SUMMARY
All plants are inhabited internally by diverse microbial communities comprising bacterial, archaeal, fungal, and protistic taxa. These microorganisms showing endophytic lifestyles play crucial roles in plant development, growth, fitness, and diversification. The increasing awareness of and information on endophytes provide insight into the complexity of the plant microbiome. The nature of plant-endophyte interactions ranges from mutualism to pathogenicity. This depends on a set of abiotic and biotic factors, including the genotypes of plants and microbes, environmental conditions, and the dynamic network of interactions within the plant biome. In this review, we address the concept of endophytism, considering the latest insights into evolution, plant ecosystem functioning, and multipartite interactions.
INTRODUCTION
Endophytes are microorganisms that spend at least parts of their life cycle inside plants. Endophyte definitions have changed in the past years and expectedly will evolve further over the coming years. The term “endophyte” has commonly been used for fungi living inside plants, but later researchers realized that interior parts of plants could be colonized by bacteria as well (1, 2). Plants do not live alone as single entities but closely associate with the microorganisms present in their neighborhood, and especially with those living internally. The emergence of the concept of the “plant microbiome,” i.e., the collective genomes of microorganisms living in association with plants, has led to new ideas on the evolution of plants where selective forces do not act merely on the plant genome itself but rather on the whole plant, including its associated microbial community. Lamarckian concepts of acquired heritable traits may be explained via the hologenome concept by vertical transmission of valuable traits provided by endophytes to plants (3).
The most common definition of endophytes is derived from the practical description given in 1997 by Hallmann and coauthors (2), who stated that endophytes are “…those (bacteria) that can be isolated from surface-disinfested plant tissue or extracted from within the plant, and that do not visibly harm the plant.” This definition has been valid for cultivable species in most laboratories in the world over the past 2 decades. However, due to the suspected lack of adequate elimination of nucleic acids after disinfection of plant surfaces, this definition appeared to be less suitable for noncultured species upon the introduction of molecular detection techniques in endophyte research (4).
Conceptual aspects related to the nature of endophytes are under dispute. For instance, must plant pathogens be considered endophytes or not, even when they have lost their virulence (5)? Recently, a typical bacterial group of endophytes beneficial to plants, the group of fluorescent pseudomonads, turned out to be detrimental to leatherleaf ferns under specific conditions (6). This indicates that potential plant mutualists can become deleterious for their hosts. Endophytes should not be harmful to the plant host, but what about harmfulness to other species, for instance, when particular bacteria that colonize internal compartments of plants are harmful to humans (7)?
The most common endophytes are typed as commensals, with unknown or yet unknown functions in plants, and less common ones are those shown to have positive (mutualistic) or negative (antagonistic) effects on plants (2). However, these properties are often tested in a single plant species or within groups of closely related plant genotypes, but rarely over a taxonomically wide spectrum of plant species. Also, the environmental conditions wherein plant-endophyte interactions are studied are often rather narrow. Furthermore, interactions between members of the endophyte community have rarely been investigated. A few studies demonstrated that interactions between taxonomically related microbial endophytes can shift whole populations inside the plant (8, 9). Bacterial and fungal endophytic communities are commonly investigated separately, but the interaction between both groups inside plants can become a fascinating new field in endophyte research (10).
Studies of plant-endophyte interactions are commonly based on controlled, optimized conditions for growth of host plants and seldom based on variable, field-realistic conditions. Effects ascribed to endophytes in healthy plants might change when host plants are grown under less favorable, or even stressful, conditions. In conclusion, our current understanding of endophytes is built on a rather small set of experimental conditions, and more varied experimental settings would be required for deeper insight into endophyte functioning. Because of this and the general preference to investigate microbial species that are relatively easy to cultivate, our knowledge of the ecology and interactions of endophytes in plants is still biased.
New developments in high-throughput technologies, such as next-generation sequencing, permit the investigation of complex microbiomes and will facilitate larger sample sizes and encourage deeper analyses of microbial communities (11). The new “omics” approaches are valuable tools for exploring, identifying, and characterizing the contributions of genetic and metabolic elements involved in the interactions between host plants and endophytes. For instance, metagenome sequencing has revealed important functions required for survival of bacterial endophytes inside plants (12), and metabolome analysis demonstrated the effects of beneficial endophytes on primary metabolites of plants (13). The combination of cultivation-independent and improved cultivation technologies will allow the exploration of hitherto uncultured groups living in association with plants (14, 15). In addition, the locations of endophytes in different plant compartments are disputable (16), but powerful image analyses can provide information about the exact colocalization within plant tissues and about physical contacts between different microbial groups (17–19). We are reaching a pivotal point in our perception of endophytes, and we expect that technical innovations in microbial detection will soon drastically change our concepts of endophytes as living entities colonizing internal plant compartments.
In this paper, we present a historical overview of the endophyte research leading to the current understanding of endophytes. The state of science for defined groups of endophytes is described in succeeding sections, based on the vast number of peer-reviewed publications on endophytes, which have been growing exponentially over the last 3 decades. Furthermore, we elaborate the expected impacts of novel technologies on endophyte research. It is our purpose to revisit current concepts on endophytes and to assess directions for new research on microbial endophytes based on the latest technological developments.
HISTORY OF ENDOPHYTE DEFINITIONS
The German botanist Heinrich Friedrich Link was the first to describe endophytes, in 1809 (20). At that time, they were termed “Entophytae” and were described as a distinct group of partly parasitic fungi living in plants. Since then, many definitions have evolved; for a long time, they mostly addressed pathogens or parasitic organisms, primarily fungi (21–23). Only Béchamp described so-called microzymas in plants, referring to microorganisms (24). Generally, in the 19th century, the belief was that healthy or normally growing plants are sterile and thus free of microorganisms (postulated by Pasteur; cited in reference 25). Nevertheless, Galippe reported the occurrence of bacteria and fungi in the interior of vegetable plants and postulated that these microorganisms derive from the soil environment and migrate into the plant, where they might play a beneficial role for the host plant (26, 27). Other studies in the late 19th century and the beginning of the 20th century confirmed the occurrence of beneficial microorganisms within plants (28, 29). Nevertheless, contrasting views on the existence of plant-beneficial endophytes prevailed at that time (28, 30–34). Nowadays, it is a well-established fact that plants are hosts for many types of microbial endophytes, including bacteria, fungi, archaea, and unicellular eukaryotes, such as algae (35) and amoebae (36).
An important discovery was made in 1888 by the Dutch microbiologist Martinus Willem Beijerinck, who isolated root nodule bacteria in pure culture from nodules of Leguminosae plants and showed that these isolates, which were later classified as Rhizobium leguminosarum (37), were capable of fixing atmospheric nitrogen (38). At the same time, Hermann Hellriegel and Hermann Wilfarth reported mineral N independence of leguminous plants, as well as the importance of symbiotic nitrogen fixation by rhizobia (39). Albert Bernhard Frank reported another important mutualistic symbiosis, i.e., the living together of unlike organisms (40), between roots of trees and underground fungi (41). He coined the term “mycorrhiza” to describe the interaction, which literally means “fungus roots.”
More recently, in 1991, Orlando Petrini defined endophytes as “all organisms inhabiting plant organs that at some time in their life cycle can colonize internal plant tissues without causing apparent harm to their host” (42). Since then, many definitions have been formulated (2, 43–48), essentially all pertaining to microorganisms which for all or part of their life cycle invade tissues of living plants without causing disease. Although this endophyte definition has been the basis of many studies and might be a pragmatic approach to distinguish between endophytes and pathogens, it has some drawbacks and raises some questions.
First, this definition is more suitable for cultivated endophytes, as only with those is it possible to assess phytopathogenicity. However, in most cases, pathogenicity assays are not performed, or they are performed with only one plant species, although pathogenicity might occur with a different plant genotype or under different conditions. Second, it is well known that some bacteria may live as latent pathogens within plants and become pathogenic under specific conditions (6) or are pathogens of other plants. Third, it has been shown that bacterial strains belonging to a well-known pathogenic species of a specific plant host may even have growth-promoting effects on another plant (49, 50). These findings demonstrate that it is not trivial to clearly distinguish a nonpathogenic endophyte from a pathogen and that properties such as pathogenicity or mutualism may depend on many factors, including plant and microbial genotype, microbial numbers, and quorum sensing or environmental conditions. With cultivation-independent analyses, it is now even more difficult to assess the pathogenicity of individual microbiome members. In conclusion, we question the currently applied definition of endophytes and claim that the term “endophyte” should refer to habitat only, not function, and therefore that the term should be more general and include all microorganisms which for all or part of their lifetime colonize internal plant tissues.
PLANT-MICROBE SYMBIOSES
Different groups of bacteria and fungi interact with higher plants. Genetic links between the association of plants with arbuscular mycorrhizal fungi (AMF) and root nodule symbioses have been found (51–53), suggesting that at least segments of bacterial and fungal endophytic populations coevolved with each other and with their host. Mutualistic interactions leading to adaptive benefits for both partners occasionally evolved to even more complex forms, in which more than two partners were involved (10).
Evolution of Plant-Fungus Symbioses
Plant-fungus symbioses are known to have occurred during early colonization of land by terrestrial plants (54). The fungal group Glomeromycota has for a long time been the prime candidate for interaction with the first terrestrial plants, in the Ordovician era, but members of the Mucoromycotina are also speculated to have had symbiotic interactions with the first terrestrial plants (55). The association between AMF and plants evolved as a symbiosis, facilitating the adaption of plants to the terrestrial environment (56). The oldest known fossils representing terrestrial fungi with properties similar to those of AMF were collected from dolomite rocks in Wisconsin and are estimated to be 460 million years old, originating from the Ordovician period (54). It was therefore assumed that terrestrial AMF already existed at the time when bryophyte-like, “lower” plants covered the land. All other plant-AMF interaction types, such as ectomycorrhiza and orchid and ericoid mycorrhiza, appeared later and are considered to be derived from the first interactions between AMF and the first terrestrial plants (57).
It is assumed that no tight interactions between plants and fungi occurred initially but that, due to nutritional limitations, interactions between both partners evolved (57). It is still unknown whether the first AMF were already mutualistic symbionts or whether mutualistic lifestyles evolved from pathogenic forms. The internal spaces of plants became important habitats for plant-colonizing fungi. Specific tissue layers, such as the endodermis and exodermis, evolved, forming the borders of cortex cells surrounding fungi internalized in the roots (57). This finally resulted in the formation of arbuscules, which are typical structures in plant-AMF interactions. AMF became more dependent on their host for energy sources and adopted an obligate life cycle. On the other hand, intraradical hyphae increased the total root surface area of the host plant, allowing substantially more nutrient (P) uptake from the soil environment. As evolution progressed, more extreme forms of plant-fungus interactions appeared, such as mycoheterotrophic plants, i.e., plants that fully exploit their fungal counterparts during interaction (58).
Evolution of Plant-Bacterium Symbioses
The best-described plant-bacterium interaction is the one between leguminous plants and rhizobia. The interactions of nitrogen-fixing bacteria belonging to the genera Azorhizobium, Bradyrhizobium, Ensifer, Mesorhizobium, Rhizobium, and Sinorhizobium (collectively called “rhizobia”; for a full list of genera, see http://www.rhizobia.co.nz/taxonomy/rhizobia) are capable of inducing differentiation in root nodule structure, as demonstrated in Fabaceae and Parasponia plants (60). Typical symptoms in roots of leguminous plants infected by rhizobia are curling of root hairs and the appearance of infection threads and, finally, nodule primordia in the inner root layers—these are all processes mediated by signal exchange between plants and rhizobia (for a review, see reference 61). In primordium cells, the bacteria become surrounded by the plant membrane, and together, the bacteria and plant structure form the symbiosome, in which atmospheric nitrogen is fixed and transferred in exchange for carbohydrates (62). Symbiosomes have a structure similar to that of mycorrhizal arbuscules, which are also surrounded by a plant membrane. It is interesting that a number of legume-nodulating rhizobial strains form endophytic associations with monocotyledonous plants, such as rice (63), maize (64), and sugarcane (65), and dicotyledonous plants, such as sweet potato (66). Although nodule primordia were not observed, rhizobial nifH transcripts were found inside roots of rice and sugarcane plants (12, 65). The contribution of rhizobium-assimilated nitrogen to the total nitrogen pool in nonleguminous plants is still a matter of debate (67).
Recent studies revealed that the nature of the association of both AMF and rhizobia with host plant species can be mutualistic, parasitic, or nonsymbiotic (68, 69). A meta-analysis demonstrated that the plant response to AMF depends on various factors, most importantly the host plant type and N fertilization (69). Apart from mutualistic rhizobia, parasitic strains which infect legumes but fix little or no nitrogen have been reported (68). The rhizobium-legume symbiosis seems to be characterized by a continuum of different types of symbiotic interactions, in many cases dependent on the presence of symbiotic genes, frequently located on plasmids, needed for the mutualistic interaction (70).
ENDOPHYTE DIVERSITY
Prokaryotic Endophytes
We present an overview of prokaryotic endophytes reported to date, based on a curated database (see Data Sets S1 and S2 in the supplemental material) comprising all currently available 16S rRNA gene sequences assigned to endophytes (published in peer-reviewed journals indexed to the PubMed or Web of Science databases and deposited in the International Nucleotide Sequence Database Collaboration repository, as of 1 March 2014). Only sequences longer than 300 bp and from studies that applied well-established surface sterilization procedures, such as the application of sodium hypochlorite (NaOCl) or mercury chloride (HgCl2), were included. The database comprises 4,146 16S rRNA gene sequences from isolates (56%) and 3,202 16S rRNA gene sequences from uncultured organisms (44%). Sequences from earlier next-generation high-throughput sequencing technologies (e.g., 454 pyrosequencing) were able to produce only relatively short nucleotide stretches (i.e., <300 bp), which limits the discriminatory power for classification of different taxonomic groups, and thus were not included in our database.
Prokaryotic endophytes considered in this database are diverse and comprise 23 recognized and candidate phyla (2 from Archaea and 21 from Bacteria) (Table 1; see Fig. S1 in the supplemental material). Despite this remarkable diversity, more than 96% of the total number of endophytic prokaryotic sequences (n = 7,348) are distributed among four bacterial phyla (54% Proteobacteria, 20% Actinobacteria, 16% Firmicutes, and 6% Bacteroidetes). These phyla have also been reported to be dominant in the plant environment (71, 72). The database comprises only a few (n = 29) sequences from Archaea, which were mainly detected in coffee cherries (73), rice and maize roots (74, 75), and the arctic tundra rush Juncus trifidus (76).
TABLE 1.
Summary of the endophytic data set from all peer-reviewed publications with prokaryotic 16S rRNA gene sequencesa
| Phylogenetic affiliationb | No. of sequences | % of sequences |
|---|---|---|
| Bacteria | 7,319 | |
| Acidobacteria | 53 | 0.72 |
| Actinobacteria | 1,461 | 19.88 |
| Armatimonadetes | 6 | 0.08 |
| Bacteroidetes | 462 | 6.29 |
| GOUTA4c | 1 | 0.01 |
| ODc | 6 | 0.08 |
| TM7c | 2 | 0.03 |
| Chlamydiae | 8 | 0.11 |
| Chlorobi | 5 | 0.07 |
| Chloroflexi | 3 | 0.04 |
| Cyanobacteria | 102 | 1.39 |
| Deinococcus-Thermus | 7 | 0.1 |
| Elusimicrobia | 1 | 0.01 |
| Firmicutes | ||
| Bacilli | 1,132 | 15.41 |
| Clostridia | 68 | 0.93 |
| Fusobacteria | 3 | 0.04 |
| Nitrospirae | 3 | 0.04 |
| Planctomycetes | 5 | 0.07 |
| Proteobacteria | ||
| Alpha- | 1,337 | 18.2 |
| Beta- | 736 | 10.02 |
| Delta- | 26 | 0.35 |
| Epsilon- | 3 | 0.04 |
| Gamma- | 1,878 | 25.56 |
| Spirochaetae | 3 | 0.04 |
| Tenericutes | 2 | 0.03 |
| Verrucomicrobia | 6 | 0.08 |
| Archaea | 29 | |
| Euryarchaeota | 23 | 0.31 |
| Thaumarcheota | 6 | 0.08 |
| Total | 7,348 |
Endophytic sequences with >300 bp were retrieved from peer-reviewed manuscripts available in the ISI Web of Science and PubMed databases (as of 1 March 2014).
Based on comparison with the small-subunit rRNA SILVA database (version 115) (372) by using the SINA aligner (364).
Candidate division phyla.
Most of the prokaryotic endophytes (26%) could be assigned to the Gammaproteobacteria, including 56 recognized and 7 unidentified genera as well as the “Candidatus Portiera” genus (see Data Set S1 and Fig. S2 in the supplemental material). It should be noted that Gammaproteobacteria also comprise a large number of genera and species which are known as phytopathogens (77, 78). Endophytic Gammaproteobacteria are largely represented by a few genera: Pseudomonas, Enterobacter, Pantoea, Stenotrophomonas, Acinetobacter, and Serratia (>50 sequences each) (see Fig. S2). Members of the genus Enterobacter associate with diverse organisms, and their ecological relationships range from mutualism to pathogenesis. For instance, four species of Enterobacter in plants have been described as opportunistic pathogens, whereas many others (at least five) are beneficial to the host (79), including a monophyletic clade that was recently named Kosakonia (80). The nature of the interactions of other members of the Gammaproteobacteria, including Pseudomonas, Pantoea, and Stenotrophomonas species, is similar to that for Enterobacter, with few species described as plant pathogens and many others described as plant mutualists. Similarly, the Alphaproteobacteria encompass a large number (18%) of endophytic sequences, belonging to 57 recognized and 14 unidentified genera as well as the “Candidatus Liberibacter” genus (see Data Set S1 and Fig. S3). Most of the sequences can be assigned to the genera Rhizobium and Bradyrhizobium, known for their N2-fixing symbioses with legumes, and Methylobacterium and Sphingomonas (>50 sequences each) (see Fig. S3). Methylobacterium is capable of growth on methanol as the sole source of carbon and energy and has been hypothesized to potentially dominate the phyllosphere environment (81). The Betaproteobacteria sequences (10%) comprise 53 recognized and 10 unidentified genera of endophytes (see Data Set S1), mainly belonging to Burkholderia, Massilia, Variovorax, and Collimonas (≥40 sequences each) (see Fig. S4). Burkholderia strains have the potential to colonize a wide range of hosts and environments (82), suggesting a great metabolic and physiological adaptability of endophytes belonging to this genus.
Among Gram-positive endophytes, the class Actinobacteria (20%) comprises diverse endophytes belonging to 107 recognized and 15 unidentified genera (see Data Set S1 in the supplemental material). Most of the sequences group with the genera Streptomyces, Microbacterium, Mycobacterium, Arthrobacter, and Curtobacterium (>50 sequences each) (see Fig. S5). Members of the genus Streptomyces are well known for their capacity to synthesize antibiotic compounds (83). The class Bacilli (15%) comprises 25 recognized and 2 unidentified genera of endophytes (see Data Set S1). The genera Bacillus, Paenibacillus, and Staphylococcus have more than 100 sequences assigned to them (see Fig. S6). Within the genus Bacillus, the species Bacillus thuringiensis is well known for its production of parasporal crystal proteins with insecticidal properties (84).
Overall, most bacterial endophytes belong to mainly four phyla, but they encompass many genera and species. Their functions cannot be assigned clearly to taxonomy and seem to depend on the host and environmental parameters.
Eukaryotic Endophytes
A data set of eukaryotic endophytic full-length internal transcribed spacer (ITS) regions was also built for this study (see Data Set S3 in the supplemental material). A total of 8,439 sequences were retrieved from the National Center for Biotechnology Information (NCBI) nucleotide database (Table 2 shows the details of data retrieval and analysis; data were current as of 1 August 2014). Endophytes mainly belong to the Glomeromycota (40%), Ascomycota (31%), Basidiomycota (20%), unidentified phyla (8%), and, to a lesser extent, Zygomycota (0.1%) (Table 2).
TABLE 2.
Summary of the endophytic data set from all peer-reviewed eukaryotic full-length ITS sequences (as of 1 August 2014)a
| Taxonomic assignmentb | No. of sequences | % of sequences |
|---|---|---|
| Total | 8,439 | |
| Ascomycota | 2,610 | 30.92 |
| Archaeorhizomycetes | 2 | 0.02 |
| Dothideomycetes | 1,272 | 15.07 |
| Eurotiomycetes | 54 | 0.64 |
| Incertae sedis | 2 | 0.02 |
| Lecanoromycetes | 5 | 0.06 |
| Leotiomycetes | 171 | 2.03 |
| Orbiliomycetes | 0 | 0 |
| Pezizomycetes | 112 | 1.33 |
| Saccharomycetes | 11 | 0.13 |
| Sordariomycetes | 785 | 9.30 |
| Unidentified | 196 | 2.32 |
| Basidiomycota | 1,712 | 20.3 |
| Agaricomycetes | 1,560 | 18.49 |
| Atractiellomycetes | 26 | 0.31 |
| Cystobasidiomycetes | 3 | 0.04 |
| Exobasidiomycetes | 0 | 0 |
| Microbotryomycetes | 23 | 0.27 |
| Pucciniomycetes | 1 | 0.01 |
| Tremellomycetes | 30 | 0.36 |
| Ustilaginomycetes | 0 | 0 |
| Unidentified | 69 | 0.82 |
| Glomeromycota | 3,390 | 40.17 |
| Glomeromycetes | 3,294 | 39.03 |
| Unidentified | 96 | 1.14 |
| Zygomycota | ||
| Incertae sedis | 5 | 0.06 |
| Unidentified | 722 | 8.56 |
Fungal ITS sequences were retrieved from the NCBI nucleotide database by using the following search strings for the endophytic data set: “Endophyt*[ALL] AND nuccore_PubMed[Filter] AND internal[Title]” and “Mycorrhiza*[ALL] AND nuccore_PubMed[Filter] AND internal[Title].” Full-length ITS (ITS1, 5.8S, and ITS2 regions) sequences were extracted using ITSx (365) and assigned to operational taxonomic units (OTUs; definition set at 97% sequence similarity) with UCLUST (366), using the QIIME pipeline (367).
The phylum Glomeromycota only comprises endophytes known as arbuscular mycorrhizal fungi (AMF) (85) (see Data Set S3 in the supplemental material). Most of the eukaryotic endophytes (39%) can be assigned to the class Glomeromycetes. All members of this class form ubiquitous endosymbioses with most land plants and are of undeniable ecological and economic importance (86–88). AMF of the genera Glomus and Rhizophagus form obligate symbioses with a wide variety of host plants from the subkingdom Embryophyta (86). Among the Ascomycota, a large number of endophytes are identified in the class Dothideomycetes (15%). Besides endophytes, many members of the Dothideomycetes class are necrotrophic plant-pathogenic fungi, which are remarkable because of their production of host-specific toxins, such as phytotoxic metabolites and peptides that are biologically active only against a particular plant species (89–92). Overall, this class contains many species of the genera Alternaria and Epicoccum comprising endophytes (see Data Set S3). Although Alternaria brassicae is considered an opportunistic plant pathogen (93), it is frequently detected in high abundance in healthy plants (94, 95). Many members of the class Sordariomycetes (9%) are endophytes, such as species of the genera Balansia, Epichloë, Nemania, Xylaria, and Colletotrichum, but this class is also well known for phytopathogenic members, such as Cryphonectria parasitica (the causal agent of chestnut blight), Magnaporthe grisea (rice blast), Ophiostoma ulmi and Ophiostoma novo-ulmi (Dutch elm disease), and Fusarium, Verticillium, and Rosellinia species (96).
Among the Basidiomycota (Table 2), the class Agaricomycetes (18%) contains a large number of endophytes, mainly mushroom-forming (basidiome) fungi causing wood decay, white and brown rot saprotrophs, and the beneficial ectomycorrhiza (EMC) symbionts (97). Furthermore, members of the order Sebacinales form mycorrhizal symbioses with a broad range of plants, including woody plants and members of the families Orchidaceae and Ericaceae and the division Marchantiophyta (98). Additional assigned classes containing endophytes are Atractiellomycetes, Cystobasidiomycetes, Microbotryomycetes, and Tremellomycetes (see Data Set S3 in the supplemental material). Similar to the case for bacterial endophytes, various taxa comprise known phytopathogens and strains without known pathogenic effects, indicating that the functions of endophytic fungi also cannot necessarily be linked to taxonomy.
LIFESTYLES OF ENDOPHYTES
Degrees of Intimacy between Plants and Endophytes
Microorganisms can be strictly bound to plants and complete a major part or even their entire life cycle inside plants. Microorganisms requiring plant tissues to complete their life cycle are classified as “obligate.” Well-documented examples of obligate endophytes are found among mycorrhizal fungi and members of the fungal genera Balansia, Epichloë, and Neotyphodium, from the family Clavicipitaceae (Ascomycota) (99, 100). On the other extreme are “opportunistic” endophytes that mainly thrive outside plant tissues (epiphytes) and sporadically enter the plant endosphere (101). Among these are rhizosphere-competent colonizers, such as bacteria of the genera Pseudomonas and Azospirillum and fungi of the genera Hypocrea and Trichoderma (102–105). It is interesting that endophytes, which are transmitted vertically via seeds, are often recovered as epiphytes, suggesting that various endophytes might also colonize surrounding host plant environments (106, 107). Between these two extremes is an intermediate group, which comprises the vast majority of endophytic microorganisms, the so-called “facultative” endophytes. Whether facultative endophytes use the plant as a vector for dissemination or are actively selected by the host is still a matter of debate (107–110). However, facultative endophytes consume nutrients provided by plants, which would in fact reduce the ecological fitness of the host plant. This point is therefore often used as an argument that the so-called facultative endophytes must be mutualists in plants, even if the details of the interaction are unclear.
Overlaps exist between these three groups; thus, these categories must be considered “marking points” within the continuum of colonization strategies existing among endophytes. Independent of class, the microbial species thriving inside plant tissues are ecologically fit to survive and to proliferate under the local conditions of the plant interior, and aspects of survival are discussed later.
COLONIZATION OF THE ENDOSPHERE
Colonization Behavior of Fungal Endophytes
Successful colonization by endophytes depends on many variables, including plant tissue type, plant genotype, the microbial taxon and strain type, and biotic and abiotic environmental conditions. Different colonization strategies have been described for clavicipitaceous and nonclavicipitaceous endophytes (111, 112). Species of the Clavicipitaceae, including Balansia spp., Epichloë spp., and Claviceps spp., establish symbioses almost exclusively with grass, rush, and sledge hosts (47, 113), in which they may colonize the entire host plant systemically. They proliferate in the shoot meristem, colonizing intercellular spaces of the newly forming shoots, and can be transmitted vertically via seeds (113). Some Neotyphodium and Epichloë species may also be transmitted horizontally via leaf fragments falling on the soil (114). At the stage of inflorescence development, the mycelium of Neotyphodium can also colonize ovules and be present during infructescence development in the scutellum and the embryo, as demonstrated for Lolium perenne (115). When the inflorescence of the grass host develops, Epichloë can also grow over the developing inflorescence and form stromata, which can be differentiated sexually with the help of Botanophila flies (116).
Based on colonization characteristics, Rodriguez et al. (117) classified clavicipitaceous endophytes as class 1 fungal endophytes. Fungi colonizing above- and below-ground plant tissues, i.e., the rhizosphere, endorhiza, and aerial tissues (118), and being horizontally and/or vertically transmitted (119) were grouped as class 2 fungal endophytes (117). Class 3 endophytes were defined to contain mostly members of the Dikaryomycota (Ascomycota or Basidiomycota), which are particularly well studied in trees, but also in other plant taxa and in various ecosystems (120–126). Members of this class are mostly restricted to aerial tissues of various hosts and are horizontally transmitted (127, 128). Class 4 endophytes comprise dark, septate endophytes, which, similar to mycorrhizal fungi, are restricted to roots, where they reside inter- and/or intracellularly in the cortical cell layers (129).
