Abstract
The Wilms’ tumor 1 protein (WT1) is a transcriptional regulator that can either activate or repress genes controlling cell growth, apoptosis and differentiation. The transcriptional corepressor BASP1 interacts with WT1 and mediates WT1’s transcriptional repression activity. BASP1 is contained within large complexes, suggesting that it works in concert with other factors. Here we report that the transcriptional repressor prohibitin is part of the WT1-BASP1 transcriptional repression complex. Prohibitin interacts with BASP1, colocalizes with BASP1 in the nucleus, and is recruited to the promoter region of WT1 target genes to elicit BASP1-dependent transcriptional repression. We demonstrate that prohibitin and BASP1 cooperate to recruit the chromatin remodeling factor BRG1 to WT1-responsive promoters and that this results in the dissociation of CBP from the promoter region of WT1 target genes. As seen with BASP1, prohibitin can associate with phospholipids. We demonstrate that the recruitment of PIP2 and HDAC1 to WT1 target genes is also dependent on the concerted activity of BASP1 and prohibitin. Our findings provide new insights into the function of prohibitin in transcriptional regulation and uncover a BASP1-prohibitin complex that plays an essential role in the PIP2-dependent recruitment of chromatin remodeling activities to the promoter.
Keywords: WT1, BASP1, Prohibitin, transcription
Introduction
The Wilms’ tumor 1 protein (WT1) plays an important role in development of several organs and is mutated or aberrantly expressed in different cancers where it acts as an oncogene or a tumor suppressor. 1-3 As a transcriptional regulator WT1 activities are complex, resulting in either transcriptional activation or repression of numerous target genes involved in disparate biological activities. 4 We identified BASP1 as a WT1 cofactor that converts WT1 from a transcriptional activator to a repressor. 5, 6 Since WT1 and BASP1 are co-expressed at numerous sites in the developing embryo, this suggests a role for BASP1 in regulating the function of WT1 during development. 6
BASP1 can localize to the nucleus through a bipartite nuclear localization sequence (NLS) and binds to WT1 at the promoters of several target genes. 6-11 BASP1 can also inhibit cellular transformation by the v-myc oncogene and blocks the regulation of myc target genes.12 Moreover, BASP1 expression is downregulated in hepatocellular carcinomas and several leukemia’s, which is attributed to silencing of the BASP1 gene through methylation. 13, 14 Taken together, these recent studies suggest a significant tumor suppressor role for BASP1.
How BASP1 acts as a transcriptional corepressor is not clear. We recently demonstrated that transcriptional repression by the WT1-BASP1 complex requires the N-terminal myristoylation of BASP1 to provide a platform for the recruitment of the phospholipid PIP2 to the promoter. The BASP1-PIP2 interaction is critical for the assembly of HDAC1 to mediate transcriptional repression. 11 Although our understanding of the transcription function of BASP1 has increased significantly in recent years, it is still not clear how BASP1 acts in concert with other components of the transcription machinery.
Previous gel filtration analyses revealed that BASP1 is contained within large complexes within the nucleus. 8 Here we report that BASP1 interacts with the transcriptional corepressor and tumor suppressor prohibitin. Prohibitin acts as a corepressor for several transcription factors including E2F, 15-20 Rb, 21estrogen receptor, ER 22-24 and androgen receptor AR 25, 26. We demonstrate that prohibitin forms an integral component of the WT1-BASP1 repressor complex and that it functions to recruit ATP-dependent chromatin remodeling complexes to WT1-dependent promoters. Furthermore BASP1 and prohibitin cooperate through PIP2 to recruit histone deacetylase activity. Our findings uncover prohibitin as a key component that regulates the activity of the WT1-BASP1 complex in a multi-faceted mechanism of transcriptional repression.
Results
Prohibitin interacts with and colocalizes with BASP1 in the nucleus
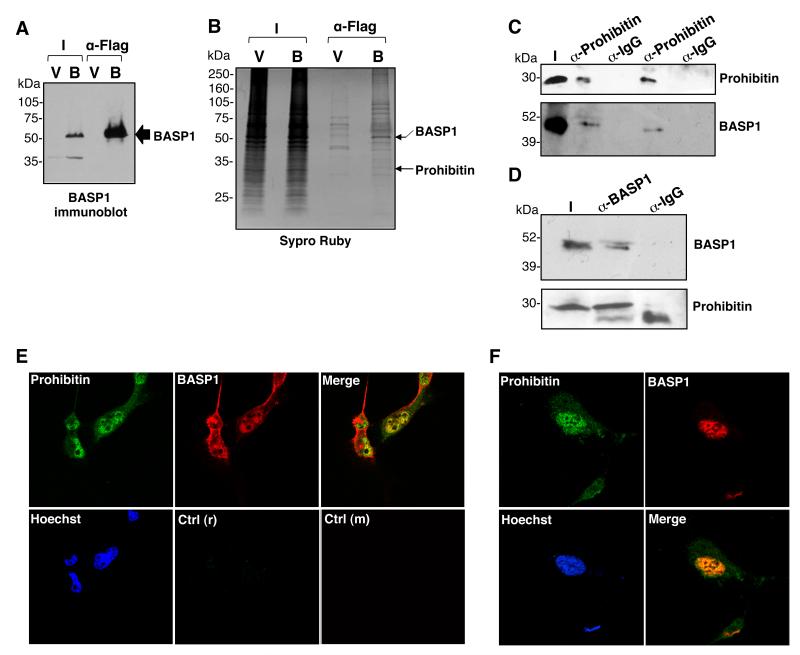
Our previous studies demonstrated that BASP1 is contained within large complexes (up to 1MDa) in nuclear extracts. 8 We therefore sought to identify proteins that coimmunoprecipitate with BASP1 from nuclear extracts. K562 cells do not normally express BASP1 and we have shown previously that the stable introduction of BASP1 into K562 cells leads to robust transcriptional repression of WT1 target genes. 6, 10-11 We employed these stable K562 cell line derivatives, that contain either pcDNA3 vector (V-K562 cells; V) or the same vector driving expression of a BASP1 derivative containing a C-terminal FLAG tag (BASP1-K562; B). Nuclear extracts were prepared from V-K562 and BASP1-K562 cells and immunoprecipitation performed with anti-FLAG antibodies. We confirmed that the anti-FLAG antibodies efficiently immunoprecipitated BASP1 from nuclear extracts prepared from BASP1-K562 cells but not the control V-K562 cells (Figure 1A). Analysis of the eluates by Sypro ruby staining revealed several proteins specific to the FLAG immunoprecipitate from BASP1-K562 cells (Figure 1B). We identified one of the proteins as prohibitin (indicated).
Figure 1. BASP1 associates with prohibitin in the nucleus.
(A) Nuclear extracts from V-K562 (V) cells and BASP1-K562 (B) cells were used in immunoprecipitation with anti-FLAG antibodies. The immunoprecipitates were immunoblotted with anti-BASP1 antibodies. (B) FLAG immunoprecipitates from part A were resolved by electrophoresis and the gel was stained with Sypro Ruby. BASP1 and prohibitin were identified by MALDI-TOF mass spectrometry. (C) HeLa cell nuclear extracts were subjected to immunoprecipitation with control IgG or anti-prohibitin antibodies and immunoblotted with anti-BASP1 antibodies. I is input. (D) Same as in part C except that the immunoprecipitation was performed with anti-BASP1 antibodies and the samples immunoblotted with anti-prohibitin antibodies. (E) MCF7 cells were used to perform coimmunoflurescence with anti-rabbit BASP1 antibodies and anti-mouse prohibitin antibodies and visualized with confocal microscopy. Hoechst was used to label the nucleus. Immunofluorescence with control anti-rabbit (r) and anti-mouse (m) antibodies are shown in the bottom row. (F) Same as in part (E) except that U2OS cells were used.
