Abstract
Background
Immunoglobulin E (IgE) antibodies play a role in allergic disease.
Results
IgE has a bent conformation in solution that becomes more bent upon binding to the FcεRI receptor, but less bent upon binding the anti-IgE omalizumab.
Conclusions
Conformational change is critical for FcεRI-mediated IgE activity.
Significance
The bent structure provides a molecular rationale for the susceptibility of IgE-FcεRI complexes to allergenic stimulation.
Keywords: antibody, omalizumab, CD23, FRET biosensor, homoFRET
The incidence of allergic disease is on the increase world-wide, and this includes a range of conditions from seasonal hayfever to asthma and fatal anaphylactic shock triggered by allergens such as peanuts. The key mediator between the allergen and the cells of the immune system is the antibody immunoglobulin E (IgE) (1). In contrast to the comparatively high levels of the other immunoglobulin isotypes (~2 mg/ml for IgA and 13 mg/ml for IgG), IgE is only found at low levels (~50–200 ng/ml) in serum. Nevertheless, IgE binds with high affinity (Ka ~ 1010 M−1) through its Fc region to the receptor FcεRI expressed on the surface of mast cells and basophils, and this high affinity and long lifetime of receptor-bound IgE in tissue (dissociation t1/2 ~14 days) (2, 3) accounts for the allergic sensitization and rapid activation of these cells upon allergen binding. Degranulation of these cells and the release of inflammatory mediators, triggered by often innocuous allergens, leads to the physiological responses associated with allergic reactions (1, 4), although the primary function of IgE may be to respond to multicellular parasitic pathogens such as helminths (5).
In IgG, the two Fab (antigen-binding) arms are connected to the Fc (a dimer of Cγ2 and Cγ3 domains) by a disulfide-linked hinge region. IgE however has the (Cε2)2 domain pair in place of the hinge, and thus IgEFc comprises six domains (a dimer of Cε2, Cε3 and Cε4, disulphide-linked between the Cε2 domains). The crystal structure of IgEFc revealed the (Cε2)2 domain pair bent back against the Cε3 domains and even contacting one of the Cε4 domains (6). This structure was even more acutely bent than that proposed in an earlier FRET study with a chimaeric IgE (7) or an X-ray and neutron scattering study of IgEFc in solution (8). Comparative studies of the kinetics of binding to the soluble extracellular domains of the IgE-binding α-chain of FcεRI (sFcεRIα) of both IgEFc and a sub-fragment lacking the Cε2 domains (IgEFc3-4), demonstrated that the Cε2 domains were in part responsible for the high affinity and slow dissociation rate (9). Although an “unbending” of the IgEFc was proposed to allow engagement of the Cε2 domains with FcεRI (4), IgEFc was found to remain bent – in fact the bend is enhanced – in the crystal structure of the IgEFc·sFcεRIα complex (10).
In order to discover whether the conformations and conformational changes seen in the crystal structures occur in solution, we generated an IgEFc biosensor to monitor conformational change in this region of the molecule. Previous solution-based fluorescence depolarization studies of human and murine IgE provided evidence only of flexibility of the Fabs relative to Fc (11-13); this was also observed by electron microscopy (14). FRET studies using recombinant mouse chimaeric IgE (with Cε4 replaced by Cγ3 of IgG) did reveal a small change in energy transfer on binding to rat FcεRI (7,15), but the donor fluorophore was placed in the Fab antigen-binding site rather than in Fc, making it difficult to distinguish between movements within Fc from those of the Fabs relative to Fc. We report here a conformational change within human IgEFc monitored by FRET between biologically encoded probes attached to the N- and C- termini of the molecule. Both hetero- and homo-energy transfer were measured using a combination of steady-state and excited-state decay techniques. These measurements were made in the presence and absence of soluble forms of the “high-affinity” mast cell receptor FcεRIα, and the “low-affinity” B cell receptor CD23 (FcεRII), as well as the Fab of the therapeutic anti-IgE antibody omalizumab (Xolair™). The results reveal not only that IgEFc is bent in solution, but also that the bend is enhanced upon binding to FcεRIα. CD23 binding has no effect upon the bend, while the anti-IgE omalizumab causes an “unbending”. Finally, we modeled the disposition of the Fabs relative to FcεRI-bound IgEFc and discovered a significant difference compared with receptor-bound IgG, which may have important implications for cross-linking of receptor-bound IgE and the activation of mast cells and basophils by allergens.
EXPERIMENTAL PROCEDURES
Cloning, expression and purification of recombinant proteins
cDNA sources were as follows: pRY 24 (16) for IgEFc (including secretion tag); pEGFP-N1 C (Clontech) for the minimal eGFP residues 1-230 (17); pRSET mRFP1 (kind gift of Roger Tsien (18) for mRFP). For expression the cDNAs were inserted into the pCEP4 vector (Invitrogen). In total four vectors were made: eGFP-IgEFc, an IgEFc labeled N-terminally with eGFP; mRFP-IgEFc, an IgEFc labeled N-terminally with mRFP; mRFP-IgEFc-eGFP, a dual labeled IgEFc with mRFP at the N-terminus and eGFP at the C-terminus; IgEFc-eGFP, an IgEFc C-terminally labeled with eGFP (see Supplemental Fig. S1). Each construct was individually transfected into HEK 293E cells using polyethyleneimine (PEI) (19). Transfection positive cells were selected using hygromycin (according to instructions provided with the pCEP4 vector) and maintained in triple layered tissue culture flasks (Nunc). Supernatants were harvested at regular intervals and active IgE fusion protein purified by affinity chromatography using a sFcεRIα receptor column (sFcεRIα-IgG4Fc fusion protein) (20). Monomeric protein was isolated by size-exclusion chromatography (21). Omalizumab Fabs were produced by papain digestion of whole antibody (kind gift of Gerald Dubois, Novartis Pharmaceuticals) using the Pierce Fab preparation kit (Thermo Scientific) followed by anion-exchange chromatography. sFcεRIα, the IgEFc3-4 fragment, whole IgE, IgEFc, derCD23 and eGFP were produced as described previously (16, 20, 22-24).
Biochemical and biophysical characterization of fluorescently labeled IgEFc
Analytical size exclusion chromatography was used to test the “bindability” of stored protein preparations as previously described (25) and also on material prior, and subsequent to all FRET assays. All fluorescent fusion proteins were compared to whole IgE for their affinity and kinetics of binding to sFcεRIα, by surface plasmon resonance using a Biacore 3000 as described previously (21). Dual polarization interferometry (DPI) measurements (26) were made using an Analight Instrument (Farfield). All experiments were carried out at 20°C. An active FcεRI surface was generated by amine coupling sFcεRIα-IgG4Fc fusion protein (20) to the chip at 100 μg/ml in HEPES buffered saline (HBS), after pre-activation of the chip surface with sulpho-GMBS cross-linker (Thermo Lifesciences). The surface was exposed to protein for ~8 minutes before remaining reactive esters were blocked using 1M ethanolamine, pH 8, for 2 minutes. All experiments were carried out and analyzed according to the manufacturer’s instructions.
Steady-state energy transfer measurements of fluorescently labeled IgEFc
Spectra were collected using a Spex Fluoromax (Jobin-Yvon) fluorimeter with polarizers set to vertical on excitation and magic angle (54.7°) on emission, to remove any polarization bias in the collected data. Measurements were made in a 50μl fluorescence cuvette maintained at 20°C in a thermostatted cell holder with concentration-matched (200 nM) samples. Comparisons were made on the basis of acceptor mRFP fluorescence at 610nm (monochromator bandwidth of 2 nm) with excitation wavelengths from 350-600 nm (bandwidth 2 nm). All spectra were corrected for the intensity of the source. Relevant spectra were also corrected for bleed-through of emission seen at 610nm as a result of excitation of eGFP (always minimal); this was detected using IgEFc constructs labeled with eGFP alone. FRET efficiency (E) was calculated according to Lakowicz (27) based on the published extinction co-efficient (ε) for the acceptor, mRFP, at the peak excitation wavelength for the eGFP donor , and the extinction coefficient for eGFP at the same wavelength (24), together with the intensities of the free acceptor at the peak emission wavelength for the acceptor and donor in the presence of acceptor at this wavelength using equation 1.
| (Eq. 1) |
Fluorescence lifetime measurements of IgEFc constructs
Measurements were made using an Edinburgh Instruments Lifespec fitted with a cooled MCP-PMT, emission polarizer and monochromator set at 510 nm (bandwidth 4 nm) for eGFP fluorescence, and a 468nm polarized pulsed laser diode light source. The polarizer settings were the same as for the steady-state measurements. Sample preparation was identical to that used for steady-state measurements, with all experiments carried out in a 50μl fluorescence cuvette at 20°C maintained in a thermostatted holder. Data were analyzed using TRI2 (28) updated with a single transient module. The photophysics of eGFP are complex, such that there are at least two electronic ground states and two excited states, for example (29-32). Together with the biexponential decay kinetics, this precludes an entirely unambiguous measurement of the separation between the donor and acceptor fluorophores as the photophysical decay cannot be definitively modeled. Instead, the true average lifetimes () were calculated using equation 2, where αn are the relative amplitudes (normalized such that Σn αn = 1) and τn are the lifetimes for the nth exponential.
| (Eq. 2) |
For the purpose of calculating FRET efficiencies, the excitation-weighted average lifetime (〈τ〉) was calculated using equation 3.
