Abstract
Homozygous truncating mutations in the helix-loop-helix transcription factor PTF1A are a rare cause of pancreatic and cerebellar agenesis. The correlation of Ptf1a dosage with pancreatic phenotype in a mouse model suggested the possibility of finding hypomorphic PTF1A mutations in patients with pancreatic agenesis or neonatal diabetes but no cerebellar phenotype. Genome wide SNP typing in two siblings with neonatal diabetes from a consanguineous pedigree revealed a large shared homozygous region (31 Mb) spanning PTF1A. Sanger sequencing of PTF1A identified a novel missense mutation, p.P191T. Testing of 259 additional patients using a targeted next generation sequencing assay for 23 neonatal diabetes genes detected one additional proband and an affected sibling with the same homozygous mutation. All 4 cases were diagnosed with diabetes at birth and are insulin treated. Two of the 4 had exocrine pancreatic insufficiency requiring replacement but none of the affected individuals have neurodevelopmental delay. Transient transfection assays of the mutant protein demonstrated a 75% reduction in transactivation activity. This study shows that the functional severity of a homozygous mutation impacts on the severity of clinical features found in patients.
Keywords: neonatal diabetes, exocrine pancreatic insufficiency, cerebellar agenesis, PTF1A, hypomorphic mutation
Introduction
Homozygous truncating mutations in the basic helix-loop-helix transcription factor PTF1A cause pancreatic agenesis and very severe neurodevelopmental problems including central hypoventilation and total cerebellar agenesis [1–3]. All 6 affected individuals from 4 families show a high degree of phenotypic concordance and none survived for more than 4 months.
A combined linkage, genome sequencing and epigenomic annotation strategy recently identified homozygous mutations in a novel enhancer located 25 kb downstream from the PTF1A gene [4]. Patients with biallelic mutations in this enhancer have isolated pancreatic agenesis without cerebellar involvement. Sparing of the cerebellum suggests that the enhancer is tissue-specific to the pancreas.
A mouse model relating Ptf1a dosage to pancreatic phenotype [5] resulted in pancreatic hypoplasia and glucose intolerance in a dosage-dependent manner. The pancreatic phenotype consisted of reduced pancreatic bud size, mis-specification of pancreatic progenitors, reduced branching morphogenesis of the exocrine pancreas and a reduction in the ratio of beta to non-beta cells in pancreatic islets. In this model system, Ptf1a RNA levels were correlated with the endocrine and exocrine pancreatic phenotype.
Biallelic mutations in a second transcription factor gene, PDX1, also result in non-syndromic pancreatic agenesis and have been identified in three unrelated cases [6–8]. Hypomorphic mutations result in neonatal diabetes in the absence of exocrine pancreatic insufficiency [9,10]. A hypomorphic mutation in the transcription factor gene Pdx1 in mice has also been modelled [11] and shown to play a role in the transition from pancreatic progenitor to endocrine progenitor, with a reduction in the number of endocrine lineages.
To date, no PTF1A coding hypomorphic mutations have been identified. We now report four individuals from two separate sibships in each of whom the same novel PTF1A coding mutation, p.P191T, was identified. All four individuals were diagnosed with neonatal diabetes, but cerebellar pathology was absent. We performed functional studies on this mutation to investigate our hypothesis that p.P191T is a hypomorphic PTF1A mutation.
Methods
Genetics
Homozygosity mapping
Homozygosity mapping was carried out as described previously [12] in one patient with pancreatic agenesis of unknown aetiology and their sibling with neonatal diabetes. Pancreatic agenesis was defined as neonatal diabetes requiring insulin treatment and exocrine pancreatic insufficiency requiring enzyme replacement therapy [13].
Next generation sequencing assay
A targeted next generation sequencing assay was used to sequence PTF1A and 22 other genes in which mutations have been reported to cause neonatal diabetes [14]. We sequenced DNA samples from 259 probands with neonatal diabetes diagnosed before 6 months and no known genetic aetiology. Nine of these patients had pancreatic agenesis. Mutations in ABCC8, KCNJ11 and INS had previously been excluded by Sanger sequencing. We had also excluded EIF2AK3 mutations in patients born to consanguineous parents.
