Abstract
Purpose
Understanding resistance to selective FGFR inhibitors is crucial to improve the clinical outcomes of patients with FGFR2-driven malignancies.
Experimental Design
We analyzed sequential ctDNA, +/- WES or targeted NGS on tissue biopsies from patients with tumors harboring activating FGFR2 alterations progressing on pan-FGFR-selective inhibitors, collected in the prospective UNLOCK program. FGFR2::BICC1 Ba/F3 and patient-derived xenografts (PDX) models were used for functional studies.
Results
Thirty-six patients were included. In cholangiocarcinoma, at resistance to both reversible inhibitors (e.g. pemigatinib, erdafitinib) and the irreversible inhibitor futibatinib, polyclonal FGFR2 kinase domain mutations were frequent (14/27 patients). Tumors other than cholangiocarcinoma shared the same mutated FGFR2 residues, but polyclonality was rare (1/9 patients). At resistance to reversible inhibitors, 14 residues in the FGFR2 kinase domain were mutated; after futibatinib, only the molecular brake N550 and the gatekeeper V565. Off-target alterations in PI3K/mTOR and MAPK pathways were found in 11 patients, often together with on-target mutations. At progression to a first FGFR inhibitor, 12 patients received futibatinib or lirafugratinib (irreversible inhibitors), with variable clinical outcomes depending on previous resistance mechanisms. Two patients with TSC1 or PIK3CA mutations benefitted from everolimus. In cell viability assays on Ba/F3 and in pharmacologic studies on PDX, irreversible inhibitors retained better activity against FGFR2 kinase domain mutations, with lirafugratinib active against the recalcitrant V565L/F/Y.
Conclusions
At progression to FGFR inhibitors, FGFR2-driven malignancies are characterized by high intra- and inter-patient molecular heterogeneity, particularly in cholangiocarcinoma. Resistance to FGFR inhibitors can be overcome by sequential, molecularly-oriented treatment strategies across FGFR2-driven tumors.
Keywords: Cholangiocarcinoma, FGFR, resistance mechanisms, mutations, futibatinib, lirafugratinib
Introduction
Molecular alterations of fibroblast growth factor receptor family members (FGFR1/2/3/4) are frequent across cancers (1,2). FGFR amplifications are the most frequent alterations observed, yet their inconsistent oncogenic potential raises questions about their suitability as targets for selective inhibition (3). FGFR2 gene fusions occur in 10-15% of intrahepatic cholangiocarcinoma cases, while activating mutations in the extracellular domain account for only a minor fraction of this malignancy (4–6). Recent research has highlighted the importance of deletions in the extracellular domain and truncations in the intracellular C-terminal domain of FGFR2 as key drivers and therapeutic targets in intrahepatic cholangiocarcinoma (6,7). Of note, the same molecular alterations can be found, with a lower incidence, across a variety of solid tumors, of almost any histology (7).
The development and availability of selective FGFR inhibitors for treating FGFR2-driven intrahepatic cholangiocarcinoma are transforming the therapeutic landscape for patients with this molecular subtype (8–10). Selective FGFR inhibitors can be categorized into reversible (e.g. infigratinib, pemigatinib, erdafitinib, derazantinib, zoligratinib) and irreversible (e.g. futibatinib, lirafugratinib), based on their binding to the tyrosine kinase domain. Erdafitinib has demonstrated effectiveness in inhibiting FGFR3 in urothelial cancer (11,12), and shows activity across various FGFR2-driven tumors, reflecting the concept of molecularly-driven, tumor-agnostic targeted therapy (13). Similarly, pemigatinib and the two irreversible FGFR inhibitors, initially developed for FGFR2-driven intrahepatic cholangiocarcinoma, have shown efficacy across multiple tumor types in dose-expansion cohorts of phase I/II clinical trials (14–17).
Studies have identified frequent on-target, polyclonal mutations in the tyrosine kinase domain of FGFR2 as common mechanisms of resistance to selective FGFR inhibitors in FGFR2-driven cholangiocarcinoma (18,19). These findings are supported by functional validation of specific FGFR2 kinase domain mutations that confer resistance, with irreversible FGFR inhibitors designed to be effective against these mutations (20–26). Recently, off-target resistance mechanisms have been explored; pathogenic variants in the MAPK and PI3K/mTOR pathways have been detected at progression on selective FGFR inhibitors in patients with cholangiocarcinoma (21,23,27).
Importantly, resistance to FGFR inhibitors in the setting of FGFR2-driven disease has been predominantly reported in cholangiocarcinoma, except in four cases of FGFR2-driven tumors that progressed on pemigatinib (15,28). The broader application of FGFR inhibitors across different histologies underscores the need to identify and address resistance mechanisms in non-cholangiocarcinoma tumors, potentially offering universal strategies to counteract resistance.
In this study, we report on- and off-target resistance mechanisms to reversible and irreversible FGFR inhibitors across FGFR2-driven tumor types, validated through functional studies. Furthermore, the outcomes from sequential molecular treatments, applied to one-third of the patients with longitudinal monitoring of molecular alterations, provide insights into strategies to overcome resistance across a spectrum of FGFR2-driven solid tumors.
Methods
Patients and treatments
UNLOCK is an institutional program which aims to decipher mechanisms of action and resistance to innovative drugs
To be included in this cohort of the UNLOCK program, patients had to satisfy the following criteria: 1) Diagnosis of an advanced solid tumor requiring systemic treatment; 2) Molecular detection of an activating alteration (i.e. fusions/rearrangements, mutations) in the FGFR2 gene; 3) Having received a selective FGFR inhibitor, either reversible (pemigatinib, erdafitinib, infigratinib, derazantinib, zoligratinib, rogaratinib, fexagratinib) or irreversible (futibatinib or lirafugratinib); 4) having post-progression molecular analyses performed on circulating tumor DNA (ctDNA) and/or tissue biopsies. In two cases showing primary resistance to futibatinib (ST4455 and MR719), in the lack of the availability of post-progression samples, pre-treatment tissue biopsy and ctDNA were analyzed.
The molecular analyses were performed within four institutional studies at Gustave Roussy, whose aim is the molecular characterization of tumors: MATCH-R (NCT02517892) (29), MOSCATO (NCT01566019)(30), STING (NCT04932525), and CTC (NCT02666612).
Patients were treated in the setting of clinical trials or compassionate use programs allowing treatment with FGFR inhibitors on the basis of molecular selection. Disease response was measured according to RECIST 1.1, and progression-free survival (PFS) was calculated from the date of targeted inhibitor start to the day of radiological evidence of progression.
All patients participating in the mentioned studies were fully informed and signed a written informed consent. The studies have been approved by ethics committees in France (French National Agency for Medicines and Health Products Safety - ANSM), and are being conducted in accordance with the Declaration of Helsinki.
Molecular analyses
Post-progression tissue biopsies, when possible, underwent whole exome sequencing (WES), with or without concomitant RNA sequencing (RNA-seq). The lower limit for WES performance was a proportion of tumor cells ≥ 30% in the tissue sample. In cases with a proportion of tumor cells between 10% and 30%, molecular analyses with targeted next-generation sequencing (NGS) panels (Mosc-4, Oncomine v3) were performed. For WES, the mean coverage was 140X.
With regard to ctDNA analyses, they were performed with GuardantHealth, Illumina, Foundation Medicine or Integragen liquid biopsy panels. For each patient with longitudinal ctDNA assessment, only analyses performed with the same platform were reported.
Among the findings of the molecular reports, only molecular events potentially implicated in resistance were reported in the present study. FGFR2 kinase domain mutations were reported according to reference transcript NM_001144913.1, as previously reported by our group and others (20,21,31,32).
Site-directed mutagenesis
Lentiviral vectors expressing FGFR2:BICC1 fusions were created using the pLenti6/V5 directional TOPO Cloning Kit (#K495510, Thermo Fisher Scientific) according to the manufacturer's instructions. Point mutations in the FGFR2 kinase domain of the FGFR2::BICC1 fusion were introduced using the QuickChange XL Site-Directed Mutagenesis Kit (#200516, Agilent) according to manufacturer's protocol.
Cell lines
Ba/F3 cells were infected with lentiviral constructs, as reported previously (33), to express the FGFR2::BICC1 fusion, this latter with or without FGFR2 kinase domain mutations. Ba/F3 cells harboring the fusion were selected in the presence of blasticidin (14 mg/mL) and IL-3 (0.5 ng/mL) until recovery, and a second selection by culturing the cells in the absence of IL-3. FGFR2 fusion and FGFR2 kinase domain mutations were confirmed on the established cell lines by Sanger sequencing. The cells were not tested for Mycoplasma contamination, but cells were not maintained in culture for more than two months after establishment or thawing.
Cell viability assays were performed in 96-well plates using the CellTiter Glo Luminescent Cell Viability Assay (G7570, Promega). We seeded 4000 cells/well and we treated cells for 48h. Half-maximal inhibitory concentration (IC50) values were derived using GraphPad Prism software.
Reagents
Lirafugratinib was provided by Relay Therapeutics. Erdafitinib, infigratinib, fexagratinib, zoligratinib, derazantinib and futibatinib were purchased from Selleck Chemicals. Pemigatinib and rogaratinib were purchased from MedChemExpress.
