Abstract
Numerous past studies have shown members of the genus Nitrospira to be the predominant nitrite-oxidizing bacteria (NOB) in nitrifying wastewater treatment plants (WWTPs). Only recently, the novel NOB “Candidatus Nitrotoga arctica” was identified in permafrost soil and a close relative was enriched from activated sludge. Still, little is known about diversity, distribution, and functional importance of Nitrotoga in natural and engineered ecosystems. Here we developed Nitrotoga 16S rRNA-specific PCR primers and FISH probes, which were applied to screen activated sludge samples from 20 full-scale WWTPs. Nitrotoga-like bacteria were detected by PCR in 11 samples and reached abundances detectable by FISH in seven sludges. They coexisted with Nitrospira in most of these WWTPs, but constituted the only detectable NOB in two systems. Quantitative FISH revealed that Nitrotoga accounted for nearly 2% of the total bacterial community in one of these plants, a number comparable to Nitrospira abundances in other WWTPs. Spatial statistics revealed that Nitrotoga co-aggregated with ammonia oxidizing bacteria, strongly supporting a functional role in nitrite oxidation. This activity was confirmed by FISH-MAR, which revealed nitrite-dependent autotrophic carbon fixation by Nitrotoga in situ. Correlation of presence or absence with WWTP operational parameters indicated low temperatures as a main factor supporting high Nitrotoga abundances, although in incubation experiments these NOB remained active over an unexpected range of temperatures, and also at different ambient nitrite concentrations. In conclusion, this study demonstrates that Nitrotoga can be functionally important nitrite oxidizers in WWTPs and can even represent the only known NOB in engineered systems.
Keywords: Nitrotoga, nitrite-oxidizing bacteria, nitrification, activated sludge, wastewater treatment
Introduction
The anthropogenic release of large amounts of nitrogen has detrimental effects on the environment such as eutrophication of inland and coastal water bodies, leading to algal blooms and hypoxia (Diaz and Rosenberg 2008). As high concentrations of ammonia and nitrite are also toxic for many organisms, their discharge into the environment must be regulated to protect ecosystems and drinking water supplies (Camargo and Alonso 2006, Conley et al. 2009). Besides agriculture, human sewage is one of the largest sources of inorganic nitrogen, in particular ammonia from urea degradation. Therefore, WWTPs designed for nutrient removal are essential for protecting aquatic ecosystems from nitrogen contamination. Most WWTPs exploit nitrifying and denitrifying microbes to aerobically oxidize ammonia via nitrite to nitrate (nitrification) and to subsequently reduce the produced nitrate to gaseous dinitrogen under anaerobic conditions (denitrification). Complete nitrification depends on the mutualistic interaction of ammonia-oxidizing microbes and nitrite-oxidizing bacteria (NOB) and thus both groups have been intensively studied (for reviews see Gujer 2010, Wagner et al. 2002).
Before cultivation-independent molecular methods became available, members of the alphaproteobacterial genus Nitrobacter were thought to be mainly responsible for nitrite oxidation in sewage treatment because the isolation of these NOB from activated sludge was straightforward (e.g., Henze et al. 1997). This view changed radically when molecular tools revealed that Nitrobacter occurs in many WWTPs only in small numbers close to or even below the detection limit of microscopy-based methods such as fluorescence in situ hybridization (FISH; Wagner et al. 1996). Instead, yet uncultured Nitrospira turned out to be the key NOB in most engineered systems (Daims et al. 2001b, Juretschko et al. 1998, Schramm et al. 1998). Since this discovery, research on NOB in WWTPs has focused mainly on Nitrospira, which belong to the distinct bacterial phylum Nitrospirae, display a considerable phylogenetic diversity, and possess genetic and physiological features that clearly distinguish them from other known NOB (Daims et al. 2001a, Foesel et al. 2008, Lücker et al. 2010, Maixner et al. 2006, Maixner et al. 2008, Schramm et al. 1999).
However, novel nitrite oxidizers are still being discovered (e.g., Schott et al. 2010). Recently, Alawi et al. (2007) enriched the novel nitrite-oxidizing betaproteobacterium “Candidatus Nitrotoga arctica” from permafrost soil. N. arctica only grows at low temperatures between 4 to 17 °C. At only 0.3 mM (Alawi et al. 2007), its nitrite concentration optimum is approximately one order of magnitude below the nitrite concentrations usually applied to cultivate Nitrospira isolates (Lebedeva et al. 2008) and even two orders of magnitude below the nitrite concentrations used to grow Nitrobacter (Prosser 1989). Interestingly, a closely related Nitrotoga strain was also enriched from a full-scale WWTP (Alawi et al. 2009), fuelling speculations that these novel NOB might be relevant for nitrite oxidation in engineered systems. This question cannot be answered by cultivation-based approaches because very few Nitrotoga cells could be sufficient as inoculum for a successful enrichment culture, whereas their in situ numbers may be low and thus irrelevant for the nitrification process in the system.
In this study we therefore applied the full-cycle rRNA approach (Amann et al. 1995) to investigate whether Nitrotoga are hitherto overlooked key nitrifiers in full-scale sewage treatment systems. Based on in-depth phylogenetic analyses of the new candidate genus Nitrotoga, cultivation-independent molecular tools for the specific detection of Nitrotoga-like bacteria were developed. These were then applied to detect, visualize, and quantify Nitrotoga in nitrifying full-scale WWTPs and to investigate their spatial distribution patterns relative to ammonia-oxidizing bacteria (AOB) within activated sludge flocs. Furthermore, FISH in combination with microradiography (FISH-MAR) was used to test for the chemolithoautotrophic capacity of Nitrotoga-like NOB across a range of nitrite concentrations and temperatures.
Material and Methods
Activated sludge sampling and fixation
Activated sludge samples were obtained from full-scale sequencing batch reactors (SBRs) operated with or without differential internal cycling (DIC) (Holm 2003), from conventional activated sludge basins, fixed bed reactors, and a membrane filtration plant. The selected WWTPs are located in Germany and Switzerland and treat municipal wastewater, which in some cases is mixed with industrial sewage or animal rendering waste (Table 1). In addition, highly enriched N. arctica was grown according to Alawi et al. (2007) and used for probe and primer evaluation.
Table 1. Characteristics of the analyzed wastewater treatment plants.
| WWTP | Reactor typea | Type of treated sewage | Detection of Nitrotogab |
Nitrospira
sublineagec,d |
Temp.e [°C] | Influente [mg/l] | Effluente[mg/l] | Sampling date (2007) | |||
|---|---|---|---|---|---|---|---|---|---|---|---|
| PCR | FISH | NH4+ | NH4+ | NO2− | NO3− | ||||||
| Altmannstein | SBR | municipal | + | + | I + II | 7 | 54.7 | 9.18 | 0.48 | 0.72 | March 24 |
| Ampfing | SBR | municipal, slaughter, and dairy waste | − | − | II | 13 | nd | 0.1 | 0.04 | 3.24 | March 26 |
| Bad Zwischenahn | DIC-SBR | municipal and industrial | + | + | − | 16 | 60 | 0.25 | 0.15 | 6.5 | May 23 |
| Bruchmühlen | DIC-SBR | municipal | + | + | I | 15 | 36 | 0.53 | 0.09 | 4.53 | May 22 |
| Deuz | DIC-SBR | municipal | + | + | I + II | 13 | nd | 0.33 | 0.09 | 3.46 | May 21 |
| Hettstedt | DIC-SBR | municipal and external activated sludge | − | − | I + II | 15 | 56 | 12.35 | 0.24 | 3 | May 24 |
| Huntlosen | DIC-SBR | municipal | − | − | I + II | 17 | 68 | 0.13 | 0.03 | 2.2 | May 23 |
| Ingolstadt | SBR | activated sludge drainage | − | − | I + II | 27 | 856 | 0.3 | <0.1 | 20.4 | May 09 |
| Kraftisried | single-stage activated sludge basin | animal rendering | − | − | I + II | 7 | 397.5 | 35.3 | 6.2 | 17.4 | January 29 |
| Langenzenn | SBR | municipal | + | + | − | 9 | 21.25 | 7.96 | 0.42 | 3.1 | March 14 |
| Lyss (ARA) | fixed bed reactor | municipal | + | − | I | 12 | 20 | 1 | 0.1 | 18 | January 29 |
| Lyss (GZM) | membrane filtration plant | animal rendering | − | − | I + II | 30 | 700 | <1 | <0.5 | 14 | January 29 |
| Oberding | fixed bed reactor | animal rendering | − | − | I + II | 26 | 450 | <1 | <0.5 | 4 | January 29 |
| Plattling | two-stage activated sludge basin | animal rendering | − | − | I + II | 30 | 750 | 1 | <0.5 | 3 | January 29 |
| Radeburg | DIC-SBR | municipal | + | − | I | 14 | nd | 0 | 0.05 | 3.3 | May 24 |
| Rosenheim | SBR | municipal | − | − | I + II | 36 | 970 | nd | nd | nd | May 30 |
| Seefeld | SBR | municipal | + | − | I | nd | 8.32 | 1.59 | nd | 1.73 | March 28 |
| Spenge | DIC-SBR | municipal | + | + | I | 14 | 24 | <0.2 | 0.05 | 1.38 | May 22 |
| Waldsassen | SBR | municipal and industrial | + | + | I | 9 | 18.5 | <0.1 | nd | 3.45 | March 27 |
| Weisstal | DIC-SBR | municipal | + | − | I + II | nd | nd | 0 | 0.02 | 4.4 | May 21 |
SBR, sequencing batch reactor; DIC-SBR, differential internal cycling SBR.