Sanger sequencing
Mutations were confirmed by PCR/Sanger sequencing and tested in other relatives.
Functional work
Cell Transfections
The reporter plasmids Ela1p.luc, with a minimal promoter directing the luciferase gene of pGL3 basic, and 3Rbpjl.Ela1p.luc, with 3 tandem repeats of the proximal PTF1 binding site of the Rbpjl gene upstream of Ela1p.luc, have been described [15]. The p.P191T mutation was introduced into the human PTF1A coding sequence by site-directed mutagenesis as previously described [16]. DNA was introduced into the human embryonic kidney cell line HEK 293 (American Type Culture Collection CRL-1573) with FuGene6 (Roche, Indianapolis, INS, USA) according to the manufacturer’s instructions. All transfections were normalized based on the β-galactosidase activity of a cotransfected reporter plasmid, pCMVbeta (Clontech, Mountain View, CA, USA).
Electrophoretic Mobility Shift Assays (EMSA)
The subunits of PTF1 were synthesized by in vitro transcription and translation (ivtt) using a TnT Reticulocyte Lysate System (Promega, Madison, WI, USA). The wild type and p.P191T mutant PTF1A plasmids described above were also used for in vitro protein synthesis. The plasmid bearing human RBPJ has previously been described [16]. The expression plasmid bearing a partial human E12/TCF3 cDNA in pCITE2a was a gift from Eric Olson, UT Southwestern, and was originally derived from E12R (20). The products of ivtt were quantified by [35S]methionine incorporation and adjustment according to the number of methionine residues in each protein.
EMSAs were performed by the method of Sawada and Littman [17] with slight modifications. The double-stranded oligonucleotide probe encompassed the proximal PTF1 binding site of the mouse Rbpjl gene [15] and was 5’-end labeled with 32P. The sequence of the top strand is GACACCTGCTGGGCAGATGTAGGCTTCCCACGG. ImageQuant software was used to analyze PhosphorImager scans of the EMSA gels (both, GE Healthcare, Life Sciences, Pittsburg, PA, USA).
Results
Homozygosity analysis of genome wide SNPs identified a homozygous region (31 Mb) on chromosome 10 which was shared between two affected siblings born to consanguineous parents. Sanger sequencing of PTF1A identified a novel homozygous missense mutation, p.P191T (c.571C>A; p.Pro191Thr) in both siblings. Analysis of PTF1A by targeted next generation sequencing of 9 patients with pancreatic agenesis identified one additional proband with the p.P191T mutation. This patient’s affected sibling was also homozygous for p.P191T. No additional PTF1A mutations were identified in a further 250 patients with neonatal diabetes.
P191T mutation characteristics
The p.P191 residue is located in the highly conserved helix loop helix domain which is critical for dimerisation and DNA binding (Figure 1). The p.P191T missense variant is not present in any public variant databases (1000 Genomes, NHLBI Exome Sequencing Project Exome Variant Server (EVS) or the ExAC browser). In silico analysis by SIFT and PolyPhen predicts the variant to be pathogenic. All four affected patients were homozygous for the p.P191T mutation and their parents were heterozygous carriers. The patients are from Saudi Arabia and Kuwait, raising the possibility of a founder mutation. Analysis of microsatellite markers flanking PTF1A showed a shared haplotype of 8 Mb (data not shown), consistent with a founder mutation segregating in the two families.
