Abstract
Loss of iRhom2, a catalytically inactive rhomboid-like protein, blocks maturation of TACE/ADAM17 in macrophages, resulting in defective shedding of the cytokine TNF. Apart from the resulting inflammatory defects, iRhom2-null mice appear normal: they do not exhibit the multiple defects seen in TACE knockouts, suggesting that TACE maturation is independent of iRhom2 in cells other than macrophages. Here we show that the physiological role of iRhoms is much broader. iRhom1 knockout mice die within 6 weeks of birth. They exhibit a severe phenotype, with defects in multiple tissues including highly penetrant brain haemorrhages. The non-overlapping phenotypes imply that iRhom 1 and 2 have distinct physiological roles, although at a cellular level both promote the maturation of TACE (but not other ADAM proteases). Both iRhoms are co-expressed in many contexts where TACE acts. We conclude that all TACE activity, constitutive and regulated, requires iRhom function. iRhoms are therefore essential and specific regulators of TACE activity, but our evidence also implies that they must have additional physiologically important clients.
Introduction
iRhoms are non-protease members of the rhomboid-like superfamily of polytopic membrane proteins. They are endoplasmic reticulum proteins that regulate the trafficking and fate of membrane proteins in a variety of contexts (Zettl et al., 2011; Adrain et al., 2012; McIlwain et al., 2012). Mouse iRhom2 is the best understood; it is a myeloid-specific regulator of the activation of TACE, an important shedding protease that releases proteins from the cell surface (Gooz, 2010). TACE (also called ADAM17) was first discovered as the enzyme responsible for the release of active TNF (Peschon et al., 1998), the primary inflammatory cytokine. It has now been shown to have many other substrates, including ligands of the epidermal growth factor receptor (EGFR) and therefore to control a wide range of physiologically and medically significant functions (Blobel, 2005).
Because of its role in macrophage TACE activation, iRhom2 knockout mice have specific inflammatory defects: they fail to release active TNF in response to immune activation by LPS or specific pathogens (Adrain et al., 2012; McIlwain et al., 2012; Siggs et al., 2012). We discovered that the cellular cause of this phenotype was a failure of TACE maturation in iRhom2 mutant macrophages (Adrain et al., 2012). In the absence of iRhom2, TACE is unable to leave the ER, thereby preventing it from being trafficked through the Golgi apparatus. Consequently, its inhibitory prodomain cannot be removed by furin, and TACE remains fully inactive. We concluded that iRhom2 acts in macrophages to promote the ER exit of TACE (Adrain et al., 2012).
Beyond the inflammatory defects of iRhom2 loss, the mutant mice appear healthy: there is no sign of the wide spectrum defects associated with loss of TACE activity in other tissues (Peschon et al., 1998). This implies that, aside from in macrophages, where iRhom2 is expressed at high levels, TACE trafficking in other cell types does not depend on iRhom2. Significantly though, there are two iRhoms in mammals (Lemberg and Freeman, 2007). Mouse iRhom1 is 57% identical and 69% similar to iRhom2 and is expressed more broadly than the latter. Here we report that the role of iRhom1 and 2 in mouse physiology and development is distinct but, at a cellular level, both participate in TACE trafficking. We also show that that iRhoms are required not only for constitutive TACE activity but also for stimulus induced shedding. Our genetic and cellular data strongly suggest that all TACE activity in all mouse tissues depends on iRhom function. Moreover, the iRhom double knockout phenotype, which is lethal during mid-embryogenesis, indicates that TACE is not the only client for iRhoms.
Results and Discussion
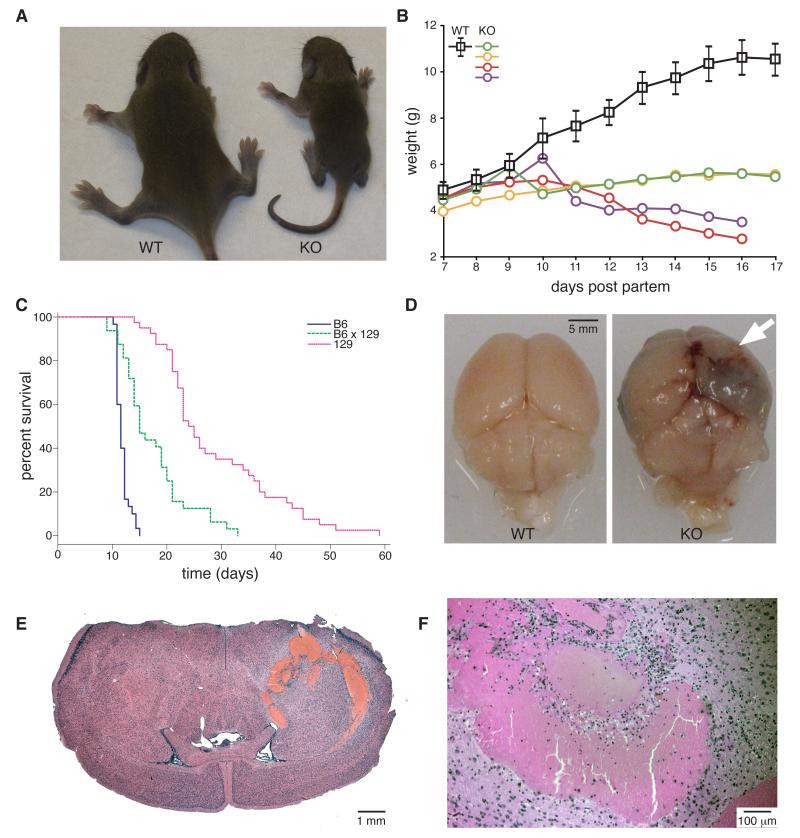
We targeted the mouse iRhom1 gene by homologous recombination to generate a loss of function mutation (Fig. S1). Homozygous knockout mice were born in a Mendelian ratio and were indistinguishable from littermates at birth but failed to gain weight, becoming obviously small and malnourished by about 6-14 days after birth (Fig. 1A, B); heterozygotes showed no phenotype. Homozygotes had no detectable white fat deposits (not shown). The rate of deterioration was dependent on genetic background (Fig 1C), but all homozygous mutants died between 9 days (background strain C57BL/6J) and 6 weeks (background strain 129S6/SvEvTac) after birth (Fig. 1C). Subsequently, knockout animals from both backgrounds were sacrificed when weight loss approached 20%.
