Abstract
Purpose
To determine if patient survival and mechanisms of right ventricular (RV) failure in pulmonary hypertension (PH) could be predicted using supervised machine learning of three-dimensional patterns of systolic cardiac motion.
Materials and methods
The study was approved by a research ethics committee and participants gave written informed consent. 256 patients (143 females, mean age 63 ± 17) with newly-diagnosed PH underwent cardiac MR imaging, right heart catheterization (RHC) and six-minute walk testing (6MWT) with a median follow-up of 4.0 years. Semi-automated segmentation of short axis cine images was used to create a three-dimensional model of right ventricular motion. Supervised principal components analysis identified patterns of systolic motion which were most strongly predictive of survival. Survival prediction was assessed by the difference in median survival time and the area under the curve (AUC) using time-dependent receiver operator characteristic for one-year survival.
Results
At the end of follow-up 33% (93/256) died and one underwent lung transplantation. Poor outcome was predicted by a loss of effective contraction in the septum and freewall coupled with reduced basal longitudinal motion. When added to conventional imaging, hemodynamic, functional and clinical markers, three-dimensional cardiac motion improved survival prediction (area under the curve 0.73 vs 0.60, p<0.001) and provided greater differentiation by difference in median survival time between high- and low- risk groups (13.8 vs 10.7 years, p<0.001).
Conclusion
Three-dimensional motion modeling with machine learning approaches reveal the adaptations in function that occur early in right heart failure and independently predict outcomes in newly-diagnosed PH patients.
Introduction
Pulmonary hypertension (PH) is a heterogeneous group of diseases defined by a resting mean pulmonary artery pressure of at least 25mmHg (1). PH may follow a rapidly progressive clinical course with impaired exercise tolerance and dyspnea associated with right ventricular (RV) hypertrophy, right heart dilatation and ultimately cardiac failure (2). Outcome prediction in PH has been extensively investigated using invasive hemodynamic data, serum biomarkers, exercise testing and cardiac imaging. These markers consistently demonstrate that survival is related to functional adaptation of the right ventricle (3). RV ejection fraction (RVEF) is a measure of global systolic function which predicts survival in PH patients (4), though the complex shape and contraction pattern of the right ventricle make this an insensitive assessment of early cardiac decompensation (5).
Patients investigated for PH routinely undergo cardiac magnetic resonance (MR) imaging which provides an accurate assessment of cardiac functional status, but to realize the full predictive potential of cardiac imaging requires methods able to select the most relevant and meaningful prognostic features (6). Computational image analysis coupled with machine learning could enable discovery of the complex functional adaptations that predict eventual right heart failure and death. Unlocking the full potential of diagnostic imaging in this way has recently become feasible with advances in computational modeling of the structure and function of the heart (7–9). The purpose of the present study was to determine if patient survival and mechanisms of RV failure in PH could be predicted using supervised machine learning of three-dimensional patterns of systolic cardiac motion.
Materials and methods
Study population
This study was part of a continuous prospective research program into the prognosis of PH patients using conventional clinical and imaging biomarkers. The program had ethics committee approval and all patients gave written informed consent. Our computational analysis was retrospectively performed on the data for the UK Digital Heart Project (http://digital-heart.org). Patients referred to the National Pulmonary Hypertension Service at Imperial College Healthcare NHS Trust for routine diagnostic assessment and cardiac imaging between May 2004 and October 2013 were included in the study with end of follow-up in September 2014. Criteria for inclusion included a documented diagnosis of PH by right heart catheterization (RHC) with a resting mean pulmonary artery pressure ≥25 mmHg. Clinical classification was according to European guidelines (1) and patients with congenital shunts, arrhythmias preventing cardiac gating, or more than three months between baseline investigations were excluded. All patients were treated with standard therapy in accordance with current guidelines and NHS England treatment policy (10).
Right heart catheterization
RHC was performed by certified interventionists with a balloon-tipped, flow-directed Swan-Ganz catheter (Baxter Healthcare Corp., Irvine, California) to derive cardiac output, cardiac index, mean pulmonary artery pressure (mPAP), pulmonary capillary wedge pressure and pulmonary vascular resistance (PVR). Six minute walk distance (6MWD) was measured according to the American Thoracic Society guidelines (11).
MR imaging protocol
Cardiac MR imaging was performed at a single site on a 1.5T Philips Achieva (Best, Netherlands) and a standard clinical protocol was followed according to published international guidelines (12). Ventricular function was assessed using balanced-steady state free precession (b-SSFP) cine images acquired in conventional cardiac short- and long- axis planes with typical parameters of - repetition time msec/echo time msec, 3.2/1.6; voxel size, 1.5 x 1.5 x 8 mm; flip angle, 60°; sensitivity encoding factor (SENSE), 2.0; bandwidth, 962 Hz/pixel; and temporal resolution 29 msec. Reproducibility was assessed in 20 subjects undergoing repeat studies on the same day. Images were stored on an open-source database (MRIdb, Imperial College London).
Quantification of right ventricular function
Volumetric analysis of cine images was performed using Philips ViewForum (Best, Netherlands) by one reader with 3 years of experience (TJWD) manually defining the RV endocardial borders at end-diastole and end-systole using a standard published protocol (13). Reference to the position of the pulmonary and tricuspid valves on long-axis images was made to ensure correct placement of the contours. Papillary muscles and trabeculae were included in the RV volume.
Three-dimensional assessment of ventricular physiology
Atlas-based approaches for segmenting the right ventricle enable a three-dimensional model of RV structure and function to be constructed (14). To ensure a fair comparison manual volumetry and computational analysis used the same standard cardiac MR images.