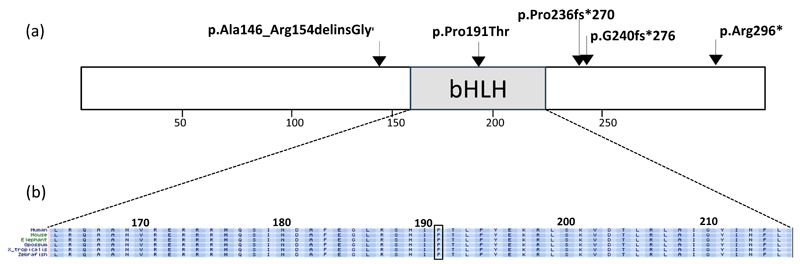
Figure 1.
(a) Schematic representation of the PTF1A protein and location of the mutations identified in all previously reported cases [1–3], and the present case. The p.P191T is the only missense mutation identified to date. It is located at a conserved residue within the bHLH domain (highlighted). (b) Amino acid conservation in the bHLH domain. The location of the p.P191T mutation found in the 4 patients us shown. ¥ The p.Ala146ArgdelinsGly mutation was previously reported as c.437_460del, p.Ala146_Arg154delfsX115 (3).
Clinical characteristics of patients with homozygous P191T mutation
The male proband in family I, (I-1), was the first child born to first cousins of Arabic descent with a birth weight of 1.98kg at 38 weeks’ gestation. He was noted to be hyperglycemic (350 mg/dl) on the first day of life while on a glucose infusion which was discontinued and an insulin infusion commenced for 20 hours. On Day 6, when on no treatment, his plasma glucose remained high (240 mg/dl) with low insulin of 0.1 MU/L (N 1.4-14) and C peptide of 0.1 ng/ml (N 0.9-4.3) confirming a diagnosis of neonatal diabetes. Insulin was recommend at 2 months of age as most glucose values were 200-300 mg/dl and his HbA1c was 7%. This has continued since then with Hba1c now 8.1%. The baby continued to have poor weight gain despite recommencing insulin and steatorrhoea was observed suggesting pancreatic insufficiency which was confirmed by elevated fecal fat, elevated stool chymotrypsin and low serum trypsinogen (Table 1). Exocrine pancreatic replacement therapy was initiated at 3 months of age. Other clinical features were a small patent ductus arteriosus, with a small atrial septal defect which were detected by echo after a cardiac murmur was noted; these did not require any intervention.
Table 1. Pancreatic clinical characteristics of patients studied.
| Patient | I-1 | I-2 | II-1 | II-2 |
| Present age | 12 years | 9 years | Deceased at 12 weeks due to sepsis | 2 years |
| Sex | Male | Female | Male | Female |
| Country of origin | Saudi Arabia | Saudi Arabia | Kuwait | Kuwait |
| Birth weight Percentile |
1980 g 0.4 |
2000 g 2 |
1275 g 0.1 |
1400 g <0.1 |
| Gestational age | 38 weeks | 37 weeks | 34 weeks | 36 weeks |
| Age at diabetes diagnosis/ Age of permanent Insulin therapy | 1 Day /2 months | 1 day/4 months | 1 day/1 day | 8 days/ 8 days |
| Diabetes treatment | Insulin | Insulin | Insulin | Insulin |
| Exocrine pancreatic insufficiency requiring replacement? | Yes | No | Yes | Yes |
| Clinical basis of exocrine insufficiency | Steatorrhoea, failure to thrive | No symptoms of exocrine insufficiency | Failure to thrive | Steatorrhoea, failure to thrive |
| Biochemical basis of exocrine insufficiency | Fecal fat: 3 g/24hr stool Stool chymotrypsin: 1 U/g (4-10U/g) Serum trypsinogen: 4 μg/l (15-25 μg/l) | Fecal fat: 0.5 g/24hr stool Stool chymotrypsin: 5 U/g (4-10U/g) Serum trypsinogen: 18 μg/l (15-25 μg/l) | Fecal elastase: undetectable (>200 μg elastase/g stool) | |
| Exocrine pancreatic replacement regimen | 10 000 U/day of lipase | None | 15 000 U/day of lipase | |
| Age at initiation of exocrine pancreatic replacement | 12 weeks | N/A | N/A | 10 weeks |
| Clinical neurocognitive function | Normal neurocognitive development with some disruption of eye movements on tracking. | Normal neurocognitive development | Normal neurocognitive development | |
| Brain imaging | Not performed | Not performed | Not performed | Normal (MRI imaging) |
His younger sister (I-2) was born at 37 weeks’ gestation, birth weight 2.00 kg. Diabetes was diagnosed on the first day of life with a glucose of 300mg/dl and treated with insulin for 4 days. Insulin was commenced at four months due to consistently high blood glucose values (> 200mg/dl) and HbA1c of 7% and has continued since then with HbA1c values 7-8%. Exocrine pancreatic supplementation has not been required and biochemical parameters (fecal fat, stool chymotrypsin and serum trypsinogen) are within normal limits, albeit at the lower limit of the normal range (see Table 1).