Figure 1.
Phenotype of iRhom1 knockout mice
(A) Photograph of 11 day-old WT versus iRhom1 KO pups from a mixed C57/B6J × 129S6/SvEvTac background.
(B) Growth curve of WT versus iRhom1 KO pups from a mixed B6/129 background. The WT curve represents mean weight +/− SD of seven WT or heterozygous animals, compared to four individual KO littermates.
(C) Kaplan Meier survival plot of iRhom1 KO animals from pure B6 (blue), mixed B6×129 (green) or pure 129 (red) background.
(D) Photograph of brains from 129 mice illustrating an intracerebral haemorrhage in the KO animals. The site of haemorrhage is indicated by an arrow.
(E, F) Haematoxylin and eosin (H & E)-stained brain sections from an iRhom1 KO mouse (5×, 50× magnification).
Attempts to establish a possible primary defect by looking for the earliest phenotypes in the most severely affected mice proved inconclusive due to the pleiotropic phenotype. We therefore focused on knockouts on the milder 129S6/SvEvTac background, which normally survived 25-40 days before the end point of 20% weight loss was reached. Post-mortem investigation of these mice revealed that all had pronounced intracerebral haemorrhages (Fig 1D-F); no other gross brain abnormalities were apparent. Brain haemorrhages were also found in 100% of animals culled at 25 days, prior to exhibiting severe weight loss (8/8 animals), suggesting that this defect might be a primary consequence of iRhom1 loss.
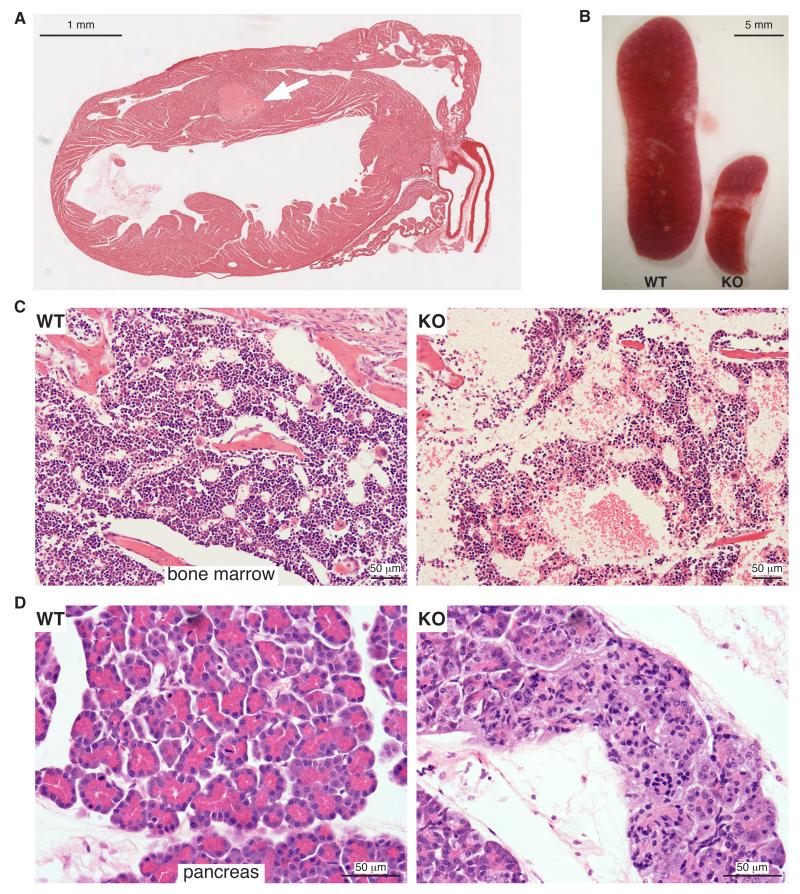
Beyond the fully penetrant brain haemorrhages on the129S6/SvEvTac background, we also found a range of other organ defects in the mixed (C57BL/6J × 129S6/SvEvTac) genetic background. There was evidence of cardiac infarction, and histological analysis indicated formation of thrombi in and around the ventricular septum close to the site of infarction (Fig 2A; 4/4 hearts examined). The spleen (Fig 2B) and thymus (not shown) were disproportionately small; the bone marrow was hypocellular, although erythroid, myeloid and megakaryocyte maturation looked normal (Fig 2C). The exocrine part of the pancreas was defective: the acini appeared small and pale and zymogen granules were absent (Fig 2D). Knockouts on both backgrounds also showed behavioural abnormalities consistent with neurological defects: B6 mice were uncoordinated and ataxic, with the hind legs being particularly affected. In 129 mice, in addition to exhibiting splayed legs and a mild transient lack of coordination around the age of 14-18 days, the main observation was obsessive eating around the time of weaning (13/13 animals examined).