To confirm that prohibitin is a bona fide BASP1-interacting protein, we performed coimmunoprecipitation with nuclear extracts prepared from HeLa cells, which contain endogenous BASP1 and prohibitin. Anti-prohibitin immunoprecipitates contained BASP1 (Figure 1C), and prohibitin was also retained in anti-BASP1 immunoprecipitates (Figure 1D). We confirmed that endogenous BASP1 and prohibitin colocalize in the nucleus of MCF7 cells using coimmunofluorescence (Figure 1E). Comparable results were obtained when BASP1 and prohibitin were analyzed in U2OS cells (Figure 1F). Taken together, the data in Figure 1 demonstrate that BASP1 and prohibitin associate in the cell nucleus.
Prohibitin mediates BASP1-dependent transcriptional repression of WT1 target genes
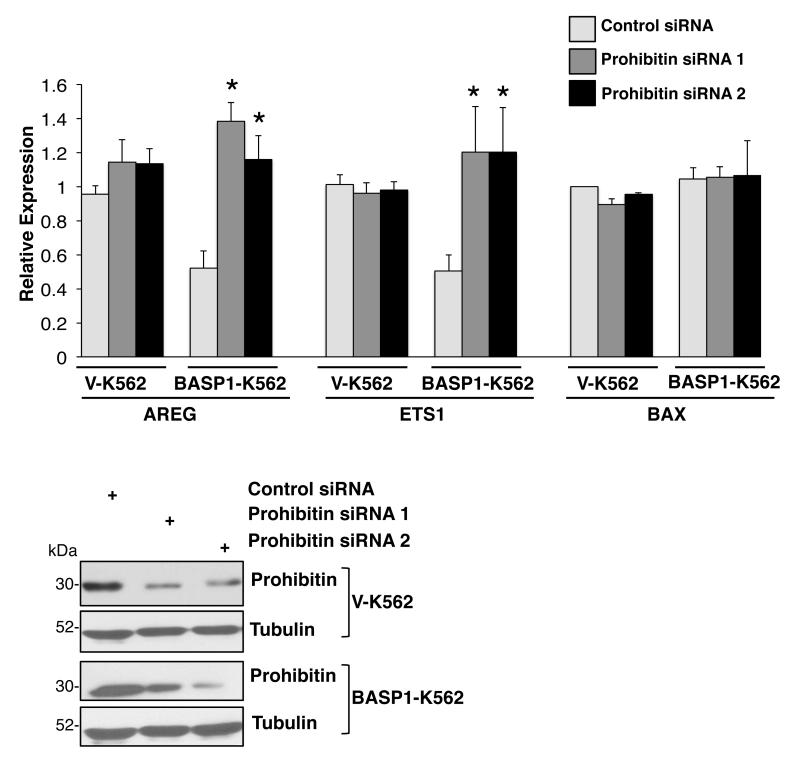
Since prohibitin had previously been identified as a transcriptional corepressor 27 we considered that it might also be required to elicit BASP1-mediated transcriptional repression of WT1 target genes. We analyzed the effect of prohibitin depletion on the expression of WT1 target genes in V-K562 and BASP1-K562 cells. V-K562 cells and BASP1-K562 cells were transfected with a control small interfering RNA (siRNA) or siRNAs targeting two different regions of prohibitin (prohibitin siRNA 1 and prohibitin siRNA 2). Immunoblotting of whole-cell extracts with anti-prohibitin antibodies confirmed the depletion of prohibitin by both siRNAs (Figure 2). 48 hours after transfection, cDNA was produced and the expression levels of the WT1 target genes AREG and Ets-1, along with Bax as a control non-WT1 target gene, was measured (Figure 2). As we reported before, expression of BASP1 in K562 cells resulted in reduced expression of AREG and Ets-1 but not Bax. 10, 11 Depletion of prohibitin in BASP1-K562 cells with either of the two different siRNAs caused elevation of AREG and Ets-1 mRNA in BASP1-K562 cells but not the control gene Bax. Conversely, in the V-K562 cells (that do not express BASP1), AREG and Ets-1 mRNA did not change. These data demonstrate that prohibitin is required for the BASP1-dependent transcriptional repression of WT1 target genes.
Figure 2. Prohibitin mediates BASP1-dependent transcriptional repression of WT1 target genes.
V-K562 cells and BASP1-K562 cells were transfected with control siRNA or each of two different siRNAs against prohibitin (prohibitin siRNA 1 and prohibitin siRNA 2). 48 hours later cDNA was prepared. qPCR was performed to detect GAPDH, AREG, Ets-1 and BAX mRNA. Error bars are obtained from SDM of three independent experiments; *p<0.05 obtained by Student’s t-test. Below, whole-cell extracts prepared in parallel after the transfection were immunoblotted with anti-prohibitin or anti-tubulin antibodies.
Prohibitin recruitment at promoters is BASP1 and WT1-dependent
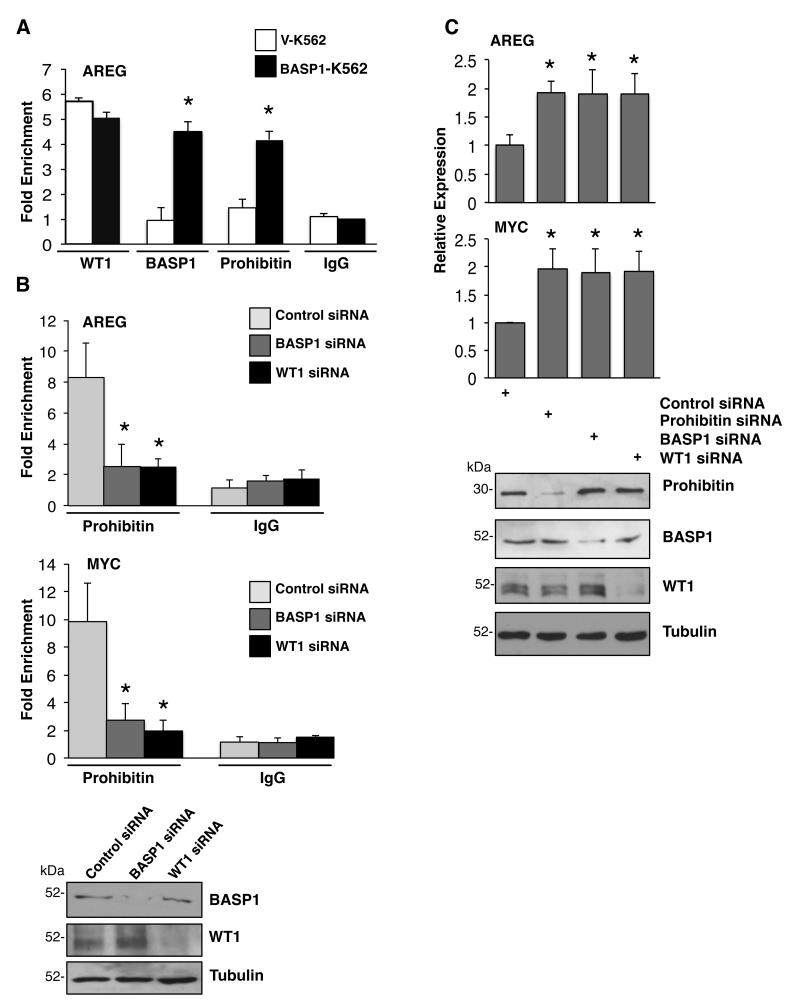
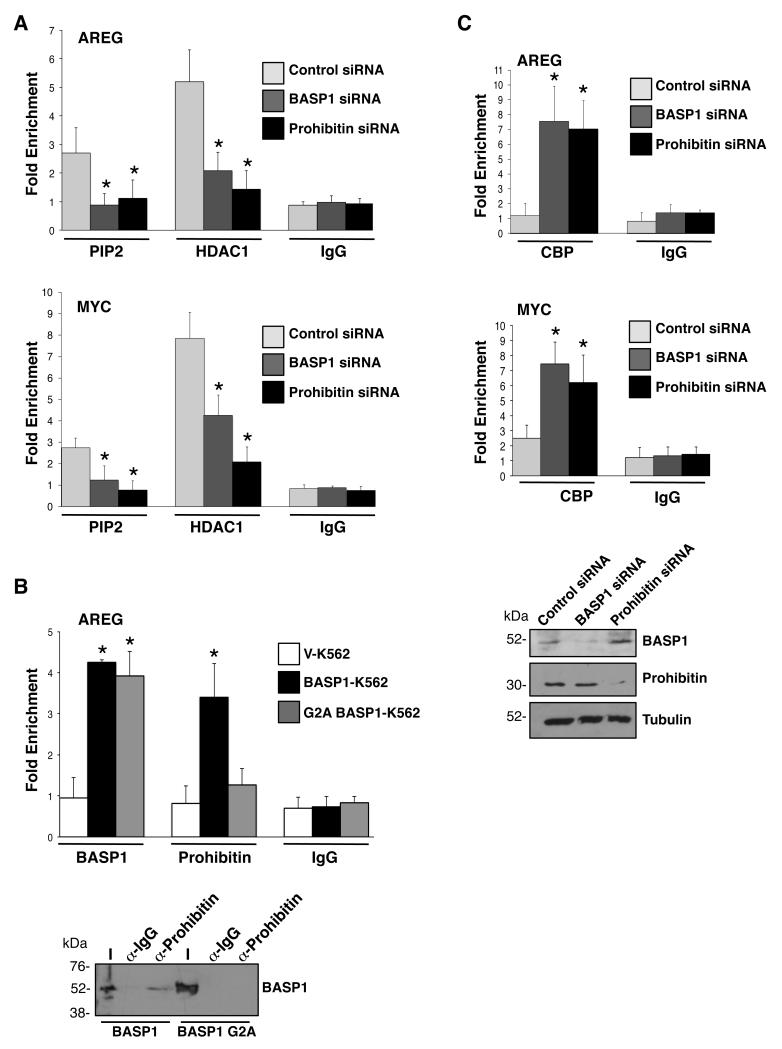
To further explore the functional role that prohibitin plays in BASP1-dependent transcriptional repression, we next used chromatin immunoprecipitation (ChIP) to determine if prohibitin is recruited to the AREG promoter, a well defined WT1 target gene. 28 ChIP was performed with control, anti-WT1, anti-BASP1 or anti-prohibitin antibodies using fragmented cross-linked chromatin from V-K562 cells and BASP1-K562 cells (Figure 3A). As previously reported by us, WT1 occupied the AREG promoter in both V-K562 cells and BASP1-K562 cells. 10, 11 BASP1 occupied the AREG promoter only in BASP1-expressing K562 cells (Figure 3A). ChiP analysis with anti-prohibitin antibodies demonstrated significant recruitment of prohibitin to the AREG promoter in BASP1-K562 cells but not the control V-K562 cells (Figure 3A). Thus, prohibitin is recruited to the AREG promoter only when BASP1 is expressed.
