Abstract
Cells couple growth with division and regulate size in response to nutrient availability. In rod-shaped fission yeast, cell size control occurs at mitotic commitment. An important regulator is the DYRK-family kinase Pom1, which forms gradients from cell poles and inhibits the mitotic activator Cdr2, itself localized at the medial cortex. Where and when Pom1 modulates Cdr2 activity is unclear as Pom1 medial cortical levels remain constant upon cell elongation. Here we show that Pom1 re-localizes to cell sides upon environmental glucose limitation, where it strongly delays mitosis. This re-localization is caused by severe microtubule destabilization upon glucose starvation, with microtubules undergoing catastrophe and depositing the Pom1 gradient nucleator Tea4 at cell sides. Microtubule destabilization requires PKA/Pka1 activity, which negatively regulates the microtubule rescue factor CLASP/Cls1/Peg1, reducing CLASP’s ability to stabilize microtubules. Thus, PKA signalling tunes CLASP’s activity to promote Pom1 cell side localization and buffer cell size upon glucose starvation.
Introduction
Cell size is a fundamental attribute, critical for cell fitness 1, 2, 3. Cell size is regulated by homeostatic mechanisms and in response to nutrient availability, with cells reducing their target size in conditions of poor nutrients. In rod-shaped fission yeast cells, size control occurs at mitotic commitment. Cyclin-dependent kinase 1 (CDK1) activation for mitotic entry is regulated by the balanced activities of the inhibiting kinase Wee1 and the activating phosphatase Cdc25 4. In response to poor nitrogen source, the target of rapamycin (TOR) and mitogen-activated protein kinase (MAPK) stress response pathways modify this balance to advance mitotic commitment and reduce cell size at division 5, 6, 7. When glucose is limiting, cells also reduce their size 8, though the mechanisms remain largely unknown. Glucose is primarily signaled by a cyclic-AMP/Protein Kinase A (cAMP/PKA) pathway, which inhibits the transcription of gluconeogenesis and sexual differentiation factors, and also modulates cell cycle progression 9, 10, 11.
In steady-state conditions, cell-intrinsic sizing mechanisms coordinate cell growth with division. Recent studies have focused on the DYRK-family kinase Pom1 and its substrate, the Wee1-inhibitory kinase Cdr2 12, 13. Pom1, which forms concentration gradients from cell poles, restricts Cdr2 localization to the cell middle 14, 15 and phosphorylates Cdr2 to inhibit its activation by the Ca2+/calmodulin-dependent protein kinase kinase (CaMKK) Ssp1 and delay mitotic commitment 14, 16. Pom1 was proposed to coordinate growth and division by inhibiting Cdr2 until attainment of sufficient cell length 12, 13. Indeed, Pom1 is highly dose-dependent, and its forced localization to cell sides delays mitosis 12, 13, 14. However, it has been unclear where and when it naturally inhibits Cdr2, as medial Pom1 concentration does not substantially vary during cell extension 14, 17. In addition, the observation that cells lacking pom1 remain homeostatic, though at a reduced cell size 18, has raised question about whether Pom1 acts as a cell length sensor.
The position of Pom1 gradients is dictated by microtubules, depositing a phosphatase-regulatory complex, Tea1-Tea4, at cell poles 19, 20, 21, 22. Tea4 associates with a phosphatase 1 catalytic subunit, which dephosphorylates Pom1 to trigger membrane binding 23, 24, 25. Pom1 concentration then decays towards the cell middle through diffusion and autophosphorylation-dependent membrane detachment 25, 26. Microtubules form antiparallel bundles nucleated from the nuclear envelope with dynamic plus-ends that grow towards cell poles 27. Microtubule plus-end dynamics – growth, shrinkage, catastrophe and rescue – are regulated by a host of microtubule-associated factors (MAPs). These include the +TIP complex Mal3/EB1-Tip1/CLIP-170-Tea2/kinesin, the Alp14/XMAP215 polymerase and the Klp5-6/kinesin-8 28, 29, 30, 31, 32, 33, which promote microtubule sliding along cell sides and restrict catastrophe events to cell poles for local Tea4 release. Microtubules are maintained in antiparallel bundles by the MAP65/PRC1-family protein Ase1, which localizes to the zones of microtubule overlap, where it recruits the CLASP Cls1/Peg1 (Cls1 below) 34, 35, 36. CLASPs are conserved microtubule stabilizers 37, initially identified as CLIP-associated proteins on microtubule plus-end in animal cells 38. By contrast, the sole fission yeast CLASP does not track microtubule plus-ends, and localizes prominently to zones of antiparallel microtubule overlap, where it is essential for microtubule rescue 34, 39.
Here, we describe that Pom1 re-localizes to cell sides and that microtubule dynamics and organization are dramatically altered upon glucose starvation. These findings lead us to uncover a novel PKA-dependent regulation of microtubule dynamics, in which PKA signaling negatively regulates the microtubule rescue factor CLASP to promote microtubule catastrophe, Tea4 delivery and Pom1 re-localization at cell-sides, where Pom1 buffers cell size upon glucose starvation.
Results
Pka1-dependent re-localization of Pom1 in limited glucose
We serendipitously observed that, in contrast to the polar gradients formed in exponentially growing cells, Pom1 is detected around the medial cortex in saturated cultures (Supplementary Fig. 1A), suggesting that starvation triggers Pom1 re-localization. Depletion of nitrogen or shift from a good to a poor nitrogen source did not modify Pom1 distribution (Supplementary Fig. 1C). By contrast, glucose starvation to levels similar to those measured in the saturated cultures resulted in Pom1 re-localization (Fig 1A, C-D; Supplementary Fig. 1B). Pom1 was almost uniform around the cell periphery in 0.03% glucose, in which cells grow very little, and was also less confined to cell tips in 0.08% glucose, in which cells proliferate at very similar rates as in 2% glucose 8. Pom1 re-localization occurred quickly, within 10 minutes, and reversibly, with no major change in global Pom1 levels as seen by both imaging and western blots (Fig 1A; Supplementary Fig. 1D). Other stresses, such as temperature (36°C) or osmotic stress (1M sorbitol) did not affect Pom1 localization. Thus, Pom1 localization responds to changes in external glucose levels.
Figure 1. Pka1-dependent reversible re-localization of Pom1 to cell sides upon glucose limitation.
(A) Sum of 5 medial spinning disk confocal images taken over 30 seconds of Pom1-tdTomato in wild-type cells grown in 2% or 0.08% or 0.03% glucose (G) for 1h. Arrowheads indicate Pom1 presence at cell sides. The last panel shows polar Pom1 upon 2% glucose replenishment for 10min.
(B) Pom1-tdTomato in pka1Δ cells as in (A).
(C) Distribution of cortical Pom1 from one cell tip to the other (0 = cell middle) in wild-type cells obtained with the Cellophane plugin. Average of 31, 27 and 35 profiles for 2%, 0.08% and 0.03%G, respectively, in 8μm-long cells. Profiles obtained from other cell lengths are similar.
(D) Box and whisker plot of cortical Pom1 fluorescence intensity in the middle 2μm region in both wild-type (n=31, 27 and 35) and pka1Δ cells (n=34, 24 and 42) in 2%, 0.08% and 0.03% glucose respectively. Experiments were performed thrice and quantification of one is shown.
(E) Left: Schematic representation of glucose detection by the PKA pathway. Right: Medial spinning disk confocal images of Pom1-tdTomato in mutants of the PKA pathway. Representative images from 2 independent experiments are shown.
Arrowheads indicate Pom1 presence at cell sides. Scale bars are 5μm.
Glucose is sensed by a dedicated G-protein-coupled receptor at the plasma membrane, Git3, which signals PKA activation through a G alpha protein, Gpa2, and the adenylate cyclase, Cyr1 11. In pka1Δ cells grown to saturation, Pom1 remained restricted to cell tips (Supplementary Fig. 1A), indicating that the PKA pathway may be required for Pom1 side-localization upon glucose limitation. Indeed, in git3Δ, gpa2Δ, cyr1Δ and pka1Δ cells, Pom1 failed to re-localize upon shift to low glucose (Fig 1B, D-E). By contrast, cells deleted for the regulatory subunit of PKA, cgs1, which exhibit constitutive Pka1 activity 40, displayed normal tip-localized Pom1 in glucose-rich conditions and Pom1 re-localization in low glucose (Fig 1E). Deletion or mutation in the two other major pathways that transduce information about nutrient availability, MAPK and TOR, 41, 42 did not prevent Pom1 re-localization in low glucose (Supplementary Fig. 1E-F), though Pom1 re-localization was less severe in sty1Δ cells or in cells treated with the MAPK inhibitor SP600125 43. Interestingly activation of the MAPK pathway using the hyperactive wis1DD allele 44 showed a drastic reduction in global Pom1 levels (Supplementary Fig. 1E). Thus, the cAMP-PKA pathway regulates the re-localization of Pom1 upon glucose limitation with some contribution from the MAPK pathway. In this study, we focused on the role of PKA signaling on Pom1 localization.
We note that the Pom1 substrate Cdr2 also distributed more widely around the cell cortex under limited glucose, in a pka1-dependent manner, but remained dependent on Pom1 for exclusion from the cell pole (Supplementary Fig. 1G-H), consistent with Pom1 regulating its cortex-binding 15.
Pka1 is active in low glucose to signal Pom1 re-localization
Previous data showed that nuclear-localized Pka1 represses transcription in presence of glucose, with de-repression observed upon glucose depletion45, 46, 47. However, the cortical, fast reversible dynamics of Pom1 re-localization upon glucose depletion suggest this may not be a transcriptional response.