All image processing was performed in Matlab (Mathworks, Natick, MA). We took the short-axis cines for each PH patient and automatically aligned each set of end-diastolic and end-systolic images image by minimizing the intensity differences between each slice (15). The segmentation process was then initialized by a reader (TJWD) who placed six pre-defined anatomical landmarks on the target images (left ventricular apex, mitral annulus and lateral wall; the RV freewall; and the superior and inferior RV insertion points - Supplementary Figure 1). These landmarks were also defined on each labeled atlas. Manually-annotated cardiac atlases at end-diastole and end-systole were derived from 47 PH patients and included in the Digital Heart Project population dataset for analysis of both shape and motion (16). Each voxel in the PH atlases was manually labeled as LV/RV cavity or myocardium using freely available software (ITKsnap, National Library of Medicine’s Insight Segmentation and Registration Toolkit). A multi-atlas approach utilized the entire dataset of labeled atlases rather than relying on a model-based average representation (17). An approximate graph search was performed to find correspondences between small cubic regions, or patches, in the image to be segmented and the database of labeled atlases. Spectral embedding using a multi-layered graph of the images was used to capture global shape properties. Finally, we estimated anatomical patch correspondences based on a joint spectral representation of the image and atlases (14, 18). The final segmentations were co-registered to an average template surface mesh, where vertex density was determined by curvature at each sampling point, allowing cardiac shape or function within the population to be compared in a common space (freely available at https://github.com/UK-Digital-Heart-Project).
Three-dimensional motion reconstruction was performed using temporal sparse free-form registration to estimate cardiac motion between the two time points at end-diastole and end-systole (Figure 1)(19). For each endocardial vertex in the right ventricle (~30,000 data points) we calculated the direction and magnitude of systolic excursion, ie the maximal displacement from the initial position in end-diastole. Each vector was then resolved into three perpendicular components (longitudinal, circumferential, and radial) relative to a long-axis defined between the tricuspid orifice and RV apex. The resulting co-registered three-dimensional dataset represented the systolic motion of the endocardial surface of the right ventricle and septum in the PH cohort. Patterns of three-dimensional motion associated with survival were then assessed with supervised principal components analysis. Accuracy was assessed by comparing manual and semi-automated segmentations using leave-one-out cross-validation, and repeatability by measuring the agreement between two studies.
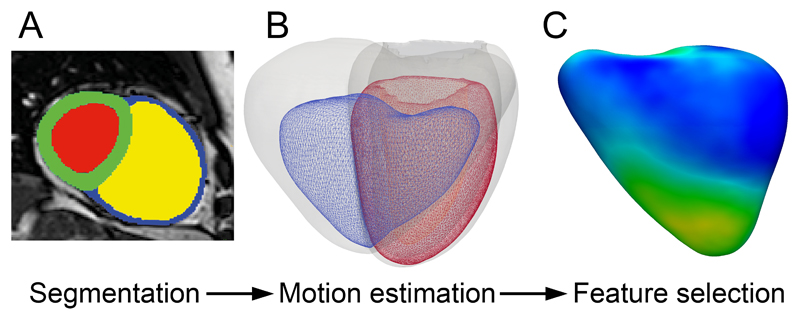
Fig 1.
An example of computational modeling is shown for a patient with idiopathic pulmonary arterial hypertension. (a) Cine magnetic resonance images were segmented using prior knowledge from a set of disease-specific atlases. Here the intensity image in the short-axis of the heart is overlaid with labels for left ventricular blood pool (red), myocardium (green), right ventricular blood pool (yellow) and free-wall (blue). (b) A three-dimensional model at end-diastole (grey) and end-systole (blue – right ventricle and red – left ventricle) was used to determine the direction and magnitude of systolic excursion at each corresponding anatomical point in the mesh using a deformable motion model. (c) A statistical model of right ventricular endocardial motion is used for feature selection to determine functional patterns that are associated with survival (relative weightings shown for the right ventricular freewall).
Statistical analysis
Data were analyzed in R using RStudio Server version 0.98 (Boston, MA). Categorical variables were expressed as percentages. Continuous variables were expressed as mean ± standard deviation (SD) or median ± inter-quartile range (IQR) for non-normal variables. Baseline anthropometric data were compared by unpaired t-tests and Mann-Whitney U tests pending normality (continuous variables) and Fisher’s Exact (nominal variables) and Cochran-Armitage tests (ordinal variables). Inter-group age differences were controlled for by linear regression.
Standard principal components analysis (PCA) is commonly used to summarize data into components that account for most of the variance in the observed data, but these components may not relate to markers of interest such as outcome or survival. Supervised PCA is a supervised learning approach that is effective for regression and classification problems using complex input data (20).
The performance of each marker was tested as a predictor of survival in the whole dataset and significant univariate predictors (p<0.05) were carried forward to three nested models designed to test the incremental benefit of groups of predictors. Model 1 included the clinical, hemodynamic and functional predictors found to be significant in univariate prediction. Model 2 additionally included markers of MR volumetry (RVEF, RVESVI, RVEDVI and SV/RVESV). Lastly, model 3 included the predictors in both models 1 and 2 as well as 3D motion. Models were optimized in the training data and then assessed in the unseen validation data using eight-fold cross-validation (Figure 2) (21). In each fold, 224 cases provided the training data for a predictive model which was then evaluated on the held-out set of 32 cases until every patient had been in a validation set exactly once. Supervised PCA proceeded as follows: i) fit a Cox proportional hazards model for each predictor, ii) perform feature selection by selecting predictors with coefficients exceeding an absolute threshold (established by cross-validation of the log-likelihood ratios), iii) use the first principal component of the reduced data matrix as a prognostic marker. The value of this principal component in unseen subjects was used to predict whether subjects would be alive or dead at censoring and to fit a Cox proportional hazards model for subject survival from which model fit was measured.