Development of patient-derived xenografts and in vivo pharmacologic studies
All animal procedures and studies have been approved by the French Ministry of “Enseignement supérieur, de la Recherche et de l’Innovation” (APAFIS#2790-2015112015055793 and APAFIS#2328-2015101914074846). Fresh tumor fragments were implanted in the subrenal capsule of 6-week-old female NOD/SCID gamma (NSG) mice obtained from Charles River Laboratories. Patient-derived xenografts (PDX)-bearing NSG mice were treated with the indicated doses of pemigatinib, erdafitinib, futibatinib and lirafugratinib. Eight mice per group were treated for up to 50 days, and tumor volume and mouse weight were measured twice weekly.
Results
Patient population and molecular treatments
We studied 36 patients with advanced solid tumors driven by FGFR2, all of whom were progressing on selective FGFR inhibitors (Supplementary Table S1). This cohort included 27 patients with intrahepatic cholangiocarcinoma and nine patients with various other tumor types: two with high-grade serous ovarian cancer, and one each with lung adenocarcinoma, urothelial cancer, triple-negative breast cancer, duodenal cancer, pancreatic ductal adenocarcinoma, adrenocortical carcinoma, and cancer of unknown primary. The majority, 31 patients, had tumors harboring FGFR2 fusions, while five had tumors driven by FGFR2 mutations located in the extracellular domain (specifically, three with FGFR2 C383R, one with FGFR2 S267P, and one with FGFR2 Y376C). FGFR2 fusion partners included BICC1 in five cases (all in intrahepatic cholangiocarcinomas), TACC2 in three cases (n = 1 intrahepatic cholangiocarcinoma, n = 2 other tumor types), STRN4 and CCSSER2 in two cases, one from each cohort. Other unique fusion partners were found in the remaining 19 tumors, detailed in Supplementary Tables S2-3.
Twenty-three patients received a reversible FGFR inhibitor (n = 13 pemigatinib, n = 8 erdafitinib, n = 1 derazantinib, n = 1 zoligratinib), and 13 the irreversible inhibitor futibatinib (Supplementary Table S1). In the cholangiocarcinoma group, patients treated with reversible inhibitors and futibatinib showed 61% and 67% objective response rate, with median PFS of 8.7 and 11.1 months, respectively (Supplementary Table S4). Given the diversity in tumor origin, detailed clinical data of the non-cholangiocarcinoma patients is provided in Supplementary Table S3.
All patients underwent post-progression ctDNA analysis. Twenty-one of them had further molecular analyses performed on post-progression tissue biopsies, 16 via whole exome sequencing (WES) with or without RNA sequencing (RNA-seq), and five via targeted next-generation sequencing (NGS).
After disease progression on the first FGFR inhibitor, 12 patients received sequential targeted treatments. Eight patients with cholangiocarcinoma received futibatinib following a reversible inhibitor, with three receiving the mTOR inhibitor everolimus based on molecular findings. Two patients with cholangiocarcinoma and two with other tumor types, progressing respectively on pemigatinib and futibatinib, were switched to the FGFR2-selective inhibitor lirafugratinib (Supplementary Table S1).
Molecular alterations observed at resistance to selective FGFR inhibitors
In order to better approach the specificities of resistance mechanisms to reversible inhibitors versus the irreversible inhibitor futibatinib, and between intrahepatic cholangiocarcinoma and other tumor types, we separated below the different groups of patients analyzed at progression after a first FGFR inhibitor.
Intrahepatic cholangiocarcinoma on reversible inhibitors
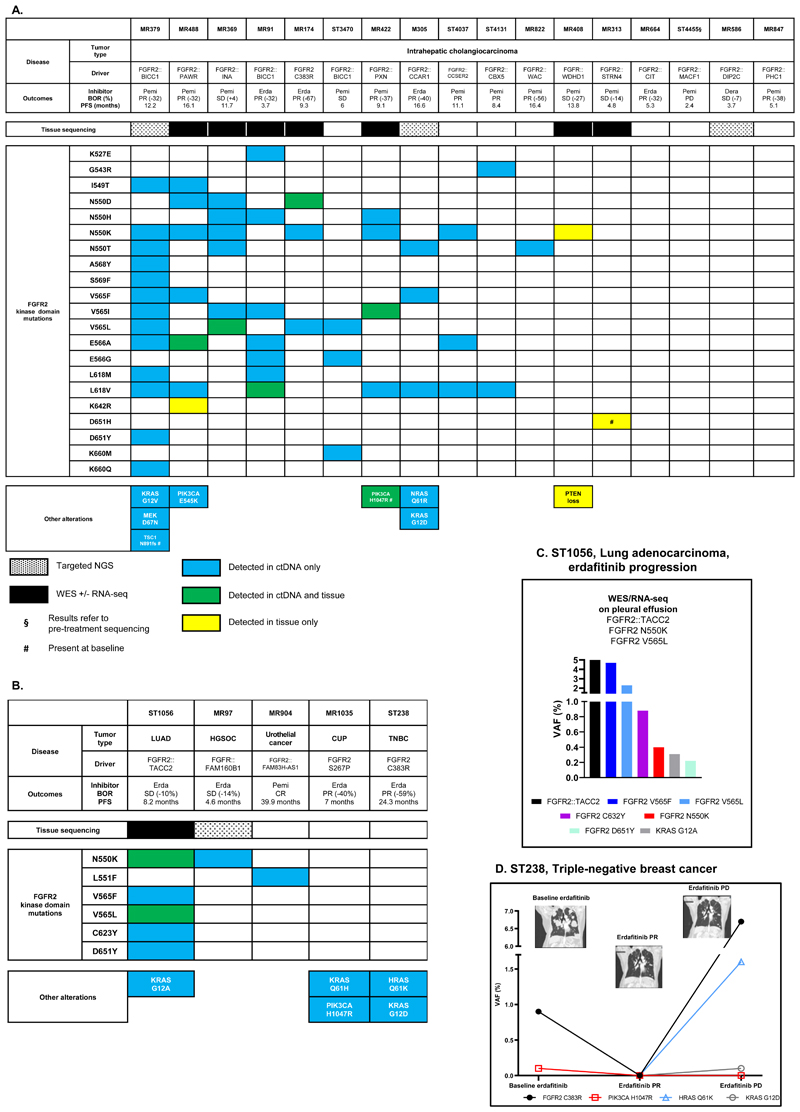
The molecular alterations detected in 17 patients with intrahepatic cholangiocarcinoma progressing on reversible inhibitors are reported in Figure 1A. Polyclonal kinase domain mutations (≥ 2 FGFR2 mutations in the same blood sample) were detected in 10 of these patients (59%).
Figure 1. Molecular findings at resistance to reversible FGFR inhibitors.
A: Patients suffering from intrahepatic cholangiocarcinoma. B: Patients suffering from other tumor types. C: Molecular findings of patient ST1056, suffering from a lung adenocarcinoma harboring a FGFR2::TACC2 fusion, at acquired progression to erdafitinib. D: Clinico-radiological and molecular evolution of patient ST238, suffering from a FGFR2 C383R-driven triple-negative breast cancer.
The ctDNA findings are reported as variant allele frequency (VAF, %).
BOR: Best objective response; PFS: Progression-free survival; PR: Partial response: SD: Stable disease; PD: Progressive disease; Pemi: Pemigatinib; Erda: Erdafitinib; Dera: Derazantinib; LUAD: Lung adenocarcinoma; HGSOC: High-grade serous ovarian cancer; CUP: Cancer of unknown primary; TBNC: Triple-negative breast cancer; CR: Complete response.
In three additional patients, a single FGFR2 mutation was detected either in the tissue biopsy (MR408 and MR313) or in ctDNA (MR822; FGFR2 N550T). Specifically, FGFR2 D651H was detected in both pre- and post-treatment biopsies of one patient (MR313). Another unique case (MR488) had two concurrent FGFR2 kinase domain mutations in a single tissue biopsy (E566A and K642R). Concurrent pathogenic alterations in the MAPK and PI3K/mTOR pathways, suggesting off-target resistance mechanisms, were observed in five patients.
Other tumor types on reversible inhibitors
Five patients with non-cholangiocarcinoma FGFR2-driven tumors exhibited diverse resistance patterns after initial tumor shrinkage (Figure 1B).
FGFR2 kinase domain mutations were detected in three of these patients, with one exhibiting polyclonal mutation. The latter (patient ST1056), was a FGFR2::TACC2 rearranged lung adenocarcinoma that progressed on erdafitinib with FGFR2 N550K, V565L/F, C632Y, D651Y mutations, as well as KRAS G12A (Figure 1C, Supplementary Figure S1).
In two patients, no FGFR2 kinase domain mutations aroused at progression to erdafitinib (MR1035, cancer of unknown primary; ST238, triple negative breast cancer, Figure 1D), but KRAS/PIK3CA and HRAS/KRAS mutations were detected in ctDNA at progression, respectively (Figure 1B).
