Abstract
Parkinson’s disease (PD) has a number of known genetic risk factors. Clinical and epidemiological studies have suggested the existence of intermediate factors that may be associated with additional risk of PD. We construct genetic risk profiles for additional epidemiological and clinical factors using known genome-wide association studies (GWAS) loci related to these specific phenotypes to estimate genetic comorbidity in a systematic review. We identify genetic risk profiles based on GWAS variants associated with schizophrenia and Crohn’s disease as significantly associated with risk of PD. Conditional analyses adjusting for SNPs near loci associated with PD and schizophrenia or PD and Crohn’s disease suggest that spatially overlapping loci associated with schizophrenia and PD account for most of the shared comorbidity, while variation outside of known proximal loci shared by PD and Crohn’s disease accounts for their shared genetic comorbidity. We examine brain methylation and expression signatures proximal to schizophrenia and Crohn’s disease loci to infer functional changes in the brain associated with the variants contributing to genetic comorbidity. We compare our results with a systematic review of epidemiological literature, while the findings are dissimilar to a degree; marginal genetic associations corroborate the directionality of associations across genetic and epidemiological data. We show a strong genetically defined level of comorbidity between PD and Crohn’s disease as well as between PD and schizophrenia, with likely functional consequences of associated variants occurring in brain.
INTRODUCTION
Parkinson’s disease (PD) is recognized to be associated with a number of genetic susceptibility factors, including variability at the loci SNCA, LRRK2, MAPT, BST1, GAK, HLA-DR, ACMSD, STK39, MCCC1/LAMP3, SYT11, CCDC62/HIP1R, PARK16/1q32, STX1B/16p11, FGF20/8p22, STBD1/4q21, GPNMB/7p15, among others, which have been identified in genome-wide association studies (GWAS). It is likely that other additional genetic risk factors also contribute. PD has been reported to be associated with a number of clinical comorbidities and altered laboratory values, such as affective disorders and serum urate levels. Several of these have themselves been associated with genetic susceptibility factors, for which information is available from the NHGRI GWAS catalog (http://www.genome.gov/26525384, (1)).
This offers the opportunity to examine genetic risk profiles associated with these clinical phenotypes as risk factors for PD. Genetic risk profiles may be thought of as the cumulative genetic load of risk alleles related to a particular disease or trait, explaining more of the attributable genetic risk associated with this disorder than a single SNP itself. While many epidemiological studies examine associations between intermediate phenotypes and a specific outcome, in this study we sought to examine how the genetic risk profiles associated with these intermediate phenotypes may be associated with PD risk themselves. We sought to identify genetic comorbidities of PD by testing associations with genetic risk profiles previously associated with intermediate phenotypes of interest. Any disease or trait with a significant risk profile score associated with PD essentially would share some genetic factors in common.
In this study, we identify PD genetic comorbidities such as Crohn’s disease and schizophrenia. We also attempt to dissect the individual contributions of these associated loci in their contributions to PD risk. Additionally, we utilize expression and methylation data to infer functional genetic consequences in brain tissues associated with these genetic comorbidities.
RESULTS
We used a large sample series of >5000 PD cases and 9000 controls with genome-wide genotyping data (Table 1). We defined a list of clinical factors that are potentially associated with PD risk or comorbidity. We then examined how SNPs associated with these clinical factors might be associated with PD itself using a measure of the cumulative effect of all SNPs related to that particular factor (Supplementary Material, Tables S1 and S2). We identified 18 clinical factors suggested by the literature and clinical observation to be possible factors associated with PD for which there were high-quality GWAS results in the NHGRI catalog (http://www.genome.gov/26525384, (1)). These include: serum amyloid, bipolar disorder, caffeine intake, Crohn’s disease, hypertension, inflammation, serum iron, melanoma, obesity, PD, psoriasis, rheumatoid arthritis, schizophrenia, smoking, type 2 diabetes, ulcerative colitis and serum urate (2-68). As a note, PD was included as a positive control. For these phenotypes of interest, alleles associated with an increase in risk for binary phenotypes and/or alleles associated with increase in the level of continuous phenotypes were summed to create the genetic risk profiles.
Table 1. Descriptive statistics of cohorts contributing to analyses.
| Cases | Controls | |||||
|---|---|---|---|---|---|---|
| Cohort | Age at onset, mean (SD) in years | % Female | n | Age at enrollment, mean (SD) in years | % Female | n |
| France | 57.548 (13.086) | 41.523 | 985 | 73.736 (5.373) | 42.843 | 1984 |
| Germany | 55.715 (11.549) | 39.595 | 740 | 47.416 (12.376) | 47.987 | 944 |
| Netherlands | 55.649 (11.826) | 36.446 | 771 | 55.721 (5.771) | 56.028 | 2024 |
| NIA—USA | 57.812 (13.156) | 40.235 | 937 | 61.933 (11.599) | 24.525 | 1896 |
| UK | 64.167 (12.434) | 42.051 | 1648 | 53 (0) | 48.388 | 2699 |
NIA denotes cohorts with data generated at the Laboratory of Neurogenetics at the National Institute on Aging, National Institutes of Health, Bethesda, MD, USA.
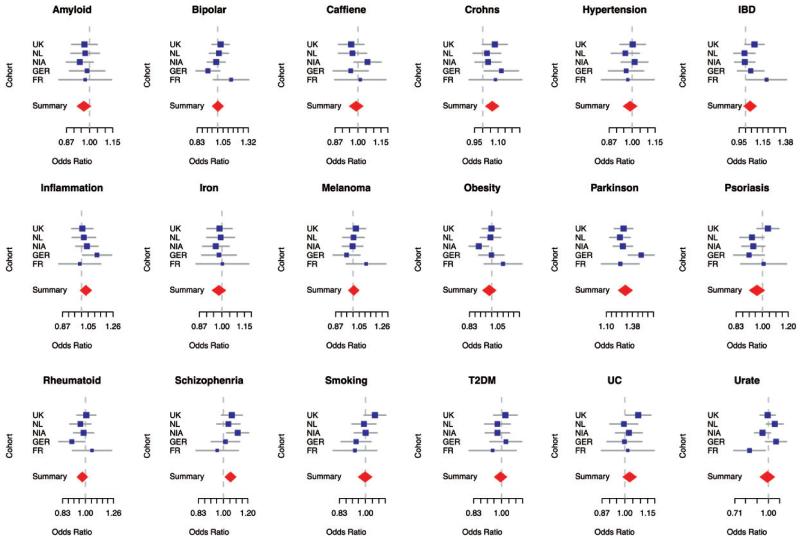
Within each of the five IPDGC cohorts participating in this analysis, each risk profile comprising a number of SNPs was tested using logistic regression for an association with PD status, adjusting for sex, age at onset/exam and population substructure. Summary statistics from each cohort were combined through meta-analysis under random effects to account for possible heterogeneity (see Figure 1 and Table 2). This resulted in three genetic risk profiles being significantly associated with PD. These include profiles based on SNPs taken from studies of Crohn’s disease, schizophrenia and PD. A single standard deviation increase in the genetic risk profile for PD, equivalent to roughly a 34% increase in the genetic burden associated with known PD risk alleles within a population, was associated with an odds ratio (OR) of 1.29 (95% confidence interval 1.22–1.38, P-value,<2E–16). Using the same burden scale, a single standard deviation increase from population means in the allelic burdens derived from Crohn’s disease and schizophrenia risk profiles was significantly associated with PD status at an OR of 1.06 (95% confidence interval 1.02–1.11, P-value 0.005) and 1.05 (95% confidence interval 1.01–1.10, P-value 0.012), respectively.
Figure 1. Forest plots from random effects meta-analyses of profiled phenotypes.
Cohort-specific OR associated with PD are shown in blue, the size of the square is proportional to the size of the study. Confidence intervals of the summary OR per phenotype are shown as red diamonds, with the centerline of each diamond representing the summary OR for that particular phenotype. The dependent variable in each model is the standardized count of disease risk or quantitative measure increasing alleles per sample associated with either disease status or continuous measures of interest. Abbreviations include Bipolar for bipolar disorder, IBD for inflammatory bowel disease, T2DM for type 2 diabetes, Crohn’s for Crohn’s disease, UC for ulcerative colitis, Parkinson’s for Parkinson’s disease, Rheumatoid for Rheumatoid arthritis, Smoking for smoking history/rate, Caffeine for caffeine or coffee consumption levels, amyloid for serum amyloid levels, iron for serum iron levels and urate for serum urate levels.
