Abstract
Tumour cells can use strategies that make them resistant to nutrient deprivation to outcompete their neighbours. A key integrator of the cell’s responses to starvation and other stresses is amino acid-dependent mechanistic Target of Rapamycin Complex 1 (mTORC1). Activation of mTORC1 on late endosomes and lysosomes is facilitated by amino acid transporters within the Solute-Linked Carrier (SLC)36 and SLC38 families. Here we analyse the functions of SLC36 family member, SLC36A4, otherwise known as Proton-assisted Amino Acid Transporter 4 (PAT4), in colorectal cancer. We show that independent of other major pathological factors, high PAT4 expression is associated with reduced relapse-free survival after colorectal cancer surgery. Consistent with this, PAT4 promotes HCT116 human colorectal cancer cell proliferation in culture and tumour growth in xenograft models. Inducible knockdown in HCT116 cells reveals that PAT4 regulates a form of mTORC1 with two distinct properties: firstly, it preferentially targets eukaryotic translation initiation factor 4E-binding protein 1 (4E-BP1) and secondly, it is resistant to rapamycin treatment. Furthermore, in HCT116 cells two non-essential amino acids, glutamine and serine, which are often rapidly metabolised by tumour cells, regulate rapamycin-resistant mTORC1 in a PAT4-dependent manner. Overexpressed PAT4 is also able to promote rapamycin resistance in HEK-293 human embryonic kidney cells. PAT4 is predominantly associated with the Golgi apparatus in a range of cell types and in situ proximity ligation analysis shows PAT4 interacts with both mTORC1 and its regulator Rab1A on the Golgi. These findings, together with other studies, suggest differentially localised intracellular amino acid transporters contribute to activation of alternate forms of mTORC1. Furthermore, our data predict that colorectal cancer cells with high PAT4 expression will be more resistant to depletion of serine and glutamine, allowing them to survive and outgrow neighbouring normal and tumorigenic cells, and potentially providing a new route for pharmacological intervention.
Keywords: SLC36A4, mTOR, proton-assisted amino acid transporter, Rab1A, glutamine/serine, transceptor
INTRODUCTION
During cancer growth, tumour cell adaptation is driven by adverse microenvironmental conditions, such as hypoxia and starvation (1). Mechanistic Target of Rapamycin Complex 1 (mTORC1) responds to both local nutrient status and growth factor signalling through phosphatidylinositol 3-kinase (PI3K) to regulate protein synthesis and cellular homeostasis, thereby modulating cancer cell growth, metabolism and metastasis (2-4). However, attempts to block tumour growth using the allosteric mTOR inhibitor rapamycin or its analogues have met with limited success (5). While these drugs strongly reduce signalling to one of the two well-characterised mTORC1 targets, ribosomal protein p70-S6 kinase 1 (S6K1), they often have more limited effects on the other, eukaryotic translation initiation factor 4E-binding protein 1 (4E-BP1), a negative regulator of eukaryotic initiation factor 4E (eIF4E) (6, 7) implicated in metastatic growth (8, 9). This resistance can sometimes be circumvented by using adenosine triphosphate (ATP)-competitive mTOR inhibitors (5-7), which also block the other mTOR kinase-containing complex, mTORC2. Nonetheless, how changes in mTOR structure (10) or mTOR regulators modulate rapamycin sensitivity remains of considerable interest.
Members of the Proton-assisted Amino acid Transporter (PAT) or SLC36 family (11) were identified as positive regulators of growth and mTORC1 signalling through an in vivo genetic overexpression screen in flies (12, 13). These effects were shown to be conserved by characterisation of the two ubiquitously transcribed human PATs, PAT1 (SLC36A1) and PAT4 (SLC36A4) (14). The prototypic PAT family member, PAT1, is a lysosomal amino acid transporter [AAT] (15, 16). In rapidly growing cells, it is located at the surface of nutrient-rich late endosomal and lysosomal (LEL) compartments (13), where mTOR accumulates in response to amino acid stimulation. The recruitment of mTOR requires assembly of a multi-protein complex, which includes Raptor, a heterodimeric pair of Ras-related Rag GTPases, the pentameric Ragulator, and the Vacuolar-H+-ATPase (V-ATPase) proton pump at the compartment surface, reviewed in (2, 3). PAT1 also interacts with this complex to promote mTOR localisation on LELs and subsequent mTORC1 signalling. Amino acid sensing by the PATs may involve transport or signalling via a so-called ‘transceptor’ mechanism (4, 13, 17).
Recent studies have identified an AAT in the related SLC38 family, SLC38A9, which also interacts on LELs with the mTORC1-regulatory machinery, potentially in response to arginine (18, 19), suggesting that different LEL-located, mTORC1-regulatory AATs may sense different amino acids. Furthermore, the identification of molecules like Golgi-localised Rab1A (20), and ADP ribosylation factor Arf1 (21) and phospholipase D (22, 23) as regulators of Rag-independent, mTORC1 activation, suggests that other amino acid sensing mechanisms remain to be discovered.
Here we investigate PAT4 function in colorectal cancer. Colorectal cancers are frequently rapamycin-resistant (6) and often metastatic, seriously impacting on clinical outcome (24, 25). We show that PAT4 up-regulation is associated with cancer progression. By employing an inducible PAT4 shRNA knockdown in HCT116 colorectal cancer cells, we find that PAT4 responds to two rapidly metabolised, non-essential amino acids, glutamine and serine (26, 27), to drive rapamycin-resistant, mTORC1-mediated cell proliferation. Furthermore, we provide evidence that PAT4 interacts with Rab1A and mTORC1 on the Golgi, suggesting a role in amino acid sensing from this compartment.
RESULTS
Validation of a novel PAT4 monoclonal antibody
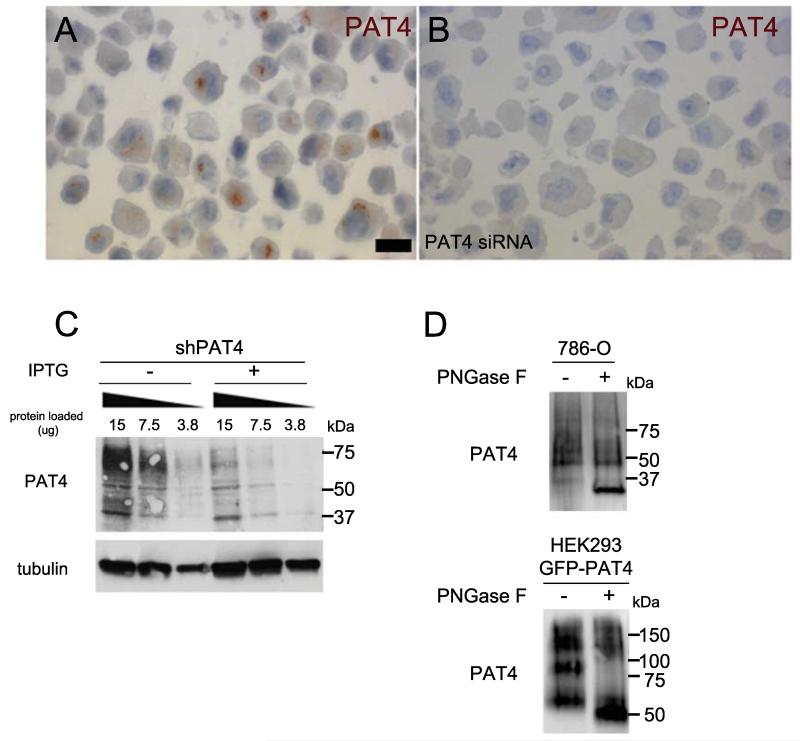
We generated a highly specific mouse monoclonal antibody against PAT4 (antibody Pat4/9/H10). Staining with this antibody revealed that PAT4 was localised to an asymmetric perinuclear region in formalin-fixed, paraffin-embedded 786-O renal cancer cells, which express high PAT4 levels, and lost in 786-O cells transfected with PAT4 siRNA (Fig. 1A and B). Bands of 60-75 kDa molecular weight were observed on western blots of cell lysates (Fig. 1C) and strongly reduced after PAT4 knockdown, confirming antibody specificity. This smear resolved into a band of approximately 30 kDa after pre-treatment of lysates with the glycosidase, Peptide-N-Glycosidase F (PNGase F), smaller than the predicted 55 kDa molecular weight (Fig. 1D).
Figure 1.
Validation of an ‘in house’-generated anti-PAT4 monoclonal antibody. A and B, formalin-fixed, paraffin-embedded 786-O cells incubated with PAT4 monoclonal antibody (Pat4/9/H10) and visualised with 3,3′-Diaminobenzidine (DAB). There is an obvious perinuclear region of staining visible in most cells treated with the scrambled control siRNA (A). This staining is absent when the PAT4 transcript is knocked down using an siRNA against PAT4 (si435 [14]; B). C, western blot analysis of serial dilutions of cell lysates (15, 7.5 and 3.8 μg of protein) produced from pools of 786-O cells carrying an IPTG-inducible shPAT4 (43587; shPAT4) probed with PAT4 monoclonal antibody Pat/9/H10. This revealed a set of bands from 60-75 kDa that was strongly reduced by IPTG-induced PAT4 knockdown (+IPTG), suggesting that they are PAT4-specific. Western blots were also probed with an anti-tubulin antibody as a loading control. D, western blot of cell lysates from 786-O cells and from a GFP-PAT4-overexpressing HEK-293 cell line treated with PNGase F prior to electrophoresis to remove glycosyl groups. This resolved the cross-reacting molecules seen in untreated cell lysates into more specific bands migrating at approximately 30 and 50 kDa respectively, smaller than the predicted molecular weights of 55 kDa (PAT4) and 85 kDa (GFP-PAT4), a phenomenon also reported for other transmembrane proteins (43).
