Abstract
The peroxisome proliferator-activated receptor γ co-activator 1α (PGC-1α) coordinates the transcriptional network response to promote an improved endurance capacity in skeletal muscle, e.g. by co-activating the estrogen-related receptor α (ERRα) in the regulation of oxidative substrate metabolism. Despite a close functional relationship, the interaction between these two proteins has not been studied on a genomic level. We now mapped the genome-wide binding of ERRα to DNA in a skeletal muscle cell line with elevated PGC-1α and linked the DNA recruitment to global PGC-1α target gene regulation. We found that, surprisingly, ERRα co-activation by PGC-1α is only observed in the minority of all PGC-1α recruitment sites. Nevertheless, a majority of PGC-1α target gene expression is dependent on ERRα. Intriguingly, the interaction between these two proteins is controlled by the genomic context of response elements, in particular the relative GC and CpG content, monomeric and dimeric repeat binding site configuration for ERRα, and adjacent recruitment of the transcription factor SP1. These findings thus not only reveal a novel insight into the regulatory network underlying muscle cell plasticity, but also strongly link the genomic context of DNA response elements to control transcription factor – co-regulator interactions.
Key terms: skeletal muscle, co-activator, transcription factor, PGC-1α, ERRα, transcriptional regulation, ChIP-Seq
Introduction
Skeletal muscle cells have an enormous capacity to respond to external stimuli, e.g. altered levels of physical activity, temperature, oxygen, nutrient composition and supply, by modulating metabolic and contractile properties (1,2). Accordingly, skeletal muscle cell plasticity entails a biological program with an enormous complexity. Thus, not surprisingly, the molecular mechanisms that control this program are still largely elusive. In recent years however, the peroxisome proliferator-activated receptor γ coactivator 1α (PGC-1α) has emerged as a regulatory nexus in the phenotypic adaptation of skeletal muscle to endurance training (3). The expression of individual or groups of target genes is positively or negatively affected by specific interactions of PGC-1α with a substantial repertoire of different transcription factors (TFs) (4). The dynamism and flexibility of a coactivator-controlled transcriptional network could therefore provide an explanation regarding how PGC-1α expression in muscle is not only sufficient to induce a high endurance phenotype in this tissue (5,6), but also to control related processes such as angiogenesis (7) or post- and pre-synaptic neuromuscular junction plasticity (8).
The estrogen-related receptor α (ERRα, NR3B1) plays a prominent role in regulating cellular metabolism that is highly reminiscent of the function of PGC-1α to boost mitochondrial biogenesis and oxidative substrate utilization (9). Indeed, a close relationship between ERRα and PGC-1α in the regulation of the expression of metabolic and other genes has been described in muscle and other tissues (10,11). Unbiased motif prediction in promoters of genes that exhibit PGC-1α-dependent changes in expression furthermore implied co-activation of ERRα by PGC-1α as a central regulatory paradigm in the control of mitochondrial oxidative phosphorylation (OXPHOS) gene expression (12). Intriguingly, at least in some crystal structures, the ligand-binding pocket of ERRα is almost completely occupied by bulk amino acid side chains and thereby, binding of putative endogenous ligands in the ligand binding pocket might be almost impossible (13). Instead, fluorescence polarization-based binding assay of the ERRα ligand binding domain (LBD) together with a coactivator peptide from PGC-1α revealed that these two partners exhibit a particularly high interaction affinity, as well as a change of the ERRα LBD into a transcriptionally active conformation in a ligand-independent manner (13). These data imply a “special relationship” between ERRα and PGC-1α to constitute the mechanistic core of PGC-1α- and ERRα-controlled gene expression whereby PGC-1α could act as the effective “ligand” of ERRα (14).
Recently, we have investigated the global DNA recruitment pattern of PGC-1α to the mouse genome in muscle cells related to PGC-1α-controlled gene transcription (4). To our surprise, a computational analysis of regulatory sites in positively regulated PGC-1α target genes not only suggested ERRα as an important TF in the regulation of direct, but also to be involved in the induction of indirect PGC-1α target genes, implying a role for ERRα in the absence of co-activation (4). To rule out the possibility of false positive computational prediction or spurious assignment of different nuclear receptor binding sites as ERRα response elements, we now studied genome-wide binding of endogenous ERRα to the mouse genome in muscle cells upon activation of PGC-1α. As in our previous study, cultured muscle cells were chosen based on their low expression of endogenous PGC-1α and hence a high signal-to-noise ratio upon adenoviral overexpression of this coactivator. Furthermore, exogenous expression of PGC-1α allowed the introduction of an epitope tag, which not only further enhances the selectivity of the immunoprecipitation, but also circumvents the problem of the currently existing low affinity antibodies that hamper an analysis of endogenous, untagged PGC-1α in cells or muscle tissue in vivo. Thus, by comparing genomic loci bound by endogenous ERRα in muscle cells that overexpress PGC-1α with those occupied by PGC-1α using chromatin immuno-precipitation followed by deep sequencing (ChIP-Seq), we aimed at identifying shared and individual recruitment sites for these two proteins in the context of PGC-1α-controlled muscle gene expression in the same cellular context. We now experimentally confirmed a role for ERRα in the regulation of PGC-1α-mediated transcription, thus after overexpression of PGC-1α, in the absence of PGC-1α co-recruitment. Importantly, we identified several parameters describing the genomic context of DNA response elements that differentiate between ERRα/PGC-1α coactivation and exclusive ERRα DNA binding. In particular, monomeric/dimeric DNA binding site configuration for ERRα, GC and CpG content of the binding region and co-recruitment of the specificity protein 1 (SP1) predict the interaction between PGC-1α and ERRα. Collectively, these findings not only significantly expand our insights into the regulation of the PGC-1α-controlled transcriptional network involved in muscle cell plasticity, but at the same time provide distinctive molecular links between genomic elements and TF – coregulator interactions.
Materials and Methods
Cell culture, shRNA knockdown of ERRα and RNA isolation
C2C12 cell culture, shRNA-mediated knockdown and RNA isolation were performed as described (4). The adenoviral vectors for the modulation of ERRα were a generous gift from Prof. A. Kralli from the Scripps Research Institute in La Jolla, California, USA. For the ERRα knockdown gene expression arrays, the RNAs from the following three conditions were used: AV-shGFP + AV-GFP + vehicle (0.02% DMSO); AV-shGFP + AV-flag-PGC-1α + vehicle (0.02% DMSO); AV-shERRα + AV-flag-PGC-1α + 2µM XCT-790. Briefly, myoblasts were differentiated into myotubes for 4 days, infected with adenoviral constructs and treated with XCT-790 for 2 additional days with daily medium change before harvesting. XCT-790 was used in the experiment to inhibit residual ERRα activity since the AV-shERRα knockdown alone was incomplete (at approx. 20% control levels, data not shown). Since modulation of PGC-1α and ERRα could potentially affect the myogenic program, the degree of differentiation of the cells was visually assessed before each experiment. Affymetrix Mouse Genome 430 2.0 arrays were used for the gene expression analysis.
ChIP and ChIP-Seq
The ERRα ChIPseq was done in cells overexpressing PGC-1α using the exact same conditions and methodology as described for the PGC-1α ChIPseq experiments (4) and the ChIP-Seq data for PGC-1α was used from previous work (4) to assess DNA binding of ERRα in the context of PGC-1α-regulated gene expression. For the immunoprecipitation of ERRα, magnetic beads (Dynabeads Protein G, Invitrogen) were coated with the monoclonal anti-ERRα antibody (ERRα Rabbit Monoclonal Antibody, Clone ID: EPR46Y, Epitomics). For the ChIP of SP1, the magnetic beads were coated with the polyclonal anti-SP1 antibody (ChIPAb+ Sp1 Rabbit Polyclonal Antibody, #17-601, Millipore).
High-throughput sequencing, read mapping and peak calling
The ERRα ChIP-Seq experiment in C2C12 cells undergoing PGC-1α over-expression was performed at the joint Quantitative Genomics core facility of the University of Basel and the Department of Biosystems Science and Engineering (D-BSSE) of the ETH Zurich in Basel on a Illumina HiSeq2000 sequencer as described (4).
The sequenced reads underwent a quality filter which retained all reads having Phred score >= 20, read length >= 25 bps and ambiguous nucleotides (Ns) per read <= 2. The reads that passed the filter were used as input for Bowtie version 0.12.7 (15) and aligned to the UCSC mm9 mouse genome assembly. Moreover, to avoid PCR amplification error, which might have arisen during sample preparation, we removed redundant reads mapping to the same location with the same orientation and we kept at most one read per position. Consequently, we obtained 2’155’507 covered positions for the IP and 84’175’472 covered positions for the WCE. Peak calling was performed as described using sliding windows (4). For the ERRα ChIP-Seq experiment, all consecutive windows having a Z-score greater than 3.5 were merged and the top scoring one from each window cluster was considered as the peak summit and used for further analyses.
TF binding site over-representation and principal component analysis
Analysis of TF binding site over-representation and principal component analysis was done as described (4). Briefly, TF binding site occurrence was compared to a randomized background set of regions and overrepresentations of TF binding sites calculated based on occurrence in peaks vs. that in the shuffled, randomized background. The principal component analysis (PCA) was based on an input matrix N containing the total number of predicted TF binding sites in each of the peaks for the 190 mammalian regulatory motifs that were defined.