Intrahepatic cholangiocarcinoma on futibatinib
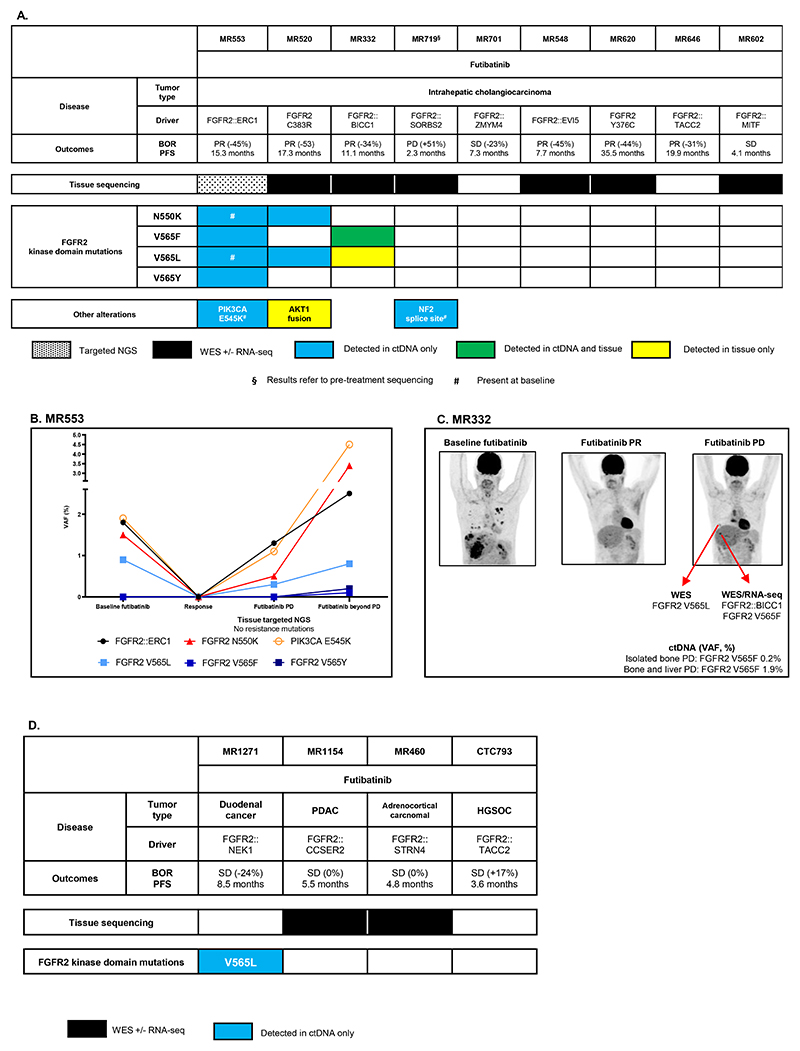
Among nine patients with intrahepatic cholangiocarcinoma progressing on futibatinib, fewer FGFR2 kinase domain mutations were observed compared to those on reversible inhibitors, mainly involving key regions like the molecular brake (N550) and gatekeeper (V565) (Figure 2A). Only three patients had polyclonal mutations, which were limited to N550K and V565F/L/Y.
Figure 2. Molecular findings at resistance to the irreversible FGFR inhibitor futibatinib.
A: Patients suffering from intrahepatic cholangiocarcinoma. B: Molecular evolution of patient MR553, suffering from an intrahepatic cholangiocarcinoma harboring a FGFR2::ERC1 fusion. C: Clinico-radiological and molecular evolution of patient MR332, suffering from an intrahepatic cholangiocarcinoma driven by a FGFR2::BICC1 fusion. D: Patients suffering from other tumor types.
The ctDNA findings are reported as variant allele frequency (VAF, %).
BOR: Best objective response; PFS: Progression-free survival; PR: Partial response: SD: Stable disease; PD: Progressive disease; PDAC: Pancreatic ductal adenocarcinoma; HGSOC: High-grade serous ovarian cancer.
One patient (MR553) showed pre-treatment FGFR2 kinase domain mutations that disappeared during response and re-emerged at progression (Figure 2B, Supplementary Figure S2).
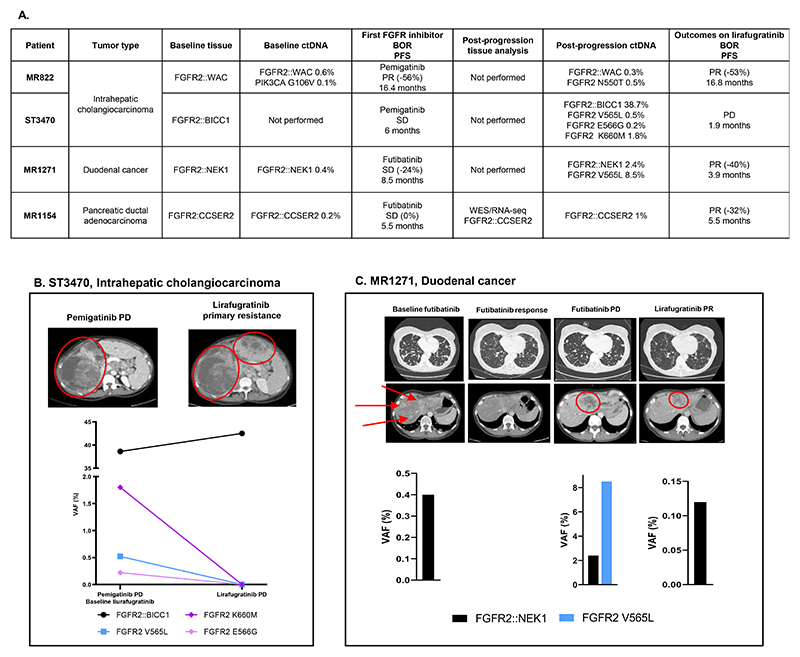
Interestingly, the patient MR332 firstly experienced an isolated bone progression, whose biopsy revealed a FGFR2 V565L mutation (not detectable in blood), followed by a hepatic progression harboring a FGFR2 V565F mutation (Figure 2C).
Other tumor types on futibatinib
Four patients with various tumor types showed resistance mechanisms to futibatinib (Figure 2D), including a duodenal cancer patient (MR1271) who exhibited a monoclonal FGFR2 V565L mutation concurrent with progression in the lung and liver (see Figure 5C).
Global analysis of candidate resistance mechanisms
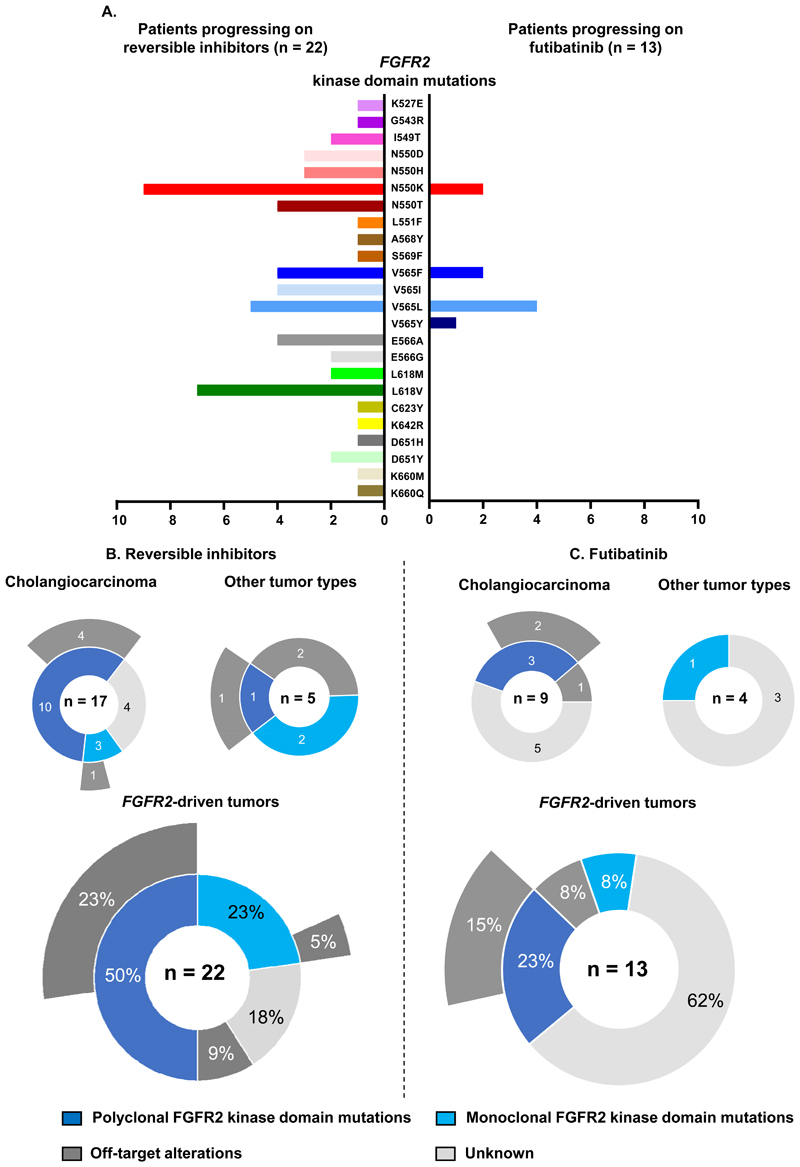
Comparing the spectrum of putative resistance mechanisms occurring in FGFR2-driven cholangiocarcinoma or other tumor types, we hypothesized that the two entities converged towards overlapping ways to escape targeted FGFR2 inhibition. We therefore pooled the molecular data of the two populations to allow a global view on resistance to a first FGFR inhibitor among FGFR2-driven tumor types (Figure 3).
Figure 3. Global view on candidate resistance mechanisms to FGFR inhibitors across FGFR2-driven malignancies.
A: Spectrum of FGFR2 kinase domain mutations detected across patients progressing to reversible inhibitors and futibatinib. B: Overview of the molecular alterations found at progression to reversible inhibitors in cholangiocarcinoma and other tumor types (upper panels), and pooled across all cases. C: Overview of the molecular alterations found at progression to the irreversible inhibitor futibatinib in cholangiocarcinoma and other tumor types (upper panels), and pooled across all cases.
