Abstract
Transposases are important tools in genome engineering, and there is considerable interest in engineering more efficient ones. Here we seek to understand the factors determining their activity using the Sleeping Beauty transposase. Recent work suggests that protein co-evolutionary information can be used to classify groups of physically connected, co-evolving residues into elements called ‘sectors’, which have proven useful for understanding the folding, allosteric interactions, and enzymatic activity of proteins. Using extensive mutagenesis data, protein modeling and analysis of folding energies, we show that 1) The Sleeping Beauty transposase contains two sectors, which span across conserved domains, and are enriched in DNA-binding residues, indicating that the DNA binding and endonuclease functions of the transposase coevolve; 2) Sector residues are highly sensitive to mutations, and most mutations of these residues strongly reduce transposition rate; 3) Mutations with a strong effect on free energy of folding in the DDE domain of the transposase significantly reduce transposition rate. 4) Mutations that influence DNA and protein-protein interactions generally reduce transposition rate, although most hyperactive mutants are also located on the protein surface, including residues with protein-protein interactions. This suggests that hyperactivity results from the modification of protein interactions, rather than the stabilization of protein fold.
Introduction
Recent findings identified a structural organization of protein domains that is distinct from their known hierarchical organization into secondary and tertiary structural elements. These structures, termed ‘sectors’1 form physically connected networks of coevolving residues within proteins, and span across secondary structural elements. Sectors are identified using multiple alignments with a procedure called Statisctical Coupling Analysis (SCA), which uses the covariance matrix of amino acid variability at different positions of the alignment, and their conservation1. It has been noticed that the residues that show correlated evolution in the alignments have a block structure in the SCA matrix: they can be partitioned into clusters of residues, which show correlated evolution within the cluster, but are essentially uncorrelated with residues of other clusters. These groups of coevolving residues were termed ‘sectors’, in analogy to financial sectors1,2. Several important biological properties of proteins are determined by sectors: although they typically make up only 10-30% of the residues of a protein, they were shown to significantly contribute to the specification of protein folds3, allosteric communication in proteins4, and evolution of novel functions5. Since it is possible to engineer functional artificial protein folds based purely on sector information6, or modify their functions using sector residues5 (at least in small domains), sectors are of considerable importance also for protein engineering. However, most work to date on the architecture of sectors, functions and importance of sectors have focused on relatively few single-domain proteins, often with only a single sector1,4,5,7, and the number of studies with multidomain and multisector proteins is low1,8. Thus, it is unclear to what degree the current findings can be generalized, and whether sectors are of similar importance in more complex multi-domain structures as in small proteins2.
Most DNA transposons contain a single gene encoding the transposase protein, which is flanked by terminal inverted repeats (TIRs). Transposons ‘jump’ by a cut-and-paste mechanism, during which the transposase moves the sequence flanked by TIRs to a new genomic location. Since transposases require only the TIRs, and any sequence flanked by TIRs can be moved by externally supplied transposases, they can be used for gene transfer 9. In consequence, transposons are popular tools that are widely used for genome engineering, including cancer gene identification by insertional mutagenesis10, germline transgenesis 11, somatic gene transfer for gene therapy 9, or cellular reprogramming 12. Their primary advantage over viral vectors for gene therapy is that they have considerably fewer side effects, including low immunogenicity and genotoxicity, while, at least for some applications, they provide stable transgene expression levels with efficiency matching viral vectors 9. Several transposon systems are currently applied as genome engineering tools, including the piggyBac, Tol2 and Sleeping Beauty transposons 13–18. The first DNA transposon tool capable for gene transfer in vertebrates was Sleeping Beauty (SB), which was reconstructed from extinct Tc1/mariner transposons in fish 19. Sleeping Beauty, and especially its hyperactive variant 20 is still one of the most widely used transposon tool, and it is the only transposon vector being currently in human clinical trials 21,22.
In this work, using our extensive mutagenesis data available for the Sleeping Beauty transposase, we investigate the structural elements that are the most sensitive to mutations, with particular emphasis on protein sectors. We show that sectors are enriched in DNA-binding residues and are highly sensitive to mutations, which cannot be explained by positional conservation. In addition, our analysis suggests that hyperactivity results from the modification of protein-protein interactions, rather than improved protein folding. Wild-type transposases are not optimal for practical use, because they evolved to transpose at relatively low frequency, as high transposition rates harm their host. As a consequence, modifying their activity or insertion patterns through point mutations is of considerable practical importance, and our results may aid their optimization by identifying mutations that are likely to result in transposases with reduced transposition rate.
Results
Determination of the tertiary structure of SB transposase and protein core
The amino acid sequence of the Sleeping Beauty transposase was obtained from Ivics et al.19. Experimentally determined protein structures are available for the DDE domain of the transposase23 and the N-terminal HTH motif of the DNA-binding domain24, but not for the entire transposase. Thus we predicted the tertiary structure of Sleeping Beauty with the I-TASSER molecular modeling platform 25,26, which uses threading and also ab-initio modeling for structure prediction. Additionally, we used the coordinates of the existing experimental structures (see above) as constraints (Figure S1A). Due to the availability of high quality templates, a high quality structure prediction was possible: the estimated template modeling (TM) score 27 of the predicted tertiary structure with an experimentally determined structure is 0.86 (± 0.1). Models of this quality can be successfully used in mutagenesis studies and stability analyses 28. The most similar structure in the Protein Data Bank (PDB, http:/www.rcsb.org) to the predicted structure (supplementary SB.pdb file) is the Mos1 transposase 29, which was also the highest ranking template used by I-TASSER (see Figure S1B and C for structural alignments between the Mos1 transposase and the predicted structure, and Figure S2 for a Ramachandran plot of the predicted SB transposase using PROCHECK 30). Transposases typically function in a dimeric 29,31 or tetrameric enzyme complex32,33 (and the N-terminal domain of SB was reported to be able to form tetramers in vitro32), but the high structural and mechanistic similarity of the monomer to Mos1 strongly suggests that the active core unit of the complex is a very similar dimer as the one seen for Mos1. (Nevertheless tetramers may exist and may even be the functional state, for example during assembly.) Thus the monomer produced by I-TASSER was used to build a dimer, using the Mos1 (3HOT) transposase as a template (Figure S1D.) DNA nucleotides were replaced with Chimera34, to match the inverted repeats of SB; next the SB transposases were superposed over the Mos1 dimer (3HOT) with TMalign35, followed by correction of clashes and minimization. Severe atomic overlaps (e.g. rings penetrated by other groups) in the initial complex model were manually corrected. The model was then subjected to energy minimization in vacuo by the steepest descent algorithm in GROMACS5 36 using the CHARMM27 force field. The minimization converged to machine precision with no remaining overlaps between atoms. Visualizations of the protein structures were made with Chimera.
As buried residues in proteins are known to be less tolerant of mutations than exposed residues 37, we determined relative solvent accessibility of each residue of the structure with DSSP38 (see methods). The 75 residues with relative solvent accessibility <= 0.1 were assumed to form the protein core. Residues that take part in protein-protein interactions were determined using the difference in solvent accessible surface areas (SASA) of the monomeric and dimeric form of SB: all residues that have different SASA in the dimer and monomer were assumed to take part in protein-protein interactions. DNA-protein interactions were determined with the SNAP tool of the 3DNA package39.