Table 2. Summary statistics from random effects meta-analysis of profile phenotypes.
| Profiled phenotype | OR | Lower limit of the OR 95% confidence interval | Upper limit of the OR 95% confidence interval | Beta | SE | P-value | I 2 | Heterogeneity P-value |
|---|---|---|---|---|---|---|---|---|
| Amyloid | 0.966 | 0.927 | 1.007 | −0.034 | 0.021 | 0.106 | 0 | 0.970 |
| Bipolar | 1.002 | 0.953 | 1.053 | 0.002 | 0.025 | 0.949 | 0.270 | 0.241 |
| Caffiene | 0.989 | 0.949 | 1.031 | −0.011 | 0.021 | 0.613 | 0 | 0.406 |
| Crohns | 1.061 | 1.018 | 1.106 | 0.059 | 0.021 | 0.005 | 0 | 0.688 |
| Hypertension | 0.988 | 0.948 | 1.030 | −0.012 | 0.021 | 0.569 | 0 | 0.867 |
| IBD | 1.038 | 0.987 | 1.092 | 0.038 | 0.026 | 0.143 | 0.284 | 0.232 |
| Inflammation | 1.032 | 0.991 | 1.076 | 0.032 | 0.021 | 0.131 | 0 | 0.535 |
| Iron | 0.982 | 0.942 | 1.024 | −0.018 | 0.021 | 0.388 | 0 | 0.986 |
| Melanoma | 1.005 | 0.965 | 1.048 | 0.005 | 0.021 | 0.796 | 0 | 0.583 |
| Obesity | 0.980 | 0.927 | 1.036 | − 0.020 | 0.028 | 0.471 | 0.416 | 0.144 |
| Parkinson | 1.298 | 1.222 | 1.380 | 0.261 | 0.031 | <2E – 16 | 0.466 | 0.112 |
| Psoriasis | 0.962 | 0.912 | 1.015 | − 0.039 | 0.027 | 0.155 | 0.359 | 0.182 |
| Rheumatoid | 0.975 | 0.934 | 1.019 | − 0.025 | 0.022 | 0.259 | 0.065 | 0.370 |
| Schizophrenia | 1.055 | 1.012 | 1.100 | 0.053 | 0.021 | 0.012 | 0 | 0.424 |
| Smoking | 0.997 | 0.949 | 1.047 | − 0.003 | 0.025 | 0.906 | 0.259 | 0.249 |
| T2DM | 0.993 | 0.953 | 1.036 | − 0.007 | 0.021 | 0.756 | 0 | 0.734 |
| UC | 1.030 | 0.988 | 1.073 | 0.029 | 0.021 | 0.168 | 0 | 0.595 |
| Urate | 0.989 | 0.918 | 1.066 | −0.011 | 0.038 | 0.772 | 0.665 | 0.018 |
The dependent variable in each model is the standardized count of disease risk or quantitative measure increasing alleles per sample associated with either disease status or continuous measures of interest. OR and beta coefficients correspond to a single standard deviation of change in the genetic profile for each phenotype based on allele counts associated with an increase in disease risk or the level of quantitative phenotype. Abbreviations include IBD for inflammatory bowel disease, T2DM for type 2 diabetes, UC for ulcerative colitis, SE denotes standard error of the beta coefficient from logistic regression and I2 is the index of heterogeneity.
All 96 SNPs within these three risk profiles (Crohn’s disease, PD and schizophrenia) were tested independently for association with PD using identical statistical models and meta-analytic methods as described for the analyses of risk profiles (Supplementary Material, Table S3). Two SNPs not previously known to be associated with PD passed Bonferroni correction for 99 tests. The first was rs11191580, a SNP in the NT5C2 gene known to be associated with schizophrenia. This SNP reached genome-wide significance in our testing (risk associated with reference allele T at OR 1.35, 95% confidence interval 1.21–1.50, P-value 3.98E–8, T frequency 91.2%). The region surrounding rs11191580 was suggested to be associated with PD initially in a previous publication by Simon-Sanchez et al. using a portion of the IPDGC data (42). However, the association at this locus was never definitively replicated in a European ancestry population and was therefore not included in the PD genetic risk profiles we have constructed in this report (42). The other SNP passing Bonferroni correction was rs11564258, a SNP near the LRRK2 risk locus from the Crohn’s disease risk profile, at an OR of 0.69 associated with the G reference allele (95% confidence interval 0.58–0.83, P-value 5.49E–5, G frequency 97.5%) showing possible LD with the LRRK2 PD risk locus (49).
To assess the independence of the risk profile associations outside of possible spatial overlaps with PD loci, risk profile score associations were recalculated adjusting for SNPs near PD risk loci as covariates. Keller et al.’s summary of PD loci was used, in addition to data mined from the GWAS catalog to further scrutinize putative PD loci (69). To define SNPs used as covariates, they must be within 1 mb of a PD risk locus described in either the downloaded GWAS catalog, Keller et al. or Simon-Sanchez et al. (1,42,69). When adjusting for SNPs near PD risk loci as covariates, the schizophrenia risk profile association was reevaluated adjusting for rs11191580 and rs7914558 in NT5C2 and rs3131296 near the HLA-DRA risk locus as additional covariates. The Crohn’s disease risk profile association was adjusted for rs11564258 and rs11175593 near LRRK2, rs17309827 near HLA-DRA and rs1736135 near the suspected PD risk locus at USP25. After meta-analyzing the cohort-specific summary statistics adjusted for these SNPs, the association between the Crohn’s disease’s risk profile remained significantly associated with PD risk (OR 1.05, 95% confidence interval 1.01–1.09, P-value 0.03), while the schizophrenia association was completely attenuated (OR 0.99, 95% confidence interval 0.92–1.06, P-value 0.76). This suggests that there are additional genetic factors outside of known PD loci overlap that contribute to the genetic comorbidity shared by PD and Crohn’s disease, while the genetic comorbidity shared by schizophrenia and PD may simply be due to possible overlapping loci, although the association at NT5C2 in PD remains to be definitively replicated.
Systematic review data were extracted for overlapping phenotypes from Noyce et al. (1) to evaluate how genetic risk may be reflected in epidemiological data (Table 3). Neither Crohn’s disease or schizophrenia associations were significantly replicated in the epidemiological data, although the directionality of effect for psychosis aspects of schizophrenia and Crohn’s disease mirror genetic risk estimates associated with PD. The directionality of effect in our study of genetic factors relating to PD comorbidity leads support to the effects described in the statistically significant epidemiologically evaluated comorbidities of smoking status, coffee drinking and hypertension as described in the systematic review.
Table 3. Systematic review of epidemiological literature for select traits of interest.
| Factor | Study/studies (n) | Cases (n) | Controls (n) | OR | Lower limit of the OR 95% confidence interval | Upper limit of the OR 95% confidence interval | SE | P-value | I 2 | Heterogeneity P-value |
|---|---|---|---|---|---|---|---|---|---|---|
| Ever versus never smokinga | 67 | 19518 | 1 053 664 | 0.64 | 0.6 | 0.69 | 0.0347 | <0.001 | 49.60% | <0.001 |
| Drinking versus non-drinking coffeea | 19 | 5801 | 723 072 | 0.67 | 0.58 | 0.76 | 0.0684 | <0.001 | 42.90% | 0.025 |
| Hypertension preceding PDa | 12 | 5993 | 187226 | 0.74 | 0.61 | 0.9 | 0.0989 | 0.003 | 76.50% | <0.001 |
| Diabetes preceding PDa | 13 | 20 025 | 303 543 | 0.91 | 0.72 | 1.15 | 0.1189 | 0.423 | 70.90% | <0.001 |
| Melanoma preceding PDb | 6 | – | – | 1.07 | 0.62 | 1.84 | – | – | 49.30% | 0.079 |
| Serum ironc | 10 | 520 | 711 | NS | NS | NS | – | – | 93.40% | <0.0000 |
| Bipolar disorder preceding PD | Shiba et al. (71, 83) | 196 | 196 | 1 | 0.1 | 16 | – | – | NA | NA |
| Psoriasis preceding PD | Rugbjerg et al. (72,84) | 13 695 | 68 445 | 1.25 | 0.51 | 3.06 | – | – | NA | NA |
| Ulcerative colitis preceding PD | Rugbjerg et al. (72,84) | 13 695 | 68 445 | 1.3 | 0.9 | 1.8 | – | – | NA | NA |
| Crohn’s disease preceding PD | Rugbjerg et al. (72,84) | 13 695 | 68 445 | 1.06 | 0.54 | 2.1 | – | – | NA | NA |
| Rheumatoid arthritis preceding PD | Rugbjerg et al. (72,84) | 13 695 | 68 445 | 0.7 | 0.5 | 0.9 | – | – | NA | NA |
| Blood inflammatory markers preceding PDd | Chen et al. (73, 85) | 84 | 165 | 3.4 | 1.1 | 10.5 | NA | NA | ||
| Obesity preceding PDa | 3/7 studies described significant associations with obesity preceding PD through a variety of different measures (RR/OR 2.8f, 2.03f, 0.43f, 0.9, 0.8, 0.86, 0.99) | |||||||||
| Urate level preceding PDa | 3/5 studies described significant associations with elevate durate preceding PD using various cut-offs (ORs 0.4f, 0.6f, 0.71f, 0.43, 1.33) and 1 with preceding gout (OR 0.69f) | |||||||||
| Schizophrenia preceding PD$e | Two studies (Shiba et al. (71, 83): schizophrenia OR 1.0 (CI 0.1–7.0); Rajput et al. (74, 86): psychosis RR 1.5 (CI 0.3–6.7)) | |||||||||
| Serum amyloid | No known epidemiological studies comparing serum amyloid levels between PD and controls | |||||||||
NS, non-significant but figures not provided; NA, not available; RR, relative risk.
Summary data from meta-analysis undertaken by Noyce et al. (1).
