Abstract
Mitochondria are involved in ageing and their function requires coordinated action of both mitochondrial and nuclear genes. Epistasis between the two genomes can influence lifespan but whether this also holds for reproductive senescence is unclear. Maternal inheritance of mitochondria predicts sex differences in the efficacy of selection on mitonuclear genotypes, which should result in differences between females and males in mitochondrial genetic effects. Mitonuclear genotype of a focal individual may also indirectly affect trait expression in the mating partner. We tested these predictions in the seed beetle Callosobruchus maculatus, using introgression lines harbouring distinct mitonuclear genotypes. Our results reveal both direct and indirect sex-specific effects of mitonuclear epistasis on reproductive ageing. Females harbouring coadapted mitonuclear genotypes showed higher lifetime fecundity due to slower senescence relative to novel mitonuclear combinations. We found no evidence for mitonuclear coadaptation in males. Mitonuclear epistasis affected age-specific ejaculate weight, but also influenced male age-dependent indirect effects on traits expressed by their female partners (fecundity, egg size, longevity). These results demonstrate important consequences of sex-specific mitonuclear epistasis for both mating partners, consistent with a role for mitonuclear genetic constraints upon sex-specific adaptive evolution.
Introduction
Ageing (senescence) is a near universal characteristic of living organisms and is manifested as decreasing performance and increasing probability of death with age, due to physiological deterioration (Monaghan et al, 2008). Mitochondrial malfunction has been implicated in ageing across species and is apparent as structural abnormalities of mitochondria, decreased electron transport chain enzyme activity in the oxidative phosphorylation (OXPHOS) pathway, increased production of reactive oxygen species (ROS) and accumulation of mutations on mitochondrial DNA (mtDNA) (Tower, 2014). Mitochondrial function relies on the coordinated expression of over a thousand nuclear genes and a few dozen mitochondrial genes (Calvo and Mootha, 2010). This suggests that ageing phenotypes may commonly be subject to a complex set of epistatic interactions between the nuclear and the mitochondrial genomes. Mitonuclear interactions affect lifespan through redox balance involving nuclear-encoded antioxidants that defend against oxidative stress (Finkel and Holbrook, 2000), retrograde signalling (Liu and Butow, 2006), and mitonuclear protein imbalance via activation of the mitochondrial unfolded protein response (Houtkooper et al, 2013).
Some studies have found effects of mitochondrial polymorphism or mitonuclear epistasis on longevity and survival (Camus et al, 2012; Maklakov et al, 2006; Rand et al, 2006; Zhu et al, 2014) but their role in reproductive ageing is less well studied (Yee et al, 2015). The influence of mitochondria in general, and of mitonuclear interactions in particular, may be even larger for reproductive senescence compared to actuarial ageing (i.e. longevity and survival) (Dobler et al, 2014; Maklakov and Lummaa, 2013). This is partly because reproduction is sensitive to cellular malfunctioning, and there is a general decline in mitochondrial performance with age (Lee and Wei, 2012; Tower, 2014), but also because sexual asymmetries in inheritance and selection should interact to sustain mitonuclear epistasis. Males and females often optimise their reproductive scheduling in distinct ways and selection should thus generate sex-differences in the pattern of reproductive and actuarial ageing (Bonduriansky et al, 2008). Such sex-specific selection predicts that mitonuclear genetic effects on the pattern of ageing may also differ between males and females. For example, the involvement of mitochondria places evolutionary constraints on male-specific optimisation of reproduction and somatic maintenance, because mtDNA can only respond to selection acting on females. This represents an irreconcilable form of intra-locus sexual conflict (Bonduriansky and Chenoweth, 2009) that can lead to the accumulation of a male-specific mutation load, a phenomenon known as Mother’s curse (Gemmell et al, 2004). One of the key predictions of the Mother’s curse is that male-harming mitochondrial mutations should select for counter-adaptations in nuclear genes interacting with the mitochondria, leading to mitonuclear coevolution (Gallach and Betran, 2011; Wade, 2014). However, it has been estimated that the rate of nuclear compensatory evolution is less than 1/4 that of male-harming mitochondrial mutations (Wade, 2014), making mitonuclear coevolution less efficient in males. Concordantly, many studies on male-specific mitonuclear effects find irregular epistasis rather than increased mitochondrial performance in coadapted mitonuclear combinations (Dobler et al, 2014). In females, selection on mitonuclear genotype would be more efficient in terms of generating mitonuclear coadaptation, simply because nuclear genes and mtDNA cosegregate in females (Rand et al, 2004; Wade, 2014). Sex-specific mitonuclear coevolution may also be constrained by sexually antagonistic selection on the nuclear genes (Rand et al, 2001). Although mitonuclear epistatic effects have been demonstrated in both sexes, very few studies have studied both sexes simultaneously (Dobler et al, 2014).
An important but overlooked aspect in the study of sex-specific reproductive senescence derives from the fact that reproductive traits in one sex, affected by mitochondrial genetic variation (Dowling et al, 2007b; Yee et al, 2013), may have important effects on reproductive phenotypes in the other sex. Reproduction is an outcome of molecular interactions between the sexes and research over the last few decades has clearly demonstrated that male traits influence female reproductive performance (Arnqvist and Rowe, 2005). It is thus possible that mitochondrial genetic effects may extend beyond focal males (i.e. direct genetic effects) to also affect their mating partners (i.e. indirect genetic effects) (Moore et al, 1997). Such effects are most likely mediated by seminal fluid proteins that are transferred to females during mating, with manifold effects on female reproduction (Avila et al, 2011). Male reproductive senescence could thus influence female traits via male seminal fluid biosynthesis and could be affected by mitonuclear epistasis through the role that mitochondria play in metabolic senescence.
Here, we explicitly test for mitonuclear epistasis on several reproductive and life-history characteristics of males and females throughout their life, using Callosobruchus maculatus seed beetles with distinct mitonuclear genotypes created by introgression between naturally occurring mtDNA haplotypes and nuclear genotypes from three populations. Our main objectives are to test the following predictions: (1) mitonuclear epistasis influences reproductive senescence, (2) such effects are sex-specific and show differences regarding mitonuclear coadaptation and (3) mitonuclear genetic variation in males is associated with indirect genetic effects on traits expressed by their mates.
Materials and Methods
Study organism
The seed beetle C. maculatus occurs in subtropical and tropical regions throughout the world, and is a major pest of legume plants. Females lay eggs on beans and the 1st instar larvae bore into them within 2-4 days. The larvae develop for approximately three weeks, and copulate soon after emergence from beans as adults. Both sexes can mate multiply. This species is facultatively aphagous and can complete its reproductive cycle with the resources acquired during the larval stage (Fox, 1993). However, adult feeding increases both longevity and productivity of the beetles (Fox, 1993). Previous work has documented mitonuclear genetic effects on basal metabolic rate in pupae (Arnqvist et al, 2010) and in mated males (Immonen et al, 2015), juvenile growth rate (Dowling et al, 2007a), behavioural syndromes (Lovlie et al, 2014) and sperm characteristics (Dowling et al, 2007b).