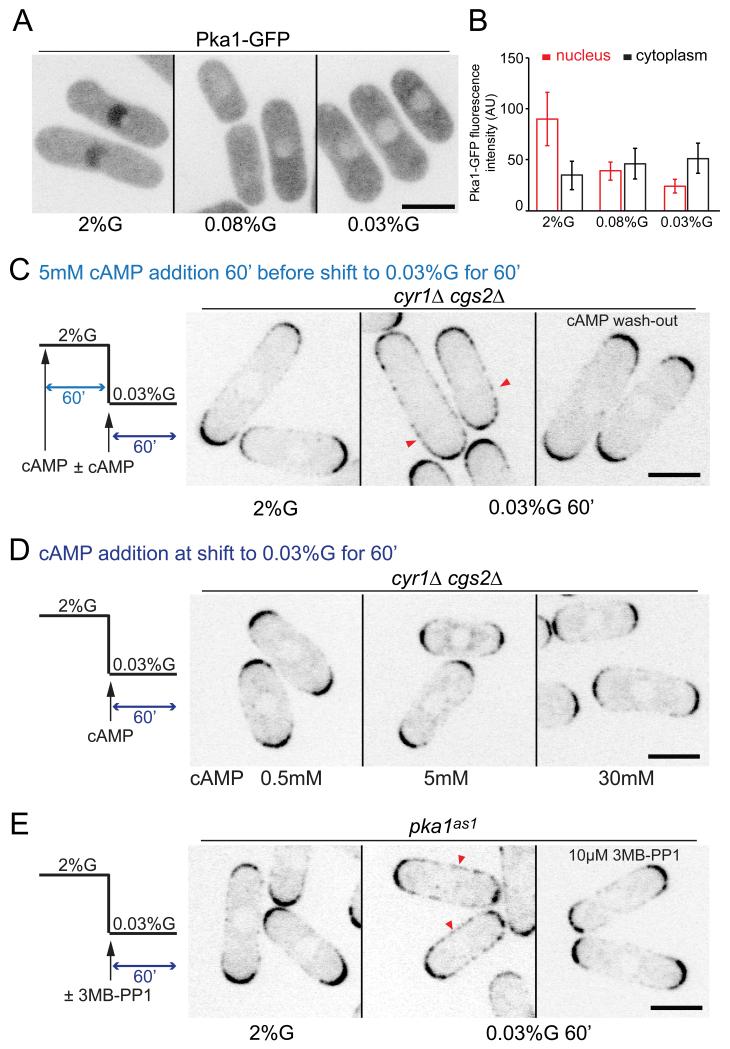
Consistent with previous data 48, 49, Pka1 localized to the nucleus in presence of 2% glucose, but was also present in the cytosol, where its concentration increased as it exited the nucleus upon glucose starvation (Fig 2A-B).48 We found that in cgs1Δ cells, nuclear and cytosolic levels of Pka1-GFP remained constant in all glucose-conditions tested (Supplementary Fig. 2A-B). As these cells showed a wild type-like Pom1 distribution (Fig 1E), we conclude that Pka1 may signal Pom1 re-localization from the cytosol, but that an increase in cytosolic Pka1 cannot be the trigger for Pom1 re-localization.
Figure 2. Pka1 is active in low glucose to promote Pom1 side-localization.
(A) Maximum intensity spinning disk projection of Pka1-GFP in wild-type cells grown in 2% or 0.08% or 0.03% glucose for 1h.
(B) Measurement of cytoplasmic and nuclear Pka1-GFP levels in cells as in (A) (n>20). Experiments were performed thrice and quantification of one is shown
(C) Medial spinning disk confocal section of Pom1-tdTomato in cyr1Δcgs2Δ cells incubated with 5mM cAMP in 2% glucose for 1h (left) and shifted to 0.03% glucose for 1h with the same amount of cAMP (middle) or without cAMP (right). Arrowheads indicate Pom1 at cell sides.
(D) Medial spinning disk confocal section of Pom1-tdTomato in cyr1Δcgs2Δ cells grown in 2% glucose and incubated with increasing amounts of cAMP at the time of shift 0.03% glucose for 1h.
(E) Medial spinning disk confocal section of Pom1-tdTomato in pka1-as1 cells grown in 2% glucose (left) and shifted to 0.03% glucose without (middle) or with 10μM 3MB-PP1 (right).
Arrowheads indicate Pom1 at cell sides. Scale bars are 5μm. Error bars are standard deviations. For C-E, representative images from 2 independent experiments are shown.
We addressed when Pka1 activity is required in two ways. First, we used cells lacking cAMP, and thus PKA activity, due to deletion of the adenylate cyclase and cAMP phospho-diesterase genes (cyr1Δ cgs2Δ). As expected, cyr1Δ cgs2Δ cells failed to re-localize Pom1, but addition of cAMP before and throughout glucose limitation restored PKA activity and yielded a uniform Pom1 localization over the entire cell cortex. However, if cAMP was washed out at the time of shift to low glucose, Pom1 failed to re-localize, suggesting PKA activity is required in low glucose (Fig 2C; Supplementary Fig. 2D). We note that up to 30mM cAMP addition only at the time of shift to low glucose was not sufficient to trigger Pom1 re-localization (Fig 2D). Control pka1Δ cgs2Δ cells treated the same way retained polar Pom1, confirming that the action of cAMP occurs through PKA activation (Supplementary Fig. 2C-D). Thus, Pka1 is active and required when glucose is limiting to signal Pom1 re-localization.
Second, we constructed an analog-sensitive Pka1 mutant (pka1as1), whose activity could be selectively inhibited by addition of an ATP-analogue 50. Untreated pka1as1 cells behaved largely as wild-type cells, re-localizing Pom1 in low glucose (Fig 2E), though these cells were somewhat shorter than wild-type, suggesting Pka1as1 is not fully functional (Supplementary Fig. 2E). Prolonged treatment with 10μM 3MB-PP1 mimicked a pka1 deletion (Supplementary Fig. 2F). Selective Pka1as1 inhibition only upon glucose limitation also blocked Pom1 re-localization, confirming that PKA is active to promote Pom1 side-localization in the glucose-limiting conditions (Fig 2E). Together, these data suggest that cytosolic Pka1 is active during glucose limitation and signals Pom1 re-localization
Mechanism of Pom1 re-localization upon glucose limitation
We considered three possible mechanisms of Pom1 re-localization: first, glucose limitation may lead to Pom1 inactivation, causing its delocalization as observed for a Pom1KD (kinase-dead) allele 25, 51; second, changes in membrane composition or potential may increase the intrinsic affinity of Pom1 for the plasma membrane; third, the delivery to cell ends of Tea4, required for Pom1 dephosphorylation and membrane-binding, may be altered. Several lines of evidence – in particular FRAP analysis, Tea4-dependency, and the fact that Pom1 controls cell length in low glucose (see below) – indicated that Pom1 remains active upon glucose limitation, and that glucose limitation only modestly affects the intrinsic affinity of Pom1 for the plasma membrane (see Supplementary Fig. 3 for details).
By contrast, the localization of the Pom1 gradient nucleator Tea4 was drastically changed upon glucose limitation (Fig 3A, C): in 0.08% glucose, there was an increase in the cortical levels of Tea4 in the middle of the cell; in 0.03% glucose, Tea4 dots were present all around the cell periphery and microtubule tracks were not detected. Tea4 re-localization reversibly occurred within 10 minutes of shift to low glucose and was pka1-dependent (Fig 3B, D). Tea4 was also present on cell sides in saturated cultures (Supplementary Fig. 3D). As localization of Tea4 on cell sides is sufficient to recruit Pom1 24, 25, we conclude that, upon glucose limitation, Pka1 regulates Tea4 localization, which in turn recruits Pom1.
Figure 3. Pka1-dependent reversible Tea4 re-localization around the cell cortex upon glucose limitation.
(A) Sum of 5 medial spinning disk confocal images taken over 30 seconds of Tea4-tdTomato in wild-type cells grown in 2% glucose or 0.03% glucose for 1h. Arrows indicate Tea4 tracks on microtubules. Arrowheads indicate Tea4 dots at cell sides.
(B) Localization of Tea4-tdTomato in pka1Δ cells as in (A).
(C) Distribution of cortical Tea4 from one tip to the other (0 = cell middle) in wild-type cells obtained with the Cellophane plugin. Average of 45, 31 and 64 profiles in 2%, 0.08% and 0.03% glucose in 8μm-long cells. Profiles obtained from other cell lengths are similar.
(D) Box and whisker plot of cortical Tea4 fluorescence intensity in the middle 2μm region in both wild-type (n=45, 31 and 64) and pka1Δ cells (n=71,66 and 62) in 2%, 0.08% and 0.03% glucose, respectively.
Scale bars represent 5μm. Experiments were performed thrice and quantification of one is shown.
Pka1 controls microtubule stability
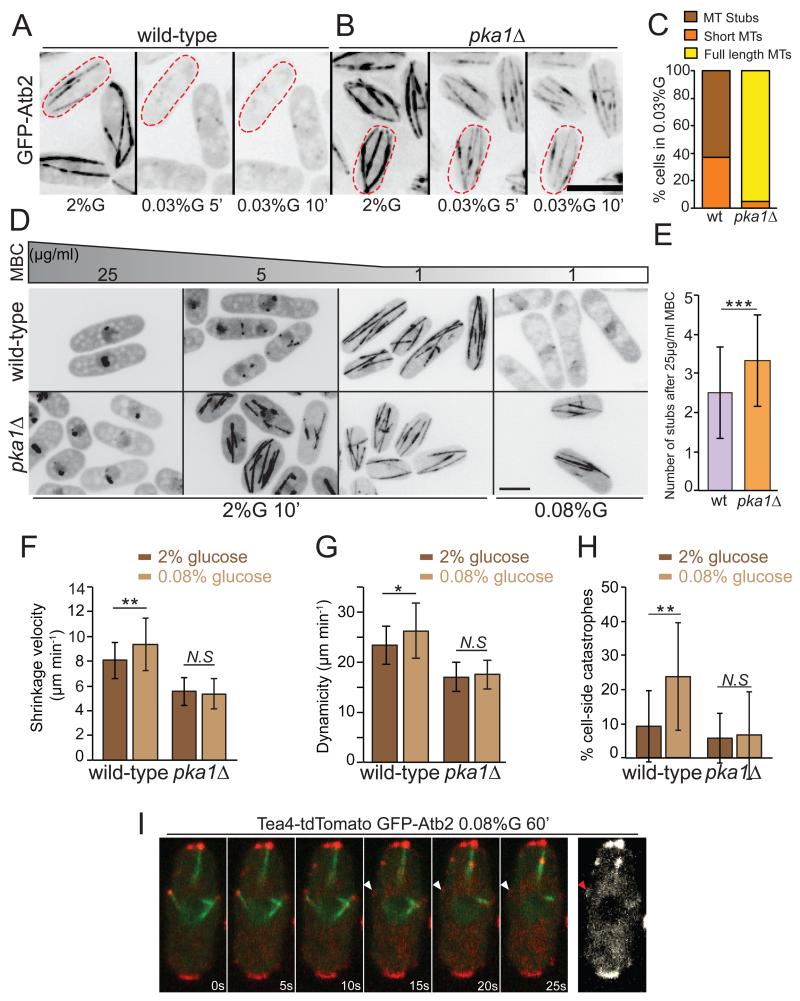
To understand the mechanism of Tea4 re-localization, we probed microtubule behavior under limited glucose conditions, using flow-chambers. A shift to 0.03% glucose caused a dramatic, rapid, reversible loss of long interphase microtubules in less than 5 minutes, with 37% cells exhibiting short dynamic microtubules and 63% displaying only microtubule stubs (stable regions of microtubule overlap) (Fig 4A, C). As medium exchange in the flow-chamber takes about 3 min, the effect occurred within 2 min of glucose deprivation. Mitotic spindles were resistant and kept elongating under these conditions. Short dynamic microtubules were observed in cells kept in 0.03% glucose for up to 20h, after which microtubules disappeared completely from all cells (Supplementary Movie 1).
