of hair cells with a reduction in the nerve fibers around inner hair cells and a dispersed distribution of nerve fibers towards outer hair cells. Minar2tm1b mutant mice show reduction in the number of crossings of type II nerves at P14 (Fig. 7). We subsequently examined synapses of inner hair cells using anti-CtBP2 antibody to label presynaptic ribbons and anti-GluR2 antibody to mark the AMPA receptor subunit R2, a part of the post-synaptic density. Ribbon synapses look qualitatively normal at both ages tested, P14 and P30, and the number of synapses per inner hair cell in homozygous mutants was not significantly different from numbers in heterozygotes or wildtype controls at P14, indicating that the raised ABR thresholds observed at that age in mutants are not due to the loss of ribbon synapses (Fig. 5 A and B). However, by P30-P32 Minar2tm1b mutant mice did show a significant decrease in the number of synapses compared to wildtype mice (Fig. 5 A and C), and this may contribute to the progression of HL.
Figure 7. Confocal maximum intensity projection images from 24 kHz region of the whole-mount cochlea.
(A and B) Total innervation with anti-2H3 Neurofilament label (green) for P1 and P14 (N=3 each group) wild type and mutant mice; scale bar A and B =20 µm (C) Schematic diagram of whole mount cochlea demonstrating the innervation pattern of type II nerves at P1 and P14 wild type and mutant mice. (D) The total number of crossings of type II nerves is show as histogram (n=3; *** P<0.001.)
Overexpression of MINAR2 suppresses angiogenesis and MINAR2 variants abolish this effect
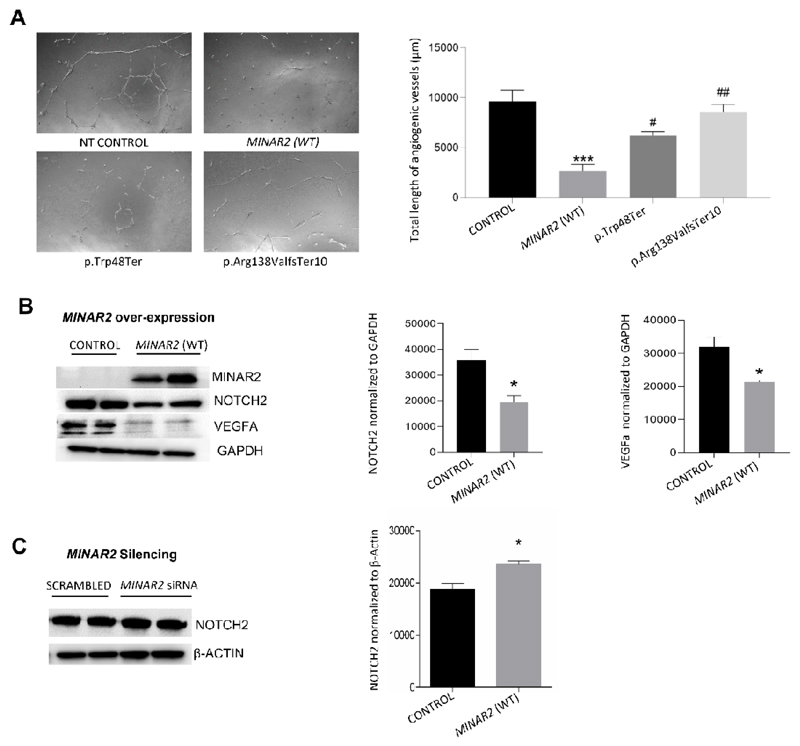
As MINAR1 has been shown to inhibit angiogenesis (20), we investigated whether MINAR2 shows the same effect on angiogenesis in human umbilical vein endothelial cells (HUVEC). These studies reveal that transient overexpression of MINAR2 suppresses angiogenesis (Fig. 8A). In addition, overexpressing each MINAR2 variant detected in Families 1 and 2 in HUVEC reduces suppression of angiogenesis, suggesting that they are loss-of-function variants (Fig. 8A).
Figure 8. Effects of MINAR2 on angiogenesis, NOTCH2 and VEGFA.