Mitonuclear lines
In order to disentangle the effects of mitochondrial and nuclear genetic variation as well as their epistatic interaction we introgressed three distinct naturally occurring mitochondrial genomes into three nuclear backgrounds. Sequencing of two mitochondrial genes (1005 bp of cytochrome oxidase subunit I [COI] and 473 bp of cytochrome b [Cyt-b]) has documented substantial genetic variation across these three mtDNA haplotypes (Arnqvist et al, 2010). Approximately 2-3% of all mtDNA sites are polymorphic, and some sites show non-synonymous differences. A detailed description of the construction of the introgression lines can be found in Kazancioglu and Arnqvist (Kazancioglu and Arnqvist, 2013). Briefly, three outbred populations of C. maculatus (Brazil [BRA], California [CA] and Yemen [YEM]), which carry distinct mtDNA haplotypes (Kazancioglu and Arnqvist, 2013), were used to generate nine fully crossed combinations of distinct cytoplasmic and nuclear lineages. These populations were selected from a larger set of potential populations as previous research has shown that mitochondrial genetic variation across these particular populations affects life history phenotypes (Arnqvist et al, 2010; Dowling et al, 2007a; Dowling et al, 2007b; Immonen et al, 2015). A single randomly selected virgin female from each of the populations (ensured to carry a distinct mtDNA haplotype by sequencing, see above) was mated to a male from the same population, and the full-sib daughters of these “mitochondrial Eves” were subsequently mated to males from one of the three populations. This orthogonal scheme resulted in nine possible mitonuclear combinations, and each particular cross was replicated twice starting with a different female (carrying the same mtDNA haplotype) (i.e. total of 18 introgression lines) to allow assessment of variation within mitonuclear combination. Within each cross, virgin daughters in every generation were backcrossed to virgin males from their father’s population, thus disassociating the mitochondrial genome from the nuclear genome with which it was originally coexpressed. Backcrossing was repeated for 16 generations, which replaces 99.998% of the original nuclear genome with the paternal nuclear genome. Replication of cross types (see above) allowed us to assess effects of this incomplete replacement of the nuclear genome. Fertility, hatching rate and adult viability was not depressed during introgression. Subsequently, the 18 introgression lines were maintained as separate populations in large glass jars seeded with 100g of Vigna unguiculata black-eyed beans, at 29°C and 12:12 h light:dark cycle, with 50% relative humidity. The beetles were maintained under aphagy and the lines were backcrossed once again to the outbred stock populations at generation 17. We validated the mitochondrial integrity of the introgression lines immediately after our experiments, by amplification and sequencing of a diagnostic mtDNA marker (COI), following Kazancioglu and Arnqvist (Kazancioglu and Arnqvist, 2013). Potential cytoplasmic endosymbionts (e.g. Wolbachia), that could confound cytoplasmic effects, have carefully been screened for in many C. maculatus populations, including the ones used here, but have never been detected (e.g. Tuda et al, 2006). Nevertheless, we treated all introgression lines with an antibiotic treatment between generations 13 and 14 (see Kazancioglu and Arnqvist, 2013 for details), to preclude the possibility that the lines may have harboured cytoplasmic bacterial infections. Moreover, mtDNA sequence divergence across these haplotypes is quantitatively associated with phenotypic divergence across haplotypes (Arnqvist et al, 2010).
Experimental procedures
The reproductive performance of the males and females from the introgression lines were recorded by mating them to individuals from an outbred standard reference population (SI USA) at fixed intervals through their reproductive life. The reference population was maintained on mung beans (Vigna radiata) under the same abiotic condition as the focal introgression lines. Virgin focal and reference individuals were collected by isolating beans with larvae in chambers from which hatching adults were collected. Virgins were then isolated in 1.5mL Eppendorf tubes (with lids punctured for air) until used in the experiments.
To ensure that beetles would not become dehydrated or resource limited with age, which could result in starvation effects rather than senescence in physiological performance, we provided both focal and reference individuals food ad libitum throughout their lifespan (baker’s yeast and 5% sucrose-solution; renewed every four days) (Arnqvist et al, 2010).
Females
To characterize the pattern of senescence in females from the introgression lines we recorded age-specific fecundity (N = 912), fertility (the proportion of hatched eggs, N= 912), larval survival (the proportion of emerged adult offspring out of all the hatched eggs, N= 629) as well as longevity (N=165). To ensure the females would not become sperm limited over time, we allowed them to mate for up to four times (to reference virgin males of age 24-48h) at fixed intervals (Figure 1). We observed all matings and separated the pairs immediately after copulation. After the first mating (at the age of 48h), the females were allowed to individually lay eggs on petri dishes with an excess of black-eyed beans as an egg laying substrate, for a fixed interval, after which they were either first provided a re-mating opportunity or directly transferred into a new egg-laying dish (Figure 1). Altogether we recorded female reproduction from six age intervals, capturing the entire reproductive age span from 2 to 26 days. We counted the hatched and un-hatched eggs from each dish after ensuring that all eggs had sufficient time to hatch, and subsequently allowed the eggs to develop into adult offspring. For the post-reproductive period (age 26 days onwards) we transferred the females individually into dishes (with only a few beans and food) where their longevity was recorded. The females were weighed upon emergence to the nearest 0.00001 g (Sartorius® Genius ME 235P) to account for the effect of weight on fecundity and longevity.
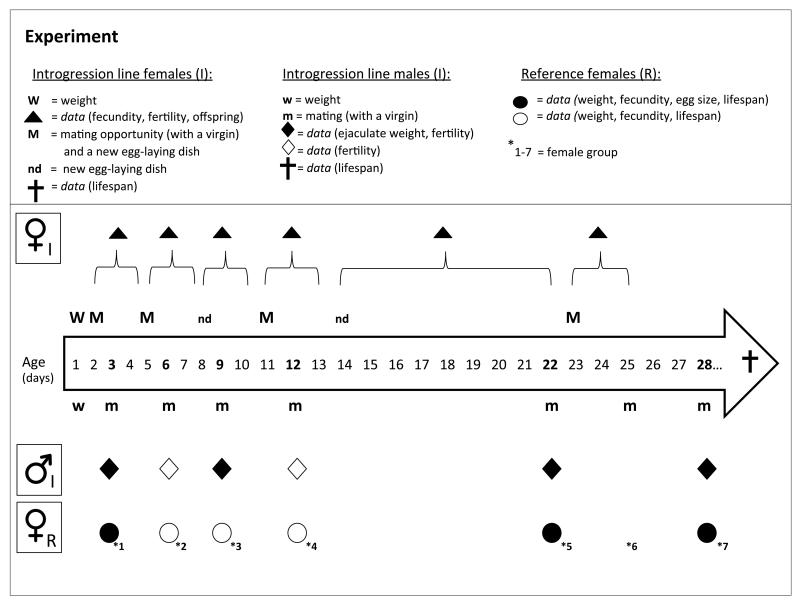
Figure 1.
Summary of the experimental design. We recorded data of age-specific reproductive performance and lifespan from mitonuclear introgression lines (I) and a reference population females (R) mated to the introgression line males. For the focal females, provided with a mating opportunity with four sets of reference virgin males (1-2 days old virgin), we collected data from six age intervals. For the focal males and their reference female mates (1-2 days old virgin), we collected data from six male age points. The symbols indicate data collected from each experimental group at each time point (open and closed symbols show which variables were recorded at a given time).
Males
We characterized effects of senescence in males from the introgression lines on both traits expressed by themselves and their reference female mating partners (i.e. direct and indirect genetic effects). The direct effects measured in males included ejaculate weight (N = 615) (measured as the difference in body mass before and after mating, Edvardsson and Tregenza, 2005) and male longevity (N = 173).
The indirect effects of male age and genotype on the expression of female traits were measured for fecundity (N = 621), egg size (N = 7971) and fertility of the eggs (the proportion of hatched eggs, N = 954). We also recorded longevity of the female mates (N = 945).
We mated males seven times to 24-48 h old virgin reference females at fixed intervals (male reproductive age span from 3 to 28 days, Figure 1). We observed every mating and separated the pairs immediately after copulation. Controlling mating rate allowed us to focus on effects of male physiological senescence and its consequences on his reproductive performance in the context of mating, without confounding effects of variation in male harassment or mating frequency. After mating, the reference females were kept individually in petri dishes, with an excess of mung beans, for 18 days (to allow plenty of time for all the eggs to hatch but no adult offspring), after which the females were transferred in isolation to new petri dishes (with food and a few beans) where their longevity was recorded. The reference females and focal males were weighed to the nearest 0.00001 g (Sartorius® Genius ME 235P). We measured the ejaculate weight from four matings (ages 3, 9, 22 and 28 days, Figure 1). To measure egg size, we randomly sampled 20 eggs per female, from three matings of male ages 3, 22 and 28 days (Figure 1), and photographed them using a Lumenera Infinity 2-2 digital camera mounted on a dissection microscope (Leica MZ8). Egg width, as a proxy for egg size, was measured using ImageJ (v. 1.48).
Statistical analyses
The crossed design allows the use of linear effects mixed models to evaluate our results. All of our inferential mixed effects models contain a random effect structure that takes the mitonuclear line replication into account (i.e. by fitting the line as a random factor) and, where necessary, the repeated measures taken across age (i.e. by fitting individual ID as a random factor). Models were fitted with R (version 3.0.1, see package information below) (RDevelopmentCoreTeam, 2011). All models include mitochondrial lineage (“mito”), nuclear lineage (“nuc”) and their interaction as factorial fixed effects, and weight at emergence (of the focal individual) as a covariate.