Identification of sectors of the SB transposase
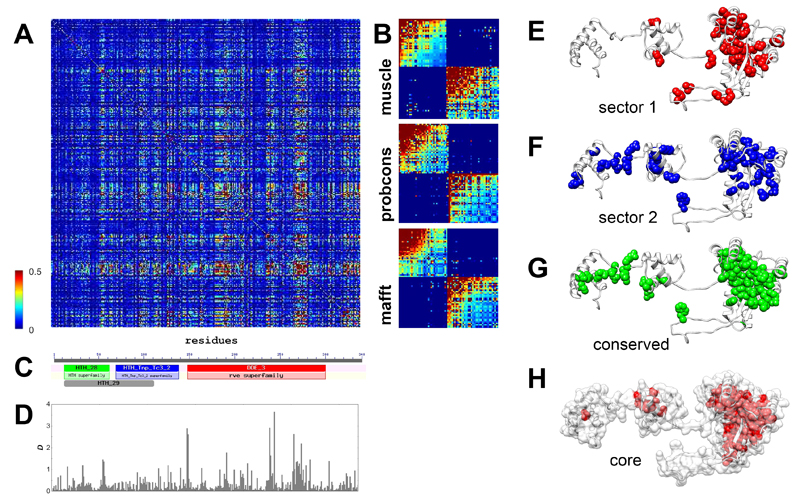
To identify sectors in SB, multiple alignments were made with three different state-of-art tools: muscle40, probcons41 and mafft42 (see methods). Using the three alignments, statistical coupling analyses were performed to identify protein sectors, with the method described by Halabi et al 1, using a modified MATLAB script provided by the same study. SCA tests whether the conservation of an amino acid at any position in the sequence alignment is correlated with the conservation of any other residue of the protein 4, i.e. identifies residues that coevolve. First, it builds a weighted correlation matrix of coevolving amino acids for all residues in the alignment (Figure 1A), and this matrix is subsequently cleaned from statistical noise with a randomization method 1. The analysis of eigenvalue spectra identified three significant eigenvalues for all three alignments (after the exclusion of the largest one), indicating that there might be up to three sectors in the protein. However, after examining residue weights along eigenvectors 2-4 we could identify only two sectors along eigenvector 2 (see Figure S3) that had similar residue compositions irrespectively of the alignment used (Table S3-S4). Due to the different spatial pattern of the residue weights of the three alignments, attempts to identify a third sector resulted in a poorly defined sector, which had different residues depending on the alignment used, and was also strongly correlated with the other two sectors. In consequence, we use only the two sectors that could be consistently defined in all three alignments, which together contain 72-78 residues, depending on the alignment. The cleaned SCA matrices of all three alignments show that the two sectors are essentially independent (Figure 1B), i.e. the correlations between the residues of a sector are much stronger than the correlations between sectors.
Figure 1. Identification of sectors and conserved domains in the Sleeping Beauty (SB) transposase.
A) Statistical Coupling Analysis (SCA) matrix for the muscle alignment of 289 homologous sequences present in RepBase (+SB). The matrix represents correlations between amino acid frequencies at each position of the alignment, i.e. residue pairs that coevolve. B) Cleaned SCA matrices for three alignments made with muscle, probcons, and mafft aligners, containing the residues of the two sectors. Residues within sectors show correlated evolution, while there is almost no correlation between sectors. C) The transposase contains three Pfam conserved domains; two HTH domains with DNA binding functions, and a DDE domain with endonuclease activity. D) The distribution of conservation scores (D) across the sequence. E-F) The location of the two sectors identified with the muscle alignment in the tertiary structure of the SB transposase. The sectors are located across secondary structure elements, and are less compact than the ones reported so far, possibly due to the low sequence similarity in the alignments. Both sectors have residues in multiple conserved domains; most notably sector 2, which has residues in all three Pfam domains of the protein. G) The location of the conserved residues (D > 0.5, muscle alignment) of the transposase. H) The residues of the protein core. All residues with relative solvent accessibility below 0.1 are highlighted with red.
The location of the sectors in the transposase structure is somewhat different from the pattern observed in smaller proteins 1 (Figure 1E-F). Residues of both sectors are located in more than one conserved domain, and in the case of the second sector, residues are present in all three Pfam 43 domains of the transposase (Figure 1C,F), indicating that the division to conserved domains does not strictly correspond with the units of the protein that actually coevolve. Sectors (but also conserved residues) are enriched in DNA-binding residues: their fraction is 29%, as opposed to the 17% observed for the entire protein (p< 0.05 for all three alignments, tests of proportions), but there is no significant difference between the two sectors. The residues of sectors are physically less tightly connected than in most small proteins examined so far, which may arise from the low sequence conservation of the alignments: inaccuracies in the alignments due to the low sequence similarity result in noise, which reduces correlations among residues, and in consequence SCA may fail to detect certain residues as sector residues. To a lesser degree, minor inaccuracies in the transposon sequences themselves may contribute to such noise, as many transposon sequences – including Sleeping Beauty – are reconstructions of extinct repeats.
The dependence of transposition rate on sectors, protein core and conservation
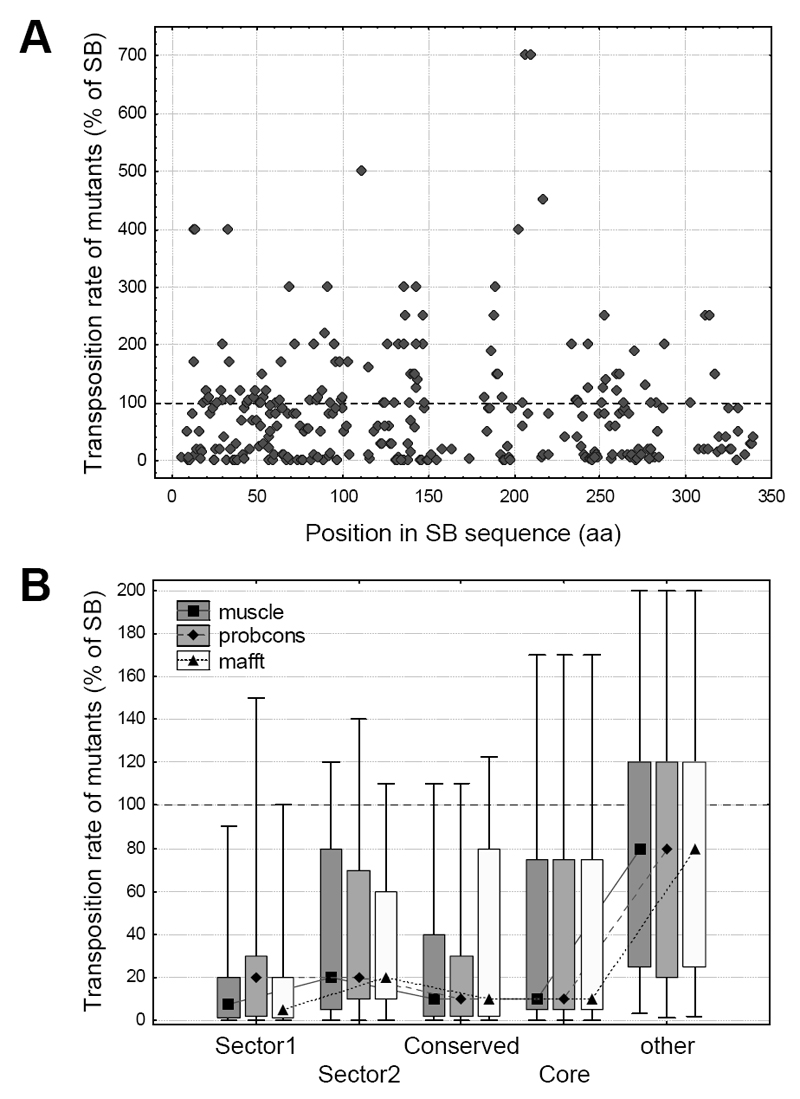
To examine the effect of different residues and protein regions on transposition rate, we used transposition rate measurements of 286 SB mutants, which represent a compilation of all Sleeping Beauty point mutations known to us and also unpublished mutants (see Methods). The distribution of 286 point mutations is approximately uniform across the SB transposase sequence (Figure 2A); however their amino acid distribution is not, as the majority of mutants were alanine replacements (Figure S4). In general, the transposition rates of mutants vary significantly, from completely inactivating the transpsosase to significantly increasing the transposition rate (Figure 2A). The location of the residues in the protein structure have a large influence on their effect: mutations in protein sectors, conserved residues (D > 0.5, see methods) and the protein core result in a significantly larger reduction in transposition rate in comparison with the residues that do not belong to any of these groups (Figure 2B, both sectors, conserved residues and the core are significantly different from other residues, p << 0.05, pairwise comparisons with Mann-Whitney U tests).