Ultrasound scanning of the abdomen of the two siblings in family I was performed in the first week of life and showed normal liver and contracted gallbladder, but failed to identify the pancreas. Parents declined further imaging of the abdomen.
Both siblings in family I underwent detailed neurodevelopmental assessment by a neurodevelopmental pediatrician at the age of 12 years in the older and at 9 years in the younger sibling. Evaluation revealed normal neurological examination apart from searching eye movements with horizontal nystagmus in patient 1. There are no other clinical abnormalities referable to cerebellar function. Parents declined MRI brain imaging. The rest of the clinical neurodevelopmental assessment was age-appropriate including vision, hearing, expressive and receptive language, gross motor and fine motor skills, social skills, and school performance. Their clinical neurodevelopmental assessment is similar to their unaffected siblings. However, formal neuropsychological testing was not performed.
The male proband in family II (II-1) was the first child born to consanguineous parents of Arabic descent. Birth weight was 1.275 kg at 34 weeks’ gestation. Neonatal diabetes was diagnosed on the first day of life and treated with insulin. There was severe failure to thrive gaining only 250g in the first 12 weeks of life but biochemical tests for pancreatic malabsorption were not performed. The patient died of necrotising enterocolitis and overwhelming sepsis at the age of 12 weeks. No further clinical details are available.
His younger sibling is a female child born at 36 weeks’ gestation with birth weight 1.40 kg. Neonatal diabetes was diagnosed at the age of 8 days and treated with insulin. She failed to thrive and fecal elastase was undetectable, confirming a diagnosis of exocrine pancreatic insufficiency. Exocrine replacement therapy was initiated at 10 weeks’ of age. Neurocognitive development and neurological examination are normal at the age of 2 years. Brain and cerebellar MRI scan was normal aged 3 months.