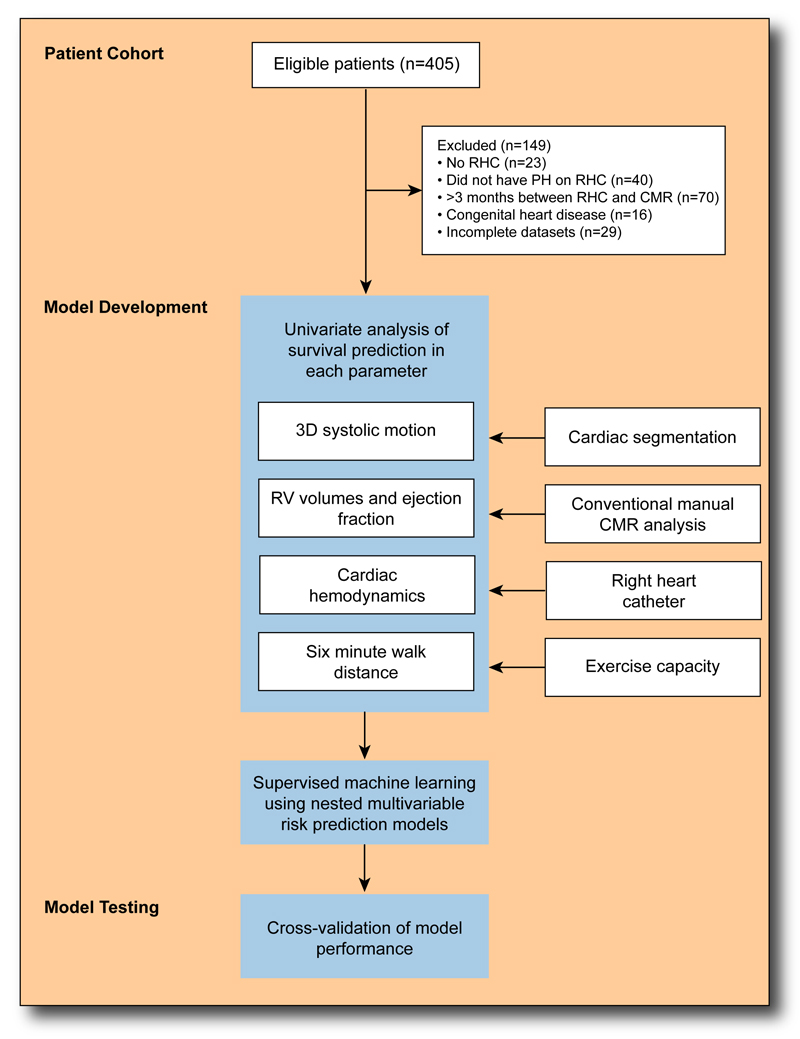
Fig. 2.
A flow diagram for recruitment and analysis of pulmonary hypertension patients. 256 eligible patients had their cardiac MR images segmented and analyzed. In the training phase supervised machine learning was used to discover patterns of right ventricular function that were associated with outcome. Predictive performance of multivariable risk models was assessed using 8-fold cross-validation to demonstrate the incremental value of computational phenotyping.
Survival was taken as the time between enrolment and death from any cause. Survival prediction for the Cox proportional hazards model was assessed by the hazards ratio, R2 for each model and the area under the curve (AUC) using time-dependent receiver operating characteristics (ROC) for one-year survival (22). The model was bootstrapped (1,000 bootstraps) to estimate performance metrics which were then compared by analysis of variance (ANOVA) with post-hoc Tukey’s testing. Models’ performance was compared to the null hypothesis by permutation testing (1,000 permutations) with a p-value <0.05 taken as significant.
This study was conducted according to the “Transparent Reporting of a multivariable prediction model for Individual Prognosis or Diagnosis” (TRIPOD) guidelines.
Results
Study Population Characteristics
In total 405 consecutive patients referred for investigation to the Centre were evaluated for eligibility and 256 subjects with confirmed PH were enrolled. Of these 33% (93/256) died and one underwent lung transplantation during follow-up, which was of median length 4.0 years (inter-quartile range (IQR): 2.0–5.7 years). In the cohort 6% (16/256) of patients were unable to do the 6MWD test. Median patient age was 67 (52-75) years and 56% were female (anthropometric, hemodynamic and cardiac MR data are given in Supplementary Table 1). Additional data stratified by PH sub-group are given in Supplementary Tables 2, 3 and 4.
Accuracy and reproducibility of segmentation and motion tracking
Mean Hausdorff distance between corresponding semi-automated and manual segmentations of the right ventricle was 3.0±1.2mm. Computational analysis showed good inter-study agreement by intraclass correlation coefficient (ICC) in determining the longitudinal, radial and circumferential positions of corresponding points in each orthogonal coordinate (ICCs: 0.98/0.98/0.91 respectively, all p<0.001) and for assessment of cardiac motion (ICCs: 0.90/0.81/0.82 respectively, all p<0.001). A three-dimensional representation of the spatial errors in segmentation is given in Supplementary Figure 2.
Survival Prediction
Univariate Cox regression analyses indicated that age, sex and race (“clinical markers”), mRAP and RV end-diastolic pressure (RVEDP) (“hemodynamic markers”), functional class and 6MWD (“functional markers”) and all baseline cardiac MR measurements (“volumetric markers”) were significantly associated with survival (Supplementary Table 1). Univariate standardized hazards ratios for each predictor and Kaplan-Meier estimates of survival comparing RVEF to 3D motion analysis are shown in Figure 4.
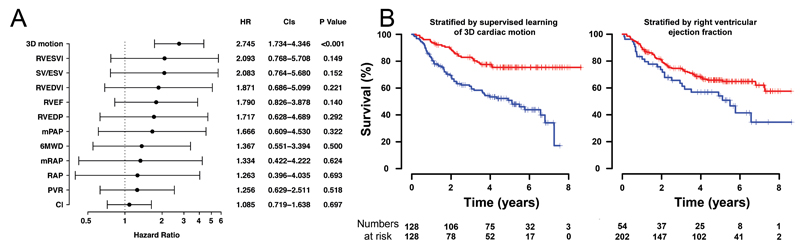
Fig. 4.