Summary data from meta-analys is undertaken by Liu et al. (86). NB: After exclusion of the only negative study OR 1.44 (CI 1.06–1.96); association of existing PD with melanoma OR 3.61 (CI 1.49–8.77).
Summary data from meta-analysis undertaken by Mariani et al. (87); data are for patients with established PD; Mariani et al. also report reduced transferrin and transferrin saturation.
OR refers to highest quintile ofIL-6 compared with the lowest (P for trend 0.03). Associations with high-sensitivity c-reactive protein (CRP), fibrinogen and tumor necrosis factor were non-significant. NB: In established PD, Song et al. (75, 87) reported increased high-sensitivity CRP compared with controls OR 2.04 (CI 1.18– 3.52).
Risk of bias through drug-induced parkinsonism due to neuroleptic treatment.
Statistically significant result (where confidence interval not given).
Mining of brain tissue to infer possible biological functionality at loci associated with PD, Crohn’s disease and schizophrenia, we examined regional methylation and expression data within ±1 mb of SNPs of interest in a large series of neurologically normal frontal cortex and cerebellar tissues. While we identified a number of loci significantly associated with methylation and expression changes in the brain tissue samples, our primary interest was to focus on SNPs from overlapping regions identified as risk loci for PD and Crohn’s or PD and schizophrenia (Table 4). SNPs in the HLA region used to construct the Parkinson’s and schizophrenia profiles were significantly associated with changes in regional methylation status in both the frontal and cerebellar tissue samples, allowing us to infer functional consequence in brain tissues associated with these proximal risk SNPs. Alleles at this locus associated with risk of PD and schizophrenia were concurrently associated with decreased methylation in the frontal cortex (49). We also show that a Crohn’s disease-associated SNP near the LRRK2 PD risk locus is also significantly associated with methylation changes in the frontal cortex. This suggests that genetic variants associated with Crohn’s disease and schizophrenia (in addition to PD) may alter brain function to some degree.
Table 4. Local changes in mRNA expression and CpG methylation are associated with SNPs of interest for Crohn’s disease, PD and schizophrenia.
| SNP name | Chromosome | SNP position (bp, build 37) | Major allele, minor allele | MAF | Imputation quality (RSQ from MiniMach) | Effect, based on minor allele dosage | Standard error | P-value | CpG or mRNA probe | Assay | Tissue sampled | Distance from SNP to probe (bp) | Probe-associated gene | Phenotype |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| rs2274910 | 1 | 160852046 | C,T | 0.2776 | 1 | −0.744 | 0.09 | 1.05E–16 | cg10106388 | CpG methylation | CRBLM | 19384 | CD244 | Crohns |
| rs10181042 | 2 | 61224259 | C,T | 0.4224 | 0.9946 | 0.414 | 0.074 | 1.82E–08 | ILMN_1754501 | mRNA expression | FCTX | 166006 | NA | Crohns |
| rs9858542 | 3 | 49701983 | G,A | 0.3273 | 0.8245 | − 0.485 | 0.095 | 3.05E–07 | cg12788313 | CpG methylation | FCTX | 24144 | MST1 | Crohns |
| rs9858542 | 3 | 49701983 | G,A | 0.322 | 0.6 | − 0.553 | 0.099 | 2.06E–08 | ILMN 1690454 | mRNA expression | CRBLM | 140324 | FAM212A | Crohns |
| rs2188962 | 5 | 131770805 | C,T | 0.4244 | 1 | 0.388 | 0.087 | 8.62E–06 | cg20512303 | CpG methylation | CRBLM | 177896 | PDLIM4 | Crohns |
| rs2188962 | 5 | 131770805 | C,T | 0.4244 | 1 | − 0.388 | 0.087 | 8.67E–06 | cg22598563 | CpG methylation | CRBLM | 206884 | P4HA2 | Crohns |
| rs359457 | 5 | 173279842 | T,C | 0.4171 | 0.999 | 0.463 | 0.074 | 4.26E–10 | ILMN_1722025 | mRNA expression | CRBLM | 105114 | CPEB4 | Crohns |
| rs359457 | 5 | 173279842 | T,C | 0.4171 | 0.999 | 0.672 | 0.075 | 2.31E–19 | ILMN_1722025 | mRNA expression | FCTX | 105114 | CPEB4 | Crohns |
| rs13194053 | 6 | 27143883 | T,C | 0.1683 | 0.9872 | 0.516 | 0.109 | 2.26E–06 | cg14345882 | CpG methylation | FCTX | 779090 | BTN3A2 | Schizophrenia |
| rs2021722 | 6 | 30174131 | C,T | 0.2031 | 0.9775 | 0.429 | 0.107 | 5.96E–05 | cg17475918 | CpG methylation | CRBLM | 283056 | HLA-E | Schizophrenia |
| rs3131296 | 6 | 32172993 | C,T | 0.1409 | 0.9875 | − 0.597 | 0.126 | 2.13E–06 | cg25764570 | CpG methylation | FCTX | 234246 | HLA-DRA | Schizophrenia |
| rs2395163 | 6 | 32387809 | T,C | 0.2251 | 1 | 0.42 | 0.102 | 3.46E–05 | cg07363637 | CpG methylation | FCTX | 540863 | SLC44A4 | Parkinson |
| rs2395163 | 6 | 32387809 | T,C | 0.2251 | 1 | 0.428 | 0.102 | 2.46E–05 | cg21415604 | CpG methylation | FCTX | 439376 | C4B | Parkinson |
| rs3129882 | 6 | 32409530 | A,G | 0.4151 | 1 | 0.473 | 0.089 | 1.00E–07 | cg25764570 | CpG methylation | FCTX | 2291 | HLA-DRA | Parkinson |
| rs2301436 | 6 | 167437988 | C,T | 0.4308 | 0.8426 | − 0.35 | 0.081 | 1.50E–05 | ILMN_1671565 | mRNA expression | CRBLM | 94917 | RNASET2 | Crohns |
| rs4077515 | 9 | 139266496 | C,T | 0.4247 | 0.9333 | − 0.901 | 0.076 | 1.67E–32 | ILMN 1811301 | mRNA expression | CRBLM | 56817 | INPP5E | Crohns |
| rs4077515 | 9 | 139266496 | C,T | 0.4247 | 0.9333 | − 0.904 | 0.076 | 6.37E–33 | ILMN 1811301 | mRNA expression | FCTX | 56817 | INPP5E | Crohns |
| rs7914558 | 10 | 104775908 | G,A | 0.3991 | 0.9932 | − 0.696 | 0.086 | 5.20E–16 | cg00035347 | CpG methylation | FCTX | 177700 | NT5C2 | Schizophrenia |
| rs7914558 | 10 | 104775908 | G,A | 0.3993 | 0.8939 | − 0.439 | 0.08 | 3.36E–08 | ILMN_1682165 | mRNA expression | CRBLM | 72030 | NT5C2 | Schizophrenia |
| rs102275 | 11 | 61557803 | T,C | 0.3279 | 0.9763 | − 0.423 | 0.075 | 1.70E–08 | ILMN 1670134 | mRNA expression | CRBLM | 9609 | FADS1 | Crohns |
| rs102275 | 11 | 61557803 | T,C | 0.3279 | 0.9763 | −0.315 | 0.075 | 2.57E–05 | ILMN_1670134 | mRNA expression | FCTX | 9609 | FADS1 | Crohns |
| rs694739 | 11 | 64097233 | A,G | 0.4073 | 0.8502 | −0.911 | 0.081 | 2.75E–29 | ILMN_1772208 | mRNA expression | CRBLM | 27286 | CCDC88B | Crohns |
| rs11175593 | 12 | 40601940 | C,T | 0.0189 | 0.9679 | 1.324 | 0.299 | 9.86E–06 | cg04575343 | CpG methylation | FCTX | 102678 | SLC2A13 | Crohns |
| rs12817488 | 12 | 123296294 | G,A | 0.4249 | 0.8039 | − 0.368 | 0.093 | 7.46E–05 | cg05384917 | CpG methylation | CRBLM | 95341 | GPR109B | Parkinson |
| rs2942168 | 17 | 43714850 | G,A | 0.2538 | 0.9091 | 0.522 | 0.104 | 5.73E–07 | cg19832721 | CpG methylation | CRBLM | 534966 | KIAA1267 | Parkinson |
| rs2942168 | 17 | 43714850 | G,A | 0.247 | 0.3054 | − 0.87 | 0.149 | 5.30E–09 | ILMN_1709549 | mRNA expression | CRBLM | 201405 | PLEKHM1 | Parkinson |
| rs2942168 | 17 | 43714850 | G,A | 0.247 | 0.3054 | − 2.287 | 0.164 | 3.66E–44 | ILMN_2393693 | mRNA expression | CRBLM | 130367 | PLEKHM1 | Parkinson |
| rs393152 | 17 | 43719143 | A,G | 0.2551 | 0.997 | 0.497 | 0.099 | 5.76E–07 | cg19832721 | CpG methylation | CRBLM | 530673 | KIAA1267 | Parkinson |
| rs393152 | 17 | 43719143 | A,G | 0.2483 | 0.335 | − 0.829 | 0.142 | 5.25E–09 | ILMN_1709549 | mRNA expression | CRBLM | 205698 | PLEKHM1 | Parkinson |
| rs393152 | 17 | 43719143 | A,G | 0.2483 | 0.335 | −2.18 | 0.156 | 3.41E–44 | ILMN_2393693 | mRNA expression | CRBLM | 134660 | PLEKHM1 | Parkinson |
| rs8070723 | 17 | 44081064 | A,G | 0.2547 | 1 | 0.496 | 0.099 | 5.77E–07 | cg19832721 | CpG methylation | CRBLM | 168752 | KIAA1267 | Parkinson |
| rs8070723 | 17 | 44081064 | A,G | 0.2592 | 0.946 | − 0.509 | 0.083 | 8.29E–10 | ILMN_1709549 | mRNA expression | CRBLM | 567619 | PLEKHM1 | Parkinson |
| rs8070723 | 17 | 44081064 | A,G | 0.2592 | 0.946 | − 1.265 | 0.091 | 1.07E–43 | ILMN_2393693 | mRNA expression | CRBLM | 496581 | PLEKHM1 | Parkinson |
| rs199533 | 17 | 44828931 | G,A | 0.2386 | 0.8935 | 0.487 | 0.108 | 5.91E–06 | cg19832721 | CpG methylation | CRBLM | 579115 | KIAA1267 | Parkinson |
| rs199533 | 17 | 44828931 | G,A | 0.2449 | 0.7188 | − 0.4 | 0.099 | 5.37E–05 | ILMN_1698680 | mRNA expression | FCTX | 234735 | ARL17A | Parkinson |
| rs281379 | 19 | 49214274 | A,G | 0.4943 | 0.9789 | 0.484 | 0.086 | 1.71E–08 | cg13881341 | CpG methylation | CRBLM | 44941 | FUT1 | Crohns |
| rs281379 | 19 | 49214274 | A,G | 0.4943 | 0.9789 | 0.389 | 0.086 | 5.73E–06 | cg16155702 | CpG methylation | CRBLM | 45936 | FGF21 | Crohns |
| rs4809330 | 20 | 62349586 | G,A | 0.3435 | 1 | − 0.388 | 0.077 | 4.23E–07 | ILMN_1794643 | mRNA expression | CRBLM | 17792 | ZGPAT | Crohns |
