Abstract
Multiple differentially methylated sites and regions associated with adiposity have now been identified in large-scale cross sectional studies. We tested for replication of associations between previously identified CpG sites at HIF3A and adiposity in ~1,000 mother-offspring pairs from the Avon Longitudinal Study of Parents and Children. Availability of methylation and adiposity measures at multiple time points, as well as genetic data, allowed us to assess the temporal associations between adiposity and methylation and to make inferences regarding causality and directionality.
Overall, our results were discordant with those expected if HIF3A methylation has a causal effect on BMI and provided more evidence for causality in the reverse direction i.e. an effect of BMI on HIF3A methylation. These results are based on robust evidence from longitudinal analyses and were also partially supported by Mendelian randomization analysis, although this latter analysis was underpowered to detect a causal effect of BMI on HIF3A methylation. Our results also highlight an apparent long-lasting inter-generational influence of maternal BMI on offspring methylation at this locus, which may confound associations between own adiposity and HIF3A methylation. Further work is required to replicate and uncover the mechanisms underlying both the direct and inter-generational effect of adiposity on DNA methylation.
Keywords: ALSPAC, ARIES, causal inference, DNA methylation, BMI, Mendelian randomization
Introduction
The notion that epigenetic processes are linked to variation in adiposity is well established.(1) Genome-wide quantification of site-specific DNA methylation has led to the identification and validation of multiple adiposity-associated differentially methylated sites and regions.(2-8)
A large-scale epigenome-wide association study (EWAS) of body mass index (BMI), undertaken using the Illumina Infinium HumanMethylation450 BeadChip array, found robust associations between BMI and DNA methylation at three neighbouring probes in intron 1 of HIF3A which were confirmed in two additional independent cohorts.(6) Since then, the site locus has also been associated with adiposity in four further studies.(7-10) Furthermore, HIF3A methylation has been found to be associated with weight but not height, and methylation at this locus in adipose tissue has been found to be strongly associated with BMI (6; 7) indicating that methylation at this locus might be related to some component of adiposity.
HIF3A and other hypoxia inducible transcription factors (HIF) regulate cellular and vascular responses to decreased levels of oxygen, and studies in mice suggest they may play key roles in metabolism, energy expenditure and obesity.(11-14) This lends support for a role of this gene in the development of obesity and its consequent comorbidities. However, it is also possible that greater BMI induces changes in HIF3A methylation as the direction of the effect is difficult to discern in these cross-sectional studies.(6)
Further research is required to determine the directionality of the association and strengthen inference regarding causality. A large-scale longitudinal design is warranted to investigate the temporal relationship between baseline adiposity and follow-up methylation, and vice versa.(15-17)
Mendelian randomization uses genetic variants as instrumental variables (IVs) to investigate the causal relationship between an exposure and outcome of interest.(18-21) The assumptions of this approach are that the instrumental variable is: robustly related to the exposure; related to the outcome only through its robust association with the exposure of interest; and not related to confounding factors for the exposure-outcome association and not influenced by the development of the outcome. If these assumptions are true then any association observed between the IV and outcome is best explained by a true causal effect of the exposure on the outcome.(22) It has been shown that genetic variants are not likely to be related to confounding factors that explain non-genetic associations and are unaffected by disease, (23) and therefore may be used to strengthen causal inference.
In the context of methylation, Mendelian randomization may be facilitated by the strong cis-effects which allow the isolation of specific loci influencing methylation (24) and has been applied elsewhere to assess causal effects.(25; 26) In the study that identified differential methylation at HIF3A,(6) cis-genetic variants robustly associated with DNA methylation at this locus were used as causal anchors in a pseudo Mendelian randomization approach to assert no causal effect of methylation at HIF3A on adiposity. However, no attempt was made to investigate causality in the reverse direction i.e. the causal effect of adiposity on HIF3A methylation. Bidirectional Mendelian randomization may be used to elucidate the causal direction between HIF3A and adiposity, using valid IVs for each trait.(21; 27; 28)
Investigating a possible inter-generational intra-uterine effect of maternal BMI on offspring methylation could further strengthen causal inference since it is plausible that maternal BMI could influence offspring methylation through intra-uterine effects independent of offspring’s own BMI.(29) Indeed, a recent study postulated and found some evidence for a confounding effect of the prenatal environment on the association between adiposity and HIF3A methylation through an assessment of birth weight.(9) Alternatively, confounding by familial socio-economic and lifestyle characteristics may explain the observed associations between adiposity on HIF3A methylation and this was not fully assessed in the previous study.(6)
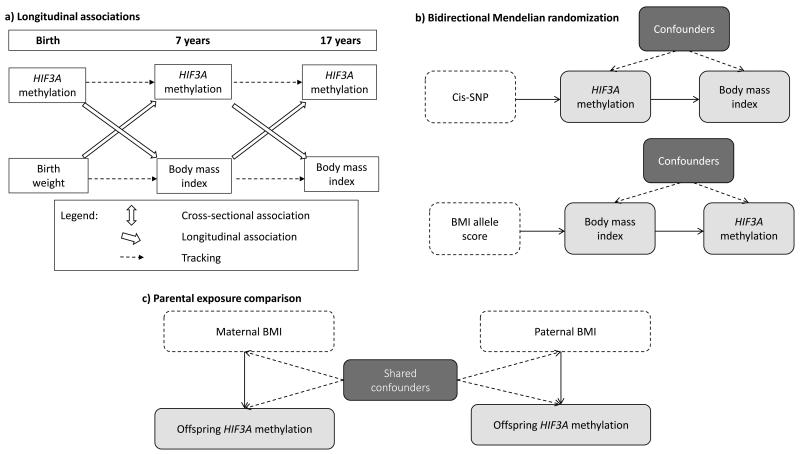
We aimed to investigate associations between methylation at HIF3A and BMI at different ages using data from the Avon Longitudinal Study of Parents and Children as part of the Accessible Resource for Integrated Epigenomics Studies (ARIES) project. We first estimated effect sizes for the three previously identified probes in HIF3A, with and without adjustment for a number of potential confounding factors. Given evidence of an association between HIF3A methylation and components of adiposity specifically, we also investigated associations between methylation at HIF3A and fat mass index (FMI).(6; 7) To further investigate the dominant direction of causality in any observed associations we undertook the following additional analyses: a) investigating longitudinal associations between BMI and methylation b) performing bidirectional Mendelian randomization analysis c) determining whether there is an inter-generational effect of parental BMI on offspring methylation, either through an intra-uterine effect of maternal BMI or a postnatal effect of paternal/maternal BMI through shared familial lifestyle or genetic factors (Figure 1). The results of the various analyses which would be expected under the different hypotheses being tested are outlined in Supplementary Table 1.
Figure 1.
Schematic diagrams of the causal inference methods being implemented in this study
a) Investigating longitudinal associations between BMI and HIF3A methylation
b) Investigating the dominant direction of causality in the association between BMI and HIF3A methylation with the use of bidirectional Mendelian randomization analysis
c) Investigating the intra-uterine effect of maternal smoking on offspring DNA methylation with the use of a parental comparison design.
Research Design and Methods
Participants
ALSPAC is a large, prospective birth cohort study based in the South West of England. 14,541 pregnant women resident in Avon, UK with expected dates of delivery 1st April 1991 to 31st December 1992 were recruited and detailed information has been collected on these women and their offspring at regular intervals.(30; 31) The study website contains details of all the data that is available through a fully searchable data dictionary (http://www.bris.ac.uk/alspac/researchers/data-access/data-dictionary/).
As part of the ARIES (Accessible Resource for Integrated Epigenomic Studies) project,(32) the Illumina Infinium® HumanMethylation450K (HM450) BeadChip (Illumina Inc., CA, USA)(33) has been used to generate epigenetic data on 1,018 mother-offspring pairs in the ALSPAC cohort (v1. Data release 2014). A web portal has been constructed to allow openly accessible browsing of aggregate ARIES DNA methylation data (ARIES-Explorer, http://www.ariesepigenomics.org.uk/).
The ARIES participants were selected based on availability of DNA samples at two time points for the mother (antenatal and at follow-up when the offspring were adolescents) and three time points for the offspring (neonatal, childhood (mean age 7.5 years) and adolescence (mean age 17.1 years)). We focused our analyses on offspring in the ARIES study who have more detailed longitudinal and parental exposure data available. Therefore, this project uses methylation data from the three time points in the offspring. A detailed description of the data available in ARIES is available in a Data Resource Profile for the study.(32)
Written informed consent has been obtained from all ALSPAC participants. Ethical approval for the study was obtained from the ALSPAC Ethics and Law Committee and the Local Research Ethics Committees.
Methylation assay – laboratory methods, quality control and pre-processing
We examined DNA methylation in relation to body mass index using methylation data from the Infinium HM450 BeadChip.(33) The Infinium HM450 Beadchip assay detects the proportion of molecules methylated at each CpG site on the array. For the samples, the methylation level at each CpG site was calculated as a beta value (β), which is the ratio of the methylated probe intensity and the overall intensity and ranges from 0 (no cytosine methylation) to 1 (complete cytosine methylation).(34; 35) All analyses of DNA methylation used these beta values.
Cord blood and peripheral blood samples (whole blood, buffy coats or blood spots) were collected according to standard procedures and the DNA methylation wet-lab and pre-processing analyses were performed as part of the ARIES project, as previously described.(32) In brief, samples from all time points in ARIES were distributed across slides using a semi-random approach to minimise the possibility of confounding by batch effects. The main batch variable was found to be the bisulphite conversion plate number. Samples failing QC (average probe p-value >= 0.01, those with sex or genotype mismatches) were excluded from further analysis and scheduled for repeat assay and probes that contained <95% of signals detectable above background signal (detection p-value <0.01) were excluded from analysis. Methylation data were pre-processed using in R (version 3.0.1), with background correction and subset quantile normalisation performed using the pipeline described by Touleimat and Tost.(36) In the offspring, 914 samples at birth, 973 samples at follow-up in childhood and 974 samples at follow-up in adolescence passed the QC.
Anthropometry
In childhood (mean age 7.5) and adolescence (mean age 17.1), offspring attended follow-up clinics where weight and height were measured with the participant in light clothing and without shoes. Weight was measured to the nearest 0.1kg with Tanita scales and height to the nearest 0.1cm using a Harpenden stadiometer. Body mass index (kg/m2) was then calculated. At the adolescent clinic, fat mass (kg) and lean mass (kg) were also assessed by Lunar Prodigy dual energy x-ray absorptiometry (DXA) scanner (GE Medical Systems Lunar, Madison, WI). The scans were visually inspected and realigned where necessary. Once complete, the tester examined the scan to ensure its quality and if necessary repeated the scan. The fat mass index (FMI; kg/m2) was calculated.
After recruitment, mothers were asked to report their height and pre-pregnancy weight in a questionnaire administered at 12 weeks gestation, which were then used to calculate pre-pregnancy maternal BMI. Reported weight was highly correlated with the first antenatal clinic measure of weight (Pearson’s correlation coefficient 0.95). Partners reported their own heights and weights in questionnaires at 12 weeks gestation, which were used to determine paternal BMI. For this study, data for partners who were not confirmed as being the biological father of the child by the mothers’ report were excluded.
