Abstract
The human microbiome influences the efficacy and safety of a wide variety of commonly prescribed drugs. Designing precision medicine approaches that incorporate microbial metabolism would require strain-and molecule-resolved, scalable computational modelling. Here, we extend our previous resource of genome-scale reconstructions of human gut microbes with a greatly expanded version. AGORA2 accounts for 7,302 strains, includes microbial drug degradation and biotransformation, and was extensively curated based on comparative genomics and literature searches. It performs very well against three independently assembled experimental data with an accuracy of 0.72 to 0.84, and predicts known microbial drug transformations with an accuracy of 0.81. We demonstrate that AGORA2 enables personalised, strain-resolved modelling by predicting the drug conversion potential of the gut microbiomes from 616 colorectal cancer patients and controls, which greatly varied between individuals and correlated with age, sex, BMI, and disease stages. AGORA2 serves as a knowledge base for the human microbiome and paves the way to personalised, predictive analysis of host-microbiome metabolic interactions.
Introduction
Trillions of microbes inhabit the human gastrointestinal tract, with a high interindividual variation depending on factors such as sex, age, ethnicity, lifestyle, and health status1. The gut microbiota synthesises bioactive metabolites, such as short-chain fatty acids, hormones, and neurotransmitters2, and participates in the metabolism of commonly prescribed drugs3, resulting in drug inactivation, activation, detoxification, or re-toxification4. Human gut microbes have been shown to metabolise 176 of 271 tested drugs5, with activity varying between individuals6. Consequently, precision medicine interventions that take diet, genetics, and the microbiome into account have been proposed7. Predicting such personalised treatments would require detailed knowledge of the distribution of drug transformation reactions across human microbial taxa as well as the stoichiometry of such transformations.
A mechanistic systems biology approach that includes a detailed stoichiometric representation of metabolism is constraint-based reconstruction and analysis (COBRA)8. COBRA relies on genome-scale reconstructions of the target organism that are often manually curated based on the available literature8. These reconstructions can be converted into predictive computational models through the application of condition-specific constraints9, including (meta-) omics and nutritional data, and linked together to interrogate strain-resolved, personalised microbiome models10, 11. Hence, the COBRA approach is well-suited for the exploration of metabolic human-microbiome co-metabolism12, 13. To facilitate the genome-scale reconstruction of the thousands of known species inhabiting humans14, semi-automated reconstruction tools, such as CarveMe15, MetaGEM16, MIGRENE17, and gapseq18, have been published. Despite their many advantages, these tools provide limited support for curation against manually refined genome annotations and experimental data from peer-reviewed literature. Both are crucial for the inclusion of not yet routinely annotated (e.g., drug metabolism) and/or species-specific pathways9. To overcome these limitations, we have developed a semi-automated curation pipeline guided by manually assembled comparative genomic analyses and experimental data19, which previously enabled the generation of AGORA, a resource of 773 genome-scale reconstructions of human gut microbe strains, representing 605 species and 14 phyla20.
Here, we present an expansion in scope and coverage of AGORA, called AGORA2, consisting of microbial reconstructions for 7,302 strains, 1,738 species, and 25 phyla. AGORA2 summarises the knowledge and experimental data obtained through manual comparative genomics analyses and literature and textbook reviews, and demonstrates high accuracy against three independently collected experimental datasets. AGORA2 has been expanded by manually formulated molecule- and strain-resolved drug biotransformation and degradation reactions covering over 5,000 strains, 98 drugs, and 15 enzymes, some of which were validated against independent experimental data. The AGORA2 reconstructions are fully compatible with the generic21 and the organ-resolved, sex-specific, whole-body human metabolic reconstructions22. We demonstrate the use of AGORA2 for the prediction of personalised gut microbial drug metabolism for a cohort of 616 individuals. Taken together, the AGORA2 reconstructions can be used independently or together for investigating microbial metabolism and host-microbiota co-metabolism in silico.
Results
Data-driven reconstruction of diverse human microbes
To build the reconstructions of the 7,302 gut microbial strains in the AGORA2 compendium (Table S1), we substantially revised and expanded (Methods) a previously developed20 data-driven reconstruction refinement pipeline, deemed DEMETER19. Overall, the DEMETER workflow consists of data collection, data integration, draft reconstruction generation, translation of reactions and metabolites into the Virtual Metabolic Human (VMH)23 name space, and simultaneous iterative refinement, gap-filling and debugging19. Reconstruction refinement follows standard operating procedures for generating high-quality reconstructions9 and is continuously verified through a test suite19 (Table S2, Supplemental Note 2).
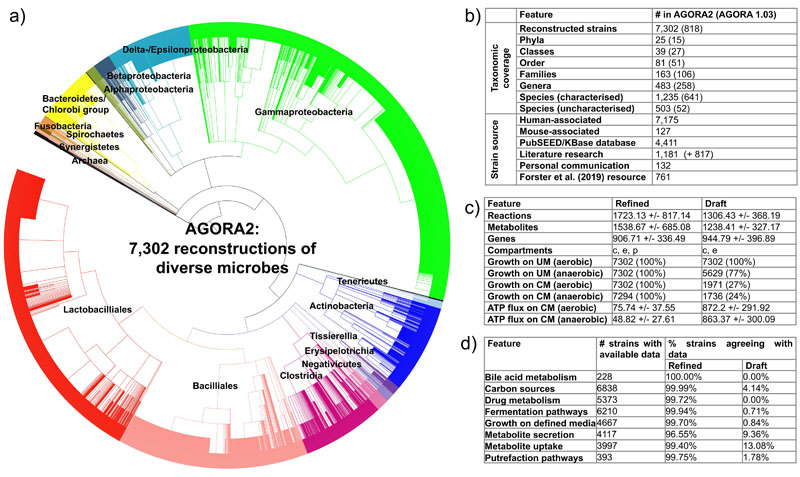
After expanding the taxonomic coverage (Figure 1a-b, Table S1, Supplemental Note 1) and retrieving the corresponding genome sequences, we generated automated draft reconstructions through the online platform KBase24, which were subsequently refined and expanded through the DEMETER pipeline19 (Methods). As a lack of accurate genome annotations is a source of uncertainty in the predictive potential of genome-scale reconstructions25, we manually validated and improved the annotations of 446 gene functions across 35 metabolic subsystems for 5,438/7,302 (74%) genomes using PubSEED26 (Table S3a-d). To further ensure accurate representation of species-specific metabolic capabilities, we performed an extensive, manual literature search spanning 732 peer-reviewed papers and two microbial reference textbooks yielding information for 6,971/7,302 strains (95%) (Methods). For the remaining 331 strains, either no experimental data was available or all biochemical tests reported in the literature were negative. The performed extensive refinement driven by the collected data resulted in average in the addition and removal of 685.72 (standard deviation (±): 620.83) and 147.79 (±93.18) reactions, respectively, per reconstruction (Figure S1). The biomass reactions provided in the draft reconstructions were curated, and reactions were placed in a periplasm compartment where appropriate (Supplemental Note 3). Moreover, we retrieved the metabolic structures for 1,838/3,613 (51%) metabolites and provide atom-atom mapping for 5,583 of the overall 8,637 (65%) enzymatic and transport reactions captured across AGORA2 (Methods). Owing to these extensive curation efforts, the metabolic models derived from the refined reconstructions showed a clear improvement in their predictive potential over models derived from the KBase draft reconstructions (Figure 1c, d, Supplemental Note 2). As an additional assessment of reconstruction quality, we generated an unbiased quality control report for all reconstructions (Methods) resulting in an average score of 73%.
Figure 1. Features of AGORA2.
a) Taxonomic coverage and sources of reconstructed strains. b) Taxonomic distribution of the included 7,302 strains. c) Features of the AGORA2 reconstructions and KBase draft reconstructions. c = cytosol, e = extracellular space, p= periplasm. Growth rates on Western diet (WD) and unlimited medium (UM) (Methods) are given in 1/hr. ATP production potential on WD is given in mmol/gdry weight/hr. Shown are averages across all models +/- standard deviations. d) Number of reconstructions with available positive findings from comparative genomics and literature, and percentage of curated and draft reconstructions agreeing with the findings for the respective organism. N/A = not applicable as the pathway was absent in draft reconstructions.
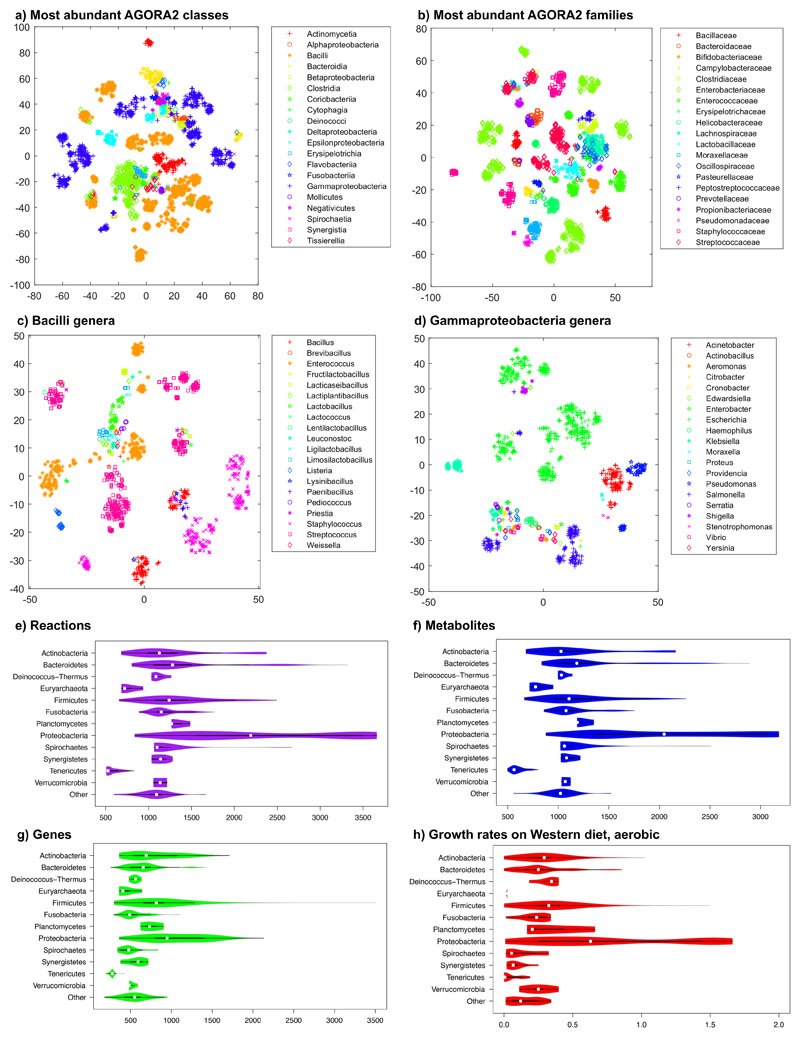
We then clustered the content of the AGORA2 reconstructions by taxonomic distribution. Overall, AGORA2 reflects the diversity of the captured strains as they clustered by class and family according to their reaction coverage (Figures 2a-b, S3a, Supplemental Note 4). Several genera in the Bacilli and Gammaproteobacteria classes formed subgroups illustrating important metabolic differences between them (Figures 2c-d, S2a-b, Supplemental Note 4, Kruskal-Wallis test: p=0.0001). Cross-phylum metabolic differences also translated to differences in reconstruction sizes and predicted growth rates (Figure 2e-h) and in their potential to consume and secrete metabolites (Figure S3a-b). Taken together, the models derived from AGORA2 capture taxon-specific metabolic traits of the reconstructed microbes.