| rs181359 | 22 | 21928641 | G,A | 0.2118 | 0.8549 | − 0.698 | 0.111 | 3.49E–10 | cg22262140 | CpG methylation | CRBLM | 58335 | FLJ36046 | Crohns |
| rs181359 | 22 | 21928641 | G,A | 0.2118 | 0.8549 | − 0.481 | 0.111 | 1.51E–05 | cg22262140 | CpG methylation | FCTX | 58335 | FLJ36046 | Crohns |
The table shows only significantly associated SNP-probe pairs after Bonferroni correction for multiple testing. Abbreviations include CRBLM for cerebellum, FCTX for frontal cortex and bp for base pair.
DISCUSSION
In this study, we identified Crohn’s disease and schizophrenia as genetic comorbidities of PD. The genetic risk profiles based on GWAS identified loci for these two diseases showed significant risk associated with PD. The genetic comborbidity associated with schizophrenia was almost entirely accounted for by SNPs in the PD associated loci near NT5C2 and HLA-DRA. On the other hand, Crohn’s disease’s risk profile and PD remained significantly associated even when adjusting for overlapping SNPs at loci near known PD risk loci USP25, HLA-DRA and LRRK2. This suggests that additional risk loci associated with Crohn’s disease are also connected to PD risk, outside of those already near known PD loci. In the near future, deep sequencing of large population based studies should help clarify further genetic comorbidities and provide greater insight into the mechanisms of multiple related diseases.
There seems to be functional connectivity between these diseases based on our analyses of expression and methylation data. SNPs associated with both schizophrenia and Crohn’s disease cause significant changes in proximal methylation and expression levels in the brain, allowing for the inference of functional changes occurring in the brain related to these genetic comorbidities. The associated changes in methylation and expression status surrounding these SNPs mirrors their genomic context in previous analyses related to PD (49,70). Future studies with single cell capture designs, deep sequencing and additional brain tissue regions sampled may better quantify possible functional genetic consequences in PD etiology.
We recognize that a lack of clinical data on many of the participants in the study pose problems. In particular, we are concerned about the issue of drug-induced parkinsonism in this study and in other GWAS of both schizophrenia and PD. There is a long history of drug-induced parkinsonism that is often acute in onset (76), and misdiagnosis could affect the power and validity of our current study. However, as part of the Queen Square Brain Bank diagnostic criteria utilized to diagnose a majority of samples contributed to the IPDGC consortium effort, we believe that the possible effect of drug-induced PD is minimal, as a concurrent diagnosis of schizophrenia was an exclusionary criteria at study inception.
We recognize that there are some shortcomings in this analysis, first and foremost is the lack of available data relating to intermediate phenotypes in our PD consortium. The IPDGC datasets contain little to no information regarding the 18 intermediate exposures that would be necessary to conduct a true Mendelian randomization (7, 77-80). A previous study of PD risk focused on serum iron levels as the intermediate exposure has attempted to extrapolate risk estimates from non-overlapping populations and conduct an indirect Mendelian randomization based on summary statistics from disparate cohorts in a complex and well-conducted study, although this was logistically prohibitive in this survey of multiple outcomes due to data availability and standardization issues across intermediate exposures (81). Due to logistic constraints, we opted to use this risk profiling method instead, which could be applied within our consortium. While this approach does not infer causality, it does elucidate comorbid factors, and the associations between intermediate factors are directly comparable using this method. In addition, this approach can successfully identify comorbid intermediate factors associated with PD, as this study has >95% power to detect an effect at the magnitude of the schizophrenia and Crohn’s disease associations (OR ~1.05, α< 0.05). On the other hand, this method cannot definitively show a true negative effect for the other 16 intermediate risk factors investigated in this manuscript (particularly those with robust epidemiological associations such as smoking and caffeine intake) as the genetic contributions of known associated SNPs may be so small, and then further diluted in their biological relationship to PD risk, that these effects may be of virtually undetectable magnitude at this current period in genetic analyses.
MATERIALS AND METHODS
Data mining of PD-related traits from GWAS
The entire NHGRI GWAS catalog was downloaded on August 15, 2012. Phenotypes for each GWAS were indexed describing the diseases and quantitative traits associated with these studies. Based on a recently published systematic review of PD-related factors and consultation, we assembled a list of possible comorbidities of interest from the GWAS catalog using the disease/trait index (Supplementary Material, Table S1). From these indexed disease/trait descriptions, we extracted all keywords described in Supplementary Material, Table S1 by text mining to identify studies of relevant phenotypes for this analysis.
To extract relevant SNPs from only high-quality studies, a series of filters were used for SNP inclusion in the profiles based on data from the GWAS catalog. Minimum sample sizes for included GWAS SNPs had to be at least 1000 cases and 1000 controls for binary phenotypes in both discovery and replication analyses, and at least 1000 samples for continuous phenotypes. In addition, since all PDGWAS samples were of European ancestry, these sample size filters were based on European ancestry participants within the reported GWAS only. These SNPs must also have achieved a discovery P-value of ≤5E–8 as reported in the GWAS catalog. The contributing GWAS study must also have made data available regarding allelic direction of effect for the most significant SNP per locus.
Once all high-quality studies and their reported SNPs were extracted, additional quality control was necessary prior to constructing phenotype-specific risk scores. For all GWAS SNPs associated with a specific phenotype of interest, duplicate SNPs were excluded, and SNPs within 250 kb of another SNP associated with the same phenotype were excluded if the r2 between the two SNPs was >0.5. For SNPs in linkage disequilibrium related to the same phenotype, the more significant SNP based on discovery phase P-value was kept for construction of the risk profile scores. r2 estimates of linkage disequilibrium were based on European ancestry samples from the most recent 1000 Genomes Project data freeze (Phase 1 alpha, version 3 available from (1)). In addition, all SNPs had to be successfully imputed in the IPDGC cohorts participating in this analysis, see below for details.
IPDG datasets
Genotyping data from five IPDGC cohorts with genome-wide genotyping were extracted for use in this study. These cohorts have been described in Table 1, and in greater detail in previous publications (49,70). In brief, standard genome-wide association quality control was used including inclusion criteria such as: minimum 95% call rate per sample, Hardy–Weinberg equilibrium P-values >1E–6, minor allele frequency >1%, missingness per SNP <5%. Additional quality control parameters for GWAS samples included European ancestry consistent with HapMap3 samples based on multidimensional scaling, X chromosome heterogeneity reflecting self-reported gender per sample, and the exclusion of cryptically related samples at the first cousin level or closer relation. In addition, SNPs were removed for palindromic alleles (A/T or G/C combinations), differential missingness between cases and controls at P-value of <1E–4 and differential missingness by haplotype P-value of <1E–4. All quality control of raw genotype data was conducted using PLINK and R (6, R Development Core Team, R: A Language and Environment for Statistical Computing, R Foundation for Statistical Computing, Vienna, Austria, URL http://www.R-project.org, 2006).