Figure 4. Pka1 negatively regulates microtubule stability.
(A) Epifluorescence deconvolved maximum intensity projection images of wild-type cells expressing GFP-Atb2 grown in microfluidic chambers in 2% glucose and 5 and 10 min after shift to 0.03% glucose.
(B) GFP-Atb2 in pka1Δ cells as in (A) (2% glucose and 0.03% glucose after 10 min). Representative images from 3 independent experiments are shown.
(C) Percentage of wild-type (n=68) and pka1Δ (n=59) cells with indicated microtubule organization in 0.03% glucose for 10min in microfluidic chambers.
(D) Maximum projection of spinning disk confocal images of wild-type and pka1Δ cells expressing GFP-Atb2 treated with the indicated concentrations of MBC for 10min in 2% or 0.08% glucose. Representative images from 2 independent experiments are shown.
(E) A graph showing the number of microtubule stubs left after 10min 25μg/ml MBC treatment (n>45 cells). p=0.001.
(F) Mean microtubule shrinkage velocity in wild-type and pka1Δ cells grown under 2% glucose or 0.08% glucose for 1h (n>27 microtubules). (p= 0.008; p=0.56)
(G) Mean microtubule dynamicity in wild-type and pka1Δ cells grown under 2% glucose or 0.08% glucose for 1h (n>27 microtubules) (p=0.031; p=0.54)
(H) Percentage of microtubule catastrophes occurring at the cell-sides in wild-type and pka1Δ cells (n>100 catastrophe events in >16 cells) grown as in (E). (p=0.002, p=0.926)
(I) Time-lapse imaging of Tea4-tdTomato and GFP-Atb2 in wild-type cells shifted to 0.08% glucose for 1h acquired on the spinning disk confocal microscope. The first 6 images are maximum projections of 2 Z-sections. The last image is a projection of the 3 timepoints shown after microtubule catastrophe. Arrowheads indicate Tea4 presence at the lateral cell cortex after microtubule catastrophe. Representative images from 4 independent experiments are shown.
Scale bars represent 5μm. Error bars are standard deviation. Statistical significance was derived using student’s t-test.
In 0.08% glucose, microtubules remained long but were also destabilized. First, they were de-polymerized by sub-optimal methyl benzimidazol-2-yl-carbamate (MBC) levels (1μg/ml) that did not affect microtubules in 2% glucose (Fig 4D). Second, they showed increased shrinkage rate and dynamicity (length change of microtubules in any direction over time), as compared to cells grown in 2% glucose (Fig 4F-G). Finally, 23.8% of microtubule catastrophes occurred upon contact at the lateral cortex (vs. only 9.2 % in 2% glucose; Fig 4H). These catastrophes at cell sides led to sustained Tea4 contact and temporary deposition at the lateral cortex (Fig 4I). Thus, frequent microtubule catastrophes at cell sides may promote Tea4 and thus Pom1 re-localization upon glucose starvation.
Microtubule destabilization upon glucose limitation was entirely dependent on Pka1: in pka1Δ cells, microtubules remained long when cells were shifted to 0.03% glucose for up to 30h, after which microtubules disappeared completely as in the wild-type situation (Fig 4B-C; Supplementary Movie 2); in 0.08% glucose, they were not depolymerized by sub-optimal dosage of MBC (Fig 4D), and dynamic parameters and location of catastrophes at cell sides were not altered as compared to pka1Δ cells grown in 2% glucose (Fig 4F-G); in 2% glucose, microtubules were also significantly more stable in pka1Δ than wild-type cells, displaying higher resistance to MBC treatment, and slower shrinkage rate and dynamicity (Fig 4D-G). Conversely, Pka1 overexpression caused faster microtubule shrinkage rates, higher dynamicity and 30.2% of catastrophes occurring along the lateral cell cortex (Supplementary Fig. 4A; Table1). As pka1 overexpression causes cell elongation, we controlled for the effect that cell length may have on microtubule dynamics by using elongated cdc25-22 cells. In these cells, microtubules showed largely wild-type dynamic parameters, with 20% side catastrophes, a percentage significantly lower than that observed upon pka1 overexpression (Supplementary Fig. 4A; Table1). We conclude that Pka1 negatively regulates microtubule stability.
Table1. Microtubule dynamic parameters.
| %G | Genotype | Shrinkage Rate (μm min−1) |
Growth Rate (μm min−1) |
Freq. Catastrophe (min−1) |
Freq. Rescue (min−1) |
Dynamicity (μm min−1) |
% side catastrophes |
|
|---|---|---|---|---|---|---|---|---|
| 25°C | 2 | WT | 8.06±1.47 | 3.3±0.57 | 0.53±0.13 | 0.51±0.14 | 23.4±3.84 | 9.17±10.5 |
| 0.08 | WT | 9.34±2.1 | 2.9±0.83 | 0.63±0.28 | 0.61±0.2 | 26.2±5.5 | 23.80±15.7 | |
| 2 | pka1 Δ | 5.55±1.13 | 2.95±0.56 | 0.53±0.13 | 0.6±0.19 | 17.06±2.9 | 5.80±7.3 | |
| 0.08 | pka1 Δ | 5.36±1.23 | 3.06±0.70 | 0.36±0.12 | 0.36±0.14 | 17.53±2.9 | 6.60±12.9 | |
| 36°C | 2 | WT | 11.2 ± 3.04 | 4.6 ± 1.37 | 0.59 ± 0.2 | 0.58 ± 0.15 | 31.83±8.25 | 3.07±4.2 |
| 0.08 | WT | 12.2 ± 3.0 | 26.8±7.3 | |||||
| 2 | pka1 Δ | 6.03 ± 1.25 | 3.8 ± 0.68 | 0.41 ± 0.09 | 0.47 ± 0.11 | 19.63±2.14 | 2.4±3.9 | |
| 0.08 | pka1 Δ | 6.19 ± 1.7 | 4.9±8.1 | |||||
| 2 | cls1-36 | 11.13 ± 2.9 | 4.45 ± 0.8 | 0.47 ± 0.16 | 0.44 ± 0.15 | 31.89±6.16 | 3.1±4.3 | |
| 0.08 | cls1-36 | 11.7 ± 2.5 | 32.4±28.8 | |||||
| 2 | cls1-36 pka1 Δ | 11.19 ± 2.7 | 4.09 ± 0.9 | 0.5 ± 0.14 | 0.48 ± 0.12 | 31.33±6.59 | 3.58±4.58 | |
| 0.08 | cls1-36 pka1 Δ | 11.4 ± 2.6 | 28.1±16.05 | |||||
| 25°C | 2 | Pka1-OE | 13.1±2.1 | 3.59±0.45 | 0.71±0.15 | 0.7±0.17 | 32.1±8.1 | 30.2±24.3 |
| cdc25-22 | 8.35±1.3 | 3.93±0.98 | 0.37±0.2 | 0.39±0.17 | 25.2±4.2 | 20.1±16.9 | ||
| mal3 Δ | 6.09±1.45 | |||||||
| mal3Δ pka1Δ | 4.5±0.85 | |||||||
| tea2 Δ | 10.09±2.23 | |||||||
| tea2Δ pka1Δ | 6.98±1.26 | |||||||
| tip1 Δ | 7.74±1.03 | |||||||
| tip1Δ pka1Δ | 5.98±0.83 | |||||||
| tip1Δ tea2Δ mal3Δ | 8.4±2.0 | |||||||
| tip1Δ tea2Δ mal3Δ pka1Δ | 5.0±0.18 | |||||||
| alp14 Δ | 4.7±0.58 | |||||||
| alp14Δ pka1Δ | 3.7±0.58 | |||||||
| ase1 Δ | 8.3±1.67 | |||||||
| ase1Δ pka1Δ | 5.7±1.24 | |||||||
| dhc1 Δ | 8.0±1.23 | |||||||
| dhc1 Δ pka1 Δ | 4.62±1.12 |
Remarkably, microtubule destabilization was sufficient to trigger both Tea4 and Pom1 side-relocation. Indeed, Tea4 and Pom1 partly re-localized to cell sides in glucose-starved pka1Δ cells when microtubules were destabilized either by a 10 min MBC treatment (Supplementary Fig. 4C-D), or by deletion of the +TIP complex (Supplementary Fig.4E). Similarly, Pom1 localized along cell sides in pka1-overexpressing cells even in 2% glucose, but not in cdc25-22 mutant cells (Supplementary Fig.4B). Thus, Pka1 promotes microtubule destabilization, which causes Tea4 and thus Pom1 re-localization to cell sides.
Pka1 regulates microtubule dynamics through CLASP
To dissect the mechanism by which Pka1 destabilizes microtubules, we screened for genetic interactions between pka1Δ and mutations in known MAPs in 2% glucose, initially measuring microtubule shrinkage rates. We reasoned that pka1Δ would not modify the phenotype of a MAP deletion if this was regulated by PKA, but would show additive effects to other MAP mutants. Although single or even triple deletions of the +TIP complex, in which microtubules are destabilized 28, 52, 53, restored Pom1 side-localization to glucose-starved pka1Δ cells (Supplementary Fig. 4E), all deletions showed additive effects with pka1Δ for microtubule shrinkage velocities (Table 1). Similarly, deletion of the TOG-domain protein Alp14/XMAP215, shown to accelerate microtubule assembly 32, was additive with pka1Δ. These additive phenotypes indicate that Pka1 does not modulate microtubule stability through these MAPs.