Figure 2. Taxonomically related strains are similar in their AGORA2 reconstruction content.
a-d) Clustering through t-distributed stochastic neighbour embedding (t-SNE)52 of reaction presence across all pathways per reconstruction. Coordinates were statistically different across taxonomic units (Kruskal-Wallis test, p=0.0001 in all cases). a) Members of the largest classes. b) Members of the largest families. c) Members of the Bacilli class by genus. d) Members of the Gammaproteobacteria class by genus. e-h) Features of all AGORA2 reconstructions across phyla: e) Number of reactions. f) Number of metabolites. g) Number of genes, and h) growth rate in 1/hr on aerobic Western Diet.
AGORA2 is predictive against three independent datasets
While automated draft reconstructions can be rapidly generated, they still require subsequent curation efforts to be predictive27. Several (semi-)automated reconstruction tools bridge the gap between automated draft and fully manually curated reconstructions including CarveMe15, gapseq18, and MIGRENE17. To further access the quality of AGORA2 and the DEMETER pipeline, we compared AGORA2’s predictive potential and model properties with other resources of microbial genome-scale reconstructions. For this purpose, we retrieved 8,075 reconstructions built through gapseq18, 1,333 reconstructions built through MIGRENE, deemed MAGMA17, as well as 72 manually curated genome-scale reconstructions deposited in the BiGG database28. Additionally, we built CarveMe15 reconstructions for 7,279 AGORA2 strains and gapseq18 reconstructions for a subset of 1,767 AGORA2 strains (Methods).
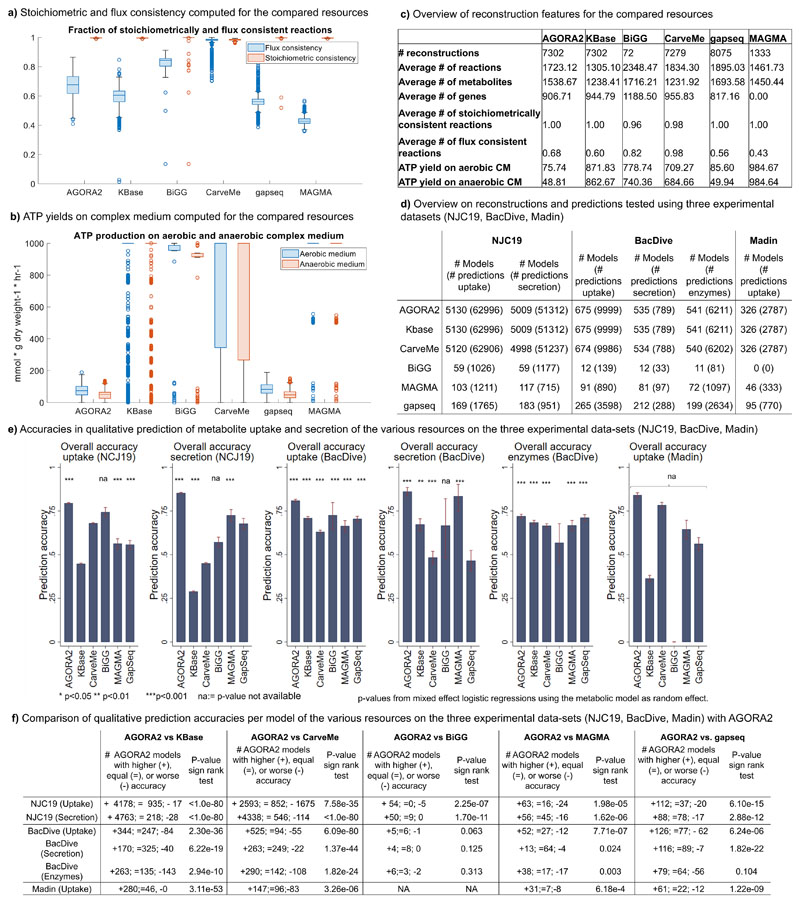
For an unbiased assessment of reconstruction quality, we first determined the fraction of flux consistent reactions29 in each resource. Only the manually curated reconstructions from BiGG and reconstructions built through CarveMe had a higher fraction of flux consistent reactions than AGORA2 (Figure 3a-b, p<1e-30, Wilcoxon rank-sum test). Note that our reconstructions represent knowledge bases, thus, if genetic or biochemical evidence exists for a gene or reaction, it will be included in the reconstruction. In contrast, CarveMe by design removes all flux inconsistent reactions from a metabolic reconstruction15. Compared with the KBase draft reconstructions, AGORA2 had a significantly higher percentage of flux consistent reactions despite being larger in metabolic content, as well as a significantly higher flux consistency than gapseq and MAGMA (Figure 3a,c, p<1e-30, Wilcoxon rank-sum test). It was also observed that all resources except AGORA2 and gapseq produced very high amounts of ATP (up to 1000 mmol/gdry weight/hr) on the complex medium for at least a subset of models (Figure 3b,c). Hence, in these models, the ATP production flux was only limited by the upper bounds on reactions, which generally indicates the existence of futile cycles9.
Figure 3. Comparison of AGORA2 refined reconstructions, draft reconstructions, and three other reconstructions resources.
Compared were the 7,302 AGORA2 and KBase draft reconstructions, 72 manually curated reconstructions from the BiGG database28, 5,587 reconstructions built through CarveMe15, 8,075 reconstructions built through gapseq18, and 1,333 MAGMA reconstructions17. a) Fraction of reactions that are stoichiometrically and flux consistent as defined in29 for each model derived from the five compared resources. Exchange and demand reactions, which are stoichiometrically inconsistent by definition, were excluded. b) Aerobic and anaerobic ATP production on complex medium (mmol/gdry weight/hr) by each model derived from the five compared resources. c) Overview of reconstruction properties for the compared resources. d) Overview on number of models and number of predictions tested in validating AGORA2, KBase, BiGG, CarveMe, gapseq, and MAGMA against three independent experimental datasets30, 32, 33. e) Bar plots with 95%-confidence intervals of overall accuracies of the five resources in predicting uptake and secretion in the three experimental datasets. Significance of prediction accuracy was determined by mixed effect logistic regressions using the metabolic model as random effect variable to account for the statistical dependence of predictions stemming from the same model. NA indicates a missing p-value due to empty categories (e.g., no true negatives detected). f) Comparison of accuracies per model of the various resources on the three experimental datasets. P-values were derived by sign rank tests.
The most crucial aspect of a genome-scale reconstruction is its accuracy in capturing known biochemical or physiological traits of the target organism9, i.e., its potential to make biologically plausible predictions. Hence, we set out to determine the predictive potential of AGORA2. For an unbiased assessment, we retrieved organism-specific experimental data from three separate sources (Methods). First, we retrieved species-level positive and negative metabolite uptake and secretion data for 455 species (5,319 strains) in AGORA2 from the NJC19 resource30. Note that a precursor of NJC19, NJS1631, containing only positive data, had been used to refine AGORA2. Next, we mapped species-level positive metabolite uptake data, retrieved from Madin et al32, for 185 species (328 strains) in AGORA2 (“Madin” data). Finally, we retrieved strain-resolved positive and negative metabolite uptake and secretion data for 676 AGORA2 strains as well as positive and negative enzyme activity data for 881 AGORA2 strains from the BacDive database33. Neither the Madin dataset nor BacDive had been used during the refinement of AGORA2. For metabolite uptake and secretion, the AGORA2 reconstructions captured the known capabilities of the target organisms very well (overall accuracy against NJC19, BacDive, and Madin of 0.82, 0.81, and 0.84, respectively, Figure 3e, Table S4d). For enzyme activity, a slightly lower accuracy of 0.72 was achieved (Figure 3e, Table S4d). AGORA2 had a lower specificity than the other resources on NJC19. However, the majority of observed false positives in AGORA2 concerned glutamate uptake in Escherichia coli (Table S4c), which was a negative finding in the NJC19 dataset based on a report for a single E. coli strain.
We then compared the predictive potential of AGORA2 with the other four resources where possible. Of the 7,302 reconstructed AGORA2 strains, 7,279 had been reconstructed through CarveMe, 451 overlapped with reconstructions built through gapseq, and 60 overlapped with reconstructed strains available at the BiGG database (Table S4a). No strains overlapped with MAGMA as it consists of pan-species reconstructions built from metagenome-assembled genomes17 but 216 reconstructions could be mapped at the species level (Table S4a). For the four resources and for each dataset, we then computed the predictive potential for the organisms overlapping with AGORA2 (Figure 3d-f, Table S4b-d). While MAGMA and AGORA2 achieved significant prediction accuracies for secretion and uptake on the NJC19 and the BacDive datasets, KBase failed to perform better than chance for metabolite uptake and secretion in NJC19, and CarveMe failed to predict significantly secretion in the NJC19 dataset (Figure 3e, Table S4d). The gaqseq reconstructions built in the present study for the subset of AGORA2 strains performed comparably to the set of gapseq reconstructions that had been published by the authors (Table S4b).
To compare the performance of AGORA2 with KBase, CarveMe, gapseq, BiGG, and MAGMA directly, we calculated the accuracy per model separately for uptake and secretion. We then compared the accuracies on models in the overlap of AGORA2 and each resource via a non-parametric sign-rank test. AGORA2 was significantly better than all other methods on all three datasets, except for BiGG on the BacDive data, where the overlap in models was too small to achieve sufficient statistical power, and gapseq on the BacDive enzyme data where it performed comparably to AGORA 2 (71% versus 72%, Figure 3e, f).
Taken together, the AGORA2 reconstructions capture the known traits of the respective organisms very well, surpassing other semi-automatedly generated reconstructions and being comparable to manually curated reconstructions. These results demonstrate the value of the extensive curation efforts refinement, guided by species-species experimental data, performed during the development of AGORA2 as outlined above. Accordingly, AGORA2 performed particularly well for metabolite uptake and secretion data, which requires curation based on experimental data, compared with enzyme activity data, which can be curated based on genome annotations. Remaining false positive and false negative predictions (Table S4c) will be addressed in future efforts following the iterative curation philosophy9. Flux inconsistent reactions, indicating they contain dead-end metabolites29, may serve as the starting point for gap-filling efforts thereby enabling biological discovery34.