High PAT4 expression is associated with poor outcome in colorectal cancer patients
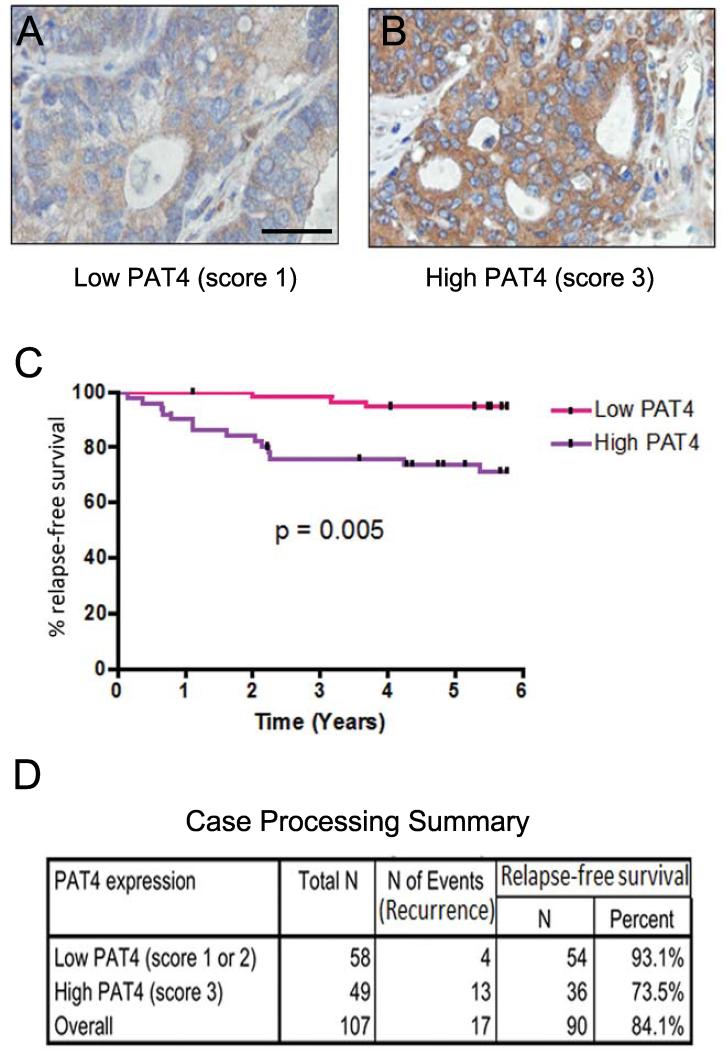
To test whether PAT4 expression is altered in human colorectal cancer, we stained primary tumour tissue microarrays from 107 patients, who had been treated by surgical resection only. The intensity of cytoplasmic staining was scored by a pathologist (CS) into three categories (Fig. 2A and B; see Materials and Methods), all of which were higher than normal colorectal epithelium. Statistical analysis showed no association between high PAT4 expression and standard clinical or pathological variables, including site of tumour, tumour stage, nodal or distal metastases, age, lymphatic, vascular or neural invasion, differentiation or gender (Supplementary Table S1).
Figure 2.
High expression of PAT4 predicts poorer relapse-free survival in colon cancer. 107 patients with primary colonic carcinoma were assessed for expression of PAT4 in their tumours and stratified according to PAT4 expression. A, B representative images of low and high PAT4 expression levels as determined by immunohistochemistry. Brown staining with diamino-benzidine indicates immunoreactivity. C, Kaplan-Meier curves compare high versus low levels of expression. P-value is the result of a log-rank test (Mantel-Cox). D, case processing summary for high and low PAT4-expressing patients. The scale bar in A is 50 μm.
In univariate analyses, high PAT4 levels (P=0.01) as well as high tumour stage (P<0.01), tumour stage score (P<0.01, the presence of bowel perforation (P=0.02), neural invasion (P<0.01), nodal (P<0.01) and synchronous metastasis (P<0.01) significantly correlated with shorter relapse-free survival (Supplementary Table S2). Patients with cancers that had higher PAT4 expression had a significantly shorter mean relapse-free survival than those with lower levels (P<0.01; Fig. 2C and D). Additionally, higher PAT4 levels showed statistical significance in multivariate survival analysis (P<0.01; Supplementary Table S3). The multivariate model included all variables significantly associated with relapse in univariate analysis, apart from overall stage, since this is calculated from tumour stage (T), nodal metastases (N) and distant metastases (M) stages. We conclude that increased PAT4 levels are associated with worse prognosis in patients with colorectal cancer.
PAT4 regulates HCT116 cell proliferation
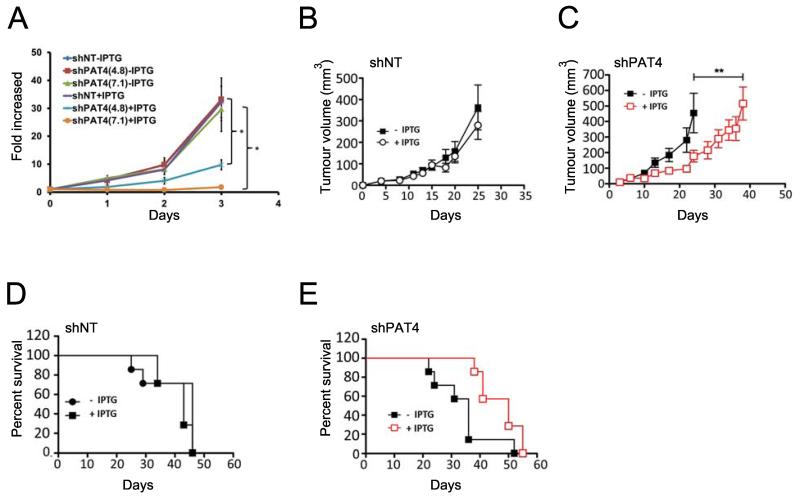
To analyse PAT4 function in HCT116 colorectal cancer cells, we generated stably transduced cell lines, each carrying one of three different lentiviral constructs expressing a PAT4 short hairpin (sh)RNA under isopropyl β-D-1-thiogalactopyranoside (IPTG)-inducible control. For each construct, pooled cells representing many individual transduction events had reduced PAT4 transcript levels, as determined by quantitative real-time PCR (qRT-PCR; Supplementary Fig. S1A). Two shRNAs, 49384 and 49387, were selected for further study. To induce a uniformly strong PAT4 knockdown, single cell clones were isolated from another shRNA transduction, and named shPAT4(4.8) and shPAT4(7.1) respectively (Supplementary Fig. S1B). In vitro culture of these clones together with HCT116 cells containing an IPTG-inducible, non-targeting shRNA gene (shNT) revealed that IPTG induction specifically inhibited proliferation of shPAT4-expressing cells (P<0.05; Fig. 3A) with no significant effect on cell death (Supplementary Table S4).
Figure 3.
PAT4 regulates the growth of HCT116 cells in vitro and in vivo. A, proliferation of clones of HCT116 cells stably transduced with one of two independent IPTG-inducible shRNA constructs targeting PAT4, namely shPAT4(4.8) and shPAT4(7.1), or the IPTG-inducible non-targeting control construct (shNT) was measured in the presence and absence of IPTG (n=3). B and C, mean growth curves (± SEM) of human HCT116 tumour xenografts in immunodeficient mice carrying pools of cells transduced with shNT (B) with (empty circles) or without (filled squares) IPTG induction, or shPAT4-transduced HCT116 cells (C) with (empty squares, outlined in red) or without (filled squares) IPTG induction (n=7). Data in (B, C) were analysed by unpaired two-tailed independent Student’s t-test. D and E, Kaplan-Meier survival curves of seven animals with and without IPTG induction of shNT- (D) and shPAT4-containing (E) HCT116 tumours; Log-rank (Mantel-Cox) Test: P=0.019 for (E). For shPAT4-inducible cells, all but one of the seven non-induced mice (filled squares) needed to be sacrificed within 36 days (median survival time of 36 days), while all seven induced mice (empty squares, outlined in red) were sacrificed from 38 days onwards with a median survival time of 50 days. Cell proliferation experiment was repeated three times. *P<0.05.
PAT4 promotes human tumour growth in xenograft models
To assess the role of PAT4 in tumour growth, pooled clones of HCT116 cells carrying shPAT4(49387), were employed in xenograft experiments. We reasoned that the variable level of PAT4 knockdown in these cells might better model changes taking place in heterogeneous tumours expressing different levels of PAT4. The effect on tumour growth of IPTG-induced PAT4 knockdown in shPAT4 HCT116 cells was assessed in mice over a 60 day period. Immunodeficient mice were provided with IPTG in their drinking water to induce shRNA expression. IPTG did not alter the size of tumours formed from shNT cells (Fig. 3B). However, IPTG-induced shPAT4 expression reduced tumour growth significantly compared to non-induced shPAT4 controls (Fig. 3C; P<0.01). In addition, shPAT4 induction extended median survival time of mice from 36 to 50 days (Ratio: 0.72, 95% CI of ratio: 0.36 to 1.07; Gehan-Breslow-Wilcoxon Test, P=0.008; Fig. 3D and E). Induction of PAT4 knockdown in a second experiment also significantly reduced mean tumour volume (P<0.05) and improved animal survival (P<0.05), demonstrating that PAT4 promotes HCT116 tumour growth in vivo.