Figure 3. WT1-BASP1 dependent recruitment of Prohibitin to the promoters of WT1 target genes.
(A) Vector control K562 cells (V-K562) and BASP1-K562 cells were subjected to ChIP assay with control IgG, WT1, BASP1 or prohibitin antibodies. The data at the AREG promoter are presented as fold-enrichment relative to a control gene region. Error bars are obtained from standard deviation from the mean (SDM) of three independent experiments; *p<0.05 obtained by Student’s t-test. (B) U2OS cells were transfected with control siRNA, BASP1 siRNA or WT1 siRNA and 48 hr later ChIP was performed with control IgG or anti-prohibitin antibodies. The data are presented as fold enrichment at the AREG and c-myc promoters relative to a control gene region. Error bars are obtained from SDM of three independent experiments; *p<0.05 obtained by Student’s t test. Whole cell extracts were prepared parallel and immunoblotted with anti-prohibitin or anti-tubulin antibodies. (C) U2OS cells were transfected with a control siRNA, prohibitin siRNA, BASP1 siRNA or WT1 siRNA. 48 hr later RNA and cDNA were prepared and qPCR was performed to quantify GAPDH, AREG and c-myc mRNA. The data are presented relative to GAPDH mRNA. Values are the mean for three independent experiments with SDM and asterisk is student’s t test of p<0.05. Whole-cell extracts were prepared in parallel and blotted with anti-prohibitin, anti-BASP1, anti-WT1 or anti-tubulin antibodies.
We sought to confirm and extend these findings by performing ChIP analysis in U2OS cells, which express endogenous WT1 and BASP1 (Figure 3B). 11, 29 U2OS cells were transfected with control siRNA, or siRNAs targeting WT1 or BASP1, and then ChIP analysis was performed with anti-prohibitin and control IgG antibodies. Immunoblotting confirmed the successful ablation BASP1 and WT1 by their respective siRNAs (Figure 3B, at bottom). ChIP analysis with anti-prohibitin antibodies revealed a significant loss of prohibitin recruitment to the AREG and c-myc promoters in U2OS cells when either BASP1 or WT1 were depleted (Figure 3B). Comparable results were obtained when we analyzed the recruitment of prohibitin to the AREG and c-myc promoters in MCF7 cells transfected with either BASP1 or WT1 siRNA (Supplementary Figure 1). These results independently confirm that the recruitment of prohibitin to the promoters of WT1 target genes is both BASP1- and WT1-dependent. To further validate the role of prohibitin as a corepressor of WT1-BASP1 complex, U2OS cells were transfected with either control siRNA or siRNA directed to prohibitin, BASP1 or WT1. Western blotting confirmed successful depletion of prohibitin, BASP1 and WT1 expression by the respective siRNAs (Figure 3C, bottom). Quantitative PCR (qPCR) was used to analyze the effect of Prohibitin, BASP1 or WT1 depletion on expression of the WT1 target genes amphiregulin (AREG) and c-myc. SiRNAs targeting prohibitin, BASP1 or WT1 caused elevation of both AREG and c-myc mRNA suggesting that each factor is required to mediate transcriptional repression of these WT1 target genes (Figure 3C). Comparable results were obtained when we analyzed the effect of transfection of prohibitin, BASP1 and WT1 siRNA on the expression of AREG and c-myc in MCF7 cells (Supplementary Figure 2).