Microbial drug metabolism guided by genome and bibliome
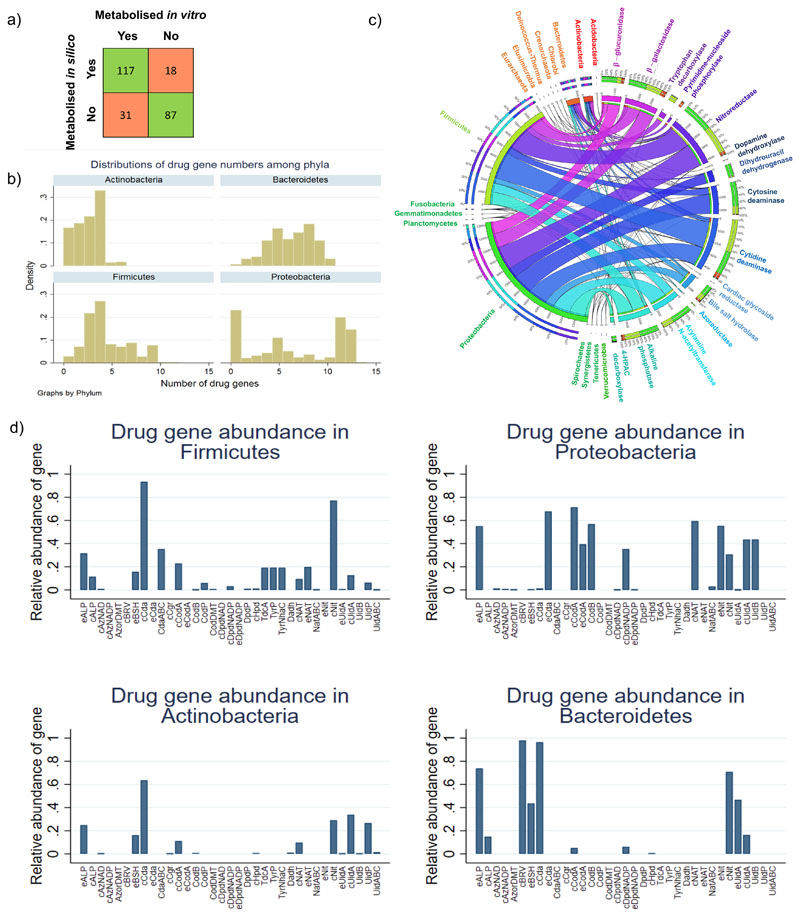
Microbes can directly or indirectly influence drug activity and toxicity through degradation (e.g., hydrolysis) and biotransformation (e.g., reduction)3, 4 is only captured to a limited extent by genome annotation pipelines and no systematic comparative genomic analysis of drug-metabolising enzymes has previously been performed. Hence, microbial drug transformations are not yet captured by any existing genome-scale reconstruction resources. To fill this gap, we performed an extensive, manual comparative genomic analysis for 25 drug genes, encoding for 15 enzymes shown to directly or indirectly affect drug metabolism (Table S5a), their subcellular locations, and 12 genes encoding for drug transporters (Table S3b). All 5,438 analysed strains carried at least one drug-metabolising enzyme (Table S3c). As these enzymes are also involved in central metabolism, e.g., nucleoside metabolism, this high coverage was expected. We then carried out a thorough literature and database review of metabolite structures, formulas, and charges for 98 frequently prescribed drugs belonging to ten drug groups and 32 subgroups (Table 5b). We formulated 1,440 drug-related reactions containing 363 metabolites (Table S6a-b) and added, on average, 188 drug-related reactions and 111 metabolites to the reconstructions depending on the genomic evidence. We validated, with an accuracy of 0.81 (sensitivity: 0.87, specificity: 0.74, Fisher’s exact test: p=2.01e-23, mixed effect logistic regression accounting for stochastic dependencies from predictions stemming from the same model: p=1.209e-07), the drug-metabolising predictions against independent published experimental data for 253 drug-microbe pairs (Table S7, Figure 4a). The 18 false positive predictions may indicate non-functional genes or regulatory mechanisms, whereas the 31 false negative predictions could be due to incompleteness of genomes or non-orthologous displacement in complete genomes, or a currently unaccounted for homolog encoding the reaction.
Figure 4. Overview of reconstructed drugs and annotated drug enzymes present in AGORA2.
a) Overlap between independent, experimentally demonstrated activity of drug-metabolising enzymes and predictions by models derived from the AGORA2 reconstructions for 253 drug-microbe pairs (Table S7). b) Distribution of the number of strains carrying each drug enzyme over the 14 analysed phyla. c) Fraction of strains carrying each gene encoding drug enzymes or transport proteins in the four main phyla in the human microbiome. d) Distribution of the number of drug genes per strain for the four main phyla. For the list of abbreviations, see Table S3b.
Taxonomic distribution of drug-metabolising capabilities
We next analysed the taxonomic distribution of the annotated drug and transport genes (Figure 4b-d, Table S3c). At least one strain in each of the 14 analysed phyla encoded for genes involved in drug metabolism (Figure 4f). The most widespread drug-metabolising enzymes were cytidine deaminase and nitroreductase, which were found in 12 and 13 phyla, respectively (Figure S7a-b). Another central metabolic enzyme, the pyrimidine-nucleoside phosphorylase, was also widely distributed, but the monophyletic branch specific for the metabolism of brivudine and sorivudine35 was only found in the Bacteroidetes phylum (Figures 4c-d, S7c). Many drugs are detoxified by the liver through the addition of glucuronic acid, a modification that is reversed by microbial β-glucuronidase4. This enzyme was in >99% of analysed Escherichia coli strains and was also widely distributed across Bacteroidetes and Firmicutes strains (Figures 4c-d, S7d), consistent with previous analyses36. E. coli was the species most enriched in drug metabolism with >99% of all analysed strains carrying seven to ten drug enzymes (Table S3c). Taken together, drug-metabolising enzymes, and transporters, are widely distributed but important phyla-specific and strain specific differences exist. To elucidate the potential benefits that these drug-metabolising capabilities could confer to the microbes, we computed the strain-specific energy, carbon, and nitrogen yields of drug degradation. This analysis revealed that many strains spread across phyla were capable of using drugs as a source of energy, carbon, and/or nitrogen (Figure S4, Table S8).
Personalised modelling of drug-metabolising capacities
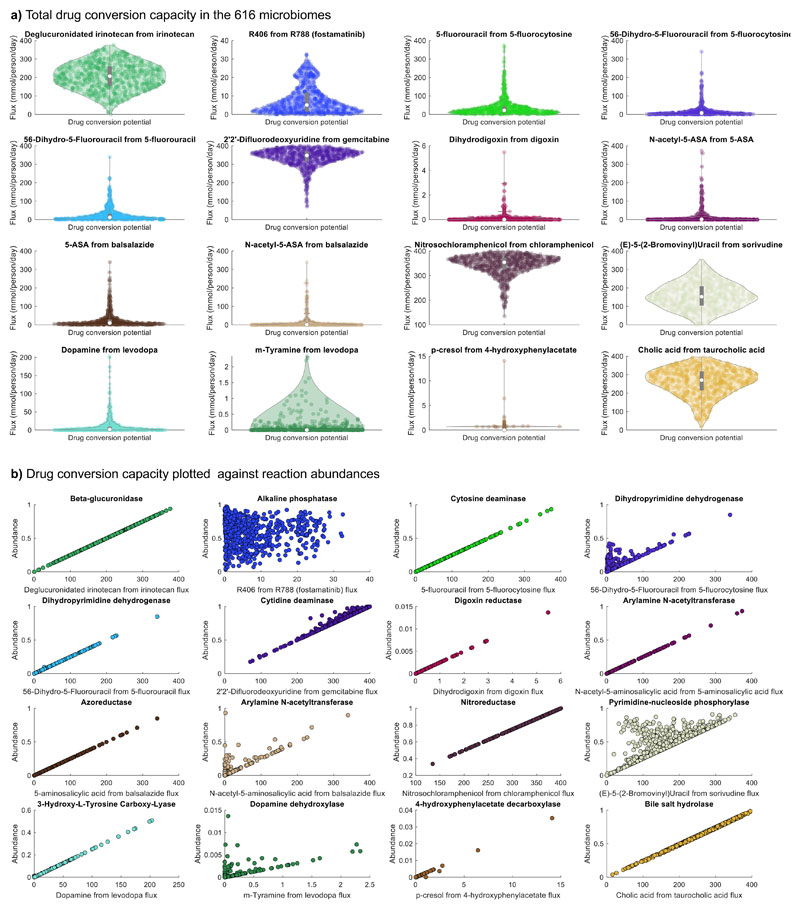
As human microbes do not exist in isolation, we addressed the important question on how the total drug-metabolising capacities may differ between individual gut microbiomes. A previously developed community modelling framework10 allows for the scalable, tractable computation of community-wide metabolic capabilities as well as organism-resolved contributions to faecal metabolite levels37. We used a metagenomic data set from a Japanese cohort of 365 colorectal cancer (CRC) patients and 251 healthy controls38 that had previously allowed us to interrogate the metabolic capabilities of each gut microbiome and validate the fluxes against metabolomic data37. A total of 97% of the named species could be mapped onto the AGORA2 (compared to 72% for AGORA). For each individual’s gut microbiome, we built and interrogated a community model (Methods), resulting in the prediction of total drug-metabolising potential (Figure 5a, Table S9). For some enzymes, e.g., dihydropyrimidine dehydrogenase and dopamine dehydroxylase, the drug conversion potential only showed limited correlation with the total abundance of the corresponding drug-metabolising reactions indicating flux-limiting metabolic bottlenecks (Figure 5b). Analysing such bottlenecks would require the simulation of enzymatic functions in their metabolic context. Shadow price analysis (Methods) revealed that, in two-step reactions, such as levodopa degradation to m-tyramine, the drug conversion potential for the second step was limited by the species abundance carrying out the first step (Supplemental Note 5, Figure S6, Table S10). Levodopa degradation is known to be a two-step pathway carried out by different species39 (Figure S6).
Figure 5. Drug conversion capacity of 616 microbiomes.
a) Drug conversion potential in the microbiomes of 616 Japanese colorectal cancer patients and controls on the Average Japanese diet. The violin plots show the distribution of drug metabolite flux in mmol/person/day. b) Drug conversion potential (mmol/person/day) plotted against the total relative abundance of the reaction producing the shown drug metabolite in the 616 microbiomes. See Table S5a for a description of each drug-metabolising enzyme.
While most drugs could be qualitatively metabolised in silico by at least 95% of the microbiomes, only 53% of the microbiomes presented the capacity to metabolise digoxin, and levodopa could be metabolised by 86% of the investigated microbiomes into dopamine and by 46% into m-tyramine (Figure 5a). Both digoxin transformation and the second step of levodopa degradation strictly depended on the presence of Eggerthella lenta (Figure S8), and are known to reduce bioavailability of the drugs4, 39. Moreover, while all but three microbiomes could activate the anti-inflammatory bowel disease prodrug Balsalazide through the azoreductase activity, the highest secretion flux of the active form of Balsalazide (5-aminosalicylic acid) achieved by any microbiome was 339.81 mmol/day/person, while the average was 25.47 +/ 40.84 mmol/day/person (Figure 5a). This variation may be of high clinical relevance, as it indicates that not all microbiomes can equally activate Balsalazide. As a sensitivity analysis, we recomputed drug-metabolising capacities using an average European diet instead of the Japanese diet and found that the drug-metabolising capacities were virtually unaltered for all drugs and, hence, highly robust towards diet constraints (Figure S7).
Microbiome-level fluxes are sensitive to clinical parameters
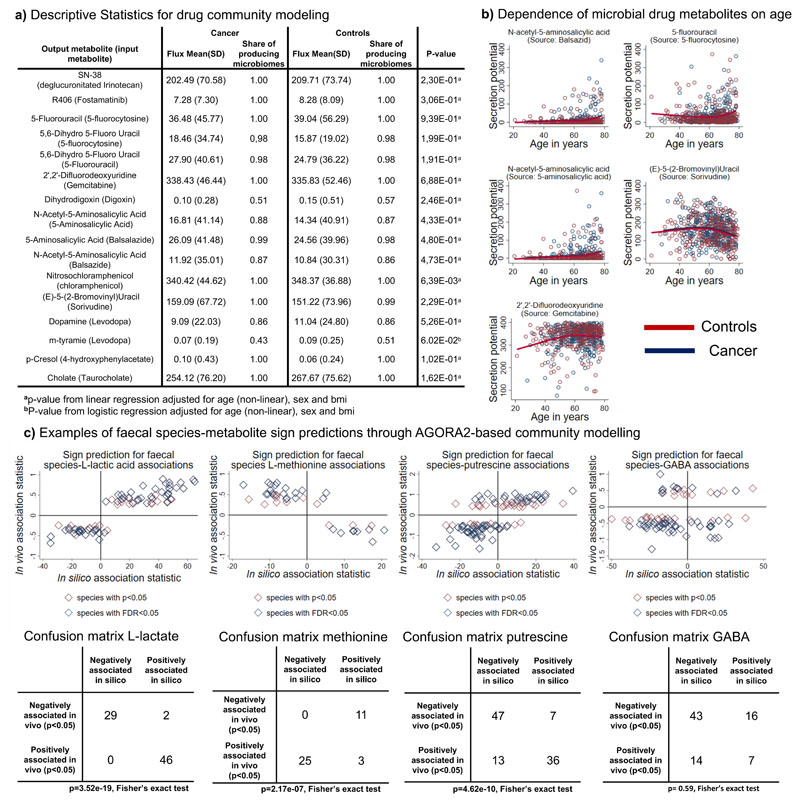
Next, we investigated whether drug-metabolising capacities were associated with CRC. For neither of the drugs, including cancer drugs, neither qualitative nor quantitative differences in drug-metabolising capacities were found after correction for multiple testing, despite the reported enrichment in 29 species in CRC metagenomes40. On a nominal level (p<0.05), nitrosochloramphenicol was increased in cancer cases (Figure 6a). Nonetheless, drastic individual differences in drug-metabolising potential, regardless of disease status, due to distinct microbiota composition existed (Figure 5a).
