Summary
Background
Trigger finger and carpal tunnel syndrome are the two most common non-traumatic connective tissue disorders of the hand. Both of these conditions frequently co-occur, often in patients with rheumatoid arthritis. However, this phenotypic association is poorly understood. Hypothesising that the co-occurrence of trigger finger and carpal tunnel syndrome might be explained by shared germline predisposition, we aimed to identify a specific genetic locus associated with both diseases.
Methods
In this genome-wide association study (GWAS), we identified 2908 patients with trigger finger and 436579 controls from the UK Biobank prospective cohort. We conducted a case-control GWAS for trigger finger, followed by co-localisation analyses with carpal tunnel syndrome summary statistics. To identify putative causal variants and establish their biological relevance, we did fine-mapping analyses and expression quantitative trait loci (eQTL) analyses, using fibroblasts from healthy donors (n=79) and tenosynovium samples from patients with carpal tunnel syndrome (n=77). We conducted a Cox regression for time to trigger finger and carpal tunnel syndrome diagnosis against plasma IGF-1 concentrations in the UK Biobank cohort.
Findings
Phenome-wide analyses confirmed a marked association between carpal tunnel syndrome and trigger finger in the participants from UK Biobank (odds ratio [OR] 11·97, 95% CI 11·1–13·0; p<1 × 10–300). GWAS for trigger finger identified five independent loci, including one locus, DIRC3, that was co-localised with carpal tunnel syndrome and could be fine-mapped to rs62175241 (0·76, 0·68–0·84; p=5·03 × 10–13). eQTL analyses found a fibroblast-specific association between the protective T allele of rs62175241 and increased DIRC3 and IGFBP5 expression. Increased plasma IGF-1 concentrations were associated with both carpal tunnel syndrome and trigger finger in participants from UK Biobank (hazard ratio >1·04, p<0·02).
Interpretation
In this GWAS, the DIRC3 locus on chromosome 2 was significantly associated with both carpal tunnel syndrome and trigger finger, possibly explaining their co-occurrence. The disease-protective allele of rs62175241 was associated with increased expression of long non-coding RNA DIRC3 and its transcriptional target, IGBP5, an antagonist of IGF-1 signalling. These findings suggest a model in which IGF-1 is a driver of both carpal tunnel syndrome and trigger finger, and in which the DIRC3-IGFBP5 axis directly antagonises fibroblastic IGF-1 signalling.
Funding
Wellcome Trust, National Institute for Health Research, Medical Research Council.
Introduction
Trigger finger (also known as stenosing flexor tenosynovitis) and carpal tunnel syndrome are the two most common non-traumatic hand disorders, with a lifetime prevalence of 2–10%1 for trigger finger and 3–10% for carpal tunnel syndrome.2 Trigger finger is caused by impaired gliding of the flexor tendons through the first annular (A1) pulley and manifests as painful clicking of the digit during flexion and extension, with progressive stiffness, locking, and loss of function. Carpal tunnel syndrome is a compression neuropathy of the median nerve that manifests as paresthesia, pain, numbness, and weakness of the hand. Both conditions are associated with marked functional impairment and reduced quality of life. Carpal tunnel syndrome is associated with an estimated loss of 78 375 disease-adjusted life years annually in the USA, and a total disease burden of US$2·7 to $4·8 billion per year.3,4
Observational studies have provided evidence to suggest that there is a link between carpal tunnel syndrome and trigger finger: a prospective study found that 43% of patients diagnosed with trigger finger had clinical signs or symptoms of carpal tunnel syndrome,4 and another study of patients with carpal tunnel syndrome or trigger finger found that, on clinical examination, 61% of patients had both carpal tunnel syndrome and trigger finger.5 63% of patients with trigger finger in another study had neurophysiological evidence of carpal tunnel syndrome compared with only 8% of controls.6 Although some studies have argued that the risk of trigger finger is increased after carpal tunnel syndrome surgical decompression, which could lead to confounding of the observed association, a retrospective longitudinal study from 2019 showed no temporal correlation between ipsilateral trigger finger and carpal tunnel syndrome surgery.7 Both conditions are characterised by synovitis and fibrosis around the flexor tendons8 and share multiple risk factors, including repetitive movements, diabetes, obesity, pregnancy, and rheumatoid arthritis.1,9 Furthermore, trigger finger and carpal tunnel syndrome can both be treated with a corticosteroid injection and surgical decompression.
The association between carpal tunnel syndrome and trigger finger is incompletely characterised. We hypothesised that the co-occurrence of these conditions might be explained by a common genetic risk locus. Although the genetic architecture of carpal tunnel syndrome has been investigated through a genome-wide association study (GWAS) using data from UK Biobank,10 much less is known about the genetic basis of trigger finger. The only published GWAS for trigger finger included 942 patients and 24 472 controls, and identified a single non-replicated locus on chromosome 13.11 In this GWAS, we aimed to leverage the UK Biobank cohort to identify a specific genetic locus associated with both conditions that could potentially explain their co-occurrence.
Methods
Study design and participants
Patients with carpal tunnel syndrome (n=16 294) were defined as previously described.10 To maximise specificity, patients with trigger finger were defined by the intersection of patients who had International Classification of Diseases(ICD)-10 codes for trigger finger (M65.3, M65.30-39), and patients who had Office of Population Censuses and Surveys Classification of Surgical Operations and Procedures (OPCS) codes for tendon release (T723; figure 1A). In sensitivity analyses, we explored including patients who had either ICD-10 or OPCS codes alone (termed the extended cohort). We also included patients with self-reported trigger finger symptoms (UK Biobank self-reported illness code 1619), termed the mixed cohort. Further information for cohort definitions is provided in the appendix (p 2). To define a control group for all subsequent genome-wide analyses, we selected 488 263 participants from UK Biobank with imputed genomic data, excluding 31657 participants who did not pass genomic quality control and 20 027 participants who had coding of any nature for either carpal tunnel syndrome of trigger finger (appendix p 12).
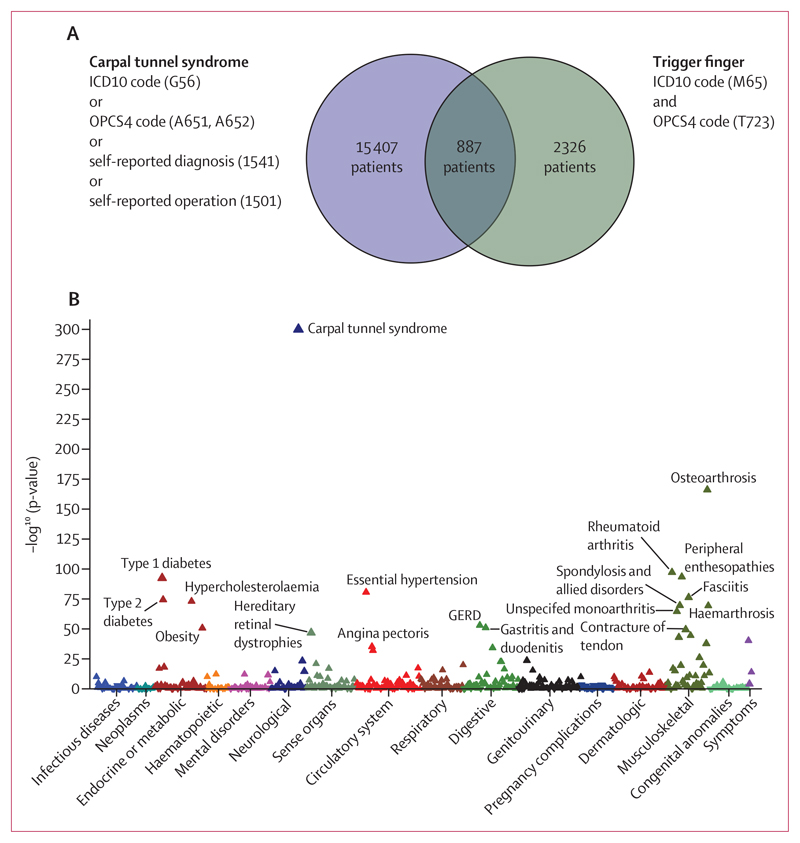
Figure 1. Association between trigger finger and carpal tunnel syndrome.