+, Nitrotoga detected; −, Nitrotoga not detected.
detection of Nitrospira by FISH: I, Nitrospira sublineage I; II, Nitrospira sublineage II; −, Nitrospira not detected.
Nitrobacter, Nitrococcus, Nitrolancea, and Nitrospina-like NOB were not detected by FISH in any sample
Temperature was measured on sampling date; nd, not determined.
For FISH analysis, activated sludge and enrichment culture samples were fixed with paraformaldehyde (PFA) according to Daims et al. (2005). Fixed biomass was stored at −20°C. Unfixed samples for DNA extraction were harvested by centrifugation (13,000 × g, 10 min, 4°C) and stored at −20°C.
Probe and primer design and evaluation
16S rRNA-targeted FISH probes and 16S rRNA gene-targeted PCR primers were designed and evaluated using the probe design and probe match functions of ARB (Ludwig et al. 2004) and a manually curated SILVA 16S rRNA database (version SSURef_NR99_115) (Quast et al. 2013). In short, the 16S rRNA sequence database was updated by importing (i) all high quality, near full-length 16S rRNA sequences (≥1250 nucleotides, pintail quality scores ≥75%) included in the SILVA SSU r117 web release and classified as Candidatus Nitrotoga (http://www.arb-silva.de/browser/), and (ii) all sequences with an identity >96% to N. arctica strain 6680 (DQ839562) from the NCBI nr database. Sequences with a pintail value <75% were regarded as potential chimeras (Ashelford et al. 2005) and excluded from further analyses. For the purpose of this study, the genus Nitrotoga was defined on the basis of phylogenetic analyses (see below) and only included sequences that formed a stable monophyletic group with N. arctica. The definition of the family Gallionellaceae was based on SILVA classification. High quality sequences of family members were extracted from the SILVA guide tree and comprised the genera Nitrotoga, Gallionella, and Sideroxydans, as well as a large number of sequences of unresolved affiliation within the family.
Optimal PCR conditions for the new Nitrotoga-targeting primer pairs were determined by temperature gradient PCR. Optimal hybridization conditions for newly developed FISH probes were determined as described previously (Daims et al. 1999). Non-target organisms were not included, as they were not available in pure culture or contained in our clone libraries for Clone-FISH (Schramm et al. 2002). The probes were thus used at the highest hybridization stringency (i.e., formamide concentration in the hybridization buffer) that still yielded bright fluorescence signals for the target organisms and in combination with unlabeled competitor probes specific for the non-target organisms with the fewest number of mismatches at the respective probe binding site. Moreover, in all experiments at least two probes labeled with different fluorochromes were used in combination to unambiguously identify the target organisms according to the multiple probe concept (Ludwig et al. 1998). Probes used for FISH were 5′-labeled with the dyes 5(6)-carboxyfluorescein-N-hydroxysuccinimide ester (FLUOS), Cy3, or Cy5. Labeled probes, unlabeled competitors, and PCR primers were obtained from Thermo Scientific (Thermo Fisher Scientific Inc., Waltham, MA, USA). All FISH probes used in this study are listed in Tables 2 and S1.
Table 2. FISH probes designed in this study.
| Probe full namea | Short name | Sequence 5′-3′ | Binding positionb | FA%c | Target group | Coveraged | |
|---|---|---|---|---|---|---|---|
| Probe | Comb. | ||||||
| S-G-Ntoga-0122-a-A-19 | Ntoga122 | TCC GGG TAC GTT CCG ATA T | 122 - 140 | 40 | genus Nitrotoga | 98.5% | 98.5% |
| cS-G-Ntoga-0122-a-A-19 e | c1Ntoga122 | TCW GGG TAC GTT CCG ATA T | 122 - 140 | − | − | ||
| cS-G-Ntoga-0122-b-A-19 e | c2Ntoga122 | TCY GGG TAC GTT CCG ATG T | 122 - 140 | − | − | ||
| S-F-Gall-0178-a-A-18 | FGall178 | TCC CCC TYA GGG CAT ATG | 178 - 195 | 30 | family Gallionellaceae | 98.9% | 98.9% |
| cS-F-Gall-0178-a-A-18e | cFGall178 | TCC CCC TYA GGG CKT ATG | 178 - 195 | − | − | ||
| S-F-Gall-0221-a-A-18 | FGall221a | TAT CGG CCA CTC CGA AAG | 221 - 238 | 30 | family Gallionellaceae | 71.6% | 90.2% |
| cS-F-Gall-0221-a-A-18 e | c1FGall221a | TAT CGG CCA CTC CTA AAG | 221 - 238 | − | − | ||
| S-F-Gall-0221-b-A-18f | FGall221b | TAT CGG CCG CTC CGA AAA | 221 - 238 | 30 | genus Nitrotoga | 97.9% | |
| family Gallionellaceae | 18.6% | ||||||
| cS-F-Gall-0221-b-A-18 | cFGall221b | CAT CGG CCG CTC CGA AAG | 221 - 238 | − | − | ||
| S-*-Gall-0438-a-A-18 | FGall438 | GTT TTC TTT CCG GCT GAA | 438 - 455 | 25 | genus Nitrotoga | 89.4% | 89.4% |
| cS-*-Gall-0438-a-A-18e | c1FGall438 | GAT TTC TTT CCG GCT GAA | 438 - 455 | Zoogloea spp. | − | ||
| cS-*-Gall-0438-b-A-18e | c2FGall438 | GTT TTC TTC CCG GCT GAA | 438 - 455 | Thaurea/Dechloromonas spp. | − | ||
| cS-*-Gall-0438-c-A-18e | c3FGall438 | GTT TTC TTT CCG TCT GAA | 438 - 455 | Azoarcus spp. | − | ||
| S-G-Ntoga-1424-a-A-18 | Ntoga1424 | CTA GCT GCT TCT GGT AGA A | 1424 - 1442 | 20 | genus Nitrotoga | 82.2% | 82.2% |
| cS-G-Ntoga-1424-a-A-18e | c1Ntoga1424 | CTA ACT GCT TCT GGT AGA A | 1424 - 1442 | Sterolibacterium/Dechlorosoma spp. | − | ||
| cS-G-Ntoga-1424-b-A-18e | c2Ntoga1424 | CTA GCT GCT TCT GGT ACA A | 1424 - 1442 | Acidothiobacillus spp. | − | ||
Probe nomenclature according to (Alm et al. 1996).
Probe binding position according to E. coli 16S rRNA gene numbering.