Other variables
Age, sex, birth weight, gestational age, maternal education, household social class, maternal smoking and alcohol consumption in pregnancy and own smoking and alcohol were also considered potential confounders. Sex, gestational age and infant birth weight were recorded in the delivery room and abstracted from obstetric records and/or birth notifications. Gestational age was based on the date of the mother’s last menstrual period, clinical records or ultrasounds. Based on questionnaire responses in pregnancy, the highest occupation of the mother or their partner was used to define family social class as either manual or non-manual (using the 1991 British Office of Population and Census Statistics (OPCS) classification). Highest educational qualification for the mother was collapsed into whether they had achieved a university degree or not. Mothers were asked about their smoking during pregnancy and these data were used to generate a binary variable of any smoking during pregnancy. In addition, mothers were asked whether they had drunk any alcohol during the first trimester and these data were used to generate a binary variable: never or ever drank alcohol during the first trimester. Offspring smoking was obtained from a questionnaire administered at the clinic when DNA was extracted for methylation at age 15-17 years, and this was categorised into never/less than weekly, weekly and daily. Adolescent alcohol intake was obtained from the same questionnaires and categorised into whether they consumed alcohol at least weekly or not.
Genotypes
ALSPAC offspring were genotyped using the Illumina HumanHap550 quad genome-wide SNP genotyping platform (Illumina Inc., San Diego, CA, USA) by the Wellcome Trust Sanger Institute (Cambridge, UK) and the Laboratory Corporation of America (Burlington, NC, US), with support from 23andMe. ALSPAC mothers were genotyped on the Illumina 660K quad chip at the Centre National de Genotypage, Paris. DNA extraction, quality control, SNP genotyping and imputation were carried out separately in the ALSPAC mothers and offspring and have been described in detail elsewhere.(37; 38)
Statistical analysis
Cross-sectional analysis
We performed multivariable regression analysis of log-transformed BMI with concurrently measured methylation level (beta values) at each of the 3 CpG sites in HIF3A identified (6), in both mothers and offspring in ARIES. Main models were adjusted for age, sex and bisulphite conversion batch in the analyses of offspring childhood BMI, and age, sex, smoking status and bisulphite conversion batch in the analyses of offspring adolescent BMI and maternal BMI. All covariates, including bisulphite conversion batch, were included as fixed effects. BMI was treated as the outcome variable by Dick et al. and so we present coefficients as percentage change per 0.1 increase in methylation so as to be able to compare the magnitude of the observational estimates directly with those reported (6). DXA-measured fat mass index (FMI) was also investigated as the outcome variable in a secondary analysis of the individuals at adolescence, which was similarly log-transformed.
Secondary models were adjusted for age, sex, bisuphite conversion batch, birth weight, gestational age, maternal education, household social class, maternal smoking and alcohol consumption in pregnancy and own smoking and alcohol . In addition, it has been demonstrated that differences in methylation can arise as a result of variability of cell composition in whole blood.(39) In order to ensure that the results are not influenced by variation in cell type fraction between samples, we estimated the fraction of CD8+T, CD4+T, NK and B cells, monocytes and granulocytes in the samples using the estimateCellCounts function in the minfi Bioconductor package implemented in R.(40) (41) This approach uses as a reference a dataset presented by Reinius and colleagues which identified differentially methylated regions which could discriminate cell types in flow-sorted leukocytes from six adult samples. (39) Analyses were repeated adjusting for cell composition by including each blood cell fraction as a covariate in the multivariable linear regression.
Additional analyses
To further investigate the dominant direction of causality in any observed associations we undertook the following additional analyses (Figure 1):
Longitudinal analysis
Multiple linear regression models were next used to establish the association of methylation with future adiposity and of adiposity with future methylation in the offspring, with adjustments made for sex, age and batch, and baseline adiposity or methylation respectively. Specifically, BMI in adolescence was regressed on childhood methylation, and methylation in adolescence on childhood BMI. Childhood methylation was also regressed on birth weight, and childhood BMI on cord blood methylation at birth. Secondary models were adjusted for age, sex, batch, baseline adiposity or methylation, birth weight (where birth weight or methylation at birth was not the outcome or main exposure), gestational age, maternal education, household social class, maternal smoking and alcohol consumption in pregnancy and own smoking and alcohol.
Mendelian randomization analysis
It is now well established that genetic factors regulate variation in methylation (42) and two single nucleotide polymorphisms (SNPs), rs8102595 and rs3826795, were found to have strong cis-effects on methylation at HIF3A.(6) These same SNPs were not associated with BMI in either the previous study cohorts or in a large-scale meta-analysis of genome-wide association studies (GWAS) for BMI,(43) implying that increasing methylation at the HIF3A CpG sites does not have a causal effect on BMI. We aimed to perform formal Mendelian randomization analysis to establish a causal effect of methylation at HIF3A on BMI using these previously identified cis-SNPs combined in a weighted allele score, using the weights from a meta-analysis of the discovery and replication cohorts in Dick et al (6) as a proxy for methylation levels.
We also performed reciprocal Mendelian randomization analysis to investigate whether there was evidence of a causal effect of BMI on HIF3A methylation using genetic variants found to be robustly associated with BMI in large-scale GWAS.(43; 44) For this, a weighted allele score was created from 97 SNPs that have been shown to be reliably associated with BMI (44) and was used as a genetic instrument for adiposity. The dose of the effect allele at each locus was weighted by the effect size of the variant in this independent meta-analysis and these doses were summed to reflect the average number of BMI-increasing alleles carried by an individual. Analyses were performed using a standardised allele score.
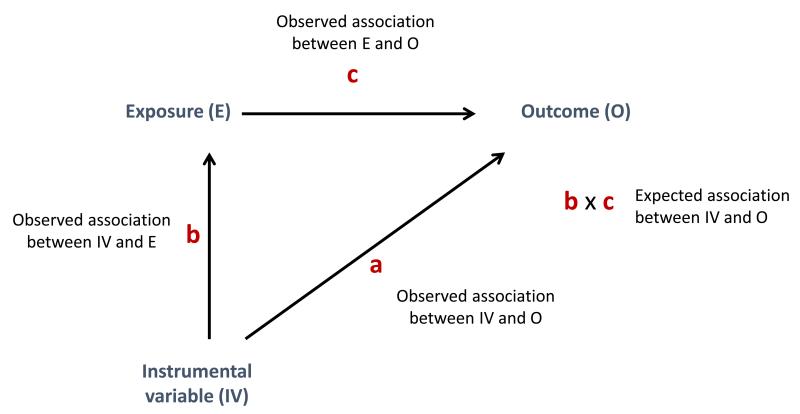
For the Mendelian randomization analyses we used the approach of “triangulation”.(45-47). This approach involves a comparison of the observed association between the instrument and the outcome with the association which would be expected if the observed exposure-outcome association were causal (Figure 2). The expected association is calculated by multiplying the observed instrument-exposure association with the observed exposure-outcome association while the standard error for the expected effect size is calculated using a second order Taylor series expansion of the product of two means, where the covariance of the estimated parameters was estimated using a bootstrapping procedure with 200 replications (48).
Figure 2.
Triangulation approach for instrumental variable analyses used in this study
The observed association between the instrumental variable and the outcome (a) is compared to that expected given the association between the instrumental variable and the exposure (b) and the association between the exposure and the outcome (c).
Here we estimated the expected effect of the instrument-outcome association based on the effect estimates for the instrument-exposure and exposure-outcome associations and compared this with the observed association of instrument with outcome (DNA methylation), performing a z-test for difference between the observed and expected estimates, where again the covariance of the estimated parameters was estimated using a bootstrapping procedure. Where the observed and expected estimates are consistent this suggests that there is unlikely to be marked residual confounding in the association between exposure and outcome (i.e. it supports a causal effect); assuming there is adequate statistical power for this comparison. The only covariate included in the main model was bisulphite conversion batch.
Inter-generational analysis
We next performed multivariable linear regression analysis to investigate associations between log-transformed maternal pre-pregnancy BMI and offspring HIF3A methylation at birth, childhood and adolescence. These models adjusted for maternal age at delivery, maternal smoking status in pregnancy, offspring sex and bisulphite conversion batch. Analyses assessing the association of maternal BMI with childhood and adolescent methylation at HIF3A were also adjusted for offspring’s age at methylation measurement.
Primarily we were interested in the direction of any causal effect and this inter-generational design effectively rules out an effect of offspring methylation on maternal BMI. Should any robust associations of maternal BMI with offspring DNA methylation at HIF3A be identified, we planned to use causal inference strategies to investigate whether these associations were likely to be caused by an intra-uterine effect of maternal BMI or rather by confounding due to shared familial lifestyles and/or genetic factors.
Specifically, these strategies were a negative control design and Mendelian randomization. In the negative control design, associations of maternal exposure and paternal exposure (the negative control) with the offspring outcome are compared. If these are similar it suggests that confounding by shared familiar factors, shared epigenetic inheritance or parental genotypes is likely, whereas a stronger maternal-offspring association (even after adjustment for paternal exposure) would provide support for a causal intra-uterine effect.( 49; 50) Associations of maternal pre-pregnancy BMI and offspring methylation at HIF3A were therefore compared, visually and formally using incremental F-tests, to associations of paternal BMI and offspring methylation, with and without mutual adjustment.
For the Mendelian randomization analysis, genetic variants in the mothers were used to create a weighted allele score for maternal BMI and the instrumental variable approach of triangulation was applied to infer a causal effect on offspring DNA methylation at HIF3A. However, in this case an obvious violation of the instrumental variable assumption is the relationship of maternal genotype to offspring (fetal) genotype which could provide a pathway from the instrument (maternal genotype) to outcome (offspring DNA methylation at HIF3A) that is not via the exposure of interest (maternal BMI) and hence would bias our findings.(51) Therefore, the analysis was adjusted for offspring’s BMI allele score. All analyses were also adjusted for bisulphite conversion batch.
All statistical analyses were performed in R (version 3.0.1).
Results
Basic characteristics
Methylation data were available for 973 children at the mean age of age 7.5 (S.D. 0.1) years and 974 adolescents at the mean age of 17.1 (1.0) years, with 940 individuals having data at both of these time points. For the three HIF3A probes identified previously, mean methylation levels were lower in adolescence than in childhood (Table 1). Methylation in childhood was positively associated with methylation in the same individuals assessed in adolescence (Pearson’s correlation coefficients: 0.72, 0.57, 0.68 at cg22891070, cg27146050 and cg16672562, respectively). R2 values for regressions of methylation in adolescence on methylation in childhood showed that childhood methylation explained 52.3%, 32.4% and 46.8% of variation in methylation in adolescence at cg22891070, cg27146050 and cg16672562 respectively
Table 1.
Characteristics of ARIES participants included in analyses
| ARIES participants* | ||
|---|---|---|
| Childhood (N=970) | Adolescence (N= 845) | |
| Age (years) | 7.5 (0.1) | 17.1 (1.0) |
| % males | 485 (49.8%) | 474 (48.7%) |
| Height (m) | 1.26 (0.05) | 1.72 (0.09) |
| Weight (kg) | 25.9 (4.6) | 66.2 (9.1) |
| Body mass index (kg/m2) | 16.2 (2.1) | 22.3 (3.9) |
| Fat mass index (kg/m2) | - | 5.9 (3.5) |
| % fat mass | - | 25.1% (11.0%) |
| % smoke at least weekly | - | 130 (15.2%) |
| Methylation of cg22891070 (β value) | 0.664 (0.102, 0.281-0.918) | 0.578 (0.120, 0.200-0.884) |
| Methylation of cg27146050 (β value) | 0.182 (0.035, 0.080-0.538) | 0.167 (0.033, 0.083-0.399) |
| Methylation of cg16672562 (β value) | 0.660 (0.131, 0.200-0.930) | 0.536 (0.147, 0.122-0.925) |
Data are mean (SD), n (%), or mean (SD, range)
Cross-sectional analysis
There were no cross-sectional associations between methylation at cg22891070 and cg16672562 and BMI in childhood or adolescence (Table 2). There was also no robust association between methylation at cg27146050 and childhood BMI (Table 2), although there was some suggestive evidence of association between and methylation across the HIF3A region and childhood BMI (Supplementary Figure 1). An association between methylation at cg27146050 and BMI in adolescence withstood Bonferroni correction; a 0.1 increase in methylation β value at cg27146050 was associated with a 4.7% (95% CI 1.0, 8.3; P=0.012) increase in BMI, which is in line with previously reported adult BMI effect estimates.(6)
Table 2.