Across the 36 patients, 14 residues in the FGFR2 kinase domain (K527, G543, I549, N550, L551, A568, S569, V565, E566, L618, C623, K642, D651, K660) were found mutated at progression. For six of them (N550, V565, E566, L618, D651 and K660), at least two possible substitutions were observed, thus representing 24 possible mutations (Figure 3A). FGFR2 C623Y and L551F were the only mutations found exclusively in non-cholangiocarcinoma (Figure 1B). FGFR2 C623Y has not previously been reported, while L551F has been described in the setting of cholangiocarcinoma progressing on infigratinib (19).
Polyclonal kinase domain mutations were detected in half (11/22) of the patients progressing on reversible inhibitors, almost exclusively with cholangiocarcinoma, whereas only three patients (23%) revealed polyclonal FGFR2 mutations after futibatinib (Figure 3B-C). When assessable due to amplicon sizes, the polyclonal FGFR2 kinase domain mutations were always detected in trans (i.e. on different alleles).
Overall, 61 FGFR2 kinase domain mutations were detected after reversible inhibitors, while only nine mutations were observed after futibatinib (Figure 3A). The most frequently mutated residues were the molecular brake N550 and the gatekeeper V565.
In 13/22 (59%) of the patients progressing on reversible inhibitors, at least one mutation affecting either of these two residues was found, whereas the two residues were co-mutated in eight cases (36%). N550 and V565 were also the residues with the highest number of different substitutions, as N550 D/H/K/T and V565F/I/L. L618V/M occurred in nine cases, followed by E566 (E566A/G) that was mutated in six patients.
In contrast, the N550 and V565 residues were the unique sites of FGFR2 kinase domain mutations found at progression to futibatinib, namely N550K (n = 2), V565F (n = 2), V565L (n = 4), and V565Y (n = 1) (Figure 2A, 3A). Of note, the mentioned molecular brake and gatekeeper mutations have been previously reported, with the exception of FGFR2 V565Y (34). This mutation is a novel entity, emerging from a double-base substitution in the corresponding Valine codon GTT, for which we hypothesize the sequential occurrence of single nucleotide substitutions, from V565F (F being coded by the codon TTT) to V565Y (Y being coded by TAT).
Off-target mutations that are potentially implicated in resistance, such as those affecting the MAPK (i.e. HRAS, KRAS, NRAS, MEK) and PI3K/mTOR pathways (i.e. PIK3CA, PTEN, TSC1), were found in 8/22 (36%) and 3/13 (23%) cases progressing on reversible inhibitors and futibatinib, respectively (Figure 1A-B, 2A, 3B-C). In eight patients (23%), these mutations co-occurred with FGFR2 kinase domain mutations. In two patients with other tumor types progressing on erdafitinib, they emerged without concomitant on-target alterations (Figure 1B).
Pooling the molecular data obtained at progression, we observed molecular candidates for resistance in 77% and 38% of the patients progressing on reversible inhibitors and futibatinib, respectively (Figure 3B-C).
Sequential treatment strategies
Within our study cohort, 33% of the patients (n = 12, including 10 with intrahepatic cholangiocarcinoma and one each from pancreatic and duodenal cancers) underwent sequential targeted therapy regimens (with longitudinal sampling) that included irreversible FGFR inhibitors and the mTOR inhibitor everolimus.
Sequential treatments including futibatinib and everolimus
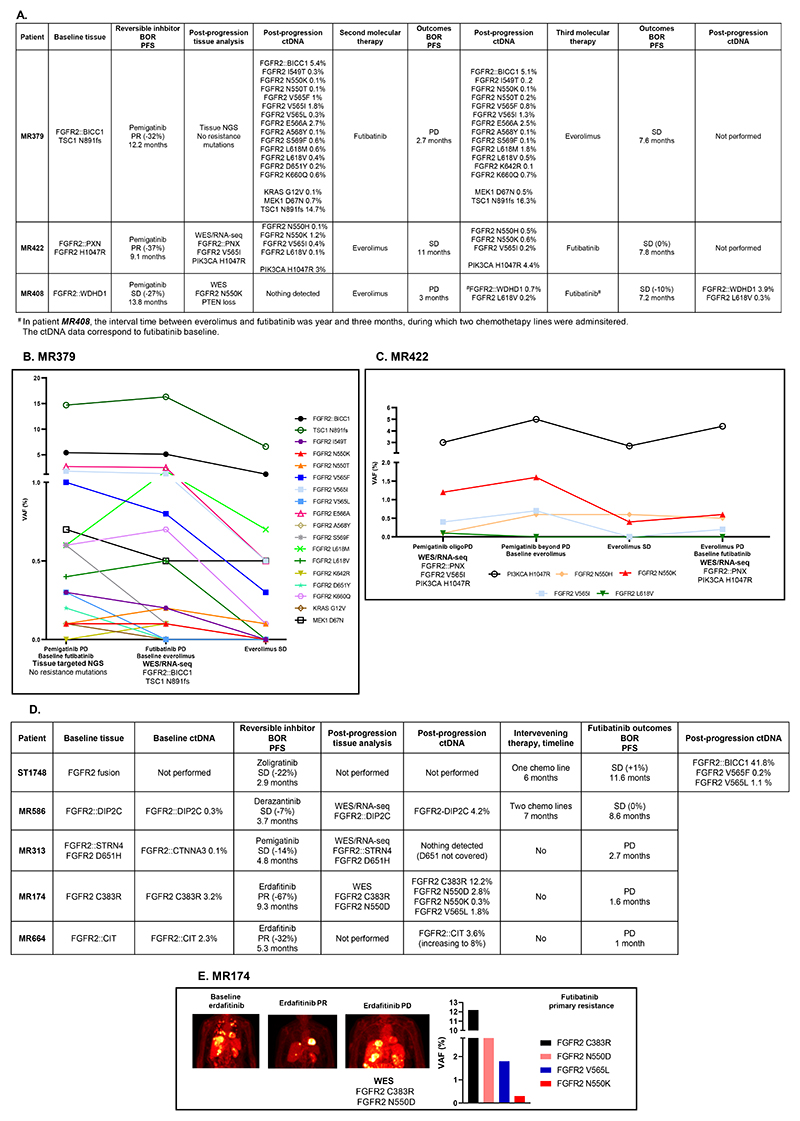
After progressing on reversible inhibitors, three patients with intrahepatic cholangiocarcinoma received a sequential treatment with futibatinib and everolimus based on specific molecular findings (Figure 4A):
Figure 4. Clinical and molecular evolution of patients with FGFR2-driven intrahepatic cholangiocarcinoma receiving sequential targeted treatments including futibatinib and everolimus.
A: Three patients received a sequential treatment of futibatinib and everolimus, this latter administered given the molecular finding of alterations in the PI3K/mTOR pathway. B: Molecular evolution of patient MR379, suffering from an FGFR2::BICC1 driven disease, with a concomitant pathogenic TSC1 frameshift mutation. C: Molecular evolution of patient MR422, suffering from a FGFR2-rearranged disease, with a concomitant PIK3CA H1047R mutation. D: Additional five patients received a sequence of reversible FGFR inhibitor followed by futibatinib. E: Clinico-radiological and molecular evolution of patient MR174, suffering from a FGFR2 C383R driven disease.
The ctDNA findings are reported as variant allele frequency (VAF, %).
BOR: Best objective response; PFS: Progression-free survival; PR: Partial response; SD: Stable disease; PD: Progressive disease; Chemo: Chemotherapy.
Patient MR379 suffered from a tumor harboring both FGFR2::BICC1 fusion and a TSC1 frameshift mutation in the baseline tissue sample. At progression to pemigatinib, the two alterations were found in ctDNA together with 13 different FGFR2 kinase domain mutations, KRAS and MEK1 mutations (Figure 4B). Futibatinib did not induce any clinical benefit (stable disease; PFS 2.7 months), while everolimus, administered due to the TCS1 loss-of-function alteration, led to stable disease with a PFS of 7.6 months. The clinical benefit was accompanied by the reduction of the allele frequencies of all the alterations in ctDNA (Figure 4B).
Patient MR422 experienced oligo-progression while on pemigatinib. A liver biopsy revealed a new FGFR2 V565I mutation alongside pre-existing FGFR2::PNX fusion and PIK3CA H1047R mutation (Figure 4A, C). Despite continued progression, pemigatinib treatment was extended, resulting in increased variant allele frequency (VAF) of PIK3CA H1047R, FGFR2 V565I, N550H and N550K mutations. Everolimus, initiated due to the PIK3CA mutation, provided stable disease for 11 months, corresponding to a decrease in the VAF of the documented mutations. Their VAF increased again at everolimus progression, and subsequent futibatinib administration allowed the achievement of disease stabilization with a PFS of 7.8 months.
Patient MR408 progressed to pemigatinib with an isolated lung nodule showing a FGFR2 N550K mutation and PTEN loss. Although everolimus showed no clinical activity, subsequent futibatinib treatment led to tumor shrinkage and a PFS of 7.2 months, despite the baseline documentation of a FGFR2 L618V mutation (Supplementary Figure S3).
Sequential treatments with reversible FGFR inhibitors followed by futibatinib
Five additional patients with FGFR2-driven cholangiocarcinoma were treated with futibatinib after experiencing resistance to reversible FGFR inhibitors (Figure 4D).
Two patients showed clinical benefit from futibatinib (MR586 and ST1748). Upon progression on futibatinib, FGFR2 V565F/L mutations were identified in patient ST1748, consistent with mutations typically seen in FGFR inhibitor-naïve patients. Three other patients experienced primary resistance to futibatinib after acquired resistance to reversible inhibitors. Of notice, FGFR2 N550D/K and V565L were present at futibatinib baseline in MR174, and could explain its lack of benefit (Figure 4E).