PAT4 regulates a rapamycin-resistant form of mTORC1
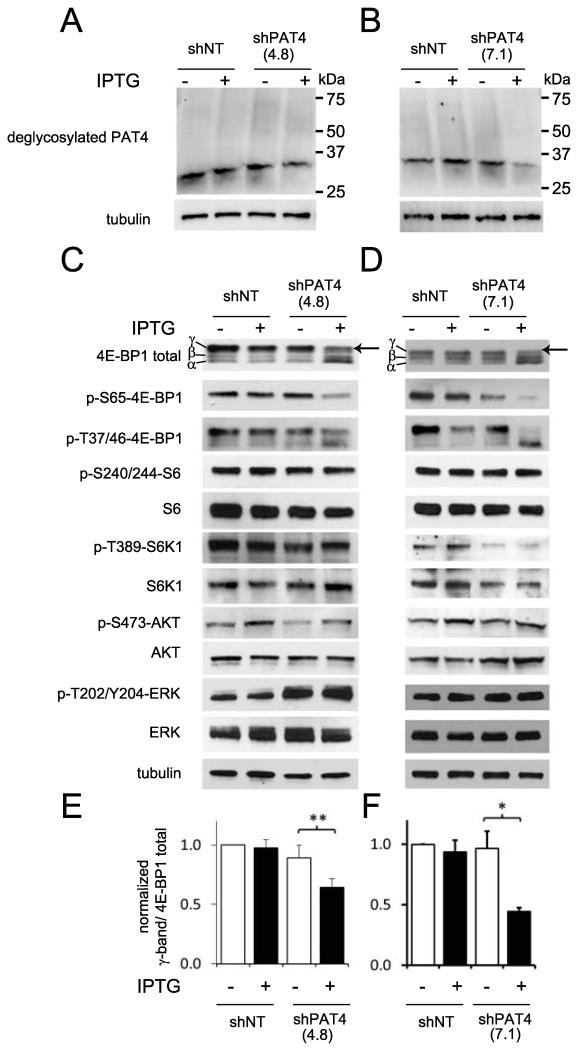
To determine how PAT4 knockdown might inhibit tumour growth, we analysed mTORC1 signalling in stably transfected HCT116 clones carrying inducible-PAT4 shRNAs, shPAT4(4.8) and shPAT4(7.1), and in the non-targeting clone, shNT. PAT4 protein is expressed at much lower levels in HCT116 cells compared to 786-O cells, so lysates from large pools of HCT116 cells were pre-treated with PNGase F to detect PAT4, which resolved into a specific 30 kDa band that was clearly reduced after IPTG addition in knockdown cells (Fig. 4A and B).
Figure 4.
PAT4 selectively affects the mTORC1 target 4E-BP1. Western blots of protein extracts from HCT116 cells carrying either IPTG-inducible shNT or shPAT4(4.8) (A,C) or shNT and shPAT4(7.1) constructs (B,D) cultured in the presence and absence of IPTG. A and B, PAT4 knockdown reduced the level of PAT4 protein detected following incubation of cell lysates with PNGase F prior to electrophoresis. C-F, PAT4 knockdown significantly reduced the level of the most phosphorylated γ-form of 4E-BP1, visualised both with an anti-phospho-Ser65-4E-BP1 (p-S65-4E-BP1) antibody and also as the upper band with a pan-4E-BP1 antibody (arrow; quantified in three independent experiments as a proportion of total 4E-BP1 staining in histograms E and F respectively). Phospho-S6K (p-T389-S6K1), phospho-S6 (p-S240/244-S6), phospho-Akt (p-S473-AKT) and phospho-ERK (p-T202/Y204-ERK) levels were essentially unaffected by PAT4 knockdown. Blots were probed with an anti-tubulin antibody as a loading control. Effects were reproduced in three separate experiments. *P<0.05; **P<0.01.
IPTG-induced PAT4 knockdown selectively reduced the most highly phosphorylated form of 4E-BP1, designated the γ-band on western blots (28), while levels of less phosphorylated forms increased (Fig. 4C-F). Human 4E-BP1 has at least eight phosphorylation sites (28). Phosphorylation of 4E-BP1 at Ser65, a key residue for eIF4E binding (29), is typically required to form the γ-band. An anti-phospho-Ser65-4E-BP1 antibody verified that phosphorylation of this residue (p-S65-4E-BP1) was strongly decreased by PAT4 knockdown. In contrast, overall phosphorylation of p-T37/46-4E-BP1 was maintained, but distributed between multiple 4E-BP1 bands after knockdown. PAT4 knockdown either had no effect on S6K1 (p-T389-S6K1) and S6 (p-S240/244-S6) phosphorylation (Fig. 4C and D), or sometimes led to a modest reduction in p-S240/244-S6 (eg., Fig. 5A and Supplementary Fig. S2C).
Figure 5.
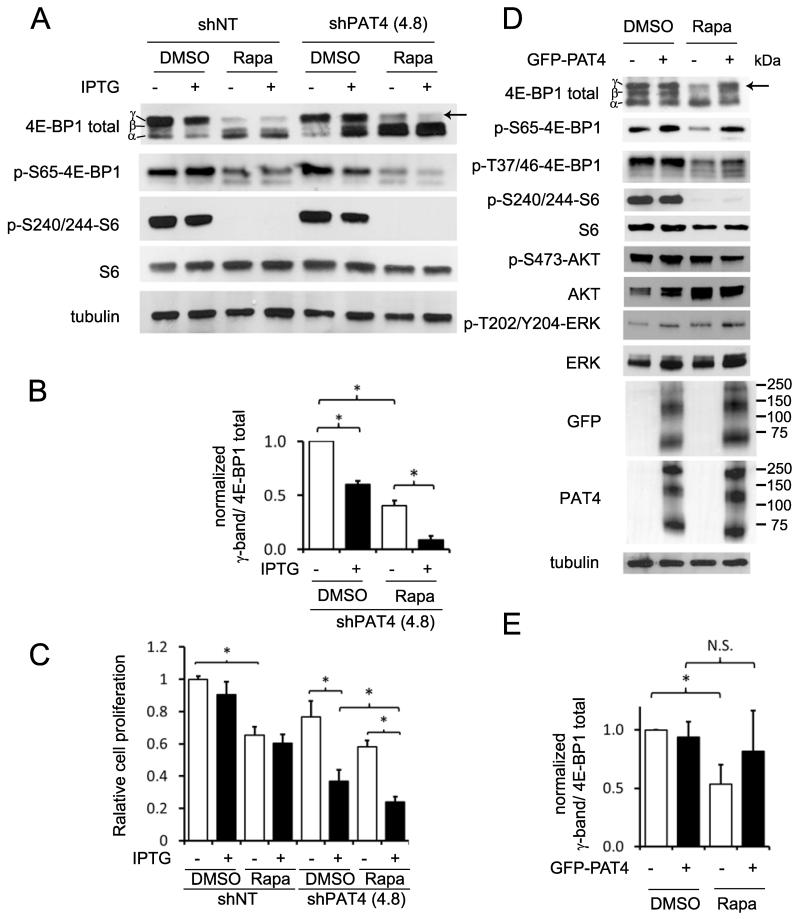
PAT4 regulates a rapamycin-resistant form of mTORC1 in HCT116 cells and increases rapamycin resistance in HEK-293 cells. A and B, clones of HCT116 cells carrying the IPTG-inducible PAT4 shRNA construct, shPAT4(4.8), and the IPTG-inducible non-targeting control construct, shNT, were cultured for five days in the absence or presence of IPTG and, if required, treated with rapamycin for the last 24 hours. Rapamycin (3 nM; see Supplementary Fig. S2A and S2B) strongly reduces phospho-S6 levels (p-S240/244-S6), and partially affects the Ser65-phosphorylated 4E-BP1 (p-S65-4E-BP1) γ-band (arrow). This rapamycin-resistant phospho-4E-BP1 γ–band is almost completely lost after PAT4 knockdown (B). C, shPAT4(4.8) cells and shNT controls were cultured for eight days in the absence or presence of IPTG and treated with rapamycin for three days. Cells were then counted revealing reduced proliferation for both cell lines in the presence of rapamycin, and also for shPAT4(4.8) in the presence of IPTG (n=3). The combination of both rapamycin and IPTG leads to further reduction in the cell number of shPAT4(4.8). D and E, normal HEK-293 cells or cells stably transfected with a constitutively expressed GFP-PAT4 construct were cultured for 24 hours in the presence or absence of 100 nM rapamycin (see Supplementary Fig. S2F), and then cell lysates analysed by western analysis. GFP-PAT4 expression reduces the effect of rapamycin on the γ-phosphorylated 4E-BP1 band (E), but not phospho-S6 (p-S240/244-S6), phospho-Akt (p-S473-AKT) or phospho-ERK (p-T202/Y204-ERK). All blots were probed with an anti-tubulin antibody as a loading control. (*P<0.05; n=3). The cell proliferation experiment was repeated three times.
To confirm that PAT4 knockdown was not altering 4E-BP1 phosphorylation by inhibiting upstream PI3K/Akt signalling, levels of phosphorylated mTORC2-regulated Akt (p-S473-AKT) were assessed; no change was observed (Fig. 4C and D). Phosphorylation of ERK MAPK (p-T202/Y204-ERK) by oncogenic forms of KRAS has also been associated with resistance to mTOR kinase inhibitors in colorectal cancer (30). HCT116 cells carry an oncogenic KRAS-G13D allele, but PAT4 knockdown did not reduce ERK phosphorylation (Fig. 4C and D), suggesting that it does not act through ERK to regulate mTORC1.