Figure 6. Descriptive statistics for the modelled drug metabolites and faecal species-metabolite associations.
a) Overview on descriptive statistics for the modelled drug metabolites. b) Scatter plots (red: controls; blue cancer) of various drug metabolites in dependence on age with non-linear regression lines for cases and controls. Regression lines were estimated with restricted cubic splines. All regression models had p<0.0001 (FDR<0.05) and regression coefficients were virtually the same for cases and controls. c) Faecal species metabolite sign prediction for L-lactic acid, L-methionine, and gamma-aminobutyrate. Upper panel represents scatter plots of in silico change in microbial community net secretion flux derived from community modelling against the change in measured faecal concentration in dependence on microbial species presence. Each dot represents one microbial species having an effect on metabolite concentration with at least p<0.05. Lower panel depicts the confusion matrix of sign prediction through in silico modelling. P-values derived from Fisher’s exact test should be treated with care due to species-species and metabolite-metabolite interdependencies.
Lastly, we investigated the statistical association pattern of age, sex, and body mass index (BMI) to the drug-metabolising capacities of the microbiome (Figure 6b, Figure S9). Five predicted secretion potentials of drug metabolites were clearly associated with age (Figure 6b), although the effect sizes were small to medium (explained variances <8%). For example, the conversion of Sorivudine into a toxic byproduct showed a nonlinear association with age, where secretion capacities declined from 60 years on (Figure 6b, R2 = 0.047 p=7.17e-06). Women had significantly higher taurocholate metabolising capability, while slightly, but significantly lower conversion potential of the chemotherapy drug Gemcitabine (Figure S9a). In conclusion, our analysis enabled the investigation into clinical parameters that were associated with drug-metabolising capacities of the gut microbiome.
Community models predict species-metabolite associations
As a last step of validation, we tested whether AGORA2-based community modelling is capable of predicting the sign of statistical associations between microbial species presence and faecal metabolite concentrations in the CRC sample, following procedures established before41. We calculated the faecal net secretion rate for 52 AGORA metabolites (Methods), for which faecal metabolomics data from the same Japanese cohort was available38. As these metabolomics data were not used in constructing the AGORA2-based community models, this procedure represents an independent validation.
After correction for multiple testing, AGORA2-based community modelling was predictive for the sign of significant species-metabolite associations in 24 of 52 metabolites (Figure 6c, Table S11) with p<0.05 and 19 with false discovery rate (FDR)<0.05. Particularly well covered were amino acids, known fermentation products (e.g., L-lactate, butyrate) as well as amines (Table S11). Notably, for certain metabolites, e.g., methionine (Figure 6c), in vivo association statistics were consistently inverse to the corresponding in silico association statistics. These latter results may correspond to net uptake of the metabolites by the microbial community. The non-significant sign prediction, as exemplarily depicted in Figure 6c for gamma-aminobutyrate, can have multiple reasons, ranging from host factors dominating the variation in faecal concentration to incomplete community models or missing confounders in the statistical models, leading to false positive in vivo associations. In conclusion, AGORA2-based community models could predict the direction of species metabolite associations for a broad range of metabolites, highlighting their general predictive nature.
Discussion
Here, we introduced AGORA2, a resource of 7,302 genome-scale reconstructions for human-associated microbes with unprecedented coverage, scope, and curation effort. AGORA2 follows the quality standards developed by the systems biology research community9, 42, accurately captures biochemical and physiological traits of the target organisms, surpassing other reconstruction resources, and includes manually refined, strain-resolved drug-metabolising capabilities. It enables personalised modelling of human microbial metabolism through a dedicated computational pipeline10, which had recently been improved upon in terms of computational efficiency and implemented features43. Hence, personalised microbiome modelling using AGORA2 can be performed in a reasonable timeframe on a standard personal computer (Methods).
Computational modelling of microbial consortia is increasingly recognised as a complementary method to in vitro and in vivo experiments and can generate experimentally testable hypotheses13, 44. Our knowledge about gut microbes remains limited and thus, any in silico reconstruction will be inherently incomplete and require regular updates45. For instance, a recent study has found that 176 of 271 tested drugs could be metabolised by human bacteria, and for a subset of these drugs, transformations could be linked to specific gene functions5. Through future comparative genomics and metabolite and reaction formulation efforts, AGORA2 may be expanded by these drug transformations to further broaden its coverage of prescription drug metabolism. As AGORA2 uses the same metabolite and reaction nomenclature23 as the human metabolic reconstruction21 and the whole-body metabolic reconstructions22, it could be used to predict host-microbiome co-metabolism, up to their potential contribution to human organ-level metabolism22.
To date, AGORA has enabled nearly 50 studies that modelled microbe-microbe, host-microbe, and microbiome interactions46, and, together with available software tools10, 47, contributed substantially to recent advances in size and scope of constraint-based modelling of multispecies interactions46. However, AGORA was to an extent hampered by its limited taxonomic coverage, which mainly included the westernised gut microbiome20. In contrast, AGORA2 also captures microbes commonly found in skin, oral, and vaginal microbiomes, includes many uncharacterised taxa as well as those from non-Westernised microbiomes, and has a high overlap with species found in several resources of isolates and metagenome-assembled genomes of the human microbiome (Supplemental Note 1). Together, this extension increases the prediction fidelity of microbiome-level models included for non-gut and non-Westernised microbiomes.
We reported associations between CRC patient-specific microbial drug conversion capabilities and clinical parameters, such as age and BMI (Figure 6). The example of Balsalazide, an anti-inflammatory drug utilised in treating inflammatory bowel disease (IBD), showcases how AGORA2 could be used to inform clinical research, and potentially facilitate personalisation of treatment. Balsalazide has high numbers need to treat (NNT) metrics for inducing remission (NNT:10) and maintenance (NNT:6) in ulcerative colitits48, indicating that most patients do not profit from the drug. Consistently, the Balsalazide activation potential varied strongly in the investigated CRC cohort microbiomes (Figure 5a) indicating that not all individuals would profit equally from Balsalazide treatment. Consequently, we propose that AGORA2 in conjunction with metagenomics could predict the stratification of inflammatory bowel disease patients into Balsalazide responders and non-responders, which could then be validated in follow-up clinical trials. The finding that drug-metabolising capabilities were associated with age-groups, BMI, and sex (Figures 6b, S9) demonstrates that AGORA2 in conjunction with community modelling can be utilised in large epidemiological cohort studies to link predicted metabolic fluxes with clinical parameters, and thereby opening new research possibilities to understand the role of the microbiome in modifying health risk and contributing to adverse health outcomes. Finally, AGORA2-based community models were able to predict the direction of species-metabolite associations for a range of metabolites (Figure 6c), demonstrating utility in delivering valid in silico markers of the microbiome’s metabolic traits.
Taken together, we present a resource of genome-scale reconstructions, AGORA2, which accurately captures organism-specific capabilities and can be used to build predictive personalised microbiome models. AGORA2 and all tools and scripts used in this study are freely available to the research community. We expect that like its predecessor, AGORA2 will be of great interest to the microbiome and constraint-based modelling communities with an even broader range of potential applications46. As a unique feature, AGORA2 captures strain-resolved microbial drug metabolism. Predicting drug response to realistic drug concentrations will require hybrid modelling approaches, e.g., integrating constrained-based modelling with physiological-based pharmacokinetic modelling49, 50. Using a constrained-based model of organ-resolved whole-body metabolism integrated with models of the gut microbiome22, and using such hybrid modelling approaches, dietary supplements, probiotics, or microbiome-targeted interventions, which have been shown to attenuate side effects of drugs4, could be predicted and validated49. Hence, AGORA2 paves the way for an integrative, multi-scale modelling approach that may enable in silico clinical trials49, 51 and contribute to precision medicine.
Material and methods
Selection of newly reconstructed organisms and retrieval of whole-genome sequences
First, we retrieved 4,185 genomes of human gut-associated strains that were available on PubSEED53 (Supplemental Note 6). To expand the species coverage, we performed an extensive literature search of species isolated from or detected in the human microbiome with available whole-genome sequences (Table S1). This search led to the addition of further 1,324 strains, which included 127 genomes of mouse-associated strains. The corresponding whole-genome sequences were retrieved in FASTA format from the National Center for Biotechnology Information (NCBI) FTP site (ftp://ftp.ncbi.nlm.nih.gov/). Moreover, we included 26 genomes of Eggerthella lenta strains54 available at https://www.ncbi.nlm.nih.gov/bioproject/PRJNA412637. Finally, we retrieved 761 human microbial genomes from the Human Gastrointestinal Bacteria Culture Collection (HBC)55 in FASTQ format from https://www.ebi.ac.uk/ena/data/view/PRJEB23845 and https://www.ebi.ac.uk/ena/data/view/PRJEB10915. Together with AGORA1.03, which was obtained from the VMH23, these combined efforts resulted in 7,302 strains and 1,738 species included in AGORA2.
Manual refinement of metabolic pathways and gene annotations through comparative genomics
Of the 7,302 analysed strains, 5,438 bacterial strains and three archaeal strains were present in the PubSEED resource53, 56 (Supplemental Note 6) and could be re-annotated for their metabolic functions through comparative genomics. A total of 34 metabolic subsystems that had been reconstructed previously for a smaller subset of gut microbial strains20, 57–60, as well as a newly created drug metabolism subsystem, were considered for the analysis (Table S3a for a comprehensive list of subsystems). All subsystems are available at the PubSEED website.
Curation of subsystems
For annotation of the genes in each subsystem, the PubSEED platform was used53. Functional roles for each subsystem were annotated based on the (1) prescribed functional role for the protein, (2) sequence similarities of the protein to proteins with previously confirmed functional role, and (3) genomic context (Supplemental Note 7).
Metabolic pathways considerations for comparative genomics analysis
Absence of gene(s) for one or more enzymes in a pathway may result in blocked reactions in a metabolic reconstruction. To avoid this, we estimated the completeness of metabolic pathways during the genome annotation. For each potentially synthesised metabolite, all the biosynthetic pathways were collected in agreement with the KEGG PATHWAY resource61 and genes of the subsystem were attributed to corresponding steps of the metabolic pathways. Absence of the consequent reactions was determined as a gap. Only pathways with no more than two gaps with gap length of no more than one step (Supplemental Note 8) were further gap-filled and used for generation of reactions.
Sequence-based gap-filling
For the gapped pathways, the bidirectional best-hit (BBH) method62 was used: (1) The gene corresponding to the gap and present in the genome for the related organisms (belonging to the same species, genus, or family) was used as a query for a BLAST search in the genome with the gap. (2) Possible BBHs were defined as homologs for that alignment with the query protein having an e-value ≤ e-50 and protein identity ≥ 50%. (3) For each possible BBH, the reverse search was done for the genome that was a source of the query protein. (4) If the query protein and its best homolog in the analysed genome formed BBH pair, the gap was filled. (5) A similar genomic context for the query protein and its ortholog was considered as an additional confirmation for orthology of the identified BBH pair.