Data from UK Biobank. (A) Overlap between trigger finger and carpal tunnel syndrome in UK Biobank, annotated with the International Classification of Diseases (ICD)-10 codes and Office of Population Censuses and Surveys Classification of Surgical Operations and Procedures (OPCS) codes that were used to define whether patients had carpal tunnel syndrome or trigger finger. (B) Phenome-wide association analysis, showing the association between trigger finger and 694 phenotypes that were derived from ICD10 coding in UK Biobank. The p-value refers to a Fisher’s exact test, and the direction of the triangle reflects the direction of the effect. GERD=gastroesophageal reflux disease.
UK Biobank has approval from the North West Multi-Centre Research Ethics Committee (11/NW/0382), and this study was conducted under UK Biobank study ID 22572. The Oxford carpal tunnel syndrome cohort was derived from two clinical studies that were approved by the National Research Ethics Service (UK): the Pain in Neuropathy Study (PiNS; 10/H07056/35), and the Molecular Genetics of Carpal Tunnel Syndrome (MGCTS) study (16/LO/1920). Written informed consent was obtained from all participants recruited to these studies. Genomic data quality control steps for the UK Biobank trigger finger cohort is summarised in the appdendix (p 12).
Phenotypic association analysis
To identify diagnoses associated with trigger finger, all UK Biobank first occurrence fields (a specific UK Biobank data resource) and cancer registry data (fields 40 005 and 40 006) were extracted, with ICD-10 codes mapped to Phecodes.12 Data entries were binarised to construct a matrix of 694 diagnosis codes (including trigger finger and carpal tunnel syndrome, as defined above) versus 502 505 participants from UK Biobank. To determine the association between trigger finger diagnosis pairs, we constructed a 2 × 2 contingency table for each pair and did a Fisher’s one-way test. Significance level was set at p<1 × 10–5 which met the appropriate Bonferroni-adjusted threshold for the 694 phenotypes tested.13
Genome-wide association analyses
Genome-wide association analyses were implemented in Regenie (version 2.2.1)14 in the European ancestry cohort using Firth approximation, with covariates including year of birth (field 34), genotyping array (which were binarised from field 22000), recruitment centre (field 54), and principal components one to ten. For phenome-wide association analysis using data from UK Biobank, summary statistics were exported from the OpenTargets Genetics Portal,15 extracting data specific to the European ancestry group and with p<1 × 10–5.
Processing of summary statistics
To identify independent signals, we conducted conditional and joint analyses, implemented in Genome-wide Complex Trait Analysis(GCTA)-Conditional and Joint analysis (COJO)16 using a linkage disequilibrium reference derived from 1000 Genomes. Linkage disequilibrium score regression was implemented in the ldsc package for R using a UK Biobank European ancestry linkage disequilibrium reference derived from the PanUK Biobank project. Lead single-nucleotide polymorphisms (SNPs) were annotated using the OpenTargets Genetics portal, which considered three annotations: the nearest coding gene, genes with a cis-expression quantitative trait loci (eQTL) variant in linkage disequilibrium (r2>0·8) with the lead SNP, and the Variant2Gene score.
Co-localisation analysis
Carpal tunnel syndrome summary statistics from our previous GWAS were used.10 To extract data that were specific to the DIRC3 locus, we filtered the merged summary statistics to a 1MB region centered at rs10203066. To extract a signed linkage disequilibrium correlation matrix for the SNPs in these regions, we used the ld_matrix_local function in the ieugwasr package in R using a linkage disequilibrium reference derived from 5000 randomly selected unrelated European participants from UK Biobank. Co-localisation analyses were implemented in the coloc package in R using the coloc. susie function, with default parameters.17 The posterior probability for hypothesis 4 (H4), reflecting the existence of a shared causal variant, was extracted. To determine the 95% credible set of co-localised variants, we extracted the posterior probabilities of each SNP, conditioned on H4 being true. These SNPs were functionally annotated using ensembl variant effect predictor in the Ensembl web server.
Replication in FinnGen cohort and meta-analysis
Summary statistics for trigger finger (M13) and carpal tunnel syndrome (G6) were downloaded from the FinnGen portal (release 4).18 The FinnGen analysis pipeline has been described previously.19 Plots were generated in LocusZoom using legacy mode and the 1000 Genomes European linkage disequilibrium reference, relative to the index SNP rs10203066. We did a sample size-weighted meta-analysis between our UK Biobank trigger finger summary statistics (core analysis) and FinnGen R4 trigger finger summary statistics, which we then implemented in METAL (version March 2011) using the analyse heterogeneity command.20
Oxford-carpal tunnel syndrome cohort sample collection
Sample collection for both the PiNS and MGCTS has been described previously.10 Briefly, patients with clinically diagnosed carpal tunnel syndrome underwent carpal tunnel decompression surgery, during which tenosynovial specimens were collected. Samples were preserved in RNAlater (Thermo Fisher, MA, USA) before extraction using the High Pure RNA Isolation Kit (Roche, Basel, Switzerland). For paired whole-genome genotyping, DNA was extracted from whole blood samples using the PureLink Genomic DNA Kit (Invitrogen, MA, USA).
RNA sequencing
RNA extraction and library preparation were done as described previously.10 Reads were aligned to the GRCh37 reference with STAR21 using the Ensembl 87 gene annotation, with gene-level counts assigned using HTSeq.22 Count-level batch correction between MGCTS and PiNS cohorts was done using ComBat-seq. To facilitate inter-sample comparisons, count-level data was Trimmed Mean of the M-values(TMM)-normalised and log-transformed to generate log-transcripts per million (log-counts per million [CPM]) data.
eQTL analysis
Harmonised summary statistics from our analysis of Genotype-Tissue Expression (GTEx) project data were downloaded from the eQTL Catalogue.23 eQTL analysis for IGFBP5 in the cohort analysed by Neavin and colleagues was done as described previously.24 Briefly, for each individual and fibroblast cluster, the quantile-normalised pseudobulk average expression was extracted, and cis-eQTL association statistics were computed using a linear model implemented in MatrixEQTL,25 with one PEER factor as covariate. eQTL analysis for DIRC3 and IGFBP5 in the Oxford-carpal tunnel syndrome cohort was implemented as a Kruskal-Wallis test for gene expression (log-CPM) against genotype.
IGF-1 in UK Biobank
IGF-1 plasma levels from the first recruitment visit were extracted from field 30770 and were normalised by Z-scoring, stratified by age (decile) and sex. For Cox regression of trigger finger syndrome-free survival or carpal tunnel syndrome-free survival against Z-scored IGF-1, the time variable used was time from blood sampling to ICD-coded diagnosis of carpal tunnel syndrome or trigger finger, or last follow-up date. Last follow-up date was determined through integration of death status, recruitment visits, and ICD coding dates. The model was adjusted for age at first recruitment visit, sex, and recruitment centre.
Role of the funding source
The funders of the study had no role in study design, data collection, data analysis, data interpretation, or writing of the report.
Results
Phenome-wide analyses were conducted across the whole UK Biobank cohort (n=502 490), which recapitulated a highly significant association between trigger finger and carpal tunnel syndrome (p<1×10−300 [odds ratio (OR) 11·97], 95% CI 11·1–13·0; figure 1B). To explore the phenotypic association between trigger finger and carpal tunnel syndrome, we quantified the overlap between trigger finger and carpal tunnel syndrome (figure 1A) and explored the clinical characteristics of patients with trigger finger, carpal tunnel syndrome, and trigger finger–carpal tunnel syndrome overlap (appendix p 3). We identified evidence for a significantly increased prevalence of type 1 diabetes and type 2 diabetes, as well as significantly increased HbA1c levels (a routinely used biomarker for glycaemic control) in patients with trigger finger–carpal tunnel syndrome overlap compared with patients with carpal tunnel syndrome alone (p<1 × 10–4) and patients with trigger finger alone (p<0·01).