The selective action of PAT4 knockdown on 4E-BP1 phosphorylation versus the S6K/S6 signalling arm of the mTORC1 pathway is complementary to the effects reported with rapamycin in these cells (6), where 4E-BP1 γ-phosphorylation is particularly resistant to this drug. Even at rapamycin concentrations that essentially blocked S6 phosphorylation, residual 4E-BP1 γ-phosphorylation was observed, as well as lower molecular weight 4E-BP1 phosphorylated on Ser65 (Fig. 5A, Supplementary Fig. S2A and B). However, when rapamycin and PAT4 knockdown were combined, the γ-form of phosphorylated 4E-BP1 and remaining p-S65-4E-BP1 bands were almost completely lost (Fig. 5A and B and Supplementary Fig. S2C and S2D). Unlike rapamycin, an ATP-kinase inhibitor of mTOR, PP242, strongly affected both 4E-BP1 γ-phosphorylation and S6K activity in HCT116 cells (Supplementary Fig. S2E). Together, these findings suggest that in HCT116 cells essentially all 4E-BP1 γ-phosphorylation is mTORC1-dependent (see also ref. 30), but that some phosphorylation is rapamycin-resistant and regulated by PAT4. Importantly, rapamycin and PAT4 knockdown also had additive effects on HCT116 cell proliferation (Fig. 5C), demonstrating they inhibit different mTORC1 complexes, which both play a role in cell proliferation.
To test the link between PAT4 expression and rapamycin resistance, we overexpressed a GFP-tagged form of PAT4 in HEK-293 cells. We have previously shown in these cells that siRNA-induced knockdown of PAT4 reduces both 4E-BP1 and S6K/S6 phosphorylation, and this was confirmed using shRNA knockdown (Supplementary Fig. S3). We observed that 4E-BP1 hyperphosphorylation shows some rapamycin-resistance in HEK-293 cells (Supplementary Fig. S2F). Strong overexpression of GFP-PAT4, which produced several protein bands resolving to a 50 kDa band on western blots after PNGase pre-treatment (Fig. 1D), increased rapamycin resistance of hyperphosphorylated, p-Ser65-4E-BP1 (Fig. 5D and E). This suggests that high PAT4 levels can induce resistance to rapamycin in multiple different cell types.
PAT4–dependent sensitivity of rapamycin-resistant mTORC1 to glutamine and serine
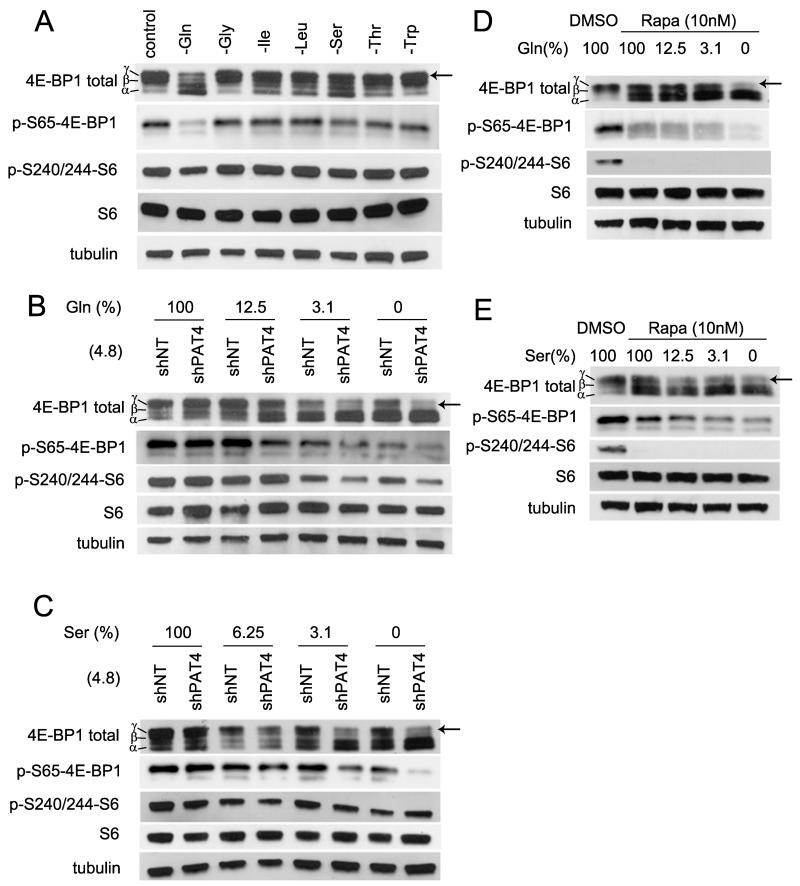
Since PATs are implicated in amino acid-dependent mTORC1 activation, we hypothesised that PAT4 might sense levels of specific amino acids that regulate rapamycin-resistant mTORC1. We starved HCT116 cells of specific amino acids, including two non-essential amino acids required for HCT116 growth. Non-essential serine is diverted into glycolysis, while glutamine fuels the tricarboxylic acid (TCA) cycle via glutaminolysis in cancer cells, including HCT116 cells (26, 27, 31). Reducing either amino acid, particularly glutamine, over four hours had a stronger inhibitory effect on 4E-BP1 hyperphosphorylation than loss of any essential amino acid (Fig. 6A).
Figure 6.
Glutamine and serine selectively regulate PAT4-dependent rapamycin-resistant mTORC1 signalling in HCT116 cells. A, HCT116 cells were starved of the essential amino acids, glycine (Gly), isoleucine (Ile), leucine (Leu), threonine (Thr) and tryptophan (Trp), and the non-essential amino acids, serine (Ser) and glutamine (Gln), for four hours in separate cultures, and then proteins extracted and subjected to western blotting. After this time, glutamine depletion produces the greatest reduction in γ-phosphorylation of 4E-BP1 (arrow); serine depletion also induces a more modest shift. B, clones of HCT116 cells carrying the IPTG-inducible PAT4 shRNA constructs, shPAT4(4.8), and the IPTG-inducible non-targeting control construct, shNT, were exposed to culture medium containing different concentrations of glutamine for four hours in the absence of IPTG. Under these conditions, shPAT4(4.8) cells express about 50% of normal PAT4 mRNA levels (Supplementary Fig. S1B) due to leaky PAT4 shRNA transcription. As glutamine concentration falls, a greater reduction in γ-phosphorylation of 4E-BP1 is apparent in shPAT4(4.8) cells. S6 phosphorylation also seems to be affected at low glutamine concentrations. C, same experiment as (B), except different levels of serine in the culture medium. Again, shPAT4(4.8) cells have lower 4E-BP1 γ-phosphorylation at reduced concentrations of serine. D and E, rapamycin-treated HCT116 cells subjected to different levels of glutamine (D) and serine starvation (E), over four hours, show a greater reduction in γ-phosphorylation of 4E-BP1.
If some of the selective effects of glutamine and serine on 4E-BP1 are mediated via a PAT4-dependent sensing mechanism, we reasoned modest changes in PAT4 levels might alter the sensitivity of HCT116 cells to glutamine and serine starvation. Our PAT4 knockdown clones displayed leaky IPTG-independent shRNA knockdown (Supplementary Fig. S1B), which normally did not affect 4E-BP1 phosphorylation (Fig. 5A) or proliferation (Fig. 3A). We tested the amino acid sensitivity of these cells in the absence of IPTG. 4E-BP1 hyperphosphorylation was more sensitive to reduction in glutamine and serine levels in both shPAT4(4.8) (Fig. 6B and C) and shPAT4(7.1) (Supplementary Fig. S4) clones, indicating that PAT4 is involved in the sensing of these metabolically important amino acids. Interestingly, S6 phosphorylation also appeared to be more sensitive to reduced glutamine and serine in these experiments, suggesting either that rapamycin-resistant mTORC1 can directly or indirectly modulate S6K activity under certain conditions or that starvation affects the specificity of PAT4 for rapamycin-resistant mTORC1. Combining glutamine or serine starvation with rapamycin treatment had a greater effect on 4E-BP1 hyperphosphorylation than either treatment alone (Fig. 6D and 6E), supporting our conclusion that these amino acids are sensed by PAT4-regulated, rapamycin-resistant mTORC1.
Golgi-localised PAT4 interacts with mTORC1
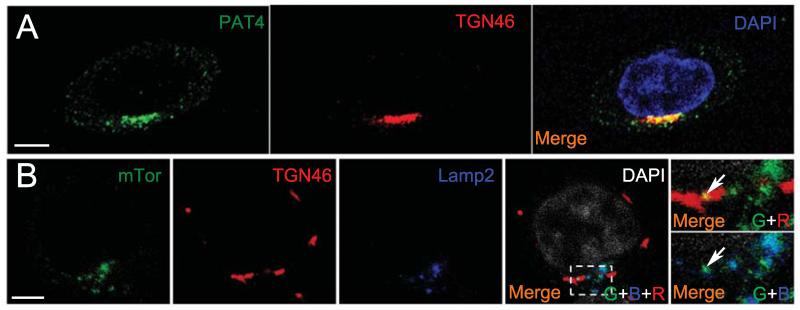
Immunofluorescence staining of HCT116 cells revealed that PAT4 is concentrated on and adjacent to the trans-Golgi network (Fig. 7A). A similar localisation was observed in 786-O cells (Supplementary Fig. S5A). A previous report also suggested that overexpressed PAT4 in HEK-293T cells is not on LELs (32). Interestingly, although under normal culture conditions, most mTOR is localised around the LELs, some colocalisation with the trans-Golgi network was also observed (Fig. 7B), raising the possibility that it might be associated with PAT4 in this compartment.