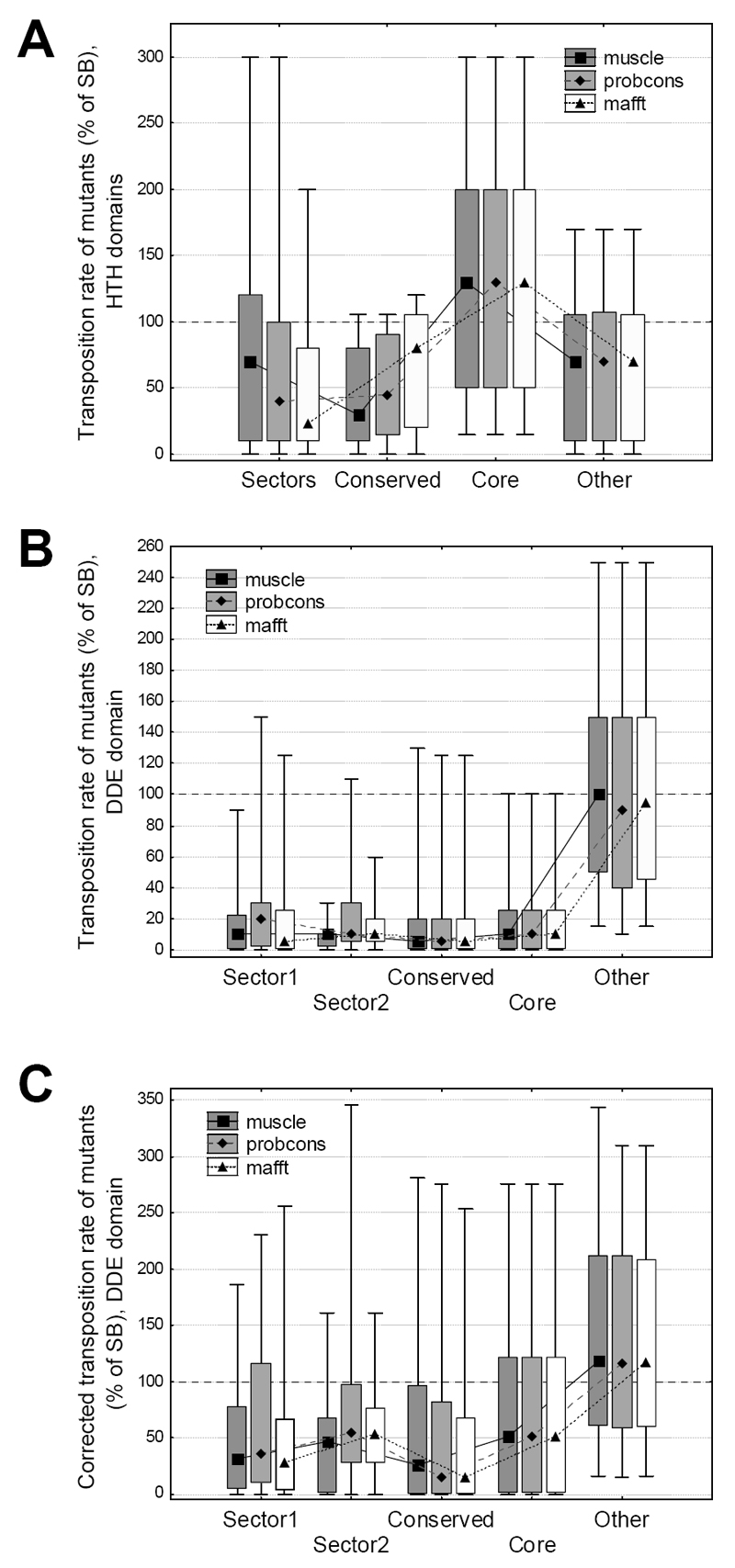
Figure 2. Effect of residue location on the transposition rate of SB mutants.
A) The location of the mutations along the SB transposase sequence, and their effect on transposition rate. The 286 mutants are distributed approximately evenly across the sequence; the majority of mutants reduces transposition rate (< 100% of SB). None of the Pfam conserved domains show a clear difference from the rest of the sequence. B) The effect of sectors, conserved residues and protein core on transposition rate (median, box: 25%-75%, whiskers: 10%-90%). Mutants in both sectors, conserved residues and residues of the protein core have significantly lower transposition rates than other residues, irrespectively of the aligner used (p < 0.05, Mann-Whitney U tests).
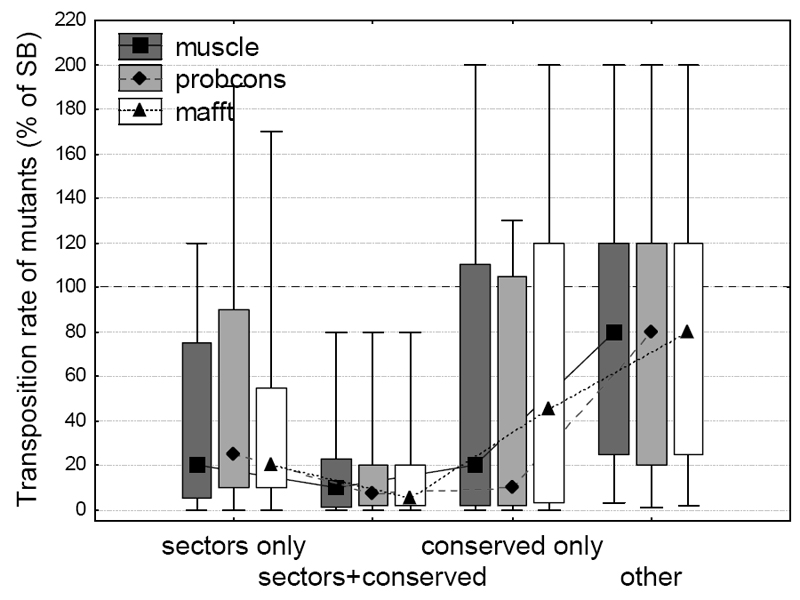
Sectors represent an extension of the traditional concept of conservation, and there is significant overlap between residues that are part of a sector and also have high positional conservation (D > 0.5). Recently it has been questioned whether the effect of sectors on transposition rate is independent from the effect of conservation2. To test this, we split sector and conserved residues into three groups: sector residues with low conservation (D < 0.5), sector residues with high conservation, and conserved residues that are not part of a sector. The comparison of these groups with residues that are neither part of sectors, nor the protein core, and are also not conserved (“other” residues, Figure 3) indicates, that the effect of sectors on transposition rate is not simply due to positional conservation, as the three groups are significantly different from the “other” residues (p < 0.05 for all comparisons except “conserved only” of the mafft alignment, Fisher post–hoc tests, ANOVA on log transformed transposition rates). There is no significant difference between non-conserved sector and non-sector conserved residues (p > 0.05 in all three alignments, Fisher post–hoc tests, ANOVA).
