Abstract
Expansions of polyglutamine (polyQ) tracts in nine different proteins cause a family of neurodegenerative disorders called polyQ diseases. Since polyQ tracts are potential therapeutic targets for these pathologies there is great interest in characterizing the conformations that they adopt and in understanding how their aggregation behavior is influenced by the sequences flanking them. We used solution NMR to study at single-residue resolution a 156-residue proteolytic fragment of the androgen receptor that contains a polyQ tract associated with the disease called spinobulbar muscular atrophy, also known as Kennedy disease. Our findings indicate that a Leu-rich region preceding the polyQ tract causes it to become α-helical and appears to protect the protein against aggregation, which represents a new mechanism by which sequence context can minimize the deleterious properties of these repetitive regions. Our results have implications for drug discovery for polyQ diseases because they suggest that the residues flanking these repetitive sequences may represent viable therapeutic targets.
Introduction
A group of nine disorders termed polyglutamine (polyQ) diseases occur as a result of the expansion of polymorphic polyQ tracts in proteins that are otherwise unrelated (1). The variable length of such tracts is due to the propensity of the CAG and GTC codon repeats that codify for them to form non-B-DNA structures that cause slippage during DNA replication (2). As a consequence of these expansions, encoded proteins with expanded polyQ tracts are formed, which often misfold, oligomerize and aggregate to form fibrillar species resembling amyloid fibrils that may play a role in the onset of polyQ diseases (3–5).
Characterizing the structural and dynamical properties of polyQ tracts is crucial for understanding the molecular basis of polyQ diseases and for developing therapeutic strategies to inhibit their aggregation (6). However, these regions of low sequence complexity are challenging targets for conventional methods of structural biology. This is due, in addition to their low solubility, to their high propensity to be intrinsically disordered, that in general hinders crystallization, and to the highly repetitive nature of their amino acid sequence, that can hamper their investigation by NMR. A number of pioneering studies have characterized the structural and dynamic properties of polyQ tracts, but to date it has not been possible to report on the conformation of one such a tract in its native context and without fusing it to a solubilizing moiety (7, 8).
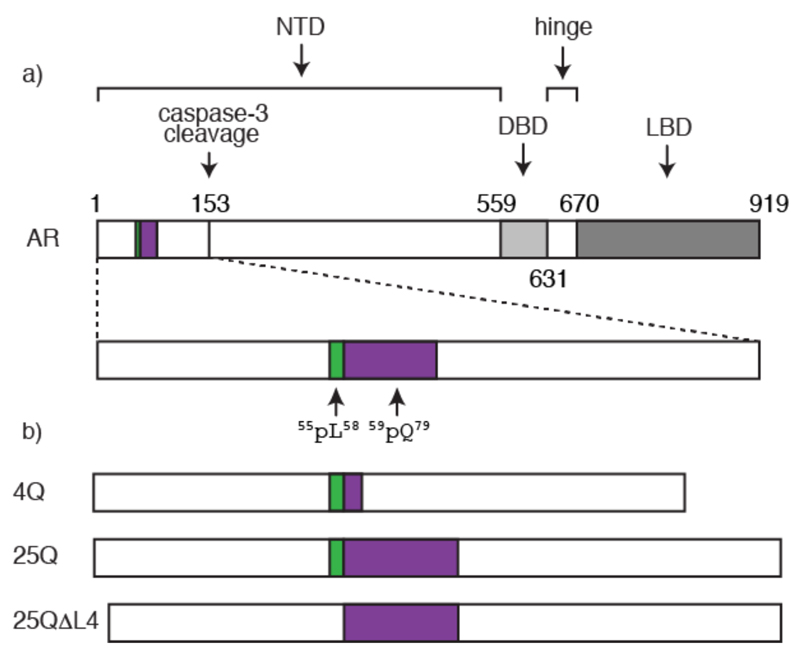
We focus here on a polyQ tract occurring in the transactivation domain of the androgen receptor (AR) which plays a key role in the onset of spinal bulbar muscular atrophy (SBMA), a rare hereditary neuromuscular polyQ disease also known as Kennedy disease (9). AR is a nuclear receptor activated by androgens such as dihydrotestosterone that regulates the expression of the male phenotype (10). The polymorphic polyQ tract in AR starts at position 59 and can be between 14 and 34 residues long in healthy individuals (Fig. 1a). Sizes over 37 residues are associated with SBMA and the length of the polyQ tract anticorrelates with the age of onset.
Figure 1.
Structural organization of AR. a) Position of the transactivation (NTD), DNA binding (DBD) and ligand binding (LBD) domains of AR with an indication of the positions of the 55LLLL58 motif, shown in green, of the polymorphic polyQ tract, shown in purple, and of the caspase 3 cleavage site. b) AR constructs used in this work (see also Fig. S1).
The polyQ tract of AR is involved in the formation of the aggregates associated with SBMA (11). However, neither its structure nor its role in the mechanism of AR aggregation are known and this acts as an important hurdle for the development of therapeutic approaches for this disease based on preventing or decreasing the rate of aggregation. To remedy this and, especially, to better understand how sequence context can influence the structural properties of polyQ tracts we used solution NMR to investigate the conformation of an N-terminal fragment of AR found in the aggregates associated with SBMA. This 156-residue fragment is the product of proteolytic cleavage by caspase 3 and plays a key role in the progression of the disease (Fig. 1a) (9).