Figure 7.
PAT4 and mTOR are located on the Golgi in HCT116 cells. A, Endogenous PAT4 (green) is expressed in compartments that include the trans-Golgi network (TGN46; red) in HCT116 cells. B, Although most mTOR (green) is located on LAMP2-positive late endosomes and lysosomes (blue) in HCT116 cells, some mTOR staining also overlaps with the trans-Golgi network (red; indicated by arrows in merged images). DAPI marks the nucleus in A (blue) and B (white). Scale bars are 5 μm. Confocal channels are indicated as follows in the merged images: green (G), blue (B) and red (R).
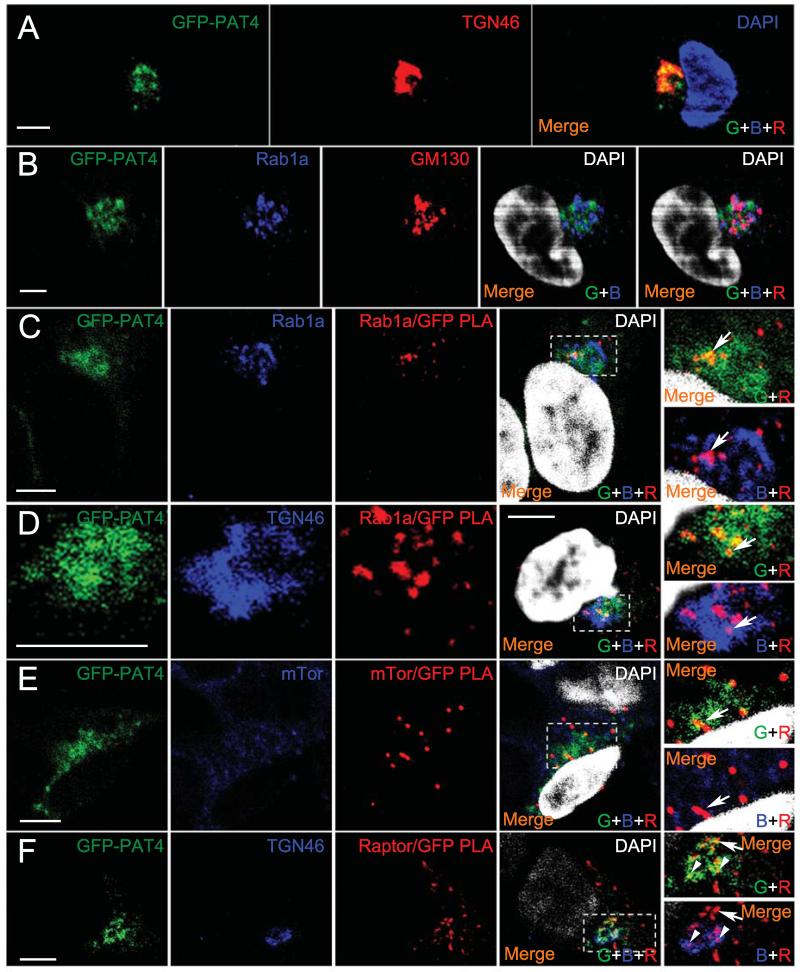
To investigate this further, we employed rapamycin-resistant HEK-293 cells expressing GFP-tagged PAT4 (Fig. 5D). GFP-PAT4 also localised on and around the trans-Golgi network (Fig 8A). Golgi-localised Rab1A, a monomeric GTPase involved in membrane trafficking events, has recently been implicated in amino acid-dependent activation of mTORC1 from the Golgi (20). We tested whether Rab1A might interact with PAT4. We used the proximity ligation assay (PLA), which detects specific protein-protein interactions in situ, when antibodies recognising these molecules are in close proximity (33). Although anti-Rab1A staining primarily localised to the cis-Golgi (Fig. 8B), Rab1A and GFP antibodies produced a PLA signal on an adjacent compartment (Fig. 8C), which was not present in cells that did not express GFP-PAT4 (Supplementary Fig, S5B). This signal frequently overlapped with GFP-PAT4 and was partly within the trans-Golgi network (Fig. 8D), suggesting that Rab1A and PAT4 can interact on the Golgi. In addition, PLA using either anti-mTOR (Fig. 8E) or anti-Raptor (Fig. 8F and Supplementary Fig. S5C) antibodies with anti-GFP also produced a specific signal primarily in GFP-PAT4-containing compartments that included the trans-Golgi network. This indicates that mTORC1 interacts with PAT4 on the Golgi, consistent with the idea that PAT4 can regulate mTORC1 activity from this compartment.
Figure 8.
PAT4 interacts with Rab1A and mTORC1 on the Golgi. A, GFP-PAT4 fusion protein (green) in HEK-293 cells shows a similar subcellular localisation to HCT116 cells on the trans-Golgi (TGN46; red). An alternative cell at higher resolution is stained with the same markers in Fig. 8D. B, Rab1A (blue) is localised primarily on the cis-Golgi, labelled by the GM130 marker (red). C, in situ proximity ligation assay (PLA; red) reveals interaction between Rab1A (blue) and GFP-PAT4 (green) primarily associated with GFP-PAT4-containing compartments (overlap between GFP-PAT4 and PLA signal is yellow in merge, top right panel; arrow). The PLA signal is also adjacent and partially overlapping with compartments on which Rab1A is concentrated in merge of Rab1A and PLA, bottom right panel. Rab1A/PAT4 PLA signals are only observed in GFP-PAT4-expressing cells (see Supplementary Fig. S5B). D, some of the GFP-PAT4/Rab1a PLA-positive interacting compartments (red) also appear to be partly or entirely labelled by trans-Golgi network marker TGN46 (blue; arrow), but not trans-Golgi regions lacking GFP-PAT4. E, PLA (red) reveals an interaction between mTOR (blue) and GFP-PAT4 (green) partially overlapping with GFP-PAT4-containing compartments (yellow in merges containing green and red channels, including magnified image in top right panel; arrow). In the low magnification merge image, the two upper cells not expressing GFP-PAT4 do not give a PLA signal, indicating that this assay specifically detects the GFP-PAT4/mTOR interaction. Blue and red channel merge (lower right-hand panel) reveals that mTOR staining is often in close proximity to PLA signal, but is also found in many other locations within the cell. F, PLA (red) reveals an interaction (red) between Raptor and GFP-PAT4 (green), in and adjacent to compartments containing GFP-PAT4 (yellow overlap in merges containing green and red channels, including magnified image in top right panel; arrow and arrowheads). Blue and red channel merge (lower right-hand panel) reveals that some PLA signals are adjacent or partially colocalise with the trans-Golgi (TGN, blue; arrowheads), while others do not (arrow). Raptor/GFP-PAT4 PLA signals are only observed in GFP-PAT4-expressing cells (see Supplementary Fig. S5C). DAPI marks the nucleus in A (blue) and B-F (white) in the merge. Confocal channels are indicated as follows in the merged images: green (G), blue (B) and red (R). Scale bars are 5 μm.
DISCUSSION
Although resistance of mTORC1 to inhibitors can be partly explained by differential in vitro sensitivity of substrate target sites (34), increasing evidence indicates there are also different mTORC1 complexes in cancer cells (20-23, 35), which may make them differentially sensitive to drugs like rapamycin. In this study, we demonstrate that PAT4 regulates rapamycin-resistant mTORC1 in HCT116 cells and can induce increased rapamycin resistance when overexpressed in HEK-293 cells. PAT4 and rapamycin-resistant mTORC1 are essential for normal cell proliferation in vitro. Furthermore, PAT4 expression levels are predictive of early relapse in colorectal cancer, suggesting a pathophysiological role in the acquisition of more aggressive tumour phenotypes.
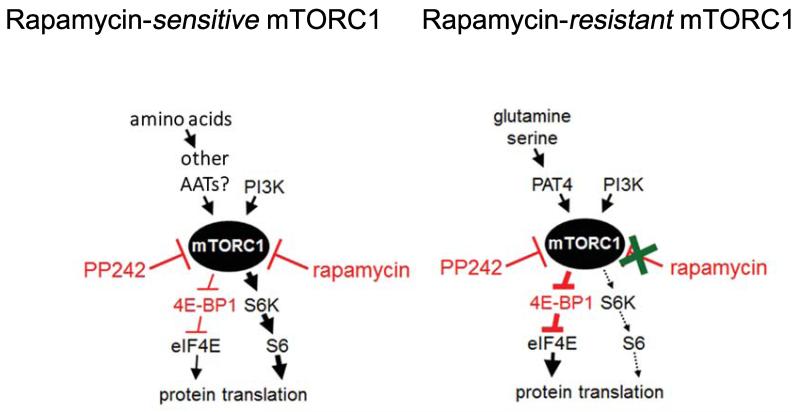
Our findings support a model in which rapamycin-resistant and rapamycin-sensitive forms of mTORC1 can be independently controlled, and provide a new genetic tool to separate these two signalling functions (Fig. 9). Rapamycin-resistant mTORC1 selectively, but not exclusively, regulates 4E-BP1 hyperphosphorylation, which is also specifically targeted by P53 activation in murine erythroleukemia cells (27), further supporting the idea that it is a distinct complex. However, this selective effect (Fig. 4 and Supplementary Fig. S2C) is not universal. In other cell types, PAT4 primarily appears to regulate an mTORC1 complex controlling 4E-BP1 and S6K (Supplementary Fig. S3; ref. 14), suggesting cell type-specific regulatory factors or PAT4 expression levels modulate this transporter’s specificity in controlling mTORC1. Indeed, it is only upon PAT4 overexpression that HEK-293 cells exhibit detectable PAT4-dependent rapamycin resistance (Fig. 5D).