The significance of terms in the models was assessed with type III sums-of-squares tested with analyses of deviance based on the chi-square distribution, as implemented in the car package (version 2.0-20) (Fox and Weisberg, 2011). Survival data was analysed using Cox Proportional Hazard mixed models, where the significance of terms was determined by model comparisons after model reductions, using Chi-square tests. Covariates in all models were standardized to a mean of zero and unit variance, with the exception of female age that was only mean centred. Model fit was validated by visual inspection and few observations, where an absolute value of the standardized residuals exceeded 3.0 were deemed as outliers and omitted. We note that, in any such occasion, the results remained qualitatively the same even if including these outliers. For the binomial models we estimated overdispersion as the sum of squared Pearson residuals divided by the residual degrees of freedom and, if overdispersed, we accounted for this by including an individual observation level random effect (ILRE) into the model (Elston et al, 2001) and validated the fit with a model comparison using AIC and log-likelihood information. Below we present the model details specific to the analyses for each sex.
Females
We examined effects on the rate of female reproductive senescence with the following models. We first investigated effects on fecundity by fitting a Zero Inflated Negative Binomial Mixed Model (ZINBMM, package “glmmADMB” version 0.8.0 (Fournier et al, 2012)), with age as a covariate and its two- and three-way interactions with the genetic factors. Because one of the egg-laying intervals was longer than others (see Figure 1), we used daily fecundity as the response variable, calculated by dividing the number of eggs with the number of days within each interval. The best model fit, based on AIC information, was achieved with “NB1” parametrization (variance = φμ). Based on the findings of this model (Figure S1) we also tested whether an observed mitonuclear × age interaction follows a pattern of mitonuclear coevolution, with an otherwise similar model (see above and Table 1) but replacing the “mito” factor with a two-level factor representing the mitonuclear coevolutionary history (i.e. potentially coadapted or not). Both models also include female lifetime mating rate as a covariate (we note that female remating rate did not differ between the genotypes, result not shown). We tested for effects on fertility and on larval survival with Generalized Linear Mixed Models (GLMM, package “lme4“, version 1.1-7, (Bates et al, 2014)) with the same model structure as for the main inferential model for fecundity, but with an additional observation-level random factor to account for a slight overdispersion.
Table 1.
Direct effects of age, mitochondrial and nuclear genetic variation on introgression line female(1) fecundity (i.e. daily rate of egg laying) and fertility, and male(2) ejaculate weight.
| Response variable: | (1) Fecundity N=912 | (1) Fertility N=912 | (2) Ejaculate weight N = 615 | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
| |||||||||||||||
| Fixed effects | Coef | se | χ 2 | Df | P | Coef | se | χ 2 | Df | P | Coef | se | χ 2 | Df | P |
| Age | −0.67 | 0.04 | 305.0 | 1 | <0.0001 | −0.81 | 0.11 | 52.5 | 1 | <0.0001 | 5.8 | 3 | 0.1198 | ||
| Body weight | 0.16 | 0.02 | 97.0 | 1 | <0.0001 | 0.01 | 0.08 | 0.002 | 1 | 0.9636 | 5.44 | 0.40 | 183.6 | 1 | <0.0001 |
| Mating rate | 0.04 | 0.02 | 6.8 | 1 | 0.0091 | 0.13 | 0.08 | 2.9 | 1 | 0.0874 | - | - | - | - | - |
| Mitochondrial lineage | 7.3 | 2 | 0.0266 | 1.8 | 2 | 0.3929 | 13.9 | 2 | 0.0010 | ||||||
| Nuclear lineage | 6.6 | 2 | 0.0373 | 0.0 | 2 | 0.9979 | 4.6 | 2 | 0.0986 | ||||||
| Mito × Nuc | 12.5 | 4 | 0.0141 | 8.1 | 4 | 0.0887 | 24.8 | 4 | <0.0001 | ||||||
| Age × Mito | 4.4 | 2 | 0.1123 | 0.3 | 2 | 0.8724 | 31.7 | 6 | <0.0001 | ||||||
| Age × Nuc | 6.0 | 2 | 0.0495 | 2.4 | 2 | 0.2964 | 24.7 | 6 | <0.0001 | ||||||
| Age × Mito × Nuc | 10.0 | 4 | 0.0422 | 2.3 | 4 | 0.6766 | 36.4 | 12 | <0.0001 | ||||||
|
| |||||||||||||||
| Random effects | Variance (σ2) | Variance (σ2) | Variance (σ2) | ||||||||||||
|
| |||||||||||||||
| Line | 2.061 × 10−9 | 0.00 | 0.08 | ||||||||||||
| ID | 2.066 × 10−9 | 0.40 | 1.08 | ||||||||||||
We examined effects on female actuarial ageing, by testing for effects on longevity, with a Linear Mixed Model (LMM, package “lme4”), including the genetic effects as factors, weight and lifetime fecundity as covariates (N = 165). We also tested for survival differences using a Cox Proportional Hazard Mixed Model (CPHMM, package “coxme” version 2.2-3 (Thernau, 2012)), with the same explanatory variables.
Males
We first assessed direct genetic and age effects on the two reproductive traits expressed by the males. We tested how male ejaculate weight (N = 615) varies with age and genotype with a LMM and how these affect male fertility (N = 954), using a GLMM, with age fitted as a factor. We also tested whether observed mitonuclear × age interactions confirm with mitonuclear coadaptation, with otherwise similar models but replacing the “mito” factor with a two-level factor representing coevolutionary history. Observation –level random factor was included in the fertility models to account for overdispersion. To examine mitonuclear genetic effects on male actuarial ageing, we tested how males vary in longevity with a LMM and in survival using a CPHMM (N = 173).
Our second aim was to assess indirect genetic and age effects of males on traits expressed by their female mating partners. We tested for effects on female fecundity with a LMM. As male ejaculate weight was clearly an important determinant, we here only present the model on the subset of the data where we have this information (N = 621, Figure 1). We note, however, that genetic and age effects where independent of the ejaculate weight effects. We also explored the effects of mitonuclear coadaptation in the same manner as described above. We tested for male effects on female egg size (N = 7971) with a LMM, including the reference female ID as an additional random factor to account for the repeated egg measures per female. To explore whether egg size covaries with female fecundity, we used the mean egg size per female as a response variable in a LMM. We assessed male effects on female survival using a CPHMM and on longevity with a LMM (N = 945), with female fecundity and weight as covariates in addition to the male age and genetic factors.