Prohibitin recruits BRG1 to WT1 target genes to mediate transcriptional repression
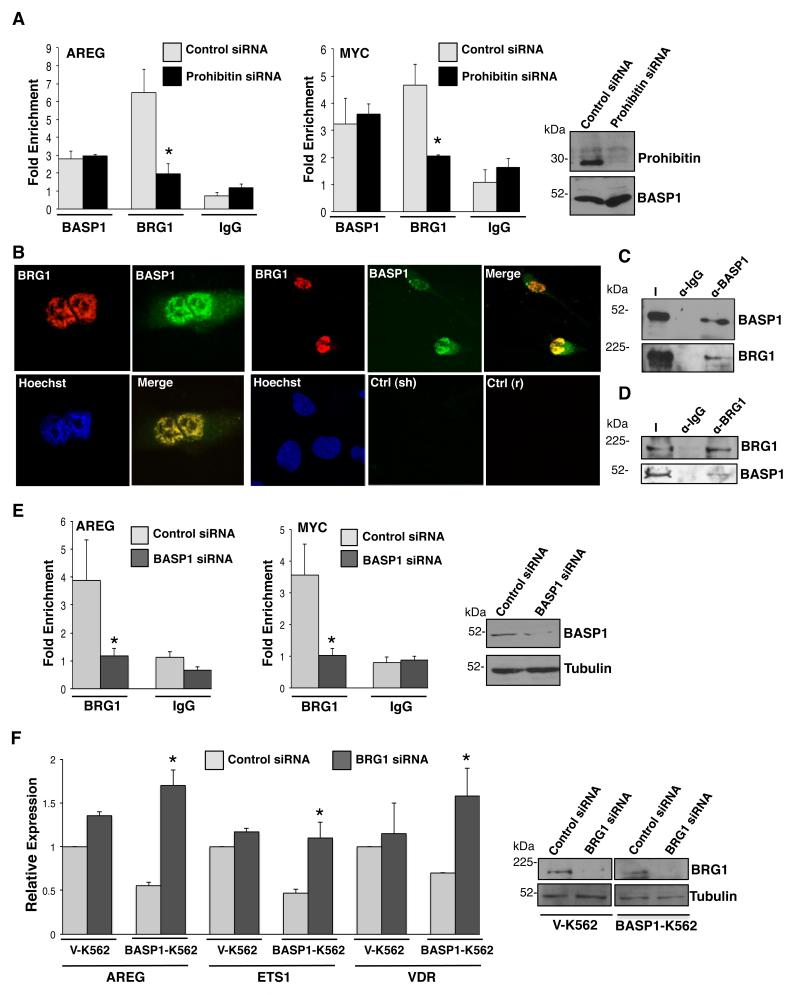
We next explored the molecular mechanism by which prohibitin directs the repression of WT1 target genes. It has been previously shown that prohibitin utilizes the SWI-SNF complex core ATPase, BRG1, to mediate repression at AR-, ER- and E2F-responsive promoters. 17, 30, 31, 26 We therefore sought to determine if BRG1 is recruited to WT1 target genes to mediate transcriptional repression. U2OS cells transfected with control siRNA or prohibitin siRNA were subjected to ChIP with control antibodies, anti-BASP1 or anti-BRG1 antibodies. Compared with the control cells, cells that were depleted of prohibitin showed a significant reduction in BRG1 recruitment to both the AREG and c-myc promoters (Figure 4A). Western blotting confirmed successful depletion of prohibitin in cells transfected with prohibitin siRNA. A reduction in BRG1 recruitment to the AREG and c-myc promoters was also observed in MCF7 cells depleted of prohibitin compared to control cells (Supplementary Figure 3). These results suggest that BRG1 is recruited to the promoters of WT1 target genes in a prohibitin-dependent manner.
Figure 4. Prohibitin and BASP1 recruit BRG1 to promoters of WT1 target genes.
(A) 48hr after transfection with prohibitin siRNA or control siRNA, U2OS cells were subjected to ChIP analysis with control IgG, anti-BASP1 or anti-BRG1 antibodies. Data are presented as fold enrichment of AREG and c-myc promoters relative to a control genome region. Error bar values are SDM for three independent experiments; *p<0.05 by Student’s t test. Immunoblotting with anti-prohibitin or anti-tubulin antibodies confirmed successful depletion of prohibitin. (B) MCF7 cells were subjected to coimmunofluorescence with sheep BASP1 antibodies and rabbit BRG1 antibodies. Hoechst was used to label the nucleus. Control anti-sheep (sh) and control anti-rabbit (r) are shown below. (C) Immunoprecipitation with control IgG antibodies or BASP1 antibodies was performed in HeLa cell nuclear extracts. The samples were then immunoblotted with BASP1 or BRG1 antibodies. I is input. (D) Immunoprecipitation with control IgG antibodies or BRG1 antibodies was performed in HeLa cell nuclear extracts and the samples were immunoblotted with either BRG1 or BASP1 antibodies. I is the input nuclear extract. (E) U2OS cells were transfected with control siRNA or BASP1 siRNA and 48 hr later were subjected to ChIP with control IgG antibodies or BRG1 antibodies. The results are presented as fold enrichments at AREG and c-myc promoters relative to a control genome region. Error bars are obtained from the SDM of three independent experiments (*p<0.05 by Student’s t test). Immunoblotting with BASP1 antibodies or tubulin antibodies confirmed successful depletion of BASP1. (F) Vector control K562 cells (V-K562) and BASP1-K562 cells were transfected with control siRNA or BRG1 siRNA and 48 hr later RNA and whole cell extracts were obtained. Samples were immunoblotted with anti-BRG1 or anti-tubulin antibodies to confirm depletion of BRG1. The expression of AREG, Ets-1 and VDR are presented as relative to GAPDH mRNA. Error bars are the SDM of three independent experiments and Student’s t test of p<0.05 was performed.
Since the association between prohibitin and BRG1 is well-established and our data suggest that BASP1 and prohibitin associate in the nucleus, we next sought to determine if there is an association between BASP1 and BRG1. Immunofluorescence with anti-BASP1 and anti-BRG1 antibodies revealed that BASP1 and BRG1 colocalize in the nucleus of MCF7 cells (Figure 4B). Furthermore, anti-BASP1 immunoprecipitates from HeLa cell nuclear extract contained BRG1 (Figure 4C) and BASP1 was detected in the corresponding anti-BRG1 immunoprecipitates (Figure 4D). Taken together, these data demonstrate that BASP1 colocalizes with and interacts with BRG1 in the nucleus and suggest that BRG1 recruitment to the promoter of WT1 target genes may be both prohibitin and BASP1-dependent. To test this possibility, U2OS cells were transfected with control siRNA or BASP1 siRNA and then subjected to ChIP assays with control antibodies or anti-BRG1 antibodies. Western blotting with anti-BASP1 antibodies confirmed depletion of BASP1 (Figure 4E). ChIP analysis revealed that BRG1 recruitment to the promoters of both the AREG and c-myc genes showed a significant decrease in BASP1-depleted cells when compared to the control U2OS cells (Figure 4E). Thus, BRG1 recruitment at promoters is BASP1- and prohibitin-dependent.
BRG1, the central catalytic subunit of numerous chromatin-modifying complexes, is a major coregulator of transcription by remodeling nucleosomal structure in an ATP-dependent manner. 31, 32 BRG1 has been implicated in both the activation and repression of gene expression depending upon the promoter context and the protein subunits comprising the complex. 33-35 Our data so far suggests that BRG1 associates with the BASP1 and prohibitin corepressor complex. To determine if BRG1 plays a role in the BASP1-dependent repression of WT1 target genes, VK562 cells and BASP1-K562 cells were transfected with control siRNA or BRG1 siRNA. Immunoblotting of whole-cell extracts with anti-BRG1 antibodies confirmed significant depletion of BRG1 in V-K562 cells and BASP1-K562 cells when compared to the cells transfected with control siRNA (Figure 4F). We then prepared cDNA and measured the expression of the WT1 target genes AREG, Ets-1 and Vitamin D receptor (VDR; Figure 4F). While expression of BRG1 was ablated successfully in both BASP1 K562 cells and V-K562 cells, AREG, Ets-1 and VDR RNA was significantly elevated only in BASP1 K562 cells and not V-K562 cells (Figure 4F). Taken together the results of Figure 4 suggest that BRG1 mediates transcriptional repression of WT1 target genes and that this requires both BASP1 and prohibitin.
Prohibitin is required for the BASP1-dependent recruitment of PIP2 and HDAC1 to the promoters of WT1 target genes
We recently reported that repression of transcription by the WT1-BASP1 complex requires the N-terminal myristoylation of BASP1 and the PIP2-dependent recruitment of HDAC1 to gene promoters. 11 Studies by others have shown that prohibitin also interacts with HDAC1 and recruits HDAC1 to repress E2F-mediated transcription. 21 Moreover, prohibitin also associates with phospholipids. 36 Thus we sought to determine if prohibitin plays a role in the recruitment of PIP2 and HDAC1 to the promoter regions of WT1 target genes. U2OS cells were transfected with BASP1 siRNA, prohibitin siRNA or control siRNA and then subjected to ChIP assays with control antibodies, anti-PIP2 antibodies, or anti-HDAC1 antibodies. Consistent with our previous results in K562 cells, 11 when the expression of BASP1 was ablated in U2OS cells by siRNA, the recruitment of PIP2 and HDAC1 to the AREG and c-myc promoters was significantly reduced (Figure 5A). siRNA-mediated depletion of prohibitin also led to a significant reduction in the recruitment of both PIP2 and HDAC1 to the AREG and c-myc promoters compared to the control cells (Figure 5A). These data suggest that prohibitin and BASP1 are both required for the recruitment of PIP2 and HDAC1 to the promoter to mediate transcriptional repression.