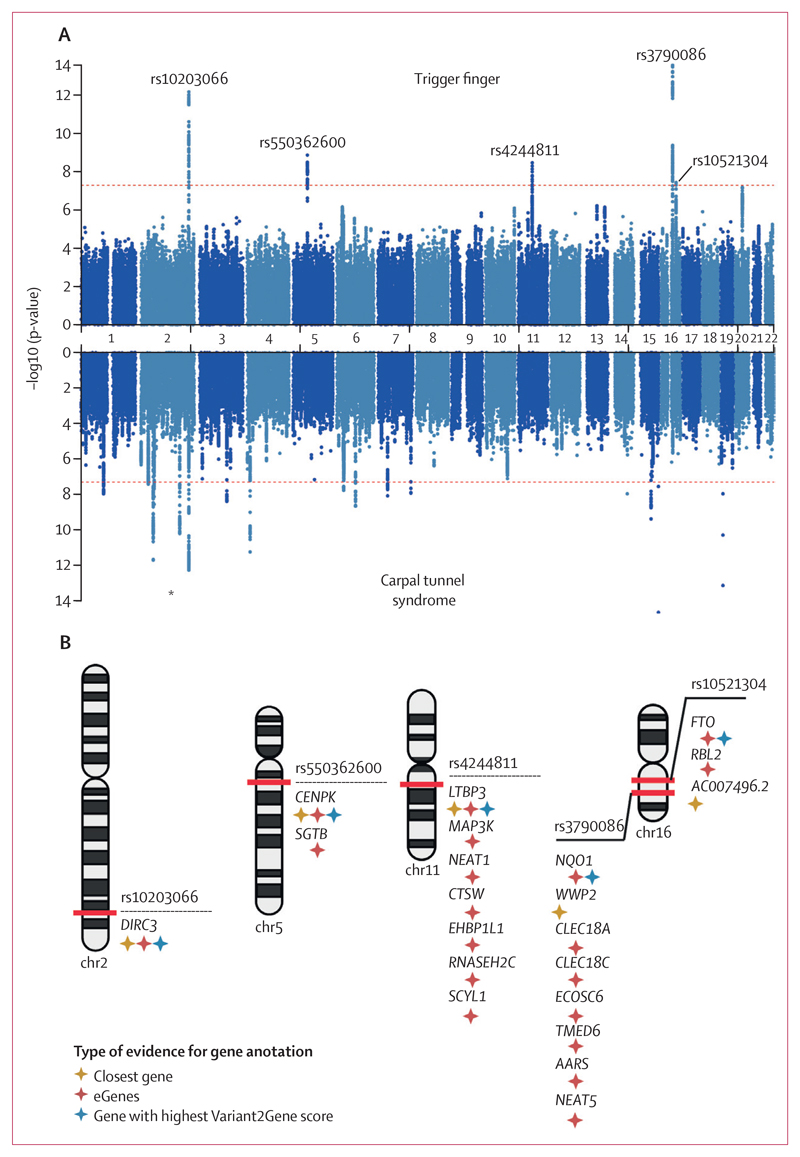
To investigate the genetic architecture of trigger finger, we conducted a GWAS in participants with European ancestry (n=456 606) from UK Biobank, incorporating 2908 patients with trigger finger (as previously defined [figure 1A]) who passed sample quality control (appendix p 12), and 436 579 controls (appendix p 2) using a mixed-model approach that accounted for unbalanced case-control ratios, population structure, and cryptic relatedness. There were insufficient numbers of patients with trigger finger in non-European ancestry groups to conduct a sufficiently powered analysis. There was also no evidence of significant confounding, with a genomic inflation factor (λGC 1·058), and we estimated the SNP-based heritability for trigger finger to be 0·8% (SE 0·1%). We identified five independent risk loci, comprising 419 variants, that met genome-wide significance (figure 2A). Conditional analyses showed no evidence of secondary signals at each locus. Using a multi-modal approach to gene-mapping, we identified 21 candidate genes at the five loci (figure 2B; appendix p 6).
Figure 2. Genome-wide association results for trigger finger and carpal tunnel syndrome.
(A) Summary statistics from trigger finger and carpal tunnel syndrome (derived from Wiberg and colleagues) genome-wide association analyses in European patients from UK Biobank. Independent loci in trigger finger summary statistics are annotated with the single-nucleotide polymorphism (SNP) identifier for the index SNP (lowest p-value) at each locus. (B) Results of gene prioritisation analysis for trigger finger trait, using multiple annotations including nearest gene to SNP, cis-eQTL associations and the Variant2Gene score from OpenTargets Genetics. AARS=alanyl-tRNA synthetase. CENPK=centromere protein K. CLEC18A=C-Type Lectin Domain Family 18 Member A. CLEC18C=C-Type Lectin Domain Family 18 Member C. CTSW=cathepsin W. DIRC3=Disrupted In Renal Carcinoma 3. EHBP1L1=EH domain binding protein 1 like 1. FTO=FTO alpha-ketoglutarate dependent dioxygenase. LTBP3=latent transforming growth factor beta binding protein 3. MAP3K=mitogen-activated protein kinase kinase kinase 1. NEAT1=nuclear paraspeckle assembly transcript 1. NEAT5=nuclear paraspeckle assembly transcript 5. NQO1=NAD(P)H quinone dehydrogenase 1. RBL2=RB transcriptional corepressor like 2. RNASEH2C=ribonuclease H2 subunit C. SCYL1=SCY1 like pseudokinase 1. SGTB=small glutamine rich tetratricopeptide repeat co-chaperone beta. TMED6=transmembrane p24 trafficking protein 6. WWP2=WW domain containing E3 ubiquitin protein ligase 2. *Signal at DIRC3 locus common to trigger finger and carpal tunnel syndrome analyses.
In support of our stringent approach to trigger finger phenotype definition, we found that conducting the GWAS by including patients with either ICD-10 or OPCS codes (ie, the extended cohort) or additionally including patients with self-reported trigger finger (ie, the mixed cohort) markedly reduced the power to detect significant associations across the five loci (appendix p 2, 6).
At the DIRC3 locus (encoding a long non-coding RNA [lncRNA]), index SNP rs10203066 (p=6·73 × 10–13 [OR 0·75]; 95% CI 0·69–0·82) was shared with our GWAS on carpal tunnel syndrome (p=2·20 × 10–12 [0·88]; 0·85–0·91; figure 3A, C).10 To confirm that this signal was not driven solely by patients with carpal tunnel syndrome in our trigger finger cohort, we undertook a further sensitivity GWAS analysis excluding any patients with carpal tunnel syndrome (appendix p 2) and confirmed that this signal retained genome-wide significance (p=1·09 × 10–10; appendix p 6). Consistent with the independent association of this locus with carpal tunnel syndrome and trigger finger, we confirmed an increase in statistical significance by merging all patients with trigger finger and carpal tunnel syndrome (p=1·28 × 10–24; appendix p 6).