(A) Angiogenic potential of non-transfected (NT) HUVEC compared to cells transfected with MINAR2 wildtype (WT), and MINAR2 p.Trp48* and p.Arg138Valfs*10 plasmid constructs. Right graph shows analysis of total length of angiogenic vessels formed after incubation of 12 hrs on Matrigel coated wells; images were analyzed with imageJ angioanalyzer and results are expressed as mean±SEM. Significant differences were shown as *** (p≤0.001) when compared to control and as ## (p ≤0.01) when compared to MINAR2(WT). (B) Left: immunoblot images showing the effect of MINAR2(WT) transient overexpression on NOTCH2 and VEGFA protein levels in HUVEC cells; Right: statistical analysis of GAPDH-normalized relative protein expressions of NOTCH2 and VEGFA. Results are expressed as mean±SEM and significant differences are shown as *(p≤0.05) when compared to control. (C) Left: immunoblot images of siRNA-induced MINAR2 silencing on NOTCH2 protein levels in PC12 cells; Right: beta-actin-normalized relative protein expression of NOTCH2. Results are expressed as mean±SEM and significant differences are shown as *(p≤0.05) when compared to control. Data analyzed by one way ANOVA with multiple comparisons.
Expression levels of MINAR2 are inversely correlated with intracellular NOTCH2 abundance
MINAR2 is named based on its structural similarity to MINAR1, which is shown to be involved in NOTCH2 signaling (20). While MINAR2 is a much smaller protein compared to MINAR1 (190 vs 917 amino acids) (20), it has recently been shown to bind NOTCH2 in a co-immunoprecipitation assay (13). Thus, we set out to explore if NOTCH2 abundance in cells is correlated with MINAR2 expression (21, 22). Furthermore, as NOTCH and VEGF signaling pathways are known to interact during angiogenesis (23), we determined the effect of MINAR2 expression on VEGF. Our studies show that overexpression of wildtype MINAR2 in HUVEC is associated with reduced abundance of NOTCH2 and VEGFA (a prominent VEGF protein in vascular endothelial cells) (Fig. 8B). Silencing of MINAR2 in PC12 cells, which endogenously express MINAR2, leads to an increase in NOTCH2 and confirms the suppressor effect of MINAR2 on NOTCH2 abundance (Fig. 8C and SI Appendix, Fig. S8).
MINAR2 is involved in MAP kinase and mTOR pathways
MINAR1 has been shown to regulate neurite outgrowth in a neuroendocrine cell line PC12 (24), in which neurite outgrowth via nerve growth factor is achieved by MAPK and PI3K pathways (25). The mTOR complex is a downstream target of PI3K, which contains mTORC1 and mTORC2 complexes in its central catalytic domain (24). We tested the effects of MINAR2 on the MAPK pathway by transiently overexpressing wildtype MINAR2 in PC12 cells and detecting levels of ERK1/2 and pERK1/2, a crucial kinase of the MAPK signaling pathway. These studies show that overexpression of wildtype MINAR2 reduces the abundance of pERK1/2, the active form of ERK1/2. On the other hand, overexpression of the two MINAR2 deafness variants in Families 1 and 2 does not show this effect, again supporting their loss-of-function (SI Appendix, Fig. S9A). When we silenced MINAR2 in PC12 cells, we observed an increase in a functional protein in mTORC1 activity, P-S6, at 12 hours (SI Appendix, Fig. S9B). However, there was no difference in pAKT, a component of mTORC2 activity (SI Appendix, Fig. S9B).