Figure 5. Prohibitin is required to recruit PIP2 and HDAC1 to the promoters of WT1 target genes.
(A) U2OS cells were transfected with control siRNA, BASP1 siRNA or prohibitin siRNA. 48hr later the cells were subjected to ChIP assay with control IgG antibodies, PIP2 antibodies and HDAC1 antibodies. The results are presented as fold enrichment at AREG and cmyc promoter relative to a control genome region. Error bars are presented as the SDM of three independent experiments and *p<0.05 by Student’s t test. (B) Control V-K562 cells, wild type BASP1-K562 cells and the mutant derivate G2A BASP1-K562 cells were subjected to ChIP with control IgG antibodies, BASP1 antibodies and prohibitin antibodies at AREG promoter relative to a control genome region. Error bars are the SDM of three independent experiments and *p<0.05 by Student’s t test. Lower panel; Nuclear extracts from BASP1-K562 cells and BASP1 G2A-K562 cells were used in immunoprecipitation with anti-prohibitin antibodies. The immunoprecipitates were immunoblotted with anti-BASP1 antibodies. (C) U2OS cells were transfected with BASP1 siRNA, prohibitin siRNA or a control siRNA and 48 hr later the cells were subjected to ChIP with control IgG antibodies and anti-CBP antibodies. The results are presents as fold enrichment at the AREG and c-myc promoter regions compared to a control genome region. Error bars are SDM of three independent experiments (*p<0.05 by Student’s t test). In parallel, whole-cell extracts were immunoblotted with anti-BASP1, anti-prohibitin or anti-tubulin antibodies to confirm knockdown of BASP1 or prohibitin.
We have previously shown that K562 cells expressing BASP1 mutant derivative G2A (which prevents the N-terminal myristoylation of BASP1) led to a reduction of PIP2 and HDAC1 recruitment at promoters compared to wild type BASP1 expressing cells and was thus defective in transcriptional repression. 11 Since our data above demonstrate that prohibitin is important for PIP2 and HDAC1 recruitment at promoters, we sought to determine if prohibitin recruitment at promoters was dependent on the N-terminal myristoylation of BASP1. Vector control K562 cells or K562 cell line derivatives expressing either wild-type BASP1 or BASP1 G2A were subjected to ChIP with control antibodies, BASP1 antibodies or prohibitin antibodies (Figure 5B). As in figure 3A, compared to the control V-K562 cells, wild-type BASP1-K562 cells showed recruitment of both BASP1 and prohibitin to the AREG promoter (Figure 5B). In contrast, cells expressing BASP1 G2A did not show efficient recruitment of prohibitin to the AREG promoter compared with control cells, even though BASP1 G2A itself was recruited (Figure 5B). Moreover, while anti-prohibitin antibodies coimmunoprecipitated wild type BASP1 from nuclear extracts prepared from BASP1-K562 cells, BASP1 G2A did not coimmunoprecipitate (Figure 5B, lower panel). Thus, the myristoylation of BASP1 is not only important for PIP2 and HDAC1 recruitment but is also required for prohibitin recruitment to the promoter to mediate transcriptional repression. 11
Prohibitin induces transcriptional repression, in part, through the removal of coactivators such as p300/CBP. 26 Since p300/CBP also acts as a coactivator for WT1, 37 we next sought to determine if prohibitin and/or BASP1 play a role in regulating the association of CBP with WT1. U2OS cells transfected with control siRNA, prohibitin siRNA or BASP1 siRNA were subjected to ChIP with control antibodies or anti-CBP antibodies. Depletion of BASP1 or prohibitin led to a significant increase in CBP recruitment to AREG and c-myc promoters compared to the control cells (Figure 5C). Thus, the BASP1/prohibitin corepressor complex blocks the association of the coactivator CBP with WT1 at the promoter.
Prohibitin is required for the BASP1-dependent diversion of the K562 differentiation program
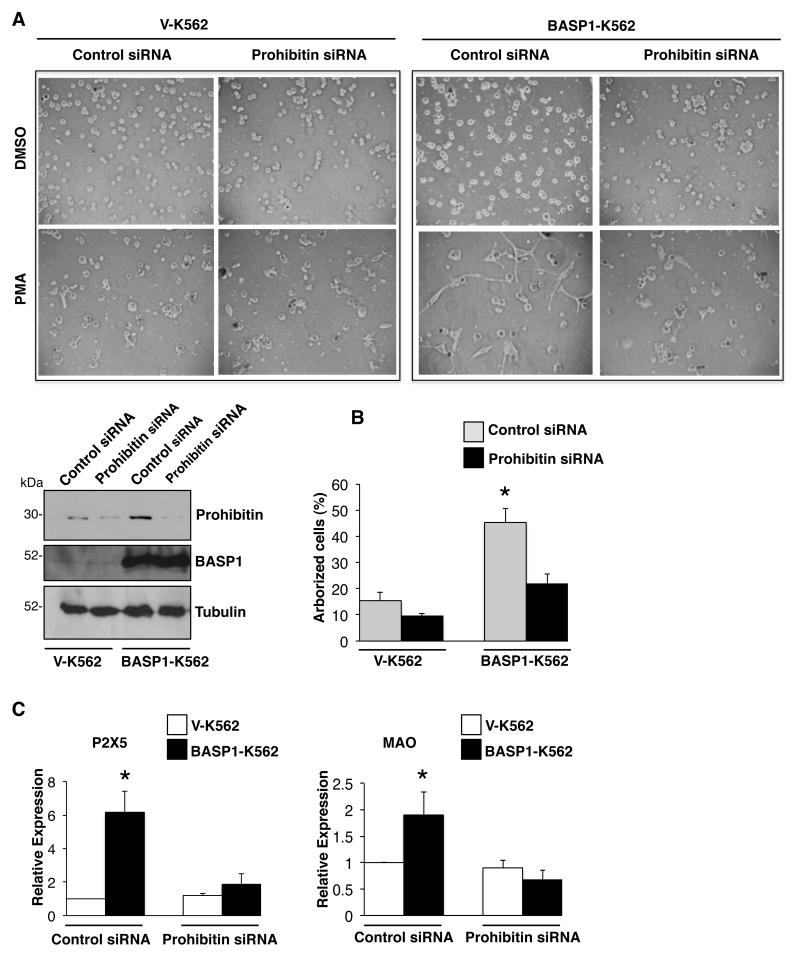
Our data so far suggest that prohibitin plays a critical role in transcriptional repression by WT1-BASP1. We next sought evidence of the functional relevance of prohibitin to the control of cell differentiation by WT1-BASP1. We have previously demonstrated that WT1 and BASP1 transcriptionally cooperate to redirect the PMA-dependent differentiation pathway of K562 cells to an arborized cell type with neuronal characteristics. 10, 11 We therefore transfected V-K562 cells or BASP1-K562 cells with control siRNA or siRNA targeting prohibitin. 48 hours after transfection, the cells were treated with PMA and allowed the cells to differentiate over 3 days. Immunoblotting confirmed that transfection of the prohibitin siRNA caused a significant reduction in prohibitin expression in both V-K562 and BASP1-K562 cells (Figure 6A). As before, BASP1-K562 cells exhibited an altered differentiation morphology, becoming highly arborized when compared to the vector control K562 cells (Figure 6A, quantitated in Figure 6B). While transfection of the prohibitin siRNA had no observable effect in PMA-induced control VK562 cells, it significantly blocked the PMA-induced arborization of BASP1-K562 cells (Figure 6A, quantitation in Figure 6B). Thus, prohibitin expression is required for the WT1- and BASP1-dependent differentiation program of BASP1 K562 cells that occurs in response to PMA treatment.