Associations between methylation at three CpG sites at HIF3A and BMI
| Childhood | Adolescence | |||||||
|---|---|---|---|---|---|---|---|---|
| Basic model (N=970)* | Adjusted model (N=918)† | Basic model (N=845)‡ | Adjusted model (N=804)† | |||||
| Percentage change in BMI (95% CI)§ | p-value | Percentage change in BMI (95% CI)§ | p-value | Percentage change in BMI (95% CI)§ | p-value | Percentage change in BMI (95% CI)§ | p-value | |
| cg22891070 | 0.44 (−0.35, 1.23) | 0.27 | 0.45 (−0.32, 0.12) | 0.25 | 0.66 (−0.31, 1.63) | 0.19 | 0.30 (−0.67, 1.28) | 0.54 |
| cg27146050 | 0.62 (−1.69, 2.93) | 0.60 | 0.34 (−1.89, 2.56) | 0.77 | 4.66 (1.04, 8.29) | 0.01 | 3.49 (−0.12, 7.10) | 0.06 |
| cg16672562 | 0.31 (−0.32, 0.93) | 0.34 | 0.32 (−0.29, 0.93) | 0.30 | 0.40 (−0.41, 1.20) | 0.34 | 0.24 (−0.56, 1.05) | 0.55 |
Childhood analyses are adjusted for age, sex and batch.
Adolescent analyses are adjusted for age, sex, smoking and batch.
Basic model additionally adjusted for smoking, alcohol, maternal education, social class, maternal smoking, maternal smoking, maternal alcohol, birthweight and gestational age.
Coefficients have been converted into percentage change in BMI for every 0.1 unit increase in methylation β value.
We investigated whether the observed association between adolescent BMI and cg27146050 methylation could be explained by additional confounding factors (Supplementary Table 2). The association between methylation at cg27146050 and BMI in adolescence was attenuated by 25% upon adjustment for these, indicating some potential confounding in the observational association (Table 2). DNA was extracted from buffy coats in adolescence. To establish the effect of correcting for buffy coat cell type, predicted cell type components were added as covariates to the main and secondary models. Evidence for association strengthened following this adjustment (Supplementary Table 3).
Effect estimates for associations between adolescent methylation and fat mass index (FMI) were consistently larger for all three of the CpG sites compared with those for BMI, particularly at cg27146050, where an increase in methylation β value of 0.1 was associated with an 11.8% (−0.1, 23.7) increase in FMI (P=0.053), however confidence intervals were wider and the p-values for the associations did not withstand Bonferroni correction. We also investigated whether the observed associations could be explained by additional confounding factors that may exist in the context of adiposity and methylation by assessing the impact of adjusting for potential confounders on the observational effect estimates. The association between methylation at cg27146050 and FMI in adolescence was similarly attenuated by 25%, indicating some potential confounding in the observational association (Supplementary Table 4).
Longitudinal associations
We next investigated the prospective associations between HIF3A methylation at birth and childhood BMI, between birth weight and childhood HIF3A methylation, between childhood HIF3A methylation and adolescent BMI and between childhood BMI and HIF3A methylation in adolescence, with and without adjustment for adiposity or methylation at the earlier time point (Table 3). We observed positive associations between birthweight and childhood methylation at all three sites, which was not attenuated with adjustment for cord blood methylation at birth (P-values ranging 0.0019 to 0.019). While there was weak evidence of inverse associations between HIF3A methylation at birth and childhood BMI, these associations were attenuated after adjusting for birth weight (Table 3).
Table 3.
Prospective associations between birthweight and childhood methylation, between cord blood methylation at birth and childhood BMI, between childhood BMI and adolescent methylation, and between childhood methylation and adolescent BMI
| Exposure | Outcome | CpG site | N | Association without adjustment for the outcome at baseline* | N | Association with adjustment for the outcome at baseline* | ||
|---|---|---|---|---|---|---|---|---|
| β (95% CI) | p-value | β (95% CI) | p-value | |||||
| Birth methylation | Childhood BMI | cg22891070 | 890 | −1.70 (−3.66, 0.30)a | 0.10 | 874 | −1.65 (−3.52, 0.26)a | 0.09 |
| cg27146050 | 890 | −0.03 (−1.04, 1.00)a | 0.96 | 874 | 0.21 (−0.77, 1.21)a | 0.67 | ||
| cg16672562 | 890 | −2.66 (−5.10, −0.16)a | 0.04 | 874 | −2.23 (−4.58, 0.17)a | 0.07 | ||
|
| ||||||||
| Birth weight | Childhood methylation | cg22891070 | 957 | 0.02 (0.01, 0.04)b | 0.01 | 871 | 0.02 (0.003, 0.035)b | 0.02 |
| cg27146050 | 957 | 0.01 (0.001, 0.012)b | 0.02 | 871 | 0.01 (0.003, 0.014)b | 0.04 | ||
| cg16672562 | 957 | 0.03 (0.01, 0.05)b | 0.01 | 871 | 0.03 (0.006, 0.046)b | 0.01 | ||
|
| ||||||||
| Childhood methylation | Adolescent BMI | cg22891070 | 922 | 0.68 (−0.40, 1.76)a | 0.22 | 919 | 0.14 (−0.64, 0.91)a | 0.73 |
| cg27146050 | 922 | 2.30 (−0.83, 5.43)a | 0.15 | 919 | 1.33 (−0.91, 3.57)a | 0.24 | ||
| cg16672562 | 922 | 0.31 (−0.54, 1.15)a | 0.48 | 919 | −0.04 (−0.64, 0.57)a | 0.90 | ||
|
| ||||||||
| Childhood BMI | Adolescent methylation | cg22891070 | 971 | 0.005(−0.002, 0.011)c | 0.17 | 937 | 0.001 (−0.004, 0.005)c | 0.78 |
| cg27146050 | 971 | 0.003 (0.001, 0.005)c | 0.001 | 937 | 0.003 (0.001, 0.004)c | 0.001 | ||
| cg16672562 | 971 | 0.005 (−0.003, 0.013)c | 0.21 | 937 | 0.002 (−0.004, 0.008)c | 0.60 | ||
Also adjusted for age at childhood/adolescence, sex, batch
Coefficients have been converted into percentage change in BMI for every 0.1 unit increase in methylation β value.
Coefficients are change in methylation per 1kg increase in birthweight.
Coefficients are change in methylation per 10% increase in BMI.
We also observed a positive association between childhood BMI and cg27146050 methylation in adolescence (0.003 (0.001, 0.005) increase in methylation β value per 10% increase in BMI; P=0.001) which was not attenuated with adjustment for childhood methylation at this site. The effect remained unchanged with adjustment for a number of potential confounders (Supplementary Table 5). However, there were no prospective associations between childhood BMI and adolescent methylation at cg22890170 or cg16672562
Mendelian randomization analysis
To investigate the potential effect of methylation at cg27146050 on BMI, we first assessed genetic associations with methylation using a score composed of two single nucleotide polymorphisms (SNPs), rs8102595 and rs3826795, found to have strong cis-effects on methylation at HIF3A in an independent study (6). There was a 0.2 (0.16, 0.25; P<10−10; R2=7.4%) increase in methylation β value at cg27146050 per unit increase in the cis-SNP score (Supplementary Table 6). Unlike for the adiposity and methylation measures, there was no strong evidence of association between the cis-SNP score and a number of potential confounding factors (Supplementary Table 8).
Given the strength of the association with methylation at cg27146050 and lack of association with confounding factors, we used the cis-SNP score as an instrument for methylation in a Mendelian randomization analysis. There was little association between the cis-SNPs and BMI compared with the expected association if methylation on BMI was causal (Table 4). However, wide confidence intervals for the observed estimates meant that there was no strong evidence of a difference between the observed and expected effect estimates (observed effect = −0.04 (−0.29, 0.22); expected effect = 0.10 (0.03, 0.17); P-for-difference= 0.30).
Table 4.
Mendelian randomization analysis for associations between adolescent BMI and methylation at cg27146050.
| Instrumental variable (IV) | Exposure (E) | Outcome (O) | Observed association between IV and O (c)* | Expected association between IV and O (a × b) | Difference between observed (c) and expected (a × b) estimates | |
|---|---|---|---|---|---|---|
| N | β (95% CI) | β (95% CI) | P-value | |||
| Adolescent cis-SNP score | Adolescent methylation at cg27146050 | Adolescent log BMI | 831 | −0.0381 (−0.2937, 0.2176) | 0.1027 (0.0315, 0.1739) | 0.30 |
| Adolescent standardised 97 SNP allele score | Adolescent log BMI | Adolescent methylation at cg27146050 | 849 | 0.0014 (−0.0009, 0.0037) | 0.0008 (0.0002, 0.0013) | 0.55 |
Analyses are adjusted for bisulphite conversion batch only
We calculated that we would need a sample of N=25,369 to confidently detect an association (at p<0.001) between the cis-SNP allele score that explained 0.1% of the variance in log BMI with 95% power. Therefore, we also tested for associations between the cis-SNPs and body mass index by performing a look-up of the SNPs in the publically-available results of the most recent GIANT consortium meta-analysis.(44) In this sample, there was no strong evidence of association between either of the SNPs and BMI (rs3826795: n=224,403, β (SE) = 0.002 (0.005), p=0·63; rs8102595: n=223,534, β (SE) = −0.002 (0.007), p=0·78), in accordance with previous findings using data from a smaller meta-analysis in GIANT. (6) In addition, we performed two-sample Mendelian randomization,(52) using SNP-methylation association estimates obtained from the ARIES data set and SNP-BMI association estimates obtained from the GIANT results to derive a Wald ratio estimate for the causal effect of methylation on BMI. Using inverse-weighted variance meta-analysis of the estimates derived using the two SNPs, a 1-unit increase in methylation was associated with a −0.021 (−0.55, 0.51); p=0.94 decrease in inverse-normally transformed BMI residuals i.e. providing further evidence against a causal effect of methylation at HIF3A on BMI (Supplementary Table 10).
To investigate the potential effect of BMI on methylation at cg27146050, we confirmed the expected association between a weighted allele score composed of 97 BMI variants identified in an independent study(44) and log-transformed BMI in our sample (β =0.036 (0.025, 0.046) P<10−10, R2=5.2%) (Supplementary Table 7). Unlike for the adiposity and methylation measures, there was no evidence of association between the BMI allele score and a number of potential confounding factors (Supplementary Table 8). Although there was some evidence for a difference in mean allele score between groups based on adolescent own smoking, this was driven by a small number of individuals in the group who smoked weekly (n= 29) and no linear trend was observed.