Figure 3. The effect of sectors on transposition rate is not a by-product of positional conservation.
Sector and conserved residues were split into three groups: sector residues with low positional conservation (D< 0.5), sector residues with high positional conservation, and conserved residues that are not part of any sector. Transposition rates of mutants (median, box: 25%-75%, whiskers: 10%-90%) in all three groups are significantly different from mutants in other residues (p << 0.05 for all comparisons, Fisher post–hoc tests, ANOVA on log transformed transposition rates), and there is no significant difference between the mutants of non-conserved sector and conserved but non-sector residues (p > 0.05 in all three alignments, Fisher post–hoc tests).
The effect of mutations on protein stability
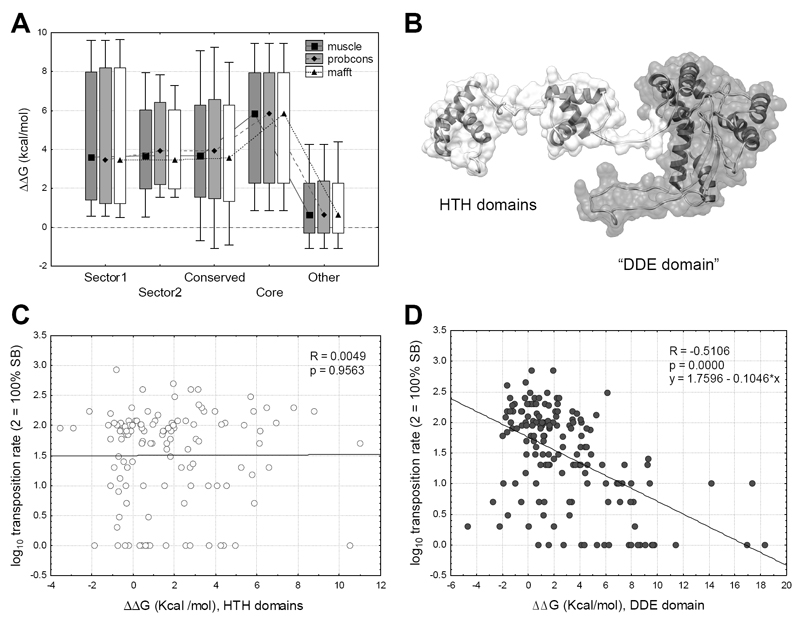
Most proteins can function only in a narrow range of folding energies 44, as unstable proteins may not fold properly and very stable ones may be too rigid to perform their functions. Mutating a residue in a protein can have significant effects on its overall stability (ΔG, the free energy of unfolding) and function, thus we tested whether the differences in transposition rate between the sectors, conserved residues and core of the protein and other residues are caused by their effect on protein stability, measured as the difference of the predicted folding energy (ΔΔG) between the wild type SB transposase and the mutants. The analysis shows that mutations in sector, conserved and core residues usually have a destabilizing effect on the structure (ΔΔG > 0, p << 0.05, t-tests; Figure 4A).
Figure 4. The effect of mutations on the change of the free energy of unfolding.
(median, box: 25%-75%, whiskers: 10%-90%). A) Mutations in sectors and the core are significantly more destabilizing (ΔΔG > 0) than mutants of other residues (p < 0.05 for all comparisons, t-tests). B) The monomer of SB. The flexible N-terminal arm of the protein containing the HTH domains (residues 1-120) is indicated with white, the globular part (residues 121-340), which contains the DDE domain, with gray. C) In the flexible arm the effect of mutations on ΔΔG is not correlated with transposition rate (p = 0.95). D) In the globular region we find a significant negative correlation (p << 0.001, R = -0.51) between ΔΔG and transposition rate.
Although three conserved domains were identified in the SB sequence, an analysis of the flexibility of the structure with the PiSQRD tool 45 and also recent analyses of the Mos1 and SB transposase 24,46 indicate that the structure can be split into two large regions; the relatively flexible N-terminal part of the protein containing the DNA binding HTH-domains (residues 1-120), and a rigid, globular region (residues 121-340) containing the DDE domain (Figure 4B, Figure 1C). Mutations have different effects on folding energies in these two regions; while we detected a clear negative correlation (p << 0.001, R = -0.51) between transposition rate and ΔΔG (Figure 4C) in the globular part of the protein, there is no such relationship (p = 0.95, R= 0.0049) in the N-terminal region containing the HTH domains (Figure 4D).
Next we tested whether mutants in the two regions have different effects on the transposition rate of SB, and we found that the two regions are markedly different. In the flexible part of the protein mutants of sector, conserved and core residues do not differ significantly from the remaining residues (p> 0.05 for all comparisons, Mann-Whitney U tests, Figure 5A), while in the region containing the DDE domain there is a highly significant difference (p << 0.001 for all comparisons, Mann-Whitney U tests, Figure 5B). Additionally, 50% of the mutants of “other” residues have higher transposition rates than the wild type.
Figure 5. The effect of residue location on transposition rate, in the two regions of the protein.
(median, box: 25%-75%, whiskers: 10%-90%). A) In the HTH-region, mutants of sectors, conserved or buried residues are not significantly different from the remaining mutants (p > 0.05 for all comparisons, Mann-Whitney U tests). B) In the DDE domain the differences are highly significant (p < 0.05 for all comparisons, Mann-Whitney U tests), even after correcting for the different effects of free energy of folding (C). Note that in the DDE domain 50% of mutants of “other” residues are characterized with higher activity than the wild type SB.
As the location of mutations has a significant effect on the free energy of folding, and in the DDE domain ΔΔG is correlated with transposition rates (Figure 4D), we tested whether the effect of sectors remains significant if we remove the effect of ΔΔG on transposition rate, i.e. we adjust all rates to ΔΔG = 0. The results show that the corrected transposition rates are still highly significantly different from other residues (p < 0.05 for all comparisons, Mann-Whitney U tests, Figure 5C), thus the biological effect of sectors and conserved residues cannot be explained with their effect on ΔΔG alone.