Annotation of the drug metabolic genes
To annotate drug-metabolising genes, we used the following pipeline. (1) Identify genes known to encode for drug-metabolising enzymes in a range of microbial organisms, from the scientific literature (Table S5a). (2) Using the amino acid sequences of these known drug-metabolising genes as queries, we performed a BLAST search for every analysed genome. (3) The resulting best BLAST hit was then used as a query for the BLAST search in the genome having known drug-metabolising gene to confirm that the known protein sequence and its best BLAST hit form a pair of best bidirectional hits (BBHs). (4) All BBHs were used for the construction of a rooted maximal-likelihood tree. (5) All previously known proteins were mapped onto the tree, and all monophyletic branches containing known drug-metabolising enzymes were determined (Figure S10). (6) All annotated proteins in these branches were considered as orthologs of the known drug-metabolising proteins. All the proteins not being in branches with known drug-metabolising proteins were considered as proteins with other functions and were excluded from further analysis. Subsequently, a tree was constructed again for orthologs of the known drug-metabolising proteins. (7) For L-tyrosine decarboxylase (TdcA, EC 4.1.1.25) and cytidine deaminase (cCda, EC 3.5.4.5), we found that genomic context is conserved between species and also analysed the genomic context. If the genomic context of a candidate gene was similar to that of a known drug-metabolising gene, the candidate was considered as an ortholog of the known protein. Otherwise, it was considered to as a false-positive prediction and excluded from further analysis (Supplemental Note 9, Figure S10). As for (6), the tree was constructed again for only the orthologs of the known proteins. (8) For each tree, including only the orthologs of the known genes, we defined the monophyletic branches containing proteins derived from only one species. For each of such species-specific branch, we predicted subcellular localisation (Supplemental Note 10), using the CELLO v.2.5 system (cello.life.nctu.edu.tw). (9) For cytoplasmic enzymes, drug transporters were predicted based on genomic context (Supplemental Note 11, Table S3b).
Tools
The PubSEED platform53, 56 was used to annotate the subsystems. To search for BBHs for previously known proteins, a BLAST algorithm63 implemented in the PubSEED platform was used. Additionally, the PubSEED platform was used for analysis of the genomic context. To analyse the protein domain structure, we searched the Conserved Domains Database (CDD)64 using the following parameters: an e-value ≤0.01 and a maximum number of hits equal to 500. For the prediction of protein subcellular localisation, the CELLO65 web tool was used. Alignments were performed using MUSCLE v.3.8.3166. For every multiple alignment, position quality scores were evaluated using Clustal X67, 68. Thereafter, all positions with a score of zero were removed from the alignment and the modified alignment was used for construction of the phylogenetic trees. Phylogenetic trees were constructed using the maximum-likelihood method with the default parameters implemented in PhyML-3.069. The obtained trees were midpoint-rooted and visualised using the interactive viewer Dendroscope, version 3.2.10, build 1970.
Literature and database searches
Biochemical and physiological characterisation papers were retrieved by entering the names of AGORA2 species into PubMed (https://www.ncbi.nlm.nih.gov/pubmed/). Information on 132 carbon sources, 30 fermentation pathways, 64 growth factors, consumption of 73 metabolites, and secretion of 51 metabolites were subsequently manually extracted on the species and/or genus level from 732 peer-reviewed papers and >8,000 pages of microbial reference textbooks71. Moreover, the traits of each reconstructed strain including taxonomy, morphology, metabolism, and genome size were retrieved through database searches. The taxonomic classification of the strains was retrieved from NCBI Taxonomy (https://www.ncbi.nlm.nih.gov/taxonomy/). Information on morphology, habitat, body site, gram status, oxygen status, metabolism, motility, and genome size were manually retrieved from the Integrated Microbial Genomes and Microbiomes72 database (https://img.jgi.doe.gov/) (Table S1). All experimental data that was used to refine AGORA2 is available at https://github.com/opencobra/COBRA.papers/tree/master/2021_demeter/input.
Generation of draft reconstructions
Draft reconstructions were generated through the KBase24 narrative interface. Genomes present in KBase were directly imported into the narrative. Otherwise, genomes in FASTA format were uploaded into the Staging Area and subsequently, imported into the narrative through the “Batch Import Assembly From Staging Area” (https://narrative.kbase.us/#catalog/apps/kb_uploadmethods/batch_import_assembly_from_staging) app. Genomes in FASTQ format were directly imported into the narrative through the “Import Paired-End Reads From Web” (https://narrative.kbase.us/#catalog/apps/kb_uploadmethods/load_paired_end_reads_from_URL) app after retrieving the links to the corresponding files from https://www.ebi.ac.uk/ena/data/view/PRJEB23845 and https://www.ebi.ac.uk/ena/data/view/PRJEB10915. The imported assemblies were annotated using RAST subsystems73 through the “Annotate Multiple Assemblies” (https://narrative.kbase.us/#appcatalog/app/RAST_SDK/annotate_contigsets) app. Draft metabolic reconstructions were generated through the “Create Multiple Metabolic Models” (https://narrative.kbase.us/#appcatalog/app/fba_tools/build_multiple_metabolic_models) app and exported in SBML format through the “Bulk Download Modelling Objects” (https://narrative.kbase.us/#appcatalog/app/fba_tools/bulk_download_modeling_objects) app.
Semi-automated, data-driven refinement pipeline
We developed a semi-automated refinement pipeline, DEMETER (Data-drivEn METabolic nEtwork Refinement)19, which had been previously used to build AGORA20. Briefly, DEMETER was developed by testing gap-filling steps in few reconstructions and propagating identified solutions to many reconstructions. Curation against experimental data is performed in DEMETER by gap-filling the appropriate reconstructions with a complete pathway for each experimentally demonstrated function. Biomass production under aerobic and anaerobic conditions and on defined media as well as biosynthesis of cell wall components are also enabled through gap-filling solutions that had been previously determined in few reconstructions. Similarly, futile cycles are solved by identifying and correcting the affected reactions in few reconstructions and propagating these changes during the development of DEMETER. More details on DEMETER are provided at Ref19. A detailed tutorial is available as part of the COBRA Toolbox47.
For the generation of AGORA2, we revised DEMETER substantially. Specifically, we (i) translated ~1,000 additional reactions and ~800 metabolites from KBase to VMH23 nomenclature; (ii) introduced additional gap-filling reactions, where needed, to enable biomass production under anoxic conditions on a complex medium with thermodynamically consistent reaction directionalities; (iii) removed futile cycles resulting in thermodynamically implausible ATP production by making the responsible reactions irreversible; (iv) ensured through gap-filling and/or deletion of appropriate reactions that all reconstructions captured the collected experimental data; and (v) adjusted biomass objective functions to account for class-specific cell membrane and cell wall structures as well as introducing a periplasm compartment (Supplemental Note 3). As described previously20, all refinement and debugging solutions were manually determined for a subset of the reconstructions and subsequently propagated to many reconstructions, as appropriate. All newly included metabolites and reactions were formulated based on literature and/or database23, 28, 74 searches, while ensuring mass and charge balance through the reconstruction tool rBioNet75. Reactions identified through comparative genomics (Table S3b-c) were added to up to 5,438 reconstructions. Non-gene associated reactions, for which the respective gene could not be found through comparative genomics, were removed from the draft reconstructions if doing so did not abolish biomass production.
Curation efforts were verified via a test suite19. Specifically, it systematically accessed whether each reconstruction (i) grew anaerobically on complex medium, (ii) had correct reconstruction structure, i.e., mass and charge balance, and correct syntax for gene-protein-reaction associations, (iii) was thermodynamically feasible, e.g., produced realistic amounts of ATP, and (iv) captured known metabolic traits of the organism according to the collected experimental and comparative genomic data. Table S2 summarises all features that are tested by the test suite.
For consistency, the existing 818 AGORA1.03 reconstructions (version 25.02.2019, available at https://www.vmh.life/files/reconstructions/AGORA/1.03/AGORA-1.03.zip) also underwent refinement through DEMETER. The AGORA1.03 reconstruction of Staphylococcus intermedius ATCC 27335 was removed since it was a duplicate of the newly reconstructed strain Streptococcus intermedius ATCC 27335. The names of eight AGORA 1.03 reconstructions were changed to correct strain determination and/or spelling (Table S1).
DEMETER has been implemented in the COBRA Toolbox47 and was run in MATLAB (Mathworks, Inc.) version R2020b.
Generation of quality control reports
The quality control reports and associated score we determined for each AGORA2 reconstruction using the MetaboReport tool in the COBRA Toolbox47. The quality checks included are consistent with the Memote42 checks, as were the calculation of the scores. All 7,302 reports can be accessed via https://metaboreport.live.
Formulation of the drug reactions
A literature search for microbial enzymes known to transform, degrade, activate, inactive, or indirectly influence commonly prescribed drugs was performed yielding 15 enzymes in total (Figure 3a, Table S5), which are encoded by 29 genes (Table S3b). To enable comparative genomic analyses, only drug transformations that could be linked to specific protein-encoding genes were considered. As described above, enzyme-encoding genes were analysed in their genomic context as outlined in76 using PubSEED subsystems26, 53. Additional information on the presence of the analysed genes were retrieved from39, 77, 78.
Literature and database searches were performed for the metabolic fate of commonly prescribed human-targeted drugs. The structures of 287 drug metabolites and drug degradation products were retrieved from 73 peer-reviewed papers, HMDB79, DrugBank79, and Transformer database80. Reactions were formulated based on the collected experimentally determined drug structures, drug downstream product metabolite structures, and reaction mechanisms. Both, cytosolic and extracellular, enzymatic reactions were formulated depending on the identified subcellular protein locations. Since at least six drugs undergoing glucuronidation in the human body have been shown to be substrates for the microbial ß-glucuronidase81, 82 (Table S6), it was assumed that all retrieved glucuronidated drug metabolites (118 in total) could serve as substrates. Additionally, ß-glucuronidase reactions were formulated for 33 glucuronidated drug metabolites from a previously reconstructed module of human drug metabolism83 and three glucuronidated hormones from Recon3D21. New metabolites and reactions were assigned VMH IDs following standards in nomenclature used for COBRA reconstructions9, and formulated while ensuring mass and charge balance through the reconstruction tool rBioNet75. In total, for 98 drugs (Figure 3b), 353 unique metabolites, 381 enzymatic reactions, 373 exchange reactions, and 710 transport reactions (Table S6a-b) were formulated.
Atom-atom mapping
The COBRA Toolbox47 function ‘generateChemicalDatabase’ was used to generate atom-atom mappings. The process to obtain the atom-atom mappings for the AGORA2 reconstructions can be summarised as follows: 1) 1,894/3,533 metabolic structures from the AGORA2 reconstructions were collected from the SMILES and InChIs associated with their metabolites and different chemical databases, such as VMH23, KEGG74, HMDB79, PubChem84, and ChEBI85 databases. The metabolic structures were standardised based on the InChI algorithm86 and can be found in the VMH database23; 2) the standardised metabolites and the reaction stoichiometry in the AGORA2 reconstructions were used to generate 5,583/7,300 MDL RXN files; 3) 5,583/7,300 AGORA2 reactions were atom mapped using the Reaction Decoder Tool algorithm87 for active transport reactions and a custom algorithm88 for passive transport reactions and coupled transport reactions. Atom-atom mappings can be found in the VMH database23 and are freely available at https://github.com/opencobra/ctf.
Simulations
All simulations were performed in MATLAB (Mathworks, Inc.) version R2020b with IBM CPLEX (IBM) as the linear and quadratic programming solver. Computations were carried out on a tower with a 2.80 GHz processor and 64 GB RAM with 12 cores dedicated to parallelisation. The simulations were carried out using functions implemented in the COBRA Toolbox47. Flux balance analysis (FBA)34 was used to simulate metabolic fluxes. All additional scripts for data generation, data analysis, and data visualisation are available at https://github.com/ThieleLab/CodeBase.