We applied this instrument to investigate the potential causal effect of BMI on HIF3A methylation (Table 4). The direction of effect observed was consistent with that expected if the effect were causal. In addition, there was little evidence of a difference between the observed and expected effect estimates (observed effect = 0.0014 (−0.0009, 0.0037); expected effect = 0.0008 (0.0002, 0.0013); P-for-difference=0.55). However, due to wide confidence intervals, no robust evidence of an association between the allele score and methylation was observed. In order to confidently detect an association between the BMI allele score and HIF3A methylation (at p<0.001) that explained 0.1% of the variance in log BMI with 95% power, we calculated that we would need a sample of N=30,523. Unfortunately, no publically-available meQTL data of this sample size are currently available to investigate this.
Inter-generational analysis
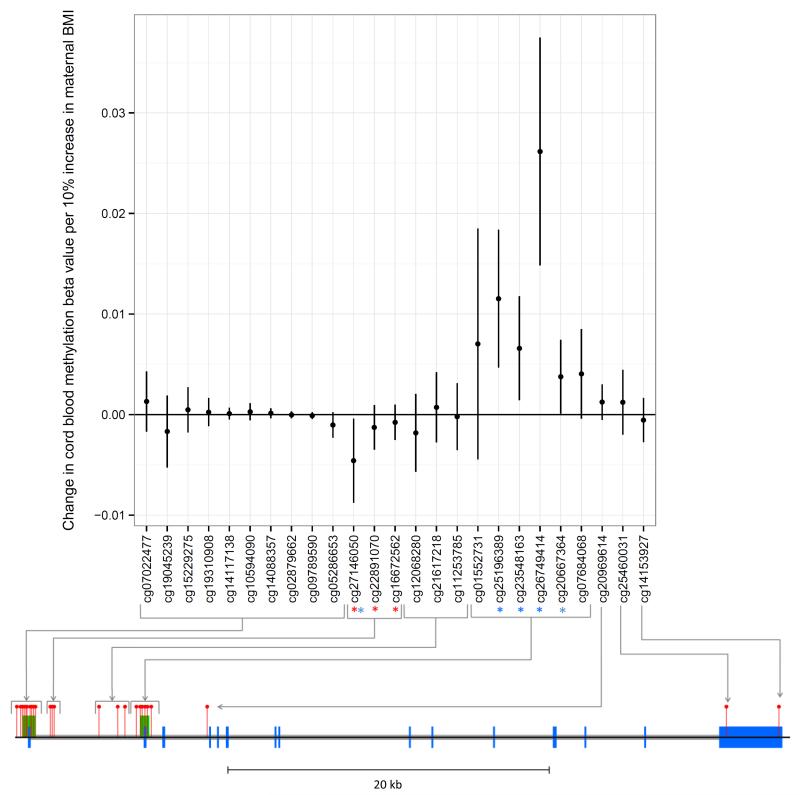
We next carried out an inter-generational analysis to investigate a potential intrauterine effect of maternal BMI on offspring methylation at cg27146050 from birth to adolescence. Maternal pre-pregnancy BMI was associated with offspring cord blood methylation at cg27146050 (P= 0.027). However, whereas own BMI was positively associated with methylation at this site, maternal BMI was inversely associated with offspring DNA methylation at cg27146050 in cord blood (−0.0048 (−0.0092, 0.0004) change in methylation per 10% increase in maternal BMI) (Figure 3).
Figure 3.
Associations between maternal BMI and offspring methylation at birth at HIF3A CpG sites
Associations of maternal BMI and offspring cord blood methylation at birth at all 25 CpG sites at the HIF3A locus (mean change in methylation per unit increase in log maternal pre-pregnancy BMI; error bars indicate 95% confidence intervals). The locations of CpG sites on the HIF3A gene are mapped on the diagram below the graph. Blue blocks are exons, grey blocks are introns, green blocks are CpG islands and red pins are CpG sites. The three sites previously identified in adult peripheral blood as associated with own BMI are highlighted with a red asterisk. All sites associated with maternal BMI with a P-value <0.05 in our analyses are highlighted with a blue asterisk.
Maternal BMI was also associated with cord blood methylation at four other CpG sites at HIF3A (cg20667364, cg26749414, cg25196389 and cg23548163; P-values ranging 7.5 × 10−6 to 4.6 × 10−2) (Figure 3). These sites in the second CpG island were found to be positively associated with maternal BMI (in contrast to cg27146050, which was negatively associated). A heatmap of the correlation between methylation β-values at HIF3A (Supplementary Figure 2) shows that the sites in the second CpG island are inversely correlated with cg27146050.
Associations between maternal BMI and offspring methylation at birth at the additional sites in the second CpG island did not persist at later ages (Supplementary Figure 3, birth n = 795, childhood n = 845, adolescence n = 851). The inverse association of maternal pre-pregnancy BMI with methylation at cg27146050 in cord blood reversed to a positive one in adolescence, in line with the association of own BMI with methylation at this site.
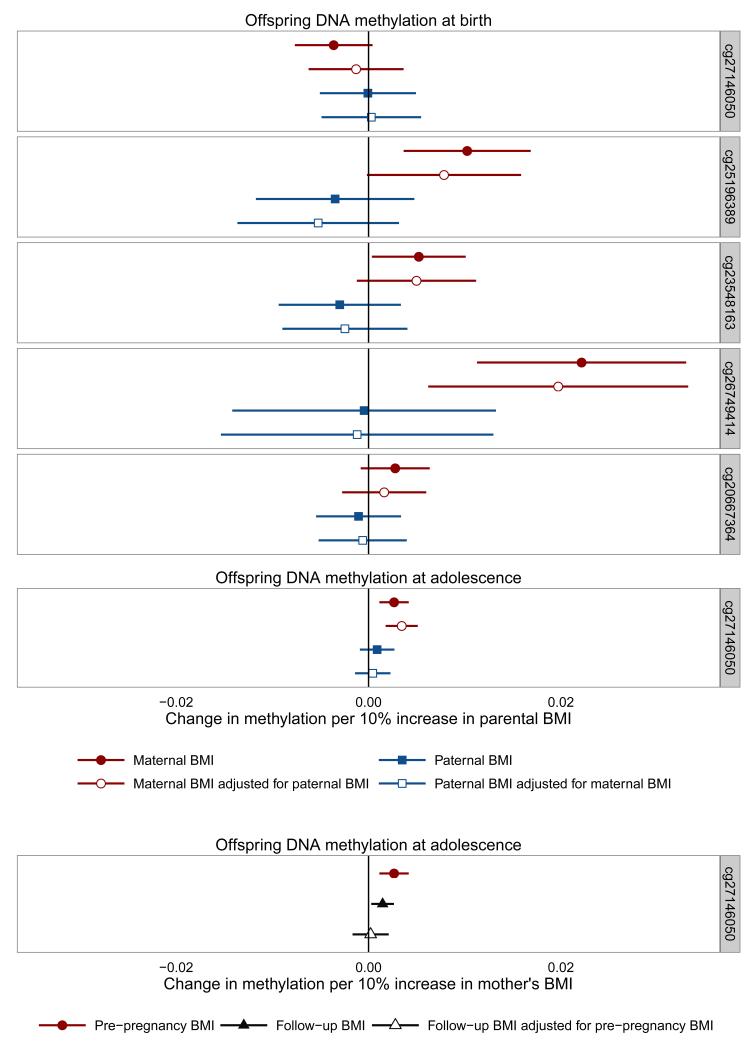
Using a negative control design, we found that the association between maternal BMI and offspring methylation at the sites identified in cord blood tended to be stronger than the association with paternal BMI (Figure 4, maternal n =797, paternal n = 655, mutually adjusted n = 625), but after mutual adjustment of maternal and paternal BMI, there was only robust evidence that they differed at cg25196389 (Wald test p-value for difference between maternal and paternal associations with mutual adjustment: 0.031 for cg25196389, all other probes > 0.05). We also found that, for cg27146050 in adolescence, the association with pre-pregnancy maternal BMI was stronger than the association with paternal BMI with and without mutual adjustment (Figure 4; Wald test p-value = 0.009), and was also stronger than the association with maternal BMI measured postnatally when their offspring were approximately age 15 (Figure 4; Wald test p-value = 0.050 in adjusted model).
Figure 4.
Associations between parental BMI and offspring DNA methylation at HIF3A
Error bars indicate 95% confidence intervals. Maternal antenatal n = 849 [birth] 904 [adolescence], paternal n = 694 [birth] 742 [adolescence], mutually adjusted n =662 [birth] 708 [adolescence], maternal at follow-up n = 819 [adolescence], maternal antenatal adjusted for maternal at follow-up n = 763 [adolescence].
In the Mendelian randomization analyses of maternal BMI on cord blood methylation (Supplementary Table 9), the observed associations between the IV and offspring methylation were stronger than the expected estimates, though 95% confidence intervals were wide and included the null value at most sites. There was little evidence that the expected and observed associations of the maternal BMI allele score with offspring methylation differed. Adjusting for offspring allelic score slightly strengthened the observed maternal allelic score –methylation relationship, but conclusions were generally the same. However, in the Mendelian randomization analysis of maternal BMI on cg27146050 methylation in adolescence, there was no observed association between maternal genotype and offspring methylation which we would expect to find if the effect of maternal BMI on offspring methylation in adolescence was causal. However, again effect estimates were imprecise. (Supplementary Table 9).
Discussion
In this study, we tested for replication of a previous investigation of the association between BMI and DNA methylation at HIF3A in childhood and adolescence in a subset of individuals from the Avon Longitudinal Study of Parents and Children.(6) Although no clear cross-sectional associations were observed between childhood BMI and methylation, we found evidence of a positive association between adolescent BMI and methylation at cg27146050 in HIF3A, with a magnitude of effect similar to that seen previously.(6)
We also examined the association between HIF3A methylation and DXA-derived FMI in adolescence and found positive associations at all three CpG sites. Effect estimates were larger than those observed in the associations with BMI, although the associations were imprecisely estimated with wide confidence intervals that included the null value.
We carried out several additional analyses to investigate the dominant direction of causality in any observed associations (Figure 1). In longitudinal analysis, we found an association between childhood BMI and methylation in adolescence, but childhood methylation was not robustly associated with BMI in adolescence, implying that the direction of any possible effect is from adiposity to methylation at this locus, rather than the other way round.
For the Mendelian randomization analysis, we confirmed associations between two cis-SNPs and methylation at HIF3A and, in line with the aforementioned study, (6) did not find associations between these SNPs and BMI, suggesting that variation in methylation at HIF3A does not causally affect BMI. This was supported by our finding that the observed effect estimate of the SNPs on BMI was different from that expected if methylation at HIF3A had a causal effect on BMI in the ARIES sample, as well as a null effect estimate for the causal effect of HIF3A methylation on BMI in the GIANT data set (44) established using a two-sample Mendelian randomization approach.
We were able to extend the analysis by using instruments for BMI to investigate causality of the reciprocal effect. We used an allele score composed of variants robustly associated with BMI in an independent GWAS (44) and assessed the magnitude of association between this score and methylation at HIF3A in adolescence. Whilst this analysis showed no robust evidence of an association between the allele score and methylation, confidence intervals were wide and here the observed effect estimate was in the same direction and exceeded the expected magnitude of a causal effect.