Colonization Behavior of Bacterial Endophytes
Many bacterial endophytes originate from the rhizosphere environment, which attracts microorganisms due to the presence of root exudates and rhizodeposits (130, 131). Mercado-Blanco and Prieto (132) suggested that the entry of bacterial endophytes into roots occurs via colonization of root hairs. To a certain extent, stem and leaf surfaces also produce exudates that attract microorganisms (130). However, UV light, the lack of nutrients, and desiccation generally reduce colonization of plant surfaces, and only adapted bacteria can survive and enter the plant via stomata, wounds, and hydathodes (130, 133). Endophytes may also penetrate plants through flowers and fruits via colonization of the anthosphere and carposphere (18, 130).
Depending on the strain, various colonization routes have been described, and specific interactions have been suggested (133, 134). Several of these routes involve passive or active mechanisms enabling bacteria to migrate from the rhizoplane to the cortical cell layer, where the plant endodermis represents a barrier for further colonization (130, 135). For bacteria that can penetrate the endodermis, the xylem vascular system is the main transport route for systemic colonization of internal plant compartments (134), whereas others colonize intercellular spaces locally. Bacteria have been shown to colonize xylem vessels, and the sizes of the holes of the perforation plates between xylem elements are sufficiently large to allow bacterial passage (130, 134, 136–138). However, vertical spread of bacteria through plants may take several weeks (139), and it is unclear why bacterial endophytes progress so slowly in the vascular system. Bacteria might even migrate to reproductive organs of Angiospermae plants and have been detected in the inner tissues of flowers (epidermis and ovary), fruits (pulp), and seeds (tegument) of grapevines (18) and in pumpkin flowers (140), as well as in the pollen of pine, a Gymnospermae plant (141). Suitable niches for colonization by bacterial endophytes have been described for different plant taxonomic groups, including Bryophytes, Pteridophytes, Gymnospermae, and Angiospermae (17, 130, 142) (Fig. 1). Overall, it is not known whether endophytes need to reach a specific organ or tissue for optimal performance of the functions which have been identified for endophytes.
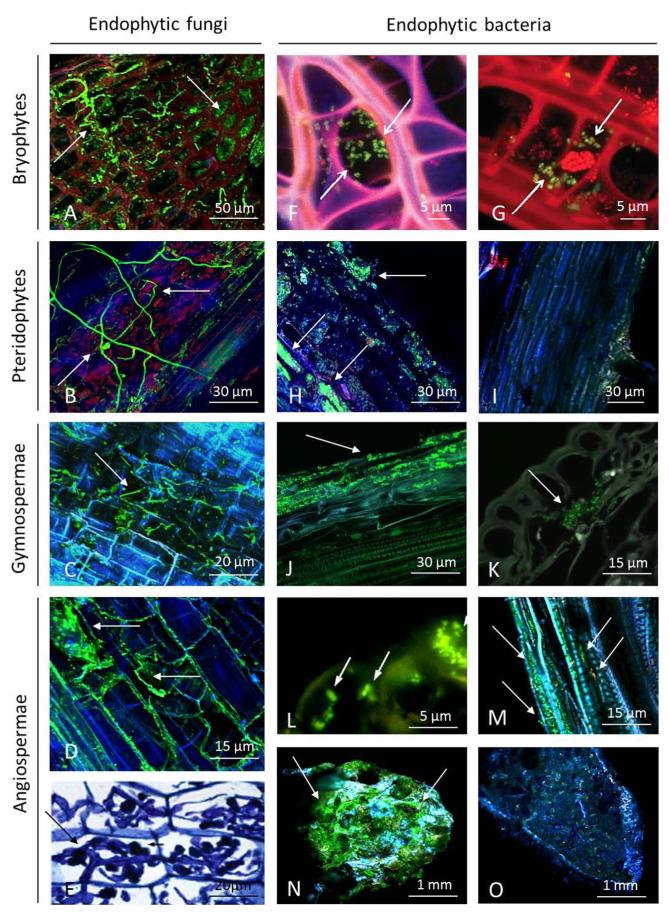
FIG 1.
Microphotographs of endophytes showing (arrows) endophytic fungi in Sphagnum sp. (Alex Fluor 488-wheat germ agglutinin [WGA]) (A), endophytic fungi in a fern stem (Alex Fluor 488-WGA) (B), endophytic fungi in a stem of a Pinus sp. (Alex Fluor 488-WGA) (C), fungal endophytes in a stolon of a Trifolium sp. (Alex Fluor 488-WGA) (D), and mycorrhiza colonizing Eleutherococcus sieboldianus (toluidine blue) (E). (F and G) Bacterial endophytes in Sphagnum magellanicum (fluorescence in situ hybridization [FISH] with probes targeting Alphaproteobacteria [F] and Planctomycetes [G]). (H and I) Bacterial endophytes in fern leaves (double labeling of oligonucleotide probes-fluorescence in situ hybridization [DOPE-FISH] with EUBMIX-FLUOS probe for all bacteria [H] and with NONEUB-FLUOS probe [I]). (J and K) Colonization of Scots pine seedling by green fluorescent protein-tagged Methylobacterium extorquens DSM13060. (L) Bacterial endophytes in flowers of grapevine plants (FISH with EUBMIX-Dylight488 and LGC-Dylight549 probes, targeting all bacteria and Firmicutes, respectively). (M) Bacterial endophytes in the xylem of grapevine plants (DOPE-FISH with EUBMIX-FLUOS and HGC69a-Cy5 probes, targeting all bacteria and Actinomycetes, respectively). (N and O) Bacterial endophytes in a nodule of Medicago lupulina (DOPE-FISH with EUBMIX-FLUOS probe targeting all bacteria [N] and with NONEUB-FLUOS probe [O]). (Panel E reprinted from reference 362. Panels F and G reprinted from reference 17 by permission from Macmillan Publishers Ltd. [copyright 2011]. Panels J and K reprinted from reference 369 with kind permission from Springer Science and Business Media. Panel L reprinted from reference 18 with kind permission from Springer Science and Business Media. Panel M reprinted from reference 363 by permission of the Society for Molecular Biology and Evolution.) All photographs show environmental samples, except those in panels J and K. Note that Alexa Fluor 488-WGA can also detect microbes other than fungi.
FUNCTIONS OF ENDOPHYTES
Some endophytes have no apparent effects on plant performance but live on the metabolites produced by the host. These are termed commensal endophytes, whereas other endophytes confer beneficial effects to the plant, such as protection against invading pathogens and (arthropod) herbivores, either via antibiosis or via induced resistance, and plant growth promotion (Fig. 2). A third group includes latent pathogens (143). Generally, endophytes can have neutral or detrimental effects to the host plant under normal growth conditions, whereas they can be beneficial under more extreme conditions or during different stages of the plant life cycle. For example, the fungus Fusarium verticillioides has a dual role both as a pathogen and as a beneficial endophyte in maize (144). The balance between these two states is dependent on the host genotype, but also on locally occurring abiotic stress factors that reduce host fitness, resulting in distortion of the delicate balance and in the occurrence of disease symptoms in the plant and production of mycotoxins by the fungus (144). However, beneficial effects have also been demonstrated, e.g., strains of the endophytic fungus F. verticillioides suppress the growth of another pathogenic fungus, Ustilago maydis, protecting their host against disease (145).
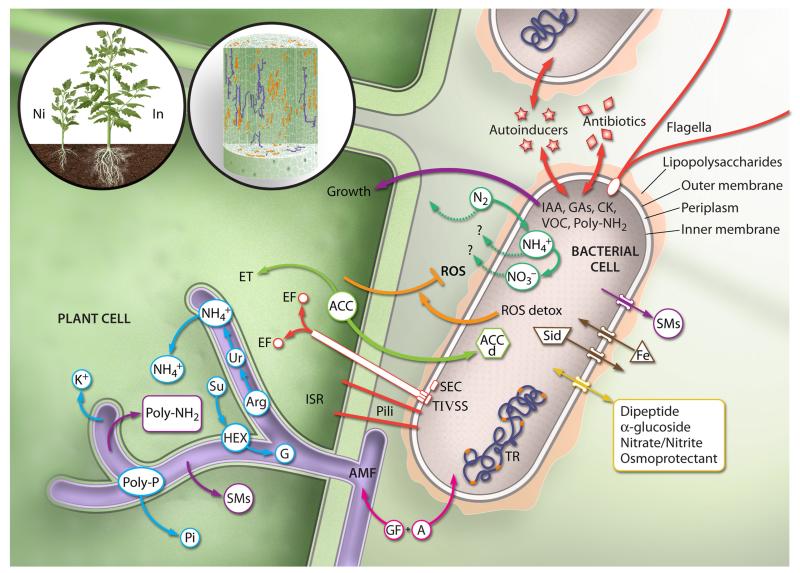
FIG 2.
Beneficial properties of endophytes. The left panel shows plants inoculated (In) with beneficial microorganisms that significantly improve plant growth compared to noninoculated (Ni) plants. Various microorganisms, in particular bacteria (orange) and fungi (purple), can colonize the internal tissues of the plant (middle panel). Once inside the plant, the endophytic bacteria and fungi interact intimately with the plant cells and with surrounding microorganisms (large panel). Endophytic fungi, represented here as arbuscular mycorrhizal fungi (AMF) (lilac), might form specialized structures, called arbuscules, where plant-derived carbon sources, mainly sucrose (Su), are exchanged for fungus-provided phosphate (Pi), nitrogen (NH4+), and potassium (K+) elements (blue). Plant cytoplasmic sucrose is transported to the periarbuscular space, where it is converted to hexose (HEX) to be assimilated by the fungus. Hexose is finally converted to glycogen (G) for long-distance transport. Phosphate and nitrogen are transported inside the fungal cytoplasm as polyphosphate granules (Poly-P), which are converted to Pi and arginine (Arg) in the arbuscule. Pi is transported to the host cytoplasm, whereas Arg is initially converted to urea (Ur) and then to ammonium (NH4+). Fungal and bacterial plant hormones, such as auxins (IAA), gibberellins (GAs), cytokinins (CKs), volatile organic compounds (VOCs), and polyamines (Poly-NH2), as well as secondary metabolites (SMs), are transferred to the host (violet). Various bacterial structures, such as flagella, pili, secretion system machineries (e.g., TIV SS and SEC), and lipopolysaccharides, as well as bacterium-derived proteins and molecules, such as effectors (EF), autoinducers, and antibiotics, are detected by the host cells and trigger the induced systemic resistance (ISR) response (red). ACC, the direct precursor of ethylene (ET), is metabolized by bacteria via the enzyme ACC deaminase (ACCd), thus ameliorating abiotic stress (light green). A range of reactive oxygen species detoxification (ROS detox) enzymes might also ameliorate the plant-induced stress (orange). Diazotrophic bacterial endophytes are capable of fixing atmospheric nitrogen (N2) and might actively transport NH4+ and nitrate (NO3−) to the host (dark green). Bacterial processes of siderophore production (Sid) and uptake (Fe) that are involved in plant growth promotion, biocontrol, and phytoremediation are shown in brown. Examples of various substrates on which the transmembrane proteins are enriched among endophytes are shown in yellow. Transcriptional regulators (TR) are also shown (orange). Communications and interactions between cells of microorganisms dwelling inside the plant tissues are promoted by growth factor (GF), antibiotic (A) (fuchsia), and autoinducer molecules.
Plant Growth Promotion and Protection against Biotic and Abiotic Stresses
ISR and production of antibiotic secondary metabolites
Carroll (111) suggested in 1988 that endophytes play a role in the defense systems of trees. Because life cycles of endophytes are considered to be much shorter than the life cycle of their host, they may evolve faster in their host, resulting in higher selection of antagonistic forms that contribute to resistance against short-living pathogens and herbivores. Later, in 1991, Carroll suggested that endophyte-mediated induced resistance occurs in Douglas fir trees (146). Endophytes may induce plant defense reactions, so-called induced systemic resistance (ISR), leading to a higher tolerance of pathogens (147, 148). There is increasing evidence that at an initial stage, interactions between beneficial microorganisms and plants trigger an immune response in plants similar to that against pathogens but that, later on, mutualists escape host defense responses and are able to successfully colonize plants (148). Bacterial strains of the genera Pseudomonas and Bacillus can be considered the most common groups inducing ISR (reviewed in references 149 and 150), although ISR induction is not exclusive to these groups (151, 152). Bacterial factors responsible for ISR induction were identified to include flagella, antibiotics, N-acylhomoserine lactones, salicylic acid, jasmonic acid, siderophores, volatiles (e.g., acetoin), and lipopolysaccharides (152, 153) (Fig. 2). The shoot endophyte Methylobacterium sp. strain IMBG290 was shown to induce resistance against the pathogen Pectobacterium atrosepticum in potato, in an inoculum-density-dependent manner (151). The observed resistance was accompanied by changes in the structure of the innate endophytic community. Endophytic community changes were shown to correlate with disease resistance, indicating that the endophytic community as a whole, or just fractions thereof, can play a role in disease suppression (151). In contrast to bacterial endophytes, fungal endophytes have less frequently been reported to be involved in protection of their hosts via ISR (154–156).
Fungal endophytes are better known for their capacity to produce compounds that have growth-inhibitory activities toward plant pathogens and herbivores. These compounds comprise alkaloids, steroids, terpenoids, peptides, polyketones, flavonoids, quinols, phenols, and chlorinated compounds (157–159) (Fig. 2). Alkaloids produced by the clavicipitaceous fungi of grasses are among the best-described compounds produced by endophytes. For example, the neurotoxic indole-diterpenoid alkaloids, so-called lolitrems, are responsible for intoxication of cattle grazing on the endophyte-infected grass (160, 161). Some of these compounds, as well as some other alkaloids, are important for protection of the plant against insect herbivores (162, 163). Also, several reports have been published on the production of antiviral, antibacterial, antifungal, and insecticidal compounds by fungal endophytes, and most of these endophytes are transmitted horizontally, forming local infections in their hosts (157, 164). Not all horizontally transmitted fungal endophytes produce protective compounds, and due to the often small window of opportunity for contact with plant pathogens, in both time and space, their role in host protection against plant pathogens is still under dispute. A study made with cacao plants indicated that pathogens commonly colonize tree leaves but that infection does not always result in the occurrence of disease, and even that they can act as beneficial or harmless endophytes in their host (165–167). A recent report supported this finding and further demonstrated that production of endophytic antimicrobial compounds by endophytes can be induced by the presence of a pathogen (168).
Bacterial endophytes also produce antimicrobial compounds (Fig. 2). For example, the endophyte Enterobacter sp. strain 638 produces antibiotic substances, including 2-phenylethanol and 4-hydroxybenzoate (169). Generally, endophytic actinomycetes are the best-known examples of antimicrobial compound producers, and compounds discovered so far include munumbicins (170), kakadumycins (171), and coronamycin (172). Recently, multicyclic indolosesquiterpenes with antibacterial activity were identified in the endophyte Streptomyces sp. HKI0595, isolated from the mangrove tree (Kandelia candel) (173), and spoxazomicins A to C, with antitrypanosomal activity, were found to be produced by Streptosporangium oxazolinicum strain K07-0450T, isolated from orchid plants (174). Some of these compounds appear to be valuable for clinical or agricultural purposes (175), but their exact roles in plant-microbe interactions still need to be elucidated.
Production of additional secondary metabolites
Secondary metabolites are biologically active compounds that are an important source of anticancer, antioxidant, antidiabetic, immunosuppressive, antifungal, anti-oomycete, antibacterial, insecticidal, nematicidal, and antiviral agents (157, 175–182). In addition, endophytes produce secondary metabolites that are involved in mechanisms of signaling, defense, and genetic regulation of the establishment of symbiosis (183). Besides the production of secondary metabolite compounds, endophytes are also able to influence the secondary metabolism of their plant host (182). This was demonstrated in strawberry plants inoculated with a Methylobacterium species strain, in which the inoculant strain influenced the biosynthesis of flavor compounds, such as furanones, in the host plants (184–186). Recently, bacterial endophytes, along with bacterial methanol dehydrogenase transcripts, were localized in the vascular tissues of strawberry receptacles and in the cells of achenes, the locations where the furanone biosynthesis gene is expressed in the plant (187). Similarly, biosynthesis and accumulation of phenolic acids, flavan-3-ols, and oligomeric proanthocyanidins in bilberry (Vaccinium myrtillus L.) plants were enhanced upon interaction with a fungal endophyte, a Paraphaeosphaeria sp. strain (188).
Iron homeostasis
Some bacterial and fungal endophytes are producers of vivid siderophores (153, 189–192). Siderophores are essential compounds for iron acquisition by soil microorganisms (193, 194), but they also play important roles in pathogen-host interactions in animals (195, 196). The role of siderophores produced by endophytes in plant colonization is unknown, but it has been suggested that these compounds play a role in induction of ISR (153) (Fig. 2). Furthermore, siderophore production was shown to play an important role in the symbiosis of Epichloë festucae with ryegrass, as shown upon interruption of the siderophore biosynthesis gene cluster in E. festucae (191). It is possible that siderophores modulate iron homeostasis in E. festucae-infected ryegrass plants. Siderophores produced by endophytic Methylobacterium strains are also involved in suppression of Xylella fastidiosa, the causative agent of citrus variegated chlorosis in Citrus trees (189). A recent comparative genomic analysis of proteobacterial endophytes revealed that strains lacking the gene clusters involved in siderophore biosynthesis have a larger total number of genes encoding membrane receptors for uptake of Fe3+-siderophore complexes, hence potentially allowing them to take up siderophores produced by other endophytes (197).
Protection against biotic and abiotic stresses
Whereas most of the described endophytes protect the plant from biotic stresses, some endophytes can also protect the plant against different abiotic stresses. For example, fungal strains of Neotyphodium spp. were shown to be able to increase tolerance toward drought in grass plants by means of osmo- and stomatal regulation (198), and they protected the plants against nitrogen starvation and water stress (199). The root fungal endophyte Piriformospora indica was shown to induce salt tolerance in barley (200) and drought tolerance in Chinese cabbage plants (201). In both cases, increases in antioxidant levels were the proposed mechanisms behind elevation in stress tolerance in these plants. Colonization of fungal endophytes of the genus Trichoderma in cacao seedlings also resulted in a delay in the response to drought stress (202), and the bacterial endophyte Burkholderia phytofirmans strain PsJN elevates drought tolerance levels in maize (203) and wheat plants (204). Furthermore, fungal endophytes have been shown to interfere with cold tolerance of rice plants (205), and B. phytofirmans strain PsJN has been shown to enhance chilling tolerance in grapevine plantlets (206).
ACC deaminase is a bacterial enzyme that is often associated with alleviation of plant stress (Fig. 2). This enzyme is responsible for lowering the levels of ethylene in the plant by cleaving the plant-produced ethylene precursor 1-aminocyclopropane-1-carboxylate (ACC) to ammonia and 2-oxobutanoate, preventing ethylene signaling (207). The plant hormone ethylene acts in the germination of seeds and in response to various stresses, and it is the key regulator of colonization of plant tissue by bacteria (208). This suggests that, apart from stress alleviation, ACC deaminase supports colonization of a number of bacterial endophytes. When the ACC deaminase gene of B. phytofirmans PsJN was inactivated, the endophyte lost the ability to promote root elongation in canola seedlings (209). Another study performed on cut flowers indicated that bacterial endophytes are able to colonize the shoot and that ACC deaminase delays flower senescence (210).
Plant growth stimulation
Some endophytes are involved in plant growth promotion, despite the fact that they are promoting growth at the expense of obtaining valuable nutrients provided by the host plant (211–213). High endophyte infection loads in plants indicate that benefit-cost balances are at least neutral or positive, suggesting that most endophytes must be beneficial to their hosts. Such beneficial effects may result from interference in photosynthesis and carbon fixation processes taking place in plants. A fungal grass endophyte strain of Neotyphodium lolii was found to influence CO2 fixation but was not shown to be able to interfere with light interception, photochemistry, or net photosynthesis (214). No effect on photosynthesis, stomatal conductance, photosynthetic water use efficiency, or the maximum and operating efficiencies of photosystem II was found in poplar trees inoculated with the bacterial plant growth-promoting endophyte Enterobacter sp. 638 (215). On the other hand, inoculation of wheat with the bacterium B. phytofirmans strain PsJN increased the photosynthetic rate, CO2 assimilation, chlorophyll content, and water use efficiency under drought conditions (204).
Phytohormone production by endophytes is probably the best-studied mechanism of plant growth promotion, leading to morphological and architectural changes in plant hosts (213, 216, 217). The ability to produce auxins and gibberellins is a typical trait for root-associated endophytes (213, 216–219). It was proposed that indole-3-acetic acid (IAA), a member of the auxin class, increases colonization efficiency (220), possibly via interference with the host defense system (221), and production of this compound or related compounds may be an important property for plant colonization by endophytes (Fig. 2). Cytokinin production is commonly observed in endophytes, but on one occasion, in a root-colonizing fungal strain of Piriformospora indica, cytokinin biosynthesis was demonstrated and mutational deletions in cytokinin biosynthesis genes resulted in abortion of any plant growth-promoting effect (222).
Besides the production of plant growth hormones, additional mechanisms for plant growth promotion exist. Adenine and adenine ribosides have been identified as growth-promoting compounds in endophytes of Scots pine (223). Volatile compounds, such as acetoin and 2,3-butanediol, can stimulate plant growth (224–226) and are produced by some bacterial endophytes (227, 228). Polyamines affect plant growth and development in plant-mycorrhiza interactions (229) and are produced by the bacterium Azospirillum brasilense (230). It can be expected that additional, not yet understood mechanisms exist among plant-associated bacteria to promote plant growth.
Nitrogen fixation
Nutrient acquisition for plants via nitrogen fixation is another mechanism behind plant growth promotion. This trait is well studied in rhizobial and actinorhizal plant symbioses. Several root endophytes fix nitrogen (e.g., Acetobacter diazotrophicus, Herbaspirillum spp., and Azoarcus spp.) (231, 232), but the efficiencies of nitrogen fixation in free-living endophytes are far lower than those in root nodules of leguminous plant-rhizobium interactions (233). One exception is the relatively high nitrogen fixation efficiency observed in endophytic strains of Gluconacetobacter diazotrophicus in symbiosis with sugarcane plants (234). Other G. diazotrophicus strains were shown to be present in the microbiome of pine needles, including some potential N2-fixing strains (235). This indicates that G. diazotrophicus strains play important roles as nitrogen fixers in wider taxonomic ranges of host plants. Another example of a N2-fixing endophyte is Paenibacillus strain P22, which has been found in poplar trees (13). Strain P22 contributed to the total nitrogen pool of the host plant and induced metabolic changes in the plant. Nitrogen fixation contributes to the fitness of the host plant, especially in nitrogen-poor environments. Even if the quantities of fixed nitrogen measured in single nitrogen-fixing species are low, it remains to be clarified if the fixed N is for the endophytes’ own demands and/or for provision to the host plant (236).
In summary, various mechanisms in endophytes can explain the profound effects that endophytes have on their plant hosts. A recent report indicates that endophyte infection can also affect the gender selection of the host plant (237), which suggests that many new properties remain to be identified among endophytes.
Plant-Microbe Symbioses Leading to Improved Plant Fitness
Endophytes taxonomically differing from AMF and rhizobia were also shown to confer increased fitness to their hosts (238, 239). As an example, spotted knapweed (Centaurea stoebe) became more competitive toward bunchgrass (Koeleria macrantha) upon inoculation with the fungal endophyte Alternaria alternata (238). Stimulation of the production of secondary compounds by the endophyte played an important role in increased fitness of the host plant. However, inoculation with other Alternaria sp. endophytes did not result in increased fitness of knapweed plants (239), indicating that the endophyte-host plant interaction was strain specific. In another case, it was shown that infection of wild red fescue plants with the ergot fungus Claviceps purpurea, a seed pathogen in many grass species, resulted in decreased herbivory by sheep (240). In association with its host, C. purpurea produces alkaloids that are toxic to mammalian species, thus protecting the host from predation. From this case, it is clear that particular microorganisms or taxa showing a lifestyle typical for endophytes can be both pathogenic and beneficial for their host. It was furthermore shown that plant-endophyte interactions can shift the gender balance in the offspring of the plant host. The fungus Epichlöe elymi, an endophyte in Elymus virginicus plants, is vertically and maternally transmitted from parent to offspring plants, thereby increasing its opportunity to establish new infections in succeeding plant generations (237). Manipulation of the sex ratio in offspring is an example of how endophytes can manipulate the fitness of their hosts, in analogy to Wolbachia infection of particular insect species, indicating that manipulation of the gender balance in offspring is common among higher eukaryote-microbe interactions (241).
DECIPHERING THE BEHAVIOR OF ENDOPHYTES BY COMPARATIVE GENOMIC ANALYSIS
Comparative genomics is an important tool for identifying genes and regulons that are important for plant penetration and colonization by endophytes (242). Specific properties discriminating endophytes from closely related nonendophytic strains have been found on several occasions (169, 197, 243–246). Lateral gene transfer (e.g., by mobile elements, such as plasmids and genomic islands) plays an important role in the acquisition of properties responsible for the capacity of bacteria and fungi to colonize the endosphere of plants. As an example, the assembled genome of the obligate biotroph fungus Rhizophagus irregularis was shown to contain up to 11% transposable elements (244). No loss of metabolic complexity was detected, only a drift of genes involved in toxin synthesis and in degradation of the plant cell wall. Also, the genome sequence of the competent bacterial endophyte Enterobacter sp. 638 revealed many transposable elements, which were often flanked by genes relevant to host-bacterium interactions (e.g., amino acid/iron transport, hemolysin, and hemagglutinin genes), as well as a large conjugative plasmid important for host colonization (169).
A comparative genomic and metabolic network study revealed major differences between pathogenic (n = 36) and mutualistic (n = 28) symbionts of plants in their metabolic capabilities and cellular processes (246). Genes involved in biosynthetic processes and functions were enriched and more diverse among plant mutualists, while genes involved in degradation and host invasion were predominantly detected among phytopathogens. Pathogens seem to require more compounds from the plant cell wall, whereas plant mutualists metabolize more plant-stress-related compounds, thus potentially helping in stress amelioration. The study revealed the presence of secretion systems in pathogen genomes, probably needed to invade the host plants, while genomic loci encoding nitrogen fixation proteins and ribulose bisphosphate carboxylase/oxygenase (RubisCO) proteins were more exclusive to mutualistic bacteria (246). Bacteria carrying relatively large genomes are often able to successfully colonize a wide range of unrelated plant hosts, as well as soils, whereas strains with smaller genomes seem to have a smaller host range (247).
Comparative Genomics To Elucidate Specific Properties That Evolved in Bacterial Endophytes
To further expand on potential functional and mechanistic aspects of endophytes, we compared the genomes of 40 well-described bacterial strains which were isolated from the plant endosphere (i.e., endophytes) with those of 42 nodule-forming symbionts, 29 well-described plant bacterial pathogens, 42 strains frequently found in the rhizosphere (i.e., rhizosphere bacteria), and 49 soil bacteria (see Data Set S4 in the supplemental material). Sequences from protein-encoding genes of each genome were assigned KEGG Ortholog (KO) tags by using the Integrated Microbial Genome (IMG) comparative analysis system (248). A feature-by-sample contingency table was created, using properties with abundances of >25% and samples within each group with <98% functional similarity. The assigned KO tags were normalized by cumulative sum scaling (CSS) normalization, and then a mixture model that implements a zero-inflated Gaussian distribution was computed to detect differentially abundant properties by using the metagenomeSeq package (249). A comparison of relevant properties in the process of host colonization and establishment for each investigated group (i.e., nodule-forming symbionts, phytopathogens, and bacterial strains isolated from the rhizosphere and from soil) and for endophytes is shown in Table 3. We are aware of the fact that endophytes may colonize the rhizosphere (soil) or may even, under certain circumstances, have a phytopathogenic lifestyle (as discussed in other parts of this review). However, the aim of the comparative genomic analysis was to obtain indications of potential typical endophytic properties, which require further confirmation.