Figure 2.
Pathological features of mixed genetic background iRhom1 KO animals
(A) H & E stained heart section from 9 day-old iRhom1 KO mouse (10× magnification). An arrow indicates a thrombus within the intraventricular septum.
(B) Photograph of spleens from 15 day-old WT and iRhom1 KO littermates.
(C,D) H & E-stained sections of bone marrow (C) or pancreas (D) from WT (left) and KO (right) 9 day-old littermates (200×, 400× magnification).
The phenotype of iRhom1 knockout is much more severe than that caused by loss of iRhom2, which appears immune cell specific (Adrain et al., 2012; McIlwain et al., 2012). Loss of iRhom1 causes some similar characteristics to loss of TACE: for example, small size, reduced fat mass and brain haemorrhages (Peschon et al., 1998; Gelling et al., 2008; Canault et al., 2010). Nevertheless, iRhom1 loss does not phenocopy loss of TACE, which has specific hallmarks including eyes open at birth and hair follicle defects (Peschon et al., 1998). We therefore examined the phenotype of double knockout mice, in which iRhom1 and 2 are absent, on 129S6/SvEvTac and C57BL/6J backgrounds. The double knockout phenotype proved to be more severe than TACE mutants: loss of both iRhom1 and 2 caused embryonic lethality (note that iRhom1+/−; iRhom2−/− were viable and fertile). This contrasts with the TACE knockouts, which depending on genetic background, exhibit perinatal lethality. We examined embryos from 129S6/SvEvTac mice in utero and found no live double mutants beyond E9.5-10.5, indicating that in the absence of both iRhoms, death occurred at this stage. We conclude three things from these genetic data. First, because loss of both iRhoms causes a more severe phenotype than loss of TACE, this implies that the physiological role of iRhoms is broader than TACE regulation alone. Second, the fact that the double knockout is worse than a simple combination of the two single mutants implies that there is some redundancy between them. Third, the non-overlapping phenotypes of iRhom1 and iRhom2 single mutants implies that they have distinct physiological roles.
Both iRhoms promote trafficking and maturation of TACE
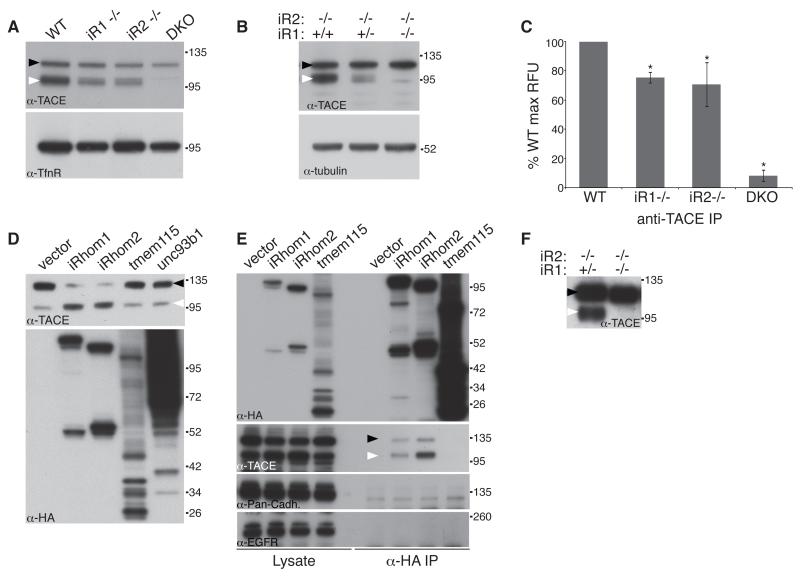
To examine whether, iRhom1 and 2 have similar molecular functions, we tested whether cellular TACE trafficking depended on both iRhoms. We isolated embryonic fibroblasts (MEFs) from iRhom1, iRhom2 and double knockout embryos. Loss of either iRhom had similar partial inhibitory effects on TACE trafficking and maturation; double knockout MEFs showed no sign of any TACE maturation (Fig 3A). This result is consistent with recent RNAi data in which knockdown of iRhom1 was shown to contribute to TACE defects in iRhom2 knockout fibroblasts (Issuree et al., 2013). Interestingly, we consistently saw that total TACE levels were reduced in iRhom KO cells. We do not understand this phenomenon but it is possible that homeostatic mechanisms prevent TACE overaccumulation when it cannot be trafficked from the ER. The conclusion that in cells other than macrophages both iRhoms contribute to TACE trafficking was further supported by a clear dependency of TACE maturation upon the levels of total iRhom (Fig 3B). The defect in TACE trafficking/maturation was mirrored in enzyme activity assays, loss of either iRhom alone reduced the proteolytic activity of TACE, but that loss of both led to complete absence of TACE mediated proteolysis (Fig 3C).