Functional work
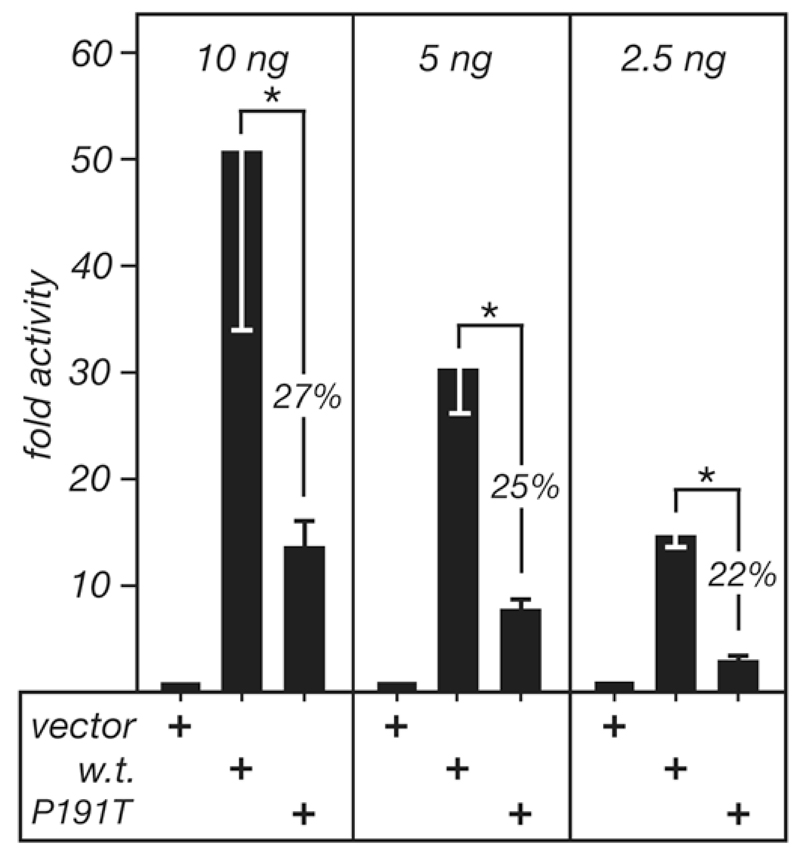
Transfection of the p.P191T mutation into HEK293 cells resulted in a 75% reduction in transcriptional activity compared to the wild type protein (Figure 2). A range in the amount of expression plasmids was tested to ensure that the activities of the wild type plasmids were proportional to the plasmid quantity and, thus, not saturated in the assay. The mean activity of the mutant PTF1A at three different concentrations within the linear range was 25 +/- 4.9% of the activity of the wild type protein (p < 0.001) (Figure 2).
Figure 2. The activity of human PTF1A-P191T in transfected cells.
The reporter plasmid 3Rbpjl.Ela1p.luc was cotransfected with expression plasmids for wild type PTF1A, PTF1A-P191T, or an insertless vector into human embryonic kidney (HEK) 293 cells. Three different amounts of each expression plasmid were used to determine whether response was proportional to plasmid quantity and, therefore, that the proteins produced were not in excess. Four independent transfections were analyzed for 10 and 5ng, and two for 2.5ng. Error bars are standard deviations. The asterisks indicate p values <0.05 for each pairwise comparison.
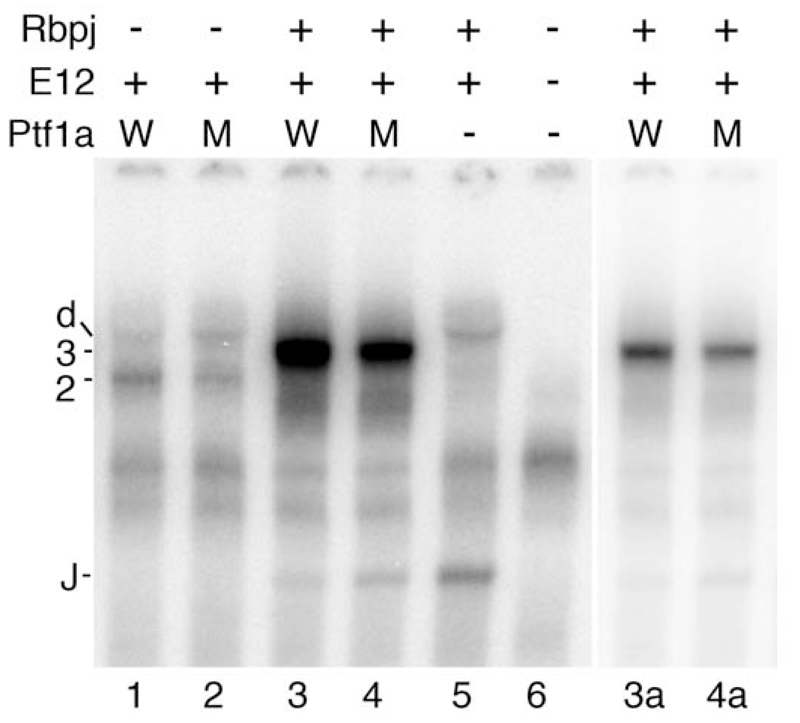
To determine the DNA binding characteristics of human PTF1A -P191T relative to wild type EMSAs were performed with a Rbpjl proximal PTF1 binding site as the radiolabeled probe (Figure 3). For the PTF1A:E12 dimer, the binding of the p.P191T mutant was 34% +/-7% of wild type (n = 3) whilst for the PTF1A:E12:Rbpj trimer, the binding of the p.P191T mutant was 88% +/-19% of wild type (n = 4).