TABLE 3.
Comparative genomics of properties relevant to plant colonization and establishmenta
| Log2 fold change in abundance in the indicated group versus endophytes |
||||
|---|---|---|---|---|
| Category and feature (gene) | Symbionts | Phytopathogens | Rhizosphere bacteria | Soil bacteria |
| Chemotaxis and motility | ||||
| Aerotaxis (aer) | −0.983*** | 0.029 | −0.259 | −0.354 |
| Serine chemotaxis (tsr) | −0.697** | 0.471* | −0.284 | 0.162 |
| Aspartate/maltose chemotaxis (tar) | −0.315 | −0.262 | −0.276* | −0.041 |
| Ribose chemotaxis (rbsB) | 1.076*** | −0.423 | −0.108 | −0.252 |
| Galactose chemotaxis (mglB) | −0.257*** | 0.030 | 0.390*** | 0.283** |
| Dipeptide chemotaxis (tap) | −0.174** | −0.172 | −0.215*** | −0.089 |
| Response regulators (cheBR) | −0.276 | −0.519*** | −0.153 | −0.280* |
| Response regulator (cheV) | −0.880*** | −0.271 | 0.143 | 0.069 |
| Response regulator (cheD) | −0.206* | −0.086 | 0.040 | 0.009 |
| Response regulator (cheC) | −0.367* | −0.861*** | 0.096 | −0.298 |
| Response regulator (cheZ) | −0.271 | −0.202 | −0.396*** | −0.155 |
| Flagellar apparatus (fliI) | −0.252** | −0.201* | −0.149 | 0.045 |
| Chemotaxis and motility (motA) | −0.555*** | −0.297* | 0.094 | −0.065 |
| Signal transduction—two-component systems | ||||
| Magnesium assimilation (phoQ-phoP) | −0.951*** | 0.052 | −0.034 | 0.042 |
| Stress (rstB-rstA) | −0.951*** | −0.022 | −0.005 | 0.070 |
| Carbon source utilization (creC-creB) | −0.726*** | 0.077 | −0.098 | −0.016 |
| Multidrug resistance (baeS-baeR) | −0.804*** | 0.032 | 0.074 | 0.000 |
| Copper efflux (cusS-cusR) | −0.821*** | −0.415 | 0.258 | 0.300 |
| Carbon storage regulator (barA-uvrY) | −0.989*** | −0.044 | −0.058 | 0.017 |
| Antibiotic resistance (evgS-evgA) | −0.868*** | −0.522*** | 0.143 | −0.288 |
| Nitrogen fixation/metabolism (ntrY-ntrX) | −0.037 | −0.233*** | −0.615*** | −0.089 |
| Type IV fimbria synthesis (pilS-pilR) | −0.902*** | 0.038 | 0.039 | 0.180 |
| Amino sugar metabolism (glrK-glrR) | −0.974*** | −0.021 | −0.061 | 0.232 |
| Twitching motility (chpA-chpB) | −0.783*** | 0.120 | 0.026 | 0.003 |
| Extracellular polysaccharide (wspE-wspR) | −0.612*** | −0.072 | 0.044 | −0.023 |
| Cell fate control (pleC-pleD) | −0.131*** | −0.255** | −0.639*** | 0.094 |
| Redox response (regB-regA) | −0.004 | −0.204* | −0.099 | −0.136 |
| Transcriptional regulators | ||||
| Nitrogen assimilation (nifA) | −0.133 | −0.757*** | −0.359*** | −0.220 |
| Carbon storage regulator (sdiA) | 0.617*** | −0.067 | −0.055 | −0.279* |
| Biofilm formation (crp) | −0.976*** | 0.036 | −0.036 | 0.068 |
| Nitric oxide reductase (norR) | −0.625*** | −0.156* | 0.193 | 0.129 |
| NAD biosynthesis (nadR) | −0.257*** | 0.012 | −0.103 | −0.079 |
| Beta-lactamase resistance (ampR) | 0.091 | 0.016 | −0.060 | −0.339*** |
| Pyrimidine metabolism (pyrR) | −0.326** | −0.051 | 0.121 | 0.015 |
| Thiamine metabolism (tenA) | 0.070 | −0.976*** | 0.109 | 0.195 |
| Stress-related enzymes | ||||
| Glutathione peroxidase (btuE) | −0.360** | −0.031 | 0.104 | −0.195 |
| Glutathione S-transferase (gst) | 0.562** | −0.435* | −0.230 | −0.351 |
| Catalase (katE) | −0.362* | −0.237 | 0.084 | 0.042 |
| Transport system | ||||
| ABC, capsular polysaccharide (kpsT) | −0.045 | −0.277*** | −0.244* | −0.221 |
| ABC, thiamine-derived products (thiY) | −0.449** | −0.958*** | 0.000 | 0.000 |
| ABC, spermidine/putrescine (potD) | 0.718*** | −0.308** | 0.092 | 0.081 |
| ABC, dipeptide (dppF) | 0.204** | −0.230*** | −0.027 | 0.09 |
| ABC, branched-chain amino acid (livK) | 0.571 | −0.884** | −0.629 | −0.734 |
| ABC, cystine (fliY) | −0.28 | −0.270* | 0.225 | −0.355* |
| ABC, methionine (metN) | −0.478*** | −0.336* | 0.031 | −0.163 |
| ABC, histidine (hisJ) | −0.302 | −0.096 | 0.349* | −0.266* |
| ABC, lysine/arginine/ornithine (argT) | 0.216 | −0.336 | 0.464 | −0.182 |
| ABC, l-arabinose (araG) | 0.145 | −0.067 | 0.066 | −0.342*** |
| ABC, rhamnose (rhaT) | −0.129 | −0.826*** | −0.043 | −0.724*** |
| PTS, cellobiose (celB) | −1.425*** | −0.947*** | −0.073 | −0.146 |
| PTS, glucose (ptsG) | −0.860*** | 0.000 | 0.000 | 0.000 |
| PTS, mannose (manY) | −0.433*** | −0.287** | −0.374*** | −0.264* |
| PTS, ascorbate (sgaA) | −0.433** | 0.003 | −0.207 | 0.158 |
| PTS, phosphocarrier (furB) | −0.317* | −0.007 | −0.065 | 0.059 |
| Others, multidrug (mdtB) | 0.076 | −0.042 | −0.217** | 0.134 |
| Other, tricarboxylic (tctA) | 0.670*** | −0.018 | 0.352* | 0.481* |
| Others, C4-dicarboxylate (dctP) | −0.123 | −0.462* | 0.382* | 0.553** |
| Others, membrane pore protein (ompC) | −1.149*** | 0.090 | −0.053 | 0.071 |
| Secretion systems | ||||
| Type I RaxAB-RaxC system (raxB) | −0.270 | 0.357 | −0.234 | −0.186 |
| Type II general pathway protein (gspD) | −0.199 | 0.213 | −0.265 | 0.064 |
| Type III secretion core apparatus (yscJ) | 0.354* | 0.263** | 0.051 | −0.181** |
| Type IV conjugal DNA protein (virB2) | 0.370 | 0.125 | −0.718*** | −0.143 |
| Type VI Imp/Vas core components (hcp) | −0.360 | −0.038 | −0.045 | 0.095 |
| Twitching motility protein (pilJ) | −0.850*** | 0.058 | 0.028 | −0.002 |
| Type I pilus assembly protein (fimA) | −0.676*** | −0.300 | 0.282 | 0.158 |
| Plant growth-promoting properties | ||||
| Nitrogenase (nifH) | 0.301** | −0.676*** | 0.226 | 0.030 |
| ACC deaminase (acdS) | 0.118 | 0.223 | 0.119 | −0.344** |
| Acetoin reductase (budC) | 0.024 | −0.259*** | −0.024 | −0.059 |
| Acetolactate decarboxylase (alsD) | −1.000*** | 0.000 | 0.000 | 0.000 |
| Butanediol dehydrogenase (butB) | −0.089 | −0.090 | 0.469** | −0.319 |
| IAA biosynthesis, IAM pathway (amiE) | 0.201 | −0.067 | −0.017 | −0.029 |
| IAA biosynthesis, IPyA pathway (ipdC) | 0.077 | −0.157 | 0.291*** | −0.043 |
| IAA biosynthesis, IAN pathway (nit) | −0.156 | 0.088 | 0.084 | −0.019 |
| IAA biosynthesis, IAN pathway (nthAB) | −0.136 | −1.054*** | −0.596*** | −0.147 |
The relative abundances of the assigned functional properties in each investigated group (symbionts [n = 42], phytopathogens [n = 29], rhizosphere bacteria [n = 42], or soil bacteria [n = 49]) compared to endophytes (n = 40) are shown as normalized log2 fold changes. Negative values are shown if the endophyte group has a higher abundance.
Significant changes were computed with a zero-inflated Gaussian mixture model, and the alpha levels, denoted by *, **, and ***, were assigned to q-value thresholds of 0.05, 0.01, and 0.001, respectively.
Motility and chemotaxis
The ability to sense and respond to environmental cues is one of the major properties driving colonization of microorganisms (249–252). Our comparative genomic analysis of properties involved in chemotaxis and motility of bacteria suggested that protein-encoding genes related to the use of aspartate/maltose (Tar) and dipeptides (Tap) are more abundant among endophytes than among strains obtained from the rhizosphere. The response regulator proteins CheBR and CheC and the flagellum biosynthesis and motility mechanisms are more abundant among endophytes than among phytopathogens (Table 3). These results indicate the specificity of aspartate and dipeptide metabolism among endophytes, whereas serine metabolism seems to be used largely by phytopathogens. In addition, endophytes might be more responsive to different environmental cues than phytopathogens and nodule-forming symbionts.
Signal transduction
Regulation of two-component response systems is essential for the process of bacterial cell communication and fundamental for the synchronization of cooperative behavior (253, 254). In this category, bacterial endophytes differ mainly from nodule-forming symbionts and only marginally from the other investigated groups. Genes putatively involved in antibiotic resistance (evgS and evgA), redox response (regB and regA), nitrogen fixation and metabolism (ntrY and ntrX), and cell fate control (pleC and pleD) are found more prominently among endophytes than among phytopathogens and rhizobacteria (for the last two). A variety of energy-generating and energy-utilizing biological processes, including photosynthesis, carbon fixation, nitrogen fixation, hydrogen oxidation, denitrification, aerobic and anaerobic respiration, electron transport, and aerotaxis mechanisms, are known to be regulated in response to cellular redox balance (255) and might assist endophytes to thrive inside the host. The transmembrane nitrogen sensor protein NtrY interacts with the regulator protein NtrX to induce the expression of nif genes (256). Under nitrogen-limiting conditions, endophytes might be better able to fix nitrogen for their own benefit than phytopathogens or rhizobacteria. Overall, these results reveal distinct characteristics that are suitable for bacteria to thrive and survive in different environmental niches and conditions.
Transcriptional regulators
Transcriptional regulators are essential for prokaryotes to rapidly respond to environmental changes, improving their adaptation plasticity, cellular homeostasis, and colonization of new niches (257). Genes putatively involved in the transcriptional regulation of nitrogen assimilation (nifA), reduction of nitric oxide (norR), regulation of carbon storage (sdiA), beta-lactamase resistance (ampR), pyrimidine metabolism (pyrR), and thiamine metabolism (tenA) are detected in significantly larger proportions among endophytes than among the other investigated groups (Table 3). Regulatory genes related to the stoichiometry of nitrogen and carbon metabolism and those involved in the metabolism of nucleotides and vitamins and in stress responses might be of great importance for a life inside plants. Nodule-forming symbionts and plant pathogens that also thrive inside plant tissues reveal mechanisms different from those in endophytes to cope with stress and metabolism of nutrients, suggesting that each group has its own regulatory set of genes required for its typical behavioral responses.
Detoxification and stress-related enzymes
Due to an abrupt burst of reactive oxygen species (ROS) and reactive nitrogen species (RNS), the internal compartments of plants are inhospitable niches for aerobic microorganisms. Therefore, enzymes with detoxification capacities are essential for plant endosphere colonization and may also function as ameliorating agents upon host-induced stresses (258). Genes encoding glutathione peroxidase (btuE), glutathione S-transferase (gst), catalase (katE), and nitric oxide reductase (norR) are enriched, according to our analysis, in endophyte genomes compared to phytopathogen or nodule-forming symbiont genomes (Table 3). These ROS- and RNS-scavenging enzymes might assist endophytes to cope with the plant oxidative burst and might also ameliorate host biotic and abiotic stresses by protecting plant cells from oxidative damages (12, 259).
Transporters
Nutrient transport is an important function for life inside plants (169, 197). The proportion of endophytes harboring genes for ATP-binding cassette (ABC), major facilitator superfamily (MFS), phosphotransferase system (PTS), solute carrier family (SLC), and other transport systems varied largely in our analysis (Table 3). Genes putatively involved in the uptake of capsular polysaccharides, organic ions, peptides, amino acids, and carbohydrates were detected more prominently among endophytes than in the other investigated groups (Table 3). These results indicate the complexity of nutrient transport systems of endophytes, which might reflect their various lifestyle strategies for acquiring nutrients inside plants.
Secretion systems
Protein secretion plays an important role in plant-bacterium interactions (67, 260). Major differences in secretion systems of endophytes and nodule-forming symbionts were observed in our analysis (Table 3). Genes putatively involved in type III secretion systems are more typical of nodule-forming symbionts and phytopathogens than of endophytes, whereas they are detected in a significantly larger proportion of endophytes than soil bacteria (Table 3). This type of secretion system is more often employed by pathogens to manipulate host metabolism (261, 262). Conversely, type IV conjugal DNA-protein transfer secretion systems were detected more prominently among endophytes than among rhizosphere bacteria (Table 3). Type IV secretion is likely to be involved in host colonization and conjugation of DNA (263–265). Protein-encoding genes involved in adhesion to the host via twitching motility and type I pilus assembly are also detected more prominently among endophytes than among nodulating symbionts. These systems might be determinants of host colonization success (266, 267).
Genes involved in plant growth promotion
The nitrogenase (nifH) gene, putatively involved in the fixation of atmospheric N2, was detected in a significantly larger proportion of endophytes than of phytopathogens (Table 3). Surprisingly, 28% of the investigated group of prokaryotic endophytes harbored this gene, indicating that it has an important function in improving plant productivity under conditions of N limitation (see above). One of the genes thought to be involved in plant stress alleviation, encoding 1-aminocyclopropane-1-carboxylate deaminase (acdS), is detected more prominently among endophytes than among soil bacteria. Recent analyses of bacterial endophyte genomes suggest that ACC deaminase is not as widely spread among endophytes as previously thought (197, 263). However, endophytes differ significantly in their favored pathways for the biosynthesis of the plant hormones acetoin, 2,3-butanediol, and IAA compared to phytopathogens or rhizosphere bacteria, thus suggesting some particular characteristics that promote plant growth.
To summarize, genomic differences can be found between the different functional groups (i.e., endophytes, nodule-forming symbionts, phytopathogens, rhizobacteria, and soil bacteria), but we have to be aware that borders between functional groups are not clear-cut. Properties that are largely discriminative for endophytes compared to the other groups are a higher responsiveness to environmental cues, nitrogen fixation, and protection against reactive oxygen and nitrogen species. Endophytes might exhibit phytopathogenic effects under certain conditions, and rhizosphere bacteria might also be able to colonize plants internally. Furthermore, the balance between mutualism and antagonism depends on multiple parameters and might depend on a very fine-tuned interaction between microbial elicitors and plant responses (268).
PATHOGENS AND ENDOPHYTES: THE BALANCE OF THE INTERACTION IS CRUCIAL
Pathogenicity: Definition and Mechanisms
Pathogenicity to humans, animals, and plants is the most acclaimed feature of microorganisms. Traditionally, pathogens have been defined as causative agents of diseases, guided by Koch’s postulates for more than a century and later advanced by making use of molecular markers (269). Next-generation sequencing-based technologies have drastically revolutionized our knowledge of the microbiome, and also of pathogens (270, 271). We learned particularly from the human microbiome that it is involved in many more diseases than recently thought and that pathogen outbreaks are associated with shifts of the whole community, including those supporting pathogens (272, 273). Recently, this was also shown for plant pathogens (140, 274).
The generally used definition of endophytes excludes pathogenic microorganisms per se. However, all recent studies have shown that plant-endophyte interactions have a much broader range reaching from beneficial to pathogenic (275, 276). Many potential human as well as plant pathogens have the capacity to colonize the plant endosphere (275, 277). Therefore, endophytes and internal pathogens share several mechanisms (277). Several studies have provided evidence that similar or even identical functions are responsible for beneficial interactions with plants and virulence in humans. For example, the involvement of siderophore uptake systems or extracellular enzymes is common to both beneficial bacteria and human pathogens (278). Dörr et al. (279) reported that type IV pili of the plant-associated Azoarcus sp. strain BH72 are responsible for adhesion to plant and fungal cells. Furthermore, the amino acid sequence of the pilus shows high similarities to those of the pili of human-associated strains of Pseudomonas aeruginosa and Neisseria gonorrhoeae. While a mutant of Pseudomonas fluorescens deficient in a lauroyl transferase involved in lipid A biosynthesis resulted in impaired root colonization (280), a similar mutant of Salmonella enterica serovar Typhimurium was limited in its ability to colonize organs of the lymphatic system of mice (281). Type III secretion systems are responsible for the introduction of effectors into eukaryotic host cells, and they have been found in pathogenic bacteria as well as plant-associated bacteria with beneficial effects on host plants (282). Genome comparisons of plant- and human-associated Stenotrophomonas strains identified many similar properties responsible for host-microbe interactions (275), but also different ones, which included factors responsible for host invasion, antibiotic resistance, and several crucial virulence factors (283, 284). Interestingly, heat shock proteins were absent and a suicide vector activated at 37°C was identified in the plant-associated Stenotrophomonas strain (283). In addition, Stenotrophomonas rhizophila DSM14405T possessed unique genes for the synthesis and transport of the plant-protective compound spermidine, plant cell wall-degrading enzymes, and high salinity tolerance (283). The role of mutation frequency in niche adaptation was identified by Turrientes et al. (285). The factors described above are important mechanisms by which harmless bacteria can behave as pathogens with a change of host or host niche, upon which their virulence potential is frequently revealed to its full extent. Stenotrophomonas maltophilia is a multiresistant pathogen, and clinical and plant-associated strains show similar levels of resistance against clinically relevant antibiotics (286). For example, quinolone resistance mediated by the efflux pump SmeDEF is important for clinical issues but also for colonization of plant roots (287). In addition, clinical Stenotrophomonas strains are still able to colonize plant environments, such as tomato roots (Fig. 3A).
FIG 3.
Colonization of endosphere tissues by clinical bacterial strains. Volume renderings are shown for confocal laser scanning micrographs for FISH analyses of stained Stenotrophomonas maltophilia cells (red signal) within the emerging lateral root of a tomato plant (beige signal) (A to C) and stained Escherichia coli cells (red signal) invading a lettuce leaf via a stoma (green signal) (D and E). (Panels A to C reprinted from reference 370 with kind permission from Springer Science and Business Media. Panel E reprinted from reference 371.)
Occurrence of Potential Pathogens in the Endosphere of Plants
The plant endosphere can be colonized by plant, animal, and human pathogens. In plant microbiome analyses, several plant pathogens were identified, although no disease symptoms were observed (277). The fungal endophyte Verticillium dahliae is an interesting example; this is a pathogen which causes large yield losses in a broad range of crops, e.g., strawberry, potato, and olive (288). On the other hand, the fungus was found in many healthy plants as a commensal endophyte, e.g., in medicinal plants, potato, and grapevine (289). Moreover, “beneficial” strains of V. dahliae were used to biologically control Ophiostoma novo-ulmi, the fungus which causes Dutch elm disease (290). Animal and human pathogens, especially Escherichia coli pathovars (7, 242), are also able to colonize endospheres (291). Figure 3D shows the invasion of E. coli cells into lettuce leaves via stomata. Opportunistic pathogens play a special role because plants, including the endosphere, are an important reservoir for emerging opportunistic pathogens (276, 277). There are many genera comprising endophytes, including Burkholderia, Enterobacter, Herbaspirillum, Mycobacterium, Ochrobactrum, Pseudomonas, Ralstonia, Serratia, Staphylococcus, Stenotrophomonas, and Xylella, that enter bivalent interactions with plant and human hosts. Several members of these genera show plant growth-promoting properties as well as excellent antagonistic properties against plant pathogens and have therefore been utilized to control pathogens or to promote plant growth (277). However, many strains of these species also successfully colonize human organs and tissues and thus cause diseases.
Enterobacteriaceae is a large family of Gram-negative bacteria that includes, along with many harmless symbionts, many of the more familiar, so-called enteric pathogens (292). Many members live in the intestines of animals, but interestingly, the plant endosphere is also a reservoir for enterobacteria (293, 294). In particular, the abundance of human enteric pathogens is enhanced after intermediate disturbances (274). Although the incidence of outbreaks of enteric pathogens associated with fresh produce in the form of raw or minimally processed vegetables and fruits has recently increased, the ecology of enteric pathogens outside their human and animal hosts is less well understood (7).
Which Functions Could Pathogens Have Inside Plants?
Plant microbiota, i.e., microbial communities associated with a particular plant, play an important role in plant growth and health. Microorganisms can support nutrient uptake and produce a broad range of phytohormones or influence the latter. Another important function is the involvement of plant-associated bacteria in pathogen defense (276). Resistance against leaf pathogens is often encoded in the plant genome but may also be mediated by plant-associated microorganisms (295). It is more difficult to find resistance genes against soilborne pathogens. Cook et al. (296) suggested that antagonistic rhizobacteria fulfil this function.
Another hypothesis is that bacteria associated with the human diet, such as Enterobacteriaceae, act as stimuli for our immune systems. Recently, Hanski et al. (297) showed that declining environmental biodiversity is associated with reduced microbial diversity on human skin and enhanced allergic disposition, as shown through significant interactions with Enterobacteriaceae. Furthermore, they showed a positive association between the abundance of Acinetobacter organisms and interleukin-10 expression in peripheral blood mononuclear cells of healthy human individuals. Interleukin-10 is an anti-inflammatory cytokine and plays a central role in maintaining immune tolerance to harmless substances (298). The endotoxin derived from Gram-negative bacteria, such as Enterobacteriaceae, is known to have allergy-protective and immune-modulatory potentials (299). If plants are a natural reservoir of Enterobacteriaceae, then these bacteria must have been a “natural” part of our diet for a long time. Taking into account how many vegetables and fruits are eaten by people worldwide, these outbreaks seem to be more an accident than the norm, particularly considering that, traditionally, food was not processed and sterilized before being eaten. Therefore, the function of the plant-associated microbiome as an immune stimulant or “natural vaccination” was suggested by Berg et al. (300).
THE PLANT BIOME AND MULTIPARTITE INTERACTIONS
Our increasing understanding of the structure and complexity of microbial communities in various environments has led to comparisons between the microbiota associated with humans and those associated with plants, particularly with roots (301, 302). Plant roots have been suggested to be analogous to the human gut, as they are the primary organs interacting with the environment and mediating signal exchange and communication between plants and microorganisms. Microbiota of both animals and plants have important functions for host health by protecting against pathogens (247, 303) as well as regulating host gene expression and nutrient uptake and providing metabolic capacities (304). The plant microbiome can be considered an extension of the host phenotype (302, 305), and the plant secretory machinery has been suggested to play an important role in establishing an extended phenotype with microbial life (306).
Determinants of Endophyte Community Structures
The composition of endophyte communities is governed by biotic and abiotic factors. Most importantly, the plant, i.e., the host genotype and developmental stage, as well as the environment from which endophytes originate (such as soil for root endophytes and air for foliar fungal endophytes), contribute to community assembly; however, the magnitudes of the effects may differ between distinct systems. Few studies have attempted to evaluate the extents to which these parameters shape the endosphere microbiome. Rasche et al. (307) investigated potato-associated bacterial endophyte communities colonizing the lower stem sections of plants grown under greenhouse conditions. Different varieties, including genetically modified plants, were grown in contrasting soil types, and plants were sampled at different vegetation stages. In addition, the experiment included a pathogen (Pectobacterium atrosepticum) treatment. Molecular community analysis showed that the soil type was the most important driver of bacterial community composition, followed by the plant developmental stage. Recently, a thorough investigation making use of next-generation sequencing technologies investigated root-associated microbiomes of eight diverse, inbred Arabidopsis accessions, cultivated in two different soil types (308). Although the developmental stage had less of an effect on structures of the microbial communities in that study, the effect of the soil environment was more pronounced than that of the plant genotype. Ding et al. (309) studied bacterial leaf endophyte communities associated with distantly related plant species grown under natural conditions. In their study, the host species was the main factor shaping the community composition, followed by sampling dates and sampling locations. Generally, genetically related plants seem to host more similar bacterial endophyte communities, although host effects have repeatedly been reported (8, 76, 307, 308, 310–313). Nevertheless, the host phylogenetic distance alone does not explain bacterial microbiota diversification (314). The host effect on bacterial communities can be explained by the fact that many or most bacterial endophytes enter the plants via roots. Different plant species and varieties are characterized by different root exudation patterns, which are likely to attract different microorganisms colonizing the rhizoplane and subsequently gaining entry into the plant. In addition, plant physiology and chemical or physical characteristics are likely to play a major role. This is evidenced by the finding of different bacterial communities in different plant tissues (315, 316).
Microbiota Associated with Plant Reproductive Organs
Seed transmission is well known for fungal endophytes. Recently, it was suggested that bacterial endophytes may also be transmitted via seeds. Johnston-Monje and Raizada (227) showed that seed endophyte diversity was conserved to a certain extent in maize seeds, from wild ancestors to modern varieties, across boundaries of evolution, ethnography, and ecology. Seed bacterial endophyte communities have been reported to be quite independent from the soil environment (317), suggesting that vertical transmission of bacterial endophytes might also contribute to the establishment and fitness of the host. The frequency of vertical transmission of bacterial endophytes along host generations is a matter of debate. Hardoim et al. (107) reported that up to 45% of the seed-borne bacterial community was transmitted vertically from two consecutive generations in rice plants. It is interesting that in insect-pollinated plants, such as apple plants, flower-associated bacterial communities were dominated by taxa that are rare in plants (318). This indicates that bacterial endophytes associated with reproductive organs of allogamous plants may have an origin different from that of bacterial endophytes associated with other organs and might derive from the air or from feeding insects. Some of these endophytes might be transmitted via seeds. The flowering and pollination properties of the host genotype likely influence the community composition of bacterial endophytes transmitted via seeds.
Grasses have very specific interactions with fungi, with many endophytes being transmitted vertically via host seeds, and communities are therefore greatly dependent on the host genotype (110, 319). Other fungal endophytes are transmitted via spores colonizing plant leaves or may derive from members of the soil environment colonizing roots (117, 285). Although the interaction between plant hosts and horizontally transmitted endophytes is less specific than the symbiosis between Neotyphodium/Epichloë and grasses, host genotype specificity has frequently been reported for horizontally transmitted endophytes (320–323). Similarly to what has been described for bacterial endophytes, specific fungal communities colonize plant tissues representing distinct niches (324, 325). Above- and below-ground plant tissues seem to obtain their fungal endophytes from different sources. Root fungal endophytes are likely to derive from the soil environment (326), whereas fungal endophytes colonizing above-ground tissues are transmitted via spores in the air (327). Bacterial and fungal endophyte communities greatly differ between different plant developmental stages (307, 324), again indicating the tight interaction between endophytes and host physiology.