A comparison of survival prediction for each parameter is shown. (a) Standardized hazards ratios (for a 1.96 standard deviation difference) with 95% confidence intervals are shown for 3D motion and conventional prognostic markers. (b) Kaplan Meier curves and numbers-at-risk for the survival of pulmonary hypertension patients comparing risk-stratification by 3D motion vs right ventricular ejection fraction.
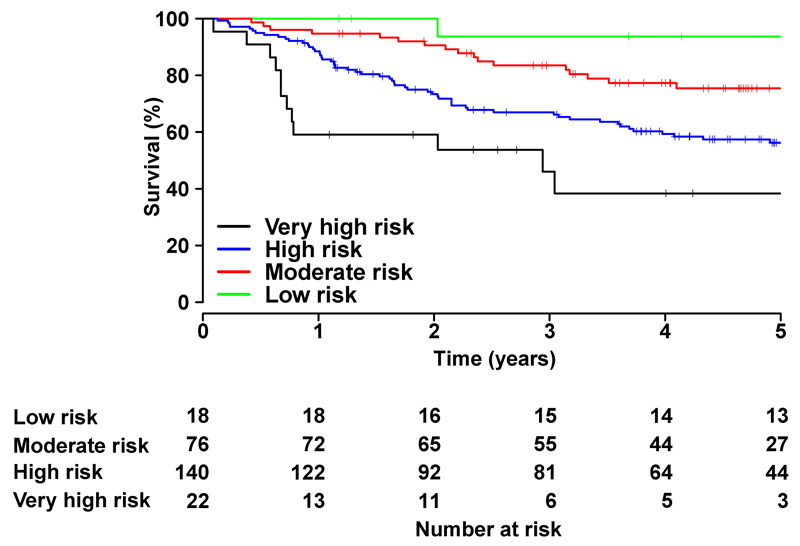
Feature extraction and supervised learning from the data is shown in Figure 3. All three nested prediction models were significant compared to the null hypothesis (all p<0.001) and there was a significant difference between the predictive power of the models (ANOVA, HR: F=80.2, p<0.001; AUC: F=94.2, p<0.001; R2: F=40.7, p<0.001). Model 3, which included three-dimensional motion, had significantly higher hazards ratio, AUC and R2, and a greater difference in median survival time (all p<0.001) (Table 1). Five-year survival from time of diagnosis by quartiles of risk predicted by Model 3 is shown in Figure 5.
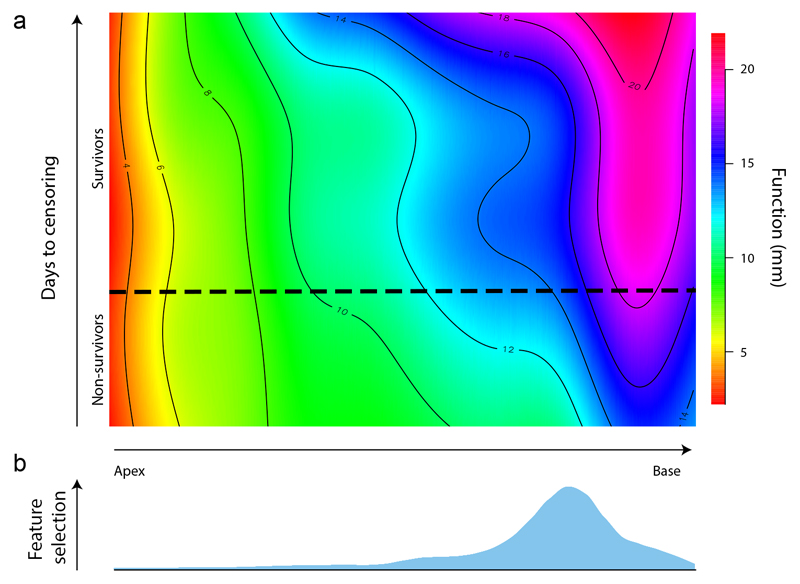
Fig 3.
An illustration of how features of right ventricular motion are automatically selected for their prognostic importance in pulmonary hypertension patients. The upper plot represents how the magnitude of systolic excursion in the right ventricle, derived from atlas-based cardiac segmentations, varies between survivors and non-survivors from basal to apical level. The lower plot shows where supervised machine learning identifies features within these motion-based data that most accurately discriminate between low risk and high risk patients. The full model used for survival prediction considered the prognostic importance of motion throughout a three-dimensional representation of the right ventricle resolved into orthogonal components.
Table 1. Survival prediction in the validation cases.