Results
Senescence in females
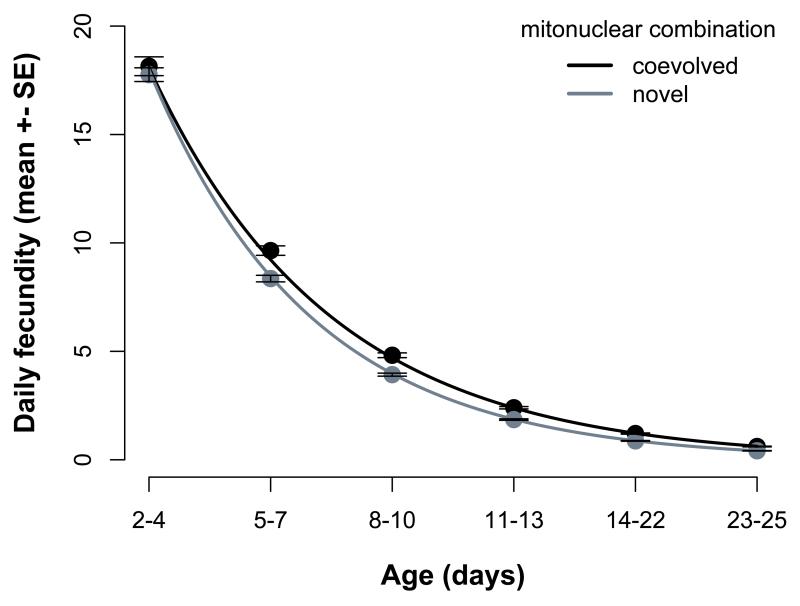
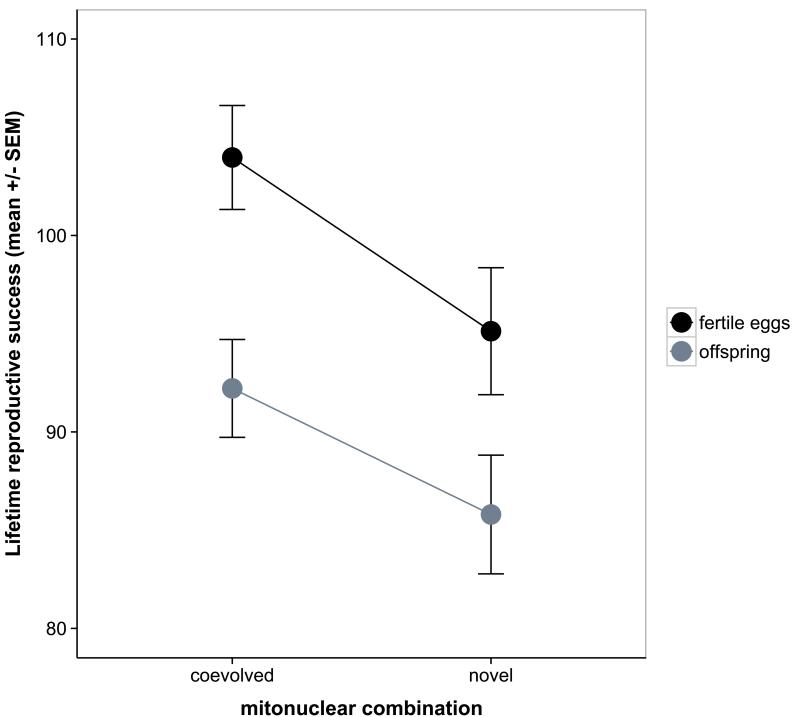
Fecundity decreased rapidly with age in all females, and the average fecundity as well as the rate of decline depended on the mitonuclear combination (Table 1 Figure S1). This epistatic interaction arose from mitonuclear coadaptation: females with a coevolved mitonuclear genotype showed a higher average fecundity (χ21=7.44, p=0.0064) and slower rate of decline in fecundity compared to females with a novel mitonuclear genotype (age × coevolution: χ21=4.4; P=0.036, Figure 2a). Consequently, the disruption of a coadapted mitonuclear combination resulted in significantly lower lifetime fecundity (All fertile eggs: χ21=5.9; P=0.015) and a marginally non-significantly lower lifetime offspring production (χ21=3.8; P=0.053, Figure 2b). We note that the lifetime fecundity difference was similar in magnitude for fertile and total number eggs (result not shown). Female fertility declined with age (Figure S2) but we found no significant effects of mitochondrial genetic variation or mitonuclear epistasis on this decline (Table 1). There were also no significant mitochondrial effects on actuarial ageing in females (i.e. longevity and survival) or on their larval offspring survival (Table S1). The average longevity of females was 38 days (±0.54 SE) with more fecund females living longer (Table S1).
Figure 2.
Daily egg-laying rate declines at a lower rate in females with a coadapted mitonuclear genotype (A), resulting in a higher lifetime reproductive success compared to females with a novel mitonuclear genotype (B). Figures show model predictions.
Senescence in males
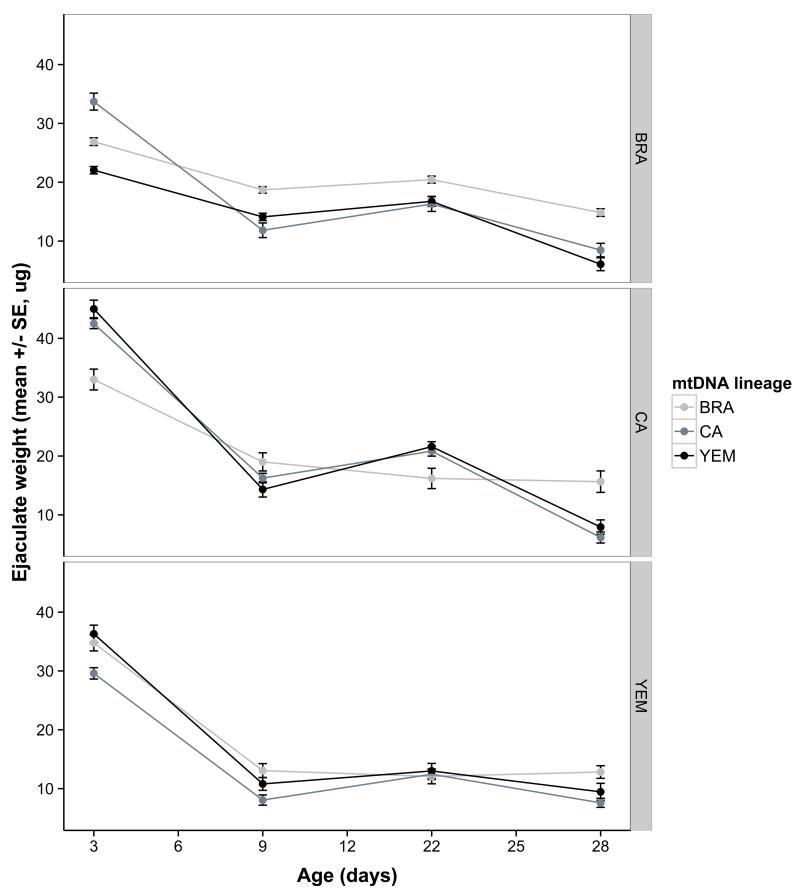
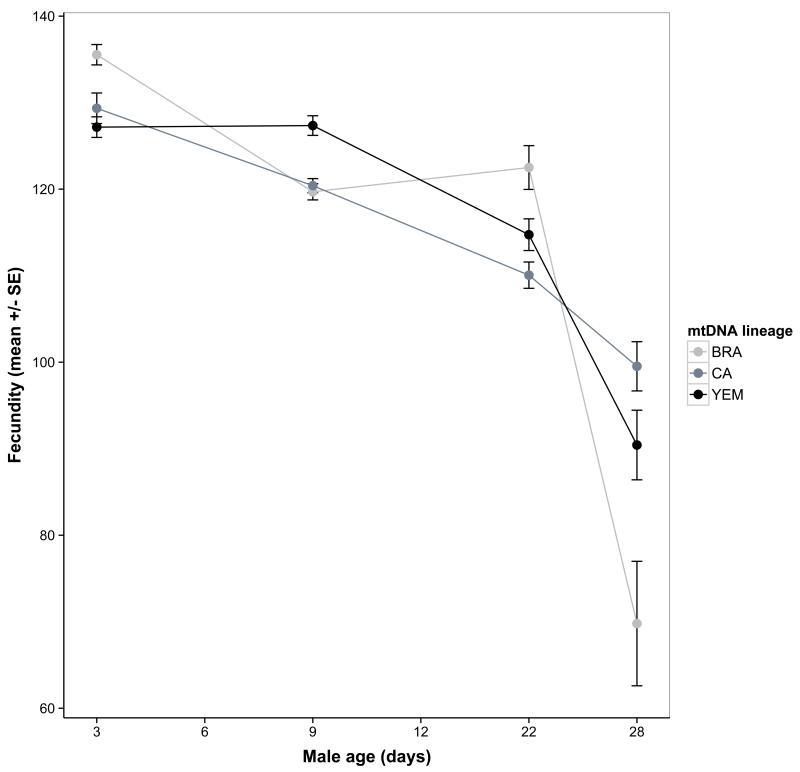
We first tested for direct mitochondrial and mitonuclear genetic effects on ageing phenotypes expressed by the males. Mitochondrial haplotype and mitonuclear combination both significantly influenced average ejaculate weight, and also its pattern of age-specific decline (Table 1, Figure 3). As for females, there were no significant direct mitochondrial genetic effects on male longevity or survival (Table S2). Males outlived the females from the introgression lines, with an average lifespan of 45 days (±1.49 SE) compared to 38 days for females.
Figure 3.
Direct mitonuclear epistatic effects on age-specific male ejaculate weight. Means represent model predictions.