Materials and Methods
Protein expression and purification
The genes codifying for 4Q, 25Q and 25QΔL (Figs. 1b and S1) were purchased from GeneArt and cloned in a pDEST-HisMBP vector (Addgene). Expression of the resulting genes led to fusion proteins containing a His6 tag and a maltose binding protein (MBP) moiety that were used, respectively, to purify and increase the solubility of the proteins. 15N labeled protein expression was carried in Rosetta E. coli cells grown in MOPS at 37°C until the value of OD600 was 0.7 and induced with 0.5 mM Isopropyl β-D-1-thiogalactopyranoside (IPTG) for 4 hours at 28°C. Cells were harvested by centrifugation and re-suspended in core buffer (50 mM sodium phosphate, 500 mM NaCl, 5% (v/v) Glycerol, 1 mM 2-Mercapto-ethanol, pH 8.0). A HisTrap HP 5 ml column (GE Healthcare) was used to purify the proteins, which were eluted by an imidazole gradient (final composition: 500 mM imidazole, 50 mM sodium phosphate, 500 mM NaCl, 5% Glycerol, 1 mM 2-mercapto-ethanol, pH 8.0), followed by a size exclusion chromatography step carried out in a Superdex HighLoad S200 26/60 column (GE Healthcare) equilibrated in a buffer with the following composition: 500 mM NaCl, 20 mM sodium phosphate, 5% (v/v) Glycerol, 1 mM DTT, pH 7.5. The pure proteins were then incubated with His6-tagged TEV protease for 16 hours at 4°C by dialysis against a buffer containing 20 mM sodium phosphate, 100 mM NaCl and 0.5 mM EDTA (Ethylenediaminetetraacetic acid), pH 8.0. The product of the proteolytic cleavage was purified by Ni2+ affinity chromatography, employing a buffer containing 8 M urea (500 mM imidazole, 50 mM sodium phosphate, 100 mM NaCl, 8 M urea, pH 8.0) to prevent the aggregation of the cleaved AR in the column. Finally, the cleaved proteins were stored at –80°C.
NMR sample preparation
Except for the experiments presented in fig. 4b the protein solutions stored at -80°C in 8M urea were thawed and dialyzed (1:1000) for 16 hours at 4°C against a buffer containing 20 mM sodium phosphate and 1 mM tris(2-carboxyethyl)phosphine (TCEP) at pH 7.4. Finally, 10% (v/v) D2O and 0.015 mM DSS (4,4-dimethyl-4-silapentane-1-sulfonic acid) were added to the samples; the concentration of these NMR samples was 400 μM. The experiments presented in fig. 4b were carried out with samples prepared with the procedure used for the CD and DLS samples (see below); the concentrations of these samples were 43 μM (25QΔL4) and 50 μM (25Q).
Figure 4.
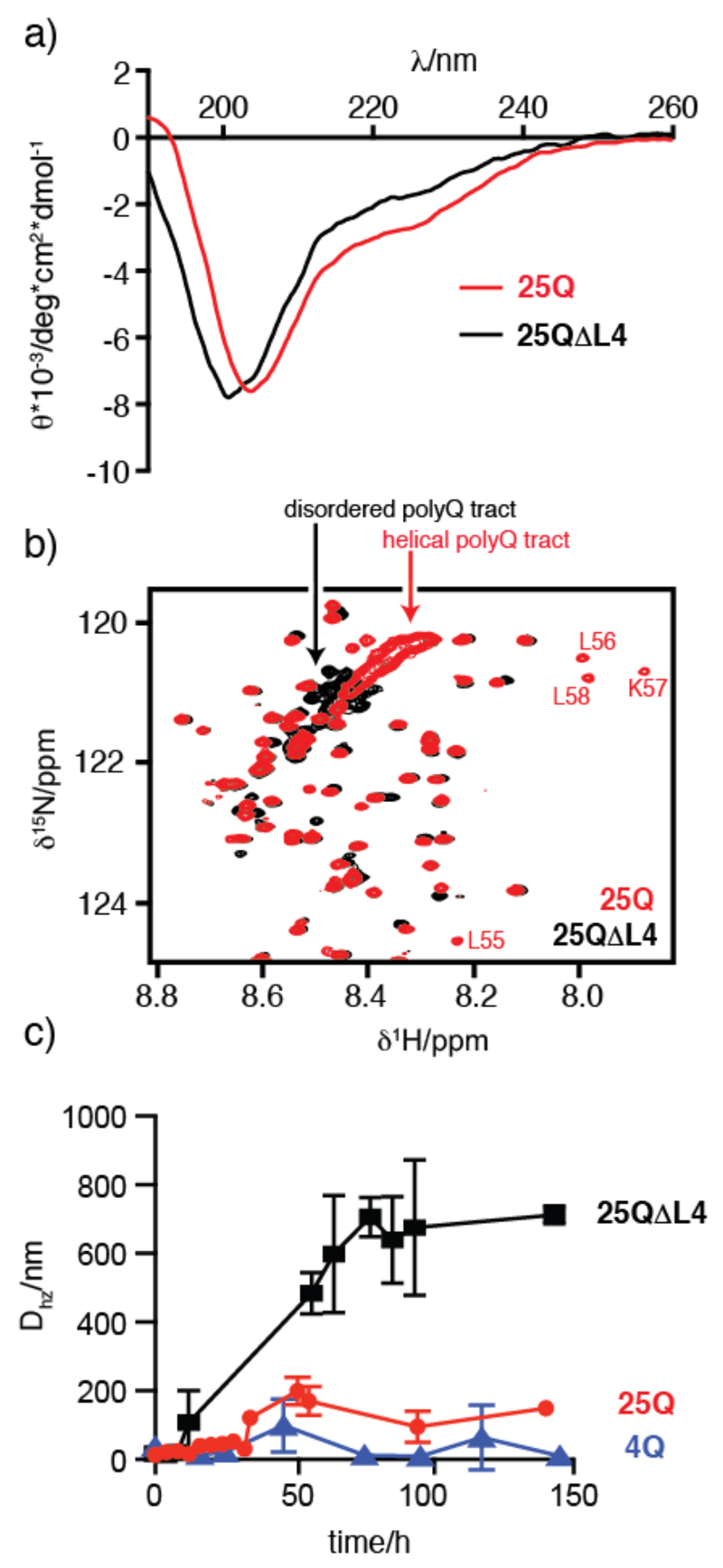
Characterization of the structural and aggregation properties of 25Q (red) and 25QΔL4 (black). a) Comparison of the CD spectra of 130 μM 25Q and 140 μM 25QΔL4 at 310K. b) Comparison of the 1H-15N HSQC NMR spectra of 50 μM 25Q and 43 μM 25QΔL4 at 278K. The cross-peaks belonging to the four Leu of 25Q immediately preceding the polyQ tract, not present in 25QΔL4, are indicated. The absence of helical propensity in 25QΔL4, which reduces the chemical shift dispersion of the resonances corresponding to the polyQ tract, is highlighted. c) Temporal evolution of the mean particle size of 4Q (blue), 25Q and 25QΔL4 at 20 μM and 310K.
DLS and CD sample preparation