Several studies have shown that maternal adiposity during pregnancy is associated with offspring DNA methylation.(53-56) We carried out inter-generational analysis and identified associations between maternal pre-pregnancy BMI and offspring cord blood methylation at cg27146050, as well as four novel CpG sites at HIF3A. Since the association of maternal BMI with offspring DNA methylation could not be explained by reverse causality, this lends further plausibility to an effect of adiposity on DNA methylation at HIF3A.
Associations of maternal BMI and offspring methylation at the novel sites at HIF3A were stronger at birth than in childhood and adolescence, suggesting that any effect of maternal BMI on neonatal DNA methylation at these sites does not persist into later life. This seemingly transient effect of maternal BMI on offspring cord methylation at HIF3A may be indicative of changes in the regulation of HIFs specific to pregnancy.(57) Meanwhile, for cg27146050, an association between maternal BMI and offspring methylation was evident at all three time points, although the direction of the association changed over time.
Some evidence for a causal intra-uterine effect of maternal BMI on offspring cord blood was supported with the use of both a parental negative control comparison analysis, where no association was seen between paternal BMI (the negative control) and offspring cord methylation, and Mendelian randomization using a BMI allele score in the mothers. For the latter, conclusions were similar even after adjustment for offspring genotype. A parental comparison analysis also provided support for a possible legacy from the intra-uterine effect of maternal BMI on offspring DNA methylation into adolescence, as has been previously identified in the case of maternal smoking in pregnancy.(58; 59) However, this could be influenced by parental differences in the proportion of environmental factors shared with offspring postnatally and, while maternal BMI in pregnancy was more strongly associated with offspring methylation than maternal BMI postnatally, Mendelian randomization did not provide strong support for a causal intra-uterine effect at this later time point.
Strengths of this analysis include the extension of a previous study, with the aim of replicating identified associations between BMI and methylation at HIF3A locus in a younger cohort. We obtained similar findings in terms of direction of effect between BMI and methylation at the identified CpG sites in HIF3A although associations were weaker, as has been found previously.(9) In addition, more thorough consideration has been given to a number of potential confounding factors and both longitudinal and Mendelian randomization analysis have been used to assess causality in the observed association.
The main limitation of this analysis was the limited power to detect a difference between the observed and expected triangulation estimates between the BMI allele score and DNA methylation and further exploration in additional large studies is warranted. Other possible limitations of Mendelian randomization include population stratification, canalization, pleiotropy and linkage disequilibrium.(18; 21; 60) Major population stratification is unlikely since this analysis was completed in unrelated individuals of European ancestry. However, a pleiotropic association of either a cis-SNP with BMI or the BMI allele score with HIF3A methylation, or linkage disequilibrium between these genotypes and a functional variant independently associated with the outcome, would violate the assumptions of the Mendelian randomization analysis.
While the genetic variants included in the cis-SNP score were found to be robustly associated with cg27146050 methylation levels, in a previous study they have been associated with methylation at the neighbouring CpG, cg22891070, implying non-specificity of these genetic instruments which instead proxy for regional HIF3A methylation levels rather than methylation at individual CpG sites. To investigate specificity of the BMI SNPs, we performed a look-up of the 97 SNPs in a large scale meQTL analysis within the ARIES data set and did not find any SNP-CpG associations which surpassed genome-wide significance, indicating that it is unlikely that the BMI SNPs have a pleiotropic influence on methylation independent of BMI.
Canalization (or developmental compensation) could potentially bias the Mendelian randomization analysis assessing causality in the adolescent BMI-methylation association but is not an issue in the inter-generational analysis since the mother’s genetic instrument will only influence the developmental environment of the offspring through the exposure of interest.(61) Nonetheless, the inter-generational Mendelian randomization estimates are potentially biased with adjustment for offspring BMI genotype, which might introduce a different pathway between maternal BMI genotype and paternal BMI genotype (a form of collider bias). However, as we have already stated it is unlikely that paternal BMI will have a direct effect on offspring methylation and adjusting for offspring BMI genotype did not substantially alter effect estimates for this MR analysis.
Further limitations of the study include missing data for BMI, FMI and some of the potential confounders which reduced the complete case sample size. It should be noted that we found no CpG sites in HIF3A that were associated with either offspring or maternal BMI with a P-value <1 × 10−7 (the widely-used Bonferroni cut-off for genome-wide significance on the HM450 array), therefore an epigenome-wide association study of own or maternal BMI in ARIES would not have identified any sites in HIF3A. However, given the existence of correlation structure and comethylation in this region, correction for multiple testing based on independent tests in an EWAS would likely be too stringent. Additionally, eight of the 25 Illumina 450k probes at HIF3A appear on a comprehensive list of probes that provide noisy or inaccurate signals.(62) This list includes two (cg22891070 and cg16672562) of the sites previously identified as being associated with own BMI, so these findings are at a high risk of being false discoveries. In addition, although not the primary focus of our analyses, we did not find strong associations between HIF3A methylation at any of the three sites and BMI in the ARIES mothers at the time of pregnancy or around 17 years later at follow-up, although the direction of effect was consistent with that found previously at these sites (6) (Supplementary Table 11).
An additional limitation is that cord blood or peripheral blood may not be the most appropriate tissues in which to study associations with BMI and a more pronounced association of BMI with HIF3A methylation has been observed in adipose tissue.(6; 7). Furthermore, this analysis was limited to blood samples with mixed cell composition. Although no differences were found in the analysis with estimated cell-type correction, it is unclear how effective the method used to correct for cell-type proportions is in these samples since the reference data sets are available only for adult peripheral blood (39).
Conclusions
Overall our results do not support a causal effect of HIF3A methylation on BMI, and are more suggestive of a causal effect in the reverse direction i.e. an effect of higher BMI on higher HIF3A methylation. Use of a range of causal inference techniques including longitudinal analysis, Mendelian randomization and a parental comparison design provided findings largely consistent with both a causal effect of own BMI on methylation at HIF3A as well as an independent intra-uterine effect of maternal BMI on offspring cord blood methylation at HIF3A (Supplementary Figure 1). Further work is required to uncover the mechanisms underlying both a direct and intra-uterine effect of adiposity on DNA methylation in this gene and to investigate its role in the downstream effects of adiposity, given that methylation changes have been shown to influence gene expression at this locus.(6)
Supplementary Material
Acknowledgments
We are extremely grateful to all the families who took part in this study, the midwives for their help in recruiting them, and the whole ALSPAC team, which includes interviewers, computer and laboratory technicians, clerical workers, research scientists, volunteers, managers, receptionists and nurses. As the guarantors for this manuscript, RCR, GCS and MEW take full responsibility for the work as a whole.
Conceived and designed the experiments: RCR, GCS, MEW, AF, DAL, GDS, CLR
Analysed the data: RCR, GCS, MEW
Wrote the paper: RCR, GCS, MEW, AF, DAL, GDS, CLR
Data production and management: OL, WM, SR, TG
Funding
This work was supported by the UK Medical Research Council Integrative Epidemiology Unit and the University of Bristol (MC_UU_12013_1, MC_UU_12013_2, MC_UU_12013_5 and MC_UU_12013_8), the Wellcome Trust (WT088806) and the United States National Institute of Diabetes and Digestive and Kidney Diseases (R01 DK10324). AF is funded by a UK Medical Research Council research fellowship (MR/M009351/1). RCR and MEW are funded by a Wellcome Trust 4-year PhD studentship (Grant Code: WT097097MF and 099873/Z/12/Z). GDS and CLR are partially supported by the ESRC (RES-060-23-0011) “The biosocial archive: transforming lifecourse social research through the incorporation of epigenetic measures” The UK Medical Research Council and the Wellcome Trust (Grant ref: 102215/2/13/2) and the University of Bristol provide core support for ALSPAC. The Accessible Resource for Integrated Epigenomics Studies (ARIES) was funded by the UK Biotechnology and Biological Sciences Research Council (BB/I025751/1 and BB/I025263/1). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. This publication is the work of the authors and RCR, GS and MEW will serve as guarantors for the contents of this paper.
Footnotes
Disclosure declaration
None to declare
References
- 1.van Dijk SJ, Molloy PL, Varinli H, Morrison JL, Muhlhausler BS, members of Epi S Epigenetics and human obesity. Int J Obes (Lond) 2015:85–97. doi: 10.1038/ijo.2014.34. [DOI] [PubMed] [Google Scholar]