Percent formamide (v/v) added to the hybridization buffer for optimal hybridization conditions.
Group coverage was calculated as the fraction of organisms with a full sequence match to the respective probe relative to the number of sequences within the respective probe target group as defined by phylogenetic analyses (see main text for details). Column “Probe” lists the coverage of single probes, column “Comb.” that of probe combinations.
Competitor probes were added to the hybridization buffer as unlabeled oligonucleotides and in equimolar amounts as the labeled probes to increase hybridization specificity.
Probe can be used alone for detection of the genus Nitrotoga or in combination with probe FGall221a for detection of the family Gallionellaceae.
DNA extraction, PCR, and cloning of 16S rRNA genes
Genomic DNA was extracted using the PowerSoil® DNA Isolation Kit (MO BIO Laboratories, Inc, Carlsbad, CA, USA) according to manufacturer’s instructions. Preliminary test PCRs were performed using the general bacterial primers 616V and 1492r to ensure that the DNA was of sufficient quantity, quality, and purity for PCR as described elsewhere (Juretschko et al. 1998, Kane et al. 1993). For the specific amplification of Nitrotoga-like 16S rRNA genes, the genus Nitrotoga-specific primer combination S-G-Ntoga-0124-a-S-19 (Ntoga124F, 5′- ATC GGA ACG TAC CCG GAA A-3′) and S-G-Ntoga-1462-a-A-18 (Ntoga1462R, 5′- CGA ACC CTA CCG TGG CAA C-3′) were used. Reaction mixtures were prepared according to the manufacturer’s recommendations in a total volume of 50 μl with 2 mM MgCl2, 0.5 μM of each primer, and 1.25 U of Taq polymerase (Fermentas, St. Leon-Rot, Germany). Additionally, 5 μg of BSA were added to circumvent PCR inhibition. PCR cycling consisted of an initial denaturation step at 94°C for 5 min, followed by 35 cycles of denaturation at 94°C for 30 s, annealing at 63°C for 30 s, and elongation at 72°C for 1 min 30 s, and was completed by a final elongation step at 72°C for 10 min. The presence and purity of amplicons were confirmed by agarose gel electrophoresis. Cloning and sequencing of amplified 16S rRNA genes were performed as described elsewhere (Juretschko et al. 1998). Cloned genes to be sequenced were selected based on different restriction fragment length polymorphism (RFLP) patterns, with 1 - 5 clones per pattern randomly chosen. For RFLP, 5 μl of M13 (Invitrogen, Carlsbad, CA, USA) PCR product were digested with 1 μl restriction enzyme MspI (Fisher Scientific, Vienna, Austria) and 1 μl buffer (universal buffer “Tango”, Fisher Scientific, Vienna, Austria) at 37°C for 3 h. Fragment patterns were separated and visualized by gel electrophoresis using a 2.5% (w/v) agarose gel.
Phylogenetic analyses
For phylogenetic tree calculations, a manually curated 16S rRNA database was created as described above (see chapter Probe and primer design and evaluation). All sequences imported into the ARB software (Ludwig et al. 2004) were automatically aligned using the tools implemented in ARB, followed by manual refinement of the alignment. Phylogenetic analyses were performed using maximum-likelihood and maximum-parsimony methods as provided by ARB, or Bayesian inference using MrBayes (Ronquist and Huelsenbeck 2003). A 50% conservation filter for the family Gallionellaceae was used (resulting in 1481 informative positions) and only near full-length sequences (>1320 nucleotides) were included in tree calculations. Bootstrap values were estimated using the maximum-likelihood and maximum-parsimony algorithms with 100 iterations, or MrBayes run with parameters stoprule=yes and stopval=0.01, causing the program to stop when the average standard deviation of the topological convergence diagnostics has reached a value <0.01.
FISH, microscopy, and digital image analysis
Aliquots of PFA-fixed biomass were spotted onto microscope slides and FISH was performed as described elsewhere (Daims et al. 2005). Probe-conferred fluorescence was recorded on a LSM 510 confocal laser scanning microscope (CLSM, Carl Zeiss AG, Oberkochen, Germany) equipped with one argon ion (450 to 514 nm) and two helium neon lasers (543 and 633 nm) for the detection of FLUOS, Cy3, and Cy5, respectively, or a Leica LSM SP8 equipped with a white light laser (Leica Microsystems, Wetzlar, Germany). For determining probe dissociation profiles (Daims et al. 1999) of newly designed FISH probes (Table 2), highly enriched N. arctica biomass was used for hybridization with the respective probe and 10 images per formamide concentration were recorded for subsequent image analysis. For quantifying relative biovolume fractions (Daims and Wagner 2007, Schmid et al. 2000), the activated sludge samples were hybridized to probe Ntoga122 and the EUB338 probe mix (Tables 2 and S1), and 40 image pairs containing each probe signal were taken at random fields of view. For analyzing spatial distribution patterns of Nitrotoga-like bacteria within activated sludge flocs (Daims et al. 2006), cells were stained by probes Ntoga122, Cluster6a192, and BET42a (Tables 2 and S1) and 40 images of these probe signals were recorded at random positions. All digital image analysis tasks were carried out by using the software daime (Daims et al., 2006).
FISH and microautoradiography
Activated sludge was sampled from two DIC-SBR reactors (Bad Zwischenahn and Deuz, Germany; Table 1) and transferred to the laboratory at 4°C within 24h. Prior to the incubations, sludge samples were diluted at a ratio of 1:5 with sterile filtered supernatant from the same reactor and pre-incubated at room temperature overnight to ensure that all endogenous ammonia and nitrite were consumed. Concentrations of these electron donors were monitored by Nessler’s reagent (Sigma-Aldrich, Vienna, Austria) and Merckoquant test stripes (Merck, Vienna, Austria), respectively. Subsequently, 5 ml sample aliquots were transferred to 100 ml glass serum bottles. All experiments were performed in duplicates and PFA-fixed biomass from the same reactor was used as dead control. To start the incubation, NaNO2 was added to the desired concentration (see below) and each vessel was supplemented with 10 μCi [14C]-H2CO3. The bottles were sealed air tight with rubber stoppers. Assuming that parallel incubations behaved similarly, nitrite consumption was followed in an additional sample for each incubation condition which contained non-radioactive bicarbonate. Nitrite was replenished when necessary with an aseptic needle without opening the radioactive culture bottles. Incubations were performed using six different nitrite concentrations (0, 0.1, 0.5, 1, 5, and 10 mM NO2−, at 14°C) and at five different temperatures (4°C, 10°C, 14°C, 20°C, and 27°C, with 0.5 mM NO2−). After 6 h, incubations were stopped by harvesting the biomass (25,500 × g, 5 min, 4°C) and performing PFA fixation as described elsewhere (Daims et al. 2005). In order to disintegrate the sludge flocs, one volume of PFA-fixed sample was diluted with four volumes 1x phosphate buffered saline (PBS), transferred to a 2 ml screw-cap reaction tube containing a ¼-inch ceramic sphere, and vortexed horizontally for 10 min at maximum speed. Following two washing steps in 1 × PBS, a small aliquot was spotted onto a cover slip and FISH-MAR was carried out as described by Lee et al. (1999) with an exposure time of 7 days.
Results and discussion
Phylogeny and environmental distribution of the candidate genus Nitrotoga
N. arctica is the proposed type strain of the new candidate genus Nitrotoga (Alawi et al. 2007), which is affiliated with the family Gallionellaceae (Skerman et al. 1980) of the order Gallionellales (Weiss et al. 2007), and contains the only known nitrite oxidizers in the Betaproteobacteria. Besides Nitrotoga, the Gallionellaceae comprise two genera of iron-oxidizing organisms, Gallionella (Garrity et al. 2005, Henrici and Johnson 1935) and Sideroxydans (Weiss et al. 2007), some of which were reported to couple Fe(II)-oxidation to nitrate reduction (Blöthe and Roden 2009).
