Abstract
The eukaryotic cortical actin cytoskeleton creates specific lipid domains, including lipid rafts, which determine the distribution of many membrane proteins. Here we show that the bacterial actin homolog MreB displays a comparable activity. MreB forms membrane associated filaments that coordinate bacterial cell wall synthesis. We noticed that the MreB cytoskeleton influences fluorescent staining of the cytoplasmic membrane. Detailed analyses combining an array of mutants, using specific lipid staining techniques and spectroscopic methods, revealed that MreB filaments create specific membrane regions with increased fluidity (RIFs). Interference with these fluid lipid domains (RIFs) perturbs overall lipid homeostasis and affects membrane protein localization. The influence of MreB on membrane organization and fluidity may explain why the active movement of MreB stimulates membrane protein diffusion. These novel MreB activities add additional complexity to bacterial cell membrane organization, and have implications for many membrane associated processes.
INTRODUCTION
The morphology of many rod-shaped bacteria is established by the coordinated incorporation of new cell wall material perpendicular to the cell axis1, 2. An essential component of this machinery is the bacterial actin homolog MreB, which polymerizes into filaments at the cell periphery3. The peripheral association of MreB is facilitated by a conserved hydrophobic membrane binding loop, which in some organisms is further supported by a membrane binding N-terminal amphipathic helix4. Upon binding, MreB forms a complex with the conserved membrane proteins MreC and MreD, and with proteins involved in peptidoglycan synthesis such as RodA, MurG, MraY, and several penicillin-binding proteins (PBPs)1, 2, 5. Interference with the MreB activity renders cells mechanically less rigid6, and in the absence of this protein cells lose their rod-shaped morphology7, 8. The prevailing model, in which helical MreB-polymers spatially direct the synthesis of new peptidoglycan and as a result determine the general shape of the cell, has recently been revised. It turned out that MreB-filaments, and the associated cell wall synthetic machinery, move around the cell in a process that is driven by peptidoglycan synthesis9-11. The MreB cytoskeleton has also been implicated in other cellular processes, including the establishment of cell polarity and chromosome segregation12, 13.
In a previous study, we have shown that the MreB cytoskeleton of Bacillus subtilis is sensitive to changes in the membrane potential, and incubation of cells with the proton ionophore CCCP results in a rapid delocalization of MreB14. The mechanism for this membrane potential -sensitivity is currently unknown. During this work, we noticed that the fluorescence of the cell membrane, when stained with the lipid dye Nile Red, shows a rapid (within one to two minutes) transformation from a uniform to a clustered signal, which indicates irregularities in the lipid membrane. Interestingly, these Nile Red foci colocalize with GFP-MreB and do not emerge in bacteria that lack MreB. MreB is a homolog of eukaryotic actin, and actin forms an intricate membrane associated network termed the cortical actin cytoskeleton15. The correlation between MreB and the lipid staining effects was intriguing since the cortical actin cytoskeleton is involved in the formation of lipid domains including lipid rafts and sphingolipid enriched domains15, 16. By applying different lipid staining techniques, and using a variety of mutant strains, we were able to show that the MreB cytoskeleton of B. subtilis is associated with fluid lipid domains, and, like the eukaryotic cortical actin cytoskeleton, is involved in the distribution of lipids and proteins. Furthermore, the active and directed movement of MreB appeared to stimulate the diffusion of proteins within the cell membrane. The consequences for membrane protein activity and cell wall synthesis are discussed.
RESULTS
Altered membrane stain upon MreB delocalization
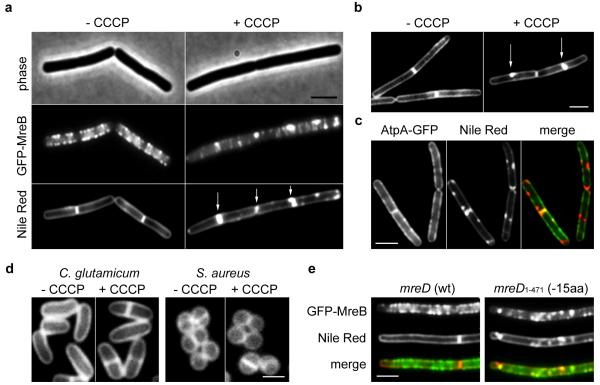
Dissipation of the membrane potential with CCCP results in delocalization of the cytoskeletal protein MreB in B. subtilis14. We noticed that such treatment also affects fluorescent membrane staining (Fig. 1a). In untreated cells, Nile Red stains cell membranes without a visible preference for certain membrane areas. A slightly stronger fluorescence signal is observed for division septa, which is caused by the presence of the two adjacent cell membranes from the newly formed daughter cells. However, in the presence of CCCP brightly fluorescent foci appear. These membrane staining effects are visible within 2 minutes after CCCP addition. CCCP is a proton carrier which abolishes the proton motive force (pmf)-driven synthesis of ATP, resulting in a rapid reduction of the cellular ATP pool14. An F1Fo ATP synthase deletion mutant is viable and generates ATP by pmf-independent means via glycolysis, and is therefore able to maintain stable ATP levels when CCCP is present for a much longer period of time14. Nevertheless, in an F1Fo ATP synthase deletion strain the addition of CCCP still results in a spotty Nile Red membrane stain, indicating that the effect is not related to changes in ATP levels (Fig. 1b). Addition of the potassium-ionophore valinomycin also gives rise to an irregular fluorescence membrane staining, whereas the proton-potassium antiporter nigericin has no effect (Supplementary Fig. 1a). Valinomycin and nigericin dissipate the membrane potential and proton gradient, respectively. Therefore, the membrane staining effects specifically occur when the membrane potential is disturbed. The presence of chloramphenicol did not prevent these effects, indicating that de novo protein synthesis is not required (Supplementary Fig. 1b). The fact that the fluorescent foci become visible within one or two minutes makes it unlikely that they are formed by local accumulation of newly synthetized membrane material. A detailed analysis of the Nile Red fluorescence spectra showed that the increased Nile Red fluorescence originates from a regular lipid bilayer environment, and is not caused by Nile Red that is bound to abnormal protein or lipid aggregates (Supplementary Fig. 2). In theory, a loss of cell turgor as a result of CCCP treatment could result in the invagination of lipid membranes through plasmolysis. However, a dissipation of the membrane potential with CCCP does not cause a rapid loss of cell turgor (Supplementary Fig. 3). Moreover, when the Nile Red membrane stain was compared with the fluorescent membrane signal of a GFP-labelled trans-membrane protein (F1Fo ATP synthase), the GFP signal remained unaffected by CCCP and did not correlate with the fluorescent Nile Red foci (Fig. 1c, Supplementary Fig. 4). For these reasons, we conclude that the increase in fluorescence signal is not explained by invagination of the cell membrane, or otherwise abnormal membrane shapes.
Figure 1. Disruption of the MreB cytoskeleton is accompanied by an aberrant membrane stain.
(a) Phase contrast images of B. subtilis cells (upper panel) expressing GFP-MreB (middle panel), and fluorescent Nile Red membrane stains (lower panel), are depicted in the absence or presence of the proton ionophore CCCP. Some of the Nile Red foci appearing with CCCP are highlighted with arrows (see Supplementary Fig. 5a for more examples). The CCCP-triggered local enrichment of MreB is not caused by the weak dimerization property of GFP, which was recently shown to stimulate protein clustering70 (Supplementary Fig. 5b), or by artificial overproduction (Supplementary Fig. 6). Strain used: B. subtilis YK405 (gfp-mreB). (b) Incubation of F1Fo ATP-synthase deficient cells with CCCP also results in a rapid (within 2 minutes) emergence of Nile Red foci, ruling out that the appearance of Nile Red foci is due to a reduction of ATP levels (see Supplementary Fig. 1c for more examples). Strain used: B. subtilis HS13 (Δatp). (c) Comparison of Nile Red foci, and the localization of the integral membrane protein F1Fo ATP synthase fused to GFP (AtpA-GFP). The lack of overlap in fluorescence signals rules out membrane invagination, or aberrantly formed septa (see arrows) as an explanation for the strong local Nile Red fluorescence (see Supplementary Fig. 4 for more examples). The reason that in some cases the Nile red stain appears to protrude into the cytoplasm is a consequence of high membrane fluorescence originating from slightly above and below the exact focal plane. Strain used: B. subtilis BS23 (atpA-gfp). (d) CCCP does not affect Nile Red fluorescent membrane stain in Staphylococcus aureus and Corynebacterium glutamicum which both lack MreB (see Supplementary Fig. 1d for more examples). Strains used: C. glutamicum RES167, and S. aureus RN4220. (e) Colocalization of GFP-MreB (upper panel) with Nile Red (middle panel) in B. subtilis strains encoding wild type MreD (wt), and a 15 amino acid C-terminal truncation of MreD (−15aa), which results in a mild delocalization of MreB in low Mg2+ medium (see Supplementary Fig. 1e for more examples). Strains used: B. subtilis HS35 (gfp-mreB mreD wt), and HS38 (gfp-mreB mreD1-471/-15aa). Scale bar, 2 μm.