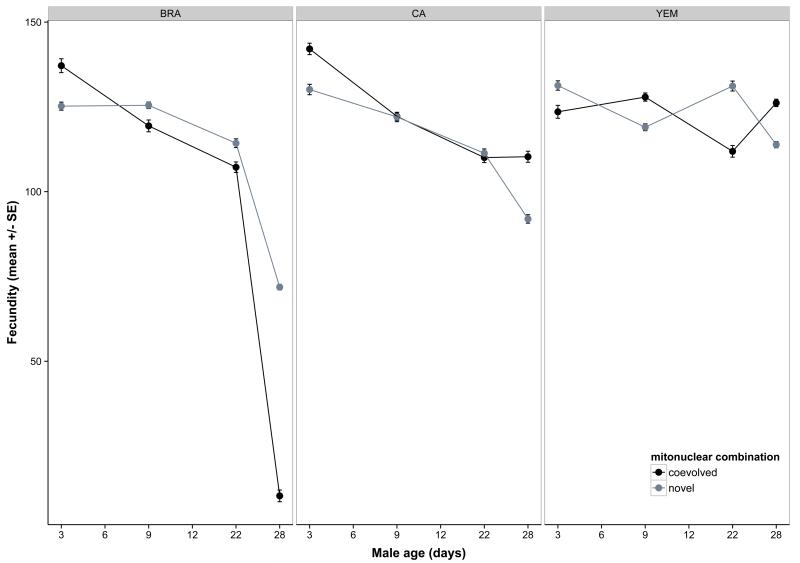
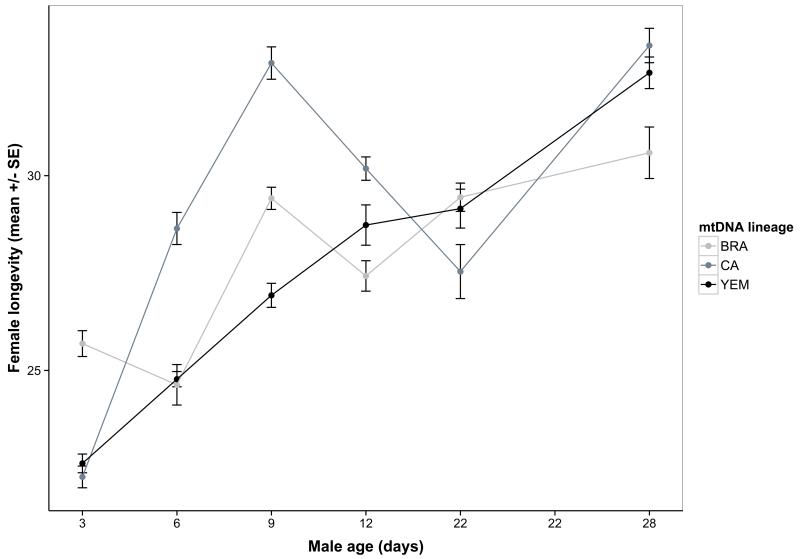
The second aim was to test for a role for mitonuclear genotype on the indirect effects of male senescence upon reproduction and/or actuarial ageing in their mates. We found widespread evidence for both. First, male ability to stimulate female fecundity declined with age, and this decline was contingent upon both mitochondrial and nuclear lineage (Table 3, Figure 4a-b). Here, variation in the two genomes primarily acted additively, as we found no significant mitonuclear interaction (Table 3). However, epistasis was apparent when modelled in terms of coevolutionary history: mitonuclear coadaptation had a strong impact on age-specific effects (coadaptation× nuc × age: χ26=23.4; P=0.0007, Figure 4b). Male ejaculate weight showed a strong positive effect on female fecundity (Table 3) but there was no interaction effect on this relationship due to the genetic factors (interactions not presented). This suggests that although ejaculate weight is an important determinant of male reproductive success and subject to mitonuclear epistasis itself, this relationship does not explain the indirect effect of male genotype on female fecundity. Second, female egg size significantly increased with male age (Table 3, Figure S3a), and this effect depended upon mitonuclear combination (Table 3, Figure S3b). When we included female fecundity into the model of egg size it removed the effect of male age (fecundity: χ21=217.8; P<0.00001; male age: χ22=4.3; P=0.12). This suggests that the male age effect on egg size was mediated by a maternal size-number trade-off, induced by the observed effect of male age on female fecundity. However, a mitonuclear interaction effect remained significant (χ24=10.5; P=0.033). Third, there was little evidence of male ageing influence on egg fertility, but the mitonuclear genotype had a marginally non-significant effect on age-dependent fertility (p=0.053, Table 2). However, we found some evidence for mito-nuclear epistasis, as there was a significant effect of mitonuclear coadaptation on age-dependent fertility that differed between the nuclear lineages (coevolution× nuc ×age: χ210=19.5; P=0.034). Fourth, increasing male age had a positive effect on female survival and there was also a marginally non-significant age × mitochondrial lineage interaction (P=0.055, Table 3, Figure S3). This interaction was significant for female longevity (P=0.047, Figure 5a), even when controlling for the strong negative effect of fecundity on female lifespan (Table 3). As for male effects on female fecundity, mitonuclear epistasis for female mates’ lifespan was evident when modelled in terms of male mitonuclear coevolutionary history: there was both a significant main effect of mitonuclear coadaptation (coevolution:χ21=4.5; P=0.0336) and an interaction with male age (coevolution × age: χ25=13.6; P=0.0183) that also depended on the nuclear lineage (coevolution × nuc × age: χ210=23.1; P=0.0105, Figure 5b).
Table 3.
The indirect effects of male age, mitochondrial and nuclear genetic variation on female actuarial ageing.
| Response variable: | Female longevity N = 945 | Female survival N = 945 | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
|
| ||||||||||
| Fixed effects | Coef | se | χ 2 | Df | P | Coef | se | χ 2 | Df | P |
| Male age | 6.9 | 5 | 0.2273 | 25.8 | 5 | <0.0001 | ||||
| Female weight | 0.36 | 0.38 | 0.9 | 1 | 0.3520 | −0.0003 | 0.0004 | 1.3 | 1 | 0.2456 |
| Lifetime fecundity | −2.07 | 0.42 | 23.9 | 1 | <0.0001 | 0.005 | 0.001 | 47.5 | 1 | <0.0001 |
| Mitochondrial lineage | 4.7 | 2 | 0.0936 | 3.3 | 2 | 0.1953 | ||||
| Nuclear lineage | 1.7 | 2 | 0.4270 | 0.3 | 2 | 0.8810 | ||||
| Mito × Nuc | 2.7 | 4 | 0.6078 | 1.4 | 4 | 0.8457 | ||||
| Age × Mito | 18.5 | 10 | 0.0466 | 18.0 | 10 | 0.0553 | ||||
| Age × Nuc | 15.7 | 10 | 0.1098 | 10.7 | 10 | 0.3836 | ||||
| Age × Mito × Nuc | 27.7 | 20 | 0.1159 | 24.6 | 20 | 0.2155 | ||||
|
| ||||||||||
| Random effects | Variance (σ2) | Variance (σ2) | ||||||||
|
| ||||||||||
| Line | 0.00 | 0.0042 | ||||||||
| Male ID | 1.03 | 0.19 | ||||||||
Figure 4.
Indirect effects of male age and mitonuclear genotype on female reproductive traits. Male age affects female fecundity but the decline over male age differs across mtDNA haplotypes (A) and depends upon an interaction between mitonuclear coadaptation and the nuclear genetic background (B). Male age also influences female egg size, an effect which differs across male mitonuclear genotypes (C). The different nuclear lineages are shown in panels. Means represent model predictions.
Table 2.