Figure 9.
Schematic diagrams of rapamycin-sensitive and -resistant mTORC1 complexes in HCT116 cells. S6K and 4E-BP1, a negative regulator of eukaryotic initiation factor (eIF4E), are the best characterised downstream targets of mTORC1. Although rapamycin treatment strongly inhibits S6K phosphorylation, it has a weaker effect on 4E-BP1 γ-band phosphorylation at Ser65. Less phosphorylated forms of 4E-BP1 bind to eIF4E leading to translational repression (8). Reducing PAT4 activity primarily affects the rapamycin-resistant form of mTORC1. This leads to a reduction in a Ser65-phosphorylated form of 4E-BP1, but has less effect on S6K phosphorylation. Other amino acid transporters in the SLC36 (PAT) and/or SLC38 family are likely to be involved in rapamycin-sensitive mTORC1 regulation, eg. PAT1 and SLC38A9. PP242 is an mTORC1 ATP kinase inhibitor that acts on both the rapamycin-sensitive and resistant forms of mTORC1. Arrows signify positive signals and cross-bars mark inhibitory signalling events.
It has been suggested that PAT4 regulates mTORC1 signalling by transporting amino acids across the plasma membrane (36). However, this seems unlikely in our cell models for three reasons: first, cell surface PAT4 protein levels are very low (Fig. 1A, 7 and 8) (32, 36); second, PAT4 appears to be a low capacity transporter (37); third, it is difficult to explain how a cell surface transporter might affect only rapamycin-resistant mTORC1. We favour a model where PAT4 interacts with mTOR on the Golgi, although it may also regulate mTORC1 from other subcellular compartments.
Our study also revealed selective sensitivity of the mTORC1 target 4E-BP1 in HCT116 cells to reduction of two non-essential amino acids, glutamine and serine, but not to the mTORC1 regulator leucine, whose intracellular levels can be modulated by glutamine (38). The Arf1 GTPase, which regulates trafficking in multiple compartments, including the Golgi, has also recently been implicated in glutamine sensing by mTORC1 (22). A key question is whether PAT4 and Arf1 are involved in a common mTORC1-regulatory mechanism. Since PAT4’s amino acid specificity is different to arginine-sensitive, mTORC1-regulatory SLC38A9 (18, 19) and PAT1 (15), it seems likely that a range of AATs may determine the sensitivity of mTORC1 to different amino acids. The unique N-terminal domain of SLC38A9 appears to bind the Ragulator complex with higher affinity than other transporters (18). SLC38A9 also interacts at lower affinity with the V-ATPase through sequences including its transmembrane domains, which share similarity with other SLC38 and SLC36 (PAT) family members. This may explain why several of these transporters can regulate mTORC1, but some, like PAT4, cannot pull down other mTORC1 super-complex components, even though PLA indicates they interact in situ.
The PAT4-dependent sensitivity of 4E-BP1 hyperphosphorylation to four hours of glutamine or serine starvation probably reflects the rapid metabolism of these two non-essential amino acids in HCT116 cells (23, 27). Despite its name, when heterologously expressed in Xenopus oocytes, PAT4 can transport amino acids via a non-proton-coupled mechanism. It appears to have a very high substrate affinity, but low capacity, for proline and tryptophan (37). Several other amino acids, including glutamine and serine, bind with lower affinity, and can compete with high affinity PAT4 substrates, although they may not be transported. This could provide a transport-independent, amino acid-sensing ‘transceptor’ mechanism, in keeping with previous suggestions for the PATs (17). Cells would become more resistant to depletion of highly metabolised amino acids by expressing more PAT4, explaining the clinical data from colorectal cancer patients.
In conclusion, our study suggests pharmacological inhibition of an upstream mTORC1 activator such as PAT4 in colorectal cancer could complement the actions of rapamycin, by blocking a rapamycin-resistant, 4E-BP1-selective pathway. PAT4 may also provide a new biomarker for more aggressive colorectal tumours that are rapamycin-resistant.
MATERIALS AND METHODS
Cell culture
HCT116 cells were cultured in McCoy’s 5A modified medium (Gibco® Invitrogen) containing 10% fetal calf serum (FCS; Gibco® Invitrogen), unless otherwise specified. 786-O and HEK-293 cells were cultured in 10% FCS-containing DMEM. Cells were incubated at 37°C in 5% CO2. Rapamycin treatment was carried out 24 hours after seeding. Cells were treated for 24 hours typically, or 72 hours for cell proliferation analysis, with 3 nM (HCT116) or 100 nM (HEK-293) rapamycin, diluted from a stock solution in DMSO (R8781; Sigma), or as specified for dose-response curves. Founder cell lines were obtained from ATCC and used within six months of resuscitation (< 25 passages). si435 used to knockdown PAT4 expression and the scrambled control siRNA (Fig. 1) were previously described in (14).
Inducible shRNA-expressing lentiviruses
Sigma Mission® lentiviral particles carrying the following clones: TRCN0000043984 5′-CCGGCCTTGATAAATGAGCAGAATT-CTCGAGAATTCTGCTCATTTATCAAGGTTTTTG-3′ referred to as shPAT4(43984), TRCN0000043985 5′-CCGG-CCTGGAGAGTAAAGTGTTTATCTCGAG-ATAAACACTTTACTCTCCAGGTTTTTG-3′ (shPAT4[43985])] and TRCN0000043987 5′-CCGGCCAGTATGTTGTCAGGAACATCTCGAG-ATGTTCCTGACAACATACTGGTTTTTG-3′ (shPAT4[43987]) and a non-targeting control construct (SHC312V; shNT) in the IPTG-inducible lentiviral vector pLKO IPTG_1×LacO were transduced into HCT116 cells. While shPAT4(43984) partially overlaps with siRNA(435), shPAT4(43985) and (43987) have no overlap with previously employed siRNAs (14). For most experiments, clone shPAT4(7.1) produced a stronger knockdown than clone shPAT4(4.8) with greater effects on proliferation and mTORC1 signalling.
Generation and induction of shRNA lines
Cells were transduced with lentiviral vectors at a multiplicity of infection of three (viral particles to cells) in the presence of polybrene. Puromycin selection began two days after transduction to generate pools of cells derived from multiple transduction events. Single cell clones were isolated soon after transduction using the limiting dilution method. 100 μM IPTG was added for induction. IPTG-treated cells were pre-induced for five days prior to plating.
Amino acid starvation media
Medium was based on DMEM (11995-065; Invitrogen), but made in-house, so that different amino acids could be omitted individually. For glutamine, a medium lacking this amino acid was already available (McCoy’s 5A without glutamine [M8403; Sigma]). Cells were starved of amino acids for four hours.
Generation of GFP-PAT4 stable cell line
The PAT4 open reading frame from IMAGE Clone ID:531323 was recombined into a pOPINE vector containing an in-frame C-terminal GFP sequence (pOPINE-GFPc; gift from J Beale and S Newstead). The following errors in the PAT4 cDNA sequence (compared to the annotated transcript sequence) were corrected using the Quickchange® site-directed mutagenesis kit (Invitrogen): I209L (ATA->CTA) H376P (CCT->CAT) I429L (ATA->CTA). The PAT4-GFP fusion was amplified by PCR and cloned into pcDNA3.1(+) at the KpnI and XbaI sites. Amplified PAT4 and GFP ORFs were recloned into pcDNA3.1(+) as a GFP-PAT4 fusion. HEK-293 cells were transfected with this construct or the empty pcDNA3.1(+) vector using Lipofectamine® 2000 (Invitrogen). Stably transfected cells were selected 48 hours later using 800 μg/ml Geneticin (Gibco®; Invitrogen), as described in (14).
Xenograft studies
All protocols were carried out under Project License 30/2771 following Home Office regulations (39) using six-seven week-old female BALB/c SCID nu/nu mice (Harlan Sprague Dawley, Inc). 2.5 × 106 HCT116 cells in 50 μl serum-free medium and 50 μl Matrigel (BD Bioscience) were subcutaneously injected into one flank (seven mice per group; animals not randomized and investigator not blinded; numbers based on previous xenograft studies). 10 mM IPTG (Carbosynth Inc) was added to drinking water of treated mice. Tumour growth was measured three times a week using callipers and volume calculated from the formula 1/6 π × length × width × height. The experiment was repeated and produced similar significant reductions in growth and effects on survival time.
Quantitative real-time PCR
RNA extraction and quantitative real-time PCR (qRT-PCR) were carried out as described previously (14), with Ct values for PAT4 normalised against the HPRT1 housekeeping gene.