Figure 6. Prohibitin is required for the PMA-dependent differentiation of BASP1 K562 cells.
(A) Vector control V-K562 cells and BASP1-K562 cells were transfected with control siRNA or prohibitin siRNA and 48 hr later were treated with DMSO as a control or treated with 100nM of PMA and then imaged 72hr later. Whole cell-extracts were prepared in parallel and immunoblotted with anti-prohibitin or anti-tubulin antibodies to confirm successful depletion of prohibitin in V-K562 cells and BASP1-K562 cells. (B) As in part A except that quantification of the percentage of arborized cells was performed. Error bars represent SDM of three independent experiments and *p<0.05 by Student’s t test. (C) As in part A except that RNA and cDNA were prepared. Expression of P2X5 and MAO were analyzed by qPCR in comparison with GAPDH. Error bars are SDM of three independent experiments (*p<0.05 by Student’s t test).
PMA treatment of BASP1-K562 cells leads to the induction of several neuronal markers (e.g. P2X5 and MAO; 10). We therefore prepared cDNA from V-K562 cells and B-K562 cells that had been transfected with control or prohibitin siRNA, and then treated with PMA for 48 hours. Ablation of prohibitin in BASP1-K562 cells blocked the induction of both of the neuronal makers, P2X5 and MAO, but did not have any significant effect on the expression of these genes in V-K562 cells (Figure 6C). Taken together the findings in Figure 6 demonstrate that the WT1-BASP1 complex requires prohibitin cooperation to elicit the PMA-induced arborization of K562 cells and the induction of neuronal markers.
Discussion
In this study we have provided evidence that prohibitin is a BASP1 cofactor that regulates the transcriptional activity of the WT1-BASP1 complex. Our findings suggest that prohibitin and BASP1 cooperate to regulate the expression of WT1 target genes by at least three mechanisms; BASP1 and prohibitin facilitate the recruitment of BRG1-containing chromatin remodeling activities to WT1 target genes. BASP1 and prohibitin cooperate through the phospholipid PIP2 to recruit HDAC1 to the promoter. Finally, BASP1 and prohibitin compete with CBP to interact with WT1 during the switch from WT1-mediated transcriptional activation to repression. Taken together, our results suggest that BASP1 and prohibitin form part of a multi-modal transcription repression mechanism that regulates WT1 target genes.
Several studies have demonstrated that prohibitin functions as a coregulator for many transcription factors. Prohibitin suppresses E2F-mediated transcription by inducing the recruitment of chromatin modifiers including HDAC1, N-CoR and BRG1/Brm. 17, 19 It is also known that prohibitin dissociates coactivators such as p300 from AR-responsive promoters to induce a repressive gene state. 26 It is not uncommon for transcriptional factors to associate at promoters along with a variety of histone-modifying enzymes, chromatin remodelers and other cofactors to repress transcription. Indeed BRG1 can associate with various multi-protein complexes to elicit transcriptional activation or repression. 35 These opposing effects are dependent upon promoter context and also the composition of the complex. 33, 34, 35 For instance, when BRG1 is found assembled in complexes with histone deacetylases, it leads to transcriptional repression. 35 Thus, this differential makeup of BRG1 found in complexes with corepressors such as prohibitin, BASP1 and histone deacetylases might be required to switch WT1 from a gene activator to a repressor. However, further work is needed to fully understand the architecture of the assembled complexes. Indeed, based on our previous gel filtration data, we anticipate that further active components of the BASP1 complex remain to be identified. 8
Repression of transcription by WT1-BASP1 requires the myristoylation of BASP1 and the PIP2-dependent recruitment of HDAC1. 11 It has also been reported that prohibitin associates with phospholipids 36 and our present study suggests that BASP1 and prohibitin are required together for the recruitment of PIP2 and HDAC1 to the promoters of WT1 target genes. We note that the myristoylation-defective BASP1 mutant derivative G2A is not able to interact with PIP2, 11 and even though it is recruited to the promoter it does not support the assembly of prohibitin or HDAC1. Thus, PIP2 appears to be critical for forming protein-protein contacts between these factors. PIP2 associates with BASP1 11 and prohibitin has a PHB domain, which a number of studies suggest has high affinity for lipids. 38 Moreover, both histone deacetylases 39-41 and ATP-dependent chromatin remodeling activities 42, 43 have been shown to require nuclear phospholipids for their activity. Indeed, a recent study demonstrated that the interaction of HDAC3 with the SMRT corepressor required the presence of inositol phosphates. 44 Furthermore, Schwabe and coworkers also revealed that the activity of HDAC1 is regulated by interaction with inositol phosphates. 45 Thus, it is tempting to speculate that several phospholipid molecules could be recruited to gene promoters through independent components of the repression complex. Indeed, a recent study reported that several components of the RNA polymerase I transcription machinery interact with PIP2 to regulate transcription of rRNA genes. 46 Taken together with previous work, our findings argue for an extensive role for nuclear phospholipids in transcriptional regulation. Since prohibitin also regulates the transcriptional activity of many transcription factors, it will be important to determine whether BASP1, prohibitin and phospholipids cooperate in the regulation of other transcriptional regulators in addition to WT1.
Our data demonstrate that the WT1-BASP1 complex requires prohibitin to transcriptionally cooperate in diversion of the PMA-induced differentiation program of K562 cells to a neuronal-like morphology. It is therefore possible that prohibitin might also be important to regulate WT1’s function during development. Prohibitin has been shown to repress ER-dependent transcription and a recent study reported that prohibitin is essential for Estrogen-dependent mouse uterine development and adult function. 47 Furthermore, it has also been demonstrated that prohibitin acts as a neural crest specifier in Xenopus development by repressing the activity of the transcriptional factor E2F1. 20 Further work is needed to determine if the WT1-BASP1-prohibitin complex plays a role in development. Moreover, considering the proposed roles of BASP1 and prohibitin as tumor suppressors, it will be important to study these factors in WT1-dependent cancers.
Materials and methods
Tissue culture, immunofluorescence and antibodies
U2OS, MCF7 and HeLa cells were cultured in Dulbecco’s modified Eagle’s medium and K562 cells were cultured in RPM1 medium. Stable K562 cell line derivatives were prepared as described before. 10 Differentiation of K562 cells with PMA and scoring of cellular arborization was performed as described previously. 10 U2OS, K562 and MCF7 cells were transfected with HiPerFect (Qiagen). For immunofluorescence, cells were grown in six-well slides, fixed with 4% paraformaldehyde, permeabilized with 0.1% TritonX-100 and washed with phosphate-buffered saline (PBS). Cells were incubated with primary antibodies for 1 h and then washed with PBS. The cells were then incubated for 30 min with DyLight 488 goat anti-mouse (1:100) and DyLight 546 goat anti-rabbit (1:100) antibodies in PBS and washed with PBS. Nuclei were counterstained with Hoechst stain (1μg/ml) in PBS.