Retrieval of reconstruction resources
Manually and semi-automatically reconstructions compared with AGORA2 were retrieved as follows: 72 fully manually curated reconstructions were downloaded from the BiGG database28 (http://bigg.ucsd.edu/). Reconstructions generated through gapseq18 (8,075 total) were downloaded from ftp://ftp.rz.uni-kiel.de/pub/medsystbio/models/EnzymaticDataTestModels.zip and exported in SBML format through the sybilSBML package in R using a custom script. MAGMA17 reconstructions (1,333 total) were downloaded from https://www.microbiomeatlas.org/data/MSP_GEM_models.zip. To enable comparability with AGORA2, exchange reactions in all retrieved reconstructions were translated to VMH23 nomenclature through custom MATLAB scripts. Moreover, an ATP demand reaction (VMH reaction ID: DM_atp_c_) was added if not already present and otherwise translated to VMH nomenclature.
Generation of reconstructions through CarveMe
Protein fasta files corresponding to 7,279 AGORA2 strains were downloaded from either NCBI (https://www.ncbi.nlm.nih.gov/assembly) or ENA (https://www.ebi.ac.uk/ena) and subsequently used to run CarveMe. The remaining 23 AGORA2 strains were excluded as a corresponding protein FASTA file was not available. Reconstructions for 7,279 strains were generated with CarveMe15 version 1.5.1 on Python 3.7.13 (retrieved from https://www.python.org/downloads/release/python-3713) and relying on DIAMOND89 version 0.9.14.
Generation of reconstructions through gapseq
Genome FASTA files retrieved as described above were used as the input for gapseq18. A total of 1,767 models were generated with gapseq 1.2, which was run in R90 version 4.1.2 on a Ubuntu 22.04 machine. The R interface of GLPK (package Rglpk) was used as the linear programming solver.
Flux and stoichiometrically consistent reactions
The subset of flux and stoichiometrically consistent reactions, as defined in29, was retrieved through the ‘findFluxConsistentSubset’ and ‘findStoichConsistentSubset’ functions implemented in the COBRA Toolbox47. The fraction of stoichiometrically and flux consistent reactions, excluding exchange and demand reactions, was subsequently determined for each AGORA2 reconstruction and corresponding KBase draft reconstruction as well as for 5,587 reconstructions generated through CarveMe15, 8,075 reconstructions generated through gapseq18, 1,333 MAGMA17 reconstructions, and 73 curated reconstructions from the BiGG database28. Briefly, the subset of stoichiometrically consistent reactions in a reconstruction includes all reactions that are mass and charge conserved, excluding exchange, demand, and sink reactions, which are by definition mass and charge imbalanced29. The subset of flux consistent reactions consists of all reactions that are stoichiometrically consistent and can carry flux under the defined set of constraints29.
Validation against three independent experimental datasets
For an independent assessment of predictive potential of genome-scale reconstructions, independent (i.e., not used for the reconstruction process) experimental data on metabolite uptake and secretion was retrieved from three sources30, 32, 33 and mapped onto the VMH23 nomenclature through custom MATLAB scripts. The experimental data included species-level positive and negative metabolite uptake and secretion data for 457 species (5,341 strains) and 269 metabolites in AGORA2 from the NJC19 resource30, and species-level positive metabolite uptake data from32 for 184 species (328 strains) and 85 metabolites in AGORA2. Moreover, strain-resolved positive and negative metabolite uptake and secretion data for 676 AGORA2 strains and 220 metabolites, and positive and negative enzyme activity data for 881 AGORA2 strains and 31 enzymes were retrieved from the BacDive database33. The enzyme data was mapped to the respective reactions in each of the compared reconstruction resources’ namespaces. Positive data indicated that the metabolite uptake or secretion capability or enzyme activity had been demonstrated in a microbe while negative data indicated that the microbe has been shown to not possess the capability. For each retrieved positive or negative data point, the capability of the respective model to take up or produce the corresponding metabolite was calculated using FBA on unlimited medium by either minimising or maximising the corresponding exchange reaction, respectively. For enzyme data, it was tested whether at least one reaction mapped to the respective enzyme was present in the model and could carry nonzero flux. If the data point was positive and the corresponding model could also take up or secrete the metabolite or produce flux through the corresponding enzymatic reactions(s), this resulted in a true positive prediction, while a false negative prediction occurred when the microbe was known to have this capability, but the corresponding model did not capture the trait. If the data point was negative and the corresponding model also could not take up or secrete the metabolite or did not produce flux through any reaction(s) mapped to the enzyme, this resulted in a true negative prediction, otherwise the prediction was a false positive.
Prediction accuracy were calculated for the three experimental datasets. For an assessment of the predictive potential of AGORA2 compared with other reconstruction resources, the analysis was repeated for the strains in KBase draft reconstructions, CarveMe reconstructions, and BiGG, gapseq, and MAGMA reconstructions that overlapped with the AGORA2 organisms with available data. To this end, the predictive value of all resources was tested via mixed effect logistic regressions with the in silico prediction as predictor and the in vivo behaviour (binary) as response variable, while introducing the model as random effect variable accounting for the stochastic dependencies of predictions for different metabolites stemming from the same model. Moreover, the accuracy per model was calculated for all resources, and then compared with the AGORA2 accuracies via non-parametric sign rank tests. The list of all strains in the compared reconstruction resources that were tested against the three datasets is shown in Table S4a. All scripts are available at https://github.com/ThieleLab/CodeBase.
Validation of drug-metabolising capacities against independent experimental data
A literature search was performed for in vitro experiments demonstrating the capabilities of human microbial strains to metabolise reconstructed drugs through the 15 annotated enzymes, resulting in 253 drug-microbe pairs (Table S7). As this data contained both positive and negative data, true positive, true negative, false positive, and false negative predictions could occur as described above. If no studies on the specific reconstructed drugs were found for the enzyme, studies on general activity of the enzyme were retrieved. If possible, the tested microbes were matched to AGORA2 models on the strain level, otherwise pan-species models were used. Subsequently, the capabilities to metabolise the drugs through the respective enzymes for the 164 AGORA2 models with available data (Table S7) were tested by computing whether the corresponding reaction could carry flux. Accuracy, sensitivity, and specificity of predictions were calculated after determining the number of true positive, true negative, false positive, and false negative predictions. P-values were calculated by Fisher’'s exact test and, for sensitivity analysis, by mixed effect logistic regression including the model as random effect variable, accounting for the stochastic dependency of predictions stemming from the same model.
Drug yields
To determine each strains’ capability to metabolise drugs, all AGORA2 were constrained with a simulated Western diet20 and the flux through the exchange reactions corresponding to each drug was minimised using FBA, corresponding to maximal uptake rate of the drug. For all AGORA2 organisms capable to take up at least one drug, the yield of ATP, carbon, and ammonia from 1 mmol of the drug gdry weight/hr was evaluated as follows. Each reconstruction was constrained to only allow the uptake of water, phosphate, and oxygen (VMH IDs: h2o, pi, o2). Demand reactions for ammonia as well as CO2 and pyruvate (as proxies for carbon sources) (VMH IDs: nh4, co2, pyr) were added, while a demand reaction for ATP (VMH ID: atp) already existed in each reconstruction. Next, the uptake of each drug metabolite (15 in total, one representative for each enzyme) was allowed one by one at an uptake rate of 1 mmo1/gdry weight/hr. For each drug metabolite, the yields of ATP, ammonia, CO2, and pyruvate from each drug metabolite were computed using flux balance analysis (FBA) by maximising the flux through the respective demand reactions. As control, yields were also computed for 1 mmo1/gdry weight/hr of glucose and without any metabolites added.
Simulation of drug metabolism by individual gut microbiomes
Previously, metagenomic sequencing from faecal samples of a cohort of 616 Japanese colorectal cancer patients and healthy controls had been performed38. Species-level abundances for this cohort, which has been determined with MetaPhIAn291, were retrieved from https://www.nature.com/articles/s41591-019-0458-7#MOESM3. Unclassified taxa on the species level, eukaryotes, and viruses were excluded. Of the remaining 517 species, 501 (97%) could be mapped onto the 1,738 AGORA2 species. Pan-species models for AGORA2 were created through the ‘createPanModels’ function. From the pan-species models, personalised microbiome models for each of the 616 samples were built through a computationally efficient pipeline43 with the species-level abundances as input data and parameterised as described elsewhere10, 60. For each individual, we integrated all microbial models having a non-zero abundance in the sample into one personalised microbiome model. To contextualise the models with appropriate diet constraints, a simulated Average Japanese Diet described previously41 (Table S12) was used. To predict the drug conversion potential of each microbiome, the faecal secretion reactions for 13 drug metabolism end products were optimised one by one using flux balance analysis34, while providing the respective precursor drug as well as oxygen at a de facto unlimited uptake rate of 1000 mmo1/gdry weight/hr.
Shadow price analysis
To determine species in microbiome models that were of importance for the microbiome’s combined potential to metabolise a drug, a shadow price analysis was performed as described previously60. Briefly, shadow prices are a feature of every flux balance analysis solution (i.e., the shadow price is the dual to the primal linear programming problem) that reflect the contribution of each metabolite in the model to the flux through the objective function8. A non-zero shadow price for a metabolite indicates that this metabolite has importance for the total flux capacity through the optimised objective function, i.e., in our case, the secretion of a drug metabolic product. A shadow price of zero indicates that increasing the availability of this metabolite would not change the flux through the objective function. To determine the species that were bottlenecks for the conversion potential of the 13 drugs in each microbiome model, nonzero shadow prices for species biomass metabolites (‘species_biomass[c]’), which reflect the contribution of the species to the community biomass reaction, were retrieved.
Statistical analysis
We analysed statistically the net production capacity of 13 drug metabolites (Figure 6a) among 252 healthy individuals and 364 CRC patients. For each drug metabolite, we calculated the mean flux and the share of microbiomes with a flux greater zero. Drug metabolites, which had in over 50% of the cases a zero flux, were dichotomised (can be produced vs. cannot be produced) and subsequently, analysed via logistic regressions. Drug metabolites with over 50% non-zero entries were analysed via linear regressions using heteroscedastic robust standard errors. First, we investigated potential effects of basic covariates (age, sex, and BMI) via generalised linear regressions (logistic or linear) with the net production capacity being the response variable (dichotomised or metric). Age and BMI were introduced into the models as restricted cubic splines92 using four knots (the 5%-percentile, the 33%-percentile, the 66%-percentile, and the 95%-percentile) resulting in three spline variables, each to test on potential non-linear relationships. Significance was then determined by testing the three spline variables belonging to age (or BMI, respectively) simultaneously on zero via the Wald test92. While for age substantial non-linearities were found, no indication for non-linear BMI effects could be identified. The final models included, therefore, only the linear BMI term. Second, we tested for potential associations of net production capacities with case control status. This test was done via generalised linear regressions (logistic or linear) with the net production capacity being the response variable (dichotomised or metric), while adjusting for age (restricted cubic splines), sex (male/female), and BMI (linear). We corrected for multiple testing using the false discovery rate, adjusting significance values for 13 tests per analyses stream. A test was considered nominal significant with p<0.05 and FDR-corrected significant if FDR<0.05. For sensitivity analysis, we recomputed the drug-metabolising capabilities using an average European diet instead of a Japanese diet. Then, we calculated Pearson correlations for each drug metabolite between the secretion potentials under Japanese and an average European diet. All statistical analyses were performed with STATA 17/MP. All scripts are available at https://github.com/ThieleLab/CodeBase.