Several experiments suggest a link between the CCCP-induced lipid staining effect and the MreB cytoskeleton. Firstly, the Nile Red-stained membrane foci colocalize with clusters of MreB (Fig. 1a, Supplementary Fig. 5). Importantly, these clusters are not caused by artificial overproduction of a GFP-fusion protein (Supplementary Fig. 6). Secondly, both the irregular Nile Red stain and the delocalization of MreB are triggered by valinomycin and not by nigericin14 (Supplementary Fig. 1a). Thirdly, when the effect of CCCP on Nile Red staining was tested in Staphylococcus aureus or Corynebacterium glutamicum, which do not contain MreB homologs, no change in the fluorescent membrane stain was observed (Fig. 1d). Finally, the localization of B. subtilis MreB depends on MreD17, and an MreD mutant that lacks the last 15 amino acids causes an irregular MreB pattern but also an uneven Nile Red stain (Fig. 1e). The latter experiment indicates that the membrane staining effect occurs upon delocalization of MreB, even when the membrane potential is not affected.
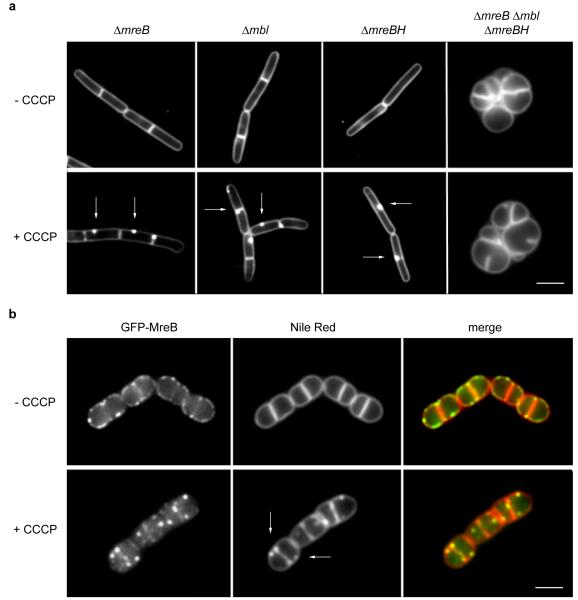
If the delocalization of MreB causes irregular fluorescent membrane signals, then the addition of CCCP should have no effect on the Nile Red membrane stain when MreB is absent. However, it appeared that CCCP still causes fluorescent Nile Red foci in the membrane of a B. subtilis mreB deletion mutant (Fig. 2a). B. subtilis encodes two other MreB homologs, Mbl and MreBH, which are known to be able to complement the activity of MreB7. Individual deletions of mbl or mreBH do not prevent the formation of CCCP-induced Nile Red foci either, but in the absence of all three genes (ΔmreB Δmbl ΔmreBH) the Nile Red membrane stain showed a smooth fluorescence signal in the presence of CCCP (Fig. 2a, see Supplementary Fig. 7 for the analysis of the foci frequency). Importantly, the absence of Nile Red foci in the triple mutant is not a consequence of the deformation of the cell shape by the inhibition of lateral cell wall synthesis, since inactivation of RodA, which is required for peptidoglycan synthesis in the lateral cell wall18, also results in round cells, yet these deformed cells still display Nile Red foci when CCCP is added (Fig. 2b). Taken together, these results support the suggestion that there is a correlation between MreB delocalization, and the irregularities in Nile Red lipid stain.
Figure 2. Distorted lipid staining requires the MreB cytoskeleton.
(a) CCCP-dependent fluorescent Nile Red foci occur in B. subtilis strains deficient for single MreB-homologues, but are absent in strains that lack all three MreB-homologues (ΔmreB, Δmbl, ΔmreBH). Some of the Nile Red foci appearing with CCCP are highlighted with arrows. See also Supplementary Fig. 7 for the analysis of the foci frequency. Strains used: B. subtilis 3728 (ΔmreB), 4261 (Δmbl), 4262 (ΔmreBH), and 4277 (ΔmreB, Δmbl, ΔmreBH). (b) Colocalization of GFP-MreB (left panels) with Nile Red (middle panels) in B. subtilis cells depleted for RodA. The MreB cytoskeleton is disrupted with CCCP. Some of the Nile Red foci appearing with CCCP are highlighted with arrows. Strain used: B. subtilis HS36 (gfp-mreB Pspac-rodA). Scale bar, 2 μm.
Involvement of specific lipids
Cardiolipin (CL), phosphatidyl-ethanolamine (PE) and phosphatidyl-glycerol (PG) are the major phospholipid species in bacteria. Both CL and PE have been reported to form domains in B. subtilis membranes19, 20. Nile Red is an uncharged fluorescent dye that partitions into lipid membranes based on its intrinsic hydrophobicity21, 22. It is not known whether Nile Red prefers certain lipid head groups but its fluorescence is sensitive to physical changes in its lipid environment23. To test whether an aberrant distribution of CL or PE is responsible for the irregular Nile Red stain, CL or PE deficient B. subtilis mutant strains were treated with CCCP. Despite the absence of CL or PE in these strains24, the fluorescent Nile Red foci still appear after depolarization of the membrane (Supplementary Fig. 8a).
Another commonly used membrane dye is FM 4-64, a styryl-dye which also partitions into lipid membrane surface based on intrinsic hydrophobicity25. Unlike Nile Red, it does carry a positive charge25 and a preference to bind negatively charged lipid species can be speculated. It has been shown that this dye exhibit a fluorescent stain in B. subtilis cells that under certain conditions weakly resembles a MreB-like helical patterns26, 27. This was interpreted as PG-enriched helical domains since PG is negatively charged, and these structures coalesce when PG is depleted26, 28. However, it turned out that this change in FM 4-64 staining pattern is caused by depolarization of the membrane potential upon PG-depletion. As in case of uncharged Nile Red, addition of CCCP readily induced FM 4-64 foci which colocalize with MreB (see Supplementary Figures 8b-e and 9 for details). Thus, we conclude that FM 4-64 is not a specific indicator for PG. The strong formation of Nile Red – foci upon depletion of PG makes it unlikely that these are specific clusters of PG. Together, these results suggest that the fluorescent membrane foci are not caused by domains enriched in lipids with specific head groups. A more probable explanation for the intense fluorescent Nile Red foci is the accumulation of lipids carrying specific fatty acid species.
Localization of fluid lipid domains
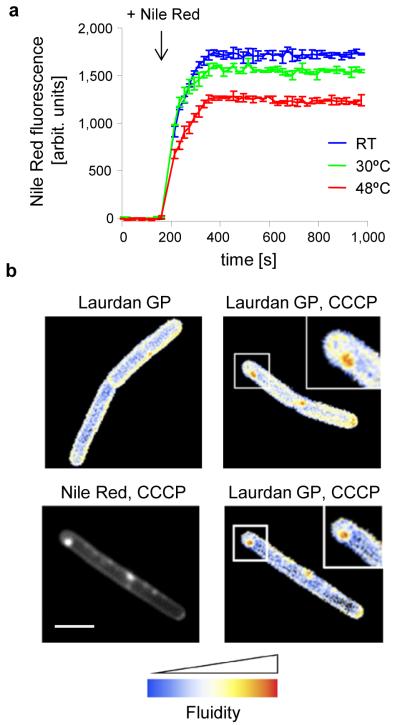
Bacterial membranes are composed of lipids that can form different conformations such as ‘gel phase’ or a more fluid ‘liquid-crystalline phase’. These distinct membrane configurations are predominantly determined by the fatty acid moiety of the lipids29, 30. The presence of unsaturated bonds or branched chains increases the space that fatty acids occupy within a bilayer31. As a result, lipids containing unsaturated or branched fatty acids assemble into a more disordered ‘liquid-crystalline phase’, characterised by a less dense packing of lipids. In contrast, lipids with saturated fatty acids favour the more compact and less fluid ‘gel-phase’. The fluidity of the membrane is known to influence the fluorescent staining of many lipid dyes, including that of Nile Red23. To test whether this is also the case in B. subtilis cells, we measured the fluorescence intensity of Nile Red using cells that differ in their membrane fluidity. In B. subtilis the fluid lipid content is greatly altered as a response to growth temperatures. In order to maintain a constant membrane fluidity at different temperatures, the fluid lipid content decreases with increasing growth temperature32. As a consequence, when cells grown at 48°C are cooled down to 30°C, their cell membrane will be less fluid compared to cells that are grown at 30°C. As shown in Fig. 3a, this results in a lower Nile Red fluorescence intensity. In contrast, if cells are grown at room temperature, and subsequently transferred to 30°C, the membrane fluidity is increased compared to cells grown at 30°C. Indeed, this leads to an increase in Nile Red fluorescence (Fig. 3a). It is therefore possible that the irregular membrane stain caused by MreB delocalization points towards local differences in membrane fluidity.