Discussion
In this study we present three loss-of-function variants in MINAR2 in four families co-segregating with non-syndromic sensorineural HL. Hearing loss started at birth or in early childhood in all 13 affected individuals. A progressive HL reaching to severe to profound degree during childhood was noted in four affected individuals. In addition to ABR thresholds showing sensorineural HL, otoacoustic emissions were absent in three children tested, suggesting dysfunction of both inner hair cell/acoustic nerve and outer hair cells. To support causality of the MINAR2 mutations in deafness and to begin understanding the mechanism of HL, we show that Minar2tm1b/tm1b mice have a severe, progressive increase in ABR thresholds from two weeks old onwards (around the time that mice start to hear), with very few responses by four weeks old. Otoacoustic emissions can be detected in the mutant mice but at raised thresholds at two weeks old, and these responses are mostly absent by four weeks old, implicating outer hair cells in the pathology. In conclusion, audiological findings both in humans and mice show that loss of MINAR2 function results in early-onset and sensorineural HL that rapidly progresses to severe to profound deafness.
The reduced number of stereocilia in the shortest row on outer hair cells would be expected to have a severe impact upon the number of transduction channel complexes available and suggests that the primary defect in these mutant mice is located at the top of the hair cell. However, we have demonstrated that the remaining MET channels are active in Minar2tm1b/tm1b mice, since the hair cells of these mice are able to uptake FM1-43 dye. Degeneration of hair cells (Fig. 6) and synaptic defects (Fig. 5) occur later in development and cannot explain the raised ABR and DPOAE thresholds at 14 days old in the Minar2tm1b homozygotes. Mutations of other genes also lead to loss of the shorter stereocilia associated with hearing impairment, for example the Baiap2l2 mutant (26) and the Clrn2 mouse mutant (27).
The Notch pathway is a highly conserved intercellular signaling cascade that is activated by the interaction of transmembrane ligands (Delta and Jagged) with Notch receptors, which are usually expressed on the surface of neighboring cells. Binding of the Notch Ligand to receptor induces cleavage of the Notch receptors intracellular domain, which binds to multiple DNA-binding proteins in the nucleus (28–30). In the initial stages of angiogenesis, Notch activation is generally repressed to allow proliferation of endothelial cells in response to VEGF stimulation, and its expression is later upregulated when endothelial cells stop proliferating and the vessels begin to stabilize (31–37). While it is possible that the effects of MINAR2 on angiogenesis may play a role in hearing, we did not observe abnormal vascular structures in the cochlea of homozygous mutant mice. Moreover, the most sensitive part of the cochlea to vascular function is the generation of EP by the stria vascularis. In this study, EP was within the normal range in homozygous mutant mice. Our results show that MINAR2 and NOTCH2 have a regulatory relationship. In the mammalian cochlea, there is no regeneration of sensory hair cells and the strictly controlled pattern of alternating hair cells and supporting cells in the organ of Corti is set up in early development. Notch signaling is believed to be a critical part of the process of cells deciding their fate as a hair cell or a supporting cell and many genes involved in notch signaling underlie hearing impairment (38, 39). As our examination of the organ of Corti of young mice shows (SI Appendix, Fig. S5), the pattern is set up correctly, ruling out an early developmental defect of cell fate or differentiation. However, notch signally may also play a role in longer-term maintenance of the organ of Corti, and our findings of progressive deterioration of this organ resulting from mutation of a gene known to be involved in notch signaling supports this suggestion.
MINAR family proteins (MINAR1 and MINAR2) have been proposed to be associated with the inhibition of cell proliferation (13, 20). In this study, we did not observe any proliferation effect in different ages of Minar2tm1b/tm1b mice. Terminal mitoses of hair cells occur early at E12-14 before birth (40). The confocal images illustrated in Fig. 6 all indicate loss of cells from the normal regular array, with no sign of any proliferation.
Our findings of an abnormal innervation pattern of hair cells as early as P1, as well as a significant decrease in the numbers of intact synapses at four weeks old, suggest that abnormal innervation of hair cells may contribute to the severe auditory dysfunction in Minar2tm1b/tm1b mice. Based on these findings, Minar2 may be playing a role in the maintenance of hair cell synapses. MINAR2 is structurally similar to MINAR1, which has been reported to be involved in controlling neurite formation during neuronal differentiation through DEP Domain Containing MTOR Interacting Protein (DEPTOR). In this study, we show that similar to MINAR1, MINAR2 downregulates mTOR signaling (SI Appendix, Fig. S9B), which may contribute to the abnormal innervation pattern observed in the organ of Corti of Minar2 mutant mice.