After quality control, all IPDGC datasets were imputed using the minimac implementation of the Markov Chain based haplotyper with reference haplotypes from the European ancestry samples available in the August 2010 release from the 1000 Genomes project (82). After imputation, imputed dosages were filtered based on minimum imputation quality of 0.30 (RSQR metric from MACH) and a minor allele frequency of 1% for each cohort.
Risk profile construction
For each of the 18 phenotypes of interest with high-quality SNPs available, risk profiles were constructed. This was accomplished by summing all imputed allele dosages from extracted SNPs for each phenotype per cohort. The allelic dosages were quantified as 0–2 dosages for each SNP relating to the specific allele that was associated in previous reports with either a risk increase for a disease phenotype or an increase in the level of a continuous phenotype. All risk profiles were relatively normally distributed and were then Z transformed so that effect estimates would be easily comparable. Z transformation was undertaken using the standard formula where individual profile scores were subtracted by the mean of the population for that score and then divided by their standard deviation.
Analyses of risk profiles
For each of the 18 profiles calculated within each of the contributing cohorts, logistic regression was used to generate summary statistics, using the profile scores as the independent variable and adjusting for age (at onset in cases and exam in controls), sex and principal components 1 and 2 derived from genotyped SNPs. The use of principal components based on an LD pruned SNP set in each cohort as a covariate allows for population substructure to be accounted for in the regression models. Summary statistics for each cohort were meta-analyzed to generate aggregate effect estimates for all profiles using random effects meta-analyses. For PD, schizophrenia and Crohn’s disease related SNPs, single SNP analyses were also performed using identical statistical methods as for the risk profile-based analyses to facilitate an in-depth examination of the individual SNPs comprising these three scores.
Expression and methylation quantitative trait analyses
Data for this aspect of analyses were made possible through collaborative efforts of the North American and United Kingdom Brain Expression Consortia (83-85). Frozen frontal cortex and cerebellar samples were obtained from >399 self-reported European ancestry samples without determinable neuropatholgical evidence of disease. Genomic DNA was extracted with phenol–chloroform. Bisulfite converted DNA and assayed at >27 000 sites on the Illumina Infinium HumanMethylation27 BeadChips. MRNA expression levels were assayed using Illumina HumanHT-12 v3 Expression Beadchips. In brief, individual probes were excluded from analyses if the P-value for detection was >0.01 or there was <95% completeness of data per probe, and samples were excluded if <95% of probes were detected. Probes were also removed if an analyzed SNP was within the probe or the probe mapped ambiguously to multiple locations in the genome. Expression data were cubic spline normalized and log 2-transformed prior to analyses.
Each tissue sample was genotyped using the Illumina HumanHap550 v3, Human610-Quad v1 or Human660W-Quad v1 Infinium Beadchips, shared SNPs were extracted prior to QC and imputation. Standard GWAS quality control was undertaken with inclusion criteria such as: minimum call rate 95% for both participants and SNPs, minor allele frequency (MAF) > 0.01, HWE > 1E–7, no first-degree relatives in the sample collection (identity by descent score <0.125 in PLINK) and European ancestry confirmed by multidimensional scaling analyses.
Data were imputed using MiniMac (http://genome.sph.umich.edu/wiki/Minimac) to the most recent data freeze of 1000 Genomes haplotypes (http://www.sph.umich.edu/csg/abecasis/MaCH/download/1000G.2012-03-14.html) using default settings. All imputed SNPs were filtered for a minimum imputation quality of 0.30. After quality control, data were available for >10 million SNPs, with expression data on 399 samples (9814 probes from the frontal cortex and 9587 probes in cerebellum) and methylation data on 292 samples (27465 CpG sites in the frontal cortex tissue samples and 27419 CpG sites in the cerebellum).
Linear regression models were utilized to estimate associations between allele dosages of per SNP and gene expression or methylation levels adjusted for covariates of gender, age at death, the first two component vectors from multidimensional scaling, postmortem interval, brain bank and batch in which preparation or hybridization were performed, using MACH2QTLv1.11 (http://www.sph.umich.edu/csg/abecasis/MaCH/download/). Analyses were carried out separately for each brain region and each array type. Only probes within ±1 mb of SNPs of interest in this study were analyzed to test only cis associations. From these analysis results, data were mined for SNPs comprising the Crohn’s disease, PD and schizophrenia risk profiles. Multiple test correction was based on simple Bonferroni correction stratified by brain region and assay type.
Systematic review of epidemiological data
Validation of the findings of associations of a genetic profile PD with genetic risk profiles for other conditions or behaviors was sought by comparison with published data from epidemiological studies on their clinical associations. A recent systematic review and meta-analysis reported the combined results from case–control and cohort studies on association of PD with preceding smoking, coffee drinking, hypertension, diabetes, raised serum urate and obesity. For full details of the search strategy, inclusion and exclusion criteria and analysis of data see Noyce et al. (1). A separate meta-analysis on association of PD with preceding as well as existing diagnosis of melanoma(86) included publications until June 2010 and no further relevant papers were published from October 2010 to March 2011; and a another meta-analysis on association of existing PD with iron levels included publications until 2011 (87). For the other factors that had not fulfilled inclusion criteria in these meta-analyses, individual publications that reported case–control or cohort studies in the general PD population up to March 31, 2011 were identified. From these publications, the number of studies, number of cases and controls, OR with 95% confidence intervals, standard errors, I2 statistic, and P-values for risk and heterogeneity (in the meta-analyses) were extracted or calculated.
Supplementary Material
ACKNOWLEDGEMENTS
We thank and acknowledge all who made this research possible, please see Supplementary Material, Text S1 for full consortia membership. This study utilized the high-performance computational capabilities of the Biowulf Linux cluster at the National Institutes of Health, Bethesda, MD (http://biowulf.nih.gov), and DNA panels, samples and clinical data from the National Institute of Neurological Disorders and Stroke Human Genetics Resource Center DNA and Cell Line Repository. People who contributed samples are acknowledged in descriptions of every panel on the repository website. We thank the French Parkinson’s Disease Genetics Study Group: Y. Agid, M. Anheim, A.-M. Bonnet, M. Borg, A. Brice, E. Broussolle, J.-C. Corvol, P. Damier, A. Destée, A. Dürr, F. Durif, S. Klebe, E. Lohmann, M. Martinez, P. Pollak, O. Rascol, F. Tison, C. Tranchant, M. Vérin, F. Viallet and M. Vidailhet. We also thank the members of the French 3C Consortium: A. Alpérovitch, C. Berr, C. Tzourio and P. Amouyel for allowing us to use part of the 3C cohort, and D. Zelenika for support in generating the genomewide molecular data. We thank P. Tienari (Molecular Neurology Programme, Biomedicum, University of Helsinki), T. Peuralinna (Department of Neurology, Helsinki University Central Hospital), L. Myllykangas (Folkhalsan Institute of Genetics and Department of Pathology, University of Helsinki) and R. Sulkava (Department of Public Health and General Practice Division of Geriatrics, University of Eastern Finland) for the Finnish controls (Vantaa85+ GWAS data).
FUNDING: For details on funding, refer Supplementary Material, Text S1.
Footnotes
Conflict of Interest statement. None declared.