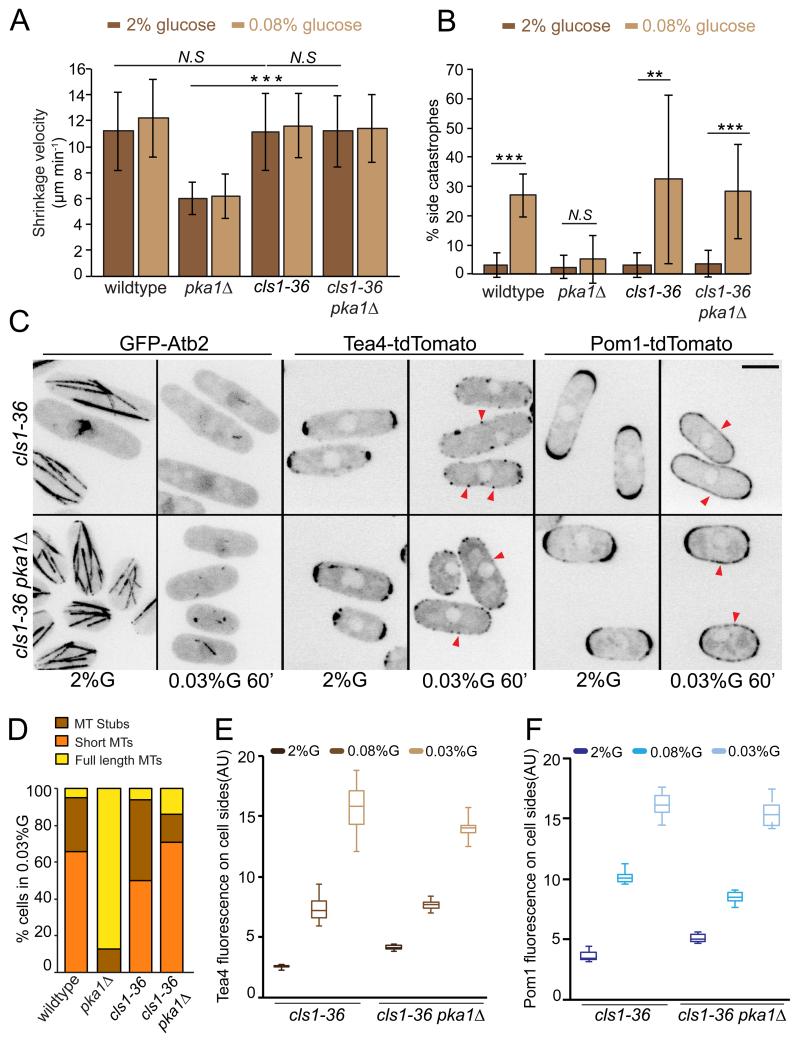
By contrast, several lines of evidence showed that Pka1 modulates microtubule dynamics through the sole fission yeast CLASP Cls1/Peg1. Consistent with previous observations 34, inactivation of Cls1, using a published temperature-sensitive mutant cls1-36 at the restrictive temperature of 36°C, revealed no or minor effect on microtubule dynamics (Table 1). Surprisingly however, cls1-36 masked the effect of pka1Δ for all microtubule dynamic parameters in both 2% and 0.08% glucose (Fig 5A). The cls1-36 mutant also restored frequent side-catastrophes in pka1Δ cells in 0.08% glucose (Fig 5B), and microtubule destabilization in 0.03% glucose (Fig 5C-D). Thus, in absence of PKA, CLASP modulates microtubule dynamics. We note though that more long microtubules were observed in double cls1-36 pka1Δ than single cls1-36 mutant in 0.03% glucose (Fig 5D), suggesting PKA may signal through additional MAPs for microtubule destabilization. CLASP inactivation further restored Tea4 and Pom1 side-localization upon glucose starvation in pka1Δ cells (Fig 5C, E-F). Conversely, cls1 overexpression, which promotes microtubule stabilization 34, led to restriction of Pom1 at cell tips upon glucose limitation, mimicking the pka1Δ phenotype (Supplementary Fig. 5). We conclude that Pka1 promotes microtubule destabilization, and thus Tea4 and Pom1 side-localization, by negatively regulating CLASP.
Figure 5. Pka1 regulates microtubule dynamics through CLASP to trigger Tea4 and Pom1 side-localization.
(A) Mean microtubule shrinkage velocity in wild-type, pka1Δ, cls1-36 and cls1-36 pka1Δ strains grown in 2% glucose or 0.08% glucose for 1h at 36°C (n>15 microtubules) (p=0.9, p=0.93, p<10−9)
(B) Percentage of microtubule catastrophes occurring at cell sides in the same strains, grown as in (A) (n>70 catastrophe events in >16 cells). Statistical significance is derived using student’s t-test. (p<10−8 p=0.46 p=0.015 p= 0.00013). Error bars are standard deviations.
(C) Maximum intensity projection of spinning disk images of GFP-Atb2, and sum of 5 medial spinning disk confocal images taken over 30 seconds of Tea4-tdTomato and Pom1-tdTomato in cls1-36 and cls1-36 pka1Δ cells grown in 2% glucose at 25°C and shifted to 36°C for 1h in either 2% glucose or 0.03% glucose. Arrowheads indicate Tea4 or Pom1 side-localization. Scale bar represents 5 μm.
(D) Percentage of wild-type (n=38), pka1Δ (n=32), cls1-36 (n=100) and cls1-36 pka1Δ (n=119) cells showing the indicated microtubule organization after shift to 0.03% glucose for 10min at 36°C.
(E) Box and whisker plot of cortical Tea4 fluorescence intensity in the middle 2μm region in both cls1-36 and cls1-36 pka1Δ cells at 36°C with cells grown in 2% glucose at 25°C and shifted to 36°C for 1h in either 2% glucose or 0.08% glucose or 0.03% glucose (n>15).
(F) Box and whisker plot of cortical Pom1 fluorescence intensity in the middle 2μm region in both cls1-36 and cls1-36 pka1Δ cells at 36°C with cells grown as in (E) (n>19). Experiments were performed thrice and quantification of one is shown.
CLASP stabilizes microtubules by binding both microtubules and tubulin directly through its S/R-rich region and N-terminal TOG domains, respectively 34, 39. In vivo, Cls1 is recruited to zones of microtubule overlaps by the microtubule bundler Ase1 34. Dynein-dependent Cls1 localization near microtubule plus-ends was also reported 54, 55. Cls1 localization was significantly changed both in low-glucose and in pka1Δ cells. In pka1Δ cells, Cls1 showed increased levels on microtubules and often formed multiple dots on a single microtubule, though global Cls1 levels were only modestly changed (Fig 6A, Supplementary Fig. 6A-C). In low-glucose, Cls1 also showed enhanced local levels and formed strong foci near the cell middle (Fig 6B). Whereas most of these foci localized on microtubule bundles in pka1Δ cells, 12-35% were at sites lacking microtubules in wild-type cells (Fig 6C). These foci that do not contain microtubules may represent less active or inactive Cls1 in agreement with the less stable microtubule bundles in these conditions. We were unable to test for the presence of Ase1 in these foci, as co-localization experiments using Ase1-mCherry and Cls1-3GFP showed synthetic effects. However, deletion of neither Ase1 nor the dynein Dhc1 masked the effect of pka1Δ on microtubule dynamics, suggesting Pka1 does not regulate Cls1 solely through these targeting factors (Table 1). Thus PKA may render Cls1 less active for microtubule rescue, leading to complete loss of the microtubule bundle. Additionally, we found that Pka1 overexpression was able to mitigate the Cls1 TOG domains-dependent microtubule stabilization (Fig 6D-E), suggesting Pka1 reduces the activity of Cls1 in microtubule stabilization.
Figure 6. Pka1 promotes microtubule destabilization by negatively regulating Cls1.
(A) Maximum intensity spinning disk projections showing time-lapse imaging of Cls1-3GFP and mCherry-Atb2 in wild-type and pka1Δ cells grown in 2% glucose. Arrowheads highlight Cls1 dots on microtubules.
(B) Maximum intensity spinning disk projections of Cls1-3GFP and mCherry-Atb2 in wild-type and pka1Δ cells grown in in2% glucose or 0.08% glucose or 0.03% glucose for 1h. Arrowheads indicate Cls1 dots present on microtubule overlaps. Arrows point to Cls1 foci not on microtubules.
(C) Percentage of Cls1 foci not on microtubules in cells as in (B) (n>96). Representative images from 3 independent experiments and quantification of one are shown.
(D) Time-lapse imaging of GFP-Atb2 in wild-type and pka1 over-expressing (pka1OE) cells transformed with pnmt1-cls1(1-500)-mCherry grown in 2% glucose and induced without thiamine for 14-16h. Arrowheads indicate stable microtubules in wild-type (upper panel) and arrows track microtubule shrinkage events in pka1OE (bottom panel) cells.
Scale bar represents 5 μm.
(E) Percentage of cells showing dynamic or at least one stabilized microtubule bundle in strains as in (D). Cells with similar range of Cls1 fluorescence levels were chosen for this analysis (n=31 for wild type, n=41 for pka1 over-expression). Representative images from 2 independent experiments and quantification of one are shown.
(F) Schematic depicting the mechanism of Pom1 side-localization under glucose limitation conditions. The left part of the cell shows the situation in high glucose, when CLASP rescues microtubules allowing them to reach cell ends. The right part of the cell shows the situation in low glucose, when PKA activity antagonizes the microtubule-stabilizer CLASP. Microtubules thus become destabilized and undergo catastrophe at the lateral cortex, depositing Tea4 there. Tea4 mediated de-phosphorylation of Pom1 leads to an increase in its levels at cell sides, promoting mitotic delay.
Pom1 re-localization buffers against excess cell shortening
We examined the physiological consequences of Pom1 re-localization in low glucose, by examining cell length at division as a proxy of cell cycle length 56. We measured cell length at division in the standard conditions of 2% glucose and in 0.08% glucose, in which cells show reliable growth and division patterns, though at shorter size 8 (Table 2).