Sequential treatments with lirafugratinib
Four patients received lirafugratinib after progressing on a previous inhibitor (pemigatinib or futibatinib), without other intervening therapies (Figure 5A).
Figure 5. Clinical and molecular evolution of patients receiving a first FGFR inhibitor followed by the irreversible, highly-FGFR2 selective inhibitor lirafugratinib.
A: Four patients harboring a FGFR2 fusion received lirafugratinib as a second FGFR inhibitor. None received intercurrent treatment between the FGFR inhibitors.
B: Clinico-radiological and molecular evolution of patient ST3470, with a cholangiorcarcinoma driven by FGFR2::BICC1 progressing on pemigatinib. C: Patient MR1271 had duodenal cancer progressing on futibatinib with emergence of FGFR2 V565L mutation, that was cleared by lirafugratinib.
The ctDNA findings are reported as variant allele frequency (VAF, %).
BOR: Best objective response; PFS: Progression-free survival; PR: Partial response; SD: Stable disease; PD: Progressive disease.
Lirafugratinib outcomes were divergent among the two patients with FGFR2-rearranged cholangiocarcinoma progressing on pemigatinib.
Patient MR822, with a prolonged initial response to pemigatinib, developed an FGFR2 N550T mutation along with a persistent driver fusion FGFR2::WAC. Lirafugratinib treatment resulted in another prolonged response, highlighting its effectiveness against this specific mutation (Supplementary Figure S4A). Patient ST3470 encountered primary progression on lirafugratinib despite no detectable FGFR2 kinase domain mutations in ctDNA, suggesting an alternative resistance mechanism (Figure 5B). Importantly, three FGFR2 mutations V565L, E566G, and K660M present before lirafugratinib were lost at progression, suggesting their sensitivity to lirafugratinib.
Two other patients suffering from tumors other than cholangiocarcinoma also benefited from lirafugratinib after futibatinib progression (Figure 5A).
Patient MR1271 suffered from a FGFR2::NEK1 driven duodenal carcinoma, who progressed on futibatinib with the acquisition of FGFR2 V565L in ctDNA, and major disease response was observed with lirafugratinib (Figure 5C).
Patient MR1154 suffered from a pancreatic carcinoma harboring FGFR2::CCSER2 fusion. No molecular events potentially implicated in resistance to futibatinib were detected, but FGFR2::CCSER2 VAF was no longer detectable three weeks after lirafugratinib initiation (Supplementary Figure S4B).
FGFR2 kinase domain mutations exert a differential spectrum of resistance according to selective FGFR inhibitors
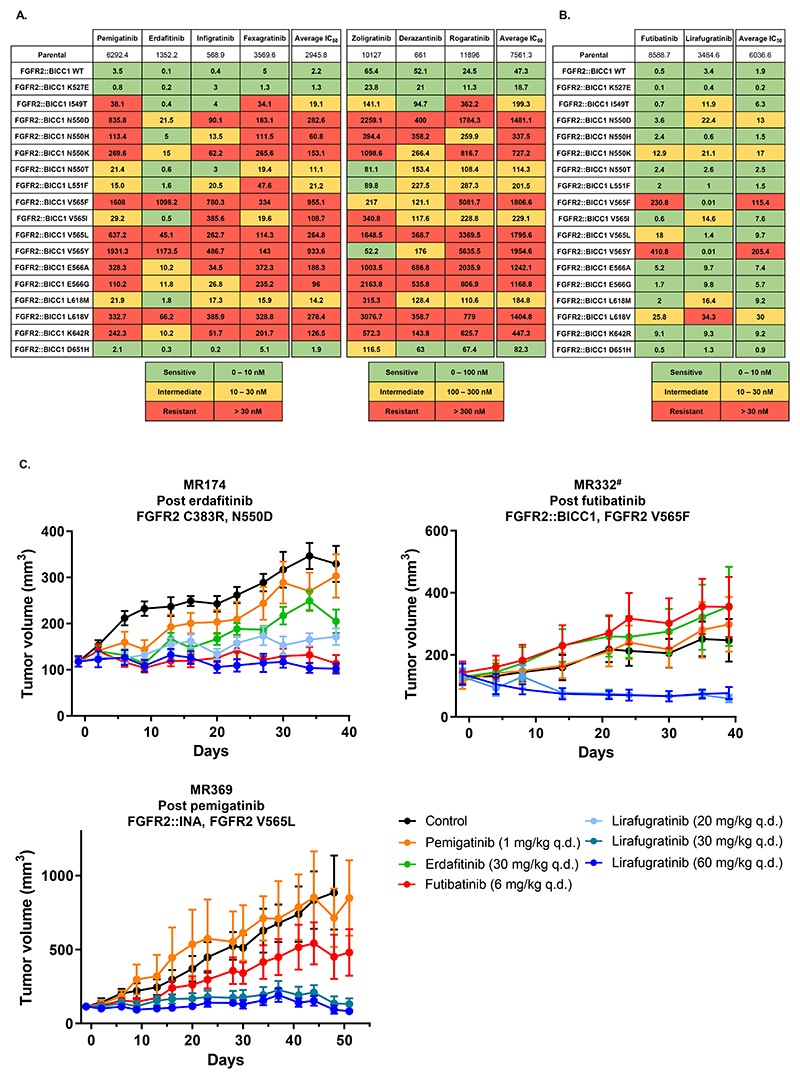
To explore how specific FGFR2 kinase domain mutations affect resistance to FGFR inhibitors, we used 18 Ba/F3 cell lines, engineered to express the FGFR2::BICC1 fusion with various secondary mutations. FGFR2::BICC1 was chosen being the most frequent fusion observed in our cohort and in other series (35). We exposed each Ba/F3 cell line to increasing concentrations of seven selective, reversible FGFR inhibitors and to the irreversible agents futibatinib and lirafugratinib, in order to establish their IC50 (Figures 6A-B, Supplementary Figure S5).
Figure 6. In vitro and in vivo evaluation of the activity of selective FGFR inhibitors against FGFR2 kinase domain mutations.
A: IC50 values of seven reversible FGFR inhibitors (and their average) against parental Ba/F3, FGFR2::BICC1 Ba/F3 (wild-type, WT), and 17 mutants. B: Graphical representation of the IC50 values of the two irreversible FGFR inhibitors futibatinib and lirafugratinib (and their average) against parental Ba/F3, FGFR2::BICC1 Ba/F3 (WT), and 17 mutants. We created two different cut-off thresholds, given the lower potency of zoligratinib, derazantinib and rogaratinib against WT FGFR2::BICC1 Ba/F3. In A and B, IC50 values (nmol/L) are reported as means of ≥ 3 independent datasets. C: Tumor growth kinetics in PDX models established from patients with cholangiocarcinoma, exposed to four FGFR inhibitors.
# MR332 PDX was established from the liver tissue biopsy harboring FGFR2 V565F, while the bone lesion harbored FGFR2 V565L (see Figure 2C).
q.d.: quaque die (i.e. daily).
In our experiments, erdafitinib emerged as the most potent inhibitor across all mutants, followed closely by infigratinib and futibatinib, achieving sub-nanomolar IC50 values against the wild-type FGFR2::BICC1 Ba/F3 cell line (Figure 6A-B).
The profiles of sensitivity and resistance conferred by individual FGFR2 kinase domain mutations matched with the spectrum of mutations emerging in patients treated with either reversible agents or futibatinib, respectively (Figure 3A, 6A-B). FGFR2 D651H did not confer resistance to any of the inhibitors, suggesting its role as a passenger event in patient MR313.
A significant finding from our study was the variable resistance patterns conferred by mutations at key residues within FGFR2, notably the molecular brake N550 and the gatekeeper V565. Mutations at these sites-N550K and V565F/L/Y-broadly conferred resistance across several reversible inhibitors, casting doubt on the effectiveness of using these drugs sequentially in patients with these mutations. However, other mutations like N550T and V565I seemed to result in a lesser degree of resistance. Interestingly, zoligratinib demonstrated relatively lower IC50 values against V565F/Y mutations compared to those against V565I/L, indicating specific interactions between the inhibitor and variant amino acids at this site.
The irreversible inhibitors generally showed superior activity compared to reversible agents in our Ba/F3 models. Both futibatinib and lirafugratinib exhibited efficacy within the 2-20 nmol/L range against various mutations, although they displayed slight differences in activity against certain mutations. Notably, lirafugratinib was particularly effective against FGFR2 V565F/Y mutations-mutations where futibatinib showed reduced activity due to steric hindrances from bulky amino acids like Phenylalanine and Tyrosine (IC50 > 200 nmol/L) (25,36). In contrast, the two irreversible inhibitors showed an opposite profile of activity against FGFR2 V565I and V565L mutants, with futibatinib being more active on V565I and lirafugratinib on V565L. The slightly difference in tridimensional structure between Leucine (V565L) and Isoleucine (V565I) likely explains the activity of the two inhibitors. In addition, FGFR2 V565I is known to increase basal activity of the kinase domain (37), and the higher potency of futibatinib could prevail in this setting.
The two irreversible inhibitors maintained an IC50 in the 10-20 nmol/L range for FGFR2 N550K, the most common mutation arising after a reversible inhibitor. Since FGFR2 N550K occurred in two patients progressing on futibatinib, and in five patients the gatekeeper FGFR2 V565L emerged (Figures 2A, 4D), we suppose that these two mutations cannot be overcome with clinically achievable concentrations of the agent.