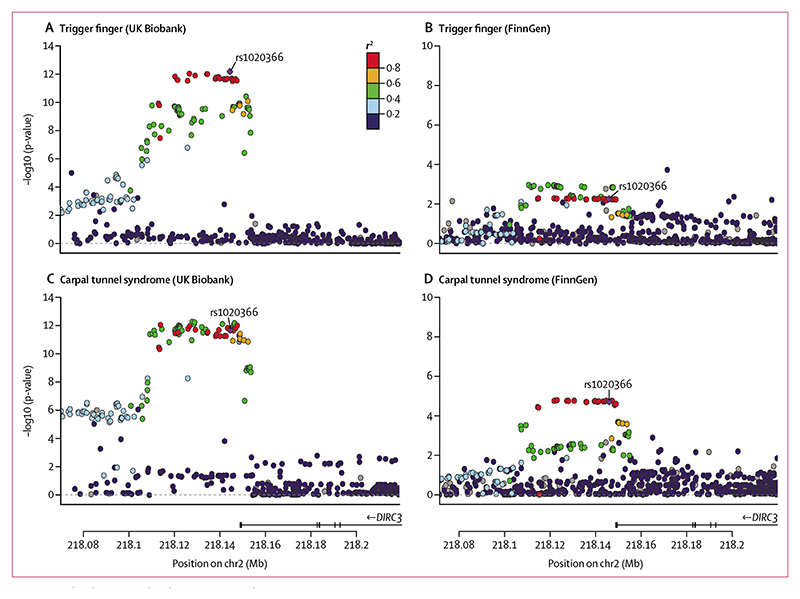
Figure 3. Co-localisation and replication at DIRC3 locus.
Single-nucleotide polymorphism(SNP)-level associations with trigger finger (A, B) and carpal tunnel syndrome (C, D) at DIRC3 locus, displayed as a LocusZoom plot, derived from UK Biobank (A, C) and FinnGen (B, D) prospectively-recruited cohorts. Index SNP at DIRC3 locus (rs10203066) is annotated in purple, and each SNP is coloured according to r2 with rs10203066, derived from the 1000 Genomes linkage disequilibrium reference. DIRC3=Disrupted In Renal Carcinoma 3. Mb=megabase.
We conducted a genetic correlation analysis using linkage disequilibrium score regression and identified evidence of a high correlation between trigger finger and carpal tunnel syndrome traits (genetic correlation estimate 0·70). To identify the loci that were driving this genetic correlation, we conducted a co-localisation analysis with a multiple causal variant assumption (SuSiE coloc). This analysis identified a high (87%) posterior probability that trigger finger and carpal tunnel syndrome share a single causal variant at this locus. To replicate the association between the DIRC3 locus and both trigger finger and carpal tunnel syndrome, we extracted summary statistics for trigger finger and carpal tunnel syndrome from the FinnGen cohort (release 4; https://finngen.gitbook.io/documentation/v/r4/). The association with both trigger finger (1485 patients and 137185 controls; p=0·0055 [OR 0·88]; 95% CI 0·81–0·96; appendix p 6) and carpal tunnel syndrome (8576 patients and 158705 controls; p=1·90 × 10–5 [0·91]; 0·88–0·95) was confirmed using a Bonferroni-corrected threshold of p<0·01 (figure 3B, D). A formal meta-analysis of UK Biobank and FinnGen summary statistics for trigger finger showed directional concordance at all loci, with low heterogeneity across three out of five loci (appendix p 7).
We leveraged the co-localisation analysis between the traits for carpal tunnel syndrome and trigger finger to fine-map the DIRC3 locus by extracting the 95% credible set (n=21) of co-localised variants (appendix pp 8–9). Next, using Ensembl Variant Effector Predictor,26 we functionally annotated the SNPs with their immediate regulatory environment. One SNP, rs62175241, had significant regulatory consequences by disrupting an enhancer site active in fibroblasts, as well as the binding motifs for a range of transcription factors, including KLF16 and KLF18 (appendix p 10).
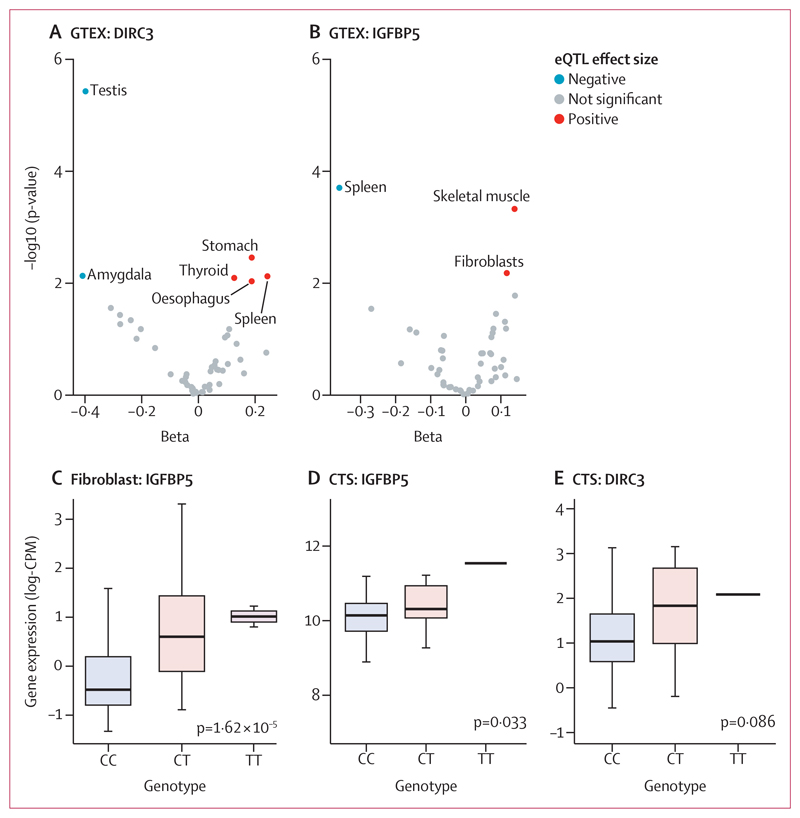
To investigate how rs62175241 (p=5·03 × 10–13 [OR 0·76]; 95% CI 0·68–0·84) might modulate the expression of DIRC3, we conducted an eQTL analysis using data from 53 tissues that were examined as part of the GTEx project.23 This analysis provided evidence that the effect of rs62175241 (T allele, which is protective for carpal tunnel syndrome and trigger finger, allele fraction 0·14) on DIRC3 expression is highly tissue-specific, with positive regulation in the stomach and spleen, and negative regulation in the testes and amygdala (figure 4A). In light of evidence that DIRC3 is able to directly activate expression of IGFBP5,27 with both genes found in the same topologically associating domain, we examined the effect of rs62175241 on IGFBP5 expression in GTEx. This analysis again provided evidence for tissue-specific eQTL associations and showed a discordant effect of rs62175241 on DIRC3 and IGFBP5 expression in the spleen (figure 4B). Considering that fibroblast proliferation is a histological feature of both trigger finger and carpal tunnel syndrome,8 we further investigated the effect of rs62175241 on IGFBP5 expression in fibroblasts. We re-analysed fibroblast single-cell eQTL data from 79 donors.24 Four of six fibroblast subtypes showed a significant positive association between the protective T allele of rs62175241 and IGFBP5 expression (appendix p 10), with the strongest association seen in HOXC6+ cluster (figure 4C). LncRNAs inherently have markedly lower abundance than mRNAs; consistent with this and the shallow depth of sequencing in single cell RNA sequencing data, DIRC3 was detected in fewer than 1% of cells, precluding further analysis.
Figure 4. rs62175241 is associated with increased RNA expression the DIRC3-IGFBP5 axis Cis-eQTL analysis for rs62175241 on DIRC3 (A) and IGFBP5 (B) gene expression in 53 tissues from the GTEx project.
Tissue-specific eQTL associations with p values <0·01 are annotated (ie, we have labelled the tissue type for significant associations). p values refer to the linear model, adjusted for PC1-6. (C) Validation of rs62175241 as an eQTL for IGFPB5 expression in the HOXC6+ fibroblast cluster, using single-cell RNA sequencing of fibroblasts from 79 donors. The p value refers to the linear model, adjusted for average expression and 1 PEER factor. Association between rs62175241 genotype and DIRC3 (D) and IGFBP5 (E) expression, derived from paired whole-genome genotyping and RNA sequencing of surgical tenosynovium samples in the Oxford-carpal tunnel syndrome cohort (n=77, including 18 patients with CT genotype and one patient with TT genotype). The p value refers to Kruskal-Wallis test. CPM=counts per million. CTS=carpal tunnel syndrome. DIRC3=Disrupted In Renal Carcinoma 3. eQTL=expression quantitative trait. GTEX=genotype-tissue expression. IGFBP5=Insulin-like growth factor-binding protein-5.