The indirect effects of male age, mitochondrial and nuclear genetic variation upon reference female fecundity, egg size and fertility.
| Response variable: | Fecundity N = 621 | Egg size N = 7971 | Fertility N = 954 | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
| |||||||||||||||
| Fixed effects | Coef | se | χ 2 | Df | P | Coef | se | χ 2 | Df | P | Coef | se | χ 2 | Df | P |
| Male age | 82.3 | 3 | <0.0001 | 11.8 | 2 | 0.0027 | 3.2 | 5 | 0.6737 | ||||||
| Female weight | 0.03 | 0.02 | 2.2 | 1 | 0.1377 | 3.21 | 0.75 | 18.3 | 1 | <0.0001 | 0.03 | 0.03 | 1.3 | 1 | 0.2525 |
| Ejaculate weight | 0.62 | 0.17 | 13.7 | 1 | <0.0001 | −1.50 | 1.24 | 1.4 | 1 | 0.2320 | - | - | - | - | - |
| Mitochondrial lineage | 3.2 | 2 | 0.2062 | 1.9 | 2 | 0.3866 | 0.3 | 2 | 0.8623 | ||||||
| Nuclear lineage | 0.3 | 2 | 0.8524 | 5.2 | 2 | 0.0750 | 5.9 | 2 | 0.0531 | ||||||
| Mito × Nuc | 4.3 | 4 | 0.3661 | 3.0 | 4 | 0.5568 | 5.8 | 4 | 0.2152 | ||||||
| Age × Mito | 27.2 | 6 | <0.0001 | 17.5 | 4 | 0.0015 | 8.3 | 10 | 0.6031 | ||||||
| Age × Nuc | 39.2 | 6 | <0.0001 | 15.9 | 4 | 0.0031 | 22.0 | 10 | 0.0150 | ||||||
| Age × Mito × Nuc | 15.4 | 12 | 0.2183 | 19.6 | 8 | 0.0117 | 31.2 | 20 | 0.0527 | ||||||
|
| |||||||||||||||
| Random effects | Variance (σ2) | Variance (σ2) | Variance (σ2) | ||||||||||||
|
| |||||||||||||||
| Line | 0.00 | 1.51 × 10−12 | 0.00 | ||||||||||||
| Male ID | 74.61 | 2.4 | 0.04 | ||||||||||||
| Female ID | - | 21.2 | - | ||||||||||||
Figure 5.
Female longevity increases with decreasing male age, however this general pattern also depends upon the male mtDNA haplotype (A) and an interaction between mitonuclear coadaptation and the nuclear genetic background (B). Means represent model predictions.
Discussion
Evolutionary theories incorporating sex-specific selection predict that males and females should age differently (Bonduriansky et al, 2008) and mitochondrial genetics should play a role in influencing these patterns (Frank and Hurst, 1996; Gemmell et al, 2004). Here we demonstrate widespread mitochondrial genetic effects on reproductive ageing in both sexes. Our results are broadly consistent with our key predictions. First, mitochondrial genetic effects were typically contingent upon the nuclear genetic background, but the nature of this mitonuclear epistasis was sex-specific. Disruption of potentially coadapted mitonuclear combinations resulted in a general increased rate of female fecundity senescence. In males, the same disruption resulted in more haphazardous effects that were contingent upon the nuclear genome, male age and the trait in question. Second, mitonuclear epistasis was associated with indirect genetic effects, such that genotypic effects on reproductive senescence in males affected their female mating partners’ fitness. Here, we discuss the implications of these patterns for the evolution of sex-specific reproductive strategies and the role of sexual conflict in ageing and maternal investment.
Sex-specific mitonuclear effects
Evolution of different optimal reproductive strategies in males and females should lead to sex-differences in the rate of senescence (Bonduriansky et al, 2008; Maklakov and Lummaa, 2013; Promislow, 2003). Empirical studies have found that sexual dimorphism in senescence is variable between species and across traits (Chen and Maklakov, 2014; Cornwallis et al, 2014; Promislow, 1992). Much of this variation likely owes to interspecific differences but rates of sex-specific senescence vary even within species. This has been suggested to result from ecological conditions, differences in age-specific intensity of sexual selection (Bonduriansky et al, 2008) or from intersexual genetic correlations constraining the evolution of sex-specific optima for life-history traits (Bonduriansky and Chenoweth, 2009). Asymmetric inheritance of mitochondrial genes imposes an extreme form of genetic correlation and our results demonstrate that sex-specific variation in ageing can arise from variation in mitonuclear genotypes (Maklakov and Lummaa, 2013). Although such effects have been considered for sex-differences in lifespan (Camus et al, 2012; Frank, 2012; Maklakov and Lummaa, 2013), to our knowledge our results are the first comprehensive report of mitochondrial and mitonuclear effects on sex-specific reproductive senescence.
The observed sex differences in mitochondrial genetic effects align well with theoretical predictions based on maternal inheritance of mitochondria (Gemmell et al, 2004). Female-specific selection should thus optimise the mitochondrial function by mitonuclear coadaptation across populations. While selection on the nuclear components of mitochondrial function may be quite strong in males (Gallach and Betran, 2011), this is a relatively weak engine of male-specific mitonuclear coadaptation (Wade, 2014). Our results show that females substantiate this prediction: across the nuclear genotypes, introgression of any novel mitochondrial haplotype hampered lifetime fecundity by accelerating ageing (Figure 2, S1). This result is also in line with an increasing number of studies that show non-neutral mitochondrial effects in females (e.g. Dobler et al, 2014; Kazancioglu and Arnqvist, 2013; Kurbalija Novicic et al, 2014; Lovlie et al, 2014; Maklakov et al, 2006). We note, however, that mitonuclear genetic variation had no significant effect on female fertility, despite the fact that fertility also showed marked ageing (Figure S2).
Several facts suggest that optimal mitochondrial function differs between the sexes (Gallach and Betran, 2011; Gemmell et al, 2004; Innocenti et al, 2011), which may help explain why male-specific mitonuclear epistasis often tends to reflect genotypic idiosyncrasies rather than general coadaptation (Dobler et al, 2014; Maklakov and Lummaa, 2013). The lack of cosegregation of mitochondrial and nuclear genes in the male germ line, coupled with sex-specific selection (Rand et al, 2001), can constrain male-specific mitonuclear coadaptation (Bonduriansky and Chenoweth, 2009; Gemmell et al, 2004; Wade, 2014) and also contribute to the maintenance of genetic variation in male phenotypes under selection. The extent to which this is true will depend on how often mitochondrial function underlies male-specific phenotypes. Our results demonstrate the existence of such pathways in C. maculatus for traits that influence male age-specific reproductive fitness such as ejaculate weight and male fecundity stimulation (Figures 3-5).
Consistent with the above, we found that the age-specific effects of particular mitonuclear combinations are rather idiosyncratic in males. For example, the mtDNA haplotype ‘BRA’ showed an elevated male ability to stimulate female fecundity early in life when coexpressed with its native nuclear genome. However, this effect declined late in life but was rescued by either of the two novel nuclear genomes (Figure 4b). It is interesting to note that this rather dramatic ageing in the male performance for the ‘BRA-RBA’ genotype is reversed for the females of the same genotype that showed decreased rate of ageing and higher relative fecundity (Figure S1a), which implies sexually antagonistic selection (Rand et al, 2001). In contrast, the ‘CA’ haplotype showed an elevated male fecundity stimulation in its native nuclear background both early and late in life, relative to the other haplotypes, whereas the ‘YEM’ nuclear lineage showed much less senescence overall. The fact that YEM nuclear lineage was capable of maintaining its performance suggests that senescence in this trait is not inevitable, but is a result of mitonuclear epistasis. Changes in ejaculate weight with age (Figure 3) do not coincide with those seen for male effects on female fecundity (Figure 4a-b), despite having an overall positive effect on female fecundity (Table 2). This suggests that age-specific male ability to stimulate female fecundity is a result of changes in ejaculate composition rather than ejaculate volume. We note that an apparent lack of mitonuclear coadaptation in males was also recently documented by Yee et al (2015), who showed that the rate of decline in male mating propensity differs across mitochondrial genotypes in Drosophila.
Although there is now ample evidence for a role of mitochondrial malfunctioning in actuarial ageing (e.g. Camus et al, 2012; Cho et al, 2011; Houtkooper et al, 2013; Lee and Wei, 2012; Zhu et al, 2014), our data revealed that mitochondrial genetic effects on longevity and survival were weak and non-significant relative to age-specific reproduction in both sexes of C. maculatus. This suggests that mitonuclear genetic effects on senescence are stronger for reproductive function than somatic maintenance governing lifespan, presumably reflecting the fact that reproduction is particularly sensitive to cellular malfunctioning. We have, however, previously found some evidence of mitonuclear epistasis on female lifespan under aphagous conditions (Dowling et al, 2010), which is in line with a recent report that lifespan is subject to complex mitonuclear-by-environment interactions (Zhu et al, 2014).