Multitrophic Interactions
The plant biome comprises the plant and multiple fungal and bacterial players, including both pathogens and mutualists, and is characterized by a dense network of multitrophic interactions, which are still poorly understood. Particularly in the case of tight interactions between the plant host and endophytes, signaling and recognition processes are highly important, inducing molecular, physiological, and morphological changes (328). However, plant-associated microorganisms may also influence plant pathways and phenotypes more generally. Quambusch et al. (329) reported distinct endophytic communities for easy- and difficult-to-propagate cherry genotypes, indicating the need for a specific microbiome, or at least specific microbiome components, for plant growth in general. The interaction of endophytes with their plant host may also affect its relationship with other microbes.
It is well known that endophytes may directly antagonize plant pathogens, which might be detectable in confrontation assays. Nevertheless, antimicrobial effects may also be induced by more sophisticated chemical communication. Combès et al. (168) demonstrated that Paraconiothyrium variabile, a fungal foliar (needle) endophyte, showed direct antagonism toward the phytopathogen Fusarium oxysporum; however, extracts of pure cultures did not show any effects. Only dual cultures of endophyte and pathogen led to competition-induced metabolite production. Oxylipins were identified as the induced metabolites, and their production was also associated with decreased mycotoxin production by the Fusarium pathogen. It is evident that chemical signaling and cross talk between endophytes and host plants are complex (330), and this example illustrates the importance of chemical communication not only between endophytes and plants but also between microorganisms. However, we are at the very beginning of understanding multitrophic metabolic interactions, which probably involve diverse chemical compounds produced by either the plant or microorganisms within the framework of their interaction (175, 183). Chemical interactions may also occur between fungal endophytes and endofungal bacteria. Hoffman and Arnold (331) reported that filamentous fungal endophytes frequently harbor diverse endohyphal bacteria, with mostly unknown importance. The same authors also recently found that such an endohyphal bacterium, a Luteibacter sp., greatly enhances IAA production from a foliar fungal endophyte, although the bacterium does not show IAA production when grown in pure culture under standard laboratory conditions (332). Another example of endofungal bacterial activity is toxin (rhizoxin) production by the fungus Rhizopus microsporus, which is responsible for rice seedling blight, but the actual toxin producer is the endofungal bacterium Burkholderia endofungorum (333, 334).
Bacteria might also play important roles in the interactions of AMF with plants and may represent examples of the evolution of multipartner associations. Representatives of the Mollicutes and “Candidatus Glomeribacter,” a group of Burkholderia-related Gram-negative species, have been demonstrated to live in hyphae and spores of AMF (335, 336). Relationships of these so-called mycorrhiza helper bacteria with AMF are close, and these bacteria most likely contribute to colonization and formation of the mycorrhizal structures in plant roots (337, 338). Another example of tripartite interactions is provided by a virus-infected fungal endophyte of Curvularia protuberata, which systemically colonizes the geothermal grass Dichanthelium lanuginosum (119, 339, 340) and increases its tolerance to high temperatures. The host plant and the endophyte can tolerate temperatures only as high as 40°C when grown separately, but in symbiosis, the plant-fungus combination is able to grow at soil temperatures as high as 65°C.
Endophytes can be prone to phage infections, and in principle, phages infecting endophytes can modulate bacterial and fungal endophytic communities (339, 341, 342). Several studies indicate that phages can play important roles in microbial community structuring (343, 344). Phages infecting endophytes of horse chestnut were more virulent for endophytes of the same trees than those of other trees, indicating selective forces on endophytic communities and that their phages can be tree specific. These examples demonstrate that neither plants nor individual endophytes act independently but that multiple organisms interact and influence the performance of the plant (biome).
Interactions between Endophytes and Pathogens/Pests
Endophytes may increase the defense against herbivores, including insects that transmit pathogens (319, 345). Deterrence of herbivores is known to be mediated via in planta production of biologically active alkaloids in grasses by endophytes, which can reduce arthropod feeding and, consequently, damage to the host (346). However, in relation to wild grasses, Faeth and Saari (347) reported that herbivore abundance and species richness may be even greater on endophyte-infected plants with high alkaloid contents than on endophyte-free plants; they argued that herbivores may develop detoxification pathways. Endophyte infection in grasses has also been tested for reducing aphid-transmitted virus infections (348). Endophyte infection and alkaloid production resulted in reduced aphid feeding, as expected, but no effect on virus titers could be observed. Nevertheless, the impact of virus infection on the host was reduced in endophyte-infected plants, indicating that the endophyte induced a host response, which was probably responsible for this effect. The interactions between plants, endophytes, aphids, and viruses were also influenced by the host and endophyte genotypes (348) as well as by abiotic factors, such as temperature (349).
The endosphere microbiome composition is affected by pathogen infection (49, 350–353), potentially leading to effects on microbial functioning. Some studies reported a reduction of bacterial (351) and fungal (353) diversity in diseased or pathogen-containing plants. Douanla-Meli et al. (353) compared culturable fungal endophyte communities of healthy and yellowing citrus leaves. The latter showed higher levels of colonization by fungal endophytes but lower levels of richness than healthy leaves. Phytoplasma infection of grapevines resulted in a reduction in diversity of bacterial endophyte communities (351). On the other hand, Reiter et al. (49) found a higher diversity of bacterial potato endophytes due to the presence of Pectobacterium atrosepticum; however, no disease symptoms were observed. Rasche et al. (307) reported that the extent to which P. atrosepticum affected the structure of bacterial endophytic communities depended on the plant genotype and on the soil environment. The various findings can be explained by the use of different plant hosts and pathogens and by the severity of disease. Pathogens induce a cascade of reactions leading to the synthesis of stress metabolites, including ROS or phytoalexins, and a range of stress signals, and these may provide a habitat with different physiological characteristics. Such an altered habitat is likely to support a differently structured endophyte community showing different functional characteristics. In addition, endophytes or rhizosphere bacteria that induce a systemic response in plants, such as Methylobacterium strains tested by Ardanov et al. (354), were reported to affect endophyte communities. The resulting bacterial community structures correlated with resistance or susceptibility to disease caused by Pectobacterium atrosepticum, Phytophthora infestans, and Pseudomonas syringae in potato and by Gremmeniella abietina in pine (354).
Interactions between Endophytes and Other Symbionts
In addition to the interactions between endophyte communities and phytopathogens, endophytes interact with other symbiotic microorganisms. Foliar fungal endophyte species composition was reported to be altered by AMF colonization (355). Wearn et al. (324) suggested that there is competition or antagonism between AMF and root endophytes, as they found negative correlations between mycorrhizal colonization and the presence of endophytes in roots of herbaceous grassland species. Some studies suggest that AMF colonization of grasses may be affected by the production of alkaloids or other allelopathic compounds by fungal endophytes (356–358). However, different AMF species or strains may behave/interact differently with plants and endophytes. For grasses, Larimer et al. (359) reported that Glomus mossae enhanced endophyte growth through increased tiller production, and in return, G. mossae showed higher colonization levels. On the other hand, colonization by another AMF species, Glomus claroideum, declined in endophyte-infected plants. In Pinus sylvestris, the interaction between a bacterial endophyte and an ectomycorrhizal fungus was shown to be species dependent, as endophytic Methylobacterium extorquens enhanced the growth of pine seedlings with one fungal species but decreased the growth when coinoculated with another ectomycorrhizal fungus (360).
In conclusion, the plant biome is characterized by multiple and complex interactions between the plant, the associated microbiota, i.e., endophytes with different functions, including pathogens, and the environment. The plant phenotype not only is determined by the response of the plant to the environment but also is regulated by the associated microbiota, the response of the microbiota to the environment, and the complex interactions between individual members.
CONCLUDING REMARKS
Technological developments, especially with respect to “-omics” technologies, will revolutionize our concepts on endosphere microbiomes. At present, we are better able to distinguish between properties specific to phytopathogens, endophytes, and other microorganisms from soil and plant habitats. This will allow us to better understand mutualists and pathogens, because from an ecological perspective, the boundaries between both groups are not always clear. Furthermore, microbial groups previously thought to be distinctive of other environments, such as human pathogens in warm-blooded animals, have been demonstrated to thrive in plants. Genomics will teach us how microbial groups from other environments adapt to plant environments and will reveal the minimal genetic requirements for successful penetration and internal colonization of plants. Novel technologies will also allow us to investigate multiple interactions between microbial groups associated with plants and the plant host itself. Nowadays, we have a better capacity to analyze impacts of invading microorganisms on the whole endophytic community composition and functioning, and vice versa. We can also better explain the resilience of plants upon invasion by potentially deleterious microorganisms by the functioning and complexity of the endophytic communities. We must learn more, however, about the still unknown roles of endophytes, particularly the so-called commensal endophytes. This group, which causes no apparent effects on plant performance but lives on the metabolites produced by the host, is presumably the most dominant functional group among endophytes by quantity (2, 361). We expect to find hidden functions within this group and to learn more about the complexity of microbial interactions within plants, including the consequences for the host plant. We also need to learn more about the interactions between endophytes and plants as well as the mechanisms employed by all partners. It will be highly relevant to elucidate the physiological conditions present in endophytes and plants during colonization, as it can be expected that an endophyte will have different characteristics inside the plant compared to growth in the soil or in the lab. Similarly, research is needed to better understand under which conditions and by which mechanisms microorganisms exhibit harmful, beneficial, or neutral effects on plant performance. By implementing new technologies and multidisciplinary approaches, our understanding of endophyte biology and ecology will consistently evolve further, leading to a better knowledge of the plant holobiome.
Supplementary Material
ACKNOWLEDGMENTS
Our presented work was conducted within the EU Cost Action “Endophytes in Biotechnology and Agriculture” (grant FA1103) and within the 7th framework project Biofector (grant 312117 awarded to L.S.V.O.). This work was also supported by grants provided by the FWF (Austrian Science Foundation) (grants P26203-B22 and P24569-B25) to A.S. and by the Portuguese FCT (Foundation for Science and Technology) (grant SFRH/BPD/78931/2011) to P.R.H.
We thank Cristiane C. P. Hardoim for performing fungal endophyte taxonomic assignments with QIIME. We thank the anonymous reviewers for many suggestions, which helped to improve the manuscript. Gobius Ciência e Comunicação is thanked for the artwork design in Fig. 2.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/MMBR.00050-14.
REFERENCES
- 1.Chanway CP. Endophytes: they’re not just fungi! Can J Bot. 1996;74:321–322. http://dx.doi.org/10.1139/b96-040. [Google Scholar]
- 2.Hallmann J, Quadt-Hallmann A, Mahaffee WF, Kloepper JW. Bacterial endophytes in agricultural crops. Can J Microbiol. 1997;43:895–914. http://dx.doi.org/10.1139/m97-131. [Google Scholar]
- 3.Rosenberg E, Sharon G, Zilber-Rosenberg I. The hologenome theory of evolution contains Lamarckian aspects within a Darwinian framework. Environ Microbiol. 2009;11:2959–2962. doi: 10.1111/j.1462-2920.2009.01995.x. http://dx.doi.org/10.1111/j.1462-2920.2009.01995.x. [DOI] [PubMed] [Google Scholar]
- 4.Garbeva P, van Overbeek LS, van Vuurde JWL, van Elsas JD. Analysis of endophytic bacterial communities of potato by plating and denaturing gradient gel electrophoresis (DGGE) of 16S rDNA based PCR fragments. Microb Ecol. 2001;41:369–383. doi: 10.1007/s002480000096. http://dx.doi.org/10.1007/s002480000096. [DOI] [PubMed] [Google Scholar]
- 5.van Overbeek LS, Bergervoet JHH, Jacobs FHH, van Elsas JD. The low-temperature-induced viable-but-nonculturable state affects the virulence of Ralstonia solanacearum biovar 2. Phytopathology. 2004;94:463–469. doi: 10.1094/PHYTO.2004.94.5.463. http://dx.doi.org/10.1094/PHYTO.2004.94.5.463. [DOI] [PubMed] [Google Scholar]
- 6.Kloepper JW, McInroy JA, Liu K, Hu C-H. Symptoms of fern distortion syndrome resulting from inoculation with opportunistic endophytic fluorescent Pseudomonas spp. PLoS One. 2013;8:e58531. doi: 10.1371/journal.pone.0058531. http://dx.doi.org/10.1371/journal.pone.0058531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.van Overbeek LS, van Doorn J, Wichers JH, van Amerongen A, van Roermund HJW, Willemsen PTJ. The arable ecosystem as battleground for emergence of new human pathogens. Front Microbiol. 2014;5:104. doi: 10.3389/fmicb.2014.00104. http://dx.doi.org/10.3389/fmicb.2014.00104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Andreote FD, de Araujo WL, de Azevedo JL, van Elsas JD, da Rocha UN, van Overbeek LS. Endophytic colonization of potato (Solanum tuberosum L.) by a novel competent bacterial endophyte, Pseudomonas putida strain P9, and its effect on associated bacterial communities. Appl Environ Microbiol. 2009;75:3396–3406. doi: 10.1128/AEM.00491-09. http://dx.doi.org/10.1128/AEM.00491-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Elasri M, Delorme S, Lemanceau P, Stewart G, Laue B, Glickmann E, Oger PM, Dessaux Y. Acyl-homoserine lactone production is more common among plant-associated Pseudomonas spp. than among soilborne Pseudomonas spp. Appl Environ Microbiol. 2001;67:1198–1209. doi: 10.1128/AEM.67.3.1198-1209.2001. http://dx.doi.org/10.1128/AEM.67.3.1198-1209.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Frey-Klett P, Burlinson P, Deveau A, Barret M, Tarkka M, Sarniguet A. Bacterial-fungal interactions: hyphens between agricultural, clinical, environmental, and food microbiologists. Microbiol Mol Biol Rev. 2011;75:583–609. doi: 10.1128/MMBR.00020-11. http://dx.doi.org/10.1128/MMBR.00020-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Knief C. Analysis of plant microbe interactions in the era of next generation sequencing technologies. Front Plant Sci. 2014;5:216. doi: 10.3389/fpls.2014.00216. http://dx.doi.org/10.3389/fpls.2014.00216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sessitsch A, Hardoim P, Döring J, Weilharter A, Krause A, Woyke T, Mitter B, Hauberg-Lotte L, Friedrich F, Rahalkar M, Hurek T, Sarkar A, Bodrossy L, van Overbeek L, Brar D, van Elsas JD, Reinhold-Hurek B. Functional characteristics of an endophyte community colonizing rice roots as revealed by metagenomic analysis. Mol Plant Microbe Interact. 2012;25:28–36. doi: 10.1094/MPMI-08-11-0204. http://dx.doi.org/10.1094/MPMI-08-11-0204. [DOI] [PubMed] [Google Scholar]
- 13.Scherling C, Ulrich K, Ewald D, Weckwerth W. A metabolic signature of the beneficial interaction of the endophyte Paenibacillus sp isolate and in vitro: grown poplar plants revealed by metabolomics. Mol Plant Microbe Interact. 2009;22:1032–1037. doi: 10.1094/MPMI-22-8-1032. http://dx.doi.org/10.1094/MPMI-22-8-1032. [DOI] [PubMed] [Google Scholar]
- 14.Nunes da Rocha U, Andreote FD, de Azevedo JL, van Elsas JD, van Overbeek LS. Cultivation of hitherto-uncultured bacteria belonging to the Verrucomicrobia subdivision 1 from the potato (Solanum tuberosum L.) rhizosphere. J Soils Sediments. 2010;10:326–339. http://dx.doi.org/10.1007/s11368-009-0160-3. [Google Scholar]
- 15.Nunes da Rocha U, Plugge CM, George I, van Elsas JD, van Overbeek LS. The rhizosphere selects for particular groups of Acidobacteria and Verrucomicrobia. PLoS One. 2013;8:e82443. doi: 10.1371/journal.pone.0082443. http://dx.doi.org/10.1371/journal.pone.0082443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.McCully ME. Niches for bacterial endophytes in crop plants: a plant biologist’s view. Aust J Plant Physiol. 2001;28:983–990. http://dx.doi.org/10.1071/PP01101. [Google Scholar]
- 17.Bragina A, Berg C, Cardinale M, Shcherbakov A, Chebotar V, Berg G. Sphagnum mosses harbour highly specific bacterial diversity during their whole lifecycle. ISME J. 2012;6:802–813. doi: 10.1038/ismej.2011.151. http://dx.doi.org/10.1038/ismej.2011.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Compant S, Mitter B, Colli-Mull JG, Gangl H, Sessitsch A. Endophytes of grapevine flowers, berries, and seeds: identification of cultivable bacteria, comparison with other plant parts, and visualization of niches of colonization. Microb Ecol. 2011;62:188–197. doi: 10.1007/s00248-011-9883-y. http://dx.doi.org/10.1007/s00248-011-9883-y. [DOI] [PubMed] [Google Scholar]
- 19.Cardinale M. Scanning a microhabitat: plant-microbe interactions revealed by confocal laser microscopy. Front Microbiol. 2014;5:94. doi: 10.3389/fmicb.2014.00094. http://dx.doi.org/10.3389/fmicb.2014.00094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Link HF. Observationes in ordines plantarum naturales, dissertatio prima, complectens anandrarum ordines Epiphytas, Mucedines, Gastromycos et Fungos. Der Gesellschaft Naturforschender Freunde zu Berlin; Berlin, Germany: 1809. [Google Scholar]
- 21.Nees von Esenbeck CG. Das System der Pilze und Schwämme. Stahelschen Buchhandlung; Würzburg, Germany: 1817. [Google Scholar]
- 22.Unger F. Die Exantheme der Pflanzen und einige mit diesen verwandten Krankheiten der Gewächse: pathogenetisch und nosographisch dargestellt. Verlag Carl Gerold; Vienna, Austria: 1833. [Google Scholar]
- 23.De Bary A. Morphologie und Physiologie der Pilze, Flechten und Myxomyceten. Verlag von Wilhelm Engelmann; Leipzig, Germany: 1866. [Google Scholar]
- 24.Béchamp A. Du rôle de la craie dans les fermentations butyrique et lactique, et des organismes actuellement vivants qu’elle contient. C R Hebd Seances Acad Sci. 1866;63:451–456. [Google Scholar]
- 25.Compant S, Sessitsch A, Mathieu F. The 125th anniversary of the first postulation of the soil origin of endophytic bacteria—a tribute to M.L.V. Galippe Plant Soil. 2012;356:299–301. http://dx.doi.org/10.1007/s11104-012-1204-9. [Google Scholar]
- 26.Galippe V. Note sur la présence de micro-organismes dans les tissus végétaux. C R Seances Soc Biol Fil. 1887;39:410–416. [Google Scholar]
- 27.Galippe V. Note sur la présence de micro-organismes dans les tissus végétaux (deuxième note) C R Seances Soc Biol Fil. 1887;39:557–560. [Google Scholar]
- 28.Laurent É . Sur l’existence de microbes dans les tissus des plantes superieures. Bull Soc R Bot Belge. 1889;1889:233–244. [Google Scholar]
- 29.Bernheim H. Die parasitären Bakterien der Ceralien. Chem Ztg. 1888;12:1321. [Google Scholar]
- 30.Smith EF. Bacteria in relation to plant diseases: history, general considerations, vascular diseases. Carnegie Institution of Washington; Washington, DC: 1911. [Google Scholar]
- 31.Fernbach A. De l’absence des microbes dans les tissus végétaux. Ann Inst Pasteur. 1888;2:567–570. [Google Scholar]
- 32.Schneider A. Mutualistic symbiosis of algae and bacteria with Cycas revoluta. Bot Gaz. 1894;19:25–32. [Google Scholar]
- 33.Schanderl H. Über die Bakteriensymbiose bei Leguminosen und Nichtleguminosen. Gartenbauwiss. 1939;13:406–440. [Google Scholar]
- 34.Schanderl H. Der derzeitige Stand in der Frage der Isolierbarkeit von Bakterien aus normalem, gesundem Pflanzengewebe. Zentralbl Bakteriol Parasitenkd. 1962;184:287–290. [Google Scholar]
- 35.Trémouillaux-Guiller J, Rohr T, Rohr R, Huss VAR. Discovery of an endophytic alga in Ginkgo biloba. Am J Bot. 2002;89:727–733. doi: 10.3732/ajb.89.5.727. http://dx.doi.org/10.3732/ajb.89.5.727. [DOI] [PubMed] [Google Scholar]
- 36.Müller P, Döring M. Isothermal DNA amplification facilitates the identification of a broad spectrum of bacteria, fungi and protozoa in Eleutherococcus sp. plant tissue cultures. Plant Cell Tissue Organ Cult. 2009;98:35–45. http://dx.doi.org/10.1007/s11240-009-9536-8. [Google Scholar]
- 37.Beijerinck MW. Cultur des Bacillus radicola aus den Knöllchen. Bot Ztg. 1888;46:740–750. [Google Scholar]
- 38.Frank BA. Ueber die Pilzsymbiose der Leguminosen. Ber Dtsch Bot Ges. 1889;7:332–346. [Google Scholar]
- 39.Hellriegel H, Wilfarth H. Untersuchungen über die Stickstoffnah-rung der Gramineen und Leguminosen. Druck von Kayssler; Berlin, Germany: 1888. [Google Scholar]
- 40.Martin BD, Schwab E. Current usage of symbiosis and associated terminology. Int J Biol. 2013;5:32–45. http://dx.doi.org/10.5539/ijb.v5n1p32. [Google Scholar]
- 41.Frank AB. Ueber die auf Wurzelsymbiose beruhende Ernährung gewisser Bäume durch unterirdische Pilze. Ber Dtsch Bot Ges. 1885;3:128–145. [Google Scholar]
- 42.Petrini O. Fungal endophytes of tree leaves. In: Andrews JH, Hirano SS, editors. Microbial ecology of leaves. Springer-Verlag; New York, NY: 1991. pp. 179–197. http://dx.doi.org/10.1007/978-1-4612-3168-4_9. [Google Scholar]
- 43.Kado CI. Plant pathogenic bacteria. In: Balows A, Tr̈uper HG, Dworkin M, Wim Harder T, Schleifer K-H, editors. The prokaryotes: a handbook on the biology of bacteria: ecophysiology, isolation, identification, application. Springer-Verlag; New York, NY: 1992. pp. 659–676. http://dx.doi.org/10.1007/978-1-4757-2191-1. [Google Scholar]
- 44.Wilson D. Endophyte—the evolution of a term, and clarification of its use and definition. Oikos. 1995;73:274–276. http://dx.doi.org/10.2307/3545919. [Google Scholar]
- 45.Di Fiore S, Gallo MD. Endophytic bacteria: their possible role in the host plant. In: Fendrik I, Gallo MD, Vanderleyden J, Miklos Zamaroczy D, editors. Azospirillum VI and related microorganisms: genetics-physiology-ecology. Springer Verlag; Berlin, Germany: 1995. pp. 169–187. http://dx.doi.org/10.1007/978-3-642-79906-8_18. [Google Scholar]
- 46.Azevedo JL. Microorganismos endofíticos. In: de Melo IS, de Azevedo JL, editors. Ecologia microbiana. Embrapa Meio Ambiente; Jaguariúna, Brazil: 1998. pp. 117–137. [Google Scholar]
- 47.Bacon CW, White J. Physiological adaptations in the evolution of endophytism in the Clavicipitaceae. In: Bacon CW, White J, editors. Microbial endophytes. Taylor & Francis; New York, NY: 2000. pp. 237–263. [Google Scholar]
- 48.Stone JK, Bacon CW, White J. An overview of endophytic microbes: endophytism defined. In: Bacon CW, White J, editors. Microbial endophytes. Taylor & Francis; New York, NY: 2000. pp. 3–29. [Google Scholar]
- 49.Reiter B, Pfeifer U, Schwab H, Sessitsch A. Response of endophytic bacterial communities in potato plants to infection with Erwinia carotovora subsp. atroseptica. Appl Environ Microbiol. 2002;68:2261–2268. doi: 10.1128/AEM.68.5.2261-2268.2002. http://dx.doi.org/10.1128/AEM.68.5.2261-2268.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Coombs JT, Franco CMM. Isolation and identification of actinobacteria from surface-sterilized wheat roots. Appl Environ Microbiol. 2003;69:5603–5608. doi: 10.1128/AEM.69.9.5603-5608.2003. http://dx.doi.org/10.1128/AEM.69.9.5603-5608.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kanamori N, Madsen LH, Radutoiu S, Frantescu M, Quistgaard EM, Miwa H, Downie JA, James EK, Felle HH, Haaning LL, Jensen TH, Sato S, Nakamura Y, Tabata S, Sandal N, Stougaard J. A nucleoporin is required for induction of Ca2+ spiking in legume nodule development and essential for rhizobial and fungal symbiosis. Proc Natl Acad Sci U S A. 2006;103:359–364. doi: 10.1073/pnas.0508883103. http://dx.doi.org/10.1073/pnas.0508883103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gherbi H, Markmann K, Svistoonoff S, Estevan J, Autran D, Giczey G, Auguy F, Péret B, Laplaze L, Franche C, Parniske M, Bogusz D. SymRK defines a common genetic basis for plant root endosymbioses with arbuscular mycorrhiza fungi, rhizobia, and Frankia bacteria. Proc Natl Acad Sci U S A. 2008;105:4928–4932. doi: 10.1073/pnas.0710618105. http://dx.doi.org/10.1073/pnas.0710618105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Markmann K, Giczey G, Parniske M. Functional adaptation of a plant receptor-kinase paved the way for the evolution of intracellular root symbioses with bacteria. PLoS Biol. 2008;6:e68. doi: 10.1371/journal.pbio.0060068. http://dx.doi.org/10.1371/journal.pbio.0060068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Redecker D, Kodner R, Graham LE. Glomalean fungi from the Ordovician. Science. 2000;289:1920–1921. doi: 10.1126/science.289.5486.1920. http://dx.doi.org/10.1126/science.289.5486.1920. [DOI] [PubMed] [Google Scholar]
- 55.Bidartondo MI, Read DJ, Trappe JM, Merckx V, Ligrone R, Duckett JG. The dawn of symbiosis between plants and fungi. Biol Lett. 2011;7:574–577. doi: 10.1098/rsbl.2010.1203. http://dx.doi.org/10.1098/rsbl.2010.1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Schüßler A. Molecular phylogeny, taxonomy, and evolution of Geosiphon pyriformis and arbuscular mycorrhizal fungi. Plant Soil. 2002;244:75–83. http://dx.doi.org/10.1023/A:1020238728910. [Google Scholar]
- 57.Brundrett MC. Coevolution of roots and mycorrhizas of land plants. New Phytol. 2002;154:275–304. doi: 10.1046/j.1469-8137.2002.00397.x. http://dx.doi.org/10.1046/j.1469-8137.2002.00397.x. [DOI] [PubMed] [Google Scholar]
- 58.Leake JR. The biology of myco-heterotrophic (‘saprophytic’) plants. New Phytol. 1994;127:171–216. doi: 10.1111/j.1469-8137.1994.tb04272.x. http://dx.doi.org/10.1111/j.1469-8137.1994.tb04272.x. [DOI] [PubMed] [Google Scholar]
- 59.Reference deleted.
