Abstract
Due to its special geological history, the New Caledonian Archipelago is a mosaic of soil types, and in combination with climatic conditions this results in a heterogeneous environment across relatively small distances. A group of over 20 endemic species of Diospyros (Ebenaceae) has rapidly and recently radiated on the archipelago after a single long-distance dispersal event. Most of the Diospyros species in the radiating group are morphologically and ecologically well differentiated, but they exhibit low levels of DNA variability. To investigate the processes that shaped the diversification of this group we employed restriction site associated DNA sequencing (RADseq). Over 8,400 filtered SNPs generally confirm species delimitations and produce a well-supported phylogenetic tree. Our analyses document local introgression, but only a limited potential for gene flow over longer distances. The phylogenetic relationships point to an early regional clustering among populations and species, indicating that allopatric speciation with respect to macrohabitat (i.e., climatic conditions) may have had a role in the initial differentiation within the group. A later, more rapid radiation involved divergence with respect to microhabitat (i.e., soil preference). Several sister species in the group show a parallel divergence in edaphic preference. Searches for genomic regions that are systematically differentiated in this replicated phenotypic divergence pointed to loci potentially involved in ion binding and cellular transport. These loci appear meaningful in the context of adaptations to soil types that differ in heavy-metal and mineral content. Identical nucleotide changes affected only two of these loci, indicating that introgression may have played a limited role in their evolution. Our results suggest that both allopatric diversification and (parapatric) ecological divergence shaped successive rounds of speciation in the Diospyros radiation on New Caledonia.
Keywords: adaptive radiation, Diospyros, hybridization, New Caledonia, RADsequencing, soil adaptation
Biogeographic regions with significant species richness (i.e., biodiversity hotspots; Willis et al. 2007), and ecological-opportunistic, explosive diversifications of one lineage into an array of new species (i.e., adaptive radiations; Gavrilets and Losos 2009) promise to provide tremendous evolutionary insights. The accumulation of biodiversity within a particular group or a geographic area opens the way to integrative studies into the drivers of evolutionary opportunity and the ecological and genetic limits of adaptation (Losos 2010). However, the complexity of evolutionary relationships in such cases poses significant challenges to phylogenetic inference (Glor 2010), which is required as starting point for further studies of the evolutionary processes shaping observed species richness.
Adaptive radiations are generally linked to ecological opportunity, often starting from long-distance dispersal events, when an alien lineage takes advantage of an array of newly formed or previously unfilled niches (Glor 2010). Mathematical models of adaptive radiation point to a rapid burst of speciation events soon after the beginning of radiations (Gavrilets and Vose 2005), in time resulting in overshooting (decreasing speciation and/or increasing extinction rates; Gavrilets and Losos 2009). However, in the case of isolated areas (e.g., oceanic islands) the initial founder population is likely to have an extremely small effective size, with only limited variation that can be selected and potentially result in novel and divergent adaptations. Due to low levels of genetic variation and small population size, the evolution of island biotas is hence intuitively expected to be shaped by neutral processes rather than natural selection (Ohta 2002). In particular for organisms with long generation time, such as trees, the initial stages of radiation are expected to be notably difficult, and the evolutionary processes shaping such radiations are little understood.
We analyse here a putative radiation of a tropical tree (Diospyros, Ebenaceae) on New Caledonia, an archipelago in the Pacific, about 1,600 km east of Australia. The continental part of New Caledonia separated from East Gondwana in the late Cretaceous (ca 80 million years ago - Ma; McLoughlin 2001). During the Paleocene and early to late Eocene (65 to 37 Ma), the Archipelago was submerged, and a thick layer of oceanic sediments accumulated over its surface (Pelletier 2006). After it re-emerged in the late Eocene, a heavy-metal rich oceanic material covered the land area, and due to later erosion around one third of the main island is still overlaid today with ultramafic substrates (i.e., heavy metal rich). Due to this complex geological history, New Caledonia displays a mosaic of soil types (Pelletier 2006; Maurizot and Vendé-Leclerc 2009), and in combination with its diverse terrain and climate this results in a heterogeneous environment across a fairly small area (i.e., roughly 60 × 400 km for the main island; Fig. 1). As a result, the Archipelago hosts an unusually concentrated level of biodiversity, and it is often cited as one of the areas with the highest rate of plant endemism (Myers et al. 2000; Mittermeier et al. 2004; Kier et al. 2009), with some genera and even families unique to these islands (Morat et al. 2012). Due to mining and agriculture, this biodiversity is under continuous human threat and New Caledonia is one of the regions with the highest predicted land loss as a result of climate change-driven increase of sea levels (Bellard 2013).
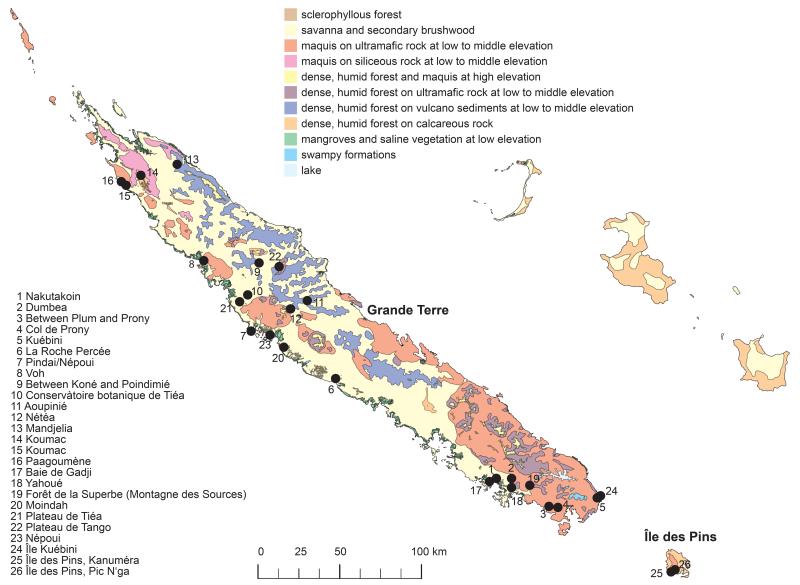
Figure1.
Map of New Caledonia indicating the 26 sampling localities for this study. Numbered dots indicate sampling sites (see also Table 1). The colours indicate the vegetation types (Jaffré et al. 2012) according to the legend insert.
Diospyros is a large pantropical genus of woody plants that colonised New Caledonia via long-distance dispersal at least four times during the last 25 million years (Duangjai et al. 2009). Three of these colonization events gave rise to only a small set of species (one to five species each; Turner et al. 2013a). However, another event gave rise to a clade of 24 closely related species that take advantage of all vegetation types on the Archipelago (except mangroves; Turner et al. 2013b). Most of these closely related species are morphologically and ecologically clearly differentiated, and only a few of them occur in local sympatry. Several of the species are point endemics. Due to extremely low levels of sequence divergence at several plastid and low-copy nuclear regions, the relationships between these species have previously proven difficult to clarify, even with extensive data sampling (Duangjai et al. 2009; Turner et al. 2013a; Turner et al. 2013b). However, delimitation of most species has been confirmed with amplified fragment length polymorphisms (AFLP; Turner et al. 2013b). Previous phylogenetic studies (Duangjai et al. 2009; Turner et al. 2013a) have shown this group to be related to species found on islands of the Indian Ocean and Hawai’i. Molecular dating based on combined plastid and low-copy nuclear DNA sequence data showed that the lineage forming this group of Diospyros species split from its sister species around nine Ma, with the majority of species diverging within the last two million years (Turner et al. 2013a). Taking into consideration that these are woody plants with generation time of ca. seven years (Verdú 2002), we can estimate there have been fewer than 1.3 million generations since the first members of the group arrived on the Archipelago. Such a time frame may be insufficient for genome-wide genetic differentiation among these species to have accumulated, especially at the scale of a relatively small area where interspecific gene flow may have been frequent.
Phenotypic novelties, reproductive isolation and adaptation to environmental conditions do not necessarily depend on large-scale genetic alterations; they can be due to divergence at only a few loci (Wu 2001; Lexer and Widmer 2008; Kane et al. 2009). Finding such relatively small differences within non-model genomes is challenging. Recently developed high-throughput DNA sequencing technologies and analysis algorithms provide an opportunity to study in detail genome-wide variability across groups of individuals, adding significant power to evolutionary investigations. In particular, restriction site associated DNA sequencing (RADseq; Baird et al. 2008) has been suggested as a powerful tool for interspecific comparisons across a large number of loci in non-model organisms (Cariou et al. 2013), delivering clear phylogenetic information even in the presence of incomplete lineage sorting (Wagner et al. 2013). We use RADseq here to attempt to resolve phylogenetic relationships among 21 (out of 24) diploid Diospyros species that recently and rapidly radiated on New Caledonia in order to identify the evolutionary processes that shaped this radiation.
Materials and Methods
Taxon Sampling and DNA isolation
Leaf material from New Caledonian Diospyros species was collected on Grande Terre and Île des Pins at 27 localities (Fig. 1, Table 1) and stored in silica gel. Herbarium vouchers are deposited in the herbaria of Nouméa (NOU), the University of Montpellier (MPU) and the University of Vienna (WU; see Table 1). Whenever possible, we aimed to investigate at least two individuals per sampling locality and a minimum of three individuals per species. Our dataset contains 84 individuals from 39 populations (Fig. 1, Table 1), including representatives of 21 species of New Caledonian Diospyros that have been previously shown to have radiated rapidly after a single long-distance dispersal event (Turner et al. 2013a). One of the studied species (collected at Île des Pins, Pic N'ga) could not be unambiguously identified - due to the absence of diagnostic reproductive organs at the time of collection -and is referred to as D. sp. Pic N'ga.
Table 1.