Figure 3. Analysis of cell membrane fluidity.
(a) Nile Red fluorescence intensity of B. subtilis cells grown in room temperature, 30°C or 48°C followed by shift to 30°C, was measured fluorometrically. To prevent rapid adaptation of membrane fluidity68, the measurements were carried out with a lipid desaturase (des) deficient strain. The curves represent average values with standard deviation of 3 replicate measurements. Strain used: B. subtilis HB5134 (Δdes). (b) Microscopic analysis of fluid membrane domains using Laurdan generalised polarisation. The images show a colour-coded Laurdan generalised polarisation (GP) intensity map in which red indicates regions of increased fluidity. Membrane fluidity is determined for untreated B. subtilis cells (left panel) and cells treated with CCCP (right panel). See also Supplementary Fig. 10 for the measurement of the average fluidity change. (c) Colocalization of Nile Red foci and Laurdan GP in CCCP-treated B. subtilis cells. Strain used: B. subtilis 168 (wild type). Scale bar, 2 μm.
To test whether the Nile Red foci are caused by the enrichment of fluid lipids, we adapted a staining technique used to visualize fluidity differences in eukaryotic membranes. The fluidity-sensitive dye Laurdan undergoes an emission wavelength shift upon changes in fluidity that can be detected microscopically33-36. As shown in Fig. 3b and Supplementary Fig. 10, incubation with CCCP generates membrane areas with a clear and statistically significant reduction in ‘Laurdan Generalised Polarisation’, which indicates an increase in local membrane fluidity. Importantly, these membrane domains colocalize with Nile Red foci (Fig. 3c).
MreB generates membrane regions with increased fluidity
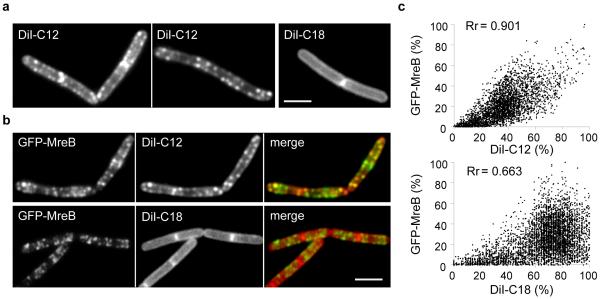
These experiments suggest a direct link between the MreB cytoskeleton and membrane Regions with Increased Fluidity, RIFs in short. So far, we have only observed RIFs when MreB coalesces into large clusters, and the question arises whether RIFs are present under conditions in which MreB is normally localized, rather that delocalized by CCCP or other means. Presumably, visualization of RIFs in normal uninhibited cells requires a lipid dye with a high affinity for membrane areas with increased fluidity. It has been shown that a lipid-mimicking dye DiI-C12 localizes with high specificity to fluid membrane areas as a consequence of its relative short acyl chains37, 38. When B. subtilis cells were stained with this dye, a distinct punctuated fluorescent pattern became visible that overlapped significantly with the GFP-MreB signal, with a Pearsons’ correlation coefficient of 0.9 (Fig. 4a-c, Supplementary Fig. 11a-b)37. As a control, we tested DiI-C18, a variant with longer acyl chains. Importantly, in this case a smooth membrane stain is observed (Fig. 4a-c, Supplementary Fig. 11a-b), indicating that the fluorescent pattern of DiI-C12 is based on differences in membrane fluidity, and not a consequence of favoured interactions between the DiI head group and certain lipids or proteins.
Figure 4. Localization of RIFs in untreated B. subtilis cells.
(a) RIFs can be visualised in normally growing untreated B. subtilis cells using the fluid membrane dye DiI-C12 (left panel). Staining with the long acyl-chain variant, DiI-C18, reveals a regular (smooth) membrane signal (right panel). Strain used: B. subtilis 168 (wild type). (b) Colocalization of DiI-C12 and GFP-MreB (see Supplementary Fig. 11 for more examples and statistical analysis of colocalization). Strain used: B. subtilis YK405 (gfp-mreB). (c) Fluorescence intensity correlation graphs are shown for the cells in panel b. The graphs display a pixel by pixel intensity correlation between DiI, and GFP-MreB fluorescence and shows the Pearson’s correlation coefficients (Rr) (see Supplementary Fig. 11 for details). It should be mentioned that some fluorescence correlation between DiI-C18 and GFP remains since both DiI-C18 and MreB are present in the cell membrane. Strain used: B. subtilis YK405 (gfp-mreB). Scale bar, 2 μm.
The ability to visualize RIFs with DiI-C12 without dissipation of membrane potential allowed us to test which of the MreBCD proteins are required for RIF formation. In the absence of all three MreB homologs (ΔmreB Δmbl ΔmreBH), staining with DiI-C12 produces a smooth fluorescent membrane signal and RIFs are no longer visible (Fig. 5a, Supplementary Fig. 11c). In contrast, mreC or mreD deletion strains that form comparably round cells still form RIFs (Fig. 5a, Supplementary Fig. 11c), indicating that the MreB-homologs rather than the integral membrane proteins MreC or MreD are essential for RIF formation. MreB can bind directly to lipid bilayers4, and when GFP-MreB is expressed in a ΔmreBCD strain, a strong colocalization of GFP-MreB clusters with DiI-C12 foci is observed (Fig. 5b). These results show that the formation and spatial organization of RIFs is a specific property of MreB.
Figure 5. DiI-C12 stained RIFs are MreB-dependent.
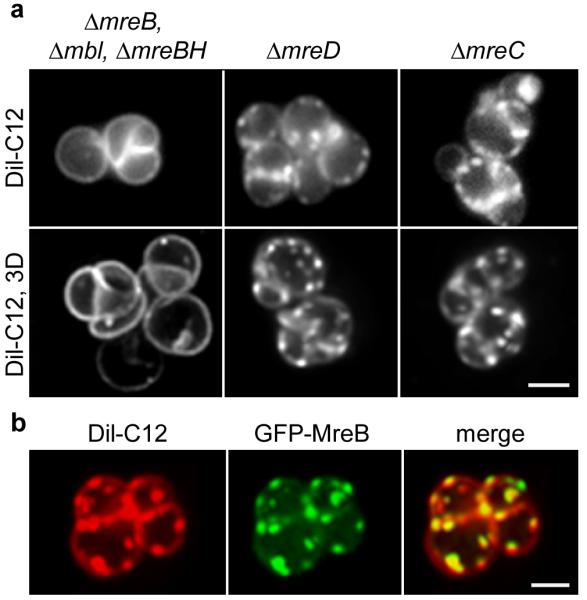
(a) DiI-C12 fluorescence of widefield (upper panel), and maximum intensity projection of a Z-stack (lower panel) of B. subtilis ΔmreC and ΔmreD cells, and cells that lack the three MreB-homologs (ΔmreB, Δmbl, ΔmreBH). See Supplementary Movie 1 for 3D-reconstruction, Supplementary Movie 2 for corresponding raw and deconvolved images, and Supplementary Fig. 11c for more examples. Strains used: B. subtilis 3481 (ΔmreC), 4311 (ΔmreD), 4277 (ΔmreB, Δmbl, ΔmreBH, ΔrsgI). (b) Colocalization of DiI-C12 and GFP-MreB in the absence of MreBCD. The maximum intensity projections of Z-stacks are depicted. See Supplementary Movie 3 for 3D-reconstruction and Supplementary Movie 4 for corresponding raw and deconvolved images. Strain used: B. subtilis HS35 (gfp-mreB ΔmreBCD). Scale bar, 2 μm.