It is notable that although the sensory hair cells are present up to four weeks old and are able to take up FM1-43 dye through transduction channels, they do not appear to be functioning normally as shown by the raised ABR and DPOAE thresholds. Most genetic and environmental causes of sensorineural HL lead to permanent loss of hair cells, reducing the chance of gene therapy or gene editing approaches. Progressive HL associated with a relatively normal appearance of hair cells in young ages makes MINAR2 a promising target for genetic therapies. Further elucidation of its role in the maintenance of hair cell synapses and stereocilia bundles may open new avenues to treat more common forms of HL resulting from similar mechanisms.
Materials and Methods
More details of Materials and Methods are in the SI Appendix.
Study approval
This study was approved by the University of Miami Institutional Review Board (20081138-USA), as well as by the Ethics Committees of Ankara University Medical School (012413-Turkey) and Rajarshi Shahu College of Pharmacy (180720-India). A signed informed consent was obtained from all participants or, in the case of a minor, from the parents.
DNA Sequencing and Bioinformatics Analysis
Genome sequencing was performed in Family 1 individuals II:1, II:2, II:3 and Family 2 II:1 (Fig. 1A and SI Appendix, Table S3) (41, 42). Reads were mapped to human genome reference (NCBI build37/hg 19 version) with Burrows Wheeler Aligner (BWA). Genome Analysis Toolkit (GATK) was used for variant calling (43–45). Copy number variants (CNVs) were called using CNVnator (46). Enlis Genome Research software (https://www.enlis.com/) was used to identify runs of homozygosity in Family 1 and Family 2 from genome sequencing data (SI Appendix, Tables S4, S5 and S6).
Illumina Infinium Global Screening Array (GSA) v2 (Illumina) kit was used for genotyping in 16 members of Family 3 and three members of Family 4 to map the shared homozygous regions in affected individuals. Additional, exome sequencing was performed on a HiSeq 2000 platform (Illumina), as described previously (47), in individuals IV:3 and IV:5 in Family 3 and IV:1 in Family 4 (SI Appendix, Table S3).
Genome sequencing data was deposited in the NCBI’s BioProject database: PRJNA623118, BioSample: SAMN27763770 (Family 1, individual II:1) and BioSample: SAMN27763769 (Family 2, individual II:1).
Ethics statement for animals
Mouse studies in the UK were carried out in accordance with UK Home Office regulations and the UK Animals (Scientific Procedures) Act of 1986 (ASPA) under UK Home Office licenses, and the study was approved by the King's College London Ethical Review Committee. Mice were culled using methods approved under these licenses to minimize any possibility of suffering. All procedures in Miami were approved by the University of Miami Institutional Animal Care and Use Committee and followed the NIH guidelines “Using Animals in Intramural Research.”
Mouse generation
Minar2tm1a mutant mice were generated at the Wellcome Sanger Institute by blastocyst microinjection of targeted ES cells on a C57BL/6N genetic background (48, 49). Minar2tm1b(KOMP)Wtsi mice (Minar2tm1b) were generated from Minar2tm1a(KOMP)Wtsi mice by exposure of pre-implantation embryos to soluble Cre recombinase (50) which leads to recombination between LoxP sites, removing the selection cassette and exon 2 to produce mice carrying the Minar2tm1b allele (SI Appendix, Fig. S3). Both tm1a and tm1b alleles are available via The European Mouse Mutant Archive (EMMA; https://www.infrafrontier.eu ).
Statistics
A one-way ANOVA with multiple comparisons was used to compare groups for count of synapses in mouse inner ear and was also used for angiogenesis, MAPK, and mTOR assays.
Supplementary Material
Significance Statement.
Molecular components of hearing in mammalians are not completely delineated. Via a genetic approach conducted in families with sensorineural hearing loss, this study presents MINAR2 as an indispensable element of hearing in humans. Similarly, disruption of Minar2 in mice leads to progressive hearing loss associated with alterations in the stereocilia of hair cells, the receptors of hearing, while hair cells remain intact until later in life. We present MINAR2 as a gene working in the inner ear that is essential for hearing in humans and mice. Degeneration of sensory epithelium is a common consequence of hereditary deafness precluding genetic therapies. The preservation of hair cells in mutant mice at young ages makes MINAR2 a good candidate for intervention.