The BASP1 antibodies used were described before. 6 Antibodies anti-WT1 (C-19), anti-BRG1 (H-88) and anti-CBP (A-22) were from Santa Cruz Biotech. Antibodies anti β-tubulin (ab6046), anti-HDAC1 (ab7028), anti-PIP2 (ab11039) and anti-prohibitin (ab28172) were from Abcam.
mRNA analysis and Chromatin Immunoprecipitation
Total RNA was generated using the QIAGEN RNeasy kit, and cDNA was prepared using the Bio-Rad cDNA synthesis kit. RNA expression primers used were; GAPDH, Forward, 5′-ACAGTCAGCCGCATCTTCTT-3′, Reverse, 5′-ACGACCAAATCCGTTGACTC-3′; AREG, Forward, 5′-TGGATTGGACCTCAATGACA-3′, Reverse, 5′- ACTGTGGTCCCCAGAAAATG-3′; Ets-1, Forward, 5′-AAACTTGCTACCATCCCGTACGT-3′, Reverse, 5′- ATGGTGAGAGTCGGCTTGAGAT-3′; VDR, Forward, 5′-CTGACCCTGGAGACTTTGAC-3′, Reverse, 5′-TTCCTCTGCACTTCCTCAT-3′: c-myc Forward 5′-CCACGTCTCCACACATCAG-3′ and Reverse, 5′-TGGTGCATTTTCGGTTGTTG-3′. siRNA to human prohibitin were purchased from Qiagen. siRNA to human BASP1, WT1 and control siRNA were purchased from Ambion.
ChIP assays were performed as described before.11, 48 ChIP primers used in this study were AREG, Forward, 5′-TTTAAGTTCCACTTCCTCTCA-3′, Reverse, 5′-GGTGTGCGAACGTCTGTA-3′ Control, Forward, 5′-CAGCTCAGTGCTGTTGGTGG-3′, Reverse, 5′-ACCATCCAACCCTGGAGATC-3′ and c-myc Forward, 5′-TCAAACAGTACTGCTACGGA-3′ Reverse 5′-AGAGCCGCATGAATTAACTA-3′.
Supplementary Material
Acknowledgements
This work was funded by the National Institute of General Medical Sciences [1R01GM098609] (to KFM and SGER) and Cancer Research UK [C1356/A6630] (to SGER). We thank Alan Siegel for help with the confocal microscopy and the facility funded by National Science Foundation [DBI 0923133].
Footnotes
Conflict of Interest The authors declare no conflict of interest.
Supplementary Information accompanies the paper on the Oncogene website (http://www.nature.com/onc)
References
- 1.Rivera MN, Haber DA. Wilms’ tumour: connecting tumorigenesis and organ development in the kidney. Nat Rev Cancer. 2005;5:699–712. doi: 10.1038/nrc1696. [DOI] [PubMed] [Google Scholar]
- 2.Hohenstein P, Hastie ND. The many facets of the Wilms’ tumour gene, WT1. Human Mol Genet. 2006;15(Spec No 2):R196–R201. doi: 10.1093/hmg/ddl196. [DOI] [PubMed] [Google Scholar]
- 3.Huff V. Wilms’ tumours: about tumour suppressor genes, an oncogene and a chameleon gene. Nat Rev Cancer. 2011;11:111–121. doi: 10.1038/nrc3002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Roberts SGE. Transcriptional regulation by WT1 in development. Curr Opin Genet Dev. 2005;15:542–547. doi: 10.1016/j.gde.2005.08.004. [DOI] [PubMed] [Google Scholar]
- 5.McKay LM, Carpenter B, Roberts SGE. Regulation of the Wilms’ tumour suppressor protein transcriptional activator domain. Oncogene. 1999;18:6546–6554. doi: 10.1038/sj.onc.1203046. [DOI] [PubMed] [Google Scholar]
- 6.Carpenter B, Hill KJ, Charalambous M, Wagner KJ, Lahiri D, James DI, Andersen JS, et al. BASP1 is a transcriptional cosuppressor for the Wilms’ tumor suppressor protein WT1. Mol Cell Biol. 2004;24:537–549. doi: 10.1128/MCB.24.2.537-549.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mosevitsky MI. Nerve ending “signal” proteins GAP-43, MARCKS, and BASP1. Int Rev Cytol. 2005;245:245–325. doi: 10.1016/S0074-7696(05)45007-X. [DOI] [PubMed] [Google Scholar]
- 8.Green LM, Wagner KJ, Campbell HA, Addison K, Roberts SGE. Dynamic interaction between WT1 and BASP1 in transcriptional regulation during differentiation. Nucleic Acids Res. 2009;37:431–440. doi: 10.1093/nar/gkn955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Essafi A, Webb A, Berry RL, Slight J, Burn SF, Spraggon L, et al. A Wt1-controlled chromatin switching mechanism underpins tissue-specific wnt4 activation and repression. Dev Cell. 2011;21:559–574. doi: 10.1016/j.devcel.2011.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Goodfellow SJ, Rebello MR, Toska E, Zeef LA, Rudd SG, Medler KF, et al. WT1 and its transcriptional cofactor BASP1 redirect the differentiation pathway of an established blood cell line. Biochem J. 2011;435:113–125. doi: 10.1042/BJ20101734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Toska E, Campbell HA, Shandilya J, Goodfellow SJ, Shore P, Medler KF, et al. Repression of transcription by WT1-BASP1 requires the myristoylation of BASP1 and the PIP2-dependent recruitment of histone deacetylase. Cell Reports. 2012;2:462–469. doi: 10.1016/j.celrep.2012.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hartl M, Nist A, Khan MI, Valovka T, Bister K. Inhibition of Myc-induced cell transformation by brain acid-soluble protein 1 (BASP1) Proc Natl Acad Sci USA. 2009;106:5604–5609. doi: 10.1073/pnas.0812101106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yeoh EJ, Ross ME, Shurtleff SA, Williams WK, Patel D, Mahfouz R, et al. Classification, subtype discovery, and prediction of outcome in pediatric acute lymphoblastic leukemia by gene expressing profiling. Cancer Cell. 2002;1:133–143. doi: 10.1016/s1535-6108(02)00032-6. [DOI] [PubMed] [Google Scholar]
- 14.Moribe T, Lizuka N, Miura T, Stark M, Tamatsukuri S, Ishitsuka H, et al. Identification of novel aberrant methylation of BASP1 and SRD5A2 for early diagnosis of hepatocellular carcinoma by genome-wide search. Int J Oncol. 2008;33:949–958. [PubMed] [Google Scholar]
- 15.Wang S, Nath N, Adlam M, Chellappan S. Prohibitin, a potential tumor suppressor, interacts with RB and regulates E2F function. Oncogene. 1999;18:3501–3510. doi: 10.1038/sj.onc.1202684. [DOI] [PubMed] [Google Scholar]
- 16.Wang S, Nath N, Fusaro G, Chellappan S. Rb and prohibitin target distinct regions of E2F1 for repression and respond to different upstream signals. Mol Cell Biol. 1999b;19:7447–7460. doi: 10.1128/mcb.19.11.7447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang S, Zhang B, Faller DV. Prohibitin requires Brg-1 and Brm for the repression of E2F and cell growth. EMBO J. 2002a;21(12):3019–28. doi: 10.1093/emboj/cdf302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Joshi B, Ko D, Ordonez-Ercan D, Chellappan SP. A putative coiled-coil domain of prohibitin is sufficient to repress E2F1-mediated transcriptional and induce apoptosis. Biochem Biophys Res Commun. 2003;312(2):459–66. doi: 10.1016/j.bbrc.2003.10.148. [DOI] [PubMed] [Google Scholar]
- 19.Choi D, Lee SJ, Hong S, Kim IH, Kang S. Prohibitin interacts with RNF2 and regulates E2F1 function via dual pathways. Oncogene. 2008;27:1716–1725. doi: 10.1038/sj.onc.1210806. [DOI] [PubMed] [Google Scholar]
- 20.Schneider M, Schambony A, Wedlich D. Prohibitin1 acts as a neural crest specifier in Xenopus development by repressing the transcription factor E2F1. Development. 2010;137(23):4073–81. doi: 10.1242/dev.053405. [DOI] [PubMed] [Google Scholar]
- 21.Wang S, Fusaro G, Padmanabhan J, Chellappan SP. Prohibitin co-localizes with Rb in the nucleus and recruits N-CoR and HDAC1 for transcriptional repression. Oncogene. 2002;21(55):8388–96. doi: 10.1038/sj.onc.1205944. [DOI] [PubMed] [Google Scholar]