| (Eq. 3) |
The lifetime FRET efficiency (Eτ) was calculated using equation 4 where 〈τDA〉 and 〈τ0D〉 are the excitation-weighted average lifetimes for the biosensor and control experiment where the acceptor was not on the same protein as the donor, respectively.
| (Eq. 4) |
Steady-state anisotropy measurements of fluorescently labeled IgEFc
Fluorescence anisotropy (r) measurements of eGFP labeled IgEFc samples were made using the same Spex Fluoromax used for intensity FRET measurements. HomoFRET measurements were made according to a method adapted from ref. 33; briefly, the emission monochromator was set at 560nm (bandwidth 2nm) and excitation was either performed at 488nm (bandwidth 2nm) and 514nm (bandwidth 2nm), or by carrying out an excitation scan from 470nm to 510nm (bandwidth 2nm). Emission anisotropies (r) were then calculated according to equation 5, using the orthogonally polarized emission intensities IVV and IVH taken with vertically polarized excitation, together with polarized emission intensities IHV and IHH taken with horizontally polarized excitation, to correct for instrumental polarization bias, (G factor; equation 6).
| (Eq. 5) |
| (Eq. 6) |
Anisotropy decay measurements of eGFP-labeled IgEFc
Anisotropy decay measurements were made using the same instrument used for the lifetime measurements. Polarized decays were collected at an emission wavelength of 510nm (bandwidth 4nm). G-factor normalization was accomplished by matching the summed counts in the VV-polarized and VH-polarized component decay curves to the separately measured steady-state anisotropies via equation 7 (27, 34) where r is the steady-state anisotropy as defined by equation 5.
| (Eq. 7) |
Generation of fusion protein models
A model of the mRFP-IgEFc-eGFP biosensor was generated based on the crystal structures of IgEFc (PDB 1O0V, (6)), and GFP (1GFL chain A, (35)). Although eGFP and mRFP show strong structural and sequence similarity in the barrel domains, the excited-state dipole transition moment vector geometry for mRFP is considerably less well defined relative to that in eGFP (36). Since this was a pre-requisite for the use of the models in the calculation of theoretical FRET efficiencies (see below), eGFP was used in the model in place of mRFP. The models were constructed by fusing eGFP N- and C-terminally to the IgEFc (1O0V) via rotational linker residues using the program FPMOD (37) to create a model of eGFP-IgEFc-eGFP simulating its genetically encoded form. The GFP units were allowed to move over 5 days computational time, and 1300 models with no clashes were generated. The procedure was repeated to produce 1300 models using the extended IgE (theoretical) model 1IGE (38).
Theoretical calculation of FRET efficiencies from the fusion protein models
Inter-probe distances (R) for FRET biosensors are often related to FRET efficiency (E) using equation 8.
| (Eq. 8) |
where kT is the rate constant for transfer, while R0 is the separation between the donor and the acceptor for which E = 50%, also known as the Förster distance, in the definition of which the orientation factor (k2) is commonly assigned its dynamic random average value of 2/3. However, this expression is inadequate for the IgEFc biosensor, in which there are two acceptors for each donor (the sum of the rate constants to each acceptor replaces the single rate constant in equation 8), and where averaging over the full multiplicity of conformers in the population exhibiting a wide range of probe separations and orientations, is necessary. Similarly, the time-dependent depolarization due to rotation coupled with homoFRET between the two eGFP moieties in each of the conformers of the C-terminal construct and of the separate N-terminal construct, is given by equation 9.
| (Eq. 9) |
where dT is the transfer depolarization factor (dT = [3/2]cos2θT – [1/2]), θT being the angle between the relevant transition moment vectors of the two eGFP fluorophores in excited-state exchange, kT is the one-way homoFRET transfer rate constant, and  is the apparent rotational correlation time. Taking into account the substitution of r0 for its limiting value of 2/5, as well as the addition of the overall multiplier representing the rotational decay , equation 9 (above) is essentially equivalent to equation 16 of ref. 39. With the additional condition of parallel absorption and emission transition moments (δ = 0), equation 9 (above) is also equivalent to equation 10 of ref. 40, with both refs. 39 and 40 deriving from the work of Tanaka and Mataga (41). Likewise with the relevant substitution r∞ = (r0 + r0dT)/2 which is the limiting anisotropy at very long (infinite) times when the excition energy is equally shared between the initially excited fluorophore and initially unexcited one, equation 9 (above) corresponds with the more recently presented equation 11 in ref. 42. Again, in the present cases, the measured depolarization will represent an ensemble average over the particular depolarization factors and homoFRET rate constants represented in the multiplicity of conformers.
is the apparent rotational correlation time. Taking into account the substitution of r0 for its limiting value of 2/5, as well as the addition of the overall multiplier representing the rotational decay , equation 9 (above) is essentially equivalent to equation 16 of ref. 39. With the additional condition of parallel absorption and emission transition moments (δ = 0), equation 9 (above) is also equivalent to equation 10 of ref. 40, with both refs. 39 and 40 deriving from the work of Tanaka and Mataga (41). Likewise with the relevant substitution r∞ = (r0 + r0dT)/2 which is the limiting anisotropy at very long (infinite) times when the excition energy is equally shared between the initially excited fluorophore and initially unexcited one, equation 9 (above) corresponds with the more recently presented equation 11 in ref. 42. Again, in the present cases, the measured depolarization will represent an ensemble average over the particular depolarization factors and homoFRET rate constants represented in the multiplicity of conformers.
The appropriate treatments for both cases are detailed in the Supplemental Information. For each of the 1300 donor-acceptor IgEFc models generated for both extended and bent forms, average intra- and cross-chain separation vectors R were calculated as the average of four vectors linking the coordinates of the donor and acceptor surrogate GFP benzylidene C1 and C2 and imidazolone N3 and C4 atoms within the fluorophores. In each case, the donor-acceptor separation was defined as being the length of vector R (i.e., the modulus, |R|). The transition moments of the donor and acceptor fluorophores are represented by the vectors D and A, respectively and were calculated for each conformation as the average of those obtained from the coordinates of the benzylidene C6 and imidazolone O5 and those of the benzylidene C3 and imidazolone N1 atoms (36, 43). The orientation factor, k2, was calculated using equation 10 (44, 45):
| (Eq. 10) |
where θT is the angle between vectors D and A, θD is the angle between vectors D and R, and θA is the angle between vectors R and A. All calculations, together with the FRET efficiencies and homoFRET depolarization courses, and the distributions of separations and of orientation factors were carried out using Mathcad 13 (Parametric Technology Corp.) as described in the Supplemental Information.
Whole antibody modeling
The same process used to create the fluorescent IgEFc fusions was used to recreate whole IgE and IgG models based on the crystal structure of the Fabs bound to an allergen β-lactoglobulin (PDB 2R56) (46). IgG models were based on crystal structures of available human IgG1-like molecules (PDB 1HZH (47) and 1IGT (48)) but with the 2R56 Fabs.
RESULTS
Fusion of fluorescent proteins to IgEFc does not perturb binding to sFcεRIα
Expressed protein was highly purified, free from aggregate and breakdown products as judged by analytical size exclusion gel filtration chromatography (see Supplemental Fig. S2). IgE and FcεRI interact with high affinity (KA ~ 1010 M−1) and the integrity of stored preparations was confirmed by a complete shift in the gel filtration elution profile of fluorescently labeled IgEFc, detected by UV and fluorescence (see Supplemental Fig. S3). In addition to confirming the “bindability” of the components, these analyses demonstrated that binding to sFcεRIα did not lead to aggregation of labeled (homo-dimeric) IgEFc. The kinetics and affinities for the binding of all labeled IgEFc constructs to sFcεRIα was compared to wild-type IgE by surface plasmon resonance (SPR, Biacore), (see Supplemental Table S1); no noteworthy differences were detected.
As well as complementing the SPR affinity measurements, dual polarization interferometry (DPI) provided a measure of the physical properties of the IgEFc·sFcεRIα complex as it formed on the biosensor surface. Initial characterization (data not shown) indicated that all constructs had the same high affinity for sFcεRIα as observed in SPR and gel filtration experiments. Upon addition of IgEFc3-4, virtually no change in layer thickness was observed; however the density increased, indicating that the fragment binds in the plane of the layer (see Supplemental Table S2). An increase in thickness was seen upon addition of IgEFc combined with a decrease in density suggesting that the Cε2 domains present in IgEFc but not IgEFc3-4 point away from the surface. This was even more pronounced for both whole IgE and the FRET pair labeled IgEFc (mRFP-IgEFc-eGFP), which are expected to have similar dimensions. The thickness change for the two larger constructs also resulted in a substantial decrease in the apparent layer density. The marked increase in thickness seen when IgE (or mRFP-IgEFc-eGFP) is bound to receptor, compared with IgEFc or IgEFc3-4, indicates that the Cε2 domains act to project the Fab region of IgE (or eGFP in the IgEFc biosensor) away from the surface (consistent with the models we derive). It appears that the biosensor mimics well the architecture and overall dimensions of the receptor-bound antibody.