Acknowledgments
We are immensely grateful to all the patients and families participating in this study. This study was supported by R01DC009645 and R01DC012836 from the National Institutes of Health/National Institute on Deafness (MT), the NIHR Biomedical Research Centre, King's College London (KPS) and the Royal National Institute for Deaf People, RNID (KPS). This research was funded in part by the Wellcome Trust. For the purpose of Open Access, the author has applied a CC BY public copyright license to any Author Accepted Manuscript (AAM) version arising from this submission. We thank the Wellcome Sanger Institute Mouse Genetics Project for generating and providing the Minar2 mouse mutant. We (GH and MD) would also like to express our sincere gratitude for all the support provided by Dr S P Jain, Principal, Rajarshi Shahu College of Pharmacy, Malvihir, Buldana, India.
Footnotes
Author Contributions: MT designed the study. GB, MD, MYB, DD, NS, GH, JN, SPJ and MT collected samples. GB, MFZ, CJS, NV, IK, SG and BC developed methods and analyzed data. MLR, NJI and KPS generated the mouse KO model, performed hearing tests and staining. JC carried out scanning electron microscopy. CA and KW performed mouse gene expression assays and protein localization. KW, JN, KPS and MT supervised experiments and analysis. GB, MFZ, KW, MLR, KPS and MT wrote the first draft. All authors reviewed the manuscript.
Competing Interest Statement: The authors have declared that no conflict of interest exists.
References
- 1.Mehl AL, Thomson V. The Colorado newborn hearing screening project, 1992-1999: on the threshold of effective population-based universal newborn hearing screening. Pediatrics. 2002;109:E7. doi: 10.1542/peds.109.1.e7. [DOI] [PubMed] [Google Scholar]
- 2.Morton CC, Nance WE. Newborn hearing screening--a silent revolution. N Engl J Med. 2006;354:2151–2164. doi: 10.1056/NEJMra050700. [DOI] [PubMed] [Google Scholar]
- 3.Van Camp G, Smith RJH. Hereditary Hearing Loss Homepage. 2021. Dec, https://hereditaryhearingloss.org .
- 4.Alford RL, et al. American College of Medical Genetics and Genomics guideline for the clinical evaluation and etiologic diagnosis of hearing loss. Genet Med. 2014;16:347–355. doi: 10.1038/gim.2014.2. [DOI] [PubMed] [Google Scholar]
- 5.Korver AM, et al. Congenital hearing loss. Nat Rev Dis Primers. 2017;3:16094. doi: 10.1038/nrdp.2016.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bademci G, et al. Comprehensive analysis via exome sequencing uncovers genetic etiology in autosomal recessive nonsyndromic deafness in a large multiethnic cohort. Genet Med. 2015 doi: 10.1038/gim.2015.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Doll J, et al. Genetic Spectrum of Syndromic and Non-Syndromic Hearing Loss in Pakistani Families. Genes (Basel) 2020;11 doi: 10.3390/genes11111329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sloan-Heggen CM, et al. Characterising the spectrum of autosomal recessive hereditary hearing loss in Iran. J Med Genet. 2015;52:823–829. doi: 10.1136/jmedgenet-2015-103389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yan D, et al. Spectrum of DNA variants for non-syndromic deafness in a large cohort from multiple continents. Hum Genet. 2016;135:953–961. doi: 10.1007/s00439-016-1697-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Atik T, Bademci G, Diaz-Horta O, Blanton S, Tekin M. Whole-exome sequencing and its impact in hereditary hearing loss. Genet Res (Camb) 2015 doi: 10.1017/S001667231500004X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ingham NJ, et al. Mouse screen reveals multiple new genes underlying mouse and human hearing loss. PLoS Biol. 2019;17:e3000194. doi: 10.1371/journal.pbio.3000194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vona B, Nanda I, Hofrichter MA, Shehata-Dieler W, Haaf T. Non-syndromic hearing loss gene identification: A brief history and glimpse into the future. Mol Cell Probes. 2015;29:260–270. doi: 10.1016/j.mcp.2015.03.008. [DOI] [PubMed] [Google Scholar]