In this study, the genus Nitrotoga was defined based on phylogenetic support and comprised only sequences that consistently formed a monophyletic cluster with N. arctica in our analyses. The affiliation of partial 16S rRNA sequences with the genus was confirmed by individually adding them to a representative tree without changing the overall tree topology. According to this conservative approach, the candidate genus Nitrotoga contains, besides the WWTP-derived sequences from this study and the enrichments described by Alawi et al. (2007, 2009), organisms thriving in diverse habitats (Figure 1). Nitrotoga-like sequences were detected by cultivation-independent approaches in different wastewater and drinking water treatment systems (Ji and Chen 2010, Kong et al. 2007, Kwon et al. 2010, Maestre et al. 2009, White et al. 2012), soil (Sattin et al. 2009), groundwater and cave-derived water (Chen et al. 2009, Flynn et al. 2013), and in lake, river, and marine water and sediment samples (Brümmer et al. 2003, Li et al. 2011, Liu et al. 2012, Martiny et al. 2011, Na et al. 2011, Percent et al. 2008, Schwarz et al. 2007, Tamminen et al. 2011). While this wide distribution of Nitrotoga-like organisms indicates that these novel NOB might contribute to nitrification in a great variety of habitat types, none of these studies provided evidence for their function in the respective ecosystem. Thus, their abundances and in situ functions in these systems, as well as their competitive success compared to other NOB such as Nitrospira and Nitrobacter, remain to be elucidated.
Figure 1. Phylogenetic analysis of the candidate genus Nitrotoga (gray box) and selected members of the family Gallionellaceae.
Displayed is a Bayesian interference tree (s.d. = 0.009882) including representative nearly full-length 16S rRNA gene sequences related to N. arctica strain 6680. Pie charts indicate statistical support of nodes based on bootstrap analysis or Bayesian inference. Bootstrap values are based on 100 iterations. Sequences obtained in this study are printed in bold. The scale bar corresponds to 5% estimated sequence divergence. MB, Bayesian inference; ML, maximum likelihood; MP, maximum parsimony; TP, Treepuzzle.
Probe design and evaluation
16S rRNA-targeted probes for the specific detection of Nitrotoga-like bacteria by FISH (Table 2) were designed according to the “multiple probe approach” (Ludwig et al. 1998), ensuring the unambiguous identification of Nitrotoga cells by phylogenetically nested probes for the candidate genus Nitrotoga and most members of the family Gallionellaceae. Probes Ntoga122 and Ntoga1424 were designed to target the candidate genus only, whereas probes FGall221b and Ntoga438 also include some sequences of uncertain affiliation within the Gallionellaceae, which have a high sequence similarity (95.8 to 97.9%) to N. arctica but did not cluster consistently with the genus Nitrotoga in our phylogenetic analyses. Probe FGall178 and the FGall221a+b probe mixture target most known members of the family Gallionellaceae for which sequence information is available in the respective region of the 16S rRNA (Table 2).
When tested on highly enriched N. arctica biomass, all probes yielded bright fluorescence signals, irrespective of the dye used for probe labeling. Only signals of probe Ntoga1424 were relatively dim, which is consistent with a low brightness of probes targeting the homologous region of the 16S rRNA in E. coli (Behrens et al. 2003). In silico evaluation indicated a good specificity of all probes, with the respective perfect-match probe binding sites found in very few non-target organisms only. Still, the new probes should always be used by combining the three genus-specific, or two genus- with one family-specific probe, all labeled in different colors. Organisms detected by all applied probes can be regarded as Nitrotoga-like bacteria with a high degree of confidence because the current 16S rRNA databases contain no non-target organism with the binding sites for three of the Nitrotoga or Gallionellaceae-specific probes. Still, as some closely related bacteria (mainly Betaproteobacteria) have few and hard to discriminate mismatches at the respective probe binding site, we recommend using the new probes in combination with the respective unlabeled competitor oligonucleotides (Table 2). In this study, unambiguous identification of Nitrotoga in the activated sludge samples was ensured by first hybridizing the sample with multiple Nitrotoga-specific probes as described above. As these experiments revealed that all applied probes bound to the same cell clusters and no cells were detected by only a subset of these probes, the new probes were subsequently also used in combination with probes targeting organisms other than Nitrotoga or Gallionellaceae (see below).
Occurrence of Nitrotoga-like NOB in WWTPs
Utilizing the new genus Nitrotoga-specific primer pair Ntoga124F/Ntoga1462R, we screened 20 full-scale WWTPs by PCR for the presence of Nitrotoga-like bacteria. With SBRs, DIC-SBRs, and conventional activated sludge systems, these plants represented different reactor types (Table 1). PCR amplicons were obtained for 11 samples, indicating that Nitrotoga-like NOB were present in as many as 55% of the screened WWTPs (Table 1). All of these 11 plants received municipal sewage, which in some cases was mixed with different amounts of industrial wastewaters. The applied PCR assay did not detect Nitrotoga-like NOB in any of the systems treating animal rendering waste.
To confirm the specificity of the primer pair, the PCR products obtained from four WWTP samples, including one plant where Nitrotoga was detectable by PCR but not by FISH (see below), were cloned and in total 61 of the cloned 16S rRNA genes were Sanger-sequenced. Indeed, none of the clone libraries contained any non-target organism. Although the sequences obtained in this study shared high sequence similarities ≥98%, they formed several sub-clusters within the candidate genus Nitrotoga in phylogenetic analyses (Figure 1). These sub-clusters might reflect microdiversity of closely related and coexisting Nitrotoga strains or the presence of multiple rrn operons with small sequence dissimilarities in Nitrotoga genomes. Alternatively, the observed sequence differences could be artifacts introduced by PCR or sequencing errors.
FISH confirmed the presence of Nitrotoga-like organisms in seven WWTPs, which all were also PCR-positive (Table 1). In these samples, the Nitrotoga cells occurred as dense clusters of heterogeneous shape located within the sludge flocs (Figure 2). The cells in these aggregates were irregularly shaped rods or cocci, resembling the morphologies described for N. arctica (Alawi et al. 2007). Interestingly, all WWTPs that harbored Nitrotoga in sufficiently high quantities for detection by FISH (Amann et al. 1995) were operated at temperatures between 7 and 16°C (Table 1). This observation is fully consistent with the optimal growth temperature range of enriched Nitrotoga cultures (Alawi et al. 2009) and temperature hence seems to be a major factor affecting the growth of Nitrotoga-like NOB in full-scale WWTPs.
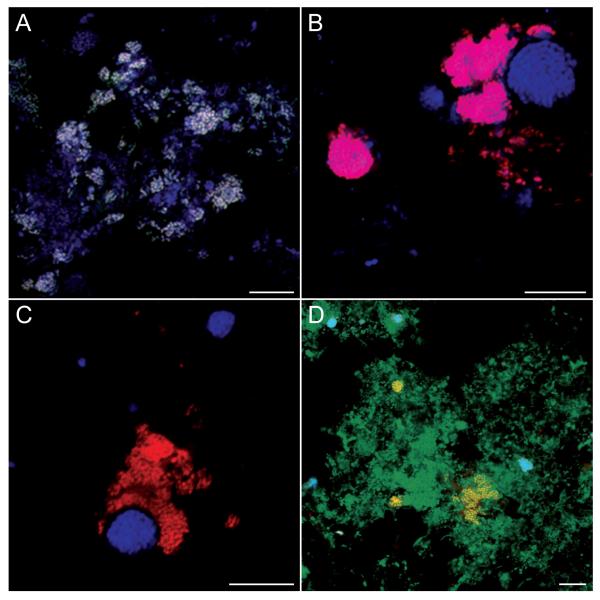
Figure 2. Confocal micrographs of FISH-stained Nitrotoga-like bacteria in activated sludge samples from WWTPs Bad Zwischenahn (A-C) and Deuz (D).