Details of the samples, populations and species included in the present study.
| Taxon | Sample ID | Sampling location1 | Voucher2 |
|---|---|---|---|
| D. calciphila F.White | BT313 BT314 BT317 |
25, littoral forest | JM6650, JM6653 (MPU, NOU, P) |
| D. cherrieri F.White | BT276 BT278 |
20, dry forest | NOU054492 NOU054008 |
| D. cherrieri | BT293 BT294 |
23, dry forest | NOU079547 |
| D. erudita F.White | BT280 BT281 |
21, dry forest | WU062858, Chambrey & Turner 22 (NOU) |
| D. flavocarpa (Vieill. ex P.Parm.) F.White | BT129 BT130 |
9, humid mountain forest | JM6625 (NOU) |
| D. flavocarpa | BT156 BT157 |
11, dense humid mountain forest | JM6632 (NOU) |
| D. glans F.White | BT093 BT094 |
5, forest near river | NOU022860 |
| D. impolita F.White | BT102 BT103 BT105 |
6, mesophyll forest near beach | NOU019538 |
| D. inexplorata F.White | BT308 BT310 BT311 |
24, littoral forest | NOU005818 |
| D. labillardierei F.White | BT122 BT125 |
9, river edge in mountain forest | JM6624 (NOU) |
| D. labillardierei | BT178 BT182 |
12, river edge | (NOU031346) |
| D. minimifolia F.White | BT131 BT135 |
10, dry forest | NOU019556 |
| D. minimifolia | BT232 BT233 |
17, mesophyll forest near beach | NOU019554 |
| D. minimifolia | BT263 BT269 |
20, dry forest | NOU079549, WU062872 NOU054493 |
| D. pancheri Kosterm. | BT028 BT031 BT035 |
3, forest near road | JM6619, JM6620 (NOU) |
| D. parviflora (Schltr.) Bakh. | BT038 BT041 BT042 |
4, wet forest | |
| D. parviflora | BT147 BT148 |
10, forest near river | JM6630 (NOU) |
| D. parviflora | BT187 | 13, mountain forest | JM6636 (NOU) |
| D. parviflora | BT250 | 19, humid forest at low elevation | tree no. 23109 |
| D. parviflora | BT289 BT290 BT291 |
22, mountain forest | NOU079550 |
| D. perplexa F.White | BT004 | 1, mesophyll forest | JM6611, JM6613 (NOU) |
| D. pustulata F.White | BT111 BT112 |
7, dry forest | |
| D. pustulata | BT137 BT140 |
10, dry forest | JM6629 (NOU) |
| D. pustulata | BT259 BT261 BT265 BT268 |
20, dry forest | WU062855 NOU079544, WU062870, NOU079548, WU062871 NOU053999 |
| D. revolutissima F.White | BT117 BT120 |
8, maquis | NOU023189 |
| D. revolutissima | BT219 BT221 |
16, maquis | JM6640 (NOU) |
| D. tridentata F.White | BT203 BT206 BT207 |
14, dry forest at low elevation | JM6639 (NOU) |
| D. trisulca F.White | BT185 BT192 BT199 BT201 |
13, mountain forest | NOU031344 JM6637 (NOU) |
| D. umbrosa F.White | BT176 BT177 |
12, dense humid forest | JM6635 (NOU) |
| D. umbrosa | BT197 | 13, mountain forest | |
| D. umbrosa | BT246 BT247 |
19, humid forest at low elevation | NOU023234 |
| D. veillonii F.White | BT224 BT226 BT227 |
17, mesophyll forest near beach | NOU019582 |
| D. vieillardii (Hiern) Kosterm. | BT025 BT026 |
2, forest near river | JM6618 (NOU) |
| D. vieillardii | BT088 BT100 |
5, forest near river | |
| D. vieillardii | BT215 BT217 |
15, maquis | NOU023242 |
| D. vieillardii | BT286 | 21, dry forest | |
| D. yahouensis (Schltr.) Kosterm. | BT238 BT239 |
18, mesophyll forest | P00057340 |
| D. sp. Pic N'ga | BT318 BT320 BT323 |
26, maquis | JM6065 (NOU) |
The identification number of sampling localities are following Figure 1.
Voucher-Codes: JMXXXX: collection number J. Munzinger; Tree N° XXX: Tree of New Caledonian Plant Inventory and Permanent Plot Network (NC-PIPPN, Ibanez et al. 2014); NOUXXXXXXX: Herbarium accession number of Noumea herbarium (NOU); WUXXXXXX: Herbarium accession number of the Herbarium of the University Vienna (WU); P: Herbarium of the Natural History Museum Paris; MPU: Herbarium of the University of Montpellier.
DNA extractions performed with a modified sorbitol/high-salt CTAB method (Tel-Zur et al. 1999) were already available from a previous study (Turner et al. 2013b). As we observed significant differences between standard Nanodrop (Thermo Scientific) and Quant-It Pico-Green (Life Technologies) quantifications of the DNA samples, these have been purified using the NucleoSpin gDNA clean-up kit (Macherey-Nagel), according to the manufacturer’s protocol.
RADseq Library Preparation
By using an average genome size in the target group of 1C = 1.9 pg (Turner et al. 2013a) and the RAD counter, available from www.wiki.ed.ac.uk/display/RADSequencing, we have estimated that 60 individually barcoded samples can be pooled together when using the SbfI restriction enzyme (New England Biolabs). Due to uneven representation of individuals within the library, a second RAD library was later prepared in order to increase the number of reads of selected samples to a minimum 1 million high-quality read pairs per individual and to add 24 additional individuals. The RAD libraries were prepared using a protocol adapted from Baird et al. (2008) with modifications. We started with 300 ng DNA per individual and used double barcoding to decrase the number of different adapters necessary. The six-base-pair P5 barcodes and, respectively, the four-base-pair P7 barcodes were chosen to differ by at least three bases from each other to avoid erroneous assignment due to sequencing error. We ligated 200 mM P5 adapters to the restricted samples overnight at 16 °C. Groups of samples barcoded with different P5 barcodes were pooled, and sheared by sonication with a Bioruptor Pico (Diagenode) to achieve an average size of ca. 400 bp, using two cycles of 55s “on” and 55s “off” at 6 °C. In order to obtain the optimal fragment sizes for Illumina sequencing, we have performed a left and right size selection with SPRIselect (Beckman Coulter), by using 0.7x and, respectively, 0.55x volume of SPRI reagent to sample. After ligating P7 adaptors, all samples were pooled after quantification, so that each sample would be equally represented. To remove unwanted primer dimers two size selections on the left side with 0.65x volume of SPRI reagent to sample have been finally performed: once before the 18 cycles PCR amplification in the Phusion Master Mix (Thermo Fischer Scientific), and again afterwards. The libraries were sequenced on an Illumina HiSeq at CSF Vienna (http://csf.ac.at/ngs/) as 100 bp paired-end reads.
Filtering SNPs from RADseq Data
The libraries were demultiplexed into individual samples according to the respective barcode combinations by allowing for single errors at the barcodes using the RADpools module of the RADtools v. 1.2.4 package (Baxter et al. 2011). The 84 individual files have been further imported in the CLC Genomic Workbench v. 6.5 (Qiagen) and trimmed/filtered to retain only full-length (i.e., 94 bp after barcode trimming) reads, free of any adaptor sequence and with all bases of a quality Phred score ≥30. The final high-quality filtered and demultiplexed data set contained over 160 million pairs of reads.
The forward reads were further used for running denovo_map.pl script of STACKS v. 1.12 (Catchen et al. 2011). To find the best settings for STACKS, we first varied the value of the minimum number of identical reads required for a stack to be formed (i.e., the setting “m”) from five to 15, by allowing one base pair difference between loci when processing one individual (i.e., the setting “M = 1”) and when building the catalogue (i.e., setting “n = 1”). We have chosen the value of m = 13 as the best for our data because it delivered the most polymorphic stacks with less than ten SNP positions (in order to avoid any pooled paralogues in the same locus) that are covered by data in at least 90% of individuals (i.e., with data present for at least 75 individuals, to avoid artificially splitting individual loci). Further, for the value of m = 13 we have run additional tests by varying the value of “M” from one to four and the value of “n” from zero to six (Supplementary Table 1; doi:10.5061/dryad.5q8r8). The final combination of settings was chosen based on the criteria above for m = 13, M = 1 and n = 1.
The deleveraging algorithm of ustacks has been left on in order to split loci merged incorrectly and remove highly repetitive sequences from further analyses. To avoid retention of any merged paralogs, the loci showing ten or more SNPs have been blacklisted in further analyses by filtering them out using the export_sql.pl script from STACKS. The SNP data have been extracted by using the populations script of STACKS. By retaining SNPs from loci covered in at least 75 individuals with a maximum of nine polymorphic nucleotide positions, we obtained a data matrix containing 8,488 concatenated SNPs (hereafter dataset D1), which has been further used for phylogenomic analyses. In an additional filtering step we retained only one SNP per locus (dataset D2 hereafter, including 1,506 SNPs) to minimize linkage in the data, and we finally filtered out all apomorphic SNPs (i.e., distinguishing only single individuals from the rest) to obtain a reduced matrix of 791 SNPs (hereafter dataset D3). Data matrices including loci with more missing data (up to 20% and, respectively, up to 50%) per locus resulted in less resolved phylogenetic trees and have been discarded (not shown).