Sign prediction of faecal metabolite-species associations using AGORA2-based community models
We utilised the publicly available metabolome dataset (n=347) from Ref.38. To test whether AGORA2-based community modelling is capable of predicting the sign of statistical associations between species presence and faecal metabolite concentrations in the CRC sample, we calculated maximal net secretion for 52 metabolites with faecal metabolome data with more than 50% of the samples having concentrations above limit of detection. Metabolite net secretion was computed using the mgPipe module in the Microbiome Modelling Toolbox10, 43 while relying on computationally efficient flux variability analysis93. Then, we calculated for each species present in at least 10% of the microbiomes and at max 90% of the microbiomes the effect of species (binary predictor: species present vs species not present) on each faecal metabolite concentration in multivariable regressions adjusting for age, sex, BMI, and study group. We then filtered for all species metabolite associations with p<0.05. Next, we calculated the effect of the species presence on the community net secretion of the corresponding metabolite in analogous regressions. Finally, we calculated for each metabolite the agreement in signs between the in vivo association statistics and the in silico association statistics. Significance was determined by Fisher’s exact test and FDR correction was applied accounting for 52 tests. Note that the p-values should be treated with care since the signs of the various association statistics may cluster due to the multivariate nature of both the metabolome and the microbiome data.
Data visualisation
The phylogenetic tree of AGORA2 organisms was constructed in PhyloT (https://phylot.biobyte.de/) and visualised in iTOL (https://itol.embl.de/)94. Violin plots were generated in BoxPlotR (http://shiny.chemgrid.org/boxplotr/). Clustering of taxa by reaction presence through t-distributed stochastic neighbour embedding (t-SNE)52 was performed using the t-SNE implementation in MATLAB with Euclidean distance, barneshut set as the algorithm, and perplexity set to 30. Taxa with fewer representatives than 0.5% of all clustered strains were excluded from the t-SNE plots. Significance of differences in coordinates across taxonomic units were determined by Kruskal-Wallis tests. Circle plots were generated using the online implementation of Circos95. Figures 6 and S9 were generated with the graphics functions of STATA 16/MP. All other data was visualised in MATLAB and R90.
Supplementary Material
Acknowledgements
We thank Prof. Peter Turnbaugh for providing genome sequences for 26 Eggerthella lenta strains, Dr. Jan Krumsiek for communicating mouse-associated microbial species, Dr. Cyrille Thinnes for valuable discussions, and Lubin Moussu and Semra Smajic for their help with the comparative genomic effort.
Funding sources
This study was funded by grants from the European Research Council (ERC) under the European Union’s Horizon 2020 research and innovation programme (grant agreement No 757922) to IT, from the National Institute on Aging grants (1RF1AG058942 and 1U19AG063744), from the European Union’s Horizon 2020 research and innovation programme under the Marie Skłodowska-Curie (grant agreement No 859890), and from the Science Foundation Ireland under Grant number 12/RC/2273-P2.
Footnotes
Author contributions
I.T. and A.H. conceived the study. D.A.R., G.A., and O.E.O. performed comparative genomic analyses. I.T., A.H., S.M., M.H., F.M., J.N.E., and C.S.H. created KBase draft reconstructions. A.H. and S.M. built the semi-automated reconstruction pipeline and the test suite. G.A. and A.H. translated reaction and metabolite identifiers to VMH nomenclature. M.H., G.A., A.H., and F.M. collected experimental data. M.H., F.M., and A.H. collected organism information. M.N. and A.H. formulated the drug reactions. A.H. performed continuous reconstruction testing and curation. A.H. performed simulations. A.H. and J.H. analysed and visualised the data. J.H. performed statistical analyses. B.N. built CarveMe and gapseq reconstructions. G. P. and R.M.T.F. performed atom-atom mappings. A.H. and J.H. drafted the paper. All authors edited the paper. I.T. supervised the study.
Competing interests
The authors declare no competing interests.
Data availability statement
The 7,302 AGORA2 reconstructions are freely available at https://www.vmh.life/. Quality control reports for all reconstructions are available at https://metaboreport.live/ and https://vmh.life/files/reconstructions/AGORA2/ for bulk download.
Code availability statement
Code and input data to reproduce the generation of the AGORA2 reconstructions and microbiome models as well as all simulations and analyses are available at https://github.com/ThieleLab/CodeBase.
References
- 1.Lynch SV, Pedersen O. The Human Intestinal Microbiome in Health and Disease. N Engl J Med. 2016;375:2369–2379. doi: 10.1056/NEJMra1600266. [DOI] [PubMed] [Google Scholar]
- 2.Nebert DW, Zhang G, Vesell ES. From human genetics and genomics to pharmacogenetics and pharmacogenomics: past lessons, future directions. Drug Metab Rev. 2008;40:187–224. doi: 10.1080/03602530801952864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tralau T, Sowada J, Luch A. Insights on the human microbiome and its xenobiotic metabolism: what is known about its effects on human physiology? Expert Opin Drug Metab Toxicol. 2015;11:411–425. doi: 10.1517/17425255.2015.990437. [DOI] [PubMed] [Google Scholar]
- 4.Spanogiannopoulos P, Bess EN, Carmody RN, Turnbaugh PJ. The microbial pharmacists within us: a metagenomic view of xenobiotic metabolism. Nat Rev Microbiol. 2016;14:273–287. doi: 10.1038/nrmicro.2016.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zimmermann M, Zimmermann-Kogadeeva M, Wegmann R, Goodman AL. Mapping human microbiome drug metabolism by gut bacteria and their genes. Nature. 2019;570:462–467. doi: 10.1038/s41586-019-1291-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Javdan B, et al. Personalized Mapping of Drug Metabolism by the Human Gut Microbiome. Cell. 2020;181:1661–1679.:e1622. doi: 10.1016/j.cell.2020.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Guthrie L, Kelly L. Bringing microbiome-drug interaction research into the clinic. EBioMedicine. 2019;44:708–715. doi: 10.1016/j.ebiom.2019.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Palsson B. Systems biology : properties of reconstructed networks. Cambridge University Press; Cambridge: 2006. [Google Scholar]
- 9.Thiele I, Palsson BØ. A protocol for generating a high-quality genome-scale metabolic reconstruction. Nature protocols. 2010;5:93–121. doi: 10.1038/nprot.2009.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Baldini F, et al. The Microbiome Modeling Toolbox: from microbial interactions to personalized microbial communities. Bioinformatics. 2019;35:2332–2334. doi: 10.1093/bioinformatics/bty941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Diener C, Gibbons SM, Resendis-Antonio O. MICOM: Metagenome-Scale Modeling To Infer Metabolic Interactions in the Gut Microbiota. mSystems. 2020;5 doi: 10.1128/mSystems.00606-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Magnusdottir S, Thiele I. Modeling metabolism of the human gut microbiome. Curr Opin Biotechnol. 2018;51:90–96. doi: 10.1016/j.copbio.2017.12.005. [DOI] [PubMed] [Google Scholar]
- 13.van der Ark KCH, van Heck RGA, Martins Dos Santos VAP, Belzer C, de Vos WM. More than just a gut feeling: constraint-based genome-scale metabolic models for predicting functions of human intestinal microbes. Microbiome. 2017;5:78. doi: 10.1186/s40168-017-0299-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lagier JC, et al. Many More Microbes in Humans: Enlarging the Microbiome Repertoire. Clin Infect Dis. 2017;65:S20–S29. doi: 10.1093/cid/cix404. [DOI] [PubMed] [Google Scholar]
- 15.Machado D, Andrejev S, Tramontano M, Patil KR. Fast automated reconstruction of genome-scale metabolic models for microbial species and communities. Nucleic Acids Res. 2018;46:7542–7553. doi: 10.1093/nar/gky537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zorrilla F, Buric F, Patil KR, Zelezniak A. metaGEM: reconstruction of genome scale metabolic models directly from metagenomes. Nucleic Acids Res. 2021 doi: 10.1093/nar/gkab815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bidkhori G, et al. The Reactobiome Unravels a New Paradigm in Human Gut Microbiome Metabolism. bioRxiv. 2021:2021.2002.2001.428114 [Google Scholar]
- 18.Zimmermann J, Kaleta C, Waschina S. gapseq: informed prediction of bacterial metabolic pathways and reconstruction of accurate metabolic models. Genome Biol. 2021;22:81. doi: 10.1186/s13059-021-02295-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Heinken A, Magnusdottir S, Fleming RMT, Thiele I. DEMETER: Efficient simultaneous curation of genome-scale reconstructions guided by experimental data and refined gene annotations. Bioinformatics. 2021 doi: 10.1093/bioinformatics/btab622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Magnusdottir S, et al. Generation of genome-scale metabolic reconstructions for 773 members of the human gut microbiota. Nat Biotechnol. 2017;35:81–89. doi: 10.1038/nbt.3703. [DOI] [PubMed] [Google Scholar]
- 21.Brunk E, et al. Recon3D enables a three-dimensional view of gene variation in human metabolism. Nat Biotechnol. 2018;36:272–281. doi: 10.1038/nbt.4072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Thiele I, et al. Personalized whole-body models integrate metabolism, physiology, and the gut microbiome. Mol Syst Biol. 2020;16:e8982. doi: 10.15252/msb.20198982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Noronha A, et al. The Virtual Metabolic Human database: integrating human and gut microbiome metabolism with nutrition and disease. Nucleic Acids Res. 2019;47:D614–D624. doi: 10.1093/nar/gky992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Arkin AP, et al. KBase: The United States Department of Energy Systems Biology Knowledgebase. Nat Biotechnol. 2018;36:566–569. doi: 10.1038/nbt.4163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bernstein DB, Sulheim S, Almaas E, Segre D. Addressing uncertainty in genome-scale metabolic model reconstruction and analysis. Genome Biol. 2021;22:64. doi: 10.1186/s13059-021-02289-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Aziz RK, et al. SEED servers: high-performance access to the SEED genomes, annotations, and metabolic models. PLoS One. 2012;7:e48053. doi: 10.1371/journal.pone.0048053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Henry CS, et al. High-throughput generation, optimization and analysis of genome-scale metabolic models. Nat Biotechnol. 2010;28:977–982. doi: 10.1038/nbt.1672. [DOI] [PubMed] [Google Scholar]
- 28.Norsigian CJ, et al. BiGG Models 2020: multi-strain genome-scale models and expansion across the phylogenetic tree. Nucleic Acids Res. 2020;48:D402–D406. doi: 10.1093/nar/gkz1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fleming RM, Vlassis N, Thiele I, Saunders MA. Conditions for duality between fluxes and concentrations in biochemical networks. Journal of theoretical biology. 2016;409:1–10. doi: 10.1016/j.jtbi.2016.06.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lim R, et al. Large-scale metabolic interaction network of the mouse and human gut microbiota. Sci Data. 2020;7:204. doi: 10.1038/s41597-020-0516-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sung J, et al. Global metabolic interaction network of the human gut microbiota for context-specific community-scale analysis. Nat Commun. 2017;8:15393. doi: 10.1038/ncomms15393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Madin JS, et al. A synthesis of bacterial and archaeal phenotypic trait data. Sci Data. 2020;7:170. doi: 10.1038/s41597-020-0497-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Reimer LC, et al. BacDive in 2019: bacterial phenotypic data for High-throughput biodiversity analysis. Nucleic Acids Res. 2019;47:D631–D636. doi: 10.1093/nar/gky879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Orth JD, Thiele I, Palsson BO. What is flux balance analysis? Nat Biotechnol. 2010;28:245–248. doi: 10.1038/nbt.1614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zimmermann M, Zimmermann-Kogadeeva M, Wegmann R, Goodman AL. Separating host and microbiome contributions to drug pharmacokinetics and toxicity. Science. 2019;363 doi: 10.1126/science.aat9931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pollet RM, et al. An Atlas of beta-Glucuronidases in the Human Intestinal Microbiome. Structure. 2017;25:967–977.:e965. doi: 10.1016/j.str.2017.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Heinken A, Hertel J, Thiele I. Metabolic modelling reveals broad changes in gut microbial metabolism in inflammatory bowel disease patients with dysbiosis. Syst Biol Appl. 2021 doi: 10.1038/s41540-021-00178-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yachida S, et al. Metagenomic and metabolomic analyses reveal distinct stage-specific phenotypes of the gut microbiota in colorectal cancer. Nat Med. 2019;25:968–976. doi: 10.1038/s41591-019-0458-7. [DOI] [PubMed] [Google Scholar]