The effect of protein-protein and protein-DNA interactions on transposition rate
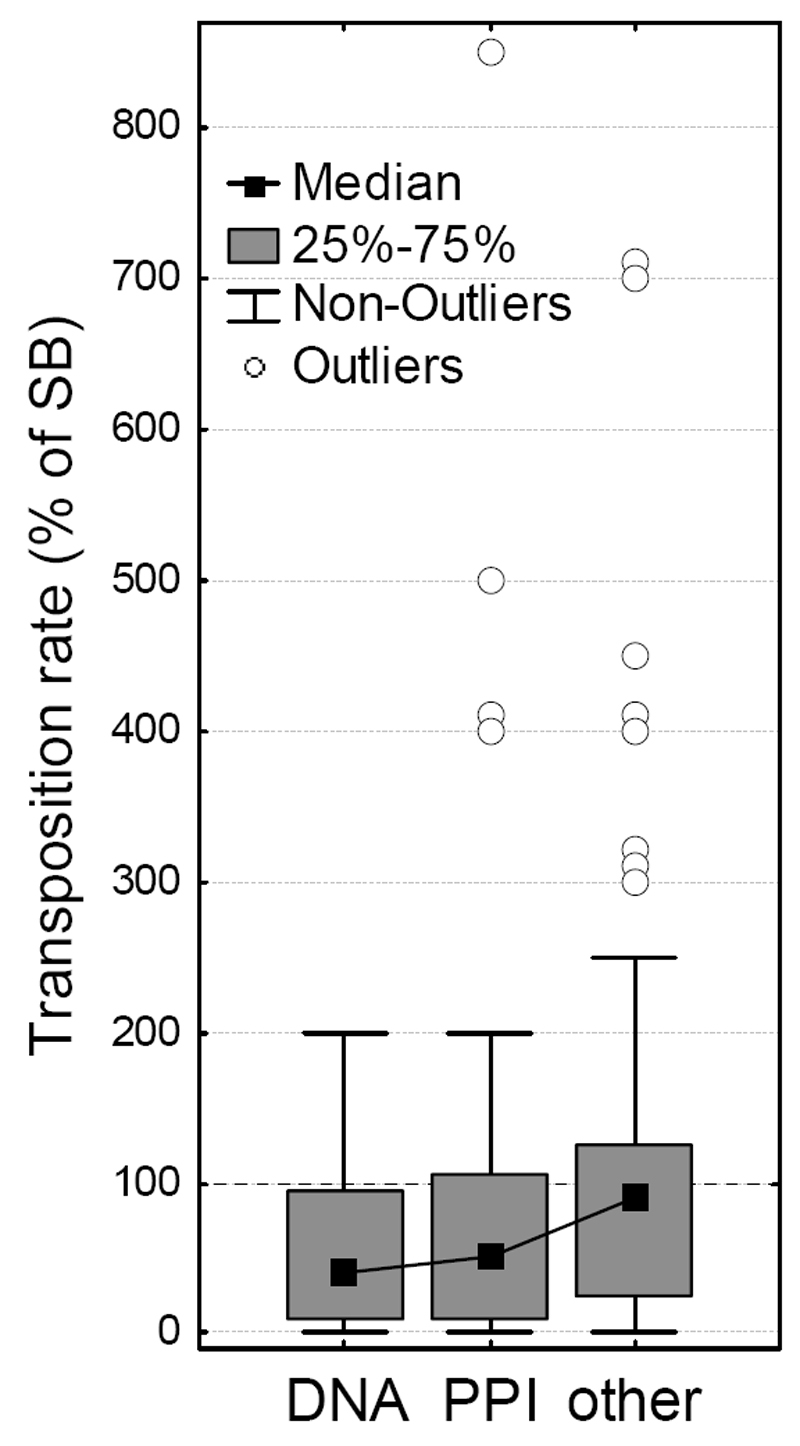
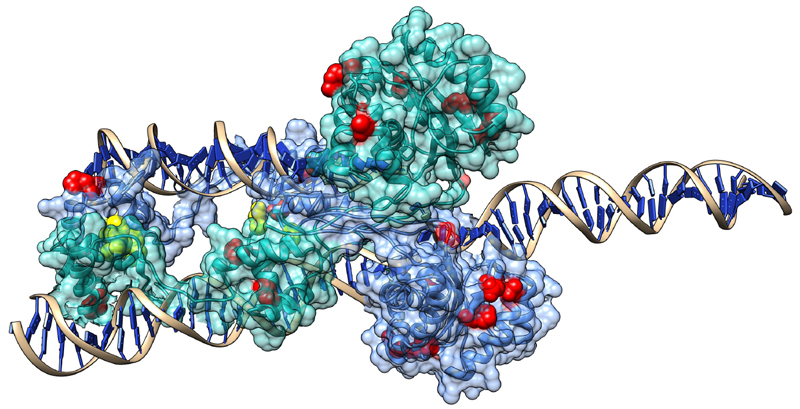
Transposases typically form protein complexes during transposition, and recent studies on mariner transposases related to Sleeping Beauty (Hsmar1 and Mos1) indicate that mutants that disrupt allosteric communication within its dimer are characterized by increased activity47–49. In particular, almost all mutants of the conserved WVPHEL motif (except P and E) of Hsmar1 transposon were hyperactive 48, most likely due to lowering the kinetic barrier to synapsis50,51. Our findings suggest that the mechanism that causes hyperactivity in SB may be comparable to Hsmar1,and probably involves the modification (or disruption) of protein–protein interactions (although the WVPHEL motif is not conserved in the SB transposase). This hypothesis is also consistent with the observation that the relationship between transposase concentration and SB activity is similar to Hsmar151. We tested whether mutants of residues taking part in protein-protein and DNA-protein interactions have different transposition rates than other residues at the protein surface. In general, when outliers are excluded, mutants of both protein and DNA-interacting residues have significantly lower transposition rates than other residues at the surface (Figure 6, p < 0.05, Kruskal-Wallis test). However, all but two of the hyperactive mutants (with 300% or higher activity) are located at the protein surface, and none are present in the core of the DDE domain. Of the 12 hyperactive surface mutations, four are in the protein-protein interfaces of the dimer (including the most active mutant), and none are in DNA-protein interfaces (see Figure 7 and supplementary Chimera visualization). Since the SB transposase can probably also form tetramers during transposition32, there are probably more residues that take part in protein-protein interactions, suggesting that modification of interactions might be a key factor responsible for hyperactivity.
Figure 6. The effect of DNA-protein and protein-protein interactions on transposition rate.
When outliers are excluded, mutants of residues interacting with DNA (“DNA”) and the other SB chain (“PPI”) have significantly lower transposition rates (p< 0.05, Mann-Whitney U test) than other residues located at the surface (RSA > 0.1). Surprisingly, 12 of the 14 hyperactive mutants (outliers, 300+% activity) are also located in the protein surface, and none in the DNA binding regions, suggesting that the modification of protein-protein interactions might be responsible for their dramatically increased activity. (Outliers with identical transposition rates were shifted by 10%, for visibility)
Figure 7. The location of the 14 hyperactive mutations in the SB dimer.
Yellow residues represent mutants in protein-protein interfaces, red residues other mutants. As SB probably forms also a tetramer in certain phases of transposition, the number of residues taking part in PPIs is probably higher. (See also the supplementary “hyperactive.py” Chimera file.)
Discussion
We performed an analysis of protein sectors in a relatively large, multidomain protein with a complex tertiary and quaternary structure, and attempt to predict the effect of mutations on transposition rate, based on their location and effect on protein stability. Although sector identification depends on the alignment used, we could identify two sectors in the SB transposase, regardless of the alignment method. Most previous studies focused on smaller, single-domain proteins 1,7,52, and one study8 identified a sector that spans two domains; our analysis indicates that sectors can span multiple conserved domains of a protein (Figure 1E-F), and, in the case of SB, are enriched in DNA binding residues. There may be at least two explanations for the observation that sectors residues are present in more than one domain: first, in some stages of transposition these residues may be in physical contact. Second, since sector identification is a purely statistical procedure which searches for coevolving residues in the entire protein sequence, in the case of two (or multi) domain proteins where both domains are necessary for the protein to function, coevolution between the domains is highly likely, and sectors that are confined to a single domain are probably present only in domains that are essentially independent.