Intra-molecular FRET occurs between eGFP and mRFP on the IgEFc biosensor
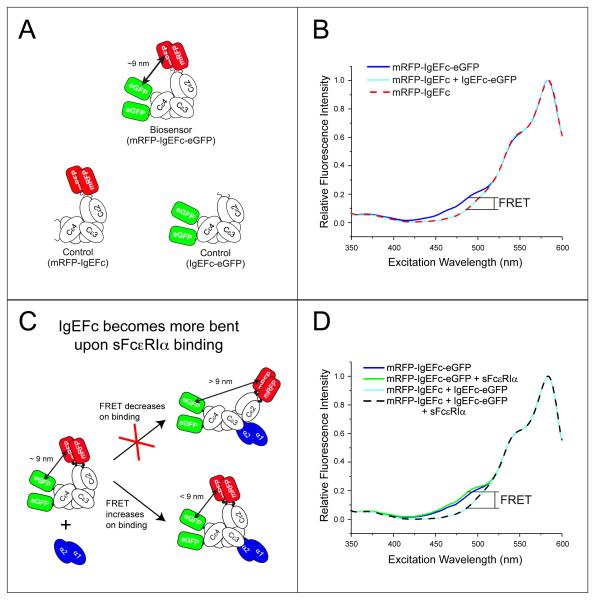
Excitation scans were collected at 610nm (bandwidth 2 nm) for both concentration-matched (200 nM) samples of both dual-labeled (mRFP-IgEFc-eGFP) and individually labeled (IgEFc-eGFP and mRFP-IgEFc) proteins (Fig. 1A), and are shown corrected for residual fluorescence emission by eGFP (Fig. 1B). Comparison of the spectra for mRFP-IgEFc alone with dual-labeled mRFP-IgEFc-eGFP shows an increase in acceptor (mRFP) emission detected at the donor (eGFP) excitation maximum, revealing that intra-molecular FRET indeed occurs with an efficiency of (9.6 ± 0.5)%. In contrast, in the control experiment in which the singly labeled mRFP-IgEFc and IgEFc-eGFP constructs were mixed together in the same cuvette, no FRET is observed (Fig. 1B).
FIG. 1.
The mRFP-IgEFc-eGFP biosensor reveals IgEFc to be bent in solution and to undergo conformational change on binding to sFcεRIα. A: Cartoons of the three fluorescent constructs. B: Steady-state fluorescence excitation scan (emission detected at 610nm) of either the FRET biosensor mRFP-IgEFc-eGFP (blue line), or control molecules mRFP-IgEFc alone (red dashed line), or with IgEFc-eGFP (cyan line). The increase in fluorescence shown here is indicative of energy transfer. C: Cartoon of the proposed conformational change model to be tested using the mRFP-IgEFc-eGFP biosensor. D: Steady-state fluorescence excitation scan (emission detected at 610nm) of either the FRET biosensor mRFP-IgEFc-eGFP alone (blue line) or saturated with sFcεRIα (green line), or control molecules mRFP-IgEFc and IgEFc-eGFP alone (cyan line) or with sFcεRIα (black dashed line). The fluorescence increases when sFcεRIα binds to the mRFP-IgEFc-GFP biosensor indicative of additional bending within IgEFc.
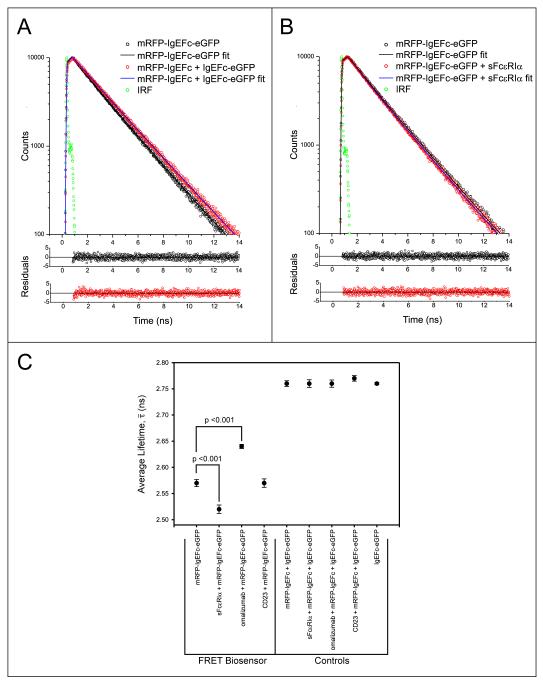
Fluorescence decay measurements of eGFP provide an additional way to quantify FRET in terms of the decrease it causes in the fluorescence lifetime of the donor. However, the complex photophysics of eGFP results in a biexponential decay, even for a single eGFP in solution (see Supplemental Fig. S4 and Table S3), as previously reported (29-32). We therefore compared the true average lifetimes (Eq. 2) for the FRET biosensor (mRFP-IgEFc-eGFP; (2.57 ± 0.01) ns), which were shorter than that in the control experiment (mRFP-IgEFc + IgEFc-eGFP; (2.76 ± 0.01) ns), providing further evidence that intra-molecular FRET is indeed occurring (Fig. 2C). To calculate the FRET efficiencies Eτ (Supplemental Table S3) from the lifetimes, the excitation-weighted average lifetimes 〈τ〉 (which are proportional to the relevant steady-state intensities) of the two exponential components (equation. 3) are employed (equation. 4) (see Fig. 2A, Supplemental Fig. S5 and Supplemental Table S3). These also qualitatively support the presence of FRET: (2.45 ± 0.01) ns for the FRET biosensor, shorter than the (2.69 ± 0.01) ns in the control, yielding a FRET efficiency of (8.9 ± 0.7)% (a summary of all the , 〈τ〉 and Eτ determined in this study is presented in Supplemental Table S3).
FIG. 2.
Fluorescence lifetime measurements for biosensor and control molecules in the presence and absence of IgE ligands. A: Fluorescence decays (excitation 468nm, emission 510nm) and bi-exponential fits (line plots) and their photon-counting weighted residuals for the mRFP-IgEFc-eGFP biosensor (open black circles), or control molecules (open red circles). Green circles are the instrument response function (IRF). Non-overlap of the fits indicates a shorter fluorescence lifetime for the mRFP-IgEFc-eGFP biosensor, and that it is therefore undergoing FRET. B: Fluorescence decays (excitation 468nm, emission 510nm) and bi-exponential fits (line plots) and their photon-counting weighted residuals, for the mRFP-IgEFc-eGFP biosensor alone (open black circles), or saturated with sFcεRIα (open red circles). Green circles are the instrument response function (IRF). Non-overlap of the fits indicates a shorter fluorescence lifetime for the mRFP-IgEFc-eGFP biosensor in the presence of sFcεRIα. C: Mean values of the results of independent experiments (n = 5) for the true average lifetimes calculated using eqn 2. Significant values were determined using a one-way ANOVA with Tukey-Kramer test. Only the biosensor mRFP-IgEFc-eGFP shows FRET, with changes in true average lifetimes on binding to both sFcεRIα and omalizumab Fab.
Modeling reveals that FRET can only occur if IgEFc has a bent conformation
In contrast to the extended conformation of the six domains in the first published model for IgEFc (37), the crystal structure (6) revealed an acutely bent structure. Calculation of inter-probe separations from FRET efficiencies for GFP-related proteins is complicated by uncertainty in the value of the orientation factor (k2) (44, 46-51), and by the biexponential decays (49, 52). To discriminate between the two extreme sets of conformations of IgEFc, we simulated the flexible movements of fluorescent proteins at the N- and C-termini of the extended (PDB 1IGE) and bent (PDB 1O0V) structures and calculated their theoretical FRET efficiencies on a simplified basis, as described above and in the Supplemental Information, where the complications above are examined in more detail. The distributions of the k2 values derived from the models are shown in Supplemental Figs. S6, and S7, and are close to random, suggesting sufficient models were produced to sample the space available to the eGFP units.
The calculated efficiency based on the extended models (0.54%) would be experimentally indistinguishable from zero; in contrast, the value based on the bent model (9.2%) is in good agreement with the experimental values of (9.6 ± 0.5)% by steady-state FRET and (8.9 ± 0.7)% by lifetime FRET. This demonstrates firstly, that the IgEFc does indeed adopt a bent conformation free in solution, and secondly, that the mRFP-IgEFc-eGFP biosensor is a useful tool for analyzing IgEFc conformation in solution.
IgEFc undergoes a conformational change on FcεRIα binding
It had been proposed that IgEFc might “unbend” upon binding to FcεRI (4, 9) whereas the crystal structure of the IgEFc·sFcεRIα complex revealed an even more acute bend (10). Unbending (see cartoon Fig. 1C) would result in a decrease in the FRET signal from the IgEFc biosensor, but the enhanced emission (Fig. 1D) and further decrease in 〈τ〉 to (2.38 ± 0.01) ns, (from (2.45 ± 0.01) ns) (Fig. 2B, and Supplemental Table S3) both point to increased intra-molecular FRET; this is consistent with a greater degree of bending upon FcεRIα binding, bringing the probes closer together (and/or, in principle, their relative orientations changing such that energy transfer between them is more favorable).
Omalizumab anti-IgE, but not CD23 (FcεRII) binding, affects IgEFc conformation
The soluble lectin “head” domain of CD23 (termed derCD23) binds to IgEFc with a 2:1 stoichiometry (20). Recombinant derCD23 was bound to mRFP-IgEFc-eGFP with no detectable change in 〈τ〉 ((2.45 ± 0.01) ns; Supplemental Table S3), implying no conformational change in IgEFc. In contrast, when omalizumab Fab was added to mRFP-IgEFc-eGFP, 〈τ〉 increased to ((2.54 ± 0.01) ns; Supplemental Table S3); the decreased FRET implies that the probes moved further apart upon omalizumab binding, in an unbending of the IgEFc (and/or their relative orientations changed) such as to reduce FRET efficiency. No effect was seen on the addition of omalizumab Fab to a control mixture of donor only (IgEFc-eGFP) and acceptor only (mRFP-IgEFc) labeled molecules (Fig. 2C and Supplemental Table S3).