The protocol was adapted from the one developed by Linse group for Aß peptide kinetic measurements (12 and Chiesa et al in preparation). After purification, the lyophilized protein was dissolved in a buffer containing 20 mM sodium phosphate, 100 mM NaCl, 6 M Guanidine Thiocyanate, pH 7.4 and 5 mM TCEP until complete reduction of the protein, as monitored by HPLC measurement with a C18 column. The solution was passed through a PD-10 column (GE Healthcare) equilibrated in 20 mM sodium phosphate, pH 7.4. The most concentrated fraction was then purified with a Superdex 75 10/300 size exclusion chromatography column and the fractions containing exclusively the monomeric protein were centrifuged at 386,000 g for 1 hour at 4°C, using an Optima MAX preparative ultracentrifuge (Beckman). Only the upper ¾ of the centrifuged solutions were used for the assays.
Dynamic light scattering (DLS)
Before the measurements, samples were centrifuged by using a tabletop centrifuge at 4°C for 10 minutes at 13,000 rpm. Measurements were taken with a Malvern Zetasizer Nano S equipped with a He-Ne of 633 nm wavelength laser. For each experiment, 20 repetitions of 20 seconds were recorded. Three measurements were performed at each time point. The concentration of the DLS samples was 20 μM and the experiments were performed at 310K.
Circular Dichroism (CD) spectroscopy
Far-UV CD measurements were performed on a JASCO 815 spectropolarimeter using a 0.01 cm length cuvette. Freshly prepares sample were diluted to the desired concentration with 20 mM sodium phosphate buffer (pH 7.4). The spectra were acquired at 0.2 nm resolution with a scan rate of 50 nm/min. For each sample, 10 spectra were collected and averaged, after subtraction the blank. The concentrations of the CD samples were 130 μM for 25Q, 120μM for 4Q and 140μM for 25QΔL4 and the experiments were performed at 310K.
NMR experiments
0.4 mM samples of 13C,15N double-labeled 4Q and 25Q in 20 mM sodium phosphate, 1 mM TCEP, pH 7.4 were prepared. 10 % (v/v) D2O was added for the lock. Identical samples, but exclusively enriched in 15N, were used to acquire 15N relaxation experiments. 13C-detected and 1H-detected NMR experiments for sequence-specific resonance assignment were acquired at 16.4 T on a Bruker Avance spectrometer operating at 700.06 MHz 1H, 176.03 MHz 13C and 70.94 MHz 15N frequencies, equipped with a cryogenically cooled probehead optimized for 13C-direct detection (TXO), and at 22.3 T on a Bruker Avance III spectrometer operating at 950.20 MHz 1H, 238.93 MHz 13C and 96.28 MHz 15N frequencies, equipped with a cryogenically cooled probehead (TCI). 15N relaxation experiments were performed at 16.4 T Bruker Avance spectrometer operating at 700.13 MHz 1H, 176.05 MHz 13C and 70.94 MHz 15N frequencies, equipped with a cryogenically cooled probehead (TXI), by measuring 15N backbone longitudinal (R1) and transverse (R2) relaxation rates and the heteronuclear 15N{15H} NOEs. The experiments presented in figure 4b were acquired at 18.8 T on a Bruker Digital Avance spectrometer operating at 800 MHz 1H, 201.20 MHz 13C and 81.08 MHz 15N frequencies, equipped with a cryogenically cooled probehead (TCI), with 15N-enriched samples. All the experiments were collected at 278 K.