Gene expression array analysis and gene ontology
Microarray probes were associated to a comprehensive collection of mouse promoters that was downloaded from the SwissRegulon database (16) as described (4). For each promoter, the log2 fold change (log2FC) was compared between the following conditions: over-expressed PGC-1α (treatment) and GFP (control); ERRα knockdown with the addition of XCT-790 (treatment) and over-expressed PGC-1α (control). The significance of the expression change was assessed by a Z score, which was computed as:
where n = 3 was the number of replicate samples, Ētreatment is the mean log2 expression across the treatment samples, Ēcontrol is the mean log2 expression across the control samples, and σ2treatment and σ2control are the variances of log2 expression levels across the replicates for the treatment and control samples, respectively. A log2FC threshold of ±0.585 (corresponding, in a more commonly used notation, to 1.5 fold change) and a Z score cutoff of ±3 were used to identify significantly up/down-regulated promoters. The criterion used to associate peaks and genes was proximity. For each gene with one or more differentially regulated promoters, we checked whether there was a peak located within 10 kb from any of the gene’s associated promoters. Gene ontology analysis was performed as described using a false discovery rate (FDR)-adjusted p-value <= 0.05 for enrichment.
Motif activity response analysis
An extended version of Motif Activity Response Analysis (ISMARA, (17)) to separately model the direct and indirect regulatory effects that ERRα and PGC-1α was performed as described (4) using the following linear model with eps denoting the log-expression of promoter p, i.e. the total log-expression of transcripts expressed from that promoter, and Npm denoting the total number of predicted TFBSs for regulatory motif m in the proximal promoter p (running from -500 to +500 relative to the transcription start site, or TSS):
In this model, cp describes the basal expression of promoter p, a sample-dependent normalization constant, and Ams is the regulatory activity of motif m in sample s, which is inferred by the model. To extend this model to now incorporate PGC-1α and ERRα binding data, we recognized that the motif activities Ams of a given regulatory motif m may be modulated by the nearby binding of PGC-1α and/or ERRα. We thus distinguished the effect Ams of a regulatory site for motif m that occurs outside of the binding of PGC-1α/ ERRα from the effect of the motif when it occurs within a binding peak of either PGC-1α or ERRα. To model our gene expression data, we applied the standard MARA model above to promoters that lacked an associated PGC-1α binding peak. The gene expression changes observed at these promoters upon knockdown of ERRα and/or over-expression of PGC-1α indicate indirect regulatory effects of ERRα, PGC-1α or both of them on the activities Ams. In contrast, for each “direct target” promoter p that has an associated binding peak (which could be an ERRα, a PGC-1α or an overlapping ERRα/PGC-1α peak) within 10 kb, we modelled its expression in terms of the predicted TFBSs in the binding peak, i.e.:
where is the number of predicted TFBSs for motif m in the peak associated with promoter p, and is the motif activity of regulator m in sample s when this motif occurs in the context of either ERRα binding, PGC-1α recruitment or both. Besides motif activities ISMARA also calculates error-bars δms for each motif m in each sample s. Using these, ISMARA calculates, for each motif m, an overall significance measure for the variation in motif activities across the samples analogous to a z-statistic:
For each motif we calculate a z-score Zm associated with its indirect activity changes, a z-score associated with its direct activity changes in the context of ERRα binding, a z-score associated with its direct activity changes in the context of PGC-1α recruitment and a z-score associated with its direct activity changes in the context of both ERRα binding and PGC-1α recruitment.
Quantitative real-time PCR and statistical analysis
Semi-quantitative real-time PCR (qPCR) was used to validate the efficiency of the ERRα knockdown in regard to gene expression and to verify that the ChIP of ERRα and the ChIP of SP1 were successful. The sequences of all primers used for qPCR are listed in Suppl. Table 1. Previously described ERRα response elements in PGC-1α target gene promoters and known ERRα/PGC-1α target genes were used as positive controls for the validation of the ChIP and gene expression, respectively. Regarding the statistical analysis of qPCR data sets, the values are presented as the mean ±SEM. Student’s t-tests were performed and a p-value < 0.05 was considered as significant. *<0.05, **<0.01, ***<0.001.
Animal experiments
Mice were housed in a conventional facility with a 12-h night/12-h day cycle with free access to chow diet pellet and water. For the experiments, male, 10- to 13-week-old skeletal muscle-specific PGC-1α knockout (MKO) mice (18) and PGC-1α muscle-specific transgenic (Tg) animals (5) were used. All experiments were performed according to the criteria outlined for the care and use of laboratory animals and with approval of the veterinary office of the Basel canton and the Swiss authorities. Injections were performed under sevoflurane (Provet, QN01AB08) anesthesia. Mice were injected intramuscularly (i.m.) with either PBS + DMSO vehicle (30μl/TA) or Mithramycin A (Cayman Chemical, 11434) (1μg/TA) dissolved in DMSO in both TA muscles. Mice were sacrificed 6 hours post-injection and the TAs isolated for further analysis.
Results
ERRα is recruited to DNA together with and independently of PGC-1α
Following up on several previous publications that implied a strong, direct co-dependence of ERRα and PGC-1α in the control of PGC-1α-regulated metabolic gene expression (10,12), we previously performed an unbiased, genome-wide analysis of PGC-1α recruitment to the mouse genome (4). The results of this study suggested a role for ERRα in controlling PGC-1α target gene expression in the absence of co-activation by PGC-1α (4). To verify these predictions and to identify all regions that are bound by this TF genome-wide in skeletal muscle cells after overexpression of PGC-1α, we performed a chromatin immunoprecipitation (ChIP) experiment followed by high-throughput sequencing (ChIP-Seq) of endogenous ERRα in differentiated C2C12 murine myotubes that overexpressed epitope-tagged PGC-1α. Thus, importantly, our experiments were not designed to map ERRα recruitment per se, but specifically the involvement of ERRα in the regulation of PGC-1α muscle target genes in the exact same cellular context as the previous mapping of PGC-1α recruitment (4). We then compared the identified ERRα binding sites with this set of PGC-1α recruitment regions that we identified previously (4). In order to identify all genomic locations significantly enriched in ERRα binding, we passed a sliding window along the genome and compared the local IP read density with the background read density from whole cell extract (WCE) for each consecutive window and quantified the significance of the enrichment by Z score. All regions with a Z score bigger than 3.5 were merged into a final total of 3225 peaks, which included binding regions in the vicinity of known ERRα target genes (Suppl. Fig. S1A), like the isocitrate dehydrogenase 3 [NAD+] alpha (Idh3a) and the pyruvate dehydrogenase lipoamide kinase isozyme 4 (Pdk4) (19). The enrichment of IP fragments from the ChIP-Seq experiment was validated for some of these ERRα target genes by quantitative real-time PCR (Suppl. Fig. S1B).
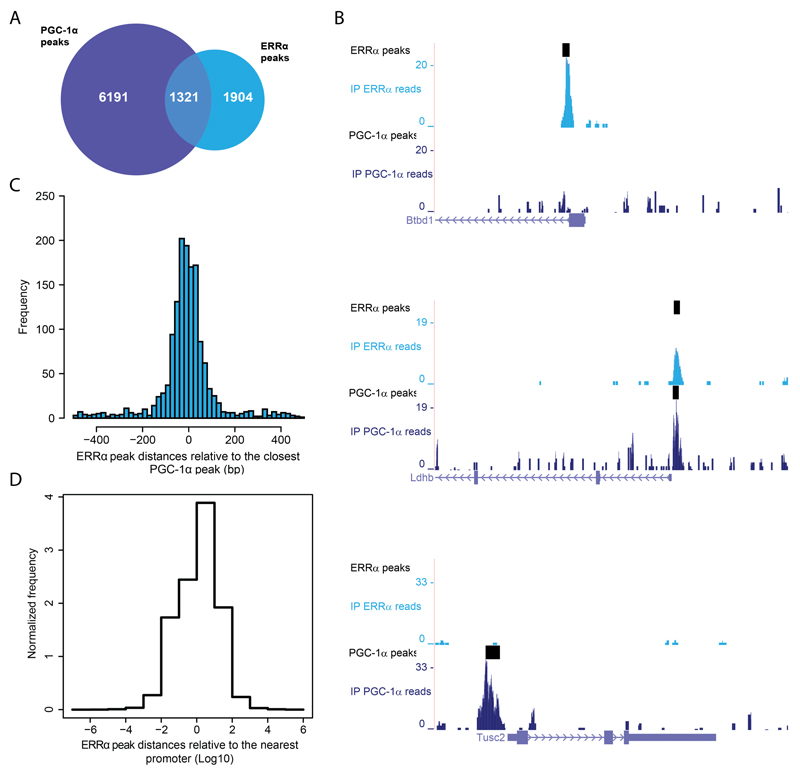
When we compared the genome-wide ERRα binding and PGC-1α DNA recruitment (Fig. 1A), we noticed that the majority of ERRα peaks (∼60%) are not overlapping a PGC-1α peak, suggesting that the so-far believed concept of symbiotic cooperation between these two proteins is in fact restricted to only a subset of their identified targets (~40% for ERRα and ~18% for PGC-1α), at least at the specific time point of analysis chosen in our experiments. It obviously is possible that the overlap between the two sets of peaks differs in a temporal manner. Moreover, the number of the PGC-1α peaks that overlap ERRα binding sites (~18%) could in part be due to the high overexpression of PGC-1α. Finally, the two ChIPseq experiments most likely differ in terms of specificity and efficacy of the antibody-antigen interaction and thus, interpretation of negative data could be hampered in the analysis. Nevertheless, the small overlap between the ERRα and PGC-1α peaks was not necessarily expected based on the literature. Some examples of the differential regulation are depicted in Fig. 1B. Of the 1321 ERRα peaks overlapping a PGC-1α site (that is, sharing at least one base pair), the vast majority of them is well centered on the closest PGC-1α peak at a distance of a couple of dozen base pairs (Fig. 1C), which could be interpreted as direct co-activation of ERRα by PGC-1α in most cases of ERRα/PGC-1α peak overlap. Notably, a larger fraction of ERRα peaks (approx. 12%) resides within 100 bp from a mouse promoter region (Fig. 1D), compared to the PGC-1α peaks (approx. 2%), which we previously found to be more distally located (4).
Figure 1. ERRα and PGC-1α are recruited to both shared and distinct sets of DNA elements and target genes.