Figure 3.
iRhoms are essential for TACE maturation
(A) Western blot of Concanavalin-A (ConA)-enriched protein extract from WT, iRhom1 KO (iR1−/−), iRhom2 KO (iR2−/−) and iRhom1/2 double KO (DKO) MEFs. Here and subsequently, immature and mature TACE are indicated by black and white arrowheads respectively.
(B) Western blot of ConA-enriched protein extract from iRhom2 KO MEFs containing two, one or no iRhom1 alleles.
(C) In vitro TACE activity assay on anti-TACE immunoprecipitates from WT, iRhom single KO, and DKO MEFs. *P<0.01, t test; pairwise comparisons between WT and each of the other treatments.
(D) Western blot showing TACE maturation in HEK293 ET cells stably expressing iRhom1, iRhom2 or the control polytopic proteins, tmem115 and Unc93b1. Expression of iRhoms, tmem115 and Unc93b1 was confirmed by anti-HA western.
(E) Western blot showing TACE binding in anti-HA immunoprecipitates from cells expressing HA-tagged iRhom1, iRhom2 or tmem115.
(F) Western blot showing TACE maturation in embryonic extracts from iRhom1−/+/iRhom2−/− versus double knockout embryos.
Overexpression of iRhom1 in human HEK cells enhanced the production of mature active TACE (Fig. 3D), implying that not only is iRhom1 necessary for TACE maturation in MEFs but it is also rate limiting: it acts as a bottleneck on the levels of mature TACE in cells, making it a potential point of regulation of TACE activity.
iRhom2 acts as a cargo receptor for TACE by direct binding (Adrain et al., 2012). We found that iRhom1 also binds specifically and strongly to TACE when co-immunoprecipitated, even under quite harsh conditions (Fig 3E). In control experiments to rule out non-specific binding between membrane proteins, we found no evidence of iRhom1 binding to other single pass transmembrane proteins including cadherin and the EGFR.
In support of the mechanistic overlap between iRhom1 and 2, we found that extracts from double knockout embryos were completely devoid of mature TACE, suggesting an essential universal role for iRhoms in the developing embryo (Fig 3F).
iRhom expression patterns explains redundancy in TACE regulation
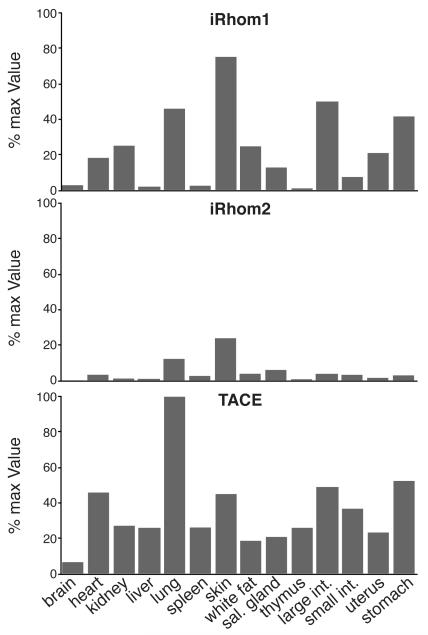
To investigate the potential extent of redundancy between iRhom1 and 2 in TACE regulation, we measured the relative expression of the two iRhoms. mRNA isolated from a panel of mouse tissues was used in real-time PCR experiments to assess the relative levels of iRhom1 and 2. These quantitative approaches showed that although iRhom2 is highly elevated in macrophages, significant levels can be found in most cell types (Fig 4). In most places, however, iRhom1 is expressed more highly than iRhom2, providing a good explanation for the more severe and widespread phenotype of iRhom1 loss. We observed that iRhoms are expressed most highly in cell types with high levels of TACE, consistent with their functional relationships (Fig 4). It is also striking that the tissues where a TACE-like phenotype is most expected (eg lung, heart, skin (Peschon et al., 1998; Jackson et al., 2003)), correspond to the tissues in which iRhom1 and 2 are significantly co-expressed (Fig 4). This provides some explanation for the lack of several characteristic TACE phenotypes in the single knockouts; in the double knockout, embryonic lethality precludes analysing later developing phenotypes.