A data set consisting of a combination of 13C-detected (4D HCBCACON (13), 4D HCBCANCO (13), 4D (HCA)CON(CA)CON (14) and 4D (HN)CON(CA)CON) (14) and 1H-detected (3D TROSY HNCO (15), 4D TROSY (H)NCO(CA)NNH (16) and 4D TROSY HN(COCA)NNH) (16) NMR experiments was used to achieve the full sequence-specific assignment of 4Q. Instead, 13C-detected 4D HCBCACON (13) and 4D (HN)CON(CA)CON (14) experiments, and 1H-detected 3D TROSY HNCO (15), 3D TROSY HN(CA)CO (17) and 4D TROSY HN(COCA)NNH (16) experiments were acquired to obtain the complete resonances assignment of 25Q. All the experiments were performed using on-grid non-uniform sampling (NUS). The o "Poisson disk” sampling scheme was chosen to generate the time schedules with the RSPack program (18). The assignments of HN, Hα, Hβ, C', Cα, Cβ and N resonances of 4Q and 25Q are reported in the BMRB (www.bmrb.wisc.edu) (19), entries 25606 and 25607. The parameters used for the acquisition of the experiments are reported in Tables S1-S4.
NMR data processing and analysis
NUS NMR data were converted with NMRPipe (20) and then processed using ToASTD (21) and reduced (22, 23) programs. Uniformly sampled NMR data were processed with TopSpin. Sparky (24) and CcpNMR Analysis (25) were used to visualize the spectra and analyze the 15N relaxation data, respectively. The secondary structure propensity from the heteronuclear chemical shifts was determined for 4Q and 25Q by using the neighbor corrected structural propensity calculator (ncSPC) method (26), available online at http://nmr.chem.rug.nl/ncSPC/. The Tamiola, Acar and Mulder random coil chemical shift library was chosen for the analyses (27) using ncSPC. The secondary structure propensities were also calculated by using the δ2D method (28), available online at http://www-mvsoftware.ch.cam.ac.uk/.
Results and Discussion
In order to investigate how the properties of the polyQ tract depend on its length we used constructs containing 4 and 25 Gln residues (4Q and 25Q, Figs. 1b and S1). Constructs featuring longer polyQ tracts could not be used in the study because they aggregate too fast with respect to the time required to perform the NMR experiments. The disordered nature of the protein and the presence of a 25 residue-long polyQ tract required an experimental strategy based on the use of recently developed 4D 13C-detected NMR experiments that lead to the complete sequence specific assignment of the fragment (13, 14). Heteronuclei were exploited to take advantage of their improved chemical shift dispersion, compared to that of protons, and of their reduced sensitivity to exchange processes (29). The extensive cross-peak overlap in the spectra was overcome by acquiring high-dimensional experiments and by exploiting non-uniform sampling (NUS) to achieve high spectral resolution in the indirect dimensions. To our knowledge, a full characterization at atomic resolution of a polyQ tract under native conditions, as the one reported here, is unprecedented.