Table 2. Mean cell length at division.
| Genotype | Mean cell length at division | Adaptation index | p value | |
|---|---|---|---|---|
| 2%G | 0.08%G | |||
| WT | 13.9±1.33 | 12.2±1.57 | 12.23 | >10−14 |
| pka1 Δ | 11.14±1.9 | 10.94±3.03 | 1.79 | 0.561090743 |
| sty1 Δ | 23.04±2.61 | 24.6±5.4 | −6.77 | 0.0002 |
| sty1Δ pka1Δ | 22.7±3.1 | 21.2±3.2 | 6.6 | >10−7 |
| pom1 Δ | 11.58±1.7 | 8.69±1.26 | 24.95 | >10−49 |
| pom1Δ pka1Δ | 9.07±1.77 | 8.64±2.17 | 4.74 | 0.116039099 |
| pom1Δ sty1Δ | 17.4±1.99 | 16.6±2.08 | 4.59 | 0.0048 |
| cdr2 Δ | 22.25±1.86 | 21.24±3.9 | 4.5 | >10−8 |
| pom1Δ cdr2Δ | 21.8±3.02 | 20.5±2.95 | 6.3 | >10−6 |
| pom16A | 19.38±1.38 | 18±3.5 | 7.12 | >10−5 |
| pom1KD | 11.5±1.5 | 8.2±1.7 | 28.69 | >10−34 |
| tea4 Δ | 15.1±1.01 | 9.6±0.9 | 36.5 | >10−82 |
| tea4RVXF* | 15.1±0.9 | 10.5±1.1 | 30.67 | >10−60 |
| cdr2T166A | 18.22±1.57 | 14.34±1.7 | 21.42 | >10−37 |
| WT (YSM2224) | 13.08±1.55 | 11.69±2.31 | 10.6 | 0.0003 |
| pom1Δ (YSM2229) | 11.25±1.38 | 8.34±1.46 | 25.8 | >10−18 |
| cdr2S755A-758A (YSM2226) | 12.46±1.08 | 11.03±2.16 | 11.47 | >10−7 |
| pom1Δ cdr2S755A-758A (YSM2234) | 12.11±2.24 | 8.52±1.24 | 20.71 | >10−31 |
The p value was calculated by using the student’s t-test comparing the cell length at division in 2% glucose vs 0.08% glucose for the same strain. A p value less than 0.05 was considered to indicate a significant difference.
All strains used are prototroph, except for the bottom four (YSM2224, YSM2226, YSM2229 and YSM2234), which are auxotroph for leucine and uracil.
Pka1 plays an ill-understood function in cell cycle progression. Indeed, pka1Δ cells are short, and pka1 overexpression yields long cells, suggesting Pka1 delays the cell cycle 9, 10. Previous work placed Pka1 upstream of MAPK signaling 57, 58. Consistently, in 2% glucose, deletion of the MAPK Sty1 was largely epistatic over Pka1, with sty1Δ pka1Δ double mutants dividing at a length similar to sty1Δ single mutants; and pom1Δ and pka1Δ were additive, with double mutants dividing at shorter size than either single mutant. By contrast, in 0.08% glucose, sty1Δ pka1Δ double mutants were significantly shorter than sty1Δ single mutants, suggesting that in these conditions Pka1 plays an additional role, not upstream of Sty1; in addition, pom1Δ pka1Δ double mutant divided at the same length as pom1Δ single mutant, suggesting Pom1 is largely epistatic over Pka1 in these conditions. We conclude that Pka1 regulates both MAPK and Pom1 pathways, with contributions depending on glucose conditions.
To better understand the changes in cell size upon glucose limitation, we devised an adaptation index representing the percent difference in cell lengths between 2% and 0.08% glucose. Wild-type cells reduced in length by about 12%. As is the case upon nitrogen limitation 5, 6, 7, MAPK likely promotes mitotic entry upon glucose limitation, as sty1Δ cells did not shorten in 0.08% glucose. Instead, these cells became longer, yielding a negative adaptation index. Remarkably, this negative adaptation was abolished by pom1 deletion. Consistently, pom1 deletion or inactivation caused hyper-adaptation with cells loosing 25% of their length in low glucose, suggesting Pom1 plays a comparatively more important role to delay mitosis in glucose-limiting conditions.
We confirmed that the newly recruited, membrane-associated Pom1 activity delays mitosis through Cdr2 regulation, as in cells grown in glucose-rich medium. First, pom1KD cells bearing a kinase-dead allele were indistinguishable from pom1Δ, indicating Pom1 kinase activity is required. Second, tea4 mutant cells, in which Pom1 is largely cytosolic 25, showed hyper-adaption to glucose limitation like pom1Δ, indicating that Pom1 signals from the plasma membrane. Third, mutation of 6 Pom1 auto-phosphorylation sites in pom16A, previously shown to delocalize active Pom16A around the cell cortex even in rich medium 25, severely dampened cell length adaption to low glucose. Fourth, cdr2Δ was epistatic to pom1Δ under limiting glucose, like in rich medium, and also showed dampened adaptation. Finally, we found that the cdr2T166A allele, carrying a mutation of the activation loop preventing phosphorylation by the activating kinase Ssp1 16, showed hyper-adaption to glucose limitation, similar to pom1Δ. This is consistent with the proposed model that Pom1 blocks activation of Cdr2 by Ssp1 16. However, mutation of the C-terminal Pom1 phosphorylation sites in cdr2S755A-758A, through which Pom1 was proposed to inhibit Cdr2 activity 14, 16, was epistatic to pom1Δ in high, but not low glucose, and did not show hyper-adaption to glucose limitation, suggesting other modes of Cdr2 regulation by Pom1 exist.
Together these data suggest that the higher levels of Pom1 along cell sides in low glucose conditions impose a stronger inhibition on Cdr2, balancing the mitosis-promoting effect of the MAPK pathway.
Discussion
We report here two physiological changes in cells grown in limiting glucose levels, a condition that cells may routinely face in the wild. First, the mitotic inhibitor Pom1 re-localizes to cell sides upon glucose limitation, where it delays division. Second, microtubule organization and dynamics are severely altered. Both changes rely on PKA signaling negatively regulating the activity of the microtubule rescue factor CLASP. Thus, when glucose is limiting, microtubules are destabilized and undergo catastrophe at cell sides, where they transiently deposit the type I phosphatase regulatory subunit Tea4. This in turn promotes Pom1 membrane binding at cell sides, which delays cell division (Fig 6F).
PKA signals microtubule destabilization. Indeed, microtubules are stabilized and insensitive to glucose levels in cells deficient in the cAMP/PKA signaling cascade, whereas Pka1 overexpression promotes microtubule destabilization. Previous investigation of PKA had shown it localizes to the nucleus in glucose-rich conditions 48, where is serves to repress transcription 46, 47. By contrast, cytosolic PKA likely controls microtubule dynamics. Indeed, the PKA-dependent destabilization of microtubules upon starvation is rapid (occurring within 2min) and reversible, and thus unlikely to be due to a transcriptional response 59. We further showed that Pka1 is active in glucose-limiting conditions and that its activity is required to signal Pom1 re-localization. These data are consistent with a cytosolic microtubule-destabilizing PKA activity in glucose-limiting conditions.
We note though that PKA is unlikely to be the sensor of glucose limitation to trigger microtubule destabilization, because cgs1Δ mutants and cls1-36 pka1Δ double mutants, which have constitutive and no PKA activity, respectively, still re-localize Pom1 to cell sides specifically upon glucose limitation. Thus, while PKA activity is necessary for microtubule destabilization, it is not sufficient for it. We deduce that an additional pathway signals microtubule destabilization and Pom1 re-localization upon glucose depletion. Because of the alterations in Pom1 levels and localization observed in sty1 and wis1 mutants (see Supplementary Fig. 1E) and given the genetic relationships of pka1 and sty1 57, 58, the stress MAPK pathway may be a prime candidate for this additional signal.
CLASP-dependent microtubule stabilization is a likely target of the PKA signal. In vitro, CLASP promotes rescue and suppresses catastrophes by recruiting tubulin dimers to the microtubules 39. In fission yeast, CLASP is recruited to zones of microtubule overlap by Ase1, where it promotes microtubule rescue and the maintenance of the microtubule bundle, but does not appear to affect microtubule plus end dynamics (34 and our data). This is in contrast to the situation in metazoan cells, where CLASP is recruited to microtubule plus-ends by CLIP-170 and protects them from catastrophe 38, or in plant cells, where CLASP promotes microtubule growth at cell edges 60. We now show that, in pka1Δ cells, CLASP loss-of-function restores microtubule depolymerization rate and catastrophes at cell sides, as well as Tea4 and Pom1 relocation, in low glucose. We conclude that CLASP also promotes microtubule growth persistence in fission yeast, but that this activity is normally inhibited by PKA signaling. Thus, tuning of CLASP activity underlies distinct microtubule organization.
PKA signaling likely antagonizes the microtubule-stabilizing function of CLASP. In glucose-starved wild-type cells, a significant number of Cls1 foci do not contain microtubules. We interpret these as sites of inactive Cls1, unable to promote microtubule rescue, from which the microtubule bundle has been lost. Contrary to this, in pka1Δ cells, most Cls1 foci contain microtubules, suggesting Cls1 is functional for microtubule rescue. In addition, Pka1 overexpression antagonizes the microtubule-stabilizing effect of the CLASP TOG domains, which directly bind tubulin dimers. PKA signaling may also antagonize CLASP microtubule binding, as CLASP localization on microtubule overlaps is enhanced in pka1Δ cells. We suggest that, similar to in vitro observations 39, in absence of PKA, direct binding by a few CLASP molecules near a microtubule plus end leads to rescue events promoting further growth of microtubules contacting cell sides.
We note however that CLASP is unlikely to be a direct PKA substrate, because mutation of five predicted PKA phosphorylated sites in Cls1 N-terminus did not alter Cls1’s ability to stabilize microtubules in vivo, and immuno-precipitated Pka1-GFP failed to phosphorylate Cls1 fragments in vitro. PKA may also indirectly modulate microtubules by regulating the cell energy levels. Indeed, recent data in the budding yeast has shown that cells lacking PKA activity exhibit higher ATP stores 61. The complete loss of microtubules we observe after long-term starvation in 0.03% glucose may reflect a loss of cellular energy, which indeed happens later in the pka1 deleted cells. Thus, PKA may regulate microtubule dynamics in direct and indirect ways.