Figure 4.
iRhom and TACE expression
Quantitative PCR analysis of RNA isolated from tissues from WT 129 mice. iRhom and TACE expression levels were normalized against TBP mRNA. Then, the most abundant signal (TACE in lung) was fixed at 100% and all other signals expressed as a proportion of this (Y axis). The individual tissues from which the RNA was obtained are indicated on the X axis. A representative result from three replicate experiments is shown.
iRhom specificity
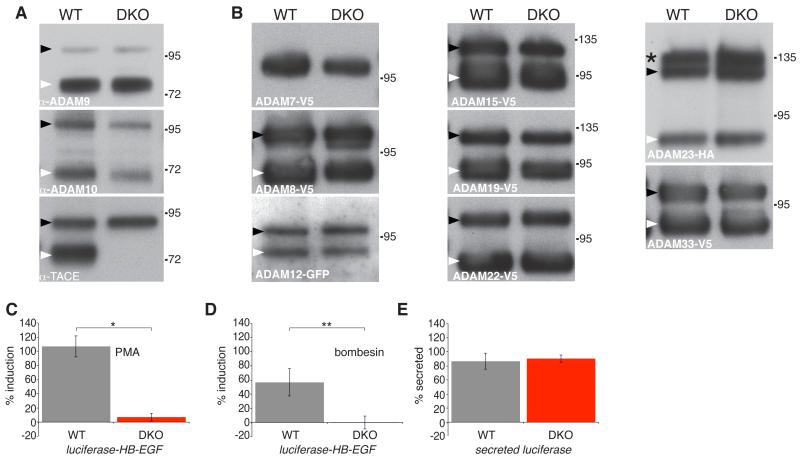
The severity of the iRhom1/2 double knockout phenotype compared to the loss of TACE indicates that iRhoms regulate more than just TACE. An obvious possibility is that other members of the ADAM family of shedding proteases might also be dependent on iRhoms. To test this we examined the trafficking and maturation of a panel of other members of the ADAM family in double knockout fibroblasts. None were affected (Fig.5 A,B, Fig S2). We also tested the trafficking of a variety of other type 1 transmembrane proteins (amphiregulin, betacellulin, EGF, EGFR, ErbB2, epiregulin, neuregulin 4, and TGFa) and, again, found none to be affected by the complete absence of both iRhoms (data not shown). Overall, we conclude that, beyond TACE, iRhoms act on additional unknown clients but that they nevertheless show a very high level of client specificity.
Figure 5.
iRhoms are specific to TACE and are essential for stimulated TACE shedding
(A) Western blot of endogenous ADAM9, ADAM10 and TACE from ConA-enriched lysates from WT or iRhom DKO MEFs. Immature and mature ADAMs are indicated by black and white arrowheads, respectively.
(B) Western blot showing tagged ADAMs −7, −8, −12, −15, −19, −22, −23, and −33 expressed in WT and iRhom DKO MEFs. Immature and mature ADAMs are indicated by black and white arrowheads, respectively. The immature and mature forms of ADAM7 can only be revealed by deglycosylation (see Fig S2A). The panel for ADAM23 shows three bands: the upper band (asterisk) is the Golgi species with prodomain intact, the middle band (black arrowhead) is the immature ER form; the lower band is mature ADAM23 lacking the prodomain (white arrowhead).
(C) Shedding of luciferase-HB-EGF after stimulation with 250 nM PMA for 1 hour. *P< = 0.01, t test.
(D) Shedding of luciferase-HB-EGF after stimulation with 1 μM bombesin for 1 hour. **P <0.05, t test.
(E) Shedding of unmodified secreted luciferase by WT versus iRhom DKO MEFs.
iRhoms are required for stimulated TACE activity
An important feature of TACE regulation, both pharmacologically and pathophysiologically, is its ability to be activated by a variety of external stimuli (Gooz, 2010). For example, TACE activity is acutely stimulated by phorbol esters and some GPCR signalling. Indeed, TACE mediates the process of transactivation, in which EGF receptor signalling is triggered indirectly by GPCR agonists, a major physiological pathway of growth factor control (Prenzel et al., 1999; Lappano and Maggiolini, 2011; Liebmann, 2011). We investigated whether such external activation of TACE was able to bypass the need for iRhom function. To test this, we fused luciferase to the TACE substrate HB-EGF and measured the ability of wild type or double knockout MEFs to shed this substrate when stimulated with PMA (Fig 5C) or the GPCR agonist, bombesin (Fig 5D). Although the double knockout cells were capable of releasing unmodified secreted luciferase as efficiently as the WT cells (Fig 5E), implying no general secretory defects, they were refractile to stimulated release of HB-EGF by either PMA or bombesin. This provides a mechanistic explanation of a previous observation that silencing of iRhom1 reduces EGFR transactivation in a specific squamous cancer cell line (Zou et al., 2009). Overall, we conclude that iRhoms are essential regulators of TACE activation not only constitutively, in all cell types we have examined, but also in response to physiological and pathological stimuli.