Absence of MreB changes membrane properties
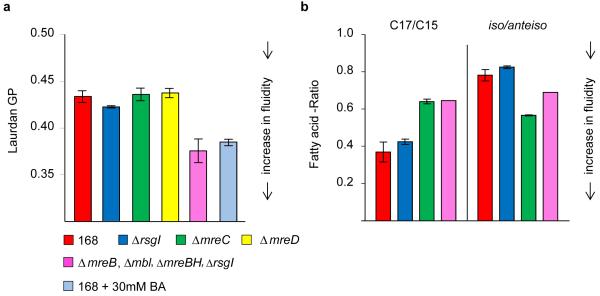
Since MreB creates local membrane areas with increased fluidity, it might be that this activity affects the general fluidity of the cell membrane. Indeed, when the overall membrane fluidity was measured using Laurdan generalised polarisation (GP), it turned out that the general membrane fluidity is significantly elevated when MreB homologs are absent (Fig. 6a). This increase in fluidity is comparable to the addition of 30 mM of the membrane fluidizer benzyl alcohol39 (Fig. 6a). It should be added that growth in the absence of all three MreB-homologs is only possible when the anti-sigma factor rsgI is deleted40, however, an ΔrsgI mutation by itself does not result in altered membrane fluidity (Fig. 6a). ΔmreC or ΔmreD strains show identical changes in cell shape and inhibition of lateral cell wall synthesis as the triple mreB mbl mreBH deletion mutant but still contain RIF clusters. Importantly, the overall membrane fluidity of these strains is comparable to wild type cells.
Figure 6. Membrane fluidity increases in the absence of MreB.
(a) Overall membrane fluidity of several cytoskeletal mutant strains was measured as Laurdan generalised polarisation (GP) of logarithmically growing B. subtilis cells. As a positive control, an increase in fluidity (= decrease in Laurdan GP) was established by adding the membrane fluidizer benzyl alcohol (BA; 30 mM) to wild type cells. The diagram depicts the average values and standard deviation of 3 independent measurements. (b) Ratios between chain lengths of the major fatty acids (C17 and C15), and ratios between the iso and anteiso forms of fatty acids are depicted for the different strains (see supplementary table 1 for the detailed composition). High C17/C15 or high iso/anteiso -ratios indicate a reduced fluidity. The diagram depicts the average values and standard deviation of 2 independent analyses. Strains used: B. subtilis 168 (wild type), 4264 (ΔrsgI), 3481 (ΔmreC), 4277 (ΔmreB, Δmbl, ΔmreBH, ΔrsgI).
The Laurdan experiments suggest that RIFs formed by MreB affect the overall fluidity. However, it is possible that the absence of MreB influences the actual lipid composition. To investigate this, the fatty acid compositions of the different strains were analysed using gas chromatography (Fig. 6b, Supplementary Table 1). Both the ΔmreC mutant and mreB triple mutant (ΔmreB/mbl/mreBH) increase their overall fatty acid chain length (less C15 and more C17 species), which normally lowers the membrane fluidity (Fig. 6b, Supplementary Table 1). The ΔmreC, and to a lesser extent the mreB triple mutant, also increases the fraction of ‘anteiso’ fatty acids compared to ‘iso’ fatty acid species. Anteiso fatty acids species carry a methyl group attached to the third last carbon of the chain and are significantly more fluid than iso species, which carry a methyl group attached to the second last carbon41. In case of ΔmreC this increase of anteiso fatty acids seems to compensate for the effect of increased fatty acid chain length since the overall membrane fluidity of this ΔmreC mutant is the same as for wild type (Fig. 6a). Importantly, this increase of anteiso fatty acids is lower in the mreB triple mutant compared to the ΔmreC mutant (Fig. 6b, Supplementary Table 1) and yet the overall fluidity of the MreB triple mutant is significantly higher (Fig. 6a). The strong increase of membrane fluidity of the mreB triple mutant can therefore not be explained by an altered fatty acid composition. This suggests that the MreB protein complex concentrates fluid lipids into RIFs, which results in lower overall membrane fluidity. These experiments provide further evidence for the specific role of MreB-homologs in maintaining a proper lipid homeostasis in the cell membrane.
MreB is important for membrane protein distribution
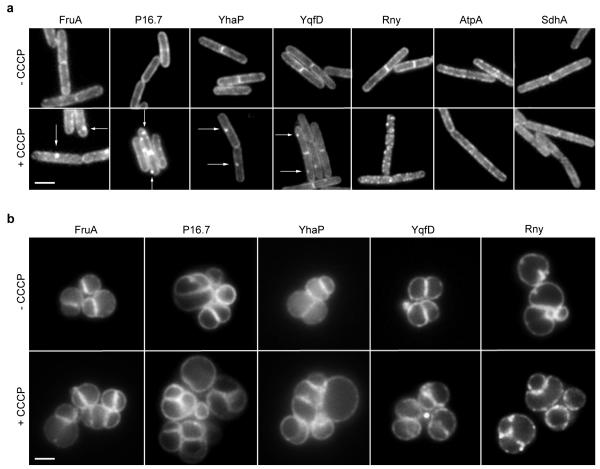
The cortical actin cytoskeleton is involved in segregation of the eukaryotic cell membrane into domains that differ in fluidity, including the formation of lipid rafts15, 16, 42-45. This activity of actin influences the localization and diffusion of many membrane proteins46-50. Since MreB is an actin homolog and creates distinct membrane domains, the question arises whether the MreB cytoskeleton also influences the distribution of membrane proteins. To test this, the MreB cytoskeleton was disrupted with CCCP, and the localization of several integral membrane proteins was analysed. Indeed, incubation with CCCP induces clustering of several membrane proteins including Fructose permease (FruA), P16.7, YhaP and YqfD, and RNase Y (Rny) (Fig. 7a). Other proteins, such as Succinate dehydrogenase (SdhA) and F1Fo ATP synthase (AtpA), showed no apparent difference in localization pattern (Fig. 7a). The proteins FruA, YhaP, P16.7 and YqfD show a clear clustering with Nile Red foci in CCCP-treated cells (Supplementary Fig. 12). To test whether this clustering depends on MreB, and is not caused by CCCP itself, we analysed the localization of these proteins in the mreB triple mutant strain. Indeed, in this strain background, the localization of these proteins was unaffected by CCCP (Fig. 7b). Thus, the MreB-induced fluid lipid domains can influence the distribution of membrane proteins. It should be added that the localization pattern of RNase Y is still influenced by dissipation of membrane potential in MreB triple mutant (Fig. 7b), suggesting that in this case the effect is MreB independent.
Figure 7. Aberrant localization of different membrane proteins.
(a) The localization of several membrane proteins (FruA, P16.7, YhaP, YqfD, and Rny) is changed after the MreB cytoskeleton was disturbed by addition of CCCP. Some of the foci appearing with CCCP are highlighted with arrows. The localization of other proteins (AtpA and SdhA) remains unaffected. See Supplementary Fig. 12 for more examples, and colocalization with Nile Red. Strains used: B. subtilis FruA-GFP, 110WA (p16.7-gfp), yhaP-gfp, yqfD-gfp, 3569 (rny-gfp), BS23 (atpA-gfp), BS112 (sdhA-gfp). (b) The CCCP-induced clustering of membrane proteins FruA, P16.7, YhaP and YqfD is absent in strains deficient for MreB-homologs. Strains used B. subtilis HS43 (rny-gfp, ΔmreB*), HS44 (fruA-gfp, ΔmreB*), HS45 (P16.7-gfp, ΔmreB*), HS46 (yhaP-gfp, ΔmreB*), HS47 (yqfD-gfp, ΔmreB*). mreB* designates (ΔmreB, Δmbl, ΔmreBH, ΔrsgI). Scale bar, 2 μm.
MreB stimulates membrane protein diffusion
The MreB-complexes of B. subtilis move rapidly along the cell membrane, driven by the cell wall synthesis machinery. This movement can be slowed down by inhibiting peptidoglycan synthesis with vancomycin9, 10. Since MreB affects the localization of lipids and membrane proteins, it is possible that the active movement of MreB also influences the diffusion dynamics of membrane proteins. To test this, the diffusion of several membrane proteins was followed using high-speed total internal reflection (TIRF) microscopy. As shown in the kymographs of Fig. 8a, the diffusion dynamics of FruA and F1Fo ATP synthase changed significantly when the movement of MreB was stopped with vancomycin. The same behaviour was observed for two other tested transmembrane proteins (Supplementary Fig. 13a, see also Supplementary Fig. 13b for quantification based on fluorescence intensity fluctuation).
Figure 8. MreB-movement influences membrane protein diffusion.