The cAMP/PKA pathway modulates the cell cycle 9, 10, and we show its effect depends on glucose levels. When glucose is in excess, PKA acts principally through the stress MAPK pathway, by ways that remain to be characterized 57, 58. By contrast, when glucose is limiting, PKA signals through Pom1 in addition, by modifying its localization. The predicted net effect is an alteration of the balance between Cdc25 and Wee1 activities in favor of Wee1, to delay mitotic commitment. This counterbalances the effect of MAPK activation helping cells to attain a critical size.
Mitotic entry is exquisitely sensitive to Pom1 dosage, but natural changes in Pom1 amounts at the cell middle in steady-state growing conditions had so far not been observed 12, 13, 14, 17, 25. We now show that modulation of Pom1 concentration at the medial cortex is a physiological cellular response to glucose levels. Pom1 side-enrichment in 0.08% glucose has important effect on cell size at division by regulating Cdr2. This is especially noticeable in absence of the MAPK Sty1, when Pom1 causes an increase in cell size. By contrast, these localization changes do not have major effect on medial division, consistent with the lower sensitivity of division site placement on Pom1 levels 14, 24. Thus, one important function of the mitotic inhibitor Pom1 is to buffer cell size during glucose-starvation. Future work should reveal whether this is due to the modification of a homeostatic sizer system.
Methods
Strain construction, media and growth conditions
All S.pombe strains used are listed in Supplementary Table 1. All plasmids used are listed in Supplementary Table 2. Strains were made by either tetrad dissection or random spore analysis and replication on appropriate antibiotic plates. Gene tagging and deletions were done using a PCR-based approach and confirmed by diagnostic PCR 62. Transformations were done using the lithium-acetate-DMSO method. Pka1-overexpressing strains were obtained as follows: For KanMX-3nmt1-GFP-Pka1 strain (Supplementary Fig. 4A-B), the first 547 bp of Pka1-ORF and the last 500 bp of Pka1-promoter were cloned in a pFA6a-KanMX6-P3nmt1-GFP plasmid using BamHI/SalI sites in frame after GFP and SacII/EcoRV sites before the kanMX cassette, respectively. The plasmid was cut with SacII/SalI to excise the cassette with the homology and transformed in appropriate yeast strains, wherein the integration was checked by diagnostic PCR. For NatMX-3nmt1-Pka1 strain (Fig 6D lower panel), the kanMX6 cassette from pFA6a-kanMX6-P3nmt1 was replaced by the natMX cassette, excised from pFA6a-natMX6 with EcoRV/BglII, followed by the same strategy as above.
To generate the pka1-as1 strain, the ORF of Pka1 and 618bp of the 3′UTR were cloned in a small plasmid using BamHI/SalI sites and the gatekeeper residue aa278 (Met) was mutated to (Gly) M278G 63 by site-directed mutagenesis and checked by sequencing. The cassette was then excised using the same enzymes and transformed into a pka1::ura4+ strain which still retains the first 200bp of the Pka1 ORF. Colonies were selected on 5′FOA plates and correct integration was checked by diagnostic PCR.
Cells were grown in standard Edinburgh minimal media (EMM) with appropriate supplements adenine, leucine, uracil (ALU) when required. For glucose limitation assays, cells were grown in 2% glucose to mid-log, washed three times in either 0.08% glucose or 0.03% glucose and then incubated in the same medium before imaging. For measurement of glucose in the medium (Supplementary Fig. 1B), cells were grown in EMM-2% glucose to saturation and glucose concentration in the medium was measured using the Glucose (HK) Assay Kit (Sigma) at the indicated time-points. For nitrogen depletion experiment, cells were first grown in EMM medium containing nitrogen and then shifted to medium lacking nitrogen for the indicated time-points.
Adenosine 3′,5′-cyclic monophosphate (cAMP) (Sigma A9501), was used at final concentrations as indicated from a stock of 30mM dissolved in the same medium as used for imaging. 4-Amino-1-tert-butyl-3- (3-methyl benzyl) pyrazolo (3,4-d pyrimidine) 3MB-PP1 (Toronto Research Chemicals Inc.) was added at final concentrations as indicated from a stock of either 5mM or 1mM in MeOH (methanol). MeOH addition had no effect on cell length or Pom1 localization. In Fig4A-B cells were first grown in EMM with proper supplements, containing 2% glucose to mid-log phase. They were diluted to an O.D of 0.1-0.2, and 100μl of cells were loaded onto ONIX microfluidic chambers (Y04C plates, CellAsic). After loading, the medium flux was kept constant at 2psi, at which the chamber refresh time is just above 3 min (CellAsic Y04C specifications). These cells were first allowed to adapt to the chambers by letting them grow for 3-4 hours prior to imaging. GFP-Atb2 was imaged in these cells in 2% glucose every 5mins for 15mins, followed by a change in medium to 0.03%G-imaged every 5min for 30mins.
Methyl-benzidazole-2-yl-carbamate (MBC, Sigma, St. Louis, MO) was used at a final concentration as indicated, from a stock of 2.5 mg/ml in DMSO. To induce the expression of Pka1, from the nmt1 promoter cells were first grown in a medium containing 5μg/ml thiamine from a 2000x stock, to repress the promoter, washed three times in the same medium without thiamine and incubated for 18-20 hours prior to imaging. All temperature sensitive strains were first grown at their permissive temperature of 25°C then shifted to their restrictive temperature of 36°C in either 2%G, 0.08% glucose or 0.03% glucose and imaged using a heated objective at 36°C, except the cdc25-22 which was grown at the semi-permissive temperature of 30°C. All other strains were grown at 30°C and imaged at room temperature (around 23-25°C) unless stated otherwise. For cell-length measurements (Table 2) all strains used were prototrophs and grown in EMM, except for cdr2S755A-758A, pom1Δ cdr2S755A-758A and corresponding control strains, which were grown in EMM-ALU, with either 2% glucose or 0.08% glucose to mid-log before imaging. Calcofluor (Sigma) was added at a final concentration of 5μg/ml from a 200X stock solution. Hoechst (Sigma) was added at a concentration of 1μg/ml for about 15-20 minutes prior to imaging.
Microscopy
Microscopy was performed on live cells, either on a spinning disk confocal microscope or on a DeltaVision epifluorescence system. Spinning disk microscopy was carried out using a Leica DMI4000B inverted microscope equipped with HCX PL APO X100/1.46 (NA) oil objective and a PerkinElmer Ultraview Confocal system (including a Yokagawa CSU22 real-time confocal scanning head, and solid-state laser and a cooled 14-bit frame transfer EMCCD C9100-50 camera) 64. Stacks of z-series confocal sections were acquired at 0.3 μm intervals with the Volocity software. For quantifications of Pom1 and Tea4 using Cellophane 14, 5 images were taken over 30 seconds and summed at 1sec exposure time, binning 1. To measure microtubule dynamics (Table 1), images were acquired on the spinning-disk microscope as 3 medial Z-stacks at an interval of 0.5 μm every 5 seconds for 10 minutes and analysis was done on maximum projections. FRAP (fluorescent recovery after photobleaching) experiments were performed on the spinning disk using the PerkinElmer photokinesis module. One full tip was bleached and post-bleach images were acquired at 5 s intervals for the first 60 s followed by 10 s intervals for the next 140 s and finally 30 s intervals for the last 600 s to minimize bleaching during image acquisition 25. For Fig4A-B, wide-field fluorescence microscopy was performed on a DeltaVision platform (Applied Precision) composed of a customized Olympus IX-71 inverted microscope and a UPlan Apo 100X/1.4 NA oil objective, a CoolSNAP HQ2 camera (Photometrics), and an Insight SSI 7 color combined unit illuminator. 7 Z-stacks were acquired at an interval of 0.6μm and a maximum projection was taken and deconvolved. For cell-length measurements (Table 2), images of calcofluor-stained cells were taken using the same setup with a Plan Apo 60x/1.42 NA objective. Figures were prepared with ImageJ64 and Adobe Photoshop CS5 and assembled using Adobe Illustrator CS5.
Biochemistry
For Western blots protein extracts were prepared in CXS buffer (50 mM HEPES, pH 7.0, 20 mM KCl, 1 mM MgCl, 2 mM EDTA, pH 7.5 and protease inhibitor cocktail) from cells grown in EMM with appropriate glucose concentrations, using glass beads and a bead beater and 25μg of protein was loaded and resolved on a SDS-PAGE gel. Primary antibodies used were anti-GFP (Roche) and anti-tubulin (TAT1), secondary antibody was anti-mouse-HRP (Promega) used at a concentration of 1:3000.
Image Analysis
Image analysis for cortical quantification of Pom1, Cdr2 and Tea4 in Fig 1C-D, Fig 3C-D, and Supplementary Fig. 1G and 4D was done using the manual mode of the ImageJ plugin-Cellophane 14. A line was drawn around the cortex to measure the fluorescence intensity and the plugin also extracts values of the cytoplasmic signal and the background. The profiles were aligned and further analyzed using the R-software. The profiles depicted span from one cell tip to the other. The mean fluorescence intensity measured in the medial 2μm region from all the cells was used for the bar and whisker plots as shown in the panels. Cortical Pom1 and Tea4 measurements in Fig 5E-F were done manually using the segmented line tool plugin of ImageJ on images taken as for Cellophane analysis. A 3-pixel line was drawn along the cell periphery from one tip to the other for 20 cells and the fluorescence intensities were obtained using the Plot profile tool. The intensities were corrected for background obtained from profiles of the same line dragged just outside the cell and the corrected intensities in the middle of each cell was selected. The mean fluorescence intensity of the medial 2μm region of 20 cells was used for the bar and whisker plots shown.
To calculate global fluorescence intensities (Supplementary Fig. 1D and 6A), a sum projection of 14 z-stacks on the spinning disk confocal microscope was done for each cell. The polygon tool of ImageJ was used to draw a line manually around the cell periphery and the mean fluorescence intensity inside the cell was obtained using the Measure tool. The fluorescence intensity was corrected for background intensity taken just outside the cell and for cells not expressing GFP or tdTomato. To measure the ratio of cls1 over microtubules in Supplementary Fig. 6B, a sum intensity projection of the 14 z-stacks was done on individual cells imaged on the spinning disk. A 5-pixel line was drawn along a microtubule, using the Straight Line tool in ImageJ and the intensities were obtained using the Plot profile tool. The maximum intensity for each cell was obtained for >27 cells for both the GFP and RFP channels and a ratio of GFP/RFP was calculated. The mean of the ratios is shown on the graph.