Figure 3. The DNA binding of wild type PTF1A and PTF1A-P191T as part of the PTF1A:E12 dimer or PTF1A:E12:Rbpj trimer in EMSA.
The relative abilities of wild type and p.P191T mutant Ptf1a to form complexes with E12 and Rbpj on the PTF1 binding site of the Rbpjl gene was tested in vitro. W, Ptf1a wild type; M, p.P191T mutant; 2, Ptf1a:E12 dimer; 3, Ptf1a:E12:Rbpj trimer; d, E12 homodimer; J, Rbpj monomer. Lanes 3a and 4a show lanes 3 and 4 with an expanded gray scale such that the PTF1 trimeric complex bands are not saturated. Ptf1a alone does not bind DNA.
Discussion
We report a novel hypomorphic homozygous PTF1A mutation in 4 affected individuals from two families who are likely to have inherited the mutation from a common distant ancestor. The clinical phenotype is of neonatal diabetes with reduced exocrine function and normal neurological function and appearance. This clinical phenotype is distinct from the previously reported syndrome caused by homozygous PTF1A truncating mutations where all 6 patients (from 4 families) died in the first 4 months of life [1–3] of severe neurological complications as well as having pancreatic and cerebellar agenesis. In vitro functional studies are consistent with the p.P191T missense mutation being a hypomorphic mutation.
The main clinical feature of homozygosity for the p.P191T missense mutation is pancreatic aplasia/hypoplasia. All 4 patients had neonatal diabetes and 2 of the 4 have clinical and biochemical evidence of clinically significant exocrine dysfunction of the pancreas requiring replacement therapy. Another patient had clinical features suggestive of exocrine insufficiency but died of sepsis at 12 weeks and the fourth patient had diabetes requiring insulin treatment but no clinical features of exocrine failure, albeit with pancreatic enzyme levels close to the lower limit of the normal range.
All four patients had normal neurological development and function. This is in contrast to the patients with homozygous null PTF1A mutations who had very severe neurological developmental problems, central hypoventilation and total cerebellar agenesis. One patient had some disrupted eye movements but the phenotype was not consistent with significant cerebellar dysfunction. A normal cerebellum was seen in the patient who had brain imaging.
The moderate phenotypic variability seen in these 4 patients is also seen in patients who have other pancreatic transcription factor mutations affecting the pancreatic stem cell. Different phenotypes due to the same mutation are seen in patients with mutations in GATA6 [18], and in HNF1B [19]. Phenotypic variability is also seen in patients with mutations in the homeodomain transcription factor PDX1: biallelic mutations in this gene have been reported in patients with pancreatic agenesis [6–8]; neonatal diabetes with biochemical but not clinical evidence of exocrine insufficiency [9,10], and neonatal diabetes with normal exocrine function, both clinically and biochemically [10].
The p.P191T missense mutation is located in the highly conserved helix loop helix domain of PTF1A, a region which is critical for dimerisation with a common bHLH E-protein such as E12/E47 and DNA binding. Proline 191 terminates helix 1, and substitution of threonine at that position could favor the extension of the first α-helix, almost a full turn and thereby shorten the loop region, mis-position the second α-helix, and make binding to the E-protein partner less favourable. Indeed, in vitro functional studies demonstrated an effect on DNA binding and transactivation activity was reduced by 75% compared to the wild-type protein. This residual activity is consistent with p.P191T being a hypomorphic mutation.