(A) Venn diagram depicting the number of ChIP-Seq binding peaks for PGC-1α (blue) and for ERRα (cyan). (B) PGC-1α and ERRα read densities around the TSS of the genes Btbd1 (only ERRα peak), Ldhb (overlapping ERRα/PGC-1α peaks) and Tusc2 (only PGC-1α peak) obtained from the UCSC Genome Browser. (C) Distribution of ERRα peaks relative to their closest PGC-1α peaks. (D) Distribution of all ERRα peaks from the nearest mouse promoter region.
ERRα function is required for the regulation of many PGC-1α target genes
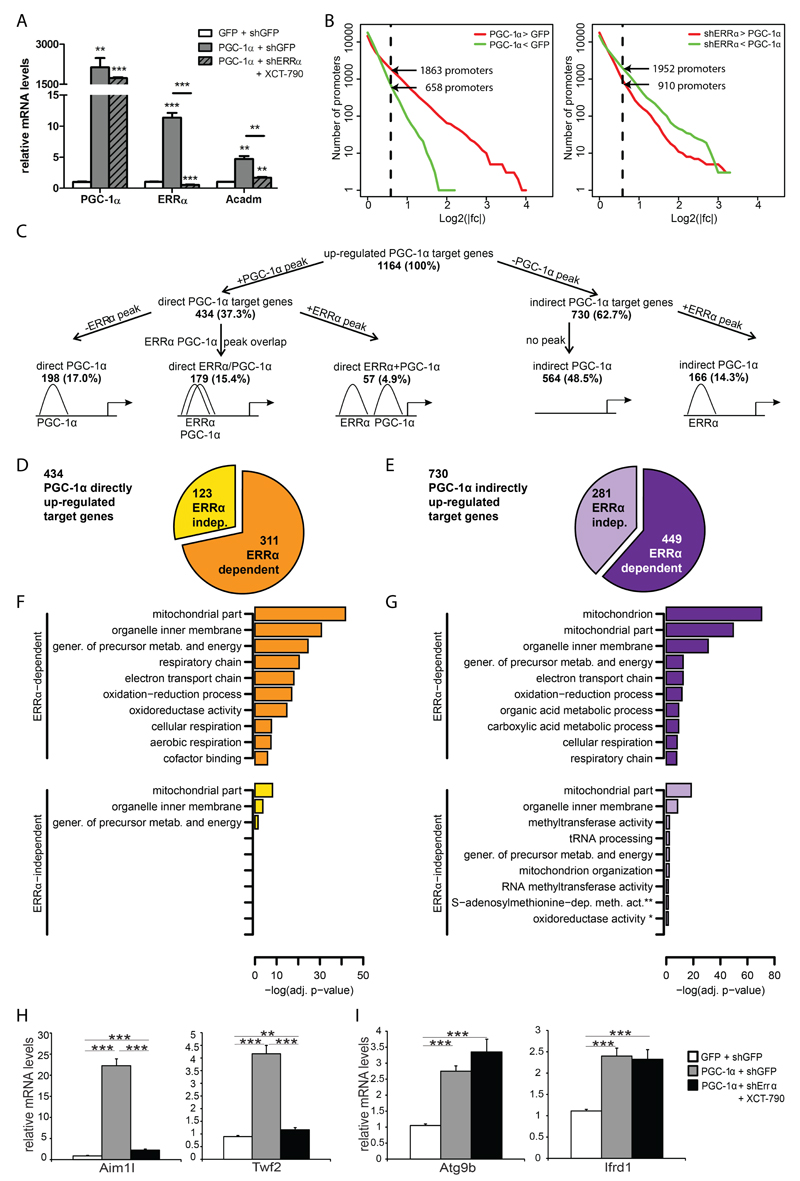
Based on DNA binding data alone, we cannot estimate how many of the non-PGC-1α overlapping ERRα peaks are non-functional. Therefore, to integrate the results obtained from the ChIP-Seq experiment with functional data in terms of PGC-1α-dependent gene expression, we further analyzed the impact of ERRα on gene expression changes downstream of PGC-1α in differentiated muscle cells using the following conditions: (i) shGFP-transfected control cells expressing small hairpin RNA (shRNA) targeted at green fluorescent protein (GFP); (ii) shGFP-transfected cells expressing PGC-1α; (iii) shERRα-transfected cells expressing PGC-1α in addition to shRNA against ERRα combined with the ERRα inverse agonist XCT-790 (12) to completely abolish ERRα activity. By comparing conditions (i) and (ii), we are able to identify gene expression changes downstream of PGC-1α induction, while comparing conditions (ii) with (iii) allows us to quantify the impact of ERRα on PGC-1α-mediated gene expression: for example, we observed a strong reduction of PGC-1α-controlled induction of Acadm, a known ERRα/PGC-1α target gene, in cells with abolished ERRα activity (Fig. 2A). After mapping the microarray probes to known transcripts and, through these, to a reference set of mouse promoters (16), we noticed that more promoters were significantly up-regulated (1863, corresponding to 1164 genes) than down-regulated (658, corresponding to 468 genes) following PGC-1α overexpression; in contrast, we observed the opposite effect in the ERRα knockdown cells: 910 promoters (corresponding to 597 genes) were significantly induced whereas 1952 promoters (corresponding to 1203 genes) were repressed, demonstrating a strong role for ERRα in PGC-1α-mediated up-regulation of gene expression (Fig. 2B). Then, a region of +/- 10kb distance from each promoter was chosen to assign peaks to promoters and hence divide target genes into direct (harboring at least on peak within this region) vs. indirect (without a peak within this region) genes, which obviously underestimates more long-range regulatory interactions. This stratification of the positively regulated PGC-1α target genes in terms of presence and absence of PGC-1α and ERRα peaks revealed several interesting findings: first, of the up-regulated PGC-1α target genes with a PGC-1α peak within 10 kb from any of their associated promoters, which constitute roughly 40% of all up-regulated PGC-1α targets, the number of genes with an overlap of ERRα and PGC-1α peaks (179 peaks, 15.4% of all up-regulated target genes) is lower than that of genes with only a PGC-1α peak (198 genes with only a PGC-1α peak and 57 genes that harbor distinct ERRα and PGC-1α peaks, thus combined representing 255 or 22% of all up-regulated genes) (Fig. 2C). Importantly, ERRα recruitment is observed in a significant number of indirectly up-regulated PGC-1α target genes (166 genes, corresponding to 22.7% of all indirect PGC-1α targets). These data suggest that, based on DNA binding, ERRα indeed plays a substantial role in PGC-1α target gene regulation, both when co-activated by PGC-1α, but equally significant when binding in the absence of this co-activator. Notably, the PGC-1α-mediated down-regulation of gene expression is almost exclusively indirect (439 out of 468 down-regulated PGC-1α target genes, corresponding to 93.8%), and the DNA binding of ERRα seems to likewise play a minor role in this process with ERRα peaks occurring in only 17 of down-regulated genes (3.6%) (Suppl. Fig. S2A). Of note, 62% of the 1321 overlapping PGC-1α/ERRα peaks (Fig. 1A) where not associated to any gene within a distance of +/-10 kb of the TSS while 25% of these peaks were linked to non-changing genes.
Figure 2. PGC-1α directly up-regulates both in an ERRα-dependent and -independent manner.
(A) qPCR analysis of PGC-1α, ERRα and Acadm mRNA levels in response to PGC-1α over-expression (OV) and shERRα knockdown (KD) + XCT-790. Data are normalized to mRNA levels in GFP infected cells. Error bars represent ± SEM. *p < 0.05; **p < 0.01; ***p < 0.001. (B) Reverse cumulative distribution of log2 fold changes for all mouse promoters in the PGC-1α OV condition versus GFP control (left panel) and in the PGC-1α OV + shERRα KD + XCT-790 versus PGC-1α OV (right panel). Promoters are colored in red (up-regulation) when their fold change is bigger than 1.5 and in green (down-regulation) when their fold change is smaller than -1.5 (obtained by taking the inverse of the linear binding ratio). (C) Tree diagram of all PGC-1α up-regulated target genes, distinguished in different subgroups according to peak presence/absence. (D) Pie-chart representing the classification of directly up-regulated PGC-1α target genes in ERRα-dependent (orange) and ERRα-independent (yellow) targets. (E) Pie-chart representing the classification of indirectly up-regulated PGC-1α target genes in ERRα-dependent (violet) and ERRα-independent (lilac) targets. (F-G) Subset of the top significantly enriched GO terms identified for ERRα-dependent and ERRα-independent PGC-1α directly (F) or indirectly (G) induced target genes. Abbreviations: gener., generation; metab. metabolites; * oxidoreductase activity, acting on a sulfur group, disulfide as acceptor; ** S-adenosylmethionine-dependent methyltransferase activity. (H-I) qPCR analysis of two ERRα-dependent (H) or ERRα-independent (I) PGC-1α target genes, in response to PGC-1α OV and shERRα KD + XCT-790. Data are normalized to mRNA levels in GFP infected cells. Error bars represent ± SEM. *p < 0.05; **p < 0.01; ***p < 0.001.
DNA recruitment of TFs or co-regulators typically only partially correlates with transcriptional changes, e.g. as indicated by a large number of PGC-1α peaks that were not assigned to regulated genes (4). Inversely, gene regulation can be brought about in an indirect manner and, therefore, might not require a peak adjacent to the gene promoter region, as seen for 48.5% of up-regulated PGC-1α target genes without a PGC-1α or ERRα peak, respectively (Fig. 2C). We classified genes that exhibit up-regulation in response to PGC-1α induction into four categories based on whether they were associated with a PGC-1α binding peak, i.e. direct versus indirect PGC-1α targets, and whether the up-regulation was dependent on ERRα. According to this classification, approximately two thirds of the up-regulated PGC-1α-controlled genes were dependent on the presence of functional ERRα protein, irrespective of whether they were direct or indirect targets of PGC-1α (Figs 2D and 2E).