| Model | Predictors | HR (95% CIs) † | Model comparison | AUC (95% CIs) † | Model comparison | R2 (95% CIs) † | Model comparison | Survival (years) (95% CIs) † | Model comparison | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Clinical | RHC | Function | MR | 3D | |||||||||
| 1 | ✓ | ✓ | ✓ | 1.71 (1.24-2.17) | 0.64 (0.49-0.76) | 0.09 (0.03 – 0.19) | 9.5 (3.3 - 22.0) | ||||||
| 2 | ✓ | ✓ | ✓ | ✓ | 1.66 (1.33-2.02) | Model 2 vs 1: P=0.06 | 0.60 (0.49-0.74) | Model 2 vs 1: P=0.33 | 0.08 (0.03 – 0.15) | Model 2 vs 1: P=0.09 | 10.7 (5.0 - 22.6) | Model 2 vs 1: P=0.52 | |
| 3 | ✓ | ✓ | ✓ | ✓ | ✓ | 1.99 (1.59-2.69) | Model 3 vs 2: P<0.001 |
0.73 (0.58-0.81) | Model 3 vs 2: P<0.001 |
0.13 (0.07 – 0.20) | Model 3 vs 2: P<0.001 |
13.8 (8.0 - 25.3) | Model 3 vs 2: P<0.001 |
Clinical markers: age, gender, race and PH subtype; RHC (right heart catheterization) hemodynamic markers: right atrial pressure, right ventricular end-diastolic pressure; Functional markers: NYHA functional class, six-minute walk distance; MR: right ventricular end-diastolic volume index, right ventricular end-systolic volume index, right ventricular ejection fraction and stroke volume / right ventricular end-systolic volume; 3D: three-dimensional motion. Receiver operating characteristics’ (ROC) area under the curve (AUC) indicates the performance of each marker in one-year survival prediction. In subgroups with >50% survival at censoring, median survival values are extrapolated. 3D, three-dimensional. All models were significantly better predictors than the null (p<0.001). †Confidence intervals.
Fig 5.
Observed 5-year survival from time of diagnosis according to predicted risk strata obtained using Model 3 as described in Table 1.
Functional contributions to survival and ventricular function
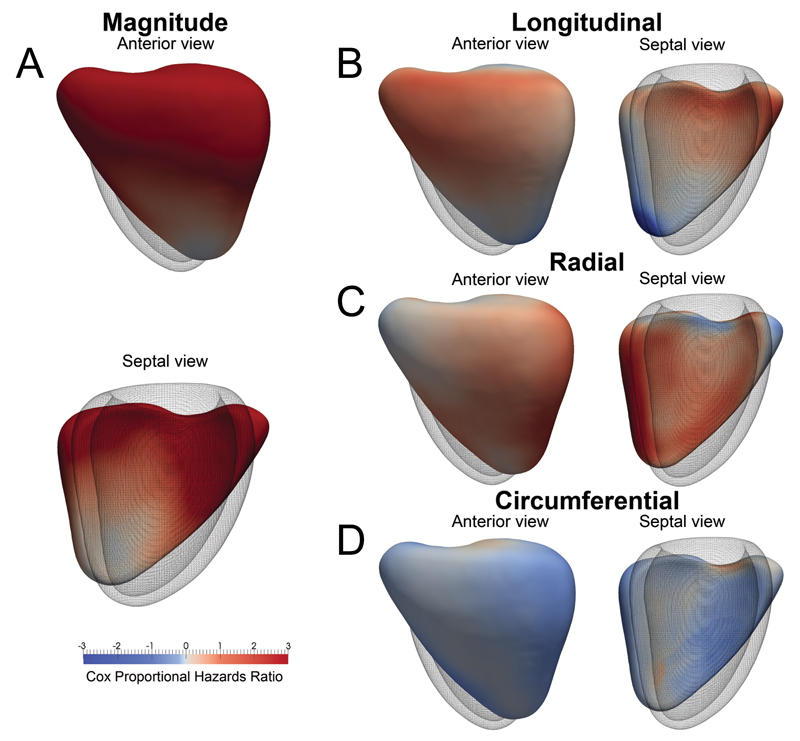
Systolic function throughout most of the right ventricle and septum was influential in patient survival (Figure 6). Reduced longitudinal excursion throughout the basal and mid-ventricular regions was association with poor outcome. A decrease in radial contraction in the septum and freewall also carried prognostic significance. Mortality was also predicted by a global increase in circumferential function. Machine learning analysis from the motion datasets predicted survival in all three PH subgroups (all p<0.001) and the variation in cardiac function between groups is shown in Supplementary Figure 3.
Fig. 6.
Three-dimensional model of the right ventricle showing the regional contributions to survival prediction in 256 pulmonary hypertension patients. The models show where reduced (red) or increased (blue) systolic motion is associated with death. This is shown by the (a) magnitude of excursion as well as each directional component (b, c, d). The right ventricle is shown in anterior and septal views with the left ventricle as a mesh. A reduction in both longitudinal basal motion and transverse bellows contraction are associated with death, as is an increase in circumferential contraction.
Discussion
Semi-automated analysis of cine cardiac MR images in PH patients is feasible, accurate and reproducible. Supervised machine learning of the patterns of cardiac motion indicates that survival in PH patients is predicted by a loss of effective contractile motion in anatomically distinct but functionally synergistic regions of the right ventricle. A machine learning survival model that includes cardiac motion has incremental prognostic power compared to conventional parameters.
Computational modeling provides a platform for improving our understanding of the heart, and the integration of experimental and clinical data is now bringing computational models closer to use in routine clinical practice (23). Advances in both cardiac imaging and analytic models offer a wealth of biological data that contribute to the search for novel biomarkers of cardiac dysfunction. Multimodal techniques can now offer biventricular electromechanical modeling, fluid-solid mechanical models and luminal flow-streamlines coupled with myocardial displacement (24, 25). Atlas-based analysis in the heart has been applied to describing shape variation amongst asymptomatic adults, in identifying persisting effects of preterm delivery on ventricular geometry and in demonstrating patterns of remodeling after myocardial infarction (9, 26, 27). The potential for this technique lies not just in shape analysis but as a means to understand how the integrated function of the heart contributes to survival in large clinical cohorts as the data from each subject is co-registered. While other approaches to cardiac segmentation are more widely available, atlas based methods maintain the anatomic correspondence of the three-dimensional mesh between patients. Machine learning, both supervised and unsupervised, can be applied to clinical data sets for the purpose of developing robust risk models and redefining patient classes (28). In this study we took a complex three-dimensional model of cardiac displacement and applied a machine learning algorithm to identify recurring patterns within this high-dimensional dataset that most strongly predicted outcomes. From standard diagnostic imaging a mathematical model of cardiac function’s relationship to survival can be generated.