- 13.Ho RX, et al. Loss of MINAR2 impairs motor function and causes Parkinson's disease-like symptoms in mice. Brain Commun. 2020;2:fcaa047. doi: 10.1093/braincomms/fcaa047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Karczewski KJ, et al. The mutational constraint spectrum quantified from variation in 141,456 humans. Nature. 2020;581:434–443. doi: 10.1038/s41586-020-2308-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Oza AM, et al. Expert specification of the ACMG/AMP variant interpretation guidelines for genetic hearing loss. Hum Mutat. 2018;39:1593–1613. doi: 10.1002/humu.23630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Richards S, et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015;17:405–424. doi: 10.1038/gim.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Elkon R, et al. RFX transcription factors are essential for hearing in mice. Nat Commun. 2015;6:8549. doi: 10.1038/ncomms9549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Orvis J, et al. gEAR: Gene Expression Analysis Resource portal for community-driven, multi-omic data exploration. Nat Methods. 2021 doi: 10.1038/s41592-021-01200-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gale JE, Marcotti W, Kennedy HJ, Kros CJ, Richardson GP. FM1-43 dye behaves as a permeant blocker of the hair-cell mechanotransducer channel. J Neurosci. 2001;21:7013–7025. doi: 10.1523/JNEUROSCI.21-18-07013.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ho RX, et al. MINAR1 is a Notch2-binding protein that inhibits angiogenesis and breast cancer growth. J Mol Cell Biol. 2018;10:195–204. doi: 10.1093/jmcb/mjy002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lanford PJ, et al. Notch signalling pathway mediates hair cell development in mammalian cochlea. Nat Genet. 1999;21:289–292. doi: 10.1038/6804. [DOI] [PubMed] [Google Scholar]
- 22.Pan W, Jin Y, Stanger B, Kiernan AE. Notch signaling is required for the generation of hair cells and supporting cells in the mammalian inner ear. Proc Natl Acad Sci U S A. 2010;107:15798–15803. doi: 10.1073/pnas.1003089107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Siekmann AF, Covassin L, Lawson ND. Modulation of VEGF signalling output by the Notch pathway. Bioessays. 2008;30:303–313. doi: 10.1002/bies.20736. [DOI] [PubMed] [Google Scholar]
- 24.Zhang H, et al. UBTOR/KIAA1024 regulates neurite outgrowth and neoplasia through mTOR signaling. PLoS Genet. 2018;14:e1007583. doi: 10.1371/journal.pgen.1007583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vaudry D, Stork PJ, Lazarovici P, Eiden LE. Signaling pathways for PC12 cell differentiation: making the right connections. Science. 2002;296:1648–1649. doi: 10.1126/science.1071552. [DOI] [PubMed] [Google Scholar]
- 26.Carlton AJ, et al. Loss of Baiap2l2 destabilizes the transducing stereocilia of cochlear hair cells and leads to deafness. J Physiol. 2021;599:1173–1198. doi: 10.1113/JP280670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dunbar LA, et al. Clarin-2 is essential for hearing by maintaining stereocilia integrity and function. EMBO Mol Med. 2019;11:e10288. doi: 10.15252/emmm.201910288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Andersson ER, Sandberg R, Lendahl U. Notch signaling: simplicity in design, versatility in function. Development. 2011;138:3593–3612. doi: 10.1242/dev.063610. [DOI] [PubMed] [Google Scholar]
- 29.Chillakuri CR, et al. Structural analysis uncovers lipid-binding properties of Notch ligands. Cell Rep. 2013;5:861–867. doi: 10.1016/j.celrep.2013.10.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lu FM, Lux SE. Constitutively active human Notch1 binds to the transcription factor CBF1 and stimulates transcription through a promoter containing a CBF1-responsive element. Proc Natl Acad Sci U S A. 1996;93:5663–5667. doi: 10.1073/pnas.93.11.5663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Henderson AM, Wang SJ, Taylor AC, Aitkenhead M, Hughes CC. The basic helix-loop-helix transcription factor HESR1 regulates endothelial cell tube formation. J Biol Chem. 2001;276:6169–6176. doi: 10.1074/jbc.M008506200. [DOI] [PubMed] [Google Scholar]