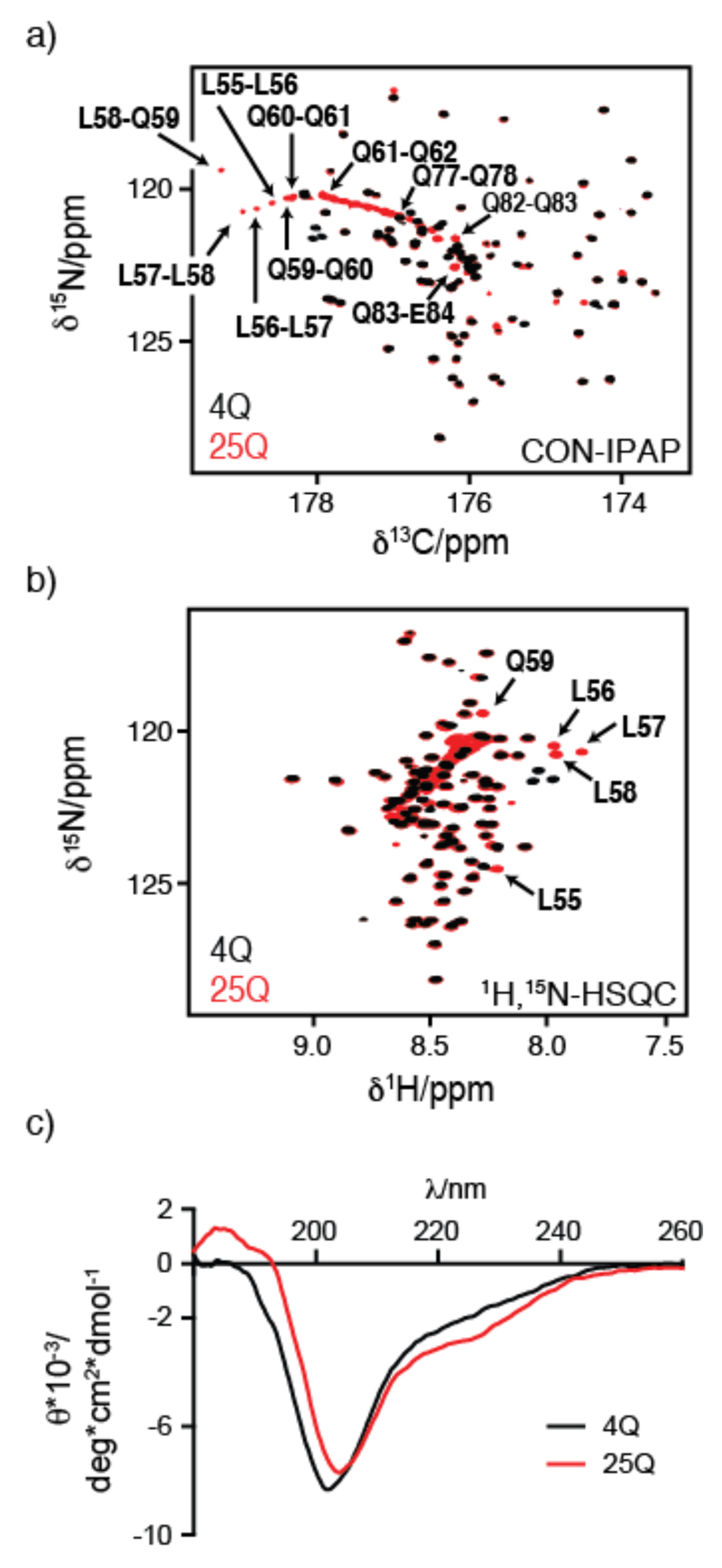
As shown in Figure 2, in which we present the central regions of the 2D 1H-15N HSQC and 2D CON-IPAP spectra of 4Q and 25Q, the spectral differences between these two constructs are limited to the resonances of the polyQ tract and of the 4 Leu residues immediately preceding it i.e. the 55LLLL58 motif. An interesting feature of the spectra of 25Q is the presence of pseudo-diagonals, a characteristic fingerprint, consisting of the cross peaks corresponding to the residues of the polyQ tract, which have 1Hn, 15N and 13C' chemical shifts that correlate with their position in the sequence of the fragment (Fig. S2 in the supplemental material).
Figure 2.
Comparison of the NMR (a,b) and CD (c) spectra of 4Q and 25Q. Central region of (a) the CON-IPAP spectrum and (b) the 1H-15N HSQC spectrum of 400 μM 4Q (black) and 25Q (red) at 278 K with an indication, in panel b, of the resonances which experience the largest chemical shift variations upon increasing the length of the polyQ tract. A close up of the CON-IPAP spectrum of 25Q with the full assignment of the polyQ tract is provided as supplemental material (Fig. S2). c) CD spectrum of 120 μM 4Q and 130 μM 25Q at 310 K.
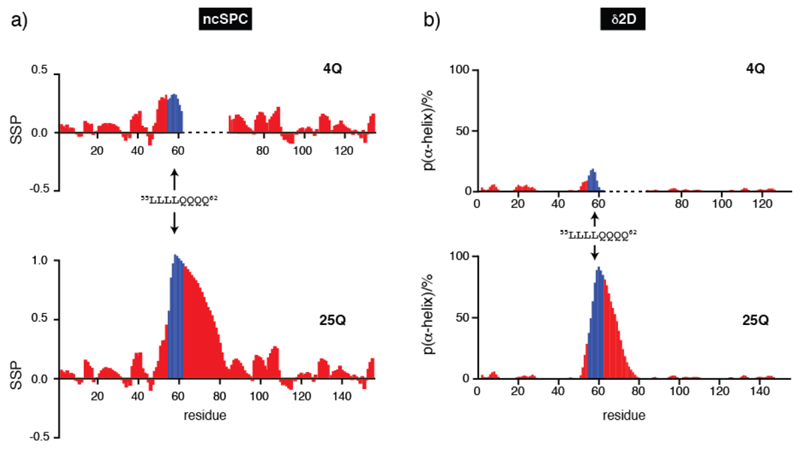
We analyzed the NMR signals chemical shifts to derive the conformational properties of 4Q and 25Q (Fig. 3) in terms of secondary structural propensities by using the algorithms ncSPC (27, 30) and δ2D (28). The values obtained with both methods indicate the absence of persistent secondary structure in the polypeptide except for the polyQ tract and for the 4 Leu residues preceding it, which show some α-helical propensity, and a dramatic increase in helical propensity of the polyQ tract upon expansion from 4 to 25 residues. Although similar observations have been reported (31) this is a surprising result considering that polyQ tracts are in general disordered or, in the fibrillar state, in an extended conformation (8, 32). The fact that the helical propensity of the polyQ tract increases with its length was confirmed by circular dichroism (CD) analysis (Fig. 2c) and 15N relaxation measurements (Figs. S3). Interestingly, the helicity was found to be most pronounced at the beginning of the polyQ tract and to decrease gradually towards its end.
Figure 3.
Secondary structure propensity of 4Q and 25Q obtained by using ncSPC (a) and population of α-helix obtained by using δ2D (b). To facilitate the comparison, values for residues of 4Q which follow the polyQ tract are shifted to the right by 21 units. Values for residues 55 to 62, corresponding to the 55LLLL58 motif and the first 4 Gln of the polyQ tract, are shown in blue to highlight the variation of the structural properties of the protein due to the different length of the polyQ tract.