- 60.Provorov NA, Vorobyov NI. Host plant as an organizer of microbial evolution in the beneficial symbioses. Phytochem Rev. 2009;8:519–534. http://dx.doi.org/10.1007/s11101-009-9140-x. [Google Scholar]
- 61.Kondorosi E, Mergaert P, Kereszt A. A paradigm for endosym-biotic life: cell differentiation of Rhizobium bacteria provoked by host plant factors. Annu Rev Microbiol. 2013;67:611–628. doi: 10.1146/annurev-micro-092412-155630. http://dx.doi.org/10.1146/annurev-micro-092412-155630. [DOI] [PubMed] [Google Scholar]
- 62.Pawlowski K. Prokaryotic symbionts in plants. Springer-Verlag; Dordrecht, Netherlands: 2009. http://dx.doi.org/10.1007/978-3-540-75460-2. [Google Scholar]
- 63.Yanni YG, Rizk RY, Corich V, Squartini A, Ninke K, Philip-Hollingsworth S, Orgambide G, de Bruijn F, Stoltzfus J, Buckley D, Schmidt TM, Mateos PF, Ladha JK, Dazzo FB. Natural endophytic association between Rhizobium leguminosarum bv. trifolii and rice roots and assessment of its potential to promote rice growth. Plant Soil. 1997;194:99–114. http://dx.doi.org/10.1023/A:1004269902246. [Google Scholar]
- 64.Gutiérrez-Zamora ML, Martinez-Romero E. Natural endophytic association between Rhizobium etli and maize (Zea mays L.) J Biotechnol. 2001;91:117–126. doi: 10.1016/s0168-1656(01)00332-7. http://dx.doi.org/10.1016/S0168-1656(01)00332-7. [DOI] [PubMed] [Google Scholar]
- 65.Burbano CS, Liu Y, Roesner KL, Reis VM, Caballero-Mellado J, Reinhold-Hurek B, Hurek T. Predominant nifH transcript phylotypes related to Rhizobium rosettiformans in field-grown sugarcane plants and in Norway spruce. Environ Microbiol Rep. 2011;3:383–389. doi: 10.1111/j.1758-2229.2010.00238.x. http://dx.doi.org/10.1111/j.1758-2229.2010.00238.x. [DOI] [PubMed] [Google Scholar]
- 66.Reiter B, Burgmann H, Burg K, Sessitsch A. Endophytic nifH gene diversity in African sweet potato. Can J Microbiol. 2003;49:549–555. doi: 10.1139/w03-070. http://dx.doi.org/10.1139/w03-070. [DOI] [PubMed] [Google Scholar]
- 67.Reinhold-Hurek B, Hurek T. Living inside plants: bacterial endophytes. Curr Opin Plant Biol. 2011;14:435–443. doi: 10.1016/j.pbi.2011.04.004. http://dx.doi.org/10.1016/j.pbi.2011.04.004. [DOI] [PubMed] [Google Scholar]
- 68.Kiers ET, Rousseau RA, West SA, Denison RF. Host sanctions and the legume-rhizobium mutualism. Nature. 2003;425:78–81. doi: 10.1038/nature01931. http://dx.doi.org/10.1038/nature01931. [DOI] [PubMed] [Google Scholar]
- 69.Hoeksema JD, Chaudhary VB, Gehring CA, Johnson NC, Karst J, Koide RT, Pringle A, Zabinski C, Bever JD, Moore JC, Wilson GWT, Klironomos JN, Umbanhowar J. A meta-analysis of context-dependency in plant response to inoculation with mycorrhizal fungi. Ecol Lett. 2010;13:394–407. doi: 10.1111/j.1461-0248.2009.01430.x. http://dx.doi.org/10.1111/j.1461-0248.2009.01430.x. [DOI] [PubMed] [Google Scholar]
- 70.Denison RF, Kiers ET. Lifestyle alternatives for rhizobia: mutualism, parasitism, and forgoing symbiosis. FEMS Microbiol Lett. 2004;237:187–193. doi: 10.1016/j.femsle.2004.07.013. http://dx.doi.org/10.1111/j.1574-6968.2004.tb09695.x. [DOI] [PubMed] [Google Scholar]
- 71.Bulgarelli D, Schlaeppi K, Spaepen S, van Themaat EVL, Schulze-Lefert P. Structure and functions of the bacterial microbiota of plants. Annu Rev Plant Biol. 2013;64:807–838. doi: 10.1146/annurev-arplant-050312-120106. http://dx.doi.org/10.1146/annurev-arplant-050312-120106. [DOI] [PubMed] [Google Scholar]
- 72.Vorholt JA. Microbial life in the phyllosphere. Nat Rev Microbiol. 2012;10:828–840. doi: 10.1038/nrmicro2910. http://dx.doi.org/10.1038/nrmicro2910. [DOI] [PubMed] [Google Scholar]
- 73.Oliveira MNV, Santos TMA, Vale HMM, Delvaux JC, Cordero AP, Ferreira AB, Miguel PSB, Totola MR, Costa MD, Moraes CA, Borges AC. Endophytic microbial diversity in coffee cherries of Coffea arabica from southeastern Brazil. Can J Microbiol. 2013;59:221–230. doi: 10.1139/cjm-2012-0674. http://dx.doi.org/10.1139/cjm-2012-0674. [DOI] [PubMed] [Google Scholar]
- 74.Chelius MK, Triplett EW. The diversity of archaea and bacteria in association with the roots of Zea mays L. Microb Ecol. 2001;41:252–263. doi: 10.1007/s002480000087. http://dx.doi.org/10.1007/s002480000087. [DOI] [PubMed] [Google Scholar]
- 75.Sun L, Qiu F, Zhang X, Dai X, Dong X, Song W. Endophytic bacterial diversity in rice (Oryza sativa L.) roots estimated by 16S rDNA sequence analysis. Microb Ecol. 2008;55:415–424. doi: 10.1007/s00248-007-9287-1. http://dx.doi.org/10.1007/s00248-007-9287-1. [DOI] [PubMed] [Google Scholar]
- 76.Nissinen RM, Mannisto MK, van Elsas JD. Endophytic bacterial communities in three arctic plants from low arctic fell tundra are cold-adapted and host-plant specific. FEMS Microbiol Ecol. 2012;82:510–522. doi: 10.1111/j.1574-6941.2012.01464.x. http://dx.doi.org/10.1111/j.1574-6941.2012.01464.x. [DOI] [PubMed] [Google Scholar]
- 77.Bull CT, de Boer SH, Denny TP, Firrao G, Fischer-Le Saux M, Saddler GS, Scortichini M, Stead DE, Takikawa Y. Comprehensive list of names of plant pathogenic bacteria, 1980 –2007. J Plant Pathol. 2010;92:551–592. [Google Scholar]
- 78.Bull CT, de Boer SH, Denny TP, Firrao G, Saux MF-L, Saddler GS, Scortichini M, Stead DE, Takikawa Y. List of new names of plant pathogenic bacteria (2008–2010) J Plant Pathol. 2012;94:21–27. http://dx.doi.org/10.4454/jpp.fa.2011.003. [Google Scholar]
- 79.Hardoim PR, Nazir R, Sessitsch A, Elhottova D, Korenblum E, van Overbeek LS, van Elsas JD. The new species Enterobacter oryziphilus sp nov and Enterobacter oryzendophyticus sp nov are key inhabitants of the endosphere of rice. BMC Microbiol. 2013;13:164. doi: 10.1186/1471-2180-13-164. http://dx.doi.org/10.1186/1471-2180-13-164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Brady C, Cleenwerck I, Venter S, Coutinho T, de Vos P. Taxonomic evaluation of the genus Enterobacter based on multilocus sequence analysis (MLSA): proposal to reclassify E. nimipressuralis and E. amnigenus into Lelliottia gen. no. as Lelliottia nimipressuralis comb. nov. and Lelliottia amnigena comb. nov., respectively, E. gergoviae and E. pyrinus into Pluralibacter gen. nov. as Pluralibacter gergoviae comb. nov. and Pluralibacter pyrinus comb. nov., respectively, E. cowanii, E. radicincitans, E. oryzae and E. arachidis into Kosakonia gen. nov. as Kosakonia cowanii comb. nov., Kosakonia radicincitans comb. nov., Kosakonia oryzae comb. nov. and Kosakonia arachidis comb. nov., respectively, and E. turicensis, E. helveticus and E. pulveris into Cronobacter as Cronobacter zurichensis nom. nov., Cronobacter helveticus comb. nov. and Cronobacter pulveris comb. nov., respectively, and emended description of the genera Enterobacter and Cronobacterv. Syst Appl Microbiol. 2013;36:309–319. doi: 10.1016/j.syapm.2013.03.005. http://dx.doi.org/10.1016/j.syapm.2013.03.005. [DOI] [PubMed] [Google Scholar]
- 81.Sy A, Timmers ACJ, Knief C, Vorholt JA. Methylotrophic metabolism is advantageous for Methylobacterium extorquens during colonization of Medicago truncatula under competitive conditions. Appl Environ Microbiol. 2005;71:7245–7252. doi: 10.1128/AEM.71.11.7245-7252.2005. http://dx.doi.org/10.1128/AEM.71.11.7245-7252.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Coenye T, Vandamme P. Diversity and significance of Burkholderia species occupying diverse ecological niches. Environ Microbiol. 2003;5:719–729. doi: 10.1046/j.1462-2920.2003.00471.x. http://dx.doi.org/10.1046/j.1462-2920.2003.00471.x. [DOI] [PubMed] [Google Scholar]
- 83.Liu G, Chater KF, Chandra G, Niu G, Tan H. Molecular regulation of antibiotic biosynthesis in Streptomyces. Microbiol Mol Biol Rev. 2013;77:112–143. doi: 10.1128/MMBR.00054-12. http://dx.doi.org/10.1128/MMBR.00054-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Schnepf E, Crickmore N, van Rie J, Lereclus D, Baum J, Feitelson J, Zeigler DR, Dean DH. Bacillus thuringiensis and its pesticidal crystal proteins. Microbiol Mol Biol Rev. 1998;62:775–806. doi: 10.1128/mmbr.62.3.775-806.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Schüßler A, Schwarzott D, Walker C. A new fungal phylum, the Glomeromycota: phylogeny and evolution. Mycol Res. 2001;105:1413–1421. http://dx.doi.org/10.1017/S0953756201005196. [Google Scholar]
- 86.Smith SE, Read DJ. Mycorrhizal symbiosis. Vol. 3. Elsevier Science; London, United Kingdom: 2008. [Google Scholar]
- 87.Casieri L, Lahmidi NA, Doidy J, Veneault-Fourrey C, Migeon A, Bonneau L, Courty P-E, Garcia K, Charbonnier M, Delteil A, Brun A, Zimmermann S, Plassard C, Wipf D. Biotrophic transportome in mutualistic plant-fungal interactions. Mycorrhiza. 2013;23:597–625. doi: 10.1007/s00572-013-0496-9. http://dx.doi.org/10.1007/s00572-013-0496-9. [DOI] [PubMed] [Google Scholar]
- 88.Jeffries P, Gianinazzi S, Perotto S, Turnau K, Barea JM. The contribution of arbuscular mycorrhizal fungi in sustainable maintenance of plant health and soil fertility. Biol Fertil Soils. 2003;37:1–16. http://dx.doi.org/10.1007/s00374-002-0546-5. [Google Scholar]
- 89.Stergiopoulos I, Collemare J, Mehrabi R, de Wit PJGM. Phytotoxic secondary metabolites and peptides produced by plant pathogenic Dothideomycete fungi. FEMS Microbiol Rev. 2013;37:67–93. doi: 10.1111/j.1574-6976.2012.00349.x. http://dx.doi.org/10.1111/j.1574-6976.2012.00349.x. [DOI] [PubMed] [Google Scholar]
- 90.Schoch CL, Crous PW, Groenewald JZ, Boehm EWA, Burgess TI, de Gruyter J, de Hoog GS, Dixon LJ, Grube M, Gueidan C, Harada Y, Hatakeyama S, Hirayama K, Hosoya T, Huhndorf SM, Hyde KD, Jones EBG, Kohlmeyer J, Kruys A, Li YM, Lucking R, Lumbsch HT, Marvanová L, Mbatchou JS, McVay AH, Miller AN, Mugambi GK, Muggia L, Nelsen MP, Nelson P, Owensby CA, Phillips AJL, Phongpaichit S, Pointing SB, Pujade-Renaud V, Raja HA, Plata ER, Robbertse B, Ruibal C, Sakayaroj J, Sano T, Selbmann L, Shearer CA, Shirouzu T, Slippers B, Suetrong S, Tanaka K, Volkmann-Kohlmeyer B, Wingfield MJ, Wood AR, Woudenberg JH, Yonezawa H, Zhang Y, Spatafora JW. A class-wide phylogenetic assessment of Dothideomycetes. Stud Mycol. 2009;64:1S10–15S10. doi: 10.3114/sim.2009.64.01. http://dx.doi.org/10.3114/sim.2009.64.01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Friesen TL, Faris JD, Solomon PS, Oliver RP. Host-specific toxins: effectors of necrotrophic pathogenicity. Cell Microbiol. 2008;10:1421–1428. doi: 10.1111/j.1462-5822.2008.01153.x. http://dx.doi.org/10.1111/j.1462-5822.2008.01153.x. [DOI] [PubMed] [Google Scholar]
- 92.Hane JK, Lowe RGT, Solomon PS, Tan K-C, Schoch CL, Spatafora JW, Crous PW, Kodira C, Birren BW, Galagan JE, Torriani SFF, McDonald BA, Oliver RP. Dothideomycete-plant interactions illuminated by genome sequencing and EST analysis of the wheat pathogen Stagonospora nodorum. Plant Cell. 2007;19:3347–3368. doi: 10.1105/tpc.107.052829. http://dx.doi.org/10.1105/tpc.107.052829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Thomma BPHJ. Alternaria spp.: from general saprophyte to specific parasite. Mol Plant Pathol. 2003;4:225–236. doi: 10.1046/j.1364-3703.2003.00173.x. http://dx.doi.org/10.1046/j.1364-3703.2003.00173.x. [DOI] [PubMed] [Google Scholar]
- 94.Jumpponen A, Jones KL. Massively parallel 454 sequencing indicates hyperdiverse fungal communities in temperate Quercus macrocarpa phyllosphere. New Phytol. 2009;184:438–448. doi: 10.1111/j.1469-8137.2009.02990.x. http://dx.doi.org/10.1111/j.1469-8137.2009.02990.x. [DOI] [PubMed] [Google Scholar]
- 95.Pan JJ, Baumgarten AM, May G. Effects of host plant environment and Ustilago maydis infection on the fungal endophyte community of maize (Zea mays) New Phytol. 2008;178:147–156. doi: 10.1111/j.1469-8137.2007.02350.x. http://dx.doi.org/10.1111/j.1469-8137.2007.02350.x. [DOI] [PubMed] [Google Scholar]
- 96.Zhang N, Castlebury LA, Miller AN, Huhndorf SM, Schoch CL, Seifert KA, Rossman AY, Rogers JD, Kohlmeyer J, Volkmann-Kohlmeyer B, Sung G-H. An overview of the systematics of the Sordariomycetes based on a four-gene phylogeny. Mycologia. 2006;98:1076–1087. doi: 10.3852/mycologia.98.6.1076. http://dx.doi.org/10.3852/mycologia.98.6.1076. [DOI] [PubMed] [Google Scholar]
- 97.Veneault-Fourrey C, Plett JM, Martin F. Who is controlling whom within the ectomycorrhizal symbiosis: insights from genomic and functional analyses. In: de Bruijn FJ, editor. Molecular microbial ecology of the rhizosphere. Vol. 1. John Wiley & Sons, Inc.; Singapore: 2013. pp. 501–512. Singapore. http://dx.doi.org/10.1002/9781118297674.ch47. [Google Scholar]
- 98.Weiss M, Selosse MA, Rexer KH, Urban A, Oberwinkler F. Sebacinales: a hitherto overlooked cosm of heterobasidiomycetes with a broad mycorrhizal potential. Mycol Res. 2004;108:1003–1010. doi: 10.1017/s0953756204000772. http://dx.doi.org/10.1017/S0953756204000772. [DOI] [PubMed] [Google Scholar]
- 99.Parniske M. Arbuscular mycorrhiza: the mother of plant root endosymbioses. Nat Rev Microbiol. 2008;6:763–775. doi: 10.1038/nrmicro1987. http://dx.doi.org/10.1038/nrmicro1987. [DOI] [PubMed] [Google Scholar]
- 100.Schardl CL, Leuchtmann A, Spiering MJ. Symbioses of grasses with seedborne fungal endophytes. Annu Rev Plant Biol. 2004;55:315–340. doi: 10.1146/annurev.arplant.55.031903.141735. http://dx.doi.org/10.1146/annurev.arplant.55.031903.141735. [DOI] [PubMed] [Google Scholar]
- 101.Hardoim PR, van Overbeek LS, van Elsas JD. Properties of bacterial endophytes and their proposed role in plant growth. Trends Microbiol. 2008;16:463–471. doi: 10.1016/j.tim.2008.07.008. http://dx.doi.org/10.1016/j.tim.2008.07.008. [DOI] [PubMed] [Google Scholar]
- 102.Gnanamanickam SS, Immanuel JE. Epiphytic bacteria, their ecology and functions. In: Gnanamanickam SS, editor. Plant-associated bacteria. Springer; Dordrecht, Netherlands: 2007. pp. 131–153. http://dx.doi.org/10.1007/978-1-4020-4538-7_4. [Google Scholar]
- 103.Steenhoudt O, Vanderleyden J. Azospirillum, a free-living nitrogen-fixing bacterium closely associated with grasses: genetic, biochemical and ecological aspects. FEMS Microbiol Rev. 2000;24:487–506. doi: 10.1111/j.1574-6976.2000.tb00552.x. http://dx.doi.org/10.1111/j.1574-6976.2000.tb00552.x. [DOI] [PubMed] [Google Scholar]
- 104.Bashan Y, de-Bashan LE. How the plant growth-promoting bacterium Azospirillum promotes plant growth: a critical assessment. In: Sparks DL, editor. Advances in agronomy. Vol. 108. Elsevier; San Diego, CA: 2010. pp. 77–136. http://dx.doi.org/10.1016/S0065-2113(10)08002-8. [Google Scholar]
- 105.Druzhinina IS, Seidl-Seiboth V, Herrera-Estrella A, Horwitz BA, Kenerley CM, Monte E, Mukherjee PK, Zeilinger S, Grigoriev IV, Kubicek CP. Trichoderma: the genomics of opportunistic success. Nat Rev Microbiol. 2011;9:749–759. doi: 10.1038/nrmicro2637. http://dx.doi.org/10.1038/nrmicro2637. [DOI] [PubMed] [Google Scholar]
- 106.Nelson EB. Microbial dynamics and interactions in the spermo-sphere. Annu Rev Phytopathol. 2004;42:271–309. doi: 10.1146/annurev.phyto.42.121603.131041. http://dx.doi.org/10.1146/annurev.phyto.42.121603.131041. [DOI] [PubMed] [Google Scholar]
- 107.Hardoim PR, Hardoim CCP, van Overbeek LS, van Elsas JD. Dynamics of seed-borne rice endophytes on early plant growth stages. PLoS One. 2012;7:e30438. doi: 10.1371/journal.pone.0030438. http://dx.doi.org/10.1371/journal.pone.0030438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Tyler HL, Triplett EW. Plants as a habitat for beneficial and/or human pathogenic bacteria. Annu Rev Phytopathol. 2008;46:53–73. doi: 10.1146/annurev.phyto.011708.103102. http://dx.doi.org/10.1146/annurev.phyto.011708.103102. [DOI] [PubMed] [Google Scholar]
- 109.Kale SD, Tyler BM. Entry of oomycete and fungal effectors into plant and animal host cells. Cell Microbiol. 2011;13:1839–1848. doi: 10.1111/j.1462-5822.2011.01659.x. http://dx.doi.org/10.1111/j.1462-5822.2011.01659.x. [DOI] [PubMed] [Google Scholar]
- 110.Saikkonen K, Wäli PR, Helander M. Genetic compatibility determines endophyte-grass combinations. PLoS One. 2010;5:e11395. doi: 10.1371/journal.pone.0011395. http://dx.doi.org/10.1371/journal.pone.0011395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Carroll GC. Fungal endophytes in stems and leaves: from latent pathogen to mutualistic symbiont. Ecology. 1988;69:2–9. http://dx.doi.org/10.2307/1943154. [Google Scholar]
- 112.Stone JK, Polishook JD, White JRJ. Endophytic fungi. In: Foster MS, Bills GF, Mueller GM, editors. Biodiversity of fungi: inventory and monitoring methods. Elsevier Science; Burlington, MA: 2004. pp. 241–270. [Google Scholar]
- 113.Saikkonen K, Ion D, Gyllenberg M. The persistence of vertically transmitted fungi in grass metapopulations. Proc Biol Sci. 2002;269:1397–1403. doi: 10.1098/rspb.2002.2006. http://dx.doi.org/10.1098/rspb.2002.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Tadych M, Bergen M, Dugan FM, White JF. Evaluation of the potential role of water in spread of conidia of the Neotyphodium endophyte of Poa ampla. Mycol Res. 2007;111:466–472. doi: 10.1016/j.mycres.2007.02.002. http://dx.doi.org/10.1016/j.mycres.2007.02.002. [DOI] [PubMed] [Google Scholar]
- 115.Philipson MN, Christey MC. The relationship of host and endophyte during flowering, seed formation, and germination of Lolium perenne. N Z J Bot. 1986;24:125–134. [Google Scholar]
- 116.Steinebrunner F, Twele R, Francke W, Leuchtmann A, Schiestl FP. Role of odour compounds in the attraction of gamete vectors in endophytic Epichloë fungi. New Phytol. 2008;178:401–411. doi: 10.1111/j.1469-8137.2007.02347.x. http://dx.doi.org/10.1111/j.1469-8137.2007.02347.x. [DOI] [PubMed] [Google Scholar]
- 117.Rodriguez RJ, White JF, Arnold AE, Redman RS. Fungal endophytes: diversity and functional roles. New Phytol. 2009;182:314–330. doi: 10.1111/j.1469-8137.2009.02773.x. http://dx.doi.org/10.1111/j.1469-8137.2009.02773.x. [DOI] [PubMed] [Google Scholar]
- 118.Gao K, Mendgen K. Seed-transmitted beneficial endophytic Stagonospora sp can penetrate the walls of the root epidermis, but does not proliferate in the cortex, of Phragmites australis. Can J Bot. 2006;84:981–988. http://dx.doi.org/10.1139/b06-056. [Google Scholar]
- 119.Redman RS, Sheehan KB, Stout RG, Rodriguez RJ, Henson JM. Thermotolerance generated by plant/fungal symbiosis. Science. 2002;298:1581. doi: 10.1126/science.1072191. http://dx.doi.org/10.1126/science.1078055. [DOI] [PubMed] [Google Scholar]
- 120.Carroll GC, Carroll FE. Studies on the incidence of coniferous needle endophytes in the Pacific Northwest. Can J Bot. 1978;56:3034–3043. http://dx.doi.org/10.1139/b78-367. [Google Scholar]
- 121.Petrini O. Taxonomy of endophytic fungi of aerial plant tissues. In: Fokkema NJ, van den Heuvel J, editors. Microbiology of the phyllosphere. Cambridge University Press; Cambridge, United Kingdom: 1986. pp. 175–187. [Google Scholar]
- 122.Gamboa MA, Bayman P. Communities of endophytic fungi in leaves of a tropical timber tree (Guarea guidonia: Meliaceae) Biotropica. 2001;33:352–360. http://www.jstor.org/stable/2663841. [Google Scholar]
- 123.Davis EC, Franklin JB, Shaw AJ, Vilgalys R. Endophytic Xylaria (Xylariaceae) among liverworts and angiosperms: phylogenetics, distribution, and symbiosis. Am J Bot. 2003;90:1661–1667. doi: 10.3732/ajb.90.11.1661. http://dx.doi.org/10.3732/ajb.90.11.1661. [DOI] [PubMed] [Google Scholar]
- 124.Higgins KL, Arnold AE, Miadlikowska J, Sarvate SD, Lutzoni F. Phylogenetic relationships, host affinity, and geographic structure of boreal and arctic endophytes from three major plant lineages. Mol Phylogenet Evol. 2007;42:543–555. doi: 10.1016/j.ympev.2006.07.012. http://dx.doi.org/10.1016/j.ympev.2006.07.012. [DOI] [PubMed] [Google Scholar]
- 125.Murali TS, Suryanarayanan TS, Venkatesan G. Fungal endophyte communities in two tropical forests of southern India: diversity and host affiliation. Mycol Prog. 2007;6:191–199. http://dx.doi.org/10.1007/s11557-007-0540-2. [Google Scholar]
- 126.Davis EC, Shaw AJ. Biogeographic and phylogenetic patterns in diversity of liverwort-associated endophytes. Am J Bot. 2008;95:914–924. doi: 10.3732/ajb.2006463. http://dx.doi.org/10.3732/ajb.2006463. [DOI] [PubMed] [Google Scholar]
- 127.Barengo N, Sieber TN, Holdenrieder O. Diversity of endophytic mycobiota in leaves and twigs of pubescent birch (Betula pubescens) Sydowia. 2000;52:305–320. [Google Scholar]
- 128.Kumar DSS, Hyde KD. Biodiversity and tissue-recurrence of endophytic fungi in Tripterygium wilfordii. Fungal Divers. 2004;17:69–90. [Google Scholar]
- 129.O’Dell TE, Massicotte HB, Trappe JM. Root colonization of Lupinus latifolius Agardh. and Pinus contorta Dougl. by Phialocephala fortinii Wang & Wilcox. New Phytol. 1993;124:93–100. http://dx.doi.org/10.1111/j.1469-8137.1993.tb03800.x. [Google Scholar]
- 130.Compant S, Clement C, Sessitsch A. Plant growth-promoting bacteria in the rhizo- and endosphere of plants: their role, colonization, mechanisms involved and prospects for utilization. Soil Biol Biochem. 2010;42:669–678. http://dx.doi.org/10.1016/j.soilbio.2009.11.024. [Google Scholar]
- 131.Philippot L, Raaijmakers JM, Lemanceau P, van der Putten WH. Going back to the roots: the microbial ecology of the rhizosphere. Nat Rev Microbiol. 2013;11:789–799. doi: 10.1038/nrmicro3109. http://dx.doi.org/10.1038/nrmicro3109. [DOI] [PubMed] [Google Scholar]
- 132.Mercado-Blanco J, Prieto P. Bacterial endophytes and root hairs. Plant Soil. 2012;361:301–306. http://dx.doi.org/10.1007/s11104-012-1212-9. [Google Scholar]
- 133.Hallmann J. Plant interactions with endophytic bacteria. In: Jeger MJ, Spence NJ, editors. Biotic interactions in plant-pathogen associations. CABI Publishing; Wallingford, United Kingdom: 2001. pp. 87–119. http://dx.doi.org/10.1079/9780851995120.0000. [Google Scholar]
- 134.James EK, Gyaneshwar P, Mathan N, Barraquio QL, Reddy PM, Iannetta PPM, Olivares FL, Ladha JK. Infection and colonization of rice seedlings by the plant growth-promoting bacterium Herbaspirillum seropedicae Z67. Mol Plant Microbe Interact. 2002;15:894–906. doi: 10.1094/MPMI.2002.15.9.894. http://dx.doi.org/10.1094/MPMI.2002.15.9.894. [DOI] [PubMed] [Google Scholar]
- 135.Mercado-Blanco J, Lugtenberg BJJ. Biotechnological applications of bacterial endophytes. Curr Biotechnol. 2014;3:60–75. [Google Scholar]
- 136.James EK, Olivares FL, de Oliveira ALM, dos Reis FB, da Silva LG, Reis VM. Further observations on the interaction between sugar cane and Gluconacetobacter diazotrophicus under laboratory and greenhouse conditions. J Exp Bot. 2001;52:747–760. doi: 10.1093/jexbot/52.357.747. http://dx.doi.org/10.1093/jexbot/52.357.747. [DOI] [PubMed] [Google Scholar]
- 137.Bell CR, Dickie GA, Harvey WLG, Chan JWYF. Endophytic bacteria in grapevine. Can J Microbiol. 1995;41:46–53. http://dx.doi.org/10.1139/m95-006. [Google Scholar]
- 138.Compant S, Duffy B, Nowak J, Clement C, Barka EA. Use of plant growth-promoting bacteria for biocontrol of plant diseases: principles, mechanisms of action, and future prospects. Appl Environ Microbiol. 2005;71:4951–4959. doi: 10.1128/AEM.71.9.4951-4959.2005. http://dx.doi.org/10.1128/AEM.71.9.4951-4959.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Compant S, Kaplan H, Sessitsch A, Nowak J, Ait Barka E, Clement C. Endophytic colonization of Vitis vinifera L. by Burkholderia phytofirmans strain PsJN: from the rhizosphere to inflorescence tissues. FEMS Microbiol Ecol. 2008;63:84–93. doi: 10.1111/j.1574-6941.2007.00410.x. http://dx.doi.org/10.1111/j.1574-6941.2007.00410.x. [DOI] [PubMed] [Google Scholar]