Phylogenomic Analyses
To study the phylogenetic relationships between the sampled Diospyros individuals, we used maximum parsimony (MP), maximum likelihood (ML) and Bayesian inference (BI) on dataset D1. For BI and molecular dating, the program BEAST v1.7.5 (Drummond et al. 2012) was run on the CIPRES Science Gateway (http://www.phylo.org/portal2/; Miller et al. 2010). Estimation of evolutionary models was conducted with jModelTest v2.1.4 (Darriba et al. 2012) that indicated the transversional model (TVMef; Posada 2003) with equal frequencies modelled with a gamma distribution and a proportion of invariable sites (TVMef+Γ+I) to be the best fit to our data. We used a relaxed uncorrelated log-normal clock model to keep the age estimations flexible (Drummond et al. 2006). Because of the relatively young age of the investigated Diospyros group, and therefore a low proportion of lineage extinction expected, we opted for a simple Yule speciation model (Yule 1925; Gernhard 2008). Priors for substitution rates between bases (gamma shape 10), alpha (gamma shape 10), and p-inv (uniform) were inferred by jModelTest v2.1.4 (Darriba et al. 2012). Two independent Metropolis-coupled Markov chain Monte Carlo (MCMC) analyses each with 20 million generations were run sampling every 1,000th generation. The initial 10% of trees obtained from each MCMC run were removed as burn in; the remaining trees of both runs were used to calculate a maximum clade credibility tree. With BEAST we also conducted molecular dating. Because no fossils pertaining to this New Caledonian Diospyros group are available, dating estimates were obtained by secondary calibration: we took into account the age of the split between D. vieillardii and the rest of the group (7.2 Ma) so that it conforms to a previous date obtained for this split (Turner et al. 2013a). We constructed the priors (log normal shape) by defining this age as a minimum (i.e., no fixed upper limit), to accommodate the uncertainty of this estimate.
Parsimony analyses were run with PAUP* v4b10 (Swofford 2003) on dataset D1 using a heuristic search with stepwise addition, random sequence addition (1,000 replicates) and tree-bisection-reconnection. To estimate clade support, bootstrapping with 1,000 replicates was performed. We rooted the tree obtained with D. vieillardii, according to earlier results (Turner et al. 2013a).
Trees were also estimated using dataset D1 within a maximum likelihood framework using RAxML 8.1.3 (Stamatakis 2014). We used the BFGS method to optimize GTR rate parameters, the gamma model of rate heterogeneity and 1,000 rapid bootstrap inferences with a subsequent thorough ML search. The results were visualized with FigTree 1.4 (available from http://tree.bio.ed.ac.uk/software/figtree/).
Finally, we inferred a species tree on dataset D2 using the Bayesian coalescent method implemented in SNAPP (Bryant et al. 2012). As SNAPP is designed to deal with bi-allelic unlinked data, we extracted one SNP at random per locus. Given the average density of our RAD loci (less than 1/Mb), the probability of any of those SNPs to be in linkage is low. Nevertheless, a small amount of linkage among markers is not expected to bias the analysis (Bryant et al. 2012). For the SNAPP analyses, the individuals were a priori grouped to species, apart from those species that were shown to form multiple clusters in the MP/ML/BI trees, where the grouping used followed sampling localities. Two localities where only single individuals were available were excluded from the analyses (i.e., D. perplexa from L1 and D. parviflora from L13). We set a prior for θ = 4μNe as a gamma distribution with α = 1.15 and β = 10000 (mean = 1.15·10−4). We ran two independent MCMC chains for 5·106 generations, each sampling every 100th generation. Burn-in was visually determined and convergence among different chains checked in Tracer 1.6. We also checked that all ESSs were > 100 across combined runs, with most of them > 200. Trees resulting from the analysis were then visualized in DensiTree (Bouckaert 2010). We summarized all trees in a set of consensus trees, where each consensus tree is calculated from the subset of trees sharing the same topology averaging the branch length over this subset. A large number of consensus trees denotes a high degree of uncertainty in topology.
Genetic Clustering and Patterns of Reticulation
Within closely related groups, representation of relationships as networks rather than bifurcating trees appears to be more appropriate (Huson and Scornavacca 2011) to account for the potential presence of hybridization. We used SplitsTree v.4.12.6 (Huson and Bryant 2006) to create a network based on Hamming distances (Hamming 1950) and dataset D1. The simple calculation method of Hamming distance was considered appropriate for the RADseq derived SNPs dataset that lacks indels after processing with STACKS.
To investigate the higher-level clustering of the included individuals and potential admixture between different groups we used Structure v2.3.3 (Pritchard et al. 2000; Hubisz et al. 2009) on dataset D3. We ran the program separately for i) all individuals analysed and ii) the individuals forming the larger cluster in the first analysis (i.e., the RR clade as defined in Results). Both analyses have been performed at the Lifeportal of the University of Oslo for K = 1 to 10, each with ten replicates, and a model based on admixture and independent allelic frequencies. Each run had one million iterations with 10% additional burn in. The calculation of ΔK (Evanno et al. 2005; Supplementary Fig. 3) was performed with Harvester (Earl and vonHoldt 2012). To avoid any stochastic aspect of the process, we have produced a permuted matrix from the ten replicates of the best K value with Clumpp v1.1.2 (Jakobsson and Rosenberg 2007) in the greedy algorithm and 1,000 repeats. The graphical display of Structure results was prepared with Distruct v1.1 (Rosenberg 2004).
Historical relationships inferred with a population graph analysis that allows both population splits and migration events were constructed with TreeMix 1.12 (Pickrell and Pritchard 2012) on dataset D2, but including only populations from the RR group, plus D. cherrieri, used as outgroup (Supplementary Fig. 4). The individuals were grouped according to sampling localities and species (excluding localities represented by single individuals, except for D. parviflora L19 which was pooled together with individuals from the same species from L4 according to above presented results). We have added migration edges stepwise with up to ten events and inspected the results for consistency between the different runs.
Patterns of Niche Divergence
To investigate how niche specialization contributed to diversification, we attempted to reconstruct ancestral preference for habitats along the inferred Bayesian phylogeny. We employed for this purpose maximum-likelihood inferences of niche evolution with the Lagrange dispersal, extinction and cladogenesis (DEC; Ree and Smith 2008) model implemented in RASP v.3.02 (Yu et al. 2015). We carried out two analyses: one for the evolution of climatic niches (i.e., humid versus dry) and one for edaphic niches (i.e., ultramafic, volcanic, limestone, schist, and, respectively, serpentine). We did not apply any constraints on the connectivity between the different niches. As all extant taxa are confined to a single soil type, and for simplicity, we allowed for ancestral ranges combining a maximum of two edaphic types.
The phylogenetic trees obtained pointed to several sister clades comprising individuals with divergent preference for ultramafic versus volcanic soils: D. flavocarpa/D. umbrosa; D. parviflora (L10)/D. parviflora (L22); D. yahouensis + D. perplexa /D. pancheri; D. minimifolia (L17)/D. sp. Pic N'ga. We searched for RAD regions that contained SNPs with pairwise FST values over 0.5 at least for two pairs of sister species with divergent soil preferences. We then made use of the paired-end RADseq data and assembled mini-contigs of the candidate stacks by extracting a list of reads for each locus with Sort_read_pairs.pl from STACKS, sorting reads from FastQ files with Fastq.filter.pl (L.M. Rodriguez unpublished, available from http://enveomics.blogspot.co.at/2013/04/fastqfilterpl.html) and assembling each set in the CLC Genomic Workbench (QIAGEN), with automatic optimization of the word and bubble sizes and updating the contigs after mapping back the reads. We finally performed functional annotation analyses for the obtained contigs using Blast2GO v. 3.1.3 (BioBam; Götz et al. 2008) with default settings (i.e., blastx searches against nr) and integrating GO (www.geneontology.org), KEGG (www.genome.jp/kegg) and InterProScan (www.ebi.ac.uk/Tools/InterProScan) information in our results. The biological meaning of the set of sequences has been investigated with the combined graph option of Blast2Go by visualizing only categories with a node score ≥ 2.00. Further, a Fisher’s exact test has been run with Blast2Go to investigate the significance of any GO term enrichment in the potential adaptive loci against a reference group of 1,000 RADseq loci assembled in a similar way.
Results
Phylogenomic Analyses
After demultiplexing, trimming and filtering the raw 100 bp reads from two RADseq libraries, we retained on average 1.9 +/− 0.7 million high-quality pairs of reads per individual. After parameter optimization as described above, the de novo assembly pipeline of STACKS produced 37,336 loci (excluding any tags present in only one individual).
The MP (Supplementary Fig. 1), ML (Supplementary Fig. 2) and BI (Fig. 2) analyses resulted in generally consistent topologies, pointing to two major groups of species: one older grade of more slowly radiating species (SR hereafter) that are more divergent, and a younger clade of more rapidly radiating species (RR hereafter). The backbones of the other trees are slightly less supported than that of the BI (Fig. 2).
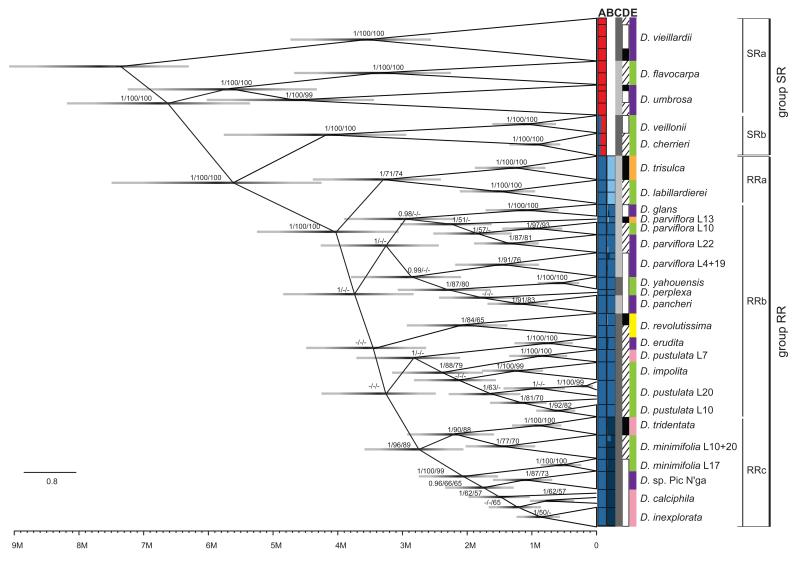
Figure 2.