HomoFRET occurs between the eGFPs attached at each terminus
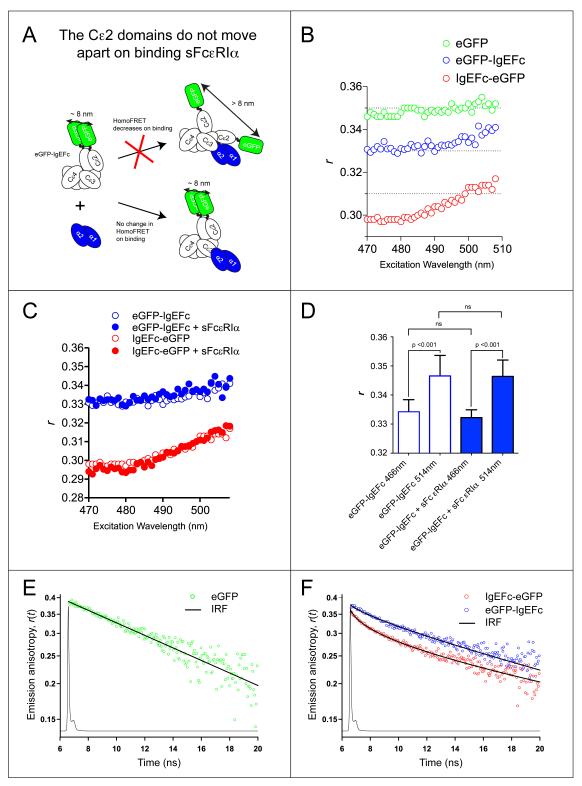
Since IgEFc is a dimer it carries two copies of each biologically encoded label (see cartoons in Figs. 1A, 1C and 3A), and homoFRET may occur between these identical fluorescent molecules, since their excitation and emission spectra overlap. GFP homoFRET is excitation wavelength dependent and detected through a “red-edge” effect in the anisotropy values, because the shape of the emission spectrum is sensitive to excitation wavelength (33, 53). Both N- and C-terminal IgEFc eGFP fusions had lower anisotropies than eGFP alone, and the measured anisotropy was excitation wavelength dependent (Fig. 3B) indicative of homoFRET. The anisotropy values measured for the C-terminal fused IgEFc were substantially lower than those for the N-terminal construct (Fig. 3B), indicating that the eGFP molecules in this construct were closer (as expected from the IgEFc crystal structure) and/or the difference in their relative orientations more extreme on average.
FIG. 3.
HomoFRET in eGFP-IgEFc and IgEFc-eGFP does not change on binding to sFcεRIα. A: Cartoon illustrating how homoFRET could be used to detect independent movements of polypeptide chains in the IgEFc homodimer on binding to sFcεRIα. B: Anisotropy excitation scan of either eGFP alone (green circles), eGFP-IgEFc (blue circles) or IgEFc-eGFP (red circles). HomoFRET occurs in both constructs as shown by the significantly lower emission anisotropy (r) at lower excitation wavelengths and the increase in r with excitation wavelength. C: sFcεRIα binding has no effect on the anisotropy excitation scans of either eGFP-IgEFc (blue circles) or IgEFc-eGFP (red circles). D: Repeat measurements of steady-state anisotropies of eGFP-IgEFc at two wavelengths. Data represents the average and standard deviations for n = 9 independent measurements. Significant values were determined using a one-way ANOVA with Tukey-Kramer test. The absence of any change on binding sFcεRIα confirms that there is no significant change in orientation between the Cε2 domains. E: Semi-log plot showing the anisotropy decay r(t) for eGFP alone, with raw data 5-point averaged for display purposes only (green circles). The fit line passing through the decay data is the best-fit impulse response for eGFP rotation (see Supplemental Information for details). The overlaid line graph shows the instrument response function (IRF). The anisotropy decay of eGFP (in the absence of homoFRET) is monoexponential and thus appears linear in this semi-log plot. F: Semi-log plot showing the anisotropy decays for eGFP-IgEFc (blue circles) and IgEFc-eGFP (red circles). Raw data was 5-point averaged for display purposes only with the lines passing through the data points being the theoretically calculated anisotropy decays based on the models (for a detailed description see Supplementary Information). The pronounced curvature of the decays in this semi-log plot demonstrates the expected deviation from monoexponential decay kinetics due to homoFRET.
For homoFRET pairs, the overlapping absorption and emission spectra are identical for each partner. Hence, given close enough separation and favorable relative orientations, there will be transfer of the excitation energy of one of them to the other, and (because the same conditions apply to the newly excited partner) back again, and so on. This leads to sharing of the initial excitation energy between them such that homoFRET is reversible, with identical transfer rate constants in both directions. The extent of completeness of this sharing is determined by the relative rates of transfer and deactivation. Since the excited-state behavior of the two partners is identical, homoFRET leads to no change in the overall emission spectrum, nor – in contrast with heteroFRET – of the intensity (quantum yield) or decay kinetics, of the exchanging pair as compared with an isolated partner (see Fig. 2 and Supplemental Table S3). HomoFRET is only observed experimentally when the emission transition dipoles of the pair (only the case of parallel absorption and emission dipoles is considered here) are not aligned parallel to one another. This leads to depolarization of the overall emission, relative to that observed for an isolated partner, the extent of depolarization depending on both the angle between the emission dipoles and the fraction of emission originating from the initially unexcited partner. This depolarization occurs on top of any rotational depolarization, converting the monoexponential anisotropy decay due to rotational depolarization observed if there were no homoFRET, to biexponential decay kinetics in its presence (see equation 9 above and ref. 54). Thus, whereas the anisotropy of monomeric eGFP decays monoexponentially as a result of depolarization due to rotational tumbling, with a harmonic mean correlation time 〈 〉 = (19.8 ± 0.2) ns (see Fig. 3E, Supplemental Fig. S8, Supplemental table S4) in good agreement with other values in the literature (see ref. 55 for example), the presence of homoFRET in the eGFP-IgEFc and IgEFc-eGFP constructs induces an additional anisotropy decay component (see Fig. 3F). The magnitude of this component of the biexponential decay depends on the angular separation of the transition dipole moments of the partners, and has a rate constant [2kT + (1/
〉 = (19.8 ± 0.2) ns (see Fig. 3E, Supplemental Fig. S8, Supplemental table S4) in good agreement with other values in the literature (see ref. 55 for example), the presence of homoFRET in the eGFP-IgEFc and IgEFc-eGFP constructs induces an additional anisotropy decay component (see Fig. 3F). The magnitude of this component of the biexponential decay depends on the angular separation of the transition dipole moments of the partners, and has a rate constant [2kT + (1/ )] (see equation 9) involving the rate constant for homoFRET. It is thus always greater, and may be very much greater than the rotational rate constant. In the present cases, where many different conformers contribute differently, the overall average anisotropy decays are expected to be highly multiexponential with a wide range of correlation times. This is seen clearly for the C-terminal IgEFc-eGFP fusion (see Fig 3F, Supplemental Fig. S8C, 〈
)] (see equation 9) involving the rate constant for homoFRET. It is thus always greater, and may be very much greater than the rotational rate constant. In the present cases, where many different conformers contribute differently, the overall average anisotropy decays are expected to be highly multiexponential with a wide range of correlation times. This is seen clearly for the C-terminal IgEFc-eGFP fusion (see Fig 3F, Supplemental Fig. S8C, 〈 〉 = (54.5 ± 0.7) ns). It is less clearly seen for the N-terminal eGFP-IgEFc construct (see Fig. 3F, Supplemental Fig. S8B, 〈
〉 = (54.5 ± 0.7) ns). It is less clearly seen for the N-terminal eGFP-IgEFc construct (see Fig. 3F, Supplemental Fig. S8B, 〈 〉 = (38.6 ± 0.3) ns), where the distribution of separations of the partner fluorophores in the various conformers is less favorable to FRET (see Supplemental Information). Good fits to the experimental decays were obtained (see Fig. 3F and Supplemental Fig. S8) by theoretical calculation of the homoFRET depolarization coupled to rotational depolarization for the 1300 bent-configuration conformers. Details of the calculations are described in the Supplemental Information, with derived rotational depolarization parameters shown in Supplemental Table S4, together with an additional analysis of the effects of using excitation averaged lifetimes and dynamic average orientation factors. The close to random distributions for the individual model homoFRET depolarization factors are shown in Supplemental Figs. S9 and S10. The calculated decays were averaged across the 1300 models, weighted for their widely different extents of depolarization which result in a large range of individual homoFRET anisotropy decays (Supplemental Fig. S11). Despite the same approximations as for the heteroFRET modeling, this confirms the presence of homoFRET, and strongly supports the validity of the modeling of heteroFRET efficiency.