Concluding remarks
Our genetic data reveal that the two iRhoms have widespread and essential physiological functions in mammals. Not only do these results make clear the central role of iRhoms in TACE activation, but our observation that the double iRhom1/2 knockout is more severe than a TACE knockout implies that iRhoms have additional biological roles. The recognition of the central position of iRhoms in regulating TACE, and the crucial role of TACE in cytokine and growth factor signalling, provides a strong incentive to understand iRhom mechanisms and to consider them as potential new pharmacological targets.
Specific point mutations in iRhom2 have recently been shown to cause the inherited condition tylosis, characterised by palmoplantar hyperkeratosis and oesophageal cancer (Blaydon et al., 2012; Saarinen et al., 2012). Furthermore, iRhoms have also been genetically implicated in several other human diseases including ovarian cancer (Yan et al., 2008; Zou et al., 2009; Galvan et al., 2011; Wojnarowicz et al., 2012). It now becomes a major goal to identify which iRhom clients – TACE or others – mediate these pathological roles. Evidently the other clients are not most of the other ADAM proteases: we show that a wide selection, most notably ADAM10, the closest homologue of TACE (Saftig and Reiss, 2010), are unaffected by iRhoms. It is an attractive possibility that, during their evolution from active rhomboid proteases (which recognize and cleave TMDs), the iRhoms have retained the ability to recognise TMDs with high selectivity. If this evolutionary logic is correct, it implies that the unknown iRhom clients are likely to be other single pass transmembrane domains.
We do not yet understand the unifying functional theme of the rhomboid-like superfamily (Lemberg and Freeman, 2007; Adrain and Freeman, 2013), but given the range of functions we already know, they could widely regulate trafficking, maturation, protein stability, or degradation of membrane proteins. Another possible function might be to protect transmembrane domains from inappropriate or premature heterotypic or homotypic interactions. Finally, our observation that iRhoms mediate the activation of TACE by important physiological stimuli, including GPCRs, implies that they are potential new targets to regulate and manipulate TACE regulated inflammatory and growth factor signalling pathways.
Materials and Methods
Details of reagents and antibodies, and generation of iRhom1 KO mice are described in Supplemental Methods and outlined in Fig S1A. Co-immunoprecipitations and analysis of TACE activity in cells were performed as described (Adrain et al., 2012). To analyse TACE in embryos, E10.5 embryos were obtained following timed matings between iRhom1+/−/iRhom2−/− animals. Embryos were disrupted in a pellet pestle homogenizer (Kontes) in TX-100 lysis buffer (see reagents in Suplemental Methods). Lysates were then enriched for glycoproteins using concanavalin A-sepharose (ConA) (Sigma) as described below.
Glycoprotein enrichment using ConA
To improve the detection of TACE and other ADAMs, cells were lysed in TX-100 buffer supplemented with 1 mM EDTA, 1 mM MnCl2, 1 mM CaCl2 and glycoproteins were captured using ConA. Beads were washed twice in the same buffer and eluted into SDS-PAGE buffer supplemented with 15% sucrose.
Generation and immortalization of MEFs
For iRhom1 and iRhom2 single knockout embryos, embryonic fibroblasts were generated from E14.5 embryos. As iRhom1/2 double knockout embryos died in utero, embryonic fibroblasts were generated from E10.5 embryos. All MEFs were immortalised using lentiviral transduction of SV40 virus large T antigen (Ef1a_Large T-antigen_Ires_Puro, Addgene plasmid 18922 (Mali et al., 2008)).
Deglycosylation analysis
Lysates from MEFs generated as described above were denatured at 65oC and treated with endoglycosidase H or PNGase F according to the manufacturer’s instructions (New England Biolabs). Note that for all assays requiring western blot analysis, lysis buffer was supplemented with 10 mM 1,10-phenanthroline to prevent autoproteolysis of TACE (Schlondorff et al., 2000).
Lentiviral/retroviral transduction and expression of ADAMs in MEFs
Lentivirus was produced as described previously (Adrain et al., 2012). 293 ET cells were transduced with either pLEX.puro empty vector or pLEX.puro containing mouse iRhom1, iRhom2, TMEM115 and Unc93b1, all with a C-terminal HA tag. Cells were selected in 5 μg/ml puromycin. For retroviral transduction of MEFs, the cDNAs of ADAMs −7, −8, −15, −19,− 22 and −33 were fused to a C-terminal V5 tag and cloned into pM6P.BLAST (kind gift of F.Randow). MEFs were selected in 4 μg/ml blasticidin. The cDNAs of ADAMs −18, −21 and −28 were fused to a C-terminal V5 tag, cloned into pcDNA6 and transfected into MEFs as described in the section below. ADAM12-GFP was a gift of Marie Kveiborg and ADAM23-HA was a gift of S. Cal.
Transient transfection of MEFs
For shedding assays and assessment of EGFR ligand and ADAM trafficking, MEFs were plated at a density of 0.15 × 106 per well in 6-well plates. The following day they were transfected with 1 μg DNA and 4.5 μl of fugene-6 (Promega).