We next investigated whether the different classes of PGC-1α targets were over-represented for genes from different functional categories. As expected, most of the enriched categories for ERRα-dependent up-regulated target genes were related to mitochondria and oxidative energy metabolism (Figs. 2F and 2G). Notably, as we observed previously (4), the same functional categories show enrichment regardless of direct or indirect PGC-1α involvement. Moreover, similar gene ontology terms were found when using the ERRα-independent PGC-1α targets as input for FatiGO (Figs. 2F and 2G). The different categories of PGC-1α target genes were confirmed by qPCR showing two ERRα-dependent (Aim1l and Twf2) and two ERRα-independent (Atg9b and Ifrd1) PGC-1α target genes (Figs. 2H and 2I, respectively).
Finally, we also checked dependency of transcriptional regulation on functional ERRα for PGC-1α down-regulated targets. Peak-gene association clearly indicates that the majority of genes whose transcription is repressed by PGC-1α lack peaks for either PGC-1α or ERRα within 10 kb of the gene promoters (approx. 94% of all down-regulated PGC-1α target genes) (Suppl. Fig. S2A). Out of these 439 indirectly down-regulated genes, about 23% (101 down-regulated, indirect PGC-1α targets) were dependent on ERRα meaning that PGC-1α-mediated repression was significantly alleviated by ERRα knockdown (Suppl. Fig. S2B). Thus, ERRα markedly contributes to boost an indirect inhibitory mechanism that is involved in PGC-1α-controlled transcriptional repression. Nevertheless, however, the majority of PGC-1α-mediated inhibition of gene expression is ERRα-independent and thus using alternative mediators, such as, for example, the indirect inhibition of the nuclear factor κB (NFκB) (20).
Coactivation specificity of monomeric vs. dimeric ERRα binding elements
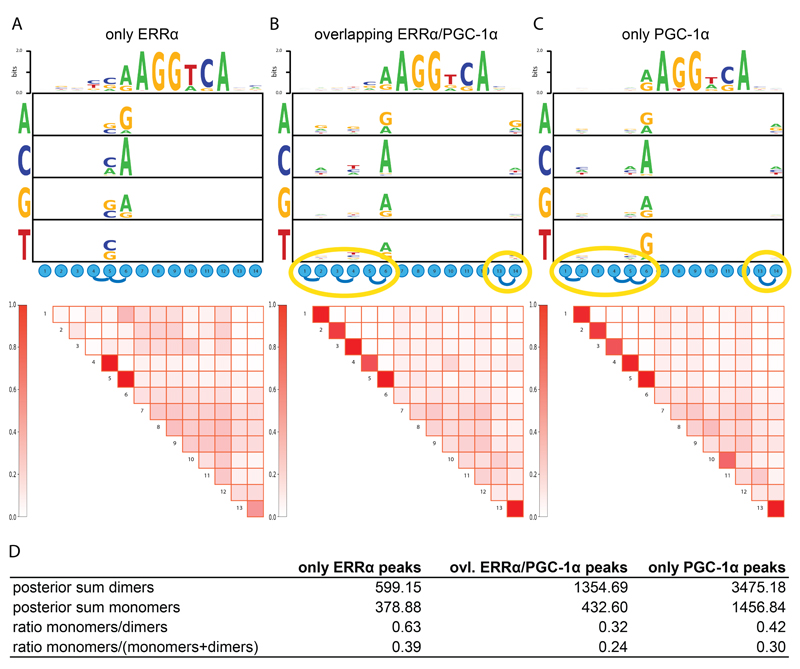
In light of the postulated intimate relationship between ERRα and PGC-1α, our data depicting a high degree of independence of these two proteins in the regulation of PGC-1α target genes in muscle cells are quite surprising. In particular, it is unclear by what molecular mechanisms PGC-1α is recruited to ERRα binding sites at some genomic loci, but not to others. ERRα can bind to a nine nucleotide-long element with the consensus sequence TNAAGGTCA called an estrogen-related receptor response element (ERRE) (21). In addition, binding of ERRα to repeats of ERREs and potentially other response elements has also been proposed (22). In both cases, ERRα has been proposed to bind as homo- or heterodimer, even to single ERREs (23). Importantly, data based on in vitro experiments implied that the base at the N position of the ERRE controls co-activation by PGC-1α with a preference for PGC-1α to interact with ERRα on ERREs with a T at the N position (TTAAGGTCA) whereas a C (TCAAGGTCA) favors reduced co-activation by PGC-1α (22). Since these in vitro studies were severely limited in terms of scope, we now investigated whether similar sequence variations can be detected in a genome-wide analysis of ERRα DNA binding elements identified by ChIP-Seq. We therefore split the ERRα and PGC-1α peaks into three distinct groups: “only ERRα”, “overlapping ERRα/PGC-1α” and “only PGC-1α” peak regions and computationally derived separate binding motifs for each set of regions. Instead of inferring standard position-specific weight matrix motifs, we employed a novel approach, recently developed in our group (Omidi, van Nimwegen et al., personal communication), which extends position-specific weight matrix models to so-called dinucleotide weight tensors, which allow arbitrary dependencies between the positions within the binding sites.
First, both the “only ERRα” and the “overlapping ERRα/PGC-1α” peak-associated motifs exhibited a more determined 5’ extension of the hexamer half-site as expected for an ERRE compared to the “only PGC-1α” peak regions (Fig. 3A-C). Intriguingly, the “only ERRα” motif harbors a stronger preference for C at position 5 when compared to the “overlapping ERRα/PGC-1α” peaks, even though the preference for this nucleotide is relatively small. However, even more strikingly, we noticed that although there are internal dependencies between the nucleotides at positions 4, 5, and 6 in every peak group, the dependencies between the initial and final positions (1-2 and 13-14) of the motif are only observed for “overlapping ERRα/PGC-1α” and “only PGC-1α” peaks, but not for “only ERRα” peaks (Fig. 3A-C).
Figure 3. In the absence of a direct coactivation by PGC-1α, ERRα prefers to bind to monomeric DNA elements.
(A-C) Motif logo showing the interdependencies between the different positions of the ESRRA weight matrix identified in “only ERRα”, “overlapping ERRα/PGC-1α” and “only PGC-1α”. Dependencies between positions are indicated by a blue curved line, while yellow ellipses highlight the dependencies which are in “overlapping ERRα/PGC-1α” and “only PGC-1α” peaks, but not in “only ERRα” peaks. (D) Table showing the posterior sum and the fraction of nuclear receptor hexamer half-site monomers and dimers across our three peak sets.
Dependencies at the ends of the motif could imply that the TF is more often binding DNA as a dimer at these sites, suggesting that ERRα binding site repeats may be more likely to recruit co-activation by PGC-1α than monomeric, extended half-sites. To test whether these motifs indeed differ in terms of hexamer repeat configuration, we next used the core recognition motif “AGGTCA” of the ESRRA weight matrix to identify nuclear receptor dimers in direct, everted or inverted configurations with a variable spacing between half-sites that ranged from 1 to 10 nucleotides around the core motif in the different peak groups. Remarkably, we found a striking difference in the relative occurrence of monomers and dimers of nuclear receptor hexamer half-sites between the “only ERRα” and the “overlapping ERRα/PGC-1α” peak sets (Fig. 3D). In the first group, the ratio of monomers to dimers was markedly higher compared to the “overlapping ERRα/PGC-1α” peaks (0.63 vs. 0.32), further supporting that dimeric ERRα binding sites are more likely to enable co-activation by PGC-1α. Furthermore, even when the number of monomers is normalized to the sum of monomers and dimers in each peak set, the “only ERRα” peaks showed the highest fraction of nuclear receptor monomers (39%) of the three groups (Fig. 3D). It should be noted, however, that despite these differences, the presence of a monomeric half-site in a ERRα peak is only a weak predictor of PGC-1α co-recruitment, as in both groups only a marginally higher proportion of “only ERRα” peaks contain a monomer compared to “overlapping ERRα/PGC-1α” (“only ERRα”: 440 out of 1904 peaks corresponding to 23.1%; “overlapping ERRα/PGC-1α”: 266 out of 1321 peaks in total corresponding to 20.1%). It is therefore very likely that the sequence specificity and the monomeric/dimeric configuration favor, but by themselves are not sufficient to entirely control co-activation of ERRα by PGC-1α.