Phylogenetic tree of the radiating Diospyros species on New Caledonia derived from Bayesian inference. For simplicity, individuals are collapsed to species or population level wherever possible. Node bars indicate the 95% confidence interval for the age of the corresponding node. A time scale is given at the bottom of the figure. Numbers at nodes detail BI posterior probabilities greater than 0.95/ML confidence over 50%/bootstrap support values over 50%. The vertical bars indicate: A) Structure results for all individuals for K=2; B) Structure results for the subset of individuals from group RR, for K = 3; C) the preference for climate type, generally following a E-W separation - light grey for humid, dark grey for dry; D) the geography, generally following a N-S separation - black for north, hashed area for centre and white bars for south; and E) for substrate type preference - lemon green for volcanic rock, dark purple for ultramafic rock, orange for schist, pink for limestone and yellow for serpentine. L. refers to sampling location given in Fig. 1 and Table 1.
Apart from D. minimifolia, D. parviflora and D. pustulata, all other species form highly supported clusters in the BI phylogeny. The individuals of the same population always group together and are well supported, except for one population of D. pustulata (location 20). Further, the sampled individuals of D. calciphila form a paraphyletic group, embedding D. inexplorata individuals despite growing on different islands. We do not observe any major grouping related to ecological factors like soil type or climate, but sister species often show divergent ecological preferences (Fig. 2). The molecular clock analysis resulted in a slightly older age for the split of D. vieillardii from the rest of the group, estimated at 7.4 Ma, with a wide 95% confidence interval of 2.7 Ma. The next divergence (i.e., D. flavocarpa/D. umbrosa from the rest of the species) took place around 6.6 Ma. The lineage forming D. cherrieri and D. veillonii separated from the rest around 5.6 Ma. The RR group started to diversify around 4 Ma. The RRc clade is a young group, about 2.7 million years old. Most speciation events seem to have taken place in this group between 3.5 and 1.5 Ma.
The MP analysis resulted in 31 equally parsimonious trees (Supplementary Fig. 1). The phylogenetic relationships between the earlier diverged lineages (group SR) are the same as in the BI genealogy. The relationships between the clades within group RR differ slightly between MP (Supplementary Fig. 1) and BI (Fig. 2). The general topology of the ML tree (Supplementary Fig. 2) inferred with RAxML is also similar to that of the BI.
The SNAPP results (Fig. 3a) reveal a species tree of similar major topology to the BI tree, but with several marked differences. Generally, the SNAPP analyses seems to have less resolving power than the BI, most probably due to more informative characters present in the matrix used for the latter. Whereas the composition of the SR and RR groups was found to be constant between the two analyses, SNAPP unexpectedly identifies D. yahouensis as the sister species to the rest of the RR group. It also points to a fairly significant time gap between any earlier split and the recent rapid radiation of the RR group. In fact, the relative age of the radiation of the RR group in comparison to the time of divergence since D. vieillardii split from the rest of the species, seems even younger in the SNAPP analyses in comparison to BI. Due to short internal branch lengths, the relationships between the species of the RR group remain generally unclear and resemble a rapid star-like radiation. However, the RRa and RRc groups that received good support in the BI, MP and ML analyses are also present in the SNAPP results.
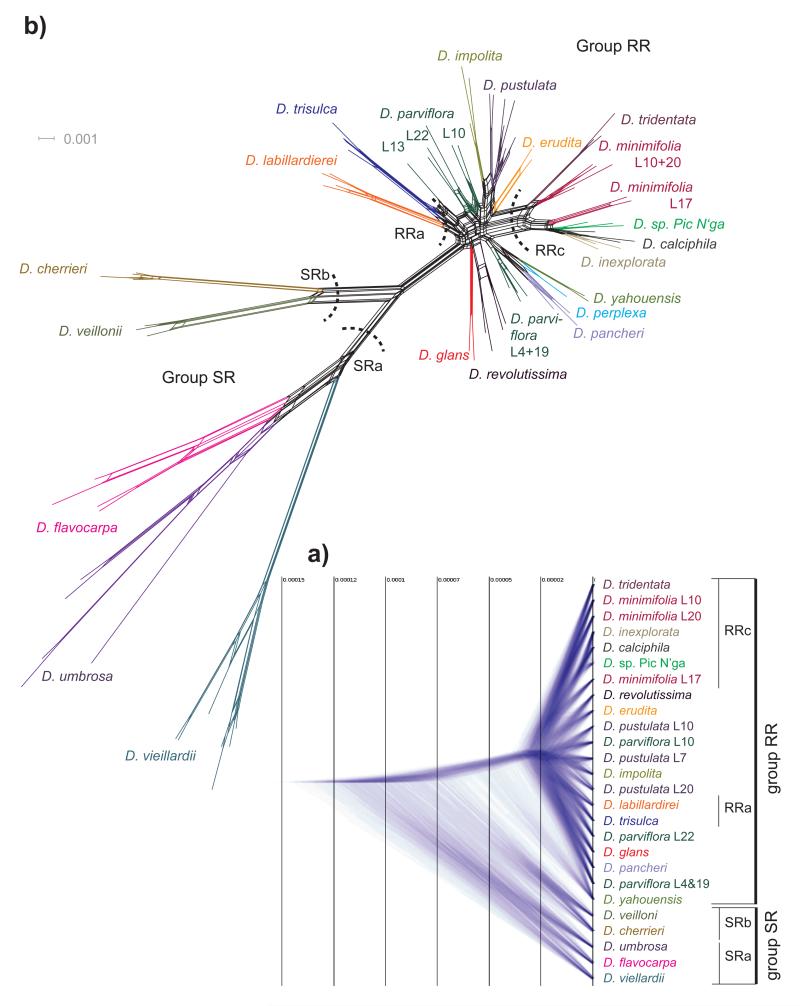
Figure 3.
a) Species trees generated by SNAPP analysis. The distances on the horizontal axis are relative measures of substitutions per site. The nodes are positioned as a star tree to highlight the timing of species radiation. Time on the grid is scaled to the number of mutations. b) NeighbourNet based on Hamming distance. Each species is shown with a different colour. L. refers to sampling locations given in Fig. 1 and Table 1.
Genetic Clustering and Patterns of Reticulation
The SplitsTree network (Fig. 3b) mirrors the general pattern evident in the phylogenetic analyses. The branches within group SR and between SR and RR groups are significantly longer than those within the RR group, in a ratio more similar to the SNAPP results rather than for BI. The RR group has a reticulate history, indicative of hybridization and/or incomplete lineage sorting. Conflicting information is also apparent at the level of deeper relationships within the SR group.
When including all individuals, Structure identifies two major clades (Fig. 2 column A; Supplementary Fig. 3a), roughly corresponding to the SR and RR groups. However, it shows the presence of potential admixture between the two groups in D. veillonii and D. cherrieri (SRb subgroup). When run only on the subset of individuals from the RR group, Structure further separates three subgroups (Fig. 2 column B; Supplementary Fig. 3b) with few admixed individuals between them.
We further used ancestry graphs implemented in TreeMix (Pickrell and Pritchard 2012) to identify patterns of divergence and migration within the RR group only, including D. cherrieri as an outgroup. The stepwise addition of migration events in the analysis resulted in an incremental, consistent inclusion of additional events up to the 6th migration. With the 7th migration, the events changed significantly, so we considered as final the result including six migrations (Supplementary Fig. 4). Those six migration events link populations from different climatic conditions (three of six), different soil types (four of six events) and from different regions (five of six events - connecting populations from the centre of the island with those in the north or in the south).
Patterns of Adaptive Divergence
Lagrange analyses (Supplementary Fig. 5) suggest the most recent common ancestor of all members of this Diospyros group that radiated on New Caledonia was most likely a generalist for the amount of humidity, and it might have occupied both ultramafic and volcanic substrates. The generalist lineage had specialist descendants with regard to climate type relatively early, resulting in large clades of extant taxa sharing the same preference for humidity. In contrast, edaphic specialists evolved in general more recently. Surprisingly, apart from multiple transitions from edaphic generalists to specialists, several shifts from specialists to generalists have also been inferred. In addition, specialists to particular substrates are often not each other’s closest relatives, but they are rather related to sister taxa adapted to different substrates.
The phylogenetic trees obtained provide evidence that substrate differences exist for several pairs of species, which suggests that ecologically driven isolation could have made a major contribution in driving speciation across the radiating group. We aimed to test if any particular genomic regions (but not necessarily the same exact SNP position) have systematically been affected as a result of positive selection or genetic hitchhiking in divergences between four replicated pairs of sister species (for details see ‘Material and Methods’) with distinct preference for ultramafic versus volcanic substrates. We therefore searched for RAD regions that contained SNPs with high fixation index values (FST > 0.5) at least for two pairs of sister species with such replicated divergent soil preferences. We have identified 50 RAD regions (GenBank accession numbers KT587203 - KT587252) showing parallel patterns, including four DNA regions that have been found to be significantly different in three of the four pairs of species (see Methods). The majority of the identified SNPs (74.6%) are not fixed differences between species, potentially indicative of polygenic adaptation, as expected for ecological traits (Savolainen et al. 2013). Using the paired-end read information, we have assembled de novo larger DNA regions of between 390 and 909 bp for the 50 candidate loci and performed functional annotations for them. Only 26 of the loci received at least one blast hit, and 15 of them could be successfully annotated (Table 2). The combined graph analysis of Blast2Go and a Fisher’s exact test against a reference group of RADseq loci indicates an enrichment for regions related to transport (i.e., amide transport, peptide transport, and nitrogen compound transport, all at p < 0.05) and ion binding (at p < 0.05) (Table 2; Supplementary Fig. 6).