- 140.Fürnkranz M, Lukesch B, Mueller H, Huss H, Grube M, Berg G. Microbial diversity inside pumpkins: microhabitat-specific communities display a high antagonistic potential against phytopathogens. Microb Ecol. 2012;63:418–428. doi: 10.1007/s00248-011-9942-4. http://dx.doi.org/10.1007/s00248-011-9942-4. [DOI] [PubMed] [Google Scholar]
- 141.Madmony A, Chernin L, Pleban S, Peleg E, Riov J. Enterobacter cloacae, an obligatory endophyte of pollen grains of Mediterranean pines. Folia Microbiol (Praha) 2005;50:209–216. doi: 10.1007/BF02931568. http://dx.doi.org/10.1007/BF02931568. [DOI] [PubMed] [Google Scholar]
- 142.Rosenblueth M, Martinez-Romero E. Bacterial endophytes and their interactions with hosts. Mol Plant Microbe Interact. 2006;19:827–837. doi: 10.1094/MPMI-19-0827. http://dx.doi.org/10.1094/MPMI-19-0827. [DOI] [PubMed] [Google Scholar]
- 143.Scortichini M, Loreti S. Occurrence of an endophytic, potentially pathogenic strain of Pseudomonas syringae in symptomless wild trees of Corylus avellana L. J Plant Pathol. 2007;89:431–434. [Google Scholar]
- 144.Bacon CW, Glenn AE, Yates IE. Fusarium verticillioides: managing the endophytic association with maize for reduced fumonisins accumulation. Toxin Rev. 2008;27:411–446. http://dx.doi.org/10.1080/15569540802497889. [Google Scholar]
- 145.Estrada AER, Jonkers W, Kistler HC, May G. Interactions between Fusarium verticillioides, Ustilago maydis, and Zea mays: an endophyte, a pathogen, and their shared plant host. Fungal Genet Biol. 2012;49:578–587. doi: 10.1016/j.fgb.2012.05.001. http://dx.doi.org/10.1016/j.fgb.2012.05.001. [DOI] [PubMed] [Google Scholar]
- 146.Carroll GC. Fungal associates of woody plants as insect antagonists in leaves and stems. In: Barbosa P, Krischik VA, Jones CG, editors. Microbial mediation of plant-herbivore interactions. Wiley; New York, NY: 1991. pp. 253–271. [Google Scholar]
- 147.Robert-Seilaniantz A, Grant M, Jones JDG. Hormone crosstalk in plant disease and defense: more than just jasmonate-salicylate antagonism. Annu Rev Phytopathol. 2011;49:317–343. doi: 10.1146/annurev-phyto-073009-114447. http://dx.doi.org/10.1146/annurev-phyto-073009-114447. [DOI] [PubMed] [Google Scholar]
- 148.Zamioudis C, Pieterse CMJ. Modulation of host immunity by beneficial microbes. Mol Plant Microbe Interact. 2012;25:139–150. doi: 10.1094/MPMI-06-11-0179. http://dx.doi.org/10.1094/MPMI-06-11-0179. [DOI] [PubMed] [Google Scholar]
- 149.Chanway CP. Bacterial endophytes: ecological and practical implications. Sydowia. 1998;50:149–170. [Google Scholar]
- 150.Kloepper JW, Ryu CM. Bacterial endophytes as elicitors of induced systemic resistance. In: Schulz BJE, Boyle CJC, Sieber TN, editors. Microbial root endophytes. Springer-Verlag; Berlin, Germany: 2006. pp. 33–52. http://dx.doi.org/10.1007/3-540-33526-9_3. [Google Scholar]
- 151.Ardanov P, Ovcharenko L, Zaets I, Kozyrovska N, Pirttilä AM. Endophytic bacteria enhancing growth and disease resistance of potato (Solanum tuberosum L.) Biol Control. 2011;56:43–49. http://dx.doi.org/10.1016/j.biocontrol.2010.09.014. [Google Scholar]
- 152.Bordiec S, Paquis S, Lacroix H, Dhondt S, Ait Barka E, Kauffmann S, Jeandet P, Mazeyrat-Gourbeyre F, Clement C, Baillieul F, Dorey S. Comparative analysis of defence responses induced by the endophytic plant growth-promoting rhizobacterium Burkholderia phytofirmans strain PsJN and the non-host bacterium Pseudomonas syringae pv. pisi in grapevine cell suspensions. J Exp Bot. 2011;62:595–603. doi: 10.1093/jxb/erq291. http://dx.doi.org/10.1093/jxb/erq291. [DOI] [PubMed] [Google Scholar]
- 153.van Loon LC, Bakker PAHM, van der Heijdt WHW, Wendehenne D, Pugin A. Early responses of tobacco suspension cells to rhizobacterial elicitors of induced systemic resistance. Mol Plant Microbe Interact. 2008;21:1609–1621. doi: 10.1094/MPMI-21-12-1609. http://dx.doi.org/10.1094/MPMI-21-12-1609. [DOI] [PubMed] [Google Scholar]
- 154.Blodgett JT, Eyles A, Bonello P. Organ-dependent induction of systemic resistance and systemic susceptibility in Pinus nigra inoculated with Sphaeropsis sapinea and Diplodia scrobiculata. Tree Physiol. 2007;27:511–517. doi: 10.1093/treephys/27.4.511. http://dx.doi.org/10.1093/treephys/27.4.511. [DOI] [PubMed] [Google Scholar]
- 155.Vu T, Hauschild R, Sikora RA. Fusarium oxysporum endophytes induced systemic resistance against Radopholus similis on banana. Nematology. 2006;8:847–852. http://dx.doi.org/10.1163/156854106779799259. [Google Scholar]
- 156.Bae H, Roberts DP, Lim H-S, Strem MD, Park S-C, Ryu C-M, Melnick RL, Bailey BA. Endophytic Trichoderma isolates from tropical environments delay disease onset and induce resistance against Phytophthora capsici in hot pepper using multiple mechanisms. Mol Plant Microbe Interact. 2011;24:336–351. doi: 10.1094/MPMI-09-10-0221. http://dx.doi.org/10.1094/MPMI-09-10-0221. [DOI] [PubMed] [Google Scholar]
- 157.Gunatilaka AAL. Natural products from plant-associated micro-organisms: distribution, structural diversity, bioactivity, and implications of their occurrence. J Nat Prod. 2006;69:509–526. doi: 10.1021/np058128n. http://dx.doi.org/10.1021/np058128n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Higginbotham SJ, Arnold AE, Ibañez A, Spadafora C, Coley PD, Kursar TA. Bioactivity of fungal endophytes as a function of endophyte taxonomy and the taxonomy and distribution of their host plants. PLoS One. 2013;8:e73192. doi: 10.1371/journal.pone.0073192. http://dx.doi.org/10.1371/journal.pone.0073192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Tejesvi MV, Segura DR, Schnorr KM, Sandvang D, Mattila S, Olsen PB, Neve S, Kruse T, Kristensen HH, Pirttilä AM. An antimicrobial peptide from endophytic Fusarium tricinctum of Rhododendron tomentosum Harmaja. Fungal Divers. 2013;60:153–159. http://dx.doi.org/10.1007/s13225-013-0227-8. [Google Scholar]
- 160.Fletcher LR, Harvey IC. An association of a Lolium endophyte with ryegrass staggers. N Z Vet J. 1981;29:185–186. doi: 10.1080/00480169.1981.34839. http://dx.doi.org/10.1080/00480169.1981.34839. [DOI] [PubMed] [Google Scholar]
- 161.Gallagher RT, Hawkes AD, Steyn PS, Vleggaar R. Tremor-genic neurotoxins from perennial ryegrass causing ryegrass staggers disorder of livestock—structure elucidation of lolitrem-B. J Chem Soc Chem Commun (Camb) 1984;9:614–616. http://dx.doi.org/10.1039/C39840000614. [Google Scholar]
- 162.Bush LP, Cornelius PL, Buckner RC, Varney DR, Chapman RA, Burrus PB, Kennedy CW, Jones TA, Saunders MJ. Association of N-acetyl loline and N-formyl loline with Epichloë typhina in tall fescue. Crop Sci. 1982;22:941–943. http://dx.doi.org/10.2135/cropsci1982.0011183X002200050010x. [Google Scholar]
- 163.Siegel MR, Latch GCM, Bush LP, Fannin FF, Rowan DD, Tapper BA, Bacon CW, Johnson MC. Fungal endophyte-infected grasses: alkaloid accumulation and aphid response. J Chem Ecol. 1990;16:3301–3315. doi: 10.1007/BF00982100. http://dx.doi.org/10.1007/BF00982100. [DOI] [PubMed] [Google Scholar]
- 164.Tejesvi MV, Kajula M, Mattila S, Pirttilä AM. Bioactivity and genetic diversity of endophytic fungi in Rhododendron tomentosum Harmaja. Fungal Divers. 2011;47:97–107. http://dx.doi.org/10.1007/s13225-010-0087-4. [Google Scholar]
- 165.Arnold AE, Mejia LC, Kyllo D, Rojas EI, Maynard Z, Robbins N, Herre EA. Fungal endophytes limit pathogen damage in a tropical tree. Proc Natl Acad Sci U S A. 2003;100:15649–15654. doi: 10.1073/pnas.2533483100. http://dx.doi.org/10.1073/pnas.2533483100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Arnold AE. Understanding the diversity of foliar endophytic fungi: progress, challenges, and frontiers. Fungal Biol Rev. 2007;21:51–66. http://dx.doi.org/10.1016/j.fbr.2007.05.003. [Google Scholar]
- 167.Clay K. Fungi and the food of the gods. Nature. 2004;427:401–402. doi: 10.1038/427401a. http://dx.doi.org/10.1038/427401a. [DOI] [PubMed] [Google Scholar]
- 168.Combès A, Ndoye I, Bance C, Bruzaud J, Djediat C, Dupont J, Nay B, Prado S. Chemical communication between the endophytic fungus Paraconiothyrium variabile and the phytopathogen Fusarium oxysporum. PLoS One. 2012;7:e47313. doi: 10.1371/journal.pone.0047313. http://dx.doi.org/10.1371/journal.pone.0047313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Taghavi S, van der Lelie D, Hoffman A, Zhang YB, Walla MD, Vangronsveld J, Newman L, Monchy S. Genome sequence of the plant growth promoting endophytic bacterium Enterobacter sp 638. PLoS Genet. 2010;6:e1000943. doi: 10.1371/journal.pgen.1000943. http://dx.doi.org/10.1371/journal.pgen.1000943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Castillo UF, Strobel GA, Ford EJ, Hess WM, Porter H, Jensen JB, Albert H, Robison R, Condron MAM, Teplow DB, Stevens D, Yaver D. Munumbicins, wide-spectrum antibiotics produced by Streptomyces NRRL 30562, endophytic on Kennedia nigriscans. Microbiology. 2002;148:2675–2685. doi: 10.1099/00221287-148-9-2675. [DOI] [PubMed] [Google Scholar]
- 171.Castillo U, Harper JK, Strobel GA, Sears J, Alesi K, Ford E, Lin J, Hunter M, Maranta M, Ge HY, Yaver D, Jensen JB, Porter H, Robison R, Millar D, Hess WM, Condron M, Teplow D. Kakadumycins, novel antibiotics from Streptomyces sp NRRL 30566, an endophyte of Grevillea pteridifolia. FEMS Microbiol Lett. 2003;224:183–190. doi: 10.1016/S0378-1097(03)00426-9. http://dx.doi.org/10.1016/S0378-1097(03)00426-9. [DOI] [PubMed] [Google Scholar]
- 172.Ezra D, Castillo UF, Strobel GA, Hess WM, Porter H, Jensen JB, Condron MAM, Teplow DB, Sears J, Maranta M, Hunter M, Weber B, Yaver D. Coronamycins, peptide antibiotics produced by a verticillate Streptomyces sp. (MSU-2110) endophytic on Monstera sp. Microbiology. 2004;150:785–793. doi: 10.1099/mic.0.26645-0. http://dx.doi.org/10.1099/mic.0.26645-0. [DOI] [PubMed] [Google Scholar]
- 173.Ding L, Maier A, Fiebig H-H, Lin W-H, Hertweck C. A family of multicyclic indolosesquiterpenes from a bacterial endophyte. Org Biomol Chem. 2011;9:4029–4031. doi: 10.1039/c1ob05283g. http://dx.doi.org/10.1039/c1ob05283g. [DOI] [PubMed] [Google Scholar]
- 174.Inahashi Y, Iwatsuki M, Ishiyama A, Namatame M, Nishihara-Tsukashima A, Matsumoto A, Hirose T, Sunazuka T, Yamada H, Otoguro K, Takahashi Y, Omura S, Shiomi K. Spoxazomicins A-C, novel antitrypanosomal alkaloids produced by an endophytic actinomycete, Streptosporangium oxazolinicum K07-0460(T) J Antibiot. 2011;64:303–307. doi: 10.1038/ja.2011.16. http://dx.doi.org/10.1038/ja.2011.16. [DOI] [PubMed] [Google Scholar]
- 175.Brader G, Compant S, Mitter B, Trognitz F, Sessitsch A. Metabolic potential of endophytic bacteria. Curr Opin Biotechnol. 2014;27:30–37. doi: 10.1016/j.copbio.2013.09.012. http://dx.doi.org/10.1016/j.copbio.2013.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Strobel G, Daisy B. Bioprospecting for microbial endophytes and their natural products. Microbiol Mol Biol Rev. 2003;67:491–502. doi: 10.1128/MMBR.67.4.491-502.2003. http://dx.doi.org/10.1128/MMBR.67.4.491-502.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Strobel G, Daisy B, Castillo U, Harper J. Natural products from endophytic microorganisms. J Nat Prod. 2004;67:257–268. doi: 10.1021/np030397v. http://dx.doi.org/10.1021/np030397v. [DOI] [PubMed] [Google Scholar]
- 178.Verma VC, Kharwar RN, Strobel GA. Chemical and functional diversity of natural products from plant associated endophytic fungi. Nat Prod Commun. 2009;4:1511–1532. [PubMed] [Google Scholar]
- 179.Tan RX, Zou WX. Endophytes: a rich source of functional metabolites. Nat Prod Rep. 2001;18:448–459. doi: 10.1039/b100918o. http://dx.doi.org/10.1039/b100918o. [DOI] [PubMed] [Google Scholar]
- 180.Aly AH, Debbab A, Kjer J, Proksch P. Fungal endophytes from higher plants: a prolific source of phytochemicals and other bioactive natural products. Fungal Divers. 2010;41:1–16. http://dx.doi.org/10.1007/s13225-010-0034-4. [Google Scholar]
- 181.Aly AH, Debbab A, Proksch P. Fungal endophytes: unique plant inhabitants with great promises. Appl Microbiol Biotechnol. 2011;90:1829–1845. doi: 10.1007/s00253-011-3270-y. http://dx.doi.org/10.1007/s00253-011-3270-y. [DOI] [PubMed] [Google Scholar]
- 182.Zhang HW, Song YC, Tan RX. Biology and chemistry of endophytes. Nat Prod Rep. 2006;23:753–771. doi: 10.1039/b609472b. http://dx.doi.org/10.1039/b609472b. [DOI] [PubMed] [Google Scholar]
- 183.Schulz B, Boyle C. The endophytic continuum. Mycol Res. 2005;109:661–686. doi: 10.1017/s095375620500273x. http://dx.doi.org/10.1017/S095375620500273X. [DOI] [PubMed] [Google Scholar]
- 184.Zabetakis I, Holden MA. Strawberry flavour: analysis and biosyn-thesis. J Sci Food Agric. 1997;74:421–434. http://dx.doi.org/10.1002/(SICI)1097-0010(199708)74:4<421::AID-JSFA817>3.0.CO;2-6. [Google Scholar]
- 185.Koutsompogeras P, Kyriacou A, Zabetakis I. The formation of 2,5-dimethyl-4-hydroxy-2H-furan-3-one by cell-free extracts of Methylobacterium extorquens and strawberry (Fragaria × ananassa cv. Elsanta) Food Chem. 2007;104:1654–1661. doi: 10.1021/jf0516033. http://dx.doi.org/10.1016/j.foodchem.2007.03.025. [DOI] [PubMed] [Google Scholar]
- 186.Verginer M, Siegmund B, Cardinale M, Mueller H, Choi Y, Miguez CB, Leitner E, Berg G. Monitoring the plant epiphyte Methylobacterium extorquens DSM 21961 by real-time PCR and its influence on the strawberry flavor. FEMS Microbiol Ecol. 2010;74:136–145. doi: 10.1111/j.1574-6941.2010.00942.x. http://dx.doi.org/10.1111/j.1574-6941.2010.00942.x. [DOI] [PubMed] [Google Scholar]
- 187.Nasopoulou C, Pohjanen J, Koskimaki JJ, Zabetakis I, Pirttilä AM. Localization of strawberry (Fragaria × ananassa) and Methylobacterium extorquens genes of strawberry flavor biosynthesis in strawberry tissue by in situ hybridization. J Plant Physiol. 2014;171:1099–1105. doi: 10.1016/j.jplph.2014.03.018. http://dx.doi.org/10.1016/j.jplph.2014.03.018. [DOI] [PubMed] [Google Scholar]
- 188.Koskimäki JJ, Hokkanen J, Jaakola L, Suorsa M, Tolonen A, Mattila S, Pirttilä AM, Hohtola A. Flavonoid biosynthesis and degradation play a role in early defence responses of bilberry (Vaccinium myrtillus) against biotic stress. Eur J Plant Pathol. 2009;125:629–640. http://dx.doi.org/10.1007/s10658-009-9511-6. [Google Scholar]
- 189.Araújo WL, Lacava PT, Andreote FD, Azevedo JL. Interaction between endophytes and plant host: biotechnological aspects. In: Barka EA, Cĺement C, editors. Plant-microbe interactions. Research Sign-post; Kerala, India: 2008. pp. 95–115. [Google Scholar]
- 190.Kajula M, Tejesvi MV, Kolehmainen S, Makinen A, Hokkanen J, Mattila S, Pirttilä AM. The siderophore ferricrocin produced by specific foliar endophytic fungi in vitro. Fungal Biol. 2010;114:248–254. doi: 10.1016/j.funbio.2010.01.004. http://dx.doi.org/10.1016/j.funbio.2010.01.004. [DOI] [PubMed] [Google Scholar]
- 191.Johnson LJ, Koulman A, Christensen M, Lane GA, Fraser K, Forester N, Johnson RD, Bryan GT, Rasmussen S. An extracellular siderophore is required to maintain the mutualistic interaction of Epichloë festucae with Lolium perenne. PLoS Pathog. 2013;9:e1003332. doi: 10.1371/journal.ppat.1003332. http://dx.doi.org/10.1371/journal.ppat.1003332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Rosconi F, Davyt D, Martínez V, Martínez M, Abin-Carriquiry JA, Zane H, Butler A, de Souza EM, Fabiano E. Identification and structural characterization of serobactins, a suite of lipopeptide siderophores produced by the grass endophyte Herbaspirillum seropedicae. Environ Microbiol. 2013;15:916–927. doi: 10.1111/1462-2920.12075. http://dx.doi.org/10.1111/1462-2920.12075. [DOI] [PubMed] [Google Scholar]
- 193.Schippers B, Bakker AW, Bakker P. Interactions of deleterious and beneficial rhizosphere microorganisms and the effect of cropping practices. Annu Rev Phytopathol. 1987;25:339–358. http://dx.doi.org/10.1146/annurev.py.25.090187.002011. [Google Scholar]
- 194.Powell PE, Cline GR, Reid CPP, Szaniszlo PJ. Occurrence of hydroxamate siderophore iron chelators in soils. Nature. 1980;287:833–834. http://dx.doi.org/10.1038/287833a0. [Google Scholar]
- 195.Bearden SW, Perry RD. The Yfe system of Yersinia pestis transports iron and manganese and is required for full virulence of plague. Mol Microbiol. 1999;32:403–414. doi: 10.1046/j.1365-2958.1999.01360.x. http://dx.doi.org/10.1046/j.1365-2958.1999.01360.x. [DOI] [PubMed] [Google Scholar]
- 196.Schrettl M, Bignell E, Kragl C, Joechl C, Rogers T, Arst HN, Haynes K, Haas H. Siderophore biosynthesis but not reductive iron assimilation is essential for Aspergillus fumigatus virulence. J Exp Med. 2004;200:1213–1219. doi: 10.1084/jem.20041242. http://dx.doi.org/10.1084/jem.20041242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Mitter B, Petric A, Shin MW, Chain PSG, Hauberg-Lotte L, Reinhold-Hurek B, Nowak J, Sessitsch A. Comparative genome analysis of Burkholderia phytofirmans PsJN reveals a wide spectrum of endophytic lifestyles based on interaction strategies with host plants. Front Plant Sci. 2013;4:120. doi: 10.3389/fpls.2013.00120. http://dx.doi.org/10.3389/fpls.2013.00120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Bacon CW, Hill NS. Symptomless grass endophytes: products of coevolutionary symbioses and their role in the ecological adaptations of grasses. In: Redlin SC, Carris LM, editors. Endophytic fungi in grasses and woody plants. American Phytopathological Society Press; St. Paul, MN: 1996. pp. 155–178. [Google Scholar]
- 199.Ravel C, Courty C, Coudret A, Charmet G. Beneficial effects of Neotyphodium lolii on the growth and the water status in perennial rye-grass cultivated under nitrogen deficiency or drought stress. Agronomie. 1997;17:173–181. http://dx.doi.org/10.1051/agro:19970304. [Google Scholar]
- 200.Baltruschat H, Fodor J, Harrach BD, Niemczyk E, Barna B, Gullner G, Janeczko A, Kogel K-H, Schaefer P, Schwarczinger I, Zuccaro A, Skoczowski A. Salt tolerance of barley induced by the root endophyte Piriformospora indica is associated with a strong increase in anti-oxidants. New Phytol. 2008;180:501–510. doi: 10.1111/j.1469-8137.2008.02583.x. http://dx.doi.org/10.1111/j.1469-8137.2008.02583.x. [DOI] [PubMed] [Google Scholar]
- 201.Sun C, Johnson J, Cai D, Sherameti I, Oelmüeller R, Lou B. Piriformospora indica confers drought tolerance in Chinese cabbage leaves by stimulating antioxidant enzymes, the expression of drought-related genes and the plastid-localized CAS protein. J Plant Physiol. 2010;167:1009–1017. doi: 10.1016/j.jplph.2010.02.013. http://dx.doi.org/10.1016/j.jplph.2010.02.013. [DOI] [PubMed] [Google Scholar]
- 202.Bae H, Sicher RC, Kim MS, Kim S-H, Strem MD, Melnick RL, Bailey BA. The beneficial endophyte Trichoderma hamatum isolate DIS 219b promotes growth and delays the onset of the drought response in Theobroma cacao. J Exp Bot. 2009;60:3279–3295. doi: 10.1093/jxb/erp165. http://dx.doi.org/10.1093/jxb/erp165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Naveed M, Mitter B, Reichenauer TG, Wieczorek K, Sessitsch A. Increased drought stress resilience of maize through endophytic colonization by Burkholderia phytofirmans PsJN and Enterobacter sp FD17. Environ Exp Bot. 2014;97:30–39. http://dx.doi.org/10.1016/j.envexpbot.2013.09.014. [Google Scholar]
- 204.Naveed M, Hussain MB, Zahir ZA, Mitter B, Sessitsch A. Drought stress amelioration in wheat through inoculation with Burkholderia phytofirmans strain PsJN. Plant Growth Regul. 2014;73:121–131. http://dx.doi.org/10.1007/s10725-013-9874-8. [Google Scholar]
- 205.Redman RS, Kim YO, Woodward CJDA, Greer C, Espino L, Doty SL, Rodriguez RJ. Increased fitness of rice plants to abiotic stress via habitat adapted symbiosis: a strategy for mitigating impacts of climate change. PLoS One. 2011;6:e14823. doi: 10.1371/journal.pone.0014823. http://dx.doi.org/10.1371/journal.pone.0014823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Ait Barka E, Nowak J, Clement C. Enhancement of chilling resistance of inoculated grapevine plantlets with a plant growth-promoting rhizobacterium, Burkholderia phytofirmans strain PsJN. Appl Environ Microbiol. 2006;72:7246–7252. doi: 10.1128/AEM.01047-06. http://dx.doi.org/10.1128/AEM.01047-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Glick BR, Penrose DM, Li JP. A model for the lowering of plant ethylene concentrations by plant growth-promoting bacteria. J Theor Biol. 1998;190:63–68. doi: 10.1006/jtbi.1997.0532. http://dx.doi.org/10.1006/jtbi.1997.0532. [DOI] [PubMed] [Google Scholar]
- 208.Iniguez AL, Dong YM, Carter HD, Ahmer BMM, Stone JM, Triplett EW. Regulation of enteric endophytic bacterial colonization by plant defenses. Mol Plant Microbe Interact. 2005;18:169–178. doi: 10.1094/MPMI-18-0169. http://dx.doi.org/10.1094/MPMI-18-0169. [DOI] [PubMed] [Google Scholar]
- 209.Sun Y, Cheng Z, Glick BR. The presence of a 1-aminocyclopro-pane-1-carboxylate (ACC) deaminase deletion mutation alters the physiology of the endophytic plant growth-promoting bacterium Burkholderia phytofirmans PsJN. FEMS Microbiol Lett. 2009;296:131–136. doi: 10.1111/j.1574-6968.2009.01625.x. http://dx.doi.org/10.1111/j.1574-6968.2009.01625.x. [DOI] [PubMed] [Google Scholar]
- 210.Ali S, Charles TC, Glick BR. Delay of flower senescence by bacterial endophytes expressing 1-aminocyclopropane-1-carboxylate deaminase. J Appl Microbiol. 2012;113:1139–1144. doi: 10.1111/j.1365-2672.2012.05409.x. http://dx.doi.org/10.1111/j.1365-2672.2012.05409.x. [DOI] [PubMed] [Google Scholar]
- 211.Singh LP, Gill SS, Tuteja N. Unraveling the role of fungal symbionts in plant abiotic stress tolerance. Plant Signal Behav. 2011;6:175–191. doi: 10.4161/psb.6.2.14146. http://dx.doi.org/10.4161/psb.6.2.14146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Clay K. Fungal endophytes of grasses: a defensive mutualism between plants and fungi. Ecology. 1988;69:10–16. http://dx.doi.org/10.2307/1943155. [Google Scholar]
- 213.Long HH, Schmidt DD, Baldwin IT. Native bacterial endophytes promote host growth in a species-specific manner; phytohormone manipulations do not result in common growth responses. PLoS One. 2008;3:e2702. doi: 10.1371/journal.pone.0002702. http://dx.doi.org/10.1371/journal.pone.0002702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Spiering MJ, Greer DH, Schmid J. Effects of the fungal endophyte, Neotyphodium lolii, on net photosynthesis and growth rates of perennial ryegrass (Lolium perenne) are independent of in planta endophyte concentration. Ann Bot. 2006;98:379–387. doi: 10.1093/aob/mcl108. http://dx.doi.org/10.1093/aob/mcl108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Rogers A, McDonald K, Muehlbauer MF, Hoffman A, Koenig K, Newman L, Taghavi S, van der Lelie D. Inoculation of hybrid poplar with the endophytic bacterium Enterobacter sp 638 increases bio-mass but does not impact leaf level physiology. Glob Change Biol Bioenergy. 2012;4:364–370. http://dx.doi.org/10.1111/j.1757-1707.2011.01119.x. [Google Scholar]
- 216.Shi Y, Lou K, Li C. Promotion of plant growth by phytohormone-producing endophytic microbes of sugar beet. Biol Fertil Soils. 2009;45:645–653. http://dx.doi.org/10.1007/s00374-009-0376-9. [Google Scholar]