(A) Nitrotoga cell aggregates hybridized to probes Ntoga122 (green), FGall221b (red), and EUB338mix (blue). Nitrotoga appear white due to overlay of all probe signals. (B) Nitrotoga detected by probes FGall221b (red) and EUB338mix (blue) at high magnification. Nitrotoga appear magenta. (C) Simultaneous detection of Nitrotoga and AOB cell clusters by probes FGall221b (red) and Cluster6a192 (blue) at high magnification. Note the close vicinity of Nitrotoga and AOB, reflecting their metabolic interaction. (D) Simultaneous detection of Nitrotoga and Nitrospira by probes Ntoga122 (red), Ntspa662 (blue), and EUB338mix (green). Nitrotoga appear yellow, Nitrospira cyan. For probe details refer to Tables 2 and S1. The scale bar in all micrographs equals 10 μm.
Positive PCR results could not be confirmed by FISH for three of the SBRs and for one fixed-bed reactor (Table 1). Possible reasons for this discrepancy may be a cellular ribosome content of Nitrotoga below the detection limit of the applied standard FISH protocol (1400 ribosomes per cell; Hoshino et al. 2008) or PCR-amplification of DNA from lysed cells or extracellular DNA. Considering that PCR is at least tenfold more sensitive than FISH (Amann et al. 1995), the abundance of Nitrotoga most likely was too low for FISH detection in these activated sludges, implying that Nitrotoga were not functionally important and probably were allochthonous organisms unable to establish large stable populations in these WWTPs.
Quantification of Nitrotoga-like bacteria in activated sludge
In most of the analyzed WWTPs containing Nitrotoga-like bacteria, they were of low abundance (<1% of the total bacterial biomass) and coexisted with NOB of the genus Nitrospira (Table 1). Intriguingly, however, no known NOB except Nitrotoga were detected in the WWTPs Langenzenn and Bad Zwischenahn. The sludge from Langenzenn harbored only few Nitrotoga cell clusters (<1% of total bacterial biomass), and high ammonia concentrations were measured in the effluent of this plant at the time of sampling, although AOB related to Nitrosomonas oligotropha were present in this sample (data not shown). On the contrary, the activated sludge from Bad Zwischenahn contained comparably large amounts of Nitrotoga-like bacteria, which according to quantitative FISH constituted between 1% and 2% of the total bacterial biovolume (measured in four technical replicates). The low concentrations of ammonia and nitrite in the effluent of this WWTP (Table 1) imply complete nitrification. As Nitrotoga-like bacteria were the only detected NOB in this plant, this finding suggests that Nitrotoga can be solely responsible for nitrite oxidation in full-scale WWTPs.
Spatial co-localization of Nitrotoga-like NOB with AOB
AOB and NOB are partners in a mutualistic symbiosis where AOB oxidize ammonia to nitrite, which then serves as substrate for NOB whose activity prevents the accumulation of nitrite that could otherwise be toxic to AOB (Stein and Arp 1998). The strong interdependence of the two functional groups is often reflected by a close spatial co-aggregation of AOB and NOB in nitrifying activated sludge and biofilm samples, as observed frequently for Nitrospira with various AOB (Juretschko et al. 1998, Maixner et al. 2006, Okabe et al. 1999, Schramm et al. 1999). The spatial arrangement patterns of microbial populations in complex samples can be analyzed by a combination of FISH, image analysis, and spatial statistics (Daims et al. 2006) and already confirmed the co-localization of Nitrospira and AOB in WWTPs (Daims et al. 2006, Maixner et al. 2006).
To test whether Nitrotoga-like bacteria also co-localize with AOB, their spatial distribution patterns in two activated sludge samples were analyzed by the aforementioned method. The first WWTP addressed by this approach was Bad Zwischenahn, where no other known NOB except Nitrotoga had been detected (Table 1). In this sludge sample, all known AOB were detected by probe Cluster6a192 as shown in test hybridizations in combination with the AOB probe mix (Table S1). Visual observation already indicated that most Nitrotoga cell clusters occurred in close vicinity of AOB within the sludge flocs, sometimes even enclosing the AOB cell aggregates (Figure 2). Quantitative analysis confirmed a pronounced co-aggregation of AOB and Nitrotoga-like bacteria at distances below 50 μm between the cell clusters (Figure 3, panel A), suggesting that Nitrotoga preferably grew in the close vicinity of AOB in this sludge. The degree of clustering was highest at distances between 2 and 40 μm with two local maxima, one at 12 and a more pronounced one at 26 μm. These two peaks might reflect the presence of at least two Nitrotoga subpopulations, which would be in line with the apparent microdiversity of Nitrotoga found in the clone libraries (including clones from WWTP Bad Zwischenahn, Figure 1). Such slightly different spatial distribution patterns relative to AOB were also observed for NOB belonging to sublineage I and II of the genus Nitrospira, which have different nitrite concentration optima and thus occurred at different distances from AOB, which are the source of nitrite (Maixner et al. 2006). At distances larger than 50 μm no co-aggregation of Nitrotoga-like bacteria and AOB was detected and the pair cross-correlation function was not significantly different from one (indicating a random distribution pattern) or was even below this threshold. However, it should be noted that low pair cross-correlation values at large distances can also result from the absence of nitrifiers outside the activated sludge flocs, a bias that occurs if the analyzed distances are close to or exceed the average floc size (Daims et al. 2006).
Figure 3. Statistical analyses of the spatial arrangement patterns of NOB and AOB in the activated sludge samples from the Bad Zwischenahn (A-C) and Deuz (D, E) WWTPs.
(A-C) Spatial arrangement of Nitrotoga-like bacteria relative to (A) AOB, (B) all other Betaproteobacteria excluding AOB, and (C) all other Betaproteobacteria including AOB. (D, E) Spatial arrangement of (D) Nitrospira-like and (E) Nitrotoga-like bacteria relative to AOB. Black circles depict the mean pair cross-correlation function, and the upper and lower lines delimit 95% confidence intervals. Values >1 indicate co-aggregation, values <1 repulsion, and values =1 (dashed horizontal line) random distribution at the corresponding distance (Daims et al. 2006).
To confirm the quantified co-aggregation of Nitrotoga and AOB, the analysis was repeated with a negative control that was artificially derived from the recorded FISH images. First, the Nitrotoga and AOB probe signals (both groups are affiliated with the Betaproteobacteria) were digitally subtracted from the BET42a probe signal (targeting all Betaproteobacteria), resulting in images containing only other betaproteobacterial cells. These cells did not belong to any known nitrifying population and thus were not expected to have any specific functional link to Nitrotoga. Subsequently, the spatial arrangement pattern of Nitrotoga relative to the non-nitrifying Betaproteobacteria was quantified, resulting in pair cross-correlation values close to one over the whole range of tested distances (Figure 3, panel B). This result indicates a random distribution of Nitrotoga relative to non-nitrifying Betaproteobacteria in the sludge flocs. When only the Nitrotoga probe signal was subtracted from the BET42a signal, the presence of AOB within the remaining BET42a probe-defined population restored the observed co-aggregation pattern at distances between 6 and 50 μm (Figure 3, panel C). Thus, the co-aggregation between Nitrotoga and AOB was specific and most likely caused by a direct physiological interaction between these organisms.
The spatial distribution of Nitrotoga-like NOB relative to AOB was quantified also in WWTP Deuz, where in addition to Nitrotoga also Nitrospira had been detected by FISH (Table 1). While the in situ analyses of the sludge from WWTP Bad Zwischenahn had suggested that Nitrotoga can functionally replace Nitrospira, it remained to be shown if Nitrotoga-like bacteria can also compete with Nitrospira for niches in the close neighborhood of AOB when both NOB groups co-occur. Indeed, spatial arrangement analyses showed that both Nitrotoga and Nitrospira co-aggregated with AOB in WWTP Deuz. Intriguingly, however, the quantified co-aggregation patterns where distinctly different. For the more abundant Nitrospira, a strong co-aggregation signal at distances ranging from 5 to 35 μm was obtained (Figure 3, panel D). Nitrotoga-like bacteria also strongly co-aggregated with AOB, but the distance range was much more narrow with a clear peak around 10 μm distance (Figure 3, panel E). This outcome strongly suggests that both NOB groups are functionally linked to AOB, but apparently show ecological niche partitioning. Nitrospira-like bacteria appear to be more flexible regarding the symbiosis with AOB, for example because they could be adapted to a relatively broad range of nitrite concentrations found at different distances from AOB, or due to the presence of several sublineages (Maixner et al. 2006). In contrast, Nitrotoga seemed to inhabit a narrower niche where they could successfully compete for nitrite or might be involved in other yet uncharacterized biological interactions with AOB. Taken together, our analyses confirmed that in two different WWTPs Nitrotoga-like NOB specifically co-aggregated with AOB at short distances and thus strongly supported the hypothesis that Nitrotoga grow by oxidizing nitrite in full-scale WWTPs.