Next, we analysed the association of rs62175241 on DIRC3 and IGFBP5 expression in diseased tenosynovium samples from patients with carpal tunnel syndrome (n=77). We confirmed that the protective T allele was associated with significantly increased IGFBP5 expression (p=0·033, allele fraction 0·13; figure 4D). DIRC3 expression levels were again low and did not show allele-specific differential expression (p=0·086, figure 4E). Because IGFBP5 is a secreted protein, we investigated whether this variant might alter plasma concentration. We analysed a publicly available plasma proteomic GWAS dataset that was obtained from the SomaLogic platform,28 and found that this variant (tagged by rs10203066, A allele, r2=0·99) was associated with a non-significant increased plasma IGFBP5 (beta=0·017; p=0·11).
As IGFBP5 is known to directly antagonise IGF-1 signalling,29–31 with evidence that exogenous growth hormone treatment can cause carpal tunnel syndrome,29 we hypothesised that higher IGF-1 plasma levels would be associated with significantly increased risk of both trigger finger and carpal tunnel syndrome in UK Biobank participants. We identified significant associations between plasma concentrations of IGF-1 and trigger finger (hazard ratio [HR] per 1 SD 1·04 [95% CI 1·01–1·07]; p=0·02) and carpal tunnel syndrome (HR per 1 SD 1·04 [1·02–1·05]; p=4·23 × 10–6), which was concordant with another carpal tunnel syndrome-specific analysis in UK Biobank.32 If the protective effect of rs62175241 was mediated via antagonism of IGF-1 signalling, we hypothesised that this variant would be associated with the attenuation of growth hormone-regulated phenotypes such as height and lean body mass. To investigate this hypothesis, we extracted growth phenotype summary statistics from UK Biobank and selected traits that met phenome-wide significance (p<1 × 10–5). This process identified 22 growth-related traits that were significantly associated with rs62175241 (appendix p 11), all of which had a negative beta, including standing height (p=3·57 × 10–18), weight (p=8·28 × 10–6), forced vital capacity (p=1·14 × 10–6), and appendicular lean mass (p=4·90 × 10–33). Altogether, the available data suggest that IGF-1 is associated with increased risk of both trigger finger and carpal tunnel syndrome, and that the T allele of rs62175241 might act to directly attenuate IGF-1 signalling, thus explaining its protective effect for trigger finger and carpal tunnel syndrome.
Discussion
This GWAS for trigger finger identified five risk loci, one of which was also identified in our previous GWAS of carpal tunnel syndrome.10 Hypothesising that a single genetic variant might contribute to the pathogenesis of both diseases, we fine-mapped this locus to a single putative causal SNP, rs62175241. Our single-cell eQTL analysis provided evidence to show that this variant was associated with tissue-specific modulation of the expression of DIRC3 and its known downstream effector, IGFBP5. Bulk RNA sequencing analysis of surgically resected tenosynovium samples from patients with carpal tunnel syndrome showed that this protective variant was associated with enhanced expression of IGFBP5. Considering that IGFBP5 is an antagonist of IGF-1, we found that both trigger finger and carpal tunnel syndrome were positively associated with IGF-1 levels. These findings are important because they provide a direct biological insight into the shared pathophysiological mechanisms contributing to trigger finger and carpal tunnel syndrome (figure 5). Furthermore, our findings provide a starting point for investigating non-surgical interventions for these two common conditions.
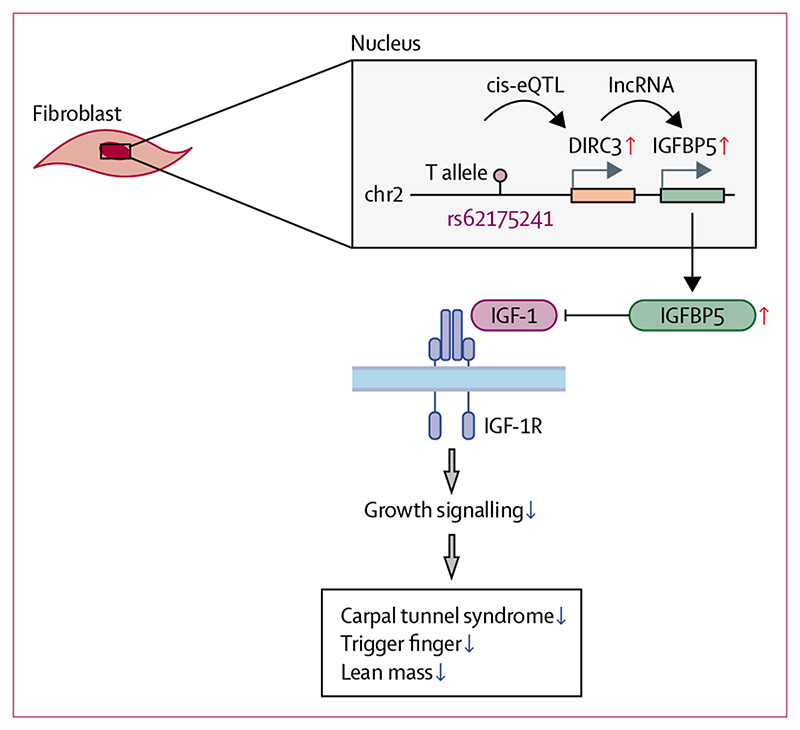
Figure 5. Proposed model for the role of DIRC3 in carpal tunnel syndrome and trigger finger.
The disease-protective T allele of rs62175241 (DIRC3 locus) is associated with increased expression of long non-coding RNA (lncRNA) DIRC3 and increased RNA expression of its direct transcriptional target IGFBP5. IGFBP5 protein is a secreted antagonist of IGF-1 signalling that acts to suppress growth signalling. Increased plasma IGF-1 is associated with both carpal tunnel syndrome and trigger finger, suggesting that IGF-1 signalling is a driver of both conditions. We propose that the association between the T allele of rs62175241 with carpal tunnel syndrome, trigger finger, and growth phenotypes (eg, lean mass) is mediated by increased antagonist of fibroblast IGF-1 signalling, altogether directly implicating IGF-1 signalling in carpal tunnel syndrome and trigger finger pathophysiology. DIRC3=Disrupted In Renal Carcinoma 3. eQTL=expression quantitative trait. IGF-1=insulin-like growth factor-1. IGFBP5=insulin-like growth factor-binding protein-5.
To the best of our knowledge, the co-localised locus that mapped to the DIRC3 gene has not been previously described in association with trigger finger. One previous GWAS11 was conducted to identify risk loci associated with trigger finger, finding a single genome-wide locus within KLHL1 that did not replicate. KLHL1 is an actin-binding protein, and the authors of this study speculated that this variant might lead to fibrocartilaginous metaplasia in tenocytes.
In this GWAS, the putative causal SNP, rs62175241, is located 3731 base pairs from the canonical transcription start site of the DIRC3 gene. The DIRC3 locus spans 450 kilobases between the IGFBP5 and TNS1 genes. By mapping the chromatin structure of the IGFBP5-DIRC3-TNS1 gene territory in human keratinocytes, Coe and colleagues27 showed that DIRC3 and IGFBP5 are located within the same topologically-associated domain. They also found two DNA looping interactions between the DIRC3 locus and IGFBP5 promoter. In their study, DIRC3 levels positively correlated with IGFBP5 in melanoma RNA sequencing samples and they discovered that DIRC3 acts in cis to control expression of IGFBP5.