ERRα binding regions without PGC-1α recruitment are enriched for SP1 binding
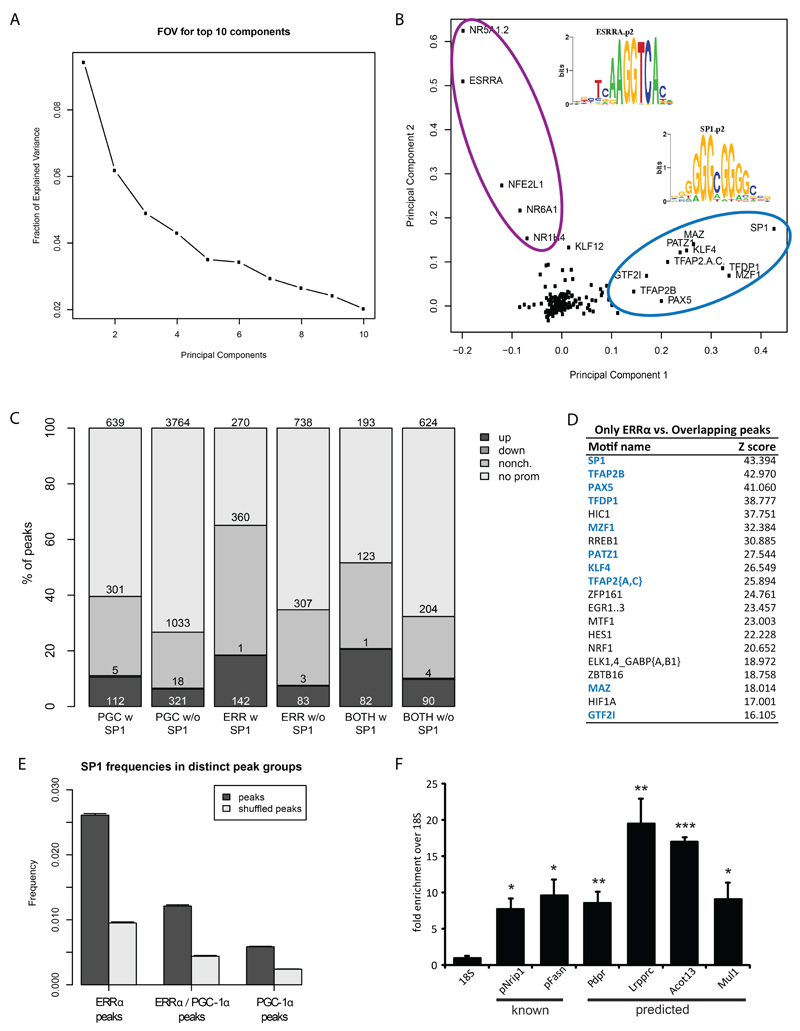
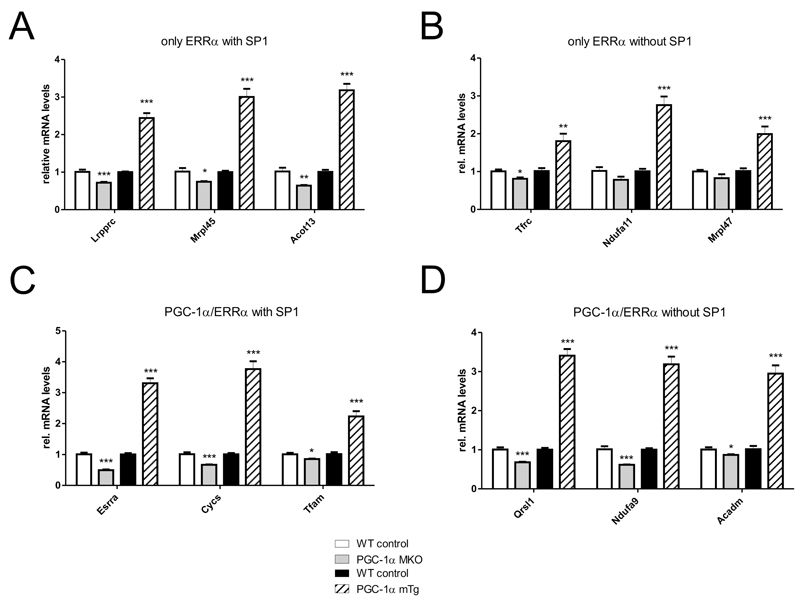
To identify additional predictors of the ERRα/PGC-1α interaction, we next analyzed the occurrence of TF DNA-binding motifs within all of the ERRα peaks. We used the software MotEvo to predict TF binding sites (TFBSs) for a set of 190 known mammalian regulatory motifs (24). In order to explain most of the binding site variation observed across the ERRα peaks, we then applied principal component analysis (PCA) to a site-count matrix N, whose elements Npm represent the number of predicted TFBSs for each motif m in each ERRα peak region p. Out of a total of 190, the first component was accounting for ~10% of the total variation in the dataset (Fig. 4A). The distribution of motif projections on the first two principal components clearly indicates two distinct clusters of motifs that are associated with variation along the first and second principal components (Fig. 4B). The first group includes ESRRA and other nuclear receptors which have binding motifs that are very similar to that of the ERRα motif. This cluster reflects the most abundant sites which can be found within the ERRα binding regions. Interestingly, besides these expected nuclear receptor motifs, the second group of motifs consists of GC-rich motifs which often are found in the proximity of transcriptional start sites. The motif with the highest score along the first principal component describes binding elements of SP1. The activity of this protein can be significantly affected by post-translational modifications, resulting in SP1 to either act as an activator or as a repressor (25). Moreover, a functional link between the occurrence of SP1 binding sites and ERRα activity, albeit without consideration of co-activation by PGC-1α, has been proposed previously (26). We thus subsequently investigated the activity of SP1 in the context of PGC-1α target gene regulation. The different classes of peaks (“only ERRα”, “only PGC-1α”, “overlapping ERRα/PGC-1α”) were therefore combined with the regulation of their assigned promoters (“up”, “down”, “non-changing”, “no promoter assigned”) as shown in Fig. 4C. Strikingly, whenever a site for SP1 is present within a peak, it is more likely for the assigned promoter to be up-regulated, strongly suggesting that in the context of PGC-1α over-expression, SP1 plays a role as an activator. This effect is particularly enhanced when SP1 is found in an ERRα peak compared to the PGC-1α peaks. Similarly, when analyzing TFBS predictions that differ between the “only ERRα” and the “overlapping ERRα/PGC-1α” groups, SP1 emerges as the top-scoring motif and thus strongly associates with “only ERRα” peaks (Fig. 4D). The specific enrichment of SP1 motifs in the “only ERRα” group was also confirmed by comparing the enrichment of predicted SP1 binding sites, relative to its occurrence in a set of randomized peak sequences, in “only ERRα” peaks with the enrichment in “overlapping ERRα/PGC-1α” and “only PGC-1α” peaks. Although SP1 sites are more frequent in all peak sets relative to randomized regions, the enrichment is by far strongest in “only ERRα” peaks (Fig. 4E). Next, we experimentally validated the presence of SP1 both at the promoters of the known target genes RIP140/Nrip1 and Fasn (27,28) and in ERRα peaks with an adjacent predicted SP1 binding site in the proximity of four distinct genes by ChIP (Fig. 4F). Finally, we studied the functional consequence of SP1 on muscle target gene expression of endogenous and overexpressed PGC-1α in gain- and loss-of-function animal models in vivo (Suppl. Figure S3). First, we validated a set of target genes belonging to all four binding categories (genes with only ERRα recruitment with SP1 motifs, only ERRα recruitment without SP1 binding sites, PGC-1α/ERRα overlapping peaks with SP1 motifs and PGC-1α/ERRα overlapping peaks without SP1 binding sites). As shown in Fig. 5, the expression of genes from all four categories was reduced in skeletal muscle-specific PGC-1α knockout (MKO) animals and elevated in skeletal muscle-specific PGC-1α transgenic (mTg) mice. Thus, at least these genes are not only regulated by overexpressed PGC-1α in cultured muscle cells, but also by endogenous and overexpressed PGC-1α in mouse muscle in vivo. Subsequently, we aimed at testing the functional involvement of SP1 in the predicted subcategories of PGC-1α target genes using the specific pharmacological SP1 inhibitor Mithramycin A (MitA) (29). First, efficacy of SP1 inhibition was demonstrated by the reduction of the known SP1 target genes Sp1 and Vegfa (Suppl. Fig. S4). Surprisingly however, MitA not only reduced the ability of PGC-1α to induce target genes that harbor an SP1 motif, but also those without a predicted SP1 binding site (Suppl. Fig. S4). Most likely, the expected selectivity of the functional involvement of SP1 is lost due to an inhibition of endogenous and transgenic PGC-1α expression by MitA (Suppl. Fig. S4). Similarly, siRNA-mediated knockdown of SP1 in cultured muscle cells likewise reduced the expression of PGC-1α (data not shown). Indeed, putative SP1 binding sites were found both in the proximal as well as in the distal/alternative promoter regions of PGC-1α (Suppl. Fig. S4). Thus, even though we found a significant functional involvement of SP1 in the regulation of PGC-1α target gene expression in mouse muscle in vivo, we were unable to validate our prediction based on the presence of SP1 motifs in a subset of these genes, most likely due to the observation of PGC-1α itself being an SP1 target.
Figure 4. SP1 is the top transcription factor partner for ERRα in skeletal muscle.
(A) Fraction of explained variance of the top 10 PCA components. (B) PCA analysis of the 3225 ERRα peaks. The names of the motifs with the largest projections on the first two principal components are indicated. Purple and light blue ellipses highlight motif clusters, as identified by PC1, of nuclear hormone receptor-like motifs and SP1-like motifs, respectively. (C) Bar chart representing the different classes of peaks (“only ERRα”, “only PGC1α”, “overlapping ERRα and PGC1α”) together with the regulation of their associated promoters (“up”, “down”, “non-changing”, “no promoter assigned”). Numbers shown on top of each box represent the absolute peak counts. (D) Top scoring results of motif search obtained by comparing the TFBSs predictions within the “only ERRα peaks” with those in the “overlapping ERRα/PGC-1α”. The motifs corresponding to the SP-1 group in the PCA are colored in blue. (E) TFBSs posterior sum for SP1 in “only ERRα”, “overlapping ERRα/PGC-1α” and “only PGC-1α” peaks. For each dataset, TFBS occurrences were compared against binding site predictions performed on the corresponding background set of shuffled peaks. (F) qPCR validation of the ChIP enrichment measured at the promoter of a set of SP1 known target genes and around the predicted SP1 site within the ERRα peaks associated to the genes Pdpr, Lrpprc, Acot13 and Mul1. Bars represent fold enrichment over that of the 18S rRNA gene, error bars represent SEM. *p < 0.05; **p < 0.01; ***p < 0.001.
Figure 5. Target genes of all four binding categories are regulated by endogenous and overexpressed PGC-1α in mouse muscle in vivo.
(A-D) The expression of PGC-1α target genes with only ERRα DNA binding with (A) and without (B) adjacent SP1 motifs as well as of PGC-1α target genes with overlapping PGC-1α and ERRα peaks with (C) and without (D) SP1 binding sites was validated in skeletal muscle-specific PGC-1α knockout (MKO) and transgenic (mTG) mice compared to the respective wildtype littermate controls.