Quantitative RT PCR measurements of iRhom and TACE mRNA levels in mouse tissues
qPCR was performed as described previously (Adrain et al., 2012) with probes for mouse TBP (Mm01277045_m1), iRhom1/RHBDF1 (Mm00711711_m1), iRhom2/RHBDF2 (Mm00553469_m1) and TACE (Mm00456428_m1). The levels of iRhom1 or iRhom2 were normalized relative to the TBP mRNA levels in each sample. After this normalisation step, the most abundant signal (TACE in lung mRNA) was set as 100% and all other signals were expressed as a percentage of this.
Shedding assays
Shedding assays were based on a chimeric form of the TACE substrate HB-EGF, lacking the first 84 amino acids, fused to Gaussia luciferase. Further details are given in Supplemental Methods.
Statistical analysis
Where indicated, values are expressed as means −/+ standard deviation. Unpaired, two-tailed Student T tests were used for statistical analysis.
Supplementary Material
Acknowledgements
We thank Dr Madhuri Warren of Pathology Diagnostics for her help with histology, and the staff of MRC animal facilities for their excellent animal care. Alan Warren and Angela Moncada-Pazos provided valuable advice. This work was supported by the Medical Research Council programme number U105178780.
References
- Adrain C, Freeman M. New lives for old: evolution of pseudoenzyme function illustrated by iRhoms. Nat Rev Mol Cell Biol. 2012;13:489–498. doi: 10.1038/nrm3392. [DOI] [PubMed] [Google Scholar]
- Adrain C, Freeman M. Regulation of receptor tyrosine kinase ligand processing. Cold Spring Harb Perspect Biol. 2013 doi: 10.1101/cshperspect.a008995. doi: 10.1101/cshperspect.a008995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adrain C, Zettl M, Christova Y, Taylor N, Freeman M. Tumor necrosis factor signaling requires iRhom2 to promote trafficking and activation of TACE. Science. 2012;335:225–228. doi: 10.1126/science.1214400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blaydon DC, Etheridge SL, Risk JM, Hennies HC, Gay LJ, Carroll R, Plagnol V, McRonald FE, Stevens HP, Spurr NK, Bishop DT, Ellis A, Jankowski J, Field JK, Leigh IM, South AP, Kelsell DP. RHBDF2 mutations are associated with Tylosis, a familial esophageal cancer syndrome. Am J Hum Genet. 2012;90:340–346. doi: 10.1016/j.ajhg.2011.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blobel CP. ADAMs: key components in EGFR signalling and development. Nat Rev Mol Cell Biol. 2005;6:32–43. doi: 10.1038/nrm1548. [DOI] [PubMed] [Google Scholar]
- Canault M, Certel K, Schatzberg D, Wagner DD, Hynes RO. The lack of ADAM17 activity during embryonic development causes hemorrhage and impairs vessel formation. PLoS ONE. 2010;5:e13433. doi: 10.1371/journal.pone.0013433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DasGupta S, Murumkar PR, Giridhar R, Yadav MR. Current perspective of TACE inhibitors: a review. Bioorg Med Chem. 2009;17:444–459. doi: 10.1016/j.bmc.2008.11.067. [DOI] [PubMed] [Google Scholar]
- Galvan A, Skorpen F, Klepstad P, Knudsen AK, Fladvad T, Falvella FS, Pigni A, Brunelli C, Caraceni A, Kaasa S, Dragani TA. Multiple loci modulate opioid therapy response for cancer pain. Clin Cancer Res. 2011;17:4581–4587. doi: 10.1158/1078-0432.CCR-10-3028. [DOI] [PubMed] [Google Scholar]
- Gelling RW, Yan W, Al-Noori S, Pardini A, Morton GJ, Ogimoto K, Schwartz MW, Dempsey PJ. Deficiency of TNFalpha converting enzyme (TACE/ADAM17) causes a lean, hypermetabolic phenotype in mice. Endocrinology. 2008;149:6053–6064. doi: 10.1210/en.2008-0775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gooz M. ADAM-17: the enzyme that does it all. Crit Rev Biochem Mol Biol. 2010;45:146–169. doi: 10.3109/10409231003628015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenblatt EJ, Olzmann JA, Kopito RR. Making the cut: intramembrane cleavage by a rhomboid protease promotes ERAD. Nat Struct Mol Biol. 2012;19:979–981. doi: 10.1038/nsmb.2398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Issuree PD, Maretzky T, McIlwain DR, Monette S, Qing X, Lang PA, Swendeman SL, Park-Min KH, Binder N, Kalliolias GD, Yarilina A, Horiuchi K, Ivashkiv LB, Mak TW, Salmon JE, Blobel CP. iRHOM2 is a critical pathogenic mediator of inflammatory arthritis. J Clin Invest. 2013;123:928–932. doi: 10.1172/JCI66168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson LF, Qiu TH, Sunnarborg SW, Chang A, Zhang C, Patterson C, Lee DC. Defective valvulogenesis in HB-EGF and TACE-null mice is associated with aberrant BMP signaling. EMBO J. 2003;22:2704–2716. doi: 10.1093/emboj/cdg264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lappano R, Maggiolini M. G protein-coupled receptors: novel targets for drug discovery in cancer. Nat Rev Drug Discov. 2011;10:47–60. doi: 10.1038/nrd3320. [DOI] [PubMed] [Google Scholar]