Conventional cardiac MR studies in PH have shown that deteriorating RV function is associated with poor outcome despite therapeutic reductions in PVR (4). Imaging can be used to assess RV systolic function in several ways, typically by global measures of pump function or regional systolic excursion (29). Our models indicate that survival is linked to a combined failure of basal longitudinal shortening and transverse contraction of the septum and freewall. The importance of these individual components of motion to pump function has been previously proposed in physiologic studies (30), and here we demonstrate their combined influence on outcome. The modeling also reveals the risk associated with adaptations of circumferential function which tends to increase in PH as global failure develops (31). Raised afterload and RV dilatation are associated with fiber reorientation towards the circumferential direction (32), and our data indicate that such adverse structural remodeling independently contributes to survival. The predictive performance of ML is independent of PH sub-type but the models identify some prognostic variations in RV geometry and function that may reflect differing responses to altered loading conditions. These integrative models of right heart physiology show that cardiac decompensation is not simply a global decline in function, but instead results from a loss of effective contractile motion in anatomically distinct but functionally synergistic regions.
Innovation in biomarker discovery and personalized medicine requires a cultural change in how clinical data are exploited (6). Here we have demonstrated how it is possible to maximize the potential of existing imaging resources for outcome prediction using computational models that require minimal human intervention. As well as accurately risk classifying individual patients the models also inform clinicians about the functional mechanisms which underlie RV failure. The potential for such computational simulations lies not only in risk stratification but also in designing trials for new therapies that have a direct effect on RV contractility. Future work will be directed at improving the depth of phenotyping using time-resolved segmentations throughout the cardiac cycle to model three-dimensional strain tensors, evaluating ML predictions on an independent validation cohort benchmarked against conventional multivariate risk models, and exploring the potential of deep learning architectures for hierarchical feature recognition (33, 34).
Our study has limitations. The pragmatic study design, which included all non-congenital cases of PH and all treatment regimens, may limit applicability in selective groups, but demonstrates that the methods are effective across a spectrum of disease and treatments. We classified our patients according to international guidelines, but it is recognized that PH patients have multifactorial disease with overlap between categories. Study endpoint and censoring were confined to all-cause mortality to avoid bias in the classification of cause of death, though markers’ performance will also be affected by variations in therapy and those selected for surgery. The accuracy of our segmentation compares well to previously published results (14), however the uncertainty in both end-diastolic and end-systolic segmentation will propagate to the uncertainty in displacement estimation. Our models are also currently limited to measuring excursion rather than contractility and deriving strain tensors may add additional prognostic data (34). Human intervention is limited to landmark placement but newer techniques offer a means to also automate this step (35). We considered only the first supervised principal component of the three-dimensional functional model as this explains the most variance in data as possible and allows a fair comparison with other single parameter predictors. The cumulative variance of subsequent components may further improve prediction although later components are increasingly influenced by noise. Contemporaneous echocardiography and cardiopulmonary exercise testing were not available for each case. Tricuspid annular plane systolic excursion offers a simple to measure prognostic indicator, and although we confirm the importance of basal longitudinal excursion to survival, the models show that regional contraction in the circumferential and radial directions also contribute to prognosis.
In conclusion, applying machine learning of complex motion phenotypes obtained from cardiac MR imaging allows more accurate prediction of patient outcomes in pulmonary hypertension.
Supplementary Material
Advances in Knowledge
A disease-specific cardiac atlas can be used to create accurate (Hausdorff distance 3.0 ± 1.2mm) and reproducible (intraclass correlation coefficients for agreement of position in each axis: 0.98/0.98/0.91, all p<0.001) segmentations of the heart in pulmonary hypertension from conventional cardiac MR images.
A supervised machine learning survival model that includes three-dimensional cardiac motion provides incremental prognostic benefit compared to conventional imaging, hemodynamic, functional and clinical markers (area under the curve 0.73 vs 0.60, p<0.001; difference in median survival time between high- and low- risk groups (13.8 vs 10.7 years, p<0.001).
Implications for patient care
Computational analysis of right ventricular motion in pulmonary hypertension can be used for risk stratification and reveals early prognostic signs of dysfunction.
Machine learning using cardiac MR should be evaluated as a tool to guide patient management.
Summary statement
Applying machine learning of complex motion phenotypes obtained from cardiac MR imaging allows more accurate prediction of patient outcomes in pulmonary hypertension.
Acknowledgments
The study was supported by the Medical Research Council, UK; National Institute for Health Research (NIHR) Biomedical Research Centre based at Imperial College Healthcare NHS Trust and Imperial College London, UK; British Heart Foundation grants PG/12/27/29489 and SP/10/10/28431, and a Wellcome Trust-GSK Fellowship Grant (100211/Z/12/Z).
Contributor Information
Timothy J.W. Dawes, MRC Clinical Sciences Centre (CSC), Du Cane Road, London W12 0NN, UK; Division of Experimental Medicine, Department of Medicine, Imperial College London, London, UK
Antonio de Marvao, MRC Clinical Sciences Centre (CSC), Du Cane Road, London W12 0NN, UK.
Wenzhe Shi, MRC Clinical Sciences Centre (CSC), Du Cane Road, London W12 0NN, UK; Department of Computing, Imperial College London, South Kensington Campus, London, UK.
Tristan Fletcher, MRC Clinical Sciences Centre (CSC), Du Cane Road, London W12 0NN, UK; Division of Experimental Medicine, Department of Medicine, Imperial College London, London, UK.
Geoffrey M.J. Watson, Division of Experimental Medicine, Department of Medicine, Imperial College London, London, UK
John Wharton, Division of Experimental Medicine, Department of Medicine, Imperial College London, London, UK.