ERRα peaks without PGC-1α co-recruitment exhibit higher GC and CpG content
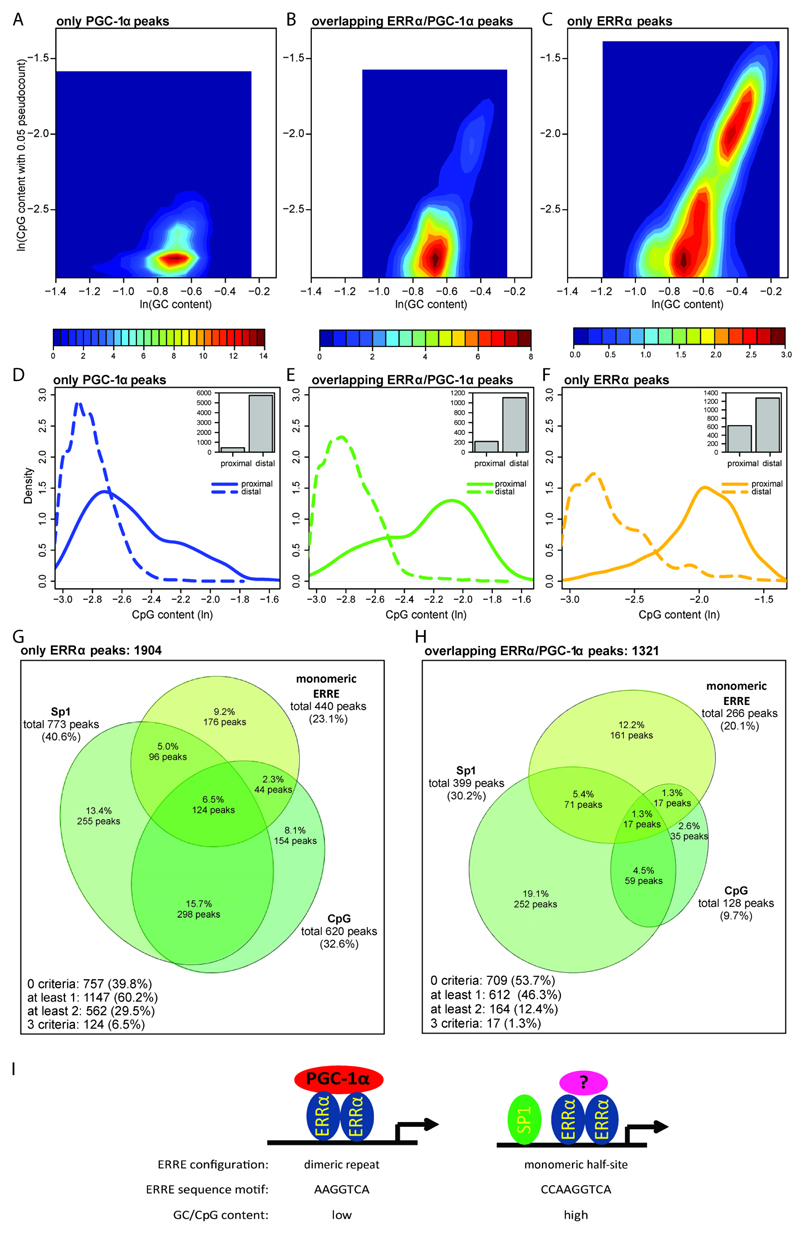
Intriguingly, the amount of predicted SP1 TFBSs (in terms of posterior sum) was much lower in PGC-1α randomized (shuffled) peaks compared to the ERRα shuffled peak dataset (Fig. 4E). Since SP1 is known to bind GC-rich regions, these results might reflect a different nucleotide composition between the peak sets. Accordingly, we analyzed the GC and CpG content of all ERRα and PGC-1α peaks. Interestingly, in contrast to the “overlapping ERRα/PGC-1α” peaks, and even more to the “only PGC-1α” peaks, the “only ERRα” peaks separated into two distinct populations, one with high and the second with lower GC content (Fig. 6A-C). Even more strikingly, these two populations in the “only ERRα” peak group also differed in the CpG content and therefore potential CpG islands. Subsequently, each peak set was further subdivided into proximal and distal binding regions, where “proximal” referred to peaks within 1kb from their associated gene promoter and “distal” to peaks located farther away. As clearly shown in Fig. 6D-F, the “only ERRα” peaks host more CpG dinucleotides with respect to “only PGC-1α” peaks; moreover, the fraction of “only ERRα” proximal peaks is much higher (~1/3) than the corresponding fraction of “only PGC-1α” peaks (~1/10). Importantly, while most of this difference stems from the CpG content in proximal peaks, even the more distal ERRα peak distribution curve exhibits shoulders towards higher CpG content that are completely missing in the PGC-1α peaks. These results suggest a preference for high GC and CpG content in ERRα DNA recruitment sites, whereas PGC-1α in the absence of ERRα is bound to response elements with a relatively lower GC and CpG content. Importantly, the “overlapping ERRα/PGC-1α” peaks behave in an intermediary manner (Fig. 6E).
Figure 6. “Only ERRα” peaks prefer to occur as ERRE monomers and to bind high CpG content regions.
(A-C) Two-dimensional histogram (shown as a heat map) of the GC base content (horizontal axis) and CpG dinucleotide content (vertical axis) of “only PGC-1α” (A), “overlapping ERRα/PGC-1α” (B) and “only ERRα” peaks (C). The values shown on both axes are expressed as logarithms. (D-F) Density plots of the CpG content of “only ERRα” (D), “overlapping ERRα/PGC-1α” (E) and “only PGC-1α” (F) peaks, located either proximally (≤ 1 kb) or distally (> 1 kb) from the closest promoter. Each inset shows the bar plot of the number of “proximal” and “distal” peaks. (G-H) Euler diagram of “only ERRα peaks” (G) and of “overlapping ERRα/PGC-1α” peaks (H). Peaks were subdivided according to three different criteria: presence of SP1 binding sites, presence of monomers and high CpG content (defined as GC content >= 50% and CpG content >= 65%). (I) Model of ERRα regulation of PGC-1α target genes in muscle cells. A combination of SP1 co-recruitment, monomeric vs. dimeric ERRα binding site configuration, nucleotide preference of the ERRE, and GC/CpG content affect co-activation of ERRα by PGC-1α in the regulation of PGC-1α target genes in muscle cells.
Strikingly, the combination of all three parameters, monomeric binding, high CpG content and presence of an SP1 binding site, synergize in discriminating between “only ERRα” and “overlapping ERRα/PGC-1α” peaks. For example, as depicted in Figure 6G-H, the percentage of peaks harboring at least two features are 2 fold more frequent in the “only ERRα” compared to the “overlapping ERRα/PGC-1α” group, while those with all three features are even 5 times more frequent. Notably, SP1 co-occurrence with high CpG content is particularly enriched in the “only ERRα” group with 15.7% of peaks, as opposed to only 4.5% in the “overlapping ERRα/PGC-1α” peak group. Similarly, the combination of all the three criteria accounts for 6.5% of “only ERRα” peaks, whereas they are found in only 1.3% of “overlapping ERRα/PGC-1α” peaks. Indeed, the CpG content, which is present in 32.6% of the “only ERRα” peaks (i.e. 3 fold higher than in the other dataset), is the feature which determines the biggest fraction of overlap among the three criteria that we focused on.
Discussion
Control of complex biological programs by co-regulator proteins has emerged as a regulatory paradigm in higher organisms in recent years. For example, the three members of the steroid receptor co-activator family SRC-1, -2 and -3 play a major role in modulating systems metabolism (30). Co-regulator control of biological programs exhibits several advantages over individual TFs (31,32): by binding to and modulating the activity of several different TFs, co-regulators usually have a broader repertoire in target gene transcriptional regulation (33). Second, the possibility of coordinating the regulation of genes within a specific transcriptional program provides kinetic advantages to accelerate the output of specific pathways beyond the possibilities of individual gene regulation (34). Furthermore, transcriptional regulation, transcript variants and a myriad of posttranslational modifications allow a combinatorial control of co-regulator stability and specificity and thereby enable dynamic control of complex cellular plasticity in a highly context-dependent manner (35). Many of these mechanistic principles are illustrated by the regulation and function of PGC-1α in the control of cellular energy homeostasis. However, mechanistic insights into the dynamic TF – co-regulator interactions remain rudimentary. Following our previous report predicting ERRα activity both in the presence and absence of direct PGC-1α coactivation based on motif representation (4), we now provide experimental and computational evidence for a contribution of the genomic context of DNA response elements to control the co-recruitment of PGC-1α and ERRα in the context of PGC-1α-controlled muscle gene expression. Our findings are particularly surprising since historically, ERRα has been thought to strongly rely on PGC-1α co-activation to regulate PGC-1α target gene expression (10,12). Interestingly, the DNA binding of PGC-1α and ERRα have been analyzed in a previous study by Charos and colleagues (36). Notably, several important differences compared to our experimental system exist: for example, Charos et al. analyzed human proteins in the human hepatoma cell line HepG2 and studied ERRα DNA binding in the absence of activated/elevated PGC-1α. Nevertheless, in both studies, a similar number of ERRα peaks were found (3786 by Charos compared to 3225 reported here), and even more importantly, the overlap between PGC-1α and ERRα peaks was likewise small: of the 3193 and 1741 multiple regulatory factor binding regions (multi-RFBRs) of ERRα and PGC-1α, respectively, only 535 were shared between these two factors (36).