- 32.Itoh F, et al. Synergy and antagonism between Notch and BMP receptor signaling pathways in endothelial cells. EMBO J. 2004;23:541–551. doi: 10.1038/sj.emboj.7600065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Leong KG, et al. Activated Notch4 inhibits angiogenesis: role of beta 1-integrin activation. Mol Cell Biol. 2002;22:2830–2841. doi: 10.1128/MCB.22.8.2830-2841.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liu Z, et al. Notch1 loss of heterozygosity causes vascular tumors and lethal hemorrhage in mice. J Clin Invest. 2011;121:800–808. doi: 10.1172/JCI43114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Noseda M, et al. Notch activation induces endothelial cell cycle arrest and participates in contact inhibition: role of p21Cip1 repression. Mol Cell Biol. 2004;24:8813–8822. doi: 10.1128/MCB.24.20.8813-8822.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Taylor KL, Henderson AM, Hughes CC. Notch activation during endothelial cell network formation in vitro targets the basic HLH transcription factor HESR-1 and downregulates VEGFR-2/KDR expression. Microvasc Res. 2002;64:372–383. doi: 10.1006/mvre.2002.2443. [DOI] [PubMed] [Google Scholar]
- 37.Williams SE, Beronja S, Pasolli HA, Fuchs E. Asymmetric cell divisions promote Notch-dependent epidermal differentiation. Nature. 2011;470:353–358. doi: 10.1038/nature09793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Driver EC, Kelley MW. Development of the cochlea. Development. 2020;147 doi: 10.1242/dev.162263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Brown R, Groves AK. Hear, Hear for Notch: Control of Cell Fates in the Inner Ear by Notch Signaling. Biomolecules. 2020;10 doi: 10.3390/biom10030370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ruben RJ. Development of the inner ear of the mouse: a radioautographic study of terminal mitoses. Acta Otolaryngol. 1967;(Suppl 220):221–244. [PubMed] [Google Scholar]
- 41.Bademci G, et al. FOXF2 is required for cochlear development in humans and mice. Hum Mol Genet. 2019;28:1286–1297. doi: 10.1093/hmg/ddy431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li C, et al. Dysfunction of GRAP, encoding the GRB2-related adaptor protein, is linked to sensorineural hearing loss. Proc Natl Acad Sci U S A. 2019;116:1347–1352. doi: 10.1073/pnas.1810951116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.DePristo MA, et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet. 2011;43:491–498. doi: 10.1038/ng.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li H, Durbin R. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics. 2010;26:589–595. doi: 10.1093/bioinformatics/btp698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.McKenna A, et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Abyzov A, Urban AE, Snyder M, Gerstein M. CNVnator: an approach to discover, genotype, and characterize typical and atypical CNVs from family and population genome sequencing. Genome Res. 2011;21:974–984. doi: 10.1101/gr.114876.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jelani M, et al. A mutation in the major autophagy gene, WIPI2, associated with global developmental abnormalities. Brain. 2019;142:1242–1254. doi: 10.1093/brain/awz075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Skarnes WC, et al. A conditional knockout resource for the genome-wide study of mouse gene function. Nature. 2011;474:337–342. doi: 10.1038/nature10163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.White JK, et al. Genome-wide generation and systematic phenotyping of knockout mice reveals new roles for many genes. Cell. 2013;154:452–464. doi: 10.1016/j.cell.2013.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ryder E, et al. Rapid conversion of EUCOMM/KOMP-CSD alleles in mouse embryos using a cell-permeable Cre recombinase. Transgenic Res. 2014;23:177–185. doi: 10.1007/s11248-013-9764-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.