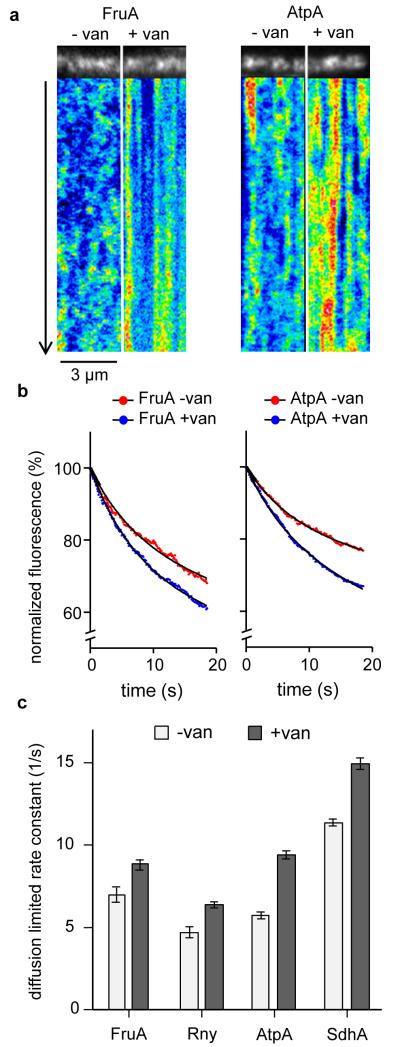
(a) Kymographs visualizing the diffusion of Fructose permease (FruA) and F1Fo ATP synthase (AtpA) in the absence and presence of vancomycin (van), which blocks MreB movement. Time lapse series with 20 s length and 100 ms time resolution were acquired using TIRF microscopy. See Supplementary Movie 5 for corresponding raw image series. (b) In TIRF microscopy, only proteins within the vicinity of the evanescent light (<200 nm distance from the coverslip surface) are subject to fluorophore (GFP) bleaching. The graphs depict the averaged, normalised, and background-subtracted fluorescent signals of Fructose permease (FruA) and F1Fo ATP synthase (AtpA) in the presence (− van) and absence (+ van) of MreB movement. A significant increase in bleaching is observed when movement of MreB is inhibited with vancomycin, which indicates a reduced diffusion of proteins in and out of the range of the evanescent light. The curve fits were performed using a two phase exponential decay model composed of a rapid bleaching of GFP within the range of the evanescent wave, and slower (diffusion limited) bleaching of the whole cell fluorescence (see Supplementary Fig. 14 for details). (c) Rate constants and standard errors of slow (diffusion limited) bleaching kinetics for Fructose permease (FruA), RNaseY (Rny), F1Fo ATP synthase (AtpA), and Succinate dehydrogenase (SdhA) in the absence and presence of vancomycin. Increased rate constant indicates a reduced exchange of protein between the TIRF-illuminated area, and rest of the cell surface. The number of analysed cells, goodness-of-fit, and standard errors are provided in supplementary figure 14. Strains used: B. subtilis BS23 (atpA-gfp), BS112 (sdhA-gfp), FruA-GFP, and 3569 (rny-gfp).
In TIRF microscopy, a major determinant is the diffusion of GFP in and out of the range of the evanescent light. The reduction in membrane protein diffusion should thus increase photo bleaching. Indeed, for all tested proteins a significant increase in diffusion-limited GFP bleaching was observed in the presence of vancomycin (Fig. 8b-c, Supplementary Fig. 14). Thus, the active movement of the MreB cytoskeleton stimulates diffusion of membrane proteins.
DISCUSSION
Despite their small size, the cell membrane of bacteria exhibits a remarkably complex organization. There are lipid species that accumulate at cell division sites and cell poles, and most membrane proteins are not uniformly distributed over the bacterial cell membrane30, 51, 52. In addition, it has been reported that B. subtilis can form low fluidity “lipid rafts”53-55. The capacity of the MreB cytoskeleton to induce fluid lipid domains (RIFs) that determine the distribution of membrane proteins adds a new dimension to this complexity.
The mechanism through which MreB organizes fluid lipids remains hypothetical. Peripheral membrane proteins have a capacity to induce lipid domains56, 57. This activity is related to the binding of lipid bilayers by means of an intercalating amphipathic helix, but these domains are absent in MreB-homologs from Gram-positive bacteria4. However, it has also been postulated that peripheral membrane proteins can directly induce fluid lipid domains by reducing the ordering effect of electrostatic attraction and repulsion between charged lipid head groups58. Although this mechanism is in good agreement with the electrostatic membrane binding mode of MreB4, we cannot rule out the involvement of other MreB-interacting proteins in the formation of RIFs.
Because of the various cellular functions of the cytoplasmic membrane, an organization into membrane areas that differ in their physical properties has a clear advantage. The optimal fluid state of the membrane is a compromise between different factors. On the one hand, high membrane fluidity promotes protein activity by reducing viscosity, and thus stimulating catalytic activity and diffusion of integral membrane proteins59. A higher membrane elasticity of fluid membranes also allows the lipid bilayer to optimally embed membrane proteins60, 61. On the other hand, high membrane fluidity has the disadvantage that it increases proton permeability62, 63, resulting in a less efficient energy and ion homeostasis. The MreB-dependent formation of fluid lipid domains (RIFs) could therefore enable the cell to maintain an optimal condition for membrane protein activity in limited areas of increased fluidity without compromising the proton permeability of the whole membrane.
In a recent study, it was shown that the random motion of chromosomes in bacterial and yeast cells is faster than can be explained by simple Brownian motion. This increased molecular diffusion rate is caused by ATP-dependent processes such as RNA synthesis64. This bears some similarity with our observation that the diffusion of membrane proteins is also stimulated by an active process, in this case peptidoglycan synthesis, and the subsequent movement of RIFs. It is tempting to speculate that the continuous mixing of the cell membrane by active RIF movement promotes diffusion of proteins in a viscous membrane environment, thereby stimulating protein-protein and protein-substrate interactions.
Inactivation of MreB prevents lateral cell wall synthesis resulting in deformed round cells. This effect can be explained by the inability to recruit the necessary proteins to the proper site of synthesis. However, the fluid membrane environment created by MreB might also be required for optimal cell wall synthesis since Lipid II, the membrane-anchored precursor for peptidoglycan synthesis, has a strong preference for fluid membranes65. Possibly, RIFs are enriched with this essential precursor. In fact, the delocalization of MreB with CCCP does coincide with a loss of lateral Lipid II localization, based on staining with fluorescently-labelled vancomycin, whereas staining of the cell division septum remains unaffected (Supplementary Fig. 15).
With its ability to organize lipids and proteins in the bacterial cell membrane, MreB bears a functional resemblance to the eukaryotic cortical actin cytoskeleton15, 42, 44, 45. There are, however, differences. The actin cytoskeleton forms a relatively static meshwork at the membrane periphery66, yet the bacterial counterpart is more fragmented9-11. In eukaryotic cells the membrane anchored actin creates fences that restrict diffusion of proteins66, whereas the active movement of the MreB cytoskeleton stimulates diffusion. Interestingly, it was recently shown that short and highly dynamic actin polymers, which influence clustering of membrane proteins, also exist in eukaryotic cells48. Thus, despite some differences, the ability of MreB and actin to induce and organize membrane domains appears comparable, and may explain why actin and MreB are evolutionarily linked.
MATERIALS AND METHODS
The growth conditions and construction of strains are described in the supplementary materials and methods. The used strains and conditions for gene induction are listed in Supplementary Table 2.
General fluorescence microscopy
For fluorescence microscopy, cells were grown to exponential growth phase at 30 °C if not stated otherwise. The cells were immobilized on microscope slides covered with a thin film of 1.2 % agarose in water. For the dissipation of membrane potential with CCCP, the mounting medium was supplemented with 0.5 % DMSO or 100 μM CCCP dissolved in DMSO (0.5 % final concentration of DMSO). Imaging was carried out within 2 min after addition of ionophores. Membranes were visualised with Nile Red or FM 4-64 (0.5 μg ml−1). Standard fluorescence microscopy was carried out using Zeiss Axiovert 200M (Zeiss Plan-Neofluar 100×/1.30 Oil Ph3 objective), Nikon Eclipse Ti (Nikon Plan Fluor 100×/1.30 Oil Ph3 DLL objective), and Applied Precision DeltaVision RT (Zeiss Plan-Neofluar 100×/1.30 Oil Ph3) microscopes. The images were acquired with Metamorph 6 (Molecular Devices, Inc), softWoRx Suite (Applied Precision), and further analysed using ImageJ v.1.38 (National Institutes of Health). Deconvolution was carried from optical sections using Huygens Essentials v.3.3 (Scientific Volume Imaging).