- 22.Montano MM, Ekena K, Delage-Mourroux R, Chang W, Martini P, Katzenellenbogen BS. An estrogen receptor-selective coregulator that potentiates the effectiveness of aniestrogenes and represses the activity of estrogens. Proc Natl Acad Sci USA. 1999;96:6947–6952. doi: 10.1073/pnas.96.12.6947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Delage-Mourroux R, Martini PG, Choi I, Kraichely DM, Hoeksema J, Katzenellenbogen BS. Analysis of estrogen receptor interaction with a repressor of estrogen receptor activity (REA) and the regulation of estrogen receptor transcriptional activity by REA. J Biol Chem. 2000;275(46):35848–56. doi: 10.1074/jbc.M001327200. [DOI] [PubMed] [Google Scholar]
- 24.He B, Feng Q, Mukherjee A, Lonard DM, DeMayo FJ, Katzenellenbogen BS, Lydon JP, et al. A repressive role for prohibitin in estrogen signaling. Mol Endocrinol. 2008;22:344–360. doi: 10.1210/me.2007-0400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gamble SC, Chotai D, Odontiadis M, Dart DA, Brooke GN, Powell SM, et al. Prohibitin, a protein downregulated by androgens, represses androgen receptor activity. Oncogene. 2007;26:1757–1768. doi: 10.1038/sj.onc.1209967. [DOI] [PubMed] [Google Scholar]
- 26.Dai Y, Ngo D, Jacob J, Forman LW, Faller DV. Prohibitin and the SW1/SNF ATPase subunit BRG1 are required for effective androgen antagonist-mediated transcriptional repression of androgen receptor regulated genes. Carcinogenesis. 2008;29(9):1725–1733. doi: 10.1093/carcin/bgn117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mishra S, Murphy LC, Murphy LJ. The prohibitins: emerging roles in diverse functions. J Cell Mol Med. 2006;10(2):353–363. doi: 10.1111/j.1582-4934.2006.tb00404.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kim HS, Kim MS, Hancock AL, Harper JCP, Park JY, Poy G, et al. Identification of novel Wilms’ tumor suppressor gene target genes implicated in kidney development. The Journal of Biological Chemistry. 2007;282:16278–1628. doi: 10.1074/jbc.M700215200. [DOI] [PubMed] [Google Scholar]
- 29.Hartkamp J, Carpenter B, Roberts SGE. The Wilms’ tumor suppressor protein WT1 is processed by the serine protease HtrA2/Omi. Mol Cell. 2010;37:159–171. doi: 10.1016/j.molcel.2009.12.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang S, Zhang B, Faller DV. BRG1/BRM and prohibitin are required for growth suppression by estrogen antagonists. EMBO J. 2004;23:2293–303. doi: 10.1038/sj.emboj.7600231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang B, Chambers Kj, Faller DV, Wang S. (2007) Reprogramming of the SWI/SNF complex for coactivation or co-repression in prohibitin-mediated estrogen receptor regulation. Oncogene. 2007;26(50):7153–7. doi: 10.1038/sj.onc.1210509. [DOI] [PubMed] [Google Scholar]
- 32.Havas K, Flaus A, Phelan M, Kingston R, Wade PA, Lilley DM, et al. Generation of superhelical torsion by ATP-dependent chromatin remodeling activities. Cell. 2000;103:1133–1142. doi: 10.1016/s0092-8674(00)00215-4. [DOI] [PubMed] [Google Scholar]
- 33.Kadam S, Emerson BM. Transcriptional specificity of human SWI/SNF BRG1 and BRM chromatin remodeling complexes. Mol Cell. 2003;11:377–89. doi: 10.1016/s1097-2765(03)00034-0. [DOI] [PubMed] [Google Scholar]
- 34.Sif S, Saurin Aj, Imbalzano AN, Kingston RE. Purification and characterization of mSin3A-containing Brg1 and hBrm chromatin remodeling complexes. Genes Dev. 2001;15:603–18. doi: 10.1101/gad.872801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Trotter KW, Archer TK. The BRG1 transcriptional coregulator. Nuclear Receptor Signaling. 2008;6:e004. doi: 10.1621/nrs.06004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ande SR, Mishra S. Prohibitin interacts with phosphatidylinositol 3,4,5-triphosphate (PIP3) and modulates insulin signaling. Biochem and Biophysical Research Communications. 2009;390:1023–1028. doi: 10.1016/j.bbrc.2009.10.101. [DOI] [PubMed] [Google Scholar]
- 37.Wang W, Lee SB, Palmer R, Eillisen LW, Haber DA. A functional interaction with CBP contributes to transcriptional activator by the Wilms’ tumor suppressor WT1. J Biol Chem. 2001;276(20):16810–6. doi: 10.1074/jbc.M009687200. [DOI] [PubMed] [Google Scholar]
- 38.Morrow IC, Parton RG. Flotillins and the PHB domain protein family: rafts, worms and anaesthetics. Traffic. 2005;6(9):725–40. doi: 10.1111/j.1600-0854.2005.00318.x. [DOI] [PubMed] [Google Scholar]
- 39.Gozani O, Karuman P, Jones DR, Ivanov D, Cha J, Lugovskoy AA, et al. The PHD finger of the chromatin-associated protein ING2 functions as a nuclear phosphoinositide receptor. Cell. 2003;114(1):99–111. doi: 10.1016/s0092-8674(03)00480-x. [DOI] [PubMed] [Google Scholar]
- 40.Hait NC, Allegood J, Maceyka M, Strub GM, Harikumar KB, Singh SK, et al. Regulation of histone acetylation in the nucleus by sphingosine 1-phosphate. Science. 2009;325:1254–1257. doi: 10.1126/science.1176709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Han BK, Emr SD. Phosphoinositide [PI (3,5)P2] lipid-dependent regulation of the general transcriptional regulator Tup1. Genes Dev. 2011;25:984–995. doi: 10.1101/gad.1998611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhao K, Wang W, Rando OJ, Xue Y, Swiderek K, Kuo A, et al. Rapid and phosphoinositol-dependent binding of the SWI-SNF-like BAF complex to chromatin after T lumphocyte receptor signaling. Cell. 1998;95:625–636. doi: 10.1016/s0092-8674(00)81633-5. [DOI] [PubMed] [Google Scholar]
- 43.Steger DJ, Haswell ES, Miller AL, Wente SR, O’Shea EK. Regulation of chromatin remodeling by inositol polyphosphates. Science. 2003;299:114–116. doi: 10.1126/science.1078062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Watson PJ, Fairall L, Santos GM, Schwabe JWR. Structure of HDAC3 bound to co-repressor and inositol tetraphosphate. Nature. 2012;481:335–340. doi: 10.1038/nature10728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Millard CJ, Watson PJ, Celardo I, Gordiyenko Y, Cowley SM, Robinson CV, et al. Class I HDACs share a common mechanism of regulation by inositol phosphates. Mol Cell. 2013;51:1–11. doi: 10.1016/j.molcel.2013.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yildirim S, Castano E, Sobol M, Philimonenko VV, Dzijak R, Venit T, et al. (2013) Involvement of phosphatidylinositol 4,5-bisphosphate in RNA polymerase 1 transcription. Journal of Cell Science. 2013;126:2730–2739. doi: 10.1242/jcs.123661. [DOI] [PubMed] [Google Scholar]
- 47.He B, Kim TH, Kommagani R, Feng Q, Lanz RB, Jeong JW, et al. Estrogen-regulated prohibitin is required for mouse uterine development and adult function. Endrocrinology. 2012;152(3):1047. doi: 10.1210/en.2010-0732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shandilya J, Wang Y, Roberts SGE. TFIIB dephosphorylation links transcription inhibition with the p53-dependent DNA damage response. Proc Natl Acad Sci USA. 2012;10:1073. doi: 10.1073/pnas.1207483109. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.