Indirect genetic effects on female reproduction
Post-copulatory sexual selection in males has selected for a suite of male traits that influence female reproductive physiology and fitness, and many female reproductive traits may thus be subject to indirect genetic effects through males (Clark et al, 1999; Edward et al, 2014). To our knowledge, our results are first to demonstrate that such indirect genetic effects can involve mtDNA. We found that mitochondrial genetic variation interacted with male age to influence female fecundity and investment into eggs (i.e., egg size, Figure S3). Females mated to young males traded a high fecundity against a lower investment per egg and vice versa for mates of older males. In Drosophila, male seminal fluid proteins have well-established effects on female egg laying rate (Avila et al, 2011; Chapman et al, 2001). We have recently identified >100 seminal fluid proteins in C. maculatus, several of which affects female fecundity (Goenaga et al. in review; Yamane et al, 2015). It is likely that the effect of male age on female fitness seen in our experiments result from changes in the seminal fluid composition of male ejaculates. Because mitonuclear genotype influences the metabolic pathways that fuel biosynthesis of seminal fluid molecules (Immonen et al, 2015) the indirect genetic effects seen in our experiments likely reflect differences across mitonuclear genotypes in age-related changes in seminal fluid composition of males. Interestingly, we also found evidence that female cost of reproduction was influenced by male mitochondrial genetic variation in interaction with age: male age affected female lifespan and this effect differed across mtDNA haplotypes (Figures 5a and S2), even when controlling for female reproductive effort. This lends further support for the interpretation that mitochondrial function affects ejaculate mediated indirect genetic effects on females. Seminal fluid molecules influence female lifespan as a toxic side effect (Avila et al, 2011), an effect that may change with male age due to deterioration in accessory gland function.
Taken together, our results suggest that ageing in C. maculatus represents both a cause and a consequence of sexual conflict. Both males and females suffer reduced reproductive output when mating with an older partner, but female ageing is further under the influence of male traits, some of which also manipulate female reproduction (fecundity and egg size investment). The mitonuclear genetic effects seen here suggest that the underlying genetic architecture of the traits involved is complex and likely to constrain male-specific adaptive evolution. This may not only depress male but also the female mating partner’s reproductive success (Figure 4).
Conclusions
Our results demonstrate that mitonuclear epistatic effects are sex-specific and affect reproductive, but not actuarial, ageing in this species. Mitonuclear coadaptation decelerated reproductive senescence in females, resulting in higher female fitness, whereas coexpression of mitochondrial and nuclear genomes with a shared evolutionary history had more variable and idiosyncratic effects in males. Intriguingly, we found age-specific indirect genetic effects of male mtDNA haplotype on female fecundity, egg size and longevity. This suggests an important role for mitochondrial pathways for both female fecundity and male reproduction, which may be mediated through male seminal fluid substances. More generally, our results illustrate the importance of considering reproductive phenotypes in both sexes as well as the nuclear genetic background in studies examining the role of mitochondria in ageing.
Supplementary Material
Acknowledgments
We are grateful to Anna Qvarnström, Alexei Maklakov, Arild Husby and Björn Rogell for helpful comments on this manuscript. We also thank Ana Che Guevara for assistance with the data collection. This work was funded by the European Research Council (AdG-294333) and the Swedish Research Council (621-2010-5266) for G. Arnqvist.
Footnotes
Conflict of interest
The authors declare no conflicts of interest.
Data availability
The data will be deposited to Dryad (http://datadryad.org) upon acceptance.
References
- Arnqvist G, Dowling DK, Eady P, Gay L, Tregenza T, Tuda M, et al. Genetic architecture of metabolic rate: environment specific epistasis between mitochondrial and nuclear genes in an insect. Evolution. 2010;64(12):3354–3363. doi: 10.1111/j.1558-5646.2010.01135.x. [DOI] [PubMed] [Google Scholar]
- Arnqvist G, Rowe L. Sexual Conflict. Princeton University Press; New Jersey: 2005. [Google Scholar]
- Avila FW, Sirot LK, LaFlamme BA, Rubinstein CD, Wolfner MF. Insect seminal fluid proteins: identification and function. Annu Rev Entomol. 2011;56:21–40. doi: 10.1146/annurev-ento-120709-144823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bates D, Maechler M, Bolker BM, Walker S. lme4: Linear mixed-effects models using Eigen and S4. R package. 1.1-7. 2014 http://CRAN.R-project.org/package=lme4.
- Bonduriansky R, Chenoweth SF. Intralocus sexual conflict. Trends Ecol Evol. 2009;24(5):280–288. doi: 10.1016/j.tree.2008.12.005. [DOI] [PubMed] [Google Scholar]
- Bonduriansky R, Maklakov A, Zajitschek F, Brooks R. Sexual selection, sexual conflict and the evolution of ageing and life span. Funct Ecol. 2008;22(3):443–453. [Google Scholar]
- Calvo SE, Mootha VK. The mitochondrial proteome and human disease. Annu Rev Genom Hum Genet. 2010;11:25–44. doi: 10.1146/annurev-genom-082509-141720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camus MF, Clancy DJ, Dowling DK. Mitochondria, maternal inheritance, and male aging. Curr Biol. 2012;22(18):1717–1721. doi: 10.1016/j.cub.2012.07.018. [DOI] [PubMed] [Google Scholar]
- Chapman T, Herndon LA, Heifetz Y, Partridge L, Wolfner MF. The Acp26Aa seminal fluid protein is a modulator of early egg hatchability in Drosophila melanogaster. Proc R Soc Biol Sci Ser B. 2001;268(1477):1647–1654. doi: 10.1098/rspb.2001.1684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen HY, Maklakov AA. Condition dependence of male mortality drives the evolution of sex differences in longevity. Curr Biol. 2014;24(20):2423–2427. doi: 10.1016/j.cub.2014.08.055. [DOI] [PubMed] [Google Scholar]
- Cho J, Hur JH, Walker DW. The role of mitochondria in Drosophila aging. Exp Gerontol. 2011;46(5):331–334. doi: 10.1016/j.exger.2010.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark AG, Begun DJ, Prout T. Female × male interactions in Drosophila sperm competition. Science. 1999;283(5399):217–220. doi: 10.1126/science.283.5399.217. [DOI] [PubMed] [Google Scholar]
- Cornwallis CK, Dean R, Pizzari T. Sex-specific patterns of aging in sexual ornaments and gametes. Am Nat. 2014;184(3):E66–78. doi: 10.1086/677385. [DOI] [PubMed] [Google Scholar]
- Dobler R, Rogell B, Budar F, Dowling DK. A meta-analysis of the strength and nature of cytoplasmic genetic effects. J Evol Biol. 2014;27(10):2021–2034. doi: 10.1111/jeb.12468. [DOI] [PubMed] [Google Scholar]
- Dowling DK, Abiega KC, Arnqvist G. Temperature-specific outcomes of cytoplasmic-nuclear interactions on egg-to-adult development time in seed beetles. Evolution. 2007a;61(1):194–201. doi: 10.1111/j.1558-5646.2007.00016.x. [DOI] [PubMed] [Google Scholar]
- Dowling DK, Meerupati T, Arnqvist G. Cytonuclear interactions and the economics of mating in seed beetles. Am Nat. 2010;176(2):131–140. doi: 10.1086/653671. [DOI] [PubMed] [Google Scholar]
- Dowling DK, Nowostawski AL, Arnqvist G. Effects of cytoplasmic genes on sperm viability and sperm morphology in a seed beetle: implications for sperm competition theory? J Evol Biol. 2007b;20(1):358–368. doi: 10.1111/j.1420-9101.2006.01189.x. [DOI] [PubMed] [Google Scholar]