Table 2.
Details of the 26 regions with blast hits, which are hypothesized to be recurrently involved in divergent edaphic adaptation.
| GenBank | SNP bp | SNP pos.3 | Sequence Description | GOs |
|---|---|---|---|---|
| KT587204 | 7/66/72 | out | protein aluminium sensitive 3 | C:plasma membrane; P:response to aluminium ion |
| KT587205 | 77 | 1st | udp-glycosyltransferase 73c3-like | F:transferase activity, transferring glycosyl groups |
| KT587206 | 15/48 | out | translation machinery-associated protein 22 | P:translational initiation; F:translation initiation factor activity |
| KT587208 | 49/55 | 2nd/2nd | probable calcium-binding protein cml23 | F:ion binding |
| KT587250 | 51/62 | out | duf616 family protein | - |
| KT587212 | 12/78 | 3rd/3rd | copia-like retrotransposable | C:membrane; F: nucleic acid binding; F: zinc ion binding; F: anion binding; P:DNA integration; P: ammonium transport |
| KT587213 | 6/51 | 3rd/3rd | trihelix transcription factor asil2-like | C:vacuole; F:chromatin binding; C:nucleus |
| KT587214 | 6/45 | out | snf2 domain-containing family protein | F:DNA binding; F:ion binding; F:helicase activity |
| KT587216 | 34/59 | out | protein nrt1 ptr family -like | P:transport |
| KT587248 | 33/63 | out | myosin-2-like isoform x1 | C:cytoskeleton; F:ion binding; C:protein complex; F:cytoskeletal protein binding |
| KT587249 | 52/79 | - | uncharacterized locus | F:ion binding; P:tRNA metabolic process; P:cellular amino acid metabolic process; F:ligase activity; P:translation |
| KT587217 | 39/47 | out | ribonuclease 3 | C:plastid |
| KT587218 | 43/60/90 | out | gag-pol polyprotein | F:nucleic acid binding; P:DNA integration; F:zinc ion binding; C:nucleus |
| KT587219 | 12/84 | - | e3 ubiquitin-protein ligase pub24-like | F:ubiquitin-protein transferase activity; P:protein ubiquitination |
| KT587220 | 42/58 | out | r- recognition motif | - |
| KT587223 | 61/92 | out | cbs domain-containing protein mitochondrial | F:adenyl nucleotide binding; P:metabolic process; F:catalytic activity |
| KT587224 | 35/80 | 1st/1st | eh domain-containing protein 1 | C:cell; F:ion binding |
| KT587225 | 38/64 | out | type ii inositol -trisphosphate 5-phosphatase fra3 isoform x1 | P:lipid metabolic process; F:phosphatase activity; F:ion binding |
| KT587226 | 28/31 | 2nd/2nd | Uncharacterized protein TCM_034686 | - |
| KT587227 | 48/72 | out | hypothetical protein VITISV_008807 | P:oxidation-reduction process; F:heme oxygenase (decyclizing) activity; P:heme oxidation; F:transferase activity; P:metabolic process; F:sulfotransferase activity |
| KT587244 | 8/43 | 3rd/2nd | hypothetical protein PRUPE_ppa021982mg | - |
| KT587247 | 24/40 | 2nd/3rd | saur-like auxin-responsive protein | - |
| KT587234 | 20/52/86 | out/3rd/1st | uncharacterized loc101212813 | C:nucleus |
| KT587231 | 68 | - | abc transporter b family member 19-like | C:integral component of membrane; F:ATP binding; F:xenobiotic-transporting ATPase activity; P:transmembrane transport; F:transmembrane transporter activity; F:ion binding; P:peptide transport; P:xenobiotic transport |
| KT587237 | 68/91 | out | mitogen-activated protein kinase 19 | P:signal transduction; F:ion binding; P:cellular protein modification process; P:cellular amino acid metabolic process; F:signal transducer activity; F:kinase activity |
| KT587245 | 69/75 | 2nd/2nd | (+)-neomenthol dehydrogenase-like isoform x1 | F:oxidoreductase activity; P:oxidation-reduction process |
SNP position refers to the position with respect of the reading frame, inferred from the blast result: out - out of ORF, 1st, 2nd and 3rd refer to codon positions.
Discussion
Inferring Shallow Phylogenetic Relationships
To resolve the phylogenetic relationships within a rapidly radiating Diospyros group on New Caledonia, we employed thousands of SNPs derived from RAD loci assembled de novo from Illumina reads. This allowed us to infer a much more completely resolved tree than previous studies using multiple DNA loci (Duangjai et al. 2009; Turner et al. 2013a) and genome-wide fingerprinting analyses (AFLP; Turner et al. 2013b). An increase in phylogenetic resolution when using RADseq in comparison with more traditional methods has already been shown for various organisms, for example, in the case of the adaptive radiation of cichlid fishes in Lake Victoria (Keller et al. 2013; Wagner et al. 2013) and the radiation of surfperches in the northern Pacific (Longo and Bernardi 2015). Despite the relatively large number of SNPs we obtained, relationships inferred here for Disopyros are not always well supported. The reason for this may lie in the limited number of generations since the extreme bottleneck associated with the initial long-distance dispersal event to New Caledonia. This renders a significant portion of the SNPs to have minor allelic variants present in only one to a handful of individuals (i.e., fairly recently evolved variants). This pattern is clearly visible in the phylogenetic trees we obtained, with well-supported terminal clades corresponding to species or just populations.
Additional processes may have blurred the phylogenetic signal in this rapidly radiating group, in particular introgression, which could have been common during some episodes of speciation in this group. Its effects are visible in the SplitsTree network (Fig. 3b) as reticulations, the Structure results (Supplementary Fig. 3) as the presence of admixed individuals, and the migration events inferred with TreeMix (Supplementary Fig. 4). Incomplete sorting of ancestral polymorphism requires a rich ancestral genetic pool (van Oppen et al. 2001; Maddison and Knowles 2006; Glor 2010; Lerner et al. 2011). Therefore we regard the latter process as less likely to have significantly affected, on a genome-wide scale, the phylogenetic patterns within this group, which radiated fairly recently after passing through the extreme bottleneck associated with arrival on New Caledonia (Duangjai et al. 2009). The results obtained are also much too structured by taxon and geography to represent this sort of distortion of phylogenetic relationships.
Early Divergence with Respect to Macrohabitat and Geographic Distance
Grande Terre, the main island of New Caledonia, is split by a mountain range into humid southeastern (2000-4000 mm precipitation per year) and dry northwestern parts (1000 mm precipitation per year) with prevailing winds and rain coming from the southeast (Maitrepierre 2012). The inferred phylogenetic relationships and the analyses of niche evolution point to an initial, but fairly slow, divergence with respect to these climatic conditions along major clades (i.e., the deep splits on the backbone of the phylogenetic tree; Fig. 2 column C and Supplementary Fig. 5A). Modelling studies have suggested that divergence with respect to the macrohabitat is indeed the first expected stage for rapid radiations (Gavrilets and Losos 2009; Glor 2010), although up to now the empirical evidence for this expectation was limited. In Diospyros, this appears to be the result of climatic specialization from a generalist lineage that, however, persists over several cladogenesis events (Supplementary Fig. 5A). This may argue against a significant contribution of climatic specialization to diversification in this particular group (Fine et al. 2014). In Diospyros, this pattern could also be merely the result of allopatric differentiation, with isolation by distance promoted by a geographic barrier between the dry and wet areas that is difficult to cross. A limited dispersal potential of New Caledonian species of Diospyros is supported by the fact that most populations cluster separately within each species (see e.g., the MP tree in Supplementary Fig. 1, or the SplitsTree network in Fig. 3b). Very little is known about pollinators and fruit dispersal in these species; most species are dioecious and have fleshy fruits. The fruits of other Diospyros species present on the island are eaten by birds - for example, the fruits of D. fasciculosa (of similar size as the fruits in the radiating group) are dispersed by the red-bellied fruit-dove, Ptilinopus greyii (Tassin et al. 2010). We assume the seeds of the radiating group are also dispersed by birds, but the genetic evidence is consistent with limited gene flow between populations.
We further observe that several intermediate-level splits follow a regional separation (Fig. 2 column D), with relatively large groups of species/populations either confined to the northern, the central or the southern portion of the island. For example, the related D. erudita, D. impolita and D. pustulata (from the RRb group) are all found in the middle western part of Grande Terre in dry vegetation (maquis or sclerophyllous forests). A similar regional clustering pattern is observed for D. pancheri, D. yahouensis, D. perplexa and part of D. parviflora ; all were collected in southern part of Grande Terre. These examples support the idea that dispersal over long geographic distances is inefficient in this group, which results in a pattern of isolation by distance.
Late Divergence with Respect to Microhabitat
In New Caledonia, Diospyros species are found in all kinds of vegetation (Fig. 1) except mangroves. However, the species that are elements of similar vegetation types are in general not closely related, a phenomenon that is characterized as phylogenetic niche convergence (Webb et al. 2002). Such a pattern can be the result of niche filling (Jakob et al. 2010), with close relatives adapting to different habitats to avoid highly similar competitors, whereas taxa from distinct clades are different enough to explore diverse resources even in sympatry. For example, typical littoral vegetation along the New Caledonian coasts is represented by species such as D. calciphila, D. inexplorata (both RRc in Fig. 2), D. impolita (RRb), and D. veillonii (SRb). On the other hand, D. cherrieri (SRb), D. minimifolia, D. tridentata (both RRc), D. perplexa, and D. pustulata (both RRb) are found in sclerophyll forests on the dry western coast of Grande Terre. Especially when considering multi-species localities (e.g., L10, L13 and L20; Table 1), sympatric species are only distantly related.