- 217.Khan AL, Hamayun M, Kang S-M, Kim Y-H, Jung H-Y, Lee J-H, Lee I-J. Endophytic fungal association via gibberellins and indole acetic acid can improve plant growth under abiotic stress: an example of Paecilomyces formosus LHL10. BMC Microbiol. 2012;12:3. doi: 10.1186/1471-2180-12-3. http://dx.doi.org/10.1186/1471-2180-1112-1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Bastián F, Cohen A, Piccoli P, Luna V, Baraldi R, Bottini R. Production of indole-3-acetic acid and gibberellins A(1) and A(3) by Acetobacter diazotrophicus and Herbaspirillum seropedicae in chemically-defined culture media. Plant Growth Regul. 1998;24:7–11. http://dx.doi.org/10.1023/A:1005964031159. [Google Scholar]
- 219.Merzaeva OV, Shirokikh IG. The production of auxins by the endophytic bacteria of winter rye. Appl Biochem Microbiol. 2010;46:44–50. http://dx.doi.org/10.1134/S0003683810010072. [PubMed] [Google Scholar]
- 220.Suzuki S, He YX, Oyaizu H. Indole-3-acetic acid production in Pseudomonas fluorescens HP72 and its association with suppression of creeping bentgrass brown patch. Curr Microbiol. 2003;47:138–143. doi: 10.1007/s00284-002-3968-2. http://dx.doi.org/10.1007/s00284-002-3968-2. [DOI] [PubMed] [Google Scholar]
- 221.Navarro L, Dunoyer P, Jay F, Arnold B, Dharmasiri N, Estelle M, Voinnet O, Jones JDG. A plant miRNA contributes to antibacterial resistance by repressing auxin signaling. Science. 2006;312:436–439. doi: 10.1126/science.1126088. http://dx.doi.org/10.1126/science.1126088. [DOI] [PubMed] [Google Scholar]
- 222.Vadassery J, Ritter C, Venus Y, Camehl I, Varma A, Shahollari B, Novak O, Strnad M, Ludwig-Mueller J, Oelmueller R. The role of auxins and cytokinins in the mutualistic interaction between Arabidopsis and Piriformospora indica. Mol Plant Microbe Interact. 2008;21:1371–1383. doi: 10.1094/MPMI-21-10-1371. http://dx.doi.org/10.1094/MPMI-21-10-1371. [DOI] [PubMed] [Google Scholar]
- 223.Pirttilä AM, Joensuu P, Pospiech H, Jalonen J, Hohtola A. Bud endophytes of Scots pine produce adenine derivatives and other compounds that affect morphology and mitigate browning of callus cultures. Physiol Plant. 2004;121:305–312. doi: 10.1111/j.0031-9317.2004.00330.x. http://dx.doi.org/10.1111/j.0031-9317.2004.00330.x. [DOI] [PubMed] [Google Scholar]
- 224.Ryu CM, Farag MA, Hu CH, Reddy MS, Wei HX, Pare PW, Kloepper JW. Bacterial volatiles promote growth in Arabidopsis. Proc Natl Acad Sci U S A. 2003;100:4927–4932. doi: 10.1073/pnas.0730845100. http://dx.doi.org/10.1073/pnas.0730845100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Ryu CM, Farag MA, Hu CH, Reddy MS, Kloepper JW, Pare PW. Bacterial volatiles induce systemic resistance in Arabidopsis. Plant Physiol. 2004;134:1017–1026. doi: 10.1104/pp.103.026583. http://dx.doi.org/10.1104/pp.103.026583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Ryu CM, Hu CH, Locy RD, Kloepper JW. Study of mechanisms for plant growth promotion elicited by rhizobacteria in Arabidopsis thaliana. Plant Soil. 2005;268:285–292. http://dx.doi.org/10.1007/s11104-004-0301-9. [Google Scholar]
- 227.Johnston-Monje D, Raizada MN. Conservation and diversity of seed associated endophytes in Zea across boundaries of evolution, ethnography and ecology. PLoS One. 2011;6:e20396. doi: 10.1371/journal.pone.0020396. http://dx.doi.org/10.1371/journal.pone.0020396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Taghavi S, Garafola C, Monchy S, Newman L, Hoffman A, Weyens N, Barac T, Vangronsveld J, van der Lelie D. Genome survey and characterization of endophytic bacteria exhibiting a beneficial effect on growth and development of poplar trees. Appl Environ Microbiol. 2009;75:748–757. doi: 10.1128/AEM.02239-08. http://dx.doi.org/10.1128/AEM.02239-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Niemi K, Julkunen-Tiitto R, Haggman H, Sarjala T. Suillus variegatus causes significant changes in the content of individual polyamines and flavonoids in Scots pine seedlings during mycorrhiza formation in vitro. J Exp Bot. 2007;58:391–401. doi: 10.1093/jxb/erl209. http://dx.doi.org/10.1093/jxb/erl209. [DOI] [PubMed] [Google Scholar]
- 230.Perrig D, Boiero ML, Masciarelli OA, Penna C, Ruiz OA, Cassan FD, Luna MV. Plant-growth-promoting compounds produced by two agronomically important strains of Azospirillum brasilense, and implications for inoculant formulation. Appl Microbiol Biotechnol. 2007;75:1143–1150. doi: 10.1007/s00253-007-0909-9. http://dx.doi.org/10.1007/s00253-007-0909-9. [DOI] [PubMed] [Google Scholar]
- 231.Baldani JI, Caruso L, Baldani VLD, Goi SR, Dobereiner J. Recent advances in BNF with non-legume plants. Soil Biol Biochem. 1997;29:911–922. http://dx.doi.org/10.1016/S0038-0717(96)00218-0. [Google Scholar]
- 232.Reinhold-Hurek B, Hurek T. Life in grasses: diazotrophic endophytes. Trends Microbiol. 1998;6:139–144. doi: 10.1016/s0966-842x(98)01229-3. http://dx.doi.org/10.1016/S0966-842X(98)01229-3. [DOI] [PubMed] [Google Scholar]
- 233.Dalla Santa OR, Hernandez RF, Alvarez GLM, Ronzelli P, Soccol CR. Azospirillum sp inoculation in wheat, barley and oats seeds greenhouse experiments. Braz Arch Biol Technol. 2004;47:843–850. http://dx.doi.org/10.1590/S1516-89132004000600002. [Google Scholar]
- 234.Dong ZM, Canny MJ, McCully ME, Roboredo MR, Cabadilla CF, Ortega E, Rodes R. A nitrogen-fixing endophyte of sugarcane stems: a new role for the apoplast. Plant Physiol. 1994;105:1139–1147. doi: 10.1104/pp.105.4.1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Carrell AA, Frank AC. Pinus flexilis and Picea engelmannii share a simple and consistent needle endophyte microbiota with a potential role in nitrogen fixation. Front Microbiol. 2014;5:333. doi: 10.3389/fmicb.2014.00333. http://dx.doi.org/10.3389/fmicb.2014.00333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Okon Y, Labandera-Gonzalez CA. Agronomic applications of Azospirillum: an evaluation of 20 years worldwide field inoculation. Soil Biol Biochem. 1994;26:1591–1601. http://dx.doi.org/10.1016/0038-0717(94)90311-5. [Google Scholar]
- 237.Gorischek AM, Afkhami ME, Seifert EK, Rudgers JA. Fungal symbionts as manipulators of plant reproductive biology. Am Nat. 2013;181:562–570. doi: 10.1086/669606. http://dx.doi.org/10.1086/669606. [DOI] [PubMed] [Google Scholar]
- 238.Aschehoug ET, Callaway RM, Newcombe G, Tharayil N, Chen S. Fungal endophyte increases the allelopathic effects of an invasive forb. Oecologia. 2014;175:285–291. doi: 10.1007/s00442-014-2891-0. http://dx.doi.org/10.1007/s00442-014-2891-0. [DOI] [PubMed] [Google Scholar]
- 239.Newcombe G, Shipunov A, Eigenbrode S, Raghavendra AK, Ding H, Anderson CL, Menjivar R, Crawford M, Schwarzlander M. Endophytes influence protection and growth of an invasive plant. Commun Integr Biol. 2009;2:29–31. doi: 10.4161/cib.2.1.7393. http://dx.doi.org/10.4161/cib.2.1.7393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Wäli PP, Wäli PR, Saikkonen K, Tuomi J. Is the pathogenic ergot fungus a conditional defensive mutualist for its host grass? PLoS One. 2013;8:e69249. doi: 10.1371/journal.pone.0069249. http://dx.doi.org/10.1371/journal.pone.0069249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Engelstädter J, Hurst GDD. The ecology and evolution of microbes that manipulate host reproduction. Annu Rev Ecol Evol Syst. 2009;40:127–149. http://dx.doi.org/10.1146/annurev.ecolsys.110308.120206. [Google Scholar]
- 242.Wright KM, Chapman S, McGeachy K, Humphris S, Campbell E, Toth IK, Holden NJ. The endophytic lifestyle of Escherichia coli O157: H7: quantification and internal localization in roots. Phytopathology. 2013;103:333–340. doi: 10.1094/PHYTO-08-12-0209-FI. http://dx.doi.org/10.1094/PHYTO-08-12-0209-FI. [DOI] [PubMed] [Google Scholar]
- 243.Amadou C, Pascal G, Mangenot S, Glew M, Bontemps C, Capela D, Carrere S, Cruveiller S, Dossat C, Lajus A, Marchetti M, Poinsot V, Rouy Z, Servin B, Saad M, Schenowitz C, Barbe V, Batut J, Medigue C, Masson-Boivin C. Genome sequence of the β-rhizobium Cupriavidus taiwanensis and comparative genomics of rhizobia. Genome Res. 2008;18:1472–1483. doi: 10.1101/gr.076448.108. http://dx.doi.org/10.1101/gr.076448.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Tisserant E, Malbreil M, Kuo A, Kohler A, Symeonidi A, Balestrini R, Charron P, Duensing N, Frey NFD, Gianinazzi-Pearson V, Gilbert LB, Handa Y, Herr JR, Hijri M, Koul R, Kawaguchi M, Krajinski F, Lammers PJ, Masclauxm FG, Murat C, Morin E, Ndikumana S, Pagni M, Petitpierre D, Requena N, Rosikiewicz P, Riley R, Saito K, Clemente HS, Shapiro H, Van Tuinen D, Becard G, Bonfante P, Paszkowski U, Shachar-Hill YY, Tuskan GA, Young PW, Sanders IR, Henrissat B, Rensing SA, Grigoriev IV, Corradi N, Roux C, Martin F. Genome of an arbuscular mycorrhizal fungus provides insight into the oldest plant symbiosis. Proc Natl Acad Sci U S A. 2013;110:20117–20122. doi: 10.1073/pnas.1313452110. http://dx.doi.org/10.1073/pnas.1313452110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Tian CF, Zhou YJ, Zhang YM, Li QQ, Zhang YZ, Li DF, Wang S, Wang J, Gilbert LB, Li YR, Chen WX. Comparative genomics of rhizobia nodulating soybean suggests extensive recruitment of lineagespecific genes in adaptations. Proc Natl Acad Sci U S A. 2012;109:8629–8634. doi: 10.1073/pnas.1120436109. http://dx.doi.org/10.1073/pnas.1120436109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Karpinets TV, Park BH, Syed MH, Klotz MG, Uberbacher EC. Metabolic environments and genomic features associated with pathogenic and mutualistic interactions between bacteria and plants. Mol Plant Microbe Interact. 2014;27:664–677. doi: 10.1094/MPMI-12-13-0368-R. http://dx.doi.org/10.1094/MPMI-12-13-0368-R. [DOI] [PubMed] [Google Scholar]
- 247.Mitter B, Brader G, Afzal M, Compant S, Naveed M, Trognitz F, Sessitsch A. Advances in elucidating beneficial interactions between plants, soil, and bacteria. In: Sparks DL, editor. Advances in agronomy. Vol. 121. Elsevier; San Diego, CA: 2013. pp. 381–445. http://dx.doi.org/10.1016/B978-0-12-407685-3.00007-4. [Google Scholar]
- 248.Markowitz VM, Chen IMA, Palaniappan K, Chu K, Szeto E, Grechkin Y, Ratner A, Jacob B, Huang J, Williams P, Huntemann M, Anderson I, Mavromatis K, Ivanova NN, Kyrpides NC. IMG: the integrated microbial genomes database and comparative analysis system. Nucleic Acids Res. 2012;40:D115–D122. doi: 10.1093/nar/gkr1044. http://dx.doi.org/10.1093/nar/gkr1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249.Paulson JN, Stine OC, Bravo HC, Pop M. Differential abundance analysis for microbial marker-gene surveys. Nat Methods. 2013;10:1200–1212. doi: 10.1038/nmeth.2658. http://dx.doi.org/10.1038/nmeth.2658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Wadhams GH, Armitage JP. Making sense of it all: bacterial chemotaxis. Nat Rev Mol Cell Biol. 2004;5:1024–1037. doi: 10.1038/nrm1524. http://dx.doi.org/10.1038/nrm1524. [DOI] [PubMed] [Google Scholar]
- 251.Hartmann A, Schmid M, van Tuinen D, Berg G. Plant-driven selection of microbes. Plant Soil. 2009;321:235–257. http://dx.doi.org/10.1007/s11104-008-9814-y. [Google Scholar]
- 252.Porter SL, Wadhams GH, Armitage JP. Signal processing in complex chemotaxis pathways. Nat Rev Microbiol. 2011;9:153–165. doi: 10.1038/nrmicro2505. http://dx.doi.org/10.1038/nrmicro2505. [DOI] [PubMed] [Google Scholar]
- 253.Waters CM, Bassler BL. Quorum sensing: cell-to-cell communication in bacteria. Annu Rev Cell Dev Biol. 2005;21:319–346. doi: 10.1146/annurev.cellbio.21.012704.131001. http://dx.doi.org/10.1146/annurev.cellbio.21.012704.131001. [DOI] [PubMed] [Google Scholar]
- 254.Keller L, Surette MG. Communication in bacteria: an ecological and evolutionary perspective. Nat Rev Microbiol. 2006;4:249–258. doi: 10.1038/nrmicro1383. http://dx.doi.org/10.1038/nrmicro1383. [DOI] [PubMed] [Google Scholar]
- 255.Elsen S, Swem LR, Swem DL, Bauer CE. RegB/RegA, a highly conserved redox-responding global two-component regulatory system. Microbiol Mol Biol Rev. 2004;68:263–279. doi: 10.1128/MMBR.68.2.263-279.2004. http://dx.doi.org/10.1128/MMBR.68.2.263-279.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256.Pawlowski K, Klosse U, Debruijn FJ. Characterization of a novel Azorhizobium caulinodans ORS571 2-component regulatory system, NtrY/NtrX, involved in nitrogen-fixation and metabolism. Mol Gen Genet. 1991;231:124–138. doi: 10.1007/BF00293830. http://dx.doi.org/10.1007/BF00293830. [DOI] [PubMed] [Google Scholar]
- 257.Balleza E, Lopez-Bojorquez LN, Martinez-Antonio A, Resendis-Antonio O, Lozada-Chavez I, Balderas-Martinez YI, Encarnacion S, Collado-Vides J. Regulation by transcription factors in bacteria: beyond description. FEMS Microbiol Rev. 2009;33:133–151. doi: 10.1111/j.1574-6976.2008.00145.x. http://dx.doi.org/10.1111/j.1574-6976.2008.00145.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258.Wu X, Monchy S, Taghavi S, Zhu W, Ramos J, van der Lelie D. Comparative genomics and functional analysis of niche-specific adaptation in Pseudomonas putida. FEMS Microbiol Rev. 2011;35:299–323. doi: 10.1111/j.1574-6976.2010.00249.x. http://dx.doi.org/10.1111/j.1574-6976.2010.00249.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 259.Mastouri F, Bjoerkman T, Harman GE. Seed treatment with Trichoderma harzianum alleviates biotic, abiotic, and physiological stresses in germinating seeds and seedlings. Phytopathology. 2010;100:1213–1221. doi: 10.1094/PHYTO-03-10-0091. http://dx.doi.org/10.1094/PHYTO-03-10-0091. [DOI] [PubMed] [Google Scholar]
- 260.Downie JA. The roles of extracellular proteins, polysaccharides and signals in the interactions of rhizobia with legume roots. FEMS Microbiol Rev. 2010;34:150–170. doi: 10.1111/j.1574-6976.2009.00205.x. http://dx.doi.org/10.1111/j.1574-6976.2009.00205.x. [DOI] [PubMed] [Google Scholar]
- 261.Abramovitch RB, Anderson JC, Martin GB. Bacterial elicitation and evasion of plant innate immunity. Nat Rev Mol Cell Biol. 2006;7:601–611. doi: 10.1038/nrm1984. http://dx.doi.org/10.1038/nrm1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 262.Xiang T, Zong N, Zou Y, Wu Y, Zhang J, Xing W, Li Y, Tang X, Zhu L, Chai J, Zhou J-M. Pseudomonas syringae effector AvrPto blocks innate immunity by targeting receptor kinases. Curr Biol. 2008;18:74–80. doi: 10.1016/j.cub.2007.12.020. http://dx.doi.org/10.1016/j.cub.2007.12.020. [DOI] [PubMed] [Google Scholar]
- 263.Frank AC. The genomes of endophytic bacteria. In: Pirttilä AM, Frank AC, editors. Endophytes of forest trees: biology and applications. Springer; Dordrecht, Netherlands: 2011. pp. 107–136. http://dx.doi.org/10.1007/978-94-007-1599-8_7. [Google Scholar]
- 264.Pitzschke A, Hirt H. New insights into an old story: Agrobacterium-induced tumour formation in plants by plant transformation. EMBO J. 2010;29:1021–1032. doi: 10.1038/emboj.2010.8. http://dx.doi.org/10.1038/emboj.2010.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 265.Salomon D, Kinch LN, Trudgian DC, Guo X, Klimko JA, Grishin NV, Mirzaei H, Orth K. Marker for type VI secretion system effectors. Proc Natl Acad Sci U S A. 2014;111:9271–9276. doi: 10.1073/pnas.1406110111. http://dx.doi.org/10.1073/pnas.1406110111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 266.Böhm M, Hurek T, Reinhold-Hurek B. Twitching motility is essential for endophytic rice colonization by the N2-fixing endophyte Azoarcus sp strain BH72. Mol Plant Microbe Interact. 2007;20:526–533. doi: 10.1094/MPMI-20-5-0526. http://dx.doi.org/10.1094/MPMI-20-5-0526. [DOI] [PubMed] [Google Scholar]
- 267.Meng YZ, Li YX, Galvani CD, Hao GX, Turner JN, Burr TJ, Hoch HC. Upstream migration of Xylella fastidiosa via pilus-driven twitching motility. J Bacteriol. 2005;187:5560–5567. doi: 10.1128/JB.187.16.5560-5567.2005. http://dx.doi.org/10.1128/JB.187.16.5560-5567.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 268.Sheibani-Tezerji R, Naveed M, Jehl M-A, Sessitsch A, Rattei T, Mitter B. The genomes of closely related Pantoea ananatis maize seed endophytes having different effects on the host plant differ in secretion system genes and mobile genetic elements. Front Microbiol. 2015;6:440. doi: 10.3389/fmicb.2015.00440. http://dx.doi.org/10.3389/fmicb.2015.00440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 269.Fredricks DN, Relman DA. Sequence-based identification of microbial pathogens: a reconsideration of Koch’s postulates. Clin Microbiol Rev. 1996;9:18–33. doi: 10.1128/cmr.9.1.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 270.Jansson JK, Neufeld JD, Moran MA, Gilbert JA. Omics for understanding microbial functional dynamics. Environ Microbiol. 2012;14:1–3. doi: 10.1111/j.1462-2920.2011.02518.x. http://dx.doi.org/10.1111/j.1462-2920.2011.02518.x. [DOI] [PubMed] [Google Scholar]
- 271.Berg G, Zachow C, Müller H, Philipps J, Tilcher R. Next-generation bio-products sowing the seeds of success for sustainable agriculture. Agronomy. 2013;3:648–656. http://dx.doi.org/10.3390/agronomy3040648. [Google Scholar]
- 272.Clemente JC, Ursell LK, Parfrey LW, Knight R. The impact of the gut microbiota on human health: an integrative view. Cell. 2012;148:1258–1270. doi: 10.1016/j.cell.2012.01.035. http://dx.doi.org/10.1016/j.cell.2012.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 273.Blaser M, Bork P, Fraser C, Knight R, Wang J. The microbiome explored: recent insights and future challenges. Nat Rev Microbiol. 2013;11:213–217. doi: 10.1038/nrmicro2973. http://dx.doi.org/10.1038/nrmicro2973. [DOI] [PubMed] [Google Scholar]
- 274.Erlacher A, Cardinale M, Grosch R, Grube M, Berg G. The impact of the pathogen Rhizoctonia solani and its beneficial counterpart Bacillus amyloliquefaciens on the indigenous lettuce microbiome. Front Microbiol. 2014;5:175. doi: 10.3389/fmicb.2014.00175. http://dx.doi.org/10.3389/fmicb.2014.00175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 275.Ryan RP, Monchy S, Cardinale M, Taghavi S, Crossman L, Avison MB, Berg G, van der Lelie D, Dow JM. The versatility and adaptation of bacteria from the genus Stenotrophomonas. Nat Rev Microbiol. 2009;7:514–525. doi: 10.1038/nrmicro2163. http://dx.doi.org/10.1038/nrmicro2163. [DOI] [PubMed] [Google Scholar]
- 276.Mendes R, Garbeva P, Raaijmakers JM. The rhizosphere micro-biome: significance of plant beneficial, plant pathogenic, and human pathogenic microorganisms. FEMS Microbiol Rev. 2013;37:634–663. doi: 10.1111/1574-6976.12028. http://dx.doi.org/10.1111/1574-6976.12028. [DOI] [PubMed] [Google Scholar]
- 277.Berg G, Eberl L, Hartmann A. The rhizosphere as a reservoir for opportunistic human pathogenic bacteria. Environ Microbiol. 2005;7:1673–1685. doi: 10.1111/j.1462-2920.2005.00891.x. http://dx.doi.org/10.1111/j.1462-2920.2005.00891.x. [DOI] [PubMed] [Google Scholar]
- 278.Tan MW, Rahme LG, Sternberg JA, Tompkins RG, Ausubel FM. Pseudomonas aeruginosa killing of Caenorhabditis elegans used to identify P. aeruginosa virulence factors. Proc Natl Acad Sci U S A. 1999;96:2408–2413. doi: 10.1073/pnas.96.5.2408. http://dx.doi.org/10.1073/pnas.96.5.2408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 279.Dörr J, Hurek T, Reinhold-Hurek B. Type IV pili are involved in plant-microbe and fungus-microbe interactions. Mol Microbiol. 1998;30:7–17. doi: 10.1046/j.1365-2958.1998.01010.x. http://dx.doi.org/10.1046/j.1365-2958.1998.01010.x. [DOI] [PubMed] [Google Scholar]
- 280.Dekkers LC, van der Bij AJ, Mulders IHM, Phoelich CC, Wentwoord RAR, Glandorf DCM, Wijffelman CA, Lugtenberg BJJ. Role of the O-antigen of lipopolysaccharide, and possible roles of growth rate and of NADH:ubiquinone oxidoreductase (nuo) in competitive tomato root-tip colonization by Pseudomonas fluorescens WCS365. Mol Plant Microbe Interact. 1998;11:763–771. doi: 10.1094/MPMI.1998.11.8.763. http://dx.doi.org/10.1094/MPMI.1998.11.8.763. [DOI] [PubMed] [Google Scholar]
- 281.Jones BD, Nichols WA, Gibson BW, Sunshine MG, Apicella MA. Study of the role of the htrB gene in Salmonella typhimurium virulence. Infect Immun. 1997;65:4778–4783. doi: 10.1128/iai.65.11.4778-4783.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 282.Preston GM, Bertrand N, Rainey PB. Type III secretion in plant growth-promoting Pseudomonas fluorescens SBW25. Mol Microbiol. 2001;41:999–1014. doi: 10.1046/j.1365-2958.2001.02560.x. http://dx.doi.org/10.1046/j.1365-2958.2001.02560.x. [DOI] [PubMed] [Google Scholar]
- 283.Alavi P, Starcher MR, Thallinger GG, Zachow C, Muller H, Berg G. Stenotrophomonas comparative genomics reveals genes and functions that differentiate beneficial and pathogenic bacteria. BMC Genomics. 2014;15:482. doi: 10.1186/1471-2164-15-482. http://dx.doi.org/10.1186/1471-2164-15-482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 284.Berg G, Martinez JL. Friends or foes: can we make a distinction between beneficial and harmful strains of the Stenotrophomonas maltophilia complex? Front Microbiol. 2015;6:241. doi: 10.3389/fmicb.2015.00241. http://dx.doi.org/10.3389/fmicb.2015.00241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 285.Turrientes MC, Baquero MR, Sánchez MB, Valdezate S, Escudero E, Berg G, Cantón R, Baquero F, Galán JC, Martínez JL. Polymorphic mutation frequencies of clinical and environmental Stenotrophomonas maltophilia populations. Appl Environ Microbiol. 2010;76:1746–1758. doi: 10.1128/AEM.02817-09. http://dx.doi.org/10.1128/AEM.02817-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 286.Berg G, Roskot N, Smalla K. Genotypic and phenotypic relationships between clinical and environmental isolates of Stenotrophomonas maltophilia. J Clin Microbiol. 1999;37:3594–3600. doi: 10.1128/jcm.37.11.3594-3600.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 287.García-León G, Hernández A, Hernando-Amado S, Alavi P, Berg G, Martínez JL. A function of the major quinolone resistance determinant of Stenotrophomonas maltophilia SmeDEF is the colonization of the roots of the plants. Appl Environ Microbiol. 2014;80:4559–4565. doi: 10.1128/AEM.01058-14. http://dx.doi.org/10.1128/AEM.01058-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 288.Jiménez-Gasco MM, Malcolm GM, Berbegal M, Armengol J, Jiménez-Díaz RM. Complex molecular relationship between vegetative compatibility groups (VCGs) in Verticillium dahliae: VCGs do not always align with clonal lineages. Phytopathology. 2014;104:650–659. doi: 10.1094/PHYTO-07-13-0180-R. http://dx.doi.org/10.1094/PHYTO-07-13-0180-R. [DOI] [PubMed] [Google Scholar]
- 289.Köberl M, Schmidt R, Ramadan EM, Bauer R, Berg G. The microbiome of medicinal plants: diversity and importance for plant growth, quality and health. Front Microbiol. 2013;4:400. doi: 10.3389/fmicb.2013.00400. http://dx.doi.org/10.3389/fmicb.2013.00400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 290.Solla A, Gil L. Evaluating Verticillium dahliae for biological control of Ophiostoma novo-ulmi in Ulmus minor. Plant Pathol. 2003;52:579–585. http://dx.doi.org/10.1046/j.1365-3059.2003.00921.x. [Google Scholar]
- 291.Yousaf S, Bulgari D, Bergna A, Pancher M, Quaglino F, Casati P, Campisano A. Pyrosequencing detects human and animal pathogenic taxa in the grapevine endosphere. Front Microbiol. 2014;5:327. doi: 10.3389/fmicb.2014.00327. http://dx.doi.org/10.3389/fmicb.2014.00327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 292.Brandl MT. Fitness of human enteric pathogens on plants and implications for food safety. Annu Rev Phytopathol. 2006;44:367–392. doi: 10.1146/annurev.phyto.44.070505.143359. http://dx.doi.org/10.1146/annurev.phyto.44.070505.143359. [DOI] [PubMed] [Google Scholar]