Laurdan Generalised Polarisation (GP) microscopy
For the microscopic analysis of membrane fluidity, B. subtilis cells grown in LB were incubated for 5 min with 100 μM Laurdan, washed and then resuspended in fresh pre-warmed LB. No inhibitory effect on growth was detected with 100 μM Laurdan. Laurdan was exited at 360 ± 20 nm and fluorescence emission was captured at 528 ± 19 nm (exposure time: 500 ms) followed by a second image at 457 ± 25 nm (exposure time: 500 ms). Image analysis and generation of a colour coded generalised polarisation (GP) map was performed with Wolfram Mathematica 7 (Wolfram Research). Briefly, the background fluorescence was subtracted and GP values were calculated for each pixel, based on intensities values of the separate images, as GP = (I457 − I528)/(I457 + I528), whereby I457 represents the emission intensity at 457 ± 25 nm and I528 emission intensity at 528 ± 19 nm. To remove signals not associated with cells, pixel pairs were regarded as insignificant when one of the channel intensities were below a threshold of 10 % of the maximal values found in the membrane. Insignificant pixel pairs were assigned a GP of zero. Finally, GP pixel values were represented as a linear colour coded GP map whereby blue represents high GP values (low fluidity) and red represents low GP values (high fluidity). Insignificant pixels (GP = 0) are shown in black. The full Wolfram Mathematica-script can be obtained from the authors upon request. The ability of the microscope-based assay to detect changes in membrane fluidity was verified by measuring changes in membrane fluidity upon cold shock or incubation with 30 mM benzyl alcohol67 (Supplementary Fig. 16a). For the measurement of mean GP values, the membrane fluorescence signal intensities at both emission channels were extracted as maximum values from a fluorescence intensity line scan perpendicular to the membrane plane. The values were measured for 70 individual cells, background subtracted, and the Laurdan GP values were calculated as described above. To prevent fast adaptation of membrane fluidity, a B. subtilis strain deficient for lipid desaturase (Δdes) was used in this case68. Des itself has no influence on the appearance of RIFs upon incubation with CCCP (Supplementary Fig. 16b).
Laurdan Generalised Polarisation (GP) spectroscopy
For the measurement of membrane fluidity in batch cultures, cells were grown in LB supplemented with 0.1 % glucose to an OD600 ~0.5, followed by 5 min incubation with 10 μM Laurdan. Subsequently, cells were washed three times with pre-warmed buffer containing 50 mM Na2HPO4/NaH2PO4 pH 7.4, 0.1 % glucose, and 150 mM NaCl with and without the membrane fluidizer benzyl alcohol (30 mM). In case of the different Mre-mutant strains, the media and buffers were supplemented with 20 mM MgCl2. The Laurdan fluorescence intensities were measured at 435 ± 5 nm and 490 ± 5 nm upon excitation at 350 ± 10 nm, using a Tecan Infinite 200M fluorometer. The Laurdan GP was calculated using the formula GP = (I435 − I490)/(I435 + I490). In both microscopic and spectroscopic measurements, the Laurdan GP assay was able to reliably detect general changes in membrane fluidity, although, due to different excitation and emission wavelengths used, the absolute values obtained with microscopy slightly differed from the normal batch fluidity measurements (Supplementary Fig. 16a). The fluorescence emission spectrum of Laurdan was recorded using 10 μM Laurdan embedded in liposomes upon excitation with 350 nm light using Tecan Infinite 200M fluorometer. The liposomes were formed from E. coli polar lipids using detergent dialysis method. In brief, the lipid extract was solved in chloroform followed by evaporation under dry argon stream. The lipids were resolubilized in 25 mM Tris/HCl pH 7.4 and 1.5% octylglycoside (25 mg ml−1 lipids), followed by dialysis against 25 mM Tris/HCl pH 7.4. At last, the formed liposomes were diluted to 1:5 in H20 and sized by extrusion through a 0.1-μm membrane.
DiI-C12 staining of fluid lipid domains
For staining of B. subtilis cells with DiI-C12, an overnight culture was diluted 1:100 in medium supplemented with 5 μg ml−1 DiI-C12 and 1% DMSO followed by growth for 2.5 h at 30 °C. Due to the high fluorescence of unbound dye, the stained cells were washed three times in pre-warmed medium supplemented with 1% DMSO, followed by microscopy. The staining of B. subtilis with DiI-C18, which is insoluble in water, was carried out by suspending small amounts of solid DiI-C18 crystals in culture medium followed by 2.5 h incubation at 30 °C, based on a method described in Kim et al. (2007)69. For microscopy, the crystals were removed by a brief low speed centrifugation. No growth inhibitory effect of the Dil-dyes was observed (Supplementary Fig. 17). For colocalization studies, the concentration of DiI-C12 was reduced to 2.5 μg ml−1. The reason for this was the extensive overlap of the excitation spectra between DiI-C12 and GFP, which results in weak GFP-fluorescence. The emission spectra of GFP and Dil-C12 are well separated, and no significant bleach-through of the emitted fluorescence was detected for DiI-C12 in the GFP-channel, and vice versa.
TIRF time lapse microscopy
The analysis of membrane protein diffusion was carried out using a Nikon Eclipse Ti equipped with, Nikon CFI APO TIRF 100×/1.49 Oil objective, TIRF illumination module, Andor Xion X3 EMCCD-camera, and 488 nm solid state laser. The cells were imaged for 20 sec with 100 ms time resolution in a continuous illumination mode. The image analysis including the generation of kymographs was carried out using ImageJ v.1.38 (National Institutes of Health) and Multiple Kymograph -plugin (J. Rietdorf and A. Seitz).
TIR-CP analysis
For the analysis of GFP bleaching kinetics upon TIRF illumination, the average fluorescence intensity of cells was measured using an automated detection of cell area based on above background fluorescence intensity (ImageJ v.1.38, National Institutes of Health). In brief, the cell detection was performed using the first image frame, and subsequently used to measure the average fluorescence intensity for each image of the time series. The background fluorescence signals were extracted from the same images as the average intensity of the remaining image area not associated with cells. The background subtracted and normalised intensity values were used to fit the GFP photo bleach kinetics as mono- or biexponential decays using GraphPad Prism 5 (GraphPad Software Inc.). The fixation of GFP, applied to inhibit diffusion was performed by incubation of the cell samples with paraformaldehyde for 5 min at RT, followed by saturation of formaldehyde crosslinking by the addition of 100 mM Glycine.
Spectral imaging fluorescence microscopy
The analysis of the emission wavelength spectrum for Nile Red-stained B. subtilis cells was carried out with Nikon A1R point scanning confocal microscope using Nikon Plan Apo VC 60× NA 1.4 oil objective, and 561 nm excitation laser. The emission spectrum was recorded with a 4.8 nm window size for the total range of 570-749 nm. Image analysis was carried out with NIS-Elements 4.0 (Nikon).
Membrane fluidity dependent Nile Red fluorescence intensity
Nile Red exhibits low fluorescence in a polar environment such as LB, but emits strongly when incorporated into hydrophobic cell membranes. This property was used to measure the intensity of Nile Red membrane fluorescence in B. subtilis cells grown in LB at different growth temperatures. For this, growing cultures were diluted to an optical density of OD600 = 0.5. Subsequently, the temperature of the cultures was rapidly adjusted to 30 °C, and the fluorescence increase was measured after addition of 0.5 μg ml−1 Nile Red. The measurements were carried out at 30°C using BMG Fluostar Optima fluorometer, and 584 nm excitation, and 620 nm emission filters. Background fluorescence was subtracted using signals measured from Nile Red in LB. In order to prevent rapid adaptation of membrane fluidity68, the measurements were carried out with a lipid desaturase deficient strain (B. subtilis Δdes).
Analysis of fatty acid composition
The fatty acid composition of B. subtilis wild type cells and the cytoskeletal mutants was determined from cells grown at 30 °C in LB supplemented with 20 mM MgCl2 that were harvested when the cultures reached an OD600 of approximately 0.5. Fatty acids were analysed as fatty acid methyl esters (FAME) using gas chromatography. All analyses were carried out in duplicates by the Identification Service of the DSMZ, Braunschweig, Germany.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Richard Daniel for strains and discussion, Alex Laude for help with spectral imaging microscopy, Darren Wilkinson and Daniel Klose for help with TIR-CP analysis, and Kursad Turgay, Stephan Gruber, Jeremy Lakey, and Tanneke den Blaauwen for critical reading of the manuscript. Funding for this research was provided by a Biotechnology and Biological Sciences Research Council (BBSRC) grant BB/I01327X/1 for L.W.H., and by the Wellcome Trust ‘Value in People’-program for H.S.
Footnotes
CONFLICT OF INTEREST
The authors declare that they have no competing financial interests.