IGFBP5 expression is altered in several fibrotic disease states. In lung tissue from patients with idiopathic pulmonary fibrosis, IGFBP5 was upregulated and exogenous IGFBP5 stimulated extracellular matrix secretion by idiopathic pulmonary fibrosis pulmonary fibroblasts.33,34 Furthermore, IGFBP5 was upregulated in skin fibroblasts from patients with systemic sclerosis.35 In the present study, the protective T allele at our putative causal SNP had tissue-specific effects on expression of DIRC3 and IGFBP5. In both skin fibroblasts and operative tenosynovium samples from patients with carpal tunnel syndrome, the protective allele was associated with enhanced expression of IGFBP5. IGFBP5 is a highly conserved and multifunctional secreted protein that binds to IGF and can have complex and varying effects on IGF signalling depending on the tissue type and context. In bone, IGFBP5 inhibits IGF-1 signalling36 by modulating binding to the IGF-1 receptor.30 Similarly, in mammary tissue, IGFBP5 regulates involution by inhibiting IGF-1 signalling.31
Consistent with the hypothesis that overactive IGF-1 signalling is important in trigger finger and carpal tunnel syndrome, we discovered that IGF-1 plasma concentrations were positively associated with both conditions. Furthermore, despite carpal tunnel syndrome generally being associated with decreased height, the protective allele at our putative causal locus was associated with decreased height, suggesting a distinct pathophysiological mechanism.
Several other lines of evidence support the role of IGF-1 signalling in trigger finger and carpal tunnel syndrome. The prevalence of carpal tunnel syndrome37 and trigger finger38 is increased in patients with acromegaly, for whom raised IGF-1 levels are characteristic. By normalising levels of IGF-1, either through pituitary resection or somatostatin analogues, increased tendon thickness at the A1 pulley can be reversed, and symptoms of trigger finger ameliorated.38 In healthy patients who do not have acromegaly, administering exogenous growth hormone stimulates a rise in IGF-1, and patients can subsequently develop carpal tunnel syndrome.29 Exogenous growth hormone is also known to increase the risk of type 2 diabetes, which, along with type 1 diabetes, we found to be significantly enriched in patients with both carpal tunnel syndrome and trigger finger39 (appendix p 3). Somatostatin analogues work not only by reducing pituitary growth hormone secretion but also by stimulating IGFBP5 secretion40 which, in turn, inhibits IGF-1 signalling. Of note, our phenotypic analysis highlighted a significant association between trigger finger and both rheumatoid arthritis and osteoarthritis, which was even stronger in the trigger finger-carpal tunnel syndrome overlap cohort (appendix pp 3–4). IGF-1 signalling has been implicated in synovitis in both osteoarthritis41 and rheumatoid arthritis,42 which would accord with our proposed pathophysiological mechanism underlying the observed phenotypic associations.
We recognise several limitations of the present study. Firstly, our GWAS of patients with trigger finger only included patients of European ancestry. It would be helpful to further characterise the genetic architecture of trigger finger in non-European groups, who are under-represented in GWASs. Although our GWAS had a greater power (81% power) to detect a hypothetical variant with a minor allele fraction of 50% and an OR of 1·2 than a previous study (3% power),13 our study was still relatively underpowered, especially for low-frequency variants, meaning that other relevant risk loci might not have met our pre-defined significance threshold. Although we were able to replicate our co-localised DIRC3 locus in patients with trigger finger and patients with carpal tunnel syndrome from the FinnGen cohort, we were unable to replicate all five trigger finger loci. This might be partly explained by the size of the replication cohort (1485 patients with trigger finger) and the use of a different case definitions in FinnGen, but might also be consistent with the so-called winner’s curse phenomenon43 in GWASs. Regardless, these validation data strongly support a role for the DIRC3 locus in both trigger finger and carpal tunnel syndrome. We have identified a biologically relevant mechanism that is likely to underpin the association between our co-localised risk locus and trigger finger and carpal tunnel syndrome. However, we recognise that the association between a protective haplotype at DIRC3, IGFBP5 and IGF-1 are all correlative, and further studies are required to dissect these mechanisms and provide evidence for a causative effect.
In conclusion, we provide evidence for a phenotypic association between trigger finger and carpal tunnel syndrome in patients from the UK Biobank cohort. We identified a putative causal variant in our GWAS of trigger finger that overlaps with carpal tunnel syndrome, which possibly accounts for some of the phenotypic overlap between these two conditions. Through multimodal expression eQTL analyses, we directly linked a protective causal variant in DIRC3 to increased expression of IGFBP5, an IGF-1 antagonist.30 Finally, we provide evidence to show a significant association between plasma IGF-1 concentrations and both trigger finger and carpal tunnel syndrome, altogether implicating IGF-1 signalling in the pathophysiology of both conditions. Collectively, our findings indicate that the protective variant at rs62175241 acts by enhancing expression of IGFBP5, via DIRC3 which, in turn, inhibits IGF-1 signalling (figure 5). Further studies are required to fully characterise this pathway and delineate whether it might be a valid target for pharmacological management of trigger finger and carpal tunnel syndrome.
Research in context.
Evidence before this study
We did a literature review using Medline to determine the co-occurrence of trigger finger and carpal tunnel syndrome. We searched terms “trigger finger” and “carpal tunnel syndrome”, and included all studies published up to March 28, 2022. We included original research studies in all languages that quantitatively assessed co-occurrence of trigger finger and carpal tunnel syndrome, and excluded individual case reports. We identified three prospective observational studies and 24 retrospective studies. Studies report that trigger finger and carpal tunnel syndrome occur together more often than would be expected by chance.
Added value of this study
Our study is, to the best of our knowledge, the largest genome-wide association study (GWAS) for trigger finger. We leveraged the results of this GWAS to identify a single locus (DIRC3) that is significantly associated with both trigger finger and carpal tunnel syndrome. We used multi-modal expression quantitative trait analysis to trace the mechanism by which DIRC3 modifies risk for these conditions, and provide evidence to show that IGF-1 signalling has a role in disease pathophysiology.
Implications of all the available evidence
The long-established co-occurrence of trigger finger and carpal tunnel syndrome might be at least partly explained by a shared germline predisposition, which acts to increase IGF-1 signalling in fibroblasts. Further research should determine whether this pathway might be a valid target for pharmacological management of trigger finger and carpal tunnel syndrome.
Acknowledgments
This study was conducted using the UK Biobank resource (application number 22572). We gratefully acknowledge the participants and investigators of the FinnGen study. We thank all patients and their families who volunteered to participate in this clinical research. We also thank the Oxford Genomics Centre at the Wellcome Centre for Human Genetics (funded by Wellcome Trust grant reference 203141/Z/16/Z) for the generation and initial processing of RNA sequencing data. We also thank our clinical collaborators at the Nuffield Department of Orthopaedics, Rheumatology and Musculoskeletal Sciences Hand Research Group for the collection of surgical specimens (https://www.ndorms.ox.ac.uk/research-groups/collaborative-hand-research-group). SOK is supported by the Starr Centennial Scholarship at the Cold Spring Harbor Laboratory School of Biological Sciences. Computational analyses at Cold Spring Harbor Laboratory were conducted with assistance from the US National Institutes of Health (grant S10OD028632-01). AW is supported by a National Institute for Health Research (NIHR) Clinical Lectureship and a grant from the Medical Research Council Doctoral Training Partnership (MR/N013468/1). DF is supported by the NIHR Biomedical Research Centre, Oxford (Biomedical Research Centre). AS is supported by a Wellcome Trust Clinical Career Development Fellowship (222101/Z/20/Z) and the NIHR Oxford Biomedical Research Centre. W-U-RA is supported by the NIHR Oxford Biomedical Research Centre, Aziz Foundation, and Royal College Surgeons of Edinburgh. The views expressed are those of the authors and not necessarily those of the National Health Service, the NIHR, or the Department of Health.
Footnotes
Contributors
BP, DF, and AW conceived and designed the study. BP constructed the statistical analysis plan and extracted case and control cohorts. SOK conducted the computational analyses. GB assimilated and processed raw RNA sequence data. W-U-RA extracted DNA for genotyping, and MN conducted quality control and imputation of the genotype data. ABS and DLB recruited and phenotyped the Pain in Neuropathy Study (PiNS) cohort, and conducted the RNA extraction with AW. DN completed single-cell analyses with supervision from JP. DF and AW supervised the project and guided all data analyses. SOK, BP, and AW accessed and verified all data reported in the manuscript. BP, SOK, DF, and AW wrote the manuscript with input from all authors. All coauthors approved the final version of the manuscript.