The fact that the residues of the 55LLLL58 motif, which precedes the polyQ tract, undergo very substantial chemical shift changes upon expansion of the latter (Fig. 2 and Fig. S3) indicates a certain degree of cooperativity in the conformational transition caused by expansion of the polyQ tract and suggests that they induce the formation of the α-helix. To verify this hypothesis, we analyzed by CD and NMR a mutant of 25Q in which the motif 55LLLL58 had been removed (25QΔL4, Fig. 1b and S1). The results obtained indicate a loss of α-helical propensity (Fig. 4a), and a decrease in the chemical shift dispersion in the resonances corresponding to the polyQ tract (Fig. 4b). Both observations confirm the hypothesis that the motif 55LLLL58 induces a helical conformation in the polyQ tract and that in its absence the tract adopts a conformation equivalent to that of synthetic polyQ peptides (11).
To investigate the effect of the 4 Leu residues on the kinetics of aggregation of the AR fragment, we compared the temporal evolution of 20 μM solutions of 4Q, 25Q and 25QΔL4 by dynamic light scattering (DLS), a technique that is well-suited to characterize the early stages of protein aggregation (Fig. 4c). The results show that the particle size (Dhz) increases much faster and reaches higher values for 25QΔL4 than for 25Q (Fig. 4c), indicating that the presence of the motif 55LLLL58 modifies the aggregation behavior of the protein. 4Q, as expected (33), was found to hardly aggregate in the timescale probed in this experiment. The effect of the 55LLLL58 motif on the aggregation properties of the AR fragment is conceptually related to that obtained for huntingtin, the protein bearing the polyQ tract involved in Huntington’s disease. In that case a polyproline flanking region was found to decrease the aggregation rate by altering, albeit in a different way, the structural properties of the polyQ tract (34).
It is important to emphasize that the mechanism by which the helical motif 55LLLL58 alters the aggregation behavior of the AR fragment is fundamentally different from the mechanism by which previously described helical flanking regions modulate the aggregation tendencies of polyQ tracts. The N-terminal 17-residue helical region of huntingtin, for example, greatly accelerates the rate of aggregation of this protein not by modifying the structural properties of the polyQ tract but, rather, by establishing intermolecular interactions, with the same region of other monomers, that increase the local concentration of polyQ tracts and therefore facilitate their oligomerization (35); a similar behavior has been observed also for other proteins bearing polyQ tracts (36).
The high resolution investigation of a protein bearing a polyQ tract achieved here by using a recently developed NMR strategy in combination with complementary techniques (CD, DLS) has revealed a new mechanism by which protein flanking regions influence the aggregation behavior of such tracts. Our findings reveal how a Leu-rich motif flanking a polyQ tract at its N-terminus can induce helical structure in the latter and in turn modify the mechanism of aggregation by adding a helix unfolding step to it. This is so because the intra-molecular hydrogen bonds that stabilize the compact α-helical secondary structure must break before the cross-β structure, rather elongated and stabilized by intermolecular hydrogen bonds, can form (37).
These results also show that the influence of flanking regions on the secondary structure of polyQ tracts spans a limited range, in our case ca 20 residues (Fig. 3) and that it may cease to be effective for the longer tracts which cause polyQ diseases (11, 38). In this scenario the potential protective role played by the induction of the helical structure would only be efficient for polyQ tracts of limited length, contributing to explaining why the protein becomes toxic only when the number of Gln residues exceeds a certain threshold. Sequence context would in this way be a contributing factor to the disease and protein-specific nature of the threshold that triggers protein aggregation and the onset of polyQ disease.
Conclusion
Our findings provide new insights into the structural properties of polyQ tracts and offer a compelling example of how flanking regions can alter the aggregation behavior and, therefore, the formation of potentially toxic fibrillar species by proteins harboring these peculiar sequences. Our increased understanding of how sequence context influences the properties of polyQ tracts opens up new avenues for the development of therapeutic strategies for polyQ diseases based on targeting residues in the flanking regions rather than the polyQ tract themselves.
Supplementary Material
Acknowledgments
This work was funded by BioNMR (EC contract no. 261863), IRB, ICREA, Obra Social “la Caixa” (B.E.), AGAUR (G.C.), MICINN (CTQ2009-08850 to X.S.), MINECOÓ (BIO2012-31043 to X.S.), Maratό de TV3 (102030 to X.S.) and the ERC (CONCERT, 648201 to X.S.). D.M. was a recipient of an Erasmus placement fellowship from the University of Florence (Italy).
Footnotes
Author Contributions
R.P., I.C.F. and X.S. conceived the project and designed the experiments; B.E., A.P., G.C. D.M. and J.G. performed the experiments and analyzed the data; all authors contributed to writing the paper.