- 293.Rastogi G, Sbodio A, Tech JJ, Suslow TV, Coaker GL, Leveau JHJ. Leaf microbiota in an agroecosystem: spatiotemporal variation in bacterial community composition on field-grown lettuce. ISME J. 2012;6:1812–1822. doi: 10.1038/ismej.2012.32. http://dx.doi.org/10.1038/ismej.2012.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 294.Leff JW, Fierer N. Bacterial communities associated with the surfaces of fresh fruits and vegetables. PLoS One. 2013;8:e59310. doi: 10.1371/journal.pone.0059310. http://dx.doi.org/10.1371/journal.pone.0059310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 295.Innerebner G, Knief C, Vorholt JA. Protection of Arabidopsis thaliana against leaf-pathogenic Pseudomonas syringae by Sphingomonas strains in a controlled model system. Appl Environ Microbiol. 2011;77:3202–3210. doi: 10.1128/AEM.00133-11. http://dx.doi.org/10.1128/AEM.00133-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 296.Cook RJ, Thomashow LS, Weller DM, Fujimoto D, Mazzola M, Bangera G, Kim D. Molecular mechanisms of defense by rhizo-bacteria against root disease. Proc Natl Acad Sci U S A. 1995;92:4197–4201. doi: 10.1073/pnas.92.10.4197. http://dx.doi.org/10.1073/pnas.92.10.4197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 297.Hanski I, von Hertzen L, Fyhrquist N, Koskinen K, Torppa K, Laatikainen T, Karisola P, Auvinen P, Paulin L, Makela MJ, Vartiainen E, Kosunen TU, Alenius H, Haahtela T. Environmental biodiversity, human microbiota, and allergy are interrelated. Proc Natl Acad Sci U S A. 2012;109:8334–8339. doi: 10.1073/pnas.1205624109. http://dx.doi.org/10.1073/pnas.1205624109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 298.Lloyd CM, Hawrylowicz CM. Regulatory T cells in asthma. Immunity. 2009;31:438–449. doi: 10.1016/j.immuni.2009.08.007. http://dx.doi.org/10.1016/j.immuni.2009.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 299.Doreswamy V, Peden DB. Modulation of asthma by endotoxin. Clin Exp Allergy. 2011;41:9–19. doi: 10.1111/j.1365-2222.2010.03628.x. http://dx.doi.org/10.1111/j.1365-2222.2010.03628.x. [DOI] [PubMed] [Google Scholar]
- 300.Berg G, Erlacher A, Grube M. The edible plant microbiome: importance and health issues. In: Lugtenberg B, editor. Principles of plant-microbe interaction. Springer International Publishing; Cham, Switzerland: 2015. pp. 419–426. http://dx.doi.org/10.1007/978-3-319-08575-3. [Google Scholar]
- 301.Berendsen RL, Pieterse CM, Bakker PA. The rhizosphere microbiome and plant health. Trends Plant Sci. 2012;17:478–486. doi: 10.1016/j.tplants.2012.04.001. http://dx.doi.org/10.1016/j.tplants.2012.04.001. [DOI] [PubMed] [Google Scholar]
- 302.Kristin A, Miranda H. The root microbiota—a fingerprint in the soil? Plant Soil. 2013;370:671–686. http://dx.doi.org/10.1007/s11104-013-1647-7. [Google Scholar]
- 303.Sekirov I, Russell SL, Antunes LCM, Finlay BB. Gut microbiota in health and disease. Physiol Rev. 2010;90:859–904. doi: 10.1152/physrev.00045.2009. http://dx.doi.org/10.1152/physrev.00045.2009. [DOI] [PubMed] [Google Scholar]
- 304.Ramírez-Puebla ST, Servín-Garcidueñas LE, Jímenez-Marín B, Bolaños LM, Rosenblueth M, Martínez J, Rogel MA, Ormeño-Orrillo E, Martínez-Romero E. Gut and root microbiota commonalities. Appl Environ Microbiol. 2013;79:2–9. doi: 10.1128/AEM.02553-12. http://dx.doi.org/10.1128/AEM.02553-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 305.Dawkins R, Dennett D. The extended phenotype: the long reach of the gene. Oxford University Press; Oxford, United Kingdom: 1999. [Google Scholar]
- 306.Bednarek P, Kwon C, Schulze-Lefert P. Not a peripheral issue: secretion in plant-microbe interactions. Curr Opin Plant Biol. 2010;13:378–387. doi: 10.1016/j.pbi.2010.05.002. http://dx.doi.org/10.1016/j.pbi.2010.05.002. [DOI] [PubMed] [Google Scholar]
- 307.Rasche F, Velvis H, Zachow C, Berg G, Van Elsas JD, Sessitsch A. Impact of transgenic potatoes expressing anti-bacterial agents on bacterial endophytes is comparable with the effects of plant genotype, soil type and pathogen infection. J Appl Ecol. 2006;43:555–566. http://dx.doi.org/10.1111/j.1365-2664.2006.01169.x. [Google Scholar]
- 308.Lundberg DS, Lebeis SL, Paredes SH, Yourstone S, Gehring J, Malfatti S, Tremblay J, Engelbrektson A, Kunin V, del Rio TG, Edgar RC, Eickhorst T, Ley RE, Hugenholtz P, Tringe SG, Dangl JL. Defining the core Arabidopsis thaliana root microbiome. Nature. 2012;488:86–90. doi: 10.1038/nature11237. http://dx.doi.org/10.1038/nature11237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 309.Ding T, Palmer MW, Melcher U. Community terminal restriction fragment length polymorphisms reveal insights into the diversity and dynamics of leaf endophytic bacteria. BMC Microbiol. 2013;13:1. doi: 10.1186/1471-2180-13-1. http://dx.doi.org/10.1186/1471-2180-13-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 310.van Overbeek LS, van Elsas JD. Effects of plant genotype and growth stage on the structure of bacterial communities associated with potato (Solanum tuberosum L.) FEMS Microbiol Ecol. 2008;64:283–296. doi: 10.1111/j.1574-6941.2008.00469.x. http://dx.doi.org/10.1111/j.1574-6941.2008.00469.x. [DOI] [PubMed] [Google Scholar]
- 311.Manter DK, Delgado JA, Holm DG, Stong RA. Pyrosequencing reveals a highly diverse and cultivar-specific bacterial endophyte community in potato roots. Microb Ecol. 2010;60:157–166. doi: 10.1007/s00248-010-9658-x. http://dx.doi.org/10.1007/s00248-010-9658-x. [DOI] [PubMed] [Google Scholar]
- 312.Hardoim PR, Andreote FD, Reinhold-Hurek B, Sessitsch A, van Over-beek LS, van Elsas JD. Rice root-associated bacteria: insights into community structures across 10 cultivars. FEMS Microbiol Ecol. 2011;77:154–164. doi: 10.1111/j.1574-6941.2011.01092.x. http://dx.doi.org/10.1111/j.1574-6941.2011.01092.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 313.Ferrando L, Mañay JF, Scavino AF. Molecular and culture-dependent analyses revealed similarities in the endophytic bacterial community composition of leaves from three rice (Oryza sativa) varieties. FEMS Microbiol Ecol. 2012;80:696–708. doi: 10.1111/j.1574-6941.2012.01339.x. http://dx.doi.org/10.1111/j.1574-6941.2012.01339.x. [DOI] [PubMed] [Google Scholar]
- 314.Schlaeppi K, Dombrowski N, Oter RG, van Themaat EVL, Schulze-Lefert P. Quantitative divergence of the bacterial root microbiota in Arabidopsis thaliana relatives. Proc Natl Acad Sci U S A. 2014;111:585–592. doi: 10.1073/pnas.1321597111. http://dx.doi.org/10.1073/pnas.1321597111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 315.Ottesen AR, Peña AG, White JR, Pettengill JB, Li C, Allard S, Rideout S, Allard M, Hill T, Evans P, Strain E, Musser S, Knight R, Brown E. Baseline survey of the anatomical microbial ecology of an important food plant: Solanum lycopersicum (tomato) BMC Microbiol. 2013;13:114. doi: 10.1186/1471-2180-13-114. http://dx.doi.org/10.1186/1471-2180-13-114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 316.Bodenhausen N, Horton MW, Bergelson J. Bacterial communities associated with the leaves and the roots of Arabidopsis thaliana. PLoS One. 2013;8:e56329. doi: 10.1371/journal.pone.0056329. http://dx.doi.org/10.1371/journal.pone.0056329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 317.van Overbeek LS, Franke AC, Nijhuis EHM, Groeneveld RMW, da Rocha UN, Lotz LAP. Bacterial communities associated with Chenopodium album and Stellaria media seeds from arable soils. Microb Ecol. 2011;62:257–264. doi: 10.1007/s00248-011-9845-4. http://dx.doi.org/10.1007/s00248-011-9845-4. [DOI] [PubMed] [Google Scholar]
- 318.Shade A, McManus PS, Handelsman J. Unexpected diversity during community succession in the apple flower microbiome. mBio. 2013;4:e00602–12. doi: 10.1128/mBio.00602-12. http://dx.doi.org/10.1128/mBio.00602-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 319.Clay K, Schardl C. Evolutionary origins and ecological consequences of endophyte symbiosis with grasses. Am Nat. 2002;160:S99–S127. doi: 10.1086/342161. http://dx.doi.org/10.1086/342161. [DOI] [PubMed] [Google Scholar]
- 320.Unterseher M, Gazis R, Chaverri P, Guarniz CFG, Tenorio DHZ. Endophytic fungi from Peruvian highland and lowland habitats form distinctive and host plant-specific assemblages. Biodivers Conserv. 2013;22:999–1016. http://dx.doi.org/10.1007/s10531-013-0464-x. [Google Scholar]
- 321.Todd D. The effects of host genotype, growth-rate, and needle age on the distribution of a mutualistic, endophytic fungus in Douglas-fir plantations. Can J For Res. 1988;18:601–605. http://dx.doi.org/10.1139/x88-087. [Google Scholar]
- 322.Ahlholm JU, Helander M, Henriksson J, Metzler M, Saikkonen K. Environmental conditions and host genotype direct genetic diversity of Venturia ditricha, a fungal endophyte of birch trees. Evolution. 2002;56:1566–1573. doi: 10.1111/j.0014-3820.2002.tb01468.x. http://dx.doi.org/10.1111/j.0014-3820.2002.tb01468.x. [DOI] [PubMed] [Google Scholar]
- 323.Rajala T, Velmala SM, Tuomivirta T, Haapanen M, Muller M, Pennanen T. Endophyte communities vary in the needles of Norway spruce clones. Fungal Biol. 2013;117:182–190. doi: 10.1016/j.funbio.2013.01.006. http://dx.doi.org/10.1016/j.funbio.2013.01.006. [DOI] [PubMed] [Google Scholar]
- 324.Wearn JA, Sutton BC, Morley NJ, Gange AC. Species and organ specificity of fungal endophytes in herbaceous grassland plants. J Ecol. 2012;100:1085–1092. http://dx.doi.org/10.1111/j.1365-2745.2012.01997.x. [Google Scholar]
- 325.Jin H, Yan Z, Liu Q, Yang X, Chen J, Qin B. Diversity and dynamics of fungal endophytes in leaves, stems and roots of Stellera chamaejasme L. in northwestern China. Antonie Van Leeuwenhoek. 2013;104:949–963. doi: 10.1007/s10482-013-0014-2. http://dx.doi.org/10.1007/s10482-013-0014-2. [DOI] [PubMed] [Google Scholar]
- 326.Domsch KH, Gams W, Anderson TH. Compendium of soil fungi. 2nd ed IHW-Verlag; London, United Kingdom: 2007. [Google Scholar]
- 327.Herre EA, Mejía LC, Kyllo DA, Rojas E, Maynard Z, Butler A, van Bael SA. Ecological implications of anti-pathogen effects of tropical fungal endophytes and mycorrhizae. Ecology. 2007;88:550–558. doi: 10.1890/05-1606. http://dx.doi.org/10.1890/05-1606. [DOI] [PubMed] [Google Scholar]
- 328.Friesen ML, Porter SS, Stark SC, von Wettberg EJ, Sachs JL, Martinez-Romero E. Microbially mediated plant functional traits. Annu Rev Ecol Evol Syst. 2011;42:23–46. http://dx.doi.org/10.1146/annurev-ecolsys-102710-145039. [Google Scholar]
- 329.Quambusch M, Pirttilä AM, Tejesvi MV, Winkelmann T, Bartsch M. Endophytic bacteria in plant tissue culture: differences between easy- and difficult-to-propagate Prunus avium genotypes. Tree Physiol. 2014;34:524–533. doi: 10.1093/treephys/tpu027. http://dx.doi.org/10.1093/treephys/tpu027. [DOI] [PubMed] [Google Scholar]
- 330.Saikkonen K, Gundel PE, Helander M. Chemical ecology mediated by fungal endophytes in grasses. J Chem Ecol. 2013;39:962–968. doi: 10.1007/s10886-013-0310-3. http://dx.doi.org/10.1007/s10886-013-0310-3. [DOI] [PubMed] [Google Scholar]
- 331.Hoffman MT, Arnold AE. Diverse bacteria inhabit living hyphae of phylogenetically diverse fungal endophytes. Appl Environ Microbiol. 2010;76:4063–4075. doi: 10.1128/AEM.02928-09. http://dx.doi.org/10.1128/AEM.02928-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 332.Hoffman MT, Arnold AE. Geographic locality and host identity shape fungal endophyte communities in cupressaceous trees. Mycol Res. 2008;112:331–344. doi: 10.1016/j.mycres.2007.10.014. http://dx.doi.org/10.1016/j.mycres.2007.10.014. [DOI] [PubMed] [Google Scholar]
- 333.Partida-Martinez LP, Hertweck C. Pathogenic fungus harbours endosymbiotic bacteria for toxin production. Nature. 2005;437:884–888. doi: 10.1038/nature03997. http://dx.doi.org/10.1038/nature03997. [DOI] [PubMed] [Google Scholar]
- 334.Lackner G, Partida-Martinez LP, Hertweck C. Endofungal bacteria as producers of mycotoxins. Trends Microbiol. 2009;17:570–576. doi: 10.1016/j.tim.2009.09.003. http://dx.doi.org/10.1016/j.tim.2009.09.003. [DOI] [PubMed] [Google Scholar]
- 335.Bonfante P, Anca I-A. Plants, mycorrhizal fungi, and bacteria: a network of interactions. Annu Rev Microbiol. 2009;63:363–383. doi: 10.1146/annurev.micro.091208.073504. http://dx.doi.org/10.1146/annurev.micro.091208.073504. [DOI] [PubMed] [Google Scholar]
- 336.Naumann M, Schussler A, Bonfante P. The obligate endobacteria of arbuscular mycorrhizal fungi are ancient heritable components related to the Mollicutes. ISME J. 2010;4:862–871. doi: 10.1038/ismej.2010.21. http://dx.doi.org/10.1038/ismej.2010.21. [DOI] [PubMed] [Google Scholar]
- 337.Frey-Klett P, Garbaye J, Tarkka M. The mycorrhiza helper bacteria revisited. New Phytol. 2007;176:22–36. doi: 10.1111/j.1469-8137.2007.02191.x. http://dx.doi.org/10.1111/j.1469-8137.2007.02191.x. [DOI] [PubMed] [Google Scholar]
- 338.Garbaye J. Helper bacteria—a new dimension to the mycorrhizal symbiosis. New Phytol. 1994;128:197–210. doi: 10.1111/j.1469-8137.1994.tb04003.x. http://dx.doi.org/10.1111/j.1469-8137.1994.tb04003.x. [DOI] [PubMed] [Google Scholar]
- 339.Márquez LM, Redman RS, Rodriguez RJ, Roossinck MJ. A virus in a fungus in a plant: three-way symbiosis required for thermal tolerance. Science. 2007;315:513–515. doi: 10.1126/science.1136237. http://dx.doi.org/10.1126/science.1136237. [DOI] [PubMed] [Google Scholar]
- 340.Rodriguez RJ, Henson J, Van Volkenburgh E, Hoy M, Wright L, Beckwith F, Kim Y-O, Redman RS. Stress tolerance in plants via habitat-adapted symbiosis. ISME J. 2008;2:404–416. doi: 10.1038/ismej.2007.106. http://dx.doi.org/10.1038/ismej.2007.106. [DOI] [PubMed] [Google Scholar]
- 341.Herrero N, Sanchez Marquez S, Zabalgogeazcoa I. Mycoviruses are common among different species of endophytic fungi of grasses. Arch Virol. 2009;154:327–330. doi: 10.1007/s00705-008-0293-5. http://dx.doi.org/10.1007/s00705-008-0293-5. [DOI] [PubMed] [Google Scholar]
- 342.Bao X, Roossinck MJ. Multiplexed interactions: viruses of endophytic fungi. Adv Virus Res. 2013;86:37–58. doi: 10.1016/B978-0-12-394315-6.00002-7. http://dx.doi.org/10.1016/B978-0-12-394315-6.00002-7. [DOI] [PubMed] [Google Scholar]
- 343.Blanquart F, Gandon S. Time-shift experiments and patterns of adaptation across time and space. Ecol Lett. 2013;16:31–38. doi: 10.1111/ele.12007. http://dx.doi.org/10.1111/ele.12007. [DOI] [PubMed] [Google Scholar]
- 344.Koskella B. Phage-mediated selection on microbiota of a long-lived host. Curr Biol. 2013;23:1256–1260. doi: 10.1016/j.cub.2013.05.038. http://dx.doi.org/10.1016/j.cub.2013.05.038. [DOI] [PubMed] [Google Scholar]
- 345.Hartley SE, Gange AC. Impacts of plant symbiotic fungi on insect herbivores: mutualism in a multitrophic context. Annu Rev Entomol. 2009;54:323–342. doi: 10.1146/annurev.ento.54.110807.090614. http://dx.doi.org/10.1146/annurev.ento.54.110807.090614. [DOI] [PubMed] [Google Scholar]
- 346.Clay K. Fungal endophytes of grasses. Annu Rev Ecol Syst. 1990;21:275–297. http://dx.doi.org/10.1146/annurev.es.21.110190.001423. [Google Scholar]
- 347.Faeth SH, Saari S. Fungal grass endophytes and arthropod communities: lessons from plant defence theory and multitrophic interactions. Fungal Ecol. 2012;5:364–371. http://dx.doi.org/10.1016/j.funeco.2011.09.003. [Google Scholar]
- 348.Rúa MA, McCulley RL, Mitchell CE. Fungal endophyte infection and host genetic background jointly modulate host response to an aphid-transmitted viral pathogen. J Ecol. 2013;101:1007–1018. http://dx.doi.org/10.1111/1365-2745.12106. [Google Scholar]
- 349.Rúa MA, McCulley RL, Mitchell CE. Climate drivers, host identity and fungal endophyte infection determine virus prevalence in a grassland ecosystem. J Ecol. 2014;102:690–699. http://dx.doi.org/10.1111/1365-2745.12238. [Google Scholar]
- 350.Araújo WL, Marcon J, Maccheroni W, van Elsas JD, van Vuurde JWL, Azevedo JL. Diversity of endophytic bacterial populations and their interaction with Xylella fastidiosa in citrus plants. Appl Environ Microbiol. 2002;68:4906–4914. doi: 10.1128/AEM.68.10.4906-4914.2002. http://dx.doi.org/10.1128/AEM.68.10.4906-4914.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 351.Bulgari D, Casati P, Crepaldi P, Daffonchio D, Quaglino F, Brusetti L, Bianco PA. Restructuring of endophytic bacterial communities in grapevine yellows-diseased and recovered Vitis vinifera L. plants. Appl Environ Microbiol. 2011;77:5018–5022. doi: 10.1128/AEM.00051-11. http://dx.doi.org/10.1128/AEM.00051-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 352.Bulgari D, Bozkurt AI, Casati P, Caglayan K, Quaglino F, Bianco PA. Endophytic bacterial community living in roots of healthy and ‘Candidatus Phytoplasma mali’-infected apple (Malus domestica, Borkh) trees. Antonie Van Leeuwenhoek. 2012;102:677–687. doi: 10.1007/s10482-012-9766-3. http://dx.doi.org/10.1007/s10482-012-9766-3. [DOI] [PubMed] [Google Scholar]
- 353.Douanla-Meli C, Langer E, Mouafo FT. Fungal endophyte diversity and community patterns in healthy and yellowing leaves of Citrus limon. Fungal Ecol. 2013;6:212–222. http://dx.doi.org/10.1016/j.funeco.2013.01.004. [Google Scholar]
- 354.Ardanov P, Sessitsch A, Häggman H, Kozyrovska N, Pirttilä AM. Methylobacterium-induced endophyte community changes correspond with protection of plants against pathogen attack. PLoS One. 2012;7:e46802. doi: 10.1371/journal.pone.0046802. http://dx.doi.org/10.1371/journal.pone.0046802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 355.Eschen R, Hunt S, Mykura C, Gange AC, Sutton BC. The foliar endophytic fungal community composition in Cirsium arvense is affected by mycorrhizal colonization and soil nutrient content. Fungal Biol. 2010;114:991–998. doi: 10.1016/j.funbio.2010.09.009. http://dx.doi.org/10.1016/j.funbio.2010.09.009. [DOI] [PubMed] [Google Scholar]
- 356.Chu-Chou M, Guo B, An ZQ, Hendrix JW, Ferriss RS, Siegel MR, Dougherty CT, Burrus PB. Suppression of mycorrhizal fungi in fescue by the Acremonium coenophialum endophyte. Soil Biol Biochem. 1992;24:633–637. http://dx.doi.org/10.1016/0038-0717(92)90041-U. [Google Scholar]
- 357.Antunes PM, Miller J, Carvalho LM, Klironomos JN, Newman JA. Even after death the endophytic fungus of Schedonorus phoenix reduces the arbuscular mycorrhizas of other plants. Funct Ecol. 2008;22:912–918. http://dx.doi.org/10.1111/j.1365-2435.2008.01432.x. [Google Scholar]
- 358.Mack KML, Rudgers JA. Balancing multiple mutualists: asymmetric interactions among plants, arbuscular mycorrhizal fungi, and fungal endophytes. Oikos. 2008;117:310–320. http://dx.doi.org/10.1111/j.2007.0030-1299.15973.x. [Google Scholar]
- 359.Larimer AL, Bever JD, Clay K. Consequences of simultaneous interactions of fungal endophytes and arbuscular mycorrhizal fungi with a shared host grass. Oikos. 2012;121:2090–2096. http://dx.doi.org/10.1111/j.1600-0706.2012.20153.x. [Google Scholar]
- 360.Pohjanen J, Koskimäki JJ, Sutela S, Ardanov P, Suorsa M, Niemi K, Sarjala T, Häggman H, Pirttilä AM. Interaction with ectomycorrhizal fungi and endophytic Methylobacterium affects nutrient uptake and growth of pine seedlings in vitro. Tree Physiol. 2014;34:993–1005. doi: 10.1093/treephys/tpu062. http://dx.doi.org/10.1093/treephys/tpu062. [DOI] [PubMed] [Google Scholar]
- 361.Sturz AV. The role of endophytic bacteria during seed piece decay and potato tuberization. Plant Soil. 1995;175:257–263. http://dx.doi.org/10.1007/BF00011362. [Google Scholar]
- 362.Döring M. Untersuchungen zur Vermehrung und Mykorrhiza von Eleutherococcus Maxim. (Araliaceae) Philipps-Universität Marburg; Marburg, Germany: 2007. Ph.D. thesis. [Google Scholar]
- 363.Campisano A, Ometto L, Compant S, Pancher M, Antonielli L, Yousaf S, Varotto C, Anfora G, Pertot I, Sessitsch A, Rota-Stabelli O. Interkingdom transfer of the acne-causing agent, Propionibacterium acnes, from human to grapevine. Mol Biol Evol. 2014;31(5):1059–1065. doi: 10.1093/molbev/msu075. http://dx.doi.org/10.1093/molbev/msu075. [DOI] [PubMed] [Google Scholar]
- 364.Pruesse E, Peplies J, Gloeckner FO. SINA: accurate high-throughput multiple sequence alignment of ribosomal RNA genes. Bioinformatics. 2012;28:1823–1829. doi: 10.1093/bioinformatics/bts252. http://dx.doi.org/10.1093/bioinformatics/bts252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 365.Bengtsson-Palme J, Ryberg M, Hartmann M, Branco S, Wang Z, Godhe A, De Wit P, Sanchez-Garcia M, Ebersberger I, de Sousa F, Amend AS, Jumpponen A, Unterseher M, Kristiansson E, Abarenkov K, Bertrand YJK, Sanli K, Eriksson KM, Vik U, Veldre V, Nilsson RH. Improved software detection and extraction of ITS1 and ITS2 from ribosomal ITS sequences of fungi and other eukaryotes for analysis of environmental sequencing data. Methods Ecol Evol. 2013;4:914–919. http://dx.doi.org/10.1111/2041-210X.12073. [Google Scholar]
- 366.Edgar RC. Search and clustering orders of magnitude faster than BLAST. Bioinformatics. 2010;26:2460–2461. doi: 10.1093/bioinformatics/btq461. http://dx.doi.org/10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
- 367.Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Pena AG, Goodrich JK, Gordon JI, Huttley GA, Kelley ST, Knights D, Koenig JE, Ley RE, Lozupone CA, McDonald D, Muegge BD, Pirrung M, Reeder J, Sevinsky JR, Tumbaugh PJ, Walters WA, Widmann J, Yatsunenko T, Zaneveld J, Knight R. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7:335–336. doi: 10.1038/nmeth.f.303. http://dx.doi.org/10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 368.Koljalg U, Nilsson RH, Abarenkov K, Tedersoo L, Taylor AFS, Bahram M, Bates ST, Bruns TD, Bengtsson-Palme J, Callaghan TM, Douglas B, Drenkhan T, Eberhardt U, Duenas M, Grebenc T, Griffith GW, Hartmann M, Kirk PM, Kohout P, Larsson E, Lindahl BD, Luecking R, Martin MP, Matheny PB, Nguyen NH, Niskanen T, Oja J, Peay KG, Peintner U, Peterson M, Poldmaa K, Saag L, Saar I, Schuessler A, Scott JA, Senes C, Smith ME, Suija A, Taylor DL, Telleria MT, Weiss M, Larsson K-H. Towards a unified paradigm for sequence-based identification of fungi. Mol Ecol. 2013;22:5271–5277. doi: 10.1111/mec.12481. http://dx.doi.org/10.1111/mec.12481. [DOI] [PubMed] [Google Scholar]
- 369.Pohjanen J, Koskimaki J, Pirttilä AM. Interactions of meristem-associated endophytic bacteria. In: Verma VC, Gange AC, editors. Advances in endophytic research. 1st ed. Springer India; New Delhi, India: 2014. pp. 103–113. http://dx.doi.org/10.1007/978-81-322-1575-2_5. [Google Scholar]
- 370.Cardinale M, Berg G. Visualization of plant-microbe interactions. In: Lugtenberg B, editor. Principles of plant-microbe interactions. Springer International Publishing; Cham, Switzerland: 2015. pp. 299–306. http://dx.doi.org/10.1007/978-3-319-08575-3_31. [Google Scholar]
- 371.Erlacher A, Cardinale M, Grube M, Berg G. Biotic stress shifted structure and abundance of Enterobacteriaceae in the lettuce micro-biome. PLoS One. 2015;10:e0118068. doi: 10.1371/journal.pone.0118068. http://dx.doi.org/10.1371/journal.pone.0118068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 372.Quast C, Pruesse E, Yilmaz P, Gerken J, Schweer T, Yarza P, Peplies J, Glockner FO. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 2013;41:D590–D596. doi: 10.1093/nar/gks1219. http://dx.doi.org/10.1093/nar/gks1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.