REFERENCES
- 1.Noyce AJ, Bestwick JP, Silveira-Moriyama L, Hawkes CH, Giovannoni G, Lees AJ, Schrag A. Meta-analysis of early nonmotor features and risk factors for Parkinson disease. Ann. Neurol. 2012;72:893–901. doi: 10.1002/ana.23687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bradfield JP, Taal HR, Timpson NJ, Scherag A, Lecoeur C, Warrington NM, Hypponen E, Holst C, Valcarcel B, Thiering E, et al. A genome-wide association meta-analysis identifies new childhood obesity loci. Nat. Genet. 2012;44:526–531. doi: 10.1038/ng.2247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Duerr RH, Taylor KD, Brant SR, Rioux JD, Silverberg MS, Daly MJ, Steinhart AH, Abraham C, Regueiro M, Griffiths A, et al. A genome-wide association study identifies IL23R as an inflammatory bowel disease gene. Science. 2006;314:1461–1463. doi: 10.1126/science.1135245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Strange A, Capon F, Spencer CCA, Knight J, Weale ME, Allen MH, Barton A, Band G, Bellenguez C, Bergboer JGM, et al. A genome-wide association study identifies new psoriasis susceptibility loci and an interaction between HLA-C and ERAP1. Nat. Genet. 2010;42:985–990. doi: 10.1038/ng.694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Scott LJ, Mohlke KL, Bonnycastle LL, Willer CJ, Li Y, Duren WL, Erdos MR, Stringham HM, Chines PS, Jackson AU, et al. A genome-wide association study of type 2 diabetes in Finns detects multiple susceptibility variants. Science. 2007;316:1341–1345. doi: 10.1126/science.1142382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Festen EAM, Goyette P, Green T, Boucher G, Beauchamp C, Trynka G, Dubois PC, Lagacé C, Stokkers PCF, Hommes DW, et al. A meta-analysis of genome-wide association scans identifies IL18RAP, PTPN2, TAGAP, and PUS10 as shared risk loci for Crohn’s disease and celiac disease. PLoS Genet. 2011;7:e1001283. doi: 10.1371/journal.pgen.1001283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Timpson NJ, Lindgren CM, Weedon MN, Randall J, Ouwehand WH, Strachan DP, Rayner NW, Walker M, Hitman GA, Doney ASF, et al. Adiposity-related heterogeneity in patterns of type 2 diabetes susceptibility observed in genome-wide association data. Diabetes. 2009;58:505–510. doi: 10.2337/db08-0906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dehghan A, Köttgen A, Yang Q, Hwang S-J, Kao WL, Rivadeneira F, Boerwinkle E, Levy D, Hofman A, Astor BC, et al. Association of three genetic loci with uric acid concentration and risk of gout: a genome-wide association study. Lancet. 2008;372:1953–1961. doi: 10.1016/S0140-6736(08)61343-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hamza TH, Zabetian CP, Tenesa A, Laederach A, Montimurro J, Yearout D, Kay DM, Doheny KF, Paschall J, Pugh E, et al. Common genetic variation in the HLA region is associated with late-onset sporadic Parkinson’s disease. Nat. Genet. 2010;42:781–785. doi: 10.1038/ng.642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Purcell SM, Wray NR, Stone JL, Visscher PM, O’Donovan MC, Sullivan PF, Sklar P. Common polygenic variation contributes to risk of schizophrenia and bipolar disorder. Nature. 2009;460:748–752. doi: 10.1038/nature08185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Raychaudhuri S, Remmers EF, Lee AT, Hackett R, Guiducci C, Burtt NP, Gianniny L, Korman BD, Padyukov L, Kurreeman FAS, et al. Common variants at CD40 and other loci confer risk of rheumatoid arthritis. Nat. Genet. 2008;40:1216–1223. doi: 10.1038/ng.233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Imielinski M, Baldassano RN, Griffiths A, Russell RK, Annese V, Dubinsky M, Kugathasan S, Bradfield JP, Walters TD, Sleiman P, et al. Common variants at five new loci associated with early-onset inflammatory bowel disease. Nat. Genet. 2009;41:1335–1340. doi: 10.1038/ng.489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stefansson H, Ophoff RA, Steinberg S, Andreassen OA, Cichon S, Rujescu D, Werge T, Pietiläinen OPH, Mors O, Mortensen PB, et al. Common variants conferring risk of schizophrenia. Nature. 2009;460:744–747. doi: 10.1038/nature08186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Benyamin B, Ferreira MAR, Willemsen G, Gordon S, Middelberg RPS, McEvoy BP, Hottenga J-J, Henders AK, Campbell MJ, Wallace L, et al. Common variants in TMPRSS6 are associated with iron status and erythrocyte volume. Nat. Genet. 2009;41:1173–1175. doi: 10.1038/ng.456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shi J, Levinson DF, Duan J, Sanders AR, Zheng Y, Pe’er I, Dudbridge F, Holmans PA, Whittemore AS, Mowry BJ, et al. Common variants on chromosome 6p22.1 are associated with schizophrenia. Nature. 2009;460:753–757. doi: 10.1038/nature08192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lill CM, Roehr JT, McQueen MB, Kavvoura FK, Bagade S, Schjeide B-MM, Schjeide LM, Meissner E, Zauft U, Allen NC, et al. Comprehensive research synopsis and systematic meta-analyses in Parkinson’s disease genetics: The PDGene database. PLoS Genet. 2012;8:e1002548. doi: 10.1371/journal.pgen.1002548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Spencer CCA, Plagnol V, Strange A, Gardner M, Paisan-Ruiz C, Band G, Barker RA, Bellenguez C, Bhatia K, Blackburn H, et al. Dissection of the genetics of Parkinson’s disease identifies an additional association 5′ of SNCA and multiple associated haplotypes at 17q21. Hum. Mol. Genet. 2011;20:345–353. doi: 10.1093/hmg/ddq469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Elliott P, Chambers JC, Zhang W, Clarke R, Hopewell JC, Peden JF, Erdmann J, Braund P, Engert JC, Bennett D, et al. Genetic Loci associated with C-reactive protein levels and risk of coronary heart disease. JAMA. 2009;302:37–48. doi: 10.1001/jama.2009.954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Qi L, Cornelis MC, Kraft P, Stanya KJ, Linda Kao WH, Pankow JS, Dupuis J, Florez JC, Fox CS, Paré G, et al. Genetic variants at 2q24 are associated with susceptibility to type 2 diabetes. Hum. Mol. Genet. 2010;19:2706–2715. doi: 10.1093/hmg/ddq156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ehret GB, Munroe PB, Rice KM, Bochud M, Johnson AD, Chasman DI, Smith AV, Tobin MD, Verwoert GC, Hwang S-J, et al. Genetic variants in novel pathways influence blood pressure and cardiovascular disease risk. Nature. 2011;478:103–109. doi: 10.1038/nature10405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Saxena R, Voight BF, Lyssenko V, Burtt NP, de Bakker PIW, Chen H, Roix JJ, Kathiresan S, Hirschhorn JN, Daly MJ, et al. Genome-wide association analysis identifies loci for type 2 diabetes and triglyceride levels. Science. 2007;316:1331–1336. doi: 10.1126/science.1142358. [DOI] [PubMed] [Google Scholar]
- 22.Stuart PE, Nair RP, Ellinghaus E, Ding J, Tejasvi T, Gudjonsson JE, Li Y, Weidinger S, Eberlein B, Gieger C, et al. Genome-wide association analysis identifies three psoriasis susceptibility loci. Nat. Genet. 2010;42:1000–1004. doi: 10.1038/ng.693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Amin N, Byrne E, Johnson J, Chenevix-Trench G, Walter S, Nolte IM, Vink JM, Rawal R, Mangino M, Teumer A, et al. Genome-wide association analysis of coffee drinking suggests association with CYP1A1/CYP1A2 and NRCAM. Mol. Psychiatry. 2012;17:1116–1129. doi: 10.1038/mp.2011.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Barrett JC, Hansoul S, Nicolae DL, Cho JH, Duerr RH, Rioux JD, Brant SR, Silverberg MS, Taylor KD, Barmada MM, et al. Genome-wide association defines more than 30 distinct susceptibility loci for Crohn’s disease. Nat. Genet. 2008;40:955–962. doi: 10.1038/NG.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McGovern DPB, Gardet A, Törkvist L, Goyette P, Essers J, Taylor KD, Neale BM, Ong RTH, Lagacé C, Li C, et al. Genome-wide association identifies multiple ulcerative colitis susceptibility loci. Nat. Genet. 2010;42:332–337. doi: 10.1038/ng.549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Scuteri A, Sanna S, Chen W-M, Uda M, Albai G, Strait J, Najjar S, Nagaraja R, Orrú M, Usala G, et al. Genome-wide association scan shows genetic variants in the FTO gene are associated with obesity-related traits. PLoS Genet. 2007;3:e115. doi: 10.1371/journal.pgen.0030115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Meyre D, Delplanque J, Chèvre J-C, Lecoeur C, Lobbens S, Gallina S, Durand E, Vatin V, Degraeve F, Proença C, et al. Genome-wide association study for early-onset and morbid adult obesity identifies three new risk loci in European populations. Nat. Genet. 2009;41:157–159. doi: 10.1038/ng.301. [DOI] [PubMed] [Google Scholar]
- 28.Franke A, Balschun T, Sina C, Ellinghaus D, Häsler R, Mayr G, Albrecht M, Wittig M, Buchert E, Nikolaus S, et al. Genome-wide association study for ulcerative colitis identifies risk loci at 7q22 and 22q13 (IL17REL) Nat. Genet. 2010;42:292–294. doi: 10.1038/ng.553. [DOI] [PubMed] [Google Scholar]