References
- 1.Zoghbi HY, Orr HT. Glutamine repeats and neurodegeneration. Annu Rev Neurosci. 2000;23:217–47. doi: 10.1146/annurev.neuro.23.1.217. [DOI] [PubMed] [Google Scholar]
- 2.Mirkin SM. Expandable DNA repeats and human disease. Nature. 2007;447:932–940. doi: 10.1038/nature05977. [DOI] [PubMed] [Google Scholar]
- 3.Liu K-Y, Shyu Y-C, Barbaro Ba, Lin Y-T, Chern Y, Thompson LM, James Shen C-K, Marsh JL. Disruption of the nuclear membrane by perinuclear inclusions of mutant huntingtin causes cell-cycle re-entry and striatal cell death in mouse and cell models of Huntington’s disease. Hum Mol Genet. 2014;24:1602–1616. doi: 10.1093/hmg/ddu574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bates GP, Dorsey R, Gusella JF, Hayden MR, Kay C, Leavitt BR, Nance M, Ross Ca, Scahill RI, Wetzel R, Wild EJ, et al. Huntington disease. Nat Rev Dis Prim. 2015:15005. doi: 10.1038/nrdp.2015.5. [DOI] [PubMed] [Google Scholar]
- 5.Sahl SJ, Weiss LE, Duim WC, Frydman J, Moerner WE. Cellular inclusion bodies of mutant huntingtin exon 1 obscure small fibrillar aggregate species. Sci Rep. 2012;2:895. doi: 10.1038/srep00895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nalavade R, Griesche N, Ryan DP, Hildebrand S, Krauss S. Mechanisms of RNA-induced toxicity in CAG repeat disorders. Cell Death Dis. 2013;4:e752. doi: 10.1038/cddis.2013.276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li P, Huey-Tubman KE, Gao T, Li X, West AP, Jr, Bennett MJ, Bjorkman PJ. The structure of a polyQ-anti-polyQ complex reveals binding according to a linear lattice model. Nat Struct Mol Biol. 2007;14:381–387. doi: 10.1038/nsmb1234. [DOI] [PubMed] [Google Scholar]
- 8.Masino L, Kelly G, Leonard K, Trottier Y, Pastore A. Solution structure of polyglutamine tracts in GST-polyglutamine fusion proteins. FEBS Lett. 2002;513:267–272. doi: 10.1016/s0014-5793(02)02335-9. [DOI] [PubMed] [Google Scholar]
- 9.Merry DE, Kobayashi Y, Bailey CK, Taye aa, Fischbeck KH. Cleavage, aggregation and toxicity of the expanded androgen receptor in spinal and bulbar muscular atrophy. Hum Mol Genet. 1998;7:693–701. doi: 10.1093/hmg/7.4.693. [DOI] [PubMed] [Google Scholar]
- 10.Gelmann EP. Molecular Biology of the Androgen Receptor. J Clin Oncol. 2002;20:3001–3015. doi: 10.1200/JCO.2002.10.018. [DOI] [PubMed] [Google Scholar]
- 11.Chen S, Berthelier V, Yang W, Wetzel R. Polyglutamine aggregation behavior in vitro supports a recruitment mechanism of cytotoxicity. J Mol Biol. 2001;311:173–82. doi: 10.1006/jmbi.2001.4850. [DOI] [PubMed] [Google Scholar]
- 12.Hellstrand E, Boland B, Walsh DM, Linse S. Amyloid ß-protein aggregation produces highly reproducible kinetic data and occurs by a two-phase process. ACS Chem Neurosci. 2010;1:13–18. doi: 10.1021/cn900015v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bermel W, Bertini I, Felli IC, Gonnelli L, Koźmiński W, Piai A, Pierattelli R, Stanek J. Speeding up sequence specific assignment of IDPs. J Biomol NMR. 2012;53:293–301. doi: 10.1007/s10858-012-9639-0. [DOI] [PubMed] [Google Scholar]
- 14.Bermel W, Felli IC, Gonnelli L, Koåmiński W, Piai A, Pierattelli R, Zawadzka-Kazimierczuk A. High-dimensionality 13C direct-detected NMR experiments for the automatic assignment of intrinsically disordered proteins. J Biomol NMR. 2013;57:353–361. doi: 10.1007/s10858-013-9793-z. [DOI] [PubMed] [Google Scholar]
- 15.Kay LE, Bax A. New Methods for the Measurement of NH- & H Constants in “ N-Labeled Proteins Coupling. J Magn Reson. 1990;86:110–126. [Google Scholar]
- 16.Piai A, Hošek T, Gonnelli L, Zawadzka-Kazimierczuk A, Koźmiński W, Brutscher B, Bermel W, Pierattelli R, Felli IC. “CON-CON” assignment strategy for highly flexible intrinsically disordered proteins. J Biomol NMR. 2014;60:209–18. doi: 10.1007/s10858-014-9867-6. [DOI] [PubMed] [Google Scholar]
- 17.Clubb RT, Thanabal V, Wagner G. A constant-time threedimensional triple-resonance pulse scheme to correlate intraresidue 1HN, 15N, and 13C′ chemical shifts in 15N- 13C-labelled proteins. J Magn Reson. 1992;97:213–217. [Google Scholar]
- 18.Kazimierczuk K, Zawadzka A, Koźmiński W. Optimization of random time domain sampling in multidimensional NMR. J Magn Reson. 2008;192:123–130. doi: 10.1016/j.jmr.2008.02.003. [DOI] [PubMed] [Google Scholar]
- 19.Ulrich EL, Akutsu H, Doreleijers JF, Harano Y, Ioannidis YE, Lin J, Livny M, Mading S, Maziuk D, Miller Z, Nakatani E, et al. BioMagResBank. Nucleic Acids Res. 2008;36:D402–8. doi: 10.1093/nar/gkm957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. NMRPipe: A multidimensional spectral processing system based on UNIX pipes. J Biomol NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 21.Kazimierczuk K, Zawadzka A, Koźmiński W, Zhukov I. Random sampling of evolution time space and Fourier transform processing. J Biomol NMR. 2006;36:157–68. doi: 10.1007/s10858-006-9077-y. [DOI] [PubMed] [Google Scholar]