FRAP quantifications were done as follows, the mean fluorescence intensities were measured over time in three regions: (1) the photo-bleached region (tip), (2) the background, and (3) another non-bleached cell. For each time point, the intensities of the bleached region and that of the non-bleached cell were adjusted by subtracting background signal. To correct for loss of signal due to imaging, the adjusted bleached region intensity was then divided by the adjusted intensity of the other cell. For each experiment, all values were normalized so that the prephoto-bleaching value equals 1 65. The data was plotted and fit to an exponential and t1/2 derived using the Igor Pro 6 software (Wavemetrics).
For measurements of microtubule dynamics, the ImageJ Manual Tracking plugin was used to measure the growth and shrinkage velocities. Microtubule dynamicity was calculated as follows 66, 67. Briefly growth and shrinkage rates were calculated as the change in microtubule length (growth or shrinkage) in time and averaged. Frequency of catastrophe and rescue were measured on a per microtubule basis over 5 min of imaging, and averaged. Dynamicity was calculated by using the following formula 66: Dynamicity = [(Growth rate/Freq. catastrophe) + (Shrinkage rate/Freq. rescue)] × (Freq. catastrophe + Freq. rescue)
For cell length measurements done on calcofluor stained cells in Supplementary Fig.2C and Table 2, a line was drawn manually across the length of septated cells from the middle of one tip to the other and the length measured using the ImageJ Measure tool. To derive the adaptation index (Table 2) the difference between the length in 2% glucose and 0.08% glucose was normalized to the length in 2% glucose. All the graphs represent mean values and the error bars indicate standard deviations except for Fig 1D, 3D, 5E-F and Supplementary Fig. 4D where box and whisker plots are shown. The middle line in the box represents the median value. The upper and lower edges of the box show the upper and lower quartile respectively. The whiskers/bars represent the greatest and the lowest value in the given dataset.
Statistical analysis
Student t-test was used throughout to test statistical significance of differences in pairwise comparisons. The level of significance is shown with asterisks with * indicating p< 0.05, ** p < 0.01 and *** p< 0.001. N.S indicates not significant (p>0.05). The exact p values are stated in the figure legends. Sample size (n) in each experiment was chosen as a number sufficient for statistical analysis, but manageable in terms of analysis time. The exact sample size is indicated in the figure and/or figure legend. No samples were excluded from the analysis and no randomization was used.
Supplementary Material
Acknowledgements
We thank Charles Hoffman, Steven Marcus, Fred Chang, Mitsuhiro Yanagida, Iain Hagan, Takashi Toda for strains and reagents, Micha Hersch for help with Cellophane profile alignments, Varun Sreenivasan for help with Igor and Richard Benton, Felipe Bendezu, Laura Merlini, Payal Bhatia and Pranav Ullal for comments on the manuscript. This work was supported by an ERC Starting grant (260493) and a Swiss National Science Foundation research grant (31003A_138177) to SGM.
Footnotes
The authors declare they have no conflict of interest.
References
- 1.Turner JJ, Ewald JC, Skotheim JM. Cell size control in yeast. Curr Biol. 2012;22:R350–359. doi: 10.1016/j.cub.2012.02.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jorgensen P, Tyers M. How cells coordinate growth and division. Curr Biol. 2004;14:R1014–1027. doi: 10.1016/j.cub.2004.11.027. [DOI] [PubMed] [Google Scholar]
- 3.Lloyd AC. The regulation of cell size. Cell. 2013;154:1194–1205. doi: 10.1016/j.cell.2013.08.053. [DOI] [PubMed] [Google Scholar]
- 4.Navarro FJ, Weston L, Nurse P. Global control of cell growth in fission yeast and its coordination with the cell cycle. Curr Opin Cell Biol. 2012;24:833–837. doi: 10.1016/j.ceb.2012.10.015. [DOI] [PubMed] [Google Scholar]
- 5.Petersen J, Hagan IM. Polo kinase links the stress pathway to cell cycle control and tip growth in fission yeast. Nature. 2005;435:507–512. doi: 10.1038/nature03590. [DOI] [PubMed] [Google Scholar]
- 6.Petersen J, Nurse P. TOR signalling regulates mitotic commitment through the stress MAP kinase pathway and the Polo and Cdc2 kinases. Nat Cell Biol. 2007;9:1263–1272. doi: 10.1038/ncb1646. [DOI] [PubMed] [Google Scholar]
- 7.Shiozaki K, Russell P. Cell-cycle control linked to extracellular environment by MAP kinase pathway in fission yeast. Nature. 1995;378:739–743. doi: 10.1038/378739a0. [DOI] [PubMed] [Google Scholar]
- 8.Pluskal T, Hayashi T, Saitoh S, Fujisawa A, Yanagida M. Specific biomarkers for stochastic division patterns and starvation-induced quiescence under limited glucose levels in fission yeast. FEBS J. 2011;278:1299–1315. doi: 10.1111/j.1742-4658.2011.08050.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Maeda T, Watanabe Y, Kunitomo H, Yamamoto M. Cloning of the pka1 gene encoding the catalytic subunit of the cAMP-dependent protein kinase in Schizosaccharomyces pombe. J Biol Chem. 1994;269:9632–9637. [PubMed] [Google Scholar]
- 10.Yu G, Li J, Young D. The Schizosaccharomyces pombe pka1 gene, encoding a homolog of cAMP-dependent protein kinase. Gene. 1994;151:215–220. doi: 10.1016/0378-1119(94)90659-9. [DOI] [PubMed] [Google Scholar]
- 11.Hoffman CS. Glucose sensing via the protein kinase A pathway in Schizosaccharomyces pombe. Biochem Soc Trans. 2005;33:257–260. doi: 10.1042/BST0330257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Martin SG, Berthelot-Grosjean M. Polar gradients of the DYRK-family kinase Pom1 couple cell length with the cell cycle. Nature. 2009;459:852–856. doi: 10.1038/nature08054. [DOI] [PubMed] [Google Scholar]
- 13.Moseley JB, Mayeux A, Paoletti A, Nurse P. A spatial gradient coordinates cell size and mitotic entry in fission yeast. Nature. 2009;459:857–860. doi: 10.1038/nature08074. [DOI] [PubMed] [Google Scholar]
- 14.Bhatia P, et al. Distinct levels in Pom1 gradients limit Cdr2 activity and localization to time and position division. Cell Cycle. 2013;13 doi: 10.4161/cc.27411. [DOI] [PubMed] [Google Scholar]
- 15.Rincon SA, et al. Pom1 regulates the assembly of Cdr2-Mid1 cortical nodes for robust spatial control of cytokinesis. J Cell Biol. 2014 doi: 10.1083/jcb.201311097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Deng L, Baldissard S, Kettenbach AN, Gerber SA, Moseley JB. Dueling kinases regulate cell size at division through the SAD kinase Cdr2. Curr Biol. 2014;24:428–433. doi: 10.1016/j.cub.2014.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pan KZ, Saunders TE, Flor-Parra I, Howard M, Chang F. Cortical regulation of cell size by a sizer cdr2p. eLife. 2014;3:e02040. doi: 10.7554/eLife.02040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wood E, Nurse P. Pom1 and cell size homeostasis in fission yeast. Cell Cycle. 2013;12 doi: 10.4161/cc.26462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Feierbach B, Verde F, Chang F. Regulation of a formin complex by the microtubule plus end protein tea1p. J Cell Biol. 2004;165:697–707. doi: 10.1083/jcb.200403090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mata J, Nurse P. tea1 and the microtubular cytoskeleton are important for generating global spatial order within the fission yeast cell. Cell. 1997;89:939–949. doi: 10.1016/s0092-8674(00)80279-2. [DOI] [PubMed] [Google Scholar]
- 21.Martin SG, McDonald WH, Yates JR, 3rd, Chang F. Tea4p links microtubule plus ends with the formin for3p in the establishment of cell polarity. Dev Cell. 2005;8:479–491. doi: 10.1016/j.devcel.2005.02.008. [DOI] [PubMed] [Google Scholar]
- 22.Tatebe H, Shimada K, Uzawa S, Morigasaki S, Shiozaki K. Wsh3/Tea4 is a novel cell-end factor essential for bipolar distribution of Tea1 and protects cell polarity under environmental stress in S. pombe. Curr Biol. 2005;15:1006–1015. doi: 10.1016/j.cub.2005.04.061. [DOI] [PubMed] [Google Scholar]
- 23.Alvarez-Tabares I, Grallert A, Ortiz JM, Hagan IM. Schizosaccharomyces pombe protein phosphatase 1 in mitosis, endocytosis and a partnership with Wsh3/Tea4 to control polarised growth. J Cell Sci. 2007;120:3589–3601. doi: 10.1242/jcs.007567. [DOI] [PubMed] [Google Scholar]
- 24.Kokkoris K, Castro DG, Martin SG. Tea4-phosphatase I landmark promotes local growth by dual Cdc42 GEF recruitment and GAP exclusion. J Cell Sci. 2014 doi: 10.1242/jcs.142174. [DOI] [PubMed] [Google Scholar]
- 25.Hachet O, Berthelot-Grosjean M, Kokkoris K, Vincenzetti V, Moosbrugger J, Martin SG. A phosphorylation cycle shapes gradients of the DYRK family kinase Pom1 at the plasma membrane. Cell. 2011;145:1116–1128. doi: 10.1016/j.cell.2011.05.014. [DOI] [PubMed] [Google Scholar]
- 26.Saunders TE, et al. Noise reduction in the intracellular pom1p gradient by a dynamic clustering mechanism. Dev Cell. 2012;22:558–572. doi: 10.1016/j.devcel.2012.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sawin KE, Tran PT. Cytoplasmic microtubule organization in fission yeast. Yeast. 2006;23:1001–1014. doi: 10.1002/yea.1404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Brunner D, Nurse P. CLIP170-like tip1p spatially organizes microtubular dynamics in fission yeast. Cell. 2000;102:695–704. doi: 10.1016/s0092-8674(00)00091-x. [DOI] [PubMed] [Google Scholar]