In situ chemolithoautotrophic activity of Nitrotoga-like NOB
FISH and spatial arrangement analyses of activated sludge samples already strongly indicated that Nitrotoga-like organisms grow by nitrite oxidation in full-scale WWTPs, but a physiological proof of this lifestyle was lacking. Thus, activated sludge from two different WWTPs was incubated in the presence or absence of nitrite, and nitrite-dependent inorganic carbon fixation by the autotrophic Nitrotoga-like bacteria was monitored at the single-cell level by FISH-MAR. In one of the plants analyzed (WWTP Bad Zwischenahn; Figures 4 and S1) Nitrotoga constituted the only known NOB, whereas in the second plant (WWTP Deuz; Figure S2) they coexisted with Nitrospira. Indeed, in both samples Nitrotoga-like bacteria readily incorporated carbon from [14C]-H2CO3 when the activated sludge was incubated at a low nitrite concentration (0.5 mM NO2−) and temperature (14°C), conditions reported optimal for growth of N. arctica and a closely related WWTP isolate (Alawi et al. 2007, Alawi et al. 2009). Hence, these FISH-MAR data clearly demonstrate the role of uncultured Nitrotoga-like bacteria as novel NOB in the two WWTPs analyzed. Moreover, they turned out to be active over a broad range of nitrite concentrations and temperatures. They readily incorporated 14C-labeled bicarbonate with as little as 0.1 mM nitrite and still remained active in presence of 10 mM nitrite (Figures S1 and S2), which was far above the tolerance limit of 1.2 mM nitrite reported for N. arctica (Alawi et al. 2007).Furthermore, they actively fixed carbon at temperatures from 4°C up to 27°C (Figures S1 and S2), an unexpected broad range of incubation temperatures as Nitrotoga isolates were reported to proliferate at low temperatures only (Alawi et al. 2007, Alawi et al. 2009). Similarly, carbon incorporation by the coexisting Nitrospira-like NOB in WWTP Deuz could be detected with all nitrite concentrations and at all temperatures tested (Figure S2). Altogether, our FISH-MAR experiments revealed that the uncultured Nitrotoga-like NOB were able to fix inorganic carbon across a much greater span of environmental conditions than previously anticipated based on the behavior of cultured representatives. However, it should be noted that the metabolic activity observed in our short-term incubations not necessarily shows growth, but rather that Nitrotoga can remain active under conditions that may be less favorable for them. Such flexibility is certainly beneficial in fluctuating natural environments or in WWTPs where the operational conditions change frequently, like in SBRs or in regions with pronounced daily temperature shifts. Since the Nitrotoga strains cultured so far were outcompeted by other NOB in long-term enrichments at high nitrite concentrations or temperatures (Alawi et al. 2009), and as we found high Nitrotoga abundances by FISH only in low-temperature WWTPs (Table 1), we expect differences between short-term activity and long-term ecological success of Nitrotoga-like NOB. Furthermore, the influence on their growth rates of additional factors like presence of organic substrates and salts remains to be investigated in future research.
Figure 4. Confocal micrographs of FISH-stained Nitrotoga-like bacteria with corresponding MAR signal.

Shown are representative images from the incubation of activated sludge from WWTP Bad Zwischenahn with 0.5 mM NO2− at 14°C. (A) Nitrotoga cell aggregates hybridized to probes Ntoga122 (red), FGall221b (green), and EUB338mix (blue); (B) corresponding DIC image showing silver grain deposition above radioactively labeled bacteria; (C) overlay of FISH and MAR signal. The scale bar in all images corresponds to 25 μm.
Conclusions
This study demonstrates that the recently discovered Nitrotoga are functionally important nitrite oxidizers in full-scale WWTPs, where they often co-exist with Nitrospira but occasionally represent the only known NOB populations. Thus, Nitrotoga should be included in studies of nitrification in WWTPs in addition to Nitrospira, Nitrobacter, and Nitrolancea (Sorokin et al. 2014). With the newly developed Nitrotoga-specific PCR primers and FISH probes, we provide molecular tools for the reliable in situ detection, visualization, and quantification of Nitrotoga-like bacteria. Clearly, to achieve encompassing insights into the microbiology of nitrification we will need further research on Nitrotoga reaching from environmental distribution surveys to functional, genomic, and post-genomic analyses. Especially in the context of wastewater treatment, future work should determine key ecophysiological parameters such as the nitrite oxidation kinetics of Nitrotoga-like bacteria, and should address their competition and co-existence with other NOB. A highly interesting topic will be the sensitivity or resilience of Nitrotoga to disturbances during reactor operation. Such information is urgently needed for all NOB relevant in WWTPs because problems and failures of the nitrification process in engineered systems are still frequently encountered, but the causes are mostly unknown and a reliable strategy to prevent such events has not been developed yet.
Supplementary Material
Acknowledgments
We thank Anneliese Müller and Christian Baranyi for excellent technical assistance. Niels Holm, Dieter Schreff, Uwe Temper, and the technical staff at the analyzed WWTPs are acknowledged for providing the activated sludge samples and for helpful discussions. This work was supported by the Austrian Science Fund (FWF) grant P24101-B22, the Vienna Science and Technology Fund (WWTF) grant LS09-040, and the German Research Foundation (DFG) grant SP 667/3-1.
Footnotes
Conflict of Interest Statement: The authors declare no conflict of interest.
Data deposition: 16S rRNA gene sequence data have been submitted to the GenBank database under accession numbers KM017111 - KM017129.