- 29.Macgregor S, Montgomery GW, Liu JZ, Zhao ZZ, Henders AK, Stark M, Schmid H, Holland EA, Duffy DL, Zhang M, et al. Genome-wide association study identifies a new melanoma susceptibility locus at 1q21.3. Nat. Genet. 2011;43:1114–1118. doi: 10.1038/ng.958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schizophrenia Psychiatric Genome-Wide Association Study (GWAS) Consortium Genome-wide association study identifies five new schizophrenia loci. Nat. Genet. 2011;43:969–976. doi: 10.1038/ng.940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wallace C, Newhouse SJ, Braund P, Zhang F, Tobin M, Falchi M, Ahmadi K, Dobson RJ, Marçano ACB, Hajat C, et al. Genome-wide association study identifies genes for biomarkers of cardiovascular disease: serum urate and dyslipidemia. Am. J. Hum. Genet. 2008;82:139–149. doi: 10.1016/j.ajhg.2007.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bishop DT, Demenais F, Iles MM, Harland M, Taylor JC, Corda E, Randerson-Moor J, Aitken JF, Avril M-F, Azizi E, et al. Genome-wide association study identifies three loci associated with melanoma risk. Nat. Genet. 2009;41:920–925. doi: 10.1038/ng.411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Barrett JH, Iles MM, Harland M, Taylor JC, Aitken JF, Andresen PA, Akslen LA, Armstrong BK, Avril M-F, Azizi E, et al. Genome-wide association study identifies three new melanoma susceptibility loci. Nat. Genet. 2011;43:1108–1113. doi: 10.1038/ng.959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Marzi C, Albrecht E, Hysi PG, Lagou V, Waldenberger M, Tönjes A, Prokopenko I, Heim K, Blackburn H, Ried JS, et al. Genome-wide association study identifies two novel regions at 11p15.5-p13 and 1p31 with major impact on acute-phase serum amyloid A. PLoS Genet. 2010;6:e1001213. doi: 10.1371/journal.pgen.1001213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bergen SE, O’Dushlaine CT, Ripke S, Lee PH, Ruderfer DM, Akterin S, Moran JL, Chambert KD, Handsaker RE, Backlund L, et al. Genome-wide association study in a Swedish population yields support for greater CNV and MHC involvement in schizophrenia compared with bipolar disorder. Mol. Psychiatry. 2012;17:880–886. doi: 10.1038/mp.2012.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stahl EA, Raychaudhuri S, Remmers EF, Xie G, Eyre S, Thomson BP, Li Y, Kurreeman FAS, Zhernakova A, Hinks A, et al. Genome-wide association study meta-analysis identifies seven new rheumatoid arthritis risk loci. Nat. Genet. 2010;42:508–514. doi: 10.1038/ng.582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chen DT, Jiang X, Akula N, Shugart YY, Wendland JR, Steele CJM, Kassem L, Park J-H, Chatterjee N, Jamain S, et al. Genome-wide association study meta-analysis of European and Asian-ancestry samples identifies three novel loci associated with bipolar disorder. Mol. Psychiatry. 2011 doi: 10.1038/mp.2011.157. doi:10.1038/mp.2011.157. [DOI] [PubMed] [Google Scholar]
- 38.Wellcome Trust Case Control Consortium Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447:661–678. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Levy D, Ehret GB, Rice K, Verwoert GC, Launer LJ, Dehghan A, Glazer NL, Morrison AC, Johnson AD, Aspelund T, et al. Genome-wide association study of blood pressure and hypertension. Nat. Genet. 2009;41:677–687. doi: 10.1038/ng.384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Padmanabhan S, Melander O, Johnson T, Di Blasio AM, Lee WK, Gentilini D, Hastie CE, Menni C, Monti MC, Delles C, et al. Genome-wide association study of blood pressure extremes identifies variant near UMOD associated with hypertension. PLoS Genet. 2010;6:e1001177. doi: 10.1371/journal.pgen.1001177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Barrett JC, Lee JC, Lees CW, Prescott NJ, Anderson CA, Phillips A, Wesley E, Parnell K, Zhang H, Drummond H, et al. Genome-wide association study of ulcerative colitis identifies three new susceptibility loci, including the HNF4A region. Nat. Genet. 2009;41:1330–1334. doi: 10.1038/ng.483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Simón-Sánchez J, Schulte C, Bras JM, Sharma M, Gibbs JR, Berg D, Paisan-Ruiz C, Lichtner P, Scholz SW, Hernandez DG, et al. Genome-wide association study reveals genetic risk underlying Parkinson’s disease. Nat. Genet. 2009;41:1308–1312. doi: 10.1038/ng.487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tobacco and Genetics Consortium Genome-wide meta-analyses identify multiple loci associated with smoking behavior. Nat. Genet. 2010;42:441–447. doi: 10.1038/ng.571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Franke A, McGovern DPB, Barrett JC, Wang K, Radford-Smith GL, Ahmad T, Lees CW, Balschun T, Lee J, Roberts R, et al. Genome-wide meta-analysis increases to 71 the number of confirmed Crohn’s disease susceptibility loci. Nat. Genet. 2010;42:1118–1125. doi: 10.1038/ng.717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Paternoster L, Evans DM, Nohr EA, Holst C, Gaborieau V, Brennan P, Gjesing AP, Grarup N, Witte DR, Jørgensen T, et al. Genome-wide population-based association study of extremely overweight young adults – the GOYA study. PLoS ONE. 2011;6:e24303. doi: 10.1371/journal.pone.0024303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nair RP, Duffin KC, Helms C, Ding J, Stuart PE, Goldgar D, Gudjonsson JE, Li Y, Tejasvi T, Feng B-J, et al. Genome-wide scan reveals association of psoriasis with IL-23 and NF-kappaB pathways. Nat. Genet. 2009;41:199–204. doi: 10.1038/ng.311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pichler I, Minelli C, Sanna S, Tanaka T, Schwienbacher C, Naitza S, Porcu E, Pattaro C, Busonero F, Zanon A, et al. Identification of a common variant in the TFR2 gene implicated in the physiological regulation of serum iron levels. Hum. Mol. Genet. 2011;20:1232–1240. doi: 10.1093/hmg/ddq552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kugathasan S, Baldassano RN, Bradfield JP, Sleiman PMA, Imielinski M, Guthery SL, Cucchiara S, Kim CE, Frackelton EC, Annaiah K, et al. Loci on 20q13 and 21q22 are associated with pediatric-onset inflammatory bowel disease. Nat. Genet. 2008;40:1211–1215. doi: 10.1038/ng.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.International Parkinson Disease Genomics Consortium. Nalls MA, Plagnol V, Hernandez DG, Sharma M, Sheerin U-M, Saad M, Simón-Sánchez J, Schulte C, Lesage S, et al. Imputation of sequencevariantsfor identification of genetic risks for Parkinson’s disease: a meta-analysis of genome-wide association studies. Lancet. 2011;377:641–649. doi: 10.1016/S0140-6736(10)62345-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Psychiatric GWAS Consortium. Bipolar Disorder Working Group Large-scale genome-wide association analysis of bipolar disorder identifies a new susceptibility locus near ODZ4. Nat. Genet. 2011;43:977–983. doi: 10.1038/ng.943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Liu JZ, Tozzi F, Waterworth DM, Pillai SG, Muglia P, Middleton L, Berrettini W, Knouff CW, Yuan X, Waeber G, et al. Meta-analysis and imputation refines the association of 15q25 with smoking quantity. Nat. Genet. 2010;42:436–440. doi: 10.1038/ng.572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Anderson CA, Boucher G, Lees CW, Franke A, D’Amato M, Taylor KD, Lee JC, Goyette P, Imielinski M, Latiano A, et al. Meta-analysis identifies 29 additional ulcerative colitis risk loci, increasing the number of confirmed associations to 47. Nat. Genet. 2011;43:246–252. doi: 10.1038/ng.764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zeggini E, Scott LJ, Saxena R, Voight BF, Marchini JL, Hu T, de Bakker PIW, Abecasis GR, Almgren P, Andersen G, et al. Meta-analysis of genome-wide association data and large-scale replication identifies additional susceptibility loci for type 2 diabetes. Nat. Genet. 2008;40:638–645. doi: 10.1038/ng.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Dehghan A, Dupuis J, Barbalic M, Bis JC, Eiriksdottir G, Lu C, Pellikka N, Wallaschofski H, Kettunen J, Henneman P, et al. Meta-analysis of genome-wide association studies in >80 000 subjects identifies multiple loci for C-reactive protein levels. Circulation. 2011;123:731–738. doi: 10.1161/CIRCULATIONAHA.110.948570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pankratz N, Beecham GW, DeStefano AL, Dawson TM, Doheny KF, Factor SA, Hamza TH, Hung AY, Hyman BT, Ivinson AJ, et al. Meta-analysis of Parkinson’s disease: identification of a novel locus, RIT2. Ann. Neurol. 2012;71:370–384. doi: 10.1002/ana.22687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yang Q, Köttgen A, Dehghan A, Smith AV, Glazer NL, Chen M-H, Chasman DI, Aspelund T, Eiriksdottir G, Harris TB, et al. Multiple genetic loci influence serum urate levels and their relationship with gout and cardiovascular disease risk factors. Circ. Cardiovasc. Genet. 2010;3:523–530. doi: 10.1161/CIRCGENETICS.109.934455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gregersen PK, Amos CI, Lee AT, Lu Y, Remmers EF, Kastner DL, Seldin MF, Criswell LA, Plenge RM, Holers VM, et al. REL, encoding a member of the NF-kappaB family of transcription factors, is a newly defined risk locus for rheumatoid arthritis. Nat. Genet. 