REFERENCES
- 1.Graumann PL. Cytoskeletal elements in bacteria. Annu Rev Microbiol. 2007;61:589–618. doi: 10.1146/annurev.micro.61.080706.093236. [DOI] [PubMed] [Google Scholar]
- 2.Margolin W. Sculpting the bacterial cell. Curr Biol. 2009;19:812–822. doi: 10.1016/j.cub.2009.06.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Eraso JM, Margolin W. Bacterial cell wall: thinking globally, actin locally. Curr Biol. 2011;21:628–630. doi: 10.1016/j.cub.2011.06.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Salje J, van den Ent F, de Boer P, Lowe J. Direct membrane binding by bacterial actin MreB. Mol Cell. 2011;43:478–487. doi: 10.1016/j.molcel.2011.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mohammadi T, et al. The essential peptidoglycan glycosyltransferase MurG forms a complex with proteins involved in lateral envelope growth as well as with proteins involved in cell division in Escherichia coli. Mol Microbiol. 2007;65:1106–1121. doi: 10.1111/j.1365-2958.2007.05851.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang S, Arellano-Santoyo H, Combs PA, Shaevitz JW. Actin-like cytoskeleton filaments contribute to cell mechanics in bacteria. Proc Natl Acad Sci U S A. 2010;107:9182–9185. doi: 10.1073/pnas.0911517107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Formstone A, Errington J. A magnesium-dependent mreB null mutant: implications for the role of mreB in Bacillus subtilis. Mol Microbiol. 2005;55:1646–1657. doi: 10.1111/j.1365-2958.2005.04506.x. [DOI] [PubMed] [Google Scholar]
- 8.Leaver M, Errington J. Roles for MreC and MreD proteins in helical growth of the cylindrical cell wall in Bacillus subtilis. Mol Microbiol. 2005;57:1196–1209. doi: 10.1111/j.1365-2958.2005.04736.x. [DOI] [PubMed] [Google Scholar]
- 9.Dominguez-Escobar J, et al. Processive movement of MreB-associated cell wall biosynthetic complexes in bacteria. Science. 2011;333:225–228. doi: 10.1126/science.1203466. [DOI] [PubMed] [Google Scholar]
- 10.Garner EC, et al. Coupled, circumferential motions of the cell wall synthesis machinery and MreB filaments in B. subtilis. Science. 2011;333:222–225. doi: 10.1126/science.1203285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.van Teeffelen S, et al. The bacterial actin MreB rotates, and rotation depends on cell-wall assembly. Proc Natl Acad Sci U S A. 2011;108:15822–15827. doi: 10.1073/pnas.1108999108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shaevitz JW, Gitai Z. The structure and function of bacterial actin homologs. Cold Spring Harb Perspect Biol. 2010;2:a000364. doi: 10.1101/cshperspect.a000364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Carballido-Lopez R. The bacterial actin-like cytoskeleton. Microbiol Mol Biol Rev. 2006;70:888–909. doi: 10.1128/MMBR.00014-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Strahl H, Hamoen LW. Membrane potential is important for bacterial cell division. Proc Natl Acad Sci U S A. 2010;107:12281–12286. doi: 10.1073/pnas.1005485107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chichili GR, Rodgers W. Cytoskeleton-membrane interactions in membrane raft structure. Cell Mol Life Sci. 2009;66:2319–2328. doi: 10.1007/s00018-009-0022-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Frisz JF, et al. Direct chemical evidence for sphingolipid domains in the plasma membranes of fibroblasts. Proc Natl Acad Sci U S A. 2013;110:E613–622. doi: 10.1073/pnas.1216585110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Defeu Soufo HJ, Graumann PL. Bacillus subtilis actin-like protein MreB influences the positioning of the replication machinery and requires membrane proteins MreC/D and other actin-like proteins for proper localization. BMC Cell Biol. 2005;6:10. doi: 10.1186/1471-2121-6-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.de Pedro MA, Donachie WD, Holtje JV, Schwarz H. Constitutive septal murein synthesis in Escherichia coli with impaired activity of the morphogenetic proteins RodA and penicillin-binding protein 2. J Bacteriol. 2001;183:4115–4126. doi: 10.1128/JB.183.14.4115-4126.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mileykovskaya E, Dowhan W. Cardiolipin membrane domains in prokaryotes and eukaryotes. Biochim Biophys Acta. 2009;1788:2084–2091. doi: 10.1016/j.bbamem.2009.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Matsumoto K, Kusaka J, Nishibori A, Hara H. Lipid domains in bacterial membranes. Mol Microbiol. 2006;61:1110–1117. doi: 10.1111/j.1365-2958.2006.05317.x. [DOI] [PubMed] [Google Scholar]
- 21.Mukherjee S, Raghuraman H, Chattopadhyay A. Membrane localization and dynamics of Nile Red: effect of cholesterol. Biochim Biophys Acta. 2007;1768:59–66. doi: 10.1016/j.bbamem.2006.07.010. [DOI] [PubMed] [Google Scholar]
- 22.Greenspan P, Fowler SD. Spectrofluorometric studies of the lipid probe, nile red. J Lipid Res. 1985;26:781–789. [PubMed] [Google Scholar]
- 23.Kucherak OA, et al. Switchable nile red-based probe for cholesterol and lipid order at the outer leaflet of biomembranes. J Am Chem Soc. 2010;132:4907–4916. doi: 10.1021/ja100351w. [DOI] [PubMed] [Google Scholar]
- 24.Salzberg LI, Helmann JD. Phenotypic and transcriptomic characterization of Bacillus subtilis mutants with grossly altered membrane composition. J Bacteriol. 2008;190:7797–7807. doi: 10.1128/JB.00720-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wu Y, Yeh FL, Mao F, Chapman ER. Biophysical characterization of styryl dye-membrane interactions. Biophys J. 2009;97:101–109. doi: 10.1016/j.bpj.2009.04.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Barak I, Muchova K, Wilkinson AJ, O’Toole PJ, Pavlendova N. Lipid spirals in Bacillus subtilis and their role in cell division. Mol Microbiol. 2008;68:1315–1327. doi: 10.1111/j.1365-2958.2008.06236.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Muchova K, Wilkinson AJ, Barak I. Changes of lipid domains in Bacillus subtilis cells with disrupted cell wall peptidoglycan. Fems Microbiol Lett. 2011;325:92–98. doi: 10.1111/j.1574-6968.2011.02417.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hashimoto M, et al. Induction of extracytoplasmic function sigma factors in Bacillus subtilis cells with membranes of reduced phosphatidylglycerol content. Genes Genet Syst. 2009;84:191–198. doi: 10.1266/ggs.84.191. [DOI] [PubMed] [Google Scholar]
- 29.Zhang YM, Rock CO. Membrane lipid homeostasis in bacteria. Nat Rev Microbiol. 2008;6:222–233. doi: 10.1038/nrmicro1839. [DOI] [PubMed] [Google Scholar]
- 30.Vanounou S, Pines D, Pines E, Parola AH, Fishov I. Coexistence of domains with distinct order and polarity in fluid bacterial membranes. Photochem Photobiol. 2002;76:1–11. doi: 10.1562/0031-8655(2002)076<0001:codwdo>2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 31.Hazel JR, Williams EE. The role of alterations in membrane lipid composition in enabling physiological adaptation of organisms to their physical environment. Prog Lipid Res. 1990;29:167–227. doi: 10.1016/0163-7827(90)90002-3. [DOI] [PubMed] [Google Scholar]
- 32.van de Vossenberg JLCM, Driessen AJM, da Costa MS, Konings WN. Homeostasis of the membrane proton permeability in Bacillus subtilis grown at different temperatures. Biochim Biophys Acta. 1999;1419:97–104. doi: 10.1016/s0005-2736(99)00063-2. [DOI] [PubMed] [Google Scholar]
- 33.Parasassi T, De Stasio G, d’Ubaldo A, Gratton E. Phase fluctuation in phospholipid membranes revealed by Laurdan fluorescence. Biophys J. 1990;57:1179–1186. doi: 10.1016/S0006-3495(90)82637-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tyler KM, Wheeler G. Widefield microscopy for live imaging of lipid domains and membrane dynamics. Biochim Biophys Acta. 2011;1808:634–641. doi: 10.1016/j.bbamem.2010.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mercier R, Domínguez-Cuevas P, Errington J. Crucial role for membrane tluidity in proliferation of primitive cells. Cell Rep. 2012;1:417–23. doi: 10.1016/j.celrep.2012.03.008. [DOI] [PubMed] [Google Scholar]
- 36.Bach JN, Bramkamp M. Flotillins functionally organize the bacterial membrane. Mol Microbiol. 2013;88:1205–1217. doi: 10.1111/mmi.12252. [DOI] [PubMed] [Google Scholar]