References
- Alawi M, Lipski A, Sanders T, Pfeiffer EM, Spieck E. Cultivation of a novel cold-adapted nitrite oxidizing betaproteobacterium from the Siberian Arctic. ISME J. 2007;1:256–264. doi: 10.1038/ismej.2007.34. [DOI] [PubMed] [Google Scholar]
- Alawi M, Off S, Kaya M, Spieck E. Temperature influences the population structure of nitrite-oxidizing bacteria in activated sludge. Environ Microbiol Rep. 2009;1:184–190. doi: 10.1111/j.1758-2229.2009.00029.x. [DOI] [PubMed] [Google Scholar]
- Amann RI, Ludwig W, Schleifer KH. Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol Rev. 1995;59:143–169. doi: 10.1128/mr.59.1.143-169.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashelford KE, Chuzhanova NA, Fry JC, Jones AJ, Weightman AJ. At least 1 in 20 16S rRNA sequence records currently held in public repositories is estimated to contain substantial anomalies. Appl Environ Microbiol. 2005;71:7724–7736. doi: 10.1128/AEM.71.12.7724-7736.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behrens S, Ruhland C, Inacio J, Huber H, Fonseca A, Spencer-Martins I, et al. In situ accessibility of small-subunit rRNA of members of the domains Bacteria, Archaea, and Eucarya to Cy3-labeled oligonucleotide probes. Appl Environ Microbiol. 2003;69:1748–1758. doi: 10.1128/AEM.69.3.1748-1758.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blöthe M, Roden EE. Composition and Activity of an Autotrophic Fe(II)-Oxidizing, Nitrate-Reducing Enrichment Culture. Appl Environ Microbiol. 2009;75:6937–6940. doi: 10.1128/AEM.01742-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brümmer IH, Felske A, Wagner-Dobler I. Diversity and seasonal variability of Betaproteobacteria in biofilms of polluted rivers: Analysis by temperature gradient gel electrophoresis and cloning. Appl Environ Microbiol. 2003;69:4463–4473. doi: 10.1128/AEM.69.8.4463-4473.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camargo JA, Alonso Á . Ecological and toxicological effects of inorganic nitrogen pollution in aquatic ecosystems: A global assessment. Environ Int. 2006;32:831–849. doi: 10.1016/j.envint.2006.05.002. [DOI] [PubMed] [Google Scholar]
- Chen Y, Wu L, Boden R, Hillebrand A, Kumaresan D, Moussard H, et al. Life without light: Microbial diversity and evidence of sulfur- and ammonium-based chemolithotrophy in Movile Cave. ISME J. 2009;3:1093–1104. doi: 10.1038/ismej.2009.57. [DOI] [PubMed] [Google Scholar]
- Conley DJ, Paerl HW, Howarth RW, Boesch DF, Seitzinger SP, Havens KE, et al. ECOLOGY: Controlling Eutrophication: Nitrogen and Phosphorus. Science. 2009;323:1014–1015. doi: 10.1126/science.1167755. [DOI] [PubMed] [Google Scholar]
- Daims H, Bruhl A, Amann R, Schleifer KH, Wagner M. The domain-specific probe EUB338 is insufficient for the detection of all Bacteria: Development and evaluation of a more comprehensive probe set. Syst Appl Microbiol. 1999;22:434–444. doi: 10.1016/S0723-2020(99)80053-8. [DOI] [PubMed] [Google Scholar]
- Daims H, Nielsen JL, Nielsen PH, Schleifer KH, Wagner M. In situ characterization of Nitrospira-like nitrite-oxidizing bacteria active in wastewater treatment plants. Appl Environ Microbiol. 2001a;67:5273–5284. doi: 10.1128/AEM.67.11.5273-5284.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daims H, Purkhold U, Bjerrum L, Arnold E, Wilderer PA, Wagner M. Nitrification in sequencing biofilm batch reactors: Lessons from molecular approaches. Water Sci Technol. 2001b;43:9–18. [PubMed] [Google Scholar]
- Daims H, Stoecker K, Wagner M. Fluorescence in situ hybridization for the detection of prokaryotes. In: Osborn AM, Smith CJ, editors. Molecular Microbial Ecology. Taylor & Francis Group; New York: 2005. pp. 213–239. [Google Scholar]
- Daims H, Lücker S, Wagner M. daime, a novel image analysis program for microbial ecology and biofilm research. Environ Microbiol. 2006;8:200–213. doi: 10.1111/j.1462-2920.2005.00880.x. [DOI] [PubMed] [Google Scholar]
- Daims H, Wagner M. Quantification of uncultured microorganisms by fluorescence microscopy and digital image analysis. Appl Microbiol Biotechnol. 2007;75:237–248. doi: 10.1007/s00253-007-0886-z. [DOI] [PubMed] [Google Scholar]
- Diaz RJ, Rosenberg R. Spreading Dead Zones and Consequences for Marine Ecosystems. Science. 2008;321:926–929. doi: 10.1126/science.1156401. [DOI] [PubMed] [Google Scholar]
- Flynn TM, Sanford RA, Ryu H, Bethke CM, Levine AD, Ashbolt NJ, et al. Functional microbial diversity explains groundwater chemistry in a pristine aquifer. BMC Microbiol. 2013;13:146. doi: 10.1186/1471-2180-13-146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foesel BU, Gieseke A, Schwermer C, Stief P, Koch L, Cytryn E, et al. Nitrosomonas Nm143-like ammonia oxidizers and Nitrospira marina-like nitrite oxidizers dominate the nitrifier community in a marine aquaculture biofilm. FEMS Microbiol Ecol. 2008;63:192–204. doi: 10.1111/j.1574-6941.2007.00418.x. [DOI] [PubMed] [Google Scholar]
- Garrity GM, Bell JA, Lilburn T. Family III. Gallionellaceae Henrici and Johnson 1935b. In: Brenner DY, Krieg NR, Staley JT, editors. Bergey’s Manual of Systematic Bacteriology, Second Edition. Springer; New York: 2005. pp. 880–886. [Google Scholar]
- Gujer W. Nitrification and me - A subjective review. Water Res. 2010;44:1–19. doi: 10.1016/j.watres.2009.08.038. [DOI] [PubMed] [Google Scholar]
- Henrici AT, Johnson DE. Studies of Freshwater Bacteria: II. Stalked Bacteria, a New Order of Schizomycetes. J Bacteriol. 1935;30:61–93. doi: 10.1128/jb.30.1.61-93.1935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henze M, Harremoes P, Jansen JLC, Arvin E. Wastwater Treatment - Biological and Chemical Processes. 2nd ed. Springer; Berlin: 1997. [Google Scholar]
- Holm N. In: Process for the discontinuous purification and installation for carrying out this process. Office EP, editor. Germany: 2003. p. 13. [Google Scholar]
- Hoshino T, Yilmaz LS, Noguera DR, Daims H, Wagner M. Quantification of target molecules needed to detect microorganisms by fluorescence in situ hybridization (FISH) and catalyzed reporter deposition-FISH. Appl Environ Microbiol. 2008;74:5068–5077. doi: 10.1128/AEM.00208-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji Z, Chen Y. Using sludge fermentation liquid to improve wastewater short-cut nitrification-denitrification and denitrifying phosphorus removal via nitrite. Environ Sci Technol. 2010;44:8957–8963. doi: 10.1021/es102547n. [DOI] [PubMed] [Google Scholar]
- Juretschko S, Timmermann G, Schmid M, Schleifer KH, Pommerening-Roser A, Koops HP, et al. Combined molecular and conventional analyses of nitrifying bacterium diversity in activated sludge: Nitrosococcus mobilis and Nitrospira-like bacteria as dominant populations. Appl Environ Microbiol. 1998;64:3042–3051. doi: 10.1128/aem.64.8.3042-3051.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kane MD, Poulsen LK, Stahl DA. Monitoring the enrichment and isolation of sulfate-reducing bacteria by using oligonucleotide hybridization probes designed from environmentally derived 16S rRNA sequences. Appl Environ Microbiol. 1993;59:682–686. doi: 10.1128/aem.59.3.682-686.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong Y, Xia Y, Nielsen JL, Nielsen PH. Structure and function of the microbial community in a full-scale enhanced biological phosphorus removal plant. Microbiology. 2007;153:4061–4073. doi: 10.1099/mic.0.2007/007245-0. [DOI] [PubMed] [Google Scholar]
- Kwon S, Kim TS, Yu GH, Jung JH, Park HD. Bacterial community composition and diversity of a full-scale integrated fixed-film activated sludge system as investigated by pyrosequencing. J Microbiol Biotechnol. 2010;20:1717–1723. [PubMed] [Google Scholar]
- Lebedeva EV, Alawi M, Maixner F, Jozsa PG, Daims H, Spieck E. Physiological and phylogenetic characterization of a novel lithoautotrophic nitrite-oxidizing bacterium, “Candidatus Nitrospira bockiana”. Int J Syst Evol Microbiol. 2008;58:242–250. doi: 10.1099/ijs.0.65379-0. [DOI] [PubMed] [Google Scholar]