In contrast, most sister species relationships seem to have been shaped by ecologically driven isolation, in particular with respect to edaphic specialization (Fig. 2 column E). Indeed, we find a plenitude of independent edaphic shifts (Supplementary Fig. 5b) that occur regionally (Fig. 2 columns D and E), in the absence of obvious geographic barriers. In this context substrate specialization appears to have fostered diversification (Fine et al. 2005, 2014) through parapatric speciation events during the evolution of this New Caledonian Diospyros group. Edaphic adaptation has been proposed as one of the main types of parapatric speciation in plants (Coyne and Orr 2004), and has been suggested to have played a key role, for example, in the diversification of Amazonian trees (Fine et al. 2005, 2013, 2014).
Grande Terre is a mosaic of soil types (Maurizot and Vendé-Leclerc 2009) and taking the climatic and geological factors together, the island is unusually rich in ecological niches. This likely imposes selection pressures that drive adaptation and isolation, potentially even in the presence of gene flow. In light of our results, the phenotypic characteristics related to substrate adaptation seem evolutionary labile and/or the number of underlying loci for these adaptations is small to allow for relatively frequent niche shifts, but further research is needed to support such conclusions However, the heterogeneous edaphic conditions have been proposed as the main reason for the high level of endemism found in New Caledonia (Pillon et al. 2010). Isolation by environment has been previously found to be a prevailing evolutionary force across environmental gradients in a wide range of organisms (Sexton et al. 2014), for example in the case of the flora of Lord Howe Island (Papadopulos et al. 2014).
Within the phylogenetic tree of the radiating species of Diospyros we have found in different subclades evidence for replicated phenotypic divergence with respect to substrate preference (i.e., ultramafic versus volcanic soils). Other examples of similar ecological specialists that evolved repeatedly include Anolis lizards on the islands of the Greater Antilles (Losos 2010) and African cichlid fishes (Brawand et al. 2014). To further investigate if natural selection affected a convergent set of loci during parallel evolution (Purugganan 2014), we searched for RAD regions containing SNPs that show high differentiation between two or more sister taxa with the same divergent soil-type preference. Most (92%) of the loci that were successfully annotated indicated that different alleles of the same genes were recruited by natural selection or swept along in the different splits (Table 2). At only two of the 26 loci with blast hits did we find that collateral evolution (sensu Stern 2013) through sorting of ancestral polymorphism may have played a role in the convergent adaptation, as the same variants were found across different splits. This could also indicate that introgression played only a limited adaptive role across these replicated speciation events. Half of the annotated loci were affected by divergence outside of ORF regions, potentially indicative of regulatory divergence. A significant portion (35%) of the annotated loci has molecular functions related to ion binding (Supplementary Fig. 6; Table 2); another important category plays a role in transport of elements within or between cells. As the New Caledonian soil types are different in heavy-metal content and availability of minerals, these specific loci appear potentially adaptive and therefore involved in edaphic specialization, and not simply linked to regions under selection. It is, however, difficult to argue that this differentiation is responsible for particular speciation events, or if they have evolved subsequently (Glor 2010). A similarly limited number of genomic regions on which positive selection might have acted has also been found in Hawaiian species of Schiedea (Kapralov et al. 2013), which exhibit, like the New Caledonian Diospyros species, great morphological and ecological variation.
The Effect of Introgression
It is generally accepted that species divergence can occur in the face of persistent and genome-wide admixture over long periods of time, as shown for example in Heliconius butterflies (Martin et al. 2013). Furthermore, it has been proposed that the high speciation rates in radiations may be fuelled by frequent hybridization with transgressive segregation (e.g., Seehausen 2004; Gavrilets and Losos 2009; Glor 2010; Losos 2010; Keller et al. 2013).
The SplitsTree and Structure analyses provide evidence for considerable admixture among the Diospyros species (Fig. 3b and Supplementary Fig. 3). The grouping of different populations of the non-monophyletic species like D. minimifolia and D. parviflora has some relationship to provenance. For example, the population of D. minimifolia from Gadji (L17, Fig. 1) is genetically divergent from the rest of the individuals of this species found in central Grande Terre. This population from Gadji clusters with species from Île des Pins (L25 and L26, Fig. 1) and Île Kuebini (L24, Fig. 1), which are all in the south of New Caledonia. The SplitsTree network (Fig. 3b) clearly indicates regional hybridization as the reason for the split between the D. minimifolia populations from the south and those from central portions of the island, which cluster together with northern D. tridentata accessions (see also Fig. 2). Furthermore, the accessions of D. parviflora collected in the central region of New Caledonia are positioned together with the northern accession from L13, closer to other individuals sampled from the northern and central part of the main island. In contrast, the southern populations of D. parviflora (i.e., L4 and L19) exhibit evidence of hybridization with southern populations of other species in the SplitsTree network (Fig. 3b). This phenomenon of individuals grouping with co-occurring species rather than with populations of the same species from other localities is also found in other organisms (e.g., Martin et al. 2013) and may be indicative of genome-wide admixture over long periods of time in many parts of their porous genomes (Gavrilets and Losos 2009; Kane et al. 2009). Since most neutral loci are either unlinked or only loosely linked to loci that are under divergent selection, they will be subjected to homogenization by gene flow across closely related sympatric species (Feder et al. 2012). However, given the obvious morphological and ecological differentiation between most of these species we hypothesize as unlikely the swamping of one species with genes of the others (Rieseberg 2009; Butlin et al. 2014). Rather, in this context hybridization might have had a positive effect of increasing biodiversity by promoting formation of cryptic species, as evidenced in other groups present in New Caledonia (Pillon et al. 2009, 2014; Swenson et al. in press).
Concluding Remarks
Altogether, the species-rich New Caledonian Diospyros clade is the result of a rapid adaptive radiation resulting in more than 20 morphologically and ecologically diversified species with low genetic divergence. After surviving a bottleneck associated with the long-distance dispersal event that brought the lineage to the Archipelago, accumulation of genetic variation happened relatively slowly, producing a small effective population size for the new lineage for millions of years. During the first phase of the radiation, this situation resulted in relatively low speciation rates, mainly triggered by allopatry or divergent preference to macrohabitat related to general climatic conditions. After a considerable amount of time (i.e, ca. 6 million years, Fig. 2), there was a burst of speciation events, mainly with respect to microhabitat (i.e., edaphic) specialization. Such a delayed burst of speciation is atypical for adaptive radiations, but could be a characteristic of long-generation organisms. Our results suggest that both allopatric diversification (i.e., resulting in regional patterns of diversity) and sympatric/parapatric ecological divergence shaped successive rounds of speciation in the Diospyros radiation.
Supplementary Material
Acknowledgements
This work was supported by the Austrian Science Fund [FWF P22159-B16 to R.S.]. In addition O.P. was partly funded from the Austrian Science Fund [FWF Y661-B16 to O.P.]. We thank G. Schneeweiss, F. Balao, S. Duangjai, F. E. Anderson, S. Renner and two anonymous reviewers for their contribution to the research presented here. lllumina sequencing was performed at the CSF NGS Unit, Vienna (http://csf.ac.at/ngs). Specimens are deposited in the herbaria of Noumea (NOU), University of Montpellier II (MPU) and the University of Vienna (WU), staff of these herbaria are acknowledged for various help. Collecting permits were provided by Environmental Services of North and South Provinces of New Caledonia.
Footnotes
Supplementary material, including datasets D1, D2 and D3, plus one Supplementary Table and six Supplementary Figures, can be found in the Dryad data repository (doi:10.5061/dryad.5q8r8).