A significant effect of mutations on transposition rate could be demonstrated in sectors, the protein core, conserved residues, protein-protein and protein-DNA interface: mutating these residues typically resulted in transposases with low transposition rates. Recently Teşileanu et al. suggested that depending on the method used for sector identification, any biological effect of the first sector may be the consequence of sequence conservation alone 2. Since we used the method of Halabi et al.1 for sector identification, which does not use the first eigenvector of the SCA correlation matrix, their concerns do not apply for our results. However, as a significant fraction of sector residues are conserved, we also analyzed sector and conserved residues independently, which shows that mutations of not conserved sector residues have a similar effect on transposition rate as mutations of conserved but non-sector residues (Figure 3), thus the biological functions of sectors cannot be explained with conservation alone.
In comparison with smaller proteins5, the influence of sectors on protein function appears to be more complicated, and depend on the tertiary structure. In the globular part of the protein, we could detect a clear effect of sectors, conservation and core on transposition rate, even when the effect of the free energy of folding was excluded (Figures 5). However, in the flexible part of the protein containing the HTH-domains we found no effect of sectors, nor a correlation between transposition rate and ΔΔG (Figure 4), which indicates that further studies are needed to evaluate the importance of sectors in non-globular (including disordered and coiled coil) proteins.
While we didn’t find a “recipe” for making hyperactive mutants of SB, our analysis allows prioritizing residues for targeted mutagenesis. Half of the residues in the DDE domain that are not part of sectors, conserved residues or protein core have increased transpositional activity. In addition, 12 out of the 14 hyperactive mutants (mutants with at least 3x increased activity compared to the wild type) are located in the protein surface, and 4 of them are in the protein-protein interfaces of the dimer, suggesting that similarly to the Hsmar1, the disruption of self-regulating protein-protein interactions may be an important factor in generating hyperactive mutants. In contrast, no hyperactive mutants are present in DNA-protein interfaces or in the buried residues (core) of the DDE domain. Since mutations of these regions typically strongly reduce the rate of transposition, this suggests that despite the fact that SB is a reconstructed sequence and it is most likely inaccurate to some degree, both DNA binding and folding are close to optimal.
Materials and Methods
Identification of SB homologs and making of multiple alignments
Transposase sequences homologous to Sleeping Beauty were identified in the 6-frame translated RepBase database (v17.12) 53, the main database of eukaryotic transposable elements, using the jackhmmer tool of the HMMER 3.0 package 54, with bit score cutoff 27. We excluded from the hits all matches that show homology only to a short fragment of SB, and kept only those hits that span at least from residues 50 to 290 of the SB transposase, thus covering more than 70% of the sequence. Next, to remove sequences with high similarity (>90%), the homologous sequences were clustered with uclust 55. The determination of protein sectors depends on multiple sequence alignments, but in the case of SB the average pairwise sequence similarity between the homologous sequences is low (19%), and in this low range of sequence similarity only approximately 50-80% of the residues can be aligned correctly with current methods56. This means that the choice of the aligner may influence the results significantly (i.e. the determination of sector and conserved residues), and to estimate the biases introduced by different alignment methods, we used three different alignment tools: muscle 40, probcons 41 and mafft42. After aligning the sequences, the alignments were trimmed to the 340 residues of SB transposase, i.e. we removed all columns with gaps in the SB sequence. All three alignments are available for download as Supplementary Data.
Determination of conservation and SCA calculations
Conservation (D, Kullback-Leibler entropy) at any given position of the sequence was defined as the divergence of the observed frequency from the background frequency of the most frequent residue at the position, and was calculated with the following equation: D = f ln(f/q) + (1–f) ln[(1-f)/(1-q)] 1, where f denotes the frequency of the amino acid at a given position of the sequence, and q represents its background frequency. We used the same background frequencies as in 1, and conserved residues were defined as residues with D > 0.5. Both for SCA and D calculations, we excluded all positions from the alignments, where the frequency of gaps was higher than 30%. SCA calculations (calculating the correlation matrix, spectral cleaning, randomization of the alignments) were performed with a modified Matlab script provided by the Halabi et al.1. Sectors were determined by a visual examination of residue weights of eigenvectors 2-4 (see Halabi et al. for details), sector 1 was defined as residues with weights < -0.05 along eigenvector 2, sector 2 as residues with weights > 0.05 along eigenvector 2 (see Figure S3).
Construction of SB mutants and determining their transposition rate
The mutants were partly obtained from published studies20,57–60, and partly (~80 mutants) represent unpublished material. Site-directed mutagenesis of the transposase gene was done by PCR following the QuikChange (Stratagene) principle of site-directed mutagenesis. The mutants were tested against the corresponding wild-type SB transposase in cell-based transposition assays, as originally described by Ivics et al.19.
Stability calculations and in-silico mutagenesis
The free energy of unfolding (ΔG) of the SB transposase, and its changes were calculated with the FoldX tool (version 4) 61,62, using the predicted structure of the SB complex. First, the structure produced by I-TASSER was optimized with the RepairPDB function to correct torsion angles, van der Waals clashes and total energies. Next we calculated the effect of the mutations on the ΔG of the structure for the 286 mutants. The difference in ΔG between the ‘wild type’ SB and its mutants is given as ΔΔG; its positive values indicate destabilizing, negative values stabilizing mutations.
Supplementary Material
Supplementary material is available in the online version of the paper: Supplementary figures 1-4; Supplementary Tables 1-4; the predicted structure of the Sleeping Beauty transposase (SB.pdb); Chimera visualizations of the transposase dimer showing the hyperactive (300+% activity) mutants (hyperactive.py) and the mutants marked as “other” (other.py); and the alignments used to determine protein sectors. A list with 229 SB mutants is available in the attached mutations.txt file. The remaining 57 mutants are available from Wang Y., Nagy E.E, Pryputniewicz-Dobrinska D. et al.: Regulated Complex Assembly Safeguards the Fidelity of Sleeping Beauty Transposition (Submitted).
Acknowledgments
We thank Mark A. Kay for support, and the referees for useful comments and suggestions. György Abrusán was supported by the Hungarian Scientific Research Fund (OTKA grant PD83571) and the Medical Research Council (grant MR/M02122X/1 to J.M.). András Szilágyi was supported by the OTKA grant K105415. Zoltán Ivics was supported by the European Union (EU FP5 JUMPY and EU FP6 INTHER grants), and also by grants from the Volkswagen Stiftung and the Bundesministerium für Bildung und Forschung (NGFN-2). Zsuzsanna Izsvák was supported by the EU FP6 INTHER grant. Joseph A. Marsh was supported by the Medical Research Council (MR/M02122X/1). Orsolya Barabás was supported by the EMBL.