〉 = (38.6 ± 0.3) ns), where the distribution of separations of the partner fluorophores in the various conformers is less favorable to FRET (see Supplemental Information). Good fits to the experimental decays were obtained (see Fig. 3F and Supplemental Fig. S8) by theoretical calculation of the homoFRET depolarization coupled to rotational depolarization for the 1300 bent-configuration conformers. Details of the calculations are described in the Supplemental Information, with derived rotational depolarization parameters shown in Supplemental Table S4, together with an additional analysis of the effects of using excitation averaged lifetimes and dynamic average orientation factors. The close to random distributions for the individual model homoFRET depolarization factors are shown in Supplemental Figs. S9 and S10. The calculated decays were averaged across the 1300 models, weighted for their widely different extents of depolarization which result in a large range of individual homoFRET anisotropy decays (Supplemental Fig. S11). Despite the same approximations as for the heteroFRET modeling, this confirms the presence of homoFRET, and strongly supports the validity of the modeling of heteroFRET efficiency.
The Cε2 domains move as a unit upon FcεRI binding
The presence of two eGFP moieties in each of the N- and C-terminal IgEFc fusions enables the following question to be addressed: do the domains within the (Cε2)2 or (Cε4)2 pairs change their relative orientation upon FcεRI binding? Gould et al. (4) speculated that the individual Cε2 domains might be displaced upon FcεRI binding (see cartoon, Fig. 3A), although the subsequent IgEFc·sFcεRIα crystal structure (10) showed no evidence of this. We therefore measured the homoFRET for both eGFP-IgEFc and IgEFc-eGFP before and after sFcεRIα binding. No change was observed in steady-state anisotropy (Figs. 3C and D) or anisotropy decay for either protein measurements (see Supplemental Figs. S8B and C) indicating that even in solution, the separation and relative orientation of the two probes at either terminus remain unchanged upon FcεRIα binding. The (Cε2)2 pair thus moves as a rigid unit when the IgEFc becomes more bent upon receptor binding, and as expected, the (Cε4)2 pair also behaves as a unit.
The bent IgEFc allows the Fabs to move freely when bound to FcεRI
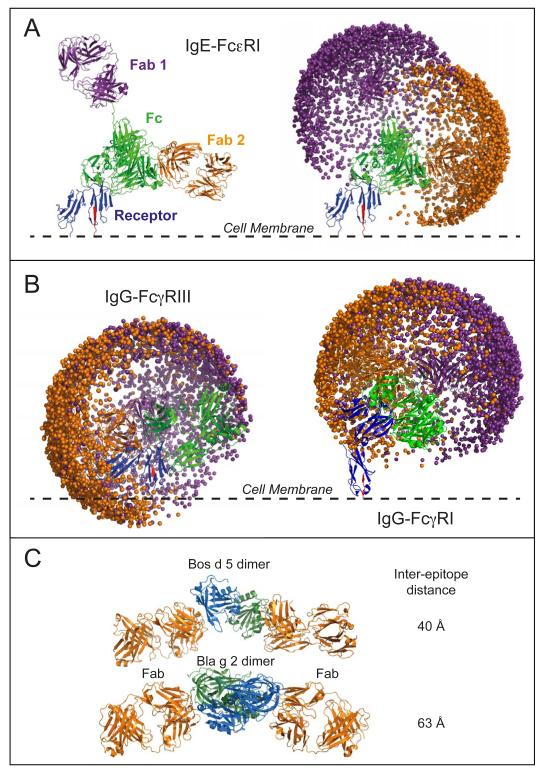
The close agreement between the observed FRET efficiencies and those calculated from the modeled structures of the IgEFc FRET biosensor suggests that modeling could also produce a realistic approximation of the relative movements of the Fabs relative to the Fc. Since the Fabs, like the probes, are connected to IgEFc by flexible linkers, 2,000 models were generated for both a whole IgE and a whole IgG in order to assess the impact of the bent IgEFc on the movement of the Fab arms (Fig.4 and supplemental video). When the IgE models (generated in the absence of sFcεRIα) were overlaid with the crystal structure of the IgEFc·sFcεRIα complex (PDB 2Y7Q (10)) only one of the 2,000 models clashed with the receptor. The models also show (Fig. 4A, right hand panel) that the Fabs occupy two essentially independent hemispheres of space pointing away from each other. These conclusions, with the Fab arms pointing away from the cell membrane, are consistent with the DPI results that showed the thickness of the IgE·sFcεRIα layer to be substantially greater than that of the IgEFc·sFcεRIα layer. The models are also consistent with the 140° inter-Fab angle seen in EM (14), and the average distance between the antigen binding sites of the models (110-130Å) agrees well with estimates from small molecule binding studies (56).
FIG. 4.
Molecular modeling of the space occupied by Fab arms in whole IgE and IgG. Example models of whole IgE (A) and IgG (B) are overlaid with their respective receptor complexes (see also supplemental video). The receptor α-chains are shown in blue with the C-terminal β-strand leading into the cell membrane in red. Fc domains are shown in green and the Fabs in purple and orange. Plots showing the spatial distributions of the Fab paratope residue Arg101 are shown for 2000 models. C: Arrangement of Fabs and allergen dimer in the Fab-allergen-allergen-Fab crystal structures for Bos d 5 (PDB 2R56 (46)) and Bla g 2 (PDB 2NR6 (67)). Images of the structures were produced using PyMOL (68).
The IgG models show two distinct differences to those of IgE. Firstly, the regions accessible to the two Fabs interpenetrate (Fig. 4B) and therefore the Fabs cannot move totally independently. In IgE the Cε2 domains direct the Fabs apart, whereas in IgG they are held closer together by the much smaller hinge region. Secondly, when the IgG models are overlaid onto the crystal structure of the IgEFc·sFcγRIIIα complex (PDB 1T83 (57)), almost a fifth of the models (392 out of 2000) clash with the receptor (Fig. 4B). It is unclear whether the bent hinge seen in whole IgG and some IgGFc crystal structures (and used in the modeling) reflects the natural arrangement in solution, but without the bend, far more IgG-receptor clashes would occur. Fig. 4B also shows that the addition of the D3 domain in FcγRIα, prevents clashes with the cell membrane that would inevitably occur in FcγRIIIα.
The range of orientations that the Fabs can adopt relative to the receptor and cell membrane are very different for IgE and IgG (see Fig. 4), and the greater range of inter-Fab angles for IgG is consistent with its enhanced flexibility as seen spectroscopically (11) and by EM (14). In IgE, the models further show that while one of the Fabs points upwards, away from the cell membrane, the other lies predominantly parallel to the cell membrane (Fig. 4A); this may have consequences for allergen recognition.
DISCUSSION
Previous efforts to investigate IgE flexibility and conformation focused on the whole antibody. Anisotropy decay measurements on fluorescently labeled murine IgE revealed less flexibility than IgG (11). Steady-state fluorescence anisotropy data obtained using the same murine IgE showed no change in flexibility on FcεRI binding (12), although later time-resolved analyses distinguished two components, one that was unaffected by receptor binding and abrogated when the Fab arms were cross-linked, and another, longer time-scale movement that was affected by receptor binding (13). Together with FRET measurements (58) this implied a rigidly bent IgE molecule bound to FcεRI with “wagging” Fabs directed away from the cell surface (13). In further studies with a chemically FRET-labeled chimaeric murine IgE/IgG, a bent conformation was confirmed, which became less flexible and slightly more bent when bound to FcεRI on cells (7, 15). This was also consistent with the “cusp-like” structure of rat myeloma IgE inferred from X-ray solution scattering (59). A compact structure for human IgEFc was also shown by solution scattering (8), and the crystal structures of both free (6) and receptor-bound (10) IgEFc reveal asymmetrically and acutely bent conformations, with a more acute bend in the complex.
In the present study we show that the asymmetrically bent IgEFc conformation exists in solution by quantitatively modeling heteroFRET and homoFRET energy transfer and depolarization processes within an ensemble of 1300 models. Rigid-body simulations that sample the conformational space of the fluorescent protein (FP) domains within the IgEFc biosensor allowed detailed calculation of distances between the FRET partners, as well as their relative orientations. These factors enabled us to determine for each conformer the degree of depolarization in homoFRET, and to avoid inappropriately assuming the problematic 2/3 value for orientation factors (Eq. 10) needed in both heteroFRET and homoFRET calculations. This assures optimally reliable predictions of the population average heteroFRET efficiency in the mRFP-IgEFc-eGFP construct, and of the population average homoFRET-induced fluorescence anisotropy decays of the C-terminal and the N-terminal eGFP constructs, to compare with the experimental measurements. A remarkable degree of correspondence is found.
Upon binding to sFcεRIα, the FRET efficiency of the mRFP-IgEFc-eGFP biosensor increases, as measured by a decrease in the average eGFP fluorescence lifetime, suggesting that the bent structure of the IgEFc becomes more bent upon sFcεRIα binding in solution (equation 8). Increased bending of IgEFc within the IgEFc·sFcεRIα complex was also observed by crystallography (10), and we show this is not an artifact of crystal packing. Additionally, the homoFRET results show that during this conformational change the (Cε2)2 and the (Cε4)2 domain pairs appear to behave as rigid units.
The IgEFc biosensor undergoes no measurable change in FRET efficiency upon binding to CD23 (FcεRII). Two molecules of derCD23 (the IgE-binding lectin domain) can bind to IgEFc (20) at locations that are distinct from the FcεRI binding site (as indicated by mutagenesis (61)). Clearly the binding mechanisms also differ, and IgEFc can bind to derCD23 in its naturally bent conformation without an apparent measurable change in the bend angle.