In order to further validate our in vitro preclinical analyses on more clinically relevant models in vivo, we established PDXs from biopsies of patients with cholangiocarcinoma, collected at the time of acquired resistance to FGFR inhibitors. We treated three PDX models with pemigatinib, erdafitinib, futibatinib and lirafugratinib (Figure 6C). The FGFR2 N550D mutation in MR174 PDX, established at progression to erdafitinib, could be overcome by futibatinib and lirafugratinib. In addition, a dose-effect was noticed for lirafugratinib, with tumor growth abrogated only at the dose of 60 mg/kg (and not 20 mg/kg), which is in line with the IC50 observed in the Ba/F3 models. MR369 PDX, established at progression to pemigatinib, harbored a FGFR2 V565L mutation, while MR332 PDX, established at futibatinib resistance, harbored FGFR2 V565F (see Figure 2C). In both cases, only lirafugratinib (even at low doses) was able to prevent tumor growth in this in vivo model, confirming our suggestion that gatekeeper mutations FGFR2 V565L/F can be difficult to overcome with futibatinib in the clinical setting, while retaining sensitivity to lirafugratinib.
Of note, two of the PDX models (MR174 and MR369) were established from patients progressing with polyclonal FGFR2 kinase domain mutations detected in ctDNA, but only one mutation was found in the corresponding tissue biopsy and PDX (Figure 6C). These observations underscore the limitation of tissue biopsies to fully recapitulate the molecular spectrum of heterogeneity observed in patients at resistance.
Discussion
FGFR inhibition in FGFR2-driven malignancies marks a significant advance in precision oncology, emphasizing the need to understand molecular mechanisms behind drug resistance to develop new treatment strategies. Our study integrates extensive clinical and molecular data, along with in vitro and in vivo validation assays, to explore resistance mechanisms in patients with FGFR2-driven cancers, including cholangiocarcinoma and other tumor types.
Our findings confirm that polyclonality of FGFR2 kinase domain mutations is commonly observed in ctDNA from patients at progression on reversible inhibitors, particularly in cholangiocarcinoma. (18–20). Indeed, FGFR2 kinase domain mutations were undetectable or found as isolated entities in tissue analyses, compared to multiple alterations in ctDNA, highlighting the "polyclonal" nature of tumor progression and the fundamental role of liquid over tissue biopsy. Interestingly, such mutations were less common after treatment with the irreversible inhibitor futibatinib, which primarily affected the molecular brake N550 and gatekeeper V565 residues.
Recently, Wu and colleagues gathered data on resistance mechanisms in patients with FGFR2-driven cholangiocarcinoma, pooling evidence from published papers and meeting abstracts (21). In our study, we were able to differentiate between resistance to reversible inhibitors and futibatinib, providing clinical proof to their functional observations. As predicted by their evaluation of clinically achievable doses of futibatinib, in our cohort FGFR2 N550K frequently emerged at progression to the irreversible agent. Interestingly, we did not detected any mutation in the binding site for irreversible inhibitors (FGFR2 C492), in line with the reduced cellular fitness caused by these mutations, which somehow suggests their limited frequency of occurrence, such as the FGFR2 C492F found in the patient reported by Berchuck and colleagues (32). On the other hand, FGFR2 V565L, labeled by Wu and colleagues as sensitive to futibatinib, was the mutation most frequently observed at progression to the irreversible inhibitor in our cohort.
Similarly, resistance mechanisms in tumors other than cholangiocarcinoma mirrored those observed in cholangiocarcinoma, involving known FGFR2 residues and off-target resistance mechanisms. If considering the nine patients with other tumor types in our cohort, together with the report from Nicolò and colleagues (FGFR2 V565L detected at pemigatinib progression in a patient with breast cancer) (28), the emergence of polyclonal FGFR2 mutations was limited to only one patient with a lung adenocarcinoma progressing on erdafitinib (Figure 1C). More recently nevertheless, Rodón and colleagues detected polyclonal FGFR2 mutations in two patients with non-cholangiocarcinoma tumors progressing on pemigatinib (15). Whatsoever, the overall small number of patients with FGFR2-driven other tumor types evaluated at resistance challenges the conclusion that the propensity of developing polyclonal FGFR2 mutations is a feature more common in FGFR2-driven cholangiocarcinoma.
Further, in concomitance with FGFR2 kinase domain mutations or not, alterations in genes of the PI3K/mTOR pathway were frequently present at progression to reversible inhibitors and futibatinib, in patients suffering from cholangiocarcinoma or from other tumor types. Interestingly, in three patients, clinical benefit was obtained from reversible FGFR inhibitors or futibatinib, despite the presence of PI3K/mTOR alterations at baseline (MR379, MR422 and MR553), which were maintained at progression (Figure 1A and 2A). In line with our clinical observations, Wu and colleagues recently reported that the PI3KCA E545K mutation does not impact futibatinib sensitivity in the context of FGFR2-driven cholangiocarcinoma cell lines (21).
Here, the mTOR inhibitor everolimus provided clinical benefit in two patients with alterations in TSC1 or PIK3CA at progression to FGFR inhibition. The reduction in VAF of concomitant FGFR2 kinase domain mutations during everolimus treatment suggests that the on-target alterations probably emerged as an early event in a tumor clone already harboring the corresponding TSC1 or PI3KCA mutations (Figure 4B-C). It therefore seems that the loss of function of TSC1 and the activation of PIK3CA do not represent the bona fide molecular mechanisms responsible for resistance to FGFR inhibitors, still explaining the clinical benefit from everolimus.
The enrichment in MAPK pathway alterations in a setting similar to ours has been recently reported by DiPeri and colleagues in cholangiocarcinoma (27). Whether these mutations can be overcome by combination treatment in the clinical setting is still to be proven, as according to their data, only an in vitro synergistic effect of FGFR/MEK inhibition was achieved, with no meaningful effect in the in vivo model.
In the present study, irreversible inhibitors were also administered after progression to reversible ones in one third of the patients. We integrated the case-by-case analysis of the clinical response in presence of precise FGFR2 kinase domain mutations, with the dynamics of resistance mutations in ctDNA during treatment sequencing, and exploring functional data in Ba/F3 cellular models and matched PDX models. Nevertheless, the unique complexity of FGFR2-driven tumors at progression to reversible inhibitors, in terms of high levels of molecular heterogeneity, hampers the definition of precise patterns of resistance suitable for the sequential treatment with an irreversible agent. This is in contrast with single on-target mutations in EGFR- and ALK- driven lung cancer, overcome by the respective third generation inhibitors in the clinical setting (38,39). In our cohort and in line with other reports (6,40), in case of progression to a first FGFR inhibitor (reversible or futibatinib) mediated by a unique FGFR2 kinase domain mutation, the clinical activity profiles of futibatinib and lirafugratinib corresponded well to our functional assessment. We were indeed able to overcome resistance to reversible inhibitors and futibatinib due to mutations occurring in the FGFR2 gatekeeper residue (FGFR2 V565F/L). In line with the initial proofs from Subbiah and colleagues (25), the FGFR2-selective inhibitor lirafugratinib was active in our Ba/F3 cellular models, PDX and in patient MR1271 (FGFR2 V565L).
On the other hand, objective responses to futibatinib and lirafugratinib were observed even in cases with polyclonal FGFR2 kinase mutations (20,25). The relative abundance of each individual resistance mutation at baseline of the irreversible inhibitors, is suspected to influence the clinical response on a systemic scale. Emblematic in this sense is the evolution of our patient ST3470, experiencing primary resistance to lirafugratinib despite clearance of three FGFR2 kinase domain mutations (Figure 5B). Considering their better on-target activity, it is possible that progression to irreversible FGFR inhibitors occurs without detectable FGFR2 kinase domain mutations or off-target alterations, suggesting the implication of additional mechanisms, as indicated by the resistance study to futibatinib in our cohort (Figure 3C).
Given the unpredictability of resistance mechanisms and the corresponding activity of irreversible inhibitors administered in a sequential way, their administration as first anti-FGFR agents seems appropriate, in particular considering the outcomes of clinical activity reported in clinical trials in this setting (9,10). As showed here in two cases (Figure 5), switching from an irreversible inhibitor to another can also be a suitable therapeutic option.
This study, however, is not without limitations. Primarily, it relies on genomic analyses, potentially overlooking non-genetic factors like epithelial-mesenchymal transition or activation of alternate resistance pathways, as recently reported for EGFR in FGFR2-driven cholangiocarcinoma (41,42). Moreover, our focus is mainly on on-target resistance mechanisms, with less emphasis on proving the role of off-target events such as MAPK and PI3K/mTOR alterations. The lack of systematic tissue biopsy and ctDNA analysis at multiple timepoints for all patients may also have constrained the depth of our insights into resistance mechanisms. Finally, the lack of clinical data of resistance to lirafugratinib limits our observations of resistance to irreversible FGFR inhibitors.
In summary, the present work provides a global approach to apprehend resistance mechanisms to FGFR inhibitors across FGFR2-driven diseases, a clinical entity of major current interest given the development of active targeted agents. Our clinical and molecular findings were corroborated by functional analyses of FGFR2 kinase domain mutations in conferring resistance to different FGFR inhibitors. The additional clinical experience with sequential treatment with FGFR inhibitors or everolimus, together with the concomitant longitudinal study on resistance mechanisms, provided further valuable information both on the potential clinical management of patients and on the molecular correlates of resistance in this setting.
Supplementary Material
Statement of Translational Relevance.