- 2.Wang XL, Zhu HD, Snieder H, Su SY, Munn D, Harshfield G, Maria BL, Dong YB, Treiber F, Gutin B, Shi HD. Obesity related methylation changes in DNA of peripheral blood leukocytes. BMC Medicine. 2010;8:87. doi: 10.1186/1741-7015-8-87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Feinberg AP, Irizarry RA, Fradin D, Aryee MJ, Murakami P, Aspelund T, Eiriksdottir G, Harris TB, Launer L, Gudnason V, Fallin MD. Personalized Epigenomic Signatures That Are Stable Over Time and Covary with Body Mass Index. Science Translational Medicine. 2010;2 doi: 10.1126/scitranslmed.3001262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Almen MS, Jacobsson JA, Moschonis G, Benedict C, Chrousos GP, Fredriksson R, Schioth HB. Genome wide analysis reveals association of a FTO gene variant with epigenetic changes. Genomics. 2012;99:132–137. doi: 10.1016/j.ygeno.2011.12.007. [DOI] [PubMed] [Google Scholar]
- 5.Xu XJ, Su SY, Barnes VA, De Miguel C, Pollock J, Ownby D, Shi HD, Zhu HD, Snieder H, Wang XL. A genome-wide methylation study on obesity Differential variability and differential methylation. Epigenetics. 2013;8:522–533. doi: 10.4161/epi.24506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dick KJ, Nelson CP, Tsaprouni L, Sandling JK, Aissi D, Wahl S, Meduri E, Morange PE, Gagnon F, Grallert H, Waldenberger M, Peters A, Erdmann J, Hengstenberg C, Cambien F, Goodall AH, Ouwehand WH, Schunkert H, Thompson JR, Spector TD, Gieger C, Tregouet DA, Deloukas P, Samani NJ. DNA methylation and body-mass index: a genome-wide analysis. Lancet. 2014;383:1990–1998. doi: 10.1016/S0140-6736(13)62674-4. [DOI] [PubMed] [Google Scholar]
- 7.Agha G, Houseman EA, Kelsey KT, Eaton CB, Buka SL, Loucks EB. Adiposity is associated with DNA methylation profile in adipose tissue. Int J Epidemiol. 2014 doi: 10.1093/ije/dyu236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Demerath EW, Guan W, Grove ML, Aslibekyan S, Mendelson M, Zhou YH, Hedman AK, Sandling JK, Li LA, Irvin MR, Zhi D, Deloukas P, Liang L, Liu C, Bressler J, Spector TD, North K, Li Y, Absher DM, Levy D, Arnett DK, Fornage M, Pankow JS, Boerwinkle E. Epigenome-wide association study (EWAS) of BMI, BMI change and waist circumference in African American adults identifies multiple replicated loci. Hum Mol Genet. 2015 doi: 10.1093/hmg/ddv161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pan H, Lin X, Wu Y, Chen L, Teh AL, Soh SE, Lee YS, Tint MT, MacIsaac JL, Morin AM, Tan KH, Yap F, Saw SM, Kobor MS, Meaney MJ, Godfrey KM, Chong YS, Gluckman PD, Karnani N, Holbrook JD, Group GS HIF3A association with adiposity: the story begins before birth. Epigenomics. 2015;1:13. doi: 10.2217/epi.15.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Huang T, Zheng Y, Qi Q, Xu M, Ley SH, Li Y, Kang JH, Wiggs J, Pasquale LR, Chan AT, Rimm EB, Hunter DJ, Manson JE, Willett WC, Hu FB, Qi L. DNA methylation variants at HIF3A locus, B vitamins intake, and long-term weight change: gene-diet interactions in two US cohorts. Diabetes. 2015 doi: 10.2337/db15-0264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Park YS, David AE, Huang Y, Park JB, He H, Byun Y, Yang VC. In vivo delivery of cell-permeable antisense hypoxia-inducible factor 1alpha oligonucleotide to adipose tissue reduces adiposity in obese mice. J Control Release. 2012;161:1–9. doi: 10.1016/j.jconrel.2012.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang H, Zhang G, Gonzalez FJ, Park SM, Cai D. Hypoxia-inducible factor directs POMC gene to mediate hypothalamic glucose sensing and energy balance regulation. PLoS Biol. 2011;9:e1001112. doi: 10.1371/journal.pbio.1001112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jiang C, Qu A, Matsubara T, Chanturiya T, Jou W, Gavrilova O, Shah YM, Gonzalez FJ. Disruption of hypoxia-inducible factor 1 in adipocytes improves insulin sensitivity and decreases adiposity in high-fat diet-fed mice. Diabetes. 2011;60:2484–2495. doi: 10.2337/db11-0174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shin MK, Drager LF, Yao Q, Bevans-Fonti S, Yoo DY, Jun JC, Aja S, Bhanot S, Polotsky VY. Metabolic consequences of high-fat diet are attenuated by suppression of HIF-1alpha. PLOS One. 2012;7:e46562. doi: 10.1371/journal.pone.0046562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Relton CL, Davey Smith G. Epigenetic Epidemiology of Common Complex Disease: Prospects for Prediction, Prevention, and Treatment. PLOS medicine. 2010;7:e1000356. doi: 10.1371/journal.pmed.1000356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Relton CL, Groom A, St Pourcain B, Sayers AE, Swan DC, Embleton ND, Pearce MS, Ring SM, Northstone K, Tobias JH, Trakalo J, Ness AR, Shaheen SO, Davey Smith G. DNA Methylation Patterns in Cord Blood DNA and Body Size in Childhood. PLOS One. 2012;7:e31821. doi: 10.1371/journal.pone.0031821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ng JW, Barrett LM, Wong A, Kuh D, Davey Smith G, Relton CL. The role of longitudinal cohort studies in epigenetic epidemiology: challenges and opportunities. Genome Biol. 2012;13:246. doi: 10.1186/gb-2012-13-6-246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Davey Smith G, Ebrahim S. 'Mendelian randomization': can genetic epidemiology contribute to understanding environmental determinants of disease? Int Journal Epidemio. 2003;32:1–22. doi: 10.1093/ije/dyg070. [DOI] [PubMed] [Google Scholar]
- 19.Didelez V, Sheehan N. Mendelian randomization as an instrumental variable approach to causal inference. Statistical Methods in Medical Research. 2007;16:309–330. doi: 10.1177/0962280206077743. [DOI] [PubMed] [Google Scholar]
- 20.Lawlor DA, Harbord RM, Sterne JAC, Timpson N, Davey Smith G. Mendelian randomization: Using genes as instruments for making causal inferences in epidemiology. Statistics in Medicine. 2008;27:1133–1163. doi: 10.1002/sim.3034. [DOI] [PubMed] [Google Scholar]
- 21.Davey Smith G, Hemani G. Mendelian randomization: genetic anchors for causal inference in epidemiological studies. Hum Mol Genet. 2014;23(R1):R89–R98. doi: 10.1093/hmg/ddu328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Didelez V, Meng S, Sheehan NA. Assumptions of IV Methods for Observational Epidemiology. Statistical Science. 2010;25:22–40. [Google Scholar]
- 23.Davey Smith G, Lawlor DA, Harbord R, Timpson N, Day I, Ebrahim S. Clustered environments and randomized genes: a fundamental distinction between conventional and genetic epidemiology. PLOS medicine. 2007;4:e352. doi: 10.1371/journal.pmed.0040352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Relton CL, Davey Smith G. Two-step epigenetic Mendelian randomization: a strategy for establishing the causal role of epigenetic processes in pathways to disease. Int J Epidemiol. 2012;41:161–176. doi: 10.1093/ije/dyr233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liang L, Willis-Owen SA, Laprise C, Wong KC, Davies GA, Hudson TJ, Binia A, Hopkin JM, Yang IV, Grundberg E, Busche S, Hudson M, Ronnblom L, Pastinen TM, Schwartz DA, Lathrop GM, Moffatt MF, Cookson WO. An epigenome-wide association study of total serum immunoglobulin E concentration. Nature. 2015;520:670–674. doi: 10.1038/nature14125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Allard C, Desgagne V, Patenaude J, Lacroix M, Guillemette L, Battista MC, Doyon M, Menard J, Ardilouze JL, Perron P, Bouchard L, Hivert MF. Mendelian randomization supports causality between maternal hyperglycemia and epigenetic regulation of leptin gene in newborns. Epigenetics : official journal of the DNA Methylation Society. 2015;10:342–351. doi: 10.1080/15592294.2015.1029700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Timpson NJ, Nordestgaard BG, Harbord RM, Zacho J, Frayling TM, Tybjaerg-Hansen A, Davey Smith G. C-reactive protein levels and body mass index: elucidating direction of causation through reciprocal Mendelian randomization. Int J Obesity. 2011;35:300–308. doi: 10.1038/ijo.2010.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Welsh P, Polisecki E, Robertson M, Jahn S, Buckley BM, de Craen AJM, Ford I, Jukema JW, Macfarlane PW, Packard CJ, Stott DJ, Westendorp RGJ, Shepherd J, Hingorani AD, Davey Smith G, Schaefer E, Sattar N. Unraveling the Directional Link between Adiposity and Inflammation: A Bidirectional Mendelian Randomization Approach. JCEM. 2010;95:93–99. doi: 10.1210/jc.2009-1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lawlor DA, Relton C, Sattar N, Nelson SM. Maternal adiposity--a determinant of perinatal and offspring outcomes? Nat Rev Endocrinol. 2012;8:679–688. doi: 10.1038/nrendo.2012.176. [DOI] [PubMed] [Google Scholar]
- 30.Boyd A, Golding J, Macleod J, Lawlor DA, Fraser A, Henderson J, Molloy L, Ness A, Ring S, Davey Smith G. Cohort Profile: the 'children of the 90s'--the index offspring of the Avon Longitudinal Study of Parents and Children. Int J Epidemiol. 2013;42:111–127. doi: 10.1093/ije/dys064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fraser A, Macdonald-Wallis C, Tilling K, Boyd A, Golding J, Davey Smith G, Henderson J, Macleod J, Molloy L, Ness A, Ring S, Nelson SM, Lawlor DA. Cohort Profile: the Avon Longitudinal Study of Parents and Children: ALSPAC mothers cohort. Int J Epidemiol. 2013;42:97–110. doi: 10.1093/ije/dys066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Relton CL, Gaunt T, McArdle W, Ho K, Duggirala A, Shihab H, Woodward G, Lyttleton O, Evans DM, Reik W, Paul YL, Ficz G, Ozanne SE, Wipat A, Flanagan K, Lister A, Heijmans BT, Ring SM, Davey Smith G. Data Resource Profile: Accessible Resource for Integrated Epigenomic Studies (ARIES) Int J Epidemiol. 2015 doi: 10.1093/ije/dyv072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dedeurwaerder S, Defrance M, Calonne E, Denis H, Sotiriou C, Fuks F. Evaluation of the Infinium Methylation 450K technology. Epigenomics. 2011;3:771–784. doi: 10.2217/epi.11.105. [DOI] [PubMed] [Google Scholar]