References
- Baird NA, Etter PD, Atwood TS, Currey MC, Shiver AL, Lewis ZA, Selker EU, Cresko WA, Johnson EA. Rapid SNP discovery and genetic mapping using sequenced RAD markers. PLoS One. 2008;3:e3376. doi: 10.1371/journal.pone.0003376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baxter SW, Davey JW, Johnston JS, Shelton AM, Heckel DG, Jiggins CD, Blaxter ML. Linkage mapping and comparative genomics using next-generation RAD sequencing of a non-model organism. PLoS One. 2011;6:e19315. doi: 10.1371/journal.pone.0019315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellard C, Leclerc C, Courchamp F. Potential impact of sea level rise on French islands worldwide. Nat. Conservat. 2013;5:75–86. [Google Scholar]
- Bouckaert RR. DensiTree: making sense of sets of phylogenetic trees. Bioinformatics. 2010;26:1372–1373. doi: 10.1093/bioinformatics/btq110. [DOI] [PubMed] [Google Scholar]
- Brawand D, Wagner CE, Li YI, Malinsky M, Keller I, Fan S, Simakov O, Ng AY, Lim ZW, Bezault E, Turner-Maier J, Johnson J, Alcazar R, Noh HJ, Russell P, Aken B, Alfoldi J, Amemiya C, Azzouzi N, Baroiller J-F, Barloy-Hubler F, Berlin A, Bloomquist R, Carleton KL, Conte MA, D'Cotta H, Eshel O, Gaffney L, Galibert F, Gante HF, Gnerre S, Greuter L, Guyon R, Haddad NS, Haerty W, Harris RM, Hofmann HA, Hourlier T, Hulata G, Jaffe DB, Lara M, Lee AP, MacCallum I, Mwaiko S, Nikaido M, Nishihara H, Ozouf-Costaz C, Penman DJ, Przybylski D, Rakotomanga M, Renn SCP, Ribeiro FJ, Ron M, Salzburger W, Sanchez-Pulido L, Santos ME, Searle S, Sharpe T, Swofford R, Tan FJ, Williams L, Young S, Yin S, Okada N, Kocher TD, Miska EA, Lander ES, Venkatesh B, Fernald RD, Meyer A, Ponting CP, Streelman JT, Lindblad-Toh K, Seehausen O, Di Palma F. The genomic substrate for adaptive radiation in African cichlid fish. Nature. 2014;513:375–381. doi: 10.1038/nature13726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryant D, Bouckaert R, Felsenstein J, Rosenberg NA, RoyChoudhury A. Inferring species trees directly from biallelic genetic markers: Bypassing gene trees in a full coalescent analysis. Mol. Biol. Evol. 2012;29:1917–1932. doi: 10.1093/molbev/mss086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butlin RK, Saura M, Charrier G, Jackson B, André C, Caballero A, Coyne JA, Galindo J, Grahame JW, Hollander J, Kemppainen P, Martínez-Fernández M, Panova M, Quesada H, Johannesson K, Rolán-Alvarez E. Parallel evolution of local adaptation and reproductive isolation in the face of gene flow. Evolution. 2014;68:935–949. doi: 10.1111/evo.12329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cariou M, Duret L, Charlat S. Is RAD-seq suitable for phylogenetic inference? An in silico assessment and optimization. Ecol. Evol. 2013;3:846–852. doi: 10.1002/ece3.512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catchen JM, Amores A, Hohenlohe P, Cresko W, Postlethwait JH. Stacks: Building and genotyping loci de novo from short-read sequences. G3: Genes, Genomes, Genet. 2011;1:171–182. doi: 10.1534/g3.111.000240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coyne JA, Orr HA. Speciation. Sinauer Associates; Sunderland, MA: 2004. [Google Scholar]
- Darriba D, Taboada GL, Doallo R, Posada D. jModelTest 2: more models, new heuristics and parallel computing. Nat. Methods. 2012;9:772. doi: 10.1038/nmeth.2109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummond AJ, Ho SYW, Phillips MJ, Rambaut A. Relaxed phylogenetics and dating with confidence. PLoS Biol. 2006;4:699–710. doi: 10.1371/journal.pbio.0040088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummond AJ, Suchard MA, Xie D, Rambaut A. Bayesian phylogenetics with BEAUti and the BEAST 1.7. Mol. Biol. Evol. 2012;29:1969–1973. doi: 10.1093/molbev/mss075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duangjai S, Samuel R, Munzinger J, Forest F, Wallnöfer B, Barfuss MHJ, Fischer G, Chase MW. A multi-locus plastid phylogenetic analysis of the pantropical genus Diospyros (Ebenaceae), with an emphasis on the radiation and biogeographic origins of the New Caledonian endemic species. Mol. Phylogenet. Evol. 2009;52:602–620. doi: 10.1016/j.ympev.2009.04.021. [DOI] [PubMed] [Google Scholar]
- Earl DA, vonHoldt BM. STRUCTURE HARVESTER: a website and program for visualizing STRUCTURE output and implementing the Evanno method. Conserv. Genet. Res. 2012;4:359–361. [Google Scholar]
- Evanno G, Regnaut S, Goudet J. Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol. Ecol. 2005;14:2611–2620. doi: 10.1111/j.1365-294X.2005.02553.x. [DOI] [PubMed] [Google Scholar]
- Feder JL, Egan SP, Nosil P. The genomics of speciation-with-gene-flow. Trends Genet. 2012;28:342–350. doi: 10.1016/j.tig.2012.03.009. [DOI] [PubMed] [Google Scholar]
- Fine PVA, Daly DC, Villa Muñoz G, Mesones I, Cameron KM. The contribution of edaphic heterogeneity to the evolution and diversity of Burseraceae trees in the western Amazon. Evolution. 2005;59:1464–1478. [PubMed] [Google Scholar]
- Fine PVA, Zapata F, Daly DC. Investigating processes of neotropical rain forest tree diversification by examining the evolution and historical biogeography of the Protieae (Burseraceae) Evolution. 2014;68:1988–2004. doi: 10.1111/evo.12414. [DOI] [PubMed] [Google Scholar]
- Fine PVA, Zapata F, Daly DC, Mesones I, Misiewicz TM, Cooper HF, Barbosa CEA. The importance of environmental heterogeneity and spatial distance in generating phylogeographic structure in edaphic specialist and generalist tree species of Protium (Burseraceae) across the Amazon Basin. J. Biogeogr. 2013;40:646–661. [Google Scholar]
- Gavrilets S, Losos JB. Adaptive radiation: contrasting theory with data. Science. 2009;323:732–737. doi: 10.1126/science.1157966. [DOI] [PubMed] [Google Scholar]
- Gavrilets S, Vose A. Dynamic patterns of adaptive radiation. Proc. Natl. Acad. Sci. USA. 2005;102:18040–18045. doi: 10.1073/pnas.0506330102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gernhard T. The conditioned reconstructed process. J. Theor. Biol. 2008;253:769–788. doi: 10.1016/j.jtbi.2008.04.005. [DOI] [PubMed] [Google Scholar]
- Glor RE. Phylogenetic insights on adaptive radiation. Annu. Rev. Ecol. Evol. Syst. 2010;41:251–270. [Google Scholar]
- Götz S, García-Gómez JM, Terol J, Williams TD, Nagraj SH, Nueda MJ, Robles M, Talón M, Dopazo J, Conesa A. High-throughput functional annotation and data mining with the Blast2GO suite. Nucleic Acids Res. 2008;36:3420–3435. doi: 10.1093/nar/gkn176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamming RW. Error detecting and error correcting codes. The Bell System Tech. J. 1950;29:147–160. [Google Scholar]
- Hubisz MJ, Falush D, Stephens M, Pritchard JK. Inferring weak population structure with the assistance of sample group information. Mol. Ecol. Res. 2009;9:1322–1332. doi: 10.1111/j.1755-0998.2009.02591.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huson DH, Bryant D. Application of phylogenetic networks in evolutionary studies. Mol. Biol. Evol. 2006;23:254–267. doi: 10.1093/molbev/msj030. [DOI] [PubMed] [Google Scholar]
- Huson DH, Scornavacca C. A survey of combinatorial methods for phylogenetic networks. Genome Biol. Evol. 2011;3:23–35. doi: 10.1093/gbe/evq077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibanez T, Munzinger J, Dagostini G, Hequet V, Rigault F, Jaffré T, Birnbaum P. Structural and floristic diversity of mixed tropical rain forest in New Caledonia: New data from the New Caledonian plant inventory and permanent plot network (NC-PIPPN) Appl. Veg. Sci. 2014;17:386–397. [Google Scholar]
- Jaffré T, Rigault F, Munzinger J. La végétation. In: Bonvallot J, Gay J-C, Habert É, editors. Atlas de la Nouvelle-Calédonie. IRD-Congrès de la Nouvelle-Calédonie; Marseille-Nouméa: 2012. pp. 77–80. [Google Scholar]
- Jakob SS, Heibl C, Rödder D, Blattner FR. Population demography influences climatic niche evolution: evidence from diploid American Hordeum species (Poaceae) Mol. Ecol. 2010;19(7):1423–1438. doi: 10.1111/j.1365-294X.2010.04582.x. 2010. [DOI] [PubMed] [Google Scholar]
- Jakobsson M, Rosenberg NA. CLUMPP: a cluster matching and permutation program for dealing with label switching and multimodality in analysis of population structure. Bioinformatics. 2007;23:1801–1806. doi: 10.1093/bioinformatics/btm233. [DOI] [PubMed] [Google Scholar]
- Kane NC, King MG, Barker MS, Raduski A, Karrenberg S, Yatabe Y, Knapp SJ, Rieseberg LH. Comparative genomic and population genetic analyses indicate highly porous genomes and high levels of gene flow between divergent Helianthus species. Evolution. 2009;63:2061–2075. doi: 10.1111/j.1558-5646.2009.00703.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapralov MV, Votintseva AA, Filatov DA. Molecular adaptation during a rapid adaptive radiation. Mol. Biol. Evol. 2013;30:1051–1059. doi: 10.1093/molbev/mst013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller I, Wagner CE, Greuter L, Mwaiko S, Selz OM, Sivasundar A, Wittwer S, Seehausen O. Population genomic signatures of divergent adaptation, gene flow and hybrid speciation in the rapid radiation of Lake Victoria cichlid fishes. Mol. Ecol. 2013;22:2848–2863. doi: 10.1111/mec.12083. [DOI] [PubMed] [Google Scholar]
- Kier G, Kreft H, Lee TM, Jetz W, Ibisch PL, Nowicki C, Mutke J, Barthlott W. A global assessment of endemism and species richness across island and mainland regions. Proc. Natl. Acad. Sci. USA. 2009;106:9322–9327. doi: 10.1073/pnas.0810306106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lerner HRL, Meyer M, James HF, Hofreiter M, Fleischer RC. Multilocus resolution of phylogeny and timescale in the extant adaptive radiation of Hawaiian honeycreepers. Curr. Biol. 2011;21:1838–1844. doi: 10.1016/j.cub.2011.09.039. [DOI] [PubMed] [Google Scholar]