References
- 1.Halabi N, Rivoire O, Leibler S, Ranganathan R. Protein sectors: evolutionary units of three-dimensional structure. Cell. 2009;138:774–786. doi: 10.1016/j.cell.2009.07.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Teşileanu T, Colwell LJ, Leibler S. Protein sectors: statistical coupling analysis versus conservation. PLoS Comput Biol. 2015;11:e1004091. doi: 10.1371/journal.pcbi.1004091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Socolich M, Lockless SW, Russ WP, Lee H, Gardner KH, Ranganathan R. Evolutionary information for specifying a protein fold. Nature. 2005;437:512–518. doi: 10.1038/nature03991. [DOI] [PubMed] [Google Scholar]
- 4.Süel GM, Lockless SW, Wall MA, Ranganathan R. Evolutionarily conserved networks of residues mediate allosteric communication in proteins. Nat Struct Biol. 2003;10:59–69. doi: 10.1038/nsb881. [DOI] [PubMed] [Google Scholar]
- 5.McLaughlin RN, Jr, Poelwijk FJ, Raman A, Gosal WS, Ranganathan R. The spatial architecture of protein function and adaptation. Nature. 2012;491:138–142. doi: 10.1038/nature11500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Russ WP, Lowery DM, Mishra P, Yaffe MB, Ranganathan R. Natural-like function in artificial WW domains. Nature. 2005;437:579–583. doi: 10.1038/nature03990. [DOI] [PubMed] [Google Scholar]
- 7.Reynolds KA, McLaughlin RN, Ranganathan R. Hot spots for allosteric regulation on protein surfaces. Cell. 2011;147:1564–1575. doi: 10.1016/j.cell.2011.10.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Smock RG, Rivoire O, Russ WP, Swain JF, Leibler S, Ranganathan R, et al. An interdomain sector mediating allostery in Hsp70 molecular chaperones. Mol Syst Biol. 2010;6:414. doi: 10.1038/msb.2010.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ivics Z, Izsvák Z. Nonviral gene delivery with the sleeping beauty transposon system. Hum Gene Ther. 2011;22:1043–1051. doi: 10.1089/hum.2011.143. [DOI] [PubMed] [Google Scholar]
- 10.Mann MB, Jenkins NA, Copeland NG, Mann KM. Sleeping Beauty mutagenesis: exploiting forward genetic screens for cancer gene discovery. Curr Opin Genet Dev. 2014;24:16–22. doi: 10.1016/j.gde.2013.11.004. [DOI] [PubMed] [Google Scholar]
- 11.Ammar I, Izsvák Z, Ivics Z. The Sleeping Beauty transposon toolbox. Methods Mol Biol. 2012;859:229–240. doi: 10.1007/978-1-61779-603-6_13. [DOI] [PubMed] [Google Scholar]
- 12.Grabundzija I, Wang J, Sebe A, Erdei Z, Kajdi R, Devaraj A, et al. Sleeping Beauty transposon-based system for cellular reprogramming and targeted gene insertion in induced pluripotent stem cells. Nucleic Acids Res. 2013;41:1829–1847. doi: 10.1093/nar/gks1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Grabundzija I, Irgang M, Mátés L, Belay E, Matrai J, Gogol-Döring A, et al. Comparative analysis of transposable element vector systems in human cells. Mol Ther. 2010;18:1200–1209. doi: 10.1038/mt.2010.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Abe G, Suster ML, Kawakami K. Tol2-mediated transgenesis, gene trapping, enhancer trapping, and the Gal4-UAS system. Methods Cell Biol. 2011;104:23–49. doi: 10.1016/B978-0-12-374814-0.00002-1. [DOI] [PubMed] [Google Scholar]
- 15.Kawakami K. Tol2: a versatile gene transfer vector in vertebrates. Genome Biol. 2007;8(Suppl 1):S7. doi: 10.1186/gb-2007-8-s1-s7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Di Matteo M, Mátrai J, Belay E, Firdissa T, Vandendriessche T, Chuah MKL. PiggyBac toolbox. Methods Mol Biol. 2012;859:241–254. doi: 10.1007/978-1-61779-603-6_14. [DOI] [PubMed] [Google Scholar]
- 17.Li X, Burnight ER, Cooney AL, Malani N, Brady T, Sander JD, et al. piggyBac transposase tools for genome engineering. Proc Natl Acad Sci USA. 2013;110:E2279–2287. doi: 10.1073/pnas.1305987110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yusa K, Zhou L, Li MA, Bradley A, Craig NL. A hyperactive piggyBac transposase for mammalian applications. Proc Natl Acad Sci USA. 2011;108:1531–1536. doi: 10.1073/pnas.1008322108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ivics Z, Hackett PB, Plasterk RH, Izsvák Z. Molecular reconstruction of Sleeping Beauty, a Tc1-like transposon from fish, and its transposition in human cells. Cell. 1997;91:501–510. doi: 10.1016/s0092-8674(00)80436-5. [DOI] [PubMed] [Google Scholar]
- 20.Mátés L, Chuah MKL, Belay E, Jerchow B, Manoj N, Acosta-Sanchez A, et al. Molecular evolution of a novel hyperactive Sleeping Beauty transposase enables robust stable gene transfer in vertebrates. Nat Genet. 2009;41:753–761. doi: 10.1038/ng.343. [DOI] [PubMed] [Google Scholar]
- 21.Guerrero AD, Moyes JS, Cooper LJN. The human application of gene therapy to reprogram T-cell specificity using chimeric antigen receptors. Chin J Cancer. 2014;33:421–433. doi: 10.5732/cjc.014.10100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Singh H, Huls H, Kebriaei P, Cooper LJN. A new approach to gene therapy using Sleeping Beauty to genetically modify clinical-grade T cells to target CD19. Immunol Rev. 2014;257:181–190. doi: 10.1111/imr.12137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Voigt F, Wiedemann L, Zuliani C, Querques I, Sebe A, Mátés L, et al. Sleeping Beauty transposase structure allows rational design of hyperactive variants for genetic engineering. Nat Commun. 2016;7:11126. doi: 10.1038/ncomms11126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Carpentier CE, Schreifels JM, Aronovich EL, Carlson DF, Hackett PB, Nesmelova IV. NMR structural analysis of Sleeping Beauty transposase binding to DNA. Protein Sci. 2014;23:23–33. doi: 10.1002/pro.2386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang Y. I-TASSER server for protein 3D structure prediction. BMC Bioinformatics. 2008;9:40. doi: 10.1186/1471-2105-9-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Roy A, Kucukural A, Zhang Y. I-TASSER: a unified platform for automated protein structure and function prediction. Nat Protoc. 2010;5:725–738. doi: 10.1038/nprot.2010.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang Y, Skolnick J. Scoring function for automated assessment of protein structure template quality. Proteins. 2004;57:702–710. doi: 10.1002/prot.20264. [DOI] [PubMed] [Google Scholar]
- 28.Zhang Y. Protein structure prediction: when is it useful? Curr Opin Struct Biol. 2009;19:145–155. doi: 10.1016/j.sbi.2009.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Richardson JM, Colloms SD, Finnegan DJ, Walkinshaw MD. Molecular architecture of the Mos1 paired-end complex: the structural basis of DNA transposition in a eukaryote. Cell. 2009;138:1096–1108. doi: 10.1016/j.cell.2009.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Laskowski RA, MacArthur MW, Moss DS, Thornton JM. PROCHECK: a program to check the stereochemical quality of protein structures. Journal of Applied Crystallography. 1993;26:283–291. [Google Scholar]