2009;41:820–823. doi: 10.1038/ng.395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zeggini E, Weedon MN, Lindgren CM, Frayling TM, Elliott KS, Lango H, Timpson NJ, Perry JRB, Rayner NW, Freathy RM, et al. Replication of genome-wide association signals in UK samples reveals risk loci for type 2 diabetes. Science. 2007;316:1336–1341. doi: 10.1126/science.1142364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Thorgeirsson TE, Gudbjartsson DF, Surakka I, Vink JM, Amin N, Geller F, Sulem P, Rafnar T, Esko T, Walter S, et al. Sequence variants at CHRNB3-CHRNA6 and CYP2A6 affect smoking behavior. Nat. Genet. 2010;42:448–453. doi: 10.1038/ng.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sulem P, Gudbjartsson DF, Geller F, Prokopenko I, Feenstra B, Aben KKH, Franke B, den Heijer M, Kovacs P, Stumvoll M, et al. Sequence variants at CYP1A1-CYP1A2 and AHR associate with coffee consumption. Hum. Mol. Genet. 2011;20:2071–2077. doi: 10.1093/hmg/ddr086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Parkes M, Barrett JC, Prescott NJ, Tremelling M, Anderson CA, Fisher SA, Roberts RG, Nimmo ER, Cummings FR, Soars D, et al. Sequence variants in the autophagy gene IRGM and multiple other replicating loci contribute to Crohn’s disease susceptibility. Nat. Genet. 2007;39:830–832. doi: 10.1038/ng2061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Döring A, Gieger C, Mehta D, Gohlke H, Prokisch H, Coassin S, Fischer G, Henke K, Klopp N, Kronenberg F, et al. SLC2A9 influences uric acid concentrations with pronounced sex-specific effects. Nat. Genet. 2008;40:430–436. doi: 10.1038/ng.107. [DOI] [PubMed] [Google Scholar]
- 63.Perry JRB, Voight BF, Yengo L, Amin N, Dupuis J, Ganser M, Grallert H, Navarro P, Li M, Qi L, et al. Stratifying type 2 diabetes cases by BMI identifies genetic risk variants in LAMA1 and enrichment for risk variants in lean compared to obese cases. PLoS Genet. 2012;8:e1002741. doi: 10.1371/journal.pgen.1002741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Li S, Sanna S, Maschio A, Busonero F, Usala G, Mulas A, Lai S, Dei M, Orrù M, Albai G, et al. The GLUT9 gene is associated with serum uric acid levels in Sardinia and Chianti cohorts. PLoS Genet. 2007;3:e194. doi: 10.1371/journal.pgen.0030194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Plenge RM, Seielstad M, Padyukov L, Lee AT, Remmers EF, Ding B, Liew A, Khalili H, Chandrasekaran A, Davies LRL, et al. TRAF1-C5 as a risk locus for rheumatoid arthritis – a genomewide study. N. Engl. J. Med. 2007;357:1199–1209. doi: 10.1056/NEJMoa073491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Voight BF, Scott LJ, Steinthorsdottir V, Morris AP, Dina C, Welch RP, Zeggini E, Huth C, Aulchenko YS, Thorleifsson G, et al. Twelve type 2 diabetes susceptibility loci identified through large-scale association analysis. Nat. Genet. 2010;42:579–589. doi: 10.1038/ng.609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Scherag A, Dina C, Hinney A, Vatin V, Scherag S, Vogel CIG, Müller TD, Grallert H, Wichmann H-E, Balkau B, et al. Two new Loci for body-weight regulation identified in a joint analysis of genome-wide association studies for early-onset extreme obesity in French and german study groups. PLoS Genet. 2010;6:e1000916. doi: 10.1371/journal.pgen.1000916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Silverberg MS, Cho JH, Rioux JD, McGovern DPB, Wu J, Annese V, Achkar J-P, Goyette P, Scott R, Xu W, et al. Ulcerative colitis-risk loci on chromosomes 1p36 and 12q15 found by genome-wide association study. Nat. Genet. 2009;41:216–220. doi: 10.1038/ng.275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Keller MF, Saad M, Bras J, Bettella F, Nicolaou N, Simón-Sánchez J, Mittag F, Büchel F, Sharma M, Gibbs JR, et al. Using genome-wide complex trait analysis to quantify ‘missing heritability’ in Parkinson’s disease. Hum. Mol. Genet. 2012;21:4996–5009. doi: 10.1093/hmg/dds335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.International Parkinson’s Disease Genomics Consortium (IPDGC) Wellcome Trust Case Control Consortium 2 (WTCCC2) A two-stage meta-analysis identifies several new loci for Parkinson’s disease. PLoS Genet. 2011;7:e1002142. doi: 10.1371/journal.pgen.1002142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Shiba M, Bower JH, Maraganore DM, McDonnell SK, Peterson BJ, Ahlskog JE, Schaid DJ, Rocca WA. Anxiety disorders and depressive disorders preceding Parkinson’s disease: a case-control study. Mov. Disord. 2000;15:669–677. doi: 10.1002/1531-8257(200007)15:4<669::aid-mds1011>3.0.co;2-5. [DOI] [PubMed] [Google Scholar]
- 72.Rajput AH, Offord KP, Beard CM, Kurland LT. A case-control study of smoking habits, dementia, and other illnesses in idiopathic Parkinson’s disease. Neurology. 1987;37:226–232. doi: 10.1212/wnl.37.2.226. [DOI] [PubMed] [Google Scholar]
- 73.Chen H, O’Reilly EJ, Schwarzschild MA, Ascherio A. Peripheral inflammatory biomarkers and risk of Parkinson’s disease. Am. J. Epidemiol. 2008;167:90–95. doi: 10.1093/aje/kwm260. [DOI] [PubMed] [Google Scholar]
- 74.Rugbjerg K, Friis S, Ritz B, Schernhammer ES, Korbo L, Olsen JH. Autoimmune disease and risk for Parkinson disease: a population-based case-control study. Neurology. 2009;73:1462–1468. doi: 10.1212/WNL.0b013e3181c06635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Song I-U, Kim J-S, Chung S-W, Lee K-S. Is there an association between the level of high-sensitivity C-reactive protein and idiopathic Parkinson’s disease? A comparison of Parkinson’s disease patients, disease controls and healthy individuals. Eur. Neurol. 2009;62:99–104. doi: 10.1159/000222780. [DOI] [PubMed] [Google Scholar]
- 76.Barbeau A. The ‘pink spot’, 3,4-dimethoxyphenylethylamine and dopamine. Relationship to Parkinson’s disease and to schizophrenia. Rev. Can. Biol. 1967;26:55–79. [PubMed] [Google Scholar]
- 77.Smith GD, Ebrahim S. ‘Mendelian randomization’: can genetic epidemiology contribute to understanding environmental determinants of disease? Int. J. Epidemiol. 2003;32:1–22. doi: 10.1093/ije/dyg070. [DOI] [PubMed] [Google Scholar]
- 78.Nitsch D, Molokhia M, Smeeth L, DeStavola BL, Whittaker JC, Leon DA. Limits to causal inference based on Mendelian randomization: a comparison with randomized controlled trials. Am. J. Epidemiol. 2006;163:397–403. doi: 10.1093/aje/kwj062. [DOI] [PubMed] [Google Scholar]
- 79.Verduijn M, Siegerink B, Jager KJ, Zoccali C, Dekker FW. Mendelian randomization: use of genetics to enable causal inference in observational studies. Nephrol. Dial. Transplant. 2010;25:1394–1398. doi: 10.1093/ndt/gfq098. [DOI] [PubMed] [Google Scholar]
- 80.Didelez V, Sheehan N. Mendelian randomization as an instrumental variable approach to causal inference. Stat. Methods Med. Res. 2007;16:309–330. doi: 10.1177/0962280206077743. [DOI] [PubMed] [Google Scholar]
- 81.Pichler I, Del Greco MF, Gögele M, Lill CM, Bertram L, Do CB, Eriksson N, Foroud T, Myers RH, PD GWAS Consortium et al. Serum iron levels and the risk of Parkinson disease: a Mendelian randomization study. PLoS Med. 2013;10:e1001462. doi: 10.1371/journal.pmed.1001462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Howie B, Fuchsberger C, Stephens M, Marchini J, Abecasis GR. Fast and accurate genotype imputation in genome-wide association studies through pre-phasing. Nat. Genet. 2012;44:955–959. doi: 10.1038/ng.2354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Gibbs JR, van der Brug MP, Hernandez DG, Traynor BJ, Nalls MA, Lai S-L, Arepalli S, Dillman A, Rafferty IP, Troncoso J, et al. Abundant quantitative trait loci exist for DNA methylation and gene expression in human brain. PLoS Genet. 2010;6:e1000952. doi: 10.1371/journal.pgen.1000952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Trabzuni D, Ryten M, Walker R, Smith C, Imran S, Ramasamy A, Weale ME, Hardy J. Quality control parameters on a large dataset of regionally dissected human control brains for whole genome expression studies. J. Neurochem. 2011;119:275–282. doi: 10.1111/j.1471-4159.2011.07432.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Trabzuni D, Wray S, Vandrovcova J, Ramasamy A, Walker R, Smith C, Luk C, Gibbs JR, Dillman A, Hernandez DG, et al. MAPT expression and splicing is differentially regulated by brain region: relation to genotype and implication for tauopathies. Hum. Mol. Genet. 2012;21:4094–4103. doi: 10.1093/hmg/dds238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Liu R, Gao X, Lu Y, Chen H. Meta-analysis of the relationship between Parkinson disease and melanoma. Neurology. 2011;76:2002–2009. doi: 10.1212/WNL.0b013e31821e554e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Mariani S, Ventriglia M, Simonelli I, Donno S, Bucossi S, Vernieri F, Melgari J-M, Pasqualetti P, Rossini PM, Squitti R. Fe and Cu do not differ in Parkinson’s disease: a replication study plus meta-analysis. Neurobiol. Aging. 2013;34:632–633. doi: 10.1016/j.neurobiolaging.2012.05.015. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.