The binding of the Fab fragment of omalizumab (an anti-IgE therapeutic IgG), caused a decrease in the FRET signal (as measured by an increase in the average donor fluorescence lifetime), indicative of an unbending of the IgEFc. Again, the precise location of the epitope recognized by omalizumab is not known, but it acts by preventing the binding of free IgE molecules to FcεRI (62). This mechanism may thus be indirect and allosteric, as a result of bending the IgE in the opposite direction to that required for receptor engagement.
Changes in FRET efficiency could potentially arise for reasons other than changes in inter-probe separation, but these can be discounted. (i) sFcεRIα binding could distort the eGFP fluorophore and/or its close environment alterating the distribution between its different photophysical states, so that the average lifetime changes for a reason other than FRET. This appears unlikely, however, since sFcεRIα binding has no effect on the donor alone control (see Supplemental Table 3). (ii) A structural change in the eGFP protein could lead to a change in the steady-state spectra (60), but this is not observed upon sFcεRIα binding (Fig. 1D). Furthermore, the modeling also showed no steric clashes between sFcεRIα and the FP domains. (iii) The change in FRET efficiency could be solely due to a change in the relative orientations of the FPs in the IgEFc, without any appreciable change in distances between them. This can be discounted, firstly because an unfeasibly large (30-40%) change in the population-average k2 would be required, in contrast to only a 4-5% change in distance. Secondly, and perhaps more importantly, other experimental evidence indicates that the distribution of orientations of the fluorescent protein probes is unchanged upon sFcεRIα binding. This is provided by steady-state fluorescence anisotropy (Fig. 3C and D) and anisotropy decay measurements (Supplemental Fig. 8B and C) where no change is observed. Furthermore, analysis of the biosensor models showed that there was a close to random distribution of k2, irrespective of the inter-probe distance (see Supplemental Figs. S6 and S7), suggesting that, even if the space available to the probes is more constrained upon sFcεRIα binding, the randomness of their orientations is not likely to be affected.
The good agreement between the observed FRET efficiencies and those calculated from the modeled IgEFc biosensor suggested that the flexibility of the Fab arms might also be modeled reliably. A comparison between the range of conformations available to the Fab arms in IgE and IgG revealed marked differences (Fig. 4, Supplemental video). In IgG, not only do the regions accessible to the two Fab arms overlap extensively, but when mapped onto the crystal structure of an IgG·FcγRIII complex, there is considerable clashing with the receptor and also with the membrane. Remarkably, the Fab arms of IgE occupy distinct regions (Fig. 4A), and show virtually no clashing with either the receptor or membrane. The rigid (Cε2)2 domain pair appears to act as a steric insulator and presents - and conformationally restricts - the Fab arms in a manner unique to IgE. In part, this difference between IgG and IgE may relate to a difference in biological function: IgG·antigen complexes generally form in solution and then interact with their low affinity receptors, whereas most IgE is receptor-bound before encountering antigen (allergen). If IgE had an IgG-like structure, the Fabs would be much closer to both the cell membrane and the receptor, which might inhibit antigen recognition. However, the manner in which the bent IgE molecule and its (Cε2)2 domains present the Fab arms may have important implications for allergen recognition and effector cell activation.
The crystal structure of the high affinity FcγRI, which has an additional domain compared with FcγRII and FcγRIII, is instructive in this context. The biological roles of the high-affinity IgG·FcγRI and IgE·FcεRI complexes have common features: (1) they are both found on dendritic cells where the high affinity results in their action as scavenger receptors that bind and internalize antigens; (2) they are both found on cells that combat infecting organisms – macrophages and bacterial infections for IgG·FcγRI, and mast cells and helminth infections for IgE·FcεRI. The FcγRI crystal structure (PDB 3RJD (63)) shows that the additional extracellular domain (D3), despite the predictions from mutagenesis (64), does not contact IgG. Instead, it acts as a spacer pushing the D1 and D2 domains (that do contact IgG) away from the membrane (Fig. 4B) Therefore it is a similar adaptation to the Cε2 domains in IgE (only this time in the receptor) that allows the Fabs to move freely without clashing with the membrane – an important ability for a scavenging receptor.
As depicted in Fig. 4A, one of the IgE Fabs predominantly points away from the membrane, while the other is oriented parallel to the membrane. The optimal inter-IgE epitope distance for FcεRI activation has been shown to be 40-60Å using rigid DNP-labeled molecular spacers (65, 66). Distances shorter than this were not assayed, but the efficiency of activation decreased rapidly at greater separations. A key feature of allergens is the spatial clustering of more than one IgE epitope, and for small allergens or those presenting only a single epitope such as β-lactoglobulin (Bos d 5), this frequently occurs through oligomerization. Two crystal structures of dimerized allergens with bound Fabs have been solved - Bos d 5 (2R56 (46) and Bla g 2 (2NR6 (67)), and are both shown in Fig. 4C. These two complexes are particularly instructive, for not only do they reveal that the allergen epitopes are optimally spaced for receptor activation (40 and 63Å respectively), but they also both display an approximately co-linear arrangement - consistent with IgE Fabs on adjacent receptors oriented parallel to the membrane. The acutely bent receptor-bound IgEFc and conformationally constrained Fab arms may therefore predispose IgE-sensitized cells to respond to small antigens with only a particular arrangement of epitopes; this may represent a structural determinant of protein allergenicity – the understanding of which has hitherto proved elusive.
Supplementary Material
SUMMARY.
IgE binding to its high affinity receptor FcεRI on mast cells and basophils is a key step in the mechanism of allergic disease and a target for therapeutic intervention. Early indications that IgE adopts a bent structure in solution have been confirmed by recent X-ray crystallographic studies of IgEFc, which further showed that the bend – contrary to expectation – is enhanced in the crystal structure of the complex with receptor. To investigate the structure of IgEFc and its conformational changes that accompany receptor binding in solution, we created a Förster resonance energy transfer (FRET) biosensor using biologically encoded fluorescent proteins fused to the N- and C-terminal IgEFc domains (Cε2 and Cε4, respectively), together with the theoretical basis for quantitating its behavior. This revealed not only that the IgEFc exists in a bent conformation in solution, but also that the bend is indeed enhanced upon FcεRI binding. No change in the degree of bending was seen upon binding to the B cell receptor for IgE, CD23 (FcεRII), but in contrast, binding of the anti-IgE therapeutic antibody omalizumab decreases the extent of the bend, implying a conformational change that opposes FcεRI engagement. HomoFRET measurements further revealed that the (Cε2)2 and (Cε4)2 domain pairs behave as rigid units flanking the conformational change in the Cε3 domains. Finally, modeling of the accessible conformations of the two Fab arms in FcεRI-bound IgE revealed a mutual exclusion not seen in IgG, and Fab orientations relative to the membrane that may predispose receptor-bound IgE to cross-linking by allergens.
Acknowledgments
We would like to thank members of the Sutton and Beavil groups past and present for useful discussions. S.A-B would like to acknowledge his collaborators in the development of the TRI2 software.
ABBREVIATIONS
- DNP
dinitrophenol
- DPI
dual polarization interferometry
- FRET
Förster resonance energy transfer;
- sFcεRIα
soluble fragment of the high affinity IgE receptor FcεRI α-chain
- SPR
surface plasmon resonance
- sulpho-GMBS
N-[γ-maleimidobutyryloxy]sulfosuccinimide ester.
Footnotes
This work was supported by a grant from Asthma UK, and the Medical Research Council (UK).