Reduced Ptf1a dosage in a mouse model [5] resulted in pancreatic hypoplasia and glucose intolerance in a dosage-dependent manner. In hypomorphic mutant mice, pancreatic bud size was small and substantial proportions of pancreatic progenitors were mis-specified to the common bile duct and duodenal cells. Exocrine pancreatic branching morphogenesis was reduced, and there was a reduction in the ratio of beta cells to non-beta cells in pancreatic islets. In this model system, Ptf1a RNA levels could be correlated with the endocrine and exocrine pancreatic phenotype. We have clearly shown a similar correlation between mutation severity and phenotype in humans.
We conclude that hypomorphic PTF1A missense mutations can cause isolated pancreatic agenesis or neonatal diabetes. Hypomorphic mutations in this gene should be considered in patients presenting with diabetes mellitus in the neonatal period.
Acknowledgements
The authors thank the families for participating in this study. We are grateful to Annet Damhuis and Anna-Maria Bussell and the Research Center, College of Medicine, King Saud University for their technical assistance and to Ward Coats for helpful discussions regarding PTF1A structure. ATH and SE are the recipients of a Wellcome Trust Senior Investigator award and this funded the genetic/clinical part of this study (WT 098395). ATH is employed as a core member of staff within the NIHR funded Exeter Clinical Research Facility and is a NIHR senior investigator. GHS and RJM were supported by NIH grant R01-DK061220.
Footnotes
Authors’ contributions
SE, JALH, SEF, RC & EdF designed, performed and interpreted the genetic studies. AH, CS-S, KH, SM, & MA designed and performed the clinical studies. RJM and GHS designed, performed and interpreted the functional studies. JALH and GHS wrote the first draft of the paper with input from SE, ATH and RJM. All co-authors reviewed and commented on the draft manuscript and reviewed the submitted manuscript. S.E. is the guarantor of this work and, as such, had full access to all the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis.
References
- 1.Sellick GS, et al. Mutations in PTF1A cause pancreatic and cerebellar agenesis. Nature genetics. 2004;36:1301–5. doi: 10.1038/ng1475. [DOI] [PubMed] [Google Scholar]
- 2.Tutak E et al. A Turkish newborn infant with cerebellar agenesis/neonatal diabetes mellitus and PTF1A mutation. Genetic counseling. 2009;20:147–52. [PubMed] [Google Scholar]
- 3.Al-Shammari M, Al-Husain M, Al-Kharfy T, Alkuraya FS. A novel PTF1A mutation in a patient with severe pancreatic and cerebellar involvement. Clinical genetics. 2011;80:196–8. doi: 10.1111/j.1399-0004.2010.01613.x. [DOI] [PubMed] [Google Scholar]
- 4.Weedon MN, Cebola I, Patch AM, Flanagan SE, De Franco E, Caswell R, Rodríguez-Seguí SA, Shaw-Smith C, Cho CH, Allen HL, Houghton JA, et al. Recessive mutations in a distal PTF1A enhancer cause isolated pancreatic agenesis. Nat Genet. 2013 Nov 10; doi: 10.1038/ng.2826. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fukuda A, Kawaguchi Y, Furuyama K, Kodama S, Horiguchi M, Kuhara T, Kawaguchi M, Terao M, Doi R, Wright CV, Hoshino M, et al. Reduction of Ptf1a gene dosage causes pancreatic hypoplasia and diabetes in mice. Diabetes. 2008 Sep;57(9):2421–31. doi: 10.2337/db07-1558. Epub 2008 Jun 30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Stoffers DA, et al. Pancreatic agenesis attributable to a single nucleotide deletion in the human IPF1 gene coding sequence. Nat Genet. 1997;15(1):106–10. doi: 10.1038/ng0197-106. [DOI] [PubMed] [Google Scholar]