Cell proliferation analysis
Cell proliferation experiments, each using a minimum of three wells, were repeated three times. Where statistically significant results were found, these were observed on all occasions. Cells were counted according to the methodology described in (14)
Western blots
Protein lysates were routinely loaded on 10% polyacrylamide gels. For immunoblotting the following primary antibodies were used at recommended dilutions: phospho-S6K1 Thr308 (Cell Signalling Technology [CST #9205]), S6K1 (CST #9202), phospho-Ser240/244-ribosomal S6 protein (Cell Signalling Technology [CST #2215]), S6 ribosomal protein (CST #2217), phospho-Ser65-4E-BP1 rabbit polyclonal (CST #9451), 4E-BP1 (CST #9644), phospho-Ser473-Akt (CST #4060), Akt (CST #9272), phospho-Thr202/Tyr204-p44/42 MAPK (Erk1/2; CST #4370), p44/42 MAPK (Erk1/2; CST #4695) and α-tubulin (Sigma #T6199). They were detected with secondary antibodies (Promega #W401B, #W402B) and visualised using the SuperSignal® West Pico Chemiluminescent Substrate kit (Thermoscientific). PAT4 was deglycosylated with PNGase F (New England Biolabs; #P0704) according to manufacturer’s instructions.
Generation of PAT4 monoclonal antibody
A monoclonal antibody against PAT4 was created by immunising mice with a keyhole limpet haemocyanin- (KLH−) conjugated, cysteine-coupled peptide (Severn Biotech Ltd) based on an antigenic amino acid sequence within the N-terminus of PAT4 (REELDMDVMRPLINEC).
Immunohistochemistry
The following commercial primary antibodies were used: mTOR (CST#2983; 1/100), LAMP2 mouse monoclonal (Abcam, ab25631: 1/100), TGN46 sheep polyclonal (Novus Biologicals NB110-40767: 1/200), Rab1a rabbit monoclonal (CST#13075: 1/100), GFP mouse monoclonal (Abcam ab1218: 1/100), GM130 mouse polyclonal (Novus Biologicals H00002801-B01P: 1/25), Raptor rabbit polyclonal (Merck Millipore, 09-217: 1/100) and secondary antibodies raised in donkey (Jackson ImmunoResearch: 1/500). Cells were processed and imaged as described in (13, 40). Proximity ligation assays were performed using the Duolink® In Situ Orange Starter Mouse/Rabbit kit (DUO92102, Sigma) according to manufacturer’s instructions.
For patient samples, slides were de-waxed and rehydrated with antigen retrieval performed using citrate buffer pH 6.0 (Sigma-Aldrich) and a pressurised De-cloaking Chamber (Biocare Medical). The intensity of cytoplasmic staining was assessed by a pathologist (CS) on a semi-quantitative scale from 0-3. High levels of expression in tumour sections were scored three, lower levels of expression scored zero, one or two.
Immunohistochemistry was carried out as previously described (41). Paraffin-embedded tissue blocks from formalin-fixed tumour samples were sectioned, de-waxed, and rehydrated using standard techniques and 4 μm sections.
Patient material
Formalin-fixed and paraffin-embedded tissue was obtained with informed consent from 111 patients with colon cancer treated by surgical resection at the John Radcliffe Hospital, Oxford from 1997 to 2000. The sample size was limited to 107 by availability of tissue with full clinicopathological data, follow-up data and ethical approval. Use of tissue in this project was approved by the Oxford Centre for Histopathological Research Panel (Project number 12/A172) and the local Research Ethics Committee (C02.216). No patients received pre-operative chemotherapy. The average age at time of surgery was 71 years (range 37-96 years), 64 patients were male (58%) and the average follow-up time was 4.1 years (as of January 2009). Based on the tumour, nodal metastases and distant metastases (TNM), cancer stage of patients at resection was as follows; 10 patients (9%) Stage 1; 55 patients (49%) Stage 2; 35 patients (32%) Stage 3 and 11 patients (10%) Stage 4. All resections resulted in clear margins. Relapse-free survival was defined as time between tumour resection and the first-documented recurrence of tumour at any site. Patients who died from unrelated causes were excluded. Tissue microarrays were assembled as previously described (42) using two representative cores of tumour and two representative cores of adjacent normal colonic mucosal epithelium for each patient.
Statistics
All data (means ± standard error of mean [xenografts] or standard deviation [in vitro]) were analysed using Excel or GraphPad Prism 5. For tumour growth analysis, a parametric generalised linear model was performed using GraphPad Prism 4.0b software. For western blot analysis, cell proliferation and qRT-PCR, statistical significance was determined using the Kruskal-Wallis one-way analysis of variance. All in vitro experiments were replicated on at least three occasions. For patient data, the χ2 test was used to determine association between PAT4 expression and categorical clinical variables (Supplementary Table S1). Cox-regression analysis was employed to determine prognostic factors in univariate (Supplementary Table S2) and multivariate survival models (Supplementary Table S3). The log rank (Mantel-Cox) test was used to assess the significance of differences in relapse-free survival between Kaplan-Meier curves. Statistical analyses were performed using SPSS Statistics (Version 21.0).
Supplementary Material
ACKNOWLEDGMENTS
We thank Alan McIntyre, Elena Favaro, Thomas Roberts, Aaron Leiblich and Margret Ögmundsdóttir for advice and technical help during this study, Katharine Carr, Kristie McCormick and Dan Stevens for critical reading of the manuscript, and are especially grateful to Richard Boyd for facilitating key collaborative interactions. This work was supported by Cancer Research UK (C191591/A6181, C19591/A9093, C7713/A6174) and C38302/A12278 grants through the Cancer Research UK Oxford Centre Development Fund, the Wellcome Trust (WT093326MA), the BBSRC (BB/L007096/1), a Nuffield Medical Fellowship to CS, funding from the Studienstiftung des deutschen Volkes and the Max Weber Programm Bayern to SH, and a studentship from The British Province of the Society of Jesus (626791) to SMWP.
Footnotes
CONFLICT OF INTEREST
The authors declare no conflict of interest.
Supplementary Information accompanies the paper on the Oncogene website (http://www.nature.com/onc)
REFERENCES
- 1.Schulze A, Harris AL. How cancer metabolism is tuned for proliferation and vulnerable to disruption. Nature. 2012;491:364–73. doi: 10.1038/nature11706. [DOI] [PubMed] [Google Scholar]
- 2.Bar-Peled L, Sabatini DM. Regulation of mTORC1 by amino acids. Trends Cell Biol. 2014;24:400–6. doi: 10.1016/j.tcb.2014.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Malik AR, Urbanska M, Macias M, Skalecka A, Jaworski J. Beyond control of protein translation: What we have learned about the non-canonical regulation and function of mammalian target of rapamycin (mTOR) Biochim Biophys Acta. 2012;1834:1434–48. doi: 10.1016/j.bbapap.2012.12.010. [DOI] [PubMed] [Google Scholar]
- 4.Jewell JL, Guan KL. Nutrient signaling to mTOR and cell growth. Trends Biochem Sci. 2013;38:233–42. doi: 10.1016/j.tibs.2013.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zaytseva YY, Valentino JD, Gulhati P, Evers BM. mTOR inhibitors in cancer therapy. Cancer Lett. 2012;319:1–7. doi: 10.1016/j.canlet.2012.01.005. [DOI] [PubMed] [Google Scholar]
- 6.Zhang Y, Zheng XF. mTOR-independent 4E-BP1 phosphorylation is associated with cancer resistance to mTOR kinase inhibitors. Cell Cycle. 2012;11:594–603. doi: 10.4161/cc.11.3.19096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hsieh AC, Ruggero D. Targeting eukaryotic translation initiation factor 4E (eIF4E) in cancer. Clin Cancer Res. 2010;16:4914–20. doi: 10.1158/1078-0432.CCR-10-0433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hsieh AC, Liu Y, Edlind MP, Ingolia NT, Janes MR, Sher A, Shi EY, Stumpf CR, Christensen C, Bonham MJ, Wang S, Ren P, Martin M, Jessen K, Feldman ME, Weissman JS, Shokat KM, Rommel C, Ruggero D. The translational landscape of mTOR signalling steers cancer initiation and metastasis. Nature. 2012;485:55–61. doi: 10.1038/nature10912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Thoreen CC, Chantranupong L, Keys HR, Wang T, Gray NS, Sabatini DM. A unifying model for mTORC1-mediated regulation of mRNA translation. Nature. 2012;485:109–13. doi: 10.1038/nature11083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Grabiner BC, Nardi V, Birsoy K, Possemato R, Shen K, Sinha S, Jordan A, Beck AH, Sabatini DM. A diverse array of cancer-associated mTOR mutations are hyperactivating and can predict rapamycin sensitivity. Cancer Discov. 2014;4:554–63. doi: 10.1158/2159-8290.CD-13-0929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Thwaites DT, Anderson CM. The SLC36 family of proton-coupled amino acid transporters and their potential role in drug transport. Br J Pharmacol. 2011;164:1802–16. doi: 10.1111/j.1476-5381.2011.01438.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Goberdhan DC, Meredith D, Boyd CA, Wilson C. PAT-related amino acid transporters regulate growth via a novel mechanism that does not require bulk transport of amino acids. Development. 2005;132:2365–75. doi: 10.1242/dev.01821. [DOI] [PubMed] [Google Scholar]