The clinical benefit generated by selective FGFR inhibitors in patients with FGFR2-driven cancer is hampered by the inevitable occurrence of resistance. We analyzed post-progression ctDNA and tissue biopsies from 36 patients suffering from FGFR2-driven cholangiocarcinoma or other tumor types, at resistance to FGFR inhibitors. We were therefore able to recognize molecular traits, characteristics of resistance to reversible inhibitors versus the irreversible agent futibatinib, especially in terms of variety of FGFR2 kinase domain mutations involved in resistance. Compared to FGFR2-driven cholangiocarcinomas, polyclonal FGFR2 kinase domain mutations were less frequent in other tumor types. Twelve patients were treated with an additional FGFR irreversible inhibitor (futibatinib or lirafugratinib), and two derived benefit from everolimus. We integrated longitudinal molecular data with the clinical outcomes on these sequential targeted treatments, with functional evidence using Ba/F3 cellular models and patient-derived xenografts, aiming to propose molecular treatment strategies to overcome resistance.
Acknowledgments
We thank Illumina and Guardant Health for performing ctDNA sequencing analysis in the setting of an academic collaboration. The work of F. Facchinetti is supported by a grant from Philanthropia–Lombard Odier Foundation. The work of L. Friboulet is supported by an ERC Consolidator Grant (agreement number 101044047). Financial support was provided by Incyte, Debiopharm and Relay Therapeutics.
Footnotes
Conflicts of interest statement
FF received honoraria from BeiGene for advisory board.
YL received honoraria from AstraZeneca, Astellas, BMS, Gilead, Janssen, Merck KGaA, MSD and Pfizer, and received payments for travel and accommodations from Astellas, BMS, Janssen, Merck KGaA, MSD, Pfizer and Roche.
DV served as speaker for Roche and AstraZeneca.
SM declares consulting or advisory role for Biophytis, Servier, Yuhan, Roche, Kedrion Biopharma, IQvia.
AI reports research grants from Bayer, BMS, MSD, AstraZeneca, Roche, Novartis, Merck.
BB declares advisory boards (to institution): Abbvie, Biontech SE, BristolMyerSqibb, Chugai pharmaceutical, CureVac AG, Daiichi Sankyo, F. Hoffmann-La Roche Ltd, Pharmamar, Regeneron, Sanofi aventis, Turning Point Therapeutics; Conseil (to institution): Abbvie, Eli Lilly, Ellipses pharma Ltd, F.Hoffmann-La Roche Ltd, Genmab, Immunocore, Janssen, MSD, Ose Immunotherapeutics, Owkin, Taiho oncology; Steering committee (to institution): Astrazeneca, Beigene, GENMAB A/S, GlaxoSmithKline, Janssen, MSD, Ose Immunotherapeutics, Pharmamar, Roche-Genentech, Sanofi, Takeda; Speaker (to institution): Abbvie AstraZeneca, Chugai pharmaceutical, Daichii Sankyo, Hedera Dx, Janssen, MSD, Roche, Sanofi Aventis Springer Healthcare Ltd.
FA declares research grants (to institution): AstraZeneca, Daiichi Sankyo, Roche, Lilly, Pfizer, Owkin, Novartis, Guardant Health; Advisory board or speaker (to institution): AstraZeneca, Daiichi Sankyo, Roche, Lilly, Pfizer, Owkin, Novartis, Guardant Health, N-Power Medicine, SERVIER, Gilead, Boston Pharmaceutics; Advisory board (personal): Lilly.
AH received honoraria from Amgen, AstraZeneca, Debiopharm, Eli Lilly and Company, Incyte Corporation, QED Therapeutics.
LF declares research funding from Debiopharm, Incyte, Relay Therapeutics, Sanofi and Nuvalent.
Data availability
WES/RNA-seq raw data files from this study are deposited at the European Genome–phenome Archive (EGA) using the accession code EGAD50000000439. Access to this shared dataset is controlled by the institutional Data Access Committee, and requests for access can be sent to the corresponding author. Further information about EGA can be found at https://ega-archive.org/. Any additional information required to reanalyze the data reported in this article is available upon request from the corresponding author.
References
- 1.Helsten T, Elkin S, Arthur E, Tomson BN, Carter J, Kurzrock R. The FGFR Landscape in Cancer: Analysis of 4,853 Tumors by Next-Generation Sequencing. Clin Cancer Res. 2016;22:259–67. doi: 10.1158/1078-0432.CCR-14-3212. [DOI] [PubMed] [Google Scholar]
- 2.Murugesan K, Necchi A, Burn TC, Gjoerup O, Greenstein R, Krook M, et al. Pan-tumor landscape of fibroblast growth factor receptor 1-4 genomic alterations. ESMO Open. 2022;7:100641. doi: 10.1016/j.esmoop.2022.100641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pearson A, Smyth E, Babina IS, Herrera-Abreu MT, Tarazona N, Peckitt C, et al. High-Level Clonal FGFR Amplification and Response to FGFR Inhibition in a Translational Clinical Trial. Cancer Discov. 2016;6:838–51. doi: 10.1158/2159-8290.CD-15-1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Graham RP, Barr Fritcher EG, Pestova E, Schulz J, Sitailo LA, Vasmatzis G, et al. Fibroblast growth factor receptor 2 translocations in intrahepatic cholangiocarcinoma. Hum Pathol. 2014;45:1630–8. doi: 10.1016/j.humpath.2014.03.014. [DOI] [PubMed] [Google Scholar]
- 5.Arai Y, Totoki Y, Hosoda F, Shirota T, Hama N, Nakamura H, et al. Fibroblast growth factor receptor 2 tyrosine kinase fusions define a unique molecular subtype of cholangiocarcinoma. Hepatology. 2014;59:1427–34. doi: 10.1002/hep.26890. [DOI] [PubMed] [Google Scholar]
- 6.Cleary JM, Raghavan S, Wu Q, Li YY, Spurr LF, Gupta HV, et al. FGFR2 Extracellular Domain In-Frame Deletions Are Therapeutically Targetable Genomic Alterations That Function as Oncogenic Drivers in Cholangiocarcinoma. Cancer Discov. 2021;11:2488–505. doi: 10.1158/2159-8290.CD-20-1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zingg D, Bhin J, Yemelyanenko J, Kas SM, Rolfs F, Lutz C, et al. Truncated FGFR2 is a clinically actionable oncogene in multiple cancers. Nature. 2022;608:609–17. doi: 10.1038/s41586-022-05066-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Abou-Alfa GK, Sahai V, Hollebecque A, Vaccaro G, Melisi D, Al-Rajabi R, et al. Pemigatinib for previously treated, locally advanced or metastatic cholangiocarcinoma: a multicentre, open-label, phase 2 study. Lancet Oncol. 2020;21:671–84. doi: 10.1016/S1470-2045(20)30109-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Goyal L, Meric-Bernstam F, Hollebecque A, Valle JW, Morizane C, Karasic TB, et al. Futibatinib for FGFR2-Rearranged Intrahepatic Cholangiocarcinoma. N Engl J Med. 2023;388:228–39. doi: 10.1056/NEJMoa2206834. [DOI] [PubMed] [Google Scholar]
- 10.Hollebecque A. LBA12 - Efficacy of RLY-4008, a highly selective FGFR2 inhibitor in patients (pts) with an FGFR2-fusion or rearrangement (f/r), FGFR inhibitor (FGFRi)-naïve cholangiocarcinoma (CCA): ReFocus trial. 2022 [Google Scholar]
- 11.Loriot Y, Necchi A, Park SH, Garcia-Donas J, Huddart R, Burgess E, et al. Erdafitinib in Locally Advanced or Metastatic Urothelial Carcinoma. N Engl J Med. 2019;381:338–48. doi: 10.1056/NEJMoa1817323. [DOI] [PubMed] [Google Scholar]
- 12.Loriot Y, Matsubara N, Park SH, Huddart RA, Burgess EF, Houede N, et al. Erdafitinib or Chemotherapy in Advanced or Metastatic Urothelial Carcinoma. N Engl J Med. 2023;389:1961–71. doi: 10.1056/NEJMoa2308849. [DOI] [PubMed] [Google Scholar]
- 13.Pant S, Schuler M, Iyer G, Witt O, Doi T, Qin S, et al. Erdafitinib in patients with advanced solid tumours with FGFR alterations (RAGNAR): an international, single-arm, phase 2 study. Lancet Oncol. 2023;24:925–35. doi: 10.1016/S1470-2045(23)00275-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Subbiah V, Iannotti NO, Gutierrez M, Smith DC, Féliz L, Lihou CF, et al. FIGHT-101, a first-in-human study of potent and selective FGFR 1-3 inhibitor pemigatinib in pan-cancer patients with FGF/FGFR alterations and advanced malignancies. Ann Oncol. 2022;33:522–33. doi: 10.1016/j.annonc.2022.02.001. [DOI] [PubMed] [Google Scholar]
- 15.Rodón J, Damian S, Furqan M, García-Donas J, Imai H, Italiano A, et al. Pemigatinib in previously treated solid tumors with activating FGFR1-FGFR3 alterations: phase 2 FIGHT-207 basket trial. Nat Med. 2024 doi: 10.1038/s41591-024-02934-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Meric-Bernstam F, Bahleda R, Hierro C, Sanson M, Bridgewater J, Arkenau H-T, et al. Futibatinib, an Irreversible FGFR1-4 Inhibitor, in Patients with Advanced Solid Tumors Harboring FGF/FGFR Aberrations: A Phase I Dose-Expansion Study. Cancer Discov. 2022;12:402–15. doi: 10.1158/2159-8290.CD-21-0697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schram AM, Subbiah V, Liao C-Y (Andy), Moreno V, Desamparados R, Ponz-Sarvise M, et al. Abstract IA006: Clinical activity of lirafugratinib (RLY-4008), a highly selective FGFR2 inhibitor, in patients with advanced FGFR2-altered solid tumors: The ReFocus study. Molecular Cancer Therapeutics. 2023;22:IA006 [Google Scholar]