- Lexer C, Widmer A. The genic view of plant speciation: recent progress and emerging questions. Philos. T. R. Soc. B. 2008;363:3023–3026. doi: 10.1098/rstb.2008.0078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longo G, Bernardi G. The evolutionary history of the embiotocid surfperch radiation based on genome-wide RAD sequence data. Mol. Phylogenet. Evol. 2015;88:55–63. doi: 10.1016/j.ympev.2015.03.027. [DOI] [PubMed] [Google Scholar]
- Losos JB. Adaptive radiation, ecological opportunity, and evolutionary determinism. Am. Nat. 2010;175:623–639. doi: 10.1086/652433. [DOI] [PubMed] [Google Scholar]
- Maddison WP, Knowles LL. Inferring phylogeny despite incomplete lineage sorting. Syst. Biol. 2006;55:21–30. doi: 10.1080/10635150500354928. [DOI] [PubMed] [Google Scholar]
- Maitrepierre L. Les types de temps et les cyclones, les éléments du climat. In: Bonvallot J, Gay J-C, Habert É, editors. Atlas de la Nouvelle-Calédonie. IRD-Congrès de la Nouvelle-Calédonie; Marseille-Nouméa: 2012. pp. 53–60. [Google Scholar]
- Martin SH, Dasmahapatra KK, Nadeau NJ, Salazar C, Walters JR, Simpson F, Blaxter M, Manica A, Mallet J, Jiggins CD. Genome-wide evidence for speciation with gene flow in Heliconius butterflies. Genome Res. 2013;23:1817–1828. doi: 10.1101/gr.159426.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maurizot P, Vendé-Leclerc M. New Caledonia geological map, scale 1/500000. Direction de l’Industrie, des Mines et de l’Energie - Service de la Géologie de Nouvelle-Calédonie, Bureau de Recherches Géologiques et Minières; New Caledonia: 2009. [Google Scholar]
- McLoughlin S. The breakup history of Gondwana and its impact on pre-cenozoic floristic provincialism. Aust. J. Bot. 2001;49:271–300. [Google Scholar]
- Miller MA, Pfeiffer W, Schwarz T. Proceedings of the Gateway Computing Environments Workshop (GCE) New Orleans, Louisiana: 2010. Creating the CIPRES Science Gateway for inference of large phylogenetic trees; pp. 1–8. [Google Scholar]
- Mittermeier RA, Robles-Gil P, Hoffmann M, Pilgrim J, Brooks T, Goettsch-Mittermeier C, Lamoreux J, da Fonseca GAB. Hotspots revisited: Earth’s biologically richest and most endangered terrestrial ecoregions. CEMEX; Mexico City: 2004. [Google Scholar]
- Morat P, Jaffré T, Tronchet F, Munzinger J, Pillon Y, Veillon J-M, Chalopin M. Le référentiel taxonomique florical et les caractéristiques de la flore vasculaire indigène de la Nouvelle-Calédonie. Adansonia. 2012;34:177–219. [Google Scholar]
- Myers N, Mittermeier RA, Mittermeier CG, da Fonseca GAB, Kent J. Biodiversity hotspots for conservation priorities. Nature. 2000;403:853–858. doi: 10.1038/35002501. [DOI] [PubMed] [Google Scholar]
- Ohta T. Near-neutrality in evolution of genes and gene regulation. Proc. Natl. Acad. Sci. USA. 2002;99:16134–16137. doi: 10.1073/pnas.252626899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papadopulos AST, Kaye M, Devaux C, Hipperson H, Lighten J, Dunning LT, Hutton I, Baker WJ, Butlin RK, Savolainen V. Evaluation of genetic isolation within an island flora reveals unusually widespread local adaptation and supports sympatric speciation. Philos. T. R. Soc. B. 2014;369:20130342. doi: 10.1098/rstb.2013.0342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelletier B. Geology of the New Caledonian region and its implications for the study of the New Caledonia biodiversity. In: Payri C, Richer de Forges B. Compendium of marine species from New Caledonia. Vol. II 7 IRD; Nouméa, Nouvelle-Caledonié: 2006. [Google Scholar]
- Pickrell JK, Pritchard JK. Inference of population splits and mixtures from genome-wide allele frequency data. PLoS Genet. 2012;8:e1002967. doi: 10.1371/journal.pgen.1002967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pillon Y, Hopkins HCF, Munzinger J, Amir H, Chase MW. Cryptic species, gene recombination and hybridization in the genus Spiraeanthemum (Cunoniaceae) from New Caledonia. Bot. J. Linn. Soc. 2009;161:137–152. [Google Scholar]
- Pillon Y, Hopkins HCF, Rigault F, Jaffré T, Stacy EA. Cryptic adaptive radiation in tropical forest trees in New Caledonia. New Phytol. 2014;202:521–530. doi: 10.1111/nph.12677. [DOI] [PubMed] [Google Scholar]
- Pillon Y, Munzinger J, Amir H, Lebrun M. Ultramafic soils and species sorting in the flora of New Caledonia. J. Ecol. 2010;98:1108–1116. [Google Scholar]
- Posada D. Using MODELTEST and PAUP* to select a model of nucleotide substitution. Curr. Protoc. Bioinformatics. 2003 doi: 10.1002/0471250953.bi0605s00. 10.1002/0471250953.bi0605s00. [DOI] [PubMed] [Google Scholar]
- Pritchard JK, Stephens M, Donnelly P. Inference of population structure using multilocus genotype data. Genetics. 2000;155:945–959. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purugganan MD. An evolutionary genomic tale of two rice species. Nat. Genet. 2014;45:931–932. doi: 10.1038/ng.3071. [DOI] [PubMed] [Google Scholar]
- Ree RH, Smith SA. Maximum likelihood inference of geographic range evolution by dispersal, local extinction, and cladogenesis. Syst. Biol. 2008;57:4–14. doi: 10.1080/10635150701883881. [DOI] [PubMed] [Google Scholar]
- Rieseberg LH. Evolution: replacing genes and traits through hybridization. Curr. Biol. 2009;19:R119–R122. doi: 10.1016/j.cub.2008.12.016. [DOI] [PubMed] [Google Scholar]
- Rosenberg NA. DISTRUCT: a program for the graphical display of population structure. Mol. Ecol. Notes. 2004;4:137–138. [Google Scholar]
- Savolainen O, Lascoux M, Merilä J. Ecological genomics of local adaptation. Nat. Rev. Genet. 2013;14:807–820. doi: 10.1038/nrg3522. [DOI] [PubMed] [Google Scholar]
- Seehausen O. Hybridization and adaptive radiation. Trends Ecol. Evol. 2004;19:198–207. doi: 10.1016/j.tree.2004.01.003. [DOI] [PubMed] [Google Scholar]
- Sexton JP, Hangartner SB, Hoffmann AA. Genetic isolation by environment or distance: which pattern of gene flow is most common? Evolution. 2014;68:1–15. doi: 10.1111/evo.12258. [DOI] [PubMed] [Google Scholar]
- Stamatakis A. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 2014;30:1312–1313. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stern DL. The genetic causes of convergent evolution. Nat. Rev. Genet. 2013;14:751–764. doi: 10.1038/nrg3483. [DOI] [PubMed] [Google Scholar]
- Swenson U, Munzinger J, Lowry PP, II, Cronholm B, Nylinder S. Island life -classification, speciation, and cryptic species of Pycnandra (Sapotaceae) in New Caledonia. Bot. J. Linn. Soc. 2015;179:57–77. [Google Scholar]
- Swofford DL. PAUP*: Phylogenetic analysis using parsimony (*and other methods) Sinauer Associates; Sunderland, MA: 2003. [Google Scholar]
- Tassin J, Boissenin M, Barré N. Can Ptilinopus greyii (Columbidae) disperse seeds in New Caledonia's dry forests? Pacific Sci. 2010;64:527–532. [Google Scholar]
- Tel-Zur N, Abbo S, Myslabodski D, Mizrahi Y. Modified CTAB procedure for DNA isolation from epiphytic cacti of genera Hylocereus and Selenicereus (Cactaceae) Pl. Molec. Biol. Report. 1999;17:249–254. [Google Scholar]
- Turner B, Munzinger J, Duangjai S, Temsch EM, Stockenhuber R, Barfuss MHJ, Chase MW, Samuel R. Molecular phylogenetic of New Caledonian Diospyros (Ebenaceae) using plastid and nuclear markers. Mol. Phylogenet. Evol. 2013a;69:740–763. doi: 10.1016/j.ympev.2013.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner B, Paun O, Munzinger J, Duangjai S, Chase MW, Samuel R. Analyses of amplified fragment length polymorphisms (AFLP) indicate rapid radiation of Diospyros species (Ebenaceae) endemic to New Caledonia. BMC Evol. Biol. 2013b;13:269. doi: 10.1186/1471-2148-13-269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Oppen MJH, McDonald BJ, Willis B, Miller DJ. The evolutionary history of the coral genus Acropora (Scleractinia, Cnidaria) based on a mitochondrial and a nuclear marker: reticulation, incomplete lineage sorting, or morphological convergence? Mol. Biol. Evol. 2001;18:1315–1329. doi: 10.1093/oxfordjournals.molbev.a003916. [DOI] [PubMed] [Google Scholar]
- Verdú M. Age at maturity and diversification in woody angiosperms. Evolution. 2002;56:1352–1361. doi: 10.1111/j.0014-3820.2002.tb01449.x. [DOI] [PubMed] [Google Scholar]
- Wagner CE, Keller I, Wittwer S, Selz OM, Mwaiko S, Greuter L, Sivasundar A, Seehausen O. Genome-wide RAD sequence data provide unprecedented resolution of species boundaries and relationships in the Lake Victoria cichlid adaptive radiation. Mol. Ecol. 2013;22:787–798. doi: 10.1111/mec.12023. [DOI] [PubMed] [Google Scholar]
- Webb CO, Ackerly DD, McPeek MA, Donoghue MJ. Phylogenies and community ecology. Annu. Rev. Ecol. Syst. 2002;33:475–505. [Google Scholar]
- Willis KJ, Gillson L, Knapp S. Biodiversity hotspots through time: an introduction. Philos. T. R. Soc. B. 2007;362:169–174. doi: 10.1098/rstb.2006.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C-I. The genic view of the process of speciation. J. Evol. Biol. 2001;14:861–865. [Google Scholar]
- Yu Y, Harris AJ, Blair C, He X. RASP (Reconstruct Ancestral State in Phylogenies): A tool for historical biogeography. Mol. Phylogenet. Evol. 2015;87:46–49. doi: 10.1016/j.ympev.2015.03.008. [DOI] [PubMed] [Google Scholar]
- Yule GU. A mathematical theory of evolution, based on the conclusions of Dr. J. C. Willis, F.R.S. Philos. T. R. Soc. B. 1925;213:21–87. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.