- 31.Nesmelova IV, Hackett PB. DDE transposases: Structural similarity and diversity. Adv Drug Deliv Rev. 2010;62:1187–1195. doi: 10.1016/j.addr.2010.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Izsvák Z, Khare D, Behlke J, Heinemann U, Plasterk RH, Ivics Z. Involvement of a bifunctional, paired-like DNA-binding domain and a transpositional enhancer in Sleeping Beauty transposition. J Biol Chem. 2002;277:34581–34588. doi: 10.1074/jbc.M204001200. [DOI] [PubMed] [Google Scholar]
- 33.Montaño SP, Pigli YZ, Rice PA. The μ transpososome structure sheds light on DDE recombinase evolution. Nature. 2012;491:413–417. doi: 10.1038/nature11602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, et al. UCSF Chimera--a visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 35.Zhang Y, Skolnick J. TM-align: a protein structure alignment algorithm based on the TM-score. Nucleic Acids Res. 2005;33:2302–2309. doi: 10.1093/nar/gki524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pronk S, Pall S, Schulz R, Larsson P, Bjelkmar P, Apostolov R, et al. GROMACS 4.5: a high-throughput and highly parallel open source molecular simulation toolkit. Bioinformatics. 2013;29:845–854. doi: 10.1093/bioinformatics/btt055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bowie JU, Reidhaar-Olson JF, Lim WA, Sauer RT. Deciphering the message in protein sequences: tolerance to amino acid substitutions. Science. 1990;247:1306–1310. doi: 10.1126/science.2315699. [DOI] [PubMed] [Google Scholar]
- 38.Touw WG, Baakman C, Black J, te Beek TAH, Krieger E, Joosten RP, et al. A series of PDB-related databanks for everyday needs. Nucleic Acids Res. 2015;43:D364–368. doi: 10.1093/nar/gku1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lu X-J, Olson WK. 3DNA: a versatile, integrated software system for the analysis, rebuilding and visualization of three-dimensional nucleic-acid structures. Nat Protoc. 2008;3:1213–1227. doi: 10.1038/nprot.2008.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Do CB, Mahabhashyam MSP, Brudno M, Batzoglou S. ProbCons: Probabilistic consistency-based multiple sequence alignment. Genome Res. 2005;15:330–340. doi: 10.1101/gr.2821705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Katoh K, Standley DM. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 2013 doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Finn RD, Mistry J, Tate J, Coggill P, Heger A, Pollington JE, et al. The Pfam protein families database. Nucleic Acids Res. 2010;38:D211–222. doi: 10.1093/nar/gkp985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.DePristo MA, Weinreich DM, Hartl DL. Missense meanderings in sequence space: a biophysical view of protein evolution. Nat Rev Genet. 2005;6:678–687. doi: 10.1038/nrg1672. [DOI] [PubMed] [Google Scholar]
- 45.Aleksiev T, Potestio R, Pontiggia F, Cozzini S, Micheletti C. PiSQRD: a web server for decomposing proteins into quasi-rigid dynamical domains. Bioinformatics. 2009;25:2743–2744. doi: 10.1093/bioinformatics/btp512. [DOI] [PubMed] [Google Scholar]
- 46.Cuypers MG, Trubitsyna M, Callow P, Forsyth VT, Richardson JM. Solution conformations of early intermediates in Mos1 transposition. Nucleic Acids Res. 2013;41:2020–2033. doi: 10.1093/nar/gks1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Claeys Bouuaert C, Walker N, Liu D, Chalmers R. Crosstalk between transposase subunits during cleavage of the mariner transposon. Nucleic Acids Res. 2014;42:5799–5808. doi: 10.1093/nar/gku172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Liu D, Chalmers R. Hyperactive mariner transposons are created by mutations that disrupt allosterism and increase the rate of transposon end synapsis. Nucleic Acids Res. 2014;42:2637–2645. doi: 10.1093/nar/gkt1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dornan J, Grey H, Richardson JM. Structural role of the flanking DNA in mariner transposon excision. Nucleic Acids Res. 2015;43:2424–2432. doi: 10.1093/nar/gkv096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Claeys Bouuaert C, Liu D, Chalmers R. A simple topological filter in a eukaryotic transposon as a mechanism to suppress genome instability. Mol Cell Biol. 2011;31:317–327. doi: 10.1128/MCB.01066-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Claeys Bouuaert C, Lipkow K, Andrews SS, Liu D, Chalmers R. The autoregulation of a eukaryotic DNA transposon. Elife. 2013;2:e00668. doi: 10.7554/eLife.00668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lockless SW, Ranganathan R. Evolutionarily conserved pathways of energetic connectivity in protein families. Science. 1999;286:295–299. doi: 10.1126/science.286.5438.295. [DOI] [PubMed] [Google Scholar]
- 53.Jurka J, Kapitonov VV, Pavlicek A, Klonowski P, Kohany O, Walichiewicz J. Repbase Update, a database of eukaryotic repetitive elements. Cytogenet Genome Res. 2005;110:462–467. doi: 10.1159/000084979. [DOI] [PubMed] [Google Scholar]
- 54.Eddy SR. A new generation of homology search tools based on probabilistic inference. Genome Inform. 2009;23:205–211. [PubMed] [Google Scholar]
- 55.Edgar RC. Search and clustering orders of magnitude faster than BLAST. Bioinformatics. 2010;26:2460–2461. doi: 10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
- 56.Nuin PAS, Wang Z, Tillier ERM. The accuracy of several multiple sequence alignment programs for proteins. BMC Bioinformatics. 2006;7:471. doi: 10.1186/1471-2105-7-471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Geurts AM, Yang Y, Clark KJ, Liu G, Cui Z, Dupuy AJ, et al. Gene transfer into genomes of human cells by the sleeping beauty transposon system. Mol Ther. 2003;8:108–117. doi: 10.1016/s1525-0016(03)00099-6. [DOI] [PubMed] [Google Scholar]
- 58.Zayed H, Izsvák Z, Walisko O, Ivics Z. Development of hyperactive sleeping beauty transposon vectors by mutational analysis. Mol Ther. 2004;9:292–304. doi: 10.1016/j.ymthe.2003.11.024. [DOI] [PubMed] [Google Scholar]
- 59.Yant SR, Park J, Huang Y, Mikkelsen JG, Kay MA. Mutational analysis of the N-terminal DNA-binding domain of sleeping beauty transposase: critical residues for DNA binding and hyperactivity in mammalian cells. Mol Cell Biol. 2004;24:9239–9247. doi: 10.1128/MCB.24.20.9239-9247.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Baus J, Liu L, Heggestad AD, Sanz S, Fletcher BS. Hyperactive transposase mutants of the Sleeping Beauty transposon. Mol Ther. 2005;12:1148–1156. doi: 10.1016/j.ymthe.2005.06.484. [DOI] [PubMed] [Google Scholar]
- 61.Guerois R, Nielsen JE, Serrano L. Predicting changes in the stability of proteins and protein complexes: a study of more than 1000 mutations. J Mol Biol. 2002;320:369–387. doi: 10.1016/S0022-2836(02)00442-4. [DOI] [PubMed] [Google Scholar]
- 62.Schymkowitz JWH, Rousseau F, Martins IC, Ferkinghoff-Borg J, Stricher F, Serrano L. Prediction of water and metal binding sites and their affinities by using the Fold-X force field. Proc Natl Acad Sci USA. 2005;102:10147–10152. doi: 10.1073/pnas.0501980102. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.