- 13.Ogmundsdottir MH, Heublein S, Kazi S, Reynolds B, Visvalingam SM, Shaw MK, Goberdhan DC. Proton-assisted amino acid transporter PAT1 complexes with Rag GTPases and activates TORC1 on late endosomal and lysosomal membranes. PloS One. 2012;7:e36616. doi: 10.1371/journal.pone.0036616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Heublein S, Kazi S, Ogmundsdottir MH, Attwood EV, Kala S, Boyd CA, Wilson C, Goberdhan DC. Proton-assisted amino-acid transporters are conserved regulators of proliferation and amino-acid-dependent mTORC1 activation. Oncogene. 2010;29:4068–79. doi: 10.1038/onc.2010.177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sagne C, Agulhon C, Ravassard P, Darmon M, Hamon M, El Mestikawy S, Gasnier B, Giros B. Identification and characterization of a lysosomal transporter for small neutral amino acids. Proc Natl Acad Sci USA. 2001;98:7206–11. doi: 10.1073/pnas.121183498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wreden CC, Johnson J, Tran C, Seal RP, Copenhagen DR, Reimer RJ, Edwards RH. The H+-coupled electrogenic lysosomal amino acid transporter LYAAT1 localizes to the axon and plasma membrane of hippocampal neurons. J Neurosci. 2003;23:1265–75. doi: 10.1523/JNEUROSCI.23-04-01265.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Goberdhan DC. Intracellular amino acid sensing and mTORC1-regulated growth: new ways to block an old target? Curr Opin Investig Drugs. 2010;11:1360–7. [PMC free article] [PubMed] [Google Scholar]
- 18.Wang S, Tsun ZY, Wolfson RL, Shen K, Wyant GA, Plovanich ME, Yuan ED, Jones TD, Chantranupong L, Comb W, Wang T, Bar-Peled L, Zoncu R, Straub C, Kim C, Park J, Sabatini BL, Sabatini DM. Metabolism. Lysosomal amino acid transporter SLC38A9 signals arginine sufficiency to mTORC1. Science. 2015;347:188–94. doi: 10.1126/science.1257132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rebsamen M, Pochini L, Stasyk T, de Araújo ME, Galluccio M, Kandasamy RK, Snijder B, Fauster A, Rudashevskaya EL, Bruckner M, Scorzoni S, Filipek PA, Huber KV, Bigenzahn JW, Heinz LX, Kraft C, Bennett KL, Indiveri C, Huber LA, Superti-Furga G. SLC38A9 is a component of the lysosomal amino acid sensing machinery that controls mTORC1. Nature. 2015;19:477–81. doi: 10.1038/nature14107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Thomas JD, Zhang YJ, Wei YH, Cho JH, Morris LE, Wang HY, Zheng XF. Rab1A is an mTORC1 activator and a colorectal oncogene. Cancer Cell. 2014;26:754–69. doi: 10.1016/j.ccell.2014.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jewell JL, Kim YC, Russell RC, Yu FX, Park HW, Plouffe SW, Tagliabracci VS, Guan KL. Metabolism. Differential regulation of mTORC1 by leucine and glutamine. Science. 2015;347:194–8. doi: 10.1126/science.1259472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xu L, Salloum D, Medlin PS, Saqcena M, Yellen P, Perrella B, Foster DA. Phospholipase D mediates nutrient input to mammalian target of rapamycin complex 1 (mTORC1) J Biol Chem. 2011;286:25477–86. doi: 10.1074/jbc.M111.249631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yoon MS, 1, Du G, Backer JM, Frohman MA, Chen J. Class III PI-3-kinase activates phospholipase D in an amino acid-sensing mTORC1 pathway. J Cell Biol. 2011;195:435–47. doi: 10.1083/jcb.201107033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Siegel R, Desantis C, Jemal A. Colorectal cancer statistics, 2014. CA Cancer J Clin. 2014;64:104–17. doi: 10.3322/caac.21220. [DOI] [PubMed] [Google Scholar]
- 25.Kim DD, Eng C. The promise of mTOR inhibitors in the treatment of colorectal cancer. Expert Opin Investig Drugs. 2012;21:1775–88. doi: 10.1517/13543784.2012.721353. [DOI] [PubMed] [Google Scholar]
- 26.Jiang P, Du W, Mancuso A, Wellen KE, Yang X. Reciprocal regulation of p53 and malic enzymes modulates metabolism and senescence. Nature. 2013;493:689–93. doi: 10.1038/nature11776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Maddocks OD, Berkers CR, Mason SM, Zheng L, Blyth K, Gottlieb E, Vousden KH. Serine starvation induces stress and p53-dependent metabolic remodelling in cancer cells. Nature. 2013;493:542–6. doi: 10.1038/nature11743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Constantinou C, Clemens MJ. Regulation of the phosphorylation and integrity of protein synthesis initiation factor eIF4GI and the translational repressor 4E-BP1 by p53. Oncogene. 2005;24:4839–50. doi: 10.1038/sj.onc.1208648. [DOI] [PubMed] [Google Scholar]
- 29.Karim MM, Hughes JM, Warwicker J, Scheper GC, Proud CG, McCarthy JE. A quantitative molecular model for modulation of mammalian translation by the eIF4E-binding protein 1. J Biol Chem. 2001;276:20750–7. doi: 10.1074/jbc.M011068200. [DOI] [PubMed] [Google Scholar]
- 30.Ducker GS, Atreya CE, Simko JP, Hom YK, Matli MR, Benes CH, Hann B, Nakakura EK, Bergsland EK, Donner DB, Settleman J, Shokat KM, Warren RS. Incomplete inhibition of phosphorylation of 4E-BP1 as a mechanism of primary resistance to ATP-competitive mTOR inhibitors. Oncogene. 2014;33:1590–600. doi: 10.1038/onc.2013.92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhao Y, Butler EB, Tan M. Targeting cellular metabolism to improve cancer therapeutics. Cell Death Dis. 2013;4:e532. doi: 10.1038/cddis.2013.60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zoncu R, Bar-Peled L, Efeyan A, Wang S, Sancak Y, Sabatini DM. mTORC1 senses lysosomal amino acids through an inside-out mechanism that requires the vacuolar H(+)-ATPase. Science. 2011;334:678–83. doi: 10.1126/science.1207056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Söderberg O, Gullberg M, Jarvius M, Riddersträle K, Leuchowius K-J, Jarvius J, Wester K, Hydbring P, Bahram F, Larsson L-G, Landegren U. Direct observation of individual endogenous protein complexes in situ by proximity ligation. Nat. Methods. 2006;3:995–1000. doi: 10.1038/nmeth947. [DOI] [PubMed] [Google Scholar]
- 34.Kang SA, Pacold ME, Cervantes CL, Lim D, Lou HJ, Ottina K, Gray NS, Turk BE, Yaffe MB, Sabatini DM. mTORC1 phosphorylation sites encode their sensitivity to starvation and rapamycin. Science. 2013;341:1236566. doi: 10.1126/science.1236566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li L, 1, Kim E, Yuan H, Inoki K, Goraksha-Hicks P, Schiesher RL, Neufeld TP, Guan KL. Regulation of mTORC1 by the Rab and Arf GTPases. J Biol Chem. 2010;285:19705–9. doi: 10.1074/jbc.C110.102483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Matsui T, Fukuda M. Rab12 regulates mTORC1 activity and autophagy through controlling the degradation of amino-acid transporter PAT4. EMBO Rep. 2013;14:450–7. doi: 10.1038/embor.2013.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pillai SM, Meredith D. SLC36A4 (hPAT4) is a high affinity amino acid transporter when expressed in Xenopus laevis oocytes. J Biol Chem. 2011;286:2455–60. doi: 10.1074/jbc.M110.172403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nicklin P, Bergman P, Zhang B, Triantafellow E, Wang H, Nyfeler B, Yang H, Hild M, Kung C, Wilson C, Myer VE, MacKeigan JP, Porter JA, Wang YK, Cantley LC, Finan PM, Murphy LO. Bidirectional transport of amino acids regulates mTOR and autophagy. Cell. 2009;136:521–34. doi: 10.1016/j.cell.2008.11.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li JL, Sainson RC, Oon CE, Turley H, Leek R, Sheldon H, Bridges E, Shi W, Snell C, Bowden ET, Wu H, Chowdhury PS, Russell AJ, Montgomery CP, Poulsom R, Harris AL. DLL4-Notch signaling mediates tumor resistance to anti-VEGF therapy in vivo. Cancer Res. 2011;71:6073–83. doi: 10.1158/0008-5472.CAN-11-1704. [DOI] [PubMed] [Google Scholar]
- 40.Sancak Y, Bar-Peled L, Zoncu R, Markhard AL, Nada S, Sabatini DM. Ragulator-Rag Complex Targets mTORC1 to the Lysosomal Surface and Is Necessary for Its Activation by Amino Acids. Cell. 2010;141:290–303. doi: 10.1016/j.cell.2010.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li JL, Sainson RC, Shi W, Leek R, Harrington LS, Preusser M, Biswas S, Turley H, Heikamp E, Hainfellner JA, Harris AL. Delta-like 4 Notch ligand regulates tumor angiogenesis, improves tumor vascular function, and promotes tumor growth in vivo. Cancer Res. 2007;67:11244–53. doi: 10.1158/0008-5472.CAN-07-0969. [DOI] [PubMed] [Google Scholar]
- 42.Bubendorf L, Nocito A, Moch H, Sauter G. Tissue microarray (TMA) technology: miniaturized pathology archives for high-throughput in situ studies. J Pathol. 2001;195:72–9. doi: 10.1002/path.893. [DOI] [PubMed] [Google Scholar]
- 43.Rath A, Glibowicka M, Nadeau VG, Chen G, Deber CM. Detergent binding explains anomalous SDS-PAGE migration of membrane proteins. Proc Natl Acad Sci U S A. 2009;106:1760–5. doi: 10.1073/pnas.0813167106. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.