Declaration of interests
DLB reports grants from Lilly and AstraZeneca, has acted as a consultant on behalf of Oxford Innovation for Amgen, Biointervene, Bristows, LatigoBio, GSK, Ionis, Lilly, Olipass, Orion, Regeneron, and Theranexus, and has a patent application “a method for the treatment or prevention of pain, or excessive neuronal activity, or epilepsy” (16/337,428). GB reports consultancy fees from Biocoding and Ivy Farm. JP is chair of Oz Single Cells and has stock or stock options in Sonic Healthcare. All other authors declare no competing interests.
Contributor Information
Benjamin Patel, Department of Plastic and Reconstructive Surgery, Southmead Hospital, North Bristol NHS Trust, Bristol, UK.
Sam O Kleeman, Cold Spring Harbor Laboratory, New York, NY, USA.
Drew Neavin, Garvan-Weizmann Centre for Cellular Genomics, Garvan Institute of Medical Research, Sydney, NSW, Australia.
Prof Joseph Powell, Garvan-Weizmann Centre for Cellular Genomics, Garvan Institute of Medical Research, Sydney, NSW, Australia; UNSW Cellular Genomics Futures Institute, University of New South Wales, NSW, Australia.
Georgios Baskozos, Nuffield Department of Clinical Neurosciences, John Radcliffe Hospital, University of Oxford, Oxford, UK.
Michael Ng, Nuffield Department of Orthopaedics, Rheumatology and Musculoskeletal Sciences, Botnar Research Centre, University of Oxford, Oxford, UK.
Waheed-Ul-Rahman Ahmed, Nuffield Department of Orthopaedics, Rheumatology and Musculoskeletal Sciences, Botnar Research Centre, University of Oxford, Oxford, UK.
Prof David L Bennett, Nuffield Department of Clinical Neurosciences, John Radcliffe Hospital, University of Oxford, Oxford, UK.
Prof Annina B Schmid, Nuffield Department of Clinical Neurosciences, John Radcliffe Hospital, University of Oxford, Oxford, UK.
Prof Dominic Furniss, Nuffield Department of Orthopaedics, Rheumatology and Musculoskeletal Sciences, Botnar Research Centre, University of Oxford, Oxford, UK.
Akira Wiberg, Nuffield Department of Orthopaedics, Rheumatology and Musculoskeletal Sciences, Botnar Research Centre, University of Oxford, Oxford, UK.
Data sharing
RNA sequencing data from the Pain in Neuropathy Study (PiNS) cohort has been reported previously and is available at accession GEO108023 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE108023). The relevant raw data (count matrix and genotype calls at rs62175241) and code that are necessary to replicate analyses in the expanded Oxford-CTS RNA sequencing cohort are provided on Github (https://github.com/samkleeman1/cts_tf/). A Jupyter Notebook summarising the genotype data quality control and genome-wide association analysis implemented in Regenie is provided on Github (https://github.com/samkleeman1/cts_tf/). Single-cell fibroblast expression quantitative trait (eQTL) summary statistics and applicable source data are published alongside the original manuscript.19 UK Biobank data can be requested through the application process detailed at https://www.ukbiobank.ac.uk/. Summary statistics for the primary trigger finger GWAS (patients with both International Classification of Diseases[ICD]-10 and Office of Population Censuses and Surveys Classification of Surgical Operations and Procedures[OPCS]-4 codes for trigger finger) have been uploaded to the GWAS Catalog (accession code GCST90104907). Summary statistics for the sensitivity analyses described in the manuscript are available from the corresponding author on request.
References
- 1.Stahl S, Kanter Y, Karnielli E. Outcome of trigger finger treatment in diabetes. J Diabetes Complications. 1997;11:287–90. doi: 10.1016/s1056-8727(96)00076-1. [DOI] [PubMed] [Google Scholar]
- 2.Atroshi I, Gummesson C, Johnsson R, Ornstein E, Ranstam J, Rosén I. Prevalence of carpal tunnel syndrome in a general population. JAMA. 1999;282:153–58. doi: 10.1001/jama.282.2.153. [DOI] [PubMed] [Google Scholar]
- 3.Hubbard ZS, Law TY, Rosas S, Jernigan SC, Chim H. Economic benefit of carpal tunnel release in the Medicare patient population. Neurosurg Focus. 2018;44:E16. doi: 10.3171/2018.1.FOCUS17802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kumar P, Chakrabarti I. Idiopathic carpal tunnel syndrome and trigger finger: is there an association? J Hand Surg Eur Vol. 2009;34:58–59. doi: 10.1177/1753193408096015. [DOI] [PubMed] [Google Scholar]
- 5.Rottgers SA, Lewis D, Wollstein RA. Concomitant presentation of carpal tunnel syndrome and trigger finger. J Brachial Plex Peripher Nerve Inj. 2009;4:13. doi: 10.1186/1749-7221-4-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Garti A, Velan GJ, Moshe W, Hendel D. Increased median nerve latency at the carpal tunnel of patients with “trigger finger”: comparison of 62 patients and 13 controls. Acta Orthop Scand. 2001;72:279–81. doi: 10.1080/00016470152846628. [DOI] [PubMed] [Google Scholar]
- 7.Zhang D, Collins J, Earp BE, Blazar P. Relationship of carpal tunnel release and new onset trigger finger. J Hand Surg Am. 2019;44:28–34. doi: 10.1016/j.jhsa.2018.10.003. [DOI] [PubMed] [Google Scholar]
- 8.Ettema AM, Amadio PC, Zhao C, Wold LE, An K-N. A histological and immunohistochemical study of the subsynovial connective tissue in idiopathic carpal tunnel syndrome. J Bone Joint Surg Am. 2004;86:1458–66. doi: 10.2106/00004623-200407000-00014. [DOI] [PubMed] [Google Scholar]
- 9.Moore JS. Flexor tendon entrapment of the digits (trigger finger and trigger thumb) J Occup Environ Med. 2000;42:526–45. doi: 10.1097/00043764-200005000-00012. [DOI] [PubMed] [Google Scholar]
- 10.Wiberg A, Ng M, Schmid AB, et al. A genome-wide association analysis identifies 16 novel susceptibility loci for carpal tunnel syndrome. Nat Commun. 2019;10:1030. doi: 10.1038/s41467-019-08993-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sood RF, Westenberg RF, Winograd JM, Eberlin KR, Chen NC. Genetic risk of trigger finger: results of a genomewide association study. Plast Reconstr Surg. 2020;146:165e–76e. doi: 10.1097/PRS.0000000000006982. [DOI] [PubMed] [Google Scholar]
- 12.Patrick W, Gifford A, Meng X, et al. Mapping ICD-10 and ICD-10-CM codes to phecodes: workflow development and initial evaluation. JMIR Med Inform. 2019;7:e14325. doi: 10.2196/14325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bastarache L, Denny JC, Roden DM. Phenome-Wide Association Studies. JAMA. 2022;327:75–76. doi: 10.1001/jama.2021.20356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mbatchou J, Barnard L, Backman J, et al. Computationally efficient whole-genome regression for quantitative and binary traits. Nat Genet. 2021;53:1097–103. doi: 10.1038/s41588-021-00870-7. [DOI] [PubMed] [Google Scholar]
- 15.Ghoussaini M, Mountjoy E, Carmona M, et al. Open Targets Genetics: systematic identification of trait-associated genes using large-scale genetics and functional genomics. Nucleic Acids Res. 2021;49:D1311–20. doi: 10.1093/nar/gkaa840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yang J, Ferreira T, Morris AP, et al. Conditional and joint multiple-SNP analysis of GWAS summary statistics identifies additional variants influencing complex traits. Nat Genet. 2012;44:369–75.:S1–3. doi: 10.1038/ng.2213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wallace C. A more accurate method for colocalisation analysis allowing for multiple causal variants. PLoS Genet. 2021;17:e1009440. doi: 10.1371/journal.pgen.1009440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.FinnGen. Introduction. [accessed June 29, 2022]. https://finngen.gitbook.io/documentation/v/r4/