- 29.Busch KE, Brunner D. The microtubule plus end-tracking proteins mal3p and tip1p cooperate for cell-end targeting of interphase microtubules. Curr Biol. 2004;14:548–559. doi: 10.1016/j.cub.2004.03.029. [DOI] [PubMed] [Google Scholar]
- 30.Busch KE, Hayles J, Nurse P, Brunner D. Tea2p kinesin is involved in spatial microtubule organization by transporting tip1p on microtubules. Dev Cell. 2004;6:831–843. doi: 10.1016/j.devcel.2004.05.008. [DOI] [PubMed] [Google Scholar]
- 31.Bieling P, et al. Reconstitution of a microtubule plus-end tracking system in vitro. Nature. 2007;450:1100–1105. doi: 10.1038/nature06386. [DOI] [PubMed] [Google Scholar]
- 32.Al-Bassam J, Kim H, Flor-Parra I, Lal N, Velji H, Chang F. Fission yeast Alp14 is a dose-dependent plus end-tracking microtubule polymerase. Mol Biol Cell. 2012;23:2878–2890. doi: 10.1091/mbc.E12-03-0205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tischer C, Brunner D, Dogterom M. Force- and kinesin-8-dependent effects in the spatial regulation of fission yeast microtubule dynamics. Molecular systems biology. 2009;5:250. doi: 10.1038/msb.2009.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bratman SV, Chang F. Stabilization of overlapping microtubules by fission yeast CLASP. Dev Cell. 2007;13:812–827. doi: 10.1016/j.devcel.2007.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Loiodice I, Staub J, Setty TG, Nguyen NP, Paoletti A, Tran PT. Ase1p organizes antiparallel microtubule arrays during interphase and mitosis in fission yeast. Mol Biol Cell. 2005;16:1756–1768. doi: 10.1091/mbc.E04-10-0899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yamashita A, Sato M, Fujita A, Yamamoto M, Toda T. The roles of fission yeast ase1 in mitotic cell division, meiotic nuclear oscillation, and cytokinesis checkpoint signaling. Mol Biol Cell. 2005;16:1378–1395. doi: 10.1091/mbc.E04-10-0859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Al-Bassam J, Chang F. Regulation of microtubule dynamics by TOGdomain proteins XMAP215/Dis1 and CLASP. Trends Cell Biol. 2011;21:604–614. doi: 10.1016/j.tcb.2011.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Akhmanova A, et al. Clasps are CLIP-115 and -170 associating proteins involved in the regional regulation of microtubule dynamics in motile fibroblasts. Cell. 2001;104:923–935. doi: 10.1016/s0092-8674(01)00288-4. [DOI] [PubMed] [Google Scholar]
- 39.Al-Bassam J, Kim H, Brouhard G, van Oijen A, Harrison SC, Chang F. CLASP promotes microtubule rescue by recruiting tubulin dimers to the microtubule. Dev Cell. 2010;19:245–258. doi: 10.1016/j.devcel.2010.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.DeVoti J, Seydoux G, Beach D, McLeod M. Interaction between ran1+ protein kinase and cAMP dependent protein kinase as negative regulators of fission yeast meiosis. Embo J. 1991;10:3759–3768. doi: 10.1002/j.1460-2075.1991.tb04945.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shiozaki K. Nutrition-minded cell cycle. Science signaling. 2009;2:pe74. doi: 10.1126/scisignal.296pe74. [DOI] [PubMed] [Google Scholar]
- 42.Yanagida M, Ikai N, Shimanuki M, Sajiki K. Nutrient limitations alter cell division control and chromosome segregation through growth-related kinases and phosphatases. Philos Trans R Soc Lond B Biol Sci. 2011;366:3508–3520. doi: 10.1098/rstb.2011.0124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hartmuth S, Petersen J. Fission yeast Tor1 functions as part of TORC1 to control mitotic entry through the stress MAPK pathway following nutrient stress. J Cell Sci. 2009;122:1737–1746. doi: 10.1242/jcs.049387. [DOI] [PubMed] [Google Scholar]
- 44.Shiozaki K, Shiozaki M, Russell P. Heat stress activates fission yeast Spc1/StyI MAPK by a MEKK-independent mechanism. Mol Biol Cell. 1998;9:1339–1349. doi: 10.1091/mbc.9.6.1339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Byrne SM, Hoffman CS. Six git genes encode a glucose-induced adenylate cyclase activation pathway in the fission yeast Schizosaccharomyces pombe. J Cell Sci. 1993;105(Pt 4):1095–1100. doi: 10.1242/jcs.105.4.1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hoffman CS, Winston F. Glucose repression of transcription of the Schizosaccharomyces pombe fbp1 gene occurs by a cAMP signaling pathway. Genes Dev. 1991;5:561–571. doi: 10.1101/gad.5.4.561. [DOI] [PubMed] [Google Scholar]
- 47.Higuchi T, Watanabe Y, Yamamoto M. Protein kinase A regulates sexual development and gluconeogenesis through phosphorylation of the Zn finger transcriptional activator Rst2p in fission yeast. Mol Cell Biol. 2002;22:1–11. doi: 10.1128/MCB.22.1.1-11.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Matsuo Y, McInnis B, Marcus S. Regulation of the subcellular localization of cyclic AMP-dependent protein kinase in response to physiological stresses and sexual differentiation in the fission yeast Schizosaccharomyces pombe. Eukaryot Cell. 2008;7:1450–1459. doi: 10.1128/EC.00168-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gupta DR, Paul SK, Oowatari Y, Matsuo Y, Kawamukai M. Multistep regulation of protein kinase A in its localization, phosphorylation and binding with a regulatory subunit in fission yeast. Curr Genet. 2011;57:353–365. doi: 10.1007/s00294-011-0354-2. [DOI] [PubMed] [Google Scholar]
- 50.Bishop AC, Buzko O, Shokat KM. Magic bullets for protein kinases. Trends Cell Biol. 2001;11:167–172. doi: 10.1016/s0962-8924(01)01928-6. [DOI] [PubMed] [Google Scholar]
- 51.Bähler J, Nurse P. Fission yeast Pom1p kinase activity is cell cycle regulated and essential for cellular symmetry during growth and division. Embo J. 2001;20:1064–1073. doi: 10.1093/emboj/20.5.1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Browning H, Hayles J, Mata J, Aveline L, Nurse P, McIntosh JR. Tea2p is a kinesin-like protein required to generate polarized growth in fission yeast. J Cell Biol. 2000;151:15–28. doi: 10.1083/jcb.151.1.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Beinhauer JD, Hagan IM, Hegemann JH, Fleig U. Mal3, the fission yeast homologue of the human APC-interacting protein EB-1 is required for microtubule integrity and the maintenance of cell form. J Cell Biol. 1997;139:717–728. doi: 10.1083/jcb.139.3.717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chiron S, Bobkova A, Zhou H, Yaffe MP. CLASP regulates mitochondrial distribution in Schizosaccharomyces pombe. J Cell Biol. 2008;182:41–49. doi: 10.1083/jcb.200712147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Grallert A, et al. S. pombe CLASP needs dynein, not EB1 or CLIP170, to induce microtubule instability and slows polymerization rates at cell tips in a dynein-dependent manner. Genes Dev. 2006;20:2421–2436. doi: 10.1101/gad.381306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Fantes PA. Control of cell size and cycle time in Schizosaccharomyces pombe. J Cell Sci. 1977;24:51–67. doi: 10.1242/jcs.24.1.51. [DOI] [PubMed] [Google Scholar]
- 57.Navarro FJ, Nurse P. A systematic screen reveals new elements acting at the G2/M cell cycle control. Genome Biol. 2012;13:R36. doi: 10.1186/gb-2012-13-5-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Stettler S, Warbrick E, Prochnik S, Mackie S, Fantes P. The wis1 signal transduction pathway is required for expression of cAMP-repressed genes in fission yeast. J Cell Sci. 1996;109(Pt 7):1927–1935. doi: 10.1242/jcs.109.7.1927. [DOI] [PubMed] [Google Scholar]
- 59.Coulon A, Chow CC, Singer RH, Larson DR. Eukaryotic transcriptional dynamics: from single molecules to cell populations. Nature reviews Genetics. 2013;14:572–584. doi: 10.1038/nrg3484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ambrose C, Allard JF, Cytrynbaum EN, Wasteneys GO. A CLASP-modulated cell edge barrier mechanism drives cell-wide cortical microtubule organization in Arabidopsis. Nat Commun. 2011;2:430. doi: 10.1038/ncomms1444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Aoh QL, Hung CW, Duncan MC. Energy metabolism regulates clathrin adaptors at the trans-Golgi network and endosomes. Mol Biol Cell. 2013;24:832–847. doi: 10.1091/mbc.E12-10-0750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bähler J, et al. Heterologous modules for efficient and versatile PCR-based gene targeting in Schizosaccharomyces pombe. Yeast. 1998;14:943–951. doi: 10.1002/(SICI)1097-0061(199807)14:10<943::AID-YEA292>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- 63.Bishop AC, et al. A chemical switch for inhibitor-sensitive alleles of any protein kinase. Nature. 2000;407:395–401. doi: 10.1038/35030148. [DOI] [PubMed] [Google Scholar]
- 64.Bendezu FO, Martin SG. Cdc42 Explores the Cell Periphery for Mate Selection in Fission Yeast. Curr Biol. 2013;23:42–47. doi: 10.1016/j.cub.2012.10.042. [DOI] [PubMed] [Google Scholar]
- 65.Martin SG, Chang F. Dynamics of the formin for3p in actin cable assembly. Curr Biol. 2006;16:1161–1170. doi: 10.1016/j.cub.2006.04.040. [DOI] [PubMed] [Google Scholar]
- 66.Fu C, Jain D, Costa J, Velve-Casquillas G, Tran PT. mmb1p binds mitochondria to dynamic microtubules. Curr Biol. 2011;21:1431–1439. doi: 10.1016/j.cub.2011.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Walker RA, et al. Dynamic instability of individual microtubules analyzed by video light microscopy: rate constants and transition frequencies. J Cell Biol. 1988;107:1437–1448. doi: 10.1083/jcb.107.4.1437. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.