Christopher J. Rhodes, Division of Experimental Medicine, Department of Medicine, Imperial College London, London, UK
Luke S.G.E. Howard, Department of Cardiology, National Pulmonary Hypertension Service, Imperial College Healthcare NHS Trust, London, UK
J. Simon R. Gibbs, Department of Cardiology, National Pulmonary Hypertension Service, Imperial College Healthcare NHS Trust, London, UK; National Heart and Lung Institute, Imperial College London, London, UK
Daniel Rueckert, Department of Computing, Imperial College London, South Kensington Campus, London, UK.
Stuart A. Cook, MRC Clinical Sciences Centre (CSC), Du Cane Road, London W12 0NN, UK; National Heart Centre Singapore, Singapore and Duke-NUS Graduate Medical School, Singapore
Martin R. Wilkins, Division of Experimental Medicine, Department of Medicine, Imperial College London, London, UK
Declan P O’Regan, MRC Clinical Sciences Centre (CSC), Du Cane Road, London W12 0NN, UK.
References
- 1.Galie N, Humbert M, Vachiery JL, et al. 2015 ESC/ERS Guidelines for the diagnosis and treatment of pulmonary hypertension: The Joint Task Force for the Diagnosis and Treatment of Pulmonary Hypertension of the European Society of Cardiology (ESC) and the European Respiratory Society (ERS): Endorsed by: Association for European Paediatric and Congenital Cardiology (AEPC), International Society for Heart and Lung Transplantation (ISHLT) Eur Respir J. 2015;46(4):903–75. doi: 10.1183/13993003.01032-2015. [DOI] [PubMed] [Google Scholar]
- 2.Benza RL, Miller DP, Gomberg-Maitland M, et al. Predicting survival in pulmonary arterial hypertension: insights from the Registry to Evaluate Early and Long-Term Pulmonary Arterial Hypertension Disease Management (REVEAL) Circulation. 2010;122(2):164–72. doi: 10.1161/CIRCULATIONAHA.109.898122. [DOI] [PubMed] [Google Scholar]
- 3.Vonk-Noordegraaf A, Haddad F, Chin KM, et al. Right heart adaptation to pulmonary arterial hypertension: physiology and pathobiology. J Am Coll Cardiol. 2013;62(25 Suppl):D22–33. doi: 10.1016/j.jacc.2013.10.027. [DOI] [PubMed] [Google Scholar]
- 4.van de Veerdonk MC, Kind T, Marcus JT, et al. Progressive right ventricular dysfunction in patients with pulmonary arterial hypertension responding to therapy. J Am Coll Cardiol. 2011;58(24):2511–9. doi: 10.1016/j.jacc.2011.06.068. [DOI] [PubMed] [Google Scholar]
- 5.Naeije R, Ghio S. More on the right ventricle in pulmonary hypertension. Eur Respir J. 2015;45(1):33–5. doi: 10.1183/09031936.00209414. [DOI] [PubMed] [Google Scholar]
- 6.Scruggs SB, Watson K, Su AI, et al. Harnessing the Heart of Big Data. Circulation Research. 2015;116(7):1115–9. doi: 10.1161/CIRCRESAHA.115.306013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fonseca CG, Backhaus M, Bluemke DA, et al. The Cardiac Atlas Project--an imaging database for computational modeling and statistical atlases of the heart. Bioinformatics. 2011;27(16):2288–95. doi: 10.1093/bioinformatics/btr360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.de Marvao A, Dawes TJ, Shi W, et al. Population-based studies of myocardial hypertrophy: high resolution cardiovascular magnetic resonance atlases improve statistical power. J Cardiovasc Magn Reson. 2014;16:16. doi: 10.1186/1532-429X-16-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Medrano-Gracia P, Cowan BR, Ambale-Venkatesh B, et al. Left ventricular shape variation in asymptomatic populations: the Multi-Ethnic Study of Atherosclerosis. J Cardiovasc Magn Reson. 2014;16:56. doi: 10.1186/s12968-014-0056-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.NHS England. Clinical Commissioning Policy: National policy for targeted therapies for the treatment of pulmonary hypertension in adults. 2014 [Google Scholar]
- 11.A. T. S. Committee on Proficiency Standards for Clinical Pulmonary Function Laboratories. ATS statement: guidelines for the six-minute walk test. Am J Respir Crit Care Med. 2002;166(1):111–7. doi: 10.1164/ajrccm.166.1.at1102. [DOI] [PubMed] [Google Scholar]
- 12.Kramer CM, Barkhausen J, Flamm SD, Kim RJ, Nagel E. Standardized cardiovascular magnetic resonance (CMR) protocols 2013 update. J Cardiovasc Magn Reson. 2013;15:91. doi: 10.1186/1532-429X-15-91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schulz-Menger J, Bluemke DA, Bremerich J, et al. Standardized image interpretation and post processing in cardiovascular magnetic resonance: Society for Cardiovascular Magnetic Resonance (SCMR) board of trustees task force on standardized post processing. J Cardiovasc Magn Reson. 2013;15:35. doi: 10.1186/1532-429X-15-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Petitjean C, Zuluaga MA, Bai W, et al. Right ventricle segmentation from cardiac MRI: a collation study. Med Image Anal. 2015;19(1):187–202. doi: 10.1016/j.media.2014.10.004. [DOI] [PubMed] [Google Scholar]
- 15.Hill DL, Batchelor PG, Holden M, Hawkes DJ. Medical image registration. Phys Med Biol. 2001;46(3):R1–45. doi: 10.1088/0031-9155/46/3/201. [DOI] [PubMed] [Google Scholar]
- 16.Bai W, Shi W, de Marvao A, et al. A bi-ventricular cardiac atlas built from 1000+ high resolution MR images of healthy subjects and an analysis of shape and motion. Med Image Anal. 2015;26(1):133–45. doi: 10.1016/j.media.2015.08.009. [DOI] [PubMed] [Google Scholar]