- 19.Kurki MI, Karjalainen J, Palta P, et al. FinnGen: unique genetic insights from combining isolated population and national health register data. medRxiv. 2022 doi: 10.1101/2022.03.03.22271360. published online March 6. (preprint) [DOI] [Google Scholar]
- 20.Willer CJ, Li Y, Abecasis GR. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics. 2010;26:2190–91. doi: 10.1093/bioinformatics/btq340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dobin A, Davis CA, Schlesinger F, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Anders S, Pyl PT, Huber W. HTSeq--a Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31:166–69. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Aguet F, Brown AA, Castel SE, et al. Genetic effects on gene expression across human tissues. Nature. 2017;550:204–13. doi: 10.1038/nature24277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Neavin D, Nguyen Q, Daniszewski MS, et al. Single cell eQTL analysis identifies cell type-specific genetic control of gene expression in fibroblasts and reprogrammed induced pluripotent stem cells. Genome Biol. 2021;22:76. doi: 10.1186/s13059-021-02293-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shabalin AA. Matrix eQTL: ultra fast eQTL analysis via large matrix operations. Bioinformatics. 2012;28:1353–58. doi: 10.1093/bioinformatics/bts163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McLaren W, Gil L, Hunt SE, et al. The ensembl variant effect predictor. Genome Biol. 2016;17:122. doi: 10.1186/s13059-016-0974-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Coe EA, Tan JY, Shapiro M, et al. The MITF-SOX10 regulated long non-coding RNA DIRC3 is a melanoma tumour suppressor. PLoS Genet. 2019;15:e1008501. doi: 10.1371/journal.pgen.1008501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Suhre K, Arnold M, Bhagwat AM, et al. Connecting genetic risk to disease end points through the human blood plasma proteome. Nat Commun. 2017;8:14357. doi: 10.1038/ncomms14357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cohn L, Feller AG, Draper MW, Rudman IW, Rudman D. Carpal tunnel syndrome and gynaecomastia during growth hormone treatment of elderly men with low circulating IGF-I concentrations. Clin Endocrinol (Oxf) 1993;39:417–25. doi: 10.1111/j.1365-2265.1993.tb02388.x. [DOI] [PubMed] [Google Scholar]
- 30.Mukherjee A, Rotwein P. Insulin-like growth factor-binding protein-5 inhibits osteoblast differentiation and skeletal growth by blocking insulin-like growth factor actions. Mol Endocrinol. 2008;22:1238–50. doi: 10.1210/me.2008-0001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ning Y, Hoang B, Schuller AG, et al. Delayed mammary gland involution in mice with mutation of the insulin-like growth factor binding protein 5 gene. Endocrinology. 2007;148:2138–47. doi: 10.1210/en.2006-0041. [DOI] [PubMed] [Google Scholar]
- 32.Papier K, Knuppel A, Perez-Cornago A, et al. Circulating insulin-like growth factor-I and risk of 25 common conditions: outcome-wide analyses in the UK Biobank Study. Eur J Epidemiol. 2022;37:25–34. doi: 10.1007/s10654-021-00811-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pilewski JM, Liu L, Henry AC, Knauer AV, Feghali-Bostwick CA. Insulin-like growth factor binding proteins 3 and 5 are overexpressed in idiopathic pulmonary fibrosis and contribute to extracellular matrix deposition. Am J Pathol. 2005;166:399–407. doi: 10.1016/S0002-9440(10)62263-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nguyen X-X, Muhammad L, Nietert PJ, Feghali-Bostwick C. IGFBP-5 promotes fibrosis via increasing its own expression and that of other pro-fibrotic mediators. Front Endocrinol (Lausanne) 2018;9:601. doi: 10.3389/fendo.2018.00601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Feghali CA, Wright TM. Identification of multiple, differentially expressed messenger RNAs in dermal fibroblasts from patients with systemic sclerosis. Arthritis Rheum. 1999;42:1451–57. doi: 10.1002/1529-0131(199907)42:7<1451::AID-ANR19>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- 36.Schneider MR, Zhou R, Hoeflich A, et al. Insulin-like growth factor-binding protein-5 inhibits growth and induces differentiation of mouse osteosarcoma cells. Biochem Biophys Res Commun. 2001;288:435–42. doi: 10.1006/bbrc.2001.5785. [DOI] [PubMed] [Google Scholar]
- 37.Vouzouneraki K, Esposito D, Mukka S, et al. Carpal tunnel syndrome in acromegaly: a nationwide study. Eur J Endocrinol. 2021;184:209–16. doi: 10.1530/EJE-20-0530. [DOI] [PubMed] [Google Scholar]
- 38.Tagliafico A, Resmini E, van Holsbeeck MT, Derchi LE, Ferone D, Martinoli C. Sonographic depiction of trigger fingers in acromegaly. J Ultrasound Med. 2009;28:1441–46. doi: 10.7863/jum.2009.28.11.1441. [DOI] [PubMed] [Google Scholar]
- 39.Cutfield WS, Wilton P, Bennmarker H, et al. Incidence of diabetes mellitus and impaired glucose tolerance in children and adolescents receiving growth-hormone treatment. Lancet. 2000;355:610–13. doi: 10.1016/S0140-6736(99)04055-6. [DOI] [PubMed] [Google Scholar]
- 40.Ruan W, Fahlbusch F, Clemmons DR, et al. SOM230 inhibits insulin-like growth factor-I action in mammary gland development by pituitary independent mechanism: mediated through somatostatin subtype receptor 3? Mol Endocrinol. 2006;20:426–36. doi: 10.1210/me.2005-0283. [DOI] [PubMed] [Google Scholar]
- 41.Lloyd ME, Hart DJ, Nandra D, et al. Relation between insulin-like growth factor-I concentrations, osteoarthritis, bone density, and fractures in the general population: the Chingford study. Ann Rheum Dis. 1996;55:870–74. doi: 10.1136/ard.55.12.870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Suzuki S, Morimoto S, Fujishiro M, et al. Inhibition of the insulin-like growth factor system is a potential therapy for rheumatoid arthritis. Autoimmunity. 2015;48:251–58. doi: 10.3109/08916934.2014.976631. [DOI] [PubMed] [Google Scholar]
- 43.Zöllner S, Pritchard JK. Overcoming the winner’s curse: estimating penetrance parameters from case-control data. Am J Hum Genet. 2007;80:605–15. doi: 10.1086/512821. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
RNA sequencing data from the Pain in Neuropathy Study (PiNS) cohort has been reported previously and is available at accession GEO108023 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE108023). The relevant raw data (count matrix and genotype calls at rs62175241) and code that are necessary to replicate analyses in the expanded Oxford-CTS RNA sequencing cohort are provided on Github (https://github.com/samkleeman1/cts_tf/). A Jupyter Notebook summarising the genotype data quality control and genome-wide association analysis implemented in Regenie is provided on Github (https://github.com/samkleeman1/cts_tf/). Single-cell fibroblast expression quantitative trait (eQTL) summary statistics and applicable source data are published alongside the original manuscript.19 UK Biobank data can be requested through the application process detailed at https://www.ukbiobank.ac.uk/. Summary statistics for the primary trigger finger GWAS (patients with both International Classification of Diseases[ICD]-10 and Office of Population Censuses and Surveys Classification of Surgical Operations and Procedures[OPCS]-4 codes for trigger finger) have been uploaded to the GWAS Catalog (accession code GCST90104907). Summary statistics for the sensitivity analyses described in the manuscript are available from the corresponding author on request.