- 18.Goyal L, Saha SK, Liu LY, Siravegna G, Leshchiner I, Ahronian LG, et al. Polyclonal Secondary FGFR2 Mutations Drive Acquired Resistance to FGFR Inhibition in Patients with FGFR2 Fusion-Positive Cholangiocarcinoma. Cancer Discov. 2017;7:252–63. doi: 10.1158/2159-8290.CD-16-1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Varghese AM, Patel J, Janjigian YY, Meng F, Selcuklu SD, Iyer G, et al. Noninvasive Detection of Polyclonal Acquired Resistance to FGFR Inhibition in Patients With Cholangiocarcinoma Harboring FGFR2 Alterations. JCO Precis Oncol. 2021;5:PO.20.00178. doi: 10.1200/PO.20.00178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Goyal L, Shi L, Liu LY, Fece de la Cruz F, Lennerz JK, Raghavan S, et al. TAS-120 Overcomes Resistance to ATP-Competitive FGFR Inhibitors in Patients with FGFR2 Fusion-Positive Intrahepatic Cholangiocarcinoma. Cancer Discov. 2019;9:1064–79. doi: 10.1158/2159-8290.CD-19-0182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wu Q, Ellis H, Siravegna G, Michel AG, Norden BL, Fece de la Cruz F, et al. Landscape of Clinical Resistance Mechanisms to FGFR Inhibitors in FGFR2-Altered Cholangiocarcinoma. Clin Cancer Res. 2024;30:198–208. doi: 10.1158/1078-0432.CCR-23-1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Krook MA, Bonneville R, Chen H-Z, Reeser JW, Wing MR, Martin DM, et al. Tumor heterogeneity and acquired drug resistance in FGFR2-fusion-positive cholangiocarcinoma through rapid research autopsy. Cold Spring Harb Mol Case Stud. 2019;5:a004002. doi: 10.1101/mcs.a004002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Krook MA, Lenyo A, Wilberding M, Barker H, Dantuono M, Bailey KM, et al. Efficacy of FGFR Inhibitors and Combination Therapies for Acquired Resistance in FGFR2-Fusion Cholangiocarcinoma. Mol Cancer Ther. 2020;19:847–57. doi: 10.1158/1535-7163.MCT-19-0631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sootome H, Fujita H, Ito K, Ochiiwa H, Fujioka Y, Ito K, et al. Futibatinib Is a Novel Irreversible FGFR 1-4 Inhibitor That Shows Selective Antitumor Activity against FGFR-Deregulated Tumors. Cancer Res. 2020;80:4986–97. doi: 10.1158/0008-5472.CAN-19-2568. [DOI] [PubMed] [Google Scholar]
- 25.Subbiah V, Sahai V, Maglic D, Bruderek K, Touré BB, Zhao S, et al. RLY-4008, the First Highly Selective FGFR2 Inhibitor with Activity across FGFR2 Alterations and Resistance Mutations. Cancer Discov. 2023;13:2012–31. doi: 10.1158/2159-8290.CD-23-0475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Balasooriya ER, Wu Q, Ellis H, Zhen Y, Norden BL, Corcoran RB, et al. The irreversible FGFR inhibitor KIN-3248 overcomes FGFR2 kinase domain mutations. Clin Cancer Res. 2024 doi: 10.1158/1078-0432.CCR-23-3588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.DiPeri TP, Zhao M, Evans KW, Varadarajan K, Moss T, Scott S, et al. Convergent MAPK pathway alterations mediate acquired resistance to FGFR inhibitors in FGFR2 fusion-positive cholangiocarcinoma. J Hepatol. 2024;80:322–34. doi: 10.1016/j.jhep.2023.10.041. [DOI] [PubMed] [Google Scholar]
- 28.Nicolò E, Munoz-Arcos L, Vagia E, D’Amico P, Reduzzi C, Donahue J, et al. Circulating Tumor DNA and Unique Actionable Genomic Alterations in the Longitudinal Monitoring of Metastatic Breast Cancer: A Case of FGFR2-KIAA1598 Gene Fusion. JCO Precis Oncol. 2023;7:e2200702. doi: 10.1200/PO.22.00702. [DOI] [PubMed] [Google Scholar]
- 29.Recondo G, Mahjoubi L, Maillard A, Loriot Y, Bigot L, Facchinetti F, et al. Feasibility and first reports of the MATCH-R repeated biopsy trial at Gustave Roussy. NPJ Precis Oncol. 2020;4:27. doi: 10.1038/s41698-020-00130-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Massard C, Michiels S, Ferté C, Le Deley M-C, Lacroix L, Hollebecque A, et al. High-Throughput Genomics and Clinical Outcome in Hard-to-Treat Advanced Cancers: Results of the MOSCATO 01 Trial. Cancer Discov. 2017;7:586–95. doi: 10.1158/2159-8290.CD-16-1396. [DOI] [PubMed] [Google Scholar]
- 31.Facchinetti F, Hollebecque A, Braye F, Vasseur D, Pradat Y, Bahleda R, et al. Resistance to Selective FGFR Inhibitors in FGFR-Driven Urothelial Cancer. Cancer Discov. 2023;13:1998–2011. doi: 10.1158/2159-8290.CD-22-1441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Berchuck JE, Facchinetti F, DiToro DF, Baiev I, Majeed U, Reyes S, et al. The clinical landscape of cell-free DNA alterations in 1671 patients with advanced biliary tract cancer. Ann Oncol. 2022;33:1269–83. doi: 10.1016/j.annonc.2022.09.150. [DOI] [PubMed] [Google Scholar]
- 33.Friboulet L, Li N, Katayama R, Lee CC, Gainor JF, Crystal AS, et al. The ALK inhibitor ceritinib overcomes crizotinib resistance in non-small cell lung cancer. Cancer Discov. 2014;4:662–73. doi: 10.1158/2159-8290.CD-13-0846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Valery M, Vasseur D, Fachinetti F, Boilève A, Smolenschi C, Tarabay A, et al. Targetable Molecular Alterations in the Treatment of Biliary Tract Cancers: An Overview of the Available Treatments. Cancers (Basel) 2023;15:4446. doi: 10.3390/cancers15184446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Silverman IM, Hollebecque A, Friboulet L, Owens S, Newton RC, Zhen H, et al. Clinicogenomic Analysis of FGFR2-Rearranged Cholangiocarcinoma Identifies Correlates of Response and Mechanisms of Resistance to Pemigatinib. Cancer Discov. 2021;11:326–39. doi: 10.1158/2159-8290.CD-20-0766. [DOI] [PubMed] [Google Scholar]
- 36.Schönherr H, Ayaz P, Taylor AM, Casaletto JB, Touré BB, Moustakas DT, et al. Discovery of lirafugratinib (RLY-4008), a highly selective irreversible small-molecule inhibitor of FGFR2. Proc Natl Acad Sci U S A. 2024;121:e2317756121. doi: 10.1073/pnas.2317756121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Byron SA, Chen H, Wortmann A, Loch D, Gartside MG, Dehkhoda F, et al. The N550K/H mutations in FGFR2 confer differential resistance to PD173074, dovitinib, and ponatinib ATP-competitive inhibitors. Neoplasia. 2013;15:975–88. doi: 10.1593/neo.121106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mok TS, Wu Y-L, Ahn M-J, Garassino MC, Kim HR, Ramalingam SS, et al. Osimertinib or Platinum-Pemetrexed in EGFR T790M-Positive Lung Cancer. N Engl J Med. 2017;376:629–40. doi: 10.1056/NEJMoa1612674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shaw AT, Solomon BJ, Besse B, Bauer TM, Lin C-C, Soo RA, et al. ALK Resistance Mutations and Efficacy of Lorlatinib in Advanced Anaplastic Lymphoma Kinase-Positive Non-Small-Cell Lung Cancer. J Clin Oncol. 2019;37:1370–9. doi: 10.1200/JCO.18.02236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rengan AK, Denlinger CS. Robust Response to Futibatinib in a Patient With Metastatic FGFR-Addicted Cholangiocarcinoma Previously Treated Using Pemigatinib. J Natl Compr Canc Netw. 2022:1–6. doi: 10.6004/jnccn.2021.7121. [DOI] [PubMed] [Google Scholar]
- 41.Gainor JF, Dardaei L, Yoda S, Friboulet L, Leshchiner I, Katayama R, et al. Molecular Mechanisms of Resistance to First-and Second-Generation ALK Inhibitors in ALK-Rearranged Lung Cancer. Cancer Discov. 2016;6:1118–33. doi: 10.1158/2159-8290.CD-16-0596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wu Q, Zhen Y, Shi L, Vu P, Greninger P, Adil R, et al. EGFR Inhibition Potentiates FGFR Inhibitor Therapy and Overcomes Resistance in FGFR2 Fusion-Positive Cholangiocarcinoma. Cancer Discov. 2022;12:1378–95. doi: 10.1158/2159-8290.CD-21-1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
WES/RNA-seq raw data files from this study are deposited at the European Genome–phenome Archive (EGA) using the accession code EGAD50000000439. Access to this shared dataset is controlled by the institutional Data Access Committee, and requests for access can be sent to the corresponding author. Further information about EGA can be found at https://ega-archive.org/. Any additional information required to reanalyze the data reported in this article is available upon request from the corresponding author.