- Lee N, Nielsen PH, Andreasen KH, Juretschko S, Nielsen JL, Schleifer KH, et al. Combination of fluorescent in situ hybridization and microautoradiography - a new tool for structure-function analyses in microbial ecology. Appl Environ Microbiol. 1999;65:1289–1297. doi: 10.1128/aem.65.3.1289-1297.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li D, Qi R, Yang M, Zhang Y, Yu T. Bacterial community characteristics under long-term antibiotic selection pressures. Water Res. 2011;45:6063–6073. doi: 10.1016/j.watres.2011.09.002. [DOI] [PubMed] [Google Scholar]
- Liu Z, Huang S, Sun G, Xu Z, Xu M. Phylogenetic diversity, composition and distribution of bacterioplankton community in the Dongjiang River, China. FEMS Microbiol Ecol. 2012;80:30–44. doi: 10.1111/j.1574-6941.2011.01268.x. [DOI] [PubMed] [Google Scholar]
- Lücker S, Wagner M, Maixner F, Pelletier E, Koch H, Vacherie B, et al. A Nitrospira metagenome illuminates the physiology and evolution of globally important nitrite-oxidizing bacteria. PNAS. 2010;107:13479–13484. doi: 10.1073/pnas.1003860107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludwig W, Amann R, Martinez-Romero E, Schönhuber W, Bauer S, Neef A, et al. rRNA based identification and detection systems for Rhizobia and other bacteria. Plant Soil. 1998;204:1–19. [Google Scholar]
- Ludwig W, Strunk O, Westram R, Richter L, Meier H, Yadhukumar, et al. ARB: a software environment for sequence data. Nucleic Acids Res. 2004;32:1363–1371. doi: 10.1093/nar/gkh293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maestre JP, Rovira R, Gamisans X, Kinney KA, Kirisits MJ, Lafuente J, et al. Characterization of the bacterial community in a biotrickling filter treating high loads of H2S by molecular biology tools. Water Sci Technol. 2009;59:1331–1337. doi: 10.2166/wst.2009.111. [DOI] [PubMed] [Google Scholar]
- Maixner F, Noguera DR, Anneser B, Stoecker K, Wegl G, Wagner M, et al. Nitrite concentration influences the population structure of Nitrospira-like bacteria. Environ Microbiol. 2006;8:1487–1495. doi: 10.1111/j.1462-2920.2006.01033.x. [DOI] [PubMed] [Google Scholar]
- Maixner F, Wagner M, Lücker S, Pelletier E, Schmitz-Esser S, Hace K, et al. Environmental genomics reveals a functional chlorite dismutase in the nitrite-oxidizing bacterium “Candidatus Nitrospira defluvii”. Environ Microbiol. 2008;10:3043–3056. doi: 10.1111/j.1462-2920.2008.01646.x. [DOI] [PubMed] [Google Scholar]
- Martiny JBH, Eisen JA, Penn K, Allison SD, Horner-Devine MC. Drivers of bacterial β-diversity depend on spatial scale. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:7850–7854. doi: 10.1073/pnas.1016308108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Na H, Kim OS, Yoon SH, Kim Y, Chun J. Comparative approach to capture bacterial diversity of coastal waters. J Microbiol. 2011;49:729–740. doi: 10.1007/s12275-011-1205-z. [DOI] [PubMed] [Google Scholar]
- Okabe S, Satoh H, Watanabe Y. In situ analysis of nitrifying biofilms as determined by in situ hybridization and the use of microelectrodes. Appl Environ Microbiol. 1999;65:3182–3191. doi: 10.1128/aem.65.7.3182-3191.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Percent SF, Frischer ME, Vescio PA, Duffy EB, Milano V, McLellan M, et al. Bacterial community structure of acid-impacted lakes: What controls diversity? Appl Environ Microbiol. 2008;74:1856–1868. doi: 10.1128/AEM.01719-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prosser JI. Autotrophic nitrification in bacteria. Adv Microb Physiol. 1989;30:125–181. doi: 10.1016/s0065-2911(08)60112-5. [DOI] [PubMed] [Google Scholar]
- Quast C, Pruesse E, Yilmaz P, Gerken J, Schweer T, Yarza P, et al. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 2013;41:D590–D596. doi: 10.1093/nar/gks1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronquist F, Huelsenbeck JP. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19:1572–1574. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
- Sattin SR, Cleveland CC, Hood E, Reed SC, King AJ, Schmidt SK, et al. Functional shifts in unvegetated, perhumid, recently-deglaciated soils do not correlate with shifts in soil bacterial community composition. J Microbiol. 2009;47:673–681. doi: 10.1007/s12275-009-0194-7. [DOI] [PubMed] [Google Scholar]
- Schmid M, Twachtmann U, Klein M, Strous M, Juretschko S, Jetten M, et al. Molecular evidence for genus level diversity of bacteria capable of catalyzing anaerobic ammonium oxidation. Syst Appl Microbiol. 2000;23:93–106. doi: 10.1016/S0723-2020(00)80050-8. [DOI] [PubMed] [Google Scholar]
- Schott J, Griffin BM, Schink B. Anaerobic phototrophic nitrite oxidation by Thiocapsa sp. strain KS1 and Rhodopseudomonas sp. strain LQ17. Microbiology. 2010;156:2428–2437. doi: 10.1099/mic.0.036004-0. [DOI] [PubMed] [Google Scholar]
- Schramm A, De Beer D, Wagner M, Amann R. Identification and activities in situ of Nitrosospira and Nitrospira spp. as dominant populations in a nitrifying fluidized bed reactor. Appl Environ Microbiol. 1998;64:3480–3485. doi: 10.1128/aem.64.9.3480-3485.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schramm A, de Beer D, van den Heuvel JC, Ottengraf S, Amann R. Microscale distribution of populations and activities of Nitrosospira and Nitrospira spp. along a macroscale gradient in a nitrifying bioreactor: Quantification by in situ hybridization and the use of microsensors. Appl Environ Microbiol. 1999;65:3690–3696. doi: 10.1128/aem.65.8.3690-3696.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schramm A, Fuchs BM, Nielsen JL, Tonolla M, Stahl DA. Fluorescence in situ hybridization of 16S rRNA gene clones (Clone-FISH) for probe validation and screening of clone libraries. Environ Microbiol. 2002;4:713–720. doi: 10.1046/j.1462-2920.2002.00364.x. [DOI] [PubMed] [Google Scholar]
- Schwarz JI, Eckert W, Conrad R. Community structure of Archaea and Bacteria in a profundal lake sediment Lake Kinneret (Israel) Syst Appl Microbiol. 2007;30:239–254. doi: 10.1016/j.syapm.2006.05.004. [DOI] [PubMed] [Google Scholar]
- Skerman VBD, McGowan V, Sneath PHA. Approved lists of bacterial names. Int J Syst Bacteriol. 1980;30:225–420. [PubMed] [Google Scholar]
- Sorokin DY, Vejmelkova D, Luecker S, Streshinskaya GM, Rijpstra I, Sinninghe Damsté J, et al. Nitrolancea hollandica gen. nov., sp. nov., a chemolithoautotrophic nitrite-oxidizing bacterium from a bioreactor belonging to the phylum Chloroflexi. Int J Syst Evol Microbiol. 2014;64:1859–1865. doi: 10.1099/ijs.0.062232-0. doi 10.1099/ijs.1090.062232-062230. [DOI] [PubMed] [Google Scholar]
- Stein LY, Arp DJ. Loss of ammonia monooxygenase activity in Nitrosomonas europaea upon exposure to nitrite. Appl Environ Microbiol. 1998;64:4098–4102. doi: 10.1128/aem.64.10.4098-4102.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamminen M, Karkman A, Corander J, Paulin L, Virta M. Differences in bacterial community composition in Baltic Sea sediment in response to fish farming. Aquaculture. 2011;313:15–23. [Google Scholar]
- Wagner M, Rath G, Koops H-P, Flood J, Amann R. In situ analysis of nitrifying bacteria in sewage treatment plants. Water Sci Technol. 1996;34:237–244. [Google Scholar]
- Wagner M, Loy A, Nogueira R, Purkhold U, Lee N, Daims H. Microbial community composition and function in wastewater treatment plants. Antonie Leeuwenhoek. 2002;81:665–680. doi: 10.1023/a:1020586312170. [DOI] [PubMed] [Google Scholar]
- Weiss J, Rentz J, Plaia T, Neubauer S, Merrill-Floyd M, Lilburn T, et al. Characterization of Neutrophilic Fe(II)-Oxidizing Bacteria Isolated from the Rhizosphere of Wetland Plants and Description of Ferritrophicum radicicola gen. nov. sp. nov., and Sideroxydans paludicola sp. nov. Geomicrobiol J. 2007;24:559–570. [Google Scholar]
- White CP, Debry RW, Lytle DA. Microbial survey of a full-scale, biologically active filter for treatment of drinking water. Appl Environ Microbiol. 2012;78:6390–6394. doi: 10.1128/AEM.00308-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.