- 7.Schwitzgebel VM, et al. Agenesis of human pancreas due to decreased half-life of insulin promoter factor 1. The Journal of clinical endocrinology and metabolism. 2003;88(9):4398–406. doi: 10.1210/jc.2003-030046. [DOI] [PubMed] [Google Scholar]
- 8.Thomas IH, et al. Neonatal diabetes mellitus with pancreatic agenesis in an infant with homozygous IPF-1 Pro63fsX60 mutation. Pediatric Diabetes. 2009;10(7):492–6. doi: 10.1111/j.1399-5448.2009.00526.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.De Franco E, Shaw-Smith C, Flanagan SE, Edghill EL, Wolf J, Otte V, Ebinger F, Varthakavi P, Vasanthi T, Edvardsson S, Hattersley AT, et al. Biallelic PDX1 (insulin promoter factor 1) mutations causing neonatal diabetes without exocrine pancreatic insufficiency. Diabet Med. 2013 May;30(5):e197–200. doi: 10.1111/dme.12122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nicolino M, Claiborn KC, Senee V, Boland A, Stoffers DA, Julier C. A novel hypomorphic PDX1 mutation responsible for permanent neonatal diabetes with subclinical exocrine deficiency. Diabetes. 2010;59:733–740. doi: 10.2337/db09-1284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Oliver-Krasinski JM, Kasner MT, Yang J, Crutchlow MF, Rustgi AK, Kaestner KH, Stoffers DA. The diabetes gene Pdx1 regulates the transcriptional network of pancreatic endocrine progenitor cells in mice. J Clin Invest. 2009 Jul;119(7):1888–98. doi: 10.1172/JCI37028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rubio-Cabezas O, Minton JA, Kantor I, et al. Homozygous mutations in NEUROD1 are responsible for a novel syndrome of permanent neonatal diabetes and neurological abnormalities. Diabetes. 2010;59:2326–2331. doi: 10.2337/db10-0011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Allen HL, et al. GATA6 haploinsufficiency causes pancreatic agenesis in humans. Nat Genet. 2012;44(1):20–2. doi: 10.1038/ng.1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ellard S, Lango Allen H, De Franco E, Flanagan SE, Hysenaj G, Colclough K, Houghton JA, Shepherd M, Hattersley AT, Weedon MN, Caswell R. Improved genetic testing for monogenic diabetes using targeted next-generation sequencing. Diabetologia. 2013 Sep;56(9):1958–6314. doi: 10.1007/s00125-013-2962-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Masui T, Long Q, Beres TM, Magnuson MA, MacDonald RJ. Early pancreatic development requires the vertebrate Suppressor of Hairless (RBPJ) in the PTF1 bHLH complex. Genes Dev. 2007 Oct 15;21(20):2629–43. doi: 10.1101/gad.1575207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Beres TM, Masui T, Swift GH, Shi L, Henke RM, MacDonald RJ. PTF1 is an organ-specific and Notch-independent basic helix-loop-helix complex containing the mammalian Suppressor of Hairless (RBP-J) or its paralogue, RBP-L. Mol Cell Biol. 2006 Jan;26(1):117–30. doi: 10.1128/MCB.26.1.117-130.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sawada S, Littman DR. A heterodimer of HEB and an E12-related protein interacts with the CD4 enhancer and regulates its activity in T-cell lines. Mol Cell Biol. 1993 Sep;13(9):5620–8. doi: 10.1128/mcb.13.9.5620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.De Franco E, Shaw-Smith C, Flanagan SE, Shepherd MH, International NDM Consortium. Hattersley AT, Ellard S. GATA6 mutations cause a broad phenotypic spectrum of diabetes from pancreatic agenesis to adult-onset diabetes without exocrine insufficiency. Diabetes. 2013 Mar;62(3):993–7. doi: 10.2337/db12-0885. Epub 2012 Dec 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Edghill EL, Stals K, Oram RA, Shepherd MH, Hattersley AT, Ellard S. HNF1B deletions in patients with young-onset diabetes but no known renal disease. Diabet Med. 2013 Jan;30(1):114–7. doi: 10.1111/j.1464-5491.2012.03709.x. [DOI] [PubMed] [Google Scholar]