Intriguingly, the decision between ERRα co-activation by PGC-1α and distinct DNA binding is to a certain extent determined by several aspects of the DNA composition of the enhancer and promoter regions (Fig. 6I). In particular, the ERRα binding element configuration as a monomeric half-site, adjacent recruitment of SP1 and a high CpG content appear to discourage co-recruitment of PGC-1α. Assuming that ERRα activity seems largely determined by coactivator action due to the small ligand-binding pocket observed in some crystallographic studies (13), the context of PGC-1α-regulated gene expression implies that separate ERRα DNA binding not only precludes association of PGC-1α, but instead favors co-activation by other co-regulators. Indeed, the transcriptional activity of hERR1, the human ortholog of the murine ERRα, is enhanced in a ligand-independent manner by the activator of thyroid and retinoic acid receptors (ACTR), the glucocorticoid receptor interacting protein 1 (GRIP1), and SRC-1 (37). Whether any of these co-activators are involved in ERRα-dependent muscle gene regulation by PGC-1α remains to be investigated. Intriguingly, such a shift in co-activator preference from PGC-1α towards binding of GRIP1 to the glucocorticoid receptor could be achieved by using pharmacological means (38). Furthermore, an inverse agonist was discovered to specifically reduce the interaction between PGC-1α and ERRα, but not other TF binding partners (12,39). However, future studies will have to aim at determining how the genomic context translates into conformational changes in a TF that then affects interaction with distinct co-regulators. Importantly, at least part of this genomic context might be amenable to dynamic regulation, for example by the overall availability or posttranslational control of the activity of SP1. Unfortunately, due to the potent effect of SP1 on PGC-1α transcription, we were unable to validate our predictions of increased presence of SP1 binding sites in ERRα only regulated PGC-1α target genes. Second, the cytosines in CpG sites are potential targets for DNA methylation and thereby mediate epigenetic regulation of gene expression (40). Even though our conclusions rely to a large extent on computational prediction and therefore, future experiments will have to further validate and expand these findings, it is intriguing to speculate that DNA methylation may not only generally repress transcription by limiting TF binding, but maybe in a more fine-tuned manner also modulate TF – co-regulator interactions. Moreover, based on the reports of epigenetic modifications in exercise, including DNA hypomethylation of the PGC-1α promoter itself (41), it thus will be interesting to study how exercise-induced epigenetic changes affect not only the expression, but also the DNA recruitment and TF coactivation pattern of this key regulator of endurance exercise adaptation in muscle.
Besides the more general implication of our results on the mechanistic aspects of genomic context, TF binding and co-regulator recruitment, a second highly surprising finding emerged from the data related to the function of ERRα and PGC-1α in muscle cells. Specifically, ERRα was described as the central partner for PGC-1α in the regulation of mitochondrial oxidative phosphorylation gene expression (10,12). Our results however now reveal a much more diverse manner by which PGC-1α regulates the expression of these and other, related metabolic pathways. Ontological analysis of the PGC-1α target genes devoid of an ERRα and PGC-1α peak demonstrate that other TFs also significantly contribute to the regulation of genes encoding enzymes in the same metabolic pathways. Importantly, in light of the close similarity of DNA binding elements and target gene activation, it is possible that some of the predicted ERREs could also be activated by ERRγ. Moreover, as implied by the prediction of TF binding motifs to be associated with PGC-1α-dependent transcriptional regulation, there might be a number of additional TFs that work with PGC-1α in controlling muscle cell plasticity, many of which have not been studied in the context of PGC-1α-mediated transcriptional control so far. Intriguingly, a certain degree of functional redundancy seems to exist: for example, inhibition of ERRα reduces the PGC-1α-induced expression of the vascular endothelial growth factor (VEGF) gene (7). Likewise however, siRNA-mediated knockdown of components of the AP-1 TF complex or of SP1 also decreases the ability of PGC-1α to increase VEGF gene expression (4). Thus, PGC-1α-controlled muscle cell plasticity might combine two mechanistic principles: on the one hand, a “regulon” to tightly coordinate the concurrent expression of genes that belong to a specific transcriptional program while on the other hand, providing a more distributed transcriptional network using a variety of different TFs, both directly as well as indirectly, to add regulatory robustness as well as flexibility to control the expression of these genes in different cellular contexts. ERRα most likely is the central factor for PGC-1α to control a bioenergetic regulon using several modulators including SP1 and potentially others such as Prox1 (42) to affect ERRα-PGC-1α interactions. Inversely, AP-1 and other TFs could complement the action of ERRα, for example by triggering muscle vascularization in different contexts such as local tissue hypoxia for AP-1 as opposed to altered metabolic demand for ERRα (4).
In conclusion, we elucidated to what extent the nuclear receptor ERRα contributes to PGC-1α target gene expression in a muscle cell line. Even though our experiments were restricted to the analysis of endogenous ERRα in the context of overexpressed PGC-1α in cultured muscle cells, several interesting mechanistic findings emerged. Intriguingly, despite a relatively low overlap in DNA binding, ERRα is crucial for the regulation of a majority of PGC-1α target genes in a muscle cell line. Moreover, the genome-wide DNA binding patterns of ERRα and PGC-1α demonstrated that coactivation of this TF by PGC-1α depends on different aspects of the genomic context of the DNA response element. Importantly however, the postulated criteria do not provide a binary distinction between co-activation and non-coactivation. Parameters with a higher predictive power might be identified in a temporal analysis of PGC-1α and ERRα DNA recruitment to PGC-1α target genes in muscle cells. Nevertheless, these findings not only provide important mechanistic insights into the regulation of complex biological programs by co-regulator proteins, but could also help to specifically modulate such networks in order to selectively address dysregulation of genes in pathological settings. In the future, studies on endogenous proteins in murine and human contexts in vivo will help to further unravel the complex mechanisms of co-activator-controlled transcriptional networks.
Supplementary Material
Acknowledgments
We would like to thank Dr. A. Kralli for the generous gift of control and shERRα adenoviral vectors. This project was funded by the ERC Consolidator grant 616830-MUSCLE_NET, the Swiss National Science Foundation, SystemsX.ch, the Swiss Society for Research on Muscle Diseases (SSEM), the Neuromuscular Research Association Basel (NeRAB), the Gebert-Rüf Foundation “Rare Diseases” Program, the “Novartis Stiftung für medizinisch-biologische Forschung”, the University of Basel, and the Biozentrum.
Footnotes
Data access
The Gene Expression Omnibus (GEO) SuperSeries accession number for the ChIP-Seq and gene expression array data reported in this paper is GSE80522.
Disclosure summary: The authors have nothing to disclose.
References
- 1.Hoppeler H, Baum O, Lurman G, Mueller M. Molecular mechanisms of muscle plasticity with exercise. Compr Physiol. 2011;1(3):1383–1412. doi: 10.1002/cphy.c100042. [DOI] [PubMed] [Google Scholar]
- 2.Blaauw B, Schiaffino S, Reggiani C. Mechanisms modulating skeletal muscle phenotype. Compr Physiol. 2013;3(4):1645–1687. doi: 10.1002/cphy.c130009. [DOI] [PubMed] [Google Scholar]
- 3.Pérez-Schindler J, Handschin C. New insights in the regulation of skeletal muscle PGC-1alpha by exercise and metabolic diseases. Drug Discov Today Dis Models. 2013;10(2):e79–85. [Google Scholar]
- 4.Baresic M, Salatino S, Kupr B, van Nimwegen E, Handschin C. Transcriptional network analysis in muscle reveals AP-1 as a partner of PGC-1alpha in the regulation of the hypoxic gene program. Mol Cell Biol. 2014;34(16):2996–3012. doi: 10.1128/MCB.01710-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lin J, Wu H, Tarr PT, Zhang CY, Wu Z, Boss O, Michael LF, Puigserver P, Isotani E, Olson EN, Lowell BB, et al. Transcriptional co-activator PGC-1 alpha drives the formation of slow-twitch muscle fibres. Nature. 2002;418(6899):797–801. doi: 10.1038/nature00904. [DOI] [PubMed] [Google Scholar]
- 6.Handschin C, Chin S, Li P, Liu F, Maratos-Flier E, Lebrasseur NK, Yan Z, Spiegelman BM. Skeletal muscle fiber-type switching, exercise intolerance, and myopathy in PGC-1alpha muscle-specific knock-out animals. J Biol Chem. 2007;282(41):30014–30021. doi: 10.1074/jbc.M704817200. [DOI] [PubMed] [Google Scholar]
- 7.Arany Z, Foo SY, Ma Y, Ruas JL, Bommi-Reddy A, Girnun G, Cooper M, Laznik D, Chinsomboon J, Rangwala SM, Baek KH, et al. HIF-independent regulation of VEGF and angiogenesis by the transcriptional coactivator PGC-1alpha. Nature. 2008;451(7181):1008–1012. doi: 10.1038/nature06613. [DOI] [PubMed] [Google Scholar]