- 17.Aljabar P, Heckemann RA, Hammers A, Hajnal JV, Rueckert D. Multi-atlas based segmentation of brain images: atlas selection and its effect on accuracy. Neuroimage. 2009;46(3):726–38. doi: 10.1016/j.neuroimage.2009.02.018. [DOI] [PubMed] [Google Scholar]
- 18.Shi W, Lombaert H, Bai W, et al. Multi-atlas spectral PatchMatch: application to cardiac image segmentation. Med Image Comput Comput Assist Interv. 2014;17(Pt 1):348–55. doi: 10.1007/978-3-319-10404-1_44. [DOI] [PubMed] [Google Scholar]
- 19.Wang H, Shi W, Zhuang X, et al. Automatic Cardiac Motion Tracking Using Both Untagged and 3D Tagged MR Images. In: Camara O, Konukoglu E, Pop M, Rhode K, Sermesant M, Young A, editors. Statistical Atlases and Computational Models of the Heart Imaging and Modelling Challenges: Second International Workshop, STACOM 2011, Held in Conjunction with MICCAI 2011, Toronto, ON, Canada, September 22, 2011. Berlin, Heidelberg: Springer Berlin Heidelberg; 2012. pp. 45–54. Revised Selected Papers. [Google Scholar]
- 20.Bair E, Tibshirani R. Semi-supervised methods to predict patient survival from gene expression data. PLoS Biol. 2004;2(4):E108. doi: 10.1371/journal.pbio.0020108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Simon RM, Subramanian J, Li MC, Menezes S. Using cross-validation to evaluate predictive accuracy of survival risk classifiers based on high-dimensional data. Brief Bioinform. 2011;12(3):203–14. doi: 10.1093/bib/bbr001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Heagerty P, Lumley T, Pepe M. Time-dependent ROC Curves for Censored Survival Data and a Diagnostic Marker. Biometrics. 2000;56:337–44. doi: 10.1111/j.0006-341x.2000.00337.x. [DOI] [PubMed] [Google Scholar]
- 23.Chabiniok R, Wang VY, Hadjicharalambous M, et al. Multiphysics and multiscale modelling, data-model fusion and integration of organ physiology in the clinic: ventricular cardiac mechanics. Interface Focus. 2016;6(2):20150083. doi: 10.1098/rsfs.2015.0083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sermesant M, Chabiniok R, Chinchapatnam P, et al. Patient-specific electromechanical models of the heart for the prediction of pacing acute effects in CRT: a preliminary clinical validation. Med Image Anal. 2012;16(1):201–15. doi: 10.1016/j.media.2011.07.003. [DOI] [PubMed] [Google Scholar]
- 25.McCormick M, Nordsletten DA, Kay D, Smith NP. Simulating left ventricular fluid–solid mechanics through the cardiac cycle under LVAD support. Journal of Computational Physics. 2013;244:80–96. [Google Scholar]
- 26.Lewandowski AJ, Augustine D, Lamata P, et al. Preterm heart in adult life: cardiovascular magnetic resonance reveals distinct differences in left ventricular mass, geometry, and function. Circulation. 2013;127(2):197–206. doi: 10.1161/CIRCULATIONAHA.112.126920. [DOI] [PubMed] [Google Scholar]
- 27.Zhang X, Cowan BR, Bluemke DA, et al. Atlas-based quantification of cardiac remodeling due to myocardial infarction. PLoS One. 2014;9(10):e110243. doi: 10.1371/journal.pone.0110243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Deo RC. Machine Learning in Medicine. Circulation. 2015;132(20):1920–30. doi: 10.1161/CIRCULATIONAHA.115.001593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Forfia PR, Fisher MR, Mathai SC, et al. Tricuspid Annular Displacement Predicts Survival in Pulmonary Hypertension. Am J Respir Crit Care Med. 2006;174(9):1034–41. doi: 10.1164/rccm.200604-547OC. [DOI] [PubMed] [Google Scholar]
- 30.Kind T, Mauritz GJ, Marcus JT, van de Veerdonk M, Westerhof N, Vonk-Noordegraaf A. Right ventricular ejection fraction is better reflected by transverse rather than longitudinal wall motion in pulmonary hypertension. J Cardiovasc Magn Reson. 2010;12:35. doi: 10.1186/1532-429X-12-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Buckberg G, Mahajan A, Saleh S, Hoffman JI, Coghlan C. Structure and function relationships of the helical ventricular myocardial band. J Thorac Cardiovasc Surg. 2008;136(3):578–89. 89 e1–11. doi: 10.1016/j.jtcvs.2007.10.088. [DOI] [PubMed] [Google Scholar]
- 32.Pettersen E, Helle-Valle T, Edvardsen T, et al. Contraction pattern of the systemic right ventricle shift from longitudinal to circumferential shortening and absent global ventricular torsion. J Am Coll Cardiol. 2007;49(25):2450–6. doi: 10.1016/j.jacc.2007.02.062. [DOI] [PubMed] [Google Scholar]
- 33.LeCun Y, Bengio Y, Hinton G. Deep learning Nature. 2015;521(7553):436–44. doi: 10.1038/nature14539. [DOI] [PubMed] [Google Scholar]
- 34.Auger DA, Zhong X, Epstein FH, Spottiswoode BS. Mapping right ventricular myocardial mechanics using 3D cine DENSE cardiovascular magnetic resonance. J Cardiovasc Magn Reson. 2012;14:4. doi: 10.1186/1532-429X-14-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Oktay O, Bai W, Guerrero R, et al. Stratified Decision Forests for Accurate Anatomical Landmark Localization. IEEE Transactions on Medical Imaging. 2016 doi: 10.1109/TMI.2016.2597270. Epub ahead of print. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.