- Lemberg MK. Sampling the membrane: function of rhomboid-family proteins. Trends Cell Biol. 2013;23:210–217. doi: 10.1016/j.tcb.2013.01.002. [DOI] [PubMed] [Google Scholar]
- Lemberg MK, Freeman M. Functional and evolutionary implications of enhanced genomic analysis of rhomboid intramembrane proteases. Genome Res. 2007;17:1634–1646. doi: 10.1101/gr.6425307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liebmann C. EGF receptor activation by GPCRs: an universal pathway reveals different versions. Mol Cell Endocrinol. 2011;331:222–231. doi: 10.1016/j.mce.2010.04.008. [DOI] [PubMed] [Google Scholar]
- Liu P, Jenkins NA, Copeland NG. A highly efficient recombineering-based method for generating conditional knockout mutations. Genome Res. 2003;13:476–484. doi: 10.1101/gr.749203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mali P, Ye Z, Hommond HH, Yu X, Lin J, Chen G, Zou J, Cheng L. Improved efficiency and pace of generating induced pluripotent stem cells from human adult and fetal fibroblasts. Stem Cells. 2008;26:1998–2005. doi: 10.1634/stemcells.2008-0346. [DOI] [PubMed] [Google Scholar]
- McIlwain DR, Lang PA, Maretzky T, Hamada K, Ohishi K, Maney SK, Berger T, Murthy A, Duncan G, Xu HC, Lang KS, Haussinger D, Wakeham A, Itie-Youten A, Khokha R, Ohashi PS, Blobel CP, Mak TW. iRhom2 regulation of TACE controls TNF-mediated protection against Listeria and responses to LPS. Science. 2012;335:229–232. doi: 10.1126/science.1214448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peschon JJ, Slack JL, Reddy P, Stocking KL, Sunnarborg SW, Lee DC, Russell WE, Castner BJ, Johnson RS, Fitzner JN, Boyce RW, Nelson N, Kozlosky CJ, Wolfson MF, Rauch CT, Cerretti DP, Paxton RJ, March CJ, Black RA. An essential role for ectodomain shedding in mammalian development. Science. 1998;282:1281–1284. doi: 10.1126/science.282.5392.1281. [DOI] [PubMed] [Google Scholar]
- Prenzel N, Zwick E, Daub H, Leserer M, Abraham R, Wallasch C, Ullrich A. EGF receptor transactivation by G-protein-coupled receptors requires metalloproteinase cleavage of proHB-EGF. Nature. 1999;402:884–888. doi: 10.1038/47260. [DOI] [PubMed] [Google Scholar]
- Saarinen S, Vahteristo P, Lehtonen R, Aittomaki K, Launonen V, Kiviluoto T, Aaltonen LA. Analysis of a Finnish family confirms RHBDF2 mutations as the underlying factor in tylosis with esophageal cancer. Fam Cancer. 2012;11:525–528. doi: 10.1007/s10689-012-9532-8. [DOI] [PubMed] [Google Scholar]
- Saftig P, Reiss K. The “A Disintegrin And Metalloproteases” ADAM10 and ADAM17: Novel drug targets with therapeutic potential? Eur J Cell Biol. 2010;90:527–535. doi: 10.1016/j.ejcb.2010.11.005. [DOI] [PubMed] [Google Scholar]
- Schlondorff J, Becherer JD, Blobel CP. Intracellular maturation and localization of the tumour necrosis factor alpha convertase (TACE) Biochem J. 2000;347:131–138. [PMC free article] [PubMed] [Google Scholar]
- Siggs OM, Xiao N, Wang Y, Shi H, Tomisato W, Li X, Xia Y, Beutler B. iRhom2 is required for the secretion of mouse TNF alpha. Blood. 2012;119:5769–5771. doi: 10.1182/blood-2012-03-417949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wojnarowicz PM, Provencher DM, Mes-Masson AM, Tonin PN. Chromosome 17q25 genes, RHBDF2 and CYGB, in ovarian cancer. Int J Oncol. 2012;40:1865–1880. doi: 10.3892/ijo.2012.1371. [DOI] [PubMed] [Google Scholar]
- Yan Z, Zou H, Tian F, Grandis JR, Mixson AJ, Lu PY, Li LY. Human rhomboid family-1 gene silencing causes apoptosis or autophagy to epithelial cancer cells and inhibits xenograft tumor growth. Mol Cancer Ther. 2008;7:1355–1364. doi: 10.1158/1535-7163.MCT-08-0104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zettl M, Adrain C, Strisovsky K, Lastun V, Freeman M. Rhomboid family pseudoproteases use the ER quality control machinery to regulate intercellular signaling. Cell. 2011;145:79–91. doi: 10.1016/j.cell.2011.02.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou H, Thomas SM, Yan ZW, Grandis JR, Vogt A, Li LY. Human rhomboid family-1 gene RHBDF1 participates in GPCR-mediated transactivation of EGFR growth signals in head and neck squamous cancer cells. FASEB J. 2009;23:425–432. doi: 10.1096/fj.08-112771. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.