REFERENCES
- 1.Gould HJ, Sutton BJ. IgE in allergy and asthma today. Nat. Rev. Immunol. 2008;8:205–217. doi: 10.1038/nri2273. [DOI] [PubMed] [Google Scholar]
- 2.Wank SA, DeLisi C, Metzger H. Analysis of the rate-limiting step in a ligand-cell receptor interaction: the IgE system. Biochemistry (USA) 1983;22:954–959. doi: 10.1021/bi00273a038. [DOI] [PubMed] [Google Scholar]
- 3.Miller L, Blank U, Metzger H, Kinet J. Expression of high-affinity binding of human immunoglobulin E by transfected cells. Science. 1989;244:334–337. doi: 10.1126/science.2523561. [DOI] [PubMed] [Google Scholar]
- 4.Gould HJ, Sutton BJ, Beavil AJ, Beavil RL, Mccloskey N, Coker HA, Fear D, Smurthwaite L. The biology of IgE and the basis of allergic disease. Annu. Rev. Immunol. 2003;21:579–628. doi: 10.1146/annurev.immunol.21.120601.141103. [DOI] [PubMed] [Google Scholar]
- 5.Soussi Gounni A, Lamkhioued B, Ochiai K, Tanaka Y, Delaporte E, Capron A, Kinet J-P, Capron M. High-affinity IgE receptor on eosinophils is involved in defence against parasites. Nature. 1994;367:183–186. doi: 10.1038/367183a0. [DOI] [PubMed] [Google Scholar]
- 6.Wan T, Beavil RL, Fabiane SM, Beavil AJ, Sohi MK, Keown M, Young RJ, Henry AJ, Owens RJ, Gould HJ, Sutton BJ. The crystal structure of IgE Fc reveals an asymmetrically bent conformation. Nat. Immunol. 2002;3:681–686. doi: 10.1038/ni811. [DOI] [PubMed] [Google Scholar]
- 7.Zheng Y, Shopes B, Holowka D, Baird B. Conformations of IgE bound to its receptor FcεRI and in solution. Biochemistry (USA) 1991;30:9125–9132. doi: 10.1021/bi00102a002. [DOI] [PubMed] [Google Scholar]
- 8.Beavil AJ, Young RJ, Sutton BJ, Perkins SJ. Bent domain structure of recombinant human IgE-Fc in solution by X-ray and neutron scat tering in conjunction with an automated curve fitting procedure. Biochemistry (USA) 1995;34:14449–14461. doi: 10.1021/bi00044a023. [DOI] [PubMed] [Google Scholar]
- 9.McDonnell JM, Calvert R, Beavil RL, Beavil AJ, Henry AJ, Sutton BJ, Gould HJ, Cowburn D. The structure of the IgE Cε2 domain and its role in stabilizing the complex with its high-affinity receptor FcεRIα. Nat. Struct. Biol. 2001;8:437–441. doi: 10.1038/87603. [DOI] [PubMed] [Google Scholar]
- 10.Holdom MD, Davies AM, Nettleship JE, Bagby SC, Dhaliwal B, Girardi E, Hunt J, Gould HJ, Beavil AJ, McDonnell JM, Owens RJ, Sutton BJ. Conformational changes in IgE contribute to its uniquely slow dissociation rate from receptor FcεRI. Nat. Struct. Mol. Biol. 2011;18:571–576. doi: 10.1038/nsmb.2044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Oi VT, Vuong TM, Hardy R, Reidler J, Dangle J, Herzenberg LA, Stryer L. Correlation between segmental flexibility and effector function of antibodies. Nature. 1984;307:136–140. doi: 10.1038/307136a0. [DOI] [PubMed] [Google Scholar]
- 12.Slattery J, Holowka D, Baird B. Segmental flexibility of receptor-bound immunoglobulin E. Biochemistry (USA) 1985;24:7810–7820. doi: 10.1021/bi00347a046. [DOI] [PubMed] [Google Scholar]
- 13.Holowka D, Wensel T, Baird B. A nanosecond fluorescence depolarization study on the segmental flexibility of receptor-bound immunoglobulin E. Biochemistry (USA) 1990;29:4607–4612. doi: 10.1021/bi00471a015. [DOI] [PubMed] [Google Scholar]
- 14.Roux KH, Strelets L, Brekke OH, Sandlie I, Michaelsen TE. Comparisons of the ability of human IgG3 hinge mutants, IgM, IgE, and IgA2, to form small immune complexes: a role for flexibility and geometry. J. Immunol. 1998;161:4083–4090. [PubMed] [Google Scholar]
- 15.Zheng Y, Shopes B, Holowka D, Baird B. Dynamic conformations compared for IgE and IgG1 in solution and bound to receptors. Biochemistry (USA) 1992;31:7446–7456. doi: 10.1021/bi00148a004. [DOI] [PubMed] [Google Scholar]
- 16.Young RJ, Owens RJ, Mackay GA, Chan CMW, Shi J, Hide M, Francis DM, Henry AJ, Sutton BJ, Gould HJ. Secretion of recombinant human IgE-Fc by mammalian cells and biological activity of glycosylation site mutants. Protein Eng. Des. Sel. 1995;8:193–199. doi: 10.1093/protein/8.2.193. [DOI] [PubMed] [Google Scholar]
- 17.Li X, Zhang G, Ngo N, Zhao X, Kain SR, Huang C-C. Deletions of the Aequorea victoria green fluorescent protein define the minimal domain required for fluorescence. J. Biol. Chem. 1997;272:28545–28549. doi: 10.1074/jbc.272.45.28545. [DOI] [PubMed] [Google Scholar]
- 18.Campbell RE, Tour O, Palmer AE, Steinbach PA, Baird GS, Zacharias DA, Tsien RY. A monomeric red fluorescent protein. Proc. Natl. Acad. Sci. U. S. A. 2002;99:7877–7882. doi: 10.1073/pnas.082243699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Durocher Y, Perret S, Kamen A. High-level and high-throughput recombinant protein production by transi ent transfection of suspension-growing human 293-EBNA1 cells. Nucleic Acids Res. 2002;30:E9. doi: 10.1093/nar/30.2.e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shi J, Ghirlando R, Beavil RL, Beavil AJ, Keown MB, Young RJ, Owens RJ, Sutton BJ, Gould HJ. Interaction of the low-affinity receptor CD23/FcεRII lectin domain with the Fcε3-4 fragment of human immunoglobulin E. Biochemistry (USA) 1997;36:2112–2122. doi: 10.1021/bi961231e. [DOI] [PubMed] [Google Scholar]
- 21.Hunt J, Bracher MG, Shi J, Fleury S, Dombrowicz D, Gould HJ, Sutton BJ, Beavil AJ. Attenuation of IgE affinity for FcεRI radically reduces the allergic response in vitro and in vivo. J. Biol. Chem. 2008;283:29882–29887. doi: 10.1074/jbc.M804742200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bruggemann M, Williams GT, Bindon CI, Clark MR, Walker MR, Jefferis R, Waldmann H, Neuberger MS. Comparison of the effector functions of human immunoglobulins using a matched set of chimeric antibodies. J. Exp. Med. 1987;166:1351–1361. doi: 10.1084/jem.166.5.1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cook JPD, Henry AJ, McDonnell JM, Owens RJ, Sutton BJ, Gould HJ. Identification of contact residues in the IgE binding site of human FcεRIα. Biochemistry (USA) 1997;36:15579–15588. doi: 10.1021/bi9713005. [DOI] [PubMed] [Google Scholar]
- 24.Peter M, Ameer-Beg SM, Hughes MKY, Keppler MD, Prag S, Marsh M, Vojnovic B, Ng T. Multiphoton-FLIM quantification of the EGFP-mRFP1 FRET pair for localization of membrane receptor-kinase interactions. Biophys. J. 2005;88:1224–1237. doi: 10.1529/biophysj.104.050153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Keown MB, Ghirlando R, Young RJ, Beavil AJ, Owens RJ, Perkins SJ, Sutton BJ, Gould HJ. Hydrodynamic studies of a complex between the Fc fragment of human IgE and a soluble fragment of the FcεRIα chain. Proc. Natl. Acad. Sci. U. S. A. 1995;92:1841–1845. doi: 10.1073/pnas.92.6.1841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Swann MJ, Peel LL, Carrington S, Freeman NJ. Dual-polarization interferometry: an analytical technique to measure changes in protein structure in real time, to determine the stoichiometry of binding events, and to differentiate between specific and nonspecific interactions. Anal. Biochem. 2004;329:190–198. doi: 10.1016/j.ab.2004.02.019. [DOI] [PubMed] [Google Scholar]
- 27.Lakowicz JR. Principles of Fluorescence Spectroscopy. 3rd ed. Springer; 2006. [Google Scholar]
- 28.Barber PR, Ameer-Beg SM, Gilbey J, Carlin LM, Keppler M, Ng TC, Vojnovic B. Multiphoton time-domain fluorescence lifetime imaging microscopy: practical application to protein–protein interactions using global analysis. J. R. Soc. Lond. Interface. 2009;6:S93–S105. [Google Scholar]
- 29.Suhling K, Siegel J, Phillips D, French PMW, Lévêque-Fort S, Webb SED, Davis DM. Imaging the environment of green fluorescent protein. Biophys. J. 2002;83:3589–3595. doi: 10.1016/S0006-3495(02)75359-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Heikal AA, Hess ST, Webb WW. Multiphoton molecular spectroscopy and excited-state dynamics of enhanced green fluorescent protein (EGFP): acid–base specificity. Chem. Phys. 2001;274:37–55. [Google Scholar]
- 31.Uskova MA, Borst J-W, Hink MA, van Hoek A, Schots A, Klyachko NL, Visser AJWG. Fluorescence dynamics of green fluorescent protein in AOT reversed micelles. Biophys. Chem. 2000;87:73–84. doi: 10.1016/s0301-4622(00)00184-8. [DOI] [PubMed] [Google Scholar]
- 32.Cotlet M, Hofkens J, Maus M, Gensch T, Van der Auweraer M, Michiels J, Dirix G, Van Guyse M, Vanderleyden J, Visser AJWG, De Schryver FC. Excited-state dynamics in the enhanced green fluorescent protein mutant probed by picosecond time-resolved single photon counting spectroscopy. J. Phys. Chem. B. 2001;105:4999–5006. [Google Scholar]
- 33.Squire A, Verveer PJ, Rocks O, Bastiaens PIH. Red-edge anisotropy microscopy enables dynamic imaging of homo-FRET between green fluorescent proteins in cells. J. Struct. Biol. 2003;147:62–69. doi: 10.1016/j.jsb.2003.10.013. [DOI] [PubMed] [Google Scholar]