- 22.Kazimierczuk K, Zawadzka-Kazimierczuk A, Koźmiński W. Non-uniform frequency domain for optimal exploitation of non-uniform sampling. J Magn Reson. 2010;205:286–92. doi: 10.1016/j.jmr.2010.05.012. [DOI] [PubMed] [Google Scholar]
- 23.Kazimierczuk K, Zawadzka A, Koźmiński W. Narrow peaks and high dimensionalities: exploiting the advantages of random sampling. J Magn Reson. 2009;197:219–28. doi: 10.1016/j.jmr.2009.01.003. [DOI] [PubMed] [Google Scholar]
- 24.Kneller DG, Kuntz ID. UCSF Sparky: An NMR Display, Annotation and Assignment Tool. J Cell Biochem. 1993;53(S17C):254. [Google Scholar]
- 25.Vranken WF, Boucher W, Stevens TJ, Fogh RH, Pajon A, Llinas M, Ulrich EL, Markley JL, Ionides J, Laue ED. The CCPN data model for NMR spectroscopy: development of a software pipeline. Proteins. 2005;59:687–96. doi: 10.1002/prot.20449. [DOI] [PubMed] [Google Scholar]
- 26.Tamiola K, Acar B, Mulder Faa. Sequence-specific random coil chemical shifts of intrinsically disordered proteins. J Am Chem Soc. 2010;132:18000–18003. doi: 10.1021/ja105656t. [DOI] [PubMed] [Google Scholar]
- 27.Tamiola K, Mulder Faa. Using NMR chemical shifts to calculate the propensity for structural order and disorder in proteins. Biochem Soc Trans. 2012;40:1014–20. doi: 10.1042/BST20120171. [DOI] [PubMed] [Google Scholar]
- 28.Camilloni C, De Simone A, Vranken WF, Vendruscolo M. Determination of secondary structure populations in disordered states of proteins using nuclear magnetic resonance chemical shifts. Biochemistry. 2012;51:2224–2231. doi: 10.1021/bi3001825. [DOI] [PubMed] [Google Scholar]
- 29.Gil S, Hošek T, Solyom Z, Kümmerle R, Brutscher B, Pierattelli R, Felli IC. NMR spectroscopic studies of intrinsically disordered proteins at near-physiological conditions. Angew Chem Int Ed Engl. 2013;52:11808–12. doi: 10.1002/anie.201304272. [DOI] [PubMed] [Google Scholar]
- 30.Marsh Ja, Singh VK, Jia Z, Forman-Kay JD. Sensitivity of secondary structure propensities to sequence differences between alpha- and gamma-synuclein: implications for fibrillation. Protein Sci. 2006;15:2795–2804. doi: 10.1110/ps.062465306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Davies P, Watt K, Kelly SM, Clark C, Price NC, McEwan IJ. Consequences of poly-glutamine repeat length for the conformation and folding of the androgen receptor amino-terminal domain. J Mol Endocrinol. 2008;41:301–314. doi: 10.1677/JME-08-0042. [DOI] [PubMed] [Google Scholar]
- 32.Buchanan LE, Carr JK, Fluitt AM, Hoganson AJ, Moran SD, de Pablo JJ, Skinner JL, Zanni MT. Structural motif of polyglutamine amyloid fibrils discerned with mixed-isotope infrared spectroscopy. Proc Natl Acad Sci U S A. 2014;111:5796–5801. doi: 10.1073/pnas.1401587111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen S, Ferrone FA, Wetzel R. Huntington’s disease age-of-onset linked to polyglutamine aggregation nucleation. Proc Natl Acad Sci U S A. 2002;99:11884–11889. doi: 10.1073/pnas.182276099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bhattacharyya A, Thakur AK, Chellgren VM, Thiagarajan G, Williams AD, Chellgren BW, Creamer TP, Wetzel R. Oligoproline effects on polyglutamine conformation and aggregation. J Mol Biol. 2006;355:524–35. doi: 10.1016/j.jmb.2005.10.053. [DOI] [PubMed] [Google Scholar]
- 35.Thakur AK, Jayaraman M, Mishra R, Thakur M, Chellgren VM, Byeon I-JL, Anjum DH, Kodali R, Creamer TP, Conway JF, Gronenborn AM, et al. Polyglutamine disruption of the huntingtin exon 1 N terminus triggers a complex aggregation mechanism. Nat Struct Mol Biol. 2009;16:380–389. doi: 10.1038/nsmb.1570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ellisdon AM, Thomas B, Bottomley SP. The two-stage pathway of ataxin-3 fibrillogenesis involves a polyglutamine-independent step. J Biol Chem. 2006;281:16888–16896. doi: 10.1074/jbc.M601470200. [DOI] [PubMed] [Google Scholar]
- 37.Calloni G, Lendel C, Campioni S, Giannini S, Gliozzi A, Relini A, Vendruscolo M, Dobson CM, Salvatella X, Chiti F. Structure and dynamics of a partially folded protein are decoupled from its mechanism of aggregation. J Am Chem Soc. 2008;130:13040–50. doi: 10.1021/ja8029224. [DOI] [PubMed] [Google Scholar]
- 38.Scherzinger E, Sittler A, Schweiger K, Heiser V, Lurz R, Hasenbank R, Bates GP, Lehrach H, Wanker EE. Self-assembly of polyglutamine-containing huntingtin fragments into amyloid-like fibrils: implications for Huntington’s disease pathology. Proc Natl Acad Sci U S A. 1999;96:4604–9. doi: 10.1073/pnas.96.8.4604. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.