- 8.Arnold AS, Gill J, Christe M, Ruiz R, McGuirk S, St-Pierre J, Tabares L, Handschin C. Morphological and functional remodelling of the neuromuscular junction by skeletal muscle PGC-1alpha. Nat Commun. 2014;5:3569. doi: 10.1038/ncomms4569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Eichner LJ, Giguere V. Estrogen related receptors (ERRs): a new dawn in transcriptional control of mitochondrial gene networks. Mitochondrion. 2011;11(4):544–552. doi: 10.1016/j.mito.2011.03.121. [DOI] [PubMed] [Google Scholar]
- 10.Schreiber SN, Emter R, Hock MB, Knutti D, Cardenas J, Podvinec M, Oakeley EJ, Kralli A. The estrogen-related receptor alpha (ERRalpha) functions in PPARgamma coactivator 1alpha (PGC-1alpha)-induced mitochondrial biogenesis. Proc Natl Acad Sci U S A. 2004;101(17):6472–6477. doi: 10.1073/pnas.0308686101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huss JM, Kopp RP, Kelly DP. Peroxisome proliferator-activated receptor coactivator-1alpha (PGC-1alpha) coactivates the cardiac-enriched nuclear receptors estrogen-related receptor-alpha and -gamma. Identification of novel leucine-rich interaction motif within PGC-1alpha. J Biol Chem. 2002;277(43):40265–40274. doi: 10.1074/jbc.M206324200. [DOI] [PubMed] [Google Scholar]
- 12.Mootha VK, Handschin C, Arlow D, Xie X, St Pierre J, Sihag S, Yang W, Altshuler D, Puigserver P, Patterson N, Willy PJ, et al. Erralpha and Gabpa/b specify PGC-1alpha-dependent oxidative phosphorylation gene expression that is altered in diabetic muscle. Proc Natl Acad Sci U S A. 2004;101(17):6570–6575. doi: 10.1073/pnas.0401401101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kallen J, Schlaeppi JM, Bitsch F, Filipuzzi I, Schilb A, Riou V, Graham A, Strauss A, Geiser M, Fournier B. Evidence for ligand-independent transcriptional activation of the human estrogen-related receptor alpha (ERRalpha): crystal structure of ERRalpha ligand binding domain in complex with peroxisome proliferator-activated receptor coactivator-1alpha. J Biol Chem. 2004;279(47):49330–49337. doi: 10.1074/jbc.M407999200. [DOI] [PubMed] [Google Scholar]
- 14.Handschin C, Mootha VK. Estrogen-related receptor alpha (ERRalpha): A novel target in type 2 diabetes. Drug Discov Today Ther Strateg. 2005;2(2):151–156. [Google Scholar]
- 15.Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10(3):R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pachkov M, Balwierz PJ, Arnold P, Ozonov E, van Nimwegen E. SwissRegulon, a database of genome-wide annotations of regulatory sites: recent updates. Nucleic Acids Res. 2013;41(Database issue):D214–220. doi: 10.1093/nar/gks1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Balwierz PJ, Pachkov M, Arnold P, Gruber AJ, Zavolan M, van Nimwegen E. ISMARA: automated modeling of genomic signals as a democracy of regulatory motifs. Genome Res. 2014;24(5):869–884. doi: 10.1101/gr.169508.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Perez-Schindler J, Summermatter S, Santos G, Zorzato F, Handschin C. The transcriptional coactivator PGC-1alpha is dispensable for chronic overload-induced skeletal muscle hypertrophy and metabolic remodeling. Proc Natl Acad Sci U S A. 2013;110(50):20314–20319. doi: 10.1073/pnas.1312039110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang Y, Ma K, Sadana P, Chowdhury F, Gaillard S, Wang F, McDonnell DP, Unterman TG, Elam MB, Park EA. Estrogen-related receptors stimulate pyruvate dehydrogenase kinase isoform 4 gene expression. J Biol Chem. 2006;281(52):39897–39906. doi: 10.1074/jbc.M608657200. [DOI] [PubMed] [Google Scholar]
- 20.Eisele PS, Salatino S, Sobek J, Hottiger MO, Handschin C. The peroxisome proliferator-activated receptor gamma coactivator 1alpha/beta (PGC-1) coactivators repress the transcriptional activity of NF-kappaB in skeletal muscle cells. J Biol Chem. 2013;288(4):2246–2260. doi: 10.1074/jbc.M112.375253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sladek R, Bader JA, Giguere V. The orphan nuclear receptor estrogen-related receptor alpha is a transcriptional regulator of the human medium-chain acyl coenzyme A dehydrogenase gene. Mol Cell Biol. 1997;17(9):5400–5409. doi: 10.1128/mcb.17.9.5400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Barry JB, Laganiere J, Giguere V. A single nucleotide in an estrogen-related receptor alpha site can dictate mode of binding and peroxisome proliferator-activated receptor gamma coactivator 1alpha activation of target promoters. Mol Endocrinol. 2006;20(2):302–310. doi: 10.1210/me.2005-0313. [DOI] [PubMed] [Google Scholar]
- 23.Horard B, Vanacker JM. Estrogen receptor-related receptors: orphan receptors desperately seeking a ligand. J Mol Endocrinol. 2003;31(3):349–357. doi: 10.1677/jme.0.0310349. [DOI] [PubMed] [Google Scholar]
- 24.Arnold P, Erb I, Pachkov M, Molina N, van Nimwegen E. MotEvo: integrated Bayesian probabilistic methods for inferring regulatory sites and motifs on multiple alignments of DNA sequences. Bioinformatics. 2012;28(4):487–494. doi: 10.1093/bioinformatics/btr695. [DOI] [PubMed] [Google Scholar]
- 25.Chu S. Transcriptional regulation by post-transcriptional modification--role of phosphorylation in Sp1 transcriptional activity. Gene. 2012;508(1):1–8. doi: 10.1016/j.gene.2012.07.022. [DOI] [PubMed] [Google Scholar]
- 26.Castet A, Herledan A, Bonnet S, Jalaguier S, Vanacker JM, Cavailles V. Receptor-interacting protein 140 differentially regulates estrogen receptor-related receptor transactivation depending on target genes. Mol Endocrinol. 2006;20(5):1035–1047. doi: 10.1210/me.2005-0227. [DOI] [PubMed] [Google Scholar]
- 27.Nichol D, Christian M, Steel JH, White R, Parker MG. RIP140 expression is stimulated by estrogen-related receptor alpha during adipogenesis. J Biol Chem. 2006;281(43):32140–32147. doi: 10.1074/jbc.M604803200. [DOI] [PubMed] [Google Scholar]
- 28.Samson SL, Wong NC. Role of Sp1 in insulin regulation of gene expression. J Mol Endocrinol. 2002;29(3):265–279. doi: 10.1677/jme.0.0290265. [DOI] [PubMed] [Google Scholar]
- 29.Malek A, Nunez LE, Magistri M, Brambilla L, Jovic S, Carbone GM, Moris F, Catapano CV. Modulation of the activity of Sp transcription factors by mithramycin analogues as a new strategy for treatment of metastatic prostate cancer. PLoS One. 2012;7(4):e35130. doi: 10.1371/journal.pone.0035130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stashi E, York B, O’Malley BW. Steroid receptor coactivators: servants and masters for control of systems metabolism. Trends Endocrinol Metab. 2014;25(7):337–347. doi: 10.1016/j.tem.2014.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mouchiroud L, Eichner LJ, Shaw RJ, Auwerx J. Transcriptional coregulators: fine-tuning metabolism. Cell Metab. 2014;20(1):26–40. doi: 10.1016/j.cmet.2014.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dasgupta S, Lonard DM, O’Malley BW. Nuclear receptor coactivators: master regulators of human health and disease. Annu Rev Med. 2014;65:279–292. doi: 10.1146/annurev-med-051812-145316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Handschin C, Spiegelman BM. Peroxisome proliferator-activated receptor gamma coactivator 1 coactivators, energy homeostasis, and metabolism. Endocr Rev. 2006;27(7):728–735. doi: 10.1210/er.2006-0037. [DOI] [PubMed] [Google Scholar]
- 34.Spiegelman BM, Heinrich R. Biological control through regulated transcriptional coactivators. Cell. 2004;119(2):157–167. doi: 10.1016/j.cell.2004.09.037. [DOI] [PubMed] [Google Scholar]
- 35.Lonard DM, O’Malley BW. Nuclear receptor coregulators: judges, juries, and executioners of cellular regulation. Mol Cell. 2007;27(5):691–700. doi: 10.1016/j.molcel.2007.08.012. [DOI] [PubMed] [Google Scholar]
- 36.Charos AE, Reed BD, Raha D, Szekely AM, Weissman SM, Snyder M. A highly integrated and complex PPARGC1A transcription factor binding network in HepG2 cells. Genome Res. 2012;22(9):1668–1679. doi: 10.1101/gr.127761.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Xie W, Hong H, Yang NN, Lin RJ, Simon CM, Stallcup MR, Evans RM. Constitutive activation of transcription and binding of coactivator by estrogen-related receptors 1 and 2. Mol Endocrinol. 1999;13(12):2151–2162. doi: 10.1210/mend.13.12.0381. [DOI] [PubMed] [Google Scholar]
- 38.Coghlan MJ, Jacobson PB, Lane B, Nakane M, Lin CW, Elmore SW, Kym PR, Luly JR, Carter GW, Turner R, Tyree CM, et al. A novel antiinflammatory maintains glucocorticoid efficacy with reduced side effects. Mol Endocrinol. 2003;17(5):860–869. doi: 10.1210/me.2002-0355. [DOI] [PubMed] [Google Scholar]
- 39.Willy PJ, Murray IR, Qian J, Busch BB, Stevens WC, Jr, Martin R, Mohan R, Zhou S, Ordentlich P, Wei P, Sapp DW, et al. Regulation of PPARgamma coactivator 1alpha (PGC-1alpha) signaling by an estrogen-related receptor alpha (ERRalpha) ligand. Proc Natl Acad Sci U S A. 2004;101(24):8912–8917. doi: 10.1073/pnas.0401420101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Blattler A, Farnham PJ. Cross-talk between site-specific transcription factors and DNA methylation states. J Biol Chem. 2013;288(48):34287–34294. doi: 10.1074/jbc.R113.512517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Barres R, Yan J, Egan B, Treebak JT, Rasmussen M, Fritz T, Caidahl K, Krook A, O’Gorman DJ, Zierath JR. Acute exercise remodels promoter methylation in human skeletal muscle. Cell Metab. 2012;15(3):405–411. doi: 10.1016/j.cmet.2012.01.001. [DOI] [PubMed] [Google Scholar]
- 42.Charest-Marcotte A, Dufour CR, Wilson BJ, Tremblay AM, Eichner LJ, Arlow DH, Mootha VK, Giguere V. The homeobox protein Prox1 is a negative modulator of ERR{alpha}/PGC-1{alpha} bioenergetic functions. Genes Dev. 2010;24(6):537–542. doi: 10.1101/gad.1871610. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.