- 34.Du P, Zhang X, Huang CC, Jafari N, Kibbe WA, Hou L, Lin SM. Comparison of Beta-value and M-value methods for quantifying methylation levels by microarray analysis. BMC bioinformatics. 2010;11:587. doi: 10.1186/1471-2105-11-587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bibikova M, Barnes B, Tsan C, Ho V, Klotzle B, Le JM, Delano D, Zhang L, Schroth GP, Gunderson KL, Fan JB, Shen R. High density DNA methylation array with single CpG site resolution. Genomics. 2011;98:288–295. doi: 10.1016/j.ygeno.2011.07.007. [DOI] [PubMed] [Google Scholar]
- 36.Touleimat N, Tost J. Complete pipeline for Infinium((R)) Human Methylation 450K BeadChip data processing using subset quantile normalization for accurate DNA methylation estimation. Epigenomics. 2012;4:325–341. doi: 10.2217/epi.12.21. [DOI] [PubMed] [Google Scholar]
- 37.Paternoster L, Zhurov AI, Toma AM, Kemp JP, St Pourcain B, Timpson NJ, McMahon G, McArdle W, Ring SM, Davey Smith G, Richmond S, Evans DM. Genome-wide association study of three-dimensional facial morphology identifies a variant in PAX3 associated with nasion position. Am J Hum Genet. 2012;90:478–485. doi: 10.1016/j.ajhg.2011.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Evans DM, Zhu G, Dy V, Heath AC, Madden PA, Kemp JP, McMahon G, St Pourcain B, Timpson NJ, Golding J, Lawlor DA, Steer C, Montgomery GW, Martin NG, Davey Smith G, Whitfield JB. Genome-wide association study identifies loci affecting blood copper, selenium and zinc. Hum Mol Genet. 2013;22:3998–4006. doi: 10.1093/hmg/ddt239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Reinius LE, Acevedo N, Joerink M, Pershagen G, Dahlen SE, Greco D, Soderhall C, Scheynius A, Kere J. Differential DNA methylation in purified human blood cells: implications for cell lineage and studies on disease susceptibility. PLoS One. 2012;7:e41361. doi: 10.1371/journal.pone.0041361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jaffe AE, Irizarry RA. Accounting for cellular heterogeneity is critical in epigenome-wide association studies. Genome Biology. 2014;15(2):R31. doi: 10.1186/gb-2014-15-2-r31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Houseman EA, Kelsey KT, Wiencke JK, Marsit CJ. Cell-composition effects in the analysis of DNA methylation array data: a mathematical perspective. BMC bioinformatics. 2015;16:95. doi: 10.1186/s12859-015-0527-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang D, Cheng L, Badner JA, Chen C, Chen Q, Luo W, Craig DW, Redman M, Gershon ES, Liu C. Genetic control of individual differences in gene-specific methylation in human brain. Am J Hum Genet. 2010;86:411–419. doi: 10.1016/j.ajhg.2010.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Speliotes EK, Willer CJ, Berndt SI, Monda KL, Thorleifsson G, Jackson AU, Lango Allen H, Lindgren CM, Luan J, Magi R, Randall JC, Vedantam S, Winkler TW, Qi L, Workalemahu T, Heid IM, Steinthorsdottir V, Stringham HM, Weedon MN, Wheeler E, Wood AR, Ferreira T, Weyant RJ, Segre AV, Estrada K, Liang L, Nemesh J, Park JH, Gustafsson S, Kilpelainen TO, Yang J, Bouatia-Naji N, Esko T, Feitosa MF, Kutalik Z, Mangino M, Raychaudhuri S, Scherag A, Smith AV, Welch R, Zhao JH, Aben KK, Absher DM, Amin N, Dixon AL, Fisher E, Glazer NL, Goddard ME, Heard-Costa NL, Hoesel V, Hottenga JJ, Johansson A, Johnson T, Ketkar S, Lamina C, Li S, Moffatt MF, Myers RH, Narisu N, Perry JR, Peters MJ, Preuss M, Ripatti S, Rivadeneira F, Sandholt C, Scott LJ, Timpson NJ, Tyrer JP, van Wingerden S, Watanabe RM, White CC, Wiklund F, Barlassina C, Chasman DI, Cooper MN, Jansson JO, Lawrence RW, Pellikka N, Prokopenko I, Shi J, Thiering E, Alavere H, Alibrandi MT, Almgren P, Arnold AM, Aspelund T, Atwood LD, Balkau B, Balmforth AJ, Bennett AJ, Ben-Shlomo Y, Bergman RN, Bergmann S, Biebermann H, Blakemore AI, Boes T, Bonnycastle LL, Bornstein SR, Brown MJ, Buchanan TA, Busonero F, Campbell H, Cappuccio FP, Cavalcanti-Proenca C, Chen YD, Chen CM, Chines PS, Clarke R, Coin L, Connell J, Day IN, den Heijer M, Duan J, Ebrahim S, Elliott P, Elosua R, Eiriksdottir G, Erdos MR, Eriksson JG, Facheris MF, Felix SB, Fischer-Posovszky P, Folsom AR, Friedrich N, Freimer NB, Fu M, Gaget S, Gejman PV, Geus EJ, Gieger C, Gjesing AP, Goel A, Goyette P, Grallert H, Grassler J, Greenawalt DM, Groves CJ, Gudnason V, Guiducci C, Hartikainen AL, Hassanali N, Hall AS, Havulinna AS, Hayward C, Heath AC, Hengstenberg C, Hicks AA, Hinney A, Hofman A, Homuth G, Hui J, Igl W, Iribarren C, Isomaa B, Jacobs KB, Jarick I, Jewell E, John U, Jorgensen T, Jousilahti P, Jula A, Kaakinen M, Kajantie E, Kaplan LM, Kathiresan S, Kettunen J, Kinnunen L, Knowles JW, Kolcic I, Konig IR, Koskinen S, Kovacs P, Kuusisto J, Kraft P, Kvaloy K, Laitinen J, Lantieri O, Lanzani C, Launer LJ, Lecoeur C, Lehtimaki T, Lettre G, Liu J, Lokki ML, Lorentzon M, Luben RN, Ludwig B, Magic, Manunta P, Marek D, Marre M, Martin NG, McArdle WL, McCarthy A, McKnight B, Meitinger T, Melander O, Meyre D, Midthjell K, Montgomery GW, Morken MA, Morris AP, Mulic R, Ngwa JS, Nelis M, Neville MJ, Nyholt DR, O'Donnell CJ, O'Rahilly S, Ong KK, Oostra B, Pare G, Parker AN, Perola M, Pichler I, Pietilainen KH, Platou CG, Polasek O, Pouta A, Rafelt S, Raitakari O, Rayner NW, Ridderstrale M, Rief W, Ruokonen A, Robertson NR, Rzehak P, Salomaa V, Sanders AR, Sandhu MS, Sanna S, Saramies J, Savolainen MJ, Scherag S, Schipf S, Schreiber S, Schunkert H, Silander K, Sinisalo J, Siscovick DS, Smit JH, Soranzo N, Sovio U, Stephens J, Surakka I, Swift AJ, Tammesoo ML, Tardif JC, Teder-Laving M, Teslovich TM, Thompson JR, Thomson B, Tonjes A, Tuomi T, van Meurs JB, van Ommen GJ, Vatin V, Viikari J, Visvikis-Siest S, Vitart V, Vogel CI, Voight BF, Waite LL, Wallaschofski H, Walters GB, Widen E, Wiegand S, Wild SH, Willemsen G, Witte DR, Witteman JC, Xu J, Zhang Q, Zgaga L, Ziegler A, Zitting P, Beilby JP, Farooqi IS, Hebebrand J, Huikuri HV, James AL, Kahonen M, Levinson DF, Macciardi F, Nieminen MS, Ohlsson C, Palmer LJ, Ridker PM, Stumvoll M, Beckmann JS, Boeing H, Boerwinkle E, Boomsma DI, Caulfield MJ, Chanock SJ, Collins FS, Cupples LA, Davey Smith G, Erdmann J, Froguel P, Gronberg H, Gyllensten U, Hall P, Hansen T, Harris TB, Hattersley AT, Hayes RB, Heinrich J, Hu FB, Hveem K, Illig T, Jarvelin MR, Kaprio J, Karpe F, Khaw KT, Kiemeney LA, Krude H, Laakso M, Lawlor DA, Metspalu A, Munroe PB, Ouwehand WH, Pedersen O, Penninx BW, Peters A, Pramstaller PP, Quertermous T, Reinehr T, Rissanen A, Rudan I, Samani NJ, Schwarz PE, Shuldiner AR, Spector TD, Tuomilehto J, Uda M, Uitterlinden A, Valle TT, Wabitsch M, Waeber G, Wareham NJ, Watkins H, Procardis C, Wilson JF, Wright AF, Zillikens MC, Chatterjee N, McCarroll SA, Purcell S, Schadt EE, Visscher PM, Assimes TL, Borecki IB, Deloukas P, Fox CS, Groop LC, Haritunians T, Hunter DJ, Kaplan RC, Mohlke KL, O'Connell JR, Peltonen L, Schlessinger D, Strachan DP, van Duijn CM, Wichmann HE, Frayling TM, Thorsteinsdottir U, Abecasis GR, Barroso I, Boehnke M, Stefansson K, North KE, McCarthy MI, Hirschhorn JN, Ingelsson E, Loos RJ. Association analyses of 249,796 individuals reveal 18 new loci associated with body mass index. Nat Genet. 2010;42:937–948. doi: 10.1038/ng.686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Locke AE, Kahali B, Berndt SI, Justice AE, Pers TH, Day FR, Powell C, Vedantam S, Buchkovich ML, Yang J, Croteau-Chonka DC, Esko T, Fall T, Ferreira T, Gustafsson S, Kutalik Z, Luan J, Magi R, Randall JC, Winkler TW, Wood AR, Workalemahu T, Faul JD, Smith JA, Hua Zhao J, Zhao W, Chen J, Fehrmann R, Hedman AK, Karjalainen J, Schmidt EM, Absher D, Amin N, Anderson D, Beekman M, Bolton JL, Bragg-Gresham JL, Buyske S, Demirkan A, Deng G, Ehret GB, Feenstra B, Feitosa MF, Fischer K, Goel A, Gong J, Jackson AU, Kanoni S, Kleber ME, Kristiansson K, Lim U, Lotay V, Mangino M, Mateo Leach I, Medina-Gomez C, Medland SE, Nalls MA, Palmer CD, Pasko D, Pechlivanis S, Peters MJ, Prokopenko I, Shungin D, Stancakova A, Strawbridge RJ, Ju Sung Y, Tanaka T, Teumer A, Trompet S, van der Laan SW, van Setten J, Van Vliet-Ostaptchouk JV, Wang Z, Yengo L, Zhang W, Isaacs A, Albrecht E, Arnlov J, Arscott GM, Attwood AP, Bandinelli S, Barrett A, Bas IN, Bellis C, Bennett AJ, Berne C, Blagieva R, Bluher M, Bohringer S, Bonnycastle LL, Bottcher Y, Boyd HA, Bruinenberg M, Caspersen IH, Ida Chen YD, Clarke R, Daw EW, de Craen AJ, Delgado G, Dimitriou M, Doney AS, Eklund N, Estrada K, Eury E, Folkersen L, Fraser RM, Garcia ME, Geller F, Giedraitis V, Gigante B, Go AS, Golay A, Goodall AH, Gordon SD, Gorski M, Grabe HJ, Grallert H, Grammer TB, Grassler J, Gronberg H, Groves CJ, Gusto G, Haessler J, Hall P, Haller T, Hallmans G, Hartman CA, Hassinen M, Hayward C, Heard-Costa NL, Helmer Q, Hengstenberg C, Holmen O, Hottenga JJ, James AL, Jeff JM, Johansson A, Jolley J, Juliusdottir T, Kinnunen L, Koenig W, Koskenvuo M, Kratzer W, Laitinen J, Lamina C, Leander K, Lee NR, Lichtner P, Lind L, Lindstrom J, Sin Lo K, Lobbens S, Lorbeer R, Lu Y, Mach F, Magnusson PK, Mahajan A, McArdle WL, McLachlan S, Menni C, Merger S, Mihailov E, Milani L, Moayyeri A, Monda KL, Morken MA, Mulas A, Muller G, Muller-Nurasyid M, Musk AW, Nagaraja R, Nothen MM, Nolte IM, Pilz S, Rayner NW, Renstrom F, Rettig R, Ried JS, Ripke S, Robertson NR, Rose LM, Sanna S, Scharnagl H, Scholtens S, Schumacher FR, Scott WR, Seufferlein T, Shi J, Vernon Smith A, Smolonska J, Stanton AV, Steinthorsdottir V, Stirrups K, Stringham HM, Sundstrom J, Swertz MA, Swift AJ, Syvanen AC, Tan ST, Tayo BO, Thorand B, Thorleifsson G, Tyrer JP, Uh HW, Vandenput L, Verhulst FC, Vermeulen SH, Verweij N, Vonk JM, Waite LL, Warren HR, Waterworth D, Weedon MN, Wilkens LR, Willenborg C, Wilsgaard T, Wojczynski MK, Wong A, Wright AF, Zhang Q, LifeLines Cohort S, Brennan EP, Choi M, Dastani Z, Drong AW, Eriksson P, Franco-Cereceda A, Gadin JR, Gharavi AG, Goddard ME, Handsaker RE, Huang J, Karpe F, Kathiresan S, Keildson S, Kiryluk K, Kubo M, Lee JY, Liang L, Lifton RP, Ma B, McCarroll SA, McKnight AJ, Min JL, Moffatt MF, Montgomery GW, Murabito JM, Nicholson G, Nyholt DR, Okada Y, Perry JR, Dorajoo R, Reinmaa E, Salem RM, Sandholm N, Scott RA, Stolk L, Takahashi A, Tanaka T, Van't Hooft FM, Vinkhuyzen AA, Westra HJ, Zheng W, Zondervan KT, Consortium AD, Group A-BW. Consortium CAD. Consortium CK. Glgc, Icbp. Investigators M. Mu TC, Consortium MI. Consortium