- 34.Blumberg WE, Dale RE, Eisinger J, Zuckerman DM. Energy transfer in tRNAphe (yeast). The solution structure of transfer RNA. Biopolymers. 1974;13:1607–1620. doi: 10.1002/bip.1974.360130808. [DOI] [PubMed] [Google Scholar]
- 35.Yang F, Moss LG, Phillips GN. The molecular structure of green fluorescent protein. Nat. Biotechnol . 1996;14:1246–1251. doi: 10.1038/nbt1096-1246. [DOI] [PubMed] [Google Scholar]
- 36.Wan S, Liu S, Zhao G, Chen M, Han K, Sun M. Photoabsorption of green and red fluorescent protein chromophore anions in vacuo. Biophys. Chem. 2007;129:218–223. doi: 10.1016/j.bpc.2007.06.003. [DOI] [PubMed] [Google Scholar]
- 37.Pham E, Chiang J, Li I, Shum W, Truong K. A computational tool for designing FRET protein biosensors by rigid-body sampling of their conformational space. Structure. 2007;15:515–523. doi: 10.1016/j.str.2007.03.009. [DOI] [PubMed] [Google Scholar]
- 38.Padlan EA, Davies DR. A model of the Fc of immunoglobulin E. Mol. Immunol. 1986;23:1063–1075. doi: 10.1016/0161-5890(86)90005-2. [DOI] [PubMed] [Google Scholar]
- 39.Gautier I, Tramier M, Durieux C, Coppey J, Pansu RB, Nicolas J-C, Kemnitz K, Coppey-Moisan M. Homo-FRET microscopy in living cells to measure monomer-dimer transition of GFP-tagged proteins. Biophys. J. 2001;80:3000–3008. doi: 10.1016/S0006-3495(01)76265-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bastiaens PI, van Hoek A, Benen JA, Brochon JC, Visser AJ. Conformational dynamics and intersubunit energy transfer in wild-type and mutant lipoamide dehydrogenase from Azotobacter vinelandii. A multidimensional time-resolved polarized fluorescence study. Biophys. J. 1992;63:839–853. doi: 10.1016/S0006-3495(92)81659-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tanaka F, Mataga N. Theory of time-dependent photo-selection in interacting fixed systems. Photochem. Photobiol. 1979;29:1091–1097. [Google Scholar]
- 42.Yeow EKL, Clayton AHA. Enumeration of o ligomerization states of membrane proteins in living cells by homo-FRET spectroscopy and microscopy: theory and application. Biophys. J. 2007;92:3098–3104. doi: 10.1529/biophysj.106.099424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shi X, Basran J, Seward HE, Childs W, Bagshaw CR, Boxer SG. Anomalous negative fluorescence anisotropy in yellow fluorescent protein (YFP 10C): quantitative analysis of FRET in YFP dimers. Biochemistry (USA) 2007;46:14403–14417. doi: 10.1021/bi701575n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dale RE, Eisinger J, Blumberg WE. The orientational freedom of molecular probes. The orientation factor in intramolecular energy transfer. Biophys. J. 1979;26:161–193. doi: 10.1016/S0006-3495(79)85243-1. corrigenda (1980) ibid. 30, 365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Förster Th. Fluoreszenz organischer Verbindungen. Vandenhoeck & Ruprecht; Göttingen: 1951. [Google Scholar]
- 46.Niemi M, Jylhä S, Laukkanen M-L, Söderlund H, Mäkinen-Kiljunen S, Kallio JM, Hakulinen N, Haahtela T, Takkinen K, Rouvinen J. Molecular interactions between a recombinant IgE antibody and the β-lactoglobulin allergen. Structure. 2007;15:1413–1421. doi: 10.1016/j.str.2007.09.012. [DOI] [PubMed] [Google Scholar]
- 47.Saphire EO, Stanfield RL, Max Crispin MD, Parren PWHI, Rudd PM, Dwek RA, Burton DR, Wilson IA. Contrasting IgG structures reveal extreme asymmetry and flexibility. J. Mol. Biol. 2002;319:9–18. doi: 10.1016/S0022-2836(02)00244-9. [DOI] [PubMed] [Google Scholar]
- 48.Harris LJ, Larson SB, Hasel KW, McPherson A. Refined Structure of an intact IgG2a monoclonal antibody. Biochemistry (USA) 1997;36:1581–1597. doi: 10.1021/bi962514+. [DOI] [PubMed] [Google Scholar]
- 49.Seward HE, Bagshaw CR. The photochemistry of fluorescent proteins: implications for their biological applications. Chem. Soc. Rev. 2009;38:2842–2851. doi: 10.1039/b901355p. [DOI] [PubMed] [Google Scholar]
- 50.van der Meer BW. Kappa-squared: from nuisance to new sense. Rev. Mol. Biotechnol. 2002;82:181–196. doi: 10.1016/s1389-0352(01)00037-x. [DOI] [PubMed] [Google Scholar]
- 51.Clegg RM. In: Fluorescence resonance energy transfer and nucleic acids. David MJ, Lilley JED, editors. Academic Press; 1992. pp. 353–388. (Methods Enzymol., vol. 211). [DOI] [PubMed] [Google Scholar]
- 52.Hink MA, Griep RA, Borst JW, van Hoek A, Eppink MH, Schots A, Visser AJ. Structural dynamics of green fluorescent protein alone and fused with a single chain Fv protein. J. Biol. Chem. 2000;275:17556–17560. doi: 10.1074/jbc.M001348200. [DOI] [PubMed] [Google Scholar]
- 53.Vogel SS, Thaler C, Blank PS, Koushik SV. Time Resolved Fluorescence Anisotropy. In: Periasamy A, Clegg RM, editors. FLIM Microscopy in Biology and Medicine. CRC Press; 2010. pp. 245–320. [Google Scholar]
- 54.Thaler C, Koushik SV, Puhl HL, Blank PS, Vogel SS. Structural rearrangement of CaMKII α catalytic domains encodes activation. Proc. Natl. Acad. Sci. U. S. A. 2009;106:6369–6374. doi: 10.1073/pnas.0901913106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Suhling K, Davis DM, Philips D. The influence of solvent viscosity on the fluorescence decay and time-resolved anisotropy of green fluorescent protein. J. Fluores. 2002;12:91–95. [Google Scholar]
- 56.Schweitzer-Stenner R, Licht A, Lüscher I, Pecht I. Oligomerization and ring closure of immunoglobulin E class antibodies by divalent haptens. Biochemistry (USA) 1987;26:3602–3612. doi: 10.1021/bi00386a053. [DOI] [PubMed] [Google Scholar]
- 57.Radaev S, Motyka S, Fridman W-H, Sautes-Fridman C, Sun PD. The structure of a human type III Fcγ receptor in complex with Fc. J. Biol. Chem. 2001;276:16469–16477. doi: 10.1074/jbc.M100350200. [DOI] [PubMed] [Google Scholar]
- 58.Baird B, Holowka D. Structural mapping of Fc receptor bound immunoglobulin E: proximity to the membrane surface of the antibody combining site and another site in the Fab segments. Biochemistry (USA) 1985;24:6252–6259. doi: 10.1021/bi00343a032. [DOI] [PubMed] [Google Scholar]
- 59.Davis KG, Glennie M, Harding SE, Burton DR. A model for the solution conformation of rat IgE. Biochem. Soc. Trans. 1990;18:935–936. doi: 10.1042/bst0180935. [DOI] [PubMed] [Google Scholar]
- 60.Kirchhofer A, Helma J, Schmidthals K, Frauer C, Cui S, Karcher A, Pellis M, Muyldermans S, Casas-Delucchi CS, Cardoso MC, Leonhardt H, Hopfner K-P, Rothbauer U. Modulation of protein properties in living cells using nanobodies. Nat. Struct. Mol. Biol. 2010;17:133–138. doi: 10.1038/nsmb.1727. [DOI] [PubMed] [Google Scholar]
- 61.Sayers I, Housden JEM, Spivey AC, Helm BA. The importance of Lys-352 of human immunoglobulin E in Fc ε RII/CD23 recognition. J. Biol. Chem. 2004;279:35320–35325. doi: 10.1074/jbc.M404575200. [DOI] [PubMed] [Google Scholar]
- 62.Holgate S, Casale T, Wenzel S, Bousquet J, Deniz Y, Reisner C. The anti-inflammatory effects of omalizumab confirm the central role of IgE in allergic inflammation. J. Allergy Clin. Immunol. 2005;115:459–465. doi: 10.1016/j.jaci.2004.11.053. [DOI] [PubMed] [Google Scholar]
- 63.Lu J, Ellsworth JL, Hamacher N, Oak SW, Sun PD. Crystal Structure of Fcγ Receptor I and Its Implication in High Affinity γ-Immunoglobulin Binding. J. Biol. Chem. 2011;286:40608–40613. doi: 10.1074/jbc.M111.257550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Harrison PT, Allen JM. High affinity IgG binding by Fcγ RI (CD64) is modulated by two distinct IgSF domains and the transmembrane domain of the receptor. Protein Eng. 1998;11:225–232. doi: 10.1093/protein/11.3.225. [DOI] [PubMed] [Google Scholar]
- 65.Paar JM, Harris NT, Holowka D, Baird B. Bivalent ligands with rigid double-stranded DNA spacers reveals tructural constraints on signaling by FcεRI. J. Immunol. 2002;169:856–864. doi: 10.4049/jimmunol.169.2.856. [DOI] [PubMed] [Google Scholar]
- 66.Sil D, Lee JB, Luo D, Holowka D, Baird B. Trivalent ligands with rigid DNA spacers reveal structural requirements for IgE receptor signaling in RBL mast cells. ACS Chem. Biol. 2007;2:674–684. doi: 10.1021/cb7001472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Li M, Gustchina A, Alexandratos J, Wlodawer A, Wünschmann S, Kepley CL, Chapman MD, Pomés A. Crystal structure of a dimerized cockroach allergen Bla g 2 complexed with a monoclonal antibody. J. Biol. Chem. 2008;283:22806–22814. doi: 10.1074/jbc.M800937200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Schrödinger, LLC . The PyMOL Molecular Graphics System, Version 1.5.0. 2011. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.