- Edvardsson M, Tregenza T. Why do male Callosobruchus maculatus harm their mates? Behav Ecol. 2005;16(4):788–793. [Google Scholar]
- Edward DA, Poissant J, Wilson AJ, Chapman T. Sexual conflict and interacting phenotypes: A quantitative genetic analysis of fecundity and copula duration in Drosophila Melanogaster. Evolution. 2014;68(6):1651–1660. doi: 10.1111/evo.12376. [DOI] [PubMed] [Google Scholar]
- Elston DA, Moss R, Boulinier T, Arrowsmith C, Lambin X. Analysis of aggregation, a worked example: numbers of ticks on red grouse chicks. Parasitology. 2001;122(Pt 5):563–569. doi: 10.1017/s0031182001007740. [DOI] [PubMed] [Google Scholar]
- Finkel T, Holbrook NJ. Oxidants, oxidative stress and the biology of ageing. Nature. 2000;408(6809):239–247. doi: 10.1038/35041687. [DOI] [PubMed] [Google Scholar]
- Fournier DA, Skaug HJ, Ancheta J, Ianelli J, Magnusson A, Maunder MN, et al. AD Model Builder: using automatic differentiation for statistical inference of highly parameterized complex nonlinear models. Optim Meth Softw. 2012;27(2):233–249. [Google Scholar]
- Fox CW. Multiple mating, lifetime fecundity and female mortality of the Bruchid beetle, Callosobruchus maculatus (Coleoptera, Bruchidae) Funct Ecol. 1993;7(2):203–208. [Google Scholar]
- Fox J, Weisberg S. An {R} Companion to Applied Regression. Second edn. Sage; 2011. [Google Scholar]
- Frank SA. Evolution: mitochondrial burden on male health. Curr Biol. 2012;22(18):R797–799. doi: 10.1016/j.cub.2012.07.066. [DOI] [PubMed] [Google Scholar]
- Frank SA, Hurst LD. Mitochondria and male disease. Nature. 1996;383(6597):224. doi: 10.1038/383224a0. [DOI] [PubMed] [Google Scholar]
- Gallach M, Betran E. Intralocus sexual conflict resolved through gene duplication. Trends Ecol Evol. 2011;26(5):222–228. doi: 10.1016/j.tree.2011.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gemmell NJ, Metcalf VJ, Allendorf FW. Mother's curse: the effect of mtDNA on individual fitness and population viability. Trends Ecol Evol. 2004;19(5):238–244. doi: 10.1016/j.tree.2004.02.002. [DOI] [PubMed] [Google Scholar]
- Houtkooper RH, Mouchiroud L, Ryu D, Moullan N, Katsyuba E, Knott G, et al. Mitonuclear protein imbalance as a conserved longevity mechanism. Nature. 2013;497(7450):451–457. doi: 10.1038/nature12188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Immonen E, Rönn J, Watson C, Berger D, Arnqvist G. Complex mitonuclear interactions and metabolic costs of mating in male seed beetles. J Evol Biol. 2015 doi: 10.1111/jeb.12789. Accepted. [DOI] [PubMed] [Google Scholar]
- Innocenti P, Morrow EH, Dowling DK. Experimental evidence supports a sex-specific selective sieve in mitochondrial genome evolution. Science. 2011;332(6031):845–848. doi: 10.1126/science.1201157. [DOI] [PubMed] [Google Scholar]
- Kazancioglu E, Arnqvist G. The maintenance of mitochondrial genetic variation by negative frequency-dependent selection. Ecol Lett. 2013;17(1):22–27. doi: 10.1111/ele.12195. [DOI] [PubMed] [Google Scholar]
- Kurbalija Novicic Z, Immonen E, Jelic M, AnEthelkovic M, Stamenkovic-Radak M, Arnqvist G. Within-population genetic effects of mtDNA on metabolic rate in Drosophila subobscura. J Evol Biol. 2014 doi: 10.1111/jeb.12565. [DOI] [PubMed] [Google Scholar]
- Lee HC, Wei YH. Mitochondria and aging. Adv Exp Med Biol. 2012;942:311–327. doi: 10.1007/978-94-007-2869-1_14. [DOI] [PubMed] [Google Scholar]
- Liu Z, Butow RA. Mitochondrial retrograde signaling. Annu Rev Genet. 2006;40:159–185. doi: 10.1146/annurev.genet.40.110405.090613. [DOI] [PubMed] [Google Scholar]
- Lovlie H, Immonen E, Gustavsson E, Kazancioglu E, Arnqvist G. The influence of mitonuclear genetic variation on personality in seed beetles. P Roy Soc B-Biol Sci. 2014;281(1796):20141039. doi: 10.1098/rspb.2014.1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maklakov AA, Friberg U, Dowling DK, Arnqvist G. Within-population variation in cytoplasmic genes affects female life span and aging in Drosophila melanogaster. Evolution. 2006;60(10):2081–2086. [PubMed] [Google Scholar]
- Maklakov AA, Lummaa V. Evolution of sex differences in lifespan and aging: causes and constraints. Bioessays. 2013;35(8):717–724. doi: 10.1002/bies.201300021. [DOI] [PubMed] [Google Scholar]
- Monaghan P, Charmantier A, Nussey DH, Ricklefs RE. The evolutionary ecology of senescence. Funct Ecol. 2008;22(3):371–378. [Google Scholar]
- Moore AJ, Brodie ED, Wolf JB. Interacting phenotypes and the evolutionary process .1. Direct and indirect genetic effects of social interactions. Evolution. 1997;51(5):1352–1362. doi: 10.1111/j.1558-5646.1997.tb01458.x. [DOI] [PubMed] [Google Scholar]
- Promislow D. Mate choice, sexual conflict, and evolution of senescence. Behav Genet. 2003;33(2):191–201. doi: 10.1023/a:1022562103669. [DOI] [PubMed] [Google Scholar]
- Promislow DEL. Costs of sexual selection in natural-populations of mammals. P Roy Soc B-Biol Sci. 1992;247(1320):203–210. [Google Scholar]
- Rand DM, Clark AG, Kann L. Sexually antagonistic cytonuclear fitness interactions in Drosophila melanogaster. Genetics. 2001;159:173–187. doi: 10.1093/genetics/159.1.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rand DM, Fry A, Sheldahl L. Nuclear-mitochondrial epistasis and drosophila aging: introgression of Drosophila simulans mtDNA modifies longevity in D. melanogaster nuclear backgrounds. Genetics. 2006;172(1):329–341. doi: 10.1534/genetics.105.046698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rand DM, Haney RA, Fry AJ. Cytonuclear coevolution: the genomics of cooperation. Trends Ecol Evol. 2004;19(12):645–653. doi: 10.1016/j.tree.2004.10.003. [DOI] [PubMed] [Google Scholar]
- RDevelopmentCoreTeam R: a language and environment for statistical computing. 2011.
- Thernau T. coxme: Mixed effects cox models. R package. 2.2-3. 2012 http://CRAN.R-project.org/package=coxme.
- Tower J. Mitochondrial maintenance failure in aging and role of sexual dimorphism. Arch Biochem Biophys. 2014;576:17–31. doi: 10.1016/j.abb.2014.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuda M, Ronn J, Buranapanichpan S, Wasano N, Arnqvist G. Evolutionary diversification of the bean beetle genus Callosobruchus (Coleoptera: Bruchidae): traits associated with stored-product pest status. Mol Ecol. 2006;15(12):3541–3551. doi: 10.1111/j.1365-294X.2006.03030.x. [DOI] [PubMed] [Google Scholar]
- Wade MJ. Paradox of mother's curse and the maternally provisioned offspring microbiome. CSH Persp Biol. 2014;6(10):a017541. doi: 10.1101/cshperspect.a017541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamane T, Goenaga J, Ronn JL, Arnqvist G. Male seminal fluid substances affect sperm competition success and female reproductive behavior in a seed beetle. PLoS ONE. 2015;10(4):e0123770. doi: 10.1371/journal.pone.0123770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yee WK, Rogell B, Lemos B, Dowling DK. Intergenomic interactions between mitochondrial and Y-linked genes shape male mating patterns and fertility in Drosophila melanogaster. Evolution. 2015 doi: 10.1111/evo.12788. doi: 10.1111/evo.12788.[Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- Yee WKW, Sutton KL, Dowling DK. In vivo male fertility is affected by naturally occurring mitochondrial haplotypes. Curr Biol. 2013;23(2):R55–R56. doi: 10.1016/j.cub.2012.12.002. [DOI] [PubMed] [Google Scholar]
- Zhu CT, Ingelmo P, Rand DM. G×G×E for lifespan in Drosophila: mitochondrial, nuclear, and dietary interactions that modify longevity. PLoS Genet. 2014;10(5):e1004354. doi: 10.1371/journal.pgen.1004354. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.