P. ReproGen C, Consortium G. International Endogene C. Heath AC. Arveiler D, Bakker SJ, Beilby J, Bergman RN, Blangero J, Bovet P, Campbell H, Caulfield MJ, Cesana G, Chakravarti A, Chasman DI, Chines PS, Collins FS, Crawford DC, Cupples LA, Cusi D, Danesh J, de Faire U, den Ruijter HM, Dominiczak AF, Erbel R, Erdmann J, Eriksson JG, Farrall M, Felix SB, Ferrannini E, Ferrieres J, Ford I, Forouhi NG, Forrester T, Franco OH, Gansevoort RT, Gejman PV, Gieger C, Gottesman O, Gudnason V, Gyllensten U, Hall AS, Harris TB, Hattersley AT, Hicks AA, Hindorff LA, Hingorani AD, Hofman A, Homuth G, Hovingh GK, Humphries SE, Hunt SC, Hypponen E, Illig T, Jacobs KB, Jarvelin MR, Jockel KH, Johansen B, Jousilahti P, Jukema JW, Jula AM, Kaprio J, Kastelein JJ, Keinanen-Kiukaanniemi SM, Kiemeney LA, Knekt P, Kooner JS, Kooperberg C, Kovacs P, Kraja AT, Kumari M, Kuusisto J, Lakka TA, Langenberg C, Le Marchand L, Lehtimaki T, Lyssenko V, Mannisto S, Marette A, Matise TC, McKenzie CA, McKnight B, Moll FL, Morris AD, Morris AP, Murray JC, Nelis M, Ohlsson C, Oldehinkel AJ, Ong KK, Madden PA, Pasterkamp G, Peden JF, Peters A, Postma DS, Pramstaller PP, Price JF, Qi L, Raitakari OT, Rankinen T, Rao DC, Rice TK, Ridker PM, Rioux JD, Ritchie MD, Rudan I, Salomaa V, Samani NJ, Saramies J, Sarzynski MA, Schunkert H, Schwarz PE, Sever P, Shuldiner AR, Sinisalo J, Stolk RP, Strauch K, Tonjes A, Tregouet DA, Tremblay A, Tremoli E, Virtamo J, Vohl MC, Volker U, Waeber G, Willemsen G, Witteman JC, Zillikens MC, Adair LS, Amouyel P, Asselbergs FW, Assimes TL, Bochud M, Boehm BO, Boerwinkle E, Bornstein SR, Bottinger EP, Bouchard C, Cauchi S, Chambers JC, Chanock SJ, Cooper RS, de Bakker PI, Dedoussis G, Ferrucci L, Franks PW, Froguel P, Groop LC, Haiman CA, Hamsten A, Hui J, Hunter DJ, Hveem K, Kaplan RC, Kivimaki M, Kuh D, Laakso M, Liu Y, Martin NG, Marz W, Melbye M, Metspalu A, Moebus S, Munroe PB, Njolstad I, Oostra BA, Palmer CN, Pedersen NL, Perola M, Perusse L, Peters U, Power C, Quertermous T, Rauramaa R, Rivadeneira F, Saaristo TE, Saleheen D, Sattar N, Schadt EE, Schlessinger D, Slagboom PE, Snieder H, Spector TD, Thorsteinsdottir U, Stumvoll M, Tuomilehto J, Uitterlinden AG, Uusitupa M, van der Harst P, Walker M, Wallaschofski H, Wareham NJ, Watkins H, Weir DR, Wichmann HE, Wilson JF, Zanen P, Borecki IB, Deloukas P, Fox CS, Heid IM, O'Connell JR, Strachan DP, Stefansson K, van Duijn CM, Abecasis GR, Franke L, Frayling TM, McCarthy MI, Visscher PM, Scherag A, Willer CJ, Boehnke M, Mohlke KL, Lindgren CM, Beckmann JS, Barroso I, North KE, Ingelsson E, Hirschhorn JN, Loos RJ, Speliotes EK. Genetic studies of body mass index yield new insights for obesity biology. Nature. 2015;518:197–206. doi: 10.1038/nature14177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Freathy RM, Timpson NJ, Lawlor DA, Pouta A, Ben-Shlomo Y, Ruokonen A, Ebrahim S, Shields B, Zeggini E, Weedon MN, Lindgren CM, Lango H, Melzer D, Ferrucci L, Paolisso G, Neville MJ, Karpe F, Palmer CNA, Morris AD, Elliott P, Jarvelin MR, Davey Smith G, McCarthy MI, Hattersley AT, Frayling TM. Common variation in the FTO gene alters diabetes-related metabolic traits to the extent expected given its effect on BMI. Diabetes. 2008;57:1419–1426. doi: 10.2337/db07-1466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.De Silva NM, Freathy RM, Palmer TM, Donnelly LA, Luan J, Gaunt T, Langenberg C, Weedon MN, Shields B, Knight BA, Ward KJ, Sandhu MS, Harbord RM, McCarthy MI, Davey Smith G, Ebrahim S, Hattersley AT, Wareham N, Lawlor DA, Morris AD, Palmer CN, Frayling TM. Mendelian randomization studies do not support a role for raised circulating triglyceride levels influencing type 2 diabetes, glucose levels, or insulin resistance. Diabetes. 2011;60:1008–1018. doi: 10.2337/db10-1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fall T, Hagg S, Magi R, Ploner A, Fischer K, Horikoshi M, Sarin AP, Thorleifsson G, Ladenvall C, Kals M, Kuningas M, Draisma HH, Ried JS, van Zuydam NR, Huikari V, Mangino M, Sonestedt E, Benyamin B, Nelson CP, Rivera NV, Kristiansson K, Shen HY, Havulinna AS, Dehghan A, Donnelly LA, Kaakinen M, Nuotio ML, Robertson N, de Bruijn RF, Ikram MA, Amin N, Balmforth AJ, Braund PS, Doney AS, Doring A, Elliott P, Esko T, Franco OH, Gretarsdottir S, Hartikainen AL, Heikkila K, Herzig KH, Holm H, Hottenga JJ, Hypponen E, Illig T, Isaacs A, Isomaa B, Karssen LC, Kettunen J, Koenig W, Kuulasmaa K, Laatikainen T, Laitinen J, Lindgren C, Lyssenko V, Laara E, Rayner NW, Mannisto S, Pouta A, Rathmann W, Rivadeneira F, Ruokonen A, Savolainen MJ, Sijbrands EJ, Small KS, Smit JH, Steinthorsdottir V, Syvanen AC, Taanila A, Tobin MD, Uitterlinden AG, Willems SM, Willemsen G, Witteman J, Perola M, Evans A, Ferrieres J, Virtamo J, Kee F, Tregouet DA, Arveiler D, Amouyel P, Ferrario MM, Brambilla P, Hall AS, Heath AC, Madden PA, Martin NG, Montgomery GW, Whitfield JB, Jula A, Knekt P, Oostra B, van Duijn CM, Penninx BW, Davey Smith G, Kaprio J, Samani NJ, Gieger C, Peters A, Wichmann HE, Boomsma DI, de Geus EJ, Tuomi T, Power C, Hammond CJ, Spector TD, Lind L, Orho-Melander M, Palmer CN, Morris AD, Groop L, Jarvelin MR, Salomaa V, Vartiainen E, Hofman A, Ripatti S, Metspalu A, Thorsteinsdottir U, Stefansson K, Pedersen NL, McCarthy MI, Ingelsson E, Prokopenko I, European Network for G. Genomic Epidemiology c The role of adiposity in cardiometabolic traits: a Mendelian randomization analysis. PLOS medicine. 2013;10:e1001474. doi: 10.1371/journal.pmed.1001474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Thomas DC, Lawlor DA, Thompson JR. Re: Estimation of bias in nongenetic observational studies using “Mendelian triangulation” by Bautista et al. Ann Epidemiol. 2007;17:511–513. doi: 10.1016/j.annepidem.2006.12.005. [DOI] [PubMed] [Google Scholar]
- 49.Davey Smith G. Negative control exposures in epidemiologic studies. Epidemiology. 2012;23:350–351. doi: 10.1097/EDE.0b013e318245912c. author reply 351-352. [DOI] [PubMed] [Google Scholar]
- 50.Richmond RC, Al-Amin A, Davey Smith G, Relton CL. Approaches for drawing causal inferences from epidemiological birth cohorts: A review. Early human development. 2014;90:769–780. doi: 10.1016/j.earlhumdev.2014.08.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lawlor DA, Timpson NJ, Harbord RM, Leary S, Ness A, McCarthy MI, Frayling TM, Hattersley AT, Davey Smith G. Exploring the developmental overnutrition hypothesis using parental-offspring associations and FTO as an instrumental variable. PLOS medicine. 2008;5:e33. doi: 10.1371/journal.pmed.0050033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pierce BL, Burgess S. Efficient design for Mendelian randomization studies: subsample and 2-sample instrumental variable estimators. AJE. 2013;178:1177–1184. doi: 10.1093/aje/kwt084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Guenard F, Tchernof A, Deshaies Y, Cianflone K, Kral JG, Marceau P, Vohl MC. Methylation and expression of immune and inflammatory genes in the offspring of bariatric bypass surgery patients. J Obes. 2013;2013:492170. doi: 10.1155/2013/492170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Liu X, Chen Q, Tsai HJ, Wang G, Hong X, Zhou Y, Zhang C, Liu C, Liu R, Wang H, Zhang S, Yu Y, Mestan KK, Pearson C, Otlans P, Zuckerman B, Wang X. Maternal preconception body mass index and offspring cord blood DNA methylation: exploration of early life origins of disease. Environ Mol Mutagen. 2014;55:223–230. doi: 10.1002/em.21827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Morales E, Groom A, Lawlor DA, Relton CL. DNA methylation signatures in cord blood associated with maternal gestational weight gain: results from the ALSPAC cohort. BMC Res Notes. 2014;7:278. doi: 10.1186/1756-0500-7-278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sharp G, Lawlor DA, Richmond RC, Fraser A, Shihab HA, Lyttleton O, McArdle W, Ring S, Gaunt T, Davey Smith G, Relton C. Maternal pre-pregnancy BMI and gestational weight gain, offspring DNA methylation and later offspring adiposity: Findings from the Avon Longitudinal Study of Parents and Children. Int J Epidemiology. 2015 doi: 10.1093/ije/dyv042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rajakumar A, Conrad KP. Expression, ontogeny, and regulation of hypoxia-inducible transcription factors in the human placenta. Biol Reprod. 2000;63:559–569. doi: 10.1095/biolreprod63.2.559. [DOI] [PubMed] [Google Scholar]
- 58.Lee KW, Richmond R, Hu P, French L, Shin J, Bourdon C, Reischl E, Waldenberger M, Zeilinger S, Gaunt T, McArdle W, Ring S, Woodward G, Bouchard L, Gaudet D, Davey Smith G, Relton C, Paus T, Pausova Z. Prenatal Exposure to Maternal Cigarette Smoking and DNA Methylation: Epigenome-Wide Association in a Discovery Sample of Adolescents and Replication in an Independent Cohort at Birth through 17 Years of Age. Environ Health Perspect. 2015;123.2:193. doi: 10.1289/ehp.1408614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Richmond RC, Simpkin AJ, Woodward G, Gaunt TR, Lyttleton O, McArdle WL, Ring SM, Smith AD, Timpson NJ, Tilling K, Davey Smith G, Relton CL. Prenatal exposure to maternal smoking and offspring DNA methylation across the lifecourse: Findings from the Avon Longitudinal Study of Parents and Children (ALSPAC) Hum Mol Genet. 2014 doi: 10.1093/hmg/ddu739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Davey Smith G, Ebrahim S. Mendelian randomization: prospects, potentials, and limitations. Int J Epidemiology. 2004;33:30–42. doi: 10.1093/ije/dyh132. [DOI] [PubMed] [Google Scholar]
- 61.Davey Smith G. Use of genetic markers and gene-diet interactions for interrogating population-level causal influences of diet on health. Genes and Nutrition. 2011;6:27–43. doi: 10.1007/s12263-010-0181-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Naeem H, Wong NC, Chatterton Z, Hong MK, Pedersen JS, Corcoran NM, Hovens CM, Macintyre G. Reducing the risk of false discovery enabling identification of biologically significant genome-wide methylation status using the HumanMethylation450 array. BMC Genomics. 2014;15:51. doi: 10.1186/1471-2164-15-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.