- 37.Baumgart T, Hunt G, Farkas ER, Webb WW, Feigenson GW. Fluorescence probe partitioning between Lo/Ld phases in lipid membranes. Biochim Biophys Acta. 2007;1768:2182–2194. doi: 10.1016/j.bbamem.2007.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhao J, Wu J, Veatch Sarah L. Adhesion stabilizes robust lipid heterogeneity in supercritical membranes at physiological temperature. Biophys J. 2013;104:825–834. doi: 10.1016/j.bpj.2012.12.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Konopásek I, Strzalka K, Svobodová J. Cold shock in Bacillus subtilis: different effects of benzyl alcohol and ethanol on the membrane organisation and cell adaptation. Biochim Biophys Acta. 2000;1464:18–26. doi: 10.1016/s0005-2736(99)00240-0. [DOI] [PubMed] [Google Scholar]
- 40.Schirner K, Errington J. The cell wall regulator σI specifically suppresses the lethal phenotype of mbl mutants in Bacillus subtilis. J Bacteriol. 2009;191:1404–1413. doi: 10.1128/JB.01497-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.de Mendoza D, Schujman GS, Aguilar PS. Biosynthesis and function of membrane lipids. In: Sonenshein AL, Hoch JA, Losick R, editors. Bacillus subtilis and its relatives: from genes to cells. ASM Press; Washington, D.C: 2001. pp. 43–55. [Google Scholar]
- 42.Liu AP, Fletcher DA. Actin polymerization serves as a membrane domain switch in model lipid bilayers. Biophys J. 2006;91:4064–4070. doi: 10.1529/biophysj.106.090852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.McMahon HT, Gallop JL. Membrane curvature and mechanisms of dynamic cell membrane remodelling. Nature. 2005;438:590–596. doi: 10.1038/nature04396. [DOI] [PubMed] [Google Scholar]
- 44.Petrov EP, Ehrig J, Schwille P. Near-critical fluctuations and cytoskeleton-assisted phase separation lead to subdiffusion in cell membranes. Biophys J. 2011;100:80–89. doi: 10.1016/j.bpj.2010.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Veatch SL, Machta BM,BB, Papanikolaou S, Sethna JP. Minimal model of plasma membrane heterogeneity requires coupling cortical actin to criticality. Biophys J. 2011;100:1668–1677. doi: 10.1016/j.bpj.2011.02.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lucero HA, Robbins PW. Lipid rafts-protein association and the regulation of protein activity. Arch Biochem Biophys. 2004;426:208–224. doi: 10.1016/j.abb.2004.03.020. [DOI] [PubMed] [Google Scholar]
- 47.Derivery E, Helfer E, Henriot V, Gautreau A. Actin polymerization controls the organization of WASH domains at the surface of endosomes. Plos One. 2012;7:e39774. doi: 10.1371/journal.pone.0039774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gowrishankar K, et al. Active remodeling of cortical actin regulates spatiotemporal organization of cell surface molecules. Cell. 2012;149:1353–1367. doi: 10.1016/j.cell.2012.05.008. [DOI] [PubMed] [Google Scholar]
- 49.Gomez-Llobregat J, Buceta J, Reigada R. Interplay of cytoskeletal activity and lipid phase stability in dynamic protein recruitment and clustering. Sci Rep. 2013;3:2608. doi: 10.1038/srep02608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lavi Y, Gov N, Edidin M, Gheber LA. Lifetime of major histocompatibility complex class-I membrane clusters is controlled by the actin cytoskeleton. Biophys J. 2012;102:1543–1550. doi: 10.1016/j.bpj.2012.01.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Vanounou S, Parola AH, Fishov I. Phosphatidylethanolamine and phosphatidylglycerol are segregated into different domains in bacterial membrane. Study with pyrene-labeled phospholipids. Biophys J. 2002;82:343a–343a. doi: 10.1046/j.1365-2958.2003.03614.x. [DOI] [PubMed] [Google Scholar]
- 52.Johnson AS, van Horck S, Lewis PJ. Dynamic localization of membrane proteins in Bacillus subtilis. Microbiology. 2004;150:2815–2824. doi: 10.1099/mic.0.27223-0. [DOI] [PubMed] [Google Scholar]
- 53.Lopez D, Kolter R. Functional microdomains in bacterial membranes. Genes Dev. 2010;24:1893–1902. doi: 10.1101/gad.1945010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Donovan C, Bramkamp M. Characterization and subcellular localization of a bacterial flotillin homologue. Microbiology. 2009;155:1786–1799. doi: 10.1099/mic.0.025312-0. [DOI] [PubMed] [Google Scholar]
- 55.Dempwolff F, Moller HM, Graumann PL. Synthetic motility and cell shape defects associated with deletions of flotillin/reggie paralogs in Bacillus subtilis and interplay of these proteins with NfeD proteins. J Bacteriol. 2012;194:4652–4661. doi: 10.1128/JB.00910-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Garcia-Saez AJ, Chiantia S, Schwille P. Effect of line tension on the lateral organization of lipid membranes. J Biol Chem. 2007;282:33537–33544. doi: 10.1074/jbc.M706162200. [DOI] [PubMed] [Google Scholar]
- 57.Cornell RB, Taneva SG. Amphipathic helices as mediators of the membrane interaction of amphitropic proteins, and as modulators of bilayer physical properties. Curr Protein Pept Sc. 2006;7:539–552. doi: 10.2174/138920306779025675. [DOI] [PubMed] [Google Scholar]
- 58.Mbamala EC, Ben-Shaul A, May S. Domain formation induced by the adsorption of charged proteins on mixed lipid membranes. Biophys J. 2005;88:1702–1714. doi: 10.1529/biophysj.104.048132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lee AG. How lipids affect the activities of integral membrane proteins. Biochim Biophys Acta. 2004;1666:62–87. doi: 10.1016/j.bbamem.2004.05.012. [DOI] [PubMed] [Google Scholar]
- 60.Andersen OS, Koeppe RE. Bilayer thickness and membrane protein function: An energetic perspective. Annu Rev Bioph Biom. 2007;36:107–130. doi: 10.1146/annurev.biophys.36.040306.132643. [DOI] [PubMed] [Google Scholar]
- 61.Cybulski LE, de Mendoza D. Bilayer hydrophobic thickness and integral membrane protein function. Curr Protein Pept Sc. 2011;12:760–766. doi: 10.2174/138920311798841681. [DOI] [PubMed] [Google Scholar]
- 62.Rossignol M, Thomas P, Grignon C. Proton permeability of liposomes from natural phospholipid mixtures. Biochim Biophys Acta. 1982;684:195–199. doi: 10.1016/0005-2736(82)90005-0. [DOI] [PubMed] [Google Scholar]
- 63.van de Vossenberg JL, Driessen AJ, da Costa MS, Konings WN. Homeostasis of the membrane proton permeability in Bacillus subtilis grown at different temperatures. Biochim Biophys Acta. 1999;1419:97–104. doi: 10.1016/s0005-2736(99)00063-2. [DOI] [PubMed] [Google Scholar]
- 64.Weber SC, Spakowitz AJ, Theriot JA. Nonthermal ATP-dependent fluctuations contribute to the in vivo motion of chromosomal loci. Proc Natl Acad Sci U S A. 2012;109:7338–7343. doi: 10.1073/pnas.1119505109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ganchev DN, Hasper HE, Breukink E, de Kruijff B. Size and orientation of the lipid II headgroup as revealed by AFM imaging. Biochemistry. 2006;45:6195–6202. doi: 10.1021/bi051913e. [DOI] [PubMed] [Google Scholar]
- 66.Kusumi A, et al. Paradigm shift of the plasma membrane concept from the two-dimensional continuum fluid to the partitioned fluid: High-speed single-molecule tracking of membrane molecules. Annu Rev Bioph Biom. 2005;34:351–354. doi: 10.1146/annurev.biophys.34.040204.144637. [DOI] [PubMed] [Google Scholar]
- 67.Konopasek I, Strzalka K, Svobodova J. Cold shock in Bacillus subtilis: different effects of benzyl alcohol and ethanol on the membrane organisation and cell adaptation. Biochim Biophys Acta. 2000;1464:18–26. doi: 10.1016/s0005-2736(99)00240-0. [DOI] [PubMed] [Google Scholar]
- 68.Weber MH, Klein W, Muller L, Niess UM, Marahiel MA. Role of the Bacillus subtilis fatty acid desaturase in membrane adaptation during cold shock. Mol Microbiol. 2001;39:1321–1329. doi: 10.1111/j.1365-2958.2001.02322.x. [DOI] [PubMed] [Google Scholar]
- 69.Kim BG, Dai HN, McAtee M, Vicini S, Bregman BS. Labeling of dendritic spines with the carbocyanine dye DiI for confocal microscopic imaging in lightly fixed cortical slices. J Neurosci Methods. 2007;162:237–243. doi: 10.1016/j.jneumeth.2007.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Landgraf D, Okumus B, Chien P, Baker TA, Paulsson J. Segregation of molecules at cell division reveals native protein localization. Nature methods. 2012;9:480–498. doi: 10.1038/nmeth.1955. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.