- 39.Maini Rekdal V, Bess EN, Bisanz JE, Turnbaugh PJ, Balskus EP. Discovery and inhibition of an interspecies gut bacterial pathway for Levodopa metabolism. Science. 2019;364 doi: 10.1126/science.aau6323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wirbel J, et al. Meta-analysis of fecal metagenomes reveals global microbial signatures that are specific for colorectal cancer. Nat Med. 2019;25:679–689. doi: 10.1038/s41591-019-0406-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hertel J, Heinken A, Martinelli F, Thiele I. Integration of constraint-based modeling with fecal metabolomics reveals large deleterious effects of Fusobacterium spp. on community butyrate production. Gut Microbes. 2021;13:1–23. doi: 10.1080/19490976.2021.1915673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lieven C, et al. MEMOTE for standardized genome-scale metabolic model testing. Nat Biotechnol. 2020;38:272–276. doi: 10.1038/s41587-020-0446-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Heinken A, Thiele I. Microbiome Modelling Toolbox 2.0: Efficient, tractable modelling of microbiome communities. Bioinformatics. 2022 doi: 10.1093/bioinformatics/btac082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sen P, Oresic M. Metabolic Modeling of Human Gut Microbiota on a Genome Scale: An Overview. Metabolites. 2019;9 doi: 10.3390/metabo9020022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Monk JM, et al. iML1515, a knowledgebase that computes Escherichia coli traits. Nat Biotechnol. 2017;35:904–908. doi: 10.1038/nbt.3956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Heinken A, Basile A, Thiele I. Advances in constraint-based modelling of microbial communities. Current Opinion in Systems Biology. 2021 [Google Scholar]
- 47.Heirendt L, et al. Creation and analysis of biochemical constraint-based models using the COBRA Toolbox v.3.0. Nat Protoc. 2019;14:639–702. doi: 10.1038/s41596-018-0098-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bebb JR, Scott BB. How effective are the usual treatments for ulcerative colitis? Aliment Pharmacol Ther. 2004;20:143–149. doi: 10.1111/j.1365-2036.2004.02018.x. [DOI] [PubMed] [Google Scholar]
- 49.Thiele I, Clancy CM, Heinken A, Fleming RMT. Quantitative systems pharmacology and the personalized drug-microbiota-diet axis. Curr Opin Syst Biol. 2017;4:43–52. doi: 10.1016/j.coisb.2017.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Krauss M, et al. Integrating cellular metabolism into a multiscale whole-body model. PLoS Comput Biol. 2012;8:e1002750. doi: 10.1371/journal.pcbi.1002750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Heinken A, Basile A, Hertel J, Thinnes C, Thiele I. Genome-Scale Metabolic Modeling of the Human Microbiome in the Era of Personalized Medicine. Annu Rev Microbiol. 2021 doi: 10.1146/annurev-micro-060221-012134. [DOI] [PubMed] [Google Scholar]
- 52.van der Maaten L, Hinton G. Viualizing data using t-SNE. Journal of Machine Learning Research. 2008;9:2579–2605. [Google Scholar]
- 53.Overbeek R, et al. The subsystems approach to genome annotation and its use in the project to annotate 1000 genomes. Nucleic Acids Res. 2005;33:5691–5702. doi: 10.1093/nar/gki866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bisanz JE, et al. A Genomic Toolkit for the Mechanistic Dissection of Intractable Human Gut Bacteria. Cell Host Microbe. 2020 doi: 10.1016/j.chom.2020.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Forster SC, et al. A human gut bacterial genome and culture collection for improved metagenomic analyses. Nat Biotechnol. 2019;37:186–192. doi: 10.1038/s41587-018-0009-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Disz T, et al. Accessing the SEED genome databases via Web services API: tools for programmers. BMC Bioinformatics. 2010;11:319. doi: 10.1186/1471-2105-11-319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ravcheev DA, Thiele I. Systematic genomic analysis reveals the complementary aerobic and anaerobic respiration capacities of the human gut microbiota. Front Microbiol. 2014;5:674. doi: 10.3389/fmicb.2014.00674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Magnusdottir S, Ravcheev D, de Crecy-Lagard V, Thiele I. Systematic genome assessment of B-vitamin biosynthesis suggests co-operation among gut microbes. Front Genet. 2015;6:148. doi: 10.3389/fgene.2015.00148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ravcheev DA, Thiele I. Genomic Analysis of the Human Gut Microbiome Suggests Novel Enzymes Involved in Quinone Biosynthesis. Front Microbiol. 2016;7:128. doi: 10.3389/fmicb.2016.00128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Heinken A, et al. Personalized modeling of the human gut microbiome reveals distinct bile acid deconjugation and biotransformation potential in healthy and IBD individuals. Microbiome. 2019;7:75. [Google Scholar]
- 61.Kanehisa M, Furumichi M, Tanabe M, Sato Y, Morishima K. KEGG: new perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Res. 2017;45:D353–D361. doi: 10.1093/nar/gkw1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wolf YI, Koonin EV. A tight link between orthologs and bidirectional best hits in bacterial and archaeal genomes. Genome Biol Evol. 2012;4:1286–1294. doi: 10.1093/gbe/evs100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Altschul SF, et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Marchler-Bauer A, et al. CDD: conserved domains and protein three-dimensional structure. Nucleic Acids Res. 2013;41:D348–352. doi: 10.1093/nar/gks1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yu CS, Chen YC, Lu CH, Hwang JK. Prediction of protein subcellular localization. Proteins. 2006;64:643–651. doi: 10.1002/prot.21018. [DOI] [PubMed] [Google Scholar]
- 66.Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Larkin MA, et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 68.Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Research. 1997;25:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Guindon S, et al. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Systematic biology. 2010;59:307–321. doi: 10.1093/sysbio/syq010. [DOI] [PubMed] [Google Scholar]
- 70.Huson DH, et al. Dendroscope: An interactive viewer for large phylogenetic trees. BMC Bioinformatics. 2007;8:460. doi: 10.1186/1471-2105-8-460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Krieg N, et al. Bergey’s Manual® of Systematic Bacteriology. 2010 [Google Scholar]
- 72.Chen IA, et al. IMG/M v.5.0: an integrated data management and comparative analysis system for microbial genomes and microbiomes. Nucleic Acids Res. 2019;47:D666–D677. doi: 10.1093/nar/gky901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Aziz RK, et al. The RAST Server: Rapid Annotations using Subsystems Technology. BMC genomics. 2008;9:75. doi: 10.1186/1471-2164-9-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kanehisa M, Sato Y, Furumichi M, Morishima K, Tanabe M. New approach for understanding genome variations in KEGG. Nucleic Acids Res. 2019;47:D590–D595. doi: 10.1093/nar/gky962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Thorleifsson SG, Thiele I. rBioNet: A COBRA toolbox extension for reconstructing high-quality biochemical networks. Bioinformatics (Oxford, England) 2011;27:2009–2010. doi: 10.1093/bioinformatics/btr308. [DOI] [PubMed] [Google Scholar]
- 76.Osterman A, Overbeek R. Missing genes in metabolic pathways: a comparative genomics approach. Curr Opin Chem Biol. 2003;7:238–251. doi: 10.1016/s1367-5931(03)00027-9. [DOI] [PubMed] [Google Scholar]
- 77.Zou L, et al. Bacterial metabolism rescues the inhibition of intestinal drug absorption by food and drug additives. Proc Natl Acad Sci U S A. 2020;117:16009–16018. doi: 10.1073/pnas.1920483117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Koppel N, Bisanz JE, Pandelia ME, Turnbaugh PJ, Balskus EP. Discovery and characterization of a prevalent human gut bacterial enzyme sufficient for the inactivation of a family of plant toxins. Elife. 2018;7 doi: 10.7554/eLife.33953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Wishart DS, et al. HMDB 4.0: the human metabolome database for 2018. Nucleic Acids Res. 2018;46:D608–D617. doi: 10.1093/nar/gkx1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Hoffmann MF, et al. The Transformer database: biotransformation of xenobiotics. Nucleic Acids Res. 2014;42:D1113–1117. doi: 10.1093/nar/gkt1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Wallace BD, et al. Alleviating cancer drug toxicity by inhibiting a bacterial enzyme. Science. 2010;330:831–835. doi: 10.1126/science.1191175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Saitta KS, et al. Bacterial beta-glucuronidase inhibition protects mice against enteropathy induced by indomethacin, ketoprofen or diclofenac: mode of action and pharmacokinetics. Xenobiotica. 2014;44:28–35. doi: 10.3109/00498254.2013.811314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Sahoo S, Haraldsdottir H, Fleming RM, Thiele I. Modeling the effects of commonly used drugs on human metabolism. FEBS Journal. 2015;282:297–317. doi: 10.1111/febs.13128. [DOI] [PubMed] [Google Scholar]
- 84.Kim S, et al. PubChem 2019 update: improved access to chemical data. Nucleic Acids Res. 2019;47:D1102–D1109. doi: 10.1093/nar/gky1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Hastings J, et al. The ChEBI reference database and ontology for biologically relevant chemistry: enhancements for 2013. Nucleic Acids Res. 2013;41:D456–463. doi: 10.1093/nar/gks1146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Heller SR, McNaught A, Pletnev I, Stein S, Tchekhovskoi D. InChI, the IUPAC International Chemical Identifier. J Cheminform. 2015;7:23. doi: 10.1186/s13321-015-0068-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Rahman SA, et al. Reaction Decoder Tool (RDT): extracting features from chemical reactions. Bioinformatics. 2016;32:2065–2066. doi: 10.1093/bioinformatics/btw096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Preciat Gea. Genome-scale metabolic modelling and 13C metabolic flux analysis in midbrain neurons (in preparation)
- 89.Buchfink B, Xie C, Huson DH. Fast and sensitive protein alignment using DIAMOND. Nat Methods. 2015;12:59–60. doi: 10.1038/nmeth.3176. [DOI] [PubMed] [Google Scholar]
- 90.Team, R.C. A Language and Environment for Statistical Computing. R Foundation for Statistical Computing; Vienna: 2013. [Google Scholar]
- 91.Truong DT, et al. MetaPhlAn2 for enhanced metagenomic taxonomic profiling. Nat Methods. 2015;12:902–903. doi: 10.1038/nmeth.3589. [DOI] [PubMed] [Google Scholar]
- 92.Harrell FE. Regression modeling strategies : with applications to linear models, logistic regression, and survival analysis. Springer; New York: 2001. [Google Scholar]
- 93.Gudmundsson S, Thiele I. Computationally efficient flux variability analysis. BMC Bioinformatics. 2010;11:489. doi: 10.1186/1471-2105-11-489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Letunic I, Bork P. Interactive Tree Of Life (iTOL) v4: recent updates and new developments. Nucleic Acids Res. 2019;47:W256–W259. doi: 10.1093/nar/gkz239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Krzywinski M, et al. Circos: an information aesthetic for comparative genomics. Genome Res. 2009;19:1639–1645. doi: 10.1101/gr.092759.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The 7,302 AGORA2 reconstructions are freely available at https://www.vmh.life/. Quality control reports for all reconstructions are available at https://metaboreport.live/ and https://vmh.life/files/reconstructions/AGORA2/ for bulk download.
Code and input data to reproduce the generation of the AGORA2 reconstructions and microbiome models as well as all simulations and analyses are available at https://github.com/ThieleLab/CodeBase.